- 1School of Life Sciences, University of Warwick, Coventry, United Kingdom
- 2School of Agriculture, Food and the Environment, Royal Agricultural University, Gloucestershire, United Kingdom
- 3College of Life and Environmental Sciences, Institute for Microbiology and Infection, University of Birmingham, Birmingham, United Kingdom
Dichelobacter nodosus is the essential pathogen in ovine footrot, an important cause of lameness in sheep that reduces productivity and welfare. The aim of this study was to investigate the feasibility of using multiple locus variable number tandem repeat analysis (MLVA) developed to investigate isolates to understand the molecular epidemiology of Dichelobacter nodosus in ovine footrot by investigation of communities of strains. MLVA sensitivity was improved by optimizing PCR conditions to 100% specificity for D. nodosus. The improved MLVA scheme was used to investigate non-cultured DNA purified from swabs (swab DNA) and cultured DNA from isolates (isolate DNA) from 152 foot and 38 gingival swab samples from 10 sheep sampled on four occasions in a longitudinal study. Isolate DNA was obtained from 6/152 (3.9%) feet and 5/6 yielded complete MLVA profiles, three strains were detected. Two of the three isolate strains were also detected in isolate DNA from 2 gingival crevice cultures. Complete MLVA profiles were obtained from swab DNA from 39 (25.7%) feet. There were 22 D. nodosus community types that were comprised of 7 single strain and 15 multi-strain communities. Six community types were detected more than once and three of these were detected on the same four sheep and the same two feet over time. There were a minimum of 17 and a maximum of 25 strain types of D. nodosus in the study. The three isolate strain types were also the most frequently detected strain types in swab DNA. We conclude that the MLVA from swab DNA detects the same strains as culture, is much more sensitive and can be used to describe and differentiate communities and strains on sheep, feet and over time. It is therefore a sensitive molecular tool to study D. nodosus strains directly from DNA without culture.
Introduction
Footrot (FR) is the most common cause of lameness in sheep in the UK and it is a health and welfare concern in sheep flocks globally (1–4). Footrot reduces productivity and sustainability of sheep farming (5, 6), costing the UK industry £20–£80 million per annum (6, 7).
There are two clinical presentations of footrot, interdigital dermatitis (ID), characterized by inflammation of the interdigital skin, and severe footrot (SFR), characterized by separation of the hoof horn from underlying tissues (8). The essential pathogen in footrot is the fastidious gram negative, aerotolerant, anaerobic bacterium Dichelobacter nodosus (9, 10) which is key in initiation of ID and in progression to SFR (11–13). D. nodosus is present in >90% of UK sheep flocks and causes ~70 % of lameness (4).
In cross-sectional studies, D. nodosus has been detected on healthy and diseased feet (11, 14–18), and in the gingival cavity (19).
Russell et al. (20) developed an MLVA assay as a strain-typing tool for cultured D. nodosus isolates based on four polymorphic loci (DNTR02, 09, 10, and 19). The assay was used by Smith et al. (21) to investigate within-flock population dynamics of strains of D. nodosus. They reported that D. nodosus strains clustered within sheep and were transmitted between ewes over time. D. nodosus isolation is challenging and time consuming, because of the organism's fastidious and anaerobic nature and direct PCR from swab DNA is more sensitive than culture (15, 22).
Muzafar et al. (23) used the MLVA developed by Russell et al. (20) to analyse D. nodosus directly from DNA extracted from interdigital skin swabs of healthy and footrot affected feet using only DNTR10 and DNTR19. They did not use DNTR09 due to poor amplification nor DNTR02 due to non-specific amplification.
Therefore, the aim of this study was to optimize and validate the full Dichelobacter nodosus MLVA for isolate DNA and community DNA and investigate its value in a pilot longitudinal study of persistence of D. nodosus in sheep.
Materials and Methods
Sample Collection
In 2014, swab samples were collected in a longitudinal study of 10 sheep (5 ewes, 5 lambs) from a UK flock with footrot. All sheep were sampled on four occasions at 2-week intervals from May to June. On each occasion, all four feet and the gingival crevice were swabbed with two swabs each, one for DNA analysis and one for culture. In addition, foot lesions were scored for ID and SFR using two 5-point scales (15). A total of 152 foot and 38 mouth swabs were collected.
Isolation and Detection of Dichelobacter nodosus
Swabs for culture were inoculated onto 4% hoof agar (HA) followed by subculture onto a 2% HA (15). Plates were incubated under anaerobic conditions at 30°C for 4–5 days (MACS-MG-1000 anaerobic workstation, Don Whitley Scientific, Shipley, UK, 80% N2, 10% CO2, 10% H2). Isolate DNA was extracted with a DNeasy Blood and Tissue Kit (Qiagen Ltd., Manchester, United Kingdom) according to the manufacturer's instructions with a lysis time of 1 h. DNA was extracted directly from swabs using the hydroxyapatite spin-column method (24) using only 0.5 ml of the sodium phosphate extraction buffers. D. nodosus was detected in DNA extracted from the foot and mouth swabs using a D. nodosus-specific rpoD-targeted qPCR (25).
