- 1Department of Microbiology, School of Natural Sciences and Ryan Institute, National University of Ireland Galway, Galway, Ireland
- 2Department of Biochemistry and Organic Chemistry, Institute of Chemistry, São Paulo State University, Araraquara, Brazil
- 3Department of Civil Engineering, School of Engineering, College of Science and Engineering, National University of Ireland Galway, Galway, Ireland
This research assessed the microbiological suitability of oleate degradation coupled to sulfidogenesis by enriching communities from anaerobic sludge treating dairy products with S0, , , and S2 as electron acceptors. The limiting factor hampering highly efficient oleate degradation was investigated in batch reactors. The best sulfidogenic performance coupled to specialization of the enriched bacterial community was obtained for S0- and S2-reducing enrichments, with 15.6 (± 0.2) and 9.0 (± 0.0) mM of sulfide production, respectively. Microbial community analyses revealed predominance of Enterobacteraceae (50.6 ± 5.7%), Sulfurospirillum (23.1 ± 0.1%), Bacteroides (7.5 ± 1.5%) and Seleniivibrio (6.9 ± 1.1%) in S0-reducing cultures. In S2-reducing enrichments, the genus Desulfurella predominated (49.2 ± 1.2%), followed by the Enterobacterales order (20.9 ± 2.3%). S0-reducing cultures were not affected by oleate concentrations up to 5 mM, while S2-reducing cultures could degrade oleate in concentrations up to 10 mM, with no significant impact on sulfidogenesis. In sequencing batch reactors operated with sulfide stripping, the S0-reducing enrichment produced 145.8 mM sulfide, precipitating Zn as ZnS in a separate tank. The S2 fed bioreactor only produced 23.4 mM of sulfide precipitated as ZnS. The lower sulfide production likely happened due to sulfite toxicity, an intermediate of thiosulfate reduction. Therefore, elemental sulfur reduction represents an excellent alternative to the currently adopted approaches for LCFA degradation. To the best of our knowledge, this is the first report of oleate degradation with the flux of electrons totally diverted toward sulfide production for metal precipitation, showing great efficiency of LCFA degradation coupled to high levels of metals precipitated as metal sulfide.
Introduction
Anaerobic reactors have been widely used for the treatment of lipid-rich wastewater, and, due to the high energy content of lipids, they are generally coupled to the production of biogas (Alves et al., 2009; Dasa et al., 2016). However, when no suitable feeding strategy is adopted and the accumulation of long-chain fatty acids (LCFAs) is not controlled, the efficiency of lipid-fed anaerobic digesters has been shown unpredictable (Hawkes et al., 1995; Hwu et al., 1998; Alves et al., 2009).
The release of LCFAs from fat, oil and grease (FOG)-rich wastewaters occurs through the activity of extracellular lipases that hydrolyze lipids into LCFA (Palatsi et al., 2009). Oleate (C18:1), stearate (C18:0), and palmitate (C16:0) are LCFAs commonly present in anaerobic digesters fed with lipids, from which oleate shows the highest abundance in wastewater systems (Komatsu et al., 1991), reasonably good solubility and high toxicity to anaerobic digestion communities (Lalman and Bagley, 2002; Zhang et al., 2011). Proton-reducing acetogenic bacteria present in the system degrade such fatty acids by performing the cyclic β-oxidation pathway, in which a recurrent cleavage of 2-carbon fragments occurs with concomitant release of acetyl-CoA (Nelson and Cox, 2005).
For biogas production, a syntrophic association of LCFA-degrading bacteria with hydrogen-utilizing microorganisms is required to guarantee a low partial pressure of hydrogen in the system (Sousa et al., 2007). Furthermore, when major products of β-oxidation accumulate to thermodynamically-limiting levels, further oxidation of LCFA and propionate is hampered, inhibiting the digestion to proceed (Labatut et al., 2014). In this scenario, the presence of LCFA has been reported to impact the activity of hydrolytic, acidogenic, acetogenic bacteria, and methanogenic archaea (Lalman and Bagley, 2002; Pereira et al., 2005). The archaeal community, however, is more resilient to increased LCFA concentrations compared to the bacterial community (Lalman and Bagley, 2002; Zhang et al., 2011; Ma et al., 2015), with partial inhibition of archaeal activity due to a reversible mineralization of LCFA (Pereira et al., 2005). Despite the reversibility of inhibition, the biogas production from the degradation of lipidic matter has yet several trammels on its way.
The difficulties related to LCFA-rich wastewaters for methane production raise the question whether other bioprocesses could be more suitable for handling this type of effluent. Sulfur cycle microorganisms, for example, are possible syntrophic partners, as they can generally use hydrogen as electron donor (Sánchez-Andrea et al., 2014; Florentino et al., 2016b). Besides, several microorganisms able to reduce sulfurous compounds are also able to directly oxidize LCFAs, without the requirement of syntrophic relationships for the complete degradation of these substrates (Bonch-Osmolovskaya et al., 1990; Miroshnichenko et al., 1999; Florentino et al., 2016a). In this case, the final product is hydrogen sulfide. The versatility of sulfidogenic microorganisms allows for many combinations of electron donors and sulfur sources, and for a wide range of operational conditions for the process.
The theoretical high levels of sulfide produced from the degradation of LCFAs can be utilized for the precipitation of heavy metals in solution, which is of great relevance from the biotechnological point of view, considering that the release of heavy metals in the environment is a major environmental problem (Akcil and Koldas, 2006; Moodley et al., 2017). Once sulfide is released, it can bind to divalent heavy metals in solution, such as Cu2+, Zn2+, and Ni2+, precipitating as insoluble metal sulfides, consequently generating a metal-free effluent. Therefore, it represents not only a strategy to alleviate metal-related pollution, but it also avails metal recycling (Johnson, 2014; Işildar et al., 2019).
In this study, we aimed to assess the microbiological suitability of LCFA degradation coupled to sulfidogenic bioconversions by enriching communities from anaerobic sludge treating dairy products. The performance of the enrichments was analyzed using different electron acceptors, and their tolerance to increasing concentrations of LCFA was studied. The limiting factors hampering efficient LCFA degradation were assessed. To the best of our knowledge, this is the first report of LCFA degradation with the flow of electrons totally diverted toward sulfide production for metal precipitation, with great efficiency of degradation yielding high levels of metal sulfides.
Materials and Methods
Inoculum Source, Media, and Screening Setup
Anaerobic sludge from a dairy industry in Mitchelstown (Co. Cork, Ireland) was used as source of inoculum. An aliquot (1 mL) of the homogenized sludge was added to 120 mL serum bottles with 50 mL sterile anoxic basal medium, prepared as described elsewhere (Stams et al., 1993). Briefly, the composition of the medium (g L−1) was: 0.41 KH2PO4; 0.53 Na2HPO4·2H2O; 0.3 NH4Cl; 0.3 NaCl; 0.1 MgCl2·6H2O; 0.11 CaCl2·2H2O; and 1 mL L−1 of acid and alkaline trace elements solution; 0.2 mL L−1 vitamins; 0.1 g L−1 yeast extract (Alfa Aesar, Ward Hill, MA) and 1 mL L−1 resazurin sodium salt solution (Fischer Scientific, Hampton, NH). The initial pH of the medium was set to 6.8 by addition of HCl or NaOH. Bicarbonate-buffer was not added to the medium, in order to monitor the pH values over time and to understand the impact of the LCFA degradation on the pH in the enrichments. Serum bottles were sealed with butyl rubber stoppers (Ochs Laborbedarf, Bovenden, Germany) and flushed with a 1.5 atm N2 headspace. Enrichments were incubated statically in the dark at 30°C.
Supplementary Figure 1 shows the experimental procedure flow chart. Four sets of transfers were performed to assess the oleate degradation coupled to sulfidogenic activity with elemental sulfur (S0), sulfite (), sulfate (), and thiosulfate (S2) as electron acceptors. The mentioned compounds were added to the enrichments to a final concentration of 25 mM. Sodium oleate 82% (Sigma-Aldrich, St. Louis, MI) was added as electron donor and carbon source from sterile anaerobic stock solutions to a final concentration of 1 mM. Two series of controls were performed without any external electron donors or electron acceptors. Activity was monitored every 4 days by sulfide production, volatile fatty acids and hydrogen profile, and pH dynamics. Screening cultivation experiments were performed in triplicates.
Four sets of serial dilutions were performed (Supplementary Figure 1). The serial dilutions were performed to eliminate the influence of organic matter present in the sludge, which would play the role of electron donor instead of the added LCFA. After the 4th transfer, as the endogenous organic matter was completely depleted in the cultures (no peaks or organic acids or sugars chromatographically detected), the sulfide production could thus be correlated solely to LCFA degradation. Results in this manuscript report the data of the 4th transfer only.
Sequencing Batch Reactors
Screening enrichments presenting the best LCFA-degrading performance and with the highest selective sulfur-metabolizing microbial community after the 4th transfer were selected for further analyses in a sequencing batch reactor (SBR). Selected enrichments were used as inoculum source (10%, v/v) in 1020 mL glass-made SBR, with 500 mL working-volume and 520 mL headspace. Bioreactors were flushed with N2 for 15 min and the final pressure was set at 1.5 atm N2 to ensure an O2-free headspace. The SBR were maintained at 30°C with the aid of thermostatic recirculation bath and submitted to an intermittent upflow recirculation flow rate of 46.5 mL min−1, and upflow velocity of 0.05 cm min−1 for 15 min every 6 h. The SBR were supplied with elemental sulfur (in excess) and thiosulfate (50 mM) as electron acceptors, and 2 mM sodium oleate 82% (Sigma-Aldrich, St. Louis, MI) as electron donor and carbon source. Activity of the biomass was monitored every 4 days by sulfide production, volatile fatty acids (VFAs) and pH dynamics.
