- 1Department of Oral and Molecular Microbiology, Osaka University Graduate School of Dentistry, Suita, Japan
- 2Department of Oral and Maxillofacial Surgery II, Osaka University Graduate School of Dentistry, Suita, Japan
Streptococcus pneumoniae is a major cause of pneumonia, sepsis, and meningitis. Previously, we identified a novel virulence factor by investigating evolutionary selective pressure exerted on pneumococcal choline-binding cell surface proteins. Herein, we focus on another pneumococcal cell surface protein. Cell wall-anchoring proteins containing the LPXTG motif are conserved in Gram-positive bacteria. Our evolutionary analysis showed that among the examined genes, nanA and bgaA had high proportions of codon that were under significant negative selection. Both nanA and bgaA encode a multi-functional glycosidase that aids nutrient acquisition in a glucose-poor environment, pneumococcal adherence to host cells, and evasion from host immunity. However, several studies have shown that the role of BgaA is limited in a mouse pneumonia model, and it remains unclear if BgaA affects pneumococcal pathogenesis in a mouse sepsis model. To evaluate the distribution and pathogenicity of bgaA, we performed phylogenetic analysis and intravenous infection assay. In both Bayesian and maximum likelihood phylogenetic trees, the genetic distances between pneumococcal bgaA was small, and the cluster of pneumococcal bgaA did not contain other bacterial orthologs except for a Streptococcus gwangjuense gene. Evolutionary analysis and BgaA structure indicated BgaA active site was not allowed to change. The mouse infection assay showed that the deletion of bgaA significantly reduced host mortality. These results indicated that both nanA and bgaA encode evolutionally conserved pneumococcal virulence factors and that molecular evolutionary analysis could be a useful alternative strategy for identification of virulence factors.
Introduction
Streptococcus pneumoniae is one of most frequently isolated bacteria from community acquired pneumonia, sepsis, and bacterial meningitis (Ishiguro et al., 2013; CDC, 2019). The phylogenetic relationship of both 16S rRNA and a core set of 136 genes indicated that this pathogen belongs to mitis group of Streptococcus (Kawamura et al., 1995; Richards et al., 2014). S. pneumoniae is capable of importing various genes including antimicrobial resistance genes via horizontal transfer from related species (Salvadori et al., 2019). In the United States of America, more than 30% of clinically isolated pneumococcal bacteria are resistant to one or more antibiotics (CDC, 2019). Currently, 23-valent pneumococcal polysaccharide and 13-valent pneumococcal-conjugated vaccines are in use in various countries. These vaccines prevent pneumococcal infections caused by vaccine-targeted serotype strains and inhibit the spread of drug resistant strains (CDC, 2019). On the other hand, the selective pressure imposed by vaccination has increased the emergence of non-vaccine serotype strains (Golubchik et al., 2012).
Recently, we applied a combination of evolutionary analysis and laboratory-based approaches to evaluate the functional significance of putative virulence factors (Yamaguchi et al., 2016, 2019a; Yamaguchi, 2018). As mutations in non-essential but important genes promote the selection of bacterial lineages in single species, genes that undergo considerable negative selection would be important for the survival and/or success of the species in its host and/or the environment. Thus, a molecular evolutionary approach enables us to estimate the contributions of bacterial proteins to species success throughout its life cycle. We have previously focused on the evolutionary selective pressures on choline-binding proteins (CBPs) in S. pneumoniae (Yamaguchi et al., 2019a). CBPs are localized to the cell surface by binding to phosphoryl choline on the cell wall. As cell surface proteins are easily and directly accessible to the external environment, these proteins could represent attractive antigen candidates for vaccine development. Our analysis revealed that CbpJ contributes to evasion of host neutrophil-mediated killing in pneumococcal pneumonia. This is surprising as CbpJ has no known functional domains apart from signal sequences and choline-binding repeats. While there are also other types of pneumococcal cell surface proteins, their degree of evolutionary conservation remains unknown.
For this study, we focused on different motif involved in pneumococcal cell surface localization, LPXTG, which is associated with cell wall-anchoring (Lofling et al., 2011). LPXTG-containing proteins are covalently attached to the cell wall, and at least some of these proteins have been identified as multi-functional proteins (Yamaguchi et al., 2008; Uchiyama et al., 2009; Dalia et al., 2010; Lofling et al., 2011). Our evolutionary analysis indicated that the nanA and bgaA genes are under considerable negative selection pressure. The nanA gene encodes pneumococcal cell surface-localized exo-α-sialidase (NanA) that hydrolyzes α2-3-, α2-6-, and α2-8-linkages of N-acetylneuraminic acid residues, and bgaA encodes exo-β-galactosidase (BgaA) that hydrolyzes β1-4-linkages of galactose residues, respectively (Hobbs et al., 2018). These glycosidases contribute to biofilm formation in glucose-poor but galactose-rich environments such as the mouse nasopharynx (Blanchette et al., 2016). The glycosidases also disrupt complement deposition and reduce opsonophagocytic killing through catalytic activities (Dalia et al., 2010). In mouse intravenous infection, NanA contributes to pneumococcal invasion into the host central nervous system by aiding penetration through the blood–brain barrier (Uchiyama et al., 2009). However, whether BgaA functions as a virulence factor in vivo remains unknown. Thus, we performed a phylogenetic analysis and mouse intravenous infection assay to address this question.
Materials and Methods
Phylogenetic and Evolutionary Analyses
The tBLASTn search was used to identify homologs and orthologs of genes that encode cell wall-anchoring proteins (Gertz et al., 2006). Phylogenetic and evolutionary analyses were performed as previously described, with minor modifications (Yamaguchi et al., 2016, 2017, 2019a). Briefly, the sequences were aligned by codon using Phylogear2 (Tanabe, 2008), MAFFT v.7.221 with an L-INS-i strategy (Katoh and Standley, 2013), and Jalview (Waterhouse et al., 2009). Conserved common codons were used for further phylogenetic analysis. The best-fitting codon evolutionary models for MrBayes and RAxML analyses were determined using Kakusan4 (Tanabe, 2011). Bayesian Markov chain Monte Carlo analyses were performed using MrBayes v.3.2.5 or v.3.2.6 (Ronquist et al., 2012), and 2–8 × 106 generations were sampled. To validate phylogenetic inferences, maximum likelihood phylogenetic trees with bootstrap values were generated with RAxML v.8.1.20 (Stamatakis, 2014). Phylogenetic trees were visualized using FigTree v.1.4.4 (Rambaut, 2018). Evolutionary analyses were performed based on aligned orthologous regions of genes that encode cell wall-anchoring proteins and Bayesian phylogenetic trees with a two-rate fixed-effects likelihood function in the HyPhy software package (Pond et al., 2005). For the evolutionary analyses, the level of statistical significance was set at P < 0.1 with the HyPhy default setting.
TIGR4 BgaA protein structure was visualized using PyMOL 2.41. The PDB ID is 4CU6 (Singh et al., 2014). The domain structures were identified using MOTIF and Pfam (Kanehisa and Goto, 2000; Finn et al., 2014).
