- 1State Key Laboratory of Materials-Oriented Chemical Engineering, College of Biotechnology and Pharmaceutical Engineering, Nanjing Tech University, Nanjing, China
- 2Jiangsu National Synergetic Innovation Center for Advanced Materials (SICAM), Nanjing Tech University, Nanjing, China
Microbial consortia are ubiquitous in nature and exhibit several attractive features such as sophisticated metabolic capabilities and strong environmental robustness. This study aimed to decipher the metabolic and ecological characteristics of synergistic interactions in acetamiprid-degrading consortia, suggesting an optimal scheme for bioremediation of organic pollutants. The microbial consortium ACE-3 with excellent acetamiprid-degrading ability was enriched from the soil of an acetamiprid-contaminated site and characterized using high-throughput sequencing (HTS). Consortium ACE-3 was able to completely degrade 50 mg⋅L–1 acetamiprid in 144 h, and was metabolically active at a wide range of pH values (6.0–8.0) and temperatures (20–42°C). Furthermore, plausible metabolic routes of acetamiprid biodegradation by the consortium were proposed based on the identification of intermediate metabolites (Compounds I, II, III and IV). The findings indicated that the consortium ACE-3 has promising potential for the removal and detoxification of pesticides because it produces downstream metabolites (Compounds I and II) that are less toxic to mammals and insects than acetamiprid. Finally, Illumina HTS revealed that β Proteobacteria were the dominant group, accounting for 85.61% of all sequences at the class level. Among the more than 50 genera identified in consortium ACE-3, Sphingobium, Acinetobacter, Afipia, Stenotrophomonas, and Microbacterium were dominant, respectively accounting for 3.07, 10.01, 24.45, and 49.12% of the total population.
Introduction
As one of the most important chloropyridinyl neonicotinoids, acetamiprid plays a major role in crop protection, but its use inevitably causes environmental pollution (Carra et al., 2016; Sun et al., 2017). In fact, acetamiprid residues have often been detected in natural bodies of ground-, surface- and drinking water due to its excessive use to enhance crop production (Cruz-Alcalde et al., 2017). Moreover, although acetamiprid pesticide has been classified as an “unlikely” human carcinogen by the US EPA, it still possesses generalized, non-specific toxicity to mammals. Moreover, acetamiprid residues in soil or on plants have a marked negative impact on non-target species, including livestock, honeybees and humans (Chakroun et al., 2016). It is therefore urgently needed to find an efficient route for the biodegradation of acetamiprid residues, reducing possible ecological risks to the natural environment.
In view of sustainable development goals, indigenous microorganisms are usually used to degrade various environmental pollutants without producing secondary pollution. Accordingly, microbes from different genera were isolated and identified as acetamiprid-biodegraders (Watanabe, 2001; Ju and Zhang, 2014). Examples include Stenotrophomonas maltophilia CGMCC 1.1788 (Chen et al., 2008), Rhodotorula sp. IM-2 (Dai et al., 2010), Pigmentiphaga sp. strain D-2, (Yang et al., 2013), Pigmentiphaga sp. strain AAP-1 (Wang et al., 2013a), Pseudoxanthomonas sp. AAP-7 (Wang et al., 2013c), Ochrobactrum sp. D-12 (Wang et al., 2013b), Rhodococcus sp. BCH2 (Phugare and Jadhav, 2015), and Fusarium sp. CS-3 (Shi et al., 2018). Among these degraders, strains of Stenotrophomonas sp. and Pigmentiphaga sp. were respectively able to degrade 58.9% of 570 mg⋅L–1 acetamiprid in 8 days and completely degrade 50 mg⋅L–1 acetamiprid after 72 h, which was more rapid than other pure bacterial cultures, enabling more efficient bioremediation. However, pure acetamiprid-biodegrading cultures are often unable to perform in situ or on-site bioremediation with practical value. One of the most important reasons is that pure bacterial cultures cannot effectively mineralize these pesticides due to the formation of derived toxic or inhibitory intermediate metabolites (Copley, 2009). Furthermore, many environmental factors still affect microbial activity (e.g., water, pH, redox potential), leading to possible failure of operation (Liu et al., 2015; Dangi et al., 2019). Therefore, it is urgent to develop reliable bioaugmentation methods for achieving bioremediation of acetamidine in the open environment.
In the open environment, 99% of microorganisms exist in the form of microbial consortia (Ding et al., 2016; Singh and Kashmir, 2016). Compared with pure cultures, microbial consortia possess more flexible and adaptive capabilities to face complex environmental stresses due to combined interactions of diverse species, performing complicated enzymatic catalysis (e.g., intestinal food digestion, wastewater purification, or lignocellulose degradation) (Festa et al., 2013; Dvořák et al., 2017; Mnif et al., 2017). According to Fida et al. (2017) and Su et al. (2019), microbial cells that exhibit low metabolic activity for long-term survival in the viable but non-culturable (VBNC) state still play a crucial role in rapid in situ biodegradation of acetamiprid in soil. Therefore, the most appropriate strategy for allowing the emergence of ecologically stable consortia for biodegradation is the development of acclimatized mixed cultures. More recently, increasing numbers of studies are exploring the role of microbial consortia and unculturable microbes in the biodegradation of pollutants. Perruchon et al. (2017b) enriched a bacterial consortium to evaluate their biodegradation capacity for neonicotinoid insecticides under various conditions (pH values, temperatures, and different pollutant concentrations), which indicated that the consortium was more suitable for biodegradation of pollutants in an unstable environment. Similarly, Sharma S. et al. (2014) explored the biodegradation of acetamiprid by another consortium from two soil samples, which achieved excellent degradation performance and realized complete mineralization without producing complex secondary pollutants. This result also indicated that the synergistic interactions between different bacterial strains enabled them to completely mineralize the pollutant.
Here, the microbial consortium ACE-3 was serially enriched from an acetamiprid-contaminated soil. It was able to utilize acetamiprid as the sole carbon source for microbial growth. In addition, the metabolic route of acetamiprid biodegradation by consortium ACE-3 was proposed based on the identification of intermediate metabolites, and the community structure of consortium ACE-3 was determined using high-throughput sequencing (HTS) technology. This study uncovered why and how community structure of mixed consortia affects their biodegradation performance, suggesting an ecologically stable strategy for optimal acetamiprid biodegradation.
Based on these results, we have tested the hypothesis that the community structure of mixed consortia affects their biodegradation performance which suggests an ecologically stable strategy for optimal acetamiprid biodegradation. This was achieved by serial enrichment of the microbial consortium ACE-3 from an acetamiprid-contaminated soil. The consortium was able to utilize acetamiprid as the sole carbon source for microbial growth. In addition, the metabolic route of acetamiprid biodegradation by consortium ACE-3 was proposed based on the identification of intermediate metabolites, and the community structure of consortium ACE-3 was determined using high-throughput sequencing (HTS) technology.
Materials and Methods
Chemicals and Media
Chromatography-grade methanol and acetonitrile were purchased from Sigma Aldrich (United States). All molecular biology reagents were purchased from TaKaRa Co., Ltd. (Dalian, China). All other chemicals and reagents were commercially available and were of analytical grade, unless stated otherwise. Acetamiprid (>99% purity) was provided by Jiangsu Academy of Agricultural Sciences, China. The stock solution of acetamiprid (1%, w/v) was prepared in acetone, followed by filtration through a sterile membrane with 0.22 μm pore size. Minimal salt medium (MSM; K2HPO4 1.5 g⋅L–1, KH2PO4 0.5 g⋅L–1, NH4NO3 1 g⋅L–1, MgSO4⋅7H2O 0.1 g⋅L–1, and NaCl 1 g⋅L–1, pH7.0) was used to enrich the acetamiprid-degrading culture and for the biodegradation tests.
