- 1Department of Agricultural Biotechnology, Research Institute of Agriculture and Life Science, Seoul National University, Seoul, South Korea
- 2School of Food Science and Biotechnology, Kyungpook National University, Daegu, South Korea
- 3Department of Environment Science and Biotechnology, Jeonju University, Jeonju, South Korea
- 4Department of Food Science, Purdue University, West Lafayette, IN, United States
- 5Department of Food Science and Human Nutrition, University of Illinois at Urbana-Champaign, Urbana, IL, United States
- 6Department of Biotechnology, Graduate School, Korea University, Seoul, South Korea
Being a microbial host for lignocellulosic biofuel production, Saccharomyces cerevisiae needs to be engineered to express a heterologous xylose pathway; however, it has been challenging to optimize the engineered strain for efficient and rapid fermentation of xylose. Deletion of PHO13 (Δpho13) has been reported to be a crucial genetic perturbation in improving xylose fermentation. A confirmed mechanism of the Δpho13 effect on xylose fermentation is that the Δpho13 transcriptionally activates the genes in the non-oxidative pentose phosphate pathway (PPP). In the current study, we found a couple of engineered strains, of which phenotypes were not affected by Δpho13 (Δpho13-negative), among many others we examined. Genome resequencing of the Δpho13-negative strains revealed that a loss-of-function mutation in GCR2 was responsible for the phenotype. Gcr2 is a global transcriptional factor involved in glucose metabolism. The results of RNA-seq confirmed that the deletion of GCR2 (Δgcr2) led to the upregulation of PPP genes as well as downregulation of glycolytic genes, and changes were more significant under xylose conditions than those under glucose conditions. Although there was no synergistic effect between Δpho13 and Δgcr2 in improving xylose fermentation, these results suggested that GCR2 is a novel knockout target in improving lignocellulosic ethanol production.
Introduction
Lignocellulosic biofuels are renewable liquid-fuel alternatives owing to abundant feedstock availability and substantial CO2 emission reduction (Lynd, 2017). Saccharomyces cerevisiae plays an essential role in the production of lignocellulosic biofuels by fermenting lignocellulosic sugars, mainly glucose and xylose, which requires engineering of the yeast via a heterologous xylose pathway (Kim et al., 2013c; Richa et al., 2019). Current efforts on the metabolic engineering of yeast remain focused on improving the xylose fermentation yield and productivity under multiple stress conditions of lignocellulosic biomass hydrolyzates (Jeong et al., 2020; Qin et al., 2020).
Previously, an efficient xylose-fermenting strain of S. cerevisiae (SR8) was developed through the introduction of a heterologous xylose pathway, optimization of its expression levels, and adaptive evolution, which resulted in a loss-of-function mutation in PHO13 (Kim et al., 2013d; Jeong et al., 2020). Continued efforts have discovered that the deletion of PHO13 (Δpho13) resulted in transcriptional and metabolic changes favorable to xylose and other C5 sugar fermentation (Kim et al., 2015; Xu et al., 2016; Ye et al., 2019). Although PHO13 was discovered as a knockout target to improve xylose fermentation a decade ago (Ni et al., 2007; Van Vleet et al., 2008), detailed molecular mechanisms remain unelucidated. To date, the most advanced finding is that Δpho13 results in the transcriptional activation of non-oxidative pentose phosphate pathway (PPP) genes, which therefore facilitates xylose metabolism (Xu et al., 2016).
The present study found that the Δpho13 effect on xylose fermentation was not observed in a couple of strains (the YSX3 and DA24 strains; the Δpho13-negative phenotype), and a loss-of-function mutation in GCR2 was responsible. Gcr2, as a transcriptional regulator of glycolysis, enhances the CT box-dependent transcriptional activation of a Rap1-Gcr1 complex required for the expression of glycolytic genes (Huie et al., 1992). Rap1 and Gcr1 provide specific DNA-binding and activation of glycolytic and ribosomal genes, respectively (Uemura and Fraenkel, 1990; Sasaki et al., 2005). Gcr2 functions as an activation domain for the Gcr1p–Gcr2p complex mediating high level of glycolytic gene expression (Uemura and Jigami, 1992; Cha et al., 2020). However, it is unknown how the Gcr2-associated regulatory systems would function when new pathways, such as those involving the heterologous xylose-assimilating genes, are introduced.
The aim of the current study is to discover genetic factors associated with the Δpho13-negative phenotype that do not affect xylose fermentation regardless of the Δpho13 and to identify its molecular mechanisms. The results of the present study suggest that a native transcriptional regulator Gcr2 contributed to the suboptimal xylose fermentation of engineered S. cerevisiae.
Materials and Methods
Strain Construction
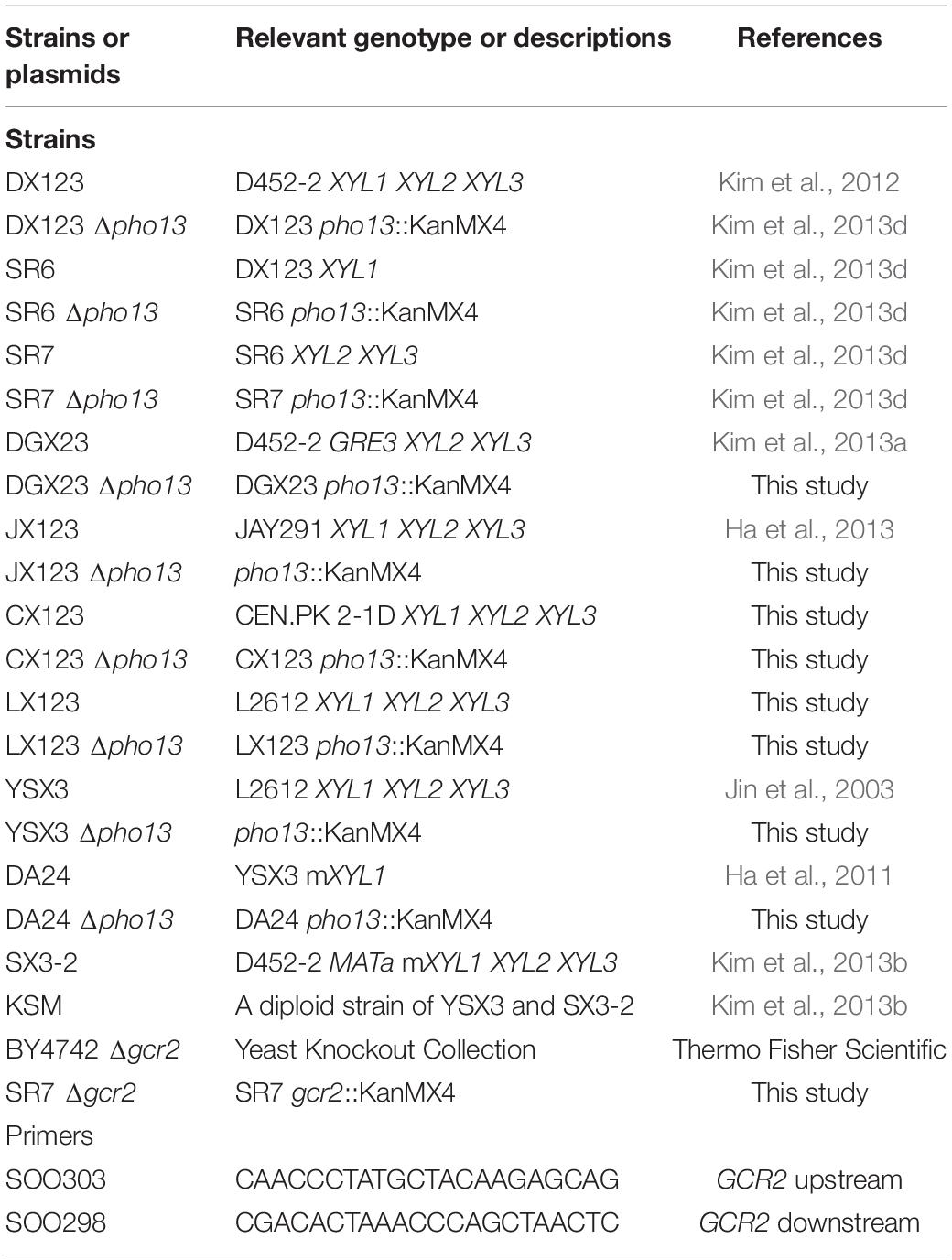
All S. cerevisiae strains used in the present study are listed in Table 1. To construct xylose-fermenting strains, the linear expression cassette of Scheffersomyces stipitis XYL1, XYL2, and XYL3 was used as previously described (Kim et al., 2013d). Parental strains (D452-2, JAY291, CENPK. 2-1D, and L2612) and their derivatives expressing the xylose assimilation pathway were tested for the pho13 effect. To delete PHO13 (GenBank accession number NC_001136) in the xylose-fermenting strains, the pho13:KanMX4 cassette was used as previously described (Kim et al., 2013d). To isolate spores from the KSM diploid strain, tetrad dissection was performed as previously described (Kim et al., 2017). To delete GCR2 (GenBank accession number NC_001146), the gcr2::KanMX4 cassette was amplified from the genomic DNA of the BY4742 Δgcr2 strain (clone ID: 12013) of the Yeast Knockout Collection (Thermo Fisher Scientific, United States) by polymerase chain reaction (PCR) using SOO303/298 primers. The PCR product was purified and integrated into the genome of SR7 strain using the LiAc transformation method (Gietz and Schiestl, 2007). The resulting deletion mutant was selected on an agar medium containing 10 g/L yeast extract, 20 g/L peptone, 20 g/L glucose (YPD), 15 g/L agar, and 300 mg/mL G418 sulfate (GoldBio, St. Louis, MO, United States).
