- Department of Microbiology, Faculty of Medicine Siriraj Hospital, Mahidol University, Bangkok, Thailand
Candidemia, a bloodstream infection caused by genus Candida, has a high mortality rate. Candida albicans was previously reported to be the most common causative species among candidemia patients. However, during the past 10 years in Thailand, Candida tropicalis has been recovered from blood more frequently than C. albicans. The cause of this shift in the prevalence of Candida spp. remains unexplored. We conducted in vitro virulence studies and antifungal susceptibility profiles of 48 C. tropicalis blood isolates collected during 2015–2017. To compare to global isolates of C. tropicalis, multilocus sequence typing (MLST), a minimum spanning tree, and an eBURST analysis were also conducted. C. tropicalis and C. albicans were the most (47–48.7%) and second-most (21.5–33.9%) common species to be isolated from candidemia patients, respectively. Of the C. tropicalis blood isolates, 29.2, 0, 100, and 93.8% exhibited proteinase activity, phospholipase activity, hemolytic activity, and biofilm formation, respectively. Moreover, 20.8% (10/48) of the isolates were resistant to voriconazole and fluconazole, and also showed high minimum inhibitory concentrations (MICs) to posaconazole and itraconazole. In contrast, most of the isolates were susceptible to anidulafungin (97.9%), micafungin (97.9%), and caspofungin (97.9%), and showed low MICs to amphotericin B (100%) and 5-flucytosine (100%). The MLST identified 22 diploid sequence types. Based on the eBURST analysis and minimum spanning tree, 9 out of 13 members (69.2%) of an eBURST group 3 were resistant to voriconazole and fluconazole, and also showed high MICs to posaconazole and itraconazole. Association analysis revealed the eBURST group 3 was significantly associated with the four triazole resistance (p < 0.001). In conclusion, the eBURST group 3 was associated with the triazole resistance and resistance to many antifungal drugs might be collectively responsible for the prevalence shift.
Introduction
Candida species is a cause of mild superficial to serious invasive infections in humans worldwide. The invasive infection often leads to significant morbidity and mortality, particularly with immunocompromised patients (Brown et al., 2012). Candida spp. was ranked as the fourth most frequent cause of bloodstream infections with a high mortality rate (Wisplinghoff et al., 2004). Several predisposing factors have been identified for candidemia in hospitalized patients, including parental nutrition, central venous catheterization, organ transplantation, usage of broad-spectrum antibiotics, and longer intensive care unit hospitalization (Kibbler, 1996; Conde-Rosa et al., 2010; Yapar, 2014).
Previously, Candida albicans was globally reported to be the most common causative species isolated from the blood of candidemia patients (Guinea, 2014). A large surveillance study in 1997–2016 reported that 46.3–57.4% of all candidemia cases were caused by C. albicans whereas only 8.3–10.7% were caused by Candida tropicalis (Pfaller et al., 2019). Although considered to be a less prevalent candidemia-causing species, bloodstream infections due to C. tropicalis are continuing to rise globally (Guinea, 2014). In the Asian-Pacific and Latin American regions, C. tropicalis has been ranked as the first and second most prevalent pathogenic Candida species, respectively (Tan et al., 2015; Doi et al., 2016). The mechanism for the increasing prevalence of C. tropicalis remains unclear. However, one report revealed that C. tropicalis has a higher rate of fluconazole resistance than C. albicans (Khairat et al., 2019). An investigation in Southern India, C. tropicalis a predominant species (54.3%), also demonstrated that the ability to develop rapid resistance to fluconazole involves the increasing prevalence (YeSudhaSon and MohanraM, 2015).
Resistance to antifungal agents and strong virulence phenotypes of C. tropicalis have been reported (Kothavade et al., 2010; Deorukhkar et al., 2014; Fan et al., 2017). Although Candida spp. is generally susceptible to most antifungal drugs, the SENTRY Antimicrobial Surveillance Program reported a high fluconazole resistance rate (9.2%) among C. tropicalis isolates in Asia-Pacific (Pfaller et al., 2019). Remarkable increasing trends of triazole resistance have also been found in China and Taiwan (Chou et al., 2007; Fan et al., 2017). As a human pathogen, C. tropicalis secretes hydrolytic enzymes, namely, proteinase and phospholipase, that digest the host cell membrane, resist phagocytosis, and invade tissues (Naglik et al., 2003). In fact, one study showed a strong proteinase and phospholipase production by C. tropicalis isolated from various clinical specimens (Deorukhkar et al., 2014). Hemolysin is another group of enzymes in which C. tropicalis can secrete to lyse human red blood cells. The hemoglobin released from the lysed red blood cells is later used as an iron source to facilitate hyphal penetration and yeast dissemination (Furlaneto et al., 2015). A previous report demonstrated that all C. tropicalis isolated from the bloodstream and urinary tract infections were able to express hemolytic activity (Negri et al., 2010).
Biofilm formation is also found to play a major role in Candida pathogenesis by protecting the pathogenic yeast from the host immune cells and causing resistance to antifungal treatment. Moreover, the biofilm formation also facilitates Candida adherence to indwelling medical devices such as vascular catheters, artificial joints, and cardiac devices (Cavalheiro and Teixeira, 2018). Evidence of C. tropicalis biofilm formation was provided by a Spain study in which 72.7% of the C. tropicalis isolates from both sterile and non-sterile sites were found to be high biofilm producers (Guembe et al., 2017).
Multilocus sequence typing (MLST) has recently been utilized to analyze the pattern of genetic variation of C. tropicalis (Tavanti et al., 2005). To date, 543 allele types (AT) and 914 diploid sequence types (DST) have been deposited in the C. tropicalis MLST database1. Studies of C. tropicalis in Kuwait and China have identified more than 59 DSTs of 63 isolates and 94 DSTs of 116 isolates, respectively (Al-Obaid et al., 2017; Wu et al., 2017). Moreover, associations of MLST genotypes with virulence phenotypes/antimicrobial susceptibility patterns have been reported. For example, a study in China showed a significant association between biofilm formation and MLST groups (Yu et al., 2017). Molecular epidemiology in Taiwan has also found a correlation between an MLST cluster and fluconazole resistance (Chou et al., 2007). Furthermore, the relationship between epidemiological data and microsatellite markers illustrated that an increase of C. tropicalis resistance to 5-flucytosine was brought by widespread of a flucytosine-resistant clone among hospitalized patients in Paris (Desnos-Ollivier et al., 2008). However, despite the high burden of C. tropicalis infections, information on the molecular epidemiology of C. tropicalis in Asia is fairly limited.
As a mechanism of the increased prevalence of C. tropicalis in Thailand remains controversial, we investigated the molecular epidemiology of C. tropicalis recovered from blood cultures at a tertiary care hospital in Thailand. The virulence characteristics and antifungal susceptibilities were also reported. The relationships between MLST-based genetic clusters and antimicrobial susceptibility testing were also described.
Materials and Methods
Isolates Collection
Upon approval by the Siriraj Institutional Review Board (COA number: SI 091/2016), information on all Candida isolates from positive hemocultures during 2015–2017 were collected from the Department of Microbiology, Faculty of Medicine Siriraj Hospital, Mahidol University, Bangkok, Thailand. To reduce selection bias, two-three C. tropicalis blood isolates per month were randomly collected between August 2015 and May 2017 from a culture collection of the diagnostic microbiology laboratory, Department of Microbiology, Faculty of Medicine, Siriraj Hospital. Finally, a total of viable 48 isolates of the C. tropicalis blood isolates was included for a molecular epidemiological study. Species identification of Candida was primarily performed by using CHROMagar Candida chromogenic media (Oxoid, Basingstoke, United Kingdom) and Remel RapIDTM Yeast Plus System (Thermo Fisher Scientific, Waltham, MA, United States). To confirm the species-level identification, internal transcribed spacer (ITS) regions of the 48 studied isolates were amplified and analyzed as previously described (Pharkjaksu et al., 2018).
