- 1State Key Laboratory of Bioreactor Engineering, East China University of Science and Technology, Shanghai, China
- 2Department of Applied Biology, East China University of Science and Technology, Shanghai, China
Lincomycin is one of the most important antibiotics in clinical practice. To further understand the regulatory mechanism on lincomycin biosynthesis, we investigated a pleiotropic transcriptional regulator AdpAlin in the lincomycin producer Streptomyces lincolnensis NRRL 2936. Deletion of adpAlin (which generated ΔadpAlin) interrupted lincomycin biosynthesis and impaired the morphological differentiation. We also found that putative AdpA binding sites were unusually scattered in the promoters of all the 8 putative operons in the lincomycin biosynthetic gene cluster (BGC). In ΔadpAlin, transcript levels of structural genes in 8 putative operons were decreased with varying degrees, and electrophoretic mobility shift assays (EMSAs) confirmed that AdpAlin activated the overall putative operons via directly binding to their promoter regions. Thus, we speculated that the entire lincomycin biosynthesis is under the control of AdpAlin. Besides, AdpAlin participated in lincomycin biosynthesis by binding to the promoter of lmbU which encoded a cluster sited regulator (CSR) LmbU of lincomycin biosynthesis. Results of qRT-PCR and catechol dioxygenase activity assay showed that AdpAlin activated the transcription of lmbU. In addition, AdpAlin activated the transcription of the bldA by binding to its promoter, suggesting that AdpAlin indirectly participated in lincomycin biosynthesis and morphological differentiation. Uncommon but understandable, AdpAlin auto-activated its own transcription via binding to its own promoter region. In conclusion, we provided a molecular mechanism around the effect of AdpAlin on lincomycin biosynthesis in S. lincolnensis, and revealed a cascade regulation of lincomycin biosynthesis by AdpAlin, LmbU, and BldA.
Introduction
Lincomycin is a naturally occurring antibiotic isolated from soil sample, and it was first introduced into clinical practice in 1963 (Macleod et al., 1964). Lincomycin and its derivatives belong to lincosamide antibiotics and exhibit biological activities against anaerobic and some protozoal infections by inhibiting protein synthesis in sensitive targets (Spizek and Rezanka, 2017). Clindamycin is a semi-synthetic chlorinated derivative of lincomycin and it is marked by being one of the 20 most important antibiotics (Spizek and Rezanka, 2004a). Given the extensive clinical application of lincomycin, multiple attempts have been taken into industrial practice to increase the production yields of lincomycin or to optimize the products (Spizek and Rezanka, 2004b; Li et al., 2007). Genetic manipulations are also adopted as complement to enhance the production of lincomycin (Pang et al., 2015; Xu et al., 2018). Though the pathway of lincomycin biosynthesis was assembled mainly within the recent 10 years (Neusser et al., 1998; Novotna et al., 2004; Sasaki et al., 2012; Lin et al., 2014; Pang et al., 2015; Zhao et al., 2015; Jiraskova et al., 2016), studies on the regulation mechanism of lincomycin biosynthesis are prompted quite slowly. Thus, various methods have limited effect on yield improvement of lincomycin.
Biosynthesis of antibiotics is controlled by elaborate regulatory mechanisms. Hormone-like signaling molecules, for example γ-butyrolactone (Takano et al., 2000; Hsiao et al., 2009; Du et al., 2011), serve as stimuli that interact with their receptor proteins to prelude the secondary metabolism (Niu et al., 2016). Global regulators and/or pleiotropic regulators then deliver these signals to CSRs which directly control the onset of antibiotic biosynthesis. In addition, researches on the secondary metabolism in Streptomyces is expanded to the post-transcriptional regulation. For example, BldA, a rare tRNA in Streptomyces, has significant importance on morphological differentiation and antibiotic biosynthesis (Hackl and Bechthold, 2015). As for lincomycin biosynthesis, only limited researches contribute to decipher the regulatory network. Lu et al. (2008) found that LmbU contributes to lincomycin biosynthesis. Hou et al. (2018a, 2019) demonstrated that LmbU, as a CSR, positively regulates lincomycin biosynthesis by controlling the transcription of lmbA, lmbC, lmbJ, lmbK, lmbW, and lmbU itself, and subsequently solved the subtle mechanism of LmbU regulon. Besides, Hou et al. (2018b) also found that BldA functions as a global regulator on both morphological differentiation and lincomycin biosynthesis at the level of translation, and genes lmbB2, lmbY, and lmbU, which all contain TTA rare codon, get involved in the regulon. Meng et al. (2017) revealed the regulatory network between nitrate metabolism and lincomycin biosynthesis where GlnR activates the transcription of lmrA, the lincomycin exporter gene. Lately, a TetR-type regulator SLCG_2919 has been identified as a repressor of lincomycin biosynthesis that controls the transcription of lmbA, lmbC, lmbE, lmbG, lmbK, lmbR, lmbV, and lmbW (Xu et al., 2019). However, to complete the regulatory network of lincomycin biosynthesis, there are lots of gaps remained.
AdpA was previously found to be an A-factor dependent regulator and repressed by ArpA (Kato et al., 2007). AdpA amplifies the A-factor signal and thus participates in morphological differentiation and secondary metabolism. By means of chromatin affinity precipitation (ChAP) and chromatin immunoprecipitation (ChIP), Higo et al. (2012) found that AdpA controls more than 500 genes in Streptomyces griseus. Afterward AdpA is considered to be a regulator of great importance in Streptomyces. In S. chattanoogensis, AdpAch controls the expression of wblAch, and thus participates in the regulation of natamycin biosynthesis (Yu et al., 2014). In S. roseosporus, AdpA controls the expression of atrA and indirectly control daptomycin biosynthesis (Mao et al., 2015). In a recent research, AdpA interacts with the two-component system PhoRP and both of them contribute to the transcription of atrA (Zheng et al., 2019). Effects of AdpA on antibiotic biosynthesis is always a popular topic. AdpA always serves as an activator of antibiotic biosynthesis, a classic case is that AdpA activates the transcription of strR, which encodes the CSR of streptomycin biosynthesis. Therefore, AdpA regulates streptomycin biosynthesis positively and indirectly (Tomono et al., 2005). Similarly, for the biosynthesis of grixazone (Higashi et al., 2007), nikkomycin (Pan et al., 2009), and natamycin (Yu et al., 2018), AdpA activates the transcription of CSRs in their BGCs and indirectly regulates antibiotic biosynthesis. On the other hand, AdpA has a negative impact on oviedomycin biosynthesis in S. ansochromogenes by repressing the transcription of CSR (Xu et al., 2017). Very recently, it was reported that AdpA from S. xiamenensis 318 negatively regulates morphological differentiation as well as polycyclic tetramate macrolactams (PTMs) production, while positively regulates xiamenmycin production by activating the transcription of two of the structural genes ximA and ximB (Bu et al., 2019). As we can speculate from previous studies, AdpA typically controls the CSR and/or some structural genes in an antibiotic BGC. So, we scanned the lincomycin BGC and found that putative AdpA binding sites were extraordinarily scattered upstream all of the 8 putative operons with different amounts and locations. Thus, we focused on the pleiotropic regulator AdpA (GenBank accession no. ANS65440.1) and attempted to investigate its regulatory mechanism of lincomycin biosynthesis in Streptomyces lincolnensis in this study.
Materials and Methods
Bacterial Strains, Plasmids, and Culture Conditions
Bacterial strains and plasmids used in this study are listed in Table 1. S. lincolnensis NRRL 2936 which served as wild type (WT) and its mutants were incubated at 28°C on mannitol soya flour (MS) medium (Kieser et al., 2000) for 3–5 days for routine cultivation, phenotype observation, and strain preservation, and then cultivated at 28°C in YEME liquid medium [10 g/L yeast extract (OXOID, United States), 5 g/L polypeptone (Nihon Pharmaceutical, Japan), 10 g/L glucose (Lingfeng, China), 3 g/L maltose (Generay, China), 5 mM MgCl2⋅2H2O (Lingfeng, China), 340 g/L sucrose (Titan, China), dissolved in dH2O] with shaking (210 rpm) for 3–5 days for routine cultivation, total DNA extraction, and sporeless strain preservation. Fermentation medium FM1 [20 g/L lactose (SCRC, China), 20 g/L glucose, 10 g/L polypeptone, 10 g/L corn steep liquor (Aladdin, China), dissolved in dH2O] is used for primary cultivation, and FM2 [20 g/L lactose, 20 g/L glucose, 10 g/L polypeptone, 10 g/L corn steep liquor, 4 g/L CaCO3 (Lingfeng, China), dissolved in dH2O] is used for dry cell weight determination and lincomycin production assays. ISP4 medium [10 g/L soluble starch (Lingfeng, China), 1 g/L K2HPO4 (Lingfeng, China), 5 g/L MgSO4⋅7H2O (Lingfeng, China), 1 g/L NaCl (Titan, China), 2 g/L (NH4)2SO4 (Lingfeng, China), 2 g/L CaCO3, 15 g/L Agar (Shize, China), 0.001 g/L FeSO4⋅7H2O, 0.001 g/L MnCl2⋅4H2O (Lingfeng, China), 0.001 g/L ZnSO4⋅7H2O (Lingfeng, China), 0.02 mol/L MgCl2, dissolved in dH2O] was used for conjugation of Escherichia coli and S. lincolnensis. Antibiotics were supplemented on demand with the following final concentration: 20 μg/mL apramycin (Sangon Biotech, China), 20 μg/mL kanamycin (Kinglyuan, China), 12 μg/mL chloramphenicol (Sigma-Aldrich, United States), and/or 20 μg/mL nalidixic acid (Aladdin, China).
Escherichia coli strains were cultivated at 37°C in Luria-Bertani (LB) liquid medium with shaking (180 rpm) or on LB solid media. Antibiotics were supplemented on demand with the following final concentration: 50 μg/mL apramycin, 50 μg/mL kanamycin, and/or 30 μg/mL chloramphenicol.
Micrococcus luteus 28001 were cultivated at 37°C in medium III (5 g/L polypeptone, 1.5 g/L beef extract (SCRC, China), 3 g/L yeast extract, 3.5 g/L NaCl, 3.68 g/L K2HPO4, 1.32 g/L KH2PO4 (Lingfeng, China), 1 g/L glucose, 18 g/L agar) for 16 to 18 h.
