- 1CAS Key Laboratory of Biogeography and Bioresource in Arid Land, Xinjiang Institute of Ecology and Geography, Ürümqi, China
- 2Department of Medicine, College of Kashgar Vocational Technology, Kashgar, China
- 3Faculty of Biology, National University of Uzbekistan, Tashkent, Uzbekistan
- 4Department of Environmental Protection, Faculty of Environmental Agricultural Sciences, Arish University, Arish, Egypt
- 5Xinjiang Laboratory of Resources Microbiology, College of Life Sciences and Technology, Xinjiang University, Ürümqi, China
- 6State Key Laboratory of Biocontrol and Guangdong Provincial Key Laboratory of Plant Resources, School of Life Sciences, Sun Yat-sen University, Guangzhou, China
We report for the first time the isolation of endophytic actinobacteria associated with wild populations of the Chinese medicinal herb Thymus roseus Schipcz obtained from the arid land in Ili and Tacheng of the Xinjiang Province, China. Strains were isolated by special pretreatment of plant tissues and identified based on their 16S rRNA gene sequences, and their antimicrobial activities in vitro were evaluated. A total of 126 endophytic actinobacteria belonging to two classes, eight orders, 14 families, and 24 genera were isolated from different organs at the Ili and Tacheng sites. In addition, the diversity of culturable endophytic actinobacteria genera was higher at Tacheng site (n = 71, 56.35%) than the Ili site (n = 55, 43.65%). A neighbor-joining tree of 126 isolated actinobacteria showing the phylogenetic relationships based on 16S rRNA gene sequences and the genus Streptomyces was the most dominant isolate. The number of endophytic actinobacteria genera obtained from root tissues (n = 54, 42.86%) was higher compared to stem (n = 35, 27.78%) and leaf tissue (n = 37, 29.36%). Among 126 endophytic actinobacteria, 54 strains were antagonistic against at least one or more indicator organisms in vitro. Notably, most strains of Streptomyces proved antagonistic activities. For example, strain T4SB028, namely Streptomyces polyantibioticus, showed the highest inhibition ratio reached 67.06, 64.20, and 70.55% against Alternaria solani, Valsa malicola, and Valsa mali, respectively. The results demonstrate that about 30.95%, 23.01% of the tested endophytic actinobacteria were capable of producing siderophores and chitinase, respectively. Additionally, the results of the amplification of biosynthetic genes polyketide synthetase (PKS-I) and non-ribosomal peptide synthetase (NRPS) indicated that at least one antibiotic biosynthetic gene was detected in 27 (50%) of the tested strains. Our result emphasizes that the endophytic actinobacteria communities are different based on the plant tissues and the geographical environment of the sampled area. Thus, we conclude that T. roseus Schipcz. provided a rich source of endophytic actinobacteria that exhibited a broad-spectrum antimicrobial agent.
Introduction
Endophytes are microorganisms that inhabit healthy plant tissues at specific growth stages, or whole stages of their life cycle, and establish a symbiotic relationship with the host without causing any apparent disease symptoms (Petrini, 1991; Duan et al., 2013; Castro et al., 2014; Egamberdieva et al., 2017b, 2018; Li et al., 2018; Liu et al., 2019). Endophytic actinobacteria are considered to be a vital resource of microbial biodiversity (Matsumoto and Takahashi, 2017). A few reports have investigated a wide range of taxonomic status of endophytic actinobacteria associated with medicinal plants (Qin et al., 2009; Zhao et al., 2011; Matsumoto and Takahashi, 2017).
Endophytic actinobacteria exploit an unusual habitat may contribute to plant natural defenses by preventing herbivores and promoting the biocontrol of pathogens and pests due to their ability to produce the same or similar compounds with the host plants (Shimizu et al., 2009). Therefore, great attention has been paid by researchers toward their potential applications in pharmaceutical, agricultural, and food industries (Strobel and Daisy, 2003; Singh and Gaur, 2016). For instance, a total of 150 endophytic actinobacteria were isolated from three medicinal plant species (Annonaceae squamosa, Camptotheca acuminata, and Taxus chinen), and 72.4% of them showed inhibition against more than one indicator microorganism, including Bacillus subtilis, Escherichia coli, Staphylococcus aureus, Candida albicans, and Aspergillus niger (Wu et al., 2009). In addition, endophytic actinobacteria were antagonistic against Pythium aphanidermatum and significantly promoted plant growth and reduced the crown and root rot of cucumber that was caused by the Pythium aphanidermatum in vitro condition (El-Tarabily et al., 2009).
Medicinal plants in China have an ethnomedicinal history, and there are about 128,000 kinds of crude medicine resources (Cai et al., 2013). In other words, the vast majority of traditional Chinese medicine and natural chemical products had been derived from medicinal plants due to their effectiveness in various components for disease prevention and treatment. Thymus roseus Schipcz is one of the traditional Chinese medicinal herbs belonging to the Labiatae family, and it has a strong adaptive capability for a variety of particular habitats, such as arid lands, saline-alkali soils, and desertified soils (Ren et al., 2010). Furthermore, modern pharmacology has demonstrated that Thymus and thyme essential oils possess of anti-inflammatory, antibiosis, antiviral, antioxidant, anticancer, and antithrombus properties (Liang et al., 2014; Bobis et al., 2015; Jariæ et al., 2015; Hosseinzadeh et al., 2015). There is high value in medical treatment and chemical use of T. roseus S.; thus, the study of endophytic actinobacteria and their interaction with the host plants Thymus and its adaptive properties to arid lands is of great import within arid land ecology. This has the potential to significantly better our understanding of its diversity and antimicrobial activities as well as to further investigate its biotechnological applications potentials. However, the selection rationale of host plants, locations, and pretreatment methods for the isolation of endophytes is essential in biodiversity research (Strobel and Daisy, 2003; Zhang et al., 2016). Besides, endophytic resources of arid land are most likely to possess unique and diverse functions, such as degrading chitin, producing siderophore, encoding antimicrobial and antibiotic biosynthetic genes, because of their high ability to adapt the extremely arid environment (Qin et al., 2009; Cho et al., 2015; Egamberdieva et al., 2016, 2017c; Li et al., 2018). For instance, our research group reported that the vast majority of endophytic isolates from Chinese traditional medicinal plants Ferula sinkiangensis, Glycyrrhiza uralensis, and Ferula songorica in Xinjiang arid land were capable of producing both antifungal and plant growth-promoting traits (Liu et al., 2016, 2017; Li et al., 2018).
To the best of our knowledge, to date, there is no report on the diversity and antimicrobial potential of endophytic actinobacteria associated with Thymus. Thus, the objectives of our study were as follows: (1) to isolate and identify endophytic actinobacteria associated with Chinese traditional medicinal plant Thymus roseus Schipcz by a culture-dependent method; (2) to analyze the species richness and distribution pattern among the different plant tissues and locations; (3) to compare the effectiveness of plant tissue pretreatment on the isolation of endophytic actinobacteria; and (4) to determine their antimicrobial capability and to screen the isolates with the antibiotic biosynthetic genes in in vitro conditions.
Materials and Methods
Site Selection and Plant Sampling
Healthy medicinal plants of T. roseus Schipcz. were randomly collected from their natural arid habitats in the Xinjiang province of China. The study sites were located at Ili (43°21′56′′ N, 84°21′51′′ E; 2116.00 m.a.s.l., belonging to continental semi-dry climate.) and Tacheng (46°55′23′′ N; 83°16′59′′ E; 1035.80 m.a.s.l., belonging to temperate continental arid climate). On August 10th, 2016 (Figure 1), the healthy plants were brought to the laboratory inside a sterile paper bag and used to isolate for endophytic actinobacteria within 48 h of collection.
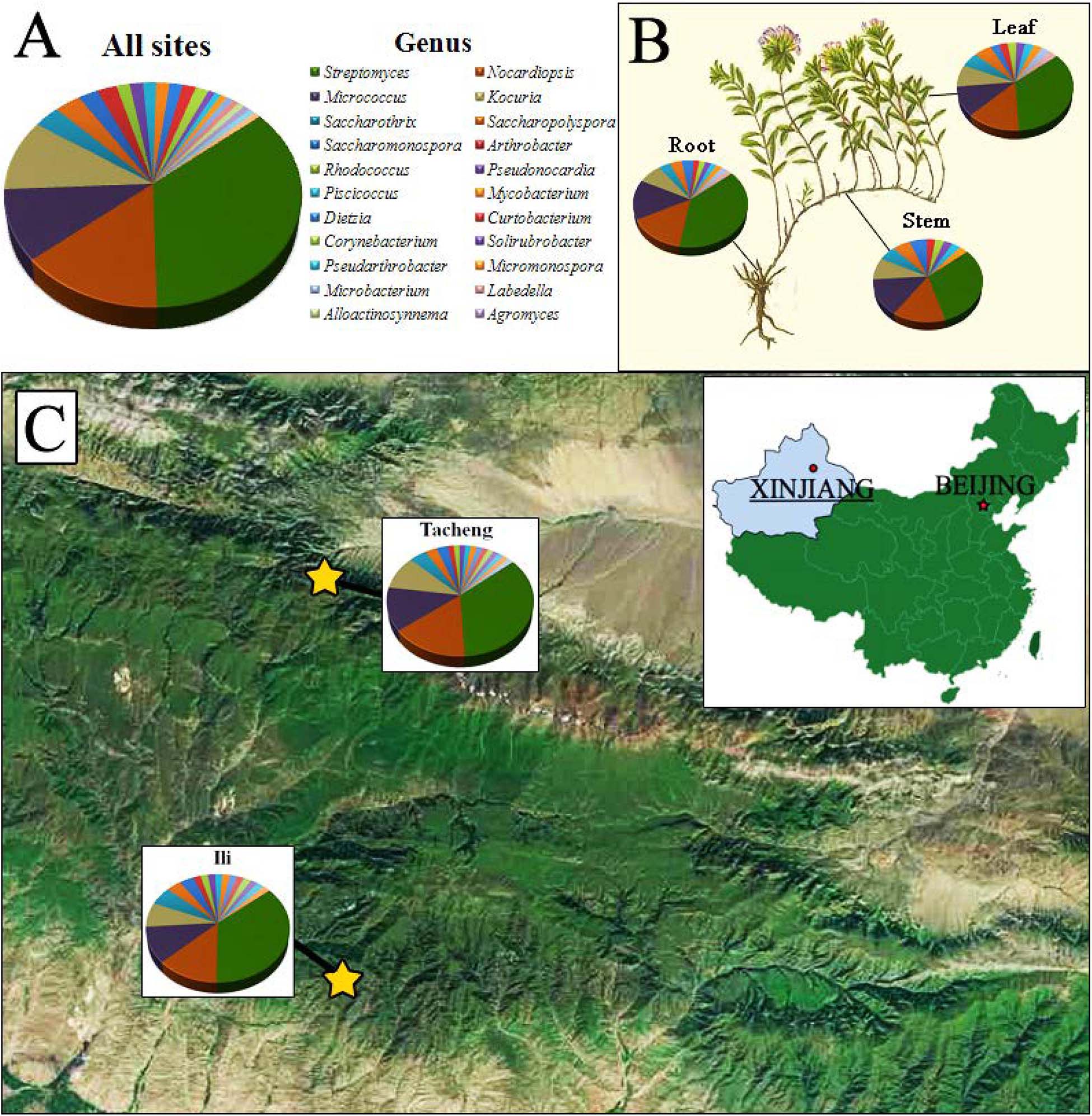
Figure 1. The distribution and identity of 126 culturable endophytic actinomycetes from T. roseus Schipcz. based on 16S rRNA gene sequences. (A) A summary of genera present at all sites. (B) Genus assignments for isolates from different tissues. (C) Genus assignments according to location and showing the high diversity of isolates from the most arid site, Tacheng.
Surface Sterilization, Pretreatment, and Isolation of Endophytic Actinobacteria
Each plant sample was washed with running tap water to remove surface clays and adhering epiphytes completely. The plant samples were separated into stem, root, and leaf segments by using sterile scissors. Subsequently, samples were washed by sonification for 30 min at 45 kHz to remove the dislodge soil and organic matter. The surface sterilization protocol included five steps: plant segments were rinsed in 0.1% Tween 20 for 1 min, sequentially immersed in 5% sodium hypochlorite (NaOCl) for 4 min leaf segments and 6 min for stem and root segments, washed for 10 min in 2.5% Na2S2O3, soaked in 70% ethanol for 4 min (leaves) and 6 min (stems and roots), and finally rinsed in 10% NaHCO3 for 10 min to disrupt the growth of fungi. After each treatment, samples were rinsed three times in sterile water. To confirm the success of the sterilization protocol, 0.2 m L–1 of the sterile distilled water from the final wash process of each tissue were placed onto International Streptomyces Project medium ISP\ 2 (Shirling and Gottlieb, 1966) and incubated at 28°C for 1 week. Finally, the samples were placed on sterilized filter paper and thoroughly dried in a sterile Petri dish under the laminar flow chamber for 24 h. After drying under sterile conditions, the sterilized samples were crushed in a sterilized commercial blender (Joyoung, JYL-C012) for 20 s (leaves) and 30 s for (stems and roots) (Qin et al., 2008; Li et al., 2018).
