Abstract
Microalgae, due to their complex metabolic capacity, are being continuously explored for nutraceuticals, pharmaceuticals, and other industrially important bioactives. However, suboptimal yield and productivity of the bioactive of interest in local and robust wild-type strains are of perennial concerns for their industrial applications. To overcome such limitations, strain improvement through genetic engineering could play a decisive role. Though the advanced tools for genetic engineering have emerged at a greater pace, they still remain underused for microalgae as compared to other microorganisms. Pertaining to this, we reviewed the progress made so far in the development of molecular tools and techniques, and their deployment for microalgae strain improvement through genetic engineering. The recent availability of genome sequences and other omics datasets form diverse microalgae species have remarkable potential to guide strategic momentum in microalgae strain improvement program. This review focuses on the recent and significant improvements in the omics resources, mutant libraries, and high throughput screening methodologies helpful to augment research in the model and non-model microalgae. Authors have also summarized the case studies on genetically engineered microalgae and highlight the opportunities and challenges that are emerging from the current progress in the application of genome-editing to facilitate microalgal strain improvement. Toward the end, the regulatory and biosafety issues in the use of genetically engineered microalgae in commercial applications are described.
Introduction
The proficient photosynthetic microorganisms including green microalgae, diatoms, and cyanobacteria offer remarkable advantage over the terrestrial plants as a rich source of various biomolecules to be used for food, feed, and fuel applications. In addition to the faster growth rate, higher biomass productivity, and ability to synthesize complex metabolites with minimal resources are some of their key advantages. The wide taxonomic and inherent biochemical diversity among the microalgal species makes them suitable resource of abundant biomolecules with industrial and biomedical importance. Owing to this, microalgae have been continuously exploited for the production of biomolecules such as lipids, proteins, and carbohydrates. Apart from the production of secondary metabolites, microalgae have also been targeted for various applications in nutraceuticals, pharmaceuticals, dietary supplements, and personal care products. Microalgae are also utilized for concomitant CO2 sequestration, wastewater treatment, and biomass production for high-volume low-value products (Yadav et al., 2014; Mehar et al., 2019). In the last few years, owing to the high lipid content in microalgae (20–70% of dry cell weight), various start-up companies in the sector of clean energy production have attempted for commercialization of microalgae derived biofuels (Mata et al., 2010; Chisti, 2013). According to a global market research, the market for algal products across various segments is expected to grow at a compound annual growth rate of 4.2% from 2018 to 2025 and will have a total market value of more than 3.4 billion USD (https://www.alliedmarketresearch.com/algae-products-market).
Even though the commercial potential of microalgae along with its market portfolio is well-known, challenges pertaining to its economic feasibility still remain to be addressed. High biomass production along with the desired metabolite(s), cost-efficient dewatering and harvesting of biomass, green and efficient process for product extraction are some of the broad challenges to further improve the microalgal process economics. Among all these, the robust and highly efficient strain with desired characteristics can substantially improve the economics of upstream processing. Though various nutritional-, environmental-, and physiological-alteration-based cultivation have been attempted for improved microalgal productivities, commercial success remains limited (Pierobon et al., 2018). This is mainly due to the fact that these biotechnological amendments in the cultivation processes could not enhance the inherent metabolic capacity of the microalgae to hyperaccumulate the desired metabolite(s). For example, triggering the lipid accumulation in microalgae through nutrient deprivation inevitably lowers the cell division, thereby making it difficult to simultaneously achieve high lipid accumulation and high growth rate, thus decreasing the final lipid productivity (Lenka et al., 2016).
In this context, the genetic engineering of microalgae can help to overcome the inherent limitation of metabolic capacity for higher accumulation of desired biomolecules, thus eventually improving the economic feasibility of the production process. Though the wide taxonomic and genetic diversity among the microalgae offer several opportunities for genetic modifications, the scarcity of genomic resources and genetic tools limits the progress in algal bioengineering. For instance, the information of genome sequence, metabolic pathway maps, and the other genetic resources that are the key to identify target gene(s) is available only for the limited (mostly model) microalgal strains. However, despite the available genome sequence information, the annotation, and the gene functional studies related to the microalgae are still very limited. Since many of the microalgal genome sequences will be studied in near future, the computational biology and the bioinformatics may play an important role in precise genome assembly and its annotation. In addition, the multiomics datasets for microalgae can also be explored to improve the biorefinery capabilities and the quality of the microalgal bioproducts (Fayyaz et al., 2020). Moreover, the functional genetic screening through genome scale mutant libraries and their high-throughput screening may help to make robust strategies for microalgal strain improvement. Therefore, such information is extremely essential for purpose-specific bioengineering of microalgal strains. The typical strategic path from the integration of different datasets to the microalgal strain improvement is illustrated in Figure 1. In the process of genetic-engineering-based strain improvement, the molecular tools for stable transformation, selective screening, and precise gene targeting are extremely important to accomplish the genetic modification. Unlike other microorganisms, such as bacteria, yeast, and fungi, the microalgal bioengineering suffer the lack of efficient genetic tools and techniques.
Figure 1
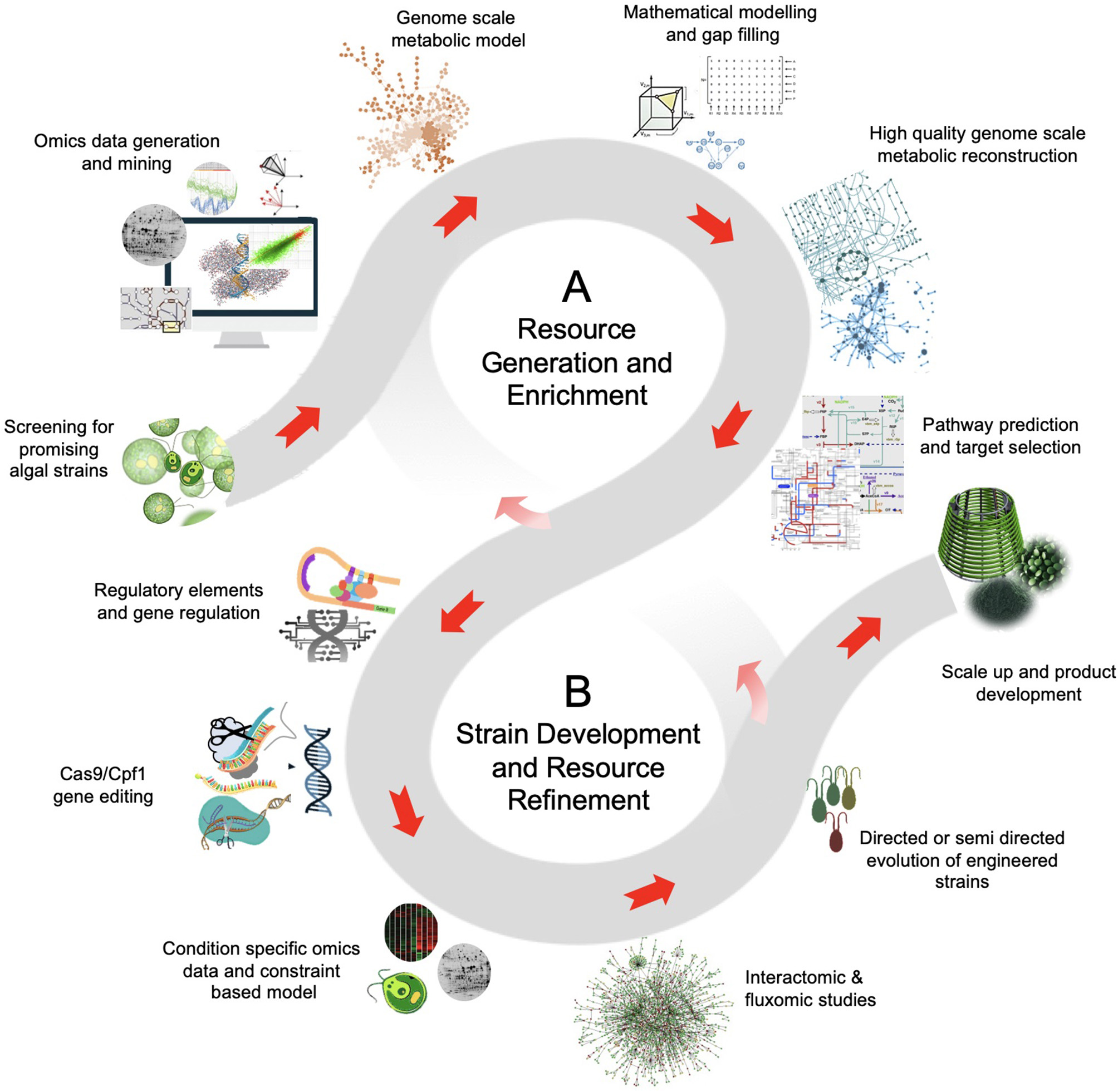
The schematic of data and resource driven strategy for microalgal bioengineering. (A)Resource generation and enrichment: The high-throughput technologies, intense computation and bioinformatic analysis, and the extensive research interest on microalgae can generate high-quality curated data. The genomic and transcriptomic data of model organisms provides a basic understanding of the biosynthetic pathway. This imperative information is aided by proteomics and metabolomics that offers functional insights for bioproduct discovery in microalgae. Also, the metabolomic data can be implemented to novel microbial isolates with limited genomic and transcriptomic information. (B)Strain development and resource refinement: The leads from metabolic models and the use of state-of-the-art technologies, such as genome-editing and high-throughput variant selection can be used for microalgae strain development. Often the metabolic flux shifts of the mutants implies an organism's evolution to optimize flux rearrangement. The objective of the flux balance shift can be biomass production or enhanced production of desired product. Moreover, the information obtained from fine-tuned modeling and genomic-editing experiments create resource avenues for further discoveries.
Considering these shortcomings, in this review, we have thoroughly mapped the information regarding the evolution of genetic modification strategies from the conventional to the emerging genome-editing tools and their implication in microalgae bioengineering. Although the bioengineering of microalgae holds the great potential to improve process economics, the risk assessment, biosafety, and regulatory issues pertaining to the use of genetically engineered microalgae must be considered and are summarized in this review. We attempt to comprehensively describe the resources for microalgae bioengineering, including omics resources, mutant resources, and their high throughput screening methodologies, transformation methods, selective markers, and precise gene-editing tools. We have also illustrated the applications of genetic engineering in the key areas of microalgal research, such as production of biomass, lipids, and bioactive molecules with the help of case studies along with the strategies used till date for the improvement of algal strains.
Advancement in the Resources for Microalgal Research
Omics Resources
Genomic and Transcriptomic Resources
Until 2008, only three microalgal species, namely Chlamydomonas reinhardtii, Thalassiosira pseudonana, and Phaeodactylum tricornutum, had been sequenced (Fu et al., 2019). In the last decade, revolution in “next-generation sequencing” technologies has led to the swift increase in the available number of draft as well as completed genomes of algal species (Table 1). Recently, Fu et al. (2019) have reviewed the efforts to sequence the genome of diverse group of microalgal species. The three sequencing projects, including one transcriptome sequencing and two genome sequencing projects, have been undertaken to generate the genetic resource for algal species. The transcriptome sequencing project named Marine Microbial Eukaryote Transcriptome Sequencing Project aimed to sequence nearly 700 marine microbial species of 17 phyla (Keeling et al., 2014). The sequence information of this dataset is available at iMicrobe Project (www.imicrobe.us/#/projects/104) and Sequence Read Archive (SRA) (BioProject PRJNA231566). Among the other sequenced transcriptomes, 140 are of marine microalgae species. Most of these sequenced species are culturable and taxonomically well-defined. Therefore, unambiguously the dataset has bias toward the gene prediction of relatively selected group of culturable isolates. Indeed, this transcriptomic data is still very helpful because it provides the extensive reference dataset for novel gene discovery and construction of computation-based metabolic models. One of the two genome sequencing projects, the ALG-ALL-CODE, was launched at NYU Abu Dhabi (lassb.abudhabi.nyu.edu/algallcode.php) and aimed at sequencing over 120 genomes of algal isolates belonging to several evolutionarily distinct phylum. Till date, the draft genome assemblies for 21 isolates are available in public domain, while the draft genome assemblies for 106 isolates will be available in near future. The other recently launched genome sequencing project is the 10KP, which aimed to generate genomic resource for 10,000 plants and eukaryotic microbes. Among the 10,000 genomes, at least 1,000 green algae (microalgae and macroalgae), and 3,000 photosynthetic and heterotrophic protists (majority will be of microalgae) are expected to be sequenced in 10KP genome sequencing initiative (Cheng et al., 2018). At present, around 60 algal accessions have been sequenced and their complete or draft genomes are available at “Phytozome” (phytozome.jgi.doe.gov) and “The Greenhouse” (greenhouse.lanl.gov). The complete or near to complete genome sequences for microalgae are summarized in Table 1. Altogether, these genome sequencing projects will generate a huge genetic resource for the microalgal species, which remained untapped due to the lack of information of their metabolic pathways, regulatory networks, and genetic potentials. In addition, there are three web-based resources available for algal genomics. The first database, pico-PLAZA, contains the genome information and other intuitive tools for functional genomics of 16 photosynthetic algal species (http://bioinformatics.psb.ugent.be/pico-plaza/) (Vandepoele et al., 2013). The second database is AlgaePath (http://algaepath.itps.ncku.edu.tw) that provides the details of gene expression based metabolic pathway prediction in Chlamydomonas reinhardtii and Neodesmus sp. UTEX 2219-4 (Zheng et al., 2014). The third one holds the information of gene co-expression data for two algal species (Chlamydomonas reinhardtii and Cyanidioschyzon merolae) and is available at ALCOdb (http://alcodb.jp) (Aoki et al., 2016). In addition, the random information of complete and draft genome sequence is available at JGI Genome Portal (https://genome.jgi.doe.gov) and Phytozome (https://phytozome.jgi.doe.gov). Besides the availability of robust computational methods, the complementation of the genome datasets with other omics datasets is indeed required for rational use of synthetic biology approach. For instance, the advantage of different omics datasets (genomics, proteomics, and metabolomics) and their integration for biological research is recently exemplified by sulfur-metabolic capacity of 14 diverse and representative strains of microalgae from different clades and habitats (Nelson et al., 2019).
Table 1
| Organism (strain used for genome sequencing) | Genome size (Mb) | Conditions or aim ofomicsstudies | Focus | Accession numbers and references | ||
|---|---|---|---|---|---|---|
| Transcriptomic studies | Proteomic studies | Metabolomic studies/ metabolic models | ||||
| Auxenochlorella protothecoides (0710) | 22.92 | Response to temperature and phosphate stress; trophic growth conditions; oil accumulation | Response to temperature, nitrogen and phosphorus starvation, ionizing radiation; trophic growth conditions, oil accumulation, | Response to temperature and, phosphate and nitrogen starvation, copper stress; oil accumulation, glycome profiling, trophic growth conditions / Genome scale and core metabolic model | Biofuel | PRJNA428835, PRJNA484804 (Li et al., 2013, 2014b; Gao et al., 2014a; Sibi et al., 2014; Wu et al., 2015; Park and Choi, 2018; Park et al., 2018; Vogler et al., 2018; Xing et al., 2018) |
| Bathycoccus prasinos (RCC 1105) | 15.07 | Normal growth conditions | - | - | Comparative analysis | PRJNA231566, https://www.imicrobe.us/#/projects/104 |
| Bigelowiella natans (CCMP2755) | 91.41 | High light stress and small RNA profiling | Profiling of proteins targeted to plastid and peri-plastid space | - | Model Organism | GSE124831, GSE115762 (Hopkins et al., 2012) |
| Botryococcus braunii (Showa) | 184.32 | Response to nitrogen deprivation, high salt, cobalt enrichment, NaHCO3, salicylic acid, methyl jasmonate, and acetic acid | - | Response to different nutrients, growth phases; tetraterpenoid and hydrocarbons analysis / Genome scale metabolic model | Hydrocarbons and biofuels | FY358876, GES71296, SRP161189, GSE96585 (Molnar et al., 2012; Cornejo-Corona et al., 2016; Thapa et al., 2016; Blifernez-Klassen et al., 2018) |
| Chlamydomonas debaryana (NIES-2212) | 120.36 | - | - | Oxylipin analysis, lipid profiling in response to different light and CO2 levels | de los Reyes et al., 2014; Toyoshima and Sato, 2015, 2018; Yoshitomi et al., 2019 | |
| Chlamydomonas reinhardtii (CC-503 cw92 mt+) | 120.4 | Response to nutrient starvation, oxidative and heat stress, high light intensity, diurnal cycle; ciliogenesis; lipid accumulation | Response to nitrogen and sulfur starvation; exposure to high salinity, high CO2, dark and anoxic conditions; lipid mutant, lipid droplet proteins | Response to nitrogen starvation, dark and anoxic conditions / Genome scale and core metabolic model | Model organism | GSE17970, PRJNA379963 (May et al., 2009; Chen et al., 2010; Baba et al., 2011; Nguyen et al., 2011; Longworth et al., 2012; Mastrobuoni et al., 2012; Choi et al., 2013; Chaiboonchoe et al., 2014; Wase et al., 2014; Sithtisarn et al., 2017; Salguero et al., 2019) |
| Chlorella pyrenoidosa (FACHB-9) | 56.99 | Response to CO2 deprivation, bisphenol A, salt stress, high light stress, glucose starvation and hydroxyl radical; trophic growth conditions | Dried biomass, exposure to inhibitor of mitochondrial respiratory electron transport | Lipid profiling under copper stress and different nitrate levels / Core metabolic model | Biofuels | SRX399080, GSE40028, GSE69816, PRJNA292642, PRJNA526277 (Yang et al., 2000; Sibi et al., 2014; Liu et al., 2018b; Wan et al., 2018; Zhang et al., 2018; Duan et al., 2019) |
| Chlorella sorokiniana (1230) | 58.53 | Response to nitrogen deprivation, different pH, and high CO2 | Response to inoculum sizes, light intensity and glucose concentrations, nitrogen starvation; bioactive peptide analysis | Response to high-density cultivation and UV radiation; fatty acid profiling | Biofuels | GAPD00000000, GSE98781, GCUV00000000 (Lu et al., 2013; Ma et al., 2013; Rosenberg et al., 2014; Li et al., 2015a; Chen et al., 2017; Kumar et al., 2018; Tejano et al., 2019) |
| Chlorella variabilis (NC64A) | 46.16 | Response to early phase of Chlorella virus-1 infection | - | Nitrogen deprivation and long-chain alkenes/Genome scale metabolic model | Biofuels | SRP026413 (Juneja et al., 2016; Sorigue et al., 2016) |
| Chlorella vulgaris (NJ-7) | 39.08 | Response to nitrogen starvation and salt stress | Response to nitrogen depletion and repletion, heterotrophic and Na induced lipid accumulation, S-nitrosylated proteome in nitrogen deplete and replete condition | Lipid profiling under copper stress, effect of graphene oxide nanomaterial, N-glycan profiling / Core metabolic model | Biofuels | LDKB00000000 (Guarnieri et al., 2011, 2013; Sibi et al., 2014; Li et al., 2015b; Ouyang et al., 2015; Henard et al., 2017; Zuñiga et al., 2018; Mocsai et al., 2019) |
| Chloroidium sp. (CF) | 54.31 | - | Normal growth conditions / Genome scale metabolic model | Ecological importance | Nelson et al., 2017, 2019 | |
| Chromochloris zofingiensis (SAG 211-14) | 58 | Response to nitrogen deprivation, high light; heterotrophic conditions, different growth conditions | Lipid droplets analysis | Lipid and carotenoid profiling in response to glucose | Carotenoids and fatty acids | SRP067324, GSE92515 (Wang et al., 2019c; Zhang et al., 2019) |
| Coccomyxa sp. (LA000219) | 48.54 | Response to arsenic treatment | - | Response to arsenic treatment | Model organism and biofuels | Koechler et al., 2016 |
| Coccomyxa subellipsoidea (C-169) | 48.83 | Response to CO2 supplementation; miRNA profiling | - | Response to nitric oxide, cadmium stress, carbon source, nitrogen starvation, phytohormones | Biofuels | GSE76638PRJNA428141 (Kováčik et al., 2015; Allen et al., 2017; Liu et al., 2018a; Wang et al., 2019e) |
| Cyanidioschyzon merolae (10D) | 16.55 | Response to diurnal cycle, different CO2 level, blue and red light, UV irradiance | Response to low temperature acclimatization; photosystem II proteins | Response to different CO2 level, diurnal cycle; hydrocarbon and lipid profiling in response to cyanobacterial Acyl-ACP Reductase overexpression | Model organism | GSE37673, GSE83828, GSE100372 (Krupnik et al., 2013; Rademacher et al., 2016; Nikolova et al., 2017; Miyagishima et al., 2019) |
| Dunaliella salina (CCAP 19/18) | 343.7 | Response to osmotic and oxidative stress, nitrogen depletion, salinity, high light; different growth phases | Response to arsenate, high salinity, high light and high bicarbonate ion level; flagella composition | Response to nitrogen starvation | Halophile, Biofuels, β-carotene and glycerol production | Katz et al., 2007; Jia et al., 2009, 2016; Gu et al., 2014; Ge et al., 2016; Lv et al., 2016; Zhao et al., 2016; Wei et al., 2017b; Wang et al., 2019d |
| Emiliana huxleyi (CCMP1516) | 167.68 | Response to nitrogen, sulfate and phosphorus starvation, calcium concentrations, elevated temperature and CO2 | Response to different calcium concentration | Response to host-virus (E. huxleyi virus) interaction, phosphorus and nitrogen starvation; lipidomic | Coccolithophore | GSE24341, E-MTAB-2274, SRP017794, SRX756940 (Benner et al., 2013; Rokitta et al., 2014; Hunter et al., 2015; McKew et al., 2015; Wördenweber et al., 2018) |
| Fistulifera solaris (JPCC DA0580) | 49.74 | Response of nutrient depleted and replete conditions on lipid accumulation and its degradation | Lipid droplet proteins | - | Biofuels | DRA002404 (Nonoyama et al., 2019) |
| Fragilariopsis cylindrus (CCMP1102) | 80.54 | Response to temperature, high CO2, prolonged darkness, and nitrogen and iron limitation; small RNA profiling | Response to temperature, salinity stress, prolonged darkness, high CO2, iron starvation | Response to different growth phases | Psychrophile | E-MTAB-5024, GSE57987 (Lyon et al., 2011; Boroujerdi et al., 2012; Kennedy et al., 2019) |
| Galdieria sulphuraria (074W) | 13.71 | Response to cold acclimation | Photosystem-II analysis | - | Extremophile | PRJNA487158, GSE89169 (Thangaraj et al., 2010) |
| Guillardia theta (CCMP2712) | 87.15 | Small RNA profiling under light and dark conditions, mRNA splicing analysis | Response to different light intensities | - | Eukaryote endosymbiosis | GSE124831, SRR747855 (Kieselbach et al., 2018) |
| Haematococcus pluvialis (SAG 192.80) | 365.78 | Response to high light, salinity, iron, acetate, salicylic acid and jasmonic acid, nitrogen depletion and repletion, photooxidative stress; distinct growth phases | Cell wall protein, astaxanthin accumulation, response to high light stress, salicylic acid, and jasmonic acid | Lipid analysis, pigments and protein profiling, live single-cell analysis | Carotenoids | Wang et al., 2004; Tran et al., 2009; Peled et al., 2011; Gu et al., 2014; Recht et al., 2014; Su et al., 2014; Gao et al., 2016; Baumeister et al., 2019; Luo et al., 2019 |
| Helicosporidium sp. (ATCC 50920) | 12.37 | Transition from free-living organism to obligate intracellular parasite | - | - | Parasite | Pombert et al., 2014 |
| Klebsormidium nitens (NIES-2285) | 104.21 | Response to auxin treatment and cold stress | - | Response to cold stress | Tolerance to UV and harsh conditions | PRJDB4958, PRJNA500592 (Nagao et al., 2008) |
| Micromonas commoda (RCC299) | 21.11 | Response to different light regimes and ultra-violet light stress | Response to chronic phosphate limitation and subsequent relief, high light and UV-radiation | - | Marine phytoplankton | Cuvelier et al., 2017; Guo et al., 2018 |
| Micromonas pusilla (CCMP1545) | 21.96 | Response to phycodnavirus MpV-SP1 infection, phosphate deplete and replete, day-night cycle | Phosphate deplete and replete condition, day-night cycle | Response to phosphate deplete and replete condition; different growth phases, | Marine phytoplankton | PRJNA422663 (van Baren et al., 2016; Waltman et al., 2016; Kujawinski et al., 2017) |
| Micromonas sp. (ASP10-01a) | 19.58 | Normal growth conditions | - | - | Marine phytoplankton | van Baren et al., 2016 |
| Monoraphidium neglectum (SAG 48.87) | 69.71 | Nitrogen deprivation | - | - | Biofuels | PRJNA221625 (Jaeger et al., 2017) |
| Nannochloropsis gaditana (CCMP1894) | 30.86 | Response to light intensity regimes and nitrogen replete and deplete condition | Fresh and atomized biomass | Response to light intensity regimes and nitrogen deprivation / Genome scale metabolic model | Biofuels | Radakovits et al., 2012; Sorigue et al., 2016; Ajjawi et al., 2017; Shah et al., 2017; Fernandez-Acero et al., 2019; Patelou et al., 2020 |
| Nannochloropsis limnetica (CCMP505) | 33.51 | - | - | Nitrogen deprivation | Biofuels | Sorigue et al., 2016 |
| Nannochloropsis oceanica (LAMB2011) | 29.26 | Response to different CO2 levels, phosphorus and nitrogen limitation, light and dark cycle, fresh water acclimation; transition from quiescence to autotrophy | Response to long-term nitrogen starvation, low CO2; single-cell-level phenotypic heterogeneity | Response to osmotic downshift and nitrogen depletion | Biofuels | Dong et al., 2013; Pal et al., 2013; Sorigue et al., 2016; Poliner et al., 2018; Chen et al., 2019; Wei et al., 2019 |
| Nannochloropsis oculate (CCMP525) | 26.27 | - | Nitrogen deprivation, cadmium stress | Nitrogen deprivation | Lipids and protein content | Kim et al., 2005; Sorigue et al., 2016; Tran et al., 2016 |
| Ostreococcus lucimarinus (CCE9901) | 13.2 | - | - | Genome scale metabolic model | Small genome | Krumholz et al., 2012 |
| Ostreococcus tauri (RCC4221) | 13.03 | Response to OtV5 virus infection, light and dark cycle, iron limitation, and high light; life cycle stages | Phosphoproteome in response to casein kinase 2, light dark cycle | Glycerolipid profiling under nutrient deprived condition, diurnal variations, nitrogen deprivation / Genome Scale metabolic model | Small genome | Krumholz et al., 2012; Martin et al., 2012; Hindle et al., 2014; Le Bihan et al., 2015; Lelandais et al., 2016; Sorigue et al., 2016; Degraeve-Guilbault et al., 2017; Hirth et al., 2017 |
| Parachlorella kessleri (NIES-2152) | 59.18 | Response to salt stress and sulfur deplete and replete | Salt stress | Nitrogen, sulfur and phosphorus deprivation | Ota et al., 2016a,b; Shaikh et al., 2019; You et al., 2019 | |
| Phaeodactylum tricornutum (CCAP 1055/1) | 27.45 | Response to nitrogen, iron, carbon and phosphorus deprivation, cadmium stress, mixotrophic growth, grazing stress, different light intensities, and regimes, salicylic acid; non-coding microRNA | Response to nitrogen limitation, oxidative and dark stress; phosphoproteomics under high light, nitrogen, and iron deficiency | Response to blue and red light, nitrogen and phosphorus deprivation; glycerolipid profile; mixotrophic growth / Genome scale and core metabolic model | Model organism | PRJEB11970, SRX648639 (Chen et al., 2014; Ge et al., 2014; Jungandreas et al., 2014; Rosenwasser et al., 2014; Yang et al., 2014; Abida et al., 2015; Alipanah et al., 2015; Feng et al., 2015; Bai et al., 2016; Longworth et al., 2016; Sorigue et al., 2016; Yoneda et al., 2016; Villanova et al., 2017; Remmers et al., 2018; Smith et al., 2019) |
| Picochlorum sp. (SENEW3 / DOE 101) | 13.39 / 15.25 | Response to salinity stress and high temperature | - | - | Biofuels | PRJNA245752, PRJNA389600 |
| Scenedesmus sp. (ARA3, ARA) | 93.24 | Response to phosphorus and nitrogen starvation, lipid accumulation | Response to salinity stress; lipid accumulation | Response to salinity and arsenic stress; lipid accumulation | Biofuels | PRJNA428298 (Chu et al., 2011; Arora et al., 2018, 2019; Wang et al., 2019b) |
| Scenedesmus obliquus (UTEX393) | 107.72 | Response to diurnal changes and nC60; wild type and starch less mutant comparison | Thylakoid membrane proteome, toxicity of silver nanoclusters | Response to nC60 and silver nanoparticles; different photoperiod and growth phases | Lipid and biomass | E-MTAB-7009 (Kantzilakis et al., 2007; Du et al., 2017; Zhang et al., 2017; Vendruscolo et al., 2019; Wang et al., 2019a) |
| Symbiodinium minutum (Mf 1.05b.01) | 609.48 | Diurnal cycle, cultured, and freshly isolated cells | - | Response to acidification | Coral symbiont | PRJNA544863 (Jiang and Lu, 2019) |
| Symbiodinium microadriaticum (CCMP2467) | 808.2 | Response to different temperature, dark, and salinity stress; normal growth conditions, miRNA profiling | - | Response to environmental variation | Coral symbiont | GSE47373, GSE47372 (Klueter et al., 2015; Aranda et al., 2016) |
| Tetraselmis striata (LANL1001) | 227.95 | Normal growth | - | - | PRJNA231566, https://www.imicrobe.us/#/projects/104 | |
| Thalassiosira oceanica (CCMP1005) | 92.18 | Response to iron and copper | Response to iron and copper; extracellular superoxide production | - | Model organism | PRJNA382002, SRA045825 (Lommer et al., 2012; Diaz et al., 2019) |
| Thalassiosira pseudonana (CCMP1335) | 32.44 | Response to nitrogen and phosphorus deprivation, salinity, light intensity, triphenyltin chloride, silicon, CO2 levels, source of light, and nitrogen | Response to nitrogen and phosphorus starvation, light intensity, salinity, triphenyltin chloride, CO2 levels, silicon, micronutrients deficiency, benzo(a)pyrene, K. brevis allelopathy; composition of nano- and micropatterned biosilica cell wall, mitochondrial and plastid proteome | Response to phosphate deplete and replete condition, cobalamin scarcity; K. brevis allelopathy | Model organism | Carvalho and Lettieri, 2011; Dyhrman et al., 2012; Du et al., 2014; Kettles et al., 2014; Kustka et al., 2014; Luo et al., 2014; Poulson-Ellestad et al., 2014; Yi et al., 2014; Jian et al., 2017; Kujawinski et al., 2017; Chen et al., 2018; Heal et al., 2019; Schober et al., 2019 |
| Trebouxia gelatinosa (LA000220) | 61.73 | Response to dehydration and subsequent rehydration | - | - | Colonization through symbiosis | PRJNA213702 |
| Volvox carteri f. magariensis (Eve) | 137.68 | Response to low dose of UV-B radiation; somatic and reproductive cells | - | - | Multicellular alga, model organism | E-MTAB-5691 and GSE104835 |
| Yamagishiella unicocca (NIES-3982) | 134.24 | Normal growth condition | - | - | Multicellular alga, model organism | PRJNA532307 |
List of microalgae and diatoms with complete or near to complete genome, and the overview of reported omics studies.
