- 1School of Life Science and Biotechnology, Dalian University of Technology, Dalian, China
- 2Institute of Food Science and Engineering, Jinzhou Medical University, Jinzhou, China
- 3College of Life Science, Dalian Minzu University, Dalian, China
In this study, attention has been focused on the ecology of yeasts during the spontaneous and inoculated fermentation processes of Vidal blanc icewine in northeast China, which is very important for screening autochthonous yeast strains, understanding the roles of these strains, and managing fermentation. The strategies were to conduct spontaneous and inoculated laboratory-scale fermentation processes simultaneously and to analyze the samples taken at different fermentation stages by culture-dependent and -independent methods. Three hundred and thirty-eight yeast strains were isolated and twelve genera were identified by sequencing. During the spontaneous fermentation process, non-Saccharomyces yeasts were predominant in the initial and middle stages, whereas Saccharomyces dominated in the later stages; Candida was preponderant in the whole process, and its abundance in the final stages was only lower than Saccharomyces. The inoculated fermentation was characterized by a predominance of Saccharomyces throughout the fermentation process; non-Saccharomyces yeasts were observed in the early stage. The internal transcribed spacer (ITS) 2 region gene was firstly used to analyze the yeast diversity in the samples during the icewine fermentation processes by high-throughput sequencing (HTS), and a more complex mycobiota was revealed. Moreover, the dynamics of other major fungi (mainly Davidiella and Alternaria) during icewine fermentation processes were also revealed, which have never been reported in icewine before.
Introduction
Icewine originated in Germany more than 200 years ago. It is a typical dessert wine made from grapes that have naturally frozen on the vine under cold weather conditions (below −8°C) (OIV, 2008). Due to the specific environmental conditions required for planting and harvesting ice grapes, icewine is manufactured only in a few countries where winter conditions are cold enough. Canada, Germany, and Austria are the world’s major producers of icewine (Tian et al., 2009). In recent years, the icewine industry in China has developed rapidly, and China has become an icewine production country that cannot be ignored (Alessandria et al., 2013; Ma et al., 2017; Huang et al., 2018). Vidal blanc grapes have a high level of winter hardiness because of thick-skinned berries (Bowen and Reynolds, 2015); it is the main cultivar for icewine production in the northeast China. Vidal blanc icewine products are considered to be of high quality because of their appealing aroma and attractive flavor (Nurgel et al., 2004; Crandles et al., 2015).
Alcoholic fermentation of grape juice is a complex biochemical process carried out by a dynamically changing microbiota, in which yeast strains play an important role. Icewine yeasts need to face many challenges during the fermentation process, e.g., high sugar concentrations, high acid concentrations and low fermentation temperature (Zhang et al., 2018). During the process of conversion of ice grape juice into icewine, the composition changes dramatically under the action of Saccharomyces and non-Saccharomyces yeasts. Several studies have shown that non-Saccharomyces yeast contributes positively to the aroma and flavor of wine (Fleet, 2003; Ciani et al., 2010; Lencioni et al., 2016; Englezos et al., 2018) and icewine (Zhang et al., 2018). A few non-Saccharomyces yeast genera were reported to adequately produce hydrolytic enzymes, which can facilitate the release of aromatic precursors (Pérez et al., 2011; Maturano et al., 2012).
Due to the easy operation and stability of product quality, commercial yeast strains are currently used to inoculate icewine fermentation in many wineries. Another strategy is to favor the screening and utilization of autochthonous yeast strains, because these yeasts can adapt well to the micro-conditions of the wine region and generate territorial characteristics in icewine products (Zhang et al., 2018). Therefore, the knowledge of the yeast diversity and dynamics during inoculated and spontaneous fermentations is very important for controlling fermentation, screening autochthonous yeast strains and understanding the role of these yeast strains in the process. However, most studies have focused on the aroma compounds and flavor characteristics of icewine (Bowen and Reynolds, 2015; Ma et al., 2017; Huang et al., 2018), and the knowledge of the microbiota involved in the fermentation of icewine is very limited.
Thus far, yeast diversity and dynamic changes during the fermentation process have been researched by culture-dependent and -independent methods. The culture-dependent approach using the Wallerstein laboratory (WL) nutrient agar medium has been employed to monitor and isolate the yeasts during grape juice fermentation based on the colors and morphologies of the colonies (Cavazza et al., 1992; Urso et al., 2008; Alessandria et al., 2013). For further identification of yeast species, the 5.8S-ITS region gene or the D1/D2 domain of the 26S rDNA gene has been sequenced (Brysch-Herzberg and Seidel, 2015). In recent years, PCR-DGGE (Alessandria et al., 2013; Bučková et al., 2018) and HTS (Andorrà et al., 2010; De Filippis et al., 2017; Mendoza et al., 2017) as culture-independent methods have been used for detecting yeast population diversity. In general, in terms of supporting the results of the culture-dependent methods, culture-independent methods have detected higher microbial diversity (Van Hijum et al., 2013). HTS has been shown to have better potential for analyzing the microbial diversity of fermented foods as compared to PCR-DGGE (Połka et al., 2015; Portillo and Mas, 2016). However, in this decade, studies on the microbiota involved in the fermentation of icewine were only carried out by using PCR-DGGE as a culture-independent method by Alessandria et al. (2013) and Bučková et al. (2018).
The main objective of this study was to comprehensively analyze yeast population diversity and dynamics during the spontaneous and inoculated fermentation processes of Vidal blanc icewine of northeast China by culture-dependent (WL nutrient agar) and culture-independent (HTS) approaches.
Materials and Methods
Laboratory-Scale Fermentation Processes and Sampling
The ice grape berries of Vidal blanc used in this study were harvested manually from the vineyard of “Sun Valley” winery (NE China, 41°45′24.97″N 123°62′20.58″E) in December 2016. Healthy ice grapes were randomly collected, and the peduncles and leaves were fully removed. The frozen grapes were then pressed to obtain ice grape juice for fermentative production of icewine. The ice grape juice was immediately placed in sterile buckets (at ≤ 0°C) and transported to the laboratory. The ice grape juice was named SZ and found to have pH 3.89 and sugar content of approximately 27.3° Brix. Laboratory-scale spontaneous and inoculated fermentation processes were carried out simultaneously. Spontaneous fermentation processes were carried out with 600 mL of the ice grape juice (containing 50 mg/L SO2) in 1000-mL sterile flasks in triplicate at 18°C under static conditions. Inoculated fermentations were performed in a similar way, although in addition commercial active dried yeast (ST; LAFFORT, Bordeaux, France; initial inoculum of approximately 106 cells/mL final concentration) was added to the ice grape juice. Samples consisted of longitudinal icewine fermentation samples, and sampling was carried out at 0, 1, 2, 4, 7, 14, 21, and 30 days during the spontaneous and inoculated fermentation processes. The samples of the spontaneous fermentation were named SSFN-[1, 2, 3] (N represented sequential sampling points, N = A, B…G), while samples from the inoculated fermentation were designated SJFN-[1, 2, 3] (N = A, B…G). Microbial cultivation and DNA extraction procedures were performed for each sample.
At the end of fermentation, the means of pH, ethanol, and sugar content in the icewines produced by the spontaneous and inoculated fermentation processes were determined. The pH was determined using a pH-meter (pHS-3C; WEIYE, Shanghai, China), and the sugar content was determined using OIV methods (OIV, 2008). The ethanol was quantified using gas chromatograph with a flame ionization detector (GC 9790 plus; Fuli, Zhejiang, China), and the system was equipped with a KB-5 capillary column (30 m × 320 μm × 0.25 μm; Kromat Corporation, Bordentown, NJ, United States); the column temperature was 50°C, the injection port was 250°C, and the detector temperature was 250°C; ethanol (GC quality standard reagents; Aladdin, Shanghai, China) as a standard was used; each sample of 0.2 μL was injected into the injection port, the carrier gas was nitrogen with the flow rate of 1 mL/min, and the injection split ratio was 50:1.
Yeast Isolation and Cultivation
Each sample was serially diluted in a sterile physiological solution (with ratios of 1:10 to 1:106) and spread-plated on WL Nutrient agar (Haibo, Qingdao, China). All plates were incubated at 28°C for 5 days. The colonies on each plate were counted, and mean values were calculated. At least 20 colonies with different colors and morphologies were selected and isolated from each plate as representatives at each sampling point of the fermentations. All pure isolates were cultured in YPD medium (yeast extract 10 g/L, peptone 20 g/L, dextrose 20 g/L; Haibo, Qingdao, China) at 28°C for 2 days and stored at −80°C after addition of glycerol (30% v/v).
Molecular Identification of the Isolates
The stored isolate was activated in the YPD medium at 28°C for 2 days, and genomic DNA was extracted by means of the yeast genomic DNA extraction kit (DP307-02; TIANGEN, Beijing, China). DNA was quantified on an ND-1000 spectrophotometer (NanoDrop, Wilmington, DE, United States), and concentration was standardized to 100 ng/μL. The ITS region (ITS1-5.8S-ITS2) was amplified with primers ITS1 (5′-TCC GTA GGT GAA CCT GCG G-3′) and ITS4 (5′-TCC TCC GCT TAT TGA TAT GC-3′) (Brysch-Herzberg and Seidel, 2015). PCR was carried out in a final volume of 25 μL, and the composition of the PCR mixture was as follows: 2.5 U Taq DNA polymerase, 2.5 μL 10 × Taq buffer, 2 μL 25 mM MgCl2, 0.5 μL of each dNTP (10 mM), 1 μL of 10 nM forward primer and 1 μL of 10 nM reverse primer, and 1 μL of the template DNA. Thermal cycling was performed as follows: initial denaturation at 95°C for 5 min; 35 cycles (95°C for 30 s, 55°C for 30 s, 72°C for 1 min); and a final elongation at 72°C for 7 min. When necessary, the 26S rDNA D1/D2 domain sequences of the isolates were additionally sequenced and analyzed (Fell et al., 2000). This region was amplified with the universal primer pairs NL1 (5′-GCATATCAATAAGCGGAGGAAAAG-3′) and NL4 (5′-GGTCCGTGTTTCAAGACGG-3′) (Brysch-Herzberg and Seidel, 2015; Graça et al., 2015); in addition, the amplification reaction and the PCR conditions were identical to those described for the ITS region (Santo et al., 2012). All PCR products of extracted DNA samples were sent to Sangon Biotech Co., Ltd (Shanghai, China) for sequencing. For identification of the isolates, these sequences were edited in the MEGA software (version 6.06) (Tamura et al., 2013) and compared with the present in the NCBI database (National Centre for Biotechnology Information1) using BLAST (Basic Local Alignment Search Tool) search to determine the closest known relatives.
