- 1Division of Pulmonary, Allergy and Critical Care Medicine, Department of Medicine, University of Pittsburgh, Pittsburgh, PA, United States
- 2Department of Microbiology and Molecular Genetics, University of Pittsburgh, Pittsburgh, PA, United States
Pseudomonas aeruginosa is an opportunistic human pathogen with a complex respiratory chain. The bacterium is predicted to express three NADH:ubiquinone oxidoreductases (NDH-1, NDH-2 and Nqr). We created deletions strains of the predicted NADH:ubiquinone oxidoreductases alone, and in combination to determine the respective roles of the NADH dehydrogenases in growth and virulence. NDH-1 and NDH-2 were largely redundant under aerobic conditions. Aerobic NADH dehydrogenase enzymatic activity assay was lost with deletion of both NDH-1 and NDH-2. Under anaerobic conditions, NDH-1 was required for robust growth, and overexpression of NDH-2 rescued the NDH-1 anaerobic growth defect in rich media. There was not compensatory upregulation of NDH-2 under anaerobic conditions in NDH-1 deletion strains. To test which genes were required for in vivo virulence, we used both an insect and plant disease model. In the Galleria mellonella model, neither deletion of NDH-1 nor NDH-2 led to a change in median lethal dose, although death occurred more slowly in the NDH-1 deletion infections. In a lettuce model of virulence, loss of NDH-1 caused a decrease in recovered viable bacteria and a decrease in visual tissue damage. The compound deletion of NDH-1/NDH-2 causes a severe growth defect, both under aerobic and anaerobic conditions, and was avirulent in a lettuce model. Together, these results demonstrate the redundancy of the P. aeruginosa respiratory chain at the NADH dehydrogenase level in aerobic growth and virulence.
Introduction
Pseudomonas aeruginosa is an opportunistic pathogen that causes pneumonia, chronic airway infections, and urinary tract infections (Magill et al., 2014; Crull et al., 2018). The bacterium is also ubiquitous in both soil and water environments. To subsist in this wide range of environments, P. aeruginosa is able to metabolize a wide range of carbon sources. The organism has a highly branched respiratory chain with multiple terminal oxidases and can respire both with oxygen and nitrogen oxides (Arai, 2011; Arai et al., 2014). Additionally, there are up to 17 predicted dehydrogenases that may be coupled to the quinone pool, though the physiological role of these enzymes remains poorly understood (Williams et al., 2007). The P. aeruginosa genome encodes at least three bioinformatically predicted NADH dehydrogenases (NADH:quinone oxidoreductases). NDH-1, encoded by the nuoA-N operon (PA2637-2649), is homologous to the mitochondrial complex I, and has a fused nuoCD subunit (Spero et al., 2016). NDH-1 enzymes both translocate protons and oxidize NADH to NAD+ (Williams et al., 2007). Previously, it had been reported that the nuo operon was required for anaerobic growth through two transposon screens (Filiatrault et al., 2006; Schurek et al., 2008). Mutations within the nuo operon are also found in association with aminoglycoside resistance, presumably due to the energy dependency of aminoglycoside uptake (Amini et al., 2011; Ezraty et al., 2013; Yen and Papin, 2017).
In addition to NDH-1, P. aeruginosa is also predicted to express one type-II NADH dehydrogenase (NDH-2), which is encoded by the ndh gene (PA4538). Type-II bacterial NADH dehydrogenases use FAD as a cofactor to oxidize NADH without translocating protons. NDH-2 enzymes have been a recent focus for antimicrobial development because many microbial organisms lack NDH-1 (Cook et al., 2014; Heikal et al., 2014). The roles of NDH-2 in P. aeruginosa growth and virulence are not known. Finally, the nqrA-F operon (PA2994-2999) has been experimentally demonstrated to be a unique sodium regulated, proton pumping NADH dehydrogenase in P. aeruginosa (Raba et al., 2018). Again, the physiologic role of NQR in P. aeruginosa is unclear. During chronic infection, anaerobic growth conditions are common, and core bacterial metabolism during infection is incompletely understood (Alvarez-Ortega and Harwood, 2007). Thus, the purpose of this paper was to determine the respective roles of the NADH: quinone oxidoreductase enzymes in P. aeruginosa physiology and in vivo virulence.
Materials and Methods
Reagents
Lysogeny broth (LB) with 5% NaCl, LB agar, KCN, sodium succinate, sodium acetate, sodium citrate, L-proline, L-glutamine, glucose and potassium nitrate were purchased from Sigma. Other reagents include Muller Hinton Broth Agar (Fluka), M9 powdered salts (BD), gentamicin (Atlanta Biologicals), Breath-Easy Membranes (Diversified BD Biotech), XTT (2,3-Bis-2-Methoxy-4-Nitro-5-Sulfophenl-2-Tetrazolium-5-Carboxanilide from Caymen chemicals) and nicotinamide adenine dinucleotide (NADH from Caymen chemicals).
Strain Construction
Deletion mutants were made using two-step allelic exchange as described by Hmelo in the PAO1 and PA14 backgrounds (Hmelo et al., 2015). Strains, primers, and plasmids are found in Supplementary Table 1. The following protocol modifications were made: the plasmid pMQ30 was used for allelic replacement (Shanks et al., 2006). 500–1000 bp upstream and downstream of the region to be deleted were amplified by PCR, joined through sequential overlap extension PCR for the allelic replacement construct (ARC) for the NADH dehydrogenase deletion mutants. The ΔroxSR ARC was made using NEB Assembly per the kit instructions (New England Biolabs). Ligation mixtures were transformed into Escherichia coli DH5α, colonies were screened for the inserts by PCR, and plasmid was isolated from colonies by Miniprep (Qiagen). Inserts were sequence confirmed (Eurofins) and mated into the P. aeruginosa recipient strain. Complementation was done by inserting the ndh or nuoIJ sequence of interest into the pUC18-mini-Tn7 plasmid containing the constitutive PnptII promoter and sequence confirmed prior to insertion into P. aeruginosa (Choi and Schweizer, 2006; Melvin et al., 2017). Transposon insertion mutants were from the PA14 library1. Strains were grown overnight in 5 ml of LB on a roller drum prior to experiments except for the ΔnuoIJΔndh and ΔnuoIJΔndhΔnqr strains which required 48 h to achieve sufficient culture density for experimentation.
Sequencing Analysis
The initial deletion was identified through whole genome sequencing on a NextSeq 500 System (Illumina) using 2 × 150 bp libraries. Breseq version 0.28.1 was used for variant calling with alignment to the PAO1 genome (Deatherage and Barrick, 2014). After identification of the initial suppressor, all subsequent deletion strains were subjected to whole genome sequencing.
Microarray Analysis
Published microarrays were analyzed using the GEO2R tool on the Gene Expression Omnibus (GEO) website (Edgar, 2002). Default settings were used, including the use of the Benjamini and Hochberg false discovery rate method and log transformation of data.
