- 1Research and Innovation Centre, Fondazione Edmund Mach, San Michele all’Adige, Italy
- 2Bi-PA nv, Londerzeel, Belgium
- 3Department of Induced Resistance and Plant Bioprotection, University of Reims, Reims, France
- 4Center Agriculture Food Environment (C3A), University of Trento, San Michele all’Adige, Italy
Tagatose is a rare sugar with no negative impacts on human health and selective inhibitory effects on plant-associated microorganisms. Tagatose inhibited mycelial growth and negatively affected mitochondrial processes in Phytophthora infestans, but not in Phytophthora cinnamomi. The aim of this study was to elucidate metabolic changes and transcriptional reprogramming activated by P. infestans and P. cinnamomi in response to tagatose, in order to clarify the differential inhibitory mechanisms of tagatose and the species-specific reactions to this rare sugar. Phytophthora infestans and P. cinnamomi activated distinct metabolic and transcriptional changes in response to the rare sugar. Tagatose negatively affected mycelial growth, sugar content and amino acid content in P. infestans with a severe transcriptional reprogramming that included the downregulation of genes involved in transport, sugar metabolism, signal transduction, and growth-related process. Conversely, tagatose incubation upregulated genes related to transport, energy metabolism, sugar metabolism and oxidative stress in P. cinnamomi with no negative effects on mycelial growth, sugar content and amino acid content. Differential inhibitory effects of tagatose on Phytophthora spp. were associated with an attempted reaction of P. infestans, which was not sufficient to attenuate the negative impacts of the rare sugar and with an efficient response of P. cinnamomi with the reprogramming of multiple metabolic processes, such as genes related to glucose transport, pentose metabolism, tricarboxylic acid cycle, reactive oxygen species detoxification, mitochondrial and alternative respiration processes. Knowledge on the differential response of Phytophthora spp. to tagatose represent a step forward in the understanding functional roles of rare sugars.
Introduction
Rare sugars have been defined as monosaccharides and their derivatives that rarely exist in nature, such as tagatose, allose, sorbose, xylulose, and xylitol (Granström et al., 2004). The potential functional properties of rare sugars are underestimated due to their limited availability in nature (Li et al., 2013). However, the implementation of novel enzymatic and microbial processes lowered the cost of rare sugar synthesis and extended their use in various industrial and scientific fields, such as agriculture, human nutrition and medicine (Granström et al., 2004; Izumori, 2006; Li et al., 2013). Among rare sugars, tagatose is a ketohexose that was found at low concentrations in many foods, such as apple, pineapple, orange and milk (Vastenavond et al., 2011). Since tagatose does not have negative impacts on human health, it was ‘generally recognized as safe’ by the Food and Drug Administration and it is used as low-calorie sweetener in several countries (Levin, 2002; Vastenavond et al., 2011). Tagatose can be utilized as a carbohydrate source by only a restricted number of microorganisms, such as Exiguobacterium spp., Lactobacillus spp., and Lactococcus spp. (Raichand et al., 2012; Martinussen et al., 2013; Van der Heiden et al., 2013; Wu and Shah, 2017). On the other hand, tagatose is not catabolised by some human-associated microorganisms, such as Bacillus cereus, Escherichia coli, Listeria monocytogenes, Staphylococcus aureus, and Yersinia enterocolitica (Bautista et al., 2000), indicating selective nutritional or anti-nutritional properties for specific microbial taxa. For example, tagatose inhibited the growth of Streptococcus mutans (Hasibul et al., 2018) and Salmonella enterica serovar Typhimurium (Lobete et al., 2017) and decreased the gene expression (e.g., glucosyltransferase, fructosidase, and phosphotransferase encoding genes) and the enzymatic activity (e.g., glucosyltransferase) of sugar metabolism in S. mutans (Hasibul et al., 2018). Conversely, tagatose enhanced the growth of Lactobacillus rhamnosus and upregulated genes associated with sugar metabolism (e.g., phosphotransferase encoding genes, phosphofructokinase and tagatose-6-phosphate kinase) (Koh et al., 2013), suggesting specific impacts of rare sugars on the microbial metabolism.
Tagatose inhibited the growth of several causal agents of plant diseases, such as Phytophthora infestans (tomato and potato late blight) (Chahed et al., 2020), Plasmopara viticola (grapevine downy mildew) and Erysiphe necator (grapevine powdery mildew) (Perazzolli et al., 2020), Hyaloperonospora parasitica (cabbage downy mildew) and Oidium violae (tomato powdery mildew) (Mochizuki et al., 2020; Mijailovic et al., 2021). Tagatose showed also possible prebiotic effects on the phyllosphere microbiota and shifted the proportions of potential beneficial and potential pathogenic microorganisms by a selective nutritional or anti-nutritional effect on plant-associated microorganisms (Perazzolli et al., 2020). Differential effects were also found on Trichoderma spp. growth, where tagatose supported the growth of T. harzianum and T. pleuroticola, but not that of T. pleurotum (Komon-Zelazowska et al., 2007), confirming the selective effect of tagatose on plant-associated microorganisms. In particular, tagatose inhibited mycelial growth with severe mitochondrial alterations and stress-related gene modulation in P. infestans, but not in P. cinnamomi (Chahed et al., 2020), indicating species-specific responses to this rare sugar. Further studies are therefore required, in order to better understand the metabolic and transcriptional reprogramming responsible for the differential inhibitory effects of tagatose on plant-associated microorganisms.
Members of the Phytophthora genus are widespread causal agents of destructive diseases for different plant species (Kamoun, 2000). In particular, P. infestans and P. cinnamomi are two of the most important phytopathogenic oomycetes (Kamoun et al., 2015) and are responsible for severe economic losses in agricultural, horticultural and forest production (Hardham, 2005; Fry et al., 2015; Kamoun et al., 2015). The control of phytopathogenic oomycetes requires the use of chemical fungicides (Judelson and Blanco, 2005) with potential negative effects on the environment and human health (Fantke et al., 2012). Tagatose could represent a valid alternative to chemical fungicides, thanks to its inhibitory properties against P. infestans (Chahed et al., 2020) and the absence of negative impacts on human health (Ohara et al., 2008), but further studies are needed to clarify its impacts on metabolic and transcriptional regulations of Phytophthora spp. For example, the fungicide ethylicin severely affected amino acid and sugar metabolic processes in P. infestans (Zhang et al., 2020). Likewise, changes in the sugar, amino acid and organic acid content were observed in P. infestans isolates resistant to the fungicide metalxyl, indicating that fungicide resistance pathways are linked to the modulation of fatty acid biosynthesis and glycerophospholipid metabolism (Maridueña-Zavala et al., 2017). Transcriptional reprogramming in Phytophthora spp. exposed to biofungicides was also found and the natural product melatonin inhibited P. infestans growth by the downregulation of genes related to amino acid and sugar metabolism (Zhang et al., 2017). Similarly, the growth inhibition of P. infestans caused by Lysobacter capsici AZ78 (Tomada et al., 2017) and that of Phytophthora sojae caused by Bacillus amyloliquefaciens JDF3 and B. subtilis RSS (Liu et al., 2019) was associated with the downregulation of genes related to protein and sugar metabolism, indicating that complex metabolic responses are activated in Phytophthora spp. in response to biological or chemical growth inhibitors. The aim of this study was to elucidate the metabolic changes and transcriptional reprogramming activated by Phytophthora spp. in response to tagatose incubation, in order to clarify the differential inhibitory mechanisms of tagatose and the molecular determinants of the species-specific reaction to this rare sugar.
Materials and Methods
Phytophthora spp. Growth Conditions and Tagatose Incubation
Phytophthora infestans strain VB3 and P. cinnamomi strain CBS 144.22 were stored in glycerol at −80°C in the fungal collection of the Fondazione Edmund Mach and they are available upon request. P. infestans and P. cinnamomi were grown in Petri dishes on pea agar medium (PAM; 12.5% frozen peas and 1.2% agar in distilled water) at 18 and 25°C in dark conditions, respectively (Chahed et al., 2020). The stock solution (50 g/L in distilled water) of tagatose (Bi-PA, Londerzeel, Belgium) was filter sterilized and added at the final concentration of 5 g/L on PAM shortly before Phytophthora spp. inoculation, as reported by Chahed et al. (2020).
Phytophthora spp. growth was assessed as previously described (Chahed et al., 2020). Briefly, a plug (5 mm diameter and 2 mm height) of a 14 days-old colony was placed at the center of each dish (90 mm diameter) on PAM in the absence (control) or presence of 5 g/L tagatose (tagatose-incubated). The radial growth of P. infestans and P. cinnamomi was assessed at 4 and at 10 days after incubation (DAI) at 18 and 25°C, respectively, calculated as the average of the two perpendicular diameters of the colony, minus the plug diameter and the result divided by two. Ten replicates (dishes) were used for each treatment and the experiment was carried out twice.
For metabolic and transcriptomic analyses, a plug (5 mm diameter and 2 mm height) of a 14 days-old colony was placed on PAM covered with sterile cellophane layers in the absence (control) and presence of 5 g/L tagatose. P. infestans and P. cinnamomi samples were collected at 4 and at 10 DAI after incubation at 18 and 25°C, respectively, as previously described (Chahed et al., 2020). Briefly, mycelium samples were collected with a sterile forceps, transferred in a sterile 50 mL-tube, immediately frozen in liquid nitrogen and stored at −80°C. Samples were crushed using a mixer mill disruptor (MM200, Retsch, Haan, Germany) at 25 Hz for 45 s with sterile steel jars and beads refrigerated in liquid nitrogen.
Sugar and Amino Acid Quantification
Ground Phytophthora spp. mycelium (500 mg) or uninoculated PAM (500 mg; Supplementary Table 1) were dissolved in 25 mL of ultrapure water, filtered through a 0.22 μm PTFE membrane (Sartorius, Göttingen, Germany) and used for sugar and amino acid quantification (Chemistry Unit at Fondazione Edmund Mach).
The sugar content (arabinose, fructose, glucose, isomaltose, lactose, maltose, mannose, melibiose, rhamnose, ribose, tagatose, trehalose, and xylose) was assessed by ion chromatography (Cataldi et al., 2000) and it was expressed as quantity of each sugar per unit of mycelium fresh weight (mg/kg), using a calibration curve of each pure sugar (Sigma-Aldrich, Merc, Kenilworth, NJ, United States) dissolved in ultrapure water within a range between 0.4 and 40 μg/mL. Briefly, Phytophthora spp. mycelium samples, uninoculated PAM samples and calibration curves of each pure sugar were analysed with an ionic chromatograph ICS 5000 (Dionex-Thermo Scientific, Waltham, MA, United States), equipped with an autosampler, a quaternary gradient pump, a column oven and a pulsed amperometric detector with a gold working electrode and a palladium counter electrode. The separation was obtained by injecting 5 μL of each Phytophthora spp. mycelium sample, uninoculated PAM sample or pure sugar onto a CarboPac PA200 3 mm × 250 mm analytical column (Dionex, Thermo Scientific, Waltham, MA, United States), preceded by a CarboPac PA200 3 mm × 50 mm guard column (Dionex, Thermo Scientific), with a NaOH gradient (from 1 to 30 mM) at 0.4 mL/min flow rate.
The amino acid content (aminobutyric acid, ethanolamine, glycine, glutamic acid, aspartic acid, alanine, arginine, asparagine, citrulline, phenylalanine, glutamine, isoleucine, histidine, leucine, lysine, ornithine, serine, tyrosine, threonine, valine, tryptophan, and methionine) was assessed by high performance liquid chromatography (HPLC) (Hill et al., 1979) and it was expressed as quantity of each amino acid per unit of mycelium fresh weight (mg/kg), using a calibration curve of each pure amino acid (Sigma-Aldrich, Merck) dissolved in ultrapure water within a range between 0.1 and 50 μg/mL. Briefly, Phytophthora spp. mycelium samples, uninoculated PAM samples and calibration curves of each amino acid were analyzed with a HPLC instrument (1100 series, Agilent Technologies, Santa Clara, CA, United States) equipped with a degasser, a quaternary gradient pump, an autosampler, a thermostatted column oven and a Fluorimetric Detector (FLD). The chromatographic separation of amino acids was obtained by injecting 5 μL of each Phytophthora spp. mycelium sample, uninoculated PAM sample or pure amino acid onto a Chromolith Performance RP-18e column (100 mm × 4.6 mm) and a guard RP-18e column (10 mm × 4.6 mm; Merck, Kenilworth, NJ, United States) kept at 40°C with an eluent flow of 2.0 mL/min. The FLD was set at 336 nm as excitation and at 445 nm as emission wavelength.
For sugar and amino acid quantification, three replicates (each replicate obtained as a pool of ten dishes) were analyzed for each Phytophthora spp., treatment and time point, and the experiment was carried out twice.