Illumina HTS
The total DNA of each sample was directly extracted with the E.Z.N.A. ®Soil DNA Kit (D5625, Omega, Inc., United States). DNA extracts were sent to LC-Bio Technology Co., Ltd. (Hangzhou, Zhejiang Province, China) for PCR amplification and HTS. The internal transcribed spacer2 (ITS2) region of high-quality DNAs of the samples was selected (Pinto et al., 2015) and amplified with slightly modified versions of primers fITS7 (5′-GTGARTCATCGAATCTTTG-3′) and ITS4 (5′-TCCTCCGCTTATTGATATGC-3′). The 5′ ends of the primers were tagged with specific barcodes and sequencing universal primers. PCR was carried out in a total volume of 25 μL, containing 12.5 μL of the PCR Premix, 2.5 μL of the forward and reverse primers, 50 ng of the template DNA, and PCR-grade water to adjust the volume. Thermal cycling was as follows: initial denaturation at 98°C for 30 s; then 35 cycles of denaturation at 98°C for 10 s, annealing at 54°C for 30 s, and extension at 72°C for 45 s; followed by a final extension at 72°C for 10 min. The amplicons were analyzed by electrophoresis on 2% agarose gels and were purified by means of AMPure XT beads (Beckman Coulter Genomics, Danvers, MA, United States). The Quant-iT PicoGreen dsDNA Assay Kit was then employed. The library was quantified on a Qubit®2.0 fluorometer (Invitrogen, Carlsbad, CA, United States). The amplicon pools were prepared for sequencing, and size and quantity of the amplicon library were evaluated on an Agilent Bioanalyzer 2100 system with the Library Quantification Kit for Illumina (Kapa Biosciences, Woburn, MA, United States). The MiSeq instrument (Illumina, San Diego, CA, United States) was used to sequence 250-bp paired-end reads in two separate runs.
Data Analysis
Paired-end reads were assigned to samples based on the unique barcodes, and the barcode and primer sequence were then removed. Paired-end reads were merged in the PEAR software (Paired-End reAd mergeR, Version 0.9.6). Raw reads were filtered to obtain high-quality clean reads under specific conditions of filtration via the FastQC (Version 0.10.1). Reads were scanned by the sliding window method, and the default value was 6 bp. In cases where the average phred quality score in the window was less than 20, sequences were truncated from the beginning of the window to the 3′ end. After truncation, reads shorter than 100 bp and with more than 5% ambiguous bases were excluded from the analysis. Moreover, the chimeric sequences were filtered by means of the VerSeach software (Version 2.3.4). Sequences were clustered into operational taxonomic units (OTUs; defined by 97% similarity) in VerSeach. Taxonomic data were assigned to the representative sequences (each OTU) using the RDP (Ribosomal Database Project) classifier. PyNAST software was used for multiple sequence alignment and to study the phylogenetic relationship between different OTUs. The α-Diversity that included two indices (Shannon and Chao1) was employed for the analysis of species diversity, and all indices were calculated in the QIIME software (Version 1.8.0). Excel 2007 (Microsoft Corp., United States) was used for graphics of the icewine yeast species abundance (mean values). The raw fungal ITS sequence data were deposited in the Sequence Read Archive (SRA) of the National Center for Biotechnology Information (NCBI) database under accession number PRJNA503950.
Results
Basic Chemical Analysis of the Composition of the Icewines
The spontaneous and inoculated fermentation processes were simultaneously terminated on day 30, and the basic chemical composition of the icewines was determined. The pH was 3.98, ethanol content was 9.8% (v/v) and residual sugar contents were 132.6 g/L in the icewine produced by spontaneous fermentation, whereas pH, ethanol, and sugar contents were 3.95, 12.6% (v/v), and 85.2 g/L, respectively, in the icewine produced by inoculated fermentation. The ethanol yields were clearly higher in the icewines produced by inoculated fermentation than those produced by spontaneous fermentation.
Yeast Population Diversity and Dynamic Changes During the Fermentation Processes by the Culture-Dependent Method
A total of 338 strains were isolated at different stages of the spontaneous and inoculated fermentation processes and were differentiated based on colony color and morphology on WL nutrient agar (Cavazza et al., 1992); 21 different morphotypes were observed. The ITS region and the 26S rDNA D1/D2 domain of the representative strains were then sequenced for species identification. Twelve genera were detected, including Aureobasidium, Candida, Cryptococcus, Filobasidium, Hanseniaspora, Pichia, Rhodotorula, Saccharomyces, Sporidiobolus, Torulaspora, Zygoascus, and Zygosaccharomyces. The species found included Aureobasidium pullulans, Candida californica, Candida railenensis, Candida salmanticensis, Candida smithsonii, Candida carpophila, Candida zemplinina, Cryptococcus flavescens, Cryptococcus magnus, Filobasidium floriforme, Hanseniaspora uvarum, Pichia kudriavzevii, Pichia occidentalis, Rhodotorula babjevae, Rhodotorula glutinis, Saccharomyces uvarum, Saccharomyces cerevisiae, Sporidiobolus pararoseus, Torulaspora delbrueckii, Zygoascus meyerae, and Zygosaccharomyces parabailii. Table 1 provides an overview of the isolated representative strains, their ITS region and D1/D2 domain fragment sizes, and their GenBank accession numbers.
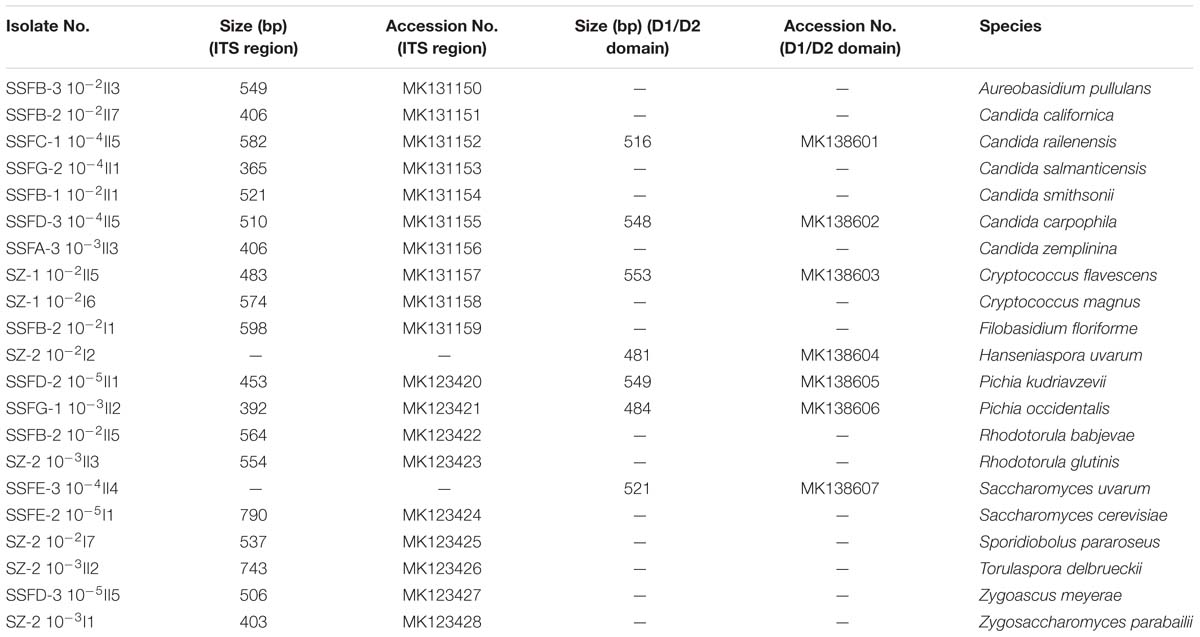
Table 1. ITS region and D1/D2 domain fragment sizes of the representative isolates and their GenBank accession numbers.
Figures 1, 2 show changes in the isolated representative strains at genus and species levels during the spontaneous and inoculated fermentation processes. During the spontaneous fermentation, Candida was present during the whole process, and the relative abundance of the genus changed dramatically. The relative abundance gradually increased and then decreased, peaking at 61.6% on day 7 (SSFD), and accounted for 25.0% of the population on day 30 (SSFG). In the genus Candida, C. zemplinina was detectable at all sampling points (8.7–23.5%), representing 20.0% of the population at the end of the spontaneous fermentation (SSFG). C. carpophila and C. californica were detected on days 1–7 and days 2–21, respectively. The relative abundance of both species initially increased and then decreased, with their highest abundance levels being 28.0% (SSFC, day 4) and 18.8% (SSFE, day 14), respectively. Pichia (P. kudriavzevii and P. occidentalis) was detected from day 4 (SSFC; 8%) to day 30 (SSFG; 15%) and increased initially and then decreased, with the highest abundance being 31.3% (SSFE, day 14). Cryptococcus (Cry. flavescens and Cry. magnus) was the predominant genus at the beginning (SZ; 39.1%) and gradually dropped to 8.0% (SSFC, day 4), and eventually became undetectable. Similar to Cryptococcus, Filobasidium (F. floriforme) and Zygosaccharomyces (Zygos. parabailii) were found in the early fermentative stage (days 0–4), with their highest abundance levels being detected on day 2 (SSFB) and representing 21.9 and 12.5% of the population, respectively. A. pullulans, Spo. pararoseus and Rhodotorula (R. babjevae and R. glutinis) were all detectable at the first three sampling points (SZ, SSFA, and SSFB) and were absent later. Moreover, H. uvarum and T. delbrueckii were found only on day 0 (SZ) at low abundance (4.3%). Saccharomyces was first observed (7.7%) on day 7 (SSFD), and gradually increased in abundance to become the dominant genus in the later fermentative stages (days 21 and 30). The highest abundance was 60.0% (S. cerevisiae accounted for 45.0%) at the end of the spontaneous fermentation (SSFG).