Isolation of the Acetamiprid-Degrading Natural Microbial Consortium
Fresh soil (0–15 cm depth) from an acetamiprid-rich environment contaminated for several years was collected at a pesticide factory located in Weifang (Shandong, China). The soil sample was used for enrichment and isolation of the acetamiprid-degrading consortium. A biodegradation system was built in an Erlenmeyer flask (500 mL) containing 5 g soil (dry weight) and 100 mL MSM supplemented with 30 mg⋅L–1 acetamiprid as the sole carbon and energy source. The enrichment culture was incubated in a rotary shaker at 180 rpm and 30°C for 6 days. After the degradation efficiency was determined, 10% of the consortium was transferred into fresh MSM containing 50 mg⋅L–1 acetamiprid, followed by incubation for another 6 days. The cultivation with increasingly concentrated media was repeated 6 times until the concentration of acetamiprid reached 100 mg⋅L–1. The consortium with the highest acetamiprid degradation efficiency was named ACE-3 and used for further study.
Growth and Biodegradation Assay
When 100 mg⋅L–1 acetamiprid was decomposed to a degree of 50%, the cells in the degradation system were collected by centrifugation at 2124 × g and 4°C, washed twice with sterile phosphate buffered saline (PBS, pH 7.0), and re-suspended in sterilized MSM, which was used as inoculum for further study. MSM containing 50 mg⋅L–1 acetamiprid was inoculated at a rate of 1%, and un-inoculated MSM was used as the control. Four milliliter samples were drawn at intervals of 24 h to measure the OD600, and the remaining acetamiprid concentration in the sample was analyzed by HPLC.
Effects of Environmental Factors on Acetamiprid Degradation
To optimize the degradation conditions for the ACE-3 consortium, the effects of different factors on acetamiprid degradation were investigated by varying the temperature (20, 25, 30, 37, and 42°C), initial pH (4.0, 5.0, 6.0, 7.0, 8.0, and 10.0), inoculum size (1, 3, 5, and 10%) and initial concentration of acetamiprid (25, 50, 100, 150, and 200 mg⋅L–1). All samples were extracted with dichloromethane immediately before HPLC analysis, and the concentration of residual acetamiprid was calculated by comparing the integrated area with that of the reference standard with known concentration. All treatments were performed in triplicate.
Identification of Metabolites
The consortium ACE-3 was seeded into a 250 mL Erlenmeyer flask containing 100 mL of MSM supplemented with 50 mg⋅L–1 of acetamiprid. The samples (acetamiprid degradation percentages of 30, 50, and 70%) were collected by centrifugation at 12,580 × g for 10 min at 4°C. Then, the supernatants were freeze-dried and re-dissolved in 1 mL of chromatography-grade methanol for HPLC-MS analysis. A separation column (internal diameter, 4.6 mm; length, 25 cm) filled with Thermo100-5C18 was used. The mobile phase consisted of 50% water and 50% acetonitrile at a flow rate of 0.6 mL⋅min–1. The injection volume was 20 μL, and the column elution was monitored at 247 nm using a UV-900 spectrophotometric detector. The concentration of acetamiprid was calculated by comparing the area under the peak with that of the standard solution (Wang et al., 2013c). The LOD (limit of detection) and LOQ (limit of quantification) of the methods were 1.0 × 10–7 mg and 0.002 mg⋅L–1, respectively (R2 = 0.9982). The metabolites were identified by MS (G6410B Triple Quad Mass; Agilent, United States), with electrospray ionization in positive ion mode, and scanned in the normal mass range from 30 m/z (mass to charge ratio) to 300 m/z. Characteristic fragment ions were detected using second-order MS as described before (Zhou et al., 2014).
Analysis of the Microbial Community by High-Throughput Sequencing
The metagenomic DNA of consortium ACE-3 was extracted using the FastDNA Spin Kit for Soil (MP Biomedicals, United States). The DNA samples were stored at -20°C for subsequent PCR amplification. The universal primers 515F (5′-CCTACGGGAGGCAGCAG-3′) and 907R (5′-TTACCGCGGCTGCTGGC-3′) with different barcodes were used to amplify the V4-V5 region of the 16S rRNA gene (Dong et al., 2017). The 50 μL PCR reaction mixtures consisted of 1 μL DNA template, 25 μL Primer Star Max, 1 μL of each primer (515F and 907R), and 22 μL of double distilled water. The PCR program encompassed an initial denaturation at 94°C for 2 min, followed by 30 cycles of 30 s at 94°C, 30 s at 55°C, and 45 s at 72°C each, and a final extension at 72°C for 10 min. The PCR products were recovered using the AxyPrep DNA gel Recovery Kit (AXYGEN, China), eluted with Tris-HCl, and visualized via 2% agarose gel electrophoresis. The resulting final high-quality sample was used for HTS.
Sequencing and Analysis
The PCR products were sent to Novogene Co., Ltd. (Beijing, China) for HTS of the 16S rDNA. According to the sequencing requirements of two samples, the Illumina PE250 library was constructed and sequenced. The PE reads obtained by Illumina PE250 sequencing were first spliced according to overlaps, after which the quality of the sequences was controlled and filtered to achieve the requirements of sequence analysis. The raw sequencing data was deposited in the NCBI SRA database under the accession number PRJNA606871.
The UPARSE-OTU algorithm (version 7.1) of the Usearch software platform was used to allocate the operational taxonomic units (OTUs) to High Identity with Tolerance (CD-HIT) groups based on 97% sequence similarity using the cluster database. PCR-generated chimeras were detected and removed using the Uchime method (version 4.2.40; Edgar, 2013). Based on the results of OTU analysis, we calculated the alpha diversity of single samples, including the Chao, Shannon and Simpson indices, using Mothur software (version 1.34.0; Schloss and Westcott, 2011). Classification was conducted using the RDP-II classifier of the ribosome database project (RDP) and the National Center for Biotechnology Information (NCBI) BLAST algorithm at http://www.ncbi.nlm.nih.gov/blast/. The construction of phylogenetic trees was conducted using MEGA software version 7.0 (Kumar et al., 2016). Bootstrap analysis with 1000 replicates was applied to assign confidence levels to the nodes in the phylogenetic trees.
Results and Discussion
Degradation of Acetamiprid by the Microbial Consortium ACE-3
The consortium ACE-3 was successfully enriched by serial selection from acetamiprid-contaminated soil after seven rounds of culture with increasing concentrations of acetamiprid as the sole carbon and energy source. As most bacteria cannot be cultured on conventional bacteriological media because they enter into the VBNC state under harsh environmental conditions (Fida et al., 2017), the growth curve was determined to ensure that the microbial consortium enriched for continuous degradation of acetamiprid was culturable. As shown in Figure 1, consortium ACE-3 was able to degrade 96% of 50 mg⋅L–1 acetamiprid within 144 h, while the cell density (OD600) increased roughly 3-fold (0.12–0.23, approximately 1.6 × 106–4.8 × 106 cfu⋅mL–1). Interestingly, degradation of acetamiprid by consortium ACE-3 was initially slow, with a degradation ratio of 12% within 24 h, possibly due to toxicity of acetamiprid for the microbial population. Subsequently, the rates of acetamiprid degradation and cell growth increased once consortium ACE-3 was well-adapted to the acetamiprid-containing medium. As anticipated, the controls without inoculation with consortium ACE-3 or the addition of acetamiprid resulted in no significant degradation of acetamiprid and nearly negligible bacterial growth, respectively. These results confirmed that consortium ACE-3 possesses the ability to degrade and utilize acetamiprid as the sole carbon source for microbial growth.
