- 1Heilongjiang Key Laboratory for Animal Disease Control and Pharmaceutical Development, Department of Preventive Veterinary Medicine, College of Veterinary Medicine, Northeast Agricultural University, Harbin, China
- 2Department for Radiological Protection, Heilongjiang Province Center for Disease Control and Prevention, Harbin, China
Bovine rotavirus (BRV), bovine parvovirus (BPV), and bovine viral diarrhea virus (BVDV) are the pathogens that cause diarrhea primarily in newborn calves. A mixed infection of BRV, BPV, and BVDV makes clinical diagnosis difficult. In this study, we designed dual-priming oligonucleotide (DPO) primers the VP6 gene of BRV, VP2 gene of BPV, and 5′UTR gene of BVDV and synthesized gold nanoparticles (GNPs) with an average diameter of 10 nm. We combined the DPOs with the GNPs to develop a DPO-nanoPCR assay for detecting BRV, BPV, and BVDV. The annealing temperature, primer concentration, and GNP concentration were optimized for this assay. Compared to a conventional PCR assay, the DPO-nanoPCR assay allowed the use of a wider range of annealing temperatures (41–65°C) to effectively amplify target genes. PCR amplification was the most efficient at 56.2°C using conventional primers. The optimal volume of all the primers (10 μM) was 1.0 μL. The optimal volume of GNPs (10 nM) for all the reactions was 0.5 μL. The detection limits of DPO-nanoPCR for pMD19-T-VP6, pMD19-T-VP2, and pMD19-T-5′UTR were 9.40 × 102 copies/μL, 5.14 × 103 copies/μL, and 4.09 × 101 copies/μL, respectively; and those using conventional PCR were 9.40 × 104 copies/μL, 5.14 × 105 copies/μL, and 4.09 × 104 copies/μL, respectively. The sensitivity of DPO-nanoPCR was at least 100-fold higher than that of conventional PCR. The specificity detection showed that the DPO-nanoPCR was able to specifically detect BRV, BPV, and BVDV. Use of clinical samples indicated that target viruses can be detected accurately. Thus, DPO-nanoPCR is a new powerful, simple, specific, and sensitive tool for detecting mixed infections of BRV, BPV, and BVDV.
Introduction
Bovine rotavirus (BRV) belongs to the genus Rotavirus in the family Reoviridae (Swiatek et al., 2010). BRV infection primarily occurs in calves between 15 and 45 days of age. The clinical symptoms include depression, loss of appetite, diarrhea, and dehydration. The rotavirus particle core has 11 double-stranded RNA segments (Crawford et al., 2017). Each segment encodes a different protein. The six structural proteins that have been identified are: VP1, VP2, VP3, VP4, VP6, and VP7. VP6 accounts for ∼51% of the total viral protein content. VP6 from group A rotavirus is highly conserved between the different serotypes with >90% amino acid sequence homology that enables it to be the major diagnostic antigen (Dennehy, 2015; Shepherd et al., 2018).
Bovine parvovirus (BPV) is a member of the Bocaparvovirus genus in the Parvoviridae family (Qiu et al., 2017). BPV infection mainly causes reproductive dysfunction in pregnant cows and respiratory and gastrointestinal diseases in newborn calves. The genome of BPV consists of three open reading frames (ORF): ORF1 encodes the non-structural protein NS1; ORF2 encodes the phosphorylated protein NP1; and ORF3 encodes the structural proteins VP1, VP2, and VP3. VP2 is the main structural protein of BPV that accounts for ∼80% of the total structural protein content (Dudleenamjil et al., 2010; Luo et al., 2013; Kailasan et al., 2015).
Bovine viral diarrhea virus (BVDV) is a globally well-distributed pathogen that infects cows leading to great economic losses (Yarnall and Thrusfield, 2017; Reichel et al., 2018). BVDV belongs to the Pestivirus genus under family Flaviviridae (Charoenlarp et al., 2018; Quintero Barbosa et al., 2019). In addition to causing respiratory, gastroenteric, and reproductive diseases, intrauterine infection with BVDV can also result in a persistent infection, thereby generating a state of immunotolerance (Lanyon et al., 2014; Khodakaram-Tafti and Farjanikish, 2017). The genome length of BVDV is ∼12.3 kb and consists of a 5′ UTR, ORF, and 3′ UTR. The 5′ UTR sequence is highly conserved in various strains of BVDV and is often used as a marker for diagnosis or classification of BVDV (Chernick and Frank, 2017; Yeşilbaǧ et al., 2017; Zoccola et al., 2017).
Bovine rotavirus, bovine parvovirus, and bovine viral diarrhea virus all cause intestinal infections. Owing to the similarity of their clinical manifestations and infection routes, mixed infections often occur. It is necessary to develop a method that simultaneously detects the three pathogens, which can save time and labor and has a huge advantage in clinical detection.
Dual-priming oligonucleotide primers were first proposed in 2007. DPO-based PCR is practical, reliable, and quick in detecting pathogens (Chun et al., 2007). The DPO primers comprise two separate initiation regions – a longer 5′ end and a shorter 3′ end (stabilizer and determinant) joined by a polyhypoxanthine (poly I) linker. Due to the special structure, DPO primers are difficult to form secondary structures. The 3′ end (6–12 base pairs long with a 40–80% GC-content) determines the specific extension of the target sequence and blocks subsequent false positive results (Kommedal et al., 2012; Ito and Suzaki, 2017). Studies have shown that mismatches of 3 or more bases in the 5′ and 3′ regions of the primer will not allow template extension. While the 5′ end (18–25 base pairs long with Tm >65°C) enables the use of a wide range of annealing temperatures (Lee H. R. et al., 2010), it is not necessary to screen primers, and optimize annealing temperatures (Lee H. J. et al., 2010). Therefore, DPO primers are perfect for developing multiplex PCR assays that can be used for amplifying multiple genes at multiple annealing temperatures (Yeh et al., 2011; Xu et al., 2015).
Nanoparticle-assisted PCR (nanoPCR) is an advanced PCR technique in which solid gold nanoparticles (GNPs) (1–100 nm) form a colloidal nanofluid to increase thermal conductivity and rapidly attain the target temperature (Li H. K. et al., 2005; Li M. et al., 2005; Rehman et al., 2015). The efficiency and sensibility of this assay are improved by shortening the time of amplification at non-target temperatures. The susceptibility and latency of BRV, BPV, and BVDV can lead to persistent infections in cows; and, the potential risks of shedding and dispersal lead to serious economic losses in the cattle industry. NanoPCR is particularly suitable for detecting samples with low viral titers in early and latent infection, which is of great significance for the prevention and control of diseases caused by BRV, BPV, and BVDV.
In this study, we combined DPO primers with nanoPCR to develop a multiplex DPO-nanoPCR system for the simultaneous detection of BRV, BPV, and BVDV. Compared to conventional PCR, DPO-nanoPCR saves time and effort and is very sensitive and specific. This is a new approach for diagnosing early and latent infections of BRV, BPV, and BVDV.
Materials and Methods
DPO Primer Design and Preparation of Recombinant Plasmids
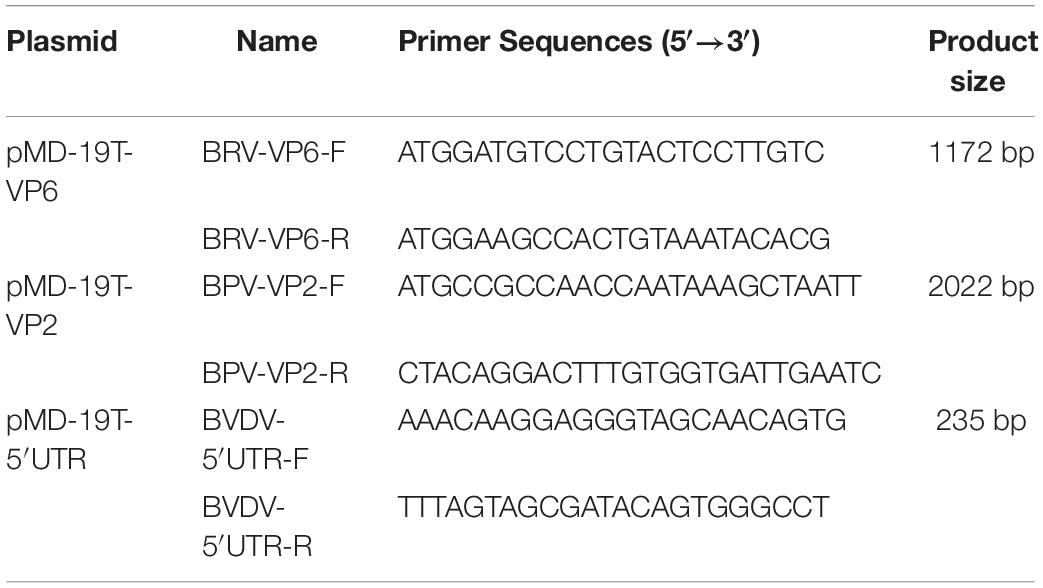
Genes VP6 from BRV (NCDV strain), VP2 from BPV (ATCC strain VR-767) and 5′ UTR gene from BVDV (BA strain) were chosen based on the sequences in the GenBank database (GenBank accession numbers for VP6, VP2, and 5′ UTR are JF693031.1, NC_001540.1, and KC695814.1, respectively). The primers for VP6 (1,172 bp), VP2 (2,022 bp), and 5′ UTR (235 bp) were designed using the Oligo6.0 software (Table 1). VP6, VP2, and 5′ UTR were amplified and inserted into the pMD19-T vector (TaKaRa Bio Inc., Dalian, China) using standard cloning procedures. The recombinant plasmids were transformed into E. coli TG1. The plasmids were purified using the TIANprep Mini Plasmid Kit (TIANGEN Biotech, Beijing, China) and stored at −20°C.