A batch cycle consisted of 35 days for the thiosulfate-reducing reactor and 39 days for the sulfur-reducing reactor. At the end of each cycle, 20% (v/v) of the medium fed with substrates were replenished to avoid limitation of substrates or other nutrients. To prevent massive biomass loss, medium replenishment was always performed 5.5 h after the last recirculation cycle.
To release the potential negative impact of sulfide accumulation in the system, hydrogen sulfide was stripped every 9 and 17 days for, respectively, thiosulfate- and elemental sulfur-fed SBR. Hydrogen sulfide stripping was performed by flushing the SBR with N2 for 5 min (200 mL N2 min−1). N2 was flushed through a porous stone installed at the bottom of the SBR to ensure smooth mass transfer. At the end of the flushing procedure, the reactor headspace was put under 1.5 atm pressure. The hydrogen sulfide stripped from the bioreactors can be used for the precipitation of Zn2+ as ZnS. To assess metal precipitation with sulfide generated by the SBR, the gas stripped from the reactor was directed to a vessel containing 100 mL of a 5% ZnCl2 solution (w/v). The ZnS precipitates were left settling overnight. Zinc and sulfide concentrations in the supernatant and in the precipitates were then determined.
Sacrificial Assays
To assess the profile of oleate degradation in S0 and S2 -fed biomass, 56 serum bottles (30 mL) were filled with the above-described anoxic basal medium and inoculated with the enriched biomass (10%, v/v) present in the reactors sampled on the 50th day of operation. Triplicate bottles were sacrificed twice a week and the whole content of the bottles was used for pH, sulfide, VFA, and LCFA measurements.
Oleate Tolerance Tests
To determine the inhibitory oleate concentration to the enriched microorganisms, elemental sulfur- and thiosulfate-reducing enrichments (using the 4th transfer biomass inoculated to the bioreactors) were incubated with sodium oleate 82% (Sigma-Aldrich, St. Louis, MI) in a concentration range from 1 to 10 mM, exceeding the reported inhibitory concentrations for anaerobic microorganisms (Hwu and Lettinga, 1997; Sousa et al., 2013; Silva et al., 2016). Experiments were conducted in triplicates and the bottles were incubated statically at 30°C. Activity was monitored every 4 days by sulfide production and pH dynamics.
Sulfite Tolerance Tests
To investigate the impact of sulfite on the thiosulfate-reducing SBR biomass, incubations with initial concentrations of sodium sulfite of 2.5, 5, 7.5, 10, and 25 mM were performed with 1 mM sodium oleate 82% (Sigma-Aldrich, St. Louis, MI) added as electron donor and carbon source. A bioreactor sample from the 58th day of operation was used as inoculum. Experiments were conducted in triplicates and cultures were incubated statically at 30°C. Activity was monitored every 4 days by sulfide production and pH dynamics.
Transmission Electron Microscopy (TEM)
Reactors were sampled on day 50th for TEM analysis. Fixation and imaging of cells were performed at the Center for Microscopy and Imaging at the National University of Ireland Galway. To cross-link cellular components and to preserve and stabilize cellular structures, samples were fixed in 2% (v/v) glutaraldehyde and 2% (w/v) paraformaldehyde in 0.1 M sodium cacodylate buffer/HCl pH 7.2 for 2 h, followed by fixation in 1% (w/v) osmium tetroxide in a 0.1 M cacodylate buffer/HCl (pH 7.2) for 2 h. Osmium-fixed samples were dehydrated in graded alcohol solutions, and pure ethanol was washed out by acetone replacement. Dehydrated samples were embedded in Agar Low Viscosity Resin (Agar Scientific, Stansted, UK). Complete polymerization of the resin occurred in an oven at 65°C for 48 h. Survey sections of 500 nm were cut using a glass knife and transferred to a glass slide, stained with toluidine blue and viewed using a light microscope. When a region of interest was identified, ultra-thin sections (70–90 nm) were cut with a diamond knife, filled on copper grids, stained with uranyl acetate and lead citrate to enhance organic matter contrast on a Leica EM AC20 automatic contrasting system (Leica, Wetzlar, DE), and examined in a Hitachi H7000 transmission electron microscope (Hitachi, Tokyo, JA).
DNA Extraction
Cultures enriched in the screening were considered for microbial community analyses. Cells were harvested at the early stationary phase by centrifuging samples at 10,000 × g for 5 min. Triplicate harvested cells were used for DNA extraction with the DNeasy PowerSoil Kit (QIAGEN, Hilden, Germany), following the instructions of the manufacturer. Purity and concentration of the extracted DNA were checked in a NanoDrop 2000C (ThermoFisher, Scientific Walthan, MA). DNA concentrations were about 25 ng μL−1, with 260/280 ratios of ~1.8. Extracted DNA samples were stored at −20°C until sequencing. No-template-controls and extraction blanks were performed and resulted in no amplification when universal primers for bacteria and archaea were employed.
Sequencing and Microbial Community Analysis
Frozen DNA samples were dispatched to Eurofins Genomics (Germany), where PCR was performed followed by sequencing on the Illumina MiSeq platform. Bacterial 16S rDNA genes were amplified using bacterial universal primers sequencing the hypervariable regions V3-V4 of the gene. Once the amplicons were obtained, all the reads with ambiguous bases (“N”) were removed, and chimeric reads were identified and removed following the UCHIME algorithm (Edgar et al., 2011) as implemented in the VSEARCH package for sequence analysis (Rognes et al., 2016). The remaining set of high-quality reads, the operational taxonomic unit (OTU)-picking strategy, was processed using minimum entropy decomposition—MED (Eren et al., 2013, 2015). The MED procedure identified and filtered sequences with a very low abundance (< ≈ 0.02% of the average sample size). DC-MEGABLAST alignments of cluster representative sequences to the sequence database (NCBI_nt—release 2019-01-05) were performed to assign taxonomic information to each OTU. The most specific taxonomic assignment for each OTU was then transferred from the set of best-matching reference sequences. The minimum requirement for a reference sequence was an identity of 70% across at least 80% of the representative sequence. Further processing of OTUs and taxonomic assignments was performed using the QIIME software package (version 1.9.1, http://qiime.org/). Abundances of bacterial taxonomic units were normalized using lineage-specific copy numbers of the relevant marker genes to improve estimates (Angly et al., 2014). The processed Illumina Miseq reads were deposited in the Sequence Read Archive of NCBI under accession number PRJNA622591.
LCFA Extraction
Extraction and transmethylation of LCFAs were performed as previously described by Guihéneuf et al. (2014). Briefly, freeze-dried samples - about 10 mg dry weight (Lyovapor L-200, Buchi Labortechnik AG, Flawil, CH) received 2 mL of 2% H2SO4 in dry methanol in 15 mL glass vials with teflon sealed caps. 10 μL of the internal standard, pentadecanoic acid (5 mg L−1), were added to the solution under N2 flow, and heated to 80°C for 1.5 hour with continuous stirring. When at room temperature, 1 mL of H2O was added to terminate the transmethylation reaction, followed by the addition of 1 mL hexane to extract the fatty acid methyl esters (FAMEs), concentrated in the upper layer.
Analytical Methods
The partial pressures of hydrogen and methane were measured in an Agilent 7890A gas chromatograph (Agilent Technologies, Santa Clara, CA), equipped with a 13803-U stainless-steel column (Sigma-Aldrich, St. Louis, MI) and a thermal conductivity detector. Temperatures of the oven and detector were set to 90 and 200°C, respectively. Argon was used as carrier gas at a flow rate of 24 mL min−1. VFAs were quantified using a 1260 Infinity II Liquid Chromatograph system (Agilent Technologies, Santa Clara, CA), equipped with a Hi-plex H column, a refractive index detector, and 14 mM H2SO4 eluent at a flow rate of 0.4 mL min−1.
Sulfide concentration in solution was determined by the photometric method using methylene blue as described previously by Cline (1969), after preservation of the sample by addition of 5% ZnCl2 solution (w/v). Sulfate, sulfite and thiosulfate concentrations were quantified using a Dionex Aquion ion chromatograph equipped with an IonPac AS14A 4 × 250 mm column, an AG14A 4 × 50 mm guard column, and a suppressed conductivity detector (ThermoFisher Scientific, Waltham, MA). The eluent was prepared as a mixture of carbonate and bicarbonate solutions to a final ratio of 3.03 mM / 0.97 mM, at a flow rate of 1 mL min−1.
LCFA concentrations were quantified as FAMEs, using an Agilent 7890A GC equipped with a flame ionization detector and a fused silica capillary column DB-WAXETR (Agilent Technologies, Santa Clara, CA). The measuring conditions followed the protocol proposed by Guihéneuf et al. (2014). Zinc analyses were performed with an ICP-OES (ThermoFisher, Scientific Walthan, MA). The device was operated at RF power: 1,300 W, argon plasma flow rate: 8 L.min−1, auxiliary argon flow rate: 0.3 L min−1, nebulizer argon flow rate: 0.80 L min−1 and sample flow: 1.0 L min−1. Zinc was read on axial mode at 213.857 nm.
Results
Enrichments
Sulfidogenic Activity
The degradation of oleate by the enrichments (4th transfer) was accompanied by sulfide production with all the electron acceptors used (Figure 1A). Acetate (Figure 1B) and CO2 were the other products detected in the incubations. Traces of hydrogen (0-0.01 mM) were detected in the headspace of all the 4th transfer cultures. Methane was not detected in the headspace of any enrichment. The pH values in S0- and S2-reducing enrichments ranged from 5.5 to 7.0, while there was stability around pH 6–6.4 in -reducing enrichments, and a variation from pH 6–8 in -reducing cultures.
