- 1School of Agriculture and Food Science, The University of Queensland, Brisbane, QLD, Australia
- 2Guangxi Crop Genetic Improvement and Biotechnology Key Lab, Guangxi Academy of Agricultural Sciences, Nanning, China
- 3Department of Agriculture and Fisheries, Maroochy Research Facility, Nambour, QLD, Australia
- 4Department of Primary Industry and Resources, Northern Territory Government, Darwin, NT, Australia
- 5School of Biological Sciences, The University of Western Australia, Perth, WA, Australia
Fusarium oxysporum f. sp. cubense (Foc) has severely curtailed banana production in the tropical regions of the world. The tropical race 4 (TR4) of Foc was detected in Australia in the 1990s and it is virulent to all Cavendish type banana cultivars, which represents the majority of banana production in Australia. Genetic resistance to Foc race 4 is urgently needed. To characterize sources of resistance, we have assessed the Foc resistance response of 34 Musa cultivars with plants grown under controlled settings. Amongst diploid banana cultivars carrying the AA genome, resistance is found in Musa acuminata sub-species including malaccensis ‘Pahang’ and burmannica ‘Calcutta4.’ In the polyploid group, the hybrids such as ‘FHIA-18’ and ‘FHIA-25’ are highly resistant against both Foc-TR4 and subtropical race 4 (Foc-STR4). Interestingly, ‘FHIA-2’ and ‘CAM020’ appear to be resistant to Foc-TR4 but susceptible to Foc-STR4, suggesting potential differences in the resistance mechanisms against the different race 4 strains. Using a GFP tagged Foc-STR4 strain challenged onto both resistant and susceptible M. a. malaccensis lines, a high inoculum dosage rapidly induced vascular wilt in the susceptible M. a. malaccensis lines at 2.5 weeks. This was associated with an accumulation of micro-conidia in the rhizome and the movement of the fungus through the xylem vessels. In contrast, the fungal movement was restrained in the rhizome of the resistant M. a. malaccensis lines and no sporulation was observed. Overall, this research suggests that the resistance response is dependent to an extent on inoculum dosage and that the plant host’s response, in the rhizome, plays an important role in inhibiting the fungus from spreading to the rest of the plant. Identifying race 4 resistant accessions can help to understand mechanisms of resistance and provide banana breeders with the genetic resources to integrate resistance genes into commercial varieties.
Introduction
Banana and plantain (Musa spp.) serve as important sources of staple food and fruit around the world and collectively are considered the world’s leading fruit crop, with a production value reaching over 100 million tons per annum (FAO, 2019). As a staple food, banana is an important export commodity in Africa and Asia, ensuring food security for millions of people (Aurore et al., 2009). One of the major constraints in the global production of banana is the disease, Fusarium wilt. It is also known as Panama disease, which is caused by Fusarium oxysporum f. sp. cubense (Foc) (Ploetz, 2006).
Foc gains entry into the plant host via roots. Once inside, it colonizes the rhizome and travels up the pseudostem, where it blocks the water-conducting xylem vessels and thus prevents the transport of water and nutrients to the aerial parts of the plant. External symptoms of Fusarium wilt start with the yellowing and wilting of the older leaves and progress to the younger leaves until the plant dies. Internally, the plants show brown discoloration and necrosis of xylem vessels in the rhizomes and stems. The disease incidence varies depending on the cultivar, the environment and the level of inoculum, but can extend to total crop loss in heavily infested fields (Moore et al., 2001).
Foc is a soil-borne pathogen that produces chlamydospores enabling its survival in the soil in the absence of the host. It is also known to survive on weed hosts in a non-pathogenic manner (Hennessy et al., 2005). Once the soil is infested with Foc, it generally becomes unsuitable for replanting for many years thereafter (Stover, 1990). Furthermore, unlike the black Sigatoka leaf spot caused by Pseudocercospora fijiensis, the other major fungal pathogen affecting the banana industry worldwide, Fusarium wilt cannot be controlled by fungicides (Ploetz, 2006).
Outbreaks of Fusarium wilt decimated a banana industry primarily based on the cultivar ‘Gros Michel’ (AAA) in Central America in the 1950s. The pathogen was subsequently named Foc race 1, and the outbreak forced the industry to shift production to the Foc race 1 resistant cultivars of the Cavendish (AAA) subgroup (Stover, 1990). Cavendish now accounts for >40% of world banana production, with export markets amounting to 15% of the total production (FAO, 2019).
Within the forma specialis (f. sp.) cubense, which include all isolates pathogenic to Musa spp. (Snyder and Hansen, 1940), there is significant pathogenic variation; members of this f. sp. are divided into four races based on their host range (Stover, 1990). Race 1 is pathogenic to ‘Gros Michel’ (AAA) and a range of other cultivars carrying the AAB genome. Race 2 targets those race 1-susceptible cultivars, as well as the hybrid triploid ‘Bluggoe’ (AAB). Race 3 affects Heliconia species and is no longer considered to be part of the cubense race structure. Race 4 is pathogenic to all race 1 and race 2 susceptible cultivars plus the Cavendish subgroup (AAA) (Su et al., 1986). Race 4 is further divided into two groups: tropical (TR4) and subtropical (STR4) race 4. Foc-STR4 isolates cause disease in Cavendish in the subtropics, whereas Foc-TR4 isolates are pathogenic to Cavendish under both tropical and subtropical conditions (Buddenhagen, 2009).
Foc isolates can also be grouped according to vegetative compatibility, which is the ability of an isolate to anastomose and form a stable heterokaryon (Moore et al., 1993). Isolates that are vegetatively compatible with one another form a VCG and typically share common biological, physiological and pathological traits (Caten and Jinks, 1966). At least 21 different VCGs of Foc have been characterized, with the majority of the groups present in Asia, where the pathogen is thought to have evolved (Fourie et al., 2009). Foc-TR4 isolates are designated as VCG 01213 (or VCG 01216, which is a different designation for the same VCG) (Bentley et al., 1998). Foc-STR4 isolates are designated as VCGs 0120, 0121, 0122, 0129, and 01211 (Buddenhagen, 2009). Since its appearance in Southeast Asia in the 1990s, Foc-TR4 (VCGs 01213 or 01216) has caused severe damage to Cavendish plantations in Malaysia, Indonesia, China, the Philippines and the Northern Territory and Queensland in Australia as well as Mozambique (Ploetz, 2006, 2015; Buddenhagen, 2009). The virulence and epidemic nature of Foc-TR4 is due to its potent pathogenicity and wide host range within the genus Musa.
Genetic resistance can offer a long-term means of control of Fusarium wilt, but no truly Foc-TR4 resistant commercially viable cultivar is available. Efforts have been made to identify Foc-TR4 resistant cultivars (Zhang et al., 2018; Zuo et al., 2018) but transferring these favorable alleles into a commercially viable cultivar with good agronomic traits has been a challenge (Dita et al., 2018). Somaclonal variants of Cavendish known as the ‘giant Cavendish tissue culture variants’ or ‘GCTCVs’ have been generated in Taiwan through tissue culturing and field trials and have shown that they possess some improved level of tolerance to Foc-TR4 (Hwang and Ko, 2004). However, these lines vary in their levels of tolerance and are considered by some to contain undesirable agronomic traits (Ploetz, 2015). Therefore, genotypic screening of cultivated and wild germplasm is extremely important in characterizing existing or novel sources of Foc race 4 resistance, which could assist phenotypic selection in breeding programs or facilitate the isolation of gene(s) underlying resistance for the purpose of engineering resistance in commercial cultivars via gene technologies.
In cultivated banana, four genome types have been identified and they include M. acuminata (A), M. balbisiana (B), M. schizocarpa (S) and those of the Australimusa section (T) (D’Hont et al., 2000). So far, most of the cultivated banana plants are diploid or triploid, originating from intra- or inter-specific hybridizations between the diploid A and the B genomes (Perrier et al., 2011). Most of the commercial cultivars are seedless and produce fruit by parthenocarpy, resulting in limited selection process and mono-culture productions. Most of these cultivars are therefore susceptible to biotic and abiotic stress, indicating a limited gene pool (D’Hont et al., 2012). Wild relatives of commercial varieties and other cultivated diploids such as ‘Pisang Jari Buaya,’ produce seeds and are considered good sources of genetic diversity worthy of exploration for improving resistance to Fusarium wilt. Indeed, resistance gene analog 2 (RGA2) was isolated from Musa acuminata ssp. malaccensis and has recently been shown to confer resistance to TR4 in field trials conducted in Australia (Peraza-Echeverria et al., 2008; Dale et al., 2017).
In this study, we assessed the resistance level of 34 banana cultivars against Foc-TR4 and Foc-STR4. These include diploids (AA and BB groups, and wild relatives) and intra- or inter-specific hybrids such as Cavendish banana ‘GCTCVs’ and plantains from the FHIA. We used a pot based bio-assay method to assess the level of resistance in each accession grown under glasshouse conditions. We characterized several Foc race 4 resistant diploids as a source of resistance; these sources may have already been incorporated into the resistant hybrids through conventional breeding. We also show that resistance levels vary amongst the genotypes and resistance is likely of quantitative nature across a spectrum. The level of observed resistance under controlled environmental conditions can be determined by factors such as inoculum dosage, maturity of plants and inoculation technique. Using a GFP tagged Foc-STR4 strain, we show that the rhizome plays an important role as a barrier to the pathogen preventing it from migrating toward the rest of the plant. We identified a diploid wild relative that exhibits a strong resistance response to Foc manifested by reduced fungal penetration in root cells and containment of the fungi in the rhizome. This is therefore, a potential novel source of resistance to Foc race 4 types.
Materials and Methods
Fusarium Strains
The monoconidial Foc-TR4 strain (NTPDc 35673) was originally collected from the Coastal Plains Research Farm at in the Northern Territory, Australia in the early 2000s. In 2016, VCG testing performed on this strain by Department of Agriculture and Fisheries (DAF, Nambour, QLD, Australia) confirmed that it is VCG 01213/16.
Three monoconidial isolates of Foc VCG0120 were obtained from the Queensland Plant Pathology Herbarium (BRIP; 63488, 43781, and 42331) at the Queensland Department of Agriculture and Fisheries. Foc BRIP 23598 from VCG 0120 was previously transformed with GFP (Henceforth, GFP-Foc-STR4) and stored at -80°C (Forsyth, 2006).
All Foc isolates were cultured on half-strength potato dextrose agar (PDA) Difco (Becton, Dickson and Co., Sparks, MD, United States) for 7 days at 25°C. The GFP-Foc-STR4 isolate was regenerated on the same media but with the addition of 50 mg L-1 of hygromycin B.