MLVA Optimization and Protocol
The sensitivity of the MLVA primers (Supplementary Table 1) was determined using DNA from D. nodosus isolates and swab samples. Improvements in sensitivity were made by changing the PCR Master mix (From Promega x2 PCR Master Mix to Bioline MyTaq™ Red Mix), increasing primer concentration (from 10 pmoles of each primer in a 50 μl reaction to 10 pmoles of each primer in a 25 μl reaction), DNA template concentration (from 1 μl/50 μl reactions to 1 μl/25 μl reactions) and the number of PCR cycles (from 30 to 40 cycles). The final protocol was, in 25 μl reactions; 12.5 μl MyTaq™ Red Mix (Bioline, London, United Kingdom), 1 μl of each primer (10 μM stock concentration; Supplementary Table 1), 1 μl bovine serum albumin (20 mg ml−1) (Sigma Aldrich, Dorset, United Kingdom) and 1 μl of DNA template. DNA from D. nodosus strain 4303 LBV and nuclease free H2O were used as positive and negative controls, respectively. All PCR reactions were carried out on an Eppendorf Mastercycler ep gradient machine (Eppendorf, Hamburg, Germany) using the following cycling conditions: One cycle of 95°C for 2 min, 40 cycles of 95°C for 1 min, 59°C for 30 s and 72°C for 1 min with a final extension of 72°C for 2 min. PCR products were visualized by ethidium bromide-stained agarose gel electrophoresis and imaged using a Gene Flash imager (Syngene Bio Imaging, Cambridge, United Kingdom).
Determination of VNTR Amplicon Size Using Fragment Analysis
MLVA amplicon size was determined using fragment analysis. Forward primers for the four loci were labeled with different fluorescent dyes (Supplementary Table 1) and amplicons for each locus were submitted separately for fragment analysis (DNA Sequencing and Services, University of Dundee, Scotland). PCR products from D. nodosus isolates were diluted 1:100 and products originating from swabs were diluted either 1:20 or 1:100 depending in the PCR band intensity seen on agarose gel. 1200 Liz dye (Applied Biosystems, Warrington, United Kingdom) was used as a size standard and data were analyzed using Peak Scanner? Software (Applied Biosystems, Warrington, United Kingdom). The bin range was set to 4bp (fragment size ± 2 bp) and minimum fragment length cut off values 500, 500, 400, and 550 bp for DNTR02, 09, 10, and 19, respectively, based on the length of each fragment without repeats.
To provide additional information on the accuracy of the assay, the 4 loci were amplified from D. nodosus strain 1703A (GenBank Accession number CP000513) and submitted for fragment analysis and Sanger sequencing. The size of the loci from fragment analysis was compared with the published 1703A sequences for the 4 loci (Genebank Accession numbers KC676717, KC676718, KC676719, and KC676720 for DNTR02, 09, 10, and 19, respectively). The sequenced VNTR loci from D. nodosus strain 1703A were analyzed using tandem repeat (TR) finding software (26) to determine the numbers of repeats in the sequence.
MLVA analysis of two isolates resulted in a primary peak and a number of small secondary peaks ≤ 20% the height of the primary peak at expected TR intervals (Supplementary Figure 1). To investigate secondary peaks, 14 more D. nodosus isolates were analyzed (Supplementary Table 2). Secondary peaks were observed in some, but not all, isolates. The secondary peaks might have been due to non-axenic D. nodosus cultures. However, this is unlikely because multiple peaks were detected at all 4 loci (Supplementary Figure 1) which would indicate many isolate strains in each culture. Other explanations for secondary peaks might be rapid, minor, within-strain variation or an artifact in the PCR process. It was not possible to adjust the PCR to prevent the formation of these small secondary peaks, therefore it was decided to consider secondary peaks ≤ 20% the height of the primary peak as artifacts (Supplementary Figure 2).
Validation of the MLVA PCR and Testing on Model Communities
Primer specificity was tested by MLVA analysis of DNA from non-target organisms previously detected on sheep feet or present in soil or feces. These were Streptococcus uberis, Staphylococcus epidermis, Staphylococcus intermedius, Staphylococcus aureus, Staphylococcus hyicus, Staphylococcus chromogenis, Streptococcus dysgalactidae, Streptococcus agalactidae, Mannheimia spp., Fusobacterium necrophorum, Pseudomonas aeruginosa, Escherichia coli, and Mycobacterium tuberculosis.
To confirm specificity PCR products from 10 D. nodosus positive samples from foot swabs and 1 gingival swab were analyzed using MLVA and the resulting VNTR amplicons were submitted for Sanger sequencing (GATC Biotech AG, Cologne, Germany). Sequences were assessed for quality using CodonCode Aligner version 6.0.2. and analyzed using BLAST (27).
MLVA sensitivity was investigated by assessing its limit of detection of D. nodosus load. D. nodosus strain 4303 LBV was incubated on Eugon Agar (28) for 5 days. After incubation 1 ml of PBS, pH 7.4, was added to the D. nodosus plates, creating a cell suspension. Suspended cells were quantified using a Petroff-Hausser counting chamber (Hausser Scientific, PA, USA) and serially diluted. Sterile swabs were spiked with 50 μl of the serially diluted cell suspension resulting in 1.07 × 106 to 1.07 D. nodosus cells per swab (equivalent to rpoD copies per sample). DNA was extracted from these swabs as above. Samples were then screened for D. nodosus using the MLVA assay. To test whether detection could be improved further, all samples were submitted to a second round of the MLVA PCR assay using the same cycling conditions.
The feasibility of using MLVA to analyse multistrain communities was investigated using model communities created from eight D. nodosus isolates with different MLVA profiles. Two model communities (1 and 2) were created, each with 4 isolates (Table 1). DNA from each isolate was standardized to a concentration of 15 ng/μl and 5 μl of DNA from each isolate mixed in a 1:1:1:1 ratio. A third model community was created with the isolates in community 1, with one isolate (D. nodosus 1703A) diluted 5-fold to investigate whether non-dominant isolates could be detected. These model communities were amplified and analyzed using the MLVA protocol above.
