- Wellcome Trust Centre for Cell Biology, School of Biological Sciences, University of Edinburgh, Scotland, United Kingdom
Protein kinases are major regulators of mitosis, with over 30% of the mitotic proteome phosphorylated on serines, threonines and tyrosines. The human genome encodes for 518 kinases that have a structurally conserved catalytic domain and includes about a dozen of cell division specific ones. Yet each kinase has unique structural features that allow their distinct substrate recognition and modes of regulation. These unique regulatory features determine their accurate spatio-temporal activation critical for correct progression through mitosis and are exploited for therapeutic purposes. In this review, we will discuss the principles of mitotic kinase activation and the structural determinants that underlie functional specificity.
Introduction
Protein phosphorylation is a key regulatory mechanism influencing various cellular processes such as cell growth, cell motility, cell differentiation and cell division. Most notably, protein phosphorylation peaks during mitosis and the timing coincides with the cell division-related chromosomal and cytoskeletal reorganization (Dephoure et al., 2008; Olsen et al., 2010). Consequently, mitotic protein kinases are considered central players orchestrating the mitotic progression and accurate spatio-temporal regulation of their activity becomes essential for error-free chromosome segregation. Many mitotic kinases are well characterized in terms of their structure and function. The list includes but is not limited to CDK (cyclin-dependent kinase; CDK1 and CDK2), Aurora (Aurora-A and Aurora-B) and Plk (Polo-like kinase; Plk1, Plk) families, Bub1, Haspin and Mps1 (see also a focused review on NIMA family of kinase by Fry et al, in this issue). Though mitotic kinases share significant structural similarities, their cellular localization, enzymatic activity and substrate specificity are determined by diverse mechanisms.
In this review, we will summarize our understanding of how structurally conserved and distinct features determine correct spatio-temporal regulation of kinase activity during mitosis.
Generally Conserved Structural Features of a Kinase Catalytic Domain
Most protein kinases have a bilobal catalytic domain of ≈290 amino acids with an N-terminal lobe made of a β-sheet and one α-helix (known as the C-helix) and an α-helical C-terminal lobe (reviewed in Johnson et al., 1996; Bayliss et al., 2012). The active site that transfers the γ-phosphate of ATP to the substrate is buried at the interface between the two lobes (Figure 1). When the kinase is catalytically active, the C-helix packs against the N-terminal lobe (Kobayashi et al., 1992; Brown et al., 1995; Johnson et al., 1996; Bayliss et al., 2003; Sessa et al., 2005; Petri et al., 2007; Endicott et al., 2012). The ATP binding pocket is largely conserved across kinases and is surrounded by relatively less conserved pockets often exploited for inhibitor design (Noble et al., 2004). The peptide backbone of the hinge region connecting the N- and C-terminal lobes makes hydrogen bonds to the adenine ring of the ATP, while nonpolar aliphatic residues lining the pocket interact with the purine structure. A glutamate from the C-helix and a lysine from the N-terminal lobe make a salt bridge (a conserved feature of an active kinase achieved by the packing of the C-helix against the N-terminal lobe) and stabilize the α- and β-phosphate groups of ATP. Note that when generating a kinase dead mutant, this lysine usually is targeted. The glycine-rich loop in the N-terminal lobe also stabilizes the ATP β- and γ-phosphate groups, in addition to its important regulatory role in controlling the access of the substrate to the kinase active site. Finally two magnesium ions in the ATP binding pocket coordinate the phosphate groups to ensure transfer of the γ-phosphate to the hydroxyl acceptor group (Johnson et al., 1996; Bayliss et al., 2012; Endicott et al., 2012).
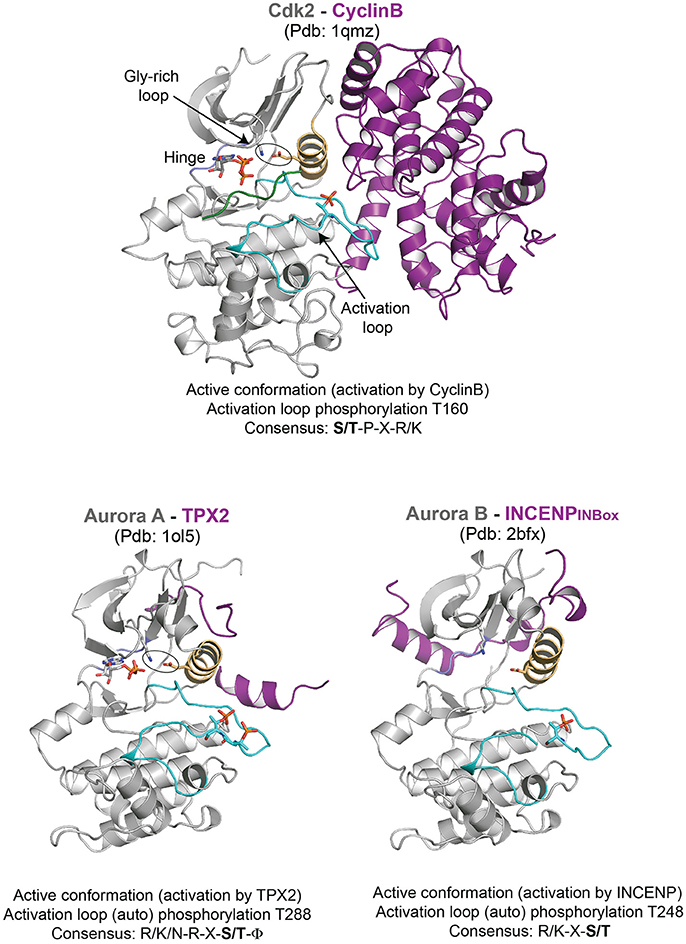
Figure 1. Representative structures of mitotic kinases I. Key structural features are highlighted: C-helix (light orange), hinge region involved in ATP binding (blue), and activation segment (cyan). The characteristic structural feature of an active enzyme, the salt bridge between a Lys and Glu from the N-terminal lobe and the C-helix is highlighted in stick representation.
The C-terminal lobe contains an activation segment that varies in length and sequence but is defined by the well-conserved amino acid motifs DFG and APE (D-[L/Y]-G and [S/P]-P-E are uncommon variations) at each end (Bayliss et al., 2012; Endicott et al., 2012). This activation segment is usually remodeled during kinase activation through phosphorylation, although intra- or inter-molecular protein binding can also trigger activation segment remodeling (Bayliss et al., 2012; Endicott et al., 2012). When non-phosphorylated, the activation segment is disordered and auto-inhibits the kinase by obstructing the substrate binding site. Phosphorylation of the activation segment on a phosphoacceptor serine or threonine typically activates the kinase by stabilizing the substrate-binding site. The negatively charged phosphate group is engaged by the basic residues contributed by the C-helix, the N-terminal lobe and the activation segment to stabilize the substrate-binding platform and hence the active conformation of the kinase (Endicott et al., 2012).