Preparation of Millet Inoculum
To prepare millet grain for Foc-STR4 inoculum, millet (Pennisetum glaucum) seed was washed in tap water, covered with distilled water, and then soaked overnight in a suitable container. Excess water was drained from the seed using a sieve, then the grain was rinsed a second time in distilled water to remove leached carbohydrate. The grain was then placed into Erlenmeyer flasks or other suitable containers and autoclaved twice for 20 min at 120°C on consecutive days. Once cooled, the grain was inoculated with approximately five 1 cm squared mycelial plugs cut from Foc cultures grown on half strength PDA. Flasks were shaken daily to distribute Foc evenly. When the millet was fully colonized by the Fusarium, it was used as plant inoculum.
The Foc-TR4 inoculum was prepared with the following modifications to allow an up-scale. Batches of 1.5 kg of millet were placed in small autoclave bags. After the addition of 500 mL of RO water, the bags were sealed and autoclaved twice over consecutive days. Half of a PDA plate containing Foc-TR4 was added to each bag of autoclaved millet. The bags were shaken every second day.
For GFP studies, four to five mycelial agar plugs of GFP-Foc-STR4 were added to half strength potato dextrose broth (PDB, Difco) containing 50 mg L-1 hygromycin B and shaken gently at 28°C for 5 days. The culture was processed using a previously described method (Li et al., 2011) to extract viable spores and a final concentration of 2 × 106 conidia mL-1 was prepared for root dipping. All work involving GFP transformed strains of Foc, was conducted under conditions of an NLRD (notifiable low risk dealing) permit according to the Office and Gene Technology Regulator, Australia.
Plant Materials and Growth
Wild relatives of the cultivated banana are known to harbor resistance against Foc. So far, field studies have identified resistant genotypes against Foc race 4 and those include diploids (AA genome) from Musa accuminata ssp. malaccensis, Musa accuminata ssp. Banksii, and Musa accuminata ssp. burmannica (Table 1). In this study, we selected 34 accessions of various ploidy levels to assess their resistance response to Foc-STR4 and Foc-TR4 in pot trials (Table 1). The diploids used include M. a. malaccensis from Sumatra, Indonesia (‘Ma846,’ ‘Ma848,’ ‘Ma850,’ ‘Ma851,’ ‘Ma852’) and from Malaysia, ‘Pahang’ and M. a. malaccensis of ITC0250. We also examined ‘Calcutta4’ (M. a. burmannica) and ‘Pisang Jari Buaya’ (AA) that have been shown to carry race 4 resistance in field trials (Table 1). Other diploid cultivars tested include ‘SH3217,’ ‘SH3362,’ and ‘SH3142’ that have been used extensively in breeding programs such as FHIA. The hybrids with polyploid genomes tested include ‘FHIA-1,’ ‘FHIA-2,’ ‘FHIA-3,’ ‘FHIA-18,’ ‘FHIA-23,’ ‘FHIA-25,’ ‘FHIA-26.’ We also looked at the two ‘GCTCVs’ lines (119 and 218) of Taiwan origin that exhibited improved tolerance against Foc-TR4 in the field (Hwang and Ko, 2004). Lines from breeding programs were also examined. These include ‘M61 Guadeloupe,’ an elite bred diploid from the Jamaican breeding program and ‘CAM020’ which is an F1 individual from a cross between ‘Calcutta 4’ and ‘Banksii Madang.’ It is part of the ‘AFCAM20’ population developed by INIBAP (International Network for the Improvement of Banana and Plantain).
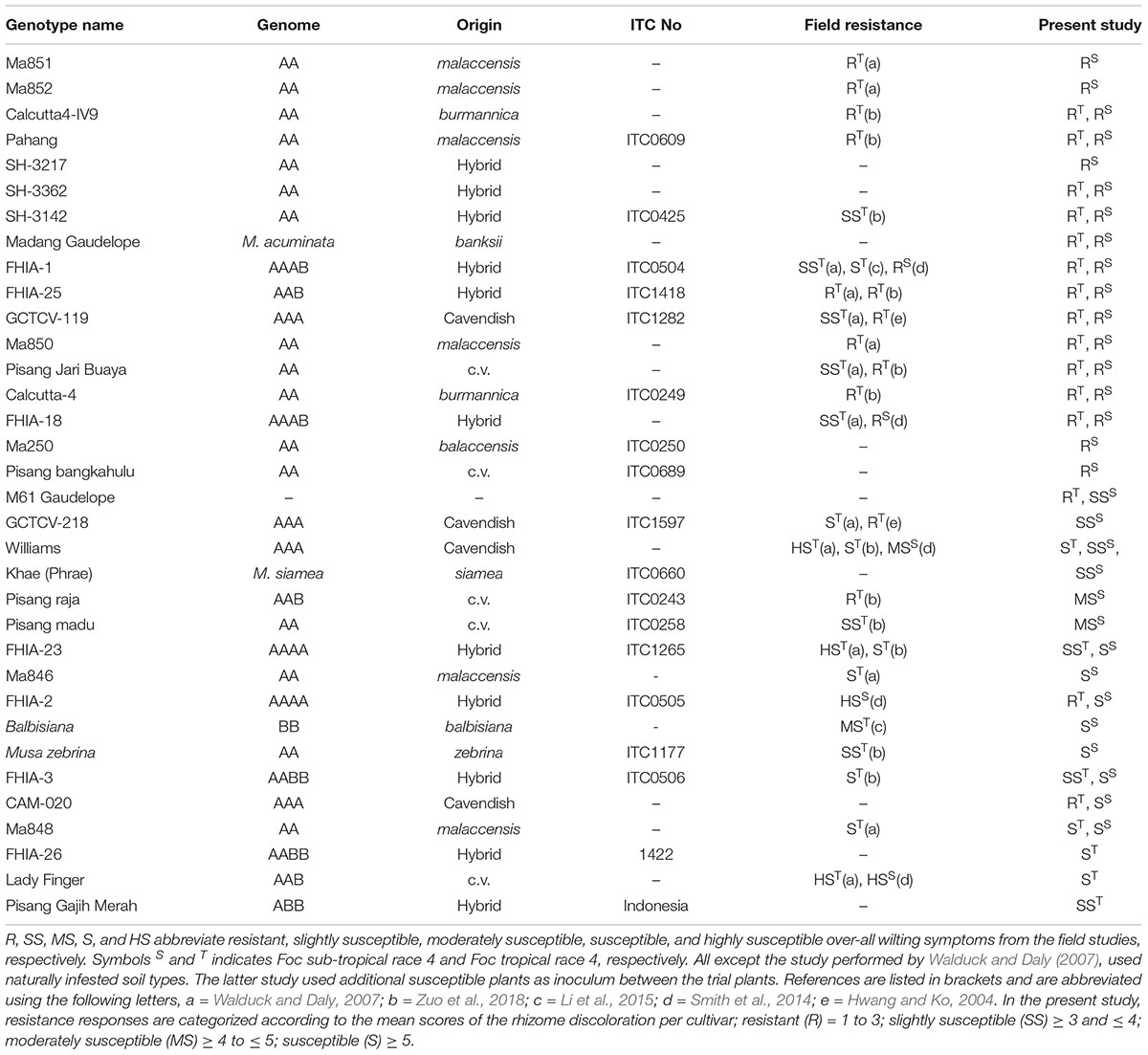
Table 1. List of genotypes presented in this study, and the available corresponding field studies that determined their resistance responses against Foc race 4 types.
Banana germplasm was kindly supplied by the Maroochy Research Facility at the Queensland Department of Agriculture and Fisheries. Germplasm is listed in Table 1 and those with known corresponding ITC numbers are indicated. Six to eight clones of each accession from Table 1 were micro-propagated and maintained in vitro as per previous study (Smith and Hamill, 1993). Seedlings approximately 10 cm in height with three to four true leaves were de-flasked into 30 well seedling trays to harden off in the laboratory under constant LED lights. The seedlings were transferred into 200 mm diameter pots in designated spaces for disease screening. Foc-TR4 is strictly quarantined in Australia and pathogen screening was instead carried out in a screen house at the Coastal Plains Research Station (Department of Primary Industry and Resources, Northern Territory) in Darwin, Australia. The average daily temperature in Darwin ranged from 30 to 35°C during the August to November months when the pot trials were conducted. Foc-STR4 screening was performed in the University of Queensland (UQ) glasshouse facility with temperature control set at 25 to 28°C. The potting mix contained 70% composted pink bark and 30% coco peat at a pH range of 5.5 to 6.5.
Plant Inoculation
Once the pseudostem of most plants reached 30 cm in height and each plant carried five to six true leaves, the plants were inoculated with either Foc-STR4 or Foc-TR4 infested millet or spores of GFP-Foc-STR4 at pre-determined concentrations. A total of 40 g of Foc-STR4 or Foc-TR4 infested millet was used as inoculum per 200 mm diameter pot. For inoculation, plants were briefly removed from the pots and 20 g of millet was added to the bottom of the pots before plants were re-potted. Another 20 g of millet was then spread evenly on the surface soil layer and finally covered with additional soil.
Assessment of Symptoms
Plants inoculated with Foc-STR4 or Foc-TR4 infested millet were scored for internal symptoms 10 to 12 weeks post-inoculation. The rhizome was vertically cut in halves to score the extent of discoloration. The rating scale for rhizome discoloration ranges from 1 to 8, with 1 being no discoloration and 8 indicating the entire rhizome is discolored and the plant dead (Mak et al., 2004). The mean score from three to eight plants was used to quantify the plant response.
Confocal Microscopy
Confocal microscopy was used to visualize GFP-Foc-STR4 in two separate experiments. Firstly, ‘Ma851’ self-derived progeny ‘p168’ (resistant) and ‘p248’ (susceptible) were examined for the presence of GFP-Foc-STR4 in the rhizome. Plants were inoculated with GFP-Foc-STR4 infested millet. Assessment was performed at 3 months post-inoculation. In the second experiment, the parent ‘Ma851’ (resistant) and ‘Ma848’ (susceptible) were used. Plant roots were dipped in a GFP-Foc-STR4 spore suspension containing 2 × 10-6 spores per mL for 2 h. Inoculated plants were then re-potted using soil containing 50,000 GFP-Foc-STR4 conidia per g of soil which is 5–10-fold of what is typically used in this type of assay (Li et al., 2012). The pathogen infection processes in these plants were observed at 1, 2, 3, 4, 7, and 14 dpi.
Transverse and longitudinal sections were hand-prepared to visualize pathogen development on the root surface and in the vascular bundles and the corms. Sliced sections were counter-stained with propidium iodide (PI, Sigma Aldrich) for 5 min at a concentration of 10 μg mL-1. A Zeiss 700 laser scanning microscope was used to visualize and acquire the confocal images. The GFP and the PI were detected using the 488 and 555 nm lasers, respectively. Z-stack acquisition mode was used to obtain 3D images consisting of 10–20 optical slices taken at intervals of 1–5 μm. T-PMT (transmission detector setting) was used to obtain the light images of the sectioned tissue.