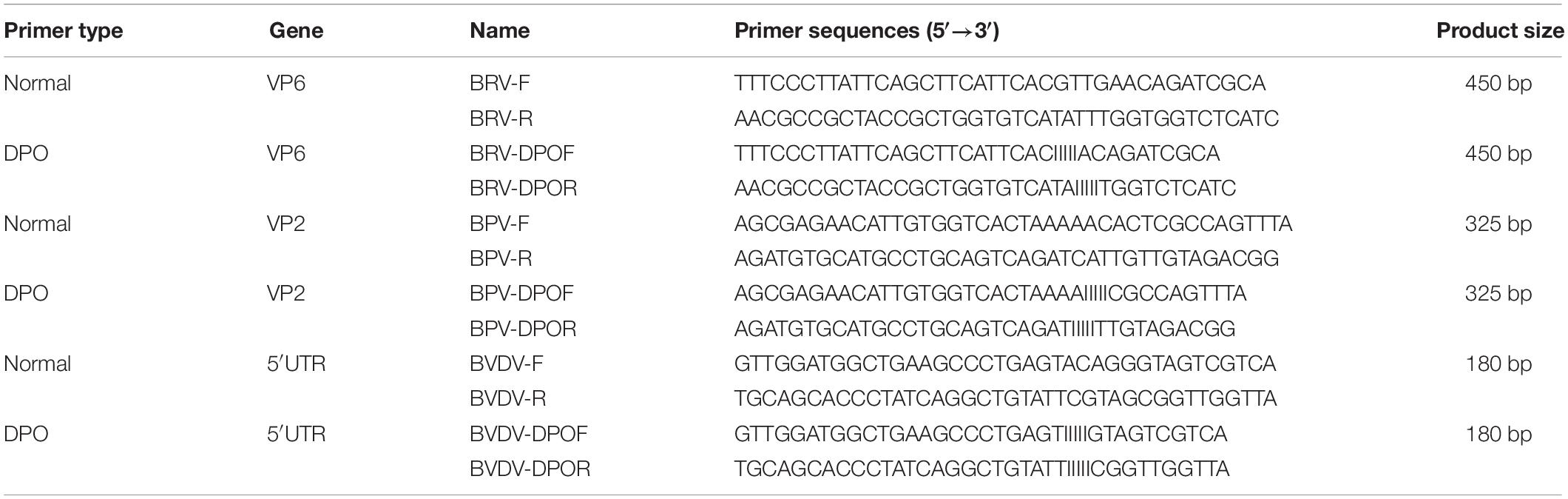
Dual-priming oligonucleotides are composed of two unequal regions (a long 5′-segment and a short 3′-segment) linked with 5 poly (I) stretches (Chun et al., 2007). The primers for DPO-nanoPCR and conventional PCR were designed using the Oligo6.0 software based on the conserved regions of VP6, VP2, and 5′ UTR (Table 2) and the amplicon sizes were 450 bp, 325 bp, and 180 bp, respectively.
Sample Collection and RNA/DNA Isolation
We collected 269 fecal samples from the Heilongjiang, Jilin, Liaoning, and Inner Mongolia provinces. BRV (strain NCDV), BPV (ATCC strain VR-767), BVDV (strain BA), bovine respiratory syncytial virus (BRSV; strain 391-2), PoRV (strain JL94), PPV (strain TJ), TGEV (strain TH98), and PEDV (strain LJB/03) were stored in our lab. BRV (strain NCDV), BPV (ATCC strain VR-767), and BVDV (strain BA) were propagated using the MA-104, BT, and MDBK cell lines, respectively.
We extracted nucleic acids from the viruses using magnetic beads (PuriMag Biotechnology Ltd., Xiamen, China) with modifications of previously described protocols (Smerkova et al., 2013; Clark et al., 2019; Li et al., 2019; Oberacker et al., 2019). First, 500 μL of phosphate-buffered saline was added to 0.2 g of feces in a centrifuge tube, shaken for 2 min, and frozen and thawed three times. After centrifugation at 12,000 r/min for 10 min, 250 μL of the supernatant was transferred into an EP tube. 500 μL of the lysis buffer [4 M guanidinium thiocyanate, 0.5 M Tris–HCl, 0.015 M sodium citrate, 5% (w/v) sodium dodecyl sulfate, and 0.1 M EDTANa2] was then added and mixed for 2 min. The mixture was incubated on ice for 10 min and shaken for 1 min followed by the addition of 20 μL of the magnetic bead suspension and mixed. The mixture was incubated at 4°C for 10 min and placed on a magnetic frame for 5 min. The aqueous phase was carefully transferred to a new EP tube following which an equal volume of cold isopropanol was added, mixed, and the mixture was incubated at −20°C for 2 h and placed on a magnetic frame for 5 min. The supernatant was discarded and 1 mL of cold 75% ethanol was subsequently added. After one wash, the tubes were placed on a magnetic frame and incubated for 5 min. The supernatants were discarded and samples were air-dried. Subsequently, 50 μL of diethylpyrocarbonate-treated water was added to dissolve the DNA/RNA pellet. The TransScript Fly First-Strand cDNA Synthesis SuperMix kit (TransGen Biotech Co., Beijing, China) was used to synthesize cDNA from the samples. The DNA and cDNA samples were stored at −20°C.
Preparation and Characterization of Gold Nanoparticles (GNPs)
Gold nanoparticles with an average diameter of 10 nm were synthesized using the Turkevich and Frens synthesis method (Emmanuel et al., 2018). One hundred milliliters of 0.01% gold chloride was boiled for 3 min following which 5 mL of 1% trisodium citrate solution (preheated to 37°C) was rapidly added while stirring with a glass rod. The solution was boiled again for 5–8 min until it turned wine red. The GNPs were visualized using a transmission electron microscope after cooling at room temperature and then stored at 4°C.
Optimization of the Conditions for DPO-nanoPCR Assay
We optimized the annealing temperature, primer concentration, and GNP concentration for the DPO-nanoPCR assay. The recombinant plasmids (pMD19-T-VP6, pMD19-T-VP2, and pMD19-T-5′ UTR) were used as templates. Conventional PCR primers usually require a specific annealing temperature. Theoretically, DPO primers are very specific over a wide range of annealing temperatures owing to their structural features (Chun et al., 2007; Xu et al., 2017). Therefore, different annealing temperatures were used to verify whether the detection can be affected with varying annealing temperatures. We used temperatures between 41 and 65°C that were chosen randomly. The reactions were performed at different annealing temperatures using DPO primers, and conventional primers were used as controls. Since reagents are limited in the multiple PCR amplification system, each pair of primers compete for the reagents in the reaction. To enhance amplification, the volume and ratios of the three pairs of primers need to be optimized (Luo et al., 2015; Wang et al., 2015). The volumes of all the primers (10 μM) used ranged from 0.1 to 1.0 μL in increments of 0.1 μL. To test the effects of GNPs on amplification, the volumes of GNPs (10 nM) used were between 0.1 and 1.0 μL in increments of 0.1 μL.
The reaction conditions were as follows: 95°C for 5 min followed by 30 cycles of 94°C for 30 s, 41–65°C for 30 s, and 72°C for 30 s and a final extension at 72°C for 10 min. Products were visualized on 2% agarose gels.
