- 1State Key Laboratory of Microbial Technology, Marine Biotechnology Research Center, Shandong University, Qingdao, China
- 2Laboratory for Marine Biology and Biotechnology, Qingdao National Laboratory for Marine Science and Technology, Qingdao, China
- 3College of Marine Life Sciences, Ocean University of China, Qingdao, China
- 4Department of Molecular Biology, Qingdao Vland Biotech Inc., Qingdao, China
The biocontrol fungus Trichoderma longibrachiatum SMF2 secretes a large quantity of peptaibols that have been shown to have a range of biological activities and therefore great application values. However, the mechanism of the regulatory expression of peptaibols is still unclear. The putative methyltransferase LaeA/LAE1 is a global regulator involved in the biosynthesis of some secondary metabolites in filamentous fungi. In this study, we demonstrated that the ortholog of LaeA/LAE1 in the biocontrol fungus T. longibrachiatum SMF2, TlLAE1, plays an important role in the regulation of peptaibols production. Deletion of Tllae1 resulted in a slight negative impact on mycelial growth, and a significant defect in conidial production. Deletion of Tllae1 also compromised the production of peptaibols to a large degree. Further analyses indicated that this defect occurred at the transcriptional level of the two synthetases-encoding genes, tlx1 and tlx2, which are responsible for peptaibols production. By contrast, constitutive expression of Tllae1 in T. longibrachiatum SMF2 led to 2-fold increased peptaibols production, suggesting that this is a strategy to improve peptaibols production in Trichoderma fungi. These results demonstrate the important role of LAE1 in the regulation of peptaibols production in T. longibrachiatum SMF2.
Introduction
Peptaibols are a group of small peptides containing 5–20 amino acid residues, some of which are non-proteinogenic amino acids. They are characterized as having a high content of α-Aminoisobutyric acid (Aib) residues and generally have both N-terminal (mainly acetyl groups) and C-terminal modifications in the form of amino alcohol groups rather than amino acids. Peptaibols are ecologically and commercially important due to their antimicrobial and anticancer properties as well as their ability to induce systemic resistance in plants against microbial invasion (Wiest et al., 2002; Viterbo et al., 2007; Reino et al., 2008).
Fungal species of the genus Trichoderma, such as the well characterized T. atroviride, T. virens, and T. longibrachiatum, are excellent producers of peptaibols (Marik et al., 2019). Of these fungal producers, T. longibrachiatum SMF2 produces a large quantity of peptaibols named as Trichokonins (TKs) that are mainly classified into 20-aa Trichokonins A (TKA) and 12-aa Trichokonins B (TKB) (Song et al., 2006; Xie et al., 2014). TKs produced by T. longibrachiatum SMF2 have been demonstrated to have broad-spectrum antimicrobial activities (Song et al., 2006; Shi et al., 2012) as well as other biological functions including the elicitation of systemic resistance in tobacco and Chinese cabbage (Luo et al., 2010; Li et al., 2014) and the induction of programmed cell death in tumor cells (Shi et al., 2010).
Fungal peptaibols are naturally synthesized by large multi-module protein complexes known as non-ribosomal peptide synthetases (NRPSs), in which each module catalyzes the incorporation of a single proteinogenic or non-proteinogenic amino acid (El-Bondkly, 2014). Combining genome sequencing and targeted gene deletion analyses, the two NRPS encoding genes, tlx1 and tlx2, responsible for 20-aa TKA and 12-aa TKB, have been identified in T. longibrachiatum SMF2 (Xie et al., 2015; Zhou et al., 2019), which is consistent with the reports that two peptaibol synthases are present in the genomes of T. longibrachiatum and other 11 different Trichoderma spp. (Kubicek et al., 2019). We recently demonstrated that the stp1 gene, encoding a putative glucose sensor in T. longibrachiatum SMF2, represses peptaibols production, and stp1 deletion led to significantly enhanced expression of tlx1 and tlx2, and therefore remarkably increased peptaibols production (Zhou et al., 2019). Despite this, our understanding of the regulatory mechanism of peptaibols production in T. longibrachiatum is much limited, which hampers efficient genetic engineering for construction of hyperproducing strains.
The putative S-adenosylmethionine-dependent methyltransferase LaeA (stands for “loss of aflR expression-A”), which forms a trimeric protein complex with VeA and VelB, has been demonstrated to be involved in the biosynthesis of a large number of secondary metabolites in many filamentous fungi, such as Aspergillus nidulans, Penicillium chrysogenum, Fusarium fujikuroi, and Cochliobolus heterostrophus (Bok and Keller, 2004; Bok et al., 2006; Kale et al., 2008; Hoff et al., 2010; Wiemann et al., 2010; Oda et al., 2011; Wu et al., 2012). LaeA is also involved in the regulation of conidiation in many filamentous fungi (Sugui et al., 2007; Bayram et al., 2010; Hoff et al., 2010; Wiemann et al., 2010; Chang et al., 2012; Jiang et al., 2012; Wu et al., 2012) and fruiting body formation in Aspergillus species (Amaike and Keller, 2009). LaeA orthologs, named LAE1, have been found in Trichoderma species including T. reesei and T. atroviride. LAE1 regulates the expression of cellulases and polysaccharide hydrolases in T. reesei (Seiboth et al., 2012), which is an excellent cellulolytic fungus widely applied in industry (Bischof et al., 2016; Druzhinina and Kubicek, 2016). Transcriptome analyses on lae1-null and -overexpressing T. reesei strains were further performed to assess the role of LAE1 in genome-wide gene expression. Genes significantly regulated by LAE1 include those encoding ankyrin proteins, iron uptake, heterokaryon incompatibility proteins, PTH11-receptors, and oxidases/monooxygenases (Karimi-Aghcheh et al., 2013). In addition, 7 of 17 polyketide or non-ribosomal peptide synthase encoding genes are positively regulated by LAE1 (Karimi-Aghcheh et al., 2013). Notably, one NRPS synthetase encoding gene responsible for synthesis of paracelsin, one kind of peptaibol secreted by T. reesei (Neuhof et al., 2007), is significantly up-regulated in the lae1 null mutant but also lae1-overexpressing mutant (Karimi-Aghcheh et al., 2013). Moreover, similar to orthologs in other filamentous fungi, LAE1 in T. reesei and T. atroviride are found to be positively involved in conidiation (Aghcheh et al., 2013; Karimi-Aghcheh et al., 2013).
In this study, we presented that the ortholog of LaeA/LAE1 in the biocontrol fungus T. longibrachiatum SMF2, TlLAE1, is involved in the regulation of peptaibols production. The results showed that targeted deletion of Tllae1 resulted in a significant defect in peptaibols production, whereas constitutive overexpression of Tllae1 evidently enhanced the yield of peptaibols in T. longibrachiatum SMF2.
Materials and Methods
Strains, Media and Cultivation Conditions
Trichoderma longibrachiatum SMF2 (CCTCC No. 209031) stocked in our laboratory was used as the wild type (WT) strain. T. longibrachiatum SMF2 strains were routinely maintained on potato dextrose agar (PDA) plates. Escherichia coli DH5α was used for plasmid construction and cloning.
