- 1DARWIN21, Biological and Environmental Sciences and Engineering Division, King Abdullah University of Science and Technology, Thuwal, Saudi Arabia
- 2Computational Bioscience Research Center, King Abdullah University of Science and Technology, Thuwal, Saudi Arabia
- 3BioScience Core Lab, King Abdullah University of Science and Technology, Thuwal, Saudi Arabia
- 4Department of Biology, University of Fribourg, Fribourg, Switzerland
- 5Max F. Perutz Laboratories, University of Vienna, Vienna, Austria
Salinity stress is a major challenge to agricultural productivity and global food security in light of a dramatic increase of human population and climate change. Plant growth promoting bacteria can be used as an additional solution to traditional crop breeding and genetic engineering. In the present work, the induction of plant salt tolerance by the desert plant endophyte Cronobacter sp. JZ38 was examined on the model plant Arabidopsis thaliana using different inoculation methods. JZ38 promoted plant growth under salinity stress via contact and emission of volatile compounds. Based on the 16S rRNA and whole genome phylogenetic analysis, fatty acid analysis and phenotypic identification, JZ38 was identified as Cronobacter muytjensii and clearly separated and differentiated from the pathogenic C. sakazakii. Full genome sequencing showed that JZ38 is composed of one chromosome and two plasmids. Bioinformatic analysis and bioassays revealed that JZ38 can grow under a range of abiotic stresses. JZ38 interaction with plants is correlated with an extensive set of genes involved in chemotaxis and motility. The presence of genes for plant nutrient acquisition and phytohormone production could explain the ability of JZ38 to colonize plants and sustain plant growth under stress conditions. Gas chromatography–mass spectrometry analysis of volatiles produced by JZ38 revealed the emission of indole and different sulfur volatile compounds that may play a role in contactless plant growth promotion and antagonistic activity against pathogenic microbes. Indeed, JZ38 was able to inhibit the growth of two strains of the phytopathogenic oomycete Phytophthora infestans via volatile emission. Genetic, transcriptomic and metabolomics analyses, combined with more in vitro assays will provide a better understanding the highlighted genes’ involvement in JZ38’s functional potential and its interaction with plants. Nevertheless, these results provide insight into the bioactivity of C. muytjensii JZ38 as a multi-stress tolerance promoting bacterium with a potential use in agriculture.
Introduction
The dramatic increase in the human population and the increasingly negative effects of climate change pose a serious threat to global food security, with a number of severe social and economic problems (FAO, 2017). Abiotic stresses, including soil salinity, nutrient deficiency, drought and heat, can cause extensive losses to agricultural yields worldwide (Boyer, 1982; Bray et al., 2000). Some of these abiotic stresses also become more intense due to global warming and climate change. For example, models have predicted a significant decrease in the percentage of soil moisture and an increase in drought-experiencing surfaces on Earth by the end of the 21st century (Burke et al., 2006; Dai, 2012). Another major abiotic stress that adversely affects plant growth and crop yields is soil salinity which affects approx. 20% of irrigated land (FAO, 2008).
Biological inoculants containing beneficial microbes are emerging as part of the 2nd green revolution technology to tackle biotic and abiotic stresses in a sustainable, environmentally-friendly and chemical-free manner and as a more rapid and cost-efficient alternative to time-consuming crop breeding (Baez-Rogelio et al., 2017; de Zélicourt et al., 2018). These beneficial microbes, including plant growth promoting bacteria (PGPB), can establish symbiotic associations and protect plants against biotic stresses, such as fungal pathogens, or promote tolerance to abiotic stresses, such as salinity or drought stress (Hardoim et al., 2008; Eida et al., 2019). PGPB may serve as biofertilizers for promoting plant growth through several mechanisms, such as increasing nutrient availability by solubilization of e.g., phosphate and zinc or sequestration of e.g., iron by siderophores (Rodriguez et al., 2004; Rodríguez et al., 2006). Other mechanisms include changes in phytohormones levels, e.g., by the production of indole-acetic acid (IAA) or modulation of ethylene levels by ACC deaminases (Idris et al., 2007; Wang et al., 2016). The production of exopolysaccharides does not only function in biofilm formation but also in water retention (Ashraf et al., 2004). Additionally, bacteria can produce volatiles (e.g., volatile organic compounds/VOCs, sulfur-containing compounds, or indole) that are used to communicate with other microbes (Weisskopf et al., 2016) or to promote growth and/or stress tolerance (Bailly et al., 2014; Liu and Zhang, 2015; Ledger et al., 2016).
Desert plants offer a promising pool of endophytic microbes from which beneficial bacteria can be isolated and exploited for their plant growth promoting potential, especially for inducing tolerance to abiotic stresses experienced in deserts (Bokhari et al., 2019). For example, we previously demonstrated the ability of the endophyte Enterobacter sp. SA187 isolated from the nodules of Indigofera argentea, a legume native to the Saudi Arabian desert, to enhance the yield of alfalfa crops using saline irrigation in the field (de Zélicourt et al., 2018). Recently, a number of desert plant species native to the Jizan region in Saudi Arabia were collected resulting in the isolation of 116 bacterial root endophyte strains of these plants (Eida et al., 2018). Here, we characterize the endophytic bacterium Cronobacter muytjensii JZ38 from this collection displaying significant growth promoting ability of the model plant Arabidopsis thaliana under salinity stress conditions in contact or contactless settings. A comprehensive genome sequence analysis of JZ38, supplemented with biochemical and phenotypic characterization, provided important insights into the pathways for conferring abiotic stress tolerance and antimicrobial activity to plants, making JZ38 a good candidate for application in agriculture.
Materials and Methods
Plant Assays
Arabidopsis thaliana wild-type Columbia (Col-0) seeds were surface sterilized by shaking for 10 min in 70% ethanol + 0.05% sodium dodecyl sulfate (SDS), then washed twice with 99% ethanol and once with sterilized ddH2O. The seeds were then sown on square Petri dishes (12 × 12 cm) containing half-strength Murashige and Skoog Basal Salt Mixture pH 5.8, 0.9% agar (1/2 MS) (Murashige and Skoog, 1962) (M5524, Sigma Aldrich). The plates were stored in the dark for 2 days at 4°C for seed stratification and then incubated vertically (∼75° angle) in growth chambers (Percival Scientific Inc.) at 22°C with a photoperiod of 16/8 h (light/dark, 100 μmol.m–2.s–1) for germination. Five-day old seedlings (∼1.0–1.5 cm in root length) were then gently transferred to fresh 1/2MS agar plates supplemented with 100 mM NaCl as a salt stress (five seedlings/plate). For contact (in-plate) assays, seeds were sown on 1/2MS plates containing JZ38 (2 × 105 CFU/mL) prior to stratification (100 μL of pure LB was added as a control) (de Zélicourt et al., 2018). For contactless assays, sterile 15 mL Falcon tube caps (VWR) were placed at the bottom of each square Petri dish before pouring the 1/2MS media. Then, 2 mL LB agar were poured into the tube caps. Once the LB agar solidified, 50 μL (5 × 106 CFU) of JZ38 were spot inoculated onto the LB agar (50 μL of pure LB was used as a control). Growth assays were performed in Percival incubators located at the KAUST Greenhouse Core Labs. Both contact and contactless assays were performed with three biological replicates.
Shoot and Root Phenotypic Measurements
The number of lateral roots was counted under a light microscope and primary root lengths (PRL) were measured using image analysis software ImageJ1. Lateral root density (LRD) was calculated by dividing the number of lateral roots by the primary root length. Fresh weight (FW) was measured using a sensitive analytical balance. All measurements were taken 16 days after inoculation (DAI). For dry weight (DW) measurements, plant material was dried for 2 days at 65°C.
Growth Conditions and Genomic DNA Isolate
Isolate JZ38 was regularly grown in LB broth (Lennox L Broth Base, Invitrogen) or on LB agar plates at 28°C. Fresh, pure bacterial cultures were used for total genomic DNA extraction using Sigma’s GenElute bacterial genomic DNA kit (Sigma Aldrich) following the manufacturer’s protocol. DNA quality and quantity were assessed by 0.7% agarose gel electrophoresis (35 V, 12 h), NanoDrop 2000 (Thermo Fisher Scientific) and Qubit dsDNA BR assay kit (Thermo Fisher Scientific).
PacBio Library Preparation and Genome Sequencing
DNA was size selected to 10 kb using the BluePippin Size-Selection System (Sage Science), following the “High-PassTM DNA Size Selection of ∼20kb SMRTbellTM Templates” manual. The SMRTbell template library was prepared according to the instructions from Pacific Biosciences’s “Procedure & Checklist - 20 kb Template Preparation using BluePippin Size-Selection System” guide. The SMRT cells were run at the KAUST Bioscience Core Labs on the PacBio RSII (Pacific Biosciences) sequencing platform using P6-C4 chemistry. One SMRT cell was run, taking one 360 min movie and generating 63,220 reads with a mean insert read length of 12,322 bp and an estimated genome coverage of 170X.
Genome Assembly
Raw data from PacBio’s platform was assembled into a draft assembly using the Hierarchical Genome Assembly Process v3 (HGAP3) (Chin et al., 2013) from PacBio’s SMRT Analysis pipeline v2.3.0.140936 patch 5. The assembly workflow can be broken down to three main steps: a preassembly step that mapped single pass reads to seed reads to generate consensus reads that were then quality trimmed. De novo assembly was done using the overlap layout consensus approach. The final step is consensus polishing using Quiver to reduce indels and base substitution using quality scores embedded in the raw data. To determine whether assembled contigs are circular, dot plots were generated using Gepard (Krumsiek et al., 2007) for detecting overlaps at the peripheries. Overlaps were collapsed and genome was closed using Minimus2 (Sommer et al., 2007). Finally, additional polishing rounds were performed using Quiver by applying quality scores from raw data to correct for indels and base substitutions where the output from one round is inputted to the next.
Genome Annotation
Genome annotation was carried out using the Automatic Annotation of Microbial Genomes (AAMG) which is an integrated module in the in-house INDIGO-Desert v1.1 pipeline (Alam et al., 2013). Briefly, gene prediction was done using prodigal v2.6.1 (Hyatt et al., 2010). Functional annotation was done using a multitude of tools and databases. InterProScan (Jones et al., 2014) was used to assign domain information, Gene Ontology (GO) terms and KEGG pathways. Predicted genes were compared using BLAST against UniProt2 for generic annotations and cross-references to Cluser of Orthologous Genes (COGs). For annotation of gene function, genes were compared to KEGG database (Functional Kyoto Encyclopedia of Genes and Genomes) (Kanehisa et al., 2016). RPS-BLAST (Marchler-Bauer et al., 2002) was used to identify conserved domains and COG (Clusters of Orthologous Groups). Predicted genes were also BLAST-ed against NCBI-nr, UniProt and KEGG. Ribosomal RNAs (rRNAs), transfer RNAs (tRNAs) and other non-coding RNAs (ncRNAs) were predicted using RNAmmer 1.2 (Lagesen et al., 2007), tRNAscan-SE 2.0 (Lowe and Eddy, 1997), and Infernal software (Nawrocki and Eddy, 2013), respectively. Prophage sequences were identified, annotated and graphically displayed using PHAST (Zhou et al., 2011). Function and pathway analysis was also performed using BlastKOALA web tool of KEGG database (Kanehisa et al., 2016). Toxin-antitoxin (T/A) systems were retrieved by using TA finder (Shao et al., 2011). Identification of gene clusters responsible for the biosynthesis of secondary metabolites was performed using antiSMASH v.4.2.0 (Weber et al., 2015). Chromosome and plasmid maps were generated using DNAPlotter release 18.0.2 (Carver et al., 2008).
Fatty Acid Analysis
The fatty analysis was done in the BCCM/LMG services, University of Gent, Belgium. In brief, isolate JZ38 and type strain Cronobacter sakazakii LMG 5740T were grown for 24 h at 28°C under aerobic conditions on LMG medium. Inoculation and harvesting of the cells, the extraction and analysis performed conform to the recommendations of the commercial identification system MIDI (Microbial Identification System, Inc., DE, United States). The whole-cell fatty acid composition was determined by gas chromatographically on an Agilent Technologies 6890N gas chromatograph (Santa Clara, CA, United States). The peak naming table MIDI TSBA 5.0 was used. The experiment was performed once as a standard method at BCCM/LMG.
Phylogenetic Analysis
16S rRNA-Based Phylogenetic Tree
The 16S rRNA gene sequences of JZ38 were predicted using RNAmmer 1.2 and the most common and identical sequences of the six copies were compared to known sequences listed in NCBI’s GenBank using BLASTn (Altschul et al., 1997). The sequences with the highest similarity in terms of sequence identity and query coverage, along with other type-strains from similar and distant genera were used for the phylogenetic tree construction. Multiple alignment of the nucleotide sequences was performed using MUSCLE (Edgar, 2004). The phylogenetic tree was constructed by the Neighbor-Joining method (Saitou and Nei, 1987), based on the Kimura 2-parameter model (Kimura, 1980), with bootstrap analysis (1,000 replications) using the software MEGA version 7 (Kumar et al., 2016).
Genome-Based Phylogenetic Tree
Using NCBI’s taxonomy website, a list of representative strains (Supplementary Table S1) belonging to different species from the Cronobacter genus were compiled. To help root the tree, Mangrovibacter plantisponsor DSM 19579 was used as an out-group. To minimize bias due to differences in annotation pipelines, all genomes were re-annotated using AAMG with identical parameters while keeping all plasmids in the analysis. Using AAMG annotations, OrthoFinder (Letunic and Bork, 2016) was used with default settings to disentangle the pan genome of the selected genomes in the form of a set of orthologous groups. Briefly, an all-vs.-all BLASTp analysis (Altschul et al., 1990) was done for the initial allocation of gene pairs. The gene pairs were then screened based on the length-normalized BLAST bitscores to generate a gene pair graph for all-vs.-all species. Next, the MCL tool v14.137 (Li et al., 2003) was used to infer orthogroups from the graph.
A gene tree was generated for each orthogroup that has at least two genomes present using the alignment-free tool DendroBlast (Kelly and Maini, 2013) and FastMe v2.1.10 (Lefort et al., 2015). A species tree was then built from the consensus of all gene trees using STAG v1.0.03 and rooted based on duplication events using STRIDE v1.0.0 (Emms and Kelly, 2017). Support values were calculated based on supporting consensus from the gene trees.
For distinguishing between Cronobacter strains at species level, Digital DNA-DNA Hybridization (dDDH) and Average Nucleotide Identity (ANI) calculations were performed. Pairwise BLAST-based Average Nucleotide Identity values (ANIb) were obtained using JSpecies (Richter et al., 2015). The genome sequence data were uploaded to the Type (Strain) Genome Server (TYGS), a free bioinformatics platform available under https://tygs.dsmz.de, for a whole genome-based taxonomic analysis (Meier-Kolthoff and Göker, 2019).
Determination of PGP Traits and Bioassays
Phenotypic classification was performed using API 20E (Biomérieux) strips for identification of Enterobacteriaceae according to manufacturer’s instructions. Cytochrome oxidase activity was tested using oxidase strips (Sigma Aldrich). Solubilization of phosphate and zinc, production of siderophores, evaluation of growth in different NaCl and PEG concentrations and antibiotic resistance were performed as previously described by Andrés-Barrao et al. (2017) with the slight modifications. Siderophore production and phosphate solubilization were assessed using the Blue Agar CAS assay (as described by Louden et al., 2011) and Pikovskaya’s (PVK) agar plates (M520, Himedia), respectively, by spot inoculation of 10 μL of 107 CFU/mL on the respective agar surface and incubated at 28°C for 2-3 days. Evaluation of growth in salinity, osmotic stress (PEG) and pH were performed in LB broth, by inoculation into 48-well plates at starting OD600 of 0.01, and PEG 8000 (Fisher Scientific) was used. Growth in heavy metal stress was performed in MM9 (g/L: 25.6, Na2HPO4; 6.0, KH2PO4; 1.0, NaCl; 2.0, NH4Cl and separately 40 mL, 20% glucose solution; 4 mL, 1 M MgSO4; 200 μL, 1 M CaCl2; 200 μL, 0.5% thiamine) supplemented with 100 mg/L of Manganese (Mn), Cadmium (Cd), Copper (Cu), Cobalt (Co), and Nickel (Ni) using MnCl2, CdCl2.2.5H2O, CuCl2.2H2O, CoCl2.6H2O and NiSO4, respectively. Carbohydrate utilization was performed using API 50 CH (Biomérieux) strips according to manufacturer’s instructions.
Indole-3-acetic acid (IAA) and indole production was quantitatively determined by inoculating 105 cells in LB broth supplemented with or without 2.5 mM L-Tryptophan (Sigma Aldrich) and incubated at 28°C with shaking at 210 rpm for 24 h. Cells were centrifuged and the supernatant was used for determination of IAA (or its precursors) and indole production. For IAA quantification, Salkowski reagent (2% of 0.5 M FeCl3 in 35% HClO4 solution) was added to supernatant in 2:1 ratio followed by 30 min incubation in the dark. For indole quantification, Kovac’s reagent (Sigma Aldrich) was added to supernatant in 1:1 ratio followed by 30 min incubation in the dark. The concentration of IAA and indole produced by the bacteria was estimated using standard curve of pure IAA (Sigma Aldrich) and indole (Sigma Aldrich) and spectrophotometric measurement at 530 and 570 nm, respectively.
Production of volatile hydrogen sulfide (H2S) and indole were determined by using strips impregnated with lead (II) acetate [Pb (CH3COO)2] for H2S and Kovac’s reagent (for indole) (Sigma Aldrich) for indole. The production was tested by inoculation of culture into falcon tube containing LB broth to starting OD600 of 0.01 and inserting the strips on the top of the tube. Blackening of the strips was indicative of H2S production while color change from yellow to pink was indicative of indole production.
Motility assay was performed by spot inoculation of 10 μL of 107 CFU/mL culture on LB agar plates containing 0.3% (swimming) and 0.6% (swarming) agar, and the plates were incubated at 28°C for 2 days. Antibiotic susceptibility tests were performed using antibiotic disks (BD BBL Sensi-Disc; Hardy Diagnostics HardyDisk AST) (Supplementary Table S2) and observation of halo formation to evaluate antibiotic resistance. All aforementioned bioassays were performed with at least three biological replicates.
Electron Microscopy
Pure colonies were picked and grown in LB broth at 28°C overnight, sub-cultured the next day to OD600 of 0.1. Cells from the exponential phase were harvested by centrifugation at 3000 × g, washed twice and resuspended in 0.1 M phosphate buffer saline (PBS). Bacterial cells were then fixed with 2.5% glutaraldehyde in cacodylate buffer (0.1 M, pH 7.4) overnight and rinsed with 0.1 M cacodylate buffer. Fixed samples were post-fixed with 2% osmium tetroxide, 1.5% potassium ferrocyanide in 0.1 M cacodylate buffer for 1h and then washed with water. Samples were then dehydrated in ethanol series and embedded in Epon epoxy resin. Contrasting sections were stained with uranyl acetate and lead citrate and imaging was performed using a Titan 80-300 S/TEM (Titan Cryo Twin; FEI Company) operating at 300 kV. The sample preparation and imaging were performed at the KAUST Imaging and Characterization Core Labs.
Gas Chromatography–Mass Spectrometry (GC/MS)
One colony from Luria-Bertani (LB) agar plate was resuspended in 3 mL of LB broth, incubated 24 h at 28°C and shaken at 190 rpm. The bacterial culture was adjusted to a density of OD595 = 1.0 in LB broth and inoculating 100 μL of this cell suspension were spread on LB agar medium poured into 5 cm glass Petri dishes. LB broth inoculated on LB-agar glass plates were used as controls. Isolate JZ38 was grown overnight at room temperature before collecting the volatiles during 48h using closed-loop-stripping analysis (CLSA) as described by Hunziker et al. (2015). Experiments were repeated at least three times. Trapped volatiles were extracted from the charcoal filter by rinsing the filter three times with 25 μL dichloromethane (≥99.8%, VWR). The experiment was repeated three times. The volatiles were analyzed by gas chromatography/mass spectrometry (GC/MS). The samples were injected in a HP6890 gas chromatography connected to a HP5973 mass selective detector fitted with an HP-5 ms fused silica capillary column (30 m; 0.25-mm inside diameter; 0.25-μm film; Agilent Technologies). Conditions were as follows: inlet pressure, 67 kPa; He, 15 ml/min; injection volume, 2 μL; transfer line, 300°C; injector, 250°C; electron energy, 70 eV. The gas chromatograph was programmed as follows: 5 min at 50°C, then increasing 5°C/min to 320°C and hold 1 min. The retention time and the relative abundance were determined by using OPENCHROM program (open source software) and the compounds were identified by comparison of mass spectra to database spectra (Wiley 275 mass spectral library).