Gentamicin Minimum Inhibitory Concentration (MIC)
Agar dilution MIC assays using Muller-Hinton Broth agar were performed as described (Wiegand et al., 2008), with the modification that gentamicin concentrations were varied by 0.1 to 0.3 mg/L for quantitative MICs and by 50–100 mg/L for strains containing aacC1 cassettes.
Aerobic Growth Curves
Strains were diluted in fresh LB, M9 with glucose or M9 with succinate and inoculated in 3–6 replicates in a 96 well plate. The plates were incubated at 37°C with a gas permeable membrane covering the top of the plate. The plate was shaken for 3 s prior to each OD600 measurement on a SpectraMax M2 plate reader (Molecular Devices). Experiments were done 3–6 times.
Anaerobic Growth Assays
Overnight cultures were grown in LB, mixed well, diluted 1:500 and spotted in 5 μl drops on plates. For anaerobic growth experiments, the following solid media was used supplemented with 1% KNO3: LB agar, M9 with sodium succinate, glucose, sodium citrate, ethanol, L-proline, L-glutamine, sodium acetate. Plates were grown in GasPak (BD Biosystems) jars at 37°C for 1–7 days as indicated. Oxygen detection strips were included to insure anaerobic conditions, and in some experiments Δanr strains were included as negative growth controls. For anaerobic growth curves, strains were diluted to an optical density of 0.0005 in media and sealed in Wheaton serum vials. At indicated intervals, vials were sampled aseptically with a needle and syringe and the optical density was measured on a plate reader at 600 nm (OD600). Lysogeny broth, M9 with glucose and M9 with succinate were used in these experiments, all supplemented with 1% KNO3 to support anaerobic respiration.
Transcriptional Reporter Assays
We followed the protocol described by Melvin et al. (2017) with the following modifications. The 300 base pair intergenic region upstream of ndh was inserted into the transcriptional fusion reporter plasmid pAG4 using NEBAssembly (New England Biolabs). The sequenced plasmid was inserted into a single site on the P. aeruginosa chromosome (Glassing and Lewis, 2015). Plasmid with no insert was used as a negative control, and the PntpII promoter was used as a positive control. For aerobic assays, overnight cultures of reporter strains were diluted in fresh Lysogeny Broth, grown to mid-log phase and luminescence was measured. For anaerobic studies, overnight cultures were grown on a roller drum and then diluted 1:50 in LB with 1%KNO3. The ΔnuoIJ and Δanr strains were diluted 1:10 so that similar culture density would result at the end of the assay. Subcultures were incubated in a GasPak jar for 6 h, at least 2 h after the indicator strips indicated oxygen depletion. Luminescence was measured using a Synergy 2 plate reader (BioTek), and measurements were normalized to OD600 and empty vector signal intensity for each day. The experiment was done in triplicate.
Enzymatic Activity Assays
Fifty milliliter overnight cultures were grown in LB with shaking. Bacteria were pelleted and sonicated in 0.1 M Tris buffer pH 7.5. Cell debris was removed via low speed centrifugation, and the supernatant was centrifuged at 32,000 × g for 10 min to pellet the membrane fraction. NADH was measured spectrophotometrically at 340 nm. Results were normalized to protein concentration. The assay was performed 10 times, and results are shown normalized to the parental strain activity for each replicate with 95% confidence intervals. The NADH-Ubiquinone oxidoreductase assay was used with 1 μM antimycin A, 5 mM KCN, and XTT (a formazan dye) as a terminal electron acceptor. Membranes were incubated with these reagents in 0.1 M Tris buffer, pH 7.5 at 37°C and formation of formazan dye product was monitored spectrophotometrically over 30 min. Equal protein was loaded for both assays. Results shown as means with 95% confidence intervals.
Galleria Virulence Model
The protocol described by Jander was used with stationary phase cultures (Jander et al., 2000). G. mellonella larvae were obtained from Grubco (Fairfield, OH) and kept at room temperature. Overnight bacterial cultures were rinsed in 10 mM MgSO4, diluted, and injected into the larvae using a 30-gauge needle. For survival curves, live worms were counted every 2 h during the period frequent deaths were occurring, and then every 4–12 h. For LD50 determinations, worms were injected with varying inocula and live worms were counted at 72 h. Sterility controls were included in all experiments. Median lethal infections inocula were determined through probit analysis.
Lettuce Virulence Model
The protocol described by Starkey was used (Starkey and Rahme, 2009). Romaine lettuce was purchased the morning of the experiment. Overnight cultures were rinsed in 10 mM MgSO4, diluted to an optical density at 600 nm (OD600) of 0.2 and injected into the rib of the lettuce leaf. Leaves were incubated at room temperature and assessed daily for visual signs of infection. After 4 days, the infected portion of leaf was excised, homogenized with a bead beater, and colony forming units (CFU) were plated by serial dilution. Experiments were performed 3–4 times with at least 3 replicate leaves per strain in each experiment.
Statistics
Prism version 7.0 software (GraphPad Software, Inc.) was used to display and analyze data. Number of replicates for experiments is described in figure legends, and at least three replicates were done for each experiment. Tests are indicated in figure legends. When applicable, one-way ANOVA tests were used with post hoc Dunnett test to compare multiple groups. For assays that were normalized to control values, a one-way t-test was done comparing to the theoretical mean of 1.0. Survival analysis was used for the G. mellonella experiment, with the log-rank test used to compare survival curves. Probit analysis was used to determine LD50.
Results
Role of NDH-1 in Anaerobic Growth and Aminoglycoside Resistance
We initially encountered a large deletion in the nuo operon (NDH-1) while constructing a deletion mutant of roxSR (PA4493-4494) using two-step allelic exchange with gentamicin as the selection agent for an unrelated study (Hmelo et al., 2015). The two-component sensor kinase RoxSR regulates aerobic terminal oxidase expression, however it was not reported to regulate pathways required for anaerobic growth (Kawakami et al., 2010). The resulting ΔroxSR strain exhibited an anaerobic growth defect that could not be complemented despite multiple approaches (Figure 1A). Because complementation failed, we subjected the strain to whole genome sequencing and found a 759 bp deletion spanning nuoIJ, in addition to the expected deletion of roxSR. The strain was reconstructed, and the reconstructed ΔroxSR strain did not have an anaerobic growth defect, which suggested that the nuoIJ deletion caused the anaerobic growth defect. It has been previously reported that transposon insertion within the nuo operon gives an anaerobic growth defect, and we were able to replicate these results using strains from the PA14 ordered transposon mutant library (Figure 1B; Filiatrault et al., 2006). NDH-1 in prokaryotes consists of a two protein arms: membrane and peripheral. The nuoIJ deletion spans both arms and is thus predicted to disrupt protein function. We used a ΔnuoIJ strain throughout due to the difficulty in complementing such a large operon in its entirety. The nuoIJ deletion was encountered while using gentamicin as a selection agent, and because the nuo operon has previously been reported to be under genetic pressure after exposure to aminoglycosides, we tested if deletion of nuoIJ resulted in an increase in gentamicin MIC using agar dilution (Yen and Papin, 2017). The MIC of PA14 was 0.4 mg/L and the MIC of PA14 ΔnuoIJ increased 3-fold to 1.2 mg/L (Figure 1C). The complemented strain was not tested because it contained a gentamicin resistance cassette. Similar results were obtained in the PAO1 background. Notably, the gentamicin MICs for the Δndh, ΔnqrA-F, and Δndh ΔnqrA-F strains were within 0.3 mg/L of the parental strain, and the ΔnuoIJΔnqrA-F strain MIC was within 0.3 mg/L of the ΔnuoIJ strain (data not shown). The slow growth of the ΔnuoIJΔndh and ΔnuoIJΔndhΔnqrA-F strains complicated interpretation of the MIC assays, however the MIC for the strains was at least as high as the ΔnuoIJ strain. The increase in gentamicin resistance imparted by ΔnuoIJ was additive to the aacC1 cassette that is frequently used for genetic manipulations in P. aeruginosa, increasing the MIC of PAO1-aacC1 from 100 to 300 mg/L (Figure 1D). In summary, we encountered an incidental 759 bp deletion in the nuo operon that caused an anaerobic growth defect and increased resistance to gentamicin. Because the genome of P. aeruginosa contains multiple putative NADH dehydrogenases, we then determined the role of each in growth and virulence.