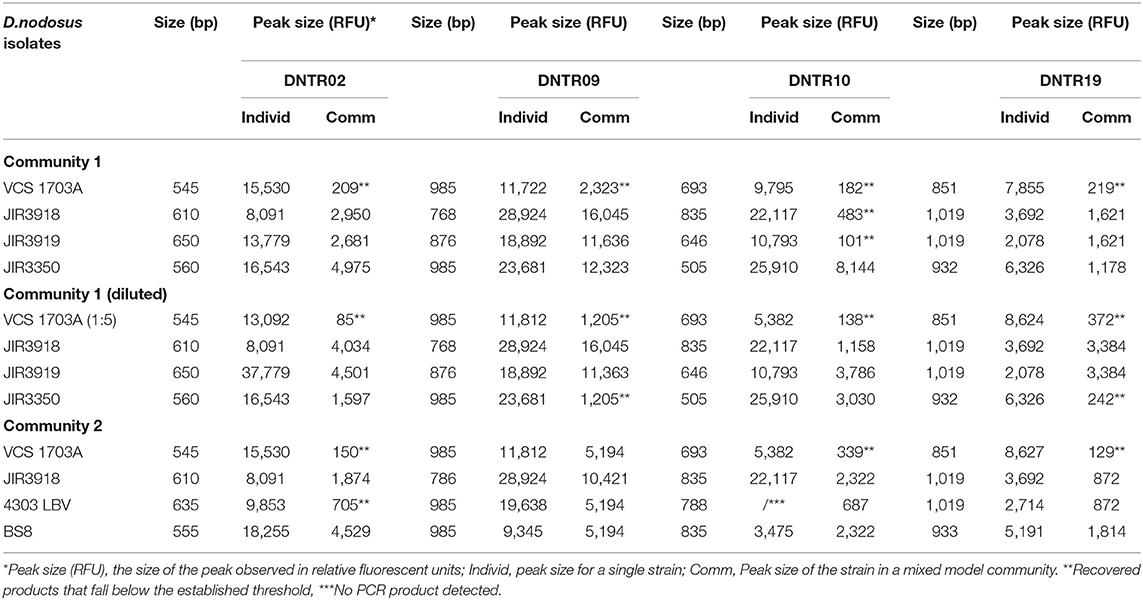
Table 1. Input and recovery of MLVA PCR amplicons and peak sizes in Dichelobacter nodosus isolates and model communities.
Assessment of the MLVA Assay on DNA Extracted From Foot and Gingival Swab Samples
Swab DNA and isolate DNA from feet and gingivae from the longitudinal study that were positive for D. nodosus by rpoD qPCR were analyzed using MLVA and fragment analysis. Secondary peaks ≤ 20% the height of the primary peak were excluded (Supplementary Figure 2). Distinct community fingerprints were obtained that could be compared visually (Figure 1). In addition, strain types within a community could be defined as “definitely present” when a community had only one variant at each locus or when more than 1 variant was detected at one locus only and one variant at the other three loci. When there were 2 or more variants at 2 or more loci strains were defined as “possibly present.” The minimum and maximum number of strain types in a community were calculated using the formulae:
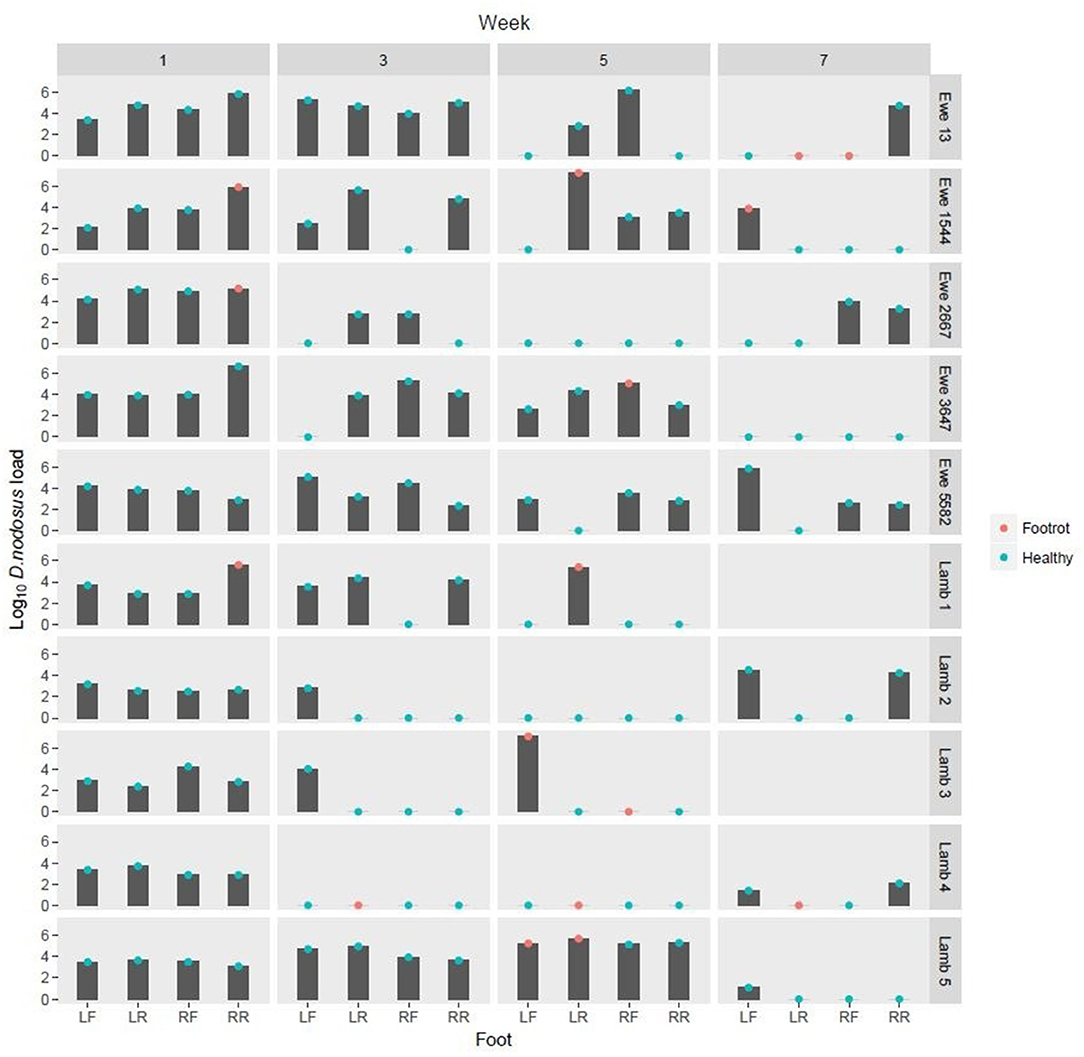
Figure 1. Log10 Dichelobacter nodosus load on DNA swabs from feet from weeks 1, 3, 5, and 7. Green dots: Healthy foot (Interdigital dermatitis score 0 or 1, footrot score 0) Red dots: Foot classed as diseased with footrot (Interdigital dermatitis score >1 and/or footrot score >0) LF, Left front; LR, left rear; RF, Right front; RR, Right Rear.
Results
Validation and Optimisation of the MLVA Scheme
The number of TRs at the four loci for strain 1703A was the same as reported by Russell et al. (20). The MLVA assay was specific for D. nodosus with no amplification in any of the 4 loci in non-target species (Supplementary Figure 3). DNA sequencing of the amplified products from foot swabs were 99–100% similar to their target sequences confirming specificity of the MLVA PCR. The mouth swab DNA yielded products for three MLVA loci, DNTR02, 09 and 10, which were 96–97% similar to their target