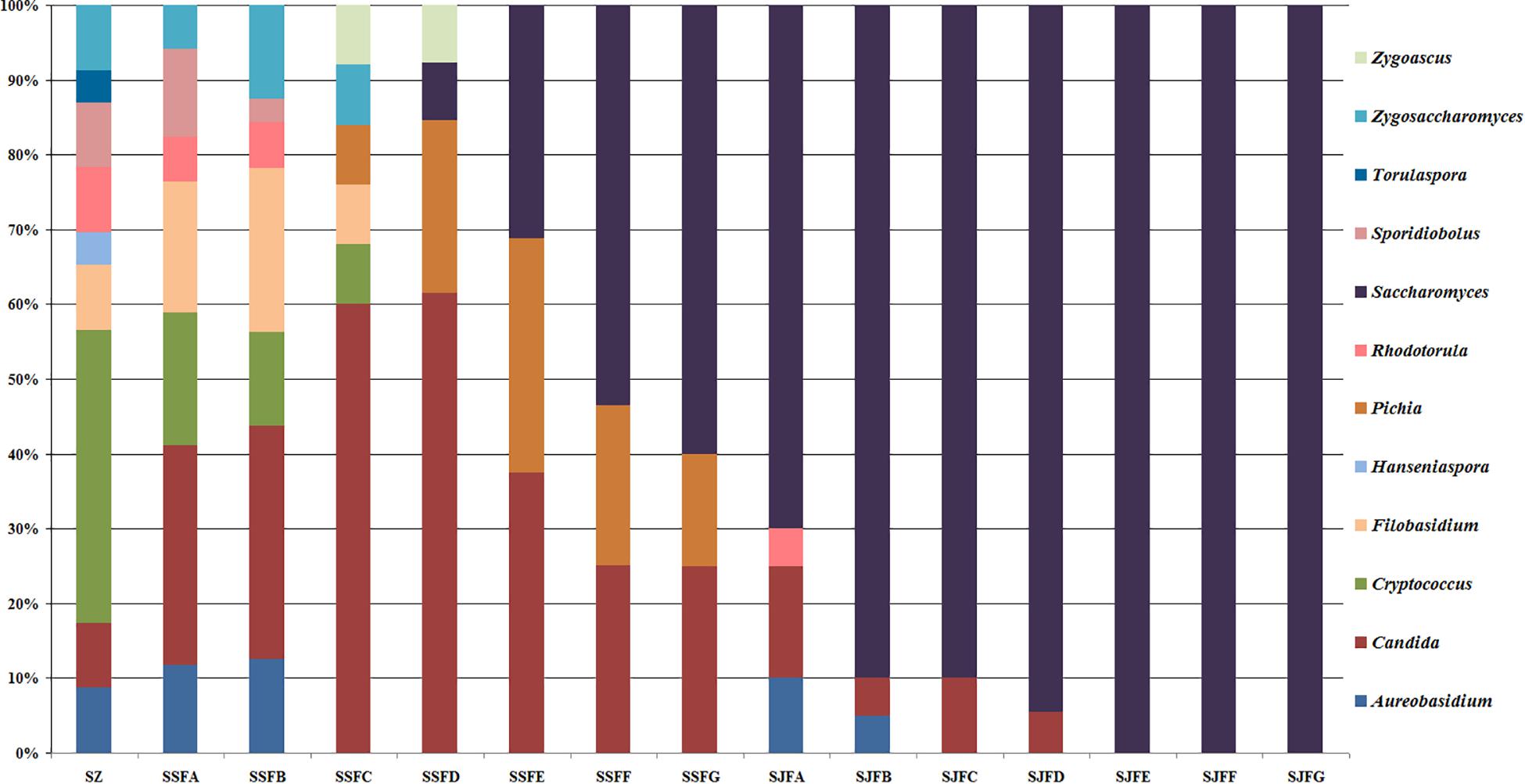
Figure 1. Histograms with the relative abundance of the yeasts isolated at each sampling point during spontaneous and inoculated fermentation processes (at the genus level).
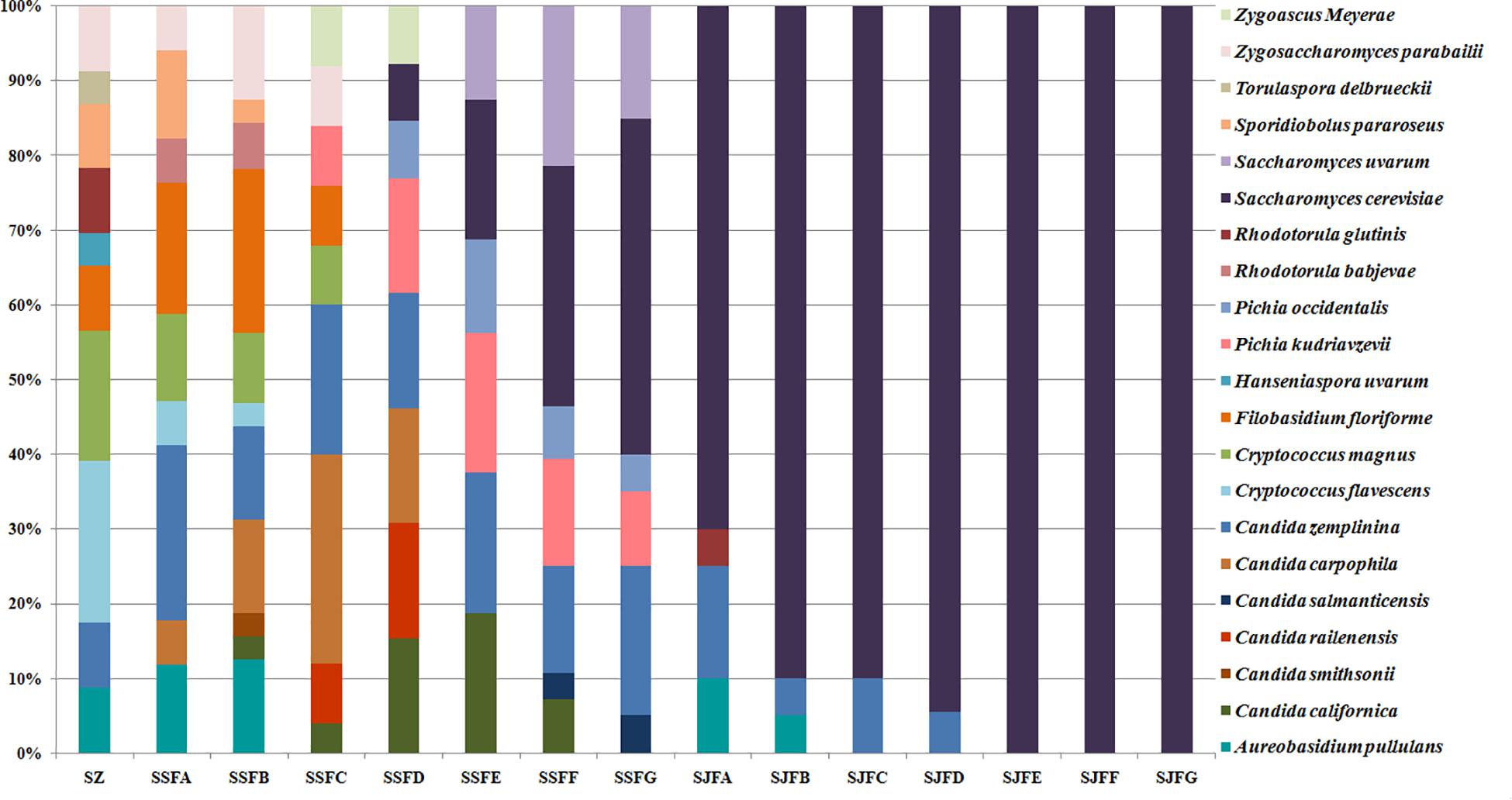
Figure 2. Histograms with the relative abundance of the yeasts isolated at each sampling point during spontaneous and inoculated fermentation processes (at the species level).
During the inoculated fermentation, A. pullulans, C. zemplinina, and R. glutinis were observed and their abundance levels decreased in the early stage. In the middle and later stages, other species were not detectable, with the exception of S. cerevisiae. From day 1 to 30, the abundance of S. cerevisiae gradually increased from 70.0% to 100.0%. Notably, S. cerevisiae was the predominant species during the whole process.
The yeast cell count in ice grape juice (SZ) was approximately 9 × 103 CFU/mL. During the spontaneous and inoculated fermentation processes, the yeast cell count initially increased and then decreased. Table 2 presents the changes of yeast cell count and residual sugar content during the spontaneous and inoculated fermentation processes. In the spontaneous fermentation process, tiny bubbles were observed on the surface on day 4 (SSFC); while in the inoculated fermentation process, obvious bubbles appeared a day after inoculated commercial yeasts (SJFA).
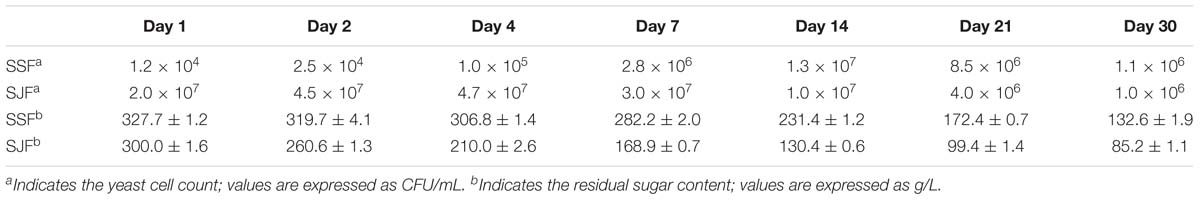
Table 2. Changes of the yeast cell count and the residual sugar content during the spontaneous and inoculated fermentation processes.
HTS Data Analysis
HTS of the ITS2 region genes was conducted to analyze fungal community structure during the spontaneous and inoculated fermentation processes. Table 3 presents the summary of HTS data. The highest numbers of raw and clean reads were detected on day 4 of the spontaneous fermentation, and were 60,214 and 58,849 (SSFC), respectively. In contrast, the lowest numbers of raw and clean reads were 14,013 and 13,564 (SJFC), and these were observed on day 4 of the inoculated fermentation. The highest and lowest Shannon diversity indices were observed in SSFB (4.02) and SJFF (0.33) samples, respectively. The Chao 1 richness index was the highest and lowest for SSFE (71.78) and SJFD (17.25) samples, respectively.
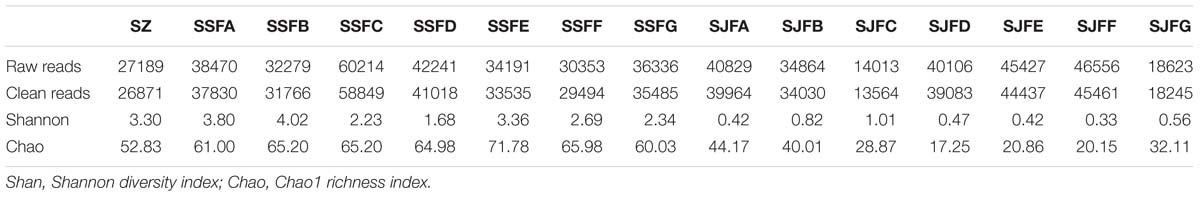
Table 3. Summary of HTS data, including the number of raw and clean reads, Shannon diversity index and Chao1 richness index for fungal ITS rDNA libraries from samples at different stages of the spontaneous and inoculated fermentation processes.
