- Key Laboratory of Animal Virology of Ministry of Agriculture and Institute of Preventive Veterinary Medicine, College of Animal Sciences, Zhejiang University, Hangzhou, China
Porcine torovirus (PToV) is a potential enteric swine pathogen, found at especially high rates in piglets with diarrhea. It was first reported in the Netherlands in 1998 and has emerged in many countries around the world. Infections are generally asymptomatic and have not directly caused large economic losses, though co-infections with other swine pathogens and intertype recombination may lead to unpredictable outcomes. This review introduces progress in PToV research regarding its discovery, relationship with other Toroviruses, virion morphological characteristics, genetic structure and variation, recent epidemiology, diagnostic methods, and possibilities for future research.
History
Toroviruses (ToV) (order Nidovirales; suborder Tornidovirineae; family Tobaniviridae; subfamily Torovirinae; genus Torovirus; subgenus Renitovirus) are responsible for infections leading to gastroenteritis in animals and humans (1–5). There are three recognized species in the Renitovirus subgenus: porcine torovirus (PToV); bovine torovirus (BToV), and equine torovirus (EToV) (Virus Taxonomy: 2018b Release (MSL #34); https://talk.ictvonline.org/taxonomy/). Human torovirus (HToV) is used to be the member of the Torovirus genus, according to the 2017 Release of Virus Taxonomy (MSL #32). Neutralizing activity against EToV has been found in the sera of other animals (cattle, goats, sheep, swine, rabbits, mice), providing serological evidence for the existence of ToVs in other animals (5).
EToV (also called Berne virus [BEV]) was the first to be discovered, and is the prototype species of the genus Torovirus. EToV was initially isolated in 1972 (but not reported until 1983) from a rectal swab taken from a horse in Berne, Switzerland which showed pseudomembranous enteritis, miliary granulomas and necrosis in the liver at necropsy. The isolated pathogenic agent couldn't be neutralized by antibodies against equine viruses known at the time. By electron microscopy, the virions appeared pleomorphic, mostly with smooth surfaces and spherical, though some were C-shaped, and some particles in damaged membranes had a “sausage-like” internal structure with transverse striations (1).
Another unclassified virus, BToV (also called Breda virus [BRV]) was discovered in 1982 in calves with diarrhea in Breda, Iowa, and confirmed to have antigenic differences from known diarrhea-related bovine viruses. The isolate was infectious when orally inoculated into gnotobiotic and conventionally reared calves (2).
In 1984, a similar virus was found in the feces of diarrheic patients (mainly children under 5 years-old) in England and the United States, with virions about 100 nm in diameter and a 7–9 nm capsule on the edge (3).
Previously, ToV-like agents had been detected in swine in many countries by electron microscopy or neutralization assays performed with EToV antibodies (5–8). In 1998, PToV was first detected and characterized in fecal samples from piglets in the Netherlands (4). Immunoelectron microscopy showed the elongated, 120- by 55-nm particles in fresh material. By sequence analysis, the N protein gene of PToV only has 68% sequence identity with BToV and EToV, which share 88% between them (4). PToV has since been reported in Canada, the United States, South Africa, China, Korea, and many European countries, such as the United Kingdom, Italy, Belgium, Hungary, and Spain (5–14).
Etiology
PToV Structure
ToVs are enveloped viruses with positive-sense single-stranded RNA genomes. PToV particles appear elongated in fresh samples, about 120 nm long and 55 nm wide, as observed by EM. After repeated freezing and thawing, virions have multiple forms, including round, kidney- and torus-shaped particles (4). The nucleocapsid is formed by N protein and viral RNA, surrounded by an envelope that contains the triple-spanning membrane (M) protein (10). On the surface of particles, two kinds of projections have been identified: longer protrusions (about 19 nm) with a drumstick or petal shape, considered to be spike (S) proteins; and shorter ones (6 nm in length) speculated to be the hemagglutinin-esterase (HE) (4, 15).
PToV Genome
ToV genomes are nonsegmented, polyadenylated, and 25–30 kb in size, with similar organization to other coronaviruses (16, 17). Both the 5′ and 3′ termini of the genome have a non-translated region (NTR), and the 5′ two-thirds of the ToV genome contains two large, overlapping open reading frames (ORF1a [13,254 bp] and ORF1b [6,875 bp]) connected by a frameshift (Figure 1), which encode two replicase polyproteins. There are four smaller ORFs downstream encoding the structural proteins (S [4,722 bp]; M [702 bp]; HE [1,284 bp]; and N [492 bp]), which are expressed through a 3′-coterminal nested set of subgenomic mRNAs (17–21). The first genomic sequence of PToV from Shanghai, China (strain SH1; GenBank accession no. JQ860350) was 28,301 bp in length, and had 79% sequence identity with BToV (21).