Proteomic Resources
The quantitative data of protein expression under different experimental conditions is advantageous for better understanding of regulatory pathways, which differ at the post-transcriptional level. Since, several studies failed to give a high correlation between transcriptomic and proteomic data (Haider and Pal, 2013), the availability of quantitative proteomic and transcriptomic data under defined experimental condition will provide strategic insights for strain improvement in microalgae. In particular, several analyses have been performed to identify the proteome dynamics and the corresponding transcriptome analysis. However, this was mainly focused to understand the lipid metabolism in model and/or oleaginous microalgae with potential of biofuel production (Table 1). The literature mining shows that the majority of proteomics studies were performed under experimental conditions, including nitrogen starvation, copper deprivation, light intensity regimes, heterotrophic cultivation, and salt stress (Table 1). The majority of differentially abundant proteins were found to have functions in metabolic pathways related to fatty acid and lipid metabolism, carbohydrate metabolism, photosynthesis, and cell structure integrity and maintenance. In addition, the large numbers of algal proteins have been predicted through genomic sequence analysis and the information is available at the Uniprot (https://www.uniprot.org) and Protein Data Bank archive (https://www.rcsb.org). In an attempt to comprehensively cumulate the structural, physicochemical, and functional information of algal proteome, the non-redundant protein database of 31 algal species was developed and is available at Algal Protein Annotation Suite (Alga-PrAS) (Kurotani et al., 2017).
Metabolomics and Metabolic Models
The metabolites are the intermediate or end products of the cellular regulatory processes that are implicated through the transcriptome and proteome, and thus represent the cellular response to the stimulus. Some metabolites are also involved in the regulation of cellular responses by regulating the activity of enzymes involved (Wegner et al., 2015). Thus, information of metabolic profile in response to the experimental conditions may help to target the processes or pathways, which could be helpful in metabolic-engineering of microalgal strains. The quantitative and qualitative analysis of metabolites is now fairly possible even though they have a wide variation in chemical properties, such as polarity, charge, solubility, volatility, and molecular weight. This has become possible due to the advances in non-targeted metabolite profiling and its platforms, such as capillary electrophoresis-mass spectrometry, gas chromatography-mass spectrometry, liquid chromatography-mass spectrometry, Fourier transform ion cyclotron resonance-mass spectrometry, and nuclear magnetic resonance spectroscopy. Similar to the transcriptome and proteomic studies in microalgae, the majority of metabolomic (though only few untargeted metabolic studies reported) studies were also focused on lipid metabolism under various environmental conditions (Table 1). Recently, the potential of single-probe mass spectrometry technology has been demonstrated for near in-situ analyses of single cell of Scrippsiella trochoidea under nitrogen starving and light vs. dark conditions to analyze the lipid content and lipid profile (Sun et al., 2018). This single-cell-targeted metabolomics may prove to be instrumental in the future algal research, since it reduces the chances of experimental artifacts and confounds, thereby minimizing the cell to cell metabolic variability. Unlike genome and transcriptome databases, unfortunately, no dedicated database is available for the microalgal metabolomics. Although the attempts were made to reconstruct genome-scale metabolic models at system level, they are based on the information of the genome, transcriptome, and scarcely available experimental data. For organisms like C. reinhardtii, Chlorella spp., P. tricornutum, and some blue-green algae (cyanobacteria), the genome-scale metabolic models are available. The core metabolic models and genome-scale system-level metabolic networks available for different microalgal species are given in Table 1. In addition, some databases, such as KEGG (https://www.genome.jp/kegg/pathway.html), Reactome (https://reactome.org), and Metacyc (https://metacyc.org) contain predicted and experimentally proven metabolic network information and can be explored for predictive and integrative biology in microalgae. The information available through the genetic characterization of cellular pathways, and high throughput genome-scale studies under different experimental conditions, is contributing toward the refinement of metabolic models for system-level analysis of biological processes.
Mutant Resources for Microalgae
The mutant library for an organism is the best available tool to accelerate the functional characterization of enormous set of uncharacterized genes for better understanding of fundamental biological processes. The potential of such mutant libraries has been exemplified by those that are available for organisms such as Saccharomyces cerevisiae (www.sequence.stanford.edu/group/yeast_deletion_project/deletions3) and Arabidopsis thaliana (www.arabidopsis.org/portals/mutants/index.jsp). The mutant libraries are instrumental in the reverse genetic studies. However, generating such libraries for microalgae is limited by the lack of efficient transformation and genetic manipulation protocols (discussed in later sections). The insertional mutagenesis through random non-homologous end-joining is the method of choice to generate the mutant libraries. Till date, only two genomewide random insertion mutant libraries have been generated for C. reinhardtii using the insertional mutagenesis approach. The first collaborative project, Chlamydomonas Library Project (CLiP) was launched in 2010 by Jonikas (now at Princeton University, USA) and Grossman at Carnegie Institution for science (USA), Fitz-Gibbon (University of California Los Angeles, USA), and Lefebvre (University of Minnesota, USA) to generate the genome-scale insertional mutant library for C. reinhardtii. The mutants from this library have been released for the research community and other stakeholders on periodic basis. The complete library featuring more than 62,000 mutants that covers 83% of nuclear protein-encoding genes is now available at Chlamydomonas Resource Center (www.chlamycollection.org/products/clip-strains). Importantly, the mutants in this library are fully mapped for insertion sites and indexed with unique DNA barcode for high-throughput screening of pooled mutants for a particular trait or biological process (Li et al., 2016b, 2019). Similarly, the Huang group at Institute of Hydrobiology, China, generated another insertional mutant library of C. reinhardtii with ~150,000 insertional mutants (Cheng et al., 2017). Although this library contains higher number of mutants than that of CLiP, the list of mutants and their mapping information is not available in public domain. In addition, a non-indexed insertional mutant library of C. reinhardtii with ~49,000 mutants was also developed and is available for the scientific community at Chlamydomonas Resource Center (http://chlamycollection.org). The potential utility of these mutant libraries can be attributed to the discovery of novel candidate genes involved in biological and physiological processes, such as photosynthesis, lipid biosynthesis, and intraflagellar transport in microalgae (Dent et al., 2015; Li et al., 2016b, 2019).
In addition to the insertional mutagenesis, the mutagenic agents are being regularly used to generate the mutant strains with desired traits. Several attempts have been made using forward genetic approach to characterize the genes involved in the molecular pathways targeting a desired trait. Since the C. reinhardtii is considered as premier reference organism for understanding the basic algal metabolism and biological processes, most of the forward genetic screens have been performed in this model organism. These forward genetic screening in C. reinhardtii and some other model microalgae have been performed mostly to identify the genetic factors responsible for desirable traits, such as higher biomass and cell culture density (Thung et al., 2018), enhanced lipid content (Cagnon et al., 2013; Lee et al., 2014), or to understand the basic cellular processes such as photosynthesis (Dent et al., 2015; Li et al., 2019), non-photochemical quenching (Schierenbeck et al., 2015), lipid metabolism (Li et al., 2016b; Schulz-Raffelt et al., 2016; Cheng et al., 2017), and flagellar responses (Hilton et al., 2016; Cheng et al., 2017). In an integrative approach, the P. tricornutum mutants with enhanced carotenoid biosynthesis were subjected to genome-scale metabolic network simulation to identify the metabolic reactions that are highly correlated with the carotenoid biosynthesis (Yi et al., 2018). This study exemplified the use of system-biology approach to target the key pathway(s) that should be considered during bioengineering in diatoms. Recently, using a modified approach named as bulked mutant analysis and bulked mutant RNA sequencing, the single nucleotide polymorphisms and indels were identified that are associated to the growth-related genes in Nannochloropsis oceanica (Liang et al., 2019). These methods of forward genetic screen have the potential to facilitate the genetic investigation of diverse microalgae with various desirable traits.
High-Throughput Screening Methodologies for Microalgae
The previous section reviewed various genomic and mutant resources that are available for the microalgae research. The resources for microalgal forward genetics have the potential to revolutionize the identification of mutants with desired traits, however limited to availability of the rapid screening methods. Moreover, the screening of microalgae natural pools to identify functional components is low due to the lack of effective rapid and high-throughput analysis tools (Lee et al., 2013). In addition, this also limits our capacity for the real-time monitoring of process for target compound production using microalgae. To enrich the mutants capable of accumulating high lipid content, Sharma et al. (2015) developed and validated a high-throughput work flow strategy based on in-situ analysis of lipid bodies using confocal Raman microscopy combined with fluorescence activated cell sorting (FACS). A precise and efficient Raman platform was developed to distinguish the contrasting features of lipids such as chain length and saturation level in lipid-expressing cells generated through UV mutagenesis. Terashima et al. (2015) introduced another high-throughput advanced technique, named Chlamydomonas high-lipid sorting (CHiLiS), which enables to isolate mutants with high lipid content. CHiLiS is based on the fact that Nile Red (lipid detecting dye)-stained lipid pools were enriched by using FACS. In this method, the staining extent was raised to a certain level for increasing the enrichment tendency without interfering with the cell's viability. These high-throughput methods have the potential of selecting the mutant strains that can be used either for the understanding of molecular basis of high lipid accumulation or engineering of microalgae for maximizing the production of lipids. Based on the staining of lipid bodies with fluorescent dyes, several high throughput systems are available commercially. Semi-automated QPixTM 400 Series system from Molecular Devices is one such example (https://www.moleculardevices.com/sites/default/files/en/assets/brochures/biologics/qpix-400-systems). The Fourier transform infrared spectroscopy also demonstrated its sensitivity to screen mutants of C. reinhardtii for variation in their lipid and carbohydrate profile under specific nutrient stress conditions (Bajhaiya et al., 2016). Based on this screening, nutrient starvation response genes (PSR1, SNRK2.1, and SNRK2.2) with possible role in lipid and starch accumulation were identified.
In an another approach, to isolate the algal cells with superior photosynthetic activity, the high-throughput microfluidics were used in the microalgal selection process (Kim et al., 2016). This system used the strong positive relationship between phototaxis and photosynthetic efficiency, where the competitive phototactic response was employed to isolate the highly photosynthetic efficient strains at the single-cell level using a microfluidic system. Also, the putative candidate genes related to the transcriptional regulation (JGI Chlre4, protein ID: 525919, 516641, 513996), cellular metabolism (519327, 523869, 515661) signal transduction (516786), flagellar function (518826), and membrane transport (protein ID; 516748, 516786, 513005, 520695, 512634) were identified, that might have some role in enhanced photosynthetic activity and phototactic response in mutant strains. The putative candidate genes identified in this study may be cataloged for their use in microalgal strain engineering strategies. Even after the identification of photosynthetic efficient microalgal strains, optimization of the light conditions remains critical to augment system efficiency. Recently, a novel high-throughput screening system was developed by Sivakaminathan et al. (2018), which simulates fluctuating light regimes in mass cultures. This high-throughput miniaturized light system is capable of screening up to 18 different combinations of light regime and up to 1,728 conditions to evaluate species-specific light conditions for maximum photosynthetic efficiency and productivity.
For the screening of biopigments accumulation, a 96-well microplate-based high-throughput assay was developed to identify P. tricornutum mutants with high carotenoid content (Yi et al., 2018). The assay was based on the fact that fluorescence intensity of chlorophyll a and neutral lipids (stained with fluorescence dye) has a significant correlation with the carotenoid content during exponential growth phase of P. tricornutum. Generally, the in-situ optical detection-based methods fail to provide detailed information on the pigment composition in microalgae because of the possible overlapping of absorbance and emission spectra of various pigments. In such cases, the extraction and subsequent detection is the only method of choice. However, the extraction of a particular pigment type is a time-consuming multi-step process that also required a suitable extraction solvent to effectively extract the pigment. A rapid and reliable microwave-assisted extraction and subsequent detection of microalgal pigment using relevant method could be helpful in developing high-throughput screening platform for microalgal pigments (Pasquet et al., 2011). An enzyme-linked immunosorbent assay (ELISA) on microtiter platform was developed by Jirásková et al. (2009) to detect the presence of phytohormones, such as abscisic acid, indole-3-acetic acid, cis- and trans-zeatin, and isopentenyladenosine in microalgae. This high-throughput application of ELISA-based microtiter platform can be extrapolated to the other bioactive compounds if suitable antibodies and/or antigens are available. Likewise, a simple and inexpensive high-throughput bioassay was developed to screen the algal mutants or isolates producing high H2 under saturating light intensity (Wecker and Ghirardi, 2014). The screening assay used the agar overlay of Rhodobacter capsulatus bacteria carrying a green fluorescent protein that responds to H2 produced by single algal colony. Among the other high-throughput screening methods, the phenotype microarray technologies have also shown promise to screen-defined metabolic activities in response to array of different drugs, chemicals, and metabolites (www.biolog.com).
Genetic Engineering in Microalgae
Transformation Technologies and Selectable Markers
The first nuclear transformation of C. reinhardtii using polyethylene glycol or poly-L-ornithine was demonstrated in early 1980's. Here, the complementation of arginine-requiring, cell-wall deficient mutant was performed through successful integration of yeast arg4 locus (Rochaix and Dillewijn, 1982). In the late 1980's, the successful stable nuclear transformation in C. reinhardtii was demonstrated using the biolistic transformation approach to deliver the native genes to complement auxotrophic growth in mutants (Debuchy et al., 1989; Kindle et al., 1989; Mayfield and Kindle, 1990). Later in the 1990's, the success of glass bead agitation and electroporation were demonstrated, where the later was found to be the most efficient method to transform the nuclear genome of C. reinhardtii (Kindle, 1990). The droplet electroporation on microfluidic chip was found to have threefold higher transformation efficiency than the electroporation cuvettes (Qu et al., 2012). In addition, the use of other methods by employing silicon carbide whiskers (Dunahay, 1993), Agrobacterium tumefaciens (Kumar et al., 2004), and nanoparticles (Kim et al., 2014) have been also demonstrated to successfully transform the nuclear genome of C. reinhardtii. The methods for the nuclear transformation in other microalgal species such as Phaeodactylum, Nannochloropsis, Dunaliella, and Haematococcus are available (Table 2). The various transformation techniques and the selectable markers used for the screening of transformants, and mainly includes the use of antibiotic, herbicide resistance, and auxotrophic markers are listed in Table 2. The evolutionary divergence of the cellular machinery in microalgae, however, limits the use of existing plant or other microbe-based selectable markers for selection purposes. For instance, the trait stacking in the industrially important microalgae “Nannochloropsis” through genetic engineering is mostly limited by the availability of selectable markers (Verruto et al., 2018). The use of auxotrophic selection marker is mostly a desirable trait; however, a pre-requisite that the strain to be transformed must be auxotrophic mutant for the selectable marker, which may sometimes interfere with the experimental setup.
Table 2
| Organism | Genetic tools and techniques | References | ||
|---|---|---|---|---|
| Transformation | Genetic manipulation | Selectable markers | ||
| Botryococcus braunii | Electroporation | Gene integration and expression | Antibiotic: aphVIII | Berrios et al., 2016 |
| Chlamydomonas reinhardtii | Biolistic, glass bead agitation, electroporation, and agrobacterium-mediated | Gene expression, RNA interference, gene-editing using ZFNs and CRISPR | Antibiotic: aphVIII, aphVII, nptII, addA, tetX, hph, and ble. Autotrophic: arg and trp. Herbicide: 2-fluoroadenin resistance | Kim and Cerutti, 2009; Greiner et al., 2017; Mini et al., 2018 |
| Chlorella pyrenoidosa | Electroporation | Gene integration and expression | Antibiotic: nptII | Run et al., 2016 |
| Chlorella sorokiniana | Biolistic | Gene integration and expression | Autotrophic: nr | Dawson et al., 1997 |
| Chlorella vulgaris | Electroporation, glass bead agitation, and agrobacterium-mediated, | Gene integration and expression | Antibiotic: nptII and aphVII | Cha et al., 2012; Muñoz et al., 2018 |
| Chromochloris zofingiensis | Biolistic | Gene integration and expression | Herbicide: norflurazon-resistance | Liu et al., 2014 |
| Coccomyxa sp. | Biolistic and electroporation | Gene integration and expression, gene-editing using CRISPR | Autotrophic: umps | Kasai et al., 2018; Yoshimitsu et al., 2018 |
| Coccomyxa subellipsoidea | Electroporation | Gene integration and expression | Antibiotic: hptII | Kania et al., 2019 |
| Cyanidioschyzon merolae | PEG-mediated | Gene integration and expression, RNAi | Antibiotic: cat Auxotrophic: ura | Ohnuma et al., 2009; Sumiya et al., 2015; Fujiwara et al., 2017 |
| Dunaliella salina | Electroporation, biolistic, glass beads agitation, and agrobacterium-mediated | Gene integration and expression | Antibiotic: aphVII and nptII Herbicide: bar Auxotrophic: nr | Li et al., 2007; Radakovits et al., 2010; Srinivasan and Gothandam, 2016 |
| Fistulifera solaris | Biolistic | Gene integration and expression | Antibiotic: nptII | Muto et al., 2013 |
| Gonium pectorale | Biolistic | Gene integration and expression | Antibiotic: aphVIII | Lerche and Hallmann, 2009 |
| Haematococcus pluvialis | Biolistic | Gene integration and expression | Antibiotic: aadA Herbicide: norflurazon resistance | Steinbrenner and Sandmann, 2006; Yuan et al., 2019 |
| Monoraphidium neglectum | Electroporation | Gene integration and expression | Antibiotic: aphVII | Jaeger et al., 2017 |
| Nannochloropsis gaditana | Electroporation | Gene integration and expression, gene-editing using CRISPR | Antibiotic: aphVII, nptII and BSD | Ajjawi et al., 2017 |
| Nannochloropsis limnetica | Electroporation | Gene integration and expression | Antibiotic:aphVII and nptII | Chen and Hu, 2019 |
| Nannochloropsis oceanica | Electroporation | Gene integration and expression, RNAi, gene-editing using CRISPR | Antibiotic: sh ble and nptII | Li et al., 2014a; Poliner et al., 2018; Osorio et al., 2019 |
| Nannochloropsis oculata | Electroporation | Gene integration and expression | Antibiotic: sh ble | Li et al., 2014a |
| Ostreococcus tauri | Electroporation and PEG-based | Gene integration and expression | Antibiotic:nptII and neo | van Ooijen et al., 2012; Sanchez et al., 2019 |
| Parachlorella kessleri | Biolistic and agrobacterium-mediated | Gene integration and expression | Antibiotic: nptII and aadA | Rathod et al., 2013 |
| Phaeodactylum tricornutum | Biolistic, electroporation, and bacterial conjugation | Gene integration and expression, gene-editing using MNs, CRISPR, and TAELNs | Antibiotic: nat, sat-1, addA, sh ble and cat Autotrophic: ura Herbicide: 5-fluoroorotic acid and 2-fluoroadenine resistance | Daboussi et al., 2014; Serif et al., 2018; Sharma et al., 2018 |
| Scenedesmus obliquus | Electroporation | Gene integration and expression | Antibiotic: cat | Guo et al., 2013 |
| Symbiodinium microadriaticum | silicon carbide whiskers | Gene integration and expression | Antibiotic:nptII and hpt | Te et al., 1998 |
| Thalassiosira pseudonana | Biolistic and bacterial conjugation | Gene integration and expression, gene-editing using CRISPR | Antibiotic: nat and sat-1 | Karas et al., 2015 |
| Volvox carteri f. magariensis | Biolistic | Gene integration and expression, gene-editing using CRISPR | Antibiotic: hpt and BSD Autotrophic: nr | Ortega-Escalante et al., 2019a,b |
List of microalgae and the molecular tool and techniques available for their genetic engineering.
