- 1Center for Marine Research, Ruđer Bošković Institute, Rovinj, Croatia
- 2Department of Functional and Evolutionary Ecology, University of Vienna, Vienna, Austria
- 3Department of Marine Microbiology and Biogeochemistry, Royal Netherlands Institute for Sea Research (NIOZ), Utrecht University, Den Burg, Netherlands
Surfaces of marine macrophytes are inhabited by diverse microbial communities. Most studies focusing on epiphytic communities of macrophytes did not take into account temporal changes or applied low sampling frequency approaches. The seasonal dynamics of epiphytic microbial communities was determined in a meadow of Cymodocea nodosa invaded by Caulerpa cylindracea and in a monospecific settlement of C. cylindracea at monthly intervals. For comparison the ambient prokaryotic picoplankton community was also characterized. At the OTU level, the microbial community composition differed between the ambient water and the epiphytic communities exhibiting host-specificity. Also, successional changes were observed connected to the macrophyte growth cycle. Taxonomic analysis, however, showed similar high rank taxa (phyla and classes) in the ambient water and the epiphytic communities, with the exception of Desulfobacterota, which were only found on C. cylindracea. Cyanobacteria showed seasonal changes while other high rank taxa were present throughout the year. In months of high Cyanobacteria presence the majority of cyanobacterial sequences were classified as Pleurocapsa. Phylogenetic groups present throughout the year (e.g., Saprospiraceae, Rhodobacteraceae, members without known relatives within Gammaproteobacteria, Desulfatitalea, and members without known relatives within Desulfocapsaceae) constituted most of the sequences, while less abundant taxa showed seasonal patterns connected to the macrophyte growth cycle. Taken together, epiphytic microbial communities of the seagrass C. nodosa and the macroalga C. cylindracea appear to be host-specific and contain taxa that undergo successional changes.
1. Introduction
Marine macrophytes (seagrasses and macroalgae) are important ecosystem engineers forming close associations with microorganisms belonging to all three domains of life (Egan et al., 2013; Tarquinio et al., 2019). Microbes can live within macrophyte tissue as endophytes or form epiphytic communities on surfaces of leaves, roots, rhizomes, and thalli (Egan et al., 2013; Hollants et al., 2013; Aires et al., 2015; Tarquinio et al., 2019). Epiphytic and endophytic microbial communities exhibit a close functional relationship with the macrophyte host. It has been proposed that this close relationship constitutes a holobiont, an integrated community where the macrophyte organism and its symbiotic partners support each other (Margulis, 1991). In addition, as suggested by the hologenome theory endophytic microbes play a critical role in the adaptation and evolution of the host species (Aires et al., 2015).
Biofilms of microbial epiphytes can contain diverse taxonomic groups and harbor cell abundances from 102 to 107 cells cm-2 (Armstrong et al., 2000; Bengtsson et al., 2010; Burke et al., 2011b). In such an environment a number of positive and negative interactions between the macrophyte and the colonizing microorganisms have been described (Egan et al., 2013; Hollants et al., 2013; Tarquinio et al., 2019). Macrophytes can promote growth of associated microbes by nutrient exudation (Wood and Hayasaka, 1981), while in return microorganisms may support macrophyte performance through improved nutrient availability (Nielsen et al., 2001; de Oliveira et al., 2012), phytohormone production (Matsuo et al., 2003; Celdran et al., 2012), and protection from toxic compounds (Küsel et al., 2006), oxidative stress (Sanchez-Amat et al., 2010), biofouling organisms (Dobretsov and Qian, 2002), and pathogens (Penesyan et al., 2009). Besides these positive interactions, macrophytes can negatively impact the associated microbes by producing reactive oxygen species (Weinberger, 2007) and secondary metabolites (Saha et al., 2011).
All these ecological roles are carried out by a taxonomically diverse community of microorganisms. At higher taxonomic ranks (phyla and classes) microbial taxa, such as Alphaproteobacteria, Gammaproteobacteria, Bacteroidota, and Cyanobacteria, have been associated with surfaces of seagrass leaves and macroalgal thalli (Crump and Koch, 2008; Tujula et al., 2010; Lachnit et al., 2011; Egan et al., 2013; Tarquinio et al., 2019; Ugarelli et al., 2019). While similar high rank taxa have been found on surfaces of different macrophyte species, in order to describe new ecological patterns it is also necessary to focus on lower taxonomic ranks (genus and OTUs) which tend to be host-specific (Lachnit et al., 2011; Hollants et al., 2013; Roth-Schulze et al., 2016). While the microbial community composition can vary between host species, metagenomic analyses revealed that the majority of microbial functions are conserved, showing that different epiphytic microbial species could be functionally similar (Burke et al., 2011a; Roth-Schulze et al., 2016; Cúcio et al., 2018). This discrepancy between the microbial taxonomic and functional composition might be explained by the lottery hypothesis (Sale, 1976). It postulates that an initial random colonization step takes place from a set of functionally equivalent taxonomic groups resulting in taxonomically different epiphytic communities sharing a core set of functional genes (Burke et al., 2011a; Stratil et al., 2013; Schmidt et al., 2015; Roth-Schulze et al., 2016).
Seagrasses are known to form close relationships with microbial communities associated with the surfaces of leaves, roots, and rhizomes (Cúcio et al., 2016; Crump et al., 2018; Ugarelli et al., 2019; Ettinger and Eisen, 2020; Wang et al., 2020). For different seagrass species a distinct microbial community from ambient seawater or bulk sediment has been reported, however no species specific communities have been found (Cúcio et al., 2016; Crump et al., 2018; Ugarelli et al., 2019). It seems that seagrasses are selecting the associated microbial community but these microbes have not coevolved with their seagrass plant host. Similar to seagrasses, siphonous macroalgae of the genus Caulerpa are also closely associated with their microbial communities (Aires et al., 2013, 2015; Rizzo et al., 2016b; Stabili et al., 2017; Morrissey et al., 2019). While some studies have found similar culturable bacterial groups associated with the surface of a Caulerpa species from different geographic locations (Stabili et al., 2017), other have reported large compositional differences that were mainly attributed to different host species of this genus, biogeography, and nutrient levels (Morrissey et al., 2019) again raising the question to which extent are associated communities host-specific.
Since marine macrophytes in temperate zones are exhibiting seasonal changes in growth and physiology (Agostini et al., 2003; Najdek et al., 2020) it is important to verify if and how surface associated microbial communities are affected by these changes. The majority of studies describing macrophyte epiphytic microbial communities have not included possible seasonal changes (Crump and Koch, 2008; Lachnit et al., 2009; Burke et al., 2011b; Roth-Schulze et al., 2016; Ugarelli et al., 2019). If seasonal changes have been taken into account, low temporal frequency, applied methodologies, and/or limited number of analyzed host species did not allow for a detailed taxonomic analysis (Tujula et al., 2010; Lachnit et al., 2011; Bengtsson et al., 2012; Michelou et al., 2013; Miranda et al., 2013; Mancuso et al., 2016). In the present study we performed a descriptive analysis of seasonal bacterial and archeal community dynamics on the surfaces of the seagrass Cymodocea nodosa, an abundant seagrass species in the Mediterranean (Short et al., 2001), and siphonous macroalga Caulerpa cylindracea, one of the most invasive macroalgal species (Klein and Verlaque, 2008; Boudouresque et al., 2009). Bacterial and archeal epiphytes were sampled in a meadow of C. nodosa invaded by the invasive C. cylindracea and in a locality of only C. cylindracea located in the proximity of the seagrass meadow. For comparison, the microbial community of the ambient seawater was also characterized. The presence of both macrophytes in the same area enabled (i) the assessment of differences in the bacterial and archeal communities between host species and settlements of C. cylindracea and (ii) the evaluation of differences between surface associated and free living (ambient seawater) communities. In addition, these differences were evaluated on a monthly scale providing insight into seasonal changes (iii).
2. Materials and Methods
2.1. Sampling
Sampling was performed in the Bay of Funtana, northern Adriatic Sea (45°10′39′′ N, 13°35′42′′ E). The sea floor in the bay is partly covered by the invasive macroalga C. cylindracea that can be found in a monospecific settlement or mixed with the seagrass C. nodosa (Figure 1). C. nodosa leaves were retrieved from a meadow of C. nodosa invaded by the invasive C. cylindracea (mixed settlement; depth, 2–2.5 m), while C. cylindracea thalli were sampled in the same invaded meadow (mixed settlement; depth, 2–2.5 m) and in a monospecific settlement (depth, 1–1.5 m) of C. cylindracea located in the proximity (20–50 m) of the invaded meadow at approximately monthly intervals from November 2017 to October 2018 (Supplementary Table 1). Leaves and thalli were collected by diving and transported to the laboratory in containers placed on ice and filled with seawater collected at the sampling site. Upon arrival to the laboratory, C. nodosa leaves were cut into sections of 1–2 cm, while C. cylindracea thalli were cut into 5–8 cm long sections. Leaves and thalli were washed three times with sterile artificial seawater (ASW) to remove loosely attached microbial cells. Ambient seawater was collected in 10 L containers by diving and transported to the laboratory where 10–20 L were filtered through a 20 μm net. The filtrate was further sequentially filtered through 3 and 0.2 μm polycarbonate membrane filters (Whatman, United Kingdom) using a peristaltic pump. Filters were briefly dried at room temperature and stored at -80°C. Seawater samples were also collected approximately monthly from July 2017 to October 2018.
Figure 1. Location of the mixed (C. nodosa and C. cylindracea) and monospecific (C. cylindracea) settlement in the Bay of Funtana, northern Adriatic Sea (© OpenStreetMap contributors, www.openstreetmap.org/copyright).
2.2. DNA Isolation
DNA from surfaces of C. nodosa and C. cylindracea was isolated from a pool of leaves (1 g wet weight) or thalli (2 g wet weight) on the sampling day using a previously modified and adapted protocol that allows for a selective epiphytic DNA isolation (Massana et al., 1997; Korlević et al., 2021). Briefly, leaves and thalli were incubated in a lysis buffer and treated with lysozyme and proteinase K. Following the incubations, the mixture containing lysed epiphytic cells was separated from the leaves and thalli and extracted using phenol-chloroform. Finally, the extracted DNA was precipitated using isopropanol. DNA from seawater picoplankton was extracted from 0.2 μm polycarbonate filters according to Massana et al. (1997) with a slight modification. Following the phenol-chloroform extraction, 1/10 of 3 M sodium acetate (pH 5.2) was added. DNA was precipitated by adding 1 volume of chilled isopropanol, incubating the mixtures overnight at -20°C and centrifuging at 20,000 × g and 4°C for 20 min. The pellet was washed twice with 500 μl of chilled 70% ethanol and centrifuged after each washing step at 20,000 × and 4°C for 5 min. Dried pellets were re-suspended in 50–100 μl of deionized water. One DNA sample originating from seawater picoplankton was obtained per each sampling point.
2.3. Illumina 16S rRNA Sequencing
Illumina MiSeq sequencing of the V4 region of the 16S rRNA gene was performed as described previously (Korlević et al., 2021). The V4 region of the 16S rRNA gene was amplified using a two-step PCR procedure. In the first PCR, the 515F (5′-GTGYCAGCMGCCGCGGTAA-3′) and 806R (5′-GGACTACNVGGGTWTCTAAT-3′) primers from the Earth Microbiome Project (https://earthmicrobiome.org/protocols-and-standards/16s/) were used (Caporaso et al., 2012; Apprill et al., 2015; Parada et al., 2016). These primers contained on their 5′ end a tagged sequence. Purified PCR products were sent for Illumina MiSeq sequencing at IMGM Laboratories, Martinsried, Germany. Prior to sequencing at IMGM, the second PCR amplification of the two-step PCR procedure was performed using primers targeting the tagged region incorporated in the first PCR. In addition, these primers contained adapter and sample-specific index sequences. Beside samples, a positive and negative control for each sequencing batch was sequenced. The negative control comprised PCR reactions without DNA template, while for a positive control a mock community composed of evenly mixed DNA material originating from 20 bacterial strains (ATCC MSA-1002, ATCC, USA) was used. Sequences obtained in this study have been deposited in the European Nucleotide Archive (ENA) at EMBL-EBI under the accession number PRJEB37267 (https://www.ebi.ac.uk/ena/browser/view/PRJEB37267).
