- 1Stem Cell Fate Laboratory, Institute of Genetics and Biophysics “A. Buzzati-Traverso,” CNR, Naples, Italy
- 2Molecular Medicine Laboratory, International Centre for Genetic Engineering and Biotechnology, Trieste, Italy
Skeletal muscle regeneration mainly depends on satellite cells, a population of resident muscle stem cells. Despite extensive studies, knowledge of the molecular mechanisms underlying the early events associated with satellite cell activation and myogenic commitment in muscle regeneration remains still incomplete. Cripto is a novel regulator of postnatal skeletal muscle regeneration and a promising target for future therapy. Indeed, Cripto is expressed both in myogenic and inflammatory cells in skeletal muscle after acute injury and it is required in the satellite cell compartment to achieve effective muscle regeneration. A critical requirement to further explore the in vivo cellular contribution of Cripto in regulating skeletal muscle regeneration is the possibility to overexpress Cripto in its endogenous configuration and in a cell and time-specific manner. Here we report the generation and the functional characterization of a novel mouse model for conditional expression of Cripto, i.e., the Tg:DsRedloxP/loxPCripto-eGFP mice. Moreover, by using a satellite cell specific Cre-driver line we investigated the biological effect of Cripto overexpression in vivo, and provided evidence that overexpression of Cripto in the adult satellite cell compartment promotes myogenic commitment and differentiation, and enhances early regeneration in a mouse model of acute injury.
Introduction
The responses of skeletal muscle tissue following acute or chronic damages are highly complex and coordinated processes, involving many different cell populations that interact each other to promote muscle regeneration, inflammation and angiogenesis, until full regeneration of the tissue and its functional recovery. This process is tightly controlled by signals released by the damaged fibers, which lead to the activation of the quiescent satellite cells, i.e., the myogenic stem cell population that is among the major players responsible for the regeneration of skeletal muscle (Collins et al., 2005). Following an injury, the satellite cells are activated and proliferate as myogenic progenitors that migrate to the damaged site, differentiate and fuse each other to form new myofibers (Hawke and Garry, 2001; Chargé and Rudnicki, 2004). This sequential process is correlated with the finely regulated expression of the Myogenic Regulatory Factors (MRFs). Indeed, quiescent satellite cells express Pax7 and, upon activation, upregulate the myogenic transcription factor MyoD. Upon commitment to differentiation these transient amplifying cells (Pax7+/MyoD+) downregulate Pax7 and upregulate differentiation genes, as Myogenin and MRF4, while a subset of these cells downregulate MyoD retaining Pax7 expression to replenish the pool of quiescent satellite cells (Zammit et al., 2004; Olguin et al., 2007; Tajbakhsh, 2009). Beside satellite cells, regeneration process is also orchestrated by the crosstalk between heterogeneous cell populations that are recruited/activated after damage, such as the inflammatory cells that are always associated with tissue regeneration, supporting myogenic progression by interacting with satellite cells (Kharraz et al., 2013; Saclier et al., 2013). Besides extensive studies, knowledge of the mechanisms underlying this highly orchestrated process and how the different cell populations establish their fate is still far to be fully elucidated, and much less is known on the extrinsic regulation of this process (Bentzinger et al., 2010). We have recently demonstrated that Cripto, a key regulator of early vertebrate embryogenesis (Shen and Schier, 2000; Minchiotti et al., 2001), is a new player in this process (Guardiola et al., 2012). Cripto is a GPI-anchored protein (Minchiotti et al., 2001) and a developmental factor required for correct orientation of the anterior–posterior axis in the vertebrate embryos (Chu et al., 2005; Minchiotti, 2005). Despite its well-characterized role in embryo development and embryonic stem cell differentiation (Minchiotti, 2005), the role of Cripto in adult life has been poorly investigated also due to the fact that Cripto knockout mice are embryonic lethal (Ding et al., 1998; Xu et al., 1999), and that its expression is almost absent in adult physiological conditions. Indeed, Cripto expression is undetectable in skeletal muscles under baseline conditions. However, it becomes rapidly and transiently re-expressed after acute injury, both in myogenic and inflammatory cells, and it is required in the myogenic compartment to achieve an efficient regeneration (Guardiola et al., 2012). Interestingly, a soluble form of the protein (sCripto) is able to rescue the effect of genetic inactivation of Cripto, thus recapitulating the function of endogenous membrane form of the protein (Guardiola et al., 2012). Yet, the cellular contribution of Cripto in skeletal muscle repair remains to be further clarified, and its knowledge is limited also by the absence of mouse models for conditional and tissue–specific Cripto expression. Here we report the generation and characterization of novel transgenic mice for conditional expression of Cripto in its endogenous configuration, which allowed us to study the biological effect of satellite cell-specific Cripto overexpression on muscle regeneration and myogenic cell fate determination.
Results
Generation of Conditional Cripto Gain of Function Transgenic Mice
To get insight into the cellular contribution of Cripto in skeletal muscle regeneration, and to finely modulate Cripto expression in vivo, we generated a novel transgenic mouse line for conditional Cripto expression based on the Cre-loxP strategy. To generate the pDsRedloxP/loxPCripto targeting vector, a Cripto-IRES-eGFP cassette was inserted downstream of the DsRed gene sequence followed by three termination sequences, and flanked by two LoxP sites (see Materials and Methods for details; Figure 1A). The effectiveness of the pDsRedloxP/loxPCripto vector was first evaluated in vitro. To this end, HEK-293T cells were transfected with the pCMV-Cre, expressing the Cre recombinase, and the pDsRedloxP/loxPCripto plasmids, either alone or in combination, and Cripto protein expression was evaluated. We first verified that eGFP expression was induced in cells cotransfected with pDsRedloxP/loxPCripto and pCMV-Cre (Figure S1A). Accordingly, Cripto protein was specifically induced (Figure 1B) and, as expected, it localized at the cell membrane (Minchiotti et al., 2000) of eGFP expressing cells (Figure S1B). Following the in vitro validation of the targeting vector, transgenic mice were generated by pronuclear injection, and the presence of the transgene in the offspring was assessed by PCR genotyping of tail biopsies (Figures 1C,D). One out of three transgenic mice obtained gave germline transmission and carried two copies of the transgene that segregated independently in the offspring (Figure 1E). Two founder lines were thus established and bred to FVB/N wild type mice to generate the Tg:DsRedloxP/loxPCripto-eGFP(A) and Tg:DsRedloxP/loxPCripto-eGFP(B) colonies (from now onwards named Tg:Cripto(A) and Tg:Cripto(B), respectively).