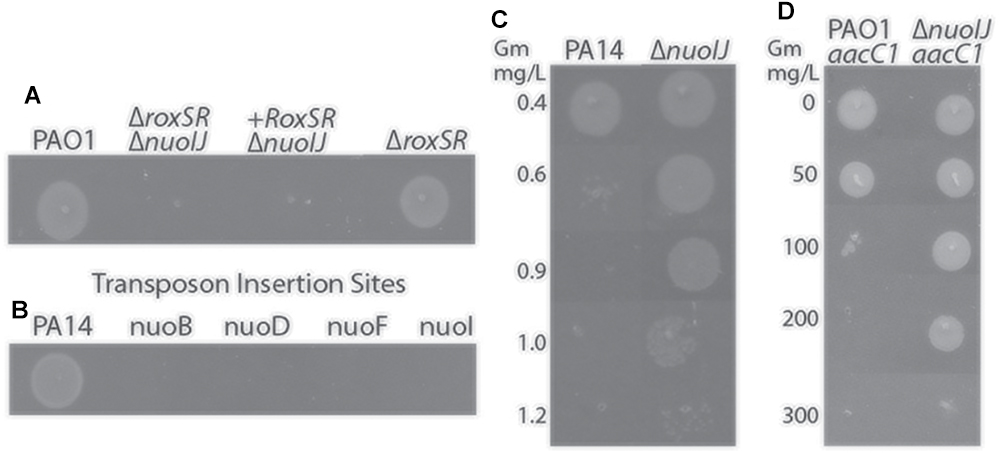
Figure 1. (A) Deletion of roxSR caused an anaerobic growth defect on LB with 1% KNO3 that failed to complement. When sequencing revealed deletion of nuoIJ, the reconstructed ΔroxSR strain grows anaerobically. (B) Transposon mutants within the nuo operon fail to grow anaerobically. (C) The gentamicin MIC for PA14 is 0.6 mg/L, while the MIC for PA14 ΔnuoIJ is increased to 1.2 mg/L. (D) The aac1 cassette inserted to a single copy chromosomal site in combination with nuoIJ deletion increases the MIC from 100 to 300 mg/L.
Role of NADH Dehydrogenase Genes in Growth
We determined the role of the NADH dehydrogenase enzymes in aerobic growth. Single deletions of nuoIJ, ndh, and nqrA-F did not affect log phase growth rate in rich media, minimal media (M9) succinate, or M9 with glucose as carbon sources (Figures 2A,C,D,F,G,I). However, the ΔnuoIJ strain grew to slightly lower stationary phase density in LB (PAO1 parental 8.3 ± 6.7 × 109CFU/ml vs. ΔnuoIJ 2.5 ± 2.7 × 109 CFU/ml, p < 0.05 by two-tailed t-test). Strains expressing nqr only (ΔnuoIJΔndh) or no known NADH dehydrogenases (ΔnuoIJΔndhΔnqrA-F, triple ko) had a growth defect that varied in severity by carbon source. In rich media and M9-succinate, the ΔnuoIJΔndh strain (nqr only) had a 30–50% decrease in log phase growth rate compared to the parental strain, and deletion of all three dehydrogenases ΔnuoIJΔndhΔnqrA-F (triple ko) did not result in a further decrease in growth (Figures 2B,E). However, with glucose as a carbon source, either NDH-1 or NDH-2 was required for growth, with no growth seen in strains expressing only NQR (Figure 2H).
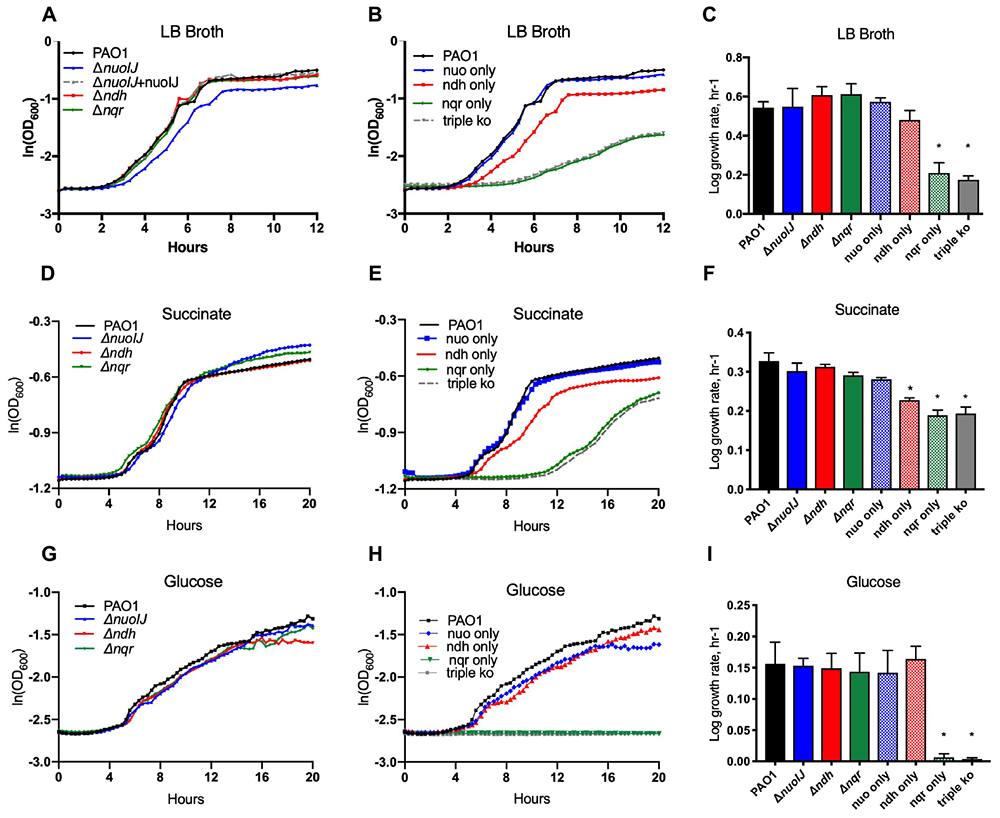
Figure 2. Indicated strains were grown in a microtiter dish on LB, M9 with glucose or succinate as carbon sources. (A) In LB, deletion of any single NADH dehydrogenase does not change log phase growth rate. (B) The compound ΔnuoIJ Δndh (“nqr only”) and ΔnuoIJ Δndh Δnqr (“triple ko”) strains grow slowly in LB. (C) Maximal log phase growth rate in LB (mean ± SD). (D) With succinate as a carbon source, deletion of any individual NADH dehydrogenase did not affect aerobic growth. (E) With succinate as a carbon source, the compound ΔnuoIJ Δndh (“nqr only”) and ΔnuoIJ Δndh Δnqr (“triple ko”) strains grew slowly, and there was a slight decrease in growth rate in a strain expressing only ndh (ΔnuoIJ Δnqr). (F) Maximal log phase growth rate in succinate (mean ± SD). (G) With glucose as a carbon source, deletion of any individual NADH dehydrogenase did not affect aerobic growth. (H) With glucose as a carbon source, the compound ΔnuoIJ Δndh (“nqr only”) and ΔnuoIJ Δndh Δnqr (“triple ko”) strains showed now growth within 20 h. (I) Maximal log phase growth rate in glucose. ∗ indicates p < 0.01 by one-way ANOVA followed by Dunnett’s multiple comparison test. At least 3 replicates were done for all experiments and representative growth curves are shown.
