- 1Optipharm, Inc., Cheongju-Si, South Korea
- 2Optipharm Animal Disease Diagnostic Center, Cheongju-Si, South Korea
Porcine circovirus type 2 (PCV2), the causative agent of porcine circovirus-associated diseases (PCVAD), poses a serious economic threat for the swine industry. Currently, PCV2 is classified into five major genotypes: PCV2a, PCV2b, PCV2c, PCV2d, and PCV2e. The aim of this study is to evaluate the performance of two commercially available methods, multiplex real-time PCR assay and PCR-reverse blot hybridization assay (REBA), for the rapid detection of PCV2 and direct identification of PCV2 genotypes from clinical samples as well as to compare the results with that of sequence analysis. Molecular diagnostic methods were used to evaluate a total of 180 samples, including tissues and blood samples from pigs that were suspected of PCVAD infection. The results of this study showed that the detection rate for positive PCV2 was 48.3% (n = 87) in both multiplex real-time PCR and PCR-REBA methods. Using sequence analysis, which is the gold standard, and multiplex real time PCR assay, the sensitivity, specificity, positive predictive value, and negative predictive value of PCV2 genotyping were found to be 97.1% (n = 67, 95% CI 0.894–0.998, p < 0.001), 100% (n = 93, 95% CI 0.966–1.000, p < 0.001), 100% (95% CI 0.953–1.000, p < 0.001), 97.9% (95% CI 0.921–0.998, p < 0.001), respectively. The results of PCR-REBA were found to be consistent with those of sequence analysis for all the samples and showed good agreement (κ = 1). The most prevalent genotypes detected in this study were PCV2d (n = 53, 60.9%), followed by PCV2a (n = 17, 19.5%), PCV2b (n = 14, 16.1%), and PCV2a/b co-infection (n = 3, 3.5%). Both the methods required ~3 h for completion. Therefore, we conclude that two molecular methods are rapid and reliable for the characterization of the causative pathogen with PCV2 genotypes.
Introduction
Different from the non-pathogenic porcine circovirus (PCV) type 1 strain (1), PCV type 2 is considered to be an important emerging pathogen that causes porcine circovirus associated diseases (PCVAD) including postweaning multisystemic wasting syndrome (PMWS), porcine dermatitis nephropathy syndrome (PDNS), porcine respiratory disease complex (PRDC), enteritis, reproductive failure (2, 3), and one of the most economically important swine diseases worldwide (4). Whenever there are outbreaks of respiratory clinical signs, wasting, and granulomatous inflammation of lymphoid tissues in pigs, PMWS is clinically suspected (5). PCV2-associated systemic infection is clinically characterized by wasting, dyspnea, and lymphadenopathy, and in some cases, might be associated with diarrhea, pallor, and jaundice (6). PCV2, belonging to the genus Circovirus of the family Circoviridae, is a small non-enveloped virus with a circular single-stranded DNA genome (7). The PCV2 genome is ~1.7 kb nucleotide long and encodes for two open reading frames (ORFs) (8, 9). In the viral genome, ORF1 codes for the replicase (Rep) protein and ORF2 for the capsid (Cap) protein. Rep is a non-structural protein and is responsible for the viral replication, while the structural Cap protein controls the immunogenicity of the virus (10–13). With emerging viral strains, PCV2 has undergone much genetic variation in recent years and has been divided into five genotypes, namely PCV2a-e strains, which are classified based on the diversity level of the ORF2 nucleotide sequences (14, 15). Continuous mutations in the PCV2 genome have made the identification of PCV more difficult, especially by traditional molecular detection methods (16, 17). It has also been demonstrated experimentally that subclinical PCV2 infection might be associated with decreased vaccine efficacy (18). Therefore, PCV2 subclinical infection is not only the most common form of infection in pigs but is also resistant to the effect of vaccines. Hence, rapid and early identification of PCV2 subclinical infection is very important for the effective prophylaxis against PCVAD (19).
Until now, commercial diagnostic tests based on ELISA (9, 12) and PCR (19) have only been developed to confirm the presence or absence of PCV2. Although it has the advantage of being able to detect PCV2 in a short time, it is required expensive antibodies for diagnostic purposes, and the PCV2 genotypes cannot be distinguished simultaneously, so most PCV2 genotypes have been identified separately using PCR-based Restriction Fragment Length Polymorphism (RFLP) (20) or sequence analysis (13, 17, 20–22). In this study, a novel diagnostic assay based on multiplex real-time PCR (Opti PCV2-genotyping; Optipharm, Osong, Republic of Korea) was developed for the rapid and accurate identification of PCV2 as well as to discriminate between the PCV2 a/e, b, and d genotypes. The PCR-based reverse blot hybridization assay (PCR-REBA, REBA PCV2-genotyping; Optipharm) was to detect PCV2 and distinguish between PCV2a, PCV2b, PCV2c, PCV2d, and PCV2e genotypes. In this study, we evaluated the clinical applicability of multiplex real-time PCR and PCR-REBA methods and compared their efficiency to that of the sequence analysis method used for detecting PCV2 and differentiating the different PCV2a-e genotypes directly from the serum and tissue samples of pigs.
Methods
Preparation of DNA Samples
To evaluate the diagnostic performance of the multiplex real-time PCR and PCR-REBA methods, a total of 180 samples suspected to be infected with PCVAD including 109 tissues and 71 bloods were provided by the Optipharm Animal Disease Diagnostic Center, which was commissioned from January to December, 2019. DNA was extracted from 200 μL of serum or 20 mg of organ tissue homogenate using a commercial automated system (Miracle-AutoXT Automated Nucleic Acid Extraction System, intronbio, Seongnam, Republic of Korea) according to the manufacturer's recommendations. To avoid cross contamination, all the samples were processed individually and stored at −20°C. The content and purity of the extracted DNA were assayed by measuring absorbance at 260 and 280 nm using an Infinite 200 NanoQuant (Tecan, Switzerland) spectrophotometer.