Mechanisms of Kinase Activation
Activation by Inter or Intra Molecular Protein Interaction
Mechanism of mitotic kinase activation was first established for the cyclin-dependent kinase (CDK) family (Evans et al., 1983; Felix et al., 1990; Kobayashi et al., 1992). Pioneering studies in yeast, sea urchin and Xenopus showed that phosphorylation and cyclins controlled the activity of CDKs to drive the cell cycle. Cyclin levels are regulated throughout cell cycle both at the transcriptional and proteolytic level, which in turn activates CDK activity temporally (Evans et al., 1983; Pines and Hunter, 1989; Felix et al., 1990). Cyclins have a cyclin box domain that binds to the N-terminal lobe and the C-helix of the kinase domain (Kobayashi et al., 1992; Brown et al., 1995; Petri et al., 2007). This triggers the packing of the C-helix to remodel the ATP binding site (Brown et al., 1999a, 2015). This mode of activation is broadly conserved in CDK1 (Cdc2 and Cdc28 in budding and fission yeast respectively), which upon cyclin B binding initiates mitosis (Santamaria et al., 2007; Gavet and Pines, 2010; Diril et al., 2012).
The Aurora family of kinases mainly Aurora A and Aurora B, are among the well characterized protein kinases (Bayliss et al., 2003; Sessa et al., 2005; Zorba et al., 2014). These exhibit basal level of kinase activity on their own, but their full activation requires specific binding partners: Aurora A, implicated in centrosome maturation and mitotic spindle stability, involves TPX2 while Aurora B activity essential for destabilizing erroneous kinetochore-microtubule attachment and cytokinesis requires INCENP (which together with Borealin and Survivin functions as the Chromosomal Passenger Complex, CPC). Though the activation loop auto-phosphorylation is sufficient for basal activity, facilitating the right conformation of the loop is required for full activation, and is achieved by the binding of TPX2 and INCENP to Aurora A and Aurora B kinases, respectively (Bayliss et al., 2003; Sessa et al., 2005). A short N-terminal fragment of TPX2 binds between the N-lobe and activation loop of Aurora A and by doing so it stabilizes the activation loop in a fully active conformation. This makes the activation loop phosphorylation (pT288) inaccessible for counteracting phosphatases and thus maintains the fully active state of Aurora A. In the case of Aurora B, the C-terminal INBOX domain of INCENP wraps around the Aurora B N-lobe interacting with the C-helix, however is not sufficient for the full activation of Aurora B (at least on the basis of the relative orientation of C-helix with respect to the N-terminal lobe as seen in the crystal structures) (Figure 1). It has been postulated that additional interactions between the phosphorylated TSS motif downstream of INBOX with Aurora B might be required for full activation (Sessa et al., 2005).
Bub1 is a kinetochore-associated protein kinase which regulates the Spindle Assembly Checkpoint (SAC) and kinetochore-microtubule attachment (Elowe, 2011). Cdc20, Mad1/2 and Histone H2A appear as major substrates, while Bub1 also plays a scaffolding role at kinetochores to recruit the pseudokinase BubR1 (Tang et al., 2004; Yamagishi et al., 2010; Suijkerbuijk et al., 2012; Overlack et al., 2015, 2017; Jia et al., 2016). Structural characterization of Bub1 kinase domain reveals the role of its N-terminal extension in positioning the C-helix and stabilizing the conformation of the activation segment (Lin et al., 2014) (Figure 2). However, the activation segment conformation seen in the crystal structure does not appear to be suitable for substrate binding and activation loop phosphorylation has been implicated to enhance kinase activity and substrate recognition (Lin et al., 2014) (Figure 2).
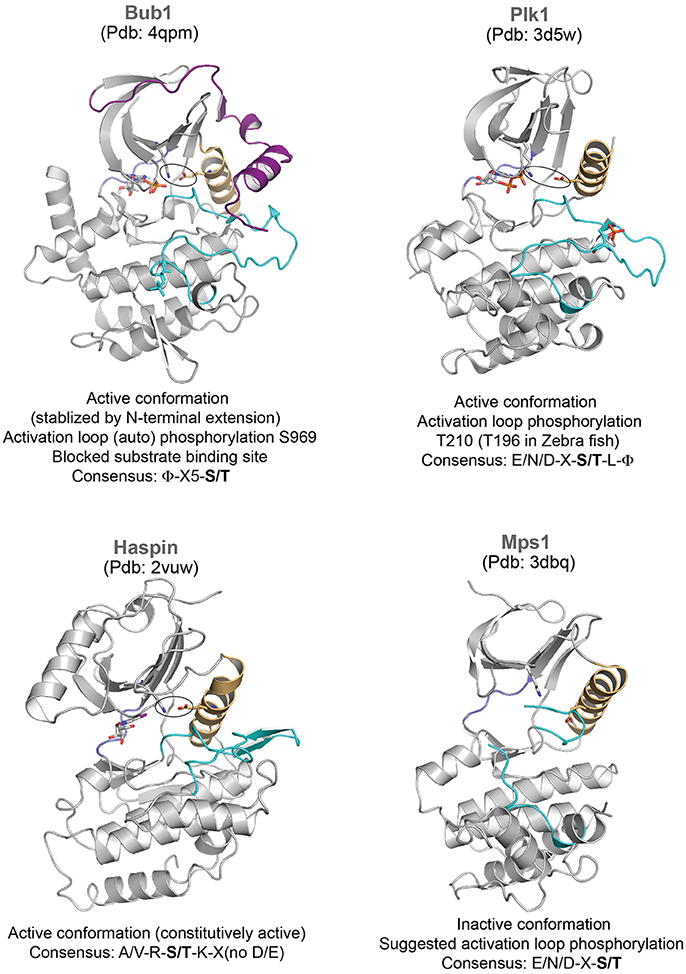
Figure 2. Representative structures of mitotic kinases II. Key structural features are highlighted: C-helix (light orange), hinge region involved in ATP binding (blue), and activation segment (cyan). The characteristic structural feature of an active enzyme, the salt bridge between a Lys and Glu from the N-terminal lobe and the C-helix is highlighted in stick representation.