Construction of T. longibrachiatum SMF2 Recombinant Strains
An overlap extension PCR method (Amaike and Keller, 2009) was used to create the fragments for targeted gene deletion. Two approximately 2.0 kb fragments upstream and downstream of Tllae1 were amplified from the genomic DNA of T. longibrachiatum SMF2 using the primer pairs ΔTllae1upF/ΔTllae1upR and ΔTllae1downF/ΔTllae1downR, respectively. The 2.4 kb hph gene (a hygromycin B phosphotransferase encoding gene) was amplified using the primer pair FhygBF/FhygBR from the plasmid pUCATPH (Lu et al., 1994). The three resulting fragments were fused and amplified using the primer pair CΔTllae1F/CΔTllae1R, and was subsequently transformed into T. longibrachiatum SMF2 to construct the Tllae1-deleted mutant ΔTllae1. To generate the complementation vector for Tllae1, three fragments named CF1, CF2, and CF3 were amplified individually and ligated into the plasmid pMD-19T. Specifically, the putative native promoter (∼1.0 kb), full-length coding sequence (∼2.2 kb), and the putative terminator of Tllae1 (∼0.5 kb) were amplified from T. longibrachiatum SMF2 with primers CpΔTllae1upF/CpΔTllae1upR to generate the fragment CF1. The second fragment CF2 (∼3 kb), the acetamidase-encoding gene (amdS) cassette, was amplified from the pALK424 vector using the primer pair CpΔTllae1-amdSF/CpΔTllae1-amdSR. The third fragment CF3 (∼1.6 kb) which is immediately downstream Tllae1 coding sequence was amplified from T. longibrachiatum SMF2 genomic DNA using the primer pair CpΔTllae1downF/CpΔTllae1downR. The pMD-19T plasmid containing the above three fragments was linearized with HindIII and applied to transform the ΔTllae1 cells to generate the Tllae1 complementation strain. To obtain the overexpression vector, the fragment including the full-length coding sequence of Tllae1 and its native terminator was amplified from T. longibrachiatum SMF2 genomic DNA with primers Tllae1CE-F/Tllae1CE-R, and was ligated into the pIG1783 plasmid (Penttilä et al., 1987), which contains the promoter sequence of the glyceraldehydes- 3-phosphate dehydrogenase encoding gene (gpd). The resultant plasmid pIG1783-Tllae1CE was linearized with HindIII, and then transformed the T. longibrachiatum SMF2 cells to generate the Tllae1 overexpression strain.
Transformation of T. longibrachiatum SMF2 was carried out essentially as previously described (Zhou et al., 2012). For southern blot hybridization, genomic DNA extracted from the WT or ΔTllae1 strain was digested using SmaI prior to hybridization. Detection of probe-hybridized DNA fragment was carried out using the DIG High Prime DNA Labeling and Detection Starter Kit I (Roche Diagnostics, Mannheim, Germany). The primers used in this study were listed in Supplementary Table S1. The illustration of recombinant strain construction and verification was shown in Supplementary Figures S1–S3.
Vegetative Growth, Conidiation and Conidial Germination Analyses
To compare the vegetative growth of the WT and mutant strains, a same-size piece (1-cm-diameter) of mycelia of each strain were inoculated on PDA plates and incubated at 28°C for 7 days. To analyze biomass accumulation, equal amounts of conidia (2 × 106) were inoculated into 100 mL of potato dextrose broth (PDB) (Hermosa et al., 2004) and cultured at 28°C. Mycelia were harvested at an interval of 24 h, and the dry weight of mycelia after dehydration at 65°C was determined. To analyze conidiation, approximately 104 conidia were spread on the 90-mm PDA plates and incubated under constant white light (31 μmol photons m–2s–1, 2200 lx) condition at 28°C for 3 days. The number of conidia was counted with a hemocytometer under an inverted optical microscope (Olympus, Tokyo, Japan). To analyze conidial germination, approximately 104 conidia were inoculated into 100 mL of PDB, and conidial germination after 12 h-cultivation was analyzed under an optical microscope.
Hydrophobicity Assay
Approximately 10 μL of the conidia suspension (106 conidia/mL) of all the tested strains was inoculated onto PDA plates and cultivated at 28°C for 7 days. Fifty μL of water was placed on a fungal colony of each strain and maintaining of the drop of water on the colony surface was monitored.
Scanning Electron Microscope Observation
To visualize the hyphal morphology difference between the mutant and WT strains, equal amounts of approximately 108 conidia were inoculated into PDB medium and cultured at 28°C for 48 h. Mycelia were fixed with 2.5% glutaraldehyde in 100 mM PBS buffer (pH 7.2) at 4°C for 12 h, and then washed with 100 mM PBS buffer three times. They were further fixed with 1% (w/v) osmium tetroxide for 2 h at room temperature, followed by wash with 100 mM PBS buffer three times, and then dehydrated with ethanol. Samples were mounted, sputter coated with 60% gold and 40% palladium, and finally viewed with a JEOL JSM-7600F scanning electron microscope.
High-Performance Liquid Chromatography (HPLC) Analyses
High-Performance Liquid Chromatography analyses were performed to quantify the yield of peptaibols production essentially as previously described (Zhou et al., 2019). Approximately equal amounts of approximately 108 conidia of the WT and mutant strains were inoculated into 500 mL Erlenmeyer flasks containing 100 mL PDB, and were cultivated at 28°C for 10 days with shaking at 160 rpm. After centrifugation at 12,000 rpm for 20 min, 30 mL of the collected culture supernatant was loaded on a Cleanert C18 SPE cartridges (500 mg/6 mL, Agela Technologies, China) and eluted with 2 mL methanol to achieve 15 times concentrated peptaibol samples. The samples (15 μL each) were subsequently subjected to HPLC to analyze the content of peptaibols using a reversed phase analytical column (Sunfire C18, 4.6 × 250 mm, Waters, Ireland). The solvent system was MeOH/H2O (84:16, v/v) at a flow rate of 1.0 mL min–1 and the chromatogram was monitored at 203 nm (Wiest et al., 2002; Mukherjee et al., 2011). The fungal mycelia after different incubation periods were collected, dried and weighted. Peptaibols collected previously (Zhou et al., 2019) were used as standards. The production yield of peptaibols was determined as the peptaibols amount per mg fungal biomass.
Quantitative Real-Time RT-PCR
Wild type, ΔTllae1, CpΔTllae1 and CETllae1 strains were cultivated in PDB at 28°C for 48 h. Total RNA was extracted from the harvested mycelia using TRIzol reagent (Invitrogen, Grand Island, NY, United States) and further treated with the TURBO DNA-free kit (Ambion, Austin, TX, United States) to remove genomic DNA according to the manufacturer’s instruction. Reverse transcription was carried out using the PrimeScript RT reagent kit (Takara, Tokyo, Japan) according to the instruction. Quantitative PCR was performed on a LightCycler 480 II thermocycler (Roche, Basel, Switzerland). Amplification reactions were performed using the SYBR Green Supermix (Takara, Tokyo, Japan) according to the manufacturer’s instructions. Data analysis was performed using the comparative CT method (Schmittgen and Livak, 2008). The endogenous tef1 gene was used as the control for normalization (Brunner et al., 2008). Primers are listed in Supplementary Table S1.
Sequence Analysis
The amino acid sequences of proteins were retrieved from NCBI or JGI databases. Sequence alignments were performed using Clustal W (Thompson et al., 2002). Phylogenetic tree was constructed with MEGA7.0 (Kumar et al., 2016) using the neighbor-joining method with 1,000 bootstraps.
Statistical Analysis
Statistical analysis was performed using the student’s t-test analysis. At least three biological replicates were performed for each analysis and the results and errors are the mean and SD, respectively, from three replicates.
Results
Identification of the LaeA/LAE1 Ortholog in T. longibrachiatum SMF2
To identify the LaeA/LAE1 ortholog in T. longibrachiatum SMF2, its genome was searched with the amino acid sequence of A. nidulans LaeA (Bok and Keller, 2004) and T. reesei LAE1 (Seiboth et al., 2012) as inquiry, respectively, and the protein-encoding gene SMF2FGGW_101365, which was hereafter named Tllae1, was retrieved. The cDNA sequence of Tllae1 has 1077 bp nucleotides in length and encodes a putative protein of 358 amino acids. The 75–260 amino acids specify the S-adenosylmethionine-dependent methyltransferase domain. Amino acid sequence comparison showed that TlLAE1 shares sequence identity of 29 and 91% with LaeA/LAE1 from A. nidulans and T. reesei, respectively. Phylogenic analysis further revealed that orthologs of TlLAE1 are widely distributed across filamentous ascomycete fungi, including several Trichoderma biocontrol fungi which produce peptaibols (Figure 1).