We then studied anaerobic growth both qualitatively on solid media (Figures 3A) or in broth quantified by optical density of the culture (Figures 3B,C). Under anaerobic conditions on LB agar supplemented with 1% KNO3, the ΔnuoIJ strain grew very slowly with scant growth visible by 7 days (Figure 3A). The growth defect was complemented by expression of either nuoIJ or ndh under the constitutive promoter PnptII (Figure 3B). Of note, this promoter increases expression of ndh approximately 40-fold as compared to baseline (data not shown). Compound mutants containing the ΔnuoIJ deletion grew poorly anaerobically, while the strain expression only nuo (ΔndhΔnqr) grew similarly to the parental strain (Figure 3C). An Δanr strain was included as a control for anaerobic conditions. ANR is the master transcriptional regulator for denitrification and is required for anaerobic growth (Zimmermann et al., 1991). In minimal media with succinate, deletion of nuoIJ resulted in a milder growth defect. Growth was clearly visible on solid media at 48 h (Figure 3D), so growth was quantitated by OD (Figure 3E). Overexpression of ndh did not complement the mild growth defect seen in the ΔnuoIJ strain (Figure 3E). That NDH-2 complemented NDH-1 on rich media but not on M9 succinate, suggested that the requirement of NADH dehydrogenases for redox balance versus proton motive force (pmf) generation may vary by substrate. Therefore, we tested the growth of these strains on several carbon sources (Figure 3D). Citrate, L-glutamine and L-proline feed into the tricarboxylic acid (TCA) cycle prior to succinate dehydrogenase. On these substrates, an anaerobic growth detect was seen in the ΔnuoIJ strain that was complemented by either expression of nuoIJ or overexpression of ndh (Figure 3D). Ethanol and acetate are not readily converted to succinate. An anaerobic growth defect was seen in the ΔnuoIJ strain on both ethanol and acetate, that was not complemented by expression of ndh (Figure 3D).
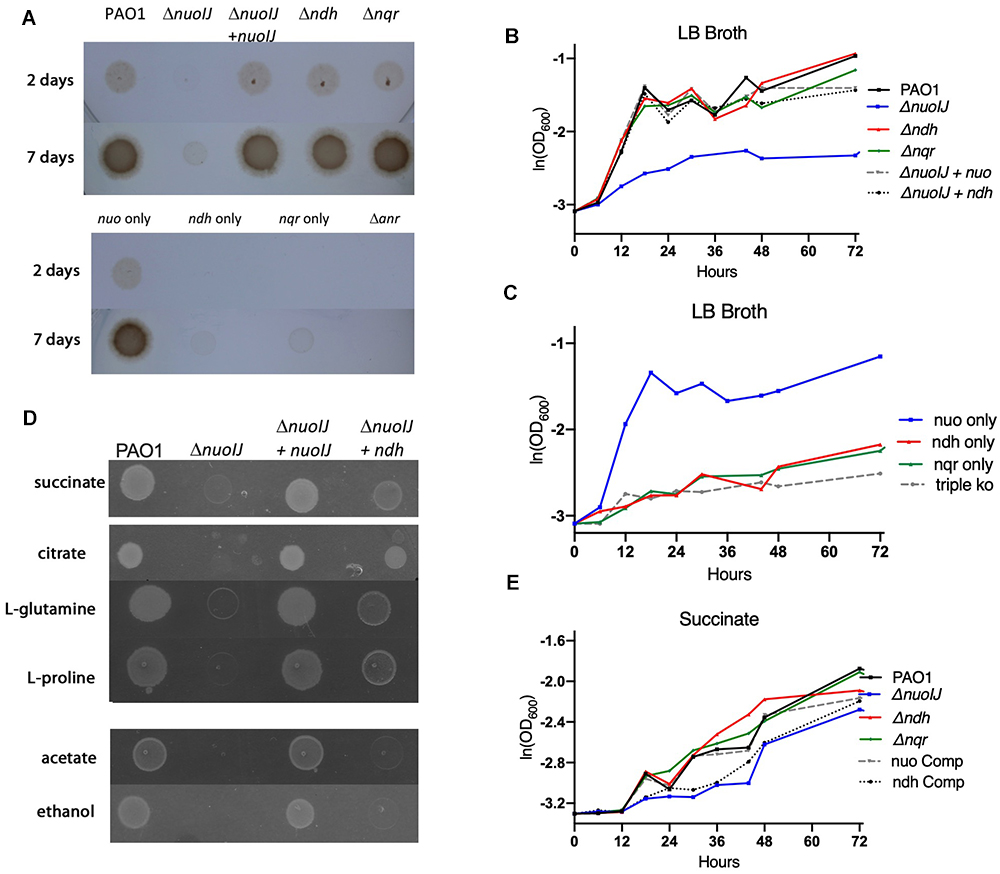
Figure 3. (A) Overnight cultures of NADH dehydrogenase mutant strains were diluted 1:500 and grown for one to seven days anaerobically on plates containing 1% KNO3. The ΔnuoIJ had scant growth at 7 days, that was complemented by nuoIJ. (B) Strains were diluted in LB broth with 1% KNO3, sealed in glass vials and incubated at 37°C. OD600 was measured at indicated times. In LB, the ΔnuoIJ strain shows little growth. The growth defect was complemented by expression of nuoIJ or overexpression of ndh. (C) In LB, the strain expressing nuo only (Δnqr Δndh) grew anaerobically, while compound deletion strains with nuoIJ deleted grew poorly. (D) M-9 agar plates with the indicated carbon sources were grown anaerobically for 48 h. The ΔnuoIJ had clearly visible growth at 48 h on succinate, faint growth on citrate, L-glutamine and L-proline and no visible growth on acetate and ethanol. Expression of nuoIJ complemented growth defects on all carbon sources. Overexpression of ndh complemented growth on citrate, L-glutamine and L-proline, but not acetate or ethanol. (E) Strains were diluted in M9-succinate containing 1% KNO3, sealed in glass vials and incubated at 37°C. OD600 was measured at indicated times. The ΔnuoIJ strain shows a mild growth defect that is not complemented by overexpression of ndh.
