- 1Service of Cardiology, Centre Hospitalier Universitaire Vaudois (CHUV), Lausanne, Switzerland
- 2Danish Stem Cell Center, University of Copenhagen, Copenhagen, Denmark
Beta cells are defined by the genes they express, many of which are specific to this cell type, and ensure a specific set of functions. Beta cells are also defined by a set of genes they should not express (in order to function properly), and these genes have been called forbidden genes. Among these, the transcriptional repressor RE-1 Silencing Transcription factor (REST) is expressed in most cells of the body, excluding most populations of neurons, as well as pancreatic beta and alpha cells. In the cell types where it is expressed, REST represses the expression of hundreds of genes that are crucial for both neuronal and pancreatic endocrine function, through the recruitment of multiple transcriptional and epigenetic co-regulators. REST targets include genes encoding transcription factors, proteins involved in exocytosis, synaptic transmission or ion channeling, and non-coding RNAs. REST is expressed in the progenitors of both neurons and beta cells during development, but it is down-regulated as the cells differentiate. Although REST mutations and deregulation have yet to be connected to diabetes in humans, REST activation during both development and in adult beta cells leads to diabetes in mice.
Introduction
REST is a transcription factor that represses numerous genes that are essential to the function of beta cells. It is thus actively excluded from beta cells and maintains beta cell genes repressed in other cell types. Similarly, REST is excluded from the majority of neurons, where it was first discovered, and represses neuronal genes in other cell types. We review the discovery of REST as well as its mechanisms of action and targets, mostly based on work with a primary focus on neurons. We also discuss more specific work uncovering how REST repression contributes to triggering beta cell differentiation and function.
REST, Discovery as a Repressor of Neuronal Traits
In 1995, Gail Mandel and David Anderson's groups discovered that the expression of a small battery of genes was restricted to neurons by the recruitment in other cell types of a zinc finger transcriptional repressor to a 21 bp DNA element (Kraner et al., 1992; Mori et al., 1992). This repressor was called RE-1 Silencing Transcription factor (REST) because it binds to a DNA element called repressor element-1 (RE-1) located in the rat Scn2a2 gene (Chong et al., 1995; Figure 1). In the context of the rat Stmn2 gene, it was called Neuron-Restrictive Silencer Factor (NRSF) and its target site was named neuron restrictive silencer element (NRSE; Schoenherr and Anderson, 1995). These breakthrough papers, followed by many others reporting the identification of novel REST target genes, described a default pathway whereby the absence of a unique factor (REST) determines part of the gene activity encoding fundamental traits of terminally differentiated neurons (RE-1-containing genes). Target genes of REST were found to be enriched in functions linked to synaptic transmission (Schoch et al., 1996; Bessis et al., 1997; Myers et al., 1998; Lietz et al., 2003; Bruce et al., 2004; Ballas et al., 2005), neurotransmitter signaling (Wood et al., 1996; Bessis et al., 1997; Bai et al., 1998; Myers et al., 1998), ion channeling (Chong et al., 1995; Yeo et al., 2009) and specifically in humans, to learning and memory (Rockowitz and Zheng, 2015), and to neuroprotection and cognitive function (Lu et al., 2014). REST expression was also found in undifferentiated neural progenitors where it prevents precocious expression of the target genes characterizing differentiated neurons (Chong et al., 1995; Schoenherr and Anderson, 1995; Ballas et al., 2005; Figure 1). REST inactivation in mice resulted in embryonic lethality starting after embryonic day (E) 9.5 with ectopic neuronal gene expression in multiple tissues but surprisingly not all (Chen et al., 1998).
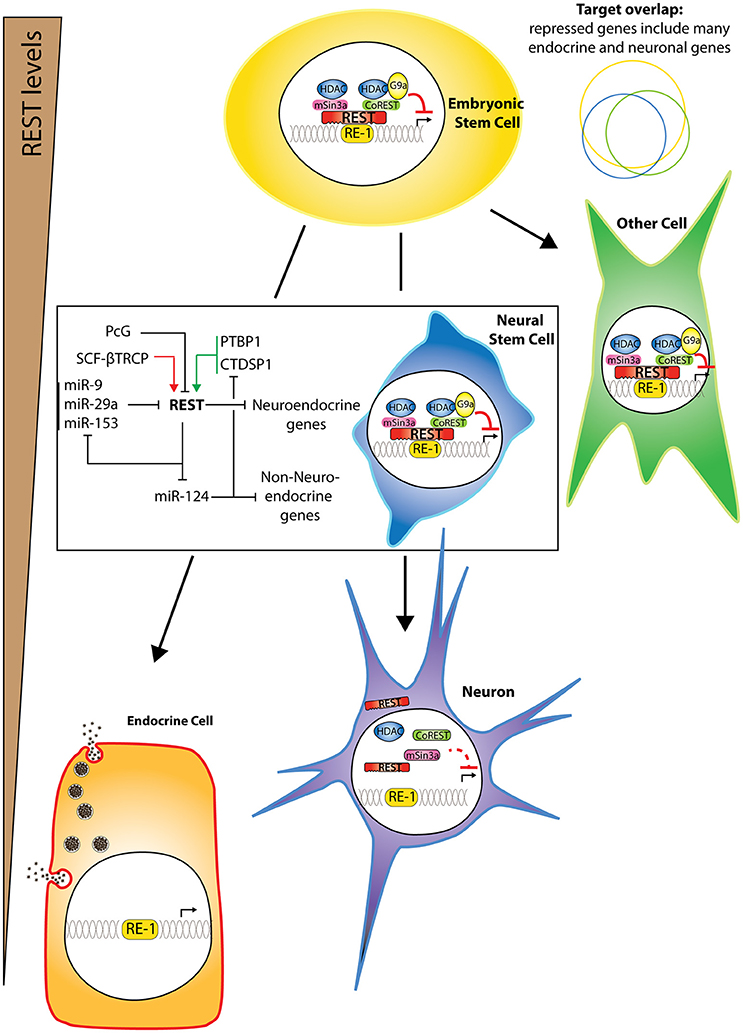
Figure 1. REST repressor complex and activity in different cell types. Different cell types express different REST levels. Endocrine cells and neurons express the lowest levels though some neurons can reactivate some REST transcripts at least in pathological scenarios. In all other cell types, there is some binding of REST to its RE-1 target sequence and repressive activity, in conjunction with co-factors. The repressed targets exhibit large but not total overlap between different cell types (with same color code as represented cells). During neuronal differentiation, REST represses neuroendocrine genes, as well as specific miRNAs that are both targets and regulators of REST, thereby forming double negative feedback loops. Imbalanced action of positive (green arrows) and negative (red arrows) regulators of REST stability might trigger a switch toward differentiation.
Several observations suggest that REST can be present and active in some neurons. Even though it is accepted that REST expression generally decreases during neuronal differentiation (Chong et al., 1995; Schoenherr and Anderson, 1995; Ballas et al., 2005), REST protein (Calderone et al., 2003; Zuccato et al., 2003; Sun et al., 2005; Gao et al., 2011; Lu et al., 2014; Schiffer et al., 2014), and mRNA (Palm et al., 1998; Calderone et al., 2003; Kuwabara et al., 2004; Lu et al., 2014) have been detected in certain mature neurons, especially those of the hippocampus. It is important to note, however, that the genuine detection of REST has been rendered questionable mainly because of a paucity of reliable antibodies or because of a lack of appropriate controls. For example, discrepancies in the molecular weight of immunoreactive bands reported as specific using western blotting may raise several concerns. While the calculated size of REST is of 121 kDa, we, and others, have always observed it around 200 kDa, in experiments including both negative (native beta cell lines) and positive controls (cells overexpressing REST full length). The numerous aspecific bands observed with many commercially available antibodies therefore render questionable observations made using immunostaining without specific controls. In some neurons, alternative splicing of REST mRNA occurs (Palm et al., 1998), giving rise to splice variants of unclear significance such as REST4, which possesses an inserted neuron-specific exon leading to translational frame shift, resulting in a truncated REST protein with four zinc fingers (Palm et al., 1998, 1999). Importantly, disturbance of REST activity in neurons has been linked to several human diseases, as discussed later in this review, suggesting that the control of REST is also important for the maintenance of neuronal function as well as for synaptic plasticity.
In non-neuronal tissues, changes in REST levels induced by pathological conditions may provide a cellular adaptation by affecting the expression of its targets. For example, down-regulated REST expression upon hypertrophic stimulus triggers the cardiac fetal gene reprogramming and correlates with increased smooth muscle cell proliferation in vascular neointimal hyperplasia (Kuwahara et al., 2003; Cheong et al., 2005). In the same spirit, it acts as a tumor suppressor in human epithelial tissues and its de-regulation promotes tumor formation (Westbrook et al., 2005, 2008). It is noteworthy that some of the targets up-regulated in these pathological scenarios are also regulated by REST in the nervous system. However, they are in several instances regulated by REST in a cell type specific manner. For example, NPPA is repressed by REST in cardiac myocytes (Kuwahara et al., 2003) but not in a REST-expressing neuronal cell line (Wood et al., 2003), while CX36 expression can be repressed by ectopic REST expression in pancreatic beta and alpha cell lines but not in a neuronal context (Hohl and Thiel, 2005).
The cell-specificity of REST target occupancy has been resolved at the genome-wide level using ChIP-seq or ChIP-chip experiments, by comparing REST regulatory networks between embryonic stem cells (ESC) and neural stem cells (NSC; Johnson et al., 2008), or between different neuronal and non-neuronal cell lines (Bruce et al., 2009) and is further discussed below.
