- 1Department of Biotechnology, Delft University of Technology, Delft, Netherlands
- 2Global Innovation and Research, HEINEKEN Supply Chain B.V., Zoeterwoude, Netherlands
Interspecies hybrids of Saccharomyces species are found in a variety of industrial environments and often outperform their parental strains in industrial fermentation processes. Interspecies hybridization is therefore increasingly considered as an approach for improvement and diversification of yeast strains for industrial application. However, current hybridization methods are limited by their reliance on pre-existing or introduced selectable phenotypes. This study presents a high-throughput phenotype-independent method for isolation of interspecies Saccharomyces hybrids based on dual dye-staining and subsequent mating of two strains, followed by enrichment of double-stained hybrid cells from a mating population by fluorescence-activated cell sorting (FACS). Pilot experiments on intra-species mating of heterothallic haploid S. cerevisiae strains showed that 80% of sorted double-stained cells were hybrids. The protocol was further optimized by mating an S. cerevisiae haploid with homothallic S. eubayanus spores with complementary selectable phenotypes. In crosses without selectable phenotype, using S. cerevisiae and S. eubayanus haploids derived from laboratory as well as industrial strains, 10 to 15% of double-stained cells isolated by FACS were hybrids. When applied to rare mating, sorting of double-stained cells consistently resulted in about 600-fold enrichment of hybrid cells. Mating of dual-stained cells and FACS-based selection allows efficient enrichment of interspecies Saccharomyces hybrids within a matter of days and without requiring selectable hybrid phenotypes, both for homothallic and heterothallic strains. This strategy should accelerate the isolation of laboratory-made hybrids, facilitate research into hybrid heterosis and offer new opportunities for non-GM industrial strain improvement and diversification.
Introduction
Saccharomyces yeasts are used in various biotechnological industries including beer brewing, wine making, biopharmaceutical protein synthesis, and biofuels production (Balat, 2011; Nielsen, 2013; Marsit and Dequin, 2015; Jansen et al., 2017; Krogerus et al., 2017a). Nine Saccharomyces species have currently been described (Hittinger, 2013; Nueno-Palop et al., 2017), which are separated by a post-zygotic barrier that causes interspecies hybrids to be sterile (Naumov et al., 2000; Greig et al., 2002; Pfliegler et al., 2012; Hou et al., 2014). Although Saccharomyces hybrids occur in natural contexts such as the guts of wasps (Stefanini et al., 2016), strains with chimeric genomes are most commonly found in domesticated environments (Almeida et al., 2014; Boynton and Greig, 2014). For instance, lager beer is brewed by S. cerevisiae × S. eubayanus hybrids, collectively indicated as S. pastorianus (Libkind et al., 2011), S. uvarum × S. eubayanus hybrids called S. bayanus are used for cider brewing among other applications (Naumov et al., 2001), and various double and triple hybrids between S. cerevisiae, S. kudriavzevii, and S. uvarum play an important role in aroma production during wine fermentation (González et al., 2006). In addition, interspecies hybridization likely contributed to the evolution of domesticated Saccharomyces strains by facilitating horizontal gene transfer (Peter et al., 2018). Genetic admixture contributed to the distinct phenotypes of, for instance, cider-fermenting S. uvarum strains and wine-fermenting S. cerevisiae strains (Naumova et al., 2011; Dunn et al., 2012).
The genomes of hybrids from different Saccharomyces species have been shown to act synergistically, a phenomenon called ‘heterosis’ or ‘hybrid vigor,’ in which a hybrid performs better than either of its parents in specific environments (Rainieri et al., 2006; Belloch et al., 2008; Querol and Bond, 2009; Tronchoni et al., 2009). Heterosis is a complex phenomenon, involving copy number effects, interactions between different dominant and recessive alleles, and epistatic interactions (Shapira et al., 2014). Hybrid physiology largely depends on the specific parental strains (Mertens et al., 2015; Krogerus et al., 2017b). While some traits such as cryotolerance or flocculation appear to be completely inherited from one of the parental strains (Coloretti et al., 2006; Hebly et al., 2015), hybrids can also show phenotypes intermediary to their parental strains, as has been demonstrated for production of flavor compounds and other metabolites (Bellon et al., 2011; Krogerus et al., 2015).
Saccharomyces hybrids have been generated in the laboratory by crossing strains from different species (Banno and Kaneko, 1989). By analogy to the chimeric hybrids used for industrial applications, laboratory hybridization can yield strains with novel or improved properties for industrial applications. For instance, laboratory-made S. cerevisiae × S. eubayanus hybrids displayed increased cold tolerance, faster oligosaccharide consumption, different flavor profiles, higher fermentation rates and higher ethanol titres than their parental strains (Steensels et al., 2014a; Hebly et al., 2015; Krogerus et al., 2015). Pioneering studies on reconstruction of naturally-occurring hybrids have inspired the generation of hybrids from novel combinations of species, such as S. cerevisiae × S. paradoxus hybrids (Bellon et al., 2011), S. cerevisiae × S. mikatae hybrids (Bellon et al., 2013; Nikulin et al., 2017), S. cerevisiae × S. arboricola hybrids (Nikulin et al., 2017), and S. cerevisiae × S. uvarum hybrids (Masneuf-Pomarède et al., 2002; Bellon et al., 2015; Lopandic et al., 2016). Their phenotypic diversity showed promise for applications ranging from the fermented beverage industry to the production of biofuels (Masneuf-Pomarède et al., 2002; Steensels et al., 2014b; Nikulin et al., 2017; Peris et al., 2017; Nikulin et al., 2018).
Analogous to intra-species mating, interspecies hybridization occurs either by mating haploid cells of opposite mating type, or by rare mating based on spontaneous mating-type switching caused by loss of heterozygosity at the MAT locus (Gunge and Nakatomi, 1972). However, interspecies hybridization occurs at a relatively low rate; reported hybridization frequencies range from 1.5 to 3.6% for mass mating of spores (Mertens et al., 2015; Krogerus et al., 2016) to frequencies as low as 1 × 10−6 to 1 × 10−8 for mass mating of cells dependent on rare mating (Gunge and Nakatomi, 1972; Krogerus et al., 2016). While the efficiency of interspecies mating can be improved by genetic modification (GM) techniques, for example by overexpression of HO-endonuclease (Alexander et al., 2016) or by the use of spore micromanipulation (Naumov, 1996), isolation of bona fide hybrids from mating cultures remains necessary.
When parental strains have different selectable phenotypes, hybrids can be isolated by transferring the mating culture to conditions requiring both phenotypes for growth. Selectable phenotypes such as auxotrophies can either occur naturally (Sato et al., 2002; Fernández-González et al., 2015; Magalhães et al., 2017), or they can be obtained by mutagenesis and/or laboratory evolution under conditions favoring auxotrophic strains (Boeke et al., 1987; Scannell et al., 2011; Pérez-Través et al., 2012; Krogerus et al., 2015). However, generation of auxotrophic mutants is time- and labor-intensive (Alexander et al., 2016) and can be further complicated by the polyploid or aneuploid nature of many industrially-relevant Saccharomyces strains (Pérez-Través et al., 2012; Gorter de Vries et al., 2017b). Alternatively, selectable phenotypes such as antibiotic resistance can be introduced using GM techniques (Jimenez and Davies, 1980; Goldstein and McCusker, 1999; Piotrowski et al., 2012; da Silva et al., 2015; Hebly et al., 2015). However, industrial strains can be resilient to GM, and customer acceptance and legislation issues still largely preclude use of GM technology for applications in the food and beverages industry (Wunderlich and Gatto, 2015; Gorter de Vries et al., 2017a).
Fluorescence-activated cell sorting (FACS) can be used to isolate fluorescent cells from populations, even if they occur at extremely low frequencies (Cormack et al., 1996). By labeling each parental strain with a fluorescent dye, FACS has previously been used to sort mated Saccharomyces cerevisiae cells from their mating culture, resulting in a threefold enrichment of mated cells (Bell et al., 1998). Although a threefold enrichment would not be sufficient to isolate interspecies hybrids from a mating culture, this early study raised the question whether it might be possible to sufficiently modify staining, mating, and FACS procedures to accomplish this goal. To address this question, we explored a method to isolate interspecies Saccharomyces hybrids based on dual fluorescent labeling of parental strains and subsequent FACS-based selection of double-stained cells, without any dependency on any selectable phenotypes. After reproducing the isolation of intra-species S. cerevisiae crosses, we optimized isolation of interspecies S. cerevisiae × S. eubayanus hybrids using strains with selectable phenotypes. The resulting method was then tested for phenotype-independent isolation of S. cerevisiae × S. eubayanus hybrids.
Materials and Methods
Strains, Media, and Cultivation
Saccharomyces cerevisiae and S. eubayanus strains used in this study are listed in Table 1. Strains were routinely grown in complex medium (YP), containing 10 g L−1 yeast extract and 20 g L−1 peptone, supplemented with 20 g L−1 glucose for YPD, and with 20 g L−1 trehalose for YPT. Synthetic medium (SM) containing 20 g L−1 glucose, 3 g L−1 KH2PO4, 5.0 g L−1 (NH4)2SO4, 0.5 g L−1 MgSO4⋅7H2O, 1 mL L−1 of a trace element solution and 1 mL L−1 of a vitamin solution, was prepared as described previously (Verduyn et al., 1992), and the pH was set to 6.0 using 2 M KOH. Presence of the KanMX marker cassette was selected for in SM+G418: SM supplemented with 0.2 g L−1 of G418 (Invitrogen, Carlsbad, CA, United States) in which (NH4)2SO4 was replaced by 1 g L−1 monosodium glutamate (Cheng et al., 2000). For solid media, 20 g L−1 agar was added to media. Strains were grown in 500 mL round-bottom shake flasks with 100 mL medium at 200 RPM in an Innova 44 incubator shaker (Eppendorf, Nijmegen, Netherlands). Cultures of S. cerevisiae and S. eubayanus were grown at 30°C and 20°C, respectively. Liquid sporulation medium contained 20 g L−1 potassium acetate and its pH set to 7.0 using acetic acid (Bahalul et al., 2010). Frozen stocks were prepared by addition of glycerol (30% v/v) to exponentially growing shake-flask cultures, after which 1-mL aliquots were aseptically stored at −80°C.