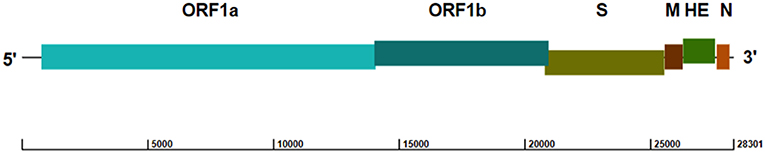
Figure 1. Genome structure of PToV strain SH1 (GenBank accession no. JQ860350). The ORFs encoding the replicase polyproteins (ORF1a and ORF1b) and structural proteins spike (S), membrane (M), hemagglutinin-esterase (HE), and nucleocapsid (N) are indicated.
PToV Structural Proteins
The mature ToV S protein contains two functional domains, S1 and S2, which mediate receptor specificity and determine viral tropism; S1 can bind independently to cellular receptors whereas S2 mediates fusion of the viral and cell membranes (22). Based on their electrophoretic mobility upon endoglycosidase F treatment, it has been demonstrated that the putative cleavage site, composed of five consecutive arginine residues, plays a role in post-translational processing in vivo. There are structural features in common with other coronavirus S proteins, such as the N-terminal signal sequence, an assumed C-terminal transmembrane anchoring domain, two heptad-repeat domains and a probable “trypsin-like” cleavage site (23). Due to its important role in host receptor binding, the S protein is the major target of neutralizing antibody responses.
The ToV M protein, previously referred to as the envelope protein, has three transmembrane α-helices in its N-terminal part. Its C-terminal part is exposed on the cytoplasmic face of the membrane, a feature of coronavirus M proteins. This protein lacks a cleavage signal sequence, so it is suspected that one of the hydrophobic transmembrane domains acts as an internal signal sequence (24).
Observed by EM, the HE protein is distributed on the virion surface as small projections. It is a class I membrane protein, and a member of the receptor destroying enzyme (RDE) protein family. There are two main domains identified in the monomer: the enzymatic acetyl-esterase region (E); and the receptor binding or lectin domain (25). The HE protein specifically but reversibly binds to mucopolysaccharides, mediating adhesion of virions to the intestinal wall. Through binding to 9-O-acetylated receptors then cleaving and rebinding to glycosylated surfaces (26, 27), virions can theoretically migrate through the mucus layer, thus promoting infection (18).
The open torus-shaped core of ToV is formed by the viral genome binding to the phosphorylated N protein, which is the only viral RNA binding protein. This 18.7 kDa protein is the most abundant protein among the ToV virions (28). Structural predictions of N protein have revealed the presence of four N-glycosylation sites [associated with antigenicity and immunogenicity (29)], two protein kinase C phosphorylation sites, one casein kinase II phosphorylation site, and an N-myristoylation site.
PToV Strain Variability
Sequencing studies around the world have shown significant variation among PToV isolates by geographic region. The genetic diversity of PToV in three Korean farms was studied in 2010, showing low nucleotide and deduced amino acid identities of partial S genes. Among the Korean PToV strains there was 73.5–94.1 and 71.4–95.0% identity in nucleotide and deduced amino acid sequences, respectively, whereas comparison with the Netherland strain Markelo revealed 74.0–93.1% and 73.2–95.5% identity, respectively. In addition, phylogenetic analysis of the N gene showed that different PToV strains were emerging in Korea, and even within the same farm (11).
The complete genome of the first PToV strain identified in the United States (PToV-NPL/2013, GenBank accession no. KM403390) contained 28,305 bp, with 92% identity to PToV-SH1. The predicted S, M, HE, and N proteins shared 94, 99, 92, and 96% amino acid identity with PToV-SH1, respectively. The gene encoding HE had 80–95% identity to other PToV strain sequences in GenBank, while the N and M genes were 95 and 93–97%, respectively, indicating that HE is more diverse among PToV strains (14).
