- 1Institute of Microbiology Paulo de Góes, Federal University of Rio de Janeiro, Rio de Janeiro, Brazil
- 2Research Center Leopoldo Américo Miguez de Mello, Rio de Janeiro, Brazil
- 3IMAM-AquaRio – Rio de Janeiro Aquarium Research Center, Rio de Janeiro, Brazil
In this study, oil spills were simulated in field-based mangrove mesocosms to compare the efficiency of bioremediation strategies and to characterize the presence of the alkB, ndo, assA, and bssA genes and the ecological structures of microbial communities in mangrove sediments at two different depths, (D1) 1–10 cm and (D2) 25–35 cm. The results indicated that the hydrocarbon degradation efficiency was higher in superficial sediment layers, although no differences in the hydrocarbon degradation rates or in the abundances of the alkB and ndo genes were detected among the tested bioremediation strategies at this depth. Samples from the deeper layer exhibited higher abundances of the analyzed genes, except for assA and bssA, which were not detected in our samples. For all of the treatments and depths, the most abundant phyla were Proteobacteria, Firmicutes and Bacteroidetes, with Gammaproteobacteria, Flavobacteriales and Clostridiales being the most common classes. The indicator species analysis (ISA) results showed strong distinctions among microbial taxa in response to different treatments and in the two collection depths. Our results indicated a high efficiency of the monitored natural attenuation (MNA) for oil consumption in the tested mangrove sediments, revealing the potential of this strategy for environmental decontamination and suggesting that environmental and ecological factors may select for specific bacterial populations in distinct niches.
Introduction
Mangrove forests are known to store a large amount of organic carbon and therefore have the potential to substantially contribute to carbon emissions when exposed to high rates of pollution, degradation and/or deforestation (Hamilton and Friess, 2018). Among the numerous sources of pollution that can affect mangroves, oil spills are one of the most important concerns because they can occur at any stage of the oil production chain and result in extensive environmental and health damage (Santos et al., 2011). Although oil spills can directly affect almost any ecosystem, coastal areas such as mangroves are among the most threatened environments (Duke, 2016). Because of their ecotonal location and structural peculiarities, mangroves typically retain oil from spillages, concentrating hydrocarbons in their surroundings and compromising their physical, chemical and biological systems for long periods of time (Lewis et al., 2011; Duke, 2016).
Several physical and chemical remediation strategies have been used for oil decontamination, but considering their high financial and environmental costs, bioremediation approaches have become increasingly useful for this purpose (Xu and Lu, 2010). Bioremediation can be applied both in situ and ex situ using different strategies, such as bioattenuation, bioaugmentation and biostimulation. Bioattenuation, also known as monitored natural attenuation (MNA), is an in situ technique that consists of the contaminant degradation by natural physical, chemical and biological processes without any human intervention, through volatilization and degradation of the toxic compounds by the autochthonous microbiota. In general, it is a slow process and requires site monitoring over a long period (Khan et al., 2004; Yu et al., 2005). Bioaugmentation consists of the injection of microorganisms grown in the laboratory, autochthonous from the contaminated site, with known ability to degrade the target contaminant. It can be used in situ and ex situ, being particularly important for in situ treatments, when the autochthonous microbiota is not presented in sufficient abundance to degrade the contaminant efficiently (Desay et al., 2010). Biostimulation, in turn, is the process by which an intentional stimulation of autochthonous microbial communities takes place through the provision of nutrients and electron acceptors/donors, pH and temperature adjustment and aeration of the substrate. This technique can be performed in situ and ex situ, but depends on the existence of an autochthonous microbiota capable of degrading the target contaminant (Khan et al., 2004; Desay et al., 2010).
Bioremediation techniques have been successfully applied worldwide in environmental oil mitigation, such as in the oil spills in Prince William Sound, Alaska, in 1989 (Bragg et al., 1994) and in the Mexican Gulf in 2010 (Atlas, 2011), and they are promising strategies for cleaning up oil-contaminated mangrove sediments (Santos et al., 2011; Duke, 2016). However, in contrast to most soils, mangrove sediments are predominantly anaerobic, which should be considered when attempting to develop a bioremediation strategy (Ghizelini et al., 2012).
Some studies have examined the microbial degradation of pollutants to address questions related to details of the bioremediation process, such as transcriptional information and kinetic behavior associated with enzymes involved in pollutant degradation (Peixoto et al., 2011a; Thatoi et al., 2013). The results of these studies have provided significant insights for improving bioremediation processes. However, most studies analyzing the effect of hydrocarbon pollution on subtidal sediments have disregarded the differential distribution of bacteria throughout the depth profile, and sediment samples have generally been analyzed in bulk or sampling has been primarily focused in the upper layer (0–5 cm), where metabolism is primarily aerobic. Other assays, such as the characterization of syntrophic pollutant-degrading microorganisms and the detection of genes that are potentially involved in the bioremediation processes of microbial communities, are still of great importance. For oil bioremediation purposes, some bacterial groups have already been recognized for their ability to metabolize oil hydrocarbons, such as members of the genera Acinetobacter, Alcaligenes, Paenibacillus, and Pseudomonas (do Carmo et al., 2011; Kostka et al., 2011; Santos et al., 2015). Some genes associated with the outset of hydrocarbon metabolism have also been identified, such as alkB (encoding alkane monooxygenase) and ndo (encoding naphthalene dioxygenase), which are active under aerobic conditions to degrade alkanes and polycyclic aromatic hydrocarbons (PAHs), respectively (Beilen et al., 1994; Gomes et al., 2007), as well as assA (encoding alkylsuccinate synthase A) and bssA (encoding benzylsuccinate synthase A), which are active under anaerobic conditions to degrade alkanes and PAHs, respectively (Andrade et al., 2012; Netzer et al., 2013).
Mangroves are permanently vulnerable to potential oil contamination. Previous studies have shown that the microbiome of these ecosystems can be heterogeneously distributed throughout the mangrove (Peixoto et al., 2011b) and is affected by the concentration of oil (Santos et al., 2010, 2011; Fasanella et al., 2012), presenting great metabolic versatility with enzymatic potential for the degradation of hydrocarbons (Santos et al., 2010). The use of native microbial species or consortia for mangrove bioremediation can represent a promising and efficient approach for the recovery of these ecosystems. However, the deposition and adsorption of oil in the subsurface layers of mangrove sediments is a problem that current bioremediation approaches do little to help solve. Therefore, the utilization of a microbial consortium containing facultative anaerobic bacteria has been suggested as a potential strategy to overcome this limitation (Andrade et al., 2012).
In our study, we selected and assembled an oil degrading bacterial consortium containing local mangrove native aerobic and facultative anaerobic bacteria and developed an innovative in situ mesocosm system in a mangrove located in the state of Rio de Janeiro, Brazil. This innovative mesocosm system was simultaneously safe against environmental contamination and dynamic to generate a realistic system that included tidal influence. After a local and controlled oil spill simulation in the mesocosms, we evaluated: (i) the presence of different hydrocarbon degradation genes during the oil contamination and bioremediation processes; (ii) oil degradation in response to different bioremediation processes; (iii) differences in bacterial diversity patterns along the sediment depth at this site.
Materials and Methods
Experimental Design
Mesocosm Construction
A mesocosm experiment was developed (Figure 1 and Supplementary Figure S1) at the mangrove of Restinga da Marambaia, Rio de Janeiro, Brazil (23°03′18.7″S/43°34′19.7′′W; Permitted by SisGen AC72B83, according to the Brazilian legislation on access to biodiversity (Law 13,123/15 and Decree 8.772/16) to test bioremediation techniques against the MF (marine fuel) 380 oil (composed of mixtures of medium and heavy fractions of fuel oil, with paraffinic and aromatic hydrocarbons of different molecular weights). Aluminum cylindrical tubes (100 cm in length and 10 cm in diameter) were used as mesocosms (Figures 1A,B and Supplementary Figure S1) and vertically inserted into mangrove sediments. Twenty-four tubes were used in this study that were divided in four groups, one for each treatment, and arranged in a linear configuration, with six tubes per group. Each group was surrounded by oil absorption barriers, and the total experimental area was physically separated from the rest of the mangrove by oil contention barriers (Supplementary Figure S1). To ensure the efficiency of the barriers despite tidal hydrodynamics, these structures were affixed to iron brackets with a length of 4 m (calculated based on the maximum tidal range) using metallic rings to provide them vertical movement. This movement allowed them to follow water levels, improving the protection efficiency in the experimental area, improving protection efficiency in the experimental area.
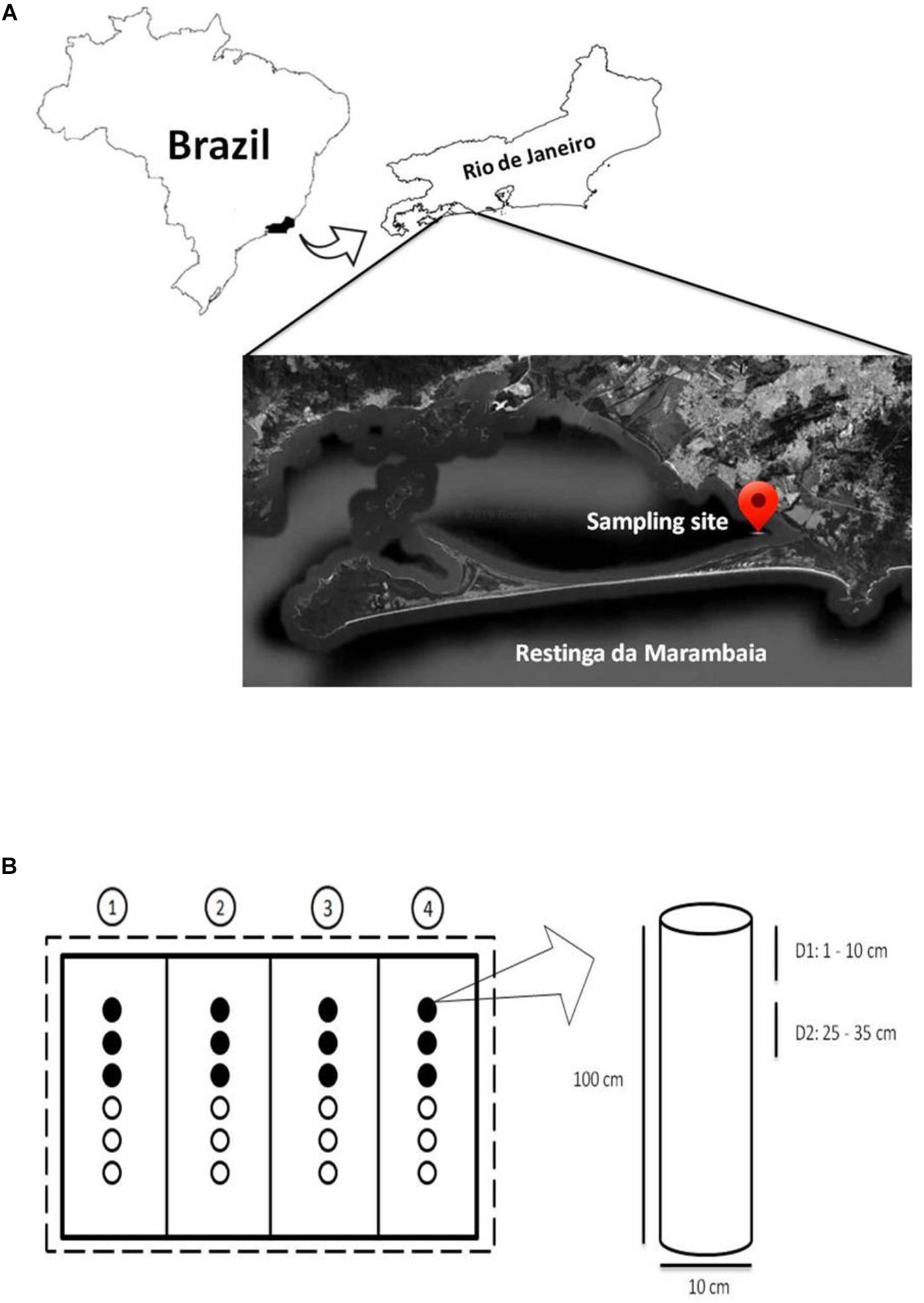
Figure 1. (A) Sampling location (modified from Google Earth – view from 72 km altitude). (B) Experimental design outline. Dashed line: contention barrier; solid lines: absorption barriers; circles: mesocosms. The numbers in circles represent the bioremediation treatments applied in each set of mesocosms: 1, Monitored Natural Bioattenuation; 2, Bioaugmentation; 3, Biostimulation; 4, Bioaugmentation + Biostimulation. Black and white circles represent samples collected after 45 and 90 days of bioremediation treatments application, respectively. The cylinder represents a mesocosm with the two collection depths: depth 1 (D1) 1–10 cm and depth 2 (D2) 25–35 cm. Not drawn to scale.