Analyzing the Sensitivity and Reproducibility of DPO-nanoPCR
To analyze the sensitivity of DPO-nanoPCR, the pMD19-T-VP6, pMD19-T-VP2, and pMD19-T-5′ UTR plasmids were purified using the TIANprep Mini Plasmid Kit (TIANGEN Biotech, Beijing, China) and quantified by UV spectroscopy (Thermo Scientific NanoDrop 2000 Spectrophotometer, Thermo Fisher Scientific, United States). Plasmid copy number (copies/μL) was calculated according to the following equation: [6.02 × 1023 (copy/mol) × DNA amount (g) × 10–9)/(DNA length (dp) × 660 (g/mol/dp)] (Kang et al., 2018). Ten-fold serial dilutions of the recombinant plasmids were used to analyze the sensitivity of DPO-nanoPCR compared to conventional PCR. Conventional PCRs were performed using the same primers and reaction conditions. The amplicons were analyzed by 2% agarose gel electrophoresis. All experiments were repeated and validated by multiple trials.
The reproducibility of DPO-nanoPCR was determined using three different concentrations of standard plasmid. Each dilution was analyzed in three independent experiments performed by two different operators on different days in accordance with MIQE guidelines (Bustin et al., 2009).
Analyzing the Specificity of DPO-nanoPCR
DNA and cDNA samples from BRV, BPV, BVDV, BRSV, PoRV, PPV, TGEV, and PEDV were used to assess the specificity of DPO-nanoPCR. A mixture of the BRV, BPV, and BVDV cultures was used as a positive control. The amplicons were analyzed on a 2% agarose gel.
Detection of Clinical Samples
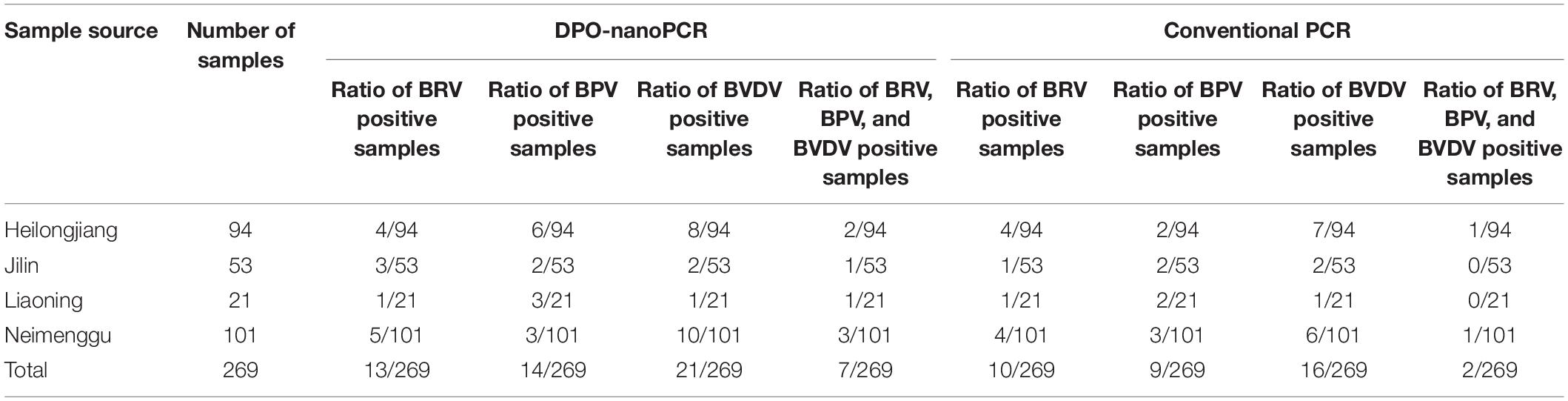
A total of 269 clinical samples were tested by the DPO-nanoPCR developed in this study. These results were compared with those from the conventional PCR assay using 2% agarose gel electrophoresis.
DNA Sequencing
The DPO-nanoPCR amplicons were sent to Kumei Company (Changchun, China) for sequencing. The sequences obtained were confirmed by the DNAStar software and BLAST of GenBank.
Results
Characterization of the Synthesized Gold Nanoparticles
The synthesized GNPs appeared wine red. Transmission electron micrographs showed that these GNPs were relatively regular in size and spherical morphology and had uniform particle size without impurities and agglutination (Figure 1).
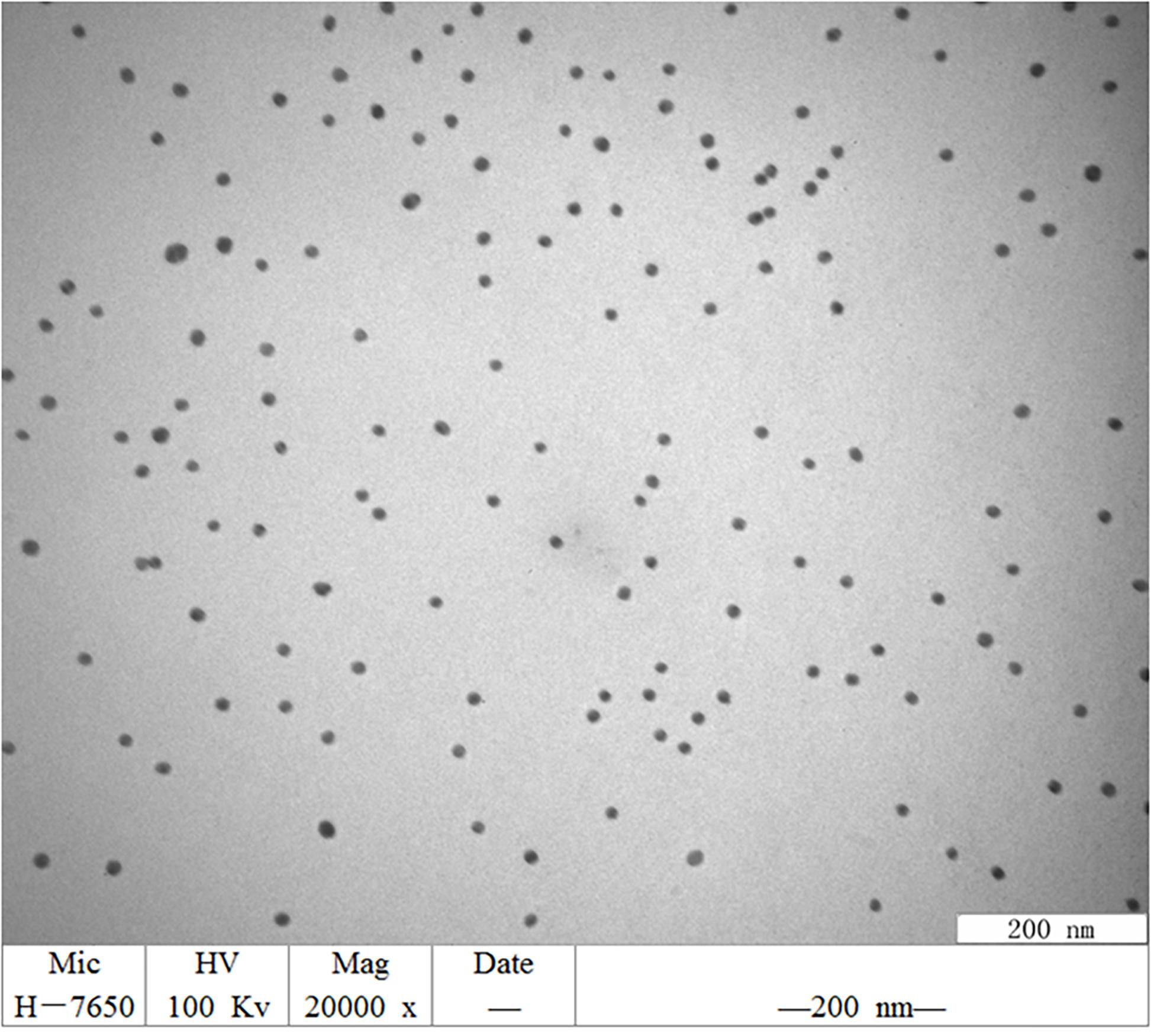
Figure 1. TEM image of GNPs synthesized. GNPs had a relatively regular size and spherical morphology, uniform particle size, no impurity fragments, and no agglutination.
Optimizing the DPO-nanoPCR Assay
Recombinant plasmids (pMD19-T-VP6, pMD19-T-VP2, and pMD19-T-5′ UTR) were used as templates to optimize the DPO-based nanoPCR assay. The annealing temperature used ranged between 41 and 65°C. Figure 2 shows that PCR using DPO primers could efficiently amplify the target under conditions of varying annealing temperatures. However, PCR amplification was most efficient at 56.2°C using conventional primers. As shown in Figures 3, 4, the optimal volume of all the primers (10 μM) was 1.0 μL and that of the GNPs (10 nM) for all the reactions was 0.5 μL.