Koch’s Postulates
Primary isolations were performed on three resistant (104A, 3A, 18A) and one susceptible (96B) self-derived progeny of ‘Ma851’ to investigate the presence or absence of GFP-Foc-STR4. Fungal strains were isolated 3 months post-inoculation with GFP-Foc-STR4 infected millet. Five regions were isolated per plant, which included the upper stem just below the first leaf petiole, mid-point of the stem, stem just above the rhizome, the central cylinder of the rhizome and the outer layer of the rhizome connected to the cortex. Four pieces of tissues were isolated in each region. These sectioned pieces were surface sterilized for 1 min in 1% hypochlorite solution, then rinsed twice in distilled water for 30 s each time. Each piece was air-dried and plated onto water agar plates containing 100 mg L-1 streptomycin for 5 days at 24°C. Segments showing Fusarium-like growth under a dissecting microscope were further isolated and then hyphal tipped to generate monoconidial isolates on half strength potato dextrose agar (PDA) plates containing 50 mg per L hygromycin B. Each isolate was then transferred to 20 mL of potato dextrose broth containing 50 mg L-1 hygromycin B (PDB) and incubated at 26°C on a platform rotating at 160 rpm for 4 days. GFP fluorescence of the isolates was visualized under Zeiss 700 confocal microscope to confirm the presence of the GFP-Foc-STR4 strain.
Results
Foc-STR4 Screening Trial
Thirty-four genotypes were tested for their response to Foc-STR4 under glasshouse conditions (Table 1). Following inoculation, the rhizome discoloration was scored using a predetermined scale (Mak et al., 2004) which is presented in Table 2. A rhizome score of 1 or 2 indicates no discoloration in the rhizome 3 months after inoculation and is associated with the AA diploids ‘Ma851,’ ‘Ma852,’ ‘Calcutta4-IV9,’ ‘SH-3217,’ ‘SH-3362,’ ‘SH-3142,’ as well as the hybrids ‘FHIA-1’ (Gold Finger), ‘FHIA-25’ and ‘GCTCV-119’ (Figure 1). Accessions that had a clear rhizome but had little or some discoloration around the junctions of roots joining the rhizome are categorized by a score of 2 to 3 (≤5% discoloration) and include the diploids ‘Ma850,’ ‘Pisang Jari Buaya,’ ‘Calcutta-4,’ ‘FHIA-18,’ ‘Ma250,’ and ‘Pisang bangkahulu.’ In this group, some of these lines, namely ‘Ma850,’ ‘Pisang Jari Buaya,’ ‘FHIA-18,’ and ‘Ma250’ had large phenotypic variation in the degree of rhizome discoloration amongst individual clones, possibly due to somaclonal variations of genetic or epigenetic origin (Figure 1 and Supplementary Figure 1). These lines also exhibited a good level of resistance against Foc-TR4 in previous field studies (Table 1). Accessions that display up to 20% rhizome discoloration fall into the slightly susceptible (SS) group. These include‘ ‘M61 Gaudelope,’ ‘GCTCV218,’ ‘Williams,’ ‘Khae’ (Figure 1). ‘Pisang raja’ and ‘Pisang madu’ showed discoloration in the 21 to 50% range and are hence considered moderately susceptible (Figure 1). The accessions that showed more than 50% discoloration are considered susceptible (S) to Foc-STR4. These include ‘FHIA-2,’ ‘FHIA-23,’ ‘Ma846,’ Musa balbisiana, Musa accuminata ssp. zebrina, ‘FHIA-3,’ ‘CAM020,’ and ‘Ma848’ (Figure 1). ‘CAM-020’ and ‘Ma-848’ were extremely susceptible to Foc-STR4; half of the clonal plants were dead at the time of assessment.

Table 2. Rhizome discoloration index as per previously described (Mak et al., 2004).
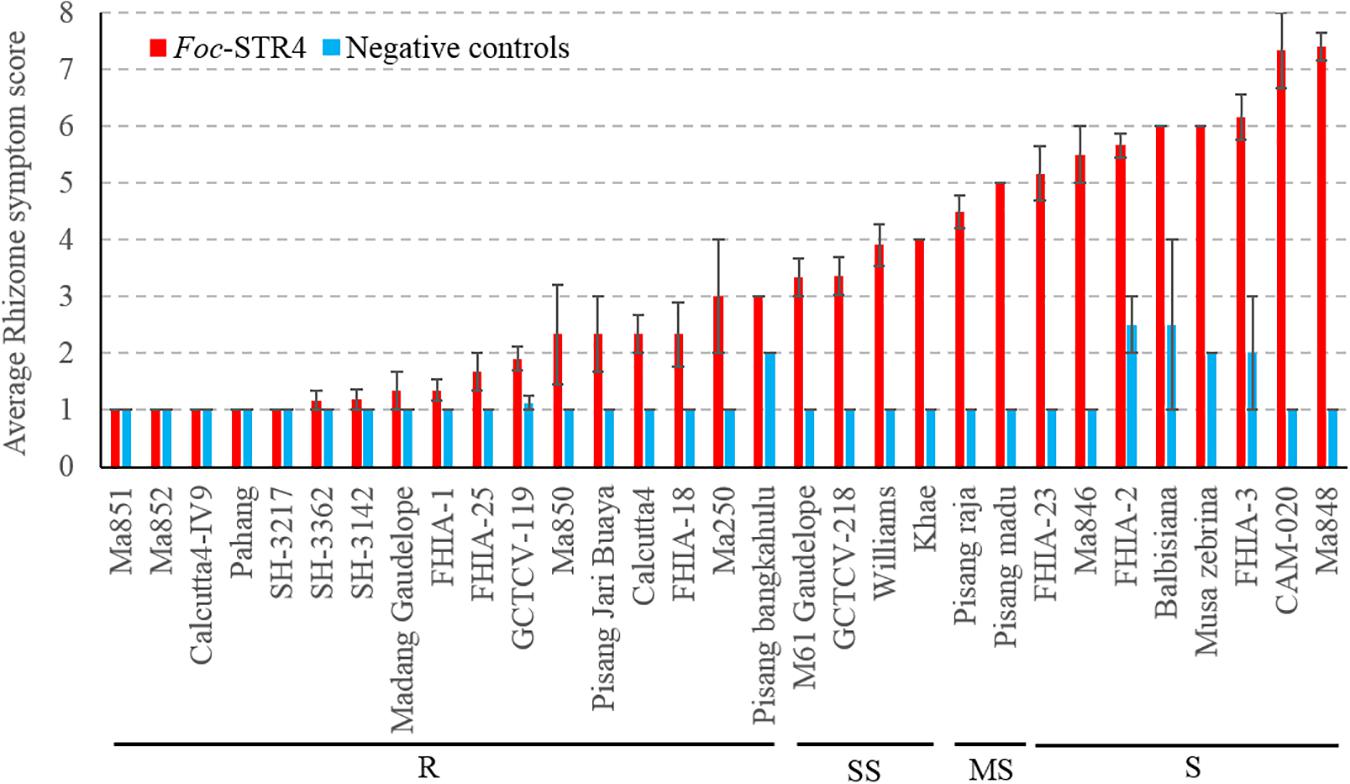
Figure 1. Sensitivity of different banana genotypes to Foc-STR4 in glasshouse pot trials. Internal symptoms were assessed by using a scoring system based on the percentage of rhizome discoloration (Mak et al., 2004). Millet was inoculated separately using three Foc-STR4 isolates BRIP63488, BRIP43781, and BRIP42331 of VCG 0120. Fully colonized millet from each strain was mixed in equal amounts and 40 g of inoculum was applied to each 200 mm diameter pot. Clean autoclaved millet was used as negative controls. Error bars indicate standard deviations of the mean derived from three to eight individual plants. Genotypes that have rhizome symptom scores between 1 and 3 (≤5% rhizome discoloration) are categorized in the resistant group (R); those with a score between 3 and 4 (>5 to ≤20% discoloration) are categorized in the slightly susceptible group (SS); those that score between 4 and 5 (>21 to ≤50%) are categorized as susceptibles (MS); those that score greater than 5 (>50%) is categorized as susceptible (S).
Foc-TR4 Screening Trial
Due to logistical constraints, only a subset of accessions was selected for testing against Foc-TR4 in the Northern Territory, Australia under shade house conditions. At the time of the assessment, the susceptible plants did not show noticeable external symptoms, however, internal symptoms were present. Internally, the diploids M. a. malaccensis (AA) ‘Pahang’ and ‘Ma850’ displayed clean rhizomes that suggests a high level of resistance against Foc-TR4 (Figure 2). Other accessions that produced resistant (R) rhizome phenotypes include ‘FHIA-1,’ ‘FHIA-2,’ ‘FHIA-18,’ ‘FHIA-25,’ ‘Calcutta4,’ ‘Calcutta4-IV9,’ ‘SH-3142,’ ‘SH-3362,’ ‘Pisang Jari Buaya,’ ‘Madang Guadelope,’ ‘M61 Guadelope,’ ‘CAM-020,’ and ‘GCTCV-119’ (Figure 2). The SS group includes ‘Pisang Gajih Merah,’ ‘FHIA-3,’ and ‘FHIA-23.’ All three cultivars displayed a high level of phenotypic variation in clonal plants, possibly owing to somaclonal variations (Figure 2 and Supplementary Figure 2). ‘Lady Finger,’ ‘Williams,’ ‘FHIA-26,’ and ‘Ma848’ all showed severe discoloration in their rhizomes and hence are considered as susceptible (S) to Foc-TR4 (Figure 2).