Investigation of in vitro Virulence Factors
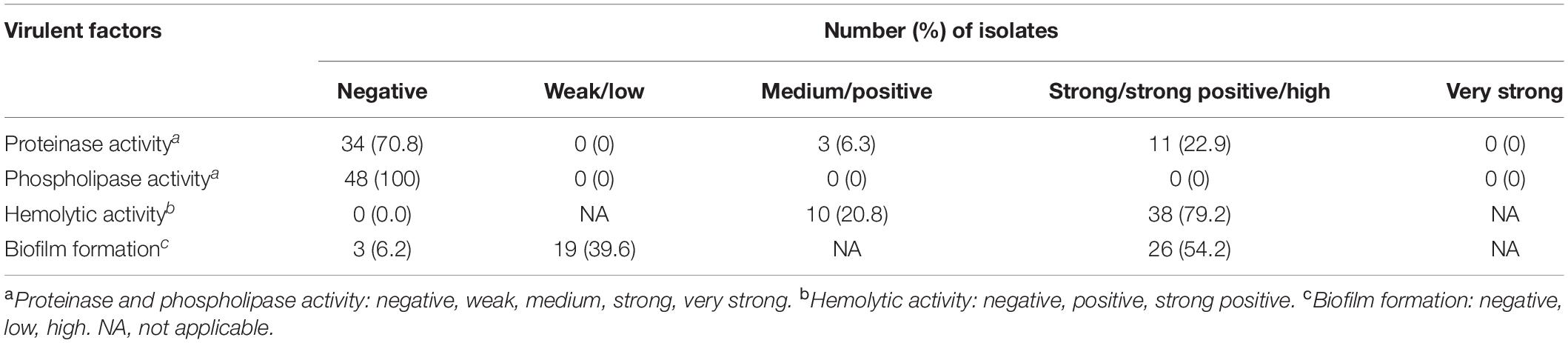
Four virulence factors, comprising phospholipase production, proteinase secretion, hemolysin production, and biofilm formation, were measured in triplicates (Pham et al., 2019). The phospholipase, proteinase, and hemolysin productions were tested by inoculating yeast cells onto an egg yolk medium, a bovine serum albumin (BSA) medium, and a sugar-enriched sheep blood medium, respectively (Sachin et al., 2012). First, the yeast isolates were pregrown on Sabouraud dextrose agar (SDA) at 30°C for 48 h before being suspended in a phosphate buffer to make a yeast suspension of 108 CFU/ml. Five microliters of the suspension was spotted onto the egg yolk medium (1.48% w/v CaCl2.2H2O, 11.7% w/v NaCl, 13% w/v SDA, and 10% v/v egg yolk), the BSA medium (0.01% w/v yeast extract, 0.2% w/v BSA, 1.17% w/v yeast carbon base, and 2% w/v agar), and the sugar-enriched sheep blood medium (1% w/v peptone, 1% w/v yeast extract, 5% w/v sheep blood, 7% w/v dextrose, and 2% w/v agar), and then incubated at 37°C for 48 h. Production of extracellular phospholipase and proteinase was observed by a precipitation zone around the colony and a clear halo zone surrounding the colony, respectively. Production of proteinase and phospholipase was classified into level by the ratio of colony diameter to the diameter of precipitation or clear zone (Pz) as follows: Pz ≤ 0.69, very strong; 0.70 ≤ Pz < 0.79, strong; 0.80 ≤ Pz < 0.89, medium; 0.90 ≤ Pz < 0.99, weak; and Pz = 1, negative. For the hemolysin production, a distinct translucent halo zone around the colony indicated hemolytic activity. Hemolytic activity was measured based on the ratio of the colony diameter to the diameter of the translucent halo zone (Pz) as follows: Pz < 0.64, strongly positive; 0.64 ≤ Pz < 1.00, positive; and Pz = 1, negative.
Biofilm formation of C. tropicalis was measured by an XTT [2,3-bis(2-methoxy-4-nitro-5-sulfophenyl)-2H-tetrazolium-5- carboxanilide] reduction assay (Loures and Levitz, 2015). C. tropicalis, pregrown on SDA at 30°C for 48 h, was used to prepare a cell suspension of 108 CFU/ml in a yeast extract peptone dextrose broth (1% w/v yeast extract, 2% w/v peptone, and 1% w/v dextrose). One hundred microliters of the suspension were seeded into each well of a flat-bottomed, 96-well plate and incubated at 37°C for 48 h. Non-adherence cells were removed by washing twice with phosphate buffer. The remaining adhered cells were measured for biofilm formation by adding 50 μl of XTT and phenazine methosulfate mixture. After further incubation for 2 h in the dark, the quantity of the biofilm layer was assessed by absorbance at 490 nm. Finally, biofilm formation was classified as: O.D. value > GM (geometric mean) = high biofilm formation, O.D. value ≤ GM = low biofilm formation, and O.D. value < 0.10 = negative biofilm formation (Li et al., 2003).