Mechanisms of Transcriptional Repression and Epigenetic Modifications
REST is a zinc finger protein related to members of the Gli-Kruppel family of transcriptional repressors. The protein contains a cluster of eight zinc fingers, which is required for binding to the RE-1 element, and two repressor domains at the N- and C-terminus (Figure 1). Even though a number of reports showed that the sole ectopic expression of REST is able to mediate target gene repression, REST activity relies on the coordinated recruitment of multiple transcriptional and epigenetic regulators at target loci. This combinatorial recruitment and interplay of co-repressors and chromatin-modifying enzymes defines the complexity and context-dependency of the regulatory landscape controlled by REST. The N-terminus of REST recruits mSin3A (Huang et al., 1999; Grimes et al., 2000), which serves as a scaffold for histone deacetylases (HDACs; Huang et al., 1999; Grimes et al., 2000). The C-terminus interacts with CoREST (Andrés et al., 1999), which also recruits HDACs in addition to the histone H3K4 demethylase, LSD1 (Shi et al., 2004), the H3K9 methyltransferase, G9a (Roopra et al., 2004), the methyl CpG binding protein MeCP2 (Lunyak et al., 2002; Ballas et al., 2005) and BRG-1 and associated factors (Battaglioli et al., 2002; Figure 1). Among others, reviewed in (Ooi and Wood, 2007; Qureshi and Mehler, 2009; Bithell, 2011), REST also interacts with components or effectors of the RNA polymerase II machinery, including the anti-neural small C-terminal domain phosphatase 1 (CTDSP1; Yeo et al., 2005; Visvanathan et al., 2007), Mediator subunits (Ding et al., 2009) and in human cells with the H3K4 demethylase SMCX (Tahiliani et al., 2007) and the corepressor C-terminal binding protein (CtBP; Garriga-Canut et al., 2006; Figure 1). In addition, REST acts in parallel with another key epigenetic system, the polycomb repressive complexes (PRC), which shapes developmental stage-specific regulatory networks by imprinting H3K27 trimethylation at target loci. However, divergent views emerge as to whether the two systems cooperate to drive gene repression at common loci. REST binding motif associates with EZH2-enriched chromatin regions (Ku et al., 2008) and might therefore influence the localization of PRC2-mediated chromatin marks at promoter regions containing RE-1 motifs. However, this crosstalk was not found to translate into transcriptional changes (Arnold et al., 2013). Contrasted observations concluded that REST has context-dependent functions for PRC1- and PRC2-recruitment (Ren and Kerppola, 2011; Dietrich et al., 2012) or that Polycomb and REST complexes direct independent epigenetic modifications controlling early neural fate decisions vs. terminal neural traits acquisition (McGann et al., 2014). Interestingly, the involvement of non-coding RNAs might explain in certain contexts how this cooperative shaping of chromatin states is orchestrated between REST and Polycomb complexes, as it appears that the LncRNA HOTAIR provides a scaffold for collective recruitment of both modifying complexes at the chromatin (Tsai et al., 2010).
Repertoire of REST-Regulated Genes
Identifying the repertoire of REST target loci has unraveled an additional level of complexity in the mechanisms dictated by REST to generate cell type-specific and developmental stage-specific gene activity programs (Bithell, 2011). The unusual length of the RE-1 motif has allowed in silico identification of putative REST targets across human and murine genomes (Bruce et al., 2004; Mortazavi et al., 2006; Wu and Xie, 2006) and accession to deep-sequencing-associated chromatin immunoprecipitation (ChIP) techniques has increased the number of genes expected to be under REST regulation to the scale of several thousands (Johnson et al., 2007; Otto et al., 2007; Bruce et al., 2009; Rockowitz and Zheng, 2015).
By comparing chromatin occupancy, chromatin modifications, and gene expression in various neuronal and non-neuronal cell types, several studies have provided novel findings. They reveal the existence of different combinations of RE-1 half sites, resulting in a variety of non-canonical RE-1 motifs with varying binding affinities for REST (Otto et al., 2007). Few REST targets were ascribed to non-neuronal functions, such as immune/inflammatory response and cell adhesion, suggesting that REST may not merely be a repressor of neuronal traits (Otto et al., 2007). Another study shows that the number of bound targets decreases as neurons differentiate: among all the targets bound in ESCs and NSCs, 45% are solely bound in ESCs while 50% are shared, and very few are specifically bound in NSC (Figure 1). Many of the ESC-specific REST targets are involved in pluripotency and are commonly targeted by key pluripotency factors OCT4, NANOG, and SOX2, including Nanog itself (Johnson et al., 2008). Therefore, REST appears to form an autoregulatory circuit which is connected to the autoregulatory circuit of pluripotency factors. Activating and repressive transcriptional signals thus control ESC pluripotency. When comparing eight neuronal and non-neuronal cell lines, 90% of all REST targets show cell-specificity in REST recruitment. The level of REST protein and the sequence variations in the RE-1 motif govern this modular association of REST to its repertoire of target loci across the different lines (Bruce et al., 2009). REST directly down-regulates a large number of genes at the transcriptional level, but also probably indirectly activates the expression of other genes at the post-transcriptional level via the repression of many noncoding targets (Conaco et al., 2006; Mortazavi et al., 2006; Wu and Xie, 2006; Visvanathan et al., 2007; Singh et al., 2008; Johnson et al., 2009), including several micro RNAs (miRNAs) considered to be brain-specific (such as miR9, miR124, miR132, miR135, miR139, and miR153; Figure 1). Importantly, REST itself appears to be a predicted target of miR-153 (Mortazavi et al., 2006; Wu and Xie, 2006), miR-9 and miR-29a (Wu and Xie, 2006). Furthermore, miR-124 and other neuronal-specific miRNAs target various components of the REST complex, including CTDSP1 and CoREST (Xue et al., 2013). Together, these reports describe a double-negative feedback loop between REST and brain-related miRNAs in controlling neuronal gene expression.
Altogether, these reports integrating transcriptomic and epigenomic data have contributed to a great extent toward the deciphering of the complex mechanisms by which REST shapes regulatory landscapes and controls gene expression in a developmental stage- and cell-specific manner.
REST is Disallowed for Normal Adult Pancreatic Beta Cell Function and Maintenance
Pancreatic beta cells and neurons are derived from different germ layers, the endoderm and ectoderm layers, respectively, yet they share a large number of functional similarities. Beta cells and neurons are both electrically excitable and respond to hormonal stimuli and glucose by depolarization and exocytosis in a process similar to neurotransmitter release. At the molecular level, there are many overlapping patterns of gene expression between the two cell types (van Arensbergen et al., 2010). The shared proteins include enzymes implicated in the synthesis of neurotransmitters, receptors for growth factors and amino acids, neurofilaments, hormones (Atouf et al., 1997), proteins involved in the machinery of exocytosis of synaptic vesicles (Burgoyne and Morgan, 2003), and many transcription factors such as NEUROD/BETA2 (Naya et al., 1995). In 1997, the group of Scharfmann was the first to suggest that, in addition to sharing common transactivators, beta cell lines and neurons are devoid of REST expression. This enables the expression of important functional genes harboring RE-1 motifs including the N-methyl-D-aspartate (NMDA) receptor, the dopamine β-hydroxylase, Scg10, Synapsin, and the nicotinic acetylcholine receptor β2 subunit (Atouf et al., 1997; Figure 2). The list of REST-regulated genes expressed in beta cells was further extended with the characterization of RE-1 motifs associated with Mapk8ip1/Ib1 and Gjd2/Cx36 (Abderrahmani et al., 2001, 2004; Martin et al., 2003). MAPK8iP1, identified as a genetic factor associated with type 2 diabetes (Waeber et al., 2000), is a scaffold protein that protects beta cells against apoptosis via the interaction with the c-Jun N-terminal kinase (JNK) signaling pathway (Haefliger et al., 2003) and GJD2, a gap junction-forming protein that controls insulin secretion via cell to cell communication (Haefliger et al., 2003; Head et al., 2012).
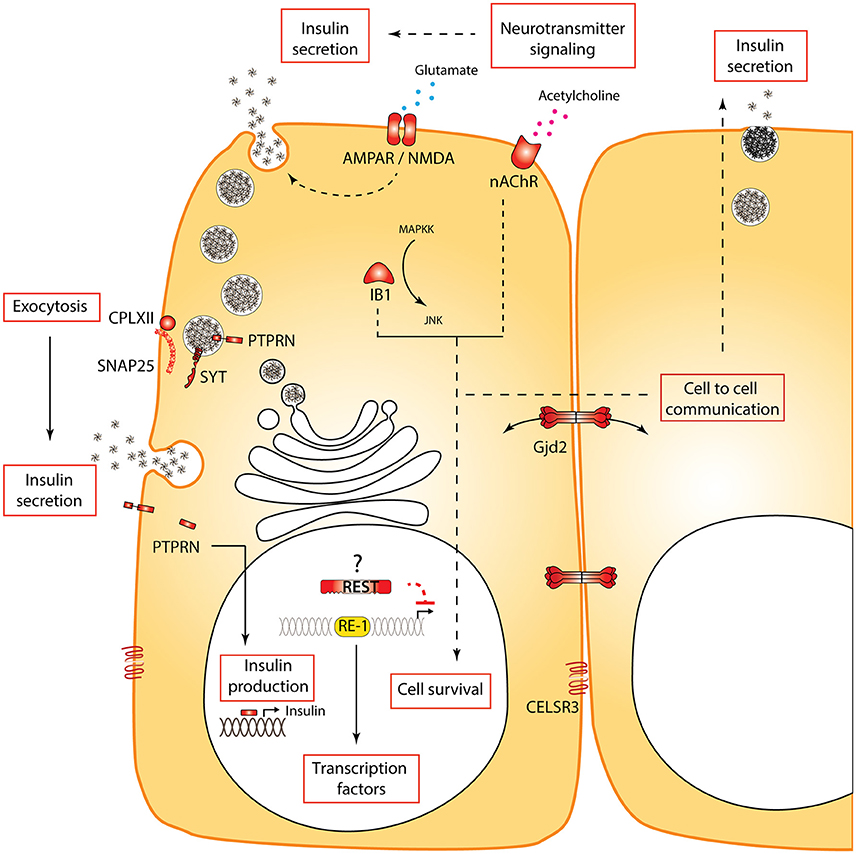
Figure 2. Processes and specific genes regulated by REST in beta cells. REST regulates multiple functional processes (boxes) in beta cells. Some non-exhaustive examples of genes and encoded proteins regulated by REST are shown in red. Several of these genes are regulated by REST not only in the functional adult beta cell but also during development. Dotted lines indicate a hypothetical or indirect link. The question mark next to REST indicates that although REST re-expression in beta cells in physiological or pathological conditions was not reported, its re-expression in neurons suggests this may be expected.
