- Departments of Genetic Medicine and Pediatrics, The Johns Hopkins University, Baltimore, MD, United States
Mammals compensate for sex differences in the number of X chromosomes by inactivating all but one X chromosome. Although they differ in the details of X inactivation, all mammals use long non-coding RNAs in the silencing process. By transcribing XIST RNA, the human inactive X chromosome has a prime role in X-dosage compensation. Yet, the autosomes also play an important role in the process. Multiple genes on human chromosome 1 interact with XIST RNA to silence the future inactive Xs. Also, it is likely that multiple genes on human chromosome 19 prevent the silencing of the single active X – a highly dosage sensitive process. Previous studies of the organization of chromosomes in the nucleus and their genomic interactions indicate that most contacts are intra-chromosomal. Co-ordinate transcription and dosage regulation can be achieved by clustering of genes and mingling of interacting chromosomes in 3D space. Unlike the genes on chromosome 1, those within the critical eight MB region of chromosome 19, have remained together in all mammals assayed, except rodents, indicating that their proximity in non-rodent mammals is evolutionarily conserved. I propose that the autosomal genes that play key roles in the process of X inactivation are non-randomly distributed in the genome and that this arrangement facilitates their coordinate regulation.
When female mammals compensate for sex differences in the dosage of X linked genes by inactivating X chromosomes, the X chromosome(s) that is silenced has a major role in the process. In all mammals, a non-coding RNA, encoded by the X, is essential to its being inactivated by epigenetic factors (Grant et al., 2012). Clearly, the bi-directional spread of Xist RNA from its locus in the middle of the X chromosome initiates the inactivation process in eutherian mammals (Brockdorff et al., 1992; Brown et al., 1992). In addition, the other long non coding RNAs, implicated in the process, i.e., the potential Xist repressors, rodent-specific Tsix (Lee and Lu, 1999), and the primate specific XACT (Vallot et al., 2017), are also encoded by the X chromosome. Once coated with enough Xist RNA, the future inactive X moves toward the nuclear lamina, where its chromatin is transformed from euchromatin to heterochromatin (McHugh et al., 2015; Moindrot and Brockdorff, 2016).
