- 1Marine Biology Research Division, Scripps Institution of Oceanography, University of California, San Diego, La Jolla, CA, United States
- 2Center for Marine Biotechnology and Biomedicine, Scripps Institution of Oceanography, University of California, San Diego, La Jolla, CA, United States
- 3Division of Biological Sciences, University of California, San Diego, La Jolla, CA, United States
Various marine gamma-proteobacteria produce omega-3 polyunsaturated fatty acids, such as eicosapentaenoic acid (20:5, EPA) and docosahexaenoic acid (22:6, DHA), which are incorporated into membrane phospholipids. Five genes, designated pfaABCDE, encode the polyketide/fatty acid synthase necessary for production of these long-chain fatty acids. In addition to de novo biosynthesis of EPA and DHA, the “Pfa synthase” is also involved with production of a long-chain polyunsaturated hydrocarbon product (31:9, PUHC) in conjunction with the oleABCD hydrocarbon biosynthesis pathway. In this work, we demonstrate that OleA mediates the linkage between these two pathways in vivo. Co-expression of pfaA-E along with oleA from Shewanella pealeana in Escherichia coli yielded the expected product, a 31:8 ketone along with a dramatic ∼10-fold reduction in EPA content. The decrease in EPA content was independent of 31:8 ketone production as co-expression of an OleA active site mutant also led to identical decreases in EPA content. We also demonstrate that a gene linked with either pfa and/or ole operons in diverse bacterial lineages, herein designated pfaT, plays a role in maintaining optimal production of Pfa synthase derived products in Photobacterium and Shewanella species.
Introduction
The pathway for biosynthesis of omega-3 polyunsaturated fatty acids (PUFAs) in certain marine bacteria occurs via a polyketide synthase type mechanism encoded by five genes pfaABCDE (Metz et al., 2001; Allen and Bartlett, 2002; Shulse and Allen, 2011; Yoshida et al., 2016; Allemann and Allen, 2018). The “Pfa synthase” has been identified in several bacterial lineages and shown to synthesize a variety of PUFA products, most notably the long-chain omega-3 PUFAs, such as eicosapentaenoic acid (20:5n-3, EPA) and docosahexaenoic (22:6n-3) acids (Shulse and Allen, 2011; Yoshida et al., 2016; Allemann and Allen, 2018). The Pfa synthase multienzyme complex contains all of the required enzymatic domains for lipid biosynthesis and these activities reside on either multi-domain or stand-alone proteins (Figure 1). Recombinant production of EPA or DHA in Escherichia coli has been demonstrated by heterologous expression of pfaA-E from various marine bacterial strains (Metz et al., 2001; Okuyama et al., 2007; Amiri-Jami and Griffiths, 2010). Akin to canonical fatty acid biosynthesis, PUFAs are synthesized from 2C malonyl extender units (Metz et al., 2001) and the final product is incorporated into membrane phospholipids of the producing strain (Yazawa, 1996; Yoshida et al., 2016).
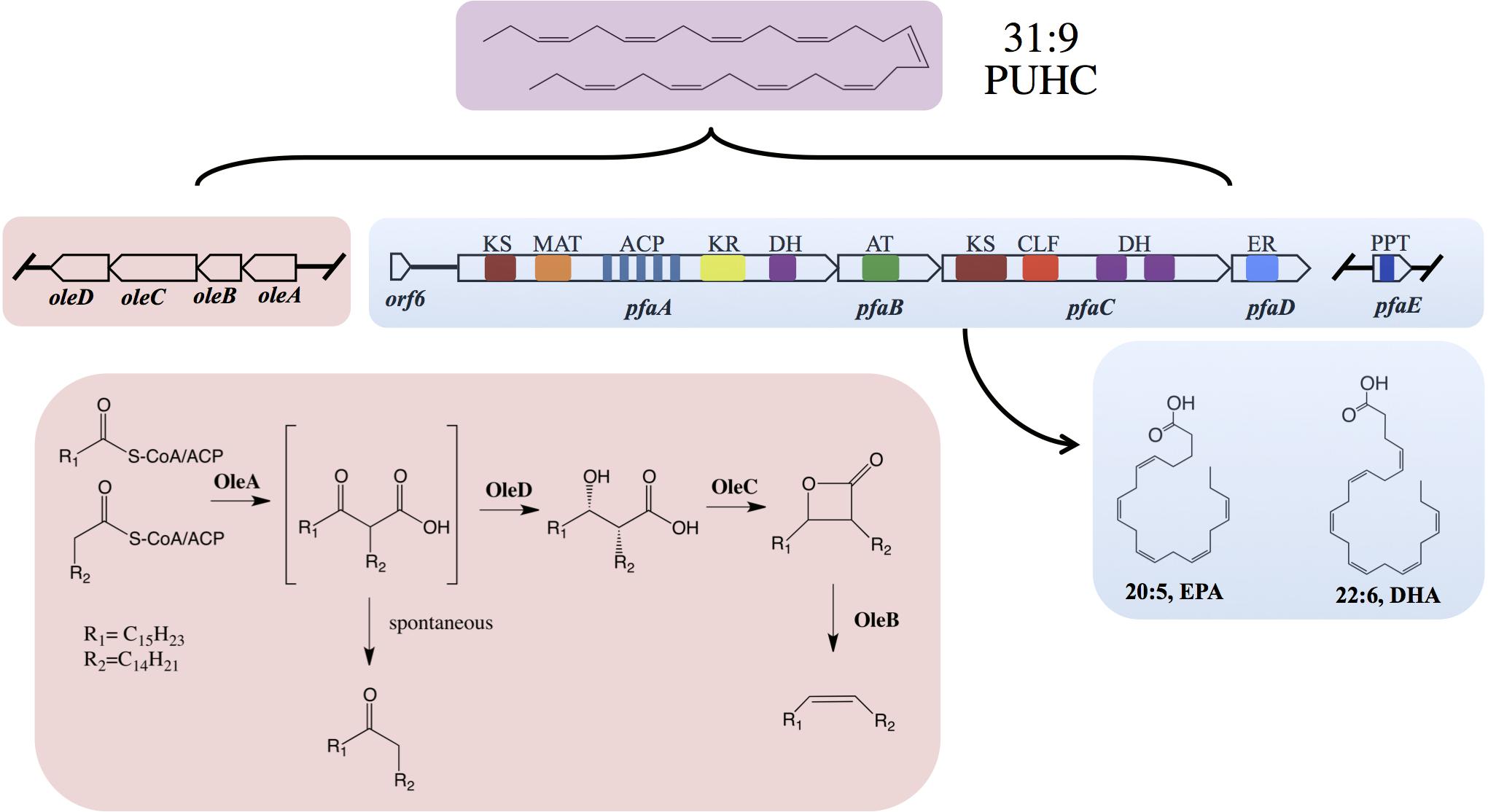
Figure 1. Biochemical and genetic aspects of PUFA and PUHC biosynthetic pathways. Domain designations within the Pfa synthase are; phosphopantetheinyl transferase (PPT), β-ketoacyl synthase (KS), malonyl-CoA:ACP transacylase (MAT), acyl-carrier protein (ACP), ketoacyl reductase (KR), dehydratase/isomerase (DH), acyltransferase (AT), chain-length factor (CLF), and enoyl reductase (ER). Both pfaE and/or oleABCD can be clustered with the pfa operon or found elsewhere in the genome depending on the host organism.