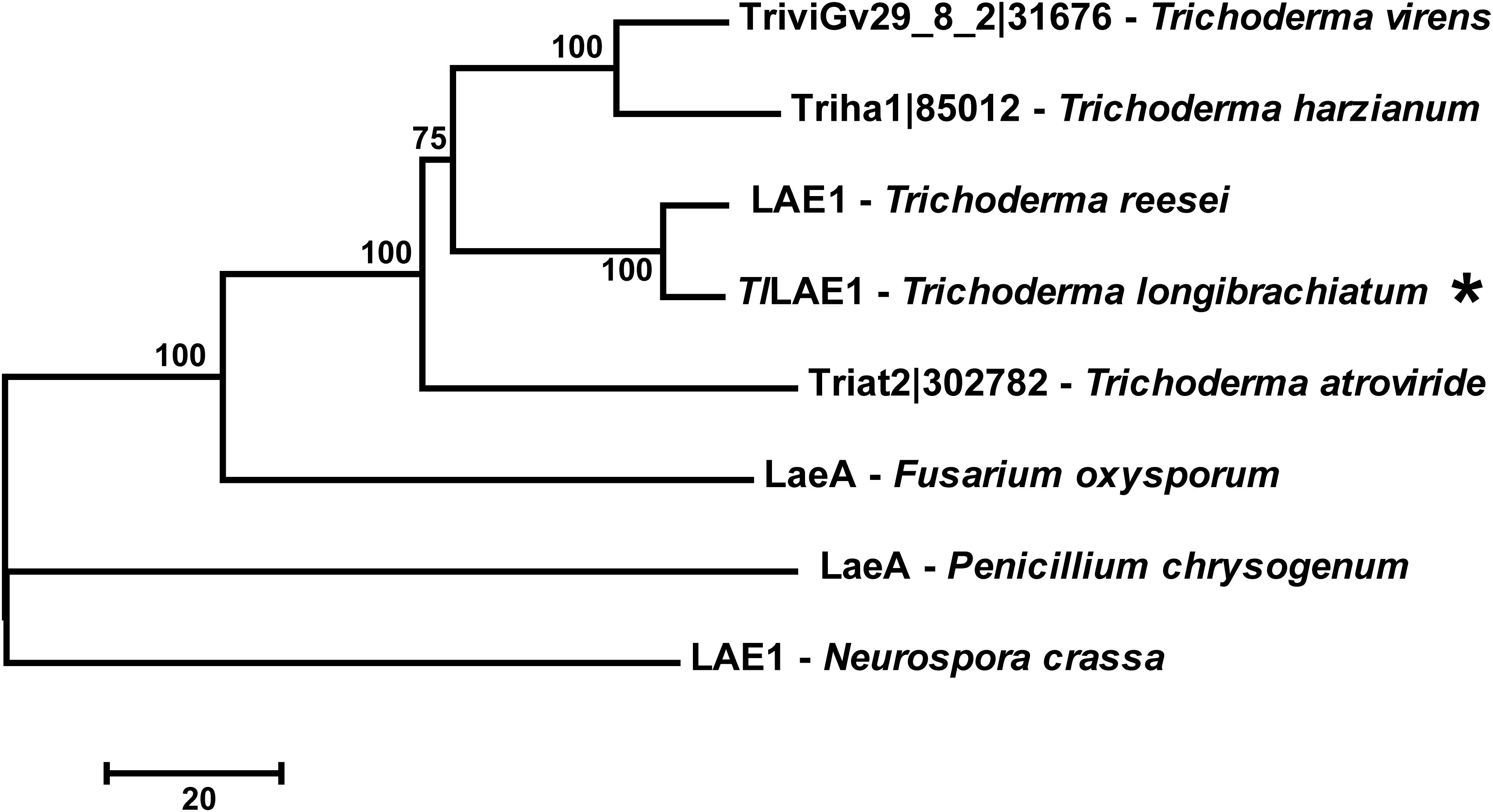
Figure 1. Phylogenetic analyses of TlLAE1 and its orthologs. Sequence alignments were performed with Clustal W (Thompson et al., 2002), and the neighbor-joining tree was generated with Mega 7.0 (Kumar et al., 2016). Numbers on the tree branches represent the bootstrap support calculated per 1000 bootstrap replicates. JGI entry numbers of TlLAE1 orthologs were shown. TlLAE1 is indicated by an asterisk.
Deletion of Tllae1 Resulted in a Slight Impact on Vegetative Growth but Significantly Compromised Conidiation of T. longibrachiatum SMF2
Targeted deletion of Tllae1 in T. longibrachiatum SMF2 was performed by gene replacement of Tllae1 coding region with an expression cassette of the hygromycin B phosphotransferase encoding gene, hph, and therefore, the mutant ΔTllae1 was constructed. To analyze the effect of the TlLAE1 absence on the vegetative growth of T. longibrachiatum SMF2, ΔTllae1 and WT strains were cultured in PDB, and their biomass was determined. As shown in Figure 2A, ΔTllae1 displayed a slight defect in biomass accumulation, as demonstrated by a 20–30% decrease in biomass accumulation. This slight growth defect was almost completely rescued by the in situ complementation of Tllae1, whose expression was driven under its native promoter (Supplementary Figure S2). In line with the slight difference in biomass accumulation, no evident difference was observed in the mycelial morphology between the mutant and the WT strain (Figure 2B), suggesting that the absence of TlLAE1 did not have a significant impact on fungal growth.
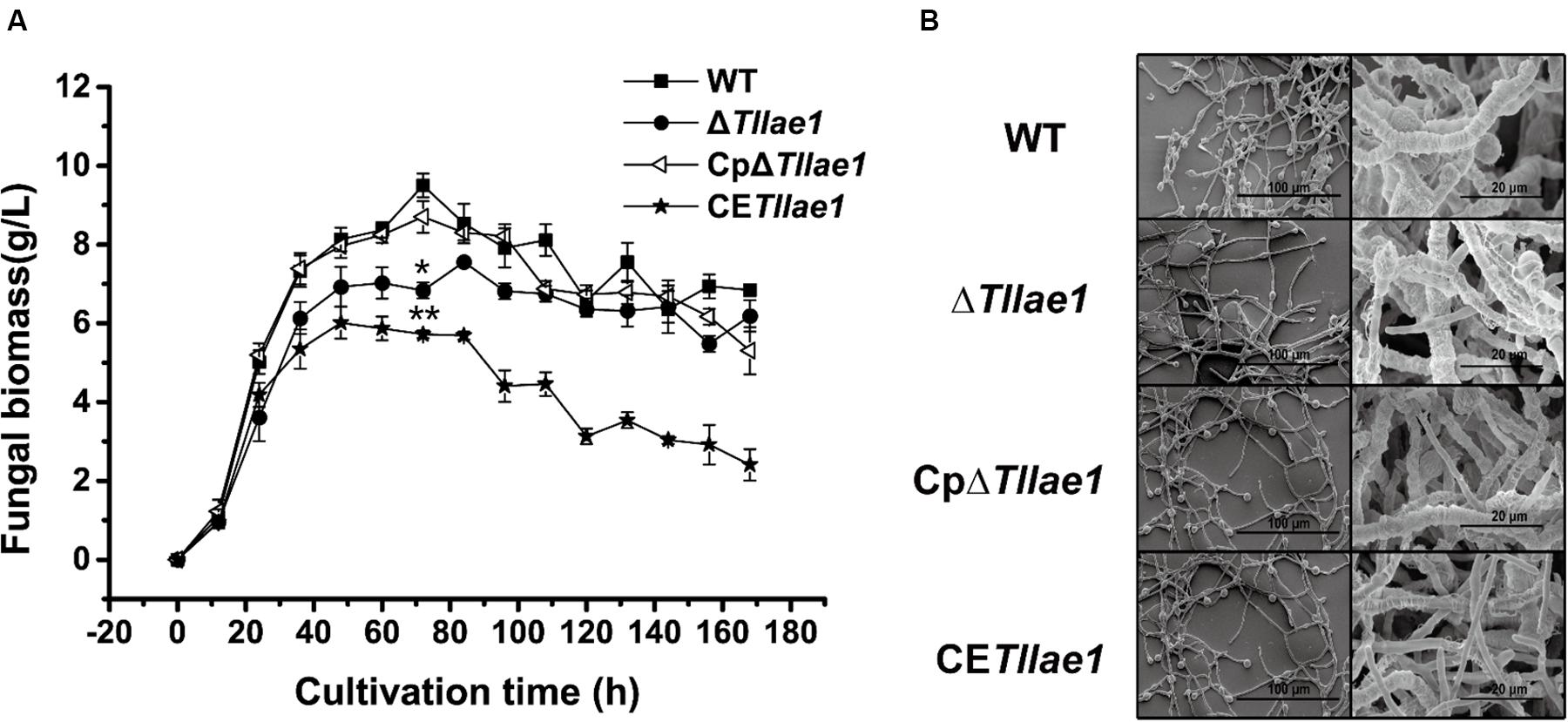
Figure 2. Growth analyses of T. longibrachiatum SMF2 and its mutants. (A) Determination of biomass accumulation of WT, ΔTllae1, CpΔTllae1 and CETllae1 strains that were cultured in PDB at 28°C. Significant differences (T-test *P < 0.05, **P < 0.01) were observed in the biomass accumulation between WT and ΔTllae1, and CETllae1. Values represent the mean of three biological replicates. Error bars are the SD from these replicates. (B) Scanning electron microscope observation of hyphal morphology of WT, ΔTllae1, CpΔTllae1 and CETllae1 strains that were cultured in PDB at 28°C for 48 h. The data shown in the graph are representative of results of triplicate experiments with three independent transformants.