Culture Conditions
A colony of yeast cells was pre-cultured in 5 mL YPD for 24 h at 30°C and 250 rpm. The culture containing 25 or 2.5 mg cells was centrifuged at 15,000 rpm for 1 min at 4°C. The cells were resuspended in 50 mL YPD (40 g/L glucose) or YPX (40 g/L xylose) in a 250 mL Erlenmeyer flask, and the culture with an initial dry cell weight of 0.5 or 0.05 g/L was incubated at 30°C and 80 rpm. All experiments were performed in triplicates.
Fermentation Profiles
Cell growth was monitored at 600 nm using a spectrophotometer (OD600). The concentrations of substrates and metabolites were determined by high-performance liquid chromatography (Agilent Technologies 1260 Series, Santa Clara, CA, United States) equipped with a refractive index detector using the Rezex ROA-Organic Acid H+ (8%) column (Phenomenex, Inc., Torrance, CA, United States). The column was eluted with 0.005 N H2SO4 at a flow rate of 0.6 mL/min and 50°C.
Genome Sequencing
For single nucleotide polymorphism (SNP) discovery in the Δpho13-negative strain (YSX3), the genome of the YSX3 strain and its parental strain (L2612) were resequenced as previously described (Kim et al., 2013d). Briefly, genomic DNA was prepared using the YeaStar Genomic DNA Kit (Zymo Research), and the DNA quality was evaluated on a 1% agarose gel. The genomic DNA samples were then sequenced using an Illumina HiSeq 2500 system, and the sequencing results were analyzed using the CLC Genomic Workbench (version 6.5) program for SNP detection.
RNA-seq
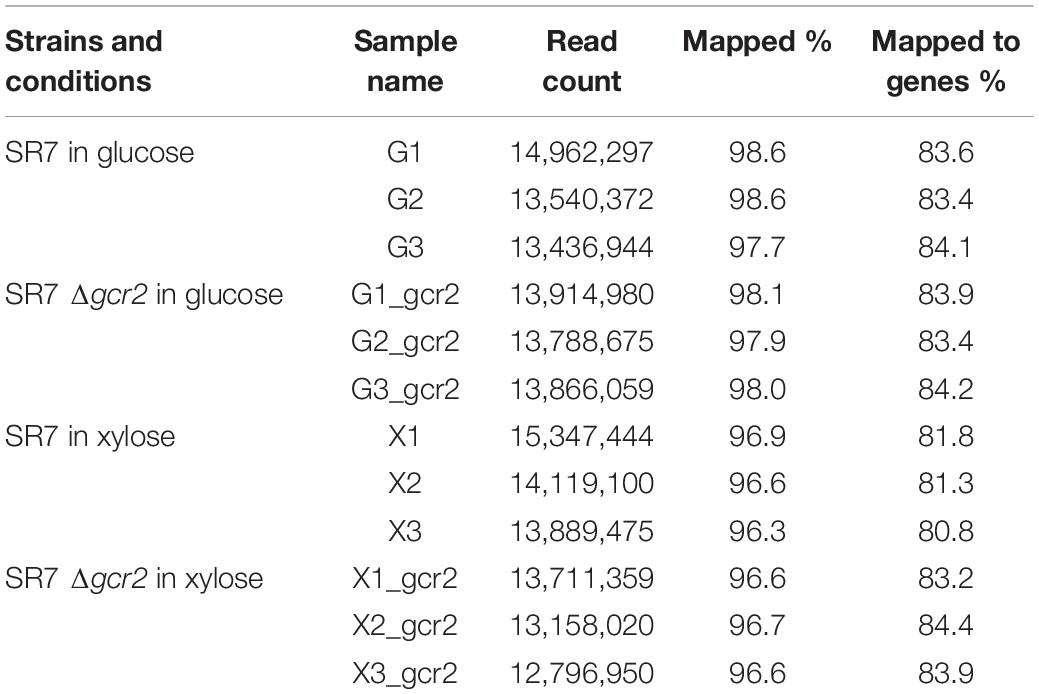
For transcriptomic analysis, RNA-seq was performed as previously described (Kim et al., 2015). Briefly, RNA was extracted from exponentially growing 0.5 mg cells of the control strain (SR7) and Δgcr2 mutant (SR7 Δgcr2) on glucose or xylose using the Qiagen RNeasy Mini Kit, and the RNA quality was evaluated using a Bioanalyzer RNA chip. The samples with high-quality total RNA were sequenced using an Illumina HiSeq 2000 system. The sequencing results were then analyzed using the CLC Genomic Workbench (version 6.5) to investigate RNA-seq quality, differentially expressed (DE) genes, and gene set enrichment analysis (GSEA). GSEA is a computational method determining statistical significance and concordant differences of defined sets of genes (Subramanian et al., 2005). Fold changes were calculated based on the total number of exon reads per kilobase of exon length per million mapped reads (RPKM) between SR7 and SR7 Δgcr2 strains.
Data Availability
Whole genome sequencing data of S. cerevisiae L2612 and YSX3 strains are available under NCBI BioProject PRJNA689417 and BioSample accessions (SAMN17207998 and SAMN17207999). RNA-seq data of S. cerevisiae SR7 and SR7 Δgcr2 strains are available under NCBI BioProject PRJNA689538 and BioSample accessions (SAMN17208951–SAMN17208954).