Figure 3 depicts a group of Venn diagrams highlighting the comparison of the OTUs at the same sampling point during the spontaneous and inoculated fermentation processes, and the OTUs obtained dynamically changed. On day 1, there were 64 OTUs in the spontaneous fermentation (SSFA) and 61 in the inoculated fermentation (SJFA), whereas 53 OTUs were identical between the two fermentation types. The number of OTUs present in SJFA but not in SSFA was 8, representing 11.1% of the total of OTUs in SSFA and SJFA. In SSFB and SJFB, there were 72 and 66 OTUs, respectively; the OTUs in the inoculated fermentation but not in the spontaneous fermentation accounted for 10% of all OTUs on day 2. From day 4 to day 14, the number of OTUs in the spontaneous fermentation increased from 78 (SSFC) to 84 (SSFE), while that in the inoculated fermentation decreased from 47 (SJFC) to 41 (SJFE); moreover, the percentage of the OTUs in the inoculated fermentation but not in the spontaneous fermentation gradually decreased from 11.4 to 3.4%. In the later fermentative stages (days 21–30), the number of OTUs in the spontaneous (SSFF and SSFG) and inoculated (SJFF and SJFG) fermentations was 45 and 40; it was notable that the number of OTUs in the inoculated fermentation but not in the spontaneous fermentation was 0. In summary, the number of OTUs increased initially and then decreased in the spontaneous fermentation, eventually reaching a peak at 84 on day 14. In the inoculated fermentation, the number of OTUs increased initially and then decreased gradually, but the highest value of OTUs was 66 on day 2. The percentage of the OTUs in the inoculated fermentation but not in the spontaneous fermentation decreased and was 0 from day 21 to the last sampling day (day 30).
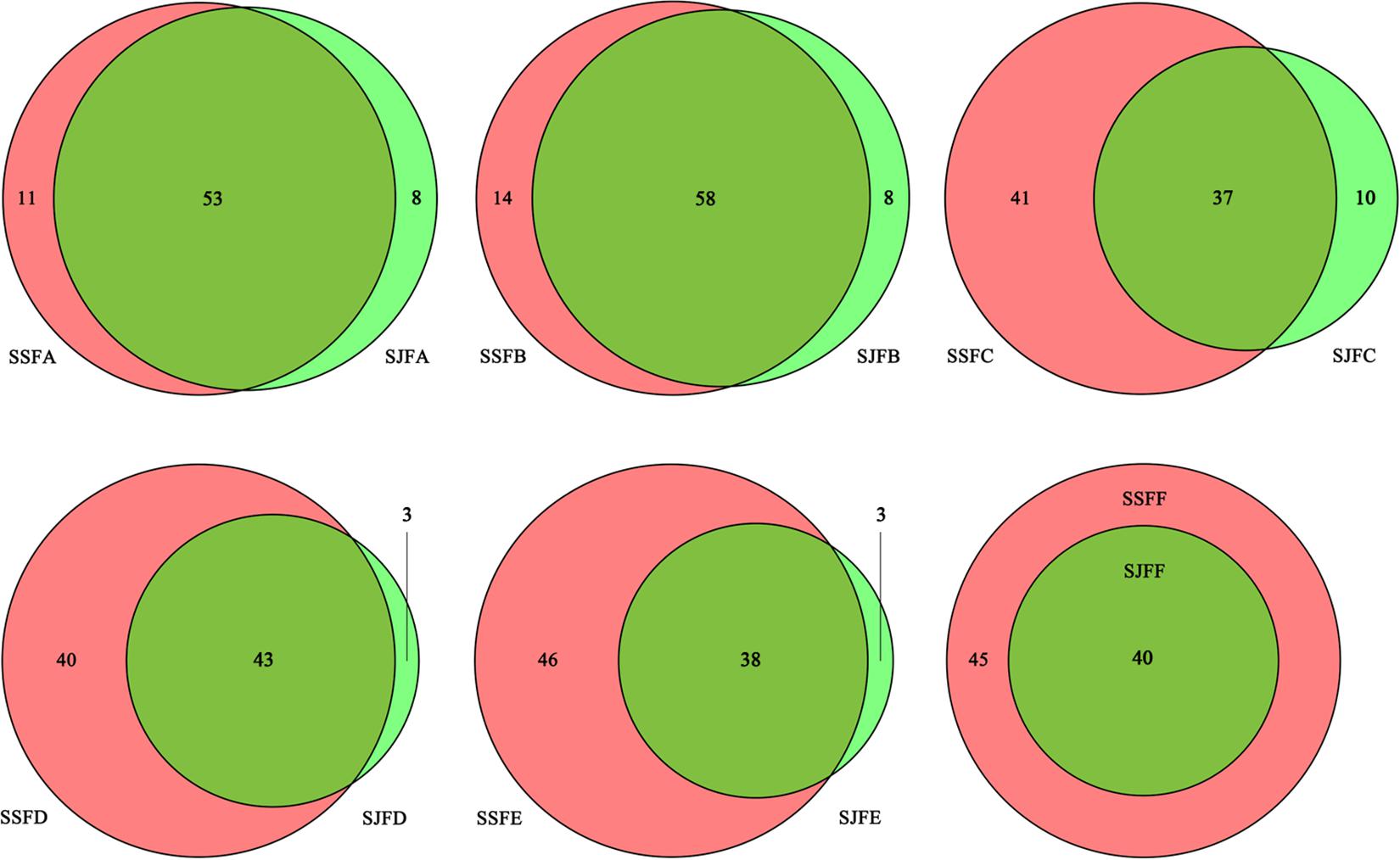
Figure 3. Venn diagrams of the number and relation of OTUs at each sampling point during the inoculated and spontaneous fermentation processes.
In the fermentation processes, the presence of three phyla along with unidentified and unclassified ones was suggested; 36 genera along with unidentified and unclassified ones; 56 species with unidentified and unclassified ones. The HTS method revealed a more complex mycobiota than the culture-dependent method did in the fermentations. E.g., Figure 4 depicts the genera detected in ice grape juice (SZ) by the culture-dependent and HTS methods; except for Aureobasidium, Candida, Cryptococcus, Filobasidium, and Zygosaccharomyces, more genera were detected by the HTS method.
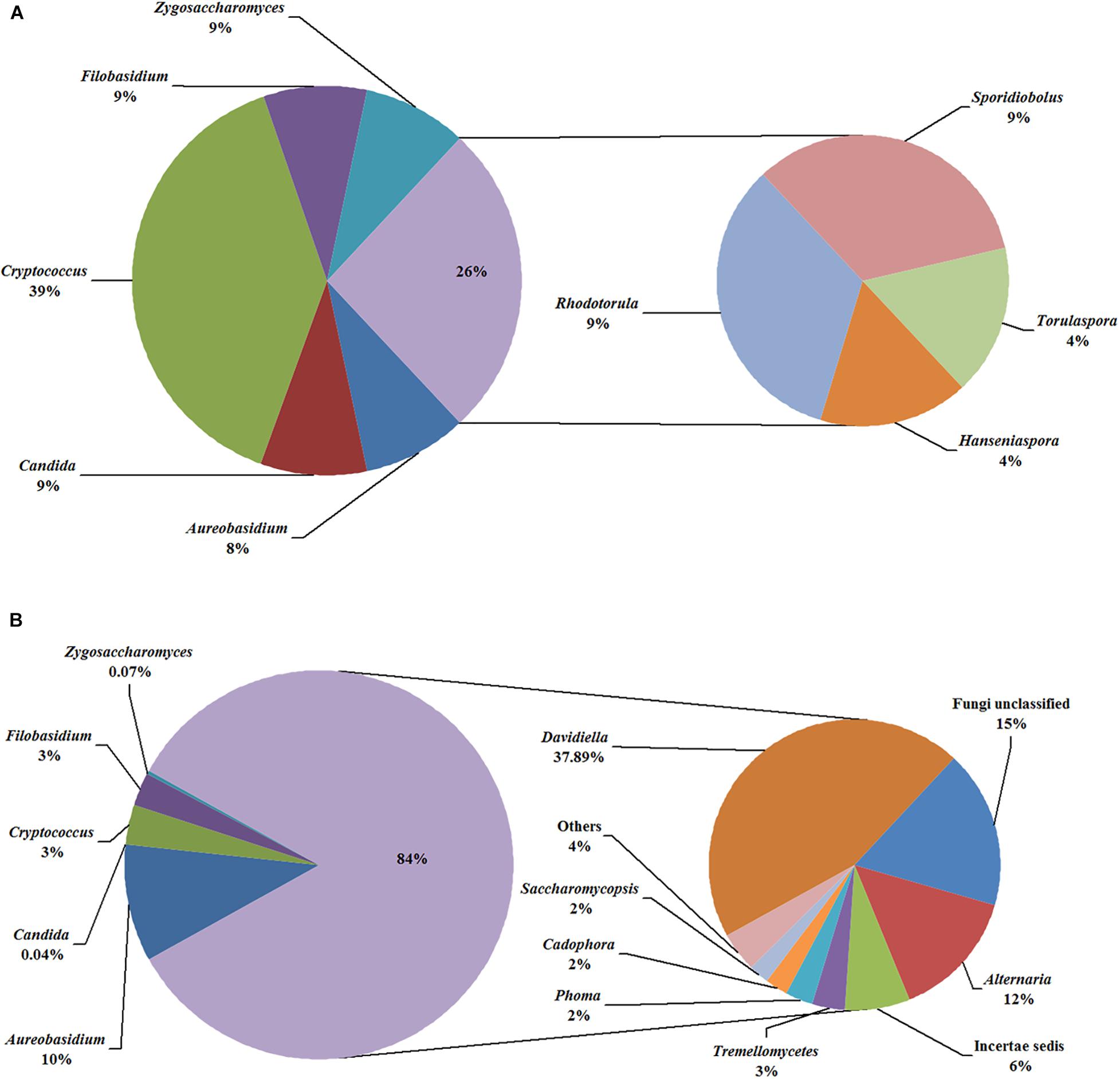
Figure 4. Relative abundance of the genera detected in the ice grape must (SZ) by culture-dependent (A) and HTS (B) methods. The “others” group includes Hanseniaspora, Rhodotorula, and so on.
An obvious dynamic change in the yeast population was observed by the HTS method during the spontaneous and inoculated fermentation procedures. Figures 5–7 show fungal diversity and community changes at phylum, genus, and species levels (top 20) during the spontaneous and inoculated fermentation processes. A high abundance of Ascomycota was detected at all sampling points. This was the dominant phylum, as it reached the highest value of 94.30% on day 7 (SSFD) and the lowest value of 51.33% on day 1 (SSFA) in the spontaneous fermentation; it represented over 90% in all samples of the inoculated fermentation. The highest (24.22%) and lowest (1.89%) abundance levels of Basidiomycota were detected on days 2 (SSFB) and 30 (SSFG) of the spontaneous fermentation, respectively. Moreover, a low abundance of Zygomycota was detected (Figure 5).