Using qPCR and Western blot, we showed that REST was also excluded from adult primary beta cells in mice (Martin et al., 2008, 2015). Importantly, while there are concerns about the specificity of available antibodies (as discussed above in the context of neurons), we provided a western blot analysis including bona fide negative (native beta cell line extracts) and positive controls (beta cells overexpressing murine REST). This allowed to identify the correct immunoreactive band and to unequivocally demonstrate the absence or extremely low levels of production of REST in murine islet cells. Previous transcriptome studies of the pancreas using next generation sequencing on whole islets or purified islet cells showed no or insignificant levels of Rest expression in mouse (Ku et al., 2012; Benner et al., 2014) and in human endocrine pancreas (Nica et al., 2013; Blodgett et al., 2015). The advent of single-cell transcriptomic analyses coupled to unbiased in silico identification of different cell types recently reaffirmed the concept that Rest is expressed in duct and in acinar cells, while totally absent from human endocrine cell types (Li et al., 2016; Muraro et al., 2016; Segerstolpe et al., 2016). We investigated the functional importance of REST target genes in pancreatic beta cells by ectopic expression of REST in beta cells using RIP-REST transgenic mice. REST mis-expression in beta cells led to impaired glucose homeostasis due to a decrease in both phases of glucose-stimulated insulin secretion. Several defects were noticed that likely accounted for this depressed insulin release: (1) pancreatic insulin content was decreased by half in RIP-REST mice, (2) beta cell counts were 30% lower than in control mice, (3) a selective battery of genes involved in exocytosis of large dense core vesicles, including Snap25, Syt4, Syt7, Syt9, and Cplx2 were under the control of functional RE-1 motif and were down-regulated (Martin et al., 2008; Figure 2). Altogether, our results indicated that besides their implication in insulin secretion, a subset of the RE-1-containing genes may be important for beta-cell turnover. This was confirmed in another line of RIP-REST animals featuring higher levels of REST transgene expression, which developed diabetes due to a dramatic loss in beta-cell mass. Genes relevant to beta cell survival were also found to be under REST control in vivo, including Gjd2, Ib1, Ptprn, and Cdk5r2 (Martin et al., 2012; Figure 2). Because of the wide range of naturally occurring combinations of RE-1 half sites in the genome (see above), it is likely that greater expression of REST led to a greater number of bound genes, especially those bearing non-canonical RE-1 motif with suboptimal binding affinity. Mis-expression in adult beta cells using an inducible transgene PDX-tTA; TetO-REST also led to diabetes, as a combined effect of REST mis-expression over both differentiation and maintenance of beta cells (Martin et al., 2015).
Taken together, our work and others' (reviewed in Thiel et al., 2015) show that REST activity needs to be repressed in adult beta cells, qualifying REST as a so-called “disallowed/forbidden” gene (Quintens et al., 2008; Pullen and Rutter, 2013; Lemaire et al., 2016). This classification, however, might be tempered by the fact that the strict definition would imply that Rest shows its lowest level of expression in pancreatic beta cells as compared to all other cell types. REST is also absent in other pancreatic endocrine cells as well as most neurons and does thus not strictly qualify the definition. We may need to categorize such genes if they become more numerous. Comprehensive analysis of the chromatin and transcriptional modifications triggered by the mis-expression of REST in adult beta cells would provide valuable insights into the number and role of all REST-controlled genes that constitute this beta-cell-specific sub-signature.
Role of Rest in Beta Cell Development
The exclusion of REST from beta cells (Atouf et al., 1997; Martin et al., 2008), the role of REST as a gatekeeper of neuronal differentiation (Ballas et al., 2005; Gao et al., 2011; Mao et al., 2011; Xue et al., 2013; Love and Prince, 2015) and the similarities between neuron and endocrine cell differentiation suggested that REST may also restrict endocrine cell differentiation during development. Accordingly, we found that REST is expressed in progenitors, and is down-regulated in differentiating alpha and beta cells during development (Martin et al., 2015). Several lines of evidence suggest that REST is repressed already in NEUROG3+ endocrine precursors (Figure 3). First, Rest transcript is decreased 5.8 times more in beta cells emerging from NEUROG3+ precursors than in young NEUROG3+ cells (http://www.betacell.org/resources/data/studies/view/study_id/3100 and ArrayExpress E-CBIL-48) (Soyer et al., 2010). Second, transcriptome analysis after NEUROG3 in vivo gain-of-function at E11.25 (Johansson et al., 2007) shows a 1.9 fold decrease of Rest in the pancreatic bud after NEUROG3 over-expression (http://www.betacell.org/resources/data/studies/view/study_id/3733) (Cortijo et al., 2012). The Rest gene acquires a Polycomb-mediated H3K27me3 repressive mark after the pancreatic progenitor stage, which can be interpreted as an outcome of the repression of REST expression and/or as a mechanism that ensures its repression (van Arensbergen et al., 2010). ChIP sequencing analysis has detected several REST binding sites in the vicinity of genes that promote endocrine development such as NeuroD, Neurog3, Onecut1, or Hnf4α (Johnson et al., 2007), which suggests that REST in progenitors represses endocrine differentiation (Figure 3). Hence, gain-of-function in pancreatic progenitors using the inducible transgenic line PDX-tTA; TetO-REST (Martin et al., 2015) severely decreases the formation of NEUROG3+ precursors and of differentiated endocrine cells. Moreover, conditional inactivation of REST in pancreas progenitors inhibits the expression of important factors of endocrine differentiation, leading to the formation of endocrine cells in appropriate numbers but in a partially differentiated state. The observation that REST depletion is not sufficient to fully promote endocrine cell formation suggests that there are multiple gates in the differentiation, and that either other repressors or activators are acting parallel to REST. The control over a subprogram is in agreement with what was observed in the context of neuronal differentiation in Rest-deficient mice (Aoki et al., 2012) and zebrafish (Kok et al., 2012). In the neural retina, Rest deletion in ganglion progenitor cells led to an increased number of retinal ganglion cells, via the up-regulation of crucial regulators of this cell type (Mao et al., 2011). A change in differentiation can be difficult to detect if complex feedback mechanisms are involved. For example, in the adult brain, loss of REST only triggers a transient increase in neurogenesis and depletion of the hippocampal stem cells (Gao et al., 2011).
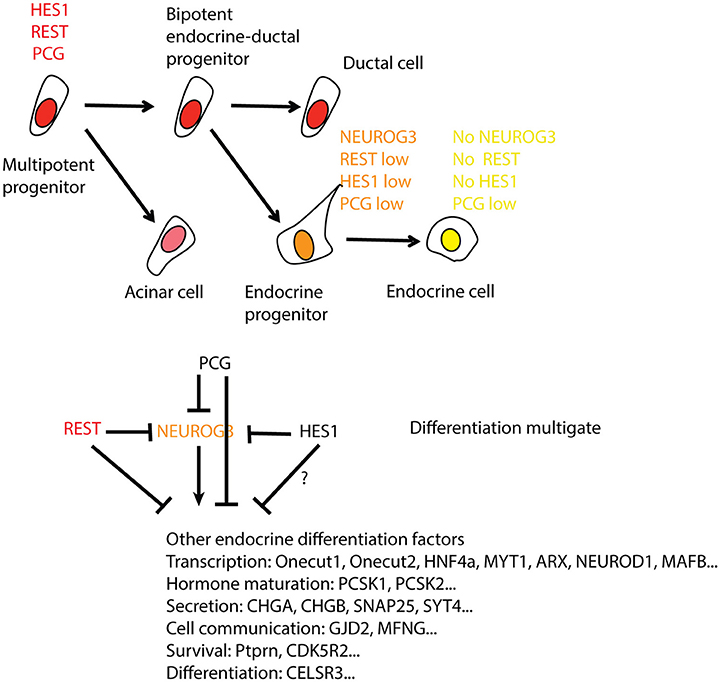
Figure 3. REST regulation and activity in the differentiation from pancreas progenitors to endocrine cells. The top panel shows the level of REST/REST activity in the endocrine lineage (red is high, orange medium, and yellow low/absent). REST levels are paralleled by polycomb (PcG) and the Notch target gene HES1. As these transcripts decrease, there is a transient peak of NEUROG3 as progenitors differentiate into endocrine progenitors. The endocrine cells derived from them exhibit low/absent expression of all four. There are multiple parallel blocks on endocrine differentiation by HES1, REST, and PCG at two stages of differentiation, firstly in the progression to endocrine precursors and secondly in their further differentiation into endocrine cells. How endocrine progenitors escape the multiple block to turn on NEUROG3 is unclear. NEUROG3 then promotes the differentiation program. The interactions between REST, HES1, and PCG remain to be deciphered at the level of targets and on their own regulatory sequences.