Bioremediation Strategies
The oil used in this study was the marine fuel MF-380, which has the following characteristics: density (0.9817), 12°API, viscosity of 5313 cP at 20°C, boiling point range from 174 to 750°C (26%, w/w residue), BTEX content of 1.56 mg.g–1 and 50 mg.g–1 PAHs. Four different bioremediation techniques were tested to mitigate the MF-380 oil spill simulation: Bioattenuation (BT), Bioaugmentation (BA), Biostimulation (BS) and Bioaugmentation + Biostimulation (BB). For the BA and BB strategies, a bacterial consortium was selected and assembled. This consortium was composed of native hydrocarbon-utilizing bacteria isolated under anaerobic and aerobic conditions. The aerobic bacteria used in this study were previously obtained by do Carmo et al. (2011) in the exactly same experimental area and have been classified at the molecular level as Acinetobacter sp., Alcaligenes sp., Azospirillum sp., Paenibacillus sp., Pseudomonas sp., Pseudomonas pseudoalcaligenes, and Pseudomonas stutzeri. The sequences from the aerobic bacteria are available at the NCBI Sequence Read Archive under the following accession numbers: HQ020607, HQ020582, HQ020562, HQ020564, HQ020609, HQ020568, and HQ020576, respectively. The anaerobic strains used in this study were isolated from mangrove sediments at the same sampling site (23°03′18.7′′S/43°34′19.7′′W). Sediment samples were collected at a depth of 15–75 cm in the syzygy tidal zone with a cylindrical sampler (7.5 cm in diameter and 75 cm in length). Subsequently, the these samples were blended, and 12.5 g was homogenized in 125 mL of a sterile reducing buffer in an anaerobic culture system. Serial dilutions (10–1 to 10–4) were generated and 100 μL aliquots were used to inoculate Bushnell-Hass broth (MgSO4, CaCl2, KH2PO4, K2HPO4, (NH4)(NO3), and FeCl3) that were cultured in triplicate under the following conditions: pH 6, 7, and 8, and salinity of 0.5 and 2%. The bacterial cultures were incubated at 27°C. All media were supplemented with MF 380 oil as the sole carbon source. After 30 days, these cultures were inoculated into Petri dishes containing marine agar medium (Marine Agar Zobell 2216, Himedia Laboratories, Mumbai, India) and incubated at 27°C. The cultured strains were selected and identified using molecular techniques before becoming part of the consortia. For this step, genomic DNA was extracted using a Wizard® Genomic DNA Purification Kit (Promega, Madison, United States) following the manufacturer’s instructions. The resulting DNA was used for PCR targeting the 16S rRNA gene with the primers 27F (AGA GTT TGA TCA TGC CTC AG), and 1492R (GTT TAC CTT GTT ACG ACT T). The PCR products were purified, sequenced, analyzed using the Ribosomal Database Project (RDP) (Cole et al., 2009) and compared with sequences deposited at the National Center for Biotechnology Information (NCBI)1 using the BLAST tool (Basic Local Alignment Search Tool) for taxonomic classification. Antagonistic tests were performed between all strains according to Giambiagi-Marval et al. (1990). These tests are important to identify antagonistic strains that should not be used simultaneously in a given consortium. Strains that did not present antagonism against each other were selected to be part of the consortium. The number of cells of each individual strain was estimated by counting the number of colony forming units (CFUs) over time using serial dilutions (103–107) in triplicate. For each dilution, 200 μL was spread onto Petri dishes containing marine Agar, and the resulting colonies were counted using a colony counter. Subsequently, the aerobic and anaerobic strains were inoculated into 300 mL of Luria–Bertani broth and Baar broth medium (Na3C6H5O7, MgSO4, CaSO4, NH4Cl, K2HPO4, sodium lactate, yeast extract and Fe(NH4)2(SO4)2), respectively, and were incubated for 48 h. The aerobic strains were incubated at 150 rpm, whereas the anaerobic strains were incubated without rotation to avoid aeration of the medium. The bacterial cultures were centrifuged at 5000 rpm for 30 min, and the resulting cell pellets were resuspended in 60 mL of NaCl 0.9% to obtain a density of 107 viable cells mL–1. Equal volumes of each bacterial isolate suspension were mixed, yielding a consortium containing 107 cells mL–1 of each selected strain. To evaluate the bioremediation strategies, each tube of the mesocosm was vertically contaminated with 90 mL of MF 380 oil (1.4%) using a syringe manufactured specifically for this purpose (Supplementary Figure S2A). The syringe had a piston with a length of 1.20 m and a cylinder of 1.10 m, with a capacity of 90 mL. The widest and narrowest points of the syringe were 10 and 4 mm across, respectively. To ensure that oil would be equally distributed in all sediment layers, we developed a isosceles triangle tool that had the largest intersection lines measuring 1.35 m, a base of 0.65 m, and a height of 1.30 m. To apply the oil, the triangle tool was placed above each mesocosm, and the syringe containing the oil was inserted into the metallic tubes until the piston fit perfectly to the upper vertex of the triangle tool. With the piston in the upper triangle vertex, the syringe was slowly removed from the mesocosm (Supplementary Figure S2B).
Four months after oil contamination, the bioremediation strategies were applied. In each tube, 90 mL of each treatment was vertically applied using the same strategy used for oil contamination. The treatments consisted of the following strategies: (1) Monitored Natural Bioatenuation: the application of no treatment to allow the catabolic activity of local microorganisms, with no other stimulation other than those from the natural environment and monitoring; (2) Bioaugmentation: the application of 45 mL of the bacterial oil-degrading consortium (107 cell/mL) under aerobic and anaerobic conditions plus 45 mL of 0.9% sterile saline; (3) Biostimulation: the application of 45 mL of a nitrogen, phosphorous and potassium (NPK) solution in a 18:2:10 (w/w) proportion (Ramsay et al., 2010) plus 45 mL of 0.9% sterile saline; (4) Bioaugmentation + Biostimulation: 45 mL of the bacterial oil-degrading consortium under aerobic and anaerobic conditions (107 cell/mL) plus 45 mL of NPK in a 18:2:10 (w/w) proportion. After 45 and 90 days of the application of the different treatments, three tubes were collected from each treatment and processed. The sediment columns were sectioned in two depth fractions, 1–10 cm (depth 1; D1) and 25–35 cm (depth 2; D2), and each fraction was manually homogenized and used for further analyses.
Physical and Chemical Characterization of Sediment
Sediment samples were characterized for their granulometry and carbon and nitrogen contents. Sediment granulometry was determined using the aerometer method after chemical dispersion (EMBRAPA, 2011). Carbon content was evaluated via the oxidation of wet organic matter with potassium dichromate in sulfuric medium using the heat released from the sulfuric acid as a heat source. The excess dichromate after oxidation was titrated with a standard solution of ammoniacal ferrous sulfate (EMBRAPA, 2011). To determine the total nitrogen content, N was converted to (NH4)2SO4 by oxidation with a mixture of CuSO4, H2SO4 and Na2SO4 or K2SO4. Subsequently, the (NH4)2SO4 was placed in an alkaline medium in a diffusion chamber, where it released ammonia that complexed with a boric acid solution containing a mixed indicator and then was measured by acidimetry (with H2SO4 or HCl) (EMBRAPA, 2011).
Evaluation of Hydrocarbon Degradation
The efficiency of the bioremediation strategies was tested by evaluating hydrocarbon degradation. The saturated hydrocarbon content (n-alkanes) was evaluated by gas chromatography and flame ionization detection (GC/FID) following the United States EPA Standard Methods for the Examination of Water and Wastewater (846-8015). The detection of polyaromatic hydrocarbons was performed according to the United States EPA Standard Method 846-8270, which describes the use of gas chromatography for semi-volatile organic compounds and mass spectrometry (GC/MS). Hydrocarbon analysis was performed at Oceanus Hidroquímica facility2.
DNA Extraction
Total DNA was extracted from sediment samples (0.5 g) using a Power SoilTM DNA Isolation kit (MoBioTM, United States) following the manufacturer’s instructions. The DNA was quantified using a fluorometer (QuBit® Fluorometric Quantification, Life TechnologiesTM, United States) and visualized on a 1% (w/v) agarose gel with SYBR-Safe DNA gel stain (Invitrogen, Life TechnologiesTM, United States) to assess its integrity prior to PCR and qPCR analysis.
qPCR for Hydrocarbon-Degrading Genes
qPCR was performed to assay the hydrocarbon-degrading alkB, ndo, assA, bssA, and 16S rRNA genes in each sample. qPCR was performed in a 20-μL reaction with 2 × Master Mix (Fermentas), 5 μM of each primer and 2–5 ng of template. In the ndo reaction, 1% of dimethyl sulfoxide was added. qPCR was performed under previously described conditions for each set of primers (Supplementary Table S1). At the end of each reaction, we obtained a denaturation curve by heating the products at 95°C for 15 s, followed by a cooling step of 60°C for 1 min and subsequent heating at 95°C for 15 s. All amplifications were performed in duplicate using a 7500 Real-Time PCR System (Applied Biosystems, Germany).
