- 1Longping Branch, Graduate School of Hunan University, Changsha, China
- 2Hunan Academy of Agricultural Sciences, Changsha, China
- 3National Agro-Tech Extension and Service Center, Beijing, China
It is well documented that the canonical function of NIa-protease (NIa-Pro) of the potyviruses is responsible for cleaving the viral polyprotein into functional proteins. Although NIa-Pro is vital for the infection cycle of potyviruses, the function of NIa-Pro in the interaction of the potyvirus host is not clear. In this study, NIa-Pro is ectopically expressed from a potato virus X (PVX) vector and infiltrates Nicotiana benthamiana wild type and 16-TGS. The pathogenicity and inhibition of host transcriptional gene silencing (TGS) are characterized. Ectopic expression of NIa-Pro from a PVX vector resulted in severe mosaic symptoms followed by a hypersensitive-like response in N. benthamiana. Furthermore, PepMoV NIa-Pro was able to reverse established TGS of a green fluorescent protein transgene by reducing methylation of promoter sequences in N. benthamiana and possessed the capacity to interfere with the global methylation of N. benthamiana. Taken together, the results of this study likely suggest that PepMoV NIa-Pro is a pathogenicity determinant and a potent suppressor of host TGS and suggest that NIa-Pro may employ novel mechanisms to suppress host antiviral defenses. To the best of our knowledge, this is the first report of a plant RNA virus modulating host TGS in a novel manner by interfering with the establishment of the methylation step of the plant DNA methylation pathway.
Introduction
Potyviruses (genus Potyvirus, family Potyviridae) constitute the largest group of plant-infecting RNA viruses affecting major crops worldwide. Potyviruses encode a large polyprotein that is subsequently cleaved by three virus-encoded proteinases to yield up to 10 mature proteins (Shukla et al., 1994). Additionally, a P3N–PIPO is a fusion of the N-terminal part of P3 and the PIPO protein-coding regions by polymerase slippage (Rodamilans et al., 2015; Untiveros et al., 2016). The main viral proteinase, NIa, is a polyprotein consisting of two domains, viral protein genome-linked (VPg) and protease (NIa-Pro), separated by an inefficiently utilized self-proteolytic site. NIa-Pro encoded by potyviruses plays many roles during the virus infecting cycle. The canonical function of NIa-Pro is the main protease of the virus responsible for the cleavage of most sites in the polyprotein (Sun et al., 2010). Tobacco etch potyvirus encoding NIa-Pro can bind NIb, which is critical for tobacco etch virus (TEV) genome replication (Daros et al., 1999). Further proteomic studies demonstrated that TEV NIa-Pro could interact with 76 host proteins involved in response to host stress, metabolism, and photosynthesis (Martínez et al., 2016). Additionally, NIa-Pro induced vein necrosis in tobacco by potato virus Y (PVY) isolates (Faurez et al., 2012), which indicated that NIa-Pro of the potyviruses may be a pathogenicity determinant.
Cytosine DNA methylation, one of the fundamental aspects of stable epigenetic modification, modulates coding genes that involve various cellular biological processes and molecular functions to counter virus infection (Wang et al., 2019). The cytosines in the genome of DNA viruses could be directly methylated through plant DNA methylation; for example, geminivirus transcription is usually disrupted by host DNA methylation, which occurs in the viral genome through RNA-directed DNA methylation (RdDM) (Coursey et al., 2018). Although RNA viruses cannot be directly methylated by plant DNA methylation owing to the absence of DNA in the replication process, the 2b protein encoded by cucumber mosaic virus (CMV) could bind the AGO4-related small RNAs and impairs AGO4 activities to release the DNA methylation of Nicotiana benthamiana, suggesting the modulation of RNA virus replication by plant DNA methylation (Hamera et al., 2012).
To counter the suppression of viral transcription mediated by plant DNA methylation, DNA viruses, mainly from geminiviruses, encode DNA methylation suppressor proteins. The C2 and C4 proteins encoded by tomato leaf curl Yunnan virus (TLCYnV), the V2 protein encoded by Tomato yellow leaf curl virus (TYLCV), and the βC1 and the Rep proteins encoded by geminiviruses can inhibit host transcriptional gene silencing (TGS) and modulate the plant DNA methylation pathway (Yang et al., 2011; Zhang et al., 2011; Rodrıguez-Negrete et al., 2013; Xie et al., 2013; Wang et al., 2014). However, the DNA methylation suppressor proteins encoded by plant RNA viruses have little been documented until now (Hamera et al., 2012).
PepMoV was first identified as an atypical species of potyvirus (Nelson and Wheeler, 1972) and is known to cause serious symptoms in many solanaceous plants (Han et al., 2006). Although most proteins encoded by potyviruses have been well characterized, the functions of PepMoV-encoded proteins are mostly annotated through comparison with other species of potyviruses and remain to be identified.
In this study, the functions of PepMoV-encoded NIa-Pro were characterized by ectopic expression of NIa-Pro from a potato virus X (PVX) vector, and the results demonstrated that PepMoV NIa-Pro is a pathogenicity determinant and a potent suppressor of host TGS and suggest that NIa-Pro may employ novel mechanisms to suppress host antiviral defenses.
Materials and Methods
Plant Materials
Wild-type and 16-TGS lines (Buchmann et al., 2009) of Nicotiana benthamiana Domin were used in this study. Plants were grown inside a culture room set at 25 ± 2°C and 60% relative humidity with a 16-h light/8-h dark illumination. The third or fourth leaves of 6- to 8-week-old plants were infiltrated individually with one of the agrobacterium cultures containing the plasmid made in this study.
Plasmid Constructs and Agroinfiltration
The NIa-Pro gene sequence was cloned from PepMoV isolate HN (GenBank accession No. LN832375) (Zhang et al., 2019) and inserted into a PVX-based vector (pGR106) between the ClaI and SalI sites. The recombinant pGR106 vectors expressing the NIa-Pro gene or βC1 gene [as a positive control, cloned from tomato yellow leaf curl China virus betasatellite (TYLCCNB) and described previously] (Buchmann et al., 2009) were transformed into Agrobacterium tumefaciens GV3101. The Agrobacterium cultures were cultured in Luria broth with appropriate antibiotic selection at 28°C for 16 h. The cells were centrifugated and resuspended in infiltration medium (10 mM of MgCl2 and 20 mM of acetosyringone) to an OD600 0.4 to 0.6 and incubated at room temperature for 3 h before injection into leaves. The underside of the third or fourth leaf of N. benthamiana was infiltrated with syringe (Rajamaki and Valkonen, 2009).
H2O2 Detection
The production and accumulation of H2O2 was detected visually in leaves using the 3,3′-diaminobenzidine (DAB)–HCl (Sigma) uptake method as described previously (Sharma et al., 2010).