Despite the recent advancement in the transformation technologies, the microalgae transformation is still facing the problem of low efficiency, except in Chlamydomonas when compared to the plant system. In an advancement, the development of nuclear episomal vector to transform diatoms via conjugation-based method that directly deliver the vector from E. coli to diatom provides an efficient method for diatom transformation (Karas et al., 2015). This method offers several advantages over the conventional transformation methods such as capacity to deliver large DNA fragments (may be multiple genes from a pathway), stable self-replication of episomal vector (due to presence of yeast-derived regulatory sequences, CEN/ARS), loss of transgene upon removal of selection pressure, and low possibility of positional or epigenetic effects (Doron et al., 2016). Recently, the application of conjugation-based method in CRISPR/cas9 mediated genome-editing of Pt MYBR1 gene in P. tricornutum (Sharma et al., 2018) and nitrate reductase gene (NR) in Nannochloropsis oceanica (Poliner et al., 2019) has been demonstrated to generate transgene-free mutants. Although 20–100 times higher transformation efficiency and rapid transformant appearance was observed in the conjugation-based method, there was a significant delay in the appearance of mutants in the positive transformants (Sharma et al., 2018). The plausible explanation for this delayed mutant appearance was attributed to the lower Cas9 expression due to higher rate of cell division in conjugatively transformed cells. In addition, the episomal vector system adapted for diatom was able to transform the green oleaginous microalgae Acutodesmus obliquus and Neochloris oleoabundans through bacterial conjugation (Muñoz et al., 2019). Although the transformation efficiency was sufficiently higher as compared to the biolistic-based vector delivery system, this application of diatom adapted episomal vector system in other microalgae has some limitations that are discussed in the following section.
Genome-Editing
Over the years, significant progress has been made to improve the catalog of available tools for genetic engineering in microalgae, with the ultimate aim to improve the feasibility of microalgae as a model organism for scientific and/or industrial applications. In the past decade, the gene-editing tools such as zinc-finger nucleases (ZFNs), meganucleases (MNs), transcription activator-like effector nucleases (TALEN), and clustered regularly interspaced short palindromic repeats (CRISPR/Cas9) have been emerged as the efficient tools for genome-editing in many organisms (Razzaq et al., 2019). All these tools are able to introduce a double-strand break at targeted DNA sequence that can be further repaired via either non-homologous end-joining (may disrupt gene through mutations) or homologous recombination (may insert or replace gene with exogenous donor DNA) (Jeon et al., 2017). The CRISPR is often used interchangeably to the term genome-editing; however, the ZFNs and TALENs were among the first molecular tools available for the genome-editing. The applicability of these tools largely depends on the factors such as cost, complexity, and ability to cause multiple edits simultaneously. Among the others, the CRISPR/cas9-mediated genome-editing system now became the state-of-the-art tool due to its simplicity and versatility.
The initial reports of gene-editing in microalgae were from ZFN-mediated genome-editing. Sizova et al. (2013) and Greiner et al. (2017) used engineered ZFNs to target the COP3 and COP4 genes in C. reinhardtii. However, the efficiency of the ZFNs was only observed in the tailored model strain of C. reinhardtii. In addition, it was also suggested that ZFNs prefer the homology-directed repair when supplied with larger donor DNA (>750 bp) for the clean and predictable gene modification. Beside the recent developments in the ZFN technology, the most challenging task is to create unique ZFNs with high specificity and affinity toward the target sites. This requires the validation of ZFNs using gene targeting selection system before conducting the actual experiment (Sizova et al., 2013). Meanwhile, the use of MNs and TALENs was also demonstrated to target the uridyl diphosphate (UDP)-glucose pyrophosphorylase in P. tricornutum for enhanced lipid accumulation (Daboussi et al., 2014). The use of TALENs for the disruption of urease gene through homologous recombination has been successfully achieved in P. tricornutum (Weyman et al., 2015). Similarly, in an attempt to evaluate the use of uridine monophosphate (UMP) synthase as an endogenous positive selectable marker for DNA-free genome editing, Serif et al. (2018) used TALEN to generate knock-out mutants of UMP synthase gene in P. tricornutum. Although the efficiency of the gene disruption using TALENs was quite low (only 16%), the applicability of TALENs for gene-editing in microalgae has been established. However, though the use of TALENs for gene-editing has been exemplified in several organisms, no report has been observed till date in Chlamydomonas. The functioning of transcription activator-like effectors (TALEs) has been established in Chlamydomonas to induce the expression of endogenous genes, ARS1 and ARS2 through the binding of gene-specific artificially designed TALEs to the promoter region of the targeted genes (Gao et al., 2014b). This study indicates that the TALEs coupled to nuclease(s) can (TALENs) be used as one of the approaches to target the gene-editing in Chlamydomonas.
The successful use of CRISPR/cas9 system in microalgae species was first demonstrated by Jiang et al. in C. reinhardtii (Jiang et al., 2014). In this study, four genes were successfully edited through the expression of codon-optimized Cas9 gene and corresponding single guide RNA (sgRNA). However, the constitutive expression of Cas9 shows cytotoxic effect in C. reinhardtii that reduce the cell viability of transformants (Ng et al., 2020). Therefore, the transient delivery of in-vitro assembled Cas9/sg RNA ribonucleoprotein (RNP) complex via electroporation is a promising methodology to efficiently edit genes in C. reinhardtii without the cytotoxic effect of Cas9, and this approach was established recently (Shin et al., 2016a; Baek et al., 2016a). The use of Cas9/sgRNA-RNP-complex-mediated approach could exempt the genome-edited microalgae from the regulations of genetically modified organism (GMO) regulations, since it does not involve the integration of foreign DNA (cas9 gene) in the host genome. In addition, the Cas9/sgRNA RNP complex further reduces the off-target effects and is less cytotoxic to the cells because of transient expression of cas9, thus improving the efficiency of gene-editing. In an effort to improve the efficiency of CRISPR/Cas9 system in C. reinhardtii, Jiang and Weeks (2017) employed gene-within-a-gene methodology that uses hybrid Cas9 gene containing an artificial intron having sgRNA gene. Although the hybrid cas9 system was functional in Chlamydomonas, the improvement in the efficiency of gene editing was only marginal. A higher editing-efficiency of up to 9 and 3.3% in Chlamydomonas was observed by Greiner et al. (2017) after using a Cas9 gene from Staphylococcus aureus and S. pyogenes, respectively. Recently, Guzmán-Zapata et al. (2019) used transient expression of S. pyogenes cas9 to disrupt the atp9 gene in Chlamydomonas with efficiency of up to 30% on preselected 2-fluoroadenine resistant colonies. This approach of pre-selection based on the editing of selectable marker gene could also be used for the multiplexed editing. In another approach, an ortholog of cas9, Cpf1 was used in single-step co-delivery of CRISPR/Cpf1 RNP complex along with single-stranded DNA repair template, and this approach resulted in ~10% efficiency for precise gene-editing in C. reinhardtii (Ferenczi et al., 2017). Using dcas9 (dead cas9, nuclease defense), the functioning of a variant of CRISPR, named as CRISPRi (CRISPR interference) was also established in C. reinhardtii through downregulation of PEPC1 expression to enhance the lipid content (Kao and Ng, 2017).
Besides Chlamydomonas, the adaptability of the CRISPR system was also successfully demonstrated for another model marine microalgae P. tricornutum. Using codon-optimized S. pyogenes cas9, the disruption of P. tricornutum CpSRP54 gene with 31% efficiency indicates that, unlike Chlamydomonas, the Cas9 constitutive expression is not likely to be toxic for diatoms (Nymark et al., 2016). Recently, Sharma et al. (2018) compared the effect of constitutive and transient expression of cas9 on editing frequency and stability of mutant lines generated through biolistic and bacterial conjugation, respectively. Although the efficiency of CRISPR-induced targeted mutations were similar for both methods, the use of conjugation-based episomal CRISPR/Cas9 system is capable of avoiding re-editing of mutant lines caused by constitutive expression of Cas9 in the progeny (Sharma et al., 2018; Slattery et al., 2018). Intriguingly, the simultaneous knock-out of multiple genes has also been demonstrated in P. tricornutum through the delivery of Cas9/sgRNA RNP complex (Serif et al., 2018). In addition, the CRISPR/cas9 system was also able to edit urease gene in another diatom, T. pseudonana with up to more than 60% of disruption efficiency (Hopes et al., 2016). The application of CRISPR system on industrially important oleaginous marine microalgae N. oceanica was first demonstrated through the disruption of nitrate reductase gene (Wang et al., 2016), however with a very low efficiency of nearly 1%. Later, the cas9 editor line of N. gaditana was developed that constitutively expressed the cas9 and was used for editing of targeted transcription factor genes with high efficiency range of up to 78% (Ajjawi et al., 2017). In the recent past, various strategies have been successfully applied for gene-editing in several microalgal species. However, the above literature shows the inconsistency in the editing-efficiency of CRISPR system across the microalgal species and is still a concern. Thus, the identification of novel or optimized nucleases that may prove to be useful in gene-editing in microalgae is required. Moreover, the constitutive expression of Cas9 (or other nucleases) may sometime induce the undesired re-editing of the mutant lines (Slattery et al., 2018). In this context, the episomal-vector system has the advantage of transient Cas9 (or other nucleases) expression that can prevent re-editing of mutant lines, which is a common complication associated with the constitutive expression of Cas9. Moreover, the elimination of episomal CRISPR/Cas9 vector from the host upon removal of selection pressure makes the mutant lines be considered as non-transgenic. In contrast to diatom, using similar vector system, recently Muñoz et al. (2019) were not able to rescue the episomal plasmids from positive transformants of green oleaginous microalgae Acutodesmus obliquus and Neochloris oleoabundans. Moreover, the continuous subculturing in the selection-free medium was not sufficient to remove the episomal vector. This indicates the possible chromosomal integration event even in the bacterial conjugation-based episomal vector delivery. Therefore, the episomal maintenance of delivered plasmids in microalgae other than diatoms through diatom-adapted, yeast-derived centromeric sequences (CEN/ARS) is not possible yet. Rather, episomal maintenance is a function of species-specific adaptation of yeast centromeric regions that should be optimized before the wider application of episomal vectors in microalgal bioengineering.
Case Studies for Genetic Engineering in Microalgae
The previous sections reviewed key resources that can augment the bioengineering in microalgae. This section describes the various algal bioengineering research such as: the enhancement of (1) photosynthesis and biomass production, (2) lipid production, (3) the production of biomolecules, and other value-added products.
Photosynthetic Efficiency and Biomass Production
Enhanced CO2 fixation through augmentation of photosynthetic efficiency is the key process to improve microalgae biomass production, a pre-requisite to develop microalgae as the next-generation feed-stock. The carbon fixation is dependent on multiple factors, where selectivity and velocity of RuBisCo enzyme remains one of the major factors. RuBisCo is capable to fix CO2 as well as O2 into 3-phosphoglycerate and 2-phosphoglycolate, where 2-phosphoglycolate is undesirable and toxic to the cells. The phenomenon called photorespiration occurs in the mitochondrion and peroxisome, which uses 2-phosphoglycolate to release CO2. These futile side reactions ultimately hamper the photosynthetic activity. Therefore, attempts have been made to simultaneously improve the selectivity and catalytic rate of RuBisCO through genetic engineering, though with limited success (Du et al., 2003; Spreitzer et al., 2005). Alternatively, the problem of RuBisCO selectivity can be mitigated by controlling the design consideration of cultivation system to enrich the CO2 supply. Nevertheless, in order to improve the catalytic rate of RuBisCO, its genetic modification is preferable mode than to select efficient RuBisCO from the diverse pool of natural variants. In one such effort, the small subunit of RuBisCO enzyme of Chlamydomonas has been replaced with that of Arabidopsis, spinach, and sunflower to enhance the carboxylation catalytic efficiency and CO2/O2 specificity (Genkov et al., 2010). Although the hybrid RuBisCO enzyme had 3–11% increase in specificity, the velocity of the enzyme remained same. Likewise, several amino-acid residues have been identified in the conserved region of small subunit of the RuBisCO that can be the potential target for engineering RuBisCo to improve its catalytic efficiency (Du et al., 2000; Spreitzer et al., 2001; Genkov et al., 2006; Genkov and Spreitzer, 2009). In another approach through regulation of RuBisCo activity, the photosynthetic biomass production in N. oceanica was substantially enhanced upon overexpression of RuBisCO activase (Wei et al., 2017a). Beside RuBisCO the other relatively low abundant enzymes of Calvin cycle regeneration phase, such as fructose−1,6-bisphosphatase (FBPase), fructose 1,6-bisphosphate aldolase (FBA), and sedoheptulose 1,7-bisphosphatase (SBPase) are the prime target to manipulate the photosynthetic activity. Recently the engineering of Calvin cycle through the overexpression of cyanobacterial FBA was found to enhance the photosynthetic capacity of C. vulgaris (Yang et al., 2017). Similarly, the overexpression of Chlamydomonas SBPase was reported to improve the photosynthetic activity in Dunaliella bardawil (Fang et al., 2012). The FBPase was found to enhance the photosynthetic efficiency upon overexpression in higher plants (Tamoi et al., 2006). However, its overexpression in Chlamydomonas had detrimental effect on growth and photosynthetic activity under high CO2 photoautotrophic conditions. This was mainly due to the reduced amount of glyceraldehyde−3-phaspahte because there was enhanced conversion of fructose−1,6-bisphosphate into fructose−6- phosphate (Dejtisakdi and Miller, 2016). This indicates that in microalgae the reaction catalyzed by the FBPase is not a rate limiting one that can be targeted to improve the photosynthetic efficiency and concomitant biomass accumulation.
The photosynthetically efficient microorganisms operate the CO2-concentrating mechanisms (CCMs) to increase the CO2 concentration in the proximity of RuBisCO, which eventually reduce the photorespiration and promote carboxylation. In comparison to the terrestrial plants, the green microalgae have efficient CCM because of sequestration of the enzymes of photosynthetic machinery in the pyrenoid or peroxisome (Mackinder, 2018; Hennacy and Jonikas, 2020). Several functional and regulatory factors have been identified, which are responsible to facilitate the carboxylation reaction of RuBisCO through CCM. Among these factors CIA5, transporter of inorganic carbon (Ci) and carbonic anhydrases (CA) are considered as the targets for manipulation to increase the photosynthetic performance and eventually biomass yield (Moroney et al., 2011; Wang et al., 2015; Yamano et al., 2015; Gee and Niyogi, 2017). However, there are no such reports on successful engineering of CCM components in microalgae, and thus it remains a challenge to enhance the carbon fixation process.
On the other hand, the cultivation of microalgae at high cell density often encounters a problem of photo-limitation because of light shading. The high light intensity at the surface cell layers saturates the photosynthetic process and causes photoinhibition, whereas excess energy is dissipated through non-photochemical quenching. Meanwhile, the low-light intensity at the lower layer of cells compels them to perform photorespiration instead of photosynthesis. This uneven distribution of the light intensity results in suboptimum photosynthetic efficiency that eventually reduces the biomass yield. Reducing the size of antenna or light-harvesting complex is one of the approaches that has the potential to improve the light transmission and light absorption capacity. For instance, the reduction of chlorophyll b content and consequent reduction of antenna size in Chlamydomonas through RNAi-mediated silencing of chlorophyllide a oxygenase, resulted in enhanced photosynthetic activity and higher growth rate as compared to chlorophyll b mutant under saturating light conditions (Perrine et al., 2012). Similarly, the C. vulgaris mutant with truncated antenna size and reduced chlorophyll a and b content, generated through random mutagenesis of chloroplast signal recognition particle (CpSRP43), exhibit enhanced photosynthetic efficiency associated with reduced non-photochemical quenching and higher biomass yield (Shin et al., 2016b, 2017). The engineering of photosystem II protein D1 isoform in Chlamydomonas showed enhanced photosynthetic efficiency under saturating light conditions (Vinyard et al., 2014). In another novel approach, the diatom P. tricornutum was engineered to establish a concept of intracellular spectral recompositioning for improved light absorption and consequent higher biomass production (Fu et al., 2017). In this case, the overexpressed green fluorescent protein absorbs the excess blue light energy from incident light and subsequently emits energy as green light that can be harvested by accessory pigments. Thus, spectral recompositioning eventually improves the light absorption and reduces the non-photochemical quenching and may mitigate the problem of photoinhibition at high cell density cultures through deeper penetration of emitted green light. A similar ecological mechanism has been observed in the coral-algae symbionts to acclimatize deep water light environment by facilitating homogenous distribution of available light energy (Smith et al., 2017). Although significant information is availed through genetic engineering to get insight into the photosynthetic efficiency, most of these leads are from the model algal systems. Moreover, this information is yet to be applied for large scale applications. In addition, the design consideration of cultivation system has significant effect on the photosynthetic efficiency and eventually on productivity. Therefore, there is a need of more comprehensive and cumulative approach, such as fine tuning the flux balance of Calvin cycle toward enhanced CO2 fixation or perturbation of multiple targets at once to get a synergistic effect. The various strategies to improve photosynthetic efficiency and biomass production are illustrated in Figure 2.
Figure 2
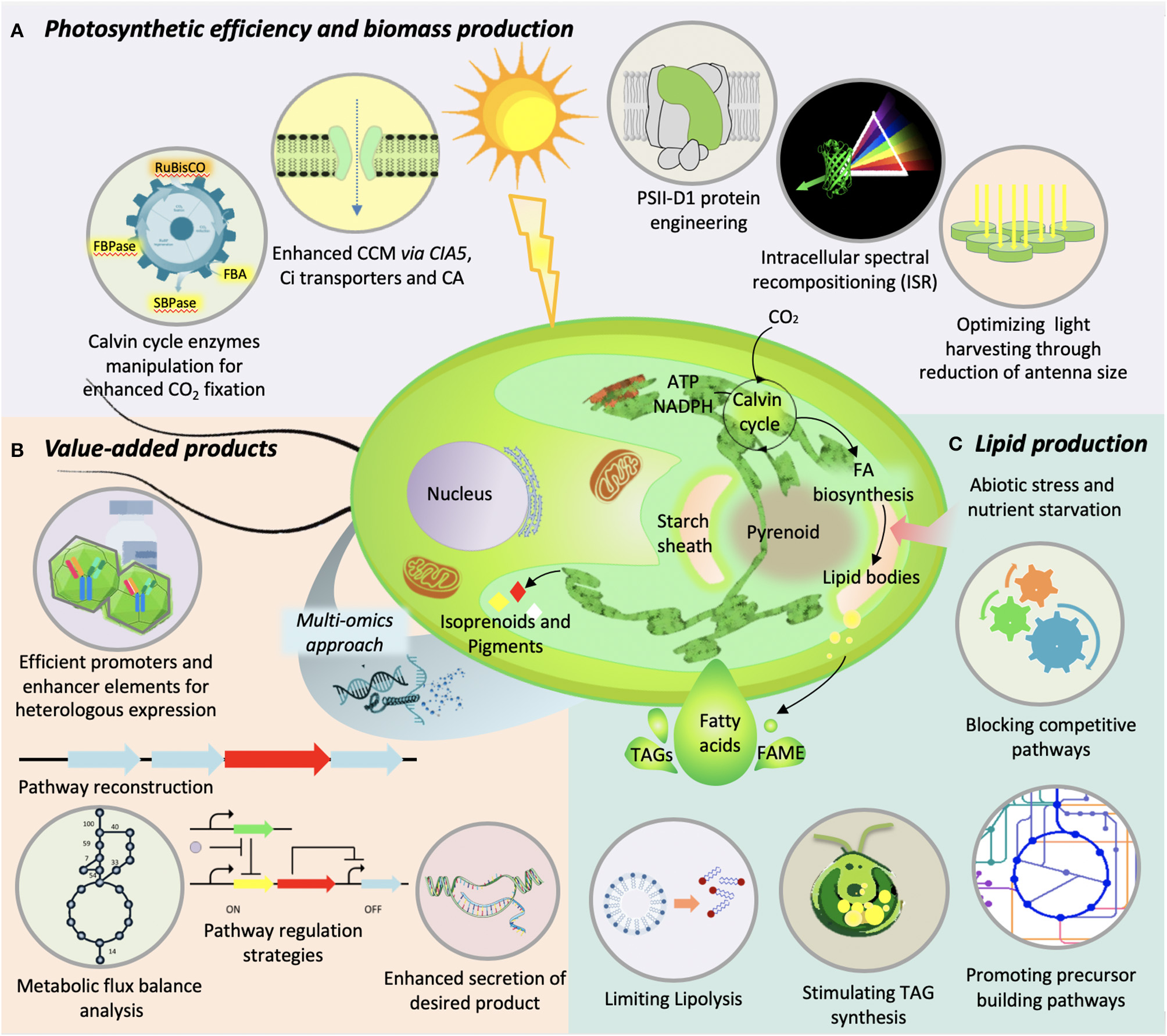
Illustration of various genetic-engineering strategies used in microalgae to improve (A) photosynthetic efficiency and biomass production, (B) value-added product synthesis, and (C) lipid production.
Lipid Production
Lipids from microalgae are at the center of attention due to their yield and nutraceutical importance. The quantity, quality, and the type of lipids synthesized by microalgae not only help in diversifying their application but influence the biodiesel properties if chosen for the fuel purpose (Shekh et al., 2016). For researchers working in this area, lipid productivity remains a key parameter for strain selection. In fact, the kind of lipids a microalga accumulates plays a key role in its commercial utilization for food, feed, or fuel purpose. Over the years, a trade-off between enhancing microalgal lipid content by various means without compromising the lipid productivity was targeted. Various augmentations in environmental, nutritional, and physiological conditions for cultivation of microalgae, as well as genetic manipulations, have been attempted for enhanced lipid production (Figure 2). However, genetic engineering of the robust strains for enhanced lipid production remains one of the most viable options to improve the process economics. In the recent past, various genes involved in lipid biosynthesis were knocked-out or overexpressed to examine their effects on lipid accumulation. Acetyl-CoA Carboxylase (ACCase), which encodes enzyme for fatty acid synthesis, was overexpressed for the first time in 1996 by Dunahay et al. (1996). Even though the overexpression of ACCase was characterized by 2- to 3-fold increase in ACCase activity, it could not lead to increased lipid accumulation (Sheehan et al., 1998). However, upregulation of ACCase in tandem with malic enzyme, which catalyzes malate to pyruvate conversion, was effective in enhanced lipid accumulation in D. salina (Talebi et al., 2014). Overexpression of diacyl glycerol acyl transferase, which catalyzes the final step in TAG synthesis, is often-used strategy, which also resulted in lipid enhancement (Niu et al., 2013; Iwai et al., 2014; Li et al., 2016a). Also, the enhanced expression of pyruvate dehydrogenase, acetyl-CoA synthase, phosphoenolpyruvate carboxylase, NAD(H) kinase, and glycerol kinase has resulted in hyperaccumulation of lipids in various microalgal species. Simultaneous expression of multiple acyl transferases from S. cerevisiae and Yarrowia lipolytica in Chlorella minutissima resulted in twofold lipid accumulation (Hsieh et al., 2012). Overexpression of RuBisCO activase in N. oceanica has resulted in an increase in the productivity, thereby increasing lipid accumulation (Wei et al., 2017a). Inhibiting the expression of a multifunctional lipase/phospholipase/acyltransferase in T. pseudonana resulted in enhanced lipid accumulation without compromising the growth (Trentacoste et al., 2013). On the other hand, it is known that the transcriptional regulation can influence metabolomic flux of the system as transcription factors can target multiple regulatory points in a metabolic pathway. Overexpression/knockdown of transcription factors targeting the upregulation of lipid biosynthesis genes may accumulate higher lipids. In one of the efforts, knockdown of a single transcription regulator ZnCys in N. gaditana resulted in twofold increase in lipid content (Ajjawi et al., 2017). Strategies to prevent the degradation of synthesized lipids were also studied to improve lipid yields. A knock-out mutant of the phospholipase A2 gene (C. reinhardtii) had the total lipid content increased up to 64.25% (Shin et al., 2019). In another study, a 10-fold increase in TAG was reported upon silencing the cht7 gene encoding a TAG lipase (Tsai et al., 2014). Most recently, CRISPR/Cas9-based technology for gene manipulation in C. vulgaris was used wherein a fragment of Cas9 with sgRNA designed on omega-3 fatty acid desaturase (fad3) gene was constructed. This has resulted in 46% (w/w) higher accumulation of lipid content (Lin and Ng, 2020). Even though various studies to genetically engineer microalgae for enhanced lipid accumulation have been attempted, they are mostly restricted to model and/or selected microalgae strains. The recent advancements in gene-editing technologies especially CRISPR/Cas9 may allow the gene manipulations in commercially important oleaginous strains so as to improve the process economics.