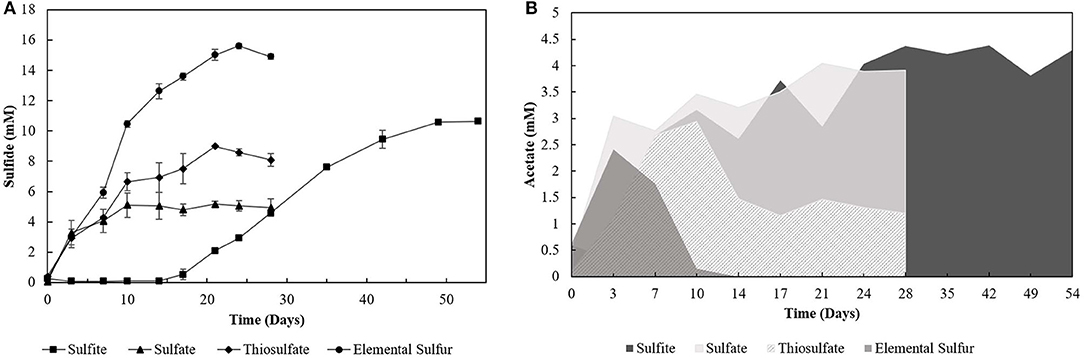
Figure 1. (A) Sulfide production and (B) acetate profile of the 4th transfer enrichments oxidizing oleate with sulfur compounds as electron acceptors. Measurements of biological triplicates were averaged, and the standard deviation is shown.
Incubations with S0 and yielded the highest production of sulfide, 15.60 (± 0.17) mM and 10.62 (± 0.12) mM, respectively. However, -reducing enrichments presented a lag phase of 14 days, and acetate accumulated at 4.29 (± 0.28) mM, while it got completely depleted within 14 days of incubation in the S0 -reducing enrichment. Enrichments incubated with as electron acceptor produced maximally 5.16 (± 0.19) mM sulfide, with an accumulation of 4.05 (± 0.14) mM of acetate. The concentration of sulfate decreased by 7.44 (± 1.60) mM. Enrichments growing on S2 produced up to 8.99 (± 0.02) mM of sulfide, and acetate accumulated at 1.20 (± 0.09) mM with a S2 consumption of 5.57 (± 1.16) mM. In control groups, sulfide production was detected in the first transfer (4.07 ± 0.18 mM) due to the endogenous electron donors present in the inoculum sludge. In further transfers, sulfide production in control groups was no longer detected.
Microbial Communities
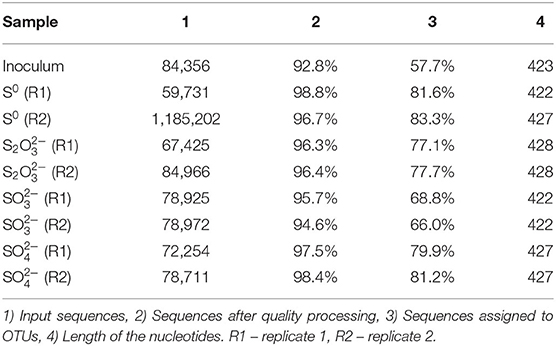
Extracted DNA from the enrichments showed amplification only with bacterial primers, indicating that archaeal communities did not develop in the cultures. Enriched cultures were analyzed in duplicate per electron acceptor used. Summarized statistics of the 16S rDNA gene amplicon analysis of the inoculum source and enriched cultures are given in Table 1. After quality processing, sequences from the inoculum assigned to OTUs represented 21 known classes, with 79 correlated genera. The three most representative classes were Deltaproteobacteria (35.7% of the sequences), Bacteroidia (32.2% of the sequences), and Spirochaetia (10.3% of the sequences) (Figure 2A). About 2.8% of the total number of sequences could not be identified at the class level and were classified at the phylum level.
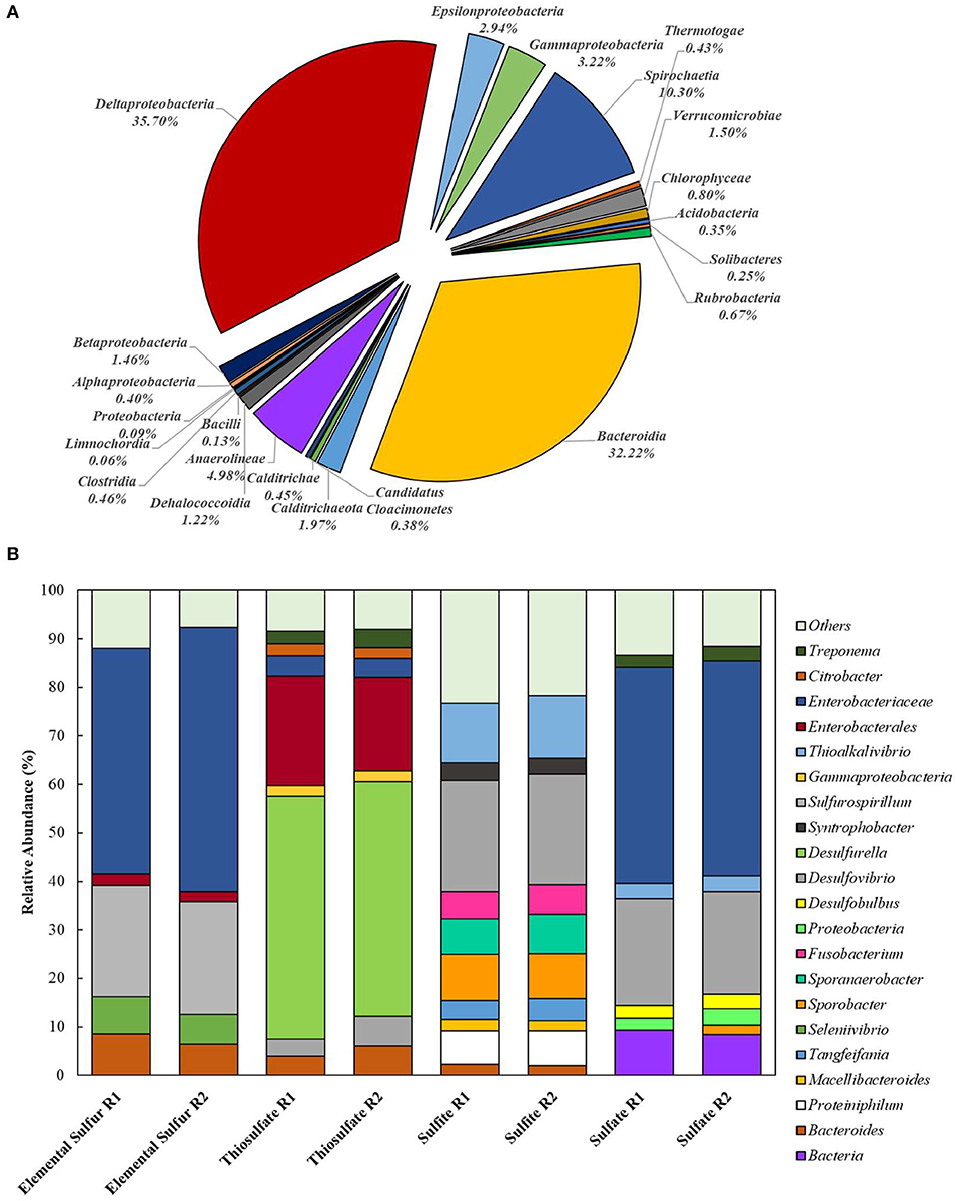
Figure 2. Bacterial diversity in the (A) inoculum (at the class level) and in the (B) 4th transfer enrichments on early stationary phase (at the genus level). Groups that could not be identified at the mentioned level were classified at the next highest possible resolution level. OTUs with less than 2% sequence reads were combined in the group “Others.” R1 – replicate 1; R2 – replicate 2.
In enriched cultures, sequences that could not be identified at the genus level were classified at the next highest possible resolution level. In S0-reducing cultures, there was a predominance of Enterobacteraceae (50.6 ± 5.7%), Sulfurospirillum (23.1 ± 0.1%), Bacteroides (7.4 ± 1.5%), and Seleniivibrio (6.9 ± 1.1%). In S2-reducing enrichments, the genus Desulfurella predominated in about 49.2 (± 1.2) % of the total number of sequences, followed by 20.9 (± 2.3)% of Enterobacterales. -reducing cultures presented the highest diversity, with 10 OTUs representing more than 2% of the total number of sequences. From these, 22.9 (± 0.1) % were represented by the genus Desulfvibrio, followed by 12.6 (± 0.5) % of Thioalkalivibrio. In cultures that received sulfate as electron acceptor, Enterobacteraceae dominated the samples with 44.4 (± 0.2) % of the sequences, followed by Desulfovibrio with 21.5 (± 0.6) % of the sequences.
Oleate Tolerance Tests
The two conditions with the best combined sulfidogenic performance, with the minimal acetate accumulation and with a more specialized microbial community (S0- and S2-reducing enrichments) were selected for further analyses. Increasing concentrations of oleate in the medium affected the sulfide production by S0-reducing cultures (Figure 3A). Concentrations of oleate of 1 and 2 mM did not impact the sulfide production by the enrichments. However, when it was added to a final concentration of 5 mM, the sulfidogenic activity of the cultures decreased by more than 50%, and triplicates receiving 7.5 mM of oleate did not show significant sulfide production when compared to the initial concentrations.
