- Graduate School of Frontier Biosciences, Osaka University, Suita, Japan
Gap junctions are intercellular channels that allow passage of ions and small molecules between adjacent cells. Gap junctions in vertebrates are composed of connexons, which are an assembly of six proteins, connexins. Docking of two connexons on the opposite cell surfaces forms a gap junction between the cytoplasm of two neighboring cells. Connexins compose a family of structurally related four-pass transmembrane proteins. In mammals, there are ~20 connexins, each of which contributes to unique permeability of gap junctions, and mutations of some connexin-encoding genes are associated with human diseases. Zebrafish has been predicted to contain 39 connexin-encoding genes; the high number can be attributed to gene duplication during fish evolution, which resulted in diversified functions of gap junctions in teleosts. The determination of body shapes and skin patterns in animal species is an intriguing question. Mathematical models suggest principle mechanisms explaining the diversification of animal morphology. Recent studies have revealed the involvement of gap junctions in fish morphological diversity, including skin pattern formation and body shape determination. This review focuses on connexins in teleosts, which are integrated in the mathematical models explaining morphological diversity of animal skin patterns and body shapes.
Introduction
Gap junctions are intercellular channels that mediate the transfer of small molecules between adjacent cells (Kumar and Gilula, 1996). Because of low size selectivity of molecules transferred through gap junctions (<1,000 Da), it is difficult to determine the biological functions of gap junctions in the organisms. Gap junctions are composed of two hemichannels formed by four-pass transmembrane proteins: connexins and innexins (Figures 1A,B; Baranova et al., 2004). Connexins are vertebrate-specific gap junction proteins, whereas innexins are expressed in invertebrates. Connexins as well as innexins form both hemichannels and gap junctions, while pannexins expressed in vertebrates but homologous to invertebrate innexins predominantly exist as hemichannels connecting the intracellular and extracellular space, rather than gap junctions connecting adjacent cells. Curiously, there is no evolutional relationship between connexins and pannexins, although both are expressed in vertebrates. In summary, connexins and innexins are functional homologs, while innexins and pannexins are evolutionary homologs (Figure 1A; Scemes et al., 2007).
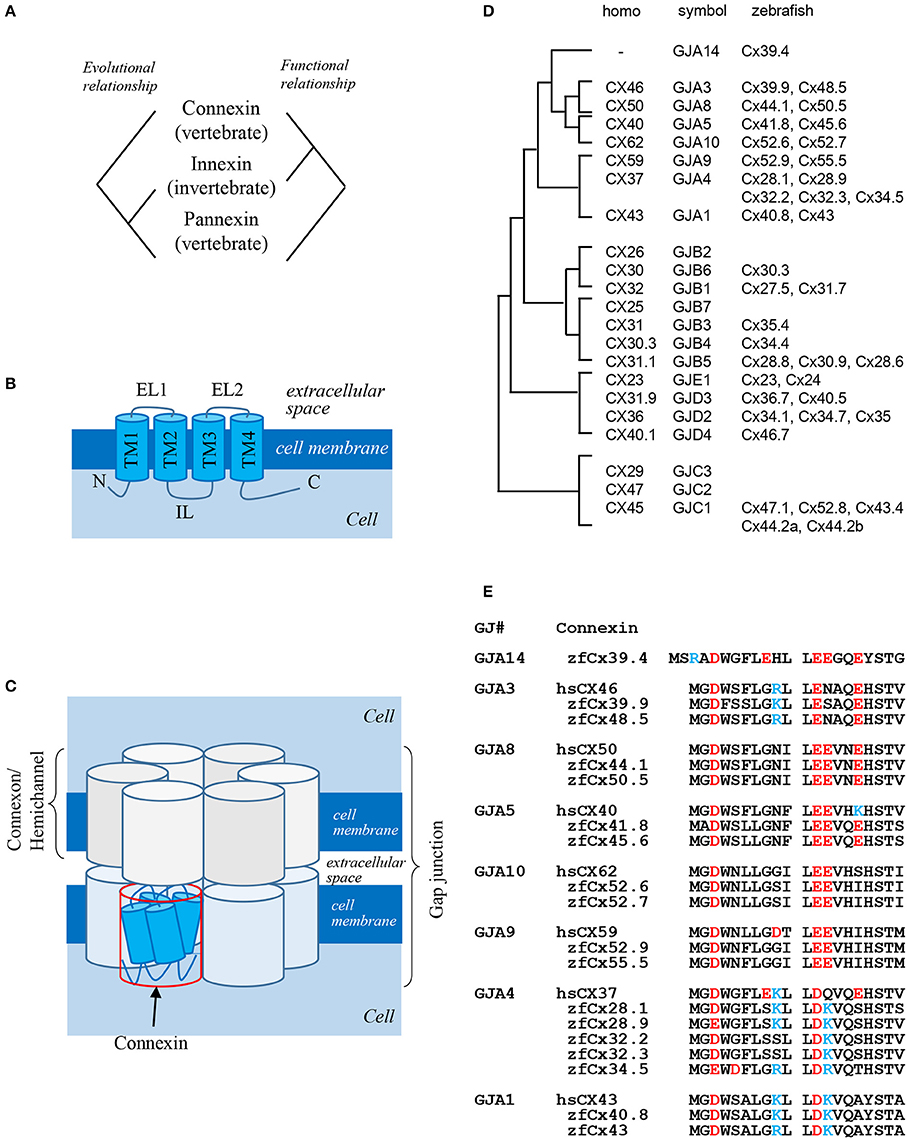
Figure 1. Gap junction proteins. (A) Relationship among three types of gap junction proteins: connexins, innexins, and pannexins (Baranova et al., 2004). (B) Structure of connexin proteins. N: N-terminus; TM: transmembrane domain; EL: extracellular loop; IL: intracellular loop; C: C-terminus (Kumar and Gilula, 1996). (C) Schematic presentation of a gap junction (Kumar and Gilula, 1996). (D) Phylogenic relationship between human and zebrafish connexins (Eastman et al., 2006; Cruciani and Mikalsen, 2007). (E) Sequence alignment of N-terminal domains of human and zebrafish alpha-type connexins (Connexin sequences were obtained from genome data base in Sanger Institute, http://www.sanger.ac.uk/).