Results
Δpho13-Negative Phenotype Was Found in a Few Xylose-Fermenting Engineered Strains
As previously reported, Δpho13 improves the xylose fermentation capability of engineered strains of S. cerevisiae (Ni et al., 2007; Van Vleet et al., 2008), and other studies have confirmed this with different strain backgrounds (Fujitomi et al., 2012; Kim et al., 2013d; Jeong et al., 2020). Although the detailed molecular mechanism remains unknown, Δpho13 causes transcriptional activation of the genes involved in non-oxidative PPP (Kim et al., 2015; Ye et al., 2019) and reduction of the dephosphorylation product of sedoheptulose-7-phosphate, suggesting the phosphatase activity of Pho13 (Xu et al., 2016). To further explore Δpho13-mediated metabolic regulation, the Δpho13 effect was tested with a broader range of strains that we have constructed, as listed in Figure 1A and Table 1. When a plasmid expressing heterologous xylose pathway (XYL1, XYL2, and XYL3 derived from S. stipitis) was introduced to four different strain backgrounds, all the resulting strains (DX123, JX123, CX123, and LX123) and their derivatives (SR6, SR7, and DGX23) showed an improved xylose consumption rate by Δpho13. However, two strains, YSX3 and its derivative (DA24) did not show any improvement in xylose consumption rate by Δpho13. The LX123 and YSX3 strains have been engineered to express a heterologous xylose pathway using S. cerevisiae L2612 as a parental strain. Thus, unlike LX123, the Δpho13-negative phenotype of YSX3 can be considered independent of the strain background. Because the YSX3 strain underwent multiple transformations and vigorous screening processes for strain optimization previously (Jin et al., 2003), we hypothesized that some spontaneous mutations were acquired in YSX3, which might be responsible for its Δpho13-negative phenotype.
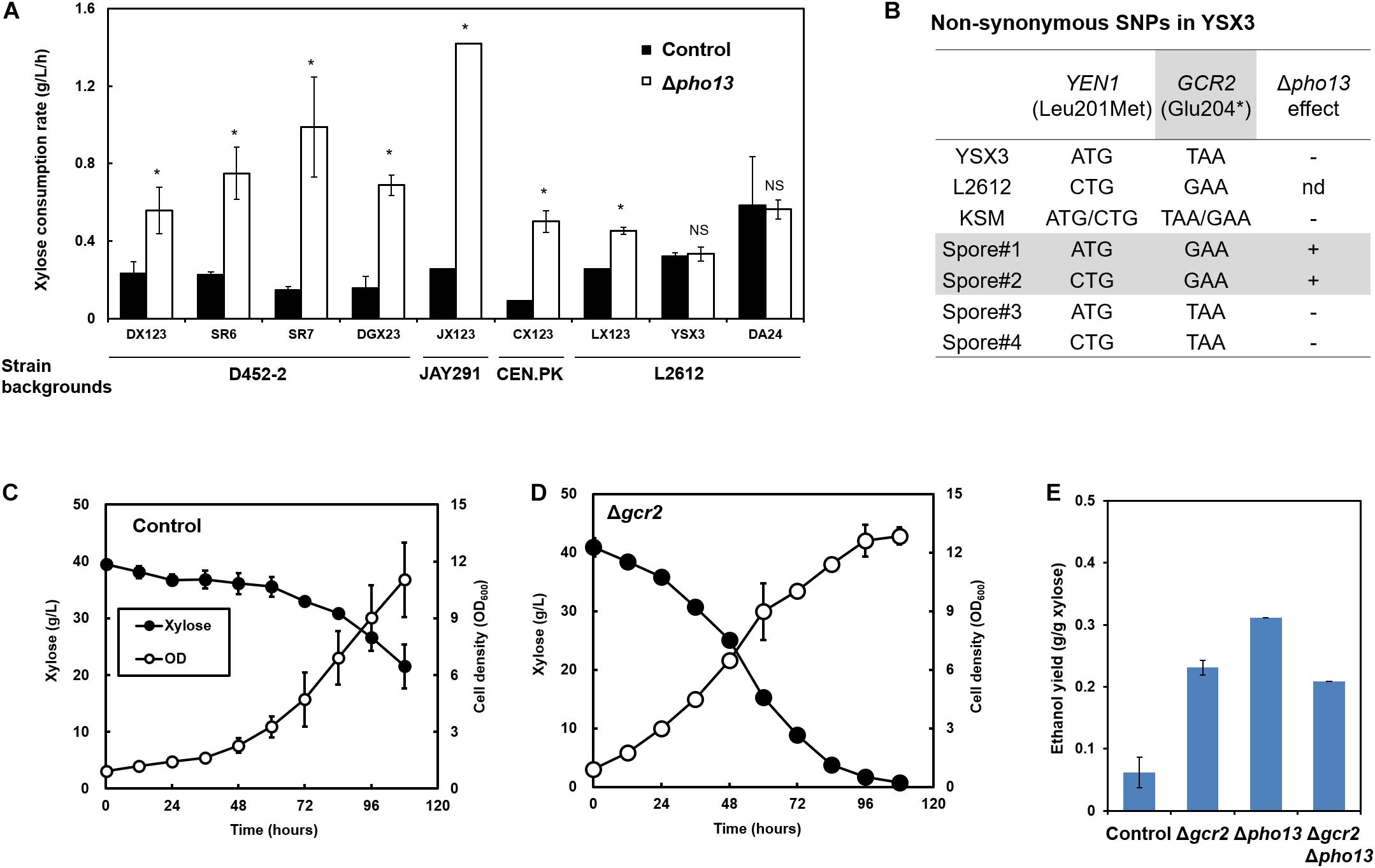
Figure 1. Loss-of-function mutation in GCR2 is responsible for the Δpho13-negative phenotype in xylose-fermenting S. cerevisiae strains. (A) Improved xylose consumption rates by the deletion of PHO13 (Δpho13) in engineered strains with different strain backgrounds, except for YSX3 and DA24 strains. (B) Genome sequencing results of the YSX3 and L2612 strains and Sanger sequencing results of haploid spores derived from the KSM diploid (YSX3 × a derivative of D452-2). (C–E) Xylose fermentation profiles by the SR7 strain (control) and its gene deletion mutants (Δgcr2, Δpho13, and Δgcr2/Δpho13). Fermentation was performed with an initial cell density of 0.5 g/L in YP medium containing 40 g/L xylose under microaerobic conditions. *p < 0.05; NS, not statistically significant; nd, not determined.
Loss-of-Function Mutation in GCR2 Is Responsible for the Δpho13-Negative Phenotype
To identify the molecular mechanism of the Δpho13-negative phenotype of the YSX3 strain, genome sequencing of the YSX3 strain and its parental strain (L2612) was performed. Among the 44 non-synonymous SNPs identified (Supplementary Table 1), only 2 SNPs in YEN1 (YER041W, 601C > A substitution) and GCR2 (YNL199C, 610G > T substitution) had a high frequency (>98%) with deep coverage (172 copies), suggesting that the two SNPs are not likely sequencing errors. Sanger sequencing of the PCR-amplified genes confirmed that the two SNPs are present in YSX3 but not in L2612 (Figure 1B).
To further identify the necessity of the two SNPs in the Δpho13-negative phenotype, the KSM diploid was constructed using two haploids, YSX3 (Δpho13-negative) and SX3. The SX3 strain is a D452-2 derivative, in which Δpho13 improves its phenotypes on xylose (Δpho13-positive). From tetrad dissection of the KSM strain, four spores were obtained. Based on xylose fermentation phenotypes of the spores, it was noted that two spores had the Δpho13-positive phenotype, whereas the other two spores had the Δpho13-negative phenotype. Sanger sequencing of the spores revealed that both Δpho13-positive spores had wild-type GCR2, whereas Δpho13-negative spores had the mutant GCR2. Therefore, it was concluded that the Δpho13-negative phenotype is strongly associated with the mutant GCR2 (YNL199C, 610G > T substitution).