Through phylogenetic analysis, the M gene sequences of 19 novel PToV strains from Sichuan, China and 21 ToV strains in GenBank could be classified into two genotypes (I & II). All of the novel Sichuan strains belonged to genotype I along with two Korean sequences (GU-07-56-11 and GU-07-56-22), whereas all other representative Korean, Netherlands and Canadian strains from GenBank belonged to genotype II. Putative amino acid sequence identities of the M gene were 99.1–99.6% among the 19 Sichuan strains and 97.4–99.6% between Sichuan and foreign strains, demonstrating high conservation of the M gene (30). Since the M gene sequences available were highly conserved, genotyping of PToV based on the M gene may be meaningless.
The sequence differences between the four Torovirus species range from 30 to 40%, which makes it easy to distinguish between them. However, evidence of intertype recombination was found in 2003, where BToV variants had emerged from recombination with PToV at the 3′ end of the HE/N genes and the 3′-NTR (31). Similar recombination events have also been found in ORF1a, ORF1b, and the 5′-NTR (32). Furthermore, some PToV and BToV variants have been isolated with chimeric HE genes, speculatively from recombination with some unknown ToVs (31). This ability for recombination underlines the potential for cross-species transmission and even the risk of zoonosis.
Inter-order Recombination Between ToV and Enterovirus in Swine
Interestingly, evidences of inter-order recombination between ToV (order Nidovirales) and the order of Picornavirales were discovered in different geographical locations most recently. The porcine enterovirus G (EV-G) genome with an insertion of ToV papain-like cysteine protease gene (PLCP) at the 2C/3A junction was detected in the feces of diarrheal pigs from farms in the United States, Belgium, Japan, China, and South Korea (33–38). The inserted PLCP sequences had lengths varied in strains from different geographical locations, ranging from 194 to 223 amino acids, which form a separate cluster distantly related to those of PToV, BToV, and EToV, suggesting that these viruses might have a common ancester. Since a mutant virus without the insertion by reverse genetics produced impaired growth and higher expression levels of innate immune genes in infected cells (36), the PLCP sequence might help the EV-G-PLCP strains establish a new host immune evasion strategy and, in some cases, determine its pathogenic potential.
Diagnostic Assays
Immunoelectron microscopy was used to detect PToV in the early years. However, this method was costly, time-consuming, and not suitable for processing a large number of samples. In addition, the polymorphism of ToV particles (4) decreases the specificity and accuracy of this detection method, and the fact that viral particles are not shed in the feces for a long duration decreases the likelihood of detection.
Neutralization assays using EToV virions obtained from cell culture (39) and an indirect ELISA method using BToV virions obtained from infected gnotobiotic calves (40) have been applied to serological diagnosis of PToV by detecting cross-reacting antibodies. However, the difficulty of virion purification and lost sensitivity resulting from use of heterologous antigens make these assays inappropriate for widespread use. The lack of a PToV culture system and infection model makes it difficult to obtain a large number of virions, hampering development of diagnostic methods and epidemiology studies. The only method available for serological diagnosis of PToV is an indirect ELISA based on a recombinant His-tagged N protein expressed in a baculovirus system. Serum samples (n = 15) collected from 6 to 8 week-old healthy piglets from a farm located in Galicia, Spain, tested by this method obtained a positive rate of 100%, whereas the positive rates when tested by western blot and neutralization assay of EToV virions were 60% (9/15) and 93% (14/15), respectively (10). It could be seen that this ELISA method had a high sensitivity, but its specificity may be slightly worse. The indirect ELISA based on PToV-HE protein is less sensitive and may have false negative results because of the diversity of HE proteins and the antigenic differences between HE lineages (14, 25). Development of other ELISA diagnostic methods based upon the PToV S protein is needed, according to experiences from porcine coronavirus such as porcine epidemic diarrhea virus (PEDV) (41).
Currently, PCR is the most widely used diagnostic assay, with the advantages of being low-cost, convenient, and highly sensitive and specific. Apart from conventional reverse transcription PCR (RT-PCR) (4, 10, 13, 31), quantitative RT-PCR (qRT-PCR) methods with primer pairs derived from the N gene (9, 39) have also been established to detect PToV and investigate epidemiology, since the N protein is the most abundant protein in the virus and has a high sequence conservation. A one-step SYBR Green qRT-PCR based on the PToV M gene was shown to be more sensitive than conventional RT-PCR and nested PCR (42). A multiplex RT-PCR method was developed for simultaneous detection of porcine kobuvirus (PKV), porcine astrovirus (PAstV) and PToV using primers based on their M gene sequences (43). Furthermore, an RT-LAMP method based on 4 specific primers from the N gene was developed for the quick detection of PToV (44).