Six connexin proteins form a hexamer called connexon, which functions as a hemichannel. After docking of two connexons on neighboring cell membranes, a gap junction is formed (Figure 1C). Connexins consist of several structural domains: the N-terminus, transmembrane region, extracellular and intracellular loops, and C-terminus (Figure 1B; Maeda et al., 2009). The N-terminal domain functions as a plug providing closure of gap junctions (Oshima et al., 2007) and as a voltage sensor of membrane potential (Verselis et al., 1994). The C-terminal domain has several phosphorylation sites which transmit signals to control the opening and closing of gap junctions and are also implicated in other biological pathways (Hebert and Stains, 2013), while extracellular loops are responsible for docking of hemichannels (Kumar and Gilula, 1996).
The number of connexin genes differs depending on animal species. In the human genome, there are 21 connexin genes, whereas in the zebrafish genome, 39 connexin genes are predicted (Hebert and Stains, 2013). Connexin proteins are named according to their molecular weight; for example, connexin 43 (Cx43) is a 43-kDa protein. This system sometimes causes confusion because orthologous genes belonging to different species have different gene symbols; thus, human CX46 and zebrafish cx39.9 are orthologous genes (Figure 1D). There is also other classification system based on gap junctions which are divided into five families, from gap junction alpha (GJA) to gap junction epsilon (GJE), and all connexins are named according to a specific subfamily (Figure 1D, symbol). In this system, orthologous genes in different animal species belong to the same gap junction subfamily. Although this classification makes it easier to understand the relationship between connexins and gap junctions, it has faults, because some connexins have a potential to form gap junctions with different connexin proteins. Thus, gap junctions composed of the same hemichannels consisting of two or more connexins are called heteromeric, while those composed of different hemichannels are called heterotypic. Although the formation of heteromeric-heterotypic gap junctions has been extensively examined in in vitro experiments, its role in vivo remains largely unknown.
Connexins in Teleosts
There are more connexin-encoding genes in zebrafish than in humans (39 and 21, respectively; Eastman et al., 2006; Cruciani and Mikalsen, 2007). Here, I analyzed the number of connexin genes in six teleost species: zebrafish, herring, catfish, fugu, tilapia, and medaka using Genome Database (Ensembl, Sanger Institute). Zebrafish, herring, and catfish form one sister group, and the other three species form another one (Figure 2A). The results show that ~40 connexin genes exist in the genomes of the examined teleost species, although it is not known whether all the genes are expressed and functional. The duplication of connexin-encoding genes may have occurred in the ancestor of the teleost lineage through chromosome duplication events. After counting the number of connexin genes and categorizing them into the GJA–GJE subfamilies, it appeared that the examined teleost species had similar gene numbers in each subfamily (Figure 2B). Figure 2C shows the number of connexin genes belonging to the GJA subfamily. For example, cx39.4 belongs to the GJA14 subfamily and all six species examined have one cx39.4 ortholog in their genome. In the fugu-tilapia-medaka lineage, connexins of the GJA3 subfamily were duplicated and cx40.8 and cx50.5 orthologs were lost in their common ancestor. On the other hand, cx55.5 might have been lost in catfish and tilapia independently. Connexins belonging to the GJA4 subfamily have three or more paralogous genes. The high copy numbers of GJA4 paralogous genes may be explained by local chromosomal duplications occurred in the ancestor. In addition to the high copy number, the cx39.4 gene is teleost-specific and is not detected in human, chicken, lizard, or Xenopus genomes (Figures 1D,E).