The biosynthesis of long-chain fatty acid-derived olefin hydrocarbons has been previously shown to be the result of the oleABCD pathway in various marine and non-marine bacterial lineages (Beller et al., 2010; Sukovich et al., 2010a,b). In this pathway, hydrocarbons are generated by the condensation of two fatty acyl chains via a “head-to-head” Claisen condensation mechanism, resulting in an intermediate β-keto acid product (Figure 1; Frias et al., 2011; Goblirsch et al., 2016). After condensation, the β-keto acid produced by OleA is processed by OleD, a NADPH-dependent reductase, to form a β-keto alcohol (Bonnett et al., 2011). OleC, a β-lactone synthetase, then generates a β-lactone moiety (Christenson et al., 2017b) which is subsequently de-carboxylated by OleB, a β-lactone decarboxylase, yielding the final olefin product (Christenson et al., 2017c). Recent work has demonstrated that OleBCD together form a large multimeric enzyme complex that processes the β-keto intermediate formed by OleA activity (Christenson et al., 2017a). Weak interactions between the OleBCD complex and OleA in vitro suggests that OleA condenses the precursor acyl groups and possibly transfers the β-keto acid product directly to the OleBCD complex for further processing (Christenson et al., 2017a). In the absence of downstream processing by OleBCD, the OleA catalyzed β-keto acid intermediate undergoes spontaneous decarboxylation to form a ketone product (Sukovich et al., 2010b; Frias et al., 2011; Figure 1).
The precursor fatty acids for olefin biosynthesis are derived from the cellular fatty acid biosynthesis pathway(s) in the producing host (Sukovich et al., 2010a). In strains such as Xanthomonas campestris and Stenotrophomonas maltophilia, the C27–C31 olefin products are derived from the core Type II fatty acid synthase (FAS), which produces saturated, monounsaturated, or branched chain fatty acids (Beller et al., 2010; Sukovich et al., 2010a). However, marine bacteria that synthesize EPA and/or DHA via the Pfa synthase mechanism produce a unique polyunsaturated hydrocarbon (Nichols et al., 1995), hentriacontanonaene (31:9, PUHC), via the OleABCD pathway (Sugihara et al., 2010; Sukovich et al., 2010a,b). Previous genetic experiments in Shewanella oneidensis MR-1 verified that PUHC biosynthesis is dependent on the Pfa synthase, as mutations in the pfa operon led to loss of PUHC production (Sukovich et al., 2010b). Interestingly, despite the presence of the Type II FAS in PUFA-producing bacteria, OleA exclusively condenses a predicted 16:4n-3 acyl chain derived from the Pfa synthase in these species.
Given the biosynthetic linkage of the ole and pfa pathways, it is not surprising that in some strains such as Photobacterium profundum SS9, the two operons are located adjacent to one another on the chromosome (Allen and Bartlett, 2002; Vezzi et al., 2005). Intriguingly, a previously characterized gene encoding an acyl-CoA thioesterase, previously designated orf6, sits between the two operons in P. profundum SS9. Previous structural and biochemical characterization of Orf6 revealed a “hot dog” fold topology with thioesterase activity on various long-chain acyl-CoA substrates in vitro (Rodríguez-Guilbe et al., 2013). Based on its activity and its conservation among EPA/DHA producing bacteria, it was speculated that this thioesterase may be involved with product release from the Pfa synthase (Rodríguez-Guilbe et al., 2013). However, modest rate enhancement of thioesterase activity in vitro raised doubts as to the role of Orf6 in product release (Rodríguez-Guilbe et al., 2013).
In this work we establish the linkage between PUFA and PUHC biosynthesis and show that OleA is responsible for mediating the linkage between the two pathways. Our results indicate that OleA can interact with the Pfa synthase directly, most likely with the acyl carrier protein (ACP) domains that shuttle acyl intermediates, including the 16:4n-3 PUHC precursor molecules, among catalytic domains during de novo PUFA biosynthesis. We also investigated the in vivo role of the orf6 gene in P. profundum SS9 and S. oneidensis MR-1, demonstrating that it is required for optimal biosynthesis of EPA or PUHC in each strain, respectively. Given our results, we have re-designated orf6 as pfaT (pfa-associated thioesterase). Together, these results provide new insight into the genetic and enzymatic determinants involved in the bacterial synthesis of long-chain fatty acid and hydrocarbon products of biotechnological interest.
Materials and Methods
Bacterial Strains and Growth Conditions
A list of strains used in this study is shown in Table 1. E. coli and S. oneidensis MR-1 strains were cultured in Luria Bertani media at 37 and 30°C, respectively, unless noted otherwise. P. profundum SS9R, a rifampin-resistant derivative of wild-type SS9, and Shewanella pealeana strains were cultured in 75% strength 2216 marine broth media (BD Difco, 28 g/L) at 15°C unless noted otherwise. For high hydrostatic pressure growth studies, SS9R strains were grown in heat-sealed bulbs as described previously (Chi and Bartlett, 1993). For analysis of heterologous production of EPA in E. coli, relevant strains were grown at 15°C for 48 h. For solid medias, agar was included at 15 g/L. The antibiotics kanamycin (50 μg/ml for E. coli and S. oneidensis MR-1; 200 μg/ml for P. profundum), chloramphenicol (15 μg/ml), carbenicillin (100 μg/ml), and rifampicin (100 μg/ml) were used as required.
Gene Disruption Mutagenesis
To generate in-frame deletions of genes, an allelic exchange approach was used similar to previous work (Eloe et al., 2008). Briefly, upstream and downstream regions of the gene of interest were amplified with the appropriate primer combinations (5′O, 5′I) and (3′O, 3′I), respectively (Supplementary Table 1). Purified PCR products were assembled using overlap PCR and subsequently amplified with 5′O and 3′O primers. Assembled fragments were then cloned into the suicide vector pRE118 (Edwards et al., 1998) using standard methods (Sambrook et al., 1989). Colony PCR and subsequent DNA sequencing were used to verify constructs.
For conjugation into P. profundum, biparental matings were performed using the E. coli donor strain S17-1λpir containing the desired plasmid to be mobilized into SS9R. Selection for exconjugants was performed on 2216 agar containing rifampicin and kanamycin as described previously (Lauro et al., 2005; Allemann and Allen, 2018). After colony purification of exconjugants, colonies were grown without selection and dilutions plated onto 2216 agar supplemented with 5% sucrose to select for a second recombination event. Colony PCR was used to screen sucrose resistant clones for the targeted deletion.
For conjugation into S. oneidensis MR-1, biparental matings were performed using an E. coli diaminopimelic acid (DAP) auxotroph, WM3064, on LB agar supplemented with DAP, as described previously (Yang et al., 2015). Exconjugants were selected on LB plates containing kanamycin without DAP. Colony purified exconjugants were grown in LB without NaCl for several generations and subsequently plated onto LB supplemented with 5% sucrose for counter-selection. Sucrose resistant colonies were screened for kanamycin sensitivity and colony PCR was used to screen kanamycin sensitive clones for the targeted deletion.
Cloning/Expression Procedures
The pfa operon from S. pealeana was cloned into the pre-linearized pCC2FOS vector following manufacturer guidelines (Epicentre, Madison, WI, United States). Briefly, a fosmid library was constructed from SwaI digested genomic DNA from S. pealeana. Colony PCR using primers listed in Supplementary Table 1 specific to pfaD and pfaE was used to screen clones for presence of the pfa operon. A single clone, which contained the entire cluster was identified and designated 1F12R. Other plasmids were constructed by PCR amplification of indicated genes using the appropriate primer pair containing restriction sites (i.e., pBAD24 SS9 orf 6 F/R) listed in Supplementary Table 1. PCR products were cloned into various plasmids at restriction sites given in Table 1 using standard procedures (Sambrook et al., 1989). Site-directed mutagenesis of OleA was accomplished using PCR mutagenesis primers listed in Supplementary Table 1. Briefly, the plasmid pMA63 was used as a template for PCR using mutagenesis primer pair “OleA C123A mut F/R.” A restriction digest using DpnI removed the original plasmid template and the resulting DNA was transformed into competent cells. DNA sequencing was used to verify the introduced mutation. For expression of genes cloned onto pBAD24 (Guzman et al., 1995) L-arabinose was added. Expression of OleA and Orf6 (PfaT) homologs was confirmed by SDS-PAGE of whole cell lysates followed by Coomassie staining (Sambrook et al., 1989).
Fatty Acid/Neutral Lipid Extraction and Analysis
Late log phase cultures were harvested by centrifugation and cell pellets rinsed once with 50% Sigma Sea Salts solution (16 g/L) and stored at -80°C. Cell pellets were lyophilized prior to fatty acid or hydrocarbon analysis.