Cumulatively, we saw that loss of nuoIJ had minimal effects on aerobic growth in multiple media types, with ndh being able to support growth in most situations. Anaerobically loss of nuoIJ resulted in a severe growth defect on most carbon sources tested, but a smaller growth defect on succinate. Overexpression of ndh was able to complement ΔnuoIJ completely in rich media, citrate, L-glutamine and L-proline.
Analysis of Gene Expression and Enzymatic Activity
One explanation for the growth defect seen with deletion of NDH-1 under anaerobic conditions is that NDH-2 is not expressed anaerobically. The GEO Database contains two microarray data sets comparing gene expression under aerobic and anaerobic conditions (Alvarez-Ortega and Harwood, 2007; Trunk et al., 2010). We examined both data sets using the GEO2R tool, and none of the NADH dehydrogenase operons were regulated in response to oxygen in these data sets. The sensitivity of these data sets using microarray technology may be limited for subtle changes, so we created transcriptional reporter strains fusing the putative ndh promoter upstream of luciferase (lux) in the plasmid pAG4 (Glassing and Lewis, 2015). Overall aerobic reporter expression was low but reliably detectable at 2-fold as compared to baseline (Figure 4A). In the parental strain there was a modest, but statistically significant, increase under anaerobic conditions. Aerobically, reporter expression was unchanged in the ΔnuoIJ as compared to parental, and the increased expression seen anaerobically in the parental strain was blunted (Figure 4A). We then measured NADH dehydrogenase enzymatic activity directly by following loss of NADH spectrophotometrically in the presence of isolated membranes. The ΔnuoIJΔndh and ΔnuoIJΔndhΔnqrA-F strains did not have measurable enzymatic activity, while the ΔnuoIJ and Δndh strains did not show a statistically significant decrease in total NADH dehydrogenase activity after ten replicates (Figure 4B). These results strongly indicate that NDH-1 and NDH-2 account for the total NADH dehydrogenase enzymatic activity under the condition tested. Regarding the absence of effect with deletion of NDH-1 or NDH-2 alone, the NADH assay may be limited by downstream capacity of the respiratory chain to accept electrons, thus we were not able to determine if loss of NDH-1 or NDH-2 resulted in decreased total NADH dehydrogenase activity. We then used the chromogenic formazan dye XTT combined with potassium cyanide and antimycin A to block transfer of electrons to the distal respiratory chain to measure NADH dehydrogenase activity. Deletion of either nuoIJ or ndh resulted in decreased NADH enzymatic activity in this assay (Figure 4C). From these data we concluded that ndh is not upregulated under anaerobic conditions in the ΔnuoIJ strains, but it does continue to be expressed. NDH-1 and NDH-2 supply the NADH dehydrogenase activity present during growth in rich media.
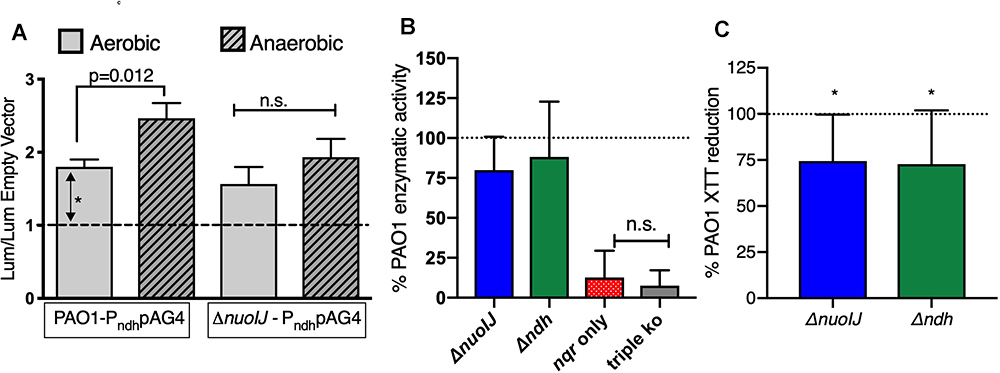
Figure 4. (A) Transcriptional reporter strains were made with the 300 bp intergenic region upstream of ndh inserted upstream of lux. Results shown normalized to empty vector luminescence and optical density of culture. Three replicates shown. p-values indicated from one-way ANOVA followed by Dunnett’s test. (B) NADH dehydrogenase assay, normalized to parental strain for each day. Ten replicates shown, and p < 0.001 for the strain expressing nqr only (ΔnuoIJ Δndh) and the triple knockout strain compared to PAO1 by one-way ANOVA. Mean ± 95% CI shown. (C) XTT assay in the presence of antimycin and cyanide. Deletion of either ΔnuoIJ or Δndh results in decreased XTT reduction. p < 0.05 by one-way t-test for each strain. Eight replicates shown. Mean ± 95% CI shown.
In vivo Roles of NADH Dehydrogenases in Plant and Insect Infections
We tested the effects of the deletion of NADH dehydrogenases on virulence in an insect and plant model. In the Galleria mellonella waxworm model of insect virulence, the median lethal dose for PAO1, ΔnuoIJ and Δndh were identical (1.3–1.5 infecting bacteria/larva). The compound deletion ΔnuoIJΔndh caused a small increase in LD50 to 8.5 bacteria/larva. While the LD50 was unchanged with loss of NDH-1, the kinetics of killing was significantly slower (p < 0.001) in the ΔnuoIJ strain as compared to PAO1 (Figure 5A and Table 1). Expression of either nuoIJ or ndh in the ΔnuoIJ strain was sufficient to return killing kinetics to that of the parental strain. We did not test killing kinetics in the ΔnuoIJΔndh strain because the LD50 was lower than PAO1. Deletion of ndh or nqr, alone or in combination, had no effect (Figure 5B).
