- Department of Neurology, Xinqiao Hospital, Army Medical University, Chongqing, China
RNA-binding proteins (RBPs) have been shown to be involved in posttranscriptional regulation, which plays an important role in the pathophysiology of intracerebral hemorrhage (ICH). Peroxiredoxin 1 (Prdx1), an RBP, plays an important role in regulating inflammation and apoptosis. On the basis that inflammation and apoptosis may contribute to ICH-induced brain injury, in this study, we used ICH models coupled with in vitro experiments, to investigate the role and mechanism of Prdx1 in regulating inflammation and apoptosis by acting as an RBP after ICH. We first found that Prdx1 was significantly up-regulated in response to ICH-induced brain injury and was mainly expressed in astrocytes and microglia in ICH rat brains. After overexpressing Prdx1 by injecting adeno-associated virus (AAV) into the striatum of rats at 3 weeks, we constructed ICH models and found that Prdx1 overexpression markedly reduced inflammation and apoptosis after ICH. Furthermore, RNA immunoprecipitation combined with high-throughput sequencing (RIP-seq) in vitro revealed that Prdx1 affects the stability of inflammation- and apoptosis-related mRNA, resulting in the inhibition of inflammation and apoptosis. Finally, overexpression of Prdx1 significantly alleviated the symptoms and mortality of rats subjected to ICH. Our results show that Prdx1 reduces ICH-induced brain injury by targeting inflammation- and apoptosis-related mRNA stability. Prdx1 may be an improved therapeutic target for alleviating the brain injury caused by ICH.
Introduction
Intracerebral hemorrhage (ICH) is a common type of stroke, with increasing incidence and mortality worldwide (Krishnamurthi et al., 2013). Increasing evidence shows that inflammation and apoptosis contribute to the brain injury seen in ICH, which is closely related to the severity of the patient’s symptoms and prognosis (Zheng et al., 2016). However, although the treatments of these therapeutic targets are effective in animal models (Chang et al., 2018; Fang et al., 2019), there are still no effective drugs used in the clinic (Chang et al., 2017). Therefore, improved therapeutic targets that could inhibit both inflammation and apoptosis need to be developed for the treatment of ICH.
Accessing posttranscriptional regulation is important to influence the occurrence and development of diseases, such as ICH (Dykstra-Aiello et al., 2015, 2016; Stamova et al., 2019). Posttranscriptional regulation includes various processes, such as alternative splicing (AS) (Paschalis et al., 2018), alternative polyadenylation (APA) (Tian and Manley, 2017), gene expression regulation (Roundtree et al., 2017), and RNA methylation (Zhao et al., 2017). Among the components of these regulative processes, RNA-binding proteins (RBPs) are critical for posttranscriptional regulation because they can influence the other regulative processes by binding to RNA and because they play vital roles in RNA modification (Hentze et al., 2018). Moreover, posttranscriptional regulation can be controlled via RBPs (Zhang et al., 2015; Fei et al., 2017). Therefore, targeting posttranscriptional regulation via RBPs may alleviate the brain injury caused by ICH, as the progression of ICH has been shown to be closely regulated by posttranscriptional regulation (Dykstra-Aiello et al., 2015, 2016; Stamova et al., 2019).
Although Prdx1, an RBP (Baltz et al., 2012; Castello et al., 2012; Kim et al., 2012; Kwon et al., 2013; Sebestyén et al., 2016), has been shown to be elevated after ICH (Nakaso et al., 2000), its roles in brain injury after ICH are still largely unknown. Prdx1 is a member of the peroxiredoxin family (Wood et al., 2003) and has been shown to be involved in regulating many pathological processes, such as redox reactions (Yamaguchi et al., 1993), inflammatory responses, apoptosis (Liu et al., 2014, 2018; Min et al., 2018; Lu et al., 2019), and tumorigenesis (Hoshino et al., 2005; Cao et al., 2009). These findings suggest that Prdx1 may also play important roles in regulating the above-mentioned pathological processes to influence the brain injury caused by ICH.
In this study, we thus created ICH models and coupled them with in vitro experiments to investigate the role that Prdx1 might play in regulating inflammation and apoptosis by acting as an RBP as well as the underlying mechanism. We found that elevated expression of Prdx1 can significantly reduce inflammation and apoptosis after ICH, and that the protective effects of Prdx1 against brain injury may be related to the binding of inflammation- and apoptosis-related mRNAs, as revealed by RIP-seq technology.
Materials and Methods
Animals
Male Sprague-Dawley (SD) rats (8–10 weeks old, 250–300 g) were purchased from the Animal Center of Daping Hospital, Third Medical Military University, Chongqing, China. All rats were raised in a clean environment and maintained at 25 ± 2°C under a 12 h light/12 h dark cycle and free access to food and water. All procedures and animal experiments were performed in agreement with the Provision and General Recommendation of Chinese Experimental Animals Administration Legislation and the Animal Management Committee of the Third Military Medical University.
ICH Model
The ICH model was established as described in previous studies (Xu et al., 2019). Briefly, autologous blood (70 μL) without anticoagulant or the same volume of saline was injected into the right striatum (0.2 mm anterior and 3.5 mm lateral of bregma at a depth of 5.5 mm) with a syringe pump (KD Scientific, Holliston, MA, United States), at a rate of 10 μL/min. The needle was left in for 10 min after the injection and then slowly withdrawn, and the skin was sutured. The success rate of the model was 90%; failed models and dead rats were excluded from this study.
Construction of the Prdx1 Overexpression Rat Model
The Prdx1-overexpressed adeno-associated virus (AAV2/9-r-Prdx1-3 × flag-mCherry) was named Prdx1-OE-AAV, and vector (AAV2/9-CMV-mCherry) was purchased from Hanbio Biotechnology (Hanbio, China). Five microliters (1.2 × 1012 vg/mL) of Prdx1-OE-AAV or the same volume of vector was injected at 1 μL/min into the right striatum (0.2 mm anterior and 3.5 mm lateral of bregma at a depth of 5.5 mm). Three weeks after the operation, the ICH model was established. The expression effect of the virus was verified by western blot analysis and quantitative real-time PCR (qRT-PCR).
Immunofluorescence Staining
As described in our previous report (Xiong et al., 2016), the brain tissue was fixed with 4% paraformaldehyde. After gradient dehydration, the tissue was embedded and cut into 30 μm thick sections. The sections were permeated at 37°C for 1 h with 0.5% Triton-X-100, blocked with 5% BSA for 2 h, and then incubated with the following primary antibodies overnight at 4°C: anti-NeuN (1:500, Abcam, Cambridge, United Kingdom), anti-GFAP (1:200, Abcam), anti-Iba1(1:200, Abcam) and anti-Prdx1 (1:200, Abcam). The sections were then incubated with fluorescent secondary antibody for 2 h at 37°C. Fluorescent secondary antibodies including Alexa Fluor 647 (1:200, donkey anti-goat), Alexa Fluor 647 (1:200, donkey anti-mouse), and Alexa Fluor 488 (1:200, donkey anti-rabbit), were all obtained from Invitrogen (Carlsbad, CA, United States). Next, 4′,6-diamidino-2-phenylindole (DAPI, 1:3000) was applied for 10 min, and samples were rinsed in PBS. All images were captured using a confocal fluorescence microscope (Leica TCS Sp5, Mannheim, Germany). Number of positive cells were counted using ImageJ software and analyzed in three different arbitrary units that can be defined as the average number of positive cells.
Immunohistochemistry Staining
Following our previous methods (Fang et al., 2014), the brain tissue was fixed with 4% paraformaldehyde, embedded in paraffin and cut into 3.5 μm thick sections. The tissues were dewaxed in xylene, rehydrated in alcohol, placed in 100°C sodium citrate buffer for antigen retrieval for 20 min, and immersed in 3% hydrogen peroxide at 37°C for 10 min to block peroxidase activity. The sections were blocked with 5% BSA for 2 h, and then incubated with the anti-Prdx1 (1:200, Abcam) overnight at 4°C. The sections were then incubated with a rabbit polyclonal secondary antibody (Maravai LifeSciences, San Diego, CA, United States) at 37°C for 2 h, incubated with SABC (Maravai) at 37°C for 1 h, and the color reaction was developed with diaminobenzidine (Zsbio, Beijing, China). All images were captured using a light microscope (BX51, Olympus, Tokyo, Japan). Numbers of positive cells were calculated using ImageJ software and analyzed in three different arbitrary units that can be defined as the average number of positive cells.
Quantitative Real-Time PCR
Using our previously described method to isolate total RNA and perform qRT-PCR (Meng et al., 2017), tissue and cellular RNA were extracted using TRIzol (Invitrogen, Carlsbad, CA) and RNA was reverse transcribed into cDNA (Takara, Dalian, China). Quantitative RT-PCR was performed on an ABI PCR instrument (ABI, CA, United States) using iQ SYBR Green reagent. Primers were synthesized by Shanghai Shenggong Biotechnology Co., Ltd. The primer sequences used in this study are shown in Supplementary Table 1. Relative gene expression levels were determined using the 2–ΔΔCT method.
Western Blot Analyses
As described in our previous report (Wang et al., 2014), protein was extracted and separated on 12% SDS-PAGE (Beyotime Biotechnology, Shanghai, China), and then transferred onto polyvinylidene fluoride membranes (Merck Millipore, Temecula, CA, United States) by electroblotting. The membranes were blocked with 5% BSA for 2 h at room temperature and incubated with the following primary antibodies overnight at 4°C: prdx1 (1:1000, Bosterbio, Pleasanton, CA, United States), Bcl2 (1:1000, Abcam), Bax (1:1000, Abcam), and β-actin (1:1000, Santa Cruz Biotechnology, Dallas, TX, United States). The membranes were washed with TBS-T washing buffer and incubated with HRP-conjugated goat anti-rabbit secondary antibodies (1:10000, Zsbio, Beijing, China) or anti-mouse secondary antibodies (1:10000, Zsbio) at 25°C for 2 h. Bound antibodies were visualized using an enhanced chemiluminescence (ECL) substrate and gray values were evaluated with ImageJ software.
Brain Water Content
We demonstrated in our previous study (Xiong et al., 2016) that the brain water content (BWC) reflects the degree of brain edema. The rats were anesthetized by intraperitoneal injection with pentobarbital, and cerebral tissues were removed, and the hematoma and contralateral brain tissue were preserved by heating to 100°C for 24 h. The BWC was calculated using the formula ((wet weight – dry weight)/wet weight) × 100%.