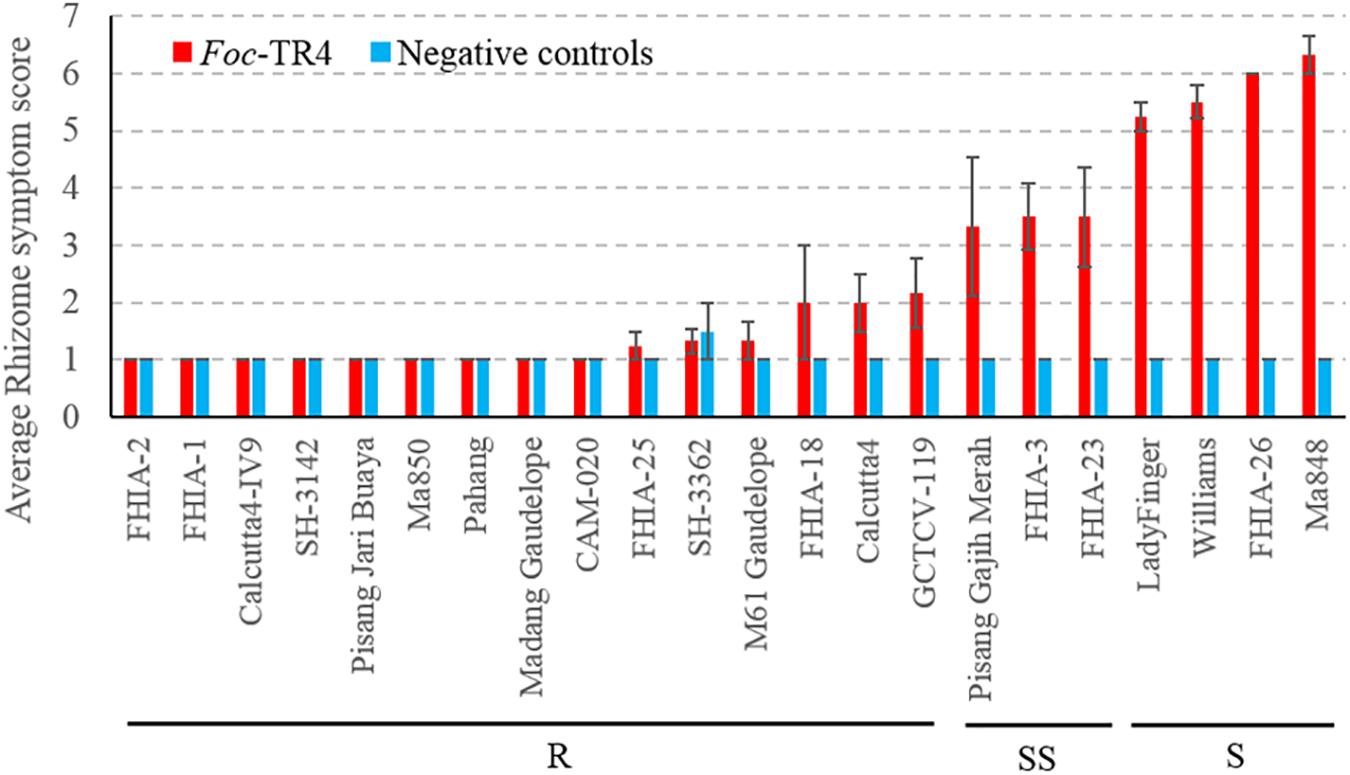
Figure 2. Sensitivity of different banana genotypes to Foc-TR4 in the shade house pot trial. Internal symptoms were assessed by using a scoring system based on the percentage of rhizome discoloration (Mak et al., 2004). Millet infested with Foc-TR4 strains carrying VCG 01213/16 was used as the inoculum. For each 200 mm diameter pot, 40 g of the inoculum was applied. Clean autoclaved millet grains were used in uninoculated control treatments. Error bars indicate standard deviations of the mean derived from three to eight individual plants. Abbreviations and categories of resistance are the same as per Figure 1.
Comparison Between Foc-STR4 and Foc-TR4 Induced Responses
Most accessions showed consistent responses between the two race 4 types (Table 1). ‘Williams,’ which belongs to the Cavendish subgroup, is the dominant variety used in commercial production in Australia. In our study, it was highly susceptible to Foc-TR4 but was only partially susceptible against Foc-STR4 when tested under glasshouse conditions (Figures 1, 2). Our results are consistent with the resistance response of ‘Williams’ to race 4 types detected in the field (Table 1). In these studies, Cavendish types (AAA) have generally shown high susceptibility against Foc race 4 but improved resistance has been detected in Cavendish variants in the field (Bhagwat and Duncan, 1998; Hwang and Ko, 2004). The giant Cavendish type somatic mutant ‘GCTVC119’ showed resistance against both Foc-STR4 and Foc-TR4 in our study (Figures 1, 2). However, the variant ‘GCTCV-218’ showed slight susceptibility against Foc-STR4. Foc-TR4 resistance levels of ‘GCTCV-218’ does not appear to be consistent as shown by field results (Table 1), possibly suggesting the presence of genotype-environment interactions at the different trial locations. ‘M61 Gaudelope,’ ‘FHIA-3,’ ‘FHIA-23,’ ‘Pisang raja’ all showed a relatively higher level of susceptibility to Foc-STR4 than Foc-TR4 (Table 1). Interestingly, ‘FHIA-2’ and ‘CAM-020’ were both resistant to Foc-TR4 but showed highly susceptible rhizome phenotypes to Foc-STR4 (Figures 1, 2).
Pathogen Infection Process
Fluorescence produced by GFP was visualized to assess the extent of the spread of the GFP-Foc-STR4 inside the rhizome of plants that had been inoculated with millet infested with the fungus. Resistant ‘p168’ and susceptible ‘p248’ progeny of ‘Ma851’ were compared. ‘P168’ and ‘p248’ were the self-pollinated progeny between the clonal plants of the parent ‘Ma851.’ Previously, segregation analysis using the self-derived progeny of ‘Ma851’ showed that they segregated for both Foc race 4 type resistance at a 3 : 1 (resistance : susceptibility) ratio indicating the respective presence of a single dominant resistance gene in ‘Ma851’ (Fraser-Smith et al., 2016). Three months after the initial inoculation, ‘p168’ showed minimal leaf symptoms, no stem splitting and a healthy rhizome (Figures 3a–c). In contrast, ‘p248’ showed necrotic lesions and the wilting of old leaves, a split stem and moderately discolored rhizome (Figures 3d–f). Confocal imagery of the inoculated resistant ‘p168’ line showed that the GFP was associated with the vascular bundles in the central cylinder toward the lower part of the rhizome (Figures 4A,B). GFP fluorescing hyphae was present in the cortex region near the xylem perforation plates (Figure 4B). Furthermore, patches of mycelial networks were detected at a low rate in the cortex region of ‘p168’ rhizome (Figure 4C). In contrast, mycelia associated with a strong GFP signal was detected in the mid and lower regions of the ‘p248’ rhizome where black discoloration was observed (Figures 4D,E).

Figure 3. Self-derived F2 progeny of the resistant parental line ‘Ma851’ showing contrasting resistance response against GFP-Foc-STR4. Plants were inoculated with GFP-Foc-STR4 infested millet and photographs were taken 3 months after inoculation. (A) The resistant progeny plant ‘p168’ showing no leaf wilting symptoms and was relatively healthy. (B) The base of the pseudostem just above the soil showing no signs of stem splitting, which is a manifestation of the wilting disease. (C) The rhizome of ‘p168,’ cut in half longitudinally, showed no traces of brown discoloration in the lower and center regions. (D) The susceptible progeny plant ‘p248’ derived from the same F2 cross displayed wilting of the old leaves. (E) Stem splitting at the base of ‘p248’ was clearly visible just above the soil level. (F) The rhizome of ‘p248’ rhizome developed extensive brown discoloration which is typically associated with high Foc susceptibility. Arrows indicate the sites of external and internal symptoms at the time of assessment. w = wilting of old leaves, ps = pseudostem splitting, rd = rhizome discoloration, pd = pseudostem discoloration.
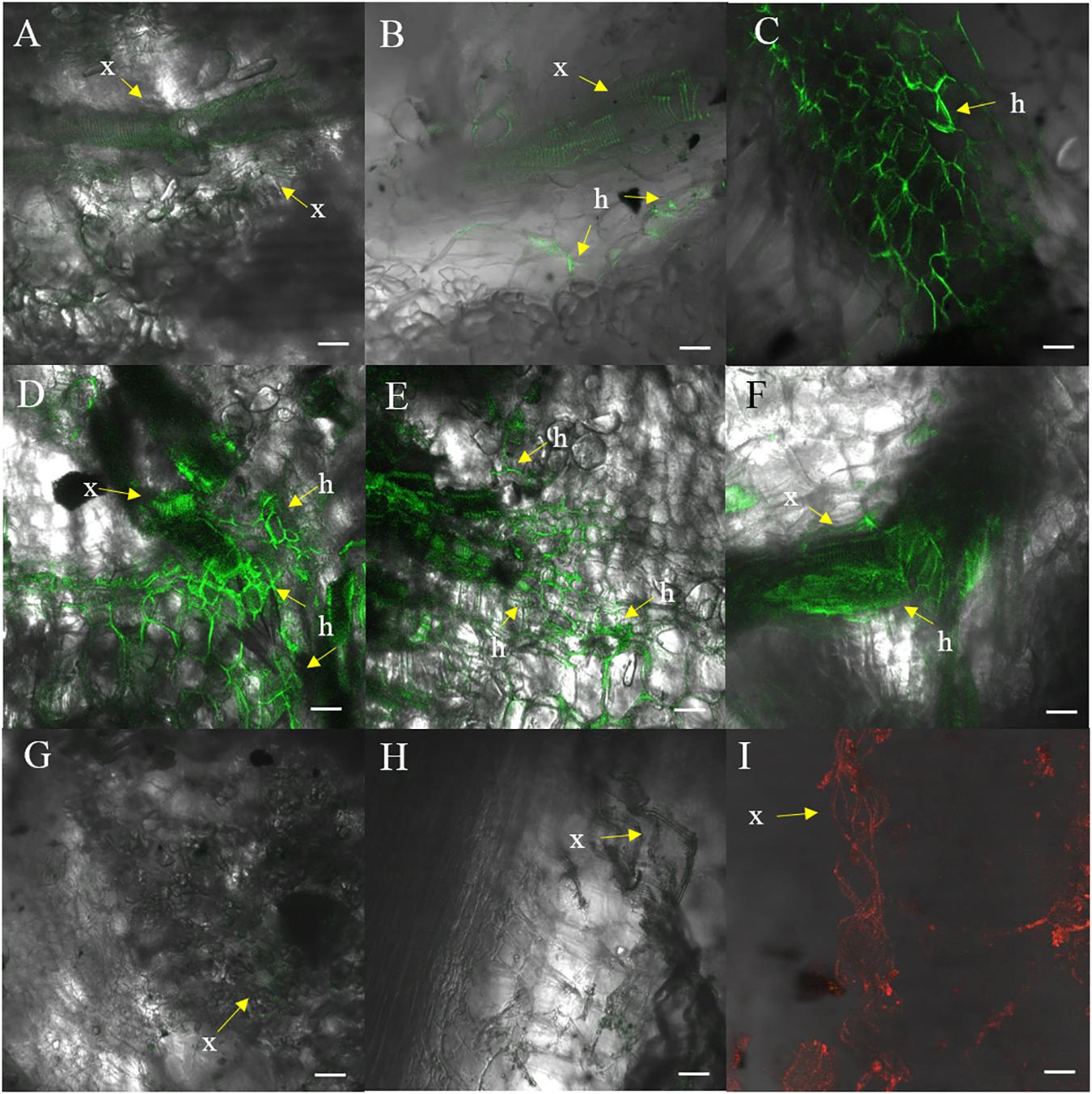
Figure 4. Confocal images of ‘p168’ (resistant) and ‘p248’ (susceptible) at 3 months post-inoculation performed using millet and GFP-Foc-STR4. (A) Xylem perforation plates observed containing GFP observed in the lower region of the ‘p168’ rhizome. (B) GFP linked to the perforation plates and the hyphae structure in the lower region of the rhizome bordering the cortex cells in ‘p168.’ (C) Presence of GFP tagged mycelial networks in the lower region of the rhizome in ‘p168.’ (D) A cross section of the lower region of the ‘p248’ rhizome showing xylem perforation plates and the expansion of mycelial networks associated with GFP. (E) A longitudinal section of the lower region of the ‘p248’ rhizome showing GFP associated with perforation plates and the expanded mycelial networks. (F) Mycelial structures establishing near the boundary between the central region of the rhizome and the cortex area in ‘p248.’ (G) Confocal showing the absence of GFP in the lower rhizome of the un-inoculated control ‘p4-19’ derived from ‘Ma851.’ (H) Xylem perforation plates with no associated fluorescence in the non-inoculated controls of ‘p4-19.’ (I) Xylem perforation plates visualized under a 555 nm laser using non-inoculated ‘p4-19’ rhizome stained with propidium iodide. GFP was visualized using the 488 nm laser. Light images are created using the T-PMT (transmitted light detector) setting. Scaled bars represent a 50 μm unit. Arrows indicate the presence of xylem vessels (x) and hyphae (h).
