- 1Department of Science, University Roma Tre, Rome, Italy
- 2National Institute of Nuclear Physics, Roma Tre Section, Rome, Italy
The emergence of antibiotic resistant bacterial pathogens is increasing at an unprecedented pace, calling for the development of new therapeutic options. Small molecules interfering with virulence processes rather than growth hold promise as an alternative to conventional antibiotics. Anti-virulence agents are expected to decrease bacterial virulence and to pose reduced selective pressure for the emergence of resistance. In the opportunistic pathogen Pseudomonas aeruginosa the expression of key virulence traits is controlled by quorum sensing (QS), an intercellular communication process that coordinates gene expression at the population level. Hence, QS inhibitors represent promising anti-virulence agents against P. aeruginosa. Virtual screenings allow fast and cost-effective selection of target ligands among vast libraries of molecules, thus accelerating the time and limiting the cost of conventional drug-discovery processes, while the drug-repurposing approach is based on the identification of off-target activity of FDA-approved drugs, likely endowed with low cytotoxicity and favorable pharmacological properties. This study aims at combining the advantages of virtual screening and drug-repurposing approaches to identify new QS inhibitors targeting the pqs QS system of P. aeruginosa. An in silico library of 1,467 FDA-approved drugs has been screened by molecular docking, and 5 hits showing the highest predicted binding affinity for the pqs QS receptor PqsR (also known as MvfR) have been selected. In vitro experiments have been performed by engineering ad hoc biosensor strains, which were used to verify the ability of hit compounds to decrease PqsR activity in P. aeruginosa. Phenotypic analyses confirmed the impact of the most promising hit, the antipsychotic drug pimozide, on the expression of P. aeruginosa PqsR-controlled virulence traits. Overall, this study highlights the potential of virtual screening campaigns of FDA-approved drugs to rapidly select new inhibitors of important bacterial functions.
Francesca D’Angelo, Institut Pasteur, Paris, France
Introduction
The long-term use of antibiotics has dramatically accelerated the emergence of multi-drug and even pan-drug resistant bacterial pathogens worldwide, leading to an alarming increase of difficult-to-treat infections. This worrying scenario especially concerns the ESKAPE pathogens (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter species), a group of bacteria that “escape” the action of almost all available antibiotics (Rice, 2008; Boucher et al., 2009). The trend toward antibiotic resistance is even more alarming if considering that only a handful of new antibiotics have been approved by the U.S. Food and Drug Administration (FDA) in the last decade, with many companies considering the R&D for new antibiotics a less attractive asset compared to more rewarding therapeutic areas. Indeed, de novo antibiotic development requires large investments that might not grant an economic reward due to the short commercial lifespan of antibiotics, caused by the rapid emergence of resistance (Ventola, 2015; Mohr, 2016; Luepke et al., 2017).
The awareness about the risk of antibiotic resistance for human health has increased in parallel with our comprehension of bacterial pathobiology, so that virulence mechanisms are now recognized as molecular targets for the development of novel anti-virulence drugs targeting the infection process rather than bacterial growth (Rasko and Sperandio, 2010). Although resistance mechanisms to anti-virulence drugs have been described (Zhu et al., 1998; Hung et al., 2005; Maeda et al., 2012; Imperi et al., 2019), targeting virulence rather than growth is expected to pose a reduced selective pressure for the emergence of resistance (Allen et al., 2014). In particular, in vitro evolution experiments indicate that drug resistant clones are counterselected if “public goods” that are shared among members of a bacterial population are targeted (e.g., toxins, exo-proteases, and siderophores) (Mellbye and Schuster, 2011; Vale et al., 2016). Since drug-resistant strains are likely to emerge only if they gain a “private” advantage over the susceptible population, quorum sensing (QS) is recognized as an ideal target for the development of anti-virulence agents. Indeed, QS is an intercellular communication system based on the production, secretion and reception of signal molecules that coordinate the expression of secreted virulence factors in different bacterial pathogens (Rampioni et al., 2014; Kalia et al., 2019).
The ESKAPE pathogen P. aeruginosa is a model organism for the development of anti-virulence drugs targeting QS (Soukarieh et al., 2018b). This Gram-negative bacterium, that is one of the most dreaded nosocomial pathogens and the main cause of death in cystic fibrosis (CF) patients, has recently been included by the World Health Organization in the list of pathogens for which new therapeutic options are urgently needed (Priority 1: Critical)1. The ability of P. aeruginosa to cause both acute and chronic infections in different districts of the human body mainly relies on its capacity to adapt to the host by fine-tuning the expression of a wide array of virulence factors, many of which are QS-controlled. As a consequence, numerous anti-virulence drugs targeting the P. aeruginosa QS circuitry have been identified in recent years, and their ability to reduce P. aeruginosa pathogenicity has been confirmed both in vitro and in vivo (Rampioni et al., 2014; Soukarieh et al., 2018b). Unfortunately, the majority of the QS inhibitors identified to date are not suitable as lead-like compounds for further drug development, mainly due to their cytotoxicity and unfavorable pharmacological properties (Maura et al., 2016; Soukarieh et al., 2018b).
With the aim to identify bioavailable and safe QS inhibitors that can faster move into clinical trials or serve as leads for drug optimization programs, our group recently undertook whole-cell biosensor-based screening campaigns of libraries of FDA-approved drugs. This drug-repurposing approach led to the identification of niclosamide, an anthelmintic drug, and clofoctol, an antibiotic active against Gram-positive bacteria, as potent and safe QS inhibitors targeting the acyl-homoserine lactones (AHL)-based and the 2-alkyl-4(1H)quinolone (AQ)-based QS systems of P. aeruginosa, respectively (Imperi et al., 2013; D’Angelo et al., 2018). These FDA-approved drugs effectively reduced P. aeruginosa pathogenic potential in animal models of infection, hence representing promising candidates for preclinical studies.
In the last decades in silico approaches have been proved as valid aids to conventional drug-discovery programs. In particular, virtual screens carried out through molecular docking simulations allow to preselect promising drug candidates in vast libraries of molecules, so that only a reduced number of predicted hits have to be validated by means of in vitro experiments. In this way, time and costs associated to conventional screening campaigns are reduced. In addition, docking simulations allow to predict the likely binding mode of candidate hits onto the selected target, providing a molecular basis for their optimization in terms of binding affinity (Reuter et al., 2015).
On this basis, the present study aims at combining the advantages of drug-repurposing and virtual screening approaches to identify FDA-approved drugs targeting the pqs QS system of P. aeruginosa via in silico molecular docking.
In P. aeruginosa the pqs QS system is based on the AQs 2-heptyl-4-hydroxyquinoline (HHQ) and 2-heptyl-3-hydroxy-4(1H)-quinolone (PQS) as signal molecules. HHQ is synthesized by the enzymes coded by the pqsABCDE-phnAB operon, and is converted to PQS by the monooxygenase PqsH. Both HHQ and PQS can bind to and activate the transcriptional regulator PqsR (also known as MvfR), that in the active form binds to the PpqsA promoter region and promotes pqsABCDE-phnAB transcription. Hence, HHQ and PQS act as autoinducers to accelerate their own synthesis (Bredenbruch et al., 2005; Heeb et al., 2011; Dulcey et al., 2013; Drees and Fetzner, 2015). While the main role of HHQ is to trigger this PqsR-dependent positive feedback loop, the signal molecule PQS and the protein PqsE (the latter coded by the fifth gene of the pqsABCDE-phnAB operon) are the main effectors of the pqs QS system. Besides activating PqsR, PQS acts as an iron chelator, is required for the biogenesis of outer membrane vesicles, and promotes the expression of virulence genes via a PqsR-independent pathway (Bredenbruch et al., 2005; Mashburn and Whiteley, 2005; Diggle et al., 2007; Rampioni et al., 2016; Lin et al., 2017). PqsE is a multifunctional protein that participates in the synthesis of HHQ and positively controls the expression of multiple virulence factors independently of AQs, likely by activating the transcriptional regulator RhlR via the production of an uncharacterized signal molecule that links the pqs and rhl QS systems (Drees and Fetzner, 2015; Hazan et al., 2010; Rampioni et al., 2010; Rampioni et al., 2016; Mukherjee et al., 2018). Overall, the production of PQS and the expression of PqsE require activated PqsR, and consequently PqsR-inhibitors have been shown to attenuate P. aeruginosa virulence both in vitro and in animal models of infection (Klein et al., 2012; Ilangovan et al., 2013; Zender et al., 2013; Lu et al., 2014; Starkey et al., 2014; Maura and Rahme, 2017; Maura et al., 2017; D’Angelo et al., 2018; Soukarieh et al., 2018a). Since the three-dimensional structure of the PqsR domain that interacts with HHQ and PQS (co-inducer binding domain; CBD) has recently been solved (Ilangovan et al., 2013; Kitao et al., 2018), this QS transcriptional regulator now constitutes an ideal target for the identification of new P. aeruginosa anti-virulence drugs via molecular docking simulations.
In this study, a virtual screening approach has been used to predict PqsR ligands in a library of 1,467 FDA-approved drugs. The ability of the best 5 hits to decrease PpqsA activity and AQs level has been tested in wild type P. aeruginosa and in ad hoc engineered strains. This process led to the identification of the antipsychotic drug pimozide as a specific PqsR inhibitor. Phenotypic assays showed that pimozide hampers the expression of PqsR-controlled virulence traits, such as the production of the virulence factor pyocyanin, swarming motility and biofilm formation, and docking simulations suggest a possible competition with native AQs for PqsR binding. These results provide a proof-of-concept that the drug-repurposing and virtual screening approaches can be combined to accelerate the selection of anti-QS molecules among FDA-approved drugs.
Materials and Methods
Bacterial Strains, Media and Chemicals
Bacterial strains used in this study are reported in Table 1. Bacterial strains were routinely grown at 37°C in Luria-Bertani (LB) broth in shaking conditions, or in LB supplemented with 15 g/L agar.
When required, tetracycline (Tc; 200 μg/mL), isopropyl β-D-1-thiogalactopyranoside (IPTG), dimethyl sulfoxide (DMSO), or synthetic PQS were added to the medium. IPTG, DMSO and synthetic PQS were used at the concentrations indicated in the text. Synthetic PQS stock solution was prepared in MeOH at 20 mM concentration (synthetic PQS was kindly provided by Paul Williams and Miguel Càmara – University of Nottingham, United Kingdom). Ergotamine and pimozide were available in our laboratory as drugs of the PHARMAKON library (10 mM stock solutions in DMSO). Dutasteride, eltrombopag and conivaptan were purchased from Sigma-Aldrich, Carbosynth Ltd., and MCE Medchem Express, respectively, and dissolved in DMSO at 10 mM concentration. Pimozide was also purchased from Sigma-Aldrich for further analyses, and dissolved in DMSO at 40 mM concentration.
Virtual Screening via Molecular Docking
Molecular docking simulations were carried out using DockingApp (Di Muzio et al., 2017), a user friendly interface to the molecular docking program AutoDock Vina (Trott and Olson, 2010), on 1,467 FDA-approved molecules extracted from the DrugBank Database and provided in ready-to-dock format as part of the DockingApp package. DockingApp is a freely available platform-independent application to perform docking simulations and virtual screening using AutoDock Vina. An intuitive graphical user interface facilitates the input phase while an embedded JMol applet allows to visualize and analyse the results. The application comes with the DrugBank set of ready-to-dock FDA-approved drugs for virtual screening and drug-repurposing purposes. In all simulations, the search space (docking grid) included the whole PqsR co-inducer binding domain (CBD) structure, in order to carry out “blind” predictions of the “hit” compounds binding sites. Simulations were first carried out on the apo form of the protein (PDB ID: 4JVC) (Ilangovan et al., 2013), by keeping all protein residues rigid. The ten best-ranking compounds, according to the AutoDock Vina scoring function, were then selected for a refinement round in which molecular docking simulations were carried out allowing flexibility of the residues building up the PqsR binding pocket (i.e., Ile149, Ala168, Val170, Ile186, Leu189, Leu207, Leu208, Phe221, Ile236, Tyr258, Asp264, and Thr265) (Ilangovan et al., 2013). The results of docking simulations were analyzed using the molecular graphics program UCSF-Chimera, version 1.12 (Pettersen et al., 2004).
Bioluminescence Assay
Analyses of PqsR activity in the presence of potential inhibitors has been performed by using ad hoc engineered reporter systems in which bioluminescence emission is proportional to PqsR activity.
