- 1Department of Cell and Molecular Biology, Faculty of Biotechnology and Biomolecular Sciences, Universiti Putra Malaysia, Selangor, Malaysia
- 2Halal Products Research Institute, Putra Infoport, Universiti Putra Malaysia, Selangor, Malaysia
- 3Department of Microbiology, Faculty of Biotechnology and Biomolecular Sciences, Universiti Putra Malaysia, Selangor, Malaysia
- 4Mass Spectrometry Technology Section, Malaysia Genome Institute, National Institute of Biotechnology Malaysia, Kajang, Malaysia
- 5Bioprocessing and Biomanufacturing Research Centre, Faculty of Biotechnology and Biomolecular Sciences, Universiti Putra Malaysia, Selangor, Malaysia
- 6Chancellory, Universiti Teknikal Malaysia Melaka, Malacca, Malaysia
The inhibitory properties of novel antimicrobial proteins against food-borne pathogens such as Listeria monocytogenes offer extensive benefits to the food and medical industries. In this study, we have identified antimicrobial proteins from a milk curd–derived bacterial isolate that exhibits antilisterial activity using genome mining and mass spectrometry analysis. The analysis of the draft genome sequence identified the isolate as Paenibacillus polymyxa Kp10, and predicted the presence of antimicrobial paenibacillin, paenilan, paeninodin, sactipeptides, thiazole-oxazole modified microcin, and histone-like DNA binding protein HU encoded in its genome. Interestingly, nanoLC-MS/MS analysis identified two histone-like DNA binding proteins HU as predicted in silico earlier, exhibiting antilisterial activity. Additionally, translation initiation factor IF-1 and 50S ribosomal protein L29 were also discovered by the mass spectrometry in the active fractions. The antilisterial activity of the four proteins was verified through heterologous protein expression and antimicrobial activity assay in vitro. This study has identified structural regulatory proteins from Paenibacillus possessing antilisterial activity with potential future application in the food and medical industries.
Introduction
Food spoilage caused by pathogenic microbes such as Listeria monocytogenes (L. monocytogenes) is a common problem, imposing serious detrimental economic effects. The pathogen L. monocytogenes has become part of most foodborne outbreaks due to its ubiquitous nature and ability to survive in extreme conditions (Angelo et al., 2017). Its ability to form a biofilm on the surface of food processing equipment and its development of resistance to the standard sterilizing procedure have imposed further burdens on the food industry (Skowron et al., 2018). Common food preservatives to eradicate pathogen contamination include acetic acid, potassium acetate, benzoic acid, benzoates, and nitrites and nitrates. However, chemically derived preservatives have been shown to compromise human health in long-term use. For instance, the cytotoxic and mutagenic activity of sodium benzoate, a common preservative in dairy products, has been found in lymphocytes (Pongsavee, 2015). In addition, the reaction of nitrites in the form of sodium and potassium nitrite with secondary amines has been postulated to contribute to the formation of carcinogenic nitrosamines molecule (Carocho et al., 2014). Antimicrobial proteins are considered greener and safer, as they are biologically produced (Bagenda and Yamazaki, 2007). Therefore, the use of antimicrobial proteins in food and medical industries has increased in the past decades due to their inhibitory activity against food-borne pathogens and multidrug-resistant bacteria.
Bacteriocin, a ribosomally synthesized antimicrobial peptide, is a common antimicrobial protein used in the food and medical fields (Bagenda and Yamazaki, 2007; Yang et al., 2014; Alvarez-Sieiro et al., 2016; Ahmad et al., 2017). Bacteriocins are classified according to their common characteristics (Alvarez-Sieiro et al., 2016). Different post-translational modification enzymes and accessory proteins encoded along with the structural antimicrobial protein in the gene cluster of the bacterial genome are required for the biosynthesis of different classes of biologically active bacteriocins (Perez et al., 2014). The conservation of post-translational modification enzymes and accessory proteins in the biosynthetic gene clusters, as well as conserved motifs in the leader peptide of bacteriocin, allow for the discovery of novel bacteriocin encoded in the bacterial genome using genome mining (Scheffler et al., 2013; Boddy, 2014). Although less common, certain proteins and peptides without bacteriocin characteristic are also produced by bacteria, such as histone-like DNA binding and ribosomal proteins (Carvalho et al., 2010). Currently, only two bacteriocins have been approved by the Food and Drug Administration for human use, namely nisin and pediocin, which are produced by Lactococcus lactis subsp. lactis and Pediococcus acidilactici, respectively. Therefore, extensive research is required to discover new antimicrobial proteins. The screening and identification of new antilisterial proteins can be conducted using methods such as genome mining and mass spectrometry.
Genome mining involves the in silico identification of biosynthetic gene cluster of antimicrobial peptides in the whole or draft genome sequence of the microbes using bioinformatics tools, followed by in vitro analysis of the gene products (Scheffler et al., 2013). Recombinant protein expression followed by in vitro antimicrobial analysis can confirm the activity of target putative proteins, complementing any in silico approach (Scheffler et al., 2013).
The current study aims to identify potential antimicrobial proteins from a local milk curd isolate through genome mining and mass spectrometry. Several potential antimicrobial proteins contributing to the antimicrobial activity were discovered. The partially purified active fractions were tested against L. monocytogenes, and fractions displaying antilisterial activity were then subjected to mass spectrometry-based protein identification. The verification of antilisterial activity of the putative proteins was conducted via heterologous protein expression in Escherichia coli (E. coli) and characterizations of the recombinant proteins in vitro.
Materials and Methods
Bacterial Strain and Culture Condition
An unidentified pure bacterial culture exhibiting antilisterial activity previously isolated from milk curd was obtained from the Bioprocessing and Biomanufacturing Research Centre, Faculty of Biotechnology and Biomolecular Sciences, Universiti Putra Malaysia. It was grown in M17 broth at 37°C overnight. Repeated streaking and sub-culturing were carried out 10 times on M17 agar to obtain a single colony.
Additionally, E. coli T7 Express LysY/Iq cells (New England BioLabs Inc., Ipswich, Massachusetts, USA) were grown on Luria Bertani (LB) medium by overnight shaking (200–300 rpm) at 37°C in an incubator shaker (Heidolph Inkubator 1000). L. monocytogenes ATCC 15313 was cultured in Brain Heart Infusion (BHI) broth at 37°C overnight.
Morphological and Biochemical Characterizations of Isolate
The isolate was subjected to Gram staining and biochemical tests. Catalase, oxidase, indole, nitrate reduction, and Voges Proskauer tests were carried out according to accredited methods at Bacteriology Laboratory of Veterinary Laboratory Service Unit, Faculty of Veterinary, Universiti Putra Malaysia.
Identification of Bacterial Isolate Using Sanger Sequencing of Partial 16S rDNA Sequence
Species identification of the isolate was initially carried out using partial 16S rDNA sequence analysis. Partial 16S rDNA sequence was amplified using 27F and 1490R primers in 1X TopTaq Master Mix (Qiagen, Hilden, Germany). PCR was conducted using the following condition 95°C 3 min, 35 cycles of 95°C 1 min, 51°C 30 s, and 72°C 1 min, and a final extension at 72°C for 10 min was used on Mastercycler Gradient (Eppendorf, Hamburg, Germany). The amplicon was electrophoresed on 1% agarose gel with 1X Tris/Borate/EDTA (TBE) as a running buffer at 100 V for 20 min. ExactMark 1 kb DNA ladder 250–10,000 bp (Axil Scientific Pte Ltd, Singapore) was used as a marker. Upon visualization using FluorChem Gel Documentation System (Alpha Innotech, San Leandro, California, USA), the amplicon was sent for bidirectional Sanger sequencing (Axil Scientific Pte Ltd) using 27F and 1490R primers. The partial 16S rRNA gene sequence was analyzed against available non-redundant sequences in National Center for Biotechnology Information (NCBI) database using the Basic Local Alignment Search Tool Nucleotide (BLASTn). Default configuration was used for analysis.