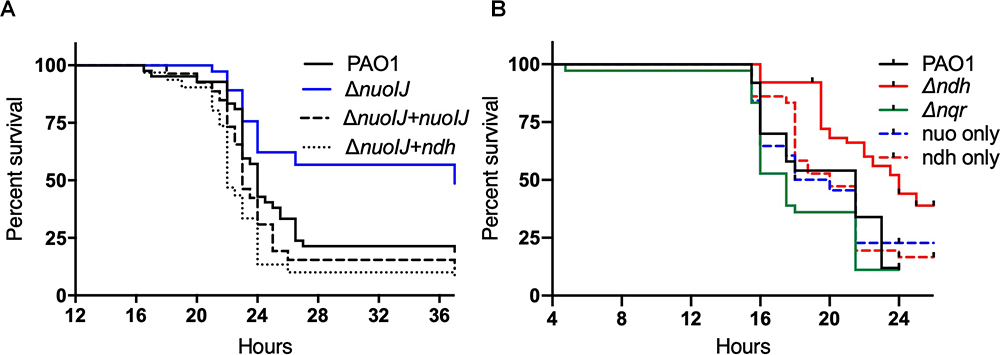
Figure 5. Survival curves for Galleria mellonella injected with 10 bacteria/larvae of indicated bacterial strains. (A) ΔnuoIJ strain has increased survival compared to parental PAO1 or ΔnuoIJ complemented with either nuoIJ or ndh (p = 0.002 by Log-Rank test). (B) Deletion of ndh or nqr did not change survival (alone or in combination).
We then used a lettuce model to test the virulence of the NDH deletion strains (Starkey and Rahme, 2009). Loss of NDH-1 caused a slight decrease in visual tissue damage (Figure 6A). The ΔnuoIJ strain had a 1-log decrease in recoverable CFUs at day 4, which was complemented. Deletion of both NDH-1 and NDH-2 caused a larger decrease in virulence (p < 0.05). Deletion of ndh or nqr, alone or in combination, had no effect (Figure 6B). Together, these data suggest that at least one NDH is required for full virulence in the lettuce model, with NDH-1 being the preferred enzyme. Overall, loss of NDH-1 caused a small but measurable decrease in virulence, while deletion of NDH-2 or NQR had no detectable effect.
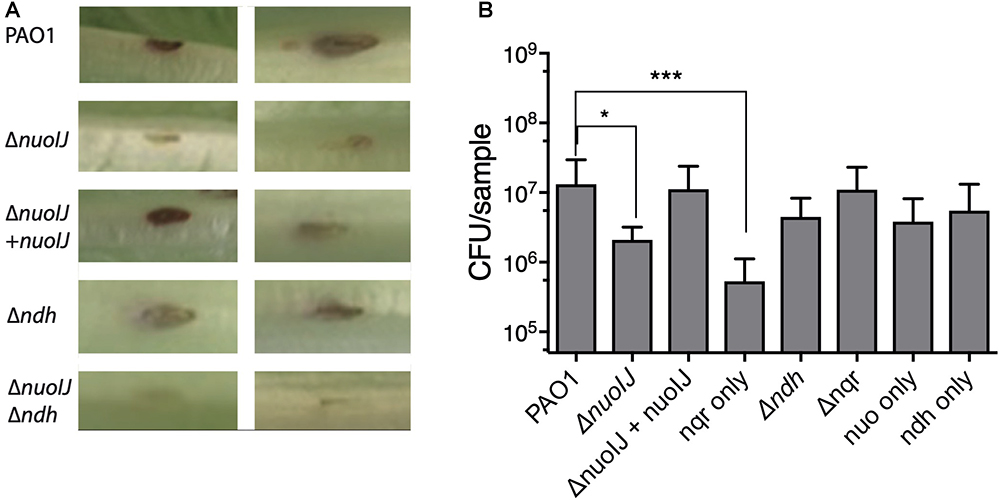
Figure 6. (A) Representative images from lettuce virulence model. Infected area appears in brown, and two representative replicates are shown for each strain. (B) Quantification of CFU from infected leaf areas. 3–4 independent experiments pooled. Brackets indicate p < 0.05 by one-way ANOVA followed by Dunnett’s test.
Discussion
In this study, we observed that of the NADH dehydrogenases in P. aeruginosa, NDH-1 (encoded by the nuo operon) is required for anaerobic growth in rich broth and glucose, as well as full virulence in plant and insect pathogenesis models. In vivo, loss of NDH-1 led to slower killing of G. mellonella larvae and less tissue damage in the lettuce infection model. We did not identify a clear physiologic role for NDH-2 in these studies. The deletion of both NDH-1 and NDH-2 leads to loss of NADH enzymatic activity demonstrating that NDH-2 is biochemically functional and expressed.
We initially encountered a large deletion within nuo while using gentamicin as a selection agent during allelic recombination. We saw a 3-fold increase in gentamicin MIC with loss of NDH-1, similar to what has been reported in E. coli and Salmonella. Aminoglycoside uptake is strongly dependent on proton motive force, to which NDH-1 contributes, thus it is perhaps not surprising that the nuo operon was the target for a suppressor while using gentamicin to genetically manipulate P. aeruginosa. We did not exclude alternative mechanisms for increased gentamicin resistance such as increased efflux. Mutations in nuo are also found during laboratory evolution experiments with aminoglycosides in P. aeruginosa, and the operon was identified as a contributor to low-level aminoglycoside resistance through transposon screening (Schurek et al., 2008; Amini et al., 2011; Yen and Papin, 2017). Apart from the change in aminoglycoside resistance, loss of NDH-1 causes a very subtle aerobic phenotype in rich media, and these results demonstrate the utility of affordable, whole genome sequencing as confirmation during the creation of genetically manipulated bacterial strains.
Redundancy of NDH Enzymes in Aerobic Growth
During aerobic growth any of the individual NADH dehydrogenases are dispensable for log phase growth on glucose, succinate or rich media. As with many studies using deletion strains, the caution that the physiologic role of any enzyme may not be revealed due to compensatory changes due to the deletions themselves applies. Strains possessing only NDH-1 or only NDH-2 grew well in all conditions tested. Strains expressing only NQR grew poorly in all conditions; and we did not detect a difference in growth between the strains expressing only nqrA-F and those with all three NADH dehydrogenase operons deleted. Compound deletion of NDH-1 and NDH-2 leads to undetectable NADH dehydrogenase activity under the conditions tested. NQR was recently reported to be a proton pumping NADH dehydrogenase in P. aeruginosa, however the physiologic role of the enzyme remains uncertain as compound deletion of NDH-1 and NDH-2 abolished NADH dehydrogenase activity under the conditions studied (Raba et al., 2018). Directly measuring NADH spectrophotometrically, we were unable to detect a change in NADH consumption with deletion of NDH-1 or NDH-2 alone. The NADH assay requires that the rest of the respiratory chain not be limiting, and it is possible that with NDH-2 alone provides enough excess NADH dehydrogenase activity compared to more limiting steps that a reduction was not detected. Using a tetrazolium dye as a terminal electron acceptor with cyanide and antimycin blocking transfer of electrons through cytochrome bc1 to the terminal oxidases, we detected a modest decrease in activity in with deletion of either NDH-1 or NDH-2. Despite these limitations, it is clear that NDH-1 and NDH-2 are the main NADH dehydrogenases required for aerobic growth.