The GFP signal of GFP-Foc-STR4 is strongly associated with the xylem perforation plates. Hyphal structures containing GFP were clearly observed in the cortex cells surrounding the vascular bundles (Figures 4D,E). The GFP-Foc-STR4 was strictly localized to the vascular bundles of the xylem and in the surrounding region (Figure 4F) suggesting that the fungus moves via the xylem. The cortex region of the uninoculated control showed little to no GFP fluorescence (Figure 4G). The xylem vessels of the uninoculated control were observed under the transmitted light setting (Figure 4H) and when stained with propidium iodide, they were associated with red-fluorescence at 555 nm (Figure 4I).
Although no internal symptoms were detected in the resistant genotype ‘p168,’ the presence of GFP-Foc-STR4 in its rhizome suggests that resistance does not inhibit the pathogen from gaining entry into its roots. To further evaluate the resistance response, we used two of the Musa acuminata ssp. malaccensis parental lines, one that has been shown to be resistant, ‘Ma851,’ and one susceptible, ‘Ma848,’ against both Foc-STR4 and Foc-TR4 (Fraser-Smith et al., 2016). Progeny of ‘Ma851’ segregates for Foc resistance (Figure 3) whereas ‘Ma848’ showed an extremely susceptible phenotype indicative of the absence of any resistance genes (Figures 1, 2). In this assay, a Foc spore suspension and a root dipping inoculation method was used which has been reported previously to study the onset of the infection process (Li et al., 2011; Zhang et al., 2018). The inoculation rapidly induced wilting in the susceptible ‘Ma848’ lines with the first signs of wilting observed at 2–3 weeks post-inoculation (Supplementary Figure 3). ‘Ma848’ plants were observed to be dead at 4 weeks (Figure 5A). Inoculated ‘Ma851’ and un-inoculated controls of both lines did not show any external symptoms at 4 weeks (Figures 5A,B). When cut in halves, the rhizomes of ‘Ma848’ showed extensive discoloration as typically associated with necrotic lesions of vascular vessels (Figure 5C). The symptoms also include the discoloration of the pseudostem and a root structure of reduced size (Figures 5D,E). While ‘Ma851’ showed some noticeable discoloration in the lower part of the rhizome, discoloration in the pseudostem was not observed (Figures 5C,D).
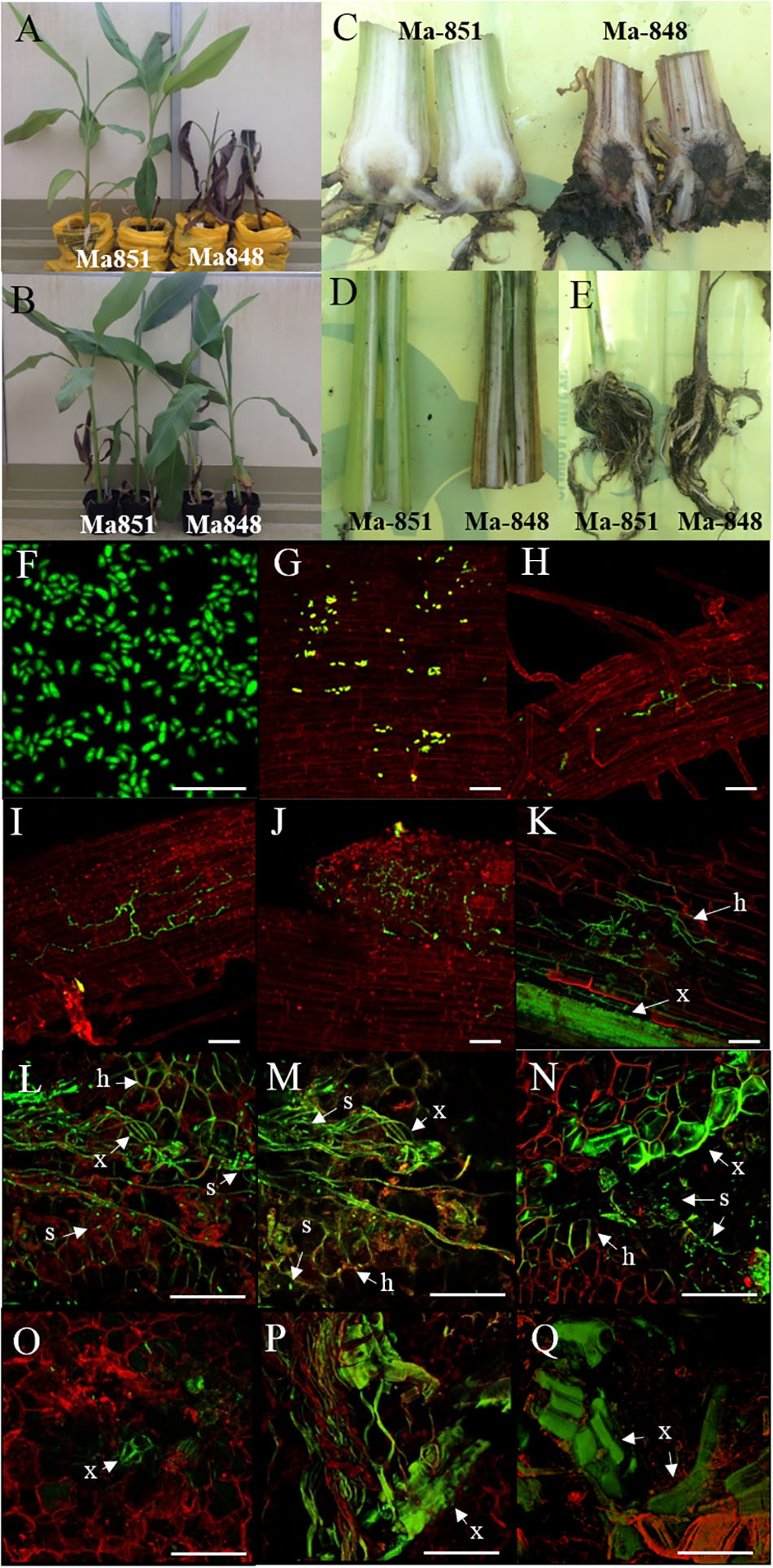
Figure 5. Characterization of Foc-race 4 resistance from Musa acuminata subsp malaccensis using a GFP tagged Foc-STR4 isolate (BRIP 23598, VCG 0120). (A) Plant phenotypes of the resistant ‘Ma851’ and susceptible ‘Ma848’ M. a malaccensis plants at 28 days post-inoculation (dpi). (B) Non-inoculated controls of ‘Ma851’ plants (left) and ‘Ma848’ plants (right). Longitudinal sections of (C) rhizomes, (D) pseudostem and (E) entire roots of ‘Ma851’ and ‘Ma848’ at 28 days dpi, respectively. (F) Spores collected from suspension culture of BRIP23598 after 5 days of growth in half strength PDB containing 50 mg per L of hygromycin B. (G–K) Visualization of the GFP protein on ‘Ma848’ under a confocal microscope. (G) Attachment of spores to the lateral root surface at 1 dpi. (H) Movement of hyphae in the epidermis layer of a lateral root at 2 dpi. (I) A mycelial network established on the epidermis at 4 dpi. (J) The apical meristem region of a root tip completely colonized by GFP-Foc-STR4 at 7 dpi. (K) Mycelial networks are established in the cortex and vascular bundles of the xylem at 14 dpi. The (L) upper, (M) mid and (N) lower sections of the rhizome from a ‘Ma848’ plant at 14 days dpi. The (O) upper, mid (P) and lower (Q) sections of the rhizome from a ‘Ma851’ plant at 14 days dpi. White bars indicate a 50 μm scale. GFP fluorescence was detected at 488 nm wavelength using a Zeiss 700 laser scanning microscope. The tissues were stained with propidium iodide to produce a red fluorescence which was detected at 555 nm wavelength. x = xylem vessels, s = individual or clumps of spores, h = hyphae.
To study the infection process, the presence of GFP-Foc-STR4 in ‘Ma848’ and ‘Ma851’ was visualized with confocal microscopy. The GFP tagged Foc-STR4 spores were confirmed to be constitutively fluorescing prior to the start of the experiment (Figure 5F). Adhesion of GFP-Foc-STR4 spores to the lateral roots of ‘Ma848’ was observed at 1 day post-inoculation (dpi, Figure 5G). The presence of hyphae on the epidermis of lateral roots of ‘Ma848’ was first observed at 2 dpi (Figure 5H). A longitudinal section of a lateral root shows the elongation of hyphae and the establishment of a mycelial network on the epidermis of lateral roots of ‘Ma848’ at 4 dpi (Figure 5I). Complete colonization of a lateral root tip by GFP-Foc-STR4 was observed in ‘Ma848’ at 7 dpi (Figure 5J). The presence of GFP-Foc-STR4 was not detected on the main roots of ‘Ma848’ during the first 7 dpi. A single hypha was observed attempting to penetrate the epidermis cell layer of the lateral roots in ‘Ma848’ at 7 dpi (Supplementary Figure 4A). Presence of hyphae was also detected on the epidermal cell layer of the fine roots in ‘Ma848’ (Supplementary Figure 4B). The presence of GFP-Foc-STR4 was not detected in the roots of the resistant line ‘Ma851’ (Supplementary Figures 4C–E). At 14 dpi, GFP tagged mycelium was observed in the root cortex region of ‘Ma848’ (Figure 5K). The xylem vessels also showed a strong GFP signal indicating the presence of the fungus within the xylem. The rhizome was sectioned into three parts, which include the upper region closer to the pseudostem, the middle region which is the central cylinder and the lower region that connects to the cortex and the roots. At 14 dpi, all three regions of the ‘Ma848’ rhizome showed the abundant presence of spores near the xylem perforation plates which were also infected with GFP-Foc-STR4 (Figures 5L–N). GFP-Foc-STR4 can be seen colonizing the cortex cells around the xylem vessels in ‘Ma848.’ In contrast, no spores were observed in the rhizome of the inoculated resistant ‘Ma851’ plants (Figures 5O–Q). Movement of hypha was also not observed. However, strong GFP fluorescence was observed in the xylem vessels, indicating the likely presence of GFP-Foc-STR4 in these vessels (Figures 5O–Q). These data suggest that the frequency of fungal penetration via the roots was greatly induced in ‘Ma851.’