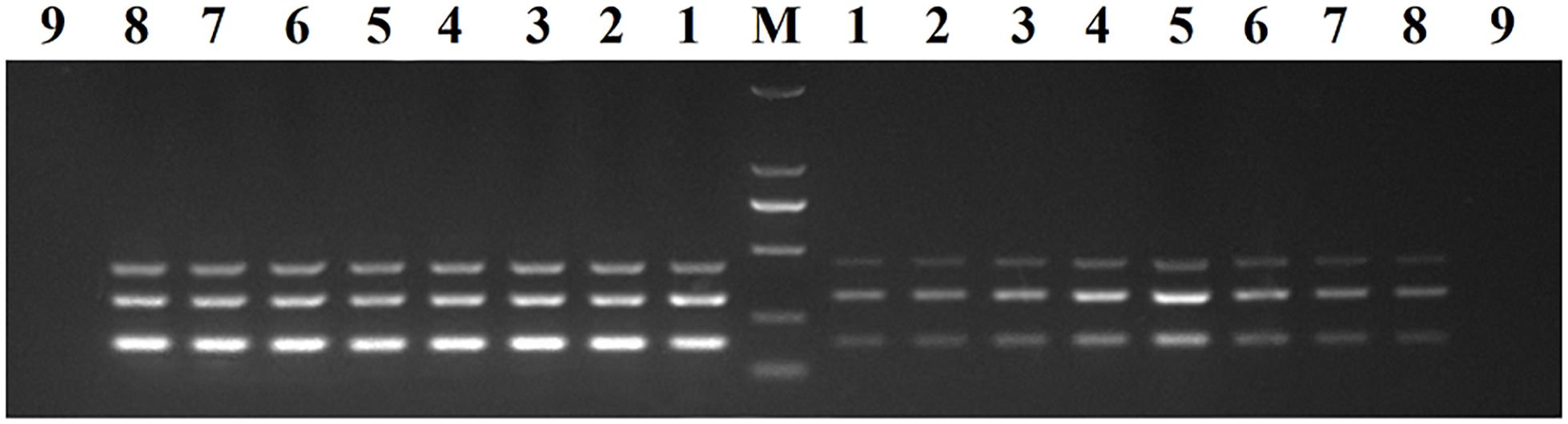
Figure 2. Optimization of the annealing temperature. The results of DPO-nanoPCR were on the left and the results of conventional PCR were on the right. Lane M, DL2000 DNA marker; Lane 1–8, 41.7°C, 45.6°C, 48.8°C, 52.6°C, 56.2°C, 60.9°C, 63.4°C, and 64.1°C; Lane 9, negative control.
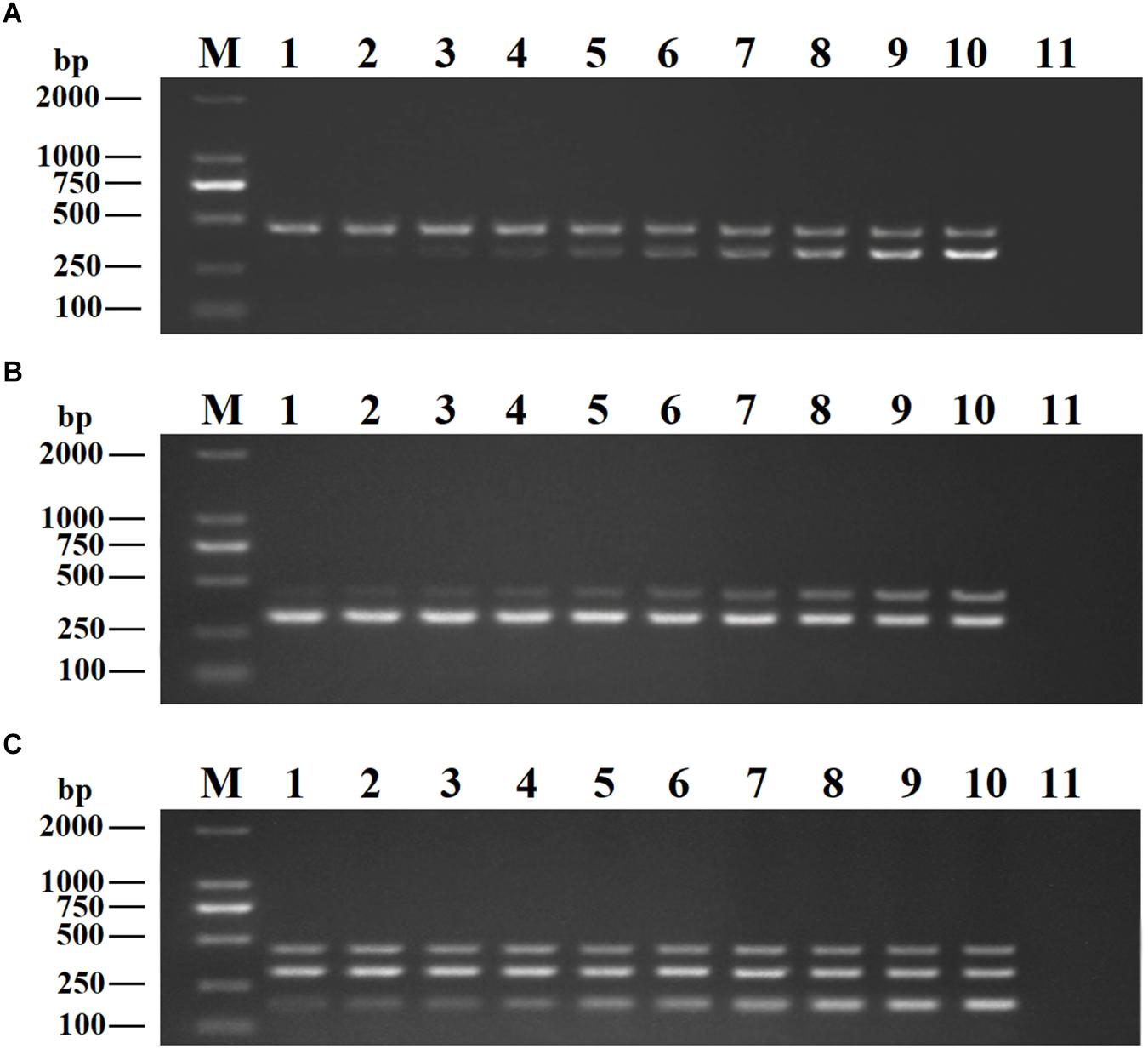
Figure 3. Optimization of primer concentrations. The recombinant plasmids (pMD19-T-VP6, pMD19-T-VP2, and pMD19-T-5′UTR) were used as templates. (A) When the volumes of BRV-DPOF (10 μM) and BRV-DPOR (10 μM) were 1.0 μL, the volumes of BPV-DPOF (10 μM), and BPV-DPOR (10 μM) were tested ranging from 0.1 to 1.0 μL in increments of 0.1 μL. (B) When the volumes of BPV-DPOF (10 μM) and BPV-DPOR (10 μM) were 1.0 μL, the volumes of BRV-DPOF (10 μM), and BRV-DPOR (10 μM) were tested ranging from 0.1 to 1.0 μL in increments of 0.1 μL. (C) When the volumes of BRV-DPOF (10 μM), BRV-DPOR (10 μM), BPV-DPOF (10 μM), and BPV-DPOR (10 μM) were all 1.0 μL, the volumes of BVDV-DPOF (10 μM) and BVDV-DPOR (10 μM) were tested ranging from 0.1 to 1.0 μL in increments of 0.1 μL. Lane M, DL2000 DNA maker; Lane 1–10, 0.1 μL, 0.2 μL, 0.3 μL, 0.4 μL, 0.5 μL, 0.6 μL, 0.7 μL, 0.8 μL, 0.9 μL and 1.0 μL, respectively; Lane 11, negative control.