In the pancreas, NEUROG3, which appears to be a direct target of REST by Chip-Seq is not induced in the knock-out and may be independently repressed by HES1 (Jensen et al., 2000; Figure 3). It is also possible that, in spite of REST binding NEUROG3 regulatory sequences, the chromatin at this locus may not be poised to activation. This scheme is suggested by the observation that REST and Polycomb repressions act in parallel in neuronal differentiation (McGann et al., 2014) as well as in pancreatic beta-cells (van Arensbergen et al., 2010). Indeed, in pancreatic progenitors, PcG marks a wide number of disallowed genes for beta cells, including regulators of alternate developmental fate that will never be activated in the differentiation path, while REST binds in ES cells to genes in which PcG repression disappears in beta cells (Figure 3).
The targets activated upon REST depletion are expected to affect a variety of functions including transcription factors that control differentiation (Onecut1, Onecut2, Hnf4a, Myt1, Arx, Neurod1, Mafb), proteins acting in insulin maturation (Pcsk1, Pcsk2), insulin secretion (Chga, Chgb, Syt4), cell to cell communication (Gjd2, Mfng), anti-apoptotic activity (Ptprn, Cdk5r2) and differentiation functions (Celsr3; Figure 3). Although the subset of REST targets in the pancreas containing Celsr3 (Jia et al., 2014) and Syt4 is also under REST repression in the nervous system, many targets are specific to one or the other tissue, suggesting that transcriptional repression by REST functions in a cell type-specific manner, as discussed above. This may be accounted for by the level of REST protein, the sequence variations in the RE-1 motif or the presence of co-factors (Johnson et al., 2008; Bruce et al., 2009).
REST Down-Regulation as a Switch for Neuronal and Endocrine Differentiation
The examples discussed above illustrate how a tight regulation of REST leads to its expression in most cells and its general exclusion from beta cells and neurons. The mechanisms regulating REST during normal development, adult life and pathological scenarios are still largely enigmatic. Three promoters, six enhancers and two repressor regions with differences of activity in different cell types regulate REST transcription but the transcription factors regulating their activity are unclear (Koenigsberger et al., 2000). The REST gene contains several RE-1 sites, indicating a putative negative autoregulatory feedback on its own expression (Johnson et al., 2007, 2008).
REST is down-regulated as neurons or pancreatic endocrine cells differentiate, but what triggers this is unclear. In the pancreas, it is accompanied by the acquisition of an epigenetic PcG-mediated H3K27me3 repressive mark occurring after the pancreatic progenitor stage, which coincides with the activation of a core beta-cell derepression program (van Arensbergen et al., 2010). During ES cell differentiation into cortical neurons, most of the regulation is initially post-transcriptional while REST transcription is eventually decreased (Ballas et al., 2005). REST abundance and stability are influenced by coordinated kinase, phosphatase, and ubiquitin ligase activities. On the one hand, the CTDSP1 phosphatase activity targets a phosphorylation site in REST and stabilizes it in stem cells. (Yeo et al., 2005; Figure 1). On the other hand, because CTDSP1 expression decreases with differentiation (Visvanathan et al., 2007), the ERK-dependent phosphorylation at the same site, together with the peptidylprolyl isomerase Pin1 activity, promote REST degradation in neural progenitors (Nesti et al., 2014). It appears that phosphorylation of the motif allows recruitment of SCFβTRCP, an E3 ubiquitin ligase that is induced during neuronal differentiation and is responsible for REST degradation and subsequent neuronal differentiation (Westbrook et al., 2008). Decreased expression of the deubiquitylase HAUSP during neuronal differentiation further destabilizes REST (Huang et al., 2011). The activity of the REST repressive complex during neuronal differentiation is also tightly controlled at the transcriptional level, and includes a HDAC-dependent regulation triggered by retinoic acid (Ballas et al., 2005). However, the most documented mechanism is post-transcriptional and involves a double negative feedback loop involving miRNAs. REST regulates the expression of miRNAs and is itself regulated by them, including miR-153 (Mortazavi et al., 2006; Wu and Xie, 2006), miR-9 and miR-29a (Wu and Xie, 2006; Figure 1). miR-124, another component of this double feedback loop, has reciprocal activity by inhibiting non neuronal transcripts (Conaco et al., 2006). REST represses miR-124, an miRNA that targets various REST components including CTDSP1 and CoREST (Xue et al., 2013). This system of large autoregulatory loops is controlled by another feedforward loop that involves the polypyrimidine-tract-binding (PTBP1) protein, which secures this system by competing at miR-124 targets (Xue et al., 2013; Figure 1).
It would be of interest to identify which signals, in addition to that directed by PcG complex, dictate REST extinction along the pancreatic developmental pathway.
Up-Regulation of REST Activity in Neurons Promotes Neurological Diseases. Could Pathological REST Activation Trigger Beta Cell Failure?
Several studies have documented that aberrant induction of REST expression in neurons or progenitors plays a role in neurodegenerative and neurodevelopmental diseases. The example studied in most depth is Rett syndrome, which is a neurodevelopmental disorder caused by mutations in the methyl CpG binding protein 2 gene (MeCP2). This causes an increase in expression of REST and CoREST, and as a consequence the downregulation of their target BDNF (Abuhatzira et al., 2007) and neuron-specific K(+)-Cl(−) cotransporter2 (KCC2; Tang et al., 2016). A few cases of type 1 diabetes have been associated to Rett syndrome, but the number of cases is still too low to determine whether this is mere serendipity or if they were caused by MeCP2 mutations and downstream REST activity (Kurtoglu et al., 2005; Rekik et al., 2010; Akin et al., 2012). De-repression of REST in neurons has also been shown to occur in response to ischemia, leading to down-regulation of REST target genes, notably the GRIA2 subunit of the glutamate receptor regulating Ca2+ permeability and miR-29c, that promoted neuronal death (Calderone et al., 2003; Noh et al., 2012; Pandi et al., 2013). Kainic acid, a glutamate analog also promotes REST expression but it is unclear whether this occurs at physiological levels to modulate neuronal function or only upon massive glutamate release (Palm et al., 1998). REST activation has also been reported in the brain of Huntington's disease patients and involves a cytoplasmic sequestering of REST by wild type huntingtin, which is lost with the mutant protein (Zuccato et al., 2003, 2007; Schiffer et al., 2014). Increased REST activity in neurons or progenitor cells is also associated with Down syndrome (Bahn et al., 2002; Lepagnol-Bestel et al., 2009).
Up-regulation of Rest mRNA and protein is also a prominent feature of a subset of human glioblastomas and medulloblastomas (Lawinger et al., 2000; Su et al., 2006; Conti et al., 2012; Kamal et al., 2012; Wagoner and Roopra, 2012). These pediatric malignant brain tumors are thought to arise from undifferentiated neural progenitors, and REST may be promoting tumor formation by preventing their differentiation (Su et al., 2006). However, the mechanism underlying REST induction in these tumors remains unknown.
REST induction in the brain may not always be detrimental. REST is promoted by BMPs during astrocyte differentiation from progenitors and is likely to enforce programs divergent from their neuronal sister cells (Kohyama et al., 2010). This appears to be direct and is mediated by a smad-binding site in REST promoter, but whether BMP also promotes REST in other contexts remains to be explored. In addition, it has been proposed that Wnt-driven REST expression increases in aging neurons of the cortex and hippocampus and may play a protective role against Alzheimer's disease and fronto-temporal dementia (Lu et al., 2014).
REST expression is forbidden in pancreatic endocrine cells and violation of this disallowance can provoke diabetes in mice (Martin et al., 2012, 2015). Its up-regulation in humans may also cause or contribute to type 2 diabetes, neonatal diabetes, or maturity onset diabetes of the young. The latter two are caused by mutations in one gene, usually key to pancreatic endocrine development or beta cell biology. Though REST gain-of-function mutations have not yet been reported as causes of monogenic diabetes, the 22 known mutations account for 80% of cases (De Franco et al., 2015). About 20% of the cases therefore remain of unknown genetic etiology.
REST Repression as a Tool for Reprogramming Cells into Neurons or Endocrine Cells
Elucidating the role of REST in development and lineage specification uncovered new roles for this factor in embryonic stem cell (ESC) pluripotency. REST is expressed at high levels in mouse ESC (Ballas et al., 2005) and a series of genome-wide analyses demonstrated that in both mouse and human ESC, REST is a target and partner of multiple factors controlling pluripotency, including NANOG, OCT4, and SOX2 (Boyer et al., 2005; Loh et al., 2006; Wang et al., 2006; Kim et al., 2008). Moreover, a subset of the REST regulatory network is shared with these three factors (Johnson et al., 2008). However, the role of REST as a pluripotency factor remains controversial. Although loss of REST in mouse ESC may decrease the expression of pluripotency factors and their self-renewal capacity via activation of miR-21 (Singh et al., 2008, 2012, 2015), other reports showed that REST loss-of-function did not restrict ESC self-renewal or their multiple lineage potential (Buckley et al., 2009; Jørgensen et al., 2009; Yamada et al., 2010). REST may be important, however, for proper timing and acquisition of the primitive endodermal fate by repressing NANOG (Johnson et al., 2008; Yamada et al., 2010).
Recent evidence suggests that depleting non-neuronal cells of REST can promote reprogramming. Fresh mouse cortical astrocytes can be reprogrammed into neurons when transduced with ASCL1 or NEUROG2. However, the conversion of astrocytes cultured for several days is possible only with the inactivation of REST and the activation of either ASCL1 or NEUROG2. Prolonged culture increases the level of H4K20me3 at the NeuroD4 promoter, a key reprogramming factor, and promotes a local chromatin environment favorable to the repressive complex REST. In these conditions REST outcompetes NEUROG2 in terms of binding to the NeuroD4 promoter (Masserdotti et al., 2015).