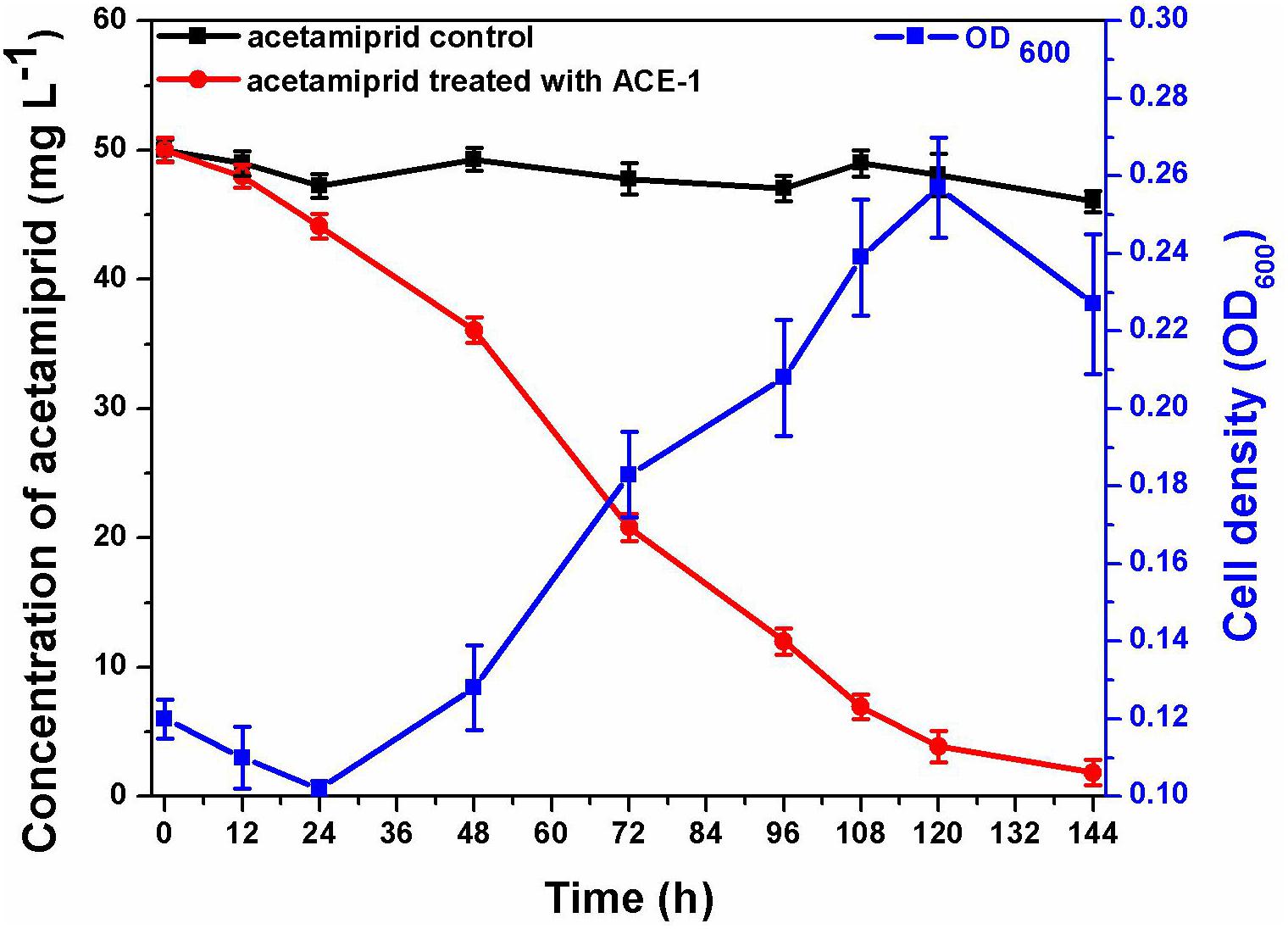
Figure 1. Degradation of acetamiprid by consortium ACE-3 and the corresponding growth curves. The data represent the means ± standard deviations of biological triplicates.
Although many acetamiprid-degrading microorganisms have been isolated and studied, cultures of single strains are usually not ecologically stable in practice. Table 1 lists the pure and mixed bacterial cultures used to degrade acetamiprid in earlier studies. In terms of degradation efficiency, both pure and mixed bacteria can achieve efficient biodegradation of acetamiprid. However, from the perspective of the degradation pathway, it is difficult for a single species of bacteria to fully mineralize the secondary metabolites. Therefore, mixed bacterial cultures are more effective for the degradation of environmental pollutants due to synergistic interactions that enable complete mineralization (Chakraborty et al., 2012; Wang et al., 2016; Mori et al., 2017). Application of natural microbial consortia has already shown promise for in situ bioremediation of gentamicin, nonyl-phenol, and atrazine-contaminated sites (Baghapour et al., 2013; Bai et al., 2017). This aimed to characterize an acetamiprid-degrading microbial consortium enriched and acclimatized to direct selection pressure for growth on acetamiprid, suggesting a novel operation strategy that can be applied for in situ bioremediation of pesticide-contaminated environments.
Degradation Characteristics
The temperature and pH are significant operation parameters that influence the rate of xenobiotic degradation. As an important parameter, the specific degradation rate (SDR) is often used to evaluate the kinetic process of pollutant degradation over time. The SDR is calculated based on degradation curve fitting, and is used to characterize the amount of acetamiprid degradation per unit of time, representing the degradation rate. As can be seen in Figure 2A, the consortium ACE-3 could degrade acetamiprid at the maximal efficiency at temperatures between 25 and 37°C, while the degradation efficiency was decreased by half at 20 or 42°C. This result indicated that 25–37°C was the optimum temperature range for the consortium to degrade acetamiprid. Similarly, an overly acidic or alkaline environment was not suitable for the biodegradation of acetamiprid by this consortium. The optimum pH for acetamiprid degradation was between 5.0 and 7.0, and the degradation rate decreased drastically at acidic pH values below 4.0 or alkaline pH above 10.0 (Figure 2B). These results indicated that the consortium ACE-3 was ecologically stable, with stable high degradation efficiency and degradation rate over a wide range of environmental changes.
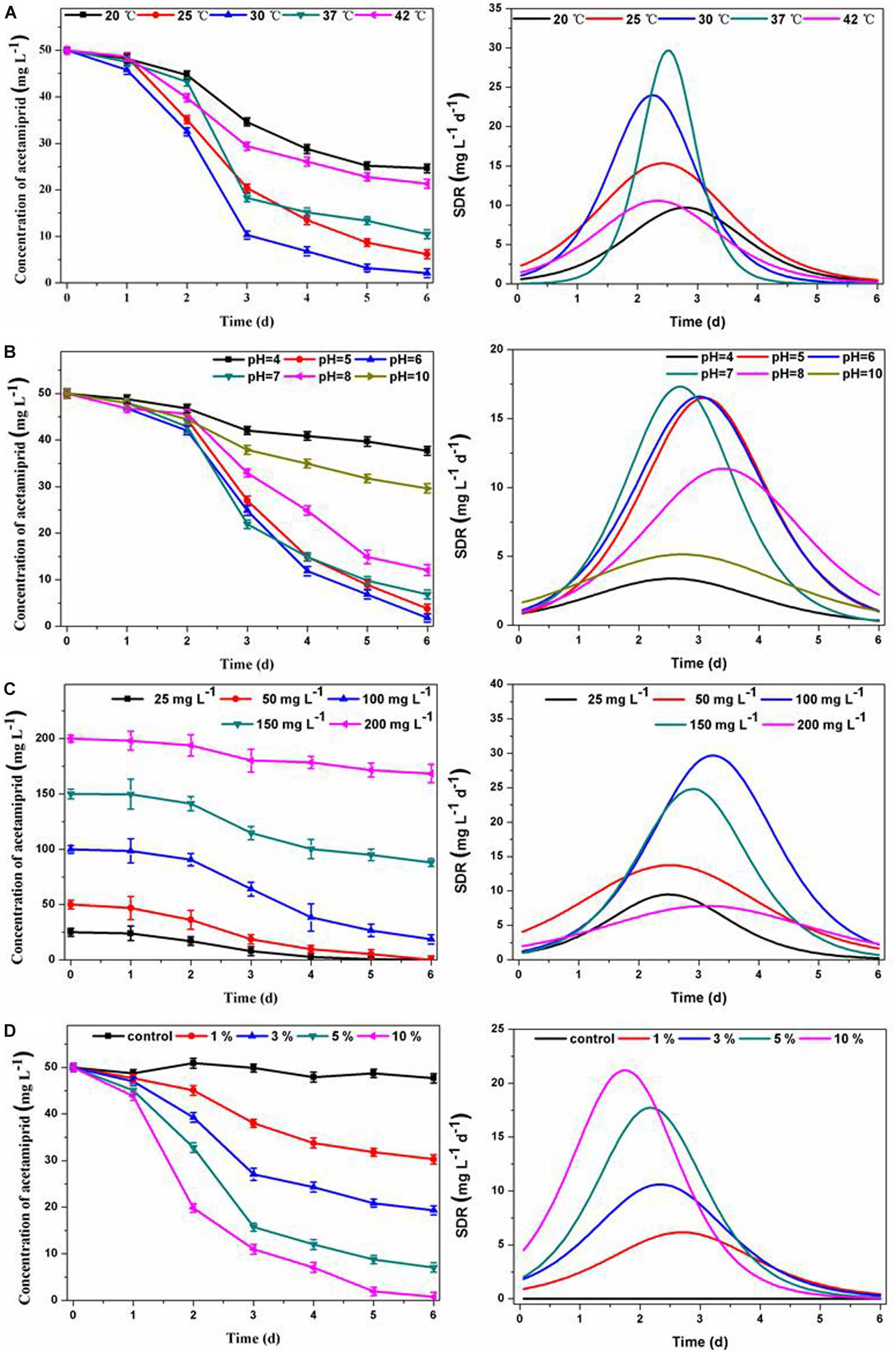
Figure 2. Effects of physicochemical conditions on acetamiprid biodegradation by consortium ACE-3 and degradation kinetics curve of SDR with time-course. (A) Temperature; (B) pH; (C) acetamiprid concentration; (D) inoculum size. The data represent the means ± standard deviations of triplicate experiments.