Deletion and Complementation of AdpAlin
To construct AdpAlin disruption strain ΔadpA in S. lincolnensis, DNA fragments of upstream and downstream region of AdpAlin were amplified separately using primers ad-F1/R1 and ad-F2/R2. Then digested, respectively, with restriction enzyme Hind III/Xba I and BamHI/EcoR I (Thermo Fisher Scientific, United States) and ligated into the E. coli-Streptomyces shuttle plasmid pMJ1 to generate plasmid pADNU. Then E. coli ET12567/pUZ8002 was used to introduce pADNU into S. lincolnensis NRRL 2936 by conjugal transfer (Hou et al., 2018b). As a result of homologous recombination ΔadpA was constructed. DNA sequencing with primers IDad-F1/IDneo-R1 and IDneo-F2/IDad-R2 was adopted for further identification.
To construct adpA complementation strain ΔadpA:adpA, a DNA fragment covering AdpAlin was amplified by PCR with primers ad-C-F/R and then digested with Nde I/EcoR I (Thermo Fisher Scientific, United States). Then ligated into the corresponding sites of the integrative vector pSET152. The resulting plasmid pADC was introduced into ΔadpA by conjugal transfer and integrated into the chromosome to generate ΔadpA:adpA where the complemented AdpAlin was under the control of the promoter ermE∗p. DNA sequencing with primers 152-F/R was adopted for further identification.
All primers used in this study are listed in Supplementary Table S1, and synthesized by Genewiz (China).
Scanning Electron Microscope (SEM)
Scanning electron microscope assay referred to a previously established method (Hou et al., 2018b) with some optimizations. S. lincolnensis NRRL 2936, ΔadpA, and ΔadpA:adpA were cultured on SMA medium at 28°C for about 5 days. Equivalent areas of the lawn were harvested and placed in 2.5% glutaraldehyde solution overnight. Dehydrated by vacuum freezing drying and sprayed with platinum by Gatan ALTO 1000E (Gatan, United States). Then observed with Hitachi S-3400N scanning electron microscopy (Hitachi, Japan).
Dry Cell Weight Determination and Lincomycin Bioassay Analysis
Streptomyces lincolnensis NRRL 2936, ΔadpA, and ΔadpA:adpA were inoculated from SMA medium into FM1 at 28°C with shaking (210 rpm) for 3 days and then inoculated into FM2 and cultured for 6 days at 28°C with shaking (210 rpm). Precipitate of each sample was harvested every 24 h and dried at 55°C for 24 h. Then the weights of the dried precipitates represent the dry cell weights. Meanwhile, to analyze the bioassay of lincomycin, supernatant of each sample was harvested at the same time, and previously mentioned method (Pharmacopoeia of the People’s Republic of China [PPRC], 1990; Hou et al., 2018a) with some modifications was adopted. M. luteus 28001, used as indicator, was cultured on medium III at 37°C for 16–18 h and the lawn was washed off with 0.9% NaCl and suspended readily to use. Lincomycin standard solutions (4, 6, 8, 10, 12, 14, and 16 μg/mL) were used for the standard curve and internal control. Diameters of inhibition zone were linearized with the logarithmic values of the concentrations of the lincomycin standard solutions. Concentration of each sample was calculated on the basis of the standard curves. All assays in this section were performed in duplication and standard errors of the mean were calculated. The software GraphPad Prism 7.00 was used to draw the line graph of dry cell weight and histogram of lincomycin bioassay.
RNA Extraction and Quantitative Real-Time PCR (qRT-PCR)
Streptomyces lincolnensis NRRL 2936, and ΔadpA cultured on the second day in FM2 medium were used to extract total RNA. Precipitate of samples were ground in liquid nitrogen (Liu et al., 2013) and followed by the method using TRIzol (Thermo Fisher Scientific, United States) (Setinova et al., 2017). After reacting with Recombinant DNase I (Takara, Japan) to remove the trace amount of DNA, 800 ng of RNA samples (analyzed by NanoDrop 2000, Thermo Fisher Scientific, United States) were reverse transcribed to cDNA using Reverse Transcriptase M-MLV (Takara, Japan). SYBR green PCR master mix (ToYoBo, Japan) was used and qRT-PCR was performed in triplication for each transcript. qRT-PCR conditions were mentioned previously (Hou et al., 2018a). To detect the transcript level of AdpAlin targets, primers qbl-F/R, qU-F/R, qA-F/R, qC-F/R, qD-F/R, qJ-F/R, qK-F/R, qV-F/R, and qW-F/R in Supplementary Table S1 were used. And primers qhrdB-F/R were used to detect the transcript level of hrdB which served as an internal control. qRT-PCR was performed with samples in triplication and data were treated with the threshold cycle (2–ΔΔCT) method (Livak and Schmittgen, 2001) and standard errors of the mean were calculated. GraphPad Prism 7.00 was used to draw the histogram of relative expression level of each AdpAlin target.
Catechol Dioxygenase Activity Analysis
DNA fragment covering reporter gene xylTE was amplified by PCR with primers xyl-F/R. Promoters of different AdpAlin targets were amplified separately by PCR with primers adp-xyl-F/R (for adpAp, from −610 to +4), blp-xyl-F/R (for bldAp, from −799 to +52), Up-xyl-F/R (for lmbUp, from −730 to +17), Ap-xyl-F/R (for lmbAp, from −533 to +3), Cp-xyl-F/R (for lmbCp, from −513 to −1), Dp-xyl-F/R (for lmbDp, from −581 to +3), Jp-xyl-F/R (for lmbJp, from −391 to +3), Kp-xyl-F/R (for lmbKp, from −896 to +3), Vp-xyl-F/R (for lmbVp, from −364 to −1), and Wp-xyl-F/R (for lmbWp, from −456 to +4). In respect to bldAp, +1 represents the start point of mature bldA. As for other promoters, +1 represents the translation starting point of the genes controlled by them. Promoter fragment and xylTE fragment were inserted into Pvu II site of the integrative vector pSET152 using Super Efficiency Fast Seamless Cloning kits (DoGene, China) (Hou et al., 2018b) to construct reporter plasmids pADPX, pBLPX, pUPX, pAPX, pCPX, pDPX, pJPX, pKPX, pVPX, pWPX, and pADPX2. Then introduced into S. lincolnensis NRRL 2936 or ΔadpA respectively, to investigate the effects of AdpAlin on these targets. Referred to the method optimized by Hou et al. (2018b), Catechol dioxygenase activity analysis was carried out in triplication. Standard errors of the mean were calculated and the software GraphPad Prism 7.00 was used to draw the histogram of catechol dioxygenase activity.
Electrophoretic Mobility Shift Assays (EMSAs)
The AdpAlin gene was amplified by PCR with primers ad-C-F/R and digested with Nde I/EcoR I. DNA fragment was cloned into corresponding sites of pET-28a (+) vector (Novagen, United States), and the resulting plasmid pADH was transformed into E. coli BL21 (DE3). Overexpression and purification of recombinant protein refer to the procedures described previously (Hou et al., 2018a). According to our experience, DNA probes with length around 200 bp are appropriate for EMSAs with AdpAlin. For the first round of amplification, primers adp-A-F/R, adp-B-F/R, adp-B-1-F/R, adp-B-F/adp-B-2-R, adp-B-F/madp-B-2-R, blp-A-F/R, blp-B-F/R, blp-N-F/R, mblp-B-F/blp-B-R, Up-A-F/R, Up-B-F/R, Up-C-F/R, mUp-B-F/R, Ap-A-F/R, mAp-F/R, Cp-A-F/R, Cp-B-F/R, Cp-C-F/R, Dp-A-F/R, Jp-A-F/R, mJp-A-F/R, Jp-B-F/R, Kp-B-F/R, mKp-B-F/R, and Vp-A-F/R in Supplementary Table S1 were used to amplify DNA probes with putative AdpA binding sites, and primers nad-F/R were used to amplify DNA probe with no AdpA binding site as a negative control. Genes lmbV and lmbW share the same DNA probe. For the second round, amplified DNA fragments are used as templates with primer Biotin-linker∗ to harvest DNA probes with biotin labeled at 5′ terminal. EMSAs were performed as previously described (Liao et al., 2015) using chemiluminescent EMSA kits (Beyotime Biotechnology, China). AdpAlin of different concentrations (0, 1.6, 3.2, and/or 6.4 μM, respectively) interacted with 2.5 nM biotin labeled DNA probe in binding buffer TGB [20 mM Tris–HCl (Shize, China), 5% glycerol (Titan, China), and 0.1% BSA (Sangon, China), pH 7.5], and 200-folds excess of unlabeled probes were added as competitive assays.
Results
AdpAlin Positively Regulates Both Lincomycin Biosynthesis and Morphological Differentiation in S. lincolnensis
It has been shown that AraC/XylS family regulators control various metabolic pathways including antibiotic biosynthesis (Ibarra et al., 2008). There are about 30 AraC/XylS family regulators in S. lincolnensis, among which, AdpA is the most famous one. Based on this, we investigated the effects of AdpAlin on lincomycin biosynthesis and attempted to propose some innovative idea on this classic regulator. Alignment of AdpA from 26 Streptomyces species (Supplementary Figures S1A,B) showed that AdpA retained the conserved N-terminal ThiJ/PfpI/DJ-1-like (also referenced as GATase-1) dimerization domain and C-terminal AraC/XylS-type DNA-binding domain (DBD) (Ohnishi et al., 2005). The first 340 amino acids possessed an ortholog with over 90% identities, and the main diversity occurred at the tail end of C-terminus with a length of no more than 90 amino acids, after the conserved DNA binding domain (Supplementary Figure S1A). To infer the evolutionary history of AdpA, phylogenetic analysis was performed using a maximum likelihood method. The results showed that AdpAlin possessed an ortholog with 89% amino acid identity to AdpAsg, and thus we classified AdpAlin to be one member of the AraC/XylS family.
To investigate the effects of AdpAlin on lincomycin biosynthesis, AdpAlin null mutant was constructed and named as ΔadpA. Lincomycin biosynthesis was significantly influenced by the non-functional AdpAlin. In medium FM2, lincomycin started to be produced on the second day in WT, reached a maximum bioassay of 30.10 μg/mL between the second and the fourth day, and maintained thereafter. However, bioassay of lincomycin in ΔadpA remained undetectable throughout the entire 6 days (Figure 1A). Furthermore, the lawns of ΔadpA on SMA medium exhibited a bald phenotype distinct from WT (Figure 1B). Deletion of AdpAlin blocked the sporulation and caused long, extended aerial hyphae when detected by SEM (Figure 1C). Complementation of AdpAlin in ΔadpA strain (ΔadpA:adpA) restored both lincomycin biosynthesis and sporulation as expected (Figure 1) though lincomycin production in complemented strain did not restore to WT level, probably due to using the promoter ermE∗p. Moreover, the biomasses of WT, ΔadpA, and ΔadpA:adpA were measured at all the four detected days. The data showed that biomasses of the three strains have no significant differences at days 1 and 6, while at day 2, ΔadpA had decreased biomass compared to WT and ΔadpA:adpA, and at day 4, ΔadpA:adpA had increased biomass compared to WT and ΔadpA. These data suggested that AdpAlin is an important regulator of lincomycin biosynthesis in S. lincolnensis.