Bacterial Strains and Construction of Mutant Strains
S. pneumoniae strains were cultured at 37°C in Todd-Hewitt broth (BD Biosciences, Franklin Lakes, NJ, United States) supplemented with 0.2% yeast extract (THY; BD Biosciences). Spectinomycin (Wako Pure Chemical Industries, Osaka, Japan) was added to the medium to a concentration of 120 μg/mL for mutant selection and maintenance. The S. pneumoniae TIGR4 isogenic bgaA (ΔbgaA) mutant strain was generated as previously described (Mori et al., 2012; Yamaguchi et al., 2019b). Briefly, the upstream region of bgaA, an aad9 cassette, and the downstream region of bgaA were combined by PCR using the primers summarized in Supplementary Table 1. The PCR product was transformed with synthesized CSP2 to construct the mutant strains by double-crossover recombination (Bricker and Camilli, 1999). The mutation was confirmed by site-specific PCR with isolated genomic DNA from the mutant strains. For growth measurement, overnight cultures of each strain were back-diluted 3:100 into fresh THY and grown at 37°C. Growth was monitored by measuring the OD600 values every 30 min. The starting point was set at an OD600 value of approximately 0.1. The experiment was repeated three times and the data is provided as Supplementary Data 1.
Mouse Intravenous Infection Assays
All mouse experiments were conducted in accordance with animal protocols approved by the Animal Care and Use Committee of Osaka University Graduate School of Dentistry (28-002-0). The mouse infection assay was performed as previously described (Hirose et al., 2018; Yamaguchi et al., 2019a). Briefly, CD-1 mice (Slc:ICR, 6 weeks, female) were infected by tail vein injection with 1 × 106 CFUs of S. pneumoniae. Mouse survival was checked twice daily for 14 days. The experiment was repeated three times and the data is provided as Supplementary Data 2. The pooled data for the three experiments was compared using a log-rank test. Statistical analysis was performed using Prism v.7.0d or v.8.4.2 software (GraphPad, Inc., La Jolla, CA, United States). The level of significance for differences between groups was set at P < 0.05.
Results
Evolutionary Selective Pressures on Genes Encoding Cell Wall-Anchoring Proteins
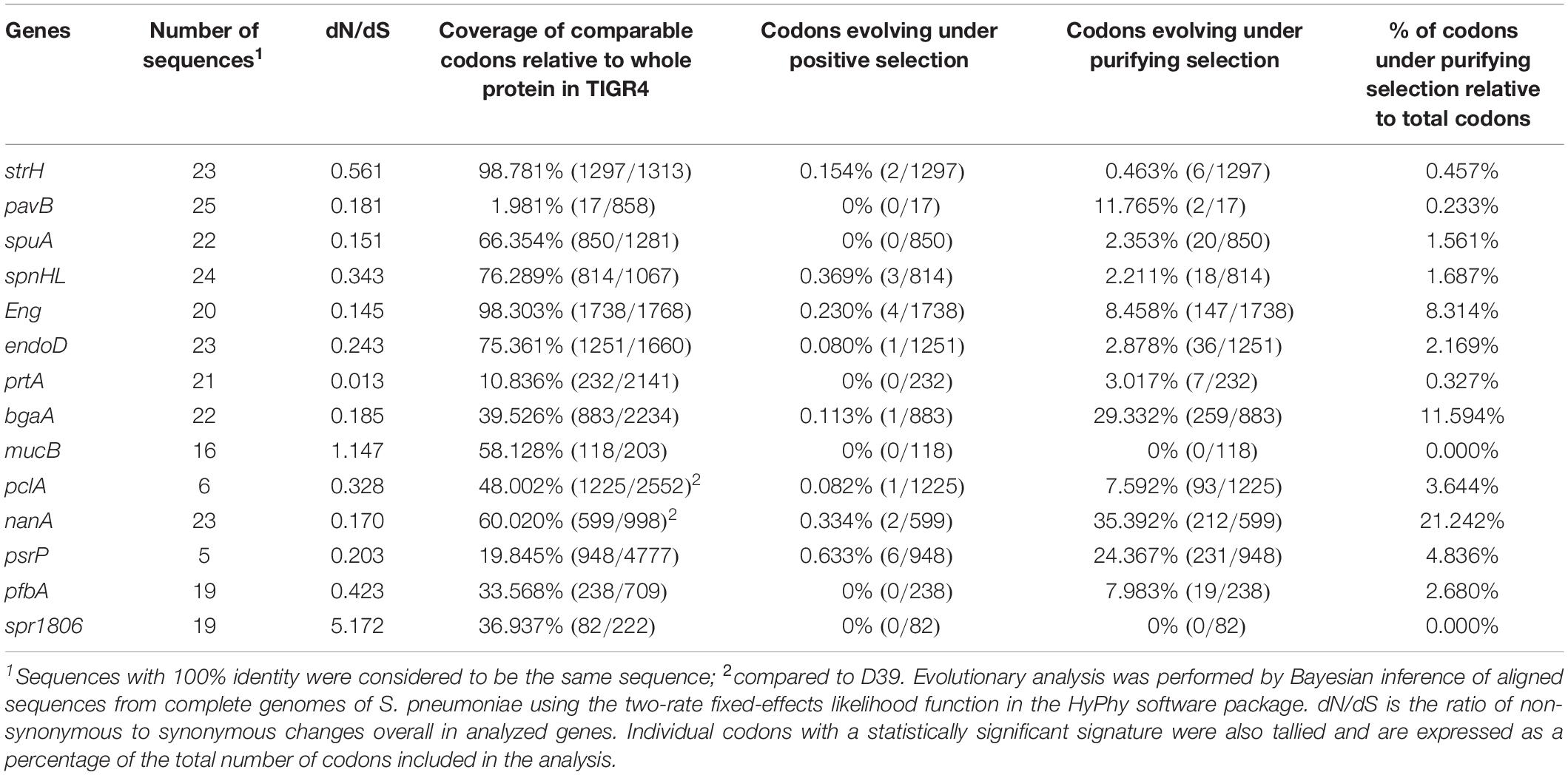
The tBLASTn function was used to search pneumococcal genomes for genes encoding cell wall-anchoring proteins (Figure 1 and Supplementary Table 2). While no genes were found to be conserved as intact open reading frames in all strains, all genes, save for pclA and psrP, were present in most strains. In particular, although the nanA gene in the TIGR4 strain contains a frameshift mutation in C-terminal region, TIGR4 NanA shows sialidase activity, and is assumed to be secreted into milieu instead of being anchored to the cell wall (Gut et al., 2011).