In vitro Antimicrobial Activity Assay
An in vitro antimicrobial activity assay was conducted using agar well diffusion method (Abbasiliasi et al., 2012) against L. monocytogenes ATCC 15313 as the indicator strain. The assay was performed in triplicate. Inhibition zones (in mm) were measured, and the value was used to calculate antimicrobial activity (AU) of the sample. Antimicrobial activity (AU) is expressed as the unit area of inhibition zone per unit volume of sample loaded into the well (mm2/mL) (Abbasiliasi et al., 2012).
Whole-Genome Sequencing of Paenibacillus polymyxa Kp10
Genomic DNA was extracted from overnight-grown Paenibacillus polymyxa Kp10 (P. polymyxa Kp10) culture using the Wizard® Genomic DNA Purification Kit (Promega, Madison, USA) as per the manufacturer's protocol. An indexed paired-end library was constructed using 1 ng DNA of P. polymyxa Kp10 and Nextera XT DNA Library Prep Kit (Illumina Inc., San Diego, California, USA) according to the manufacturer's protocol. There were 151 base-pair paired-end (2 × 151 bp) sequencing reads obtained using MiSeq™ system (Illumina Inc., San Diego, California, USA) and were scanned for adapter sequences and low-quality sequences using BBDuk (BBTools version 36) (Bushnell and Brian, 2014). De novo genome assembly was carried out using SPAdes version 3.9.0 (Bankevich et al., 2012). NCBI Prokaryotic Genome Annotation Pipeline (Tatusova et al., 2016) was used to annotate the genome.
Species Identification
The similarity of complete 16S rRNA gene sequence of the isolate obtained from the whole genome sequencing was analyzed against the available non-redundant sequences in the National Center for Biotechnology Information (NCBI) database using Basic Local Alignment Search Tool Nucleotide (BLASTn). Default configuration was used for the analysis. Additionally, the draft genome sequence of the isolate was analyzed using TrueBac™ID genome-based bacterial identification system (ChunLab Inc., Seoul, Korea) (Ha et al., 2019) to estimate the overall genome relatedness index (OGRI) against genome sequence of bacterial species that share more than 98.7% identity with the 16S rRNA gene sequence of the isolate.
Mining the Draft Genome Sequence of P. polymyxa Kp10 for Antimicrobial Genes Using Bioinformatic Analysis
The draft genome sequence of P. polymyxa Kp10 was analyzed using BAGEL4 (Van Heel et al., 2018) and antiSMASH 5.0 (Medema et al., 2011) using default parameters. A Conserved Domain (CD)-Search (Marchler-Bauer et al., 2017) of putative biosynthetic proteins of the predicted antimicrobial proteins was carried out to verify their function based on the conserved domain. Amino acid sequences of <10 kDa proteins were also subjected to CD-Search using the default configuration to identify proteins with histone-like DNA binding protein domains. Accession numbers of contigs and locus tags of putative biosynthetic proteins were obtained from the Kp10's draft genome sequence data deposited in NCBI Genbank (PRJNA449134), which was updated as of April 2020.
Partial Purification of Antilisterial Proteins From the Cell-Free Culture Supernatant of P. polymyxa Kp10
A 1 L overnight culture of P. polymyxa Kp10 was centrifuged (10,000 × g, 24°C for 5 min) to obtain cell-free culture supernatant (CFCS). Total proteins from CFCS were precipitated with the gradual addition of 790 g ammonium sulfate at 24°C. Following overnight incubation at 4°C, the precipitated protein was sedimented through centrifugation (12,000 × g, 4°C for 30 min) and dissolved in 50 mL sterile deionized distilled water. High molecular weight proteins were removed using Amicon® Ultra-15 Centrifugal Filter Unit with Nominal Molecular Weight Limit (NMWL) of 10 kDa (Merck, USA) through centrifugation (4,000 × g at 25°C for 10 min). The eluent (lower fraction) was then concentrated using Amicon® Ultra-15 Centrifugal Filter Unit with NMWL of 3,000 Da (Merck, USA) through centrifugation (4,000 × g at 25°C for 45 min). The resulting upper fraction was desalted using Hi Trap Desalting column before being further purified using Hi Trap SP HP cation exchange column on ÄKTA Purifier System (GE Healthcare, Chicago, Illinois, USA). The column was equilibrated with Tris-Cl buffer (pH 7.5) at a flow rate of 1 mL/min, and the proteins were eluted in 1 mL fractions using a sodium chloride gradient (0% to 100% of 1 M) in Tris-Cl buffer (pH 7.5). The protein concentrations of each fraction were monitored at 215 nm throughout purification.
Protein concentrations of all unbound and eluted fractions were determined using NanoOrange Protein Quantitation Kit (Invitrogen, Paisley, UK) according to the manufacturer's protocol. In vitro antilisterial activity of all unbound and eluted fractions was determined using the agar well diffusion method (Abbasiliasi et al., 2012). Partial purification, protein concentration determination, and in vitro antilisterial activity was carried out in triplicates.
Sodium Dodecyl Sulfate-Polyacrylamide Gel Electrophoresis
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) of the resulting protein fractions was performed using a precast 16.5% Mini-PROTEAN® Tris-Tricine Gel (BioRad, Hercules, USA). A 15 μL aliquot of Precision Plus Protein™ Dual Xtra Prestained Protein Standards (Bio-Rad, Hercules, USA) was used as molecular weight marker. An amount of 15 μg of protein samples was dissolved in 100 μL tricine sample buffer (200 mM Tris-HCl, pH 6.8, 40% glycerol, 2% SDS, 0.04% Coomassie Blue G-250) containing 2% fresh β-mercaptoethanol. The sample mixture was heated at 95°C for 5 min. Electrophoresis was conducted using tricine running buffer (100 mM Tris, 100 mM Tricine, 0.1% SDS, pH 8.3) at 100 V for 100 min. The SDS-PAGE gel was fixed using a fixative solution (40% methanol, 10% acetic acid) for 30 min, followed by staining using 0.025% (w/v) Coomassie Blue G-250 in 10% acetic acid for 1 h. The gel was then destained for 15 min in 10% acetic acid. The gel was viewed using a GS-800™ Calibrated Imaging Densitometer (Bio-Rad, Hercules, USA).