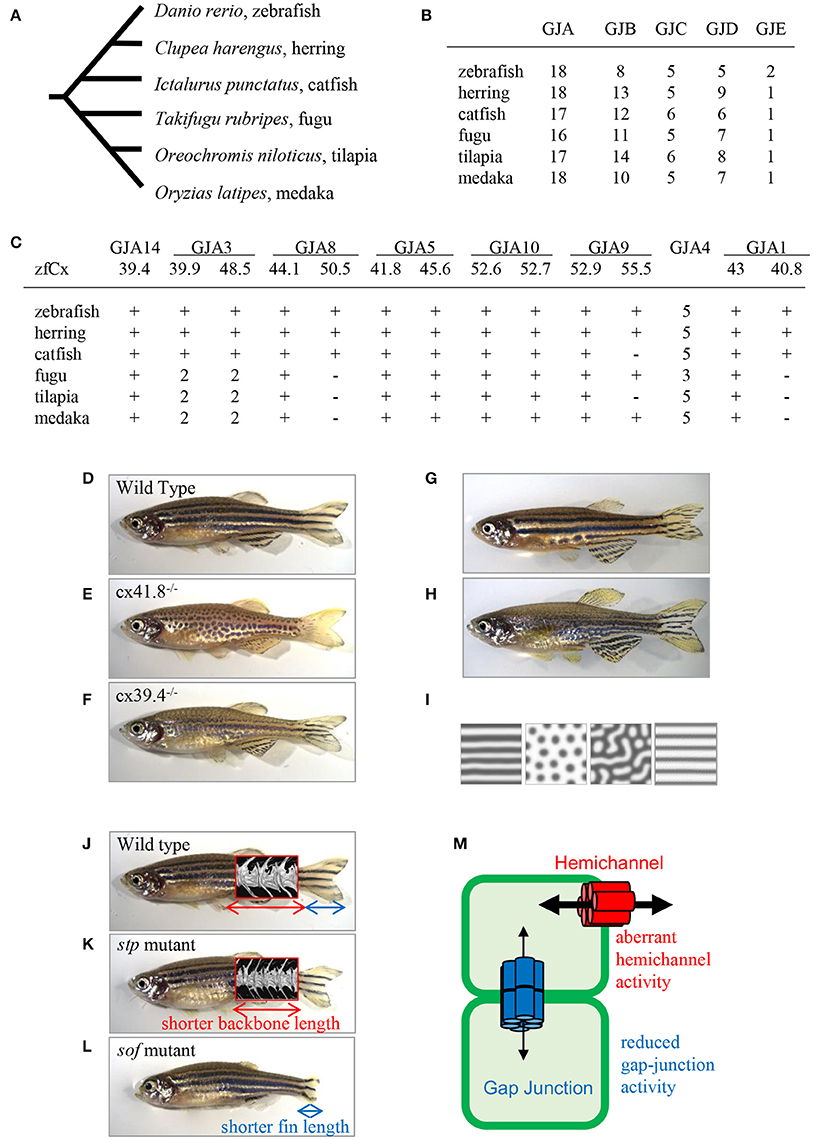
Figure 2. Connexins in teleosts. (A) Phylogenic relationship among six teleost species (Chen et al., 2004). (B) The number of connexin-encoding genes in six teleost species. (C) The number of genes encoding connexins of the alpha family gap junctions. “zfCx” indicates zebrafish connexins; “+” and “−” indicate the existence or absence, respectively, of an ortholog. If more than one orthologous gene was found, gene numbers are indicated (B,C; Connexin sequences were obtained from genome data base in Sanger Institute, http://www.sanger.ac.uk/). (D–I) Connexins in zebrafish pigment patterns. Wild-type zebrafish (D; Watanabe and Kondo, 2012); leopard mutant (E; Watanabe and Kondo, 2012); luchs mutant (F; Irion et al., 2014; Watanabe et al., 2016); transgenic zebrafish Tg(mitfa-cx41.8) >> leopard (G; Watanabe and Kondo, 2012); transgenic zebrafish Tg(mitfa-cx41.8M7) >> wild-type (H; Watanabe and Kondo, 2012); reaction-diffusion (R-D) patterns (I; Watanabe and Kondo, 2012). (J–M) Connexins in zebrafish bones; micro-CT images of vertebrae are superimposed. Wild-type zebrafish (J; Misu et al., 2016), stp mutant (K; Misu et al., 2016), sof mutant (L; Iovine et al., 2005; Misu et al., 2016). Schematic presentation of gap junction and hemichannel functions in zebrafish mutants (M; Misu et al., 2016). Red font, functional activity of hemichannels in the stp-Cx43 mutant; blue font, functional activity of gap junctions in the sof-Cx43 mutant.