In contrast to mycelial growth, ΔTllae1 displayed a severe defect in conidiation, as evidenced by the significantly decreased formation of conidia compared to that of the WT strain (Figures 3A,B). In filamentous fungi, small secreted proteins known as hydrophobins are mostly found in conidia, making the conidia surface hydrophobic (Linder et al., 2005). We therefore examined the hydrophobicity of the mutant and WT colonies, respectively, and found that water droplets dispersed immediately on the mutant colony (Figure 3C), indicating that the mutant colony was less hydrophobic than the WT colony. This is consistent with the notion that the ΔTllae1 cells produced much less conidia than the WT cells. The defect in conidia production was rescued in the complemented strain, CpΔTllae1 (Figure 3C). However, although the conidia production was significantly compromised, conidial germination was slightly impacted in the ΔTllae1 mutant (Figure 3D).
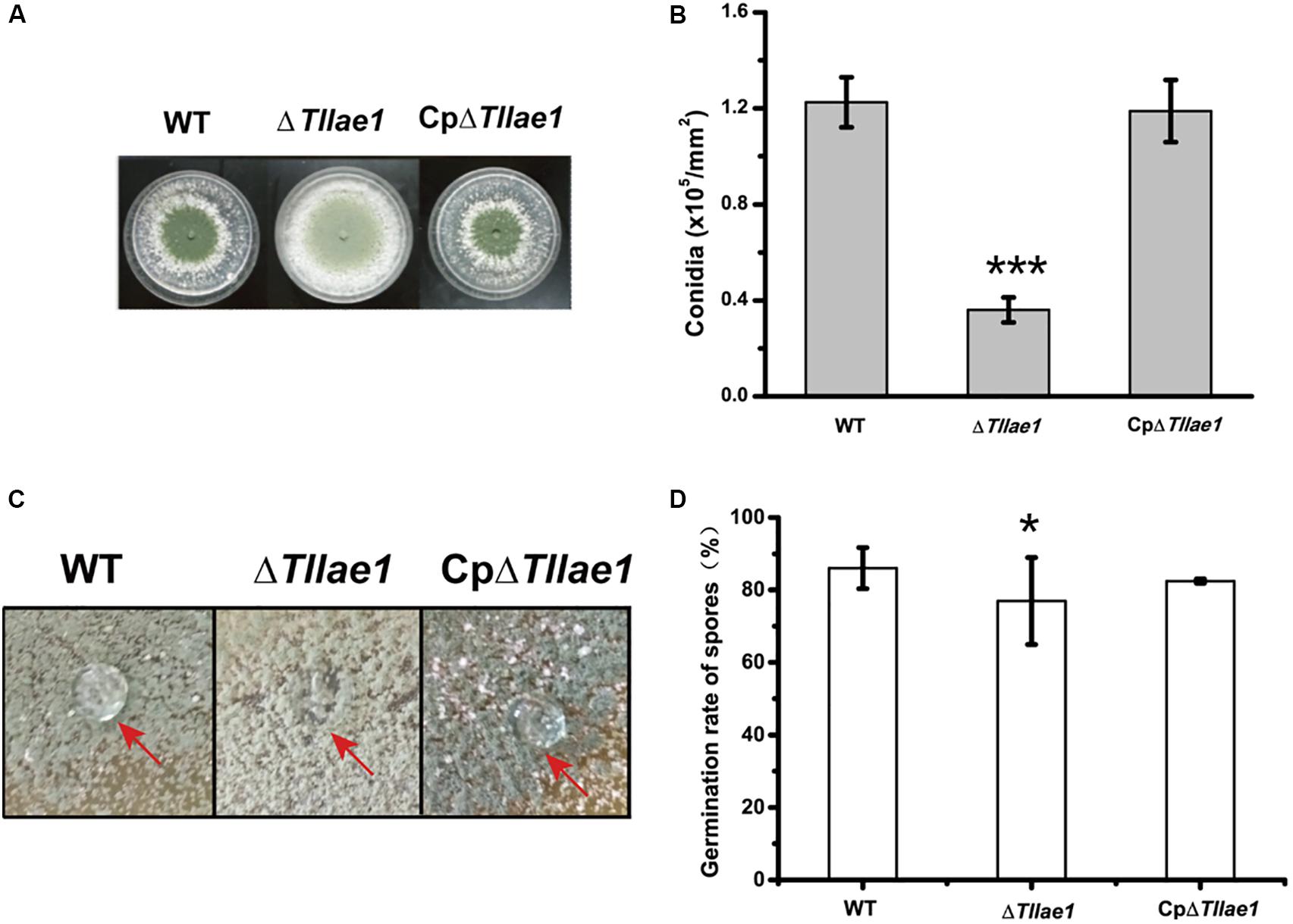
Figure 3. Effect of deletion of Tllae1 on conidial production and conidial germination of T. longibrachiatum SMF2 and its mutants. (A) Phenotypic analyses of conidial production of WT, ΔTllae1, and CpΔTllae1 strains cultured on PDA plates at 28°C for 3 days. (B) Quantitation of conidia produced by WT, ΔTllae1, and CpΔTllae1 strains as shown in panel (A). (C) Hydrophobic phenotypes of conidia produced by mycelial colonies of WT, ΔTllae1, and CpΔTllae1 strains cultured on PDA plates at 28°C for 5 days. Fifty μL of water was pipetted on the colony surface and photographs were taken 5 min later. (D) Analyses of conidial germination from WT, ΔTllae1, and CpΔTllae1 strains after being inoculated into PDB and cultured at 28°C for 12 h. Values represent the mean of three biological replicates. Error bars are the SD from these replicates. Significant differences (T-test *P < 0.05, ***P < 0.001) were observed in conidia production and germination between WT and ΔTllae1, and CETllae1.
Deletion of Tllae1 Significantly Compromised Peptaibols Production of T. longibrachiatum SMF2
In order to study the role of TlLAE1 in the regulation of peptaibols synthesis, we measured the extracellular amount of peptaibols in the ΔTllae1 and WT strains that were cultured in PDB for 10 days. As shown in Figure 4, deletion of Tllae1 resulted in a significant defect in the production of extracellular peptaibols, compared to that of the WT strain. We further analyzed the amount of the two major peptaibols, 20-aa TKA and 12-aa TKB, and found that their synthesis was both significantly compromised, with a more severe impact on TKA. To analyze whether this effect occurred at the transcriptional level, quantitative RT-PCR was performed to determine the relative transcripts of the two peptaibol synthesase encoding genes, tlx1 and tlx2, that are responsible for synthesizing TKA and TKB, respectively (Xie et al., 2014; Zhou et al., 2019). The results indicated that the relative transcription of tlx1 and tlx2 were both significantly decreased, with a more severe impact on that of tlx1 (Figure 5), which is consistent with the observation that extracellular TKA production was more affected than that of TKB. The significant defect in peptaibols production resulted from Tllae1 deletion is almost completely rescued in the complemented strain (Figures 4, 5). As a whole, the above results suggested that TlLAE1 is involved in the regulation of peptaibols production.