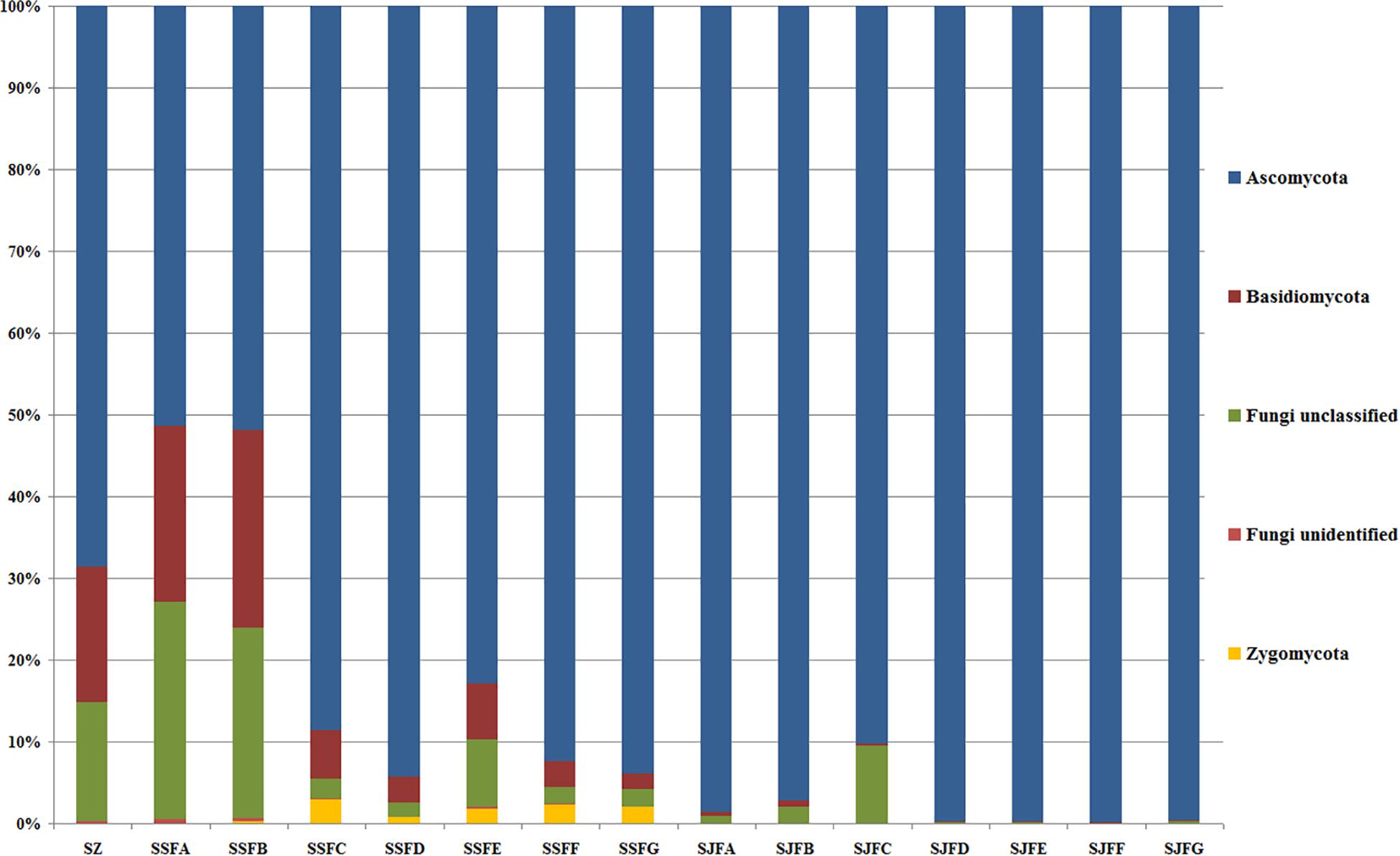
Figure 5. Histograms with the relative abundance of the yeasts during spontaneous and inoculated fermentation processes (at the phylum level) according to HTS analysis.
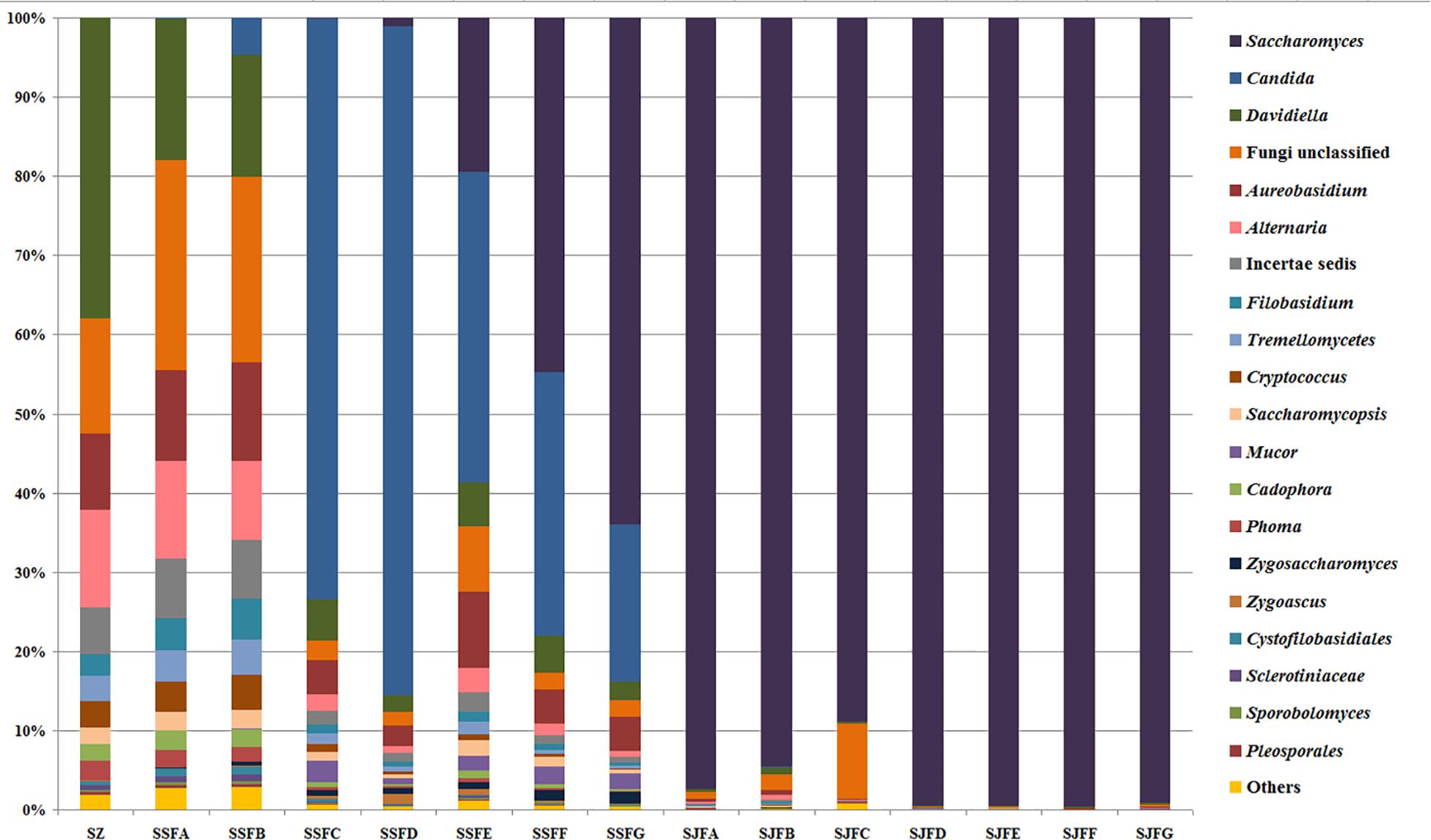
Figure 6. Histograms with the relative abundance of the yeasts during spontaneous and inoculated fermentation processes (at the genus level) according to HTS analysis.
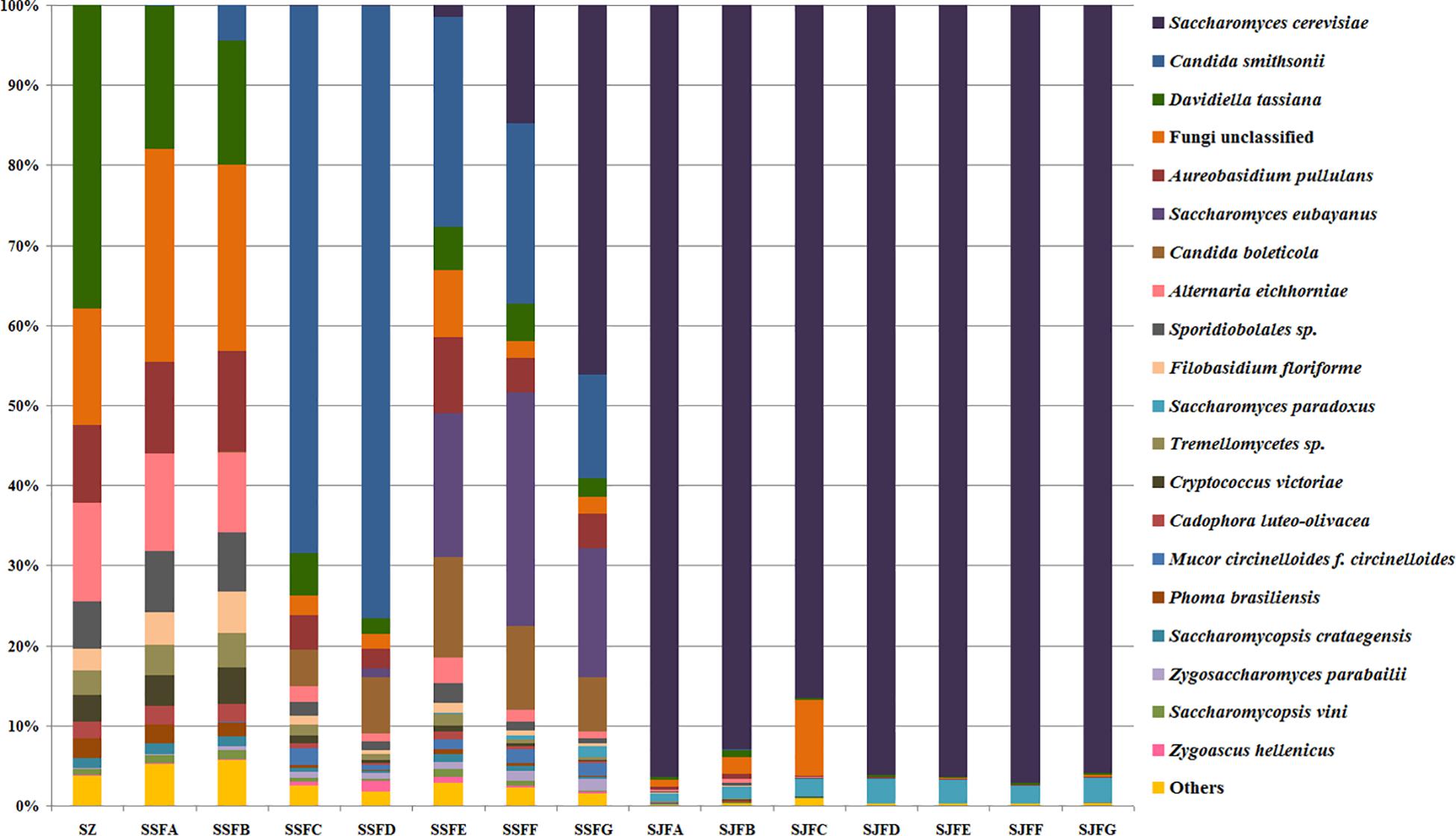
Figure 7. Histograms with the relative abundance of the yeasts during spontaneous and inoculated fermentation processes (at the species level) according to HTS analysis.
