- 1School of Applied Biosciences, Kyungpook National University, Daegu, South Korea
- 2Department of Biology, College of Natural Sciences, Kyungpook National University, Daegu, South Korea
- 3Research Institute of Ulleung-Do & Dok-Do, Kyungpook National University, Daegu, South Korea
- 4School of Life Sciences and Biotechnology, BK21 Plus KNU Creative BioResearch Group, Kyungpook National University, Daegu, South Korea
Bacterial and algal floc formation was induced by inoculating three species of wastewater-derived bacteria (Melaminivora jejuensis, Comamonas flocculans, and Escherichia coli) into algal cultures (Chlorella sorokiniana). Bacterial and algal flocs formed in algal cultures inoculated with M. jejuensis and C. flocculans, and these flocs showed higher sedimentation rates than pure algal culture. The floc formed by M. jejuensis (4988.46 ± 2589.81 μm) was 10-fold larger than the floc formed by C. flocculans (488.60 ± 226.22 μm), with a three-fold higher sedimentation rate (M. jejuensis, 91.08 ± 2.32% and C. flocculans, 32.55 ± 6.33%). Biomass and lipid productivity were improved with M. jejuensis inoculation [biomass, 102.25 ± 0.35 mg/(L·day) and 57.80 ± 0.20 mg/(L·day)] compared with the productivity obtained under pure algal culture conditions [biomass, 78.00 ± 3.89 mg/(L·day) and lipids, 42.26 ± 2.11 mg/(L·day)]. Furthermore, the fatty acid composition of the biomass produced under pure algal culture conditions was mainly composed of C16:0 (43.67%) and C18:2 (45.99%), whereas the fatty acid composition of the biomass produced by M. jejuensis was mainly C16:0 (31.80%), C16:1 (24.45%), C18:1 (20.23%), and C18:2 (16.11%). These results suggest the possibility of developing an efficient method for harvesting microalgae using M. jejuensis and provide information on how to improve biomass productivity using floc-forming bacteria.
Introduction
Microalgae are a source of bioenergy raw material that can be used to produce biofuels. This unique bioresource has been proposed as a solution to combat energy shortages and alleviate problems associated with global warming (Morales et al., 2019; Tan et al., 2019). Compared to terrestrial plants, microalgae have greater potential as a bioenergy raw material (Chen et al., 2020) with greater biomass productivity (Cooney et al., 2009; Chen et al., 2020). Typically, 10–20% of the biomass derived from microalgae consists of fatty acids that can be used as raw materials for bioenergy (Sajjadi et al., 2018). However, some limitations for industrial applications of microalgae bioenergy remain (Lee, 2001). The biomass produced through microalgae cultivation is harvested using processes such as centrifugation and filtration (Dassey and Theegala, 2013; Farooq et al., 2013). Significant losses and production costs are incurred during harvest (Dassey and Theegala, 2013; Farooq et al., 2013). Therefore, solutions to reduce the losses and production costs associated with harvesting processes are essential (Jonker and Faaij, 2013).
To reduce microalgae biomass production costs, methods have been developed to utilize wastewater to culture strains with high sedimentation rates (Cheng et al., 2019; Leite and Daniel, 2020). Multicellular strains with high sedimentation rates, such as those belonging to the Pediastrum genus, were proposed for this application (DA LUZ et al., 2002); however, culturing Pediastrum requires more time than that required for unicellular strains with lower sedimentation rates and biomass productivity, such as members of the Chlorella genus (Duygu et al., 2019; Stone et al., 2019). In addition, while the sedimentation rates of multicellular strains are relatively high, it takes a long time to harvest the sedimented biomass (Su et al., 2012). As a result of these sedimentation rate limitations, wastewater-based cultivation methods have also been investigated in an attempt to reduce the production costs associated with cultivation (Cheng et al., 2019). Wastewater contains inorganic materials required for microalgal growth such as nitrogen and phosphorous (Benítez et al., 2019). To purify wastewater, nitrogen, and phosphorous must be removed; thus, the cultivation of microalgae using wastewater can be used to simultaneously purify wastewater and produce biomass at a low cost (Benítez et al., 2019; Cheng et al., 2019). Furthermore, bacteria can be cultured together with microalgae during cultivation with wastewater (Kwon et al., 2019). Depending on the characteristics of the bacteria being cultured, bacteria-derived floc can often form. This bacteria-derived floc that forms contains microalgal cells, and the sedimentation properties of this floc are greater than those of the pure microalgal biomass. Therefore, the floc formed by bacteria can be applied to microalgal biomass harvesting (Wieczorek et al., 2015).
In this study, three bacterial strains [Melaminivora jejuensis, Korean Collection for Type Cultures (KCTC) 32230; Comamonas flocculans, KCTC 62943; Escherichia coli, DH5α] were inoculated into medium to form a floc with the microalgal strain Chlorella sorokiniana KNUA114. The floc-forming abilities of the inoculated strains were investigated and the sedimentation rates of the resulting biomass were compared. Gas chromatography/mass spectrometry (GC/MS) was used to determine the composition of fatty acid in biomass contained within each floc as a bioenergy raw material. Here, we demonstrated the application of floc-forming bacteria in microalgal harvesting and established the benefits for bioenergy development.
Materials and Methods
Whole Genome Analysis
The whole-genome analysis was applied to confirm the composition of bacterial floc-forming-related genes and the distribution of EPS genes that are expected to be related to flocculation with microalgae (Lee et al., 2013). Melaminivora jejuensis KCTC 32230 whole-genome sequencing data were assembled with PacBio SMRT Link 7.0.1 using the HGAP4 protocol (Pacific Biosciences, USA). PacBio RS II sequencing data were assembled with SMRT Portal 2.3.0 using the HGAP2 protocol (Pacific Biosciences, USA). The resulting contigs were circularized using Circlator 1.4.0 (Sanger Institute). Gene-finding and functional annotation pipeline of whole-genome assemblies were performed using the EzBioCloud genome database. Protein-coding sequences (CDSs) were predicted by Prodigal 2.6.2 (Hyatt et al., 2010). The CDSs were classified based on their roles, with reference to orthologous groups (EggNOG 4.5; http://eggnogdb.embl.de) (Powell et al., 2014). For more functional annotation, the predicted CDSs were compared with the Swissprot (Consortium, 2015), KEGG (Kanehisa et al., 2014), and SEED (Overbeek et al., 2005) databases using the UBLAST program (Edgar, 2010). The whole-genome shotgun BioProject accession number is PRJNA663428 with the locus tag prefix IDM45 (https://www.ncbi.nlm.nih.gov/bioproject/?term=txid1267217[Organism:~noexp]).
Collection and Screening of Floc-Forming Bacterial Strains
Based on a previous study, we tried to select floc-forming bacteria, which are expected to form floc with microalgae, among strains derived from wastewater (Jimoh et al., 2019; Petrini et al., 2020). C. flocculans KCTC 62943 was isolated from a livestock wastewater treatment plant wastewater sample (52-109, Gyebaek-ro 499beon-gil, Chaeun-myeon, Nonsan-si, Chungcheongnam-do, 36°10′ 36.6″ N, 127°03′ 10.4″ E; Kim et al., 2019, 2020). Melaminivora jejuensis KCTC 32230, which was originally isolated from swine waste on Jeju Island, Republic of Korea (Kim et al., 2018), was obtained from the KCTC at the Korea Research Institute of Bioscience and Biotechnology, Korea. The isolation process was performed by streaking using R2A medium agar plates (with 15 g/L agarose; Reasoner and Geldreich, 1985). The isolated strains were identified using 16S rRNA gene sequencing (Wang and Qian, 2009). The floc-forming ability of the identified strains was screened; the flocs formed were observed and measured using an optical microscope (model BX53F; Olympus).