Volatile-Mediated Effects of JZ38 on Phytophthora infestans and Other Phytopathogenic Fungi
Split Petri dishes were used to analyze the volatile-mediated effect of JZ38 on two Phytophthora infestans strains (88069 and Rec01). LB agar was poured in one half and V8 agar in the other half. A plug of mycelium (5 mm) was inoculated on the V8 medium and five drops of 10 μl (5 × 106 CFU) of overnight bacterial culture were spot inoculated on the LB medium (bacteria-free LB drops were used for control plates) on the same day. Plates were sealed with Parafilm, incubated at 23°C in the dark and photographed at different time points (10 days for 88069, 13 days for Rec01) from below with a reflex camera mounted on a stand. The obtained pictures were analyzed with the digital imaging software ImageJ. The mycelium area was determined with the freehand area measurement tool of ImageJ. This growth value was then compared to that measured in control plates (targets exposed to LB only), and a percentage was calculated. These dual culture assays were performed with four biological replicates (Petri dishes). This is a protocol modified from Hunziker et al. (2015). For the other phytopathogens, experiments were performed as described earlier, however, area of mycelial growth was determined 4 days after inoculation for the fungi (Rhizoctonia solani, Fusarium culmorum, and Botrytis cinerea) and 11 days after inoculation for the oomycete (P. infestans Rec01).
Data Availability
The data for the bacterial genome assembly and sequencing was deposited in NCBI/DDBJ/EMBL database under the accession numbers CP017662, CP017663 and CP017664, biosample number SAMN05828177. The annotations obtained by in-house INDIGO pipeline are available through the KAUST library repository4.
Statistical Analysis
The data were subjected to non-parametric one-way ANOVA, or Kruskal–Wallis H test (Kruskal and Wallis, 1952). Data were expressed as the mean ± standard error of the mean (SEM). The differences among the various treatment means were compared and the values with a p-value of ≤0.05 were considered statistically significant. Statistical analysis was done using DEVELVE statistical software5.
Results and Discussion
JZ38 Promotes Arabidopsis Growth Under Salinity Stress
Previously, JZ38 was highlighted as a plant growth promoting bacteria from a collection isolated from the root endosphere of the desert plant Tribulus terrestris (Eida et al., 2018). Here, the exhibited phenotype and salinity stress tolerance promoting (SSTP) ability of JZ38 was quantitatively confirmed on Arabidopsis thaliana as a model plant. Growth promotion of plants under salinity stress (100 mM NaCl) was observed using two different inoculation methods: contact (bacteria inoculated in-plate during germination) and contactless (bacteria grown on LB caps without contact to with the plants) (Figure 1 and Supplementary Table S3). The contactless assay can be used to study the effects of volatile compounds on plant biomass and root systems at late stages of growth (Supplementary Figure S1) when compared to circular Petri dish set-ups.
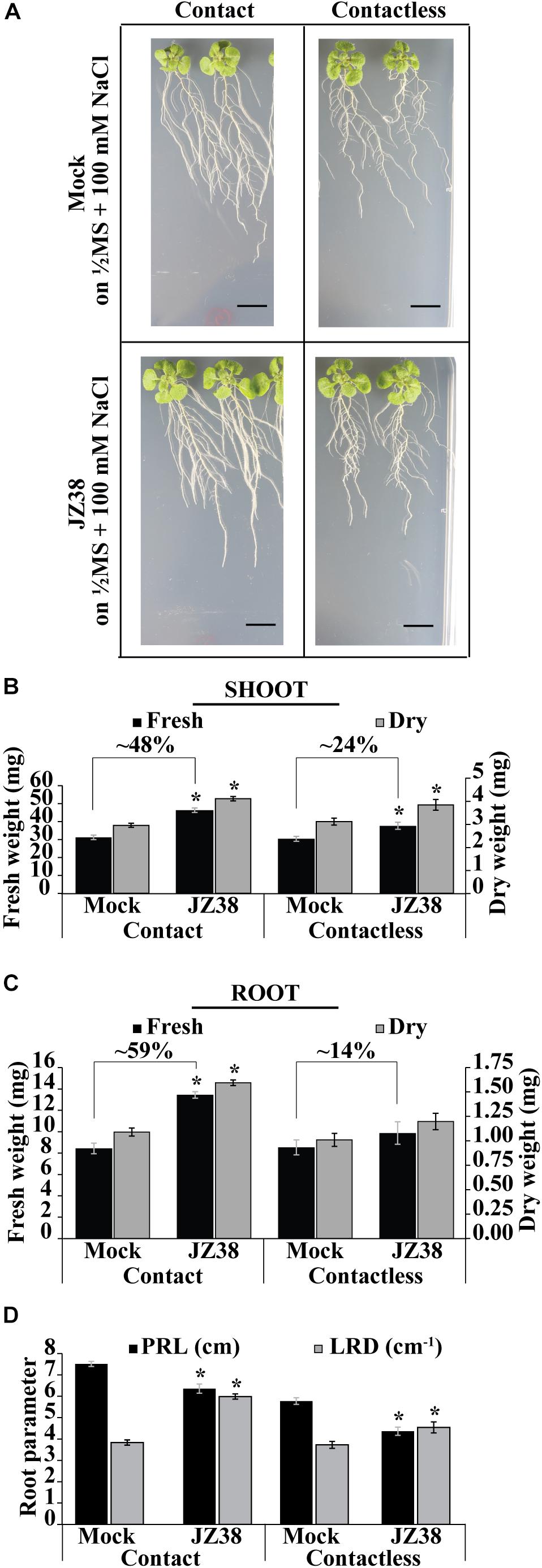
Figure 1. Effects of JZ38 on Arabidopsis growth under salinity stress using different inoculation methods. Representative images of A. thaliana plants inoculated with JZ38 or mock (bacteria-free LB) growing on 1/2MS + 100 mM NaCl, collected at 16 DAI (days after inoculation) (A). Fresh and dry weight measurements of shoots (B) and roots (C) and different root parameters (PRL, primary root length; LRD, lateral root density) collected at 16 DAI (D). Data are means of 3 biological replicates of 10 plants per treatment. Error bars represent standard error of the mean (SEM). Asterisks indicate significant differences between mock and bacteria-inoculated plants (Kruskal–Wallis test, p ≤ 0.05). Black bars correspond to 1 cm.
Under contact setting, JZ38 increased the fresh weight of shoot (Figure 1B) and roots (Figure 1C) by 48 and 59%, respectively (Supplementary Table S3). Under contactless setting, JZ38-mediated volatiles resulted in an increase of 24% in shoot fresh weight but no statistically significant effect on root fresh weight was observed. The dry weight measurements also exhibited similar results. JZ38 also induced changes in the root system architecture by reducing primary root growth (shorter primary root length) and increasing the lateral root density of plants under salinity stress using both inoculation settings. Previously isolated endophytes of the Enterobacteriaceae family (SA187, JZ29) also exhibited a similar pattern of growth promotion with higher LRDs compared to mock plants (de Zélicourt et al., 2018; Eida et al., 2019). The ability of JZ38 to promote growth in the contactless set-up suggested that volatile compounds are emitted by JZ38 which help plants to grow under saline conditions.
Cronobacter sp. JZ38 Genome Features
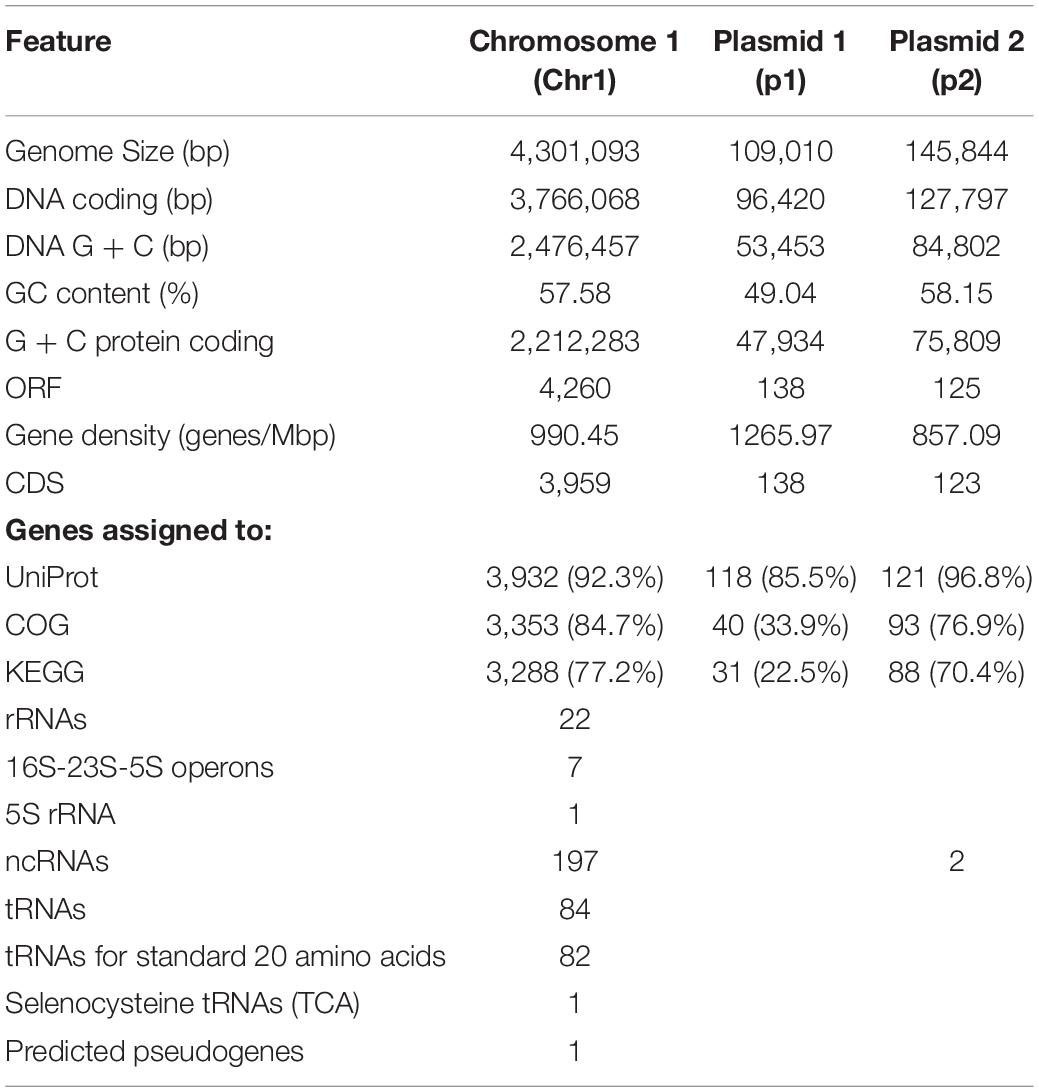
Following the confirmation of the plant growth promoting ability of JZ38 under salt stress conditions, whole genome sequence analysis of JZ38 was performed in order to obtain a reliable taxonomic classification and identify genes or pathways that could potentially contribute to the plant growth promoting effects. The genome of JZ38 was found to consist of three circular contigs (Figure 2): a single circular chromosome of ∼4.3 Mbp (Chr1) and two circular plasmids of ∼109 kbp (p1) and ∼145 kbp (p2) with an average GC content of 57.58, 49.04, and 58.15%, respectively (Table 1). A clear GC skew transition was observed at positions 270,964 and 2,425,765 of chromosome Chr1 corresponding to the origin of replication (oriC) and replication termination (terC) sites, respectively (Figure 2). The oriC and terC are predicted solely based on the GC skew transitions, while the oriC in Chr1 is confirmed by conservation of gyrB-recF-dnaN-dnaA-rpmH-rnpA gene organization in that region (Ogasawara et al., 1985).
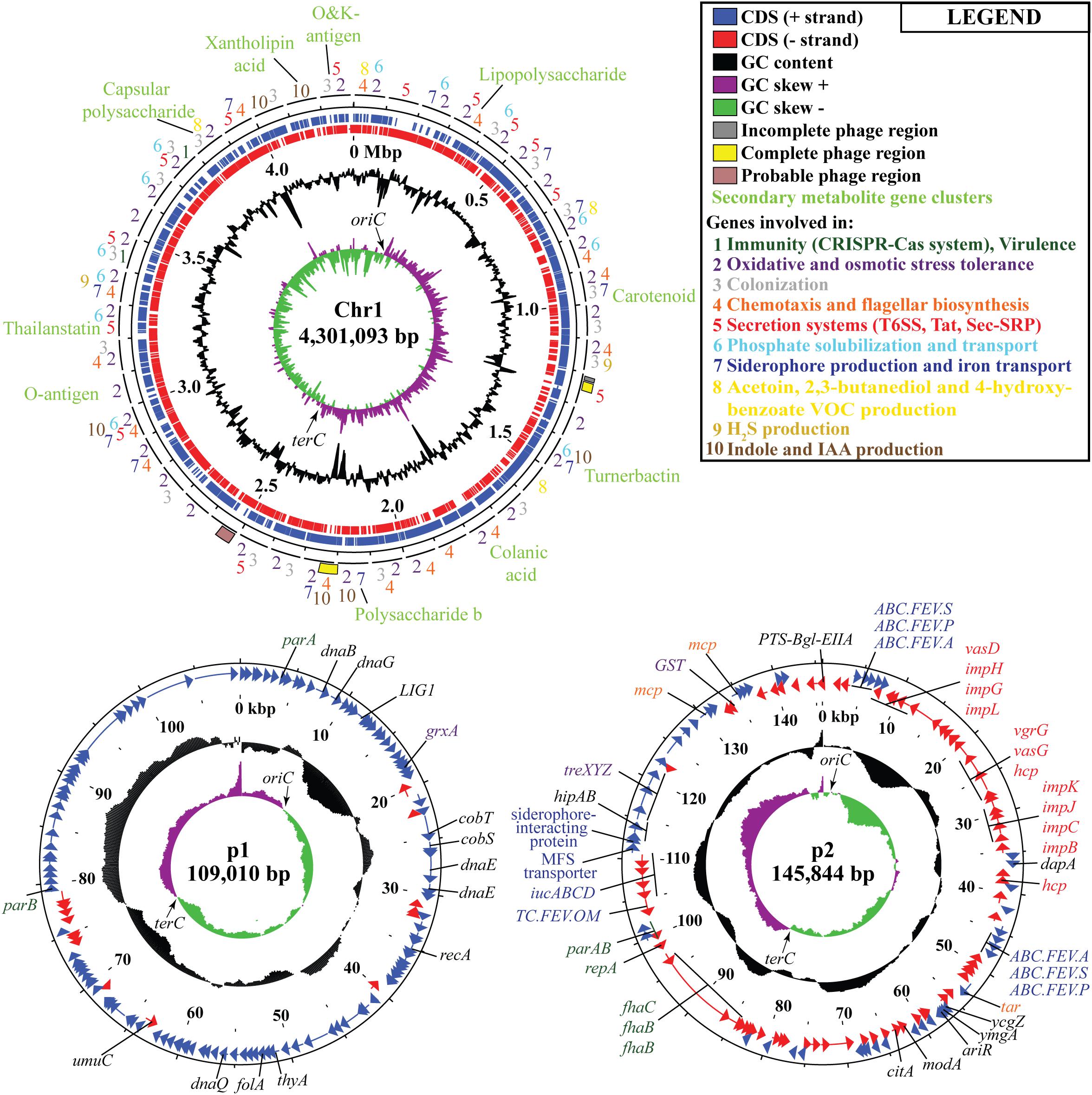
Figure 2. Circular representation of chromosome Chr1 and plasmids p1 and p2 in JZ38. The tracks from inside to outside: GC skew [(G-C/(G + C)] positive (purple) and negative (green),% GC content (black), coordinates in mega base pair (Mbp) for Chr1 and kilo base pair (kbp) for p1 and p2, reverse-strand CDSs (red), forward-strand CDSs (blue), phages predicted and genes present in chromosome or plasmids or the categories they belong to (number- or color-coded according to legend). Arrows on the GC skews indicate the origin of replication (oriC) and replication termination (terC) regions where the shifts occur. Maps generated using DNAPlotter release 18.0.2.
A total of 4,523 ORFs were predicted in the genome of which 4,220 (3,959 in the chromosome) were putative protein-coding DNA sequences (CDS). These numbers are quite similar to those reported for other Cronobacter species (Chase et al., 2017; Gopinath et al., 2018; Jang et al., 2018). In addition, a total of 197 ncRNA, 7 complete rRNA operons (16S-23S-5S), one additional 5S rRNA and 84 tRNA-coding genes (82 encoding standard amino acids, 1 selenocysteine, 1 pseudogene) were predicted in the chromosome sequence, while only 2 ncRNAs were found in plasmid p2. The presence of several copies of the rrn operon is characteristic of soil microbes and is thought to assist in the adaptation to changing environmental or growth conditions (Klappenbach et al., 2000; Bussema et al., 2008).
Taxonomic Affiliation of JZ38
Isolate JZ38 was first classified as Cronobacter sp. based on 16S rRNA gene sequence (Eida et al., 2018). Identification strip kit for Enterobacteriaceae (API 20E) phenotypically verified isolate JZ38 as a member of the genus Cronobacter (Supplementary Table S7). Cronobacter (C.) species, formerly known as Enterobacter sakazakii, are a group of gram-negative, rod-shaped bacteria that belong to the family Enterobacteriaceae (Iversen et al., 2007). The genus Cronobacter consists of seven species: C. condiment, C. dublinensis, C. malonaticus, C. muytjensii, C. sakazakii, C. turicensis, and C. universalis (Iversen et al., 2007; Iversen et al., 2008; Joseph et al., 2012).
Cronobacter sakazakii is an opportunistic foodborne pathogen that is classified as a public health issue due to neonatal meningitis outbreaks in powdered infant formula (Himelright et al., 2002; FAO/WHO, 2004; Drudy et al., 2006). However, there are indications that the virulence of C. sakazakii is due to the presence of several components found on plasmids such as pESA3 and pCTU1 (Franco et al., 2011a, b). These components include genes encoding iron acquisition systems (eitCBAD, iucABCD/iutA), type 6 secretion systems (T6SS), filamentous hemagglutinin (fhaB) and its transporter (fhaC), a RepFIB-like origin of replication (repA) and chromosome/plasmid partitioning proteins (parAB) or outer membrane serine proteases (cpa, plasminogen activator). A BLAST search of the sequences of the two plasmids against JZ38 revealed alignment of pESA3 (NC_009780.1) and pCTU1 (NC_013283.1) with plasmid p2 at 68 and 63% coverage, respectively, and 87% identity. Analysis of JZ38 genome revealed the presence of iucABCD, as part of the T6SS, two copies of fhaB and one copy of fhaC, parAB, and repA in plasmid p2, in addition to a complete T6SS and one copy of fhaB in the chromosome (Supplementary Table S6). However, the eitCBAD operon and the cpa gene were absent.
The fatty acid profiles can be used to discriminate between bacterial species (von Wintzingerode et al., 1997; Ehrhardt et al., 2010). A fatty acid profile was therefore obtained from JZ38 and the type strain of C. sakazakii LMG 5740T. The C12:0, C14:0, C16:0, C17:0cyclo, C18:1 ω7c, together with summed features 2 and 3 (Table 2), were the most abundant and present FAME in the two strains. C17:0 was also present in both strains, but at a lower percentage. Isolate JZ38 exhibited a similar FAME profile to C. sakazakii, indicating that they belong to the same genus. The presence of C18:0 in JZ38 differentiates it from C. sakazakii, indicating that the two strains belong to different species.
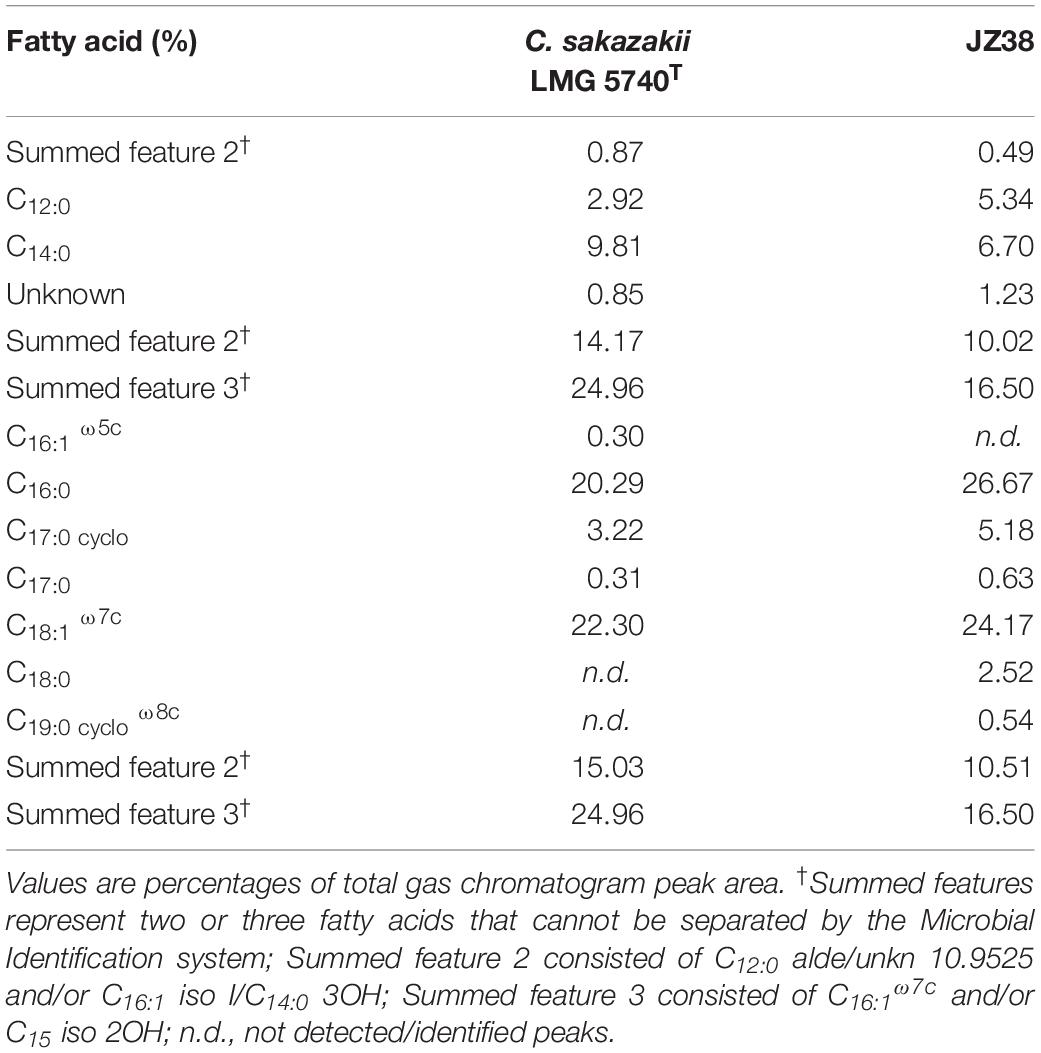
Table 2. Comparison of cellular fatty acid compositions of JZ38 and Cronobacter sakazakii LMG 5740T.