Isolation of GFP-Foc-STR4 Using Koch’s Postulates
Susceptible ‘p96’ and resistant ‘p3,’ ‘p18,’ ‘p104’ self-derived progeny of ‘Ma851’ were inoculated with GFP-Foc-STR4 and grown in a glasshouse for 3 months. At harvest, internal and external symptoms of Fusarium wilt were visible on the ‘p96’ plants (Supplementary Figures 5, 6). Extensive leaf wilting, corm rot and stem splitting were present in the susceptible ‘96’ plant but not in the resistant plants. Some slight discoloration of the corm was evident in the resistant plants (Supplementary Figure 5).
The presence of GFP-Foc-STR4 was detected in most of the regions isolated from the susceptible ‘p96’ plant, which include the throat and the mid-stem region of stem, and the mid-corm and cortex region of the corm (Supplementary Figures 7, 8 and Table 1). GFP-Foc-STR4 was not detected at all in two resistant ‘p3’ and ‘p18’ plants. However, it was isolated with the presence of the GFP confirmed in the regions of the throat in the third resistant ‘p104’ plant (Supplementary Figures 7, 8 and Table 1).
Discussion
Fusarium wilt is one of the major threats confronting the banana industry today. An effective long-term solution to this problem would be to identify genetic resistance from the untapped gene pools in the wild relatives of the cultivated banana and introduce the identified sources of resistance loci back into commercially viable varieties. Resistance screens are traditionally performed in the field, which is labor-intensive owing to the long growth cycle and the large size of banana plants. In this study, a glasshouse screen method was adopted to assess resistance level in relatively young plants grown in pots. This type of assay can have a short turn-over time due to the up-scaling of the number of plants that can be tested at a given time under controlled conditions with minimized cross infection by other micro-organisms (Smith et al., 2008; Li et al., 2015; Zuo et al., 2018). In addition, high inoculum dosage can be applied to identify highly resistant germplasm.
Resistance against F. oxysporum f. sp. cubense was assessed in a collection of 34 genotypes of diploid and polyploid banana plants in the glasshouse (Table 1) and their resistance levels were ranked using the internal discoloration of the rhizome. A wide range of disease responses from completely resistant, to partially resistant and highly susceptible, was revealed in the plant rhizomes against Foc race 4 types (Figures 1, 2). Resistance levels of the genotypes tested were mostly consistent with previously published field data (Table 1). This response shows that resistance to Foc race 4 is mainly of a quantitative nature. Quantitative resistance has been detected in other pathosystems including Pisum sativum and Medicago truncatula against Fusarium oxysporum f. sp. pisi and Fusarium oxysporum f. sp. medicaginis, respectively (Bani et al., 2012; Rispail and Rubiales, 2014). In this study, wilt resistance can be detected in each of the diploid, triploid and tetraploid genome groups and in different Musa accuminata ssp. malaccensis, Musa accuminata ssp. burmannica, Musa accuminata ssp. banksia.
The M. acuminata ssp. malaccenis line ‘Ma851’ appears to carry strong wilting resistance to Foc. Wild M. acuminata diploids are highly diversified and have been heavily integrated in the breeding of edible banana cultivars (Perrier, 2009; Perrier et al., 2011; Li et al., 2013; Christelova et al., 2017). Geographically M. acuminata sub-species shows a wide-spread pattern of distribution in East Asia with each sub-species localized in distinct regions (Perrier et al., 2011). This suggests that some of these diploids may have evolved different mechanisms of resistance independently of one another. Similar findings were obtained in other studies, particularly in Medicago truncatula and Pisum sativum, which showed that quantitative resistance in their respective collections corresponds to accessions originating from multiple locations (Grünwald et al., 2003; Rispail and Rubiales, 2014). Future work will aim to identify QTLs controlling resistance in M. a. malaccensis lines.
The screen for Foc race 4 resistance revealed that the collection of Musa spp. contained sufficient genetic variations for the resistance responses to be detected. Cultivars shown to be highly susceptible to Foc race 4 in the field showed strong necrotic lesions in the rhizomes under our conditions (Table 1). In our study, the extent of the discoloration also depends on the inoculation technique used as it may influence the inoculum dosage and thereby the rate of the infection process. This observation is supported by studies that show a positive correlation between the amount of F. oxysporum detected within plant tissue and the resistance level in several species, including pea, tomato, watermelon, and chickpea (Gao et al., 1995; Zhou and Everts, 2004; Jimenez-Fernandez et al., 2011; Rispail and Rubiales, 2014).
In banana, contrasting resistance responses against Foc race 4 types have not been previously reported. In the present study, two cultivars, namely ‘FHIA-2’ and ‘CAM020’ showed excellent resistance to Foc-TR4 but were highly susceptible to Foc-STR4 (Figures 1, 2). This indicates that resistance to the two race 4 type VCGs might be differentially regulated. One possible explanation is that both cultivars lack a gene(s) or a component of the PAMPs (pathogen-associated molecular patterns) triggered immunity (PTI) or the effector-triggered immunity (ETI) that is specifically required for Foc-STR4 mediated resistance in these lines. Mechanisms of PTI and ETI are discussed in detail in recent reviews (Jie and Zhou, 2010; Petit-Houdenot and Fudal, 2017). Furthermore, both Foc race 4 type resistance can potentially be controlled by a single gene. For example, it has been shown in tomato that the immune receptor Ve1 mediates resistance against multiple pathogens by recognizing not only the Verticillium effector Ave1, but also related homologs of Ave1, from Fusarium oxysporum f. sp. lycopersici and Cercospora beticola (de Jonge et al., 2012).
In soil borne plant diseases, such as those caused by Foc, where the pathogen gains entry via plant roots, the state of the rhizome is a good indicator of the severity of infection. Necrotic lesions in the rhizome are the consequences of Foc colonizing the vascular tissues to cause senescence in a localized manner. This was evident in the rhizomes of the susceptible banana plants used in this study (Figures 3, 4). As shown using GFP tagged Foc-STR4, mycelial networks typically migrated along the xylem vessels and expanded outwards from the pits in susceptible plants. Our results are consistent with the behavior and the strong virulence of Foc race 4 observed in susceptible banana cultivars (Li et al., 2011, 2017; Zhang et al., 2018). A high level of sporulation further supports this observation in the present study (Figure 4).
In our study, the resistant plants typically showed no discoloration when millet was used as the inoculum (Figures 1–3). Furthermore, fungal isolation performed on rhizome and stems tissues of the resistant plants 3 months post-millet inoculation also failed to recover the GFP-STR4 strain (Supplementary Table 1). This suggests that the frequency of fungal colonization was too low to be detected in these lines at the respective early and late stages. This finding is consistent with a previous study which showed that Foc was not detectable at an early stage of infection in resistant banana plants (Li et al., 2011). A possibility could be that the growth of Foc was inhibited. Root exudates can potentially inhibit spore germination and fungal growth. In pea, phytoalexin pisatin, one of the metabolites detected in the root exudate extracts of pea, negatively correlated with the extent of Fusarium oxysporum f. sp. pisi germination (Bani et al., 2018a). It is of note that the millet inoculation technique minimized root wounding, whereas artificially induced wounding can enhance the infection rate in the roots and cause GFP tagged Foc to be detected in the rhizome of resistant banana cultivar Pahang (Zhang et al., 2018). However, in our study, Foc was never-the-less inhibited from traveling further up the pseudo-stem of the plant.
When a relatively more invasive method, root dipping, and concentrated micro-conidia were applied, GFP signals were not detected in the roots of resistant ‘Ma851’ plants at 7 dpi (Supplementary Figure 4). However, necrotic lesions were observed in the rhizome of the resistant plants at 18 dpi (Supplementary Figure 3). At this time, GFP fluorescence was associated with the xylem perforation plates. However, no sporulation or mycelial networks were observed (Figure 4). This suggests that the colonization by the fungus was mainly contained in the rhizome of ‘Ma851.’ Furthermore, no GFP-Foc-STR4 was detected in the stems of ‘Ma851’ suggesting that the xylem was likely uninfected (Supplementary Table 1). A similar pattern of restricted colonization in Dianthus caryophyllus by Fusarium oxysporum f. sp. dianthi has been reported and further characterization revealed that the infected regions of the xylem became compartmentalized by cell wall thickening and hyperplasia of parenchyma cells (Ouellette et al., 1999).
This type of resistance mechanisms against vascular pathogens have been characterized in several plant species (Beckman et al., 1982; Bishop and Cooper, 1983a,b; Baayen et al., 1989; Tessier et al., 1990; Pereira et al., 2013; Bani et al., 2018b). Plant hosts develop physical and chemical barriers to block pathogen progressions at different stages during the infection process. These include cell wall strengthening by lignification and suberization, formation of papillae at penetration sites, the accumulation of tyloses inside cells and production of antifungal compounds (Beckman et al., 1982; Bishop and Cooper, 1983a; Baayen and Elgersma, 1985; Charchar and Kraft, 1989; Ouellette et al., 1999; Grayer and Kokubun, 2001; Yadeta and Thomma, 2013; Pouralibaba et al., 2017; Bani et al., 2018b). Beckman et al. (1982) showed that F. oxysporum triggered callose deposition in the parenchyma cells of tomato plants and that the rate of deposition was faster in the resistant than the susceptible plants. In a separate experiment, Robb et al. (1991) showed that Verticillium infected tomato petioles induced suberization in the membranes of the pits and the intercellular spaces around the vascular vessels. Furthermore, the level of vascular coating positively correlated with resistance and negatively correlated with the frequencies of pathogen penetration of pit membranes in alfalfa (Newcombe and Robb, 1988). One or any of these mechanisms could potentially explain the resistance mechanism observed in this study.
In the present study, we have assessed a collection of 34 banana cultivars for resistance against F. oxysporum f. sp. cubense race 4 types and identified a range of resistance responses in the rhizomes. The rhizome appears to be a key factor in preventing the fungus from further spreading to other parts of the plant. We characterized diploid wild relative M. a. malaccensis lines that exhibit strong wilt resistance to Foc by inhibiting fungal growth in its rhizome. They are potential sources of ‘complete’ resistance to Foc race 4. Furthermore, contrasting resistance responses to different Foc race 4 types were observed. Phenotypic methods used in this study can help accelerate the efforts in breeding programs. Overall, this study paves the way for further characterizations in the defense mechanisms of Foc resistance at the cellular and molecular level in this important plant species.