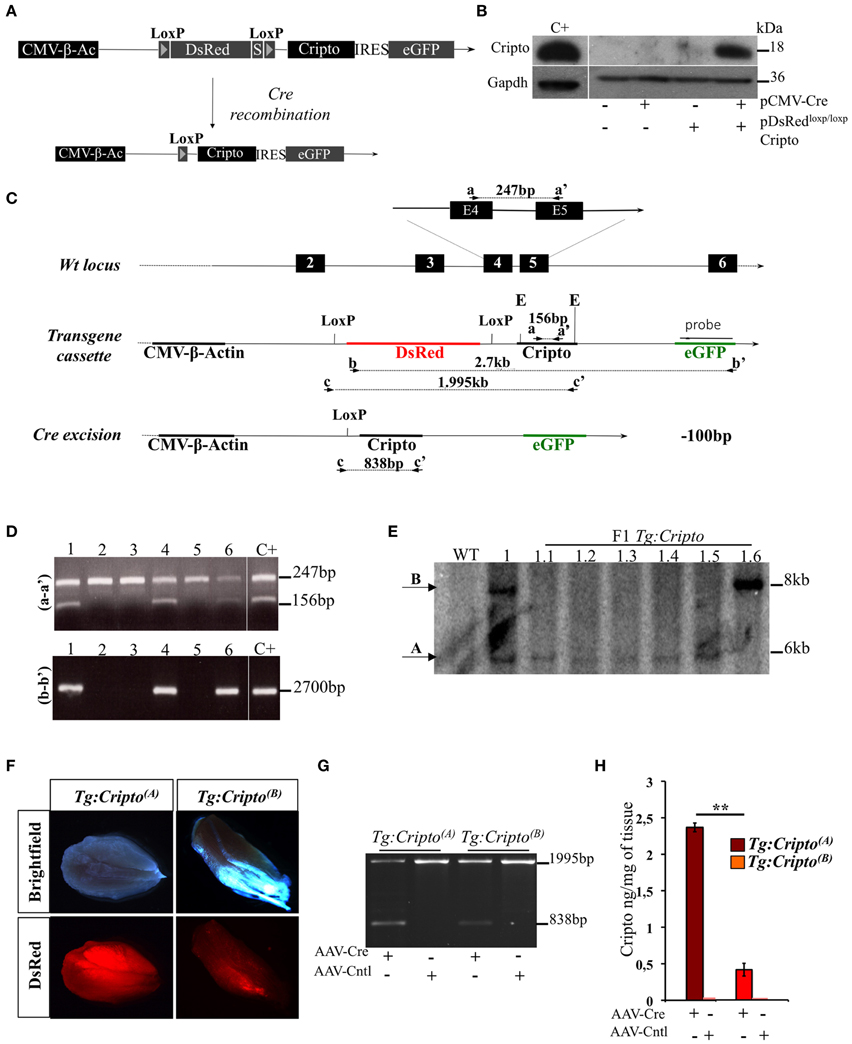
Figure 1. (A) Schematic representation of the pDsRedloxP/loxPCripto targeting vector. The nuclear DsRed gene flanked by two loxP sites and followed by three termination sequences (S) is transcribed by a constitutive CMV-β-Actin (CMV-β-Ac) promoter. Upon cre- mediated recombination, the DsRed sequence is excised and the Cripto-IRES-eGFP cassette is transcribed from the CMV/β-actin promoter. (B) Western blot analysis using anti-Cripto antibody on total protein extracts of HEK-293T cells transfected with pDsRedloxP/loxPCripto vector either alone or in combination with pCMV-Cre vector. Gapdh was used as a loading control. Protein extracts from undifferentiated mouse embryonic stem cells (ESCs) were used as positive control (C+). (C) Schematic representation of the genotyping strategy. Location of PCR primers (black arrowheads) and the probe (black bar) used for genotyping are indicated. Numbered boxes represent wild-type exons (black). E, EcoRI. (D) Representative PCR screening on tail genomic DNA from mice generated after oocyte injection. Primers a-a′ (top panel) amplified a 247-bp fragment of the wild-type Cripto locus and a 156-bp fragment of the Cripto transgene. Primers b-b' (bottom panel) amplified a 2700-bp fragment spanning the DsRed-IRES-Cripto cassette. (E) Genotyping by Southern blot analysis. Genomic DNA from wild type (WT), founder (1) and F1 offspring mice (from 1.1 to 1.6) was digested with EcoRI and hybridized with eGFP probe shown in (C). The sizes of hybridized fragments are indicated in kilobases. (F) Representative pictures of direct fluorescence of freshly isolated skeletal muscle from Tg:Cripto(A) and Tg:Cripto(B) mice, showing different levels of DsRed expression. (G) PCR screening of biopsy from Tg:Cripto(A) and Tg:Cripto(B) TA muscles infected with either AAV-Cre or AAV-Control (AAV-Cntl). Primers c-c' (C) amplified a 1995-bp fragment of the transgenic allele and a 838-bp fragment of the recombining allele. (H) ELISA assay of Cripto protein levels expressed as ng/mg of tissue in Tg:Cripto(A) and Tg:Cripto(B) muscles infected with AAV-Cre and AAV-Cntl (2.36 ± 0.06 ng/mg in Tg:Cripto(A) vs. 0.43 ± 0.09 ng/mg in Tg:Cripto(B); **P ≤ 0.005).
Functional Characterization of the Tg:Cripto Transgenic Lines
Different studies have shown that significative differences exist in the expression level of the same transgene between individual founder siblings, due to different integration loci, and the influence of the genomic sequences flanking the integration site (Palmiter et al., 1982; Overbeek et al., 1986). To characterize Tg:Cripto(A) and Tg:Cripto(B) transgenic lines, we first assessed DsRed expression in freshly isolated muscles by direct fluorescence and found a stronger DsRed signal in Tg:Cripto(A) compared to Tg: Cripto(B) muscles (Figure 1F). We thus evaluated whether Cripto was expressed upon Cre-mediated recombination in vivo. To this end, TA muscles of both transgenic lines were injected with Adeno Associated Virus 9 (Inagaki et al., 2006) encoding Cre recombinase (AAV-Cre) or empty vector (AAV-Cntl) as control. Ten days after infection, TA muscles were explanted and genotyped by PCR. As expected, a 838-bp band characterizing the recombining allele was specifically detected in the AAV-Cre injected muscles (Figure 1G), and Cripto protein was induced at different levels as determined by ELISA-based assay (2.36 ± 0.06 ng/mg in Tg:Cripto(A) vs. 0.43 ± 0.09 ng/mg in Tg:Cripto(B); **P = 0.005) (Figure 1H).
All together these data demonstrate that Cripto expression is regulated upon Cre-mediated recombination and is induced at different levels in the two transgenic lines.