The primary screening for potential PqsR inhibitors was performed as previously described (D’Angelo et al., 2018). Briefly, the screening was based on the co-culture of P. aeruginosa PAO1 wild type (PAO1) and the reporter strain PAO1 ΔpqsA PpqsA::lux (AQ-Rep). PAO1 and AQ-Rep were grown for 16 h at 37°C with shaking (200 rpm) in LB broth or in LB broth supplemented with 200 μg/L Tc, respectively. After growth, PAO1 and AQ-Rep were washed with sterile saline and mixed into LB broth to a final OD600 of 0.03 and 0.1, respectively (wild type/reporter ratio ≈ 1/3). Two-hundred μL aliquots of the diluted co-cultures were dispensed into 96-wells black clear-bottom microtiter plates. All compounds used in the primary screening were dissolved in DMSO to 10 mM concentration. The compounds were added to the microtiter plates containing the co-cultures at the final concentrations of 20 and 200 μM. As untreated controls, the same amount of DMSO alone as in the treated samples was added to the microtiter wells containing the co-culture.
For further analysis with pimozide from Sigma-Aldrich, 100 μL LB-grown aliquots of the PAO1/AQ-Rep co-culture (OD600 = 0.06 and 0.2, respectively) or of other reporter systems indicated in the text (OD600 = 0.02) were dispensed into 96-wells black clear-bottom microtiter plates, and 100 μL of pimozide diluted in LB at concentrations ranging from 50 to 400 μM were added to each well. Also in this case, DMSO alone was used as a control.
For all light emission assays, plates were incubated at 37°C with shaking (120 rpm) for 5 h, and then light emission (RLU) and cell density (OD600) of the reporter system were recorded by using an automated Spark 10 M luminometer-spectrophotometer (Tecan). Reporter activity was evaluated as Relative Light Units (RLU) normalized to cell density (OD600). Alteration in promoter activity induced by the tested compounds was determined by comparing the promoter activity of the specific biosensor system in untreated and treated samples.
Quantification of AQs
AQ signal molecules in P. aeruginosa PAO1 culture supernatants were quantified as previously described (Fletcher et al., 2007). PAO1 wild type cultures were grown at 37°C in 96-well microtiter plates with shaking (120 rpm) in LB broth supplemented with the tested compounds or solvent vehicle (i.e., DMSO) as a control. After 7 h of incubation, cell-free supernatants of PAO1 wild type cultures were collected and 5 μL were added to 195 μL of the AQ-Rep biosensor (OD600 = 0.1) dispensed into 96-wells black clear-bottom microtiter plates. Plates were incubated for 5 h at 37°C with shaking (120 rpm), and light emission (RLU) and cell density (OD600) of the cultures were recorded by using an automated Spark 10 M luminometer-spectrophotometer (Tecan). A calibration curve was generated by growing the AQ-Rep biosensor strain with synthetic PQS at concentrations ranging from ∼ 45 nM to 300 μM. The resulting dose-response curve was used as a landmark to determine the concentration of the AQs in each culture supernatant.
Pyocyanin Production, Swarming Motility and Biofilm Formation Assays
The assay for pyocyanin extraction and quantification has been performed as previously described (Essar et al., 1990) on PAO1 wild type and PAO1 ΔpqsR strains. Bacterial strains were grown for 16 h at 37°C with shaking (200 rpm) in LB broth in the presence of 100 μM pimozide or 0.25% (v/v) DMSO (solvent vehicle control).
Swarming motility assays were performed on PAO1 wild type and PAO1 ΔpqsR by using swarming plates [0.8% (w/v) nutrient broth N.2, 0.5% (w/v) glucose, 0.5% (w/v) bacteriological agar] (Rampioni et al., 2009). Plates were supplemented with 100 μM pimozide or 0.25% (v/v) DMSO (solvent vehicle control). Swarming motility was directly observed at the air-agar interface after 16 h of incubation at 37°C.
The biofilm formation assay was performed in eight-well chamber slides as previously described (Jurcisek et al., 2011; D’Angelo et al., 2018), with minor modifications. Briefly, PAO1 wild type and PAO1 ΔpqsR constitutively expressing GFP via the pMRP9-1 plasmid (Davies et al., 1998) were inoculated in an eight-well chamber slide at an OD600 of 0.02 in 700 μL of M9 minimal medium supplemented with 20 mM glucose as carbon source, in the presence of 0.25% (v/v) DMSO (solvent vehicle control) or 100 μM pimozide. Bacterial cultures were incubated at 30°C for 24 h. Planktonic cells were gently removed and the wells of the chamber slide were rinsed with sterile saline before confocal microscope (Leica TCS SP5) imaging of the bacterial cells adhered to the glass surface.
Statistical Analysis
For statistical analysis the software GraphPad Prism 5 was used; one-way analysis of variance (ANOVA) followed by Tukey-Kramer multiple comparison tests were performed. Differences having a p value < 0.05 were considered statistically significant.
Results
Virtual Screening for the Identification of FDA-Approved PqsR Ligands
We performed a virtual screening to select possible PqsR ligands in a library of 1,467 FDA-approved compounds extracted from the DrugBank database2 and already provided in ready-to-dock format as part of the DockingApp software package (see section “Materials and Methods,” for details).
PqsR is a multi-domain transcriptional regulator composed by a N-terminal helix-turn-helix DNA-binding domain and a C-terminal co-inducer binding domain (CBD). Since the three-dimensional structure of full-length PqsR is unavailable, molecular docking simulations were performed based on the crystal structure of PqsR-CBD. The CBD of PqsR has been crystallized in the apo form (PDB ID: 4JVC) or as a complex with the native AQ ligand 2-nonyl-4-hydroxy-quinoline (NHQ) (PDB ID: 4JVD) or with the quinazolinone (QZN) inhibitor 3-NH2-7Cl-C9-QZN (PDB ID: 4JVI) (Ilangovan et al., 2013). It has been shown that both the native ligand and the inhibitor bind to the CBD of PqsR in a site consisting of two adjacent pockets: the quinolone ring is accommodated in pocket B, while the aliphatic chain makes hydrophobic interactions with pocket A (Ilangovan et al., 2013). More recently, the benzamide-benzimidazole inhibitor M64 was shown to bind to PqsR-CBD in a similar way as NHQ and 3-NH2-7Cl-C9-QZN, with its benzimidazole group bound in pocket B and the phenoxy group occupying pocket A (PDB ID: 6B8A; Kitao et al., 2018). To avoid selection bias, molecular docking simulations were performed on the apo form of PqsR-CBD. In addition, to increase the reliability of the simulations, the docking search space was not restricted to the A and B binding pockets, but extended to the entire PqsR-CBD, i.e., a “blind” docking procedure was carried out. To speed up the process, the initial screening of the 1,467 FDA-approved compounds was carried out by keeping all amino acid residues rigid.
Possible PqsR ligands were ranked based on the predicted binding affinity calculated with the AutoDock Vina scoring function. In the case of multiple ligands with the same binding affinity, these were prioritized based on the size-independent ligand efficiency (SILE) coefficient. SILE is a normalized parameter derived from the ligand efficiency (LE), a predictive measure of the per-atom binding affinity of a ligand to its binding partner (Kuntz et al., 1999).
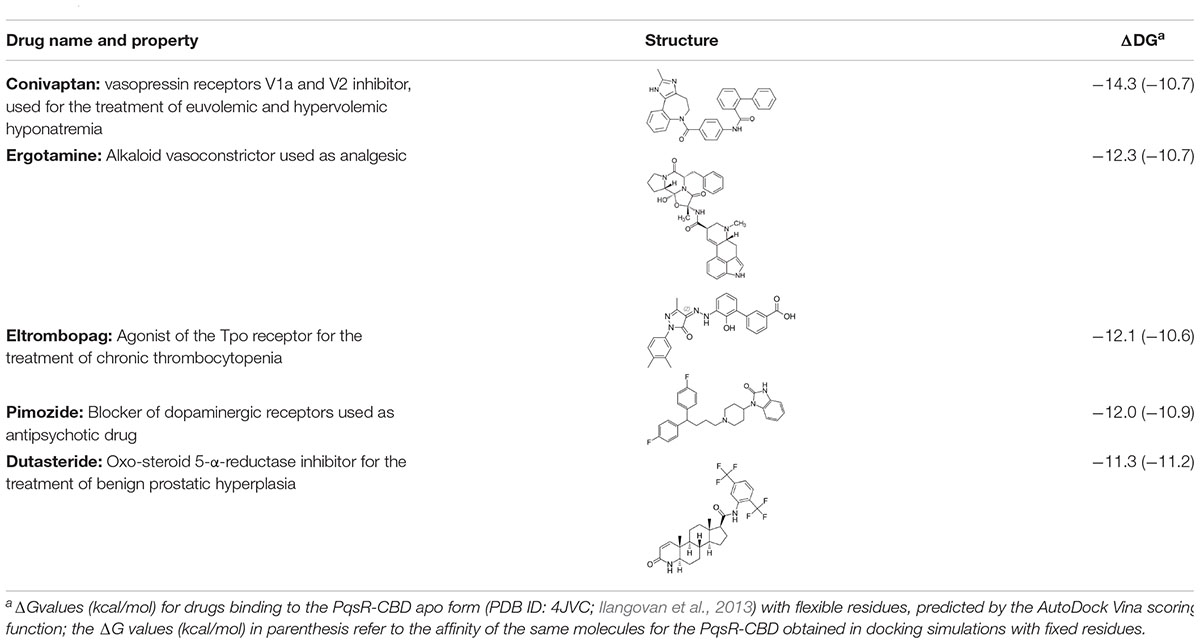
Following the above procedure, the ten best-ranking putative PqsR ligands (predicted binding affinity ranging from -11.2 to -10.0 kcal/mol) were selected for a second round of molecular docking simulations in which residues previously reported to be involved in the binding of the natural ligand NHQ to the PqsR-CBD (Ilangovan et al., 2013) were considered flexible (see section “Materials and Methods,” for details). The five molecules predicted to display the highest affinity to PqsR-CBD with fixed residues ranked in the first five positions also in the analysis with flexible residues, and are listed in Table 2. Conivaptan is an non-peptide inhibitor of the vasopressin receptor subtypes V1a and V2, commonly used to treat euvolemic and hypervolemic hyponatremia (Ferguson-Myrthil, 2010); ergotamine is an alkaloid acting as a serotonin agonist with vasoconstrictor and analgesic properties (Saxena and De Deyn, 1992); eltrombopag, an agonist of the thrombopoietin physiological target, is used for the treatment of thrombocytopenia (McCormack, 2015); pimozide is an antipsychotic drug used to treat schizophrenia, chronic psychosis, Tourette’s syndrome, and resistant tics (Tueth and Cheong, 1993); dutasteride is a 5α-reductase inhibitor used to treat benign prostatic hyperplasia (Azzouni and Mohler, 2012).
Superimposition of the five PqsR-ligand molecular complexes obtained by docking simulations with the crystal structure of the PqsR-NHQ complex (PDB ID: 4JVD) (Ilangovan et al., 2013) predicts that all the five ligands bind in the NHQ binding site, interacting with residues building up the A and B pockets of PqsR-CDB, as observed for the natural ligand NHQ (Figure 1).