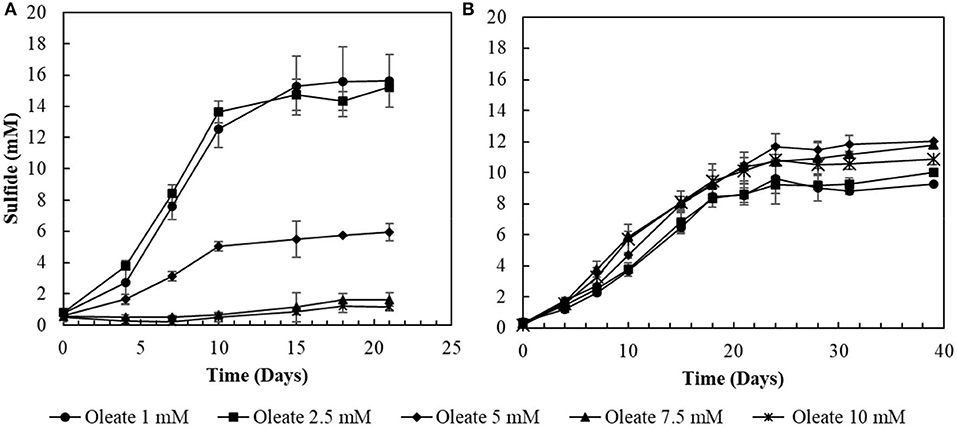
Figure 3. Toxicity profiles of oleate to (A) elemental sulfur- and (B) thiosulfate-reducing cultures. Measurements of biological triplicates were averaged, and the standard deviation is shown.
In S2-reducing cultures, the increase in oleate concentrations in the medium initially promoted an improvement in sulfide production, from 9.3 (± 0.2) mM in 1 mM oleate condition to 12.0 (± 0.6) mM in the 5 mM oleate condition. Although oleate concentrations exceeding 5 mM still promoted sulfide concentrations higher than at 1 mM, there was a decrease in comparison to the optimum obtained with 5 mM of sodium oleate (Figure 3B). In the 7.5 mM oleate culture, the production of sulfide reached 11.8 (± 0.2) mM, and, when oleate was supplied to a final concentration of 10 mM, the sulfide production decreased to 10.9 (± 0.3) mM (Figure 3B).
LCFA Degradation Profile
Sacrificial experiments were performed to investigate the degradation profile of oleate by the enrichments (4th transfer) in the presence of S0 and S2. The S0-reducing enrichments degraded 0.9 (± 0.1) mM of oleate in 22 days, with no accumulation of palmitate or any other LCFA analyzed (Figure 4A). The acetate concentration peaked at 2.0 (± 0.0) mM in 6 days of experiment but got completely depleted within 22 days. The sulfide production reached 15.8 (± 1.3) mM (Figure 5). In S2-reducing enrichments, 0.8 (± 0.2) mM of oleate was consumed in 14 days, while 0.5 (± 0.3) mM of palmitate was produced. The concentration of myristate did not significantly change along the sacrificial experiment (Figure 4B). Laureate was not detected at any point of the growth curve. In these S2 -fed enrichments, 10.0 (± 0.9) mM of sulfide was produced, while a peak of acetate was observed on day 6 (2.0 ± 0.0 mM) and decreased to 1.1 (± 0.1) mM at the end of the incubation (Figure 5).
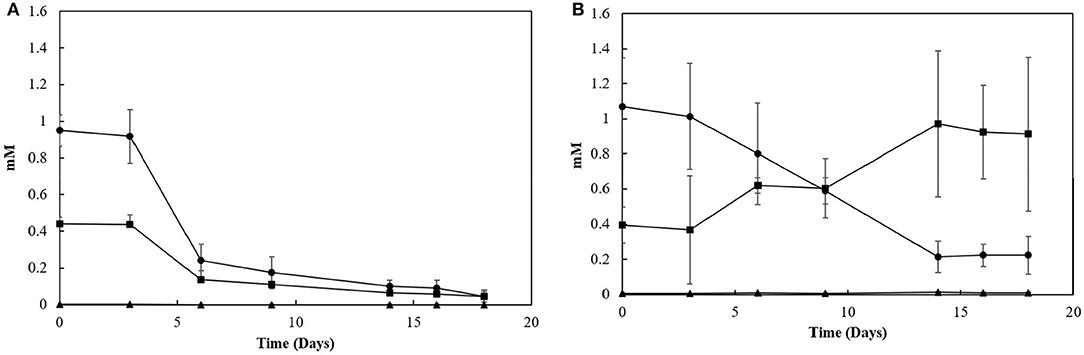
Figure 4. LCFA profiles in sacrificial experiments with cultures growing with (A) elemental sulfur or (B) thiosulfate as electron acceptors. Measurements of biological triplicates were averaged, and the standard deviation is shown. • oleate, ■ palmitate, ▴ myristate.
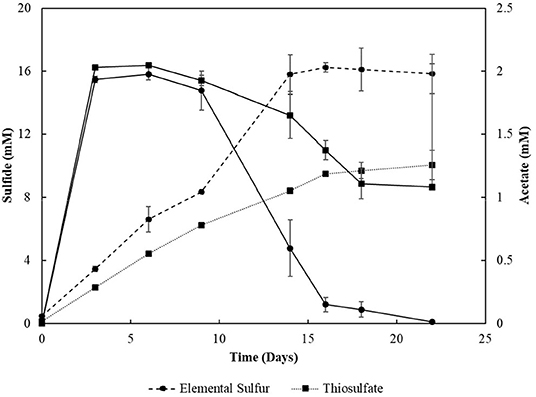
Figure 5. Sulfide production and acetate profile of the enrichments oxidizing oleate with elemental sulfur or thiosulfate as electron acceptors in sacrificial experiments. Dashed lines correlate to sulfide, and continuous lines correlate to acetate concentrations. Measurements of biological triplicates were averaged, and the standard deviation is shown.
Performance of Sequencing Batch Reactors
The elemental sulfur reactor was operated for 104 days (Figure 6A). Every ± 17 days, when sulfide reached the plateau phase (10.0 ± 0.6 mM), it was stripped from the system. Its production was subsequently restored, coupled to the degradation of oleate, replenished every about 39 days to a concentration of 2 mM. Acetate was transiently detected in the reactor mixed liquor and its concentration reached a maximum of 0.4 mM. The sulfide accumulated till day 39 was stripped and introduced to an acidic ZnCl2 solution, where about 73.4 mM of zinc precipitated as ZnS. The sulfide accumulated during the second cycle before replenishment of the medium (day 92) resulted in a total of 51.4 mM of zinc precipitated as ZnS.
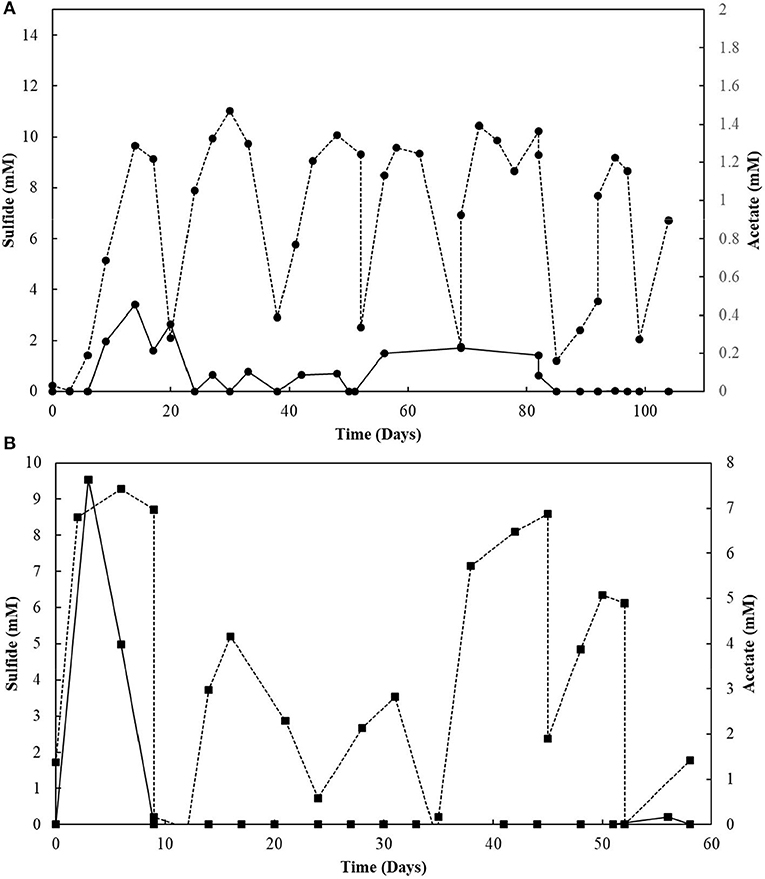
Figure 6. (A) Elemental sulfur and (B) Thiosulfate reactors performance operating in sequencing batch with sulfide stripping. Dashed lines correlate to sulfide, and continuous lines to acetate concentrations.
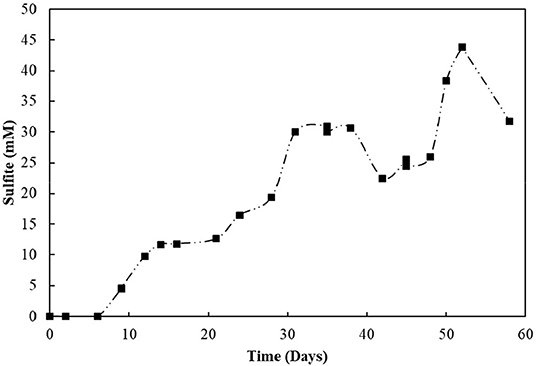
The S2-reducing bioreactor was operated for 58 days (Figure 6B). Sulfide production reached 9.3 mM in the first 6 days of reactor operation. However, after the first gas stripping, the sulfide production suffered a significant decrease, reaching maximally 5.2 mM in the next 30 days of operation, after which it increased up to 8.6 mM on day 45, and decreased again. The acetate concentration peaked at 7.6 mM within 3 days of operation, with full depletion in 9 days. After 51 days of operation, the acetate concentration slightly raised in the reactor. From the initial 50 mM of thiosulfate fed to the sequencing batch reactor, 17.2 mM was left after 58 days of operation, and the sulfite concentration of the reactor mixed liquor increased up to 43.8 mM within 52 days (Figure 7). The accumulated sulfide stripped precipitated a total of 23.4 mM of Zn as ZnS in the first cycle of sulfide stripping, while the cumulative sulfide in the second cycle precipitated as 49.4 mM ZnS.
