- 1Department of Microbiology and Biotechnology, Universität Hamburg, Hamburg, Germany
- 2Technology Platform Next Generation Sequencing, Heinrich Pette Institut, Hamburg, Germany
- 3Department of Microbiology, IWWR, Radboud University, Nijmegen, Netherlands
- 4PROBIEN (CONICET-UNCo), Departamento de Química, Facultad de Ingeniería, Universidad Nacional del Comahue, Neuquén, Argentina
Nitrification is a key process for N-removal in engineered and natural environments, but recent findings of novel nitrifying microorganisms with surprising features revealed that our knowledge of this functional guild is still incomplete. Especially nitrite oxidation – the second step of nitrification – is catalyzed by a phylogenetically diverse bacterial group, and only recently bacteria of the phylum Chloroflexi have been identified as thermophilic nitrite-oxidizing bacteria (NOB). Among these, Nitrolancea hollandica was isolated from a laboratory-scale nitrifying bioreactor operated at 35°C with a high load of ammonium bicarbonate. However, its distribution remains cryptic as very few closely related environmental 16S rRNA gene sequences have been retrieved so far. In this study, we demonstrate how such thermophilic NOB can be enriched using modified mineral media inoculated with samples from a wastewater side-stream reactor operated at 39.5°C. Distinct cultivation conditions resulted in quick and reproducible high enrichment of two different strains of Nitrolancea, closely related to Nl. hollandica. The same cultivation approach was applied to a complex nitrite-oxidizing pre-enrichment at 42°C inoculated with biomass from a geothermal spring in the Copahue volcano area in Neuquen, Argentina. Here, an additional distinct representative of the genus Nitrolancea was obtained. This novel species had 16S rRNA and nitrite oxidoreductase alpha subunit (nxrA) gene sequence identities to Nl. hollandica of 98.5% and 97.2%, respectively. A genomic average nucleotide identity between the Argentinian strain and Nl. hollandica of 91.9% indicates that it indeed represents a distinct species. All Nitrolancea cultures formed lancet-shaped cells identical to Nl. hollandica and revealed similar physiological features, including the capability to grow at high nitrite concentrations. Growth was optimal at temperatures of 35–37°C and was strongly enhanced by ammonium supplementation. Genomic comparisons revealed that the four Nitrolancea strains share 2399 out of 3387 orthologous gene clusters and encode similar key functions. Our results define general growth conditions that enable the selective enrichment of Nitrolancea from artificial and natural environments. In most natural habitats these NOB apparently are of low abundance and their proliferation depends on the balanced presence of nitrite and ammonium, with an optimal incubation temperature of 37°C.
Introduction
As important part of the global nitrogen cycle, nitrification converts reduced nitrogen species into oxidized forms, namely ammonium (−III) to nitrite (+III) and further to nitrate (+V). In wastewater treatment plants, nitrification is essential to remove N-compounds like ammonium and nitrite, and to provide nitrate as electron acceptor for denitrification. In agricultural, but also natural systems, nitrification causes nitrogen loss due to leaching of negatively charged nitrate ions and emission of gaseous N-compounds produced by denitrifying bacteria.
Both steps of nitrification are performed by distinct groups of bacteria or archaea (Koops and Pommerening-Röser, 2005; Spieck and Bock, 2005; Stieglmeier et al., 2014), but the traditional differentiation into ammonia and nitrite oxidizers has been overturned with the discovery of comammox Nitrospira, which can perform the complete oxidation of ammonia to nitrate (Daims et al., 2015; van Kessel et al., 2015).
Nitrite-oxidizing bacteria (NOB) from six genera belonging to four different phyla have been isolated so far. These are Nitrobacter, Nitrotoga and Nitrococcus within the Alpha-, Beta- and Gammaproteobacteria, respectively (Spieck and Bock, 2005; Alawi et al., 2007), Nitrospira and Nitrospina, which belong to separate phyla (Ehrich et al., 1995; Lücker et al., 2013), and the moderate thermophilic NOB Nitrolancea, which is phylogenetically affiliated with the Chloroflexi (Sorokin et al., 2012, 2014). The latter phylum also contains novel thermophilic nitrite oxidizers that have been highly enriched from hot springs at Yellowstone National Park (Spieck et al., 2020). Indications for further marine NOB have been obtained by metagenomics studies (Lüke et al., 2016; Ngugi et al., 2016; Sun et al., 2019).
Nitrolancea hollandica has been isolated from a lab-scale nitrifying bioreactor receiving a high load of ammonium bicarbonate and operating at 35°C (Vejmelkova et al., 2012). It has a high tolerance against nitrite as typical for r-strategists among the NOB (Andrews and Harris, 1986; Nowka et al., 2015), which appears to correlate with the cytoplasmic localization of the key enzyme nitrite oxidoreductase (NXR) (Spieck et al., 1998; Lücker et al., 2010). The genomic organization of the nxr genes in Nitrolancea is very similar to those in Nitrobacter, Nitrococcus (Sorokin et al., 2012) and the recently described Candidatus (Ca.) Nitrocaldera (Spieck et al., 2020). In contrast, NOB containing a periplasmic NXR like Nitrospira (Spieck et al., 1996) are mostly inhibited by high substrate concentrations (Off et al., 2010). This distinct Nitrospira-type NXR is closely related to that of anaerobic ammonium-oxidizing (anammox) bacteria (Lücker et al., 2010), while the cytoplasmic Nitrobacter-type NXR is closely related to dissimilatory nitrate reductases (NAR) of nitrate-reducing microorganisms (Kirstein and Bock, 1993).
The low substrate affinity of Nl. hollandica (Km (nitrite) = 1 mM; Sorokin et al., 2012) posed the question of its natural niche. Nitrite is an intermediate of nitrification, denitrification and nitrate reduction to ammonium. It can accumulate when these processes are imbalanced, an effect that can be exaggerated by abiotic factors like pH, temperature or certain operational modes in reactor systems (Philips et al., 2002).
Temperature is a main factor shaping the community structure of nitrite-oxidizing bacteria and variations of this incubation parameter have resulted in the discovery of novel NOB (Alawi et al., 2007; Lebedeva et al., 2008). Nitrite oxidation at elevated temperature was found to be performed by Nitrospira (Lebedeva et al., 2005, 2011; Courtens et al., 2016), a common NOB in geothermal habitats (Marks et al., 2012; Edwards et al., 2013), or novel NOB of the Chloroflexi phylum (Spieck et al., 2020).
Based on the observed properties, the natural habitat of Nl. hollandica has been proposed to feature high temperatures as well as high ammonium and nitrite concentrations, as present for instance in compost or dung (Sorokin et al., 2012). Nitrolancea-like 16S rRNA gene sequences have only rarely been detected in specialized incubations of water or soil samples (Sorokin et al., 2018). These include soil columns used for aquifer treatment (Friedman et al., 2018), 1,3-dichloropropene fumigated soil (Fang et al., 2019), and manure composting (Zhang et al., 2016). A closely related 16S rRNA gene sequence (JN087902) has been detected in a SHARON (single reactor system for high activity ammonium removal over nitrite) reactor in Korea running at elevated temperature, a system with conditions comparable to the source of Nl. hollandica. Furthermore, recent surveys revealed the presence of this NOB in four Danish enhanced biological phosphorus removal (EBPR) systems and an N-removal plant in China (Speirs et al., 2019). Additionally, Nitrolancea-like 16S rRNA gene sequences have been found in reactors used for partial nitrification (Yu et al., 2018, 2020) and in free nitrous acid (FNA) treated wastewater (Wang et al., 2020). However, so far it is unclear what the lower temperature limit for proliferation of this moderate thermophilic NOB is and if Nitrolancea is present and active in full-scale nitrifying sludge systems especially as activated sludge does not contain ammonium or nitrite in the high concentrations apparently required by Nitrolancea to be competitive.
Apart from engineered systems, 16S rRNA gene sequences distantly related to Nitrolancea were found in warm natural environments, like the Atacama Desert (Orlando et al., 2010, EU603380) and the Kalahari (Elliott et al., 2014). These findings raise the question whether geothermal habitats might be suitable habitats for Nitrolancea, too. Therefore, in this study the geothermal area Las Máquinas in Argentina was investigated for the presence of nitrifying microorganisms. This region belongs to the Copahue-Caviahue system, which is mostly located in the Neuquen Province in Northern Patagonia and is characterized by a high biodiversity (Chiacchiarini et al., 2010; Urbieta et al., 2012, 2015a,b; Giaveno et al., 2013; Willis Poratti et al., 2016).
In the present study, three new representatives of Nitrolancea were selectively enriched under comparable conditions, and two pure cultures were obtained. Two of the enrichment cultures originated from a full-scale centrate treatment reactor at a wastewater treatment plant (WWTP) in Hamburg, Germany, and one from the geothermal Las Máquinas area in Argentina. The obtained species were compared to the type strain Nl. hollandica with regard to their genomic, phenotypic and physiological features.
Materials and Methods
Sampling
WWTP Hamburg-Dradenau, Germany
On May 22, 2017, a centrate treatment side-stream reactor (3300 m3) at the full-scale WWTP Hamburg-Dradenau was sampled (sample Z2; Supplementary Figure S1A). The centrate originated from centrifuged digested sludge and contained high ammonium (38 mM) and nitrite (44 mM), and low nitrate (9 mM) concentrations, and had a temperature of 39.5°C and a pH of 6.5–6.9 on the sampling day. In the treatment reactor, this liquid was mixed with activated sludge from the nitrification stage (approx. 30% v/v) and was supplied with 2 mg O2 l–1. The HRT was 1 day. The solids content and chemical oxygen demand (COD) amounted to 550 mg l–1 and 1750 mg l–1, respectively. Subsequent to this “Store and treat” (SAT) procedure, patent-registered by the HSE (Hamburger Stadtentwässerung), the reactor effluent was applied for anammox treatment.
Las Máquinas, Argentina
The Copahue volcano (37°51′S, 71°10.2′W; 2977 m a.s.l), the Rio Agrio (pH 0.5–7) and Caviahue Lake (pH 2.1–3.7) are geographical landmarks that frame a geothermal field, which is composed of five hydrothermal sites: Las Máquinas, Las Maquinitas, Anfiteatro, Termas de Copahue (in Argentina) and Chancho-Co (in Chile). The Las Máquinas area contains many thermal fluid emission systems, consisting of fumaroles, boiling-bubbling water and mud pools with wide ranges of pH (2–7) and temperature (30–90°C). The chemical composition of the fumaroles, which are mainly fed by meteoric water (Chiodini et al., 2015), showed that the main emitted gas is CO2, followed by minor concentrations of N2, H2, CH4, and H2S. CO and He concentrations are <1 ppm. Ammonium as a substrate for nitrification was detected in concentrations between 0 and 20 mM at different sampling sites of Las Máquinas, whereas nitrite and nitrate occurred between 0 and 6 μM (unpublished results).
In December 2008 (sample E2) and in March 2011 (sample A4), two mud pools in the geothermal area Las Máquinas, Argentina, were sampled. Both sampling sites were characterized by high sulfate concentrations (Supplementary Table S1). For sample E2 (Supplementary Figure S1B), red to brownish sediment was removed at a site that had a temperature of 40°C, a pH of 2.5–3.0 and an ammonium content of 0.6 mM. The sampling site of A4 (Supplementary Figure S1C) had a temperature of 45°C, a pH of 6.0 and ammonium was below the detection limit. Sample aliquots were transported at ambient temperature by courier to the University of Hamburg, Germany. Temperature, pH, and electric conductivity were measured in situ in each sample point using specific electrodes (Thermo Scientific Orion 5-Star Plus benchtop multifunction meter, electrode Orion 9142BN, Waltham, MA, United States). Water samples for metal analysis were filtered using 0.2 μm membrane filters (Cellulose acetate, Sartorius, Göttingen, Germany) and their concentrations (Supplementary Table S1) determined by atomic absorption spectrophotometry (AAS; Merodio and Martínez, 1985). To quantify anions, a standard turbidimetric assay for the determination of sulfate with BaCl2 (Kolmert et al., 2000) and a titration of chloride ions with mercuric nitrate solution using diphenyl carbazide indicator were used (APHA et al., 1992).
Cultivation Media
The enrichments were performed in 0.05 to 0.15 l batch cultures in mineral salts medium with the following composition: 0.007 g l–1 CaCO3, 0.5 g l–1 NaCl, 0.05 g l–1 MgSO4 × 7 H2O, and 0.15 g l–1 KH2PO4 dissolved in distilled water. If not stated otherwise, 0.02–0.2 g l–1 NaNO2 (0.29 – 2.9 mM) were supplied as energy source. The trace element solution (1 ml l–1) contained 33.8 mg l–1 MnSO4 × H2O, 49.4 mg l–1 H3BO3, 43.1 mg l–1 ZnSO4 × 7 H2O, 37.1 mg l–1 (NH4)6Mo7O24, 973.0 mg l–1 FeSO4 × 7 H2O, and 25.0 mg l–1 CuSO4 × 5 H2O. The pH was adjusted to 8.0 and changed to 7.4–7.6 2 days after autoclaving. In order to enrich Nitrolancea, NH4Cl was added in concentrations between 0.5 and 10 mM (Supplementary Table S2). When bicarbonate (0.5 mM) was included, the pH had to be adjusted to 6.7 to reach a final pH of 7.4–7.6 after autoclaving. For growth of the centrate-derived cultures, and the Argentinian cultures from 2018 on, trace elements according to Widdel and Bak (1991) were added to the medium (1 ml l–1): 100 mg l–1 MnCl2 × 4 H2O, 30 mg l–1 H3BO3, 144 mg l–1 ZnSO4 × 7 H2O, 36 mg l–1 Na2MoO4 × 2 H2O, 2.1 g l–1 FeSO4 × 7 H2O, 2 mg l–1 CuCl2 × 2 H2O, 190 mg l–1 CoCl2 × 6 H2O, and 24 mg l–1 NiCl2 × 6 H2O.
Enrichment
WWTP Hamburg-Dradenau
Two batches of mineral NOB medium (see above) containing 3 mM NaNO2 and 3 mM NH4Cl were inoculated (1% v/v) with the centrate sample Z2 on June 22, 2017, and incubated without agitation at 42°C and 46°C, respectively. A transfer was performed on October 23, 2017, and the incubation temperature was lowered to 37°C. In the following, the cultures grew very well and nitrite concentrations supplied to the medium were continuously increased (Supplementary Table S2).