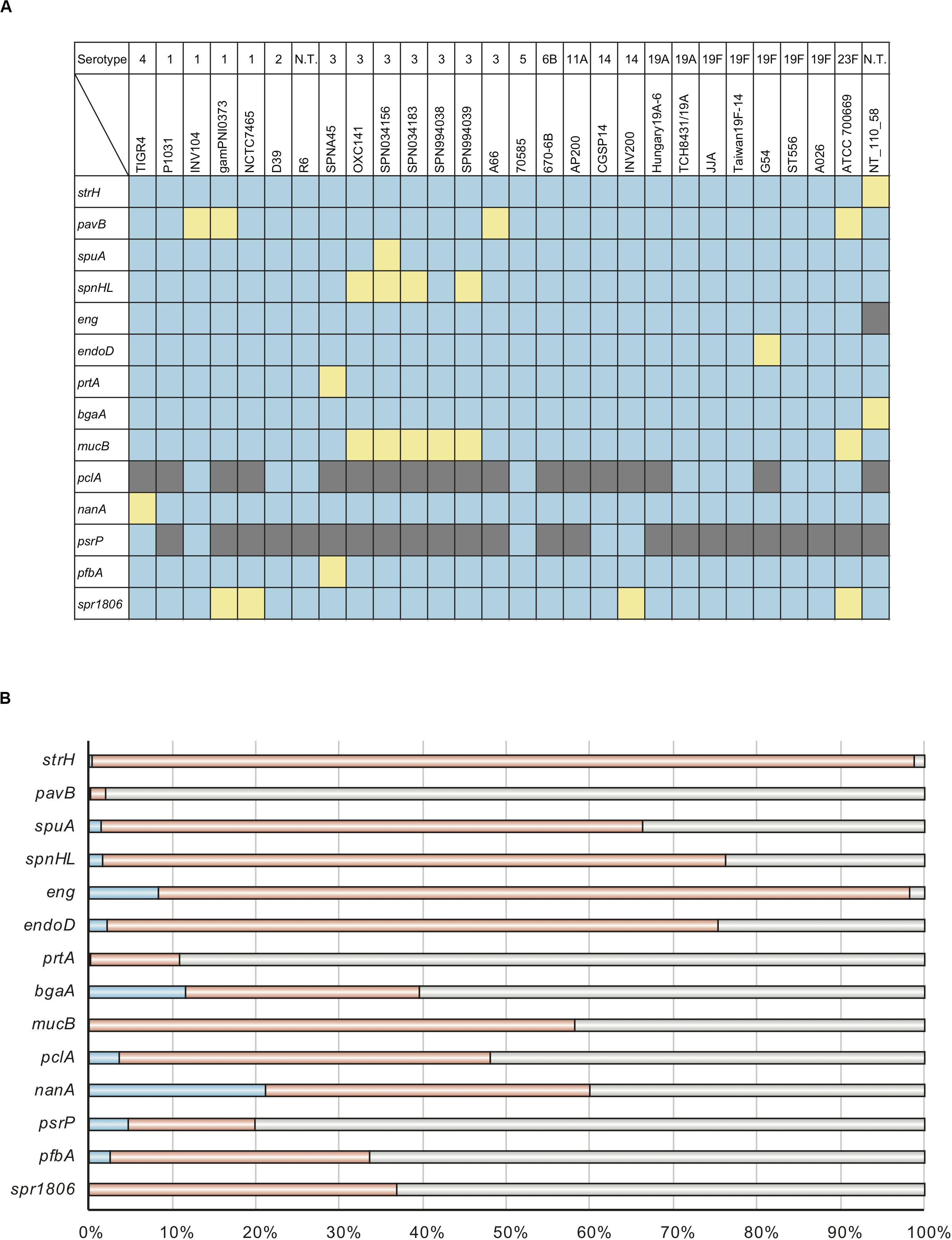
Figure 1. Distribution of genes that encode cell wall-anchoring proteins and percentage of codons that are evolutionarily conserved for these genes. (A) Distribution of genes that encode cell wall-anchoring proteins from pneumococcal strains. The gene locus tag numbers are summarized in Supplementary Table 2. Blue, yellow, and gray represent the presence, pseudogenization, and absence of genes, respectively. (B) Codons of genes encoding cell wall-anchoring proteins evolving under purifying selection were identified using HyPhy software with phylogenetic trees and aligned sequences. Blue, orange, and gray represent the percentage of codons under purifying selection, comparable common codons, and incomparable codons, respectively. The actual numbers and other parameters are listed in Table 1.
To evaluate the degree of evolutionary conservation in cell wall-anchoring proteins, we performed molecular evolutionary calculations based on each phylogenetic relationship and the DNA sequences aligned by codon. The calculated selective pressure for each gene is summarized in Table 1. The percentage of codons that are negatively selected for is visualized in Figure 1B. There was negative selection for over 11% of total codons in nanA and bgaA. This contrasts with less than 5% of total codons in most other genes, indicating that these two genes play an important role in the success of S. pneumoniae species. This same tendency was observed in our previous analysis on pneumococcal CBPs using the same genome sequences. Specifically, in the previous study, more than 13% of codons in the top two genes, cbpJ and lytA, were negatively selected (Yamaguchi et al., 2019a). Further, we previously reported that the pfbA gene showed high specificity to S. pneumoniae species and had a low level of sequence diversity (Yamaguchi et al., 2019b). Interestingly, our evolutionary analysis also indicated that the pfbA gene is under relaxed selective pressure.
Phylogenetic Relationships of the bgaA Gene
Evolutionary analysis indicated that the top two genes, nanA and bgaA, had high percentage of codons that were under negative selection pressure. We have previously reported that streptococcal nanA orthologs diverged into two major groups, with one group consisting of Streptococcus mitis, Streptococcus intermedius and S. pneumoniae, and the other consisting of Streptococcus agalactiae and the Streptococcus iniae groups (Yamaguchi et al., 2016). However, the phylogenetic relationships of bgaA have not been previously described. We used tBLASTn to search for bgaA-homologs, and performed Bayesian and maximum likelihood phylogenetic analyses (Figure 2 and Supplementary Figure 1). The tBLASTn search of the NCBI Nucleotide collection database showed that the bgaA gene homologs were identified in various streptococcal species and other Gram-positive bacteria of the phylum Firmicutes, including genus Clostridium and Bacillus. The β-galactosidase genes of these Gram-positive bacteria were used to root the phylogenetic trees. Bayesian and maximum likelihood phylogenetic analyses produced almost identical trees. The bgaA genes in S. pneumoniae and S. pseudopneumoniae had small genetic distances and formed an independent cluster within a cluster of streptococcal strains. In contrast, orthologous genes in other streptococcal species were genetically diverse. One of the bgaA orthologs from the Streptococcus gwangjuense strain ChDC B345 belonged to the pneumococcal clusters. As this gene is distinct from other S. gwangjuense genes, there is a possibility that it had been obtained from S. pneumoniae via horizontal gene transfer.
