- 1The Second Affiliated Hospital and Yuying Children’s Hospital, Wenzhou Medical University, Wenzhou, China
- 2Key Laboratory of Medical Genetics of Zhejiang Province, Key Laboratory of Laboratory Medicine, Ministry of Education of China, School of Laboratory Medicine and Life Sciences, Wenzhou Medical University, Wenzhou, China
- 3Institute of Biomedical Informatics, Wenzhou Medical University, Wenzhou, China
- 4Institute of Translational Medicine, Baotou Central Hospital, Baotou, China
Yokenella regensburgei, a member of the family Enterobacteriaceae, is usually isolated from environmental samples and generally resistant to early generations of cephalosporins. To characterize the resistance mechanism of Y. regensburgei strain W13 isolated from the sewage of an animal farm, whole genome sequencing, comparative genomics analysis and molecular cloning were performed. The results showed that a novel chromosomally encoded class C β-lactamase gene with the ability to confer resistance to β-lactam antibiotics, designated blaYOC–1, was identified in the genome of Y. regensburgei W13. Kinetic analysis revealed that the β-lactamase YOC-1 has a broad spectrum of substrates, including penicillins, cefazolin, cefoxitin and cefotaxime. The two functionally characterized β-lactamases with the highest amino acid identities to YOC-1 were CDA-1 (71.69%) and CMY-2 (70.65%). The genetic context of the blaYOC–1-ampR-encoding region was unique compared with the sequences in the NCBI nucleotide database. The plasmid pRYW13-125 of Y. regensburgei W13 harbored 11 resistance genes (blaOXA–10, blaLAP–2, dfrA14, tetA, tetR, cmlA5, floR, sul2, ant(3″)-IIa, arr-2 and qnrS1) within an ∼34 kb multidrug resistance region; these genes were all related to mobile genetic elements. The multidrug resistance region of pYRW13-125 shared the highest identities with those of two plasmids from clinical Klebsiella pneumoniae isolates, indicating the possibility of horizontal transfer of these resistance genes between bacteria of various origins.
Introduction
Yokenella regensburgei is a species of Enterobacteriaceae originally identified by Kosako et al. (1984) through DNA hybridization and biochemical tests in 1984. In 1985, Hickman-Brenner et al. (1985) proposed the name Koserella trabulsii for a new group of Enterobacteriaceae they had discovered that was previously called Enteric Group 45. However, it was subsequently hown by DNA hybridization that Y. regensburgei and K. trabulsii were the same species (Kosako et al., 1987). In 1991, the Centers for Disease Control and Prevention acknowledged that Y. regensburgei had priority upon the basis of prior publication; therefore, the use of K. trabulsii was discontinued in 1991 (McWhorter et al., 1991). Y. regensburgei has been found in a variety of environmental samples (Jain et al., 2013; Chi et al., 2017). This organism has also been sporadically isolated from clinical samples, thus demonstrating its ability to act as an opportunistic pathogen (Abbott and Janda, 1994; Bhowmick and Weinstein, 2013; Jain et al., 2013; Chi et al., 2017; Palmisano et al., 2019; Wright et al., 2019). Intriguingly, Y. regensburgei closely resembles Hafnia alvei and thus has possibly been misidentified as the latter by automated systems. It has been hypothesized that infections caused by Y. regensburgei were underestimated due to misidentification of the bacterium (Bhowmick and Weinstein, 2013).
Many studies have reported that Y. regensburgei possesses an ampC gene and that the expression of this gene is highly inducible (Chi et al., 2017). However, to date, there has been no detailed study of the AmpC β-lactamase in Y. regensburgei, and no complete genome sequence of Y. regensburgei is available in the public NCBI genome database. Y. regensburgei appears to be intrinsically resistant to colistin (Hickman-Brenner et al., 1985), azithromycin and some β-lactam antibiotics, and this resistance is likely related to chromosome-mediated AmpC (Jachymek et al., 1999). In addition to the production of ESBLs, resistance to third-generation cephalosporins can also be mediated by chromosomal and plasmid-encoded AmpCs. In general, AmpCs exhibit broad substrate specificity, including penicillins, narrow-spectrum cephalosporins, cephamycins and aztreonam (variable), and their expression can confer resistance to all these compounds. AmpCs are characterized by their greater capability of hydrolyzing cephalosporins and resistance to β-lactam-based β-lactamase inhibitors, such as clavulanate, sulbactam and tazobactam (Jacoby, 2009; Meini et al., 2019).
The acquisition of exogenous resistance genes is often associated with conjugative plasmids, which may carry multiple resistance genes (De Toro et al., 2015; Medaney et al., 2016). Studies have reported that plasmids are abundant in Enterobacteriaceae, such as Escherichia coli and Klebsiella pneumoniae (Decre et al., 2002; Doi et al., 2002; Gao et al., 2016). However, except for sporadic clinical case reports available in the literature, there have been no reports of the molecular characteristics of the Y. regensburgei genome. In this study, for the first time, we determined the complete genome sequence of the Y. regensburgei strain W13 isolated from the sewage of an animal farm. Based on sequence analysis, we identified and characterized a novel chromosomally encoded AmpC β-lactamase gene, blaYOC–1, and a conjugative plasmid, pYRW13-125, carrying multiple resistance genes. The identification of the novel chromosome-encoded blaYOC–1 gene in this environmental bacterium can provide detailed information on the intrinsic resistance mechanism of this unusual opportunistic pathogen.
Materials and Methods
Bacterial Strains
Y. regensburgei W13 was isolated from sewage discharged from an animal farm in Wenzhou, southeastern China. The strain was first identified using the bioMérieux VITEK 2 Compact Instrument (bioMérieux, Marcy L’etoile, France) and then verified by analysis of the 16S rRNA gene sequences and average nucleotide identity (ANI). The strains and plasmids used in this work are listed in Table 1.
Antimicrobial Susceptibility Testing
The minimum inhibitory concentrations (MICs) were determined using the agar dilution method following the guidelines of the Clinical and Laboratory Standards Institute (CLSI), and the susceptibility patterns were interpreted according to the CLSI breakpoint criteria (CLSI, 2019). E. coli ATCC 25922 was used as a reference strain for quality control.
Whole-Genome Sequencing (WGS) and Functional Annotation of the Genome Sequence
Bacterial DNA was extracted using the Generay Genomic DNA Miniprep kit (Shanghai Generay Biotech Co., Ltd., Shanghai, China) from a single colony subcultured in brain heart infusion broth at 37°C for approximately 16 h. Genomic DNA was sequenced on a PacBio RS II instrument (Pacific Biosciences). The PacBio long reads were initially assembled by Canu v1.8 (Koren et al., 2017), and then two FASTQ sequence files generated using the Illumina HiSeq 2500 platform were mapped onto the primary assembly to control assembly quality and to correct possible misidentified bases by using Bwa (Li and Durbin, 2009) and the Genome Analysis Toolkit (McKenna et al., 2010). Genes were predicted and annotated by using Prokka v1.14.0 (Seemann, 2014); furthermore, the predicted proteins were searched against the NCBI nonredundant and Swiss-Prot databases using DIAMOND (Buchfink et al., 2015) with an e-value threshold of 1e-5. Annotation of the resistance genes was performed using ResFinder (Zankari et al., 2012) and Resistance Gene Identifier (RGI) software of Comprehensive Antibiotic Resistance Database version 4.0.31 (McArthur et al., 2013). GView was used to construct basic genomic features (Petkau et al., 2010). Annotation of MGEs was performed using ISfinder (Siguier et al., 2006) and INTEGRALL (Moura et al., 2009). Average nucleotide identity was calculated using OrthoANI version 0.93.1 (Lee et al., 2016). Easyfig v2.2.2 (Sullivan et al., 2011) software was used to generate the figure showing structural comparisons and the nucleotide identities of the different MDR regions. The molecular weight and pI value of YOC-1 were predicted using ProtParam2. The putative signal peptide cleavage site of YOC-1 was identified by SignalP 5.03. Amino acid alignment and neighbor-joining phylogenetic tree construction of YOC-1 with other AmpCs were performed using MEGAX (Kumar et al., 2018).
