- 1Stony Brook University, Stony Brook, NY, United States
- 2University of Florida, Gainesville, FL, United States
- 3Duraspace, Beaverton, OR, United States
- 4Oregon Health and Science University, Portland, OR, United States
- 5Cornell University, Ithaca, NY, United States
- 6Feinberg School of Medicine, Northwestern University, Chicago, IL, United States
- 7Digital Science, London, United Kingdom
- 8University of Iowa, Iowa, IA, United States
OpenVIVO is a free and open-hosted semantic web platform that anyone can join and that gathers and shares open data about scholarship in the world. OpenVIVO, based on the VIVO open-source platform, provides transparent access to data about the scholarly work of its participants. OpenVIVO demonstrates the use of persistent identifiers, the automatic real-time ingest of scholarly ecosystem metadata, the use of VIVO-ISF and related ontologies, the attribution of work, and the publication and reuse of data—all critical components of presenting, preserving, and tracking scholarship. The system was created by a cross-institutional team over the course of 3 months. The team created and used RDF models for research organizations in the world based on Digital Science GRID data, for academic journals based on data from CrossRef and the US National Library of Medicine, and created a new model for attribution of scholarly work. All models, data, and software are available in open repositories.
Transparency in Scholarship
Scholarship requires knowledge of previous work. The growth of scholarship worldwide and the proliferation of scholarly output types—from papers and monographs to preprints, conference papers, datasets, posters, and presentation slides—have fundamentally changed the scholarly ecosystem from an environment dependent on libraries to one that is dependent on the electronic resources made available by libraries to support discovery and knowledge transfer. This shift clearly drives a need for the representation of scholarly works using standard metadata formats to facilitate indexing and discovery.
For scholars to have knowledge of previous work, the work must be indexed and discoverable via electronic systems. Metadata regarding the work must be available. The metadata regarding scholarly work is typically not copyrighted or restricted by license. Many commercial systems are available to collect such open metadata, index it, and provide discovery tools powered by the metadata to scholars. However, the cost of such systems can be a barrier to many institutions.
Systems and data regarding scholarship should provide scholars with knowledge of previous work in a transparent manner, providing data regarding work that has been done. A number of recent efforts have contributed toward increased transparency. VIVO (Börner et al., 2012) provides an open data model for the representation of scholarship and an open API for accessing data in VIVO systems. ORCID (2016b) provides an open API to data marked open by users. Figshare (2016a), PubMed (US National Library of Medicine, 2016c), Crossref (2016), SHARE (2016), and DataCite (2016) each provide open APIs that can be used to identify and collect metadata regarding scholarly works.
The VIVO platform served as the framework for the study described herein. VIVO provides the means for organizations to host data regarding their scholarship, integrating VIVO with their internal systems to provide detailed, open information automatically from institutional repositories and enterprise systems, for their scholars, regarding their scholarship. Using VIVO ontological models for representing scholarship, open APIs, and persistent identifiers, OpenVIVO provides transparent access to the metadata of the scholarly works of its participants.
Ontological Models
OpenVIVO is a VIVO system and uses classes, object properties, and data properties from the 20 ontologies that are used by VIVO (Börner et al., 2012) to represent the scholarship of its participants. OpenVIVO is a native semantic web application using a triple store to record all information. Triples conform to the VIVO Integrated Semantic Framework (VIVO-ISF) (OpenRIF, 2016) and its associated ontologies. The OpenVIVO team used the VIVO-ISF to develop data models for all entities represented in OpenVIVO. Figure 1 shows a portion of the OpenVIVO ontological model centered on the person class with data properties associated with each class and object properties associating the person with other entities such as scholarly works, positions held, and contact information.

Figure 1. OpenVIVO Person Model showing relationships to contact information, profile photograph, positions, academic works, events, and research areas.
Working with Digital Science, the OpenVIVO team developed an ontological model using VIVO-ISF to represent the GRID (2016) research organizational data, which is openly available. A new data property was added to contain the GRID identifier, and a model was created to represent Figshare (2016a) data; figshare subsequently added VIVO RDF as an output format to the figshare open API. Additional models were constructed from existing VIVO classes, object properties, and data properties to represent journals and date times. These models were used to develop processes for loading data into OpenVIVO and to provide datasets that others can use.
The OpenVIVO System
OpenVIVO is a hosted VIVO platform that anyone can join. It is provided freely to anyone who would like to have an OpenVIVO profile.
Project Goals
OpenVIVO was developed as a demonstration of VIVO (Börner et al., 2012) for the Force16 (FORCE11, 2016) conference. The project team consisted of 16 volunteers from 10 institutions. The team established six project goals which were realized over the course of the 3-month project leading to the debut of OpenVIVO at the FORCE16 Conference, held in Portland, OR, USA, April 17–19, 2016:
1. Provide a VIVO experience for everyone, a demonstration of VIVO, a platform for experimentation, and an ownership experience for the VIVO team.
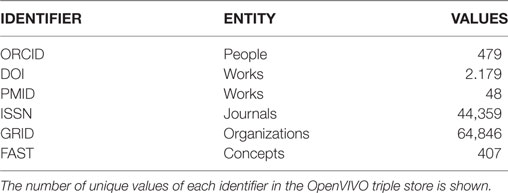
2. Use open persistent identifiers for all entities. The identifiers come from trusted sources: people (ORCID), works [Digital Object Identifiers (DOIs) and PMID], organizations (GRID), journals (ISSN), and concepts (FAST). The team wanted to understand if the platform could be entirely populated by using persistent identifiers for all entities; this project clearly demonstrated that this is indeed possible.
3. Automatic, real-time ingest of metadata via public APIs.
4. Attribution of works by scholars using the contribution role ontology (CRO), to indicate their roles in works or projects.