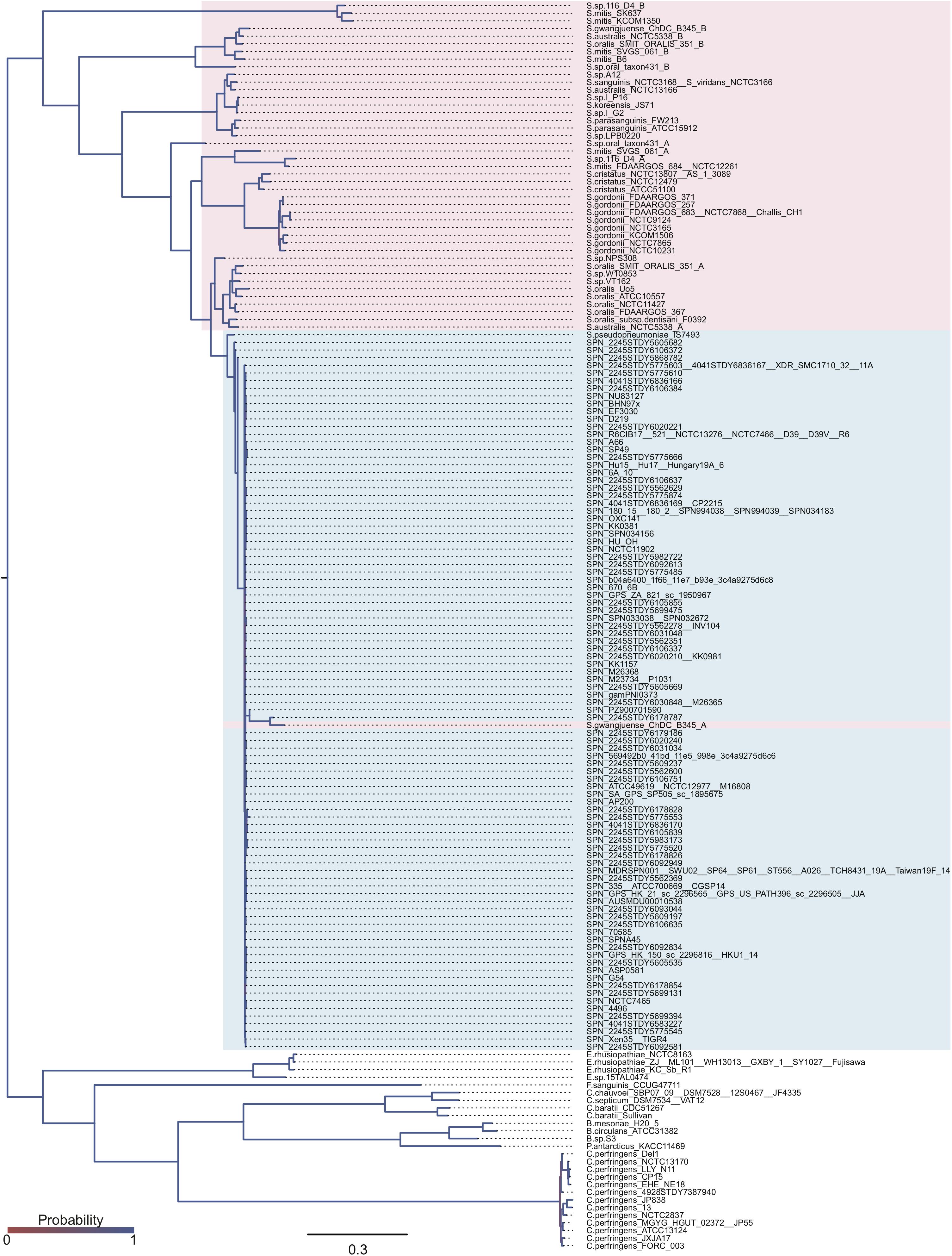
Figure 2. Bayesian phylogenetic analysis of the bgaA gene. The codon-based Bayesian phylogenetic relationship was calculated using the MrBayes program. Strains with identical sequences are listed on the same branch. S. pneumoniae and S. pseudopneumoniae bgaA genes are shaded in cyan. Other streptococcal bgaA ortholog genes are shaded in magenta. The color gradation of phylogenetic tree represents posterior probability. The scale bar indicates nucleotide substitutions per site.
Evolutionarily Conserved Catalytic Residues Contribute to the Conformation of BgaA Active Site
The domain structures and amino acid residues of TIGR4 BgaA that are under negative selection are shown in Figures 3A–D and Supplementary Figure 2. BLAST search showed that the TIGR4 BgaA amino acid sequence did not have high similarity with human proteins (Supplementary Table 3). BgaA contains glycosyl hydrolase domains in its N-terminus, and most evolutionarily conserved residues were present, from the glycosyl hydrolase sugar binding domain to the first bacterial Ig-like domain (Figures 3A,B and Supplementary Figure 2). All evolutionarily conserved proline located on loop regions (Figure 3C). Since proline has restricted phi-psi space that arise from the 5-membered ring and stabilize protein structure (Morris et al., 1992), the residues may contribute to the conservation of BgaA catalytic region via stabilization of those loop structures. The active site of BgaA contains 15 catalytic residues, R288, H450, H484, E564, D599, R602, F603, Y624, W685, W708, Y713, E716, T718, H721, and F733 (Singh et al., 2014). All catalytic residues were present in comparable residues encoded by commonly conserved codons in pneumococcal species. Interestingly, eight catalytic residues shown in Figure 3D were evolutionarily conserved ones. The other seven catalytic residues were flanked (R288, H450, W685, and W708) by or located (E564, D599, and Y713) within two residues from evolutionarily conserved residues. These results indicated that evolutionarily conserved residues contribute to the conformation of the BgaA active site.
Figure 3. Evolutionarily conserved residues in the BgaA catalytic region. (A) Pictorial representation of the structure of TIGR4 BgaA catalytic region (PDB ID: 4CU6). Glycosyl hydrolase family 2 sugar binding domain, glycosyl hydrolase family 2 domain, glycosyl hydrolase family 2 TIM barrel domain, and DUF4982 are colored in yellow, green, cyan, and magenta, respectively. (B) The side chains of evolutionarily conserved residues are shown as colored stick models. Carbon, nitrogen, and oxygen are shown as green, blue, and red, respectively. (C) BgaA active site is shown as pink stick models, and evolutionary conserved proline residues are in orange. (D) The active site of BgaA. The carbon of the evolutionarily conserved catalytic residues is shown in purple (H484, R602, F603, Y624, E716, T718, H721, and F733). The carbon of the evolutionarily conserved non-catalytic residues is shown in green. Nitrogen and oxygen are shown as blue and red, respectively.
BgaA Deficiency Decreases Pneumococcal Pathogenicity in a Mouse Sepsis Model
NanA has been identified as a multi-functional virulence factor. It contributes to pneumococcal biofilm formation, adhesion to and invasion of host epithelial and endothelial cells, inducing excessive host inflammatory responses, and resistance to opsonophagocytosis (King et al., 2006; Uchiyama et al., 2009; Dalia et al., 2010; Chang et al., 2012; Blanchette et al., 2016). BgaA also has been reported to contribute to pneumococcal adherence to host epithelial cells, host immune evasion, and in vivo biofilm formation (Dalia et al., 2010; Limoli et al., 2011; Singh et al., 2014; Blanchette et al., 2016). However, several studies reported that deletion of bgaA had limited effects on S. pneumoniae in mouse colonization models (King et al., 2006; Limoli et al., 2011; Blanchette et al., 2016; Hobbs et al., 2018). It also remains unclear if the deletion of bgaA would significantly affect host survival rate in a mouse model of sepsis. Thus, we constructed a TIGR4 bgaA mutant (ΔbgaA) strain, and performed a mouse intravenous infection assay to compare host survival for TIGR4 wild type and ΔbgaA strains. These strains showed similar growth rates in THY medium (Supplementary Figure 3 and Supplementary Data 1). In this infection model, we found that ΔbgaA-infected mice had a significantly higher survival rate compared to mice infected with the TIGR4 wild type strain (Figure 4 and Supplementary Data 2). This result indicates that BgaA functions as a virulence factor in a mouse sepsis model.