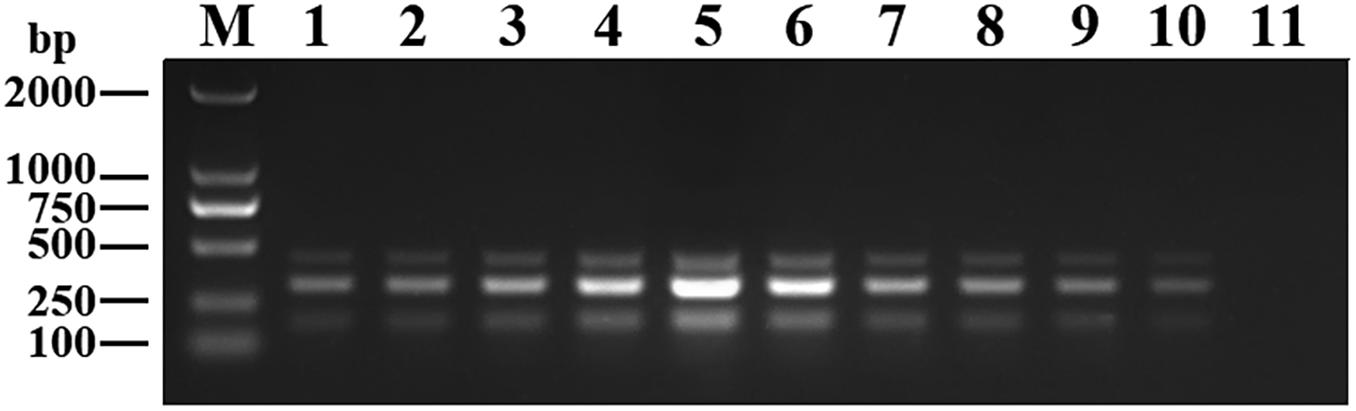
Figure 4. Optimization of GNPs concentration. The volumes of GNPs (10 nM) were tested ranging from 0.1 to 1.0 μL in increments of 0.1 μL. Lane M, DL2000 DNA maker; Lane 1–10, 0.1 μL, 0.2 μL, 0.3 μL, 0.4 μL, 0.5 μL, 0.6 μL, 0.7 μL, 0.8 μL, 0.9 μL, and 1.0 μL, respectively; Lane 11, negative control.
The reaction conditions of DPO-nanoPCR for detecting BRV, BPV, and BVDV were optimized. The DPO-nanoPCR reaction system (25 μL) comprised: 1 μL of cDNA, 0.5 μL of ExTaq (5 U/μL; TaKaRa, Dalian, China), 5 μL of 10 × ExTaq PCR Buffer (with Mg2+; 20 mM), 10 μM of the forward and reverse DPO primers each, 2.5 μL of dNTPs (2.5 mM), and 0.5 μL of the GNPs (10 nM). Nuclease-free water was used to make the volume up to 25 μL. The reaction conditions of DPO-nanoPCR were as follows: 95°C for 5 min followed by 30 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 30 s and a final extension at 72°C for 10 min. The amplicons were visualized using 2% agarose gels.
This DPO-nanoPCR assay was developed for simultaneously detecting BRV, BPV, and BVDV based on the optimized reaction system. Figure 5 shows that bands obtained by the optimized DPO-nanoPCR were clear and specific (with a clean negative control).
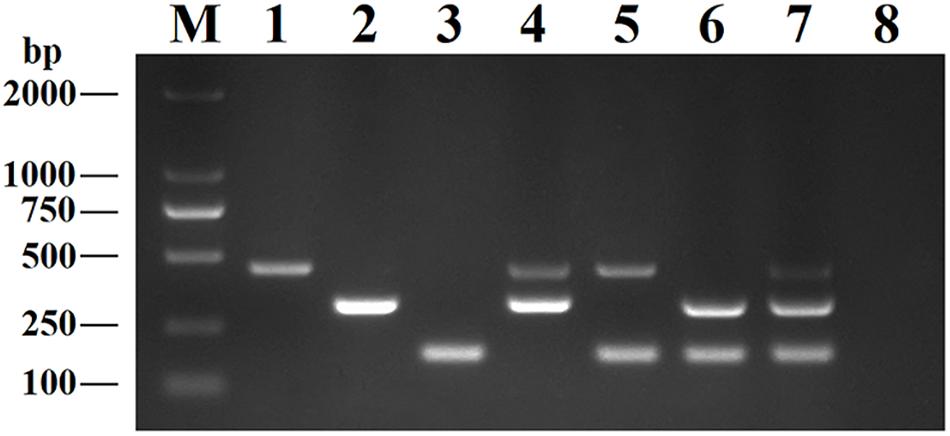
Figure 5. The DPO-nanoPCR assay for detection of BRV, BPV and BVDV. Lane M: DL2000 DNA marker; Lane 1, pMD19-T-VP6; Lane 2, pMD19-T-VP2; Lane 3, pMD19-T-5′UTR; Lane 4, pMD19-T-VP6 and pMD19-T-VP2; Lane 5, pMD19-T-VP6 and pMD19-T-5′UTR; Lane 6, pMD19-T-VP2 and pMD19-T-5′UTR; Lane 7, pMD19-T-VP6, pMD19-T-VP2 and pMD19-T-5′UTR; Lane 8, negative control.
Sensitivity and Reproducibility of the DPO-nanoPCR Assay
The purified pMD19-T-VP6, pMD19-T-VP2, and pMD19-T-5′ UTR plasmids were quantified using UV spectroscopy (Thermo Scientific NanoDrop 2000 Spectrophotometer; Thermo Fisher Scientific, United States). Ten-fold serial dilutions of the recombinant plasmids (9.40 × 1010 copies/μL of pMD19-T-VP6, 5.14 × 1010 copies/μL of pMD19-T-VP2, and 4.09 × 1011 copies/μL of pMD19-T-5′ UTR) were used to determine the sensitivity of the DPO-nanoPCR assay. The results indicated that the detection limits for pMD19-T-VP6, pMD19-T-VP2, and pMD19-T-5′ UTR were 9.40 × 102 copies/μL, 5.14 × 103 copies/μL, and 4.09 × 101 copies/μL, respectively (Figure 6B) and those using conventional PCR were 9.40 × 104 copies/μL, 5.14 × 105 copies/μL, and 4.09 × 104 copies/μL (Figure 6A), respectively. Thus, the sensitivity of DPO-nanoPCR was at least 100-fold higher than that of conventional PCR. The reproducibility of DPO-nanoPCR assay was evaluated by testing different concentrations of standard plasmids. Detection results were identical (Supplementary Figure S1). The results indicated satisfied reproducibility for DPO-nanoPCR assay.
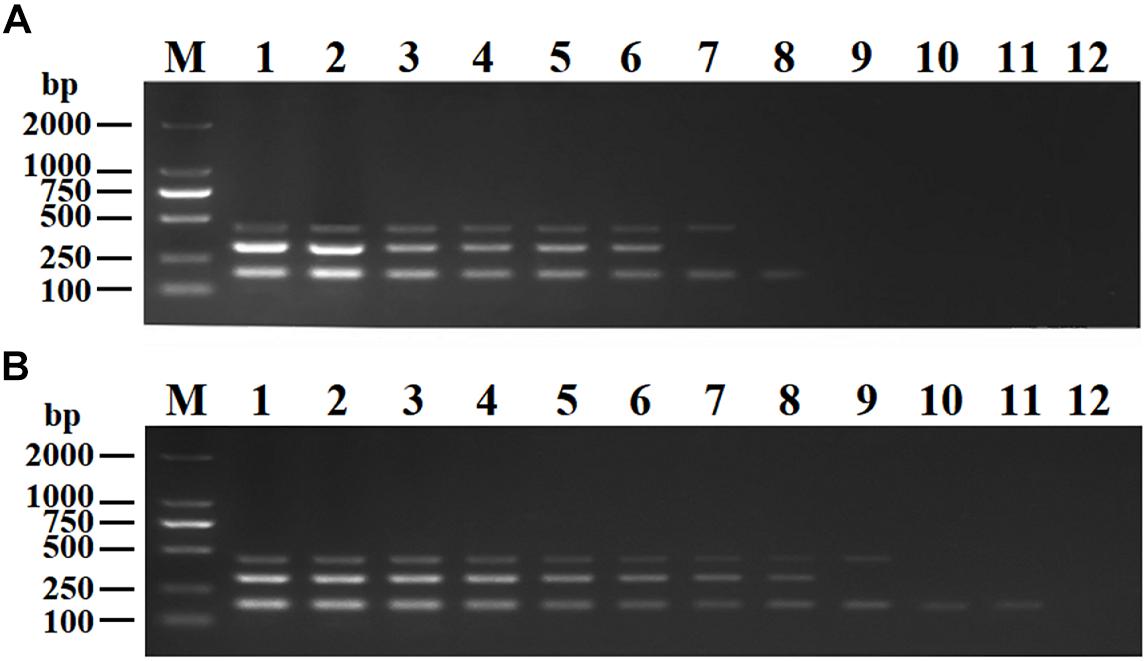
Figure 6. Sensitivity of conventional PCR (A) and DPO-nanoPCR (B). A serial 10-fold diluted plasmid mixture was used. Lane M, DL2000 DNA marker; Lane 1–11, pMD19-T-VP6 concentrations ranging from 9.40 × 1010copies/μL to 9.40 × 100 copies/μL, pMD19-T-VP2 concentrations ranging from 5.14 × 1010 copies/μL to 5.14 × 100 copies/μL, and pMD19-T-5′UTR concentrations ranging from 4.09 × 1011 to 4.09 × 101 copies/μL; Lane 12, negative control.