RNA Extraction, Sequencing and Mapping to Reference Genomes
Total RNA was extracted from ground Phytophthora spp. mycelium (100 mg) using the Spectrum Plant total RNA kit (Sigma-Aldrich, Merck) with an on-column DNase treatment using the RNase-Free DNase Set (Qiagen, Hilden, Germany). Total RNA was quantified using a Qubit (Thermo Fisher Scientific) and RNA quality was checked using a Bioanalyzer 2100 (Agilent Technologies). Four replicates (each replicate obtained as a pool of 10 dishes) were analyzed for each Phytophthora spp., treatment and time point. RNA samples were subjected to RNA-Seq library construction, using the TruSeq Stranded Total RNA Library Prep Plant protocol (Illumina, San Diego, CA, United States) with rRNA depletion with the Ribo-Zero Plant kit (Tomada et al., 2017) according to the manufacturer’s instructions. Paired-end reads of 150 bases were obtained using a HiSeq 2500 (Illumina) at the Institute of Applied Genomics (Udine, Italy).
Raw Illumina reads were cleaned and filtered using Trimmomatic program version 0.36 (Bolger et al., 2014) and low-quality bases with an average Phred quality score lower than 15 in a sliding window of four base were removed. Reads shorter than 36 bases in length were removed from the analysis and quality check of raw reads was performed using Fast QC version 0.11.7. Read pairs of P. infestans samples were aligned to the P. infestans ASM14294v1 genome1 using the STAR V2.7 program (Dobin et al., 2013), while those of P. cinnamomi samples were aligned to P. cinnamomi V1.0 genome2. Counts of unambiguously mapped read pairs were obtained during the alignment with the STAR V2.7 program. Putative orthologous genes of P. infestans and P. cinnamomi (orthologous genes) were identified by reciprocal best BLAST hit using the BLAST+ 2.7.1 program (Camacho et al., 2009) with a threshold of 70% identity and 50% alignment length on amino acid sequences, in order to better analyze the differential Phytophthora spp. response.
Identification and Functional Annotation of Differentially Expressed Genes
Differentially expressed genes (DEGs) were identified with the Limma-Voom package (Law et al., 2014) which estimates the mean–variance relationship of log counts, generating a precision weight for each observation that is fed into the Limma empirical Bayes analysis pipeline (Smyth, 2004). Pearson correlation analysis and Volcano Plot analysis (Patterson et al., 2006) were carried out using the Python programming language and the matplotlib package (Hunter, 2007) and a double cutoff on P-value (P ≤ 0.01) and minimum Log2 fold change (FC) of one [Log2 (FC) ≥ 1 or Log2 (FC) ≤ −1] was used to select DEGs, as previously reported (Tomada et al., 2017; Shen et al., 2020). Four pairwise comparisons were analyzed, in order to identify DEGs in each Phytophthora spp.: tagatose-incubated vs. control of P. infestans at 4 DAI, tagatose-incubated vs. control of P. infestans at 10 DAI, tagatose-incubated vs. control of P. cinnamomi at 4 DAI, tagatose-incubated vs. control of P. cinnamomi at 10 DAI. The distribution of P. infestans and P. cinnamomi DEGs was summarized using Venn diagram3 and DEGs were grouped in upregulated genes [P ≤ 0.01 and Log2 (FC) ≥ 1] or downregulated genes [P ≤ 0.01 and Log2 (FC) ≤ −1] at both time points (4 and 10 DAI cluster) and exclusively at 4 DAI (4 DAI cluster) or at 10 DAI (10 DAI cluster) for each Phytophthora spp. The heat map diagram of Log2-transformed FC values of Phytophthora spp. DEGs was visualized using the Java Treeview tool (Saldanha, 2004).
On the list of Phytophthora orthologous genes (8,908 in total), orthologous DEGs were identified imposing a P-value lower than 0.01 and minimum Log2 FC of one using a Volcano Plot (Patterson et al., 2006) with the Python programming language and the matplotlib package (Hunter, 2007) in the pairwise comparisons between tagatose-incubated samples and control samples at 4 DAI and at 10 DAI for each Phytophthora spp. Contrasts were defined in order to identify genes that respond differently in P. infestans compared to P. cinnamomi in the comparison between control and tagatose-incubated samples at 4 and at 10 DAI. Orthologous DEGs were then grouped in 16 clusters (defined by a four-letter code) based on their upregulation (U) or downregulation (D) in tagatose-incubated samples compared to control samples in the two Phytophthora spp. at 4 and at 10 DAI (e.g., UUDD cluster includes genes upregulated in P. infestans at 4 and 10 DAI and downregulated in P. cinnamomi at 4 and 10 DAI).
A principal component analysis (PCA) was performed using the Python programming language and the scikit.learn Python package4 on Limma-normalized expression values. Gene sequences of P. infestans and P. cinnamomi were aligned against the UniProtKB database (downloaded on May 20195) using a BLAST-X search and the three or five best protein hits with an E-value lower than 1 × 10–5 were selected for functional annotation. In particular, DEGs were annotated based on the Phytophthora spp. protein description resulted by BLAST-X search and grouped into 17 functional categories according to the previous literature. Phytophthora spp. genes were further annotated using ARGOT2 function prediction tool for the Gene Ontology functional annotation (Falda et al., 2012). Gene ontology (GO) terms significantly overrepresented (P ≤ 0.05, Benjamin and Hochberg FDR correction) in the DEG lists in comparison to the whole transcriptome of the respective Phytophthora spp. were identified using the Biological Networks Gene Ontology (BiNGO) tool (Maere et al., 2005) and biological networks were visualized with Cytoscape version 3.7.2 (Shannon et al., 2003).
Gene Expression Analysis by Quantitative Real-Time RT-PCR
Phytophthora spp. genes were selected for quantitative real-time PCR (qPCR) analysis (Supplementary Table 2). A primer pair compatible for the P. infestans and P. cinnamomi sequence was designed on conserved coding regions in the case of orthologous genes. The first strand cDNA was synthesized from 1 μg of DNase-treated RNA using Superscript III (Invitrogen, Thermo Fisher Scientific) and the oligo-dT primer. qPCR reactions were carried out with Platinum SYBR Green qPCR SuperMix-UDG (Invitrogen, Thermo Fisher Scientific) and specific primers using the Light Cycler 480 (Roche Diagnostics, Mannheim, Germany), as previously described (Chahed et al., 2020). Briefly, the PCR conditions were: 50°C for 2 min and 95°C for 2 min as initial steps, followed by 50 cycles of 95°C for 15 s and 60°C for 1 min. Each sample was examined in three technical replicates and dissociation curves were analyzed to verify the specificity of each amplification reaction. Light Cycler 480 SV1.5.0 software (Roche) was used to extract Ct values based on the second derivative calculation and LinReg software was used to calculate reaction efficiencies (Ruijter et al., 2009). The relative expression level (FC) of each gene was calculated according to the Pfaffl equation (Pfaffl, 2001) for tagatose-incubated samples compared to the respective control samples (calibrator) for each Phytophthora spp. and time point. The gene encoding β-tubulin (tub-b) was used as constitutive gene for the expression level normalization (Yan and Liou, 2006), because its expression was not significantly affected by the treatments (Supplementary Table 2). Four replicates (each replicate obtained as a pool of ten dishes) were analyzed for each Phytophthora spp., treatment and time point.
Statistical Analysis
Mycelial growth, sugar and amino acid data were analyzed with Statistica 13.1 software (TIBCO Software, Palo Alto, CA, United States). Normal distribution (Kolmogorov–Smirnov test, P > 0.05) and variance homogeneity of the data (Levene’s tests, P > 0.05) were checked and parametric tests were used when both assumptions were respected. Each experimental repetition was analyzed singularly and a two-way analysis of variance (two-way ANOVA) was used to demonstrate non-significant differences between the two experiments (P > 0.05). Data from the two experimental repetitions were pooled and significant differences between tagatose-incubated samples and control samples were assessed with the Student’s t-test (P ≤ 0.05) for each Phytophthora spp. and time point.
Results
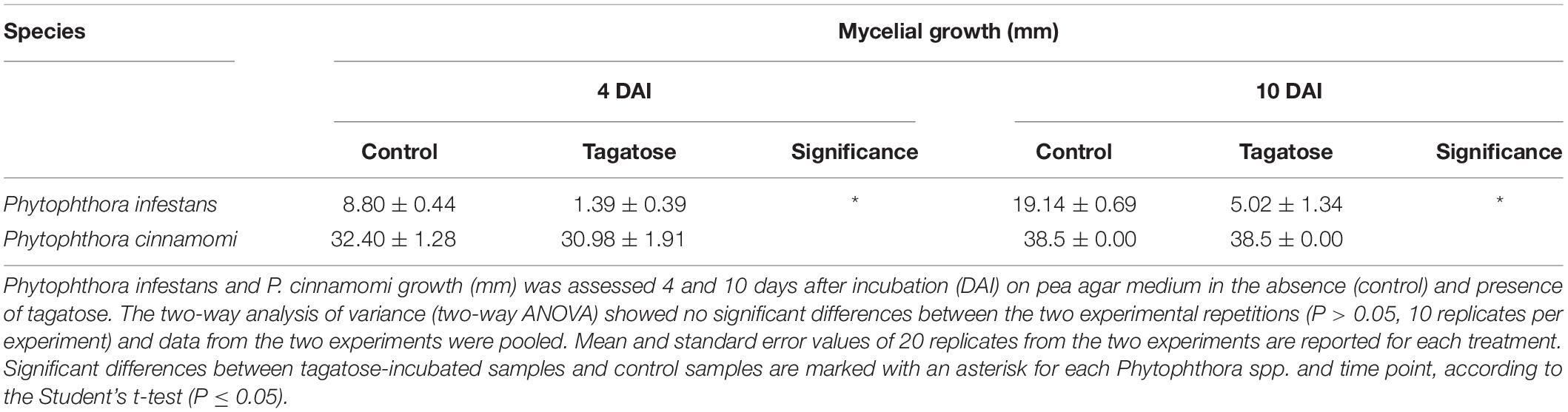
Tagatose Affects the Mycelial Growth, Sugar Content and Amino Acid Content in Phytophthora infestans, but Not in P. cinnamomi
Tagatose inhibited the growth of P. infestans and not that of P. cinnamomi (Table 1 and Supplementary Figure 1) at 4 and at 10 DAI on PAM. In order to investigate impacts of tagatose on Phytophthora spp. metabolism, sugar content and amino acid content of P. infestans and P. cinnamomi mycelium were analyzed at 4 and at 10 DAI on PAM in the absence (control) and presence of tagatose (tagatose-incubated) by ion chromatography and HPLC, respectively (Figure 1 and Supplementary Figure 2). In P. infestans, tagatose incubation decreased the glucose, mannose and ribose content at 4 and at 10 DAI and the xylose content at 10 DAI compared to the respective control samples (Figures 1A,B). Conversely, the fructose content of P. infestans was higher in tagatose-incubated samples compared to control samples at 4 and at 10 DAI. In P. cinnamomi, the content of each sugar was comparable in tagatose-incubated samples and control samples at 4 and at 10 DAI (Figures 1A,B). As expected, tagatose was found only in tagatose-incubated samples of P. infestans and P. cinnamomi at both time points, but not in the respective control samples. The sugar content in control samples differed between P. infestans and P. cinnamomi in terms of glucose, maltose and mannose content at 4 and at 10 DAI, fructose content at 4 DAI, ribose and xylose content at 10 DAI.