Multiplex Real-Time PCR Assay
Detection of PCV2 and identification of genotypes in clinical samples was performed with Opti PCV2-genotyping (Optipharm), a quantitative multiplex real-time PCR-based assay, using the CFX-96 real-time PCR system (Bio-Rad, Hercules, CA, USA) for thermocycling and fluorescence detection. Both the detection and genotype identification of PCV2 can be performed in a single tube using this assay [PCV2 (Cy5), PCV2a/e (FAM), PCV2b/d (CAL Flour Red 610), and PCV2d (HEX)] by incorporating specific TaqMan probes labeled with different fluorophores. Real-time PCR amplification was performed in a total reaction volume of 20 μL containing 10 μL of 2 × Thunderbird probe qPCR mix (Toyobo, Osaka, Japan), 5 μL of a mixture of primer and TaqMan probe that were labeled with different fluorophores, and 5 μL template DNA. The real-time PCR kits consisted of an internal control (IC) DNA, which was used to indicate successful nucleic acid extraction, the quality of the sample and to check for the presence of PCR inhibitors in the reaction. The IC DNA is designed to have minimal sequence similarity with the target gene and also facilitates detection of false negatives. Therefore, it does not directly compete with the amplification of the species-specific target in multiplex real-time PCR. Positive (Plasmid DNA with mixed PCV2a, b, d genotypes) and negative controls consisting of molecular grade (DNAse/RNAse-free) water (Ultra pure water; Welgene, Gyeongsan, Republic of Korea) without template DNA were included in each assay and the assay was performed under the following conditions: 95°C for 3 min followed by 40 cycles of 95°C for 20 s and 55°C for 40 s. Each sample was tested in duplicate by running the PCR cycles twice. The viral load was quantified by determining the cycle threshold (CT), and the number of PCR cycles required for the fluorescence to exceed a value significantly higher than the background fluorescence. Positive result was indicated when the CT value was <38.
PCR-Reverse Blot Hybridization Assay (PCR-REBA)
Oligonucleotide primers corresponding to both strands of the ORF2 region of PCV2 (Figure 1) were designed by Primer3Plus (http://www.bioinformatics.nl/cgi-bin/primer3plus/primer3plus.cgi). The primers were made as probes corresponding to the complementary strand and were used exclusively thereafter. To validate the efficiency of the selected probes, target DNA samples amplified from the PCV2 strains were applied to the REBA membrane strips and spotted with the selected probes. Two types of DNA samples (PCV2c and PCV2e) were synthesized (Bioneer, Daejeon, Republic of Korea) and amplified with custom PCR primers (PCV2c, F-5′-TAAGTGGGGGGTCTTTAAGA-3′ and R-5′-TCCTCCGCCGCCGCCCCTGG-3′; PCV2e, F-5′-TAAGTGGGGGGTCTTTAA-3′ and R-5′-CTTGGCCATATCCTCCGCC-3′), resulting in amplicons of 630 and 640 bp, respectively. The resultant products were mutagenized after subcloning into the pBHA vector. Two plasmids were extracted from the transformants, and the mutated sequences were confirmed by sequence analysis (CosmoGenetech, Daejeon, Republic of Korea). PCR was performed using a 20 μL reaction mixture (GeNet Bio, Daejeon, Republic of Korea) containing 2 × master mix (10 μL), 2 μL of primer mixture, 5 μL sample DNA, and 3 μL Ultra pure water (Welgene) to make up the final volume. The reactions were run on a Verity thermocycler (Applied Biosystems, CA, USA) under the following conditions: one cycle at 94°C for 5 min, followed by 35 cycles of 94°C for 30 s, annealing 60°C for 30 s, initial extension at 72°C for 30 s, and a final extension of 72°C for 10 min to complete the synthesis of all strands. The amplified target was visualized as a single band corresponding to a length of 620 bp using the ChemiDoc system (Vilber Lourmat, Eberhardzell, Germany).
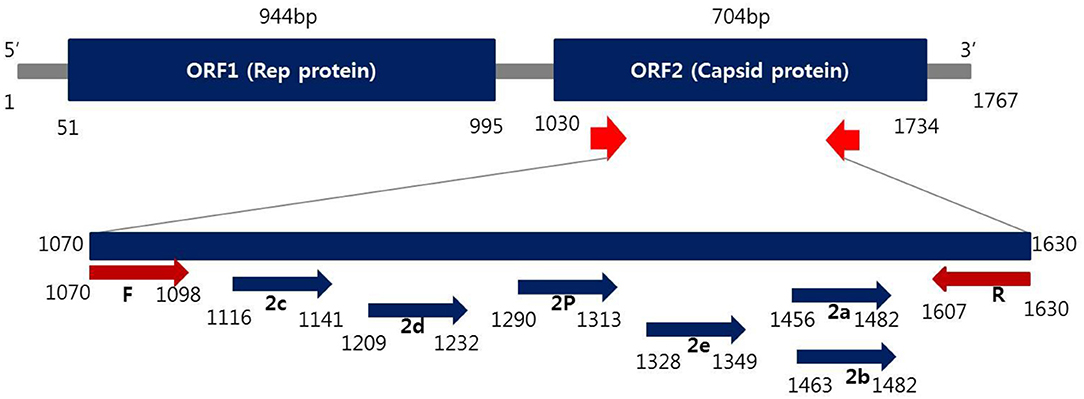
Figure 1. Schematic representation of the ORF2 gene to distinguish PCV2 genotyping from positions of primer and probes used in this study.