Author’s Note
Plants used in this study were generated from the Australian in vitro banana cultivar collection that is maintained in the Quality Banana Approved Nursery (QBAN) scheme accredited Plant Biotechnology Laboratory located at the Maroochy Research Facility, Department of Agriculture and Fisheries, Nambour, Queensland, Australia. The cultivars were directly sourced from the owners or institutions under verbal or written agreement and where plants were directly collected, such as Musa acuminata ssp. malaccensis lines, they were sourced prior to the development of International treaties for germplasm. All cultivars were sourced under agreements allowing that they are able to be used for research purposes. No ownership is claimed for those cultivars sourced from owners or other institutions. The plants were destructively sampled for the purposes of this experiment and will not be further propagated.
Author Contributions
AC, SH, JB, and EA proposed, organized, and planned the experiments. AC, JS, AM, LA-E, NC, SM, and LT-N carried out and performed the experiments. AC wrote the manuscript draft. All authors commented and contributed to the preparation of the final manuscript.
Funding
This project has been funded by the International Institute of Tropical Agriculture (IITA) through the project ‘Improvement of Banana for Smallholder Farmers in the Great Lakes Region of Africa,’ and Hort Innovation, using Fusarium wilt Tropical Race 4 research program BA14014 and contributions from the Australian Government. Hort Innovation is the grower-owned, not-for-profit research and development corporation for Australian Horticulture. While JS was a visiting scholar at the University of Queensland, she was supported by the National Natural Science Foundation of China, through the project ‘31660560.’
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Mr. Dean Beasley (BRIP, Queensland Department of Agriculture and Fisheries) for providing the BRIP STR4 isolates, Dr. Leanne Forsyth for generating the GFP-Foc-STR4 isolate, Dr. Wayne O’Neill (Biosecurity Queensland, Queensland Department of Agriculture and Fisheries) for providing useful advice on the millet inoculation technique, Ms. Samantha Cullen for the millet preparation and assisting with setting up the pot trial in Darwin, Mr. Chris Kelly and Mr. Kenneth Haycock for plant maintenance at the Coastal Plains Research Farm in the Northern Territory, Ms Gabrielle Crawford for helping with processing the plants for confocal microscopy. We also thank Dr. Rebecca Lyons and Prof. Kathryne Everts for critical reading of the manuscript; Mr. Ken Hayes, Mr. Daniel Schwartz and Ms. Jodie Smith for glasshouse maintenance at the University of Queensland.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.01062/full#supplementary-material
Abbreviations
dpi, days post-inoculation; Foc, Fusarium oxysporum f sp. cubense; FHIA, Fundacion Hondurena de Investigación Agrícola; GFP, green fluorescent protein; Ma, Musa accuminata ssp. malaccensis; SH, synthetic hybrid; STR4, subtropical race 4; TR4, tropical race 4; VCG, vegetative compatibility group.
References
Aurore, G., Parfait, B., and Fahrasmane, L. (2009). Bananas, raw materials for making processed food products. Trends Food Sci. Tech. 20, 78–91. doi: 10.1016/j.tifs.2008.10.003
Baayen, R. P., and Elgersma, D. M. (1985). Colonization and histopathology of susceptible and resistant carnation cultivars infected with Fusarium oxysporum f.sp. dianthi. Neth. J. Plant Pathol. 91, 119–135. doi: 10.1007/bf01976386
Baayen, R. P., Vaneijk, C., and Elgersma, D. M. (1989). Histology of roots of resistant and susceptible carnation cultivars from soil infested with Fusarium oxysporum f. sp. dianthi. Neth. J. Plant Pathol. 95, 3–13. doi: 10.1007/bf02000875
Bani, M., Cimmino, A., Evidente, A., Rubiales, D., and Rispail, N. (2018a). Pisatin involvement in the variation of inhibition of Fusarium oxysporum f. sp pisi spore germination by root exudates of Pisum spp. germplasm. Plant Pathol. 67, 1046–1054. doi: 10.1111/ppa.12813
Bani, M., Perez-De-Luque, A., Rubiales, D., and Rispail, N. (2018b). Physical and chemical barriers in root tissues contribute to quantitative resistance to Fusarium oxysporum f. sp pisi in pea. Front. Plant Sci. 9:199. doi: 10.3389/fpls.2018.00199
Bani, M., Rubiales, D., and Rispail, N. (2012). A detailed evaluation method to identify sources of quantitative resistance to Fusarium oxysporum f. sp pisi race 2 within a Pisum spp. germplasm collection. Plant Pathol. 61, 532–542. doi: 10.1111/j.1365-3059.2011.02537.x
Beckman, C. H., Mueller, W. C., Tessier, B. J., and Harrison, N. A. (1982). Recognition and callose deposition in response to vascular infection in Fusarium wilt-resistant or susceptible tomato plants. Physiol. Plant Pathol. 20, 1–10. doi: 10.1016/0048-4059(82)90018-2
Bentley, S., Pegg, K. G., Moore, N. Y., Davis, R. D., and Buddenhagen, I. W. (1998). Genetic variation among vegetative compatibility groups of Fusarium oxysporum f. sp. cubense analyzed by DNA fingerprinting. Phytopathology 88, 1283–1293. doi: 10.1094/Phyto.1998.88.12.1283
Bhagwat, B., and Duncan, E. J. (1998). Mutation breeding of Highgate (Musa acuminata, AAA) for tolerance to Fusarium oxysporum f. sp. cubense using gamma irradiation. Euphytica 101, 143–150. doi: 10.1023/A:1018391619986
Bishop, C. D., and Cooper, R. M. (1983a). An ultrastructural study of root invasion in 3 vascular wilt diseases. Physiol. Plant Pathol. 22, 15–27. doi: 10.1016/S0048-4059(83)81034-0
Bishop, C. D., and Cooper, R. M. (1983b). An ultrastructural study of vascular colonization in 3 vascular wilt diseases. 1. Colonization of susceptible cultivars. Physiol. Plant Pathol. 23, 323–343. doi: 10.1016/0048-4059(83)90018-8
Buddenhagen, I. W. (2009). “Understanding strain diversity in Fusarium oxysporum f. sp cubense and history of introduction of ‘tropical race 4’ to better manage banana production,” in International Symposium on Recent Advances in Banana Crop Protection for Sustainable Production and Improved Livelihoods, Vol. 828, eds D. Jones and I. Van der Bergh (Leuven: International Society for Horticultural Science), 193–204.
Caten, C. E., and Jinks, J. L. (1966). Heterokaryosis: its significance in wild homothallic ascomycetes and fungi imperfecti. Br. Mycol. Soc. Trans. 49, 81–93. doi: 10.1016/S0007-1536(66)80038-4
Charchar, M., and Kraft, J. M. (1989). Response of near-isogenic pea cultivars to infection by Fusarium oxysporum f. sp. pisi races 1 and 5. Can. J. Plant Sci. 69, 1335–1346. doi: 10.4141/cjps89-161
Christelova, P., De Langhe, E., Hribova, E., Cizkova, J., Sardos, J., Husakova, M., et al. (2017). Molecular and cytological characterization of the global Musa germplasm collection provides insights into the treasure of banana diversity. Biodivers. Conserv. 26, 801–824. doi: 10.1007/s10531-016-1273-9
Dale, J., James, A., Paul, J. Y., Khanna, H., Smith, M., Peraza-Echeverria, S., et al. (2017). Transgenic cavendish bananas with resistance to Fusarium wilt tropical race 4. Nat. Commun. 8:1496. doi: 10.1038/s41467-017-01670-6
de Jonge, R., van Esse, H. P., Maruthachalam, K., Bolton, M. D., Santhanam, P., Saber, M. K., et al. (2012). Tomato immune receptor Ve1 recognizes effector of multiple fungal pathogens uncovered by genome and RNA sequencing. Proc. Natl. Acad. Sci. U.S.A. 109, 5110–5115. doi: 10.1073/pnas.1119623109
D’Hont, A., Denoeud, F., Aury, J. M., Baurens, F. C., Carreel, F., Garsmeur, O., et al. (2012). The banana (Musa acuminata) genome and the evolution of monocotyledonous plants. Nature 488, 213–217. doi: 10.1038/nature11241
D’Hont, A., Paget-Goy, A., Escoute, J., and Carreel, F. (2000). The interspecific genome structure of cultivated banana, Musa spp. Revealed by genomic DNA in situ hybridization. Theor. Appl. Genet. 100, 177–183. doi: 10.1007/s001220050024
Dita, M., Barquero, M., Heck, D., Mizubuti, E. S. G., and Staver, C. P. (2018). Fusarium wilt of banana: current knowledge on epidemiology and research needs toward sustainable disease management. Front. Plant Sci. 9:1468. doi: 10.3389/fpls.2018.01468
FAO (2019). Banana Market Review Preliminary Results for 2019. Available at: http://www.fao.org/economic/est/est-commodities/bananas/en/. (accessed 6th Mar 2019).
Forsyth, L. M. (2006). Understanding the Role of Induced Resistance in the Control of Fusarium Wilt of Banana. Ph.D.thesise, The University of Queensland, Queensland.