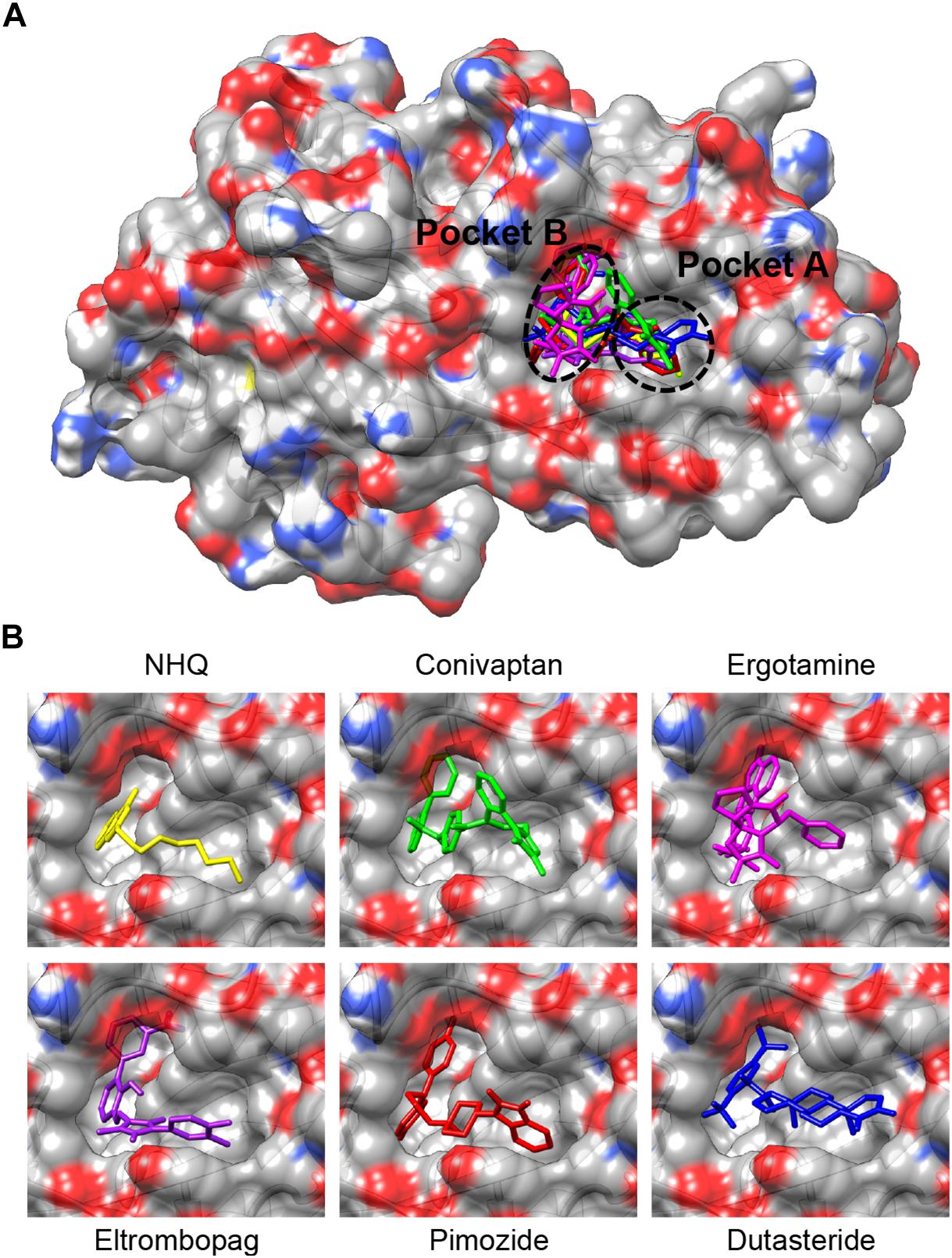
Figure 1. (A) Superimposition of the putative complexes formed by conivaptan (green), ergotamine (magenta), eltrombopag (purple), pimozide (red) or dutasteride (blue) with the PqsR-CBD, obtained by molecular docking simulations. Binding of the natural ligand NHQ (yellow) is also shown (Ilangovan et al., 2013). The binding pockets A and B are indicated. (B) Enlargement of the ligand-binding site from (A).
Anti-QS Activity of the Predicted PqsR Ligands
To validate the ability of the predicted PqsR ligands to inhibit PqsR activity in P. aeruginosa, the best 5 hits were tested for their ability to reduce bioluminescence in a co-culture system based on wild type P. aeruginosa PAO1 (Nottingham collection; herein referred to as PAO1) and isogenic P. aeruginosa PAO1 ΔpqsA PpqsA::lux (named AQ-Rep) (Fletcher et al., 2007; D’Angelo et al., 2018). The AQ-Rep biosensor strain is unable to synthesize HHQ and PQS signal molecules as a consequence of pqsA mutation, and it carries single-copy chromosomal insertion of a transcriptional fusion between the PqsR-activated promoter PpqsA and the luxCDABE operon for light emission. Therefore, in the PAO1/AQ-Rep co-culture the AQs produced by PAO1 activate PqsR in AQ-Rep, and consequently promote bioluminescence; hence, a PqsR inhibitor is expected to reduce light emission in this reporter system. The use of the PAO1/AQ-Rep co-culture recently allowed the identification of new PqsR inhibitors (D’Angelo et al., 2018), thus proving its efficacy for the selection of anti-virulence drugs targeting the pqs QS system. A co-culture of PAO1 and of a ΔpqsA derivative strain carrying the mini-CTX::lux empty vector for constitutive light emission (named C-Rep) was used to discriminate between molecules targeting PqsR and molecules affecting light emission in a PqsR-independent way.
The PAO1/AQ-Rep co-culture was incubated with the predicted PqsR-ligands identified via the preliminary virtual screening: conivaptan, ergotamine, eltrombopag, pimozide, and dutasteride. Molecules were tested at 20 μM and 200 μM concentrations. Since these drugs were dissolved in DMSO, solvent vehicle control samples in which 0.2% or 2% DMSO alone was added were also analyzed. To ensure specificity, the predicted ligands of PqsR were also analyzed by using the PAO1/C-Rep control co-culture. For each predicted ligand, the residual reporter activity (RLU/OD600) was calculated by comparing the reporter activity of the culture grown in the presence of the tested molecule to the reporter activity of the same culture grown in the presence of solvent vehicle, considered as 100%.
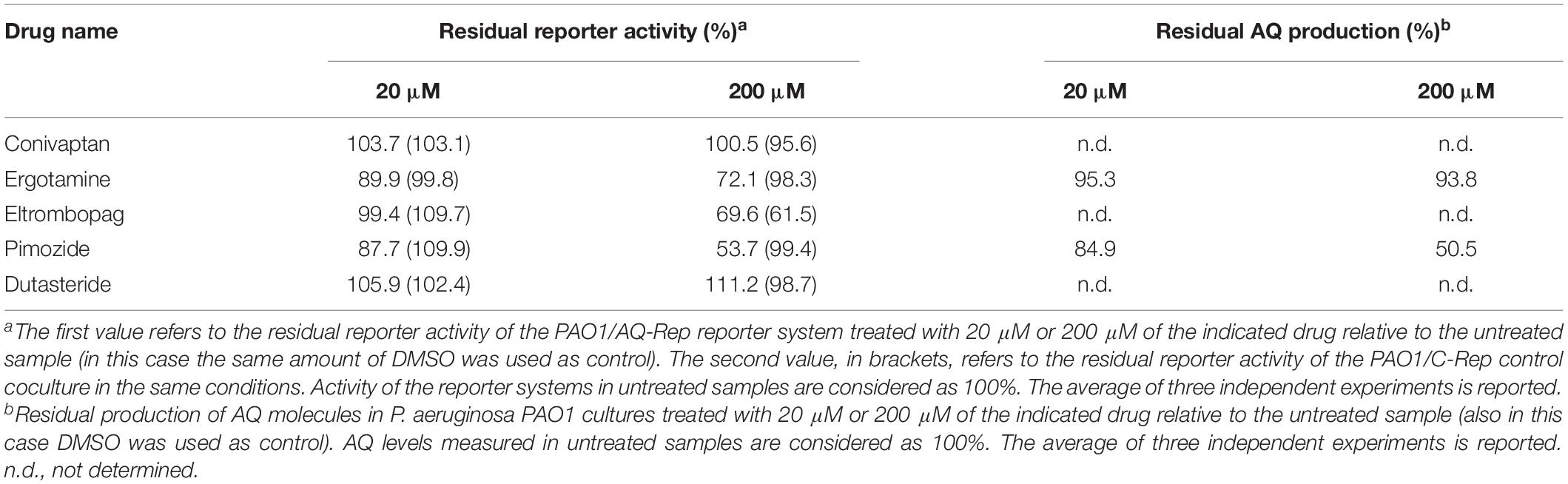
As reported in Table 3, 200 μM ergotamine, eltrombopag and pimozide were able to reduce light emission in the PAO1/AQ-Rep co-culture system by 27.9, 30.4, and 46.3%, respectively, while conivaptan and dutasteride did not affect reporter activity. However, 200 μM eltrombopag reduced light emission also in the PAO1/C-Rep control co-culture system (38.5% reduction), suggesting PqsR-independent light inhibition, while ergotamine and pimozide did not significantly alter light emission in the control samples. Notably, ergotamine and pimozide reduced reporter activity at 20 μM concentration (10.1 and 12.3% reduction, respectively), and none of the tested drugs significantly altered P. aeruginosa cell density (data not shown). Since ergotamine and pimozide were effective in specifically reducing light emission in the PAO1/AQ-Rep co-culture, without altering bacterial growth, these molecules were selected for further investigations.
A secondary screening was performed investigating the ability of ergotamine and pimozide to affect the production of the signal molecules AQs. To this end, PAO1 was grown in LB supplemented with ergotamine or pimozide (20 μM and 200 μM) or with DMSO, as a control. The amount of AQs in the corresponding cell-free supernatants was evaluated by using the AQ-Rep biosensor strain, in which light emission is proportional to the amount of AQs present in the medium. As shown in Table 3, ergotamine slightly decreased the level of AQs at both 20 μM and 200 μM (4.7 and 6.2% reduction, respectively), while pimozide reduced the production of these QS signal molecules of 15.1% and 49.5% at 20 μM and 200 μM, respectively.
Pimozide Inhibits the pqs QS System and PqsR-Controlled Virulence Traits
According to the primary and secondary screenings (Table 3) pimozide from the PHARMAKON library reduced PpqsA activity in the co-culture system PAO1/AQ-Rep and AQs production in PAO1, without altering bacterial growth. To confirm these data, experiments were replicated with pimozide purchased from a different vendor (Sigma-Aldrich).
Growth curves reported in Figure 2A show that up to 400 μM pimozide does not alter the growth profile of PAO1. Data reported in Figure 2B confirmed that pimozide significantly reduces light emission in the PAO1/AQ-Rep co-culture in a dose-dependent manner. In detail, 100 μM, 200 μM and 400 μM pimozide significantly reduced PAO1/AQ-Rep activity of 30.1, 45.2, and 64.7%, respectively (Figure 2B, white bars). Hence, the decrease in light emission from the co-culture system was comparable to what previously observed for 200 μM pimozide in the primary screening (reduction of PAO1/AQ-Rep activity = 46.3%; Table 3). Conversely, pimozide did not affect bioluminescence in the PAO1/C-Rep control system up to 400 μM (Figure 2B, gray bars).
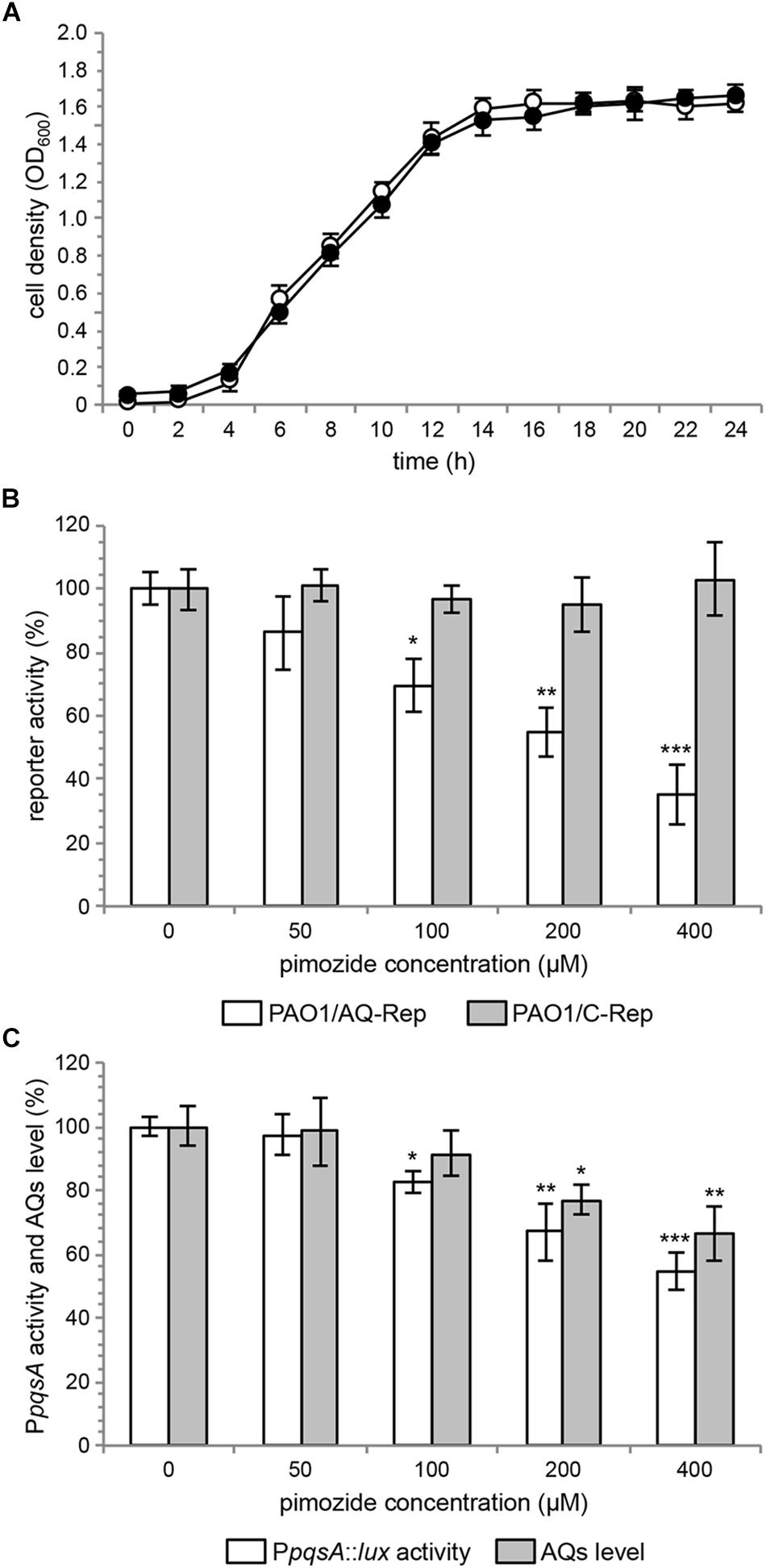
Figure 2. (A) Growth curves of PAO1 wild type incubated at 37°C in shaking conditions in LB supplemented with 400 μM pimozide (black circles) or with the corresponding amount of solvent vehicle [i.e., 1% (v/v) DMSO] (open circles). (B) Activity of the PAO1/AQ-Rep (white bars) and PAO1/C-Rep (gray bars) co-cultures treated with the indicated concentrations of pimozide. Bioluminescence of the untreated co-cultures normalized to cell density are considered as 100%. (C) PpqsA::lux activity (white bars) and AQ production (gray bars) in PAO1 treated with the indicated concentrations of pimozide. PpqsA activity and AQ level measured in untreated PAO1 normalized to cell density are considered as 100%. For (A–C), the average of three independent experiments is reported with SD. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001 (ANOVA).