DNA Methylation Analysis Using Restriction Digestion and Quantitative Real-Time PCR
Genomic DNA was extracted from plant leaves using the cetyl trimethylammonium bromide (CTAB) reagent (Tiangen, Beijing, China). DraI (TaKaRa, Kyoto, Japan), a methylation-insensitive restriction endonuclease, and McrBC (New England Biolabs, Ipswich, MA, United States). A methylation-dependent restriction endonuclease was used for digestion analysis of genomic DNA according to the manufacturer’s instructions. Briefly, genomic DNA (10 ng) was digested by 50 U of the respective enzyme in a 50-ml reaction mix. Digested genomic DNA was immediately separated by 1.5% agarose gel electrophoresis. For quantitative real-time PCR (qPCR) analysis, specific primer pairs for 35S promoter or GFP gene (Supplementary Table S1) were used, and NbActin was set as reference gene (Li et al., 2015).
RNA Extraction and Reverse Transcription Quantitative Real-Time PCR Analysis
Total RNA was isolated from infiltrated leaves of wild-type or 16-TGS N. benthamiana using TRIzol reagent (Tiangen, Beijing, China). For reverse transcription quantitative real-time PCR (RT-qPCR) analysis, specific primer pairs (Supplementary Table S1) were designed by Primer Premier 5 software or referred to published documents, and NbGADPH was set as reference gene (Li et al., 2014).
Western Blotting
Total protein was extracted from infiltrated systemic leaves as described previously (Li et al., 2014). Western blotting was performed with primary mouse monoclonal antibody [green fluorescent protein (GFP) or PVX CP protein monoclonal antibody, HuaAn Beijing, China], followed by goat anti-mouse secondary antibody conjugated to alkaline peroxidase (Bio-Rad). Blotted membranes were washed thoroughly and visualized using NBT/BCIP.
Green Fluorescent Protein Images
To visualize GFP in TGS assays, representative plants were photographed at 14 dpi with a Canon 400D digital camera with a 58-mm yellow filter. GFP fluorescence was activated by a hand-held longwave UV lamp (100 W, Black Ray model B 100 A; UV products).
Results
Phylogenetic Analyses of PepMoV-Encoded NIa-Pro
To examine the evolutionary relationships of NIa-Pro sequences from different potyviruses, a phylogenetic dendrogram of the complete NIa-Pro nucleotide sequences of 16 representative potyvirus species was generated. As shown in Figure 1A, two major subclusters are grouped. Subcluster I contains 11 of 16 potyviruses, and cluster II consists of five of 16 potyviruses. The PepMoV NIa-Pro protein is located in cluster II and shares the highest sequence identity with PVY. Sequence alignment showed that potyvirus NIa-Pro proteins are highly divergent in amino acid sequences. The structure of the NIa-Pro protein was analyzed by the Conserved Domain Database (CCD) available at the National Center for Biotechnology Information (Marchler-Bauer et al., 2011), and only one conserved domain architecture was retrieved for the NIa-Pro protein (Figure 1B). All NIa-Pro proteins from potyviruses contain this conserved domain. The presence of the same conserved position in many potyviruses suggests that NIa-Pro may play an important role during virus infection.
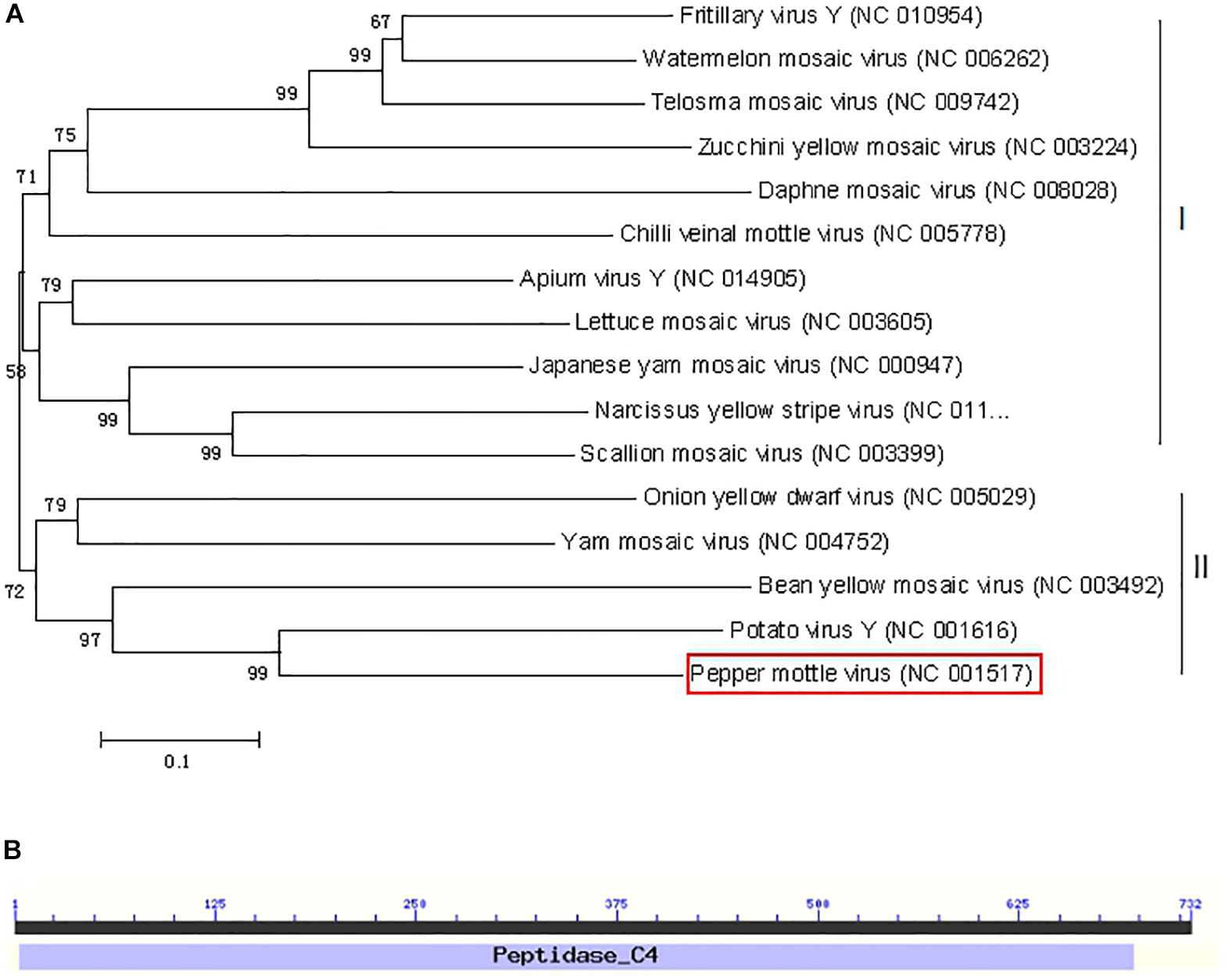
Figure 1. Phylogenetic relationships and domain structure of NIa-Pro nucleotide (nt) sequences of representative potyviruses (NIa-Pro gene of Pepper mottle virus shown in red box. (A) The nucleotide sequences of the NIa-pro gene from 16 potyvirus viruses were aligned using the neighbor-joining method with the MEGA6 program with 1,000 replications. The numbers beside each node represent the percentages for the bootstrap value (>50%). (B) One conserved peptidase domain was identified using the Conserved Domain Database (CDD).