Specificity of the DPO-nanoPCR Assay
To analyze the specificity of DPO-nanoPCR, DNA or cDNA samples from BRV, BPV, BVDV, BRSV, PoRV, PPV, TGEV, and PEDV were used. The results showed that DPO-nanoPCR could not amplify any of the other five viruses except BRV, BPV, and BVDV (Figure 7), indicating that the assay is specific.
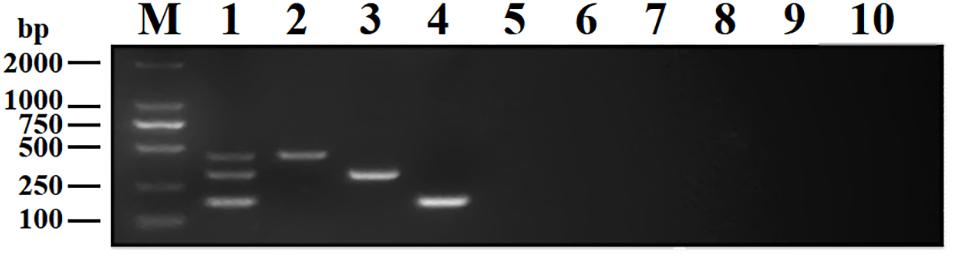
Figure 7. Specificity of the DPO-nanoPCR assay for detecting BRV, BPV, and BVDV. Lane M, DL2000 DNA marker; Lane 1, BRV, BPV, and BVDV; Lane 2, BRV; Lane 3, BPV; Lane 4, BVDV; Lane 5, BRSV; Lane 6, PoRV; Lane 7, PPV; Lane 8, PEDV; Lane 9, TGEV; Lane 10, negative control.
Detection of Clinical Samples
DPO-nanoPCR and conventional PCR were used to test 269 clinical samples. The results are shown in Table 3. Thirteen (4.8%), fourteen (5.2%), and twenty-one samples (7.8%) were positive for BRV, BPV, and BVDV, respectively using DPO-nanoPCR. Ten (3.7%), nine (3.3%), and sixteen samples (5.9%) were positive for BRV, BPV, and BVDV, respectively, using conventional PCR. Of all the clinical samples, 2.6% (7 out of 269) were positive for BRV, BPV, and BVDV by DPO-nanoPCR, and 0.7% (2 out of 269) were positive for BRV, BPV, and BVDV by conventional PCR (Table 3).
Positive results obtained by conventional PCR were consistent with DPO-nanoPCR. Three samples were positive for BRV by DPO-nanoPCR but negative by conventional PCR. Five samples were positive for BPV by DPO-nanoPCR but negative by conventional PCR. Three samples were positive for BVDV by DPO-nanoPCR but negative by conventional PCR. However, there were no samples that were found to be negative using DPO-nanoPCR but positive using conventional PCR. Sequence analysis showed high similarity (100%) between the reference sequences of the target viruses and the DPO-nanoPCR amplicons. All these results indicated that DPO-nanoPCR assay is more sensitive than conventional PCR.
Discussion
With the advances in the cattle industry, large-scale and intensive breeding has led to increased physical contact between cattle. This often enables and accelerates the spread of contagious diseases resulting in mixed infections of two or more pathogens. BRV, BPV, and BVDV are all transmitted by the fecal-oral route and shed through feces. The clinical symptoms of BRV, BPV, and BVDV infections are similar. They all primarily infect newborn calves and cause diarrhea (Rigo-Adrover et al., 2019). Mixed infections cause severe diarrhea that leads to increased mortality (Ruiz et al., 2015; Fan et al., 2017) and make clinical diagnosis difficult. Therefore, it is imperative to develop a diagnostic method that can simultaneously detect multiple pathogens resulting in enhanced epidemic surveillance. In this study, we combined DPO primers with a nanoPCR assay to establish a multiplex DPO-nanoPCR method for the simultaneous detection of BRV, BPV, and BVDV. To the best of our knowledge, this is the first report of a detection method combining DPO primers with nanoPCR.
Multiplex PCR is a rapid and economical assay that is often used for the detection of mixed infections. However, conventional primer system-based multiplex PCR using multi-primer sets often shows a lower specificity of amplification owing to primer competition, formation of primer dimers, or the different annealing temperatures used (Chun et al., 2007). DPOs have two separate primer segments – one of which is longer than the other – joined by a polydeoxyinosine [poly (I)] linker. Since DPO primers have special structures, non-specific hybridization between primers, and nucleotide sequences is prevented such that non-specific amplification can be eliminated without disrupting the amplification of target sequences (Chun et al., 2007; Fu et al., 2017; Xu et al., 2017).
We used the DPO-nanoPCR assay to detect the presence of BRV, BPV, BVDV, and other viruses. Only the target viruses were specifically detected and the presence of no other viruses were observed. Moreover, the DPO primers were not sensitive to changes in the annealing temperature. Using the 41–65°C range of annealing temperatures, there was specific amplification of the target genes. The specificity of detection was not affected upon altering the annealing temperatures within a certain range. So far, various PCR methods using DPO primers have been widely used in multiplex PCR, genotyping PCR, real-time PCR, and reverse transcription PCR for the detection of bacterial and viral pathogens (Chung et al., 2014; Xu et al., 2017). It has been reported that DPO-based PCR showed higher specificity for target sequences compared to conventional PCR (Ma et al., 2015; Chung et al., 2016). Taken together, these findings prove that DPO-based PCR is a reliable method for clinical diagnosis.
In this study, efficiency of the DPO-PCR assay was improved using nanoparticles. Nanofluids are formed upon the addition of GNPs into the PCR system. Nanofluids possess greater thermal conductivity. Thus, thermal conductivity is enhanced in nanoparticle-containing PCR systems that enables attaining the target temperature in a shorter time. Efficient heat transfer generates a larger number of amplicons, thereby improving the sensitivity of the reaction (Gabriel et al., 2018). We developed a multiplex DPO-nanoPCR assay for detecting BRV, BPV, and BVDV that is 100–1000 times more sensitive than conventional multiplex PCR. This indicates that the GNPs increase productivity by acting as modulators of PCR. Using the multiplex DPO-nanoPCR method to detect BRV, BPV, and BVDV in clinical samples showed that the target viruses could be specifically detected, and the DPO-nanoPCR assay was more sensitive than conventional PCR. It has also been reported that nanoRT-PCR exhibits a 10–100-fold higher sensitivity than conventional RT-PCR. Ma et al. (2013) established a nanoPCR method to rapidly detect and distinguish between the field and vaccine strains of the pseudorabies virus. The sensitivity of this method was 100–1000 times higher than that of conventional PCR (Ma et al., 2013). Liu et al. (2019) developed a nanoPCR assay for detecting BRSV. The sensitivity of this assay was also 10 times higher than that of conventional PCR (Liu et al., 2019). These studies prove that the addition of GNPs to PCRs effectively increase the sensitivity of PCR.
To the best of our knowledge, this is the first report of using a combination of nanoPCR with DPOs for simultaneously detecting BRV, BPV, and BVDV. Detection using DPO-nanoPCR was 100–1000 times more sensitive than conventional multiplex PCR. Thus, this DPO-nanoPCR assay is a new powerful tool that has great potential in clinical diagnoses of BRV, BPV, and BVDV.
Data Availability Statement
Nucleotide sequences generated for this study can be found in the NCBI GenBank, MN565845, MN565846, MN565847, MN565848, MN565849, MN565850, MN565851, MN565852, MN565853, MN565854, MN565855, MN565856, MN565857, MN567095, MN567096, MN567097, MN567098, MN567099, MN567100, MN567101, MN567102, MN567103, MN567104, MN567105, MN567106, MN567107, MN567108, MN565858, MN565859, MN565860, MN565861, MN565862, MN565863, MN565864, MN565865, MN565866, MN565867, MN565868, MN565869, MN565870, MN565871, MN565872, MN565873, MN565874, MN565875, MN565876, MN565877, and MN565878.
Ethics Statement
This study was carried out in accordance with the principles of the Basel Declaration and recommendations of Guidelines on Animal Experimentation, the Ethical Committee for Animal Sciences of Heilongjiang Province. The protocol was approved by the Ethical Committee for Animal Sciences of Heilongjiang Province.
Author Contributions
MW and YY developed the DPO-nanoPCR assay. YJ synthesized the gold nanoparticles. RW and LW optimized the reaction conditions. HZ collected the clinical samples. WC detected the clinical samples. XQ, YL, YX, and LT conceived the project. XQ was the grant holder and drafted the manuscript. All authors read, revised, and approved the final manuscript.