Cloning of BlaYOC–1, Expression, and Purification of YOC-1
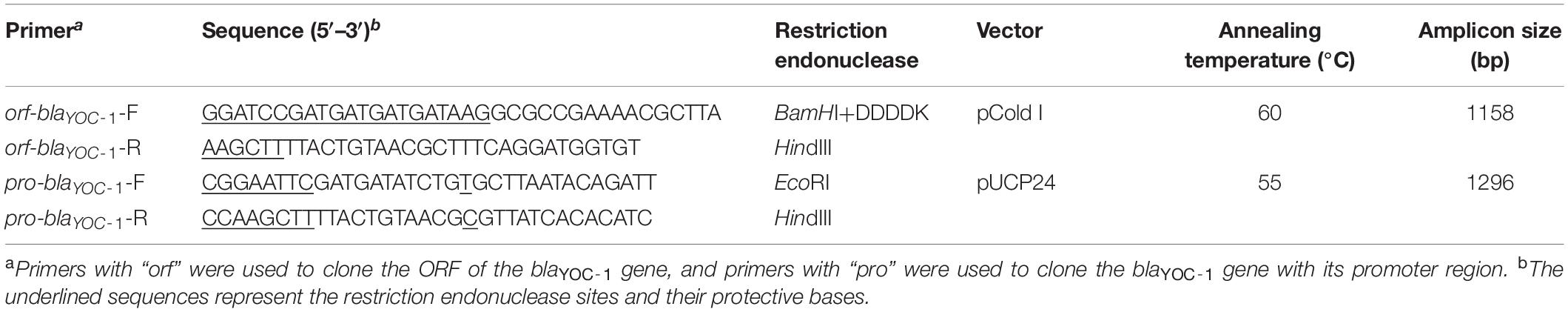
The blaYOC–1 gene sequence with its promoter region and a pair of flanking restriction endonuclease adapters (EcoRI for the forward primer pro-blaYOC–1-F and HindIII for the reverse primer pro-blaYOC–1-R, respectively, Table 2) was PCR-amplified, and the PCR product eluted from the agarose gel was digested with the corresponding restriction endonucleases and ligated into pUCP24 (digested with EcoRI and HindIII) with a T4 DNA ligase cloning kit (Takara Bio Inc., Dalian, China) (Table 2). The resulting recombinant plasmid (pUCP24-pro-blaYOC–1) was transformed into E. coli DH5α using the calcium chloride method (Suzuki et al., 2019). Transformants (pUCP24-pro-blaYOC–1/DH5α) were selected on Luria-Bertani (LB) agar plates containing 40 μg/mL gentamicin. The cloned PCR product was further confirmed by Sanger sequencing (Shanghai Sunny Biotechnology Co., Ltd., Shanghai, China). The resistance activity of the transformants containing blaYOC–1 to antibiotics was further determined.
The predicted cleavage site of the YOC-1 β-lactamase is located after the Ala-22 residue. In order to obtain the β-lactamase YOC-1, the ORF of blaYOC–1 without a signal sequence was cloned with a pair of flanking restriction endonuclease adapters (BamHI and DDDDK for the forward primer orf-blaYOC–1-F and HindIII for the reverse primer orf-blaYOC–1-R, Table 2) by similar procedures as described above. Using pCold I as a cloning vector and E. coli BL21 as the recipient, transformants (pCold I-blaYOC–1/BL21) were selected on LB agar plates containing 100 μg/mL ampicillin. The overnight culture of the recombinant strain (pCold I -blaYOC–1/BL21) was diluted 100-fold in 1 L of LB medium, incubated for 2–3 h at 37°C with orbital shaking at 250 rpm until the OD600 reached 0.6, and incubated with 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG) (Sigma Chemicals Co., St. Louis, MO, United States) for 24 h at 16°C (Choi and Geletu, 2018). The recombinant protein was purified by affinity chromatography using BeyoGold His-tag Purification Resin (Beyotime, Shanghai, China) according to the manufacturer’s instructions. The histidine tag was removed by Enterokinase (GenScript, Nanjing, China) for 24 h at 23°C.
Conjugation Experiment
E. coli C600 (MIC to rifampicin > 2048 μg/mL) was used as the recipient in conjugation experiments to detect the transferability of plasmids carried by Y. regensburgei W13 using the broth dilution method. The transconjugant was selected on a brain heart infusion plate supplemented with 2048 μg/mL rifampicin and 16 μg/mL florfenicol. The candidate transconjugant was first tested by plasmid profiling and then indole biochemical property and 16S rRNA gene sequence analyses, which could distinguish whether the candidate transconjugant was the donor, Y. regensburgei W13, or the recipient, E. coli C600 (Y. regensburgei W13 is negative but E. coli C600 is positive for the indole test) (Abbott and Janda, 1994). The plasmid of the transconjugant (pYRW13-125/EC600) was further analyzed by PCR and sequencing for the presence of resistance genes.
Enzyme Kinetic Analysis
Kinetic parameters for hydrolysis of β-lactams by the purified β-lactamase YOC-1 were examined using a UV-VIS spectrophotometer (U-3900, HITACHI, Japan) at 37°C in 10 mM phosphate buffer (pH 7.0) in a final reaction volume of 200 μL. The steady-state kinetic parameters (kcat and Km) were determined by nonlinear regression of the initial reaction rates with the Michaelis-Menten equation in Prism (version 7) software (GraphPad Software, CA, United States) (Chen et al., 2019).
β-Lactamase inhibition was studied with nitrocefin (100 μM) as the substrate. The β-lactamase inhibitors avibactam, GMP (disodium 5′-guanosine monophosphate) and clavulanic acid at various concentrations were preincubated with purified YOC-1 at 37°C for 5 min before the addition of substrate. The inhibitor concentration required to reduce the hydrolysis of 100 μM nitrocefin by 50% was determined by nonlinear regression with the log (inhibitor) vs. response – Variable slope equation in Prism (version 7) software (GraphPad Software, CA, United States) (Chen et al., 2019).
Nucleotide Sequence Accession Numbers
The complete chromosome and plasmid sequences of Y. regensburgei W13 have been submitted to GenBank, and the accession numbers of the chromosome, pYRW13-125 and pYRW13-131 are CP050811, CP050812, and CP050813, respectively.
Results and Discussion
Genome Characteristics and the Resistance Profile of Y. regensburgei W13
The 16S rRNA gene similarity analysis showed that Y. regensburgei W13 had the closest relationship with Y. regensburgei CIP 105435 (NR_104934.1) at 99.00% coverage and 98.17% identity. The average nucleotide identity (ANI) analysis results revealed that W13 shared high identity (99.05%) with Yokenella regensburgei ATCC 43003 (NZ_AGCL00000000), and this isolate was finally designated Y. regensburgei W13.