Copahue – Argentina
A 0.5 ml aliquot of sample E2 (spring water with sediment) was inoculated into a 100 ml Erlenmeyer flask with 50 ml mineral unbuffered AOB medium (Krümmel and Harms, 1982) containing 0.5 mM ammonium as substrate. When ammonium was depleted, it was replenished to 400–500 μM with sterile 5 M NH4Cl solution. Ammonium measurements were done with Quantofix test stripes (Macherey-Nagel, Düren, Germany). The pH was manually adjusted to approx. 7.4 with 5% (w/v) NaHCO3 solution. For sample A4, 1 ml was inoculated in 100 ml mineral medium for NOB with 0.3 mM nitrite as substrate in a 250 ml screw cap Schott bottle. Both cultures were grown at 42°C under static conditions and evaporation was compensated for with sterile distilled water. Both cultures were regularly transferred with 1% (v/v) inoculum into mineral NOB media containing different concentrations of ammonium and nitrite with or without addition of 0.5 mM bicarbonate, and incubated under different conditions, as detailed in Supplementary Table S2.
Isolation Attempts
Cells from the enrichment cultures Z2.3.3 and A4.5.1 (Supplementary Table S2) were manually separated by an optical tweezers system (PALM MicroTweezers, Carl Zeiss Microscopy GmbH, Munich, Germany). Cells were visualized by bright-field microscopy at 1000× magnification (Axio Observer.Z1, Carl Zeiss, Jena, Germany), trapped with an infrared laser (1064 nm, 3 W) and separated by micromanipulation (PatchMan NP2, CellTram Vario, Eppendorf, Hamburg, Germany). Separated cells were transferred to NOB medium (0.5 mM NaNO2, 0.5 mM NH4Cl and 0.5 mM NaHCO3) in 1.5 ml Eppendorf tubes, and incubated at 37°C. A total of 10 and 20 tubes for enrichments Z2.3.3 and A4.5.1, respectively, were inoculated with single cells or short chains resembling Nitrolancea by morphology. After nitrite consumption, active cultures were transferred twice to 5ml tubes (10% inoculum) and lastly into 100 ml Erlenmeyer flasks with 75 ml NOB medium. Additional attempts to eliminate accompanying heterotrophic bacteria were serial tenfold dilutions for cultures Z2.3.3 and A4.5.2 in three to four parallels in mineral NOB medium containing 0.5 mM NaNO2, 0.5 mM NH4Cl and 0.5 mM NaHCO3, and incubated at 37°C. All investigations of Nitrolancea were done on enrichment cultures.
Purity Tests
The purity of the cultures (in terms of absence of heterotrophs) was tested repeatedly by incubating culture aliquots in complex liquid medium (0.5 g l–1 bactopeptone, 0.5 g l–1 yeast extract, 0.5 g l–1 meat extract, 0.584 g l–1 NaCl in distilled water, pH 7.4), and on solid RT (2.5 g l–1 meat extract, 2.5 g l–1 casamino acids, 0.5 g l–1 yeast extract, 1.0 g l–1 KH2PO4, 0.5 g l–1 NaCl, pH 7.4, 15.0 g l–1 agar in distilled water), R2A (half-strength; 0.25 g l–1 yeast extract, 0.25 g l–1 proteose peptone, 0.25 g l–1 casamino acids, 0.25 g l–1 glucose, 0.25 g l–1 soluble starch, 0.25 g l–1 Na-pyruvate, 0.15 g l–1 K2HPO4, 0.025 g l–1 MgSO4 × 7 H2O, pH 7.4, 15.0 g l–1 agar in distilled water) or mixotrophic NOB medium (0.2 g l–1 NaNO2, 0.15 g l–1 yeast extract, 0.15 g l–1 bactopeptone, 0.055 g l–1 sodium pyruvate, trace elements [see mineral medium above], pH 8.0, 13 g l–1 agarose in distilled water).
Physiological Experiments
Growth of Nitrolancea was investigated in batch cultures, using 100 ml mineral medium in 300 ml Erlenmeyer flasks under static conditions at 37°C. Cultures were inoculated with 1% (v/v). To test for nitrite consumption as indication for growth, the Griess-Ilosvay spot test was used (Schmidt and Belser, 1994) or nitrite and nitrate were determined qualitatively with analytical test stripes (Merck KGaA, Germany). Nitrite concentrations for physiological experiments were measured by the HPLC technique.
Chemical Analyses
Nitrite and nitrate were determined quantitatively by HPLC via ion pair chromatography on a LiChrospher RP-18 column (125 × 4 mm; Merck) with UV detection in an automated system (Hitachi LaChrom Elite; VWR International GmbH, Darmstadt, Germany). Data acquisition and processing of nitrite and nitrate concentrations was performed with the integrated software EZChrom Elite 3.3.2.
Growth Characteristics
Nitrite consumption, nitrate production, cell numbers and protein content of cultures of A4.5.2 and Z2.3.4 were determined to calculate generation times (d–1) as well as protein and cell yield per mmol nitrite. Cells were counted using a Thoma cell chamber (0.02 mm depth), and protein content was quantified by fluorescence using the NanoOrange protein quantitation kit (Thermo Fisher Scientific, Waltham, MA, United States).
Activity Measurements and Oxidation Kinetics
Nitrite-dependent oxygen consumption of cells from culture of A4.5.2 (1 replicate) and culture Z2.3.4 (2 biological replicates) was measured following Nowka et al. (2015), using a micro-respiration system (Unisense AS, Denmark) consisting of a 1-channel oxygen sensor amplifier (OXY-Meter), a Clark-type oxygen microsensor (OX-MR), 2 ml glass chambers with glass stoppers, glass coated magnets and a stirring system. Cells were grown at 37°C in mineral medium as described above, with 10 mM and 35 mM (A4.5.2) or 5 mM and 15 mM (Z2.3.4) ammonium and nitrite, respectively. 12–48 h after nitrite was depleted, cells were added to the chambers, which were sealed and mounted on a stirring rack in a water bath at 37°C. The oxygen microsensor was inserted through a capillary hole inside the glass stopper. After an initial equilibration period of 15–30 min, the measurements were started by adding nitrite from stock solutions through a second capillary hole using a syringe. Nitrite oxidation kinetics were estimated from oxygen consumption rates at varying defined nitrite concentrations using the R-package “drc” (Ritz et al., 2015), which fits Michaelis-Menten kinetic to the data with the following equation: V = (Vmax × [S])/(Km + [S]), where V is activity, Vmax is maximum specific activity (μmol mg protein–1 h–1), Km is the half-saturation constant for nitrite oxidation (μM), and [S] is concentration of nitrite (μM). For cell specific Vmax, the calculated maximum activity was translated to fmol NO2– cell–1 h–1.
Microscopic Investigations
For electron microscopy, cells were collected by centrifugation (13,000 × g for 15 min), fixed with 2.5% (v/v) glutaraldehyde as well as 2% (w/v) osmium tetroxide, and embedded in a mixture of Spurr and acetone as described previously (Spieck and Lipski, 2011). Thin sections were stained with 2% (w/v) uranyl acetate and 2% (w/v) lead citrate. Microscopic examination was carried out with a transmission electron microscope (Zeiss model Leo 906E with a CCD camera model 794). For the visualization of whole cells, 3 μl concentrated biomass was pipetted on EM grids (300 mesh, Stork Veco B.V, Netherlands) and stained with 2% (w/v) uranyl acetate. 4′-6′-diamidino-2-phenylindole (DAPI) stained cells were observed using a confocal laser scanning microscope LSM 800 (Zeiss, Jena, Germany) and Plan-Apochromat 63x and 100 × 1.4 oil objectives.
Fluorescence in situ Hybridization
Cells were pelleted at 10°C and 13,000 × g for 15 min, washed with 0.9% (w/v) NaCl and fixed with 1:1 (v/v) 96% ethanol:PBS as described previously (Amann et al., 1995). After dehydration in 50, 80 and 96% ethanol for 3 min each (Manz et al., 1992), cells were hybridized overnight in hybridization buffer containing 20% formamide with the FITC-labeled universal bacterial probe EUB338 (Amann et al., 1990) and the Cy3-labeled probe Ntlc804 specific for Nitrolancea (Sorokin et al., 2012). Cells were embedded in Citifluor AF1 (Citifluor Ltd., London, United Kingdom) prior to microscopic observation at the LSM 800 (Zeiss). Signal specificity was ensured by performing negative control hybridizations using the NON-338 probe labeled in Cy3 (Wallner et al., 1993).
DNA Extraction
DNA for PCR and Illumina sequencing was extracted with the PowerSoil DNA isolation Kit (MO BIO Laboratories, Inc., Carlsbad, CA, United States) according to manufacturer’s instructions with slight modifications except for cultures E2 and A4.1: Before cell disruption via vortexing (step 5), Proteinase K (1 mg ml–1), lysozyme (4 mg ml–1) and RNase A (1 mg ml–1) were added. The vortexing step was extended to 30 min at 37°C.
PCR
The presence of Nitrolancea was tested for with specific primer sets for 16S rRNA and nxrA genes (Supplementary Table S3) and subsequent gel electrophoresis. Presence of NOB belonging to Nitrospira was checked using the nxrB-targeted primer set NxrB-169f/NxrB-638r (Pester et al., 2014). The following PCR program was used: initial denaturation at 94°C, 5 min; 32 cycles consisting of 30 s denaturation at 94°C, 30 s annealing at 55°C (16S rRNA gene), 60°C (nxrA) or 56°C (nxrB) and 3.5 min (for Nitrolancea primers) or 45 s (Nitrospira primers) elongation at 72°C; final elongation at 72°C, 20 min.
Amplicon Sequencing
For selected cultures, 10 ng of genomic DNA were used for 16S rRNA gene amplicon sequencing at MR DNA (Molecular Research LP, Shallowater, TX, United States). For the cultures E2 and A4.1, amplicons were generated using the primers 341F and 785R (Herlemann et al., 2011), followed by 454 pyrosequencing (300 bp, 3000 reads per sample). For all other cultures the primers 515F and 806R (Caporaso et al., 2011) were used with subsequent Illumina MiSeq sequencing (2 × 300 bp, 20000 reads per sample). Operational taxonomic units (OTUs) were obtained from the preprocessed sequences using the de novo OTU picking workflow in qiime (v1.9.1) (Caporaso et al., 2010), and summarized and visualized by the QIIME summarize_taxa_through_plots.py script. Illumina sequences were processed, classified and summarized by MR DNA (Dowd et al., 2008). In short, paired-end sequences were joined and depleted of barcodes, followed by removal of sequences <150 bp or with ambiguous base calls. Sequences were denoised, OTUs generated and chimeras removed. OTUs were defined by clustering at 3% divergence (97% similarity). Final OTUs were taxonomically classified using BLASTn against a curated database derived from NCBI and RDPII.1,2
Metagenomic Sequencing
DNA was fragmented using a Bioruptor (Diagenode, Seraing, Belgium). Libraries for Illumina HiSeq sequencing were generated using the NEBNext Ultra DNA Library Prep Kit for Illumina (New England Biolabs, Ipswich, MA, United States) as per manufacturer’s recommendations. Size and quality of the libraries were visualized on a Bioanalyzer High Sensitivity Chip (Agilent Technologies, Santa Clara, CA, United States). Diluted libraries were multiplex sequenced on the Illumina HiSeq 2500 instrument (Illumina, St. Diego, CA, United States) by paired-end sequencing (2 × 100 bp [metagenomes 40 and 51] or 2 × 250 bp [metagenome 35] on the Illumina MiSeq platform (Illumina, St. Diego, CA, United States). For sequencing of the metagenomes A2, Z2, and Z4, library preparation was done using the Nextera XT kit (Illumina, San Diego, CA, United States) according the manufacturer’s instructions. Tagmentation was performed starting with 1 ng of DNA, followed by incorporation of the indexed adapters and amplification of the library. After purification of the amplified library using AMPure XP beads (Beckman Coulter, Indianapolis, IA, United States), libraries were checked for quality and size distribution using the Agilent 2100 Bioanalyzer and the High sensitivity DNA kit. Quantitation of the library was performed by Qubit with the dsDNA HS Assay Kit (Thermo Fisher Scientific Inc., Waltham, MA, United States). The libraries were pooled, denatured and sequenced on a Illumina MiSeq (San Diego, CA, United States). Paired end sequencing of 2 × 300 bp was performed using MiSeq Reagent Kit v3 (San Diego, CA, United States) according to the manufacturer’s protocol.
Metagenome Assembly and Binning
Adapter removal, contaminant filtering and quality trimming of the Illumina HiSeq and MiSeq paired-end sequencing reads was performed using BBDUK (BBTOOLS version 37.76). Terminal base calls with a quality score < Q17 were trimmed and the reads with a mean quality score of ≤Q20 were discarded. The processed HiSeq reads with a length of ≥70 bp were then corrected for sequencing errors using BayesHammer (Nikolenko et al., 2013). Additional filtering parameters were applied to the MiSeq reads (k = 23, mink = 11, hdist = 1, ktrim = r, tbo = t, maxns = 0, tossjunk = t) and only reads with a length of ≥150 bp were kept. The corrected reads were assembled using metaSPAdes v3.11.1 (Nurk et al., 2017) with default settings. For the samples sequenced with HiSeq, the reads for all samples were co-assembled with iterative assemblies using kmer sizes of 21, 33, 55, 77, 99, and 127, whereas for the MiSeq reads, the assemblies were performed separately for each sample with kmer sizes of 21, 33, and 55. Trimmed HiSeq reads were mapped back to the metagenome separately for each sample using Burrows-Wheeler Aligner (BWA v0.7.17) (Li and Durbin, 2009), employing the “mem” algorithm. For samples sequenced with MiSeq, reads were mapped back to the metagenomes using the minimap2 (v2.16-r922) preset for genomic short-read mapping (Li, 2018). The sequence mapping files were handled and converted as needed using SAMtools 1.6 (Li et al., 2009). The MiSeq metagenomes were manually binned using anvi’o (v5, Eren et al., 2015) and taxonomically classified using the ‘classify workflow’ of GTDB-Tk (version 0.3.2, Parks et al., 2018). Subsequently, the Nitrolancea-like bins were annotated using Prokka (version 1.12-beta, Seemann, 2014) with the RefSeq bacterial nr protein database as a reference and the Kyoto Encyclopedia of Genes and Genomes (KEGG). The completeness and redundancy of the bins was assessed using the checkM ‘lineage’ workflow (Parks et al., 2015).