Pimozide was effective in reducing the activity of the PqsR-controlled PpqsA promoter in a dose-dependent manner also in PAO1 wild type, with reductions in bioluminescence emission of 17.6%, 33.1 and 45.5% for pimozide concentrations of 100 μM, 200 μM, and 400 μM, respectively (Figure 2C, white bars).
To validate the results of the secondary screening, PAO1 was grown in the absence or in the presence of pimozide at different concentrations, and AQ levels were measured in cell-free supernatants by means of the AQ-Rep biosensor strain. In these conditions, pimozide significantly decreased AQ production in a dose-dependent manner only at 200 μM (22.9% reduction) and 400 μM (33.8% reduction) (Figure 2C, gray bars).
Due to its ability to hamper the pqs QS signaling system, pimozide is expected to reduce the expression of pqs-controlled virulence traits, such as pyocyanin production, swarming motility and biofilm formation. As shown in Figure 3A, 100 μM pimozide reduced pyocyanin production in PAO1 (24.8% reduction of pyocyanin level in pimozide-treated cultures relative to the control sample supplemented with the solvent vehicle DMSO). Moreover, exposure to 100 μM pimozide significantly altered the swarming motility phenotype by abolishing dendrites formation (Figure 3B) and reduced biofilm formation in a PAO1 strain constitutively expressing GFP (Figure 3C). Despite exerting a milder effect, the inhibition exerted by pimozide on the tested phenotypes in wild type PAO1 mimicked pqsR deletion (ΔpqsR isogenic strain; Figure 3), thus supporting the hypothesis that PqsR is the likely target of pimozide.
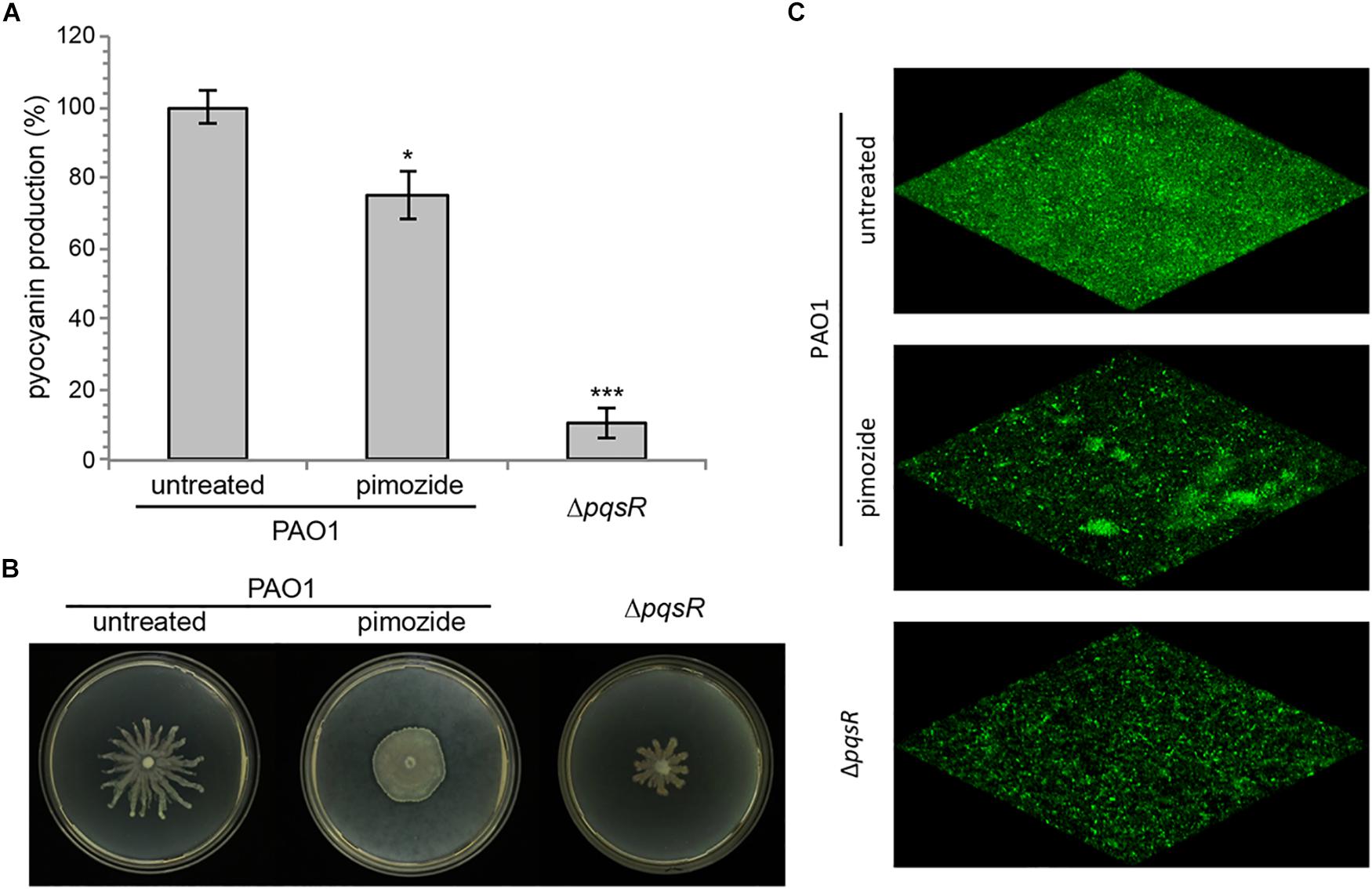
Figure 3. Effect of 100 μM pimozide on pyocyanin production (A), swarming motility (B), and biofilm formation (C) in PAO1. The same phenotypes were evaluated in the ΔpqsR mutant as a control. For pyocyanin production, the average of three independent experiments is reported with SD. ∗p < 0.05; ∗∗∗p < 0.001 (ANOVA). For swarming motility and biofilm formation, representative pictures of three independent experiments are shown.
Validation of the Molecular Mechanism of Action of Pimozide
The inhibitory activity exerted by pimozide on PpqsA activity, combined with the reduction of AQs level and attenuation of pqs-dependent virulence traits, does not allow to rule out the possibility that pimozide affects AQs biosynthesis instead of, or in addition to, AQs reception by PqsR. To tackle this issue, the effect of pimozide on PpqsA promoter activity has been tested in the AQ-Rep biosensor strain grown in the presence of 5 μM synthetic PQS. As shown in Figure 4A, pimozide reduced PpqsA activity also in this experimental setting, in which the AQ molecule PQS required to activate PqsR is not endogenously produced by PAO1. Secondly, AQs production was measured in a PAO1 ΔpqsR mutant strain carrying the pFD-pqsABCD plasmid for constitutive expression of the AQs biosynthetic enzymes. In this genetic background, in which AQ synthesis is PqsR-independent, pimozide did not reduce AQ levels (Figure 4B), indicating that this drug does not affect the activity of the AQs biosynthetic enzymes. Taken together, these experiments indicate that pimozide targets the PqsR-dependent AQs response rather than AQs biosynthesis.
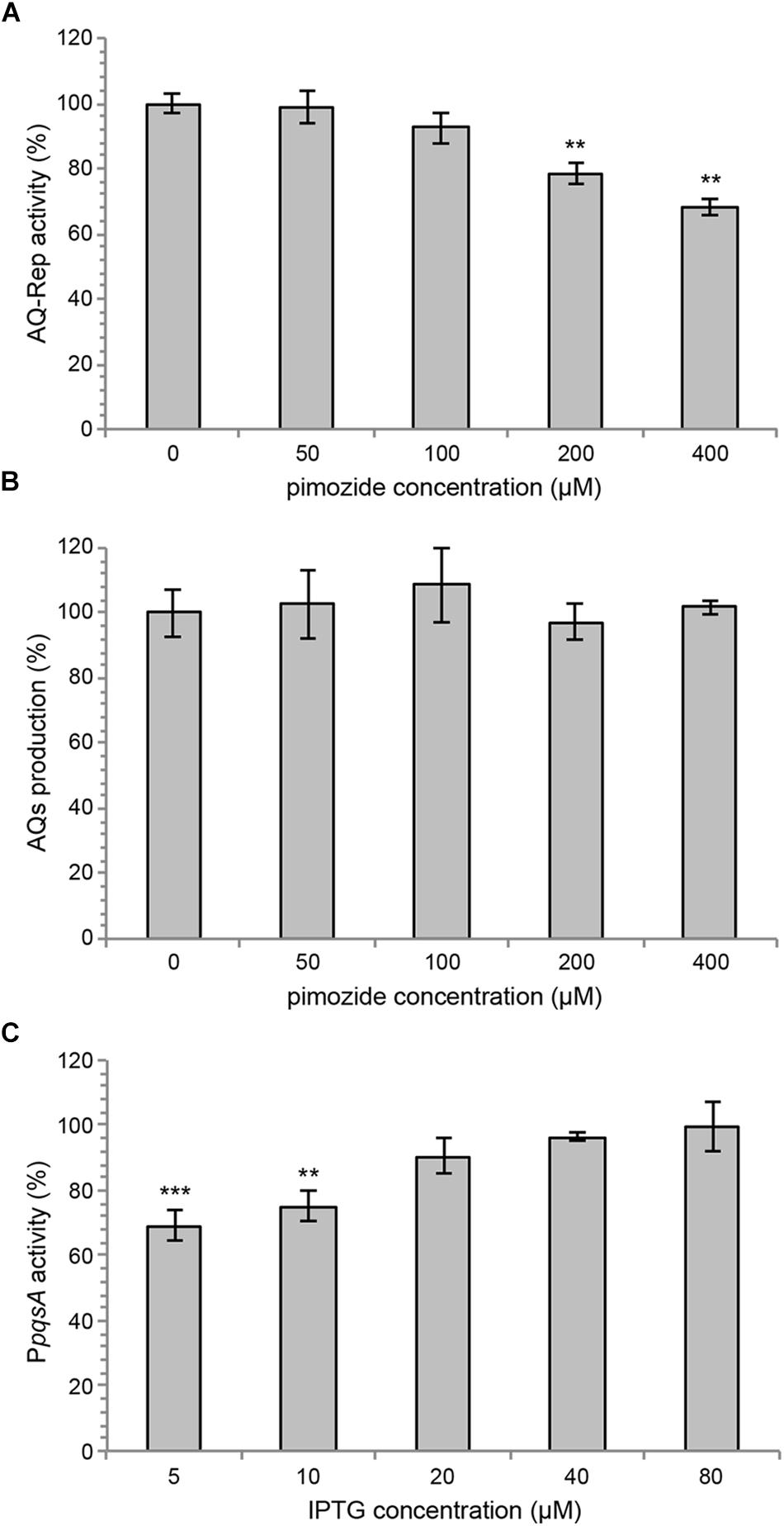
Figure 4. (A) PpqsA activity in the AQ-Rep biosensor grown at 37°C in shaking conditions in LB supplemented with 5 μM synthetic PQS and the indicated concentrations of pimozide. Biosensor activity in the untreated sample is considered as 100%. (B) Production of AQs in P. aeruginosa PAO1 ΔpqsR (pFD-pqsABCD) grown for 16 h in LB in the absence or in the presence of pimozide. The AQ level measured in the untreated sample is considered as 100%. (C) Effect of 100 μM pimozide on PpqsA::lux activity in the PAO1 ΔpqsAHR triple mutant carrying the pPqsR-6H plasmid, grown in LB supplemented with 5 μM PQS and different concentrations of IPTG, as indicated in the graph. For (A–C), the average of three independent experiments is reported with SD. ∗∗p < 0.01; ∗∗∗p < 0.001 (ANOVA).