Furthermore, the initial substrate concentration and inoculum size also significantly affected the performance of cell growth and acetamiprid degradation. As shown in Figure 2C, the degradation efficiencies were 99.2 and 97.1% at the initial concentrations of 20 and 50 mg⋅L–1, respectively. The degradation efficiency gradually decreased with increasing substrate concentration, most likely due to the toxicity of acetamiprid to bacterial cells. By comparing the maximum degradation rate, it could be seen that when the concentration of acetamiprid reached 200 mg⋅L–1, the degradation rate was significantly reduced. The most likely reason for the decrease of the degradation rate is direct inhibition of bacterial growth by the substrate. Similar observations have been reported in the biodegradation of other toxic substances. Correspondingly, when the biomass of the microbial consortium was greater, the acetamiprid degradation rate was significantly increased. As shown in Figure 2D, the acetamiprid degradation ratio at 6 days of incubation increased from 44 to 99.74% when the inoculum size was increased from 1 to 10%. More importantly, as the inoculum size increased, the time to reach the maximum degradation rate was also shorter. These results indicate that the degradation efficiency of acetamiprid is positively associated with the inoculum size of consortium ACE-3.
In summary, the results indicate that consortium ACE-3 could degrade acetamiprid with the highest efficiency under mild conditions. Similarly, the optimal conditions for the degradation of di-n-butyl phthalate (DBP) by consortium LV-1 were pH 6.0 and 30°C (Wang et al., 2017). Moreover, rapid degradation of thiabendazole by a bacterial consortium dominated by the Sphingomonas phylotype B13 was observed at slightly acidic pH (5.5–6.5) and moderate temperatures (26–37°C) (Perruchon et al., 2017a). The bacterial consortium DV-AL could degrade 1000 mg⋅L–1 of naphthalene within 24 h at an initial pH 8.0 and 37°C (Patel et al., 2012). According to the previous studies listed in Table 1, the optimal conditions of acetamiprid degradation for both pure and mixed cultures were all in the near-neutral range (pH 6 ∼ 8) and mesophilic temperatures (25∼37°C). However, the degradation efficiency changed significantly with the change of temperature and pH. As indicated by Wang et al. (2013b), the degradation process was still possible at pH 5, even if the degradation ratio was significantly reduced from over 80% at pH 7 to around 40%. Also, changes of temperature can have a similar effect. In this study, acetamiprid could be degraded by consortium ACE-3 at a broader range of temperatures and pH values, making such mixed cultures applicable to the bioremediation of various acetamiprid-contaminated environments.
Identification of Metabolites
To decipher the metabolic network of acetamiprid biodegradation, HPLC and HPLC-MS were employed to identify metabolic intermediates and propose the pathway of acetamiprid biodegradation by ACE-3. As the profile of the HPLC-MS analysis (Figure 3) indicated, acetamiprid was observed in the control sample. Mass spectrometry revealed the presence of four intermediates in the sample that underwent acetamiprid biodegradation. These correspond to the deprotonated molecules of compounds I {m/z = 163.08 [C5H3N-CH2-N(CH3)-C(O)eCH3]–, 154.91 [Cl-C5H3N- CH2-N-CH3]–, and 126.00 [Cl-C5H3N-CH2]–}, II [m/z = 126 (Cl-C5H3N-CH2)–], III [m/z = 194 (M + H-CH3), 167 (M-CH3-CN) and 126 (M-NHCCH3NCN)] and IV [m/z = 194 (M + H-CH3), 167 (M-CH3-CN) and 126 (M-NHCCH3NCN)]. Their respective molecular weights were 194.5, 152.5, 207.5, and 203.5. After comparison with the database, the detected metabolites were identified as N′-[(6-chloropyridin-3-yl) methyl]-N-methyl acetamide (Figure 3A), 1-(6-chloropyridin-3-yl)-N-methyl methaneamine (Figure 3B), (E)-N1-[(6-chloro-3-pyridyl)-methyl]-N2-cyano-acetamidine (Figure 3C) and (E)-3-(((6-chloropyridin-3yl) methyl) (methyl) amino) acrylonitrile (Figure 3D). Notably, compound III was found to be identical to the reported findings for IM 2-1 (Chen et al., 2008). Based on the identified intermediates, a plausible metabolic pathway for the biodegradation of acetamiprid was proposed as illustrated in Figure 4.
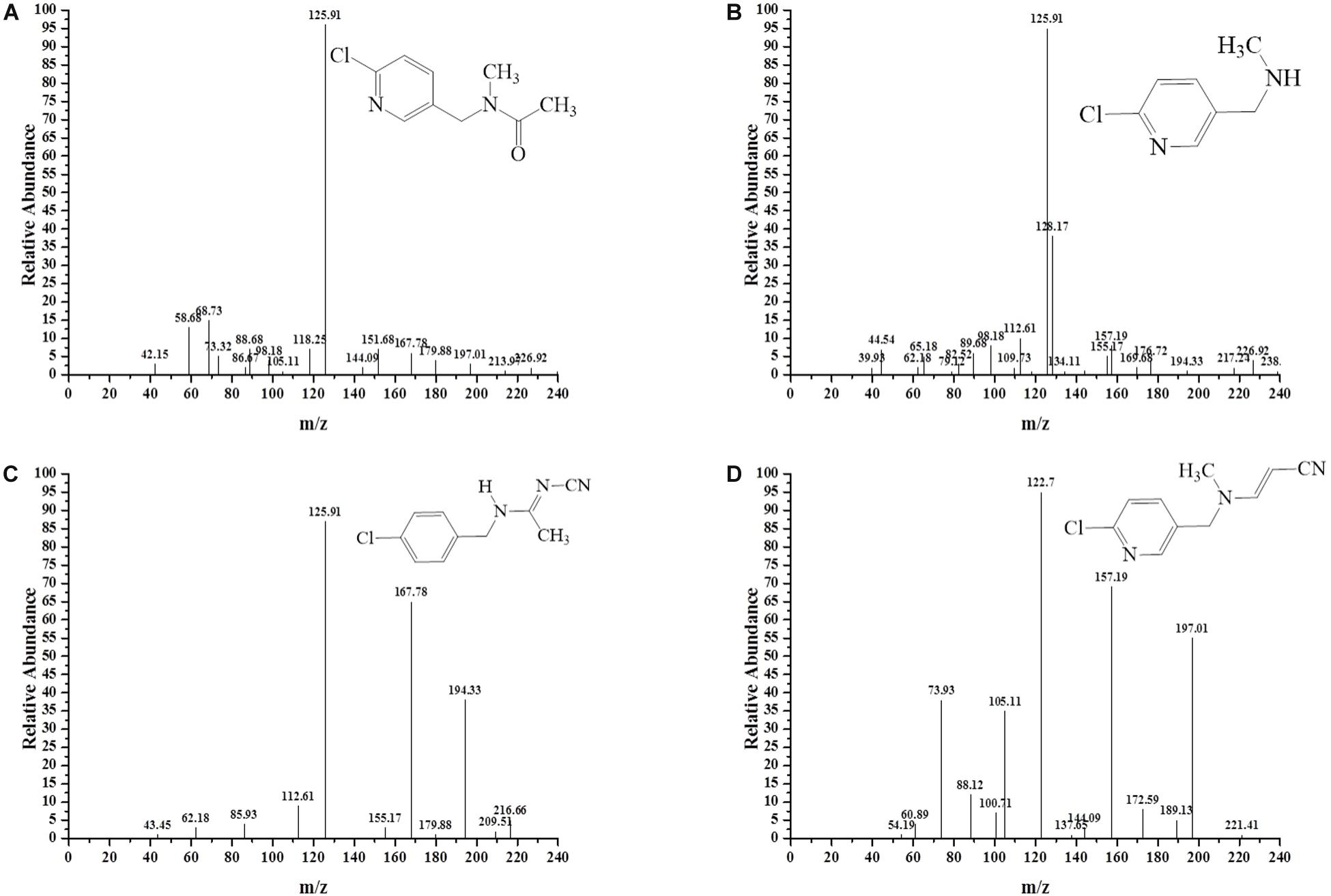
Figure 3. MS/MS identification of the metabolites produced during acetamiprid degradation. Second-order mass spectra of (A) compound I; (B) compound II; (C) compound III; (D) compound IV.