Because the mutation in GCR2 resulted in the truncation of the protein (Glu204∗), we assumed that it is a loss-of-function mutation. To test the hypothesis, the deletion of GCR2 (Δgcr2) was tested in a different strain background (SR7). Consequently, the SR7 Δgcr2 strain showed faster xylose consumption and higher ethanol production than the control SR7 strain (Figures 1C,D). Also, the GCR2 deletion improved ethanol yield of xylose fermentation from 0.06 g ethanol/g xylose (SR7) to 0.23 g/g (SR7 Δgcr2) (Figure 1E). However, the degree of improvement by the GCR2 deletion was not as great as that by the PHO13 deletion; SR7 Δpho13 showed the highest ethanol yield (0.31 g/g). Moreover, the GCR2 deletion in the SR7 Δpho13 strain did not further improve the ethanol yield. In other words, there was no synergistic effect by double deletion of GCR2 and PHO13. These results suggested that the loss-of-function mutation in GCR2 is responsible for the Δpho13-negative phenotype of the YSX3 strain. Meanwhile, it can be concluded that GCR2 is a novel deletion target to improve xylose fermentation.
Global Transcriptional Changes Induced by Δgcr2
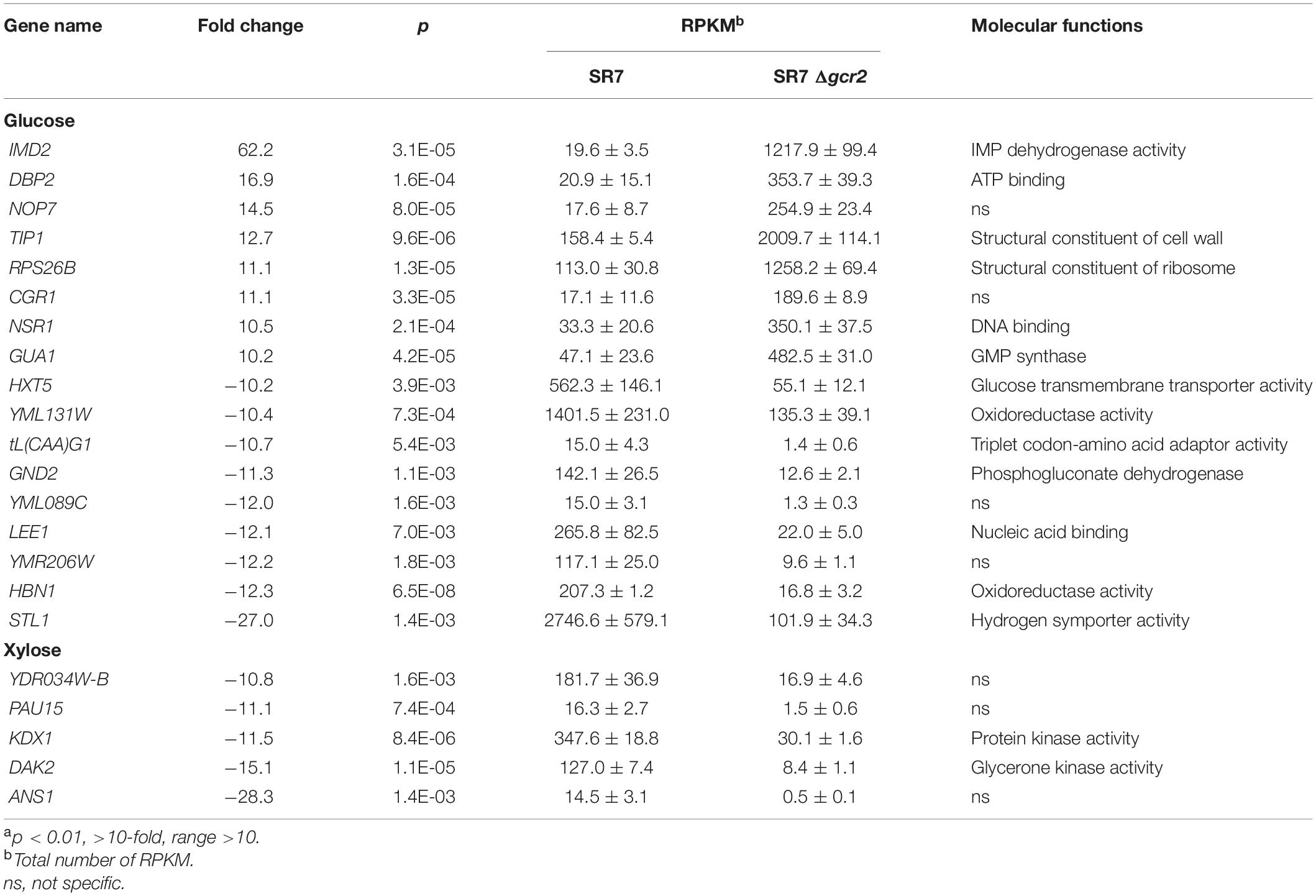
GCR2 encodes a transcriptional activator of glycolytic genes; therefore, its deletion leads to the transcriptional downregulation of the glycolytic genes and upregulation of the citric acid cycle genes during glucose metabolism (Uemura and Jigami, 1992; Sasaki and Uemura, 2005; Fendt et al., 2010). To investigate the transcriptional changes cause by Δgcr2 during xylose metabolism, S. cerevisiae SR7 and SR7 Δgcr2 strains cultured on glucose or xylose were subjected to RNA-seq, and high-quality sequencing data were obtained (Table 2). Hierarchical clustering and multivariate analysis based on Pearson’s correlation and principal component analysis, respectively, indicated that the transcriptomic profiles of glucose and xylose metabolism were the primary determinants (Figure 2). Notably, the Δgcr2 mutant samples were clustered separately from control samples on both glucose and xylose fermentation conditions, suggesting global transcriptional changes evoked by Δgcr2 regardless of the type of substrate. Meanwhile, the number of DE genes in the Δgcr2 mutant compared with those in the control (p < 0.05, >2-fold) was 1638 and 605 on glucose and xylose, respectively. Moreover, the most significant DE genes in the Δgcr2 mutant compared with those in the control (p < 0.01, >10-fold) were 17 and 5 on glucose and xylose, respectively (Table 3). The larger number of DE genes and the more significant fold changes under glucose conditions suggest that Gcr2 is responsible for higher global transcriptional regulation of glucose metabolism than that of xylose metabolism.