To further support target specificity, the effect of pimozide on PpqsA activity was evaluated in a P. aeruginosa recombinant strain with tunable levels of PqsR, named PAO1 ΔpqsAHR PpqsA::lux (pPqsR-6H). This strain carries the PpqsA::lux transcriptional fusion and deletion of the pqsA, pqsH and pqsR genes, therefore it does not synthesize AQs and does not produce the native PqsR regulator, which can be expressed upon IPTG induction via the pPqsR-6H plasmid. Therefore, in the absence of IPTG and in the presence of synthetic PQS, the PAO1 ΔpqsAHR PpqsA::lux (pPqsR-6H) strain should express basal level of active PqsR, and the effect of a PqsR inhibitor on PpqsA activity should be maximal due to target paucity. Conversely, increasing concentrations of IPTG in the presence of synthetic PQS should result in increased levels of active PqsR, thus reducing the effect of PqsR inhibitors due to increased target abundance. As shown in Figure 4C, in this recombinant strain the repressive effect exerted by 100 μM pimozide on PpqsA activity was apparent only for IPTG concentrations ≤ 10 μM, while pimozide had no significant effect on the PpqsA promoter for IPTG concentrations ≥ 20 μM. These observations support the hypothesis that pimozide is a ligand and an inhibitor of PqsR.
As shown in Figure 5, pimozide is predicted to bind to the PqsR CBD establishing mainly hydrophobic interactions that closely match those of the natural ligand NHQ (Ilangovan et al., 2013), at least as far as pocket A is concerned. Slightly different interactions are instead observed in pocket B, likely due to a rearrangement of the pocket residues needed to accommodate the bulkier bis(fluorophenyl) moiety of pimozide. Interestingly, pimozide binding to pocket A is predicted to be stabilized also by a π-stacking interaction between the drug benzimidazole group and the Tyr258 aromatic ring (Figure 5, bottom panel), an interaction that mimics the π-stacking interaction experimentally observed between the phenoxy group of the PqsR competitive inhibitor M64 and Tyr258 (Kitao et al., 2018).
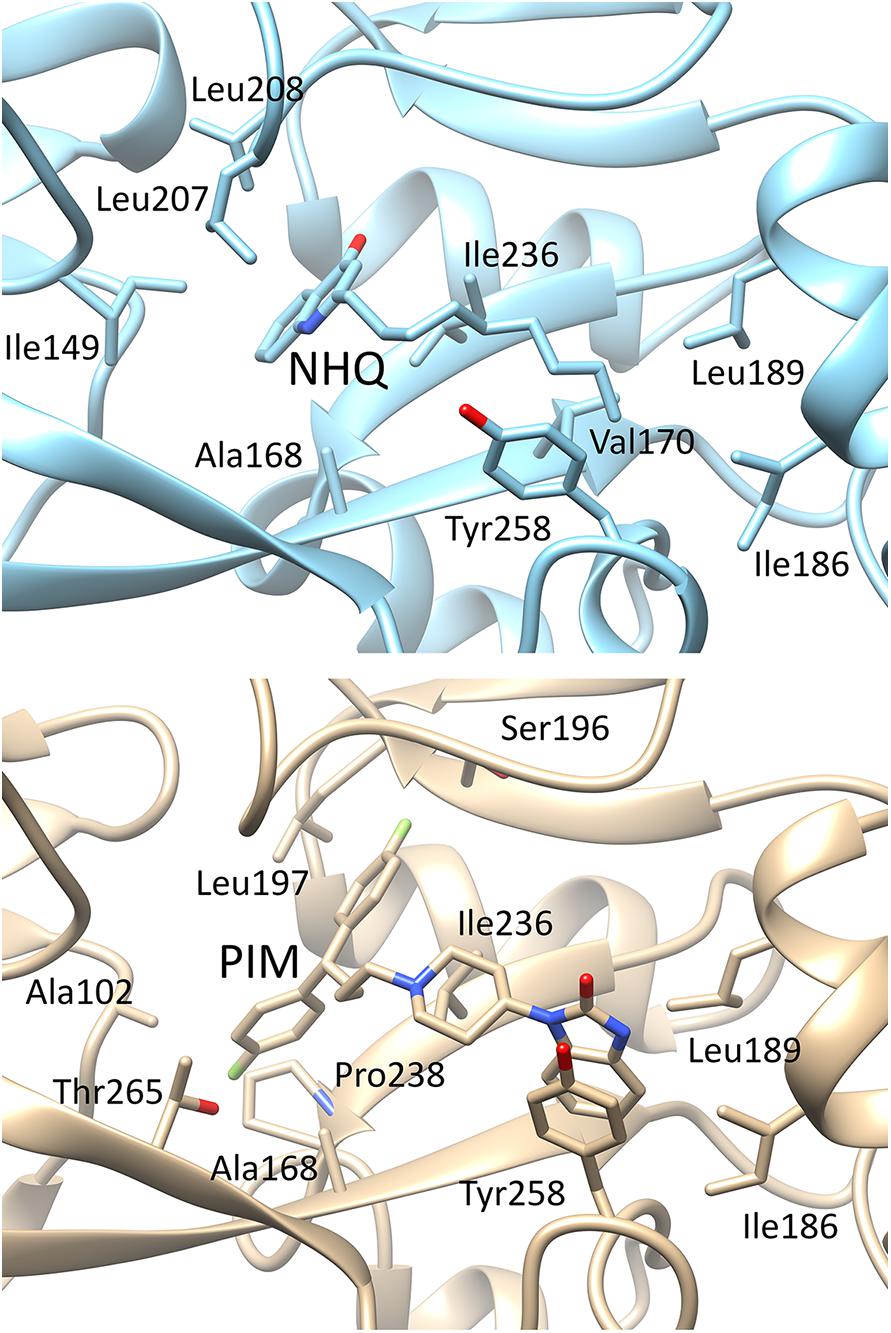
Figure 5. Details of the possible interactions of NHQ (top panel; PDB code 4JVD) and pimozide (bottom panel; predicted) with the PqsR-CBD binding site. The orientation of the macromolecule is the same as in Figure 1; pocket B is on the left side of the macromolecule and pocket A on the right side.
Discussion
Anti-virulence drugs that do not affect bacterial growth hold promise as new therapeutic agents since they are expected to decrease bacterial adaptability to the host environment and to pose a reduced selective pressure for the emergence of resistance with respect to antibiotics. Moreover, virulence mechanisms are often pathogen-specific, thus anti-virulence drugs could avoid dysbiosis usually associated to antibiotic treatments (Rampioni et al., 2014, 2017; Monserrat-Martinez et al., 2019).
The pqs QS system controls the expression of multiple virulence factors and biofilm formation, so that P. aeruginosa mutants defective in the pqs QS system display attenuated pathogenicity in different plant and animal models of infection (Cao et al., 2001; Déziel et al., 2005; Xiao et al., 2006; Lesic et al., 2007; Rampioni et al., 2010; Dubern et al., 2015). Notably, the pqs QS system is active during the infection (Collier et al., 2002; Barr et al., 2015), and while P. aeruginosa mutants impaired in the las QS system are frequently isolated from CF patients (Hoffman et al., 2009; Feltner et al., 2016), the highest proportion of P. aeruginosa strains isolated from CF lung are proficient for AQs production (Guina et al., 2003; Jiricny et al., 2014). Moreover, AQ-based QS systems have not been described in the human microbiota so far, suggesting that drugs targeting PqsR should exert a limited effect on the host microbiota. Intriguingly, recent reports indicate that the pqs QS system might contribute to the RhlR-dependent activation of virulence genes in the absence of functional LasR (Chen et al., 2019; Kostylev et al., 2019), and that this compensatory role might involve a yet uncharacterized signal molecule produced by PqsE and perceived by RhlR in addition to C4-HSL (Mukherjee et al., 2018). Therefore, by hampering PqsE expression, PqsR inhibitors would impact on virulence factors controlled by both the pqs and the rhl QS systems, and could be particularly active against las-deficient strains emerging during chronic infection in CF patients. On this basis, many inhibitors of the pqs QS system have been described in the last decade, proving the ability of anti-pqs drugs to reduce the expression of P. aeruginosa virulence traits both in vitro and in animal models of infection (Calfee et al., 2001; Lesic et al., 2007; Klein et al., 2012; Storz et al., 2012; Ilangovan et al., 2013; Sahner et al., 2013; Weidel et al., 2013; Zender et al., 2013; Lu et al., 2014; Starkey et al., 2014; Sahner et al., 2015; Ji et al., 2016; Thomann et al., 2016; Maura and Rahme, 2017; Maura et al., 2017; D’Angelo et al., 2018; Soukarieh et al., 2018a).
Despite the promise of anti-pqs agents for the treatment of P. aeruginosa infection, none of these molecules has entered clinical trials so far, likely due to poor pharmacological properties and to the lack of ADME-TOX studies required for their evaluation in humans (Maura et al., 2016; Soukarieh et al., 2018b). To overcome this limitation, we recently exploited a drug-repurposing strategy for the identification of anti-pqs drugs via whole-cell biosensor-based screening. This strategy succeeded in identifying the FDA-approved drugs clofoctol, miconazole and clotrimazole as new inhibitors of PqsR (D’Angelo et al., 2018).
Most pqs-inhibitors have been identified via costly and time-consuming biosensor-based screenings or via the rational design and experimental validation of AQ analogs or precursors based on the structure of PqsR and of AQ biosynthetic enzymes. Virtual screenings could reduce the time and costs associated to conventional drug discovery programs, hence in silico techniques have been extensively applied for the identification of molecules hampering the las QS system of P. aeruginosa (Yang et al., 2009; Skovstrup et al., 2013; Tan et al., 2013; Soheili et al., 2015; Gökalsın et al., 2017; Kalia et al., 2017; Xu et al., 2017) or QS systems in other bacteria (Zhu et al., 2012; Ali et al., 2018; Ding et al., 2018, 2019; Medarametla et al., 2018). To the best of our knowledge, only synthetic quinoline-based molecules have so far been identified as PqsR antagonists by means of in silico docking analyses (Soukarieh et al., 2018a).
In this study we combined the advantages of drug-repurposing and in silico screening approaches by exploiting recent knowledge of PqsR-CBD structure and availability of advanced molecular docking tools to identify new FDA-approved drugs with anti-pqs activity. The virtual screening led to selection of five hits for which high binding affinity for PqsR was predicted, and in vitro experiments demonstrated the anti-pqs activity of two of them, namely pimozide and ergotamine. Since pimozide showed the highest inhibitory activity, this drug was experimentally characterized. Phenotypic assays showed that exposure of P. aeruginosa PAO1 to pimozide decreased key PqsR-controlled virulence determinants, such as AQ signal molecules, pyocyanin, swarming motility and biofilm formation, without altering bacterial growth, as one would expect for an anti-virulence drug. Additional experiments performed with ad hoc engineered P. aeruginosa strains and refined in silico docking simulations suggest that pimozide competes with the natural ligands HHQ and PQS for PqsR binding, hence hampering the activity of the PpqsA promoter. Indeed, analysis of the highest ranking pimozide-PqsR docking complex indicated that the drug interacts with the binding pocket occupying the same position of the natural ligand NHQ (Figure 5), and establishes interactions experimentally demonstrated for both the natural ligand and the competitive inhibitor M64 (Ilangovan et al., 2013; Kitao et al., 2018).