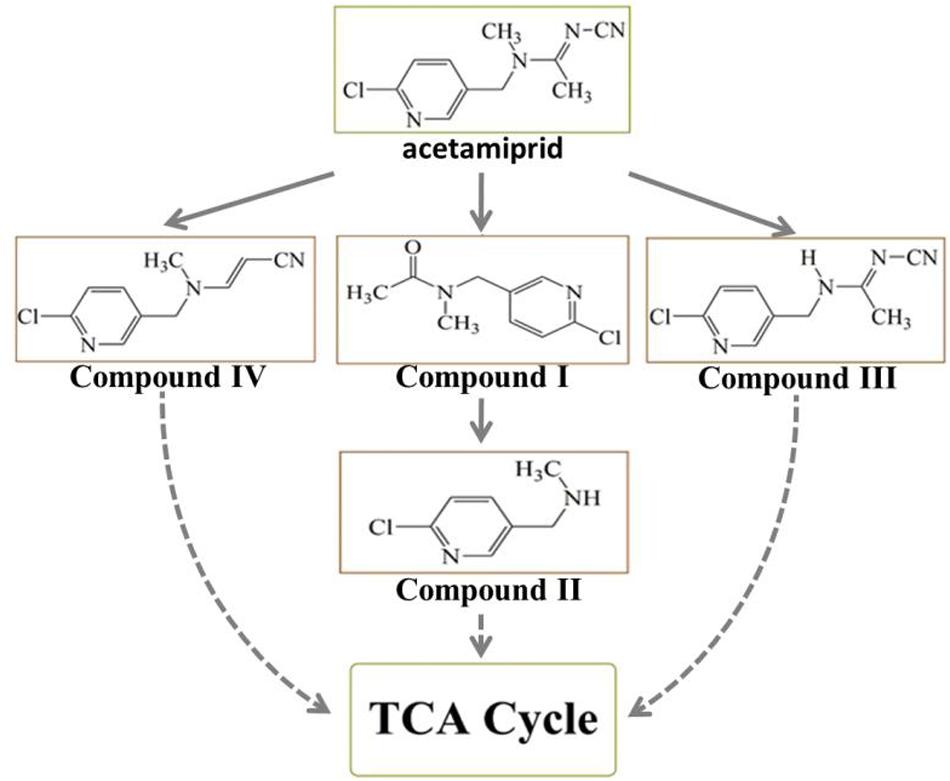
Figure 4. The proposed metabolic pathway of acetamiprid degradation by consortium ACE-3. The dotted line indicates a putative metabolic pathway.
Because the intermediate compound I was detected in the culture broth, it stands to reason that consortium ACE-3 is able to oxidatively cleave acetamiprid. The formation of compound I by the oxidative cleavage of acetamiprid is consistent with the findings for the bacterial strain Pigmentiphaga sp. D-2 (Yang et al., 2013). In addition, as Dai et al. (2010) previously reported, Rhodotorula mucilaginosa IM-2 can also transform acetamiprid into compound I. Subsequently, Compound I could be N-deacetylated to compound II, which was also identified in Stenotrophomonas sp. THZ-XP and honeybees (Brunet et al., 2005; Tang et al., 2012). The cyanoimine (= N-CN) moiety of acetamiprid gives it a much higher affinity for the nicotinic acetylcholine receptor (nAChR) of insects over that of vertebrates, leading to paralysis and death of insect pests without harming humans and livestock. Furthermore, the loss of the nitro or nitrile group can completely reverse the selective toxicity of neonicotinoids (Tang et al., 2012). Compounds I and II, which apparently lack the cyano-substituent, might be relatively less harmful to mammals or insects than acetamiprid, even if they are released back into the soil. This is obviously beneficial for the removal and detoxification of the pesticide from soil and plants.
The major metabolite compound III was also reported in earlier studies on acetamiprid degradation. Stenotrophomonas maltophilia CGMCC 1.1788 can de-methylate acetamiprid to form IM2-1, but the transformation of either the nitro or the cyano group was not observed (Chen et al., 2008). Brunet et al. (2005) also studied some associated metabolites present in honeybees exposed to the pesticide. In the process of co-metabolic biodegradation of acetamiprid by Pseudoxanthomonas sp. AAP-7, Wang et al. (2004) also identified the metabolite B, which was produced by demethylation of acetamiprid, as well as compound IV.
Analysis of the Bacterial Community
The interactive behavior of bacterial populations can affect the degradation of pesticides. Accordingly, further exploration of community diversity and phylogenetic structures of the bacterial consortium ACE-3 through HTS was carried out. After the non-repeat sequences were extracted from the optimized sequence pool, there analysis yielded 40832 valid sequences. The average length of the 16S rRNA gene sequences derived from the community of ACE-3 was found to be 376 bp. To gain an overview of the most likely acetamiprid-degrading populations, the classification and proportions of bacterial species in ACE-3 at the phylum level based on OTU sequence analysis was conducted as shown in Figure 5. In total, seven phyla were identified, whereby α Proteobacteria (6.9%) and β Proteobacteria (85.61%) were the dominant phyla in consortium ACE-3. At the class level, 98.35% of sequences were assigned, with Actinobacteria, Sphingobacteria, Cytophagia, and Flavobacteria accounting for 5.57, 0.037, 0.31, and 0.25%, respectively.