Epidemiology
The worldwide distribution of PToV has been proven, with a high infection rate in pigs. However, due to limitations of diagnostic assays and asymptomatic infections (4, 13), there are not many reports on epidemiology. In 1998, Kroneman et al. performed a neutralization assay using EToV to detect cross-reacting antibodies and found that 81.4% (96/118) of the pig serum samples collected from farms in the Netherlands contained EToV-neutralizing antibodies (4). A qRT-PCR method was applied to detect PToV in rectal swabs collected from piglets at a farm in northeastern Spain in 2010, with a positive detection rate of 39.6% (19/48) (9). A longitudinal serological and virological study of PToV in Spain detected serum antibody levels by N protein ELISA, and fecal shedding by qRT-PCR based on the N gene. Seroprevalence in one hundred and twenty piglets at 1, 3, 7, 11, and 15 weeks-of-age was 92, 58, 91, 100, and 100% positive, respectively, and the corresponding 30 sows were all seropositive, reflecting the process of maternal antibody decline and subsequent immune response. As for fecal shedding in a 36-piglet subpopulation, 92% (33/36) of piglets had detectable PToV RNA at some age (39). Another epidemiological study in Spanish farms was done in 2012, with serum samples collected from 100 farms tested by N protein ELISA, revealing a total seroprevalence of 95.7% (2550/2664) and prevalence at different ages ranging from 59.4 to 99.6%. The lowest seroprevalence was detected in 3-week-old piglets (98/165) (45).
Shin et al. examined the prevalence of PToV in Korea in 2007, revealing 6.4% (19/295) of diarrheic pig samples were positive by RT-PCR (11). Among samples from diarrheic pigs collected in Korea during 2004–2005 and 2007, 36% (31/86) were positive by SYBR Green qRT-PCR (42). RT-PCR targeting the S gene was used to test stool and intestinal samples of diarrheic piglets from 20 farms in southwest China collected in the winter of 2011, with 45% (9/20) farms positive for PToV. In addition, 7 of those 9 farms had mixed infection with other swine viruses including PEDV, PKV, porcine rotavirus group A (PRV-A), transmissible gastroenteritis virus (TGEV), PAstV and mammalian orthoreovirus (MRV) (12). In Sichuan Province in the southwest of China, 872 fecal samples collected from diarrheic swine in 2011–2013 were tested by RT-PCR based on the conserved region of the S gene. An overall positive rate of 37.96% (331/872) was found, with positive co-infection with PEDV, TGEV or PRV-A in 4.1% (36/872) of these samples. Among the different ages tested, piglets at 1–3 weeks-of-age had the highest infection rate of 42.47% (295/697) (30).
Discussion
Most of our existing knowledge about ToV is based on the study of BToV and EToV, or the members of the Coronaviridae. The lack of an adaptive culture system and infection model to grow the virus hampers the study of viral characteristics and development of diagnostic tools. On the other hand, as PToV has not caused great economic losses, people do not pay attention to it, resulting in a lack of research on treatment and prevention.
The limited studies of sequence diversity may impede development of accurate diagnostic assays and vaccine production. Especially, a lack of study of the variability of the S gene also limits our understanding of the serology of PToV. So far, though many test methods have been established, there are no commercially available diagnostic kits. The expression of structural proteins by various means is important in order to screen for antibodies against PToV, and monoclonal antibodies are needed for further research on important topics like the mechanism of pathogenesis. In particular, the sequence of structural genes, as well as their processing and modification, may affect host specificity of the virus.
Intertype recombination events that have occurred in Europe (31) and Japan (32), among other places, remind us not to underestimate the danger posed by PToV from the possibility of cross-species infection. The mechanism of pathogenesis of PToV is still unclear, and its role during co-infections with other swine enteric pathogens such as PRV A, PAstV, PEDV, TGEV, PKV and Salmonella spp. is unknown (11, 30, 46). Considering the prevalence of asymptomatic PToV infections, more research is needed to explore whether it may aggravate the diseases caused by other swine pathogens.
Author Contributions
Z-MH, Y-WH wrote the manuscript. All other co-authors edited the manuscript.
Funding
This work was supported by The National Key Research and Development Program of China (2017YFD0500103), The Key Research and Development Program of Zhejiang province (2015C02021), and The National Natural Science Foundation of China (31872488).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the professional editing service of NB Revisions for technical preparation of the text prior to submission.