Requirement of NDH-1 for Anaerobic Growth
We observed a profound anaerobic growth defect with deletion of NDH-1 on most substrates. NADH dehydrogenases serve two potential metabolic roles. During canonical anaerobic denitrification only 6 protons are pumped per 2 electrons (Chen and Strous, 2013). NDH-1 pumps 4 of these 6 protons, thus NDH-1 is predicted to significantly contribute to proton motive force generation. Additionally, on some substrates NADH dehydrogenases may be required to redox balance NADH generated by the TCA cycle or other reactions while pmf is at least partially supported through other means. On succinate the growth defect with loss of NDH-1 was milder than on rich media. Complementation studies were difficult to reliably interpret because the role of NDH-1 in growth was less. Compounds that feed into the TCA cycle prior to succinate dehydrogenase (L-proline, L-glutamine and citrate) can contribute to electron transport via complex 2. However, these energy sources also are predicted to generate NADH requiring some means of balancing the redox state of the cell. On these substrates NDH-2, which can provide redox balance but not generate pmf, was able to complement NDH-1. Substrates that cannot be readily converted to succinate (e.g., acetate and ethanol) cannot contribute to pmf via complex 2 and therefore require NDH-1. More than redox balance is required to support growth on these substrates. P. aeruginosa displays great metabolic flexibility with a highly branched respiratory chain at the terminal oxidase level. At least five aerobic terminal oxidase are expressed that have a range of oxygen affinities and proton translocation efficiencies. Branching at both the NDH and terminal oxidase levels may increase the flexibility of the chain by allowing the decoupling of pmf generation and regeneration of NAD+ (Calhoun et al., 1993; Kerscher et al., 2008; Bekker et al., 2009).
Redundancy in Virulence
NDH-1 was uniquely required for full in vivo virulence. P. aeruginosa is highly virulent in waxworms, with an LD50 of only a single bacterium. Loss of NDH-1 does not change the LD50 but causes a small delay in time to death. We saw a slightly more pronounced phenotype in the lettuce model, with less visible tissue damage and fewer recoverable CFUs with loss of NDH-1. In both the insect and lettuce models, compound deletion of NDH-1 and NDH-2 resulted in a dramatic decrease in virulence. Our results show that NADH dehydrogenase activity is required for virulence, but NDH-1 and NDH-2 may have overlapping functions in vivo. Apparent redundancy of function between NDH-1 and NDH-2 in virulence is found in a number of other bacterial species. Mycobacterium tuberculosis has one NDH-I and two NDH-2 genes, which were recently shown to be largely functionally redundant, although Δndh was attenuated in a murine model. Pseudomonas fluorescens WCS365 expresses both nuo and ndh during rhizosphere colonization, and the nuo deficient strain was less competitive than the parental strain during colonization assays (Camacho Carvajal et al., 2002). Our findings underscore the need for at least one NDH enzyme for growth and virulence in P. aeruginosa. Under aerobic conditions, these enzymes are largely functionally redundant. Anaerobically, NDH-2 is not upregulated to a degree that will support anaerobic growth in the absence of NDH-1. Loss of NDH-1 leads to a slightly decreased virulence. These data reinforce the apparent redundancy and flexibility of the respiratory chain.
Author Contributions
AT, NK, MD, and AZ acquired the data. AZ, VC, and JB analyzed and interpreted the data. All authors contributed to writing the manuscript.
Funding
This work was supported in part by the National Institutes of Health (NIH K23HL131930 to AZ and NIH R01HL123771 to JB) and the Cystic Fibrosis Foundation (ZEMKE16Q0 to AZ and BOMBER14G0 to JB). No persons employed by the funders played any role in the study, in preparation of the manuscript, or in the decision to publish.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Anthony Richardson (University of Pittsburgh) for providing helpful discussion. We also thank D. A. Hogan (Geisel School of Medicine, Dartmouth) for providing strains.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.00075/full#supplementary-material
Footnote
- ^http://ausubellab.mgh.harvard.edu/cgi-bin/pa14/home.cgi, accessed April 5, 2018.
References
Alvarez-Ortega, C., and Harwood, C. S. (2007). Responses of Pseudomonas aeruginosa to low oxygen indicate that growth in the cystic fibrosis lung is by aerobic respiration. Mol. Microbiol. 65, 153–165. doi: 10.1111/j.1365-2958.2007.05772.x
Amini, S., Hottes, A. K., Smith, L. E., and Tavazoie, S. (2011). Fitness landscape of antibiotic tolerance in pseudomonas aeruginosa biofilms. PLoS Pathog. 7:e1002298. doi: 10.1371/journal.ppat.1002298
Arai, H. (2011). Regulation and function of versatile aerobic and anaerobic respiratory metabolism in Pseudomonas aeruginosa. Front. Microbiol. 2:103. doi: 10.3389/fmicb.2011.00103
Arai, H., Kawakami, T., Osamura, T., Hirai, T., Sakai, Y., and Ishii, M. (2014). Enzymatic characterization and in vivo function of five terminal oxidases in Pseudomonas aeruginosa. J. Bacteriol. 196, 4206–4215. doi: 10.1128/JB.02176-14
Bekker, M., de Vries, S., Ter Beek, A., Hellingwerf, K. J., and de Mattos, M. J. T. (2009). Respiration of Escherichia coli can be fully uncoupled via the nonelectrogenic terminal cytochrome bd-II oxidase. J. Bacteriol. 191, 5510–5517. doi: 10.1128/JB.00562-09
Calhoun, M. W., Oden, K. L., Gennis, R. B., de Mattos, M. J., and Neijssel, O. M. (1993). Energetic efficiency of Escherichia coli: effects of mutations in components of the aerobic respiratory chain. J. Bacteriol. 175, 3020–3025. doi: 10.1128/jb.175.10.3020-3025.1993
Camacho Carvajal, M. M., Wijfjes, A. H., Mulders, I. H., Lugtenberg, B. J., and Bloemberg, G. V. (2002). Characterization of NADH dehydrogenases of Pseudomonas fluorescens WCS365 and their role in competitive root colonization. Mol. Plant Microbe Interact. 15, 662–671. doi: 10.1094/MPMI.2002.15.7.662
Chen, J., and Strous, M. (2013). Denitrification and aerobic respiration, hybrid electron transport chains and co-evolution. Biochim. Biophys. Acta Bioenerg. 1827, 136–144. doi: 10.1016/j.bbabio.2012.10.002