The silencing of the future inactive X, or Xs, is attributable to a Rube-Goldberg type of mechanism that not only brings it close to the nuclear periphery (where inactive chromatin tends to reside), but also attracts the epigenetic factors that silence it. Ultimately, the binding of Xist RNA results in expulsion of factors from the inactive X that make chromatin accessible for transcription (Jegu et al., 2019). The few active (escape) genes on that X chromosome manage to find their way out of the heterochromatic mass of inactive chromatin towards the center of the nucleus, where transcription occurs (Fraser and Bickmore, 2007). Yet, Xist RNA cannot do this alone, as autosomal gene products are essential to complete the silencing process (McHugh et al., 2015; Moindrot and Brockdorff, 2016; Patil et al., 2016).
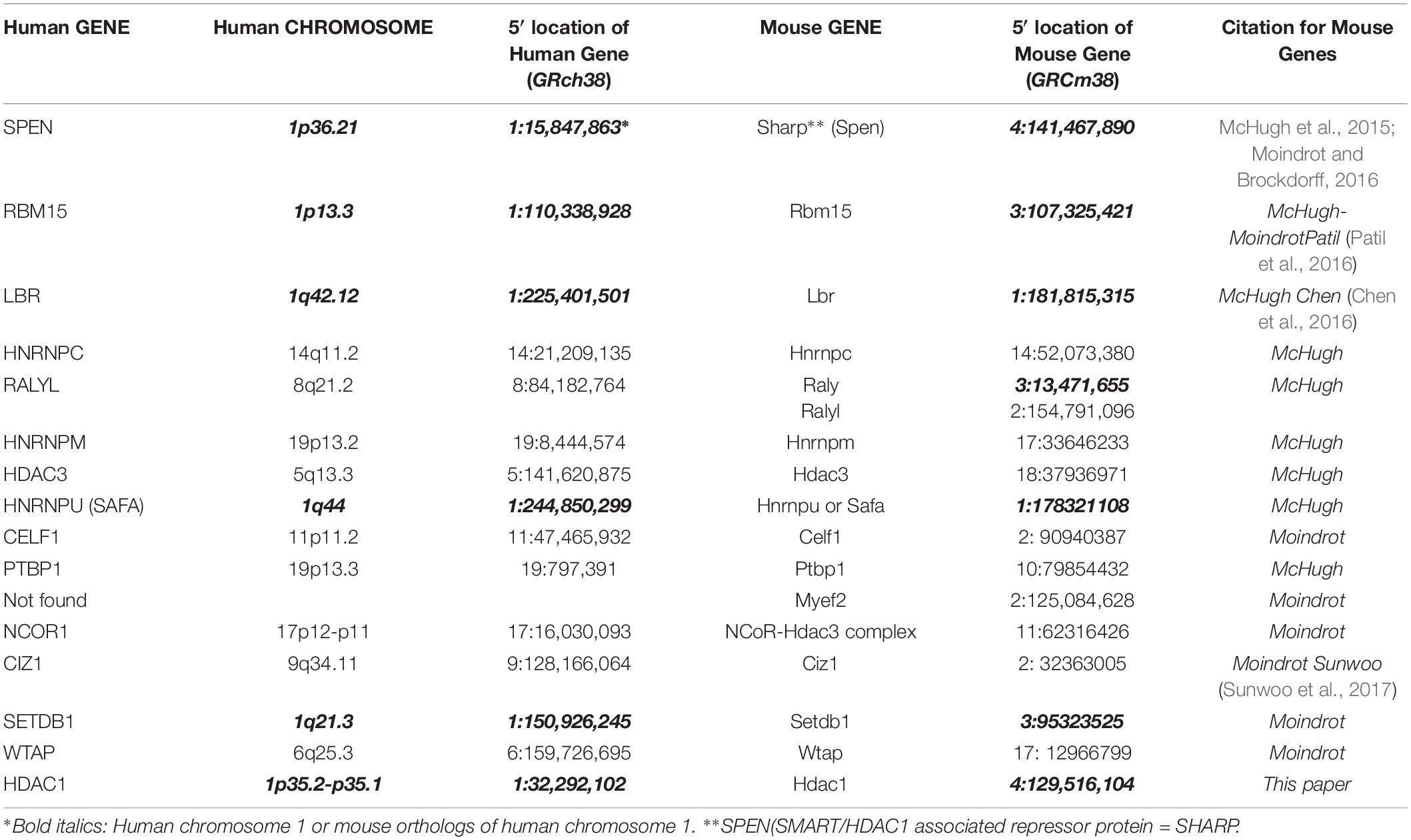
In pursuit of autosomal genes that cooperate with the X chromosome, Percec et al. (2003) used ENU chemical mutagenesis to screen for autosomal mutations involved in the initiation of X inactivation in mice. They identified regions of mouse chromosomes 5, 10, and 15, which seemed to affect the choice of the mouse inactive X. More recent studies in mice have elucidated the essential autosomal products that interact with Xist RNA to silence the chromosome (McHugh et al., 2015; Chen et al., 2016; Moindrot and Brockdorff, 2016; Sunwoo et al., 2017) (Table 1). These include the lamin B receptor (Lbr), the satellite attachment factor A (Saf-A) and Sharp (Smrt and Hdac Associated Repressor Protein, also called Spen). SPEN, LBR, and SAFA map to human chromosome 1; Lbr and Safa also map to mouse chromosome 1, whereas Sharp is on mouse chromosome 4 (orthologous to human chromosome 1). Other genes that have been implicated in the silencing process are RBM 15 and SETDB1, on human chromosome 1, and mouse chromosome 3 – also orthologous to human chromosome 1. Therefore, the genes on human chromosome 1 that play a role in silencing the future inactive X also map to mouse chromosome 1 or its orthologs (Table 1 and Figure 1A). Conceivably, genes that were on three different chromosomes in mice have evolved to be on a single human chromosome to facilitate their interaction in silencing the X.