5. Continuous publication of OpenVIVO metadata via CC-BY license.
6. Demonstrate consumption and reuse of OpenVIVO scholarly metadata.
VIVO Experience
Establishing an OpenVIVO profile is a straightforward process. Anyone with an ORCID (2016a) account can join Open VIVO by visiting the Open VIVO website (VIVO, 2016a) and logging in with their ORCID username and password. Upon logging in, OpenVIVO creates the profile, and individuals can customize their profile with their profile photograph and other information. Data from ORCID can be marked public by users; it is the data marked public that will automatically get transferred to OpenVIVO via the ORCID public API. Individuals will then have an option to add a brief overview for themselves. It takes about 30 s to make a full OpenVIVO profile.
The OpenVIVO project team chose ORCID authentication for several reasons: (1) to simplify the user experience for users of OpenVIVO, as users do not have another username and password to manage; (2) to eliminate password management for the OpenVIVO team, as forgotten passwords can be reset using existing ORCID capabilities; and (3) to provide an ORCID identifier for every person signing on to OpenVIVO, as the identifier provides a key value for using the ORCID public API (ORCID, 2016b) to populate OpenVIVO from the ORCID profile. Figure 2 shows an OpenVIVO profile. The user can add links to other websites, position and title, photograph, an overview, research areas, and more. Works are added from ORCID at each sign-on. Publication visualizations such as publications over time, coauthor networks, and a map of science are available.
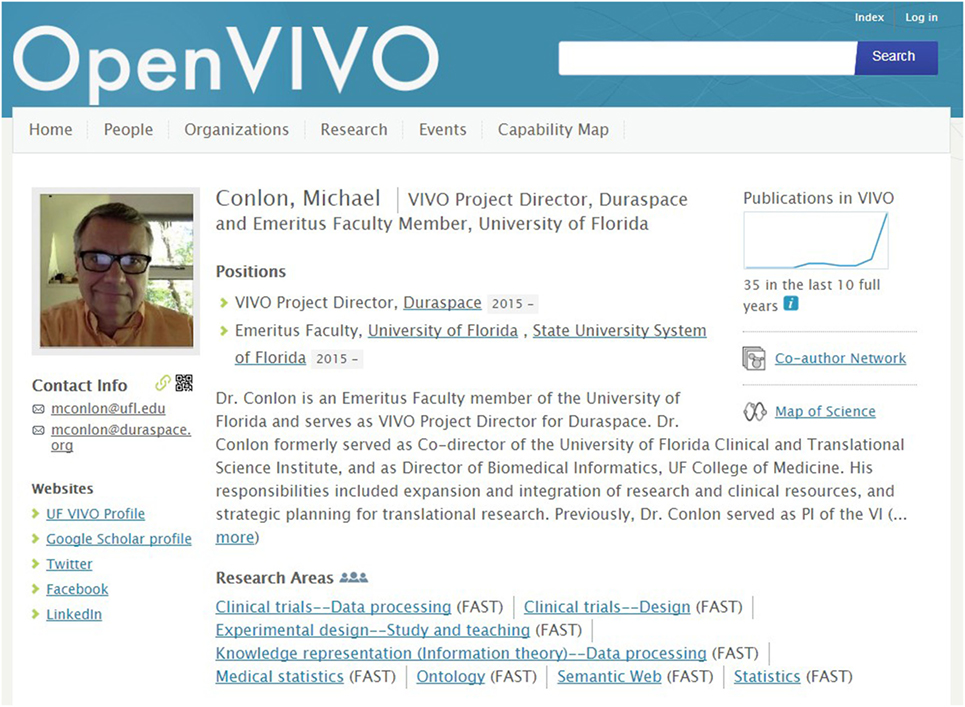
Figure 2. An OpenVIVO profile of Dr. Michael Conlon. Written and informed consent has been obtained from Dr. Michael Conlon for the publication of his identifiable image and his OpenVIVO profile. Users sign on to OpenVIVO using ORCID usernames and passwords. Users edit their overview, title, and contact information. OpenVIVO automatically populates the profile from ORCID.
The VIVO experience for OpenVIVO is different from that of VIVO hosted by an institution. Design choices were made by the OpenVIVO project team to simplify VIVO for use as an unrestricted public site. One’s profile may be edited in VIVO, to add detail regarding one’s scholarship. These self-edits are typically curated by institutional representatives to ensure that common language and formatting are used. The OpenVIVO project does not have an editorial team. As a result, self-edit was limited to those attributes that were simple data properties, such as contact information, and edits that could be made by selecting elements from force-choice drop-down menus. Over 50 self-edits available in VIVO were turned off in OpenVIVO, a change resulting in clean data without the need for manual curation—although at the cost of significant simplification in the level of detail that can be provided by the scholar to the system.
OpenVIVO provides a platform for demonstration of new VIVO features. Shortly after the debut, the capability map for interactive exploration of expertise was added to the VIVO code base and to OpenVIVO. The capability map, modeled on the Find An Expert site at the University of Melbourne (2016), is shown in Figure 3. Several months later, document viewers were added to display content from Figshare (2016a). In this way, features new to VIVO can be shown to the community before they are implemented at institutional VIVO sites. Institutional sites may wait months to schedule and upgrade their VIVO sites. OpenVIVO can be upgraded by the VIVO team. OpenVIVO provides an ownership opportunity for the VIVO team at Duraspace (2016), the nonprofit membership organization that stewards the VIVO project and the VIVO community. System administration is provided by the VIVO team. No manual effort is used to create, curate, or publish data, as these processes are completely automated.