It is not clear whether this change in chromatin at REST loci upon culture is a general principle and whether it can be exploited to reprogram other cell types. Genetic inactivation of REST does not efficiently turn fibroblasts into neurons despite the induction of some neuronal genes, suggesting that additional epigenetic or transcription factors are required (Aoki et al., 2012). A similar strategy applied to generate reprogrammed beta cells had very limited success. Indeed, forced expression of ShRest/Nrsf, shShh, and PDX1 in bone marrow-derived mesenchymal stem cells induces several markers of beta cells including insulin, but to <10 fold, which is minimal since these cells express very low levels of beta cell genes (Li et al., 2012). However, shREST was able to trigger neuronal differentiation from MEFs, potentially via a feedforward loop that involves PTBP1 and miR-124 (Xue et al., 2013). Whether the same paradigm applies to humans is unclear (Xue et al., 2016). In these attempts, one must bear in mind that depleting REST from a cell that does express it may have detrimental effects. A recent model of REST inactivation in neuronal progenitors shows that the absence of REST in proliferative cells that normally express it leads to DNA damage (Nechiporuk et al., 2016). If the affected cells are prevented from dying by p53 inactivation, some develop glioblastoma and primitive neuroectodermal tumor.
Despite the relatively large amount of information regarding the importance of REST in the regulation of the neuronal lineage, the study of REST vis-a-vis endocrine differentiation and beta cell function is in its infancy. Proof of concepts have established that REST must be excluded from beta cells but a global view of its regulation and targets during differentiation, in the embryo, in the early post-natal cells, during aging and in pathological processes is missing. Although it is becoming clear that REST is absent in human pancreatic endocrine cells, a tally of genes regulated by REST would be valuable, as well as information on its expression and role in other endocrine lineages in the intestine. Finally, most research has been restricted to mice but its targets in the human endocrine lineage and its role in diabetes will eventually be of utmost interest.
Author Contributions
DM and AB conceived the review, collected the bibliographic references and wrote the manuscript. Some of the work presented is based on their own experimental investigations.
Funding
This project was financed by a Novo Nordisk Foundation grant to AB.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Abderrahmani, A., Niederhauser, G., Plaisance, V., Haefliger, J. A., Regazzi, R., and Waeber, G. (2004). Neuronal traits are required for glucose-induced insulin secretion. FEBS Lett. 565, 133–138. doi: 10.1016/j.febslet.2004.04.002
Abderrahmani, A., Steinmann, M., Plaisance, V., Niederhauser, G., Haefliger, J. A., Mooser, V., et al. (2001). The transcriptional repressor REST determines the cell-specific expression of the human MAPK8IP1 gene encoding IB1 (JIP-1). Mol. Cell. Biol. 21, 7256–7267. doi: 10.1128/MCB.21.21.7256-7267.2001
Abuhatzira, L., Makedonski, K., Kaufman, Y., Razin, A., and Shemer, R. (2007). MeCP2 deficiency in the brain decreases BDNF levels by REST/CoREST-mediated repression and increases TRKB production. Epigenetics 2, 214–222. doi: 10.4161/epi.2.4.5212
Akin, L., Adal, E., Akin, M. A., and Kurtoglu, S. (2012). A case of diabetes mellitus associated with Rett syndrome. J. Pediatr. Endocrinol. Metab. 25, 197–198. doi: 10.1515/jpem.2011.337
Andrés, M. E., Burger, C., Peral-Rubio, M. J., Battaglioli, E., Anderson, M. E., Grimes, J., et al. (1999). CoREST: a functional corepressor required for regulation of neural-specific gene expression. Proc. Natl. Acad. Sci. U.S.A. 96, 9873–9878. doi: 10.1073/pnas.96.17.9873
Aoki, H., Hara, A., Era, T., Kunisada, T., and Yamada, Y. (2012). Genetic ablation of rest leads to in vitro-specific derepression of neuronal genes during neurogenesis. Development 139, 667–677. doi: 10.1242/dev.072272
Arnold, P., Schöler, A., Pachkov, M., Balwierz, P. J., Jørgensen, H., Stadler, M. B., et al. (2013). Modeling of epigenome dynamics identifies transcription factors that mediate Polycomb targeting. Genome Res. 23, 60–73. doi: 10.1101/gr.142661.112
Atouf, F., Czernichow, P., and Scharfmann, R. (1997). Expression of neuronal traits in pancreatic beta cells. Implication of neuron-restrictive silencing factor/repressor element silencing transcription factor, a neuron-restrictive silencer. J. Biol. Chem. 272, 1929–1934. doi: 10.1074/jbc.272.3.1929
Bahn, S., Mimmack, M., Ryan, M., Caldwell, M. A., Jauniaux, E., Starkey, M., et al. (2002). Neuronal target genes of the neuron-restrictive silencer factor in neurospheres derived from fetuses with Down's syndrome: a gene expression study. Lancet 359, 310–315. doi: 10.1016/S0140-6736(02)07497-4
Bai, G., Norton, D. D., Prenger, M. S., and Kusiak, J. W. (1998). Single-stranded DNA-binding proteins and neuron-restrictive silencer factor participate in cell-specific transcriptional control of the NMDAR1 gene. J. Biol. Chem. 273, 1086–1091. doi: 10.1074/jbc.273.2.1086
Ballas, N., Grunseich, C., Lu, D. D., Speh, J. C., and Mandel, G. (2005). REST and its corepressors mediate plasticity of neuronal gene chromatin throughout neurogenesis. Cell 121, 645–657. doi: 10.1016/j.cell.2005.03.013
Battaglioli, E., Andres, M. E., Rose, D. W., Chenoweth, J. G., Rosenfeld, M. G., Anderson, M. E., et al. (2002). REST repression of neuronal genes requires components of the hSWI.SNF complex. J. Biol. Chem. 277, 41038–41045. doi: 10.1074/jbc.M205691200
Benner, C., van der Meulen, T., Cacéres, E., Tigyi, K., Donaldson, C. J., and Huising, M. O. (2014). The transcriptional landscape of mouse beta cells compared to human beta cells reveals notable species differences in long non-coding RNA and protein-coding gene expression. BMC Genomics 15:620. doi: 10.1186/1471-2164-15-620
Bessis, A., Champtiaux, N., Chatelin, L., and Changeux, J. P. (1997). The neuron-restrictive silencer element: a dual enhancer/silencer crucial for patterned expression of a nicotinic receptor gene in the brain. Proc. Natl. Acad. Sci. U.S.A. 94, 5906–5911. doi: 10.1073/pnas.94.11.5906
Bithell, A. (2011). REST: transcriptional and epigenetic regulator. Epigenomics 3, 47–58. doi: 10.2217/epi.10.76
Blodgett, D. M., Nowosielska, A., Afik, S., Pechhold, S., Cura, A. J., Kennedy, N. J., et al. (2015). Novel observations from next-generation RNA sequencing of highly purified human adult and fetal islet cell subsets. Diabetes 64, 3172–3181. doi: 10.2337/db15-0039
Boyer, L. A., Lee, T. I., Cole, M. F., Johnstone, S. E., Levine, S. S., Zucker, J. P., et al. (2005). Core transcriptional regulatory circuitry in human embryonic stem cells. Cell 122, 947–956. doi: 10.1016/j.cell.2005.08.020
Bruce, A. W., Donaldson, I. J., Wood, I. C., Yerbury, S. A., Sadowski, M. I., Chapman, M., et al. (2004). Genome-wide analysis of repressor element 1 silencing transcription factor/neuron-restrictive silencing factor (REST/NRSF) target genes. Proc. Natl. Acad. Sci. U.S.A. 101, 10458–10463. doi: 10.1073/pnas.0401827101
Bruce, A. W., López-Contreras, A. J., Flicek, P., Down, T. A., Dhami, P., Dillon, S. C., et al. (2009). Functional diversity for REST (NRSF) is defined by in vivo binding affinity hierarchies at the DNA sequence level. Genome Res. 19, 994–1005. doi: 10.1101/gr.089086.108
Buckley, N. J., Johnson, R., Sun, Y. M., and Stanton, L. W. (2009). Is REST a regulator of pluripotency? Nature 457, E5–E6. discussion: E7. doi: 10.1038/nature07784
Burgoyne, R. D., and Morgan, A. (2003). Secretory granule exocytosis. Physiol. Rev. 83, 581–632. doi: 10.1152/physrev.00031.2002