Antifungal Susceptibility Testing
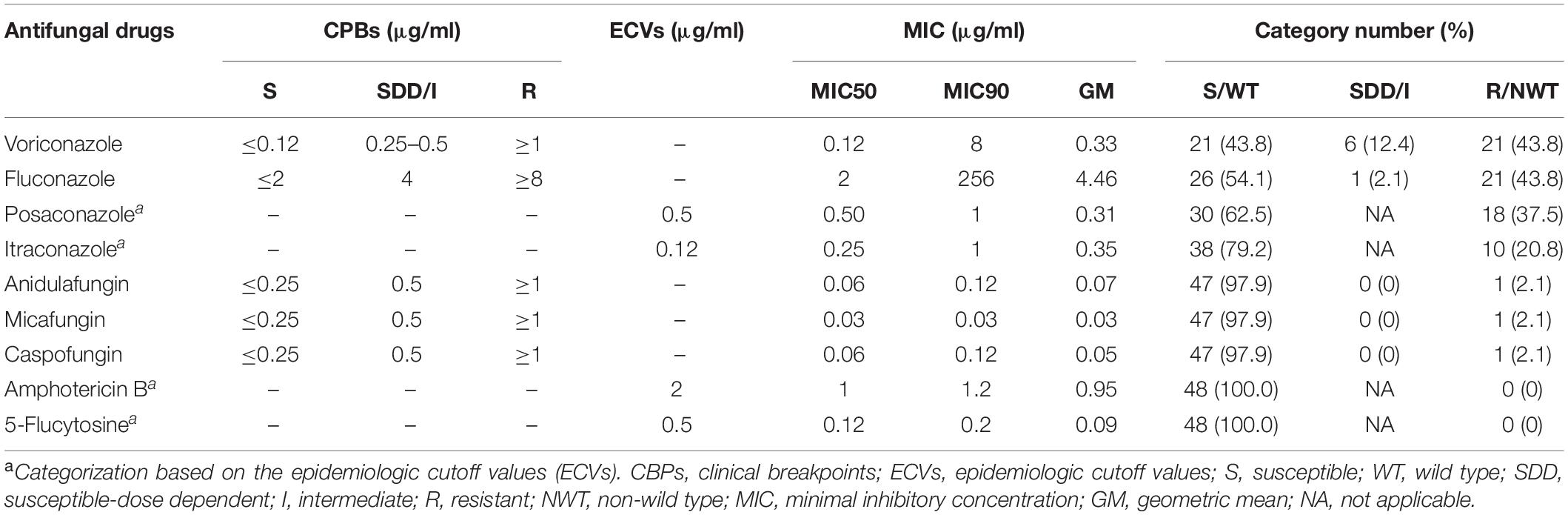
The susceptibility of C. tropicalis to nine antifungal drugs (fluconazole, voriconazole, itraconazole, posaconazole, 5-flucytosine, anidulafungin, micafungin, caspofungin, and amphotericin B) was determined by using Sensititre YeastOne YO10 (SYO; Thermo Fisher Scientific, Waltham, MA, United States), a colorimetric microdilution method, according to the manufacturer’s instructions. Briefly, 20 μl of 0.5 McFarland yeast suspension was transferred into 11 ml of YeastOne inoculum broth to obtain a concentration of 1.5–8 × 103 CFU/ml. Then, 100 μl of the inoculum was inoculated into each well of a YeastOne susceptibility plate. The concentrations of each drug ranged as follows: fluconazole, 0.12–256 μg/ml; voriconazole, 0.008–8 μg/ml; itraconazole, 0.015–16 μg/ml; posaconazole, 0.008–8 μg/ml; 5-flucytosine, 0.06–64 μg/ml; anidulafungin, 0.015–8 μg/ml; micafungin, 0.008–8 μg/ml; caspofungin, 0.008–8 μg/ml; and amphotericin B, 0.12–8 μg/ml. After 24 h of incubation at 35°C, the minimum inhibitory concentration (MIC) was determined from the change of the colorimetric growth indicator according to the manufacturer recommendations. Candida parapsilosis ATCC 20019 and Candida krusei ATCC 6258 were used as quality controls. The results were interpreted following the recommendations of Clinical Laboratory Standards Institute documents M27-S4 (Clinical and Laboratory Standards Institute, 2012): clinical breakpoints (CBPs) for fluconazole, MIC ≤ 2 μg/ml susceptible, MIC = 4 μg/ml susceptible-dose dependent, MIC ≥ 8 μg/ml resistant; voriconazole, MIC ≤ 0.12 μg/ml susceptible, MIC 0.25–0.5 μg/ml susceptible-dose dependent, MIC ≥ 1 μg/ml resistant; and anidulafungin, micafungin, and caspofungin, MIC ≤ 0.25 μg/ml susceptible, MIC 0.5 μg/ml intermediate, MIC ≥ 1 μg/ml resistant. For antifungal agents which no CBPs, epidemiological cutoff values (ECVs) assigned by CLSI document M59 were used: itraconazole = 0.5 μg/ml, posaconazole = 0.12 μg/ml, and amphotericin B = 2 μg/ml. As CLSI CBP and ECV for 5-flucytosine was not available, ECV = 0.5 μg/ml was used according to a previously published report (Cantón et al., 2012). To ensure the SYO result was not compromised, 38 of the 48 isolates (including all 32 azole-resistant isolates and 6 azole-susceptible isolates) were tested for their MIC level to fluconazole and itraconazole by the original CLSI methods. According to a previous report (Cantón et al., 2012), 82.1% of ECVs estimated by the SYO method was equal to or within one two-fold dilution of those reported for the CLSI method. Our SYO result showed that 100% of fluconazole and 97.3% of itraconazole results were equal to or within one two-fold dilution of those reported for the CLSI method. Therefore, we believed our SYO result was not compromised.
Multilocus Sequence Typing Analysis
Candida tropicalis was pregrown on SDA at 30°C for 48 h, after which genomic DNA was extracted by a standard phenol-chloroform procedure (Danesi et al., 2014). Amplifications of six housekeeping genes—ICL1, MDR1, SAPT2, SAPT4, XYR1, and ZWF1a—were performed according to a previously described protocol (Tavanti et al., 2005). Subsequently, the purified PCR products of each gene were used for bidirectional sequencing using forward and reverse primers from Axil Scientific Pte. Ltd., Singapore. The sequencing results were edited by MEGA7 software2. With each fragment, the forward and reverse sequence chromatograms were visually examined for a strong overlapping peak to define heterozygosity. Each heterozygous position was transformed into a degenerate nucleotide according to the IUPAC nucleotide code. Six sequence fragments of the housekeeping genes of each isolate were compared with information on the C. tropicalis MLST database3 to define AT and DST. The new allelic profiles and new allele combinations were submitted to a curator of the database, Hsiu-Jung Lo, to verify and assign new allele numbers and DSTs, respectively.
Phylogenetic Analysis
The edited nucleotide sequences of six fragments were concatenated into a single sequence by the MEGA7 software. Degenerate nucleotides were pre-modified for analysis as previously described (Tavanti et al., 2005). The phylogenetic analysis of the 48 C. tropicalis clinical isolates was conducted by the unweighted pair group method with arithmetic average (UPGMA) using the MEGA7 software. To illustrate the phylogenetic relatedness between our 48 isolates and global isolates deposited in the database (accessed in August 2019), a minimum spanning tree was constructed using BioNumerics software version 7.6.3 (Applied Maths, Austin, TX, United States). Each genotype cluster was identified by using the goeBURST algorithm version 1.2.14. Isolates were grouped when five out of six alleles were identical (Francisco et al., 2009).
Statistical Analysis
The statistical analysis was performed with PASW Statistic for Windows (SPSS Inc., Chicago, IL, United States). An association between eBURST group, antifungal susceptibility, and in vitro virulence phenotypes was determined using a Chi-square test. Likewise, the significance of the difference in in vitro virulence expression and antifungal susceptibility between C. albicans and C. tropicalis was also determined by the Chi-square test. Finally, post hoc analysis using nQuery Advisor (Elashoff, 2000) to ensure a sufficient sample size was performed by estimation of a type II error in the association analysis. A sufficient sample size was achieved when power for a two-sided test reached 90% at a significant level of 5%. The p-value of the Chi-square test was obtained from the web-based calculator5. A p-value of <0.05 was considered to be significant.
Results
In vitro Virulence Phenotypes Among C. tropicalis Blood Isolates
Interestingly, none of the isolates could produce phospholipase, and only 29.2% (14/48 isolates) were able to produce proteinase. In contrast, all of the isolates were able to produce hemolysin, and almost all (93.8%) of the isolates could form biofilm. For hemolysin, 79.2% (38/48 isolates) had strong hemolytic activity. A high biofilm formation was found in 54.2% (26/48 isolates) of the isolates (Table 1).
Antifungal Susceptibility of C. tropicalis Blood Isolates
It was found that 43.8, 43.8, 37.5, and 20.8% of the isolates were resistant to voriconazole and fluconazole, and also showed high MICs to posaconazole and itraconazole, respectively. The MIC90 value of fluconazole was extremely high (256 μg/ml). In contrast, none of the isolates showed high MICs to amphotericin B and 5-flucytosine (Table 2). Only a few isolates were resistant to anidulafungin (2.1%), micafungin (2.1%), and caspofungin (2.1%).