The plant tissues (roots, stems, and leaves) were introduced to three kinds of pretreatment in order to kill bacteria and isolate of endophytic actinobacteria following the method of Qin et al. (2011) with some modification: (1) surface sterilized plant tissues were directly taken to isolation; (2) freezing treatment involved plant tissues being frozen at –80 °C for 2 weeks; (3) high temperature treatment involved plant tissues drying at 100°C for 20 min.
For each pretreated sample (roots, stems, and leaves), about 1 g of tissue homogenate was weighed aseptically and macerated with a sterile mortar and pestle along with 9 mL sterile double distilled water. After precipitation for 10 min at room temperature, the supernatant was serially diluted (10–2 and 10–3), and a suspension of 0.2 ml was spread onto ten different selective isolation media (Li et al., 2018; Table 1). In order to inhibit the growth of bacteria and fungi, the isolation media were supplemented with antibiotics: nystatin 50 mg L–1, K2Cr2O7 25 mg L–1 and nalidixic acid 25 mg L–1. All agar plates were performed three replicates for each dilution and then warped with parafilm and incubated at 28°C for 45 days before being monitored every 7 days for microbial growth. The colonies were observed and selected according to their characteristics and colony morphology. The purified actinobacteria isolates were stored on the ISP2 slope medium at 4°C and in 20% glycerol at −80°C (Li et al., 2018).

Table 1. Composition of the media used for the isolation of endophytic actinomycetes from medicinal plants.
Taxonomic Characterization and 16S rRNA Gene Sequencing
Genomic DNA was extracted using the microwave thermal shock method (Orsini and Romano-Spica, 2001) and the TIANamp Bacteria DNA Kit (TIANGEN BIOTECH). The microwave thermal shock method was used: fresh bacterial biomass (50 mg) was resuspended in 35 μl lysis solution (50 mM sucrose, 50 mM Tris–HCl pH 8.0, and 20 mM EDTA-Na) and 15 μl 20% SDS and then heated in a microwave oven for 90 s. The mixture was treated with 450 μl DNA extraction solution (100 mmol.L–1 Tris, 100 mmol.L–1 EDTA, 200 mmol.L–1 NaCl, 2% PVP, 3% CTAB, and pH 9.0) and then extracted twice with phenol/chloroform/isoamyl alcohol (25:24:1 v/v/v) (Sangon Biotech) followed by precipitation with 800 μl ethanol and 80 μl sodium acetate (3 mol L–1 and pH 4.8–5.2). The DNA pellet was washed with 70% ethanol, air-dried, resuspended in 50 μl deionized distilled water, and then stored at −20°C for further study. A 16S rRNA gene sequence was amplified using universal primers 27F (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1492R (5′-GGTTACCTTGTTACGACTT-3′) (Wang et al., 2006). The PCR mixture (25 μl) contained 12.5 μl 2 × Taq PCR Master Mix procured from TIANGEN BIOTECH (Beijing, China), 2 μl of the DNA template, and 1 μl of each primer. We used an amplification protocol: denaturation took place at 95°C for 5 min, and it was followed by 35 cycles of denaturation at 94°C for 1 min, annealing at 56°C for 1.5 min, extension at 72°C for 2 min, and a final extension at 72°C for 10 min. PCR products were purified and sequenced by a commercial company, Sangon Biotech. The 16S rRNA gene sequences were compared with Ezbiocloud1 (Chun et al., 2007) and GenBank databases using BLAST software. Multiple sequence alignment was performed using the CLUSTAL X 1.83 program (Thompson et al., 1997), and a phylogenetic tree was generated by the neighbor-joining method using MEGA version 7.0 software (Kumar et al., 2016). The 16S rRNA gene sequences determined in this study were deposited in GenBank under accession numbers MN686679–MN686702(24), MN687832–MN687853(22), MN688237–MN688255(19), MN688672–MN688674(3), MN688677(1), MN688679–MN688680(2), MN686608–MN686629(22), MN686648–686678(31), and MNMN688648–688649(2). Reference sequences used are noted in the phylogenetic trees.
Antagonistic Assays of Antifungal Activities In vitro
The antifungal activity of each endophyte actinobacteria was screened for antagonism against six pathogenic fungi (provided by Key Laboratory of Biogeography and Bioresource in Arid Land, Xinjiang Institute of Ecology and Geography, Chinese Academy of Sciences)—Tomato Fusarium wilt (Fusarium oxysporum f.sp.), Tomato leaf mildew [Fulvia fulva(Cooke)Cif.], Tomato Early Blight (Alternaria solani), Cotton fusarium wilt (Fusarium oxysporum), Apple Valsa Canker (Valsa malicola), and Apple Valsa Canker (Valsa mali)—by the plate confrontation method as described by Liu et al. (2017). Briefly, a fungal disk 5 mm in diameter containing 7-day-old mycelial growth was placed at the center of a 9 cm potato dextrose agar (PDA) plates.
The four actinobacteria disks cultured on ISP2 medium for 4–7 days at 28°C were placed onto the agar surface at four equidistant points, 2.5 cm from the plate periphery (Loqman et al., 2009). Plates with pathogenic fungi alone served as a control. All agar plates were wrapped with parafilm and incubated at 28 ± 2°C for 3–5 days and observed for the inhibition of the pathogen.
The antifungal activity of isolates was determined by calculating the growth inhibition ratio. The formula of growth inhibition ratio (%) was calculated by I = (R 0–Ri)/RO × 100%, where R 0 is the radial of pathogen in control plates, and Ri is the radial of pathogen in test plates (Mohamad et al., 2018). Each experiment was conducted with three replicates.
Antagonistic Activity Against Human Pathogenic Bacteria
A slightly modified method from Nie et al. (2012) for detecting antibacterial activity was used against the three common bacteria—Staphylococcus aureus, Bacillus cereus, and Salmonella enteritidis—provided by Key Laboratory of Biogeography and Bioresource in Arid Land, Xinjiang Institute of Ecology and Geography, Chinese Academy of Sciences. The bacteria were cultured for 1 night in Luria-Bertani LB medium at 37°C. The tested bacteria and endophytes isolates were each pre-cultured overnight, and 5 m L–1 of each culture was centrifuged at 5000 rpm for 10 min. The pellets were resuspended in sterile DDH2O and density adjusted to 108 colony forming units CFU/mL by using Densicheck plus (Biomerieux, United States). A total of 100 μL of the typical bacteria cell were inoculated and evenly spread by sterile cotton swaps onto the surface of the LB medium, and then four 5 mm diameter pieces of sterile filter paper were placed on each corner of the agar plate. After this, 10 μL of each actinobacteria strain was added dropwise to the filter paper. All plates were wrapped with parafilm, incubated at 37 ± 2°C for 24 h, and observed for the inhibition of the common bacteria (Mohamad et al., 2018). Antibacterial activity was assessed by measuring the diameter of the clear zone of growth inhibition. An equivalent volume of sterile DDH2O instead of the endophytic actinobacteria was used as a negative control.
Chitinase and Siderophore Production
Chitinase production was assessed by using a Colloidal chitin-supplemented medium, as described by Agrawal and Kotasthane (2012). The protocol for preparing the colloidal chitin was such that Chitin was hydrolyzed in concentrated hydrochloric acid by stirring at 4°C for 12 h, and this was followed by extraction of colloidal chitin in 200 mL–1 of ice-cold 99% ethanol, neutralization at room temperature for another 12 h, and then centrifugation at 5000 rpm for 10 min. The pellet was washed with distilled water by centrifugation at 5000 rpm for 5 min for several times until the smell of alcohol was removed entirely and the pH was natural. Chitinase medium composed of (L–1) 2.0 g of KH2PO4, 1.0 g of citric acid monohydrate, 0.3 g of MgSO4⋅7H2O, 3.0 g of (NH4)2SO4, 15 g of agar, 200 μL of Tween-80, and 4.5 g of colloidal chitin was supplemented with 0.15 g of bromocresol purple and then autoclaved at 121°C for 20 min. All agar plates were wrapped with parafilm and three replicates for each isolate, incubated at 25 ± 2°C for 3–7 days, and were observed for the formation of a colored zone around bacterial colony. Sterile chitin agar media was used as a control for bacterial growth.
Siderophore production was evaluated by incubating endophytic actinobacteria on Chrome azurol S (CAS) agar medium at 28°C for 7 days following the methods described by Li et al. (2018). An orange halo surrounding the actinobacteria colony indicates the tested strains have the siderophore-producing ability. The experiment was conducted with three replicates.
Screening for Natural Product Biosynthetic Gene Clusters by PCR Method
Fifty-four strains showed antimicrobial activity against one or more indicator organisms and were used for screening for natural product biosynthetic gene clusters by PCR. Three sets of degenerate primers targeting biosynthetic genes were used for PCR amplification: KSF (5′-GTSCCSGTSSCRTGSSHYTCSA-3′) and KSR (5′-CGCTCCATGGAYCCSCARCA-3′), which target PKS-I KS and methyl malonyl transferase domains (Kun et al., 2010); KSαF (5′-TSGCSTGCTTGGAYGCSATC-3′) and KSαR (5′-TGGAANCCGCCGAABCCGCT-3′), which target PKS-II KSα genes (Metsä-Ketelä et al., 1999); and A3F (5′-GCSTACSYSATSTACACSTCSGG-3′) and A7R (5′-SASGTCVCCSGTSCGGTAS-3′), which target non-ribosomal peptide synthetase (NRPS) genes. A PCR amplification protocol as described by Qin et al. (2009) was followed.
Data Analysis
To study the level of significance, statistical analysis was made by one-way ANOVA using SPSS ver. 16.0 software. Mean comparison carried out by using Duncan’s multiple range tests at P < 0.05. Data are presented as means ± SD.
Results
Effectiveness of Surface Sterilization
No microbial growth was detected on the isolation media after 7 days of incubation at 28°C when the distilled water used in the final rinse of surface-sterilization was plated. This result indicated that the five-step surface sterilization protocol was successful, and all isolates were considered to be true endophytes.