Time -Dependent Effect of Cripto Overexpression in Adult Satellite Cells on Skeletal Muscle Regeneration
We have recently shown that adenovirus–mediated soluble Cripto (sCripto) overexpression accelerates muscle regeneration induced by acute injury (Guardiola et al., 2012). Nevertheless, viral mediated Cripto expression does not allow us to discriminate between the inflammatory and satellite cell contribution of Cripto overexpression. To overcome this limitation, we crossed Tg:Cripto(A) and Tg:Cripto(B) mice with the tamoxifen-inducible Tg:Pax7CreERT2 mice (Mourikis et al., 2012) and obtained the Tg:Pax7CreERT2::DsRedloxP/loxPCripto-eGFP(A) and Tg:Pax7CreERT2::DsRedloxP/loxPCripto-eGFP(B) trangenic lines (from now onwards named Tg:Pax7CT2::Cripto(A) and Tg:Pax7CT2::Cripto(B)) (Figure 2A). We first assessed the effects of satellite cell-specific Cripto overexpression on skeletal muscle regeneration. To this end, Tg:Pax7CT2::Cripto(A) and Tg:Pax7CT2::Cripto(B) adult mice and their control littermates were treated with tamoxifen once a day for 5 days; at day 4, muscle regeneration was triggered in TA muscles by local injection of cardiotoxin (CTX; Figure 2A). Genetic recombination was first confirmed by PCR analysis on TA muscle genomic DNA (Figure 2B, Figure S2A), and Cripto protein levels were quantified by ELISA assay on total protein extracts at different time points after injury. Increased Cripto protein levels were detected in both Tg:Pax7CT2::Cripto(A) and Tg:Pax7CT2::Cripto(B) mice compared to control, with Tg:Pax7CT2::Cripto(A) showing the highest levels of Cripto upon Cre-mediated recombination (Figure 2C, Figure S2B). To assess if Cripto overexpression might influence muscle regeneration, we first evaluated the expression of the embryonic Myosin Heavy Chain (eMyHC), which is a marker of newly regenerated fibers, at day 8 after injury. Both immunofluorescence and Western blot analysis clearly showed increased eMyHC protein levels in Tg:Pax7CT2::Cripto(A) mice compared to their control littermates (Figures 2D,E). In line with these findings, expression of both neonatal Myosin Heavy Chain (nMyHC) and the early muscle differentiation marker Myogenin (Myog) similarly increased in the overexpressing mice (Figures 2F,G). Furthermore, expression of Myostatin (Mstn), which is a negative regulator of muscle growth (Thomas et al., 2000), was significantly reduced in Tg:Pax7CT2::Cripto(A)mice compared to control (Figure 2H). Of note, similar results were obtained from the analysis of the other transgenic line (Tg:Pax7CT2::Cripto(B); Figures S2C–F). Morphological analysis of H&E-stained TA muscle sections at day 8 showed that the number of myofibers containing more than one nucleus (n >1) significantly increased in Cripto overexpressing mice compared to control (0.12 ± 0.02 for Tg:Pax7CT2::Cripto(A) vs. 0.06 ± 0.01 for Tg:Cripto(A) of fibers; *P ≤ 0.05; Figures 3A,B). Interestingly, while there was no significant difference in Cross Sectional Area (CSA) between the two groups at day 8 (Figures 3A,C), later on (i.e., at day 15) both CSA distribution and the relative average values significantly increased in the Tg:Pax7CT2::Cripto(A) mice compared to control (Figures 3A,D,E). To assess if the positive effect of Cripto overexpression on muscle regeneration was either transient or persistent, we extended the analysis to a later time point (i.e., at day 30). Morphometric analysis of the CSA showed no difference in the distribution of muscle fibers in the two groups at day 30 (Figures 3A,F); thus providing evidence of a time-dependent effect of satellite cell -Cripto overexpression on skeletal muscle regeneration. Most remarkably, similar experiments carried on the Tg:Pax7CT2::Cripto(B) mice gave comparable results (Figures S3A–F), thus ruling out the possibility of a position-effect of the transgene.
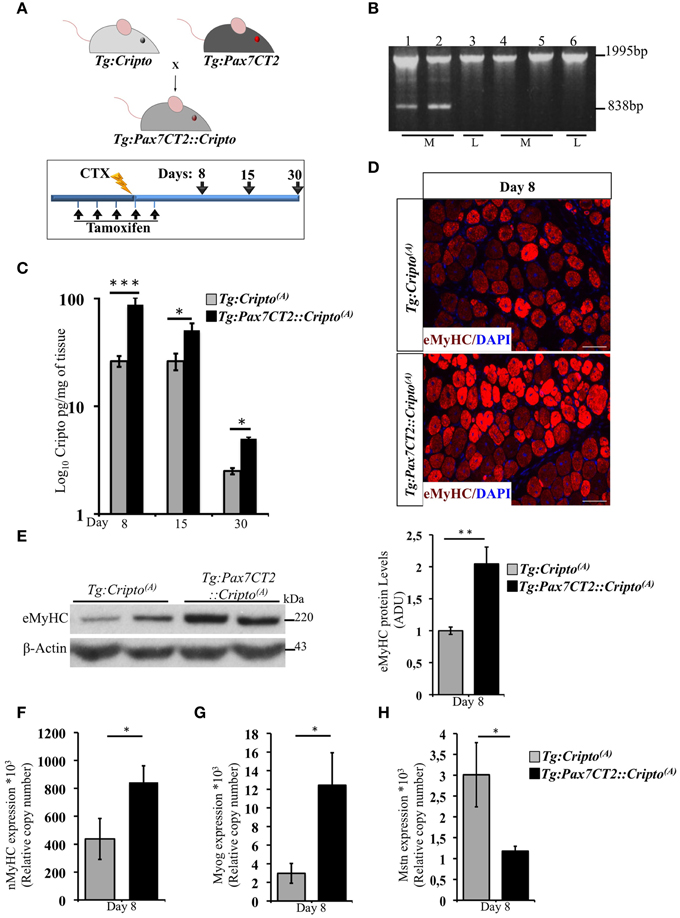
Figure 2. (A) Schematic representation of mouse breeding (upper panel) and Cripto conditional gain of function strategy in adult satellite cells (lower panel). Tamoxifen were injected i.p., in adult (5 weeks old) Tg:Pax7CT2::Cripto mice and Tg:Cripto control littermates once a day for 5 days. At day 4, regeneration was triggered by CTX injection in TA muscle of both groups, and analysis performed at the indicated time points (days 8, 15, and 30). The consecutive bars indicate the days. (B) PCR screening on muscle genomic DNA showing tamoxifen-induced recombination only in the muscle of Tg:Pax7CT2::Cripto(A). Primers c-c′ (See Figure 1C) amplified a 1995-bp fragment of the transgenic allele and a 838-bp fragment of the recombining allele. The recombining allele is detected in the muscle (M; lanes 1, 2) but not in the liver (L; lane 3) of Tg:Pax7CT2::Cripto(A) mice, and is not detected in the muscle (M; lanes 4, 5) and in the liver (L; lane 6) of Tg:Cripto(A) control littermates. (C) ELISA assay of Cripto protein levels in TA muscles at different time points after CTX injection expressed as pg/mg of muscle tissue at the indicated time points. Values are mean ± SEM of n = 5 mice/group. *P ≤ 0.05; ***P = 0.0005. (D) Representative pictures of embryonic Myosin Heavy Chain (eMyHC) Immunofluorescence on TA sections from Tg:Pax7CT2::Cripto(A) mice and Tg:Cripto(A) control littermates at day 8. Scale bars = 100 μm. (E) Representative Western blot using anti-eMyHC antibody on total protein extracts of TA muscles from Tg:Pax7CT2::Cripto(A) mice and their Tg:Cripto(A) control littermates at day 8 after CTX. β-Actin was used as a loading control. The densitometric analysis is expressed as Arbitrary Densitometric Unit (ADU) expressing the eMyHC/β-Actin ratio. Values are mean ± SEM of n = 3 mice/group. **P = 0.005. (F–H) qRT-PCR analysis of neonatal Myosin Heavy Chain (nMyHC) (F) Myogenin (Myog) (G) and Myostatin (Mstn) (H) on TA muscle total RNAs from Tg:Pax7CT2::Cripto(A) mice and the Tg:Cripto(A) control littermates at day 8 after CTX injection. Values are mean ± SEM, 5 mice/group. *P ≤ 0.05.
