- 1Department of Integrative Marine Ecology, Stazione Zoologica Anton Dohrn, Naples, Italy
- 2Department of Public Health and Infectious Diseases, Sapienza University of Rome, Rome, Italy
Studies describing the parasite fauna of sunfish species from the Mediterranean Sea are to date limited, despite information gained through parasitological examination may reveal unknown ecological and biological aspects of both hosts and parasites. Moreover, recent molecular studies on sunfish taxonomy revealed the presence of two species belonging to the genus Mola in the Mediterranean basin, namely M. mola and M. alexandrini. These two fish taxa have long been synonymized or confused among them, which implies that the majority of the studies carried out so far reported the parasites infecting both species under a single host species, generally referred to as M. mola. We hereby investigated the parasite fauna of a 43 cm long M. mola specimen from the Mediterranean Sea, whose identification was confirmed by molecular tool, and provided the first evidence of the occurrence of the nematode Anisakis simplex (s.s.) and of the cestode Gymnorhynchus isuri in Mola species anywhere. The use of helminth species as biological tags for the sunfish is also discussed.
Introduction
The sunfish genus Mola Koelreuter, 1766 has been considered, until recently, as composed of two species, namely Mola mola (Linnaeus, 1758), with a cosmopolitan distribution, and Mola ramsayi (Giglioli, 1883), restricted to the southern hemisphere (1). In the past few years, the use of molecular approaches revealed the presence of three valid species within this genus: Mola alexandrini (Ranzani, 1839) (syn. M. ramsayi) and M. mola, with a wide and sympatric distribution in the world's oceans, including the Mediterranean Sea, and Mola tecta Nyegaard, Sawai, Gemmell, Gillum, Loneragan, Yamanoue and Stewart, 2017, mostly found from temperate waters of southern hemisphere (2–4).
Mola species are well-known to host a variegate parasite fauna, and in particular several papers focused in the past on the parasite fauna of “M. mola,” whose information was reviewed by de Figueiredo et al. (5). However, since the clarification of the worldwide taxonomy of the genus Mola, only Ahuir-Baraja et al. (6) investigated the endo-parasites of a Mola individual stranded on the Valencian coast (western Mediterranean Sea), identified by molecular tools as M. alexandrini. This likely implies that the majority of the studies carried out so far reported the parasites infecting the entire genus under the single species M. mola [see (5)].
Host specificity is often considered to be a result of various factors, including phylogenetic, physiological, and ecological aspects (7, 8). Since most marine parasites exhibit at least some degree of host-specificity or host-preference, it is plausible that a degree of host-specificity may exist for parasites infecting fishes of the genus Mola. Moreover, among the several methods applied to obtain biological and ecological data of fishes, the use of parasites as biological tags in marine environment has become a useful tool in producing data for the host stock identification and for studying their migration by using molecular tools to unequivocally identify both hosts and their parasites (9–12). In fact, parasites can be used as biological tags since their geographic ranges are definite, so that a host can only become infected when entering the parasite endemic range (9, 10).
We here investigated the whole parasite fauna of a M. mola specimen obtained from the Gulf of Naples (Tyrrhenian Sea) whose identification was confirmed through molecular analysis. We provide the first evidence in Mola species of the occurrence of larval forms of two helminth parasites typically known from the Atlantic Ocean, suggesting a potential long-distance migration of the examined host fish.
Materials and Methods
Sampling
A juvenile male Mola specimen (4,190 kg in weight; 43 cm in total length) was fished at ~50 m depth on January 3, 2020 off Ischia Porto (~40.753766, 13.947723, Ischia Island, central Tyrrhenian Sea, Mediterranean) by an amateur fisherman, using a fishing line with the Mediterranean mussel (Mytilus galloprovincialis Lamarck, 1819) as bait. Once the fish was landed, it was first shown to the public and then discarded, being considered as bycatch. As one of the authors (FC) was there at that moment, and the specimen was already dead, it was collected by him and soon after frozen at −20°C for molecular and parasitological analyses.
DNA Extraction and Molecular Analysis for Host Identification
Total genomic DNA was extracted from a defrosted muscle sample, and a partial sequence of the cytochrome c oxidase subunit I (COX1) mitochondrial gene (mtDNA cox1) was amplified and sequenced following methods used in Osca et al. (13).
The sequence obtained was assembled using Sequencher v. 5.0.1 (GeneCodes Co.) and compared with reference sequences from the NCBI nucleotide (NT) database using BLASTn (14). Partial sequences of further Mola specimens were downloaded from GenBank and BOLD, together with those of Masturus lanceolatus (Liénard, 1840) to be used as outgroup (2). Nucleotide sequences were aligned with the Translator X server (15), using the MAFFT v7 (16) option with default settings. By subsequently deleting identical sequences, a total of 12 sequences of Mola specimens and of five of M. lanceolatus were used for the phylogenetic analyses (Supplementary Table 1).
Alignment format conversions were performed using the ALTER webserver (17). The best-fit model of substitution was determined using the Akaike information criterion (AIC) (18) implemented in PartitionFinder v.1.1.1 (19). Phylogenetic relationships were inferred performing maximum likelihood (ML) analyses in RAxML v.8.1.16 (20), using the rapid hill-climbing algorithm, and Bayesian inference (BI) analyses in MrBayes v.3.1.2 (21), running four simultaneous MCMC (Markov chain Monte Carlo) for 2 million generations, a sampling interval every 1,000 generations, and a burn-in of 25%. Two independent Bayesian inference runs were performed to increase the chance of adequate mixing of the Markov chains and of detecting failure to converge. Support for internal branches was evaluated by non-parametric bootstrapping (22) with 1,000 replicates (ML) and by posterior probabilities (BI). Finally, based on the results of the phylogenetic trees, diagnostic nucleotides were identified in our alignment as to further confirm the molecular identification of the specimen studied here.