Calderone, A., Jover, T., Noh, K. M., Tanaka, H., Yokota, H., Lin, Y., et al. (2003). Ischemic insults derepress the gene silencer REST in neurons destined to die. J. Neurosci. 23, 2112–2121. Available online at: http://www.jneurosci.org/content/23/6/2112.long
Chen, Z. F., Paquette, A. J., and Anderson, D. J. (1998). NRSF/REST is required in vivo for repression of multiple neuronal target genes during embryogenesis. Nat. Genet. 20, 136–142. doi: 10.1038/2431
Cheong, A., Bingham, A. J., Li, J., Kumar, B., Sukumar, P., Munsch, C., et al. (2005). Downregulated REST transcription factor is a switch enabling critical potassium channel expression and cell proliferation. Mol. Cell 20, 45–52. doi: 10.1016/j.molcel.2005.08.030
Chong, J. A., Tapia-Ramírez, J., Kim, S., Toledo-Aral, J. J., Zheng, Y., Boutros, M. C., et al. (1995). REST: a mammalian silencer protein that restricts sodium channel gene expression to neurons. Cell 80, 949–957. doi: 10.1016/0092-8674(95)90298-8
Conaco, C., Otto, S., Han, J. J., and Mandel, G. (2006). Reciprocal actions of REST and a microRNA promote neuronal identity. Proc. Natl. Acad. Sci. U.S.A. 103, 2422–2427. doi: 10.1073/pnas.0511041103
Conti, L., Crisafulli, L., Caldera, V., Tortoreto, M., Brilli, E., Conforti, P., et al. (2012). REST controls self-renewal and tumorigenic competence of human glioblastoma cells. PLoS ONE 7:e38486. doi: 10.1371/journal.pone.0038486
Cortijo, C., Gouzi, M., Tissir, F., and Grapin-Botton, A. (2012). Planar cell polarity controls pancreatic beta cell differentiation and glucose homeostasis. Cell Rep. 2, 1593–1606. doi: 10.1016/j.celrep.2012.10.016
De Franco, E., Flanagan, S. E., Houghton, J. A., Lango Allen, H., Mackay, D. J., Temple, I. K., et al. (2015). The effect of early, comprehensive genomic testing on clinical care in neonatal diabetes: an international cohort study. Lancet 386, 957–963. doi: 10.1016/S0140-6736(15)60098-8
Dietrich, N., Lerdrup, M., Landt, E., Agrawal-Singh, S., Bak, M., Tommerup, N., et al. (2012). REST-mediated recruitment of polycomb repressor complexes in mammalian cells. PLoS Genet. 8:e1002494. doi: 10.1371/journal.pgen.1002494
Ding, N., Tomomori-Sato, C., Sato, S., Conaway, R. C., Conaway, J. W., and Boyer, T. G. (2009). MED19 and MED26 are synergistic functional targets of the RE1 silencing transcription factor in epigenetic silencing of neuronal gene expression. J. Biol. Chem. 284, 2648–2656. doi: 10.1074/jbc.M806514200
Gao, Z., Ure, K., Ding, P., Nashaat, M., Yuan, L., Ma, J., et al. (2011). The master negative regulator REST/NRSF controls adult neurogenesis by restraining the neurogenic program in quiescent stem cells. J. Neurosci. 31, 9772–9786. doi: 10.1523/JNEUROSCI.1604-11.2011
Garriga-Canut, M., Schoenike, B., Qazi, R., Bergendahl, K., Daley, T. J., Pfender, R. M., et al. (2006). 2-Deoxy-D-glucose reduces epilepsy progression by NRSF-CtBP-dependent metabolic regulation of chromatin structure. Nat. Neurosci. 9, 1382–1387. doi: 10.1038/nn1791
Grimes, J. A., Nielsen, S. J., Battaglioli, E., Miska, E. A., Speh, J. C., Berry, D. L., et al. (2000). The co-repressor mSin3A is a functional component of the REST-CoREST repressor complex. J. Biol. Chem. 275, 9461–9467. doi: 10.1074/jbc.275.13.9461
Haefliger, J. A., Tawadros, T., Meylan, L., Gurun, S. L., Roehrich, M. E., Martin, D., et al. (2003). The scaffold protein IB1/JIP-1 is a critical mediator of cytokine-induced apoptosis in pancreatic beta cells. J. Cell Sci. 116(Pt 8), 1463–1469. doi: 10.1242/jcs.00356
Head, W. S., Orseth, M. L., Nunemaker, C. S., Satin, L. S., Piston, D. W., and Benninger, R. K. (2012). Connexin-36 gap junctions regulate in vivo first- and second-phase insulin secretion dynamics and glucose tolerance in the conscious mouse. Diabetes 61, 1700–1707. doi: 10.2337/db11-1312
Hohl, M., and Thiel, G. (2005). Cell type-specific regulation of RE-1 silencing transcription factor (REST) target genes. Eur. J. Neurosci. 22, 2216–2230. doi: 10.1111/j.1460-9568.2005.04404.x
Huang, Y., Myers, S. J., and Dingledine, R. (1999). Transcriptional repression by REST: recruitment of Sin3A and histone deacetylase to neuronal genes. Nat. Neurosci. 2, 867–872. doi: 10.1038/13165
Huang, Z., Wu, Q., Guryanova, O. A., Cheng, L., Shou, W., Rich, J. N., et al. (2011). Deubiquitylase HAUSP stabilizes REST and promotes maintenance of neural progenitor cells. Nat. Cell Biol. 13, 142–152. doi: 10.1038/ncb2153
Jensen, J., Heller, R. S., Funder-Nielsen, T., Pedersen, E. E., Lindsell, C., Weinmaster, G., et al. (2000). Independent development of pancreatic alpha- and beta-cells from neurogenin3-expressing precursors: a role for the notch pathway in repression of premature differentiation. Diabetes 49, 163–176. doi: 10.2337/diabetes.49.2.163
Jia, Z., Guo, Y., Tang, Y., Xu, Q., Li, B., and Wu, Q. (2014). Regulation of the protocadherin Celsr3 gene its role in globus pallidus development connectivity. Mol. Cell Biol. 34, 3895–3910. doi: 10.1128/MCB.00760-14
Johansson, K. A., Dursun, U., Jordan, N., Gu, G., Beermann, F., Gradwohl, G., et al. (2007). Temporal control of neurogenin3 activity in pancreas progenitors reveals competence windows for the generation of different endocrine cell types. Dev. Cell 12, 457–465. doi: 10.1016/j.devcel.2007.02.010
Johnson, D. S., Mortazavi, A., Myers, R. M., and Wold, B. (2007). Genome-wide mapping of in vivo protein-DNA interactions. Science 316, 1497–1502. doi: 10.1126/science.1141319
Johnson, R., Teh, C. H., Jia, H., Vanisri, R. R., Pandey, T., Lu, Z. H., et al. (2009). Regulation of neural macroRNAs by the transcriptional repressor REST. RNA 15, 85–96. doi: 10.1261/rna.1127009
Johnson, R., Teh, C. H., Kunarso, G., Wong, K. Y., Srinivasan, G., Cooper, M. L., et al. (2008). REST regulates distinct transcriptional networks in embryonic and neural stem cells. PLoS Biol. 6:e256. doi: 10.1371/journal.pbio.0060256
Jørgensen, H. F., Terry, A., Beretta, C., Pereira, C. F., Leleu, M., Chen, Z. F., et al. (2009). REST selectively represses a subset of RE1-containing neuronal genes in mouse embryonic stem cells. Development 136, 715–721. doi: 10.1242/dev.028548
Kamal, M. M., Sathyan, P., Singh, S. K., Zinn, P. O., Marisetty, A. L., Liang, S., et al. (2012). REST regulates oncogenic properties of glioblastoma stem cells. Stem Cells 30, 405–414. doi: 10.1002/stem.1020
Kim, J., Chu, J., Shen, X., Wang, J., and Orkin, S. H. (2008). An extended transcriptional network for pluripotency of embryonic stem cells. Cell 132, 1049–1061. doi: 10.1016/j.cell.2008.02.039
Koenigsberger, C., Chicca, J. J. II, Amoureux, M. C., Edelman, G. M., and Jones, F. S. (2000). Differential regulation by multiple promoters of the gene encoding the neuron-restrictive silencer factor. Proc. Natl. Acad. Sci. U.S.A. 97, 2291–2296. doi: 10.1073/pnas.050578797
Kohyama, J., Sanosaka, T., Tokunaga, A., Takatsuka, E., Tsujimura, K., Okano, H., et al. (2010). BMP-induced REST regulates the establishment and maintenance of astrocytic identity. J. Cell Biol. 189, 159–170. doi: 10.1083/jcb.200908048
Kok, F. O., Taibi, A., Wanner, S. J., Xie, X., Moravec, C. E., Love, C. E., et al. (2012). Zebrafish rest regulates developmental gene expression but not neurogenesis. Development 139, 3838–3848. doi: 10.1242/dev.080994
Kraner, S. D., Chong, J. A., Tsay, H. J., and Mandel, G. (1992). Silencing the type II sodium channel gene: a model for neural-specific gene regulation. Neuron 9, 37–44. doi: 10.1016/0896-6273(92)90218-3
Ku, G. M., Kim, H., Vaughn, I. W., Hangauer, M. J., Myung Oh, C., German, M. S., et al. (2012). Research resource: RNA-Seq reveals unique features of the pancreatic beta-cell transcriptome. Mol. Endocrinol. 26, 1783–1792. doi: 10.1210/me.2012-1176