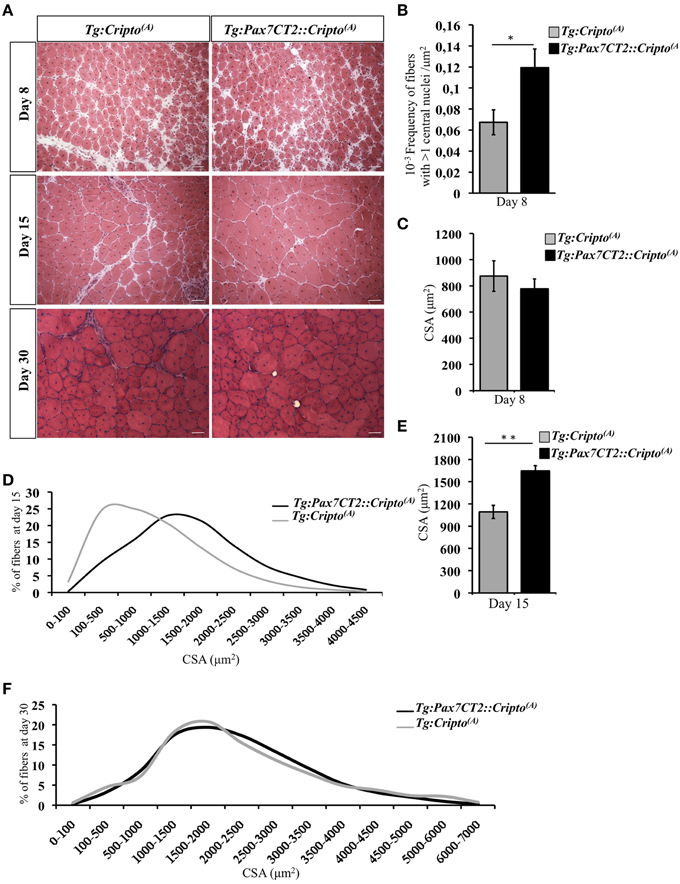
Figure 3. (A) Representative H&E staining of TA muscle sections from Tg:Pax7CT2::Cripto(A) and Tg:Cripto(A) TA muscles at the indicated time points. Scale bars = 50μm. (B) Morphological analysis on muscle sections at day 8 after injury showing the number of myofibers containing more than one central nuclei/area in each group. Values are mean ± SEM, 5 mice/group *P ≤ 0.05. (C) Average of centrally nucleated myofibers size values in TA muscle sections at day 8 after injury. Values are mean ± SEM, 5 mice/group. (D) Myofiber Cross Sectional Area distribution at 15 days after CTX injection; 5 mice/group. (E) Average of centrally nucleated myofibers size values in TA muscle sections at day 15 after injury of Tg:Pax7CT2::Cripto(A) mice and Tg:Cripto(A) control littermates. Values are mean ± SEM, 5 mice/group. **P ≤ 0.005. (F) Myofiber Cross Sectional Area distribution at 30 days after CTX injection; 4 mice/group.
Conditional Overexpression of Cripto in Satellite Cells Enhances Myogenic Differentiation
To determine how Cripto overexpression in the satellite cells impact on skeletal muscle regeneration, we first evaluated its effect on early stages of myoblast differentiation in vitro. To address this issue, we determined the fusion index (i.e., the percentage of the MF20+ cells with n ≥ 2 nuclei) of the postnatal myogenic progenitor cells (MPCs) isolated from hindlimb muscles of new-born Tg:Pax7CT2::Cripto(A) mice and their Tg:Cripto(A) control littermates at postnatal (P) day 7. Cre-mediated recombination was induced by tamoxifen injection (Figure 4A) and assessed by PCR genotyping of muscle tissues (Figure 4B). We then verified that Cripto protein was induced in Tg:Pax7CT2::Cripto(A) by ELISA assay on postnatal MPCs (70.8 ± 14 * 10−2 pg/μg for Tg:Pax7CT2::Cripto(A) vs. 32.1 ± 2.6 * 10−2 pg/μg for Tg:Cripto(A); *P ≤ 0.05; Figure 4C). Cells were maintained in culture for 24 h in proliferation medium and then shifted to differentiation medium (DM) for 24 and 72 h (Figure 4D). Quantitative analysis showed that the percentage of MF20+ cells with n ≥ 2 nuclei progressively increased in cultures from Tg:Pax7CT2::Cripto(A) mice compared to control, at both time points (29.03 ± 1.79% for Tg:Pax7CT2::Cripto(A) vs. 18.4 ± 2.75% for Tg:Cripto(A) at 24 h; 35.85 ± 2.24% for Tg:Pax7CT2::Cripto(A) vs. 26.87 ± 1.08% for Tg:Cripto(A) at 72 h; *P ≤ 0.05; Figure 4E), thus suggesting that Cripto overexpression enhanced in vitro myoblasts to myotubes differentiation. In line with these findings, the percentage of Pax−/MyoD+ cells significantly increased in Tg:Pax7CT2::Cripto(A) mice compared to control (9.92 ± 1.88% Pax−/MyoD+ for Tg:Pax7CT2::Cripto(A) vs. 3 ± 1.62% Pax−/MyoD+ for Tg:Cripto(A); *P ≤ 0.05; Figures S4A,B). Interestingly, these results are in line with data previously reported on isolated myofibers in culture in which addition of sCripto increases the number of Pax7−/MyoD+ myogenic cell population, promoting satellite cell progression into the myogenic lineage (Guardiola et al., 2012). We thus went on and extended the analysis in vivo to assess the effects of Cripto overexpression on the distribution of the satellite cell population during muscle regeneration. To this end, TA muscle sections of Tg:Pax7CT2::Cripto(A) mice and their control littermates at day 8 after injury (see Figure 2A) were double stained with anti-Pax7 and anti-MyoD antibodies (Figure 4F) and the frequency of (i) quiescent satellite cells (Pax7+/MyoD−), (ii) transient amplifying cells (Pax7+/MyoD+), and (iii) cells committed to myogenic differentiation (Pax7−/MyoD+) was evaluated. Remarkably, the frequency of Pax7−/MyoD+ cells significantly increased in Tg:Pax7CT2::Cripto(A) mice compared to control (0.32 ± 0.03 for Tg:Pax7CT2::Cripto(A) vs. 0.16 ± 0.04 for control of Pax7−/MyoD+ cells; *P ≤ 0.05; Figures 4F,G), at the expense of Pax7+/MyoD+, which conversely decreased in Tg:Pax7CT2::Cripto(A) mice (0.13 ± 0.04 for Tg:Pax7CT2::Cripto(A) vs. 0.3 ± 0.05 for control of Pax7+/MyoD+ cells; *P ≤ 0.05; Figures 4F,G); thus providing in vivo evidence that adult satellite cell specific Cripto overexpression accelerated myogenic lineage progression.