Connexins in Zebrafish
Zebrafish is a small tropical fish with a body 3–4-cm long. Because of its transparent embryo, easy breeding, generation of transgenic lines, and availability of genomic resources through advances in genome sequencing technology, zebrafish is considered an important and convenient model organism for developmental studies in vertebrates. However, knocking out of some connexin gene shows no phenotypes in zebrafish, probably because of gene redundancy or other reasons. To date, the expression of several connexin genes was identified in zebrafish mutants: cx39.9 in muscle (Hirata et al., 2012), cx52.6 and cx55.5 in retina (Klaassen et al., 2011), cx43 in fins and vertebrae (Iovine et al., 2005; Misu et al., 2016), and cx36.7 in the heart (Sultana et al., 2008), while cx41.8 and cx39.4 were found to be responsible for pigment pattern (Watanabe et al., 2006; Irion et al., 2014).
Connexins in Zebrafish Pigment Pattern
One of the famous characteristics of zebrafish is “zebra” stripe observed on the skin surface (Figure 2D), and the mechanism underlying the generation of the stripe pattern has long been an intriguing question. Sixty years ago, English mathematician Alan M. Turing proposed a mathematical model called the reaction-diffusion (R-D) model, which explained the mechanism underlying pattern formation (Turing, 1952) and which was later applied to biological phenomena (Kondo and Asai, 1995). This mathematical model represents the interaction and diffusion of two hypothetical factors, allowing, by changing the parameters in the equations, to generate various patterns in silico. Zebrafish stripes are made of two types of pigment cells, melanophores and xanthophores, and it is shown that interactions between these pigment cells satisfy the condition of the R-D model (Yamaguchi et al., 2007; Nakamasu et al., 2009). Thus, it can be assumed that the zebrafish skin pattern is generated in the R-D manner, which makes zebrafish a model organism for pattern formation studies.
One of the most famous zebrafish skin pattern mutants is leopard fish, which has spots instead of stripes (Figure 2E). This mutant was originally identified from field and several alleles were isolated from mutagenesis pools (Haffter et al., 1996). Because the spot is a representative pattern of the R-D model (Figure 2I), the leopard mutant is an important target of pattern formation studies (Asai et al., 1999). Ten years ago, our group identified the gene responsible for the leopard pattern, which encoded a gap junction protein Cx41.8 (Watanabe et al., 2006), a zebrafish ortholog of mammalian CX40 and a paralog of zebrafish Cx45.6 (Eastman et al., 2006). Knocking out cx41.8 results in a spotted skin pattern, whereas knocking out cx45.6 does not produce a skin phenotype. It should be noted that molecules functioning in cell–cell interaction should participate in stripe-to-spot changes in vivo because such changes are predicted by the mathematical model for the interaction between two hypothetical factors. This notion was confirmed when we successfully generated the R-D patterns on zebrafish skin using Cx41.8 mutants (Figures 2G–I; Watanabe and Kondo, 2012). To further investigate the role of gap junctions in skin pattern formation, we constructed transgenic fish lines in which connexin-encoding genes were ectopically expressed in pigment cells of the leopard fish (Watanabe et al., 2012). The results indicated that, in addition to cx41.8, other genes such as cx44.1, cx45.6, and cx48.5 rescued the leopard phenotype, while cx27.5, cx30.8, cx32.2, or cx43 did not. Amino acid alignment of N-terminal connexin domains revealed that connexins which rescued the leopard phenotype belonged to the GJA14, GJA3, GJA8, and GJA5 subfamilies as evidenced by the presence of the ExxxE motif (Figure 1E), a polyamine-binding site important for rectifying properties of gap junctions (Musa et al., 2004).