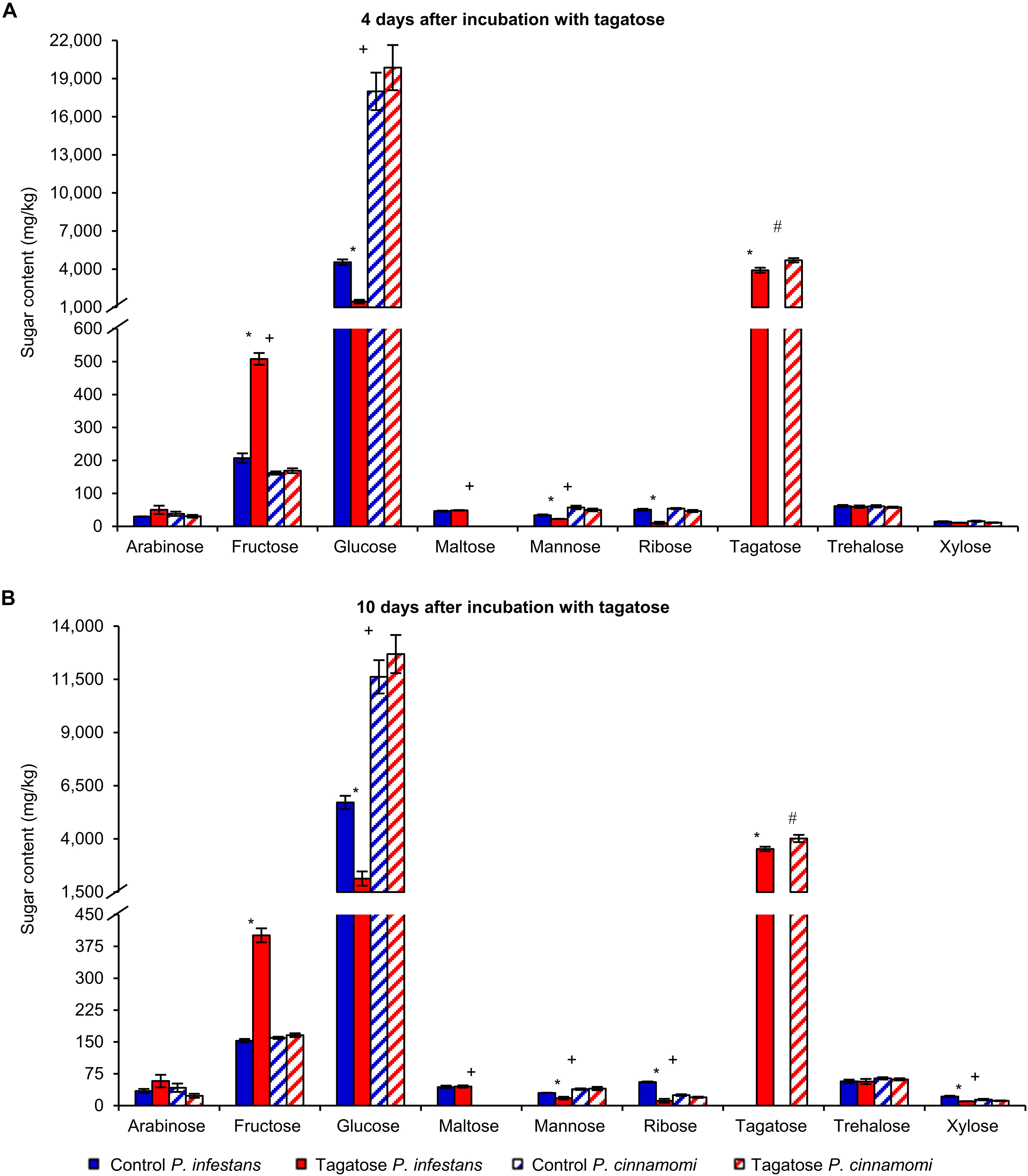
Figure 1. Effect of tagatose on Phytophthora spp. sugar content. Phytophthora infestans (solid bars) and P. cinnamomi (stripped bars) sugar content was quantified at 4 (A) and 10 (B) days after incubation (DAI) on pea agar medium in the absence (control, blue) and presence of tagatose (red). The two-way analysis of variance (two-way ANOVA) showed no significant differences between the two experimental repetitions (P > 0.05, three replicates per experiment) and data from the two experiments were pooled. Mean and standard error values of six replicates from the two experiments are presented for each treatment. For each sugar and time point, significant differences between tagatose-incubated samples and control samples are marked with an asterisk (*) for P. infestans and a hashtag (#) for P. cinnamomi, according to the Student’s t-test (P ≤ 0.05). Significant differences between P. infestans control and P. cinnamomi control for each sugar are marked with a plus sign (+), according to the Student’s t-test (P ≤ 0.05). Isomaltose, lactose, melibiose and rhamnose were not detected and omitted from the chart.
The amino acid content was affected by tagatose in P. infestans at 4 and at 10 DAI, but only slightly in P. cinnamomi at 4 DAI (Supplementary Figures 2A,B). In P. infestans, tagatose incubation decreased the content of 14 amino acids at 4 and at 10 DAI (aminobutyric acid, ethanolamine, glycine, aspartic acid, arginine, asparagine, phenylalanine, glutamine, isoleucine, leucine, ornithine, serine, tryptophan, and methionine), the content of two amino acids at 4 DAI (alanine and histidine) and the content of two amino acids at 10 DAI (lysine and tyrosine) compared to the respective control samples. Conversely, the glutamic acid content at 4 and at 10 DAI, or the histidine content at 10 DAI, was higher in tagatose-incubated samples compared to control P. infestans samples. In P. cinnamomi, the amino acid content was comparable in tagatose-incubated samples and control samples at both time points, except for the lower glutamic acid and glutamine content found in tagatose-incubated samples compared to control samples at 4 DAI.
RNA-Seq Analysis Reveals Species-Specific Response of Phytophthora spp. Genes to Tagatose Incubation
To clarify Phytophthora spp. transcriptional regulations in response to tagatose incubation, RNA-Seq analysis was carried out on control and tagatose-incubated samples of P. infestans and P. cinnamomi at 4 and at 10 DAI. Sequences of the 32 samples [two Phytophthora spp. (P. infestans and P. cinnamomi), two incubation conditions (tagatose-incubated and control), two time points (4 and 10 DAI) and four replicates] were obtained (BioProject number PRJNA622764) and the total number of read pairs that mapped uniquely to the P. infestans and P. cinnamomi genome ranged from 7,862,604 to 12,711,308 and from 4,197,875 to 9,047,846 for each replicate, respectively (Supplementary Table 3). The expression level of P. infestans and P. cinnamomi genes was assessed (Supplementary Tables 4, 5) and high Pearson correlation values among replicates were found (Supplementary Figures 3–10 and Supplementary Tables 6–8). Global effects of tagatose incubation on the Phytophthora spp. transcriptome were visualized by PCA, which discriminated P. infestans samples according to the incubation condition in the first principal component (first PC, 81.6%) and according to the time point in the second PC (5.22%; Supplementary Figure 11A). In the case of P. cinnamomi, PCA classified samples according to the time point in the first PC (54.15%) and according to the incubation condition in the second PC (7.73%; Supplementary Figure 11B). Moreover, the PCA on the putative orthologous genes of P. infestans and P. cinnamomi (8,908 orthologous genes, in total) discriminated samples according to the species in the first PC (72.48%) and highlighted changes between tagatose-incubated samples and control samples of P. infestans on the second PC (Supplementary Figure 11C).
Tagatose incubation resulted in the modulation of 3,901 in P. infestans (Supplementary Table 9) and 512 DEGs in P. cinnamomi (Supplementary Table 10), according to the Volcano Plot analysis (P ≤ 0.01 and minimum Log2 (FC) of one; Supplementary Figure 12). A large fraction (75.9%) of P. infestans DEGs and almost half (50.5%) of P. cinnamomi DEGs were downregulated by tagatose (Figure 2). DEGs were grouped in upregulated or downregulated genes at both time points (4 and 10 DAI cluster) and exclusively at 4 DAI (4 DAI cluster) or at 10 DAI (10 DAI cluster) for each Phytophthora spp. (Figure 2 and Supplementary Figure 13). Moreover, 2,172 orthologous DEGs were identified and they were grouped in 16 clusters (defined by a four-letter code) based on the upregulation or downregulation in tagatose-incubated samples compared to control samples in the two Phytophthora spp. and time points, such as modulation in P. infestans at 4 DAI (first letter); P. infestans at 10 DAI (second letter); P. cinnamomi at 4 DAI (third letter); P. cinnamomi at 10 DAI (fourth letter; Supplementary Table 11).
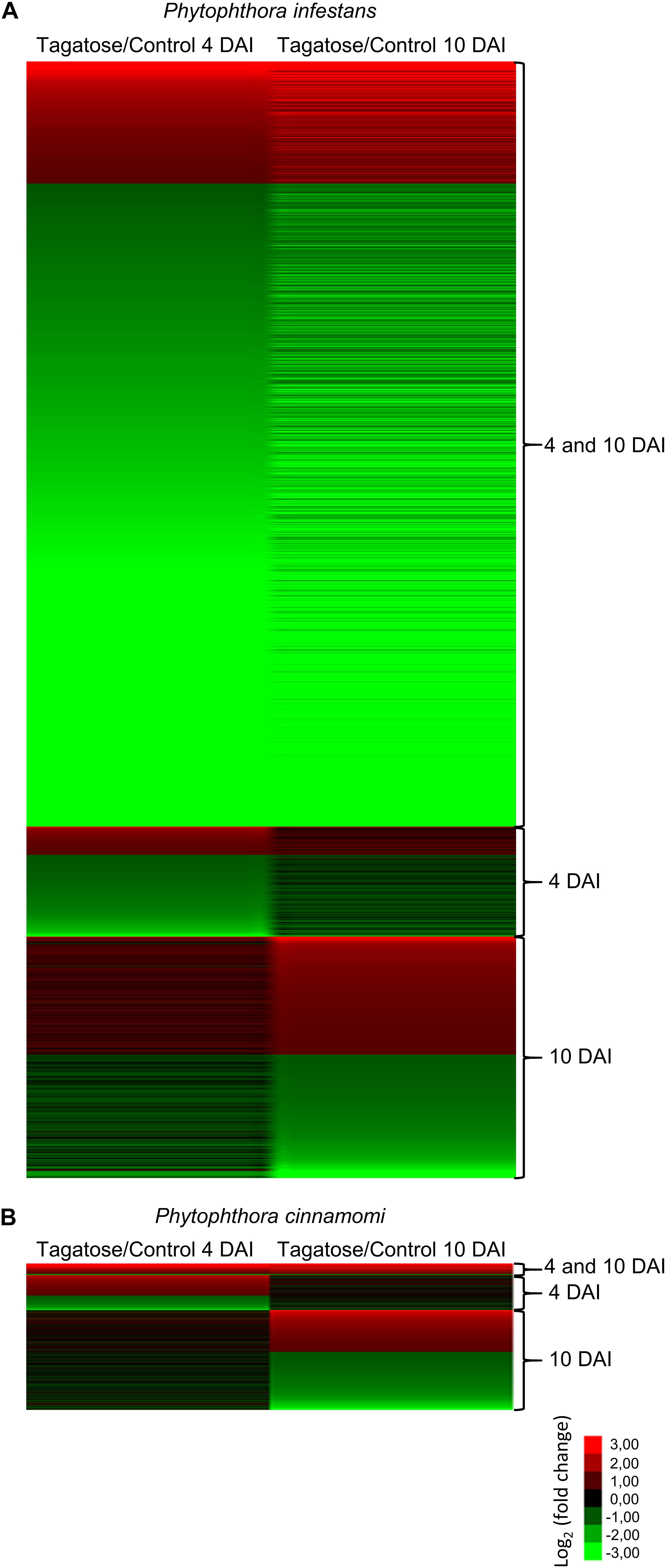
Figure 2. Clustering of genes modulated by tagatose in Phytophthora spp. Heat map diagram of fold change values of differentially expressed gens (DEGs) identified in Phytophthora infestans (A) and P. cinnamomi (B) at 4 and 10 days after incubation (DAI) on pea agar medium in the presence of tagatose compared to the respective control samples grown in the absence of tagatose. For each Phytophthora spp., DEGs were grouped in genes significantly upregulated or downregulated at both time points (4 and 10 DAI cluster) and exclusively modulated at 4 DAI (4 DAI cluster) or at 10 DAI (10 DAI cluster). The heat map diagram was visualized using Java Treeview according to the color scale legend shown.
The RNA-Seq results were validated by qPCR analysis of 14 Phytophthora spp. genes (Supplementary Table 2) that were selected according to their expression profiles (six P. infestans, six P. cinnamomi and two orthologous genes belonging to different clusters) and functional categories (e.g., sugar metabolism, oxidative stress, and transport). A close correlation (Pearson r = 0.90) between RNA-Seq and qPCR expression data was observed (Supplementary Figure 14) and expression profiles agreed completely for 12 genes and differed slightly for two genes (Supplementary Table 2).
Tagatose Incubation Causes a Severe Transcriptional Response in Phytophthora infestans
A high number of P. infestans genes was modulated at 4 and 10 DAI (2,688 DEGs: 427 upregulated and 2,247 downregulated; Figure 2A, Supplementary Figure 13A and Supplementary Table 9). The functional annotation revealed that upregulated genes of the 4 and 10 DAI cluster were mainly involved in primary metabolism (e.g., one epoxide hydrolase and two aldehyde dehydrogenases), protein metabolism, transport and energy metabolism (e.g., a fructose-bisphosphate aldolase, three glyceraldehyde-3-phosphate dehydrogenases and two phosphoglycerate mutases; Figure 3A and Supplementary Table 9). In particular, the GO biological process enrichment analysis revealed the overrepresentation of oxidation reduction category (e.g., four glutathione S-transferases and two quinone oxidoreductases) and transport-related processes [e.g., nine ATP-binding cassette (ABC) proteins and seven P-type ATPases (P-ATPase)] in the cluster of upregulated genes at 4 and 10 DAI (Figure 3B and Supplementary Table 9). Phytophthora infestans genes downregulated by tagatose at 4 and 10 DAI were mainly implicated in transport (e.g., one glucose transporter, three mitochondrial carriers and three amino acid transporters), signal transduction (e.g., three protein phosphatases and three cyclin-dependent kinases), growth and development (e.g., five myosin-like proteins, five dynein light chains and 21 kinesin-like proteins), sugar metabolism (e.g., an alpha-trehalose-phosphate synthase, a glucokinase, two lysosomal β-glucosidases, a β-glucosidase, an endo-1,4-β-xylanase and 18 glycoside hydrolases) and virulence (Figure 3A and Supplementary Table 9). Thus, the GO categories related to signaling and transport processes were overrepresented in the cluster of downregulated genes at 4 and 10 DAI (Figure 3C).