Regulation by Phosphorylation
Activation segment phosphorylation regulates kinase activity of several kinases during mitosis. Both Aurora A and Aurora B kinases undergo auto-phosphorylation within their activation segments (T288 and T248 in Aurora A and Aurora B, respectively) which is essential for their increased kinase activity (Bayliss et al., 2003; Sessa et al., 2005). Bub1 undergoes constitutive autophosphorylation in its activation loop throughout cell cycle, which is suggested to trigger reorganization in the substrate binding site and enhance kinase activity (Lin et al., 2014). Likewise, Mps1 auto-phosphorylates multiple residues in its activation loop to activate itself (Mattison et al., 2007; Wang et al., 2009). The crystal structure of Mps1 kinase dead mutant revealed that it adopts an inactive conformation (Figure 2), where the activation loop is disordered and displaces the C-helix (Chu et al., 2008; Wang et al., 2009). Auto-phosphorylation of the activation loop may reorient and allow correct positioning of the C-helix for full activity. The activation segment phosphorylations of CDK1 and Plk1 are mediated by other kinases—CAK (CDK Activating Kinase belonging to the family of CDKs) (Russo et al., 1996; Brown et al., 1999b), Aurora family of kinases (Aurora A in mammals, Jang et al., 2002; Macurek et al., 2008; Seki et al., 2008 and Aurora B in Drosophila, Carmena et al., 2012a; Xu et al., 2013; Kachaner et al., 2014), respectively.
Phosphorylation within the kinase module has also been used to inactivate kinases. For example, the wee1 kinase inhibits CDK1 by phosphorylating the Tyrosine 15 in the N-terminal glycine-rich loop and obstructing access of the substrate to the kinase active site (Russell and Nurse, 1987; Parker and Piwnica-Worms, 1992; McGowan and Russell, 1993; Welburn et al., 2007). This inhibition is reversed by the phosphatase Cdc25 (Frazer and Young, 2012).
Kinases with Constitutive Activity
Unlike most mitotic kinases, the haspin kinase responsible for the histone H3 Thr3 phosphorylation (essential for the centromere localization of Aurora B/CPC) and the budding yeast Hrr25 (Casein kinase) critical for meiosis co-orientation of kinetochore do not require the activation loop phosphorylation and their kinase domains are constitutively active (Eswaran et al., 2009; Villa et al., 2009; Ye et al., 2016). However, the intrinsic activity of the haspin kinase is inhibited during prophase by its conserved basic N terminus (haspin 380-399, human numbering) preceding the kinase domain. During prophase to metaphase transition, the concerted activities of CDK1 and Plk1 relieve the auto-inhibition through phosphorylation in the N-terminal segment, and thus restrict the haspin activity to metaphase (Ghenoiu et al., 2013).
Mechanisms for Achieving Substrate Specificity
Kinases achieve substrate specificity through multiple mechanisms in mitosis (Ubersax and Ferrell, 2007; Johnson, 2011): by specific recognition of amino acid sequence flanking the phosphorylation site or/and by employing a domain capable of recognizing substrates marked by priming phosphorylation. In addition, specific subcellular localization of kinases also facilitates substrate specificity by spatially restricting the kinases.
Consensus Phosphorylation Motifs
Kinase substrate specificity is generally determined by the architecture of the substrate binding site, which might select negatively against certain residues flanking the phosphorylation site. The identification of substrates in vitro and in vivo thus far has helped define kinase substrate specificity. Typically about 4-6 amino acids flanking the phospho-acceptor residue P can contribute to the selectivity of kinases for their substrate. The molecular basis for substrate recognition mainly comes from the structural work on CDK2/cyclin A bound to its substrate (Brown et al., 1999a). This work identified S/T-P-X-R/K as a preferred consensus phosphorylation motif for CDK. The activation segment adopts a conformation such that the carbonyl group of V164 is unusually strained and cannot form a hydrogen bond with the backbone of P+1 residue. As a result, other residues other than a proline at this position are unfavored. The preference for K/R at P+3 arises from an interaction between the phosphorylated and highly negatively charged threonine in the activation loop of CDK and the positively charged side chain of K/R (Heald and McKeon, 1990; Peter et al., 1990; Ward and Kirschner, 1990; Brown et al., 1999a).
The identification of multiple budding yeast kinetochore Aurora B kinase substrates, allowed to define an Aurora consensus phosphorylation sequences (Cheeseman et al., 2002) and subsequent identification of other Aurora substrates (Andrews et al., 2004; Lan et al., 2004; Cheeseman et al., 2006; DeLuca et al., 2006). Aurora kinases preferentially phosphorylate substrates with basic residues at P-3 and P-2 and containing a hydrophobic residue at P+1. Plk1 has a preference for an acidic residue at P-2 and a hydrophobic residue at P+1 (Toyoshima-Morimoto et al., 2001; Nakajima et al., 2003). An integrated approach combining biochemical, proteomic and structural biology methods identified substrates of the Haspin kinase, its consensus substrate recognition motif and their mode of interaction (Maiolica et al., 2014). Crystal structure analysis revealed that residues P-2 (A), P-1 (R), P (S/T) and P+1 (K) were specifically recognized within a deep pocket and thus providing a rational for the proposed consensus recognition motif A/V-R-S/T-K (Maiolica et al., 2014). In the case of Bub1, in vitro phosphorylation of kinetochore proteins by Bub1 followed by phosphorylation-directed staining and mass spectrometric analyses identified many prospective Bub1 substrates with a putative consensus motif ϕ-X5-S/T (Breit et al., 2015).
The use of peptide libraries against kinases has greatly contributed to the identification of optimal peptide sequence motifs and “anti-motifs” for kinases (Hutti et al., 2004). For example, Plk1, Aurora A and Aurora B strongly discriminate against proline at P+1 (Alexander et al., 2011). However, kinases do tolerate variations in the consensus sequence of their substrates. Proteomic studies combined with known phosphorylation consensus sites and bioinformatics represent powerful ways to uncover and validate new substrates in vivo (Dephoure et al., 2008; Mok et al., 2010; Kettenbach et al., 2011; Santamaria et al., 2011). Quantitative proteomics also inform substrate specificity of kinases not previously known. For example, such studies revealed that Mps1 and Plk1 share the same substrate preference (Dou et al., 2011; Petrone et al., 2016). Overall the substrate consensus motif plays a determining role in kinase-substrate interactions.