Parasitological Analysis
During the fish necropsy, skin, musculature, gills, mouth cavity, digestive tract, liver, heart, testes, visceral cavity, and mesenteries of the sunfish were examined for metazoan parasites. Organs and tissues were removed and placed individually in plastic Petri dishes (200 mm in diameter); the organs were then dissected and the surfaces were examined visually. After the larger helminths were removed using tweezers, organs and tissues were washed through a 100 μm mesh screen. The remaining washed material from each organ was examined under a dissecting microscope (Leica M165 C), and parasites were collected, counted, washed in physiological saline, and preserved in 70% ethanol or frozen at −20°C (23). For identification, crustacean parasites and nematodes were clarified respectively in 20% potassium hydroxide and Amman's lactophenol, and then returned to 70% ethanol; digeneans and cestodes were stained with Mayer's acid carmine and mounted in Canada balsam. Parasites were studied with a compound microscope (Leica DM1000) and identified using published identification keys (24–27).
DNA Extraction and Molecular Analysis for Selected Larval Parasites
Total genomic DNA from a cestode and a nematode larva whose morphological identification at species level proved to be uncertain, was extracted using Quick-gDNA Miniprep Kit (ZYMO RESEARCH) following the standard manufacturer-recommended protocol. DNA was quantified by a NanoDrop®TC1-E20 spectrophotometer (BioTek Synergy HT).
A cestode larva morphologically considered as belonging to the family Gymnorhynchidae Dollfus, 1935 (Trypanorhyncha) was identified to species level by sequence analysis of the complete small subunit of the ribosomal RNA gene (ssrDNA) (28) and the partial large subunit ribosomal gene (lsrDNA) (29). Complete ssrDNA was amplified using the primers WormA (5′-GCGAATGGCTCATTAAATCAG-3′) and WormB (5′-CTTGTTACGACTTTTACTTCC-3′). Partial 1srDNA was amplified using the primers ZX-1 (5′-ACCCGCTGAATTTAAGCATAT-3′) and 1500R (5′- GCTATCCTGAGGGAAACTTCG-3′). Both PCR reactions were carried out following the procedure previously reported in Palm et al. (30). The identity of the specimens was checked using the Basic Local Alignment Search Tool (Blast, www.ncbi.nih.gov/BLAST/) and the sequences of ssrDNA and 1srDNA were aligned with the Gymnorhynchidae reference sequences from the NCBI nucleotide (NT) database, using Clustal X (31). The aligned data were concatenated using sequence Matrix v.1.7.8 (32). The phylogenetic analysis of the combined ssrDNA and 1srDNA sequences were carried out by BI, using MrBayes 3.1 (21). JModeltest (33) was used to determine the best-fit substitution model for both sequences dataset (ssrDNA and 1srDNA), as implemented with Akaike's Information Criterion (AIC). BI analysis was performed using the Bayesian posterior probability analysis using the MCMC algorithm, with four chains, 0.2 as the temperature of heated chains, 2,000,000 generations, with a subsampling frequency of 500 and a burn-in fraction of 0.25. Posterior probabilities were estimated and used to assess support for each branch. The phylogenetic trees were rooted using Hepatoxylon trichiuri (Holten, 1802) Bosc, 1811 as outgroup.
The Anisakis larva was identified at the species level by sequencing the mitochondrial cytochrome c oxidase subunit II (COX2) gene (mtDNA cox2). PCR amplification was performed using the primers 211F (5′- TTTTCTAGTTATATAGATTGRTTTYAT-3′) and 210R (5′-CACCAACTCTTAAAATTATC-3′). PCR was carried out according to the procedures provided by Mattiucci et al. (34). The sequences obtained at the mtDNA cox2 for the larval nematodes were compared with those already obtained for the same gene and deposited in GenBank.
Results
Molecular and Phylogenetic Analyses for Host Identification
A 606 bp partial sequence of the mtDNA cox1 gene was obtained for the fish specimen. The ML (-lnL = 1322.1) and BI (-lnL = 1356.15 for run 1; -lnL = 1353.81 for run 2) analyses arrived at similar tree topologies. Two major clades were defined with high support for both analyses (ML and BI), in which M. tecta and M. alexandrini were placed as sister groups. The other clade, with high support for ML and maximal support for BI, included sequences of M. mola. The latter clade also included the sequence of the specimen studied here (Supplementary Figure 1). The analysis of the positions of diagnostic nucleotides in our alignment revealed that M. tecta is diagnosed by 45 (G), 474 (A), 558 (G), and 576 (T), M. alexandrini is diagnosed by 168 (G), 240 (A), 291 (T), 516 (A), 597 (A), and 603 (G), and M. mola is diagnosed by 15 (A), 150 (T), 172 (G), 186 (C), 219 (C), 225 (A), 339 (T), 387 (C), 426 (G), 429 (C), 450 (G), 456 (A), 465 (G), 498 (A), 540 (G), 543 (T), 579 (T), and 606 (T). No differences with respect to present results were obtained when analyzing the unpublished cox1 sequence of M. mola obtained here. The sequence was deposited in GenBank under the accession number MT913440.