A taxonomic classification of JZ38 based on the complete 16S rRNA gene sequence (extracted from the JZ38 genome) revealed the phylogenetic affiliation of JZ38 to C. muytjensii (Figure 3A). To obtain a more accurate taxonomic classification and confirm the affiliation of JZ38, a whole genome based phylogenetic analysis was performed using strains related to the genus Cronobacter (seven different species) (Figure 3B and Supplementary Table S1). The analysis confirmed the affiliation of JZ38 with other C. muytjensii species and separated them from other species of this genus, such as the opportunistic food-borne pathogen C. sakazakii. Finally, C. muytjensii ATCC 51329 showed the highest similarity to JZ38 with an ANIb value of 98.74% and a dDDH value of 93.3% (formula d6), which are well above the species cutoff threshold of 95 and 70%, respectively, further confirming the affiliation of JZ38 to this species.
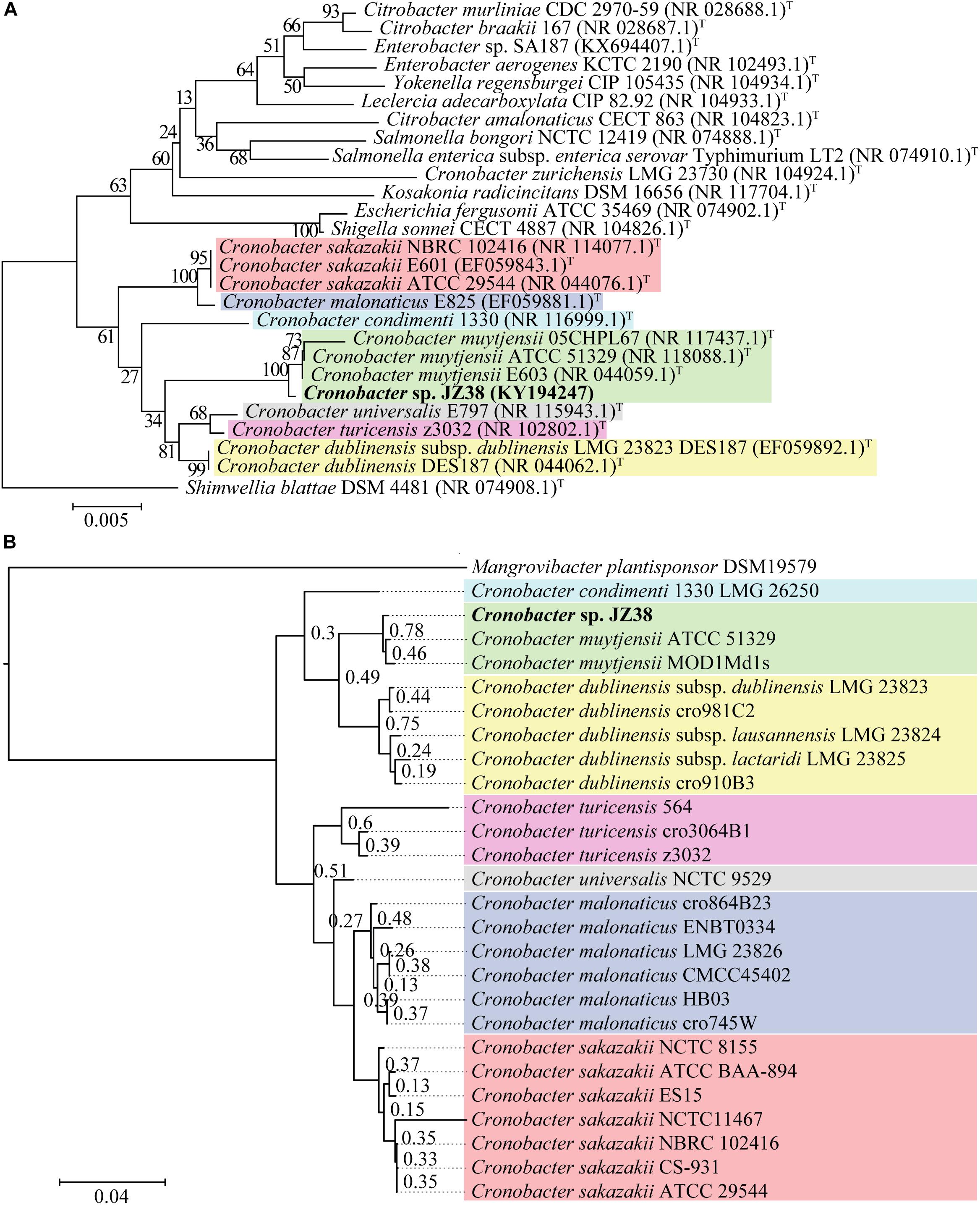
Figure 3. Taxonomic classification of JZ38. (A) 16S rRNA-based phylogenetic tree, evolutionary relationships inferred using the Neighbor-Joining method and the evolutionary distances computed using the Kimura 2-parameter method. GenBank accession numbers of isolates are presented in parentheses and type strains are indicated by a T after the parentheses. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) are shown next to the branches; (B) Genome-based phylogenetic tree, genomes of different species of Cronobacter genus were aligned using DendroBlast and FastMe and species tree was built using STAG and rooted based on duplications using STRIDE, and support values were calculated based on supporting consensus from the gene trees.
Function Analysis of Cronobacter muytjensii Strain JZ38
In order to understand the metabolic capacity and plant growth promoting potential of JZ38, a functional analysis (BlastKOALA) was performed and identified 2,678 genes (67.6% of all CDSs) with assigned functions on the chromosome Chr1, while 19 (13.8%) and 53 (43.1%) genes were assigned to plasmids p1 and p2, respectively. The JZ38 genome contains a large number of metabolic genes (such as carbohydrate and amino acid metabolism), followed by environmental information processing (such as membrane transport and signal transduction) (Table 3). AntiSMASH analysis revealed the presence of 19 clusters of genes for the biosynthesis of secondary metabolites, of which only one was present on plasmid p2 while the remaining 18 were on Chr1 (Supplementary Table S4). Most interesting are the clusters with high similarity to the biosynthesis of carotenoids, colonic acid, lipopolysaccharides and aerobactin-like and enterobactin-like (turnerbactin) siderophores. Many of these compounds may be involved in establishing symbiotic associations with plants (Danese et al., 2000; Becker et al., 2005; Bible et al., 2016). In addition to secondary metabolites, JZ38 contains genes involved in oxidative and osmotic stress tolerance, colonization, chemotaxis and flagellar assembly, secretion systems, nutrient solubilization and transport, volatile compounds and phytohormone production (Figure 2).
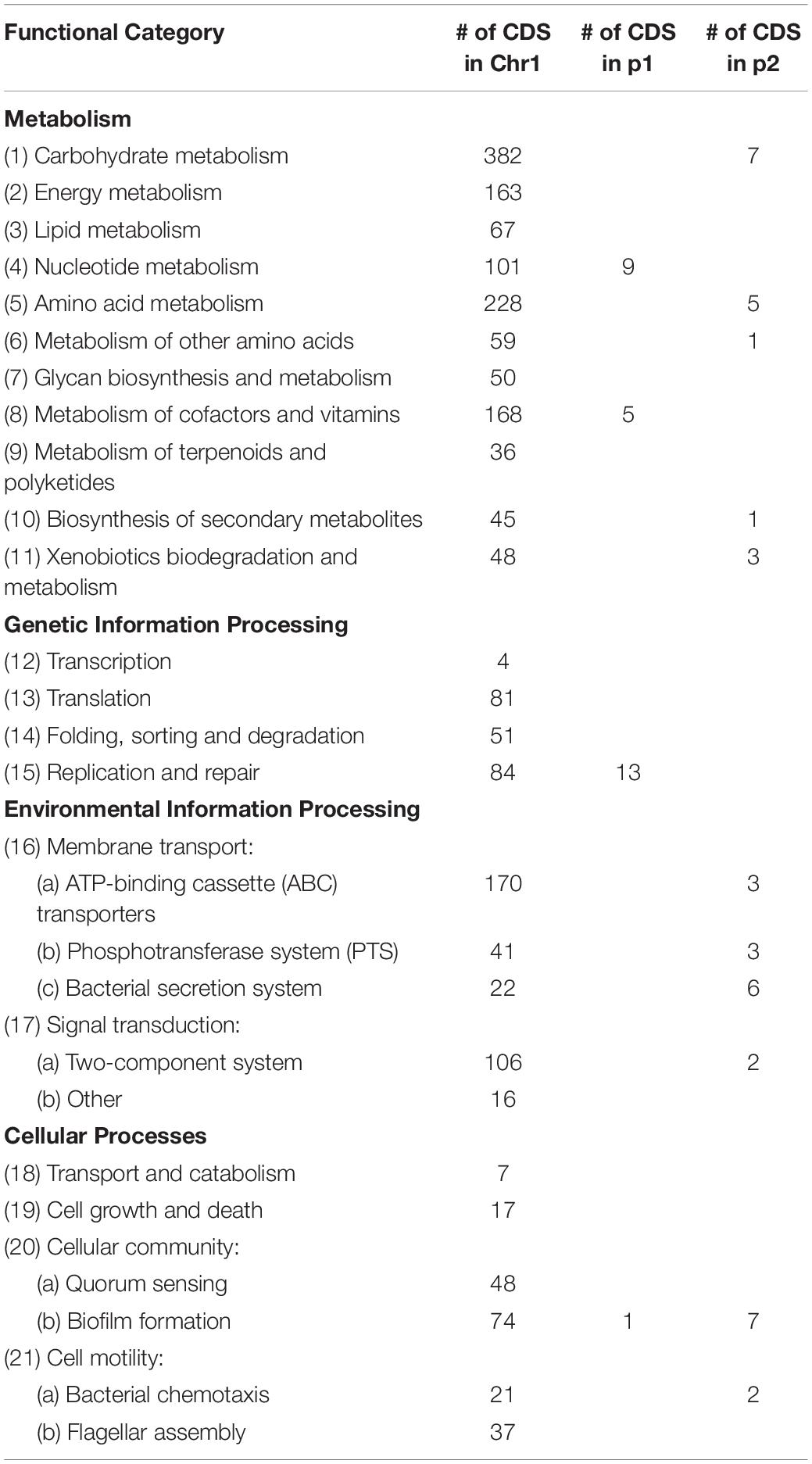
Table 3. Summary of major functional KEGG pathways annotation of predicated genes in JZ38 using BlastKOALA.
Identification of phages using the PHAST web tool revealed the presence of two intact phages (Enterobacteria phage mEp235 and Cronobacter phage phiES15) (Lee et al., 2012). Moreover, one incomplete and one possible region on Chr1 also detected phages, while plasmid p1 contained a phage that is highly similar to Salmonella phage SSU5 (Kim et al., 2012) (Supplementary Table S5). The SSU5 phage has been suggested as a possible auxiliary component of phage cocktails for biocontrol of Salmonella (Kim et al., 2014), which could be the case for JZ38. Bacteria have evolved Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) and CRISPR-associated (Cas) proteins (CRISPR-Cas systems) for providing adaptive immunity and acquired resistance against bacteriophages and plasmids (Darmon and Leach, 2014). Isolate JZ38 contains two operons encoding CRISPR-Cas systems, namely Type I (two copies of cas1, cas2, three copies of cas3), subtype I-F (csy1234) and “Cascade” complex systems (casABCDE) (Supplementary Table S6). The presence of the CRISPR system suggests its potential in protecting JZ38 from lysogenization and phage induction.
Survival Under Abiotic Stress Conditions
Isolate JZ38 previously exhibited the ability to survive under osmotic (20% polyethylene glycol/PEG) and salinity stress (855 mM NaCl) (Eida et al., 2018). Further experiments demonstrated JZ38’s ability to grow in salt concentrations of up to 1 M NaCl (Supplementary Figure S2A), in pH varying from 4 to 9 (Supplementary Figure S2B) and in the presence of heavy metals (100 mg/L of Mn, Cd, Cu, Co, and Ni ions) (Supplementary Figure S2C). Genome mining of JZ38 revealed the presence of genes related to the production of osmoprotectants, osmoregulation and abiotic stress tolerance such as heat, acidity and oxidative stress (Supplementary Table S8).
In bacteria, one of the early responses to osmotic stress involves the uptake and accumulation of K+, which along with its glutamate counter-ion, may be involved in signal transduction for secondary responses (Booth and Higgins, 1990). JZ38 contains genes encoding K+-uptake transporters, such as the Kdp-ATPase system (kdpABCDE), Trk system (trkAGE) and Kup (kup), a K+-efflux system (kefBCFG) and a K+-channel (kch) (Sleator and Hill, 2002) (Supplementary Table S8). As a secondary response to high salt concentrations, bacteria accumulate high levels of osmoprotectants (or compatible solutes), such as sugars (e.g., sucrose, trehalose), amino acids (e.g., glycine betaine, proline) or polyols (e.g., glycerol, inositol), which enable bacteria to adapt to changes in osmolarity of the external environment induced by drought or salt stress (Sleator and Hill, 2002; Reina-Bueno et al., 2012; Paul, 2013). Trehalose is synthesized via five different pathways: OtsA-OtsB, TreY-TreZ, TreS, TreT and TreP pathway (Strom and Kaasen, 1993; Paul et al., 2008). In JZ38, two copies of otsA and a copy of otsB for intracellular trehalose synthesis can be found on Chr1, while genes responsible for conversion of starch to maltodextrin (treX), maltodextrin to maltooligosyl-trehalose (treY) and, then, to trehalose (treZ) are present on plasmid p2 (Supplementary Table S8). For trehalose homeostasis, JZ38 contains genes for trehalose catabolism, by periplasmic trehalase (two copies of treA) and trehalose-6-phosphate hydrolase (treC) to glucose, trehalose uptake transporters (thuEFGK, treB) and a transcriptional regulator (treR) (Supplementary Table S8) (Strom and Kaasen, 1993).
Glycine betaine is synthesized from choline via a two-step enzymatic reaction by choline dehydrogenase (BetA) and betaine aldehyde dehydrogenase (BetB), while proline is synthesized from glutamate by the action of three enzymes (ProABC) (Mandon et al., 2003). Production of glycine betaine requires the uptake of choline by BetT, ProU and ProP transporters and ABC transporter permease OpuABC (Kappes et al., 1999; Biemans-Oldehinkel and Poolman, 2003; Ly et al., 2004). In addition to choline, ProP and ProU also plays a role in the uptake of proline and in osmotic stress tolerance (10% NaCl) (Wood, 1988). The ProU complex consists of a cytoplasmic ATPase (ProV), a transmembrane subunit (ProW) and a periplasmic binding protein (ProX) (Gul and Poolman, 2013). Furthermore, PutP and ProY have also been suggested to transport proline (Wood, 1988; Liao et al., 1997). Genome mining revealed the presence of all aforementioned genes, in addition to part of the ABC transporter complex GltIJKL involved in glutamate and aspartate uptake (gltJKL), a transcriptional repressor of bet genes (betI) and extra copies of the opuABC system and proP (Supplementary Table S8).
Genes encoding enzymes with proteolytic/hydrolytic activity against oxidative stress such as two superoxide dismutases (SOD1 and SOD2), hydrogen peroxide catalases (katE and katG), alkyl hydroperoxide reductases (ahpCF) and thiol peroxidases (tpx) were present (Supplementary Table S8) (Zhao and Drlica, 2014). In addition, genes for the detoxification of free radical nitric oxides by a flavohemoprotein nitric oxide dioxygenase (hmp), anaerobic nitrate reduction (norRVW), nitric oxide sensor (nsrR) were found (Zeidler et al., 2004). There are also five copies of glutathione S-transferases (GST), four glutathione peroxidases (gpx), a glutathione ABC transporter (gsiABCD), a gamma-glutamate-cysteine ligase (gshA), a glutathione synthetase (gshB), a glutathione reductase (gor) and hydrolase (ggt), glutaredoxins (grxABCD) and peroxiredoxins (BCP, ahpCF) which function in detoxification systems (e.g., reactive oxygen species/ROS detoxification) (Fones and Preston, 2012; Zhao and Drlica, 2014). Key regulators for controlling oxidative stresses include the stress response sigma factor (rpoS), hydrogen peroxide sensor (oxyR), regulators of the superoxide radical response (soxRS) and ferric uptake regulator (fur) (Supplementary Table S8) (Chiang and Schellhorn, 2012; Zhao and Drlica, 2014). There are also a number of universal stress proteins (uspABCEFG), some of which confer resistance to oxidative stress while others are important for motility (Nachin et al., 2005).
Polyamines, such as putrescine, cadaverine, spermidine and spermine, are also involved during oxidative, osmotic, heat and acid stress (Rhee et al., 2007; Miller-Fleming et al., 2015). JZ38 contains genes encoding polyamine transporters (potABCD, potFGHI, potE, puuP), for synthesis of putrescine from L-arginine (speAB) and L-orthinine (two copies of speC), its conversion to spermidine (speE) and N-acetyl spermidine (speG), synthesis of spermidine from S-adenosyl-L-methionine (speD), and synthesis of cadaverine from L-lysine (cadA) (Furuchi et al., 1991; Pistocchi et al., 1993; Kashiwagi et al., 1997; Kurihara et al., 2005) (Supplementary Table S8). In the case of pH, a number of genes found in JZ38 were previously shown to be involved in acid resistance, including antiporters (nhaA, nhaB, nhaP2, chaA, yrbG), molecular chaperones (groEL, dnaK), starvation inducible proteins (two copies of phoH, dps), ATPases (atpABCDEFGHI), sigma factors and regulators (rpoS, ariR, nhaR) transcriptional repressor LexA (lexA) and proteases (clpXP) (Kuhnert et al., 2004; Guazzaroni et al., 2013; Liu et al., 2016) (Supplementary Table S8). JZ38 also possesses genes encoding for resistance to heavy metals such as copper (copA, copC, copD, cueO, cueR, bhsA) and zinc/cadmium/cobalt (zntA, zntR), arsenate (arsC1) and chromate (chrR) (Supplementary Table S9). However, heavy metals are also important co-factors in bacteria, and JZ38 possesses several genes encoding transporters for zinc (znuABC, zntB, zur), nickel (nixA, hypA, hypB), manganese (mntH, mntR) and other metals (chaA, czcD). Indeed, JZ38 growth was completely unaffected by addition of 100 mg/L Cu2+ and Mn2+ ions.
The presence of genes for resistance to osmotic, oxidative, acidity and heavy metal stress suggests that JZ38 possesses the potential to grow and tolerate these stresses confirming the phenotypic assays (Supplementary Figure S2).
JZ38-Plant Interaction
An endophytic lifestyle provides access to all essential nutrients for bacterial proliferation. Consequently, the genome of JZ38 contains a multitude of genes involved in the uptake, transport and metabolism of nitrogen, sulfur and carbon-based compounds. The list of genes encoding ABC transporters, Major Facilitator Superfamily (MFS) transporters and Phosphotransferase systems (PTS) is shown in Supplementary Table S10. Genes for the transport and metabolism of carbohydrates, nitrogen and sulfur are highlighted in Supplementary Tables S11–S13, respectively. For example, JZ38 contains genes encoding several sugar transporters: arabinose (araFGH, araE), arabinogalactan (ganQP, cycB), cellobiose (PTS-Cel-EIIABC), fructose (PTS-Fru-EIIA, PTS-Fru-EIIB), galacticol (gatABC), galactose (galP), glucose (PTS-Glc-EIIABC), glycerol (glpF), glycosides (ascF/PTS-Asc-EIIB, bglF/PTS-Bgl-EIIA), inositol (iolT), lactose (three copies of lacY), maltose/maltodextrin (malGFEK, two extra copies of malK, malB and PTS-MalGlc-EIIBC, thuEFGK), mannitol (PTS-Mtl-EIIA), melibiose and raffinose (rafB, scrA/PTS-Scr-EIIBC, scrY), rhamnose (rhaSTPQ), ribose (rbsACB, rbsD), sucrose (scrY, two copies of scrA/PTS-Scr-EIIBC), trehalose (thuEFGK, treB) and xylose (xylFGH).