Azole Drug Resistance Isolates and the UPGMA-Based Cluster
According to the C. tropicalis MLST database, a total of 24 DSTs were identified. Seven DSTs (DST94, DST139, DST225, DST434, DST506, DST522, and DST754; 22 isolates) were already present in the database, while 17 DSTs (26 isolates) were newly identified in this study (Figure 1). Based on the UPGMA dendrogram, most isolates (69.2%, 9/13 isolates) with resistance to voriconazole and fluconazole, and high MICs to posaconazole and itraconazole were on the same cluster (Figure 1). Further analysis with the goeBURST program showed that the azole-resistance cluster belonged to the eBURST group 3 with a high resistance/non-wild type rate of 69.2% (9/13 isolates) while the low rate of 2.9% (1/35 isolates) was found in the non-eBURST group 3 isolates (Table 3 and Figure 1). Other eBURST groups and singletons are illustrated in the Figures 1, 2. The statistical analysis also revealed a significant association between the eBURST group 3 isolates and the four drugs resistance (Table 3; p < 0.001). The post hoc analysis indicated that the 48 studied isolates of C. tropicalis were sufficient for demonstrating the association between the four triazole resistance and eBURST cluster 3. In contrast, there was no significant correlation between the eBURST group 3 isolates with any virulence phenotypes. Other studies also reported that Asia was the origin of most eBURST group 3 isolates (Supplementary Figure S1).
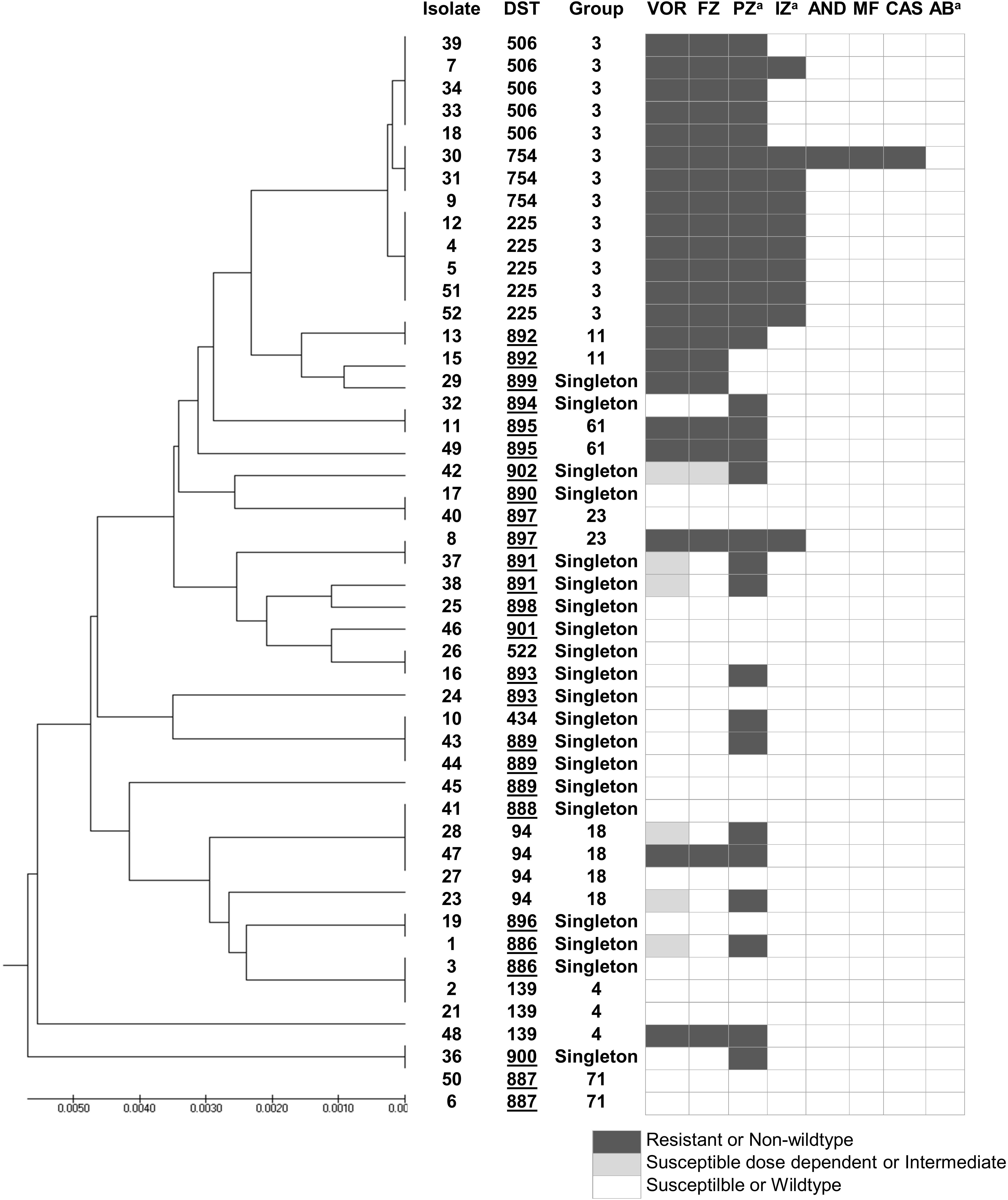
Figure 1. A UPGMA dendrogram based on MLST of six gene fragments against antifungal susceptibility pattern of 48 C. tropicalis isolates causing candidemia in Thailand. aSusceptibility categorized based on epidemiologic cutoff values; underlined DST numbers, new DST identified in this study; Group, group defined by goeBURST; black box, resistant or non-wild type; gray box, susceptible dose dependent or intermediate; white box, susceptible or wild type. DST, diploid sequence type; VOR, voriconazole; IZ, itraconazole; FZ, fluconazole; AND; anidulafungin; MF, micafungin; CAS, caspofungin; AB, amphotericin B; PZ, posaconazole.