Sporulation, Spore Isolation, and Germination
Sporulation was performed by aerobic incubation at 20°C during at least 72 h on sporulation medium. Presence of asci was verified using phase-contrast microscopy at a magnification of 400x. Spores were isolated as described by Herman and Rine (1997) with minor modifications. In short, spores were pelleted (1000 g, 5 min), resuspended in softening buffer (10 mM dithiothreitol, 100 mM Tris-SO4, pH set to 9.4 with H2SO4) and incubated at 30°C for 10 min. Cells were then washed using demineralized water, resuspended in spheroplasting buffer [2.1 M sorbitol, 10 mM KH2PO4, pH set to 7.2 with 1 M NaOH, 0.8 g L−1 zymolyase 20-T (AMS Biotechnology, Ltd., Abingdon, United Kingdom)] and incubated overnight at 30°C. After incubation, the culture was pelleted (1000 g, 10 min), washed with demineralized water and resuspended in 0.5% Triton X-100 (Sigma-Aldrich, Zwijndrecht, Netherlands). Spores were then sonicated for 15 s at 50 Hz with an amplitude of 6 μ while kept on ice using a Soniprep 150 (MSE, London, United Kingdom). During initial optimization of the protocol, a short protocol with only the zymolyase-step was also tested. Isolation of spores was confirmed by microscopic inspection as described above. For germination, spores were washed once with YPD and subsequently resuspended in 20 mL YPD to a concentration of approximately 106 cells mL−1. The germination culture was incubated in a 100 mL round bottom flask at 30°C and 200 RPM for 5 h. A protocol using different incubation times in 2% glucose medium and in YPD was tested during initial optimization of the interspecies mating.
Staining of Saccharomyces Cultures
For staining, CellTraceTM Violet, CellTraceTM CFSE and CellTraceTM Far Red fluorescent dyes (Thermo Fisher Scientific, Waltham, MA, United States) were prepared according to the manufacturers’ recommendations. Cultures were stained with 2 μL CellTraceTM dye per mL culture and incubated overnight in the dark at 12°C and 200 RPM. Stained cultures were washed twice with YP medium, as remaining unbound dye molecules would bind to the amide groups in yeast extract and peptone.
Intra-Species Mating
Heterothallic haploid parental strains were propagated until mid-exponential phase. The cultures of two parental strains were washed and diluted in sterile Isoton II (Beckman Coulter, Woerden, NL, United States) to a final cell density of approximately 106 cells mL−1, stained with CellTraceTM Violet or CellTraceTM CFSE and washed to remove unbound stain. 100 μL of each stained culture was transferred to an Eppendorf tube and centrifuged briefly (2000 g, 1 min) to increase proximity of the cells for more efficient mating. Subsequently, the mating culture was statically incubated up to 42 h at 12°C in the dark until FACS analysis. Longer incubation and higher temperatures yielded significant dilution of staining due to cell division.
Interspecies Mating and Rare Mating
Diploid parental strains and heterothallic haploid parental strains were propagated until mid-exponential phase. Haploid parental cells from homothallic strains were obtained via sporulation, spore isolation and germination. Cells were washed and diluted in sterile Isoton II (Beckman Coulter) to a final cell density of approximately 106 cells mL−1, stained with CellTraceTM Violet or CellTraceTM CFSE, and washed to remove unbound stain, as described. For rare mating, a final cell density of approximately 2 × 107 cells mL−1 was used and cells were stained with CellTraceTM Far Red or CellTraceTM CFSE. 100 μL of each stained culture was transferred to an Eppendorf tube and centrifuged briefly (2000 g, 1 min) to increase proximity of the cells for more efficient mating. Subsequently, the mating culture was statically incubated up to 30 h at 12°C in the dark until FACS analysis. Longer incubation and higher temperatures yielded significant dilution of staining due to cell division.
FACS Analysis and Sorting
Cultures for FACS analysis and sorting were diluted in sterile Isoton II and vortexed thoroughly to disrupt cell aggregates. For rare mating, 50 mM EDTA was added to disrupt cell aggregates formed by flocculation. The cultures were analyzed on a BD FACSAriaTM II SORP Cell Sorter (BD Biosciences, Franklin Lakes, NJ, United States) equipped with 355, 445, 488, 561, and 640 nm lasers and a 70 μm nozzle, and FACSFlowTM sheath fluid (BD Biosciences). Correct cytometer performance was evaluated prior to each experiment by running a Cytometer Setup & Tracking cycle using a CS&T bead kit (BD Biosciences) for calibration. Drop delay for sorting was determined by running an Auto Drop Delay cycle using Accudrop Beads (BD Biosciences). CellTraceTM Violet fluorescence was excited by the 355 nm laser and emission was detected through a 450 nm bandpass filter with a bandwidth of 50 nm. CellTraceTM CFSE was excited by the 488 nm laser and emission was detected through a 545 nm bandpass filter with a bandwidth of 30 nm. CellTraceTM Far Red was excited by the 640 nm laser and emission was detected through a 780 nm bandpass filter with a bandwidth of 60 nm. Morphology of the cells was analyzed by plotting forward scatter (FSC) against side scatter (SSC). Fluorescence of mating cultures was analyzed on either a CFSE versus Violet or a CFSE versus Far Red plot. Prior to sorting, at least 105 events were analyzed. Sorting regions (‘gates’) were set on these plots to determine the types of cells to be sorted. Gated single cells were sorted in 96-well microtiter plates containing YPD using a “single cell” sorting mask (0/32/16), and the plates were incubated at RT for 2 days. When cells with selectable phenotypes were used, the fraction of mated cells was determined by replica-plating to 96-well plates with selective medium (SM or SM+G418), using an ethanol-flame sterilized 96-pin replicator. FACS data were analyzed using FlowJo® software (version 3.05230, FlowJo, LLC, Ashland, OR, United States).
Determination of the Fraction of Growing Cells
After FACS sorting, the fraction of growing cells was determined by counting the number of wells in which growth was observed. For populations with low viabilities, up to 1000 cells were sorted per well and Poisson statistics were used to estimate the fraction of growing cells (Dube et al., 2008). The fraction of growing cells was calculated from (P), the fraction of wells containing a colony, (W) the total number of wells and (n), the total number of cells sorted into the wells (Eq. 1).
Imaging
Cells were imaged using a Zeiss Axio Imager Z1 (Carl Zeiss AG, Oberkochen, Germany). For fluorescent imaging, cells were excited with a xenon lamp. Fluorescence from CellTraceTM CFSE was imaged through a GFP filter set (Carl Zeiss AG) containing a 470 nm bandpass excitation filter with a bandwidth of 20 and 540 nm emission filter with a bandwidth of 25 nm. CellTraceTM Far Red was imaged through a Cy5 filter set (Carl Zeiss AG) containing a 640 nm bandpass excitation filter with a bandwidth of 30 nm and a 690 nm emission filter with a bandwidth of 50 nm. Images were processed using AxioVision SE64 (Rel. 4.9.1. Carl Zeiss AG) and FIJI (Schindelin et al., 2012).
Ploidy Determination by Flow Cytometry
For ploidy determination, samples were fixed using ethanol as previously described (Hebly et al., 2015). Staining of cells with SYTOX® Green Nucleic Acid Stain (Invitrogen) was performed as previously described (Haase and Reed, 2002) with some minor modifications. Cells were washed in 50 mM Tris-HCl (pH 7.5) and resuspended in 100 μL RNase solution (1 mg/mL RNase A in 50 mM Tris-HCl). 100 μL of cells was added to 1 mL of SYTOX® Green solution. When processing large numbers of samples, a high-throughput protocol in 96-well microtiter plates was used with a PIPETMAN® M multichannel electronic pipette (Gilson, Middleton, WI, United States). In this modified protocol, 100 μL sample was fixated by adding 150 μL 70% ethanol and in the final step 20 μL sample was added to 180 μL SYTOX® Green solution. An unstained control was included along with every sample. Fluorescence of the samples was measured on a BD AccuriTM C6 CSampler Flow Cytometer (BD Biosciences). The fluorophore was excited with the 488 nm laser of the flow cytometer and emission was detected through a 533 nm bandpass filter with a bandwidth of 30 nm. Ploidy data was analyzed using FlowJo® software (version 3.05230, FlowJo).