Pretreatment, Isolation Medium, and Effectiveness of Isolation
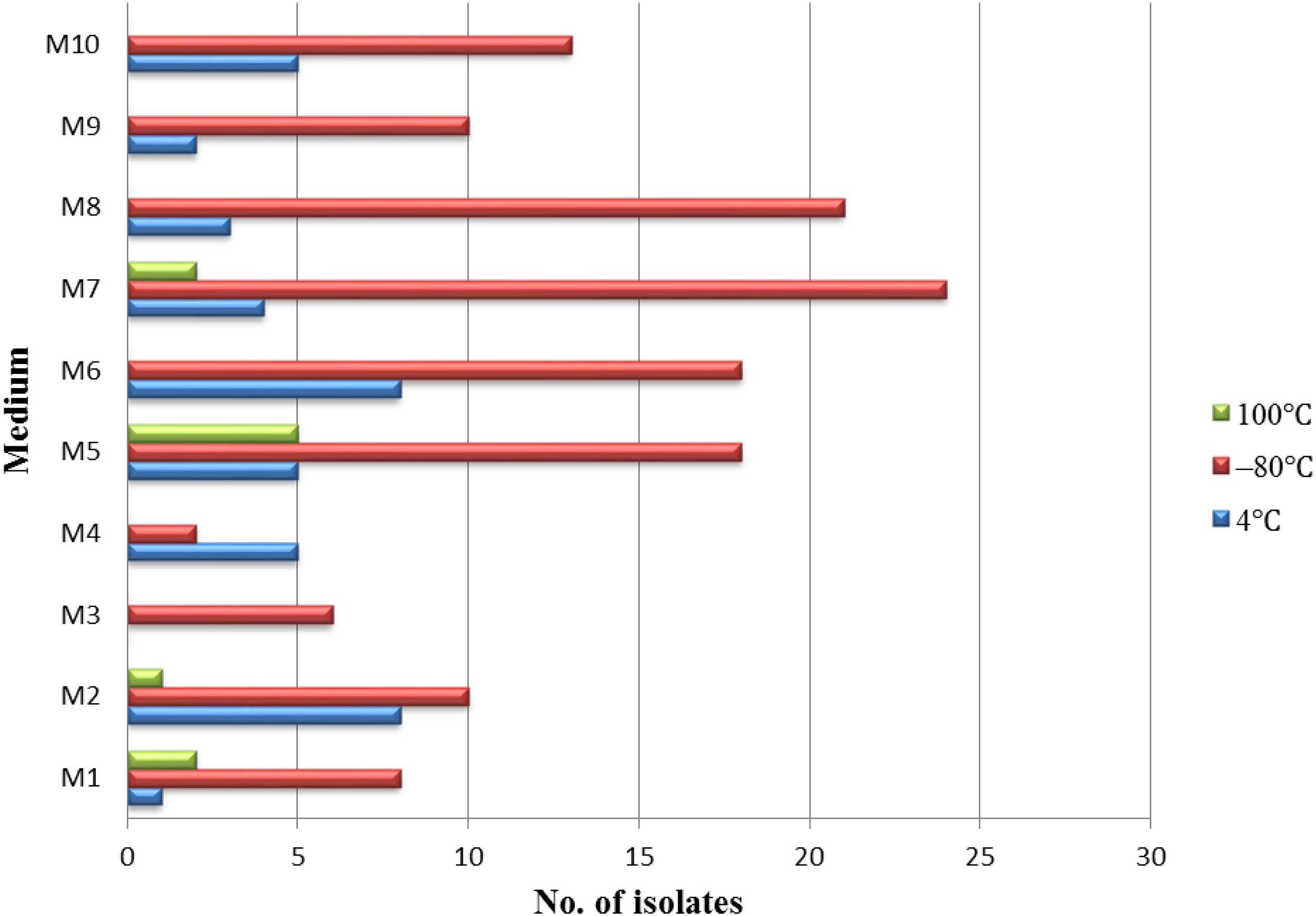
Different pretreatment and media have been used for the isolation of endophytic actinobacteria. A total of 181 endophytic actinomycetes were isolated from three types of pretreatment of medicinal plant T. roseus S tissues. The 128, 41, and 10 strains were isolated from the plant tissues treated with freezing (–80°C), and this was followed by average temperature (4°C) and high-temperature treatment (100°C). Freezing treatment showed the highest number of isolations with comparing to the other two treatments (Figure 2). The effect of different media on the isolation of endophytic actinobacteria used in this study showed dramatic results; the majority of isolations were from humic acid-vitamin (HV) medium M7, followed by sodium propionate agar medium M5, raffinose-histidine agar medium M6, and oatmeal agar medium M8 (Figure 2).
Diversity and Tissues Specificity of Endophytic Actinobacteria
A total of 126 endophytic actinobacteria isolates were obtained from the T. roseus S based on 16S rRNA gene sequences, and 55 and 71 isolates were from Ili and Tacheng sites, respectively. The diversity of culturable endophytic actinobacteria were the highest at the Tacheng site and included one class, five orders, nine families, 19 genera, and 49 species (Figure 3). While isolates from the Ili site included two classes, eight orders, 12 families, 18 genera, and 37 species (Figure 4). The endophytic actinobacteria community clearly differed in composition with regard to geography.
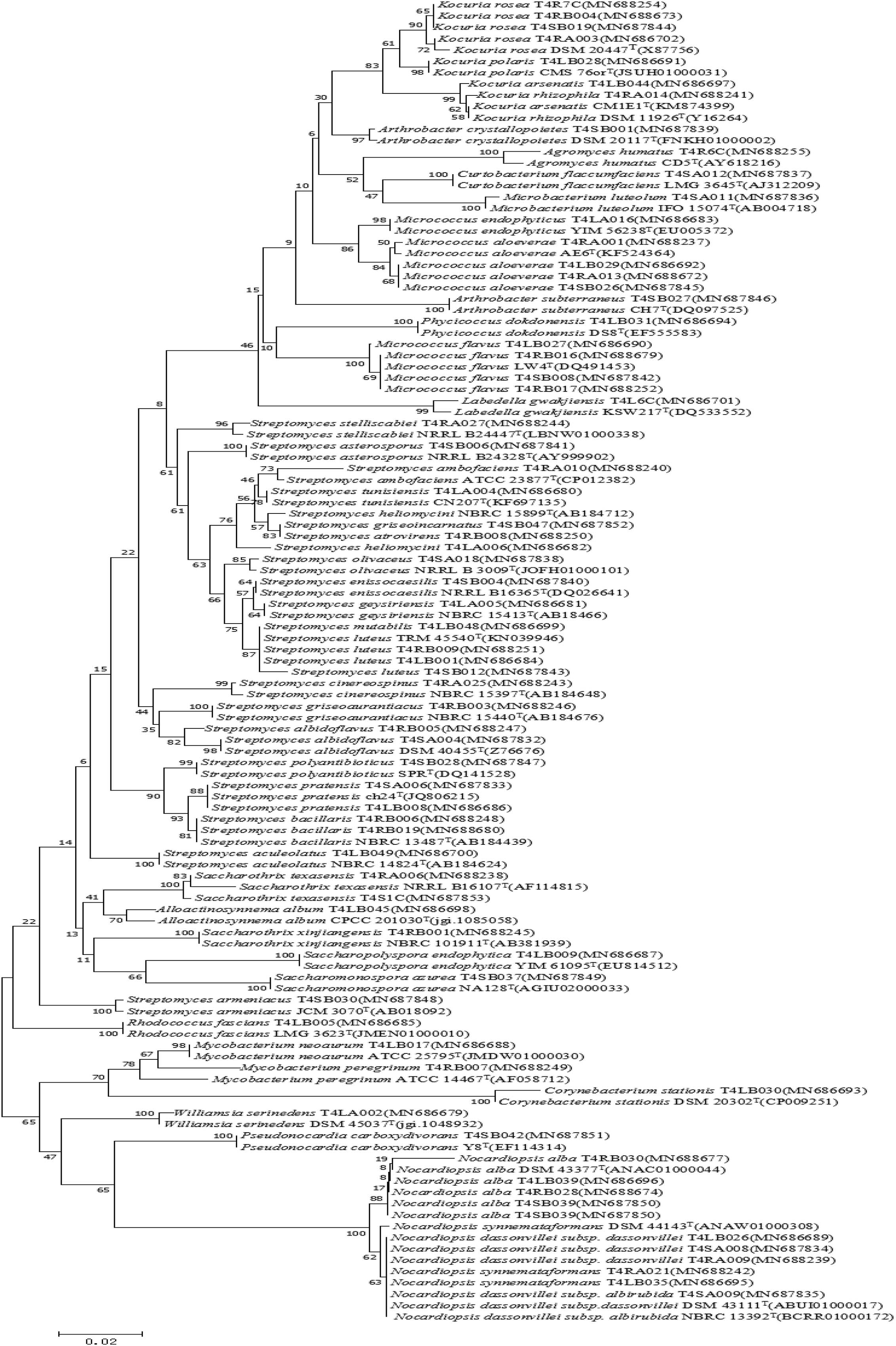
Figure 3. Neighbor-joining tree based on 16S rRNA gene sequences showing relationships between the endophytic actinomycetes (isolated from T. roseus Schipcz. in Tacheng) and the nearest type strains. The bar represents 2% sequence divergence.
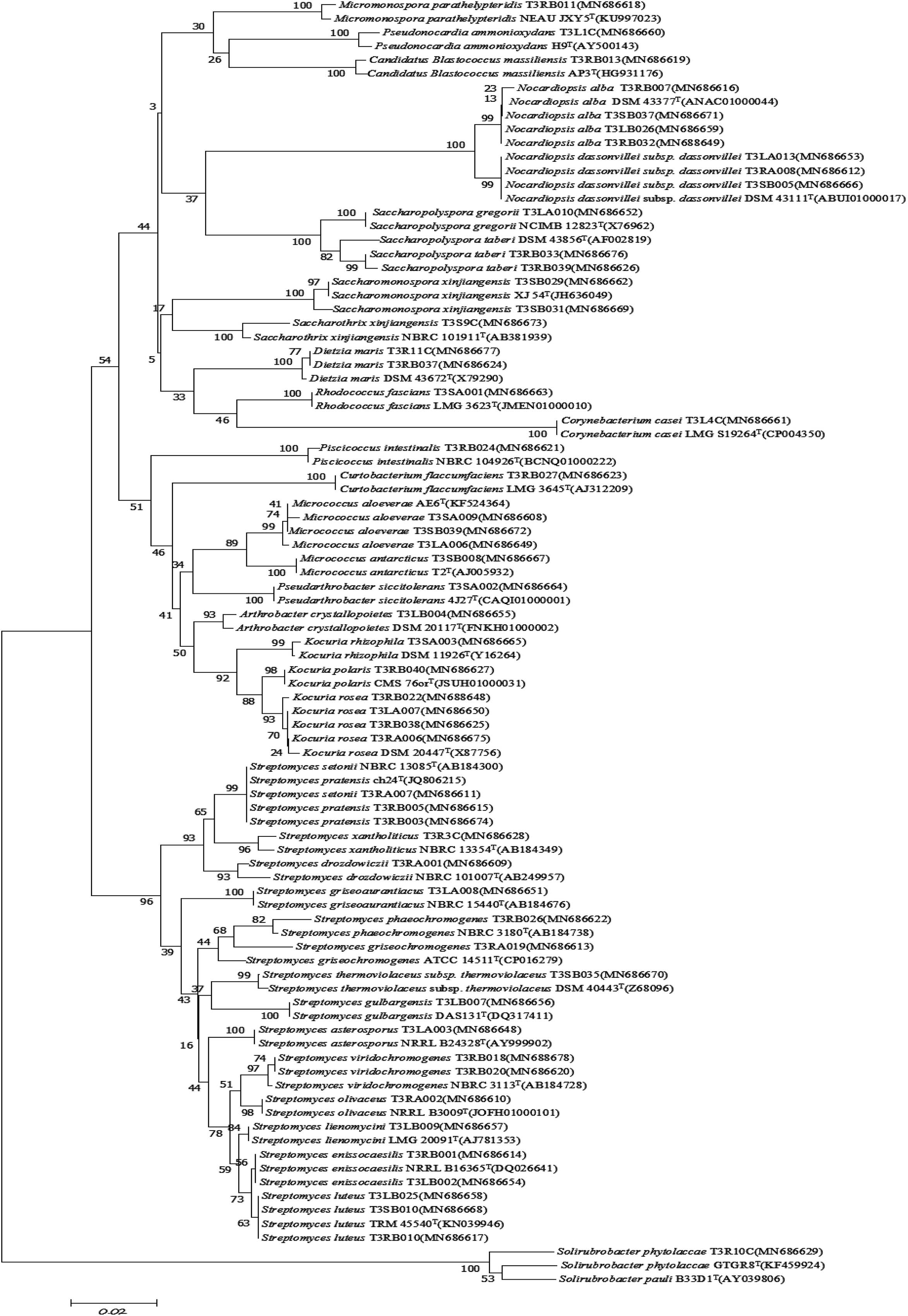
Figure 4. Neighbor-joining tree based on 16S rRNA gene sequences showing relationships between the endophytic actinomycetes (isolated from T. roseus Schipcz. in Ili) and the nearest type strains. The bar represents 2% sequence divergence.
The isolates from both sites grouped into 24 genera with a predominance of Streptomyces, followed by Nocardiopsis, Micrococcus, Kocuria, and others. Among these isolates, the number of these genera was four times as predominant from Tacheng than at the Ili site (Figure 1). Six rare genera representative 5.56 % of all genera obtained—Agromyces, Alloactinosynnema, Labedella, Microbacterium, Mycobacterium, and Williamsia—were isolated only from Tacheng, while the five rare genera representative 4.76% of all genera obtained—Blastococcus, Dietzia, Micromonospora, Pseudarthrobacter, and Solirubrobacter—were isolated only from Ili site. These results indicated that there are some differences in the composition of endophytes from the same plant species in different geographical sites (Figure 5).