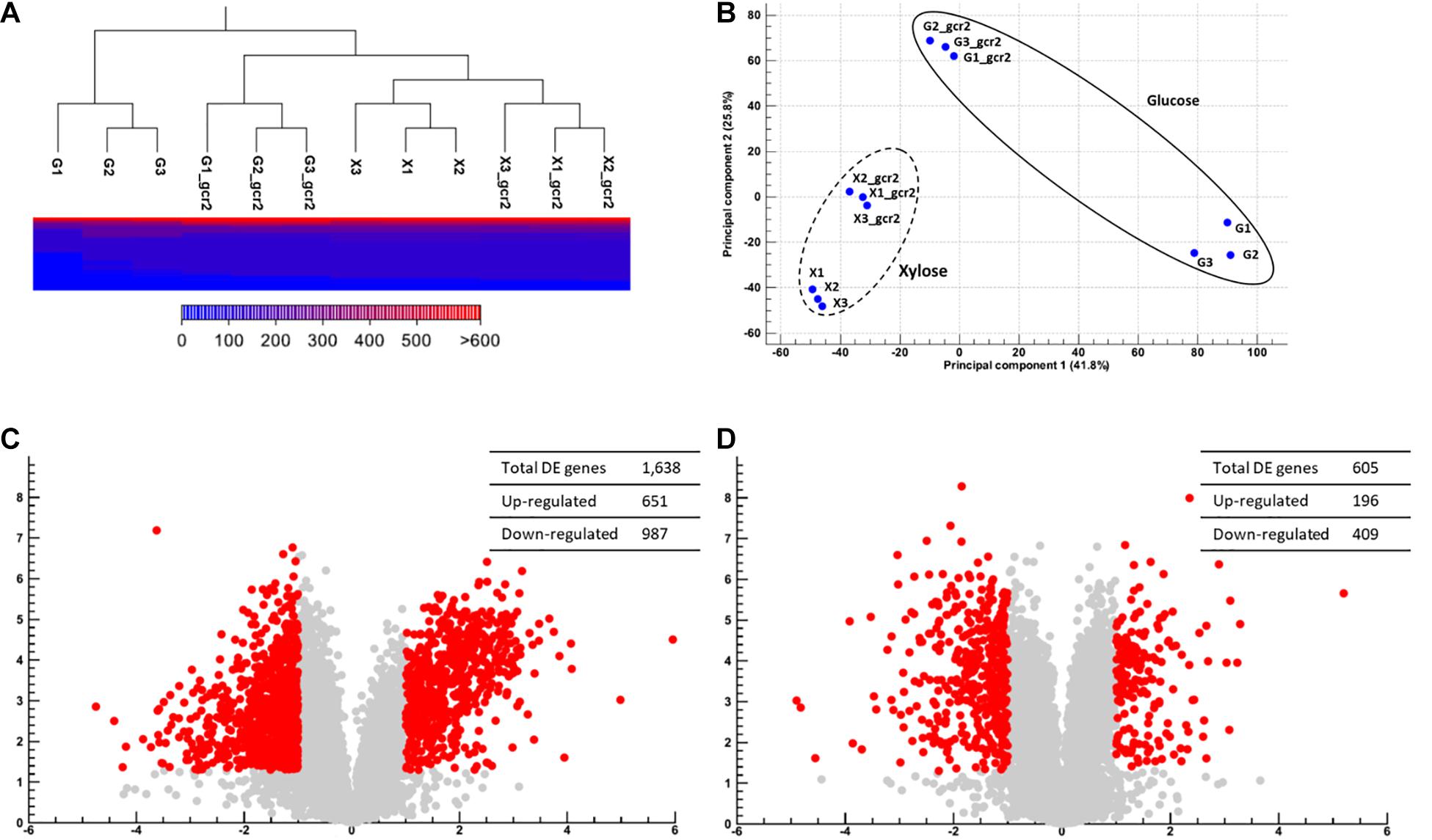
Figure 2. Global transcriptional changes induced by Δgcr2. Hierarchical clustering and multivariate analysis based on Pearson’s correlation (A) and principal component analysis (B). DE genes (p < 0.05, >2-fold) on glucose (C) and on xylose (D) were identified. RNA samples were extracted from exponentially growing SR7 and SR7 Δgcr2 strains on glucose and xylose.
GSEA of DE Genes by Δgcr2
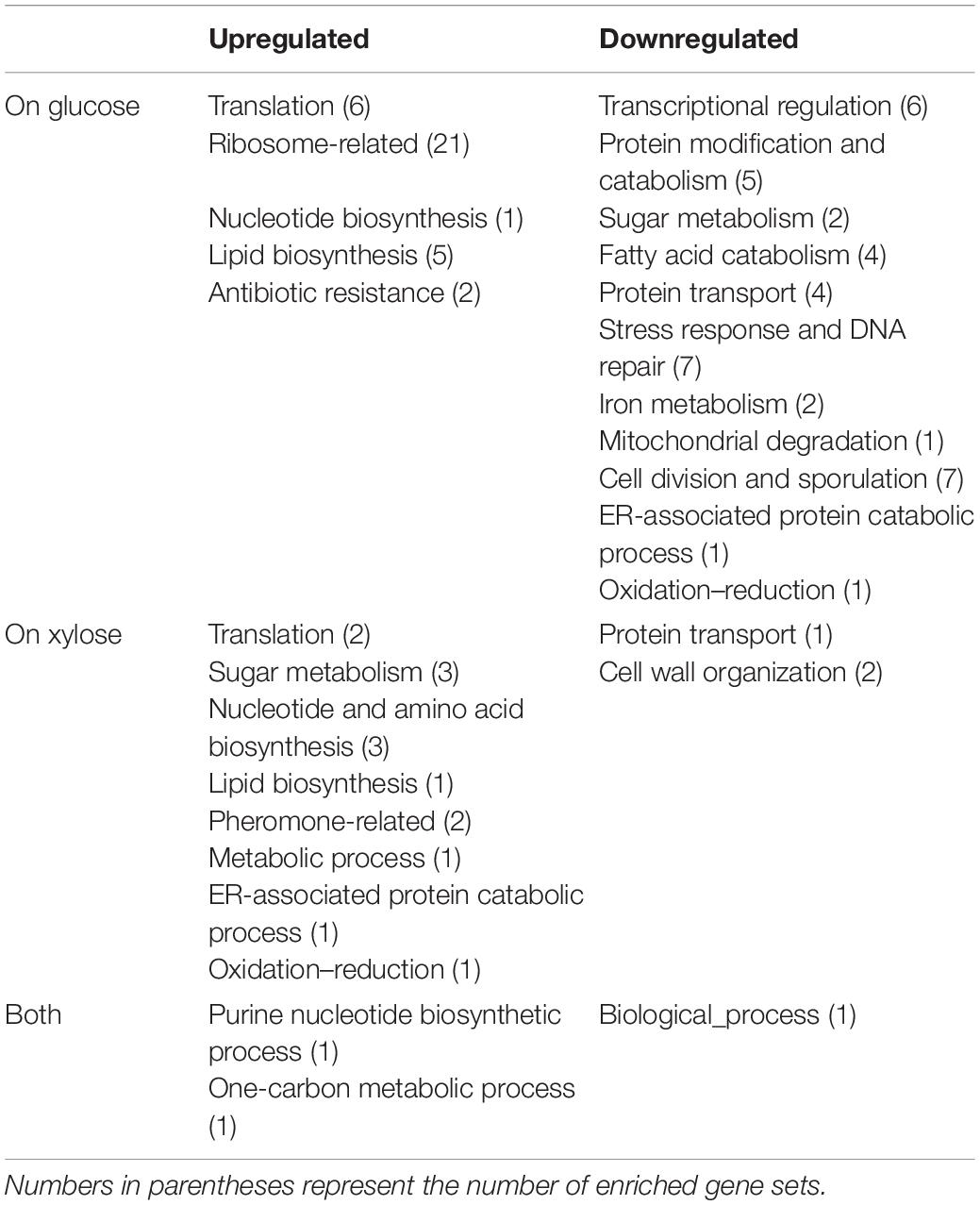
Differentially expressed genes in Δgcr2 were subjected to GSEA using Gene Ontology (GO) biological process (Table 4). Under both glucose and xylose conditions, genes associated with translation, nucleotide biosynthesis, lipid biosynthesis, and one-carbon metabolism were upregulated and those associated with protein transport were downregulated. However, the direction of the transcriptional changes by Δgcr2 in two gene sets (sugar metabolism and oxidation–reduction) were opposite depending on the type of substrates; they were upregulated with xylose as substrate but downregulated with glucose as substrate (Figure 3). For example, ALD3, encoding aldehyde dehydrogenase, is induced in response to stress; it was induced by Δgcr2 under xylose conditions but repressed under glucose conditions. Because Gcr2 is a native transcriptional activator for glucose metabolism, the heterologous xylose metabolism might interfere with the native metabolic regulation and cause some discrepancies in the direction of transcriptional regulation mediated by Gcr2.