Identification of Antilisterial Proteins Using nanoLC-MS/MS
A 1 mg/mL eluted protein displaying antilisterial activity in 0.1 M ammonium bicarbonate was digested by adding 100 μL of 0.05% Rapigest™ SF (Waters Corporation, Massachusetts, USA). After heat treatment at 80°C for 15 min, dithiothreitol with a final concentration of 100 mM was added, followed by incubation at 37°C for 30 min. Iodoacetamide with a final concentration of 200 mM was then added and incubated at room temperature for 45 min. An amount of 1 μg of trypsin powder was then added and further incubated at 37°C for overnight. A volume of 1 μL concentrated trifluoroacetic acid was added to stop digestion. The reaction mixture was centrifuged at room temperature (20,800 × g for 10 min). Then, 100 μL of supernatant was analyzed using LC Dionex 3000 Ultimate™ RSLCnano (Thermo Fisher Scientific) with A (0.1% formic acid in water) and B (0.1% formic acid in acetonitrile) as mobile phase. Peptides were separated on gradient basis: 5 to 40% B for 91 min, 2 min to 95% of B, 6 min at 95% of B, back to 5% of B in 2 min at the flow rate of 250 nL/min. EASY-Spray Column Acclaim™ PepMap™ C18 (100 Å, 2 μm particle size, 50 μm id × 15 cm) was used as separation column at 40°C. Next, 2 μL of protein sample was injected into Mass Spectrometer Orbitrap Fusion (Thermo Fisher Scientific) operated in data-dependent mode. Full scan spectra were collected (OTMS1) using the following parameters: scan range 310–1,800 m/z, resolving power of 120,000, automatic gain control (AGC) target of 4.0e5 (400,000), and maximum injection time of 50 ms. The method consisted of 3 s Top Speed Mode where precursors were selected for a maximum 3 s cycle. Only precursors with an assigned monoisotopic m/z and a charge state of 2–7 were further analyzed for MS2. All precursors were filtered using a 20 s dynamic exclusion window and an intensity threshold of 5000. The Ion Trap MS2 spectra (ITMS2) were analyzed using the following parameters: rapid scan rate with a resolving power of 60,000; AGC target of 1.0e2 (100); 1.6 m/z isolation window; and a maximum injection time of 250 ms. The precursors were fragmented by collision-induced dissociation (CID) and high energy collision dissociation (HCD) at normalized collision energy of 30 and 28%. Thermo Scientific™ Proteome Discoverer™ Software (Version 2.1) was used to analyze the data. The resulting output was used to search against the draft genome sequence of P. polymyxa Kp10 (Bioproject Accession: PRJNA449134;) with the following parameters: Missed cleavage: 2, MS1 tolerance: 10 ppm, MS2 tolerance: 0.6 Da including variable modification: Oxidation (M), deamidation of asparagine (N), and glutamine (Q) and Fixed modification: Carbamidomethyl (C). All peptides were validated using the percolator® algorithm, based on q-value <1% False Discovery Rate (FDR). The nanoLC-MS/MS analysis of the active fraction was repeated three times.
Cloning and Heterologous Expression of Putative Antilisterial Proteins in Escherichia coli T7 Express LysY/Iq
Genes encode for putative antilisterial proteins P1, P2, P3, and P4 were amplified from the genomic DNA of P. polymyxa Kp10 using the primers listed in Table S1 to incorporate enterokinase recognition site at the 3' end. NcoI and XhoI restriction sites were used to flank the amplified DNA fragment at 5' and 3' end, respectively, to allow directional cloning into pET28b+ (Novagen). Upon transformation into E. coli T7 Express LysY/Iq (New England BioLabs, Inc) according to manufacturer's protocol, the positive transformants were selected on LB agar containing 30 μg/mL of kanamycin. Positive transformants of the respective constructs were confirmed using colony PCR and sequencing. Upon verification, the positive clones were designated as pET28b_P1, pET28b_P2, pET28b_P3, and pET28b_P4.
The expression of recombinant antilisterial proteins was initiated by inoculating 10 mL LB broth containing 30 μg/mL kanamycin with single colonies of positive clones and grown at 37°C until OD600 reached 0.4. The E. coli cells containing each recombinant plasmid (pET28b_P1, pET28b_P2, pET28b_P3, and pET28b_P4) were cultured in triplicates. Expression was then induced by 0.4 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) followed by incubation at 37°C for 2 h. Proteins from the cell pellets were extracted using xTractor Buffer (Clontech Laboratories, Inc., Mountain View, USA) according to the manufacturer's protocol. The amount of total protein was estimated using the NanoOrange Protein Quantitation Kit (Invitrogen, Paisley, UK). In vitro antimicrobial activity of the recombinant antilisterial proteins was then determined using agar well diffusion assay (Abbasiliasi et al., 2012). For respective proteins, each well was loaded with the crude recombinant proteins obtained from three different overnight cultures as biological replicates. CFCS of Kp10 was used as positive control in the assay.
Results and Discussion
Biochemical and Antimicrobial Activity Characterizations of Isolate and Bacterial Identification Using Partial 16S rDNA Sequence Analysis
Besides Bacillus (Abriouel et al., 2011; Barbosa et al., 2015) and Lactobacillus (Messaoudi et al., 2013; da Silva Sabo et al., 2014), Paenibacillus (Aleti et al., 2016; Cochrane and Vederas, 2016) is another genus of bacteria with a rich resource of genes encodes for novel antimicrobial proteins and secondary metabolites. The CFCS of an unidentified pure bacterial culture isolated from milk curd has preliminarily been found to confer high antilisterial activity at 1051.84 ± 11.84 AU (Figure S1).
Biochemical characteristics of P. polymyxa Kp10 in comparison to that of other Paenibacillus species identified by other researchers are summarized in Table S2. Gram staining of the pure isolate shows purple-stained bacteria with a rod-shape morphology. The isolate was positive in catalase and acetyl-methyl carbinol production and was negative in oxidase and indole test. The positive reduction of nitrate showed that the isolate produced nitrate reductase. The biochemical characteristics of this isolate are similar to P. polymyxa OSY-DF (He et al., 2007) with the exception of acetyl-methyl carbinol production property that was not determined in the authors' study.
BLASTn analysis of the partial 16S rDNA sequence of the bacterium amplified using 27F and 1490R primers (Song et al., 2016) found that it shared 99% identity to P. polymyxa CF05 (Accession: CP009909.1), P. polymyxa SQR-21 (Accession: CP006872.1), P. polymyxa HY96-2 (Accession: CP025957.1), P. polymyxa Sb3-1 (Accession: CP010268.1), and 98% identity to Paenibacillus sp Izh-N1 (Accession: CP025696.1), with expectation value of 0.1 and query coverage of 98%. However, partial 16S rDNA sequence is insufficient to define the taxonomy of bacteria (Chun et al., 2018). A combination of the complete 16S rDNA sequence similarity and OGRI has been suggested to infer prokaryotic taxonomy (Chun et al., 2018). Hence, a high-quality draft of the bacterial genome sequence of P. polymyxa Kp10 was also obtained.
Whole-Genome Sequencing and Molecular Identification of P. polymyxa Kp10
General genome features of the isolate are summarized in Table S3. The sequencing using MiSeq (Illumina) generated 6,154,868 paired reads with length ranging from 35 to 151 nucleotides. A total of 4,554,785 paired reads were obtained after removal of low-quality reads and adapter sequences. De novo genome assembly generated 122 contigs with a total genome size of 5,653,475 bp. The longest contig length is 1,294,617 bp and the average G+C content is 45.47%. N50 value of the genome was 619,841 bp with estimated assembly coverage of 158.28X. The genome annotation of the draft genome sequence using the NCBI Prokaryotic Genome Annotation Pipeline revealed a total of 5,054 genes including 4,808 protein-coding genes, and 124 RNA genes. The Whole Genome Shotgun project has been deposited to NCBI with GenBank assembly accession no: GCA_003052505.1 and BioProject accession no: PRJNA449134. Based on the minimal standards proposed by Chun et al. (2018) for prokaryotes taxonomy, BLASTn analysis of the full length 16S ribosomal RNA sequence of the antilisterial isolate shows 100% identity with P. polymyxa CF05 (Accession: CP009909.1) and P. polymyxa HY96-2 (Accession: CP025957.1) with expectation value of 0.0 and query coverage of 100%. TrueBac™ID genome analysis successfully identified Kp10 as a genuine P. polymyxa by 98.33 and 91.3% of ANI and ANI coverage, respectively, to P. polymyxa ATCC842. This online identification tool revealed 99.8, 99.62, and 99.52% of 16S rRNA, recA, and rplC sequence similarity, respectively, to P. polymyxa. In another study, the draft genome sequence of Kp10 was included in a comparative genomic analysis with other 49 selected Paenibacillus strains. A dendrogram based on ANI matrix has disclosed that Kp10 belongs to the same cluster with P. polymyxa ATCC 842, which was classified as an original P. polymyxa species (Jeong et al., 2019). In this work, the strain designation of Kp10 was given to this bacterium to ease future reference.