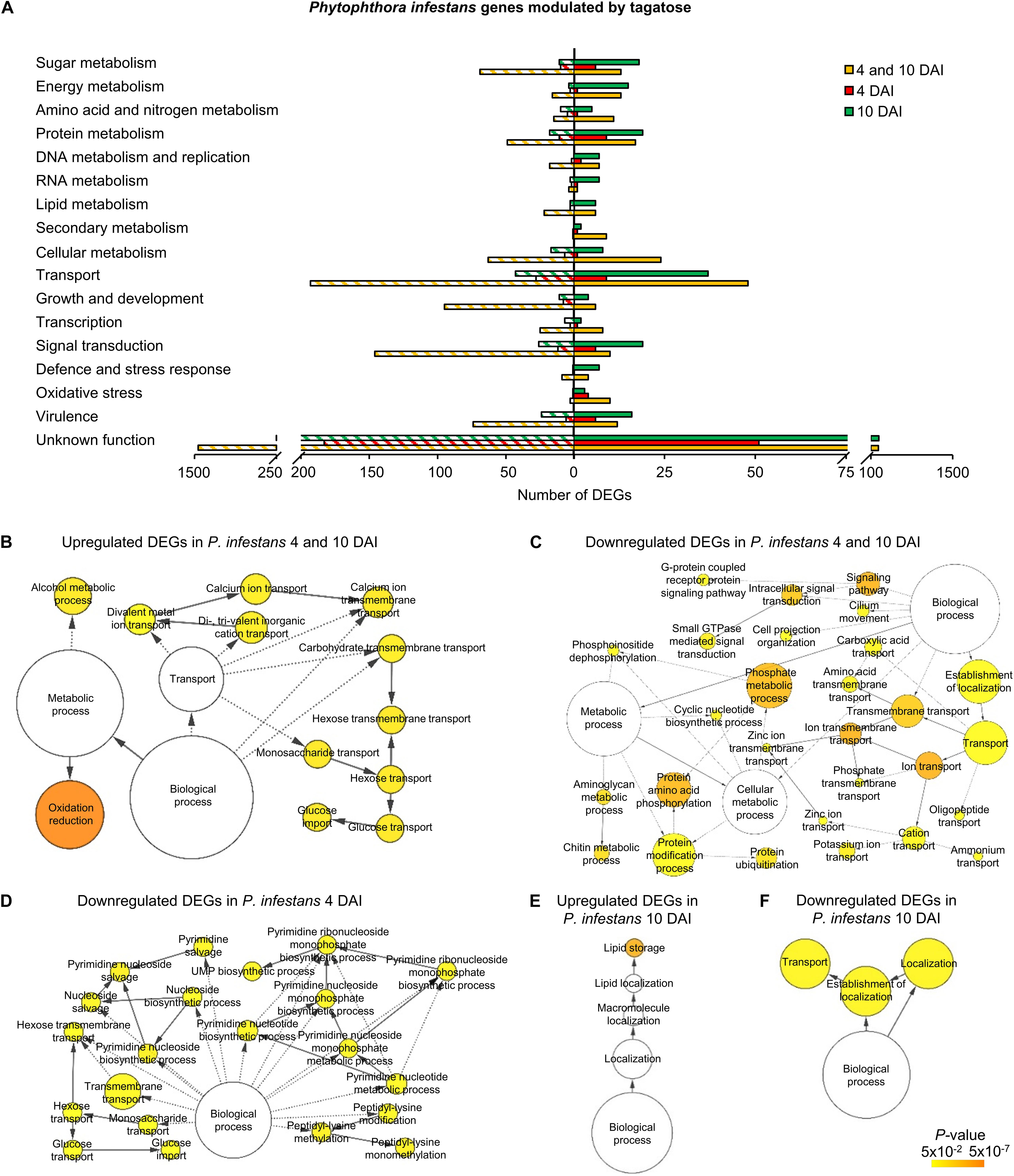
Figure 3. Functional annotation of genes modulated by tagatose in P. infestans. Differentially expressed genes (DEGs) were identified in P. infestans at 4 and 10 days after incubation (DAI) on pea agar medium in the presence of tagatose compared to the respective control samples grown in the absence of tagatose. Functional categories (A) were assigned based on the protein description of upregulated (solid bars) or downregulated (stripped bars) genes at both time points (4 and 10 DAI; orange) and exclusively at 4 DAI (red) or at 10 DAI (green; Supplementary Table 9). Biological networks of significantly enriched (P ≤ 0.05) Gene Ontology (GO) terms of P. infestans genes upregulated at 4 and 10 DAI (B), downregulated at 4 and 10 DAI (C), downregulated at 4 DAI (D), upregulated at 10 DAI (E) and downregulated at 10 DAI (F) were identified using the BiNGO tool and visualized with Cytoscape software. The color scale legend indicates the level of significance for enriched GO terms and white nodes indicate not significantly overrepresented categories. Dotted lines indicate connection between biological process categories in the GO chart, where ancestor and child are omitted for simplicity. No significant GO enrichment was found for P. infestans genes upregulated at 4 DAI.
Phytophthora infestans genes modulated at 4 DAI (4 DAI cluster) and at 10 DAI (10 DAI cluster) included 385 DEGs (99 upregulated and 286 downregulated) and 842 DEGs (411 upregulated and 431 downregulated), respectively (Supplementary Figure 13 and Supplementary Table 9). Downregulated genes at 4 DAI were mainly involved in transport (e.g., two glucose transporters and two sugar transporters), signal transduction (e.g., two protein kinases) and protein metabolism (e.g., two serine protease inhibitors and three serine proteases; Figure 3A and Supplementary Table 9). Moreover, the GO categories related to transport (e.g., glucose transport) and pyrimidine metabolism were overrepresented in the cluster of downregulated genes at 4 DAI (Figure 3D and Supplementary Table 9). Upregulated genes of the 10 DAI cluster were mainly related to protein metabolism (Figure 3A) with the overrepresentation of the lipid storage GO category (Figure 3E and Supplementary Table 9). Moreover, downregulated genes at 10 DAI were mainly involved in transport (e.g., 10 ABC proteins and three mitochondrial carriers), virulence (six crinkler family proteins, one elicitin-like protein and one cutinase) and signal transduction (Figures 3A,F and Supplementary Table 9). However, a large fraction of genes with an unknown function was modulated in the 4 and 10 DAI (228 upregulated and 1,445 downregulated), 4 DAI (51 upregulated and 183 downregulated) and 10 DAI (236 upregulated and 252 downregulated) cluster, and indicated that that several yet-to-be identified proteins may have been involved in the P. infestans response to tagatose.
In summary, P. infestans response to tagatose incubation involved the upregulation of energy metabolism and oxidative stress response. However, the P. infestans transcriptional reprogramming was characterized by the downregulation of genes of transport-, sugar metabolism-, signal transduction- and growth-related processes (Figure 4A), in agreement with the inhibition of mycelial growth and alteration of sugar and amino acid content.

Figure 4. Overview of the main processes modulated by tagatose in Phytophthora spp. Main cellular pathways affected by tagatose were identified for differentially expressed genes (DEGs) of P. infestans (A), P. cinnamomi (B), and Phytophthora spp. orthologous (C) genes at 4 and 10 days after incubation (DAI) on pea agar medium. For each P. infestans and P. cinnamomi gene, two squares represent the Log2-transformed fold change values of tagatose-incubated samples calculated as compared to control samples at 4 (left square) and 10 (right square) DAI, according to the color scale reported. Orthologous DEGs were grouped in 16 clusters (defined by a four-letter code, black circles) based on their upregulation (U) or downregulation (D) in tagatose-incubated samples [P. infestans at 4 DAI (first letter); P. infestans at 10 DAI (second letter); P. cinnamomi at 4 DAI (third letter); P. cinnamomi at 10 DAI (fourth letter)], such as genes upregulated in P. infestans and downregulated in P. cinnamomi at 4 DAI and at 10 DAI (UUDD), genes downregulated in P. infestans at 4 DAI and at 10 DAI and upregulated in P. cinnamomi at 4 DAI and at 10 DAI (DDUU), at 4 DAI (DDUD) or at 10 DAI (DDDU).
Tagatose Incubation Causes an Efficient Transcriptional Response in Phytophthora cinnamomi
The three clusters of P. cinnamomi DEGs included 41 genes modulated at 4 and 10 DAI (36 upregulated and 5 downregulated), 122 genes modulated at 4 DAI (71 upregulated and 51 downregulated) and 349 genes modulated at 10 DAI (146 upregulated and 203 downregulated; Figure 2B, Supplementary Figure 13B, and Supplementary Table 10). Although a large fraction of genes with an unknown function was found for P. cinnamomi DEGs (17, 78, and 228 in the 4 and 10 DAI, 4 and 10 DAI cluster, respectively), functional annotations revealed the upregulation of genes involved in signal transduction (e.g., one phosphoinositide-3-kinase and two magnesium-dependent phosphatases), oxidative stress (e.g., three alternative oxidases and one quinone oxidoreductase), energy metabolism (e.g., two pyruvate kinases and one glycerol-3-phosphate dehydrogenase) and transport (e.g., one stromal interaction molecule, one glucose transporter and one voltage-gated potassium channel) in the 4 and 10 DAI cluster (Figure 5A and Supplementary Table 10). As a consequence, the GO categories related to energy metabolism (e.g., glycerol 3-phosphate catabolic process and glycolosis), oxidation reduction and signaling were overrepresented in the cluster of upregulated genes at 4 and 10 DAI (Figure 5B and Supplementary Table 10).
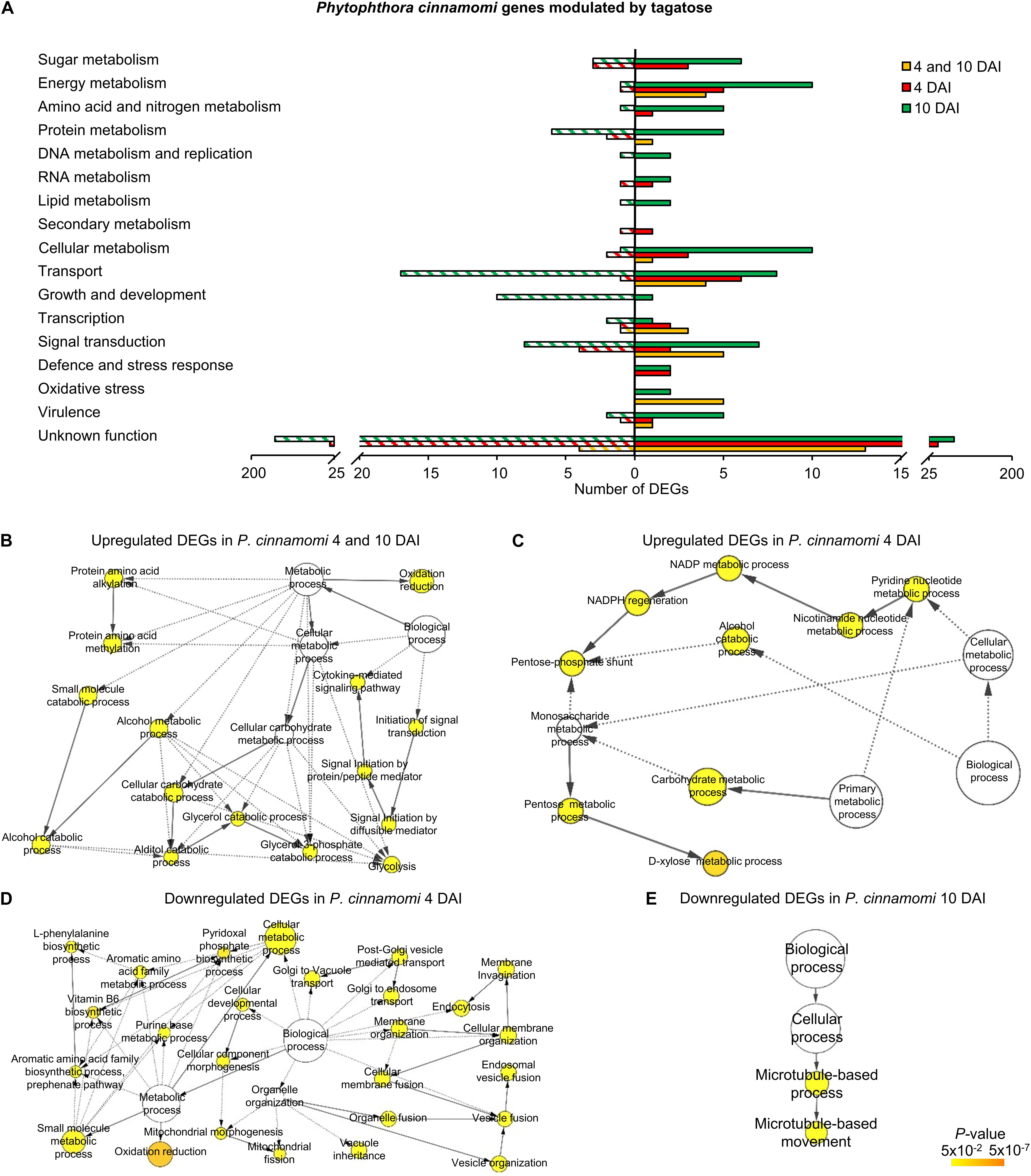
Figure 5. Functional annotation of genes modulated by tagatose in Phytophthora cinnamomi. Differentially expressed genes (DEGs) were identified in P. cinnamomi at 4 and 10 days after incubation (DAI) on pea agar medium in the presence of tagatose compared to the respective control samples grown in the absence of tagatose. Functional categories (A) were assigned based on the protein description of upregulated (solid bars) or downregulated (stripped bars) genes at both time points (4 and 10 DAI; orange) and exclusively at 4 DAI (red) or at 10 DAI (green; Supplementary Table 10). Biological networks of significantly enriched (P ≤ 0.05) Gene Ontology (GO) terms of P. cinnamomi genes upregulated at 4 and 10 DAI (B), upregulated at 4 DAI (C), downregulated at 4 DAI (D) and downregulated at 10 DAI (E) were identified using the BiNGO tool and visualized with Cytoscape software. The color scale legend indicates the level of significance for enriched GO terms and white nodes indicate not significantly overrepresented categories. Dotted lines indicate connection between biological process categories in the GO chart, where ancestor and child are omitted for simplicity. No significant GO enrichment was found for P. cinnamomi genes downregulated at 4 and 10 DAI and upregulated at 10 DAI.