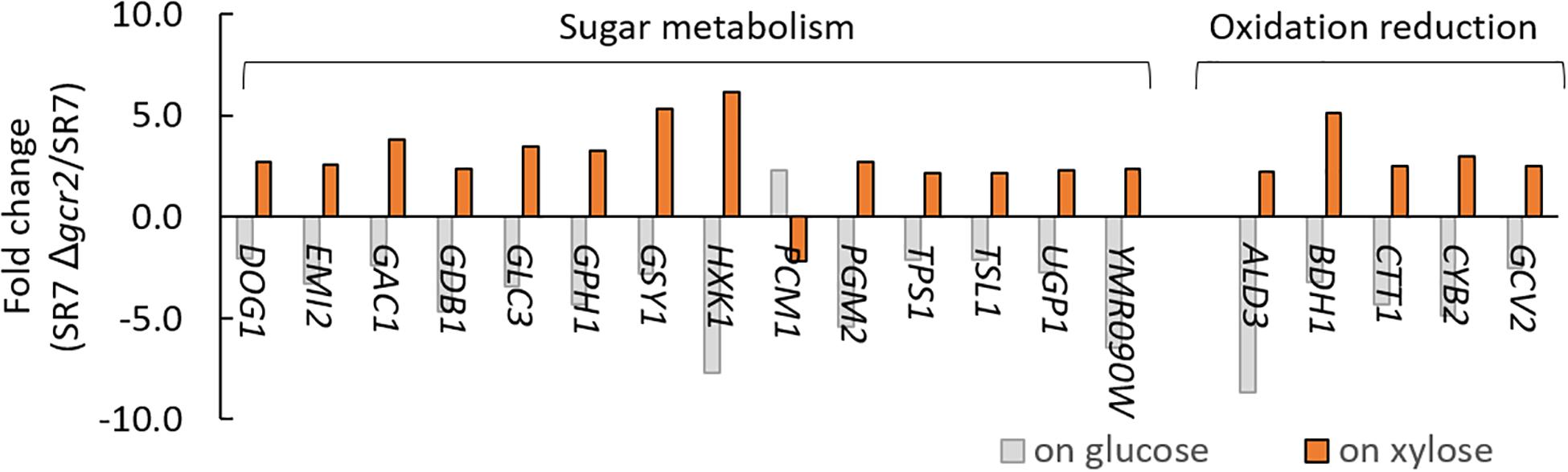
Figure 3. Gene sets and their genes that are oppositely affected by Δgcr2. RNA samples were extracted from exponentially growing SR7 and SR7 Δgcr2 strains on glucose and xylose.
Transcriptional Changes in Central Metabolic Pathways Induced by Δgcr2
To better understand the effect of Δgcr2 on xylose metabolism, the fold changes of DE genes (p < 0.05) in the glycolytic pathway, PPP, and citric acid cycle were systematically compared between glucose and xylose conditions (Figure 4). Two significant transcriptional changes were observed on both glucose and xylose conditions. First, Δgcr2 led to the downregulation of some glycolytic genes, most critically GPM1 encoding phosphoglycerate mutase, which is a key enzyme of the lower glycolic pathway. Second, Δgcr2 upregulated non-oxidative PPP genes, most critically the TAL1 gene encoding transaldolase. However, Δgcr2-mediated transcriptional changes in central metabolic pathways were more prominent during xylose metabolism. TDH2, ENO1, and CDC19 in the lower glycolytic pathway were significantly downregulated only under xylose conditions. Moreover, SOL4, GND2, and TKL2 in oxidative and non-oxidative PPP were considerably upregulated only under xylose conditions. Meanwhile, down-regulation of XYL3 was not consistent with the XYL1, of which expression were shared under the TDH3 promoter.
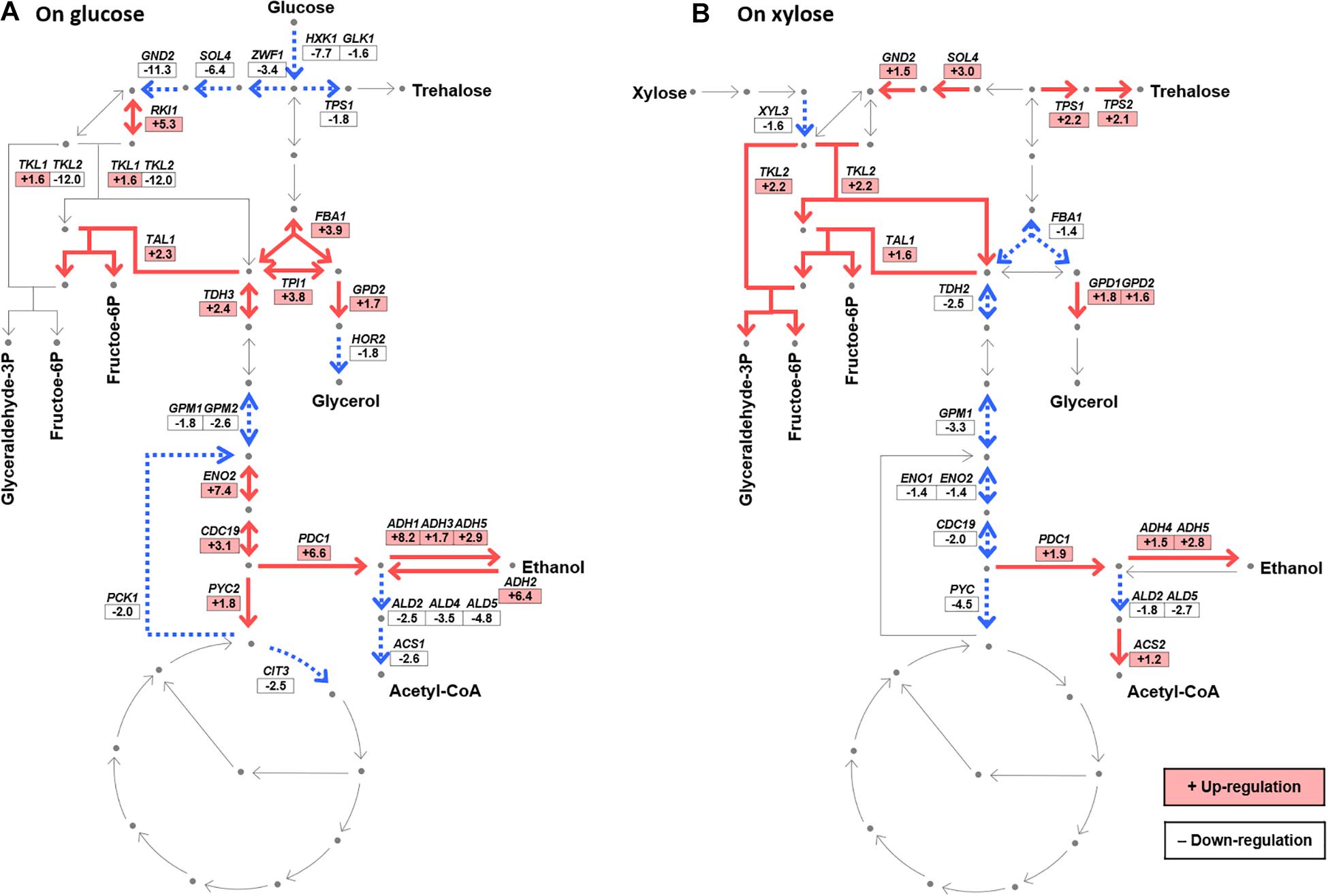
Figure 4. Transcriptional changes in the central metabolic pathways induced by Δgcr2 during glucose (A) or xylose (B) metabolism. The fold change in expression in Δgcr2 mutant relative to that in the wild-type strain is presented (p < 0.05). Glyceraldehyde-3P, glyceraldehyde-3-phosphate; fructose-6P, fructose-6-phosphate; acetyl-CoA, acetyl coenzyme A. RNA samples were extracted from exponentially growing SR7 and SR7 Δgcr2 strains on glucose and xylose.