Bacterial Community Structure Analysis
Bacterial communities were assessed by high-throughput sequencing targeting the V4 variable region of the 16S rRNA gene using the primers 515F (5′-GTG CCA GCM GCC GCG GTA A 3′) and 806R (5′ GGA CTA CHV GGG TWT CTA AT 3′) (Caporaso et al., 2012) with specific adapters. The following thermocycling conditions were used: 94°C for 3 min, followed by 30 cycles of 94°C for 30 s, 53°C for 40 s, and 72°C for 1 min, with a final elongation step at 72°C for 5 min. Equimolar amounts of the PCR products from the 48 samples were combined and sequenced via Ion Torrent Personal Genome Machine (PGM) technology following the manufacturer’s guidelines at the MrDNA facility3. Raw sequences were processed using Mothur v.1.36.1 (Schloss et al., 2009). To reduce sequencing error, all sequences that failed to comply with any of the following criteria were excluded: average quality lower than 20, length less than 200 bases, presence of ambiguities, more than 1 nucleotide mismatch to the primer and/or barcodes, or homopolymers longer than 8 nucleotides. The remaining high quality sequences were aligned using the Silva reference database (Quast et al., 2013), and chimeras that were detected using Uchime (Edgar et al., 2011) were eliminated. The resulting alignment file was used as input to construct a distance matrix and to cluster the sequences into operational taxonomic units (OTUs), with a cutoff of 3% dissimilarity. The clusters were used to calculate the species-richness estimators and Shannon diversity index. The samples were then randomly normalized to the same number of sequences. For taxonomic assignments, the 16S rRNA gene sequences were classified using the RDP database with a 50% confidence threshold (Cole et al., 2009). For our analysis, we only considered the OTUs that were represented by least three sequences. The sequences from the 48 samples are available at the NCBI Sequence Read Archive under the accession numbers SRR3984611–SRR3984659.
Data Analysis
For analytical purposes, we previously tested the distribution of the data based on assumptions of normality and homoscedasticity. When the required assumptions for parametric tests were not met, equivalent non-parametric tests were used. To compare the number of copies of the alkB, ndo, and 16S rRNA genes among the bioremediation treatments, we performed one-way analysis of variance (ANOVA). To compare the number of alkB, ndo, and 16S rRNA genes between the two sediment layers, we performed Student’s t-tests, whereas we performed ANOVA to make comparisons between depths. To evaluate the efficiency of the four bioremediation treatments with respect to total hydrocarbon and alkane degradation, we performed Kruskal-Wallis tests. To evaluate the efficiency of total hydrocarbon and alkane degradation between the two sediment layers, we performed Wilcoxon tests. We conducted these analyses in the R environment (R Core Team, 2015) using the car (Fox and Weisberg, 2011), beeswarm (Eklund, 2013), and PMCMR (Pohlert, 2014) packages. To examine differences among abiotic factors, we performed two-way ANOVA using PAST3 (Hammer et al., 2001). To evaluate samples based on their microbial compositions, we normalized the number of sequences prior to clustering. After that, the singletons and doubletons were removed, which produced samples with slight variations in the number of sequences. Most samples were close to the average of 9406 sequences (the average number of sequences per sample was, respectively, 9300, 9547, 9665, and 9204 for BA, BB, BE, and BT treatments). This shows that there was no strong sequence number bias in a specific treatment, which validates the comparison of the OTU number between the samplesnormalized the number of sequences before clustering. After that, singletons and doubletons were removed, which produced samples with small variations in the number of sequences. Then, we performed a Non-metric Multidimensional Scaling (NMDS) analysis using the Bray-Curtis similarity index, which was calculated as a function of the OTU distribution among the samples using the PC-ORD 6.04 Multivariate Analysis of Ecological Data software (McCune and Mefford, 2011). An outlier sample for the Bioaugmentation + Biostimulation (BB) treatment was retained from the NMDS analysis because it was compressing all of the other samples. Differences in community structure among treatments were tested with a two-way ANOSIM test using PAST3, and a Mantel’s test was performed to test the influence of abiotic factors on community structure with PC-ORD 6.04. To assess whether the treatments and depths had an effect on specific OTUs, we performed blocked indicator species analysis (ISA) (Dufrene and Legendre, 1997). In addition, we performed correlation analysis among the obtained OTUs and abiotic factors. For all tests, we assigned the significance level as α = 0.05.
Results
Composition of the Bacterial Consortium
Fifteen anaerobic bacterial strains were isolated in this study, but only two, from the Desulfovibrio genus, showed satisfactory growth in the presence of the other consortium strains in antagonistic tests (data not shown). Among the isolated aerobic strains, only four did not show antagonistic activity toward each other. Thus, the hydrocarbon-utilizing bacterial consortium used in the treatments with bioaugmentation conditions was composed of one strain of each of the following genera: Acinetobacter (HQ020607), Azospirillum (HQ020562), Paenibacillus (HQ020564), and Pseudomonas (HQ020609), and two isolated strains from the genus Desulfovibrio (SAMN06627399).
Sediment Characterization
Regarding the sediment granulometry, carbon and nitrogen composition/distribution, no differences were observed between treatments or over time (two-way ANOVA, p > 0.05), indicating the same physical-chemical conditions across the entire mesocosm. The carbon content was higher in the superficial layer (two-way ANOVA, F = 6.07, p = 0.01), but the nitrogen content and the C/N ratio did not differ significantly between the two depths. The granulometry of the sediment remained constant throughout the experiment, with no variations observed with respect to treatment or depth.
Efficiency of Bioremediation Strategies
The degradation percentage rate of total petroleum hydrocarbon (TPH) and alkane did not differ among the bioremediation treatments (Bioattenuation, Bioaugmentation, Biostimulation and Bioaugmentation + Biostimulation) (TPH – Kruskal–Wallis, p = 0.35; alkane – Kruskal–Wallis, p = 0.30). However, there were differences in degradation rates between the sediment layers (TPH – Wilcoxon, p = 0.03, Figure 2A; alkane – Wilcoxon test, p < 0.01, Figure 2B) when they were considered as a sum of the degradation percentage of all the treatments. The degradation rates were higher in the superficial sediment (1–10 cm) (TPH – 45.6 ± 41.8%; alkane – 68.1 ± 46.1%) than in the deeper layer (25–35 cm) (TPH – 9.2 ± 29.2%; alkane – 10.0 ± 31.6%) for both TPH and alkane.
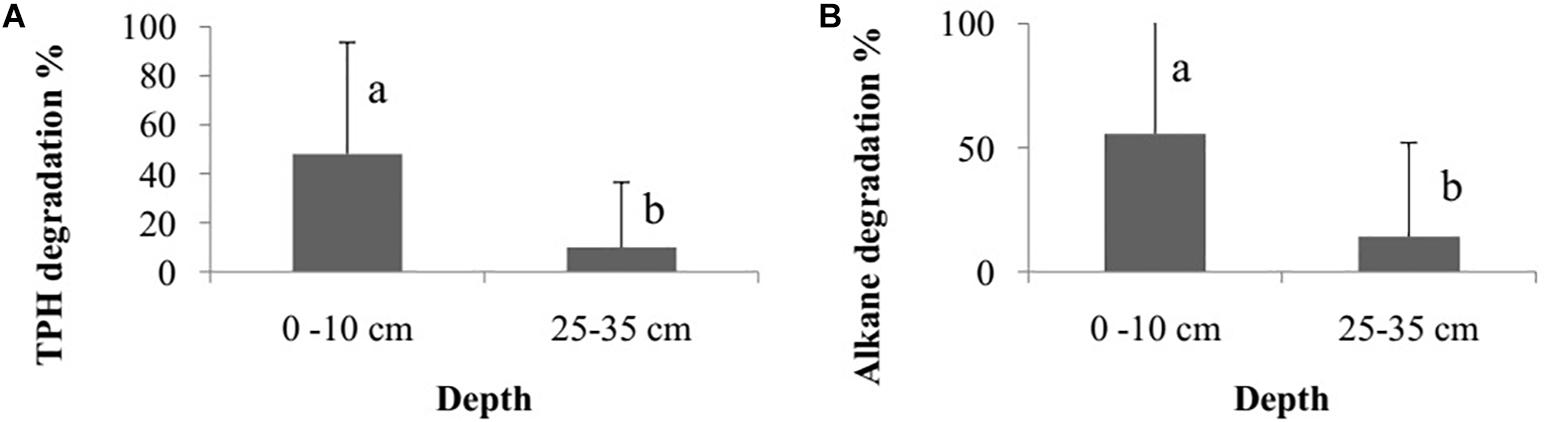
Figure 2. Degradation percentage rate of total petroleum hydrocarbon – TPH (A) and alkane (B) between different sediment layers (1–10 cm and 25–35 cm). The bars represent the arithmetic mean of all treatments for each depth. The error bars represent the standard deviation. Different letters indicate significant differences between depths.
Quantification of Hydrocarbon-Degrading Genes
The genes assA and bssA, related to anaerobic hydrocarbon degradation, were not detected in our samples. Therefore, we specifically quantified the aerobic genes alkB and ndo. The quantification of hydrocarbon-degrading genes did not differ among the treatments Bioattenuation, Bioaugmentation, Biostimulation and Bioaugmentation + Biostimulation for alkB (one-way ANOVA, p = 0.24; Figure 3A) or ndo (one-way ANOVA, p = 0.14; Figure 3B). However, the number of copies of alkB (Student’s t-test, p = 0.01; Figure 3C) and ndo (Student’s t-test, p = 0.01; Figure 3D) differed between the two sediment layers. The inferior layer had significantly more copies of alkB and ndo (alkB, 1.1E2 ± 6.3E1 copies/ng DNA; ndo, 4.4E4 ± 1.2E4 copies/ng DNA) than the superficial layer (alkB, 6.2E1 ± 2.0E1 copies/ng DNA; ndo, 3.1E4 ± 1.0E4 copies/ng DNA). We did not detect differences for either gene for the different collection times.