NIa-Pro Is a Pathogenicity Determinant in Nicotiana benthamiana
To determine whether ectopically expressed NIa-Pro is a symptom determinant in Nicotiana benthamiana, the NIa-Pro gene was inserted into a PVX-based vector (pGR106) for overexpression. As shown in Figure 2 (upper panel), the phenotype in plants infected with PVX was similar to that of plants infected with PVX-NIa-Pro at 10 dpi. At 20 dpi, PVX-infected plants showed a loss of veinal chlorosis and mild mosaic symptoms; however, continuous NIa-Pro expression resulted in symptoms of severe mosaic, chlorosis, and leaf deformity at 20 dpi (Figure 2, lower panel). In addition, pronounced necrotic symptoms were observed on the systemic leaves of PVX-βC1-infected plants, but plants inoculated with PVX-NIa-Pro failed to develop the necrotic phenotype (Figure 2, lower panel).
NIa-Pro Elicits a Hypersensitive-Like Response in Nicotiana benthamiana
To determine whether NIa-Pro elicits the hypersensitive-like response (HLR), PVX-, and PVX-NIa-Pro-infected leaves were analyzed by the DAB uptake method. DAB polymers can stain the accumulated H2O2 as a deep brown color, which can be visualized after ethanol clearing of the tissue. As anticipated, compared with PVX-infected plants, PVX-NIa-Pro-infected plants accumulated high concentrations of H2O2 in systemically infected leaves (Figure 3A). To determine whether severe symptoms in the presence of NIa-Pro are a consequence of higher accumulation of PVX, RT-qPCR and Western blotting against PVX CP protein were employed to examine PVX genomic RNA levels and accumulation of CP protein at 20 dpi (Figures 3B,C). More PVX genomic RNAs and CP proteins accumulated in PVX-NIa-Pro-infected plants compared with PVX-infected plants at 20 dpi, suggesting that NIa-Pro is a virulence factor that enhances replication of PVX in N. benthamiana. These data reveal that NIa-Pro is a symptom determinant that can elicit an HLR when expressed from PVX-NIa-Pro.
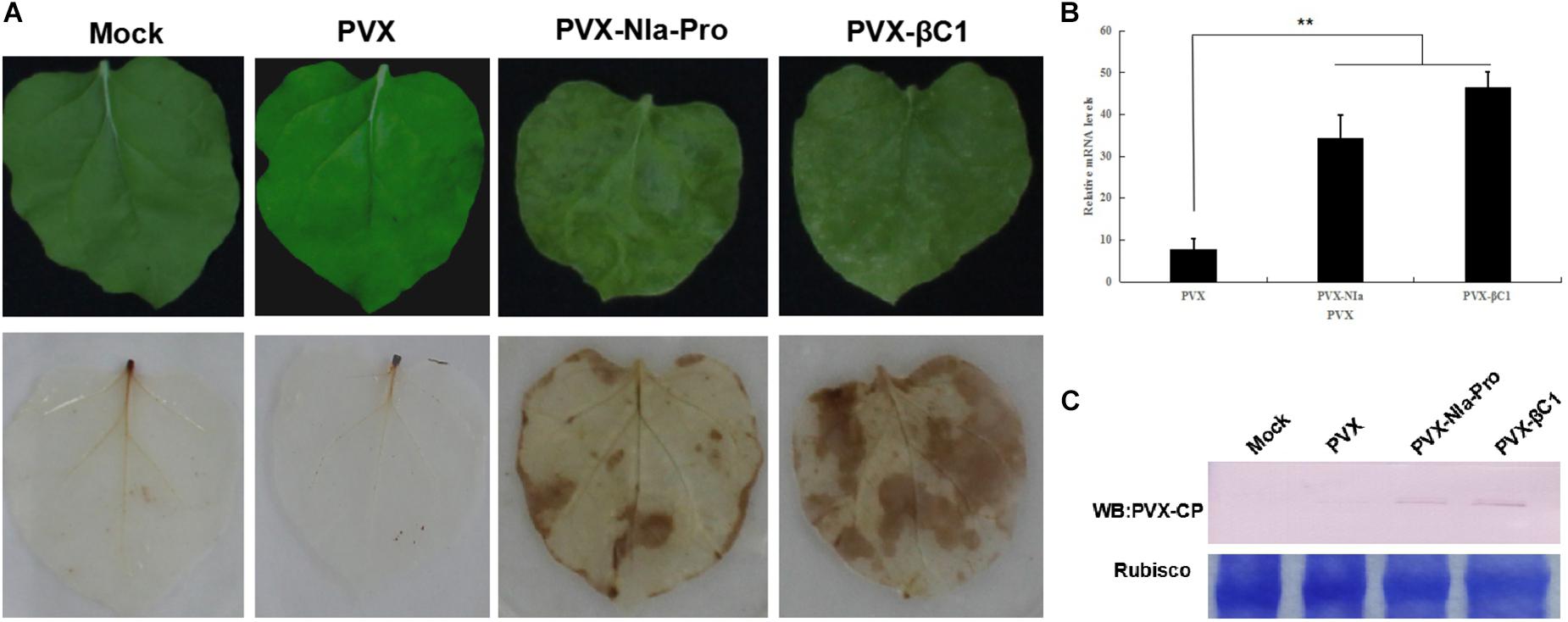
Figure 3. Symptoms exhibited H2O2 accumulation in plants inoculated with potato virus X (PVX) or PVX-NIa-Pro. (A) Severe mosaic symptoms and H2O2 accumulation induced by PVX-NIa-Pro. Nicotiana benthamiana plants were infected with PVX or PVX-NIa-Pro. Upper infected leaves with 3,3’-diaminobenzidine (DAB) staining (lower panel) were photographed directly at 20 dpi. (B) RT-qPCR analysis of PVX genomic RNA in systemic leaves from N. benthamiana plants infected with PVX or PVX-NIa-Pro at 20 dpi. Values are the mean ± SD. Highly significant differences (** p < 0.01) between samples in each pair are indicated. (C) Western blotting analysis of PVX CP protein in systemic leaves from N. benthamiana plants infected with PVX or PVX-NIa-Pro at 20 dpi.
To test if there was any association between NIa-Pro and endoplasmic reticulum (ER) stress, the expression levels of key genes involved in ER stress, including Bip (ER-localized binding protein), HSP90-2 (heat shock protein 90-2), and bZIP60 (basic leucine zipper protein 60), were quantified. qRT-PCR analysis showed that the expression of Bip and HSP90-2 was significantly induced by NIa-Pro; otherwise, the expression of bZIP60 was not influenced by NIa-Pro (Figure 4). These results suggest that NIa-Pro can induce ER stress by stabilizing proteins for correct folding or re-folding in response to ER stress but not the unfold protein response (UPR) to ER stress.