For fatty acid analysis, lipids were converted to fatty acid methyl esters (FAME) by adding 5% H2SO4 in methanol directly to lyophilized biomass and refluxing at 90°C for 90 min. After cooling, hexanes were added and non-esterified fatty acids were saponified by addition of 10% NaCl. The hexane extraction was repeated twice and pooled fractions were evaporated completely under a gentle N2 stream and re-dissolved in 1 ml of hexane. Samples were stored at -80°C until analysis.
Hydrocarbon/ketones were extracted from freeze-dried biomass by addition of a mixture of dichloromethane/methanol (2:1, vol/vol) and stirred overnight at room temperature. The crude extract was filtered using a Pasteur pipet packed with glass wool and celite and subsequently dried under a gentle stream of N2 gas. A mixture of hexanes and methanol (4:1, vol:vol) were added to the residue and allowed to form a two-phase mixture. The hexane phase was removed and two additional hexane volumes were added to the methanol phase and subsequently extracted. The hexane fractions were pooled, dried under a stream of N2 and dissolved in 1 ml of hexane. For quantitative analysis hentriacontane (31:0) (Sigma) was added as an internal standard.
Gas chromatography mass spectrometry (GC-MS) analyses were performed on an Agilent Technologies model 7890A GC connected to a 5975C VL MSL quadrupole MS(EI). Samples were separated on a 30 m HP5ms Ultra Inert Agilent GC-MS column using helium as carrier gas. Fatty acid samples were injected in splitless mode and held at 110°C for 3 min followed by a gradient of 15°C/min, and held at a final temperature of 280°C for an additional 3 min. For analysis of neutral lipid extracts, samples were injected in splitless mode and held at 100°C for 3 min followed by a gradient of 15°C/min and held at a final temperature of 300°C for an additional 10 min. Both injector and detector for the mass spectrometer were maintained at 250°C. Additional MS operating conditions were as follows: mass range 50–500 atomic mass units, 3 min solvent delay. Peak areas were quantified and mass spectra processed using ChemStation software (Agilent Technologies). FAMEs were identified by comparing MS fragmentation patterns to spectra from authentic standards or from spectra on the NIST 2008 Spectral Library.
Phylogenetic Analysis
Homolog amino acid sequences were obtained from a gene neighborhood search on the Joint Genome Institute Integrated Microbial Genome web portal (accessed on October 16, 2018). Sequences were uploaded to the Phylogeny.fr web portal and a maximum likelihood phylogenetic tree was constructed as described previously (Dereeper et al., 2008). For tree construction bootstrap values were generated from 100 re-samplings of the data.
Results
Analysis of EPA and PUHC content in S. pealeana and S. oneidensis MR-1 as a function of growth temperature displayed the trend of both compounds increasing in abundance as temperature decreased (Figure 2). This is consistent with previous findings regarding EPA (Allen et al., 1999; Okuyama et al., 2007; Yoshida et al., 2016) and PUHC (Sukovich et al., 2010b) content as a function of growth temperature. Given the relationship between the ole and pfa pathways, increased flux through the Pfa synthase is predicted to result in a concurrent increase in PUHC production. Conversely, reducing flux into the ole pathway is predicted to lead to an increase in the pfa pathway. Given the relationship between the two pathways, it was hypothesized that previously observed differences in EPA content amongst members of the Shewanella genus (Kato and Nogi, 2001), may be due to the diversion of acyl substrates from the EPA pathway into the PUHC pathway. Deletion of the entire ole operon resulted in a modest ∼twofold increase in EPA content relative to wild-type S. oneidensis MR-1 at 15°C (Supplementary Table 2).
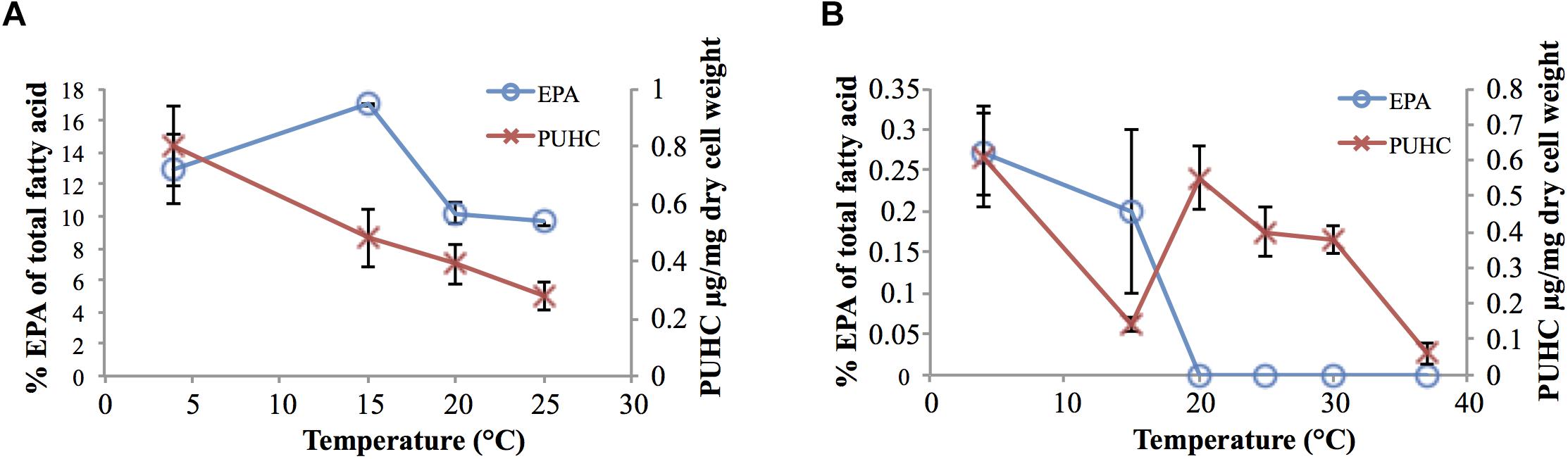
Figure 2. EPA and PUHC content as a function of temperature in (A) S. pealeana and (B) S. oneidensis MR-1. Error bars represent standard deviations based on three biological replicates.
To gain a better understanding of how the ole and pfa pathways interact, we performed combinatorial co-expression experiments in E. coli. The pfa operon was cloned from S. pealeana ATCC700345 onto pCC2FOS to yield the construct 1F12R. The pfa operon from S. pealeana was chosen due to its high EPA and PUHC production phenotypes in the native strain. Transformation of 1F12R into the E. coli strain JW1794 yielded strain MAE21 that produced EPA at ∼21% of total fatty acid when cultured at 15°C (Supplementary Table 3). The fatty acid profile of a control strain containing pCC2FOS vector only did not produce EPA. Given that OleA catalyzes the first committed step of hydrocarbon biosynthesis in the OleABCD pathway, we speculated that co-expression of OleA from S. pealeana might impact EPA production in MAE21. The OleA homolog in S. pealeana was successfully cloned under an arabinose inducible promoter (pMA63) and shown to produce a protein of the expected size (38 kDa) in MAE21 upon induction with L-arabinose (Supplementary Figure 1). Fatty acid analysis of the MAE21 strain co-expressing OleA (pMA63) indicated a dramatic ∼10-fold decrease in EPA content relative to the vector only control strain (Figure 3A). Attempts to titrate this effect with varying amounts of L-arabinose were unsuccessful (data not shown) and are most likely due to our inability to effectively titrate expression of the pBAD promoter under the growth conditions employed. Site-directed mutagenesis of the universally conserved OleA catalytic cysteine residue to alanine (C123A) was employed to determine if catalytically active OleA is required for this reduced EPA phenotype. Under identical conditions, co-expression of mutant OleAC123A (pMA70) in MAE21 yielded nearly identical results as seen for OleAWT (pMA63) (Figure 3A).