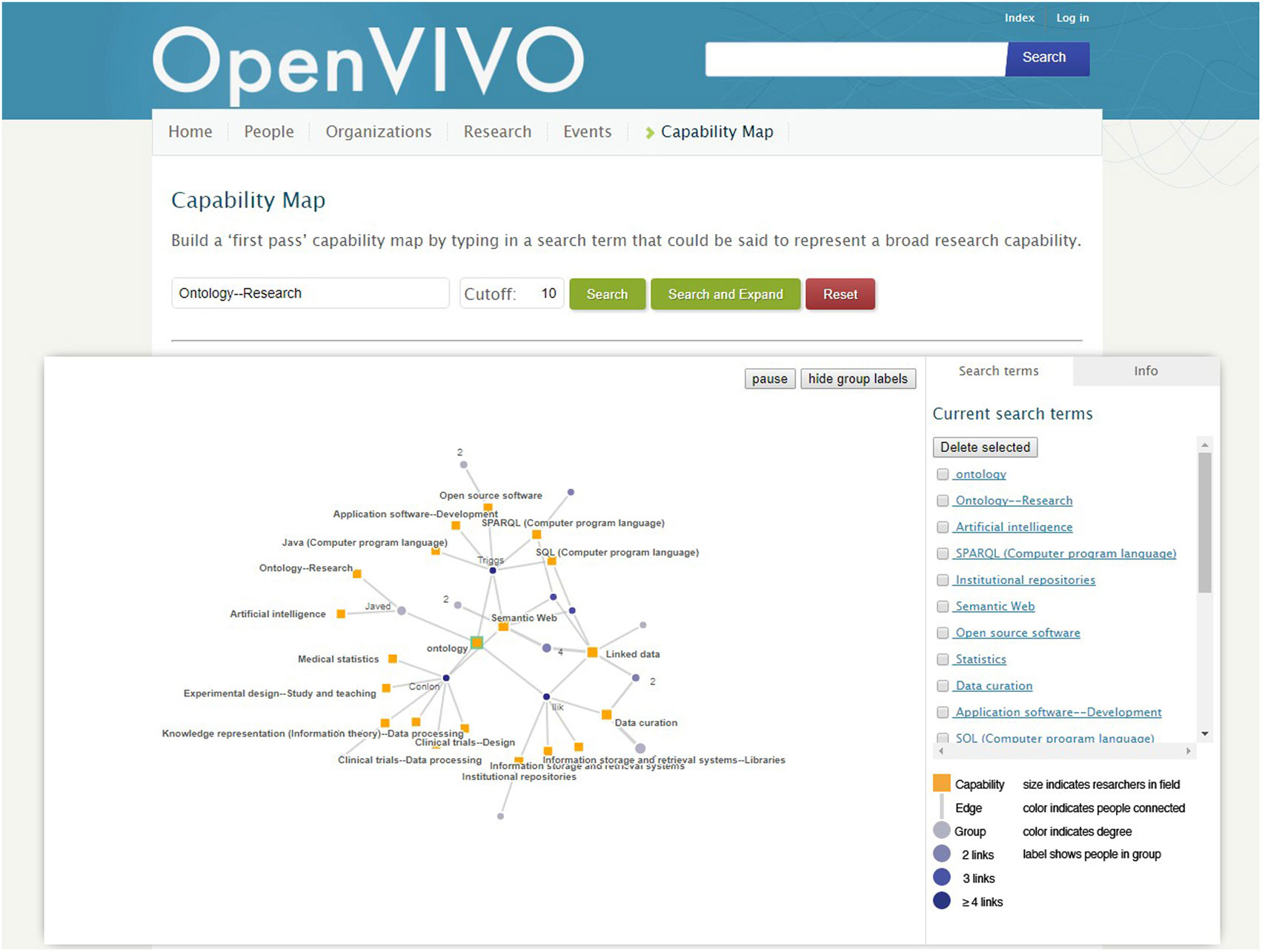
Figure 3. The capability map allows users to select concepts of interest, displaying people whose work involves those concepts. Connections between people represent coauthorship. The map can be tuned, navigated, and “drilled down” to individual investigators.
Hosting Environment
OpenVIVO is hosted on a dedicated server leased from Hetzner Online GmbH (Hetzner, 2016) and supported by VIVO members (VIVO, 2016c). The specifications include a quad core processor, 32 GB RAM and 500 GB SSD (with 6 GB/s transfer). This option was chosen for affordability and the very high storage bandwidth, which makes a significant difference to the performance of the triple store. The software environment was kept as simple and realistic to common usage as possible. As such, CentOS 6 (a community-supported Linux using the RedHat Enterprise sources) is installed, along with the following software, all from the CentOS repositories: OpenJDK 8, Apache Maven 3.0.5, Apache Tomcat 7, Apache HTTPd 2.4, and MariaDB 5.
Global Persistent Identifiers (GPIDs)
Global persistent identifiers are used in OpenVIVO to uniquely identify people, scholarly works, journals, and organizations conducting research. As a semantic web application, every entity in OpenVIVO also has a Uniform Resource Identifier (URI). GPID are attributes of the entities identified by URI. GPIDs serve to identify entities outside of OpenVIVO and support data ingest, identification of entities by humans, and data linkage between OpenVIVO and other systems. OpenVIVO takes a strict approach to the use of GPID for the entities identified by them—such entities must have a GPID attribute. In each case, the attribute value that appears is part of the entity URI.
ORCID, the Open Researcher and Contributor Identifier (ORCID, 2016a) is a 16-character string, part of the ISNI name space. URI for person entities in OpenVIVO have the format http://openvivo.org/a/ORCIDaaaa-bbbb-cccc-dddd, where aaaa-bbbb-cccc-dddd is the value of the ORCID for the person entity. Persons without ORCID identifiers cannot appear in OpenVIVO.
DOI and PubMed Unique Identifiers, DOI (International DOI Foundation, 2016), and/or PubMed Unique Identifiers (PMID) (US National Library of Medicine, 2016b) are required for each scholarly work in OpenVIVO. Works without either a DOI or a PubMed ID cannot appear in OpenVIVO. URI for works identified by DOI have the format http://openvivo.org/a/doivalue where value is the DOI value, typically beginning with 10. URI for works identified by PubMed ID has the format http://openvivo.org/a/pubmedvalue where value is the value of the PubMed ID. Works may have both a DOI and a PMID.