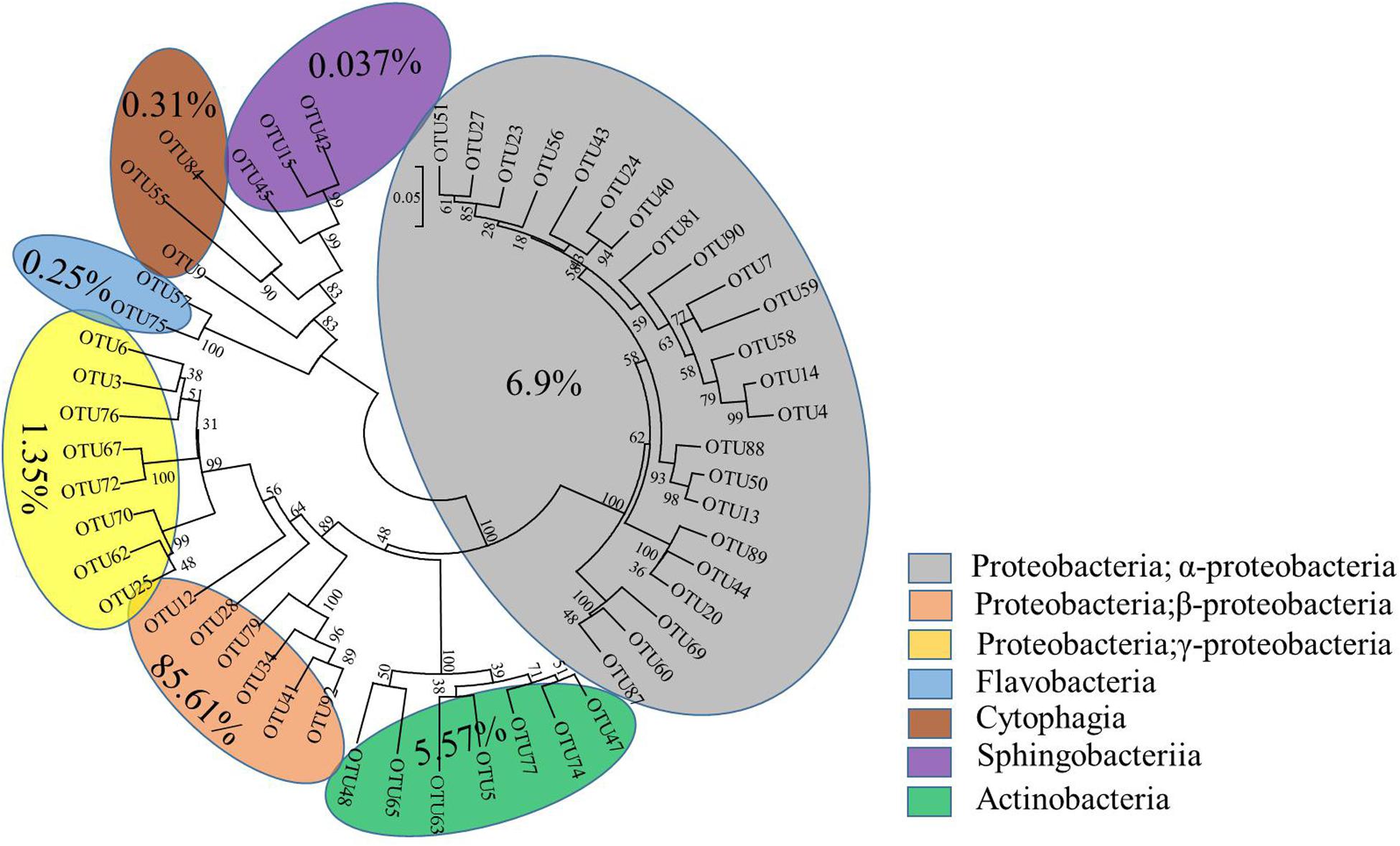
Figure 5. Phylogenetic tree based on the HTS of the acetamiprid-degrading consortium ACE-3. The scale bar indicates 0.05 substitutions per nucleotide position.
Further analysis was carried out to investigate the bacterial community abundance at the genus level, and a total of 51 genera were identified in the bacterial community of consortium ACE-3. To characterize the species diversity of the microbial community, the Shannon’s Diversity Index (H′) (Shannon, 1948) at the genus level was used as a performance index to characterize the diversity of species (OTUs) in consortium ACE-3. Shannon’s Diversity Index was calculated using the formula:
where Sobs is the total number of species (OTUs) and Pi is the fraction of the total number of individuals in a particular genotype or taxon i. The values for H’ range from 0 (low diversity) to 5 (high diversity), and can directly represent the richness of different OTUs in the bacterial community. After calculation, Shannon’s diversity index reached 3.11, which indicated that consortium ACE-3 was rich in diversity.
The dominant genera are summarized in Table 2. These included Sphingobium, Acinetobacter, Afipia, Stenotrophomonas, and Microbacterium, which respectively accounted for 3.07, 10.01, 24.45, and 49.12% of the total population. Obviously, some of these genera play a key role in pollutant degradation and can be regularly detected in various contaminated environments. For instance, Sphingobium fuliginis HC3 was identified as a novel biodegrader that can mineralize biphenyls and polychlorinated biphenyls, while no significant accumulation of derived intermediates was observed (Hu et al., 2015). Stenotrophomonas species are widely used for the biodegradation of various aromatic and chlorinated aromatic organic compounds. For example, Stenotrophomonas maltophilia D310-3 has the ability of efficiently degrading chlorimuron-ethyl, producing eight degradation products (Zang et al., 2016). Enterobacter sp. and Acinetobacter baumannii can degrade pesticides and pesticide intermediates, such as imazamox, p-hydroxyphenyl acetate, and phenol (Sharma T. et al., 2014; Liu et al., 2016; Thotsaporn et al., 2016).
In order to elucidate the mechanisms of pesticide biodegradation, the bacterial diversity of many pesticide-degrading consortia has recently been deciphered by different methods. For example, a thiabendazole-degrading consortium was found to contain the Sphingomonas phylotype B13 as the major degrading member via denaturing gradient gel electrophoresis (DGGE) analysis (Perruchon et al., 2017a). The propanoyl-degrading consortium T4 was found to contain 59 OTUs using restriction fragment length polymorphism (RFLP) (Hou et al., 2015). Interestingly, these results indicated that the species diversity of bacterial consortia capable of degrading different pesticides, which utilize xenobiotic compounds as the carbon source, became limited after serial enrichment (Zhou et al., 2012).
In recent years, biodegradation technologies have been shown to be viable processes for the complete mineralization of chemical pesticides. For the biodegradation of acetamiprid, increasing numbers of mixed cultures have been screened to study their degradability, including the consortium ACE-3 introduced in this work. However, most of these studies were carried out in the laboratory, which is quite different from the actual degradation in the open environment. Notably, the nutrient conditions in the natural environment are relatively inadequate, and cannot guarantee the efficient growth of bacteria with the desired degradation ability. Moreover, the more complex degradation conditions (e.g., swings of temperature, pH, and oxygen) may be unfavorable to the screened mixed bacteria cultures (Shi et al., 2018). In addition, the nutrient competition and even parasitism by other microorganisms may lead to instability or even the disappearance of the biodegrading bacteria (Ramanan et al., 2015). These disadvantageous conditions are both a bottleneck and a motive force of further research. These problems inspired the exploration and transformation of exciting new technologies, such as microbial modification and evolution, multicellular immobilization and microfluidic screening (Emelyanova et al., 2017; Chen et al., 2019). However, there is no doubt that the microbial consortium ACE-3 screened in this work has great application potential for acetamiprid removal in actual contaminated soil and water environments in the future.
Conclusion
The natural consortium ACE-3 was gradually acclimatized, enriched and isolated based on its ability to use acetamiprid as the sole carbon and energy source. Possible metabolic pathways for acetamiprid degradation were proposed via intermediate identification. In addition, species diversity analysis was implemented to reveal the changes of community structure and phylogenetic associations. This study provides detailed information on a novel microbial consortium with promising capability for acetamiprid degradation, offering a theoretical basis for further studies on biostimulation or bioaugmentation for in situ or on-site bioremediation of pesticide-contaminated environments.
Data Availability Statement
All datasets generated for this study are included in the article.
Author Contributions
BX, RX, and JZ performed the experiments, analyzed the data, and wrote the manuscript. XW and ZS helped to modify the graphs. FX, WZ, and MJ assisted the manuscript checking. WD provided assistance and guidance throughout the research. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Key Research and Development Program of China (2019YFA0905500 and 2018YFA0902200), the National Natural Science Foundation of China (31961133017, 31700092, and 21978129), the Jiangsu Province Natural Science Foundation for Youths (BK20170997), Jiangsu Agriculture Science and Technology Innovation Fund (CX(19)3104), and the Jiangsu Synergetic Innovation Center for Advanced Bio-Manufacture of China.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Baghapour, M. A., Nasseri, S., and Derakhshan, Z. (2013). Atrazine removal from aqueous solutions using submerged biological aerated filter. J. Environ. Health Sci. Eng. 11, 6–15.