Biomolecules and Value-Added Products
Beside lipids, microalgae are rich in biomolecules such as carotenoids with potential application in human health. The accumulation of biopigments in microalgae is known to be affected by various biotic and abiotic factors, the details of which have been recently reviewed by Saini et al. (2020). Since the carotenoid biosynthesis pathway has been extensively studied, the metabolic engineering, in addition to the mutant screening, has been applied to enhance the production of carotenoids in microalgae. Perturbing the pathway enzymes such as phytoene synthase and phytoene desaturase, microalgae have known to enhance the production of carotenoids (Steinbrenner and Sandmann, 2006; Couso et al., 2011; Dambek et al., 2012; Tran et al., 2012; Liu et al., 2014; Eilers et al., 2016; Galarza et al., 2018). In addition, several other enzymes involved in the subsequent steps of the carotenoid pathways have also been targeted. For instance, the overexpression of Haematococus pluvialis gene encoding β-carotene ketolase in Dunaliella salina resulted in production of astaxanthin (Anila et al., 2016). The downregulation of squalene epoxidase through RNAi in Chlamydomonas was found to accumulate squalene (Kajikawa et al., 2015). Similarly, the knock-out mutant of zeaxanthin epoxidase in Chlamydomonas had significantly higher zeaxanthin content than the wild type (Baek et al., 2018). However, diverting the flux toward desired metabolites is not that simple and may require perturbation of multiple genes of a pathway. In one such recent example, the overexpression of three exogenous enzymes, namely oxidosqualene cyclase (from Lotus japonicus) and cytochrome P450 along with its native reductase (from Medicago truncatula) in P. tricornutum, leads to the production of triterpenoids viz. lupeol and botulin (D'Adamo et al., 2019). Similarly, the production of sesquiterpenoids and diterpenoids through genetic engineering of Chlamydomonas has also been reported (Lauersen et al., 2016, 2018; Wichmann et al., 2018). The introduction of additional copy of gene encoding gateway enzyme of terpenoid pathway, 1-deoxy-D-xylulose 5-phosphate synthase (dxs), resulted in enhanced accumulation of fucoxanthin in P. tricornutum (Eilers et al., 2016). However, the hyperaccumulation of carotenoids or any other secondary metabolites sometimes causes feedback inhibition. Therefore, generating an additional metabolic sink (in a place other than the site of production) or expressing the flux controlling enzyme(s) that can resist the feedback inhibition could be the possible strategies. However, this strategy may get limited by the lack of information on transporters or the flux controlling enzymes. Here the genomic information can substantially improve the scenario of metabolic engineering in microalgae.
The algal nuclear or chloroplast engineering has been extensively carried out using synthetic biology approach for the production of recombinant proteins having therapeutic properties. Some of the inherent features of algae such as lack of infectious agents or toxins, efficient folding of complex proteins and scope for the development of whole algae as low-cost oral vaccine, makes them ideal platform for heterologous production and offer several advantages over the better established microbial and mammalian systems. Although, most of the chloroplast transformation attempts have been made in the model microalgae Chlamydomonas, the successful chloroplast engineering has also been demonstrated in few other microalgal species [reviewed by Siddiqui et al. (2020)]. It was reported that over 100 different recombinant proteins have been successfully expressed in algal chloroplast. Among these recombinant proteins, the vaccines, antibodies and immunotoxins, and therapeutic proteins are the major targets (Dyo and Purton, 2018). The production of whole algal cells as oral vaccines specially for farm animals, where the fusion of protein adjuvant (cholera toxin B subunit: CTB) to the N-terminus of the antigen facilitates the antigen absorption through gut epithelium, provided an alternative low-cost vaccination strategy. Moreover, this bioencapsulation of therapeutic proteins has advantages of long-term storage at room temperature and also protects them from the degradation in animal stomach (Dreesen et al., 2010; Gregory et al., 2013). Besides all these advantages, the yield of the recombinant proteins is still a major concern to adopt algae as protein production platform. Although several recombinant proteins have been successfully produced through genetic engineering of nuclear genome, a much lower success rate with production yield of only up to 0.25% of total soluble proteins was reported (Scranton et al., 2016). In comparison, the production of proteins through chloroplast engineering may reach up to 0.1–5% of total soluble protein (Dyo and Purton, 2018). Nevertheless, the nuclear expression of the protein offers some interesting features such as ability to target the protein to secretory pathway or to the specific organelles that may also allow the post-translational modification of the proteins (Lauersen et al., 2015). The various signal peptides have been used to target the proteins either to secretory pathway or to an organelle. Recently, the performance of two in-silico identified signal peptides (1,3-α-glucosidase and SAD1p derived) to efficiently secrete expressed reporter protein in C. reinhardtii has been successfully demonstrated (Molino et al., 2018). The different promoters and their respective 5' UTRs as well as their synthetic variants have been used to derive the expression of transgene in order to mitigate the constrains of inefficient transgene expression in microalgae (Coragliotti et al., 2011; Specht and Mayfield, 2013; Gimpel et al., 2015). For example, the use of strong promoter such as 16S ribosomal RNA fused to 5'UTRs of endogenous photosynthetic genes can be used to enhance the expression of transgene to some extent (Rasala et al., 2011). However, the performance of the endogenous 5′UTRs to translate the gene of interest is still the major constrain. The intrinsic features of photosynthetic genes derived 5′UTRs are also responsible for feedback regulation of translation. It does so through “control by epistasis of synthesis” that prevent overaccumulation of protein subunit in the absence of other subunits of the protein assembly (Coragliotti et al., 2011). In addition, the constitutive expression of the transgene negatively impacts the growth of the transgenic algae as an extra metabolic burden. Therefore, the use of inducible promoter to tightly regulate the expression of transgene could be the better strategy to improve the growth efficiency, and hence the productivity of desired product (Fajardo et al., 2020). The various promoters used so far in the microalgal research are given in Table 3. The advancement in the synthetic biology and our understanding on the regulation of protein synthesis in microalgae will enable us to improve the protein expression level in microalgae so as to make microalgae a feasible host system for commercial application. The various strategies to improve production of bioactive of interest in microalgae are illustrated in Figure 2.
Table 3
| Target species | Promoters | Nuclear (N)/ chloroplast (C) expression | Salient features | References |
|---|---|---|---|---|
| Ankistrodesmus convolutus | AcRbcS promoter | N | Light-regulated promoter | Thanh et al., 2012 |
| C. reinhardtii | ARG7 promoter | N | Strong promoter | Specht et al., 2015 |
| β-TUB2 promoter | N | Constitutive promoter | Crozet et al., 2018 | |
| CABII-1 | N | Light-dependent promoter | Doron et al., 2016 | |
| Cyc6 and Cpx1 promoter | N | Copper- and oxygen-dependent promoter | Quinn et al., 2000 | |
| CrGPDH3 promoter | N | Salt inducible promoter | Beltran-Aguilar et al., 2019 | |
| Fea1 promoter | N | Iron-responsive promoter | Barjona do Nascimento Coutinho et al., 2019 | |
| HSP70A-RBCS2 promoter | N | Strong hybrid promoter | Lauersen et al., 2015 | |
| HSP70A promoter | N | Strong promoter activity | Schroda et al., 2000 | |
| psaD promoter | N | Light-responsive constitutive promoter | Crozet et al., 2018 | |
| sap11 promoter | N | Synthetic strong promoter | Scranton et al., 2016 | |
| RBCS2 promoter | N | Strong promoter activity | Lumbreras et al., 1998 | |
| psaA promoter | C | Light-responsive strong promoter | Michelet et al., 2011 | |
| psbA promoter | C | Light-responsive strong promoter | Rasala et al., 2011 | |
| psbD promoter | C | Light-responsive strong promoter | Rasala et al., 2011 | |
| atpA promoter | C | Constitutive promoter activity | Rasala et al., 2011 | |
| 16S promoter-psbA 5' UTR | C | Strong promoter | Rasala et al., 2011 | |
| rbcL promoter | C | Light-responsive strong constitutive promoter | Rasala et al., 2011 | |
| Chaetoceros gracilis | Lhcr5 promoter | N | Constitutive promoter | Ifuku et al., 2015 |
| C. vulgaris | CaMV35S promoter | N | Constitutive promoter | Chow and Tung, 1999 |
| CvpsaD promoter | N | Light-responsive promoter | Kim et al., 2018 | |
| Chlorella ellipsoida | Ubi1- Ω promoter | N | Strong constitutive expression | Chen et al., 2001 |
| Cyclotella cryptica | ACCase promoter | N | Constitutive promoter | Dunahay et al., 1996 |
| Cylindrotheca fusiformis | fruα3 promoter | N | Strong constitutive expression | Fischer et al., 1999 |
| D. salina | LIP promoter | N | Light-inducible promoter | Baek et al., 2016b |
| GAPDH promoter | N | Constitutive promoter | Doron et al., 2016 | |
| Fistulifera sp. | fcpB promoter | N | Constitutive promoter | Muto et al., 2013 |
| H4 promoter | N | Constitutive promoter | Muto et al., 2013 | |
| H. pluvialis | CaMV 35S | N | Constitutive promoter | Kathiresan et al., 2009 |
| Ptub promoter | N | Strong promoter | Yuan et al., 2019 | |
| rbcL promoter | C | Light-responsive strong constitutive promoter | Gutiérrez et al., 2012 | |
| P. tricornutum | CaMV 35S promoter | N | Constitutive promoter | Chow and Tung, 1999 |
| U6 promoter | N | Constitutive promoter | Serif et al., 2018; Stukenberg et al., 2018 | |
| Lhcf promoter | N | Light-dependent promoter | Lepetit et al., 2010 | |
| NIT promoter | N | Ammonium inducible promoter | Chu et al., 2016 | |
| pPhAP1 promoter | N | Strong promoter | Lin et al., 2017 | |
| Pt211 promoter | N | Strong constitutive promoter | Zou et al., 2018 | |
| fcp promoter | N | Constitutive promoter | Watanabe et al., 2018 | |
| V-ATPase promoter | N | Strong constitutive promoter | Watanabe et al., 2018 | |
| ef2 promoter | N | Constitutive promoter | Seo et al., 2015 | |
| HASP1 promoter | N | Strong constitutive promoter | Erdene-Ochir et al., 2019 | |
| rbcL promoter | C | Light-responsive strong constitutive promoter | Xie et al., 2014 | |
| N. oceanica | β-tubulin promoter | N | Constitutive promoter | Li et al., 2014a |
| CMV viral promoter | N | Constitutive promoter | Osorio et al., 2019 | |
| ef promoter | N | Constitutive promoter | Poliner et al., 2018 | |
| Ribi promoter | N | Bidirectional strong constitutive promoter | Poliner et al., 2018 | |
| EM7 promoter | N | Constitutive promoter | Osorio et al., 2019 | |
| NIT promoter | N | Ammonium inducible promoter | Jackson et al., 2019 | |
| VCP promoter | N | Constitutive promoter | Li et al., 2014a | |
| rbcL promoter | C | Light-responsive strong constitutive promoter | Gan et al., 2018 | |
| N. gaditana | TCT promoter | N | Constitutive promoter | Ajjawi et al., 2017 |
| RPL24 promoter | N | Constitutive promoter | Ajjawi et al., 2017 | |
| 4ALL promoter | N | Constitutive promoter | Ajjawi et al., 2017 | |
| EIF3 promoter | N | Constitutive promoter | Ajjawi et al., 2017 | |
| N. oculata | HSP70A-RBCS2 promoter | N | Strong hybrid promoter | Shih et al., 2015 |
| N. salina | TUB promoter | N | Constitutive promoter | Koh et al., 2019 |
| UEP promoter | N | Constitutive promoter | Koh et al., 2019 | |
| T. pseudonana | Lcfs9 promoter | N | Constitutive promoter | Poulsen et al., 2006 |
| NIT promoter | N | Nitrate inducible promoter | Poulsen et al., 2006 | |
| Volvox carteri | LHCBM1 promoter | N | Constitutive promoter | Tian et al., 2018 |
| nitA promoter | N | Nitrate inducible promoter | von der Heyde et al., 2015 | |
| ISG promoter | N | Developmental stage (embryonic inversion) specific promoter | Hallmann and Sumper, 1994 | |
| Arylsulfate promoter | N | Sulfur starvation inducible promoter | Hallmann and Sumper, 1994 |
List of endogenous and heterologous promoters used in microalgae research.
Risk Assessment, Biosafety, and Regulatory Issues
Though genetic engineering is considered as one of the most potent tools to augment production of commercially valuable metabolites in microalgae, it inevitably invites varying opinions on the safe use of genetically modified (GM) algae for consumption and environment. On the contrary, several algal performance-improvement strategies, which could have environmental and ecological threats, are in use without much debate. In many parts of the world, strict laws/policies require transgenic/recombinant algae to undergo regulatory compliances. When research and policy complement each other, technological advances move at a rapid pace. In this case, even if various researchers across the globe are working on strain improvement for enhanced microalgae performance through genetic modifications, their commercial use is restricted. Reports indicate that the Florida-based biotechnology company named Algenol was given approval for use of GM cyanobacteria for cultivation in outdoor closed-photobioreactor. At the same time, the secretariat of the Convention on Biological Diversity in its 2015 report has raised the concerns over strict physical containment of these GM microorganisms by the company (https://www.cbd.int/ts/cbd-ts-82-en.pdf). It is arguably said that the U.S. Environmental Protection Agency (US-EPA) relies upon a regulatory regime-Toxic Substances Control Act (TSCA), which has become outdated and is incapable of assessing the novel risks arising out of the new biotechnological inventions. Under TSCA, companies are only required to file a Microbial Commercial Activity Notice for commercialization of a new GM microorganism. Till date, no outdoor cultivation of GM microalgae is reported probably due to various predictable and unexpected risks associated with its open cultivation (Nethravathy et al., 2019). Cultivation of GM microalgae possesses several risks, which includes spills that may become uncontrollable. These algae upon proliferation compete with natural species and may outgrow them. In fact, the genetically modified traits of the organisms may provide them the competitive advantage in natural ecosystem. Risks also exist for genetic contamination /interbreeding with wild-type or sexually compatible strains. Threats of harmful algal blooms, negative impacts on ecosystem, increased selection pressure, horizontal gene transfer, health and environmental impacts, unpredictable future of GM traits, loss of management control, and ethical concerns are some of the major concerns associated with cultivation of GM algae (Nethravathy et al., 2019). Apart from regulations for the use of GM algae, strict biosecurity laws are required to safeguard the importation of foreign species (GM and/or wild-type) to the local environment. Though the import and use of foreign algae strains, which are non-native to local environment, have a very little regulatory control, the associated risk of these strains dominating the local species must be seriously considered (Campbell, 2011). The concrete environmental risk due to algal spills must not only be limited to the GM aspect of the strains. Further, assessment needs to be carried out considering fitness of invading foreign species in comparison with local algal community along with intricacies and population stability characteristics of the ecological system in question (Henley et al., 2013). To further improve the situations for the use of GM algae, in-depth cost-benefit analysis of GM microalgae to society and environment must be carried out. Strict monitoring of the handling and cultivation process with health and environmental risk assessment analysis are integral to design the biosafety regulations for GM microalgae. Since GM algae are considered as one of the solutions to overcome techno-economic challenges in algal industry, it is imperative that various stakeholders including business promoters and policy makers collectively reach to a consensus on a road map for the use of GM algae in future. Various federal governments across the globe must bring in place the policies and regulations that govern the safe use of GM algae for human and environmental benefit.
Conclusion and Future Prospects
Currently, economically feasible, environmentally sustainable, and replicable microalgal processes with higher technology readiness levels are required for ease of doing algal business. To improve the economic feasibility of the algal processes, the genetic engineering of microalgae is at forefront for development of robust microalgal strains. Advances in the high-throughput technologies and molecular biology tools have facilitated the biotechnological approach to engineer the microalgal strains for performance improvement. The synergy of microalgal multi-omics datasets and the advanced molecular tools offer a rapid and predictable strategic path for the strain improvement. In this review, various microalgal resources such as genome sequence, mutant libraries, high-throughput screening methodologies, and genetic tools and techniques were summarized that holds the potential for the development of microalgae as a next-generation renewable resource. In addition, the catalog of various omics study under different conditions across the diverse microalgal species is generated (Table 1). Despite the variation in the inter- and intraspecies omics datasets, the several conserved factors can be mined to predict the biological outcomes with the comprehensive use of system biology approach. Various omics-based approaches must aim to enhance microalgal capacities to produce high value metabolites. Future research may focus on developing purpose-specific robust bioengineered strains for high photosynthetic efficiency, high CO2 fixation, and high biomass productivities. Also, targeted enhancement of low-volume, high-value metabolites of biomedical applications from microalgae must be considered using genetic engineering.
Though the genetic engineering of microalgae holds great potential to improve process economics, it is limited mainly due to the unavailability of the genetic information for robust and commercially suitable strains. In recent times, rapid advances in DNA synthesis, genetic manipulation tools and techniques, availability of functional genomes have improved the chances to better engineer microalgae with complex functions. However, the lack of genetic strain design principles is still hurting the progress in this area. Further, once the genetically improved strains are developed, safety to human health and environment will define its commercial success. Therefore, it is recommended that strict regulations and monitoring should be in place to evaluate the environmental and human health risk of using GM microalgae particularly in outdoor cultivation. Here, the recent development in precise genome editing technologies such as non-transgenic and marker-free CRISPR has the potential to revolutionize the microalgal bioengineering for the production of non-GMO algal products. The non-GMO tag to the bioengineered microalgae is expected to improve the biosafety and alleviate the regulatory issues associated with the usage of GM microalgae. In view of uncertainty within the academic and industrial community regarding the regulations for the use of GM strains, and the inadequacy of current regulations for the use of GM algae, a clear road map for regulatory regime covering the commercial use of GM microalgae is urgently required. Since the robustness of non-model microalgae species has advantages in commercial and industrial applications over model species, there is a need to develop advanced research tools for the non-model microalgal species. Moreover, to improve the economic competitiveness of algal-derived products, the development of efficient extraction methods or the use of whole cells is needed. Indeed, beside all the developments, bio-prospection for novel and robust microalgae with industrial viability must continue.
Statements
Author contributions
GK: conceptualization, writing—original draft preparation, writing—reviewing, editing, and supervision. AS: conceptualization, writing—original draft preparation, writing—reviewing, and editing. SJ: investigation—data collection and writing—original draft preparation. YS: visualization and writing—original draft preparation. RK: writing—original draft preparation. TS: writing—reviewing, editing, and supervision. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Department of Science and Technology, Government of India under INSPIRE faculty award scheme.