Mining the Draft Genome Sequence of P. polymyxa Kp10 for Antimicrobial Genes and Biosynthetic Gene Clusters Using Bioinformatic Analysis
Bioinformatic analysis using BAGEL4, antiSMASH, and CD-Search successfully predicted eight antimicrobial peptides in the genome sequence of Kp10. The amino acid sequences of the antimicrobial peptides are presented in Table 1, and their biosynthetic gene clusters and putative functions are described in Table S4. Arrangement of genes encode for structural peptides and putative biosynthetic proteins in biosynthetic gene cluster of Kp10 is illustrated in Figure 1. BAGEL4 and antiSMASH analysis detected the existence of biosynthetic gene clusters responsible for the production of six ribosomally-synthesized antimicrobial proteins, namely two lantibiotics (predicted as paenibacillin and paenilan), one lasso peptide (predicted as paeninodin), one predicted thiazole/oxazole modified microcins (TOMM) peptide, and two sactipeptides (designated as sacti5 and sacti10). The functions of the putative biosynthetic proteins encoded at the upstream and downstream of the precursor gene were further verified through conserved domain analysis using CD-Search (Table S4). In comparison to the extensively studied paenibacillin, paenilan, and paeninodin produced by Paenibacillus polymyxa OSY-DF (He et al., 2008), Paenibacillus polymyxa E681 (Park et al., 2017), and Paenibacillus dendritiformis C454 (Zhu et al., 2016), respectively, characterization and determination of biosynthesis mechanisms of TOMM and sactipeptide in other Paenibacillus polymyxa received less attention by the researchers. TOMM is featured by the presence of thiazole and oxazole heterocycles derived from modification of cysteine and serine residues (Metelev and Ghilarov, 2014).
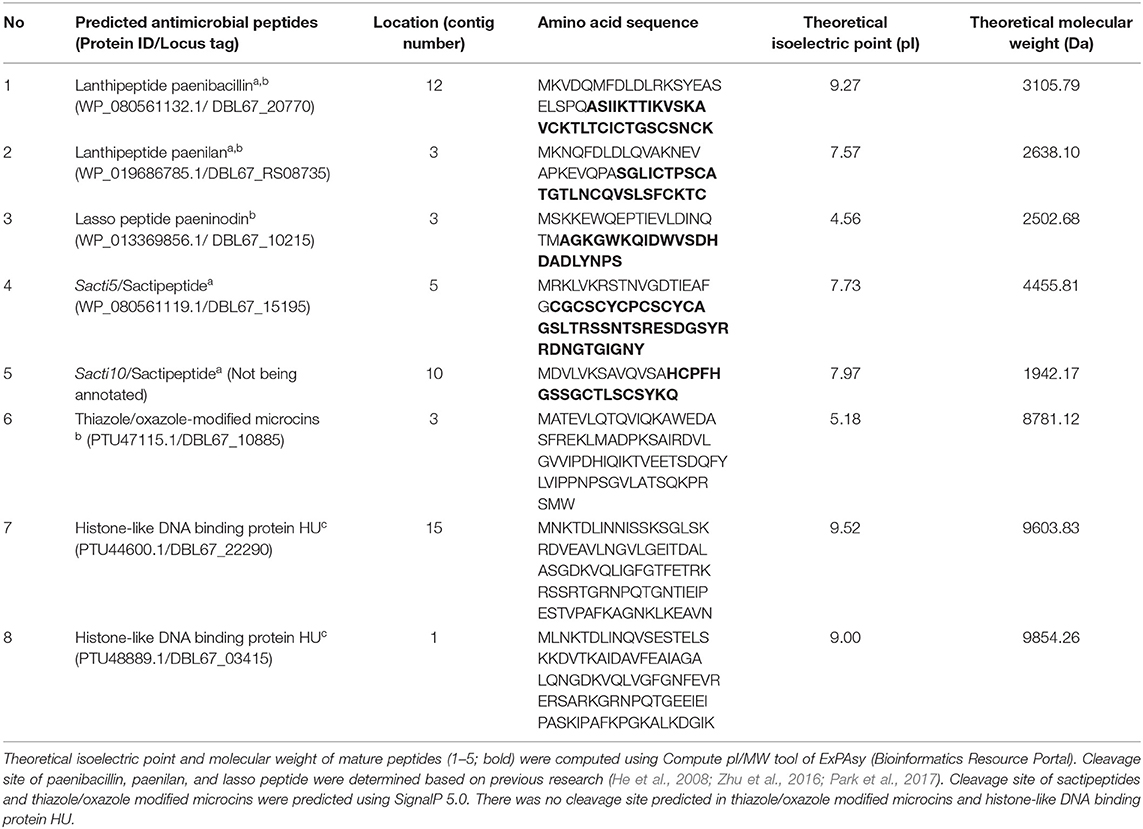
Table 1. Potential antimicrobial peptides or proteins identified in the draft genome sequence of Paenibacillus polymyxa Kp10 using BAGEL4a, antiSMASHb, and CD-Searchc.
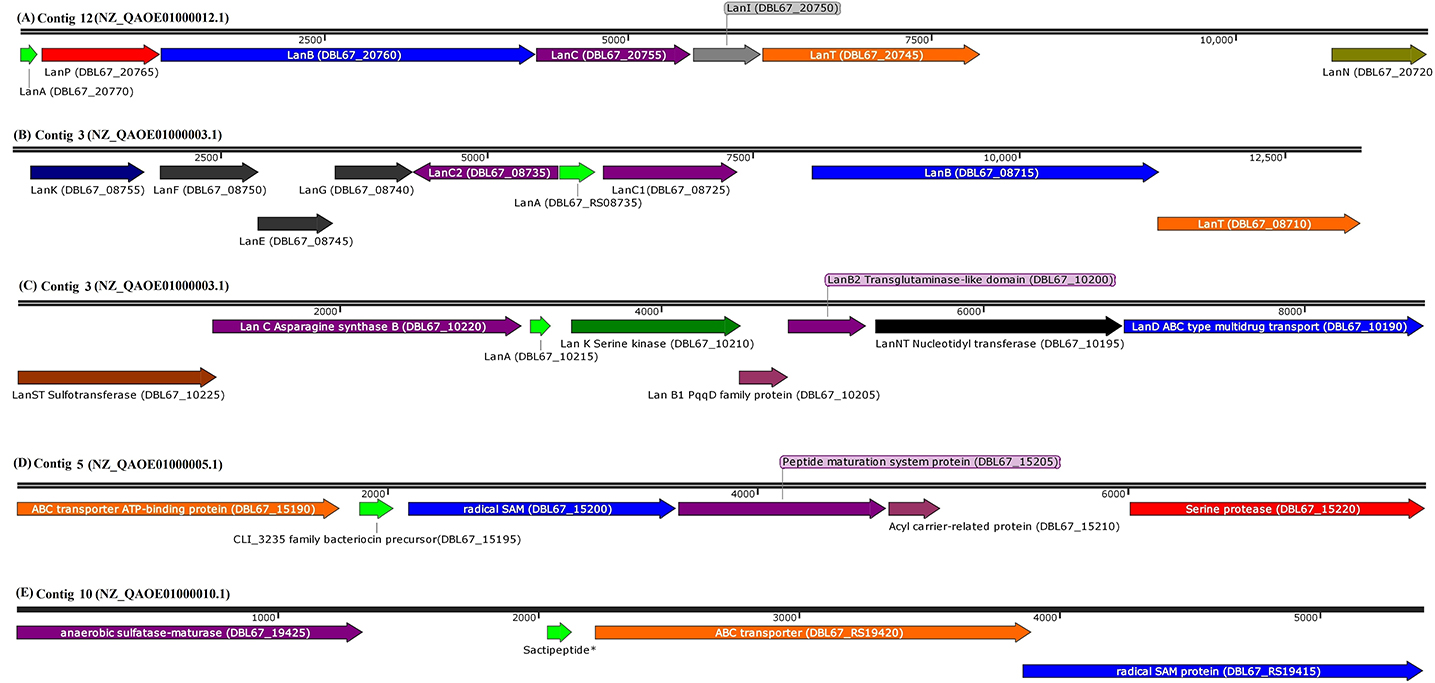
Figure 1. Arrangement of genes encode for structural peptide and putative biosynthetic proteins in biosynthetic gene cluster of (A) paenibacillin (GenBank coordinates from 63008 to 75697 in complementary strand), (B) paenilan (GenBank coordinates from 26187 to 38676 in complementary strand), (C) paeninodin (GenBank coordinates from 328300 to 337092 in complementary strand), (D) CLI_3235 family bacteriocin precursor, predicted as sactipeptide (sacti5) by BAGEL4 (Genbank coordinates from 183766 to 191369 in forward strand), and (E) sactipeptide* (GenBank coordinates from 68257 to 73648 in complementary strand) in respective contigs of P. polymyxa Kp10 genome. Sactipeptide* of the contig 10 (sacti10) has failed to be annotated by NCBI Prokaryotic Genome Annotation Pipeline. BAGEL4 analysis annotates the open reading frame as sactipeptide. The contigs with their respective accessions and locus tags of each genes/coding sequences are included in the figure. Genes are shown to scale and coordinates are relative to the length of each segment.