Bai, N., Abuduaini, R., Wang, S., Zhang, M., Zhu, X., and Zhao, Y. (2017). Nonylphenol biodegradation characterizations and bacterial composition analysis of an effective consortium NP-M2. Environ. Pollut. 220, 95–104. doi: 10.1016/j.envpol.2016.09.027
Brunet, J., Badiou, A., and Belzunces, L. P. (2005). In vivo metabolic fate of [14C]-acetamiprid in six biological compartments of the honeybee. Apis mellifera L. Pest Manag. Sci. 61, 742–748. doi: 10.1002/ps.1046
Carra, I., Sánchez Pérez, J. A., Malato, S., Autin, O., Jefferson, B., and Jarvis, P. (2016). Performance of different advanced oxidation processes for tertiary wastewater treatment to remove the pesticide acetamiprid. J. Chem. Technol. Biot. 91, 72–81. doi: 10.1002/jctb.4577
Chakraborty, R., Wu, C. H., and Hazen, T. C. (2012). Systems biology approach to bioremediation. Curr. Opin. Biotechnol. 23, 483–490. doi: 10.1016/j.copbio.2012.01.015
Chakroun, S., Ezzi, L., Grissa, I., Kerkeni, E., Neffati, F., Bhouri, R., et al. (2016). Hematological, biochemical, and toxicopathic effects of subchronic acetamiprid toxicity in Wistar rats. Environ. Sci. Pollut. R. 23, 25191–25199. doi: 10.1007/s11356-016-7650-9
Chen, D. W., Liu, S. J., and Du, W. B. (2019). Chemotactic screening of imidazolinone-degrading bacteria by microfluidic SlipChip. J. Hazard. Mater. 366, 512–519. doi: 10.1016/j.jhazmat.2018.12.029
Chen, T., Dai, Y., Ding, J., Yuan, S., and Ni, J. (2008). N-demethylation of neonicotinoid insecticide acetamiprid by bacterium Stenotrophomonas maltophilia CGMCC 1.1788. Biodegradation 19, 651–658. doi: 10.1007/s10532-007-9170-2
Copley, S. D. (2009). Evolution of efficient pathways for degradation of anthropogenic chemicals. Nat. Chem. Biol. 5, 559–566. doi: 10.1038/nchembio.197
Cruz-Alcalde, A., Sans, C., and Esplugas, S. (2017). Priority pesticides abatement by advanced water technologies: the case of acetamiprid removal by ozonation. Sci. Total Environ. 599, 1454–1461. doi: 10.1016/j.scitotenv.2017.05.065
Dai, Y., Ji, W., Chen, T., Zhang, W., Liu, Z., Ge, F., et al. (2010). Metabolism of the neonicotinoid insecticides acetamiprid and thiacloprid by the yeast Rhodotorula mucilaginosa strain IM-2. J. Agr. Food Chem. 58, 2419–2425. doi: 10.1021/jf903787s
Dangi, A. K., Sharma, B., Hill, R. T., and Shukla, P. (2019). Bioremediation through microbes: systems biology and metabolic engineering approach. Crit. Rev. Biotechn. 39, 79–98. doi: 10.1080/07388551.2018.1500997
Ding, M. Z., Song, H., Wang, E. X., Liu, Y., and Yuan, Y. J. (2016). Design and construction of synthetic microbial consortia in China. Synth. Syst. Biotechnol. 1, 230–235. doi: 10.1016/j.synbio.2016.08.004
Dong, W., Liu, K., Wang, F., Xin, F., Zhang, W., Zhang, M., et al. (2017). The metabolic pathway of metamifop degradation by consortium ME-1 and its bacterial community structure. Biodegradation 28, 181–194. doi: 10.1007/s10532-017-9787-8
Dvořák, P., Nikel, P. I., Damborský, J., and de Lorenzo, V. (2017). Bioremediation 3.0: engineering pollutant-removing bacteria in the times of systemic biology. Biotechnol. Adv. 35, 845–866. doi: 10.1016/j.biotechadv.2017.08.001
Edgar, R. C. (2013). UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10, 996–998. doi: 10.1038/nmeth.2604
Emelyanova, E. V., Souzina, N. E., Polivtseva, V. N., Reshetilov, A. N., and Solyanikova, I. P. (2017). Survival and biodegradation activity of Gordonia polyisoprenivorans 135: Basics of a biosensor receptor. Appl. Biochem. Microbiol. 53, 580–586. doi: 10.1134/s0003683817050039
Festa, S., Coppotelli, B. M., and Morelli, I. S. (2013). Bacterial diversity and functional interactions between bacterial strains from a phenanthrene-degrading consortium obtained from a chronically contaminated-soil. Int. Biodeter. Biodegr. 85, 42–51 doi: 10.1016/j.ibiod.2013.06.006
Fida, T. T., Forero, S. K. M., Breugelmans, P., Heipieper, H. J., Röling, W. F. M., and Springael, D. (2017). Physiological and transcriptome response of the polycyclic aromatic hydrocarbon degrading Novosphingobium sp. LH128 after inoculation in soil. Environ. Sci. Technol. 51, 1570–1579. doi: 10.1021/acs.est.6b03822
Hou, Y., Li, S., Dong, W., Yuan, Y., Wang, Y., Shen, W., et al. (2015). Community structure of a propanil-degrading consortium and the metabolic pathway of Microbacterium sp. strain T4-7. Int. Biodeter. Biodegr. 105, 80–89. doi: 10.1016/j.ibiod.2015.08.018
Hu, J., Qian, M., Zhang, Q., Cui, J., Yu, C., Su, X., et al. (2015). Sphingobium fuliginis HC3: a novel and robust isolated biphenyl- and polychlorinated biphenyls-degrading bacterium without dead-end intermediates accumulation. PLoS One 10:e0122740. doi: 10.1371/journal.pone.0122740
Ju, F., and Zhang, T. (2014). Novel microbial populations in ambient and mesophilic biogas-producing and phenol-degrading consortia unraveled by high-throughput sequencing. Microb. Ecol. 68, 235–246. doi: 10.1007/s00248-014-0405-6
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Liu, C., Yang, X., Lai, Y., Lu, H., Zeng, W., Geng, G., et al. (2016). Imazamox microbial degradation by common clinical bacteria: Acinetobacter baumannii IB5 isolated from black soil in China shows high potency. J. Integrated Agr. 15, 1798–1807. doi: 10.1016/s2095-3119(16)61344-8
Liu, X., Xu, X., Li, C., Zhang, H., Fu, Q., Shao, X., et al. (2015). Degradation of chiral neonicotinoid insecticide cycloxaprid in flooded and anoxic soil. Chemosphere 119, 334–341. doi: 10.1016/j.chemosphere.2014.06.016
Liu, Z., Dai, Y., Huang, G., Gu, Y., Ni, J., Wei, H., et al. (2011). Soil microbial degradation of neonicotinoid insecticides imidacloprid, acetamiprid, thiacloprid and imidaclothiz and its effect on the persistence of bioefficacy against horsebean aphid Aphis craccivora Koch after soil application. Pest Manag. Sci. 67, 1245–1252. doi: 10.1002/ps.2174
Mnif, I., Sahnoun, R., Ellouz-Chaabouni, S., and Ghribi, D. (2017). Application of bacterial biosurfactants for enhanced removal and biodegradation of diesel oil in soil using a newly isolated consortium. Process Saf. Environ. 109, 72–81. doi: 10.1016/j.psep.2017.02.002
Mori, T., Wang, J., Tanaka, Y., Nagai, K., Kawagishi, H., and Hirai, H. (2017). Bioremediation of the neonicotinoid insecticide clothianidin by the white-rot fungus Phanerochaete sordida. J. Hazard. Mater. 321, 586–590. doi: 10.1016/j.jhazmat.2016.09.049
Patel, V., Jain, S., and Madamwar, D. (2012). Naphthalene degradation by bacterial consortium (DV-AL) developed from Alang-Sosiya ship breaking yard. Gujarat, India. Bioresour. Technol. 107, 122–130. doi: 10.1016/j.biortech.2011.12.056
Perruchon, C., Chatzinotas, A., Omirou, M., Vasileiadis, S., Menkissoglou-Spiroudi, U., and Karpouzas, D. G. (2017a). Isolation of a bacterial consortium able to degrade the fungicide thiabendazole: the key role of a Sphingomonas phylotype. Appl Microbiol. Biot. 101, 3881–3893. doi: 10.1007/s00253-017-8128-5
Perruchon, C., Pantoleon, A., Veroutis, D., Blanco, S. G., Laurent, F. M., Liadaki, K., et al. (2017b). Characterization of the biodegradation, bioremediation and detoxification capacity of a bacterial consortium able to degrade the fungicide thiabendazole. Biodegradation. 28, 383–394. doi: 10.1007/s10532-017-9803-z
Phugare, S. S., and Jadhav, J. P. (2015). Biodegradation of acetamiprid by isolated bacterial strain Rhodococcus sp. BCH2 and toxicological analysis of its metabolites in silkworm (Bombax mori). 43, 296–304. doi: 10.1002/clen.201200563
Ramanan, R., Kang, Z., Kim, B. H., Cho, D. H., Jin, L., and Oh, H. M. (2015). Phycosphere bacterial diversity in green algae reveals an apparent similarity across habitats. Algal Res. 8, 140–144. doi: 10.1016/j.algal.2015.02.003
Schloss, P. D., and Westcott, S. L. (2011). Assessing and improving methods used in operational taxonomic unit-based approaches for 16S rRNA gene sequence analysis. Appl. Environ. Microb. 77, 3219–3226. doi: 10.1128/aem.02810-10
Sharma, S., Singh, B., and Gupta, V. K. (2014). Biodegradation of imidacloprid by consortium of two soil Isolated Bacillus sp. Bull. Environ. Contam. Toxicol. 93, 637–642. doi: 10.1007/s00128-014-1386-3
Sharma, T., Rajor, A., and Toor, A. P. (2014). Degradation of imidacloprid in liquid by Enterobacter sp. strain ATA1 using co-metabolism. Bioremediat. J. 18, 227–235. doi: 10.1080/10889868.2014.918575
Shi, Z., Dong, W., Xin, F., Liu, J., Zhou, X., Xu, F., et al. (2018). Characteristics and metabolic pathway of acetamiprid biodegradation by Fusarium sp. strain CS-3 isolated from soil. Biodegradation 29, 593–603. doi: 10.1007/s10532-018-9855-8
Singh, B., and Kashmir, S. (2016). Microbial degradation of herbicides. Crit. Rev. Microbiol. 42, 245–261.