ISSN, the International Standard Serial Number (2016), is used to identify journals. The OpenVIVO team assembled a set of journals with ISSN using data from Crossref (2016) and from the National Library of Medicine in the United States (US National Library of Medicine, 2016a), both publically available for download and reuse. Journals with an associated print ISSN from each set were modeled with the VIVO-ISF ontology by using the Karma data integration tool (Information Integration Group, Information Sciences Institute, University of Southern California, 2016).
Global Research Identifier Database (GRID) (GRID, 2016) is used to identify organizations conducting research. GRID is an open, curated dataset of research organizations across the world, each with a unique identifier, the GRID ID. Working with the creators of GRID, the OpenVIVO team developed an RDF model for organizations using the VIVO ontologies (Börner et al., 2012). The resulting collection of research organizations was loaded into OpenVIVO. Users of OpenVIVO can then select organizations from a list of the research organizations in the world. URI for organizations have the format http://openvivo.org/a/gridvalue, where value is the value of the GRID identifier for the organization entity. GRID updates its data monthly. OpenVIVO replicates these updates in the RDF in OpenVIVO for organizations. All organizations in OpenVIVO must have a GRID identifier.
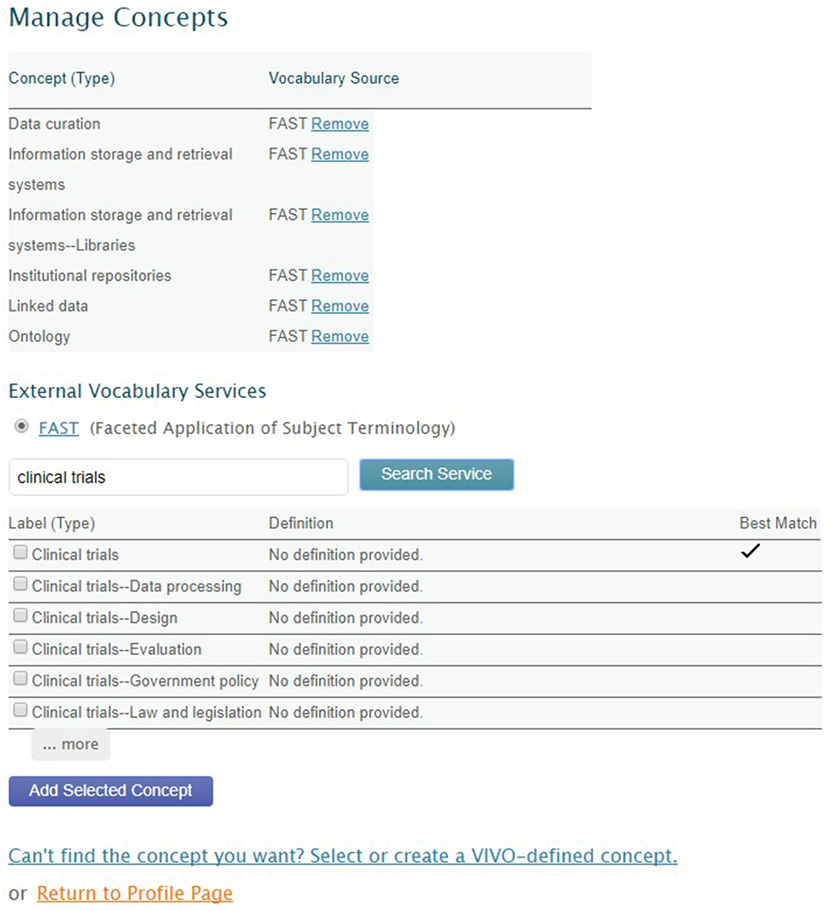
Faceted Application of Subject Terminology (FAST) (OCLC Research, 2016) provides a controlled vocabulary of research areas. OpenVIVO users can select from the FAST terminology using an external interface to FAST, enabling terms selected by users to be added to the OpenVIVO triple store. Figure 4 shows the OpenVIVO screen by which a user can search FAST for concepts and add selected concepts as research areas. The user has selected “clinical trials,” and FAST has offered a collection of possible terms. The user has indicated two concepts to add as research areas. Triples defining the selected concepts are fetched from FAST and added to the OpenVIVO triple store, along with inferred triples.
Table 1 shows the counts of various identifiers, including FAST concepts, in OpenVIVO as of the writing of this paper.
Real-Time Ingest of Metadata
For ingesting works into OpenVIVO, public APIs from various publication databases are used to retrieve metadata. By these means, we are able to acquire high-quality, consistent metadata, while users are asked to enter as little information as possible. This includes parsing a user’s ORCID profile for DOIs and using DOIs and PubMed identifiers to retrieve metadata.
ORCID. Whenever a user authenticates via ORCID, OpenVIVO queries the public API to obtain all of the data that the person has stored on their public profile. If it is the first time that somebody has logged on, then an OpenVIVO profile will be created, using their ORCID as part of the OpenVIVO URI and populating as much of the biography information (name, email, location, about, etc.) as possible. For all of the works listed with a DOI, the list of DOIs is passed to the OpenVIVO “claim your works” workflow, where the user can approve that they are associated with the work and add contribution information. On revisiting OpenVIVO, the list of works in the user’s public ORCID profile is compared with the works that they are associated with in OpenVIVO, and they are only asked to claim works with DOIs that they are not currently associated with.
CrossRef/DataCite. There are a multiple ways of retrieving metadata associated with a given DOI. For providers like CrossRef who provide their own APIs, these can be queried directly, but require custom code for each provider. Alternatively, using content negotiation, metadata can be retrieved from the DOI resolver. This provides a single, consistent interface that covers many of the DOI registries—including the two most prominent providers, CrossRef and DataCite. In OpenVIVO, we use content negotiation with the DOI resolver to retrieve metadata in a JSON serialization. This is then mapped to an intermediate format for the “claim your works” process.