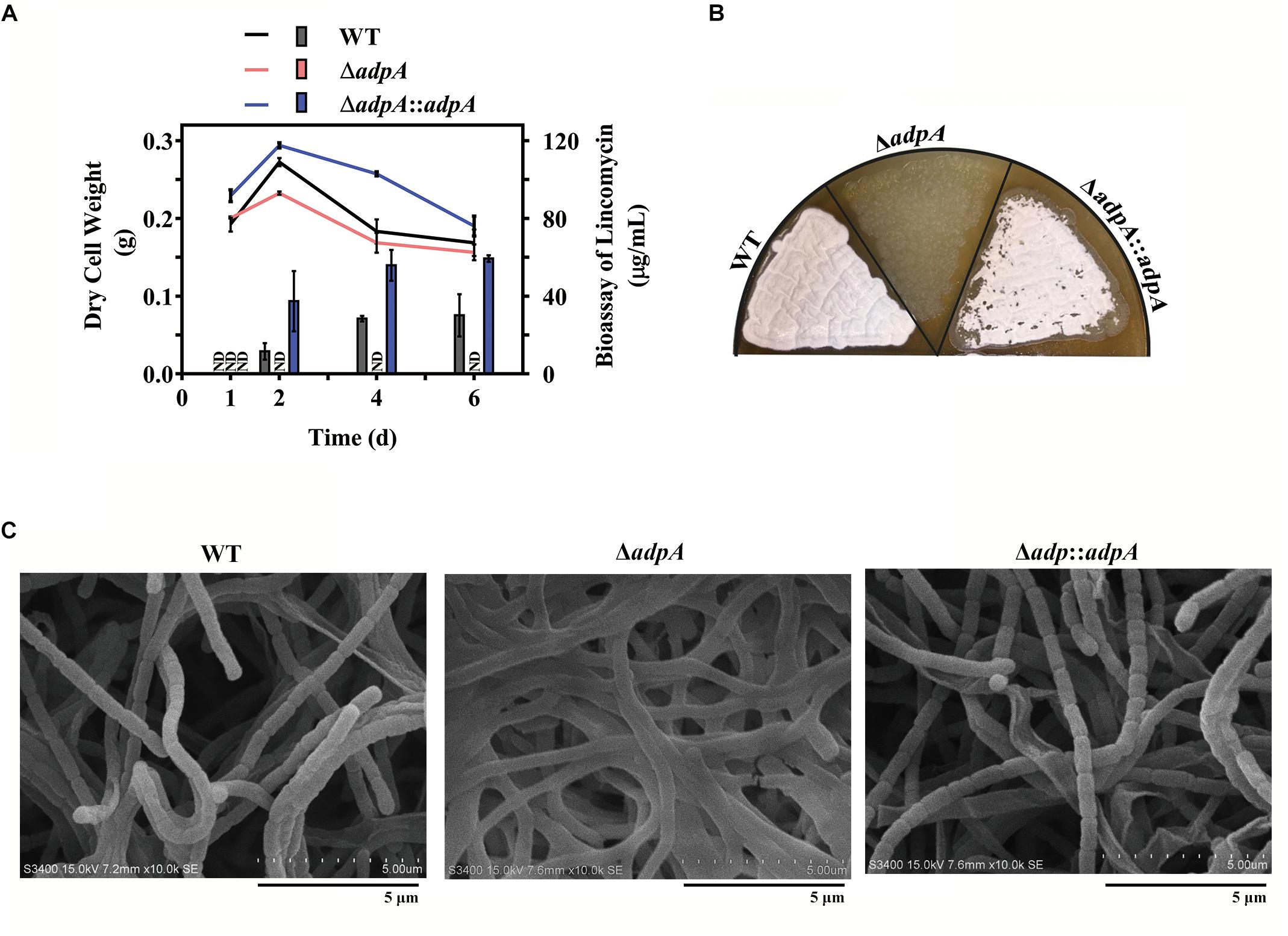
Figure 1. Effects of AdpAlin on lincomycin biosynthesis and morphological differentiation. (A) Changing of dry cell weight (line graph), which represented growth curve. And bioassay of lincomycin (histogram) of WT, ΔadpA and ΔadpA:adpA, which showed the production of lincomycin. ND represents not detected. (B,C) Growth of WT and AdpAlin mutants cultivated on SMA medium. (B) Lawns of WT and AdpAlin mutants grown on SMA medium. (C) Scanning electron micrograph with a scale bar of 5 μm.
AdpAlin Directly Activates Transcription of the Structural Genes in the lmb Cluster
AdpA regulates more than 500 genes in S. griseus (Higo et al., 2012), and AdpA binding motifs have been well studied in other Streptomyces species such as S. griseus (Yamazaki et al., 2004), S. coelicolor (Kim et al., 2005), and S. lividans (Guyet et al., 2013). Additionally, Ming et al. have solved the complex structure of AdpA-DBD and target DNA in S. griseus (Yao et al., 2013). AdpA binding site is recognized as 5′-TGGCSNGWWY-3′ (where S is G or C, W is A or T, Y is T or C, and N is any nucleotide), and G at position 2 and C at position 4 are more highly conserved than the other nucleotides in this motif (Yao et al., 2013). Lincomycin BGC was named as the lmb cluster, and the gene organization was shown in Supplementary Figure S2A. The lmb cluster contains 8 putative operons and the first genes of them are lmbA, lmbC, lmbD, lmbJ, lmbK, lmbV, lmbW, and lmbU, respectively. We looked into the nucleic acid sequence of the lmb cluster and searched with the conserved AdpA binding sequence, and no more than 3 mismatches in the last 3 bp were allowed. We found that putative AdpA binding sites were scattered in the upstream region of all the 8 putative operons (Supplementary Figures S2B–F). The numbers of putative AdpA binding sites in the upstream of lmbA, lmbC, lmbD, lmbJ, lmbK, lmbV, lmbW, and lmbU are 1, 6, 1, 3, 3, 2, 2, and 10, respectively. Therefore, we speculated that the entire biosynthesis process of lincomycin might be under the control of AdpAlin.
qRT-PCR analysis showed that the transcript level of lmbA, lmbC, lmbD, lmbJ, lmbV, and lmbW dramatically decreased in ΔadpA with fold changes 129.41, 43.64, 60.30, 215.29, 11.95, and 301.20, respectively (Figure 2A). It was suggested that in ΔadpA, lincomycin biosynthesis was blocked because of the decreased expression of structural genes lmbA, lmbC, lmbD, lmbJ, lmbV, and lmbW. Due to the low transcript level of lmbK (data not shown), we failed to calculate the relative expression in both WT and ΔadpA. To investigate the regulation between AdpAlin and lmbK, we cloned the 895 bp of DNA sequences upstream from lmbK translation starting site (TSS) and constructed reporting plasmid for catechol dioxygenase activity assay. The results demonstrated that AdpAlin got involved in regulating the transcription of lmbK (Figure 2B). Besides, transcriptions of lmbA, lmbC, lmbD, lmbJ, lmbV, and lmbW were activated by AdpAlin in catechol dioxygenase activity assay as well (Figure 2B). These data suggested that AdpAlin has primary effects on the structural genes in the lmb cluster.
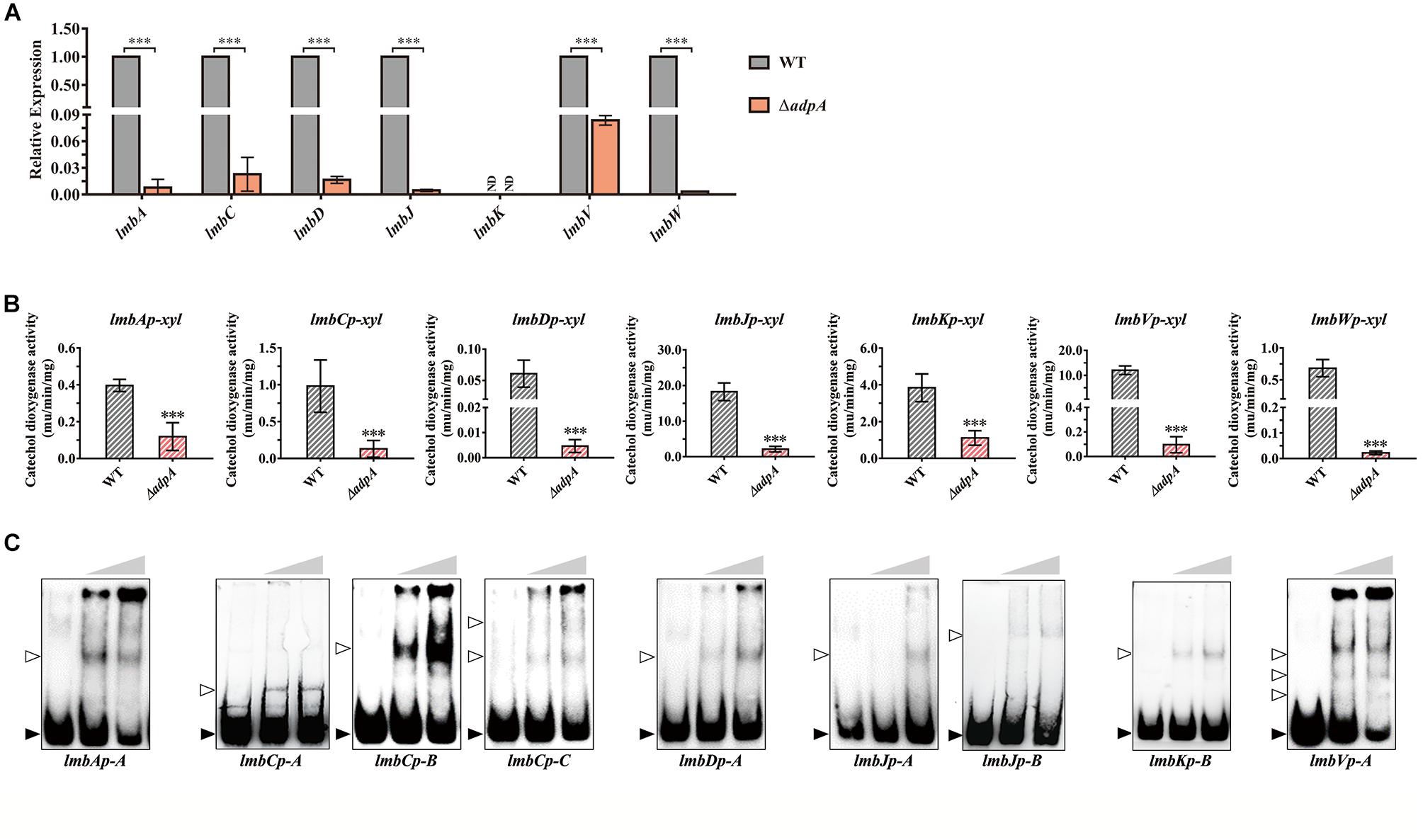
Figure 2. AdpAlin activates the transcription of structural genes in lmb cluster. (A) Relative expression of lmbA, lmbC, lmbD, lmbJ, lmbK, lmbV, and lmbW in WT and ΔadpA. ND means not detected. (B) Catechol dioxygenase activity assays of WT and ΔadpA transformed with corresponding reporter plasmids. (C) EMSAs of AdpAlin with 5′-biotin labeled probes in Supplementary Figure S2. Solid triangles point to the bands of probes and hollow triangles point to the complexes of AdpAlin and probes. Concentrations of AdpAlin are 0, 1.6, and 3.2 μM, respectively. ∗∗∗P < 0.001.