Strains and Culture Conditions
Melaminivora jejuensis KCTC 32230 (Kim et al., 2018) and C. flocculans KCTC 62943 (Kim et al., 2019, 2020), which were clearly observed to form floc, were selected as the experimental strains. E. coli, which is related to floc and biofilm, was selected as the control strain. Three flocculation bacterial strains (M. jejuensis KCTC 32230, C. flocculans KCTC 62943, and E. coli, dh5α) and the microalgal strain C. sorokiniana KNUA114 were prepared. C. sorokiniana KNUA114 was isolated from Okcheon Stream in Ulleung, Korea (Okcheon Stream, Ulleung-eup, Ulleung-gun, Gyeongsangbuk-do; 37°28′ 24.0″ N, 130°53′ 06.8″ E; Yun et al., 2020). The bacterial and algal strains were maintained at 30°C on R2A agar plates composed of the following: proteose peptone, 0.5 g/L; casamino acid, 0.5 g/L; yeast extract, 0.5 g/L; dextrose, 0.5 g/L; soluble starch, 0.5 g/L; sodium pyruvate, 0.3 g/L; K2HPO4, 0.3 g/L; and MgSO4·7H2O, 0.05 g/L. For the experiments, each strain was cultured in R2A medium for 4 days at 30°C on an orbital shaker at 160 rpm in an incubation room. Cells were collected by centrifugation (4,000 rpm, 10 min) and washed three times with sterile distilled water. The initial cell density of the C. sorokiniana culture was set to an OD of 0.3 at 680 nm (OD680). Bacterial seeds were resuspended in fresh R2A medium (OD600 = 0.3). Each prepared cell sample (15 mL) was inoculated into 150 mL fresh R2A medium in a 250 mL flask. To compare the algal yields to bacterial flocs, six experimental groups (M, M. jejuensis only; C, C. flocculans only; E, E. coli only; MC, M. jejuensis and C. sorokiniana; CC, C. flocculans and C. sorokiniana; and EC, E. coli and C. sorokiniana) and one control group (N, C. sorokiniana only) were prepared. The experimental groups were cultured in heterotrophic conditions for 4 days at 30°C on an orbital shaker at 160 rpm in an incubation room.
Flocs Size and Sedimentation Analysis
Floc size and its relationship to sedimentation rate were measured. The sizes of the floc colonies formed in the culture medium were measured using an optical microscope (model BX53F; Olympus). Samples were collected from 4-day-old cultures; 20 floc colonies were measured for each experimental condition.
To overcome the limitations of the existing sedimentation measurement using an Imhoff cone, the dry weight was used compared to separating the supernatant and the sediment (Sojka et al., 1992). For the sedimentation rate analysis, 4-day-old samples from each experimental condition were prepared, and 100 mL of each prepared sample were transferred to a glass container. The samples were shaken by hand until thoroughly mixed, and then 80 mL of supernatant was removed to obtain the lower 20 mL layer containing the sediment. The weight of cells contained in the supernatant and sediment was measured using the dry weight method (Yoo et al., 2010). In brief, a cell pellet was obtained using a pre-weighed glass-fiber filter and then dried in a drying oven at 105°C for 1 day before weighing (Yoo et al., 2010). This process was repeated for each experimental condition and measurement time.
Biomass Collection and Lipid Content Analysis
Four-day-old cells were harvested by centrifugation at 4,000 rpm for 10 min. The collected cells were freeze-dried for 7 days, then ground using a mortar. The sulfo-phospho-vanillin reaction method was used to determine the total lipid content of each sample by mixing 10 mg of ground sample with 1 mL distilled water (Mishra et al., 2014). Different volumes of the mixture (10, 20, 30, 40, and 50 μL) were transferred to glass tubes and distilled water was added to achieve a total volume of 100 μL in each tube. Sulfuric acid (2 mL) was added to each of the glass tubes, which were then heated in a water bath at 100°C for 5 min. The tubes were allowed to cool to room temperature. Once cool, 5 mL phospho-vanillin reagent was added to each of the glass tubes, which were then shaken at 200 rpm in an incubator at 37°C for 15 min. The OD530 values of the mixtures were measured and total lipid content was calculated using canola oil as a reference standard (Mishra et al., 2014). To compare the fatty acid compositions between culture conditions, fatty acids were extracted from 30 mg of ground sample using a mixture of chloroform and methanol (1:1) (Yeo et al., 2011). The chloroform was removed from the fatty acid extract using an evaporator and methanol and potassium hydroxide were added to the mixture to facilitate transesterification of any lipids. Hexane was added to the extracted fatty acid mixture to isolate fatty acid methyl esters (FAMEs). The separated mixture was heated in a water bath at 70°C for 3 h. A gas chromatograph (model 6890N, Agilent) was used to analyze the FAME composition of the pre-treated samples (Furuhashi and Weckwerth, 2013).
Statistical Analysis
We expressed the ratio of biomass present and the fatty acids contents, which we defined as 100%. We compared individual data points using Student's t-test, and a P-value of < 0.05 was considered statistically significant. All experiments were performed at least in triplicate, and the general microbiology test data were expressed as mean ± standard deviation (SD) (n = 3).
Results and Discussion
Whole-Genome Sequencing and Expected Genes for Flocculation
The characteristics of M. jejuensis are described in Tables 1, 2, and Supplementary Figure 1. The complete genome of M. jejuensis was comprised of a 3,754,826-bp chromosome, genome coverage was 195.39X, and G+C content was 67.5%. The genome contained 3,472 genes, including 3,380 protein-coding genes (CDSs), 30 pseudogenes, nine rRNA genes, and 53 tRNA genes. Total CDSs were matched to the EggNOG database (Powell et al., 2014). The M. jejuensis genome included eight aminopeptidase-coding genes, among which there was one ywaD gene (a leucyl aminopeptidase coding gene), which has been shown to control flocculation (Table 2; Geesey and Van Ommen Kloeke, 2004; Zhao et al., 2018). The genome also contained 17 polysaccharide biosynthesis protein-coding genes, including two UTP–glucose-1-phosphate uridylyltransferases (Table 2), which synthesize the EPS that form floc (Degeest and De Vuyst, 2000). Conversely, C. flocculans and E. coli each possessed only one UTP–glucose-1-phosphate uridylyltransferase-coding gene and no flocculation-controlling ywaD genes.
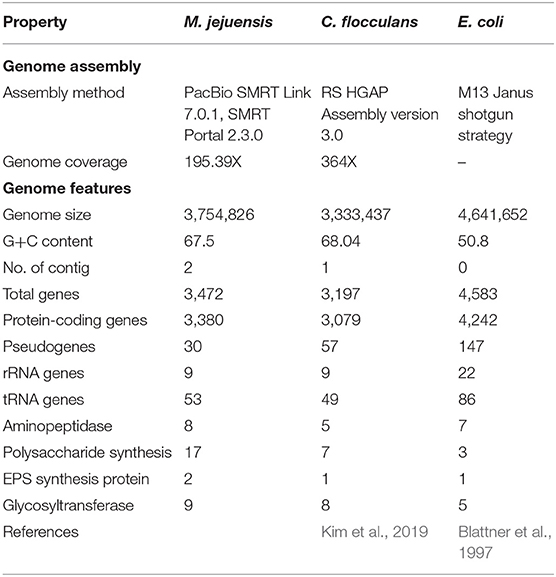
Table 1. General characteristics of M. jejuensis, C. flocculans, and E. coli, including flocculation-related genes.