Discussion
In the current study, GCR2 encoding the transcriptional activator of glycolytic genes was identified as a novel deletion target (Δgcr2) to improve xylose fermentation by S. cerevisiae, expressing a heterologous xylose pathway. RNA-seq results revealed that Δgcr2 results in not only the downregulation of glycolytic genes but also the upregulation of PPP genes, which explains the improved xylose metabolism by Δgcr2. However, manipulating the regulatory systems of the central metabolism such as glycolysis could result in some growth defects under certain conditions (Myung Hyun et al., 2020), which needs to be tested in follow-up studies.
Loss of function in GCR2 leads to the Δpho13-negative effect. Considering previous studies on Δpho13 (Kim et al., 2015; Xu et al., 2016), both Δpho13 and Δgcr2 result in the upregulation of TAL1, the essential overexpression target to improve xylose fermentation, as well as other genes in PPP. One hypothesis is that a simple activation signal of PPP was the main mechanism of improving xylose fermentation by Δgcr2 as well as Δpho13; in this scenario, because PPP is already activated in the Δgcr2 mutant, no further effect by Δpho13 might be expected.
However, some interaction between Δgcr2 and Δpho13 might be possible as well. Δgcr2 results in more global transcriptional changes than Δpho13. The number of DE genes by Δpho13 was 12 and 277 on glucose and xylose, respectively (Kim et al., 2015), which was one order of magnitude lower than that by Δgcr2. Moreover, some transcriptional changes induced by Δgcr2 were in opposite directions from those induced by Δpho13; especially, genes in the lower glycolytic pathway were repressed by Δgcr2 but activated by Δpho13 during xylose fermentation. Therefore, inhibition of the glycolytic pathway by Δgcr2 might partially counteract the Δpho13 effect during xylose fermentation, leading to imbalance of cofactor synthesis and/or energy generation required for xylose utilization.
Mutation of GCR2 triggers the upregulation of oxidative and non-oxidative PPP genes. How does the up-regulated expression of oxidative PPP relate to the xylose utilization and ethanol production? A prominent property of oxidative PPP is to generate NADPH, which subsequently can be utilized by xylose reductase encoded by XYL1 (Kwak and Jin, 2017). The increased balance between redox cofactor and the associating gene expression would promote xylose assimilation and the following non-oxidative PPP. The upregulated TAL1 and TKL2 genes expression suggests that native regulatory system such as Gcr2 acts negatively to the xylose fermentation, more specifically the PPP, ultimately providing the Δgcr2 mutant improved xylose utilization and ethanol production.
Some native regulatory systems of S. cerevisiae might act negatively to heterologous metabolism; however, it is challenging to systematically investigate all native regulatory genes to identify inhibitory ones. One of the most practical solutions for metabolic engineering is to use adaptive evolution to induce spontaneous mutations favorable to heterologous metabolism. Using omics approaches, such as genome sequencing and RNA-seq, our previous and present studies have identified spontaneous mutations in PHO13 (adaptive evolution) and GCR2 (multiple transformations and selection), which were involved in native inhibitory mechanisms against the heterologous xylose pathway.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
Y-SJ and SRK designed the experiments. MS, HP, EO, DJ, and CF carried out the experiments. SK and KK performed the statistical analysis of RNA-seq data. MS, Y-SJ, and SRK drafted and finalized the manuscript. All authors contributed to the article and read and approved the submitted version.
Funding
This work was supported by a National Research Foundation of Korea (NRF) grant (2019R1F1A1062633) funded by the Korea Government (MSIT).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer J-HS declared a shared affiliation, with no collaboration, with one of the authors MS to the handling editor at the time of review.
Acknowledgments
This article has been uploaded to a preprint server Authorea (Shin et al., 2020).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbioe.2021.654177/full#supplementary-material
Supplementary Table 1 | SNPs unique to YSX3 relative to L2612.
References
Cha, S., Hong, C. P., Kang, H. A., and Hahn, J.-S. (2020). Differential activation mechanisms of two isoforms of Gcr1 transcription factor generated from spliced and un-spliced transcripts in Saccharomyces cerevisiae. Nucleic Acids Res. 49, 745–759. doi: 10.1093/nar/gkaa1221
Fendt, S.-M., Buescher, J. M., Rudroff, F., Picotti, P., Zamboni, N., and Sauer, U. (2010). Tradeoff between enzyme and metabolite efficiency maintains metabolic homeostasis upon perturbations in enzyme capacity. Mol. Syst. Biol. 6:356. doi: 10.1038/msb.2010.11
Fujitomi, K., Sanda, T., Hasunuma, T., and Kondo, A. (2012). Deletion of the PHO13 gene in Saccharomyces cerevisiae improves ethanol production from lignocellulosic hydrolysate in the presence of acetic and formic acids, and furfural. Bioresour. Technol. 111, 161–166. doi: 10.1016/j.biortech.2012.01.161
Gietz, R. D., and Schiestl, R. H. (2007). High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat. Protoc. 2, 31–34. doi: 10.1038/nprot.2007.13
Ha, S.-J., Galazka, J. M., Rin Kim, S., Choi, J.-H., Yang, X., Seo, J.-H., et al. (2011). Engineered Saccharomyces cerevisiae capable of simultaneous cellobiose and xylose fermentation. Proc. Natl. Acad. Sci. U. S. A. 108, 504–509. doi: 10.1073/pnas.1010456108
Ha, S.-J., Kim, S. R., Kim, H., Du, J., Cate, J. H. D., and Jin, Y.-S. (2013). Continuous co-fermentation of cellobiose and xylose by engineered Saccharomyces cerevisiae. Bioresour. Technol. 149, 525–531. doi: 10.1016/j.biortech.2013.09.082
Huie, M. A., Scott, E. W., Drazinic, C. M., Lopez, M. C., Hornstra, I. K., Yang, T. P., et al. (1992). Characterization of the DNA-binding activity of GCR1: in vivo evidence for two GCR1-binding sites in the upstream activating sequence of TPI of Saccharomyces cerevisiae. Mol. Cell. Biol. 12, 2690–2700. doi: 10.1128/mcb.12.6.2690
Jeong, D., Oh, E. J., Ko, J. K., Nam, J.-O., Park, H.-S., Jin, Y.-S., et al. (2020). Metabolic engineering considerations for the heterologous expression of xylose-catabolic pathways in Saccharomyces cerevisiae. PLoS One 15:e0236294. doi: 10.1371/journal.pone.0236294