Phylogenetic Analyses
The average nucleotide identity (ANI) between the recovered Nitrolancea genomes and the genome of Nl. hollandica was calculated using orthoANI (Lee et al., 2016).
Sequences (16S rRNA and nxrA) were aligned using the multiple sequence alignment web server T-Coffee (Notredame et al., 2000). Subsequently, maximum-likelihood trees were calculated using Mega-X (Kumar et al., 2018) with 1000 bootstraps.
Genome Comparison
The orthologous gene clusters were identified using the OrthoVenn 2 web service tool (Xu et al., 2019) with an E-Value of 1e-2 and an inflation value of 1.5.
Results
Selective Enrichment of Nitrolancea
The enrichment process of Nitrolancea from WWTP centrate and geothermal mud pool samples was monitored by specific PCR, fluorescence in situ hybridization (FISH; Supplementary Table S3), and 16S rRNA gene amplicon and metagenomic sequencing. Additional details on the enrichment process can be found in the Supplementary Material.
As shown by amplicon sequencing, the microbial community of the original centrate treatment reactor sample Z2 was dominated by Proteobacteria (56%), Bacteriodetes (22%) and Firmicutes (10%). Chloroflexi were present at low amounts (2%; Supplementary Figure S2), but none of these sequences were affiliated with the genus Nitrolancea. Members of the nitrifying genera Nitrosomonas and Nitrosovibrio were detected (2.1 and 0.7%, respectively), but no known nitrite oxidizer could be identified.
Over a period of only 4 months, Nitrolancea could be selectively enriched from 1.4 to 24% (Supplementary Table S2). Different isolation attempts were performed to increase the relative abundance of Nitrolancea-like bacteria in the nitrite-oxidizing cultures. For Z2, incubations with increasing substrate concentrations (up to 70 mM nitrite) revealed a very high enrichment of lancet-shaped cells (Figure 1a) and combination with the use of an optical tweezers system or serial dilution series increased the enrichment further (Figure 1b). Two consecutive dilution series with growth up to the dilution 10–8 and 10–6, respectively, lead to a pure culture of Nitrolancea strain Z based on purity tests using four different complex media.
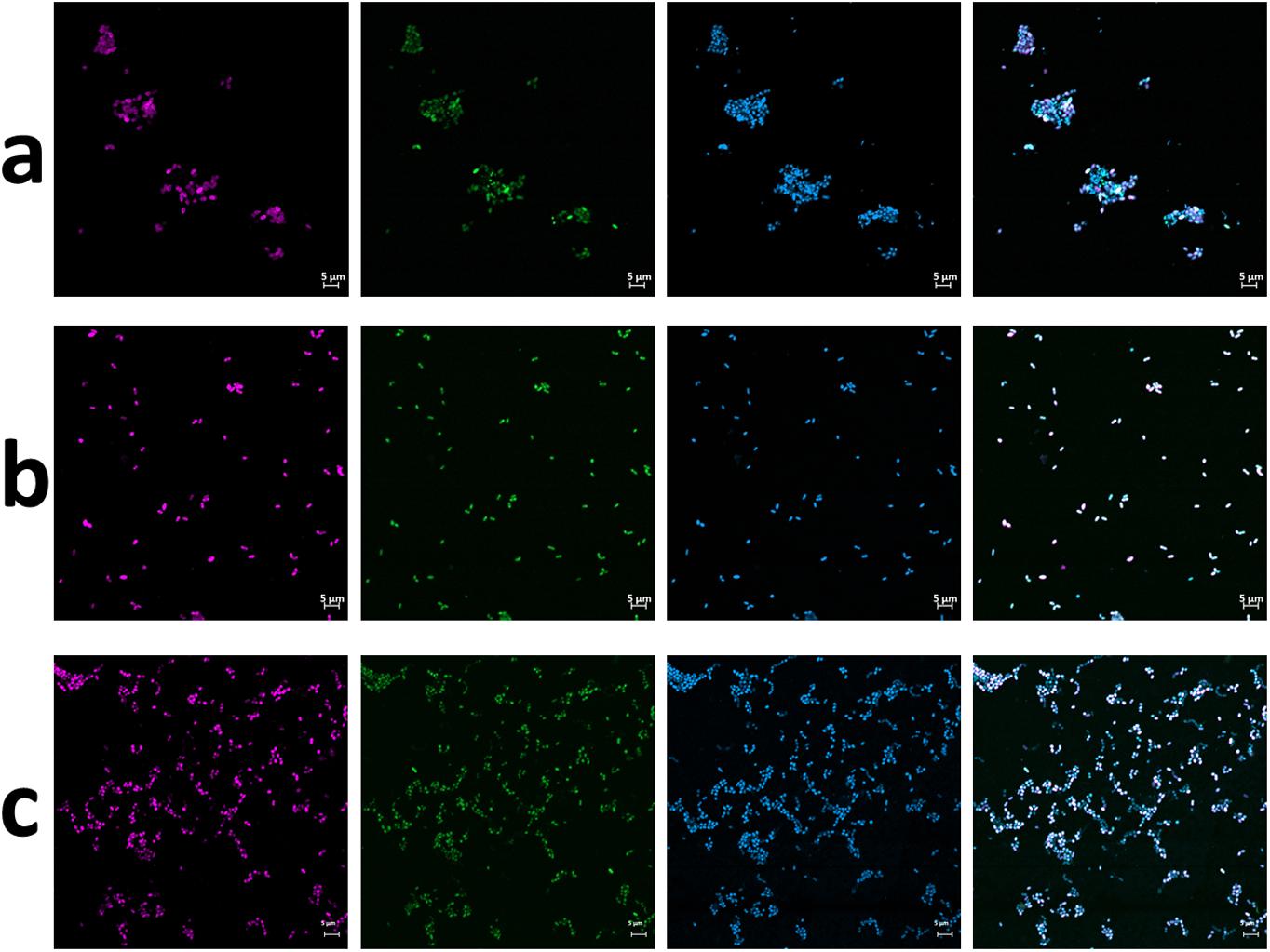
Figure 1. FISH micrographs of culture Z2.3.3 (a) before and (b) after 10−8 dilution and (c) of culture A4.5.2. From left to right: Nitrolancea-like bacteria detected with probe Ntlc804 labeled in Cy3 (red); all bacteria detected with EUB338 labeled in FITC (green); all cells stained with DAPI (blue); overlay of all three channels.
Unlike the centrate treatment reactor sample, the primary nitrite-oxidizing enrichment of Las Máquinas (A4.1) grown at 42°C was dominated by members of the phylum Chloroflexi (45%), followed by Deinococcus-Thermus (28%) and Proteobacteria (22%) according to 16S rRNA amplicon sequencing (Supplementary Figure S2). This enrichment had been supplied with 0.3 mM of nitrite without ammonium addition. Nitrolancea abundances increased to 14.1% after the culture had been repeatedly replenished with nitrite over a period of 4 years and increased further to 44% after using high substrate concentrations (30 mM) and adding ammonium (Supplementary Table S2). Manual sorting of cells from this enrichment with the optical tweezers system generated six follow-up cultures that, however, all still contained several accompanying heterotrophic bacteria (not shown). Meanwhile, the parallel culture A4.5.2, which received 40 mM nitrite in addition to the ammonium and bicarbonate also present in A4.5.1, was nearly pure as shown by inoculation of four different complex media and FISH (Figure 1c) and only tiny colonies grew on half-strength R2A plates. These heterotrophic bacteria were lost upon another serial dilution to the step 10–7 (A4.5.3) and the strain deemed isolated. The novel Nitrolancea strain was provisionally designated as Ca. Nitrolancea copahuensis based on 16S rRNA gene and whole-genome dissimilarity to Nl. hollandica (see below).
Notably, in an ammonia-oxidizing culture that had been inoculated with an earlier sample from Las Maquinas (E2) with 0.5 mM ammonium, but without nitrite addition, Nitrolancea-like NOB were also co-enriched with unknown ammonia-oxidizing microorganisms. This culture was dominated by Chloroflexi (34%), Proteobacteria (40%) and Actinobacteria (21%), while Nitrolancea-like bacteria constituted nearly 20% of the microbial community after 3.5 years of incubation at 42°C. Contrastingly, no Nitrolancea-like sequences could be detected in a parallel nitrite-oxidizing culture of E2, which was grown without ammonium addition at 42°C (not shown).
Regarding other known NOB, Nitrospira were nearly absent in all centrate treatment reactor enrichments and constituted only 0.3% of the retrieved amplicon sequences in the Argentinian cultures A4.2, A4.3 and A4.4 (Supplementary Table S2). Nitrobacter-like 16S rRNA gene sequences amounted to 0.8% in the centrate cultures Z2.1 and Z2.2 (not shown). In the Argentinian A4 cultures, Nitrobacter was undetectable, but constituted a tiny fraction in culture E2 (0.05%).
Morphology and Ultrastructure
Light and transmission electron microscopy of the nitrite-oxidizing enrichment cultures that had been inoculated with biomass from the Hamburg-Dradenau centrate treatment reactor and from Las Máquinas revealed Nitrolancea-like cells with characteristic tapered ends (Figure 2) similar to those of Nl. hollandica (Sorokin et al., 2012), but smaller in size (Table 1). The lancet shape was less pronounced in A4 cultures, where most cells had an ovoid shape (Figure 2F). Nitrolancea-like bacteria in all cultures appeared as single cells, or formed pairs and short chains. In late nitrite-oxidizing cultures, cells of Nitrolancea aggregated to roundish flocs, whereas in primary enrichments of cultures E2 and A4 tiny yellow to orange particles accumulated.
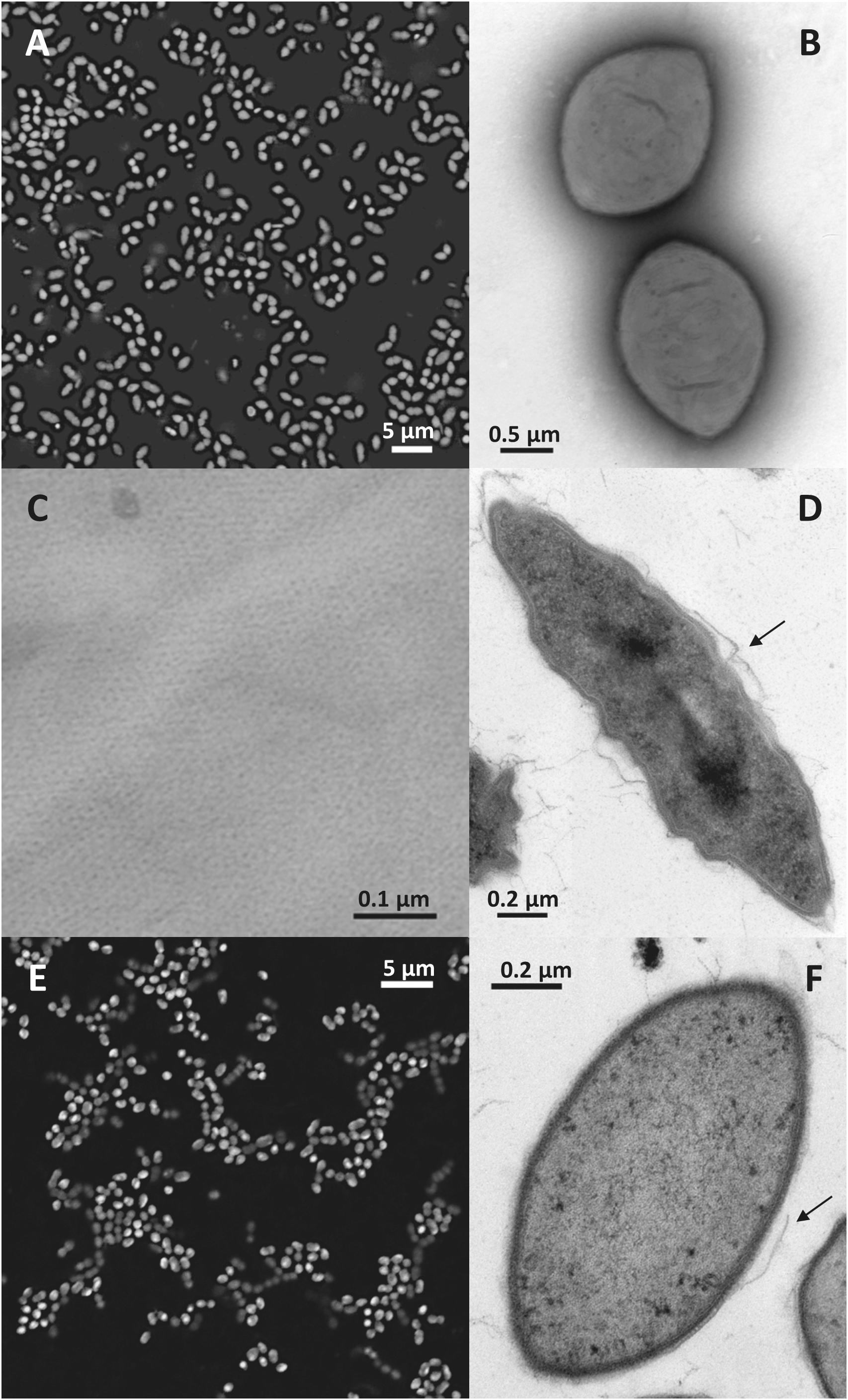
Figure 2. Nitrolancea-like cells in the nitrite-oxidizing enrichments derived from centrate (A–D; culture Z2.3.3) and of Las Máquinas (E,F). (A) Cell shape of Nitrolancea hollandica strain Z stained with DAPI. (B) Negatively stained cells showing the tapered ends in detail, stained with uranyl acetate. (C) Regular protein layer of the cell wall. (D) Electron micrograph of an ultrathin section. (E) Fluorescence micrograph of culture A4.4 stained with DAPI. (F) Ultrathin section of culture A4.3. The arrows indicate where the disrupted external protein layer is detached from the cell wall.