The inability of the predicted PqsR ligands conivaptan and dutasteride to hamper the pqs QS system in P. aeruginosa and to decrease bioluminescence in the control biosensor system may be related to drawbacks typically associated to virtual screening approaches, including cell impermeability to the selected compound or its modification/inactivation by cellular metabolism. This is in line with the notion that hits emerging from in vitro screens, as well as from screens employing heterologous organisms, may lack activity or even function as agonists when tested on the target pathogen (Galloway et al., 2011). As an example, the HHQ analog 2-heptyl-6-nitroquinolin-4(1H)-one acted as an antagonist in an Escherichia coli-based AQ-reporter strain, but as an agonist in P. aeruginosa as a consequence of metabolic modification (Lu et al., 2012). However, a subsequent synthetic modification of this molecule resulted in a strong PqsR antagonist also in P. aeruginosa, showing that agonists may still prove useful in the search for antagonists (Lu et al., 2014). More often, the inability of selected hits identified in silico or in vitro to inhibit target functionality in bacterial cell relates to a lack of internalization or to active efflux. This does not seem to be the case for eltrombopag, since the inhibitory effect exerted by this drug on both the specific and control reporter systems indicates its ability to penetrate P. aeruginosa cells, suggesting a QS-independent effect on bioluminescence. In a commentary on the use of whole-cell reporter systems for screens of QS inhibitors, the need for adequate control experiments to assess off-target effects of the tested compounds on reporter activity was emphasized (Defoirdt et al., 2013). For example, pyrogallol was reported to act as a potent inhibitor of AI-2 dependent QS in V. harveyi, but subsequent experiments revealed that the apparent inhibitory activity of pyrogallol was a side effect of its peroxide-generating activity on the reporter system, rather than on QS itself (Defoirdt et al., 2013). In our case, unspecific effects of eltrombopag on the reporter system may mask its impact on PqsR functionality. Obviously, the possibility that the virtual screening approach could select false positive hits cannot be ruled out. When molecular assays for in vitro evaluation of PqsR activity will be available, it will be possible to verify the ability of the five hits identified in this study to hamper PqsR functionality in a cell-free system.
Searching for side activities in FDA-approved drugs represents a shortcut to develop new therapeutic agents, with considerable potential for shortening the time-consuming and expensive hit-to-lead and lead-optimization phases of drug-discovery programs (Rangel-Vega et al., 2015). In the last years an increasing number of studies identified some antibacterial activity in several drugs approved for different purposes, including anticancer, antifungal, cardiovascular and antipsychotic therapies (Miró-Canturri et al., 2019). However, a possible drawback of drug-repurposing approaches relies on the primary activity of the repurposed drug. As an example, the antipsychotic activity of the dopamine antagonist pimozide, clinically used for the treatement of Tourette’s syndrome and schizophrenia (Tueth and Cheong, 1993), could limit its therapeutic use as anti-virulence drug against P. aeruginosa. In fact, it has to be considered that, although rarely, pimozide has been associated to potentially serious adverse effects, including arrhythmia, cardiac arrest, seizures, and neutropenia (Singer, 2010). Neutropenia, in particular, is a worrisome adverse effect for patients suffering a bacterial infection. In addition, the peak serum concentration of pimozide in conventional treatment as an antipsychotic drug is in the nanomolar range (Yan et al., 2010), far below the concentration required to inhibit the pqs QS system in P. aeruginosa. In spite of these limitations, the pimozide molecular scaffold could serve as the basis for chemical modifications aimed at lowering its dopamine antagonistic activity, while improving membrane permeability and affinity for the PqsR active site, in line with the selective optimization of side activity (SOSA) approach (Wermuth, 2006). However, such a hit-to-lead optimization process would partly compromise the advantage of drug reuporisng, since chemical modification of pimozide would invalidate the FDA-approval, with additional pharmacological testing being required by regulatory agencies. It must be recognized that repurposing of old drugs for new therapies can result in seamless adoption into the clinical practice only if their off-target effect overcomes their primary activity.
That said, pimozide has already been repurposed to inhibit Listeria monocytogenes virulence by decreasing cell invasion, vacuole escape and cell-to-cell spread in phagocytic host cells (Lieberman and Higgins, 2009), to inhibit the growth of the protozoan parasite Toxoplasma gondii (Dittmar et al., 2016) and for the treatment of breast cancer (Dakir et al., 2018; Elmaci and Altinoz, 2018). Notably, pimozide also inhibited Chikungunya virus (CHIKV) replication in a mouse model of Chikungunya infection when administered in combination with the fatty acid synthesis inhibitor 5-tetradecyloxy-2-furoic acid, with low toxicity in vivo (Karlas et al., 2016).
In conclusion, despite low potency of pimozide as a pqs inhibitor and predictable side-effects due to its primary antipsychotic activity, this study demonstrates for the first time the potential of virtual screening campaigns to rapidly select new FDA-approved QS inhibitors.
Data Availability Statement
All datasets generated for this study are included in the manuscript/supplementary files.
Author Contributions
FP and GR conceived the study. FP, GR, LL, and PV designed the experiments and contributed reagents, materials, and analysis tools. MM, ED, FD, VB, and SF performed the experiments. FP, GR, LL, MM, FD, ED, and VB analyzed the data. GR, FP, and MM wrote the manuscript. All authors corrected, amended the draft of the manuscript, and approved the submitted version.
Funding
This work was supported by the Italian Ministry for Education, University and Research (Futuro in Ricerca n. RBFR10LHD1_002 to GR, PRIN 2017 grant protocol 2017483NH8 to FP, and PRIN 2017 grant protocol 20177J5Y3P to PV), Italian Cystic Fibrosis Research Foundation (FFC 17/2018 to LL). The Grant of Excellence Departments, MIUR-Italy (ARTICOLO 1, COMMI 314 – 337 LEGGE 232/2016) is gratefully acknowledged.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Prof. Paul Williams and Miguel Càmara (Centre for Biomolecular Sciences, University of Nottingham, United Kingdom) for kindly providing the synthetic AQs HHQ and PQS. We are grateful to the Reviewers for constructive criticisms to the manuscript.
Footnotes
- ^ http://www.who.int/en/news-room/detail/27-02-2017-who-publishes-list-of-bacteria-for-which-new-antibiotics-are-urgently-needed
- ^ https://www.drugbank.ca/
References
Ali, F., Yao, Z., Li, W., Sun, L., Lin, W., and Lin, X. (2018). In silico prediction and modeling of the quorum sensing LuxS protein and inhibition of AI-2 biosynthesis in Aeromonas hydrophila. Molecules 23:E2627. doi: 10.3390/molecules23102627
Allen, R. C., Popat, R., Diggle, S. P., and Brown, S. P. (2014). Targeting virulence: can we make evolution-proof drugs? Nat. Rev. Microbiol. 12, 300–308. doi: 10.1038/nrmicro3232
Azzouni, F., and Mohler, J. (2012). Role of 5α-reductase inhibitors in prostate cancer prevention and treatment. Urology 79, 1197–1205. doi: 10.1016/j.urology.2012.01.024
Barr, H. L., Halliday, N., Cámara, M., Barrett, D. A., Williams, P., Forrester, D. L., et al. (2015). Pseudomonas aeruginosa quorum sensing molecules correlate with clinical status in cystic fibrosis. Eur. Respir. J. 46, 1046–1054. doi: 10.1183/09031936.00225214
Boucher, H. W., Talbot, G. H., Bradley, J. S., Edwards, J. E., Gilbert, D., Rice, L. B., et al. (2009). Bad bugs, no drugs: no ESKAPE! An update from the infectious diseases society of America. Clin. Infect. Dis. 48, 1–12. doi: 10.1086/595011
Bredenbruch, F., Nimtz, M., Wray, V., Morr, M., Müller, R., and Häussler, S. (2005). Biosynthetic pathway of Pseudomonas aeruginosa 4-hydroxy-2-alkylquinolines. J. Bacteriol. 187, 3630–3635. doi: 10.1128/jb.187.11.3630-3635.2005
Calfee, M. W., Coleman, J. P., and Pesci, E. C. (2001). Interference with Pseudomonas quinolone signal synthesis inhibits virulence factor expression by Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U.S.A. 98, 11633–11637. doi: 10.1073/pnas.201328498
Cao, H., Krishnan, G., Goumnerov, B., Tsongalis, J., Tompkins, R., and Rahme, L. G. (2001). A quorum sensing-associated virulence gene of Pseudomonas aeruginosa encodes a LysR-like transcription regulator with a unique self-regulatory mechanism. Proc. Natl. Acad. Sci. U.S.A. 98, 14613–14618. doi: 10.1073/pnas.251465298
Chen, R., Déziel, E., Groleau, M. C., Schaefer, A. L., and Greenberg, E. P. (2019). Social cheating in a Pseudomonas aeruginosa quorum-sensing variant. Proc. Natl. Acad. Sci. U.S.A. 116, 7021–7026. doi: 10.1073/pnas.1819801116
Collier, D. N., Anderson, L., McKnight, S. L., Noah, T. L., Knowles, M., Boucher, R., et al. (2002). A bacterial cell to cell signal in the lungs of cystic fibrosis patients. FEMS Microbiol. Lett. 215, 41–46. doi: 10.1016/s0378-1097(02)00937-0
Dakir, E. H., Pickard, A., Srivastava, K., McCrudden, C. M., Gross, S. R., Lloyd, S., et al. (2018). The anti-psychotic drug pimozide is a novel chemotherapeutic for breast cancer. Oncotarget 9, 34889–34910. doi: 10.18632/oncotarget.26175
D’Angelo, F., Baldelli, V., Halliday, N., Pantalone, P., Polticelli, F., Fiscarelli, E., et al. (2018). Identification of FDA-approved drugs as antivirulence agents targeting the pqs quorum-sensing system of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 62:e1296-18. doi: 10.1128/AAC.01296-18
Davies, D. G., Parsek, M. R., Pearson, J. P., Iglewski, B. H., Costerton, J. W., and Greenberg, E. P. (1998). The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science 280, 295–298. doi: 10.1126/science.280.5361.295
Defoirdt, T., Brackman, G., and Coenye, T. (2013). Quorum sensing inhibitors: how strong is the evidence? Trends Microbiol. 21, 619–624. doi: 10.1016/j.tim.2013.09.006
Déziel, E., Gopalan, S., Tampakaki, A. P., Lépine, F., Padfield, K. E., Saucier, M., et al. (2005). The contribution of MvfR to Pseudomonas aeruginosa pathogenesis and quorum sensing circuitry regulation: multiple quorum sensing-regulated genes are modulated without affecting lasRI, rhlRI or the production of N-acyl-L-homoserine lactones. Mol. Microbiol. 55, 998–1014. doi: 10.1111/j.1365-2958.2004.04448.x
Di Muzio, E., Toti, D., and Polticelli, F. (2017). DockingApp: a user friendly interface for facilitated docking simulations with AutoDock Vina. J. Comput. Aided Mol. Des. 31, 213–218. doi: 10.1007/s10822-016-0006-1
Diggle, S. P., Matthijs, S., Wright, V. J., Fletcher, M. P., Chhabra, S. R., Lamont, I. L., et al. (2007). The Pseudomonas aeruginosa 4-quinolone signal molecules HHQ and PQS play multifunctional roles in quorum sensing and iron entrapment. Chem. Biol. 14, 87–96. doi: 10.1016/j.chembiol.2006.11.014
Ding, T., Li, T., and Li, J. (2018). Identification of natural product compounds as quorum sensing inhibitors in Pseudomonas fluorescens P07 through virtual screening. Bioorg. Med. Chem. 26, 4088–4099. doi: 10.1016/j.bmc.2018.06.039
Ding, T., Li, T., and Li, J. (2019). Virtual screening for quorum-sensing inhibitors of Pseudomonas fluorescens P07 from a food-derived compound database. J. Appl. Microbiol. 127, 763–777. doi: 10.1111/jam.14333
Dittmar, A. J., Drozda, A. A., and Blader, I. J. (2016). Drug repurposing screening identifies novel compounds that effectively inhibit Toxoplasma gondii growth. mSphere 1:e042-15. doi: 10.1128/mSphere.00042-15
Drees, S. L., and Fetzner, S. (2015). PqsE of Pseudomonas aeruginosa acts as pathway-specific thioesterase in the biosynthesis of alkylquinolone signaling molecules. Chem. Biol. 22, 611–618. doi: 10.1016/j.chembiol.2015.04.012
Dubern, J. F., Cigana, C., De Simone, M., Lazenby, J., Juhas, M., Schwager, S., et al. (2015). Integrated whole-genome screening for Pseudomonas aeruginosa virulence genes using multiple disease models reveals that pathogenicity is host specific. Environ. Microbiol. 17, 4379–4393. doi: 10.1111/1462-2920.12863
Dulcey, C. E., Dekimpe, V., Fauvelle, D. A., Milot, S., Groleau, M. C., Doucet, N., et al. (2013). The end of an old hypothesis: the Pseudomonas signaling molecules 4-hydroxy-2-alkylquinolines derive from fatty acids, not 3-ketofatty acids. Chem. Biol. 20, 1481–1491. doi: 10.1016/j.chembiol.2013.09.021
Elmaci, I., and Altinoz, M. A. (2018). Targeting the cellular schizophrenia. Likely employment of the antipsychotic agent pimozide in treatment of refractory cancers and glioblastoma. Crit. Rev. Oncol. Hematol. 128, 96–109. doi: 10.1016/j.critrevonc.2018.06.004
Essar, D. W., Eberly, L., Hadero, A., and Crawford, I. P. (1990). Identification and characterization of genes for a second anthranilate synthase in Pseudomonas aeruginosa: interchangeability of the two anthranilate synthases and evolutionary implications. J. Bacteriol. 172, 884–900. doi: 10.1128/jb.172.2.884-900.1990
Feltner, J. B., Wolter, D. J., Pope, C. E., Groleau, M. C., Smalley, N. E., Greenberg, E. P., et al. (2016). LasR variant cystic fibrosis isolates reveal an adaptable quorum-sensing hierarchy in Pseudomonas aeruginosa. mBio 7:e1513. doi: 10.1128/mBio.01513-16
Ferguson-Myrthil, N. (2010). Novel agents for the treatment of hyponatremia: a review of conivaptan and tolvaptan. Cardiol. Rev. 18, 313–321. doi: 10.1097/CRD.0b013e3181f5b3b7
Fletcher, M. P., Diggle, S. P., Crusz, S. A., Chhabra, S. R., Cámara, M., and Williams, P. (2007). A dual biosensor for 2-alkyl-4-quinolone quorum-sensing signal molecules. Environ. Microbiol. 9, 2683–2693. doi: 10.1111/j.1462-2920.2007.01380.x
Galloway, W. R., Hodgkinson, J. T., Bowden, S. D., Welch, M., and Spring, D. R. (2011). Quorum sensing in Gram-negative bacteria: small-molecule modulation of AHL and AI-2 quorum sensing pathways. Chem. Rev. 111, 28–67.