Jin, Y.-S., Ni, H., Laplaza, J. M., and Jeffries, T. W. (2003). Optimal growth and ethanol production from xylose by recombinant Saccharomyces cerevisiae require moderate d-xylulokinase activity. Appl. Environ. Microbiol. 69, 495–503. doi: 10.1128/aem.69.1.495-503.2003
Kim, S. R., Ha, S.-J., Kong, I. I., and Jin, Y.-S. (2012). High expression of XYL2 coding for xylitol dehydrogenase is necessary for efficient xylose fermentation by engineered Saccharomyces cerevisiae. Metab. Eng. 14, 336–343. doi: 10.1016/j.ymben.2012.04.001
Kim, S. R., Kwee, N. R., Kim, H., and Jin, Y.-S. (2013a). Feasibility of xylose fermentation by engineered Saccharomyces cerevisiae overexpressing endogenous aldose reductase (GRE3), xylitol dehydrogenase (XYL2), and xylulokinase (XYL3) from Scheffersomyces stipitis. FEMS Yeast Res. 13, 312–321. doi: 10.1111/1567-1364.12036
Kim, S. R., Lee, K.-S., Kong, I. I., Lesmana, A., Lee, W.-H., Seo, J.-H., et al. (2013b). Construction of an efficient xylose-fermenting diploid Saccharomyces cerevisiae strain through mating of two engineered haploid strains capable of xylose assimilation. J. Biotechnol. 164, 105–111. doi: 10.1016/j.jbiotec.2012.12.012
Kim, S. R., Park, Y.-C., Jin, Y.-S., and Seo, J.-H. (2013c). Strain engineering of Saccharomyces cerevisiae for enhanced xylose metabolism. Biotechnol. Adv. 31, 851–861. doi: 10.1016/j.biotechadv.2013.03.004
Kim, S. R., Skerker, J. M., Kang, W., Lesmana, A., Wei, N., Arkin, A. P., et al. (2013d). Rational and evolutionary engineering approaches uncover a small set of genetic changes efficient for rapid xylose fermentation in Saccharomyces cerevisiae. PLoS One 8:e57048. doi: 10.1371/journal.pone.0057048
Kim, S. R., Skerker, J. M., Kong, I. I., Kim, H., Maurer, M. J., Zhang, G.-C., et al. (2017). Metabolic engineering of a haploid strain derived from a triploid industrial yeast for producing cellulosic ethanol. Metab. Eng. 40, 176–185. doi: 10.1016/j.ymben.2017.02.006
Kim, S. R., Xu, H., Lesmana, A., Kuzmanovic, U., Au, M., Florencia, C., et al. (2015). Deletion of PHO13, encoding haloacid dehalogenase type IIA phosphatase, results in upregulation of the pentose phosphate pathway in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 81, 1601–1609. doi: 10.1128/aem.03474-14
Kwak, S., and Jin, Y.-S. (2017). Production of fuels and chemicals from xylose by engineered Saccharomyces cerevisiae: a review and perspective. Microb. Cell Fact. 16:82. doi: 10.1186/s12934-017-0694-9
Lynd, L. R. (2017). The grand challenge of cellulosic biofuels. Nat. Biotechnol. 35, 912–915. doi: 10.1038/nbt.3976
Myung Hyun, N., Sanghak, C., Minsun, K., and Jung, G. Y. (2020). Recent advances in microbial cell growth regulation strategies for metabolic engineering. Biotechnol. Bioprocess Eng. 25, 810–828. doi: 10.1007/s12257-019-0511-x
Ni, H., Laplaza, J. M., and Jeffries, T. W. (2007). Transposon mutagenesis to improve the growth of recombinant Saccharomyces cerevisiae on d-Xylose. Appl. Environ. Microbiol. 73, 2061–2066. doi: 10.1128/aem.02564-06
Qin, L., Dong, S., Yu, J., Ning, X., Xu, K., Zhang, S.-J., et al. (2020). Stress-driven dynamic regulation of multiple tolerance genes improves robustness and productive capacity of Saccharomyces cerevisiae in industrial lignocellulose fermentation. Metab. Eng. 61, 160–170. doi: 10.1016/j.ymben.2020.06.003
Richa, A., Shuvashish, B., Nilesh Kumar, S., and Sachin, K. (2019). Evaluating the pathway for co-fermentation of glucose and xylose for enhanced bioethanol production using flux balance analysis. Biotechnol. Bioprocess Eng. 24, 924–933. doi: 10.1007/s12257-019-0026-5
Sasaki, H., Kishimoto, T., Mizuno, T., Shinzato, T., and Uemura, H. (2005). Expression of GCR1, the transcriptional activator of glycolytic enzyme genes in the yeast Saccharomyces cerevisiae, is positively autoregulated by Gcr1p. Yeast 22, 305–319. doi: 10.1002/yea.1212
Sasaki, H., and Uemura, H. (2005). Influence of low glycolytic activities in gcr1 and gcr2 mutants on the expression of other metabolic pathway genes in Saccharomyces cerevisiae. Yeast 22, 111–127. doi: 10.1002/yea.1198
Shin, M., Park, H., Kim, S., Oh, E. J. O., Jeong, D., Florencia C., et al. (2020). Transcriptomic changes induced by de-activation of lower glycolysis and its advantage on pentose sugar metabolism in Saccharomyces cerevisiae. Authorea [Preprint]. doi: 10.22541/au.160010788.88280741
Subramanian, A., Tamayo, P., Mootha, V. K., Mukherjee, S., Ebert, B. L., Gillette, M. A., et al. (2005). Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U. S. A. 102, 15545–15550. doi: 10.1073/pnas.0506580102
Uemura, H., and Fraenkel, D. G. (1990). gcr2, a new mutation affecting glycolytic gene expression in Saccharomyces cerevisiae. Mol. Cell. Biol. 10, 6389–6396. doi: 10.1128/mcb.10.12.6389
Uemura, H., and Jigami, Y. (1992). Role of GCR2 in transcriptional activation of yeast glycolytic genes. Mol. Cell. Biol. 12, 3834–3842. doi: 10.1128/mcb.12.9.3834
Van Vleet, J. H., Jeffries, T. W., and Olsson, L. (2008). Deleting the para-nitrophenyl phosphatase (pNPPase), PHO13, in recombinant Saccharomyces cerevisiae improves growth and ethanol production on d-xylose. Metab. Eng. 10, 360–369. doi: 10.1016/j.ymben.2007.12.002
Xu, H., Kim, S., Sorek, H., Lee, Y., Jeong, D., Kim, J., et al. (2016). PHO13 deletion-induced transcriptional activation prevents sedoheptulose accumulation during xylose metabolism in engineered Saccharomyces cerevisiae. Metab. Eng. 34, 88–96. doi: 10.1016/j.ymben.2015.12.007
Keywords: lignocellulosic biomass, yeast metabolic engineering, transcriptomics, glucose repression, GCR2, pentose phosphate pathway
Citation: Shin M, Park H, Kim S, Oh EJ, Jeong D, Florencia C, Kim KH, Jin Y-S and Kim SR (2021) Transcriptomic Changes Induced by Deletion of Transcriptional Regulator GCR2 on Pentose Sugar Metabolism in Saccharomyces cerevisiae. Front. Bioeng. Biotechnol. 9:654177. doi: 10.3389/fbioe.2021.654177
Received: 15 January 2021; Accepted: 08 March 2021;
Published: 25 March 2021.
Edited by:
Zhi-Qiang Liu, Zhejiang University of Technology, ChinaReviewed by:
Xinqing Zhao, Shanghai Jiao Tong University, ChinaJin-Ho Seo, Seoul National University, South Korea
Yong-Cheol Park, Kookmin University, South Korea
Copyright © 2021 Shin, Park, Kim, Oh, Jeong, Florencia, Kim, Jin and Kim. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yong-Su Jin, eXNqaW5AaWxsaW5vaXMuZWR1; Soo Rin Kim, c29vcmlua2ltQGtudS5hYy5rcg==
 Minhye Shin1
Minhye Shin1 Sooah Kim
Sooah Kim Eun Joong Oh
Eun Joong Oh Yong-Su Jin
Yong-Su Jin Soo Rin Kim
Soo Rin Kim