When cross-sectional TEM analysis was performed on the reactors' biomass, a distinct pattern of interaction between S0/ S2 -fed bacterial cells and LCFA can be hypothesized. S0-reducing cells likely internalized the LCFA as particles, visible as the intracellular dark dots (Figure 8A), while S2-reducing microorganisms seemed to perform the degradation on the surface of their membranes (Figure 8B). Analysis of the microbial diversity shows, as for the enrichments, a greater diversity of OTUs in the reactor operating with elemental sulfur as electron acceptor (Figure 9). In the biomass present in this reactor, sequences belonging to the family Enterobacteraceae represented 26.5% of the diversity, followed by the genera Halothiobacillus, Sulfurospirillum, and Bacteroides, with 19.7, 13.3, and 11.5%, respectively. In the reactor fed with thiosulfate as electron acceptor, the bacterial diversity was greatly dominated by the genus Desulfurella, with 41.3% of the sequences. The order Enterobacterales and the genus Bacteroides comprised the second (17.9%) and third (11.6%) OTUs better represented in the thiosulfate-fed reactor.
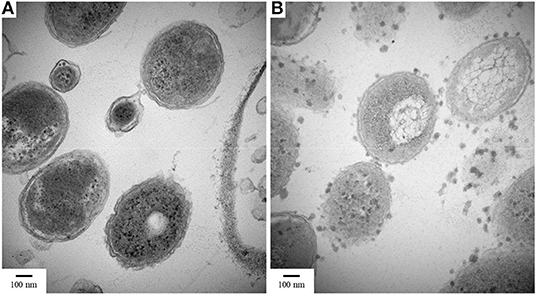
Figure 8. TEM images of cultures growing with oleate and (A) elemental sulfur or (B) thiosulfate. Samples were taken on day 50 of the reactors' operation. Magnification 80,000x.
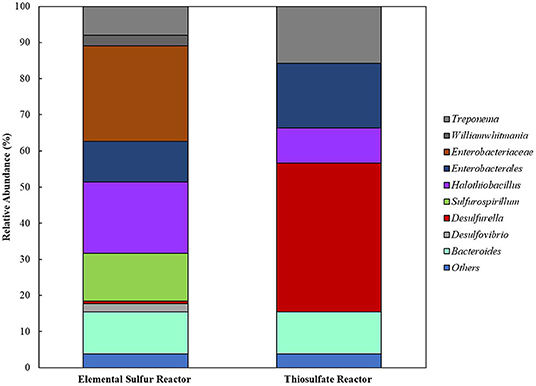
Figure 9. Bacterial diversity in the reactors oxidizing oleate with elemental sulfur or thiosulfate as electron acceptors. OTUs with <2% sequence reads were combined in the group “Others”.
Sulfite Toxicity
The decrease in sulfide production in the thiosulfate-reducing reactor motivated the investigation of sulfite toxicity to the biomass. When S2–SBR biomass was incubated with increasing sulfite concentrations, a great impact on the sulfide production was confirmed (Figure 10). When sulfite was supplied to the S2–SBR biomass in initial concentrations up to 7.5 mM, the production of sulfide was equivalent to the concentration of the electron acceptors. When 10 mM of sulfite was used, sulfide generation by the S2–SBR biomass drastically decreased, reaching maximally 1.5 (± 1.0) mM. At an initial concentration of 25 mM, sulfide was no longer produced by the S2–SBR biomass.
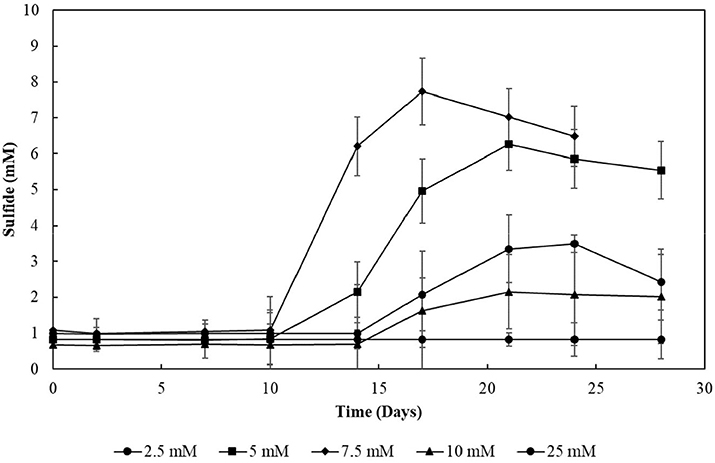
Figure 10. Evolution of sulfide production in cultures incubated with increasing sulfite concentrations. Measurements of biological triplicates were averaged, and the standard deviation is shown.
Discussion
Electron Acceptor Screening
This study showed that incubation of anaerobic sludge treating dairy wastewater with oleate and sulfur compounds diverted the fluxes of electrons from methanogenesis toward sulfide production, leading to different enrichments of sulfur compounds-metabolizing cultures. To the best of our knowledge, this is the first report in which different sulfur compounds were screened for oleate-degradation coupled to sulfidogenic capability.
Although not reaching the maximum amount of sulfide expected per mM of electron donor added, the sulfidogenic capacity of the enrichments and reactors' biomass followed the proposed stoichiometric reactions for each compound (Equations 1–4). The lag phase observed in the -reducing enrichments (Figure 1A) reflects the possible toxic effect of sulfite to the cells, in accordance with previously reported observations (Irwin et al., 2017). Together with the -reducing enrichments, they showed the highest acetate accumulation (Figure 1B), indicating an incomplete oxidation metabolism by the enriched microorganisms.
The limitation in sulfide production observed in all the enrichments might have provoked a slighter decrease in pH than expected by the above-shown equations, allowing the viability of the cells. The decrease in pH to a minimum of 5.0 is also an important feature of the enrichments, as this does not allow dissociation of acetate to acetic acid >50% (pKa = 4.75), which would acidify the medium and inhibit the microorganisms.
The addition of LCFA and sulfur compounds to anaerobic sludge was also described by Sharma and Biswas (2013), in which linoleic acid was used to inhibit methanogenic activity and enhance sulfate reduction in the sludge. Although the presence of sulfate in methanogenic environments thermodynamically favors the sulfidogenic reaction, in that study the authors observed that the diversion of electron fluxes (in more than 30%) was a function of the LCFA concentration. Salvador et al. (2019) investigated the degradation of oleate by an anaerobic culture acclimated to methane production. In the presence of a sulfonate with methanogenic-inhibiting activity (isethionate), sulfate-reducing bacteria capable of sulfonate metabolism got enriched and started a synergistic activity with oleate-degrading bacteria with conversion of the LCFA to acetate. In both studies, only sulfate was tested as electron acceptor for sulfide production.
The bacterial diversity obtained in the enrichments (Figure 2) reveals the great potential for sulfur metabolism in the anaerobic sludge treating dairy products. Besides, the metabolic flexibility of microorganisms able to reduce sulfur compounds is well-known (Plugge et al., 2011; Rabus et al., 2013; Florentino et al., 2016b). S2-reducing enrichments revealed the greatest bacterial specialization, with the predominance of members of the Desulfurella genus. This genus is known for its ability of reducing elemental sulfur as its major metabolism, coupled to the oxidation of simple and complex organic matter (Bonch-Osmolovskaya et al., 1990; Miroshnichenko et al., 1998; Florentino et al., 2016a). Although it has been reported that Desulfurella species can grow on LCFA, there are no reports of growth curves or physiological studies of this genus directly degrading LCFA for sulfide production. The second predominance was of the order Enterobacterales, which has not been reported as a group of sulfur metabolizers as major metabolism. This opens possibilities for isolation of novel microorganisms from this order with great ability for sulfide production coupled to degradation of FOG. A second possibility is, however, that members of the Enterobacterales played a role as LCFA-degrading groups (Odeyemi et al., 2013; Liu et al., 2018), not coupled to the reduction of thiosulfate, but generating acetate and hydrogen to be used as electron donors for Desulfurella members, as acetate is the best electron donor for these species (Schmitz et al., 1990; Florentino et al., 2016a).
The Enterobacteraceae family was predominant in S0-reducing enrichments, followed by the Desulfovibrio genus (Figure 2). The presence of Desulfovibrio species with elemental sulfur as electron acceptor opens possibilities for further studies, as there are no reports of extended sulfidogenesis from the reduction of sulfur coupled to the degradation of complex organic matter by this group. Escobar et al. (2007) showed the feasibility of elemental sulfur reduction by Desulfovibrio desulfuricans with hydrogen as electron donor and obtained maximal sulfide production rates of 2.1 g H2S L−1 d−1. However, Biebl and Pfennig (1977) tested D. desulfuricans and D. vulgaris and no growth on elemental sulfur was observed, but slow and definite growth was observed for D. gigas. As the Enterobacteraceae family was highly predominant, it is also reasonable to hypothesize that the LCFA degradation in those enrichments was not directly coupled to elemental sulfur reduction, but the acetate produced from the β-oxidation chain reaction was likely the actual electron donor for the Desulfovibrio species for the sulfur-reducing process.