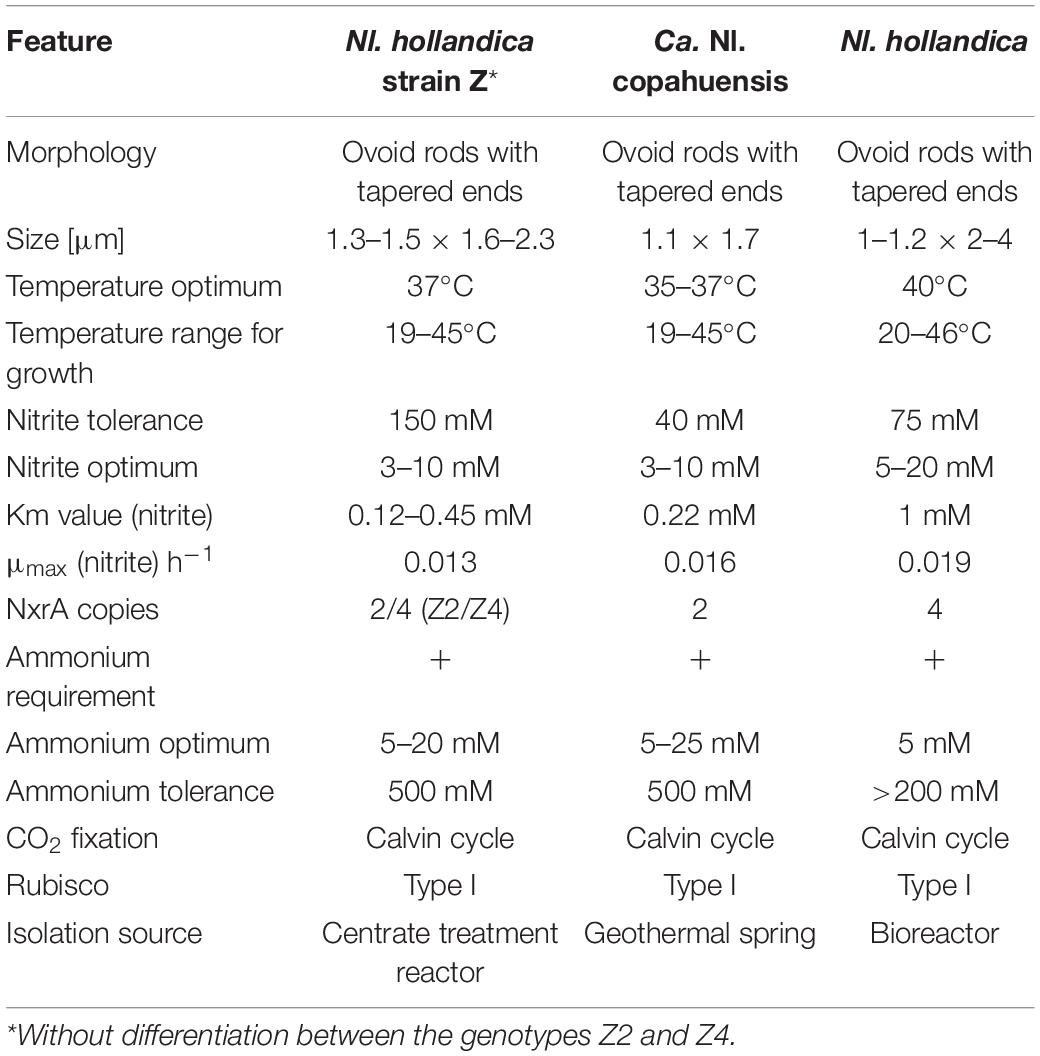
Table 1. Comparison of morphological, physiological and genomic features of the novel Nitrolancea strains and Nl. hollandica.
The Nitrolancea-like cells in both incubation series had the typical Gram-positive type complex cell wall, including a regular proteinaceous surface layer (Figure 2C), which is a common component of archaeal and bacterial cell walls (Rachel et al., 1997). This protein layer is easily disrupted during the embedding process (Figures 2D,F), resulting in loss of the typical cell morphology. As described for Nl. hollandica, the novel Nitrolancea strains did not possess carboxysomes or intracytoplasmic membrane systems. However, in some cells membrane invaginations as described by Sorokin et al. (2014) were observed at the cell poles (not shown).
Physiology
As also known for Nl. hollandica, cells of both Nitrolancea enrichments derived from the samples Z2 and A4 were able to grow lithoautotrophically with nitrite as energy source and CO2 as carbon source. Nitrite was oxidized aerobically and stoichiometrically to nitrate with a minimum generation time of 53 h (culture Z2.3.4; Figure 3A) or 43 h (culture A4.5.2; Figure 4A).
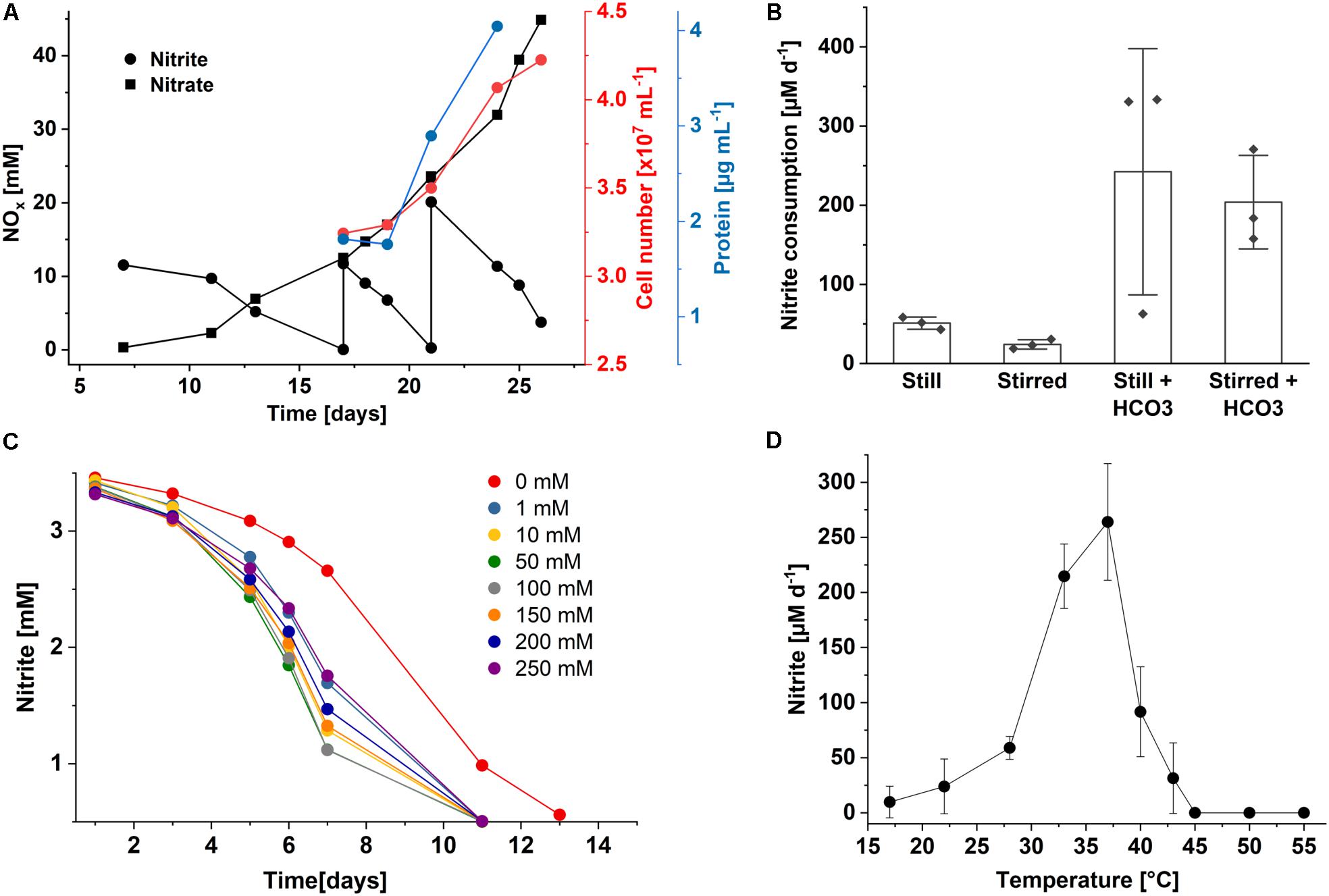
Figure 3. Growth characteristics of Nitrolancea hollandica strain Z. (A) Stoichiometric oxidation of nitrite to nitrate leading to increase in cell number and biomass. (B) Nitrite consumption of enrichment culture Z2.3.3 under different incubation conditions at 37°C: without agitation (still) or stirred at approx. 120 rpm, and with or without the addition of 0.5 mM NaHCO3. Bars represents the mean ± standard error of biological triplicates, diamonds indicate individual data points; (C) nitrite oxidation over time in medium supplemented with 3 mM nitrite and different ammonium concentrations (0 – 250 mM); (D) nitrite oxidation rates per day at different temperatures. The rates were determined for the period between day 3 and 10 after inoculation (2.5 mM initial nitrite concentration). Data represents the mean ± standard error of biological triplicates. For all incubations, medium was inoculated with 1% (v/v) enrichment culture of (A) Z2.3.4, (B,D) Z2.3.3 or (C) Z2.3.1.
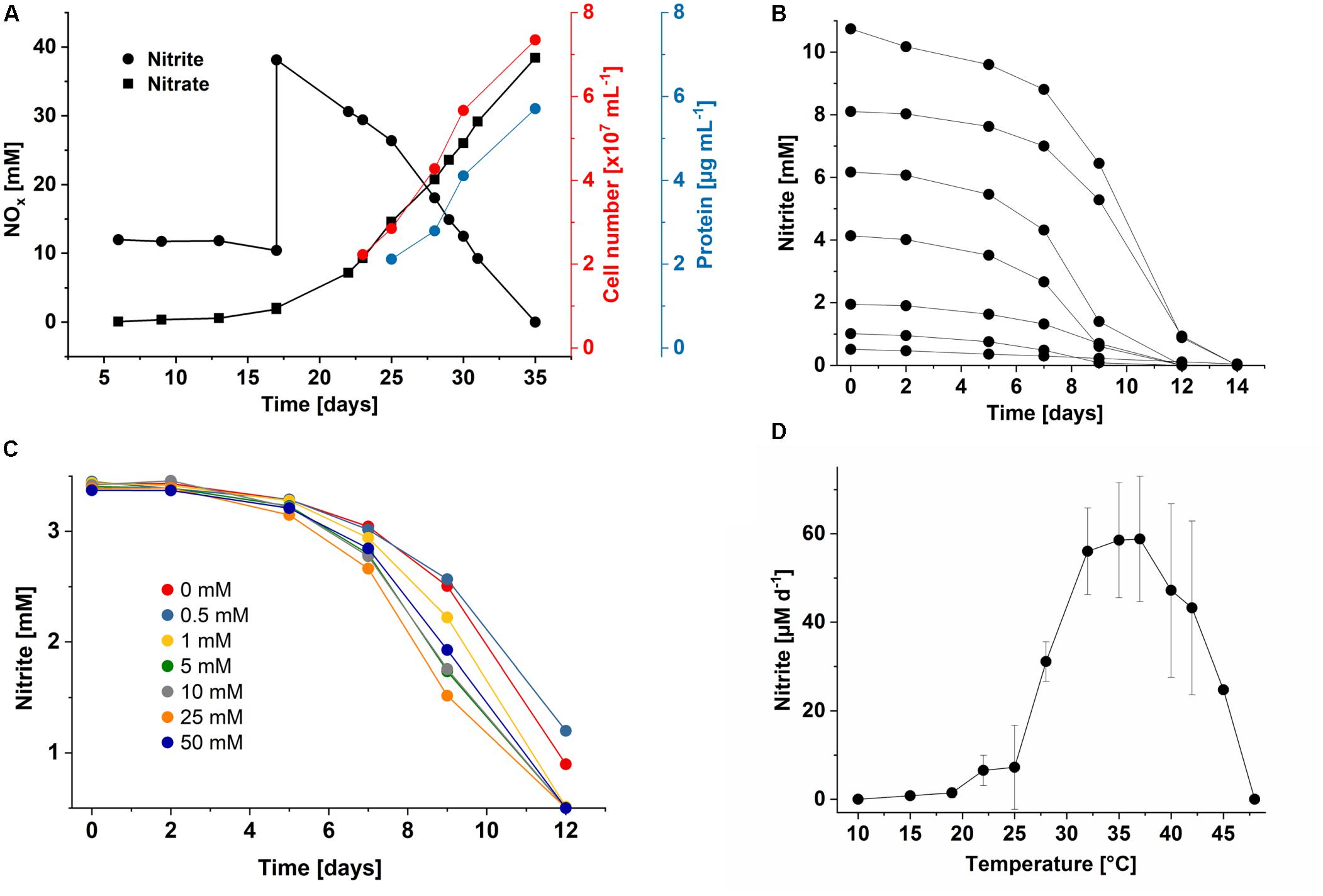
Figure 4. Growth characteristics of Ca. Nitrolancea copahuensis. (A) Stoichiometric oxidation of nitrite to nitrate leading to increase in cell number and biomass. (B) Substrate consumption test with different initial nitrite concentrations in culture A4.4, showing the rapid consumption of nitrite. Reduced nitrite oxidation was still observed at up to 40 mM nitrite (not shown). (C) Nitrite consumption of culture A4.4 in medium supplemented with different ammonium concentrations (0 to 50 mM). (D) Influence of temperature on nitrite oxidation in culture A4.5.1 with 0.5 mM nitrite. Nitrite oxidation rates were determined for the period between day 4 and 11 after inoculation. Data represents the mean ± standard error of biological triplicates. At 42°C, two biological replicates were incubated.