Polyamines, mainly putrescine, spermidine, and spermine, are small molecules important for cell proliferation and differentiation; they are known to regulate K+ inward-rectifier (Kir) channels through binding to the channel pore (Hibino et al., 2010). We have isolated Kir7.1 from a zebrafish skin pattern mutant, jaguar, and shown that the Kir7.1 channel is expressed in melanophores, where it forms resting potential (Iwashita et al., 2006); this hyperpolarization is important for the generation of a clear boundary between melanophores and xanthophores on fish skin (Inaba et al., 2012). Based on this finding, we introduced the ssat gene encoding polyamine metabolic enzyme spermidine/spermine N1-acetyltransferaseinto melanophores, and found that the ectopic expression of ssat disturbed the stripe pattern of zebrafish (Watanabe et al., 2012). Interestingly, ssat-expressing transgenic zebrafish showed a unique phenotype of large spots and wide stripes, which is an intermediate pattern between the leopard (cx41.8) and jaguar (kir7.1) mutants (Iwashita et al., 2006; Watanabe et al., 2006, 2012). Furthermore, a recent study showed that spermidine synthase was also involved in skin pattern formation as confirmed by the isolation of spermidine, but not spermine, from melanophores (Frohnhofer et al., 2016). Taken together, these data indicate that spermidine may bind both Kir7.1 and Cx41.8, and control the rectification properties of Kir7.1 as well as of Cx41.8 gap junctions in melanophores. As a result, the expected unidirectional functioning of gap junctions from xanthophores to melanophores would be provided, which is consistent with a previous observation that xanthophores are required for melanophore survival (Nakamasu et al., 2009).
Cx39.4 is another connexin protein shown to be involved in the skin pattern formation of zebrafish. Cx39.4 is the teleost lineage-specific connexin (Figures 1D,E) recently isolated from a zebrafish skin pattern mutant, luchs (Figure 2F) (Irion et al., 2014). We examined the expression of alpha-type connexins in pigment cells and found that the cx41.8 and cx39.4 genes were expressed in melanophores and xanthophores, which was also confirmed in transgenic zebrafish carrying a reporter gene under connexin promoters. To compare Cx39.4 and Cx41.8 functions in skin pattern formation, we performed complementation experiments when cx39.4 was introduced into the leopard mutant and cx41.8 into the luchs mutant (Watanabe et al., 2016). None of them was able to rescue each other phenotypes, indicating that Cx41.8 and Cx39.4 have distinct functional activities. Although Cx39.4 contains the N-terminal ExxxE motif important for pattern formation, the sequence of its N-terminus is unique because it is two residues longer than that in other alpha-type connexins and has a basic residue at the third position. Our electrophysiological analysis showed that large voltage-dependent current was absent in Cx39.4-expressing oocytes, indicating that the basic residue at the third position affected the characteristics of gap junctions and accounted for the difference of channeling properties between Cx39.4 and Cx41.8 (Watanabe et al., 2016).
Connexins in Zebrafish Bones, Fins, and Vertebrae
The variation in body shape among animal species has long been an intriguing question. A century ago, Scottish mathematician and biologist D'Arcy Thompson proposed the theory of transformations suggesting that new body shapes arise by changing angles, extending the length, or enlarging body parts, pointing out correlations between biological forms and mechanical phenomena (Thompson, 1961). Bones determine body shape in vertebrates, and zebrafish fins present a valuable model for the study of bone organogenesis and regeneration because of their rapid growth. Recent advances in transgenic techniques enable the detection of gene expression in bone-producing cells during bone growth and regeneration in live fish. Among the connexin family members expressed in zebrafish, Cx43 is known to be involved in the formation and regeneration of the fin and in determining vertebra proportions (Iovine et al., 2005; Misu et al., 2016).
Ten years ago, zebrafish Cx43 was identified in the short-of-fin (sof) mutant who has a short fin segment (Figure 2L; Iovine et al., 2005). Four sof alleles were isolated from zebrafish mutagenesis pools and amino acid substitutions causing the loss or decrease of Cx43 gap junction functional activity were identified in three mutant alleles. On the other hand, no mutation was found in the coding region of cx43 in the fourth allele; however, the downregulation of both mRNA and protein expression were detected in the carriers of this allele. Overall, these findings indicate that a decrease of Cx43 gap junction properties produces the short-fin phenotype (Figure 2L). In addition, Cx40.8, a paralog of Cx43 in zebrafish is also involved in fin development and regeneration (Gerhart et al., 2009). Interestingly, although the biological function of Cx40.8 is very similar to that of Cx43, its membrane localization is differentially controlled depending on the developmental phases and regeneration status (Gerhart et al., 2012).