Despite the great diversity observed in -reducing enrichments (Figure 2), members of the Thioalkalivibrio and Desulfovibrio genera showed relative predominance. The pronounced enrichment of Thioalkalivibrio members in the tested conditions is unexpected, as this haloalkaliphilic group, belonging to the order of Purple Sulfur Bacteria, is commonly reported as sulfide and/or elemental sulfur oxidizers (Sorokin and Kuenen, 2005; Ahn et al., 2017). Therefore, the enrichment of Thioalkalivibrio requires further investigation focused on the isolation and characterization of the potential novel species able to thrive under sulfidogenic conditions. As above-mentioned, it could be that the presence of Desulfovibrio was correlated to the release of acetate and hydrogen by the β-oxidation chain reaction performed by LCFA-degrading groups, such as Syntrophobacter, also identified in this enrichment (Figure 2). However, the accumulation of acetate in -reducing enrichment suggest that Desulfovibrio members might perform incomplete oxidation in this incubation, and therefore such members would not be able to use acetate as electron donor, only hydrogen. Further investigation is required to clarify the role played by the enriched groups in LCFA degradation.
Like in the S2-reducing enrichments, members of the family Enterobacteraceae represented the great majority of the identified groups in -reducing cultures, but, in this condition, it was followed by members of the Desulfovibrio genus. Sousa et al. (2009) also observed an enrichment of members of the genus Desulfovibrio in oleate-degrading cultures incubated with sulfate as electron acceptor. The latter authors associated the presence of the Desulfovibrio species to the release of acetate and hydrogen by the β-oxidation chain reaction performed by syntrophic bacterial groups. However, as mentioned for -reducing enrichments, acetate accumulation could be an indication of the incomplete oxidation metabolic route in the enriched Desulfovibrio species, with hydrogen as the only electron donor in this case.
LCFA Degradation in S0 vs. S2 -Fed SBRs
The sulfidogenic activity of the S0-reducing reactor biomass was affected by increasing oleate concentrations, with negative effects starting from 5 mM (Figure 3). In contrast, biomass from the thiosulfate-reducing reactors did not suffer a great impact from concentrations of oleate up to 10 mM. There are no reports about the effects of oleate on sulfidogenic pure cultures or bioreactor sludge. On methanogenic communities, however, the inhibitory effects of oleate already occur at concentrations ranging from 0.3 to 2.3 mM (Hwu and Lettinga, 1997; Sousa et al., 2013; Silva et al., 2016). The higher tolerance of the sulfidogenic biomass to oleate reveals a noticeably higher resilience of sulfidogenic communities than methanogenic communities. Such resilience makes sulfidogenic dominated biomass an approachable surrogate for methanogenic groups for the degradation of LCFA, if the sulfide produced can be properly managed, as [e.g., by using for metal precipitation (see below)].
Furthermore, although the reactors' biomass was able to oxidize oleate, its degradation profile in the reactors did not follow the same pattern (Figure 4). In the S0-reducing enrichment, the decrease in oleate concentration in the medium was not followed by a peak of palmitate, myristate or any other LCFA that can be detected by the analytical method applied. Although the electron donor was not fully consumed (residual oleate concentration after 18 days: 0.9 mM), the sulfide production (16.1 mM) did not correlate to the observed decrease in oleate concentration. However, the presence of acetate reinforces the degradation of oleate (and the residual palmitate) initially present in the medium. Therefore, it is likely that the insolubility of elemental sulfur might have interfered with the LCFA extraction method, hampering the detection of the intermediate products and consequently the oleate degradation profile.
In S2-reducing enrichments, however, the concentration of palmitate gradually increased, while no change in myristate or laureate concentration was observed (Figure 4B). Considering the amount of oleate and palmitate consumed, assuming that all the other long and short chain fatty acids were consumed and accounting the acetate left in the medium, about 3 mM of acetate was consumed by the enrichment, which would contribute to the production of ~6 mM of sulfide, less than the observed concentration (16.1 mM, Figure 5). The exceeding sulfide produced might have been produced by the hydrogen released by the β-oxidation process. However, no accurate calculation can be made.
Sulfidogenic enrichments performed on oleate did not show sulfide production any close to the stoichiometric reaction (Table 2). Two possible explanations for this observation are the maximal tolerance of the cultures to sulfide and/or the increasing concentration of palmitate in the medium as an intermediate product of oleate degradation. Although the literature reports negative effects of hydrogen sulfide on various microorganisms, including environmental bacteria (Beauchamp et al., 1984), there are only a few reports on the effects of this compound on its own producers. In general, the proposed sulfide concentration to inhibition of growth of sulfate-reducing bacteria is about 16 mM (Reis et al., 1992). Specific values for elemental sulfur- or thiosulfate-reducing cultures are not reported. As a second explanation, the accumulation of non-degraded LCFA in the cultures can also be a key factor for growth and activity limitation in methanogenic cultures (Pereira et al., 2005; Sharma et al., 2015). The adsorption of LCFA on the cell surface (Figure 8) is a possible explanation for physical inhibition of the bacterial community (Pereira et al., 2005). Besides, the differences in the composition of the cell wall might exert an influence on the sensitivity of microorganisms to the LCFA mixture present in the medium, which might play a selective role in the community (Silva et al., 2016).

Table 2. Expected and observed sulfide production from the oleate degradation coupled to elemental sulfur and thiosulfate reduction in the enriched bacterial communities during the screening experiment (4th transfer only).
Sulfide vs. Sulfite Toxicity
The removal of hydrogen sulfide from the bioreactors by gas stripping shifted the reaction toward more LCFA degradation and, consequently, more sulfide production (Figure 6A). Moreover, the total sulfide produced from 2 mM of oleate was close to that stoichiometrically expected for the conversion of oleate to palmitate in the β-oxidation chain reaction, which indicates the degradation of palmitate. Thus, our hypothesis of growth and activity inhibition due to elevated sulfide concentrations was reinforced.
This observation was less pronounced in the thiosulfate reactor (Figure 6B). Elevated initial concentrations of thiosulfate as electron acceptors for the biomass in the reactor promoted a high production of sulfite, as an intermediate product of thiosulfate reduction (Equation 5). Typically, the reductive process of thiosulfate is reported to happen in two steps. First, thiosulfate is converted to sulfide and sulfite. The sulfide formed leaves the cell, while the sulfite enters the cytoplasm and gets reduced to sulfide. Overall, one sulfide is generated during the first step, directly from thiosulfate molecule, and the other is generated in the second step, during sulfite reduction (Surkov et al., 2000; Stoffels et al., 2012; Florentino et al., 2019).
Sulfite was toxic to the enriched cultures at initial concentrations of 10 mM (Figure 10), which is in accordance with the reactor's performance: the sulfide production in this case decreased by about 80% (Figure 6B). The increased concentration of sulfite likely imposed inhibition to the S2-reducing cultures and a decreased sulfide production, motivating the sulfite toxicity experiments (discussed in previous section). It is worth noting that, in sulfidogenic reactors fed thiosulfate as electron acceptor, the thiosulfate influent concentration should thus be carefully monitored, as the production of sulfite as an intermediate depends on it and might cause damage to growth and sulfidogenic performance of the reactor.
Sulfite toxicity to cells has been widely reported (Daniel, 1986; Gardner et al., 2015; Kappler and Enemark, 2015; Kappler and Schwarz, 2016). Although this highly reactive compound is on one hand referred to damage proteins, DNA and lipids through the formation of adducts, and on the other hand sulfite-oxidizing enzymes (SOEs) are found in nearly all forms of life (Kappler and Enemark, 2015; Kappler and Schwarz, 2016), very little is known about its mechanism of toxicity toward anaerobic microorganisms.
Metal Recovery
The aquatic environmental pollution originating from heavy metals in solution is of utmost relevance due to its impact on public health, environment, and economy. Sulfidogenic reactors have become suitable alternatives for the treatment of metal-rich wastewaters (Escobar et al., 2007; Gallegos-Garcia et al., 2009), with high levels of metals precipitated as highly insoluble metal sulfides. As a proof of concept, the stripping of sulfide for heavy metal precipitation shown in this study (Figure 6) increased the possibility of degrading the highly energetic bonds in LCFA compounds, mitigating the complexity of FOGs in lipid-rich wastewater while removing and recycling metals from metallurgical emissions.
Although the precipitation of metals with sulfide is widely implemented in several reactors configurations, the majority of the studies is performed with sulfate as electron acceptor and with simple organic compounds or hydrogen as electron donors (Jong and Parry, 2003; Kaksonen et al., 2004; Sierra-Alvarez et al., 2007; Gallegos-Garcia et al., 2009). The use of LCFA by mixed microbial communities with the flux of electrons drained toward sulfide production has, thus far, not received much attention. Sharma and Biswas (2013) applied sulfate as electron acceptor in a methanogenic culture fed with glucose in the presence of linoleic acid and observed a shift in the reaction toward sulfide production. However, the authors did not perform any further investigation on the utilization of the produced sulfide, or on the microbial community shift.
The reactor operating with thiosulfate kept the bacterial diversity, with Desulfurella and Enterobacterales as the most dominant groups. In the elemental sulfur-reducing reactor, although Enterobacteraceae and Sulfurospirillum members remained with good abundance, Halothiobacillus and Bacteroides members got enriched as well. To our knowledge, the abundance and diversity of sulfidogenic microorganisms in anaerobic reactors is limited, it is thus worthy to investigate how to further improve the efficiency of metal precipitation processes by applying specific conditions to enhance the growth and metabolic activity of these specialized microorganisms.
The sulfite toxicity to S2-reducing enrichments observed in this study imposes some constraints to the broad utilization of sulfidogenic LCFA-degrading biomass for biotechnological purposes, when thiosulfate is present in elevated concentrations. The robustness and great efficiency of S0-reducing cultures, however, reveals a promising alternative to the currently applied strategies for the degradation of LCFA, with no accumulation of intermediate products and feasible destination of the end-product (H2S). It is, therefore, a good prospect for enhanced metal recovery and generation of clean industrial effluents.
Data Availability Statement
The datasets generated for this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/, PRJNA622591.