The gene encoding for biosynthetic proteins is absent both upstream and downstream of the TOMM precursor in Kp10. Nevertheless, the BLASTp analysis against non-redundant protein sequences database shows that it shares 100% identity to nitrile-hydratase leader peptide (NHLP)-related natural product precursor of P. polymyxa (WP_016821365.1). The sequence is conserved in multispecies of Paenibacillus. Quite recently, the emergence of NHLP-related peptide as a new precursor in TOMM family has been addressed (Haft et al., 2010; Melby et al., 2011; Cox et al., 2015). Post-translational modification of NHLP-related TOMM precursor is commonly carried out by dehydrogenase and cyclodehydratase, which introduces thiazole and oxazole heterocycles in the TOMM structure (Metelev and Ghilarov, 2014).
In the case of sactipeptide, BLASTp analysis shows that sacti10 does not share any similarity with protein sequences available in the database (Table S4). This suggests the novelty of sacti10 found encoded in Kp10. BLASTp of sacti5 shows that it shares 100% identity to CLI_3235 family bacteriocin precursor (Figure 1), which is a Cys-rich putative bacteriocin precursor peptides that commonly found in Clostridia. Radical SAM protein is commonly found encoded downstream of the CLI_3235 family bacteriocin precursor (Haft and Basu, 2011). The thioether bond between sulfur and α-carbon is a unique and distinctive characteristic of sactipeptide (Grove et al., 2017). Zhao et al. (2018) identified several putative biosynthetic gene clusters encode for sactipeptides in Paenibacillus larvae, Paenibacillus odorifer, Paenibacillus graminis, Paenibacillus riograndensis, and Paenibacillus sp. through bioinformatic analysis. However, the detailed amino acid sequence of the protein was not disclosed.
In addition to the two lantibiotics, one lasso peptide, one TOMM peptide, and two sactipeptides predicted by BAGEL4 and antiSMASH, CD-Search of proteins <10 kDa identified two different proteins with histone-like DNA binding protein HU domain (Table 1 and Table S4). The histone-like DNA binding proteins were not predicted by BAGEL4 and antiSMASH.
Partial Purification of Antilisterial Proteins From the CFCS of KP10 and Protein Identification Using Nano LC-MS/MS
The in silico discovery of eight potential antimicrobial proteins from Kp10 indicates the antimicrobial potential of Kp10. Hence, we hypothesize the involvement of at least one of the proteins in the antilisterial activity of Kp10. In theory, all these proteins may not necessarily involve in the antilisterial property displayed by the CFCS of the isolate. The genes found in the genome may be silenced or constitutively/inductively expressed by the bacteria. Therefore, we carried out protein identification by analyzing the active protein fractions using nanoLC-MS/MS. In the initial attempt to purify the proteins, ammonium sulfate-precipitated proteins from the CFCS of Kp10 was subjected to separation based on molecular weight using Amicon® Ultra-15 Centrifugal Filter Unit with NMWL of 10 kDa and 3 kDa. Both the retentate and eluant were analyzed for in vitro antimicrobial activity. Antilisterial activity was only detected in the eluant of 10 kDa and retentate of 3 kDa, suggesting that the antilisterial protein has a size between 3 and 10 kDa. The final cation exchange chromatography step was carried out using Tris-Cl buffer at pH 7.5. Consecutive purification steps successfully yielded a single peak that covers F2, F3, and F4 fractions (Figure 2A). Antilisterial activity was successfully detected in the mixture of these three fractions with a total activity of 2.18 × 104 ± 0 AU (Figure 2B). The purification recovery of the antilisterial proteins from each purification steps is summarized in Table S5. SDS-PAGE of the fractions' mixture yielded single electrophoretic bands between 5 and 10 kDa (Figure 2C).
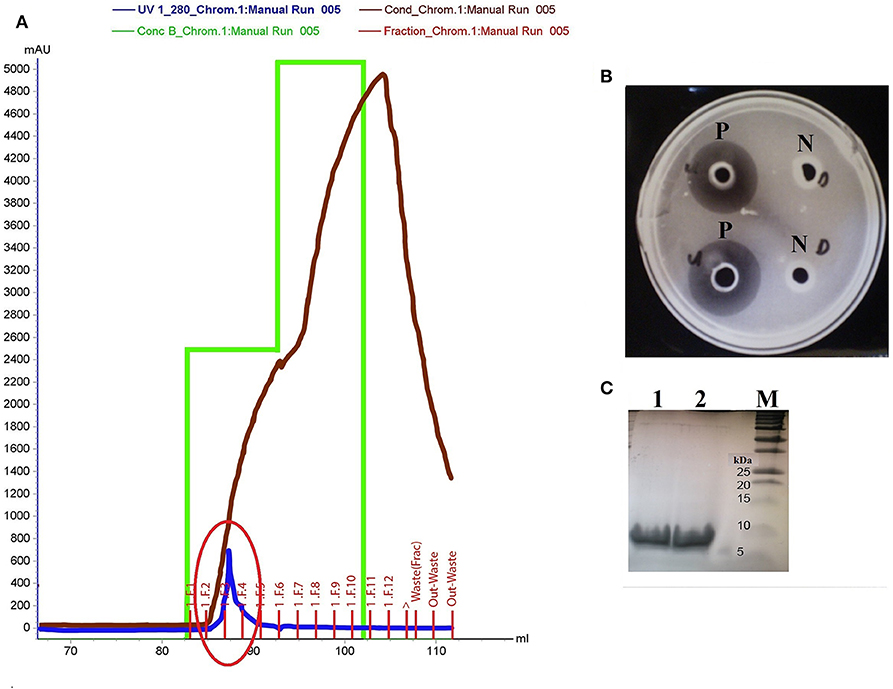
Figure 2. (A) Purification of active fraction using cation exchange chromatography (Hi Trap SP HP cation exchange column) on Akta Purifier System. The purification yields a single peak that covers F2, F3, and F4 fractions (red circle). (B) P: Antilisterial activity assay of the pooled active fractions determined using agar well diffusion method, analyzed in duplicates. N: There was no inhibition zone obtained in the control wells containing Tris-Cl buffer, pH 7.5. (C) SDS-PAGE of the active fraction mixture pooled from F2, F3, and F4 fractions analyzed in duplicates (Lane 1 and 2). Lane M: Precision Plus Protein™ Dual Xtra Standards (BioRad).