sequence. The improved detection limit of the MLVA protocol after a single round of PCR was 102,103, 102, and 103 copies μl−1 DNA template for DNTR02, DNTR09, DNTR10, and DNTR19, respectively. A second round of MLVA PCR resulted in non-specific amplification and diluting samples did not improve specificity or sensitivity (data not shown).
VNTR Amplicons From Dichelobacter nodosus Model Communities
All D. nodosus strains in the model communities were detected, including community 3 where one strain was at 5 fold dilution; 17/48 secondary peaks were ≤ 20% the height of the primary peak and excluded (Table 1); 11 of these 17 were from the VCS strain, which was only detected correctly once, suggesting that this laboratory strain is particularly difficult to detect using MLVA.
Longitudinal Study of Persistence of D. nodosus in the Epidemiology of Footrot
FR was detected at least once on the feet of 8/10 sheep (Figure 1). D. nodosus was detected by qPCR in 97/152 (63.8%) foot swab DNA samples. It was detected on both healthy and diseased feet and on all sheep, but not all weeks (Figure 1).
Out of the 97 D. nodosus positive foot swab DNA samples, 53 (54.6%) amplified all 4 MLVA loci and in 39/53 (73.6%) samples complete MLVA profiles were obtained (Figure 2). The 39 complete profiles had a total of 156 loci with 75/156 (48.1%) with secondary peaks. The total number of peaks in these 156 loci was 220. After application of the ≤ 20% threshold (Supplementary Figure 2) 106/220 (48.2%) peaks were excluded from further analysis. DNTR02 was the most variable locus with 6 TRs and DNTRs 09 and 10 were the least variable with only 2 TRs (Figure 3).
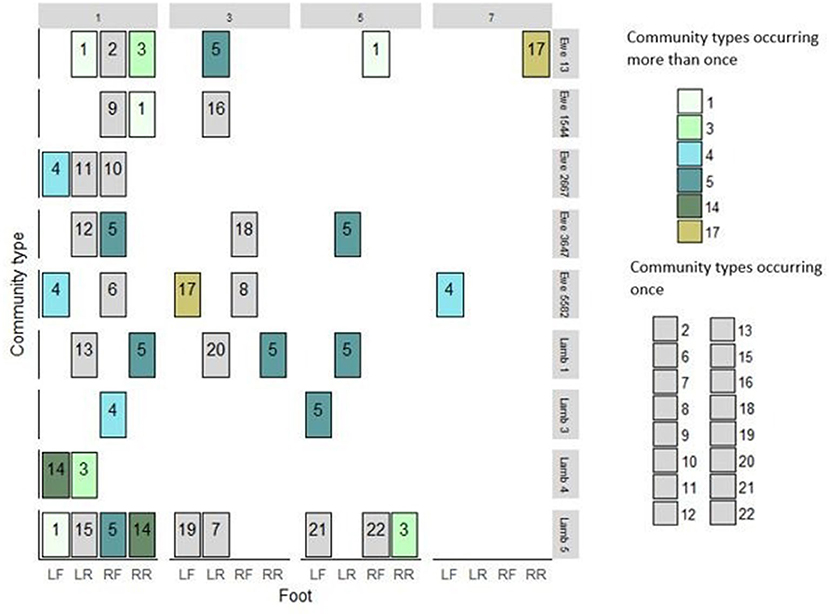
Figure 2. Dichelobacter nodosus community profiles from 39 feet of 9 sheep from weeks 1, 3, 5, and 7. Community types in green shades were detected repeatedly. Community types in gray occurred only once in the study. LF, Left front; LR, left rear; RF, Right front; RR, Right Rear.