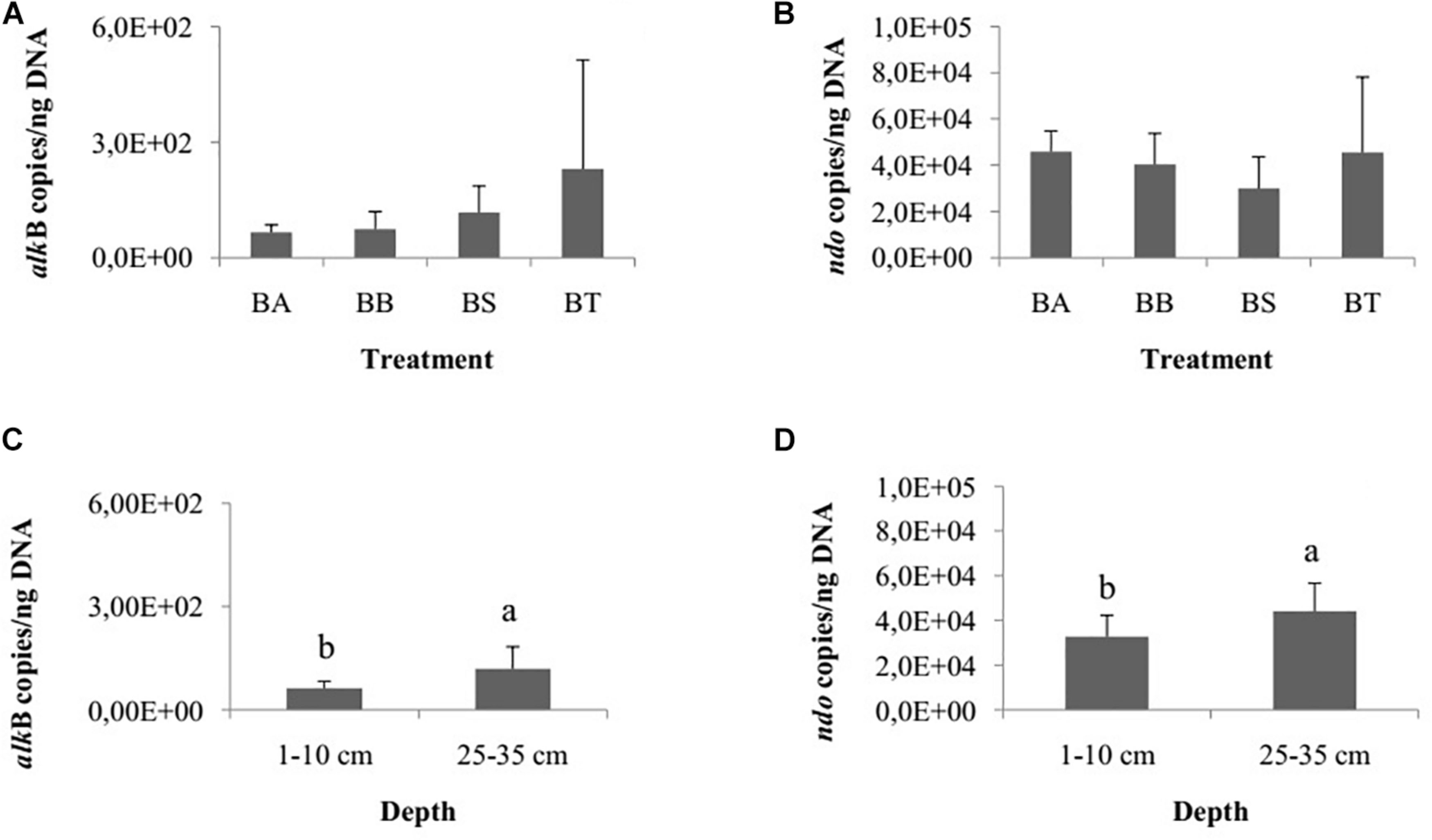
Figure 3. Number of copies of alkB (A) and ndo (B) genes/ng DNA among the bioremediation treatments (BA, Bioaugmentation; BB, Biostimulation + Bioaugmentation; BS, Biostimulation; BT, Bioattenuation). The error bars represent the standard deviation. Different letters indicate significant differences between depths. Number of copies of alkB (C) and ndo (D) genes between the different sediment layers (1–10 cm and 25–35 cm). The bars represent the arithmetic mean of all treatments for each depth. The error bars represent the standard deviation. Different letters indicate significant differences between depths.
Structure of Bacterial Communities
The number of 16S rRNA gene copies did not differ among treatments (one-way ANOVA, p = 0.62; Figure 4A) and collection times (ANOVA, p = 0.08), but they did differ between the two sediment layers, with a higher number of copies observed in the 25–35 cm layer (Wilcoxon test, p < 0.01; Figure 4B).
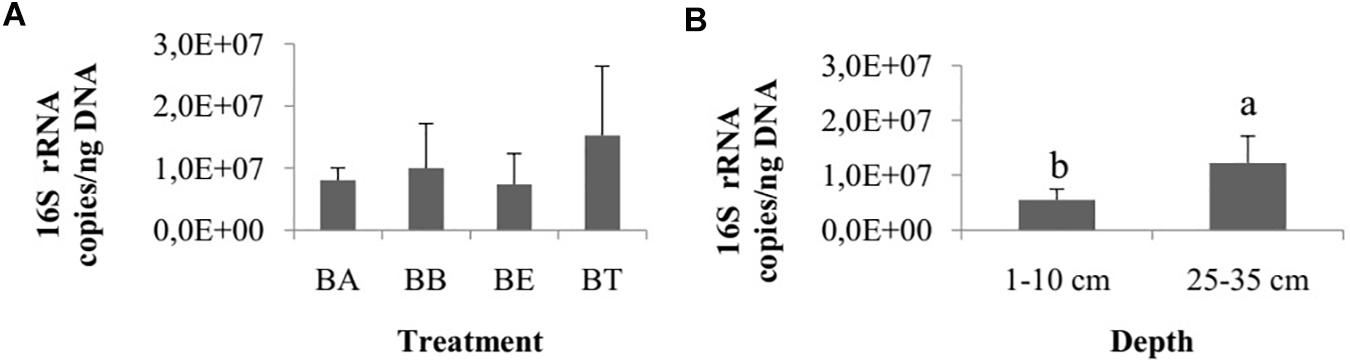
Figure 4. (A) Number of copies of 16S rRNA genes among the four bioremediation treatments (BA, Bioaugmentation; BB, Biostimulation + Bioaugmentation; BS, Biostimulation; BT – Bioattenuation.). The error bars represent the standard deviation. (B) Number of copies of 16S rRNA genes between different sediment layers. The bars represent the arithmetic mean of all treatments for each depth. The error bars represent the standard deviation. Different letters indicate significant differences.
From the four treatments, a total of 8000 sequences were obtained after quality trimming and normalization, which clustered into 11,021 OTUs that were classified into 26 phyla, 58 classes, 100 orders, 200 families, and 402 different genera. The OTU structure was significantly influenced by the bioremediation treatments (two-way ANOSIM, p < 0.01) and sediment depths (two-way ANOSIM, p < 0.01) but was not influenced by collection time (two-way ANOSIM, p = 0.50). For the two assayed depths, the OTU number was higher in the 25–35 cm layer (two-way ANOSIM, p < 0.01). In this layer, the BS treatment resulted in higher carbon and nitrogen contents (Mantel test, p < 0.05), whereas no other abiotic factors stood out for any other treatment (Mantel test, p = 0.373) (Figure 5).
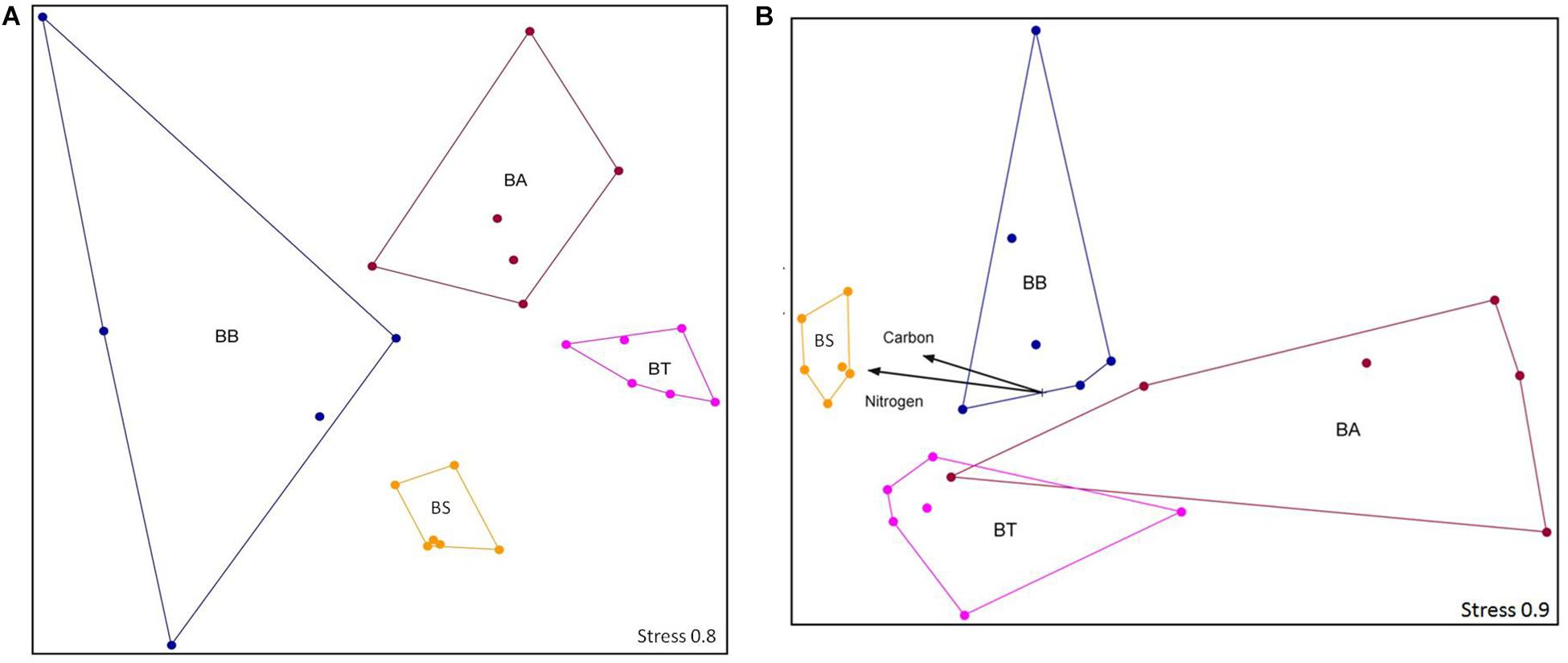
Figure 5. NMS ordination of the sequencing data at the OTU level at depths 1–10 cm (A) and 25–35 cm (B). Stress is presented on a scale from 1 to 100. BA, Bioaugmentation; BB, Biostimulation + Bioaugmentation; BS, Biostimulation; BT, Bioattenuation.
The community composition at the phylum and order levels did not differ among the treatments, whereas differences were detected between the two sediment layers in all the treatments (Figure 6). The most abundant phyla among all treatments and depths were Proteobacteria, Firmicutes and Bacteroidetes, while the most abundant orders were Gammaproteobacteria OIS∗, Flavobacteriales and Clostridiales. A large number of unclassified phyla and orders were also detected. It is worth noting that the most representative fraction of the bacterial community was unclassified at the order level.