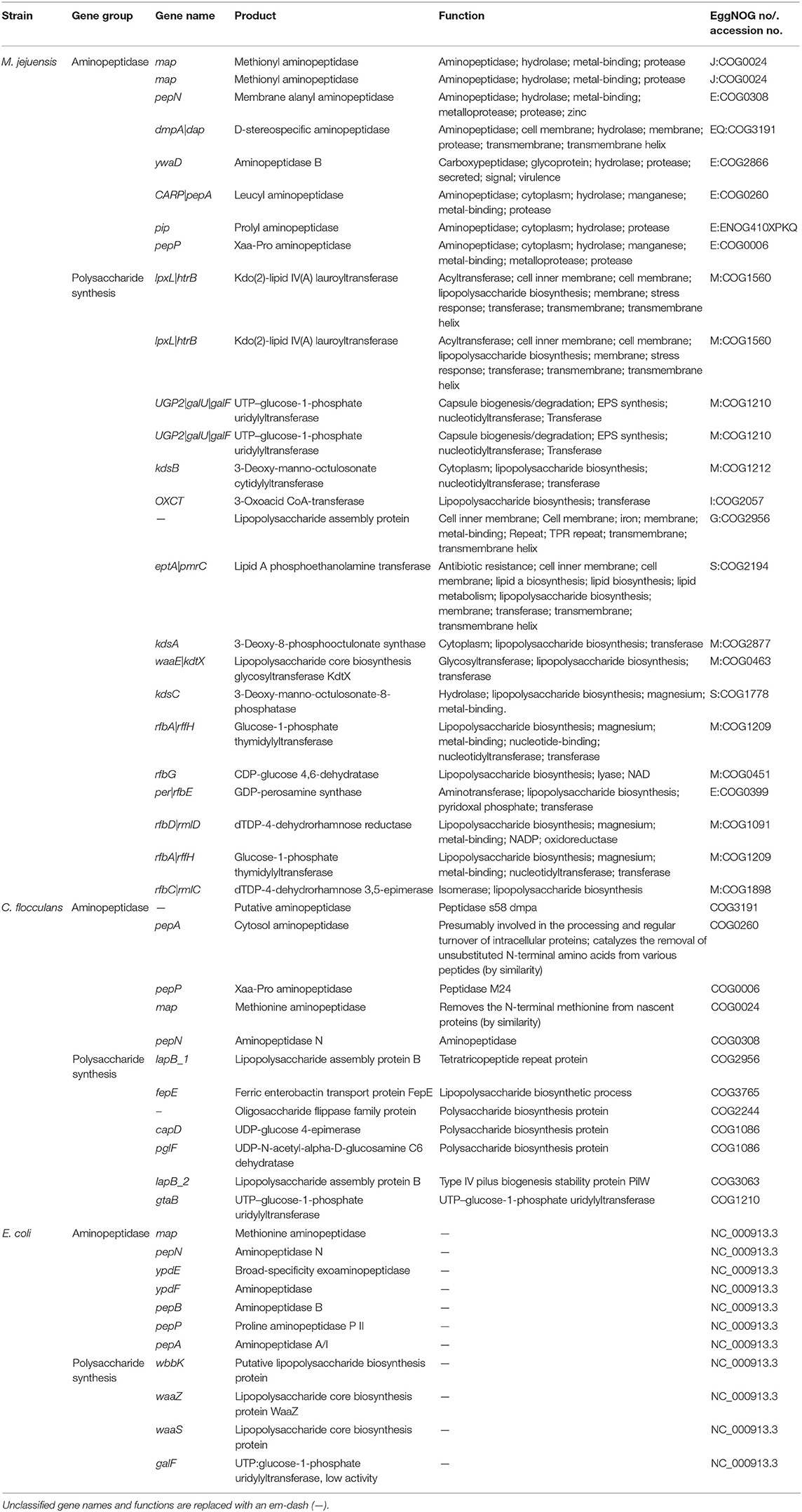
Table 2. List of predicted aminopeptidase and polysaccharide synthesis genes related to flocculation in M. jejuensis, C. flocculans, and E. coli.
Based on the whole-genome sequencing results, M. jejuensis (aminopeptidase, eight genes; polysaccharide synthesis, 17 genes) possesses more flocculation-related genes than C. flocculans (aminopeptidase, five genes; polysaccharide synthesis, seven genes; Kim et al., 2019) and E. coli (aminopeptidase, seven genes; polysaccharide synthesis, three genes; Blattner et al., 1997). Furthermore, the gene expected to regulate flocculation, i.e., the ywaD gene (leucyl aminopeptidase-coding gene), was identified in M. jejuensis only. The UTP–glucose-1-phosphate uridylyltransferase gene involved in EPS synthesis was more abundant in M. jejuensis (two genes) than in C. flocculans (one gene) and E. coli (one gene; Table 1). Therefore, we expect rapid and efficient flocculation by M. jejuensis (Lee et al., 2013). Based on these results, we suggest that M. jejuensis has the potential to flocculate more efficiently than C. flocculans and E. coli.
Flocculation and Sedimentation of Bacterial and Algal Flocs
The co-culture of bacteria and algae resulted in the formation of both bacterial and algal flocs in some culture conditions (Figure 1). In the MC condition in which M. jejuensis and C. sorokiniana were cultured together, a green-colored floc of a visually recognizable size was observed; this floc was composed of tightly connected bacteria and algae. A floc composed of bacteria and algae also formed in the CC condition, where C. flocculans and C. sorokiniana were co-cultured. However, no flocs formed in the EC condition, where E. coli and C. sorokiniana were cultured together, or in the pure culture of C. sorokiniana (N condition). Based on previous studies, the cause of the formation of floc composed of microalgae and bacteria was expected to be from bacteria-derived materials, including EPS, and it was suggested that the difference in the material produced by each bacteria would be involved in the formation and characteristic of floc (Jimoh et al., 2019).
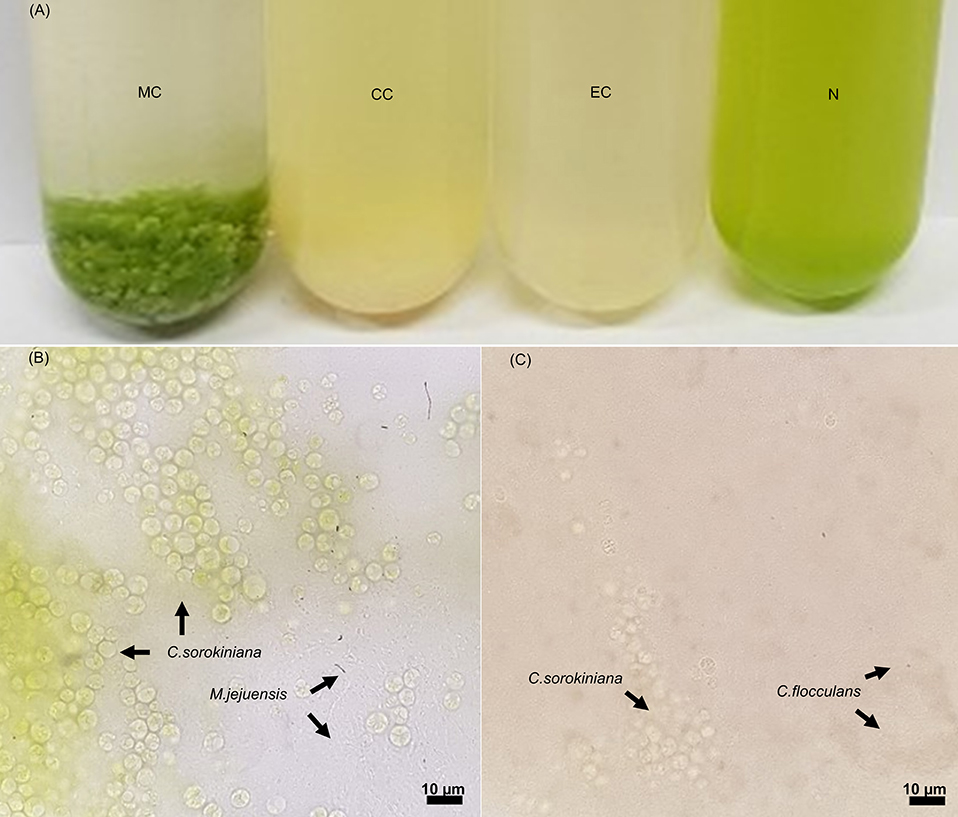
Figure 1. Images of cultures and light microscopy images of floc. In co-culturing conditions (A), floc was formed in the MC and CC conditions, but no floc was observed in the EC and N conditions. The floc formed in the MC (B) and CC (C) conditions contained bacteria and algae (scale bar, 10 μm). The location of each species is marked with an arrow. MC, M. jejuensis and C. sorokiniana; CC, C. flocculans and C. sorokiniana; EC, E. coli and C. sorokiniana; N, C. sorokiniana only, control group. Marked characteristics are annotated in Supplementary Figure 1.