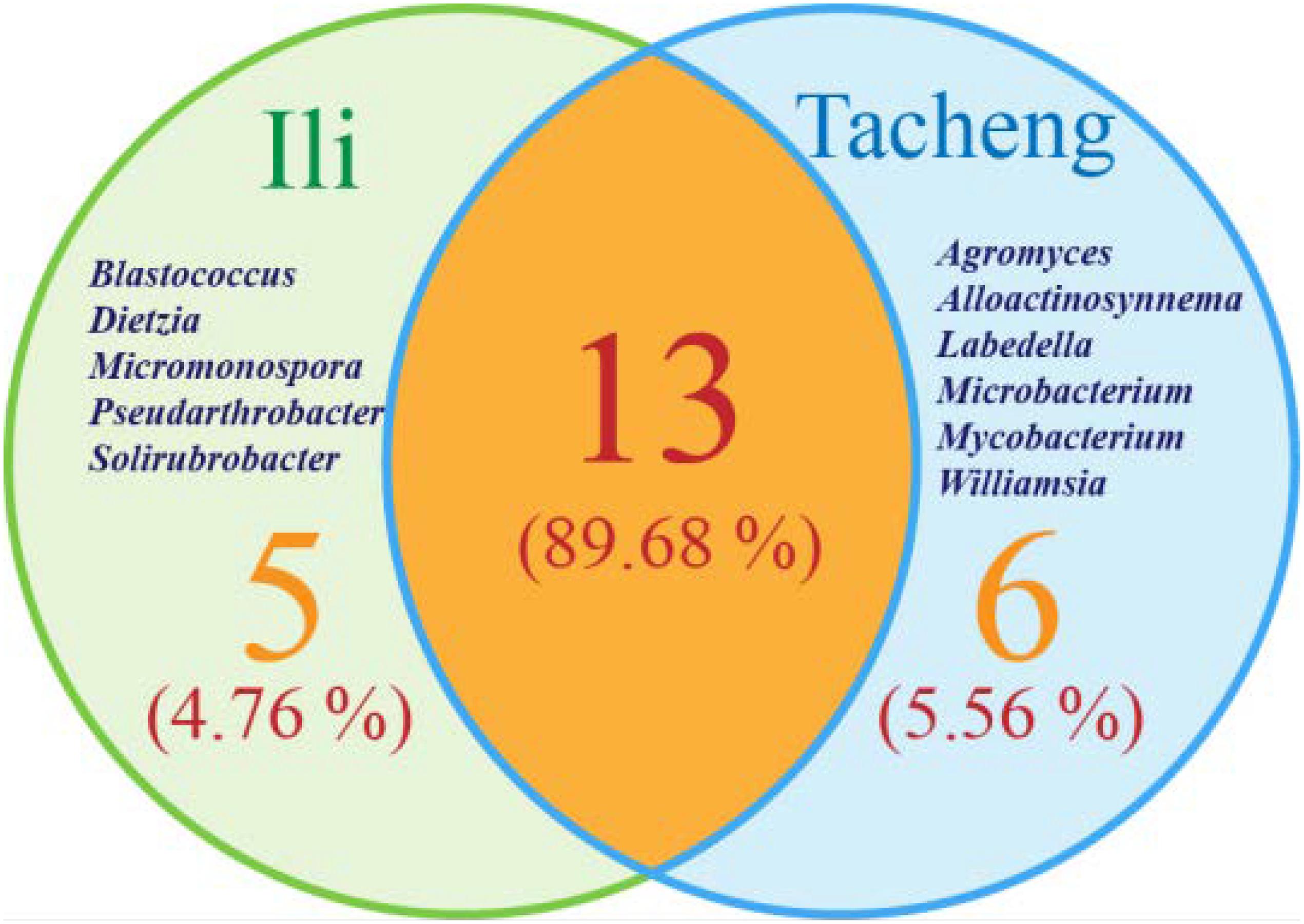
Figure 5. Venn diagram of endophytic actinomycetes in genera level from the T. roseus Schipcz. collected in two different geographical environments.
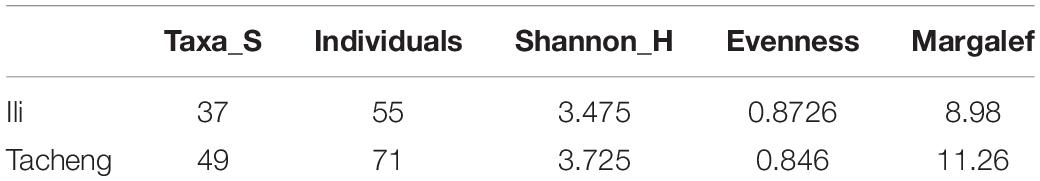
In this research, the endophytic actinomycete isolates displayed considerable diversity. The diversity parameters of each sample were different, and the values of the Shannon H indices (3.7205) and Margalf (11.26) of the species from Tacheng were higher than in the Ili site, while the Evenness (0.8460) was lower than the Ili site. All the indices clearly demonstrated that the diversity of endophytic actinomycete communities from Tacheng was more abundant than in Ili (Table 2).
Figure 1 exhibited that the genera to which strains from each sampled area and tissues belonged as well as the quantity of endophytes from each genus. The abundance of endophytic actinobacteria isolates in root tissues (n = 54, 42.86%) was significantly higher than in stems (n = 35, 27.78%) and leaves (n = 37, 29.37%), especially the isolates belonged to genera Streptomyces, Nocardiopsis, and Kocuria. However, the distribution of the genus Micrococcus was relatively similar in three kinds of tissues.
Antimicrobial Activity of the Isolates
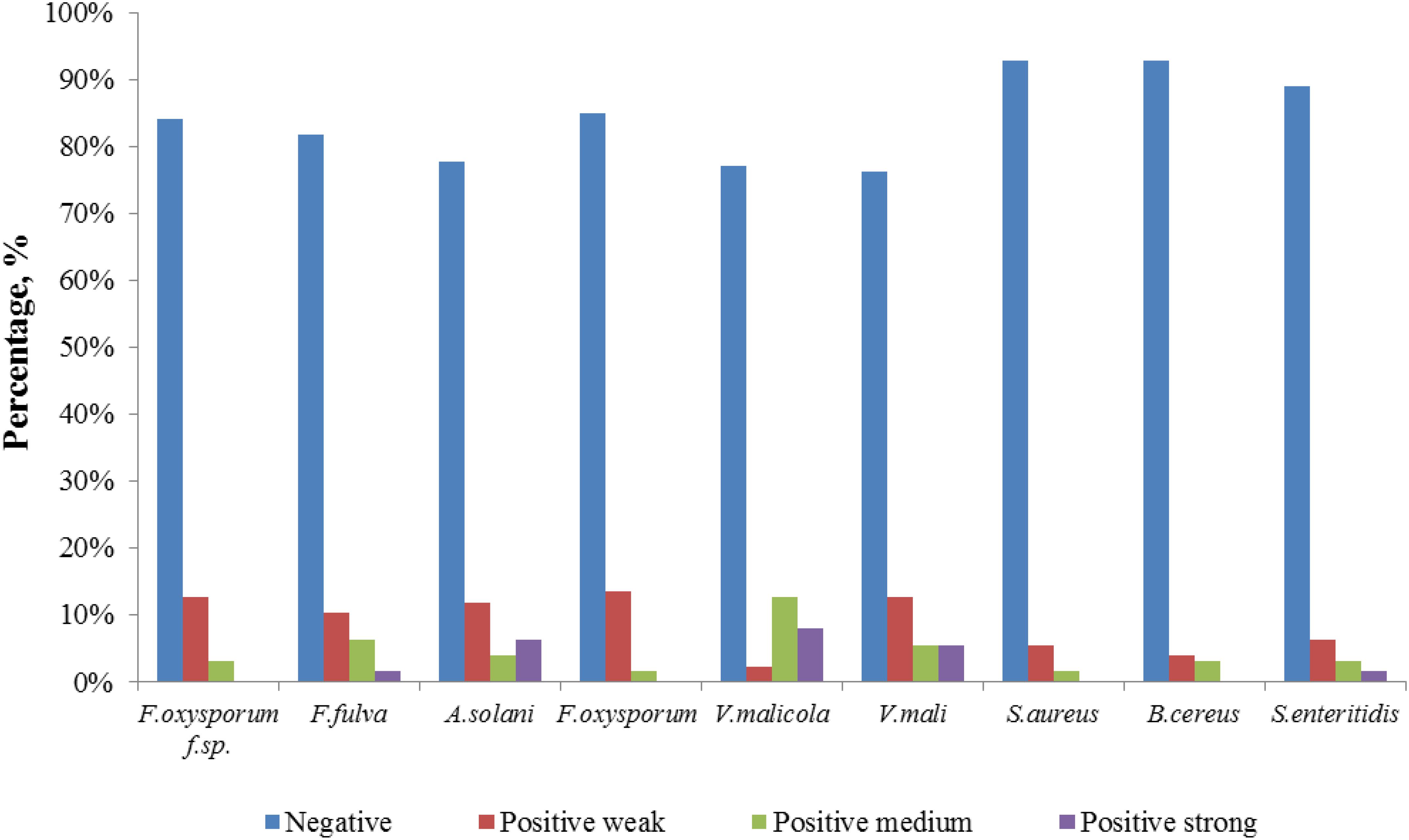
The 126 isolated endophytic actinobacteria from medicinal plant T. roseus S were tested, using plate confrontation assays, for their antibacterial and antifungal properties in in vitro conditions. Among these tested isolates, 54 strains were showed antimicrobial activity against one or more indicator pathogens (Table 3). The endophytes actinobacteria varied in their ability to inhibit the growth of the fungi, with the percentage of inhibition.
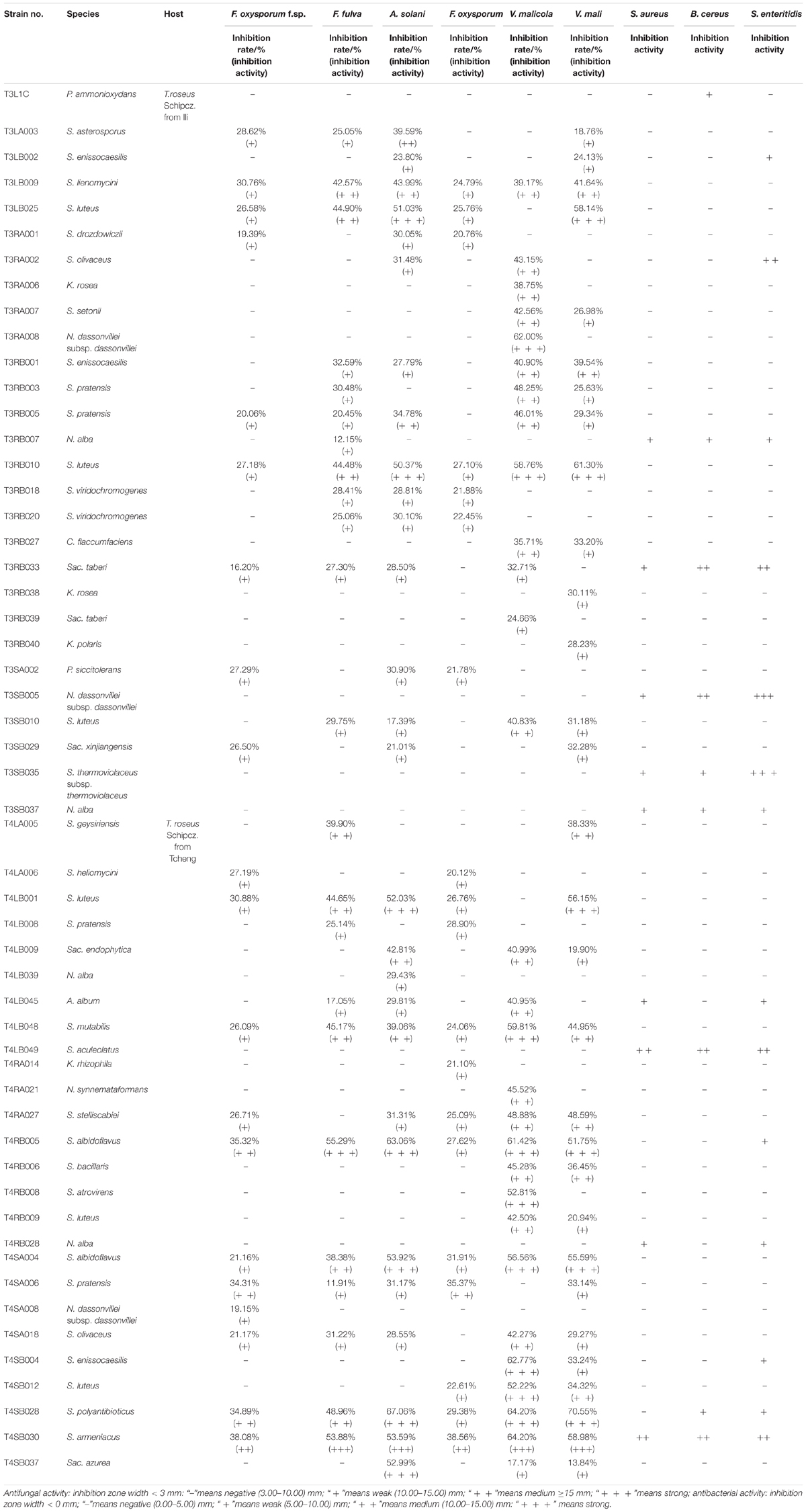
Table 3. Antimicrobial activity of 54 endophytic actinomycetes on indicated fungal and bacterial strains.