Parasitological Analysis
A total of 46 metazoan parasites were found, belonging to 10 different species. These included eight helminth taxa (one monogenean, three digeneans, three cestodes, and one nematode) and two copepods (Table 1). All parasites were found at adult stage, except for the larvae stages of the cestodes H. trichiuri and Gymnorhynchus isuri Robinson, 1959 (Figure 1) and the nematode Anisakis simplex (s.s.) (Rudolphi, 1809) (see below for the molecular identification of the selected larval parasites). Among the ectoparasites, the monogenean Capsala martinierei Bosc, 1811 and the copepods Cecrops latreillii Leach, 1816 and Lepeophtheirus nordmanni (Milne Edwards, 1836) were among those found on gills and skin.
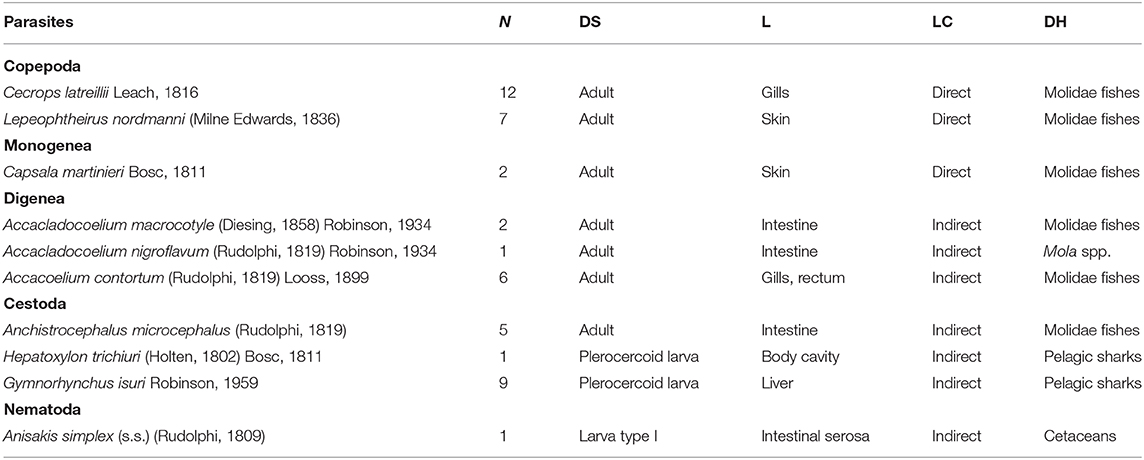
Table 1. Metazoan parasites found in a sunfish Mola mola from off Ischia Island (Tyrrhenian Sea, Mediterranean Sea), with number of individuals (N), developmental stage (DS), location in the host (L), life cycle type (LC), and known definitive host/s (DH).
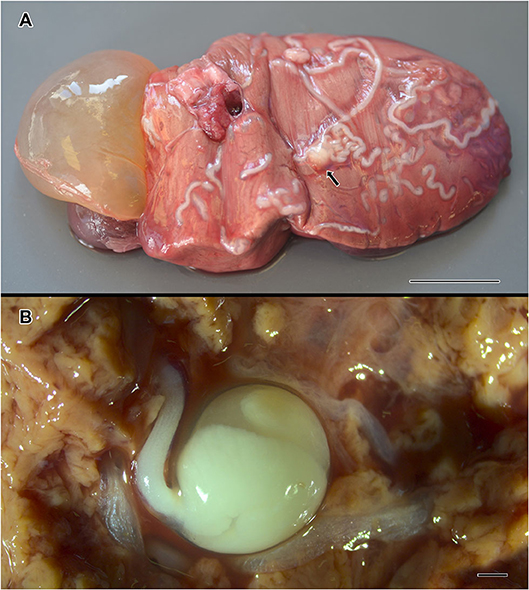
Figure 1. Gymnorhynchus isuri infection in the liver of the sunfish (M. mola). Several white larvae are embedded in the liver (A); the arrow in (A) indicates the blastocyst of a G. isuri larva enlarged in (B) after disruption of the hepatic tissue. Bar scale: (A): 2 cm; (B): 100 μm.
Molecular Analysis for Selected Larval Parasites
According to the obtained sequences at the complete small subunit (ssrDNA) and the partial large subunit (1srDNA) ribosomal RNA gene, a larva was identified as G. isuri. The lsrDNA sequences (1,300 bp) and the ssrDNA sequences (1,968 bp) matched at 99–100% with the sequences of G. isuri from GenBank (25).
The BI obtained from the combined lsrDNA and the ssrDNA sequences dataset showed that the larva of G. isuri clustered together with the reference sequence of G. isuri from GenBank (25) (Figure 2). The obtained lsrDNA and ssrDNA sequences were deposited in GenBank under the accession numbers MT667258 and MT667257, respectively.