In the beginning of the cultivation process, the Z2.2 culture was tested for its ammonium requirements. Growth was slow in ammonium-free medium and was clearly accelerated by the addition of 0.5 – 3 mM ammonium (not shown). Consequently, all mineral NOB media were prepared with the addition of ammonium. Fascinatingly, NH4Cl concentrations up to 500 mM could be tolerated, although consumption of 3 mM nitrite took 4 months. Addition of 250 mM ammonium caused slight inhibition only (Figure 3C). In accordance with Nl. hollandica, the cultures derived from the centrate were able to grow in the presence of very high nitrite concentrations and completely oxidized up to 150 mM nitrite within several months. Growth occurred between 22 and 43°C and with a delay at 19 and 45°C. Nitrite was oxidized optimally at 37°C (Figure 3D). Additionally, the influence of aeration and bicarbonate supplementation on growth and activity were tested. The highest nitrite oxidation rates were obtained when cells were incubated in Erlenmeyer flasks without agitation and were further accelerated by addition of 0.5 mM NaHCO3 (Figure 3B).
The nitrite oxidation rates of the Nitrolancea cultures obtained from Las Máquinas (A4.4) increased with increasing substrate concentrations (1–10 mM NaNO2) without a prolonged lag-phase (Figure 4B). The nitrite tolerance level was 40 mM. Nitrite was also oxidized slowly in medium without ammonium, but optimal growth depended on the addition of up to 50 mM NH4Cl (Figure 4C). The ammonium tolerance limit was 500 mM. Growth of the Argentinian Nitrolancea strain was observed between 22 and 45°C, delayed at 19°C, and with an optimum growth temperature of 35–37°C (Figure 4D). A comparison of key parameters between Nl. hollandica and the strains investigated here is shown in Table 1.
Nitrite oxidation kinetics were determined by microsensor-based oxygen consumption measurements in the different Nitrolancea cultures (Supplementary Figure S3). Both representatives revealed moderate substrate affinities with apparent half-saturation constants (Km) of 0.12–0.45 mM nitrite for Nl. hollandica strain Z and 0.22 mM for Nl. copahuensis (Supplementary Table S4). The maximum specific activity (Vmax) was relatively low (11.1–26.8 μmol nitrite mg protein–1 h–1). Growth yields ranged from 0.17 to 0.88 mg protein mmol nitrite–1.
Genomic Analyses
Biomass of cultures Z2.2, Z2.3.2, and A4.4 was used for metagenomic sequencing (Table 2). The draft genomes of the three resulting Nitrolancea strains described here (Z2, Z4, A2) could be assembled into 48 to 150 contigs. These high-quality draft genomes ranged in size from 3.19 to 3.45 Mbp and had G+C contents between 62.7 and 62.9 mol%. The number of predicted coding sequences was between 3023 and 3235, including 40 to 46 tRNAs.
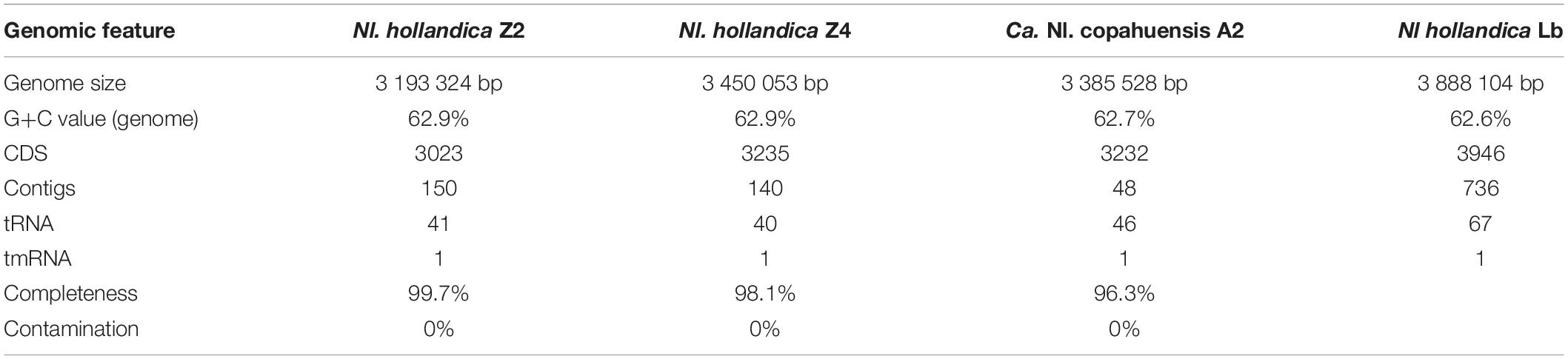
Table 2. Genome characteristics of the four Nitrolancea strains as predicted using PROKKA (Version 1.12).
After assembly and binning of the Nitrolancea strains, comparative genomics revealed that two highly similar strains (Z2 and Z4) were enriched from the centrate treatment reactor sample, while the strain A2 obtained from sample A4 was clearly distinct (see below). The comparison of these metagenome-assembled genomes (MAGs) with Nl. hollandica revealed 2399 conserved clusters out of in total 3387 orthologous gene clusters within the four Nitrolancea strains (Figure 5). The type strain Nl. hollandica exhibits 14 unique gene clusters that were absent in the genomes of the three novel strains, which, on the contrary, shared 131 of orthologous clusters. Additionally, the Argentinian strain A2 contained 6 unique gene clusters, while the two genomes obtained from the centrate enrichment cultures in total had 109 clusters not present in the other Nitrolancea strains.
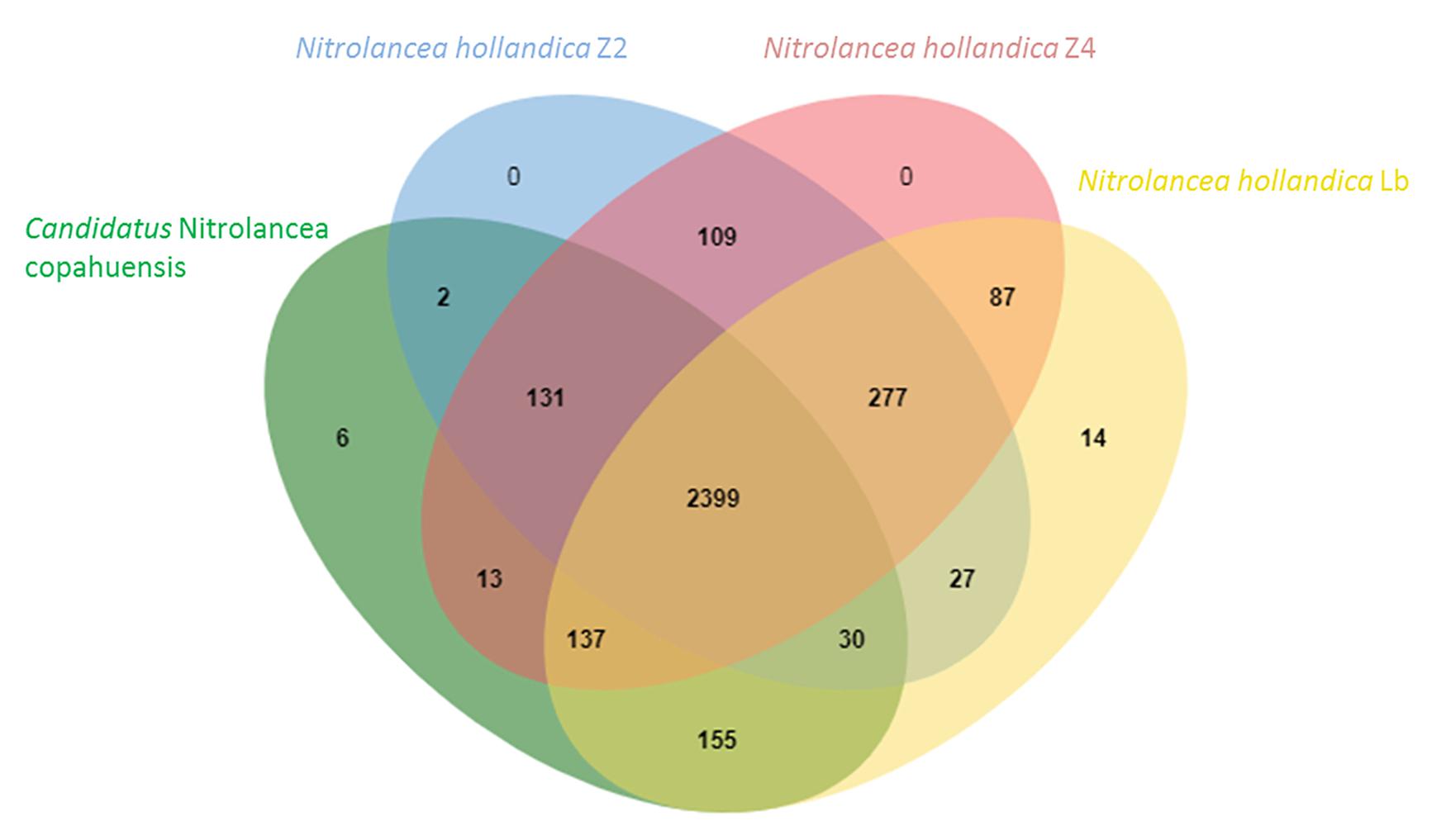
Figure 5. Venn diagram indicating the number of shared orthologous clusters among the genomes of Nitrolancea hollandica and the Nitrolancea strains obtained from the nitrite-oxidizing enrichments A4.4 (A2), Z2.2 (Z2) and Z2.3.2 (Z4). The diagram was plotted by using OrthoVenn2 (Xu et al., 2019).
Phylogenetic and sequence similarity-based analysis of the 16S rRNA genes of the novel Nitrolancea strains (Supplementary Table S5, Figure 6) revealed a close affiliation with the previously described type strain Nl. hollandica (Sorokin et al., 2014). Ca. Nitrolancea copahuensis had a 16S rRNA gene sequence identity of 98.5% to Nl. hollandica, whereas the two centrate strains were almost identical to the type strain (99.9 and 100% identity). Comparison of the NxrA sequences of the novel strains and Nl. hollandica revealed high amino acid identities (Supplementary Table S5, Supplementary Figure S4). Furthermore, the draft genome sequences of the centrate strains and Nl. hollandica had high average nucleotide identity (ANI) values of 99.2 and 99.3%, respectively, which was only 91.9% for the geothermal strain (Supplementary Table S6). The two draft genomes from the centrate enrichments were almost identical and had an ANI value of 99.96%. Thus, based on 16S rRNA gene and ANI similarity, Z2 and Z4 belong to the species Nl. hollandica, while the Argentinian strain A2 had 16S rRNA and ANI identity values below the species threshold of 98.7–99.0% (Stackebrandt and Ebers, 2006) and 95% (Rodriguez-R and Konstantinidis, 2016), respectively. Consequently, this strain represents a separate species within the genus Nitrolancea.
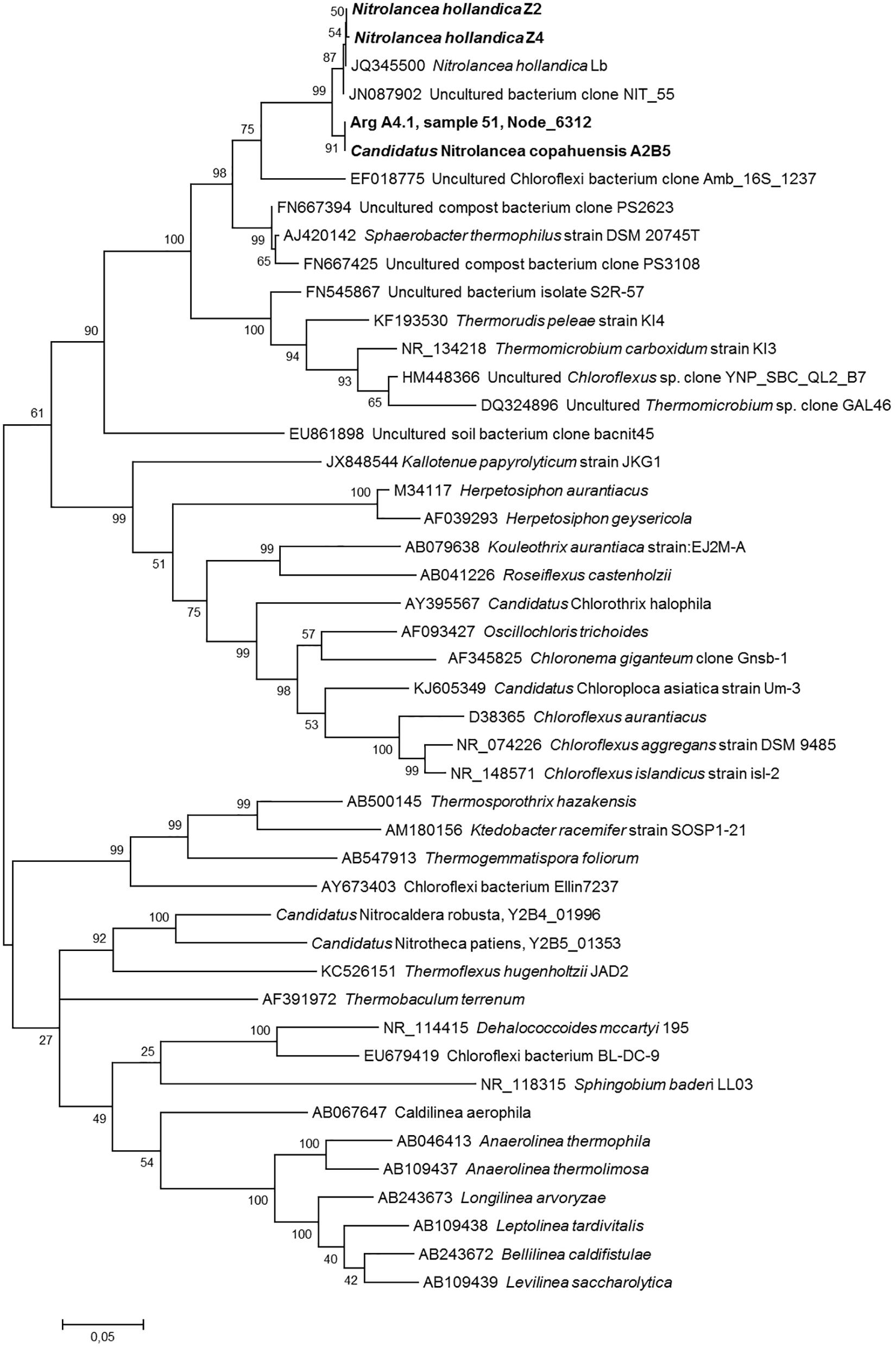
Figure 6. Phylogenetic relationship of the Nitrolancea strains based on 16S rRNA gene sequences. Maximum likelihood tree indicating the affiliation of the 16S rRNA genes identified in the centrate treatment reactor and Argentinian enrichment cultures. Statistical branching support values based on 1000 bootstraps are indicated on the nodes. The alignment contained 1703 valid positions used for tree calculation. Sequences obtained in this study are shown in bold. The scale bar represents the expected changes per nucleotide. For designation of cultures see Supplementary Table S2.