2.4. Sequence and Data Analysis
Obtained sequences were analyzed on the computer cluster Isabella (University Computing Center, University of Zagreb) using mothur (version 1.43.0; Schloss et al., 2009) according to the MiSeq Standard Operating Procedure (MiSeq SOP; https://mothur.org/wiki/MiSeq_SOP; Kozich et al., 2013) and recommendations provided by the Riffomonas project to enhance data reproducibility (http://www.riffomonas.org/). For alignment and classification of sequences the SILVA SSU Ref NR 99 database (release 138; http://www.arb-silva.de) was used (Quast et al., 2013; Yilmaz et al., 2014). Sequences were clustered into operational taxonomic units (OTUs) at a similarity level of 97%.
Pipeline data processing and visualization was done using R (version 3.6.0) (R Core Team, 2019) combined with packages vegan (version 2.5-6) (Oksanen et al., 2019), tidyverse (version 1.3.0) (Wickham, 2017; Wickham et al., 2019), and multiple other packages (Neuwirth, 2014; Xie, 2014, 2015, 2019a,b,c; Xie et al., 2018; Allaire et al., 2019; Wilke, 2019; Zhu, 2019). Observed number of OTUs, Chao1, ACE, exponential of the Shannon diversity index and Inverse Simpson index were calculated after normalization to the minimum number of reads per sample using vegan's function rrarefy to account for different sequencing depths (Oksanen et al., 2019). Chao1 and ACE estimators were calculated using vegan's function estimateR, while Shannon and Inverse Simpson diversity indices were retrieved using vegan's function diversity (Oksanen et al., 2019). To express both diversity indices in terms of effective number of OTUs the exponential of the Shannon diversity index was calculated (Jost, 2006). The proportions of shared OTUs and communities between samples and community types (seawater, C. nodosa [mixed], C. cylindracea [mixed] and C. cylindracea [monospecific]) were expressed as the Jaccard's (on presence/absence data) and Bray-Curtis similarity coefficient, respectively. The coefficients were calculated on the OTU data table using vegan's function vegdist and converted from dissimilarities to similarities (Borcard et al., 2011; Legendre and Legendre, 2012; Oksanen et al., 2019). The Principal Coordinates Analysis (PCoA) was performed on Bray-Curtis dissimilarities based on OTU abundances using the function cmdscale (Borcard et al., 2011; Legendre and Legendre, 2012). Differences between communities were tested by performing the Analysis of Similarities (ANOSIM) using the vegan's function anosim and 1,000 permutations (Oksanen et al., 2019), while differences in relative contributions or proportions of shared OTUs and communities were tested by applying the Mann–Whitney U-test using the function wilcox.test. In addition, differences between community type estimators or indices were tested by performing the Kruskal-Wallis H-test (function kruskal.test) followed by a pairwise comparison using the Mann-Whitney U-test (function pairwise.wilcox). Bonferroni correction was used to address the problem of multiple comparisons.
A total of 1.7 million sequences after quality curation and exclusion of sequences without known relatives (no relative sequences) and eukaryotic, chloroplast, and mitochondrial sequences were obtained (Supplementary Table 1). The number of reads per sample ranged between 8,408 and 77,463 sequences (Supplementary Table 1). Even when the highest sequencing effort was applied the rarefaction curves did not level off as commonly observed in high-throughput 16S rRNA amplicon sequencing approaches (Supplementary Figure 1). Following quality curation and exclusion of sequences as mentioned above reads were clustered into 28,750 different OTUs. Read numbers were normalized to the minimum number of sequences (8,408, Supplementary Table 1) using previously mentioned vegan's function rrarefy resulting in 17,201 different OTUs ranging from 352 to 2,062 OTUs per sample (Supplementary Figure 2). Based on the ATCC MSA-1002 mock community included in the analysis an average sequencing error rate of 0.01% was determined, which is in line with previously reported values for next-generation sequencing data (Kozich et al., 2013; Schloss et al., 2016). In addition, the negative controls processed together with the samples yielded only two sequences after sequence quality curation. The detailed analysis procedure including the R Markdown file is available as a GitHub repository (https://github.com/MicrobesRovinj/Korlevic_EpiphyticDynamics_FrontMicrobiol_2021).
3. Results
A total of 35 samples originating from epiphytic archeal and bacterial communities associated with surfaces of the seagrass C. nodosa and the macroalga C. cylindracea were analyzed. In addition, 18 samples (one of the samples was sequenced twice) originating from the ambient seawater were also processed for comparison. Generally, richness estimators and diversity indices showed similar trends. On average, higher values were found for C. cylindracea (mixed [Number of OTUs, 1,688.4 ± 136.6 OTUs] and monospecific [Number of OTUs, 1,750.4 ± 165.7 OTUs]) than for C. nodosa (Number of OTUs, 1,063.7 ± 210.6 OTUs) and lowest values were obtained for the microbial community of the ambient seawater (Number of OTUs, 531.0 ± 143.9 OTUs; Kruskal-Wallis, p < 0.0001) (Supplementary Figure 2 and Supplementary Tables 2, 3). Temporal changes did not reveal such large dissimilarities. C. nodosa communities showed a slow increase in all calculated richness estimators toward the end of the study, while C. cylindracea (mixed and monospecific) communities were characterized by slightly higher values in spring and summer than in autumn and winter (Supplementary Figure 2).
A clear separation between ambient seawater and surface associated communities was found (Figure 2). In addition, a separation of epiphytic bacterial and archeal communities based on host species was detected. This separation was further supported by ANOSIM (R = 0.96, p < 0.001). The highest proportion of shared OTUs and community was found between mixed and monospecific C. cylindracea (Jaccard, 0.35; Bray-Curtis, 0.77), while lower shared values were calculated between ambient seawater and epiphytic communities (Jaccard, 0.10–0.11; Bray-Curtis, 0.05–0.06). Shared proportions of OTUs and communities between C. nodosa and C. cylindracea (either mixed or monospecific) were approximately in-between the values obtained for the comparison of ambient seawater with all other communities and for the comparison of the mixed and monospecific C. cylindracea associated community. Seasonal changes of C. nodosa associated communities indicated a separation between spring, summer, and autumn/winter samples (ANOSIM, R = 0.56, p < 0.01). For C. cylindracea associated communities a separation between summer and autumn/winter/spring samples was observed that was, however, not as strong as for C. nodosa associated communities (ANOSIM, R = 0.30, p < 0.05; Figure 2). Shared proportions of OTUs between consecutive sampling points were lower for ambient seawater (19.6 ± 2.5%) than for C. nodosa (28.3 ± 5.2%) and C. cylindracea (mixed [26.3 ± 2.1%] and monospecific [27.2 ± 2.0%]) associated communities (p < 0.0001), while mean proportions of shared communities between consecutive sampling points did not show such a difference (seawater, 57.4 ± 14.7%; C. nodosa, 53.4 ± 9.3%; C. cylindracea [mixed], 55.0 ± 7.0%; C. cylindracea [monospecific], 55.1 ± 5.2%; p = 0.1), although in ambient seawater higher fluctuations could be observed (Figure 3). In addition, only 0.4–1.0% of OTUs from each surface associated community were present at all seasons. These persistent OTUs constituted a high proportion of total sequences (40.2–53.2 %) and were mainly contributing to abundant phylogenetic groups present throughout the year, e.g. the no relative Rhodobacteraceae in the case of C. nodosa or taxa within Desulfobacterota in the case of C. cylindracea (see below; Supplementary Table 4).
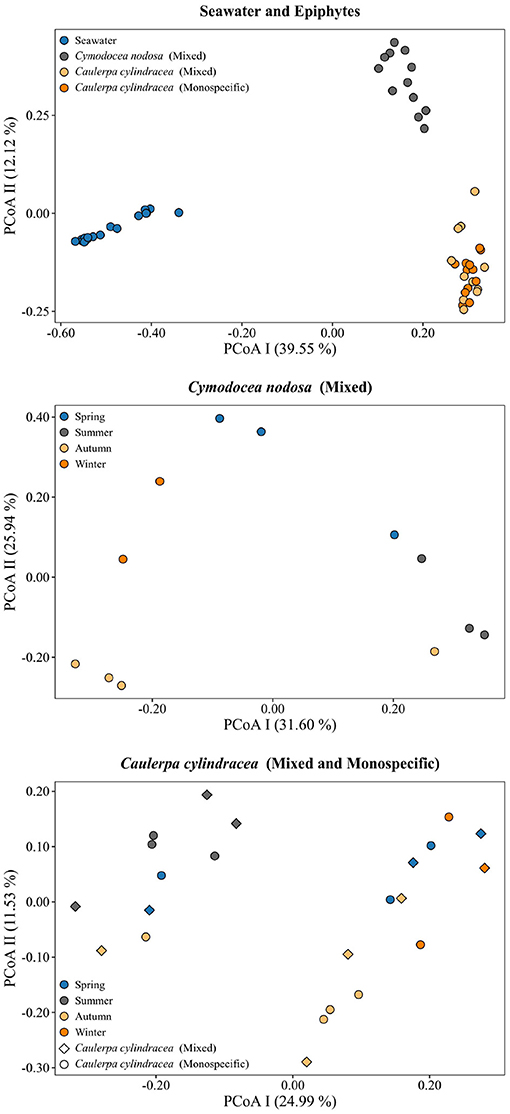
Figure 2. Principal Coordinates Analysis (PCoA) of Bray-Curtis distances based on OTU abundances of bacterial and archeal communities from the surfaces of the macrophytes C. nodosa (mixed settlement) and C. cylindracea (mixed and monospecific settlement) and in the ambient seawater. Samples from different environments or seasons are labeled in different color and shape. The proportion of explained variation by each axis is shown on the corresponding axis in parentheses.
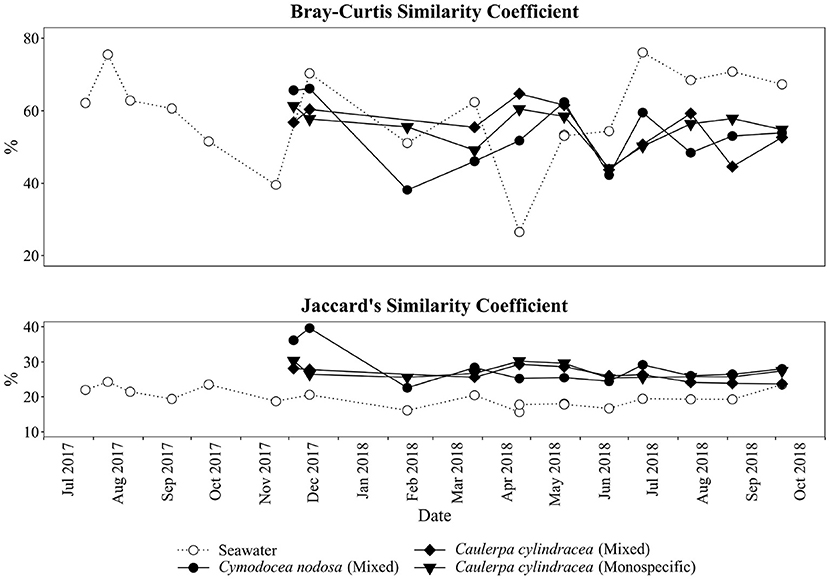
Figure 3. Proportion of shared bacterial and archeal communities (Bray-Curtis similarity coefficient) and shared bacterial and archaeal OTUs (Jaccard's similarity coefficient) between consecutive sampling dates and from the surfaces of the macrophytes C. nodosa (mixed settlement) and C. cylindracea (mixed and monospecific settlement) and in the ambient seawater.