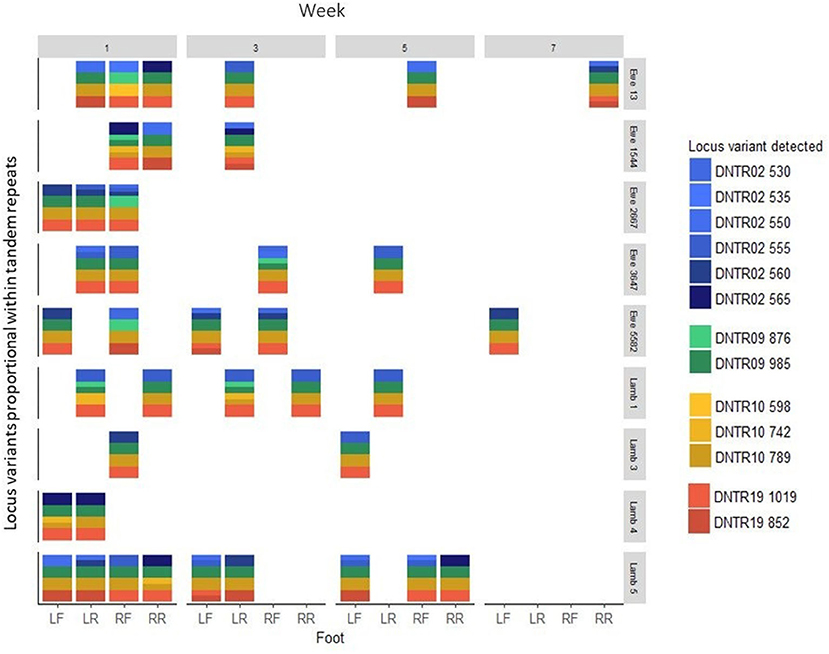
Figure 3. D. nodosus MLVA DNTR = D. nodosus tandem repeat 02, 09, 10, 19, variants proportional within TR from DNA from swab samples from 39 feet from 9 sheep in weeks 1, 3, 5, and 7. LF, Left front; LR, left rear; RF, Right front; RR, Right rear. Four colors, each of one shade, indicates a single strain.
D. nodosus was detected by qPCR in 8/38 (21.1%) gingival swab DNA samples, however, no complete MLVA profiles were obtained. Three loci were amplified in one sample and these matched loci in community types 3 and 4.
Dichelobacter nodosus Community Profiles From Foot Swab DNA
There were more MLVA positive feet in week 1, and detections declined over the 4 visits (Figure 3). There were 22 D. nodosus community types in total; seven were single strains and fifteen were multistrain; six community types were detected more than once, three of these were detected on the same four sheep and the same two feet over time (Figure 2). Community type 5, a single strain community, was most frequently detected (8/39 feet) (Figure 2). The results indicate that the MLVA differentiated D. nodosus communities spatially between feet, sheep, and over time.
Dichelobacter nodosus Strain Type Analysis From Foot Swab DNA
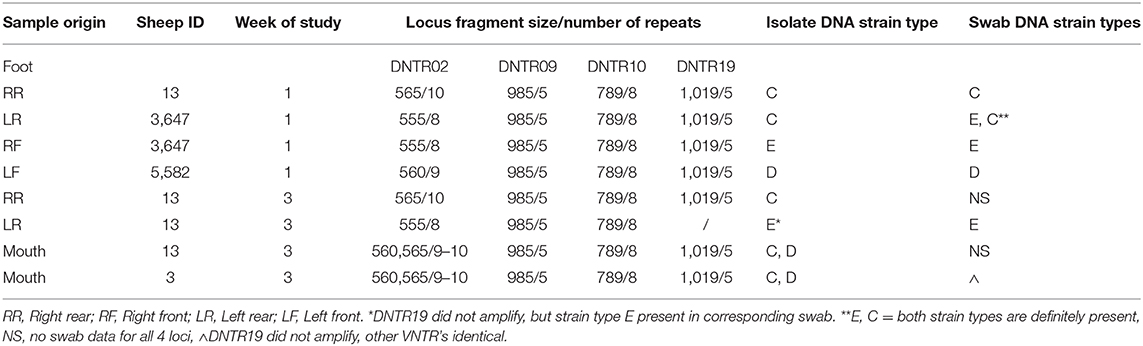
There were 17 strains definitely present and a further 8 possibly present in the 22 D. nodosus community types in foot DNA (Table 2). Seven strains were, definitely or possibly, present on more than 4 occasions and the remaining 18 strains were present on 1 to 3 occasions. The three most frequently detected single strains were C, D and E (Table 2), these were the single strain community types 3, 4, 5 (Figure 2).