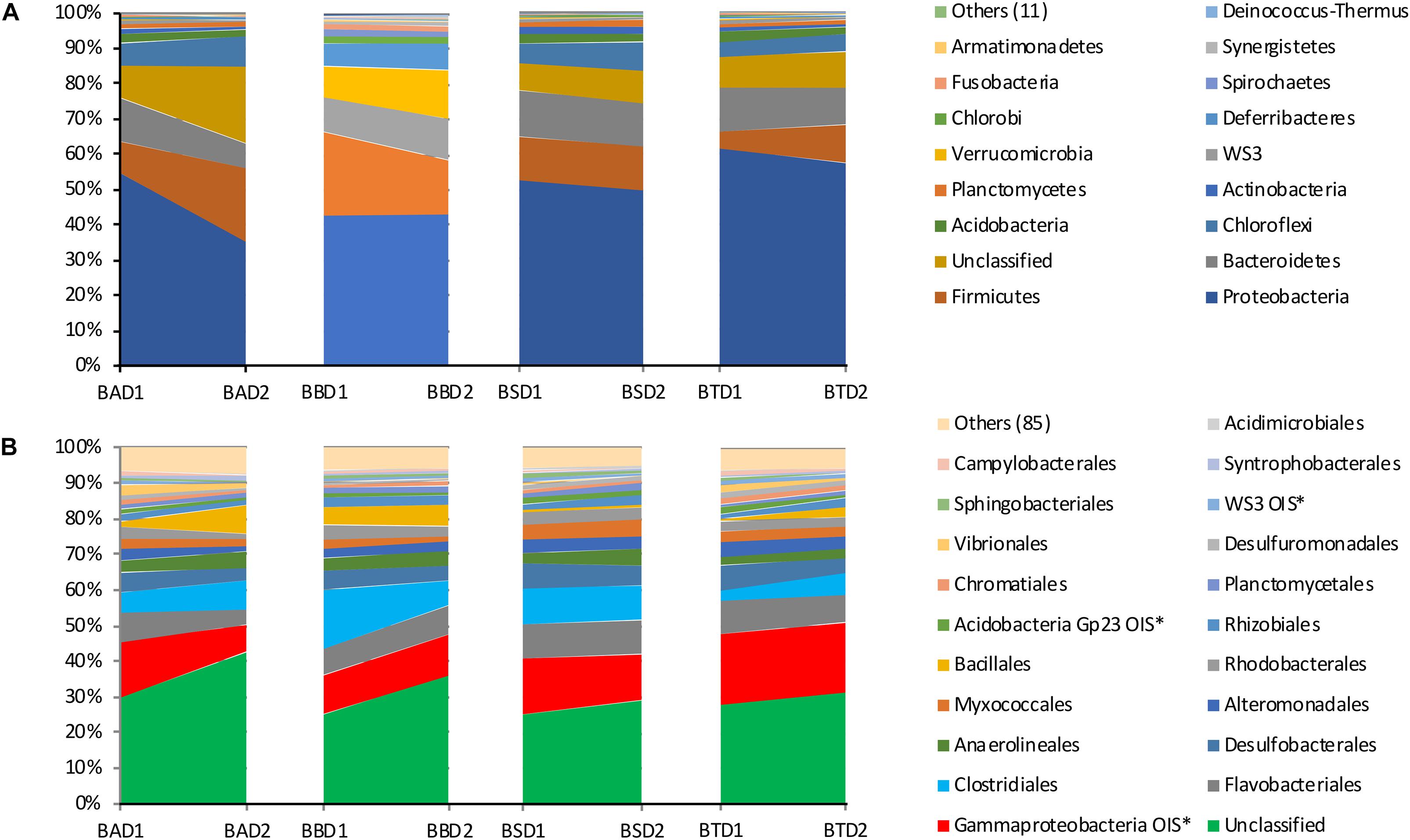
Figure 6. Relative abundance of the bacterial phyla (A) and orders (B) found in each treatment at each collection depth. BA, bioaugmentation; BB, biostimulation + bioaugmentation; BS, biostimulation; BT, bioattenuation; D1, depth 1–10 cm; D2, depth 25–35 cm.
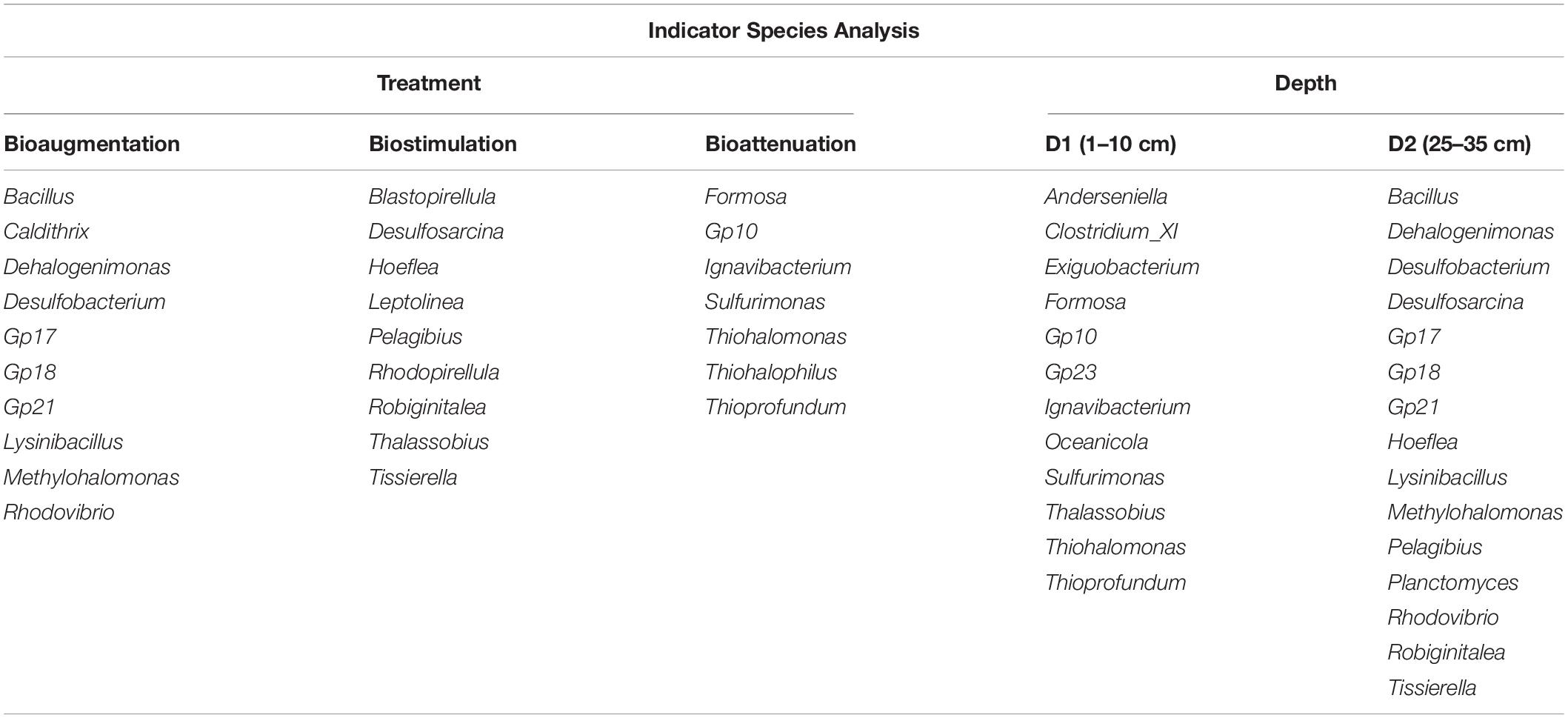
The bacterial α-diversity did not vary among significantly samples and depths, as revealed by the observed Shannon H diversity index values and the Chao and Ace richness index values. However, differences in bacterial community composition were revealed by ISA, the results of which showed that 26 different genera were significantly associated with specific bioremediation conditions, 10 of which were related to the BA conditions, nine to the BS conditions and seven to the BT conditions (Table 1). ISA also identified 12 genera that were significantly associated with the two assayed depths (1 to 10 and 15 to 25–35 cm) (Table 1).
The analysis showed that the distribution of some of the 20 most abundant OTUs correlated with some abiotic factors at D1 and D2. At D1, OTUs from the genus Desulfopila (phylum Proteobacteria and class Deltaproteobacteria) correlated positively with nitrogen content (R = 0.52, p < 0.01) and negatively with alkane (R = -0.41, p = 0.04). At D2, OTUs from the genera Actibacter (phylum Bacteroidetes and class Flavobacteriia) (R = 0.68, p < 0.01), Gp23 (phylum Acidobacteria and class Acidobacteria_Gp23) (R = 0.64, p < 0.01) and Hailea (phylum Proteobacteria and class Gammaproteobacteria) (R = 0.60, p < 0.01) and from the family Desulfobulbaceae (phylum Proteobacteria and class Deltaproteobateria) (R = 0.44, p = 0.02) and the order Myxococcales (phylum Proteobacteria and class Deltaproteobacteria) (R = 0.58, p < 0.01) correlated positively with nitrogen content.
Discussion
Hydrocarbon Degradation Efficiency of the Bioremediation Strategies
The quantified hydrocarbon rates differed between the two sediment layers, with higher degradation percentages observed at the 1–10 cm depth (D1). This result is probably related to a more anoxic condition on the deeper layers compared with the superficial ones. Generally, hydrocarbon consumption is higher and faster in the presence of oxygen than in its absence (Grishchenkova et al., 2000; Salminen et al., 2004). In a study designed with nitrate-reducing bacteria, the authors identified that, under aerobic conditions (a 10-day experiment), bacteria degraded 20–25% of the hydrocarbon provided, including up to 90–95% of all alkanes analyzed (n-C10–C35). Under anaerobic conditions (a 50-day experiment), the same microorganisms degraded 15–18% of the hydrocarbons, including alkanes (20–25%) and polycyclic aromatic hydrocarbons (15–18%) (Grishchenkova et al., 2000). Other research designed microcosms to study the hydrocarbon consumption by bacterial in an oil contaminated site undergoing a bioattenuation process. It was detected that, in the aerobic microcosms, 31% of the initial oil was removed during a 3-month incubation, while, in the anaerobic ones, on average 44% of the initial oil was removed during a 12-month anaerobic incubation, with removal of some alkanes (n-C11–C15) (Salminen et al., 2004). Despite the difference in depths, the observed oil degradation rates did not differ among the bioremediation treatments. Similar results were also observed in a previous study that used creosote as the contaminant in microcosms, where the degradation rates did not differ among the bioremediation strategies after 6 months of contamination (Simarro et al., 2013). In addition, similar results were observed in a study using crude oil, where the total petroleum hydrocarbon degradation observed using a combined bioaugmentation and biostimulation strategy did not differ from that with bioattenuation after 112 days of contamination (Wu et al., 2017). However, increases in the hydrocarbon degradation rates have also been reported in other sites over short periods of time using bioaugmentation and biostimulation strategies rather than bioattenuation (Yu et al., 2005; Meyer et al., 2014; Müeller et al., 2017). Differences in the obtained results may be explained by studies describing limitations with respect to the biostimulation and bioaugmentation strategies (Boopathy, 2000; Bouchez et al., 2000; Gough and Nielsen, 2016). Regarding biostimulation, the primary challenges are to identify indigenous microbial communities that are capable of degrading target contaminants and to deliver additives in a manner that allows them to be readily available to microorganisms (Boopathy, 2000; Adams et al., 2015). Moreover, at some sites and under some conditions, the addition of nutrients may also promote the growth of microorganisms that can outcompete the native contaminant-degrading organisms (Adams et al., 2015). With respect to the bioaugmentation strategy, the ability of microbes to survive over time depends on multiples variables, such as their attachment to surfaces, substrate availability, predator/prey interactions, competition, and other components that can be associated with the site, environmental conditions and the selected microbial consortia (Bouchez et al., 2000; Gough and Nielsen, 2016). Additionally, it has been suggested that the capacity of soil microbial communities to degrade hydrocarbons depends on their previous exposure to the contaminant (Sánchez et al., 2004; Johnsen and Karlson, 2005). Given that our studied area is a pristine mangrove, with no previous oil contamination reported, our results corroborate the thesis that previous exposure of microorganisms to oil is not needed to stimulate its degradation.