To provide deeper understanding of the molecular mechanisms supporting the theory of transformations, our group focused on a zebrafish body-shape mutant named stoepsel (stp; Figure 2K) identified 20 years ago from a zebrafish mutagenesis pool based on reduced body length. Recently, we performed precise analysis of bone shape development in this mutant fish. Micro-CT scanning images revealed that the vertebra shape of the stp mutant was almost the same as that of the wild-type fish, although the vertebra size along the anterior-posterior (A-P) axis was decreased (Figures 2J,K). Because the vertebra height along the dorsal-ventral (D-V) axis was unchanged, this fish presents a proportion mutant according to the theory of transformations. We also found that the mutant phenotype appeared 50 days post-fertilization, indicating that it is expressed in the adulthood. To disclose the underlying molecular mechanism, we performed positional cloning experiments and identified a point mutation in the cx43 coding region. Then, we asked a question why mutations occurring in same cx43 gene caused different phenotypes, i.e., short fin or short vertebra. To address this question, we compared the functions of gap junctions and hemichannels by performing dual-cell voltage clamp experiments and found that the functional activity of stp-Cx43 gap junction was decreased similar to that of the sof-Cx43 mutant (Misu et al., 2016). Measurements of fin segment length in the stp mutant revealed that it was 5% shorter than that of the wild-type fish (Misu et al., 2016), which is consistent with previous findings that shortening of the fin segment is proportional to the reduction in gap junction function (Hoptak-Solga et al., 2007). On the other hand, we detected aberrant increase of stp-Cx43 hemichannel activity, while no difference was detected between sof-Cx43 and the wild-type Cx43. These findings suggest that malfunctioning of hemichannels results in reduced backbone length, whereas a decrease in gap junction activity causes shortening of the fin segment (Figure 2M), although the underlying mechanism remains unclear (Misu et al., 2016).
The mutation in the human GJA1 gene encoding CX43 is known to be responsible for an extremely rare disease, oculodentodigital dysplasia (ODDD), manifested by small eyes, underdeveloped teeth, and malformation of fingers (Paznekas et al., 2003). As zebrafish Cx43 is an analog of human CX43, zebrafish can present a good experimental model to study ODDD and would help understand the mechanisms controlling bone formation.
Mathematical Models Predict Gap Junction Functions in Pattern Formation
It is very interesting that two independent projects which aimed to disclose the molecular mechanism hidden in mathematical models, found the involvement of gap junction proteins: Cx41.8 in the R-D model and Cx43 in the theory of transformation.
In the R-D model, positive interactions between two factors are important. As mentioned above, Cx41.8-gap junctions might be formed between xanthophores and melanophores, and control the directional flow of small molecules from a xanthophore to a melanophore depending on spermidine concentration. In addition, the R-D model predicts that Cx41.8 might be involved in melanophore differentiation. The reduction of basal level synthesis of a factor also causes pattern changes in the R-D model (Asai et al., 1999), and the reduction of the number of melanophores are observed in the leopard mutant (Figure 2E).
Regarding the theory of transformation, the stp mutant might be the simplest example. It is possible that the reduction of osteogenic activity causes the formation of shorter vertebra in the stp mutant, although the difference in the mechanisms underlying the formation of shorter vertebra in the stp fish and shorter fins in the sof fish is unclear. Recent gap junction studies have revealed that hemichannels formed by connexins and/or pannexins function as sensors of mechanical stress (Jiang et al., 2009; Thi et al., 2012), which supports the possibility that gap junctions control body shape variations, including shortening, expanding, and twisting of body frames as predicted by the theory of transformation.
Many questions still remain, including functional differences among connexin proteins, the type of transferred molecules, and the evolutionary events that have led to the acquisition of this intercellular communication system in the animal kingdom. Future mechanistic and genetic studies would address these questions.
Author Contributions