Then we carried out EMSAs to investigate the interplay between AdpAlin and promotors of above lmb structural genes. DNA fragments containing putative AdpA binding sites were labeled with biotin and incubated with purified AdpAlin. Results of EMSAs showed that AdpAlin interacted with all of the 8 promoter regions containing putative AdpA binding sites (Figure 2C). DNA probe lmbJp-A served as part of the promoter regions of both lmbJ and lmbK, and similarly, DNA probe lmbVp-A served as the promoter regions of both lmbV and lmbW. For promoters containing more than one AdpA binding sites, such as genes lmbC, lmbJ, lmbK, lmbV, and lmbW, AdpAlin interacted with different putative AdpA binding sites and generated different forms of complexes (Figure 2C). DNA fragment without AdpA binding site, i.e., probe-neg, could not interact with AdpAlin (Supplementary Figure S3). To confirm the exact binding sites of AdpAlin with promoters, we deleted the putative AdpA binding sites in lmbAp-A, lmbJp-A and lmbKp-B, and EMSAs showed AdpAlin no longer interacted with these DNA probes (Supplementary Figure S4). Thus, we speculated that AdpAlin activates all of the 8 promoters in the lmb cluster by directly binding to putative AdpA binding sites.
AdpAlin Directly Activates Transcription of the CSR Gene lmbU
LmbU was recently reported by Hou et al. (2018a, 2019) to be a novel transcriptional regulator cited in the lmb cluster and positively regulate lincomycin biosynthesis. Here, we investigated the regulatory relationship between AdpAlin and lmbU. We analyzed the 770 bp of promoter region upstream from lmbU TSS, and found 10 putative AdpA binding sites where two of them are overlapped (Supplementary Figure S2F). Relative expression of lmbU significantly decreased by 26.26 folds in ΔadpA compared with WT (Figure 3A). We cloned the 770 bp of DNA sequences upstream from lmbU TSS to construct reporter plasmid for catechol dioxygenase assay. In accordance with qRT-PCR results, catechol dioxygenase assay demonstrated that AdpAlin remarkably activated lmbU promoter (Figure 3B). As displayed in Supplementary Figure S2F, lmbUp-A, lmbUp-B, and lmbUp-C are three DNA probes containing putative AdpA binding sites in the promoter region of lmbU. EMSAs indicated that AdpAlin directly bound to lmbUp-A, lmbUp-B, and lmbUp-C, separately (Figure 3C). Then, the putative AdpA binding site in lmbUp-B was deleted, and EMSA showed that AdpAlin could not bind to this DNA probe (Supplementary Figure S4), suggesting that AdpAlin activates the transcription of lmbU by directly binding to the lmbU promoter and thus gets involved in the activation of lincomycin biosynthesis.
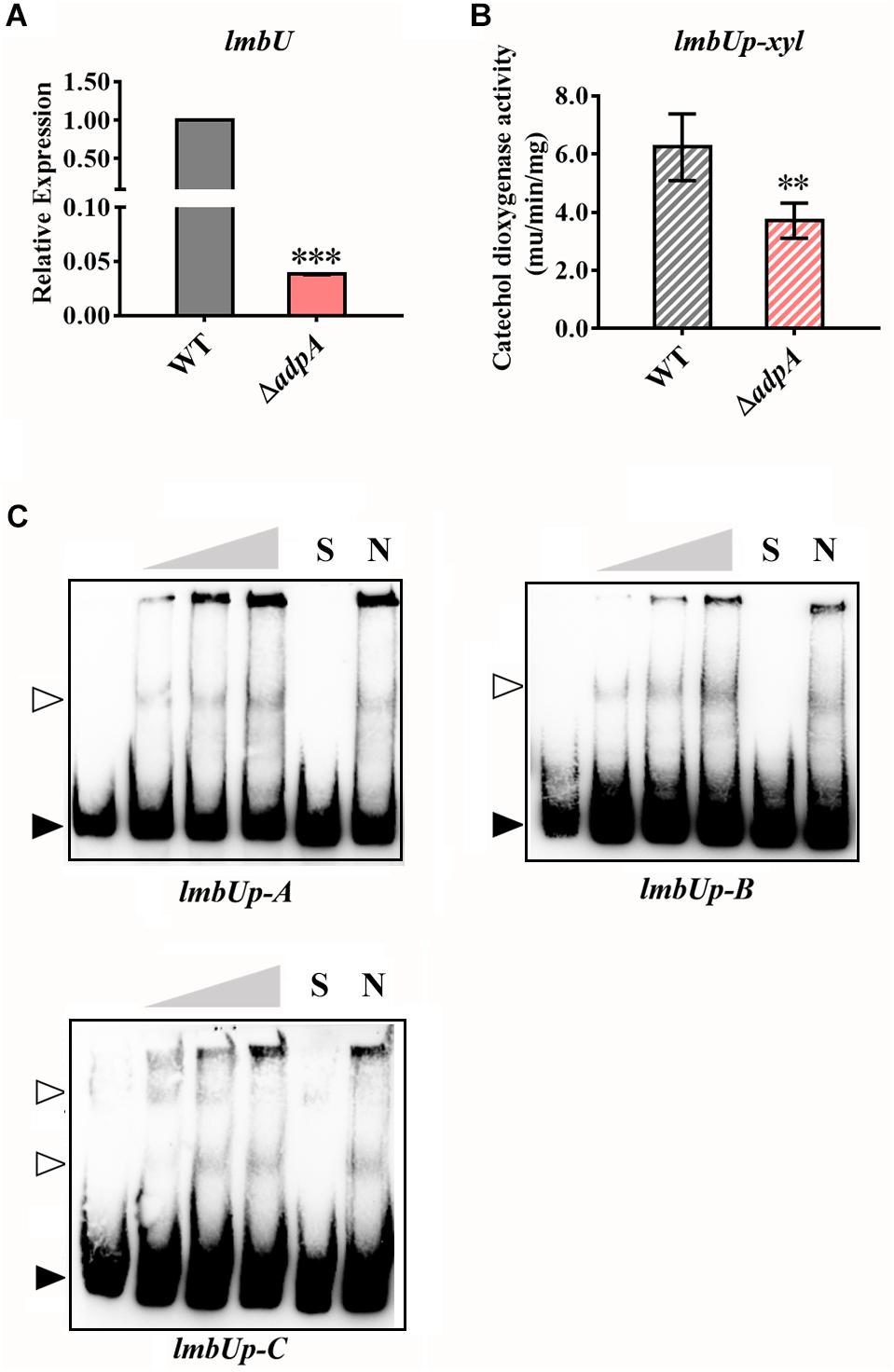
Figure 3. AdpAlin activates the transcription of the CSR gene lmbU. (A) Relative expression of lmbU in WT and ΔadpA. (B) Catechol dioxygenase activity analysis of WT and ΔadpA transformed with pUPX. (C) EMSAs of AdpAlin with 5′-biotin labeled DNA probes lmbUp-A, lmbUp-B, and lmbUp-C. Solid triangles point to the bands of probes and hollow triangles point to the complexes of AdpAlin and probes. Concentrations of AdpAlin are 0, 1.6, 3.2, 6.4, 6.4, and 6.4 μM, respectively. Competitive assays were carried out with a 200-fold excess of unlabeled specific probe lmbUp-A, lmbUp-B, or lmbUp-B (lane S) or with a 200-fold excess of unlabeled non-specific probe probe-neg (lane N). ∗∗∗P < 0.001 and ∗∗P < 0.01, respectively.
AdpAlin Directly Activates the Expression of the Global Regulator Gene bldA
As mentioned above, deletion of AdpAlin in S. lincolnensis not only blocked lincomycin biosynthesis, but also significantly impaired the generation of spores (Figures 1B,C), which suggested that AdpAlin had complicated connections with genes outside the lmb cluster. Hou et al. (2018b) previously identified that BldA regulates morphological differentiation and lincomycin biosynthesis in S. lincolnensis. We analyzed the 799 bp of promoter region upstream from the mature bldA, three putative AdpA binding sites were found (Figure 4A). Relative expression of bldA exhibited a 7.29-fold decreased in ΔadpA compared with WT (Figure 4B). The 799 bp of DNA sequences upstream from the mature bldA was cloned to construct reporter plasmid for catechol dioxygenase activity assay. And the results indicated that AdpAlin significantly activates the transcription of bldA in vivo (Figure 4C). BldAp-A and bldAp-B are two DNA probes containing putative AdpA binding sites marked in Figure 4A. We performed EMSAs of AdpAlin with bldAp-A and bldAp-B separately, and as displayed in Figure 4D, AdpAlin directly bound to DNA fragments containing putative AdpA binding sites in vitro. Thus, we speculated that AdpAlin participates in the lincomycin biosynthesis in S. lincolnensis through activating the transcription of bldA, and indirectly mediates the morphological differentiation.