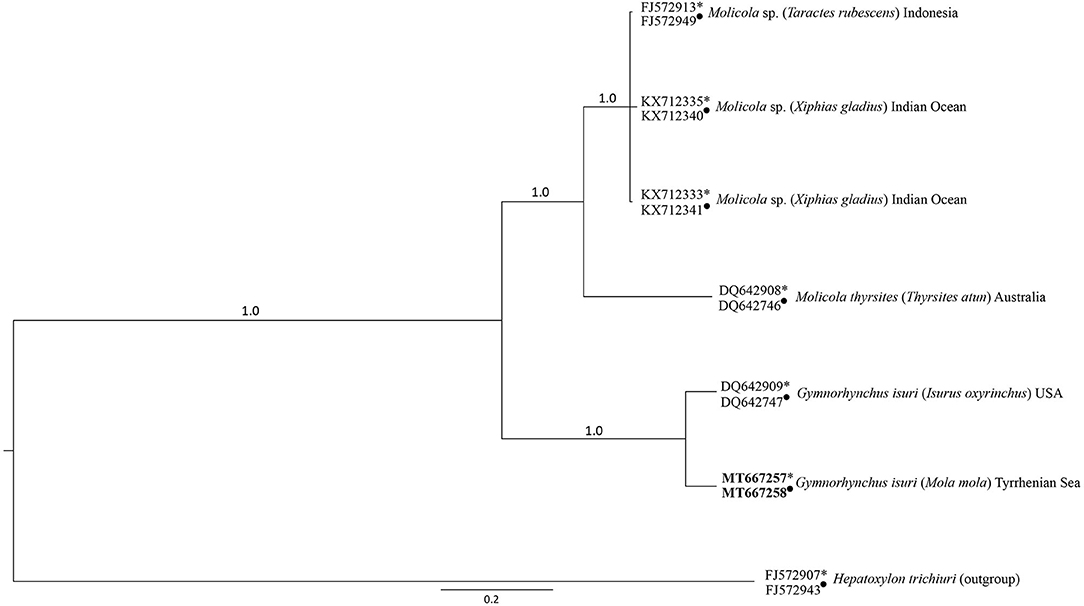
Figure 2. Phylogenetic condensed tree from Bayesian analyses based on 1srDNA and ssrDNA sequences obtained in the present study, in comparison with those available in GenBank at the same genes loci (1srDNA indicated with ∙, ssrDNA indicated with *) from cestodes of the Gymnorhynchidae Family. Hepatoxylon trichiuri was used as outgroup.
According to the sequences of 629 bp in length of the mtDNA cox2 gene locus, the Anisakis type I larva was assigned to the species A. simplex (s.s.). The obtained sequence matched 100% with the A. simplex (s.s.) sequences from GenBank. The obtained cox2 sequence was deposited in GenBank under the accession number MT667259.
Discussion
Mola mola is usually considered a highly migratory fish, characterized by daily vertical movements following zooplankton preys and generally jellyfishes (1). Its horizontal movements are linked to seasonal variations of sea surface temperature and forage availability, with migrations shown to exceed 1,000 km in the northeast and northwest Atlantic (1, 35, 36). Recent studies suggest an ontogenetic shift in diet with age, with smaller sunfishes feeding on benthic invertebrates and larger individuals feeding on gelatinous zooplankton occurring in the water column (37–39). The present finding agrees with these statements, as our juvenile sunfish specimen (43 cm total length, corresponding to ca. 15-months of age according to captive growth rates for M. mola) [see (40)] was collected with a fishing line near the seabed, thus suggesting that the sunfish was foraging in the benthic zone.
The ecto-parasites found in the specimen from off Ischia Island were considered all Mola specialists with a direct life cycle, and have been found through the whole geographical range of their hosts, while the endo-parasites were all trophically transmitted with a heteroxenous life cycle (6, 24–27, 41, 42). All parasites, except G. isuri and A. simplex (s.s.), were previously found in Mola species from the Mediterranean Sea (6, 41, 42).
In the present study, we have identified two parasite species [G. isuri and A. simplex (s.s.)] that potentially could be used as biological tags, having both some features able to producing data for fish stock identification and for a better understanding of the host migration routes [see (9–11)].
Nematodes of the genus Anisakis infect a wide range of marine hosts (with cetaceans as definitive hosts) including at least 12 species of dolphins, porpoises and whales (43). Small crustaceans (Euphausiidae), fishes, squids, and other invertebrates serve as first and second intermediate or paratenic hosts (43, 44). Anisakis larvae have been previously reported in Mola species from Spain, New Zealand, and Chile (5, 6, 42), although no molecular analyses were performed to identify those larvae to species level. Anisakis simplex (s.s.) is known from the Atlantic and Pacific Oceans, and has its southern limit of distribution in the North-Eastern Atlantic waters along the Spanish-Portuguese Atlantic coast, being never recorded in the Mediterranean Sea except from the Alboran Sea, a transitional zone between Atlantic and Mediterranean [see (43)], and in pelagic fishes like the Atlantic mackerel Scomber scombrus Linnaeus, 1758 and the Atlantic bluefin tuna Thunnus thynnus Linnaeus, 1758 in the Eastern Mediterranean (45, 46); however, it has been suggested that the latter findings were related to the large migratory routes of those intermediate/paratenic fish hosts (45, 46).
Trypanorhynchan cestodes use copepods as first and other invertebrates or small fishes as second intermediate hosts (47, 48). Species of the genera Gymnorhynchus occur as adults in pelagic sharks, with the larval stages infecting a wide range of teleosts and sharks (47, 48). The genus Gymnorhynchus includes two species, namely G. gigas and G. isuri (48). To the best of our knowledge, only G. gigas has to date been reported from the Mediterranean basin in the swordfish Xiphias gladius Linnaeus, 1758, the Ray's bream Brama brama (Bonnaterre, 1788), and the silver scabbardfish Lepidopus caudatus (Euphrasen, 1788) (49, 50). Gymnorhynchus isuri is known from the north, southwestern, and northeast Atlantic and the Tasmanian Sea in the southwestern Pacific, infecting the shortfin mako Isurus oxyrinchus Rafinesque, 1810 and the blue shark Prionace glauca (Linnaeus, 1758) (30, 51, 52), and no record exists of G. isuri in Mola species anywhere.