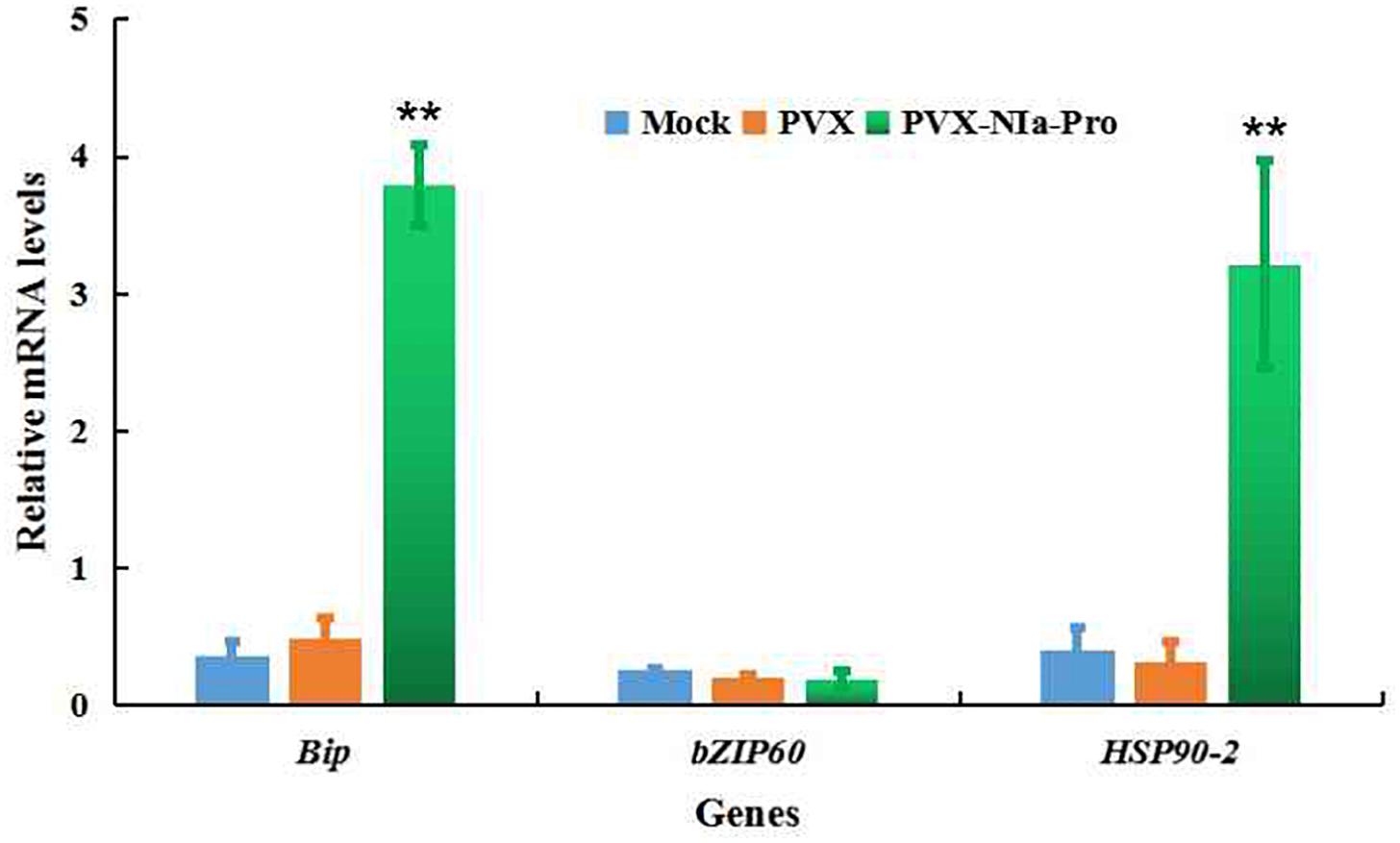
Figure 4. Relative mRNA levels of the indicated endoplasmic reticulum (ER) stress marker genes in NIa-Pro-expressing Nicotiana benthamiana plants as determined by qRT-PCR. Relative mRNA levels of N. benthamiana ER-localized binding (Bip), Heat shock 90-2 (HSP90-2), and basic leucine zipper 60 (bZIP60) were measured in potato virus X (PVX)-NIa-Pro and PVX vector-infected N. benthamiana plants at 20 days post inoculation (dpi). Values are the mean ± SD. Highly significant differences (** p < 0.01) between samples in each pair are indicated.
NIa-Pro Reverses Established Methylation-Mediated Transcriptional Gene Silencing
Plant DNA methylation modulates coding-gene expression to induce TGS (Law and Jacobsen, 2010). To determine whether NIa-Pro inhibited plant methylation-mediated TGS, transgenic N. benthamiana 16-TGS plants containing a GFP transgene downstream of a transcriptionally silenced CaMV 35S promoter were employed in this study. Previously, this assay demonstrated the TGS suppression activity of several geminivirus-encoded proteins (Raja et al., 2008; Yang et al., 2011). The mock culture (GV3101 Agrobacterium tumefaciens cultures), PVX (negative control), PVX-βC1 (positive control), or PVX-NIa-Pro 16-TGS plants were inoculated in the third and fourth leaves. By 14 dpi, mild symptoms were exhibited in systemic leaves of PVX-infected plants; however, the typical βC1-associated phenotypes, including enations and upward leaf curling, developed in systemic leaves of PVX-βC1-infected plants. The phenotypes induced by PVX and PCX-βC1 were in agreement with those described previously (Yang et al., 2011). Severe mosaic symptoms developed in plants infected with PVX-NIa-Pro. In addition, yellow-green GFP fluorescence was visible in plants infected with PVX-βC1 and PVX-NIa-Pro (Figure 5A). As anticipated, Western blot analysis verified the visible fluorescence in PVX-NIa-Pro- or PVX-βC1-inoculated plants (Figure 5B).