The genomic data was furthermore mined to reconstruct the core metabolic pathways of the three novel Nitrolancea strains (Table 3). Genes encoding all proteins of the Calvin cycle for CO2 fixation, but not for carboxysomes have been identified. The potential for nitrite and formate oxidation was also present and the genomes contained additional non-operonal nxrA copies, as has been observed for Nl. hollandica (Sorokin et al., 2012). The A2 and Z2 genomes contained only one orphan nxrA gene, while the Z4 genome encoded 3 additional NxrA copies. All other NXR subunits were not duplicated and encoded within a highly conserved operon (Supplementary Figure S5). Furthermore, genes for a NO-forming nitrite reductase (narK) and the cytoplasmic assimilatory nitrate reductase (nasAB) were detected in the three analyzed genomes, but not for assimilatory nitrite reductases (nirA or nirBD) or dissimilatory nitrite reduction to ammonium (nrfA).
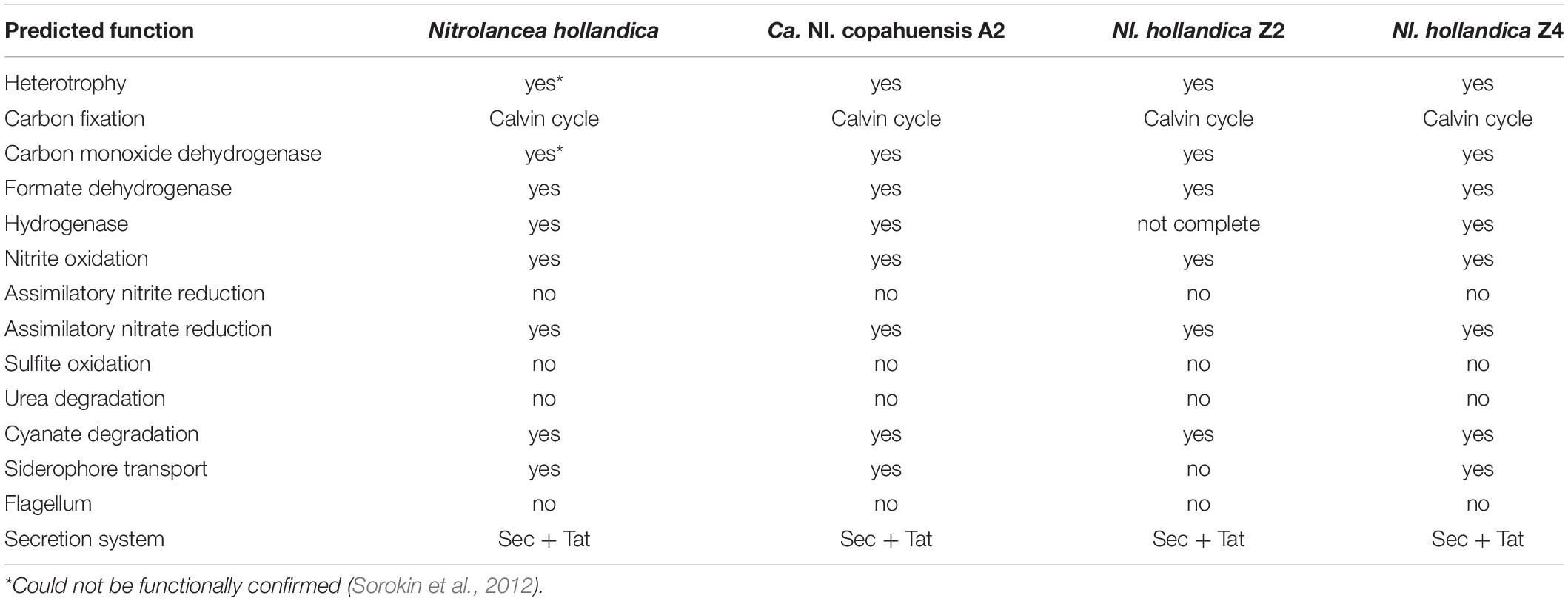
Table 3. Comparison of key functions predicted for the four Nitrolancea strains using PROKKA (Version 1.12) and the Kyoto Encyclopedia of Genes and Genomes (KEGG).
Additionally, all Nitrolancea genomes encoded an aerobic-type CO dehydrogenase, cyanase and an [NiFe]-hydrogenase. Based on the genomic data, all strains have the potential to grow heterotrophically and to use thiosulfate for energy conservation. So far, none of these phenotypes could be observed in growth experiments with Nl. hollandica (Sorokin et al., 2012). Within the sulfur metabolism, the assimilatory sulfate reduction pathway involving sulfate adenylyltransferase (sat), adenylyl-sulfate kinase (cysC), phosphoadenosine phosphosulfate reductase (cysH) and ferredoxin-dependent sulfite reductase (sir) could be identified in the three analyzed genomes, along with genes encoding thiosulfate sulfurtransferases (glpE, sseA) and a putative sulfite oxidase (sorA).
Discussion
The first isolate of Nitrolancea was only described in 2012 (Sorokin et al., 2012). Growth depended on the use of a modified NOB medium (Spieck and Lipski, 2011), as Nl. hollandica grew optimally in mineral medium containing high substrate concentrations of 20 to 50 mM KNO2 and supplementation of 5 mM NH4HCO3. Unexpectedly for a nitrite-oxidizing organism, Nl. hollandica depends on ammonium as N-source and accordingly its genome did not encode any enzymes for nitrite reduction to ammonium (Sorokin et al., 2012). This absence of assimilatory nitrite reduction pathways was recently also observed in other thermophilic NOB within the phylum Chloroflexi (Spieck et al., 2020), and some Nitrotoga species are apparently dependent on ammonium addition, too (Ishii et al., 2017; Wegen et al., 2019). In contrast, the early Nitrolancea enrichments obtained in this study were able to oxidize nitrite also without ammonium addition, although no assimilatory nitrite reductase genes were detected in their genomes. However, it is likely that these Nitrolancea strains were provided with ammonium or organic N-sources by the accompanying heterotrophs present in the cultures, as the ability for growth without ammonium addition was lost upon increased purity of the cultures.
The optimal growth conditions for the novel Nitrolancea strains were assessed by testing different incubation temperatures, as well as nitrite and ammonium concentrations. Although Nitrolancea has been described as NOB, that requires high substrate concentrations, early enrichments from Las Máquinas were done at very low nitrite availability (Supplementary Table S2). Nevertheless, once an active culture was obtained, an increase of substrate concentrations was observed to be beneficial for the selective enrichment of Nitrolancea-like NOB. High ammonium concentrations can also enhance proliferation of Nitrolancea, as was observed in culture A4.4. However, we cannot exclude that this effect was caused by growth stimulation of accompanying bacteria. After initial cultivation at 42°C, reducing the incubation temperatures to the apparent optimum of 37°C led to a reliable enrichment of Nitrolancea-like cells from both sample locations (Supplementary Table S2). Intriguingly, this cultivation strategy resulted in the successful enrichment of two novel Nl. hollandica strains from the centrate treatment reactor, even though no Nitrolancea-like 16S rRNA gene sequences were detected in the original sample. This finding indicates that these NOB are more widely distributed than previously assumed, albeit at very low numbers. The minimum temperature for growth for Nitrolancea derived from the centrate reactor as well as from the geothermal mud pool was 19°C, but related 16S rRNA gene sequences were also found in a bioreactor system operated at 12°C (Yu et al., 2018). Therefore, the environmental distribution of these NOB seems not to be restricted to thermophilic conditions.
Fast and reproducible initial enrichment of Nitrolancea spp. was achieved by designing a standard medium consisting of a mineral salt solution supplemented with 3 mM NaNO2, 3 mM NH4Cl and, as observed in later cultivation approaches, 0.5 mM NaHCO3. The positive effect of bicarbonate probably is due to resulting elevated CO2 concentrations within the cell, which might reduce the reaction of ribulose 1,5-bisphosphate carboxylase/oxygenase (RuBisCO) with the competing substrate O2 and thus increase carbon fixation efficiency in the absence of other CO2-concentrating mechanisms like carboxysomes (Price et al., 2008). In this study, a preference for a reduced oxygen supply (Figure 3B) of N. hollandica strain Z was found and culturing was done without agitation. Accordingly, Nitrolancea-like sequences have been detected in a reactor treating synthetic low-strength wastewater at low dissolved oxygen contents (Yu et al., 2020).
The proliferation of the different Nitrolancea strains at nitrite concentrations between 3 and 40 and even 70 mM, respectively, matches the extremely high half-saturation constant (Km) for nitrite determined for Nl. hollandica (Sorokin et al., 2012). The Km of this strain was found to be 1 mM nitrite, which corresponds to the lowest substrate affinity of any NOB determined so far and is in line with the high substrate concentrations present in the bioreactor Nl. hollandica was isolated from. The Nitrolancea cultures investigated here revealed a higher affinity for nitrite, although there was a great variance between values for strain Z and only one measurement was done for Nl. copahuensis. Nevertheless, our preliminary results suggest that the Km value of Nitrolancea is in the range of the different Nitrobacter species (Nowka et al., 2015). This higher substrate affinity fits with the presence of Nitrolancea in the geothermal area of Las Máquinas, where nitrite is rarely detected as usual for most natural systems. Nevertheless, high substrate concentrations are necessary for optimal growth of Nitrolancea and nitrite was oxidized slowly when present in low concentrations (Figures 4B,D).
Comparing the genomes and NxrA gene sequences from the different enrichments, it has become obvious that distinct representatives of Nitrolancea grew depending on the respective medium (for instance 3 mM vs. 60 mM nitrite for strains Z2 and Z4). Similarly, the representatives of Nitrolancea enriched from the Las Máquinas samples differed in the initial enrichments E2 and A4.4 in AOB and NOB medium, respectively (Supplementary Figure S4). The nxrA gene within the metagenome 40 of the culture E2 is non-operonal, whereas in close vicinity to the nxrA gene of the metagenome 35 a cytochrome c was found. Therefore, the initial samples must have contained a microdiversity of Nitrolancea strains, which selectively proliferate once their special demands were met.
Nitrolancea-like 16S rRNA gene sequences have only occasionally been detected in full-scale nitrifying activated sludge systems so far (Speirs et al., 2019). The Nitrolancea-typical marker fatty acid 12-methyl octadecanoic acid (18:0 12methyl) (Sorokin et al., 2012) was measured in low amounts (0.2% of total fatty acids) in activated sludge of the Hamburg-Dradenau WWTP by Kruse et al. (2013). The abundance of 12-methyl octadecanoic acid increased slightly to 0.7% after incubations with nitrite at 28°C and 32°C, which confirms the preference of Nitrolancea for elevated temperatures. However, similar glycolipids are also characteristic for other thermophilic Chloroflexi, like for instance Thermomicrobium, which has also been detected in WWTPs based on 16S rRNA gene sequences (Spieck et al., 2020). Therefore, it is still speculative if Nitrolancea also occurs in activated sludge, which was used as inoculum for the centrate reactor.
The geothermal area in Las Máquinas apparently offers beneficial conditions for Nitrolancea-like NOB, as revealed by our 16S rRNA amplicon data (Supplementary Figure S2, Supplementary Table S2) although closely related sequences have not been detected in this area previously (Urbieta et al., 2015a). Elevated ammonium or nitrite concentrations could not be measured at the sampling sites, thus posing the question if alternative substrates might have enabled proliferation of Nitrolancea. Some NOB are metabolically versatile and can use simple organic carbon compounds like formate, hydrogen, and even ammonia as additional energy sources (Koch et al., 2014, 2015; Daims et al., 2015; van Kessel et al., 2015). Furthermore, different NOB possess the genetic potential to use various sulfur compounds (Starkenburg et al., 2008; Lücker et al., 2013; Boddicker and Mosier, 2018; Kitzinger et al., 2018; Palomo et al., 2018) and the oxidation of sulfide has been demonstrated for Nitrococcus mobilis (Füssel et al., 2017). In the Las Máquinas area, a highly active sulfur cycle is present (Giaveno et al., 2013; Urbieta et al., 2014) and the ability of Nitrolancea species to oxidize thiosulfate should be tested in further studies. The ability for cyanate degradation might provide an alternative source to generate ammonia for N-assimilation.
In conclusion, we physiologically and genomically characterized three novel representatives affiliated with the genus Nitrolancea, which we enriched from a centrate treatment reactor at the WWTP Hamburg-Dradenau and the geothermal area Las Máquinas in Neuquen-Argentina. One of these strains represents a new species within the genus Nitrolancea. We provisionally refer to this strain as, Candidatus Nitrolancea copahuensis‘ (co.pa.hu.en’sis. N.L.fem.adj. copahuensis pertaining to the Copahue region in North-Patagonia, Argentina).
The strain is a mesophilic, aerobic, chemolithoautotrophic bacterium, which stoichiometrically converts nitrite to nitrate and uses carbon dioxide as carbon source. Forms short rods with tapered ends; Gram-positive. Requires ammonium supplementation and tolerates high nitrite and ammonium concentrations. Optimum growth temperature is 35–37°C. DNA G+C content is 62.7 mol%. Originated from a hot spring in the geothermal field Las Máquinas, Neuquen-Argentina.
Data Availability Statement
The datasets generated for this study are available in the European Nucleotide Archive (ENA) under accession numbers ERS4399870, ERS4399871, ERS4399872, ERS4647270-74, ERS4648393-96, and LR794103-5. Strain availability will be performed upon request.