Phenotypic characterization of sugar metabolism using API CH 50 strips demonstrated JZ38’s ability to metabolize, by fermentation or oxidation, a range of carbon sources for growth (Supplementary Table S7). These sugars included monosaccharides (L-enantiomers of arabinose and rhamnose and D-enantiomers of ribose, xylose, galactose, glucose, fructose and mannose), disaccharides (D-enantiomers of cellobiose, maltose, lactose, melibiose, saccharose/sucrose and trehalose and gentiobiose), glycosides (amygdalin, arbutin, esculin and salicin), polysaccharides (D-raffinose), polyols (glycerol, galacticol, inositol, and D-mannitol), and salts (potassium gluconate). Isolate JZ38 has the genomic potential for the metabolism of arabinose (araBAD) (Fritz et al., 2014), arabinogalactan (bgaB/ganA, ganB) (Watzlawick et al., 2016), cellulose and cellobiose (bcsZ, two copies of bglX, bglB, yliI) (Xie et al., 2007), citric acid (citABCDEFXG, citA in plasmid p2) (Martín et al., 2004), galactose (lacA, galK, galT) (Chai et al., 2012), galacticol (gatD, gatYZ, gatR) (Nolle et al., 2017), glucose (pgm, yihX, glk, pgi), glycerol (glpK, glpD) (Holmberg et al., 1990; Yang et al., 2014), glycosides (bglB, bglA/ascB, ascG) (Desai et al., 2010; Zangoui et al., 2015), inositol (iolBCDEGHI) (Morinaga et al., 2010), lactose (lacA, lacZ) (Zeng et al., 2010), maltose/matlodextrin (malP, malQ, malS, malZ, glvA) (Boos and Shuman, 1998), mannitol (mtlD) (Wisselink et al., 2004), melibiose and raffinose (rafA, rafR, scrB) (Russell et al., 1992; Hugouvieux-Cotte-Pattat and Charaoui-Boukerzaza, 2009), rhamnose (rhaBAD) (Schwartz et al., 1974; Rodionova et al., 2013), ribose (rbsK, rbsD, rbsR) (Mauzy and Hermodson, 1992; Sigrell et al., 1998), sucrose (scrB, two copies of scrK, scrR) (Hugouvieux-Cotte-Pattat and Charaoui-Boukerzaza, 2009), trehalose (two copies of treA, treC, treR) (Baker et al., 2018), and xylose (xylAB) (Shamanna and Sanderson, 1979).
Nitrogen and sulfur are essential elements in nutrients for both bacteria and plants. The genome of JZ38 lacks genes for nitrogen fixation, but contains genes encoding for ammonium (amt, glnK), urea (urtABCDE), nitrate and nitrite (nrtABC, narK) transport, nitrate (narLXKGHJI, narP) and nitrite (nirBD) reduction and nitrate assimilation (nasA, nasB annotated as nirB but in the same operon as nasA) (Supplementary Table S12). The genome also contains genes for sulfate (cysPUWA, one extra copy of cysA), taurine (tauACB) and alkanesulfonate (ssuACB) transport, metabolism of taurine (tauD), alkanesulfonate (ssuDE) and thiosulfate (glpE), and finally assimilatory sulfate reduction (cysND, cysC, cysH, cysJI) (Supplementary Table S13).
The ability of JZ38 to metabolize a diverse range of carbon sources and the presence of genes responsible for their metabolism, along with the transport and metabolism of other micronutrients confirm JZ38’s potential for a lifestyle as a plant endophyte.
A large number of genes encoding two-component systems (TCS) for rapid sensing and adjustment to changes in the external environment were present in JZ38 (Supplementary Table S14). The TCSs in JZ38 belong to the OmpR, NtrC, NarL, LytTR, CitB and chemotaxis families. The TCSs present are important for envelope stress (cpxAB, baeSR) (Batchelor et al., 2005; Leblanc et al., 2011), cell surface adhesion, biofilm formation, motility and hemolysis (cpxAB, rstAB, tctDE) (Otto and Silhavy, 2002; Huang et al., 2018; Taylor et al., 2019) and capsular polysaccharide synthesis (rcsABCD) (Cheng et al., 2010; Paczosa and Mecsas, 2016). Some were shown to be involved in carbon catabolism (creBC) (Godoy et al., 2016) and nitrogen metabolism (glnGL, glnKR, narXL) (Pahel et al., 1982; Nohno et al., 1989; Schreier et al., 1989). Others were involved in responses to antimicrobial peptides and acid pH (phoPQ) (Fields et al., 1989; Foster and Hall, 1990), osmotic stress (kdpDE), heavy metals (basRS) (Ogasawara et al., 2012) and phosphate starvation and production of acid phosphatases (phoBR, phoPQ) (Kier et al., 1979; Wanner, 1996).
Production and resistance to antibiotics and antimicrobial compounds play a role in suppressing soil-borne plant pathogens or enhancing the persistence of the producers in a highly competitive environment (Raaijmakers et al., 2002; Raaijmakers and Mazzola, 2012). JZ38 was also tested for its sensitivity to a range of antibiotics and possessed a number of genes involved in antibiotic resistance, antimicrobial compounds and defense mechanisms. Among the tested antibiotics, JZ38 displayed resistance to ampicillin, linezolid and penicillin G (Supplementary Table S15). Genome mining revealed the presence of genes involved in β-lactam, cationic antimicrobial peptide (CAMP), vancomycin and antifolate resistance (Supplementary Table S16). Most notably, the presence of a β-lactamase (ampC), a cytosolic amidase (ampD), an inner membrane permease (ampG), regulators of AmpC (ampH, nagZ), penicillin binding proteins involved in peptidoglycan biosynthesis (mrcAB, pbpC, mrdA, ftsI, dacB, three copies of dacC, pbpG) and multidrug efflux system (acrAB, tolC) may confer resistance to ampicillin and penicillin (Sauvage et al., 2008; Guérin et al., 2015) (Supplementary Table S16). JZ38 could also possess fungal antagonism abilities due the presence of a putative chitinase (Chernin et al., 1995).
Bacteria have evolved Toxin-Antitoxin (TA) systems composed of a toxin and its neutralizing antitoxin (Yamaguchi et al., 2011). The toxins are believed to slow down (dormancy) or suppress growth (cell death) in order to survive in rapidly changing environments or stress conditions (Gerdes et al., 1986; Yamaguchi and Inouye, 2009). TA systems have other diverse functions and roles, including ensuring persistence of plasmids during replication, virulence, antibiotic tolerance, phage defense and biofilm formation (Hayes, 2003; Sass et al., 2014; Shidore and Triplett, 2017). A number of genes related to different classes of TA systems were found in JZ38 (Supplementary Table S16), including an adenylate cyclase toxin (cyaA) (Cannella et al., 2017), SOS-induced toxins (tisB, symE) and their regulator (lexA) (Gerdes and Wagner, 2007), membrane stress phage shock proteins (pspABCD) (Engl et al., 2010), toxins involved in cell division (cptAB, fstZ, merB) (Masuda et al., 2012) cell death (phd, doc, clpXP) (Smith and Magnuson, 2004), promoters of persister cells (hha-tomB, hipAB in plasmid p2).
The presence of TCSs and the production of, or resistance to, antibiotics, antimicrobials and TA systems may enable JZ38 to sense their environment, compete with other microbes in the soil and rhizosphere and to persist in the microbial community under biotic and abiotic stress conditions.
Chemotaxis, Motility and Colonization
Bacterial colonization of plants begins with recognition of signals from root exudates (e.g., via TCSs) and chemotactic responses toward these signals (de Weert et al., 2002; Lugtenberg et al., 2002). Bacteria can then either flow through soil water fluxes toward the roots, or they can actively induce flagellar activity enabling internalization and colonization of different parts of the plant (van der Lelie et al., 2009). The ability to move toward plant roots, adhere and colonize the surface and systematically spread within plant tissues is an important characteristic of endophytic bacteria (Hardoim et al., 2008). Motility assays revealed the ability of JZ38 to spread on 0.3% agar (swimming) but not on 0.6% (swarming) (Supplementary Figure S3A). Transmission electron photomicrograph of JZ38 also revealed the presence of peritrichous flagella (Supplementary Figure S3B).
Indeed, genome analysis revealed the presence of genes for flagellar biosynthesis, assembly and chemotaxis (Supplementary Table S17). Genes present in the genome of JZ38 which encode for structural components of the flagellar body, important regulatory factors and determinants of chemotaxis include several operons; flgNMABCDEFGHIJKL, flhDC motAB cheAW fimCD tar cheRBYZ flhBAE, fliYZA and fliCDEFGHIJKLMNOPQRST (Supplementary Table S17) (Fitzgerald et al., 2014). In addition to extra copies of fliC and fimD, some genes encoding for parts of the type IV pilus (hofBC, hofMNOQ) system were also present. Furthermore, genes involved in chemotaxis, such as methyl-accepting and TCS chemotaxis proteins (four copies of tsr and tar, trg, aer, nine copies of mcp, and cheRBYZV), were also present on both chromosome Chr1 and plasmid p2 (Supplementary Table S17). Along with the presence of TCSs (Supplementary Table S14), which assist in signal recognition of exudates and adaptation to the environment within the plants, these results indicate that JZ38 possesses the potential to respond to nutrients as signals and, consequently, move toward and within plant roots.
Prior to internalization and systematic invasion of plants, bacteria may attach and adhere to the surface of roots and form microcolonies and biofilms. Bacteria can form aggregates in a self-produced matrix composed of water and exopolysaccharides (EPS), such as cellulose, called biofilms (Augimeri et al., 2015), which can assist in plant colonization (Laus et al., 2005; Yaron and Römling, 2014). Some surface components of bacterial cells, such as flagella, curli fibers, type I fimbriae are involved in the formation of biofilms (Beloin et al., 2008). Bacterial biofilms can be regulated by a mechanism by which small signaling molecules called autoinducers are used for cellular communication, called quorum sensing (QS), allowing bacteria to regulate gene expression in a cell-density-dependent manner (Fazli et al., 2014; Pérez-Montaño et al., 2014). The genome of JZ38 contains genes for colonization, root surface adhesion, biofilm formation, quorum sensing, and degradation of cell walls for internalization (Supplementary Table S18).
The presence of two recombinases/integrases (xerC, xerD) suggests the ability of JZ38 to colonize the rhizosphere and root surfaces (Martínez-Granero et al., 2005). The genome encodes genes involved in mediating surface adhesion by pili formation (ppdD, hofBC, ppdABC, hofMNOQ) and in lipopolysaccharides, cellulose and colanic acid biosynthesis (Dörr et al., 1998; Sauvonnet et al., 2000) (Supplementary Table S18). Cellulose and colanic acid are EPSs critical for the formation of biofilms (Danese et al., 2000). JZ38 contains the Bcs operon for the biosynthesis of cellulose (bcsABZC, one extra bcsA) (Ahmad et al., 2016). JZ38 also contains a complete gene cluster for the biosynthesis and translocation of colanic acid (wza wzb wzc wcaABCDEF gmd fcl wcaHI manCD wcaJ wzxC wcaKLM) and a transcriptional regulator (mcbR) (Supplementary Tables S4, S18) (Schmid et al., 2015). JZ38 contains enzymes (tqsA, luxS) that catalyze the synthesis of the signal precursor for autoinducer-2 mediated quorum sensing which are found in a number of Enterobacteriaceae (Rezzonico et al., 2012).
The internalization of bacteria into the plant host occurs through sites of tissue damage during growth, root hairs or intact epidermis cells requiring the active secretion of cell-wall degrading enzymes (Sprent and de Faria, 1988; Morgante et al., 2005). Cellulose, hemicelluloses and pectin are the major components of the primary cell wall. The genome of JZ38 does not contain known genes encoding cellulases or hemicellulases. However, it does contain genes for the catabolism of the hexuronate D-galacturonate, the main monomer of pectin, via the isomerase pathway (uxaABC, two copies of kdgK, kdgA) (Kuivanen et al., 2019) (Supplementary Table S18). The genome also encodes genes involved in transport (two copies of exuT, togMNAB, kdgT, kdgM), transcriptional regulation (two copies of kdgR, exuR) and catabolism of other hexuronates (uxuAB, kduID, two copies of ogl).
Carotenoids are organic pigments produced by plants, algae and several bacteria and fungi, which might contribute to the yellow color of JZ38. The production of these pigments has been reported to be important for root colonization and fitness under abiotic stresses (Johler et al., 2010; Mohammadi et al., 2012; Bible et al., 2016). Furthermore, the carotenoid zeaxanthin is a precursor of the phyothormone abscisic acid (ABA), which is involved in abiotic stress tolerance (e.g., drought, heat, salinity and UV) and, thus, carotenoids may play a role in the growth promotion of JZ38 (Vishwakarma et al., 2017). The carotenoid biosynthesis gene cluster in Enterobacteriaceae includes six enzymes: geranylgeranyl pyrophosphate synthase (CrtE), phytoene synthase (CrtB), phytoene desaturase (CrtI), lycopene β-cyclase (CrtY), 3,(3′)-β-ionone hydroxylase (CrtZ) and zeaxanthin glucosyl transferase (CrtX), which produce zeaxanthin diglucoside from farnesyl pyrophosphate (Sedkova et al., 2005). Farnesyl pyrophosphate is produced via the isoprenoid pathway catalyzed by a number of enzymes encoded by dxs, dxr, ispDEFGH, idi (isopentenyl pyrophosphate isomerase) and ispA (farnesyl pyrophosphate synthase). JZ38 contains the carotenoid gene cluster (crtE-idi-crtXYIBZ), as well as the genes in the isoprenoid pathway (dxs, dxr, ispDEFH, ispB, ispA, idi) (Supplementary Table S18).
Secretion Systems and Effector Proteins
Bacteria possess protein secretion systems that play important roles in biotic interactions, such as pathogenicity and colonization (Iniguez et al., 2005; Tseng et al., 2009). Compounds secreted include antimicrobial compounds, enzymes, ions, peptides, secondary metabolites, toxins, or effectors to the surrounding environment or into host cells, triggering defense responses, altering host cell’s physiology or structure or combating stresses (Green and Mecsas, 2016). The universal two-step Sec (general secretory pathway) and Tat (Twin arginine translocation) systems are ubiquitous systems responsible for the export of proteins across the plasma membrane into the periplasmic space, which can then be exported by other secretion systems (Natale et al., 2008). The T6SS is a versatile secretion system, found in many gram-negative bacteria, responsible for the export of multiple effector proteins, ions and toxins to the extracellular milieu or into the eukaryotic or prokaryotic target cells (Hood et al., 2010; Zoued et al., 2014; Wang et al., 2015). Genome analysis revealed the presence of genes encoding Sec, Tat and Type VI secretion systems (T6SS) on chromosome Chr1 (Supplementary Table S19). The presence of these secretion systems (including additional genes encoding some parts of the T6SS) may be associated with JZ38’s ability to participate in various physiological processes, including stress responses and colonization in planta (Weber et al., 2009; Jani and Cotter, 2010; Shidore et al., 2012; Si et al., 2017).
Plant Growth Promoting Traits and Potential for Growth Promotion: Nutrient Acquisition
JZ38 exhibited a number of plant growth promoting traits involved in nutrient acquisition and modulating plant hormone levels that could possibly be responsible for the growth promotion. Root exudates released by plants include organic acids, which can acidify the soil and result in the solubilization of Zn or mineral P, and siderophores, which increase Fe availability through chelation (Zhang et al., 1991; Ström, 1997; Dakora and Phillips, 2002). However, bacteria also possess the ability to produce organic acids and/or siderophores and, therefore, can assist in the promotion of plant growth in field applications with crops (Sahu et al., 2018). JZ38 displayed iron (Fe) sequestration or acquisition by production of siderophores and phosphate and zinc solubilization abilities (Supplementary Figure S3C).
Phosphate (P) is an essential macronutrient for growth of all living organisms as it is a key component of nucleic acids, phospholipids and ATP. It is a major limiting factor for plant growth due to inaccessible/insoluble form in soil or rhizosphere (Holford, 1997; Eida et al., 2017). Production of organic acids by P-solubilizing microbes is well documented, and the most common form responsible for solubilization of mineral P is gluconic acid (GA) (Rodriguez and Fraga, 1999). GA is synthesized via the direct oxidation of glucose by glucose-1-dehydrogenase (gcd) and its co-factor pyrrolo-quinolone quinine (PQQ) (Liu et al., 1992; de Werra et al., 2009). Genome analysis of JZ38 revealed the presence of gcd and pqqBCDEF for gluconic acid production, but a lack of ppqA. JZ38 also contains P transporters such as the high-affinity pstSCAB transporter system and low-affinity P transporter (pit), genes involved in phosphonoacetate degradation (phnA) and polyP formation (ppk) and genes encoding phosphatases (ppa, two copies of gpx-gppA) and TCSs (phoBR, phoU, two copies of phoH) (Supplementary Table S20) (Ohtake et al., 1996; Yuan et al., 2006; Agarwal et al., 2011).
Iron (Fe) is a micronutrient that plays an important role in DNA synthesis, respiration and photosynthesis. Fe is abundant in most soils but, similar to P, inaccessible to plants due to formation of insoluble forms at neutral pH (Mori, 1999). Siderophores are Fe-specific chelating compounds that can increase Fe availability. Siderophores can be classified into several groups; hydroxymates (e.g., aerobactin, ferrichrome, ferrioxamine B), catecholates (e.g., enterobactin, bacillibactin), carboxylates (e.g., petrobactin, rhizobactin) or phenolates (e.g., pyochelin) (Boukhalfa and Crumbliss, 2002; Raymond et al., 2015). Genome analysis revealed the presence of genes involved in the biosynthesis of enterobactin (entABCDEF) and aerobactin (iucABCD located in plasmid p2) (Supplementary Tables S4, S20). There are also genes responsible for enterobactin secretion (entS), Fe-enterobactin and other Fe-siderophore uptake complexes (exbBD, tonB, two copies of fepA, fhuEF), enterobactin extraction (three copies of fes) and siderophore regulator (ahpC). In the rhizosphere, siderophore production by certain bacteria may deprive Fe from potential plant-pathogenic microbes and, thus, can contribute to triggering plant immunity and protection (Bakker et al., 2013; Dellagi and Aznar, 2015). The presence of efficient Fe-uptake systems can help bacteria compete for Fe in such environments, and JZ38 possesses genes encoding two ferrous iron uptake systems (feoABC and efeUOB) and a number of ABC transporters and receptor proteins and MFS transporter (Supplementary Table S21). This suggest that JZ38 is not only able to solubilize Fe but can also import and export it to the host plant.
Zinc (Zn) is another micronutrient which is important for metabolic enzyme systems and optimal plant growth and development (Brown et al., 1993). Zn deficiency is well reported in soils around the world and is a commonly occurring problem in crop plants, leading to decreases in crop yield and nutritional quality (White and Zasoski, 1999; Mumtaz et al., 2017). Similar to P and Fe, Zn is also inaccessible to plants due to its low solubility where it is found in different forms in soil (e.g., Smithsonite/ZnCO3) and its solubility is highly dependent on pH (higher solubility at low pH) (Martínez and Motto, 2000; Vodyanitskii, 2010). In comparison to macronutrients P (720 mg/kg) and Fe (∼1665 mg/kg), the amount of the micronutrient Zn (∼32 mg/kg) in Jizan desert soil was much lower (Eida et al., 2018). Previously, JZ38 was identified to possess ZnO and ZnCO3-solubilizing abilities on agar plate assays (Eida et al., 2018). Therefore, Zn solubilization by JZ38 may have been an important trait for plant growth promotion of the T. terrestris from which it was isolated.
Secretion of organic acids, such as 5-ketogluconic acid and pentanoic acid, by bacteria is associated with acidification of soil or medium and subsequent solubilization of ZnCO3 (Saravanan et al., 2007). Indeed, Zn-solubilizing bacteria belonging to different genera of the Enterobacteriaceae family have been shown to promote the growth and Zn content of cotton (Kamran et al., 2017). The genes for the production of gluconic acid from glucose (gcd) and subsequent conversion to 5-ketogluconic acid (ghrB) were present in JZ38 (Supplementary Table S20) (De Muynck et al., 2007). In addition, a high-affinity ABC transporter for Zn import (znuABC), its regulator (zur) and efflux systems for constitutive (zitB) and regulated (zntAB, zntR) Zn export are also present (Supplementary Table S9) (Wang D. et al., 2012). These results suggest bacteria are able to solubilize Zn, import it into their cells and/or export it to the plant host.