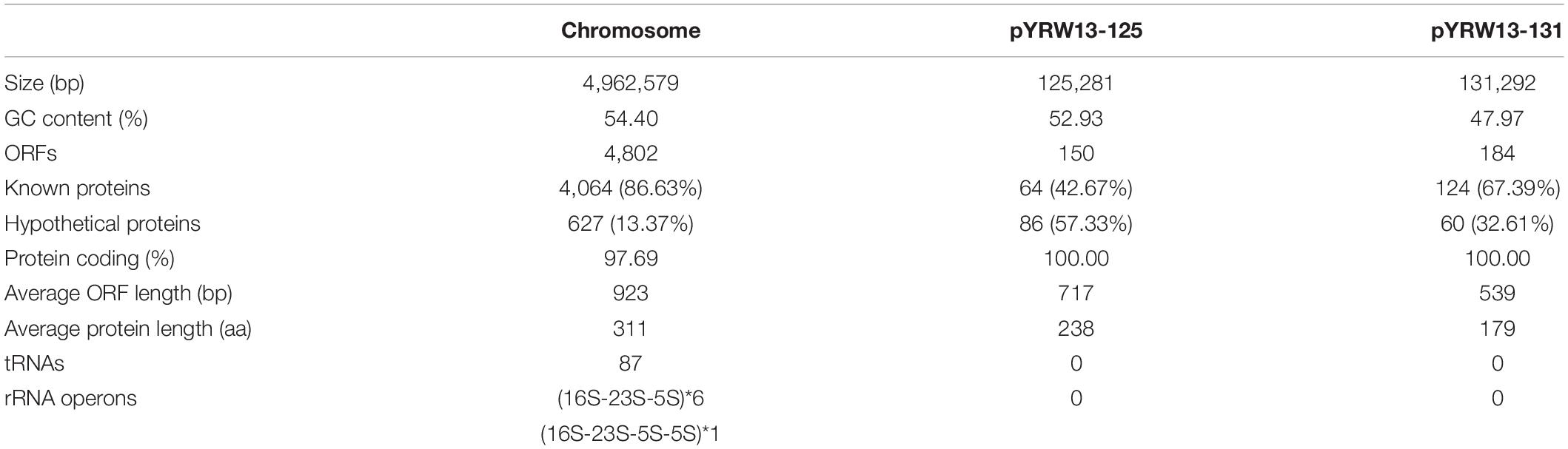
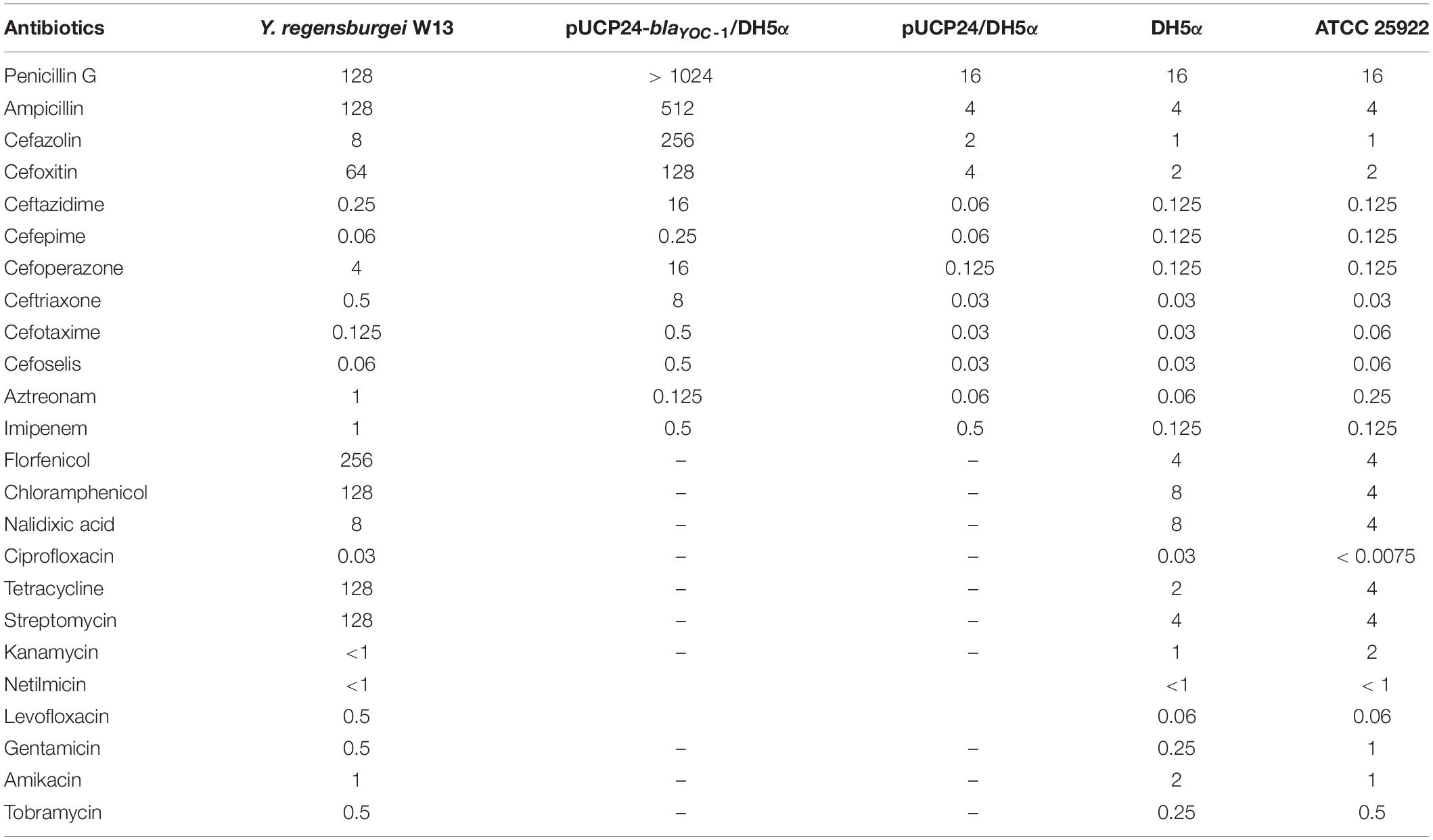
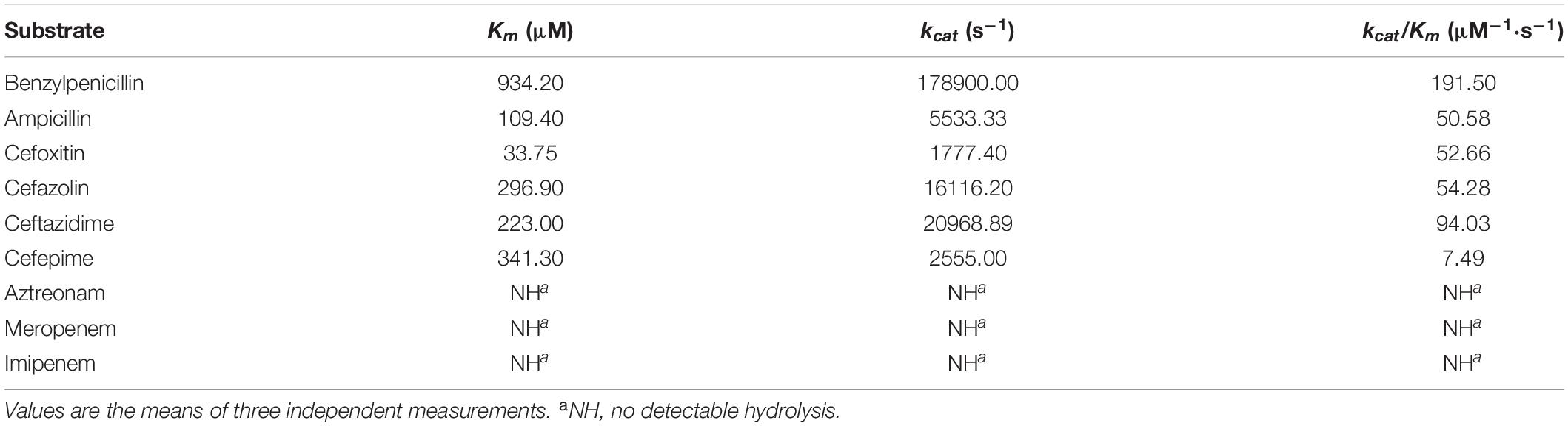
The whole genome of Y. regensburgei W13 consisted of a chromosome and two circular plasmids. The chromosome was approximately 4.96 Mb in length and encoded 4,802 ORFs. One plasmid, pYRW13-131, was 131,292 bp in length and encoded 184 ORFs, of which 124 were predicted to encode proteins with known functions (Table 3). The other plasmid, pYRW13-125, encoding eleven resistance genes against six classes of antibiotics, was 125,281 bp in length and encoded 150 ORFs (Figure 1). Similar to most Yokenella regensburgei, the in vitro susceptibility test showed that wild-type Y. regensburgei W13 exhibited resistance to penicillin G, ampicillin, cefoxitin, tetracycline and streptomycin (Table 4).