Identification of Interspecies Hybrids by PCR
The presence of genetic material from S. cerevisiae and from S. eubayanus and the mating type of potential hybrids was verified by PCR using DreamTaq PCR Mastermix (Life Technologies), as described previously (Hebly et al., 2015). To ensure that single cells isolates were tested, sorted dual-stained cells were grown in YPD and a second FACS step was used to sort a single cell from each culture. DNA from the single cell isolate cultures was released by boiling 2 μL of a liquid culture in 2 μL of NaOH for 15 min at 99°C. The S. cerevisiae-specific MEX67 gene was amplified using primers 8570 and 8571 (Supplementary Table 1) and the S. eubayanus-specific FSY1 gene was amplified using primers 8572 and 8573 (Muir et al., 2011; Pengelly and Wheals, 2013). The mating type was determined by amplifying MAT-loci, using primers 11, 12, and 13 (Huxley et al., 1990). PCR products were separated on a 2% (w/v) agarose gel in 0.5X TBE buffer (45 mM Tris-borate, 1 mM EDTA, pH 8).
Results
Isolating Intra-Species Hybrids From a Mating Culture Using FACS
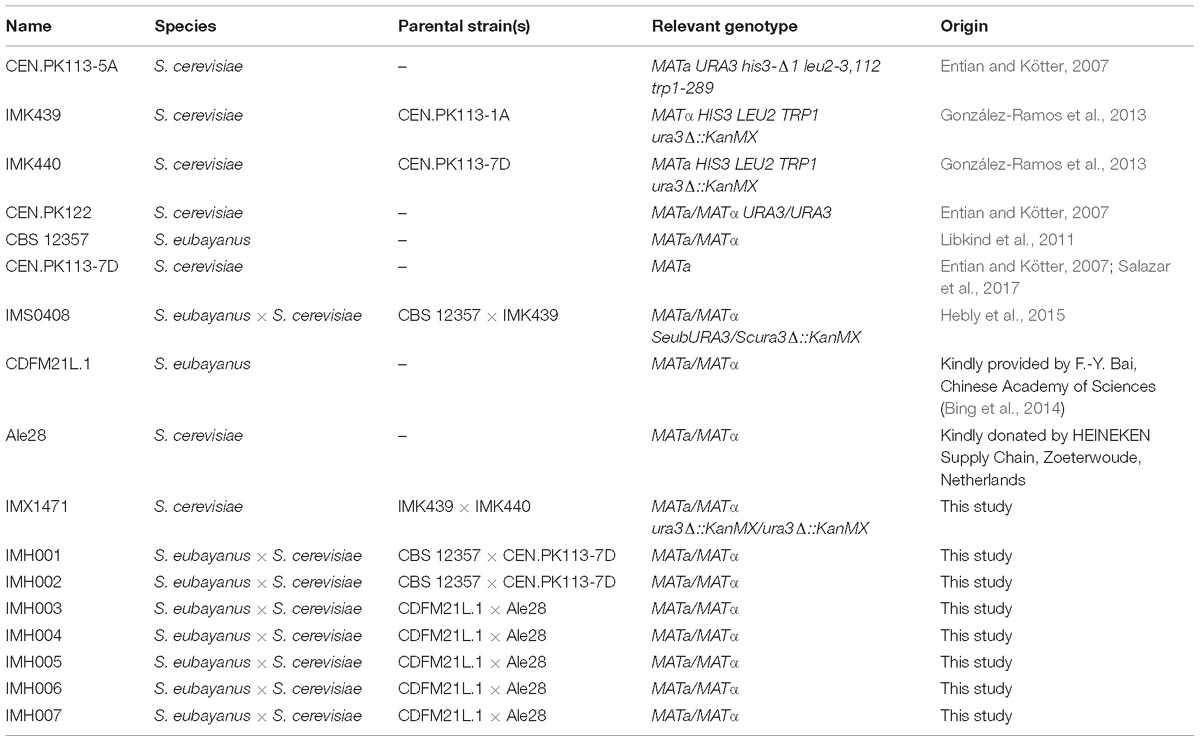
A functional protocol for dual staining of parental strains, mating and FACS-based sorting of double-stained cells was developed using the heterothallic haploid S. cerevisiae strains CEN.PK113-5A (MATa, His−, Lys−, Trp−) and IMK439 (MATα, Ura−). Due to their complementary auxotrophies, the fraction of mated cells could easily be quantified before and after FACS-based selection of double-stained cells by measuring the ability to grow on synthetic medium without histidine, lysine, tryptophan, and uracil. CEN.PK113-5A and IMK439 were stained using the commercially-available fluorescent CellTraceTM dyes CFSE and Violet, respectively. These dyes covalently bind to amine groups and thereby irreversibly label the parental cells (Filby et al., 2015). Mated cells should then be identifiable by the presence of fluorescent material from both parental strains. Efficient staining of the parental strains was confirmed for both dyes using flow cytometry (Figure 1A). To minimize dilution of the dye due to cell division, stained cells were mated by co-incubation in YPT medium at 12°C, which resulted in slow growth of S. cerevisiae. Flow cytometry of the mating culture indicated a progressive increase of the incidence of double-stained cells, from 0.90% after 18 h to 5.25% after 42 h (Figure 1A). Approximately 10% double-stained cells exhibited a morphology (Figure 1B) characteristic of Saccharomyces zygotes (Herskowitz, 1988). To determine the fraction of mated cells, cells from the total population and from the double-stained population were sorted on SM using FACS. Only 4% of the total population was able to grow on SM, while 74–82% of double-stained cells grew on this medium, indicating a 20-fold enrichment of mated cells in the double-stained population (Figure 1C).
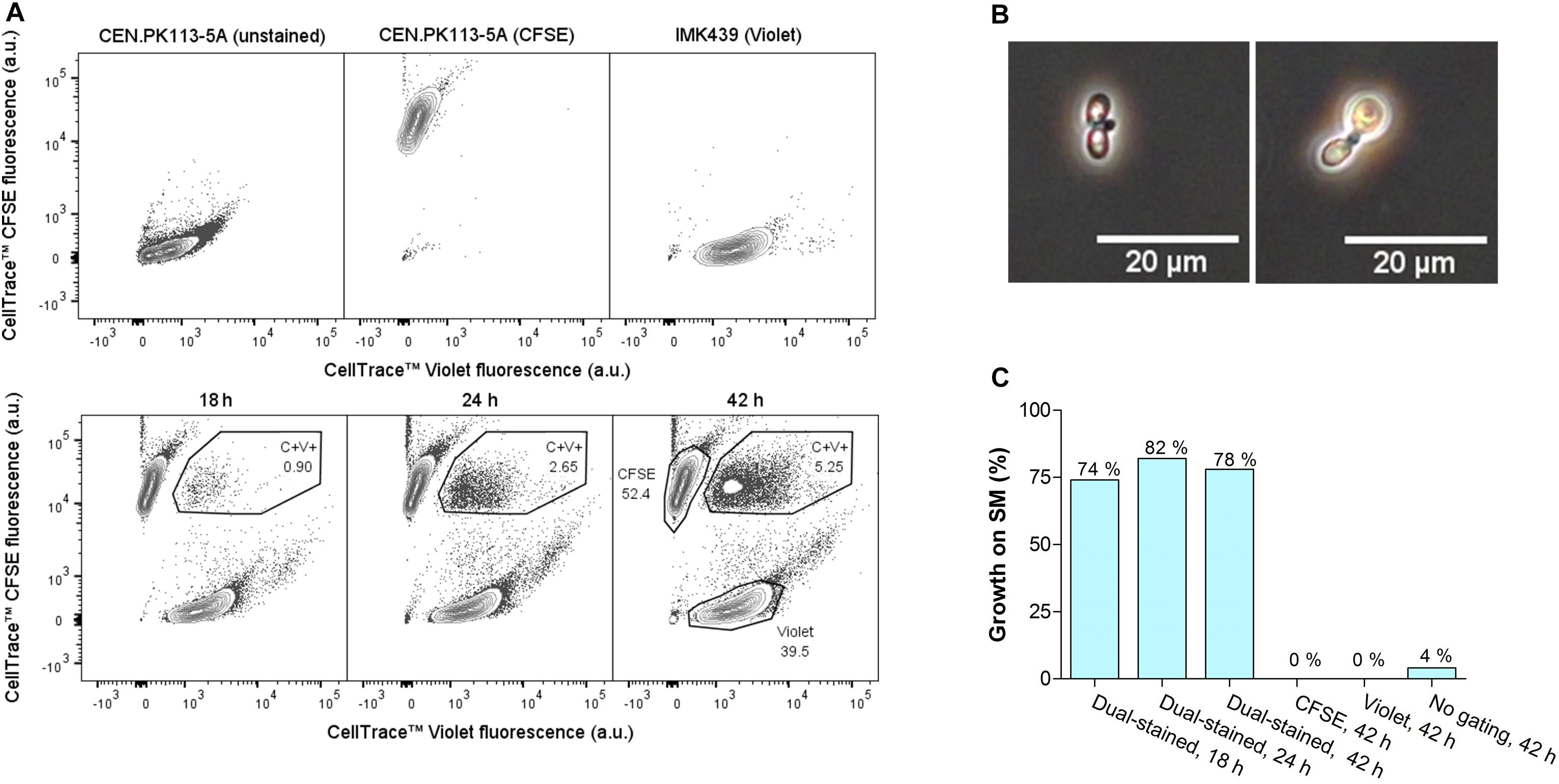
Figure 1. Intra-species mating of S. cerevisiae strains CEN.PK113-5A (MATa URA3 his3-Δ1 leu2-3,112 trp1-289) and IMK439 (MATα HIS3 TRP1 LEU2 ura3Δ::KanMX). (A) Fluorescence contour plots of unstained CEN.PK113-5A, CEN.PK113-5A stained with CellTraceTM CFSE, IMK439 stained with CellTraceTM Violet, and of the mating culture after 18, 24, and 42 h. The indicated gated areas were used for sorting cells, event rates of each gate are indicated as a percentage of total cell counts. (B) Microscope image (400×) of zygotes sorted from the double-stained population (C+V+) after 42 h of mating. (C) Percentage of cells able to grow on synthetic medium without auxotrophy-complementing supplements in different populations sorted by FACS, as indicated in (A).