PubMed. PubMed is a major biomedical publication database, which can be queried via public APIs. While CrossRef and DataCite cover a wider range of disciplines, PubMed provides a much larger historical database within its scope. Within biomedicine, PubMed identifiers are often more widely known than the DOIs. For these reasons, there were many requests to include direct support for PubMed identifiers in OpenVIVO. This was achieved in two ways. First, the PubMed APIs were used to attempt to resolve the PubMed identifiers to DOIs. For consistency, where DOIs were available, these were then used to retrieve the metadata via the DOI resolver. When a DOI is not available, the PubMed APIs are used to retrieve a JSON serialization. This is then mapped to an intermediate format for the “claim your works” process.
figshare. Using the figshare open API (Figshare, 2016b), the OpenVIVO team created a utility, figshare2vivo (Conlon, 2016), for mining figshare and adding content to OpenVIVO. figshare2vivo can collect works from figshare based on ORCID identifiers of authors, or by tags on the works. Using a tag for a conference, such as #vivo2016 for the VIVO 2016 conference, all works for a conference can be collected. Collected works are then represented in RDF using an OpenVIVO model developed in conjunction with Digital Science, the parent company of figshare. The resulting RDF can then be loaded to OpenVIVO.
Contribution Role Ontology
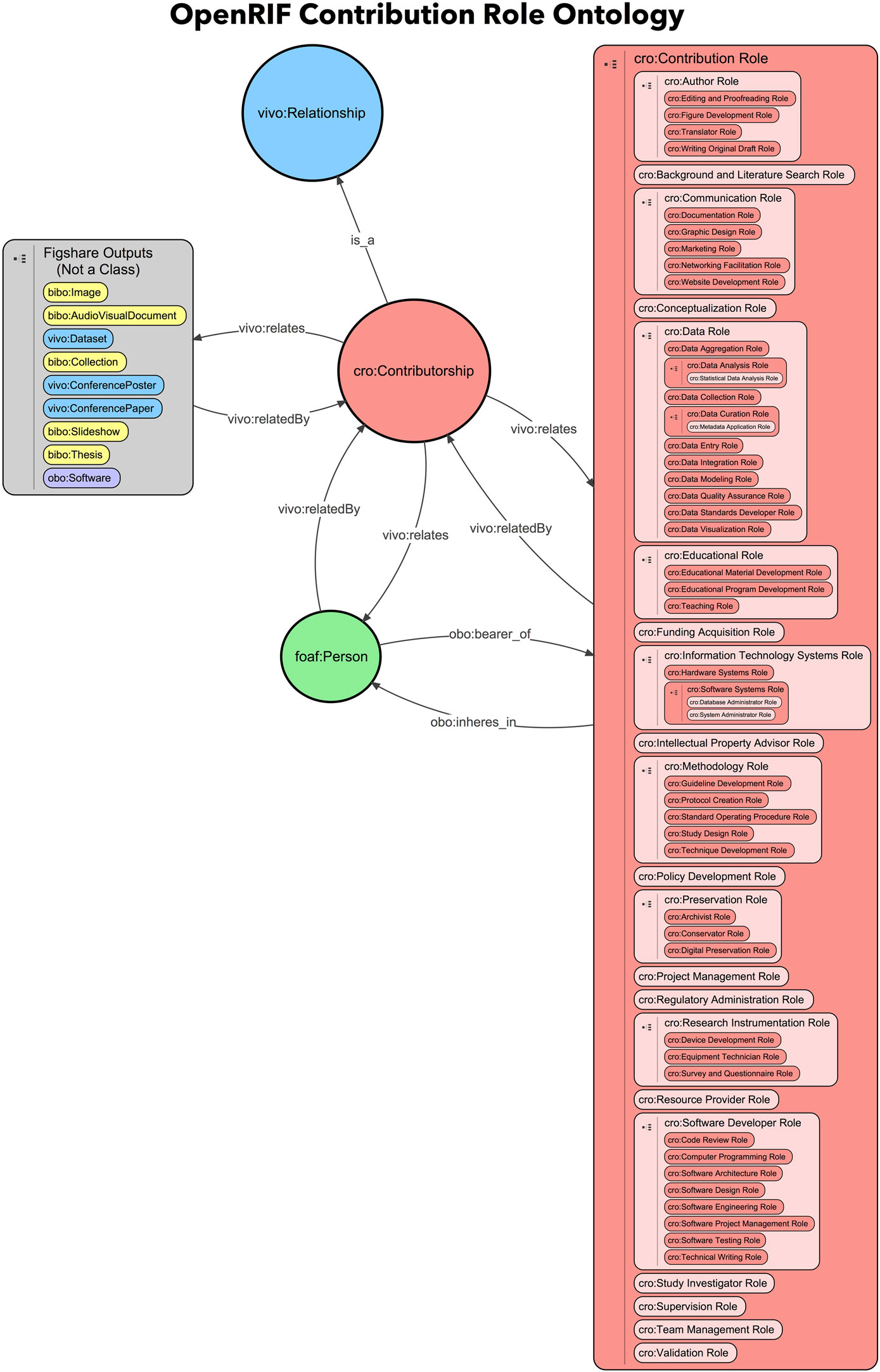
OpenVIVO utilizes the CRO developed by the FORCE11 Attribution Working Group. This ontology extends the contributions to scholarship beyond manuscript authorship to capture the broadening of researchers’ participation in scientific discoveries that have not been previously recognized by traditional measures of scholarly impact. Representation of all scholarly contributions and activities and their relationships to people and organizations is crucial to the health of the scholarly ecosystem as a whole. The FORCE11 Attribution Working Group along with the OpenVIVO Task Force members reviewed existing scholarly contribution taxonomies and extended the CRediT taxonomy (Allen et al., 2014) to create a prototype contributorship model that covers a wide selection of fields of research.