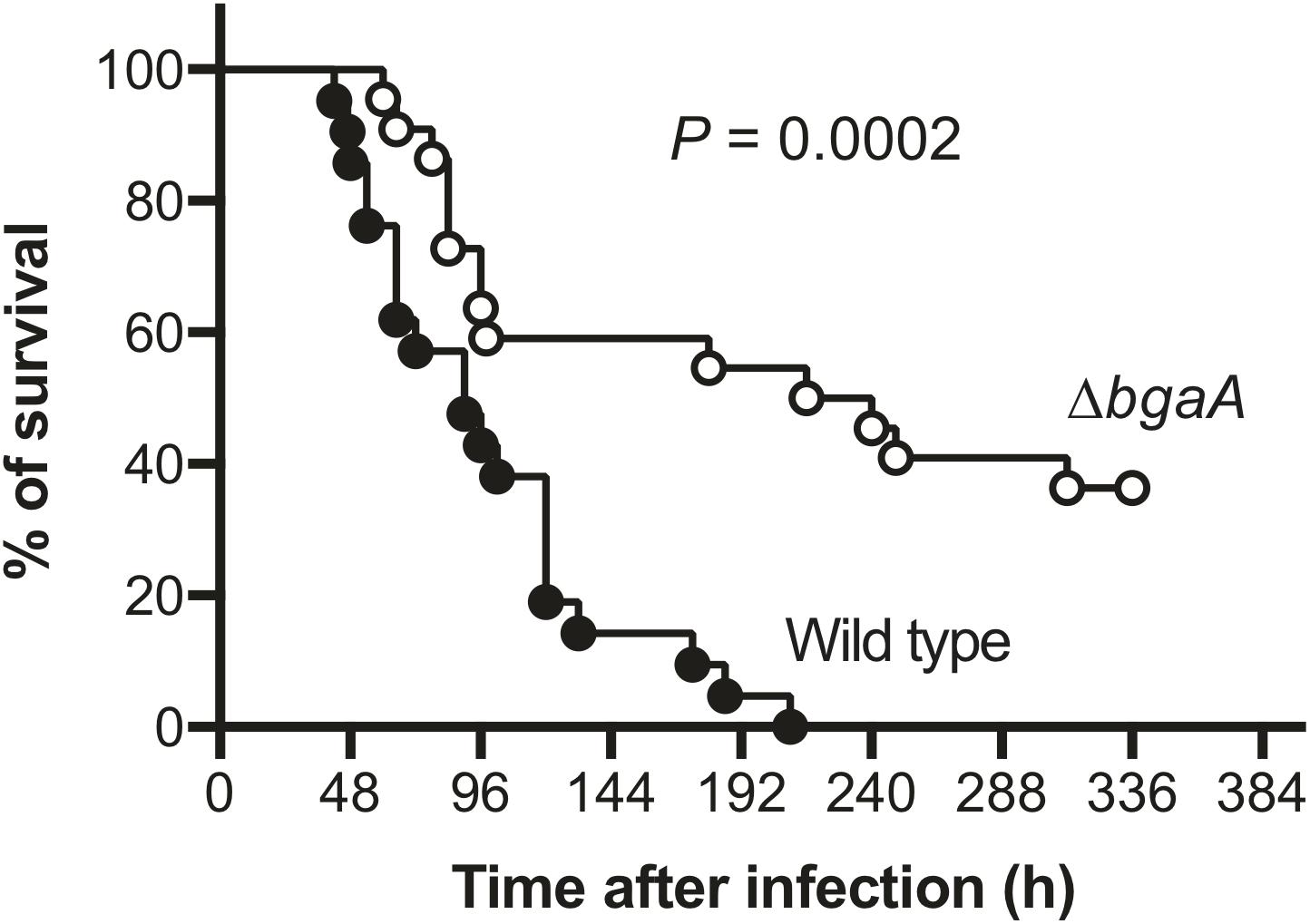
Figure 4. Deficiency of bgaA decreases pneumococcal virulence in a mouse model of sepsis. Mice were infected by intravenous injection with 1 × 106 colony forming units (CFU) of S. pneumoniae TIGR4 wild type (n = 21), or ΔbgaA (n = 22), and survival was monitored for 14 days. The difference between infected mouse groups was compared using a log-rank test. The data were obtained from three independent experiments.
Discussion
In this study, we investigated the percentage of codons in genes that encode cell wall-anchoring proteins that are under negative selection in S. pneumoniae species. Over 11% of codons in the nanA and bgaA genes had significant negative selection. As NanA is a well-studied virulence factor in S. pneumoniae, we chose to focus on the bgaA gene that encodes β-galactosidase. Phylogenetic analysis indicated that the bgaA genes in S. pneumoniae and S. pseudopneumoniae had small genetic distances and formed a distinct cluster within streptococcal bgaA orthologs. In addition, eight catalytic residues were evolutionarily conserved, and the other seven catalytic residues were located near evolutionarily conserved ones. We also demonstrated using a mouse intravenous infection model that BgaA contributes to pneumococcal pathogenesis in vivo. In combination with our previous work on pneumococcal CBPs, these results suggest that the degree of evolutionary conservation could be an effective parameter for estimating the importance of bacterial cell surface proteins (Yamaguchi et al., 2019a).
While a classical molecular microbiology approach involves analysis of a few representative strains, an evolutionary analysis approach is able to reflect importance in a species based on analysis of a few dozen to 1000s of genome sequences. For genes that encode pneumococcal CBPs, we have recently shown that more than 13% of codons in cbpJ and lytA genes are negatively selected, and CbpJ acts as a novel virulence factor in pneumococcal pneumonia both in vitro and in vivo (Yamaguchi et al., 2019a). In this study, we observed negative selective pressure for over 11% of codons in nanA and bgaA. In contrast, this figure was 3% of codons for pfbA. We have previously reported that PfbA is pneumococcal cell surface protein that forms a right-handed parallel β-helix and interacts with human fibronectin, plasmin, plasminogen, albumin, hemoglobin, and fibrinogen (Yamaguchi et al., 2008; Beulin et al., 2014, 2017; Radhakrishnan et al., 2018; Sankar et al., 2020). A BLAST search and phylogenetic analysis showed that pfbA is highly conserved in S. pneumoniae among mitis group Streptococcus (Yamaguchi et al., 2019b). In addition, in vitro assays revealed that PfbA functions as an adhesin and invasin for human epithelial cells, is a TLR2 ligand, and an anti-phagocytic factor for human neutrophils (Yamaguchi et al., 2008, 2019b). On the other hand, the deletion of pfbA in S. pneumoniae had no significant effect on host mortality in a mouse pneumonia model, and enhanced pneumococcal pathogenesis in a mouse model of sepsis (Yamaguchi et al., 2019b). With regards to pneumococcal cell surface proteins, its degree of evolutionary conservation shows good correlation with results in a mouse infection model. Although an evolutionary analysis approach has some limitations, including the fact that it is unable to identify diverse virulence factors within a species (Yamaguchi et al., 2019a), this approach could also be an effective alternative strategy for identification of common virulence factors. Furthermore, in this study, evolutionary analysis with protein structure information open the possibility that the residues under negative selection are important for the protein function and/or structure. Increasing availability and accessibility of bacterial genomic information would allow us to combine evolutionary analysis and laboratory-based approaches to study various bacterial species and proteins.
BgaA is regarded as a multi-functional protein and putative virulence factor; however, BgaA plays a limited or inconsequential role in in vivo colonization (King et al., 2006; Burnaugh et al., 2008; Dalia et al., 2010; Limoli et al., 2011; Singh et al., 2014; Blanchette et al., 2016; Hobbs et al., 2018). Here, we revealed that bgaA deficiency significantly reduced mortality in a mouse model of blood infection. As the host bloodstream is a glucose-rich environment, the ability to utilize host galactose as an alternative carbon source would not be crucial to pneumococcal survival (Blanchette et al., 2016). BgaA inhibits complement deposition, and consequently opsonophagocytosis, by cleaving N-glycans on host glycoproteins that are involved in the complement cascade (Dalia et al., 2010). As such, our observation that the ΔbgaA strain reduced host mortality in a mouse blood infection model may be explained by an inability to evade host complement deposition and phagocytosis. Although NanA, BgaA, and StrH have been reported to inhibit C3b deposition to the same extent (Dalia et al., 2010), the evolutionary approach indicated that high rates of nanA and bgaA codons are under negative selection, while strH is not. This discrepancy may arise from specificities of the glycosidases. For example, N-linked glycans are cleaved by NanA, BgaA, and StrH sequentially. NanA cleaves terminal sialic acids, after which BgaA cleaves terminal, or NanA-exposed galactose, since sialic acids are commonly linked to the C-3 or C-6 positions of galactose. Subsequently, StrH reportedly cleaves host glycans after NanA and BgaA cleavage (Hobbs et al., 2018). Therefore, one possible hypothesis is that the cleavage order of these glycosidases affects their relative importance. Specifically, StrH may function as a complementary glycosidase since NanA and BgaA may provide sufficient sialic acid, and galactose as alternative carbon sources in glucose-poor environments. Alternatively, the additional roles of NanA and BgaA may simply be more important for pneumococcal survival in the host. However, further studies are required to elucidate the precise role of BgaA in sepsis.