Candida was observed at all sampling points, and its abundance levels were dramatically changed during the spontaneous fermentation. The relative abundance of Candida initially increased and then decreased, and became the dominant genus on days 4 and 7 (SSFC and SSFD), with the highest abundance being 84.44% on day 7 (SSFD). In the genus Candida, C. smithsonii showed significant changes, and gradually increased and then decreased during the spontaneous fermentation, with the highest abundance being 76.5% (day 7, SSFD). The abundance of Candida boleticola also increased initially, but then decreased, with the highest (12.6%) abundance detected on day 14 (SSFE). A. pullulans was also found at all sampling points and reached the highest abundance of 12.55% on day 7 (SSFD) and the lowest of 2.52% on day 2 (SSFB). F. floriforme and Cryptococcus victoriae were detected at all sampling points, with their highest abundance values being 4–6% on day 2 (SSFB) and the lowest observed on day 30 (SSFG, <1%). The abundance of Saccharomyces gradually increased and it became the dominant genus in the later fermentative stages, representing 63.95% of the population at the end of the spontaneous fermentation (SSFG). In the genus Saccharomyces, S. cerevisiae, Saccharomyces eubayanus and Saccharomyces paradoxus were observed during the spontaneous fermentation. S. cerevisiae abundance gradually increased in the middle and later periods and accounted for 46.19% of the population on day 30 (SSFG). S. eubayanus was detected on days 7–30 and its abundance initially increased and then decreased, with the highest abundance detected being 29.12% on day 21 (SSFF). S. paradoxus was present on days 14–30 at low abundance levels (<2%).
During the inoculated fermentation, Saccharomyces was the predominant genus during the whole process, and its abundance levels at all sampling points were above 90%, with the exception of SJFC (88.95%). S. cerevisiae and S. paradoxus were observed, and their highest abundance levels were 95.95% (SJFG) and 3.06% (SJFD), respectively. A. pullulans was also found at all sampling points, with the highest (9.73%) and lowest (0.1%) abundance levels detected at the beginning and end of the inoculated fermentation (SZ, SJFG), respectively.
Comparison of Yeast Diversity and Dynamics Between During the Spontaneous and Inoculated Fermentation Processes
Clearly, yeast diversity in the spontaneous fermentation is much richer than that in the inoculated fermentation in both the culture-independent and HTS results. In detail, 12 genera (21 species) were detected by the culture-independent method in the spontaneous fermentation, and there were four genera (four species) in the inoculated fermentation.
During the spontaneous fermentation, non-Saccharomyces yeasts were predominant in the initial and middle stages, whereas Saccharomyces dominated in the later fermentative stages. However, Saccharomyces was the predominant genus during the whole inoculated fermentation process, and non-Saccharomyces yeasts were detectable in the early stages and then almost disappeared.
Discussion
In this study, an obvious succession of yeast community was detected during the fermentation processes. To the best of our knowledge, this is the first study on the comparison of yeast diversity and the dynamics between during the spontaneous and inoculated fermentation of icewine using the cultivation approach (WL nutrient agar) and HTS approaches. During the spontaneous fermentation, non-Saccharomyces yeasts and the non-yeast fungi decreased in abundance, whereas Saccharomyces was gradually increased. This phenomenon can be explained as follows: the increase in ethanol concentration with the increase of fermentation time prevented the growth of non-Saccharomyces yeasts and non-yeast fungi (Fleet, 2006; Mills et al., 2008). The changes of non-Saccharomyces yeasts during the inoculated fermentation can be explained that the inoculated S. cerevisiae suppressed the growth of autochthonous non-Saccharomyces yeasts and performed rapid and complete fermentation (Urso et al., 2008).
In terms of non-Saccharomyces dynamics in the spontaneous and inoculated fermentation processes, it clearly decreased from the initial to the final stages. Of particular interest was the dynamic change of Candida. C. zemplinina was detected at all sampling points of the spontaneous fermentation by the culture-dependent method, which is in agreement with our previous study (Li et al., 2018). Notably, C. zemplinina was consistently detected in the early stage (days 0–7) of the inoculated fermentation. This is because C. zemplinina can grow at high sugar concentrations, at low temperatures and in the presence of ethanol (Sipiczki, 2003, 2004); and it can participate actively in fermentative production of sweet wine (Urso et al., 2008). Moreover, in previous studies, C. zemplinina was used to participate in mixed fermentation processes with S. cerevisiae that can reduce ethanol and acetic acid content and improve the aroma profile of wine (Englezos et al., 2017, 2018). The dynamic change of C. californica in the spontaneous fermentation was obvious according to the cultivation method. C. californica had been found in different wine-growing regions before (Vaudano et al., 2018; Aplin et al., 2019), and the latest studies have revealed that C. californica, when applied to inoculated fermentation with S. cerevisiae, can produce lower concentrations of residual sugar and ethanol as compared to fermentation with only S. cerevisiae (Aplin et al., 2019). Additionally, C. carpophila and C. railenensis were present in the early stages of the spontaneous fermentation; they have been detected in wine-growing regions before (Brežná et al., 2010; Brysch-Herzberg and Seidel, 2015; Drumonde-Neves et al., 2017). In further studies, the autochthonous strains of Candida can be screened on the basis of their outstanding enzymatic activities that affect and modulate the chemical and aromatic profile; these enzymatic activities are crucial to producing icewine by “multi-species” fermentation with unique characteristics.
Moreover, Cry. victoriae was detected at all sampling points in the spontaneous fermentation by HTS, but not by cultivation. Alessandria et al. (2013) also stated that Cry. victoriae was consistently detected during the spontaneous fermentation by the PCR-DGGE method (Alessandria et al., 2013). Pichia was detected in the spontaneous fermentation by cultivation, but not by HTS; it has been found in different wine-growing regions of the world (Brysch-Herzberg and Seidel, 2015; De Filippis et al., 2017; Bučková et al., 2018). Studies have revealed that P. kudriavzevii can positively influence the aroma and flavor of wine and has the potential to be used in winemaking (Del Monaco et al., 2014). H. uvarum and T. delbrueckii were often observed on grapes and in wine fermentation (Alessandria et al., 2013; De Filippis et al., 2017; Vaudano et al., 2018), but they were found only in the ice grape juice by the culture-dependent method in this study.
Interestingly, A. pullulans was detected both in the spontaneous and inoculated fermentation processes. It is a ubiquitous fungus in the environment (Singh et al., 2008), and is the yeast-like fungus and does not have the ability to spoil wine (Alessandria et al., 2013). In this study, it was detected in the initial stages of the spontaneous and inoculated fermentation processes but not observed in the later stages by the cultivation method. Nonetheless, it was detected throughout the entire fermentation process by HTS. Alessandria et al. (2013) stated that A. pullulans was found throughout the spontaneous fermentation process of icewine at the RNA level by the PCR-DGGE method; however, A. pullulans was not detected in the inoculated fermentation process; in fact, the starter profiles were only showed in the inoculated fermentation process by PCR-DGGE method (Alessandria et al., 2013). It has been reported that PCR-DGGE is not useful for detecting species with densities below 103 CFU/mL (Portillo and Mas, 2016). Therefore, the HTS method is more comprehensive than the PCR-DGGE method for analyzing the microbial diversity of fermented foods.
Thus far, ITS-rDNA as a target amplicon was generally selected for amplification in the studies on fungal populations during wine fermentations (Bokulich et al., 2014; Pinto et al., 2015). In our previous research, the ITS1 region of the samples was selected for amplification; the notably “incertae sedis” result was obtained after HTS, and some species were detected by cultivation method but not by HTS (Li et al., 2018). In addition, the ITS2 region was suggested to be more variable than ITS1 (Bazzicalupo et al., 2013; Sommermann et al., 2018). Because of the above issues, the ITS2 region of the samples was selected for amplification in this study; however, “Fungi unclassified” and “Incertae sedis” were also observed in HTS results, and some species were found by cultivation method but not in HTS results. Regarding “Fungi unclassified” and “Incertae sedis,” that is because the databases used for fungal comparisons generally have limited coverage and need extended follow-up in the future (Ercolini, 2013).
Regarding the differences in the results between the cultivation and HTS methods, Mendoza et al. (2017) also stated that HTS results revealed a large number of yeast species not recovered by culture-dependent method; moreover, the cultivation method detected very few yeast taxa, most of them corresponding with very few or zero reads in HTS analysis (Mendoza et al., 2017). The differences in the results by the two methods have also been reported in previous studies of foods and wines (David et al., 2014; Greppi et al., 2015). In this study, it can be explained by the limitations of the two methods; the cells in a sublethal or injured physiological state can be absent in the results of the cultivation (Rantsiou et al., 2005; Mendoza et al., 2017); the bias of different primers exist, the sequencing depth of the HTS method may be insufficient, and preferential amplification of shorter fragments may occur during PCR, and so forth (Ercolini, 2013; De Filippis et al., 2017; Mendoza et al., 2017; Karst et al., 2018). Because of the above issues, both the culture-dependent and HTS methods were employed to analyze yeast diversity and dynamics during the spontaneous and inoculated fermentation processes, which is a more comprehensive strategy.