Gökalsın, B., Aksoydan, B., Erman, B., and Sesal, N. C. (2017). Reducing virulence and biofilm of Pseudomonas aeruginosa by potential quorum sensing inhibitor Carotenoid:Zeaxanthin. Microb. Ecol. 74, 466–473. doi: 10.1007/s00248-017-0949-3
Guina, T., Purvine, S. O., Yi, E. C., Eng, J., Goodlett, D. R., Aebersold, R., et al. (2003). Quantitative proteomic analysis indicates increased synthesis of a quinolone by Pseudomonas aeruginosa isolates from cystic fibrosis airways. Proc. Natl. Acad. Sci. U.S.A. 100, 2771–2776. doi: 10.1073/pnas.0435846100
Hazan, R., He, J., Xiao, G., Dekimpe, V., Apidianakism, Y., Lesic, B., et al. (2010). Homeostatic interplay between bacterial cell-cell signaling and iron in virulence. PLoS Pathog. 6:e1000810. doi: 10.1371/journal.ppat.1000810
Heeb, S., Fletcher, M. P., Chhabra, S. R., Diggle, S. R., Williams, P., and Cámara, M. (2011). Quinolones: from antibiotics to autoinducers. FEMS Microbiol. Rev. 35, 247–274. doi: 10.1111/j.1574-6976.2010.00247.x
Hoffman, L. R., Kulasekara, H. D., Emerson, J., Houston, L. S., Burns, J. L., Ramsey, B. W., et al. (2009). Pseudomonas aeruginosa lasR mutants are associated with cystic fibrosis lung disease progression. J. Cyst. Fibros. 8, 66–70. doi: 10.1016/j.jcf.2008.09.006
Hung, D. T., Shakhnovich, E. A., Pierson, E., and Mekalanos, J. J. (2005). Small-molecule inhibitor of Vibrio cholerae virulence and intestinal colonization. Science 310, 670–674. doi: 10.1126/science.1116739
Ilangovan, A., Fletcher, M., Rampioni, G., Pustelny, C., Rumbaugh, K., Heeb, S., et al. (2013). Structural basis for native agonist and synthetic inhibitor recognition by the Pseudomonas aeruginosa quorum sensing regulator PqsR (MvfR). PLoS Pathog. 9:e1003508. doi: 10.1371/journal.ppat.1003508
Imperi, F., Fiscarelli, E. V., Visaggio, D., Leoni, L., and Visca, P. (2019). Activity and impact on resistance development of two antivirulence fluoropyrimidine drugs in Pseudomonas aeruginosa. Front. Cell. Infect. Microbiol. 9:49. doi: 10.3389/fcimb.2019.00049
Imperi, F., Massai, F., Ramachandran Pillai, C., Longo, F., Zennaro, E., Rampioni, G., et al. (2013). New life for an old drug: the anthelmintic drug niclosamide inhibits Pseudomonas aeruginosa quorum sensing. Antimicrob. Agents Chemoter. 57, 996–1005. doi: 10.1128/AAC.01952-12
Ji, C., Sharma, I., Pratihar, D., Hudson, L. L., Maura, D., Guney, T., et al. (2016). Designed small-molecule inhibitors of the anthranilyl-CoA synthetase PqsA block quinolone biosynthesis in Pseudomonas aeruginosa. ACS Chem. Biol. 11, 3061–3067. doi: 10.1021/acschembio.6b00575
Jiricny, N., Molin, S., Foster, K., Diggle, S. P., Scanlan, P. D., Ghoul, M., et al. (2014). Loss of social behaviours in populations of Pseudomonas aeruginosa infecting lungs of patients with cystic fibrosis. PLoS One 9:e83124. doi: 10.1371/journal.pone.0083124
Jurcisek, J. A., Dickson, A. C., Bruggeman, M. E., and Bakaletz, L. O. (2011). In vitro biofilm formation in an 8-well chamber slide. J. Vis. Exp. 47:e2481. doi: 10.3791/2481
Kalia, M., Singh, P. K., Yadav, V. K., Yadav, B. S., Sharma, D., Narvi, S. S., et al. (2017). Structure based virtual screening for identification of potential quorum sensing inhibitors against LasR master regulator in Pseudomonas aeruginosa. Microb. Pathog. 107, 136–143. doi: 10.1016/j.micpath.2017.03.026
Kalia, V. C., Patel, S. K. S., Kang, Y. C., and Lee, J. K. (2019). Quorum sensing inhibitors as antipathogens: biotechnological applications. Biotechnol. Adv. 37, 68–90. doi: 10.1016/j.biotechadv.2018.11.006
Karlas, A., Berre, S., Couderc, T., Varjak, M., Braun, P., Meyer, M., et al. (2016). A human genome-wide loss-of-function screen identifies effective chikungunya antiviral drugs. Nat. Commun. 7:11320. doi: 10.1038/ncomms11320
Kitao, T., Lepine, F., Babloudi, S., Walte, F., Steinbacher, S., Maskos, K., et al. (2018). Molecular insights into function and competitive inhibition of Pseudomonas aeruginosa multiple virulence factor regulator. mBio 9:e02158-17. doi: 10.1128/mBio.02158-17
Klein, T., Henn, C., de Jong, J. C., Zimmer, C., Kirsch, B., Maurer, C. K., et al. (2012). Identification of small-molecule antagonists of the Pseudomonas aeruginosa transcriptional regulator PqsR: biophysically guided hit discovery and optimization. ACS Chem. Biol. 7, 1496–1501. doi: 10.1021/cb300208g
Kostylev, M., Kim, D. Y., Smalley, N. E., Salukhe, I., Greenberg, E. P., and Dandekar, A. A. (2019). Evolution of the Pseudomonas aeruginosa quorum-sensing hierarchy. Proc. Natl. Acad. Sci. U.S.A. 116, 7027–7032.
Kuntz, I. D., Chen, K., Sharp, K. A., and Kollman, P. A. (1999). The maximal affinity of ligands. Proc. Natl. Acad. Sci. U.S.A. 96, 9997–10002. doi: 10.1073/pnas.96.18.9997
Lesic, B., Lépine, F., Déziel, E., Zhang, J., Zhang, Q., Padfield, K., et al. (2007). Inhibitors of pathogen intercellular signals as selective anti-infective compounds. PLoS Pathog. 3, 1229–1239.