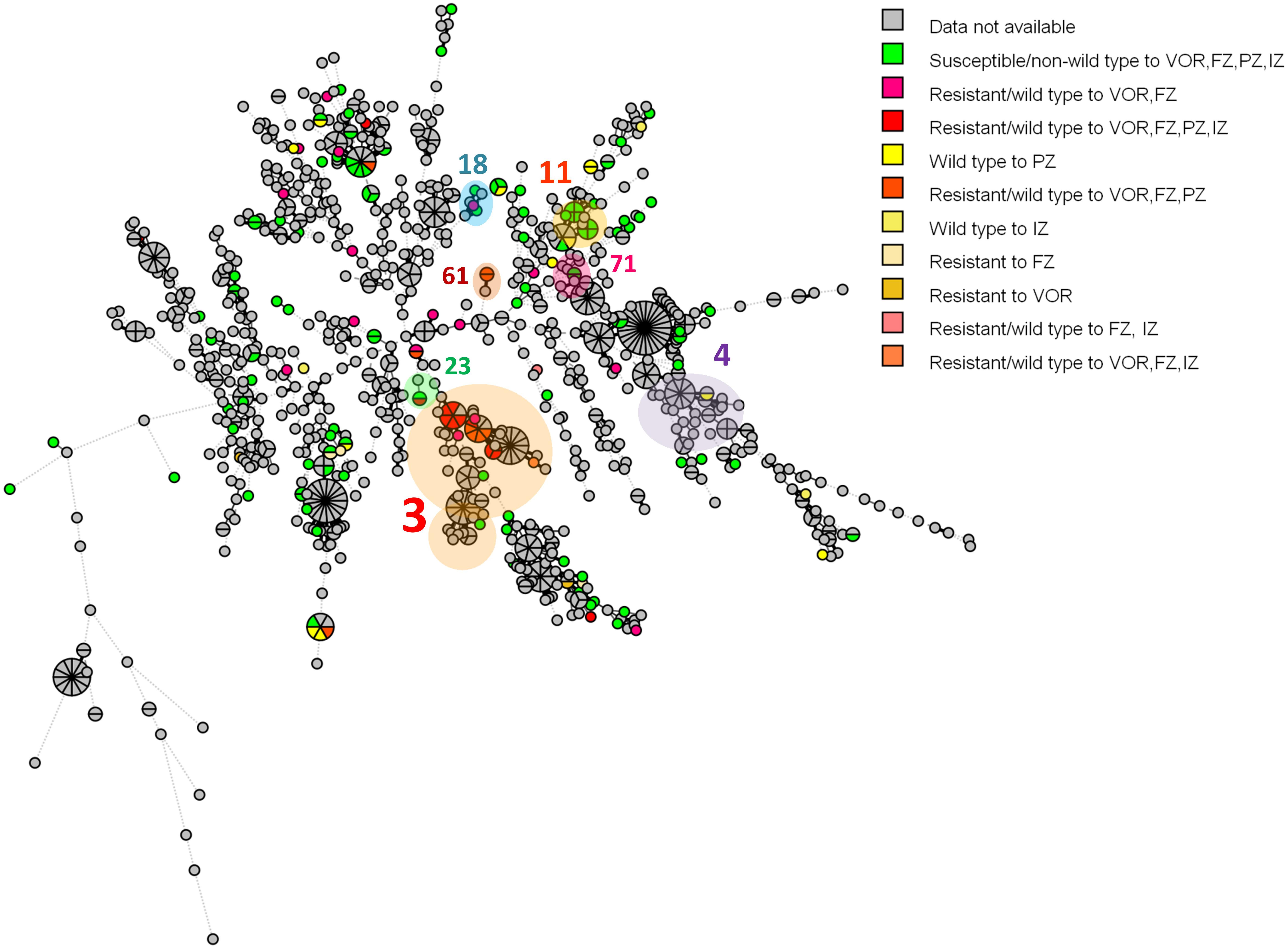
Figure 2. A minimum spanning tree illustrating the relationship between the 48 C. tropicalis isolates from Thailand and 1,019 isolates from other countries available from the C. tropicalis MLST database as of August 2019. Each circle corresponds to a unique DST; the number outside the circle indicates an eBURST cluster; the size of the circle represents the number of isolates belonging to the same DST; and the colors inside the circle represent voriconazole, fluconazole, posaconazole, and itraconazole susceptibility. VOR, voriconazole; FZ, fluconazole; PZ, posaconazole; IZ, itraconazole.
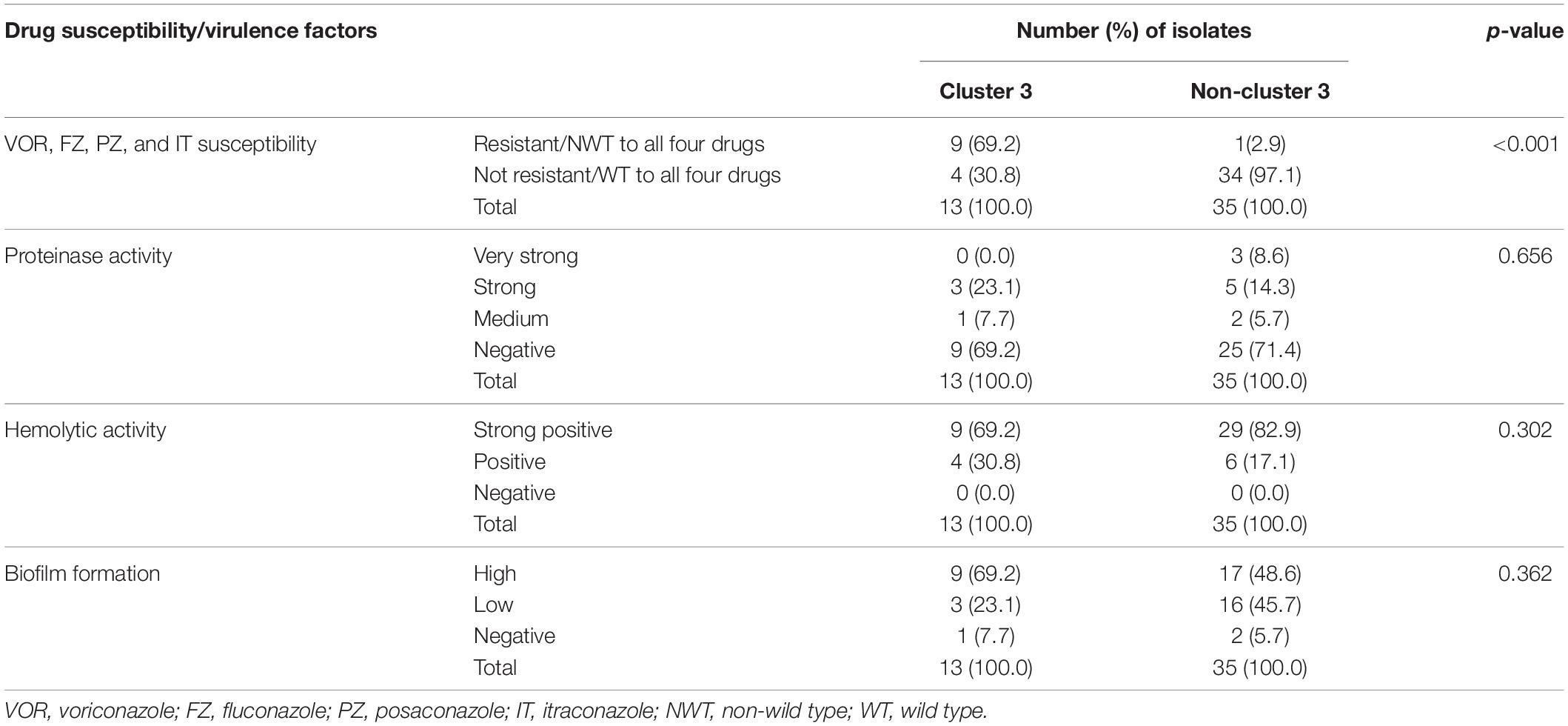
Table 3. Number of isolates in cluster 3 and non-cluster 3 in correlation to four azole drugs (voriconazole, fluconazole, posaconazole, and itraconazole) susceptibility and four virulence factors.
Discussion
During 2012–2018, a total of 2,048 Candida isolates were identified from positive blood cultures. C. tropicalis was ranked as the most common cause of candidemia (41.1–48.7%; mean 46.7%; Supplementary Figure S2). The prevalence of C. tropicalis causing candidemia among candidemia patients at Siriraj Hospital has increased considerably from 28.0% during 2006–2009 to 46.7% during 2012–2018 (Boonyasiri et al., 2013). A study reported that the prevalence in Thailand was almost 50% during 2010–2011 in Thailand (Tan et al., 2015). Compared with the average for other Asian countries (25.4%), the proportion of C. tropicalis among candidemia patients in Thailand was much higher. In fact, a large surveillance study of candidemia in Asia indicated that the proportion of C. tropicalis was higher in tropical regions (46.2%), such as India, Singapore, and Thailand, than that for temperate regions (18.9%) (Tan et al., 2015). Furthermore, our study demonstrated that C. tropicalis was the predominant cause of candidemia, followed by C. albicans. This shift in species distribution of invasive candidiasis has also been found in India and Pakistan (Farooqi et al., 2013; Chakrabarti et al., 2015).