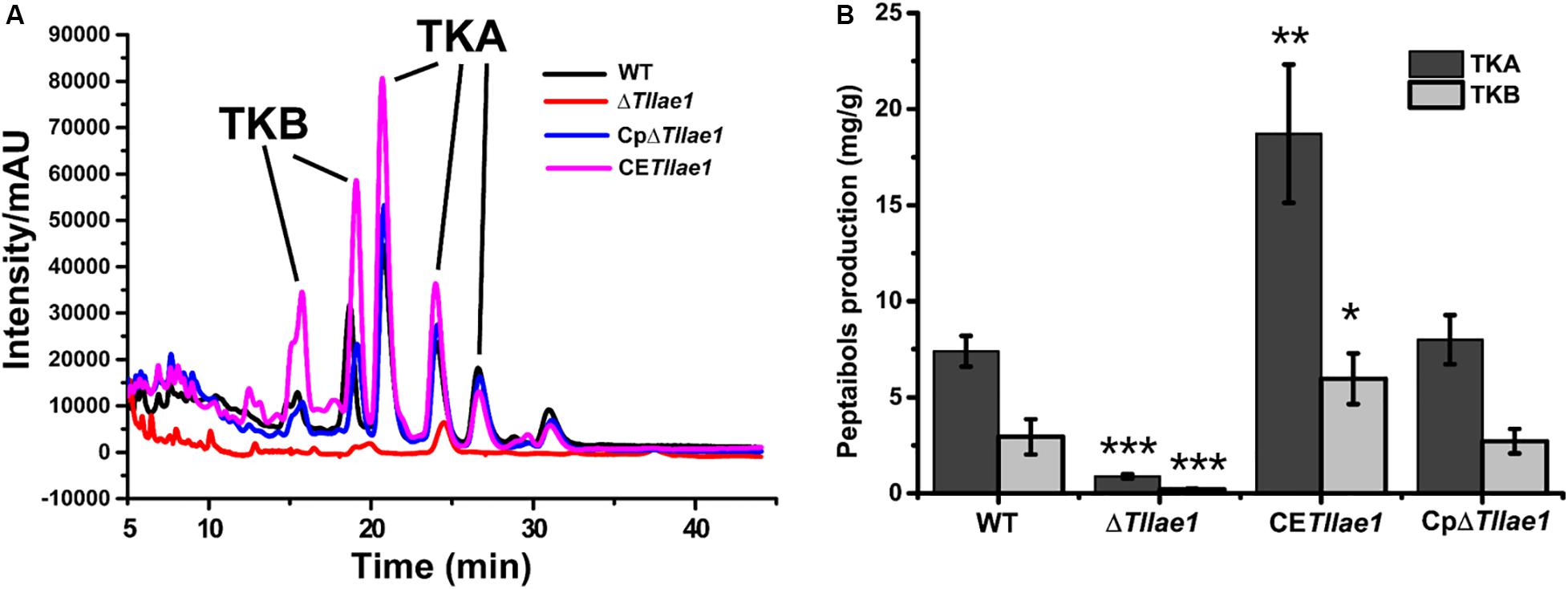
Figure 4. HPLC analyses of peptaibols production of T. longibrachiatum SMF2 and its mutants. (A) HPLC analyses of extracellular peptaibols produced by WT, ΔTllae1, CpΔTllae1, and CETllae1 strains cultured in PDB medium at 28°C for 10 days. (B) Peptaibols production of WT, CETllae1 and CpΔTllae1 strains after cultivation in PDB at 28°C. Values represent the mean of three biological replicates. Error bars are the SD from these replicates. Significant differences (T-test *P < 0.05, **P < 0.01, ***P < 0.001) were observed in production of TKA and TKB between WT and ΔTllae1, and CETllae1.
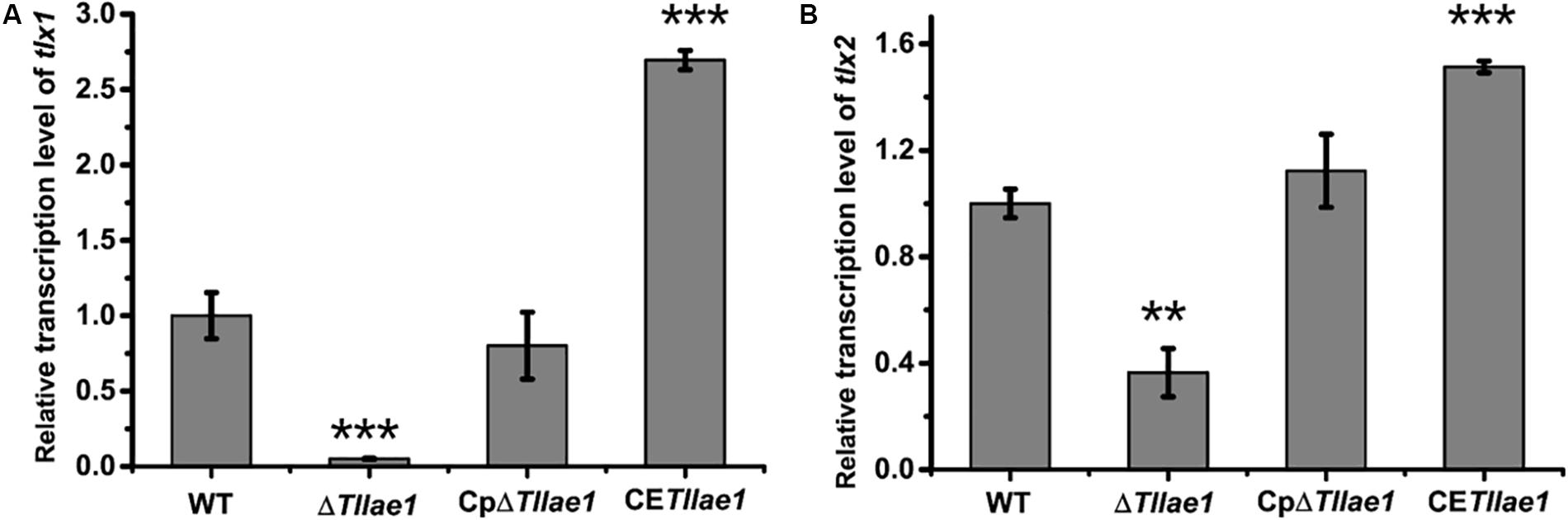
Figure 5. The effect of the absence or constitutive expression of Tllae1 on the relative transcriptional levels of the two synthetases-encoding genes, tlx1 and tlx2. (A) Relative transcription level of tlx1. (B) Relative transcription level of tlx2. All the strains were cultured at 28°C for 48 h. The gene tef1 was used as the control. Values represent the mean of three biological replicates. Error bars are the SD from these replicates. Significant differences (T-test **P < 0.01, ***P < 0.001) were observed in the relative transcriptional levels of tlx1 and tlx2 between WT and ΔTllae1, and CETllae1.
Constitutive Expression of Tllae1 Led to Enhanced Peptaibols Production
We further analyzed the effect of constitutive expression of Tllae1, under control of the constitutive gpd promoter (Punt et al., 1990; Supplementary Figure S3), on extracellular peptaibols production and mycelial growth. The overexpression of Tllae1 resulted in a compromised biomass accumulation and differing mycelial morphology (Figure 2), however, the overall peptaibols production in the overexpression strain was obviously enhanced (Figures 4, 5). Specifically, compared to those in the WT strain, production of TKA and TKB increased around 2.5- and 2-fold, respectively (Figure 4). These results indicated that increasing Tllae1 expression is an effective strategy to improve peptaibols production in T. longibrachiatum SMF2.