Isolation of Interspecies Hybrids From a Mating Culture Using FACS
The developed protocol was applied to obtain interspecies hybrids between S. eubayanus strain CBS 12357 and S. cerevisiae strain IMK439 (MATα, ura3Δ::KanMX). Hybrids of these strains can be easily identified due to combined uracil prototrophy and resistance to the antibiotic G418. As CBS 12357 is a homothallic strain it was sporulated prior to staining and mating. To limit a bias toward self-mating of sister spores, a protocol for digestion of the ascus sack was developed based on a combined treatment with the surfactant Triton X-100 and zymolyase (Herman and Rine, 1997) (Supplementary Figure 1). When staining isolated spores of S. eubayanus CBS 12357, approximately half of the population was not fluorescently labeled after staining and incubation (Supplementary Figure 2). As the dye may not be able to penetrate the spore cell wall, the observed loss of staining could be due to loss of bound fluorophores during germination, when the spore cell wall is lost. To allow for efficient germination of spores while minimizing cell division prior to staining, a 5 h incubation on YPD at 30°C was implemented (Supplementary Figure 3). Using a protocol that included these optimizations, germinated spores of S. eubayanus CBS 12357 stained with CellTraceTM CFSE were mated with haploid cells of S. cerevisiae strain IMK439 stained with CellTraceTM Violet dye (Figure 2A). As the lack of necessity for trehalose consumption would broaden the applicability of our method, we mated the cells in YPD as well as on YPT medium at 12°C. The fraction of hybridized cells was monitored during mating by sorting double-stained cells onto YPD and determining the fraction of sorted cells which could grow on selective medium (Figure 2B). After 7 h, 1% of the double-stained population of both mating cultures on YPT and YPD was hybrid (Figure 2C). In contrast to intra-species S. cerevisiae mating, mating on YPT yielded no increase in the fraction of hybrids upon prolonged incubation for interspecies mating. However, in YPD, the fraction of hybrids among the double-stained cells increased to 18% after 24 h and remained stable up to 30 h (Figure 2C). In contrast, after 30 h incubation in YPD without sorting, only 0.3% of the total population was able to grow on selective medium. These results indicated that FACS-based sorting of double-stained cells resulted in a 70-fold enrichment of interspecies hybrids by sorting.
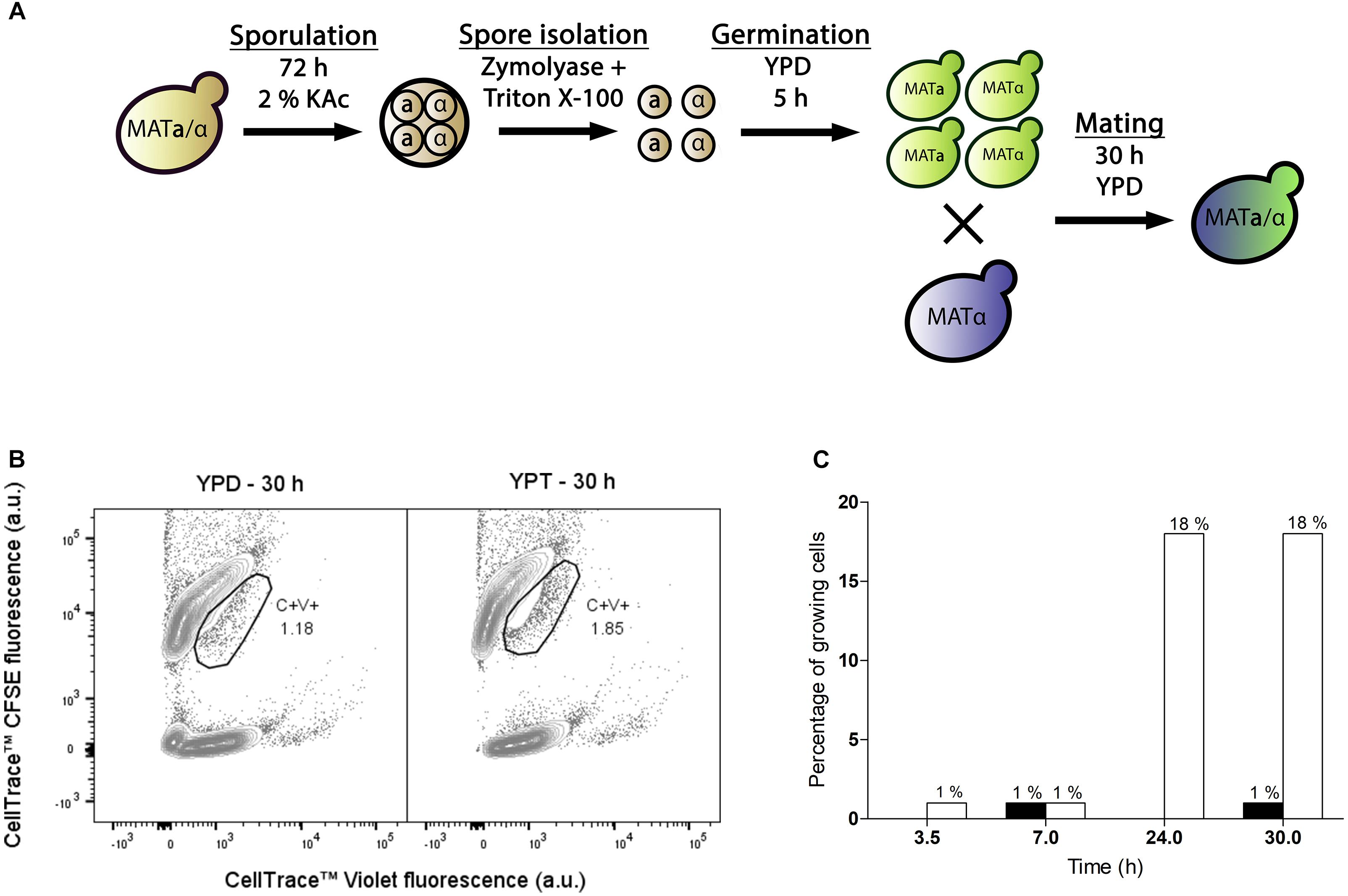
Figure 2. Optimization of interspecies hybridization between haploid S. eubayanus and S. cerevisiae strains. (A) Overview of the optimized protocol for interspecies spore-to-cell mating. (B) Fluorescence contour plots of mating cultures of stained CBS 12357 spores and IMK439 cells after 30 h of mating on YPD and YPT. The gated areas were used for sorting cells, event rates of each gate are indicated as a percentage. (C) Percentages of cells in the double-stained population able to grow on SM+G418 after 3.5, 7, 24, and 30 h of incubation on YPT (black) and YPD (white). Mating on YPT was only assessed at 7 and 30 h.
Enrichment of Interspecies Hybrids Without Selectable Phenotypes
To test applicability of the dual fluorescent staining FACS protocol for isolation of hybrids without selectable genetic markers, spores of S. eubayanus CBS 12357 (Libkind et al., 2011) were crossed with the haploid S. cerevisiae strain CEN.PK113-7D(MATa) (Entian and Kötter, 2007). In parallel, we crossed spores of the Tibetan S. eubayanus isolate CDFM21L.1 (Bing et al., 2014) with spores of the ale-brewing S. cerevisiae isolate Ale28, which was provided by HEINEKEN Supply Chain. These diploid strains were sporulated and germinated as described previously (Figure 2A). S. eubayanus parents were stained with CellTraceTM CFSE, S. cerevisiae parents with CellTraceTM Violet, and cells were co-incubated in YPD during 30 h at 12°C. Individual double-stained cells were sorted into 96 well plates containing 100 μL YPD per well, and incubated at 30°C until cultures were fully grown (Figure 3A). To eliminate false positives due to co-sorting of S. eubayanus/S. cerevisiae combinations, a single cell from each well was sorted into a second 96 well plate containing YPD. After incubation at 30°C, the presence of genetic material from both parents was verified by PCR amplification of the S. cerevisiae specific MEX67 gene and of the S. eubayanus specific FSY1 gene (Muir et al., 2011; Pengelly and Wheals, 2013). For the CBS 12357 × CEN.PK113-7D cross, a band corresponding to MEX67 and to FSY1 was observed for 2 of 22 tested single-cell isolates. These isolates were stored as IMH001 and IMH002 (Figure 3C). For the CDFM21L.1 × Ale28 cross, a band corresponding to MEX67 and to FSY1 was produced for 5 of 34 tested single-cell isolates, which were stored as IMH003-IMH007 (Figure 3C). To verify if strains IMH001-IMH007 were hybrids and not mixtures of haploid S. cerevisiae and S. eubayanus cells, the ploidy of the sorted cells was determined by DNA staining using SYTOX Green and flow cytometric analysis. The genome content of IMH001-IMH006 was diploid, whereas IMH007 was aneuploid (Figure 3B), indicating successful mating. Therefore, 9% of tested cells from the mating between CBS 12357 and CEN.PK113-7D and 15% of cells from the mating between CDFM21L.1 and Ale28 were hybrids. These results indicate that fluorescent staining and FACS enable a substantial enrichment of hybrid cells both for laboratory and industrial-relevant strains. A simple PCR protocol was sufficient to identify hybrids after enrichment.