Substrate Priming
Kinases may use docking sites or sites that are primed by another kinase to enhance their substrate selectivity. For example, to bind and to be phosphorylated by Plk1, a substrate generally needs to be primed by another kinase (Lee et al., 2008). Plk family members have two Polo-box domains (PBD), that recognize Plk1 substrates (typically containing S-S/Tp-P) primed by other kinases such as Cdk1. By doing so the PBD of Plk1 recruits the catalytic domain to the phosphorylated substrates (Cheng et al., 2003; Elia et al., 2003a,b; Barr et al., 2004). Known mitotic substrates of Plk1 are the checkpoint proteins Bub1, BubR1 and Wee1 (Watanabe et al., 2005; Qi et al., 2006; Elowe et al., 2007). There have also been evidence for Plk1 “self-priming” (reviewed in Lee et al., 2008).
CDK-cyclins also use docking sites to recognize and phosphorylate temporally their substrates. Certain cyclin partners have a hydrophobic docking patch that recognizes an “RXL” motif on substrates 40Å away from the catalytic site of the CDK active site (Schulman et al., 1998; Brown et al., 1999a). A subset of substrates have a “RXL” motif recognized by a cyclin and are phosphorylated earlier in the cell cycle because they have a higher affinity for the CDK-cyclin complex (Loog and Morgan, 2005; Koivomagi et al., 2011). Substrates that do not have this docking site are phosphorylated later during mitosis or may not be recognized by the CDK-cyclin complex (Koivomagi et al., 2011). Overall the docking site interactions increase the local concentration of the substrate and ensure accurate spatio-temporal substrate phosphorylation essential for correct mitotic progression (Brown et al., 2015).
Subcellular Localization
Many mitotic kinases rely on spatial targeting to phosphorylate their specific substrates. This restricts the activity of the kinase to generate gradients of kinase activity. The most well characterized spatially-targeted kinases are Aurora A and B kinases and they appear to share the same substrate specificity (Fu et al., 2009). However Aurora A predominantly associates with centrosomes and mitotic spindle, while Aurora B is localized at centromeres and kinetochores. Centrosome association of Aurora A is mediated by TPX2, whereas Aurora B (and hence the CPC) localization to centromeres is mediated by phosphorylation marks on Histone H3 (Thr3) and Histone H2A (Thr120) created by Haspin and Bub1, respectively (reviewed in Carmena et al., 2012b; Kitagawa and Lee, 2015). While the Survivin subunit of the CPC recognizes the histone H3 mark, Sgo1 recognizes the H2A mark (Kelly et al., 2010; Wang et al., 2010; Yamagishi et al., 2010). The CPC indirectly recognizes histone H2A via the interaction of CDK1-phosphorylated Borealin with histone H2A bound Sgo1 (Tsukahara et al., 2010). Due to their distinct subcellular localization, while Aurora B phosphorylates substrates such as histone H3, kinetochore proteins and spindle midzone proteins (Gruneberg et al., 2004; Guse et al., 2005; Nunes Bastos et al., 2013), Aurora A phosphorylates a number of centrosomal and spindle-localized substrates (Sardon et al., 2010).
The molecular basis for the recruitment of Mps1 kinase to the outer kinetochore is also well established. Multiple kinases including CDK1, Aurora B, Plk1, and Mps1 itself are implicated in Mps1 kinetochore targeting (Morin et al., 2012; Nijenhuis et al., 2013; von Schubert et al., 2015). In addition, direct interaction of Mps1 with the Ndc80 complex is crucial for its localization and function (Hiruma et al., 2015; Ji et al., 2015). While the N-terminal extension of Mps1 directly interacts with the CH-domain of Ndc80 (adjacent to the microtubule binding region), the conserved middle region of Mps1 interacts with the Nuf2 CH domain. The affinity of Mps1 is higher for Aurora B-phosphorylated Ndc80, indicating that Aurora B promotes Mps1-Ndc80 interaction in response to unattached kinetochores (Ji et al., 2015). Mps1 can then phosphorylate Knl1 to activate the spindle checkpoint.
Summary
High resolution mechanistic understanding of kinase regulation is essential not only to define how kinases achieve error-free cell division, but also to exploit the differences in their regulatory mechanisms to specifically target them in mitosis-related human disorders. Structural studies of kinases thus far have provided key insights into the similarities and differences in the modes of activation and regulation of many mitotic kinases. Although kinases possess a broadly conserved catalytic core, their level of kinase activity and substrate specificity are determined by specific inter/intra molecular interactions and phosphorylation. In this review, we summarize how key structural regulatory elements such as the relative orientation of the C-helix, activation segment conformation and spatial regulatory elements responsible for correct kinase sub-cellular localization achieve accurate kinase function. However, there are still many open questions, particularly on factors determining the graded level of kinase activation and its implications on their mitotic role. More structural analyses of kinases in complex with their regulatory binding partners with and without bound substrates, and their functional implications in cells will undoubtedly further advance our mechanistic understanding of this essential class of mitotic regulators.
Author Contributions
JW and AJ have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank M. A. Abad, B. A. Medina-Pritchard, Ben Craske, and Thibault Legal for the critical reading of the manuscript. AJ is supported by the Wellcome Senior Research Fellowship (202811). JW is supported by a CRUK Career Development Fellowship (C40377/A12840) and a Royal Society grant (RG160003). The Wellcome Trust Centre for Cell Biology is supported by core funding from the Wellcome Trust (203149).