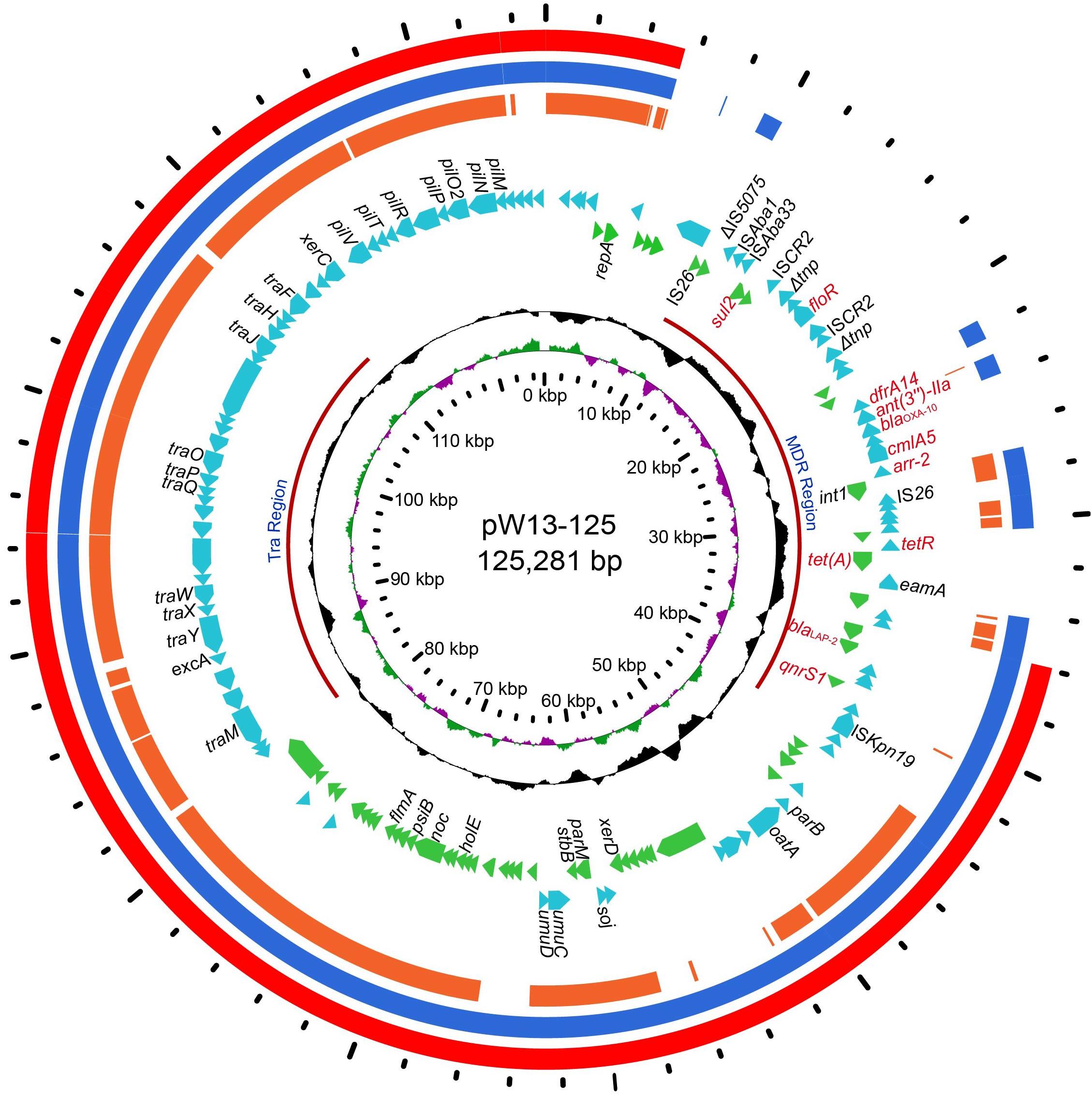
Figure 1. The circular map of pW13-125 and comparative genomics analysis with other closely related plasmids. Circle 1 (from inside to outside) shows the scale in kb. Circles 2 and 3 show the GC skew and GC content, respectively. Circle 4 shows the two different functional regions. Circles 5 and 6 present genes encoded in the forward strand and reverse strand, respectively. Circles 7–9 show the homologous regions of pOP-I (KY126370), an unnamed plasmid (CP027111) and p34399 (CP010385), respectively, and the regions without similar hits were left blank.
Homologs to YOC-1 Are Present in Other Enterobacterales Species
To investigate whether any novel β-lactamase gene was encoded in the Y. regensburgei W13 genome, we analyzed the predicted β-lactamase genes and found one containing conserved motifs of an Ambler class C β-lactamase; the predicted protein shared the highest amino acid identity (71.69%) with a functionally characterized AmpC, CDA-1 (Ammenouche et al., 2014) (Figure 2). We cloned the potential β-lactamase gene, and the results of the antibacterial susceptibility test showed that it was functional (Table 4). The novel β-lactamase gene, designated blaYOC–1, is 1,158 bp and encodes a preprotein of 385 amino acids, ca. 41.5 kDa. A signal peptide cleavage site was predicted to be between an alanine and asparagine at amino acid residues 22 and 23, respectively. Furthermore, the pI value of YOC-1 was predicted to be 8.50. Even though a hypothetical class C β-lactamase (EHM51798.1) showing the highest amino acid identity (99.48%, 383/385) with blaYOC–1 was predicted to be encoded in a draft genome of Yokenella regensburgei ATCC 43003 (NZ_AGCL00000000), the closest functionally characterized relatives to YOC-1 were β-lactamases CDA-1 (AID52933.1) and CMY-2 (X91840.1). They share only 71.69% (274/385) and 70.65% (272/385) amino acid sequence identity with YOC-1, respectively. When searching for YOC-1-like proteins (≥70% aa identity and ≥90% coverage) in the NCBI nucleotide database, it was found that more than half of the close relatives (52.04%, 51/98) were derived from Enterobacter species. YOC-1 had a close relationship with the YOC-1-like AmpCs (Figure 2).
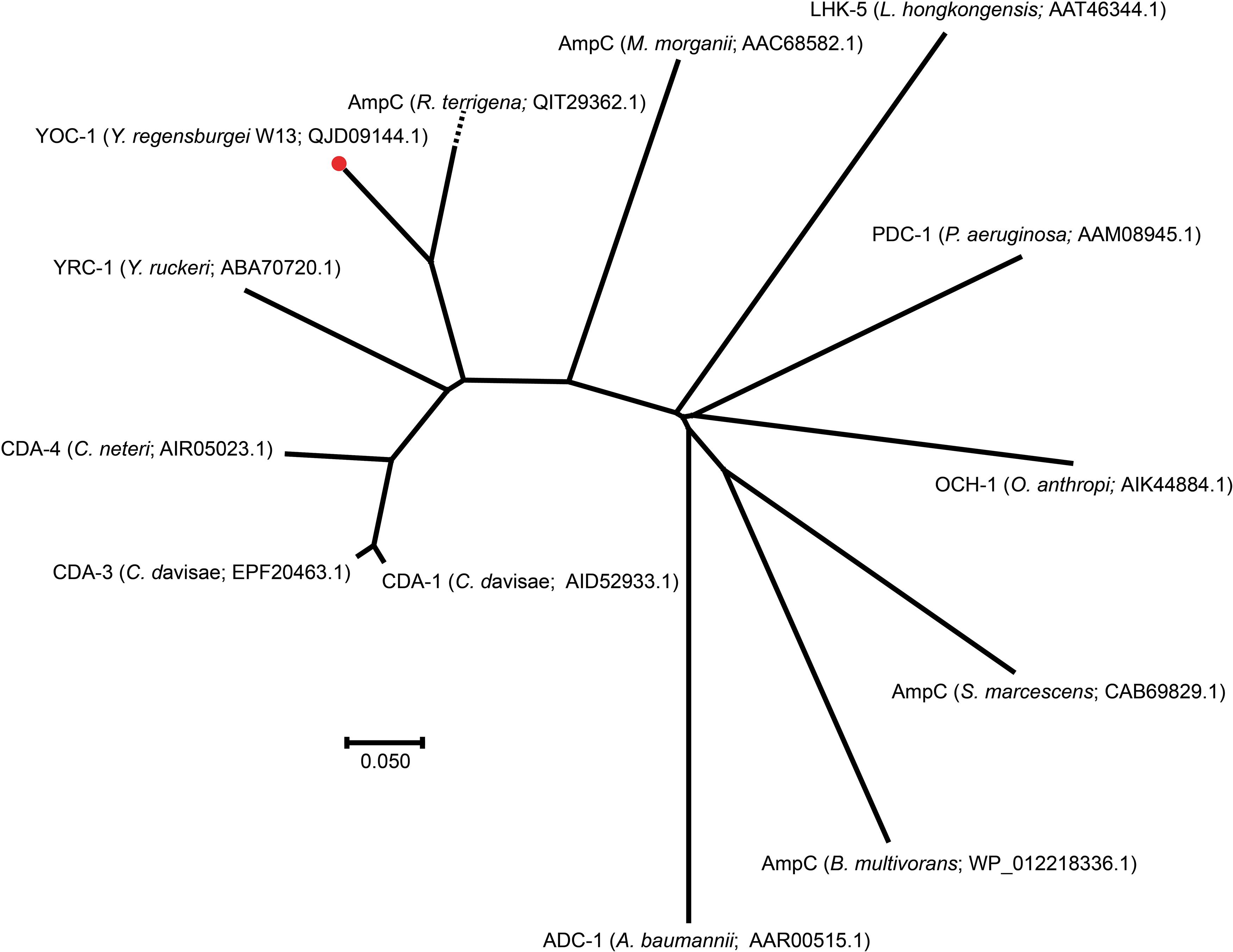
Figure 2. A phylogenetic tree showing the relationship of YOC-1 with other chromosome-borne AmpC β-lactamases. YOC-1 from our study is shown with a red dot.