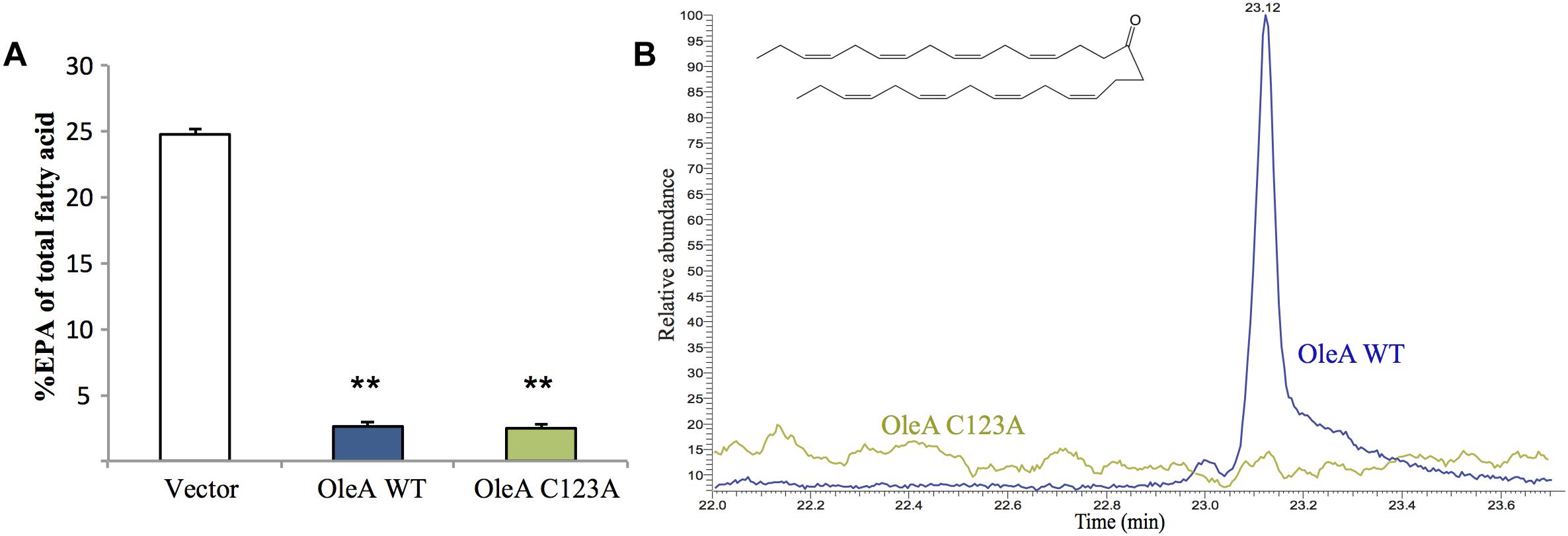
Figure 3. Expression of OleA in E. coli MAE21 strain harboring fosmid 1F12R with the pfa operon from S. pealeana leads to production of a 31:8 ketone and reduced EPA. (A) EPA content of MAE21 strains decreases upon induction of OleAWT (pMA63) and OleAC123A (pMA70) with 0.05% L-Arabinose. Bars represent averages of at least three biological replicates with error bars signifying one standard deviation (∗∗P < 0.005). (B) Total ion chromatograms (TIC) of neutral lipid extracts from MAE21 OleAWT (pMA63 – blue) and MAE21 OleAC123A (pMA70 – green). 31:8 ketone peak seen at 23.1 min.
Previous work in S. oneidensis MR-1 had shown that expression of oleA alone without oleBCD led to production of a polyunsaturated ketone (31:8), which is the result of spontaneous decarboxylation of the OleA β-keto acid product (Figure 1; Sukovich et al., 2010a,b; Frias et al., 2011). Neutral lipid extracts of MAE21 containing OleAWT (pMA63) and mutant OleAC123A (pMA70) were analyzed by GC-MS. A peak at 23.1 min corresponding to the 31:8 ketone was found in the wild-type OleA (pMA63) containing strain but not in the mutant OleAC123A (pMA70) containing strain (Figure 3B). Mass spectra associated with the peak at 23.1 min also matched the spectra of the 31:8 ketone produced by S. oneidensis Δole strain containing pOleA (Supplementary Figures 2A,B). MAE21 is a derivative of JW1794, which contains a ΔfadD::kan mutation, rendering the strain unable to produce acyl-CoA from free fatty acids (Cronan, 1997; Black and DiRusso, 2003). The appearance of the 31:8 ketone in the MAE21 strain background indicates that OleA can condense the appropriate acyl products in the absence of acyl-CoA synthetase activity.
Given its clustering with the pfa and/or ole operons in various EPA/DHA producing strains, pfaT (previously orf6) and its orthologs could be involved with one or both biosynthetic pathways. A search for homologs of pfaT was conducted and a protein sequence phylogenetic tree is shown in Figure 4. All of the included species are either known PUFA producers and/or contain both pfa and ole operons. With the exception of the Shewanella and Psychromonas species, all pfaT homologs were found to be genetically linked with either pfa or ole operons. All homologs shown in the tree contain the active site aspartate residue previously described to be essential for thioesterase activity (Rodríguez-Guilbe et al., 2013).
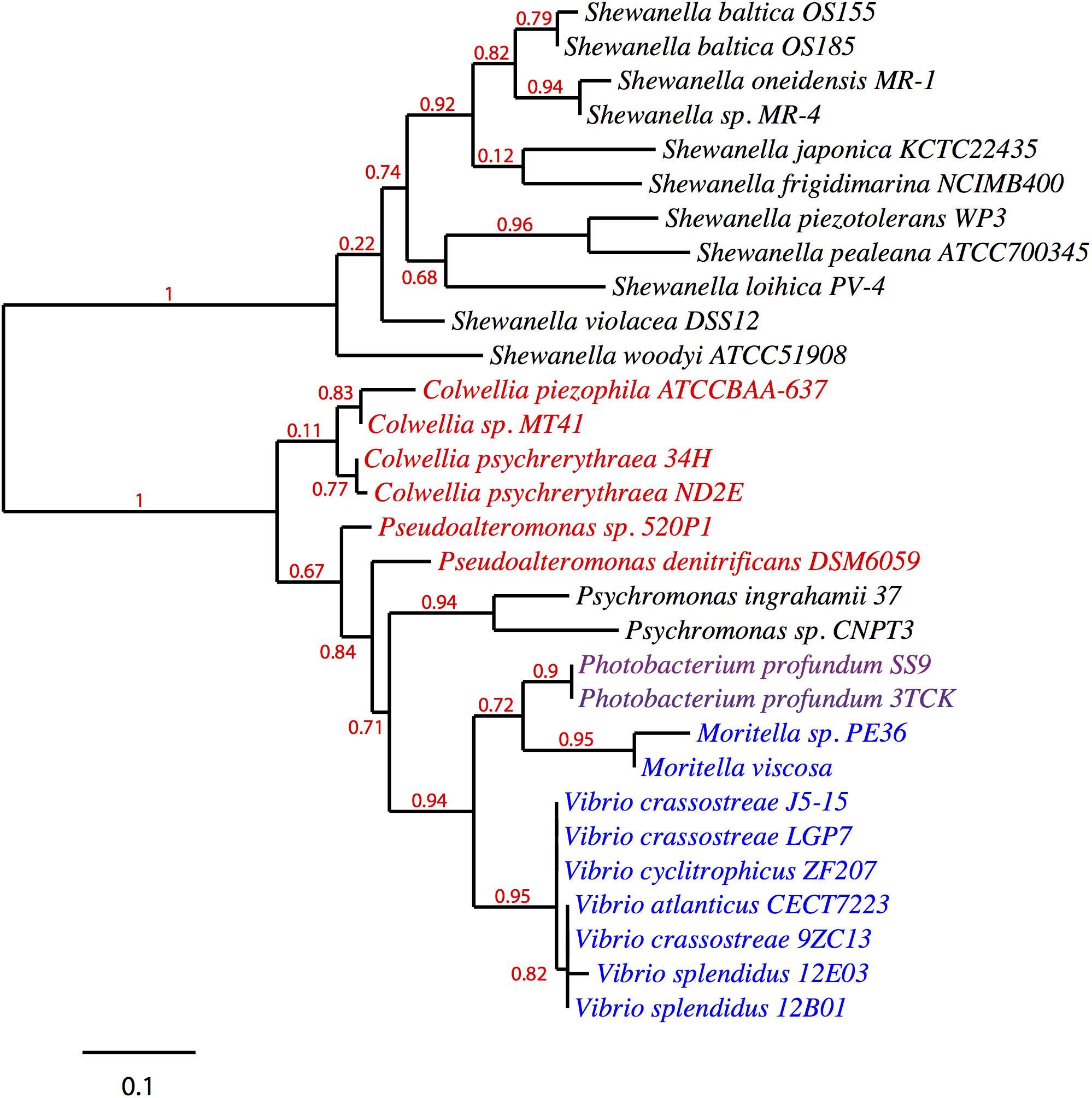
Figure 4. Phylogenetic distribution of PfaT homologs. A maximum likelihood unrooted tree of PfaT homologs found in bacteria with pfa and/or ole pathways. PfaT homologs clustered with the pfa operon are indicated in blue, homologs clustered with ole operon in red text, and clustering between the operons in purple. Numbers at branch points indicate bootstrap values based on 100 replicates.