The supernatant in the MC condition became transparent immediately after shaking, although no particular difference between the turbidity of the supernatant and the lower layer was observed in the cultures of the other conditions (CC, EC, and N), demonstrating a greater biomass sedimentation ability in the MC condition. To understand the improved sedimentation ability observed in the MC condition, we measured the floc size as a trait related to sedimentation (Stone and Krishnappan, 2003; Bainbridge et al., 2012). The average floc size in the MC condition (4988.46 ± 2589.81 μm) was about 10 times larger than that in the CC condition (488.60 ± 226.22 μm). The size of floc obtained from MC and CC conditions exceeded that of pure microalgae cells, and this size was expected to have a large influence on sedimentation (Cheng et al., 2011; Baroni et al., 2019; Zhang et al., 2019).
Furthermore, we evaluated the biomass sedimentation ability of each culture (Figure 2). Considering the biomass of the whole culture, we compared the distribution of the sedimented biomass and the biomass contained in the supernatant. The highest sedimentation rates were observed in the MC condition (91.08 ± 2.32%). Although floc formed in the CC condition (32.55 ± 6.33%), there was no remarkable difference in the sedimentation rate compared to that in the EC condition (22.36 ± 19.52%) and the N condition (25.80 ± 10.53%). Our results demonstrate that the co-culture of bacteria and algae resulted in floc formation, and the highest biomass sedimentation rate was associated with large floc size. Therefore, it is expected that the sedimentation ability of the biomass produced by co-culturing the bacterial strain M. jejuensis KCTC 32230 with C. sorokiniana KNUA114 will be improved.
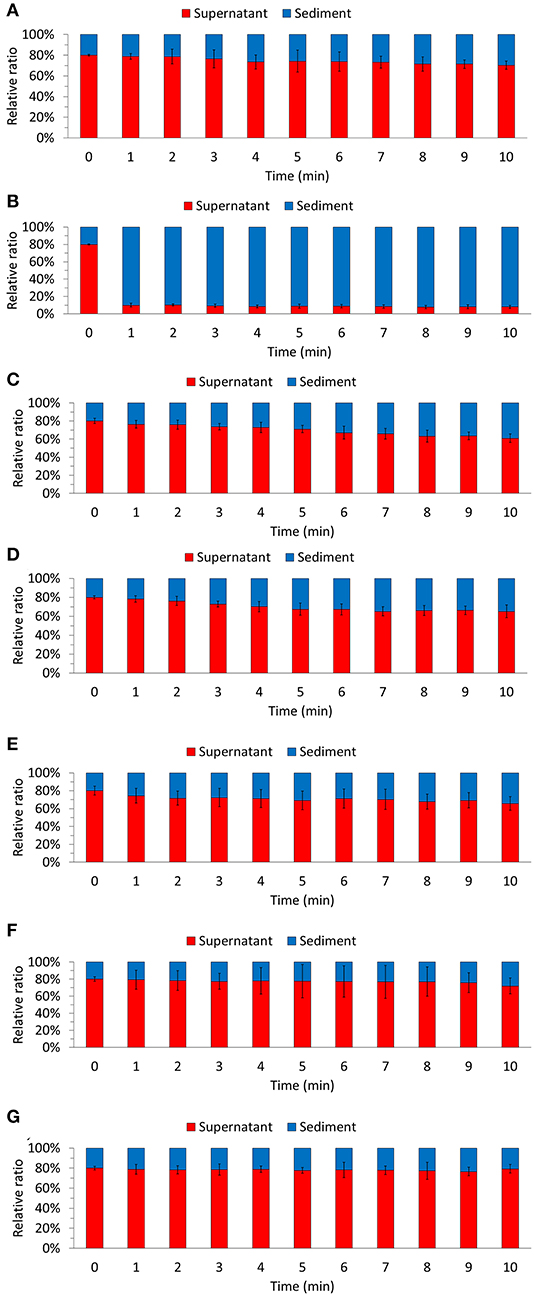
Figure 2. Biomass sedimentation ability was evaluated for each of the culture condition. N (A), MC (B), M. jejuensis (C), CC (D), C. flocculans (E), EC (F), and E. coli (G). MC, M. jejuensis and C. sorokiniana; CC, C. flocculans, and C. sorokiniana; EC, E. coli and C. sorokiniana; N, C. sorokiniana only, control group. Marked characteristics are annotated in Supplementary Figure 1. We herein expressed the ratio of biomass present, which we defined as 100%.
The floc formation by bacteria and algae improved sedimentation ability and reduced turbidity in a short period (Figure 2). Floc size is one of the main contributing factors affecting sedimentation ability (Stone and Krishnappan, 2003; Bainbridge et al., 2012). We also observed that the settleability improved in proportion to the floc size. In the MC condition, the sedimentation rate was 50 to 60% higher than in the CC condition. In addition, the floc-forming CC condition had a higher sedimentation rate (32.55 ± 6.33%) than the EC condition (22.36 ± 19.52%) and N condition (25.80 ± 10.53%), in which floc was not formed. Furthermore, when comparing the EC and N conditions, algae (C. sorokiniana), which has a larger cell size than bacteria (E. coli), exhibited a relatively high sedimentation ability. Based on the measured sedimentation ability and rate, we confirmed that floc formation affected biomass sedimentation. Therefore, our results suggest that biomass sedimentation efficiency can be improved by co-culturing bacteria and algae. Furthermore, our results support the potential application of M. jejuensis KCTC 32230, which formed the largest floc, for biomass production.
Pure Culture and Co-culture Growth Comparison
The growth patterns of co-culture and pure culture are summarized in Figure 3. The highest dry weight was measured in the MC condition (409.00 ± 1.41 mg/L). The CC condition (381.00 ± 1.41 mg/L) also had a higher dry weight than the N condition (327.00 ± 20.09 mg/L), but the EC condition (163.87 ± 16.48 mg/L) did not. Additionally, the pure algae culture (N condition) had a higher dry weight than all pure bacteria cultures (M. jejuensis, 280.00 ± 30.99 mg/L; C. flocculans, 265.00 ± 21.79 mg/L; and E. coli, 161.3 ± 10.74 mg/L). The time taken to reach the stationary phase was the shortest in the EC condition (1 day) and pure E. coli culture (1 day). The stationary phase was reached in 2 to 3 days in the rest of the conditions and strains (M. jejuensis, 2 days; C. flocculans, 3 days; N condition, 3–4 days; MC condition, 3–4 days; and CC condition, 3–4 days). These results revealed that the co-culture of bacteria and algae had higher growth than pure bacterial and algal cultures.
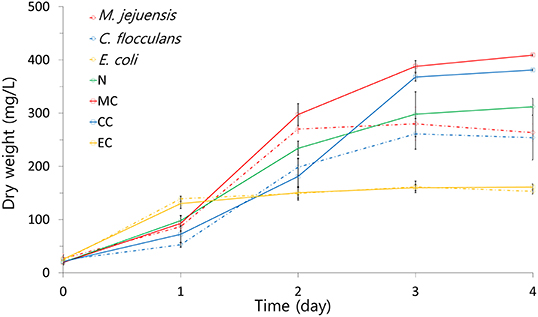
Figure 3. The growth pattern of each culture condition was visualized using dry weight. MC, M. jejuensis and C. sorokiniana; CC, C. flocculans and C. sorokiniana; EC, E. coli and C. sorokiniana; N, C. sorokiniana only, control group. Marked characteristics are annotated in Supplementary Figure 1.