Although, in the current study, we focused on the top two evolutionary conserved genes, the third gene, eng, may also serve as an important virulence factor since 8% of eng codons are negatively selected. The eng gene encodes Eng, endo-α-N-acetylgalactosamidase, which specifically cleaves core-1 O-linked glycans (Brooks and Savage, 1997; Marion et al., 2009). Although Eng reportedly contributes to the colonization of mouse airway, its specific role in pneumococcal pathogenesis, and whether it affects host mortality, remain unclear. Hence, eng is also an attractive target for further investigations.
Emerging antimicrobial resistance of S. pneumoniae and serotype replacement after the introduction of pneumococcal vaccines are serious global challenges (O’Neill, 2016; CDC, 2019). A potential solution would involve developing a novel vaccine with a common antigen. S. pneumoniae has the ability to import genes from related species and undergo recombination (Kilian et al., 2014; Kilian and Tettelin, 2019). Thus, to minimize the possibility of selective pressure-mediated antigenic variation, a multi-valent vaccine would be superior to a monovalent vaccine. Our evolutionary analysis showed that four pneumococcal cell surface proteins are evolutionarily conserved. A combination of intact or truncated LytA, CbpJ, NanA, and BgaA may be attractive antigen candidates for the development of a universal pneumococcal vaccine. Several groups have already reported that individually, NanA and LytA work as protective antigens in mouse infection models (Berry et al., 1989; Lock et al., 1992; Long et al., 2004; Tong et al., 2005; Yuan et al., 2011; Janapatla et al., 2018). On the other hand, as non-pathogenic native microflora and other S. mitis group species also contain some of these genes, it is necessary to examine the possibility that the vaccine may alter the composition of oral and/or lung microbiomes. Evaluation of potential side effects on host microbiome, alongside characterization of the immunogenicity of antigens and efficacy, would help to guide the design of a novel vaccine.
PspA is a promising vaccine candidate for pneumococcal infection, as this vaccine is generally assumed to be multi-valent since PspA is a highly variable protein under positive selection (Piao et al., 2014; Chen et al., 2015; Cornick et al., 2017; Yamaguchi et al., 2019a). Hence, host immune systems, including the humoral immune response, would select for pneumococcal PspA. This may prove advantageous for a vaccine candidate since the presence of positive selection indicates that the protein is capable of inducing host acquired immunity. At the same time, the variety of PspA reflects that S. pneumoniae evolves to evade host acquired immunity through obtaining PspA mutations. Thus, the introduction of PspA vaccine would induce novel selective pressure causing “PspA replacement,” referring to an increase in vaccine-uncovered type PspA, as well as serotype replacement by pneumococcal polysaccharide capsule vaccines. It would, therefore, be of importance to determine whether the multi-valent PspA vaccine effectively covers the selective pressure-extended PspA variety. Indeed, two different selective pressures for polysaccharide and PspA vaccines may overcome the replacement issue. Hence, during pandemics of emerging infectious diseases, such as COVID-19, an effective strategy may be to develop novel vaccines that simultaneously target conserved proteins under positive and negative selections. However, proteins under positive selection are suitable as initial targets for rapid vaccine development as they have a strong probability to elicit antibodies in humans. Meanwhile, to address the evolution of pathogens, evolutionarily conserved proteins would serve as effective targets in the development of later vaccine candidates.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics Statement
The animal study was reviewed and approved by the Animal Care and Use Committee of Osaka University Graduate School of Dentistry (28-002-0).
Author Contributions
MY and SK designed the study. MY and KH performed bioinformatics analyses. MT performed the animal experiments. MY, KG, YH, TS, MN, and SK contributed to the experimental setup. MY wrote the manuscript. MT, KH, KG, YH, TS, MN, NU, and SK contributed to the writing of the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This study was supported in part by the Japan Society for the Promotion of Science KAKENHI (grant numbers 16K15787, 17H05103, 18K19643, 19H03825, and 19K22710); SECOM Science and Technology Foundation; AMED (20wm0325001h0001); Takeda Science Foundation; MSD Life Science Foundation, Public Interest Incorporated Foundation; GSK Japan Research Grant; Asahi Glass Foundation; Kurata Memorial Hitachi Science and Technology Foundation; The Naito Foundation, and Kobayashi International Scholarship Foundation; and GlaxoSmithKline Japan. The funders had no role in the study design, data collection or analysis, decision to publish, or preparation of the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Hikaru Kodama for technical assistance.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.582437/full#supplementary-material
Footnotes
References
Berry, A. M., Lock, R. A., Hansman, D., and Paton, J. C. (1989). Contribution of autolysin to virulence of Streptococcus pneumoniae. Infect. Immun. 57, 2324–2330.
Beulin, D. S., Yamaguchi, M., Kawabata, S., and Ponnuraj, K. (2014). Crystal structure of PfbA, a surface adhesin of Streptococcus pneumoniae, provides hints into its interaction with fibronectin. Int. J. Biol. Macromol. 64, 168–173. doi: 10.1016/j.ijbiomac.2013.11.035
Beulin, D. S. J., Radhakrishnan, D., Suresh, S. C., Sadasivan, C., Yamaguchi, M., Kawabata, S., et al. (2017). Streptococcus pneumoniae surface protein PfbA is a versatile multidomain and multiligand-binding adhesin employing different binding mechanisms. FEBS J. 284, 3404–3421. doi: 10.1111/febs.14200
Blanchette, K. A., Shenoy, A. T., Milner, J. II, Gilley, R. P., McClure, E., Hinojosa, C. A., et al. (2016). Neuraminidase A-exposed galactose promotes Streptococcus pneumoniae biofilm formation during colonization. Infect. Immun. 84, 2922–2932. doi: 10.1128/IAI.00277-16
Bricker, A. L., and Camilli, A. (1999). Transformation of a type 4 encapsulated strain of Streptococcus pneumoniae. FEMS Microbiol. Lett. 172, 131–135.