References
1. Weiss M, Steck F, Horzinek MC. Purification and partial characterization of a new enveloped RNA virus (Berne virus). J General Virol. (1983) 64 (Pt 9):1849–58. doi: 10.1099/0022-1317-64-9-1849
2. Woode GN, Reed DE, Runnels PL, Herrig MA, Hill HT. Studies with an unclassified virus isolated from diarrheic calves. Vet Microbiol. (1982) 7:221–40. doi: 10.1016/0378-1135(82)90036-0
3. Beards GM, Hall C, Green J, Flewett TH, Lamouliatte F, Du Pasquier P. An enveloped virus in stools of children and adults with gastroenteritis that resembles the Breda virus of calves. Lancet. (1984) 1:1050–2. doi: 10.1016/S0140-6736(84)91454-5
4. Kroneman A, Cornelissen LA, Horzinek MC, de Groot RJ, Egberink HF. Identification and characterization of a porcine torovirus. J Virol. (1998) 72:3507–11.
5. Weiss M, Steck F, Kaderli R, Horzinek MC. Antibodies to Berne virus in horses and other animals. Vet Microbiol. (1984) 9:523–31. doi: 10.1016/0378-1135(84)90014-2
6. Durham PJ, Hassard LE, Norman GR, Yemen RL. Viruses and virus-like particles detected during examination of feces from calves and piglets with diarrhea. Can Vet J. (1989) 30:876–81.
7. Wang J, Ma S, Liu C, Yu W, Wang M, Hao G. Identification of Torovirus-like particles in diarrheal pig samples. Chin J Vet Med. (1989) 8:2–3.
8. Penrith ML, Gerdes GH. Breda virus-like particles in pigs in South Africa. J S Afr Vet Assoc. (1992) 63:102.
9. Pignatelli J, Jimenez M, Grau-Roma L, Rodriguez D. Detection of porcine torovirus by real time RT-PCR in piglets from a Spanish farm. J Virol Methods. (2010) 163:398–404. doi: 10.1016/j.jviromet.2009.10.031
10. Pignatelli J, Jimenez M, Luque J, Rejas MT, Lavazza A, Rodriguez D. Molecular characterization of a new PToV strain. Evolutionary implications. Virus Res. (2009) 143:33–43. doi: 10.1016/j.virusres.2009.02.019
11. Shin DJ, Park SI, Jeong YJ, Hosmillo M, Kim HH, Kim HJ, et al. Detection and molecular characterization of porcine toroviruses in Korea. Arch Virol. (2010) 155:417–22. doi: 10.1007/s00705-010-0595-2
12. Zhou Y, Chen L, Zhu L, Xu Z. Molecular detection of porcine torovirus in piglets with diarrhea in southwest China. Sci World J. (2013) 2013:984282. doi: 10.1155/2013/984282
13. Matiz K, Kecskemeti S, Kiss I, Adam Z, Tanyi J, Nagy B. Torovirus detection in faecal specimens of calves and pigs in Hungary: short communication. Acta Vet Hung. (2002) 50:293–6. doi: 10.1556/AVet.50.2002.3.5
14. Anbalagan S, Peterson J, Wassman B, Elston J, Schwartz K. Genome sequence of torovirus identified from a pig with porcine epidemic diarrhea virus from the United States. Genome Announc. (2014) 2:14. doi: 10.1128/genomeA.01291-14
15. Horzinek MC, Ederveen J, Kaeffer B, de Boer D, Weiss M. The peplomers of Berne virus. J General Virol. (1986) 67 (Pt 11):2475–83. doi: 10.1099/0022-1317-67-11-2475
16. Snijder EJ, Horzinek MC. The molecular biology of toroviruses. In: Siddell SG, editors. The Coronaviridae. The Viruses. Boston, MA: Springer (1995). p. 219–38.
17. Snijder EJ, Horzinek MC. Toroviruses: replication, evolution and comparison with other members of the coronavirus-like superfamily. J General Virol. (1993) 74(Pt 11):2305–16.
18. Cornelissen LA, Wierda CM, van der Meer FJ, Herrewegh AA, Horzinek MC, Egberink HF, et al. Hemagglutinin-esterase, a novel structural protein of torovirus. J Virol. (1997) 71:5277–86.