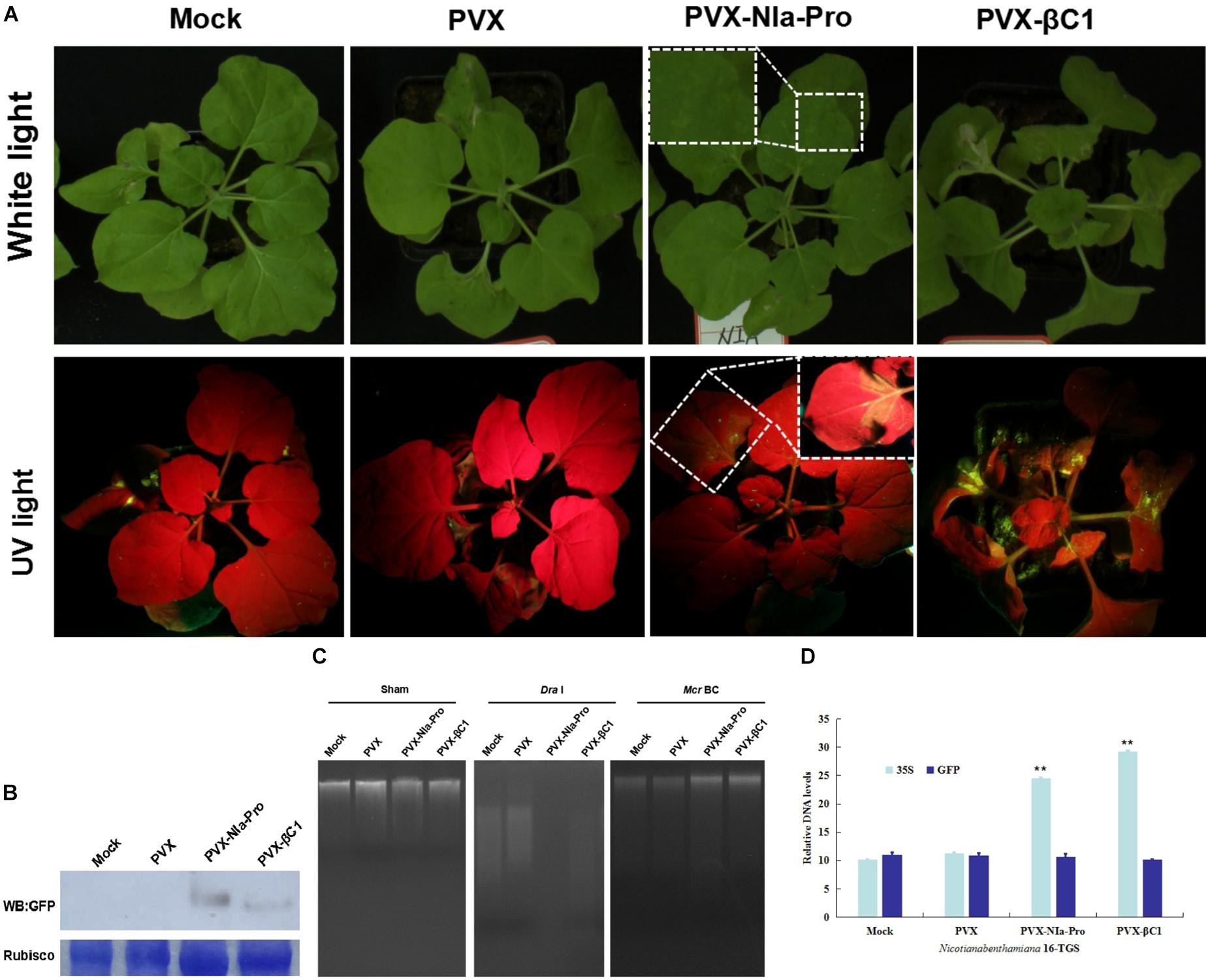
Figure 5. NIa-Pro inhibits transcriptional gene silencing (TGS) of a green fluorescent protein (GFP) transgene and suppresses DNA methylation. (A) Nicotiana benthamiana 16-TGS plants were mock-inoculated or inoculated with potato virus X (PVX), PVX-NIa-Pro, or PVX-βC1 (positive control), and leaves were photographed under white light or UV light at 14 days post inoculation (dpi). (B) Western blotting assay of GFP protein accumulation in systemically infected leaves shown in panel (A). Coomassie blue staining of the large subunit of Rubisco was used as loading controls for Western blotting. (C) DNA methylation analysis by restriction enzyme digestion in NIa-Pro-expressing N. benthamiana plants. Genomic DNA extracted from systemically infected leaves shown in (A) was digested with the methylation-dependent restriction enzyme McrBC and the methylation-insensitive enzyme DraI. “Sham” indicates a mock digestion with no enzyme included. (D) qPCR analysis of DNA levels from systemically infected leaves shown in panel (A) was digested with the methylation-dependent restriction enzyme McrBC. Values are the mean ± SD. Highly significant differences (** p < 0.01) between samples in each pair are indicated.
To determine whether NIa-Pro affects global DNA methylation, a methylation-dependent restriction enzyme (McrBC), which only cleaves methylated DNA sequences (Stewart et al., 2000), was used. Genomic DNA was extracted from leaves of N. benthamiana plants inoculated with mock culture (GV3101 A. tumefaciens cultures), PVX (negative control), PVX-βC1 (positive control), or PVX-NIa-Pro and subjected to restriction digestion. A mock digestion with no enzyme was included (sham) as a control. All four genomic DNA samples were able to digest by DraI, a methylation-independent restriction endonuclease, as indicated by the presence of a “smear” of different sized fragments on the gel (Figure 5C). However, although McrBC was able to cleave a significant portion of the genomic DNA of plants with Mock and PVX into small fragments, DNA from plants expressing NIa-Pro and βC1 was largely resistant to McrBC digestion (Figure 5C). DNA quantification by qPCR of 35S promoter and adjacent GFP gene has shown that the DNA level of 35S of PVX-NIa-Pro was relative higher than that of mock and PVX; otherwise, no difference appeared in adjacent GFP gene (Figure 5D). Thus, the expression of NIa-Pro in N. benthamiana plants likely reduced DNA methylation at a genome-wide level.
NIa-pro Expression Inhibits Transcription of NbAGO4
To provide mechanistic insights into the effects of NIa-Pro on the suppression of DNA methylation and epigenetic TGS, the expression levels of N. benthamiana DNA methyltransferases, demethylases, and key genes involved in RdDM in the presence of NIa-Pro were quantified by RT-qPCR. These genes included N. benthamiana homologs of METHYLTRANSFERASE1 (NbMET1), DRM2 (NbDRM2), CHROMOMETHYLASE3 (NbCMT3), REPRESSOR OF SILENCING1 (NbROS1), REPRESSOR OF SILENCING 2 (NbROS2), DICER3 (NbDCL3), ARGONAUTE 1-1 (NbAGO1), and ARGONAUTE 4-1 (NbAGO4). N. benthamiana plants expressing empty vector (PVX) and NIa-Pro were analyzed at 15 dpi. As shown in Figure 6, overexpression of NIa-Pro significantly induced NbAGO4 expression (32% of that of control plants; p < 0.01), and expression of NbMET1, NbDRM2, and NbCMT3 increased 46% (p < 0.01), 38% (p < 0.01), and 26% (p < 0.05), respectively (Figure 6). NbROS1 and NbROS2 expression increased by 18 and 15%, respectively, but not significantly (Figure 6). Taken together, these results demonstrate that the expression of NIa-Pro significantly suppresses the expression of NbAGO4, a gene responsible for establishing DNA methylation, which is the first step of the plant DNA methylation pathway. In addition, activation of NbROS1 and NbROS2, two DNA glycosylases and repressors of TGS, may also contribute to TGS suppression by NIa-Pro.