For REBA PCV2 genotyping, the hybridization and washing processes were performed as follows; each sample was tested in duplicate and all PCR-REBA runs were performed twice. In brief, biotinylated PCR products were denatured at 25°C for 5 min in denaturation solution and then the denatured single-stranded PCR products suspended in hybridization solution were incubated with REBA PCV2-genotyping membrane strips at 55°C with shaking at 90 rpm in a blotting tray for 30 min. The strips were then washed twice with gentle shaking in 1 ml of washing solution for 10 min at 55°C, incubated at 25°C with 1:2,000 diluted streptavidin-alkaline phosphatase (AP) conjugate (Roche Diagnostics, Mannheim, Germany) in conjugate diluent solution (CDS) for 30 min, and finally washed twice with 1 ml CDS at room temperature for 1 min. The colorimetric hybridization signals were visualized by adding a 1:50 dilution of nitro blue tetrazolium chloride/5-bromo-4-chloro-3-indolyl (NBT/BCIP) AP-mediated staining solution (Roche Diagnostics), and then incubated until a color change was detected. Finally, the band pattern was read and interpreted visually.
Sequence Analysis
To confirm the results of the two molecular diagnostic methods, the PCR amplicons of all the clinical samples were sequenced using an ABI 3730 automated DNA sequencer (Applied Biosystems, Foster City, CA, USA) and the ABI Prism BigDye Terminator (Applied Biosystems) system (CosmoGenetech, Republic of Korea). The primer set used to amplify the target ORF2 gene was 5′-TCTGAATTGTACATACATRGTTAYACGG-3′ (1070F) and 5′- TACCGYTGGAGAAGGAAAAATGG-3′ (1630R), which resulted in a 560-bp PCR product. The obtained sequences were compared with sequences in the National Center for Biotechnology Information (NCBI) GenBank database for species identification.
Results
Analytical Sensitivity and Specificity of the Multiplex Real-Time PCR and PCR-REBA Methods
Analytical sensitivity of the two molecular methods for the detection of PCV2 was determined by using 10-fold diluted (1 ng, 100 pg, 10 pg, 1 pg, 100 fg, 10 fg, and 1 fg) samples for obtaining the standard curve for the DNA extracted from PCV2 strains. The detection limit of the multiplex real-time PCR assay for PCV2-P (PCV2 species-specific probe), PCV2a/e-P (PCV2a/e genotype-specific probe), PCV2b/d-P (PCV2b/d genotype-specific probe), PCV2c (PCV2c genotype-specific probe), PCV2d-P (PCV2d genotype-specific probe), and PCV2e-P (PCV2e genotype-specific probe) ranged from 100 to 10 fg DNA per reaction. The CT values for PCV2-P, PCV2a/e, PCV2b/d, PCV2c, PCV2d, and PCV2e for each DNA concentration ranged from 17 to 36.8, 16.5 to 34.9, 16.7 to 35.1, 16.1 to 35, 16.8 to 35.3, and 15.5 to 33.1, respectively (Supplementary Figures 1A–D). The PCR-REBA detection limit for PCV2a, PCV2b, PCV2c, PCV2d, and PCV2e was ~100 fg to 10 fg DNA per reaction (Supplementary Figures 1E–J). In addition, the detection limit for mixed co-infection PCV2 subtypes in multiplex real-time PCR was 1 pg DNA per reaction, and the CT value was found to be 17.97–32.86 (Data not shown). The PCR-REBA detection limit for mixed co-infection PCV2 subtypes detected ~100 fg DNA per reaction (Supplementary Figure 1K).
To determine the specificity of the two molecular assays, primers and probes for detecting PCV2 positive genotypes were used for testing 55 DNA samples, respectively, extracted from specific pathogen-free swine serum samples and used as negative controls. The multiplex real-time PCR and PCR-REBA assay for detecting PCV2 positive genotypes yielded negative results with all strains except PCV2 strains (including mixed co-infection PCV2 subtypes), hence, the cross reactivity was not detected (Supplementary Table 1).
Detection of PCV2 DNA Using Multiplex Real-Time PCR and PCR-REBA Methods in Clinical Samples
To evaluate the performance of the multiplex real-time PCR and PCR-REBA assay, a total of 180 clinical samples including tissue (n = 109, 60.6%) and whole blood (n = 71, 39.4%) were analyzed. Of 180 clinical samples, 87 (48.3%) samples were positive for PCV2, and 93 (51.7%) samples were negative as detected by both multiplex real-time PCR and PCR-REBA (Table 1).

Table 1. Detection of porcine circovirus 2 DNA in 180 clinical samples suspected of PCVAD infection using the multiplex real-time PCR and PCR-REBA assay.
Multiplex Real-Time PCR and PCR-REBA Methods for the Detection of PCV2 Genotyping in Clinical Samples
Of the 87 PCV2 positive samples, 53 (60.9%), 17 (19.5%), 12 (13.8%), and 3 (3.5%) samples showed positive fluorescence signals for PCV2d, PCV2a, PCV2b, and PCV2a/b co-infections, respectively, as evaluated using multiplex real-time PCR assay (Figure 2A) while no PCV2 genotypes were detected in 2 cases (2.3%). All the clinical samples showed positive IC signals and the CT values of the 87 positive and 93 negative samples ranged from 23.62 to 32.7 (mean 24.89, SD ± 1) and 22.47 to 33.4 (mean 25.36, SD ± 0.47), respectively. The CT values of the PCV2d, PCV2a, PCV2b, and PCV2a/b co-infections samples ranged from 15.75 to 35.08 (mean 23.43, SD ± 5.6), 20.97 to 33.85 (mean 26.5, SD ± 4.07), 22.89 to 34.37 (mean 28.81, SD ± 5.78), and 21.2 to 30.73 (SD ± 2.4), respectively. PCR-REBA, which is another method for performing the molecular identification of PCV2 genotypes, was performed with the same clinical samples (Figure 2B). Of the 87 positive samples, the following PCV2 genotypes were identified using PCR-REBA: PCV2d was the most prevalent at 60.9% (n = 53), followed by PCV2a (n = 17, 19.5%), PCV2b (n = 14, 16.1%), and PCV2a/b co-infections (n = 3, 3.5%), respectively (Table 2).