In addition to wine yeasts, we found some other fungi in the fermentation by the HTS method, mainly Davidiella and Alternaria. They were also found in grape must or fermented wine samples of different wine-making regions (Bokulich et al., 2014; Pinto et al., 2015). Davidiella tassiana (other name is Cladosporium herbarum) is a fungal plant pathogen, which causes a common disease of grapevines (Cladosporium rot) (Latorre et al., 2011). Previous research indicates that this pathogen can reduce the yield and affect the quality of wines (Briceño et al., 2009). It is found particularly in the vines that are harvested very late (Briceño and Latorre, 2008). In the present study, the ice grapes used for making icewine were harvested in December, which was later than the harvest time of common wine grapes. Alternaria has been the main component of the wine grape mycobiota in several wine-making regions worldwide (Prendes et al., 2015; Bučková et al., 2018). Alternaria includes numerous species, and some of these species produce mycotoxins that can pose a potential health hazard for consumers. Alternaria toxins in wine, beer, and other foods have been reported (Fan et al., 2016). Alternaria alternate is the most prevalent species, which is the most important mycotoxin-producing fungus (European Food Safety Authority [EFSA], Arcella et al., 2016). Nevertheless, Alternaria alternate was not detected in our study, but Alternaria eichhorniae was found. Very few reports have studied Alternaria eichhorniae. Shabana et al. (1995) stated that Alternaria eichhorniae is a safe and effective bioherbicide for waterhyacinth (Shabana et al., 1995). In the present study, the highest abundance levels of Davidiella and Alternaria were both detected in raw ice grape juice. Thereafter, their abundance decreased clearly, and their abundance levels at the end of the spontaneous fermentation were low. The abundance decrease may be attributed to the inappropriate cultivation conditions for these species with the increase in fermentative time. Bokulich et al. (2014) stated that fungal profiles were dominated by filamentous fungi in the grape juice samples (close to 70%), and mentioned that many microorganisms inhabiting the grape surface cannot survive the low-pH, ethanolic, anaerobic conditions of wine fermentation processes, e.g., phytopathogenic fungi, but their metabolic activity can have long-ranging consequences (Bokulich et al., 2014). The abundance levels of Davidiella and Alternaria in the inoculated fermentation were all no more than 1% from day 1 to day 30. Comparison of Davidiella and Alternaria abundances during the spontaneous and inoculated fermentation processes shows that we have reason to believe that the inoculated fermentation icewine should be safer than the spontaneous fermentation icewine. To the best of our knowledge, the presence of Davidiella and Alternaria and their dynamic changes during icewine fermentation were revealed for the first time. The metabolites and mycotoxins in icewine should be researched in more detail in future works.
In this study, yeast diversity and dynamics during the spontaneous fermentation were found to be different from those in our previous research (Li et al., 2018). For example, C. californica and C. carpophila have been found in this study, but not in the previous research; Metschnikowia had been isolated from the samples of the previous research, but not in this study. Although the same grape variety was employed and the vineyards were both in northeast China in these two studies, different locations, years of sampling, and compositions of samples could explain these discrepancies. In the previous studies, different wine-growing regions were reported to maintain different microbial communities (called “microbial terroir”) (Bokulich et al., 2014; Gilbert et al., 2014).
Conclusion
The combination of a microbial cultivation method and HTS approaches is useful for estimating across-the-board microbial diversity and dynamics during the spontaneous and inoculated fermentation processes of Vidal blanc icewine. Further studies on the oenological characteristics of autochthonous yeast strains and their contribution to the sensory quality, as well as the microbial safety of the dominant yeast strains found in this study, could help us to use them in co-inoculation trials to produce icewines with territorial characteristics and particular flavors.
Data Availability
The datasets generated for this study are available in the Genbank and Sequence Read Archive (SRA) of the National Center for Biotechnology Information (NCBI) database, PRJNA503950.
Author Contributions
JL and W-ZH designed the experiments. JL conducted the experiments and analyzed the experimental data. JL, W-ZH, and Y-PX wrote the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
All samples of ice grape berries were provided by the “Sun Valley” Winery.
Abbreviations
HTS, high-throughput sequencing; ITS region, internal transcribed spacer region; PCR-DGGE, polymerase chain reaction with denaturing gradient gel electrophoresis; rDNA, ribosomal DNA; WL nutrient agar, Wallerstein laboratory nutrient agar.
Footnotes
References
Alessandria, V., Giacosa, S., Campolongo, S., Rolle, L., Rantsiou, K., and Cocolin, L. (2013). Yeast population diversity on grapes during on-vine withering and their dynamics in natural and inoculated fermentations in the production of icewines. Food Res. Int. 54, 139–147. doi: 10.1016/j.foodres.2013.06.018
Andorrà, I., Landi, S., Mas, A., Esteve-Zarzoso, B., and Guillamón, J. M. (2010). Effect of fermentation temperature on microbial population evolution using culture-independent and dependent techniques. Food Res. Int. 43, 773–779. doi: 10.1016/j.foodres.2009.11.014
Aplin, J. J., White, K. P., and Edwards, C. G. (2019). Growth and metabolism of non-Saccharomyces yeasts isolated from Washington state vineyards in media and high sugar grape musts. Food Microbiol. 77, 158–165. doi: 10.1016/j.fm.2018.09.004
Bazzicalupo, A. L., Bálint, M., and Schmitt, I. (2013). Comparison of ITS1 and ITS2 rDNA in 454 sequencing of hyperdiverse fungal communities. Fungal Ecol. 6, 102–109. doi: 10.1016/j.funeco.2012.09.003
Bokulich, N. A., Thorngate, J. H., Richardson, P. M., and Mills, D. A. (2014). Microbial biogeography of wine grapes is conditioned by cultivar, vintage, and climate. Proc. Natl. Acad. Sci. U.S.A. 111, E139–E148. doi: 10.1073/pnas.1317377110
Bowen, A. J., and Reynolds, A. G. (2015). Aroma compounds in ontario vidal and riesling icewines. I. Effects of harvest date. Food Res. Int. 76, 540–549. doi: 10.1016/j.foodres.2015.06.046
Brežná, B., Zenišová, K., Chovanová, K., Chebeňová, V., Kraková, L., Kuchta, T., et al. (2010). Evaluation of fungal and yeast diversity in Slovakian wine-related microbial communities. Antonie Van Leeuwenhoek 98, 519–529. doi: 10.1007/s10482-010-9469-6
Briceño, E. X., and Latorre, B. A. (2008). Characterization of Cladosporium rot in grapevines, a problem of growing importance in Chile. Plant Dis. 92, 1635–1642. doi: 10.1094/PDIS-92-12-1635
Briceño, E. X., Latorre, B. A., and Bordeu, E. (2009). Effect of Cladosporium rot on the composition and aromatic compounds of red wine. Span. J. Agric. Res. 7, 119–128.
Brysch-Herzberg, M., and Seidel, M. (2015). Yeast diversity on grapes in two German wine growing regions. Int. J. Food Microbiol. 214, 137–144. doi: 10.1016/j.ijfoodmicro.2015.07.034
Buèková, M., Puškárová, A., Ženišová, K., Kraková, L., Piknová, L’, Kuchta, T., et al. (2018). Novel insights into microbial community dynamics during the fermentation of Central European ice wine. Int. J. Food Microbiol. 266, 42–51. doi: 10.1016/j.ijfoodmicro.2017.11.010
Cavazza, A., Grando, M. S., and Zini, C. (1992). Rilevazione della flora microbica di mosti e vini. Vignevini 9, 17–20.
Ciani, M., Comitini, F., Mannazzu, I., and Domizio, P. (2010). Controlled mixed culture fermentation: a new perspective on the use of non-Saccharomyces yeasts in winemaking. FEMS Yeast Res. 10, 123–133. doi: 10.1111/j.1567-1364.2009.00579.x
Crandles, M., Reynolds, A. G., Khairallah, R., and Bowen, A. (2015). The effect of yeast strain on odor active compounds in riesling and vidal blanc icewines. LWT-Food Sci. Technol. 64, 243–258. doi: 10.1016/j.lwt.2015.05.049
David, V., Terrat, S., Herzine, K., Claisse, O., Rousseaux, S., Tourdot-Maréchal, R., et al. (2014). High-throughput sequencing of amplicons for monitoring yeast biodiversity in must and during alcoholic fermentation. J. Ind. Microbiol. Biot. 41, 811–821. doi: 10.1007/s10295-014-1427-2
De Filippis, F., Storia, A. L., and Blaiotta, G. (2017). Monitoring the mycobiota during Greco di Tufo and aglianico wine fermentation by 18S rRNA gene sequencing. Food Microbiol. 63, 117–122. doi: 10.1016/j.fm.2016.11.010
Del Monaco, S. M., Barda, N. B., Rubio, N. C., and Caballero, A. C. (2014). Selection and characterization of a Patagonian Pichia kudriavzevii for wine deacidification. J. Appl. Microb. 117, 451–464. doi: 10.1111/jam.12547
Drumonde-Neves, J., Franco-Duarte, R., Lima, T., Schuller, D., and Pais, C. (2017). Association between grape yeast communities and the vineyard ecosystems. PLoS One 12:e0169883. doi: 10.1371/journal.pone.0169883
Englezos, V., Giacosa, S., Rantsiou, K., Rolle, L., and Cocolin, L. (2017). Starmerella bacillaris in winemaking: opportunities and risks. Curr. Opin. Food Sci. 17, 30–35. doi: 10.1016/j.cofs.2017.08.007
Englezos, V., Rantsiou, K., Cravero, F., Torchio, F., Pollon, M., Daniela, F., et al. (2018). Volatile profile of white wines fermented with sequential inoculation of Starmerella bacillaris and Saccharomyces cerevisiae. Food Chem. 257, 350–360. doi: 10.1016/j.foodchem.2018.03.018
Ercolini, D. (2013). High-throughput sequencing and metagenomics: moving forward in the culture-independent analysis of food microbial ecology. Appl. Environ. Microbiol. 79, 3148–3155. doi: 10.1128/AEM.00256-13
European Food Safety Authority [EFSA], Arcella, D., Eskola, M., and Ruiz, J. A. G. (2016). Dietary exposure assessment to Alternaria toxins in the European population. EFSA J. 14:4654.