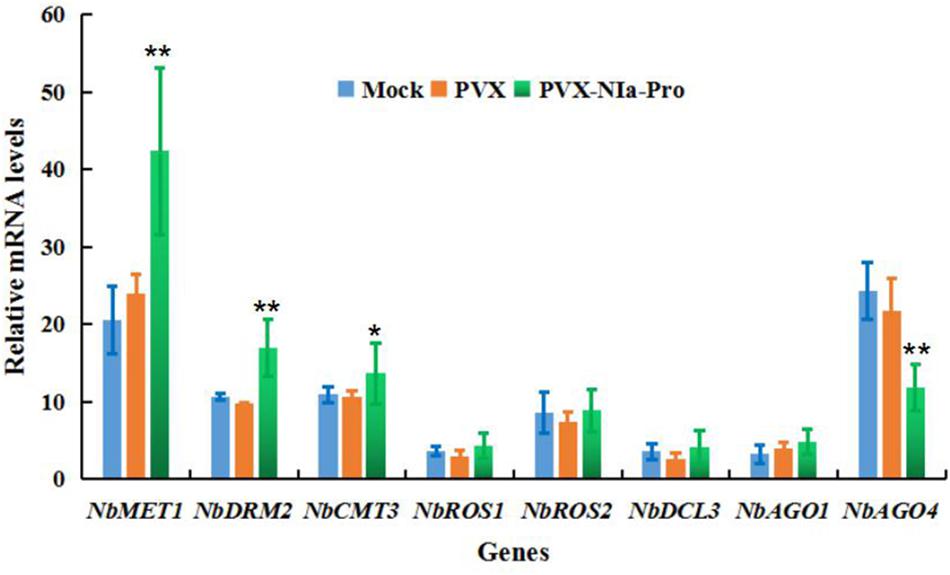
Figure 6. Overexpression of NIa-Pro represses transcription of Nicotiana benthamiana ARGONAUTE 4 (NbAGO4) mRNA. The effects of NIa-Pro on the expression of DNA methyltransferases, demethylases, and key genes involved in RNA-directed DNA methylation (RdDM) were analyzed by RT-qPCR. Relative mRNA levels of N. benthamiana DNA METHYLTRANSFERASE1 (NbMET1), NbDRM2, CHROMOMETHYLASE3 (NbCMT3), REPRESSOR OF SILENCING 1 (NbROS1), NbROS2, DICER3 (NbDCL3), ARGONAUTE 1 (NbAGO1), and NbAGO4 were measured in potato virus X (PVX)-NIa-Pro and PVX vector-infected N. benthamiana plants at 15 days post inoculation (dpi). Each mean value was derived from nine plants in three independent experiments. Student’s t-test was performed to assess differences between PVX-NIa- and PVX vector-infected plants. Values are the mean ± SD. Significant differences (* p < 0.05) and highly significant differences (** p < 0.01) between samples in each pair are indicated.
Discussion
PepMoV was first identified as an atypical isolate of the genus Potyvirus for serum differentiation and low amino acid sequence identity (Zitter, 1972). For example, PepMoV NIa-Pro exhibits 63.5% identity in sequence with PVY NIa-Pro and only 45.9% identity with TEV NIa-Pro (Mestre et al., 2000). Although the canonical function of NIa-protease (NIa-Pro) encoded by all potyviruses is responsible for cleaving the viral polyprotein into functional proteins, the other functions of NIa-Pro in the virus infecting cycle of potyviruses are inconsistent. TEV NIa-Pro could not induce a hypersensitive response (HR); however, PVY NIa-Pro could induce an HR in Ry-mediated resistance potatoes (Mestre et al., 2000). PepMoV NIa-Pro shared the highest nucleotide and amino acid sequences with PVY (Figure 1), and PVY NIa-Pro induced tobacco vein necrosis with HR (Faurez et al., 2012), suggesting PepMoV NIa-Pro would be a pathogenicity determinant. In this study, by using a PVX vector as an ectopic expression system, we found that PepMoV NIa-Pro could result in severe mosaic symptoms (Figure 2) and enhanced PVX replication (Figures 3B,C) but was not able to induce vein necrosis in leaves in Nicotiana benthamiana, which demonstrated that PepMoV NIa-Pro is a symptom determinant; however, it functioned in a different manner with PVY NIa-Pro.
Plant responses to infection by pathogens are often accompanied by a HLR, which is associated with an oxidative burst (including O2 and H2O2) (Bao and Howell, 2017). PepMoV NIa-Pro can induce H2O2 accumulation during infection with a PVX-based vector expressing NIa-Pro (Figure 3A). Additionally, PepMoV NIa-Pro could significantly activate genes expressing Bip and HSP90-2 but did not change the expression of bZIP60 (Figure 4). Bip is a chaperone gene that restores proper protein folding during ER stress (Bao and Howell, 2017), the HSP90-2 gene is involved in stabilizing proteins for correct folding or re-folding in response to ER stress (Sangster et al., 2007), and bZIP60 is the downstream transcription factor that activates genes required for coping with UPR (Arias Gaguancela et al., 2016). Therefore, PepMoV NIa-Pro likely induces N. benthamiana ER stress with a branch pathway of protein folding but does not activate the UPR.
It was well documented that DNA viruses, such as geminivirus-encoded pathogenicity determinants, also function as TGS suppressors targeting different steps of the TGS pathway. The βC1 protein of TYLCCNB directly interacts and inhibits host S-adenosyl homocysteine hydrolase (SAHH) activity to suppress host DNA methylation (Yang et al., 2011). The C2 protein of beet curly top virus (BCTV) attenuates the degradation of S-adenosyl-methionine decarboxylase 1 (SAMDC1) of Arabidopsis to inhibit host DNA methylation (Zhang et al., 2011). Intriguingly, a recent study demonstrated that the pathogenicity determinant of RNA viruses could also modulate the host methylation pathway. The 1a protein encoded by CMV, which is essential for viral infection, could recruit a novel methyltransferase from Nicotiana tabacum to methylate itself, which promotes CMV infection (Kim et al., 2008). Our results showed that PepMoV NIa-Pro can reverse the established TGS of N. benthamiana, suggesting that PepMoV NIa-Pro functioned as a host TGS suppressor. To our knowledge, this is the first report of plant RNA virus directly suppressing the plant DNA methylation pathway.