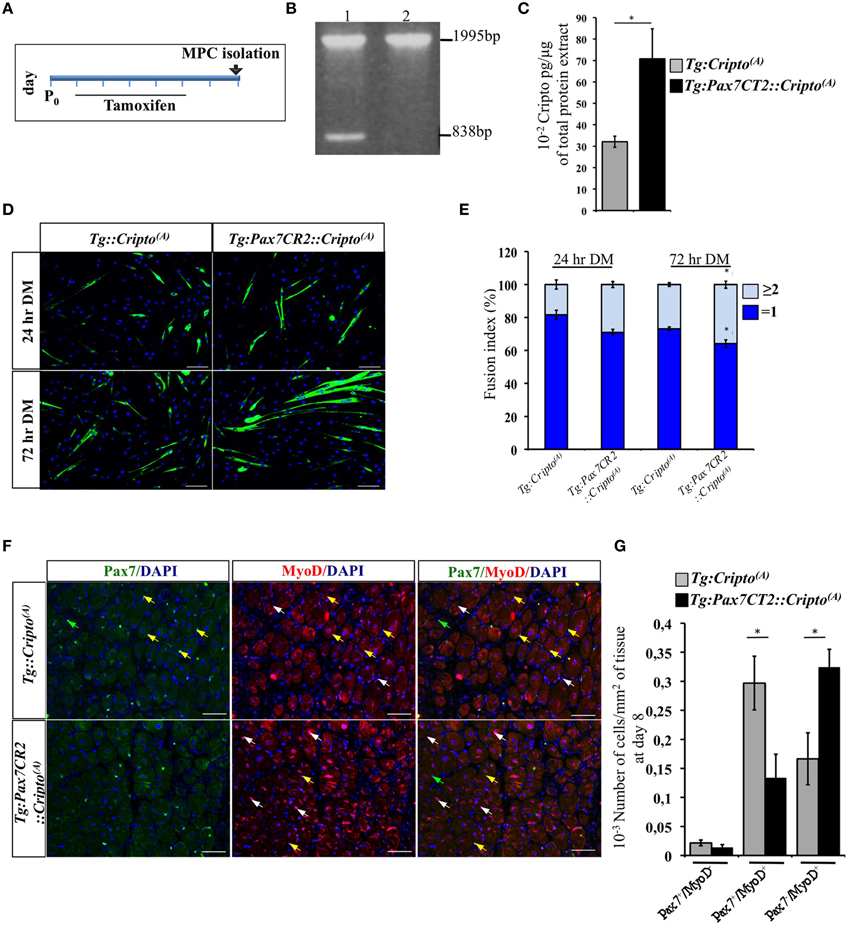
Figure 4. (A) Schematic representation of experimental strategy for myogenic progenitor cells (MPCs) isolation. Tamoxifen was injected once a day for 5 days (from P1 to P5) in new-born Tg:Pax7CT2::Cripto(A) mice and their control littermates, and MPCs were isolated at P7. (B) PCR analysis on muscle genomic DNA showing tamoxifen-induced recombination in Tg:Pax7CT2::Cripto(A) mice. Primers c-c′ (Figure 1C) amplified a 1995-bp fragment of the transgenic allele and a 838-bp fragment of the recombining allele. (C) ELISA assay of Cripto protein levels in primary myoblasts from Tg:Pax7CT2::Cripto(A) and control mice expressed as pg/μg of total protein extracts. Values are mean ± SEM of n = 3. *P ≤ 0.05. (D) Representative pictures of Tg:Pax7CT2::Cripto(A) and Tg:Cripto(A) myotubes after 24 and 72 h in differentiation medium (DM) and stained with MF20 antibody. Scale bars = 100 μm. (E) Fusion index was calculated as the percentage of MF20+ cells with n = 2 nuclei at 24 and 72 h in DM. The analysis was performed on 20 independent and random chosen microscope fields. Values are mean ± SEM of 3 independent experiments. *P = 0.05. (F) Representative pictures of double immunofluorescence with antibodies anti-Pax7 (green) and anti-MyoD (red) in TA muscles at day 8 after injury. Nuclei were counterstained with DAPI. Arrowheads indicate Pax7+/MyoD− (green), Pax7+/MyoD+ (yellow) and Pax7−/MyoD+ (white) cells. Scale bars = 100 μm. (G) Effect of satellite cells-Cripto overexpression on Pax7±/MyoD± cell distribution in vivo. The number Pax7±/MyoD±/area from Tg:Pax7CT2::Cripto(A) mice and the Tg:Cripto(A) control littermates was calculated in TA muscles and reported in the chart. Values are mean ± SEM, 5 mice/group. *P ≤ 0.05.
Taken together our results provide the first in vivo evidence that Cripto overexpression in myogenic compartment accelerates the early stages of regeneration process promoting satellite cell myogenic commitment.
Discussion
Growing evidence link adult tissue repair to reactivation of developmental gene program; nevertheless, our knowledge on the role of key regulators of early vertebrate embryogenesis in tissue regeneration is still limited also by the lack of adequate in vivo genetic tools. Indeed, transcription levels and timing of expression of key developmental genes are finely tuned, and experimental strategies to generate constitutive gain-of-function and loss-of-function mouse mutants often result in embryonic lethality. In this scenario, we have focused our attention on the developmental gene Cripto, which is a key regulator of vertebrate embryogenesis. Here we report the generation and functional characterization of a conditional Cripto gain-of-function mouse model Tg:DsRedloxP/loxPCripto-eGFP as a complementary strategy to investigate the effects of Cripto in postnatal and adult life, and potentially in all cell types. Specifically, by crossing Tg:DsRedloxP/loxPCripto-eGFP mice with a Tg:Pax7CreERT2 mouse line, we have investigated the effect of satellite cell- Cripto overexpression on skeletal muscle regeneration and overcome the limitation of the viral-mediated approach (Guardiola et al., 2012). Consistent with our previous data on isolated myofibers in culture, we provide in vivo evidence that satellite cell overexpression of membrane Cripto promotes myogenic commitment and differentiation. However, unlike the viral-mediated overexpression of soluble Cripto (sCripto; Guardiola et al., 2012), the positive effect of satellite cell Cripto overexpression on skeletal muscle regeneration is time-dependent, being restricted to the early phases of regeneration. It is unlikely that the discrepancy in the two approaches is due to protein configuration, i.e., membrane vs. soluble Cripto; indeed, sCripto is able to rescue muscle regeneration defects caused by the genetic ablation of Cripto in the satellite cells, indicating that it can recapitulate the role of the endogenous protein (Guardiola et al., 2012). Yet, it is important to consider that Cripto is expressed both in myogenic and inflammatory cells during regeneration. Thus, the different timing of regeneration in the two models might be explained by the fact that while the genetic overexpression is restricted to the satellite cells, the viral-mediated sCripto overexpression has a broader effect and likely affects both the myogenic and the inflammatory cells. Further studies will be necessary to test this hypothesis by deeply investigating the specific contribution of Cripto in inflammatory cells, as well as the autocrine and/or paracrine effects of the endogenous protein.
The positive effect of Cripto overexpression on skeletal muscle regeneration indicated by the increased CSA at day 15 is the expected consequence of its earliest effect on the satellite cell compartment. First, the number of centrally located nuclei is higher in Cripto overexpressing myofibers at day 8 after injury; thus suggesting accelerated differentiation. Accordingly, the myoblast fusion index is increased in postnatal MPCs isolated from Cripto overexpressing mice. Finally, an in depth analysis of Pax7±/MyoD± cell distribution shows that the number of Pax7−/MyoD+ cells committed to differentiation increases both in vivo and in vitro upon Cripto overexpression. We thus conclude that not only the addition of sCripto protein but also its overexpression in a more physiological context, promotes/accelerates satellite cells commitment to differentiation both in vitro and in vivo. Taken together our data provide evidence that a timely overexpression of Cripto in the satellite cells promotes myogenic commitment and fusion/differentiation and transiently accelerates regeneration after acute injury. It will be interesting to investigate in future studies the effect of sustained Cripto overexpression on satellite cell self renewal and regeneration after repeated muscle injury.
Finally, in line with the idea that early regeneration is enhanced upon Cripto overexpression, we found that the expression of Myostatin, a member of the TGFβ superfamily which inhibits myoblasts differentiation and acts as a negative regulator of skeletal muscle growth (Amthor et al., 2002; Langley et al., 2002) is downregulated in Cripto overexpressing mice. Interestingly, this Cripto/Myostatin inverse correlation is consistent with previous findings on isolated myofibers in culture showing that sCripto antagonizes Myostatin leading to increased number of myoblast population committed to differentiation (Pax7−/MyoD+) (Guardiola et al., 2012).
In conclusion, all together these findings broaden our knowledge on the role of Cripto in skeletal muscle regeneration, and add novel insights into its activity in the satellite cell compartment. We believe that these novel mouse models, which also offer the unique opportunity to study the effect of Cripto in the regulation of satellite cell quiescence, activation and self-renewal in vivo, will be instrumental to get fundamental insights into the role of Cripto in skeletal muscle homeostasis and regeneration, as an important step toward the therapeutic use of this molecule.