A possible scenario seems to suggest that the sunfish specimen here analyzed had migrated from Atlantic Ocean waters into the Mediterranean Sea or at least from Alboran Sea waters. This hypothesis is based on the finding of parasite species typically known from Atlantic waters. No data exist on migration in juvenile M. mola, nor on Molidae in general within the Mediterranean Sea or from the Atlantic Ocean to the Mediterranean and vice versa, except for a study showing movements from the Atlantic Iberian coast to the Alboran Sea (36). Published information on the parasite fauna of the target fish species highlighted assemblages characterized by species with wide geographic range. This suggests the hypothesis that host migration route may be responsible for the parasite range expansion, as suggested for other marine vertebrates, that migrate from the Atlantic to the Mediterranean sea waters and vice versa (11, 45, 46, 53).
Among trypanorhynchans, Molicola horridus (Goodsir, 1841) Dollfus, 1935 (Gymnorhynchidae) is often reported infecting the liver of Mola species (6, 41, 42). Molicola is closely related to Gymnorhynchus having a similar scolex morphology, surface ultrastructure, and tentacular armature, and members of both genera also cause similar pathological changes; however, whereas M. horridus is considered a specific parasite of Mola species, G. isuri is known from bramid fishes (48). As the plerocerci of these two genera are difficult to be differentiated by morphology only (encysted forms are often obtained damaged, dead and with tentacles retracted), there is the possibility that the two species were often mixed up or misidentified in the past literature. In fact, the most important diagnostic feature between the two genera is the arrangement of hooks on the external surface of tentacular armature, that is impossible to study in encysted dead larval stages (48). Indeed, in recent years, molecular tools for recognition of members of Gymnorhynchidae was successfully applied (30).
Regarding to the other helminths identified in the present study, the cestodes H. trichiuri (Sphyriocephalidae) and A. microcephalus (Triaenophoridae) parasitize as adults the intestine of pelagic sharks and Molidae fishes respectively (5, 6, 26, 41, 42, 47, 48). It is believed that copepods can serve as intermediate hosts for both cestode species (48, 54). Digeneans of the genera Accacoelium, and Accacladocoelium (Accacoeliidae) are specific parasites of Molidae fishes, that acquire the infection when feeding on nektonic organisms, and especially on cnidarians and ctenophores infected by accacoeliid metacercariae (24, 55).
In conclusion, traditional parasitological studies implemented by molecular tools are still needed to understand if host specificity exists for most of the parasites infecting Mola species. However, despite we have here analyzed only a single M. mola specimen, we have showed that this sunfish species serves as an intermediate host for two cestodes (H. trichiuri and G. isuri) and as a definitive host for the remaining parasites except A. simplex (s.s.), considered here as an accidental finding. In fact, due to low prevalence and intensity of infection or absence of this nematode larvae, as also reported in other studies from Mediterranean (6, 41, 42), it is reasonable to conclude that this fish ecologically likely represents a dead end for the parasite (56). For the reasons listed above, the potential use of A. simplex (s.s.) as biological tag for sunfishes remains doubtful. In contrast, the cestode G. isuri has a useful potential to be used as biological indicator and could be used to study the sunfish movements at least in the Mediterranean and adjacent areas. However, despite the present study reveals for the first time the occurrence of G. isuri in the Mediterranean Sea and indicates M. mola as a new intermediate host record for this cestode species, a molecular screening of specimen morphologically identified in the past literature and in museum collections as M. horridus could confirm if the present finding of G. isuri in the Mediterranean is a new or casual event or the species is already widespread, but undetected and/or misidentified, in the basin.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics Statement
Ethical review and approval was not required for the animal study because the sunfish was collected during amateur fishing with no additional experimental catches being performed. According to Italian law DL16/92 and European directive 2010/63/EU, this study did not require a specific permit. Procedures for this study were performed in accordance with the guide for the care and use of animals by the Italian Ministry of Health.
Author Contributions
MS: performed fish necropsy, collection, morphological identification of parasites, and wrote the paper. SM and MP: performed molecular and phylogenetic analyses of selected larval stages of parasites. DO and FC: performed molecular and phylogenetic analyses of the sunfish. All authors contributed to manuscript revision, read, and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2020.579728/full#supplementary-material
Supplementary Figure 1. Phylogenetic relationships in the genus Mola based on mtDNA cox1 partial sequences downloaded from GenBank and BOLD (ID codes as in the Supplementary Table 1), and the specimen from off Ischia Island marked in bold. Numbers above/below branches represent bootstrap values (ML) and posterior probabilities values (BI). Scale bar represents nucleotide substitution. Poor support values (<50%, 0.90) are not shown. **The putative misidentification (see Supplementary Table 1) is based on an incorrect entry in GenBank [see (2), where the same specimen is labeled as Mola sp. A = Mola alexandrini].