Recently, studies have reported the application of heterotrophic cultivation methods for Chlorella (Fan et al., 2020). Studies have described microalgal species with high sedimentation and methods for improving microalgae harvesting efficiency through flocculation (Demir et al., 2020). Our research was conducted with wastewater and heterotrophic cultivation approaches in mind. Thus, we tried to induce microalgae floc using wastewater-derived or -related bacteria. Chlorella was selected as an experimental microalgal strain due to its smaller cell size than other microalgae, contributing to low sedimentation rates (Cheng and Liu, 2020; Demir et al., 2020; Potocar et al., 2020). Low sedimentation rate is the main limitation for biomass production from Chlorella (Cheng and Liu, 2020; Demir et al., 2020; Potocar et al., 2020); therefore, improving sedimentation can increase the usefulness of Chlorella. Various approaches to improving sedimentation have been attempted (Cheng and Liu, 2020; Demir et al., 2020; Potocar et al., 2020). Furthermore, industrial-scale facilities, such as wastewater treatment plants, require floc-forming and pollutant removal capabilities to remove pollutants. Sludge can be separated efficiently through floc-forming, and water can be purified through the ability of microalgae to remove nitrogen and adsorb heavy metals. Therefore, the floc-forming ability of bacteria and algae is important for the application of industrial-scale facilities (Chatsungnoen and Chisti, 2019; Sajana et al., 2019; Kim and Kwak, 2020; Zhou et al., 2020).
Comparing the growth trends of bacteria and algae co-cultures to pure cultures, better results were observed in all co-cultures, except E. coli (Figure 3). The fact that both MC and CC conditions reached a higher dry weight than the N condition suggests that co-culturing enables more efficient biomass production than algae alone cultured in the same conditions (Cho et al., 2015). However, based on the E. coli co-culture results, not all strains are suitable for co-culture. These growth-related results support improved growth by co-culture of M. jejuensis and C. flocculans with C. sorokiniana; M. jejuensis has the greatest potential to produce biomass efficiently.
Co-culture Effects on Biomass Productivity and Lipid Quality
Biomass productivity was enhanced through the co-culture of bacteria and algae; the measurement results are summarized in Figure 4A. Among the tested conditions, MC had the highest biomass productivity [102.25 ± 0.35 mg/(L·day)]. The biomass productivity of the CC and N conditions was 95.25 ± 0.35 and 78.00 ± 3.89 mg/(L·day), respectively. Unlike the MC and CC co-cultures, which had enhanced biomass productivity compared to the N condition, the EC co-culture produced the lowest biomass of 40.25 ± 1.41 mg/(L·day). Furthermore, co-culturing improved not only biomass productivity, but also lipid productivity (Figure 4A). The MC condition showed the highest lipid productivity [57.80 ± 0.20 mg/(L·day)] in addition to the highest biomass productivity. Conversely, in the CC condition, which showed improved biomass productivity compared to that of the N condition, lipid productivity was inhibited [CC condition, 38.63 ± 0.14 mg/(L·day); N condition, 42.26 ± 2.11 mg/(L·day)]. The EC condition had the lowest lipid productivity at 22.75 ± 0.80 mg/(L·day). Therefore, the MC co-culture was the only condition in which both biomass and lipid productivity improved.
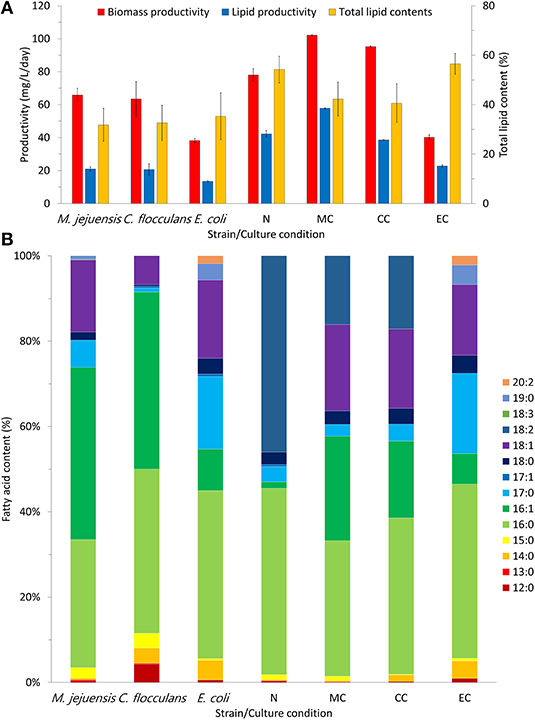
Figure 4. Productivity and fatty acid composition in each culture condition. The productivity of biomass and lipid content in each culture condition was measured (A). Through fatty acid composition analysis, we confirmed the quality of lipids that were produced in each condition (B). MC, M. jejuensis and C. sorokiniana; CC, C. flocculans and C. sorokiniana; EC, E. coli and C. sorokiniana; N, C. sorokiniana only, control group. We herein expressed the fatty acids contents, which we defined as 100%.
Additionally, we measured the changes in biomass lipid composition after improved productivity (Figure 4B). The biomass lipids obtained from co-culture (in MC and CC conditions) consisted mainly of C16:0 (MC condition, 31.80%; CC condition, 36.66%), C16:1 (MC condition, 24.45%; CC condition, 18.00%), C18:1 (MC condition, 20.23%; CC condition, 18.6%), and C18:2 (MC condition, 16.11%; CC condition, 17.14%). The biomass lipids obtained from the pure algal culture (N condition) consisted mainly of C16:0 (43.67%) and C18:2 (45.99%). Notably, the biomass lipids in the EC condition were mainly C16:0 (40.83%), C17:0 (18.80%), and C18:1 (16.57%); the composition was similar to that of pure E. coli (C16:0, 39.37%; C17:0, 17.05%; C18:1, 18.36%). Furthermore, when comparing the experimental groups (MC and CC conditions) to the respective pure bacterial biomass (M. jejuensis and C. flocculans) composed mainly of C16:0 (M. jejuensis, 30.01%; C. flocculans, 38.46%), C16:1 (M. jejuensis, 40.35%; C. flocculans, 41.51%), and C18:1 (M. jejuensis, 16.90%; C. flocculans, 6.74%), C16:1 increased and C18:1 and C18:2 decreased in the experimental group. Thus, the lipids in the co-cultured biomass showed the intermediate characteristics of the pure bacterial and algal lipid compositions.