Funding
This work was supported by grants from the National Key Research and Development Plan Project (No. 2016YFD0500904), the “Academic Backbone” Project of Northeast Agricultural University (18XG21), and the Natural Science Foundation of Heilongjiang Province (C2016028). The funding bodies played no role in the design of the study, data collection, analysis, interpretation, or in writing the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Chuang Xu from Heilongjiang Bayi Agricultural University and Jing Bai from Northeast Agricultural University for help with sampling.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.02884/full#supplementary-material
FIGURE S1 | Reproducibility test results. The reproducibility of DPO-nanoPCR assay was evaluated by testing different concentrations of standard plasmids. Each dilution was analyzed in three independent experiments performed by two different operators (A,B). A serial 10-fold diluted plasmid mixture was used. Lane M, DL2000 DNA marker. (A1,B1) Lane 1–11, pMD19-T-VP6 concentrations ranging from 9.40 × 1010 copies/μL to 9.40 × 100 copies/μL, pMD19-T-VP2 concentrations ranging from 5.14 × 1010 copies/μL to 5.14 × 100 copies/μL, and pMD19-T-5′UTR concentrations ranging from 4.09 × 1011 to 4.09 × 101 copies/μL. Lane 12, negative control. (A2,B2) Lane 1–11, pMD19-T-VP6 concentrations ranging from 9.40 × 109 copies/μL to 9.40 × 10–1 copies/μL, pMD19-T-VP2 concentrations ranging from 5.14 × 109 copies/μL to 5.14 × 10–1 copies/μL, and pMD19-T-5′UTR concentrations ranging from 4.09 × 1010 to 4.09 × 100 copies/μL. Lane 12, negative control. (A3,B3) Lane 1–11, pMD19-T-VP6 concentrations ranging from 9.40 × 108 copies/μL to 9.40 × 10–2 copies/μL, pMD19-T-VP2 concentrations ranging from 5.14 × 108 copies/μL to 5.14 × 10–2 copies/μL, and pMD19-T-5′UTR concentrations ranging from 4.09 × 109 to 4.09 × 10–1 copies/μL. Lane 12, negative control.
References
Bustin, S. A., Benes, V., Garson, J. A., Hellemans, J., Huggett, J., Kubista, M., et al. (2009). The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 55, 611–622. doi: 10.1373/clinchem.2008.112797
Charoenlarp, W., Frankena, K., Strain, S. A. J., Guelbenzu-Gonzalo, M., Graham, J., and Byrne, A. W. (2018). Spatial and risk factor analysis of bovine viral diarrhoea (BVD) virus after the first-year compulsory phase of BVD eradication programme in Northern Ireland. Prev. Vet. Med. 157, 34–43. doi: 10.1016/j.prevetmed.2018.05.011
Chernick, A., and Frank, V. D. M. (2017). Evolution of Bovine viral diarrhea virus in Canada from 1997 to 2013. Virology 509, 232–238. doi: 10.1016/j.virol.2017.06.024
Chun, J. Y., Kim, K. J., Hwang, I. T., Kim, Y. J., Lee, D. H., Lee, I. K., et al. (2007). Dual priming oligonucleotide system for the multiplex detection of respiratory viruses and SNP genotyping of CYP2C19 gene. Nucleic Acids Res. 35:e40. doi: 10.1093/nar/gkm051
Chung, W. C., Jeon, E. J., Oh, J. H., Park, J. M., Kim, T. H., Cheung, D. Y., et al. (2016). Dual-priming oligonucleotide-based multiplex PCR using tissue samples from the rapid urease test kit for the detection of Helicobacter pylori in bleeding peptic ulcers. Dig. Liver Dis. 48, 899–903. doi: 10.1016/j.dld.2016.04.012
Chung, W. C., Jung, S. H., Oh, J. H., Kim, T. H., Cheung, D. Y., Kim, B. W., et al. (2014). Dual-priming oligonucleotide-based multiplex PCR using tissue samples in rapid urease test in the detection of Helicobacter pylori infection. World J. Gastroenterol. 20, 6547–6553. doi: 10.3748/wjg.v20.i21.6547
Clark, K. D., Zhu, C., and Anderson, J. L. (2019). Maximizing ion-tagged oligonucleotide loading on magnetic ionic liquid supports for the sequence-specific extraction of nucleic acids. Anal. Chem. 91, 5945–5952. doi: 10.1021/acs.analchem.9b00350
Crawford, S. E., Ramani, S., Tate, J. E., Parashar, U. D., Svensson, L., Hagbom, M., et al. (2017). Rotavirus infection. Nat. Rev. Dis. Primers 3, 1–39. doi: 10.1038/nrdp.2017.83
Dennehy, P. H. (2015). Rotavirus infection: a disease of the past? Infect. Dis. Clin. North Am. 29, 617–635. doi: 10.1016/j.idc.2015.07.002
Dudleenamjil, E., Lin, C. Y., Dredge, D., Murray, B. K., Robison, R. A., and Johnson, F. B. (2010). Bovine parvovirus uses clathrin-mediated endocytosis for cell entry. J. Gen. Virol. 91, 3032–3041. doi: 10.1099/vir.0.024133-0
Emmanuel, A., Luca, P., Asterios, G., and Luca, M. (2018). A model for the formation of gold nanoparticles in the citrate synthesis method. Chem. Eng. Sci. 191, 318–331. doi: 10.1016/j.ces.2018.06.046
Fan, Q., Xie, Z., Xie, Z., Deng, X., Xie, L., Huang, L., et al. (2017). Development of a GeXP-multiplex PCR assay for the simultaneous detection and differentiation of six cattle viruses. PLoS One 12:e0171287. doi: 10.1371/journal.pone.0171287
Fu, W., Wei, S., Wang, C., Du, Z., Zhu, P., Wu, X., et al. (2017). A temperature-tolerant multiplex elements and genes screening system for genetically modified organisms based on dual priming oligonucleotide primers and capillary electrophoresis. Food Chem. 229, 396–402. doi: 10.1016/j.foodchem.2017.02.088
Gabriel, S., Rasheed, A. K., Siddiqui, R., Appaturi, J. N., Fen, L. B., and Khan, N. A. (2018). Development of nanoparticle-assisted PCR assay in the rapid detection of brain-eating amoebae. Parasitol. Res. 117, 1801–1811. doi: 10.1007/s00436-018-5864-0
Ito, T., and Suzaki, K. (2017). Universal detection of phytoplasmas and Xylella spp. by TaqMan singleplex and multiplex real-time PCR with dual priming oligonucleotides. PLoS One 12:e0185427. doi: 10.1371/journal.pone.0185427
Kailasan, S., Halder, S., Gurda, B., Bladek, H., Chipman, P. R., McKenna, R., et al. (2015). Structure of an enteric pathogen, bovine parvovirus. J. Virol. 89, 2603–2614. doi: 10.1128/JVI.03157-14
Kang, C. W., Lim, H. G., Yang, J., Noh, M. H., Seo, S. W., and Jung, G. Y. (2018). Synthetic auxotrophs for stable and tunable maintenance of plasmid copy number. Metab. Eng. 48, 121–128. doi: 10.1016/j.ymben.2018.05.020
Khodakaram-Tafti, A., and Farjanikish, G. H. (2017). Persistent bovine viral diarrhea virus (BVDV) infection in cattle herds. Iran J. Vet. Res. 18, 154–163.