The DNA methylation in plants is not a static pathway and is dynamically modulated with three steps of establishing DNA methylation, maintenance methylation, and demethylation. Three types of cytosine methylation have been extensively characterized in plants: symmetric CG and CHG sites and asymmetric CHH (where H = A, C, or T) sites (Law and Jacobsen, 2010). Plant DNA viruses, such as geminivirus Rep protein, are the main viral proteins that can alter the levels of expression of the plant DNA methyltransferases responsible for the maintenance of symmetric methylation (Rodrıguez-Negrete et al., 2013), and AC5 protein of mungbean yellow mosaic India virus (MYMIV) inhibits methylation of cytosine in CHH and CHG to suppress NbDRM2 expression (Li et al., 2015). These results suggest that plant DNA viruses can modulate the host DNA methylation pathway in different manners. However, evidence of plant RNA viruses interacting with the host DNA methylation pathway was observed in a study where the CMV 1a protein was methylated by a novel protein involving host DNA methylation (Kim et al., 2008); this is not direct evidence of plant RNA viruses with the capacity to modulate host DNA methylation. PepMoV NIa-Pro can reverse established TGS by suppressing NbAGO4 (Figure 6), which is critical for establishing DNA methylation in the first step of the plant DNA methylation pathway (Law and Jacobsen, 2010). NIa-pro significantly activates the expression of NbMET1, NbDRM2, and NbCMT, which catalyze CG, CHG, and CHH methylation (Law and Jacobsen, 2010), suggesting that NIa-Pro likely activates the second step of plant DNA methylation. Interestingly, NIa-Pro could also activate the expression of NbROS1 and NbROS2 (Figure 6), which has been demonstrated to demethylate DNA methylation (Kim et al., 2019). It is likely that PepMoV NIa-Pro also possesses the capacity to inhibit the plant DNA methylation pathway through activating demethylation, which is the last step of plant DNA methylation. Further studies of the molecular mechanisms of PepMoV NIa-Pro modulating the plant DNA methylation pathway would contribute to uncovering novel modulating mechanisms of plant DNA methylation by plant RNA viruses.
Overall, the observations of this study show that PepMoV NIa-Pro is a multifunctional protein, and to the best of our knowledge, this is the first report testifying that a potyvirus NIa-Pro protein can both determine the symptom and induce an HR and modulate plant DNA methylation in multiple manners.
Conclusion
In conclusion, the results of this study suggest that PepMoV NIa-Pro is a pathogenicity determinant and a potent suppressor of host TGS and suggest that NIa-Pro may employ novel mechanisms to suppress host antiviral defenses.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation, to any qualified researcher.
Author Contributions
S-BZ, X-WL, and YL designed the research and prepared the draft of the manuscript. Y-NG, R-QT, YZ, JP, and OX performed the experiments. Z-HZ, D-YZ, and HL collected the data. All authors reviewed and approved the final version of the manuscript.
Funding
This research was granted by the National Natural Science Foundation of China (317017765) and the China Agriculture Research System (CARS-23-D-02).
Conflict of Interest
HL is employed by National Agro-Tech Extension and Service Center.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to thank Prof. Fanfan Li, Chinese Academy of Agricultural Sciences, for gifting 16-TGS Nicotiana benthamiana seeds.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.00102/full#supplementary-material
References
Arias Gaguancela, O. P., Zúñiga, L. P., Arias, A. V., Halterman, D., Flores, F. J., Johansen, I. E., et al. (2016). The IRE1/bZIP60 pathway and bax inhibitor 1 suppress systemic accumulation of potyviruses and potexviruses in Arabidopsis and N. benthamiana plants. Mol. Plant Microbe Interact. 29, 750–766. doi: 10.1094/mpmi-07-16-0147-r
Bao, Y., and Howell, S. H. (2017). The unfolded protein response supports plant development and defense as well as responses to abiotic stress. Front. Plant Sci. 8:344. doi: 10.3389/fpls.2017.00344
Buchmann, R. C., Asad, S., Wolf, J. N., Mohannath, G., and Bisaro, D. M. (2009). Geminivirus AL2 and L2 proteins suppress transcriptional gene silencing and cause genome-wide reductions in cytosine methylation. J. Virol. 83, 5005–5013. doi: 10.1128/JVI.01771-08
Coursey, T., Regedanz, E., and Bisaro, D. M. (2018). Arabidopsis RNA polymerase V mediates enhanced compaction and silencing of geminivirus and transposon chromatin during host recovery from infection. J. Virol. 92:e01320. doi: 10.1128/JVI.01320-17
Daros, J. A., Schaad, M. C., and Carrington, J. C. (1999). Functional analysis of the interaction between VPg-proteinase (NIa) and RNA polymerase (NIb) of tobacco etch potyvirus, using conditional and suppressor mutants. J. Virol. 73, 8732–8740. doi: 10.1128/jvi.73.10.8732-8740.1999
Faurez, F., Baldwin, T., Tribodet, M., and Jacquot, E. (2012). Identification of new Potato virus Y (PVY) molecular determinants for the induction of vein necrosis in tobacco. Mol. Plant Pathol. 13, 948–959. doi: 10.1111/j.1364-3703.2012.00803.x
Hamera, S., Song, X. G., Su, L., Chen, X. Y., and Fang, R. X. (2012). Cucumber Mosaic virus suppressor 2b binds to AGO4-related small RNAs and impairs AGO4 activities. Plant J. 69, 104–115. doi: 10.1111/j.1365-313X.2011.04774.x
Han, J. H., Choi, H. S., Kim, D. H., Lee, H. R., and Kim, B. D. (2006). Biological, physical and cytological properties of Pepper mottle virus-SNU1 and its RT-PCR detection. Plant Pathol. J. 22, 155–160. doi: 10.5423/ppj.2006.22.2.155
Kim, M. J., Huh, S. U., Ham, B., and Paek, K. H. (2008). A novel methyltransferase methylates Cucumber mosaic virus 1a protein and promotes systemic spread. J. Virol. 82, 4823–4833. doi: 10.1128/JVI.02518-07
Kim, M. Y., Ono, A., Scholten, S., Kinoshita, T., Zilberman, D., Okamoto, T., et al. (2019). DNA demethylation by ROS1a in rice vegetative cells promotes methylation in sperm. Proc. Natl. Acad. Sci. 116, 9652–9657. doi: 10.1073/pnas.1821435116
Law, J. A., and Jacobsen, S. E. (2010). Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet. 11, 204–220. doi: 10.1038/nrg2719
Li, F., Huang, C., Li, Z., and Zhou, X. (2014). Suppression of rna silencing by a plant dna virus satellite requires a host calmodulin-like protein to repress rdr6 expression. PLoS Pathog. 10:e1003921. doi: 10.1371/journal.ppat.1003921
Li, F. F., Xu, X. B., Huang, C. J., Gu, Z. H., Cao, L. G., Hu, T., et al. (2015). The AC5 protein encoded by Mungbean yellow mosaic India virus is a pathogenicity determinant that suppresses RNA silencing-based antiviral defenses. N. Phytol. 208, 555–569. doi: 10.1111/nph.13473
Marchler-Bauer, A., Lu, S., Anderson, J. B., Chitsaz, F., Derbyshire, M. K., DeWeese-Scott, C., et al. (2011). CDD: a conserved domain database for the functional annotation of proteins. Nucleic Acids Res. 39, D225–D229. doi: 10.1093/nar/gkq1189
Martínez, F., Rodrigo, G., Aragonés, V., Ruiz, M., Lodewijk, I., Fernández, U., et al. (2016). Interaction network of tobacco etch potyvirus NIa protein with the host proteome during infection. BMC Genom. 17:87. doi: 10.1186/s12864-016-2394-y
Mestre, P., Brigneti, G., and Baulcombe, D. C. (2000). An Ry-mediated resistance response in potato requires the intact active site of the NIa proteinase from Potato virus Y. Plant J. 23, 653–661. doi: 10.1046/j.1365-313x.2000.00834.x
Nelson, M. R., and Wheeler, R. E. (1972). A new virus disease of pepper in Arizona. Plant Dis. Rep. 56, 731–735.