Comparisons of chromosomal (ACT-28 and CDA-1) and plasmidic (CMY-2 and CMY-10) AmpCs that displayed slight carbapenemase activity (Kim et al., 2006; Ammenouche et al., 2014; Kotsakis et al., 2015; Jousset et al., 2019) revealed several conserved residues in CMY-2, CMY-10, CDA-1, and ACT-28 but mutated in YOC-1 [Tyr65, Ala122, Gln231 (Ω loop), Ser244 (Ω loop), Lys274, Leu276, Met278, Asp310 (9 helix), Ala314 (9 helix) and Gly377 (11 helix)] that could lead to the susceptibility of YOC-1 to carbapenem, however, no substitution was observed at amino acid residue 370, which was reported to contribute significantly to the carbapenemase activity of the CMY-2 enzyme4. Within the deduced amino acid sequence of YOC-1, three structural elements (one with two copies) characteristic of Ambler class C β-lactamases were identified: S-X-S-K at positions 88–91, K-T-G at positions 339–341, and Y-X-N at positions 174–176 (Supplementary Figure S1). A phylogenetic tree containing YOC-1 and other chromosome-borne class C β-lactamases was constructed (Figure 2), and the results showed that YOC-1 had a close relationship with AmpC derived from Raoultella terrigena.
Functional Characterization of the YOC-1 β-Lactamase
Chromosomal AmpC enzymes, usually inducible, are often expressed at low levels and therefore may not contribute to clinical β-lactam antibiotic resistance (Wu et al., 1999). To assess the potential resistance activities to β-lactams, the coding sequence of blaYOC–1 together with its promoter region was cloned into the pUCP24 vector and then transformed into E. coli DH5α. The MICs of penicillin G, ampicillin and several first- to fourth-generation cephalosporins against the recombinant strain containing blaYOC–1 (pUCP24-blaYOC–1/DH5α) increased 4- and 256-fold compared with the control strains (DH5α and DH5α carrying the vector pUCP24). The MIC of benzylpenicillin increased more than 64-fold, the MICs of ceftriaxone, ampicillin and cefoxitin increased at least 256-, 64-, and 32-fold, respectively, and the MICs of cefazolin, ceftazidime and cefoperazone increased 128-fold (Table 4). The results showed that the increase in the MIC of ceftazidime was higher than that of cefotaxime, and ceftazidime is documented to induce AmpC cephalosporinases, which may be the cause of the phenomenon (Jones, 1998). blaYOC–1 did not confer resistance to carbapenems, which is different from CDA-1; this result may be due to some mutations in conversed residues associated with the hydrolysis of carbapenem. The antibiotic resistance spectrum of YOC-1 is consistent with most members of the family Enterobacteriaceae with an inducible chromosomal ampC gene, which are usually not resistant to extended-spectrum cephalosporins, unless the ampC gene is expressed at a very high level. The antibiotic resistance spectrum of YOC-1 is similar to that of LHK-5, a β-lactamase from a clinical L. hongkongensis isolate (Lau et al., 2005).
Similar to other chromosome- and plasmid-encoded class C β-lactamases, the catalytic efficiency (kcat/Km) of the purified β-lactamase was high against first- and second-generation cephalosporins, such as cefazolin and cefoxitin, but low against extended-spectrum cephalosporins, such as cefepime (Horii et al., 1993; Raimondi et al., 2001; Lau et al., 2005; Zioga et al., 2008). The highest catalytic efficiency was observed with penicillin G (kcat/Km rate of 191.5), which is similar to CMY-2 (Kotsakis et al., 2015). YOC-1 was more active against cefoxitin and ceftazidime than CMY-2 because of its much higher kcat, which was partially compensated for by higher Km values. The turnover rate (kcat) of YOC-1 for ceftazidime was higher than that of CDA-1, which shared 71.69% global amino acid identity with YOC-1 (Ammenouche et al., 2014; Kotsakis et al., 2015). However, the Michaelis constant (Km) of YOC-1 for cefepime was much lower than that of CDA-1. In addition, no measurable hydrolytic activities were observed for aztreonam or imipenem, which is different from CDA-1 (Ammenouche et al., 2014; Table 5).
The hydrolysis of aztreonam was not detectable for the purified enzyme, which was contradicted by the increase in the MIC value of aztreonam against the recombinant carrying blaYOC–1, which may have been an experimental error in MIC determination. This discrepancy between phenotypic and biochemical approaches could also result from the low but not zero deacylation rate of AmpC β-lactamases with those compounds. A similar phenomenon was observed for CDA-1, whose host showed resistance to ertapenem, but the β-lactamase did not have hydrolytic activity against ertapenem (Ammenouche et al., 2014).
The results of β-lactamase activity inhibition analysis, as measured by the IC50 (50% inhibitory concentration), showed that YOC-1 was poorly inhibited by clavulanic acid (IC50: 57.33 ± 0.03 μM) and GMP (IC50: 81.73 ± 0.02 μM) and was strongly inhibited by avibactam (IC50: 0.001172 ± 0.0002 μM), which is consistent with the majority of class C β-lactamases (Bonnefoy et al., 2004; Na et al., 2017), especially CMY-2 (Bauernfeind et al., 1996; Na et al., 2017), which is the most prevalent AmpC (Doi et al., 2009).
Comparative Genomic Analysis of the Genetic Context of the BlaYOC–1-Encoding Region
To compare the genetic environments of blaYOC–1 in Y. regensburgei W13 with those of the closest homologous ampC-type genes (identity > 80%), the chromosome sequences of three Raoultella spp. strains and one Klebsiella aerogenes strain were retrieved from the NCBI nucleotide database. The ampC-ampR genetic locus was conserved but located in quite different genetic environments, and there were no obvious sequence similarities between the blaYOC–1-ampR-encoding fragment and each of the other four fragments. Moreover, no sequence showing similarity with the blaYOC–1-ampR-encoding fragment among the completely sequenced bacterial genomes was retrieved from the NCBI nucleotide database. Furthermore, the region encoding pdeG-blaYOC–1-ampR-nimR-hp was flanked by a pair of perfect 7 bp direct repeats (DRs), indicating that the gene array might be mobile between strains of different species or genera through horizontal gene transfer (HGT). This exclusive gene content of the blaYOC–1-ampR-encoding region was probably ascribed to the scarcity of completely sequenced genomes of Yokenella or other closely related species. Nevertheless, it could not be ruled out that this might be caused by HGT events (Figure 3).