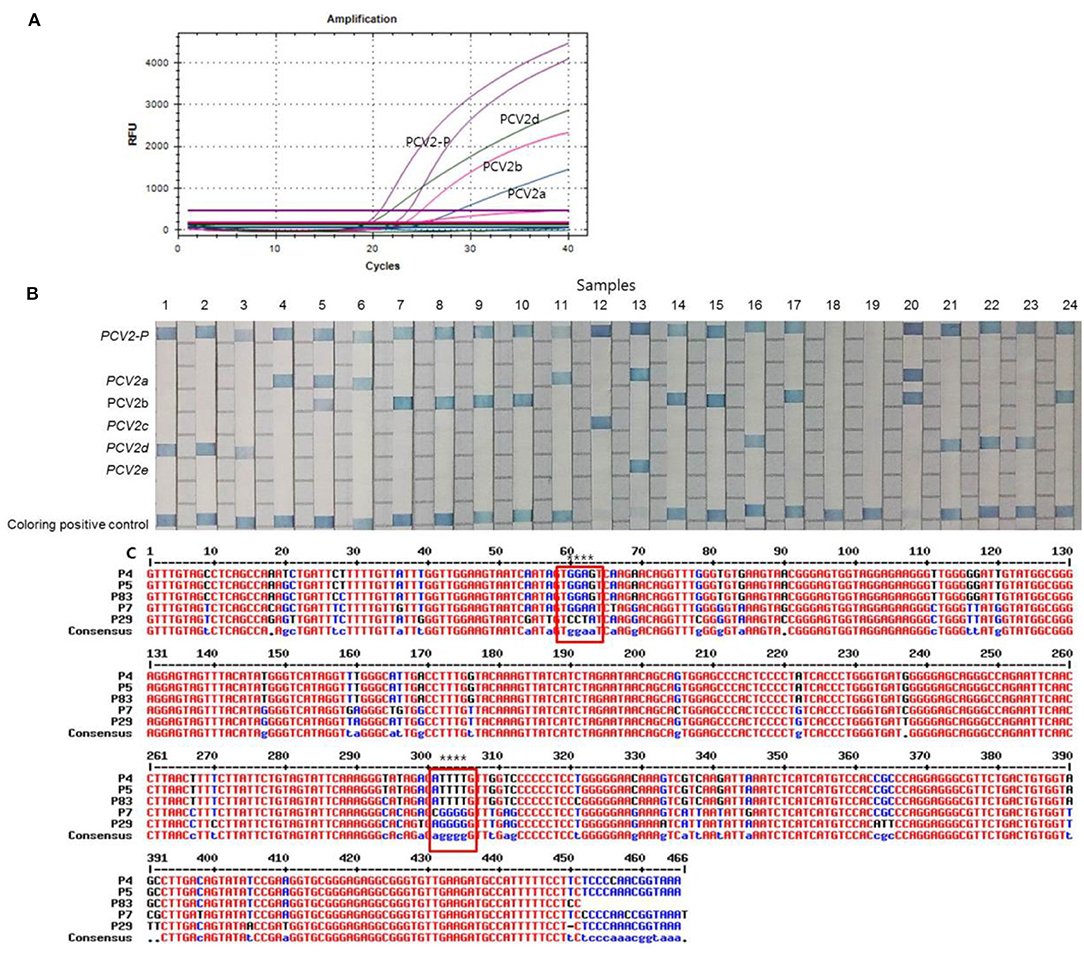
Figure 2. Typical results of the multiplex real-time PCR, PCR-REBA, and sequence analysis with clinical samples. (A) Overall results for PCV2-positive, PCV2a, PCV2b, and PCV2d. Fluorescent dyes of specific TaqMan probes for multiplex real-time PCR were used PCV2 (Cy5), PCV2a/e (FAM), PCV2b/d (CAL Flour Red 610), and PCV2d (HEX), respectively. (B) Results of PCR-REBA; Lanes 1–3, 16, 21–23: PCV2d; lanes 4, 6, and 11: PCV2a; lanes 5 and 20: PCV2a and PCV2b co-infection; lane 7–10, 14–15, 17, and 24: PCV2b; lane 12: PCV2c; lane 13: PCV2e; lane 18 and 19: negative. PCV2c and PCV2e were used to synthesize the DNA as a control. (C) Sequence alignment results of a fragment of the genomic sequence of the clinical samples; P4, PCV2a; P7, PCV2b, P29, PCV2d, P5 and P83 showed that the two samples detected as PCV2a/b co-infection positive by the multiplex real-time PCR and PCR-REBA methods were shown as only PCV2a positive by sequence analysis; The red boxes indicate the position where three genotypes (PCV2a, 2b, and 2d) can be identified.
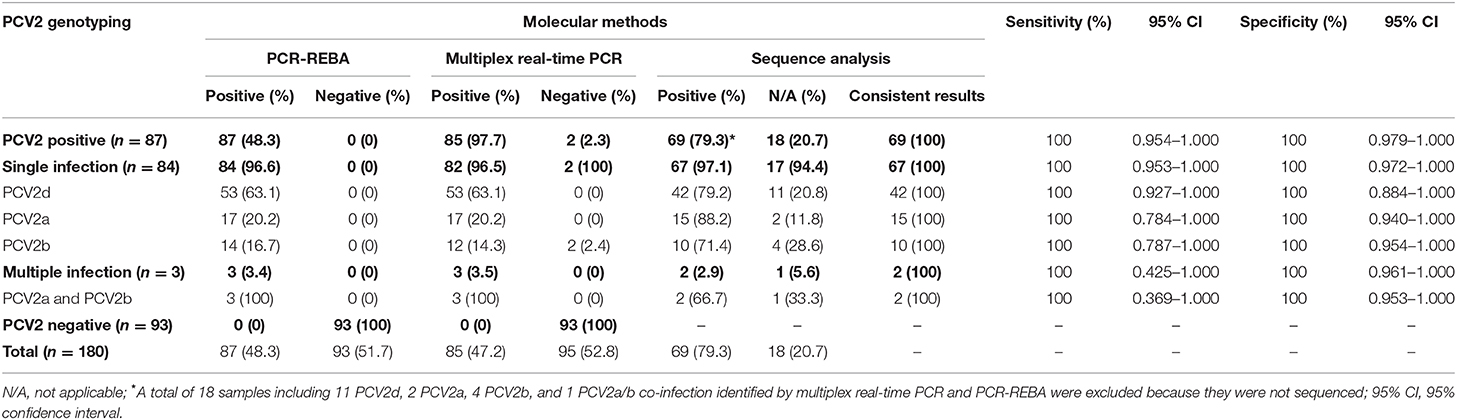
Table 2. Comparison of multiplex real-time PCR, PCR-REBA, and sequence analysis results for the detection of PCV2 genotypes in 180 clinical samples suspected of PCVAD.