Acknowledgments
We gratefully acknowledge Executive Director, National Agri-Food Biotechnology Institute (NABI), Mohali, India for the constant support. GK and AS acknowledge the Department of Science and Technology, India, for the INSPIRE Faculty award.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1
AbidaH.DolchL.-J.MeïC.VillanovaV.ConteM.BlockM. A.et al. (2015). Membrane glycerolipid remodeling triggered by nitrogen and phosphorus starvation in Phaeodactylum tricornutum. Plant Physiol.167, 118 LP−136. 10.1104/pp.114.252395
2
AjjawiI.VerrutoJ.AquiM.SoriagaL. B.CoppersmithJ.KwokK.et al. (2017). Lipid production in Nannochloropsis gaditana is doubled by decreasing expression of a single transcriptional regulator. Nat. Biotechnol.35, 647–652. 10.1038/nbt.3865
3
AlipanahL.RohloffJ.WingeP.BonesA. M.BrembuT. (2015). Whole-cell response to nitrogen deprivation in the diatom Phaeodactylum tricornutum. J. Exp. Bot.66, 6281–6296. 10.1093/jxb/erv340
4
AllenJ. W.DiRussoC. C.BlackP. N. (2017). Carbon and acyl chain flux during stress-induced triglyceride accumulation by stable isotopic labeling of the polar microalga Coccomyxa subellipsoidea C169. J. Biol. Chem.292, 361–374. 10.1074/jbc.M116.760843
5
AnilaN.SimonD. P.ChandrashekarA.RavishankarG. A.SaradaR. (2016). Metabolic engineering of Dunaliella salina for production of ketocarotenoids. Photosynth. Res.127, 321–333. 10.1007/s11120-015-0188-8
6
AokiY.OkamuraY.OhtaH.KinoshitaK.ObayashiT. (2016). ALCOdb: gene coexpression database for microalgae. Plant Cell Physiol.57:e3. 10.1093/pcp/pcv190
7
ArandaM.LiY.LiewY. J.BaumgartenS.SimakovO.WilsonM. C.et al. (2016). Genomes of coral dinoflagellate symbionts highlight evolutionary adaptations conducive to a symbiotic lifestyle. Sci. Rep.6:39734. 10.1038/srep39734
8
AroraN.DubeyD.SharmaM.PatelA.GuleriaA.PruthiP. A.et al. (2018). NMR-based metabolomic approach to elucidate the differential cellular responses during mitigation of arsenic(III, V) in a green microalga. ACS Omega3, 11847–11856. 10.1021/acsomega.8b01692
9
AroraN.KumariP.KumarA.GangwarR.GulatiK.PruthiP. A.et al. (2019). Delineating the molecular responses of a halotolerant microalga using integrated omics approach to identify genetic engineering targets for enhanced TAG production. Biotechnol. Biofuels12:2. 10.1186/s13068-018-1343-1
10
BabaM.SuzukiI.ShiraiwaY. (2011). Proteomic analysis of high-CO2-inducible extracellular proteins in the unicellular green alga, Chlamydomonas reinhardtii. Plant Cell Physiol.52, 1302–1314. 10.1093/pcp/pcr078
11
BaekK.KimD. H.JeongJ.SimS. J.MelisA.KimJ.et al. (2016a). DNA-free two-gene knockout in Chlamydomonas reinhardtii via CRISPR-Cas9 ribonucleoproteins. Nat. Publ. Gr.6:30620. 10.1038/srep30620
12
BaekK.LeeY.NamO.ParkS.SimS. J.JinE. (2016b). Introducing Dunaliella LIP promoter containing light-inducible motifs improves transgenic expression in Chlamydomonas reinhardtii. Biotechnol. J.11, 384–392. 10.1002/biot.201500269
13
BaekK.YuJ.JeongJ.SimS. J.BaeS.JinE. (2018). Photoautotrophic production of macular pigment in a Chlamydomonas reinhardtii strain generated by using DNA-free CRISPR-Cas9 RNP-mediated mutagenesis. Biotechnol. Bioeng.115, 719–728. 10.1002/bit.26499
14
BaiX.SongH.LavoieM.ZhuK.SuY.YeH.et al. (2016). Proteomic analyses bring new insights into the effect of a dark stress on lipid biosynthesis in Phaeodactylum tricornutum. Sci. Rep.6:25494. 10.1038/srep25494
15
BajhaiyaA. K.DeanA. P.DriverT.TrivediD. K.RattrayN. J. W.AllwoodJ. W.et al. (2016). High-throughput metabolic screening of microalgae genetic variation in response to nutrient limitation. Metabolomics12:9. 10.1007/s11306-015-0878-4
16
Barjona do Nascimento CoutinhoP.FriedlC.HeilmannM.BuchholzR.StuteS. C. (2019). Validated nuclear-based transgene expression regulated by the Fea1 iron-responsive promoter in the green alga Chlamydomonas reinhardtii. Mol. Biotechnol.61, 305–316. 10.1007/s12033-018-00148-0
17
BaumeisterT. U. H.ValletM.KaftanF.Svato,šA.PohnertG. (2019). Live single-cell metabolomics with matrix-free laser/desorption ionization mass spectrometry to address microalgal physiology. Front. Plant Sci.10:172. 10.3389/fpls.2019.00172
18
Beltran-AguilarA. G.Peraza-EcheverriaS.López-OchoaL. A.Borges-ArgáezI. C.Herrera-ValenciaV. A. (2019). A novel salt-inducible CrGPDH3 promoter of the microalga Chlamydomonas reinhardtii for transgene overexpression. Appl. Microbiol. Biotechnol.103, 3487−3499. 10.1007/s00253-019-09733-y
19
BennerI.DinerR. E.LefebvreS. C.LiD.KomadaT.CarpenterE. J.et al. (2013). Emiliania huxleyi increases calcification but not expression of calcification-related genes in long-term exposure to elevated temperature and pCO2. Philos. Trans. R. Soc. Lond. B. Biol. Sci.368:20130049. 10.1098/rstb.2013.0049
20
BerriosH.ZapataM.RivasM. (2016). A method for genetic transformation of Botryococcus braunii using a cellulase pretreatment. J. Appl. Phycol.28, 201–208. 10.1007/s10811-015-0596-3
21
Blifernez-KlassenO.ChaudhariS.KlassenV.WördenweberR.SteffensT.CholewaD.et al. (2018). Metabolic survey of Botryococcus braunii: impact of the physiological state on product formation. PLoS ONE13:e0198976. 10.1371/journal.pone.0198976
22
BoroujerdiA. F. B.LeeP. A.DiTullioG. R.JanechM. G.ViedS. B.BeardenD. W. (2012). Identification of isethionic acid and other small molecule metabolites of Fragilariopsis cylindrus with nuclear magnetic resonance. Anal. Bioanal. Chem.404, 777–784. 10.1007/s00216-012-6169-2
23
CagnonC.MirabellaB.NguyenH. M.Beyly-AdrianoA.BouvetS.CuinéS.et al. (2013). Development of a forward genetic screen to isolate oil mutants in the green microalga Chlamydomonas reinhardtii. Biotechnol. Biofuels6:178. 10.1186/1754-6834-6-178
24
CampbellM. L. (2011). Assessing biosecurity risk associated with the importation of non-indigenous microalgae. Environ. Res.111, 989–998. 10.1016/j.envres.2011.02.004
25
CarvalhoR. N.LettieriT. (2011). Proteomic analysis of the marine diatom Thalassiosira pseudonana upon exposure to benzo(a)pyrene. BMC Genomics12:159. 10.1186/1471-2164-12-159
26
ChaT. S.YeeW.AzizA. (2012). Assessment of factors affecting Agrobacterium-mediated genetic transformation of the unicellular green alga, Chlorella vulgaris. World J. Microbiol. Biotechnol.28, 1771–1779. 10.1007/s11274-011-0991-0
27
ChaiboonchoeA.DohaiB. S.CaiH.NelsonD. R.JijakliK.Salehi-AshtianiK. (2014). Microalgal metabolic network model refinement through high-throughput functional metabolic profiling. Front. Bioeng. Biotechnol.2, 1–12. 10.3389/fbioe.2014.00068
28
ChenC.HarstA.YouW.XuJ.NingK.PoetschA. (2019). Proteomic study uncovers molecular principles of single-cell-level phenotypic heterogeneity in lipid storage of Nannochloropsis oceanica. Biotechnol. Biofuels12:21. 10.1186/s13068-019-1361-7
29
ChenH.ZhengY.ZhanJ.HeC.WangQ. (2017). Comparative metabolic profiling of the lipid-producing green microalga Chlorella reveals that nitrogen and carbon metabolic pathways contribute to lipid metabolism. Biotechnol. Biofuels1, 1–20. 10.1186/s13068-017-0839-4
30
ChenM.ZhaoL.SunY.-L.CuiS.-X.ZhangL.-F.YangB.et al. (2010). Proteomic analysis of hydrogen photoproduction in sulfur-deprived Chlamydomonas cells. J. Proteome Res.9, 3854–3866. 10.1021/pr100076c
31
ChenX.-H.LiY.-Y.ZhangH.LiuJ.-L.XieZ.-X.LinL.et al. (2018). Quantitative proteomics reveals common and specific responses of a marine diatom Thalassiosira pseudonana to different macronutrient deficiencies. Front. Microbiol.9:2761. 10.3389/fmicb.2018.02761
32
ChenY.HuH. (2019). High efficiency transformation by electroporation of the freshwater alga Nannochloropsis limnetica. World J. Microbiol. Biotechnol.35:119. 10.1007/s11274-019-2695-9
33
ChenY.WangY.SunY.ZhangL.LiW. (2001). Highly efficient expression of rabbit neutrophil peptide-1 gene in Chlorella ellipsoidea cells. Curr. Genet.39, 365–370. 10.1007/s002940100205
34
ChenZ.YangM.LiC.WangY.ZhangJ.WangD.et al. (2014). Phosphoproteomic analysis provides novel insights into stress responses in Phaeodactylum tricornutum, a model diatom. J. Proteome Res.13, 2511–2523. 10.1021/pr401290u
35
ChengS.MelkonianM.SmithS. A.BrockingtonS.ArchibaldJ. M.DelauxP.-M.et al. (2018). 10KP: A phylodiverse genome sequencing plan. Gigascience7, 1–9. 10.1093/gigascience/giy013
36
ChengX.LiuG.KeW.ZhaoL.LvB.MaX.et al. (2017). Building a multipurpose insertional mutant library for forward and reverse genetics in Chlamydomonas. Plant Methods13:36. 10.1186/s13007-017-0183-5
37
ChistiY. (2013). Constraints to commercialization of algal fuels. J. Biotechnol.167, 201–214. 10.1016/j.jbiotec.2013.07.020
38
ChoiY.-E.HwangH.KimH.-S.AhnJ.-W.JeongW.-J.YangJ.-W. (2013). Comparative proteomics using lipid over-producing or less-producing mutants unravels lipid metabolisms in Chlamydomonas reinhardtii. Bioresour. Technol.145, 108–115. 10.1016/j.biortech.2013.03.142
39
ChowK.-C.TungW. L. (1999). Electrotransformation of Chlorella vulgaris. Plant Cell Rep.18, 778–780. 10.1007/s002990050660
40
ChuF. L.PirastruL.PopovicR.SlenoL. (2011). Carotenogenesis up-regulation in Scenedesmus sp. using a targeted metabolomics approach by liquid chromatography-high-resolution mass spectrometry. J. Agric. Food Chem. 59, 3004–3013. 10.1021/jf105005q
41
ChuL.EweD.Río BártulosC.KrothP. G.GruberA. (2016). Rapid induction of GFP expression by the nitrate reductase promoter in the diatom Phaeodactylum tricornutum. PeerJ4:e2344. 10.7717/peerj.2344
42
CoragliottiA. T.BeligniM. V.FranklinS. E.MayfieldS. P. (2011). Molecular factors affecting the accumulation of recombinant proteins in the Chlamydomonas reinhardtii chloroplast. Mol. Biotechnol.48, 60–75. 10.1007/s12033-010-9348-4
43
Cornejo-CoronaI.ThapaH. R.BrowneD. R.DevarenneT. P.Lozoya-GloriaE. (2016). Stress responses of the oil-producing green microalga Botryococcus braunii Race B. PeerJ4:e2748. 10.7717/peerj.2748
44
CousoI.VilaM.RodriguezH.VargasM. A.LeonR. (2011). Overexpression of an exogenous phytoene synthase gene in the unicellular alga Chlamydomonas reinhardtii leads to an increase in the content of carotenoids. Biotechnol. Prog.27, 54–60. 10.1002/btpr.527
45
CrozetP.NavarroF. J.WillmundF.MehrshahiP.BakowskiK.JonathanK.et al. (2018). Birth of a photosynthetic chassis : a MoClo toolkit enabling synthetic biology in the microalga Chlamydomonas reinhardtii. ACS Synth. Biol. 7, 2074–2086. 10.1021/acssynbio.8b00251
46
CuvelierM. L.GuoJ.OrtizA. C.van BarenM. J.TariqM. A.PartenskyF.et al. (2017). Responses of the picoprasinophyte Micromonas commoda to light and ultraviolet stress. PLoS ONE12:e0172135. 10.1371/journal.pone.0172135
47
DaboussiF.LeducS.MaréchalA.DuboisG.GuyotV.Perez-MichautC.et al. (2014). Genome engineering empowers the diatom Phaeodactylum tricornutum for biotechnology. Nat. Commun.5:3831. 10.1038/ncomms4831
48
D'AdamoS.Schiano di VisconteG.LoweG.Szaub-NewtonJ.BeachamT.LandelsA.et al. (2019). Engineering the unicellular alga Phaeodactylum tricornutum for high-value plant triterpenoid production. Plant Biotechnol. J.17, 75–87. 10.1111/pbi.12948
49
DambekM.EilersU.BreitenbachJ.SteigerS.BuchelC.SandmannG. (2012). Biosynthesis of fucoxanthin and diadinoxanthin and function of initial pathway genes in Phaeodactylum tricornutum. J. Exp. Bot.63, 5607–5612. 10.1093/jxb/ers211
50
DawsonH. N.BurlingameR.CannonsA. C. (1997). Stable transformation of Chlorella: rescue of nitrate reductase-deficient mutants with the nitrate reductase gene. Curr. Microbiol.35, 356–362. 10.1007/s002849900268
51
de los ReyesC.Ávila-RománJ.OrtegaM. J.de la JaraA.García-MauriñoS.MotilvaV.et al. (2014). Oxylipins from the microalgae Chlamydomonas debaryana and Nannochloropsis gaditana and their activity as TNF-α inhibitors. Phytochemistry102, 152–161. 10.1016/j.phytochem.2014.03.011
52
DebuchyR.PurtonS.RochaixJ. D. (1989). The argininosuccinate lyase gene of Chlamydomonas reinhardtii: an important tool for nuclear transformation and for correlating the genetic and molecular maps of the ARG7 locus. EMBO J.8, 2803–2809. 10.1002/j.1460-2075.1989.tb08426.x
53
Degraeve-GuilbaultC.BrehelinC.HaslamR.SayanovaO.Marie-LuceG.JouhetJ.et al. (2017). Glycerolipid characterization and nutrient deprivation-associated changes in the green picoalga Ostreococcus tauri. Plant Physiol.173, 2060–2080. 10.1104/pp.16.01467
54
DejtisakdiW.MillerS. M. (2016). Overexpression of Calvin cycle enzyme fructose 1,6-bisphosphatase in Chlamydomonas reinhardtii has a detrimental effect on growth. Algal Res.14, 116–126. 10.1016/j.algal.2016.01.003
55
DentR. M.SharifiM. N.MalnoëA.HaglundC.CalderonR. H.WakaoS.et al. (2015). Large-scale insertional mutagenesis of Chlamydomonas supports phylogenomic functional prediction of photosynthetic genes and analysis of classical acetate-requiring mutants. Plant J.82, 337–351. 10.1111/tpj.12806
56
DiazJ. M.PlummerS.HanselC. M.AndeerP. F.SaitoM. A.McIlvinM. R. (2019). NADPH-dependent extracellular superoxide production is vital to photophysiology in the marine diatom Thalassiosira oceanica. Proc. Natl. Acad. Sci. U. S. A.116, 16448 LP−16453. 10.1073/pnas.1821233116
57
DongH.-P.WilliamsE.WangD.XieZ.-X.HsiaR.JenckA.et al. (2013). Responses of Nannochloropsis oceanica IMET1 to long-term nitrogen starvation and recovery. Plant Physiol.162, 1110–1126. 10.1104/pp.113.214320
58
DoronL.SegalN.ShapiraM. (2016). Transgene expression in microalgae-from tools to applications. Front. Plant Sci.7:505. 10.3389/fpls.2016.00505
59
DreesenI. A. J.Charpin-El HamriG.FusseneggerM. (2010). Heat-stable oral alga-based vaccine protects mice from Staphylococcus aureus infection. J. Biotechnol.145, 273–280. 10.1016/j.jbiotec.2009.12.006
60
DuC.LiangJ.-R.ChenD.-D.XuB.ZhuoW.-H.GaoY.-H.et al. (2014). iTRAQ-based proteomic analysis of the metabolism mechanism associated with silicon response in the marine diatom Thalassiosira pseudonana. J. Proteome Res.13, 720–734. 10.1021/pr400803w
61
DuC.ZhangB.HeY.HuC.NgQ. X.ZhangH.et al. (2017). Biological effect of aqueous C60 aggregates on Scenedesmus obliquus revealed by transcriptomics and non-targeted metabolomics. J. Hazard. Mater.324, 221–229. 10.1016/j.jhazmat.2016.10.052
62
DuY.-C.PeddiS. R.SpreitzerR. J. (2003). Assessment of structural and functional divergence far from the large subunit active site of ribulose-1,5-bisphosphate carboxylase/oxygenase. J. Biol. Chem.278, 49401–49405. 10.1074/jbc.M309993200
63
DuY. C.HongS.SpreitzerR. J. (2000). RbcS suppressor mutations improve the thermal stability and CO2/O2 specificity of rbcL- mutant ribulose-1,5-bisphosphate carboxylase/oxygenase. Proc. Natl. Acad. Sci. U. S. A.97, 14206–14211. 10.1073/pnas.260503997
64
DuanL.ChenQ.DuanS. (2019). Transcriptional analysis of Chlorella pyrenoidosa exposed to Bisphenol A. Int. J. Environ. Res. Public Health16:81374. 10.3390/ijerph16081374
65
DunahayT. G. (1993). Transformation of chlamydomonas reinhardtii with silicon carbide whiskers. Biotechniques, 15:452–460.
66
DunahayT. G.JarvisE. E.DaisS. S.RoesslerP. G. (1996). Manipulation of microalgal lipid production using genetic engineering. Appl. Biochem. Biotechnol.57:223. 10.1007/BF02941703
67
DyhrmanS. T.JenkinsB. D.RynearsonT. A.SaitoM. A.MercierM. L.AlexanderH.et al. (2012). The transcriptome and proteome of the diatom Thalassiosira pseudonana reveal a diverse phosphorus stress response. PLoS ONE7:e33768. 10.1371/journal.pone.0033768
68
DyoY. M.PurtonS. (2018). The algal chloroplast as a synthetic biology platform for production of therapeutic proteins. Microbiology164, 113–121. 10.1099/mic.0.000599
69
EilersU.BikoulisA.BreitenbachJ.BüchelC.SandmannG. (2016). Limitations in the biosynthesis of fucoxanthin as targets for genetic engineering in Phaeodactylum tricornutum. J. Appl. Phycol.28, 123–129. 10.1007/s10811-015-0583-8
70
Erdene-OchirE.ShinB.-K.KwonB.JungC.PanC.-H. (2019). Identification and characterisation of the novel endogenous promoter HASP1 and its signal peptide from Phaeodactylum tricornutum. Sci. Rep.9:9941. 10.1038/s41598-019-45786-9
71
FajardoC.De DonatoM.CarrascoR.Martínez-RodríguezG.ManceraJ. M.Fernández-AceroF. J. (2020). Advances and challenges in genetic engineering of microalgae. Rev. Aquacult.12, 365–381. 10.1111/raq.12322
72
FangL.LinH. X.LowC. S.WuM. H.ChowY.LeeY. K. (2012). Expression of the Chlamydomonas reinhardtii sedoheptulose-1,7-bisphosphatase in Dunaliella bardawil leads to enhanced photosynthesis and increased glycerol production. Plant Biotechnol. J.10, 1129–1135. 10.1111/pbi.12000
73
FayyazM.ChewK. W.ShowP. L.LingT. C.NgI. S.ChangJ. S. (2020). Genetic engineering of microalgae for enhanced biorefinery capabilities. Biotechnol Adv. 43:107554. 10.1016/j.biotechadv.2020.107554
74
FengT.-Y.YangZ.-K.ZhengJ.-W.XieY.LiD.-W.MuruganS. B.et al. (2015). Examination of metabolic responses to phosphorus limitation via proteomic analyses in the marine diatom Phaeodactylum tricornutum. Sci. Rep.5:10373. 10.1038/srep10373
75
FerencziA.PyottD. E.XipnitouA.MolnarA. (2017). Efficient targeted DNA editing and replacement in Chlamydomonas reinhardtii using Cpf1 ribonucleoproteins and single-stranded DNA. Proc. Natl. Acad. Sci. U. S. A.114, 13567–13572. 10.1073/pnas.1710597114
76
Fernandez-AceroF. J.Amil-RuizF.Duran-PenaM. J.CarrascoR.FajardoC.GuarnizoP.et al. (2019). Valorisation of the microalgae Nannochloropsis gaditana biomass by proteomic approach in the context of circular economy. J. Proteomics193, 239–242. 10.1016/j.jprot.2018.10.015
77
FischerH.RoblI.SumperM.KrögerN. (1999). Targeting and covalent modification of cell wall and membrane proteins heterologously expressed in the diatom Cylindrotheca Fusiformis (Bacillariophyceae). J. Phycol.35, 113–120. 10.1046/j.1529-8817.1999.3510113.x
78
FuW.ChaiboonchoeA.KhraiweshB.SultanaM.JaiswalA.JijakliK.et al. (2017). Intracellular spectral recompositioning of light enhances algal photosynthetic efficiency. Sci. Adv.3:e1603096. 10.1126/sciadv.1603096
79
FuW.NelsonD. R.MystikouA.DaakourS.Salehi-ashtianiK. (2019). Advances in microalgal research and engineering development. Curr. Opin. Biotechnol.59, 157–164. 10.1016/j.copbio.2019.05.013
80
FujiwaraT.OhnumaM.KuroiwaT.OhbayashiR.HirookaS.MiyagishimaS.-Y. (2017). Development of a double nuclear gene-targeting method by two-step transformation based on a newly established chloramphenicol-selection system in the red alga Cyanidioschyzon merolae. Front. Plant Sci.8:343. 10.3389/fpls.2017.00343
81
GalarzaJ. I.GimpelJ. A.RojasV.Arredondo-VegaB. O.HenríquezV. (2018). Over-accumulation of astaxanthin in Haematococcus pluvialis through chloroplast genetic engineering. Algal Res.31, 291–297. 10.1016/j.algal.2018.02.024
82
GanQ.JiangJ.HanX.WangS.LuY. (2018). Engineering the chloroplast genome of oleaginous marine microalga Nannochloropsis oceanica. Front. Plant Sci.9:439. 10.3389/fpls.2018.00439
83
GaoC.WangY.ShenY.YanD.HeX.DaiJ.et al. (2014a). Oil accumulation mechanisms of the oleaginous microalga Chlorella protothecoides revealed through its genome, transcriptomes, and proteomes. BMC Genomics15:582. 10.1186/1471-2164-15-582
84
GaoH.WrightD. A.LiT.WangY.HorkenK.WeeksD. P.et al. (2014b). TALE activation of endogenous genes in Chlamydomonas reinhardtii. Algal Res.5, 52–60. 10.1016/j.algal.2014.05.003
85
GaoZ.MiaoX.ZhangX.WuG.GuoY.WangM.et al. (2016). Comparative fatty acid transcriptomic test and iTRAQ-based proteomic analysis in Haematococcus pluvialis upon salicylic acid (SA) and jasmonic acid (JA) inductions. Algal Res.17, 277–284. 10.1016/j.algal.2016.05.012
86
GeF.HuangW.ChenZ.ZhangC.XiongQ.BowlerC.et al. (2014). Methylcrotonyl-CoA carboxylase regulates triacylglycerol accumulation in the model diatom Phaeodactylum tricornutum. Plant Cell26, 1681–1697. 10.1105/tpc.114.124982
87
GeY.NingZ.WangY.ZhengY.ZhangC.FigeysD. (2016). Quantitative proteomic analysis of Dunaliella salina upon acute arsenate exposure. Chemosphere145, 112–118. 10.1016/j.chemosphere.2015.11.049
88
GeeC. W.NiyogiK. K. (2017). The carbonic anhydrase CAH1 is an essential component of the carbon-concentrating mechanism in Nannochloropsis oceanica. Proc. Natl. Acad. Sci. U. S. A.114, 4537 LP−4542. 10.1073/pnas.1700139114
89
GenkovT.DuY.-C.SpreitzerR. J. (2006). Small-subunit cysteine-65 substitutions can suppress or induce alterations in the large-subunit catalytic efficiency and holoenzyme thermal stability of ribulose-1,5-bisphosphate carboxylase/oxygenase. Arch. Biochem. Biophys.451, 167–174. 10.1016/j.abb.2006.04.012
90
GenkovT.MeyerM.GriffithsH.SpreitzerR. J. (2010). Functional hybrid rubisco enzymes with plant small subunits and algal large subunits: engineered rbcS cDNA for expression in Chlamydomonas. J. Biol. Chem.285, 19833–19841. 10.1074/jbc.M110.124230
91
GenkovT.SpreitzerR. J. (2009). Highly conserved small subunit residues influence rubisco large subunit catalysis. J. Biol. Chem.284, 30105–30112. 10.1074/jbc.M109.044081