References
Alexander, J., Lim, D., Joughin, B. A., Hegemann, B., Hutchins, J. R., Ehrenberger, T., et al. (2011). Spatial exclusivity combined with positive and negative selection of phosphorylation motifs is the basis for context-dependent mitotic signaling. Sci. Signal. 4:ra42. doi: 10.1126/scisignal.2001796
Andrews, P. D., Ovechkina, Y., Morrice, N., Wagenbach, M., Duncan, K., Wordeman, L., et al. (2004). Aurora B regulates MCAK at the mitotic centromere. Dev. Cell 6, 253–268. doi: 10.1016/S1534-5807(04)00025-5
Barr, F. A., Sillje, H. H., and Nigg, E. A. (2004). Polo-like kinases and the orchestration of cell division. Nat. Rev. Mol. Cell Biol. 5, 429–440. doi: 10.1038/nrm1401
Bayliss, R., Fry, A., Haq, T., and Yeoh, S. (2012). On the molecular mechanisms of mitotic kinase activation. Open Biol. 2:120136. doi: 10.1098/rsob.120136
Bayliss, R., Sardon, T., Vernos, I., and Conti, E. (2003). Structural basis of Aurora-A activation by TPX2 at the mitotic spindle. Mol. Cell 12, 851–862. doi: 10.1016/S1097-2765(03)00392-7
Breit, C., Bange, T., Petrovic, A., Weir, J. R., Muller, F., Vogt, D., et al. (2015). Role of intrinsic and extrinsic factors in the regulation of the mitotic checkpoint kinase Bub1. PLoS ONE 10:e0144673. doi: 10.1371/journal.pone.0144673
Brown, N. R., Korolchuk, S., Martin, M. P., Stanley, W. A., Moukhametzianov, R., Noble, M. E., et al. (2015). CDK1 structures reveal conserved and unique features of the essential cell cycle CDK. Nat. Commun. 6:6769. doi: 10.1038/ncomms7769
Brown, N. R., Noble, M. E., Endicott, J. A., Garman, E. F., Wakatsuki, S., Mitchell, E., et al. (1995). The crystal structure of cyclin A. Structure 3, 1235–1247. doi: 10.1016/S0969-2126(01)00259-3
Brown, N. R., Noble, M. E., Endicott, J. A., and Johnson, L. N. (1999a). The structural basis for specificity of substrate and recruitment peptides for cyclin-dependent kinases. Nat. Cell Biol. 1, 438–443. doi: 10.1038/15674
Brown, N. R., Noble, M. E., Lawrie, A. M., Morris, M. C., Tunnah, P., Divita, G., et al. (1999b). Effects of phosphorylation of threonine 160 on cyclin-dependent kinase 2 structure and activity. J. Biol. Chem. 274, 8746–8756. doi: 10.1074/jbc.274.13.8746
Carmena, M., Pinson, X., Platani, M., Salloum, Z., Xu, Z., Clark, A., et al. (2012a). The chromosomal passenger complex activates Polo kinase at centromeres. PLoS Biol. 10:e1001250. doi: 10.1371/journal.pbio.1001250
Carmena, M., Wheelock, M., Funabiki, H., and Earnshaw, W. C. (2012b). The chromosomal passenger complex (CPC): from easy rider to the godfather of mitosis. Nat. Rev. Mol. Cell Biol. 13, 789–803. doi: 10.1038/nrm3474
Cheeseman, I. M., Anderson, S., Jwa, M., Green, E. M., Kang, J., Yates, J. R. III., et al. (2002). Phospho-regulation of kinetochore-microtubule attachments by the Aurora kinase Ipl1p. Cell 111, 163–172. doi: 10.1016/S0092-8674(02)00973-X
Cheeseman, I. M., Chappie, J. S., Wilson-Kubalek, E. M., and Desai, A. (2006). The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell 127, 983–997. doi: 10.1016/j.cell.2006.09.039
Cheng, K. Y., Lowe, E. D., Sinclair, J., Nigg, E. A., and Johnson, L. N. (2003). The crystal structure of the human polo-like kinase-1 polo box domain and its phospho-peptide complex. EMBO J. 22, 5757–5768. doi: 10.1093/emboj/cdg558
Chu, M. L., Chavas, L. M., Douglas, K. T., Eyers, P. A., and Tabernero, L. (2008). Crystal structure of the catalytic domain of the mitotic checkpoint kinase Mps1 in complex with SP600125. J. Biol. Chem. 283, 21495–21500. doi: 10.1074/jbc.M803026200
DeLuca, J. G., Gall, W. E., Ciferri, C., Cimini, D., Musacchio, A., and Salmon, E. D. (2006). Kinetochore microtubule dynamics and attachment stability are regulated by Hec1. Cell 127, 969–982. doi: 10.1016/j.cell.2006.09.047
Dephoure, N., Zhou, C., Villen, J., Beausoleil, S. A., Bakalarski, C. E., Elledge, S. J., et al. (2008). A quantitative atlas of mitotic phosphorylation. Proc. Natl. Acad. Sci. U.S.A. 105, 10762–10767. doi: 10.1073/pnas.0805139105
Diril, M. K., Ratnacaram, C. K., Padmakumar, V. C., Du, T., Wasser, M., Coppola, V., et al. (2012). Cyclin-dependent kinase 1 (CDK1) is essential for cell division and suppression of DNA re-replication but not for liver regeneration. Proc. Natl. Acad. Sci. U.S.A. 109, 3826–3831. doi: 10.1073/pnas.1115201109
Dou, Z., von Schubert, C., Korner, R., Santamaria, A., Elowe, S., and Nigg, E. A. (2011). Quantitative mass spectrometry analysis reveals similar substrate consensus motif for human Mps1 kinase and Plk1. PLoS ONE 6:e18793. doi: 10.1371/journal.pone.0018793
Elia, A. E., Cantley, L. C., and Yaffe, M. B. (2003a). Proteomic screen finds pSer/pThr-binding domain localizing Plk1 to mitotic substrates. Science 299, 1228–1231. doi: 10.1126/science.1079079
Elia, A. E., Rellos, P., Haire, L. F., Chao, J. W., Ivins, F. J., Hoepker, K., et al. (2003b). The molecular basis for phosphodependent substrate targeting and regulation of Plks by the Polo-box domain. Cell 115, 83–95. doi: 10.1016/S0092-8674(03)00725-6
Elowe, S. (2011). Bub1 and BubR1: at the interface between chromosome attachment and the spindle checkpoint. Mol. Cell. Biol. 31, 3085–3093. doi: 10.1128/MCB.05326-11
Elowe, S., Hummer, S., Uldschmid, A., Li, X., and Nigg, E. A. (2007). Tension-sensitive Plk1 phosphorylation on BubR1 regulates the stability of kinetochore microtubule interactions. Genes Dev. 21, 2205–2219. doi: 10.1101/gad.436007
Endicott, J. A., Noble, M. E., and Johnson, L. N. (2012). The structural basis for control of eukaryotic protein kinases. Annu. Rev. Biochem. 81, 587–613. doi: 10.1146/annurev-biochem-052410-090317
Eswaran, J., Patnaik, D., Filippakopoulos, P., Wang, F., Stein, R. L., Murray, J. W., et al. (2009). Structure and functional characterization of the atypical human kinase haspin. Proc. Natl. Acad. Sci. U.S.A. 106, 20198–20203. doi: 10.1073/pnas.0901989106
Evans, T., Rosenthal, E. T., Youngblom, J., Distel, D., and Hunt, T. (1983). Cyclin: a protein specified by maternal mRNA in sea urchin eggs that is destroyed at each cleavage division. Cell 33, 389–396. doi: 10.1016/0092-8674(83)90420-8
Felix, M. A., Labbe, J. C., Doree, M., Hunt, T., and Karsenti, E. (1990). Triggering of cyclin degradation in interphase extracts of amphibian eggs by cdc2 kinase. Nature 346, 379–382. doi: 10.1038/346379a0