Plant Growth Promoting Traits and Potential for Growth Promotion: Phytohormone and Volatile Compound Production
Plant growth promotion can be achieved by either direct interactions between the beneficial bacteria and their host and/or antagonistic activity against plant pathogenic microbes. Plant growth and abiotic stress tolerance promotion by bacteria is well established (Dimkpa et al., 2009; Glick, 2012), but the precise mechanisms are still not fully understood. However, there are numerous indications that bacterial phytohormone production, such as IAA, can play important roles (Patten and Glick, 2002; Enebe and Babalola, 2018). Production of IAA occurs through five different pathways using tryptophan (Trp) as a precursor: indole-3-acetonitrile (IAN), indole-3-acetamide (IAM), tryptophan side-chain oxidase (TSO), tryptamine (TAM) and indole-3-pyruvate (IPyA) pathways (Patten and Glick, 1996; Carreño-Lopez et al., 2000; Spaepen et al., 2007). Biochemical analysis of JZ38 revealed its ability to produce IAA (10 μg/mL), or its precursors (IAM or IPyA) in liquid culture, with a 34% (13.4 μg/mL) increase when supplemented with 2.5 mM L-Trp (Table 4). Genomic analysis of JZ38 revealed the presence of Trp biosynthesis genes (trpABCDE), a Trp-specific importer (mtr) and IAA biosynthesis genes in the IPyA pathway (aspC, ipdC, aldA, aldB) (Supplementary Table S22) (McClerklin et al., 2018).
Indole is an important interspecies and inter-kingdom signaling molecule and can influence a number of biological functions (Kim and Park, 2015; Lee et al., 2015). It has been shown to promote root development and growth at specific concentrations, possibly by Trp-independent or conversion of indole back to Trp by the tryptophan synthase-β subunit (TSB1 and TSB2) (Last et al., 1991; Ouyang et al., 2000; Blom et al., 2011; Bailly et al., 2014). Phenotypic analysis revealed the ability of JZ38 to produce indole (46.6 μg/mL) in liquid, with a 31.5% (61.3 μg/mL) increase when supplemented with L-Trp, and as a volatile compound (Table 4). Indeed, JZ38 possesses the genes encoding tryptophanase enzyme for conversion of Trp to indole (tnaA) and excretion of indole (acrEF) (Supplementary Table S22) (Kawamura-Sato et al., 1999; Li and Young, 2013). Endophytic bacteria from the same plant as JZ38’s host, which displayed salinity stress tolerance promotion (SSTP) in Arabidopsis but belonged to different taxa, also produced IAA and/or indole (Eida et al., 2019).
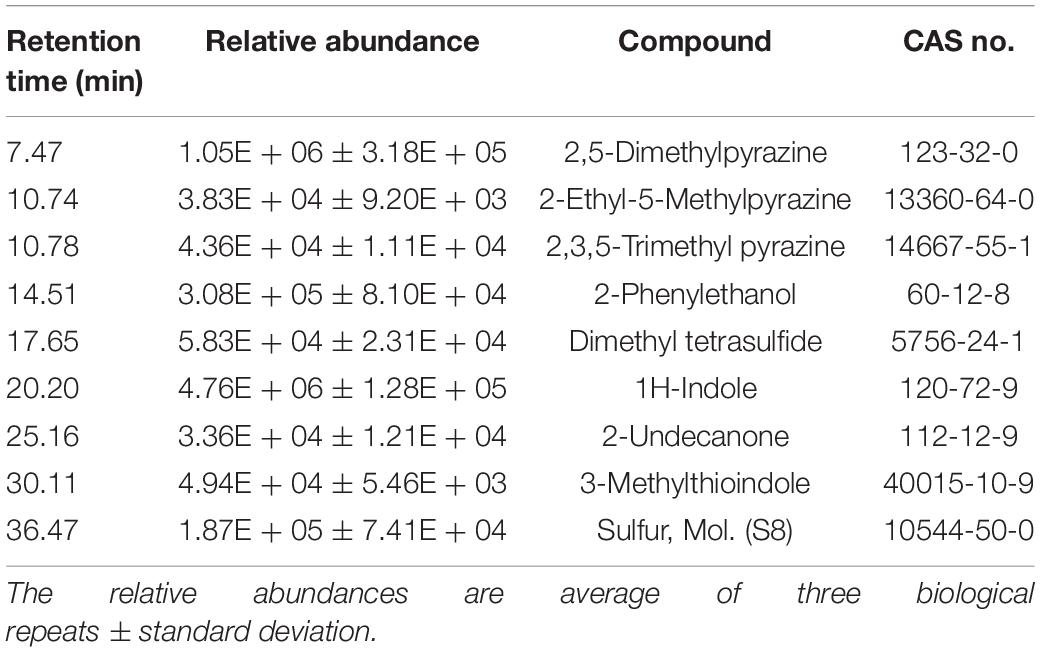
In addition to indole volatiles, SSTP endophytic bacteria from the Jizan region (Eida et al., 2019) shared a common characteristic with JZ38 to produce sulfur-containing volatile compounds, such as hydrogen sulfide (H2S) (Table 4). This gaseous compound has been shown to induce salinity stress tolerance in crop plants, such as alfalfa (Wang Y. et al., 2012), barley (Chen et al., 2015) and rice (Mostofa et al., 2015), and Arabidopsis (Li et al., 2014; Shi et al., 2015). Genes required for assimilatory sulfate reduction (H2S production) were present in JZ38 (cysND, cysC, cysH, cysJI) (Supplementary Table S22). In addition, genes encoding enzymes cystathionine β synthase (CBS) and cystathionine-γ-lyase (CTH) involved in other possible pathways for H2S production were also present (Supplementary Table S22) (Dias and Weimer, 1998; Rose et al., 2017). Bacterial production of other volatile organic compounds (VOCs), such as acetoin and 2,3-butanediol, have also been shown to promote plant growth (Ryu et al., 2003). Indeed, JZ38 produced acetoin (Supplementary Table S7) and possessed the genes for acetoin (ilvM, ilvH, three copies of ilvB, budA) and 2,3-butanediol (budC, butA) production (Supplementary Table S22) (Taghavi et al., 2010). In addition, the gene for synthesis of the VOC 4-hydroxybenzoate (ubiC) (Siebert et al., 1994), which is thought to be involved in antimicrobial activity and suppression of plant pathogens, was also found in JZ38 (Walker et al., 2003).
In line with the bioassays, the GC/MS profile revealed the presence of indole and 3-methylthioindole, an intermediate in the Gassman indole synthesis (Sundberg, 1984), in the volatiles emitted by JZ38 (Table 5 and Supplementary Table S23). Additionally, sulfur molecular (octasulfur, S8), was also detected and could indicate the presence of H2S. Some volatiles detected are involved in antagonistic activities against pathogenic microbes. For example, 2-phenylethanol has been shown to inhibit mycelial growth of the fungus Guignardia citricarpa and the oomycete P. infestans (Fialho et al., 2011; De Vrieze et al., 2015). 2-Undecanone completely inhibited the growth of the fungi R. solani and Alternaria alternata, the nematode Caenorhabditis elegans, Panagrellus redivivus, and Bursaphelenchus xylophilus flies and slight inhibition of oomycete P. infestans sporangiogenesis (Gu et al., 2007; Groenhagen et al., 2013; De Vrieze et al., 2015).
JZ38 Inhibits the Growth of Phytophthora infestans via Volatile-Emission
Since the GC/MS suggested the presence of potential antimicrobial volatile compounds, we hypothesized that JZ38 could possess volatile-mediated antagonistic activity against some phytopathogens. Therefore, the ability to inhibit the growth of two strains of the phytopathgenic oomycete P. infestans via volatile-emission by JZ38 was tested. As hypothesized, JZ38 exhibited growth inhibition activity against P. infestans via emission of volatiles (Figure 4). JZ38 inhibited the growth of strains 88069 and Rec01 by approximately 58 and 45%, respectively. However, when JZ38 was tested with phytopathogenic fungi R. solani, F. culmorum and B. cinerea, no antagonistic effect were observed (Supplementary Figure S4), indicating a degree of specificity by volatiles emitted by JZ38 toward phyotpathogens.
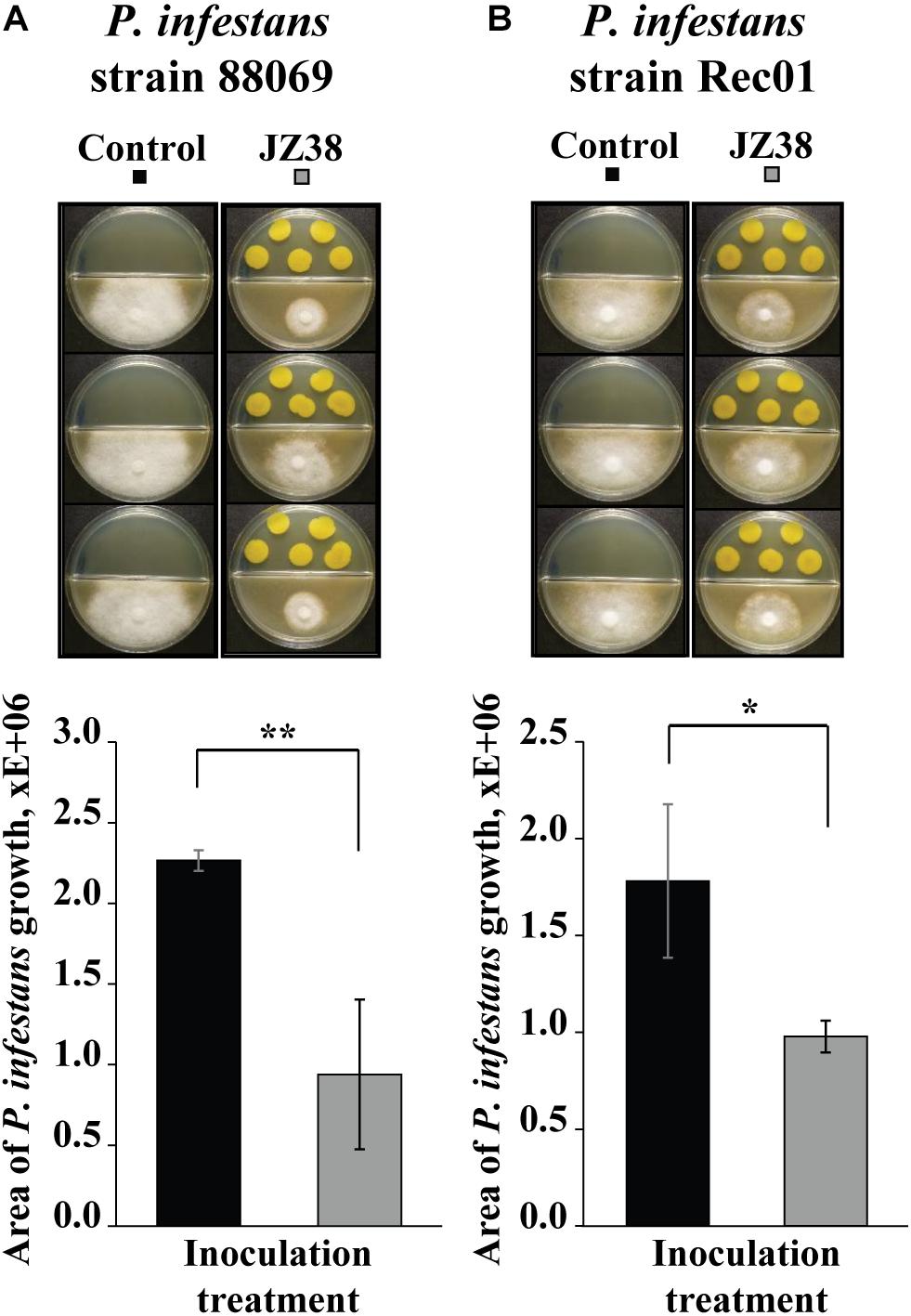
Figure 4. Volatile-mediated effects of JZ38 on mycelial growth of the phytopathogenic oomycete Phytophthora infestans. Representative images of the growth inhibition of P. infestans strains 88069 (A) and Rec1 (B) observed on V8 agar medium (lower side of split-dishes) mediated by volatiles emitted by JZ38 grown on LB agar medium (upper side). Area of mycelial growth was determined 10 and 13 days after inoculation for strains 88069 and Rec1, respectively. Significant differences from the LB-bacteria free control (Student’s t test; n = 3–5) are indicated by asterisks (*p < 0.05; **p < 0.01).
Phytophthora infestans is an oomycete that cause the late blight disease in potato, a major crop in Europe. The late blight disease costs approximately over 1 billion Euros annually (Haverkort et al., 2008). Some of the current solutions are synthetic chemical substances that are costly and have negative side effects on the environment and human health. The use of microbes with antagonistic activity against disease-causing pathogens (termed “biological control or biocontrol”) provides a sustainable, environmentally friendly solution to many plant diseases. Whether the use of JZ38 could suppress late blight in potato under field conditions needs to be tested. However, the potential to inhibit the growth of P. infestans is evident by the demonstrated plate assay.
Conclusion
The endophytic bacterial isolate JZ38 was demonstrated to possess SSTP abilities on Arabidopsis plants either by direct contact with plant tissues or indirectly via emission of volatile compounds. Genome sequencing of the bacterium and its taxonomic analysis identified JZ38 as Cronobacter muytjensii, separating it from the pathogenic C. sakazakii. Genome analysis and additional phenotypic characterization showed that JZ38 can grow under abiotic stresses in the absence and in association with the model plant Arabidopsis. JZ38 was also demonstrated to possess genes for chemotaxis, motility, plant colonization and secretion systems. JZ38 produced a mixture of volatiles, including indole and sulfur compounds that probably underlie the contact-independent SSTP activity of JZ38. Indole might be a major player in the growth-promoting effects under salinity stress conditions and high concentrations of indole have been shown to promote shoot growth and lateral root formation and to decrease the length of the primary roots under normal conditions (Bailly et al., 2014). The high levels of indole detected in the GC/MS might explain the root phenotype observed by JZ38 inoculation in the contactless assay, suggesting a crucial role of indole in promoting salinity stress tolerance in Arabidopsis. Furthermore, the detection of VOCs with possible biocontrol activity (e.g., 2-phenylethanol and 2-undecanone) may contribute to JZ38’s VOC-mediated growth inhibition of the two oomycetes P. infestans strains. To our knowledge, this is the first report on the PGP potential and SSTP ability of a C. muytjensii strain. The results of the various assays and genome analysis reveal the potential of using JZ38 as both abiotic and biotic stress tolerance promoting bacterium in agricultural applications. However, genetic, transcriptomic and metabolomics analyses are required to confirm the highlighted genes’ involvement in JZ38’s functional potentials.
Data Availability Statement
The datasets generated for this study can be found in the NCBI/DDBJ/EMBL database under the accession numbers CP017662, CP017663, and CP017664.
Author Contributions
AE performed the plant screening and phenotyping assays, gDNA extraction, bioassays, and taxonomic analysis based on 16S rRNA. SB performed genome assembly, gene prediction, annotation of genome data, and taxonomic analysis based on whole genomes. IA integrated all bioinformatics databases and facilitated all genomic annotation platforms. FL’H and LW performed the volatile-mediated pathogen assays, VOCs GC/MS profile, and the data analysis. AE, MS, and HH wrote the manuscript. MS, VB, and HH conceived the overall study.
Funding
This publication is supported by the King Abdullah University of Science and Technology (KAUST) to HH No. BAS/1/1062-01-01.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank all members of the Hirt lab, CDA management team, and KAUST Core Labs for the technical assistance and for their help in many aspects of this work. We would also like to thank Abhishek Anand for his input on the GC/MS analysis and Dr. Cristina Andrés Barrao for her input on the fatty acid analysis.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.00369/full#supplementary-material
Footnotes
- ^ https://imagej.nih.gov/ij/
- ^ https://www.uniprot.org/
- ^ https://github.com/davidemms/STAG
- ^ https://doi.org/10.25781/KAUST-17TW0
- ^ https://www.develve.net/
References
Agarwal, V., Borisova, S. A., Metcalf, W W., van der Donk, W. A., and Nair, S K. (2011). Structural and mechanistic insights into C-P bond hydrolysis by phosphonoacetate hydrolase. Chem. Biol. 18, 1230–1240. doi: 10.1016/j.chembiol.2011.07.019
Ahmad, I., Rouf, S. F., Sun, L., Cimdins, A., Shafeeq, S., Le Guyon, S., et al. (2016). BcsZ inhibits biofilm phenotypes and promotes virulence by blocking cellulose production in Salmonella enterica serovar Typhimurium. Microb. Cell Fact. 15:177. doi: 10.1186/s12934-016-0576-6
Alam, I., Antunes, A., Kamau, A. A., Ba alawi, W., Kalkatawi, M., Stingl, U., et al. (2013). INDIGO – integrated data warehouse of microbial genomes with examples from the Red Sea extremophiles. PLoS One 8:e82210. doi: 10.1371/journal.pone.0082210
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
Altschul, S. F., Madden, T. L., Schäffer, A. A., Zhang, J., Zhang, Z., Miller, W., et al. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. doi: 10.1093/nar/25.17.3389
Andrés-Barrao, C., Lafi, F. F., Alam, I., de Zélicourt, A., Eida, A. A., Bokhari, A., et al. (2017). Complete genome sequence analysis of Enterobacter sp. SA187, a plant multi-stress tolerance promoting endophytic bacterium. Front. Microbiol. 8:2023. doi: 10.3389/fmicb.2017.02023
Ashraf, M., Hasnain, S., Berge, O., and Mahmood, T. (2004). Inoculating wheat seedlings with exopolysaccharide-producing bacteria restricts sodium uptake and stimulates plant growth under salt stress. Biol. Fertility Soils 40, 157–162. doi: 10.1007/s00374-004-0766-y
Augimeri, R. V., Varley, A. J., and Strap, J. L. (2015). Establishing a role for bacterial cellulose in environmental interactions: lessons learned from diverse biofilm-producing proteobacteria. Front. Microbiol. 6:1282. doi: 10.3389/fmicb.2015.01282
Baez-Rogelio, A., Morales-García, Y. E., Quintero-Hernández, V., and Muñoz-Rojas, J. (2017). Next generation of microbial inoculants for agriculture and bioremediation. Microb. Biotechnol. 10, 19–21. doi: 10.1111/1751-7915.12448
Bailly, A., Groenhagen, U., Schulz, S., Geisler, M., Eberl, L., and Weisskopf, L. (2014). The inter-kingdom volatile signal indole promotes root development by interfering with auxin signalling. Plant J. 80, 758–771. doi: 10.1111/tpj.12666
Baker, J. L., Lindsay, E. L., Faustoferri, R. C., To, T. T., Hendrickson, E. L., He, X., et al. (2018). Characterization of the trehalose utilization operon in Streptococcus mutans reveals that the trer transcriptional regulator is involved in stress response pathways and toxin production. J. Bacteriol. 200:e00057-18. doi: 10.1128/JB.00057-18
Bakker, P. A. H. M., Doornbos, R. F., Zamioudis, C., Berendsen, R. L., and Pieterse, C. M. J. (2013). Induced systemic resistance and the rhizosphere microbiome. Plant Pathol. J. 29, 136–143. doi: 10.5423/PPJ.SI.07.2012.0111
Batchelor, E., Walthers, D., Kenney, L. J., and Goulian, M. (2005). The Escherichia coli CpxA-CpxR envelope stress response system regulates expression of the porins OmpF and OmpC. J. Bacteriol. 187, 5723–5731. doi: 10.1128/JB.187.16.5723-5731.2005
Becker, A., Fraysse, N., and Sharypova, L. (2005). Recent advances in studies on structure and symbiosis-related function of rhizobial K-antigens and lipopolysaccharides. Mol. Plant Microbe Interact. 18, 899–905. doi: 10.1094/MPMI-18-0899
Beloin, C., Roux, A., and Ghigo, J. M. (2008). “Escherichia coli biofilms,” in Bacterial Biofilms, ed. T. Romeo (Berlin: Springer), 249–289. doi: 10.1007/978-3-540-75418-3_12
Bible, A. N., Fletcher, S. J., Pelletier, D. A., Schadt, C. W., Jawdy, S. S., Weston, D. J., et al. (2016). A carotenoid-deficient mutant in Pantoea sp. YR343, a bacteria isolated from the rhizosphere of Populus deltoides, is defective in root colonization. Front. Microbiol. 7:491. doi: 10.3389/fmicb.2016.00491
Biemans-Oldehinkel, E., and Poolman, B. (2003). On the role of the two extracytoplasmic substrate−binding domains in the ABC transporter OpuA. EMBO J. 22, 5983–5993. doi: 10.1093/emboj/cdg581
Blom, D., Fabbri, C., Connor, E. C., Schiestl, F. P., Klauser, D. R., Boller, T., et al. (2011). Production of plant growth modulating volatiles is widespread among rhizosphere bacteria and strongly depends on culture conditions. Environ. Microbiol. 13, 3047–3058. doi: 10.1111/j.1462-2920.2011.02582.x
Bokhari, A., Essack, M., Lafi, F. F., Andres-Barrao, C., Jalal, R., Alamoudi, S., et al. (2019). Bioprospecting desert plant Bacillus endophytic strains for their potential to enhance plant stress tolerance. Sci. Rep. 9:18154. doi: 10.1038/s41598-019-54685-y
Boos, W., and Shuman, H. (1998). Maltose/maltodextrin system of Escherichia coli: transport, metabolism, and regulation. Microbiol. Mol. Biol. Rev. 62, 204–229. doi: 10.1128/mmbr.62.1.204-229.1998
Booth, I. R., and Higgins, C. F. (1990). Enteric bacteria and osmotic stress: intracellular potassium glutamate as a secondary signal of osmotic stress? FEMS Microbiol. Lett. 75, 239–246. doi: 10.1111/j.1574-6968.1990.tb04097.x
Boukhalfa, H., and Crumbliss, A. L. (2002). Chemical aspects of siderophore mediated iron transport. Biometals 15, 325–339. doi: 10.1023/A:1020218608266
Boyer, J. S. (1982). Plant productivity and environment. Science 218, 443–448. doi: 10.1126/science.218.4571.443
Bray, E. A., Bailey-Serres, J., and Weretilnyk, E. (2000). “Responses to abiotic stresses,” in Biochemistry and Molecular Biology of Plants, eds B. B. Buchanan, W. Gruissem, and R. L. Jones (Rockville, Md: American Society of Plant Physiologists).