Nissl and Fluoro-Jade B (FJB) Staining
We performed Nissl staining as previously described (Li Q. et al., 2018). Briefly, the tissues were dewaxed in xylene, rehydrated in alcohol, incubated with Nissl staining solution (Beyotime) at 62°C for 1 h and rinsed in PBS. The procedure also followed our previously reported method (Wang et al., 2018). Tissue was adhered to the slide and rinsed in PBS, soaked in a solution containing 1% sodium hydroxide in 80% alcohol for 5 min, and then transferred to 0.06% potassium permanganate for 10 min. The samples were incubated with Fluoro-Jade B (FJB) dye (Chemicon International, Temecula, CA, United States) at 37°C for 30 min. All images were captured using a light microscope (BX51, Olympus, Japan). The number of positive cells were calculated by ImageJ software and analyzed in three different arbitrary units that can be defined as the average number of positive cells.
RNA Immunoprecipitation, High-Throughput Sequencing (RIP-seq), and Data Analysis
The cell sample was suspended in a cell culture dish with precooled PBS, and the dish was irradiated with a UV cross-linker. Collected cells were added to the lysate at a ratio of 1:10 to lyse and digest the DNA. The lysate was incubated with the RIP antibodies overnight at 4°C for immunoprecipitation. The magnetic beads were resuspended with the antigen-antibody complex solution for 1 h, and the magnetic beads resuspended in MNase were placed on a hot mixer for 10 min and eluted. After removal of MNase, the sample was dephosphorylated with FastAP enzyme, treated with T4 PNK enzyme, and then treated with proteinase K to digest protein, and the RNA was extracted. The Illumina ScriptSeqTM v2 RNA-Seq Library Preparation Kit (Li et al., 2017; Song et al., 2018) was used for the library construction and sequencing. The quality of the RIP library was judged according to the mapping result. Quality-qualified data were entered for the downstream analysis to obtain valid reads for the genomic location distribution, peak calling, and motif analysis, which revealed the type and pattern of mRNA and ncRNA that Prdx1 binds at the genome-wide level.
Construction of cDNA Library and Data Analysis
The DNA was digested by RQ1 DNase (Promega) enzyme in total RNA. A total of 5 μg of total RNA was taken for the polyA library construction. The PCR product with a fragment size of 300–500 bp was selected for sequencing. The results of the library were sequenced using an Illumina HiSeq X Ten sequencing platform for sequencing at 150 bp. The systematic evaluation of the data included the extraction and quality assessment of effective reads, the length distribution of effective reads, sequencing saturation statistics, and base content statistics. Differentially expressed genes (DEGs) were identified in different samples using the edgeR software system (Pan et al., 2017; Sun et al., 2017) and submitted to a DAVID function GO analysis.
Survival Analysis
Following methods described in the literature (Meng et al., 2017), the number of dead rats was counted once every hour after ICH. To calculate the survival rate, the following equation was used: (number of ICH rats per group – number of dead rats per group)/number of ICH rats per group. A Kaplan-Meier survival analysis was then conducted to evaluate survival rates.
Flow Cytometry
An Annexin V/Dead Cell kit was purchased from BD Biosciences (San Jose, CA, United States), and apoptosis staining was performed according to the manufacturer’s instruction. The cells were divided into normal cells, early apoptosis cells, late apoptosis cells, and dead cells using flow cytometry, and the proportion of the four types of cells was calculated.
Hematoma Measurement
According to our previous methods (Meng et al., 2017), the rats were sacrificed at 3 d after ICH, and brain tissues were taken and frozen at −20°C for 30 min. Coronal sections of the brain were made with a thickness of 1mm. The area of hematoma in each section was determined using Image-Pro Plus 5.0 image processing software (Media Cybernetics, Bethesda, MD, United States) and the hematoma volume was measured according to the formula V = t × (A1 + A2 + ⋯ + An), where V is the hematoma volume, t is the slice thickness, and A is the hematoma area.
Modified Neurological Severity Score
The modified neurological severity score (mNSS) was calculated according to previous methods (Li et al., 2019), and the detailed scoring rules are shown in Supplementary Table 2. The score is based on an evaluation of the neurological defects, according to aspects of movement, sensation, balance, etc. Higher scores indicate more obvious neurological defects. The score in the current study were determined by three experienced laboratory researchers who were unaware of the group assignments of the rat. The final score for each rat was the average of all scores for that rat.
Primary Astrocyte Sorting
SD rats aged 8–12 weeks were anesthetized with 1% pentobarbital and perfused with pre-cooled PBS. The brain was isolated, then the brain tissue was cut into pieces of 1–3 mm with surgical scissors, and 4 mg/mL Papain(Worthington, Lakewood, United States) was added to digest the brain tissues at 37°C for 1 h; the tissues were slightly shaken every 5 min, followed by repeated pipetting of the cell suspension; then, the supernatant was used for centrifugation and cells were deposited at the bottom of the tube, using Alexa Fluor 488 GFAP antibody (1:20, BD Biosciences) combined with flow sorting to separate astrocytes for RIP.
Experimental Design
First, 67 rats were randomly divided into two groups: a sham group and an ICH group, 20 rats of each group were used to calculate mortality and sacrificed at 5 days after ICH. 12 ICH rats were sacrificed at 12, 24, 72, and 120 h, respectively (n = 3 each timepoint), and RNA were extracted to detect the Prdx1 mRNA level. Three rats in each group sacrificed at 72 h for immunofluorescence and immunohistochemistry staining, and three rats in each group sacrificed at 72 h for western blotting (Figure 1A). Second, 132 rats were randomly divided into four groups (n = 43 each group): a sham group and three ICH groups (WT, Vector, Prdx1-OE); three rats of each group were used for qRT-PCR and western blotting, three rats of each group were used for FJB and Nissl staining, six rats of each group were used to detect BWC, five rats of each group were used for functional assessments, and six rats of each group were used to calculate hematoma volume, these rats all sacrificed at 3 days after ICH. Twenty rats of each group were used to calculate mortality and sacrificed at 5 days after ICH (Figure 1A and Supplementary Figures S3A,B). Finally, for the in vitro experiments, Prdx1-overexpressing plasmids were transfected into HeLa cells, Prdx1-overexpressing HeLa cell and the cells from control group were collected for RIP-seq, RNA-seq and flow cytometry analysis. The primary astrocytes were extracted and subjected to RIP-qRT-PCR, to verify the RIP-seq data (Figure 1B).
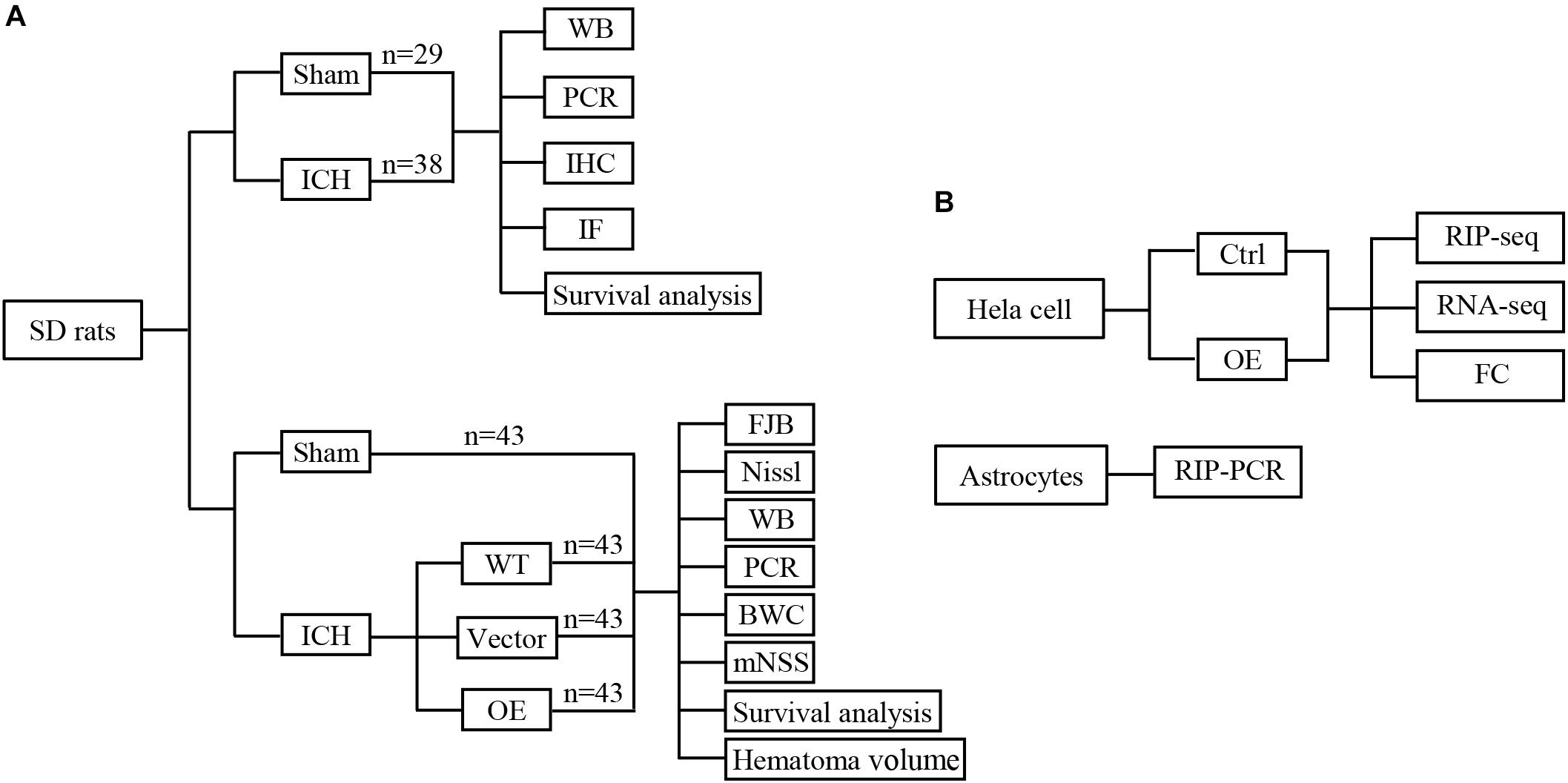
Figure 1. Conceptual illustrations of the experimental design. Flow diagram of in vivo (A) and in vitro (B) experiments.
Statistical Analysis
All data are expressed as the mean ± SEM or percentage. Student’s t-tests were used to evaluate the differences between independent samples, and comparisons among multiple groups were examined using one-way ANOVA. Two-way ANOVA was used to evaluate the differences in BWC between groups. A Kaplan Meier survival analysis was applied to evaluate survival rates. Differences were considered to be significant at P < 0.05.