Discussion
LaeA was first identified as a regulator of secondary metabolism in Aspergillus, and later on was demonstrated to be required for the biosynthesis of many secondary metabolites in fungi (Bok and Keller, 2004; Bok et al., 2006; Kale et al., 2008; Hoff et al., 2010; Wiemann et al., 2010; Oda et al., 2011; Wu et al., 2012). Moreover, LaeA/LAE1 has been found to be involved in the regulation of conidiation and morphological development (Sugui et al., 2007; Amaike and Keller, 2009; Bayram et al., 2010; Wiemann et al., 2010; Chang et al., 2012; Jiang et al., 2012; Wu et al., 2012; Karimi-Aghcheh et al., 2013). Therefore, LaeA/LAE1 is considered to act as a global regulator. In this study, we identified the ortholog of A. nidulans LaeA and T. reesei LAE1, TlLAE1, in the biocontrol fungus T. longibrachiatum SMF2 that produces large quantities of peptaibols. Our results showed that TlLAE1 is involved in the regulation of conidiation and peptaibols production in T. longibrachiatum SMF2. These results are consistent with the previous reports regarding the function of LaeA/LAE1 in conidiation and secondary metabolism (Bok and Keller, 2004; Bok et al., 2006; Kale et al., 2008; Hoff et al., 2010; Wiemann et al., 2010; Oda et al., 2011; Wu et al., 2012).
It is not uncommonly observed that overexpression of LaeA results in enhanced production of several secondary metabolites in fungi. For example, several PKS (polyketide synthetase) and NRPS encoding genes have been shown to be upregulated in the lae1-overexpressing T. reesei (Karimi-Aghcheh et al., 2013); overexpression of LaeA in A. fumisynnematus significantly increased the production of cyclopiazonic acid, which was previously known as mycotoxin (Hong et al., 2015); and in P. citrinum, overexpression of LaeA enhanced mevastatin production (Zheng et al., 2014). More strikingly, overexpression of LaeA has even led to synthesis of new metabolites that have not been detected in the WT strain (Karimi-Aghcheh et al., 2013; Hong et al., 2015; Jiang et al., 2016; Soukup et al., 2016). In our study, constitutive overexpression of Tllae1 markedly enhanced the peptaibols yield in T. longibrachiatum SMF2. The yield of the two main 20-aa TKA and 12-aa TKB were increased 2.5- and 2-fold, respectively, as a result of constitutive overexpression of Tllae1. These findings support that overexpression of LaeA/LAE1 serves as an effective strategy to improve production of biologically active secondary metabolites and even to discover new metabolites via activating silent gene clusters in fungi (Soukup et al., 2016), which are rich resources for valuable secondary metabolites that have great potentials in agriculture, industry, and pharmaceutics.
LaeA in Aspergillus spp. is postulated to serve as a histone methyltransferase to modify chromatin structure thereby to regulate transcription of target genes in a larger genomic region. Strauss and Reyes-Dominguez (2011) provided evidence to show that LaeA somehow counteracts the trimethylation of H3K9 and the binding of heterochromatin protein to this repressive chromatin mark (Reyes-Dominguez et al., 2010). However, in the filamentous fungus T. reesei, no correlation between the LAE1-modulated expression of genes and changes in histone methylation has been observed (Seiboth et al., 2012; Karimi-Aghcheh et al., 2013). This might be caused by the less conservation between LaeA from Aspergillus spp. and LAE1 from T. reesei, as demonstrated by the observation that functional T. reesei LAE1 does not complement an A. nidulans ΔlaeA strain (Karimi-Aghcheh et al., 2013). Currently, the direct target substrates of LaeA/LAE1 are unknown, although LaeA/LAE1 was demonstrated to be a bona fide methyltransferase that methylates itself (Patananan et al., 2013) and is localized to the nucleus (Reyes-Dominguez et al., 2010). It is therefore speculated that LaeA/LAE1 may bind to other proteins and exert its function indirectly to control gene expression.
TlLAE1 of T. longibrachiatum SMF2 shares high sequence identity of 90% with T. reesei LAE1, suggesting that these two proteins are highly conserved. Nevertheless, difference in the effect of LAE1 perturbation or overexpression on the expression of peptaibols synthetase encoding genes has been observed between T. reesei and T. longibrachiatum SMF2. One of the two peptaibol synthetase encoding genes in T. reesei (Trire2:23171) is up-regulated in response to LAE1 perturbation or overexpression when cells were cultivated with lactose as the sole carbon source (Karimi-Aghcheh et al., 2013), whereas the two peptaibol synthetase encoding genes in T. longibrachiatum SMF2 were significantly down-regulated in lae1-null mutant and exhibited contrasting expression pattern in lae1-overexpressing mutant. This discrepancy may be caused by the intrinsic contrasting functions of LAE1 in controlling the expression of the NRPS encoding genes responsible for peptaibol synthesis. However, given that the regulatory effect of LAE1 in T. reesei on secondary metabolism including peptaibols production appears to be growth rate dependent (Fekete et al., 2014), possibility could not be excluded that different cultivation conditions and thus different growth rates of T. reesei (with lactose as the sole carbon source) and T. longibrachiatum SMF2 (with glucose as the sole carbon source) result in this discrepancy.
We previously found that peptaibols produced by T. longibrachiatum SMF2 exhibited antimicrobial activity against a range of Gram-positive bacterial and fungal phytopathogens (Song et al., 2006), so it could be assumed that the elimination of LAE1 in this fungus, which caused a significant defect in peptaibols production, should remarkably affect the ability of this fungus to repress plant pathogens that is contributed by peptaibols. Moreover, given that LAE1/LaeA is considered to be a global regulator in many filamentous fungi, elimination of TlLAE1 in T. longibrachiatum SMF2 would have broad effects on expression of a number of genes, probably including cellulase encoding genes, as the case in the well-known cellulolytic fungus, T. reesei (Seiboth et al., 2012). These relevant issues would be investigated in the future.
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material.
Author Contributions
J-CS, W-LS, and Y-RZ performed the experiments. X-YS, W-XZ, X-LC, Y-ZZ, and XZ performed the data analysis. X-YS, Y-ZZ, and X-LC designed and supervised the project. X-LC, W-XZ, Y-RZ, and XZ wrote the manuscript. All the authors approved the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (grants 31570042, 31971535, and 31670038), the Key Research Project of Shandong Province (2017GSF21106), the Program of Shandong for Taishan Scholars (tspd20181203), AoShan Talents Cultivation Program Supported by Qingdao National Laboratory for Marine Science and Technology (2017ASTCP-OS14), and the Major Scientific and Technological Innovation Project (MSTIP) of Shandong Province (2019JZZY010817).
Conflict of Interest
XZ was employed by Qingdao Vland Biotech Inc.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Marja Paloheimo at Roal Oy (Rajamäki, Finland) for kindly providing the pALK424 plasmid.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.01267/full#supplementary-material
References
Aghcheh, R. K., Druzhinina, I. S., and Kubicek, C. P. (2013). The putative protein methyltransferase LAE1 of Trichoderma atroviride is a key regulator of asexual development and mycoparasitism. PLoS One 8:e67144. doi: 10.1371/journal.pone.0067144
Amaike, S., and Keller, N. P. (2009). Distinct roles for VeA and LaeA in development and pathogenesis of Aspergillus flavus. Eukaryotic Cell 8, 1051–1060. doi: 10.1128/ec.00088-09
Bayram, ÖS., Bayram, Ö, Valerius, O., Park, H. S., Irniger, S., Gerke, J., et al. (2010). LaeA control of velvet family regulatory proteins for light-dependent development and fungal cell-type specificity. PLoS Genet. 6:e1001226. doi: 10.1371/journal.pgen.1001226
Bischof, R. H., Ramoni, J., and Seiboth, B. (2016). Cellulases and beyond: the first 70 years of the enzyme producer Trichoderma reesei. Microbial Cell Fact. 15:106.