With the CRO, scholars are able to annotate the record with their specific contribution to scholarly works beyond authorship for manuscripts. The prototype CRO ontology contains 60 contribution roles which allow scholars to select the exact role they had on each of their scholarly works. These roles were a mix from the 42 roles collected from the FORCE 2016 Conference participants at the point of depositing their work in the official digital repository for the conference proceedings, the roles collected during the Attribution Working Group workshop at the FORCE 2015 Conference, the CRediT taxonomy, and the MARC code list of relator terms developed by the Library of Congress that designates the relationship between an agent and a bibliographic resource (Library of Congress, 2016). For example, in the production of a paper, each scholar can claim the exact role they played, which means that one can claim that they had a conceptualization role, data analysis role, or data aggregation role, just to name a few (see Figure 5).
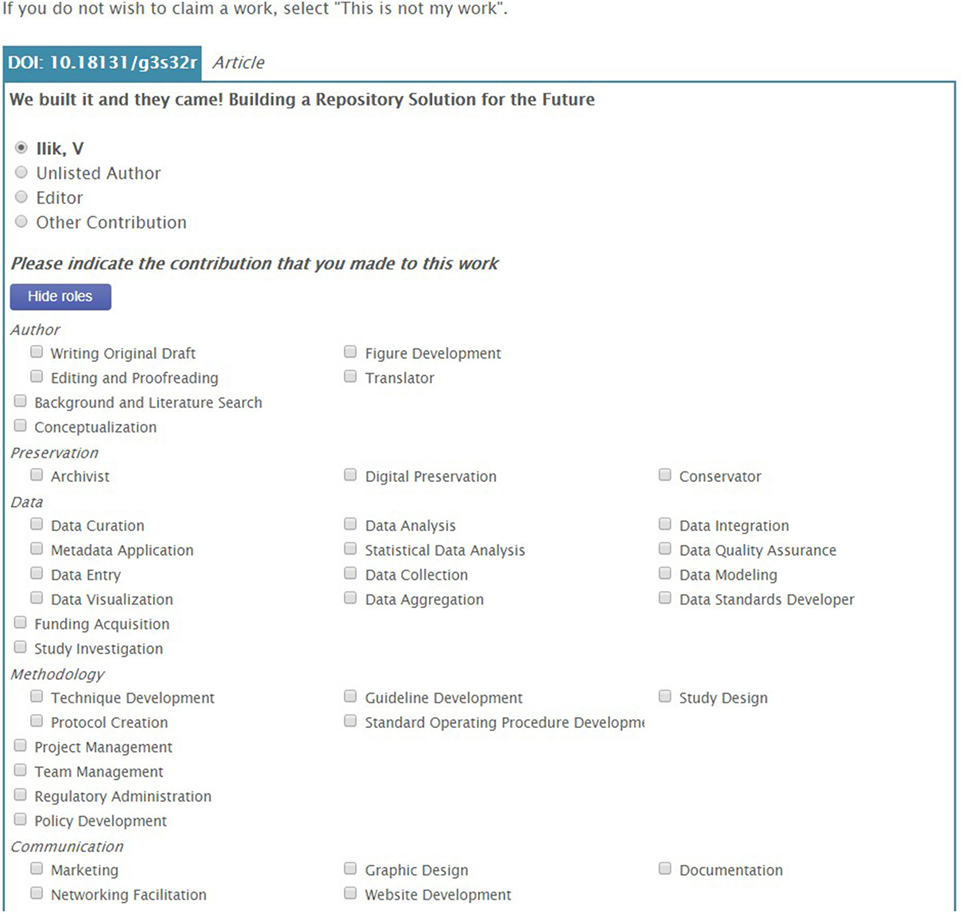
Figure 5. The claim process. Once metadata have been retrieved for an identified work, the user is asked to confirm their author position—OpenVIVO attempts to guess from the metadata—and/or assign any contribution roles.
The CRO can be found under the umbrella of Open Research Information Framework (OpenRIF) (OpenRIF Research Identification Framework, 2016), an open-source community devoted to representing scholarly research and expertise ecosystems. OpenRIF works on developing and promoting interoperable and extensible semantic infrastructure, such as the VIVO-ISF, an ontology for representing people, works, and the relationships between them; federated databases modeled on PARDI, the Portfolio Analysis and Reporting Data Infrastructure, for research impact and evaluation; and eagle-i, which aims to make research resources discoverable via a semantic search interface and represents their relationships to scholarly activities.
The image of the contribution role hierarchy is located on the OpenRIF’s GitHub repository: https://github.com/openrif/contribution-ontology/tree/reorganized-hierarchy (see Figure 6). OpenVIVO has an open text box for entering new contribution roles. The roles that are added as text will eventually be examined and evaluated by the ontology team for inclusion in the CRO.
Publication and Reuse of Data
The data are published openly and hourly (VIVO, 2016b) which points to the fact that the OpenVIVO data are freely available and open for anyone to use. OpenVIVO is not just about linked data it is about linked open data. The data in OpenVIVO is open and it is aggregated into one platform to showcase the scholarship.
In order for the data to be reusable, the contents of the asserted and inferred graphs are exported from the triple store in an N-Triple format. These exports occur hourly, and to automate this, we reused a VIVO tool (VIVO-Harvester) that can transfer data between serialized triple files and triples stores. This tool is usually used as part of an automated process to extract, transform, and load data from external systems into VIVO. By switching the input to be a graph from the triple store and the output an N-Triple serialization, the export can be scripted without writing an additional utility.
The N-Triple serializations, along with full SQL backups of the Jena SDB triple store and copies of all the uploaded thumbnail images, are placed in an export directory, replacing the previous exports. Apache HTTPd is then configured to serve this directory, allowing anyone to download all of the data simply by visiting http://openvivo.org/data/. Once downloaded, the data can be used and reused in any way. The readme file has information about the files and how to best use them.