Author Contributions
AG performed the sampling. ES, SK, and SL designed the research. SH worked on the Nitrolancea enrichments. ES and KS isolated the nitrite oxidizers and performed FISH. SK did physiological experiments. DI performed genome sequencing. MS, LK, and SL analyzed the data. ES, KS, SK, LK, AG, and SL wrote the manuscript. All authors read and agreed on the final manuscript.
Funding
This work was funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation grant SP 667/7-1+2, grant SP 667/10-1+2, and grant SP 667/11-1) and Netherlands Organization for Scientific Research (Grants 016.Vidi.189.050 and SIAM Gravitation Grant 024.002.002).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the Government of the Province of Neuquen for allowing sampling in the protected natural area Copahue geothermal park and Edgardo Donati and Wolfgang Sand for providing samples from Las Máquinas. HSE is appreciated for the support in getting samples from Hamburg-Dradenau. Elke Woelken is acknowledged for excellent technical help in electron microscopy and Theo van Alen for sequencing. We are grateful that Katja Wendt, Kristina Detzel, Katrin Petersen as well as Laura Nissen did initial cultivation and laboratory work. We also thank Malik Alawi, Lia Burkhardt, and Kerstin Reumann for sequencing assistance.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.01522/full#supplementary-material
Footnotes
References
Alawi, M., Lipski, A., Sanders, T., and Eva-Maria-Pfeiffer Spieck, E. (2007). Cultivation of a novel cold-adapted nitrite oxidizing betaproteobacterium from the Siberian Arctic. ISME J. 1, 256–264. doi: 10.1038/ismej.2007.34
Amann, R. I., Krumholz, L., and Stahl, D. A. (1990). Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J. Bacteriol. 172, 762–770. doi: 10.1128/jb.172.2.762-770.1990
Amann, R. I., Ludwig, W., and Schleifer, K. H. (1995). Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59, 143–169. doi: 10.1128/mmbr.59.1.143-169.1995
Andrews, J. H., and Harris, R. F. (1986). “r- and K-selection and microbial ecology,” in Advances in Microbial Ecology, ed. K. C. Marshall, (Boston, MA: Springer US), 99–147. doi: 10.1007/978-1-4757-0611-6_3
APHA, AWWA, and WPCF. (1992). Métodos Normalizados Para el Análisis de Aguas Potables y Residuales. Madrid: Díaz de Santos, 1715.
Boddicker, A. M., and Mosier, A. C. (2018). Genomic profiling of four cultivated Candidatus Nitrotoga spp. predicts broad metabolic potential and environmental distribution. ISME J. 12, 2864–2882. doi: 10.1038/s41396-018-0240-248
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., and Costello, E. K. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Met. 7, 335–336. doi: 10.1038/nmeth.f.303
Caporaso, J. G., Lauber, C. L., Walters, W. A., Berg-Lyons, D., Lozupone, C. A., and Turnbaugh, P. J. (2011). Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. U.S.A. 108(Suppl. 1), 4516–4522. doi: 10.1073/pnas.1000080107
Chiacchiarini, P., Lavalle, L., Giaveno, A., and Donati, E. (2010). First assessment of acidophilic microorganisms from geothermal Copahue-Caviahue system. Hydrometallurgy 104, 334–341. doi: 10.1016/j.hydromet.2010.02.020
Chiodini, G., Cardellini, C., Lamberti, M. C., Agusto, M., Caselli, A., Liccioli, C., et al. (2015). Carbon dioxide diffuse emission and thermal energy release from hydrothermal systems at Copahue-Caviahue volcanic complex (Argentina). J. Volcanol. Geoth. Res. 304, 294–303. doi: 10.1016/j.jvolgeores.2015.09.007
Courtens, E. N., Spieck, E., Vilchez-Vargas, R., Bodé, S., Boeckx, P., Schouten, S., et al. (2016). A robust nitrifying community in a bioreactor at 50°C opens up the path for thermophilic nitrogen removal. ISME J. 10, 2293–2303. doi: 10.1038/ismej.2016.8
Daims, H., Lebedeva, E. V., Pjevac, P., Han, P., Herbold, C., Albertsen, M., et al. (2015). Complete nitrification by Nitrospira bacteria. Nature 528, 504–509. doi: 10.1038/nature16461
Dowd, S. E., Sun, Y., Secor, P. R., Rhoads, D. D., Wolcott, B. M., James, G. A., et al. (2008). Survey of bacterial diversity in chronic wounds using pyrosequencing, DGGE, and full ribosome shotgun sequencing. BMC Microbiol. 8:43. doi: 10.1186/1471-2180-8-43
Edwards, T. A., Calica, N. A., Huang, D. A., Manoharan, N., Hou, W., Huang, L., et al. (2013). Cultivation and characterization of thermophilic Nitrospira species from geothermal springs in the US Great Basin, China, and Armenia. FEMS Microbiol. Ecol. 85, 283–292. doi: 10.1111/1574-6941.12117
Ehrich, S., Behrens, D., Lebedeva, E., Ludwig, W., and Bock, E. (1995). A new obligately chemolithoautotrophic, nitrite-oxidizing bacterium, Nitrospira moscoviensis sp. nov. and its phylogenetic relationship. Arch. Microbiol. 164, 16–23. doi: 10.1007/bf02568729
Elliott, D. R., Thomas, A. D., Hoon, S. R., and Sen, R. (2014). Niche partitioning of bacterial communities in biological crusts and soils under grasses, shrubs and trees in the Kalahari. Biodivers. Conserv. 23, 1709–1733. doi: 10.1007/s10531-014-0684-688
Eren, A. M., Esen, O. C., Quince, C., Vineis, J. H., Morrison, H. G., Sogin, M. L., et al. (2015). Anvi’o: An advanced analysis and visualization platform for ‘omics data. PeerJ 2015, e1319. doi: 10.7717/peerj.1319
Fang, W., Yan, D., Huang, B., Ren, Z., Wang, X., Liu, X., et al. (2019). Biochemical pathways used by microorganisms to produce nitrous oxide emissions from soils fumigated with dimethyl disulfide or allyl isothiocyanate. Soil Biol. Biochem. 132, 1–13. doi: 10.1016/j.soilbio.2019.01.019
Friedman, L., Mamane, H., Avisar, D., and Chandran, K. (2018). The role of influent organic carbon-to-nitrogen (COD/N) ratio in removal rates and shaping microbial ecology in soil aquifer treatment (SAT). Wat. Res. 146, 197–205. doi: 10.1016/j.watres.2018.09.014
Füssel, J., Lücker, S., Yilmaz, P., Nowka, B., van Kessel, M. A. H. J., Bourceau, P., et al. (2017). Adaptability as the key to success for the ubiquitous marine nitrite oxidizer Nitrococcus. Sci. Adv. 3:e1700807. doi: 10.1126/sciadv.1700807
Giaveno, M. A., Urbieta, M. S., Ulloa, J. R., Toril, E. G., and Donati, E. R. (2013). Physiologic versatility and growth flexibility as the main characteristics of a novel thermoacidophilic Acidianus strain isolated from Copahue geothermal area in Argentina. Microb. Ecol. 65, 336–346. doi: 10.1007/s00248-012-0129-124
Herlemann, D. P., Labrenz, M., Jürgens, K., Bertilsson, S., Waniek, J. J., and Andersson, A. F. (2011). Transitions in bacterial communities along the 2000 km salinity gradient of the Baltic Sea. ISME J. 5, 1571–1579. doi: 10.1038/ismej.2011.41
Ishii, K., Fujitani, H., Soh, K., Nakagawa, T., Takahashi, R., and Tsuneda, S. (2017). Enrichment and physiological characterization of a cold-adapted nitrite-oxidizing Nitrotoga sp. from an eelgrass sediment. Appl. Environ. Microbiol. 83:e00549-17. doi: 10.1128/AEM.00549-17
Kirstein, K., and Bock, E. (1993). Close genetic relationship between Nitrobacter hamburgensis nitrite oxidoreductase and Escherichia coli nitrate reductases. Arch. Microbiol. 160, 447–453. doi: 10.1007/BF00245305
Kitzinger, K., Koch, H., Lücker, S., Sedlacek, C. J., Herbold, C., Schwarz, J., et al. (2018). Characterization of the first “Candidatus Nitrotoga” isolate reveals metabolic versatility and separate evolution of widespread nitrite-oxidizing bacteria. mBio 9:e01186-18. doi: 10.1128/mBio.01186-18
Koch, H., Galushko, A., Albertsen, M., Schintlmeister, A., Gruber-Dorninger, C., Lücker, S., et al. (2014). Microbial metabolism: Growth of nitrite-oxidizing bacteria by aerobic hydrogen oxidation. Science 345, 1052–1054. doi: 10.1126/science.1256985
Koch, H., Lücker, S., Albertsen, M., Kitzinger, K., Herbold, C., Spieck, E., et al. (2015). Expanded metabolic versatility of ubiquitous nitrite-oxidizing bacteria from the genus Nitrospira. Proc. Natl. Acad. Sci. U.S.A. 112, 11371–11376. doi: 10.1073/pnas.1506533112
Kolmert, A., Wikström, P., and Hallberg, K. (2000). A fast and simple turbidimetric method for the determination of sulfate in sulfate-reducing bacterial cultures. J. Microbiol. Meth. 41, 179–184. doi: 10.1016/S0167-7012(00)00154-158
Koops, H.-P., and Pommerening-Röser, A. (2005). “The lithoautotrophic ammonia-oxidizing bacteria,” in Bergey’s Manual of Systematic Bacteriology, eds D. J. Brenner, N. R. Krieg, J. T. Staley, and G. M. Garrity, (Boston, MA: Springer), 778–811. doi: 10.1007/0-387-28021-9_18
Krümmel, A., and Harms, H. (1982). Effect of organic matter on growth and cell yield of ammonia-oxidizing bacteria. Arch. Microbiol. 133, 50–54. doi: 10.1007/bf00943769
Kruse, M., Zumbrägel, S., Bakker, E., Spieck, E., Eggers, T., and Lipski, A. (2013). The nitrite-oxidizing community in activated sludge from a municipal wastewater treatment plant determined by fatty acid methyl ester-stable isotope probing. Syst. Appl. Microbiol. 36, 517–524. doi: 10.1016/j.syapm.2013.06.007
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Lebedeva, E. V., Alawi, M., Fiencke, C., Namsaraev, B., Bock, E., and Spieck, E. (2005). Moderately thermophilic nitrifying bacteria from a hot spring of the Baikal rift zone. FEMS Microbiol. Ecol. 54, 297–306. doi: 10.1016/j.femsec.2005.04.010
Lebedeva, E. V., Alawi, M., Maixner, F., Jozsa, P. G., Daims, H., and Spieck, E. (2008). Physiological and phylogenetic characterization of a novel lithoautotrophic nitrite-oxidizing bacterium, “Candidatus Nitrospira bockiana. Int. J. Syst. Evol. Microbiol. 58, 242–250. doi: 10.1099/ijs.0.65379-65370
Lebedeva, E. V., Off, S., Zumbrägel, S., Kruse, M., Shagzhina, A., Lücker, S., et al. (2011). Isolation and characterization of a moderately thermophilic nitrite-oxidizing bacterium from a geothermal spring. FEMS Microbiol. Ecol. 75, 195–204. doi: 10.1111/j.1574-6941.2010.01006.x
Lee, I., Kim, Y. O., Park, S. C., and Chun, J. (2016). OrthoANI: an improved algorithm and software for calculating average nucleotide identity. Int. J. Syst. Evol. Microbiol. 66, 1100–1103. doi: 10.1099/ijsem.0.000760
Li, H. (2018). Minimap2: Pairwise alignment for nucleotide sequences. Bioinformatics 34, 3094–3100. doi: 10.1093/bioinformatics/bty191
Li, H., and Durbin, R. (2009). Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760. doi: 10.1093/bioinformatics/btp324
Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homer, N., et al. (2009). The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079. doi: 10.1093/bioinformatics/btp352
Lücker, S., Nowka, B., Rattei, T., Spieck, E., and Daims, H. (2013). The genome of Nitrospina gracilis illuminates the metabolism and evolution of the major marine nitrite oxidizer. Front. Microbiol. 4:27. doi: 10.3389/fmicb.2013.00027
Lücker, S., Wagner, M., Maixner, F., Pelletier, E., Koch, H., Vacherie, B., et al. (2010). A Nitrospira metagenome illuminates the physiology and evolution of globally important nitrite-oxidizing bacteria. Proc. Natl. Acad. Sci. U.S.A. 107, 13479–13484. doi: 10.1073/pnas.1003860107
Lüke, C., Speth, D. R., Kox, M. A. R., Villanueva, L., and Jetten, M. S. M. (2016). Metagenomic analysis of nitrogen and methane cycling in the Arabian Sea oxygen minimum zone. PeerJ 4:e1924. doi: 10.7717/peerj.1924
Manz, W., Amann, R., Ludwig, W., Wagner, M., and Schleifer, K. H. (1992). Phylogenetic oligodeoxynucleotide probes for the major subclasses of proteobacteria: problems and solutions. Syst. Appl. Microbiol. 15, 593–600. doi: 10.1016/s0723-2020(11)80121-9
Marks, C. R., Stevenson, B. S., Rudd, S., and Lawson, P. A. (2012). Nitrospira-dominated biofilm within a thermal artesian spring: a case for nitrification-driven primary production in a geothermal setting. Geobiology 10, 457–466. doi: 10.1111/j.1472-4669.2012.00335.x
Merodio, J. C., and Martínez, J. M. (1985). Análisis químico de componentes mayoritarios en rocas silicatadas. Rev. Asoc. Argentina Mineral. Petrol. 16, 7–16.