92
GimpelJ. A.HyunJ. S.SchoeppN. G.MayfieldS. P. (2015). Production of recombinant proteins in microalgae at pilot greenhouse scale. Biotechnol. Bioeng.112, 339–345. 10.1002/bit.25357
93
GregoryJ. A.TopolA. B.DoernerD. Z.MayfieldS. (2013). Alga-produced Cholera toxin-Pfs25 fusion proteins as oral vaccines. Appl. Environ. Microbiol.79, 3917–3925. 10.1128/AEM.00714-13
94
GreinerA.KelterbornS.EversH.KreimerG.SizovaI.HegemannP. (2017). Targeting of photoreceptor genes in Chlamydomonas reinhardtii via zinc-finger nucleases and CRISPR/Cas9. Plant Cell29, 2498–2518. 10.1105/tpc.17.00659
95
GuW.LiH.ZhaoP.YuR.PanG.GaoS.et al. (2014). Quantitative proteomic analysis of thylakoid from two microalgae (Haematococcus pluvialis and Dunaliella salina) reveals two different high light-responsive strategies. Sci. Rep.4:6661. 10.1038/srep06661
96
GuarnieriM. T.NagA.SmolinskiS. L.DarzinsA.SeibertM.PienkosP. T. (2011). Examination of triacylglycerol biosynthetic pathways via de novo transcriptomic and proteomic analyses in an unsequenced microalga. PLoS ONE6:e25851. 10.1371/journal.pone.0025851
97
GuarnieriM. T.NagA.YangS.PienkosP. T. (2013). Proteomic analysis of Chlorella vulgaris: potential targets for enhanced lipid accumulation. J. Proteomics93, 245–253. 10.1016/j.jprot.2013.05.025
98
GuoJ.WilkenS.JimenezV.ChoiC. J.AnsongC.DannebaumR.et al. (2018). Specialized proteomic responses and an ancient photoprotection mechanism sustain marine green algal growth during phosphate limitation. Nat. Microbiol.3, 781–790. 10.1038/s41564-018-0178-7
99
GuoS.-L.ZhaoX.-Q.TangY.WanC.AlamM. A.HoS.-H.et al. (2013). Establishment of an efficient genetic transformation system in Scenedesmus obliquus. J. Biotechnol.163, 61–68. 10.1016/j.jbiotec.2012.10.020
100
GutiérrezC. L.GimpelJ.EscobarC.MarshallS. H.HenríquezV. (2012). Chloroplast genetic tools for the green microalagae Haematococcus pluvialis (Chlorophyceae, Volvocales). J. Phycol.48, 976–983. 10.1111/j.1529-8817.2012.01178.x
101
Guzmán-ZapataD.Sandoval-VargasJ. M.Macedo-OsorioK. S.Salgado-ManjarrezE.Castrejón-FloresJ. L.Oliver-SalvadorM. D. C.et al. (2019). Efficient editing of the nuclear APT reporter gene in Chlamydomonas reinhardtii via expression of a CRISPR-Cas9 module. Int. J. Mol. Sci.20, 1–13. 10.3390/ijms20051247
102
HaiderS.PalR. (2013). Integrated analysis of transcriptomic and proteomic data. Curr. Genomics14, 91–110. 10.2174/1389202911314020003
103
HallmannA.SumperM. (1994). Reporter genes and highly regulated promoters as tools for transformation experiments in Volvox carteri. Proc. Natl. Acad. Sci. U. S. A.91, 11562–11566. 10.1073/pnas.91.24.11562
104
HealK. R.KelloggN. A.CarlsonL. T.LionheartR. M.IngallsA. E. (2019). Metabolic consequences of cobalamin scarcity in the diatom Thalassiosira pseudonana as revealed through metabolomics. Protist170, 328–348. 10.1016/j.protis.2019.05.004
105
HenardC. A.GuarnieriM. T.KnoshaugE. P. (2017). The Chlorella vulgaris S-nitrosoproteome under nitrogen replete and deplete conditions. Front. Bioeng. Biotechnol.4:100. 10.3389/fbioe.2016.00100
106
HenleyW. J.LitakerR. W.Novovesk,áL.DukeC. S.QuemadaH. D.SayreR. T. (2013). Initial risk assessment of genetically modified (GM) microalgae for commodity-scale biofuel cultivation. Algal Res.2, 66–77. 10.1016/j.algal.2012.11.001
107
HennacyJ. H.JonikasM. C. (2020). Prospects for engineering biophysical CO2 concentrating mechanisms into land plants to enhance yields. Annu. Rev. Plant Biol.71, 461–485. 10.1146/annurev-arplant-081519-040100
108
HiltonL. K.MeiliF.BuckollP. D.Rodriguez-PikeJ. C.ChoutkaC. P.KirschnerJ. A.et al. (2016). A forward genetic screen and whole genome sequencing identify deflagellation defective mutants in Chlamydomonas, including assignment of ADF1 as a TRP Channel. G3 Genes, Genomes, Genet.6, 3409–3418. 10.1534/g3.116.034264
109
HindleM. M.MartinS. F.NoordallyZ. B.van OoijenG.Barrios-LlerenaM. E.SimpsonT. I.et al. (2014). The reduced kinome of Ostreococcus tauri: core eukaryotic signalling components in a tractable model species. BMC Genomics15:640. 10.1186/1471-2164-15-640
110
HirthM.LiveraniS.MahlowS.BougetF.-Y.PohnertG.SassoS. (2017). Metabolic profiling identifies trehalose as an abundant and diurnally fluctuating metabolite in the microalga Ostreococcus tauri. Metabolomics13:68. 10.1007/s11306-017-1203-1
111
HopesA.NekrasovV.KamounS.MockT. (2016). Editing of the urease gene by CRISPR-Cas in the diatom Thalassiosira pseudonana. Plant Methods12:49. 10.1186/s13007-016-0148-0
112
HopkinsJ. F.SpencerD. F.LaboissiereS.NeilsonJ. A. D.EveleighR. J. M.DurnfordD. G.et al. (2012). Proteomics reveals plastid- and periplastid-targeted proteins in the chlorarachniophyte alga Bigelowiella natans. Genome Biol. Evol.4, 1391–1406. 10.1093/gbe/evs115
113
HsiehH.-J.SuC.-H.ChienL.-J. (2012). Accumulation of lipid production in Chlorella minutissima by triacylglycerol biosynthesis-related genes cloned from Saccharomyces cerevisiae and Yarrowia lipolytica. J. Microbiol.50, 526–534. 10.1007/s12275-012-2041-5
114
HunterJ. E.FradaM. J.FredricksH. F.VardiA.Van MooyB. A. S. (2015). Targeted and untargeted lipidomics of Emiliania huxleyi viral infection and life cycle phases highlights molecular biomarkers of infection, susceptibility, and ploidy. Front. Mar. Sci.2:81. 10.3389/fmars.2015.00081
115
IfukuK.YanD.MiyaharaM.Inoue-KashinoN.YamamotoY. Y.KashinoY. (2015). A stable and efficient nuclear transformation system for the diatom Chaetoceros gracilis. Photosynth. Res.123, 203–211. 10.1007/s11120-014-0048-y
116
IwaiM.IkedaK.ShimojimaM.OhtaH. (2014). Enhancement of extraplastidic oil synthesis in Chlamydomonas reinhardtii using a type-2 diacylglycerol acyltransferase with a phosphorus starvation-inducible promoter. Plant Biotechnol. J.12, 808–819. 10.1111/pbi.12210
117
JacksonH. O.BerepikiA.BaylayA. J.TerryM. J.MooreC. M.BibbyT. S. (2019). An inducible expression system in the alga Nannochloropsis gaditana controlled by the nitrate reductase promoter. J. Appl. Phycol.31, 269–279. 10.1007/s10811-018-1510-6
118
JaegerD.HubnerW.HuserT.MussgnugJ. H.KruseO. (2017). Nuclear transformation and functional gene expression in the oleaginous microalga Monoraphidium neglectum. J. Biotechnol.249, 10–15. 10.1016/j.jbiotec.2017.03.011
119
JeonS.LimJ. M.LeeH. G.ShinS. E.KangN. K.ParkY. (2017). Current status and perspectives of genome editing technology for microalgae. Biotechnol. Biofuels. 10, 267. 10.1186/s13068-017-0957-z
120
JiaY.XueL.LiJ.LiuH. (2009). Isolation and proteomic analysis of the halotolerant alga Dunaliella salina flagella using shotgun strategy. Mol. Biol. Rep.37:711. 10.1007/s11033-009-9563-x
121
JiaY.-L.ChenH.ZhangC.GaoL.-J.WangX.-C.QiuL.-L.et al. (2016). Proteomic analysis of halotolerant proteins under high and low salt stress in Dunaliella salina using two-dimensional differential in-gel electrophoresis. Genet. Mol. Biol.39, 239–247. 10.1590/1678-4685-gmb-2015-0108
122
JianJ.ZengD.WeiW.LinH.LiP.LiuW. (2017). The combination of RNA and protein profiling reveals the response to nitrogen depletion in Thalassiosira pseudonana. Sci. Rep.7:8989. 10.1038/s41598-017-09546-x
123
JiangJ.LuY. (2019). Metabolite profiling of Breviolum minutum in response to acidification. Aquat. Toxicol.213:105215. 10.1016/j.aquatox.2019.05.017
124
JiangW.BrueggemanA. J.HorkenK. M.PlucinakT. M.WeeksD. P. (2014). Successful transient expression of Cas9 and single guide RNA genes in Chlamydomonas reinhardtii. Eukaryot Cell13, 1465–1469. 10.1128/EC.00213-14
125
JiangW. Z.WeeksD. P. (2017). A gene-within-a-gene Cas9/sgRNA hybrid construct enables gene editing and gene replacement strategies in Chlamydomonas reinhardtii. Algal Res.26, 474–480. 10.1016/j.algal.2017.04.001
126
JiráskováD.PoulíčkováA.NovákO.SedlákováK.HradeckáV.StrnadM. (2009). High-throughput screening technology for monitoring phytohormone production in microalgae. J. Phycol.45, 108–118. 10.1111/j.1529-8817.2008.00615.x
127
JunejaA.ChaplenF. W. R.MurthyG. S. (2016). Genome scale metabolic reconstruction of Chlorella variabilis for exploring its metabolic potential for biofuels. Bioresour. Technol.213, 103–110. 10.1016/j.biortech.2016.02.118
128
JungandreasA.Schellenberger CostaB.JakobT.von BergenM.BaumannS.WilhelmC. (2014). The acclimation of Phaeodactylum tricornutum to blue and red light does not influence the photosynthetic light reaction but strongly disturbs the carbon allocation pattern. PLoS ONE9:e99727. 10.1371/journal.pone.0099727
129
KajikawaM.KinohiraS.AndoA.ShimoyamaM.KatoM.FukuzawaH. (2015). Accumulation of squalene in a microalga Chlamydomonas reinhardtii by genetic modification of squalene synthase and squalene epoxidase genes. PLoS ONE10:e0120446. 10.1371/journal.pone.0120446
130
KaniaK.ZienkiewiczM.DrozakA. (2019). Stable transformation of unicellular green alga Coccomyxa subellipsoidea C-169 via electroporation. Protoplasma257, 607–611. 10.1007/s00709-019-01447-2
131
KantzilakisK.AivaliotisM.KotakisC.KrasanakisF.RizosA. K.KotzabasisK.et al. (2007). A comparative approach towards thylakoid membrane proteome analysis of unicellular green alga Scenedesmus obliquus. Biochim. Biophys. Acta1768, 2271–2279. 10.1016/j.bbamem.2007.04.028
132
KaoP.-H.NgI.-S. (2017). CRISPRi mediated phosphoenolpyruvate carboxylase regulation to enhance the production of lipid in Chlamydomonas reinhardtii. Bioresour. Technol.245, 1527–1537. 10.1016/j.biortech.2017.04.111
133
KarasB. J.DinerR. E.LefebvreS. C.McQuaidJ.PhillipsA. P. R.NoddingsC. M.et al. (2015). Designer diatom episomes delivered by bacterial conjugation. Nat. Commun.6:6925. 10.1038/ncomms7925
134
KasaiY.TsukaharaT.IkedaF.IdeY.HarayamaS. (2018). Metabolic engineering using iterative self-cloning to improve lipid productivity in Coccomyxa. Sci. Rep.8:11742. 10.1038/s41598-018-30254-7
135
KathiresanS.ChandrashekarA.RavishankarG. A.SaradaR. (2009). Agrobacterium-mediated transformation in the green alga Hematococcus pluvialis (Chlorophyceae, Volvocales). J. Phycol.45, 642–649. 10.1111/j.1529-8817.2009.00688.x
136
KatzA.WaridelP.ShevchenkoA.PickU. (2007). Salt-induced Changes in the Plasma membrane proteome of the halotolerant alga Dunaliella salina as revealed by blue native gel electrophoresis and nano-LC-MS/MS analysis. Mol. Cell. Proteomics6, 1459 LP−1472. 10.1074/mcp.M700002-MCP200
137
KeelingP. J.BurkiF.WilcoxH. M.AllamB.AllenE. E.Amaral-ZettlerL. A.et al. (2014). The Marine Microbial Eukaryote Transcriptome Sequencing Project (MMETSP): illuminating the functional diversity of eukaryotic life in the oceans through transcriptome sequencing. PLoS Biol.12:e1001889. 10.1371/journal.pbio.1001889
138
KennedyF.MartinA.BowmanJ. P.WilsonR.McMinnA. (2019). Dark metabolism: a molecular insight into how the Antarctic sea-ice diatom Fragilariopsis cylindrus survives long-term darkness. New Phytol.223, 675–691. 10.1111/nph.15843
139
KettlesN. L.KoprivaS.MalinG. (2014). Insights into the regulation of DMSP synthesis in the diatom Thalassiosira pseudonana through APR activity, proteomics and gene expression analyses on cells acclimating to changes in salinity, light and nitrogen. PLoS ONE9:e94795. 10.1371/journal.pone.0094795
140
KieselbachT.CheregiO.GreenB. R.FunkC. (2018). Proteomic analysis of the phycobiliprotein antenna of the cryptophyte alga Guillardia theta cultured under different light intensities. Photosynth. Res.135, 149–163. 10.1007/s11120-017-0400-0
141
KimE.-J.CeruttiH. (2009). Targeted gene silencing by RNA interference in Chlamydomonas. Methods Cell Biol.93, 99–110. 10.1016/S0091-679X(08)93005-3
142
KimJ.LiuL.HuZ.JinE. (2018). Identification and functional analysis of the psaD promoter of Chlorella vulgaris using heterologous model strains. Int. J. Mol. Sci.19:1969. 10.3390/ijms19071969
143
KimJ. Y. H.KwakH. S.SungY. J.ChoiH.HongM. E.LimH. S.et al. (2016). Microfluidic high-throughput selection of microalgal strains with superior photosynthetic productivity using competitive phototaxis. Sci. Rep.6:21155. 10.1038/srep21155
144
KimS.LeeY.-C.ChoD.-H.LeeH. U.HuhY. S.KimG.-J.et al. (2014). A simple and non-invasive method for nuclear transformation of intact-walled Chlamydomonas reinhardtii. PLoS ONE9:e101018. 10.1371/journal.pone.0101018
145
KimY. K.YooW. I.LeeS. H.LeeM. Y. (2005). Proteomic analysis of cadmium-induced protein profile alterations from marine alga Nannochloropsis oculata. Ecotoxicology14, 589–596. 10.1007/s10646-005-0009-5
146
KindleK. L. (1990). High-frequency nuclear transformation of Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. U. S. A.87, 1228–1232. 10.1073/pnas.87.3.1228
147
KindleK. L.SchnellR. A.FernandezE.LefebvreP. A. (1989). Stable nuclear transformation of Chlamydomonas using the Chlamydomonas gene for nitrate reductase. J. Cell Biol.109, 2589–2601. 10.1083/jcb.109.6.2589
148
KlueterA.CrandallJ. B.ArcherF. I.TeeceM. A.CoffrothM. A. (2015). Taxonomic and environmental variation of metabolite profiles in marine dinoflagellates of the genus Symbiodinium. Metabolites5, 74–99. 10.3390/metabo5010074
149
KoechlerS.BertinP. N.PlewniakF.BaltenweckR.CasiotC.HeipieperH. J.et al. (2016). Arsenite response in Coccomyxa sp. Carn explored by transcriptomic and non-targeted metabolomic approaches. Environ. Microbiol.18, 1289–1300. 10.1111/1462-2920.13227
150
KohH. G.KangN. K.JeonS.ShinS.-E.JeongB.ChangY. K. (2019). Heterologous synthesis of chlorophyll b in Nannochloropsis salina enhances growth and lipid production by increasing photosynthetic efficiency. Biotechnol. Biofuels12:122. 10.1186/s13068-019-1462-3
151
KováčikJ.KlejdusB.BabulaP.HedbavnyJ. (2015). Nitric oxide donor modulates cadmium-induced physiological and metabolic changes in the green alga Coccomyxa subellipsoidea. Algal Res.8, 45–52. 10.1016/j.algal.2015.01.004
152
KrumholzE. W.YangH.WeisenhornP.HenryC. S.LibourelI. G. L. (2012). Genome-wide metabolic network reconstruction of the picoalga Ostreococcus. J. Exp. Bot.63, 2353–2362. 10.1093/jxb/err407
153
KrupnikT.KotabovaE.van BezouwenL. S.MazurR.GarstkaM.NixonP. J.et al. (2013). A reaction center-dependent photoprotection mechanism in a highly robust photosystem II from an extremophilic red alga, Cyanidioschyzon merolae. J. Biol. Chem.288, 23529–23542. 10.1074/jbc.M113.484659
154
KujawinskiE. B.LongneckerK.AlexanderH.DyhrmanS. T.FioreC. L.HaleyS. T.et al. (2017). Phosphorus availability regulates intracellular nucleotides in marine eukaryotic phytoplankton. Limnol. Oceanogr. Lett.2, 119–129. 10.1002/lol2.10043
155
KumarS. V.MisquittaR. W.ReddyV. S.RaoB. J.RajamM. V. (2004). Genetic transformation of the green alga-Chlamydomonas reinhardtii by Agrobacterium tumefaciens. Plant Sci.166, 731–738. 10.1016/j.plantsci.2003.11.012
156
KumarV.NandaM.KumarS.ChauhanP. K. (2018). The effects of ultraviolet radiation on growth, biomass, lipid accumulation and biodiesel properties of microalgae. Energy Sources A Recover. Util. Environ. Eff.40, 787–793. 10.1080/15567036.2018.1463310
157
KurotaniA.YamadaY.SakuraiT. (2017). Alga-PrAS (Algal Protein Annotation Suite): a database of comprehensive annotation in algal proteomes. Plant Cell Physiol.58:e6. 10.1093/pcp/pcw212
158
KustkaA. B.MilliganA. J.ZhengH.NewA. M.GatesC.BidleK. D.et al. (2014). Low CO2 results in a rearrangement of carbon metabolism to support C4 photosynthetic carbon assimilation in Thalassiosira pseudonana. New Phytol.204, 507–520. 10.1111/nph.12926
159
LauersenK. J.BaierT.WichmannJ.WordenweberR.MussgnugJ. H.HubnerW.et al. (2016). Efficient phototrophic production of a high-value sesquiterpenoid from the eukaryotic microalga Chlamydomonas reinhardtii. Metab. Eng.38, 331–343. 10.1016/j.ymben.2016.07.013
160
LauersenK. J.KruseO.MussgnugJ. H. (2015). Targeted expression of nuclear transgenes in Chlamydomonas reinhardtii with a versatile, modular vector toolkit. Appl Microbiol Biotechnol.99, 3491–3503. 10.1007/s00253-014-6354-7
161
LauersenK. J.WichmannJ.BaierT.KampranisS. C.PaterakiI.LindbergB.et al. (2018). Phototrophic production of heterologous diterpenoids and a hydroxy- functionalized derivative from Chlamydomonas reinhardtii. Metab. Eng.49, 116–127. 10.1016/j.ymben.2018.07.005
162
Le BihanT.HindleM.MartinS. F.Barrios-LlerenaM. E.KrahmerJ.KisK.et al. (2015). Label-free quantitative analysis of the casein kinase 2-responsive phosphoproteome of the marine minimal model species Ostreococcus tauri. Proteomics15, 4135–4144. 10.1002/pmic.201500086
163
LeeB.ChoiG. G.ChoiY. E.SungM.ParkM. S.YangJ. W. (2014). Enhancement of lipid productivity by ethyl methane sulfonate-mediated random mutagenesis and proteomic analysis in Chlamydomonas reinhardtii. Korean J. Chem. Eng.31, 1036–1042. 10.1007/s11814-014-0007-5
164
LeeT.-H.ChangJ.-S.WangH.-Y. (2013). Current developments in high-throughput analysis for microalgae cellular contents. Biotechnol. J.8, 1301–1314. 10.1002/biot.201200391
165
LelandaisG.ScheiberI.Paz-YepesJ.LozanoJ.-C.BotebolH.Pilátov,áJ.et al. (2016). Ostreococcus tauri is a new model green alga for studying iron metabolism in eukaryotic phytoplankton. BMC Genomics17:319. 10.1186/s12864-016-2666-6
166
LenkaS. K.CarbonaroN.ParkR.MillerS. M.ThorpeI.LiY. (2016). Current advances in molecular, biochemical, and computational modeling analysis of microalgal triacylglycerol biosynthesis. Biotechnol. Adv.34, 1046–1063. 10.1016/j.biotechadv.2016.06.004
167
LepetitB.VolkeD.GilbertM.WilhelmC.GossR. (2010). Evidence for the existence of one antenna-associated, lipid-dissolved and two protein-bound pools of diadinoxanthin cycle pigments in diatoms. Plant Physiol.154, 1905–1920. 10.1104/pp.110.166454
168
LercheK.HallmannA. (2009). Stable nuclear transformation of Gonium pectorale. BMC Biotechnol.9:64. 10.1186/1472-6750-9-64
169
LiD.-W.CenS.-Y.LiuY.-H.BalamuruganS.ZhengX.-Y.AlimujiangA.et al. (2016a). A type 2 diacylglycerol acyltransferase accelerates the triacylglycerol biosynthesis in heterokont oleaginous microalga Nannochloropsis oceanica. J. Biotechnol.229, 65–71. 10.1016/j.jbiotec.2016.05.005
170
LiF.GaoD.HuH. (2014a). High-efficiency nuclear transformation of the oleaginous marine Nannochloropsis species using PCR product. Biosci. Biotechnol. Biochem.78, 812–817. 10.1080/09168451.2014.905184
171
LiJ.XueL.YanH.WangL.LiuL.LuY.et al. (2007). The nitrate reductase gene-switch: a system for regulated expression in transformed cells of Dunaliella salina. Gene403, 132–142. 10.1016/j.gene.2007.08.001
172
LiT.GargouriM.FengJ.ParkJ.-J.GaoD.MiaoC.et al. (2015a). Regulation of starch and lipid accumulation in a microalga Chlorella sorokiniana. Bioresour. Technol.180, 250–257. 10.1016/j.biortech.2015.01.005
173
LiX.PatenaW.FauserF.JinkersonR. E.SaroussiS.MeyerM. T.et al. (2019). A genome-wide algal mutant library and functional screen identifies genes required for eukaryotic photosynthesis. Nat. Genet.51, 627–635. 10.1038/s41588-019-0370-6
174
LiX.ZhangR.PatenaW.GangS. S.BlumS. R.IvanovaN.et al. (2016b). An indexed, mapped mutant library enables reverse genetics studies of biological processes in Chlamydomonas reinhardtii. Plant Cell28, 367–387. 10.1105/tpc.15.00465
175
LiY.HanF.XuH.MuJ.ChenD.FengB.et al. (2014b). Potential lipid accumulation and growth characteristic of the green alga Chlorella with combination cultivation mode of nitrogen (N) and phosphorus (P). Bioresour. Technol.174, 24–32. 10.1016/j.biortech.2014.09.142
176
LiY.MuJ.ChenD.XuH.HanF.FengB.et al. (2015b). Proteomics analysis for enhanced lipid accumulation in oleaginous Chlorella vulgaris under a heterotrophic-Na(+) induction two-step regime. Biotechnol. Lett.37, 1021–1030. 10.1007/s10529-014-1758-0
177
LiY.YuanZ.MuJ.ChenD.FengB. (2013). Proteomic analysis of lipid accumulation in Chlorella protothecoides cells by heterotrophic N deprivation coupling cultivation. Energy Fuels27, 4031–4040. 10.1021/ef4000177
178
LiangS.ZhangZ.LiuH.GuoL.SunS.YangG. (2019). Identifying the growth associating genes of Nannochloropsis oceanica by bulked mutant analysis (BMA) and RNA. J. Appl. Phycol.31, 3677–3690. 10.1007/s10811-019-01867-w
179
LinH.-Y.YenS.-C.KuoP.-C.ChungC.-Y.YehK.-L.HuangC.-H.et al. (2017). Alkaline phosphatase promoter as an efficient driving element for exogenic recombinant in the marine diatom Phaeodactylum tricornutum. Algal Res.23, 58–65. 10.1016/j.algal.2017.01.007
180
LinW.-R.NgI.-S. (2020). Development of CRISPR/Cas9 system in Chlorella vulgaris FSP-E to enhance lipid accumulation. Enzyme Microb. Technol.133:109458. 10.1016/j.enzmictec.2019.109458
181
LiuJ.SunZ.GerkenH.HuangJ.JiangY.ChenF. (2014). Genetic engineering of the green alga Chlorella zofingiensis: a modified norflurazon-resistant phytoene desaturase gene as a dominant selectable marker. Appl. Microbiol. Biotechnol.98, 5069–5079. 10.1007/s00253-014-5593-y
182