Author Contributions
AF: study conception, experimental design, and drafting of manuscript. AF, RC, and YH: acquisition of data. AF and RC: analysis and interpretation of data. RC, YH, VO'F, and PL: critical revision. All authors contributed to the article and approved the submitted version.
Funding
This research was funded by the Science Foundation Ireland Research Professorship Innovative Energy Technologies for Biofuels, Bioenergy and a Sustainable Irish Bioeconomy [IETSBIO3; Grant Number 15/RP/2763], and the Research Infrastructure research grant Platform for Biofuel Analysis (Grant Number 16/RI/3401). The postdoctoral research of RC was financed by the grant 18/01524-7 from the São Paulo Research Foundation (FAPESP), Brazil.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors acknowledge the facilities and scientific and technical assistance of the Center for Microscopy and Imaging at the National University of Ireland Galway (www.imaging.nuigalway.ie).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbioe.2020.550253/full#supplementary-material
References
Ahn, A.-C., Meier-Kolthoff, J. P., Overmars, L., Richter, M., Woyke, T., Sorokin, D. Y., et al. (2017). Genomic diversity within the haloalkaliphilic genus Thioalkalivibrio. PLoS ONE 12:e0173517. doi: 10.1371/journal.pone.0173517
Akcil, A., and Koldas, S. (2006). Acid Mine Drainage (AMD): causes, treatment and case studies. J. Clean. Prod. 14, 1139–1145. doi: 10.1016/j.jclepro.2004.09.006
Alves, M. M., Pereira, M. A., Sousa, D. Z., Cavaleiro, A. J., Picavet, M., Smidt, H., et al. (2009). Waste lipids to energy: how to optimize methane production from long-chain fatty acids (LCFA). Microb. Biotechnol. 2, 538–550. doi: 10.1111/j.1751-7915.2009.00100.x
Angly, F. E., Dennis, P. G., Skarshewski, A., Vanwonterghem, I., Hugenholtz, P., and Tyson, G. W. (2014). CopyRighter: a rapid tool for improving the accuracy of microbial community profiles through lineage-specific gene copy number correction. Microbiome 2:11. doi: 10.1186/2049-2618-2-11
Beauchamp, R. O. Jr, Bus, J. S., Popp, J. A., Boreiko, C. J., and Andjelkovich, D. A. (1984). A critical review of the literature on hydrogen sulfide toxicity. Crit. Rev. Toxicol. 13, 25–97. doi: 10.3109/10408448409029321
Biebl, H., and Pfennig, N. (1977). Growth of sulfate-reducing bacteria with sulfur as electron acceptor. Archiv. Microbiol. 112, 115–117. doi: 10.1007/BF00446664
Bonch-Osmolovskaya, E. A., Sokolova, T. G., Kostrikina, N. A., and Zavarzin, G. A. (1990). Desulfurella acetivorans gen. nov. and sp. nov. – a new thermophilic sulfur-reducing eubacterium. Archiv. Microbiol. 153, 151–155. doi: 10.1007/BF00247813
Cline, J. D. (1969). Spectrophotometric determination of hydrogen sulfide in natural waters. Limnol. Oceanogr. 14, 454–458. doi: 10.4319/lo.1969.14.3.0454
Daniel, J. W. (1986). Metabolic aspects of antioxidants and preservatives. Xenobiotica. 16, 1073–8. doi: 10.3109/00498258609038984
Dasa, K. T., Westman, S. Y., Millati, R., Cahyanto, M. N., Taherzadeh, M. J., and Niklasson, C. (2016). Inhibitory effect of long-chain fatty acids on biogas production and the protective effect of membrane bioreactor. BioMed. Res. Int. 2016:7263974. doi: 10.1155/2016/5727203
Edgar, R. C., Haas, B. J., Clemente, J. C., Quince, C., and Knight, R. (2011). UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27, 2194–2200. doi: 10.1093/bioinformatics/btr381
Eren, A., Morrison, H., Lescault, P., Reveillaud, J., Vineis, J. H., and Sogin, M. L. (2015). Minimum entropy decomposition: unsupervised oligotyping for sensitive partitioning of high-throughput marker gene sequences. ISME J. 9, 968–979. doi: 10.1038/ismej.2014.195
Eren, A. M., Vineis, J. H., Morrison, H. G., and Sogin, M. L. (2013). A filtering method to generate high quality short reads using Illumina paired-end technology. PLoS ONE 8:e66643. doi: 10.1371/journal.pone.0066643
Escobar, C., Bravo, L., Hernández, J., and Herrera, L. (2007). Hydrogen sulfide production from elemental sulfur by Desulfovibrio desulfuricans in an anaerobic bioreactor. Biotechnol. Bioeng. 98, 569–577. doi: 10.1002/bit.21457
Florentino, A. P., Brienza, C., Stams, A. J. M., and Sánchez-Andrea, I. (2016a). Desulfurella amilsii sp. nov., a novel acidotolerant sulfur-respiring bacterium isolated from acidic river sediments. Int. J. Syst. Evol. Microbiol. 66, 1249–1253. doi: 10.1099/ijsem.0.000866
Florentino, A. P., Pereira, I. A. C., Boeren, S., van den Born, M., Stams, A. J. M., and Sánchez-Andrea, I. (2019). Insight into the sulfur metabolism of Desulfurella amilsii by differential proteomics. Environ. Microbiol. 21:209–225. doi: 10.1111/1462-2920.14442
Florentino, A. P., Weijma, J., Stams, A. J. M., and Sánchez-Andrea, I. (2016b). “Ecophysiology and application of acidophilic sulfur-reducing microorganisms,” in Biotechnology of Extremophiles: Grand Challenges in Biology and Biotechnology, vol 1. ed P. Rampelotto (Cham: Springer), 141–175. doi: 10.1007/978-3-319-13521-2_5
Gallegos-Garcia, M., Celis, L. B., Rangel-Méndez, R., and Razo-Flores, E. (2009). Precipitation and recovery of metal sulfides from metal containing acidic wastewater in a sulfidogenic down-flow fluidized bed reactor. Biotechnol. Bioeng. 102, 91–99. doi: 10.1002/bit.22049
Gardner, P. R., Gardner, D. P., and Gardner, A. P. (2015). Globins scavenge sulfur trioxide anion radical. J. Biol. Chem. 290, 27204–27214. doi: 10.1074/jbc.M115.679621
Guihéneuf, F., Schmid, M., and Stengel, D. B. (2014). Lipids and fatty acids in algae: extraction, fractionation into lipid classes, and analysis by gas chromatography coupled with flame ionization detector (GC-FID). Method. Mol. Biol. 1308, 173–190. doi: 10.1007/978-1-4939-2684-8_11
Hawkes, F. R., Donnelly, T., and Anderson, G. K. (1995). Comparative performance of anaerobic digesters operating on ice-cream wastewater. Water Res. 29, 525–533. doi: 10.1016/0043-1354(94)00163-2
Hwu, C.-S., Tseng, S.-K., Yuan, C.-Y., Kulik, Z., and Lettinga, G. (1998). Biosorption of long-chain fatty acids in UASB treatment process. Water Res. 32, 1571–1579. doi: 10.1016/S0043-1354(97)00352-7
Hwu, C. S., and Lettinga, G. (1997). Acute toxicity of oleate to acetate-utilizing methanogens in mesophilic and thermophilic anaerobic sludges. Enzyme Microb. Technol. 21, 297–301. doi: 10.1016/S0141-0229(97)00050-1
Irwin, S. V., Fisher, P., Graham, E., Malek, A., and Robidoux, A. (2017). Sulfites inhibit the growth of four species of beneficial gut bacteria at concentrations regarded as safe for food. PLoS ONE 12:e0186629. doi: 10.1371/journal.pone.0186629
Işildar, A., van Hullebusch, E. D., Lenz, M., Du Laing, G., Marra, A., Cesaro, A., et al. (2019). Biotechnological strategies for the recovery of valuable and critical raw materials from waste electrical and electronic equipment (WEEE)—a review. J. Hazard. Mater. 362, 467–481. doi: 10.1016/j.jhazmat.2018.08.050
Johnson, D. B. (2014). Biomining-biotechnologies for extracting and recovering metals from ores and waste materials. Curr. Opin. Biotechnol. 30, 24–31. doi: 10.1016/j.copbio.2014.04.008
Jong, T., and Parry, D. L. (2003). Removal of sulfate and heavy metals by sulfate reducing bacteria in short-term bench scale upflow anaerobic packed bed reactor runs. Water Res. 37, 3379–3389. doi: 10.1016/S0043-1354(03)00165-9
Kaksonen, A. H., Franzmann, P. D., and Puhakka, J. A. (2004). Effects of hydraulic retention time and sulfide toxicity on ethanol and acetate oxidation in sulfate-reducing metal-precipitating fluidized-bed reactor. Biotechnol. Bioeng. 86, 332–343. doi: 10.1002/bit.20061
Kappler, U., and Enemark, J. H. (2015). Sulfite-oxidizing enzymes. J. Biol. Inorg. Chem. 20, 253–264. doi: 10.1007/s00775-014-1197-3
Kappler, U., and Schwarz, G. (2016). “The sulfite oxidase family of molybdenum enzymes,” in Molybdenum and Tungsten Enzymes, eds R. Hille, C. Schulzke, and M. L. Kirk (Cambridge: Royal Society of Chemistry), 240–273. doi: 10.1039/9781782623915-00240
Komatsu, T., Hanaki, K., and Matsuo, T. (1991). Prevention of lipid inhibition in anaerobic processes by introducing a two-phase system. Water Sci. Technol. 23, 1189–1200. doi: 10.2166/wst.1991.0570
Labatut, R. A., Angenent, L. T., and Scott, N. R. (2014). Conventional mesophilic vs. thermophilic anaerobic digestion: a trade-off between performance and stability? Water Res. 53, 249–258. doi: 10.1016/j.watres.2014.01.035
Lalman, J. A., and Bagley, D. M. (2002). Effect of C18 long chain fatty acids on glucose, butyrate and hydrogen degradation. Water Res. 36, 3307–3313. doi: 10.1016/S0043-1354(02)00014-3
Liu, P., Ji, J., Wu, Q., Ren, J., Wu, G., Yu, Z., et al. (2018). Klebsiella pneumoniae sp. LZU10 degrades oil in food waste and enhances methane production from co-digestion of food waste and straw. Int. Biodeteriorat. Biodegrad. 126, 28–36. doi: 10.1016/j.ibiod.2017.09.019
Ma, J., Zhao, Q. B., Laurens, L. L., Jarvis, E. E., Nagle, N. J., Chen, S., et al. (2015). Mechanism, kinetics and microbiology of inhibition caused by long-chain fatty acids in anaerobic digestion of algal biomass. Biotechnol. Biofuels 8:141. doi: 10.1186/s13068-015-0322-z
Miroshnichenko, M. L., Rainey, F. A., Hippe, H., Chernyh, N. A., Kostrikina, N. A., and Bonch-Osmolovskaya, E. A. (1998). Desulfurella kamchatkensis sp. nov. and Desulfurella propionica sp. nov., new sulfur-respiring thermophilic bacteria from Kamchatka thermal environments. Int. J. Bacteriol. 48, 475–479. doi: 10.1099/00207713-48-2-475
Miroshnichenko, M. L., Rainey, F. A., Rhode, M., and Bonch-Osmolovskaya, E. A. (1999). Hippea maritima gen. nov., sp. nov., a new genus of thermophilic, sulfur-reducing bacterium from submarine hot vents. Int. J. Syst. Evol. Microbiol. 49, 1033–1038. doi: 10.1099/00207713-49-3-1033
Moodley, I., Sheridan, C. M., Kappelmeyer, U., and Akcil, A. (2017). Environmentally sustainable acid mine drainage remediation: research developments with a focus on waste/by-products. Miner. Eng. 126, 207–220. doi: 10.1016/j.mineng.2017.08.008
Nelson, D. L., and Cox, M. M. (2005). Lehninger Principles of Biochemistry. 4th edn. New York, NY: W. H. Freeman and Company. ISBN 978-0-7167-4339-2.