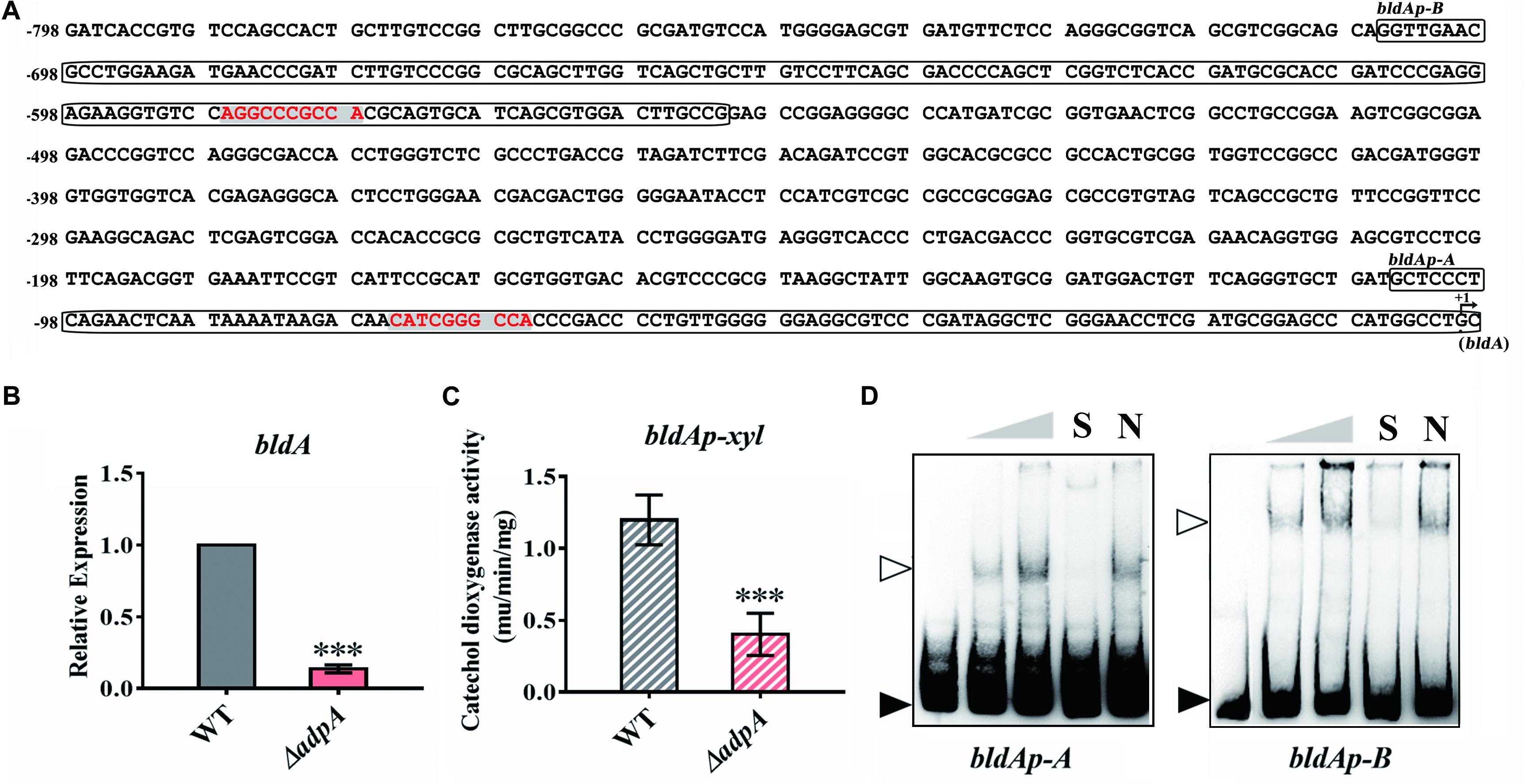
Figure 4. AdpAlin activates the transcription of bldA. (A) Nucleic acid sequence of bldA promoter region where +1 represents the start of mature bldA. Putative AdpA binding sites are marked in red color with gray background. Probes for EMSA, bldAp-A, and bldAp-B, are framed. (B) Relative expression of bldA in WT and ΔadpA. (C) Catechol dioxygenase activity analysis of WT and ΔadpA transformed with pBLPX. (D) EMSAs of AdpAlin with 5′-biotin labeled DNA probes bldAp-A and bldAp-B. Solid triangles point to the bands of probes and hollow triangles point to the complexes of AdpAlin and DNA probes. Concentrations of AdpAlin are 0, 1.6, 3.2, 3.2, and 3.2 μM, respectively. Competitive assays were carried out with a 200-fold excess of unlabeled specific probe bldAp-A or bldAp-B (lane S) or with a 200-fold excess of unlabeled non-specific probe probe-neg (lane N). ∗∗∗P < 0.001.
Since AdpA has been identified to regulate the transcription of bldA in S. griseus (Higo et al., 2011), we further analyzed the bldA promoter from S. lincolnensis and S. griseus. The data showed that putative AdpA binding site and its flanking sequence in bldAp-A was highly conserved (Supplementary Figure S5A), and EMSA of AdpAlin with bldAp-A confirmed the binding (Figure 4D). However, the sequences upstream bldAp-A are various between the two species, which contains the AdpA binding sites in S. griseus, but not in S. lincolnensis (Supplementary Figure S5A). Further EMSA demonstrated that AdpAlin cannot bind to this fragment bldAp-N (Supplementary Figure S5B), confirming that it is not a functional AdpA binding site in S. lincolnensis. Moreover, putative AdpA binding site in bldAp-B, which has not been studied before, was confirmed to be a AdpA binding site in S. lincolnensis (Supplementary Figure S5C).
AdpAlin Positively Autoregulates Its Own Transcription via Directly Binding to Its Own Promoter
We carried out a detailed analysis of 610 bp of upstream DNA sequences from AdpAlin TSS and found 6 putative AdpAlin binding sites where 4 of them are overlapped in pairs (Figure 5A). To investigate the transcriptional regulation between AdpAlin and its own transcription, we cloned the 610 bp of DNA sequence upstream from AdpAlin TSS to construct a reporter plasmid for catechol dioxygenase activity assay. The results revealed that AdpAlin slightly activated its own transcription in vivo (Figure 5B). As displayed in Figure 5A, adpAp-A and adpAp-B are two DNA probes containing putative AdpAlin binding sites, and the results of EMSAs verified that AdpAlin directly interacted with the promoter region of AdpAlin in vitro (Figure 5C).
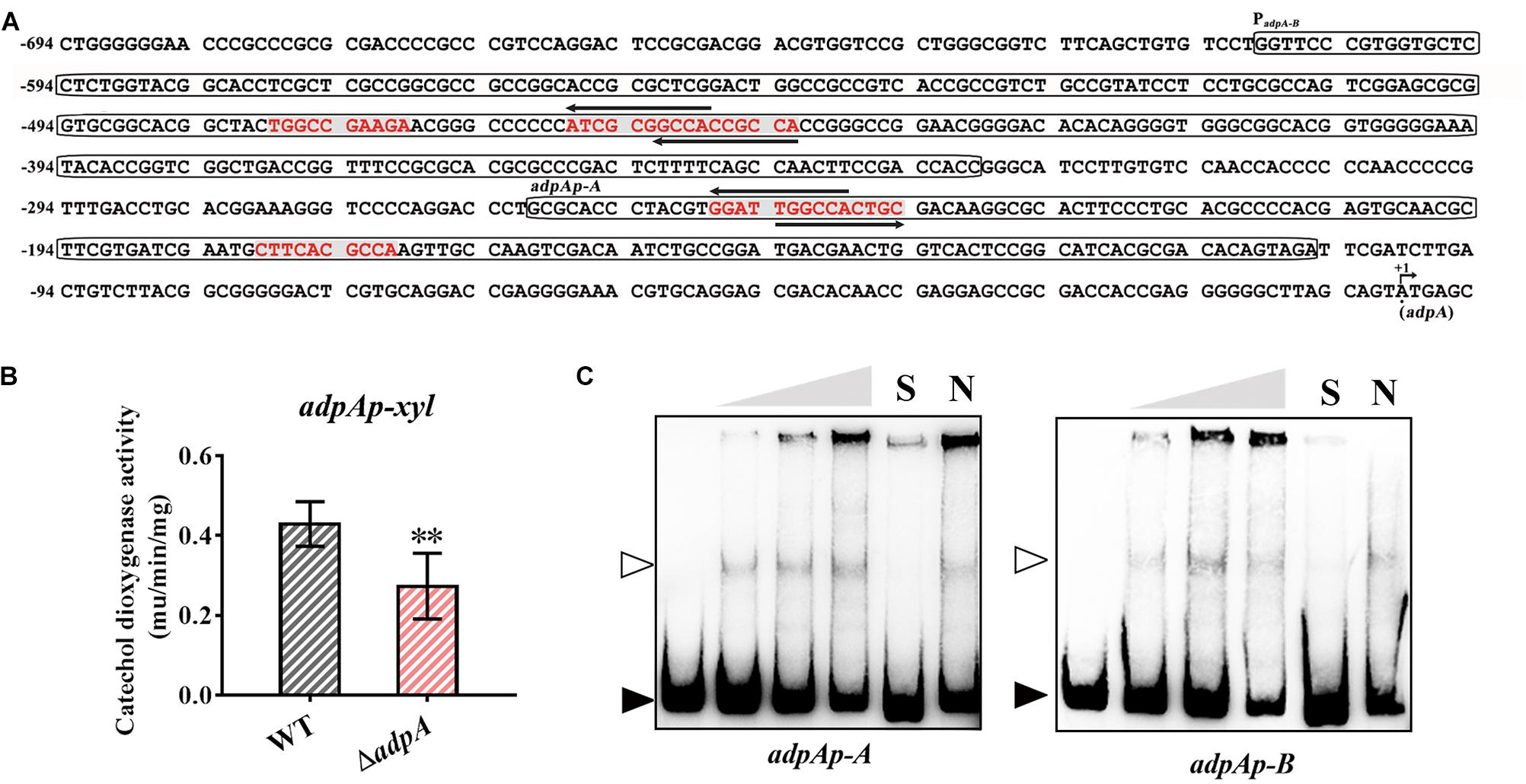
Figure 5. AdpAlin positively regulates its own transcription. (A) Nucleic acid sequence of AdpAlin promoter region where +1 represents the translation starting site (TSS) of adpA. Putative AdpA binding sites are marked in red color with gray background. Probes for EMSA, adpAp-A and adpAp-B, are framed. (B) Catechol dioxygenase activity analysis of WT and ΔadpA transformed with pADPX. ∗∗P < 0.01. (C) EMSAs of AdpAlin with 5′-biotin labeled DNA probes adpAp-A and adpAp-B. Solid triangles point to the bands of probes and hollow triangles point to the complexes of AdpAlin and DNA fragments. Concentrations of AdpAlin are 0, 1.6, 3.2, 6.4, 6.4, and 6.4 μM, respectively. Competitive assays were carried out with a 200-fold excess of unlabeled specific probe adpAp-A or adpAp-B (lane S) or with a 200-fold excess of unlabeled non-specific probe probe-neg (lane N).