The author confirms being the sole contributor of this work and approved it for publication.
Conflict of Interest Statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the Ministry of Education, Culture, Sports, Science, and Technology in Japan (KAKENHI Grant 26291049).
References
Asai, R., Taniguchi, E., Kume, Y., Saito, M., and Kondo, S. (1999). Zebrafish leopard gene as a component of the putative reaction-diffusion system. Mech. Dev. 89, 87–92. doi: 10.1016/S0925-4773(99)00211-7
Baranova, A., Ivanov, D., Petrash, N., Pestova, A., Skoblov, M., Kelmanson, I., et al. (2004). The mammalian pannexin family is homologous to the invertebrate innexin gap junction proteins. Genomics 83, 706–716. doi: 10.1016/j.ygeno.2003.09.025
Chen, W.-J., Orti, G., and Meyer, A. (2004). Novel evolutionary relationship among four fish model systems. Trends Genet. 20, 244–231. doi: 10.1016/j.tig.2004.07.005
Cruciani, V., and Mikalsen, S. O. (2007). Evolutionary selection pressure and family relationships among connexin genes. Biol. Chem. 388, 253–264. doi: 10.1515/bc.2007.028
Eastman, S. D., Chen, T. H., Falk, M. M., Mendelson, T. C., and Iovine, M. K. (2006). Phylogenetic analysis of three complete gap junction gene families reveals lineage-specific duplications and highly supported gene classes. Genomics 87, 265–274. doi: 10.1016/j.ygeno.2005.10.005
Frohnhofer, H. G., Geiger-Rudolph, S., Pattky, M., Meixner, M., Huhn, C., Maischein, H. M., et al. (2016). Spermidine, but not spermine, is essential for pigment pattern formation in zebrafish. Biol. Open 5, 736–744. doi: 10.1242/bio.018721
Gerhart, S. V, Eble, D. M., Burger, R. M., Oline, S. N., Vacaru, A., Sadler, K. C., et al. (2012). The Cx43-like connexin protein Cx40.8 is differentially localized during fin ontogeny and fin regeneration. PLoS ONE 7:e31364. doi: 10.1371/journal.pone.0031364
Gerhart, S. V, Jefferis, R., and Iovine, M. K. (2009). Cx40.8, a Cx43-like protein, forms gap junction channels inefficiently and may require Cx43 for its association at the plasma membrane. FEBS Lett. 583, 3419–3424. doi: 10.1016/j.febslet.2009.09.054
Haffter, P., Odenthal, J., Mullins, M. C., Lin, S., Farrell, M. J., Vogelsang, E., et al. (1996). Mutations affecting pigmentation and shape of the adult zebrafish. Dev. Genes Evol. 206, 260–276. doi: 10.1007/s004270050051
Hebert, C., and Stains, J. P. (2013). An intact connexin43 is required to enhance signaling and gene expression in osteoblast-like cells. J. Cell. Biochem. 114, 2542–2550. doi: 10.1002/jcb.24603
Hibino, H., Inanobe, A., Furutani, K., Murakami, S., Findlay, I., and Kurachi, Y. (2010). Inwardly rectifying potassium channels: their structure, function, and physiological roles. Physiol. Rev. 90, 291–366. doi: 10.1152/physrev.00021.2009
Hirata, H., Wen, H., Kawakami, Y., Naganawa, Y., Ogino, K., Yamada, K., et al. (2012). Connexin 39.9 protein is necessary for coordinated activation of slow-twitch muscle and normal behavior in zebrafish. J. Biol. Chem. 287, 1080–1089. doi: 10.1074/jbc.M111.308205
Hoptak-Solga, A. D., Klein, K. A., DeRosa, A. M., White, T. W., and Iovine, M. K. (2007). Zebrafish short fin mutations in connexin43 lead to aberrant gap junctional intercellular communication. FEBS Lett. 581, 3297–3302. doi: 10.1016/j.febslet.2007.06.030
Inaba, M., Yamanaka, H., and Kondo, S. (2012). Pigment pattern formation by contact-dependent depolarization. Science 335, 677. doi: 10.1126/science.1212821
Iovine, M. K., Higgins, E. P., Hindes, A., Coblitz, B., and Johnson, S. L. (2005). Mutations in connexin43 (GJA1) perturb bone growth in zebrafish fins. Dev. Biol. 278, 208–219. doi: 10.1016/j.ydbio.2004.11.005
Irion, U., Frohnhofer, H. G., Krauss, J., Çolak Champollion, T., Maischein, H. M., Geiger-Rudolph, S., et al. (2014). Gap junctions composed of connexins 41.8 and 39.4 are essential for colour pattern formation in zebrafish. Elife 3:e05125. doi: 10.7554/eLife.05125
Iwashita, M., Watanabe, M., Ishii, M., Chen, T., Johnson, S. L., Kurachi, Y., et al. (2006). Pigment pattern in jaguar/obelix zebrafish is caused by a Kir7.1 mutation: implications for the regulation of melanosome movement. PLoS Genet. 2:e197. doi: 10.1371/journal.pgen.0020197
Jiang, J. X., Siller-Jackson, A. J., and Burra, S. (2009). Roles of gap junctions and hemichannels in bone cell functions and in signal transmission of mechanical stress. Front. Biosci. 12, 1450–1462. doi: 10.2741/2159