In heterotrophic conditions, where the cultures were completely dependent on the organic carbon source in the media, a larger amount of biomass was produced in the co-culture than in the pure algal culture (Figure 4A). This phenomenon is contrary to previous reports of competition-induced growth degradation caused by limited organic carbon sources (Yadav and Archer, 1988). However, microorganisms are known to have preferred organic carbon sources, and different types of organic carbon are available depending on the environment (Brückner and Titgemeyer, 2002; Abreu et al., 2012). In addition, some studies suggest that the medium obtained by culturing certain microbial species can be recycled (Loftus and Johnson, 2017; Wang et al., 2018; Li et al., 2019). Therefore, it is expected that there has been no competition to impede growth in the use of organic carbon sources among species in conditions in which higher biomass could be produced (Loftus and Johnson, 2017; Wang et al., 2018; Li et al., 2019). Furthermore, improved lipid productivity may be related to lipid accumulation due to nutrient starvation (Chu et al., 2020; Poh et al., 2020). These results suggest that, although bacteria and algae were competing for a limited energy source, biomass production was more efficient in the co-culture than the pure algal culture (Berthold et al., 2019). However, the total lipid contents of the MC and CC conditions, which showed enhanced biomass productivity, were lower than that of the N condition. Nevertheless, the improved lipid productivity in the MC condition could be attributed to the increased biomass productivity, which improved enough to overcome the lower total lipid contents, unlike the CC condition (Figure 4A). Additionally, the co-culture biomass value was between the total lipid contents of the pure bacteria and the algae-derived biomass, which is presumed because the co-culture biomass was composed of bacterial and algae-derived biomass together (Berthold et al., 2019; Demir et al., 2020). Moreover, the improvement of lipid productivity and biomass suggests that high-quality biomass can be produced through the co-culture of bacteria and algae. Considering the biomass lipid composition results, we observed no extreme dominance between bacteria and algae in co-culture (Figure 4B). The identified biomass of the co-culture mainly consisted of C16:0, C16:1, C18:1, and C18:2, which was similar to the lipid composition of the biomass derived from pure bacteria (C16:0, C16:1, and C18:1) and algae (C16:0 and C18:2) together. The main components are expected to be derived from bacteria (C16:0, C16:1, and C18:1) and algae (C16:0 and C18:2). However, improvement in biomass and lipid productivity cannot be expected from all bacteria (Safonova and Reisser, 2005). Among the tested strains, the biomass and lipid productivity of E. coli in the EC condition was the lowest among the measured values. Moreover, the lipid composition of the EC condition was similar to that of pure E. coli. Based on this fact, it is likely that the EC condition was dominated by E. coli; we can assume that E. coli inhibited algal growth and biomass productivity (Safonova and Reisser, 2005). Therefore, to improve biomass and lipid productivity through the co-culture of bacteria and algae, it is necessary to select the appropriate species. Based on our experimental results, M. jejuensis KCTC 32230 is the most suitable bacteria for co-culture with C. sorokiniana KNUA114 among the strains tested here. Furthermore, our research demonstrates the potential for efficient biomass production through the co-culture of bacteria and algae.
Conclusion
In this study, we demonstrated the value of bacterial- and algal-derived biomass for improving the harvesting efficiency using bacterial and algal floc. Among the bacterial strains tested, M. jejuensis KCTC 32230 formed the largest floc with C. sorokiniana, with the highest sedimentation ability. Furthermore, the M. jejuensis KCTC 32230 co-culture improved biomass and lipid productivity compared with the pure algal culture. However, co-culturing with C. flocculans KCTC 62943 or E. coli dh5α did not increase the productivity of biomass or lipids. Therefore, M. jejuensis KCTC 32230 is the most suitable bacterial strain for biomass production through the co-culturing of bacteria and algae. Our research provides insights into the efficient production and harvesting of biomass through the co-culture of bacteria and algae and highlights the need for suitable strain selection.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author Contributions
Y-SK, J-GK, D-HK, and H-SY designed the project and were involved in the writing of the manuscript. Y-SK and J-GK wrote the manuscript. D-HK and H-SY carried out most of the experiments and analyzed the data. All authors contributed to the article and approved the submitted version.
Funding
This research was supported by a project for Cooperative R&D between Industry, Academy, and Research Institute funded by the Korea Ministry of SMEs and Startups in 2018 Grants No. S2597396 and the Basic Science Research Program through the National Research Foundation of Korea (NRF), funded by the Ministry of Education (2016R1A6A1A05011910 and 2018R1D1A3B07049385), South Korea and also by this research was supported by Kyungpook National University Development Project Research Fund 2018, South Korea.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Jae-Hong Park (Research Institute for Dok-do and Ulleung-do, Kyungpook National University) for supplying algae strains. We also thank Ho-Sung Yoon (Advanced Bio-resource Research Center, Kyungpook National University) for helpful discussions and assistance with the Materials and Methods.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbioe.2020.588210/full#supplementary-material
References
Abreu, A. P., Fernandes, B., Vicente, A. A., Teixeira, J., and Dragone, G. (2012). Mixotrophic cultivation of Chlorella vulgaris using industrial dairy waste as organic carbon source. Bioresour. Technol. 118, 61–66. doi: 10.1016/j.biortech.2012.05.055
Bainbridge, Z. T., Wolanski, E., Álvarez-Romero, J. G., Lewis, S. E., and Brodie, J. E. (2012). Fine sediment and nutrient dynamics related to particle size and floc formation in a Burdekin River flood plume, Australia. Mar. Pollut. Bull. 65, 236–248. doi: 10.1016/j.marpolbul.2012.01.043
Baroni, É. G., Yap, K. Y., Webley, P. A., Scales, P. J., and Martin, G. J. (2019). The effect of nitrogen depletion on the cell size, shape, density and gravitational settling of Nannochloropsis salina, Chlorella sp. (marine) and Haematococcus pluvialis. Algal. Res. 39:101454. doi: 10.1016/j.algal.2019.101454
Benítez, M. B., Champagne, P., Ramos, A., Torres, A. F., and Ochoa-Herrera, V. (2019). Wastewater treatment for nutrient removal with Ecuadorian native microalgae. Environ. Technol. 40, 2977–2985. doi: 10.1080/09593330.2018.1459874
Berthold, D. E., Shetty, K. G., Jayachandran, K., Laughinghouse, I. V. H. D., and Gantar, M. (2019). Enhancing algal biomass and lipid production through bacterial co-culture. Biomass. Bioenerg. 122, 280–289. doi: 10.1016/j.biombioe.2019.01.033
Blattner, F. R., Plunkett, G., Bloch, C. A., Perna, N. T., Burland, V., Riley, M., et al. (1997). The complete genome sequence of Escherichia coli K-12. Science 277, 1453–1462. doi: 10.1126/science.277.5331.1453