The taxonomic composition of both, macrophyte associated and ambient seawater community, was dominated by bacterial (99.1 ± 2.1%) over archeal sequences (0.9 ± 2.1%; Figure 4). Higher relative abundances of chloroplast related sequences were only observed in surface associated communities, with higher values in autumn/winter (37.2 ± 11.2%) than in spring/summer (20.9 ± 9.7%) (p < 0.0001; Supplementary Figure 3). Generally, at higher taxonomic ranks (phylum-class), epiphytic and ambient seawater microbial communities were composed of similar bacterial taxa. Ambient seawater communities were mainly comprised of Actinobacteriota, Bacteroidota, Cyanobacteria, Alphaproteobacteria, Gammaproteobacteria, and Verrucomicrobiota. Communities associated with C. nodosa consisted additionally of Planctomycetota contributing more in summer 2018 than in other seasons. In addition, communities from mixed and monospecific C. cylindracea were similar and characterized by the same groups as ambient seawater and C. nodosa communities with the addition of Desulfobacterota (Figure 4). Larger differences between environments and host species were observed at lower taxonomic ranks (Figures 5–9).
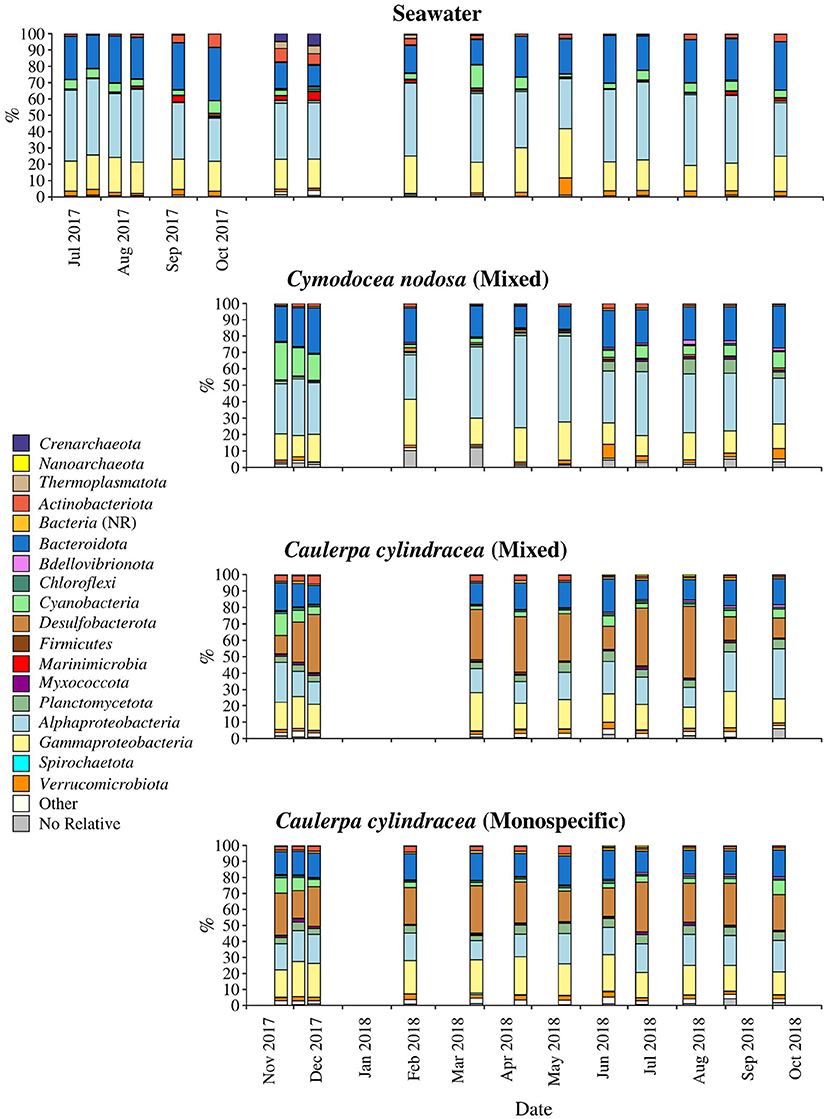
Figure 4. Taxonomic classification and relative contribution of the most abundant (≥1%) bacterial and archeal sequences on the surfaces of the macrophytes C. nodosa (mixed settlement) and C. cylindracea (mixed and monospecific settlement) and in the ambient seawater. NR, no relative (sequences without known relatives within the corresponding group).
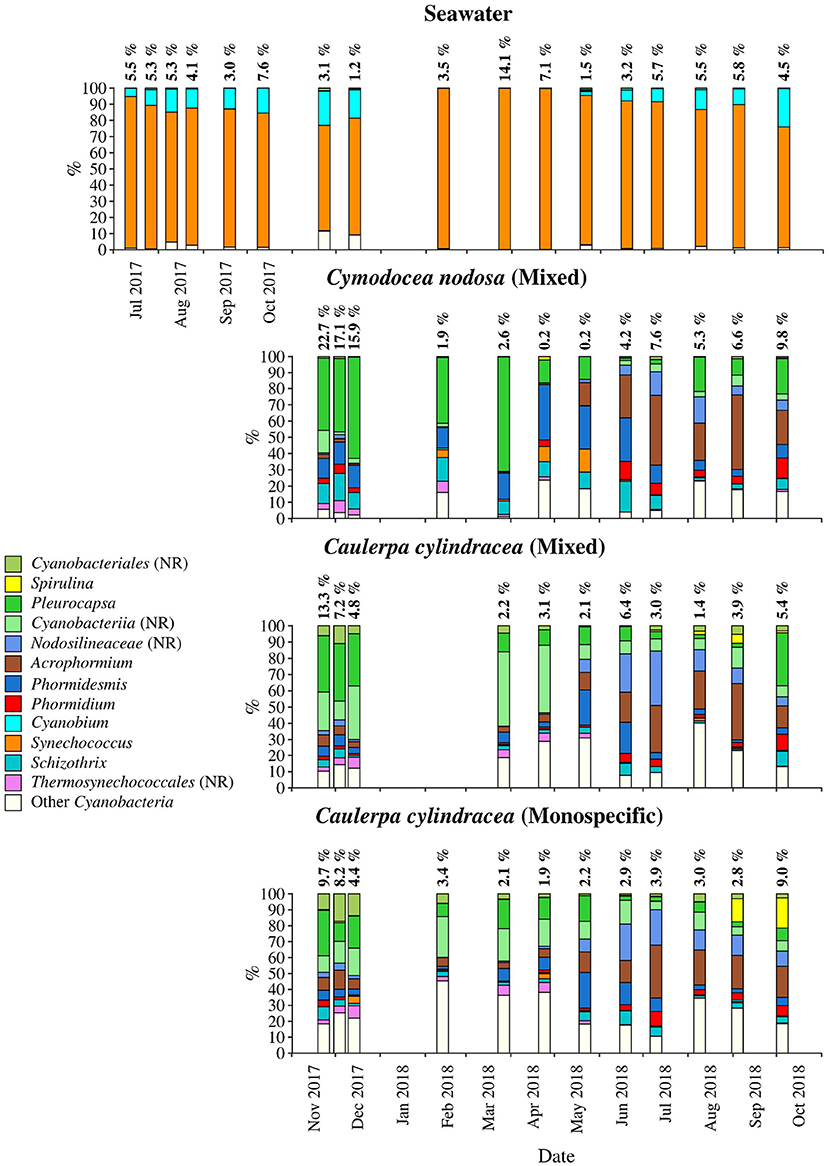
Figure 5. Taxonomic classification and relative contribution of the most abundant (≥1%) cyanobacterial sequences on the surfaces of the macrophytes C. nodosa (mixed settlement) and C. cylindracea (mixed and monospecific settlement) and in the ambient seawater. The proportion of cyanobacterial sequences in the total bacterial and archeal community is given above the corresponding bar. NR, no relative (sequences without known relatives within the corresponding group).
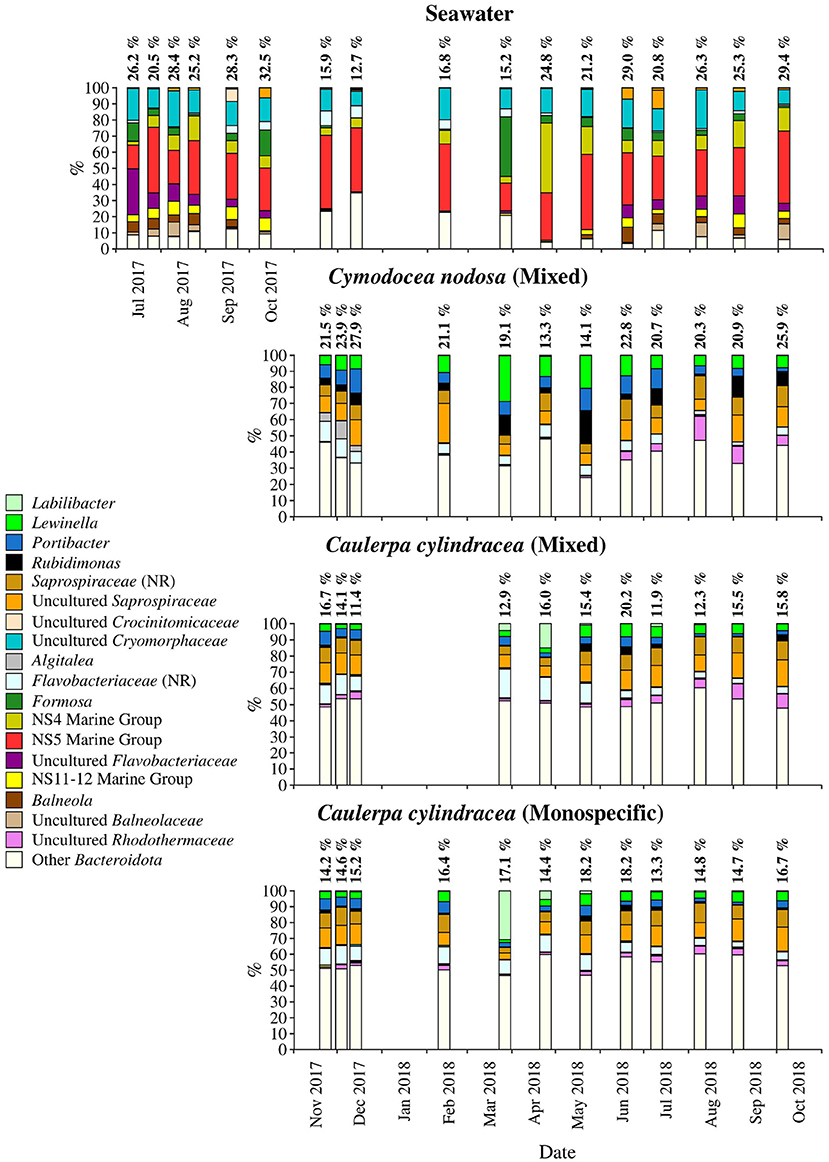
Figure 6. Taxonomic classification and relative contribution of the most abundant (≥ 2%) sequences within the Bacteroidota on the surfaces of the macrophytes C. nodosa (mixed settlement) and C. cylindracea (mixed and monospecific settlement) and in the ambient seawater. The proportion of sequences classified as Bacteroidota in the total bacterial and archaeal community is given above the corresponding bar. NR – No Relative (sequences without known relatives within the corresponding group).