The frequency of infections by C. tropicalis is increasing (Guinea, 2014). One study has reported that the disease characteristics and prior antifungal treatment are involved in the change in the prevalence of the non-albicans Candida (Makanjuola et al., 2018). Therefore, in vitro virulence and antifungal susceptibility studies of the C. tropicalis blood isolates were performed.
Previous findings have suggested that the ability to produce the two hydrolytic enzymes, to secrete the hemolytic enzyme, and to form the biofilm are key virulence factors that facilitate hematogenous infections by Candida spp. (Gokce et al., 2007; Deorukhkar et al., 2014). Although the secretion of phospholipase is considered a key contribution of host invasion, none of our studied isolates produced the enzyme. Our finding was similar to previous analysis in Candida spp. (Samaranayake et al., 1984) which reported that C. tropicalis has no ability to secrete phospholipase. Although another study reported a high percentage of C. tropicalis isolates secreted phospholipase, the isolates exhibited very low production (Galan-Ladero et al., 2010; Jiang et al., 2016). On the other hand, among non-albicans Candida spp., the highest production of phospholipase was found in C. tropicalis in another report (Deorukhkar et al., 2014). However, this might not be applicable in Thailand as a previous study showed more than 90% of the C. parapsilosis sensu stricto isolates produced phospholipase (Pharkjaksu et al., 2018). The variability of results might be brought by biological differences among isolates and sensitivity of phospholipase detection methods. In addition, previous exposure of antifungal drugs such as nystatin and amphotericin B causes a significant reduction of phospholipase activity (Anil and Samaranayake, 2003). Unfortunately, information on prior drug treatment was not evaluated in the present study.
Typically, the fluconazole-resistant rates of C. tropicalis only range from 1.1 to 2.5% in the United States and Europe. Although as high as a 10% triazole resistance rate has been reported in the Asia-Pacific region (Pfaller et al., 2019), the resistance of C. tropicalis was much higher in this study with one-fifth of isolates being resistant to voriconazole and fluconazole, and high MICs to posaconazole and itraconazole. Comparing to a previous study in Thailand during 1999–2002, the fluconazole-resistant rate of C. tropicalis substantially increased from 0 to 43.8% in this study (Foongladda et al., 2004). In contrast, a recent study reported a very low resistance/non-wild type rate of C. albicans to all antifungal agents (Pham et al., 2019). Because most antibiotics, including all the triazoles, has been available over the counter in Thailand as both topical and oral form for at least the past 40 years (Aswapokee et al., 1990; Hoge et al., 1998), the susceptible C. albicans in the skin and mucosal microbiota might be “selected out” by the overuse of triazoles. Finally, with the higher frequency of C. tropicalis isolates in skin and microbiota, the higher C. tropicalis candidemia is to be expected. However, this remains to be proven by further microbiota screening among the Thai population.
Multilocus sequence typing and phylogenetic analysis of the C. tropicalis isolates in this study showed high genetic diversity, with as many as 22 DSTs being identified from the 48 isolates in this study. As expected, an eBURST group associated with the four triazole resistance was found. As a previous study in Taiwan also reported a cluster associated with fluconazole resistance in Taiwan (Chou et al., 2007), we investigated if this would also be the case in Thailand. Interestingly, this eBURST group of the four azoles resistant isolates also included the resistant isolates from Taiwan and China identified previously (Chou et al., 2007; Fan et al., 2017). This implied that this eBURST group 3 is strongly associated with the triazole resistance of Asian C. tropicalis isolates.
Finally, the ability to express in vitro virulence and antifungal susceptibility pattern between C. albicans and C. tropicalis were compared. Information on in vitro virulence and antifungal susceptibility of 46 C. albicans blood isolates was retrieved from our recent study (Pham et al., 2019). By comparing to the C. albicans blood isolates which isolated at the same time, the C. tropicalis exhibited significantly more overall proteinase activity and stronger hemolytic activity. Moreover, the C. tropicalis blood isolates showed higher resistance to voriconazole and fluconazole, and higher MICs to posaconazole (Supplementary Tables S1, S2). In fact, a previous study of Candida spp. in Thailand in 1999–2002 reported that all C. tropicalis isolates were susceptible to fluconazole (Foongladda et al., 2004). This suggests the antifungal resistance of the C. tropicalis blood isolates did occur only recently. Unfortunately, the in vitro virulence has never been studied in Thailand before this study. However, the high hemolytic activity was reported in C. tropicalis blood isolates previously (Favero et al., 2014; Seneviratne et al., 2016) and the predominant MLST clade 17 of C. albicans blood isolates showed significant stronger hemolytic activity than the less common clade (Pham et al., 2019). These suggest in vitro virulence could somewhat influence the pathogenesis of the yeasts. Taken together, the higher in vitro virulence and antifungal drug resistance were collectively responsible for the prevalence shift of C. tropicalis among candidemia patients.
Conclusion
An increased prevalence of C. tropicalis among candidemia patients during the past 10 years has been reported. The azole resistance/high MICs isolates are strongly associated with the eBURST group 3 based on the MLST analysis. Finally, C. tropicalis blood isolates exhibited higher proteinase activity, hemolytic activity, and antifungal drug resistance/high MIC rates than the C. albicans blood isolates. These implied that these virulence phenotypes and antifungal resistance were collectively responsible for the prevalence shift of C. tropicalis. However, although the post hoc analysis confirmed sufficient isolates for the association analysis, the 48 isolates might not perfectly represent the molecular distribution of C. tropicalis in Thailand. Therefore, a further study with more isolates per time point is needed.
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material.
Ethics Statement
Upon approval by the Siriraj Institutional Review Board (COA number: SI 091/2016), information on all Candida isolates from positive hemocultures during 2012-2018 were collected from the Department of Microbiology, Faculty of Medicine Siriraj Hospital, Mahidol University, Bangkok, Thailand.
Author Contributions
PN and PC designed the study. OT and SP performed the experiments and analyzed the data. OT and PN wrote the manuscript. All authors read the manuscript.
Funding
This research project is supported by the Siriraj Research Fund, Grant Number (IO) R016033021 Faculty of Medicine Siriraj Hospital, Mahidol University.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.00934/full#supplementary-material
FIGURE S1 | Minimum spanning tree illustrating relationships between the 48 C. tropicalis isolates from Thailand and 1,019 isolates from other countries available from the C. tropicalis MLST database as of August 2019. Each circle corresponds to a unique DST; the number outside the circle indicates an eBURST cluster; the size of the circle represents the number of isolates belonging to the same DST; and the colors inside the circle represent the origin country of the C. tropicalis isolates.
FIGURE S2 | Species distribution of Candida species isolated from positive blood cultures at Siriraj Hospital during 2012–2018.
TABLE S1 | In vitro virulence comparison between C. tropicalis and C. albicans blood isolates.
TABLE S2 | Antifungal susceptibility comparison between C. tropicalis and C. albicans blood isolates.