Klaassen, L. J., Sun, Z., Steijaert, M. N., Bolte, P., Fahrenfort, I., Sjoerdsma, T., et al. (2011). Synaptic transmission from horizontal cells to cones is impaired by loss of connexin hemichannels. PLoS Biol. 9:e1001107. doi: 10.1371/journal.pbio.1001107
Kondo, S., and Asai, R. (1995). A reaction-diffusion wave on the skin of marine angelfish Pomacanthus. Nature 376, 765–768. doi: 10.1038/376765a0
Kumar, N. M., and Gilula, N. B. (1996). The gap junction communication channel. Cell 84, 381–388. doi: 10.1016/S0092-8674(00)81282-9
Maeda, S., Nakagawa, S., Suga, M., Yamashita, E., Oshima, A., Fujiyoshi, Y., et al. (2009). Structure of the connexin 26 gap junction channel at 3.5 A resolution. Nature 458, 597–602. doi: 10.1038/nature07869
Misu, A., Yamanaka, H., Aramaki, T., Kondo, S., Skerrett, I. M., Iovine, M. K., et al. (2016). Two different functions of connexin43 confer two different bone phenotypes in zebrafish. J. Biol. Chem. 291, 12601–12611. doi: 10.1074/jbc.M116.720110
Musa, H., Fenn, E., Crye, M., Gemel, J., Beyer, E. C., and Veenstra, R. D. (2004). Amino terminal glutamate residues confer spermine sensitivity and affect voltage gating and channel conductance of rat connexin40 gap junctions. J. Physiol. 557(Pt 3), 863–878. doi: 10.1113/jphysiol.2003.059386
Nakamasu, A., Takahashi, G., Kanbe, A., and Kondo, S. (2009). Interactions between zebrafish pigment cells responsible for the generation of Turing patterns. Proc. Natl. Acad. Sci. U.S.A. 106, 8429–8434. doi: 10.1073/pnas.0808622106
Oshima, A., Tani, K., Hiroaki, Y., Fujiyoshi, Y., and Sosinsky, G. E. (2007). Three-dimensional structure of a human connexin26 gap junction channel reveals a plug in the vestibule. Proc. Natl. Acad. Sci. U.S.A. 104, 10034–10039. doi: 10.1073/pnas.0703704104
Paznekas, W. A., Boyadjiev, S. A., Shapiro, R. E., Daniels, O., Wollnik, B., Keegan, C. E., et al. (2003). Connexin 43 (GJA1) mutations cause the pleiotropic phenotype of oculodentodigital dysplasia. Am. J. Hum. Genet. 72, 408–418. doi: 10.1086/346090
Scemes, E., Suadicani, S. O., Dahl, G., and Spray, D. C. (2007). Connexin and pannexin mediated cell-cell communication. Neuron Glia Biol. 3, 199–208. doi: 10.1017/S1740925X08000069
Sultana, N., Nag, K., Hoshijima, K., Laird, D. W., Kawakami, A., and Hirose, S. (2008). Zebrafish early cardiac connexin, Cx36.7/Ecx, regulates myofibril orientation and heart morphogenesis by establishing Nkx2.5 expression. Proc. Natl. Acad. Sci. U.S.A. 105, 4763–4768. doi: 10.1073/pnas.0708451105
Thi, M. M., Islam, S., Suadicani, S. O., and Spray, D. C. (2012). Connexin43 and pannexin1 channels in osteoblasts: who is the “hemichannel”? J. Membrane Biol. 245, 401–409. doi: 10.1007/s00232-012-9462-2
Turing, A. (1952). The chemical basis of morphogenesis. Philos. Trans. R. Soc. Lond. B 237, 37–72. doi: 10.1098/rstb.1952.0012
Verselis, V. K., Ginter, C. S., and Bargiello, T. A. (1994). Opposite voltage gating polarities of two closely related connexins. Nature 368, 348–351. doi: 10.1038/368348a0
Watanabe, M., Iwashita, M., Ishii, M., Kurachi, Y., Kawakami, A., Kondo, S., et al. (2006). Spot pattern of leopard Danio is caused by mutation in the zebrafish connexin41.8 gene. EMBO Rep. 7, 893–897. doi: 10.1038/sj.embor.7400757
Watanabe, M., and Kondo, S. (2012). Changing clothes easily: connexin41.8 regulates skin pattern variation. Pigment Cell Melanoma Res. 25, 326–330. doi: 10.1111/j.1755-148X.2012.00984.x
Watanabe, M., Sawada, R., Aramaki, T., Skerrett, I. M., and Kondo, S. (2016). The physiological characterization of connexin41.8 and connexin39.4, which are involved in the striped pattern formation of zebrafish. J. Biol. Chem. 291, 1053–1063. doi: 10.1074/jbc.M115.673129
Watanabe, M., Watanabe, D., and Kondo, S. (2012). Polyamine sensitivity of gap junctions is required for skin pattern formation in zebrafish. Sci. Rep. 2:473. doi: 10.1038/srep00473
Keywords: connexin, gap junction, skin pattern, bone shape, zebrafish
Citation: Watanabe M (2017) Gap Junction in the Teleost Fish Lineage: Duplicated Connexins May Contribute to Skin Pattern Formation and Body Shape Determination. Front. Cell Dev. Biol. 5:13. doi: 10.3389/fcell.2017.00013
Received: 16 December 2016; Accepted: 07 February 2017;
Published: 21 February 2017.
Edited by:
Takaaki Matsui, Nara Institute of Science and Technology, JapanReviewed by:
Igor Jakovcevski, German Center for Neurodegenerative Diseases (HZ), GermanyKoichi Nishiyama, Kumamoto University Hospital, Japan
Copyright © 2017 Watanabe. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Masakatsu Watanabe, d2F0YW5hYmUtbUBmYnMub3Nha2EtdS5hYy5qcA==
 Masakatsu Watanabe
Masakatsu Watanabe