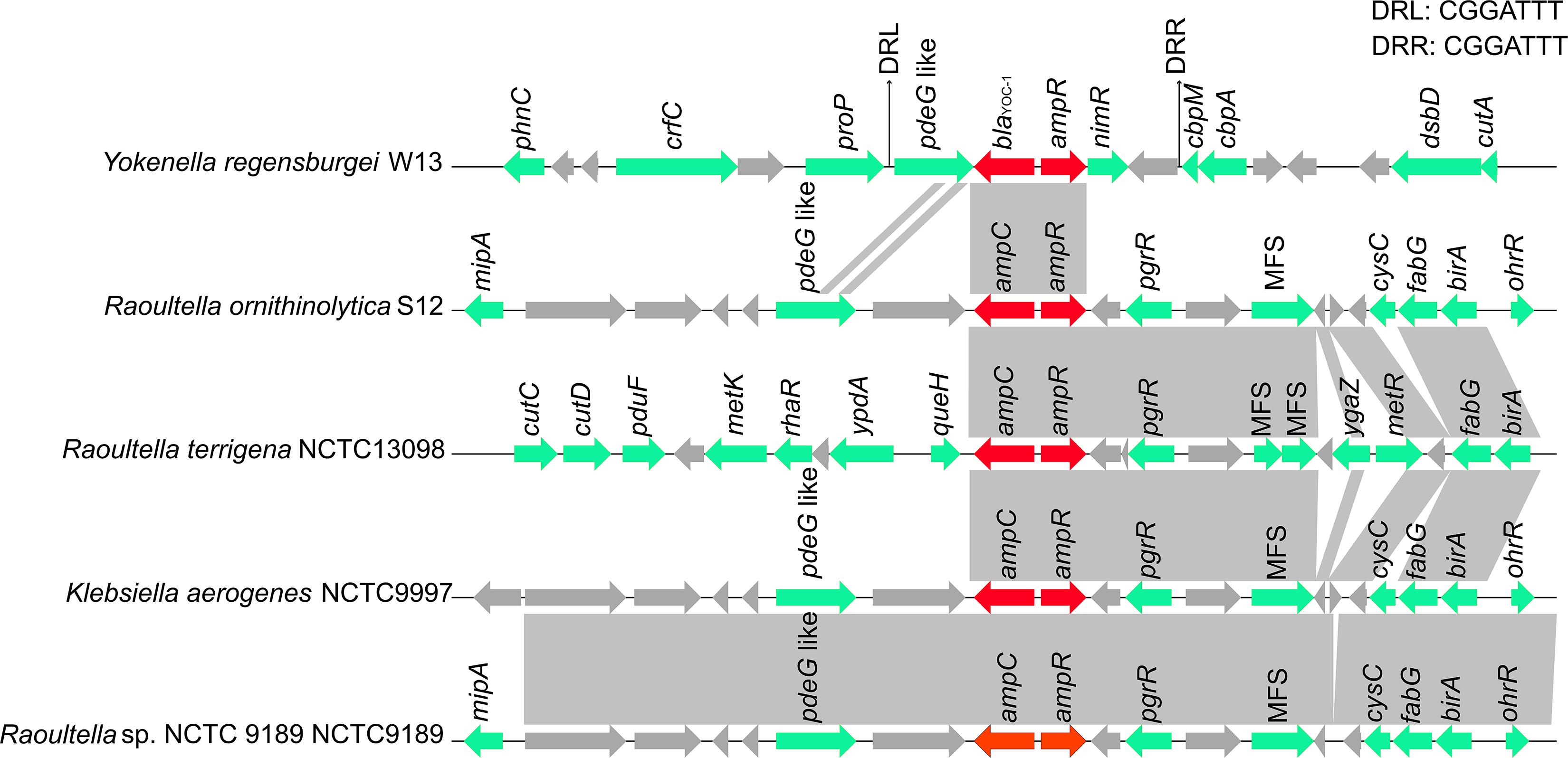
Figure 3. Comparison of the genetic environment of the blaYOC–1 gene with those carrying its homologous genes. Genes are shown as arrows and colored based on gene function classification. Genes without functional annotation are colored in gray. Shading denotes homologous regions between them. MFS represents the genes encoding major facilitator superfamily (MFS) transport proteins.
Comparative Genomics Analysis of the Plasmid Carrying Multiple Resistance Genes
In this study, 11 resistance genes related to different mobile genetic elements were identified in the plasmid pYRW13-125, including two β-lactam resistance genes (blaOXA–10 and blaLAP–2), two tetracycline resistance genes (tetA and tetR), one chloramphenicol resistance gene (cmlA5), one chloramphenicol/florfenicol resistance gene (floR), one sulfanilamide resistance gene (sul2), one aminoglycoside resistance gene (ant(3″)-IIa), one diaminopyrimidine resistance gene (dfrA14), one rifampicin resistance gene (arr-2) and one quinolone resistance gene (qnrS1) (Figure 1). The plasmid was transferable and could be successfully transferred into recipient cells (E. coli C600) through conjugation.
Comparative genome analysis revealed that pYRW13-125 shared the highest identity with three plasmids, including an unnamed plasmid of Enterobacter hormaechei BW (CP027111, 81% coverage and 99.98% identity), p34399 of Enterobacter hormaechei subsp. Xiangfangensis (CP010385, 75% coverage and 100.00% identity) and pOP-I of Enterobacter cloacae subsp. Cloacae MN201516 (KY126370, 64% coverage and 94.49% identity). The sequence differences among these four plasmids were mainly located in the multidrug resistance (MDR) region (9.70–44.58 kb) of pYRW13-125 (Figure 1).
To better characterize the MDR region, the MDR region of pYRW13-125 was comparatively analyzed with other related sequences. The results showed that the MDR region of pYRW13-125 shared the highest nucleotide sequence similarities with a plasmid from a clinical K. pneumoniae strain AR_0152 (CP021946, 99.80% identity and 84% coverage) and a plasmid from the clinical K. pneumoniae strain 130411-38618 (MK649826, 99.88% identity and 61% coverage). The MDR region of pYRW13-125 was a complex mosaic structure consisting of 3 mobile genetic element-related units (Figure 4), including a truncated IS26-blaLAP–2-qnrS1-tetA-tetR-IS26 unit, an atypical class 1 integron, an ISCR2-floR-sul2 unit. The plasmid tig00000195 harbors a complete IS26-blaLAP–2-qnrS1-tetA-tetR-IS26 unit, and the presence of TnAs1 remnant indicated that this unit might be a result of recombination of two subregions (IS26-blaLAP–2-qnrS1-IS26 unit and IS26-tetA-tetR-IS26 unit) mediated by TnAs1. Furthermore, the atypical class 1 integron (organized with a 5′-conserved segment [5′-CS: intI1], a variable region [VR: arr-2, cmlA5, blaOXA–10, ant(3″)-IIa and dfrA14]) without a 3′-CS on pYRW13-125 was identical to that on p130411-38618_1. The integron did not contain the 3′-CS (qacEΔ1/sul1) usually found in clinical class 1 integrons, suggesting that this could be a preclinical integron (Ghaly et al., 2017). The integron in pYRW13-125 was immediately flanked by a pair of IS26s, suggesting the potential of its mobility. ISCR2 was found upstream of floR and downstream of sul2 on pYRW13-125. These elements are known to move by a process called rolling-circle replication, and a function of this process is the concomitant movement of accessory sequences found upstream of their transposase genes (Toleman and Walsh, 2010). The presence of ISCR2 makes the transmission of these two genes possible. The genetic relatedness of the MDR region of pYRW13-125 with those of the two plasmids of clinical strains indicated that the environmental strain W13 might be closely related to multidrug resistant bacteria of human clinical sources.

Figure 4. Comparison of the MDR region of pW13-125 with the related regions on the other plasmids. Genes are denoted by arrows and colored according to gene function classification. Shading indicates the homologous regions. The detected direct repeat sequence is marked in the upper right corner.
Conclusion
In this study, the first complete genome sequence of Y. regensburgei was reported. Apart from a novel chromosome-encoded AmpC β-lactamase gene, blaYOC–1, the strain also harbored a plasmid (pYRW13-125) encoding multidrug resistance gene arrays that conferred resistance to a series of antibiotics, including tetracycline, amphenicols (chloramphenicol and florfenicol) and streptomycin. Among all of the recently reported functionally characterized β-lactamases, the novel β-lactamase YOC-1 shares the highest amino acid identities of 71.69 and 70.65% with CDA-1 and CMY-2, respectively. The newly identified AmpC β-lactamase encoded in Y. regensburgei W13, which carries a multidrug-resistant plasmid, will be helpful for understanding the intrinsic resistance mechanism of this unusual opportunistic pathogen and developing treatments for human (or animal) infections caused by Y. regensburgei.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/genbank/, MT271603.
Author Contributions
DZ, ZS, JL, HoL, WL, HaL, XuZ, and QL collected the strains and performed the experiments. DZ, ZS, HX, XL, HZ, TX, and KL analyzed the experimental results and performed the bioinformatics analysis. DZ, ZS, TX, KL, and QB wrote the manuscript. HZ, TX, KL, and QB designed the experiments. All authors read and approved the final manuscript.
Funding
This study was supported by the Natural Science Foundation of Zhejiang Province, China (LY19C060002 and LQ17H190001), the National Natural Science Foundation of China (81973382), and the Science & Technology Project of Wenzhou City, China (Y20170205 and 2019Y0358).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to acknowledge all study participants and individuals who contributed to the study.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.02021/full#supplementary-material
FIGURE S1 | Amino acid alignment of YOC-1 with carbapenem-resistant AmpCs. Dots indicate the absence of amino acid residues. The accession numbers of the cephalosporinases (from up to down) are as follows: QJD09144.1, AID52933.1, AIM49619.1, AAK59969.1, WP_023331841.1. The logo size indicates the degree of conservation of the amino acid residues. The amino acids in proteins with ≥80% identity are shaded in blue. The length of each AmpC protein is marked at the end of the alignment. Conserved motifs are indicated at the top of the sequence alignment. The red star indicates the unique residue in YOC-1, and the predicted signal sequence cleavage site for YOC-1 is indicated by a vertical arrow.