Comparison of the Results Between the Two Molecular Assays and Sequence Analysis for Identification of the PCV2 Genotypes in the Clinical Samples
To confirm the results obtained from the multiplex real-time PCR and PCR-REBA assay, sequence analysis was performed using the same clinical samples (Figure 2C). Only 69 (79.3%) of the 87 PCV2 positive samples could be identified for PCV2 genotyping by sequence analysis. Therefore, 18 samples including 11 PCV2d, 2 PCV2a, 4 PCV2b, and 1 PCV2a/b co-infected samples identified by multiplex real-time PCR and PCR-REBA were excluded from the comparative analysis. The results of the multiplex real-time PCR and sequence analysis methods were consistent except for two cases. In these two cases, while PCV2 genotypes were not detected using multiplex real-time PCR, the samples were identified as PCV2b-positive by sequence analysis. The results of the PCR-REBA for PCV2 genotyping of all 69 samples were consistent with the sequencing results (Table 2). The phylogenetic tree was constructed using Phylogeny.fr software (23) after alignment of the 69 sequenced results. Analysis of the phylogenetic tree indicated that the sequences could be divided into three genotypes (PCV2d, PCV2a, and PCV2b), which accounted for 60.9, 24.6, and 14.5%, respectively (Figure 3).
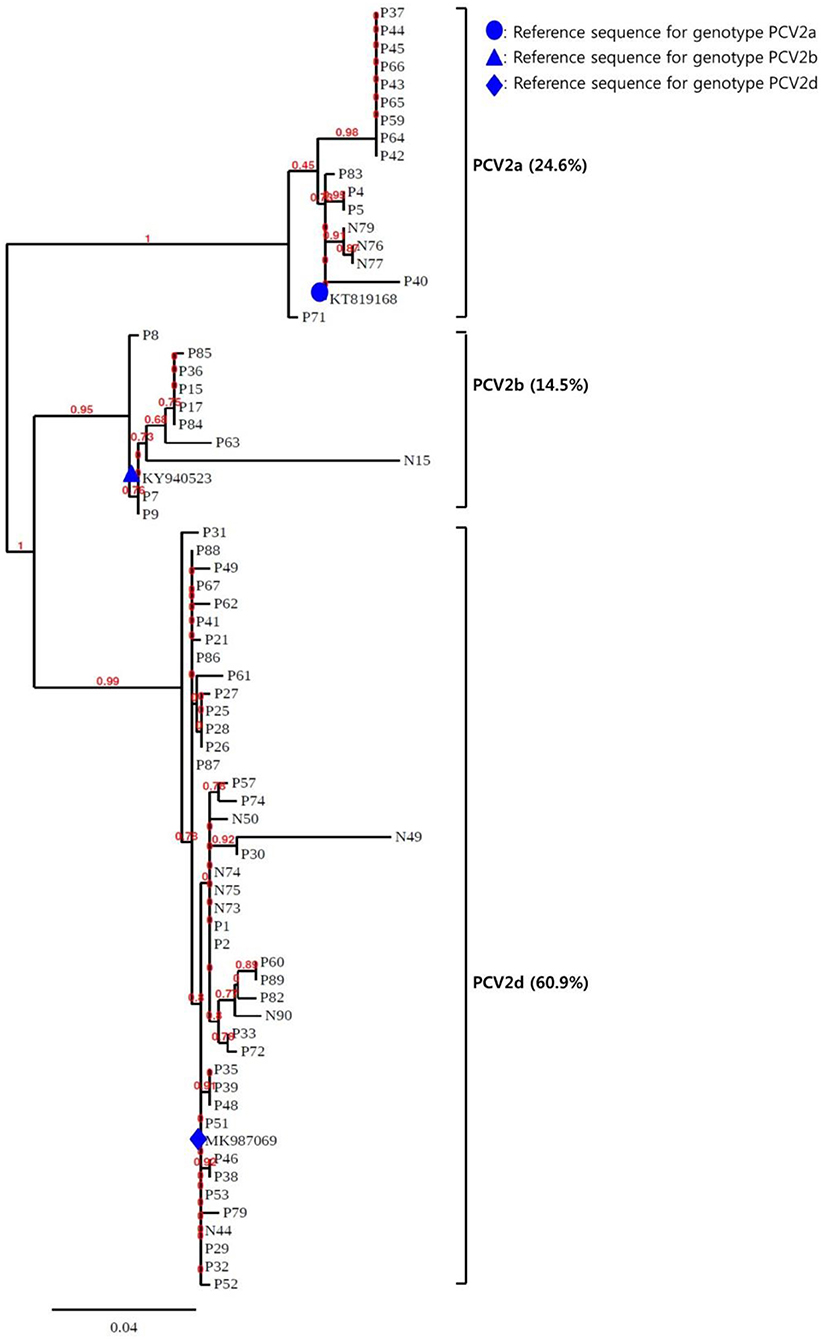
Figure 3. Phylogenetic analysis of the 69 PCV2 isolates. The phylogenetic tree was constructed using Phylogeny.fr software after alignment of the 69 sequenced results. The PCV2genomes were mainly assigned to three genotypes (PCV2a, PCV2b, and PCV2d).