Genes upregulated by tagatose at 4 DAI were mainly involved in transport (e.g., two aquaporins, one MtN3-like protein and one carboxylic acid transporter) and energy metabolism (two xylose isomerases, one xylulose kinase and one succinate dehydrogenase; Figure 5A and Supplementary Table 10) with the overrepresentation of energy-related GO categories (e.g., NADPH regeneration, pentose metabolic process and carbohydrate metabolic process; Figure 5C and Supplementary Table 10). On the other hand, downregulated genes at 4 DAI were mainly involved in signal transduction (e.g., two protein kinase and one protein phosphatase) with the overrepresentation of oxidative reduction and vesicle-mediated transport (Figures 5A,D and Supplementary Table 10). Upregulated genes of the 10 DAI cluster were mainly related to energy metabolism (e.g., two phosphoglycerate mutases, one glyceraldehyde-3-phosphate dehydrogenase and one pyruvate dehydrogenase; Figure 5A and Supplementary Table 10). Moreover, downregulated genes at 10 DAI were mainly involved in transport, growth and development (Figure 5A), as well as the GO categories of microtubule-based processes (Figure 5E and Supplementary Table 10). Thus, P. cinnamomi response to tagatose was characterized by the upregulation of genes implicated in transport-, energy metabolism- and oxidative stress-related processes (Figure 4B), possibly to adapt the cellular metabolism and minimize the alteration of the sugar content and mycelial growth.
Tagatose Differentially Modulates Orthologous Genes in Phytophthora infestans and P. cinnamomi
Although a large fraction of genes with an unknown function was found for the orthologous DEGs, functional annotations revealed that orthologous genes upregulated in both species and time points (UUUU cluster) were mainly implicated in transport (e.g., one mitochondrial carrier), amino acid and nitrogen metabolism (e.g., one glutamine amidotransferase, three aspartate aminotransferases and one histidinol-phosphate aminotransferase; Figure 6A and Supplementary Table 11). Orthologous genes upregulated in P. infestans and downregulated in P. cinnamomi (UUDD cluster) were mainly related to transport (e.g., six ABC proteins and one voltage-gated ion channel) and protein metabolism (e.g., seven ribosomal genes, one ubiquitin-specific protease and one elongation factor; Figure 6A and Supplementary Table 11) with the overrepresentation of translation and rRNA metabolic processes (Figure 6B). Moreover, orthologous genes downregulated in P. infestans and upregulated in P. cinnamomi (DDUU cluster) were mainly associated with sugar metabolism (e.g., seven glycoside hydrolases, one UDP-sugar pyrophospharylase, one sorbitol dehydrogenase, one endo-1,4-β-xylanase and one lysosomal β-glucosidase), transport (e.g., one glucose transporter, one bidirectional sugar transporter and one glycoside-cation symporter) and signal transduction (Figure 6A and Supplementary Table 11).
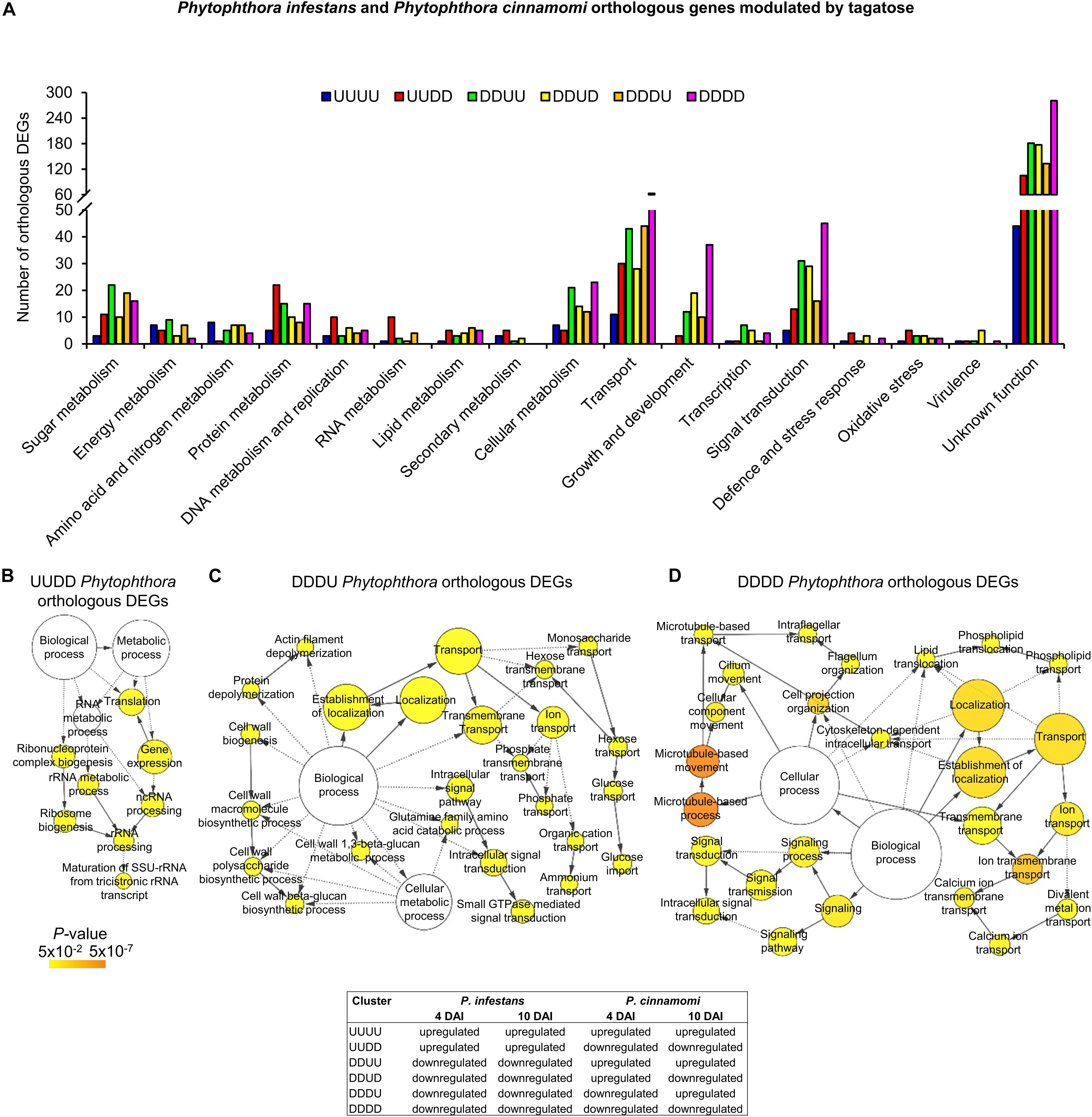
Figure 6. Functional annotation of P. infestans and P. cinnamomi orthologous genes modulated by tagatose. Orthologous genes of P. infestans and P. cinnamomi differentially expressed (orthologous DEGs) were identified at 4 and 10 days after incubation (DAI) on pea agar medium in the presence of tagatose compared to the respective control samples grown in the absence of tagatose. Orthologous DEGs were grouped in 16 clusters (defined by a four-letter code) based on their upregulation (U) or downregulation (D) in tagatose-incubated samples (Supplementary Table 11): P. infestans at 4 DAI (first letter); P. infestans at 10 DAI (second letter); P. cinnamomi at 4 DAI (third letter); P. cinnamomi at 10 DAI (fourth letter); such as the cluster of genes upregulated in P. infestans and downregulated in P. cinnamomi at 4 and 10 DAI (UUDD). Functional categories (A) of the clusters UUUU (blue), UUDD (red), DDUU (green), DDUD (yellow), DDDU (orange) and DDDD (pink) were assigned based on the protein description. Biological networks of significantly enriched (P ≤ 0.05) Gene Ontology (GO) terms of UUDD (B), DDDU (C), and DDDD (D) cluster of orthologous DEGs were identified using the BiNGO tool and visualized with Cytoscape software. The color scale legend indicates the level of significance for enriched GO terms and white nodes indicate not significantly overrepresented categories. Dotted lines indicate connection between biological process categories in the GO chart, where ancestor and child are omitted for simplicity. No significant GO enrichment was found for the UUUU, DDUU and DDUD clusters.
Orthologous genes of the DDUD cluster were mainly linked to transport [e.g., one multidrug/oligosaccharidyl-lipid/polysaccharide (MOP) flippase and two choline transporters), signal transduction, growth and development (e.g., four kinesin-like proteins, one dynein heavy chain and one myosin-like gene; Figure 6A and Supplementary Table 11). The DDDU cluster consisted of 273 orthologous DEGs that were mainly involved in transport (e.g., two phosphate transporters, a sugar transporter and a MOP flippase) and sugar metabolism (e.g., two lysosomal β-glucosidases, a maltose O-acetyltransferase and a β-glucan synthesis protein; Figure 6A and Supplementary Table 11) with the enrichment of monosaccharide transport and growth-related processes (e.g., actin filament depolymerization; Figure 6C and Supplementary Table 11). Orthologous genes downregulated in both species (DDDD cluster) were mainly related to transport, signal transduction, growth and development (Figure 6A and Supplementary Table 11) with the enrichment of the GO categories of transport, signaling and growth (e.g., microtubule-based process; Figure 6D and Supplementary Table 11). In summary, differential transcriptional regulation of Phytophthora spp. orthologous genes was found in presence of tagatose, including the upregulation of protein metabolism in P. infestans (UUDD cluster) and the upregulation of sugar metabolism, transport, signal transduction and growth-related processes in P. cinnamomi (DDUU, DDUD, and DDDU cluster; Figure 4C).
Discussion
Tagatose showed nutritional or anti-nutritional properties for specific microbial taxa (Bautista et al., 2000; Perazzolli et al., 2020) and caused differential growth inhibition in some species belonging to the same genus, such as Phytophthora (Chahed et al., 2020). In particular, the tagatose dosage showing a differential effects on Phytophthora spp. growth was previously identified [5 g/L tagatose; (Chahed et al., 2020)] and it was used to assess metabolic and transcriptional changes. Differential response to tagatose incubation was found in P. infestans and P. cinnamomi with species-specific metabolic and transcriptional changes at the time points (4 and 10 DAI) that previously showed cellular ultrastructure changes in P. infestans, but not in P. cinnamomi (Chahed et al., 2020). In particular, sugar content and amino acid content were affected by tagatose in P. infestans, but not in P. cinnamomi. Moreover, a high number of genes was modulated by tagatose in P. infestans (3,915 DEGs) compared to P. cinnamomi (512 DEGs), suggesting that tagatose incubation caused a severe transcriptional reprogramming in P. infestans, rather than in P. cinnamomi. In addition, P. infestans DEGs were mainly repressed (75.70%), unlike P. cinnamomi (50.58%), suggesting global downregulation of functional processes associated with the growth inhibition caused by tagatose in P. infestans. Metabolic and transcriptional changes were assessed for each tagatose-incubated sample in comparison to the respective control sample, in order to detect the tagatose effects for each species and time point. However, the possible contribution of the different growth rate and developmental stage to the differential response of P. infestans and P. cinnamomi to tagatose cannot be totally excluded.