As we detected high levels of hydrocarbon degradation for all treatments and found no differences of degradation rates among them all, our results suggest a high efficiency of the natural bioattenuation for oil consumption in the studied mangrove sediments, revealing the potential of the bioattenuation for environmental decontamination for this site.
Besides this, the presence of the genes alkB and ndo in all treatments and depths reinforces the idea that native microbial communities have the potential to degrade petroleum hydrocarbons. Our results showed that the number of alkB and ndo gene copies did not differ among the bioremediation treatments and collection times, although they were detected in higher abundances in the deeper sediment layer. This is a noteworthy finding, given that these genes are associated with aerobic hydrocarbon degradation. As, regarding the evaluated paremeters, the carbon content was the only one that differed between sediment layers, with higher values observed in the superficial layers, we suggest that other environmental conditions, such as dissolved oxygen, pH, temperature and the concentrations of other nutrient may affect the abundances of microorganisms containing these genes in mangrove superficial sediment layers. Furthermore, AlkB-encoding genes are widespread in nature, and although they share considerable sequence homology, the alkB gene sequence varies greatly within bacteria harboring this gene (Kuhn et al., 2009; Jurelevicius et al., 2013; Viggor et al., 2015). The same phenomenon has been observed for the ndo gene (Larkin et al., 1999; Kasai et al., 2003; Gomes et al., 2007). Thus, the design of broad-ranging alkB and ndo primers is a challenge. Despite this difficulty and the existence of other mechanisms of aerobic alkane and PHA degradation, degradation routes that involve the alkB and ndo gene products have been investigated in great detail (Bellinaso et al., 2003; Kloos et al., 2006; Gomes et al., 2007; Flocco et al., 2009; Touvora et al., 2016; Long et al., 2017) and are some of the most commonly studied routes of alkane and PHA degradation in natural environments.
We could not detect the presence of the assA and bssA genes in any of our samples. Nevertheless, this does not mean that anaerobic hydrocarbon-degrading microorganisms are not present in the studied area. In fact, hydrocarbon degradation was detected in D2, a sediment layer considered to be anaerobic, which suggest the presence of anaerobic hydrocarbon degraders in the studied mangrove. Our failure to detect the target anaerobic genes may stem from the fact that microorganisms containing these genes are present at such low abundances in this mangrove that they could not be detected by the standard PCR protocol used. Another possible explanation is that anaerobic hydrocarbon degraders may possess variants of ass/bss sequences, lacking homology to our set of primers. Indigenous microbial communities may also preferentially use other alkane and aromatic hydrocarbon degradation routes catalyzed by enzymes that are not encoded by the target genes assayed with the selected primers, such as carboxylation reactions (Andrade et al., 2012).
To gain a better comprehension of the dynamics of hydrocarbon degradation in mangrove sediments, we suggest the use of more than one set of primers for each of the target genes and that other environmental conditions that may affect microbial communities be investigated, which could improve the detection of each gene and expand knowledge of microorganisms with the potential to degrade hydrocarbon compounds.
Bacterial Community Structure
The 16S rRNA gene copy number did not differ among the treatments and collection times, whereas they did differ between the assayed depths. In contrast, the OTU number and composition were different for the various treatments and sediment stratum layers, with higher richness observed in the deeper layer. Although this is not a common pattern with respect to soil microbiology, where superficial soil layers can generally be up to one-third more diverse than deeper ones (Fierer et al., 2003; Lamontagne et al., 2003; Eilers et al., 2012; Li et al., 2012; Mendes and Tsai, 2014; Raynaud and Nunan, 2014), other studies (Li et al., 2015) have already identified this pattern in soil, with more diverse bacterial phyla associated with sediments of deep soil layers. The data ordination based on the relative abundances of bacterial groups in each treatment showed an effect of the bioremediation strategies on bacterial communities at the OTU level. Additionally, although the carbon and nitrogen contents and the C/N ratio did not differ among the treatments, and the C content was higher in D1, the bacterial communities in layer D2 from the BS treatment were associated with these factors. Chu et al. (2016) tested the dissimilarity between bacterial communities from surface and subsurface sediment layers and observed that the total carbon content and carbon to nitrogen ratio were good predictors of the microbial community distribution. Additionally, as some nutrients can be primarily retained in the soil surface, the introduction of carbon and nitrogen into the deeper sediment layers can stimulate microbial communities and promote changes in their structure, as noticed for bioremediation strategies involving nitrogen addition (Ramsay et al., 2010; Simarro et al., 2013) and across sediment depths (Mueller et al., 2015), as well as for the introduction of carbon into soil environments (Santos et al., 2011; Li et al., 2012; Simarro et al., 2013).
For all the treatments and depths, the most abundant phyla were Proteobacteria, Firmicutes and Bacteroidetes, with Gammaproteobacteria, Flavobacteriales and Clostridiales being the most common orders. Generally, these are very common and most abundant taxa in soil environments (Fierer et al., 2009; Eilers et al., 2012; Mueller et al., 2015; Nogueira et al., 2015). Furthermore, these taxa harbor several biological families and genera containing hydrocarbon-degrader microorganisms (Jaekel et al., 2013; Berdugo-Clavijo and Gieg, 2014), which reveals the potential of the mangrove bacteria to degrade these contaminants.
ISA revealed strong differences in the observed taxa in response to the different treatments and between the D1 and D2 layers. Some of the detected genera in the D1 layer, such as Clostridium and Exiguobacterium, are commonly detected in environments with extreme conditions, with additional detected genera including the sulfur-utilizing bacteria Thioprofundum and Sulfurimonas (Mori et al., 2011; Han and Perner, 2016) and the genera Formosa, Ignavibacterium, Oceanicola, and Thalassobius. In the 25–35 cm layer, the detected groups included the spore-forming bacterial genera Bacillus and Lysinibacillus; Desulfobacterium, and Desulfosarcina, which are associated with sulfur metabolism; Dehalogenimonas, which has the ability to reductively dehalogenate chlorinated alkanes (Moe et al., 2009); the methylotrophic bacterium Methylohalomonas (Sorokin et al., 2007); and Hoeflea, Pelagibius, Rhodovibrio, and Tissierella. Some of these genera, such as Bacillus, Clostridium, Dehalogenimonas, Desulfobacterium, Desulfosarcina, and Tissierella are also associated with hydrocarbon degradation (Jaekel et al., 2013; Berdugo-Clavijo and Gieg, 2014). This result, in addition to the higher abundance of OUT at D2, reveals the potential of deeper sediments microorganisms to degrade these contaminants. Besides, we detected taxa that were positively correlated to the nitrogen content at different depths. The genus Desulfopila was detected at D1, while the taxa detected at D2 included the genera Actibacter, Gp23 and Hailea; the family Desulfobulbaceae; and order Myxococcales, some members of which also have hydrocarbon degradation functions, such as members of the family Desulfobulbaceae (Laban et al., 2015).
Investigations to better understand the dynamics of bioremediation are crucial in the design of efficient, less costly and environmentally sound cleaning strategies. In this study, we describe the use of in situ mesocosms to study the effects of bioremediation strategies for petroleum hydrocarbons to evaluate changes in the microbial community during the process and assess their potential for oil degradation. We believe that closer interactions between bench and field studies would be beneficial by contributing to a better scientific understanding of microbial processes and capabilities in specific areas. Such knowledge can contribute to the development of efficient bioremediation processes at different locations.
Data Availability
The datasets generated for this study can be accessed from NCBI Sequence Read Archive, SRR3984611–SRR3984659.
Author Contributions
LM, EM, RP, and AR conceived and designed the experiments. LM, JP, and EM performed the experiments. LM, DL, CR, RP, and AR analyzed the data. JP, RP, and AR contributed to the reagents, materials, and analysis tools. LM, CR, RP, and AR wrote the manuscript.
Funding
This work was supported in part by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior, Brasil (CAPES) (Grant code 001), by grants from Fundação de Amparo à Pesquisa do Estado do Rio de Janeiro (FAPERJ), Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), and Petrobras (Grant number 0050.0055450.09.9). The funder had the following involvement with the study: provided the oil sample used in this study and funds to purchase reagents, consumables, and laboratory supplies. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. The terms of this arrangement have been reviewed and approved by the Federal University of Rio de Janeiro, in accordance with its conflict of interest policies.