Discussion
Although the severity of the economic losses caused by PCV2 infection has been mitigated by vaccination, PCVAD is still detected quite often and is an important porcine pathogen. It is important to distinguish between the PCV2 genotypes for the laboratory diagnosis of PCV2 as different PCV2 genotypes have been found in samples from pigs affected with other PCVAD (24). The currently used PCR-based methods for diagnosis, including nucleotide sequencing, PCR, nested PCR, and PCR-RFLP (25–28), are time consuming for general veterinary clinical applications; they can only be used in specialized diagnostic institutes as these methods require specialized equipment and reagents, and need to be inspected by professionals to differentiate the subtype virus. Therefore, there is an increasing requirement for diagnostic techniques that can be complemented with traditional methods in clinical diagnostic laboratories.
In the present study, two molecular assays were developed for detecting PCV2 genotypes. The purpose of the present study was to evaluate the clinical efficacy of the multiplex RT-PCR assay (Opti PCV2-genotyping) and PCR-REBA (REBA PCV2-genotyping) for rapid and accurate detection as well as identification of PCV2 genotypes based on the ORF2 regions. We also compared the results of these two molecular assays with those obtained from conventional methods such as sequence analysis. The characteristics of the multiplex real-time PCR and PCR-REBA used in this study were similar as they were both rapid (turnaround time of 2–3 h), sensitive, specific, and comparatively easy to perform without requiring any specialized laboratory equipment, other than a PCR machine and water bath.
The Opti PCV2-genotyping assay is designed to simultaneously detect PCV2 and to distinguish PCV2a/e, PCV2b, and PCV2d genotypes. Multiplex real-time PCR, a recognized technique, is faster and more effective for the rapid detection of bacterial or viral infection compared to conventional PCR and other detection methods. Multiplex real-time PCR assay is a rapid method with a turnaround time of ~1.5–2 h, which includes 30 min for DNA preparation and 1.5 h for target DNA amplification. The combination of excellent sensitivity and specificity as well as ease of handling enable rapid and simultaneous detection of multiple species, and minimizes the possibility of contamination by eliminating the need for additional post-PCR processing of the samples, which has made this technology appealing for clinical microbiology laboratory applications (29). PCR-REBA is a highly sensitive and specific probe-based method in which multiple oligonucleotide probes are immobilized on nitrocellulose strips, hybridized with biotin-labeled PCR products, and can be used to derive rapid results within 4 h (30). In addition to the time required for target DNA amplification (1.5 h), PCR-REBA is a 3-step process with a hybridization step (30 min), a washing step (20 min), and a chromogenic detection and data interpretation step (40 min). It also requires a fully automated system for the washing, hybridization, and interpretation steps. The PCR-REBA molecular diagnostic assay can be used to isolate all types of PCV2 genotypes as well as detect PCV2 directly from serum or tissue samples. In addition, PCR-REBA has the advantage of the flexibility to add more specific-probes to the membrane strip for increasing the range of PCV2 genotypes detected.
In this study, the concordance rate of the multiplex real-time PCR assay and sequence analysis was 98.8% (95% confidence interval [CI] 0.953–0.999, p < 0.001). Using sequence analysis as the gold standard, the sensitivity, specificity, and positive and negative predictive values of the PCV2 genotyping results by multiplex real-time PCR assay were 97.1% (n = 67, 95% CI 0.894–0.998, p < 0.001), 100% (n = 93, 95% CI 0.966–1.000, p < 0.001), 100% (95% CI 0.953–1.000, p < 0.001), 97.9% (95% CI 0.921–0.998, p < 0.001), respectively. The results of PCR-REBA were found to be consistent with those of sequence analysis and showed good agreement (κ = 1).
Studies have shown that the most common PCV2 genotypes detected worldwide are PCV2b (53.1%) and PCV2a (34.4%) in Taiwan (24), PCV2b (87.5%) and PCV2a (12.5%) in Mexico (31), and PCV2d (45.3%) and PCV2b (41.1%) in China (13). In this study, the most prevalent genotypes detected were PCV2d (n = 53, 60.9%), followed by PCV2a (n = 17, 19.5%), PCV2b (n = 14, 16.1 %), and PCV2a/b co-infection (n = 3, 3.5%). Co-infection of PCV2a and PCV2b in clinical samples has been suggested to be the primary cause of other PCVAD while dual heterologous infection of PCV2a and PCV2b in gnotobiotic pigs has been shown to induce severe clinical symptoms (24, 32). Therefore, rapid identification of co-infection of PCV2a and PCV2b is crucial. Generally, when identified samples from dually infected pigs were sequenced, only the predominant PCV2 genotype was detected. Our results also showed that only PCV2a could be identified by sequence analysis method in the three samples in which PCV2a/b co-infection was detected using the two molecular diagnostic methods.
There are potential limitations in this study. Firstly, the multiplex real-time PCR assay cannot distinguish between PCV2a type and PCV2e type, and does not include PCV2c type that has not yet been detected in Korea. Therefore, there should be an additional tube to include all of these genotypes. Secondly, although PCR-REBA detect all other genotypes in addition to PCV2, additional steps are required after PCR. Thirdly, the PCV2c and PCV2e genotypes were not detected in this study and further investigations of the samples are required.
Conclusions
The two recently developed molecular assays are accurate, rapid, and convenient tools for identifying PCV2. These assays can also discriminate between the PCV2 genotypes and directly detect PCV2 from clinical samples in only 2–3 h. Therefore, these two molecular assays can provide essential information that can help expedite therapeutic decisions for early and appropriate vaccinations during the acute phase of PCV2 infection. We believe that these assays can reduce the labor and time for PCV2 diagnosis in industrial animal area.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation, to any qualified researcher.
Ethics Statement
All samples used in this study were animal diagnostic samples submitted by the clients and there was no animal handling involved.
Author Contributions
HW performed evaluation of the experiments, analyzed the data, and drafted the manuscript. JS and SS provide clinical samples and clinical information. HK revised the manuscript. All authors have read and approved the final manuscript.