19. Snijder EJ, den Boon JA, Spaan WJ, Verjans GM, Horzinek MC. Identification and primary structure of the gene encoding the Berne virus nucleocapsid protein. J General Virol. (1989) 70(Pt 12):3363–70. doi: 10.1099/0022-1317-70-12-3363
20. Snijder EJ, Horzinek MC, Spaan WJ. A 3'-coterminal nested set of independently transcribed mRNAs is generated during Berne virus replication. J Virol. (1990) 64:331–8.
21. Sun H, Lan D, Lu L, Chen M, Wang C, Hua X. Molecular characterization and phylogenetic analysis of the genome of porcine torovirus. Arch Virol. (2014) 159:773–8. doi: 10.1007/s00705-013-1861-x
22. Gallagher TM, Buchmeier MJJV. Coronavirus spike proteins in viral entry and pathogenesis. (2001) 279:371–4. doi: 10.1006/viro.2000.0757
23. Snijder EJ, Den Boon JA, Spaan WJ, Weiss M, Horzinek MC. Primary structure and post-translational processing of the Berne virus peplomer protein. Virology. (1990) 178:355–63. doi: 10.1016/0042-6822(90)90332-L
24. Den Boon JA, Snijder EJ, Locker JK, Horzinek MC, Rottier PJ. Another triple-spanning envelope protein among intracellularly budding RNA viruses: the torovirus E protein. Virology. (1991) 182:655–63. doi: 10.1016/0042-6822(91)90606-C
25. Pignatelli J, Alonso-Padilla J, Rodriguez D. Lineage specific antigenic differences in porcine torovirus hemagglutinin-esterase (PToV-HE) protein. Vet Res. (2013) 44:126. doi: 10.1186/1297-9716-44-126
26. de Groot RJ. Structure, function and evolution of the hemagglutinin-esterase proteins of corona- and toroviruses. Glycoconj J. (2006) 23(1–2):59–72. doi: 10.1007/s10719-006-5438-8
27. Langereis MA, Zeng Q, Gerwig GJ, Frey B, von Itzstein M, Kamerling JP, et al. Structural basis for ligand and substrate recognition by torovirus hemagglutinin esterases. Proc Natl Acad Sci USA. (2009) 106:15897–902. doi: 10.1073/pnas.0904266106
28. Horzinek MC, Ederveen J, Weiss M. The nucleocapsid of Berne virus. J General Virol. (1985) 66(Pt 6):1287–96. doi: 10.1099/0022-1317-66-6-1287
29. Zhou T, Zhou L, Zhou Y, Liu X, Xu Z, Zhu L, et al. Cloning and bioinformatics analysis of porcine torovirus N protein gene. Chin J Vet Sci. (2015) 35:845–50. doi: 10.16303/j.cnki.1005-4545.2015.06.01
30. Zhou L, Wei H, Zhou Y, Xu Z, Zhu L, Horne J. Molecular epidemiology of Porcine torovirus (PToV) in Sichuan Province, China: 2011–2013. Virol J. (2014) 11:106. doi: 10.1186/1743-422X-11-106
31. Smits SL, Lavazza A, Matiz K, Horzinek MC, Koopmans MP, de Groot RJ. Phylogenetic and evolutionary relationships among torovirus field variants: evidence for multiple intertypic recombination events. J Virol. (2003) 77:9567–77. doi: 10.1128/JVI.77.17.9567-9577.2003
32. Ito M, Tsuchiaka S, Naoi Y, Otomaru K, Sato M, Masuda T, et al. Whole genome analysis of Japanese bovine toroviruses reveals natural recombination between porcine and bovine toroviruses. Infect Genetics Evol. (2016) 38:90–5. doi: 10.1016/j.meegid.2015.12.013
33. Wang Y, Zhang W, Liu Z, Fu X, Yuan J, Zhao J, et al. Full-length and defective enterovirus G genomes with distinct torovirus protease insertions are highly prevalent on a Chinese pig farm. Arch Virol. (2018) 163:2471–6. doi: 10.1007/s00705-018-3875-x
34. Lee S, Lee C. First detection of novel enterovirus G recombining a torovirus papain-like protease gene associated with diarrhoea in swine in South Korea. Transboundary Emerg Dis. (2018) 66:1023–8. doi: 10.1111/tbed.13073