Ku, M., Koche, R. P., Rheinbay, E., Mendenhall, E. M., Endoh, M., Mikkelsen, T. S., et al. (2008). Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS Genet. 4:e1000242. doi: 10.1371/journal.pgen.1000242
Kurtoglu, S., Atabek, M. E., Kumandas, S., and Keskin, M. (2005). Diabetes mellitus type 1: association with Rett syndrome. Pediatr. Int. 47, 90–91. doi: 10.1111/j.1442-200x.2005.02018.x
Kuwabara, T., Hsieh, J., Nakashima, K., Taira, K., and Gage, F. H. (2004). A small modulatory dsRNA specifies the fate of adult neural stem cells. Cell 116, 779–793. doi: 10.1016/S0092-8674(04)00248-X
Kuwahara, K., Saito, Y., Takano, M., Arai, Y., Yasuno, S., Nakagawa, Y., et al. (2003). NRSF regulates the fetal cardiac gene program and maintains normal cardiac structure and function. EMBO J. 22, 6310–6321. doi: 10.1093/emboj/cdg601
Lawinger, P., Venugopal, R., Guo, Z. S., Immaneni, A., Sengupta, D., Lu, W., et al. (2000). The neuronal repressor REST/NRSF is an essential regulator in medulloblastoma cells. Nat. Med. 6, 826–831. doi: 10.1038/77565
Lemaire, K., Thorrez, L., and Schuit, F. (2016). Disallowed and allowed gene expression: two faces of mature islet beta cells. Annu. Rev. Nutr. 36, 45–71. doi: 10.1146/annurev-nutr-071715-050808
Lepagnol-Bestel, A. M., Zvara, A., Maussion, G., Quignon, F., Ngimbous, B., Ramoz, N., et al. (2009). DYRK1A interacts with the REST/NRSF-SWI/SNF chromatin remodelling complex to deregulate gene clusters involved in the neuronal phenotypic traits of Down syndrome. Hum. Mol. Genet. 18, 1405–1414. doi: 10.1093/hmg/ddp047
Li, H. T., Jiang, F. X., Shi, P., Zhang, T., Liu, X. Y., Lin, X. W., et al. (2012). In vitro reprogramming of rat bone marrow-derived mesenchymal stem cells into insulin-producing cells by genetically manipulating negative and positive regulators. Biochem. Biophys. Res. Commun. 420, 793–798. doi: 10.1016/j.bbrc.2012.03.076
Li, J., Klughammer, J., Farlik, M., Penz, T., Spittler, A., Barbieux, C., et al. (2016). Single-cell transcriptomes reveal characteristic features of human pancreatic islet cell types. EMBO Rep. 17, 178–187. doi: 10.15252/embr.201540946
Lietz, M., Hohl, M., and Thiel, G. (2003). RE-1 silencing transcription factor (REST) regulates human synaptophysin gene transcription through an intronic sequence-specific DNA-binding site. Eur. J. Biochem. 270, 2–9. doi: 10.1046/j.1432-1033.2003.03360.x
Loh, Y. H., Wu, Q., Chew, J. L., Vega, V. B., Zhang, W., Chen, X., et al. (2006). The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat. Genet. 38, 431–440. doi: 10.1038/ng1760
Love, C. E., and Prince, V. E. (2015). Rest represses maturation within migrating facial branchiomotor neurons. Dev. Biol. 401, 220–235. doi: 10.1016/j.ydbio.2015.02.021
Lu, T., Aron, L., Zullo, J., Pan, Y., Kim, H., Chen, Y., et al. (2014). REST and stress resistance in ageing and Alzheimer's disease. Nature 507, 448–454. doi: 10.1038/nature13163
Lunyak, V. V., Burgess, R., Prefontaine, G. G., Nelson, C., Sze, S. H., Chenoweth, J., et al. (2002). Corepressor-dependent silencing of chromosomal regions encoding neuronal genes. Science 298, 1747–1752. doi: 10.1126/science.1076469
Mao, C. A., Tsai, W. W., Cho, J. H., Pan, P., Barton, M. C., and Klein, W. H. (2011). Neuronal transcriptional repressor REST suppresses an Atoh7-independent program for initiating retinal ganglion cell development. Dev. Biol. 349, 90–99. doi: 10.1016/j.ydbio.2010.10.008
Martin, D., Allagnat, F., Chaffard, G., Caille, D., Fukuda, M., Regazzi, R., et al. (2008). Functional significance of repressor element 1 silencing transcription factor (REST) target genes in pancreatic beta cells. Diabetologia 51, 1429–1439. doi: 10.1007/s00125-008-0984-1
Martin, D., Allagnat, F., Gesina, E., Caille, D., Gjinovci, A., Waeber, G., et al. (2012). Specific silencing of the REST target genes in insulin-secreting cells uncovers their participation in beta cell survival. PLoS ONE 7, e45844. doi: 10.1371/journal.pone.0045844
Martin, D., Kim, Y. H., Sever, D., Mao, C. A., Haefliger, J. A., and Grapin-Botton, A. (2015). REST represses a subset of the pancreatic endocrine differentiation program. Dev. Biol. 405, 316–327. doi: 10.1016/j.ydbio.2015.07.002
Martin, D., Tawadros, T., Meylan, L., Abderrahmani, A., Condorelli, D. F., Waeber, G., et al. (2003). Critical role of the transcriptional repressor neuron-restrictive silencer factor in the specific control of connexin36 in insulin-producing cell lines. J. Biol. Chem. 278, 53082–53089. doi: 10.1074/jbc.M306861200
Masserdotti, G., Gillotin, S., Sutor, B., Drechsel, D., Irmler, M., Jorgensen, H. F., et al. (2015). Transcriptional mechanisms of proneural factors and REST in regulating neuronal reprogramming of astrocytes. Cell Stem Cell 17, 74–88. doi: 10.1016/j.stem.2015.05.014
McGann, J. C., Oyer, J. A., Garg, S., Yao, H., Liu, J., Feng, X., et al. (2014). Polycomb- and REST-associated histone deacetylases are independent pathways toward a mature neuronal phenotype. Elife 3:e04235. doi: 10.7554/eLife.04235
Mori, N., Schoenherr, C., Vandenbergh, D. J., and Anderson, D. J. (1992). A common silencer element in the SCG10 and type II Na+ channel genes binds a factor present in nonneuronal cells but not in neuronal cells. Neuron 9, 45–54. doi: 10.1016/0896-6273(92)90219-4
Mortazavi, A., Leeper Thompson, E. C., Garcia, S. T., Myers, R. M., and Wold, B. (2006). Comparative genomics modeling of the NRSF/REST repressor network: from single conserved sites to genome-wide repertoire. Genome Res. 16, 1208–1221. doi: 10.1101/gr.4997306
Muraro, M. J., Dharmadhikari, G., Grun, D., Groen, N., Dielen, T., Jansen, E., et al. (2016). A single-cell transcriptome atlas of the human pancreas. Cell Syst. 3, 385–394.e383. doi: 10.1016/j.cels.2016.09.002
Myers, S. J., Peters, J., Huang, Y., Comer, M. B., Barthel, F., and Dingledine, R. (1998). Transcriptional regulation of the GluR2 gene: neural-specific expression, multiple promoters, and regulatory elements. J. Neurosci. 18, 6723–6739.
Naya, F. J., Stellrecht, C. M., and Tsai, M. J. (1995). Tissue-specific regulation of the insulin gene by a novel basic helix-loop-helix transcription factor. Genes Dev. 9, 1009–1019. doi: 10.1101/gad.9.8.1009
Nechiporuk, T., McGann, J., Mullendorff, K., Hsieh, J., Wurst, W., Floss, T., et al. (2016). The REST remodeling complex protects genomic integrity during embryonic neurogenesis. Elife 5:e09584. doi: 10.7554/eLife.09584
Nesti, E., Corson, G. M., McCleskey, M., Oyer, J. A., and Mandel, G. (2014). C-terminal domain small phosphatase 1 and MAP kinase reciprocally control REST stability and neuronal differentiation. Proc. Natl. Acad. Sci. U.S.A. 111, E3929–E3936. doi: 10.1073/pnas.1414770111
Nica, A. C., Ongen, H., Irminger, J. C., Bosco, D., Berney, T., Antonarakis, S. E., et al. (2013). Cell-type, allelic, and genetic signatures in the human pancreatic beta cell transcriptome. Genome Res. 23, 1554–1562. doi: 10.1101/gr.150706.112
Noh, K. M., Hwang, J. Y., Follenzi, A., Athanasiadou, R., Miyawaki, T., Greally, J. M., et al. (2012). Repressor element-1 silencing transcription factor (REST)-dependent epigenetic remodeling is critical to ischemia-induced neuronal death. Proc. Natl. Acad. Sci. U.S.A. 109, E962–E971. doi: 10.1073/pnas.1121568109
Ooi, L., and Wood, I. C. (2007). Chromatin crosstalk in development and disease: lessons from REST. Nat. Rev. Genet. 8, 544–554. doi: 10.1038/nrg2100
Otto, S. J., McCorkle, S. R., Hover, J., Conaco, C., Han, J. J., Impey, S., et al. (2007). A new binding motif for the transcriptional repressor REST uncovers large gene networks devoted to neuronal functions. J. Neurosci. 27, 6729–6739. doi: 10.1523/JNEUROSCI.0091-07.2007
Palm, K., Belluardo, N., Metsis, M., and Timmusk, T. (1998). Neuronal expression of zinc finger transcription factor REST/NRSF/XBR gene. J. Neurosci. 18, 1280–1296.