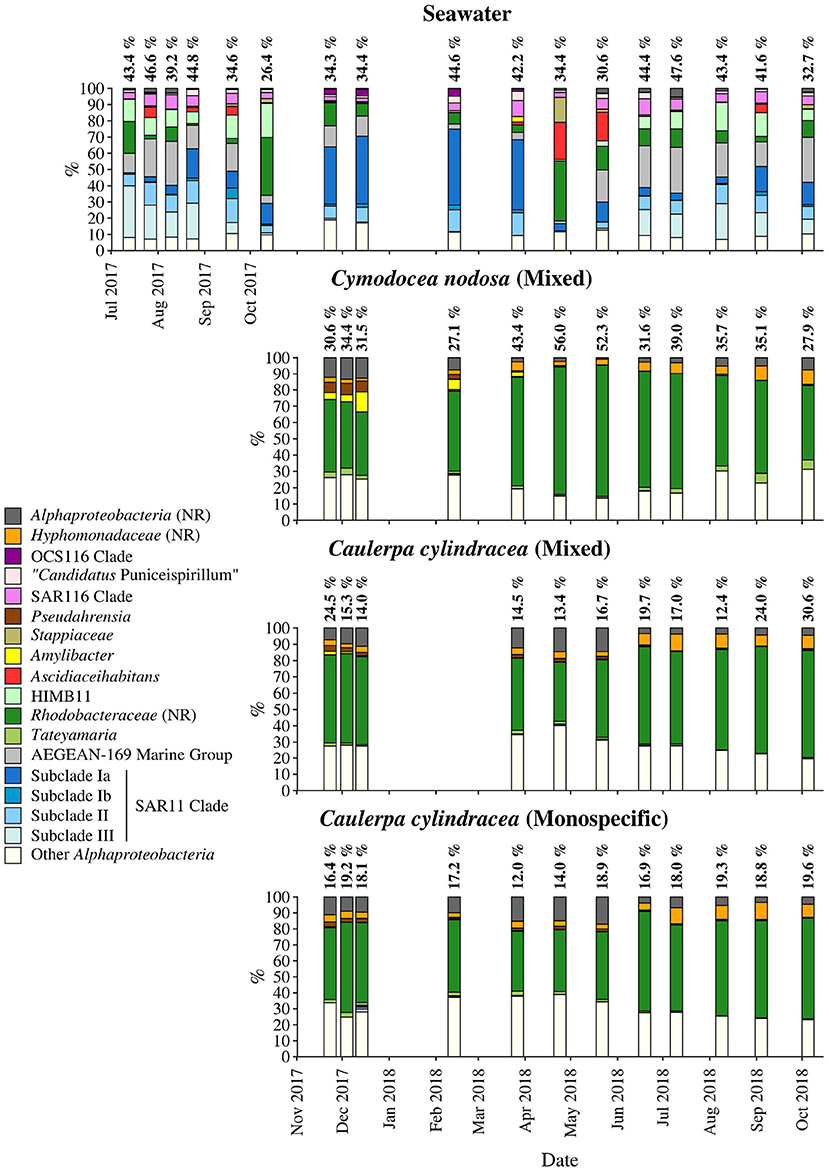
Figure 7. Taxonomic classification and relative contribution of the most abundant (≥ 2%) alphaproteobacterial sequences on the surfaces of the macrophytes C. nodosa (mixed settlement) and C. cylindracea (mixed and monospecific settlement) and in the ambient seawater. The proportion of alphaproteobacterial sequences in the total bacterial and archaeal community is given above the corresponding bar. NR – No Relative (sequences without known relatives within the corresponding group).
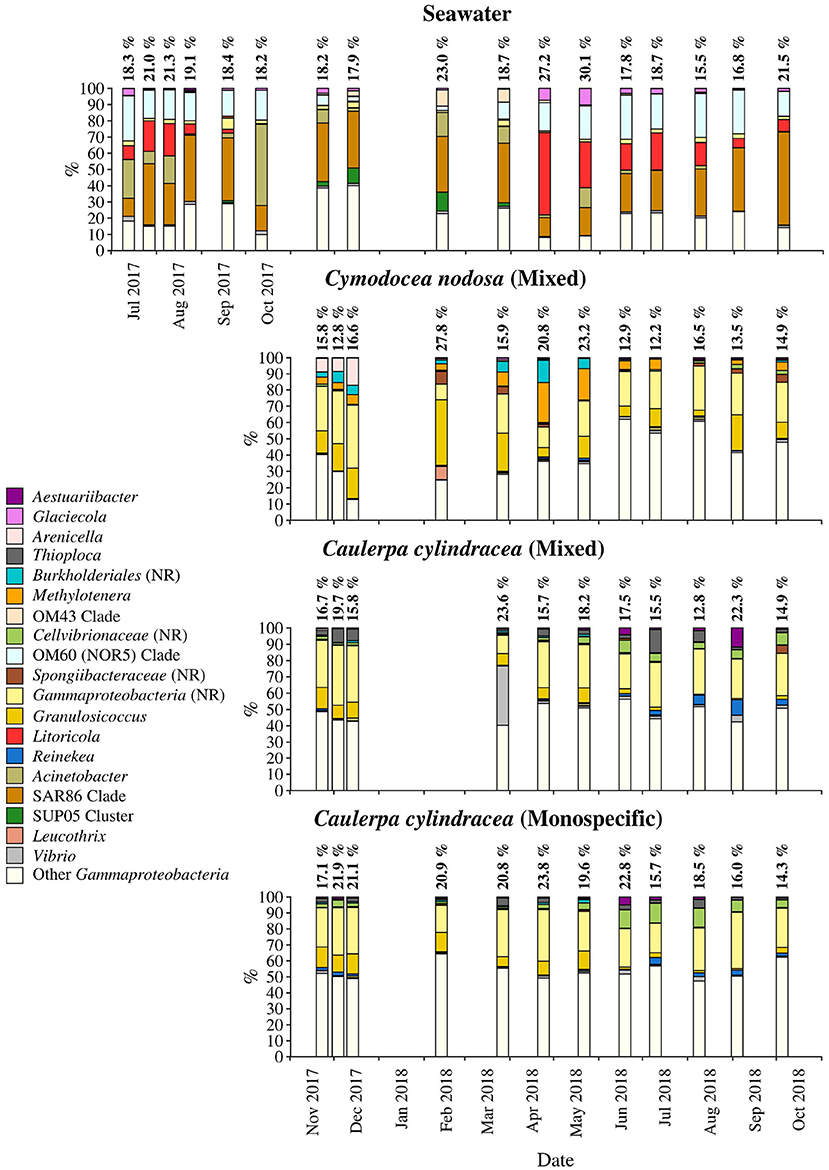
Figure 8. Taxonomic classification and relative contribution of the most abundant (≥1%) gammaproteobacterial sequences on the surfaces of the macrophytes C. nodosa (mixed settlement) and C. cylindracea (mixed and monospecific settlement) and in the ambient seawater. The proportion of gammaproteobacterial sequences in the total bacterial and archeal community is given above the corresponding bar. NR, no relative (sequences without known relatives within the corresponding group).
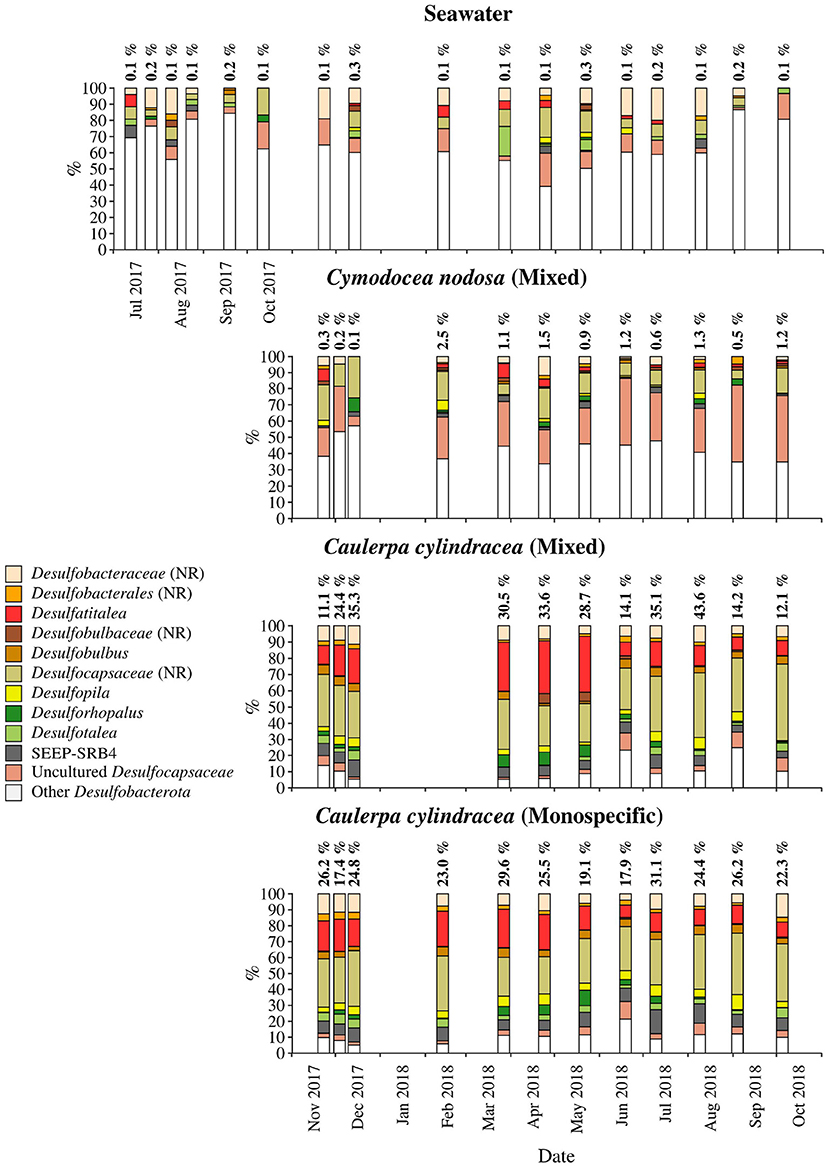
Figure 9. Taxonomic classification and relative contribution of the most abundant (≥ 1%) sequences within the Desulfobacterota on the surfaces of the macrophytes C. nodosa (mixed settlement) and C. cylindracea (mixed and monospecific settlement) and in the ambient seawater. The proportion of sequences classified as Desulfobacterota in the total bacterial and archaeal community is given above the corresponding bar. NR, no relative (sequences without known relatives within the corresponding group).
Cyanobacteria related sequences comprised, on average, 5.5 ± 4.4% of total sequences (Figure 5). Higher proportions were found for C. nodosa (16.4 ± 5.3%) and C. cylindracea mixed (7.7 ± 3.9%) and monospecific (7.8 ± 2.4%) associated communities in autumn (p < 0.0001) and for ambient seawater communities in winter (8.8 ± 7.5%). Large taxonomic differences between surface associated and ambient seawater cyanobacterial communities were observed. Ambient seawater communities were mainly comprised of Cyanobium and Synechococcus, while surface associated communities were comprised of Pleurocapsa and sequences within the class Cyanobacteriia that could not be further classified (no relative Cyanobacteriia; Figure 5). In addition, seasonal changes in surface associated cyanobacterial communities were observed with Pleurocapsa and no relative Cyanobacteriia comprising larger proportions of Cyanobacteria in autumn and winter and Acrophormium, Phormidesmis and sequences without known relatives within the Nodosilineaceae (no relative Nodosilineaceae) in spring and summer (Figure 5).