Brückner, R., and Titgemeyer, F. (2002). Carbon catabolite repression in bacteria: choice of the carbon source and autoregulatory limitation of sugar utilization. FEMS Microbiol. Lett. 209, 141–148. doi: 10.1016/S0378-1097(02)00559-1
Chatsungnoen, T., and Chisti, Y. (2019). “Flocculation and electroflocculation for algal biomass recovery,” in Biofuels from Algae (Amsterdam), 257–286. doi: 10.1016/B978-0-444-64192-2.00011-1
Chen, C., Huang, Y., Qin, S., Huang, D., Bu, X., and Huang, H. (2020). Slagging tendency estimation of aquatic microalgae and comparison with terrestrial biomass and waste. Energy 194:116889. doi: 10.1016/j.energy.2019.116889
Cheng, D., Ngo, H., Guo, W., Chang, S., Nguyen, D., and Kumar, S. (2019). Microalgae biomass from swine wastewater and its conversion to bioenergy. Bioresour. Technol. 275, 109–122. doi: 10.1016/j.biortech.2018.12.019
Cheng, H., and Liu, Z. (2020). Brachionus plicatilis culture filtrate promotes sedimentation and harvesting of Chlorella. Fung. Appl. Limnol. 193, 253–259. doi: 10.1127/fal/2020/1275
Cheng, Y. S., Zheng, Y., Labavitch, J. M., and VanderGheynst, J. S. (2011). The impact of cell wall carbohydrate composition on the chitosan flocculation of Chlorella. Process Biochem. 46, 1927–1933. doi: 10.1016/j.procbio.2011.06.021
Cho, D. H., Ramanan, R., Heo, J., Lee, J., Kim, B. H., Oh, H. M., et al. (2015). Enhancing microalgal biomass productivity by engineering a microalgal–bacterial community. Bioresour. Technol. 175, 578–585. doi: 10.1016/j.biortech.2014.10.159
Chu, F., Cheng, J., Li, K., Wang, Y., Li, X., and Yang, W. (2020). Enhanced lipid accumulation through a regulated metabolic pathway of phosphorus luxury uptake in the microalga Chlorella vulgaris under nitrogen starvation and phosphorus repletion. Acs. Sustain. Chem. Eng. 8, 8137–8147. doi: 10.1021/acssuschemeng.9b07447
Consortium, U. (2015). UniProt: a hub for protein information. Nucleic Acids Res. 43, D204–212. doi: 10.1093/nar/gku989
Cooney, M., Young, G., and Nagle, N. (2009). Extraction of bio-oils from microalgae. Sep. Purif. Rev. 38, 291–325. doi: 10.1080/15422110903327919
DA LUZ, C. F. P., Nogueira, I. D. S., Barth, O. M., and Silva, C. G. (2002). Differential sedimentation of algae Chlorococcales (Scenedesmus, Coelastrum and Pediastrum) in Lagoa de Cima, Campos dos Goitacazes municipality (Rio de Janeiro, Brazil). Pesquisas em Geociências. 29, 65–75. doi: 10.22456/1807-9806.19606
Dassey, A. J., and Theegala, C. S. (2013). Harvesting economics and strategies using centrifugation for cost effective separation of microalgae cells for biodiesel applications. Bioresour. Technol. 128, 241–245. doi: 10.1016/j.biortech.2012.10.061
Degeest, B., and De Vuyst, L. (2000). Correlation of activities of the enzymes α-phosphoglucomutase, UDP-galactose 4-epimerase, and UDP-glucose pyrophosphorylase with exopolysaccharide biosynthesis by Streptococcus thermophilus LY03. Appl. Environ. Microbiol. 66, 3519–3527. doi: 10.1128/AEM.66.8.3519-3527.2000
Demir, I., Besson, A., Guiraud, P., and Formosa-Dague, C. (2020). Towards a better understanding of microalgae natural flocculation mechanisms to enhance flotation harvesting efficiency. Water. Sci. Technol. 82, 1009–1024. doi: 10.2166/wst.2020.177
Duygu, D. Y., Erkaya, I. A., Erdem, B., and Yalcin, B. (2019). Characterization of silver nanoparticle produced by Pseudopediastrum boryanum (Turpin) E. Hegewald and its antimicrobial effects on some pathogens. Int. J. Environ. Sci. Technol. 16, 7093–7102. doi: 10.1007/s13762-019-02315-5
Edgar, R. C. (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461. doi: 10.1093/bioinformatics/btq461
Fan, J., Chen, Y., Zhang, T. C., Ji, B., and Cao, L. (2020). Performance of Chlorella sorokiniana-activated sludge consortium treating wastewater under light-limited heterotrophic condition. Chem. Eng. J. 382:122799. doi: 10.1016/j.cej.2019.122799
Farooq, W., Lee, Y. C., Han, J. I., Darpito, C. H., Choi, M., and Yang, J. W. (2013). Efficient microalgae harvesting by organo-building blocks of nanoclays. Green Chem. 15, 749–755. doi: 10.1039/c3gc36767c
Furuhashi, T., and Weckwerth, W. (2013). “Introduction to lipid (FAME) analysis in algae using gas chromatography–mass spectrometry,” in The Handbook of Plant Metabolomics (New York, NY: John Wiley & Sons, Inc.) 215–225. doi: 10.1002/9783527669882.ch11
Geesey, G. G., and Van Ommen Kloeke, F. (2004). Extracellular Enzymes Associated with Microbial flocs from Activated Sludge of Wastewater Treatment Systems. Boca Raton, FL: CRC. Press. 295–316. doi: 10.1201/9780203485330.pt3
Hyatt, D., Chen, G. L., LoCascio, P. F., Land, M. L., Larimer, F. W., and Hauser, L. J. (2010). Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11:119. doi: 10.1186/1471-2105-11-119
Jimoh, T. A., Keshinro, M. O., and Cowan, K. A. (2019). Microalgal–bacterial flocs and extracellular polymeric substances: two essential and valuable products of integrated algal pond systems. Water Air Soil Pollut. 230:95. doi: 10.1007/s11270-019-4148-3
Jonker, J., and Faaij, A. (2013). Techno-economic assessment of micro-algae as feedstock for renewable bio-energy production. Appl. Energy 102, 461–475. doi: 10.1016/j.apenergy.2012.07.053
Kanehisa, M., Goto, S., Sato, Y., Kawashima, M., Furumichi, M., and Tanabe, M. (2014). Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res. 42, D199–D205. doi: 10.1093/nar/gkt1076
Kim, D. H., Han, K. I., Kwon, H. J., Kim, M. G., Kim, Y. G., Choi, D. H., et al. (2019). Complete genome sequence of Comamonas sp. NLF-7-7 isolated from biofilter of wastewater treatment plant. Korean. J. Microbiol. 55, 309–312. doi: 10.7845/kjm.2019.9099
Kim, D. H., Han, K. I., Kwon, H. J., Kim, M. G., Kim, Y. G., Choi, D. H., et al. (2020). Comamonas flocculans sp. nov., a floc-forming bacterium isolated from livestock wastewater. Curr. Microbiol. 77, 1902–1908. doi: 10.1007/s00284-020-01940-5
Kim, J. Y., Park, S. H., Lee, D. H., Song, G., and Kim, Y. J. (2018). Melaminivora jejuensis sp. nov., isolated from Swinery waste. Int. J.Syst. Evol. Microbiol. 68, 9–13. doi: 10.1099/ijsem.0.002294
Kim, M. S., and Kwak, D. H. (2020). Auto/bio-flocculation conditions to separate algal particles without chemical coagulants for flotation and sedimentation processes. Sep. Sci. Technol. 55, 1185–1196. doi: 10.1080/01496395.2019.1579842
Kwon, G., Kim, H., Song, C., and Jahng, D. (2019). Co-culture of microalgae and enriched nitrifying bacteria for energy-efficient nitrification. Biochem. Eng. J. 152:107385. doi: 10.1016/j.bej.2019.107385
Lee, J., Cho, D. H., Ramanan, R., Kim, B. H., Oh, H. M., and Kim, H. S. (2013). Microalgae-associated bacteria play a key role in the flocculation of Chlorella vulgaris. Bioresour. Technol. 131, 195–201. doi: 10.1016/j.biortech.2012.11.130
Lee, Y. K. (2001). Microalgal mass culture systems and methods: their limitation and potential. J. Appl. Phycol. 13, 307–315. doi: 10.1023/A:1017560006941
Leite, L. d. S., and Daniel, L. A. (2020). Optimization of microalgae harvesting by sedimentation induced by high pH. Water Sci. Technol. 82, 1227–1236. doi: 10.2166/wst.2020.106