Brooks, M. M., and Savage, A. V. (1997). The substrate specificity of the enzyme endo-alpha-N-acetyl-D-galactosaminidase from Diplococcus pneumonia. Glycoconj. J. 14, 183–190. doi: 10.1023/a:1018585604073
Burnaugh, A. M., Frantz, L. J., and King, S. J. (2008). Growth of Streptococcus pneumoniae on human glycoconjugates is dependent upon the sequential activity of bacterial exoglycosidases. J. Bacteriol. 190, 221–230. doi: 10.1128/JB.01251-07
Chang, Y. C., Uchiyama, S., Varki, A., and Nizet, V. (2012). Leukocyte inflammatory responses provoked by pneumococcal sialidase. mBio 3:e00220-11. doi: 10.1128/mBio.00220-11
Chen, A., Mann, B., Gao, G., Heath, R., King, J., Maissoneuve, J., et al. (2015). Multivalent pneumococcal protein vaccines comprising pneumolysoid with epitopes/Fragments of CbpA and/or PspA elicit strong and broad protection. Clin. Vaccine Immunol. 22, 1079–1089. doi: 10.1128/CVI.00293-15
Cornick, J. E., Tastan Bishop, O., Yalcin, F., Kiran, A. M., Kumwenda, B., Chaguza, C., et al. (2017). The global distribution and diversity of protein vaccine candidate antigens in the highly virulent Streptococcus pnuemoniae serotype 1. Vaccine 35, 972–980. doi: 10.1016/j.vaccine.2016.12.037
Dalia, A. B., Standish, A. J., and Weiser, J. N. (2010). Three surface exoglycosidases from Streptococcus pneumoniae, NanA, BgaA, and StrH, promote resistance to opsonophagocytic killing by human neutrophils. Infect. Immun. 78, 2108–2116. doi: 10.1128/IAI.01125-09
Finn, R. D., Bateman, A., Clements, J., Coggill, P., Eberhardt, R. Y., Eddy, S. R., et al. (2014). Pfam: the protein families database. Nucleic Acids Res. 42, D222–D230. doi: 10.1093/nar/gkt1223
Gertz, E. M., Yu, Y. K., Agarwala, R., Schaffer, A. A., and Altschul, S. F. (2006). Composition-based statistics and translated nucleotide searches: improving the TBLASTN module of BLAST. BMC Biol. 4:41. doi: 10.1186/1741-7007-4-41
Golubchik, T., Brueggemann, A. B., Street, T., Gertz, R. E. Jr., Spencer, C. C., Ho, T., et al. (2012). Pneumococcal genome sequencing tracks a vaccine escape variant formed through a multi-fragment recombination event. Nat. Genet. 44, 352–355. doi: 10.1038/ng.1072
Gut, H., Xu, G., Taylor, G. L., and Walsh, M. A. (2011). Structural basis for Streptococcus pneumoniae NanA inhibition by influenza antivirals zanamivir and oseltamivir carboxylate. J. Mol. Biol. 409, 496–503. doi: 10.1016/j.jmb.2011.04.016
Hirose, Y., Yamaguchi, M., Goto, K., Sumitomo, T., Nakata, M., and Kawabata, S. (2018). Competence-induced protein Ccs4 facilitates pneumococcal invasion into brain tissue and virulence in meningitis. Virulence 9, 1576–1587. doi: 10.1080/21505594.2018.1526530
Hobbs, J. K., Pluvinage, B., and Boraston, A. B. (2018). Glycan-metabolizing enzymes in microbe-host interactions: the Streptococcus pneumoniae paradigm. FEBS Lett. 592, 3865–3897. doi: 10.1002/1873-3468.13045
Ishiguro, T., Takayanagi, N., Yamaguchi, S., Yamakawa, H., Nakamoto, K., Takaku, Y., et al. (2013). Etiology and factors contributing to the severity and mortality of community-acquired pneumonia. Intern. Med. 52, 317–324. doi: 10.2169/internalmedicine.52.8830
Janapatla, R. P., Chen, C. L., Hsu, M. H., Liao, W. T., and Chiu, C. H. (2018). Immunization with pneumococcal neuraminidases NanA, NanB and NanC to generate neutralizing antibodies and to increase survival in mice. J. Med. Microbiol. 67, 709–723. doi: 10.1099/jmm.0.000724
Kanehisa, M., and Goto, S. (2000). KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30.
Katoh, K., and Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Kawamura, Y., Hou, X. G., Sultana, F., Miura, H., and Ezaki, T. (1995). Determination of 16S rRNA sequences of Streptococcus mitis and Streptococcus gordonii and phylogenetic relationships among members of the genus Streptococcus. Int. J. Syst. Evol. Micrbiol. 45, 406–408.
Kilian, M., and Tettelin, H. (2019). Identification of virulence-associated properties by comparative genome analysis of Streptococcus pneumoniae, S. pseudopneumoniae, S. mitis, Three S. oralis Subspecies, and S. infantis. mBio 10:e01985-19. doi: 10.1128/mBio.01985-19
Kilian, M., Riley, D. R., Jensen, A., Bruggemann, H., and Tettelin, H. (2014). Parallel evolution of Streptococcus pneumoniae and Streptococcus mitis to pathogenic and mutualistic lifestyles. mBio 5:e01490-14. doi: 10.1128/mBio.01490-14
King, S. J., Hippe, K. R., and Weiser, J. N. (2006). Deglycosylation of human glycoconjugates by the sequential activities of exoglycosidases expressed by Streptococcus pneumoniae. Mol. Microbiol. 59, 961–974. doi: 10.1111/j.1365-2958.2005.04984.x
Limoli, D. H., Sladek, J. A., Fuller, L. A., Singh, A. K., and King, S. J. (2011). BgaA acts as an adhesin to mediate attachment of some pneumococcal strains to human epithelial cells. Microbiology 157(Pt 8), 2369–2381. doi: 10.1099/mic.0.045609-0
Lock, R. A., Hansman, D., and Paton, J. C. (1992). Comparative efficacy of autolysin and pneumolysin as immunogens protecting mice against infection by Streptococcus pneumoniae. Microb. Pathog. 12, 137–143. doi: 10.1016/0882-4010(92)90116-6
Lofling, J., Vimberg, V., Battig, P., and Henriques-Normark, B. (2011). Cellular interactions by LPxTG-anchored pneumococcal adhesins and their streptococcal homologues. Cell Microbiol. 13, 186–197. doi: 10.1111/j.1462-5822.2010.01560.x
Long, J. P., Tong, H. H., and DeMaria, T. F. (2004). Immunization with native or recombinant Streptococcus pneumoniae neuraminidase affords protection in the chinchilla otitis media model. Infect. Immun. 72, 4309–4313. doi: 10.1128/IAI.72.7.4309-4313.2004
Marion, C., Limoli, D. H., Bobulsky, G. S., Abraham, J. L., Burnaugh, A. M., and King, S. J. (2009). Identification of a pneumococcal glycosidase that modifies O-linked glycans. Infect. Immun. 77, 1389–1396. doi: 10.1128/IAI.01215-08
Mori, Y., Yamaguchi, M., Terao, Y., Hamada, S., Ooshima, T., and Kawabata, S. (2012). α-Enolase of Streptococcus pneumoniae induces formation of neutrophil extracellular traps. J. Biol. Chem. 287, 10472–10481. doi: 10.1074/jbc.M111.280321
Morris, A. L., MacArthur, M. W., Hutchinson, E. G., and Thornton, J. M. (1992). Stereochemical quality of protein structure coordinates. Proteins 12, 345–364. doi: 10.1002/prot.340120407
O’Neill, J. (2016). Tackling Drug-Resistant Infections Globally: Final Report and Recommendations. London: The Review on Antimicrobial Resistance.