Results
Prdx1 Expression Was Significantly Upregulated After ICH
Before investigating the role of Prdx1 in ICH-induced brain injury, we first detected its expression profiles in the perihematomal tissue of ICH rat models. Our results indicated that Prdx1 mRNA level increased at 24 h, peaked at 72 h and then gradually declined (Figure 2A). Because Prdx1 expression peaked at 72h, and at same timepoint, there was a statistical difference in mortality between the two groups (Figure 2B), so we chose this timepoint for the current experiments. Using western blotting, we found that Prdx1 protein levels (Figure 2C) in model rats were significantly increased compared to the level in the sham group at 3 days after ICH. Next, we performed immunohistochemical and immunofluorescence staining, and found that in the striatum, the number of Prdx1-positive cells was significantly higher in the model rats than in the sham group (Figure 2D and Supplementary Figures S1A,B), and in the cortex, there was no significant difference in two groups (Supplementary Figures S1C,D). Furthermore, immunofluorescence staining was used to explore the cellular resource of Prdx1 in normal brain tissue and perihematomal brain tissues in the striatum (Supplementary Figure S2B), and we found that Prdx1 was mainly colocalized with GFAP+ astrocytes and Iba1+ microglia (Figure 2E and Supplementary Figure S2A). Together, these results suggest that enhanced Prdx1 expression is mainly derived from astrocytes and microglia in the striatum and may be involved in the regulation of brain injury after ICH.
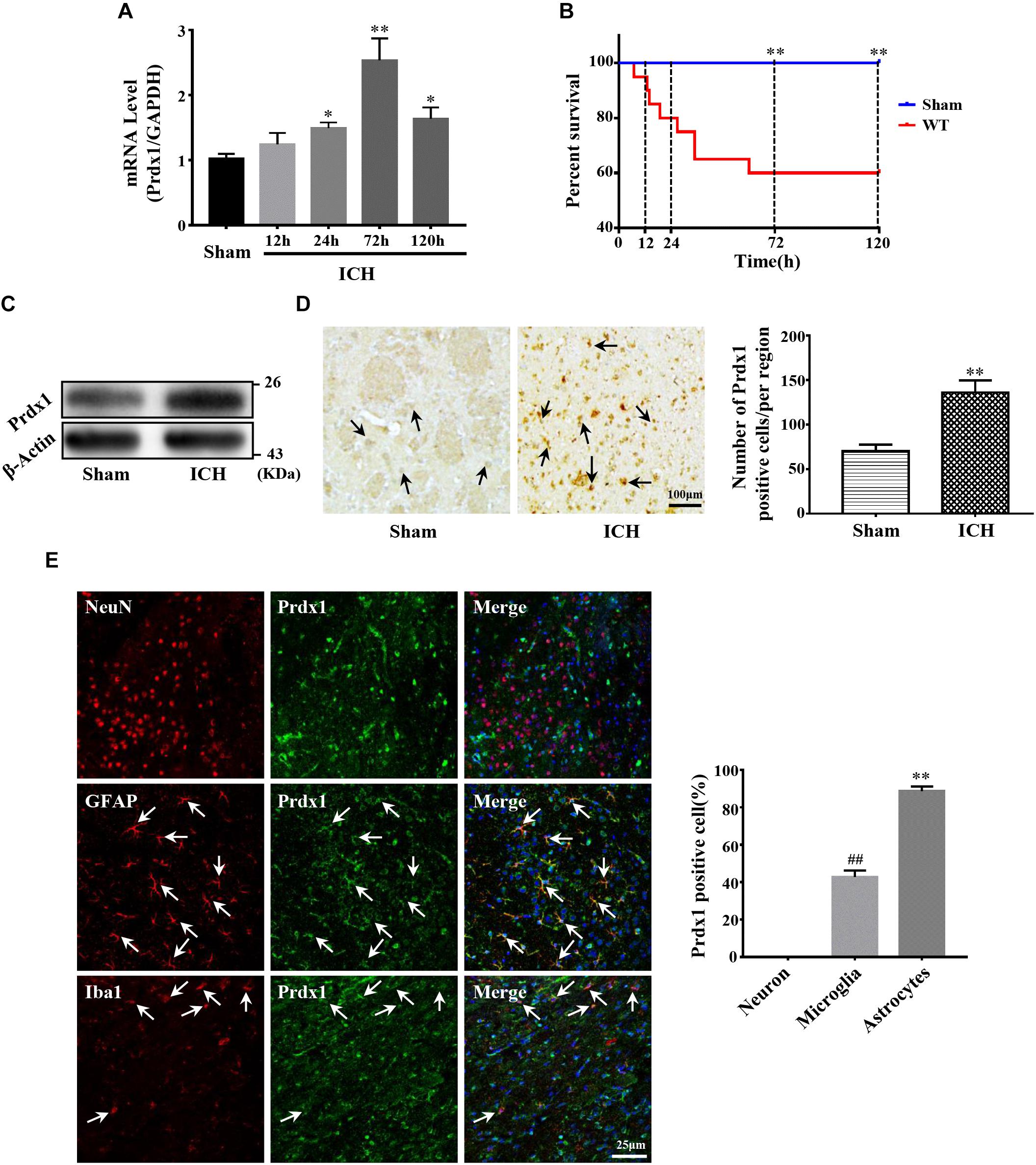
Figure 2. Prdx1 expression was significantly increased in rats after ICH. (A) Prdx1 expression in the perihematomal area was detected by qRT-PCR (df = 4, F = 35.945, ∗P < 0.05 versus sham, ∗∗P < 0.01 versus sham, n = 3). (B) Survival statistics of the sham group and WT group (χ2 = 10.457, df = 1, ∗∗P = 0.01, n = 20). (C) Prdx1 expression in the perihematomal area was detected by western blot analysis (F = 2.014, t = –3.432, ∗∗P < 0.01 versus sham, n = 3). (D) Prdx1 expression in perihematomal tissue was detected by immunohistochemistry (scale bars, 100 μm, F = 1.225, t = –7.288, ∗∗P < 0.01 versus sham, n = 3). (E) Prdx1 expression in perihematomal tissue was determined by immunofluorescence, and the percentage of positive Prdx1/NeuN, Prdx1/Ibα-1 and Prdx1/GFAP in three randomly chosen fields within the perihematomal area was counted (scale bars, 25 μm, F = 10.964, t = –11.790, ##P < 0.01 versus Prdx1/NeuN. F = 12.000, t = –60.228, ∗∗P < 0.01 versus Prdx1/NeuN, n = 3).
Prdx1 Suppressed ICH-Induced Apoptosis and Inflammation
Next, we investigated the role of Prdx1 in brain injury after ICH. Prdx1-overexpressing AAV or empty vector was injected in rats 3 weeks before ICH surgery (Supplementary Figure S3E), and the overexpression of Prdx1 in the brain was confirmed by qRT-PCR (Supplementary Figure S3C) and western blot analysis (Supplementary Figure S3D). First, we found that the mortality rate (Figure 3A) and BWC (Figure 3B) were significantly decrease in the Prdx1-OE rats compared to the WT and Vector rats after ICH. The hematoma volumes (Figure 3C) and mNSS (Figure 3D) were also significantly lower in the Prdx1-OE rats. Then, we performed Nissl staining to detect the surviving cells around the hematoma and found that the number of Nissl-positive cells in perihematomal brain tissue was higher in the Prdx1-OE group than in the WT and Vector groups (Figure 4A); in contrast, the number of damaged cells detected by FJB staining was significantly decreased in Prdx1-OE rats (Figure 4B). Furthermore, we used western blotting to investigate the expression levels of the apoptosis-related proteins Bax and Bcl2 and found that Bcl2/Bax was significantly increased in Prdx1-OE ICH rats compared with the WT and Vector groups (Figure 4C). We also assessed inflammatory factors in perihematomal brain tissue after ICH, and our results show that mRNA levels of TNF-α, IL-10, and IL-6 were markedly decreased in the Prdx1-OE group in contrast to the levels in the WT and Vector groups (Figure 4D). Together, these data indicate that Prdx1 can alleviate acute brain injury after ICH by reducing inflammation and apoptosis.
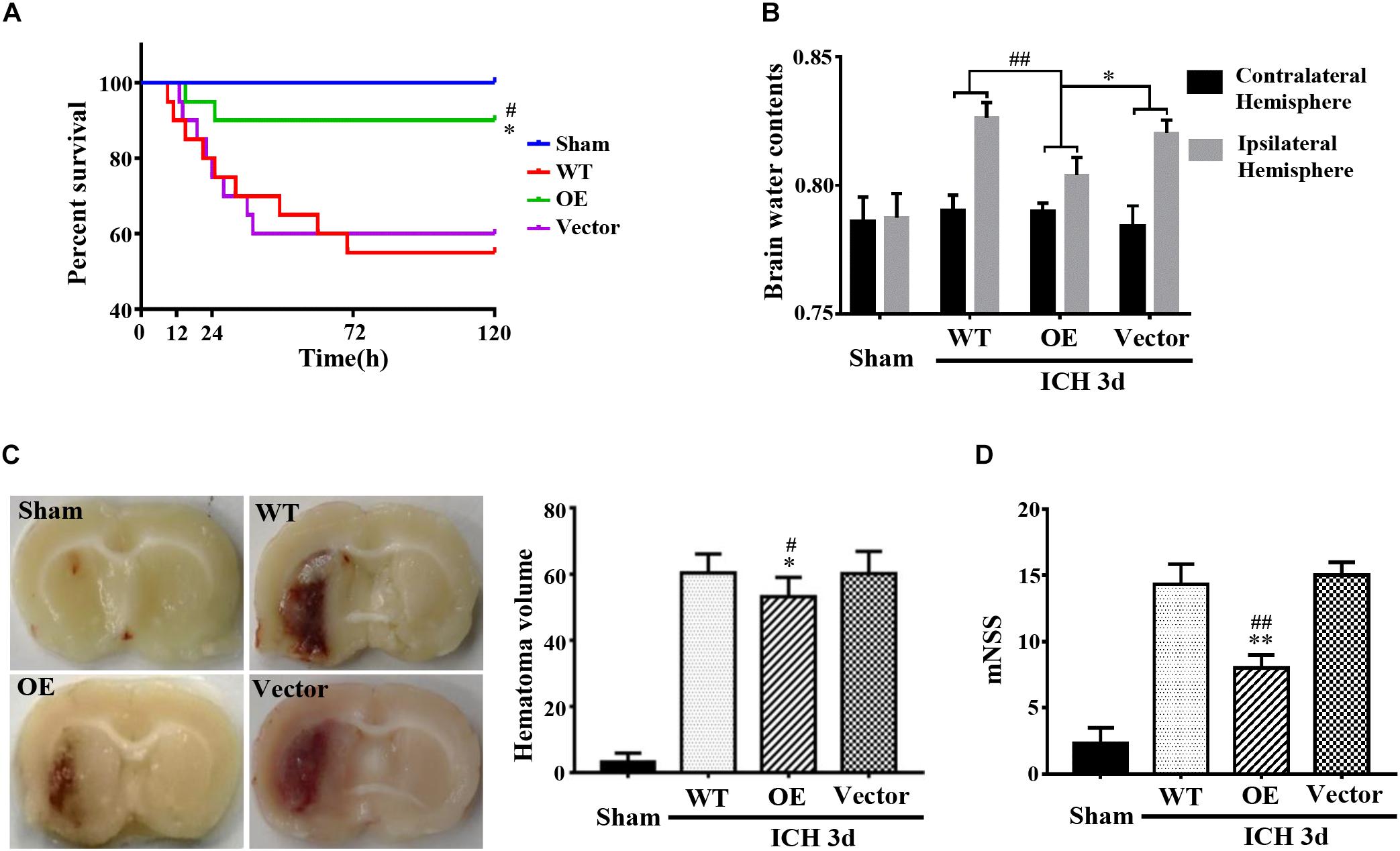
Figure 3. Prdx1 overexpression alleviated the symptoms of rats after ICH. (A) Survival statistics of the sham group, WT group, Vector group, and Prdx1-OE group (χ2 = 14.310, df = 3, ∗P < 0.05 versus Vector, #P < 0.05 versus WT, n = 20). (B) Brain water content of the four groups (df = 3, F = 9.324, ∗P < 0.05 versus Vector, ##P < 0.01 versus WT, n = 6). (C) Hematoma volume of the sham group, the WT group, the Vector group, and the Prdx1-OE group (df = 3, F = 151.467, ∗P < 0.05 versus Vector, #P < 0.05 versus WT, n = 6). (D) The mNSS was determined for four groups 3 days after ICH (df = 3, F = 75.196, ∗∗P < 0.01 versus Vector, ##P < 0.01 versus WT, n = 5).