Palm, K., Metsis, M., and Timmusk, T. (1999). Neuron-specific splicing of zinc finger transcription factor REST/NRSF/XBR is frequent in neuroblastomas and conserved in human, mouse and rat. Brain Res. Mol. Brain Res. 72, 30–39. doi: 10.1016/S0169-328X(99)00196-5
Pandi, G., Nakka, V. P., Dharap, A., Roopra, A., and Vemuganti, R. (2013). MicroRNA miR-29c down-regulation leading to de-repression of its target DNA methyltransferase 3a promotes ischemic brain damage. PLoS ONE 8:e58039. doi: 10.1371/journal.pone.0058039
Pullen, T. J., and Rutter, G. A. (2013). When less is more: the forbidden fruits of gene repression in the adult beta-cell. Diabetes Obes. Metab. 15, 503–512. doi: 10.1111/dom.12029
Quintens, R., Hendrickx, N., Lemaire, K., and Schuit, F. (2008). Why expression of some genes is disallowed in beta-cells. Biochem. Soc. Trans. 36(Pt 3), 300–305. doi: 10.1042/BST0360300
Qureshi, I. A., and Mehler, M. F. (2009). Regulation of non-coding RNA networks in the nervous system–what's the REST of the story? Neurosci. Lett. 466, 73–80. doi: 10.1016/j.neulet.2009.07.093
Rekik, N. M., Kamoun, M., Mnif, F., Charfi, N., Mnif, M. F., and Abid, M. (2010). Type 1 diabetes mellitus and Rett syndrome: is there a link? J. Endocrinol. Invest. 33, 851. doi: 10.1007/BF03350352
Ren, X., and Kerppola, T. K. (2011). REST interacts with Cbx proteins and regulates polycomb repressive complex 1 occupancy at RE1 elements. Mol. Cell. Biol. 31, 2100–2110. doi: 10.1128/MCB.05088-11
Rockowitz, S., and Zheng, D. (2015). Significant expansion of the REST/NRSF cistrome in human versus mouse embryonic stem cells: potential implications for neural development. Nucleic Acids Res. 43, 5730–5743. doi: 10.1093/nar/gkv514
Roopra, A., Qazi, R., Schoenike, B., Daley, T. J., and Morrison, J. F. (2004). Localized domains of G9a-mediated histone methylation are required for silencing of neuronal genes. Mol. Cell 14, 727–738. doi: 10.1016/j.molcel.2004.05.026
Schiffer, D., Caldera, V., Mellai, M., Conforti, P., Cattaneo, E., and Zuccato, C. (2014). Repressor element-1 silencing transcription factor (REST) is present in human control and Huntington's disease neurones. Neuropathol. Appl. Neurobiol. 40, 899–910. doi: 10.1111/nan.12137
Schoch, S., Cibelli, G., and Thiel, G. (1996). Neuron-specific gene expression of synapsin I. Major role of a negative regulatory mechanism. J. Biol. Chem. 271, 3317–3323. doi: 10.1074/jbc.271.6.3317
Schoenherr, C. J., and Anderson, D. J. (1995). The neuron-restrictive silencer factor (NRSF): a coordinate repressor of multiple neuron-specific genes. Science 267, 1360–1363. doi: 10.1126/science.7871435
Segerstolpe, A., Palasantza, A., Eliasson, P., Andersson, E. M., Andreasson, A. C., Sun, X., et al. (2016). Single-cell transcriptome profiling of human pancreatic islets in health and type 2 diabetes. Cell Metab. 24, 593–607. doi: 10.1016/j.cmet.2016.08.020
Shi, Y., Lan, F., Matson, C., Mulligan, P., Whetstine, J. R., Cole, P. A., et al. (2004). Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 119, 941–953. doi: 10.1016/j.cell.2004.12.012
Singh, S. K., Kagalwala, M. N., Parker-Thornburg, J., Adams, H., and Majumder, S. (2008). REST maintains self-renewal and pluripotency of embryonic stem cells. Nature 453, 223–227. doi: 10.1038/nature06863
Singh, S. K., Marisetty, A., Sathyan, P., Kagalwala, M., Zhao, Z., and Majumder, S. (2015). REST-miR-21-SOX2 axis maintains pluripotency in E14Tg2a.4 embryonic stem cells. Stem Cell Res. 15, 305–311. doi: 10.1016/j.scr.2015.05.003
Singh, S. K., Veo, B. L., Kagalwala, M. N., Shi, W., Liang, S., and Majumder, S. (2012). Dynamic status of REST in the mouse ESC pluripotency network. PLoS ONE 7:e43659. doi: 10.1371/journal.pone.0043659
Soyer, J., Flasse, L., Raffelsberger, W., Beucher, A., Orvain, C., Peers, B., et al. (2010). Rfx6 is an Ngn3-dependent winged helix transcription factor required for pancreatic islet cell development. Development 137, 203–212. doi: 10.1242/dev.041673
Su, X., Gopalakrishnan, V., Stearns, D., Aldape, K., Lang, F. F., Fuller, G., et al. (2006). Abnormal expression of REST/NRSF and Myc in neural stem/progenitor cells causes cerebellar tumors by blocking neuronal differentiation. Mol. Cell. Biol. 26, 1666–1678. doi: 10.1128/MCB.26.5.1666-1678.2006
Sun, Y. M., Greenway, D. J., Johnson, R., Street, M., Belyaev, N. D., Deuchars, J., et al. (2005). Distinct profiles of REST interactions with its target genes at different stages of neuronal development. Mol. Biol. Cell 16, 5630–5638. doi: 10.1091/mbc.E05-07-0687
Tahiliani, M., Mei, P., Fang, R., Leonor, T., Rutenberg, M., Shimizu, F., et al. (2007). The histone H3K4 demethylase SMCX links REST target genes to X-linked mental retardation. Nature 447, 601–605. doi: 10.1038/nature05823
Tang, X., Kim, J., Zhou, L., Wengert, E., Zhang, L., Wu, Z., et al. (2016). KCC2 rescues functional deficits in human neurons derived from patients with Rett syndrome. Proc. Natl. Acad. Sci. U.S.A. 113, 751–756. doi: 10.1073/pnas.1524013113
Thiel, G., Ekici, M., and Rossler, O. G. (2015). RE-1 silencing transcription factor (REST): a regulator of neuronal development and neuronal/endocrine function. Cell Tissue Res. 359, 99–109. doi: 10.1007/s00441-014-1963-0
Tsai, M. C., Manor, O., Wan, Y., Mosammaparast, N., Wang, J. K., Lan, F., et al. (2010). Long noncoding RNA as modular scaffold of histone modification complexes. Science 329, 689–693. doi: 10.1126/science.1192002
van Arensbergen, J., Garcia-Hurtado, J., Moran, I., Maestro, M. A., Xu, X., Van de Casteele, M., et al. (2010). Derepression of Polycomb targets during pancreatic organogenesis allows insulin-producing beta-cells to adopt a neural gene activity program. Genome Res. 20, 722–732. doi: 10.1101/gr.101709.109
Visvanathan, J., Lee, S., Lee, B., Lee, J. W., and Lee, S. K. (2007). The microRNA miR-124 antagonizes the anti-neural REST/SCP1 pathway during embryonic CNS development. Genes Dev. 21, 744–749. doi: 10.1101/gad.1519107
Waeber, G., Delplanque, J., Bonny, C., Mooser, V., Steinmann, M., Widmann, C., et al. (2000). The gene MAPK8IP1, encoding islet-brain-1, is a candidate for type 2 diabetes. Nat. Genet. 24, 291–295. doi: 10.1038/73523
Wagoner, M. P., and Roopra, A. (2012). A REST derived gene signature stratifies glioblastomas into chemotherapy resistant and responsive disease. BMC Genomics 13, 686. doi: 10.1186/1471-2164-13-686
Wang, J., Rao, S., Chu, J., Shen, X., Levasseur, D. N., Theunissen, T. W., et al. (2006). A protein interaction network for pluripotency of embryonic stem cells. Nature 444, 364–368. doi: 10.1038/nature05284
Westbrook, T. F., Hu, G., Ang, X. L., Mulligan, P., Pavlova, N. N., Liang, A., et al. (2008). SCFbeta-TRCP controls oncogenic transformation and neural differentiation through REST degradation. Nature 452, 370–374. doi: 10.1038/nature06780
Westbrook, T. F., Martin, E. S., Schlabach, M. R., Leng, Y., Liang, A. C., Feng, B., et al. (2005). A genetic screen for candidate tumor suppressors identifies REST. Cell 121, 837–848. doi: 10.1016/j.cell.2005.03.033
Wood, I. C., Belyaev, N. D., Bruce, A. W., Jones, C., Mistry, M., Roopra, A., et al. (2003). Interaction of the repressor element 1-silencing transcription factor (REST) with target genes. J. Mol. Biol. 334, 863–874. doi: 10.1016/j.jmb.2003.10.017
Wood, I. C., Roopra, A., and Buckley, N. J. (1996). Neural specific expression of the m4 muscarinic acetylcholine receptor gene is mediated by a RE1/NRSE-type silencing element. J. Biol. Chem. 271, 14221–14225. doi: 10.1074/jbc.271.24.14221
Wu, J., and Xie, X. (2006). Comparative sequence analysis reveals an intricate network among REST, CREB and miRNA in mediating neuronal gene expression. Genome Biol. 7:R85. doi: 10.1186/gb-2006-7-9-r85
Xue, Y., Ouyang, K., Huang, J., Zhou, Y., Ouyang, H., Li, H., et al. (2013). Direct conversion of fibroblasts to neurons by reprogramming PTB-regulated microRNA circuits. Cell 152, 82–96. doi: 10.1016/j.cell.2012.11.045
Xue, Y., Qian, H., Hu, J., Zhou, B., Zhou, Y., Hu, X., et al. (2016). Sequential regulatory loops as key gatekeepers for neuronal reprogramming in human cells. Nat. Neurosci. 19, 807–815. doi: 10.1038/nn.4297
Yamada, Y., Aoki, H., Kunisada, T., and Hara, A. (2010). Rest promotes the early differentiation of mouse ESCs but is not required for their maintenance. Cell Stem Cell 6, 10–15. doi: 10.1016/j.stem.2009.12.003
Yeo, M., Berglund, K., Augustine, G., and Liedtke, W. (2009). Novel repression of Kcc2 transcription by REST-RE-1 controls developmental switch in neuronal chloride. J. Neurosci. 29, 14652–14662. doi: 10.1523/JNEUROSCI.2934-09.2009
Yeo, M., Lee, S. K., Lee, B., Ruiz, E. C., Pfaff, S. L., and Gill, G. N. (2005). Small CTD phosphatases function in silencing neuronal gene expression. Science 307, 596–600. doi: 10.1126/science.1100801
Zuccato, C., Belyaev, N., Conforti, P., Ooi, L., Tartari, M., Papadimou, E., et al. (2007). Widespread disruption of repressor element-1 silencing transcription factor/neuron-restrictive silencer factor occupancy at its target genes in Huntington's disease. J. Neurosci. 27, 6972–6983. doi: 10.1523/JNEUROSCI.4278-06.2007
Keywords: transcription, diabetes, endocrinology, secretion, repressor, embryo, development, pancreas
Citation: Martin D and Grapin-Botton A (2017) The Importance of REST for Development and Function of Beta Cells. Front. Cell Dev. Biol. 5:12. doi: 10.3389/fcell.2017.00012
Received: 01 November 2016; Accepted: 07 February 2017;
Published: 24 February 2017.
Edited by:
Guy A. Rutter, Imperial College London, UKReviewed by:
Amar Abderrahmani, Lille University, FranceAlice Kong, The Chinese University of Hong Kong, Hong Kong
Copyright © 2017 Martin and Grapin-Botton. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: David Martin, ZGF2aWQubWFydGluMkBjaHV2LmNo
Anne Grapin-Botton, YW5uZS5ncmFwaW4tYm90dG9uQHN1bmQua3UuZGs=
 David Martin1*
David Martin1* Anne Grapin-Botton
Anne Grapin-Botton