LiuT.LuoF.WangZ.LiY. (2018a). The enhanced biomass and lipid accumulation in Coccomyxa subellipsoidea with an integrated treatment strategy initiated by brewery effluent and phytohormones. World J. Microbiol. Biotechnol.34:25. 10.1007/s11274-018-2408-9
183
LiuZ.LiT.HeQ.SunZ.JiangY. (2018b). role of mitochondria in regulating lutein and chlorophyll biosynthesis in Chlorella pyrenoidosa under heterotrophic conditions. Mar. Drugs16:100354. 10.3390/md16100354
184
LommerM.SpechtM.RoyA.-S.KraemerL.AndresonR.GutowskaM. A.et al. (2012). Genome and low-iron response of an oceanic diatom adapted to chronic iron limitation. Genome Biol.13:R66. 10.1186/gb-2012-13-7-r66
185
LongworthJ.NoirelJ.PandhalJ.WrightP. C.VaidyanathanS. (2012). HILIC- and SCX-based quantitative proteomics of Chlamydomonas reinhardtii during nitrogen starvation induced lipid and carbohydrate accumulation. J. Proteome Res.11, 5959–5971. 10.1021/pr300692t
186
LongworthJ.WuD.Huete-OrtegaM.WrightP. C.VaidyanathanS. (2016). Proteome response of Phaeodactylum tricornutum, during lipid accumulation induced by nitrogen depletion. Algal Res.18, 213–224. 10.1016/j.algal.2016.06.015
187
LuS.WangJ.MaQ.YangJ.LiX.YuanY.-J. (2013). Phospholipid metabolism in an industry microalga Chlorella sorokiniana: the impact of inoculum sizes. PLoS ONE8:e70827. 10.1371/journal.pone.0070827
188
LumbrerasV.StevensD. R.PurtonS. (1998). Efficient foreign gene expression in Chlamydomonas reinhardtii mediated by an endogenous intron. Plant J.14, 441–447. 10.1046/j.1365-313X.1998.00145.x
189
LuoC.-S.LiangJ.-R.LinQ.LiC.BowlerC.AndersonD. M.et al. (2014). Cellular responses associated with ROS production and cell fate decision in early stress response to iron limitation in the diatom Thalassiosira pseudonana. J. Proteome Res.13, 5510–5523. 10.1021/pr5004664
190
LuoQ.BianC.TaoM.HuangY.ZhengY.LvY.et al. (2019). Genome and transcriptome sequencing of the astaxanthin-producing green microalga, Haematococcus pluvialis. Genome Biol. Evol.11, 166–173. 10.1093/gbe/evy263
191
LvH.CuiX.WangS.JiaS. (2016). Metabolic profiling of Dunaliella salina shifting cultivation conditions to nitrogen deprivation. J. Postgenomics Drug Biomark. Dev.6, 1–9. 10.4172/2153-0769.1000170
192
LyonB. R.LeeP. A.BennettJ. M.DiTullioG. R.JanechM. G. (2011). Proteomic analysis of a sea-ice diatom: salinity acclimation provides new insight into the dimethylsulfoniopropionate production pathway. Plant Physiol.157, 1926–1941. 10.1104/pp.111.185025
193
MaQ.WangJ.LuS.LvY.YuanY. (2013). Quantitative proteomic profiling reveals photosynthesis responsible for inoculum size dependent variation in Chlorella sorokiniana. Biotechnol. Bioeng.110, 773–784. 10.1002/bit.24762
194
MackinderL. C. M. (2018). The Chlamydomonas CO2-concentrating mechanism and its potential for engineering photosynthesis in plants. New Phytol.217, 54–61. 10.1111/nph.14749
195
MartinS. F.MunagapatiV. S.Salvo-ChirnsideE.KerrL. E.Le BihanT. (2012). Proteome turnover in the green alga Ostreococcus tauri by time course 15N metabolic labeling mass spectrometry. J. Proteome Res.11, 476–486. 10.1021/pr2009302
196
MastrobuoniG.IrgangS.PietzkeM.AssmusH. E.WenzelM.SchulzeW. X.et al. (2012). Proteome dynamics and early salt stress response of the photosynthetic organism Chlamydomonas reinhardtii. BMC Genomics13:215. 10.1186/1471-2164-13-215
197
MataT. M.MartinsA. A.CaetanoN. S. (2010). Microalgae for biodiesel production and other applications: a review. Renew. Sustain. Energy Rev.14, 217–232. 10.1016/j.rser.2009.07.020
198
MayP.ChristianJ. O.KempaS.WaltherD. (2009). ChlamyCyc: an integrative systems biology database and web-portal for Chlamydomonas reinhardtii. BMC Genomics10, 1–11. 10.1186/1471-2164-10-209
199
MayfieldS. P.KindleK. L. (1990). Stable nuclear transformation of Chlamydomonas reinhardtii by using a C. reinhardtii gene as the selectable marker. Proc. Natl. Acad. Sci. U. S. A.87, 2087–2091. 10.1073/pnas.87.6.2087
200
McKewB. A.MetodievaG.RainesC. A.MetodievM. V.GeiderR. J. (2015). Acclimation of Emiliania huxleyi (1516) to nutrient limitation involves precise modification of the proteome to scavenge alternative sources of N and P. Environ. Microbiol.17, 4050–4062. 10.1111/1462-2920.12957
201
MeharJ.ShekhA. M. U NSaradaR.ChauhanV. S.et al. (2019). Automation of pilot-scale open raceway pond: a case study of CO2-fed pH control on Spirulina biomass, protein and phycocyanin production. J. CO2 Util.33, 384–393. 10.1016/j.jcou.2019.07.006
202
MicheletL.Lefebvre-LegendreL.BurrS. E.RochaixJ.-D.Goldschmidt-ClermontM. (2011). Enhanced chloroplast transgene expression in a nuclear mutant of Chlamydomonas. Plant Biotechnol. J.9, 565–574. 10.1111/j.1467-7652.2010.00564.x
203
MiniP.DemurtasO. C.ValentiniS.PallaraP.ApreaG.FerranteP.et al. (2018). Agrobacterium-mediated and electroporation-mediated transformation of Chlamydomonas reinhardtii: a comparative study. BMC Biotechnol.18:11. 10.1186/s12896-018-0416-3
204
MiyagishimaS.EraA.HasunumaT.MatsudaM.HirookaS.SumiyaN.et al. (2019). Day/night separation of oxygenic energy metabolism and nuclear DNA replication in the unicellular red alga Cyanidioschyzon merolae. MBio10, e00833–e00819. 10.1128/mBio.00833-19
205
MocsaiR.FiglR.TroschlC.StrasserR.SvehlaE.WindwarderM.et al. (2019). N-glycans of the microalga Chlorella vulgaris are of the oligomannosidic type but highly methylated. Sci. Rep.9:331. 10.1038/s41598-018-36884-1
206
MolinoJ. V. D.de CarvalhoJ. C. M.MayfieldS. P. (2018). Comparison of secretory signal peptides for heterologous protein expression in microalgae: expanding the secretion portfolio for Chlamydomonas reinhardtii. PLoS ONE13:e0192433. 10.1371/journal.pone.0192433
207
MolnarI.LopezD.WisecaverJ. H.DevarenneT. P.WeissT. L.PellegriniM.et al. (2012). Bio-crude transcriptomics: gene discovery and metabolic network reconstruction for the biosynthesis of the terpenome of the hydrocarbon oil-producing green alga, Botryococcus braunii race B (Showa). BMC Genomics13:576. 10.1186/1471-2164-13-576
208
MoroneyJ. V.MaY.FreyW. D.FusilierK. A.PhamT. T.SimmsT. A.et al. (2011). The carbonic anhydrase isoforms of Chlamydomonas reinhardtii: intracellular location, expression, and physiological roles. Photosynth. Res.109, 133–149. 10.1007/s11120-011-9635-3
209
MuñozC. F.de JaegerL.SturmeM. H. J.LipK. Y. F.OlijslagerJ. W. J.SpringerJ.et al. (2018). Improved DNA/protein delivery in microalgae—a simple and reliable method for the prediction of optimal electroporation settings. Algal Res.33, 448–455. 10.1016/j.algal.2018.06.021
210
MuñozC. F.SturmeM. H. J.D'AdamoS.WeusthuisR. A.WijffelsR. H. (2019). Stable transformation of the green algae Acutodesmus obliquus and Neochloris oleoabundans based on E. coli conjugation. Algal Res.39:101453. 10.1016/j.algal.2019.101453
211
MutoM.FukudaY.NemotoM.YoshinoT.MatsunagaT.TanakaT. (2013). Establishment of a genetic transformation system for the marine pennate diatom Fistulifera sp. strain JPCC DA0580–a high triglyceride producer. Mar. Biotechnol.15, 48–55. 10.1007/s10126-012-9457-0
212
NagaoM.MatsuiK.UemuraM. (2008). Klebsormidium flaccidum, a charophycean green alga, exhibits cold acclimation that is closely associated with compatible solute accumulation and ultrastructural changes. Plant. Cell Environ.31, 872–885. 10.1111/j.1365-3040.2008.01804.x
213
NelsonD. R.ChaiboonchoeA.FuW.HazzouriK. M.HuangZ.JaiswalA.et al. (2019). Potential for heightened sulfur-metabolic capacity in coastal subtropical microalgae. iScience11, 450–465. 10.1016/j.isci.2018.12.035
214
NelsonD. R.KhraiweshB.FuW.AlseekhS.JaiswalA.ChaiboonchoeA.et al. (2017). The genome and phenome of the green alga Chloroidium sp. UTEX 3007 reveal adaptive traits for desert acclimatization. Elife6:25783. 10.7554/eLife.25783
215
NethravathyM.MeharJ. G.MudliarS. N.ShekhA. Y. (2019). Recent advances in microalgal bioactives for food, feed, and healthcare products: commercial potential, market space, and sustainability. Compr. Rev. Food Sci. Food Saf.18, 1882–1897. 10.1111/1541-4337.12500
216
NgI.KeskinB. B.TanS. (2020). A critical review of genome editing and synthetic biology applications in metabolic engineering of microalgae and cyanobacteria. Biotechnol. J.15:1900228. 10.1002/biot.201900228
217
NguyenH. M.BaudetM.CuineS.AdrianoJ.-M.BartheD.BillonE.et al. (2011). Proteomic profiling of oil bodies isolated from the unicellular green microalga Chlamydomonas reinhardtii: with focus on proteins involved in lipid metabolism. Proteomics11, 4266–4273. 10.1002/pmic.201100114
218
NikolovaD.WeberD.ScholzM.BaldT.ScharsackJ. P.HipplerM. (2017). Temperature-induced remodeling of the photosynthetic machinery tunes photosynthesis in the thermophilic alga Cyanidioschyzon merolae. Plant Physiol.174, 35–46. 10.1104/pp.17.00110
219
NiuY.-F.ZhangM.-H.LiD.-W.YangW.-D.LiuJ.-S.BaiW.-B.et al. (2013). Improvement of neutral lipid and polyunsaturated fatty acid biosynthesis by overexpressing a type 2 diacylglycerol acyltransferase in marine diatom Phaeodactylum tricornutum. Mar. Drugs11, 4558–4569. 10.3390/md11114558
220
NonoyamaT.NojimaD.MaedaY.NodaM.YoshinoT.MatsumotoM.et al. (2019). Proteomics analysis of lipid droplets indicates involvement of membrane trafficking proteins in lipid droplet breakdown in the oleaginous diatom Fistulifera solaris. Algal Res.44:101660. 10.1016/j.algal.2019.101660
221
NymarkM.SharmaA. K.SparstadT.BonesA. M.WingeP. (2016). A CRISPR / Cas9 system adapted for gene editing in marine algae. Nat. Publ. Gr.6:24951. 10.1038/srep24951
222
OhnumaM.MisumiO.FujiwaraT.WatanabeS.TanakaK.KuroiwaT. (2009). Transient gene suppression in a red alga, Cyanidioschyzon merolae 10D. Protoplasma236, 107–112. 10.1007/s00709-009-0056-5
223
Ortega-EscalanteJ. A.JasperR.MillerS. M. (2019a). CRISPR/Cas9 mutagenesis in Volvox carteri. Plant J.97, 661–672. 10.1111/tpj.14149
224
Ortega-EscalanteJ. A.KwokO.MillerS. M. (2019b). New selectable markers for Volvox carteri transformation. Protist170, 52–63. 10.1016/j.protis.2018.11.002
225
OsorioH.JaraC.FuenzalidaK.Rey-JuradoE.VásquezM. (2019). High-efficiency nuclear transformation of the microalgae Nannochloropsis oceanica using Tn5 transposome for the generation of altered lipid accumulation phenotypes. Biotechnol. Biofuels12:134. 10.1186/s13068-019-1475-y
226
OtaS.OshimaK.YamazakiT.KimS.YuZ.YoshiharaM.et al. (2016a). Highly efficient lipid production in the green alga Parachlorella kessleri: draft genome and transcriptome endorsed by whole-cell 3D ultrastructure. Biotechnol. Biofuels9:13. 10.1186/s13068-016-0424-2
227
OtaS.YoshiharaM.YamazakiT.TakeshitaT.HirataA.KonomiM.et al. (2016b). Deciphering the relationship among phosphate dynamics, electron-dense body and lipid accumulation in the green alga Parachlorella kessleri. Sci. Rep.6:25731. 10.1038/srep25731
228
OuyangS.HuX.ZhouQ. (2015). Envelopment-internalization synergistic effects and metabolic mechanisms of graphene oxide on single-cell Chlorella vulgaris are dependent on the nanomaterial particle size. ACS Appl. Mater. Interfaces7, 18104–18112. 10.1021/acsami.5b05328
229
PalD.Khozin-GoldbergI.Didi-CohenS.SolovchenkoA.BatushanskyA.KayeY.et al. (2013). Growth, lipid production and metabolic adjustments in the euryhaline eustigmatophyte Nannochloropsis oceanica CCALA 804 in response to osmotic downshift. Appl. Microbiol. Biotechnol.97, 8291–8306. 10.1007/s00253-013-5092-6
230
ParkE.-J.ChoiJ. (2018). Resistance and proteomic response of microalgae to ionizing irradiation. Biotechnol. Bioprocess Eng.23, 704–709. 10.1007/s12257-018-0468-1
231
ParkS.-H.KyndtJ.ChouguleK.ParkJ.-J.BrownJ. K. (2018). Low-phosphate-selected Auxenochlorella protothecoides redirects phosphate to essential pathways while producing more biomass. PLoS ONE13:e0198953. 10.1371/journal.pone.0198953
232
PasquetV.ChérouvrierJ.-R.FarhatF.ThiéryV.PiotJ.-M.BérardJ.-B.et al. (2011). Study on the microalgal pigments extraction process: performance of microwave assisted extraction. Process Biochem.46, 59–67. 10.1016/j.procbio.2010.07.009
233
PatelouM.InfanteC.DardelleF.RandewigD.KouriE. D.UdvardiM. K.et al. (2020). Transcriptomic and metabolomic adaptation of Nannochloropsis gaditana grown under different light regimes. Algal Res.45:101735. 10.1016/j.algal.2019.101735
234
PeledE.LeuS.ZarkaA.WeissM.PickU.Khozin-GoldbergI.et al. (2011). Isolation of a novel oil globule protein from the green alga Haematococcus pluvialis (Chlorophyceae). Lipids46, 851–861. 10.1007/s11745-011-3579-4
235
PerrineZ.NegiS.SayreR. T. (2012). Optimization of photosynthetic light energy utilization by microalgae. Algal Res.1, 134–142. 10.1016/j.algal.2012.07.002
236
PierobonS. C.ChengX.GrahamP. J.NguyenB.KarakolisE. G.SintonD. (2018). Emerging microalgae technology: a review. Sustain. Energy Fuels2, 13–38. 10.1039/C7SE00236J
237
PolinerE.PulmanJ. A.ZienkiewiczK.ChildsK.BenningC.FarrE. M. (2018). A toolkit for Nannochloropsis oceanica CCMP1779 enables gene stacking and genetic engineering of the eicosapentaenoic acid pathway for enhanced long-chain polyunsaturated fatty acid production. Plant Biotechnol. J.16, 298–309. 10.1111/pbi.12772
238
PolinerE.TakeuchiT.DuZ.-Y.BenningC.FarreE. M. (2019). Non-transgenic marker-free gene disruption by an episomal CRISPR system in the oleaginous microalga, Nannochloropsis oceanica CCMP1779. ACS Synth. Biol.7, 962–968. 10.1021/acssynbio.7b00362
239
PombertJ.-F.BlouinN. A.LaneC.BouciasD.KeelingP. J. (2014). A lack of parasitic reduction in the obligate parasitic green alga Helicosporidium. PLOS Genet.10:e1004355. 10.1371/journal.pgen.1004355
240
PoulsenN.ChesleyP. M.KrögerN. (2006). Molecular genetic manipulation of the diatom Thalassiosira pseudonana (Bacillariophyceae). J. Phycol.42, 1059–1065. 10.1111/j.1529-8817.2006.00269.x
241
Poulson-EllestadK. L.JonesC. M.RoyJ.ViantM. R.FernandezF. M.KubanekJ.et al. (2014). Metabolomics and proteomics reveal impacts of chemically mediated competition on marine plankton. Proc. Natl. Acad. Sci. U. S. A.111, 9009–9014. 10.1073/pnas.1402130111
242
QuB.EuY.JeongW.KimD. (2012). Lab on a chip droplet electroporation in microfluidics for efficient cell transformation with or without cell wall removal. Lab Chip. 12, 4483–4488.
243
QuinnJ. M.BarracoP.ErikssonM.MerchantS. (2000). Coordinate copper- and oxygen-responsive Cyc6 and Cpx1 expression in Chlamydomonas is mediated by the same element. J. Biol. Chem.275, 6080–6089. 10.1074/jbc.275.9.6080
244
RadakovitsR.JinkersonR. E.DarzinsA.PosewitzM. C. (2010). Genetic engineering of algae for enhanced biofuel production. Eukaryot. Cell9, 486–501. 10.1128/EC.00364-09
245
RadakovitsR.JinkersonR. E.FuerstenbergS. I.TaeH.SettlageR. E.BooreJ. L.et al. (2012). Draft genome sequence and genetic transformation of the oleaginous alga Nannochloropis gaditana. Nat. Commun.3:686. 10.1038/ncomms1688
246
RademacherN.KernR.FujiwaraT.Mettler-AltmannT.MiyagishimaS.HagemannM.et al. (2016). Photorespiratory glycolate oxidase is essential for the survival of the red alga Cyanidioschyzon merolae under ambient CO2 conditions. J. Exp. Bot.67, 3165–3175. 10.1093/jxb/erw118
247
RasalaB. A.MutoM.SullivanJ.MayfieldS. P. (2011). Improved heterologous protein expression in the chloroplast of Chlamydomonas reinhardtii through promoter and 5' untranslated region optimization. Plant Biotechnol. J.9, 674–683. 10.1111/j.1467-7652.2011.00620.x
248
RathodJ. P.PrakashG.PanditR.LaliA. M. (2013). Agrobacterium-mediated transformation of promising oil-bearing marine algae Parachlorella kessleri. Photosynth. Res.118, 141–146. 10.1007/s11120-013-9930-2
249
RazzaqA.SaleemF.KanwalM.MustafaG.YousafS.Imran ArshadH. M.et al. (2019). Modern trends in plant genome editing: an inclusive review of the CRISPR/Cas9 toolbox. Int. J. Mol. Sci.20:164045. 10.3390/ijms20164045
250
RechtL.TöpferN.BatushanskyA.SikronN.GibonY.FaitA.et al. (2014). Metabolite profiling and integrative modeling reveal metabolic constraints for carbon partitioning under nitrogen starvation in the green algae Haematococcus pluvialis. J. Biol. Chem.289, 30387–30403. 10.1074/jbc.M114.555144
251
RemmersI. M.D'AdamoS.MartensD. E.de VosR. C. H.MummR.AmericaA. H. P.et al. (2018). Orchestration of transcriptome, proteome and metabolome in the diatom Phaeodactylum tricornutum during nitrogen limitation. Algal Res.35, 33–49. 10.1016/j.algal.2018.08.012
252
RochaixJ. D.DillewijnJ. (1982). Transformation of the green alga Chlamydomonas reinhardii with yeast. Nature296, 70–72. 10.1038/296070a0
253
RokittaS. D.Von DassowP.RostB.JohnU. (2014). Emiliania huxleyi endures N-limitation with an efficient metabolic budgeting and effective ATP synthesis. BMC Genomics15:1051. 10.1186/1471-2164-15-1051
254
RosenbergJ. N.KobayashiN.BarnesA.NoelE. A.BetenbaughM. J.OylerG. A. (2014). Comparative analyses of three Chlorella species in response to light and sugar reveal distinctive lipid accumulation patterns in the MicroalgaC. sorokiniana. PLoS ONE9:e92460. 10.1371/journal.pone.0092460
255
RosenwasserS.Graff van CreveldS.SchatzD.MalitskyS.TzfadiaO.AharoniA.et al. (2014). Mapping the diatom redox-sensitive proteome provides insight into response to nitrogen stress in the marine environment. Proc. Natl. Acad. Sci. U. S. A.111, 2740 LP−2745. 10.1073/pnas.1319773111
256
RunC.FangL.FanJ.FanC.LuoY.HuZ.et al. (2016). Stable nuclear transformation of the industrial alga Chlorella pyrenoidosa. Algal Res.17, 196–201. 10.1016/j.algal.2016.05.002
257
SainiD. K.ChakdarH.PabbiS.ShuklaP. (2020). Enhancing production of microalgal biopigments through metabolic and genetic engineering. Crit. Rev. Food Sci. Nutr.60, 391–405. 10.1080/10408398.2018.1533518
258
SalgueroD. A. M.Fernández-NiñoM.Serrano-BermúdezL. M.MeloD. O. P.WinckF. V.CaldanaC.et al. (2019). Development of a Chlamydomonas reinhardtii metabolic network dynamic model to describe distinct phenotypes occurring at different CO2 levels. PeerJ6:e5528. 10.7717/peerj.5528
259
SanchezF.GeffroyS.NorestM.YauS.MoreauH.GrimsleyN. (2019). Simplified transformation of Ostreococcus tauri using polyethylene glycol. Genes10:50399. 10.3390/genes10050399
260
SchierenbeckL.RiesD.RoggeK.GreweS.WeisshaarB.KruseO. (2015). Fast forward genetics to identify mutations causing a high light tolerant phenotype in Chlamydomonas reinhardtii by whole-genome-sequencing. BMC Genomics16, 1–15. 10.1186/s12864-015-1232-y
261
SchoberA. F.Ri O Bi RtulosC.BischoffA.LepetitB.GruberA.KrothP. G. (2019). Organelle studies and proteome analyses of mitochondria and plastids fractions from the diatom Thalassiosira pseudonana. Plant Cell Physiol.60, 1811–1828. 10.1093/pcp/pcz097
262
SchrodaM.BloD.BeckC. F. (2000). The HSP70A promoter as a tool for the improved expression of transgenes in Chlamydomonas. 21, 121–131. 10.1046/j.1365-313x.2000.00652.x
263
Schulz-RaffeltM.ChochoisV.AuroyP.Cuin,éS.BillonE.DauvilléeD.et al. (2016). Hyper-accumulation of starch and oil in a Chlamydomonas mutant affected in a plant-specific DYRK kinase. Biotechnol. Biofuels9, 1–12. 10.1186/s13068-016-0469-2
264
ScrantonM. A.OstrandJ. T.GeorgiannaD. R.LofgrenS. M.LiD.EllisR. C.et al. (2016). Synthetic promoters capable of driving robust nuclear gene expression in the green alga Chlamydomonas reinhardtii. Algal Res.15, 135–142. 10.1016/j.algal.2016.02.011
265
SeoS.JeonH.HwangS.JinE.ChangK. S. (2015). Development of a new constitutive expression system for the transformation of the diatom Phaeodactylum tricornutum. Algal Res.11, 50–54. 10.1016/j.algal.2015.05.012
266
SerifM.DuboisG.FinouxA.-L.TesteM.-A.JalletD.DaboussiF. (2018). One-step generation of multiple gene knock-outs in the diatom Phaeodactylum tricornutum by DNA-free genome editing. Nat. Commun.9:3924. 10.1038/s41467-018-06378-9
267
ShahA. R.AhmadA.SrivastavaS.Jaffar AliB. M. (2017). Reconstruction and analysis of a genome-scale metabolic model of Nannochloropsis gaditana. Algal Res.26, 354–364. 10.1016/j.algal.2017.08.014
268
ShaikhK. M.NesammaA. A.AbdinM. Z.JuturP. P. (2019). Molecular profiling of an oleaginous trebouxiophycean alga Parachlorella kessleri subjected to nutrient deprivation for enhanced biofuel production. Biotechnol. Biofuels12:182. 10.1186/s13068-019-1521-9
269
SharmaA. K.NymarkM.SparstadT.BonesA. M.WingeP. (2018). Transgene-free genome editing in marine algae by bacterial conjugation—comparison with biolistic CRISPR/Cas9 transformation. Sci. Rep.8:14401. 10.1038/s41598-018-32342-0
270
SharmaS. K.NelsonD. R.AbdrabuR.KhraiweshB.JijakliK.ArnouxM.et al. (2015). An integrative Raman microscopy—based workflow for rapid in situ analysis of microalgal lipid bodies. Biotechnol. Biofuels8:164. 10.1186/s13068-015-0349-1
271
SheehanJ.DunahayT.BenemannJ.RoesslerP. (1998). A look back at the US department of energy's aquatic species program: biodiesel from algae. National Renewable Energy Laboratory, 328, 1–294.