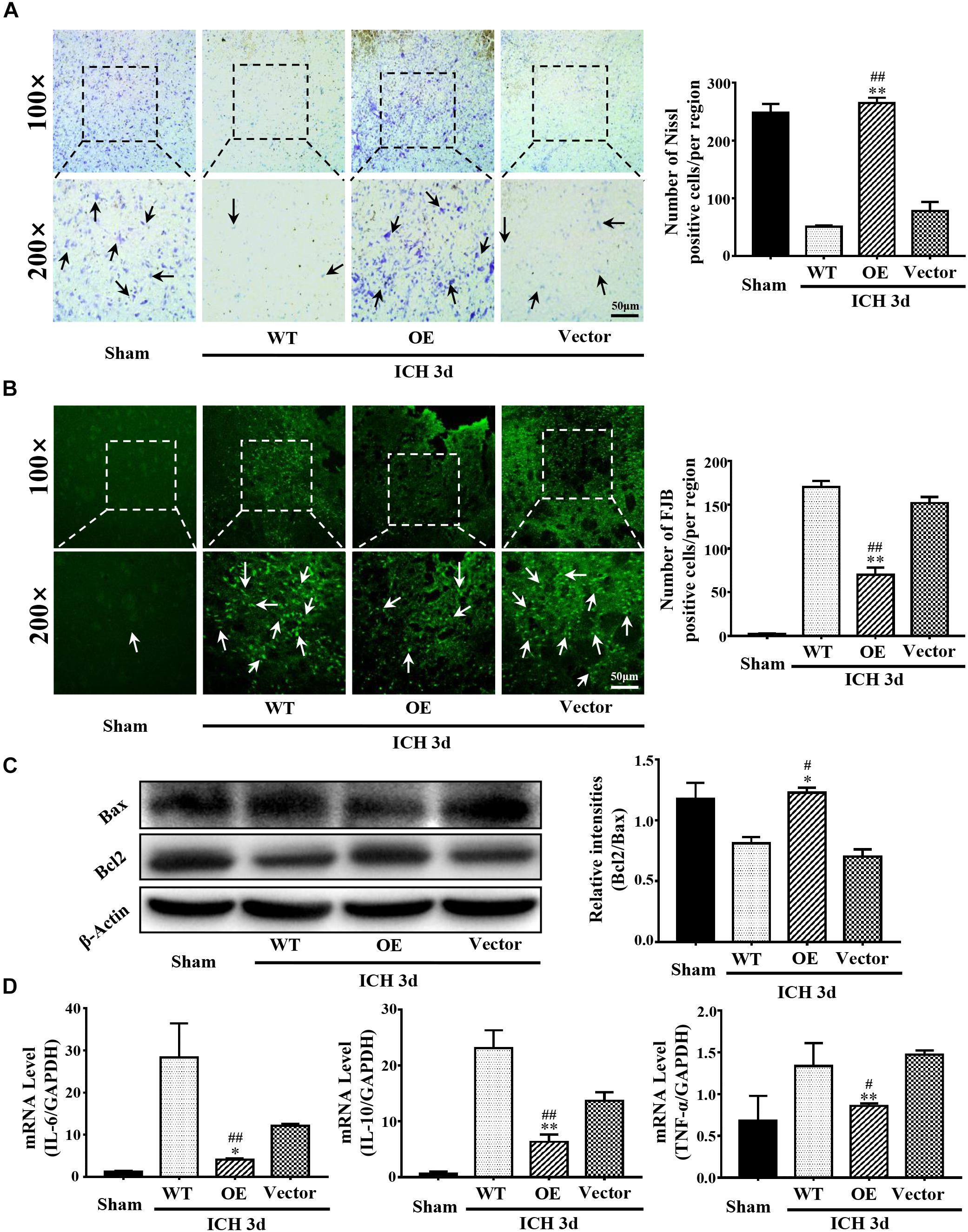
Figure 4. Prdx1 overexpression inhibited inflammation and apoptosis after ICH. (A) Representative Nissl staining in sham, WT, Prdx1-OE and Vector rats. The number of Nissl-positive cells was assessed (scale bars, 50 μm, df = 3, F = 174.206, ∗∗P < 0.01 versus Vector; ##P < 0.01 versus WT, n = 3). (B) Representative FJB staining in sham, WT, Prdx1-OE, and Vector rats. The number of FJB-positive cells was assessed (scale bars, 50 μm, df = 3, F = 435.050, ∗∗P < 0.01 versus Vector; ##P < 0.01 versus WT, n = 3). (C) Western blotting was used to detect Bcl2 and Bax expression in the four groups (df = 3, F = 32.759, ∗P < 0.05 versus Vector; #P < 0.05 versus WT, n = 3). (D) Comparison of the expression of inflammatory factors in the four groups using qRT-PCR: IL-6 (df = 3, F = 27.046, ∗P < 0.05 versus Vector; ##P < 0.01 versus WT, n = 3), IL-10 (df = 3, F = 79.041, ∗∗P < 0.01 versus Vector, ##P < 0.01 versus WT, n = 3), TNF-α (df = 3, F = 10.274, ∗∗P < 0.01 versus Vector; #P < 0.05 versus WT, n = 3).
We also injected shPrdx1 AAV into rats to generate Prdx1-knockdown rats. However, most Prdx1-knockdown rats died within 3 days after ICH surgery (data not shown), and the related experiments in Prdx1-knockdown ICH rats are difficult to perform. This also illustrates the protective effect of Prdx1 in brain injury after ICH.
Characterization of Prdx1 Binding to Target RNA in vitro
To further delineate the underlying mechanism of Prdx1 in reducing inflammation and apoptosis, we used RNA immunoprecipitation coupled with next-generation sequencing (RIP-seq) to identify all RNAs that interact with Prdx1. We chose HeLa cells for these experiments for the following reasons: studies have shown that HeLa cells are model cells for studying RBPs (D’Amico et al., 2019; Loughlin et al., 2019; Wilbertz et al., 2019) and HeLa cells are good for gene regulation studies in the central nervous system (Chu et al., 2013). In addition, Prdx1 also shows anti-apoptotic and anti-inflammatory effects in HeLa cells (Nassour et al., 2016) (Supplementary Figure S4A), suggesting that Prdx1 may play a similar role in the central nervous system. First, we tested the consistency of two biological replicates of Prdx1 RIP-seq data and found 10466 consensus genes, showing that our results were reproducible (Figure 5A,B and Supplementary Data 1). Next, we analyzed the regions of the Prdx1-bound RNAs. By comparison with the whole genome, we found the regions where the Prdx1-bound RNAs were mainly concentrated in the coding sequence (CDS) region, the 5′ UTR and the 3′ UTR (Figures 5C,E). Furthermore, de novo motif analysis showed that the Prdx1-bound motif was mainly in the AG-enriched region (Figure 5D), and GO Ontology (GO) analysis of Prdx1-bound RNAs revealed significantly enriched terms in RNA/mRNA processing and splicing (Supplementary Figure S4B), which is consistent with our prediction. Our Prdx1 RIP-seq results revealed that Prdx1 bound to mRNA transcripts was associated with the inflammatory response (BCL6 and TLR6) and the apoptotic process (PTEN and FOS) (Figure 5F), consistent with the RIP-qPCR result for these mRNAs in astrocytes (Figure 5G), suggesting that Prdx1 plays a role in inflammation and apoptosis.