Conflict of Interest
All authors were employed by Optipharm, Inc during the preparation and execution of the study.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2020.00200/full#supplementary-material
Supplementary Figure 1. Detection limits of the multiplex real-time PCR and PCR-REBA methods evaluated using 10-fold serial diluted samples. Serially diluted PCV2-P, PCV2a/e, PCV2b, and PCV2d DNA samples ranging from 1 ng to 1 fg per reaction were used to determine the detection limit of the multiplex real-time PCR and PCR-REBA methods. In the multiplex real-time PCR assay, the amplification curve of the specific probe (A) for detecting PCV2 (R2 = 0.997), PCV2a/e probe (B), PCV2b probe (C), and PCV2d probe (D), for detecting PCV2 genotypes (R2 = 0.999) are shown. The overall detection limit of this assay for the PCV2 genotypes ranged from ~100 to 10 fg DNA per reaction. CT was plotted against the input of the quantity of PCV2, 2a/e, 2b, 2c, 2d, and 2e DNA (repeated 40 times). The intensity of fluorescence is shown on the Y-axis (R2 = reporter signal/passive reference signal). RFU, relative fluorescence unit and R2, fluorescence units. Serially diluted PCV2-P (E), PCV2a (F), PCV2b (G), PCV2c (H), PCV2d (I), PCV2e (J), and 5 mixed co-infection PCV2 subtypes (K) with DNA amounts from 1 ng (lane 1), 100 pg (lane 2), 10 pg (lane 3), 1 pg (lane 4), 100 fg (lane 5), 10 fg (lane 6), and 1 fg (lane 7) were used to determine the detection limit of the PCR-REBA (E–J). N, negative control. PCV2c and PCV2e used synthesized DNA as a control.
Supplementary Table 1. Analytical specificity of the multiplex real-time PCR and PCR-REBA assay for detecting PCV2 and PCV2 genotypes with 25 strains and 30 normal serum samples, respectively, obtained from pigs. ATCC, American type culture collection, PCV2, Porcine circovirus type2.
References
1. Olvera A, Cortey M, Segalés J. Molecular evolution of porcine circovirus type 2 genomes: phylogeny and clonality. Virology. (2007) 357:175–85. doi: 10.1016/j.virol.2006.07.047
2. Opriessnig T, Meng XJ, Halbur PG. Porcine circovirus type 2 associated disease: update on current terminology, clinical manifestations, pathogenesis, diagnosis, and intervention strategies. J Vet Diagn Invest. (2007) 19:591–615. doi: 10.1177/104063870701900601
3. Qiu X, Li T, Zhang G, Cao J, Jin Y, Xing G, et al. Development of a loop-mediated isothermal amplification method to rapidly detect porcine circovirus genotypes 2a and 2b. Virol J. (2012) 9:318. doi: 10.1186/1743-422X-9-318
4. Chae C. Postweaning multisystemic wasting syndrome: a review of aetiology, diagnosis and pathology. Vet J. (2004) 168:41–9. doi: 10.1016/S1090-0233(03)00182-5
5. Rosell C, Segales J, Plana-Duran J, Balasch M, Rodriguez-Arrioja GM, Kennedy S, et al. Pathological, immunohistochemical, and in-situ hybridization studies of natural cases of postweaning multisystemic wasting syndrome (PMWS) in pigs. J Comp Pathol. (1999) 120:59–78. doi: 10.1053/jcpa.1998.0258
6. Harding JC. The clinical expression and emergence of porcine circovirus 2. Vet Microbiol. (2004) 98:131–5. doi: 10.1016/j.vetmic.2003.10.013
7. Allan GM, Ellis JA. Porcine circoviruses: a review. J Vet Diagn Invest. (2000) 12:3–14. doi: 10.1177/104063870001200102
8. Hamel AL, Lin LL, Nayar GP. Nucleotide sequence of porcine circovirus associated with postweaning multisystemic wasting syndrome in pigs. J Virol. (1998) 72:5262–7. doi: 10.1128/JVI.72.6.5262-5267.1998
9. Nainys J, Lasickiene R, Petraityte-Burneikiene R, Dabrisius J, Lelesius R, Sereika V, et al. Generation in yeast of recombinant virus-like particles of porcine circovirus type 2 capsid protein and their use for a serologic assay and development of monoclonal antibodies. BMC Biotechnol. (2014) 14:100. doi: 10.1186/s12896-014-0100-1
10. Cheung AK. The essential and nonessential transcription units for viral protein synthesis and DNA replication of porcine circovirus type 2. Virology. (2004) 313:452–9. doi: 10.1016/S0042-6822(03)00373-8
11. Mankertz A, Mankertz J, Wolf K, Buhk HJ. Identification of a protein essential for replication of porcine circovirus. J Gen Virol. (1998) 79:381–4. doi: 10.1099/0022-1317-79-2-381
12. Nawagitgul P, Harms PA, Morozov I, Thacker BJ, Sorden SD, Lekcharoensuk C, et al. Modified indirect porcine circovirus (PCV) type 2-based and recombinant capsid protein (ORF2)-based enzyme-linked immunosorbent assays for detection of antibodies to PCV. Clin Diagn Lab Immunol. (2002) 9:33–40. doi: 10.1128/CDLI.9.1.33-40.2002
13. Yao J, Qin Y, Zeng Y, Ouyang K, Chen Y, Huang W, et al. Genetic analysis of porcine circovirus type 2 (PCV2) strains between 2002 and 2016 reveals PCV2 mutant predominating in porcine population in Guangxi, China. BMC Vet Res. (2019) 15:118. doi: 10.1186/s12917-019-1859-z
14. Franzo G, Tucciarone CM, Cecchinato M, Drigo M. Porcine circovirus type 2 (PCV2) evolution before and after the vaccination introduction: a large scale epidemiological study. Sci Rep. (2016) 6:39458. doi: 10.1038/srep39458