Comparatively potent activities against Fulvia fulva (Cooke) Cif., Alternaria solani, Valsa malicola, Valsa mali, and Salmonella enteritidis were identified in 1.59, 6.35, 7.94, 5.56, and 1.59% of the isolates, respectively. In contrast, all tested strains exhibited no potent inhibition against Fusarium oxysporum f.sp. and Fusarium oxysporum, and they merely showed moderate and weak activities. Of the 126 isolates, few strains showed high activities against human pathogens (Figure 6). Strain T3SB005 revealed completely antagonistic activity against all the three human pathogens of which T3SB005 showed a sizeable clear zone of inhibition to Salmonella enteritidis (+++) (Table 3).
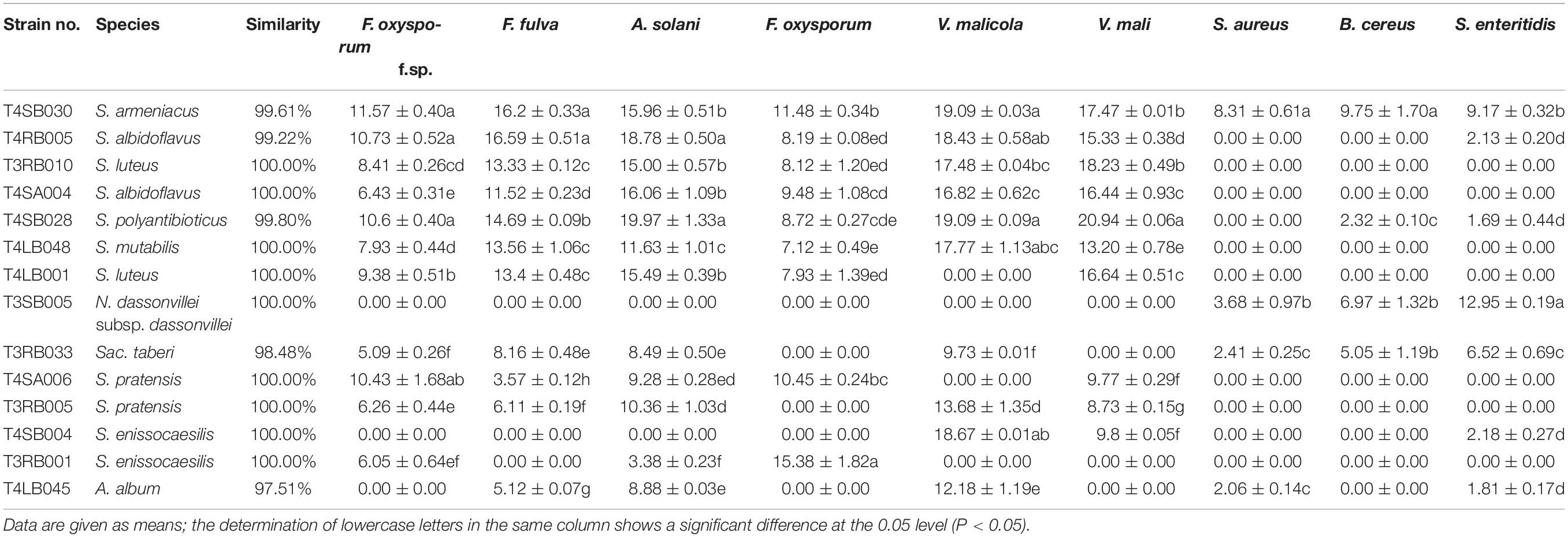
Among these positive isolates, 14 strains, including T4RB005, T3RB010, T4SA004, T4SB028, T4LB048, T4LB001, T3SB005, T3RB033, T4SA006, T3RB005, T4SB004, T3RB001, and T4LB045, showed a high and broad antimicrobial spectrum. Strain T4SB030 namely Streptomyces armeniacus had an inhibitory effect on the growth of all nine tested organisms. Moreover, strain T4SB028, namely Streptomyces polyantibioticus, showed inhibition against all the pathogens except for Staphylococcus aureus, and the inhibition ratio of T4SB028 reached 67.06, 64.20, and 70.55% against Alternaria solani, Valsa malicola, and Valsa mali, respectively. The statistical analysis revealed that the highest inhibition activity against Valsa mali was shown by T4SB028 (P < 0.05) (Tables 3, 4).
Of the 14 antagonistic strains, T3RB005 and T4LB045 displayed antagonisms against at least five indicator organisms. T3RB005 exhibited 98.48% 16S rRNA gene sequence similarity with the Saccharopolyspora taberi, while strain TLB045 displayed 97.51 % 16S rRNA gene sequence similarity with Alloactinosynnema album (Table 4). The active strains exhibited some selectivity in terms of some pathogens. Strain T4SB004 inhibited the growth of three pathogens, including Valsa malicola, Valsa mali, and Salmonella enteritidis; in a similar way, strain T3RB001 inhibited the growth of Fusarium oxysporum f.sp., Fusarium oxysporum, and Alternaria solani. Moreover, these two strains exhibited 100 % similarity with Streptomyces enissocaesilis; however, the two strains isolated from T. roseus Schipcz. in Ili and Tacheng, respectively (Tables 3, 4). This result revealed that the strains with the same taxonomy—but that are isolated from different geographic sites—showed different antagonistic activity, which can be further exemplified by the statistical analysis between strain T4SA006 and T3RB005 as well as strain T3RB010 and T4LB001 (Table 4). A representative picture illustrating the antagonistic activities is shown in Figure 7.
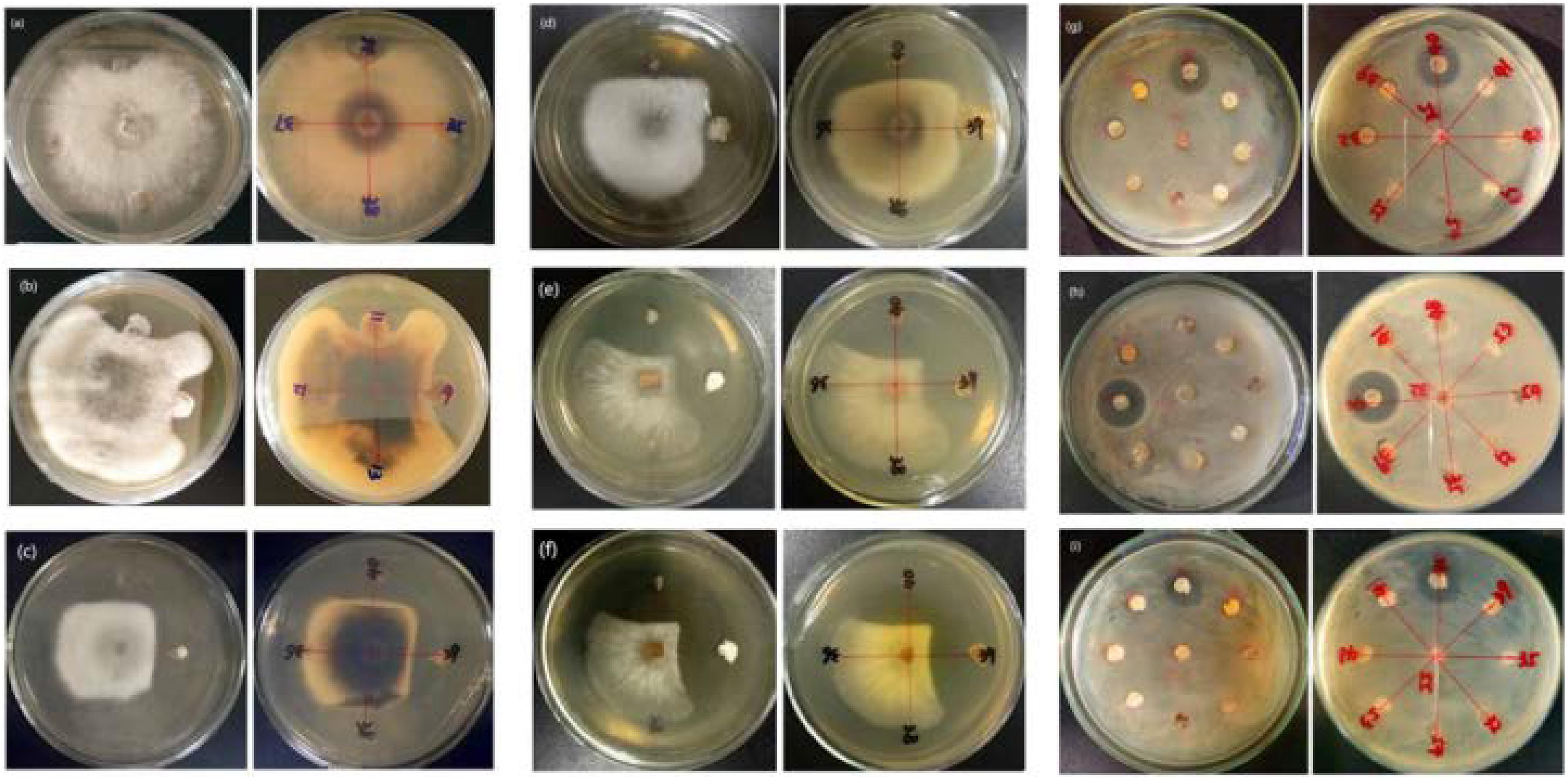
Figure 7. Representation of antimicrobial activities by the endophytic actinomycetes isolated from T. roseus Schipcz. (a) biocontrol activity against F. oxysporum f.sp; (b) biocontrol activity against F. fulva (Cooke) Cif.; (c) biocontrol activity against A. solani; (d) biocontrol activity against F. oxysporum; (e) biocontrol activity against V. malicola; (f) biocontrol activity against V. mali; (g) biocontrol activity against S. enteritidis; (h) biocontrol activity against B. cereus; (i) biocontrol activity against S. aureus; and 39 and 40 represent T4SB028 (S. armeniacus) and T4SB030 (S. polyantibioticus), respectively.
Screening of Chitinase- and Siderophore-Producing Strains
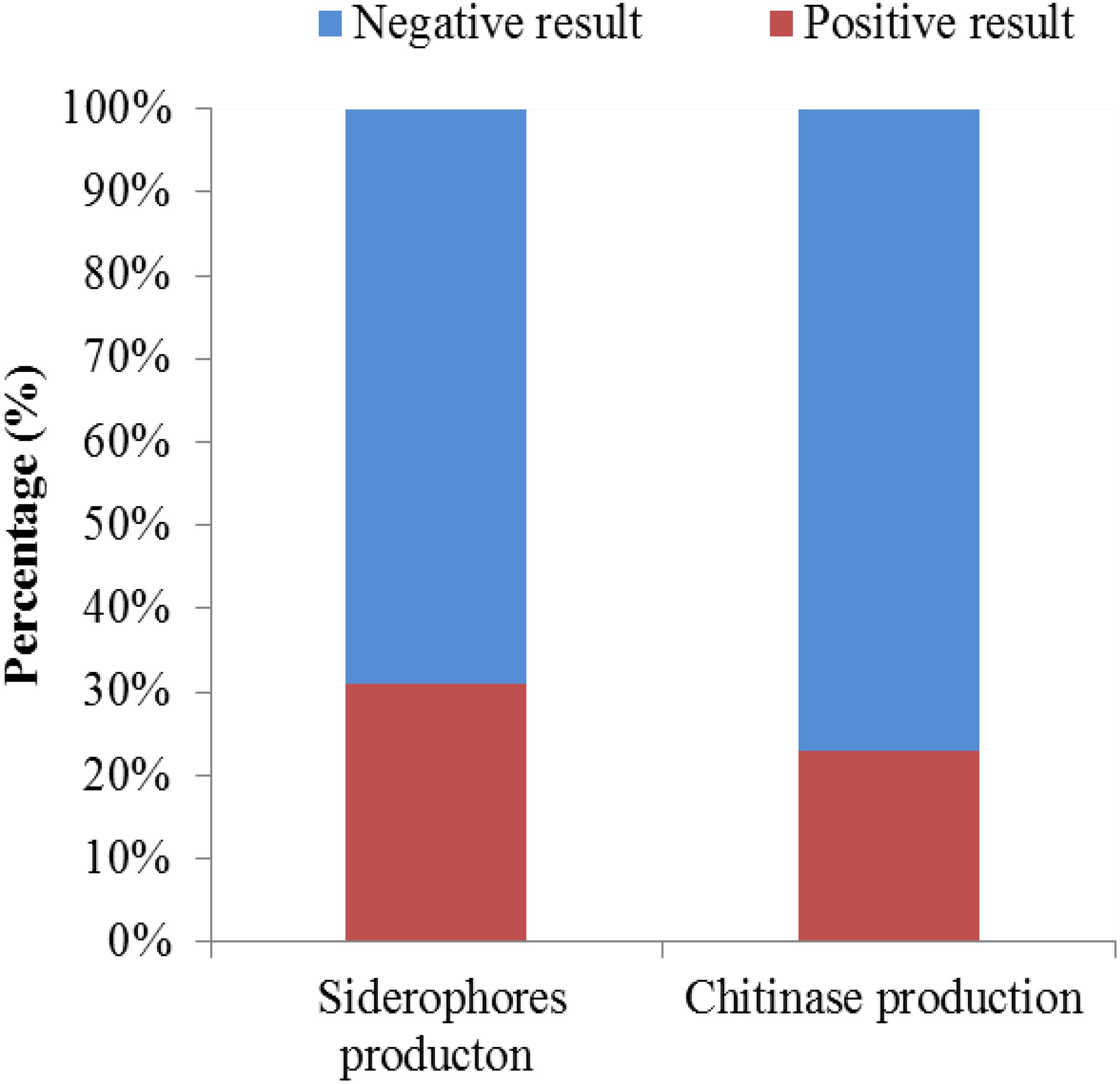
A total of 126 endophytic actinomycete isolates were screened for their siderophores and chitinase production abilities (Figure 8). Around 30.95 and 23.01% of the tested strains produced siderophores and chitinase, respectively.