Figure 3. Enrichment of interspecies hybrids without selectable phenotypes from CBS 12357 (S. eubayanus, sporulated) and CEN.PK113-7D (MATa) and from CDFM21L.1 (S. eubayanus, sporulated) and Ale28 (S. cerevisiae, sporulated). (A) Fluorescence contour plots of mating cultures between CBS 12357 (CFSE) × CEN.PK113-7D (Violet) and CDFM21L.1 (CFSE) × Ale28 (Violet). Gated areas were used for sorting cells, event rates of each gate are indicated as a percentage of the total population size. (B) Flow cytometric quantification of the genome content of constructed hybrids using SYTOX Green staining. S. cerevisiae strains CEN.PK113-7D and CEN.PK122 were used as a haploid and diploid control, respectively. (C) Multiplex PCR amplification of the S. cerevisiae specific MEX67 gene (150 bp) and the S. eubayanus specific FSY1 gene (228 bp) in single-cell isolates of the double-stained populations from CBS 12357 × CEN.PK113-7D and CDFM21L.1 × Ale28 mating cultures. For CBS 12357 × CEN.PK113-7D, 4 of the 22 tested isolates are shown. Genomic DNA of S. cerevisiae Ale26, S. eubayanus CDFM21L.1 and S. cerevisiae × S. eubayanus IMS0408 were used as controls. Hybrid isolates are indicated by arrows. L: Generuler 50 bp DNA Ladder.
Enrichment of Interspecies Hybrids by Rare Mating
Polyploidy and aneuploidy are commonly observed in industrial Saccharomyces hybrids (González et al., 2006; Querol and Bond, 2009; Peris et al., 2012), and chromosome copy number can play a key role in industrial performance (Krogerus et al., 2016; Gorter de Vries et al., 2017b). The poor sporulation efficiency of many industrial strains can preclude hybridization by conventional mating (Anderson and Martin, 1975). We explored the use of dual fluorescent staining and FACS to enrich hybrids obtained by rare mating by testing combinations of haploid and diploid S. eubayanus and S. cerevisiae strains. To evaluate low fractions of hybrid cells, we used S. eubayanus strains with uracil prototrophy and S. cerevisiae strains with uracil auxotrophy and resistance to the antibiotic G418. To obtain a diploid S. cerevisiae strain with uracil auxotrophy and resistance to the antibiotic G418, we first crossed IMK439 (MATα ura3Δ::KanMX) and IMK440 (MATa ura3Δ::KanMX) using fluorescent staining and FACS, resulting in IMX1471 (MATa/MATα, ura3Δ::KanMX/ura3Δ::KanMX). The mating types, ploidy, ability to sporulate, uracil prototrophy and G418 resistance of IMX1471 were verified (Supplementary Figure 4). Due to the anticipated low frequency of rare mating, S. eubayanus cells were stained with CellTraceTM CFSE and S. cerevisiae cells with CellTraceTM Far Red, as these dyes have little spectral overlap (Supplementary Figure 4). In total, three different crosses were made: CBS 12357 (sporulated, 1n) × IMX1471 (2n), CBS 12357 (2n) × IMK439 (1n) and CBS 12357 (2n) × IMX1471 (2n). The frequency of hybrid cells in each mating culture was assessed by plating 2 × 108 cells on SM+G418 plates and counting colonies. In parallel, the mating culture was analyzed by FACS and double-stained cells were sorted and replica-plated to SM+G418 to determine the frequency of hybrid cells after sorting. Due to the low frequency of rare mating, wells were inoculated with 1, 10, or 100 double-stained cells and the fraction of growing cells was calculated using Poisson statistics.
For the CBS 12357 (2n) × IMK439 (1n) cross, the fraction of hybrids in the total population varied between 1.6 × 10−6 and 7.2 × 10−6 between 24 and 168 h. After sorting, the fraction increased on average by a factor of 590 to between 4.3 × 10−4 and 1.3 × 10−3 (Table 2). For the CBS 12357 (1n) × IMX1471 (2n) cross, the fraction of hybrids in the total population varied between 3 × 10−7 and 1.5 × 10−6 between 24 and 168 h. Sorting only yielded a single hybrid after 96 h, at a fraction on 4.3 × 10−4, corresponding to a 540-fold enrichment. For the CBS 12357 (2n) × IMX1471 (2n) cross, a single hybrid was observed after 96 h of incubation, corresponding to a rate of 1 × 10−7, while no hybrids were identified after sorting. Overall, while rare mating was possible between the haploid and diploid strains, mated cells were present in very low frequencies both in the mating cultures and in the double-stained cells. In theory, fluorescent staining and FACS could be combined with high throughput PCR screening for hybrids in the sorted population. However, hundreds of cells would need to be screened for the diploid CBS 12357 × haploid IMK439 cross, and even more for the other crosses.
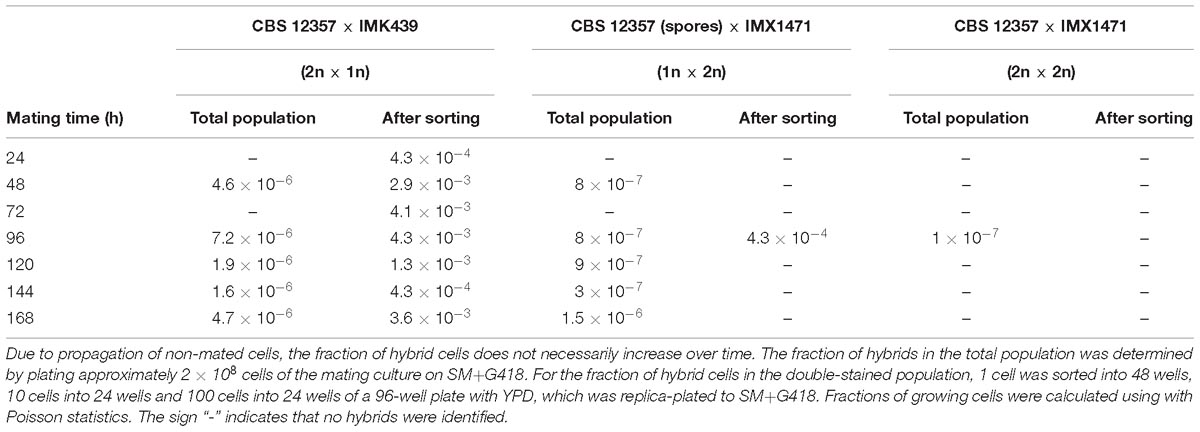
Table 2. Fraction of hybrid cells after interspecies rare mating between S. eubayanus strain CBS 12357 and S. cerevisiae strains IMK439 (1n) and IMX1471 (2n) as determined by the ability to grow on SM+G418.
Discussion
This study presents a new method for the enrichment of interspecies Saccharomyces hybrids that does not require parental strains and/or the resulting hybrids to have selectable phenotypes. By dual staining of parental cells with commercially-available fluorescent dyes prior to mating, mated cells could be enriched by up to 600-fold through sorting double-stained cells using FACS. In order to be able to mate homothallic strains, we developed a protocol for sporulation and germination prior to staining. Double-stained subpopulations selected by FACS after application of this protocol contained about 80% mated cells for intra-species crosses and 10–15% of mated cells for interspecies hybridization. By screening sorted double-stained cells using PCR, hybrids were successfully isolated from crosses of both laboratory strains and industrially relevant strains that did not have selectable phenotypes. By circumventing the need of conventional hybridization techniques for pre-existing or engineered selectable phenotypes (Pérez-Través et al., 2012; da Silva et al., 2015; Fernández-González et al., 2015; Krogerus et al., 2015), this method enables the isolation of hybrids from a wide range of strains within just a few days. Interspecies hybrids have been obtained previously by fluorescent staining and protoplast fusion (Katsuragi et al., 1994). However, while protoplast fusion is considered as a GM technique in some countries, hybridization by mating is not, making it suitable for application in the globalized food and beverage industry (Gibson et al., 2017). The use of staining is not problematic for industrial application as it is rapidly lost by dilution during subsequent cell division of the hybrid cells (Filby et al., 2015).
The isolation of interspecies hybrids using fluorescent labeling and FACS provides new opportunities for the use of laboratory-made hybrids for applications such as the production of fermented beverages and biofuels (Bellon et al., 2011, 2015; Mertens et al., 2015; Magalhães et al., 2017; Peris et al., 2017). Since hybrid physiology depends strongly on the combination of parental strains (Mertens et al., 2015; Krogerus et al., 2017b), the possibility to mate strains without any selectable phenotype could widen the phenotypic diversity of laboratory-made hybrids. Germination, staining and/or mating conditions could be optimized to account for traits of specific parental strains such as temperature optima, carbohydrate utilization, and growth kinetics. In the future, mass-mating of more than two fluorescently-labeled parental strains could broaden the phenotypic diversity of obtained laboratory-hybrids. Due to the inability of many industrial strains to produce viable spores (Steensels et al., 2014b), and due to the potential value of higher-ploidy hybrids (Krogerus et al., 2016; Gorter de Vries et al., 2017b), the isolation of interspecies hybrids by rare mating would also be valuable for industrial strain development. While fluorescent labeling and FACS did enable a 600-fold enrichment of interspecies hybrids obtained by rare mating, the isolation of hybrids would require extensive screening due to the high incidence of false positives. The presence of both stains in virtually all sorted cells, as verified by microscopy, indicated that false positives are not due to unwanted co-sorting of single stained cells. Instead, staining may be transferred between cells without resulting in hybrid cells. Due to the covalent binding of stains, such transfer would likely involve transfer of significant amounts of cell components, such as cell membrane or cytoplasm. Such transfer could result from abortive mating or cytoduction, which result in cytoplasmic fusion without nuclear fusion and would likely increase in frequency as mating efficiency decreases (Sigurdson et al., 1981). However, PCR based screening of hundreds of candidates is not impossible in industrial strain improvement programs. Moreover, the development of high throughput methods such as microfluidic lab-on-a-chip setups could further simplify screening after FACS sorting of rare hybrids (Schmitz et al., 2009; Gach et al., 2017).