AdpA also has been identified to autoregulate its own transcription in S. griseus (Kato et al., 2005), then we compared AdpAlin promoter from S. lincolnensis with adpA promoter from S. griseus. In S. lincolnensis, the putative AdpA binding site and its flanking sequence in adpAp-A was high conserved (Supplementary Figure S6A) and the result of EMSA verified the binding (Figure 5C). However, the nucleic acid sequences upstream adpAp-A were highly diverse (Supplementary Figure S6A). Then we designed two probes, adpAp-B-1 and adpAp-B-2, which contain the unconserved AdpA binding motifs from S. griseus (Kato et al., 2005) and putative AdpA binding sites from S. lincolnensis, respectively. The results of EMSAs showed that AdpAlin can bind to adpAp-B-2, but not to adpAp-B-1 (Supplementary Figures S6B,C), indicating that two new AdpA binding sites are found in S. lincolnensis, and the differences between the two probes may allow them to respond to different regulatory mechanisms.
Discussion
In this study, we elucidated the effect of AdpAlin on lincomycin biosynthesis and morphological differentiation. There are 8 putative operons in the lmb cluster. Before this study, none of a regulator has been identified to directly bind to all the eight promoters in the lmb cluster and have an overall impact on the entire lincomycin biosynthesis progress. By deciphering the regulations on the 8 putative operons, we attempted to envision the transcriptional regulatory network on lincomycin biosynthesis. Lincomycin biosynthesis contains three main parts: formation of α-methylthiolincosaminide (MTL), formation of propylproline (PPL), and condensation and final methylation (Supplementary Figure S2A). AdpAlin activates lmbKp and lmbVp, therefore we speculate that AdpAlin directly activates the transcription of lmbK, lmbR, lmbO, and lmbN, so that AdpAlin positively regulates the biosynthesis of MTL structure (Sasaki et al., 2012; Lin et al., 2014). AdpAlin activates the transcription of lmbAp, lmbWp, and lmbUp, which means lmbB1, lmbB2, lmbX and lmbY are also activated by AdpAlin, thus we speculated that AdpAlin directly regulates the biosynthesis of PPL structure (Novotna et al., 2004, 2013; Pang et al., 2015; Jiraskova et al., 2016). In addition, AdpAlin activates the transcription lmbCp, lmbDp, lmbJp, and lmbVp, which means transcriptions of lmbT, lmbE, lmbF, and lmbG are activated by AdpAlin (Hola et al., 2003; Zhao et al., 2015; Kadlcik et al., 2017; Zhong et al., 2017; Zhang et al., 2018). Therefore, we inferred that AdpAlin functions as a primary activator of lincomycin biosynthesis and regulates the entire biosynthetic process. And this is the first case that AdpA directly activates the transcription of the overall structural genes in such a complicated antibiotic biosynthetic gene cluster (BGC). In addition, EMSAs of AdpAlin with targets showed that AdpAlin binds to different binding sites with different affinities (Figure 2C). Based on these results, some strategies of genetic manipulations may be proposed for hyper-production of lincomycin, such as mutation, deletion or addition of AdpA binding sites in the promoter regions of target genes.
Besides directly participating in lincomycin biosynthesis, as a pleiotropic regulator, AdpAlin controls lincomycin biosynthesis by regulating other transcriptional regulators as well. LmbU has been identified to activate the transcription of lmbA, lmbC, and lmbJ, and repress the transcription of lmbK and lmbU itself (Hou et al., 2018a). In this study, we confirmed that the transcription of lmbU was activated by AdpAlin (Figure 3). As described by Hou et al. (2018b) there is a UUA codon in lmbU, and translation of lmbU is controlled by BldA. Existence of rare codon means very small changes of the tRNA could induce the significant change of protein amount (Chater, 2006). Besides, UUA codons also exist in lmbB2 and lmbY in the lmb cluster, indicating that LmbB2 and LmbY might be important regulatory targets during lincomycin biosynthesis. Furthermore, it has been showed that the adpA gene contains a UUA codon as well, on the other hand, transcription of bldA is regulated by AdpAlin (Figure 4), which may function as a feedback regulatory mechanism to keep the organism in balance. In this study, we speculated that AdpAlin, LmbU, and BldA formed a regulatory cascade that mediate lincomycin biosynthesis in S. lincolnensis (Figure 6). In addition, considering AdpA responds to the GBL-involved cascade regulation (Healy et al., 2009; Tan et al., 2015), bioinformatics analysis was performed and two GBL-signaling systems were found in S. lincolnensis (Figure 6). One system consists of the GBL receptor SLINC_6539 (GenBank accession number ANS68763.1) and biosynthetic enzyme SLINC_6540 (GenBank accession number ANS68764.1) which were highly homology with many receptors and enzymes in Streptomyces, whereas SLINC_6539 and SLINC_6540 had identities with ArpA (47%) and AsfA (70%) in S. griseus, respectively. The other system consists of SLINC_5093 (GenBank accession number WP_067437987.1) and SLINC_5094 (GenBank accession number WP_067437989.1) which were low similarities with other receptors and enzymes, while SLINC_5093 and SLINC_5094 had identities with ArpA (44%) and AsfA (33%) in S. griseus, respectively. But how these two GBL-signaling systems works to affect lincomycin biosynthesis will be needed further research.
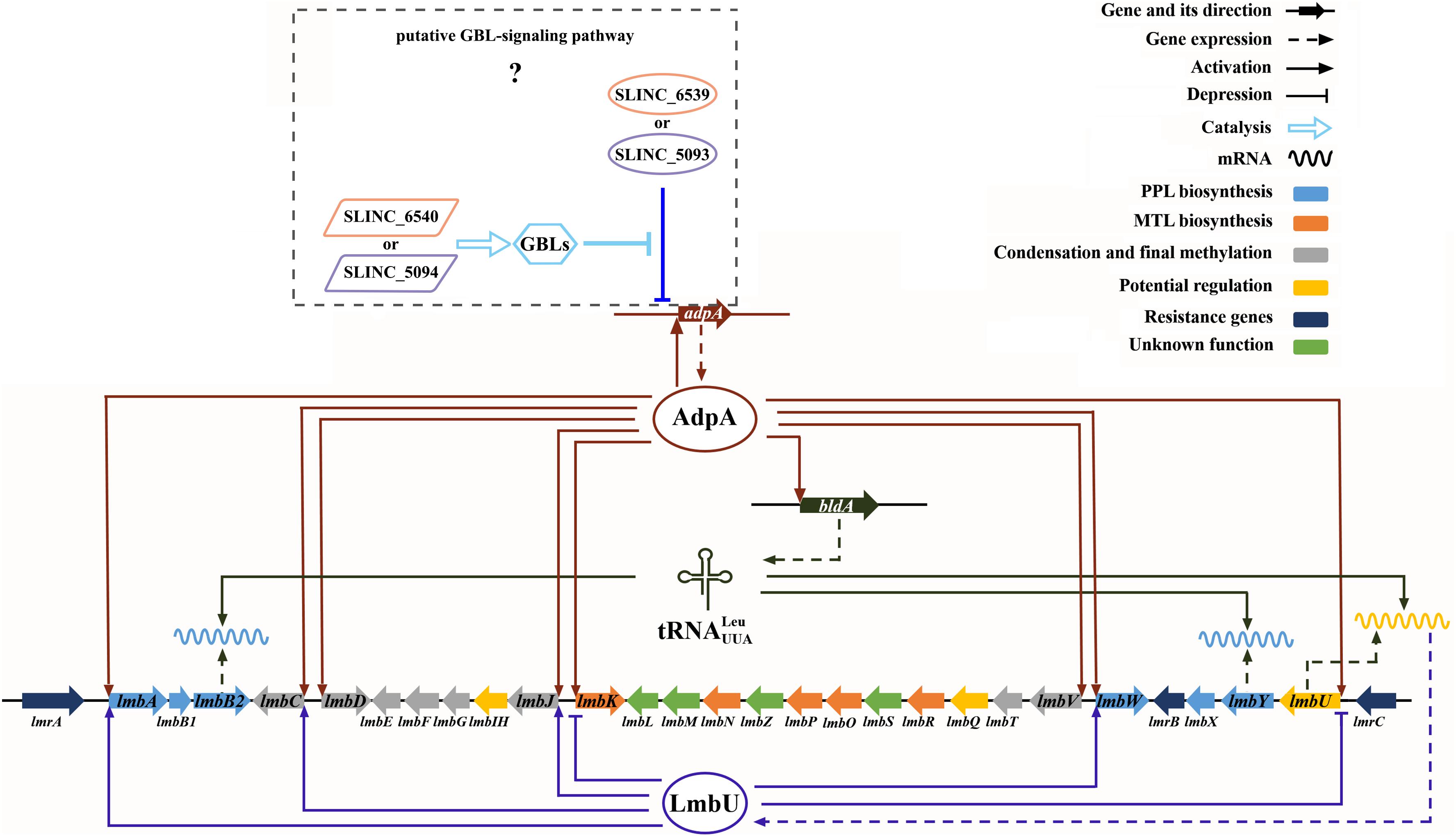
Figure 6. Cascade regulation of AdpAlin, LmbU, and BldA on lincomycin biosynthesis in S. lincolnensis.