Conflict of Interest Statement
JP was employed by company Petrobras.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank the Brazilian Army and Colonel Franco, who provided the permits and logistical support to collect samples at the Restinga da Marambaia mangrove. We would also like to thank Leandro Lobo and Regina Domingues for the valuable support with the anaerobic bacterial work, Fabiano Balieiro for the sediment characterization analysis, and Daniel Cunha Passos for the valuable contributions and for reviewing an early draft of this manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.02107/full#supplementary-material
Footnotes
References
Adams, G. O., Fufeyin, P., Okoro, S. E., and Ehinomem, I. (2015). Bioremediation, biostimulation and bioaugmentation: a review. Int. J. Environ. Bioremed. Biodegrad. 3, 28–39. doi: 10.12691/ijebb-3-1-5
Andrade, L. L., Leite, D., Ferreira, E., Ferreira, L., Paula, G. R., Maguire, M., et al. (2012). Microbial diversity and anaerobic hydrocarbon degradation potential in an oil-contaminated mangrove sediment. BMC Microbiol. 12:186. doi: 10.1186/1471-2180-12-186
Atlas, R. M. (2011). Oil biodegradation and bioremediation: a tale of the worst spills in U.S. history. Environ. Sci. Technol. 45, 6709–6715. doi: 10.1021/es2013227
Beilen, J. B., Wubbolts, M. G., and Witholt, B. (1994). Genetics of alkane oxidation by Pseudomonas oleovorans. Biodegradation 5, 161–174. doi: 10.1007/bf00696457
Bellinaso, M. D. L., Henriques, J. A. P., Gaylarde, C. C., and Greer, C. W. (2003). Genes similar to naphthalena dioxygenase genes in trifluralin-degrading bacteria. Pest Manag. Sci. 60, 474–8. doi: 10.1002/ps.835
Berdugo-Clavijo, C., and Gieg, L. M. (2014). Conversion of crude oil to methane by a microbial consortium enriched from oil reservoir production waters. Front. Microbiol. 5:197. doi: 10.3389/fmicb.2014.00197
Boopathy, R. (2000). Factors limiting bioremediation technologies. Bioresour. Technol. 74, 63–67. doi: 10.1016/s0960-8524(99)00144-3
Bouchez, T., Patureaua, D., Dabert, P., Juretschko, S., Doré, J., Delgenès, P., et al. (2000). Ecological study of a bioaugmentation failure. Environ. Microbiol. 2, 179–190. doi: 10.1046/j.1462-2920.2000.00091.x
Bragg, J. R., Prince, R. C., Harner, E. J., and Atlas, R. M. (1994). Effectiveness of bioremediation for the Exxon Valdez oil spill. Nature 368, 413–418. doi: 10.1038/368413a0
Caporaso, J. G., Lauber, C. L., Walters, W. A., Berg-Lyons, D., Huntley, J., Fierer, N., et al. (2012). Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 6, 1621–1624. doi: 10.1038/ismej.2012.8
Chu, H., Sun, H., Tripathi, B. M., Adams, J. M., Huang, R., Zhan, Y., et al. (2016). Bacterial community dissimilarity between the surface and subsurface soils equals horizontal differences over several kilometers in the western Tibetan Plateau. Environ. Microbiol. 18, 1523–1533. doi: 10.1111/1462-2920.13236
Cole, J. R., Wang, Q., Cardenas, E., Fish, J., Chai, B., Farris, R. J., et al. (2009). The ribosomal database project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 37, D141–D145. doi: 10.1093/nar/gkn879
R Core Team, (2015). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing.
Desay, C., Pathak, H., and Madamwar, D. (2010). Advances in molecular and “-omics”technologies to gauge microbial communities and bioremediation at xenobiotic/anthropogencontaminated sites. Bioresour. Technol. 101, 1558–1569. doi: 10.1016/j.biortech.2009.10.080
do Carmo, F. L., do Santos, H. F., Martins, E. F., van Elsas, J. D., Rosado, A. S., and Peixoto, R. S. (2011). Bacterial structure and characterization of plant growth promoting and oil degrading bacteria from rizhospheres of mangrove plants. J. Microbiol. 49, 535–543. doi: 10.1007/s12275-011-0528-0
Dufrene, M., and Legendre, P. (1997). Species assemblages and indicator species: the need for a flexible asymmetrical approach. Ecol. Monogr. 67, 345–366. doi: 10.1890/0012-9615(1997)067
Duke, N. C. (2016). Oil spill impacts on mangroves: recommendations for operational planning and action based on a global review. Mar. Pollut. Bull. 109, 700–715. doi: 10.1016/j.marpolbul.2016.06.082
Edgar, R. C., Hass, B. J., Clemente, J. C., Quince, C., and Knight, R. (2011). UCHIME improves sensitivy and speed of chimera detection. Bioinformatics 27, 2194–2200. doi: 10.1093/bioinformatics/btr381
Eilers, K. G., Debenport, S., Anderson, S., and Fierer, N. (2012). Digging deeper to find unique microbial communitites: the strong effect of depth on the structure of bacerial and archael communities in soil. Soil Biol. Biochem. 50, 58–65. doi: 10.1016/j.soilbio.2012.03.011
Eklund, A. (2013). Beeswarm: The Bee Swarm Plot, an Alternative to Stripchart. R Package Version 0.1.6. doi: 10.1016/j.soilbio.2012.03.011
EMBRAPA (2011). Centro Nacional de Pesquisa de Solos Manual de Métodos de Análise de Solo, 2nd Edn. Rio de Janeiro, RJ: EMBRAPA, 230.
Fasanella, C. C., Dias, A. C. F., Rigonato, J., Fiore, M. F., Soares-Jr, F. L., Melo, I. S., et al. (2012). The selection exerted by oil contamination on mangrove fungal communities. Water Air Soil Pollut. 223, 4233–4243. doi: 10.1007/s11270-012-1187-4
Fierer, N., Schimel, J. P., and Holden, P. A. (2003). Variations in microbial community composition through two soil depth profiles. Soil Biol. Biochem. 35, 167–176. doi: 10.1016/S0038-0717(02)00251-1
Fierer, N., Strickland, M. S., Liptzin, D., Bradford, M. A., and Cleveland, C. C. (2009). Global patterns in below ground communities. Ecol. Lett. 12, 1238–1249. doi: 10.1111/j.1461-0248.2009.01360.x
Flocco, C. G., Gomes, N. C. M., Cormack, W. M., and Smalla, K. (2009). Occurence, and diversity of naphthalene dioxygenase genes in soil microbial communities from the Maritime Antartic. Environ. Microbiol. 11, 700–714. doi: 10.1111/j.1462-2920.2008.01858.x
Fox, J., and Weisberg, S. (2011). An {R} Companion to Applied Regression, 2nd Edn. Thousand Oaks CA: Sage.
Ghizelini, A. M., Mendonça-Hagler, L. C. S., and Macrae, A. (2012). Microbial diversity in Brazilian mangrove sediments – a mini review. Braz. J. Microbiol. 43, 1242–1254. doi: 10.1590/S1517-83822012000400002
Giambiagi-Marval, M., Mafra, M. A., Penido, E. G. C., and Bastos, M. C. F. (1990). Distinct groups of plasmids correlated with bacteriocin production in Staphylococcus aureus. J. Gen. Microbiol. 136, 1591–1599. doi: 10.1099/00221287-136-8-1591
Gomes, N. C. N., Borges, L. R., Paranhos, R., Pinto, F. N., Krogerrecklenfort, E., Mendonça-Hagler, L. C. S., et al. (2007). Diversity of ndo genes in mangrove sediments exposed to different sources of polycyclic aromatic hydrocarbon pollution. Appl. Environ. Microbiol. 73, 7392–7399. doi: 10.1128/AEM.01099-07
Gough, H. L., and Nielsen, J. L. (2016). “Bioaugmentation,” in Hydrocarbon and Lipid Microbiology Protocols, eds T. McGenity, K. Timmis, and B. Nogales, (Berlin: Springer).
Grishchenkova, V. G., Townsend, R. T., McDonald, T. K., Autenrieth, R. L., Bonner, J. S., and Boronin, A. M. (2000). Degradation of petroleum hydrocarbons by facultative anaerobic bacteria under aerobic and anaerobic conditions. Process Biochem. 35, 889–896. doi: 10.1016/S0032-9592(99)00145-4
Hamilton, S. E., and Friess, D. A. (2018). Global carbon stocks and potential emissions due to mangrove deforestation from 2000 to 2012. Nat. Clim. Chang. 8, 240–244. doi: 10.1038/s41558-018-0090-4
Hammer, O., Harper, D. A. T., and Ryan, P. D. (2001). PAST – Paleontological Statistics sofware package for education and data analysis. Palaeo. Electron. 4, 1–9.
Han, Y., and Perner, M. (2016). Sulfide consumption in Sulfurimonas denitrificans and heterologoues expression of its three SQR homologs. J. Bacteriol. 198, 1260–1267. doi: 10.1128/JB.01021-15
Jaekel, U., Musat, N., Adam, B., Kuypers, M., Grundmann, O., and Musat, F. (2013). Anaerobic degradation of propane and butane by sulfate-reducing bacteria enriched from marine hydrocarbon cold seeps. ISME J. 7, 885–895. doi: 10.1038/ismej.2012.159
Johnsen, A. R., and Karlson, U. (2005). PAH degradation capacity of soil microbial communities – does it depende on PAH exposure? Microb. Ecol. 50, 488–495. doi: 10.1007/s00248-005-0022-5
Jurelevicius, D., Alvarez, V. M., Peixoto, R. S., Rosado, A. S., and Seldin, L. (2013). The use of a combination of alkB primers to better characterize the distribution of alkanedegrading bacteria. PLoS One 8:e66565. doi: 10.1371/journal.pone.0066565
Kasai, Y., Shindo, K., Harayama, S., and Misawa, N. (2003). Molecular characterization and substrate preference of a polycyclic aromatic hydrocarbon dioxygenase from Cycloclasticus sp. strain A5. Appl. Environ. Microbiol. 69, 6688–6697. doi: 10.1128/AEM.69.11.6688-6697.2003
Khan, F. I., Husain, T., and Hejazi, R. (2004). An overview and analysis of site remediation technologies. J. Environ. Manag. 71, 95–122. doi: 10.1016/j.jenvman.2004.02.003
Kloos, K., Munch, J. C., and Schloter, M. (2006). A new method for detection of alkane-monooxygenase homologous genes (alkB) in soils based on PCR-hybridization. J. Microbiol. Meth. 66, 486–496. doi: 10.1016/j.mimet.2006.01.014
Kostka, J. E., Prakash, O., Overholt, W. A., Green, S. J., Freyer, G., Canion, A., et al. (2011). Hydrocarbon-degrading bacteria and the bacterial community response in Gulf of Mexico beach sands impacted by the Deepwater Gorizon oil spill. Appl. Environ. Microbiol. 77, 7962–7974. doi: 10.1128/AEM.05402-11
Kuhn, E., Bellicanta, G. S., and Pellizari, V. H. (2009). New alk genes detected in Antarctic marine sediments. Environ. Microbiol. 11, 669–673. doi: 10.1111/j.1462-2920.2008.01843.x
Laban, N. A., Dao, A., and Foght, J. (2015). DNA stable-isotope probing of oil sands tailings pond enrichment cultures reveals diferent key players for toluene degradation under methanogenic and sulfidogenic conditions. FEMS Microbiol. Ecol. 91:fiv039. doi: 10.1093/femsec/fiv039
Lamontagne, M. G., Schimel, J. P., and Holden, P. A. (2003). Comparison of subsurface and surface soil bacterial communities in California grassland as assessed by terminal restriction fragment length polymorphisms of PCR-amplified 16S rRNA Genes. Microb. Ecol. 46, 216–227. doi: 10.1007/s00248-003-1006-y
Larkin, M. J., Allen, C. C. R., Kukalov, L. A., and Lipscomb, A. A. (1999). Purification and characterization of a novel naphthalene dioxygenase from Rhodococcus sp. Strain NCIMB12038. J. Bacteriol. 181, 6200–6204. doi: 10.1007/s10661-017-5798-5
Lewis, M., Pryor, R., and Wilking, L. (2011). Fate and effects of anthropogenic chemicals in mangrove ecosystems: a review. Environ. Pollut. 159, 2328–2346. doi: 10.1016/j.envpol.2011.04.027
Li, D., Sharp, J. O., and Drewes, J. E. (2015). Influence of wastewater discharge on the metabolic potential of the microbial community in river sediments. Microb. Ecol. 71, 78–86. doi: 10.1007/s00248-015-0680-x
Li, D., Sharp, J. O., Saikaly, P. E., Ali, S., Alidina, M., Alarawi, M. S., et al. (2012). Dissolved organic carbon influences microbial community composition and diversity in managed aquifer recharge systems. Appl. Environ. Microbiol. 78, 6819–6828. doi: 10.1128/AEM.01223-12
Long, H., Wang, Y., Chang, S., Liu, G., Chen, T., Huo, G., et al. (2017). Diversity of crude oil-degrading bactéria and alkane hydroxylase (alkB) genes from the Qinghai-Tibet Plateau. Environ. Monit. Assess. 189, 116.