Fan, C., Cao, X., Liu, M., and Wang, W. (2016). Determination of Alternaria mycotoxins in wine and juice using ionic liquid modified counter current chromatography as a pretreatment method followed by high-performance liquid chromatography. J. Chromatogr. A 1436, 133–140. doi: 10.1016/j.chroma.2016.01.069
Fell, J. W., Boekhout, T., Fonseca, A., Scorzetti, G., and Statzell-Tallman, A. (2000). Biodiversity and systematics of basidiomycetous yeasts as determined by large-subunit rDNA D1/D2 domain sequence analysis. Int. J. Syst. Evol. Microbiol. 50(Pt 3), 1351–1371. doi: 10.1099/00207713-50-3-1351
Fleet, G. H. (2003). Yeast interactions and wine flavour. Int. J. Food Microbiol. 86, 11–22. doi: 10.1016/s0168-1605(03)00245-9
Fleet, G. H. (2006). “The commercial and community significance of yeasts in food and beverage production,” in Yeasts in Food and Beverages, eds A. Querol and G. H. Fleet (Berlin: Springer-Verlag), 1–12. doi: 10.1007/978-3-540-28398-0_1
Gilbert, J. A., van der Lelie, D., and Zarraonaindia, I. (2014). Microbial terroir for wine grapes. Proc. Natl. Acad. Sci. U.S.A. 111, 5–6. doi: 10.1073/pnas.1320471110
Graça, A., Santo, D., Esteves, E., Nunes, C., Abadias, M., and Quintas, C. (2015). Evaluation of microbial quality and yeast diversity in fresh-cut apple. Food Microbiol. 51, 179–185. doi: 10.1016/j.fm.2015.06.003
Greppi, A., Ferrocino, I., Storia, A. L., Rantsiou, K., Ercolini, D., and Cocolin, L. (2015). Monitoring of the microbiota of fermented sausages by culture independent rRNA-based approaches. Int. J. Food Microbiol. 212, 67–75. doi: 10.1016/j.ijfoodmicro.2015.01.016
Huang, L., Ma, Y., Tian, X., Li, J. M., Li, L. X., Tang, K., et al. (2018). Chemosensory characteristics of regional Vidal icewines from China and Canada. Food Chem. 261, 66–74. doi: 10.1016/j.foodchem.2018.04.021
Karst, S. M., Dueholm, M. S., McIlroy, S. J., Kirkegaard, R. H., Nielsen, P. H., and Albertsen, M. (2018). Retrieval of a million high-quality, full-length microbial 16S and 18S rRNA gene sequences without primer bias. Nat. Biotechnol. 36, 190–195. doi: 10.1038/nbt.4045
Latorre, B. A., Briceño, E. X., and Torres, R. (2011). Increase in Cladosporium spp. populations and rot of wine grapes associated with leaf removal. Crop Prot. 30, 52–56. doi: 10.1016/j.cropro.2010.08.022
Lencioni, L., Romani, C., Gobbi, M., Comitini, F., Ciani, M., and Domizio, P. (2016). Controlled mixed fermentation at winery scale using Zygotorulaspora florentina and Saccharomyces cerevisiae. Int. J. Food Microbiol. 234, 36–44. doi: 10.1016/j.ijfoodmicro.2016.06.004
Li, J., Hu, W. Z., Huang, X. J., and Xu, Y. P. (2018). Investigation of yeast population diversity and dynamics in spontaneous fermentation of vidal blanc icewine by traditional culture-dependent and high-throughput sequencing methods. Food Res. Int. 112, 66–77. doi: 10.1016/j.foodres.2018.06.011
Ma, Y., Tang, K., Xu, Y., and Li, J. M. (2017). Characterization of the key aroma compounds in chinese vidal icewine by gas chromatography-olfactometry, quantitative measurements, aroma recombination, and omission tests. J. Agric. Food Chem. 65, 394–401. doi: 10.1021/acs.jafc.6b04509
Maturano, Y. P., Rodríguez Assaf, L. A., Toro, M. E., Nally, M. C., Vallejo, M., Castellanos, et al. (2012). Multi-enzyme production by pure and mixed cultures of Saccharomyces and non-Saccharomyces yeasts during wine fermentation. Int. J. Food Microbiol. 155, 43–50. doi: 10.1016/j.ijfoodmicro.2012.01.015
Mendoza, L. M., Neef, A., Vignolo, G., and Belloch, C. (2017). Yeast diversity during the fermentation of Andean chicha: a comparison of high-throughput approaches. Food Microbiol. 67, 1–10. doi: 10.1016/j.fm.2017.05.007
Mills, D. A., Phister, T., Neeley, E., and Johannsen, E. (2008). “Wine fermentation,” in Molecular Techniques in the Microbial Ecology of Fermented Foods, eds L. Cocolin and D. Ercolini (New York, NY: Springer), 161–192.
Nurgel, C., Pickering, G. J., and Inglis, D. L. (2004). Sensory and chemical characteristics of Canadian ice wines. J. Sci. Food Agric. 84, 1675–1684. . doi: 10.1002/jsfa.1860
Pérez, G., Fariña, L., Barquet, M., Boido, E., Gaggero, C., Dellacassa, E., et al. (2011). A quick screening method to identify β-glucosidase activity in native wine yeast strains: application of esculin glycerol agar (EGA) medium. World J. Microb. Biot. 27, 47–55. doi: 10.1007/s11274-010-0425-4
Pinto, C., Pinho, D., Cardoso, R., Custódio, V., Fernandes, J., Sousa, S., et al. (2015). Wine fermentation microbiome: a landscape from different Portuguese wine appellations. Front. Microbiol. 6:905. doi: 10.3389/fmicb.2015.00905
Połka, J., Rebecchi, A., Pisacane, V., Morelli, L., and Puglisi, E. (2015). Bacterial diversity in typical Italian salami at different ripening stages as revealed by high-throughput sequencing of 16S rRNA amplicons. Food Microbiol. 46, 342–356. doi: 10.1016/j.fm.2014.08.023
Portillo, M. D. C., and Mas, A. (2016). Analysis of microbial diversity and dynamics during wine fermentation of Grenache grape variety by high-throughput barcoding sequencing. LWT-Food Sci. Technol. 72, 317–321. doi: 10.1016/j.lwt.2016.05.009
Prendes, L. P., Merín, M. G., Andreoni, M. A., Ramírez, M. L., and Ambrosini, V. I. M. D. (2015). Mycobiota and toxicogenic Alternaria spp. strains in Malbec wine grapes from DOC San Rafael, Mendoza, Argentina. Food Control 57, 122–128. doi: 10.1016/j.foodcont.2015.03.041
Rantsiou, K., Urso, R., Iacumin, L., Cantoni, C., Cattaneo, P., Comi, G., et al. (2005). Culture-dependent and -independent methods to investigate the microbial ecology of Italian fermented sausages. Appl. Environ. Microb. 71, 1977–1986. doi: 10.1128/aem.71.4.1977-1986.2005
Santo, D. E., Galego, L., Gonçalves, T., and Quintas, C. (2012). Yeast diversity in the Mediterranean strawberry tree (Arbutus unedo L.) fruits fermentations. Food Res. Int. 47, 45–50. doi: 10.1016/j.foodres.2012.01.009
Shabana, Y. M., Charudattan, R., and Elwakil, M. A. (1995). Identification, pathogenicity, and safety of Alternaria eichorniae from Egypt as a bioherbicide agent of waterhyacinth. Biol. Control 5, 123–135. doi: 10.1006/bcon.1995.1015
Singh, R. S., Saini, G. K., and Kennedy, J. F. (2008). Pullulan: microbial sources, production and applications. Carbohyd. Polym. 73, 515–531. doi: 10.1016/j.carbpol.2008.01.003
Sipiczki, M. (2003). Candida zemplinina sp. nov., an osmotolerant and psychrotolerant yeast that ferments sweet botrytized wines. Int. J. Syst. Evol. Microbiol. 53, 2079–2083. doi: 10.1099/ijs.0.02649-0
Sipiczki, M. (2004). Species identification and comparative molecular and physiological analysis of Candida zemplinina and Candida stellata. J. Basic Microbiol. 44, 471–479. doi: 10.1002/jobm.200410449
Sommermann, L., Geistlinger, J., Wibberg, D., Deubel, A., Zwanzig, J., Babin, D., et al. (2018). Fungal community profiles in agricultural soils of a long-term field trial under different tillage, fertilization and crop rotation conditions analyzed by high-throughput ITS-amplicon sequencing. PLoS One 13:e0195345. doi: 10.1371/journal.pone.0195345
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Tian, R. R., Li, G., Wan, S. B., Pan, Q. H., Zhan, J. C., Li, J. M., et al. (2009). Comparative study of 11 phenolic acids and five flavan-3-ols in cv. Vidal: Impact of natural icewine making versus concentration technology. Aust. J. Grape Wine R. 15, 216–222. doi: 10.1111/j.1755-0238.2009.00055.x
Urso, R., Rantsiou, K., Dolci, P., Rolle, L., Comi, G., and Cocolin, L. (2008). Yeast biodiversity and dynamics during sweet wine production as determined by molecular methods. FEMS Yeast Res. 8, 1053–1062. doi: 10.1111/j.1567-1364.2008.00364.x
Van Hijum, S. A., Vaughan, E. E., and Vogel, R. F. (2013). Application of state-of-art sequencing technologies to indigenous food fermentations. Curr. Opin. Biotech. 24, 178–186. doi: 10.1016/j.copbio.2012.08.004
Vaudano, E., Quinterno, G., Costantini, A., Pulcini, L., Pessione, E., and Garcia-Moruno, E. (2018). Yeast distribution in Grignolino grapes growing in a new vineyard in Piedmont and the technological characterization of indigenous Saccharomyces spp. strains. Int. J. Food Microbiol. 289, 154–161. doi: 10.1016/j.ijfoodmicro.2018.09.016
Keywords: icewine, yeast, dynamics, spontaneous and inoculated fermentations, culture-dependent method, high-throughput sequencing
Citation: Li J, Hu W-Z and Xu Y-P (2019) Diversity and Dynamics of Yeasts During Vidal Blanc Icewine Fermentation: A Strategy of the Combination of Culture-Dependent and High-Throughput Sequencing Approaches. Front. Microbiol. 10:1588. doi: 10.3389/fmicb.2019.01588
Received: 16 April 2019; Accepted: 25 June 2019;
Published: 09 July 2019.
Edited by:
Rosanna Tofalo, University of Teramo, ItalyReviewed by:
Catarina Prista, University of Lisbon, PortugalTeresa Arroyo Casado, Instituto Madrileño de Investigación y Desarrollo Rural, Agrario y Alimentario, Spain
Copyright © 2019 Li, Hu and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wen-Zhong Hu, d2VuemhvbmdoQHNpbmEuY29t
 Jing Li
Jing Li Wen-Zhong Hu
Wen-Zhong Hu Yong-Ping Xu1
Yong-Ping Xu1