In S. griseus, AdpA auto-depresses its own transcription (Ohnishi et al., 2005), whereas in S. lincolnensis, AdpAlin has a positive impact on its own transcription (Figure 5B). Differences of AdpA binding sites in S. lincolnensis and S. griseus indicated that AdpA homologs from different resource have specialized regulatory mechanism on their own transcription. Catechol dioxygenase activity showed that AdpAlin had a negatively effect on the AdpA binding site near the AdpAlin TSS (Data not shown). Thus, we speculated that AdpAlin bound to this putative site and prevented the AdpAlin promotor from being activated. As for another putative AdpA binding sites, we presumed it might recruit RNA polymerase after interacting with AdpAlin and thus the overall effect of AdpAlin on its own promoter appeared to be positive. In the natamycin producer S. chattanoogensis, AdpAch was an activator of natamycin biosynthesis, and 6 AdpA binding sites were identified in the scnRI-scnRII intergenic region (Du et al., 2011; Yu et al., 2018). It is notable that although the general effect of AdpAch on the transcription of scnRI is positive, AdpA binding site A and B serve as repression sites. Thus, we speculated that the varying amounts and locations of AdpA binding sites in the promoter region of AdpAlin targets exhibited different effects and constituted a complicated and subtle regulatory network of AdpA regulons.
In summary, we reported a lesser-known case that AdpAlin interacted with all of the 8 putative operons and activated the transcription of structural genes in the lmb cluster. Furthermore, we deduced AdpAlin, LmbU, and BldA in cascade regulation that controlled lincomycin biosynthesis. Based on these knowledge, more efforts should be devoted to complete the regulatory mechanism of lincomycin biosynthesis and to enhance to production of lincomycin.
Data Availability Statement
All datasets generated for this study are included in the manuscript/Supplementary Files.
Author Contributions
YK, BH, and HW designed the experiments. YK, YW, and RW carried out the experiments. YK, BH, XZ, JY, and HW analyzed the data. YK, JY, and HW wrote the manuscript. JY and HZ guided the work. All authors assisted with critical reading of the manuscript.
Funding
This study was supported by the National Natural Science Foundation of China (NSFC) (31900059), the China Postdoctoral Science Foundation (2019M650079), and the Research Program of State Key Laboratory of Bioreactor Engineering.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Weihong Jiang for providing the plasmid for catechol dioxygenase activity assay and all the members of our laboratory.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.02428/full#supplementary-material
References
Bierman, M., Logan, R., O’Brien, K., Seno, E. T., Rao, R. N., and Schoner, B. E. (1992). Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene 116, 43–49. doi: 10.1016/0378-1119(92)90627-2
Bu, X. L., Weng, J. Y., He, B. B., Xu, M. J., and Xu, J. (2019). A novel AdpA homologue negatively regulates morphological differentiation in Streptomyces xiamenensis 318. Appl. Environ. Microbiol. 85:e3107-18. doi: 10.1128/AEM.03107-18
Chater, K. F. (2006). Streptomyces inside-out: a new perspective on the bacteria that provide us with antibiotics. Philos. Trans. R. Soc. Lond. B Biol. Sci. 361, 761–768. doi: 10.1098/rstb.2005.1758
Du, Y. L., Li, S. Z., Zhou, Z., Chen, S. F., Fan, W. M., and Li, Y. Q. (2011). The pleitropic regulator AdpAch is required for natamycin biosynthesis and morphological differentiation in Streptomyces chattanoogensis. Microbiology 157, 1300–1311. doi: 10.1099/mic.0.046607-0
Guyet, A., Gominet, M., Benaroudj, N., and Mazodier, P. (2013). Regulation of the clpP1clpP2 operon by the pleiotropic regulator AdpA in Streptomyces lividans. Arch. Microbiol. 195, 831–841. doi: 10.1007/s00203-013-0918-2
Hackl, S., and Bechthold, A. (2015). The gene bldA, a regulator of morphological differentiation and antibiotic production in Streptomyces. Arch. Pharm. 348, 455–462. doi: 10.1002/ardp.201500073
Healy, F. G., Eaton, K. P., Limsirichai, P., Aldrich, J. F., Plowman, A. K., and King, R. R. (2009). Characterization of gamma-butyrolactone autoregulatory signaling gene homologs in the angucyclinone polyketide WS5995B producer Streptomyces acidiscabies. J. Bacteriol. 191, 4786–4797. doi: 10.1128/JB.00437-439
Higashi, T., Iwasaki, Y., Ohnishi, Y., and Horinouchi, S. (2007). A-factor and phosphate depletion signals are transmitted to the grixazone biosynthesis genes via the pathway-specific transcriptional activator GriR. J. Bacteriol. 189, 3515–3524. doi: 10.1128/JB.00055-07
Higo, A., Hara, H., Horinouchi, S., and Ohnishi, Y. (2012). Genome-wide distribution of AdpA, a global regulator for secondary metabolism and morphological differentiation in Streptomyces, revealed the extent and complexity of the AdpA regulatory network. DNA Res. 19, 259–273. doi: 10.1093/dnares/dss010
Higo, A., Horinouchi, S., and Ohnishi, Y. (2011). Strict regulation of morphological differentiation and secondary metabolism by a positive feedback loop between two global regulators AdpA and BldA in Streptomyces griseus. Mol. Microbiol. 81, 1607–1622. doi: 10.1111/j.1365-2958.2011.07795.x
Hola, K., Janata, J., Kopecky, J., and Spizek, J. (2003). LmbJ and LmbIH protein levels correlate with lincomycin production in Streptomyces lincolnensis. Lett. Appl. Microbiol. 37, 470–474. doi: 10.1046/j.1472-765X.2003.01432.x
Hou, B., Lin, Y., Wu, H., Guo, M., Petkovic, H., Tao, L., et al. (2018a). The novel transcriptional regulator LmbU promotes lincomycin biosynthesis through regulating expression of its target genes in Streptomyces lincolnensis. J. Bacteriol. 200:e00447-e17. doi: 10.1128/JB.00447-17
Hou, B., Tao, L., Zhu, X., Wu, W., Guo, M., Ye, J., et al. (2018b). Global regulator BldA regulates morphological differentiation and lincomycin production in Streptomyces lincolnensis. Appl. Microbiol. Biotechnol. 102, 4101–4115. doi: 10.1007/s00253-018-8900-1
Hou, B., Zhu, X., Kang, Y., Wang, R., Wu, H., Ye, J., et al. (2019). LmbU, a cluster-situated regulator for lincomycin, consists of a DNA-binding domain, an auto-inhibitory domain, and forms homodimer. Front. Microbiol. 10:989. doi: 10.3389/fmicb.2019.00989
Hsiao, N. H., Nakayama, S., Merlo, M. E., de Vries, M., Bunet, R., Kitani, S., et al. (2009). Analysis of two additional signaling molecules in Streptomyces coelicolor and the development of a butyrolactone-specific reporter system. Chem. Biol. 16, 951–960. doi: 10.1016/j.chembiol.2009.08.010
Ibarra, J. A., Perez-Rueda, E., Segovia, L., and Puente, J. L. (2008). The DNA-binding domain as a functional indicator: the case of the AraC/XylS family of transcription factors. Genetica 133, 65–76. doi: 10.1007/s10709-007-9185-y
Jiraskova, P., Gazak, R., Kamenik, Z., Steiningerova, L., Najmanova, L., Kadlcik, S., et al. (2016). New concept of the biosynthesis of 4-Alkyl-l-proline precursors of lincomycin, hormaomycin, and pyrrolobenzodiazepines: could a γ-glutamyltransferase cleave the C-C bond? Front. Microbiol. 7:276. doi: 10.3389/fmicb.2016.00276
Kadlcik, S., Kamenik, Z., Vasek, D., Nedved, M., and Janata, J. (2017). Elucidation of salicylate attachment in celesticetin biosynthesis opens the door to create a library of more efficient hybrid lincosamide antibiotics. Chem. Sci. 8, 3349–3355. doi: 10.1039/c6sc04235j
Kato, J. Y., Funa, N., Watanabe, H., Ohnishi, Y., and Horinouchi, S. (2007). Biosynthesis of gamma-butyrolactone autoregulators that switch on secondary metabolism and morphological development in Streptomyces. Proc. Natl. Acad. Sci. U.S.A. 104, 2378–2383. doi: 10.1073/pnas.0607472104
Kato, J. Y., Ohnishi, Y., and Horinouchi, S. (2005). Autorepression of AdpA of the AraC/XylS family, a key transcriptional activator in the A-factor regulatory cascade in Streptomyces griseus. J. Mol. Biol. 350, 12–26. doi: 10.1016/j.jmb.2005.04.058
Kieser, T., Bibb, M. J., Buttner, M. J., Chater, K. F., and Hopwood, D. A. (2000). Practical Streptomyces Genetics. Norwich: John Innes Foundation.
Kim, D. W., Chater, K., Lee, K. J., and Hesketh, A. (2005). Changes in the extracellular proteome caused by the absence of the bldA gene product, a developmentally significant tRNA, reveal a new target for the pleiotropic regulator AdpA in Streptomyces coelicolor. J. Bacteriol. 187, 2957–2966. doi: 10.1128/JB.187.9.2957-2966.2005
Li, X. B., Zhao, G. R., Zheng, H., and Yuan, Y. J. (2007). Improved industrial fermentation of lincomycin by phosphorus feeding. Process Biochem. 42, 662–668. doi: 10.1016/j.procbio.2006.12.007
Liao, C. H., Yao, L., Xu, Y., Liu, W. B., Zhou, Y., and Ye, B. C. (2015). Nitrogen regulator GlnR controls uptake and utilization of non-phosphotransferase-system carbon sources in actinomycetes. Proc. Natl. Acad. Sci. U.S.A. 112, 15630–15635. doi: 10.1073/pnas.1508465112
Lin, C. I., Sasaki, E., Zhong, A., and Liu, H. W. (2014). In vitro characterization of LmbK and LmbO: identification of GDP-D-erythro-alpha-D-gluco-octose as a key intermediate in lincomycin a biosynthesis. J. Am. Chem. Soc. 136, 906–909. doi: 10.1021/ja412194w
Liu, F., Xu, D., Zhang, Y., Zhu, Y., Ye, J., and Zhang, H. (2015). Identification of BagI as a positive transcriptional regulator of bagremycin biosynthesis in engineered Streptomyces sp. Tu 4128. Microbiol. Res. 173, 18–24. doi: 10.1016/j.micres.2015.01.011
Liu, Y., Yan, T., Jiang, L., Wen, Y., Song, Y., Chen, Z., et al. (2013). Characterization of SAV7471, a TetR-family transcriptional regulator involved in the regulation of coenzyme a metabolism in Streptomyces avermitilis. J. Bacteriol. 195, 4365–4372. doi: 10.1128/JB.00716-13
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-ΔΔCT) method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lu, C., Yang, J., and Ye, J. (2008). Knockout and retro-complementation of a lincomycin biosynthetic gene lmbU. J. East China Univ. Sci. Techno. 34:60. doi: 10.3724/SP.J.1141.2008.00373
Macleod, A. J., Ross, H. B., Ozere, R. L., Digout, G., and Van, R. (1964). Lincomycin: a new antibiotic active against staphylococci and other gram-positive cocci: clinical and laboratory studies. Can. Med. Assoc. J. 91, 1056–1060.