Bok, J. W., and Keller, N. P. (2004). LaeA, a regulator of secondary metabolism in Aspergillus spp. Eukaryotic Cell 3, 527–535. doi: 10.1128/ec.3.2.527-535.2004
Bok, J. W., Noordermeer, D., Kale, S. P., and Keller, N. P. (2006). Secondary metabolic gene cluster silencing in Aspergillus nidulans. Mol. Microbiol. 61, 1636–1645. doi: 10.1111/j.1365-2958.2006.05330.x
Brunner, K., Omann, M., Pucher, M. E., Delic, M., Lehner, S. M., Domnanich, P., et al. (2008). Trichoderma G protein-coupled receptors: functional characterisation of a cAMP receptor-like protein from Trichoderma atroviride. Curr. Genet. 54, 283–299. doi: 10.1007/s00294-008-0217-7
Chang, P.-K., Scharfenstein, L. L., Ehrlich, K. C., Wei, Q., Bhatnagar, D., and Ingber, B. F. (2012). Effects of laeA deletion on Aspergillus flavus conidial development and hydrophobicity may contribute to loss of aflatoxin production. Fungal Biol. 116, 298–307. doi: 10.1016/j.funbio.2011.12.003
Druzhinina, I., and Kubicek, C. (2016). Familiar stranger: ecological genomics of the model saprotroph and industrial enzyme producer Trichoderma reesei breaks the stereotypes. Adv. Appl. Microbiol. 95, 69–147.
El-Bondkly, A. M. (2014). “Sequence analysis of industrially important genes from Trichoderma,” in Biotechnology and Biology of Trichoderma, Vol. 2014, eds V. Gupta, M. Schmoll, A. Herrera-Estrella, R. S. Upadhyay, I. Druzhinina, and M. Tuohy, (Amsterdam: Elsevier), 377–392. doi: 10.1016/b978-0-444-59576-8.00028-x
Fekete, E., Karaffa, L., Aghcheh, R. K., Németh, Z., Fekete, É, Orosz, A., et al. (2014). The transcriptome of lae1 mutants of Trichoderma reesei cultivated at constant growth rates reveals new targets of LAE1 function. BMC Genomics 15:447. doi: 10.1186/1471-2164-15-447
Hermosa, M. R., Keck, E., Chamorro, I., Rubio, B., Sanz, L., Vizcaíno, J. A., et al. (2004). Genetic diversity shown in Trichoderma biocontrol isolates. Mycol. Res. 108, 897–906. doi: 10.1017/s0953756204000358
Hoff, B., Kamerewerd, J., Sigl, C., Mitterbauer, R., Zadra, I., Kürnsteiner, H., et al. (2010). Two components of a velvet-like complex control hyphal morphogenesis, conidiophore development, and penicillin biosynthesis in Penicillium chrysogenum. Eukaryotic Cell 9, 1236–1250. doi: 10.1128/ec.00077-10
Hong, E. J., Kim, N. K., Lee, D., Kim, W. G., and Lee, I. (2015). Overexpression of the laeA gene leads to increased production of cyclopiazonic acid in Aspergillus fumisynnematus. Fungal Biol. 119, 973–983. doi: 10.1016/j.funbio.2015.06.006
Jiang, J., Yun, Y., Liu, Y., and Ma, Z. (2012). FgVELB is associated with vegetative differentiation, secondary metabolism and virulence in Fusarium graminearum. Fungal Genet. Biol. 49, 653–662. doi: 10.1016/j.fgb.2012.06.005
Jiang, T., Wang, M., Li, L., Si, J., Song, B., Zhou, C., et al. (2016). Overexpression of the global regulator LaeA in Chaetomium globosum leads to the biosynthesis of chaetoglobosin Z. J. Nat. Prod. 79, 2487–2494. doi: 10.1021/acs.jnatprod.6b00333
Kale, S. P., Milde, L., Trapp, M. K., Frisvad, J. C., Keller, N. P., and Bok, J. W. (2008). Requirement of LaeA for secondary metabolism and sclerotial production in Aspergillus flavus. Fungal Genet. Biol. 45, 1422–1429. doi: 10.1016/j.fgb.2008.06.009
Karimi-Aghcheh, R., Bok, J. W., Phatale, P. A., Smith, K. M., Baker, S. E., Lichius, A., et al. (2013). Functional analyses of Trichoderma reesei LAE1 reveal conserved and contrasting roles of this regulator. G3 Genes Genomes Genetics 3, 369–378. doi: 10.1534/g3.112.005140
Kubicek, C. P., Steindorff, A. S., Chenthamara, K., Manganiello, G., Henrissat, B., Zhang, J., et al. (2019). Evolution and comparative genomics of the most common Trichoderma species. BMC Genomics 20:485. doi: 10.1186/s12864-019-5680-7
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Li, H.-Y., Luo, Y., Zhang, X.-S., Shi, W.-L., Gong, Z.-T., Shi, M., et al. (2014). Trichokonins from Trichoderma pseudokoningii SMF2 induce resistance against Gram-negative Pectobacterium carotovorum subsp. carotovorum in Chinese cabbage. FEMS Microbiol. Lett. 354, 75–82. doi: 10.1111/1574-6968.12427
Linder, M. B., Szilvay, G. R., Nakari-Setälä, T., and Penttilä, M. E. (2005). Hydrophobins: the protein-amphiphiles of filamentous fungi. FEMS Microbiol. Rev. 29, 877–896. doi: 10.1016/j.femsre.2005.01.004
Lu, S., Lyngholm, L., Yang, G., Bronson, C., Yoder, O., and Turgeon, B. G. (1994). Tagged mutations at the Tox1 locus of Cochliobolus heterostrophus by restriction enzyme-mediated integration. Proc. Natl. Acad. Sci. U.S.A. 91, 12649–12653. doi: 10.1073/pnas.91.26.12649
Luo, Y., Zhang, D.-D., Dong, X.-W., Zhao, P.-B., Chen, L.-L., Song, X.-Y., et al. (2010). Antimicrobial peptaibols induce defense responses and systemic resistance in tobacco against tobacco mosaic virus. FEMS Microbiol. Lett. 313, 120–126. doi: 10.1111/j.1574-6968.2010.02135.x
Marik, T., Tyagi, C., Balázs, D., Urbán, P., Szepesi, Á, Bakacsy, L., et al. (2019). Structural diversity and bioactivities of peptaibol compounds from the Longibrachiatum Clade of the filamentous fungal genus Trichoderma. Front. Microbiol. 10:1434. doi: 10.3389/fmicb.2019.01434
Mukherjee, P. K., Wiest, A., Ruiz, N., Keightley, A., Morandiez, M. E., Mccluskey, K., et al. (2011). Two classes of new peptaibols are synthesized by a single non-ribosomal peptide synthetase of Trichoderma virens. J. Biol. Chem. 286, 4544–4554. doi: 10.1074/jbc.m110.159723
Neuhof, T., Dieckmann, R., Druzhinina, I. S., Kubicek, C. P., and von Döhren, H. (2007). Intact-cell MALDI-TOF mass spectrometry analysis of peptaibol formation by the genus Trichoderma/Hypocrea: can molecular phylogeny of species predict peptaibol structures? Microbiology 153, 3417–3437. doi: 10.1099/mic.0.2007/006692-0
Oda, K., Kobayashi, A., Ohashi, S., and Sano, M. (2011). Aspergillus oryzae laeA regulates kojic acid synthesis genes. Biosci. Biotechnol. Biochem. 75, 1832–1834. doi: 10.1271/bbb.110235
Patananan, A. N., Palmer, J. M., Garvey, G. S., Keller, N. P., and Clarke, S. G. (2013). A novel automethylation reaction in the Aspergillus nidulans LaeA protein generates S-methylmethionine. J. Biol. Chem. 288, 14032–14045. doi: 10.1074/jbc.m113.465765