Materials and Methods
Mice and Genotyping
To generate the pDsRedloxP/loxPCripto targeting vector, a mouse Cripto cDNA (750 bp) (Minchiotti et al., 2001) -IRES-eGFP cassette was cloned downstream of the Discosoma sp. red fluorescent protein (DsRed) gene into the pGapDsRedloxP/loxP vector, containing the CMV enhancer/chicken beta-actin promoter (CMV/β-Actin) and two loxP sites flanking the DsRed gene upstream of triple STOP-polyA sequences (kindly provided by Dr. Silvia Brunelli). Briefly, a Xho-AflII 2.2 kb DNA fragment spanning the Cripto-IRES-eGFP cassette was excised from the pCripto-IRES-eGFP plasmid (kindly provided by Dr. Giovanna L. Liguori), blunt ended and cloned into the backbone of the pGapDsRedloxP/loxP vector previously linearized with EcoR1 and filled in with Klenow Fragment of DNA polymerase I (New England Biolabs). Proper orientation of the inserted Cripto-IRES-eGFP DNA fragment was verified by restriction digestion and DNA sequencing. To generate the transgenic mice, a SalI DNA fragment spanning the entire CMV/β-Actin/DsRedloxP/loxPCriptoIRES-eGFP transcription unit released from the pDsRedloxP/loxPCripto targeting vector was gel purified and injected into fertilized oocytes of FVB/N mice, in collaboration with the Core Facility of Conditional Mutagenesis at San Raffaele Hospital, Milan, Italy. The resulting mice were genotyped by PCR-based screening using primers (a-a′) spanning the Cripto gene exon4-exon5 junction for the amplification of both Cripto wt (247 bp) and transgenic (156 bp) alleles. The integrity of the transgene was further evaluated by PCR using primers (b-b′) annealing on the DsRed and eGFP sequences (2700 bp). Three out of sixty mice analyzed (5%) were positive for the transgene and only one of them (mouse 1) gave germline transmission of the transgene. Two copies of the transgene, which segregate independently, were identified in the genome of the founder mouse by Southern blot analysis. After germline transmission, heterozygous Tg:DsRedloxP/loxPCripto-eGFP(A) and Tg:DsRedloxP/loxPCripto-eGFP(B) mice colonies were maintained by crossing with wild-type FVB mice and named Tg:Cripto(A) and Tg:Cripto(B), respectively.
For Southern Blot analysis, 20 ug of genomic DNA was prepared from tail biopsies, digested with EcoRI and blotted on Immobilion-Ny+ (Millipore). The 32P-labeled probe was a PCR amplified DNA fragment spanning the eGFP insert (546 bp) from the pDsRedloxP/loxPCripto vector. The heterozygous Tg:Pax7CreERT2::DsRedloxP/loxPCripto-eGFP(A)/(B) mice were generated by crossing Tg:Pax7CreERT2 (Mourikis et al., 2012) with Tg:Cripto(A)/(B) animals; the offsprings of these crosses were genotyped by double PCR analysis to identify the Tg:Cripto (213 bp) and the Tg:Pax7CreERT2 (600 bp) alleles (DsRedCripto for/rev and Pax7Cre for/rev primers). The DNA was prepared from tail biopsy samples of breeding animals. To induce genetic recombination, Tg:Pax7CreERT2::Cripto and Tg:Cripto adult mice (5 weeks old) were injected intraperitoneally (i.p.) with tamoxifen (60 μg/g of body weight per day, Sigma–Aldrich) once a day for 5 days. The genetic recombination was verified by PCR analysis (primers c-c′) to distinguish the transgenic (1995 bp) from the recombining (838 bp) allele. The DNA was obtained from tibialis anterior (TA) muscle biopsy samples. The primer sequences for PCR screening, genotyping and genetic recombination analysis are reported in Table S1.
Cell Culture and Transfection
HEK-293T cells were grown in Dulbecco Modified Eagle Medium (DMEM, Gibco) supplemented with 10% Fetal Bovine Serum (FBS, Euroclone) and 1% penicillin-streptomycin (EuroClone), and plated on six-well plates at a density of 2 × 105 cells/cm2. Twenty-four hours after plating, cells were transfected with 4 μg of DNA (pDsRedloxP/loxPCripto ± pCMV-Cre recombinase expression vector), using Lypofectamin Transfecion Reagent (Life Technologies) following manufacturer's instructions. Forty-eight hours after transfection, cells were fixed for immunofluorescence analysis or visualized by fluorescence microscopy (DMI600; Leica), and images were acquired on an Orca R2 camera (Hamamatsu).
Muscle Injections, Preparation, and Analysis
Tibialis Anterior (TA) muscles of adult Tg:Cripto(A)/(B) transgenic mice were injected with either Cre recombinase-encoding Adeno Associated Virus (AAV-Cre) or the empty vector (AAV-Cntl) at 1 × 1012 genome copies/ml in 10 μl. Muscles were collected after 10 days and total protein extracts were prepared for further analysis. The AAV vectors were prepared by the AAV Vector Unit at ICGEB Trieste (http://icgeb.org/avu-core-facility.html) by packaging AAV2 vector genome backbone into AAV capsid serotype 9, as previously described (Arsic et al., 2003). To induce muscle damage, 10 μl of cardiotoxin (CTX, 70 μM in PBS, Sigma-Aldrich) were injected in the TA muscles of adult (5 weeks old) Tg:Pax7CreERT2::Cripto and Tg:Cripto mice. For morphometric analysis, muscles were harvested at the indicated time points after damage, and frozen in isopentane-cooled liquid nitrogen for cryosection. Myofiber Cross Sectional Area (CSA) was analyzed on haematoxylin/eosin (H&E) stained muscle sections using ImageJ software (freely available software developed at the National Institutes of Health, Bethesda, Maryland, USA). The number of myofibers having more than one nucleus on total area was evaluated, as previously described (Mittal et al., 2010).
All experiments were conducted in strict accordance with the institutional guidelines for animal research and approved by the Department of Public Health, Animal Health, Nutrition and Food Safety of the Italian Ministry of Health in accordance with the law on animal experimentation.
Western Blot
Western blot analysis was performed as previously described (Parisi et al., 2003) on total protein extracts prepared from either cells or TA muscles samples. TA muscles were homogenized with TissueLyserII (Qiagen) following manufacturer's instructions. These primary antibodies were used: Cripto (1:500, R&D System), eMyHC (1:100, Developmental Studies Hybridoma Bank), Gapdh (1:40.000, Abcam) and β-Actin (1:5000, Sigma-Aldrich).
Isolation of Mouse Satellite Cells and in Vitro Differentiation
Tg:Pax7CreERT2::Cripto and Tg:Cripto new-born mice were injected i.p. with tamoxifen (10 μM, 5 μl) every day, from day 1 to day 5 after birth (P1–P5). Preparation of MPCs from new-born mice (P7) was performed as previously described (Rando and Blau, 1994). Briefly the hindlimb and forelimb were removed and separated from the bones. Muscles were mechanically minced with surgical scissors and dissociated by enzymatic digestion [600 U/ml Collagenase Type IV (Gibco) and 1.8 U/ml Dispase II (Roche) in DMEM (Gibco)] at 37°C for 2 h in the shaker. Digested muscles were spinned at 500 rpm for 5′ and satellite cells were spinned down from supernatant (1500 rpm for 10′). Cells were plated on gelatin-coated dishes in proliferation medium [1% Pen/Strep (EuroClone), 1% Glutamin (Euroclone), 20% FBS (Euroclone), 2% UltraSerum (LifeScience) in DMEM (Gibco),] and myoblast population was enriched by several pre-plating steps (Richler and Yaffe, 1970). For Pax7/MyoD cell counting, MPCs were cytospinned at a density of 50.000 cells/spot after 24 h in proliferating medium. For myogenic differentiation, myoblasts were plated in 48 wells at a density of 2 × 104 cells/cm2 in proliferation medium. After twenty-four hours, proliferation medium was replaced with differentiation medium for 24–72 h [DMEM (Gibco) containing 2% (v/v) horse serum (Gibco) and 1% pen/strep (EuroClone)] and the fusion index was determined as the ratio of the number of nuclei in myotubes (MF20+ cells with n ≥ 2 nuclei) on the total nuclei (MF20+ cells).