Supplementary Table 1. GenBank and Barcode of Life Data System mtDNA cox1 partial sequences used in the molecular systematic analyses and associated specimen data (locality obtained from web databases or relevant paper/s). *The specimen from Ischia Island (central Tyrrhenian Sea, Mediterranean) analysed in the present study. **The putative misidentification is based on an incorrect entry in GenBank (where the same specimen is labelled Mola sp. A = Mola alexandrini).
References
1. Pope E, Hays G, Thys T, Doyle T, Sims D, Queiroz N, et al. The biology and ecology of the ocean sunfish Mola mola: a review of current knowledge and future research perspectives. Rev Fish Biol Fish. (2010) 20:471–87. doi: 10.1007/s11160-009-9155-9
2. Nyegaard M, Sawai E, Gemmell N, Gillum J, Loneragan NR, Yamanoue Y, et al. Hiding in broad daylight: molecular and morphological data reveal a new ocean sunfish species (Tetraodontiformes: Molidae) that has eluded recognition. Zool J Linnean Soc. (2018) 182:631–58. doi: 10.1093/zoolinnean/zlx040
3. Sawai E, Yamanoue Y, Nyegaard M, Sakai Y. Redescription of the bump-head sunfish Mola alexandrini (Ranzani 1839), senior synonym of Mola ramsayi (Giglioli 1883), with designation of a neotype for Mola mola (Linnaeus 1758) (Tetraodontiformes: Molidae). Ichthyol Res. (2018) 65:142–60. doi: 10.1007/s10228-017-0603-6
4. Mangel JC, Pajuelo M, Pasara-Polack A, Vela G, Segura-Cobeña E, Alfaro-Shigueto J. The effect of Peruvian small-scale fisheries on sunfishes (Molidae). J Fish Biol. (2019) 94:77–85. doi: 10.1111/jfb.13862
5. de Figueiredo NC, de Lima JTAX, Freitas CI, da Silva CG. Checklist dos parasitos do peixe Lua (Mola mola: Molidae) no mundo. Pubvet. (2018) 12:1–9. doi: 10.22256/pubvet.v12n3a42.1-9
6. Ahuir-Baraja AE, Yamanoue Y, Kubicek L. First confirmed record of Mola sp. A in the western Mediterranean Sea: morphological, molecular and parasitological findings. J Fish Biol. (2017) 90:1133–41. doi: 10.1111/jfb.13247
7. Poulin R. Determinants of host-specificity in parasites of freshwater fishes. Int J Parasitol. (1992) 22:753–8. doi: 10.1016/0020-7519(92)90124-4
8. Poulin R, Krasnov BR, Mouillot D. Host specificity in phylogenetic and geographic space. Trends Parasitol. (2011) 27:355–61. doi: 10.1016/j.pt.2011.05.003
9. MacKenzie K, Abaunza P. Parasites as biological tags for stock discrimination of marine fish: a guide to procedures and methods. Fish Res. (1998) 38:45–56. doi: 10.1016/S0165-7836(98)00116-7
10. Catalano SR, Whittington ID, Donnellan SC, Gillanders BM. Parasites as biological tags to assess host population structure: guidelines, recent genetic advances and comments on a holistic approach. Int J Parasitol Parasites Wildl. (2013) 3:220–6. doi: 10.1016/j.ijppaw.2013.11.001
11. Mattiucci S, Cimmaruta R, Cipriani P, Abaunza P, Bellisario B, Nascetti G. Integrating Anisakis spp. parasites data and host genetic structure in the frame of a holistic approach for stock identification of selected Mediterranean Sea fish species. Parasitology. (2015) 142:90–108. doi: 10.1017/S0031182014001103
12. Santoro M, Di Nocera F, Iaccarino D, Cipriani P, Guadano Procesi I, Maffucci F, et al. Helminth parasites of the dwarf sperm whale Kogia sima (Cetacea: Kogiidae) from the Mediterranean Sea, with implications on host ecology. Dis Aquat Organ. (2018) 129:175-82. doi: 10.3354/dao03251
13. Osca D, Tanduo V, Tiralongo F, Giovos I, Almabruk SAA, Crocetta F, et al. The indo-pacific sergeant abudefduf vaigiensis (Quoy & Gaimard, 1825) (Perciformes: Pomacentridae) in libya, South-central mediterranean sea. J Mar Sci Eng. (2020) 8:14. doi: 10.3390/jmse8010014
14. Morgulis A, Coulouris G, Raytselis Y, Madden TL, Agarwala R, Schäffer AA. Database indexing for production MegaBLAST searches. Bioinformatics. (2008) 24:1757–64. doi: 10.1093/bioinformatics/btn322
15. Abascal F, Zardoya R, Telford MJ. TranslatorX: multiple alignment of nucleotide sequences guided by amino acid translations. Nucleic Acids Res. (2010) 38(Suppl.2):W7–13. doi: 10.1093/nar/gkq291
16. Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. (2013) 30:772–80. doi: 10.1093/molbev/mst010
17. Glez-Peña D, Gómez-Blanco D, Reboiro-Jato M, Fdez-Riverola F, Posada D. ALTER: program-oriented conversion of DNA and protein alignments. Nucleic Acids Res. (2010) 38:14–8. doi: 10.1093/nar/gkq321
18. Akaike H. Information theory and an extension of the maximum likelihood principle. In: Petrov BN, Csaki F, editors. Second International Symposium on Information Theory. Budapest: Academiai Kiado (1973). p. 267–81.