Penttilä, M., Nevalainen, H., Rättö, M., Salminen, E., and Knowles, J. (1987). A versatile transformation system for the cellulolytic filamentous fungus Trichoderma reesei. Gene 61, 155–164. doi: 10.1016/0378-1119(87)90110-7
Punt, P. J., Dingemanse, M. A., Kuyvenhoven, A., Soede, R. D., Pouwels, P. H., and van den Hondel, C. A. (1990). Functional elements in the promoter region of the Aspergillus nidulans gpdA gene encoding glyceraldehyde-3-phosphate dehydrogenase. Gene 93, 101–109. doi: 10.1016/0378-1119(90)90142-e
Reino, J. L., Guerrero, R. F., Hernández-Galán, R., and Collado, I. G. (2008). Secondary metabolites from species of the biocontrol agent Trichoderma. Phytochem. Rev. 7, 89–123. doi: 10.1007/s11101-006-9032-2
Reyes−Dominguez, Y., Bok, J. W., Berger, H., Shwab, E. K., Basheer, A., Gallmetzer, A., et al. (2010). Heterochromatic marks are associated with the repression of secondary metabolism clusters in Aspergillus nidulans. Mol. Microbiol. 76, 1376–1386. doi: 10.1111/j.1365-2958.2010.07051.x
Schmittgen, T. D., and Livak, K. J. (2008). Analyzing real-time PCR data by the comparative C T method. Nat. Protoc. 3, 1101–1108. doi: 10.1038/nprot.2008.73
Seiboth, B., Karimi, R. A., Phatale, P. A., Linke, R., Hartl, L., Sauer, D. G., et al. (2012). The putative protein methyltransferase LAE1 controls cellulase gene expression in Trichoderma reesei. Mol. Microbiol. 84, 1150–1164. doi: 10.1111/j.1365-2958.2012.08083.x
Shi, M., Chen, L., Wang, X.-W., Zhang, T., Zhao, P.-B., Song, X.-Y., et al. (2012). Antimicrobial peptaibols from Trichoderma pseudokoningii induce programmed cell death in plant fungal pathogens. Microbiology 158, 166–175. doi: 10.1099/mic.0.052670-0
Shi, M., Wang, H.-N., Xie, S.-T., Luo, Y., Sun, C.-Y., Chen, X.-L., et al. (2010). Antimicrobial peptaibols, novel suppressors of tumor cells, targeted calcium-mediated apoptosis and autophagy in human hepatocellular carcinoma cells. Mol. Cancer 9:26. doi: 10.1186/1476-4598-9-26
Song, X., Shen, Q., Xie, S., Chen, X., Sun, C., and Zhang, Y. (2006). Broad-spectrum antimicrobial activity and high stability of Trichokonins from Trichoderma koningii SMF2 against plant pathogens. FEMS Microbiol. Lett. 260, 119–125. doi: 10.1111/j.1574-6968.2006.00316.x
Soukup, A. A., Keller, N. P., and Wiemann, P. (2016). Enhancing nonribosomal peptide biosynthesis in filamentous fungi. Methods Mol. Biol. 1401, 149–160. doi: 10.1007/978-1-4939-3375-4_10
Strauss, J., and Reyes-Dominguez, Y. (2011). Regulation of secondary metabolism by chromatin structure and epigenetic codes. Fungal Genet. Biol. 48, 62–69. doi: 10.1016/j.fgb.2010.07.009
Sugui, J. A., Pardo, J., Chang, Y. C., Müllbacher, A., Zarember, K. A., Galvez, E. M., et al. (2007). Role of laeA in the regulation of alb1, gliP, conidial morphology, and virulence in Aspergillus fumigatus. Eukaryotic Cell 6, 1552–1561.
Thompson, J. D., Gibson, T. J., and Higgins, D. G. (2002). Multiple sequence alignment using ClustalW and ClustalX. Curr. Protoc. Bioinform. Chapter 2, 2.3.1–2.3.22. doi: 10.1002/0471250953.bi0203s00
Viterbo, A., Wiest, A., Brotman, Y., Chet, I., and Kenerley, C. (2007). The 18mer peptaibols from Trichoderma virens elicit plant defence responses. Mol. Plant Pathol. 8, 737–746. doi: 10.1111/j.1364-3703.2007.00430.x
Wiemann, P., Brown, D. W., Kleigrewe, K., Bok, J. W., Keller, N. P., Humpf, H. U., et al. (2010). FfVel1 and FfLae1, components of a velvet−like complex in Fusarium fujikuroi, affect differentiation, secondary metabolism and virulence. Mol. Microbiol. 77, 972–994.
Wiest, A., Grzegorski, D., Xu, B.-W., Goulard, C., Rebuffat, S., Ebbole, D. J., et al. (2002). Identification of peptaibols from Trichoderma virens and cloning of a peptaibol synthetase. J. Biol. Chem. 277, 20862–20868. doi: 10.1074/jbc.m201654200
Wu, D., Oide, S., Zhang, N., Choi, M. Y., and Turgeon, B. G. (2012). ChLae1 and ChVel1 regulate T-toxin production, virulence, oxidative stress response, and development of the maize pathogen Cochliobolus heterostrophus. PLoS Pathog. 8:e1002542. doi: 10.1371/journal.ppat.1002542
Xie, B.-B., Li, D., Shi, W.-L., Qin, Q.-L., Wang, X.-W., Rong, J.-C., et al. (2015). Deep RNA sequencing reveals a high frequency of alternative splicing events in the fungus Trichoderma longibrachiatum. BMC Genomics 16:54. doi: 10.1186/s12864-015-1251-8
Xie, B.-B., Qin, Q.-L., Shi, M., Chen, L.-L., Shu, Y.-L., Luo, Y., et al. (2014). Comparative genomics provide insights into evolution of Trichoderma nutrition style. Genome Biol. Evol. 6, 379–390. doi: 10.1093/gbe/evu018
Zheng, Y., Cao, S., Huang, Y., Liao, G., and Hu, C. (2014). Overexpression of LaeA enhances mevastatin production and reduces sporulation of Penicillium citrinum. Wei Sheng Wu Xue Bao 54, 1438–1445.
Zhou, Q., Xu, J., Kou, Y., Lv, X., Zhang, X., Zhao, G., et al. (2012). Differential involvement of β-glucosidases from Hypocrea jecorina in rapid induction of cellulase genes by cellulose and cellobiose. Eukaryotic Cell 11, 1371–1381. doi: 10.1128/ec.00170-12
Keywords: Trichoderma longibrachiatum SMF2, LAE1, conidiation, peptaibol, secondary metabolite
Citation: Shi J-C, Shi W-L, Zhou Y-R, Chen X-L, Zhang Y-Z, Zhang X, Zhang W-X and Song X-Y (2020) The Putative Methyltransferase TlLAE1 Is Involved in the Regulation of Peptaibols Production in the Biocontrol Fungus Trichoderma longibrachiatum SMF2. Front. Microbiol. 11:1267. doi: 10.3389/fmicb.2020.01267
Received: 01 February 2020; Accepted: 18 May 2020;
Published: 12 June 2020.
Edited by:
Monika Schmoll, Austrian Institute of Technology (AIT), AustriaReviewed by:
Paul Daly, Jiangsu Academy of Agricultural Sciences (JAAS), ChinaRosa Hermosa, University of Salamanca, Spain
Benjamin A. Horwitz, Technion Israel Institute of Technology, Israel
Copyright © 2020 Shi, Shi, Zhou, Chen, Zhang, Zhang, Zhang and Song. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xia Zhang, emhhbmd4QHZsYW5kZ3JvdXAuY29t; Wei-Xin Zhang, emhhbmd3eEBzZHUuZWR1LmNu; Xiao-Yan Song, eHlzb25nQHNkdS5lZHUuY24=
†These authors have contributed equally to this work
 Jin-Chao Shi1†
Jin-Chao Shi1† Xiu-Lan Chen
Xiu-Lan Chen Yu-Zhong Zhang
Yu-Zhong Zhang Wei-Xin Zhang
Wei-Xin Zhang Xiao-Yan Song
Xiao-Yan Song