Immunofluorescence Analysis
Cells were fixed for 10′ with 4% (wt/vol) paraformaldehyde (PFA) in Phosphate Buffer Saline (PBS) and blocked with 1% bovin serum albumin (BSA) in PBS. Primary antibodies used were: Cripto (Parisi et al., 2003), MF20 (1:50, Developmental Studies Hybridoma Bank), Pax7 (1:10, Developmental Studies Hybridoma Bank) and MyoD (1:50, SantaCruz). Appropriate fluorophore-conjugated secondary antibodies, Alexa Fluor 594 or 488 (1:600, Molecular Probes), biotin-conjugated goat anti-mouse (Jackson), and Cy3- (Jackson) or 488- (Molecular Probes) conjugated streptavidin were used for the visualization using DMI600 or DM600 (Leica) fluorescence microscopes.
Muscles were freshly frozen and cut in cryostat sections. Slides were fixed in 4% (wt/vol) PFA and processed as previously described (Nicolas et al., 2005). Briefly, muscle sections were permeabilized in ice-cold methanol for 6′ at −20°C, and boiled 15 min in 10 mM sodium citrate pH 6. Unbinding sites were blocked with 4% IgG-free BSA (Jackson) in PBS for 2–3 h at RT followed by a second blocking step with AffiniPure Fab Fragment goat anti-mouse IgG (Jackson). Primary antibodies were as follows: eMyHC (1:50, F1.652-s, Developmental Studies Hybridoma Bank), Pax7 (1:10, Developmental Studies Hybridoma Bank) and MyoD (1:50, SantaCruz) Appropriate fluorophore-conjugated secondary antibodies, Alexa Fluor 488 or 594 (1:600, Molecular Probes), biotin-conjugated goat anti-mouse (Jackson) and Cy3- (Jackson) or 488- (Molecular Probes) conjugated streptavidin were used for visualization. Nuclei were counterstained with DAPI (1:100 in PBS, VinciBiochem) and FluoSave Reagent (Calbiochem) was used for mounting. Labeling was visualized by epifluorescent illumination using a DM600 microscope (Leica), and images were acquired on a DFC360-FX camera (Leica).
ELISA-based Assay
ELISA–based assay was performed as previously described (Guardiola et al., 2012). Briefly ninety-six well plates were coated with 0.5 μg/ml of sheep anti-mouse Cripto Ab (R&D System, AF1538) in PBS overnight (O.N.) at 4°C. Unbinding sites were blocked with PBS-BSA 1% for 2 h at RT. After washing, protein extracts (300 μg) were added and plates were incubated O.N. at 4°C. The plates were incubated with 1 μg/ml of mouse Cripto biotinylated Ab (R&D System, BAF 1538) in PBET (0.1% BSA, 5 mM EDTA, 0.004% Tween-20 in PBS 1X pH 7,5) for 1 h at 37°C, and then for 1 h at RT. Finally, the plates were incubated for 1 h at RT with avidin/streptavidin complex conjugated with horse-radish peroxidase (Vectastain elite ABC kit, Vector Laboratories). The signals were visualized with o-phenylenediamineperoxidase substrate (OPD, Sigma-Aldrich) and the relative absorbance was read at 490 nm on a Benchmark microplate reader (Bio-Rad Laboratories).
RNA Preparation and qRT-PCR
TA muscles were homogenized with TissueLyserII (Qiagen) in TriReagent (Life Technologies) and total RNA was isolated according to the manufacturer's instructions. One microgram of total RNA was utilized for cDNA synthesis using SuperScript II Reverse Transcriptase and random hexamers (Qiagen). qRT-PCR was performed using SYBR Green PCR master mix (EuroClone). Primers are listed in Table S2.
Statistical Analysis
All values are expressed as mean ± Standard Error of the Mean (SEM). To determine significance between two groups, comparisons were made using unpaired Student t-tests. *P ≤ 0.05, **P ≤ 0.005, and ***P ≤ 0.0005 were considered statistically significant.
Author Contributions
CP, design of the work, data acquisition and analysis, data interpretation. SI, data acquisition and analysis, data interpretation. GA, LZ, FI, data acquisition and analysis. OG, conception and design of the work, data analysis, data interpretation and draft of the article. GM, conception and design of the work, data interpretation, writing and revising of the article. All the authors have read and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the Animal House and the Integrated Microscopy Facilities of IGB-CNR, and the AAV Vector Unit at ICGEB Trieste for technical assistance. The Core Facility of Conditional Mutagenesis at San Raffaele Hospital (HSR), Milan, is acknowledged for generating the Tg:Cripto transgenic mice. We are grateful to Dr. Shahragim Tajbakhsh for providing the Pax7-CT2 mice and to Dr. Silvia Brunelli and Dr. Giovanna L. Liguori for providing plasmids. This work has benefited from research funding from the European Community's Seventh Framework Programme in the project ENDOSTEM (Activation of vasculature associated stem cells and muscle stem cells for the repair and maintenance of muscle tissue, Grant agreement number 241440), the Italian Ministry of Education-University-Research (MIUR-PRIN and PON Cluster IRMI), and CARIPLO foundation to GM.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fcell.2015.00031/abstract
References
Amthor, H., Huang, R., McKinnell, I., Christ, B., Kambadur, R., Sharma, M., et al. (2002). The regulation and action of myostatin as a negative regulator of muscle development during avian embryogenesis. Dev. Biol. 251, 241–257. doi: 10.1006/dbio.2002.0812
Arsic, N., Zentilin, L., Zacchigna, S., Santoro, D., Stanta, G., Salvi, A., et al. (2003). Induction of functional neovascularization by combined VEGF and angiopoietin-1 gene transfer using AAV vectors. Mol. Ther. 7, 450–459. doi: 10.1016/S1525-0016(03)00034-0
Bentzinger, C. F., Von Maltzahn, J., and Rudnicki, M. A. (2010). Extrinsic regulation of satellite cell specification. Stem Cell Res. Ther. 1:27. doi: 10.1186/scrt27
Chargé, S., and Rudnicki, M. (2004). Cellular and molecular regulation of muscle regeneration. Physiol. Rev. 84, 209–238. doi: 10.1152/physrev.00019.2003
Chu, J., Ding, J., Jeays-Ward, K., Price, S. M., Placzek, M., and Shen, M. M. (2005). Non-cell-autonomous role for Cripto in axial midline formation during vertebrate embryogenesis. Development 132, 5539–5551. doi: 10.1242/dev.02157
Collins, C. A., Olsen, I., Zammit, P. S., Heslop, L., Petrie, A., Partridge, T. A., et al. (2005). Stem cell function, self-renewal, and behavioral heterogeneity of cells from the adult muscle satellite cell niche. Cell 122, 289–301. doi: 10.1016/j.cell.2005.05.010
Ding, J., Yang, L., Yan, Y. T., Chen, A., Desai, N., Wynshaw-Boris, A., et al. (1998). Cripto is required for correct orientation of the anterior-posterior axis in the mouse embryo. Nature 395, 702–707. doi: 10.1038/27215
Guardiola, O., Lafuste, P., Brunelli, S., Iaconis, S., Touvier, T., Mourikis, P., et al. (2012). Cripto regulates skeletal muscle regeneration and modulates satellite cell determination by antagonizing myostatin. Proc. Natl. Acad. Sci. U.S.A. 109, E3231–E3240. doi: 10.1073/pnas.1204017109
Hawke, T., and Garry, D. (2001). Myogenic satellite cells: physiology to molecular biology. J. Appl. Physiol. 1985 91, 534–551.