15. Xiao CT, Harmon KM, Halbur PG, Opriessnig T. PCV2d-2 is the predominant type of PCV2 DNA in pig samples collected in the U.S. During 2014-2016. Vet Microbiol. (2016) 197:72–7. doi: 10.1016/j.vetmic.2016.11.009
16. Eddicks M, Fux R, Szikora F, Eddicks L, Majzoub-Altweck M, Hermanns W, et al. Detection of a new cluster of porcine circovirus type 2b strains in domestic pigs in Germany. Vet Microbiol. (2005) 176:337–43. doi: 10.1016/j.vetmic.2015.01.013
17. Yang S, Yin S, Shang Y, Liu B, Yuan L, Zafar Khan MU, et al. Phylogenetic and genetic variation analyses of porcine circovirus type 2 isolated from China. Transbound Emerg Dis. (2018) 65:e383–92. doi: 10.1111/tbed.12768
18. Opriessnig T, McKeown NE, Harmon KL, Meng XJ, Halbur PG. Porcine circovirus type 2 infection decreases the efficacy of a modified live porcine reproductive and respiratory syndrome virus vaccine. Clin Vaccine Immunol. (2006) 13:923–9. doi: 10.1128/CVI.00074-06
19. Huang Y, Zhang X, Du Q, Wang F, Zhao X, Zhang W, et al. Preclinical detection of porcine circovirus type 2 infection using an ultrasensitive nanoparticle DNA probe-based PCR assay. PLoS ONE. (2014) 9:e97869. doi: 10.1371/journal.pone.0097869
20. Guo LJ, Lu YH, Wei YW, Huang LP, Liu CM. Porcine circovirus type 2 (PCV2): genetic variation and newly emerging genotypes in China. Virol J. (2010) 7:273. doi: 10.1186/1743-422X-7-273
21. Rincón Monroy MA, Ramirez-Nieto GC, Vera VJ, Correa JJ, Mogollón-Galvis JD. Detection and molecular characterization of porcine circovirus type 2 from piglets with porcine circovirus associated diseases in Colombia. Virol J. (2014) 11:143. doi: 10.1186/1743-422X-11-143
22. Xiao CT, Halbur PG, Opriessnig T. Global molecular genetic analysis of porcine circovirus type 2 (PCV2) sequences confirms the presence of four main PCV2 genotypes and reveals a rapid increase of PCV2d. J Gen Virol. (2015) 96:1830–41. doi: 10.1099/vir.0.000100
23. Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, et al. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. (2008) 36:W465–9. doi: 10.1093/nar/gkn180
24. Wang C, Pang VF, Lee F, Liao PC, Huang YL, Lin YL, et al. Development and evaluation of a loop-mediated isothermal amplification method for rapid detection and differentiation of two genotypes of porcine circovirus type 2. J Microbiol Immunol Infect. (2014) 47:363–70. doi: 10.1016/j.jmii.2013.05.003
25. Wen LB, Guo X, Yang HC. Genotyping of porcine circovirus type 2 from a variety of clinical conditions in china. Vet Microbiol. (2005) 110:141–6. doi: 10.1016/j.vetmic.2005.07.003
26. Cheung AK, Lager KM, Kohutyuk OI, Vincent AL, Henry SC, Baker RB, et al. Detection of two porcine circovirus type 2 genotypic groups in United States swine herds. Arch Virol. (2007) 152:1035–44. doi: 10.1007/s00705-006-0909-6
27. Li W, Wang X, Ma T, Feng Z, Li Y, Jiang P. Genetic analysis of porcine circovirus type 2 (PCV2) strains isolated between 2001 and 2009: genotype PCV2b predominate in postweaning multisystemic wasting syndrome occurrences in eastern China. Virus Genes. (2010) 40:244–51. doi: 10.1007/s11262-009-0438-y
28. Lyoo KS, Kim HB, Joo HS. Evaluation of a nested polymerase chain reaction assay to differentiate between two genotypes of Porcine circovirus-2. J Vet Diagn Invest. (2008) 20:283–8. doi: 10.1177/104063870802000304
29. Wang HY, Kim H, Choi EH, Lee H. Performance of the Real Fungus-ID kit based on multiplex RT-PCR assay for the rapid detection and identification of Trichophyton spp. and Microsporum spp. in clinical specimens with suspected dermatophyte infection. J Appl Microbiol. (2016) 120:234–47. doi: 10.1111/jam.12993
30. Ajbani K, Shetty A, Mehta A, Rodrigues C. Rapid diagnosis of extensively drug-resistant tuberculosis by use of a reverse line blot hybridization assay. J Clin Microbiol. (2011) 49:2546–51. doi: 10.1128/JCM.02511-10
31. Bedolla López F, Trujillo Ortega ME, Mendoza Elvira S, Quintero Ramírez V, Alonso Morales R, Ramírez-Mendoza H, et al. Identification and genotyping of porcine circovirus type II (PCV2) in Mexico. Virus Dis. (2018) 29:385–9. doi: 10.1007/s13337-018-0460-6
Keywords: porcine circovirus type 2, multiplex real-time PCR, PCR-REBA, ORF2, diagnosis
Citation: Wang H-y, Song JK, Shin S and Kim H (2020) Comparison of Multiplex Real-Time PCR and PCR-Reverse Blot Hybridization Assays for the Direct and Rapid Detection of Porcine Circovirus Type 2 Genotypes. Front. Vet. Sci. 7:200. doi: 10.3389/fvets.2020.00200
Received: 28 January 2020; Accepted: 25 March 2020;
Published: 30 April 2020.
Edited by:
Anuwat Wiratsudakul, Mahidol University, ThailandReviewed by:
Xiuqing Wang, South Dakota State University, United StatesFrancisco Rivera-Benítez, National Institute of Forestry, Agriculture and Livestock Research (INIFAP), Mexico
Copyright © 2020 Wang, Song, Shin and Kim. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hyunil Kim, aGlraW1Ab3B0aXBoYXJtLmNvLmty
 Hye-young Wang
Hye-young Wang Joong Ki Song2
Joong Ki Song2