The amino acid sequences of the predicted antimicrobial proteins obtained from the draft genome sequence of Kp10 (NZ_QAOE00000000.1) were used as a reference database in nanoLC-MS/MS analysis. Intriguingly, two proteins identified by the CD-Search of protein <10 kDa in the Kp10 genome; the histone-like DNA binding proteins were found in the active fractions. The two proteins were designated as P1 and P2 (Table 2). On the other hand, the other six predicted bacteriocins (paenibacillin, paenilan, paeninodin, TOMM, sactipeptides) previously identified in silico in the Kp10 genome were not detected in the active fractions. Besides the P1 and P2 proteins, two other proteins detected in the active fractions were translation initiation factor IF-1 (P3) and 50S ribosomal protein L29 (P4) (Table 2).
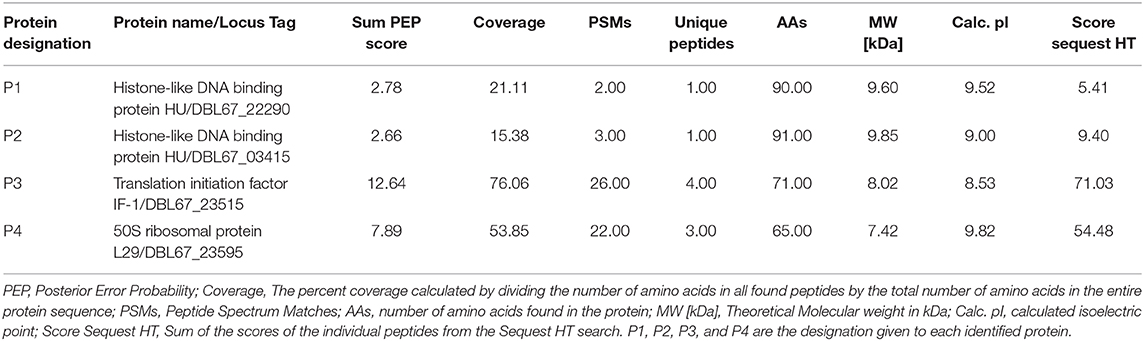
Table 2. Identification of proteins in active fraction mixture obtained from cation exchange chromatography using nanoLC-MS/MS.
Confirmation of Putative Antilisterial Proteins by Cloning and Heterologous Expression in E. coli
Further verification on the antilisterial activity of the putative antimicrobial proteins was carried out by repeating the antilisterial activity assay using the recombinant proteins. The genes encoding for P1, P2, P3, and P4 were separately cloned into pET28b+ for expression in E. coli T7 Express LysY/Iq (New England BioLabs). PCR amplification of genes encode for P1, P2, P3, and P4 obtained amplicon of 305, 287, 227, and 209 bp in size, respectively (Figure S2A). The colony PCR of the transformants (pET28b_P1, pET28b_P2, pET28b_P3, and pET28b_P4) confirmed the successful cloning of the four genes into pET28b+ (Figure S2B).
Upon induction of protein expression and extraction, in vitro antimicrobial activity assay of the total crude protein containing the recombinant proteins conferred antilisterial activity (Figure 3). Recombinant P1, P2, P3, and P4 showed inhibition zones of 14±0 mm in diameter, corresponding to 1257.14 ± 0 AU/mL. There was no antilisterial activity detected in the crude protein of empty E. coli T7 Express LysY/Iq cultures. Pidutti et al. (2018) classified the strength of antibacterial activity of protein into three groups: strong (10–15 mm inhibition zone); moderate (5–10 mm inhibition zone); and no inhibitory activity (0–5 mm inhibition zone). Based on this classification, strong antilisterial activity was observed in all recombinant proteins.
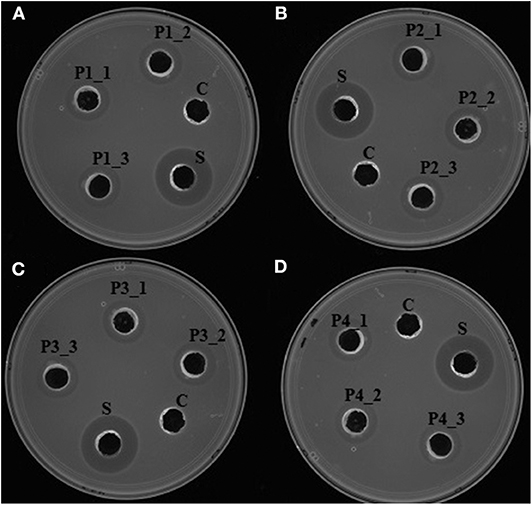
Figure 3. In vitro antimicrobial activity assay of recombinant antilisterial proteins. Each crude recombinant protein was obtained from three different cultures and assayed in triplicates. (i) (A) P1_1, P1_2, and P1_3, (B) P2_1, P2_2, P2_3, (C) P3_1, P3_2, P3_3, and (D) P4_1, P4_2, P4_3 using agar well diffusion assay. C: crude protein of untransformed E. coli T7 Express LysY/Iq (New England BioLabs, Inc). S: cell-free culture supernatant of P. polymyxa Kp10. Inhibition zones were obtained in all of the recombinant proteins, thus confirming their antilisterial activity.
The antimicrobial activity of the P1, P2, P3, and P4 proteins is not unprecedented. Carvalho et al. (2018) also identified several ribosomal proteins (30S ribosomal protein S19, S20, and S21; 50S ribosomal protein L24 and L29), one histone-like DNA binding protein HU, and one translation initiation factor IF-1 produced by Lactobacillus sakei subsp. sakei 2a with the ability to inhibit the growth of several pathogenic bacteria, including L. monocytogenes and L. innocua. In addition, Pidutti et al. (2018) discovered the antibacterial activity of 50S ribosomal protein L27 and L30 produced by Lactobacillus salivarius SGL 03 against Enterococcus faecium Sintal Group, Streptococcus pyogenes ATCC 19615, and Staphylococcus uberis ATCC700407. The antimicrobial mechanisms of these proteins have been proposed by several authors. Carvalho et al. (2018) suggested that the histone-like DNA binding protein disrupts the DNA structure of susceptible strain and halts DNA replication, in turn contributing to cell death. A study on the mechanism of action of antimicrobial histone-like DNA binding protein, translation initiation factor IF-1, and ribosomal proteins produced by Lactobacillus sakei subsp. sakei 2a found that these proteins caused dissipation of membrane potential but has no impact on pH gradient. Carvalho et al. (2010) suggest that these proteins do not cause cell leakage or cell lysis. The high isoelectric point of these proteins may trigger strong attraction towards a negatively charged phosphate group of nucleic acids at normal physiological pH. Similarly, Pidutti et al. (2018) showed that ribosomal proteins L27 and L30 of Lactobacillus salivarius SGL03 conferred bactericidal action against Streptococcus pyogenes. Due to the role of these proteins in DNA replication and the regulation of protein expression, they are constitutively expressed by all P. polymyxa strains. Hence, a dedicated secretion pathway is probably present specifically in Kp10 to allow the secretion of these proteins to the extracellular milieu, either through direct penetration or exocytosis (Le et al., 2017). Polycationic nature of these peptides could also trigger high affinity to the anionic surfaces of the bacterial cell wall. This interaction may enable the peptide to cross the lipid bilayer of the sensitive strains. However, this remains to be elucidated.