Frazer, C., and Young, P. G. (2012). Carboxy-terminal phosphorylation sites in Cdc25 contribute to enforcement of the DNA damage and replication checkpoints in fission yeast. Curr. Genet. 58, 217–234. doi: 10.1007/s00294-012-0379-1
Fu, J., Bian, M., Liu, J., Jiang, Q., and Zhang, C. (2009). A single amino acid change converts Aurora-A into Aurora-B-like kinase in terms of partner specificity and cellular function. Proc. Natl. Acad. Sci. U.S.A. 106, 6939–6944. doi: 10.1073/pnas.0900833106
Gavet, O., and Pines, J. (2010). Progressive activation of CyclinB1-CDK1 coordinates entry to mitosis. Dev. Cell 18, 533–543. doi: 10.1016/j.devcel.2010.02.013
Ghenoiu, C., Wheelock, M. S., and Funabiki, H. (2013). Autoinhibition and Polo-dependent multisite phosphorylation restrict activity of the histone H3 kinase Haspin to mitosis. Mol. Cell 52, 734–745. doi: 10.1016/j.molcel.2013.10.002
Gruneberg, U., Neef, R., Honda, R., Nigg, E. A., and Barr, F. A. (2004). Relocation of Aurora B from centromeres to the central spindle at the metaphase to anaphase transition requires MKlp2. J. Cell Biol. 166, 167–172. doi: 10.1083/jcb.200403084
Guse, A., Mishima, M., and Glotzer, M. (2005). Phosphorylation of ZEN-4/MKLP1 by aurora B regulates completion of cytokinesis. Curr. Biol. 15, 778–786. doi: 10.1016/j.cub.2005.03.041
Heald, R., and McKeon, F. (1990). Mutations of phosphorylation sites in lamin A that prevent nuclear lamina disassembly in mitosis. Cell 61, 579–589. doi: 10.1016/0092-8674(90)90470-Y
Hiruma, Y., Sacristan, C., Pachis, S. T., Adamopoulos, A., Kuijt, T., Ubbink, M., et al. (2015). CELL DIVISION CYCLE. Competition between MPS1 and microtubules at kinetochores regulates spindle checkpoint signaling. Science 348, 1264–1267. doi: 10.1126/science.aaa4055
Hutti, J. E., Jarrell, E. T., Chang, J. D., Abbott, D. W., Storz, P., Toker, A., et al. (2004). A rapid method for determining protein kinase phosphorylation specificity. Nat. Methods 1, 27–29. doi: 10.1038/nmeth708
Jang, Y. J., Ma, S., Terada, Y., and Erikson, R. L. (2002). Phosphorylation of threonine 210 and the role of serine 137 in the regulation of mammalian polo-like kinase. J. Biol. Chem. 277, 44115–44120. doi: 10.1074/jbc.M202172200
Ji, Z., Gao, H., and Yu, H. (2015). CELL DIVISION CYCLE. Kinetochore attachment sensed by competitive Mps1 and microtubule binding to Ndc80C. Science 348, 1260–1264. doi: 10.1126/science.aaa4029
Jia, L., Li, B., and Yu, H. (2016). The Bub1-Plk1 kinase complex promotes spindle checkpoint signalling through Cdc20 phosphorylation. Nat. Commun. 7:10818. doi: 10.1038/ncomms10818
Johnson, L. N. (2011). Substrates of mitotic kinases. Sci. Signal. 4:pe31. doi: 10.1126/scisignal.2002234
Johnson, L. N., Noble, M. E., and Owen, D. J. (1996). Active and inactive protein kinases: structural basis for regulation. Cell 85, 149–158. doi: 10.1016/S0092-8674(00)81092-2
Kachaner, D., Pinson, X., El Kadhi, K. B., Normandin, K., Talje, L., Lavoie, H., et al. (2014). Interdomain allosteric regulation of Polo kinase by Aurora B and Map205 is required for cytokinesis. J. Cell Biol. 207, 201–211. doi: 10.1083/jcb.201408081
Kelly, A. E., Ghenoiu, C., Xue, J. Z., Zierhut, C., Kimura, H., and Funabiki, H. (2010). Survivin reads phosphorylated histone H3 threonine 3 to activate the mitotic kinase Aurora B. Science 330, 235–239. doi: 10.1126/science.1189505
Kettenbach, A. N., Schweppe, D. K., Faherty, B. K., Pechenick, D., Pletnev, A. A., and Gerber, S. A. (2011). Quantitative phosphoproteomics identifies substrates and functional modules of Aurora and Polo-like kinase activities in mitotic cells. Sci. Signal. 4:rs5. doi: 10.1126/scisignal.2001497
Kitagawa, M., and Lee, S. H. (2015). The chromosomal passenger complex (CPC) as a key orchestrator of orderly mitotic exit and cytokinesis. Front. Cell Dev. Biol. 3:14. doi: 10.3389/fcell.2015.00014
Kobayashi, H., Stewart, E., Poon, R., Adamczewski, J. P., Gannon, J., and Hunt, T. (1992). Identification of the domains in cyclin A required for binding to, and activation of, p34cdc2 and p32CDK2 protein kinase subunits. Mol. Biol. Cell 3, 1279–1294. doi: 10.1091/mbc.3.11.1279
Koivomagi, M., Valk, E., Venta, R., Iofik, A., Lepiku, M., Morgan, D. O., et al. (2011). Dynamics of CDK1 substrate specificity during the cell cycle. Mol. Cell 42, 610–623. doi: 10.1016/j.molcel.2011.05.016
Lan, W., Zhang, X., Kline-Smith, S. L., Rosasco, S. E., Barrett-Wilt, G. A., Shabanowitz, J., et al. (2004). Aurora B phosphorylates centromeric MCAK and regulates its localization and microtubule depolymerization activity. Curr. Biol. 14, 273–286. doi: 10.1016/j.cub.2004.01.055
Lee, K. S., Park, J. E., Kang, Y. H., Zimmerman, W., Soung, N. K., Seong, Y. S., et al. (2008). Mechanisms of mammalian polo-like kinase 1 (Plk1) localization: self- versus non-self-priming. Cell Cycle 7, 141–145. doi: 10.4161/cc.7.2.5272
Lin, Z., Jia, L., Tomchick, D. R., Luo, X., and Yu, H. (2014). Substrate-specific activation of the mitotic kinase Bub1 through intramolecular autophosphorylation and kinetochore targeting. Structure 22, 1616–1627. doi: 10.1016/j.str.2014.08.020
Loog, M., and Morgan, D. O. (2005). Cyclin specificity in the phosphorylation of cyclin-dependent kinase substrates. Nature 434, 104–108. doi: 10.1038/nature03329
Macurek, L., Lindqvist, A., Lim, D., Lampson, M. A., Klompmaker, R., Freire, R., et al. (2008). Polo-like kinase-1 is activated by aurora A to promote checkpoint recovery. Nature 455, 119–123. doi: 10.1038/nature07185
Maiolica, A., de Medina-Redondo, M., Schoof, E. M., Chaikuad, A., Villa, F., Gatti, M., et al. (2014). Modulation of the chromatin phosphoproteome by the Haspin protein kinase. Mol. Cell. Proteomics 13, 1724–1740. doi: 10.1074/mcp.M113.034819
Mattison, C. P., Old, W. M., Steiner, E., Huneycutt, B. J., Resing, K. A., Ahn, N. G., et al. (2007). Mps1 activation loop autophosphorylation enhances kinase activity. J. Biol. Chem. 282, 30553–30561. doi: 10.1074/jbc.M707063200
McGowan, C. H., and Russell, P. (1993). Human Wee1 kinase inhibits cell division by phosphorylating p34cdc2 exclusively on Tyr15. EMBO J. 12, 75–85.