Kommedal, Ø, Simmon, K., Karaca, D., Langeland, N., and Wiker, H. G. (2012). Dual priming oligonucleotides for broad-range amplification of the bacterial 16S rRNA gene directly from human clinical specimens. J. Clin. Microbiol. 50, 1289–1294. doi: 10.1128/JCM.06269-11
Lanyon, S., Hill, F., Reichel, M., and Brownlie, J. (2014). Bovine viral diarrhoea: pathogenesis and diagnosis. Vet. J. 199, 201–209. doi: 10.1016/j.tvjl.2013.07.024
Lee, H. J., Choi, J., Hwang, T. S., Shong, Y. K., Hong, S. J., and Gong, G. (2010). Detection of BRAF mutations in thyroid nodules by allele-specific PCR using a dual priming oligonucleotide system. Am. J. Clin. Pathol. 133, 802–808. doi: 10.1309/AJCPO3F2ENKMDTUS
Lee, H. R., Kim, S. Y., Chang, H. E., Song, S. H., Lee, H. S., Park, K. U., et al. (2010). Novel multiplex PCR using dual-priming oligonucleotides for detection and discrimination of the Mycobacterium tuberculosis complex and M. bovis BCG. J. Clin. Microbiol. 48, 4612–4614. doi: 10.1128/JCM.00872-10
Li, H. K., Huang, J. H., Lv, J. H., An, H. J., Zhang, X. D., Zhang, Z. Z., et al. (2005). Nanoparticle PCR: nanogold-assisted PCR with enhanced specificity. Angew Chem. Int. Ed. 44, 2–5. doi: 10.1002/anie.200500403
Li, M., Lin, Y. C., Wu, C. C., and Liu, H. S. (2005). Enhancing the efficiency of a PCR using gold nanoparticles. Nucleic Acids Res. 33:e184. doi: 10.1093/nar/gni183
Li, P., Mi, R., Zhao, R., Li, X., Zhang, B., Yue, D., et al. (2019). Quantitative real-time PCR with high-throughput automatable DNA preparation for molecular screening of Nosema spp. in Antheraea pernyi. J. Invertebr. Pathol. 164, 16–22. doi: 10.1016/j.jip.2019.04.003
Liu, Z., Li, J., Liu, Z., Li, J., Li, Z., Wang, C., et al. (2019). Development of a nanoparticle-assisted PCR assay for detection of bovine respiratory syncytial virus. BMC Vet. Res. 15:110. doi: 10.1186/s12917-019-1858-0
Luo, J. G., Ge, J. W., Tang, L. J., Qiao, X. Y., Jiang, Y. P., Cui, W., et al. (2013). Development of a loop-mediated isothermal amplification assay for rapid detection of bovine parvovirus. J. Virol. Methods 191, 155–161. doi: 10.1016/j.jviromet.2012.05.002
Luo, Y., Liang, L., Zhou, L., Zhao, K., and Cui, S. (2015). Concurrent infections of pseudorabies virus and porcine bocavirus in China detected by duplex nanoPCR. J. Virol. Methods 219, 46–50. doi: 10.1016/j.jviromet.2015.03.016
Ma, X., Cui, Y., Qiu, Z., Zhang, B., and Cui, S. (2013). A nanoparticle-assisted PCR assay to improve the sensitivity for rapid detection and differentiation of wild-type pseudorabies virus and gene-deleted vaccine strains. J. Virol. Methods 193, 374–378. doi: 10.1016/j.jviromet.2013.07.018
Ma, X., Xu, H., Shi, L., Yang, P., Zhang, L., Sun, X., et al. (2015). A multiplex PCR assay for the detection of five influenza viruses using a dual priming oligonucleotide system. BMC Infect. Dis. 15:93. doi: 10.1186/s12879-015-0818-y
Oberacker, P., Stepper, P., Bond, D. M., Höhn, S., Focken, J., Meyer, V., et al. (2019). Bio-On-Magnetic-Beads (BOMB): open platform for high-throughput nucleic acid extraction and manipulation. PLoS Biol. 17:e3000107. doi: 10.1371/journal.pbio.3000107
Qiu, J., Söderlund-Venermo, M., and Young, N. S. (2017). Human Parvoviruses. Clin. Microbiol. Rev. 30, 43–113. doi: 10.1128/CMR.00040-16
Quintero Barbosa, J., Corredor Figueroa, A. P., Salas, S. S., Camargo, H., Sanchéz, A., Tobón, J., et al. (2019). High prevalence of persistently infected animals from bovine viral diarrhea in Colombian cattle. BMC Vet. Res. 15:23. doi: 10.1186/s12917-018-1769-5
Rehman, A., Sarwar, Y., Raza, Z. A., Hussain, S. Z., Mustafa, T., Khan, W. S., et al. (2015). Metal nanoparticles assisted polymerase chain reaction for strain typing of Salmonella Typhi. Analyst 140, 7366–7372. doi: 10.1039/c5an01286d
Reichel, M. P., Lanyon, S. R., and Hill, F. I. (2018). Perspectives on current challenges and opportunities for bovine viral diarrhoea virus eradication in Australia and New Zealand. Pathogens 7, 14–23. doi: 10.3390/pathogens7010014
Rigo-Adrover, M., Knipping, K., Garssen, J., Saldaña-Ruíz, S., Franch, À, Castell, M., et al. (2019). Rotavirus double infection model to study preventive dietary interventions. Nutrients 11, 131–146. doi: 10.3390/nu11010131
Ruiz, V., Mozgovoj, M. V., Dus Santos, M. J., and Wigdorovitz, A. (2015). Plant-produced viral bovine vaccines: what happened during the last 10 years? Plant Biotechnol. J. 13, 1071–1077. doi: 10.1111/pbi.12440
Shepherd, F. K., Herrera-Ibata, D. M., Porter, E., Homwong, N., Hesse, R., Bai, J., et al. (2018). Whole genome classification and phylogenetic analyses of rotavirus B strains from the United States. Pathogens 7, 44–58. doi: 10.3390/pathogens7020044
Smerkova, K., Dostalova, S., Vaculovicova, M., Kynicky, J., Trnkova, L., Kralik, M., et al. (2013). Investigation of interaction between magnetic silica particles and lambda phage DNA fragment. J. Pharm. Biomed. Anal. 86, 65–72. doi: 10.1016/j.jpba.2013.07.039
Swiatek, D. L., Palombo, E. A., Lee, A., Coventry, M. J., Britz, M. L., and Kirkwood, C. D. (2010). Characterisation of G8 human rotaviruses in Australian children with gastroenteritis. Virus Res. 148, 1–7. doi: 10.1016/j.virusres.2009.11.013
Wang, J., Cheng, Y., Zhang, M., Zhao, H., Lin, P., Yi, L., et al. (2015). Development of a nanoparticle-assisted PCR (nanoPCR) assay for detection of mink enteritis virus (MEV) and genetic characterization of the NS1 gene in four Chinese MEV strains. BMC Vet Res. 11:1. doi: 10.1186/s12917-014-0312-6
Xu, Y. G., Liu, Z. M., Guan, X. T., Cui, L. C., and Li, S. L. (2015). Dual priming oligo-nucleotide (DPO)-based multiplex PCR assay for specific detection of four diarrheagenic Escherichia coli in food. Lett. Appl. Microbiol. 61, 146–152. doi: 10.1111/lam.12426
Xu, Y. G., Sun, L. M., Wang, Y. S., Chen, P. P., Zhong, M. L., Li, Y. J., et al. (2017). Simultaneous detection of Vibrio cholerae, Vibrio alginolyticus, Vibrio parahaemolyticus and Vibrio vulnificus in seafood using dual priming oligonucleotide (DPO) system-based multiplex PCR assay. Food Control 71, 64–70. doi: 10.1016/j.foodcont.2016.06.024
Yarnall, M. J., and Thrusfield, M. V. (2017). Engaging veterinarians and farmers in eradicating bovine viral diarrhoea: a systematic review of economic impact. Vet. Rec. 181, 347–354. doi: 10.1136/vr.104370
Yeh, J. Y., Lee, J. H., Seo, H. J., Park, J. Y., Moon, J. S., Cho, I. S., et al. (2011). Simultaneous detection of Rift Valley Fever, bluetongue, rinderpest, and Peste des petits ruminants viruses by a single-tube multiplex reverse transcriptase-PCR assay using a dual-priming oligonucleotide system. J. Clin. Microbiol. 49, 1389–1394. doi: 10.1128/JCM.00710-10
Yeşilbaǧ, K., Alpay, G., and Becher, P. (2017). Variability and global distribution of subgenotypes of Bovine viral diarrhea virus. Viruses 9, 128–146. doi: 10.3390/v9060128
Keywords: dual-priming oligonucleotide, nanoparticle-assisted PCR assay, bovine rotavirus, bovine parvovirus, bovine viral diarrhea virus
Citation: Wang M, Yan Y, Wang R, Wang L, Zhou H, Li Y, Tang L, Xu Y, Jiang Y, Cui W and Qiao X (2019) Simultaneous Detection of Bovine Rotavirus, Bovine Parvovirus, and Bovine Viral Diarrhea Virus Using a Gold Nanoparticle-Assisted PCR Assay With a Dual-Priming Oligonucleotide System. Front. Microbiol. 10:2884. doi: 10.3389/fmicb.2019.02884
Received: 05 July 2019; Accepted: 29 November 2019;
Published: 12 December 2019.
Edited by:
Souvik Ghosh, Ross University School of Veterinary Medicine, Saint Kitts and NevisReviewed by:
Yashpal S. Malik, Indian Veterinary Research Institute (IVRI), IndiaSoma Chattopadhyay, Institute of Life Sciences (ILS), India
Copyright © 2019 Wang, Yan, Wang, Wang, Zhou, Li, Tang, Xu, Jiang, Cui and Qiao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xinyuan Qiao, cWlhb3hpbnl1YW5AMTI2LmNvbQ==
†These authors have contributed equally to this work
 Mengmeng Wang
Mengmeng Wang Yue Yan
Yue Yan Ruichong Wang2
Ruichong Wang2 Xinyuan Qiao
Xinyuan Qiao