Conclusion
This study determined four extracellular antilisterial proteins produced by Kp10, an isolate originating from milk curd. Eight different ribosomally-synthesized antimicrobial proteins were predicted in silico. Interestingly, two of the eight predicted proteins, namely histone-like DNA binding proteins (P1 and P2), were identified via mass spectrometry as being responsible for the antilisterial activity. In addition, one translation initiation factor IF-1 (P3) and one 50S ribosomal protein L29 (P4) were also found to contribute to the antilisterial activity of the Kp10. This was further confirmed by heterologous protein expression in E. coli and the antilisterial assay of the recombinant proteins against L. monocytogenes. Future studies on the mechanism of inhibition, factors that affect secretion, and the applicability of these proteins in food and medicine are called for. In the future, heterologous expression of the remaining antimicrobial proteins identified in silico in this study should be carried out, which increase the variability of antimicrobial protein reservoir to be explored further for food and medical industries.
Data Availability Statement
Draft genome sequence of P. polymyxa Kp10 can be found in NCBI Repository with assembly accession no: GCA_003052505.1 and BioProject accession no: PRJNA449134.
Author Contributions
The research was designed by NM, RAR, SM, and SA. The experiments were carried out by NM, AZ, SA, AAA, and RR. Data were analyzed and interpreted by NM, AM, and SA. SM, IH, AA, and RAR contributed reagents, materials and analysis tools. The manuscript was written by NM and revised by SA, AM, and IH. RAR, SM, and AM supervised and provided critical evaluation of the project. All authors had read and approved the final manuscript.
Funding
This study was supported by the funds of Ministry of Education, Malaysia and Universiti Putra Malaysia through Fundamental Research Grant Scheme (FRGS/1/2017/SKK11/UPM/01/1) and Putra Grant (GP/2017/9571800; Vote No: 9571800), respectively.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.00960/full#supplementary-material
References
Abbasiliasi, S., Tan, J. S., Ibrahim, T. A. T., Ramanan, R. N., Vakhshiteh, F., Mustafa, S., et al. (2012). Isolation of Pediococcus acidilactici Kp10 with ability to secrete bacteriocin-like inhibitory substance from milk products for applications in food industry. BMC Microbiol. 12:260. doi: 10.1186/1471-2180-12-260
Abriouel, H., Franz, C. M. A. P., Omar, N., Omar, N. B., and Galvez, A. (2011). Diversity and applications of Bacillus bacteriocins. FEMS Microbiol. Rev. 35, 201–232. doi: 10.1111/j.1574-6976.2010.00244.x
Ahmad, V., Khan, M. S., Jamal, Q. M. S., Alzohairy, M. A., Al Karaawi, M. A., and Siddiqui, M. U. (2017). Antimicrobial potential of bacteriocins: in therapy, agriculture and food preservation. Int. J. Antimicrob. Agents 49, 1–11. doi: 10.1016/j.ijantimicag.2016.08.016
Aleti, G., Antonielli, L., Corretto, E., Nikolic, B., Sessitsch, A., and Brader, G. (2016). The draft genome sequence of Paenibacillus polymyxa strain CCI-25 encompasses high potential for secondary metabolite production. Genome Announc. 4, 3–4. doi: 10.1128/genomeA.00366-16
Alvarez-Sieiro, P., Montalbán-López, M., Mu, D., and Kuipers, O. P. (2016). Bacteriocins of lactic acid bacteria: extending the family. Appl. Microbiol. Biotechnol. 100, 2939–2951. doi: 10.1007/s00253-016-7343-9
Angelo, K. M., Conrad, A. R., Saupe, A., Dragoo, H., West, N., Sorenson, A., et al. (2017). Multistate outbreak of Listeria monocytogenes infections linked to whole apples used in commercially produced, prepackaged caramel apples: United States, 2014–2015. Epidemiol. Infect. 145, 848–856. doi: 10.1017/S0950268816003083
Bagenda, D. K., and Yamazaki, K. (2007). Application of bacteriocins in food preservation and safety. Glob. Sci. Books 1, 137–148.
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Barbosa, J., Caetano, T., and Mendo, S. (2015). Class I and class II lanthipeptides produced by Bacillus spp. J. Nat. Prod. 78, 2850–2866. doi: 10.1021/np500424y
Boddy, C. N. (2014). Bioinformatics tools for genome mining of polyketide and non-ribosomal peptides. J. Ind. Microbiol. Biotechnol. 41, 443–450. doi: 10.1007/s10295-013-1368-1
Bushnell Brian (2014). BBMap: A Fast, Accurate, Splice-Aware Aligner. Available online at: https://www.osti.gov/biblio/1241166
Carocho, M., Barreiro, M. F., Morales, P., and Ferreira, I. C. F. R. (2014). Adding molecules to food, pros and cons: a review on synthetic and natural food additives. Comp. Rev. Food Sci. Food Saf. 13, 377–399. doi: 10.1111/1541-4337.12065
Carvalho, K. G., Bambirra, F. H. S., Kruger, M. F., Barbosa, M. S., Oliveira, J. S., Santos, A. M. C., et al. (2010). Antimicrobial compounds produced by Lactobacillus sakei subsp. sakei 2a, a bacteriocinogenic strain isolated from a Brazilian meat product. J. Ind. Microbiol. Biotechnol. 37, 381–390. doi: 10.1007/s10295-009-0684-y
Carvalho, K. G., Bambirra, F. H. S., Nicoli, J. R., Oliveira, J. S., Santos, A. M. C., Bemquerer, M. P., et al. (2018). Characterization of multiple antilisterial peptides produced by sakacin P-producing Lactobacillus sakei subsp. sakei 2a. Arch. Microbiol. 200, 635–44. doi: 10.1007/s00203-018-1477-3
Chun, J., Oren, A., Ventosa, A., Christensen, H., Arahal, D. R., da Costa, M. S., et al. (2018). Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int. J. Syst. Evol. Microbiol. 68, 461–466. doi: 10.1099/ijsem.0.002516
Cochrane, S. A., and Vederas, J. C. (2016). Lipopeptides from Bacillus and Paenibacillus spp.: a gold mine of antibiotic candidates. Med. Res. Rev. 36, 4–31. doi: 10.1002/med.21321
Cox, C. L., Doroghazi, J. R., and Mitchell, D. A. (2015). The genomic landscape of ribosomal peptides containing thiazole and oxazole heterocycles. BMC Genomics 16:778. doi: 10.1186/s12864-015-2008-0
da Silva Sabo, S., Vitolo, M., González, J. M. D., and de Oliveira, R. P. S. (2014). Overview of Lactobacillus plantarum as a promising bacteriocin producer among lactic acid bacteria. Food Res. Int. 64, 527–536. doi: 10.1016/j.foodres.2014.07.041
Grove, T. L., Himes, P. M., Hwang, S., Yumerefendi, H., Bonanno, J. B., Kuhlman, B., et al. (2017). Structural insights into thioether bond formation in the biosynthesis of sactipeptides. J. Am. Chem. Soc. 139, 11734–11744. doi: 10.1021/jacs.7b01283