Choi, K. H., and Schweizer, H. P. (2006). mini-Tn7 insertion in bacteria with single attTn7 sites: example Pseudomonas aeruginosa. Nat. Protoc. 1, 153–161. doi: 10.1038/nprot.2006.24
Cook, G. M., Greening, C., Hards, K., and Berney, M. (2014). Energetics of pathogenic bacteria and opportunities for drug development. Adv. Microb. Physiol. 65, 1–62. doi: 10.1016/bs.ampbs.2014.08.001
Crull, M. R., Somayaji, R., Ramos, K. J., Caldwell, E., Mayer-Hamblett, N., Aitken, M. L., et al. (2018). Changing rates of chronic Pseudomonas aeruginosa infections in cystic fibrosis: a population-based cohort study. Clin. Infect. Dis. 67, 1089–1095. doi: 10.1093/cid/ciy215
Deatherage, D. E., and Barrick, J. E. (2014). Identification of mutations in laboratory-evolved microbes from next-generation sequencing data using breseq. Methods Mol. Biol. 1151, 165–188. doi: 10.1007/978-1-4939-0554-6_12
Edgar, R. (2002). Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30, 207–210. doi: 10.1093/nar/30.1.207
Ezraty, B., Vergnes, A., Banzhaf, M., Duverger, Y., Huguenot, A., Brochado, A. R., et al. (2013). Fe-S cluster biosynthesis controls uptake of aminoglycosides in a ROS-less death pathway. Science 340, 1583–1587. doi: 10.1126/science.1238328
Filiatrault, M. J., Picardo, K. F., Ngai, H., Passador, L., and Iglewski, B. H. (2006). Identification of Pseudomonas aeruginosa genes involved in virulence and anaerobic growth. Infect. Immun. 74, 4237–4245. doi: 10.1128/IAI.02014-05
Glassing, A., and Lewis, T. A. (2015). An improved Tn7-lux reporter for broad host range, chromosomally-integrated promoter fusions in Gram-negative bacteria. J. Microbiol. Methods 118, 75–77. doi: 10.1016/j.mimet.2015.08.016
Heikal, A., Nakatani, Y., Dunn, E., Weimar, M. R., Day, C. L., Baker, E. N., et al. (2014). Structure of the bacterial type II NADH dehydrogenase: a monotopic membrane protein with an essential role in energy generation. Mol. Microbiol. 91, 950–964. doi: 10.1111/mmi.12507
Hmelo, L. R., Borlee, B. R., Almblad, H., Love, M. E., Randall, T. E., Tseng, B. S., et al. (2015). Precision-engineering the Pseudomonas aeruginosa genome with two-step allelic exchange. Nat. Protoc. 10, 1820–1841. doi: 10.1038/nprot.2015.115
Jander, G., Rahme, L. G., and Ausubel, F. M. (2000). Positive correlation between virulence of Pseudomonas aeruginosa mutants in mice and insects. J. Bacteriol. 182, 3843–3845. doi: 10.1128/JB.182.13.3843-3845.2000
Kawakami, T., Kuroki, M., Ishii, M., Igarashi, Y., and Arai, H. (2010). Differential expression of multiple terminal oxidases for aerobic respiration in Pseudomonas aeruginosa. Environ. Microbiol. 12, 1399–1412. doi: 10.1111/j.1462-2920.2009.02109.x
Kerscher, S., Dröse, S., Zickermann, V., and Brandt, U. (2008). “The three families of respiratory NADH dehydrogenases, bioenergetics,” in Results and Problems in Cell Differentiation, eds G. Schäfer and H. Penefsky (Heidelberg: Springer Berlin).
Magill, S. S., Edwards, J. R., Bamberg, W., Beldavs, Z. G., Dumyati, G., Kainer, M. A., et al. (2014). Multistate point-prevalence survey of health care-associated infections. N. Engl. J. Med. 370, 1198–1208. doi: 10.1056/NEJMoa1306801
Melvin, J. A., Gaston, J. R., Phillips, S. N., Springer, M. J., Marshall, C. W., Shanks, R. M. Q., et al. (2017). Pseudomonas aeruginosa contact-dependent growth inhibition plays dual role in host-pathogen interactions. mSphere 2, e00336-17. doi: 10.1128/mSphere.00336-17
Raba, D. A., Rosas-Lemus, M., Menzer, W. M., Li, C., Fang, X., Liang, P., et al. (2018). Characterization of the Pseudomonas aeruginosa NQR complex, a bacterial proton pump with roles in autopoisoning resistance. J. Biol. Chem. 293, 15664–15677. doi: 10.1074/jbc.RA118.003194
Schurek, K. N., Marr, A. K., Taylor, P. K., Wiegand, I., Semenec, L., Khaira, B. K., et al. (2008). Novel genetic determinants of low-level aminoglycoside resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 52, 4213–4219. doi: 10.1128/AAC.00507-08
Shanks, R. M., Caiazza, N. C., Hinsa, S. M., Toutain, C. M., and O’Toole, G. A. (2006). Saccharomyces cerevisiae-based molecular tool kit for manipulation of genes from gram-negative bacteria. Appl. Environ. Microbiol. 72, 5027–5036. doi: 10.1128/AEM.00682-06
Spero, M. A., Brickner, J. R., Mollet, J. T., Pisithkul, T., Amador-Noguez, D., and Donohue, T. J. (2016). Different functions of phylogenetically distinct bacterial complex I isozymes. J. Bacteriol. 198, 1268–1280. doi: 10.1128/JB.01025-15
Starkey, M., and Rahme, L. G. (2009). Modeling Pseudomonas aeruginosa pathogenesis in plant hosts. Nat. Protoc. 4, 117–124. doi: 10.1038/nprot.2008.224
Trunk, K., Benkert, B., Quäck, N., Münch, R., Scheer, M., Garbe, J., et al. (2010). Anaerobic adaptation in Pseudomonas aeruginosa: definition of the Anr and Dnr regulons. Environ. Microbiol. 12, 1719–1733. doi: 10.1111/j.1462-2920.2010.02252.x
Wiegand, I., Hilpert, K., and Hancock, R. E. (2008). Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat. Protoc. 3, 163–175. doi: 10.1038/nprot.2007.521
Williams, H. D., Zlosnik, J. E., and Ryall, B. (2007). Oxygen, cyanide and energy generation in the cystic fibrosis pathogen Pseudomonas aeruginosa. Adv. Microb. Physiol. 52, 1–71.
Yen, P., and Papin, J. A. (2017). History of antibiotic adaptation influences microbial evolutionary dynamics during subsequent treatment. PLoS Biol. 15:e2001586. doi: 10.1371/journal.pbio.2001586
Keywords: Pseudomonas aeruginosa, NADH dehydrogenase, NADH:ubiquinone oxidoreductases, metabolism, aminoglycoside resistance
Citation: Torres A, Kasturiarachi N, DuPont M, Cooper VS, Bomberger J and Zemke A (2019) NADH Dehydrogenases in Pseudomonas aeruginosa Growth and Virulence. Front. Microbiol. 10:75. doi: 10.3389/fmicb.2019.00075
Received: 25 September 2018; Accepted: 15 January 2019;
Published: 05 February 2019.
Edited by:
Martin G. Klotz, Washington State University, United StatesReviewed by:
Hiroyuki Arai, The University of Tokyo, JapanOscar Juarez, Illinois Institute of Technology, United States
Davide Zannoni, University of Bologna, Italy
Copyright © 2019 Torres, Kasturiarachi, DuPont, Cooper, Bomberger and Zemke. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Anna Zemke, emVta2VhQHVwbWMuZWR1
 Angela Torres1
Angela Torres1 Matthew DuPont
Matthew DuPont Vaughn S. Cooper
Vaughn S. Cooper Jennifer Bomberger
Jennifer Bomberger Anna Zemke
Anna Zemke