35. Conceicao-Neto N, Theuns S, Cui T, Zeller M, Yinda CK, Christiaens I, et al. Identification of an enterovirus recombinant with a torovirus-like gene insertion during a diarrhea outbreak in fattening pigs. Virus Evol. (2017) 3:vex024. doi: 10.1093/ve/vex024
36. Shang P, Misra S, Hause B, Fang Y. A naturally occurring recombinant enterovirus expresses a torovirus deubiquitinase. J Virol. (2017) 91:e00450–17. doi: 10.1128/JVI.00450-17
37. Tsuchiaka S, Naoi Y, Imai R, Masuda T, Ito M, Akagami M, et al. Genetic diversity and recombination of enterovirus G strains in Japanese pigs: high prevalence of strains carrying a papain-like cysteine protease sequence in the enterovirus G population. PLoS ONE. (2018) 13:e0190819. doi: 10.1371/journal.pone.0190819
38. Knutson TP, Velayudhan BT, Marthaler DG. A porcine enterovirus G associated with enteric disease contains a novel papain-like cysteine protease. J General Virol. (2017) 98:1305–10. doi: 10.1099/jgv.0.000799
39. Pignatelli J, Grau-Roma L, Jimenez M, Segales J, Rodriguez D. Longitudinal serological and virological study on porcine torovirus (PToV) in piglets from Spanish farms. Vet Microbiol. (2010) 146:260–8. doi: 10.1016/j.vetmic.2010.05.023
40. Brown DW, Beards GM, Flewett TH. Detection of Breda virus antigen and antibody in humans and animals by enzyme immunoassay. J Clin Microbiol. (1987) 25:637–40.
41. Gerber PF, Gong Q, Huang YW, Wang C, Holtkamp D, Opriessnig T. Detection of antibodies against porcine epidemic diarrhea virus in serum and colostrum by indirect ELISA. Vet J Engl. (2014) 202:33–6. doi: 10.1016/j.tvjl.2014.07.018
42. Hosmillo MD, Jeong YJ, Kim HJ, Collantes TM, Alfajaro MM, Park JG, et al. Development of universal SYBR Green real-time RT-PCR for the rapid detection and quantitation of bovine and porcine toroviruses. J Virol Methods. (2010) 168:212–7. doi: 10.1016/j.jviromet.2010.06.001
43. Gu F, Zhou Y, Huang J, Fan Y, Zhao Z, Qiao X, et al. Establishment and clinical application of a multiplex RT-PCR for detection of porcine kobuvirus, porcine astrovirus and porcine torovirus. Chin Vet Sci. (2015) 45:661–7. doi: 10.16656/j.issn.1673-4696.2015.07.001
44. Liu X, Zhou Y, Yang F, Liu P, Cai Y, Huang J, et al. Rapid and sensitive detection of porcine torovirus by a reverse transcription loop-mediated isothermal amplification assay (RT-LAMP). J Virol Methods. (2016) 228:103–7. doi: 10.1016/j.jviromet.2015.11.009
45. Alonso-Padilla J, Pignatelli J, Simon-Grife M, Plazuelo S, Casal J, Rodriguez D. Seroprevalence of porcine torovirus (PToV) in Spanish farms. BMC Res Notes. (2012) 5:675. doi: 10.1186/1756-0500-5-675
Keywords: porcine torovirus, etiology, epidemiology, diagnostic assays, recombination
Citation: Hu Z-M, Yang Y-L, Xu L-D, Wang B, Qin P and Huang Y-W (2019) Porcine Torovirus (PToV)—A Brief Review of Etiology, Diagnostic Assays and Current Epidemiology. Front. Vet. Sci. 6:120. doi: 10.3389/fvets.2019.00120
Received: 07 November 2018; Accepted: 29 March 2019;
Published: 18 April 2019.
Edited by:
Jesus Hernandez, Centro de Investigación en Alimentación y Desarrollo (CIAD), MexicoReviewed by:
Faten Abdelaal Okda, St. Jude Children's Research Hospital, United StatesTongling Shan, Shanghai Veterinary Research Institute (CAAS), China
Francisco Rivera-Benítez, Instituto Nacional de Investigaciones Forestales, Agrícolas y Pecuarias (INIFAP), Mexico
Copyright © 2019 Hu, Yang, Xu, Wang, Qin and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yao-Wei Huang, eWh1YW5nQHpqdS5lZHUuY24=
 Zhang-Min Hu
Zhang-Min Hu Yong-Le Yang
Yong-Le Yang Yao-Wei Huang
Yao-Wei Huang