Sequences classified as Bacteroidota comprised, on average, 19.2 ± 5.5% of all sequences (Figure 6). Similar to Cyanobacteria, large differences in the taxonomic composition between ambient seawater and surface associated communities were found (Figure 6). The ambient seawater community was characterized by the NS4 and NS5 marine groups, uncultured Cryomorphaceae, uncultured Flavobacteriaceae, NS11-12 marine group, Balneola, uncultured Balneolaceae and Formosa. In contrast, in surface associated communities Lewinella, Portibacter, Rubidimonas, sequences without known relatives within the Saprospiraceae (no relative Saprospiraceae), uncultured Saprospiraceae, sequences without known relatives within the Flavobacteriaceae (no relative Flavobacteriaceae) and uncultured Rhodothermaceae were found. Some groups showed minor seasonal changes such as no relative Flavobacteriaceae whose sequences were more abundant from November 2017 until June 2018. In contrast, uncultured Rhodothermaceae showed higher proportions from June 2018 until the end of the study period. Surface associated Bacteroidota communities were very diverse as observed in the high proportion of taxa clustering as other Bacteroidota (Figure 6).
On average, Alphaproteobacteria were in comparison to the other high rank taxa the largest taxonomic group, comprising 29.2 ± 12.0% of all sequences (Figure 8). In accordance to the above described taxa, large differences between ambient seawater and surface associated communities were observed. Ambient seawater communities were composed mainly of the SAR11 clade, AEGEAN-169 marine group, SAR116 clade, sequences without known relatives within the Rhodobacteraceae (no relative Rhodobacteraceae), HIMB11 and the OCS116 clade, while surface associated communities were composed mainly of no relative Rhodobacteraceae and to a lesser degree of Pseudoahrensia, Amylibacter, and sequences without known relatives within the Alphaproteobacteria (no relative Alphaproteobacteria) and Hyphomonadaceae (no relative Hyphomonadaceae). Representatives of no relative Rhodobacteraceae comprised on average 54.7 ± 11.5% of all alphaproteobacterial sequences in the epiphytic community (Figure 7). In addition, Amylibacter was detected mainly in C. nodosa from November 2017 until March 2018.
Sequences related to Gammaproteobacteria comprised on average 18.6 ± 3.9 % of all sequences (Figure 8). Similar to above mentioned taxa, large taxonomic differences between ambient seawater and surface associated communities were found. Ambient seawater communities were mainly comprised of the OM60 (NOR5) clade, Litoricola, Acinetobacter and the SAR86 clade, while epiphytic communities were mainly composed of sequences without known relatives within the Gammaproteobacteria (no relative Gammaproteobacteria) and Granulosicoccus. Beside these two groups specific to all three epiphytic communities, C. nodosa was characterized by Arenicella, Methylotenera, and sequences without known relatives within the Burkholderiales (no relative Burkholderiales), while Thioploca, Reinekea, and sequences without known relatives within Cellvibrionaceae (no relative Cellvibrionaceae) were more specific to both mixed and monospecific C. cylindracea. In addition, Arenicella was more pronounced in November and December 2017, while no relative Burkholderiales and Methylotenera were characteristic for the period from March until May 2018. For the C. cylindracea specific taxa no relative Cellvibrionaceae and Reinekea showed seasonality and were characteristic for samples originating from June to October 2018. In addition, similar to Bacteroidota, a large proportion of the surface associated community was grouped as other Gammaproteobacteria indicating high diversity within this group (Figure 8).
Desulfobacterota were specific for C. cylindracea. In the mixed and monospecific C. cylindracea communities the proportion of Desulfobacterota was 25.7 ± 11.2 and 24.0 ± 4.3%, respectively (Figure 9). In contrast, in ambient seawater and C. nodosa communities the contribution of Desulfobacterota was only 0.1 ± 0.08 and 1.0 ± 0.7%, respectively. In C. cylindracea the community consisted mainly of Desulfatitalea, Desulfobulbus, Desulfopila, Desulforhopalus, Desulfotalea, SEEP-SRB4, uncultured Desulfocapsaceae and sequences without known relatives within the Desulfobacteraceae (no relative Desulfobacteraceae), Desulfobulbaceae (no relative Desulfobulbaceae), and Desulfocapsaceae (no relative Desulfocapsaceae; Figure 9).
4. Discussion
In the present study, we applied a selective epiphytic DNA isolation procedure based on direct cellular lysis (Korlević et al., 2021) coupled with a monthly sampling and Illumina amplicon sequencing to describe in detail the bacterial and archaeal communities associated with the surfaces of two marine macrophytes, C. nodosa and C. cylindracea. Highest richness was observed for C. cylindracea (mixed and monospecific) followed by C. nodosa and lowest richness was found in ambient seawater microbial communities. Higher richness of microbial communities associated with macrophytes than in ambient seawater has been described earlier (Mancuso et al., 2016; Ugarelli et al., 2019) and could be attributed to a larger set of inhabitable microniches existing on macrophyte surfaces than in the ambient seawater. The highest richness observed for C. cylindracea might be partly due to its contact with the sediment. The stolon of C. cylindracea is attached to the sediment surface with rhizoids and thus, the stolon and rhizoids are in direct contact with the sediment. Also, studies have shown that the presence of C. cylindracea can alter the content and biochemical composition of sedimentary organic matter (Pusceddu et al., 2016; Rizzo et al., 2017, 2020) possibly further expanding the number of inhabitable microniches and thus causing the observed increase in richness. Seasonal differences in richness observed for surface attached communities indicated a slightly higher richness in spring and summer. This pattern could be explained by a more intense macrophyte growth in these two seasons than in autumn and winter (Zavodnik et al., 1998; Ruitton et al., 2005; Najdek et al., 2020). During their main growth season in spring and summer macrophytes exhibit a more dynamic chemical interaction with the surface community probably causing an increase in the number of inhabitable microniches (Borges and Champenois, 2015; Rickert et al., 2016). Proportions of shared epiphytic OTUs between consecutive sampling points were low also indicated by the proportion of OTUs ( ≤ 1.0%) present at every sampling date (Figure 3). These persistent OTUs, however, accounted for a high proportion of sequences (≥40.2%), as is often the case with similar high-frequency sampling studies (Gilbert et al., 2009, 2012). In comparison to the seawater community, higher values of shared OTUs between consecutive sampling points were observed for the macrophyte surface associated communities. It appears that macrophyte surfaces are providing more stable conditions than the ambient seawater.
We observed a strong differentiation between the surface attached and ambient seawater communities at the level of OTUs which is in agreement with most published studies (Burke et al., 2011b; Michelou et al., 2013; Mancuso et al., 2016; Roth-Schulze et al., 2016; Crump et al., 2018; Ugarelli et al., 2019; Sanders-Smith et al., 2020). This indicates that marine macrophytes are selecting microorganisms from the pool of microbial taxa present in the ambient seawater, modifying the microbial community once the macrophyte associated microbial biofilm develops (Salaün et al., 2012; Michelou et al., 2013). In addition, similar to the study of Roth-Schulze et al. (2016) seagrass and macroalgae specific microbial communities were identified, while no difference between C. cylindracea settlements was observed indicating that seagrass and macroalgae specific metabolism is involved in the selection and development of the associated biofilm. At the level of OTUs seasonal changes of C. nodosa and C. cylindracea associated communities were identified that could be linked to the growth cycle of the seagrass and macroalgae (Agostini et al., 2003; Najdek et al., 2020). C. nodosa was characterized by a spring community during maximum seagrass proliferation, a summer community during the highest standing stock of C. nodosa and an autumn/winter community during the decay of seagrass biomass. In contrast, C. cylindracea started to proliferate in late spring and was characterized only by a summer community during high growth rates and by an autumn/winter/spring community when the biomass was at the peak and decaying thereafter. Similar seasonal changes in the epiphytic community have also been described for other macroalgae (Tujula et al., 2010; Lachnit et al., 2011).
The taxonomic analysis showed higher chloroplast sequence abundances in autumn/winter than in spring/summer. This pattern is not surprising as seagrasses harbour more algal epiphytes during autumn/winter than in spring/summer (Reyes and Sansón, 2001). Furthermore, we used an adapted DNA isolation protocol that is known to partially co-extract DNA from planktonic eukaryotes (Korlević et al., 2015). In general, the taxonomic analysis identified epiphytic phylogenetic groups present throughout the year comprising most of the reads, and taxa present in lower proportions showing seasonal patterns. The first group was comprised of members of the Bacteroidota family Saprospiraceae, the alphaproteobacterial Rhodobacteraceae and Hyphomonadaceae, the gammaproteobacterial genus Granulosicoccus, sequences without known relatives within Gammaproteobacteria and various taxa within Desulfobacterota (Figures 6–9). All these groups were found on all host species, with the exception of Desulfobacterota that was characteristic for C. cylindracea. In addition, the persistence of Rhodobacteraceae in the case of C. nodosa and Desulfobacterota in the case of C. cylindracea could be observed in the taxonomic classification of OTUs present at every sampling date. Within the Bacteroidota different groups within Saprospiraceae (e.g., Lewinella, Portibacter, and Rubidimonas) were identified to be persistent. It has been suggested that members of this family are important in the hydrolysis and utilization of complex organic sources (McIlroy and Nielsen, 2014). Surface attached life style would be beneficial to these microbes as they could thrive on products of host cellular breakdown or by-products of host metabolism, so it not surprising that they are often found associated with macrophyte surfaces (Burke et al., 2011b; McIlroy and Nielsen, 2014; Crump et al., 2018). Rhodobacteraceae are often detected on macrophyte surfaces and usually are one of the most abundant groups (Burke et al., 2011b; Michelou et al., 2013; Mancuso et al., 2016). The functional association between macrophytes and members of this groups is difficult to assess based on 16S rRNA analysis as this family is phenotypically, metabolically, and ecologically very diverse (Pujalte et al., 2014). However, some interesting metabolic capacities linked to this group were described. Genomic analysis of Rhodobacteraceae strains and metatranscriptomic sequencing of seagrass microbiomes revealed the potential for biosynthesis of indole-3-acetic acid (IAA), a plant hormone (Simon et al., 2017), indicating a possible intake by seagrasses. However, another study found no effect of IAA on C. nodosa growth showing the complexity of macrophyte–microbes interactions (Muñoz, 1995). Another persistent alphaproteobacterial family was the Hyphomonadaceae, a group that contain species with stalks used to attach cells to different surfaces (Abraham and Rohde, 2014). This group has been previously associated with seagrass surfaces (Weidner et al., 2000) and it is believed that possessing stalks could be an advantage to keep the cells in the proximity of exudate excreted by the host (Weidner et al., 2000; Abraham and Rohde, 2014).