Main Cellular Processes Affected by Tagatose Incubation in Phytophthora infestans
Tagatose incubation impaired P. infestans growth and caused severe impacts on sugar content and amino acid content with the downregulation of genes related to transport, sugar metabolism and growth-related process. In particular, tagatose incubation increased the content of fructose and decreased the content of glucose, mannose and ribose in P. infestans, but not in P. cinnamomi. Tagatose is the C-4 epimer of fructose and it may inhibit fructose-metabolizing enzymes in P. infestans, as previously reported for the fructokinase in mammalian (Lu et al., 2008) and in Hyaloperonospora arabidopsidis (Mochizuki et al., 2020), with the consequent increase of the fructose content. On the other hand, the decrease in glucose content could be ascribed to the tagatose-depended inhibition of P. infestans β-glucosidase enzymes (Corneo et al., 2021), which are involved in polysaccharide and disaccharide hydrolysis (Saha et al., 1994). As corroboration, genes encoding ß-glucosidase enzymes were mainly down- and up-regulated by tagatose in P. infestans and P. cinnamomi, respectively. In agreement with these findings, previous transcriptomic studies showed that the growth inhibition of Phytophthora spp., caused by biological products (melatonin), biocontrol bacteria (L. capsici AZ78) and fungicides (dimethomorph and metalaxyl), was associated with the downregulation of genes related to sugar metabolism (Tomada et al., 2017; Zhang et al., 2017; Hao et al., 2019) and to the decrease in sugar content (Maridueña-Zavala et al., 2017), indicating a strong correlation between sugar metabolism and P. infestans growth. Likewise, sugar- and growth-related genes involved in cell wall integrity and biogenesis (e.g., trehalose synthase, glucosyltransferase, glucose 4,6-dehydratases and β-glucan synthase genes) were downregulated by tagatose in P. infestans, but not in P. cinnamomi. The impact of tagatose on oomycete cell wall was previously reported in H. arabidopsidis, where tagatose inhibited the metabolism of mannan (Mochizuki et al., 2020), which is an essential component of oomycete cell wall (Melida et al., 2013). The alteration of cell wall integrity can negatively affect P. infestans pathogenicity (Resjö et al., 2017), and genes related to virulence and infection processes, such as RxLR (Kamoun, 2006), necrosis-inducing protein (NPP1) (Qutob et al., 2002) and thrombospondin (Robold and Hardham, 2005) were downregulated in tagatose-incubated P. infestans. Although possible alterations on pathogenicity should be validated by artificial inoculations on tomato plants, transcriptional changes indicated severe impacts of tagatose on key functional processes of P. infestans.
As a possible consequence of sugar metabolism inhibition, tagatose incubation decreased amino acid content in P. infestans, but not in P. cinnamomi. Previous studies showed that some fungal species (e.g., Candida albicans) can exploit amino acids as a carbon source under sugar-limiting conditions (Ene et al., 2014), suggesting that P. infestans could catabolise amino acids in the presence of tagatose. The amino acid catabolism involves cytoplasmic transaminases to form glutamic acid (Judelson et al., 2009), which is the only amino acid to increase in tagatose-incubated P. infestans. As corroboration, genes involved in glutamic acid biosynthesis (e.g., glutamine amidotransferase and histidinol-phosphate transaminase genes) were upregulated by tagatose, suggesting that amino acid metabolism was reprogrammed in P. infestans possibly to compensate the inhibition of sugar metabolism. Likewise, the growth inhibition of Phytophthora spp. caused by biological products (melatonin) and fungicides (e.g., ethylicin and SYP-14288) was previously associated with the inhibition of amino acid metabolism and protein synthesis (Zhang et al., 2017, 2020; Cai et al., 2019). For example, arginine content was decreased in tagatose-incubated P. infestans and genes involved in arginine biosynthesis (e.g., arginino-succinate synthase) were downregulated. The decrease of amino acid content was associated also with the downregulation of amino acid transporters in tagatose-incubated P. infestans. Similarly, the downregulation of amino acid transporter genes was previously linked to P. infestans growth inhibition caused by Pseudomonas fluorescens LBUM223 (Roquigny et al., 2018), indicating strong correlations between sugar metabolism, amino acid metabolism, amino acid transport and P. infestans growth.
Processes With Opposite Modulation in Phytophthora infestans and in P. cinnamomi After Tagatose Incubation
Opposite modulation of genes involved in transport, energy metabolism, growth-related process, and oxidative stress response were found in P. infestans and P. cinnamomi, indicating species-specific reaction to tagatose incubation. In particular, tagatose incubation led to the down- and up-regulation of genes encoding sugar transporters (e.g., glucose transporters, bidirectional sugar transporters, glycoside-pentoside transporters and multidrug/oligosaccharidyl-lipid/polysaccharide flippases) in P. infestans and P. cinnamomi, respectively. Clinical studies showed that tagatose may act by attenuating glucose absorption in the human intestine (Donner et al., 1999). Likewise, the rare sugar sorbose inhibited glucose transport in Saccharomyces cerevisiae leading to growth retardation (van Uden, 1967), suggesting that the growth inhibitory effects of rare sugars may be mediated by glucose transport inhibition. Thus, the upregulation of glucose and sugar transporters in P. cinnamomi could be an efficient cellular response to mitigate tagatose inhibitory effects on sugar transporters and to maintain sufficient glucose uptake and energy production. Likewise, genes implicated in the tricarboxylic acid (TCA) cycle (e.g., malate synthase, succinate-semialdehyde dehydrogenase, succinate dehydrogenase and pyruvate dehydrogenase) and mitochondrial respiration (e.g., ubiquinone biosynthesis protein COQ7, cytochrome b5 and, NADH dehydrogenase), were down- and up-regulated by tagatose incubation in P. infestans and P. cinnamomi, respectively. In agreement with these findings, tagatose incubation decreased the content of TCA cycle intermediates in P. infestans (such as malic acid and succinic acid) (Corneo et al., 2021) and impaired normal respiration and ATP synthesis in P. infestans, but not in P. cinnamomi (Chahed et al., 2020), indicating species-specific effects of tagatose on some pathways of energy metabolism. As a possible consequence of energy limitation, genes associated with growth and development (e.g., myosin-like protein and actin-like protein) were mainly down- and up-regulated by tagatose incubation in P. infestans and P. cinnamomi, respectively. Likewise, Phytophthora capsici growth inhibition by the fungicide benzothiazole was previously associated with the downregulation of growth-related genes, such as those encoding actin and ankyrin repeat-protein (Mei et al., 2019).
Although genes involved in oxidative stress response were upregulated by tagatose incubation in both Phytophthora spp., some genes belonging to this functional category showed species-specific profiles. For example, glutathione S-transferase genes were up- and down-regulated in P. infestans and P. cinnamomi, respectively (UUDD cluster), while alternative oxidase genes showed opposite profile (DDUU cluster). Glutathione S-transferase gene is a marker of oxidative stress and it was induced by biocontrol bacteria (L. capsici AZ78 and P. fluorescens LBUM223) in P. infestans (Tomada et al., 2017; Roquigny et al., 2018). Thus, expression profiles of glutathione S-transferase genes agreed with the stronger accumulation of reactive oxygen species (ROS) previously found in P. infestans compared to P. cinnamomi during tagatose incubation (Chahed et al., 2020). Conversely, the upregulation of alternative oxidase genes in P. cinnamomi rather than P. infestans suggest a key contribution of these enzymes in the ROS detoxification, as previously found for the oxidative stress-handling machinery of Ustilago maydis (Juarez et al., 2006). Alternative oxidases are also key enzymes of the alternative respiration pathways in phytopathogenic fungi (Tian et al., 2020) and their expression was upregulated by non-fermentable carbon sources (e.g., glycerol and ethanol) in C. albicans (Huh and Kang, 2001). Thus, P. cinnamomi may upregulate alternative oxidase pathways, in order to mitigate the oxidative stress and to allow alternative respiration, in agreement with the normal ROS level and oxygen consumption rate previously found during tagatose incubation (Chahed et al., 2020). Moreover, genes related to signal transduction were mainly down- and up-regulated in P. infestans (e.g., protein phosphatases 2C and cAMP kinases) and P. cinnamomi, respectively. Phosphatase 2C genes were downregulated by oxidative stress in Nicotiana tabacum (Vranová et al., 2000) and signaling genes (e.g., cAMP domain-containing protein) were downregulated by growth inhibition in P. capsici (Mei et al., 2019), suggesting the activation of complementary signal transduction pathways in P. infestans and P. cinnamomi to modulate the response to tagatose incubation.
Main Cellular Processes Affected by Tagatose Incubation in Phytophthora cinnamomi
Mycelial growth, sugar and amino acid content were not affected by tagatose incubation in P. cinnamomi and the transcriptional response revealed the upregulation of genes related to transport, energy metabolism, sugar metabolism and oxidative stress. In particular, the response of P. cinnamomi included the upregulation of genes related to pentose metabolism, such as xylose isomerase, xylulose kinase and inositol oxygenase, indicating the upregulation of alternative sugar catabolism. Previous studies showed that blocking hexose entry into glycolysis upregulated pentose metabolism in Aspergillus nidulans (Khosravi et al., 2018) and that xylulose kinase co-expression with arabinan-degrading genes was essential for Aspergillus niger growth on xylose and arabinose (VanKuyk et al., 2001). Likewise, genes encoding sorbitol dehydrogenase and arabinan-degrading enzymes responsible for the release of pentose sugars from polysaccharides side chains (e.g., endo-1,4-beta-xylanase, arabinogalactan endo-beta-1,4-galactanase, arabinan endo-1,5-alpha-L-arabinosidase) were upregulated by tagatose incubation in P. cinnamomi. Consequently, the pentose sugars originated by these enzymatic activities can be assimilated into the pentose phosphate pathway, suggesting that P. cinnamomi metabolized alternative sugars and upregulated the pentose metabolism to alleviate metabolic impacts caused by tagatose. In addition, the P. cinnamomi response included the upregulation of a mannitol dehydrogenase gene, which was upregulated in fungicide-resistant Phytophthora isolates (Childers et al., 2015), and the upregulation of transcription factors (e.g., WRKY, zink finger proteins and MYB-like DNA-binding protein), suggesting the activation of an efficient machinery to modulate the response to tagatose incubation.
Processes Commonly Affected by Tagatose Incubation in Both Phytophthora spp.
Some oxidative stress-related genes (e.g., quinone oxidoreductase) were upregulated in P. infestans and in P. cinnamomi, indicating that both species incurred an oxidative stress after tagatose incubation. However, the efficient scavenging of reactive oxygen species found in P. cinnamomi (Chahed et al., 2020) was associated with the upregulation of additional genes (such as alternative oxidases discussed above), which were downregulated in P. infestans. In both Phytophthora spp., some genes involved in the energy metabolism of glycolysis (e.g., phosphoglycerate mutase, glyceraldehyde-3-P dehydrogenase and pyruvate kinase) were upregulated by tagatose incubation. Tagatose was patented as glycolysis inhibitor in humans (Kim et al., 2014) and showed inhibitory effects on glycolysis-related enzymes in H. arabidopsidis (fructokinase and phosphomannose isomerase) (Mochizuki et al., 2020) and Escherichia coli (fructose phosphate aldolase) (Stellmacher et al., 2016). As a possible consequence of glycolysis inhibition, P. infestans and P. cinnamomi upregulated a pyruvate phosphate dikinase gene (cluster UUUU) to generate pyruvate, as a bypass of pyruvate kinase in glycolysis reactions (Judelson et al., 2009). Likewise, genes encoding glycerol-3-phosphate dehydrogenase were upregulated in both Phytophthora spp. and they can reinforce glycolysis pathways by providing an extra source of dihydroxyacetone phosphate (Lalle et al., 2015), as an attempted reaction to alleviate metabolic impacts derived from the glycolysis inhibition. The enzymatic inhibition caused by tagatose can be ascribed to the structural similarity with fructose and to the possible interference with the substrate binding by the active site, as in the case of the mammalian fructokinase (Lu et al., 2008) and S. mutans glucosyltransferase (Hasibul et al., 2018). This competitive effect was also suggested by the fructose-dependent attenuation of tagatose effects in P. infestans (Corneo et al., 2021) and in S. mutants (Hasibul et al., 2018), but further enzymatic studies are required in order to better clarify the inhibitory effects of tagatose on Phytophthora spp. enzymes. Likewise, metabolic and transcriptomic studies at early time points in presence of different dosages of rare sugars and/or common sugars will be required, in order to investigate the early response of Phytophthora spp. to tagatose and to clarify possible dose-dependent interactions between rare sugar and common sugar metabolism.