Su, X., Xue, B., Wang, Y., Hashmi, M. Z., Lin, H., Chen, J., et al. (2019). Bacterial community shifts evaluation in the sediments of Puyang River and its nitrogen removal capabilities exploration by resuscitation promoting factor. Ecotox. Environ. Saf. 179, 188–197. doi: 10.1016/j.ecoenv.2019.04.067
Sun, S., Yang, W., Guo, J., Zhou, Y., Rui, X., Chen, C., et al. (2017). Biodegradation of the neonicotinoid insecticide acetamiprid in surface water by the bacterium Variovorax boronicumulans CGMCC 4969 and its enzymatic mechanism. RSC Adv. 7, 25387–25397. doi: 10.1039/c7ra01501a
Tang, H., Li, J., Hu, H., and Xu, P. (2012). A newly isolated strain of Stenotrophomonas sp. hydrolyzes acetamiprid, a synthetic insecticide. Process Biochem. 47, 1820–1825. doi: 10.1016/j.procbio.2012.06.008
Thotsaporn, K., Tinikul, R., Maenpuen, S., Phonbuppha, J., Watthaisong, P., Chenprakhon, P., et al. (2016). Enzymes in the p-hydroxyphenylacetate degradation pathway of Acinetobacter baumannii. J. Mol. Catal. B Enzym. 134, 353–366. doi: 10.1016/j.molcatb.2016.09.003
Wang, G., Chen, X., Yue, W., Zhang, H., Li, F., and Xiong, M. (2013a). Microbial degradation of acetamiprid by Ochrobactrum sp. D-12 isolated from contaminated soil. PLoS. One 8:e82603. doi: 10.1371/journal.pone.0082603
Wang, G., Yue, W., Liu, Y., Li, F., Xiong, M., and Zhang, H. (2013b). Biodegradation of the neonicotinoid insecticide acetamiprid by bacterium Pigmentiphaga sp. strain AAP-1 isolated from soil. Bioresour. Technol. 138, 359–368. doi: 10.1016/j.biortech.2013.03.193
Wang, G., Zhao, Y., Gao, H., Yue, W., Xiong, M., Li, F., et al. (2013c). Co-metabolic biodegradation of acetamiprid by Pseudoxanthomonas sp. AAP-7 isolated from a long-term acetamiprid-polluted soil. Bioresour. Technol. 150, 259–265. doi: 10.1016/j.biortech.2013.10.008
Wang, G., Zhu, D., Xiong, M., Zhang, H., and Liu, Y. (2016). Construction and analysis of an intergeneric fusion from Pigmentiphaga sp. strain AAP-1 and Pseudomonas sp. CTN-4 for degrading acetamiprid and chlorothalonil. Environ. Sci. Pollut. R. 23, 13235–13244. doi: 10.1007/s11356-016-6482-y
Wang, Y., Fan, Y., and Gu, J. D. (2004). Dimethyl phthalate ester degradation by two planktonic and immobilized bacterial consortia. Int. Biodeter. Biodegr. 53, 93–101. doi: 10.1016/j.ibiod.2003.10.005
Wang, Y., Li, F., Ruan, X., Song, J., Lv, L., Chai, L., et al. (2017). Biodegradation of di-n-butyl phthalate by bacterial consortium LV-1 enriched from river sludge. PLos One 12:e0178213. doi: 10.1371/journal.pone.0178213
Watanabe, K. (2001). Microorganisms relevant to bioremediation. Curr. Opin. Biotechnol. 12, 237–241. doi: 10.1016/s0958-1669(00)00205-6
Yang, H., Wang, X., Zheng, J., Wang, G., Hong, Q., Li, S., et al. (2013). Biodegradation of acetamiprid by Pigmentiphaga sp. D-2 and the degradation pathway. Int. Biodeter. Biodegr. 85, 95–102. doi: 10.1016/j.ibiod.2013.03.038
Zang, H., Yu, Q., Lv, T., Cheng, Y., Feng, L., Cheng, X., et al. (2016). Insights into the degradation of chlorimuron-ethyl by Stenotrophomonas maltophilia D310-3. Chemosphere 144, 176–184. doi: 10.1016/j.chemosphere.2015.08.073
Zhou, L., Li, K. P., Mbadinga, S. M., Yang, S. Z., Gu, J. D., and Mu, B. Z. (2012). Analyses of n-alkanes degrading community dynamics of a high-temperature methanogenic consortium enriched from production water of a petroleum reservoir by a combination of molecular techniques. Ecotoxicology 21, 1680–1691. doi: 10.1007/s10646-012-0949-5
Zhou, L., Zhang, L., Sun, S., Ge, F., Mao, S., Ma, Y., et al. (2014). Degradation of the neonicotinoid insecticide acetamiprid via the N-carbamoylimine derivate (IM-1-2) mediated by the nitrile hydratase of the nitrogen-fixing bacterium Ensifer meliloti CGMCC 7333. J. Agr. Food Chem. 62, 9957–9964. doi: 10.1021/jf503557t
Keywords: acetamiprid, microbial consortium, community structure, species diversity, degradation pathway
Citation: Xu B, Xue R, Zhou J, Wen X, Shi Z, Chen M, Xin F, Zhang W, Dong W and Jiang M (2020) Characterization of Acetamiprid Biodegradation by the Microbial Consortium ACE-3 Enriched From Contaminated Soil. Front. Microbiol. 11:1429. doi: 10.3389/fmicb.2020.01429
Received: 13 February 2020; Accepted: 02 June 2020;
Published: 08 July 2020.
Edited by:
Chaofeng Shen, Zhejiang University, ChinaReviewed by:
Xiaomei Su, Zhejiang Normal University, ChinaWei-Min Wu, Stanford University, United States
Copyright © 2020 Xu, Xue, Zhou, Wen, Shi, Chen, Xin, Zhang, Dong and Jiang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Weiliang Dong, ZHdsQG5qdGVjaC5lZHUuY24=
 Bin Xu
Bin Xu Rui Xue1
Rui Xue1 Fengxue Xin
Fengxue Xin Wenming Zhang
Wenming Zhang Weiliang Dong
Weiliang Dong Min Jiang
Min Jiang