The genome composition and stability of laboratory hybrids has been extensively researched (Sipiczki, 2018). However, the ability to generate diverse interspecies hybrids using fluorescent labeling could simplify ongoing research on, for example, hybrid-specific phenomena such as heterosis (Shapira et al., 2014; Bernardes et al., 2017), the inheritance of mitochondrial DNA in hybrids (Hsu and Chou, 2017), genome stability and loss of heterozygosity (Smukowski Heil et al., 2017) or hybrid sterility (Greig et al., 2002; Lee et al., 2008). Overall, the interspecies mating procedure presented here may be strongly accelerate industrial strain improvement programs and fundamental research into hybrid yeasts. Moreover, the described method can be complementary with subsequent strain improvement: laboratory evolution can result in admixture of parental genomes, resulting in further adaptation and potentially in improved performance, analogous to the admixed genomes of industrial chimeric strains (Pérez Través et al., 2014; Peris et al., 2017; Gorter de Vries et al., 2019).
Data Availability
The raw data supporting the conclusions of this manuscript will be made available by the authors, without undue reservation, to any qualified researcher.
Author Contributions
AG and J-MD conceived the study. AG, CK, SW, and ML designed experimental approaches. CK, SW, and AG performed experimental work. NK and J-MG provided strain ALE28 and supported the project. AG and CK wrote the manuscript with critical input from JP and J-MD. JP and J-MD supervised the study. All authors read and approved the final manuscript.
Funding
This work was performed within the BE-Basic R&D Program, which was granted an FES subsidy from the Dutch Ministry of Economic Affairs, Agriculture and Innovation (EL&I) (TKIBE01003 and TKIBE01001).
Conflict of Interest Statement
AG, CK, NK, J-MG, and J-MD have filed a patent application concerning this work. NK and J-MG are employed by Heineken Supply Chain B.V. (Zoeterwoude, Netherlands).
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Prof. Fen-Yang Bai (Chinese Academy of Sciences) for kindly providing us with CDFM21L.1 and Xavier Hakkaart (Delft University of Technology) for his expertise and support in fluorescence microscopy.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.00871/full#supplementary-material
References
Alexander, W. G., Peris, D., Pfannenstiel, B. T., Opulente, D. A., Kuang, M., and Hittinger, C. T. (2016). Efficient engineering of marker-free synthetic allotetraploids of Saccharomyces. Fungal Genet. Biol. 89, 10–17. doi: 10.1016/j.fgb.2015.11.002
Almeida, P., Gonçalves, C., Teixeira, S., Libkind, D., Bontrager, M., Masneuf-Pomarède, I., et al. (2014). A Gondwanan imprint on global diversity and domestication of wine and cider yeast Saccharomyces uvarum. Nat. Commun. 5:4044. doi: 10.1038/ncomms5044
Anderson, E., and Martin, P. (1975). The sporulation and mating of brewing yeasts. J. Inst. Brew. 81, 242–247. doi: 10.1002/j.2050-0416.1975.tb03685.x
Bahalul, M., Kaneti, G., and Kashi, Y. (2010). Ether–zymolyase ascospore isolation procedure: an efficient protocol for ascospores isolation in Saccharomyces cerevisiae yeast. Yeast 27, 999–1003. doi: 10.1002/yea.1808
Balat, M. (2011). Production of bioethanol from lignocellulosic materials via the biochemical pathway: a review. Energy Convers. Manag. 52, 858–875. doi: 10.1016/j.enconman.2010.08.013
Banno, I., and Kaneko, Y. (1989). A genetic analysis of taxonomic relation between Saccharomyces cerevisiae and Saccharomyces bayanus. Yeast 5,S373–S377.
Bell, P. J., Deere, D., Shen, J., Chapman, B., Bissinger, P. H., Attfield, P. V., et al. (1998). A flow cytometric method for rapid selection of novel industrial yeast hybrids. Appl. Environ. Microbiol. 64, 1669–1672.
Belloch, C., Orlic, S., Barrio, E., and Querol, A. (2008). Fermentative stress adaptation of hybrids within the Saccharomyces sensu stricto complex. Int. J. Food Microbiol. 122, 188–195. doi: 10.1016/j.ijfoodmicro.2007.11.083
Bellon, J. R., Eglinton, J. M., Siebert, T. E., Pollnitz, A. P., Rose, L., De Barros Lopes, M., et al. (2011). Newly generated interspecific wine yeast hybrids introduce flavour and aroma diversity to wines. Appl. Microbiol. Biotechnol. 91, 603–612. doi: 10.1007/s00253-011-3294-3
Bellon, J. R., Schmid, F., Capone, D. L., Dunn, B. L., and Chambers, P. J. (2013). Introducing a new breed of wine yeast: interspecific hybridisation between a commercial Saccharomyces cerevisiae wine yeast and Saccharomyces mikatae. PLoS One 8:e62053. doi: 10.1371/journal.pone.0062053
Bellon, J. R., Yang, F., Day, M. P., Inglis, D. L., and Chambers, P. J. (2015). Designing and creating Saccharomyces interspecific hybrids for improved, industry relevant, phenotypes. Appl. Microbiol. Biotechnol. 99, 8597–8609. doi: 10.1007/s00253-015-6737-4
Bernardes, J. P., Stelkens, R. B., and Greig, D. (2017). Heterosis in hybrids within and between yeast species. J. Evol. Biol. 30, 538–548. doi: 10.1111/jeb.13023
Bing, J., Han, P.-J., Liu, W.-Q., Wang, Q.-M., and Bai, F.-Y. (2014). Evidence for a Far East Asian origin of lager beer yeast. Curr. Biol. 24, R380–R381. doi: 10.1016/j.cub.2014.04.031
Boeke, J. D., Trueheart, J., Natsoulis, G., and Fink, G. R. (1987). 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 154, 164–175. doi: 10.1016/0076-6879(87)54076-9
Boynton, P. J., and Greig, D. (2014). The ecology and evolution of non-domesticated Saccharomyces species. Yeast 31, 449–462. doi: 10.1002/yea.3040
Cheng, T.-H., Chang, C.-R., Joy, P., Yablok, S., and Gartenberg, M. R. (2000). Controlling gene expression in yeast by inducible site-specific recombination. Nucleic Acids Res. 28:e108.
Coloretti, F., Zambonelli, C., and Tini, V. (2006). Characterization of flocculent Saccharomyces interspecific hybrids for the production of sparkling wines. Food Microbiol. 23, 672–676. doi: 10.1016/j.fm.2005.11.002
Cormack, B. P., Valdivia, R. H., and Falkow, S. (1996). FACS-optimized mutants of the green fluorescent protein (GFP). Gene 173, 33–38. doi: 10.1016/0378-1119(95)00685-0
da Silva, T., Albertin, W., Dillmann, C., Bely, M., La Guerche, S., Giraud, C., et al. (2015). Hybridization within Saccharomyces genus results in homoeostasis and phenotypic novelty in winemaking conditions. PLoS One 10:e0123834. doi: 10.1371/journal.pone.0123834
Dube, S., Qin, J., and Ramakrishnan, R. (2008). Mathematical analysis of copy number variation in a DNA sample using digital PCR on a nanofluidic device. PLoS One 3:e2876. doi: 10.1371/journal.pone.0002876
Dunn, B., Richter, C., Kvitek, D. J., Pugh, T., and Sherlock, G. (2012). Analysis of the Saccharomyces cerevisiae pan-genome reveals a pool of copy number variants distributed in diverse yeast strains from differing industrial environments. Genome Res. 22, 908–924. doi: 10.1101/gr.130310.111
Entian, K.-D., and Kötter, P. (2007). 25 Yeast genetic strain and plasmid collections. Methods Microbiol. 36, 629–666. doi: 10.1016/s0580-9517(06)36025-4
Fernández-González, M., Úbeda, J. F., and Briones, A. I. (2015). Study of Saccharomyces cerevisiae wine strains for breeding through fermentation efficiency and tetrad analysis. Curr. Microbiol. 70, 441–449. doi: 10.1007/s00284-014-0741-2
Filby, A., Begum, J., Jalal, M., and Day, W. (2015). Appraising the suitability of succinimidyl and lipophilic fluorescent dyes to track proliferation in non-quiescent cells by dye dilution. Methods 82, 29–37. doi: 10.1016/j.ymeth.2015.02.016
Gach, P. C., Iwai, K., Kim, P., Hillson, N., and Singh, A. K. (2017). Droplet microfluidics for synthetic biology. Lab Chip 17, 3388–3400. doi: 10.1039/c7lc00576h
Gibson, B., Geertman, J.-M. A., Hittinger, C. T., Krogerus, K., Libkind, D., Louis, E. J., et al. (2017). New yeasts-new brews: modern approaches to brewing yeast design and development. FEMS Yeast Res. 17:fox038. doi: 10.1093/femsyr/fox038
Goldstein, A. L., and McCusker, J. H. (1999). Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast 15, 1541–1553. doi: 10.1002/(sici)1097-0061(199910)15:14<1541::aid-yea476>3.3.co;2-b
González, S. S., Barrio, E., Gafner, J., and Querol, A. (2006). Natural hybrids from Saccharomyces cerevisiae, Saccharomyces bayanus and Saccharomyces kudriavzevii in wine fermentations. FEMS Yeast Res. 6, 1221–1234.