A marker-less in-frame deletion of pfaT was generated in P. profundum SS9 and the resulting strain, MAP1, was analyzed for EPA content under a variety of culture conditions. As shown in Figure 5A, MAP1 produced ∼fourfold less EPA compared to its parental strain SS9R at 15°C. This reduction in EPA content was also seen during growth at low temperature (4°C) and at high hydrostatic pressure (30 MPa) (Figure 5A), culture conditions that elicit increased EPA content in wild-type SS9 (Allen et al., 1999). The decreased EPA phenotype was also complemented in trans by a construct (pMA20) containing pfaT under control of its native promoter (Figure 5B). Full fatty acid profiles of SS9R and MAP1 along with the corresponding genetically complemented strains and controls are given in Supplementary Tables 4, 5, respectively. The ability to complement the pfaT mutation in strain MAP1 also confirms that the observed decrease in EPA is due to the loss of pfaT and not a polar effect on transcription of the pfa operon. Co-expression of orf6 from P. profundum SS9R (pMA47) or the homolog from S. pealeana (pMA48) in the recombinant EPA-producing E. coli strain MAE21 did not lead to alterations in EPA content or changes in overall fatty acid profile (Supplementary Table 3). Furthermore, expression of both homologs in strain MAE21 or JW1794 did not lead to accumulation of free fatty acids (data not shown).
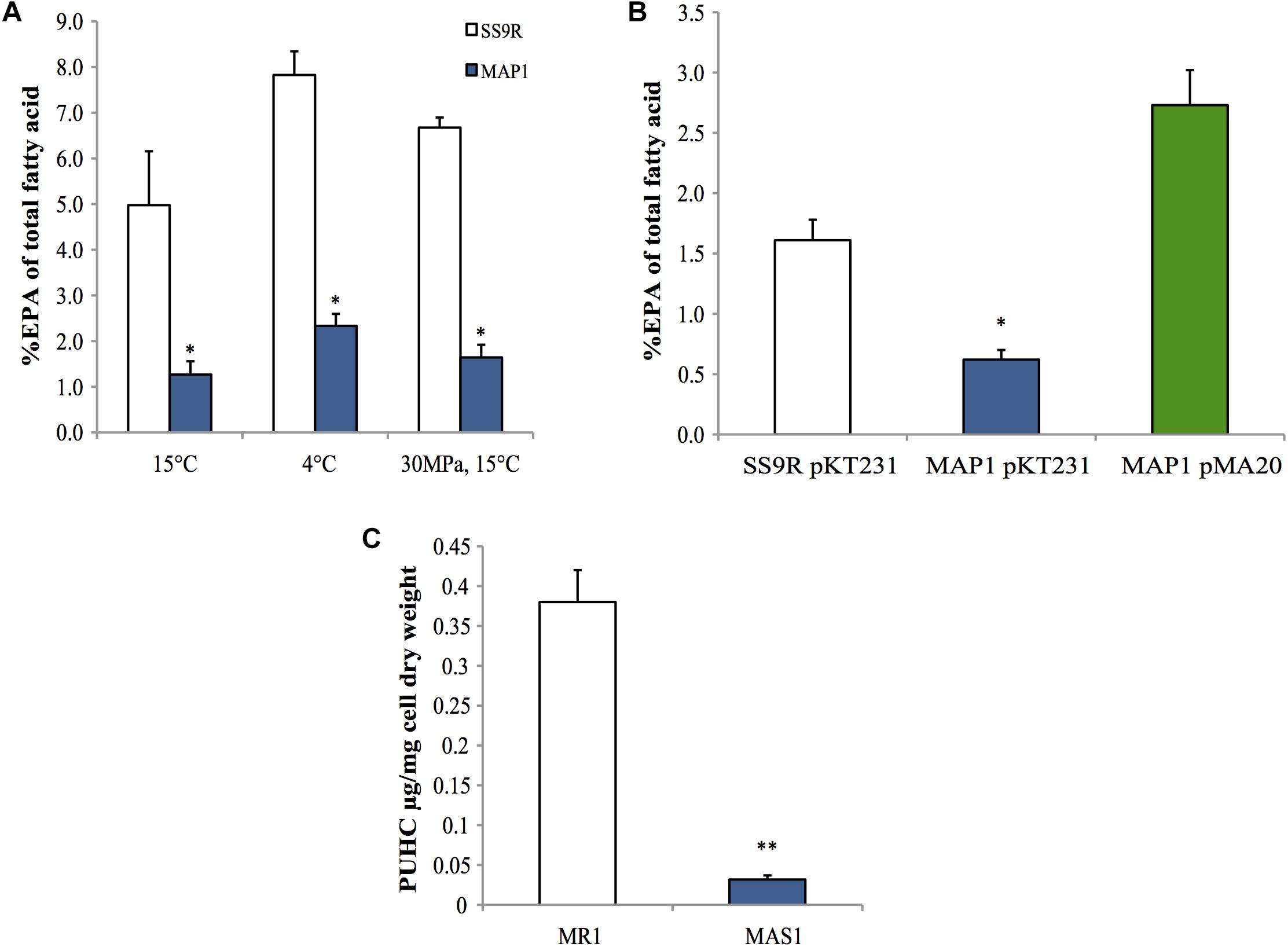
Figure 5. Phenotypes of pfaT deletion mutants in P. profundum SS9 and S. oneidensis MR-1. (A) An approximate fourfold decrease in EPA composition in P. profundum strain MAP1 harboring an in-frame deletion of pfaT relative to SS9R under various culturing conditions. (B) Complementation analysis. Comparison of the amount of EPA as a percentage of total fatty acids among parent strain SS9R harboring empty complementation plasmid pKT231, P. profundum strain MAP1 harboring empty complementation plasmid pKT231, and P. profundum strain MAP1 harboring plasmid pMA20 containing pfaT under control of its native promoter. Cells grown at 15°C as in A. Bars represent averages of at least three experimental replicates and error bars represent one standard deviation. (C) PUHC content of S. oneidensis strain MAS1 (harboring an in-frame deletion of the pfaT homolog) and parent strain S. oneidensis MR-1 at 30°C. Bars represent averages of three biological replicates and error bars represent one standard deviation (∗P < 0.05; ∗∗P < 0.005).
Surprisingly, neither MAP1 nor SS9R produced PUHC at a detectable level despite the presence of the oleABCD operon (data not shown). Given our previous characterization of S. oneidensis MR-1 and its genetic tractability, we generated an in-frame deletion of the pfaT homolog (locus tag SO1256) in S. oneidensis MR-1 (MAS1). Neutral lipid extracts of this strain indicated a drastic ∼10-fold reduction in PUHC content relative to the parental strain under identical conditions (Figure 5C). Fatty acid profiles of S. oneidensis MR-1 and MAS1 grown at 15°C are shown in Supplementary Table 2 and a similar fourfold decrease in EPA was observed.