Brown, P. H., Cakmak, I., and Zhang, Q. (1993). “Form and function of zinc plants,” in Zinc in Soils and Plants: Proceedings of the International Symposium on ‘Zinc in Soils and Plants’ Held at the University of Western Australia, 27–28 September, 1993, ed. A. D. Robson (Dordrecht: Springer), 93–106. doi: 10.1007/978-94-011-0878-2_7
Burke, E. J., Brown, S. J., and Christidis, N. (2006). Modeling the recent evolution of global drought and projections for the twenty-first century with the hadley centre climate model. J. Hydrometeorol. 7, 1113–1125. doi: 10.1175/JHM544.1
Bussema, C. III, Lee, Z. M.-P., and Schmidt, T. M. (2008). rrnDB: documenting the number of rRNA and tRNA genes in bacteria and archaea. Nucleic Acids Res. 37(Suppl. 1), D489–D493. doi: 10.1093/nar/gkn689
Cannella, S. E., Ntsogo Enguéné, V. Y., Davi, M., Malosse, C., Sotomayor Pérez, A. C., Chamot-Rooke, J., et al. (2017). Stability, structural and functional properties of a monomeric, calcium–loaded adenylate cyclase toxin, CyaA, from Bordetella pertussis. Sci. Rep. 7:42065. doi: 10.1038/srep42065
Carreño-Lopez, R., Campos-Reales, N., Elmerich, C., and Baca, B. E. (2000). Physiological evidence for differently regulated tryptophan-dependent pathways for indole-3-acetic acid synthesis in Azospirillum brasilense. Mol. Gen. Genet. 264, 521–530. doi: 10.1007/s004380000340
Carver, T., Thomson, N., Bleasby, A., Berriman, M., and Parkhill, J. (2008). DNAPlotter: circular and linear interactive genome visualization. Bioinformatics 25, 119–120. doi: 10.1093/bioinformatics/btn578
Chai, Y., Beauregard, P. B., Vlamakis, H., Losick, R., and Kolter, R. (2012). Galactose metabolism plays a crucial role in biofilm formation by Bacillus subtilis. mBio 3:e00184-12. doi: 10.1128/mBio.00184-12
Chase, H. R., Eberl, L., Stephan, R., Jeong, H., Lee, C., Finkelstein, S., et al. (2017). Draft genome sequence of Cronobacter sakazakii GP1999, sequence type 145, an epiphytic isolate obtained from the tomato’s rhizoplane/rhizosphere continuum. Genome Announc. 5:e00723-17. doi: 10.1128/genomeA.00723-17
Chen, J., Wang, W.-H., Wu, F.-H., He, E.-M., Liu, X., Shangguan, Z.-P., et al. (2015). Hydrogen sulfide enhances salt tolerance through nitric oxide-mediated maintenance of ion homeostasis in barley seedling roots. Sci. Rep. 5:12516. doi: 10.1038/srep12516
Cheng, H. Y., Chen, Y. S., Wu, C. Y., Chang, H. Y., Lai, Y. C., and Peng, H. L. (2010). RmpA regulation of capsular polysaccharide biosynthesis in Klebsiella pneumoniae CG43. J. Bacteriol. 192, 3144–3158. doi: 10.1128/jb.00031-10
Chernin, L., Ismailov, Z., Haran, S., and Chet, I. (1995). Chitinolytic Enterobacter agglomerans antagonistic to fungal plant pathogens. Appl. Environ. Microbiol. 61, 1720–1726. doi: 10.1128/aem.61.5.1720-1726.1995
Chiang, S. M., and Schellhorn, H. E. (2012). Regulators of oxidative stress response genes in Escherichia coli and their functional conservation in bacteria. Arch. Biochem. Biophys. 525, 161–169. doi: 10.1016/j.abb.2012.02.007
Chin, C.-S., Alexander, D. H., Marks, P., Klammer, A. A., Drake, J., Heiner, C., et al. (2013). Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat. Methods 10, 563–569. doi: 10.1038/nmeth.2474
Dai, A. (2012). Increasing drought under global warming in observations and models. Nat. Clim. Change 3:171. doi: 10.1038/nclimate1633
Dakora, F. D., and Phillips, D. A. (2002). Root exudates as mediators of mineral acquisition in low-nutrient environments. Plant Soil 245, 35–47. doi: 10.1023/a:1020809400075
Danese, P. N., Pratt, L. A., and Kolter, R. (2000). Exopolysaccharide production is required for development of Escherichia coli K-12 biofilm architecture. J. Bacteriol. 182, 3593–3596. doi: 10.1128/JB.182.12.3593-3596.2000
Darmon, E., and Leach, D. R. F. (2014). Bacterial genome instability. Microbiol. Mol. Biol. Rev. 78, 1–39. doi: 10.1128/MMBR.00035-13
De Muynck, C., Pereira, C. S. S., Naessens, M., Parmentier, S., Soetaert, W., and Vandamme, E. J. (2007). The genus Gluconobacter oxydans: comprehensive overview of biochemistry and biotechnological applications. Crit. Rev. Biotechnol. 27, 147–171. doi: 10.1080/07388550701503584
De Vrieze, M., Pandey, P., Bucheli, T. D., Varadarajan, A. R., Ahrens, C. H., Weisskopf, L., et al. (2015). Volatile organic compounds from native potato-associated Pseudomonas as potential anti-oomycete agents. Front. Microbiol. 6:1295. doi: 10.3389/fmicb.2015.01295
de Weert, S., Vermeiren, H., Mulders, I. H. M., Kuiper, I., Hendrickx, N., Bloemberg, G. V., et al. (2002). Flagella-driven chemotaxis towards exudate components is an important trait for tomato root colonization by Pseudomonas fluorescens. Mol. Plant Microbe Interact. 15, 1173–1180. doi: 10.1094/MPMI.2002.15.11.1173
de Werra, P., Péchy-Tarr, M., Keel, C., and Maurhofer, M. (2009). Role of gluconic acid production in the regulation of biocontrol traits of Pseudomonas fluorescens CHA0. Appl. Environ. Microbiol. 75, 4162–4174. doi: 10.1128/AEM.00295-09
de Zélicourt, A., Synek, L., Saad, M. M., Alzubaidy, H., Jalal, R., Xie, Y., et al. (2018). Ethylene induced plant stress tolerance by Enterobacter sp. SA187 is mediated by 2−keto−4−methylthiobutyric acid production. PLoS Genet. 14:e1007273. doi: 10.1371/journal.pgen.1007273
Dellagi, A., and Aznar, A. (2015). New insights into the role of siderophores as triggers of plant immunity: what can we learn from animals? J. Exp. Bot. 66, 3001–3010. doi: 10.1093/jxb/erv155
Desai, S. K., Nandimath, K., and Mahadevan, S. (2010). Diverse pathways for salicin utilization in Shigella sonnei and Escherichia coli carrying an impaired bgl operon. Arch. Microbiol. 192, 821–833. doi: 10.1007/s00203-010-0610-8
Dias, B., and Weimer, B. (1998). Conversion of methionine to thiols by Lactococci, Lactobacilli, and Brevibacteria. Appl. Environ. Microbiol. 64, 3320–3326. doi: 10.1128/aem.64.9.3320-3326.1998
Dimkpa, C., Weinand, T., and Asch, F. (2009). Plant–rhizobacteria interactions alleviate abiotic stress conditions. Plant Cell Environ. 32, 1682–1694. doi: 10.1111/j.1365-3040.2009.02028.x
Dörr, J., Hurek, T., and Reinhold-Hurek, B. (1998). Type IV pili are involved in plant–microbe and fungus–microbe interactions. Mol. Microbiol. 30, 7–17. doi: 10.1046/j.1365-2958.1998.01010.x
Drudy, D., Mullane, N. R., Quinn, T., Wall, P. G., and Fanning, S. (2006). Enterobacter sakazakii: an emerging pathogen in powdered infant formula. Clin. Infect. Dis. 42, 996–1002. doi: 10.1086/501019
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Ehrhardt, C. J., Chu, V., Brown, T., Simmons, T. L., Swan, B. K., Bannan, J., et al. (2010). Use of fatty acid methyl ester profiles for discrimination of Bacillus cereus T-strain spores grown on different media. Appl. Environ. Microbiol. 76, 1902–1912. doi: 10.1128/AEM.02443-09
Eida, A. A., Alzubaidy, H. S., de Zélicourt, A., Synek, L., Alsharif, W., Lafi, F. F., et al. (2019). Phylogenetically diverse endophytic bacteria from desert plants induce transcriptional changes of tissue-specific ion transporters and salinity stress in Arabidopsis thaliana. Plant Sci. 280, 228–240. doi: 10.1016/j.plantsci.2018.12.002
Eida, A. A., Hirt, H., and Saad, M. M. (2017). “Challenges faced in field application of phosphate-solubilizing bacteria,” in Rhizotrophs: Plant Growth Promotion to Bioremediation, ed. S. Mehnaz (Singapore: Springer), 125–143. doi: 10.1007/978-981-10-4862-3_6
Eida, A. A., Ziegler, M., Lafi, F. F., Michell, C. T., Voolstra, C. R., Hirt, H., et al. (2018). Desert plant bacteria reveal host influence and beneficial plant growth properties. PLoS One 13:e0208223. doi: 10.1371/journal.pone.0208223
Emms, D. M., and Kelly, S. (2017). STRIDE: species tree root inference from gene duplication events. Mol. Biol. Evol. 34, 3267–3278. doi: 10.1093/molbev/msx259
Enebe, M. C., and Babalola, O. O. (2018). The influence of plant growth-promoting rhizobacteria in plant tolerance to abiotic stress: a survival strategy. Appl. Microbiol. Biotechnol. 102, 7821–7835. doi: 10.1007/s00253-018-9214-z
Engl, C., Jovanovic, G., Joly, N., Buck, M., Huvet, M., Stumpf, M. P. H., et al. (2010). Managing membrane stress: the phage shock protein (Psp) response, from molecular mechanisms to physiology. FEMS Microbiol. Rev. 34, 797–827. doi: 10.1111/j.1574-6976.2010.00240.x
FAO (2008). FAO Land and Plant Nutrition Management Service. Available online at: http://www.fao.org/ag/agl/agll/spush
FAO (2017). The Future of Food and Agriculture: Trends and Challenges. Rome: Food and Agriculture Organization.
FAO/WHO (2004). Enterobacter Sakazakii and Other Microorganisms in Powdered Infant Formula: Meeting Report. Rome: Food and Agriculture Organization.
Fazli, M., Almblad, H., Rybtke, M. L., Givskov, M., Eberl, L., and Tolker-Nielsen, T. (2014). Regulation of biofilm formation in Pseudomonas and Burkholderia species. Environ. Microbiol. 16, 1961–1981. doi: 10.1111/1462-2920.12448
Fialho, M. B., Ferreira, L. F. R., Monteiro, R. T. R., and Pascholati, S. F. (2011). Antimicrobial volatile organic compounds affect morphogenesis-related enzymes in Guignardia citricarpa, causal agent of citrus black spot. Biocontrol Sci. Technol. 21, 797–807. doi: 10.1080/09583157.2011.580837
Fields, P., Groisman, E., and Heffron, F. (1989). A Salmonella locus that controls resistance to microbicidal proteins from phagocytic cells. Science 243, 1059–1062. doi: 10.1126/science.2646710
Fitzgerald, D. M., Bonocora, R. P., and Wade, J. T. (2014). Comprehensive mapping of the Escherichia coli flagellar regulatory network. PLoS Genet. 10:e1004649. doi: 10.1371/journal.pgen.1004649
Fones, H., and Preston, G. M. (2012). Reactive oxygen and oxidative stress tolerance in plant pathogenic Pseudomonas. FEMS Microbiol. Lett. 327, 1–8. doi: 10.1111/j.1574-6968.2011.02449.x
Foster, J. W., and Hall, H. K. (1990). Adaptive acidification tolerance response of Salmonella typhimurium. J. Bacteriol. 172, 771–778. doi: 10.1128/jb.172.2.771-778.1990
Franco, A. A., Hu, L., Grim, C. J., Gopinath, G., Sathyamoorthy, V., Jarvis, K. G., et al. (2011a). Characterization of putative virulence genes on the related RepFIB plasmids harbored by Cronobacter spp. Appl. Environ. Microbiol. 77, 3255–3767. doi: 10.1128/AEM.03023-10
Franco, A. A., Kothary, M. H., Gopinath, G., Jarvis, K. G., Grim, C. J., Hu, L., et al. (2011b). Cpa, the outer membrane protease of Cronobacter sakazakii activates plasminogen and mediates resistance to serum bactericidal activity. Infect. Immun. 79, 1578–1587. doi: 10.1128/IAI.01165-10
Fritz, G., Megerle, J. A., Westermayer, S. A., Brick, D., Heermann, R., Jung, K., et al. (2014). Single cell kinetics of phenotypic switching in the arabinose utilization system of E. coli. PLoS One 9:e89532. doi: 10.1371/journal.pone.0089532
Furuchi, T., Kashiwagi, K., Kobayashi, H., and Igarashi, K. (1991). Characteristics of the gene for a spermidine and putrescine transport system that maps at 15 min on the Escherichia coli chromosome. J. Biol. Chem. 266, 20928–20933.
Gerdes, K., Bech, F. W., Jørgensen, S. T., Løbner-Olesen, A., Rasmussen, P. B., Atlung, T., et al. (1986). Mechanism of postsegregational killing by the hok gene product of the parB system of plasmid R1 and its homology with the relF gene product of the E. coli relB operon. EMBO J. 5, 2023–2029. doi: 10.1002/j.1460-2075.1986.tb04459.x
Gerdes, K., and Wagner, E. G. H. (2007). RNA antitoxins. Curr. Opin. Microbiol. 10, 117–124. doi: 10.1016/j.mib.2007.03.003
Glick, B. R. (2012). Plant growth-promoting bacteria: mechanisms and applications. Scientifica 2012:963401. doi: 10.6064/2012/963401
Godoy, M. S., Nikel, P. I., Cabrera Gomez, J. G., and Pettinari, M. J. (2016). The CreC regulator of Escherichia coli, a new target for metabolic manipulations. Appl. Environ. Microbiol. 82, 244–254. doi: 10.1128/aem.02984-15
Gopinath, G. R., Chase, H. R., Gangiredla, J., Eshwar, A., Jang, H., Patel, I., et al. (2018). Genomic characterization of malonate positive Cronobacter sakazakii serotype O:2, sequence type 64 strains, isolated from clinical, food, and environment samples. Gut Pathog. 10:11. doi: 10.1186/s13099-018-0238-9
Green, E. R., and Mecsas, J. (2016). Bacterial secretion systems: an overview. Microbiol. Spectr. 4, 213–239. doi: 10.1128/microbiolspec.VMBF-0012-2015
Groenhagen, U., Baumgartner, R., Bailly, A., Gardiner, A., Eberl, L., Schulz, S., et al. (2013). Production of bioactive volatiles by different Burkholderia ambifaria strains. J. Chem. Ecol. 39, 892–906. doi: 10.1007/s10886-013-0315-y
Gu, Y.-Q., Mo, M.-H., Zhou, J.-P., Zou, C.-S., and Zhang, K.-Q. (2007). Evaluation and identification of potential organic nematicidal volatiles from soil bacteria. Soil Biol. Biochem. 39, 2567–2575. doi: 10.1016/j.soilbio.2007.05.011
Guazzaroni, M.-E., Morgante, V., Mirete, S., and González-Pastor, J. E. (2013). Novel acid resistance genes from the metagenome of the Tinto River, an extremely acidic environment. Environ. Microbiol. 15, 1088–1102. doi: 10.1111/1462-2920.12021
Guérin, F., Isnard, C., Cattoir, V., and Giard, J. C. (2015). Complex regulation pathways of AmpC-mediated β-lactam resistance in Enterobacter cloacae complex. Antimicrob. Agents Chemother. 59, 7753–7761. doi: 10.1128/aac.01729-15
Gul, N., and Poolman, B. (2013). Functional reconstitution and osmoregulatory properties of the ProU ABC transporter from Escherichia coli. Mol. Membr. Biol. 30, 138–148. doi: 10.3109/09687688.2012.754060
Hardoim, P. R., van Overbeek, L. S., and Elsas, J. D. V. (2008). Properties of bacterial endophytes and their proposed role in plant growth. Trends Microbiol 16, 463–471. doi: 10.1016/j.tim.2008.07.008
Haverkort, A. J., Boonekamp, P. M., Hutten, R., Jacobsen, E., Lotz, L. A. P., Kessel, G. J. T., et al. (2008). Societal costs of late blight in potato and prospects of durable resistance through cisgenic modification. Potato Res. 51, 47–57. doi: 10.1007/s11540-008-9089-y
Hayes, F. (2003). Toxins-antitoxins: plasmid maintenance, programmed cell death, and cell cycle arrest. Science 301, 1496–1499. doi: 10.1126/science.1088157
Himelright, I., Harris, E., Lorch, V., Anderson, M., Jones, T., Craig, A., et al. (2002). Enterobacter sakazakii infections associated with the use of powdered infant formula—Tennessee, 2001. JAMA 287, 2204–2205. doi: 10.1001/jama.287.17.2204
Holford, I. C. R. (1997). Soil phosphorus: its measurement, and its uptake by plants. Soil Res. 35, 227–240. doi: 10.1071/S96047
Holmberg, C., Beijer, L., Rutberg, B., and Rutberg, L. (1990). Glycerol catabolism in Bacillus subtilis: nucleotide sequence of the genes encoding glycerol kinase (glpK) and glycerol-3-phosphate dehydrogenase (glpD). J. Gen. Microbiol. 136, 2367–2375. doi: 10.1099/00221287-136-12-2367
Hood, R. D., Singh, P., Hsu, F., Güvener, T., Carl, M. A., Trinidad, R. R. S., et al. (2010). A type VI secretion system of Pseudomonas aeruginosa targets a toxin to bacteria. Cell Host Microbe 7, 25–37. doi: 10.1016/j.chom.2009.12.007
Huang, L., Xu, W., Su, Y., Zhao, L., and Yan, Q. (2018). Regulatory role of the RstB-RstA system in adhesion, biofilm production, motility, and hemolysis. Microbiologyopen 7:e00599. doi: 10.1002/mbo3.599
Hugouvieux-Cotte-Pattat, N., and Charaoui-Boukerzaza, S. (2009). Catabolism of raffinose, sucrose, and melibiose in Erwinia chrysanthemi 3937. J. Bacteriol. 191, 6960–6967. doi: 10.1128/JB.00594-09
Hunziker, L., Bönisch, D., Groenhagen, U., Bailly, A., Schulz, S., and Weisskopf, L. (2015). Pseudomonas strains naturally associated with potato plants produce volatiles with high potential for inhibition of Phytophthora infestans. Appl. Environ. Microbiol. 81, 821–830. doi: 10.1128/AEM.02999-14
Hyatt, D., Chen, G.-L., LoCascio, P. F., Land, M. L., Larimer, F. W., and Hauser, L. J. (2010). Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11:119. doi: 10.1186/1471-2105-11-119
Idris, E. E., Iglesias, D. J., Talon, M., and Borriss, R. (2007). Tryptophan-dependent production of indole-3-acetic acid (IAA) affects level of plant growth promotion by Bacillus amyloliquefaciens FZB42. Mol. Plant Microbe Interact. 20, 619–626. doi: 10.1094/MPMI-20-6-0619
Iniguez, A. L., Dong, Y., Carter, H. D., Ahmer, B. M. M., Stone, J. M., and Triplett, E. W. (2005). Regulation of enteric endophytic bacterial colonization by plant defenses. Mol. Plant Microbe Interact. 18, 169–178. doi: 10.1094/MPMI-18-0169
Iversen, C., Lehner, A., Mullane, N., Bidlas, E., Cleenwerck, I., Marugg, J., et al. (2007). The taxonomy of Enterobacter sakazakii: proposal of a new genus Cronobacter gen. nov. and descriptions of Cronobacter sakazakii comb. nov. Cronobacter sakazakii subsp. sakazakii, comb. nov., Cronobacter sakazakii subsp. malonaticus subsp. nov., Cronobacter turicensis sp. nov., Cronobacter muytjensii sp. nov., Cronobacter dublinensis sp. nov. and Cronobacter genomospecies 1. BMC Evol. Biol. 7:64. doi: 10.1186/1471-2148-7-64
Iversen, C., Mullane, N., McCardell, B., Tall, B. D., Lehner, A., Fanning, S., et al. (2008). Cronobacter gen. nov., a new genus to accommodate the biogroups of Enterobacter sakazakii, and proposal of Cronobacter sakazakii gen. nov., comb. nov., Cronobacter malonaticus sp. nov., Cronobacter turicensis sp. nov., Cronobacter muytjensii sp. nov., Cronobacter dublinensis sp. nov., Cronobacter genomospecies 1, and of three subspecies, Cronobacter dublinensis subsp. dublinensis subsp. nov., Cronobacter dublinensis subsp. lausannensis subsp. nov. and Cronobacter dublinensis subsp. lactaridi subsp. nov. Int. J. Syst. Evol. Microbiol. 58(Pt 6), 1442–1447. doi: 10.1099/ijs.0.65577-0