Inagaki, K., Fuess, S., Storm, T. A., Gibson, G. A., McTiernan, C. F., Kay, M. A., et al. (2006). Robust systemic transduction with AAV9 vectors in mice: efficient global cardiac gene transfer superior to that of AAV8. Mol. Ther. 14, 45–53. doi: 10.1016/j.ymthe.2006.03.014
Kharraz, Y., Guerra, J., Mann, C. J., Serrano, A. L., and Munoz-Canoves, P. (2013). Macrophage plasticity and the role of inflammation in skeletal muscle repair. Mediators Inflamm. 2013:491497. doi: 10.1155/2013/491497
Langley, B., Thomas, M., Bishop, A., Sharma, M., Gilmour, S., and Kambadur, R. (2002). Myostatin inhibits myoblast differentiation by down-regulating MyoD expression. J. Biol. Chem. 277, 49831–49840. doi: 10.1074/jbc.M204291200
Minchiotti, G. (2005). Nodal-dependant Cripto signaling in ES cells: from stem cells to tumor biology. Oncogene 24, 5668–5675. doi: 10.1038/sj.onc.1208917
Minchiotti, G., Manco, G., Parisi, S., Lago, C. T., Rosa, F., and Persico, M. G. (2001). Structure-function analysis of the EGF-CFC family member Cripto identifies residues essential for nodal signalling. Development 128, 4501–4510.
Minchiotti, G., Parisi, S., Liguori, G., Signore, M., Lania, G., Adamson, E. D., et al. (2000). Membrane-anchorage of Cripto protein by glycosylphosphatidylinositol and its distribution during early mouse development. Mech. Dev. 90, 133–142. doi: 10.1016/S0925-4773(99)00235-X
Mittal, A., Bhatnagar, S., Kumar, A., Paul, P. K., Kuang, S., and Kumar, A. (2010). Genetic ablation of TWEAK augments regeneration and post-injury growth of skeletal muscle in mice. Am. J. Pathol. 177, 1732–1742. doi: 10.2353/ajpath.2010.100335
Mourikis, P., Sambasivan, R., Castel, D., Rocheteau, P., Bizzarro, V., and Tajbakhsh, S. (2012). A critical requirement for notch signaling in maintenance of the quiescent skeletal muscle stem cell state. Stem Cells 30, 243–252. doi: 10.1002/stem.775
Nicolas, N., Marazzi, G., Kelley, K., and Sassoon, D. (2005). Embryonic deregulation of muscle stress signaling pathways leads to altered postnatal stem cell behavior and a failure in postnatal muscle growth. Dev. Biol. 281, 171–183. doi: 10.1016/j.ydbio.2005.02.022
Olguin, H. C., Yang, Z., Tapscott, S. J., and Olwin, B. B. (2007). Reciprocal inhibition between Pax7 and muscle regulatory factors modulates myogenic cell fate determination. J. Cell Biol. 177, 769–779. doi: 10.1083/jcb.200608122
Overbeek, P. A., Lai, S. P., Van Quill, K. R., and Westphal, H. (1986). Tissue-specific expression in transgenic mice of a fused gene containing RSV terminal sequences. Science 231, 1574–1577. doi: 10.1126/science.3006249
Palmiter, R. D., Brinster, R. L., Hammer, R. E., Trumbauer, M. E., Rosenfeld, M. G., Birnberg, N. C., et al. (1982). Dramatic growth of mice that develop from eggs microinjected with metallothionein-growth hormone fusion genes. Nature 300, 611–615. doi: 10.1038/300611a0
Parisi, S., D'Andrea, D., Lago, C. T., Adamson, E. D., Persico, M. G., and Minchiotti, G. (2003). Nodal-dependent Cripto signaling promotes cardiomyogenesis and redirects the neural fate of embryonic stem cells. J. Cell Biol. 163, 303–314. doi: 10.1083/jcb.200303010
Rando, T. A., and Blau, H. M. (1994). Primary mouse myoblast purification, characterization, and transplantation for cell-mediated gene therapy. J. Cell Biol. 125, 1275–1287. doi: 10.1083/jcb.125.6.1275
Richler, C., and Yaffe, D. (1970). The in vitro cultivation and differentiation capacities of myogenic cell lines. Dev. Biol. 23, 1–22. doi: 10.1016/S0012-1606(70)80004-5
Saclier, M., Cuvellier, S., Magnan, M., Mounier, R., and Chazaud, B. (2013). Monocyte/macrophage interactions with myogenic precursor cells during skeletal muscle regeneration. FEBS J. 280, 4118–4130. doi: 10.1111/febs.12166
Shen, M. M., and Schier, A. F. (2000). The EGF-CFC gene family in vertebrate development. Trends Genet. 16, 303–309. doi: 10.1016/S0168-9525(00)02006-0
Tajbakhsh, S. (2009). Skeletal muscle stem cells in developmental versus regenerative myogenesis. J. Intern. Med. 266, 372–389. doi: 10.1111/j.1365-2796.2009.02158.x
Thomas, M., Langley, B., Berry, C., Sharma, M., Kirk, S., Bass, J., et al. (2000). Myostatin, a negative regulator of muscle growth, functions by inhibiting myoblast proliferation. J. Biol. Chem. 275, 40235–40243. doi: 10.1074/jbc.M004356200
Xu, C., Liguori, G., Persico, M. G., and Adamson, E. D. (1999). Abrogation of the Cripto gene in mouse leads to failure of postgastrulation morphogenesis and lack of differentiation of cardiomyocytes. Development 126, 483–494.
Keywords: Cripto, transgenic mice, satellite cells, muscle regeneration, myogenic commitment
Citation: Prezioso C, Iaconis S, Andolfi G, Zentilin L, Iavarone F, Guardiola O and Minchiotti G (2015) Conditional Cripto overexpression in satellite cells promotes myogenic commitment and enhances early regeneration. Front. Cell Dev. Biol. 3:31. doi: 10.3389/fcell.2015.00031
Received: 06 March 2015; Accepted: 02 May 2015;
Published: 21 May 2015.
Edited by:
Pura Muñoz-Cánoves, Pompeu Fabra University, SpainReviewed by:
Hugo C. Olguín, Pontificia Universidad Católica de Chile, ChilePier Lorenzo Puri, SanfordBurnham Medical Research Intitute, USA
Copyright © 2015 Prezioso, Iaconis, Andolfi, Zentilin, Iavarone, Guardiola and Minchiotti. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ombretta Guardiola and Gabriella Minchiotti, Stem Cell Fate Laboratory, Institute of Genetics and Biophysics “A. Buzzati-Traverso,” CNR, Via Pietro Castellino 111, Naples 80131, Italy,b21icmV0dGEuZ3VhcmRpb2xhQGlnYi5jbnIuaXQ=;Z2FicmllbGxhLm1pbmNoaW90dGlAaWdiLmNuci5pdA==
 Carolina Prezioso
Carolina Prezioso Salvatore Iaconis
Salvatore Iaconis Gennaro Andolfi
Gennaro Andolfi Lorena Zentilin
Lorena Zentilin Francescopaolo Iavarone
Francescopaolo Iavarone Ombretta Guardiola
Ombretta Guardiola Gabriella Minchiotti
Gabriella Minchiotti