Ngugi, D. K., Blom, J., Stepanauskas, R., and Stingl, U. (2016). Diversification and niche adaptations of Nitrospina-like bacteria in the polyextreme interfaces of Red Sea brines. ISME J. 10, 1383–1399. doi: 10.1038/ismej.2015.214
Nikolenko, S. I., Korobeynikov, A. I., and Alekseyev, M. A. (2013). BayesHammer: Bayesian clustering for error correction in single-cell sequencing. BMC Genomics 14:S7. doi: 10.1186/1471-2164-14-S1-S7
Notredame, C., Higgins, D. G., and Heringa, J. (2000). T-coffee: A novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 302, 205–217. doi: 10.1006/jmbi.2000.4042
Nowka, B., Daims, H., and Spieck, E. (2015). Comparison of oxidation kinetics of nitrite-oxidizing bacteria: Nitrite availability as a key factor in niche differentiation. Appl. Environ. Microbiol. 81, 745–753. doi: 10.1128/AEM.02734-2714
Nurk, S., Meleshko, D., Korobeynikov, A., and Pevzner, P. A. (2017). MetaSPAdes: A new versatile metagenomic assembler. Genome Res. 27, 824–834. doi: 10.1101/gr.213959.116
Off, S., Alawi, M., and Spieck, E. (2010). Enrichment and physiological characterization of a novel Nitrospira-like bacterium obtained from a marine sponge. Appl. Environ. Microbiol. 76, 4640–4646. doi: 10.1128/AEM.03095-3099
Orlando, J., Alfaro, M., Bravo, L., Guevara, R., and Carú, M. (2010). Bacterial diversity and occurrence of ammonia-oxidizing bacteria in the Atacama Desert soil during a “desert bloom” event. Soil Biol. Biochem. 42, 1183–1188. doi: 10.1016/j.soilbio.2010.03.025
Palomo, A., Pedersen, A. G., Fowler, S. J., Dechesne, A., Sicheritz-Pontén, T., and Smets, B. F. (2018). Comparative genomics sheds light on niche differentiation and the evolutionary history of comammox Nitrospira. ISME J. 12, 1779–1793. doi: 10.1038/s41396-018-0083-83
Parks, D. H., Chuvochina, M., Waite, D. W., Rinke, C., Skarshewski, A., Chaumeil, P. A., et al. (2018). A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat. Biotechnol. 36:996. doi: 10.1038/nbt.4229
Parks, D. H., Imelfort, M., Skennerton, C. T., Hugenholtz, P., and Tyson, G. W. (2015). CheckM: Assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 25, 1043–1055. doi: 10.1101/gr.186072.114
Pester, M., Maixner, F., Berry, D., Rattei, T., Koch, H., Lücker, S., et al. (2014). NxrB encoding the beta subunit of nitrite oxidoreductase as functional and phylogenetic marker for nitrite-oxidizing Nitrospira. Environ. Microbiol. 16, 3055–3071. doi: 10.1111/1462-2920.12300
Philips, S., Laanbroek, H. J., and Verstraete, W. (2002). Origin, causes and effects of increased nitrite concentrations in aquatic environments. Rev. Environ. Sci. Biotechnol. 1, 115–141. doi: 10.1023/A:1020892826575
Price, G. D., Badger, M. R., Woodger, F. J., and Long, B. M. (2008). Advances in understanding the cyanobacterial CO2-concentrating-mechanism (CCM): functional components, Ci transporters, diversity, genetic regulation and prospects for engineering into plants. J. Exp. Bot. 59, 1441–1461. doi: 10.1093/jxb/erm112
Rachel, R., Pum, D., Šmarda, J., Šmajs, D., Komrska, J., Krzyzánek, V., et al. (1997). II. Fine structure of S-layers. FEMS Microbiol. Rev. 20, 13–23. doi: 10.1111/j.1574-6976.1997.tb00302.x
Ritz, C., Baty, F., Streibig, J. C., and Gerhard, D. (2015). Dose-response analysis using R. PLoS One 10:e0146021. doi: 10.1371/journal.pone.0146021
Rodriguez-R, L. M., and Konstantinidis, K. T. (2016). The enveomics collection: a toolbox for specialized analyses of microbial genomes and metagenomes. PeerJ 4:e1900v1.
Schmidt, E. L., and Belser, L. W. (1994). “Autotrophic nitrifying bacteria,” in Methods of Soil Analysis, eds B. P. Weaver and J. S. Angle, (Madison, WI: Soil Science Society of America), 159–177. doi: 10.2136/sssabookser5.2.c10
Seemann, T. (2014). Prokka: Rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069. doi: 10.1093/bioinformatics/btu153
Sorokin, D. Y., Lücker, S., and Daims, H. (2018). “Nitrolancea,” in Bergey’s manual of systematics of Archaea and Bacteria, eds W. B. Whitman, F. Rainey, P. Kämpfer, M. Trujillo, J. Chun, P. DeVos, et al. (Hoboken, NJ: Wiley), 1–6. doi: 10.1002/9781118960608.gbm01563
Sorokin, D. Y., Lücker, S., Vejmelkova, D., Kostrikina, N. A., Kleerebezem, R., Rijpstra, W. I. C., et al. (2012). Nitrification expanded: discovery, physiology and genomics of a nitrite-oxidizing bacterium from the phylum Chloroflexi. ISME J. 6, 2245–2256. doi: 10.1038/ismej.2012.70
Sorokin, D. Y., Vejmelkova, D., Lücker, S., Streshinskaya, G. M., Rijpstra, W. I. C., Sinninghe Damsté, J. S., et al. (2014). Nitrolancea hollandica gen. nov., sp. nov., a chemolithoautotrophic nitrite-oxidizing bacterium isolated from a bioreactor belonging to the phylum Chloroflexi. Int. J. Syst. Evol. Microbiol. 64, 1859–1865. doi: 10.1099/ijs.0.062232-62230
Speirs, L. B. M., Rice, D. T. F., Petrovski, S., and Seviour, R. J. (2019). The phylogeny, biodiversity, and ecology of the Chloroflexi in activated sludge. Front. Microbiol. 10:2015. doi: 10.3389/fmicb.2019.02015
Spieck, E., Aamand, J., Bartosch, S., and Bock, E. (1996). Immunocytochemical detection and location of the membrane-bound nitrite oxidoreductase in cells of Nitrobacter and Nitrospira. FEMS Microbiol. Lett. 139, 71–76. doi: 10.1111/j.1574-6968.1996.tb08181.x
Spieck, E., and Bock, E. (2005). “The lithoautotrophic nitrite-oxidizing bacteria,” in Bergey’s Manual of Systematic Bacteriology, eds D. J. Brenner, N. R. Krieg, J. T. Staley, and G. M. Garrity, (Boston, MA: Springer), 149–153. doi: 10.1007/0-387-28021-9_19
Spieck, E., Ehrich, S., Aamand, J., and Bock, E. (1998). Isolation and immunocytochemical location of the nitrite-oxidizing system in Nitrospira moscoviensis. Arch. Microbiol. 169, 225–230. doi: 10.1007/s002030050565
Spieck, E., and Lipski, A. (2011). “Cultivation, growth physiology, and chemotaxonomy of nitrite-oxidizing bacteria,” in Methods in Enzymology, ed. M. G. Klotz, (Burlington: Academic Press), 109–130. doi: 10.1016/b978-0-12-381294-0.00005-5
Spieck, E., Spohn, M., Wendt, K., Bock, E., Shively, J., Frank, J., et al. (2020). Extremophilic nitrite-oxidizing Chloroflexi from Yellowstone hot springs. ISME J. 14, 364–379. doi: 10.1038/s41396-019-0530-539
Stackebrandt, E., and Ebers, J. (2006). Taxonomic parameters revisited: Tarnished gold standards. Microbiol. Today 33, 152–155.
Starkenburg, S. R., Larimer, F. W., Stein, L. Y., Klotz, M. G., Chain, P. S., Sayavedra-Soto, L. A., et al. (2008). Complete genome sequence of Nitrobacter hamburgensis X14 and comparative genomic analysis of species within the genus Nitrobacter. Appl. Environ. Microbiol. 74, 2852–2863. doi: 10.1128/AEM.02311-2317
Stieglmeier, M., Alves, R. J. E., and Schleper, C. (2014). “The phylum Thaumarchaeota,” in The Prokaryotes: Other major lineages of Bacteria and the Archaea, eds E. Rosenberg, E. F. DeLong, S. Lory, E. Stackebrandt, and F. Thompson, (Berlin: Springer), 347–362. doi: 10.1007/978-3-642-38954-2_338
Sun, X., Kop, L. F. M., Lau, M. C. Y., Frank, J., Jayakumar, A., Lücker, S., et al. (2019). Uncultured Nitrospina-like species are major nitrite oxidizing bacteria in oxygen minimum zones. ISME J. 13, 2391–2402. doi: 10.1038/s41396-019-0443-447
Urbieta, M. S., González-Toril, E., Bazán, ÁA., Giaveno, M. A., and Donati, E. (2015a). Comparison of the microbial communities of hot springs waters and the microbial biofilms in the acidic geothermal area of Copahue (Neuquén, Argentina). Extremophiles 19, 437–450. doi: 10.1007/s00792-015-0729-722
Urbieta, M. S., Porati, G., Segretín, A., González-Toril, E., Giaveno, M., and Donati, E. (2015b). Copahue geothermal system: A volcanic environment with rich extreme prokaryotic biodiversity. Microorganisms 3, 344–363. doi: 10.3390/microorganisms3030344
Urbieta, M. S., González Toril, E., Aguilera, A., Giaveno, M. A., and Donati, E. (2012). First prokaryotic biodiversity assessment using molecular techniques of an acidic river in Neuquén, Argentina. Microb. Ecol. 64, 91–104. doi: 10.1007/s00248-011-9997-9992
Urbieta, M. S., Toril, E. G., Alejandra Giaveno, M., Bazán, ÁA., and Donati, E. R. (2014). Archaeal and bacterial diversity in five different hydrothermal ponds in the Copahue region in Argentina. Syst. Appl. Microbiol. 37, 429–441. doi: 10.1016/j.syapm.2014.05.012
van Kessel, M. A. H. J., Speth, D. R., Albertsen, M., Nielsen, P. H., Op Den Camp, H. J. M., Kartal, B., et al. (2015). Complete nitrification by a single microorganism. Nature 528, 555–559. doi: 10.1038/nature16459
Vejmelkova, D., Sorokin, D. Y., Abbas, B., Kovaleva, O. L., Kleerebezem, R., Kampschreur, M. J., et al. (2012). Analysis of ammonia-oxidizing bacteria dominating in lab-scale bioreactors with high ammonium bicarbonate loading. Appl. Microbiol. Biotechnol. 93, 401–410. doi: 10.1007/s00253-011-3409-x
Wallner, G., Amann, R., and Beisker, W. (1993). Optimizing fluorescent in situ hybridization with rRNA-targeted oligonucleotide probes for flow cytometric identification of microorganisms. Cytometry 14, 136–143. doi: 10.1002/cyto.990140205
Wang, B., Wang, Z., Wang, S., Qiao, X., Gong, X., Gong, Q., et al. (2020). Recovering partial nitritation in a PN/A system during mainstream wastewater treatment by reviving AOB activity after thoroughly inhibiting AOB and NOB with free nitrous acid. Environ. Int. 139:105684. doi: 10.1016/j.envint.2020.105684
Wegen, S., Nowka, B., and Spieck, E. (2019). Low temperature and neutral pH define “Candidatus Nitrotoga sp.” as a competitive nitrite oxidizer in coculture with Nitrospira defluvii. Appl. Environ. Microbiol. 85:e02569-18. doi: 10.1128/AEM.02569-2518
Widdel, F., and Bak, F. (1991). “Gram-negative mesophilic sulfate-reducing bacteria,” in The Prokaryotes: a Handbook on the Biology of Bacteria, eds A. Balows, H. G. Trüper, M. Dworkin, W. Harder, and K. H. Schleifer, (New York, NY: Springer), 3352–3379. doi: 10.1007/978-1-4757-2191-1_21
Willis Poratti, G., Yaakop, A. S., Chan, C. S., Urbieta, M. S., Chan, K. G., Ee, R., et al. (2016). Draft genome sequence of the sulfate-reducing bacterium Desulfotomaculum copahuensis strain CINDEFI1 isolated from the geothermal Copahue system, Neuquén, Argentina. Genome Announc. 4:e00870-16. doi: 10.1128/genomeA.00870-816
Xu, L., Dong, Z., Fang, L., Luo, Y., Wei, Z., Guo, H., et al. (2019). OrthoVenn2: a web server for whole-genome comparison and annotation of orthologous clusters across multiple species. Nucleic Acids Res. 47, W52–W58. doi: 10.1093/nar/gkz333
Yu, H., Meng, W., Song, Y., and Tian, Z. (2018). Understanding bacterial communities of partial nitritation and nitratation reactors at ambient and low temperature. Chem. Eng. J. 337, 755–763. doi: 10.1016/j.cej.2017.10.120
Yu, H., Tian, Z., Zuo, J., and Song, Y. (2020). Enhanced nitrite accumulation under mainstream conditions by a combination of free ammonia-based sludge treatment and low dissolved oxygen: reactor performance and microbiome analysis. RSC Adv. 10, 2049–2059. doi: 10.1039/C9RA07628J
Keywords: nitrification, nitrite oxidation, Nitrolancea, cultivation, wastewater treatment plant, geothermal springs
Citation: Spieck E, Sass K, Keuter S, Hirschmann S, Spohn M, Indenbirken D, Kop LFM, Lücker S and Giaveno A (2020) Defining Culture Conditions for the Hidden Nitrite-Oxidizing Bacterium Nitrolancea. Front. Microbiol. 11:1522. doi: 10.3389/fmicb.2020.01522
Received: 22 April 2020; Accepted: 12 June 2020;
Published: 10 July 2020.
Edited by:
Thomas E. Hanson, University of Delaware, United StatesReviewed by:
Lisa Y. Stein, University of Alberta, CanadaPetra Pjevac, University of Vienna, Austria
Copyright © 2020 Spieck, Sass, Keuter, Hirschmann, Spohn, Indenbirken, Kop, Lücker and Giaveno. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Eva Spieck, ZXZhLnNwaWVja0B1bmktaGFtYnVyZy5kZQ==
 Eva Spieck
Eva Spieck Katharina Sass
Katharina Sass Sabine Keuter
Sabine Keuter Sophia Hirschmann
Sophia Hirschmann Michael Spohn
Michael Spohn Daniela Indenbirken2
Daniela Indenbirken2 Linnea F. M. Kop
Linnea F. M. Kop Sebastian Lücker
Sebastian Lücker Alejandra Giaveno
Alejandra Giaveno