Jang, H., Woo, J., Lee, Y., Negrete, F., Finkelstein, S., Chase, H. R., et al. (2018). Draft genomes of Cronobacter sakazakii strains isolated from dried spices bring unique insights into the diversity of plant-associated strains. Stand. Genomic Sci. 13:35. doi: 10.1186/s40793-018-0339-6
Jani, A. J., and Cotter, P. A. (2010). Type VI secretion: not just for pathogenesis anymore. Cell Host Microbe 8, 2–6. doi: 10.1016/j.chom.2010.06.012
Johler, S., Stephan, R., Hartmann, I., Kuehner, K. A., and Lehner, A. (2010). Genes involved in yellow pigmentation of Cronobacter sakazakii ES5 and influence of pigmentation on persistence and growth under environmental stress. Appl. Environ. Microbiol. 76, 1053–1061. doi: 10.1128/AEM.01420-09
Jones, P., Binns, D., Chang, H.-Y., Fraser, M., Li, W., McAnulla, C., et al. (2014). InterProScan 5: genome-scale protein function classification. Bioinformatics 30, 1236–1240. doi: 10.1093/bioinformatics/btu031
Joseph, S., Cetinkaya, E., Drahovska, H., Levican, A., Figueras, M. J., and Forsythe, S. J. (2012). Cronobacter condimenti sp. nov., isolated from spiced meat, and Cronobacter universalis sp. nov., a species designation for Cronobacter sp. genomospecies 1, recovered from a leg infection, water and food ingredients. Int. J. Syst. Evol. Microbiol. 62, 1277–1283. doi: 10.1099/ijs.0.032292-0
Kamran, S., Shahid, I., Baig, D. N., Rizwan, M., Malik, K. A., and Mehnaz, S. (2017). Contribution of zinc solubilizing bacteria in growth promotion and zinc content of wheat. Front. Microbiol. 8:2593. doi: 10.3389/fmicb.2017.02593
Kanehisa, M., Sato, Y., Kawashima, M., Furumichi, M., and Tanabe, M. (2016). KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 44, D457–D462. doi: 10.1093/nar/gkv1070
Kappes, R. M., Kempf, B., Kneip, S., Boch, J., Gade, J., Meier-Wagner, J., et al. (1999). Two evolutionarily closely related ABC transporters mediate the uptake of choline for synthesis of the osmoprotectant glycine betaine in Bacillus subtilis. Mol. Microbiol. 32, 203–216. doi: 10.1046/j.1365-2958.1999.01354.x
Kashiwagi, K., Shibuya, S., Tomitori, H., Kuraishi, A., and Igarashi, K. (1997). Excretion and uptake of putrescine by the PotE protein in Escherichia coli. J. Biol. Chem. 272, 6318–6323. doi: 10.1074/jbc.272.10.6318
Kawamura-Sato, K., Shibayama, K., Horii, T., Iimuma, Y., Arakawa, Y., and Ohta, M. (1999). Role of multiple efflux pumps in Escherichia coli in indole expulsion. FEMS Microbiol. Lett. 179, 345–352. doi: 10.1111/j.1574-6968.1999.tb08748.x
Kelly, S., and Maini, P. K. (2013). DendroBLAST: approximate phylogenetic trees in the absence of multiple sequence alignments. PLoS One 8:e58537. doi: 10.1371/journal.pone.0058537
Kier, L. D., Weppelman, R. M., and Ames, B. N. (1979). Regulation of nonspecific acid phosphatase in Salmonella: phoN and phoP genes. J. Bacteriol. 138, 155–161. doi: 10.1128/jb.138.1.155-161.1979
Kim, J., and Park, W. (2015). Indole: a signaling molecule or a mere metabolic byproduct that alters bacterial physiology at a high concentration? J. Microbiol. 53, 421–428. doi: 10.1007/s12275-015-5273-3
Kim, M., Kim, S., Park, B., and Ryu, S. (2014). Core lipopolysaccharide-specific phage SSU5 as an auxiliary component of a phage cocktail for Salmonella biocontrol. Appl. Environ. Microbiol. 80, 1026–1034. doi: 10.1128/AEM.03494-13
Kim, M., Kim, S., and Ryu, S. (2012). Complete genome sequence of bacteriophage SSU5 specific for Salmonella enterica serovar Typhimurium rough strains. J. Virol. 86:10894. doi: 10.1128/JVI.01796-12
Kimura, M. (1980). A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16, 111–120. doi: 10.1007/bf01731581
Klappenbach, J. A., Dunbar, J. M., and Schmidt, T. M. (2000). rRNA operon copy number reflects ecological strategies of bacteria. Appl. Environ. Microbiol. 66, 1328–1333. doi: 10.1128/AEM.66.4.1328-1333.2000
Krumsiek, J., Arnold, R., and Rattei, T. (2007). Gepard: a rapid and sensitive tool for creating dotplots on genome scale. Bioinformatics 23, 1026–1028. doi: 10.1093/bioinformatics/btm039
Kruskal, W. H., and Wallis, W. A. (1952). Use of ranks in one-criterion variance analysis. J. Am. Stat. Assoc. 47, 583–621. doi: 10.1080/01621459.1952.10483441
Kuhnert, W. L., Zheng, G., Faustoferri, R. C., and Quivey, R. G. (2004). The F-ATPase operon promoter of Streptococcus mutans is transcriptionally regulated in response to external pH. J. Bacteriol. 186, 8524–8528. doi: 10.1128/JB.186.24.8524-8528.2004
Kuivanen, J., Biz, A., and Richard, P. (2019). Microbial hexuronate catabolism in biotechnology. AMB Express 9:16. doi: 10.1186/s13568-019-0737-1
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Kurihara, S., Oda, S., Kato, K., Kim, H. G., Koyanagi, T., Kumagai, H., et al. (2005). A novel putrescine utilization pathway involves γ-glutamylated intermediates of Escherichia coli K-12. J. Biol. Chem. 280, 4602–4608. doi: 10.1074/jbc.M411114200
Lagesen, K., Hallin, P., Rødland, E. A., Stærfeldt, H.-H., Rognes, T., and Ussery, D. W. (2007). RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 35, 3100–3108. doi: 10.1093/nar/gkm160
Last, R. L., Bissinger, P. H., Mahoney, D. J., Radwanski, E. R., and Fink, G. R. (1991). Tryptophan mutants in Arabidopsis: the consequences of duplicated tryptophan synthase beta genes. Plant Cell 3, 345–358. doi: 10.1105/tpc.3.4.345
Laus, M. C., van Brussel, A. A. N., and Kijne, J. W. (2005). Role of cellulose fibrils and exopolysaccharides of Rhizobium leguminosarum in attachment to and infection of Vicia sativa root hairs. Mol. Plant Microbe Interact. 18, 533–538. doi: 10.1094/MPMI-18-0533
Leblanc, S. K. D., Oates, C. W., and Raivio, T. L. (2011). Characterization of the induction and cellular role of the BaeSR two-component envelope stress response of Escherichia coli. J. Bacteriol. 193, 3367–3375. doi: 10.1128/JB.01534-10
Ledger, T., Rojas, S., Timmermann, T., Pinedo, I., Poupin, M. J., Garrido, T., et al. (2016). Volatile-mediated effects predominate in Paraburkholderia phytofirmans growth promotion and salt stress tolerance of Arabidopsis thaliana. Front. Microbiol. 7:1838. doi: 10.3389/fmicb.2016.01838
Lee, J.-H., Choi, Y., Shin, H., Lee, J., and Ryu, S. (2012). Complete genome sequence of Cronobacter sakazakii temperate bacteriophage phiES15. J. Virol. 86, 7713–7714. doi: 10.1128/JVI.01042-12
Lee, J.-H., Wood, T. K., and Lee, J. (2015). Roles of indole as an interspecies and interkingdom signaling molecule. Trends Microbiol. 23, 707–718. doi: 10.1016/j.tim.2015.08.001
Lefort, V., Desper, R., and Gascuel, O. (2015). FastME 2.0: a comprehensive, accurate, and fast distance-based phylogeny inference program. Mol. Biol. Evol. 32, 2798–2800. doi: 10.1093/molbev/msv150
Letunic, I., and Bork, P. (2016). Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 44, W242–W245. doi: 10.1093/nar/gkw290
Li, G., and Young, K. D. (2013). Indole production by the tryptophanase TnaA in Escherichia coli is determined by the amount of exogenous tryptophan. Microbiology 159, 402–410. doi: 10.1099/mic.0.064139-0
Li, J., Jia, H., Wang, J., Cao, Q., and Wen, Z. (2014). Hydrogen sulfide is involved in maintaining ion homeostasis via regulating plasma membrane Na+/H+ antiporter system in the hydrogen peroxide-dependent manner in salt-stress Arabidopsis thaliana root. Protoplasma 251, 899–912. doi: 10.1007/s00709-013-0592-x
Li, L., Stoeckert, C. J., and Roos, D. S. (2003). OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 13, 2178–2189. doi: 10.1101/gr.1224503
Liao, M.-K., Gort, S., and Maloy, S. (1997). A cryptic proline permease in Salmonella typhimurium. Microbiology 143, 2903–2911. doi: 10.1099/00221287-143-9-2903
Liu, S. T., Lee, L. Y., Tai, C. Y., Hung, C. H., Chang, Y. S., Wolfram, J. H., et al. (1992). Cloning of an Erwinia herbicola gene necessary for gluconic acid production and enhanced mineral phosphate solubilization in Escherichia coli HB101: nucleotide sequence and probable involvement in biosynthesis of the coenzyme pyrroloquinoline quinone. J. Bacteriol. 174, 5814–5819. doi: 10.1128/jb.174.18.5814-5819.1992
Liu, W., Wang, Q., Hou, J., Tu, C., Luo, Y., and Christie, P. (2016). Whole genome analysis of halotolerant and alkalotolerant plant growth-promoting rhizobacterium Klebsiella sp. D5A. Sci. Rep. 6:26710. doi: 10.1038/srep26710
Liu, X.-M., and Zhang, H. (2015). The effects of bacterial volatile emissions on plant abiotic stress tolerance. Front. Plant Sci. 6:774. doi: 10.3389/fpls.2015.00774
Louden, B. C., Haarmann, D., and Lynne, A. M. (2011). Use of blue agar CAS assay for siderophore detection. J. Microbiol. Biol. Educ. 12, 51–53. doi: 10.1128/jmbe.v12i1.249
Lowe, T. M., and Eddy, S. R. (1997). tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25, 955–964. doi: 10.1093/nar/25.5.955
Lugtenberg, B. J. J., Chin-A-Woeng, T. F. C., and Bloemberg, G. V. (2002). Microbe–plant interactions: principles and mechanisms. Antonie Van Leeuwenhoek 81, 373–383. doi: 10.1023/A:1020596903142
Ly, A., Henderson, J., Lu, A., Culham, D. E., and Wood, J. M. (2004). Osmoregulatory systems of Escherichia coli identification of betaine-carnitine-choline transporter family member BetU and distributions of betU and trkG among pathogenic and nonpathogenic isolates. J. Bacteriol. 186, 296–306. doi: 10.1128/jb.186.2.296-306.2004
Mandon, K., Østerås, M., Boncompagni, E., Trinchant, J. C., Spennato, G., Poggi, M. C., et al. (2003). The Sinorhizobium meliloti glycine betaine biosynthetic genes (betICBA) are induced by choline and highly expressed in bacteroids. Mol. Plant Microbe Interact. 16, 709–719. doi: 10.1094/MPMI.2003.16.8.709
Marchler-Bauer, A., Panchenko, A. R., Shoemaker, B. A., Thiessen, P. A., Geer, L. Y., and Bryant, S. H. (2002). CDD: a database of conserved domain alignments with links to domain three-dimensional structure. Nucleic Acids Res. 30, 281–283. doi: 10.1093/nar/30.1.281
Martín, M. G., Sender, P. D., Peirú, S., de Mendoza, D., and Magni, C. (2004). Acid-inducible transcription of the operon encoding the citrate lyase complex of Lactococcus lactis biovar diacetylactis CRL264. J. Bacteriol. 186, 5649–5660. doi: 10.1128/jb.186.17.5649-5660.2004
Martínez-Granero, F., Capdevila, S., Sánchez-Contreras, M., Martín, M., and Rivilla, R. (2005). Two site-specific recombinases are implicated in phenotypic variation and competitive rhizosphere colonization in Pseudomonas fluorescens. Microbiology 151, 975–983. doi: 10.1099/mic.0.27583-0
Martínez, C. E., and Motto, H. L. (2000). Solubility of lead, zinc and copper added to mineral soils. Environ. Pollut. 107, 153–158. doi: 10.1016/S0269-7491(99)00111-6
Masuda, H., Tan, Q., Yamaguchi, Y., Inouye, M., and Awano, N. (2012). A novel membrane-bound toxin for cell division, CptA (YgfX), inhibits polymerization of cytoskeleton proteins, FtsZ and MreB, in Escherichia coli. FEMS Microbiol. Lett. 328, 174–181. doi: 10.1111/j.1574-6968.2012.02496.x
Mauzy, C. A., and Hermodson, M. A. (1992). Structural and functional analyses of the repressor, RbsR, of the ribose operon of Escherichia coli. Protein Sci. 1, 831–842. doi: 10.1002/pro.5560010701
McClerklin, S. A., Lee, S. G., Harper, C. P., Nwumeh, R., Jez, J. M., and Kunkel, B. N. (2018). Indole-3-acetaldehyde dehydrogenase-dependent auxin synthesis contributes to virulence of Pseudomonas syringae strain DC3000. PLoS Pathog. 14:e1006811. doi: 10.1371/journal.ppat.1006811
Meier-Kolthoff, J. P., and Göker, M. (2019). TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat. Commun. 10:2182. doi: 10.1038/s41467-019-10210-3
Miller-Fleming, L., Olin-Sandoval, V., Campbell, K., and Ralser, M. (2015). Remaining mysteries of molecular biology: the role of polyamines in the cell. J. Mol. Biol. 427, 3389–3406. doi: 10.1016/j.jmb.2015.06.020
Mohammadi, M., Burbank, L., and Roper, M. C. (2012). Biological role of pigment production for the bacterial phytopathogen Pantoea stewartii subsp. stewartii. Appl. Environ. Microbiol. 78, 6859–6865. doi: 10.1128/AEM.01574-12
Morgante, C., Angelini, J., Castro, S., and Fabra, A. (2005). Role of rhizobial exopolysaccharides in crack entry/intercellular infection of peanut. Soil Biol. Biochem. 37, 1436–1444. doi: 10.1016/j.soilbio.2004.12.014
Mori, S. (1999). Iron acquisition by plants. Curr. Opin. Plant Biol. 2, 250–253. doi: 10.1016/S1369-5266(99)80043-0
Morinaga, T., Ashida, H., and Yoshida, K.-I. (2010). Identification of two scyllo-inositol dehydrogenases in Bacillus subtilis. Microbiology 156, 1538–1546. doi: 10.1099/mic.0.037499-0
Mostofa, M. G., Saegusa, D., Fujita, M., and Tran, L.-S. P. (2015). Hydrogen sulfide regulates salt tolerance in rice by maintaining Na+/K+ balance, mineral homeostasis and oxidative metabolism under excessive salt stress. Front. Plant Sci. 6:1055. doi: 10.3389/fpls.2015.01055
Mumtaz, M. Z., Ahmad, M., Jamil, M., and Hussain, T. (2017). Zinc solubilizing Bacillus spp. potential candidates for biofortification in maize. Microbiol. Res. 202, 51–60. doi: 10.1016/j.micres.2017.06.001
Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plant. 15, 473–497. doi: 10.1111/j.1399-3054.1962.tb08052.x
Nachin, L., Nannmark, U., and Nyström, T. (2005). Differential roles of the universal stress proteins of Escherichia coli in oxidative stress resistance, adhesion, and motility. J. Bacteriol. 187, 6265–6272. doi: 10.1128/jb.187.18.6265-6272.2005
Natale, P., Brüser, T., and Driessen, A. J. M. (2008). Sec- and Tat-mediated protein secretion across the bacterial cytoplasmic membrane—Distinct translocases and mechanisms. Biochim. Biophys. Acta 1778, 1735–1756. doi: 10.1016/j.bbamem.2007.07.015
Nawrocki, E. P., and Eddy, S. R. (2013). Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics 29, 2933–2935. doi: 10.1093/bioinformatics/btt509
Nohno, T., Noji, S., Taniguchi, S., and Saito, T. (1989). The narX and narL genes encoding the nitrate-sensing regulators of Escherichia coli are homologous to a family of prokaryotic two-component regulatory genes. Nucleic Acids Res. 17, 2947–2957. doi: 10.1093/nar/17.8.2947
Nolle, N., Felsl, A., Heermann, R., and Fuchs, T. M. (2017). Genetic characterization of the galactitol utilization pathway of Salmonella enterica serovar Typhimurium. J. Bacteriol. 199:e00595-16. doi: 10.1128/JB.00595-16
Ogasawara, H., Shinohara, S., Yamamoto, K., and Ishihama, A. (2012). Novel regulation targets of the metal-response BasS–BasR two-component system of Escherichia coli. Microbiology 158, 1482–1492. doi: 10.1099/mic.0.057745-0
Ogasawara, N., Moriya, S., von Meyenburg, K., Hansen, F. G., and Yoshikawa, H. (1985). Conservation of genes and their organization in the chromosomal replication origin region of Bacillus subtilis and Escherichia coli. EMBO J. 4, 3345–3350. doi: 10.1002/j.1460-2075.1985.tb04087.x
Ohtake, H., Wu, H., Imazu, K., Anbe, Y., Kato, J., and Kuroda, A. (1996). Bacterial phosphonate degradation, phosphite oxidation and polyphosphate accumulation. Resour. Conserv. Recy. 18, 125–134. doi: 10.1016/S0921-3449(96)01173-1
Otto, K., and Silhavy, T. J. (2002). Surface sensing and adhesion of Escherichia coli controlled by the Cpx-signaling pathway. Proc. Natl. Acad. Sci. U.S.A. 99, 2287–2292. doi: 10.1073/pnas.042521699
Ouyang, J., Shao, X., and Li, J. (2000). Indole-3-glycerol phosphate, a branchpoint of indole-3-acetic acid biosynthesis from the tryptophan biosynthetic pathway in Arabidopsis thaliana. Plant J. 24, 327–334. doi: 10.1046/j.1365-313x.2000.00883.x
Paczosa, M. K., and Mecsas, J. (2016). Klebsiella pneumoniae going on the offense with a strong defense. Microbiol. Mol. Biol. Rev. 80, 629–661. doi: 10.1128/mmbr.00078-15
Pahel, G., Rothstein, D. M., and Magasanik, B. (1982). Complex glnA-glnL-glnG operon of Escherichia coli. J. Bacteriol. 150, 202–213. doi: 10.1128/jb.150.1.202-213.1982
Patten, C. L., and Glick, B. R. (1996). Bacterial biosynthesis of indole-3-acetic acid. Can. J. Microbiol. 42, 207–220. doi: 10.1139/m96-032
Patten, C. L., and Glick, B. R. (2002). Role of Pseudomonas putida indoleacetic acid in development of the host plant root system. Appl. Environ. Microbiol. 68, 3795–3801. doi: 10.1128/aem.68.8.3795-3801.2002
Paul, D. (2013). Osmotic stress adaptations in rhizobacteria. J. Basic Microbiol. 53, 101–110. doi: 10.1002/jobm.201100288
Paul, M. J., Primavesi, L. F., Jhurreea, D., and Zhang, Y. (2008). Trehalose metabolism and signaling. Annu. Rev. Plant Biol. 59, 417–441. doi: 10.1146/annurev.arplant.59.032607.092945
Pérez-Montaño, F., Jiménez-Guerrero, I., Del Cerro, P., Baena-Ropero, I., López-Baena, F. J., Ollero, F. J., et al. (2014). The symbiotic biofilm of Sinorhizobium fredii SMH12, necessary for successful colonization and symbiosis of Glycine max cv Osumi, is regulated by quorum sensing systems and inducing flavonoids via NodD1. PLoS One 9:e105901. doi: 10.1371/journal.pone.0105901
Pistocchi, R., Kashiwagi, K., Miyamoto, S., Nukui, E., Sadakata, Y., Kobayashi, H., et al. (1993). Characteristics of the operon for a putrescine transport system that maps at 19 minutes on the Escherichia coli chromosome. J. Biol. Chem. 268, 146–152.