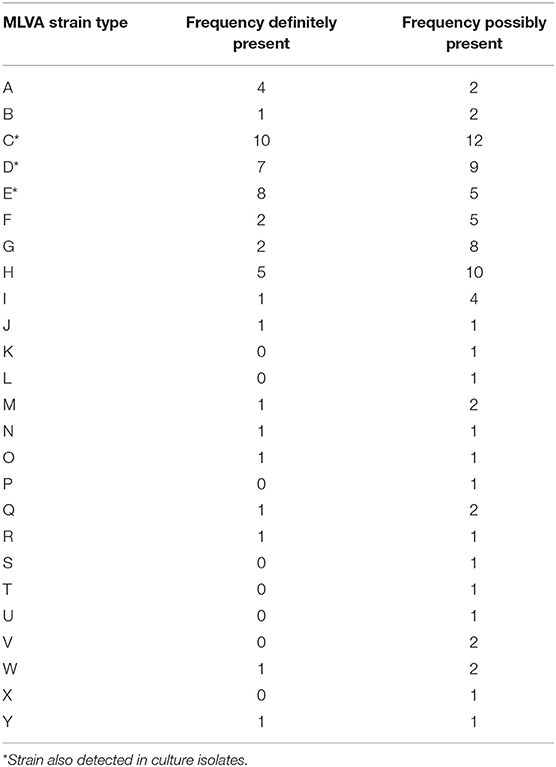
Table 2. MLVA Dichelobacter nodosus strain types from swab DNA from the feet of sheep by definitely and possibly present.
Dichelobacter nodosus Strains and Communities From Isolate DNA
D. nodosus was cultured from 6/152 (3.9%) foot swabs from 3 sheep in weeks 1, 3, and 5, with 5 complete and 1 partial MLVA profile obtained from these isolates. The three most frequently detected strains, C, D, and E (Table 2), were also detected in swab DNA from the same foot at the same time (Table 3). Complete MLVA profiles were obtained from 2 isolates from the gingival crevice and D. nodosus strains C and D (Community types 3 and 4) were detected in both samples (Table 3).
Discussion
The optimized D. nodosus MLVA (20) was developed and used successfully to investigate D. nodosus isolates and communities from DNA extracted directly from swab samples. The variability in loci and community types in the longitudinal study indicate that the improved MLVA scheme is more sensitive than previous studies (21, 23) and can be used to improve understanding of the epidemiology of communities of D. nodosus on feet over time.
Smith et al. (21) investigated transmission and persistence of D. nodosus strains on sheep's feet over a 10 months period using the original assay (20) on isolates of D. nodosus. They reported 45 animal-level repeat D. nodosus isolation events, 47% of those were isolation events of the same strain from the same foot over time. In addition, they detected and isolated the population dominant strain. In this, albeit shorter study, we obtained similar results with 4 animal-level repeat detection of D. nodosus communities and one dominant single strain community (community 5) that was detected on the same foot consecutively.
There was a high level of variability in D. nodosus communities in our study, with most variation occurring in week 1. This is possibly attributable to rainfall on and preceding the day of sampling as wet weather facilitates persistence of D. nodosus (29). A high level of D. nodosus variability was also reported by Smith et al. (21) who isolated 87 MLVA types over 10 weeks, which suggest that a large number of strains are present in footrot affected flocks and suggest that our findings represent true variability.
Our optimized MLVA was used in a subsequent longitudinal study to investigate persistence of D. nodosus strains on feet; this demonstrated that D. nodosus strains persist on the feet of diseased sheep, but not on the feet of healthy sheep (29).
Only 6 isolates were cultured in this study using published D. nodosus isolation techniques, however, the strain types from culture isolates were a subset of the strains detected from the non-culture DNA (Table 3) at the same site and time, indicating that MLVA analysis of non-cultured DNA is more sensitive than DNA from culture, as reported by others for other bacterial species (30–32). This indicates that previous studies that have compared relationships between D. nodosus using MLVA profiles of isolates (21) are incomplete and analysis would have been improved using MLVA from DNA directly.
The MLVA scheme had a limit of detection of ~103 genome copies μl−1 of extracted DNA and so there were a number of samples that were positive for D. nodosus by qPCR and negative by MLVA. Therefore, even the MLVA is not 100% sensitive. Despite this, the ability to use MLVA on non-cultured DNA to identify strains as definitely and possibly present and to produce fingerprint profiles is novel and adds to the value of MLVA as a tool to investigate strains of D. nodosus on feet over time.
This is the first occasion that a D. nodosus strain profile has been obtained from the gingival crevice, although complete strain profiles were detected from isolates and only incomplete strain profiles were obtained directly from swab DNA. Strain types C and D were present in mouths and were also among the strains most frequently detected on feet (Table 3).
The presence of regular secondary peaks in fragment analysis when testing isolate DNA has not been reported previously, although artifactual DNA extension during PCR has been reported but usually only for short sequences (33, 34). Further investigation of the secondary peaks was outside the scope of the current study and so a conservative threshold was applied to decrease the probability of artificially increasing the number of loci in a sample. As a consequence, it is possible that some non-dominant strains that were present at low abundance were classified as artifacts.
Conclusions
A sensitive and specific D. nodosus MLVA assay using four VNTR loci was validated and optimized for use on non-culture DNA. The strain types detected from isolate DNA from the same site were a subsample of those detected from swab DNA, but many more strains were present in swab DNA, indicating that it is more sensitive to analyse D. nodosus from DNA directly rather than via culture. Because the MLVA can be used to identify communities of D. nodosus on the feet of sheep over time it can be used to investigate persistence and transmission of communities of D. nodosus and so improve epidemiological understanding of D. nodosus. Other MLVA schemes may be developed for use in the non-culture based study other bacterial species.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics Statement
The animal study was reviewed and approved by Animal Welfare & Ethical Review Body (AWERB), University of Warwick.