Ha, S. M., Kim, C. K., Roh, J., Byun, J. H., Yang, S. J., Choi, S., et al. (2019). Application of the whole genome-based bacterial identification system, TRUEBAC ID, using clinical isolates that were not identified with three matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) systems. Ann. Lab. Med. 39, 530–536. doi: 10.3343/alm.2019.39.6.530
Haft, D. H., and Basu, M. K. (2011). Biological systems discovery in silico: radical S-adenosylmethionine protein families and their target peptides for posttranslational modification. J. Bacteriol. 193, 2745–2755. doi: 10.1128/JB.00040-11
Haft, D. H., Basu, M. K., and Mitchell, D. A. (2010). Expansion of ribosomally produced natural products: a nitrile hydratase- and Nif11-related precursor family. BMC Biol. 8:70. doi: 10.1186/1741-7007-8-70
He, Z., Kisla, D., Zhang, L., Yuan, C., Green-Church, K. B., and Yousef, A. E. (2007). Isolation and identification of a Paenibacillus polymyxa strain that coproduces a novel lantibiotic and polymyxin. Appl. Environ. Microbiol. 73, 168–178. doi: 10.1128/AEM.02023-06
He, Z., Yuan, C., Zhang, L., and Yousef, A. E. (2008). N-terminal acetylation in paenibacillin, a novel lantibiotic. FEBS Lett. 582, 2787–2792. doi: 10.1016/j.febslet.2008.07.008
Jeong, H., Choi, S. K., Ryu, C. M., and Park, S. H. (2019). Chronicle of a soil bacterium: Paenibacillus polymyxa E681 as a tiny guardian of plant and human health. Front. Microbiol. 10:467. doi: 10.3389/fmicb.2019.00467
Le, C.-F., Fang, C.-M., and Sekaran, S. D. (2017). Intracellular targeting mechanisms by antimicrobial peptides. Antimicrob. Agents Chemother. 61:e02340-16. doi: 10.1128/AAC.02340-16
Marchler-Bauer, A., Bo, Y., Han, L., He, J., Lanczycki, C. J., Lu, S., et al. (2017). CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 45, D200–D203. doi: 10.1093/nar/gkw1129
Medema, M. H., Kai, B., Peter, C., Victor, D. J., Piotr, Z., Michael, A. F., et al. (2011). AntiSMASH: rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucleic Acids Res. 39, W339–46. doi: 10.1093/nar/gkr466
Melby, J. O., Nard, N. J., and Mitchell, D. A. (2011). Thiazole/oxazole-modified microcins: complex natural products from ribosomal templates. Curr. Opin. Chem. Biol. 15, 369–378. doi: 10.1016/j.cbpa.2011.02.027
Messaoudi, S., Manai, M., Kergourlay, G., Prévost, H., Connil, N., Chobert, J. M., and Dousset, X. (2013). Lactobacillus salivarius: bacteriocin and probiotic activity. Food Microbiol. 36, 296–304. doi: 10.1016/j.fm.2013.05.010
Metelev, M. V., and Ghilarov, D. A. (2014). Structure, function, and biosynthesis of thiazole/oxazole-modified microcins. Mol. Biol. 48, 29–45. doi: 10.1134/S0026893314010105
Park, J. E., Kim, H. R., Park, S. Y., Choi, S. K., and Park, S. H. (2017). Identification of the biosynthesis gene cluster for the novel lantibiotic paenilan from Paenibacillus polymyxa E681 and characterization of its product. J. Appl. Microbiol. 123, 1133–1147. doi: 10.1111/jam.13580
Perez, R. H., Zendo, T., and Sonomoto, K. (2014). Novel bacteriocins from lactic acid bacteria (LAB): various structures and applications. Microb. Cell Fact. 13(Suppl. 1):S3. doi: 10.1186/1475-2859-13-S1-S3
Pidutti, P., Federici, F., Brandi, J., Manna, L., Rizzi, E., Marini, U., and Cecconi, D. (2018). Purification and characterization of ribosomal proteins L27 and L30 having antimicrobial activity produced by the Lactobacillus salivarius SGL 03. J. Appl. Microbiol. 124, 398–407. doi: 10.1111/jam.13646
Pongsavee, M. (2015). Effect of sodium benzoate preservative on micronucleus induction, chromosome break, and ala40thr superoxide dismutase gene mutation in lymphocytes. Biomed Res. Int. 2015:103512. doi: 10.1155/2015/103512
Scheffler, R. J., Colmer, S., Tynan, H., Demain, A. L., and Gullo, V. P. (2013). Antimicrobials, drug discovery, and genome mining. Appl. Microbiol. Biotechnol. 97, 969–978. doi: 10.1007/s00253-012-4609-8
Skowron, K., Hulisz, K., Gryn, G., Olszewska, H., Wiktorczyk, N., and Paluszak, Z. (2018). Comparison of selected disinfectants efficiency against Listeria monocytogenes biofilm formed on various surfaces. Int. Microbiol. 21, 23–33. doi: 10.1007/s10123-018-0002-5
Song, N. E., Cho, H. S., and Baik, S. H. (2016). Bacteria isolated from Korean black raspberry vinegar with low biogenic amine production in wine. Braz. J. Microbiol. 47, 452–460. doi: 10.1016/j.bjm.2016.01.016
Tatusova, T., Dicuccio, M., Badretdin, A., Chetvernin, V., Nawrocki, E. P., Zaslavsky, L., et al. (2016). NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 44, 6614–6624. doi: 10.1093/nar/gkw569
Van Heel, A. J., de Jong, A., Song, C., Jakob, H. V., Jan, K., and Oscar, P. K. (2018). BAGEL4: a user friendly web server to thoroughly mine RIPPs and bacteriocins. Nucleic Acids Res. 46, W278–81. doi: 10.1093/nar/gky383
Yang, S. C., Lin, C. H., Sung, C. T., and Fang, J. Y. (2014). Antibacterial activities of bacteriocins: application in foods and pharmaceuticals. Front. Microbiol. 5:683. doi: 10.3389/fmicb.2014.00683
Zhao, P., Xue, Y., Gao, W., Li, J., Zu, X., Fu, D., et al. (2018). Bacillaceae-derived peptide antibiotics since 2000. Peptides 101, 10–16. doi: 10.1016/j.peptides.2017.12.018
Keywords: antilisterial protein, genome mining, heterologous protein expression, mass spectrometry, Paenibacillus polymyxa Kp10, Listeria monocytogenes
Citation: Mokhtar NFK, Hashim AM, Hanish I, Zulkarnain A, Raja Nhari RMH, Abdul Sani AA, Abbasiliasi S, Ariff A, Mustafa S and Rahim RA (2020) The Discovery of New Antilisterial Proteins From Paenibacillus polymyxa Kp10 via Genome Mining and Mass Spectrometry. Front. Microbiol. 11:960. doi: 10.3389/fmicb.2020.00960
Received: 19 December 2019; Accepted: 22 April 2020;
Published: 02 July 2020.
Edited by:
Daniel Yero, Autonomous University of Barcelona, SpainReviewed by:
Haeyoung Jeong, Korea Research Institute of Bioscience and Biotechnology (KRIBB), South KoreaQiong Shi, Beijing Genomics Institute (BGI), China
Copyright © 2020 Mokhtar, Hashim, Hanish, Zulkarnain, Raja Nhari, Abdul Sani, Abbasiliasi, Ariff, Mustafa and Rahim. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Amalia Mohd Hashim, YW1hbGlhQHVwbS5lZHUubXk=; Sahar Abbasiliasi, c2FoYXJAdXBtLmVkdS5teQ==; cy5hYmJhc2lsaWFzaUBnbWFpbC5jb20=
 Nur Fadhilah Khairil Mokhtar
Nur Fadhilah Khairil Mokhtar Amalia Mohd Hashim
Amalia Mohd Hashim Irwan Hanish
Irwan Hanish Aisyah Zulkarnain2
Aisyah Zulkarnain2 Raja Mohd Hafidz Raja Nhari
Raja Mohd Hafidz Raja Nhari Sahar Abbasiliasi
Sahar Abbasiliasi