Discussion
In this work we have demonstrated that bacterial PUFA and PUHC biosynthesis are linked and this linkage is mediated by OleA. Our initial work with S. pealeana, which produces both PUFA and PUHC products, indicated that culture conditions which lead to increases in PUFA lead to commensurate increases in PUHC. While the physiological role of PUHC remains obscure, this result suggests that PUHC may play an additional role in adaptation to cold and/or high-pressure environments, conditions that impact membrane physical structure, e.g., fluidity. The modest EPA content in the Δole strain (<1% of total fatty acid) indicates that other factors in the biosynthesis and incorporation of EPA into membrane phospholipids are responsible for the previously observed differences in EPA production amongst members of the Shewanella genus (Kato and Nogi, 2001).
Our results also indicate that the in vivo OleA substrate is an acyl-ACP and not an acyl-CoA as described previously (Frias et al., 2011; Goblirsch et al., 2016). Our heterologous expression system in E. coli demonstrated that co-expressing oleA along with pfaABCDE was sufficient for producing the expected 31:8 ketone previously observed in S. oneidensis MR-1 mutants (Sukovich et al., 2010a). The nearly identical reduction in EPA content associated with co-expression of wild-type OleA or the OleAC123A mutant suggests that OleA interacts directly with the ACP domains of the Pfa synthase and that this reduction is not a result of acyl groups being removed from the synthase. Rather, the reduction in EPA production may instead reflect steric competition for the ACP domains between OleA and the various catalytic domains on the Pfa synthase. From a biosynthetic standpoint, such direct ACP interactions would be the most efficient method for obtaining acyl groups for OleA condensation. Other routes of diverting substrates such as thioester cleavage would require acyl-CoA synthetase activity and would consume ATP to regenerate the acyl-CoA needed for condensation by OleA. Previous work indicated that OleA and its homologs specifically act on acyl chains derived from either the Type II FAS or the Pfa synthase (Sukovich et al., 2010a; Frias et al., 2011). Protein–protein interactions between OleA and its cognate ACP may be the mechanism for this substrate specificity.
In this work the in vivo role of the previously characterized orf6 thioesterase, now designated pfaT, was investigated. While pfaT was not completely essential to EPA biosynthesis in P. profundum SS9, it was required for wild-type production levels. Similarly, the pfaT homolog in S. oneidensis MR-1 was not essential to PUHC biosynthesis; rather it was required for optimal biosynthesis of PUHC. Reduction in both end products of the Pfa synthase indicates that PfaT is not involved in a process specific to either pathway alone. Rather, these results suggest that PfaT may function as a Type II thioesterase with activity upon the Pfa synthase. Type II thioesterases serve accessory roles in removing aberrant intermediates or starter units from polyketide synthases and their genetic disruption typically leads to a reduction in the final polyketide product (Kotowska and Pawlik, 2014). Intriguingly, co-expression of various pfaT homologs in the EPA-producing E. coli strain MAE21 did not lead to modulation in EPA production. Previous studies in E. coli have described accumulation of free fatty acids in the culture media or intracellularly in response to expression of acyl-CoA/ACP thioesterases, particularly in strains in which β-oxidation is non-functional (Cho and Cronan, 1995; Lennen et al., 2011; Zhang et al., 2011). In all instances, expression of pfaT homologs in MAE21 or JW1794, did not lead to any free fatty acid accumulation in the media or the appearance of novel fatty acids. This discrepancy between results from the heterologous host E. coli, wherein omission or co-expression of PfaT led to no changes in EPA production, and native strains in which genetic disruption of PfaT led to decreases in both EPA and PUHC suggests that PfaT performs a function specific to biosynthesis of PUFA and/or PUHC in the native producing strains only. The relatively low in vitro thioesterase activity of PfaT (Orf6) reported previously (Rodríguez-Guilbe et al., 2013), along with phenotypic data presented herein is suggestive of PfaT acting as a type II thioesterase.
From the data presented, a model depicting the interaction between PUFA and PUHC biosynthesis is depicted in Figure 6. During EPA biosynthesis, a 16:4n-3 acyl group, a predicted intermediate of the biosynthetic pathway, is processed by OleA via direct interaction with the ACP domains on PfaA. After condensation, the β-keto acid product is further processed by the activities of the OleBCD complex to form the final 31:9 PUHC product (Figure 1). The absence of a significant increase in EPA production in the S. oneidensis ΔoleABCD strain relative to other members of the Shewanella genus indicates that diversion of intermediates to PUHC biosynthesis is not strictly responsible for the differences in EPA phenotypes observed previously (Kato and Nogi, 2001). The relative activities of the Pfa synthase between strains might be a possible mechanism for the observed differences in EPA production potential among Shewanella species.
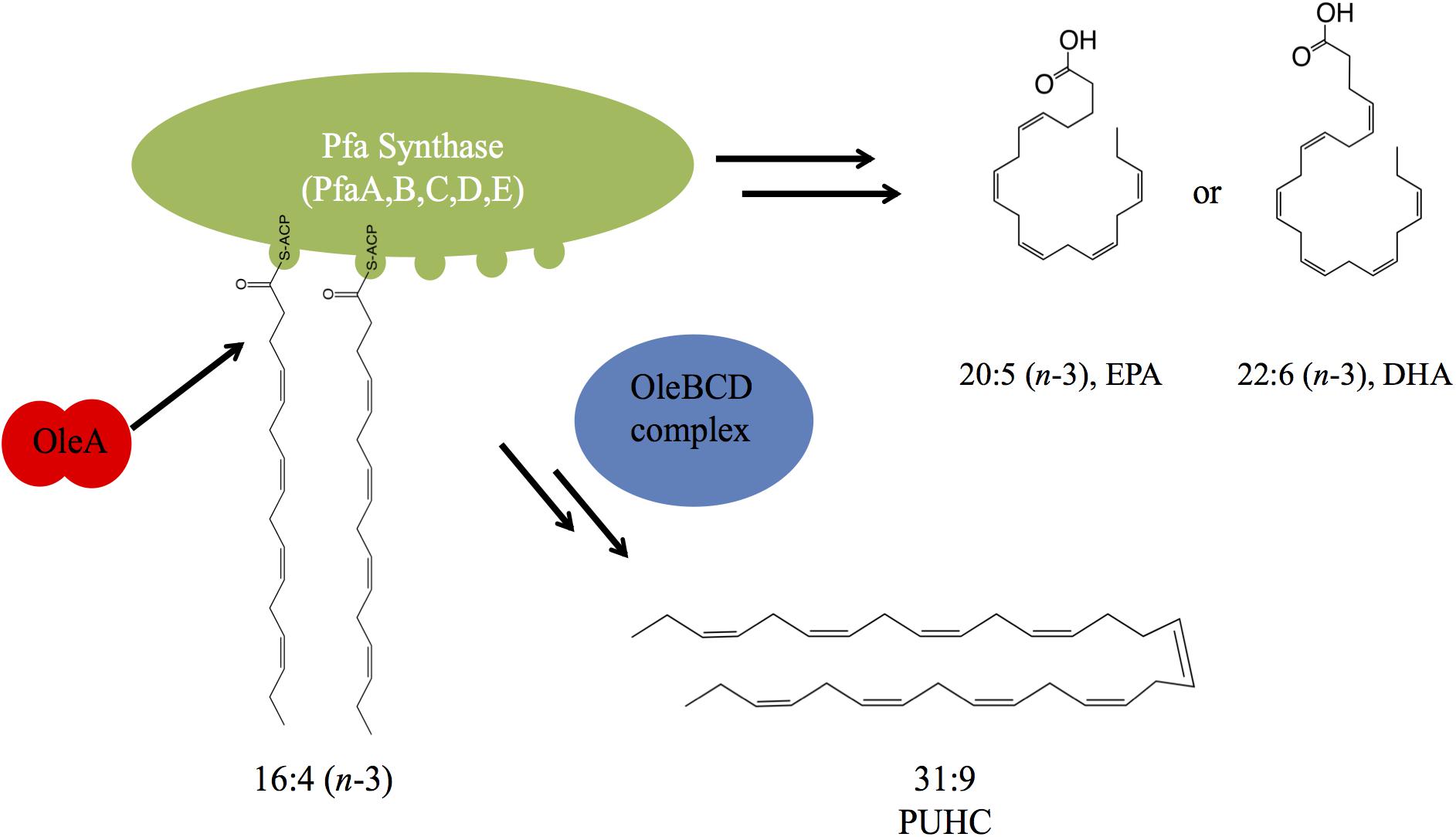
Figure 6. A model depicting the interaction between the Pfa synthase and Ole biosynthetic pathways. OleA interacts with the ACP domain(s) carrying a 16:4 acyl group. Condensation of two fatty acids is followed by further biosynthetic processing by the OleBCD complex to yield the 31:9 PUHC.