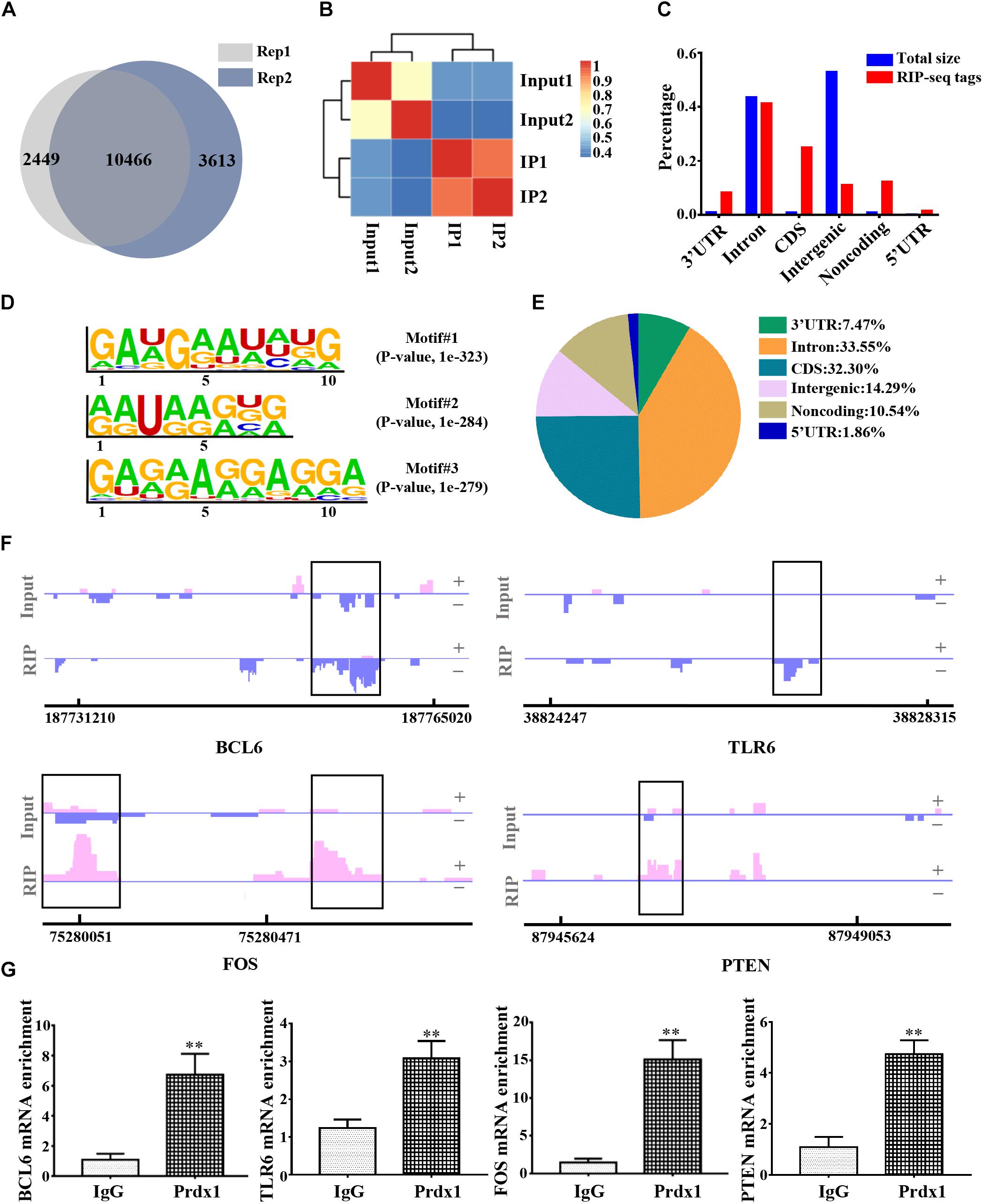
Figure 5. The genome-wide landscape of Prdx1 binding sites on RNA. (A) Venn diagram of Prdx1 RIP-seq genes from two biological replicates. (B) Heat map showing the correlation coefficient of two biological replicates. (C) The Prdx1 RIP-seq peaks are predominantly enriched in the CDS region, the 3′ UTR and the 5′ UTR. All RIP-seq peaks were classified according to their distribution on the RNA elements and compared to the genomic background. (D) De novo motif analysis identified GA repeat and GA-enriched sequences as Prdx1 binding motifs. (E) Prdx1 RIP-seq peak distribution proportion. (F) Prdx1 RIP-seq peaks are shown as track signals. The peak area is indicated by the black frame. (G) RIP-qPCR analysis of Prdx1 binding RNAs, IgG RIP was negative RIP control (BCL6: F = 3.745, t = –6.708, ∗∗P < 0.01 versus IgG, n = 3; TLR6: F = 1.668, t = –7.065, ∗∗P < 0.01 versus IgG, n = 3; FOS: F = 5.224, t = –10.432, ∗∗P < 0.01 versus IgG, n = 3; PTEN: F = 0.000, t = –6.998, ∗∗P < 0.01 versus IgG, n = 3).
Prdx1 Affects Inflammation- and Apoptosis-Related mRNA Stability
Prdx1 mainly binds to the 3′ UTR, CDS, and 5′ UTR of the RNAs, indicating that its function may be related to the stability of RNA and AS (Mayr, 2016; Yee et al., 2019). To obtain a comprehensive view of Prdx1-dependent DEGs, control or Prdx1-overexpressing vector was transfected into HeLa cells (Figure 6A and Supplementary Figure S5A) and RNA-seq was performed. The RNA-seq data showed good reproducibility (R = 0.994, Figure 6B). A total of 863 Prdx1-dependent DEGs were identified; among them, 471 were upregulated genes, and 392 were downregulated genes (Figure 6C, Supplementary Figure S5B, and Supplementary Data 2). Next, we performed GO analysis on these DEGs and found that these genes were mainly associated with redox reactions, cytokine pathways, and inflammatory responses, as well as with apoptosis and DNA repair (Figure 6D). Further, we screened for genes involved in inflammation and apoptosis in these DEGs (Figure 6E and Supplementary Data 3), verified their changing profiles using qRT-PCR, and found similar trends of these mRNAs in ICH rats when Prdx1 overexpression was compared with RNA-seq data in HeLa cells (Figure 6F and Supplementary Figure S5C).
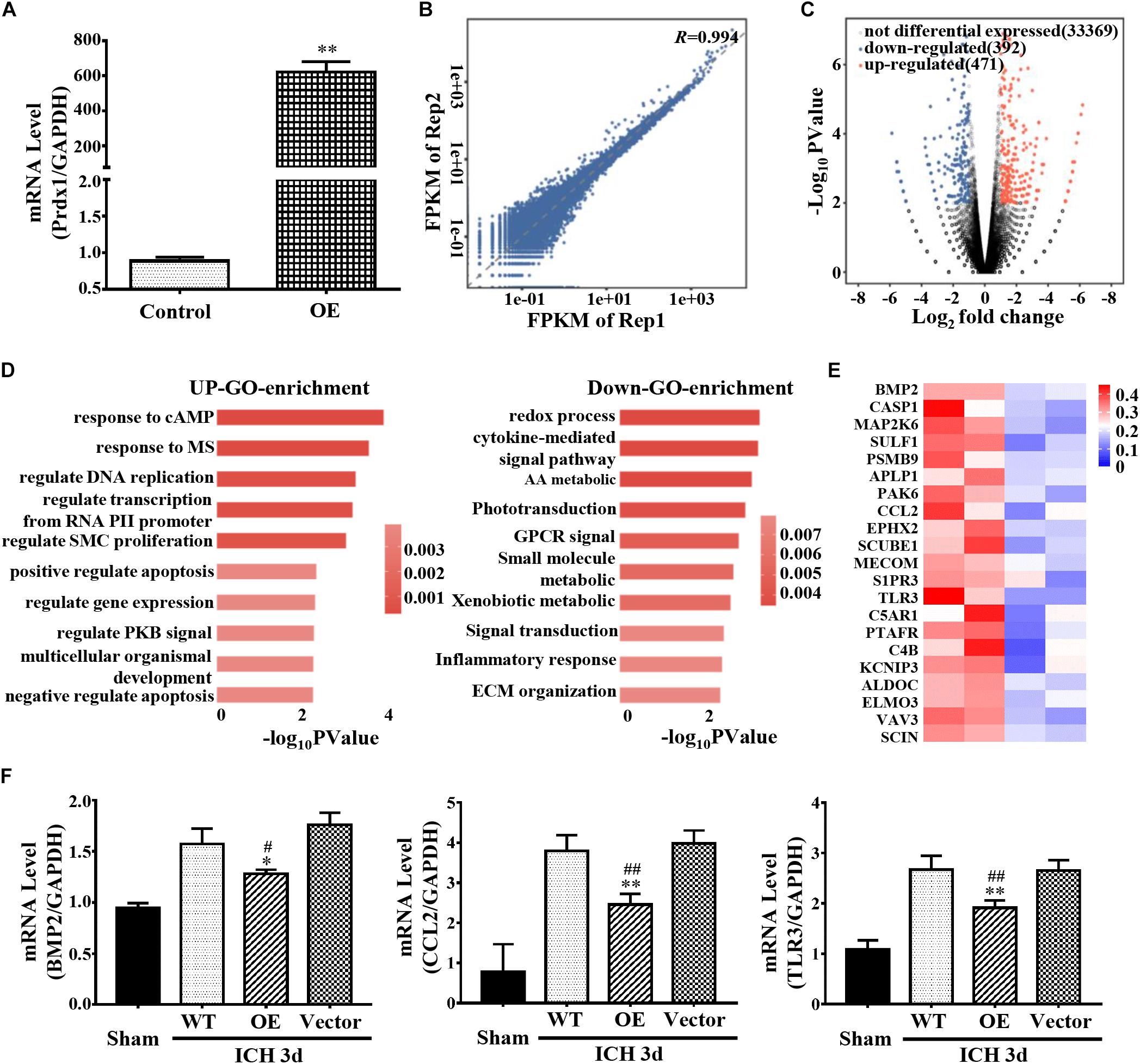
Figure 6. Prdx1 affects inflammation- and apoptosis-related mRNA stability. (A) Prdx1 expression in control HeLa cells and in the Prdx1-OE group was detected by qRT-PCR (F = 8.494, t = 40.635, ∗∗P < 0.01 versus Control). (B) Scatter plots and correlation coefficients of two biological replicates of RNA-seq. (C) Volcano map: red dot indicates a gene that is upregulated after Prdx1 overexpression, black dot indicates a gene with no significant change, and blue dot indicates a downregulated gene. (D) Gene ontology (GO) analysis of Prdx1-dependent DEGs. Significantly enriched GO terms of genes. The x-axis indicates the enrichment P-value on a −log10 scale; the y-axis indicates terms. (E) Heat map showing that apoptosis- and inflammation-related genes were significantly decreased after Prdx1 was upregulated in HeLa cells. (F) qRT-PCR was performed in four groups 3 days after ICH (BMP2: F = 8.949, df = 3, ∗P < 0.05 versus Vector, #P < 0.05 versus WT, n = 3; CCL2: F = 33.103, df = 3, ∗∗P < 0.01 versus Vector, ##P < 0.01 versus WT, n = 3; TLR3: F = 39.330, df = 3, ∗∗P < 0.01 versus Vector, ##P < 0.01 versus WT, n = 3).
Coupling RIP-seq and RNA-seq, we found that Prdx1 can combine with inflammation- and apoptosis-related RNAs, such as ANGPTL4, GADD45A, and THBS1, and cause differential expression of these mRNAs in HeLa cells (Figure 7A and Supplementary Data 2). We validated these results in astrocytes and ICH models, and found that Prdx1 also combined with these mRNAs in astrocytes (Figure 7B); similar trends of these mRNA levels were confirmed in the rat ICH model (Figure 7C).