Screening of Isolates With Antibiotics Biosynthetic Genes
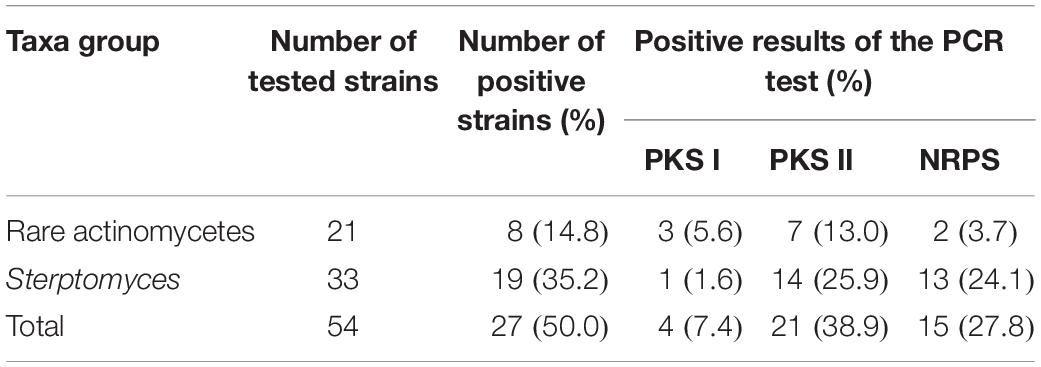
In this study, 27 out of the 54 strains possessed at least one of the test genes. Among the 54 antagonistic strains examined, 4 (7.4 %), 21 (38.9 %), and 15 (27.8 %) were identified as positive of PKS I, PKS II, and NRPS encoding genes, respectively, in the PCR reactions (Table 5). PKS-II (25.9%) and NRPS (24.1%) genes exhibited a high positive rate in the members of Streptomyces, while that of PKS-I genes was higher in rare actinobacteria (5.6%).
Discussion
Endophytic actinobacteria associated with medicinal plants as a biocontrol agent are considered to be beneficial for plant diseases management, and they spend their whole life cycle within plant tissues without causing any detectable infectious or signs of disease symptoms and may also produce the same metabolites and within the living tissues of host plant (Qin et al., 2009; Verma et al., 2009; Passari et al., 2015; Dos Santos et al., 2016). Therefore, these findings encouraged us to investigate for the first time the Chinese traditional medicinal plants T. roseus S for understanding the biodiversity and species distribution of the endophytic actinobacteria with respect to different ecological environments and their biosynthetic potential as antimicrobial agents.
The pretreatment of plant tissues is an advantageous method to promote and release rare endophytic actinobacteria from the inner parts of plant tissues. It is an essential and useful step for the isolation of endophytic actinobacteria that is depending on many factors, including host species, geographic and habitat distribution, pretreatment of plants, and selective media (Golinska et al., 2015). In this study, we isolated 126 endophytic actinobacteria from three types of pretreated plant tissues. Among different pretreatments, the number of endophytic actinobacteria with freezing treatment of plant tissues was more than the other two pretreatments. This result indicated that freezing treatment of plant tissues were more useful for the isolation of endophytic actinobacteria and helpful to release rare genera, such as strains T4LB045 and T3RB037, which belong to Alloactinosynnema and Dietzia. Freezing treatment of plant tissues could inhibit the majority of gram-negative species and the resistibility to low temperatures of spore-forming actinobacteria stronger than the non-spore-forming bacteria (Qin et al., 2011). The current findings indicated that the humic acid-vitamin (HV) medium and sodium propionate agar medium, which supplied by medicinal plant extracts of T. roseus S, was the most effective medium for isolating the endophytic actinobacteria due to the fact that the amino acids are the primary nitrogen sources in many plants. This result was in accordance with the previous study of Qin et al. (2009). This also could be due to some physiological characteristics of some actinobacteria in plant tissues being different (Kurtboke, 2003). Moreover, it is worth mentioning the combination of different pretreatment methods, especially freezing pretreatment and media influencing the isolation effectiveness of endophytic actinobacteria.
A total of 126 isolates belonging to 2 classes, 8 orders, 14 families, and 24 genera were isolated from medicinal plants T. roseus S from Ili and Tacheng sites in Xinjiang. The results showed a great diversity in populations of endophytic actinobacteria. The diversity analysis showed that the evaluated parameters from each location sample were different; the diversity from Tacheng was more abundant than the Ili site. For example, Venny of endophytic actinobacteria (Figure 3) showed that 5.56% rare genera were only from Tacheng, while 4.76% rare genera were only from the Ili site (Figure 3). Besides, the Streptomyces genus was the most abundant genus isolated from two samples (45 out of 126). These data corroborated the results obtained by Qiu et al. (2015), who isolated 119 endophytic actinobacteria 66 of them being affiliated to genus Streptomyces based on 16S rRNA gene sequence analysis. In accordance with these results, previous study has shown that 192, 146, and 159 endophytic actinobacteria were isolated from mangrove plants sampled in the three different natural protection areas Macao, Hainan, and Guangdong, respectively, and only 12 genera were isolated from Macao site (Xue et al., 2013; Li et al., 2016, 2017). In addition, in our previous investigation for isolation of endophytic bacteria associated with medicinal plants in the arid land of Xinjing, Chinam obtained similar observations regarding to geographical environment (Li et al., 2018). However, these results suggested that the select of different geographical environmental habitat to the same host plant to explore the diversity of endophytic actinobacteria resources will helps us to understand more characteristics of the symbiotic relation of host plants with respect to geographical environment of sampled area such as the type of soil and nutrient content (Qin et al., 2011; Govindasamy et al., 2014).
In the current study, Streptomyces. sp. was the most dominant in the root tissues, whereas fewer species were isolated from leaves and stem. This finding was in agreement with previous studies of Verma et al. (2009), Kaewkla and Franco (2013), and Kemung et al. (2018); they stated that plant roots provided a suitable habitat for endophytic actinobacteria and genera Streptomyces are majority in roots. This may be associated with the fact that the rhizospheric actinobacteria can move along the internal plant tissues and can colonize the roots (Golinska et al., 2015).
The antagonistic activity of the endophytic actinobacteria community associated with medicinal plant T. roseus S were screening in vitro condition by using a dual culture technique against a diverse range of phytopathogens. The results showed, substantial antagonistic activities against five pathogens, including Fulvia fulva (Cooke)Cif., Alternaria solani, Valsa malicola, Valsa mali, and Salmonella enteritidis. In contrast, all tested strains showed moderate and weak activities against Fusarium oxysporum f.sp. and Fusarium oxysporum. The antagonism of endophytic actinobacteria observed in this study was in accordance with previous reports; this includes, for example, the activity of actinobacteria isolated from tomato plants toward a diverse range of phytopathogens, such as Alternaria solani, Fusarium oxyporum f sp. lycopersici, and Rhizoctonia sp. (de Oliveira et al., 2010; Li et al., 2016). In this study, the majority of the 54 antagonistic strains belonged to Streptomyces.sp (de Oliveira et al., 2010; Kim et al., 2012; Mingma et al., 2014). Strain T3SB005 belonging to Streptomyces albidoflavus showed antagonistic activity against three human pathogens, and it strongly inhibited the growth of Salmonella enteritidis. However, Zhang et al. (2015) found that the secondary metabolites of strain Nocardiopsis dassonvillei subsp. dassonvillei showed potent inhibitory activity against Staphylococcus aureus. In addition, strain Streptomyces sp. T4SB030 and T4SB028 were able to control many bacterial and fungal pathogens. The inhibition ratio of T4SB028 against Alternaria solani, Valsa malicola, and Valsa mali was 67.06, 64.20, and 70.55%, respectively. This had accordance with the previous study (le Roes-Hill and Meyers, 2009; Fan et al., 2016; Li et al., 2016; Law et al., 2017). This result demonstrated the potential of strains Streptomyces sp. T4SB030 and T4SB028 isolated from T. roseus S. in the biocontrol of Alternaria solani, Valsa malicola, and Valsa mali. According to our work, the strain T4SB030 and T4SB028 were related to S. armeniacus, and S. polyantibioticus could be used as putative broad-spectrum biocontrol agents in future studies. Moreover, T3RB033 and T4LB045 might be a new putative antagonistic novel species isolated from T. roseus.
Interestingly, strain T4SB004 and T3RB001 with the same taxonomy based on the 16S RNA analysis and isolated from two different geographic and environmental sites exhibited different antagonistic activity. For instance, strain T4SB004 showed antagonistic activity toward the growth of Valsa malicola, Valsa mali, and Salmonella enteritidis, while strain T3RB001 inhibited the growth of Fusarium oxysporum f.sp., Alternaria solani, and Fusarium oxysporum. This indicated that active endophytic strains showed specific selectivity to some pathogens as well as further confirm the fact that the antimicrobial properties of endophytes had a relationship with the geographical site of host plants. In the present study, 30.95%, 23.00% of the tested endophytic actinobacteria were capable of producing siderophores and chitinase, respectively. It is well known that endophytic actinobacteria inhibit the growth of the phytopathogen by competing for iron in the environment (Singh et al., 2017). Besides, chitin is the crucial component of the fungal cell wall. Chitinase, as a fungal cell-wall-degrading enzyme produced by endophytic microorganisms, plays an active role in antagonism (Kalai-Grami et al., 2014; Egamberdieva et al., 2017a). Additionally, the endophytic actinomycete isolates from Achillea fragrantissima showed varying degrees of antifungal activity against fungal pathogens and produced chitinases or siderophores, according to El-Shatoury et al. (2009).
In order to understand the biosynthetic potential of the isolates, the detection of genes encoding polyketide synthases and non-ribosomal peptide synthetases, responsible for the synthesis of most biologically active polyketide and peptide compounds, has been broadly used for assessing biosynthetic potential of culturable and non-culturable microorganisms. In this study, at least one antibiotic biosynthetic gene was detected in 27 (representing 50% of tested strains) antagonistic strains. The antimicrobial genes screening results indicated that the presence of PKS and NRPS encoding genes were widespread in the tested strains. The higher proportions of PKS II (38.9 %) and NRPS (27.8 %) as well as lower detection rates of PKS I (7.4 %) these results were in agreement with the previous study (Lee et al., 2014; Qiu et al., 2015). The absence of amplification of PKS I, PKS II, and NRPS genes in some of the tested strains may be due to the absence of these genes, and the amplified primers were not suitable for screening the biosynthetic genes of some tested strains (Ayuso-Sacido and Genilloud, 2005). Moreover, we obtained the PKS and NRPS existent strains with other PKS I, PKS II, and NRPS genes primers (Wu et al., 2009). PKS-II (25.9%) and NRPS (24.1%) genes exhibited high positive rates in the members of Streptomyces, and this result was in opposition to the study of Qiu et al. (2015); they argued that the NRPS genes displayed wide distribution in the members of both Streptomyces and rare actinobacteria. Among tested endophytes, strains T3L1C, T3RB033, T3SB005, T3SB037, T4LB045, T4LB049, and T4RB005 displayed remarkable antibacterial activity in the bioactive assay, while PKS and NRPS genes were negative. According Finking and Marahiel (2004), the NRPS genes could also influences quorum-sensing of bacteria, though not in the biosynthesis of bioactive secondary metabolites.