OleA is a member of the thiolase protein family which have been shown to utilize either acyl-CoA and/or acyl-ACP substrates (Cronan and Rock, 2008; Goblirsch et al., 2012). Patent literature describing OleA from S. maltophilia demonstrated that OleA utilizes both acyl-CoA and acyl-ACP substrates in vitro (Friedman and Da Costa, 2008). Detailed examination of previously published crystal structures of OleA (Goblirsch et al., 2012, 2016) from X. campestris showed that the pantetheine channel entrance contains a cluster of positively charged residues, which form a “positive patch.” This positive patch feature is found in many fatty acid biosynthesis enzymes, which are known to interact with ACP (Zhang et al., 2001; Finzel et al., 2015). While there is no structural data corresponding to OleA homologs from any PUHC producers it is expected that a similar positive patch feature would be present.
Polyunsaturated fatty acids and PUHC biosynthesis are intrinsically linked and the results of this work have identified that OleA mediates this linkage in vivo. Notably, the results presented here suggest that OleA is capable of interacting with the Pfa synthase in vivo and that the acyl substrates are derived directly from the ACP domains of the synthase. This work has also more clearly defined the role of PfaT, not as a thioesterase for final product release, but rather as an accessory enzyme whose activity is required for optimal production of Pfa synthase derived end products including omega-3 PUFAs and long-chain olefin hydrocarbons.
Author Contributions
MA, CS, and EA conceived the idea, designed the project, and contributed to the discussion and critical writing. MA and CS carried out the experiments and collected and analyzed the data. MA wrote the manuscript.
Funding
This work was supported by National Science Foundation Division of Molecular and Cellular Biosciences grant MCB-1149552 to EA.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to Dr. Larry Wackett for providing us with the Shewanella oneidensis MR-1 ΔoleABCD strain along with the pOleA construct. We would like to thank Dr. Doug Bartlett for various strains/plasmids and advice on constructing mutants of Photobacterium profundum SS9. GC-MS analyses for Figure 2 were carried out in Dr. Lihini Aluwihare’s laboratory at Scripps Institution of Oceanography with expert advice provided by Nellie Shaul. We would also like to thank Dr. Kenneth Nealson and Andrea Cheung for providing the wild-type Shewanella oneidensis MR-1 used in this study.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.00702/full#supplementary-material
References
Allemann, M. N., and Allen, E. E. (2018). Characterization and application of marine microbial omega-3 polyunsaturated fatty acid synthesis. Methods Enzymol. 605, 3–32. doi: 10.1016/bs.mie.2018.02.018
Allen, E. E., and Bartlett, D. H. (2002). Structure and regulation of the omega-3 polyunsaturated fatty acid synthase genes from the deep-sea bacterium Photobacterium profundum strain SS9. Microbiology 148, 1903–1913. doi: 10.1099/00221287-148-6-1903
Allen, E. E., Facciotti, D., and Bartlett, D. H. (1999). Monounsaturated but not polyunsaturated fatty acids are required for growth of the deep-sea bacterium Photobacterium profundum SS9 at high pressure and low temperature. Appl. Environ. Microbiol. 65, 1710–1720.
Amiri-Jami, M., and Griffiths, M. W. (2010). Recombinant production of omega-3 fatty acids in Escherichia coli using a gene cluster isolated from Shewanella baltica MAC1. J. Appl. Microbiol. 109, 1897–1905. doi: 10.1111/j.1365-2672.2010.04817.x
Bagdasarian, M. M., Lurz, R., Rückert, B., Franklin, F. C., Bagdasarian, M. M., Frey, J., et al. (1981). Specific-purpose plasmid cloning vectors II. Broad host range, high copy number, RSF1010-derived vectors, and a host-vector system for gene cloning in Pseudomonas. Gene 16, 237–247. doi: 10.1016/0378-1119(81)90080-9
Beller, H. R., Goh, E.-B., and Keasling, J. D. (2010). Genes involved in long-chain alkene biosynthesis in Micrococcus luteus. Appl. Environ. Microbiol. 76, 1212–1223. doi: 10.1128/AEM.02312-09
Better, M., and Helinski, D. R. (1983). Isolation and characterization of the recA gene of Rhizobium meliloti. J. Bacteriol. 155, 311–316.
Black, P. N., and DiRusso, C. C. (2003). Transmembrane movement of exogenous long-chain fatty acids: proteins, enzymes, and vectorial esterification. Microbiol. Mol. Biol. Rev. 67, 454–472. doi: 10.1128/MMBR.67.3.454-472.2003
Bonnett, S. A., Papireddy, K., Higgins, S., del Cardayre, S., and Reynolds, K. A. (2011). Functional characterization of an NADPH dependent 2-alkyl-3-ketoalkanoic acid reductase involved in olefin biosynthesis in Stenotrophomonas maltophilia. Biochemistry 50, 9633–9640. doi: 10.1021/bi201096w
Chi, E., and Bartlett, D. H. (1993). Use of a reporter gene to follow high pressure signal transduction in the deep-sea bacterium Photobacterium sp.strain SS9. J. Bacteriol. 175, 7533–7540. doi: 10.1128/jb.175.23.7533-7540.1993
Cho, H., and Cronan, J. E. (1995). Defective export of a periplasmic enzyme disrupts regulation of fatty acid synthesis. J. Biol. Chem. 270, 4216–4219. doi: 10.1074/jbc.270.9.4216
Christenson, J. K., Jensen, M. R., Goblirsch, B. R., Mohamed, F., Zhang, W., Wilmot, C. M., et al. (2017a). Active multienzyme assemblies for long-chain olefinic hydrocarbon biosynthesis. J. Bacteriol. 199, 1–11. doi: 10.1128/JB.00890-16
Christenson, J. K., Richman, J. E., Jensen, M. R., Neufeld, J. Y., Wilmot, C. M., and Wackett, L. P. (2017b). β-Lactone synthetase found in the olefin biosynthesis pathway. Biochemistry 56, 348–351. doi: 10.1021/acs.biochem.6b01199
Christenson, J. K., Robinson, S. L., Engel, T. A., Richman, J. E., Kim, A. N., and Wackett, L. P. (2017c). OleB from bacterial hydrocarbon biosynthesis is a β-lactone decarboxylase that shares key features with haloalkane dehalogenases. Biochemistry 56, 5278–5287. doi: 10.1021/acs.biochem.7b00667
Cronan, J. E. (1997). In vivo evidence that acyl coenzyme a regulates DNA binding by the Escherichia coli FadR global transcription factor. J. Bacteriol. 179, 1819–1823. doi: 10.1128/jb.179.5.1819-1823.1997
Cronan, J. E., and Rock, C. O. (2008). Biosynthesis of membrane lipids. EcoSal Plus 3, 1–44. doi: 10.1128/ecosalplus.3.6.4
Dereeper, A., Guignon, V., Blanc, G., Audic, S., Buffet, S., Chevenet, F., et al. (2008). Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 36, W465–W469. doi: 10.1093/nar/gkn180
Edwards, R. A., Keller, L. H., and Schifferli, D. M. (1998). Improved allelic exchange vectors and their use to analyze 987P fimbria gene expression. Gene 207, 149–157. doi: 10.1016/S0378-1119(97)00619-7
Eloe, E. A., Lauro, F. M., Vogel, R. F., and Bartlett, D. H. (2008). The Deep-Sea Bacterium Photobacterium profundum SS9 utilizes separate flagellar systems for swimming and swarming under high-pressure conditions. Appl. Environ. Microbiol. 74, 6298–6305. doi: 10.1128/AEM.01316-08
Finzel, K., Lee, D. J., and Burkart, M. D. (2015). Using modern tools to probe the structure-function relationship of fatty acid synthases. Chembiochem 16, 528–547. doi: 10.1002/cbic.201402578
Frias, J. A., Richman, J. E., Erickson, J. S., and Wackett, L. P. (2011). Purification and characterization of OleA from Xanthomonas campestris and demonstration of a non-decarboxylative claisen condensation reaction. J. Biol. Chem. 286, 10930–10938. doi: 10.1074/jbc.M110.216127
Friedman, L., and Da Costa, B. (2008). Hydrocarbon-Producing Genes and Methods of their Use. International Patent WO 2008/147781 A2.