Conclusion
The differential inhibitory effect of tagatose on P. infestans and P. cinnamomi was associated with species-specific metabolic and transcriptional changes. In particular, an attempted response was upregulated by P. infestans, but it was not sufficient to contrast the negative effects of tagatose incubation on mycelial growth, sugar content and amino acid content. Thus, P. infestans transcriptional reprogramming was mainly characterized by the severe downregulation of genes implicated in transport, sugar metabolism, signal transduction and growth-related process. Conversely, P. cinnamomi response to tagatose incubation was characterized by the activation of processes related to transport, energy metabolism, sugar metabolism and oxidative stress-related, in order to limit negative impacts on mycelial growth, sugar content and amino acid content. In particular, P. cinnamomi was able to implement multiple pathways to modulate the cellular metabolism based on the upregulation of genes related to glucose transport, pentose metabolism, TCA cycle, ROS detoxification, mitochondrial and alternative respiration. These metabolic and transcriptional results represent a major contribution to the characterization of the species-specific mode of action of tagatose on Phytophthora spp. and might pave the way for functional genomic and biochemical studies to further characterize enzymatic reactions affected by this rare sugar.
Data Availability Statement
The sequences were deposited at the Sequence Read Archive of the National Center for Biotechnology under the BioProject number PRJNA622764. The datasets generated for this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
AC and AN carried out the experiments, sample collection, and RNA extraction. VL carried out the qPCR experiments. MM analyzed the RNA-Seq data. PEC helped to carry out the experiments and sample collection. AC, MP, EAB, GP, and IP contributed to data interpretation and the manuscript writing. MP conceived the study, designed the experiments, and coordinated all research activities. All authors revised and approved the final manuscript.
Funding
This project has received funding from the European Union’s Horizon 2020 research and innovation programme under the Marie Skłodowska-Curie grant agreement no. 722642 (project INTERFUTURE).
Conflict of Interest
AC and AN were employed by Biological Products for Agriculture (Bi-PA).
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Valerio Battaglia (Council for Agricultural Research and Agricultural Economics Analysis) for providing the Phytophthora infestans strain VB3 and Mario Malacarne of the Chemistry Unit at Fondazione Edmund Mach for metabolic analyses.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.711545/full#supplementary-material
Supplementary Figure 1 | Phytophthora spp. growth.
Supplementary Figure 2 | Effect of tagatose on Phytophthora spp. amino acid content.
Supplementary Figure 3 | Correlation matrixes of Phytophthora infestans control samples at 4 days after incubation.
Supplementary Figure 4 | Correlation matrixes of P. infestans control samples at 10 days after incubation.
Supplementary Figure 5 | Correlation matrixes of P. infestans tagatose-incubated samples at 4 days after incubation.
Supplementary Figure 6 | Correlation matrixes of P. infestans tagatose-incubated samples at 10 days after incubation.
Supplementary Figure 7 | Correlation matrixes of P. cinnamomi control samples at 4 days after incubation.
Supplementary Figure 8 | Correlation matrixes of P. cinnamomi control samples at 10 days after incubation.
Supplementary Figure 9 | Correlation matrixes of P. cinnamomi tagatose-incubated samples at 4 days after incubation.
Supplementary Figure 10 | Correlation matrixes of P. cinnamomi tagatose-incubated samples at 10 days after incubation.
Supplementary Figure 11 | Principal component analysis (PCA) of Phytophthora spp. samples.
Supplementary Figure 12 | Volcano Plot analysis for the identification of differentially expressed genes in Phytophthora spp.
Supplementary Figure 13 | Venn diagrams of genes modulated by tagatose in Phytophthora spp.
Supplementary Figure 14 | Correlation analysis of gene expression data assessed by RNA-Seq and quantitative real-time PCR.
Supplementary Table 1 | Sugar content in the pea agar medium.
Supplementary Table 2 | Primer sequences of the Phytophthora spp. genes analyzed by quantitative real-time PCR.
Supplementary Table 3 | RNA-Seq sequencing and mapping results of each replicate.
Supplementary Table 4 | Expression levels of Phytophthora infestans genes.
Supplementary Table 5 | Expression levels of P. cinnamomi genes.
Supplementary Table 6 | Pearson correlation values of P. infestans samples.
Supplementary Table 7 | Pearson correlation values of P. cinnamomi samples.
Supplementary Table 8 | Pearson correlation values of Phytophthora spp. samples.
Supplementary Table 9 | Fold change values and functional annotation of genes modulated by tagatose in P. infestans.
Supplementary Table 10 | Fold change values and functional annotation of genes modulated by tagatose in P. cinnamomi.
Supplementary Table 11 | Expression profiles and functional annotation of P. infestans and P. cinnamomi orthologous genes modulated by tagatose.
Footnotes
- ^ https://protists.ensembl.org/Phytophthora_infestans/Info/Index
- ^ https://mycocosm.jgi.doe.gov/Phyci1/Phyci1.home.html
- ^ http://bioinformatics.psb.ugent.be/webtools/Venn/
- ^ www.python.org
- ^ http://www.uniprot.org
References
Bautista, D. A., Pegg, R. B., and Shand, P. J. (2000). Effect of L-glucose and D-tagatose on bacterial growth in media and a cooked cured ham product. J. Food Prot. 63, 71–77. doi: 10.4315/0362-028X-63.1.71
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Cai, M., Wang, Z., Ni, X., Hou, Y., Peng, Q., Gao, X., et al. (2019). Insights from the proteome profile of Phytophthora capsici in response to the novel fungicide SYP-14288. PeerJ 7:e7626. doi: 10.7717/peerj.7626
Camacho, C., Coulouris, G., Avagyan, V., Ma, N., Papadopoulos, J., Bealer, K., et al. (2009). BLAST+: architecture and applications. BMC Bioinformatics 10:421. doi: 10.1186/1471-2105-10-421
Cataldi, T. R. I., Campa, C., and De Benedetto, G. E. (2000). Carbohydrate analysis by high-performance anion-exchangechromatography with pulsed amperometric detection: the potential is still growing. Fresenius J. Anal. Chem. 368, 739–758. doi: 10.1007/s002160000588
Chahed, A., Nesler, A., Navazio, L., Baldan, B., Busato, I., Ait-Barka, E., et al. (2020). The rare sugar tagatose selectively inhibits the growth of Phytophthora infestans and not Phytophthora cinnamomi by interfering with mitochondrial processes. Front. Microbiol. 11:128. doi: 10.3389/fmicb.2020.00128
Childers, R., Danies, G., Myers, K., Fei, Z., Small, I. M., and Fry, W. E. (2015). Acquired resistance to mefenoxam in sensitive isolates of Phytophthora infestans. Phytopathology 105, 342–349. doi: 10.1094/PHYTO-05-14-0148-R
Corneo, P. E., Nesler, A., Lotti, C., Chahed, A., Vrhovsek, U., Pertot, I., et al. (2021). Interactions of tagatose with the sugar metabolism are responsible for Phytophthora infestans growth inhibition. Microbil. Res. 247:126724. doi: 10.1016/j.micres.2021.126724
Dobin, A., Davis, C. A., Schlesinger, F., Drenkow, J., Zaleski, C., Jha, S., et al. (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. doi: 10.1093/bioinformatics/bts635
Donner, T. W., Wilber, J. F., and Ostrowski, D. (1999). D-tagatose, a novel hexose: acute effects on carbohydrate tolerance in subjects with and without type 2 diabetes. Diabetes Obes. Metab. 1, 285–291. doi: 10.1046/j.1463-1326.1999.00039.x
Ene, I. V., Brunke, S., Brown, A. J., and Hube, B. (2014). Metabolism in fungal pathogenesis. Cold Spring Harb. Perspect. Med. 4:a019695. doi: 10.1101/cshperspect.a019695
Falda, M., Toppo, S., Pescarolo, A., Lavezzo, E., Di Camillo, B., Facchinetti, A., et al. (2012). Argot2: a large scale function prediction tool relying on semantic similarity of weighted Gene Ontology terms. BMC Bioinformatics 13:S14. doi: 10.1186/1471-2105-13-S4-S14
Fantke, P., Friedrich, R., and Jolliet, O. (2012). Health impact and damage cost assessment of pesticides in Europe. Environ. Int. 49, 9–17. doi: 10.1016/j.envint.2012.08.001
Fry, W. E., Birch, P. R., Judelson, H. S., Grunwald, N. J., Danies, G., Everts, K. L., et al. (2015). Five reasons to consider Phytophthora infestans a reemerging pathogen. Phytopathology 105, 966–981. doi: 10.1094/PHYTO-01-15-0005-FI
Granström, T. B., Takata, G., Tokuda, M., and Izumori, K. (2004). Izumoring. J. Biosci. Bioeng. 97, 89–94. doi: 10.1016/s1389-1723(04)70173-5
Hao, K., Lin, B., Nian, F., Gao, X., Wei, Z., Luo, G., et al. (2019). RNA-seq analysis of the response of plant-pathogenic oomycete Phytophthora parasitica to the fungicide dimethomorph. Rev. Argent. Microbiol. 51, 268–277. doi: 10.1016/j.ram.2018.08.007
Hardham, A. R. (2005). Phytophthora cinnamomi. Mol. Plant Pathol. 6, 589–604. doi: 10.1111/j.1364-3703.2005.00308.x
Hasibul, K., Nakayama-Imaohji, H., Hashimoto, M., Yamasaki, H., Ogawa, T., Waki, J., et al. (2018). D-Tagatose inhibits the growth and biofilm formation of Streptococcus mutans. Mol. Med. Rep. 17, 843–851. doi: 10.3892/mmr.2017.8017
Hill, D. W., Walters, F. H., Wilson, T. D., and Stuart, J. D. (1979). High performance liquid chromatographic determination of amino acids in the picomole range. Anal. Chem 51, 1338–1341. doi: 10.1021/ac50044a055
Huh, W. K., and Kang, S. O. (2001). Characterization of the gene family encoding alternative oxidase from Candida albicans. Biochem. J. 356, 595–604. doi: 10.1042/0264-6021:3560595
Hunter, J. D. (2007). “Matplotlib: a 2D graphics environment,” in Computing in Science & Engineering, 3 Edn, eds G. K. Thiruvathukal and K. Läufer (Piscataway, NJ: IEEE), 90–95.
Izumori, K. (2006). Izumoring: a strategy for bioproduction of all hexoses. J. Biotechnol. 124, 717–722. doi: 10.1016/j.jbiotec.2006.04.016
Juarez, O., Guerra, G., Velazquez, I., Flores-Herrera, O., Rivera-Perez, R. E., and Pardo, J. P. (2006). The physiologic role of alternative oxidase in Ustilago maydis. FEBS J. 273, 4603–4615. doi: 10.1111/j.1742-4658.2006.05463.x
Judelson, H. S., and Blanco, F. A. (2005). The spores of Phytophthora: weapons of the plant destroyer. Nat. Rev. Microbiol. 3, 47–58. doi: 10.1038/nrmicro1064
Judelson, H. S., Tani, S., and Narayan, R. D. (2009). Metabolic adaptation of Phytophthora infestans during growth on leaves, tubers and artificial media. Mol. Plant Pathol. 10, 843–855. doi: 10.1111/j.1364-3703.2009.00570.x
Kamoun, S. (2000). “Phytophthora,” in Fungal Pathology, 1 Edn, ed. J. W. Kronstad (Dordrecht: Springer), 237–265.
Kamoun, S. (2006). A catalogue of the effector secretome of plant pathogenic oomycetes. Annu. Rev. Phytopathol. 44, 41–60. doi: 10.1146/annurev.phyto.44.070505.143436
Kamoun, S., Furzer, O., Jones, J. D., Judelson, H. S., Ali, G. S., Dalio, R. J., et al. (2015). The Top 10 oomycete pathogens in molecular plant pathology. Mol. Plant Pathol. 16, 413–434. doi: 10.1111/mpp.12190
Khosravi, C., Battaglia, E., Kun, R. S., Dalhuijsen, S., Visser, J., Aguilar-Pontes, M. V., et al. (2018). Blocking hexose entry into glycolysis activates alternative metabolic conversion of these sugars and upregulates pentose metabolism in Aspergillus nidulans. BMC Genomics 19:214. doi: 10.1186/s12864-018-4609-x
Kim, Y. J., Park, J. H., Kim, M. H., Kim, S. B., Hwang, S. H., and Lee, Y. M. (2014). Sweetener Composition for Preventing and Improving Obesity, Containing Glycolysis Inhibitor Ingredient. EP 2 756 764 A2.