Mok, J., Kim, P. M., Lam, H. Y., Piccirillo, S., Zhou, X., Jeschke, G. R., et al. (2010). Deciphering protein kinase specificity through large-scale analysis of yeast phosphorylation site motifs. Sci. Signal. 3:ra12. doi: 10.1126/scisignal.2000482
Morin, V., Prieto, S., Melines, S., Hem, S., Rossignol, M., Lorca, T., et al. (2012). CDK-dependent potentiation of MPS1 kinase activity is essential to the mitotic checkpoint. Curr. Biol. 22, 289–295. doi: 10.1016/j.cub.2011.12.048
Nakajima, H., Toyoshima-Morimoto, F., Taniguchi, E., and Nishida, E. (2003). Identification of a consensus motif for Plk (Polo-like kinase) phosphorylation reveals Myt1 as a Plk1 substrate. J. Biol. Chem. 278, 25277–25280. doi: 10.1074/jbc.C300126200
Nijenhuis, W., von Castelmur, E., Littler, D., De Marco, V., Tromer, E., Vleugel, M., et al. (2013). A TPR domain-containing N-terminal module of MPS1 is required for its kinetochore localization by Aurora B. J. Cell Biol. 201, 217–231. doi: 10.1083/jcb.201210033
Noble, M. E., Endicott, J. A., and Johnson, L. N. (2004). Protein kinase inhibitors: insights into drug design from structure. Science 303, 1800–1805. doi: 10.1126/science.1095920
Nunes Bastos, R., Gandhi, S. R., Baron, R. D., Gruneberg, U., Nigg, E. A., and Barr, F. A. (2013). Aurora B suppresses microtubule dynamics and limits central spindle size by locally activating KIF4A. J. Cell Biol. 202, 605–621. doi: 10.1083/jcb.201301094
Olsen, J. V., Vermeulen, M., Santamaria, A., Kumar, C., Miller, M. L., Jensen, L. J., et al. (2010). Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci. Signal. 3:ra3. doi: 10.1126/scisignal.2000475
Overlack, K., Bange, T., Weissmann, F., Faesen, A. C., Maffini, S., Primorac, I., et al. (2017). BubR1 promotes Bub3-dependent APC/C inhibition during spindle assembly checkpoint signaling. Curr. Biol. 27, 2915.e7–2927.e7. doi: 10.1016/j.cub.2017.08.033
Overlack, K., Primorac, I., Vleugel, M., Krenn, V., Maffini, S., Hoffmann, I., et al. (2015). A molecular basis for the differential roles of Bub1 and BubR1 in the spindle assembly checkpoint. Elife 4:e05269. doi: 10.7554/eLife.05269
Parker, L. L., and Piwnica-Worms, H. (1992). Inactivation of the p34cdc2-cyclin B complex by the human WEE1 tyrosine kinase. Science 257, 1955–1957. doi: 10.1126/science.1384126
Peter, M., Nakagawa, J., Doree, M., Labbe, J. C., and Nigg, E. A. (1990). Identification of major nucleolar proteins as candidate mitotic substrates of cdc2 kinase. Cell 60, 791–801. doi: 10.1016/0092-8674(90)90093-T
Petri, E. T., Errico, A., Escobedo, L., Hunt, T., and Basavappa, R. (2007). The crystal structure of human cyclin B. Cell Cycle 6, 1342–1349. doi: 10.4161/cc.6.11.4297
Petrone, A., Adamo, M. E., Cheng, C., and Kettenbach, A. N. (2016). Identification of Candidate Cyclin-dependent kinase 1 (CDK1) substrates in mitosis by quantitative phosphoproteomics. Mol. Cell. Proteomics 15, 2448–2461. doi: 10.1074/mcp.M116.059394
Pines, J., and Hunter, T. (1989). Isolation of a human cyclin cDNA: evidence for cyclin mRNA and protein regulation in the cell cycle and for interaction with p34cdc2. Cell 58, 833–846. doi: 10.1016/0092-8674(89)90936-7
Qi, W., Tang, Z., and Yu, H. (2006). Phosphorylation- and polo-box-dependent binding of Plk1 to Bub1 is required for the kinetochore localization of Plk1. Mol. Biol. Cell 17, 3705–3716. doi: 10.1091/mbc.E06-03-0240
Russell, P., and Nurse, P. (1987). Negative regulation of mitosis by wee1+, a gene encoding a protein kinase homolog. Cell 49, 559–567. doi: 10.1016/0092-8674(87)90458-2
Russo, A. A., Jeffrey, P. D., and Pavletich, N. P. (1996). Structural basis of cyclin-dependent kinase activation by phosphorylation. Nat. Struct. Biol. 3, 696–700. doi: 10.1038/nsb0896-696
Santamaria, A., Wang, B., Elowe, S., Malik, R., Zhang, F., Bauer, M., et al. (2011). The Plk1-dependent phosphoproteome of the early mitotic spindle. Mol. Cell. Proteomics 10:M110.004457. doi: 10.1074/mcp.M110.004457
Santamaria, D., Barriere, C., Cerqueira, A., Hunt, S., Tardy, C., Newton, K., et al. (2007). CDK1 is sufficient to drive the mammalian cell cycle. Nature 448, 811–815. doi: 10.1038/nature06046
Sardon, T., Pache, R. A., Stein, A., Molina, H., Vernos, I., and Aloy, P. (2010). Uncovering new substrates for Aurora A kinase. EMBO Rep. 11, 977–984. doi: 10.1038/embor.2010.171
Schulman, B. A., Lindstrom, D. L., and Harlow, E. (1998). Substrate recruitment to cyclin-dependent kinase 2 by a multipurpose docking site on cyclin A. Proc. Natl. Acad. Sci. U.S.A. 95, 10453–10458. doi: 10.1073/pnas.95.18.10453