Footnotes
- ^ http://pubmlst.org/ctropicalis, accessed in August 2019
- ^ http://www.megasoftware.net/
- ^ http://pubmlst.org/ctropicalis/
- ^ http://www.phyloviz.net/goeburst/
- ^ https://www.icalcu.com/stat/chisqtest.html
References
Al-Obaid, K., Asadzadeh, M., Ahmad, S., and Khan, Z. (2017). Population structure and molecular genetic characterization of clinical Candida tropicalis isolates from a tertiary-care hospital in Kuwait reveal infections with unique strains. PLoS One 12:e0182292. doi: 10.1371/journal.pone.0182292
Anil, S., and Samaranayake, L. (2003). Brief exposure to antimycotics reduces the extracellular phospholipase activity of Candida albicans and Candida tropicalis. Chemotherapy 49, 243–247. doi: 10.1159/000072448
Aswapokee, N., Vaithayapichet, S., and Heller, R. F. (1990). Pattern of antibiotic use in medical wards of a university hospital, Bangkok, Thailand. Rev. Infect. Dis. 12, 136–141. doi: 10.1093/clinids/12.1.136
Boonyasiri, A., Jearanaisilavong, J., and Assanasen, S. (2013). Candidemia in Siriraj hospital: epidemiology and factors associated with mortality. J. Med. Assoc. Thai. 96(Suppl. 2), S91–S97.
Brown, G. D., Denning, D. W., Gow, N. A., Levitz, S. M., Netea, M. G., and White, T. C. (2012). Hidden killers: human fungal infections. Sci. Transl. Med. 4:165rv113.
Cantón, E., Pemán, J., Hervás, D., Iñiguez, C., Navarro, D., Echeverría, J., et al. (2012). Comparison of three statistical methods for establishing tentative wild-type population and epidemiological cutoff values for echinocandins, amphotericin B, flucytosine, and six Candida species as determined by the colorimetric Sensititre YeastOne method. J. Clin. Microbiol. 50, 3921–3926. doi: 10.1128/JCM.01730-12
Cavalheiro, M., and Teixeira, M. C. (2018). Candida biofilms: threats, challenges, and promising strategies. Front. Med. 5:28. doi: 10.3389/fmed.2018.00028
Chakrabarti, A., Sood, P., Rudramurthy, S. M., Chen, S., Kaur, H., Capoor, M., et al. (2015). Incidence, characteristics and outcome of ICU-acquired candidemia in India. Intensive Care Med. 41, 285–295. doi: 10.1007/s00134-014-3603-2
Chou, H.-H., Lo, H.-J., Chen, K.-W., Liao, M.-H., and Li, S.-Y. (2007). Multilocus sequence typing of Candida tropicalis shows clonal cluster enriched in isolates with resistance or trailing growth of fluconazole. Diagn. Microbiol. Infect. Dis. 58, 427–433. doi: 10.1016/j.diagmicrobio.2007.03.014
Clinical and Laboratory Standards Institute (2012). Reference Method for Broth Dilution Antifungal Susceptibility Testing of Yeasts; Approved Standard. CLSI Document M27-A3 and Supplement S, Vol. 3. Wayne, PA: CLSI, 6–12.
Conde-Rosa, A., Amador, R., Perez-Torres, D., Colón, E., Sánchez-Rivera, C., Nieves-Plaza, M., et al. (2010). Candidemia distribution, associated risk factors, and attributed mortality at a university-based medical center. P. R. Health Sci. J. 29, 26–29.
Danesi, P., Firacative, C., Cogliati, M., Otranto, D., Capelli, G., and Meyer, W. (2014). Multilocus sequence typing (MLST) and M13 PCR fingerprinting revealed heterogeneity amongst Cryptococcus species obtained from Italian veterinary isolates. FEMS Yeast Res. 14, 897–909. doi: 10.1111/1567-1364.12178
Deorukhkar, S. C., Saini, S., and Mathew, S. (2014). Virulence factors contributing to pathogenicity of Candida tropicalis and its antifungal susceptibility profile. Int. J. Microbiol. 2014:456878. doi: 10.1155/2014/456878
Desnos-Ollivier, M., Bretagne, S., Bernède, C., Robert, V., Raoux, D., Chachaty, E., et al. (2008). Clonal population of flucytosine-resistant Candida tropicalis from blood cultures, Paris, France. Emerg. Infect. Dis. 14, 557–565. doi: 10.3201/eid1404.071083
Doi, A. M., Pignatari, A. C. C., Edmond, M. B., Marra, A. R., Camargo, L. F. A., Siqueira, R. A., et al. (2016). Epidemiology and microbiologic characterization of nosocomial candidemia from a Brazilian national surveillance program. PLoS One 11:e0146909. doi: 10.1371/journal.pone.0146909
Elashoff, J. D. (2000). nQuery Advisor (Version 4.0). User’s Guide. Los Angeles, CA: Dixon Associates.
Fan, X., Xiao, M., Liao, K., Kudinha, T., Wang, H., Zhang, L., et al. (2017). Notable increasing trend in azole non-susceptible Candida tropicalis causing invasive candidiasis in China (August 2009 to July 2014): molecular epidemiology and clinical azole consumption. Front. Microbiol. 8:464. doi: 10.3389/fmicb.2017.00464
Farooqi, J., Jabeen, K., Saeed, N., Iqbal, N., Malik, B., Lockhart, S., et al. (2013). Invasive candidiasis in Pakistan: clinical characteristics, species distribution and antifungal susceptibility. J. Med. Microbiol. 62(Pt 2), 259–268. doi: 10.1099/jmm.0.048785-0
Favero, D., Furlaneto-Maia, L., Franca, E. J., Goes, H. P., and Furlaneto, M. C. (2014). Hemolytic factor production by clinical isolates of Candida species. Curr. Microbiol. 68, 161–166. doi: 10.1007/s00284-013-0459-6
Foongladda, S., Sakulmaiwatana, P., Petlum, P., and Vanprapar, N. (2004). Candida species, genotypes and antifungal susceptibility of Candida isolates from blood samples of patients at the largest tertiary care hospital in Thailand during 1999-2002. J. Med. Assoc. Thai. 87, 92–99.
Francisco, A. P., Bugalho, M., Ramirez, M., and Carriço, J. A. (2009). Global optimal eBURST analysis of multilocus typing data using a graphic matroid approach. BMC Bioinformatics 10:152. doi: 10.1186/1471-2105-10-152
Furlaneto, M. C., Favero, D., França, E. J. G., and Furlaneto-Maia, L. (2015). Effects of human blood red cells on the haemolytic capability of clinical isolates of Candida tropicalis. J. Biomed. Sci. 22:13. doi: 10.1186/s12929-015-0120-8
Galan-Ladero, M., Blanco, M., Sacristan, B., Fernández-Calderón, M., Perez-Giraldo, C., and Gomez-Garcia, A. (2010). Enzymatic activities of Candida tropicalis isolated from hospitalized patients. Med. Mycol. 48, 207–210. doi: 10.3109/13693780902801242
Gokce, G., Cerikcioglu, N., and Yagci, A. (2007). Acid proteinase, phospholipase, and biofilm production of Candida species isolated from blood cultures. Mycopathologia 164, 265–269. doi: 10.1007/s11046-007-9053-4
Guembe, M., Cruces, R., Peláez, T., Muñoz, P., and Bouza, E., Geidi study group (2017). Assessment of biofilm production in Candida isolates according to species and origin of infection. Enferm. Infecc. Microbiol. Clin. 35, 37–40. doi: 10.1016/j.eimc.2016.04.003
Guinea, J. (2014). Global trends in the distribution of Candida species causing candidemia. Clin. Microbiol. Infect. 20, 5–10. doi: 10.1111/1469-0691.12539
Hoge, C. W., Gambel, J. M., Srijan, A., Pitarangsi, C., and Echeverria, P. (1998). Trends in antibiotic resistance among diarrheal pathogens isolated in Thailand over 15 years. Clin. Infect. Dis. 26, 341–345. doi: 10.1086/516303
Jiang, C., Li, Z., Zhang, L., Tian, Y., Dong, D., and Peng, Y. (2016). Significance of hyphae formation in virulence of Candida tropicalis and transcriptomic analysis of hyphal cells. Microbiol. Res. 192, 65–72. doi: 10.1016/j.micres.2016.06.003
Khairat, S. M., Sayed, A. M., Nabih, M., Soliman, N. S., and Hassan, Y. M. (2019). Prevalence of Candida blood stream infections among children in tertiary care hospital: detection of species and antifungal susceptibility. Infect. Drug Resist. 12, 2409–2416. doi: 10.2147/IDR.S196972
Kibbler, C. (1996). Fungaemia and Disseminated Fungal Infection. Principles and Practice of Clinical Mycology. Chichester: John Wiley & Sons Ltd, 143–164.