Fourie, G., Steenkamp, E. T., Gordon, T. R., and Viljoen, A. (2009). Evolutionary relationships among the Fusarium oxysporum f. sp cubense vegetative compatibility groups. Appl. Environ. Microb. 75, 4770–4781. doi: 10.1128/Aem.00370-09
Fraser-Smith, S., Czislowski, E., Daly, A., Meldrum, R., Hamill, S., Smith, M., et al. (2016). “Single gene resistance to Fusarium oxysporum f. sp. cubense Race 4 in the wild banana Musa acuminata subsp. malaccensis,” in XXIX International Horticultural Congress on Horticulture: Sustaining Lives, Livelihoods and Landscapes (IHC2014): IX International Symposium on Banana: ISHS-Promusa Symposium on Unravelling the Banana’s Genomic Potential, Vol. 1114, eds I. Van den Bergh, M. Smith, and C. Picq (Brisbane: International Society for Horticultural Science), 95–100. doi: 10.17660/ActaHortic.2016.1114.13
Gao, H., Beckman, C. H., and Mueller, W. C. (1995). The rate of vascular colonization as a measure of the genotypic interaction between various cultivars of tomato and various formae or races of Fusarium oxysporum. Physiol. Mol. Plant Pathol. 46, 29–43. doi: 10.1006/pmpp.1995.1003
Grayer, R. J., and Kokubun, T. (2001). Plant-fungal interactions: the search for phytoalexins and other antifungal compounds from higher plants. Phytochemistry 56, 253–263. doi: 10.1016/s0031-9422(00)00450-7
Grünwald, N. J., Coffman, V. A., and Kraft, J. M. (2003). Sources of partial resistance to Fusarium root rot in the Pisum core collection. Plant Dis. 87, 1197–1200. doi: 10.1094/PDIS.2003.87.10.1197
Hennessy, C., Walduck, G., Daly, A., and Padovan, A. (2005). Weed hosts of Fusarium oxysporum f. sp cubense tropical race 4 in northern Australia. Aust. Plant Pathol. 34, 115–117. doi: 10.1071/Ap04091
Hwang, S. C., and Ko, W. H. (2004). Cavendish banana cultivars resistant to Fusarium wilt acquired through somaclonal variation in Taiwan. Plant Dis. 88, 580–588. doi: 10.1094/Pdis.2004.88.6.580
Jie, Z., and Zhou, J. M. (2010). Plant immunity triggered by microbial molecular signatures. Mol. Plant 3, 783–793. doi: 10.1093/mp/ssq035
Jimenez-Fernandez, D., Montes-Borrego, M., Jimenez-Diaz, R. M., Navas-Cortes, J. A., and Landa, B. B. (2011). In planta and soil quantification of Fusarium oxysporum f. sp. ciceris and evaluation of Fusarium wilt resistance in chickpea with a newly developed quantitative polymerase chain reaction assay. Phytopathology 101, 250–262. doi: 10.1094/PHYTO-07-10-0190
Li, C. Q., Yang, J. H., Li, W. B., Sun, J. B., and Peng, M. (2017). Direct root penetration and rhizome vascular colonization by Fusarium oxysporum f. sp cubense are the key steps in the successful infection of Brazil Cavendish. Plant Dis. 101, 2073–2078. doi: 10.1094/Pdis-04-17-0467-Re
Li, C. Y., Chen, S., Zuo, C. W., Sun, Q. M., Ye, Q., Yi, G. J., et al. (2011). The use of GFP-transformed isolates to study infection of banana with Fusarium oxysporum f. sp cubense race 4. Eur. J. Plant Pathol. 131, 327–340. doi: 10.1007/s10658-011-9811-5
Li, C. Y., Deng, G. M., Yang, J., Viljoen, A., Jin, Y., Kuang, R. B., et al. (2012). Transcriptome profiling of resistant and susceptible Cavendish banana roots following inoculation with Fusarium oxysporum f. sp cubense tropical race 4. BMC Genomics 13:374. doi: 10.1186/1471-2164-13-374
Li, L. F., Wang, H. Y., Zhang, C., Wang, X. F., Shi, F. X., Chen, W. N., et al. (2013). Origins and domestication of cultivated banana inferred from chloroplast and nuclear genes. PLoS One 8:e80805. doi: 10.1371/journal.pone.0080502
Li, W. M., Dita, M., Wu, W., Hu, G. B., Xie, J. H., and Ge, X. J. (2015). Resistance sources to Fusarium oxysporum f. sp cubense tropical race 4 in banana wild relatives. Plant Pathol. 64, 1061–1067. doi: 10.1111/ppa.12340
Mak, C., Mohamed, A. A., Liew, K. W., and Ho, Y. W. (2004). “Early screening technique for Fusarium wilt resistance in banana micropropagated plants,” in Banana Improvement: Cellular, Molecular Biology, and Induced Mutations, eds R. Swennen and M. S. Jain (Enfield, NH: Science Publishers, Inc.),219–227.
Moore, N. Y., Pegg, K. G., Allen, R. N., and Irwin, J. A. G. (1993). Vegetative compatibility and distribution of Fusarium oxysporum f. sp cubense in Australia. Aust. J. Exp. Agric. 33, 797–802. doi: 10.1071/Ea9930797
Moore, N. Y., Pegg, K. G., Smith, L. J., Langdon, P. W., Bentley, S., and Smith, M. K. (2001). “Fusarium wilt of banana in Australia,” in Banana Fusarium Wilt Management: Towards Sustainable Cultivation. Proceedings of the International Workshop on the Banana Fusarium wilt Disease, eds A. B. Molina, N. H. Nik Masdek, and K. W. Liew (Los Baños: INIBAP), 64–75.
Newcombe, G., and Robb, J. (1988). The function and relative importance of the vascular coating response in highly resistant, moderately resistant and susceptible alfalfa infected by Verticillium Albo-Atrum. Physiol. Mol. Plant Pathol. 33, 47–58. doi: 10.1016/0885-5765(88)90042-2
Ouellette, G. B., Baayen, R. P., Simard, M., and Rioux, D. (1999). Ultrastructural and cytochemical study of colonization of xylem vessel elements of susceptible and resistant Dianthus caryophyllus by Fusarium oxysporum f.sp dianthi. Can. J. Bot. 77, 644–663. doi: 10.1139/b99-033
Peraza-Echeverria, S., Dale, J. L., Harding, R. M., Smith, M. K., and Collet, C. (2008). Characterization of disease resistance gene candidates of the nucleotide binding site (NBS) type from banana and correlation of a transcriptional polymorphism with resistance to Fusarium oxysporum f.sp cubense race 4. Mol. Breed. 22, 565–579. doi: 10.1007/s11032-008-9199-x
Pereira, A. C., Cruz, M. F. A., Paula Junior, T. J., Rodrigues, F. A., Carneiro, J. E. S., Vieira, R. F., et al. (2013). Infection process of Fusarium oxysporum f. sp phaseoli on resistant, intermediate and susceptible bean cultivars. Trop. Plant Pathol. 38, 323–328. doi: 10.1590/S1982-56762013005000022
Perrier, X. (2009). Combining biological approaches to shed light on the evolution of edible bananas. Ethnobot. Res. Appl. 7, 199–216.
Perrier, X., De Langhe, E., Donohue, M., Lentfer, C., Vrydaghs, L., Bakry, F., et al. (2011). Multidisciplinary perspectives on banana (Musa spp.) domestication. Proc. Natl. Acad. Sci. U.S.A. 108, 11311–11318. doi: 10.1073/pnas.1102001108
Petit-Houdenot, Y., and Fudal, I. (2017). Complex interactions between fungal avirulence genes and their corresponding plant resistance genes and consequences for disease resistance management. Front. Plant Sci. 8:1072. doi: 10.3389/fpls.2017.01072
Ploetz, R. C. (2006). Fusarium wilt of banana is caused by several pathogens referred to as Fusarium oxysporum f. sp cubense. Phytopathology 96, 653–656. doi: 10.1094/Phyto-96-0653
Ploetz, R. C. (2015). Fusarium wilt of banana. Phytopathology 105, 1512–1521. doi: 10.1094/Phyto-04-15-0101-Rvw
Pouralibaba, H. R., Perez-De-Luque, A., and Rubiales, D. (2017). Histopathology of the infection on resistant and susceptible lentil accessions by two contrasting pathotypes of Fusarium oxysporum f.sp. lentis. Eur. J. Plant Pathol. 148, 53–63. doi: 10.1007/s10658-016-1068-6
Rispail, N., and Rubiales, D. (2014). Identification of sources of quantitative resistance to Fusarium oxysporum f. sp medicaginis in Medicago truncatula. Plant Dis. 98, 667–673. doi: 10.1094/PDIS-03-13-0217-RE
Robb, J., Lee, S. W., Mohan, R., and Kolattukudy, P. E. (1991). Chemical characterization of stress-induced vascular coating in tomato. Plant Physiol. 97, 528–536. doi: 10.1104/pp.97.2.528
Smith, L. J., Smith, M. K., Tree, D., O’Keefe, D., and Galea, V. J. (2008). Development of a small-plant bioassay to assess banana grown from tissue culture for consistent infection by Fusarium oxysporum f. sp cubense. Aust. Plant Pathol. 37, 171–179. doi: 10.1071/Ap08006
Smith, M. K., and Hamill, S. D. (1993). Early detection of dwarf off-types from micropropagated Cavendish bananas. Aust. J. Exp. Agric. 33, 639–644. doi: 10.1071/Ea9930639
Smith, M. K., Langdon, P. W., Pegg, K. G., and Daniells, J. W. (2014). Growth, yield and Fusarium wilt resistance of six FHIA tetraploid bananas (Musa spp.) grown in the Australian subtropics. Sci. Hortic. 170, 176–181. doi: 10.1016/j.scienta.2014.02.029
Snyder, W. C., and Hansen, H. N. (1940). The species concept in Fusarium. Am. J. Bot. 27, 64–67. doi: 10.2307/2436688
Stover, R. H. (1990). Fusarium wilt of Banana: Some history and current Status of the Disease. St. Paul, MN: APS Press.
Su, H. J., Hwang, S. C., and Ko, W. H. (1986). Fusarial wilt of cavendish bananas in Taiwan. Plant Dis. 70, 814–818.
Tessier, B. J., Mueller, W. C., and Morgham, A. T. (1990). Histopathology and ultrastructure of vascular-responses in peas resistant or susceptible to Fusarium oxysporum f. sp. pisi. Phytopathology 80, 756–764. doi: 10.1094/Phyto-80-756
Walduck, G., and Daly, A. (2007). Banana Tropical race 4 Panama Disease Management. Northern Territory Department of Primary Industry, Fisheries and Mines Primary Industry. Technical report 2006-2007.
Yadeta, K. A., and Thomma, B. P. J. (2013). The xylem as battleground for plant hosts and vascular wilt pathogens. Front. Plant Sci. 4:97. doi: 10.3389/fpls.2013.00097
Zhang, L., Yuan, T. L., Wang, Y. Z., Zhang, D., Bai, T. T., Xu, S. T., et al. (2018). Identification and evaluation of resistance to Fusarium oxysporum f. sp cubense tropical race 4 in Musa acuminata Pahang. Euphytica 214:106. doi: 10.1007/s10681-018-2185-4
Zhou, X. G., and Everts, K. L. (2004). Quantification of root and stem colonization of watermelon by Fusarium oxysporum f. sp. niveum and its use in evaluating resistance. Phytopathology 94, 832–841. doi: 10.1094/PHYTO.2004.94.8.832
Keywords: Fusarium wilt, banana, Musa acuminata ssp. malaccensis, green fluorescent protein, Fusarium oxysporum f. sp. cubense
Citation: Chen A, Sun J, Matthews A, Armas-Egas L, Chen N, Hamill S, Mintoff S, Tran-Nguyen LTT, Batley J and Aitken EAB (2019) Assessing Variations in Host Resistance to Fusarium oxysporum f sp. cubense Race 4 in Musa Species, With a Focus on the Subtropical Race 4. Front. Microbiol. 10:1062. doi: 10.3389/fmicb.2019.01062
Received: 13 December 2018; Accepted: 26 April 2019;
Published: 15 May 2019.
Edited by:
Gert H. J. Kema, Wageningen University & Research, NetherlandsReviewed by:
Li-Jun Ma, University of Massachusetts Amherst, United StatesGeorge Newcombe, University of Idaho, United States
Copyright © 2019 Chen, Sun, Matthews, Armas-Egas, Chen, Hamill, Mintoff, Tran-Nguyen, Batley and Aitken. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Andrew Chen, YS5jaGVuMkB1cS5lZHUuYXU=
 Andrew Chen
Andrew Chen Jiaman Sun
Jiaman Sun Andrea Matthews
Andrea Matthews Liz Armas-Egas
Liz Armas-Egas Ning Chen1
Ning Chen1 Sharon Hamill
Sharon Hamill Sharl Mintoff
Sharl Mintoff Jaqueline Batley
Jaqueline Batley Elizabeth A. B. Aitken
Elizabeth A. B. Aitken