Within the Gammaproteobacteria, sequences without known representatives were the most pronounced group present throughout the year. Gammaproteobacteria are often a major constituent of macrophyte epiphytic communities (Burke et al., 2011b; Michelou et al., 2013; Crump et al., 2018). A study has attributed the expression of enzymes for the degradation of galactose-based algal polymers to this class indicating their possible involvement into epibiotic algal biofilm control (Crump et al., 2018). In addition, Granulosicoccus was also found in almost all samples. A species of this genus has been isolated from the leaf surface of the seagrass Zostera marina (Kurilenko et al., 2010), while sequences related to this genus have been found on the surfaces of macroalgae (Lachnit et al., 2011; Bengtsson et al., 2012), including C. cylindracea (Rizzo et al., 2016a), indicating this group preference for macrophyte surfaces. It is possible that bacteria of this genus can thrive on exudates of different macrophytes as it is known from cultivated members that they can utilize various sugars and amino acids (Ivanova and Webb, 2014). The presence of Desulfobacterota only on C. cylindracea is to be expected as part of the epiphytic community is in direct contact with the sediment. The Desulfobacterota community was comprised of known sulphate sediment groups such as the Desulfatitalea and no relative Desulfocapsaceae (Kuever, 2014; Higashioka et al., 2015). Sequences related to sulphur cycling bacterial groups have been found in Caulerpa endophytic and epiphytic communities (Aires et al., 2013). It is possible that these groups are involved into enhanced sulphate reduction rates observed in sediments underlying Caulerpa settlements causing unsuitable conditions to sulphide-sensitive seagrasses (Holmer et al., 2009).
The only high rank taxonomic group showing strong seasonal fluctuations was Cyanobacteria. Cyanobacterial sequences were more pronounced in November and December than in spring and summer. In the months of high cyanobacterial sequence abundances the majority of sequences from this group were classified as Pleurocapsa, a group known to colonize different living and non-living surfaces (Burns et al., 2004; Longford et al., 2007; Mobberley et al., 2012; Reisser et al., 2014; Kolda et al., 2020). While we observed a strong temporal pattern for this group, a study of surface sediment cyanobacterial communities did not find any seasonal dynamics for Pleurocapsa (Kolda et al., 2020), indicating a possibility that there is a reduced selection of the epiphytic community by the seagrass during periods of low photosynthetic activity (Zavodnik et al., 1998), causing leaves to become a suitable surface for non-specific colonizers. Beside all these groups comprising most of the sequences, a set of taxa present in lower proportions and showing seasonal patterns was identified. This group was comprised of e.g., Bacteroidota sequences without known relatives within Flavobacteriaceae and Rhodothermaceae, the alphaproteobacterial Amylibacter and the gammaproteobacterial Methylotenera, Reinekea and sequences without known relatives within Cellvibrionaceae (Figures 6, 8).
It is possible that Flavobacteriaceae and Rhodothermaceae are occupying similar niches with Rhodothermaceae being more adapted to higher temperatures as it is known that culturable members of this family exhibit mesophilic and thermophilic characteristics (Park et al., 2014). This would explain why we observed a higher presence of Rhodothermaceae in the warmer period of the year. A strain belonging to the Rhodobacteraceae genus Amylibacter has been isolated from the surface of a green macroalga indicating that members of this group can exhibit surface attached life style (Nedashkovskaya et al., 2016). In addition, since this is a relatively novel genus it is possible that novel taxa within Rhodobacteraceae will be described in the future elucidating the taxonomy of the currently high proportion of Rhodobacteraceae sequences without known relatives. The genus Methylotenera belongs to the methylotrophic family Methylophilaceae, a group capable of oxidizing non-methane single-carbon compounds such as methanol and methylamine (Chistoserdova and Kalyuzhnaya, 2018). Interestingly, angiosperms produce methanol during cell-wall synthesis (Nemecek-Marshall et al., 1995; Dorokhov et al., 2018), so it is not surprising that we found members of this genus only on C. nodosa and in spring, during a period of maximum seagrass proliferation. Other studies have also found Methylotenera-specific sequences associated with seagrass roots and leaves indicating that this group members are important constituents of the seagrass microbiome (Crump et al., 2018; Sanders-Smith et al., 2020). Genomic and physiological analyses of cultivated Cellvibrionaceae and Reinekea members showed the capabilities to use important algal polysaccharides (Avcı et al., 2017; Xie et al., 2017) indicating their possible involvement into the degradation of C. cylindracea polysaccharides and/or the control of its algal epiphytes.
The epiphytic microbial community associated with marine macrophytes is undergoing seasonal changes that can be attributed to the fluctuations of environmental conditions, the growth cycle of macrophytes inhabiting temperate zones or the combined effect of both. In the present study, we could identify in analyzed high rank taxa phylogenetic groups present throughout the year, comprising most of the sequences and a lower proportion of taxa showing seasonal patterns connected to the macrophyte growth cycle (Figures 4, 9). Studies focusing on functional comparisons between communities associated with different hosts showed that the majority of functions could be found in every community, indicating functional redundancy (Roth-Schulze et al., 2016). This difference between phylogenetic variability and functional stability has been explained by the lottery hypothesis assuming an initial random colonization step performed by a set of functionally equivalent taxonomic groups (Burke et al., 2011a; Roth-Schulze et al., 2016). It is possible that functional redundancy is a characteristic of high abundance taxa detected to be present throughout the year, while seasonal and/or host-specific functions are an attribute of taxa displaying successional patterns. Further studies connecting taxonomy with functional properties will be required to elucidate the degree of functional redundancy or specificity in epiphytic microbial communities.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repositories and the accession number can be found in the article.
Author Contributions
MK, GH, and MN designed the study. MK, MM, ZZ, and MN performed sampling and laboratory analysis. MK, MM, and ZZ analyzed the data. MK prepared the manuscript with editorial help from MM, ZZ, GH, and MN. All authors contributed to the article and approved the final submitted version.
Funding
This work was funded by the Croatian Science Foundation through the MICRO-SEAGRASS project (project number IP-2016-06-7118). ZZ and GH were supported by the Austrian Science Fund (FWF) project ARTEMIS (project number P28781-B21).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We would like to thank Margareta Buterer for technical support, Paolo Paliaga for help during sampling and the University Computing Center of the University of Zagreb for access to the computer cluster Isabella.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.671342/full#supplementary-material
References
Abraham, W. R., and Rohde, M. (2014). “The family Hyphomonadaceae,” in The Prokaryotes: Alphaproteobacteria and Betaproteobacteria, eds E. Rosenberg, E. F. DeLong, S. Lory, E. Stackebrandt, and F. Thompson (Berlin; Heidelberg: Springer-Verlag), 283–299.
Agostini, S., Pergent, G., and Marchand, B. (2003). Growth and primary production of Cymodocea nodosa in a coastal lagoon. Aquat. Bot. 76, 185–193. doi: 10.1016/S0304-3770(03)00049-4
Aires, T., Moalic, Y., Serrao, E. A., and Arnaud-Haond, S. (2015). Hologenome theory supported by cooccurrence networks of species-specific bacterial communities in siphonous algae (Caulerpa). FEMS Microbiol. Ecol. 91:fiv067. doi: 10.1093/femsec/fiv067
Aires, T., Serrão, E. A., Kendrick, G., Duarte, C. M., and Arnaud-Haond, S. (2013). Invasion is a community affair: Clandestine followers in the bacterial community associated to green algae, Caulerpa racemosa, track the invasion source. PLoS ONE 8:e68429. doi: 10.1371/journal.pone.0068429
Allaire, J. J., Xie, Y., McPherson, J., Luraschi, J., Ushey, K., Atkins, A., et al. (2019). rmarkdown: Dynamic Documents for R.
Apprill, A., McNally, S., Parsons, R., and Weber, L. (2015). Minor revision to V4 region SSU rRNA 806R gene primer greatly increases detection of SAR11 bacterioplankton. Aquat. Microb. Ecol. 75, 129–137. doi: 10.3354/ame01753
Armstrong, E., Rogerson, A., and Leftley, J. W. (2000). The abundance of heterotrophic protists associated with intertidal seaweeds. Estuar. Coast. Shelf Sci. 50, 415–424. doi: 10.1006/ECSS.1999.0577
Avcı, B., Hahnke, R. L., Chafee, M., Fischer, T., Gruber-Vodicka, H., Tegetmeyer, H. E., et al. (2017). Genomic and physiological analyses of ‘Reinekea forsetii' reveal a versatile opportunistic lifestyle during spring algae blooms. Environ. Microbiol. 19, 1209–1221. doi: 10.1111/1462-2920.13646
Bengtsson, M., Sjøtun, K., and Øvreås, L. (2010). Seasonal dynamics of bacterial biofilms on the kelp Laminaria hyperborea. Aquat. Microb. Ecol. 60, 71–83. doi: 10.3354/ame01409
Bengtsson, M. M., Sjøtun, K., Lanzén, A., and Øvreås, L. (2012). Bacterial diversity in relation to secondary production and succession on surfaces of the kelp Laminaria hyperborea. ISME J. 6, 2188–2198. doi: 10.1038/ismej.2012.67
Borcard, D., Gillet, F., and Legendre, P. (2011). Numerical Ecology With R 1st Edn. New York, NY: Springer-Verlag.
Borges, A. V., and Champenois, W. (2015). Seasonal and spatial variability of dimethylsulfoniopropionate (DMSP) in the Mediterranean seagrass Posidonia oceanica. Aquat. Bot. 125, 72–79. doi: 10.1016/j.aquabot.2015.05.008
Boudouresque, C. F., Bernard, G., Pergent, G., Shili, A., and Verlaque, M. (2009). Regression of Mediterranean seagrasses caused by natural processes and anthropogenic disturbances and stress: a critical review. Bot. Mar. 52, 395–418. doi: 10.1515/BOT.2009.057
Burke, C., Steinberg, P., Rusch, D., Kjelleberg, S., and Thomas, T. (2011a). Bacterial community assembly based on functional genes rather than species. Proc. Natl. Acad. Sci. U.S.A. 108, 14288–14293. doi: 10.1073/pnas.1101591108
Burke, C., Thomas, T., Lewis, M., Steinberg, P., and Kjelleberg, S. (2011b). Composition, uniqueness and variability of the epiphytic bacterial community of the green alga Ulva australis. ISME J. 5, 590–600. doi: 10.1038/ismej.2010.164
Burns, B. P., Goh, F., Allen, M., and Neilan, B. A. (2004). Microbial diversity of extant stromatolites in the hypersaline marine environment of Shark Bay, Australia. Environ. Microbiol. 6, 1096–1101. doi: 10.1111/j.1462-2920.2004.00651.x
Caporaso, J. G., Lauber, C. L., Walters, W. A., Berg-Lyons, D., Huntley, J., Fierer, N., et al. (2012). Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 6, 1621–1624. doi: 10.1038/ismej.2012.8
Celdran, D., Espinosa, E., Sánchez-Amat, A., and Marín, A. (2012). Effects of epibiotic bacteria on leaf growth and epiphytes of the seagrass Posidonia oceanica. Mar. Ecol. Prog. Ser. 456, 21–27. doi: 10.3354/meps09672
Chistoserdova, L., and Kalyuzhnaya, M. G. (2018). Current trends in methylotrophy. Trends Microbiol. 26, 703–714. doi: 10.1016/j.tim.2018.01.011
Crump, B. C., and Koch, E. W. (2008). Attached bacterial populations shared by four species of aquatic angiosperms. Appl. Environ. Microbiol. 74, 5948–5957. doi: 10.1128/AEM.00952-08
Crump, B. C., Wojahn, J. M., Tomas, F., and Mueller, R. S. (2018). Metatranscriptomics and amplicon sequencing reveal mutualisms in seagrass microbiomes. Front. Microbiol. 9:388. doi: 10.3389/fmicb.2018.00388
Cúcio, C., Engelen, A. H., Costa, R., and Muyzer, G. (2016). Rhizosphere microbiomes of european seagrasses are selected by the plant, but are not species specific. Front. Microbiol. 7:440. doi: 10.3389/fmicb.2016.00440
Cúcio, C., Overmars, L., Engelen, A. H., and Muyzer, G. (2018). Metagenomic analysis shows the presence of bacteria related to free-living forms of sulfur-oxidizing chemolithoautotrophic symbionts in the rhizosphere of the seagrass Zostera marina. Front. Mar. Sci. 5:171. doi: 10.3389/fmars.2018.00171
de Oliveira, L. S., Gregoracci, G. B., Silva, G. G. Z., Salgado, L. T., Filho, G. A., Alves-Ferreira, M., et al. (2012). Transcriptomic analysis of the red seaweed Laurencia dendroidea (Florideophyceae, Rhodophyta) and its microbiome. BMC Genomics 13:487. doi: 10.1186/1471-2164-13-487
Dobretsov, S. V., and Qian, P.-Y. (2002). Effect of bacteria associated with the green alga Ulva reticulata on marine micro- and macrofouling. Biofouling 18, 217–228. doi: 10.1080/08927010290013026
Dorokhov, Y. L., Sheshukova, E. V., and Komarova, T. V. (2018). Methanol in plant life. Front. Plant Sci. 9:1623. doi: 10.3389/fpls.2018.01623
Egan, S., Harder, T., Burke, C., Steinberg, P., Kjelleberg, S., and Thomas, T. (2013). The seaweed holobiont: understanding seaweed-bacteria interactions. FEMS Microbiol. Rev. 37, 462–476. doi: 10.1111/1574-6976.12011
Ettinger, C. L., and Eisen, J. A. (2020). Fungi, bacteria and oomycota opportunistically isolated from the seagrass, Zostera marina. PLoS ONE 15:e0236135. doi: 10.1371/journal.pone.0236135
Gilbert, J. A., Field, D., Swift, P., Newbold, L., Oliver, A., Smyth, T., et al. (2009). The seasonal structure of microbial communities in the Western English Channel. Environ. Microbiol. 11, 3132–3139. doi: 10.1111/j.1462-2920.2009.02017.x
Gilbert, J. A., Steele, J. A., Caporaso, J. G., Steinbrück, L., Reeder, J., Temperton, B., et al. (2012). Defining seasonal marine microbial community dynamics. ISME J. 6, 298–308. doi: 10.1038/ismej.2011.107
Higashioka, Y., Kojima, H., Watanabe, T., and Fukui, M. (2015). Draft genome sequence of Desulfatitalea tepidiphila S28bFT. Genome Announc. 3:e01326–15. doi: 10.1128/genomeA.01326-15
Hollants, J., Leliaert, F., De Clerck, O., and Willems, A. (2013). What we can learn from sushi: A review on seaweed-bacterial associations. FEMS Microbiol. Ecol. 83, 1–16. doi: 10.1111/j.1574-6941.2012.01446.x
Holmer, M., Marbà, N., Lamote, M., and Duarte, C. M. (2009). Deterioration of sediment quality in seagrass meadows (Posidonia oceanica) invaded by macroalgae (Caulerpa sp.). Estuar. Coast 32, 456–466. doi: 10.1007/s12237-009-9133-4
Ivanova, E. P., and Webb, H. K. (2014). “The family Granulosicoccaceae,” in The Prokaryotes: Gammaproteobacteria, eds E. Rosenberg, E. F. DeLong, S. Lory, E. Stackebrandt, and F. Thompson (Berlin; Heidelberg: Springer-Verlag), 315–317.