Mendes, L. W., and Tsai, S. M. (2014). Variations of bacterial community structure and composition in mangrove sediment at different depths in Southerastern Brazil. Diversity 6, 827–843. doi: 10.3390/d6040827
Meyer, D. D., Beker, S. A., Bücker, F., Peralba, M. C. R., Frazzon, A. P. G., Osti, J. F., et al. (2014). Bioremediation srategies for diesel and biodiesel in oxisol from southern Brazil. Int. Biodeterior. Biodegrad. 95, 356–363. doi: 10.1016/j.ibiod.2014.01.026
Moe, W. M., Yan, J., Nobre, M. F., Costa, M. S., and da Rainey, F. A. (2009). Dehalogenimonas lykanthroporepellens gen. nov., sp. nov., a reductively dehalogenating bacterium isolated from chlorinated solvent-contaminated groundwater. Int. J. Syst. Evol. Microbiol. 59, 2692–2697. doi: 10.1099/ijs.0.011502-0
Mori, K., Suzuki, K., Urabe, T., Sugihara, M., Tanaka, K., Hamada, M., et al. (2011). Thioprofundum hispidum sp. nov., an obligately chemolithoautotrophic sulfur-oxidizing gammaproteobacterium isolated from the hydrothermal field on Suiyo Seamount, and proposal of Thioalkalispiraceae fam. nov. in the order Chromatiales. Int. J. Syst. Evol. Microbiol. 61(Pt 10), 2412–2418. doi: 10.1099/ijs.0.026963-0
Müeller, J. B., Ramos, D. T., Larose, C., Fernandes, M., Lazzarin, H. S. C., Vogel, T. M., et al. (2017). Combined iron and sulfate reduction biostimulation as a novel approach to enhance BTEX and PAH source-zone biodegradation in biodiesel blend-contaminated groundwater. J. Hazard. Mat. 326, 229–236. doi: 10.1016/j.jhazmat.2016.12.005
Mueller, R. C., Belnap, J., and Kuske, C. R. (2015). Soil bacterial and fungal community responses to nitrogen addition across soil depth and microhabitat in na arid shrubland. Front. Microbiol. 6:891. doi: 10.3389/fmicb.2015.00891
Netzer, F. V., Pilloni, G., Kleindienst, S., Krüger, M., Knittel, K., Gründger, F., et al. (2013). Enhanced gene detection assays for fumarate-adding enzymes allow uncovering of anaerobic hydrocarbon degraders in terrestrial and marine systems. Appl. Environ. Microbiol. 79, 543–552. doi: 10.1128/AEM.02362-12
Nogueira, V. L. R., Rocha, L. L., Colares, G. B., Angelim, A. L., Normando, L. R. O., Cantão, M. E., et al. (2015). Microbiomes and potential metabolic pathways of pristine and anthropized Brazilian mangroves. Reg. Stud. Mar. Sci. 2, 56–64. doi: 10.1016/j.rsma.2015.08.008
Peixoto, R. S., Chaer, G. M., Carmo, F. L., Araújo, F. V., Paes, J. E., Volpon, A., et al. (2011b). Bacterial communities reflect the spatial variation in pollutant levels in Brazilian mangrove sediment. Antonie van Leeuwenhoek 99, 341–354. doi: 10.1007/s10482-010-9499-0
Peixoto, R. S., Vermelho, A. B., and Rosado, A. S. (2011a). Petroleum-degrading enzymes: bioremediation and new prospects. Enzyme Res. 2011:475193. doi: 10.4061/2011/475193
Pohlert, T. (2014). The Pairwise Multiple Comparison of Mean Ranks Package (PMCMR). Available at: http://CRAN.R-project.org/package=PMCMR (accessed January 22, 2016).
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acid Res. 41, D590–D596. doi: 10.1093/nar/gks1219
Ramsay, M. A., Swannell, R. P. J., Shipton, W. A., Duke, N. C., and Hill, R. T. (2010). Effect of bioremediation on the microbial community in oiled mangrove sediments. Mar. Pollut. Bull. 41, 413–419. doi: 10.1016/S0025-326X(00)00137-5
Raynaud, X., and Nunan, N. (2014). Spatial ecology of bacteria at microscale in soil. PLoS One 9:e87217. doi: 10.1371/journal.pone.0087217
Salminen, J. M., Tuomi, P. M., Suortti, A. M., and Jørgensen, K. S. (2004). Potential for aerobic and anaerobic biodegradation of petroleum hydrocarbons in boreal subsurface. Biodegradation 15, 29–39. doi: 10.1023/b:biod.0000009954.21526.e8
Sánchez, M. A., Vásquez, M., and González, B. (2004). A previously unexposed Forest soil microbial community degrades high levels of the pollutant 2,4,6-Trichlorophenol. Appl. Environ. Microbiol. 70, 7567–7570. doi: 10.1128/AEM.70.12.7567-70.2004
Santos, H. F., Carmo, F. L., Paes, J. E. S., Rosado, A. S., and Peixoto, R. S. (2011). Bioremediation of mangroves impacted by petroleum. Water Air Soil Pollut. 216, 329–350. doi: 10.1007/s11270-010-0536-4
Santos, H. F., Cury, J. C., Carmo, F. L., Rosado, A. S., and Peixoto, R. S. (2010). 18S rDNA sequences from microeukaryotes reveal oil indicators in mangrove sediment. PLoS One 5:e12437. doi: 10.1371/journal.pone.0012437
Santos, H. F., Duarte, G. A. S., Rachid, C. T. C. C., Chaloub, R. M., Calderon, E. N., Marangoni, L. F. B., et al. (2015). Impact of oil spills on coral reefs can be reduced by bioremediation using probiotic microbiota. Sci. Rep. 5:18268. doi: 10.1038/srep18268
Schloss, P. D., Westcott, S. L., Ryabin, T., Hall, J. R., Hartmann, M., Hollister, E. B., et al. (2009). Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75, 7537–7541. doi: 10.1128/AEM.01541-09
Simarro, R., González, N., Bautista, L. F., and Molina, M. C. (2013). Assessment of the efficiency of in situ bioremediation techniques in a creosote polluted soil: change in bacterial community. J. Hazard. Mat. 262, 158–167. doi: 10.1016/j.jhazmat.2013.08.025
Sorokin, D. Y., Trotsenko, Y. A., Doronina, N. V., Tourova, T. P., Galiski, E. A., Kolganova, T. V., et al. (2007). Methylohalomonas lacus gen. nov., sp. nov. and Methylonatrum kenyense gen. nov., sp. nov., methylotrophic gammaproteobacteria from hypersaline lakes. Int. J. Syst. Evol. Microbiol. 57(Pt 12), 2762–2769. doi: 10.1099/ijs.0.64955-0
Thatoi, H., Behera, B. C., and Mishra, R. R. (2013). Ecological role and biotechnological potential of mangrove fungi: a review. Micology 4, 54–71. doi: 10.1080/21501203.2013.785448
Touvora, T. P., Sokolova, D. S., Semenova, E. M., Shumkova, E. S., Korshunova, A. V., Babich, T. L., et al. (2016). Detection of n-alkane biodegradation genes alkB and ladA in thermophilic hydrocarbon-oxidizing bacteria of the genera Aeribacillus and Geobacillus. Microbiology 85, 693–707. doi: 10.1134/S0026261716060199
Viggor, S., Joesaar, M., Vedler, E., Kiiker, R., Parnpuu, L., and Heinaru, A. (2015). Occurrence of diverse alkane hydroxylase alkB genes in indigenous oil-degrading bacteria of Baltic Sea surface water. Mar. Pollut. Bull. 101, 507–516. doi: 10.1016/j.marpolbul.2015.10.064
Wu, M., Ye, Z., Chen, K., Li, W., Yuan, J., and Jiang, X. (2017). Bacterial community shift and hydrocarbon transformation during bioremediation of short-term petroleum-contaminated soil. Environ. Pollut. 223, 657–664. doi: 10.1016/j.envpol.2017.01.079
Xu, Y., and Lu, M. (2010). Bioremediation of crude oil-contaminated soil - comparison of different biostimulation and bioaugmentation treatements. J. Hazard. Mat. 183, 395–401. doi: 10.1016/j.jhazmat.2010.07.038
Keywords: bioremediation, mangrove, mesocosm, oil-degrading genes, bacterial diversity
Citation: Machado LF, Leite DCA, Rachid CTCC, Paes JE, Martins EF, Peixoto RS and Rosado AS (2019) Tracking Mangrove Oil Bioremediation Approaches and Bacterial Diversity at Different Depths in an in situ Mesocosms System. Front. Microbiol. 10:2107. doi: 10.3389/fmicb.2019.02107
Received: 29 March 2019; Accepted: 27 August 2019;
Published: 13 September 2019.
Edited by:
Pascal E. Saikaly, King Abdullah University of Science and Technology, Saudi ArabiaReviewed by:
Zhenmei Lu, Zhejiang University, ChinaDong Li, University of California, Santa Barbara, United States
Copyright © 2019 Machado, Leite, Rachid, Paes, Martins, Peixoto and Rosado. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Alexandre Soares Rosado, YXNyb3NhZG9AbWljcm8udWZyai5icg==
†Present address: Laís Feitosa Machado, Ecology Department, Federal University of Vale do São Francisco, Senhor do Bonfim, Brazil; Deborah Catharine de Assis Leite, Federal Technological University of Paraná, Curitiba, Brazil
 Laís Feitosa Machado
Laís Feitosa Machado Deborah Catharine de Assis Leite
Deborah Catharine de Assis Leite Caio Tavora Coelho da Costa Rachid
Caio Tavora Coelho da Costa Rachid Jorge Eduardo Paes2
Jorge Eduardo Paes2 Raquel Silva Peixoto
Raquel Silva Peixoto Alexandre Soares Rosado
Alexandre Soares Rosado