Odeyemi, A., Aderiye, B., and Bamidele, O. S. (2013). Lipolytic activity of some strains of Klebsiella, Pseudomonas and Staphylococcus spp. from restaurant wastewater and receiving stream. J. Microbiol. Res. 3, 43–52. doi: 10.5923/j.microbiology.20130301.07
Palatsi, J., Laureni, M., Andrés, M. V., Flotats, X., Nielsen, H. B., and Angelidaki, I. (2009). Recovery strategies from long-chain fatty acids inhibition in anaerobic thermophilic digestion of manure. Bioresour. Technol. 100, 4588–4596. doi: 10.1016/j.biortech.2009.04.046
Pereira, M. A., Pires, O. C., Mota, M., and Alves, M. M. (2005). Anaerobic biodegradation of oleic and palmitic acids: evidence of mass transfer limitations caused by long chain fatty acid accumulation onto the anaerobic sludge. Biotechnol. Bioeng. 92, 15–23. doi: 10.1002/bit.20548
Plugge, C. M., Zhang, W., Scholten, J. C., and Stams, A. J. (2011). Metabolic flexibility of sulfate-reducing bacteria. Front. Microbiol. 2:81. doi: 10.3389/fmicb.2011.00081
Rabus, R., Hansen, T. A., and Widdel, F. (2013). “Dissimilatory sulfate- and sulfur-reducing prokaryotes,” in The Prokaryotes, eds E. Rosenberg, E. F. DeLong, S. Lory, E. Stackebrandt, and F. Thompson (Berlin; Heidelberg: Springer), 309–404. doi: 10.1007/978-3-642-30141-4_70
Reis, M. A., Almeida, J. S., Lemos, P. C., and Carrondo, M. J. (1992). Effect of hydrogen sulfide on growth of sulfate reducing bacteria. Biotechnol. Bioeng. 40, 593–600. doi: 10.1002/bit.260400506
Rognes, T., Flouri, T., Nichols, B., Quince, C., and Mahé, F. (2016). VSEARCH: a versatile open source tool for metagenomics. PeerJ. 4:e2584. doi: 10.7717/peerj.2584
Salvador, A. F., Cavaleiro, A. J., Paulo, A., Silva, S. A., Guedes, A. P., Pereira, M. A., et al. (2019). Inhibition studies with 2-bromoethanesulfonate reveal a novel syntrophic relationship in anaerobic oleate degradation. Appl. Environ. Microbiol. 85:e01733-18. doi: 10.1128/AEM.01733-18
Sánchez-Andrea, I., Sanz, J. L., Bijmans, M. F. M., and Stams, A. J. M. (2014). Sulfate reduction at low pH to remediate acid mine drainage. J. Hazard. Mater. 269, 98–109. doi: 10.1016/j.jhazmat.2013.12.032
Schmitz, R. A., Bonch-Osmolovskaya, E. A., and Thauer, R. K. (1990). Different mechanisms of acetate activation in Desulfurella acetivorans and Desulfuromonas acetoxidans. Archiv. Microbiol. 154, 274–279. doi: 10.1007/BF00248967
Sharma, M., and Biswas, N. (2013). Diverting electron fluxes towards sulfate reduction using linoleic acid in a mixed anaerobic culture. J. Environ. Eng. Sci. 8, 587–600. doi: 10.1680/jees.2013.0062
Sharma, P., Khardenavis, A. A., and Purohit, H. J. (2015). “Metabolism of Long-Chain Fatty Acids (LCFAs) in methanogenesis,” in Microbial Factories, ed V. Kalia (New Delhi: Springer). doi: 10.1007/978-81-322-2598-0_16
Sierra-Alvarez, R., Hollingsworth, J., and Zhou, M. S. (2007). Removal of copper in an integrated sulfate reducing bioreactor-crystallization reactor system. Environ. Sci. Technol. 41, 1426–1431. doi: 10.1021/es062152l
Silva, S. A., Salvador, A. F., Cavaleiro, A. J., Pereira, M. A., Stams, A. J., Alves, M. M., et al. (2016). Toxicity of long chain fatty acids towards acetate conversion by Methanosaeta concilii and Methanosarcina mazei. Microb. Biotechnol. 9, 514–518. doi: 10.1111/1751-7915.12365
Sorokin, D. Y., and Kuenen, J. G. (2005). Haloalkaliphilic sulfur-oxidizing bacteria in soda lakes. FEMS Microbiol. Rev. 29, 685–702. doi: 10.1016/j.femsre.2004.10.005
Sousa, D. Z., Pereira, M. A., Stams, A. J. M., Alves, M. M., and Smidt, H. (2007). Microbial communities involved in anaerobic degradation of unsaturated or saturated long chain fatty acids (LCFA). Appl. Environ. Microbiol. 73, 1054–1064. doi: 10.1128/AEM.01723-06
Sousa, D. Z., Salvador, A. F., Ramos, J., Guedes, A. P., Barbosa, S., Stams, A. J. M., et al. (2013). Activity and viability of methanogens in anaerobic digestion of unsaturated and saturated long-chain fatty acids. Appl. Environ. Microbiol. 79, 4239–4245. doi: 10.1128/AEM.00035-13
Sousa, D. Z., Smidt, H., Alves, M. M., and Stams, A. J. M. (2009). Ecophysiology of syntrophic communities that degrade saturated and unsaturated long-chain fatty acids. FEMS Microbiol. Ecol. 68, 257–272. doi: 10.1111/j.1574-6941.2009.00680.x
Stams, A. J. M., Van Dijk, J. B., Dijkema, C., and Plugge, C. M. (1993). Growth of syntrophic propionate-oxidizing bacteria with fumarate in the absence of methanogenic bacteria. Appl. Environ. Microbiol. 59, 1114–1119. doi: 10.1128/AEM.59.4.1114-1119.1993
Stoffels, L., Krehenbrink, M., Berks, B. C., and Unden, G. (2012). Thiosulfate reduction in Salmonella enterica is driven by the proton motive force. J. Bacteriol. 194, 475–485. doi: 10.1128/JB.06014-11
Surkov, A. V., Bottcher, M. E., and Kuever, J. (2000). Stable sulfur isotope fractionation during the reduction of thiosulfate by Dethiosulfovibrio russensis. Arch. Microbiol. 174, 448–451. doi: 10.1007/s002030000217
Keywords: sulfidogenesis, long chain fatty acid, beta-oxidation, metal precipitation, oleate toxicity
Citation: Florentino AP, Costa RB, Hu Y, O'Flaherty V and Lens PNL (2020) Long Chain Fatty Acid Degradation Coupled to Biological Sulfidogenesis: A Prospect for Enhanced Metal Recovery. Front. Bioeng. Biotechnol. 8:550253. doi: 10.3389/fbioe.2020.550253
Received: 08 April 2020; Accepted: 25 September 2020;
Published: 23 October 2020.
Edited by:
Roland Wohlgemuth, Lodz University of Technology, PolandReviewed by:
Elizaveta Bonch-Osmolovskyaya, Winogradsky Institute of Microbiology (RAS), RussiaBo Svensson, Linköping University, Sweden
Copyright © 2020 Florentino, Costa, Hu, O'Flaherty and Lens. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Anna Patrícya Florentino, YW5uYXBhdHJpY3lhQGdtYWlsLmNvbQ==
 Anna Patrícya Florentino
Anna Patrícya Florentino Rachel Biancalana Costa
Rachel Biancalana Costa Yuansheng Hu
Yuansheng Hu Vincent O'Flaherty
Vincent O'Flaherty Piet N. L. Lens1
Piet N. L. Lens1