This result suggested that the notion of that there is no direct correlation between the occurrence of biosynthetic genes and the production of antibacterial activities.
Conclusion
In summary, the results from our pilot study revealed that medicinal plant T. roseus Schipcz. provided a rich source of endophytic actinobacteria that exhibited a broad-spectrum antimicrobial agent in in vitro conditions. Besides, the diversity of endophytes differed between the plant tissues and geographic locations. These results further confirmed the notion that the pretreatment of plant materials, using selective media and different geographic locations, is an essential step for pure culture isolation. This study showed that bacterial isolates, such as S. armeniacus and S. polyantibioticus, with antifungal activity could be used as potential biocontrol agents against different kinds of plant pathogens. Furthermore, systematic investigations are needed to explore the chemical diversity of bioactive metabolites produced by these endophytes, which can offer an opportunity to discover novel natural products for the treatment of human diseases as well.
Data Availability Statement
The 16S rRNA gene sequences determined in this study were deposited in GenBank under accession numbers MN686679–MN686702(24), MN687832–MN687853(22), MN688237–MN688255(19), MN688672–MN688674(3), MN688677(1), MN688679–MN688680(2), MN686608–MN686629(22), MN686648–686678(31), and MN688648–688649(2). Reference sequences used are noted in the phylogenetic trees.
Author Contributions
LL and W-JL designed the experiments. ZM, GA, and JM conducted the experiments. DE, YL, and OAAM analyzed the data and reviewed the manuscript. ZM, JM, and LL wrote the manuscript. All the authors read and approved the manuscript.
Funding
This research was supported by the National Key Research Project (2016YFC0501502), National Natural Science Foundation of China (Grant Nos. U1703106 and 31650110479), the Chinese Academy of Sciences “President’s International Fellowship Initiative” (Grant No. 2018VBA002S) to DE, and Xinjiang Uygur Autonomous Region regional coordinated innovation project (Shanghai Cooperation Organization Science and Technology Partnership Program) (No. 2017E01031).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
References
Agrawal, T., and Kotasthane, A. S. (2012). Chitinolytic assay of indigenous Trichoderma isolates collected from different geographical locations of Chhattisgarh in Central India. Springerplus 1:73. doi: 10.1186/2193-1801-1-73
Ayuso-Sacido, A., and Genilloud, O. (2005). New PCR primers for the screening of NRPS and PKS-I systems in actinomycetes: detection and distribution of these biosynthetic gene sequences in major taxonomic groups. Microb. Ecol. 49, 10–24. doi: 10.1007/s00248-004-0249-6
Bobis, O., Dezmirean, D., Tomos, L., Chirila, F., and Marghitas, L. A. (2015). Influence of phytochemical profile on antibacterial activity of different medicinal plants against gram-positive and gram-negative bacteria. Appl. Biochem. Microb. 51, 113–118. doi: 10.1134/S0003683815010044
Cai, S. Q., Qin, L. P., Chen, D. F., and Yin, J. (2013). Pharmacognosy, 6th Edn. Beijing: People’s Medical Publishing House, 2.
Castro, R. A., Quecine, M. C., Lacava, P. T., Batista, B. D., Luvizotto, D. M., Marcon, J., et al. (2014). Isolation and enzyme bioprospection of endophytic bacteria associated with plants of Brazilian mangrove ecosystem. Springerplus 3:382. doi: 10.1186/2193-1801-3-382
Cho, S.-T., Chang, H.-H., Egamberdieva, D., Kamilova, F., Lugtenberg, B., and Kuo, C.-H. (2015). Genome analysis of Pseudomonas fluorescens PCL1751: a rhizobacterium that controls root diseases and alleviates salt stress for its plant host. PLoS One 10:e0140231. doi: 10.1371/journal.pone.0140231
Chun, J., Lee, J.-H., Jung, Y., Kim, M., Kim, S., Kim, B. K., et al. (2007). EzTaxon: a web-based tool for the identification of prokaryotes based on 16S ribosomal RNA gene sequences. Int. J. Syst. Evol. Microbiol. 57, 2259–2261. doi: 10.1099/ijs.0.64915-0
de Oliveira, M. F., da Silva, M. G., and Van Der Sand, S. T. (2010). Anti-phytopathogen potential of endophytic actinobacteria isolated from tomato plants (Lycopersicon esculentum) in southern Brazil, and characterization of Streptomyces sp. R18 (6), a potential biocontrol agent. Res. Microbiol. 161, 565–572. doi: 10.1016/j.resmic.2010.05.008
Dos Santos, P. J. C., Savi, D. C., Gomes, R. R., Goulin, E. H., Senkiv, C. D. C., Tanaka, F. A. O., et al. (2016). Diaporthe endophytica and D. terebinthifolii from medicinal plants for biological control of Phyllosticta citricarpa. Microbiol. Res. 186, 153–160. doi: 10.1016/j.micres.2016.04.002
Duan, J.-L., Li, X.-J., Gao, J.-M., Wang, D.-S., Yan, Y., and Xue, Q.-H. (2013). Isolation and identification of endophytic bacteria from root tissues of Salvia miltiorrhiza Bge. and determination of their bioactivities. Ann. Microbiol. 63, 1501–1512. doi: 10.1007/s13213-013-0614-0
Egamberdieva, D., Jabborova, D., Wirth, S., Alam, P., Alyemeni, M. N., and Ahmad, P. (2018). Interactive effects of nutrients and Bradyrhizobium japonicum on the growth and root architecture of soybean (Glycine max L.). Front. Microbiol. 9:1000. doi: 10.3389/fmicb.2018.01000
Egamberdieva, D., Li, L., Lindström, K., and Räsänen, L. A. (2016). A synergistic interaction between salt-tolerant Pseudomonas and Mesorhizobium strains improves growth and symbiotic performance of liquorice (Glycyrrhiza uralensis Fish.) under salt stress. Appl. Microbiol. Biotech. 100, 2829–2841. doi: 10.1007/s00253-015-7147-3
Egamberdieva, D., Wirth, S., Alqarawi, A. A., Abd Allah, E. F., and Hashem, A. (2017a). Phytohormones and beneficial microbes: essential components for plants to balance stress and fitness. Front. Microbiol. 8:2104. doi: 10.3389/fmicb.2017.02104
Egamberdieva, D., Wirth, S., Behrendt, U., Ahmad, P., and Berg, G. (2017b). Antimicrobial activity of medicinal plants correlates with the proportion of antagonistic endophytes. Front. Microbiol. 8:199. doi: 10.3389/fmicb.2017.00199
Egamberdieva, D., Wirth, S. J., Shurigin, V. V., Hashem, A., and Abd Allah, E. F. (2017c). Endophytic bacteria improve plant growth, symbiotic performance of chickpea (Cicer arietinum L.) and induce suppression of root rot caused by Fusarium solani under salt stress. Front. Microbiol. 8:1887. doi: 10.3389/fmicb.2017.01887
El-Shatoury, S., Abdulla, H., El-Karaaly, O., El-Kazzaz, W., and Dewedar, A. (2006). Bioactivities of endophytic actinomycetes from selected medicinal plants in the World Heritage Site of Saint Katherine Egypt. Int. J. Bot. 2, 307–312. doi: 10.3923/ijb.2006.307.312
El-Shatoury, S., El-Kraly, O., El-Kazzaz, W., and Dewedar, A. (2009). Antimicrobial activities of actinomycetes inhabiting Achillea fragrantissima (family: compositae). Egypt. J. Nat. Toxiol. 6, 1–15.
El-Tarabily, K., Nassar, A., Hardy, G. S. J., and Sivasithamparam, K. (2009). Plant growth promotion and biological control of Pythium aphanidermatum, a pathogen of cucumber, by endophytic actinomycetes. J. Appl. Microbiol. 106, 13–26. doi: 10.1111/j.1365-2672.2008.03926.x
Fan, D., Li, Y., Zhao, L., Li, Z., Huang, L., and Yan, X. (2016). Study on interactions between the major apple Valsa Canker Pathogen Valsa mali and its biocontrol agent Saccharothrix yanglingensis Hhs. 015 Using RT-qPCR. PLoS One 11:e0162174. doi: 10.1371/journal.pone.0162174
Finking, R., and Marahiel, M. A. (2004). Biosynthesis of nonribosomal peptides. Annu. Rev. Microbiol. 58, 453–488. doi: 10.1146/annurev.micro.58.030603.123615
Golinska, P., Wypij, M., Agarkar, G., Rathod, D., Dahm, H., and Rai, M. (2015). Endophytic actinobacteria of medicinal plants: diversity and bioactivity. Antonie Van Leeuwenhoek 108, 267–289. doi: 10.1007/s10482-015-0502-7
Govindasamy, V., Franco, C. M., and Gupta, V. V. (2014). “Endophytic actinobacteria: diversity and ecology,” in Advances in Endophytic Research, eds V. Verma and A. Gange, (Cham: Springer), 27–59. doi: 10.1007/978-81-322-1575-2_2
Hayakawa, M. (1990). Selective isolation methods and distribution of soil actinomycetes (in Japanese with English abstract). Actinomycetologica 4, 103–112. doi: 10.3209/saj.4_103
Hosseinzadeh, S., Jafarikukhdan, A., Hosseini, A., and Armand, R. (2015). The application of medicinal plants in traditional and modern medicine: a review of Thymus vulgaris. Int. J. Clin. Med. 6, 635–642. doi: 10.4236/ijcm.2015.69084
Jarić, S., Mitrović, M., and Pavlović, P. (2015). Review of ethnobotanical, phytochemical, and pharmacological study of Thymus serpyllum L. Evid. Based Complementary Altern. Med. 2015:101978. doi: 10.1155/2015/101978
Kaewkla, O., and Franco, C. M. (2013). Rational approaches to improving the isolation of endophytic actinobacteria from Australian native trees. Microb. Ecol. 65, 384–393. doi: 10.1007/s00248-012-0113-z
Kalai-Grami, L., Saidi, S., Bachkouel, S., Slimene, I. B., Mnari-Hattab, M., Hajlaoui, M. R., et al. (2014). Isolation and characterization of putative endophytic bacteria antagonistic to Phoma tracheiphila and Verticillium albo-atrum. Appl. Biochem. Biotechnol. 174, 365–375. doi: 10.1007/s12010-014-1062-4
Kemung, H. M., Tan, L. T.-H., Khan, T. M., Chan, K.-G., Pusparajah, P., Goh, B.-H., et al. (2018). Streptomyces as a prominent resource of future anti-MRSA drugs. Front. Microbiol. 9:2221. doi: 10.3389/fmicb.2018.02221
Kim, T.-U., Cho, S.-H., Han, J.-H., Shin, Y. M., Lee, H. B., and Kim, S. B. (2012). Diversity and physiological properties of root endophytic actinobacteria in native herbaceous plants of Korea. J. Microbiol. 50, 50–57. doi: 10.1007/s12275-012-1417-x
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Kun, L., Zhixiang, Z., and Xiaofei, L. (2010). Screening for PKS gene from soil metagenomics library and identification of the actives against root-knot nematodes. Plant Prot. 36, 38–42.
Kurtboke, D. (2003). Selective Isolation of Rare Actinomycetes. Brisbane: Queensland Complete Printing Services.
Law, J. W.-F., Ser, H.-L., Khan, T. M., Chuah, L.-H., Pusparajah, P., Chan, K.-G., et al. (2017). The potential of Streptomyces as biocontrol agents against the rice blast fungus, Magnaporthe oryzae (Pyricularia oryzae). Front. Microbiol. 8:3. doi: 10.3389/fmicb.2017.00003
le Roes-Hill, M., and Meyers, P. R. (2009). Streptomyces polyantibioticus sp. nov., isolated from the banks of a river. Int. J. Syst. Evol. Microbiol. 59, 1302–1309. doi: 10.1099/ijs.0.006171-0
Lee, L., Zainal, N., Azman, A., Eng, S., Goh, B., Yin, W., et al. (2014). Diversity and antimicrobial activities of Actinobacteria isolated from tropical mangrove sediments in Malaysia. Sci. World J. 2014:698178. doi: 10.1155/2014/698178
Li, F. N., Pan, Z., Tuo, L., Liu, S.-W., Zuo, X., Lei, C., et al. (2017). Studies on the diversity and novelty of endophytic actinobacteria isolated from mangrove plants collected in Macao. Chin. J. Antibiot. 42, 284–293.