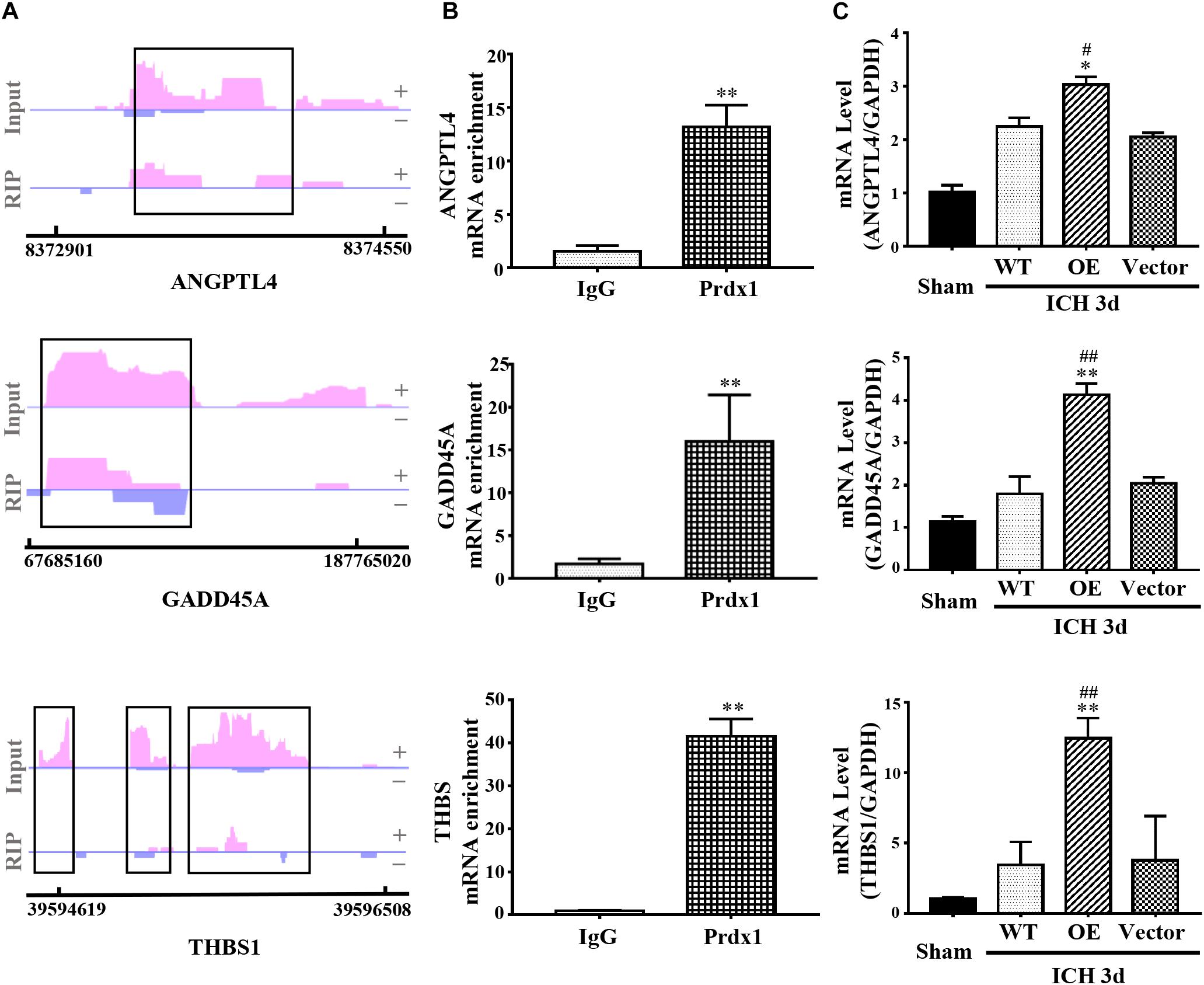
Figure 7. Prdx1 plays similar roles in vitro and in vivo. (A) Prdx1 RIP-seq peaks are shown as track signals of ANGPTL4, GADD45A and THBS1. The peak area is indicated by the black frame. (B) RIP-qPCR analysis of Prdx1 binding RNAs in astrocytes with IgG RIP as a negative RIP control (ANGPTL4: F = 4.297, t = –13.551, ∗∗P < 0.01 versus IgG, n = 3; GADD45A: F = 12.277, t = –8.909, ∗∗P < 0.01 versus IgG, n = 3; THBS: F = 4.072, t = –20.619, ∗∗P < 0.01 versus IgG, n = 3). (C) qRT-PCR was performed in four groups of ANGPTL4, GADD45A, and THBS1 mRNA (ANGPTL4: F = 87.049, df = 3, ∗P < 0.05 versus Vector, #P < 0.05 versus WT, n = 3; GADD45A: F = 73.252, df = 3, ∗∗P < 0.01 versus Vector, ##P < 0.01 versus WT, n = 3; THBS: F = 20.553, df = 3, ∗∗P < 0.01 versus Vector, ##P < 0.01 versus WT, n = 3).
Discussion
RNA-binding protein-mediated posttranscriptional regulation plays an important role in the pathophysiological processes of diseases (Wurth et al., 2016; Bar-Ziv et al., 2019; Baser et al., 2019). Recent studies have shown that posttranscriptional regulation is also associated with ICH (Dykstra-Aiello et al., 2015; Stamova et al., 2019). However, what roles RBPs play in ICH is still largely unknown. In this study, we found that Prdx1, an RBP, was significantly increased in astrocytes and microglia in response to brain injury after ICH, that is regulated inflammation- and apoptosis-related mRNA stability, and that is reduced inflammation- and apoptosis-related molecular production and release, thereby inhibiting the inflammatory response and reducing apoptosis after ICH, suggesting that Prdx1 could serve as an improved therapeutic target to inhibit both inflammation and apoptosis after ICH.
In this study, we first found that Prdx1 expression in perihematomal brain tissues was markedly increased, which in line with previous studies (Nakaso et al., 2000; Liu et al., 2014). Immunofluorescent staining revealed that elevated Prdx1 was mainly expressed in astrocytes and microglia but not in neurons in the striatum of ICH rats. Interestingly, we also found that Prdx1 colocalized with NeuN+ neurons in the cortex (Supplementary Figure S6), differential cellular localization of Prdx1 indicate that Prdx1 expression in the CNS may exert functional diversity. In addition, our results show that Prdx1 can reduce the hematoma volume after ICH, its mechanism needs further study, we speculate that Prdx1 inhibits the release of inflammatory factors such as TNF-α, thereby promoting the expression of CD36 and causing the absorption of hematomas.
While previous studies investigated the roles of Prdx1 in other diseases, our study is the first to demonstrate that Prdx1 can reduce inflammation and apoptosis after ICH. Liu et al. showed that Prdx1 exacerbate brain injury after ICH (Liu et al., 2016). However, the study did not use Prdx1 overexpression or interference models to verify the effect of Prdx1 after ICH, but only inferred its role in ICH based on the function of Prdx1 in macrophages. In fact, Prdx1 performs different and even opposite functions in different cell types, thus the study failing to fully elucidate the role and mechanisms of Prdx1 in ICH. In our study, we first overexpressed Prdx1 in rat brain and established ICH model, found that the symptoms of Prdx1-OE group was significantly alleviated compare with WT or Vector group, our study provided direct evidence of its role in ICH.
However, the roles of Prdx1 in regulating inflammation and apoptosis are currently controversial. For example, Liu et al. reported that Prdx1 can promote the release of inflammatory mediators by activating macrophages (Liu et al., 2014; He et al., 2019), and Riddell et al. declare Prdx1 could activate the TLR4/MyD88 signaling pathway (Riddell et al., 2010, 2012), thereby enhancing inflammation and apoptosis. However, Lu et al. showed that Prdx1 reduces the inflammatory response by inhibiting the activation of NF-κB and oxidative stress (Min et al., 2018; Lu et al., 2019), and studies have also shown that Prdx1 inhibits the activity of the ubiquitin ligase TRAF6 (Min et al., 2018), thereby producing an anti-inflammatory effect. Our data suggests that Prdx1 inhibits inflammation and apoptosis by regulating the posttranscriptional regulation of inflammation- and apoptosis-related mRNAs, and thus provides new evidence and insights into the anti-inflammatory effects of Prdx1.
Kim et al. (2012) first reported that Prdx1 can act as an RBP to bind RNAs, which has also been demonstrated in a number of recent studies (Baltz et al., 2012; Castello et al., 2012; Kwon et al., 2013; Sebestyén et al., 2016). However, it is still not clear what kinds of RNAs Prdx1 can combine with and what the effects of such couplings are. In the current study, we first investigated the properties of Prdx1 as an RBP from a genome-wide perspective and found that Prdx1 can regulate mRNA stability because Prdx1 is mainly combined with the CDS region, the 3′ UTR and 5′ UTR in AG-enriched RNA motifs. For example, we found that Prdx1 can combine with ANGPTL4, GADD45A, and THBS1 mRNAs, and it upregulated these mRNAs when Prdx1 was overexpressed. Previous studies showed that ANGPTL4 can significantly inhibit the inflammatory response (Guo et al., 2015) and alleviate neurological deficits and cerebral edema after ICH (Qiu et al., 2018). GADD45A is a DNA damage-inducing protein and has been shown to inhibit oxidative stress and inflammatory responses (Tanaka et al., 2017; Li F. H. et al., 2018). THBS1 is also an anti-inflammatory molecule (Cursiefen et al., 2011) and accelerates synaptogenesis (Xu et al., 2010). These earlier findings, together with our current results, demonstrate the anti-inflammatory and anti-apoptotic effects of Prdx1 and suggest Prdx1 as an improved therapeutic target to inhibit both inflammation and apoptosis after ICH.
Data Availability Statement
The datasets generated for this study can be found in the GEO, GSE134505.
Ethics Statement
The animal study was reviewed and approved by the Animal Management Committee of the Third Military Medical University.
Author Contributions
Q-WY and X-YX designed the research presented in this manuscript. G-QY and J-CH conducted the cell cultures and qRT-PCR as well as the histological and behavioral analyses. J-JY, C-XG, and QZ performed the western blot and the RIP-seq. QX, L-XX, and RC built the ICH rat model. Z-MQ, KZ, and G-HJ performed the RNA-seq. RX and QC conducted the statistical analysis.
Funding
This work was supported by the “13th Five-Year” National Key R&D Program (2016YFC1300505) and the National Natural Science Fund for Distinguished Young Scholars (81525008).