19. Lanfear R, Calcott B, Ho SYW, Guindon S. PartitionFinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol Biol Evol. (2012) 29:1695–701. doi: 10.1093/molbev/mss020
20. Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. (2014) 30:1312–3. doi: 10.1093/bioinformatics/btu033
21. Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. (2003) 19:1572–4. doi: 10.1093/bioinformatics/btg180
22. Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. (1985) 39:783–91. doi: 10.1111/j.1558-5646.1985.tb00420.x
23. Santoro M, Mattiucci S, Cipriani P, Bellisario B, Romanelli F, Cimmaruta R, et al. Parasite communities of icefish (Chionodraco hamatus) in the Ross Sea (Antarctica): influence of the host sex on the helminth infracommunity structure. PLoS ONE. (2014) 9:e88876. doi: 10.1371/journal.pone.0088876
24. Bray RA, Gibson DI. The accacoeliidae (Digenea) of fishes from the North-East Atlantic. Bull Brit Mus Nat Hist Zool. (1977) 31:53–99.
25. Kabata Z. Copepoda and branchiura. In: Margolis L, Kabata Z, editors. Guide to the parasites of fishes of Canada. Part II – Crustacea: Can Spec Publ Fish Aquat Sci. (1988). p. 3–127.
26. Khalil LF, Jones A, Bray RA. Keys to the Cestode Parasites of Vertebrates. Wallingford: CAB International. (1994). p. 768.
27. Lamothe-Argumedo R. Nuevo arreglo taxonómico de la subfamilia Capsalinae (Monogenea: Capsalinae), clave para los géneros y dos combinaciones nuevas. An Inst Biol Univ Nac Autón Mexico Ser Zool. (1997) 68:207–23.
28. Littlewood DTJ, Cribb TH, Olson PD, Bray RA. Platyhelminth phylogenetics-a key to understanding parasitism? Belg J Zool. (2001) 131:35–46.
29. van der Auwera G, Chapelle S, De Wachter R. Structure of the large ribosomal subunit RNA of Phytophthora megasperma, and phylogeny of the oomycetes. FEBS Lett. (1994) 338:133–6. doi: 10.1016/0014-5793(94)80350-1
30. Palm HW, Waeschenbach A, Olson PD, Littlewood DT. Molecular phylogeny and evolution of the trypanorhyncha Diesing, 1863 (Platyhelminthes: Cestoda). Mol Phylogenet Evol. (2009) 52:351–67. doi: 10.1016/j.ympev.2009.01.019
31. Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, et al. Clustal W and clustal X version 2.0. Bioinformatics. (2007) 23:2947–8. doi: 10.1093/bioinformatics/btm404
32. Vaidya G, Lohman DJ, Meier R. SequenceMatrix: concatenation software for the fast assembly of multi-gene datasets with character set and codon information. Cladistics. (2011) 27:171–80. doi: 10.1111/j.1096-0031.2010.00329.x
33. Posada D, Buckley TR. Model selection and model averaging in phylogenetics: advantages of Akaike information criterion and Bayesian approaches over likelihood ratio tests. Syst Biol. (2004) 53:793–808. doi: 10.1080/10635150490522304
34. Mattiucci S, Cipriani P, Webb SC, Paoletti M, Marcer F, Bellisario B, et al. Genetic and morphological approaches distinguish the three sibling species of the Anisakis simplex species complex, with a species designation as Anisakis berlandi n. sp. for A. simplex sp. C (Nematoda: Anisakidae). J Parasitol. (2014) 100:199–214. doi: 10.1645/12-120.1
35. Potter IF, Galuardi B, Howell WH. Horizontal movement of ocean sunfish, Mola mola, in the northwest Atlantic. Mar Biol. (2011) 158:531–40. doi: 10.1007/s00227-010-1578-2
36. Sousa L, Queiroz N, Mucientes G, Humphries NE, Sims DW. Environmental influence on the seasonal movements of satellite-tracked ocean sunfish Mola mola in the north-east Atlantic. Anim Biotelemetry. (2016) 4:7. doi: 10.1186/s40317-016-0099-2
37. Syväranta J, Harrod C, Kubicek L, Cappanera V, Houghton JD. Stable isotopes challenge the perception of ocean sunfish Mola mola as obligate jellyfish predators. J Fish Biol. (2012) 80:225–31. doi: 10.1111/j.1095-8649.2011.03163.x
38. Sousa, L, Xavier R, Costa V, Humphries NE, Trueman C, et al. DNA barcoding identifies a cosmopolitan diet in the ocean sunfish. Sci Rep. (2016) 6:28762. doi: 10.1038/srep28762
39. Nakamura I, Sato K. Ontogenetic shift in foraging habit of ocean sunfish Mola mola from dietary and behavioral studies. Mar Biol. (2014) 161:1263–73. doi: 10.1007/s00227-014-2416-8
40. Nakatsubo T, Hirose H. Growth of captive ocean sunfish, Mola mola. Suisan Zoshoku. (2007) 55:403–7.
41. Gustinelli A, Nardini G, Aureli G, Trentini M, Affronte M, Fioravanti ML. Parasitofauna of Mola mola (Linnaeus, 1758) from Italian seas. Biol Mar Med. (2006) 13:872–6.
42. Ahuir-Baraja AE. Estudio parasitológico del pez luna, Mola mola (l.), en el mediterráneo occidental. (PhD Thesis), Universitat de València. (2012).