Li, J., Dai, S. J., Tuo, L., Jiang, Z., Liu, S. W., Jiang, M. G., et al. (2016). Diversity and antimicrobial activity of endophytic actinobacteria isolated from mangroves collected in Dong Zhai Gang of Hainan Province. Microbiology 43, 1753–1765.
Li, L., Mohamad, O. A. A., Ma, J., Friel, A. D., Su, Y., Wang, Y., et al. (2018). Synergistic plant–microbe interactions between endophytic bacterial communities and the medicinal plant Glycyrrhiza uralensis F. Antonie Van Leeuwenhoek 111, 1735–1748. doi: 10.1007/s10482-018-1062-4
Liang, D., Li, F., Fu, Y., Cao, Y., Song, X., Wang, T., et al. (2014). Thymol inhibits LPS-stimulated inflammatory response via down-regulation of NF-κB and MAPK signaling pathways in mouse mammary epithelial cells. Inflammation 37, 214–222. doi: 10.1007/s10753-013-9732-x
Liu, Y., Guo, J., Li, L., Asem, M. D., Zhang, Y., Mohamad, O. A., et al. (2017). Endophytic bacteria associated with endangered plant Ferula sinkiangensis KM Shen in an arid land: diversity and plant growth-promoting traits. J. Arid Land 9, 432–445. doi: 10.1007/s40333-017-0015-5
Liu, Y.-H., Guo, J.-W., Salam, N., Li, L., Zhang, Y.-G., Han, J., et al. (2016). Culturable endophytic bacteria associated with medicinal plant Ferula songorica: molecular phylogeny, distribution and screening for industrially important traits. 3 Biotech 6:209. doi: 10.1007/s13205-016-0522-7
Liu, Y.-H., Wei, Y.-Y., Mohamad, O. A. A., Salam, N., Zhang, Y.-G., Guo, J.-W., et al. (2019). Diversity, community distribution and growth promotion activities of endophytes associated with halophyte Lycium ruthenicum Murr. 3 Biotech 9:144. doi: 10.1007/s13205-019-1678-8
Loqman, S., Barka, E. A., Clément, C., and Ouhdouch, Y. (2009). Antagonistic actinomycetes from Moroccan soil to control the grapevine gray mold. World J. Microbiol. Biotechnol. 25, 81–91. doi: 10.1007/s11274-008-9864-6
Matsumoto, A., and Takahashi, Y. (2017). Endophytic actinomycetes: promising source of novel bioactive compounds. J. Antibiot. 70, 514–519. doi: 10.1038/ja.2017.20
Metsä-Ketelä, M., Salo, V., Halo, L., Hautala, A., Hakala, J., Mäntsälä, P., et al. (1999). An efficient approach for screening minimal PKS genes from Streptomyces. FEMS Microbiol. Lett. 180, 1–6. doi: 10.1111/j.1574-6968.1999.tb08770.x
Mingma, R., Pathom-aree, W., Trakulnaleamsai, S., Thamchaipenet, A., and Duangmal, K. (2014). Isolation of rhizospheric and roots endophytic actinomycetes from Leguminosae plant and their activities to inhibit soybean pathogen, Xanthomonas campestris pv. glycine. World J. Microbiol. Biotechnol. 30, 271–280. doi: 10.1007/s11274-013-1451-9
Mohamad, O. A., Li, L., Ma, J. B., Hatab, S., Xu, L., Guo, J. W., et al. (2018). Evaluation of the antimicrobial activity of endophytic bacterial populations from Chinese traditional medicinal plant licorice and characterization of the bioactive secondary metabolites produced by Bacillus atrophaeus against Verticillium dahliae. Front. Microbiol. 9:924. doi: 10.3389/fmicb.2018.00924
Nie, Y., Zeng, X.-C., Yang, Y., Luo, F., Luo, X., Wu, S., et al. (2012). A novel class of antimicrobial peptides from the scorpion Heterometrus spinifer. Peptides 38, 389–394. doi: 10.1016/j.peptides.2012.09.012
Orsini, M., and Romano-Spica, V. (2001). A microwave-based method for nucleic acid isolation from environmental samples. Lett. Appl. Microbiol. 33, 17–20. doi: 10.1046/j.1472-765X.2001.00938.x
Passari, A. K., Mishra, V. K., Saikia, R., Gupta, V. K., and Singh, B. P. (2015). Isolation, abundance and phylogenetic affiliation of endophytic actinomycetes associated with medicinal plants and screening for their in vitro antimicrobial biosynthetic potential. Front. Microbiol. 6:273. doi: 10.3389/fmicb.2015.00273
Petrini, O. (1991). “Fungal endophytes of tree leaves,” in Microbial Ecology of Leaves, eds J. H. Andrews and S. S. Hirano, (New York, NY: Springer), 179–197. doi: 10.1007/978-1-4612-3168-4_9
Qin, S., Li, J., Chen, H.-H., Zhao, G.-Z., Zhu, W.-Y., Jiang, C.-L., et al. (2009). Isolation, diversity, and antimicrobial activity of rare actinobacteria from medicinal plants of tropical rain forests in Xishuangbanna, China. Appl. Environ. Microbiol. 75, 6176–6186. doi: 10.1128/AEM.01034-09
Qin, S., Wang, H.-B., Chen, H.-H., Zhang, Y.-Q., Jiang, C.-L., Xu, L.-H., et al. (2008). Glycomyces endophyticus sp. nov., an endophytic actinomycete isolated from the root of Carex baccans Nees. Int. J. Syst. Evol. Microbiol. 58, 2525–2528. doi: 10.1099/ijs.0.2008/000398-0
Qin, S., Xing, K., Jiang, J.-H., Xu, L.-H., and Li, W.-J. (2011). Biodiversity, bioactive natural products and biotechnological potential of plant-associated endophytic actinobacteria. Appl. Microbiol. Biotechnol. 89, 457–473. doi: 10.1007/s00253-010-2923-6
Qiu, P., Feng, Z.-X., Tian, J.-W., Lei, Z.-C., Wang, L., Zeng, Z.-G., et al. (2015). Diversity, bioactivities, and metabolic potentials of endophytic actinomycetes isolated from traditional medicinal plants in Sichuan, China. Chin. J. Nat. Med. 13, 942–953. doi: 10.3724/SP.J.1009.2015.00942
Ren, M., Wang, D., and Tian, C. (2010). Research of function application and composition of Thymus plants. China Brewing 12, 8–11.
Shimizu, M., Yazawa, S., and Ushijima, Y. (2009). A promising strain of endophytic Streptomyces sp. for biological control of cucumber anthracnose. J. Gen. Plant Pathol. 75, 27–36. doi: 10.1007/s10327-008-0138-9
Shirling, E. T., and Gottlieb, D. (1966). Methods for characterization of Streptomyces species. Int. J. Syst. Bacteriol. 16, 313–340. doi: 10.1099/00207713-16-3-313
Singh, S. P., Gupta, R., Gaur, R., and Srivastava, A. K. (2017). Antagonistic actinomycetes mediated resistance in Solanum lycopersicon Mill. against Rhizoctonia solani Kühn. Proc. Natl. Acad. Sci. India B Biol. Sci. 87, 789–798. doi: 10.1007/s40011-015-0651-5
Singh, S., and Gaur, R. (2016). Evaluation of antagonistic and plant growth promoting activities of chitinolytic endophytic actinomycetes associated with medicinal plants against Sclerotium rolfsii in chickpea. J. Appl. Microbiol. 121, 506–518. doi: 10.1111/jam.13176
Strobel, G., and Daisy, B. (2003). Bioprospecting for microbial endophytes and their natural products. Microbiol. Mol. Biol. Rev. 67, 491–502. doi: 10.1128/MMBR.67.4.491-502.2003
Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F., and Higgins, D. G. (1997). The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25, 4876–4882. doi: 10.1093/nar/25.24.4876
Verma, V. C., Gond, S. K., Kumar, A., Mishra, A., Kharwar, R. N., and Gange, A. C. (2009). Endophytic actinomycetes from Azadirachta indica A. Juss.: isolation, diversity, and anti-microbial activity. Microb. Ecol. 57, 749–756. doi: 10.1007/s00248-008-9450-3
Vickers, J. C., Williams, S. T., and Ross, G. W. (1984). “A taxonomic approach to selective isolation of streptomycetes from soil,” in Biological, Biochemical and Biomedical Aspects of Actinomycetes, eds L. Ortiz-Ortiz, L. F. Bojalil, and V. Yakoleff, (London: Academic Press), 553–561. doi: 10.1016/b978-0-12-528620-6.50045-8
Wang, H. (2015). Study on Biodiversity of Endophytic Bacteria Isolated from Four Chenopodiaceae Halophytes in Xinjiang and Evaluation of Their Growth-Promoting Function and Salt-Tolerance Ability. Ph.D. Dissertation, Xinjiang Institute of Ecology and Geography, Ürümqi.
Wang, L. L., Wang, E. T., Liu, J., Li, Y., and Chen, W. X. (2006). Endophytic occupation of root nodules and roots of Melilotus dentatus by Agrobacterium tumefaciens. Microb. Ecol. 52, 436–443. doi: 10.1007/s00248-006-9116-y
Wu, Y., Lu, C., Qian, X., Huang, Y., and Shen, Y. (2009). Diversities within genotypes, bioactivity and biosynthetic genes of endophytic actinomycetes isolated from three pharmaceutical plants. Curr. Microbiol. 59, 475–482. doi: 10.1007/s00284-009-9463-2
Xue, L., Xue, Q., Chen, Q., Lin, C., Shen, G., and Zhao, J. (2013). Isolation and evaluation of rhizosphere actinomycetes with potential application for biocontrol of Verticillium wilt of cotton. Crop Prot. 43, 231–240. doi: 10.1016/j.cropro.2012.10.002
Zhang, J. L., Qin, Y. L., Xiong, Z. J., Zhao, G. Z., Zhu, W. Y., Zhao, L. X., et al. (2013). The selective isolation of endophytic actinomycetes. Microbiol. China 40, 1305–1313.
Zhang, X.-Y., Han, X.-X., Chen, X.-L., Dang, H.-Y., Xie, B.-B., Qin, Q.-L., et al. (2015). Diversity of cultivable protease-producing bacteria in sediments of Jiaozhou Bay, China. Front. Microbiol. 6:1021. doi: 10.3389/fmicb.2015.01021
Zhang, Y. F., Lu, L. L., and Lu, J. (2016). Identification of actinomycetes TRM45037 in extreme environment and analysis of its secondary metabolites. Chin. J. Biochem. Mol. Biol. 32, 281–288.
Keywords: endophytic actinobacteria, Thymus roseus Schipcz, diversity, antimicrobial activity, medicinal plant, environmental microbiology
Citation: Musa Z, Ma J, Egamberdieva D, Abdelshafy Mohamad OA, Abaydulla G, Liu Y, Li W-J and Li L (2020) Diversity and Antimicrobial Potential of Cultivable Endophytic Actinobacteria Associated With the Medicinal Plant Thymus roseus. Front. Microbiol. 11:191. doi: 10.3389/fmicb.2020.00191
Received: 17 July 2019; Accepted: 27 January 2020;
Published: 12 March 2020.
Edited by:
Maria Carolina Quecine, University of São Paulo, BrazilReviewed by:
Learn-Han Lee, Monash University Malaysia, MalaysiaVincent Vineeth Leo, Mizoram University, India
Copyright © 2020 Musa, Ma, Egamberdieva, Abdelshafy Mohamad, Abaydulla, Liu, Li and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Osama Abdalla Abdelshafy Mohamad, b3NhbWFAQXJ1LkVkdS5FZw==; Wen-Jun Li, bGl3ZW5qdW4zQG1haWwuc3lzdS5lZHUuY24=; Li Li, bGlsaS5ib2JvQG91dGxvb2suY29t
†These authors have contributed equally to this work
 Zulpiya Musa
Zulpiya Musa Jinbiao Ma
Jinbiao Ma Dilfuza Egamberdieva
Dilfuza Egamberdieva Osama Abdalla Abdelshafy Mohamad
Osama Abdalla Abdelshafy Mohamad Gulsumay Abaydulla
Gulsumay Abaydulla Yonghong Liu
Yonghong Liu Wen-Jun Li
Wen-Jun Li Li Li
Li Li