Seki, A., Coppinger, J. A., Jang, C. Y., Yates, J. R., and Fang, G. (2008). Bora and the kinase Aurora a cooperatively activate the kinase Plk1 and control mitotic entry. Science 320, 1655–1658. doi: 10.1126/science.1157425
Sessa, F., Mapelli, M., Ciferri, C., Tarricone, C., Areces, L. B., Schneider, T. R., et al. (2005). Mechanism of Aurora B activation by INCENP and inhibition by hesperadin. Mol. Cell 18, 379–391. doi: 10.1016/j.molcel.2005.03.031
Suijkerbuijk, S. J., van Dam, T. J., Karagoz, G. E., von Castelmur, E., Hubner, N. C., Duarte, A. M., et al. (2012). The vertebrate mitotic checkpoint protein BUBR1 is an unusual pseudokinase. Dev. Cell 22, 1321–1329. doi: 10.1016/j.devcel.2012.03.009
Tang, Z., Shu, H., Oncel, D., Chen, S., and Yu, H. (2004). Phosphorylation of Cdc20 by Bub1 provides a catalytic mechanism for APC/C inhibition by the spindle checkpoint. Mol. Cell 16, 387–397. doi: 10.1016/j.molcel.2004.09.031
Toyoshima-Morimoto, F., Taniguchi, E., Shinya, N., Iwamatsu, A., and Nishida, E. (2001). Polo-like kinase 1 phosphorylates cyclin B1 and targets it to the nucleus during prophase. Nature 410, 215–220. doi: 10.1038/35065617
Tsukahara, T., Tanno, Y., and Watanabe, Y. (2010). Phosphorylation of the CPC by CDK1 promotes chromosome bi-orientation. Nature 467, 719–723. doi: 10.1038/nature09390
Ubersax, J. A., and Ferrell, J. E. Jr. (2007). Mechanisms of specificity in protein phosphorylation. Nat. Rev. Mol. Cell Biol. 8, 530–541. doi: 10.1038/nrm2203
Villa, F., Capasso, P., Tortorici, M., Forneris, F., de Marco, A., Mattevi, A., et al. (2009). Crystal structure of the catalytic domain of Haspin, an atypical kinase implicated in chromatin organization. Proc. Natl. Acad. Sci. U.S.A. 106, 20204–20209. doi: 10.1073/pnas.0908485106
von Schubert, C., Cubizolles, F., Bracher, J. M., Sliedrecht, T., Kops, G. J., and Nigg, E. A. (2015). Plk1 and Mps1 cooperatively regulate the spindle assembly checkpoint in human cells. Cell Rep. 12, 66–78. doi: 10.1016/j.celrep.2015.06.007
Wang, F., Dai, J., Daum, J. R., Niedzialkowska, E., Banerjee, B., Stukenberg, P. T., et al. (2010). Histone H3 Thr-3 phosphorylation by Haspin positions Aurora B at centromeres in mitosis. Science 330, 231–235. doi: 10.1126/science.1189435
Wang, W., Yang, Y., Gao, Y., Xu, Q., Wang, F., Zhu, S., et al. (2009). Structural and mechanistic insights into Mps1 kinase activation. J. Cell. Mol. Med. 13, 1679–1694. doi: 10.1111/j.1582-4934.2008.00605.x
Ward, G. E., and Kirschner, M. W. (1990). Identification of cell cycle-regulated phosphorylation sites on nuclear lamin C. Cell 61, 561–577. doi: 10.1016/0092-8674(90)90469-U
Watanabe, N., Arai, H., Iwasaki, J., Shiina, M., Ogata, K., Hunter, T., et al. (2005). Cyclin-dependent kinase (CDK) phosphorylation destabilizes somatic Wee1 via multiple pathways. Proc. Natl. Acad. Sci. U.S.A. 102, 11663–11668. doi: 10.1073/pnas.0500410102
Welburn, J. P., Tucker, J. A., Johnson, T., Lindert, L., Morgan, M., Willis, A., et al. (2007). How tyrosine 15 phosphorylation inhibits the activity of cyclin-dependent kinase 2-cyclin A. J. Biol. Chem. 282, 3173–3181. doi: 10.1074/jbc.M609151200
Xu, J., Shen, C., Wang, T., and Quan, J. (2013). Structural basis for the inhibition of Polo-like kinase 1. Nat. Struct. Mol. Biol. 20, 1047–1053. doi: 10.1038/nsmb.2623
Yamagishi, Y., Honda, T., Tanno, Y., and Watanabe, Y. (2010). Two histone marks establish the inner centromere and chromosome bi-orientation. Science 330, 239–243. doi: 10.1126/science.1194498
Ye, Q., Ur, S. N., Su, T. Y., and Corbett, K. D. (2016). Structure of the Saccharomyces cerevisiae Hrr25:Mam1 monopolin subcomplex reveals a novel kinase regulator. EMBO J. 35, 2139–2151. doi: 10.15252/embj.201694082
Keywords: mitotic kinase, mitosis, phosphorylation, structure, mechanism, substrate
Citation: Welburn JPI and Jeyaprakash AA (2018) Mechanisms of Mitotic Kinase Regulation: A Structural Perspective. Front. Cell Dev. Biol. 6:6. doi: 10.3389/fcell.2018.00006
Received: 31 October 2017; Accepted: 19 January 2018;
Published: 05 February 2018.
Edited by:
Sabine Elowe, Laval University, CanadaReviewed by:
Darja Lavogina, University of Tartu, EstoniaVictor M. Bolanos-Garcia, Oxford Brookes University, United Kingdom
Copyright © 2018 Welburn and Jeyaprakash. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Julie P. I. Welburn, anVsaWUud2VsYnVybkBlZC5hYy51aw==
A. Arockia Jeyaprakash, amV5YXByYWthc2guYXJ1bGFuYW5kYW1AZWQuYWMudWs=
 Julie P. I. Welburn
Julie P. I. Welburn A. Arockia Jeyaprakash
A. Arockia Jeyaprakash