Raja, P., Sanville, B. C., Buchmann, R. C., and Bisaro, D. M. (2008). Viral genome methylation as an epigenetic defense against geminiviruses. J. Virol. 82, 8997–9007. doi: 10.1128/JVI.00719-08
Rajamaki, M. L., and Valkonen, J. P. T. (2009). Control of nuclear and nucleolar localization of nuclear inclusion protein a of picorna-like Potato virus A in Nicotiana species. Plant Cell 21, 2485–2502. doi: 10.1105/tpc.108.064147
Rodamilans, B., Valli, A., Mingot, A., San Leon, D., Baulcombe, D., Lopez-Moya, J. J., et al. (2015). RNA polymerase slippage as a mechanism for the production of frameshift gene products in plant viruses of the Potyviridae family. J. Virol. 89, 6965–6967. doi: 10.1128/JVI.00337-15
Rodrıguez-Negrete, E., Lozano-Duran, R., Piedra-Aguilera, A., Cruzado, L., Bejarano, E. R., and Castillo, A. G. (2013). Geminivirus rep protein interferes with the plant DNA methylation machinery and suppresses transcriptional gene silencing. New Phytol. 199, 464–475. doi: 10.1111/nph.12286
Sangster, T. A., Adam, B., Amity, W., Etsuko, W., Kurt, S., Catherine, M. L., et al. (2007). Phenotypic diversity and altered environmental plasticity in Arabidopsis thaliana with reduced hsp90 levels. PLoS One 2:e648. doi: 10.1371/journal.pone.0000648
Sharma, P., Ikegami, M., Jaeger, K. E., Graf, A., Wigge, P. A., and Saitou, N. (2010). Tomato leaf curl Java virus v2 protein is a determinant of virulence, hypersensitive response and suppression of posttranscriptional gene silencing. Virology 396, 85–93. doi: 10.1016/j.virol.2009.10.012
Shukla, D. D., Ward, C. W., and Brunt, A. A. (1994). The Potyviridae. Wallingford, OX: CAB International.
Stewart, F. J., Panne, D., Bickle, T. A., and Raleigh, E. A. (2000). Methyl-specific dna binding by mcrbc, a modification-dependent restriction enzyme. J. Mol. Biol. 298, 611–622. doi: 10.1006/jmbi.2000.3697
Sun, P., Austin, B. P., Tozser, J., and Waugh, D. S. (2010). Structural determinants of tobacco vein Mottling virus protease substrate specificity. Protein Sci. 19, 2240–2251. doi: 10.1002/pro.506
Untiveros, M., Olspert, A., Artola, K., Firth, A. E., Kreuze, J., and Valkonen, J. P. (2016). A novel sweet potato potyvirus open reading frame (ORF) is expressed via polymerase slippage and suppresses RNA silencing. Mol. Plant. Pathol. 17, 1111–1123. doi: 10.1111/mpp.12366
Wang, B., Li, F., Huang, C., Yang, X., Qian, Y., Xie, Y., et al. (2014). V2 of tomato yellow leaf curl virus can suppress methylation-mediated transcriptional gene silencing in plants. J. Gen. Virol. 95, 225–230. doi: 10.1099/vir.0.055798-0
Wang, C. G., Wang, C. N., Zou, J. Z., Yang, Y. S., Li, Z. H., and Zhu, S. F. (2019). Epigenetics in the plant-virus interaction. Plant Cell Rep. 38, 1031–1038. doi: 10.1007/s00299-019-02414-0
Xie, Y., Zhao, L., Jiao, X., Jiang, T., Gong, H., Wang, B., et al. (2013). A recombinant begomovirus resulting from exchange of the C4 gene. J. Gen. Virol. 94, 1896–1907. doi: 10.1099/vir.0.053181-0
Yang, X., Xie, Y., Raja, P., Li, S., Wolf, J. N., Shen, Q., et al. (2011). Suppression of methylation-mediated transcriptional gene silencing by bC1-SAHH protein interaction during geminivirus-betasatellite infection. PLoS Pathog. 7:e1002329. doi: 10.1371/journal.ppat.1002329
Zhang, Y., Luo, X. W., Zhang, D. Y., OuYang, X., Zhang, Z. H., Li, F., et al. (2019). Genome and phylogenetic analyses of Chinese Pepper mottle virus isolates from chili pepper plants. J. Plant Pathol. 101, 559–564. doi: 10.1007/s42161-018-00235-w
Zhang, Z., Chen, H., Huang, X., Xia, R., Zhao, Q., Lai, J., et al. (2011). BSCTV C2 attenuates the degradation of SAMDC1 to suppress DNA methylation-mediated gene silencing in Arabidopsis. Plant Cell 23, 273–288. doi: 10.1105/tpc.110.081695
Keywords: NIa-Pro, DNA methylation, transcriptional gene silencing, Pepper mottle virus, pathogenicity determinant
Citation: Gong Y-N, Tang R-Q, Zhang Y, Peng J, Xian O, Zhang Z-H, Zhang S-B, Zhang D-Y, Liu H, Luo X-W and Liu Y (2020) The NIa-Protease Protein Encoded by the Pepper Mottle Virus Is a Pathogenicity Determinant and Releases DNA Methylation of Nicotiana benthamiana. Front. Microbiol. 11:102. doi: 10.3389/fmicb.2020.00102
Received: 05 December 2019; Accepted: 17 January 2020;
Published: 21 February 2020.
Edited by:
Ralf Georg Dietzgen, The University of Queensland, AustraliaReviewed by:
Adrian Alejandro Valli, Centro Nacional de Biotecnología (CNB), SpainMing-Bo Wang, Commonwealth Scientific and Industrial Research Organisation (CSIRO), Australia
Copyright © 2020 Gong, Tang, Zhang, Peng, Xian, Zhang, Zhang, Zhang, Liu, Luo and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiang-Wen Luo, bHVveHd6YnNAMTYzLmNvbQ==; Yong Liu, aGFvYXNsaXVAMTYzLmNvbQ==
 Yi-Nuo Gong1
Yi-Nuo Gong1 Song-Bai Zhang
Song-Bai Zhang Yong Liu
Yong Liu