González-Ramos, D., Van Den Broek, M., Van Maris, A. J. A., Pronk, J. T., and Daran, J.-M. G. (2013). Genome-scale analyses of butanol tolerance in Saccharomyces cerevisiae reveal an essential role of protein degradation. Biotechnol. Biofuels 6:48. doi: 10.1186/1754-6834-6-48
Gorter de Vries, A. R., Groot, P. A., Broek, M., and Daran, J.-M. G. (2017a). CRISPR-Cas9 mediated gene deletions in lager yeast Saccharomyces pastorianus. Microb. Cell Fact. 16:222. doi: 10.1186/s12934-017-0835-1
Gorter de Vries, A. R., Pronk, J. T., and Daran, J.-M. G. (2017b). Industrial relevance of chromosomal copy number variation in Saccharomyces yeasts. Appl. Environ. Microbiol. 83:e03206-16. doi: 10.1128/AEM.03206-16
Gorter de Vries, A. R., Voskamp, M., Van Aalst, A. C., Kristensen, L. H., Jansen, L., Van Den Broek, M., et al. (2019). Laboratory evolution of a Saccharomyces cerevisiae x S. eubayanus hybrid under simulated lager-brewing conditions. Front. Genet. 10:242. doi: 10.3389/fgene.2019.00242
Greig, D., Borts, R. H., and Louis, E. J. (2002). Epistasis and hybrid sterility in Saccharomyces. Proc. Biol. Sci. 269, 1167–1171. doi: 10.1098/rspb.2002.1989
Gunge, N., and Nakatomi, Y. (1972). Genetic mechanisms of rare matings of the yeast Saccharomyces cerevisiae heterozygous for mating type. Genetics 70, 41–58.
Haase, S. B., and Reed, S. I. (2002). Improved flow cytometric analysis of the budding yeast cell cycle. Cell Cycle 1, 117–121. doi: 10.4161/cc.1.2.114
Hebly, M., Brickwedde, A., Bolat, I., Driessen, M. R. M., De Hulster, E. A. F., Van Den Broek, M., et al. (2015). S. cerevisiae × S. eubayanus interspecific hybrid, the best of both worlds and beyond. FEMS Yeast Res. 15:fov005. doi: 10.1093/femsyr/fov005
Herman, P. K., and Rine, J. (1997). Yeast spore germination: a requirement for Ras protein activity during re-entry into the cell cycle. EMBO J. 16, 6171–6181. doi: 10.1093/emboj/16.20.6171
Herskowitz, I. (1988). Life cycle of the budding yeast Saccharomyces cerevisiae. Microbiol. Rev. 52, 536–553.
Hittinger, C. T. (2013). Saccharomyces diversity and evolution: a budding model genus. Trends Genet. 29, 309–317. doi: 10.1016/j.tig.2013.01.002
Hou, J., Friedrich, A., De Montigny, J., and Schacherer, J. (2014). Chromosomal rearrangements as a major mechanism in the onset of reproductive isolation in Saccharomyces cerevisiae. Curr. Biol. 24, 1153–1159. doi: 10.1016/j.cub.2014.03.063
Hsu, Y.-Y., and Chou, J.-Y. (2017). Environmental factors can influence mitochondrial inheritance in the Saccharomyces yeast hybrids. PLoS One 12:e0169953. doi: 10.1371/journal.pone.0169953
Huxley, C., Green, E. D., and Dunham, I. (1990). Rapid assessment of S. cerevisiae mating type by PCR. Trends Genet. 6:236. doi: 10.1016/0168-9525(90)90190-h
Jansen, M. L. A., Bracher, J. M., Papapetridis, I., Verhoeven, M. D., De Bruijn, H., De Waal, P. P. P., et al. (2017). Saccharomyces cerevisiae strains for second-generation ethanol production: from academic exploration to industrial implementation. FEMS Yeast Res. 17:fox044. doi: 10.1093/femsyr/fox044
Jimenez, A., and Davies, J. (1980). Expression of a transposable antibiotic resistance element in Saccharomyces. Nature 287, 869–871. doi: 10.1038/287869a0
Katsuragi, T., Kawabata, N., and Sakai, T. (1994). Selection of hybrids from protoplast fusion of yeasts by double fluorescence labelling and automatic cell sorting. Lett. Appl. Microbiol. 19, 92–94. doi: 10.1111/j.1472-765x.1994.tb00913.x
Krogerus, K., Arvas, M., De Chiara, M., Magalhães, F., Mattinen, L., Oja, M., et al. (2016). Ploidy influences the functional attributes of de novo lager yeast hybrids. Appl. Microbiol. Biotechnol. 100, 7203–7222. doi: 10.1007/s00253-016-7588-3
Krogerus, K., Magalhães, F., Vidgren, V., and Gibson, B. (2015). New lager yeast strains generated by interspecific hybridization. J. Ind. Microbiol. Biotechnol. 42, 769–778. doi: 10.1007/s10295-015-1597-6
Krogerus, K., Magalhães, F., Vidgren, V., and Gibson, B. (2017a). Novel brewing yeast hybrids: creation and application. Appl. Microbiol. Biotechnol. 101, 65–78. doi: 10.1007/s00253-016-8007-5
Krogerus, K., Seppänen-Laakso, T., Castillo, S., and Gibson, B. (2017b). Inheritance of brewing-relevant phenotypes in constructed Saccharomyces cerevisiae × Saccharomyces eubayanus hybrids. Microb. Cell Fact. 16:66. doi: 10.1186/s12934-017-0679-8
Lee, H.-Y., Chou, J.-Y., Cheong, L., Chang, N.-H., Yang, S.-Y., and Leu, J.-Y. (2008). Incompatibility of nuclear and mitochondrial genomes causes hybrid sterility between two yeast species. Cell 135, 1065–1073. doi: 10.1016/j.cell.2008.10.047
Libkind, D., Hittinger, C. T., Valério, E., Gonçalves, C., Dover, J., Johnston, M., et al. (2011). Microbe domestication and the identification of the wild genetic stock of lager-brewing yeast. Proc. Natl. Acad. Sci. U.S.A. 108, 14539–14544. doi: 10.1073/pnas.1105430108
Lopandic, K., Pfliegler, W. P., Tiefenbrunner, W., Gangl, H., Sipiczki, M., and Sterflinger, K. (2016). Genotypic and phenotypic evolution of yeast interspecies hybrids during high-sugar fermentation. Appl. Microbiol. Biotechnol. 100, 6331–6343. doi: 10.1007/s00253-016-7481-0
Magalhães, F., Krogerus, K., Vidgren, V., Sandell, M., and Gibson, B. (2017). Improved cider fermentation performance and quality with newly generated Saccharomyces cerevisiae× Saccharomyces eubayanus hybrids. J. Ind. Microbiol. Biotechnol. 44, 1203–1213. doi: 10.1007/s10295-017-1947-7
Marsit, S., and Dequin, S. (2015). Diversity and adaptive evolution of Saccharomyces wine yeast: a review. FEMS Yeast Res. 15:fov067. doi: 10.1093/femsyr/fov067
Masneuf-Pomarède, I., Murat, M.-L., Naumov, G. I., Tominaga, T., and Dubourdieu, D. (2002). Hybrids Saccharomyces cerevisiae x Saccharomyces bayanus var. uvarum having a high liberating ability of some sulfur varietal aromas of Vitis vinifera Sauvignon blanc wines. J. Int. Sci. Vigne Vin 36, 205–212.
Mertens, S., Steensels, J., Saels, V., De Rouck, G., Aerts, G., and Verstrepen, K. J. (2015). A large set of newly created interspecific Saccharomyces hybrids increases aromatic diversity in lager beers. Appl. Environ. Microbiol. 81, 8202–8214. doi: 10.1128/AEM.02464-15
Muir, A., Harrison, E., and Wheals, A. (2011). A multiplex set of species-specific primers for rapid identification of members of the genus Saccharomyces. FEMS Yeast Res. 11, 552–563. doi: 10.1111/j.1567-1364.2011.00745.x
Naumov, G. (1996). Genetic identification of biological species in the Saccharomyces sensu stricto complex. J. Ind. Microbiol. 17, 295–302. doi: 10.1007/bf01574704
Naumov, G. I., Naumova, E. S., Masneuf, I., Aigle, M., Kondratieva, V. I., and Dubourdieu, D. (2000). Natural polyploidization of some cultured yeast Saccharomyces sensu stricto: auto-and allotetraploidy. Syst. Appl. Microbiol. 23, 442–449. doi: 10.1016/s0723-2020(00)80076-4
Naumov, G. I., Nguyen, H.-V., Naumova, E. S., Michel, A., Aigle, M., and Gaillardin, C. (2001). Genetic identification of Saccharomyces bayanus var. uvarum, a cider-fermenting yeast. Int. J. Food Microbiol. 65, 163–171. doi: 10.1016/s0168-1605(00)00515-8
Naumova, E. S., Naumov, G. I., Michailova, Y. V., Martynenko, N. N., and Masneuf-Pomarède, I. (2011). Genetic diversity study of the yeast Saccharomyces bayanus var. uvarum reveals introgressed subtelomeric Saccharomyces cerevisiae genes. Res. Microbiol. 162, 204–213. doi: 10.1016/j.resmic.2010.09.023
Nielsen, J. (2013). Production of biopharmaceutical proteins by yeast: advances through metabolic engineering. Bioengineered 4, 207–211. doi: 10.4161/bioe.22856
Nikulin, J., Krogerus, K., and Gibson, B. (2017). Alternative Saccharomyces interspecies hybrid combinations and their potential for low-temperature wort fermentation. Yeast 35, 113–127. doi: 10.1002/yea.3246
Nikulin, J., Krogerus, K., and Gibson, B. (2018). Alternative Saccharomyces interspecies hybrid combinations and their potential for low-temperature wort fermentation. Yeast 35, 113–127.