Klein, J., and Verlaque, M. (2008). The Caulerpa racemosa invasion: a critical review. Mar. Pollut. Bull. 56, 205–225. doi: 10.1016/j.marpolbul.2007.09.043
Kolda, A., Ljubešiç, Z., Gavrilović, A., Jug-Dujaković, J., Pikelj, K., and Kapetanović, D. (2020). Metabarcoding Cyanobacteria in coastal waters and sediment in central and southern Adriatic Sea. Acta Bot. Croat. 79, 157–169. doi: 10.37427/botcro-2020-021
Korlević, M., Markovski, M., Zhao, Z., Herndl, G. J., and Najdek, M. (2021). Selective DNA and protein isolation from marine macrophyte surfaces. Front. Microbiol. 12:665999. doi: 10.3389/fmicb.2021.665999
Korlević, M., Pop Ristova, P., Garić, R., Amann, R., and Orli, S. (2015). Bacterial diversity in the South Adriatic Sea during a strong, deep winter convection year. Appl. Environ. Microbiol. 81, 1715–1726. doi: 10.1128/AEM.03410-14
Kozich, J. J., Westcott, S. L., Baxter, N. T., Highlander, S. K., and Schloss, P. D. (2013). Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl. Environ. Microbiol. 79, 5112–5120. doi: 10.1128/AEM.01043-13
Kuever, J. (2014). “The family Desulfobulbaceae,” in The Prokaryotes: Deltaproteobacteria and Epsilonproteobacteria, eds E. Rosenberg, E. F. DeLong, S. Lory, E. Stackebrandt, and F. Thompson (Berlin; Heidelberg: Springer-Verlag), 75–86.
Kurilenko, V. V., Christen, R., Zhukova, N. V., Kalinovskaya, N. I., Mikhailov, V. V., Crawford, R. J., et al. (2010). Granulosicoccus coccoides sp. nov., isolated from leaves of seagrass (Zostera marina). Int. J. Syst. Evol. Microbiol. 60, 972–976. doi: 10.1099/ijs.0.013516-0
Küsel, K., Trinkwalter, T., Drake, H. L., and Devereux, R. (2006). Comparative evaluation of anaerobic bacterial communities associated with roots of submerged macrophytes growing in marine or brackish water sediments. J. Exp. Mar. Biol. Ecol. 337, 49–58. doi: 10.1016/j.jembe.2006.06.004
Lachnit, T., Blümel, M., Imhoff, J. F., and Wahl, M. (2009). Specific epibacterial communities on macroalgae: phylogeny matters more than habitat. Aquat. Biol. 5, 181–186. doi: 10.3354/ab00149
Lachnit, T., Meske, D., Wahl, M., Harder, T., and Schmitz, R. (2011). Epibacterial community patterns on marine macroalgae are host-specific but temporally variable. Environ. Microbiol. 13, 655–665. doi: 10.1111/j.1462-2920.2010.02371.x
Longford, S., Tujula, N., Crocetti, G., Holmes, A., Holmström, C., Kjelleberg, S., et al. (2007). Comparisons of diversity of bacterial communities associated with three sessile marine eukaryotes. Aquat. Microb. Ecol. 48, 217–229. doi: 10.3354/ame048217
Mancuso, F. P., D'Hondt, S., Willems, A., Airoldi, L., and De Clerck, O. (2016). Diversity and temporal dynamics of the epiphytic bacterial communities associated with the canopy-forming seaweed Cystoseira compressa (Esper) Gerloff and Nizamuddin. Front. Microbiol. 7:476. doi: 10.3389/fmicb.2016.00476
Margulis, L. (1991). “Symbiogenesis and symbionticism,” in Symbiosis as a Source of Evolutionary Innovation: Speciation and Morphogenesis, eds L. Margulis and R. Fester (Cambridge, MA: The MIT Press), 1–14.
Massana, R., Murray, A. E., Preston, C. M., and DeLong, E. F. (1997). Vertical distribution and phylogenetic characterization of marine planktonic Archaea in the Santa Barbara Channel. Appl. Environ. Microbiol. 63, 50–56.
Matsuo, Y., Suzuki, M., Kasai, H., Shizuri, Y., and Harayama, S. (2003). Isolation and phylogenetic characterization of bacteria capable of inducing differentiation in the green alga Monostroma oxyspermum. Environ. Microbiol. 5, 25–35. doi: 10.1046/j.1462-2920.2003.00382.x
McIlroy, S. J., and Nielsen, P. H. (2014). “The family Saprospiraceae,” in The Prokaryotes: Other Major Lineages of Bacteria and the Archaea, eds E. Rosenberg, E. F. DeLong, S. Lory, E. Stackebrandt, and F. Thompson (Berlin; Heidelberg: Springer-Verlag), 863–889.
Michelou, V. K., Caporaso, J. G., Knight, R., and Palumbi, S. R. (2013). The ecology of microbial communities associated with Macrocystis pyrifera. PLoS ONE 8:e67480. doi: 10.1371/journal.pone.0067480
Miranda, L. N., Hutchison, K., Grossman, A. R., and Brawley, S. H. (2013). Diversity and abundance of the bacterial community of the red macroalga Porphyra umbilicalis: did bacterial farmers produce macroalgae? PLoS ONE 8:e58269. doi: 10.1371/journal.pone.0058269
Mobberley, J. M., Ortega, M. C., and Foster, J. S. (2012). Comparative microbial diversity analyses of modern marine thrombolitic mats by barcoded pyrosequencing. Environ. Microbiol. 14, 82–100. doi: 10.1111/j.1462-2920.2011.02509.x
Morrissey, K. L., Çavas, L., Willems, A., and De Clerck, O. (2019). Disentangling the influence of environment, host specificity and thallus differentiation on bacterial communities in siphonous green seaweeds. Front. Microbiol. 10:717. doi: 10.3389/fmicb.2019.00717
Muñoz, J. T. (1995). Effects of some plant growth regulators on the growth of the seagrass Cymodocea nodosa (Ucria) Ascherson. Aquat. Bot. 51, 311–318. doi: 10.1016/0304-3770(95)00481-E
Najdek, M., Korlević, M., Paliaga, P., Markovski, M., Ivančić, I., Iveša, L., et al. (2020). Effects of the invasion of Caulerpa cylindracea in a Cymodocea nodosa meadow in the Northern Adriatic Sea. Front. Mar. Sci. 7:602055. doi: 10.3389/fmars.2020.602055
Nedashkovskaya, O. I., Kukhlevskiy, A. D., Zhukova, N. V., and Kim, S. B. (2016). Amylibacter ulvae sp. nov., a new alphaproteobacterium isolated from the Pacific green alga Ulva fenestrata. Arch. Microbiol. 198, 251–256. doi: 10.1007/s00203-015-1185-1
Nemecek-Marshall, M., MacDonald, R. C., Franzen, J. J., Wojciechowski, C. L., and Fall, R. (1995). Methanol emission from leaves (enzymatic detection of gas-phase methanol and relation of methanol fluxes to stomatal conductance and leaf development). Plant Physiol. 108, 1359–1368. doi: 10.1104/pp.108.4.1359
Nielsen, L. B., Finster, K., Welsh, D. T., Donelly, A., Herbert, R. A., de Wit, R., et al. (2001). Sulphate reduction and nitrogen fixation rates associated with roots, rhizomes and sediments from Zostera noltii and Spartina maritima meadows. Environ. Microbiol. 3, 63–71. doi: 10.1046/j.1462-2920.2001.00160.x
Oksanen, J., Blanchet, F. G., Friendly, M., Kindt, R., Legendre, P., McGlinn, D., et al. (2019). vegan: Community Ecology Package.
Parada, A. E., Needham, D. M., and Fuhrman, J. A. (2016). Every base matters: Assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ. Microbiol. 18, 1403–1414. doi: 10.1111/1462-2920.13023
Park, S., Akira, Y., and Kogure, K. (2014). “The family Rhodothermaceae,” in The Prokaryotes: Other Major Lineages of Bacteria and the Archaea, eds E. Rosenberg, E. F. DeLong, S. Lory, E. Stackebrandt, and F. Thompson (Berlin; Heidelberg: Springer-Verlag), 849–856.
Penesyan, A., Marshall-Jones, Z., Holmstrom, C., Kjelleberg, S., and Egan, S. (2009). Antimicrobial activity observed among cultured marine epiphytic bacteria reflects their potential as a source of new drugs. FEMS Microbiol. Ecol. 69, 113–124. doi: 10.1111/j.1574-6941.2009.00688.x
Pujalte, M. J., Lucena, T., Ruvira, M. A., Arahal, D. R., and Macin, M. C. (2014). “The family Rhodobacteraceae,” in The Prokaryotes: Alphaproteobacteria and Betaproteobacteria, eds E. Rosenberg, E. F. DeLong, S. Lory, E. Stackebrandt, and F. Thompson (Berlin; Heidelberg: Springer-Verlag), 439–512.