Conflict of Interest
The authors declare that this study received advice from the Wuhan Life Beauty Technology Co., Ltd. The company was not involved in the study design, collection, analysis, interpretation of data, the writing of this article or the decision to submit it for publication.
Acknowledgments
We would like to thank Wuhan Life Beauty Technology Co., Ltd. for advice and suggestions on this work.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnins.2020.00181/full#supplementary-material
References
Baltz, A. G., Munschauer, M., Schwanhäusser, B., Vasile, A., Murakawa, Y., Schueler, M., et al. (2012). The mRNA-bound proteome and its global occupancy profile on protein-coding transcripts. Mol. Cell 46, 674–690. doi: 10.1016/j.molcel.2012.05.021
Bar-Ziv, R., Nguyen, N. T., and Dillin, A. (2019). Vive ut Numquam Moriturus: tweaking translational control to regulate longevity. Mol. Cell 73, 643–644. doi: 10.1016/j.molcel.2019.02.006
Baser, A., Skabkin, M., Kleber, S., Dang, Y., Gülcüler Balta, G. S., Kalamakis, G., et al. (2019). Onset of differentiation is post-transcriptionally controlled in adult neural stem cells. Nature 566, 100–104. doi: 10.1038/s41586-019-0888-x
Cao, J., Schulte, J., Knight, A., Leslie, N. R., Zagozdzon, A., Bronson, R., et al. (2009). Prdx1 inhibits tumorigenesis via regulating PTEN/AKT activity. EMBO J. 28, 1505–1517. doi: 10.1038/emboj.2009.101
Castello, A., Fischer, B., Eichelbaum, K., Horos, R., Beckmann, B. M., Strein, C., et al. (2012). Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell 149, 1393–1406. doi: 10.1016/j.cell.2012.04.031
Chang, C. F., Goods, B. A., Askenase, M. H., Hammond, M. D., Renfroe, S. C., Steinschneider, A. F., et al. (2018). Erythrocyte efferocytosis modulates macrophages towards recovery after intracerebral hemorrhage. J. Clin. Invest. 128, 607–624. doi: 10.1172/JCI95612
Chang, J. J., Kim-Tenser, M., Emanuel, B. A., Jones, G. M., Chapple, K., Alikhani, A., et al. (2017). Minocycline and matrix metalloproteinase inhibition in acute intracerebral hemorrhage: a pilot study. Eur. J. Neurol. 24, 1384–1391. doi: 10.1111/ene.13403
Chu, C. T., Ji, J., Dagda, R. K., Jiang, J. F., Tyurina, Y. Y., Kapralov, A. A., et al. (2013). Cardiolipin externalization to the outer mitochondrial membrane acts as an elimination signal for mitophagy in neuronal cells. Nat. Cell Biol. 15, 1197–1205. doi: 10.1038/ncb2837
Cursiefen, C., Maruyama, K., Bock, F., Saban, D., Sadrai, Z., Lawler, J., et al. (2011). Thrombospondin 1 inhibits inflammatory lymphangiogenesis by CD36 ligation on monocytes. J. Exp. Med. 208, 1083–1092. doi: 10.1084/jem.20092277
D’Amico, D., Mottis, A., Potenza, F., Sorrentino, V., Li, H., Romani, M., et al. (2019). The RNA-binding protein PUM2 impairs mitochondrial dynamics and mitophagy during aging. Mol. Cell 73, 775–787.e10. doi: 10.1016/j.molcel.2018.11.034
Dykstra-Aiello, C., Jickling, G. C., Ander, B. P., Shroff, N., Zhan, X., Liu, D., et al. (2016). Altered expression of long noncoding RNAs in blood after ischemic stroke and proximity to putative stroke risk loci. Stroke 47, 2896–2903. doi: 10.1161/STROKEAHA.116.013869
Dykstra-Aiello, C., Jickling, G. C., Ander, B. P., Zhan, X., Liu, D. Z., Hull, H., et al. (2015). Intracerebral hemorrhage and ischemic stroke of different etiologies have distinct alternatively spliced mRNA profiles in the blood: a pilot RNA-seq study. Transl. Stroke Res. 6, 284–289. doi: 10.1007/s12975-015-0407-9
Fang, H., Chen, J., Lin, S., Wang, P., Wang, Y., Xiong, X., et al. (2014). CD36-mediated hematoma absorption following intracerebral hemorrhage: negative regulation by TLR4 signaling. J. Immunol. 192, 5984–5992. doi: 10.4049/jimmunol.1400054
Fang, Y., Tian, Y., Huang, Q., Wan, Y., Xu, L., Wang, W., et al. (2019). Deficiency of TREK-1 potassium channel exacerbates blood-brain barrier damage and neuroinflammation after intracerebral hemorrhage in mice. J. Neuroinflammation 16:96. doi: 10.1186/s12974-019-1485-5
Fei, T., Chen, Y., Xiao, T., Li, W., Cato, L., Zhang, P., et al. (2017). Genome-wide CRISPR screen identifies HNRNPL as a prostate cancer dependency regulating RNA splicing. Proc. Natl. Acad. Sci. U.S.A. 114, E5207–E5215. doi: 10.1073/pnas.1617467114
Guo, L., Li, S., Zhao, Y., Qian, P., Ji, F. Y., Qian, L., et al. (2015). Silencing angiopoietin-like protein 4 (ANGPTL4) protects against lipopolysaccharide-induced acute lung injury via regulating SIRT1/NF-kB pathway. J. Cell. Physiol. 230, 2390–2402. doi: 10.1002/jcp.24969
He, Y., Li, S., Tang, D., Peng, Y., Meng, J., Peng, S., et al. (2019). Circulating peroxiredoxin-1 is a novel damage-associated molecular pattern and aggravates acute liver injury via promoting inflammation. Free Radic. Biol. Med. 137, 24–36. doi: 10.1016/j.freeradbiomed.2019.04.012
Hentze, M. W., Castello, A., Schwarzl, T., and Preiss, T. (2018). A brave new world of RNA-binding proteins. Nat. Rev. Mol. Cell Biol. 19, 327–341. doi: 10.1038/nrm.2017.130
Hoshino, I., Matsubara, H., Hanari, N., Mori, M., Nishimori, T., Yoneyama, Y., et al. (2005). Histone deacetylase inhibitor FK228 activates tumor suppressor Prdx1 with apoptosis induction in esophageal cancer cells. Clin. Cancer Res. 11, 7945–7952. doi: 10.1158/1078-0432.CCR-05-0840
Kim, J. H., Lee, J. M., Lee, H. N., Kim, E. K., Ha, B., Ahn, S. M., et al. (2012). RNA-binding properties and RNA chaperone activity of human peroxiredoxin 1. Biochem. Biophys. Res. Commun. 425, 730–734. doi: 10.1016/j.bbrc.2012.07.142
Krishnamurthi, R. V., Feigin, V. L., Forouzanfar, M. H., Mensah, G. A., Connor, M., Bennett, D. A., et al. (2013). Global and regional burden of first-ever ischaemic and haemorrhagic stroke during 1990-2010: findings from the Global Burden of disease study 2010. Lancet Glob. Health 1, e259–e281. doi: 10.1016/S2214-109X(13)70089-5
Kwon, S. C., Yi, H., Eichelbaum, K., Föhr, S., Fischer, B., You, K. T., et al. (2013). The RNA-binding protein repertoire of embryonic stem cells. Nat. Struct. Mol. Biol. 20, 1122–1130. doi: 10.1038/nsmb.2638
Li, F. H., Han, N., Wang, Y., and Xu, Q. (2018). Gadd45a knockdown alleviates oxidative stress through suppressing the p38 MAPK signaling pathway in the pathogenesis of preeclampsia. Placenta 65, 20–28. doi: 10.1016/j.placenta.2018.03.007
Li, P., Tang, T., Liu, T., Zhou, J., Cui, H., He, Z., et al. (2019). Systematic analysis of tRNA-derived small RNAs reveals novel potential therapeutic targets of traditional Chinese medicine (Buyang-Huanwu-Decoction) on intracerebral hemorrhage. Int. J. Biol. Sci. 15, 895–908. doi: 10.7150/ijbs.29744
Li, Q., Han, Y., Du, J., Jin, H., Zhang, J., Niu, M., et al. (2018). Alterations of apoptosis and autophagy in developing brain of rats with epilepsy: changes in LC3, P62, Beclin-1 and Bcl-2 levels. Neurosci. Res. 130, 47–55. doi: 10.1016/j.neures.2017.08.004
Li, Y., Wei, W., Feng, J., Luo, H., Pi, M., Liu, Z., et al. (2017). Genome re-annotation of the wild strawberry Fragaria vesca using extensive Illumina- and SMRT-based RNA-seq datasets. DNA Res. 25, 61–70. doi: 10.1093/dnares/dsx038
Liu, D., Mao, P., Huang, Y., Liu, Y., Liu, X., Pang, X., et al. (2014). Proteomic analysis of lung tissue in a rat acute lung injury model: identification of PRDX1 as a promoter of inflammation. Mediators Inflamm. 2014:469358. doi: 10.1155/2014/469358
Liu, D. L., Zhao, L. X., Zhang, S., and Du, J. R. (2016). Peroxiredoxin 1-mediated activation of TLR4/NF-κB pathway contributes to neuroinflammatory injury in intracerebral hemorrhage. Int. Immunopharmacol. 41, 82–89. doi: 10.1016/j.intimp.2016.10.025
Liu, W., Guo, W. J., Zhu, Y. C., Peng, S., Zheng, W., Zhang, C., et al. (2018). Targeting peroxiredoxin 1 by a curcumin analogue, AI-44, inhibits NLRP3 inflammasome activation and attenuates lipopolysaccharide-induced sepsis in mice. J. Immunol. 201, 2403–2413. doi: 10.4049/jimmunol.1700796
Loughlin, F. E., Lukavsky, P. J., Kazeeva, T., Reber, S., Hock, E. M., Colombo, M., et al. (2019). The solution structure of FUS bound to RNA reveals a bipartite mode of RNA recognition with both sequence and shape specificity. Mol. Cell 73, 490–504.e6. doi: 10.1016/j.molcel.2018.11.012