Results
OpenVIVO offers the VIVO team a platform to experiment with various features and decide which ones will eventually end up in the VIVO main code. The code and all the relevant data about OpenVIVO can be found on the GitHub repository (OpenVIVO Task Force, 2016). Persistent identifiers play an important role in OpenVIVO: works have a DOI or a PubMed ID, people have an ORCID, and organizations are identified through the GRID data available with a CC-BY open license and can be freely used and downloaded. Close to 45,000 journals are identified with ISSNs and are represented in RDF, N-Triples format, and modeled with the help of the VIVO-ISF ontology. FAST subjects/concepts are identified with their WorldCat identifiers, and all entities have OpenVIVO URIs.
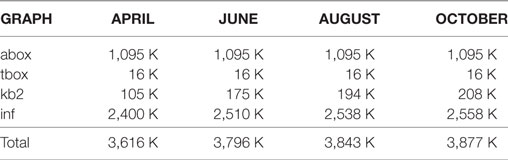
Linked data triples counts for OpenVIVO are shown in Table 2. When OpenVIVO started in April 2016 for the Force 2016 Conference (Conlon et al., 2016), there were 3,616 K triples in all four graphs. The abox and tbox are relatively fixed graphs in every VIVO instance. The triples for profile information are in the kb2 graph. The inference graph (inf) serves to collect the triples that are generated by inference. For example, if one enters the information that he or she is a librarian, the inference graph will contain the inference that the entity is a person.
OpenVIVO can be used to provide metadata in RDF regarding conferences. In collaboration with Force11 and figshare, OpenVIVO collected metadata on presentations and posters and created for Force2016 using figshare2vivo. In OpenVIVO, the metadata results in an event page for the conference (Force2016 Authors, 2016). The same approach was used to provide a complete collection of metadata regarding presentations and posters at the 2016 VIVO conference (VIVO 2016 Authors, 2016). Since the debut, OpenVIVO has also been used to provide metadata for software development (Symplectic Team, Simon Porter, 2016), provide metadata regarding works presented at conferences (Force2016 Authors, 2016; VIVO 2016 Authors, 2016), and provide metadata for aggregated search (Eichmann, 2016).
Identification of organizations, scholars, and their works remains a significant problem for the scholarly ecosystem. OpenVIVO demonstrates the utility of open API, persistent identifiers, and a common open data model for the collection and dissemination of open access to metadata regarding scholarship.
Next Steps
OpenVIVO meets a number of key needs in the scholarly ecosystem for scholars, for their organizations, and for the VIVO project, itself. In its most basic form, OpenVIVO can provide a profile of scholarly activity and roles, offering users a way to present their expertise, activities, and interest in an attractive manner. By leveraging a range of persistent identifiers, OpenVIVO makes it easy for users to populate their profiles. Moreover, the features that traditional VIVO instances enjoy, such as coauthorship and domain visualizations, feature prominently in the OpenVIVO platform. For institutions and those who have roles related to scholarly assessment, the OpenVIVO platform provides a valuable tool. The profile is not tied to a single institution, making the OpenVIVO profile something that can easily move and grow with a scholar. This offers an incredible benefit to those who are tasked with the longitudinal tracking of trainees, for example. The quality metadata from the records can be used for reporting and subsequent visualizations. Perhaps, the most promising is the discoverability of the scholarly products that can be fostered through the machine-readable data in OpenVIVO. OpenVIVO demonstrates open scholarship, the connected ecosystem, and linked open data.
OpenVIVO serves as a technology demonstration for the VIVO project, providing a way to test and demonstrate new features that may appear in the released enterprise version. As new versions of VIVO are released, OpenVIVO will be upgraded. Data packages developed for OpenVIVO, such as GRID, journals, and dates, will be made available to the larger VIVO community for local use. The CRO will also be continuously improved—especially based on our results of open text fields in OpenVIVO to expressing desired roles not already in the ontology. Additional software development could also provide the capability of extracting triples from OpenVIVO for use in a local VIVO. In this way, the institutional VIVO could contain the triples for an individual, which are asserted in OpenVIVO; cross-linking VIVO sites to consume triples in real time is also under consideration. The project team and the VIVO community expect to provide access to OpenVIVO to the scholarly community indefinitely.
Author Contributions
VI contribution roles include Conceptualization, Methodology, Software, Validation, Formal Analysis, Resources, Data Curation, Writing—Original Draft Preparation, Writing—Review & Editing, Preparation, Visualization, and Project Administration. MC contribution roles include Conceptualization, Methodology, Software, Validation, Formal Analysis, Resources, Data Curation, Writing—Original Draft Preparation, Writing—Review & Editing, Preparation, Visualization, Supervision, and Project Administration. GT contribution roles include Conceptualization, Methodology, Software, Validation, Formal Analysis, Resources, Data Curation, Writing—Original Draft Preparation, Preparation, and Visualization. MW contribution roles include Conceptualization and Methodology. MJ contribution roles include Ontology Engineering, Ontology Change Management, and Linked Data Modeling. MB contribution roles include Conceptualization and Methodology. KG contribution roles include Reviewing and Editing. SE contribution roles include Conceptualization and Methodology. PF contribution roles include Software and Visualization. SP contribution roles include Conceptualization, Software, and Resources. MS contribution roles include Conceptualization, Software, and Resources. MH contribution roles include Conceptualization, Methodology, Investigation, Supervision, and Funding Acquisition. DE contribution roles include Conceptualization, Methodology, and Testing. KH contribution roles include Conceptualization, Methodology, Resources, Writing, Supervision, and Funding Acquisition.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to acknowledge Ted Lawless, Jim Blake, John Serafin, Alexandre Rademaker for their work on this project.