Footnotes
- ^ https://card.mcmaster.ca/
- ^ https://web.expasy.org/protparam/
- ^ www.cbs.dtu.dk/services/SignalP/
- ^ http://dx.doi.org/10.1111/1574-6968.12199
References
Abbott, S. L., and Janda, J. M. (1994). Isolation of Yokenella regensburgei (“Koserella trabulsii”) from a patient with transient bacteremia and from a patient with a septic knee. J. Clin. Microbiol. 32, 2854–2855. doi: 10.1128/jcm.32.11.2854-2855.1994
Ammenouche, N., Dupont, H., and Mammeri, H. (2014). Characterization of a novel AmpC beta-lactamase produced by a carbapenem-resistant Cedecea davisae clinical isolate. Antimicrob. Agents Chemother. 58, 6942–6945. doi: 10.1128/AAC.03237-14
Bauernfeind, A., Stemplinger, I., Jungwirth, R., and Giamarellou, H. (1996). Characterization of the plasmidic beta-lactamase CMY-2, which is responsible for cephamycin resistance. Antimicrob. Agents Chemother. 40, 221–224. doi: 10.1128/aac.40.1.221
Bhowmick, T., and Weinstein, M. P. (2013). A deceptive case of cellulitis caused by a Gram-negative pathogen. J. Clin. Microbiol. 51, 1320–1323. doi: 10.1128/JCM.02975-12
Bonnefoy, A., Dupuis-Hamelin, C., Steier, V., Delachaume, C., Seys, C., Stachyra, T., et al. (2004). In vitro activity of AVE1330A, an innovative broad-spectrum non-beta-lactam beta-lactamase inhibitor. J. Antimicrob. Chemother. 54, 410–417. doi: 10.1093/jac/dkh358
Buchfink, B., Xie, C., and Huson, D. H. (2015). Fast and sensitive protein alignment using DIAMOND. Nat. Methods 12, 59–60. doi: 10.1038/nmeth.3176
Chen, Q., Zhou, W., Qian, C., Shen, K., Zhu, X., Zhou, D., et al. (2019). OXA-830, a novel chromosomally encoded extended-spectrum class D β-lactamase in Aeromonas simiae. Front. Microbiol. 10:2732. doi: 10.3389/fmicb.2019.02732
Chi, X., Liu, M., and Chen, Y. (2017). Yokenella regensburgei septicemia in a Chinese farmer immunosuppressed by HIV: a case report and literature review. Case Rep. Infect. Dis. 2017, 5962463. doi: 10.1155/2017/5962463
Choi, T. J., and Geletu, T. T. (2018). High level expression and purification of recombinant flounder growth hormone in E. coli. J. Genet. Eng. Biotechnol. 16, 347–355. doi: 10.1016/j.jgeb.2018.03.006
CLSI (2019). Performance Standards for Antimicrobial Susceptibility Testing, 29th Edn. Wayne, PA: Clinical and Laboratory Standards Institute.
De Toro, M., Garcillán-Barcia, M. P., De La Cruz, F. (2015). “Plasmid diversity and adaptation analyzed by massive sequencing of Escherichia coli plasmids,” in Plasmids, eds Tolmasky, M. E., and Alonso, J. C. 219–235. doi: 10.1128/9781555818982.ch13
Decre, D., Verdet, C., Raskine, L., Blanchard, H., Burghoffer, B., Philippon, A., et al. (2002). Characterization of CMY-type beta-lactamases in clinical strains of Proteus mirabilis and Klebsiella pneumoniae isolated in four hospitals in the Paris area. J. Antimicrob. Chemother. 50, 681–688. doi: 10.1093/jac/dkf193
Doi, Y., Paterson, D. L., Adams-Haduch, J. M., Sidjabat, H. E., O’Keefe, A., Endimiani, A., et al. (2009). Reduced susceptibility to cefepime among Escherichia coli clinical isolates producing novel variants of CMY-2 beta-lactamase. Antimicrob. Agents Chemother. 53, 3159–3161. doi: 10.1128/AAC.00133-09
Doi, Y., Shibata, N., Shibayama, K., Kamachi, K., Kurokawa, H., Yokoyama, K., et al. (2002). Characterization of a novel plasmid-mediated cephalosporinase (CMY-9) and its genetic environment in an Escherichia coli clinical isolate. Antimicrob. Agents Chemother. 46, 2427–2434. doi: 10.1128/aac.46.8.2427-2434.2002
Gao, R., Wang, Q., Li, P., Li, Z., and Feng, Y. (2016). Genome sequence and characteristics of plasmid pWH12, a variant of the mcr-1-harbouring plasmid pHNSHP45, from the multi-drug resistant E. coli. Virulence 7, 732–735. doi: 10.1080/21505594.2016.1193279
Ghaly, T. M., Chow, L., Asher, A. J., Waldron, L. S., and Gillings, M. R. (2017). Evolution of class 1 integrons: mobilization and dispersal via food-borne bacteria. PLoS One 12:e0179169. doi: 10.1371/journal.pone.0179169
Hickman-Brenner, F. W., Huntley-Carter, G. P., Fanning, G. R., Brenner, D. J., and Farmer, J. (1985). Koserella trabulsii, a new genus and species of Enterobacteriaceae formerly known as enteric group 45. J. Clin. Microbiol. 21, 39–42. doi: 10.1128/jcm.21.1.39-42.1985
Horii, T., Arakawa, Y., Ohta, M., Ichiyama, S., Wacharotayankun, R., Kato, N., et al. (1993). Plasmid-mediated AmpC-type beta-lactamase isolated from Klebsiella pneumoniae confers resistance to broad-spectrum beta-lactams, including moxalactam. Antimicrob. Agents Chemother. 37, 984–990. doi: 10.1128/aac.37.5.984
Jachymek, W., Niedziela, T., Petersson, C., Lugowski, C., Czaja, J., and Kenne, L. (1999). Structures of the O-specific polysaccharides from Yokenella regensburgei (Koserella trabulsii) strains PCM 2476, 2477, 2478, and 2494: high-resolution magic-angle spinning NMR investigation of the O-specific polysaccharides in native lipopolysaccharides and directly on the surface of living bacteria. Biochemistry 38, 11788–11795. doi: 10.1021/bi990673y
Jacoby, G. A. (2009). AmpC beta-lactamases. Clin. Microbiol. Rev. 22, 161–82,TableofContents. doi: 10.1128/CMR.00036-08
Jain, S., Gaind, R., Gupta, K. B., Dawar, R., Kumar, D., Paul, P., et al. (2013). Yokenella regensburgei infection in India mimicking enteric fever. J. Med. Microbiol. 62(Pt 6), 935–939. doi: 10.1099/jmm.0.058669-0
Jones, R. N. (1998). Important and emerging β-lactamase-mediated resistances in hospital-based pathogens: the Amp C enzymes. Diagn. Microbiol. Infect. Dis. 31, 461–466. doi: 10.1016/s0732-8893(98)00029-7
Jousset, A. B., Oueslati, S., Bernabeu, S., Takissian, J., Creton, E., Vogel, A., et al. (2019). False-positive carbapenem-hydrolyzing confirmatory tests due to ACT-28, a chromosomally encoded AmpC with weak carbapenemase activity from Enterobacter kobei. Antimicrob. Agents Chemother. 63:e02388-18.