Nueno-Palop, C., Bond, C. J., Mcghie, H., Roberts, I. N., and Delneri, D. (2017). Saccharomyces jurei sp. nov., Isolation and genetic identification of a novel yeast species from Quercus robur. Int. J. Syst. Evol. Microbiol. 67, 2046–2052. doi: 10.1099/ijsem.0.002013
Pengelly, R. J., and Wheals, A. E. (2013). Rapid identification of Saccharomyces eubayanus and its hybrids. FEMS Yeast Res. 13, 156–161. doi: 10.1111/1567-1364.12018
Pérez Través, L., Lopes, C. A., Barrio Esparducer, E., and Querol Simón, A. (2014). Stabilization process in Saccharomyces intra and interspecific hybrids in fermentative conditions. Int. Microbiol. 17, 213–224. doi: 10.2436/20.1501.01.224
Pérez-Través, L., Lopes, C. A., Barrio, E., and Querol, A. (2012). Evaluation of different genetic procedures for the generation of artificial hybrids in Saccharomyces genus for winemaking. Int. J. Food Microbiol. 156, 102–111. doi: 10.1016/j.ijfoodmicro.2012.03.008
Peris, D., Belloch, C., Lopandić, K., Álvarez-Pérez, J. M., Querol, A., and Barrio, E. (2012). The molecular characterization of new types of Saccharomyces cerevisiae× S. kudriavzevii hybrid yeasts unveils a high genetic diversity. Yeast 29, 81–91. doi: 10.1002/yea.2891
Peris, D., Moriarty, R. V., Alexander, W. G., Baker, E., Sylvester, K., Sardi, M., et al. (2017). Hybridization and adaptive evolution of diverse Saccharomyces species for cellulosic biofuel production. Biotechnol. Biofuels 10:78. doi: 10.1186/s13068-017-0763-7
Peter, J., De Chiara, M., Friedrich, A., Yue, J.-X., Pflieger, D., Bergström, A., et al. (2018). Genome evolution across 1,011 Saccharomyces cerevisiae isolates. Nature 556, 339–344. doi: 10.1038/s41586-018-0030-5
Pfliegler, W. P., Antunovics, Z., and Sipiczki, M. (2012). Double sterility barrier between Saccharomyces species and its breakdown in allopolyploid hybrids by chromosome loss. FEMS Yeast Res. 12, 703–718. doi: 10.1111/j.1567-1364.2012.00820.x
Piotrowski, J. S., Nagarajan, S., Kroll, E., Stanbery, A., Chiotti, K. E., Kruckeberg, A. L., et al. (2012). Different selective pressures lead to different genomic outcomes as newly-formed hybrid yeasts evolve. BMC Evol. Biol. 12:46. doi: 10.1186/1471-2148-12-46
Querol, A., and Bond, U. (2009). The complex and dynamic genomes of industrial yeasts. FEMS Microbiol. Lett. 293, 1–10. doi: 10.1111/j.1574-6968.2008.01480.x
Rainieri, S., Kodama, Y., Kaneko, Y., Mikata, K., Nakao, Y., and Ashikari, T. (2006). Pure and mixed genetic lines of Saccharomyces bayanus and Saccharomyces pastorianus and their contribution to the lager brewing strain genome. Appl. Environ. Microbiol. 72, 3968–3974. doi: 10.1128/aem.02769-05
Salazar, A. N., Gorter de Vries, A. R., Van Den Broek, M., Wijsman, M., De La Torre Cortés, P., Brickwedde, A., et al. (2017). Nanopore sequencing enables near-complete de novo assembly of Saccharomyces cerevisiae reference strain CEN.PK113-7D. FEMS Yeast Res. 17:fox074. doi: 10.1093/femsyr/fox074
Sato, M., Kishimoto, M., Watari, J., and Takashio, M. (2002). Breeding of brewer’s yeast by hybridization between a top-fermenting yeast Saccharomyces cerevisiae and a cryophilic yeast Saccharomyces bayanus. J. Biosci. Bioeng. 93, 509–511. doi: 10.1263/jbb.93.509
Scannell, D. R., Zill, O. A., Rokas, A., Payen, C., Dunham, M. J., Eisen, M. B., et al. (2011). The awesome power of yeast evolutionary genetics: new genome sequences and strain resources for the Saccharomyces sensu stricto genus. G3 1, 11–25. doi: 10.1534/g3.111.000273
Schindelin, J., Arganda-Carreras, I., Frise, E., Kaynig, V., Longair, M., Pietzsch, T., et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682. doi: 10.1038/nmeth.2019
Schmitz, C. H., Rowat, A. C., Köster, S., and Weitz, D. A. (2009). Dropspots: a picoliter array in a microfluidic device. Lab Chip 9, 44–49. doi: 10.1039/b809670h
Shapira, R., Levy, T., Shaked, S., Fridman, E., and David, L. (2014). Extensive heterosis in growth of yeast hybrids is explained by a combination of genetic models. Heredity 113, 316–326. doi: 10.1038/hdy.2014.33
Sigurdson, D. C., Gaarder, M. E., and Livingston, D. M. (1981). Characterization of the transmission during cytoductant formation of the 2 μm DNA plasmid from Saccharomyces. Mol. Gen. Genet. 183, 59–65. doi: 10.1007/bf00270139
Sipiczki, M. (2018). Interspecies hybridisation and genome chimerisation in Saccharomyces: combining of gene pools of species and its biotechnological perspectives. Front. Microbiol. 9:3071. doi: 10.3389/fmicb.2018.03071
Smukowski Heil, C. S., Desevo, C. G., Pai, D. A., Tucker, C. M., Hoang, M. L., and Dunham, M. J. (2017). Loss of heterozygosity drives adaptation in hybrid yeast. Mol. Biol. Evol. 34, 1596–1612. doi: 10.1093/molbev/msx098
Steensels, J., Meersman, E., Snoek, T., Saels, V., and Verstrepen, K. J. (2014a). Large-scale selection and breeding to generate industrial yeasts with superior aroma production. Appl. Environ. Microbiol. 80, 6965–6975. doi: 10.1128/AEM.02235-14
Steensels, J., Snoek, T., Meersman, E., Nicolino, M. P., Voordeckers, K., and Verstrepen, K. J. (2014b). Improving industrial yeast strains: exploiting natural and artificial diversity. FEMS Microbiol. Rev. 38, 947–995. doi: 10.1111/1574-6976.12073
Stefanini, I., Dapporto, L., Berná, L., Polsinelli, M., Turillazzi, S., and Cavalieri, D. (2016). Social wasps are a Saccharomyces mating nest. Proc. Natl. Acad. Sci. U.S.A. 113, 2247–2251. doi: 10.1073/pnas.1516453113
Tronchoni, J., Gamero, A., Arroyo-López, F. N., Barrio, E., and Querol, A. (2009). Differences in the glucose and fructose consumption profiles in diverse Saccharomyces wine species and their hybrids during grape juice fermentation. Int. J. Food Microbiol. 134, 237–243. doi: 10.1016/j.ijfoodmicro.2009.07.004
Verduyn, C., Postma, E., Scheffers, W. A., and Van Dijken, J. P. (1992). Effect of benzoic acid on metabolic fluxes in yeasts: a continuous-culture study on the regulation of respiration and alcoholic fermentation. Yeast 8, 501–517. doi: 10.1002/yea.320080703
Keywords: FACS, Saccharomyces eubayanus × Saccharomyces cerevisiae hybrids, heterosis, marker-free mating, lager beer brewing, non-GMO
Citation: Gorter de Vries AR, Koster CC, Weening SM, Luttik MAH, Kuijpers NGA, Geertman J-MA, Pronk JT and Daran J-MG (2019) Phenotype-Independent Isolation of Interspecies Saccharomyces Hybrids by Dual-Dye Fluorescent Staining and Fluorescence-Activated Cell Sorting. Front. Microbiol. 10:871. doi: 10.3389/fmicb.2019.00871
Received: 11 February 2019; Accepted: 04 April 2019;
Published: 26 April 2019.
Edited by:
Haifeng Zhao, South China University of Technology, ChinaReviewed by:
Ed Louis, University of Leicester, United KingdomMatthias Sipiczki, University of Debrecen, Hungary
Linda F. Bisson, University of California, Davis, United States
Copyright © 2019 Gorter de Vries, Koster, Weening, Luttik, Kuijpers, Geertman, Pronk and Daran. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jean-Marc G. Daran, Si5HLkRhcmFuQHR1ZGVsZnQubmw=
†These authors have contributed equally to this work
 Arthur R. Gorter de Vries
Arthur R. Gorter de Vries Charlotte C. Koster
Charlotte C. Koster Susan M. Weening
Susan M. Weening Marijke A. H. Luttik1
Marijke A. H. Luttik1 Jack T. Pronk
Jack T. Pronk Jean-Marc G. Daran
Jean-Marc G. Daran