Raaijmakers, J. M., and Mazzola, M. (2012). Diversity and natural functions of antibiotics produced by beneficial and plant pathogenic bacteria. Annu. Rev. Phytopathol. 50, 403–424. doi: 10.1146/annurev-phyto-081211-172908
Raaijmakers, J. M., Vlami, M., and de Souza, J. T. (2002). Antibiotic production by bacterial biocontrol agents. Antonie Van Leeuwenhoek 81:537. doi: 10.1023/A:1020501420831
Raymond, K. N., Allred, B. E., and Sia, A. K. (2015). Antibiotic production by bacterial biocontrol agents. Acc. Chem. Res. 48, 2496–2505. doi: 10.1021/acs.accounts.5b00301
Reina-Bueno, M., Argandoña, M., Salvador, M., Rodríguez-Moya, J., Iglesias-Guerra, F., Csonka, L. N., et al. (2012). Role of trehalose in salinity and temperature tolerance in the model halophilic bacterium Chromohalobacter salexigens. PLoS One 7:e33587. doi: 10.1371/journal.pone.0033587
Rezzonico, F., Smits, T. H. M., and Duffy, B. (2012). Detection of AI-2 receptors in genomes of Enterobacteriaceae suggests a role of type-2 quorum sensing in closed ecosystems. Sensors (Basel) 12, 6645–6665. doi: 10.3390/s120506645
Rhee, H. J., Kim, E.-J., and Lee, J. K. (2007). Physiological polyamines: simple primordial stress molecules. J. Cell. Mol. Med. 11, 685–703. doi: 10.1111/j.1582-4934.2007.00077.x
Richter, M., Rosselló-Móra, R., Oliver Glöckner, F., and Peplies, J. (2015). JSpeciesWS: a web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics 32, 929–931. doi: 10.1093/bioinformatics/btv681
Rodionova, I., Li, X., Thiel, V., Stolyar, S., Fredrickson, J., Bryant, D., et al. (2013). Comparative genomics and functional analysis of rhamnose catabolic pathways and regulons in bacteria. Front. Microbiol 4:407. doi: 10.3389/fmicb.2013.00407
Rodriguez, H., and Fraga, R. (1999). Phosphate solubilizing bacteria and their role in plant growth promotion. Biotechnol. Adv. 17, 319–339. doi: 10.1016/S0734-9750(99)00014-2
Rodríguez, H., Fraga, R., Gonzalez, T., and Bashan, Y. (2006). Genetics of phosphate solubilization and its potential applications for improving plant growth-promoting bacteria. Plant Soil 287, 15–21. doi: 10.1007/s11104-006-9056-9
Rodriguez, H., Gonzalez, T., Goire, I., and Bashan, Y. (2004). Gluconic acid production and phosphate solubilization by the plant growth-promoting bacterium Azospirillum spp. Naturwissenschaften 91, 552–555. doi: 10.1007/s00114-004-0566-0
Rose, P., Moore, P. K., and Zhu, Y. Z. (2017). H2S biosynthesis and catabolism: new insights from molecular studies. Cell. Mol. Life Sci. 74, 1391–1412. doi: 10.1007/s00018-016-2406-8
Russell, R. R., Aduse-Opoku, J., Sutcliffe, I. C., Tao, L., and Ferretti, J. J. (1992). A binding protein-dependent transport system in Streptococcus mutans responsible for multiple sugar metabolism. J. Biol. Chem. 267, 4631–4637.
Ryu, C.-M., Farag, M. A., Hu, C.-H., Reddy, M. S., Wei, H.-X., Paré, P. W., et al. (2003). Bacterial volatiles promote growth in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 100, 4927–4932. doi: 10.1073/pnas.0730845100
Sahu, A., Bhattacharjya, S., Mandal, A., Thakur, J. K., Atoliya, N., Sahu, N., et al. (2018). “Microbes: a sustainable approach for enhancing nutrient availability in agricultural soils,” in Role of Rhizospheric Microbes in Soil: Nutrient Management and Crop Improvement, Vol. 2, ed. V. S. Meena (Singapore: Springer), 47–75. doi: 10.1007/978-981-13-0044-8_2
Saitou, N., and Nei, M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425. doi: 10.1093/oxfordjournals.molbev.a040454
Saravanan, V. S., Kalaiarasan, P., Madhaiyan, M., and Thangaraju, M. (2007). Solubilization of insoluble zinc compounds by Gluconacetobacter diazotrophicus and the detrimental action of zinc ion (Zn2+) and zinc chelates on root knot nematode Meloidogyne incognita. Lett. Appl. Microbiol. 44, 235–241. doi: 10.1111/j.1472-765X.2006.02079.x
Sass, A., Nelis, H. J., Van Acker, H., Dhondt, I., and Coenye, T. (2014). Involvement of toxin–antitoxin modules in Burkholderia cenocepacia biofilm persistence. Pathog. Dis. 71, 326–335. doi: 10.1111/2049-632X.12177
Sauvage, E., Kerff, F., Terrak, M., Ayala, J. A., and Charlier, P. (2008). The penicillin-binding proteins: structure and role in peptidoglycan biosynthesis. FEMS Microbiol. Rev. 32, 234–258. doi: 10.1111/j.1574-6976.2008.00105.x
Sauvonnet, N., Gounon, P., and Pugsley, A. P. (2000). PpdD type IV pilin of Escherichia coli K-12 can be assembled into pili in Pseudomonas aeruginosa. J. Bacteriol. 182, 848–854. doi: 10.1128/JB.182.3.848-854.2000
Schmid, J., Sieber, V., and Rehm, B. (2015). Bacterial exopolysaccharides: biosynthesis pathways and engineering strategies. Front. Microbiol 6:496. doi: 10.3389/fmicb.2015.00496
Schreier, H. J., Brown, S. W., Hirschi, K. D., Nomellini, J. F., and Sonenshein, A. L. (1989). Regulation of Bacillus subtilis glutamine synthetase gene expression by the product of the glnR gene. J. Mol. Biol. 210, 51–63. doi: 10.1016/0022-2836(89)90290-8
Schwartz, N. B., Abram, D., and Feingold, D. S. (1974). L-Rhamnulose 1-phosphate aldolase of Escherichia coli. Role of metal in enzyme structure. Biochemistry 13, 1726–1730. doi: 10.1021/bi00705a026
Sedkova, N., Tao, L., Rouvière, P. E., and Cheng, Q. (2005). Diversity of carotenoid synthesis gene clusters from environmental Enterobacteriaceae strains. Appl. Environ. Microbiol. 71, 8141–8146. doi: 10.1128/AEM.71.12.8141-8146.2005
Shao, Y., Harrison, E. M., Bi, D., Tai, C., He, X., and Ou, H.-Y., et al., (2011). TADB: A web-based resource for Type 2 toxin-antitoxin loci in bacteria and archaea. Nucleic Acids Res. 39, D606–D611. doi: 10.1093/nar/gkq908
Shamanna, D. K., and Sanderson, K. E. (1979). Uptake and catabolism of D-xylose in Salmonella typhimurium LT2. J. Bacteriol. 139, 64–70. doi: 10.1128/jb.139.1.64-70.1979
Shi, H., Ye, T., Han, N., Bian, H., Liu, X., and Chan, Z. (2015). Hydrogen sulfide regulates abiotic stress tolerance and biotic stress resistance in Arabidopsis. J. Integr. Plant Biol. 57, 628–640. doi: 10.1111/jipb.12302
Shidore, T., Dinse, T., Öhrlein, J., Becker, A., and Reinhold-Hurek, B. (2012). Transcriptomic analysis of responses to exudates reveal genes required for rhizosphere competence of the endophyte Azoarcus sp. strain BH72. Environ. Microbiol. 14, 2775–2787. doi: 10.1111/j.1462-2920.2012.02777.x
Shidore, T., and Triplett, L. R. (2017). Toxin-antitoxin systems: implications for plant disease. Annu. Rev. Phytopathol. 55, 161–179. doi: 10.1146/annurev-phyto-080516-035559
Si, M., Zhao, C., Burkinshaw, B., Zhang, B., Wei, D., Wang, Y., et al. (2017). Manganese scavenging and oxidative stress response mediated by type VI secretion system in Burkholderia thailandensis. Proc. Natl. Acad. Sci. U.S.A. 114, E2233–E2242. doi: 10.1073/pnas.1614902114
Siebert, M., Severin, K., and Heide, L. (1994). Formation of 4-hydroxybenzoate in Escherichia coli: characterization of the ubiC gene and its encoded enzyme chorismate pyruvate-lyase. Microbiology 140, 897–904. doi: 10.1099/00221287-140-4-897
Sigrell, J. A., Cameron, A. D., Jones, T. A., and Mowbray, S. L. (1998). Structure of Escherichia coli ribokinase in complex with ribose and dinucleotide determined to 1.8 å resolution: insights into a new family of kinase structures. Structure 6, 183–193. doi: 10.1016/S0969-2126(98)00020-3
Sleator, R. D., and Hill, C. (2002). Bacterial osmoadaptation: the role of osmolytes in bacterial stress and virulence. FEMS Microbiol. Rev. 26, 49–71. doi: 10.1111/j.1574-6976.2002.tb00598.x
Smith, J. A., and Magnuson, R. D. (2004). Modular organization of the Phd repressor/antitoxin protein. J. Bacteriol. 186, 2692–2698. doi: 10.1128/JB.186.9.2692-2698.2004
Sommer, D. D., Delcher, A. L., Salzberg, S. L., and Pop, M. (2007). Minimus: a fast, lightweight genome assembler. BMC Bioinformatics 8:64. doi: 10.1186/1471-2105-8-64
Spaepen, S., Vanderleyden, J., and Remans, R. (2007). Indole-3-acetic acid in microbial and microorganism-plant signaling. FEMS Microbiol. Rev. 31, 425–448. doi: 10.1111/j.1574-6976.2007.00072.x
Sprent, J. I., and de Faria, S. M. (1988). Mechanisms of infection of plants by nitrogen fixing organisms. Plant Soil 110, 157–165. doi: 10.1007/BF02226795
Strom, A. R., and Kaasen, I. (1993). Trehalose metabolism in Escherichia coli: stress protection and stress regulation of gene expression. Mol. Microbiol. 8, 205–210. doi: 10.1111/j.1365-2958.1993.tb01564.x
Ström, L. (1997). Root exudation of organic acids: importance to nutrient availability and the calcifuge and calcicole behaviour of plants. Oikos 80, 459–466. doi: 10.2307/3546618
Sundberg, R. J. (1984). “Pyrroles and their benzo derivatives: synthesis and applications,” in Comprehensive Heterocyclic Chemistry, eds A. R. Katritzky and C. W. Rees (Oxford: Pergamon), 313–376. doi: 10.1016/b978-008096519-2.00056-4
Taghavi, S., van der Lelie, D., Hoffman, A., Zhang, Y.-B., Walla, M. D., Vangronsveld, J., et al. (2010). Genome sequence of the plant growth promoting endophytic bacterium Enterobacter sp. 638. PLoS Genet. 6:e1000943. doi: 10.1371/journal.pgen.1000943
Taylor, P. K., Zhang, L., and Mah, T.-F. (2019). Loss of the two-component system TctD-TctE in Pseudomonas aeruginosa affects biofilm formation and aminoglycoside susceptibility in response to citric acid. mSphere 4, e00102–e00119. doi: 10.1128/mSphere.00102-19
Tseng, T.-T., Tyler, B. M., and Setubal, J. C. (2009). Protein secretion systems in bacterial-host associations, and their description in the Gene Ontology. BMC Microbiol. 9:S2. doi: 10.1186/1471-2180-9-S1-S2
van der Lelie, D., Taghavi, S., Monchy, S., Schwender, J., Miller, L., Ferrieri, R., et al. (2009). Poplar and its bacterial endophytes: coexistence and harmony. Crit. Rev. Plant Sci. 28, 346–358. doi: 10.1080/07352680903241204
Vishwakarma, K., Upadhyay, N., Kumar, N., Yadav, G., Singh, J., Mishra, R. K., et al. (2017). Abscisic acid signaling and abiotic stress tolerance in plants: a review on current knowledge and future prospects. Front. Plant Sci. 8:161. doi: 10.3389/fpls.2017.00161
Vodyanitskii, Y. N. (2010). Zinc forms in soils (Review of publications). Eurasian Soil Sci. 43, 269–277. doi: 10.1134/S106422931003004X
von Wintzingerode, F., Rainey, F. A., Kroppenstedt, R. M., and Stackebrandt, E. (1997). Identification of environmental strains of Bacillus mycoides by fatty acid analysis and species-specific 16S rDNA oligonucleotide probe. FEMS Microbiol. Ecol. 24, 201–209. doi: 10.1111/j.1574-6941.1997.tb00437.x
Walker, T. S., Bais, H. P., Halligan, K. M., Stermitz, F. R., and Vivanco, J. M. (2003). Metabolic profiling of root exudates of Arabidopsis thaliana. J. Agric. Food Chem. 51, 2548–2554. doi: 10.1021/jf021166h
Wang, D., Hosteen, O., and Fierke, C. A. (2012). ZntR-mediated transcription of zntA responds to nanomolar intracellular free zinc. J. Inorg. Biochem. 111, 173–181. doi: 10.1016/j.jinorgbio.2012.02.008
Wang, Q., Dodd, I. C., Belimov, A. A., and Jiang, F. (2016). Rhizosphere bacteria containing 1-aminocyclopropane-1-carboxylate deaminase increase growth and photosynthesis of pea plants under salt stress by limiting Na+ accumulation. Funct. Plant Biol. 43, 161–172. doi: 10.1071/FP15200
Wang, T., Si, M., Song, Y., Zhu, W., Gao, F., Wang, Y., et al. (2015). Type VI secretion system transports Zn2+ to combat multiple stresses and host immunity. PLoS Pathog. 11:e1005020. doi: 10.1371/journal.ppat.1005020
Wang, Y., Li, L., Cui, W., Xu, S., Shen, W., and Wang, R. (2012). Hydrogen sulfide enhances alfalfa (Medicago sativa) tolerance against salinity during seed germination by nitric oxide pathway. Plant Soil 351, 107–119. doi: 10.1007/s11104-011-0936-2
Wanner, B. L. (1996). “Phosphorus assimilation and control of the phosphate regulon,” in Escherichia Coli and Salmonella: Cellular and Molecular Biology, Vol. 41, 2nd Edn, F. C. Neidhardt RCI, J. L Ingraham, E. C. C Lin, K. B Low, B. Magasanik, W. S Reznikoff, et al. (Washington, DC: ASM Press), 1357–1381.
Watzlawick, H., Morabbi Heravi, K., and Altenbuchner, J. (2016). Role of the ganSPQAB operon in degradation of galactan by Bacillus subtilis. J. Bacteriol. 198, 2887–2896. doi: 10.1128/jb.00468-16
Weber, B., Hasic, M., Chen, C., Wai, S. N., and Milton, D. L. (2009). Type VI secretion modulates quorum sensing and stress response in Vibrio anguillarum. Environ. Microbiol. 11, 3018–3028. doi: 10.1111/j.1462-2920.2009.02005.x
Weber, T., Blin, K., Duddela, S., Krug, D., Kim, H. U., Bruccoleri, R., et al. (2015). AntiSMASH 3.0—a comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic Acids Res. 43, W237–W243. doi: 10.1093/nar/gkv437
Weisskopf, L., Ryu, C.-M., Raaijmakers, J., and Garbeva, P. (2016). Editorial: smelly fumes – volatile-mediated communication between bacteria and other organisms. Front. Microbiol. 7:2031. doi: 10.3389/fmicb.2016.02031
White, J. G., and Zasoski, R. J. (1999). Mapping soil micronutrients. Field Crops Res. 60, 11–26. doi: 10.1016/S0378-4290(98)00130-0
Wisselink, H. W., Mars, A. E., van der Meer, P., Eggink, G., and Hugenholtz, J. (2004). Metabolic engineering of mannitol production in Lactococcus lactis: influence of overexpression of mannitol 1-phosphate dehydrogenase in different genetic backgrounds. Appl. Environ. Microbiol. 70, 4286–4292. doi: 10.1128/aem.70.7.4286-4292.2004
Wood, J. M. (1988). Proline porters effect the utilization of proline as nutrient or osmoprotectant for bacteria. J. Membr. Biol. 106, 183–202. doi: 10.1007/BF01872157
Xie, G., Bruce, D. C., Challacombe, J. F., Chertkov, O., Detter, J. C., Gilna, P., et al. (2007). Genome sequence of the cellulolytic gliding bacterium Cytophaga hutchinsonii. Appl. Environ. Microbiol. 73, 3536–3546. doi: 10.1128/aem.00225-07
Yamaguchi, Y., and Inouye, M. (2009). Chapter 12 mRNA Interferases, sequence-specific endoribonucleases from the toxin–antitoxin systems. Prog. Mol. Biol. Transl. Sci. 85, 467–500. doi: 10.1016/S0079-6603(08)00812-X
Yamaguchi, Y., Park, J.-H., and Inouye, M. (2011). Toxin-antitoxin systems in bacteria and archaea. Annu. Rev. Genet. 45, 61–79. doi: 10.1146/annurev-genet-110410-132412
Yang, Y., Yuan, C., Dou, J., Han, X., Wang, H., Fang, H., et al. (2014). Recombinant expression of glpK and glpD genes improves the accumulation of shikimic acid in E. coli grown on glycerol. World J. Microbiol. Biotechnol. 30, 3263–3272. doi: 10.1007/s11274-014-1753-6
Yaron, S., and Römling, U. (2014). Biofilm formation by enteric pathogens and its role in plant colonization and persistence. Microb. Biotechnol. 7, 496–516. doi: 10.1111/1751-7915.12186
Yuan, Z.-C., Zaheer, R., and Finan, T. M. (2006). Regulation and properties of PstSCAB, a high-affinity, high-velocity phosphate transport system of Sinorhizobium meliloti. J. Bacteriol. 188, 1089–1102. doi: 10.1128/jb.188.3.1089-1102.2006
Zangoui, P., Vashishtha, K., and Mahadevan, S. (2015). Evolution of aromatic β-glucoside utilization by successive mutational steps in Escherichia coli. J. Bacteriol. 197, 710–716. doi: 10.1128/jb.02185-14
Zeidler, D., Zähringer, U., Gerber, I., Dubery, I., Hartung, T., Bors, W., et al. (2004). Innate immunity in Arabidopsis thaliana: lipopolysaccharides activate nitric oxide synthase (nos) and induce defense genes. Proc. Natl. Acad. Sci. U.S.A. 101, 15811–15816. doi: 10.1073/pnas.0404536101
Zeng, L., Das, S., and Burne, R. A. (2010). Utilization of lactose and galactose by Streptococcus mutans transport, toxicity, and carbon catabolite repression. J. Bacteriol. 192, 2434–2444. doi: 10.1128/jb.01624-09
Zhang, F., Romheld, V., and Marschner, H. (1991). Release of zinc mobilizing root exudates in different plant species as affected by zinc nutritional status. J. Plant Nutr. 14, 675–686. doi: 10.1080/01904169109364234
Zhao, X., and Drlica, K. (2014). Reactive oxygen species and the bacterial response to lethal stress. Curr. Opin. Microbiol. 21, 1–6. doi: 10.1016/j.mib.2014.06.008
Zhou, Y., Liang, Y., Lynch, K. H., Dennis, J. J., and Wishart, D. S. (2011). PHAST: a fast phage search tool. Nucleic Acids Res. 39(Suppl. 2), W347–W352. doi: 10.1093/nar/gkr485
Keywords: desert bacteria, plant growth promotion, genome, abiotic stress, volatile compounds, biocontrol, Phytophthora infestans
Citation: Eida AA, Bougouffa S, L’Haridon F, Alam I, Weisskopf L, Bajic VB, Saad MM and Hirt H (2020) Genome Insights of the Plant-Growth Promoting Bacterium Cronobacter muytjensii JZ38 With Volatile-Mediated Antagonistic Activity Against Phytophthora infestans. Front. Microbiol. 11:369. doi: 10.3389/fmicb.2020.00369
Received: 12 July 2019; Accepted: 19 February 2020;
Published: 11 March 2020.
Edited by:
Brigitte Mauch-Mani, Université de Neuchâtel, SwitzerlandReviewed by:
Ben Fan, Nanjing Forestry University, ChinaKhaled Abbas El-Tarabily, United Arab Emirates University, United Arab Emirates
Copyright © 2020 Eida, Bougouffa, L’Haridon, Alam, Weisskopf, Bajic, Saad and Hirt. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Maged M. Saad, bWFnZWQuc2FhZEBrYXVzdC5lZHUuc2E=
 Abdul Aziz Eida
Abdul Aziz Eida Salim Bougouffa
Salim Bougouffa Floriane L’Haridon4
Floriane L’Haridon4 Intikhab Alam
Intikhab Alam Laure Weisskopf
Laure Weisskopf Vladimir B. Bajic
Vladimir B. Bajic Maged M. Saad
Maged M. Saad Heribert Hirt
Heribert Hirt