Goblirsch, B. R., Frias, J. A., Wackett, L. P., and Wilmot, C. M. (2012). Crystal structures of Xanthomonas campestris OleA reveal features that promote head-to-head condensation of two long-chain fatty acids. Biochemistry 51, 4138–4146. doi: 10.1021/bi300386m
Goblirsch, B. R., Jensen, M. R., Mohamed, F., Wackett, L. P., and Wilmot, C. M. (2016). Substrate trapping in crystals of the thiolase OleA identifies three channels that enable long-chain olefin biosynthesis. J. Biol. Chem. 291, 26698–26706. doi: 10.1074/jbc.M116.760892
Guzman, L. M., Belin, D., Carson, M. J., and Beckwith, J. (1995). Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177, 4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995
Heidelberg, J. F., Paulsen, I. T., Nelson, K. E., Gaidos, E. J., Nelson, W. C., Read, T. D., et al. (2002). Genome sequence of the dissimilatory metal ion-reducing bacterium Shewanella oneidensis. Nat. Biotechnol. 20, 1118–1123. doi: 10.1038/nbt749
Kato, C., and Nogi, Y. (2001). Correlation between phylogenetic structure and function: examples from deep-sea Shewanella. FEMS Microbiol. Ecol. 35, 223–230. doi: 10.1111/j.1574-6941.2001.tb00807.x
Kotowska, M., and Pawlik, K. (2014). Roles of type II thioesterases and their application for secondary metabolite yield improvement. Appl. Microbiol. Biotechnol. 98, 7735–7746. doi: 10.1007/s00253-014-5952-8
Lauro, F. M., Eloe, E. A., Liverani, N., Bertoloni, G., and Bartlett, D. H. (2005). Conjugal vectors for cloning, expression, and insertional mutagenesis in gram-negative bacteria. Biotechniques 38, 708–712. doi: 10.2144/05385BM06
Lennen, R. M., Kruziki, M. A., Kumar, K., Zinkel, R. A., Burnum, K. E., Lipton, M. S., et al. (2011). Membrane stresses induced by overproduction of free fatty acids in Escherichia coli. Appl. Environ. Microbiol. 77, 8114–8128. doi: 10.1128/AEM.05421-11
Metz, J. G., Roessler, P., Facciotti, D., Levering, C., Dittrich, F., Lassner, M., et al. (2001). Production of polyunsaturated fatty acids by polyketide synthases in both prokaryotes and eukaryotes. Science 293, 290–293. doi: 10.1126/science.1059593
Nichols, D. S., Nichols, P. D., and McMeekin, T. A. (1995). A new n-C 31:9 polyene hydrocarbon from Antarctic bacteria. FEMS Microbiol. Lett. 125, 281–285. doi: 10.1111/j.1574-6968.1995.tb07369.x
Okuyama, H., Orikasa, Y., Nishida, T., Watanabe, K., and Morita, N. (2007). Bacterial genes responsible for the biosynthesis of eicosapentaenoic and docosahexaenoic acids and their heterologous expression. Appl. Environ. Microbiol. 73, 665–670. doi: 10.1128/AEM.02270-06
Rodríguez-Guilbe, M., Oyola-Robles, D., Schreiter, E. R., and Baerga-Ortiz, A. (2013). Structure, activity, and substrate selectivity of the Orf6 thioesterase from Photobacterium profundum. J. Biol. Chem. 288, 10841–10848. doi: 10.1074/jbc.M112.446765
Saltikov, C. W., and Newman, D. K. (2003). Genetic identification of a respiratory arsenate reductase. Proc. Natl. Acad. Sci. U.S.A. 100, 10983–10988. doi: 10.1073/pnas.1834303100
Sambrook, J., Fritsch, E. F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory.
Shulse, C. N., and Allen, E. E. (2011). Widespread occurrence of secondary lipid biosynthesis potential in microbial lineages. PLoS One 6:e20146. doi: 10.1371/journal.pone.0020146
Simon, R., Priefer, U., and Pühler, A. (1983). A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. BioTechnology 1, 784–791. doi: 10.1038/nbt1183-784
Sugihara, S., Hori, R., Nakanowatari, H., Takada, Y., Yumoto, I., Morita, N., et al. (2010). Possible biosynthetic pathways for all cis-3,6,9,12,15,19,22, 25,28-hentriacontanonaene in bacteria. Lipids 45, 167–177. doi: 10.1007/s11745-009-3380-9
Sukovich, D. J., Seffernick, J. L., Richman, J. E., Gralnick, J. A., and Wackett, L. P. (2010a). Widespread head-to-head hydrocarbon biosynthesis in bacteria and role of OleA. Appl. Environ. Microbiol. 76, 3850–3862. doi: 10.1128/AEM.00436-10
Sukovich, D. J., Seffernick, J. L., Richman, J. E., Hunt, K. A., Gralnick, J. A., and Wackett, L. P. (2010b). Structure, function, and insights into the biosynthesis of a head-to-head hydrocarbon in Shewanella oneidensis strain MR-1. Appl. Environ. Microbiol. 76, 3842–3849. doi: 10.1128/AEM.00433-10
Vezzi, A., Campanaro, S., D’Angelo, M., Simonato, F., Vitulo, N., Lauro, F. M., et al. (2005). Life at depth: Photobacterium profundum genome sequence and expression analysis. Science 307, 1459–1461. doi: 10.1126/science.1103341
Yang, X. W., Jian, H. H., and Wang, F. (2015). pSW2, a novel low-temperature-inducible gene expression vector based on a filamentous phage of the deep-sea bacterium Shewanella piezotolerans WP3. Appl. Environ. Microbiol. 81, 5519–5526. doi: 10.1128/AEM.00906-15
Yazawa, K. (1996). Production of eicosapentaenoic acid from marine bacteria. Lipids 31, S297–S300. doi: 10.1007/BF02637095
Yoshida, K., Hashimoto, M., Hori, R., Adachi, T., Okuyama, H., Orikasa, Y., et al. (2016). Bacterial long-chain polyunsaturated fatty acids: their biosynthetic genes, functions, and practical use. Mar. Drugs 14:94. doi: 10.3390/md14050094
Zhang, X., Li, M., Agrawal, A., and San, K.-Y. (2011). Efficient free fatty acid production in Escherichia coli using plant acyl-ACP thioesterases. Metab. Eng. 13, 713–722. doi: 10.1016/j.ymben.2011.09.007
Keywords: hydrocarbon, thioesterase, omega-3 polyunsaturated fatty acid, Shewanella, Photobacterium
Citation: Allemann MN, Shulse CN and Allen EE (2019) Linkage of Marine Bacterial Polyunsaturated Fatty Acid and Long-Chain Hydrocarbon Biosynthesis. Front. Microbiol. 10:702. doi: 10.3389/fmicb.2019.00702
Received: 21 December 2018; Accepted: 20 March 2019;
Published: 03 April 2019.
Edited by:
Martin Koenneke, University of Bremen, GermanyReviewed by:
Tatsuo Kurihara, Kyoto University, JapanGuang Zhao, Qingdao Institute of Bioenergy and Bioprocess Technology (CAS), China
Copyright © 2019 Allemann, Shulse and Allen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Eric E. Allen, ZWFsbGVuQHVjc2QuZWR1
†Present address: Christine N. Shulse, Department of Energy, Joint Genome Institute, Walnut Creek, CA, United States
 Marco N. Allemann
Marco N. Allemann Christine N. Shulse
Christine N. Shulse Eric E. Allen
Eric E. Allen