Kothavade, R. J., Kura, M., Valand, A. G., and Panthaki, M. (2010). Candida tropicalis: its prevalence, pathogenicity and increasing resistance to fluconazole. J. Med. Microbiol. 59, 873–880. doi: 10.1099/jmm.0.013227-0
Li, X., Yan, Z., and Xu, J. (2003). Quantitative variation of biofilms among strains in natural populations of Candida albicans. Microbiology 149(Pt 2), 353–362. doi: 10.1099/mic.0.25932-0
Makanjuola, O., Bongomin, F., and Fayemiwo, S. (2018). An update on the roles of non-albicans Candida species in vulvovaginitis. J. Fungi (Basel) 4:121. doi: 10.3390/jof4040121
Naglik, J. R., Challacombe, S. J., and Hube, B. (2003). Candida albicans secreted aspartyl proteinases in virulence and pathogenesis. Microbiol. Mol. Biol. Rev. 67, 400–428. doi: 10.1128/mmbr.67.3.400-428.2003
Negri, M., Martins, M., Henriques, M., Svidzinski, T. I., Azeredo, J., and Oliveira, R. (2010). Examination of potential virulence factors of Candida tropicalis clinical isolates from hospitalized patients. Mycopathologia 169, 175–182. doi: 10.1007/s11046-009-9246-0
Pfaller, M. A., Diekema, D. J., Turnidge, J. D., Castanheira, M., and Jones, R. N. (2019). Twenty years of the SENTRY antifungal surveillance program: results for Candida species from 1997–2016. Open Forum Infect. Dis. 6(Suppl. 1), S79–S94.
Pham, L. T. T., Pharkjaksu, S., Chongtrakool, P., Suwannakarn, K., and Ngamskulrungroj, P. (2019). A predominance of clade 17 Candida albicans isolated from hemocultures in a tertiary care hospital in Thailand. Front. Microbiol. 10:1194. doi: 10.3389/fmicb.2019.01194
Pharkjaksu, S., Chongtrakool, P., Suwannakarn, K., and Ngamskulrungroj, P. (2018). Species distribution, virulence factors, and antifungal susceptibility among Candida parapsilosis complex isolates from clinical specimens at Siriraj hospital, Thailand, from 2011 to 2015. Med. Mycol. 56, 426–433. doi: 10.1093/mmy/myx058
Sachin, C., Ruchi, K., and Santosh, S. (2012). In vitro evaluation of proteinase, phospholipase and haemolysin activities of Candida species isolated from clinical specimens. Int. J. Med. Biomed. Res. 1, 153–157. doi: 10.14194/ijmbr.1211
Samaranayake, L., Raeside, J. M., and MacFarlane, T. (1984). Factors affecting the phospholipase activity of Candida species in vitro. Sabouraudia 22, 201–207. doi: 10.1080/00362178485380331
Seneviratne, C. J., Rajan, S., Wong, S. S., Tsang, D. N., Lai, C. K., Samaranayake, L. P., et al. (2016). Antifungal susceptibility in serum and virulence determinants of Candida bloodstream isolates from Hong Kong. Front. Microbiol. 7:216. doi: 10.3389/fmicb.2016.00216
Tan, B. H., Chakrabarti, A., Li, R. Y., Patel, A. K., Watcharananan, S. P., Liu, Z., et al. (2015). Incidence and species distribution of candidaemia in Asia: a laboratory-based surveillance study. Clin. Microbiol. Infect. 21, 946–953. doi: 10.1016/j.cmi.2015.06.010
Tavanti, A., Davidson, A. D., Johnson, E. M., Maiden, M. C., Shaw, D. J., Gow, N. A., et al. (2005). Multilocus sequence typing for differentiation of strains of Candida tropicalis. J. Clin. Microbiol. 43, 5593–5600. doi: 10.1128/jcm.43.11.5593-5600.2005
Wisplinghoff, H., Bischoff, T., Tallent, S. M., Seifert, H., Wenzel, R. P., and Edmond, M. B. (2004). Nosocomial bloodstream infections in US hospitals: analysis of 24,179 cases from a prospective nationwide surveillance study. Clin. Infect. Dis. 39, 309–317. doi: 10.1086/421946
Wu, J.-Y., Guo, H., Wang, H.-M., Yi, G.-H., Zhou, L.-M., He, X.-W., et al. (2017). Multilocus sequence analyses reveal extensive diversity and multiple origins of fluconazole resistance in Candida tropicalis from tropical China. Sci. Rep. 7:42537.
Yapar, N. (2014). Epidemiology and risk factors for invasive candidiasis. Ther. Clin. Risk Manag. 10, 95–105.
YeSudhaSon, B. L., and MohanraM, K. (2015). Candida tropicalis as a predominant isolate from clinical specimens and its antifungal susceptibility pattern in a tertiary care hospital in Southern India. J. Clin. Diagn. Res. 9, DC14–DC16.
Keywords: Candida tropicalis, resistance, Thailand, candidemia, prevalence, virulence factor, MLST, prevalence shift
Citation: Tulyaprawat O, Pharkjaksu S, Chongtrakool P and Ngamskulrungroj P (2020) An Association of an eBURST Group With Triazole Resistance of Candida tropicalis Blood Isolates. Front. Microbiol. 11:934. doi: 10.3389/fmicb.2020.00934
Received: 08 January 2020; Accepted: 20 April 2020;
Published: 19 May 2020.
Edited by:
Orazio Romeo, University of Messina, ItalyReviewed by:
Alexandre Alanio, Université Paris Diderot, FranceMihai Mares, Ion Ionescu de la Brad University of Agricultural Sciences and Veterinary Medicine of Iaşi, Romania
Copyright © 2020 Tulyaprawat, Pharkjaksu, Chongtrakool and Ngamskulrungroj. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Popchai Ngamskulrungroj, cG9wY2hhaS5uZ2FAbWFoaWRvbC5hYy50aA==
 Orawan Tulyaprawat
Orawan Tulyaprawat Sujiraphong Pharkjaksu
Sujiraphong Pharkjaksu Piriyaporn Chongtrakool
Piriyaporn Chongtrakool Popchai Ngamskulrungroj
Popchai Ngamskulrungroj