Lu, Y., Zhang, X. S., Zhou, X. M., Gao, Y. Y., Chen, C. L., Liu, J. P., et al. (2019). Peroxiredoxin 1/2 protects brain against HO-induced apoptosis after subarachnoid hemorrhage. FASEB J. 33, 3051–3062. doi: 10.1096/fj.201801150R
Mayr, C. (2016). Evolution and biological roles of alternative 3′ UTRs. Trends Cell Biol. 26, 227–237. doi: 10.1016/j.tcb.2015.10.012
Meng, Z. Y., Zhao, T., Zhou, K., Zhong, Q., Wang, Y. C., Xiong, X. Y., et al. (2017). A20 ameliorates intracerebral hemorrhage-induced inflammatory injury by regulating TRAF6 polyubiquitination. J. Immunol. 198, 820–831. doi: 10.4049/jimmunol.1600334
Min, Y., Kim, M. J., Lee, S., Chun, E., and Lee, K. Y. (2018). Inhibition of TRAF6 ubiquitin-ligase activity by PRDX1 leads to inhibition of NFKB activation and autophagy activation. Autophagy 14, 1347–1358. doi: 10.1080/15548627.2018.1474995
Nakaso, K., Kitayama, M., Mizuta, E., Fukuda, H., Ishii, T., Nakashima, K., et al. (2000). Co-induction of heme oxygenase-1 and peroxiredoxin I in astrocytes and microglia around hemorrhagic region in the rat brain. Neurosci. Lett. 293, 49–52. doi: 10.1016/s0304-3940(00)01491-9
Nassour, H., Wang, Z., Saad, A., Papaluca, A., Brosseau, N., Affar, E. B., et al. (2016). Peroxiredoxin 1 interacts with and blocks the redox factor APE1 from activating interleukin-8 expression. Sci. Rep. 6:29389. doi: 10.1038/srep29389
Pan, W. K., Zhang, Y. F., Yu, H., Gao, Y., Zheng, B. J., Li, P., et al. (2017). Identifying key genes associated with Hirschsprung’s disease based on bioinformatics analysis of RNA-sequencing data. World J. Pediatr. 13, 267–273. doi: 10.1007/s12519-017-0002-0
Paschalis, A., Sharp, A., Welti, J. C., Neeb, A., Raj, G. V., Luo, J., et al. (2018). Alternative splicing in prostate cancer. Nat. Rev. Clin. Oncol. 15, 663–675. doi: 10.1038/s41571-018-0085-0
Qiu, Z., Yang, J., Deng, G., Fang, Y., Li, D., and Zhang, S. (2018). Angiopoietin-like 4 attenuates brain edema and neurological deficits in a mouse model of experimental intracerebral hemorrhage. Med. Sci. Monit. 24, 880–890. doi: 10.12659/msm.907939
Riddell, J. R., Patricia, M., Sass, S. N., Moser, M. T., Foster, B. A., and Gollnick, S. O. (2012). Peroxiredoxin 1 stimulates endothelial cell expression of VEGF via TLR4 dependent activation of HIF-1α. PLoS One 7:e50394. doi: 10.1371/journal.pone.0050394
Riddell, J. R., Wang, X. Y., Minderman, H., and Gollnick, S. O. (2010). Peroxiredoxin 1 stimulates secretion of proinflammatory cytokines by binding to TLR4. J. Immunol. 184, 1022–1030. doi: 10.4049/jimmunol.0901945
Roundtree, I. A., Evans, M. E., Pan, T., and He, C. (2017). Dynamic RNA modifications in gene expression regulation. Cell 169, 1187–1200. doi: 10.1016/j.cell.2017.05.045
Sebestyén, E., Singh, B., Miñana, B., Pagès, A., and Eyras, E. (2016). Large-scale analysis of genome and transcriptome alterations in multiple tumors unveils novel cancer-relevant splicing networks. Genome Res. 26, 732–744. doi: 10.1101/gr.199935.115
Song, Y., Milon, B., Ott, S., Zhao, X., Sadzewicz, L., Shetty, A., et al. (2018). A comparative analysis of library prep approaches for sequencing low input translatome samples. BMC Genomics 19:696. doi: 10.1186/s12864-018-5066-2
Stamova, B., Ander, B. P., Jickling, G., Hamade, F., Durocher, M., Zhan, X., et al. (2019). The intracerebral hemorrhage blood transcriptome in humans differs from the ischemic stroke and vascular risk factor control blood transcriptomes. J. Cereb. Blood Flow Metab. 39, 1818–1835. doi: 10.1177/0271678X18769513
Sun, Y., Lin, X., and Li, L. (2017). Identification of biomarkers for schistosoma-associated pulmonary arterial hypertension based on RNA-seq data of mouse whole lung tissues. Lung 195, 377–385. doi: 10.1007/s00408-017-9999-z
Tanaka, N., Takahashi, S., Xiao, H., Lu, Y., Fujimori, N., Golla, S., et al. (2017). Growth arrest and DNA damage-inducible 45α protects against nonalcoholic steatohepatitis induced by methionine- and choline-deficient diet. Biochim. Biophys. Acta Mol. Basis Dis. 1863, 3170–3182. doi: 10.1016/j.bbadis.2017.08.017
Tian, B., and Manley, J. L. (2017). Alternative polyadenylation of mRNA precursors. Nat. Rev. Mol. Cell Biol. 18, 18–30. doi: 10.1038/nrm.2016.116
Wang, F. X., Yang, X. L., Ma, Y. S., Jia, W. Y., Yang, M. H., Chen, X., et al. (2018). TRIF contributes to epileptogenesis in temporal lobe epilepsy during TLR4 activation. Brain Behav. Immun. 67, 65–76. doi: 10.1016/j.bbi.2017.07.157
Wang, Y. C., Zhou, Y., Fang, H., Lin, S., Wang, P. F., Xiong, R. P., et al. (2014). Toll-like receptor 2/4 heterodimer mediates inflammatory injury in intracerebral hemorrhage. Ann. Neurol. 75, 876–889. doi: 10.1002/ana.24159
Wilbertz, J. H., Voigt, F., Horvathova, I., Roth, G., Zhan, Y., and Chao, J. A. (2019). Single-molecule imaging of mRNA localization and regulation during the integrated stress response. Mol. Cell 73, 946–958.e7. doi: 10.1016/j.molcel.2018.12.006
Wood, Z. A., Schröder, E., Harris, J. R., and Poole, L. B. (2003). Structure, mechanism and regulation of peroxiredoxins. Trends Biochem. Sci. 28, 32–40. doi: 10.1016/s0968-0004(02)00003-8
Wurth, L., Papasaikas, P., Olmeda, D., Bley, N., Calvo, G., Guerrero, S., et al. (2016). UNR/CSDE1 drives a post-transcriptional program to promote melanoma invasion and metastasis. Cancer Cell 30, 694–707. doi: 10.1016/j.ccell.2016.10.004)
Xiong, X. Y., Liu, L., Wang, F. X., Yang, Y. R., Hao, J., Wang, P. F., et al. (2016). Toll-like receptor 4/MyD88-mediated signaling of hepcidin expression causing brain iron accumulation, oxidative injury, and cognitive impairment after intracerebral hemorrhage. Circulation 134, 1025–1038. doi: 10.1161/CIRCULATIONAHA.116.021881
Xu, H., Cao, J., Xu, J., Li, H., Shen, H., Li, X., et al. (2019). GATA-4 regulates neuronal apoptosis after intracerebral hemorrhage via the NF-κB/Bax/Caspase-3 pathway both in vivo and in vitro. Exp. Neurol. 315, 21–31. doi: 10.1016/j.expneurol.2019.01.018
Xu, J., Xiao, N., and Xia, J. (2010). Thrombospondin 1 accelerates synaptogenesis in hippocampal neurons through neuroligin 1. Nat. Neurosci. 13, 22–24. doi: 10.1038/nn.2459
Yamaguchi, M., Sato, H., and Bannai, S. (1993). Induction of stress proteins in mouse peritoneal macrophages by oxidized low-density lipoprotein. Biochem. Biophys. Res. Commun. 193, 1198–1201. doi: 10.1006/bbrc.1993.1752
Yee, B. A., Pratt, G. A., Graveley, B. R., Van Nostrand, E. L., and Yeo, G. W. (2019). RBP-maps enables robust generation of splicing regulatory maps. RNA 25, 193–204. doi: 10.1261/rna.069237.118
Zhang, Y., Gu, L., Hou, Y., Wang, L., Deng, X., Hang, R., et al. (2015). Integrative genome-wide analysis reveals HLP1, a novel RNA-binding protein, regulates plant flowering by targeting alternative polyadenylation. Cell. Res. 25, 864–876. doi: 10.1038/cr.2015.77
Zhao, B. S., Roundtree, I. A., and He, C. (2017). Post-transcriptional gene regulation by mRNA modifications. Nat. Rev. Mol. Cell Biol. 18, 31–42. doi: 10.1038/nrm.2016.132
Keywords: RNA-binding protein, peroxiredoxin 1, intracerebral hemorrhage, inflammation, apoptosis, posttranscriptional regulation
Citation: Yang G-Q, Huang J-C, Yuan J-J, Zhang Q, Gong C-X, Chen Q, Xie Q, Xie L-X, Chen R, Qiu Z-M, Zhou K, Xu R, Jiang G-H, Xiong X-Y and Yang Q-W (2020) Prdx1 Reduces Intracerebral Hemorrhage-Induced Brain Injury via Targeting Inflammation- and Apoptosis-Related mRNA Stability. Front. Neurosci. 14:181. doi: 10.3389/fnins.2020.00181
Received: 26 October 2019; Accepted: 19 February 2020;
Published: 10 March 2020.
Edited by:
Jong-Min Kim, Seoul National University Bundang Hospital, South KoreaReviewed by:
Nafisa M. Jadavji, Midwestern University, United StatesAnatol Manaenko, University Hospital Erlangen, Germany
Copyright © 2020 Yang, Huang, Yuan, Zhang, Gong, Chen, Xie, Xie, Chen, Qiu, Zhou, Xu, Jiang, Xiong and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiao-Yi Xiong, eGlvbmd4eV84OUAxNjMuY29t; Qing-Wu Yang, eWFuZ3F3bWx5c0AxNjMuY29t; eWFuZ3F3bWx5c0Bob3RtYWlsLmNvbQ==
†These authors have contributed equally to this work
 Guo-Qiang Yang
Guo-Qiang Yang Jia-Cheng Huang
Jia-Cheng Huang Jun-Jie Yuan
Jun-Jie Yuan Qin Zhang
Qin Zhang Chang-Xiong Gong
Chang-Xiong Gong Qiong Chen
Qiong Chen Qi Xie
Qi Xie Le-Xing Xie
Le-Xing Xie Ru Chen
Ru Chen Zhong-Ming Qiu
Zhong-Ming Qiu Kai Zhou
Kai Zhou Rui Xu
Rui Xu Xiao-Yi Xiong
Xiao-Yi Xiong Qing-Wu Yang
Qing-Wu Yang