Lieberman, L. A., and Higgins, D. E. (2009). A small-molecule screen identifies the antipsychotic drug pimozide as an inhibitor of Listeria monocytogenes infection. Antimicrob. Agents Chemother. 53, 756–764. doi: 10.1128/AAC.00607-08
Lin, J., Zhang, W., Cheng, J., Yang, X., Zhu, K., Wang, Y., et al. (2017). A Pseudomonas T6SS effector recruits PQS-containing outer membrane vesicles for iron acquisition. Nat. Commun. 8:14888. doi: 10.1038/ncomms14888
Lu, C., Kirsch, B., Zimmer, C., de Jong, J. C., Henn, C., Maurer, C. K., et al. (2012). Discovery of antagonists of PqsR, a key player in 2-alkyl-4-quinolone-dependent quorum sensing in Pseudomonas aeruginosa. Chem. Biol. 19, 381–390. doi: 10.1016/j.chembiol.2012.01.015
Lu, C., Maurer, C. K., Kirsch, B., Steinbach, A., and Hartmann, R. W. (2014). Overcoming the unexpected functional inversion of a PqsR antagonist in Pseudomonas aeruginosa: an in vivo potent antivirulence agent targeting pqs quorum sensing. Angew. Chem. Int. Ed. Engl. 53, 1109–1112. doi: 10.1002/anie.201307547
Luepke, K. H., Suda, K. J., Boucher, H., Russo, R. L., Bonney, M. W., Hunt, T. D., et al. (2017). Past, present, and future of antibacterial economics: increasing bacterial resistance, limited antibiotic pipeline, and societal implications. Pharmacotherapy 37, 71–84. doi: 10.1002/phar.1868
Maeda, T., García-Contreras, R., Pu, M., Sheng, L., Garcia, L. R., Tomás, M., et al. (2012). Quorum quenching quandary: resistance to antivirulence compounds. ISME J. 6, 493–501. doi: 10.1038/ismej.2011.122
Mashburn, L. M., and Whiteley, M. (2005). Membrane vesicles traffic signals and facilitate group activities in a prokaryote. Nature 437, 422–425. doi: 10.1038/nature03925
Maura, D., Ballok, A. E., and Rahme, L. G. (2016). Considerations and caveats in anti-virulence drug development. Curr. Opin. Microbiol. 33, 41–46. doi: 10.1016/j.mib.2016.06.001
Maura, D., Drees, S. L., Bandyopadhaya, A., Kitao, T., Negri, M., Starkey, M., et al. (2017). Polypharmacology approaches against the Pseudomonas aeruginosa MvfR regulon and their application in blocking virulence and antibiotic tolerance. ACS Chem. Biol. 12, 1435–1443. doi: 10.1021/acschembio.6b01139
Maura, D., and Rahme, L. G. (2017). Pharmacological inhibition of the Pseudomonas aeruginosa MvfR quorum sensing system interferes with biofilm formation and potentiates antibiotic-mediated biofilm disruption. Antimicrob. Agents Chemother. 61:e01362-17. doi: 10.1128/AAC.01362-17
McCormack, P. L. (2015). Eltrombopag: a review of its use in patients with severe aplastic anaemia. Drugs 75, 525–531. doi: 10.1007/s40265-015-0363-4
Medarametla, P., Gatta, V., Kajander, T., Laitinen, T., Tammela, P., and Poso, A. (2018). Structure-based virtual screening of LsrK kinase inhibitors to target quorum sensing. ChemMedChem 13, 2400–2407. doi: 10.1002/cmdc.201800548
Mellbye, B., and Schuster, M. (2011). The sociomicrobiology of antivirulence drug resistance: a proof of concept. mBio 2:e0131-11. doi: 10.1128/mBio.00131-11
Miró-Canturri, A., Ayerbe-Algaba, R., and Smani, Y. (2019). Drug repurposing for the treatment of bacterial and fungal infections. Front. Microbiol. 10:41. doi: 10.3389/fmicb.2019.00041
Mohr, K. I. (2016). History of antibiotics research. Curr. Top. Microbiol. Immunol. 398, 237–272. doi: 10.1007/82_2016_499
Monserrat-Martinez, A., Gambin, Y., and Sierecki, E. (2019). Thinking outside the bug: molecular targets and strategies to overcome antibiotic resistance. Int. J. Mol. Sci. 20:E1255. doi: 10.3390/ijms20061255
Mukherjee, S., Moustafa, D. A., Stergioula, V., Smith, C. D., Goldberg, J. B., and Bassler, B. L. (2018). The PqsE and RhlR proteins are an autoinducer synthase-receptor pair that control virulence and biofilm development in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U.S.A. 115, E9411–E9418. doi: 10.1073/pnas.1814023115
Pettersen, E. F., Goddard, T. D., Huang, C. C., Couch, G. S., Greenblatt, D. M., Meng, E. C., et al. (2004). UCSF Chimera - a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612. doi: 10.1002/jcc.20084
Rampioni, G., Falcone, M., Heeb, S., Frangipani, E., Fletcher, M. P., Dubern, J. F., et al. (2016). Unravelling the genome-wide contributions of specific 2-alkyl-4-quinolones and PqsE to quorum sensing in Pseudomonas aeruginosa. PLoS Pathog. 12:e1006029. doi: 10.1371/journal.ppat.1006029
Rampioni, G., Leoni, L., and Williams, P. (2014). The art of antibacterial warfare: deception through interference with quorum sensing-mediated communication. Bioorg. Chem. 55, 60–68. doi: 10.1016/j.bioorg.2014.04.005
Rampioni, G., Pustelny, C., Fletcher, M. P., Wright, V. J., Bruce, M., Rumbaugh, K. P., et al. (2010). Transcriptomic analysis reveals a global alkyl-quinolone-independent regulatory role for PqsE in facilitating the environmental adaptation of Pseudomonas aeruginosa to plant and animal hosts. Environ. Microbiol. 12, 1659–1673. doi: 10.1111/j.1462-2920.2010.02214.x
Rampioni, G., Schuster, M., Greenberg, E. P., Zennaro, E., and Leoni, L. (2009). Contribution of the RsaL global regulator to Pseudomonas aeruginosa virulence and biofilm formation. FEMS Microbiol. Lett. 301, 210–217. doi: 10.1111/j.1574-6968.2009.01817.x
Rampioni, G., Visca, P., Leoni, L., and Imperi, F. (2017). Drug repurposing for antivirulence therapy against opportunistic bacterial pathogens. Emerg. Top. Life Sci. 1, 13–22. doi: 10.1042/etls20160018
Rangel-Vega, A., Bernstein, L. R., Mandujano-Tinoco, E. A., García-Contreras, S. J., and García-Contreras, R. (2015). Drug repurposing as an alternative for the treatment of recalcitrant bacterial infections. Front. Microbiol. 6:282. doi: 10.3389/fmicb.2015.00282
Rasko, D. A., and Sperandio, V. (2010). Anti-virulence strategies to combat bacteria-mediated disease. Nat. Rev. Drug Discov. 9, 117–128. doi: 10.1038/nrd3013
Reuter, K., Steinbach, A., and Helms, V. (2015). Interfering with bacterial quorum sensing. Perspect. Med. Chem. 8, 1–15.
Rice, L. B. (2008). Federal funding for the study of antimicrobial resistance in nosocomial pathogens: no ESKAPE. J. Infect. Dis. 197, 1079–1081. doi: 10.1086/533452
Sahner, J. H., Brengel, C., Storz, M. P., Groh, M., Plaza, A., Müller, R., et al. (2013). Combining in silico and biophysical methods for the development of Pseudomonas aeruginosa quorum sensing inhibitors: an alternative approach for structure-based drug design. J. Med. Chem. 56, 8656–8664. doi: 10.1021/jm401102e
Sahner, J. H., Empting, M., Kamal, A., Weidel, E., Groh, M., Börger, C., et al. (2015). Exploring the chemical space of ureidothiophene-2-carboxylic acids as inhibitors of the quorum sensing enzyme PqsD from Pseudomonas aeruginosa. Eur. J. Med. Chem. 96, 14–21. doi: 10.1016/j.ejmech.2015.04.007
Saxena, V. K., and De Deyn, P. P. (1992). Ergotamine: its use in the treatment of migraine and its complications. Acta Neurol. 14, 140–146.
Singer, H. S. (2010). Treatment of tics and tourette syndrome. Curr. Treat. Options Neurol. 12, 539–561. doi: 10.1007/s11940-010-0095-4
Skovstrup, S., Le Quement, S. T., Hansen, T., Jakobsen, T. H., Harmsen, M., Tolker-Nielsen, T., et al. (2013). Identification of LasR ligands through a virtual screening approach. ChemMedChem 8, 157–163. doi: 10.1002/cmdc.201200434
Soheili, V., Bazzaz, B. S., Abdollahpour, N., and Hadizadeh, F. (2015). Investigation of Pseudomonas aeruginosa quorum-sensing signaling system for identifying multiple inhibitors using molecular docking and structural analysis methodology. Microb. Pathog. 89, 73–78. doi: 10.1016/j.micpath.2015.08.017
Soukarieh, F., Vico Oton, E., Dubern, J. F., Gomes, J., Halliday, N., de Pilar Crespo, M., et al. (2018a). In silico and in vitro-guided identification of inhibitors of alkylquinolone-dependent quorum sensing in Pseudomonas aeruginosa. Molecules 23:E257. doi: 10.3390/molecules23020257
Soukarieh, F., Williams, P., Stocks, M. J., and Cámara, M. (2018b). Pseudomonas aeruginosa quorum sensing systems as drug discovery targets: current position and future perspectives. J. Med. Chem. 61, 10385–10402. doi: 10.1021/acs.jmedchem.8b00540
Starkey, M., Lepine, F., Maura, D., Bandyopadhaya, A., Lesic, B., He, J., et al. (2014). Identification of anti-virulence compounds that disrupt quorum-sensing regulated acute and persistent pathogenicity. PLoS Pathog. 10:e1004321. doi: 10.1371/journal.ppat.1004321
Storz, M. P., Maurer, C. K., Zimmer, C., Wagner, N., Brengel, C., de Jong, J. C., et al. (2012). Validation of PqsD as an anti-biofilm target in Pseudomonas aeruginosa by development of small-molecule inhibitors. J. Am. Chem. Soc. 134, 16143–16146. doi: 10.1021/ja3072397
Tan, S. Y., Chua, S. L., Chen, Y., Rice, S. A., Kjelleberg, S., Nielsen, T. R., et al. (2013). Identification of five structurally unrelated quorum-sensing inhibitors of Pseudomonas aeruginosa from a natural-derivative database. Antimicrob. Agents Chemother. 57, 5629–5641. doi: 10.1128/AAC.00955-13
Thomann, A., de Mello Martins, A. G., Brengel, C., Empting, M., and Hartmann, R. W. (2016). Application of dual inhibition concept within looped autoregulatory systems toward antivirulence agents against Pseudomonas aeruginosa infections. ACS Chem. Biol. 11, 1279–1286. doi: 10.1021/acschembio.6b00117
Trott, O., and Olson, A. J. (2010). AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comp. Chem. 31, 455–461. doi: 10.1002/jcc.21334
Tueth, M., and Cheong, J. (1993). Clinical uses of pimozide. South. Med. J. 86, 344–349. doi: 10.1097/00007611-199303000-00019
Vale, P. F., McNally, L., Doeschl-Wilson, A., King, K. C., Popat, R., Domingo-Sananes, M. R., et al. (2016). Beyond killing: can we find new ways to manage infection? Evol. Med. Public Health 2016, 148–157. doi: 10.1093/emph/eow012
Ventola, C. L. (2015). The antibiotic resistance crisis: part 1: causes and threats. P T 40, 277–283.
Weidel, E., de Jong, J. C., Brengel, C., Storz, M. P., Braunshausen, A., Negri, M., et al. (2013). Structure optimization of 2-benzamidobenzoic acids as PqsD inhibitors for Pseudomonas aeruginosa infections and elucidation of binding mode by SPR, STD NMR, and molecular docking. J. Med. Chem. 56, 6146–6155. doi: 10.1021/jm4006302
Wermuth, C. G. (2006). Selective optimization of side activities: the SOSA approach. Drug Discov. Today 11, 160–164. doi: 10.1016/s1359-6446(05)03686-x
Xiao, G., He, J., and Rahme, L. G. (2006). Mutation analysis of the Pseudomonas aeruginosa mvfR and pqsABCDE gene promoters demonstrates complex quorum-sensing circuitry. Microbiology 152, 1679–1686. doi: 10.1099/mic.0.28605-0
Xu, Y., Tong, X., Sun, P., Bi, L., and Lin, K. (2017). Virtual screening and biological evaluation of biofilm inhibitors on dual targets in quorum sensing system. Future Med. Chem. 9, 1983–1994. doi: 10.4155/fmc-2017-0127
Yan, M., Li, H. D., Chen, B. M., Liu, X. L., Xu, P., and Zhu, Y. G. (2010). Quantitative determination of pimozide in human plasma by liquid chromatography-mass spectrometry and its application in a bioequivalence study. J. Pharm. Biomed. Anal. 51, 1161–1164. doi: 10.1016/j.jpba.2009.11.015
Yang, L., Rybtke, M. T., Jakobsen, T. H., Hentzer, M., Bjarnsholt, T., Givskov, M., et al. (2009). Computer-aided identification of recognized drugs as Pseudomonas aeruginosa quorum-sensing inhibitors. Antimicrob. Agents Chemother. 53, 2432–2443. doi: 10.1128/AAC.01283-08
Zender, M., Klein, T., Henn, C., Kirsch, B., Maurer, C. K., Kail, D., et al. (2013). Discovery and biophysical characterization of 2-amino-oxadiazoles as novel antagonists of PqsR, an important regulator of Pseudomonas aeruginosa virulence. J. Med. Chem. 56, 6761–6774. doi: 10.1021/jm400830r
Zhu, J., Beaber, J. W., Moré, M. I., Fuqua, C., Eberhard, A., and Winans, S. C. (1998). Analogs of the autoinducer 3-oxooctanoyl-homoserine lactone strongly inhibit activity of the TraR protein of Agrobacterium tumefaciens. J. Bacteriol. 180, 5398–5405.
Keywords: Pseudomonas aeruginosa, anti-virulence strategy, quorum sensing inhibition, pimozide, in silico screening, molecular docking, new therapeutics, PqsR
Citation: Mellini M, Di Muzio E, D’Angelo F, Baldelli V, Ferrillo S, Visca P, Leoni L, Polticelli F and Rampioni G (2019) In silico Selection and Experimental Validation of FDA-Approved Drugs as Anti-quorum Sensing Agents. Front. Microbiol. 10:2355. doi: 10.3389/fmicb.2019.02355
Received: 01 August 2019; Accepted: 27 September 2019;
Published: 10 October 2019.
Edited by:
Rustam Aminov, University of Aberdeen, United KingdomReviewed by:
Natalia V. Kirienko, Rice University, United StatesFadi Soukarieh, University of Nottingham, United Kingdom
Copyright © 2019 Mellini, Di Muzio, D’Angelo, Baldelli, Ferrillo, Visca, Leoni, Polticelli and Rampioni. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fabio Polticelli, ZmFiaW8ucG9sdGljZWxsaUB1bmlyb21hMy5pdA==; Giordano Rampioni, Z2lvcmRhbm8ucmFtcGlvbmlAdW5pcm9tYTMuaXQ=
†Present address: Francesca D’Angelo, Institut Pasteur, Paris, France
 Marta Mellini
Marta Mellini Elena Di Muzio1
Elena Di Muzio1 Valerio Baldelli
Valerio Baldelli Paolo Visca
Paolo Visca Livia Leoni
Livia Leoni Fabio Polticelli
Fabio Polticelli Giordano Rampioni
Giordano Rampioni