43. Mattiucci S, Cipriani P, Levsen A, Paoletti M, Nascetti G. Molecular epidemiology of Anisakis and Anisakiasis: an ecological and evolutionary road map. Adv Parasitol. (2018) 99:93–263. doi: 10.1016/bs.apar.2017.12.001
44. Klimpel S, Palm HW, Rückert S, Piatkowski U. The life cycle of Anisakis simplex in the norwegian deep (northern North Sea). Parasitol Res. (2004) 94:1–9. doi: 10.1007/s00436-004-1154-0
45. Levsen A, Cipriani P, Mattiucci S, Gay M, Hastie LC, MacKenzie K, et al. Anisakis species composition and infection characteristics in Atlantic mackerel, Scomber scombrus, from major European fishing grounds - reflecting changing fish host distribution and migration pattern. Fish Res. (2018) 202:112–21. doi: 10.1016/j.fishres.2017.07.030
46. Mladineo I, Trumbić Z, Radonić I, Vrbatović A, Hrabar J, Bušelić I. Anisakis simplex complex: ecological significance of recombinant genotypes in an allopatric area of the Adriatic Sea inferred by genome-derived simple sequence repeats. Int J Parasitol. (2017) 47:215–23. doi: 10.1016/j.ijpara.2016.11.003
47. Campbell RA, Beveridge I. Order trypanorhyncha Diesing, 1863. In: Khalil LF, Jones A, Bray RA, editors. Keys to the Cestode Parasites of Vertebrates. Wallingford: CAB International (1994). p. 51–148.
49. Manfredi MT, Gandini G, Traldi G. Infestazione muscolare da larve di cestodi Trypanorhyncha in pesce spada (Xiphias gladius). Atti Soc Ital Sci Vet. (1993) 47:765–7.
50. Giarratana F, Muscolino D, Beninati C, Ziino G, Giuffrida A, Trapani M, et al. Gymnorhynchus gigas in Lepidopus caudatus (Actinopterygii: perciformes: trichiuridae): prevalence and related effects on fish quality. Czech J Food Sci. (2014) 32:320–5. doi: 10.17221/330/2013-CJFS
51. Knoff M, São Clemente SC, Pinto RM, Lanfredi RM, Gomes DC. Redescription of Gymnorhynchus isuri (Cestoda: Trypanorhyncha) from Isurus oxyrinchus (Elasmobranchii: Lamnidae). Folia Parasitol. (2007) 54:208–14. doi: 10.14411/fp.2007.028
52. Penadés-Suay J, Tomás J, Merchán M, Aznar FJ. Intestinal helminth fauna of the shortfin mako Isurus oxyrinchus (Elasmobranchii: Lamnidae) in the northeast Atlantic Ocean. Dis Aquat Organ. (2017) 123:45–54. doi: 10.3354/dao03081
53. Santoro M, Di Nocera F, Iaccarino D, Lawton SP, Cerrone A, Degli Uberti B, et al. Pathology and molecular analysis of Hapalotrema mistroides (Digenea: Spirorchiidae) infecting a Mediterranean loggerhead turtle Caretta caretta. Dis Aquat Organ. (2017) 124:101–8. doi: 10.3354/dao03117
54. Kuchta R, Scholz T, Bray RA. Revision of the order Bothriocephalidea Kuchta, Scholz, Brabec & Bray, 2008 (Eucestoda) with amended generic diagnoses and keys to families and genera. Syst Parasitol. (2008) 71:81–136. doi: 10.1007/s11230-008-9153-7
55. Gibson DI. Family accacoeliidae Odhner, 1911. In: Gibson DI, Jones A, Bray RA, editors. Keys to the Trematoda Volume 1. London, UK: CABI Publishing and The Natural History Museum. (2002). p. 341–7. doi: 10.1079/9780851995472.0341
56. Santoro M, Mattiucci S, Paoletti M, Liotta A, Uberti BD, Galiero G, et al. Molecular identification and pathology of Anisakis pegreffii (Nematoda: Anisakidae) infection in the Mediterranean loggerhead sea turtle (Caretta caretta). Vet Parasitol. (2010) 174:65–71. doi: 10.1016/j.vetpar.2010.08.018
Keywords: Anisakis simplex (s.s.), Molidae fishes, Tyrrhenian Sea, Gymnorhynchus isuri, metazoan parasites, Trypanorhyncha
Citation: Santoro M, Palomba M, Mattiucci S, Osca D and Crocetta F (2020) New Parasite Records for the Sunfish Mola mola in the Mediterranean Sea and Their Potential Use as Biological Tags for Long-Distance Host Migration. Front. Vet. Sci. 7:579728. doi: 10.3389/fvets.2020.579728
Received: 03 July 2020; Accepted: 11 September 2020;
Published: 19 October 2020.
Edited by:
Rachel E. Marschang, Laboklin GmbH & Co. KG, GermanyReviewed by:
Ken MacKenzie, University of Aberdeen, United KingdomMarianne Nyegaard, Auckland Museum, New Zealand
Copyright © 2020 Santoro, Palomba, Mattiucci, Osca and Crocetta. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mario Santoro, bWFyaW8uc2FudG9yb0Bzem4uaXQ=
 Mario Santoro
Mario Santoro Marialetizia Palomba
Marialetizia Palomba Simonetta Mattiucci
Simonetta Mattiucci David Osca
David Osca Fabio Crocetta
Fabio Crocetta