Pusceddu, A., Fraschetti, S., Scopa, M., Rizzo, L., and Danovaro, R. (2016). Meiofauna communities, nematode diversity and C degradation rates in seagrass (Posidonia oceanica L.) and unvegetated sediments invaded by the algae Caulerpa cylindracea (Sonder). Mar. Environ. Res. 119, 88–99. doi: 10.1016/j.marenvres.2016.05.015
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2013). The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucl. Acids Res. 41, D590–D596. doi: 10.1093/nar/gks1219
R Core Team (2019). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing.
Reisser, J., Shaw, J., Hallegraeff, G., Proietti, M., Barnes, D. K. A., Thums, M., et al. (2014). Millimeter-sized marine plastics: a new pelagic habitat for microorganisms and invertebrates. PLoS ONE 9:e100289. doi: 10.1371/journal.pone.0100289
Reyes, J., and Sansón, M. (2001). Biomass and production of the epiphytes on the leaves of Cymodocea nodosa in the Canary Islands. Bot. Mar. 44, 307–313. doi: 10.1515/BOT.2001.039
Rickert, E., Wahl, M., Link, H., Richter, H., and Pohnert, G. (2016). Seasonal variations in surface metabolite composition of Fucus vesiculosus and Fucus serratus from the Baltic Sea. PLoS ONE 11:e0168196. doi: 10.1371/journal.pone.0168196
Rizzo, L., Fraschetti, S., Alifano, P., Pizzolante, G., and Stabili, L. (2016a). The alien species Caulerpa cylindracea and its associated bacteria in the Mediterranean Sea. Mar. Biol. 163, 4. doi: 10.1007/s00227-015-2775-9
Rizzo, L., Fraschetti, S., Alifano, P., Tredici, M. S., and Stabili, L. (2016b). Association of Vibrio community with the Atlantic Mediterranean invasive alga Caulerpa cylindracea. J. Exp. Mar. Biol. Ecol. 475, 129–136. doi: 10.1016/j.jembe.2015.11.013
Rizzo, L., Pusceddu, A., Bianchelli, S., and Fraschetti, S. (2020). Potentially combined effect of the invasive seaweed Caulerpa cylindracea (Sonder) and sediment deposition rates on organic matter and meiofaunal assemblages. Mar. Environ. Res. 159:104966. doi: 10.1016/j.marenvres.2020.104966
Rizzo, L., Pusceddu, A., Stabili, L., Alifano, P., and Fraschetti, S. (2017). Potential effects of an invasive seaweed (Caulerpa cylindracea, Sonder) on sedimentary organic matter and microbial metabolic activities. Sci. Rep. 7:12113. doi: 10.1038/s41598-017-12556-4
Roth-Schulze, A. J., Zozaya-Valdés, E., Steinberg, P. D., and Thomas, T. (2016). Partitioning of functional and taxonomic diversity in surface-associated microbial communities. Environ. Microbiol. 18, 4391–4402. doi: 10.1111/1462-2920.13325
Ruitton, S., Verlaque, M., and Boudouresque, C. F. (2005). Seasonal changes of the introduced Caulerpa racemosa var. cylindracea (Caulerpales, Chlorophyta) at the northwest limit of its Mediterranean range. Aquat. Bot. 82, 55–70. doi: 10.1016/j.aquabot.2005.02.008
Saha, M., Rempt, M., Grosser, K., Pohnert, G., and Weinberger, F. (2011). Surface-associated fucoxanthin mediates settlement of bacterial epiphytes on the rockweed Fucus vesiculosus. Biofouling 27, 423–433. doi: 10.1080/08927014.2011.580841
Salaün, S., La Barre, S., Santos-Goncalvez, M. D., Potin, P., Haras, D., and Bazire, A. (2012). Influence of exudates of the kelp Laminaria digitata on biofilm formation of associated and exogenous bacterial epiphytes. Microb. Ecol. 64, 359–369. doi: 10.1007/s00248-012-0048-4
Sanchez-Amat, A., Solano, F., and Lucas-Elío, P. (2010). Finding new enzymes from bacterial physiology: a successful approach illustrated by the detection of novel oxidases in Marinomonas mediterranea. Mar. Drugs 8, 519–541. doi: 10.3390/md8030519
Sanders-Smith, R., Segovia, B. T., Forbes, C., Hessing-Lewis, M., Morien, E., Lemay, M. A., et al. (2020). Host-specificity and core taxa of seagrass leaf microbiome identified across tissue age and geographical regions. Front. Ecol. Evol. 8:605304. doi: 10.3389/fevo.2020.605304
Schloss, P. D., Jenior, M. L., Koumpouras, C. C., Westcott, S. L., and Highlander, S. K. (2016). Sequencing 16S rRNA gene fragments using the PacBio SMRT DNA sequencing system. PeerJ 4:e1869. doi: 10.7717/peerj.1869
Schloss, P. D., Westcott, S. L., Ryabin, T., Hall, J. R., Hartmann, M., Hollister, E. B., et al. (2009). Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75, 7537–7541. doi: 10.1128/AEM.01541-09
Schmidt, V. T., Smith, K. F., Melvin, D. W., and Amaral-Zettler, L. A. (2015). Community assembly of a euryhaline fish microbiome during salinity acclimation. Mol. Ecol. 24, 2537–2550. doi: 10.1111/mec.13177
Short, F. T., Coles, R. G., and Pergent-Martini, C. (2001). “Global seagrass distribution,” in Global Seagrass Research Methods, eds F. T. Short and R. G. Coles (Amsterdam: Elsevier Science B.V.), 5–30.
Simon, M., Scheuner, C., Meier-Kolthoff, J. P., Brinkhoff, T., Wagner-Döbler, I., Ulbrich, M., et al. (2017). Phylogenomics of Rhodobacteraceae reveals evolutionary adaptation to marine and non-marine habitats. ISME J. 11, 1483–1499. doi: 10.1038/ismej.2016.198
Stabili, L., Rizzo, L., Pizzolante, G., Alifano, P., and Fraschetti, S. (2017). Spatial distribution of the culturable bacterial community associated with the invasive alga Caulerpa cylindracea in the Mediterranean Sea. Mar. Environ. Res. 125, 90–98. doi: 10.1016/j.marenvres.2017.02.001
Stratil, S. B., Neulinger, S. C., Knecht, H., Friedrichs, A. K., and Wahl, M. (2013). Temperature-driven shifts in the epibiotic bacterial community composition of the brown macroalga Fucus vesiculosus. Microbiologyopen 2, 338–349. doi: 10.1002/mbo3.79
Tarquinio, F., Hyndes, G. A., Laverock, B., Koenders, A., and Säwström, C. (2019). The seagrass holobiont: understanding seagrass-bacteria interactions and their role in seagrass ecosystem functioning. FEMS Microbiol. Lett. 366:fnz057. doi: 10.1093/femsle/fnz057
Tujula, N. A., Crocetti, G. R., Burke, C., Thomas, T., Holmström, C., and Kjelleberg, S. (2010). Variability and abundance of the epiphytic bacterial community associated with a green marine Ulvacean alga. ISME J. 4, 301–311. doi: 10.1038/ismej.2009.107
Ugarelli, K., Laas, P., and Stingl, U. (2019). The microbial communities of leaves and roots associated with turtle grass (Thalassia testudinum) and manatee grass (Syringodium filliforme) are distinct from seawater and sediment communities, but are similar between species and sampling sites. Microorganisms 7:4. doi: 10.3390/microorganisms7010004
Wang, L., Tomas, F., and Mueller, R. S. (2020). Nutrient enrichment increases size of Zostera marina shoots and enriches for sulfur and nitrogen cycling bacteria in root-associated microbiomes. FEMS Microbiol. Ecol. 96:fiaa129. doi: 10.1093/femsec/fiaa129
Weidner, S., Arnold, W., Stackebrandt, E., and Pühler, A. (2000). Phylogenetic analysis of bacterial communities associated with leaves of the seagrass Halophila stipulacea by a culture-independent small-subunit rRNA gene approach. Microb. Ecol. 39, 22–31. doi: 10.1007/s002489900194
Weinberger, F. (2007). Pathogen-induced defense and innate immunity in macroalgae. Biol. Bull. 213, 290–302. doi: 10.2307/25066646
Wickham, H., Averick, M., Bryan, J., Chang, W., McGowan, L. D., François, R., et al. (2019). Welcome to the tidyverse. J. Open Source Softw. 4:1686. doi: 10.21105/joss.01686
Wood, D. C., and Hayasaka, S. S. (1981). Chemotaxis of rhizoplane bacteria to amino acids comprising eelgrass (Zostera marina L.) root exudate. J. Exp. Mar. Biol. Ecol. 50, 153–161. doi: 10.1016/0022-0981(81)90047-2
Xie, Y. (2014). “knitr: A comprehensive tool for reproducible research in R,” in Implementing Reproducible Computational Research, eds V. Stodden, F. Leisch, and R. D. Peng (New York, NY: Chapman and Hall; CRC Press), 3–32.
Xie, Y. (2015). Dynamic Documents With R and knitr, 2nd Edn. Boca Raton, FL: Chapman and Hall; CRC Press.
Xie, Y. (2019b). TinyTeX: A lightweight, cross-platform, and easy-to-maintain LaTeX distribution based on TeX Live. TUGboat 40, 30–32.
Xie, Y. (2019c). tinytex: Helper Functions to Install and Maintain ‘TeX Live', and Compile ‘LaTeX' Documents.
Xie, Y., Allaire, J. J., and Grolemund, G. (2018). R Markdown: The Definitive Guide, 1st Edn. Boca Raton, FL: Chapman and Hall; CRC Press.
Xie, Z., Lin, W., and Luo, J. (2017). Comparative phenotype and genome analysis of Cellvibrio sp. PR1, a xylanolytic and agarolytic bacterium from the Pearl River. BioMed Res. Int. 2017:6304248. doi: 10.1155/2017/6304248
Yilmaz, P., Parfrey, L. W., Yarza, P., Gerken, J., Pruesse, E., Quast, C., et al. (2014). The SILVA and "All-Species Living Tree Project (LTP)" taxonomic frameworks. Nucleic Acids Res. 42, D643–D648. doi: 10.1093/nar/gkt1209
Keywords: epiphytic microbial community, seasonal dynamics, Illumina 16S rRNA sequencing, Cymodocea nodosa, Caulerpa cylindracea
Citation: Korlević M, Markovski M, Zhao Z, Herndl GJ and Najdek M (2021) Seasonal Dynamics of Epiphytic Microbial Communities on Marine Macrophyte Surfaces. Front. Microbiol. 12:671342. doi: 10.3389/fmicb.2021.671342
Received: 23 February 2021; Accepted: 10 August 2021;
Published: 09 September 2021.
Edited by:
Nona Sheila Romualdo Agawin, University of the Balearic Islands, SpainReviewed by:
Lucia Rizzo, Zoological Station Anton Dohrn, ItalyAntonio Busquets Bisbal, University of the Balearic Islands, Spain
Fiona Tomas, Mediterranean Institute for Advanced Studies (IMEDEA), Spain
Copyright © 2021 Korlević, Markovski, Zhao, Herndl and Najdek. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Marino Korlević, bWFyaW5vLmtvcmxldmljQGlyYi5ocg==
 Marino Korlević
Marino Korlević Marsej Markovski
Marsej Markovski Zihao Zhao
Zihao Zhao Gerhard J. Herndl
Gerhard J. Herndl Mirjana Najdek
Mirjana Najdek