Li, Y., Zhang, Z., Duan, Y., and Wang, H. (2019). The effect of recycling culture medium after harvesting of Chlorella vulgaris biomass by flocculating bacteria on microalgal growth and the functionary mechanism. Bioresour. Technol. 280, 188–198. doi: 10.1016/j.biortech.2019.01.149
Loftus, S. E., and Johnson, Z. I. (2017). Cross-study analysis of factors affecting algae cultivation in recycled medium for biofuel production. Algal. Res. 24, 154–166. doi: 10.1016/j.algal.2017.03.007
Mishra, S. K., Suh, W. I., Farooq, W., Moon, M., Shrivastav, A., Park, M. S., et al. (2014). Rapid quantification of microalgal lipids in aqueous medium by a simple colorimetric method. Bioresour. Technol. 155, 330–333. doi: 10.1016/j.biortech.2013.12.077
Morales, M., Collet, P., Lardon, L., Hélias, A., Steyer, J. P., and Bernard, O. (2019). “Life-cycle assessment of microalgal-based biofuel,” in Biofuels from Algae (Amsterdam: Elsevier Science B.V.) 507–550. doi: 10.1016/B978-0-444-64192-2.00020-2
Overbeek, R., Begley, T., Butler, R. M., Choudhuri, J. V., Chuang, H. Y., Cohoon, M., et al. (2005). The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res. 33, 5691–5702. doi: 10.1093/nar/gki866
Petrini, S., Foladori, P., Beghini, F., Armanini, F., Segata, N., and Andreottola, G. (2020). How inoculation affects the development and the performances of microalgal-bacterial consortia treating real municipal wastewater. J. Environ. Manage. 26:110427. doi: 10.1016/j.jenvman.2020.110427
Poh, Z. L., Kadir, W. N. A., Lam, M. K., Uemura, Y., Suparmaniam, U., Lim, J. W., et al. (2020). The effect of stress environment towards lipid accumulation in microalgae after harvesting. Renew. Energy 154, 1083–1091. doi: 10.1016/j.renene.2020.03.081
Potocar, T., de Souza Leite, L., Daniel, L. A., Pivokonsky, M., Matoulkova, D., and Branyik, T. (2020). Cooking oil-surfactant emulsion in water for harvesting Chlorella vulgaris by sedimentation or flotation. Bioresour. Technol. 311:123508. doi: 10.1016/j.biortech.2020.123508
Powell, S., Forslund, K., Szklarczyk, D., Trachana, K., Roth, A., Huerta-Cepas, J., et al. (2014). eggNOG v4. 0: nested orthology inference across 3686 organisms. Nucleic. Acids. Res. 42, D231–D239. doi: 10.1093/nar/gkt1253
Reasoner, D. J., and Geldreich, E. (1985). A new medium for the enumeration and subculture of bacteria from potable water. Appl. Environ. Microbiol. 49, 1–7. doi: 10.1128/AEM.49.1.1-7.1985
Safonova, E., and Reisser, W. (2005). Growth promoting and inhibiting effects of extracellular substances of soil microalgae and cyanobacteria on Escherichia coll and Micrococcus luteus. Phycol. Res. 53, 189–193. doi: 10.1111/j.1440-1835.2005.tb00370.x
Sajana, T., Pandit, S., Jadhav, D. A., Abdullah-Al-Mamun, M., and Fosso-Kankeu, E. (2019). “Sediment microbial fuel cell for wastewater treatment: a new approach,” in Nano and Bio-Based Technologies for Wastewater Treatment (New York, NY: John Wiley & Sons, Inc.) 303. doi: 10.1002/9781119577119.ch9
Sajjadi, B., Chen, W. Y., Raman, A. A. A., and Ibrahim, S. (2018). Microalgae lipid and biomass for biofuel production: a comprehensive review on lipid enhancement strategies and their effects on fatty acid composition. Renew. Sust. Energ. Rev. 97, 200–232. doi: 10.1016/j.rser.2018.07.050
Sojka, R., Carter, D., and Brown, M. (1992). Imhoff cone determination of sediment in irrigation runoff. Soil. Sci. Soc. Am. J. 56, 884–890. doi: 10.2136/sssaj1992.03615995005600030034x
Stone, M., and Krishnappan, B. (2003). Floc morphology and size distributions of cohesive sediment in steady-state flow. Water. Res. 37, 2739–2747. doi: 10.1016/S0043-1354(03)00082-4
Stone, S., Adams, M., Stauber, J., Jolley, D. F., and Warne, M. S. J. (2019). Development and application of a multispecies toxicity test with tropical freshwater microalgae. Environ. Pollut. 250, 97–106. doi: 10.1016/j.envpol.2019.03.058
Su, Y., Mennerich, A., and Urban, B. (2012). Comparison of nutrient removal capacity and biomass settleability of four high-potential microalgal species. Bioresour. Technol. 124, 157–162. doi: 10.1016/j.biortech.2012.08.037
Tan, C. H., Nagarajan, D., Show, P. L., and Chang, J. S. (2019). “Biodiesel from microalgae,” in Biofuels: Alternative feedstocks and Conversion Processes for the Production of Liquid and Gaseous Biofuels, (Elsevier). 601–628. doi: 10.1016/B978-0-12-816856-1.00025-7
Wang, X., Lin, L., Lu, H., Liu, Z., Duan, N., Dong, T., et al. (2018). Microalgae cultivation and culture medium recycling by a two-stage cultivation system. Front. Env. Sci. Eng. 12:14. doi: 10.1007/s11783-018-1078-z
Wang, Y., and Qian, P. Y. (2009). Conservative fragments in bacterial 16S rRNA genes and primer design for 16S ribosomal DNA amplicons in metagenomic studies. PLoS ONE 4:e7401. doi: 10.1371/journal.pone.0007401
Wieczorek, N., Kucuker, M. A., and Kuchta, K. (2015). Microalgae-bacteria flocs (MaB-Flocs) as a substrate for fermentative biogas production. Bioresour. Technol. 194, 130–136. doi: 10.1016/j.biortech.2015.06.104
Yadav, V., and Archer, D. (1988). Specific inhibition of sulphate-reducing bacteria in methanogenic co-culture. Lett. Appl. Microbiol. 7, 165–168. doi: 10.1111/j.1472-765X.1988.tb01270.x
Yeo, I. J., Jeong, J. E., Cho, Y. J., Hong, J. W., Yoon, H. S., Kim, S. H., et al. (2011). Characterization and comparison of biodiesels made from Korean freshwater algae. B. Korean Chem. Soc. 32, 2830–2832. doi: 10.5012/bkcs.2011.32.8.2830
Yoo, C., Jun, S. Y., Lee, J. Y., Ahn, C. Y., and Oh, H. M. (2010). Selection of microalgae for lipid production under high levels carbon dioxide. Bioresour. Technol. 101, S71–S74. doi: 10.1016/j.biortech.2009.03.030
Yun, H. S., Kim, Y. S., and Yoon, H. S. (2020). Characterization of Chlorella sorokiniana and Chlorella vulgaris fatty acid components under a wide range of light intensity and growth temperature for their use as biological resources. Heliyon 6:e04447. doi: 10.1016/j.heliyon.2020.e04447
Zhang, H., Yang, L., Zang, X., Cheng, S., and Zhang, X. (2019). Effect of shear rate on floc characteristics and concentration factors for the harvesting of Chlorella vulgaris using coagulation-flocculation-sedimentation. Sci. Total. Environ. 688, 811–817. doi: 10.1016/j.scitotenv.2019.06.321
Zhao, T., Zhang, Y., Wu, H., Wang, D., Chen, Y., Zhu, M. J., et al. (2018). Extracellular aminopeptidase modulates biofilm development of Pseudomonas aeruginosa by affecting matrix exopolysaccharide and bacterial cell death. Environ. Microbiol. Rep. 10, 583–593. doi: 10.1111/1758-2229.12682
Keywords: Chlorella sorokiniana, flocculation, Melaminivora jejuensis, sedimentation, biomass harvesting
Citation: Kim D-H, Yun H-S, Kim Y-S and Kim J-G (2020) Effects of Co-culture on Improved Productivity and Bioresource for Microalgal Biomass Using the Floc-Forming Bacteria Melaminivora Jejuensis. Front. Bioeng. Biotechnol. 8:588210. doi: 10.3389/fbioe.2020.588210
Received: 30 July 2020; Accepted: 30 November 2020;
Published: 18 December 2020.
Edited by:
Fu-Li Li, Qingdao Institute of Bioenergy and Bioprocess Technology (CAS), ChinaReviewed by:
Changhong Yao, Sichuan University, ChinaApilak Salakkam, Khon Kaen University, Thailand
Copyright © 2020 Kim, Yun, Kim and Kim. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Young-Saeng Kim, a3lzbGhoMTIyOEBoYW5tYWlsLm5ldA==; Jong-Guk Kim, a2ltamdAa251LmFjLmty
†These authors have contributed equally to this work
 Dong-Hyun Kim1†
Dong-Hyun Kim1† Young-Saeng Kim
Young-Saeng Kim