Piao, Z., Akeda, Y., Takeuchi, D., Ishii, K. J., Ubukata, K., Briles, D. E., et al. (2014). Protective properties of a fusion pneumococcal surface protein A (PspA) vaccine against pneumococcal challenge by five different PspA clades in mice. Vaccine 32, 5607–5613. doi: 10.1016/j.vaccine.2014.07.108
Pond, S. L., Frost, S. D., and Muse, S. V. (2005). HyPhy: hypothesis testing using phylogenies. Bioinformatics 21, 676–679. doi: 10.1093/bioinformatics/bti079
Radhakrishnan, D., Yamaguchi, M., Kawabata, S., and Ponnuraj, K. (2018). Streptococcus pneumoniae surface adhesin PfbA and its interaction with erythrocytes and hemoglobin. Int. J. Biol. Macromol. 120(Pt A), 135–143. doi: 10.1016/j.ijbiomac.2018.08.080
Rambaut, A. (2018). FigTree ver.1.4.4. Available: https://github.com/rambaut/figtree/releases. (accessed November 26, 2018).
Richards, V. P., Palmer, S. R., Pavinski Bitar, P. D., Qin, X., Weinstock, G. M., Highlander, S. K., et al. (2014). Phylogenomics and the dynamic genome evolution of the genus Streptococcus. Genome Biol. Evol. 6, 741–753. doi: 10.1093/gbe/evu048
Ronquist, F., Teslenko, M., van der Mark, P., Ayres, D. L., Darling, A., Hohna, S., et al. (2012). MrBayes 3.2: efficient bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542. doi: 10.1093/sysbio/sys029
Salvadori, G., Junges, R., Morrison, D. A., and Petersen, F. C. (2019). Competence in Streptococcus pneumoniae and close commensal relatives: mechanisms and implications. Front. Cell Infect. Microbiol. 9:94. doi: 10.3389/fcimb.2019.00094
Sankar, S., Yamaguchi, M., Kawabata, S., and Ponnuraj, K. (2020). Streptococcus pneumoniae Surface Adhesin PfbA exhibits host specificity by binding to human serum albumin but not bovine, rabbit and porcine serum albumins. Protein J. 39, 1–9. doi: 10.1007/s10930-019-09875-y
Singh, A. K., Pluvinage, B., Higgins, M. A., Dalia, A. B., Woodiga, S. A., Flynn, M., et al. (2014). Unravelling the multiple functions of the architecturally intricate Streptococcus pneumoniae beta-galactosidase. BgaA. PLoS Pathog. 10:e1004364. doi: 10.1371/journal.ppat.1004364
Stamatakis, A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Tanabe, A. S. (2008). Phylogears2 ver. 2.0. Available online at: http://www.fifthdimension.jp/ (accessed May 10, 2019).
Tanabe, A. S. (2011). Kakusan4 and Aminosan: two programs for comparing nonpartitioned, proportional and separate models for combined molecular phylogenetic analyses of multilocus sequence data. Mol. Ecol. Resour. 11, 914–921. doi: 10.1111/j.1755-0998.2011.03021.x
Tong, H. H., Li, D., Chen, S., Long, J. P., and DeMaria, T. F. (2005). Immunization with recombinant Streptococcus pneumoniae neuraminidase NanA protects chinchillas against nasopharyngeal colonization. Infect. Immun. 73, 7775–7778. doi: 10.1128/IAI.73.11.7775-7778.2005
Uchiyama, S., Carlin, A. F., Khosravi, A., Weiman, S., Banerjee, A., Quach, D., et al. (2009). The surface-anchored NanA protein promotes pneumococcal brain endothelial cell invasion. J. Exp. Med. 206, 1845–1852. doi: 10.1084/jem.20090386
Waterhouse, A. M., Procter, J. B., Martin, D. M., Clamp, M., and Barton, G. J. (2009). Jalview Version 2–a multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189–1191. doi: 10.1093/bioinformatics/btp033
Yamaguchi, M. (2018). Synergistic findings from microbiological and evolutional analyses of virulence factors among pathogenic streptococcal species. J. Oral Biosci. 60, 36–40. doi: 10.1016/j.job.2018.02.004
Yamaguchi, M., Goto, K., Hirose, Y., Yamaguchi, Y., Sumitomo, T., Nakata, M., et al. (2019a). Identification of evolutionarily conserved virulence factor by selective pressure analysis of Streptococcus pneumoniae. Commun. Biol. 2:96. doi: 10.1038/s42003-019-0340-7
Yamaguchi, M., Hirose, Y., Takemura, M., Ono, M., Sumitomo, T., Nakata, M., et al. (2019b). Streptococcus pneumoniae evades host cell phagocytosis and limits host mortality through its cell wall anchoring protein PfbA. Front. Cell Infect. Microbiol. 9:301. doi: 10.3389/fcimb.2019.00301
Yamaguchi, M., Hirose, Y., Nakata, M., Uchiyama, S., Yamaguchi, Y., Goto, K., et al. (2016). Evolutionary inactivation of a sialidase in group B Streptococcus. Sci. Rep. 6:28852. doi: 10.1038/srep28852
Yamaguchi, M., Nakata, M., Sumioka, R., Hirose, Y., Wada, S., Akeda, Y., et al. (2017). Zinc metalloproteinase ZmpC suppresses experimental pneumococcal meningitis by inhibiting bacterial invasion of central nervous systems. Virulence 8, 1516–1524. doi: 10.1080/21505594.2017.1328333
Yamaguchi, M., Terao, Y., Mori, Y., Hamada, S., and Kawabata, S. (2008). PfbA, a novel plasmin- and fibronectin-binding protein of Streptococcus pneumoniae, contributes to fibronectin-dependent adhesion and antiphagocytosis. J. Biol. Chem. 283, 36272–36279. doi: 10.1074/jbc.M807087200
Keywords: Streptococcus pneumoniae, molecular evolutionary analysis, pneumococcal cell wall-anchoring proteins, bgaA, nanA
Citation: Yamaguchi M, Takemura M, Higashi K, Goto K, Hirose Y, Sumitomo T, Nakata M, Uzawa N and Kawabata S (2020) Role of BgaA as a Pneumococcal Virulence Factor Elucidated by Molecular Evolutionary Analysis. Front. Microbiol. 11:582437. doi: 10.3389/fmicb.2020.582437
Received: 12 July 2020; Accepted: 01 September 2020;
Published: 24 September 2020.
Edited by:
Zhan Zhou, Zhejiang University, ChinaReviewed by:
Masamitsu Kono, Wakayama Medical University, JapanAnders P. Hakansson, Lund University, Sweden
Copyright © 2020 Yamaguchi, Takemura, Higashi, Goto, Hirose, Sumitomo, Nakata, Uzawa and Kawabata. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Masaya Yamaguchi, eWFtYWd1Y2hpQGRlbnQub3Nha2EtdS5hYy5qcA==; Shigetada Kawabata, a2F3YWJhdGFAZGVudC5vc2FrYS11LmFjLmpw
†Present address: Kana Goto, Department of Pediatric Dentistry, Okayama University Graduate School of Medicine, Dentistry and Pharmaceutical Sciences, Okayama, Japan; Masanobu Nakata, Department of Oral Microbiology, Kagoshima University Graduate School of Medical and Dental Sciences, Kagoshima, Japan
 Masaya Yamaguchi
Masaya Yamaguchi Moe Takemura
Moe Takemura Kotaro Higashi
Kotaro Higashi Kana Goto1†
Kana Goto1† Yujiro Hirose
Yujiro Hirose Tomoko Sumitomo
Tomoko Sumitomo Masanobu Nakata
Masanobu Nakata Shigetada Kawabata
Shigetada Kawabata