272
ShekhA. Y.ShrivastavaP.GuptaA.KrishnamurthiK.DeviS. S.MudliarS. N. (2016). Biomass and lipid enhancement in Chlorella sp. with emphasis on biodiesel quality assessment through detailed FAME signature. Bioresour. Technol.201, 276–286. 10.1016/j.biortech.2015.11.058
273
ShihC.-H.ChenH.-Y.LeeH.-C.TsaiH.-J. (2015). Purple chromoprotein gene serves as a new selection marker for transgenesis of the microalga Nannochloropsis oculata. PLoS ONE10:e0120780. 10.1371/journal.pone.0120780
274
ShinS.-E.LimJ.-M.KohH. G.KimE. K.KangN. K.JeonS.et al. (2016a). CRISPR/Cas9-induced knockout and knock-in mutations in Chlamydomonas reinhardtii. Sci. Rep.6:27810. 10.1038/srep27810
275
ShinW.-S.LeeB.JeongB.ChangY. K.KwonJ.-H. (2016b). Truncated light-harvesting chlorophyll antenna size in Chlorella vulgaris improves biomass productivity. J. Appl. Phycol.28, 3193–3202. 10.1007/s10811-016-0874-8
276
ShinW.-S.LeeB.KangN. K.KimY.-U.JeongW.-J.KwonJ.-H.et al. (2017). Complementation of a mutation in CpSRP43 causing partial truncation of light-harvesting chlorophyll antenna in Chlorella vulgaris. Sci. Rep.7:17929. 10.1038/s41598-017-18221-0
277
ShinY. S.JeongJ.NguyenT. H. T.KimJ. Y. H.JinE.SimS. J. (2019). Targeted knockout of phospholipase A2 to increase lipid productivity in Chlamydomonas reinhardtii for biodiesel production. Bioresour. Technol.271, 368–374. 10.1016/j.biortech.2018.09.121
278
SibiG.AnuraagT. S.BafilaG. (2014). Copper stress on cellular contents and fatty acid profiles in Chlorella species. Online J. Biol. Sci.14, 209–217. 10.3844/ojbsci.2014.209.217
279
SiddiquiA.WeiZ.BoehmM.AhmadN. (2020). Engineering microalgae through chloroplast transformation to produce high-value industrial products. Biotechnol. App. Biochem.67, 30–40. 10.1002/bab.1823
280
SithtisarnS.YokthongwattanaK.MahongB.RoytrakulS.PaemaneeA.PhaonakropN.et al. (2017). Comparative proteomic analysis of Chlamydomonas reinhardtii control and a salinity-tolerant strain revealed a differential protein expression pattern. Planta246, 843–856. 10.1007/s00425-017-2734-4
281
SivakaminathanS.HankamerB.WolfJ.YarnoldJ. (2018). High-throughput optimisation of light-driven microalgae biotechnologies. Sci. Rep.8:11687. 10.1038/s41598-018-29954-x
282
SizovaI.GreinerA.AwasthiM.KateriyaS.HegemannP. (2013). Nuclear gene targeting in Chlamydomonas using engineered zinc-finger nucleases. Plant J.73, 873–882. 10.1111/tpj.12066
283
SlatteryS. S.DiamondA.WangH.TherrienJ. A.LantJ. T.JazeyT.et al. (2018). An expanded plasmid-based genetic toolbox enables Cas9 genome editing and stable maintenance of synthetic pathways in Phaeodactylum tricornutum. ACS Synth. Biol.7, 328–338. 10.1021/acssynbio.7b00191
284
SmithE. G.D'AngeloC.SharonY.TchernovD.WiedenmannJ. (2017). Acclimatization of symbiotic corals to mesophotic light environments through wavelength transformation by fluorescent protein pigments. Proceedings. Biol. Sci.284:20170320. 10.1098/rspb.2017.0320
285
SmithS. R.DupontC. L.McCarthyJ. K.BroddrickJ. T.ObornikM.HorakA.et al. (2019). Evolution and regulation of nitrogen flux through compartmentalized metabolic networks in a marine diatom. Nat. Commun.10:4552. 10.1038/s41467-019-12407-y
286
SorigueD.LegeretB.CuineS.MoralesP.MirabellaB.GuedeneyG.et al. (2016). Microalgae synthesize hydrocarbons from long-chain fatty acids via a light-dependent pathway. Plant Physiol.171, 2393–2405. 10.1104/pp.16.00462
287
SpechtE. A.MayfieldS. P. (2013). Synthetic oligonucleotide libraries reveal novel regulatory elements in Chlamydomonas chloroplast mRNAs. ACS Synth. Biol.2, 34–46. 10.1021/sb300069k
288
SpechtE. A.Nour-eldinH. H.HoangK. T. D.MayfieldS. P. (2015). An improved ARS2-derived nuclear reporter enhances the efficiency and ease of genetic engineering in Chlamydomonas. Biotechnol. J.10, 473–479. 10.1002/biot.201400172
289
SpreitzerR. J.EsquivelM. G.DuY. C.McLaughlinP. D. (2001). Alanine-scanning mutagenesis of the small-subunit beta A-beta B loop of chloroplast ribulose-1,5-bisphosphate carboxylase/oxygenase: substitution at Arg-71 affects thermal stability and CO2/O2 specificity. Biochemistry40, 5615–5621. 10.1021/bi002943e
290
SpreitzerR. J.PeddiS. R.SatagopanS. (2005). Phylogenetic engineering at an interface between large and small subunits imparts land-plant kinetic properties to algal Rubisco. Proc. Natl. Acad. Sci. U. S. A.102, 17225–17230. 10.1073/pnas.0508042102
291
SrinivasanR.GothandamK. M. (2016). Synergistic action of D-glucose and acetosyringone on Agrobacterium strains for efficient Dunaliella transformation. PLoS ONE11:e0158322. 10.1371/journal.pone.0158322
292
SteinbrennerJ.SandmannG. (2006). Transformation of the green alga Haematococcus pluvialis with a phytoene desaturase for accelerated astaxanthin biosynthesis. Appl. Environ. Microbiol.72, 7477–7484. 10.1128/AEM.01461-06
293
StukenbergD.ZaunerS.Dell'AquilaG.MaierU. G. (2018). Optimizing CRISPR/Cas9 for the Diatom Phaeodactylum tricornutum. Front. Plant Sci.9:740. 10.3389/fpls.2018.00740
294
SuY.WangJ.ShiM.NiuX.YuX.GaoL.et al. (2014). Metabolomic and network analysis of astaxanthin-producing Haematococcus pluvialis under various stress conditions. Bioresour. Technol.170, 522–529. 10.1016/j.biortech.2014.08.018
295
SumiyaN.KawaseY.HayakawaJ.MatsudaM.NakamuraM.EraA.et al. (2015). Expression of cyanobacterial Acyl-ACP reductase elevates the triacylglycerol level in the red alga Cyanidioschyzon merolae. Plant Cell Physiol.56, 1962–1980. 10.1093/pcp/pcv120
296
SunM.YangZ.WawrikB. (2018). Metabolomic fingerprints of individual algal cells using the single-probe mass spectrometry technique. Front. Plant Sci.9, 1–10. 10.3389/fpls.2018.00571
297
TalebiA. F.TohidfarM.BagheriA.LyonS. R.Salehi-AshtianiK.TabatabaeiM. (2014). Manipulation of carbon flux into fatty acid biosynthesis pathway in Dunaliella salina using AccD and ME genes to enhance lipid content and to improve produced biodiesel quality. Biofuel Res. J.1, 91–97. 10.18331/BRJ2015.1.3.6
298
TamoiM.NagaokaM.MiyagawaY.ShigeokaS. (2006). Contribution of fructose-1,6-bisphosphatase and sedoheptulose-1,7-bisphosphatase to the photosynthetic rate and carbon flow in the Calvin cycle in transgenic plants. Plant Cell Physiol.47, 380–390. 10.1093/pcp/pcj004
299
TeM. R.Lohuis MillerD. J. (1998). Genetic transformation of dinoflagellates (Amphidinium and Symbiodinium): expression of GUS in microalgae using heterologous promoter constructs. Plant J.13, 427–435. 10.1046/j.1365-313X.1998.00040.x
300
TejanoL. A.PeraltaJ. P.YapE. E. S.PanjaitanF. C. A.ChangY.-W. (2019). Prediction of bioactive peptides from Chlorella sorokiniana proteins using proteomic techniques in combination with bioinformatics analyses. Int. J. Mol. Sci.20:71786. 10.3390/ijms20071786
301
TerashimaM.FreemanE. S.JinkersonR. E.JonikasM. C. (2015). A fluorescence-activated cell sorting-based strategy for rapid isolation of high-lipid Chlamydomonas mutants. Plant J.81, 147–159. 10.1111/tpj.12682
302
ThangarajB.RyanC. M.SoudaP.KrauseK.FaullK. F.WeberA. P. M.et al. (2010). Data-directed top-down Fourier-transform mass spectrometry of a large integral membrane protein complex: photosystem II from Galdieria sulphuraria. Proteomics10, 3644–3656. 10.1002/pmic.201000190
303
ThanhT.ChiV. T. Q.OmarH.AbdullahM. P.NapisS. (2012). Sequence analysis and potentials of the native RbcS promoter in the development of an alternative eukaryotic expression system using green microalga Ankistrodesmus convolutus. Int. J. Mol. Sci.13, 2676−2691. 10.3390/ijms13032676
304
ThapaH. R.NaikM. T.OkadaS.TakadaK.MolnarI.XuY.et al. (2016). A squalene synthase-like enzyme initiates production of tetraterpenoid hydrocarbons in Botryococcus braunii Race L. Nat. Commun.7:11198. 10.1038/ncomms11198
305
ThungL.HeJ.ZhuQ.XuZ.LiuJ.ChowY. (2018). Insertional mutations exhibiting high cell-culture density HCD phenotypes are enriched through continuous subcultures in Chlamydomonas reinhardtii. Algae33, 127–141. 10.4490/algae.2018.33.2.28
306
TianY.GaoS.von der HeydeE. L.HallmannA.NagelG. (2018). Two-component cyclase opsins of green algae are ATP-dependent and light-inhibited guanylyl cyclases. BMC Biol.16:144. 10.1186/s12915-018-0613-5
307
ToyoshimaM.SatoN. (2015). High-level accumulation of triacylglycerol and starch in photoautotrophically grown Chlamydomonas debaryana NIES-2212. Plant Cell Physiol.56, 2447–2456. 10.1093/pcp/pcv163
308
ToyoshimaM.SatoN. (2018). Optimization of triacylglycerol and starch production in Chlamydomonas debaryana NIES-2212 with regard to light intensity and CO2 concentration. Microbiology164, 359–368. 10.1099/mic.0.000603
309
TranN.-A. T.PadulaM. P.EvenhuisC. R.CommaultA. S.RalphP. J.TamburicB. (2016). Proteomic and biophysical analyses reveal a metabolic shift in nitrogen deprived Nannochloropsis oculata. Algal Res.19, 1–11. 10.1016/j.algal.2016.07.009
310
TranN.-P.ParkJ.-K.HongS.-J.LeeC.-G. (2009). Proteomics of proteins associated with astaxanthin accumulation in the green algae Haematococcus lacustris under the influence of sodium orthovanadate. Biotechnol. Lett.31:1917. 10.1007/s10529-009-0095-1
311
TranP. T.SharifiM. N.PoddarS.DentR. M.NiyogiK. K. (2012). Intragenic enhancers and suppressors of phytoene desaturase mutations in Chlamydomonas reinhardtii. PLoS ONE7:e42196. 10.1371/journal.pone.0042196
312
TrentacosteE. M.ShresthaR. P.SmithS. R.GleC.HartmannA. C.HildebrandM.et al. (2013). Metabolic engineering of lipid catabolism increases microalgal lipid accumulation without compromising growth. Proc. Natl. Acad. Sci. U. S. A.110, 19748–19753. 10.1073/pnas.1309299110
313
TsaiC.-H.WarakanontJ.TakeuchiT.SearsB. B.MoelleringE. R.BenningC. (2014). The protein compromised hydrolysis of triacylglycerols 7 (CHT7) acts as a repressor of cellular quiescence in Chlamydomonas. Proc. Natl. Acad. Sci. U. S. A.111, 15833–15838. 10.1073/pnas.1414567111
314
van BarenM. J.BachyC.ReistetterE. N.PurvineS. O.GrimwoodJ.SudekS.et al. (2016). Evidence-based green algal genomics reveals marine diversity and ancestral characteristics of land plants. BMC Genomics17:267. 10.1186/s12864-016-2585-6
315
van OoijenG.KnoxK.KisK.BougetF.-Y.MillarA. J. (2012). Genomic transformation of the picoeukaryote Ostreococcus tauri. J. Vis. Exp.65:e4074. 10.3791/4074
316
VandepoeleK.Van BelM.RichardG.Van LandeghemS.VerhelstB.MoreauH.et al. (2013). pico-PLAZA, a genome database of microbial photosynthetic eukaryotes. Environ. Microbiol.15, 2147–2153. 10.1111/1462-2920.12174
317
VendruscoloR. G.FagundesM. B.MaronezeM. M.do NascimentoT. C.de MenezesC. R.BarinJ. S.et al. (2019). Scenedesmus obliquus metabolomics: effect of photoperiods and cell growth phases. Bioprocess Biosyst. Eng.42, 727–739. 10.1007/s00449-019-02076-y
318
VerrutoJ.FrancisK.WangY.LowM. C.GreinerJ.TackeS.et al. (2018). Unrestrained markerless trait stacking in Nannochloropsis gaditana through combined genome editing and marker recycling technologies. Proc. Natl. Acad. Sci. U. S. A.115, E7015–E7022. 10.1073/pnas.1718193115
319
VillanovaV.FortunatoA. E.SinghD.BoD. D.ConteM.ObataT.et al. (2017). Investigating mixotrophic metabolism in the model diatom Phaeodactylum tricornutum. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 372. 10.1098/rstb.2016.0404
320
VinyardD. J.GimpelJ.AnanyevG. M.MayfieldS. P.DismukesG. C. (2014). Engineered photosystem II reaction centers optimize photochemistry versus photoprotection at different solar intensities. J. Am. Chem. Soc.136, 4048–4055. 10.1021/ja5002967
321
VoglerB. W.StarkenburgS. R.SudasingheN.SchambachJ. Y.RollinJ. A.PattathilS.et al. (2018). Characterization of plant carbon substrate utilization by Auxenochlorella protothecoides. Algal Res.34, 37–48. 10.1016/j.algal.2018.07.001
322
von der HeydeE. L.KleinB.AbramL.HallmannA. (2015). The inducible nitA promoter provides a powerful molecular switch for transgene expression in Volvox carteri. BMC Biotechnol.15:5. 10.1186/s12896-015-0122-3
323
WaltmanP. H.GuoJ.ReistetterE. N.PurvineS.AnsongC. K.van BarenM. J.et al. (2016). Identifying aspects of the post-transcriptional program governing the proteome of the green alga Micromonas pusilla. PLoS ONE11:e0155839. 10.1371/journal.pone.0155839
324
WanX.LiT.LiuD.ChenY.LiuY.LiuB.et al. (2018). Effect of marine microalga Chlorella pyrenoidosa ethanol extract on lipid metabolism and gut microbiota composition in high-fat diet-fed rats. Mar. Drugs16:120498. 10.3390/md16120498
325
WangP.ZhangB.ZhangH.HeY.OngC. N.YangJ. (2019a). Metabolites change of Scenedesmus obliquus exerted by AgNPs. J. Environ. Sci.76, 310–318. 10.1016/j.jes.2018.05.017
326
WangQ.LuY.XinY.WeiL.HuangS.XuJ. (2016). Genome editing of model oleaginous microalgae Nannochloropsis spp. by CRISPR/Cas9. Plant J.88, 1071–1081. 10.1111/tpj.13307
327
WangS.SirbuD.ThomsenL.KuhnertN.UllrichM. S.ThomsenC. (2019b). Comparative lipidomic studies of Scenedesmus sp. (Chlorophyceae) and Cylindrotheca closterium (Bacillariophyceae) reveal their differences in lipid production under nitrogen starvation. J. Phycol.55, 1246–1257. 10.1111/jpy.12887
328
WangS.-B.HuQ.SommerfeldM.ChenF. (2004). Cell wall proteomics of the green alga Haematococcus pluvialis (Chlorophyceae). Proteomics4, 692–708. 10.1002/pmic.200300634
329
WangX.WeiH.MaoX.LiuJ. (2019c). Proteomics analysis of lipid droplets from the oleaginous alga Chromochloris zofingiensis reveals novel proteins for lipid metabolism. Genomics. Proteom. Bioinform.17, 260–272. 10.1016/j.gpb.2019.01.003
330
WangY.CongY.WangY.GuoZ.YueJ.XingZ.et al. (2019d). Identification of early salinity stress-responsive proteins in Dunaliella salina by isobaric tags for relative and absolute quantitation (iTRAQ)-based quantitative proteomic analysis. Int. J. Mol. Sci.20:30599. 10.3390/ijms20030599
331
WangY.StessmanD. J.SpaldingM. H. (2015). The CO2 concentrating mechanism and photosynthetic carbon assimilation in limiting CO2: how Chlamydomonas works against the gradient. Plant J.82, 429–448. 10.1111/tpj.12829
332
WangZ.LuoF.WangZ.ZhouR.TangY.LiY. (2019e). The potential growth and lipid accumulation in Coccomyxa subellipsoidea triggered by glucose combining with sodium acetate. World J. Microbiol. Biotechnol.35:110. 10.1007/s11274-019-2682-1
333
WaseN.BlackP. N.StanleyB. A.DiRussoC. C. (2014). Integrated quantitative analysis of nitrogen stress response in Chlamydomonas reinhardtii using metabolite and protein profiling. J. Proteome Res.13, 1373–1396. 10.1021/pr400952z
334
WatanabeY.KadonoT.KiraN.SuzukiK.IwataO.OhnishiK.et al. (2018). Development of endogenous promoters that drive high-level expression of introduced genes in the model diatom Phaeodactylum tricornutum. Mar. Genomics42, 41–48. 10.1016/j.margen.2018.06.003
335
WeckerM. S. A.GhirardiM. L. (2014). High-throughput biosensor discriminates between different algal H2 -photoproducing strains. Biotechnol. Bioeng.111, 1332–1340. 10.1002/bit.25206
336
WegnerA.MeiserJ.WeindlD.HillerK. (2015). How metabolites modulate metabolic flux. Curr. Opin. Biotechnol.34, 16–22. 10.1016/j.copbio.2014.11.008
337
WeiL.El HajjamiM.ShenC.YouW.LuY.LiJ.et al. (2019). Transcriptomic and proteomic responses to very low CO2 suggest multiple carbon concentrating mechanisms in Nannochloropsis oceanica. Biotechnol. Biofuels12:168. 10.1186/s13068-019-1506-8
338
WeiL.WangQ.XinY.LuY.XuJ. (2017a). Enhancing photosynthetic biomass productivity of industrial oleaginous microalgae by overexpression of RuBisCO activase. Algal Res.27, 366–375. 10.1016/j.algal.2017.07.023
339
WeiS.BianY.ZhaoQ.ChenS.MaoJ.SongC.et al. (2017b). Salinity-induced palmella formation mechanism in halotolerant algae Dunaliella salina revealed by quantitative proteomics and phosphoproteomics. Front. Plant Sci.8:810. 10.3389/fpls.2017.00810
340
WeymanP. D.BeeriK.LefebvreS. C.RiveraJ.MccarthyJ. K.HeubergerA. L.et al. (2015). Inactivation of Phaeodactylum tricornutum urease gene using transcription activator-like effector nuclease-based targeted mutagenesis. Plant Biotechnol. J.13, 460–470. 10.1111/pbi.12254
341
WichmannJ.BaierT.WentnagelE.LauersenK. J.KruseO. (2018). Tailored carbon partitioning for phototrophic production of (E)-alpha-bisabolene from the green microalga Chlamydomonas reinhardtii. Metab. Eng.45, 211–222. 10.1016/j.ymben.2017.12.010
342
WördenweberR.RokittaS. D.HeidenreichE.CoronaK.KirschhöferF.FahlK.et al. (2018). Phosphorus and nitrogen starvation reveal life-cycle specific responses in the metabolome of Emiliania huxleyi (Haptophyta). Limnol. Oceanogr.63, 203–226. 10.1002/lno.10624
343
WuC.XiongW.DaiJ.WuQ. (2015). Genome-based metabolic mapping and 13C flux analysis reveal systematic properties of an oleaginous microalga Chlorella protothecoides. Plant Physiol.167, 586–599. 10.1104/pp.114.250688
344
XieW.-H.ZhuC.-C.ZhangN.-S.LiD.-W.YangW.-D.LiuJ.-S.et al. (2014). Construction of novel chloroplast expression vector and development of an efficient transformation system for the diatom Phaeodactylum tricornutum. Mar. Biotechnol.16, 538–546. 10.1007/s10126-014-9570-3
345
XingG.YuanH.YangJ.LiJ.GaoQ.LiW.et al. (2018). Integrated analyses of transcriptome, proteome and fatty acid profilings of the oleaginous microalga Auxenochlorella protothecoides UTEX 2341 reveal differential reprogramming of fatty acid metabolism in response to low and high temperatures. Algal Res.33, 16–27. 10.1016/j.algal.2018.04.028
346
YadavR. R.KrishnamurthiK.ShekhA. Y.MudliarS. N.DeviS. S.ChakrabartiT. (2014). Activity enhancement of carbonic anhydrase in Chlamydomonas sp. for effective CO2 sequestration. Clean Technol. Environ. Policy16, 1827–1833. 10.1007/s10098-014-0734-7
347
YamanoT.SatoE.IguchiH.FukudaY.FukuzawaH. (2015). Characterization of cooperative bicarbonate uptake into chloroplast stroma in the green alga Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. U. S. A.112, 7315–7320. 10.1073/pnas.1501659112
348
YangB.LiuJ.MaX.GuoB.LiuB.WuT.et al. (2017). Genetic engineering of the Calvin cycle toward enhanced photosynthetic CO2 fixation in microalgae. Biotechnol. Biofuels10:229. 10.1186/s13068-017-0916-8
349
YangC.HuaQ.ShimizuK. (2000). Energetics and carbon metabolism during growth of microalgal cells under photoautotrophic, mixotrophic and cyclic light-autotrophic/dark-heterotrophic conditions. Biochem. Eng. J.6, 87–102. 10.1016/S1369-703X(00)00080-2
350
YangZ.-K.MaY.-H.ZhengJ.-W.YangW.-D.LiuJ.-S.LiH.-Y. (2014). Proteomics to reveal metabolic network shifts towards lipid accumulation following nitrogen deprivation in the diatom Phaeodactylum tricornutum. J. Appl. Phycol.26, 73–82. 10.1007/s10811-013-0050-3
351
YiA. X.LeungP. T. Y.LeungK. M. Y. (2014). Photosynthetic and molecular responses of the marine diatom Thalassiosira pseudonana to triphenyltin exposure. Aquat. Toxicol.154, 48–57. 10.1016/j.aquatox.2014.05.004
352
YiZ.SuY.XuM.BergmannA.IngthorssonS.RolfssonO.et al. (2018). Chemical mutagenesis and fluorescence-based high-throughput screening for enhanced accumulation of carotenoids in a model marine diatom Phaeodactylum tricornutum. Mar. Drugs16:80272. 10.3390/md16080272
353
YonedaK.YoshidaM.SuzukiI.WatanabeM. M. (2016). Identification of a major lipid droplet protein in a marine diatom Phaeodactylum tricornutum. Plant Cell Physiol.57, 397–406. 10.1093/pcp/pcv204
354
YoshimitsuY.AbeJ.HarayamaS. (2018). Cas9-guide RNA ribonucleoprotein-induced genome editing in the industrial green alga Coccomyxa sp. strain KJ. Biotechnol. Biofuels11:326. 10.1186/s13068-018-1327-1
355
YoshitomiT.KaminagaS.SatoN.ToyoshimaM.MoriyamaT.YoshimotoK. (2019). Formation of spherical palmelloid colony with enhanced lipid accumulation by gel encapsulation of Chlamydomonas debaryana NIES-2212. Plant Cell Physiol.61, 158–168. 10.1093/pcp/pcz188
356
YouZ.ZhangQ.PengZ.MiaoX. (2019). Lipid droplets mediate salt stress tolerance in Parachlorella kessleri. Plant Physiol.181, 510–526. 10.1104/pp.19.00666
357
YuanG.XuX.ZhangW.ZhangW.CuiY.QinS.et al. (2019). Biolistic transformation of Haematococcus pluvialis with constructs based on the flanking sequences of its endogenous alpha tubulin gene. Front. Microbiol.10:1749. 10.3389/fmicb.2019.01749
358
ZhangL.GoswamiN.XieJ.ZhangB.HeY. (2017). Unraveling the molecular mechanism of photosynthetic toxicity of highly fluorescent silver nanoclusters to Scenedesmus obliquus. Sci. Rep.7:16432. 10.1038/s41598-017-16634-5
359
ZhangQ.YouZ.MiaoX. (2018). Variation of fatty acid desaturation in response to different nitrate levels in Auxenochlorella pyrenoidosa. R. Soc. Open Sci.5:181236. 10.1098/rsos.181236
360
ZhangY.ShiM.MaoX.KouY.LiuJ. (2019). Time-resolved carotenoid profiling and transcriptomic analysis reveal mechanism of carotenogenesis for astaxanthin synthesis in the oleaginous green alga Chromochloris zofingiensis. Biotechnol. Biofuels12:287. 10.1186/s13068-019-1626-1
361
ZhaoY.HouY.LiuZ.ChenS.ChenF. (2016). Identification of NaHCO3 stress responsive proteins in Dunaliella salina HTBS using iTRAQ-based analysis. J. Proteomics Bioinform.9, 137–143. 10.4172/jpb.1000399
362
ZhengH.ChienC.HsuB. J.LiuT.ChenC. N.ChangW. (2014). AlgaePath : comprehensive analysis of metabolic pathways using transcript abundance data from next-generation sequencing in green algae. BMC Genomics15, 1–12. 10.1186/1471-2164-15-196
363
ZouL.-G.ChenJ.-W.ZhengD.-L.BalamuruganS.LiD.-W.YangW.-D.et al. (2018). High-efficiency promoter-driven coordinated regulation of multiple metabolic nodes elevates lipid accumulation in the model microalga Phaeodactylum tricornutum. Microb. Cell Fact.17:54. 10.1186/s12934-018-0906-y
364
ZuñigaC.LeveringJ.AntoniewiczM. R.GuarnieriM. T.BetenbaughM. J.ZenglerK. (2018). Predicting dynamic metabolic demands in the photosynthetic eukaryote Chlorella vulgaris. Plant Physiol.176, 450–462. 10.1104/pp.17.00605
Summary
Keywords
microalgae, genetic engineering, omics, genome editing, regulatory issues
Citation
Kumar G, Shekh A, Jakhu S, Sharma Y, Kapoor R and Sharma TR (2020) Bioengineering of Microalgae: Recent Advances, Perspectives, and Regulatory Challenges for Industrial Application. Front. Bioeng. Biotechnol. 8:914. doi: 10.3389/fbioe.2020.00914
Received
08 April 2020
Accepted
15 July 2020
Published
03 September 2020
Volume
8 - 2020
Edited by
Xiaochao Xiong, Washington State University, United States
Reviewed by
Na Pang, Michigan State University, United States; Xinqing Zhao, Shanghai Jiao Tong University, China
Updates
Copyright
© 2020 Kumar, Shekh, Jakhu, Sharma, Kapoor and Sharma.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gulshan Kumar gulshan.ihbt@gmail.comAjam Shekh azamsheikh24@gmail.comTilak Raj Sharma trsharma1965@gmail.com
This article was submitted to Industrial Biotechnology, a section of the journal Frontiers in Bioengineering and Biotechnology
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.