Mao, X. M., Luo, S., Zhou, R. C., Wang, F., Yu, P., Sun, N., et al. (2015). Transcriptional regulation of the daptomycin gene cluster in Streptomyces roseosporus by an autoregulator. AtrA. J. Biol. Chem. 290, 7992–8001. doi: 10.1074/jbc.M114.608273
Meng, S., Wu, H., Wang, L., Zhang, B., and Bai, L. (2017). Enhancement of antibiotic productions by engineered nitrate utilization in actinomycetes. Appl. Microbiol. Biotechnol. 101, 5341–5352. doi: 10.1007/s00253-017-8292-7
Neusser, D., Schmidt, H., Spizek, J., Novotna, J., Peschke, U., Kaschabeck, S., et al. (1998). The genes lmbB1 and lmbB2 of Streptomyces lincolnensis encode enzymes involved in the conversion of L-tyrosine to propylproline during the biosynthesis of the antibiotic lincomycin A. Arch. Microbiol. 169, 322–332. doi: 10.1007/s002030050578
Niu, G., Chater, K. F., Tian, Y., Zhang, J., and Tan, H. (2016). Specialised metabolites regulating antibiotic biosynthesis in Streptomyces spp. FEMS Microbiol. Rev. 40, 554–573. doi: 10.1093/femsre/fuw012
Novotna, J., Honzatko, A., Bednar, P., Kopecky, J., Janata, J., and Spizek, J. (2004). l-3,4-Dihydroxyphenyl alanine-extradiol cleavage is followed by intramolecular cyclization in lincomycin biosynthesis. Eur. J. Biochem. 271, 3678–3683. doi: 10.1111/j.1432-1033.2004.04308.x
Novotna, J., Olsovska, J., Novak, P., Mojzes, P., Chaloupkova, R., Kamenik, Z., et al. (2013). Lincomycin biosynthesis involves a tyrosine hydroxylating heme protein of an unusual enzyme family. PLoS One 8:e79974. doi: 10.1371/journal.pone.0079974
Ohnishi, Y., Yamazaki, H., Kato, J. Y., Tomono, A., and Horinouchi, S. (2005). AdpA, a central transcriptional regulator in the A-factor regulatory cascade that leads to morphological development and secondary metabolism in Streptomyces griseus. Biosci. Biotechnol. Biochem. 69, 431–439. doi: 10.1271/bbb.69.431
Pan, Y., Liu, G., Yang, H., Tian, Y., and Tan, H. (2009). The pleiotropic regulator AdpA-L directly controls the pathway-specific activator of nikkomycin biosynthesis in Streptomyces ansochromogenes. Mol. Microbiol. 72, 710–723. doi: 10.1111/j.1365-2958.2009.06681.x
Pang, A. P., Du, L., Lin, C. Y., Qiao, J., and Zhao, G. R. (2015). Co-overexpression of lmbW and metK led to increased lincomycin a production and decreased byproduct lincomycin B content in an industrial strain of Streptomyces lincolnensis. J. Appl. Microbiol. 119, 1064–1074. doi: 10.1111/jam.12919
Pharmacopoeia of the People’s Republic of China [PPRC], (1990). Pharmacopoeia of the People’s Republic of China. Beijing: China Medico-Pharmaceutical Science & Technology Publishing House.
Sasaki, E., Lin, C. I., Lin, K. Y., and Liu, H. W. (2012). Construction of the octose 8-phosphate intermediate in lincomycin a biosynthesis: characterization of the reactions catalyzed by LmbR and LmbN. J. Am. Chem. Soc. 134, 17432–17435. doi: 10.1021/ja308221z
Setinova, D., Smidova, K., Pohl, P., Music, I., and Bobek, J. (2017). RNase III-binding-mRNAs revealed novel complementary transcripts in Streptomyces. Front. Microbiol. 8:2693. doi: 10.3389/fmicb.2017.02693
Spizek, J., and Rezanka, T. (2004a). Lincomycin, clindamycin and their applications. Appl. Microbiol. Biotechnol. 64, 455–464. doi: 10.1007/s00253-003-1545-7
Spizek, J., and Rezanka, T. (2004b). Lincomycin, cultivation of producing strains and biosynthesis. Appl. Microbiol. Biotechnol. 63, 510–519. doi: 10.1007/s00253-003-1431-3
Spizek, J., and Rezanka, T. (2017). Lincosamides: chemical structure, biosynthesis, mechanism of action, resistance, and applications. Biochem. Pharmacol. 133, 20–28. doi: 10.1016/j.bcp.2016.12.001
Takano, E., Nihira, T., Hara, Y., Jones, J. J., Gershater, C. J., Yamada, Y., et al. (2000). Purification and structural determination of SCB1, a gamma-butyrolactone that elicits antibiotic production in Streptomyces coelicolor A3(2). J. Biol. Chem. 275, 11010–11016. doi: 10.1074/jbc.275.15.11010
Tan, G. Y., Peng, Y., Lu, C., Bai, L., and Zhong, J. J. (2015). Engineering validamycin production by tandem deletion of gamma-butyrolactone receptor genes in Streptomyces hygroscopicus 5008. Metab. Eng. 28, 74–81. doi: 10.1016/j.ymben.2014.12.003
Tomono, A., Tsai, Y., Yamazaki, H., Ohnishi, Y., and Horinouchi, S. (2005). Transcriptional control by a-factor of strR, the pathway-specific transcriptional activator for streptomycin biosynthesis in Streptomyces griseus. J. Bacteriol. 187, 5595–5604. doi: 10.1128/JB.187.16.5595-5604.2005
Xu, J., Zhang, J., Zhuo, J., Li, Y., Tian, Y., and Tan, H. (2017). Activation and mechanism of a cryptic oviedomycin gene cluster via the disruption of a global regulatory gene, adpA, in Streptomyces ansochromogenes. J. Biol. Chem. 292, 19708–19720. doi: 10.1074/jbc.M117.809145
Xu, Y., Ke, M., Li, J., Tang, Y., Wang, N., Tan, G., et al. (2019). TetR-type regulator SLCG_2919 is a negative regulator of lincomycin biosynthesis in Streptomyces lincolnensis. Appl. Environ. Microbiol. 85:e02091-18. doi: 10.1128/AEM.02091-18
Xu, Y., Tan, G., Ke, M., Li, J., Tang, Y., Meng, S., et al. (2018). Enhanced lincomycin production by co-overexpression of metK1 and metK2 in Streptomyces lincolnensis. J. Ind. Microbiol. Biotechnol. 45, 345–355. doi: 10.1007/s10295-018-2029-1
Yamazaki, H., Tomono, A., Ohnishi, Y., and Horinouchi, S. (2004). DNA-binding specificity of AdpA, a transcriptional activator in the a-factor regulatory cascade in Streptomyces griseus. Mol. Microbiol. 53, 555–572. doi: 10.1111/j.1365-2958.2004.04153.x
Yao, M. D., Ohtsuka, J., Nagata, K., Miyazono, K., Zhi, Y., Ohnishi, Y., et al. (2013). Complex structure of the DNA-binding domain of AdpA, the global transcription factor in Streptomyces griseus, and a target duplex DNA reveals the structural basis of its tolerant DNA sequence specificity. J. Biol. Chem. 288, 31019–31029. doi: 10.1074/jbc.M113.473611
Yu, P., Bu, Q. T., Tang, Y. L., Mao, X. M., and Li, Y. Q. (2018). Bidirectional regulation of AdpAch in controlling the expression of scnRI and scnRII in the natamycin biosynthesis of Streptomyces chattanoogensis L10. Front. Microbiol. 9:361. doi: 10.3389/fmicb.2018.00316
Yu, P., Liu, S.-P., Bu, Q.-T., Zhou, Z.-X., Zhu, Z.-H., Huang, F.-L., et al. (2014). WblAch, a pivotal activator of natamycin biosynthesis and morphological differentiation in Streptomyces chattanoogensis L10, is positively regulated by AdpAch. Appl. Environ. Microbiol. 80, 6879–6887. doi: 10.1128/aem.01849-1814
Zhang, D., Tang, Z., and Liu, W. (2018). Biosynthesis of lincosamide antibiotics: reactions associated with degradation and detoxification pathways play a constructive role. Acc. Chem. Res. 51, 1496–1506. doi: 10.1021/acs.accounts.8b00135
Zhao, Q., Wang, M., Xu, D., Zhang, Q., and Liu, W. (2015). Metabolic coupling of two small-molecule thiols programs the biosynthesis of lincomycin A. Nature 518, 115–119. doi: 10.1038/nature14137
Zheng, Y., Sun, C. F., Fu, Y., Chen, X. A., Li, Y. Q., and Mao, X. M. (2019). Dual regulation between the two-component system PhoRP and AdpA regulates antibiotic production in streptomyces. J. Ind. Microbiol. Biotechnol. 46, 725–737. doi: 10.1007/s10295-018-02127-2125
Keywords: AdpA, lincomycin, Streptomyces lincolnensis, transcriptional regulator, cascade regulation
Citation: Kang Y, Wang Y, Hou B, Wang R, Ye J, Zhu X, Wu H and Zhang H (2019) AdpAlin, a Pleiotropic Transcriptional Regulator, Is Involved in the Cascade Regulation of Lincomycin Biosynthesis in Streptomyces lincolnensis. Front. Microbiol. 10:2428. doi: 10.3389/fmicb.2019.02428
Received: 14 August 2019; Accepted: 08 October 2019;
Published: 23 October 2019.
Edited by:
Yinhua Lu, Shanghai Normal University, ChinaReviewed by:
Xu-Ming Mao, Zhejiang University, ChinaYing Wen, China Agricultural University (CAU), China
Copyright © 2019 Kang, Wang, Hou, Wang, Ye, Zhu, Wu and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jiang Ye, eWVqQGVjdXN0LmVkdS5jbg==; Huizhan Zhang, aHVpemh6aEBlY3VzdC5lZHUuY24=
 Yajing Kang
Yajing Kang Yingying Wang
Yingying Wang Bingbing Hou
Bingbing Hou Ruida Wang
Ruida Wang Jiang Ye
Jiang Ye Xiaoyu Zhu
Xiaoyu Zhu Haizhen Wu
Haizhen Wu Huizhan Zhang1,2*
Huizhan Zhang1,2*