Kim, J. Y., Jung, H. I., An, Y. J., Lee, J. H., Kim, S. J., Jeong, S. H., et al. (2006). Structural basis for the extended substrate spectrum of CMY-10, a plasmid-encoded class C β-lactamase. Mol. Microbiol. 60, 907–916. doi: 10.1111/j.1365-2958.2006.05146.x
Koren, S., Walenz, B. P., Berlin, K., Miller, J. R., Bergman, N. H., and Phillippy, A. M. (2017). Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 27, 722–736. doi: 10.1101/gr.215087.116
Kosako, Y., Sakazaki, R., Huntley-Carter, G. P., and Farmer, J. J. I. I. I. (1987). Yokenella regensburgei and Koserella trabulsii are subjective synonyms. Int. J. Syst. Bacteriol. 37, 127–129. doi: 10.1099/00207713-37-2-127
Kosako, Y., Sakazaki, R., and Yoshizaki, E. (1984). Yokenella regensburgei gen. nov., sp. nov.: a new genus and species in the family Enterobacteriaceae. Jpn. J. Med. Sci. Biol. 37, 117–124. doi: 10.7883/yoken1952.37.117
Kotsakis, S. D., Miriagou, V., Vetouli, E. E., Bozavoutoglou, E., Lebessi, E., Tzelepi, E., et al. (2015). Increased hydrolysis of oximino-beta-lactams by CMY-107, a Tyr199Cys mutant form of CMY-2 produced by Escherichia coli. Antimicrob. Agents Chemother. 59, 7894–7898. doi: 10.1128/AAC.01793-15
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Lau, S. K., Ho, P. L., Li, M. W., Tsoi, H. W., Yung, R. W., Woo, P. C., et al. (2005). Cloning and characterization of a chromosomal class C beta-lactamase and its regulatory gene in Laribacter hongkongensis. Antimicrob. Agents Chemother. 49, 1957–1964. doi: 10.1128/AAC.49.5.1957-1964.2005
Lee, I., Ouk Kim, Y., Park, S. C., and Chun, J. (2016). OrthoANI: an improved algorithm and software for calculating average nucleotide identity. Int. J. Syst. Evol. Microbiol. 66, 1100–1103. doi: 10.1099/ijsem.0.000760
Li, H., and Durbin, R. (2009). Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760. doi: 10.1093/bioinformatics/btp324
McArthur, A. G., Waglechner, N., Nizam, F., Yan, A., Azad, M. A., Baylay, A. J., et al. (2013). The comprehensive antibiotic resistance database. Antimicrob. Agents Chemother. 57, 3348–3357. doi: 10.1128/AAC.00419-13
McKenna, A., Hanna, M., Banks, E., Sivachenko, A., Cibulskis, K., Kernytsky, A., et al. (2010). The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303. doi: 10.1101/gr.107524.110
McWhorter, A., Haddock, R., Nocon, F. A., Steigerwalt, A. G., Brenner, D., Aleksiæ, S., et al. (1991). Trabulsiella guamensis, a new genus and species of the family Enterobacteriaceae that resembles Salmonella subgroups 4 and 5. J. Clin. Microbiol. 29, 1480–1485. doi: 10.1128/jcm.29.7.1480-1485.1991
Medaney, F., Ellis, R. J., and Raymond, B. (2016). Ecological and genetic determinants of plasmid distribution in Escherichia coli. Environ. Microbiol. 18, 4230–4239. doi: 10.1111/1462-2920.13552
Meini, S., Tascini, C., Cei, M., Sozio, E., and Rossolini, G. M. (2019). AmpC beta-lactamase-producing Enterobacterales: what a clinician should know. Infection 47, 363–375. doi: 10.1007/s15010-019-01291-9
Moura, A., Soares, M., Pereira, C., Leitao, N., Henriques, I., and Correia, A. (2009). INTEGRALL: a database and search engine for integrons, integrases and gene cassettes. Bioinformatics 25, 1096–1098. doi: 10.1093/bioinformatics/btp105
Na, J. H., An, Y. J., and Cha, S. S. (2017). GMP and IMP are competitive inhibitors of CMY-10, an extended-spectrum class C beta-Lactamase. Antimicrob. Agents Chemother. 61:e00098-17. doi: 10.1128/AAC.00098-17
Palmisano, S., Campisciano, G., Silvestri, M., Guerra, M., Giuricin, M., Casagranda, B., et al. (2019). Changes in gut microbiota composition after bariatric surgery: a new balance to decode. J. Gastrointest. Surg. 24, 1736–1746. doi: 10.1007/s11605-019-04321-x
Petkau, A., Stuart-Edwards, M., Stothard, P., and Van Domselaar, G. (2010). Interactive microbial genome visualization with GView. Bioinformatics 26, 3125–3126. doi: 10.1093/bioinformatics/btq588
Raimondi, A., Sisto, F., and Nikaido, H. (2001). Mutation in Serratia marcescens AmpC beta-lactamase producing high-level resistance to ceftazidime and cefpirome. Antimicrob. Agents Chemother. 45, 2331–2339. doi: 10.1128/AAC.45.8.2331-2339.2001
Seemann, T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069. doi: 10.1093/bioinformatics/btu153
Siguier, P., Perochon, J., Lestrade, L., Mahillon, J., and Chandler, M. (2006). ISfinder: the reference centre for bacterial insertion sequences. Nucl. Acids Res. 34, D32–D36. doi: 10.1093/nar/gkj014
Sullivan, M. J., Petty, N. K., and Beatson, S. A. (2011). Easyfig: a genome comparison visualizer. Bioinformatics 27, 1009–1010. doi: 10.1093/bioinformatics/btr039
Suzuki, Y., Ida, M., Kubota, H., Ariyoshi, T., Murakami, K., Kobayashi, M., et al. (2019). Multiple beta-Lactam resistance gene-carrying plasmid harbored by Klebsiella quasipneumoniae isolated from urban sewage in Japan. mSphere 4:e00391-19. doi: 10.1128/mSphere.00391-19
Toleman, M. A., and Walsh, T. R. (2010). ISCR elements are key players in IncA/C plasmid evolution. Antimicrob. Agents Chemother. 54:3534. doi: 10.1128/aac.00383-10
Wright, W. F., Utz, J. L., Bruckhart, C., Baghli, S., and Janda, J. M. (2019). Yokenella regensburgei necrotizing fasciitis in an immunocompromised host. J. Infect. Chemother. 25, 816–819. doi: 10.1016/j.jiac.2019.03.025
Wu, S. W., Dornbusch, K., Kronvall, G., and Norgren, M. (1999). Characterization and nucleotide sequence of a Klebsiella oxytoca cryptic plasmid encoding a CMY-type β-lactamase: confirmation that the plasmid-mediated cephamycinase originated from the Citrobacter freundii AmpC β-lactamase. Antimicrob. Agents Chemother. 43, 1350–1357. doi: 10.1128/aac.43.6.1350
Zankari, E., Hasman, H., Cosentino, S., Vestergaard, M., Rasmussen, S., Lund, O., et al. (2012). Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 67, 2640–2644. doi: 10.1093/jac/dks261
Keywords: Yokenella regensburgei, β-lactamase, blaYOC–1, kinetic analysis, plasmid
Citation: Zhou D, Sun Z, Lu J, Liu H, Lu W, Lin H, Zhang X, Li Q, Zhou W, Zhu X, Xu H, Lin X, Zhang H, Xu T, Li K and Bao Q (2020) Characterization of a Novel Chromosomal Class C β-Lactamase, YOC-1, and Comparative Genomics Analysis of a Multidrug Resistance Plasmid in Yokenella regensburgei W13. Front. Microbiol. 11:2021. doi: 10.3389/fmicb.2020.02021
Received: 11 April 2020; Accepted: 30 July 2020;
Published: 20 August 2020.
Edited by:
Kristina Kadlec, Independent Researcher, Wunstorf, GermanyReviewed by:
Marco Maria D’Andrea, University of Rome Tor Vergata, ItalySun-Shin Cha, Ewha Womans University, South Korea
Copyright © 2020 Zhou, Sun, Lu, Liu, Lu, Lin, Zhang, Li, Zhou, Zhu, Xu, Lin, Zhang, Xu, Li and Bao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Teng Xu, eHV0ZW5nQHdtdS5lZHUuY24=; Kewei Li, Y3Vyd2F5bGVlQGFsaXl1bi5jb20=; Y3Vyd2F5bGVlQHdtdS5lZHUuY24=; Qiyu Bao, YmFvcXlAZ2Vub21pY3MuY24=
†These authors have contributed equally to this work
 Danying Zhou
Danying Zhou Zhewei Sun
Zhewei Sun Junwan Lu
Junwan Lu Hongmao Liu
Hongmao Liu Wei Lu
Wei Lu Hailong Lin1,2,3
Hailong Lin1,2,3 Xueya Zhang
Xueya Zhang Xi Lin
Xi Lin Hailin Zhang
Hailin Zhang Teng Xu
Teng Xu Qiyu Bao
Qiyu Bao