Koh, J. H., Choi, S. H., Park, S. W., Choi, N. J., Kim, Y., and Kim, S. H. (2013). Synbiotic impact of tagatose on viability of Lactobacillus rhamnosus strain GG mediated by the phosphotransferase system (PTS). Food Microbiol. 36, 7–13. doi: 10.1016/j.fm.2013.03.003
Komon-Zelazowska, M., Bissett, J., Zafari, D., Hatvani, L., Manczinger, L., Woo, S., et al. (2007). Genetically closely related but phenotypically divergent Trichoderma species cause green mold disease in oyster mushroom farms worldwide. Appl. Environ. Microbiol. 73, 7415–7426. doi: 10.1128/AEM.01059-07
Lalle, M., Camerini, S., Cecchetti, S., Finelli, R., Sferra, G., Muller, J., et al. (2015). The FAD-dependent glycerol-3-phosphate dehydrogenase of Giardia duodenalis: an unconventional enzyme that interacts with the g14-3-3 and it is a target of the antitumoral compound NBDHEX. Front. Microbiol. 6:544. doi: 10.3389/fmicb.2015.00544
Law, C. W., Chen, Y., Shi, W., and Smyth, G. K. (2014). voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 15:R29. doi: 10.1186/gb-2014-15-2-r29
Levin, G. V. (2002). Tagatose, the new GRAS sweetener and health product. J. Med. Food 5, 23–36. doi: 10.1089/109662002753723197
Li, Z., Gao, Y., Nakanishi, H., Gao, X., and Cai, L. (2013). Biosynthesis of rare hexoses using microorganisms and related enzymes. Beilstein J. Org. Chem. 9, 2434–2445. doi: 10.3762/bjoc.9.281
Liu, D., Li, K., Hu, J., Wang, W., Liu, X., and Gao, Z. (2019). Biocontrol and action mechanism of Bacillus amyloliquefaciens and Bacillus subtilis in soybean Phytophthora blight. Int. J. Mol. Sci. 20:2908. doi: 10.3390/ijms20122908
Lobete, M. M., Noriega, E., Batalha, M. A., De Beurme, S., Van De Voorde, I., and Van Impe, J. F. (2017). Effect of tagatose on growth dynamics of Salmonella Typhimurium and Listeria monocytogenes in media with different levels of structural complexity and in UHT skimmed milk. Food Control 73, 31–42. doi: 10.1016/j.foodcont.2016.05.049
Lu, Y., Levin, G. V., and Donner, T. W. (2008). Tagatose, a new antidiabetic and obesity control drug. Diabetes Obes. Metab. 10, 109–134. doi: 10.1111/j.1463-1326.2007.00799.x
Maere, S., Heymans, K., and Kuiper, M. (2005). BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 21, 3448–3449. doi: 10.1093/bioinformatics/bti551
Maridueña-Zavala, M. G., Freire-Peñaherrera, A., Cevallos-Cevallos, J. M., and Peralta, E. L. (2017). GC-MS metabolite profiling of Phytophthora infestans resistant to metalaxyl. Eur. J. Plant Pathol. 149, 563–574. doi: 10.1007/s10658-017-1204-y
Martinussen, J., Solem, C., Holm, A. K., and Jensen, P. R. (2013). Engineering strategies aimed at control of acidification rate of lactic acid bacteria. Curr. Opin. Biotechnol. 24, 124–129. doi: 10.1016/j.copbio.2012.11.009
Mei, X., Liu, Y., Huang, H., Du, F., Huang, L., Wu, J., et al. (2019). Benzothiazole inhibits the growth of Phytophthora capsici through inducing apoptosis and suppressing stress responses and metabolic detoxification. Pestic. Biochem. Physiol. 154, 7–16. doi: 10.1016/j.pestbp.2018.12.002
Melida, H., Sandoval-Sierra, J. V., Dieguez-Uribeondo, J., and Bulone, V. (2013). Analyses of extracellular carbohydrates in oomycetes unveil the existence of three different cell wall types. Eukaryot. Cell 12, 194–203. doi: 10.1128/EC.00288-12
Mijailovic, N., Nesler, A., Perazzolli, M., Aït Barka, E., and Aziz, A. (2021). Rare sugars: recent advances and their potential role in sustainable crop protection. Molecules 26:1720. doi: 10.3390/molecules26061720
Mochizuki, S., Fukumoto, T., Ohara, T., Ohtani, K., Yoshihara, A., Shigematsu, Y., et al. (2020). The rare sugar D-tagatose protects plants from downy mildews and is a safe fungicidal agrochemical. Commun. Biol. 3:423. doi: 10.1038/s42003-020-01133-7
Ohara, T., Ishida, Y., Kudou, R., Kakibuchi, K., Akimitsu, K., Izumori, K., et al. (2008). Plant disease Control Agent Comprising d-Tagatose as Active Ingredient, and plant Disease Control Method. EP 2 329 713 A1, 1-46.∗
Patterson, T. A., Lobenhofer, E. K., Fulmer-Smentek, S. B., Collins, P. J., Chu, T.-M., Bao, W., et al. (2006). Performance comparison of one-color and two-color platforms within the Microarray Quality Control (MAQC) project. Nat. Biotechnol. 24, 1140–1150. doi: 10.1038/nbt1242
Perazzolli, M., Nesler, A., Giovannini, O., Antonielli, L., Puopolo, G., and Pertot, I. (2020). Ecological impact of a rare sugar on grapevine phyllosphere microbial communities. Microbiol. Res. 232:126387. doi: 10.1016/j.micres.2019.126387
Pfaffl, M. W. (2001). A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res. 29, 2002–2007. doi: 10.1093/nar/29.9.e45
Qutob, D., Kamoun, M., and Gijzen, M. (2002). Expression of a Phytophthora sojae necrosis-inducing protein occurs during transition from biotrophy to necrotrophy. Plant J. 32, 361–373. doi: 10.1046/j.1365-313x.2002.01439.x
Raichand, R., Pareek, S., Singh, N. K., and Mayilraj, S. (2012). Exiguobacterium aquaticum sp. nov., a member of the genus Exiguobacterium. Int. J. Syst. Evol. Microbiol. 62, 2150–2155. doi: 10.1099/ijs.0.035790-0
Resjö, S., Brus, M., Ali, A., Meijer, H. J. G., Sandin, M., Govers, F., et al. (2017). Proteomic analysis of Phytophthora infestans reveals the importance of cell wall proteins in pathogenicity. Mol. Cell Proteomics 16, 1958–1971. doi: 10.1074/mcp.M116.065656
Robold, A. V., and Hardham, A. R. (2005). During attachment Phytophthora spores secrete proteins containing thrombospondin type 1 repeats. Curr. Genet. 47, 307–315. doi: 10.1007/s00294-004-0559-8
Roquigny, R., Novinscak, A., Arseneault, T., Joly, D. L., and Filion, M. (2018). Transcriptome alteration in Phytophthora infestans in response to phenazine-1-carboxylic acid production by Pseudomonas fluorescens strain LBUM223. BMC Genomics 19:474. doi: 10.1186/s12864-018-4852-1
Ruijter, J. M., Ramakers, C., Hoogaars, W. M., Karlen, Y., Bakker, O., Van Den Hoff, M. J., et al. (2009). Amplification efficiency: linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res. 37:e45. doi: 10.1093/nar/gkp045
Saha, B. C., Freer, S. N., and Bothast, R. J. (1994). Production, purification, and properties of a thermostable 13-glucosidase from a color variant strain of Aureobasidium pullulans. Appl. Environ. Microbiol. 60, 3774–3780. doi: 10.1128/AEM.60.10.3774-3780.1994
Saldanha, A. J. (2004). Java Treeview–extensible visualization of microarray data. Bioinformatics 20, 3246–3248. doi: 10.1093/bioinformatics/bth349
Shannon, P., Markiel, A., Ozier, O., Baliga, N. S., Wang, J. T., Ramage, D., et al. (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504. doi: 10.1101/gr.1239303
Shen, T., Wang, Q., Li, C., Zhou, B., Li, Y., and Liu, Y. (2020). Transcriptome sequencing analysis reveals silver nanoparticles antifungal molecular mechanism of the soil fungi Fusarium solani species complex. J. Hazard Mater. 388:122063. doi: 10.1016/j.jhazmat.2020.122063
Smyth, G. K. (2004). Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3:Article3. doi: 10.2202/1544-6115.1027
Stellmacher, L., Sandalova, T., Schneider, S., Schneider, G., Sprenger, G. A., and Samland, A. K. (2016). Novel mode of inhibition by D-tagatose 6-phosphate through a Heyns rearrangement in the active site of transaldolase B variants. Acta Crystallogr. D Struct. Biol. 72, 467–476. doi: 10.1107/S2059798316001170
Tian, F., Lee, S. Y., Woo, S. Y., and Chun, H. S. (2020). Alternative oxidase: a potential target for controlling aflatoxin contamination and propagation of Aspergillus flavus. Front. Microbiol. 11:419. doi: 10.3389/fmicb.2020.00419
Tomada, S., Sonego, P., Moretto, M., Engelen, K., Pertot, I., Perazzolli, M., et al. (2017). Dual RNA-Seq of Lysobacter capsici AZ78 - Phytophthora infestans interaction shows the implementation of attack strategies by the bacterium and unsuccessful oomycete defense responses. Environ. Microbiol. 19, 4113–4125. doi: 10.1111/1462-2920.13861
Van der Heiden, E., Delmarcelle, M., Lebrun, S., Freichels, R., Brans, A., Vastenavond, C. M., et al. (2013). A pathway closely related to the D-tagatose pathway of Gram-negative enterobacteria identified in the Gram-positive bacterium Bacillus licheniformis. Appl. Environ. Microbiol. 79, 3511–3515. doi: 10.1128/AEM.03918-12
van Uden, N. (1967). Transport-limited fermentation and growth of Saccharomyces cerevisiae and its competitive inhibition. Arch. Mikrobiol. 58, 155–168. doi: 10.1007/BF00406676
VanKuyk, P. A., De Groot, M. J. L., Ruijter, G. J. G., De Vries, R. P., and Visser, J. (2001). The Aspergillus niger D-xylulose kinase gene is co-expressed with genes encoding arabinan degrading enzymes, and is essential for growth on D-xylose and L-arabinose. Eur. J. Biochem. 268, 5414–5423. doi: 10.1046/j.0014-2956.2001.02482.x
Vastenavond, C., Bertelsen, H., Hansen, S. R., Laursen, R., Saunders, J., and Eriknauer, K. (2011). “Tagatose (D-tagatose),” in Alternative Sweeteners, 4 Edn, ed. L. O’brien-Nabors (New York, NY: CRC Press), 197–222.
Vranová, E., Langebartels, C., Montagu, M. V., Inzé, D., and Camp, W. V. (2000). Oxidative stress, heat shock and drought differentially affect expression of a tobacco protein phosphatase 2C. J. Exp. Bot. 51, 1763–1764. doi: 10.1093/jexbot/51.351.1763
Wu, Q., and Shah, N. P. (2017). The potential of species-specific tagatose-6-phosphate (T6P) pathway in Lactobacillus casei group for galactose reduction in fermented dairy foods. Food Microbiol. 62, 178–187. doi: 10.1016/j.fm.2016.10.027
Yan, H. Z., and Liou, R. F. (2006). Selection of internal control genes for real-time quantitative RT-PCR assays in the oomycete plant pathogen Phytophthora parasitica. Fungal Genet. Biol. 43, 430–438. doi: 10.1016/j.fgb.2006.01.010
Zhang, S., Zhang, M., Khalid, A. R., Li, L., Chen, Y., Dong, P., et al. (2020). Ethylicin prevents potato late blight by disrupting protein biosynthesis of Phytophthora infestans. Pathogens 9:E299. doi: 10.3390/pathogens9040299
Zhang, S., Zheng, X., Reiter, R. J., Feng, S., Wang, Y., Liu, S., et al. (2017). Melatonin attenuates potato late blight by disrupting cell growth, stress tolerance, fungicide susceptibility and homeostasis of gene expression in Phytophthora infestans. Front. Plant Sci. 8:1993. doi: 10.3389/fpls.2017.01993
Keywords: rare sugar, Phytophthora spp., transcriptomics, gene expression level, targeted metabolomics, transcriptional reprogramming
Citation: Chahed A, Lazazzara V, Moretto M, Nesler A, Corneo PE, Barka EA, Pertot I, Puopolo G and Perazzolli M (2021) The Differential Growth Inhibition of Phytophthora spp. Caused by the Rare Sugar Tagatose Is Associated With Species-Specific Metabolic and Transcriptional Changes. Front. Microbiol. 12:711545. doi: 10.3389/fmicb.2021.711545
Received: 18 May 2021; Accepted: 16 June 2021;
Published: 07 July 2021.
Edited by:
Alberto A. Iglesias, CONICET Coastline Agrobiotechnology Institute (IAL), ArgentinaReviewed by:
Maria Victoria Aguilar Pontes, Concordia University, CanadaSergio Adrian Guerrero, National University of the Littoral, Argentina
Copyright © 2021 Chahed, Lazazzara, Moretto, Nesler, Corneo, Barka, Pertot, Puopolo and Perazzolli. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Michele Perazzolli, bWljaGVsZS5wZXJhenpvbGxpQHVuaXRuLml0
 Abdessalem Chahed
Abdessalem Chahed Valentina Lazazzara
Valentina Lazazzara Marco Moretto
Marco Moretto Andrea Nesler
Andrea Nesler Paola Elisa Corneo
Paola Elisa Corneo Essaid Ait Barka
Essaid Ait Barka Ilaria Pertot
Ilaria Pertot Gerardo Puopolo
Gerardo Puopolo Michele Perazzolli
Michele Perazzolli