Author Contributions
The work presented in this article was conducted by the KG. LG and KP supervised the research. All authors contributed to the article and approved the submitted version.
Funding
KG was funded by a Natural Environment Research Council studentship (NE/K007491/1). LG and KP were supported by BBSRC (BB/M012980/1).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr Ruth Kennan, Monash University for providing isolates of D. nodosus and Drs Ed Smith, Emma Monaghan, Rachel Clifton, and Jessica Taylor for their assistance with collection of samples from sheep and laboratory support.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2020.581342/full#supplementary-material
References
1. Gurung RB, Tshering P, Dhungyel OP, Egerton JR. Distribution and prevalence of footrot in Bhutan. Vet J. (2006) 171:346–51. doi: 10.1016/j.tvjl.2004.11.012
2. Gilhuus M, Kvitle B, L'Abee-Lund TM, Vatn S, Jorgensen HJ. A recently introduced Dichelobacter nodosus strain caused an outbreak of footrot in Norway. Acta Vet Scand. (2014) 56:29. doi: 10.1186/1751-0147-56-29
3. Sreenivasulu D, Vijayalakshmi S, Raniprameela D, Karthik A, Wani SA. Prevalence of ovine footrot in in the tropical climate of southern India and isolation and characterization of Dichelobacter nodosus. Rev Sci Tech OIE. (2013) 32:869–77. doi: 10.20506/rst.32.2.2209
4. Winter JR, Kaler J, Ferguson E, KilBride AL, Green LE. Changes in prevalence of, and risk factors for, lameness in random samples of english sheep flocks: 2004-2013. Prev Vet Med. (2015) 122:121–8. doi: 10.1016/j.prevetmed.2015.09.014
5. Marshall DJ, Walker RI, Cullis BR, Luff MF. The effect of footrot on body-weight and wool growth of sheep. Aust Vet J. (1991) 68:45–9. doi: 10.1111/j.1751-0813.1991.tb03126.x
6. Wassink GJ, King EM, Grogono-Thomas R, Brown JC, Moore LJ, Green LE. A within farm clinical trial to compare two treatments (parenteral antibacterials and hoof trimming) for sheep lame with footrot. Prev Vet Med. (2010) 96:93–103. doi: 10.1016/j.prevetmed.2010.05.006
7. Nieuwhof GJ, Bishop SC. Costs of the major endemic diseased in sheep in Great Britain and the potential benefits of reduction in disease impact. J of Anim Sci. (2005) 81:23–9. doi: 10.1079/ASC41010023
8. Green L, Clifton R. Diagnosing and managing footrot in sheep: an update. Practice. (2018) 40:17–26. doi: 10.1136/inp.j4575
9. Beveridge WIB. Footrot in sheep: a transmissible disease due to infection with Fusiformis nodosus. Studies on its cause, epidemiology and control. CSIR. (1941) 140:1–56.
10. Kennan RM, Han X, Porter CJ, Rood JI. The pathogenesis of ovine footrot. Vet Microbiol. (2011) 153:59–66. doi: 10.1016/j.vetmic.2011.04.005
11. Witcomb LA, Green LE, Kaler J, Ul-Hassan A, Calvo-Bado LA, Medley GF, et al. A longitudinal study of the role of Dichelobacter nodosus and Fusobacterium necrophorum load in initiation and severity of footrot in sheep. Prev Vet Med. (2014) 115:48–55. doi: 10.1016/j.prevetmed.2014.03.004
12. Witcomb LA, Green LE, Calvo-Bado LA, Russell CL, Smith EM, Grogono-Thomas R, et al. First study of pathogen load and localisation of ovine footrot using fluorescence in situ hybridisation (FISH). Vet Microbiol. (2015) 176:321–32. doi: 10.1016/j.vetmic.2015.01.022
13. Atia J, Monaghan E, Kaler J, Purdy K, Green L, Keeling M. Mathematical modeling of ovine footrot in thye UK: the effect of Dichelobacter nodosus and Fusobacterium necrophorum on the disease dynamics. Epidemics. (2017) 21:13–20. doi: 10.1016/j.epidem.2017.04.001
14. Bennett G, Hickford J, Sedcole R, Zhou H. Dichelobacter nodosus, Fusobacterium necrophorum and the epidemiology of footrot. Anaerobe. (2009) 15:173–6. doi: 10.1016/j.anaerobe.2009.02.002
15. Moore LJ, Wassink GJ, Green LE, Grogono-Thomas R. The detection and characterisation of Dichelobacter nodosus from cases of ovine footrot in England and Wales. Vet Microbiol. (2005) 108:57–67. doi: 10.1016/j.vetmic.2005.01.029
16. Stäuble A, Steiner A, Frey J, Kuhnert P. Simultaneous detection and discrimination of virulent and benign Dichelobacter nodosus in sheep of flocks affected by footrot and in clinically healthy flocks by competitive real-Time PCR. J Clin Microbiol. (2014) 52:1228–31. doi: 10.1128/JCM.03485-13
17. Maboni G, Frosth S, Aspan A, Totemeyer S. Ovine footrot: new insights into bacterial colonisation. Vet Record. (2016) 176:228. doi: 10.1136/vr.103610
18. Kraft AF, Strobel H, Hilke J, Steiner A, Kuhnert O. The prevalence of Dichelobacter nodosus in clinically footrot-free sheep flocks: comparative field study on elimination strategies. BMC Vet Res. (2020) 16:21. doi: 10.1186/s12917-020-2243-8
19. Witcomb LA. The in Situ Analysis of the Microbial Community Associated in Sheep. (PhD thesis), University of Warwick, Coventry (2012).