Funding
Research reported in this publication was supported, in part, by the National Institutes of Health’s National Center for Advancing Translational Sciences, Grant Number UL1TR001422 and Grant Number 3UL1TR000128-10S1 and by the National Institute of General Medical Sciences, Grant Number 3R25GM114820-02S1. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
References
Allen, L., Brand, A., Scott, J., Altman, M., and Hlava, M. (2014). Credit where credit is due. Nature 508, 312–313. doi: 10.1038/508312a
Börner, K., Conlon, M., Corson-Rikert, J., and Ding, Y. (eds) (2012). VIVO: A Semantic Approach to Scholarly Networking and Discovery, Synthesis Lectures on the Semantic Web: Theory and Technology, Vol. 2. Morgan-Claypool, 1–178.
Conlon, M. (2016). Open VIVO Task Force: Figshare Harvester for OpenVIVO. Available at: https://github.com/OpenVIVO/figshare-rdf
Conlon, M., Triggs, G., Ilik, V., Lawless, T., Holmes, K., Blake, J., et al. (2016). “OpenVIVO: a vivo anyone can join,” in Force 2016 Conference, Portland, OR (Figshare).
Crossref. (2016). Crossref.org. Available at: http://crossref.org
DataCite. (2016). Datacite Home Page. Available at: http://datacite.org
Duraspace. (2016). Duraspace: Home. Available at: http://duraspace.org
Eichmann, D. (2016). CTSAsearch. Available at: http://research.icts.uiowa.edu/polyglot/
Figshare. (2016a). Figshare—Credit for All Your Research. Available at: http://figshare.com
Figshare. (2016b). Figshare API Documentation Home Page. Available at: https://docs.figshare.com/
FORCE11. (2016). FORCE2016 Conference. Available at: https://www.force11.org/meetings/force2016
Force2016 Authors. (2016). Force2016 Conference Presentations and Posters in OpenVIVO. Available at: http://openvivo.org/a/eventFORCE2016
GRID. (2016). GRID—Global Research Identifier Database. Available at: http://grid.ac
Hetzner. (2016). Hetzner Online GmbH. Available at: http://hetzner.de
Information Integration Group, Information Sciences Institute, University of Southern California. (2016). Karma: A Data Integration Tool. Available at: http://usc-isi-i2.github.io/karma/
International DOI Foundation. (2016). Digital Object Identifier System. Available at: http://doi.org
International Standard Serial Number. (2016). ISSN. Available at: http://issn.org
Library of Congress. (2016). MARC Code List of Relators. Available at: http://id.loc.gov/vocabulary/relators.html
OCLC Research. (2016). FAST (Faceted Application of Subject Terminology). Available at: http://www.oclc.org/research/themes/data-science/fast.html
OpenRIF. (2016). Vivo Integrated Semantic Framework Data Standard Github Repository. Available at: https://github.com/openrif/vivo-isf-ontology
OpenRIF Research Identification Framework. (2016). Welcome to OpenRIF. Available at: http://openrif.org
OpenVIVO Task Force. (2016). OpenVIVO GitHub Repository. Available at: https://github.com/OpenVIVO
ORCID. (2016a). ORCID: Connecting Research and Researchers. Available at: http://ORCID.org
ORCID. (2016b). Register a Public API Client Application. Available at: http://support.ORCID.org/knowledgebase/articles/343182
SHARE. (2016). Share Home Page. Available at: http://share.osf.io
Symplectic Team, Simon Porter. (2016). Pimp My VIVO Slides. Available at: https://github.com/OpenVIVO
University of Melbourne. (2016). Find an Expert—the University of Melbourne. Available at: https://findanexpert.unimelb.edu.au/
US National Library of Medicine. (2016a). National Library of Medicine—National Institutes of Health. Available at: http://www.nlm.nih.gov
US National Library of Medicine. (2016b). PubMed Tutorial—Building the Search—Searching by Field—PubMed Unique Identifier. Available at: https://www.nlm.nih.gov/bsd/disted/pubmedtutorial/020_830.html
US National Library of Medicine. (2016c). PubMed. Available at: https://www.ncbi.nlm.nih.gov/pubmed
VIVO. (2016a). OpenVIVO. Available at: http://openvivo.org
VIVO. (2016b). OpenVIVO Data. Available at: http://openvivo.org/data
VIVO. (2016c). VIVO Project Members. Available at: http://vivoweb.org/community/membership
VIVO 2016 Authors. (2016). VIVO 2016 Conference Presentations and Posters in OpenVIVO. Available at: http://openvivo.org/a/eventVIVO2016
Keywords: VIVO, scholarly ecosystem, transparency, data sharing, semantic web, ORCID, GRID, contribution role ontology
Citation: Ilik V, Conlon M, Triggs G, White M, Javed M, Brush M, Gutzman K, Essaid S, Friedman P, Porter S, Szomszor M, Haendel MA, Eichmann D and Holmes KL (2018) OpenVIVO: Transparency in Scholarship. Front. Res. Metr. Anal. 2:12. doi: 10.3389/frma.2017.00012
Received: 01 October 2017; Accepted: 06 December 2017;
Published: 01 March 2018
Edited by:
Henk F. Moed, Sapienza Università di Roma, ItalyCopyright: © 2018 Ilik, Conlon, Triggs, White, Javed, Brush, Gutzman, Essaid, Friedman, Porter, Szomszor, Haendel, Eichmann and Holmes. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Violeta Ilik, aWxpay52aW9sZXRhQGdtYWlsLmNvbQ==
 Violeta Ilik
Violeta Ilik Michael Conlon
Michael Conlon Graham Triggs
Graham Triggs Marijane White
Marijane White Muhammad Javed5
Muhammad Javed5 Karen Gutzman
Karen Gutzman Melissa Anne Haendel
Melissa Anne Haendel David Eichmann
David Eichmann Kristi L. Holmes
Kristi L. Holmes