20. Russell CL, Smith EM, Calvo-Bado LA, Green LE, Wellington EMH, Medley GF, et al. Multiple locus VNTR analysis highlights that geographical clustering and distribution of Dichelobacter nodosus, the causal agent of footrot in sheep, correlates with inter-country movements. Infect Genet Evol. (2014) 22:273–9. doi: 10.1016/j.meegid.2013.05.026
21. Smith EM, Gilbert A, Russell CL, Purdy KJ, Medley GF, Muzafar M, et al. Within-flock population dynamics of Dichelobacter nodosus. Front Vet Sci. (2017) 4:58. doi: 10.3389/fvets.2017.00058
22. McPherson AS, Dhungyel OP, Whittington RJ. Detection and Serogrouping of Dichelobacter nodosus infection by use of direct PCR from lesion swabs to support outbreak-specific vaccination for virulent footrot in sheep. J Clin Microbiol. (2018) 56:e01730–17. doi: 10.1128/JCM.01730-17
23. Muzafar M, Calvo-Bado LA, Green LE, Smith EM, Russell CL, Grogono-Thomas R, et al. The role of the environment in transmission of Dichelobacter nodosus between ewes and their lambs. Vet Microbiol. (2015) 179:53–9. doi: 10.1016/j.vetmic.2015.04.010
24. Purdy KJ. Nucleic acid recovery from complex environmental samples. Method Enzymol. (2005) 397:271–92. doi: 10.1016/S0076-6879(05)97016-X
25. Calvo-Bado LA, Oakley BB, Dowd SE, Green LE, Medley GF, Ul-Hassan A, et al. Ovine pedomics: the first study of the ovine foot 16S rRNA-based microbiome. ISME J. (2011) 5:1426–37. doi: 10.1038/ismej.2011.25
26. Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. (1999) 27:573–80. doi: 10.1093/nar/27.2.573
27. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. (1990) 215:403–10. doi: 10.1016/S0022-2836(05)80360-2
28. Cagatay IT, Hickford JGH. Update on ovine footrot in New Zealand: isolation, identification, and characterization of Dichelobacter nodosus strains. Vet Microbiol. (2005) 111:171–80. doi: 10.1016/j.vetmic.2005.09.010
29. Clifton R, Giebel K, Liu NLBH, Purdy KJ, Green LE. Sites of persistence of Fusobacterium necrophorum and Dichelobacter nodosus: a paradigm shift in understanding the epidemiology of footrot in sheep. Sci Rep. (2019) 9:14429. doi: 10.1038/s41598-019-50822-9
30. Vranckx K, Maes D, Calus D, Villarreal I, Pasmans F, Haesebrouck F. Multiple-locus variable-number tandem-repeat analysis is a suitable tool for differentiation of Mycoplasma hyopneumoniae strains without cultivation. J Clin Microbiol. (2011) 49:2020–3. doi: 10.1128/JCM.00125-11
31. Ceglie L, Guerrini E, Rampazzo E, Barberio A, Tilburg J, Hagen F, et al. Molecular characterization by MLVA of Coxiella burnetii strains infecting dairy cows and goats of north-eastern Italy. Microb Infect. (2015) 17:776–81. doi: 10.1016/j.micinf.2015.09.029
32. Sannö X, Jacobson M, Sterner S, Thisted-Lambertz S, Aspán A. The development of a screening protocol for Salmonella spp. and enteropathogenic Yersinia spp. in samples from wild boar (Sus scrofa) also generating MLVA–data for Y. enterocolitica and Y. pseudotuberculosis. J Microbiol Methods. (2018) 150:32–8. doi: 10.1016/j.mimet.2018.05.014
33. Tuntiwechapikul W, Salazar M. Mechanism of in vitro expansion of long DNA repeats: Effect of temperature, repeat length, repeat sequence, and DNA polymerases. Biochemistry. (2002) 41:854–60. doi: 10.1021/bi0110950
Keywords: Dichelobacter nodosus, footrot, sheep, MLVA, veterinary epidemiology, bacteriology, PCR
Citation: Giebel K, Green LE and Purdy KJ (2020) A Pilot Study to Investigate the Feasibility of a Multiple Locus Variable Number Tandem Repeat Analysis to Understand the Epidemiology of Dichelobacter nodosus in Ovine Footrot. Front. Vet. Sci. 7:581342. doi: 10.3389/fvets.2020.581342
Received: 08 July 2020; Accepted: 09 November 2020;
Published: 02 December 2020.
Edited by:
Van Giap Nguyen, Vietnam National University of Agriculture, VietnamReviewed by:
Om P. Dhungyel, The University of Sydney, AustraliaMichael James Dark, University of Florida, United States
Copyright © 2020 Giebel, Green and Purdy. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Katharina Giebel, S2F0aGFyaW5hLkdpZWJlbEByYXUuYWMudWs=
†These authors have contributed equally to this work
 Katharina Giebel
Katharina Giebel Laura E. Green
Laura E. Green Kevin J. Purdy
Kevin J. Purdy