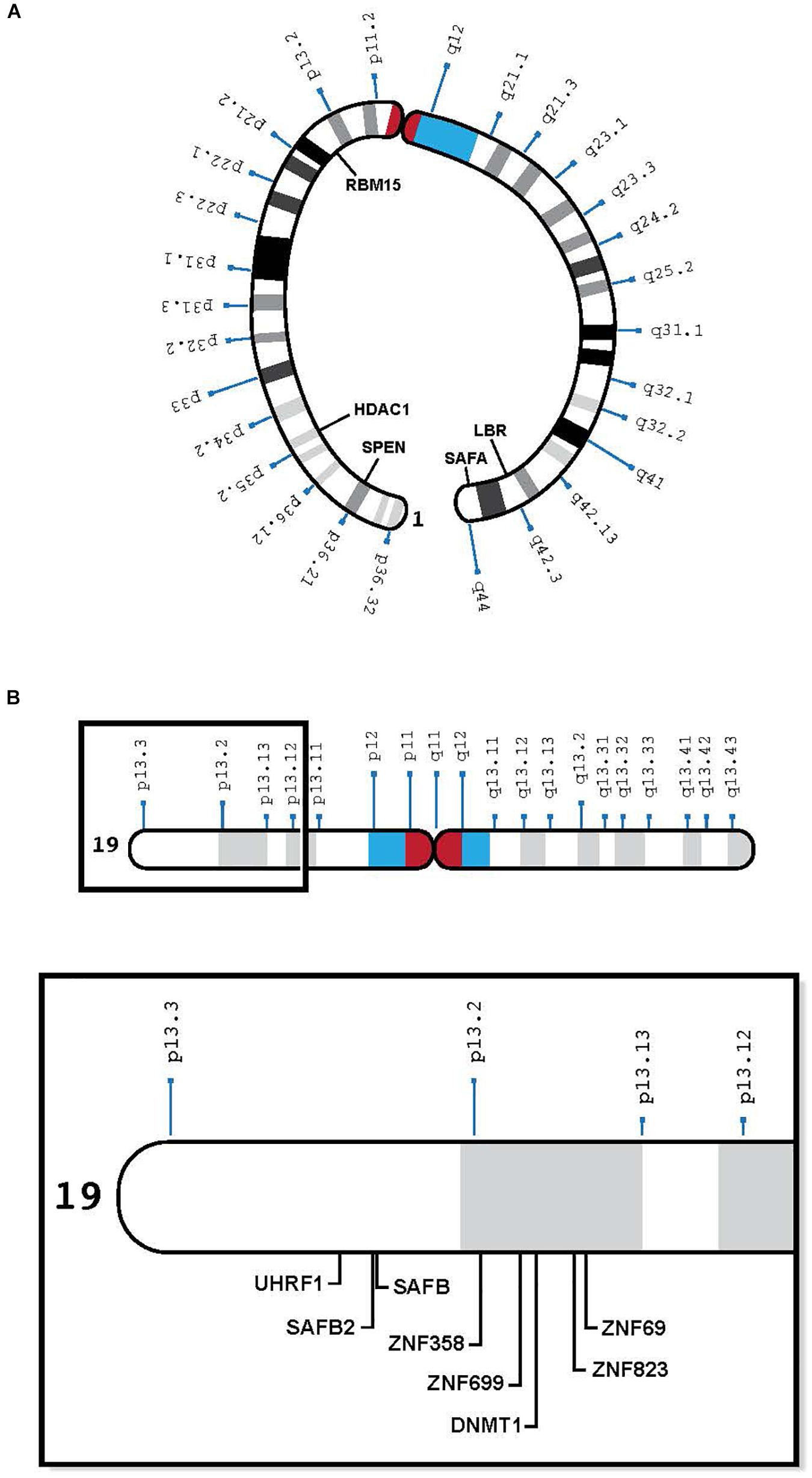
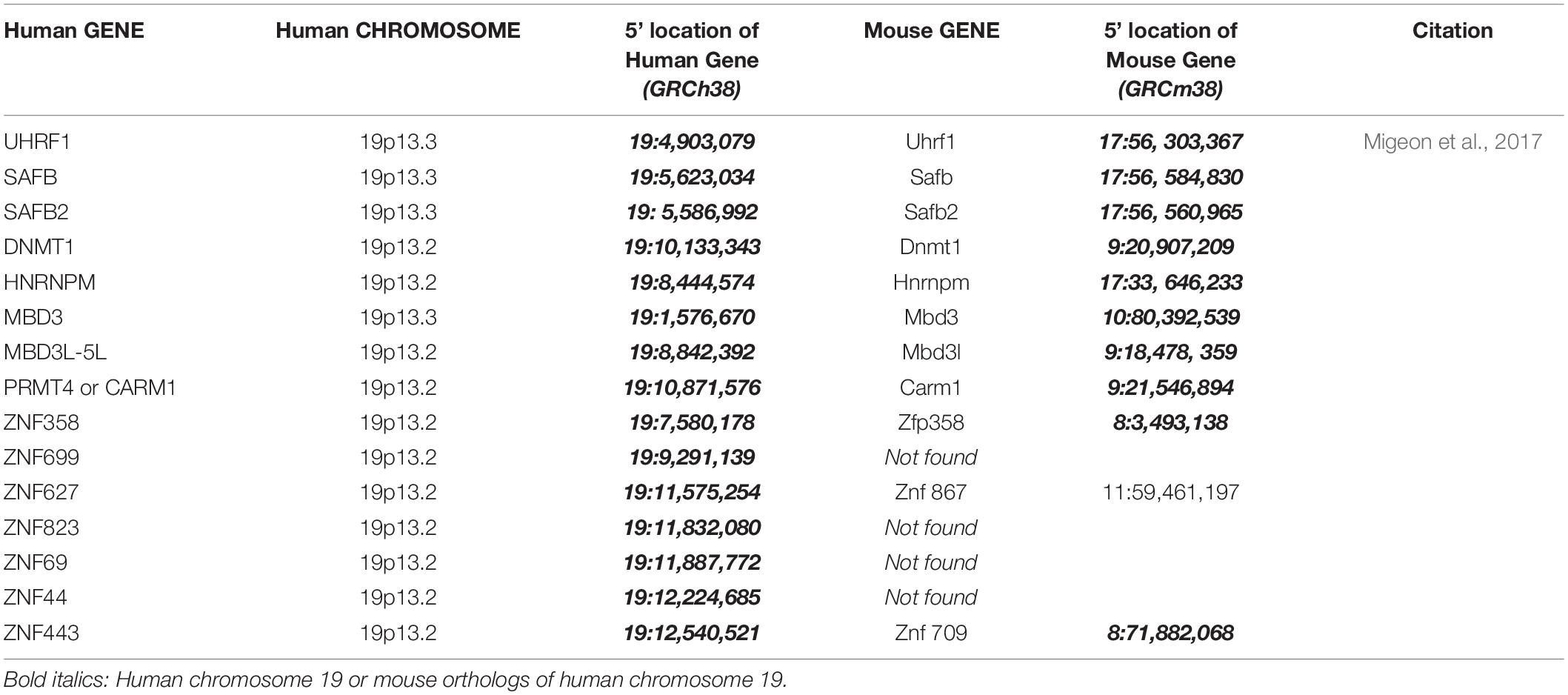
Figure 1. (A) Human chromosome 1 with relevant genes, bent to show telomeres in proximity: SPEN 1p36.2, HDAC1 1p35.2, RBM15 1p13.3, LBR 1q42.12, and SAFA 1q44 (see Table 1). (B) Human chromosome 19 insert with relevant genes, showing proximity of genes in 19p13.1-13.2: UHRF1, SAFB2, SAFB, ZNF 358, ZNF 699, DNMT1, ZNF 823, and ZNF 69 (see Table 2).
In prokaryotes, interactions between genes with a common function are facilitated because such genes are contiguous in the genome, organized into operons, with a common promoter (Jacob, 2011). On the other hand, most eukaryotic genes that interact with each other, do not share promotors, and are less well clustered (Dekker and Misteli, 2015). Yet, it has become apparent that the spatial arrangement of genes in the mammalian nucleus is non-random; chromosome folding and intermingling enable the proximity of genes that reside on the same chromosome, by looping, and even on different chromosomes, by chromosome clustering. The likely advantage of interactions between genes is coordination of their expression – perhaps in the same transcription factory, thought to occur in a discrete nuclear region (Rieder et al., 2014).
Based on HI-C studies of the human genome (Lieberman-Aiden et al., 2009), Thevenin et al. (2014) showed that a significant number of functional groups (pairs of interacting proteins, genes with common functions and those in interactive pathways) are either clustered within the same chromosome or dispersed over a relatively few chromosomes. Those on different chromosomes tend to co-localize in space. These investigators found that, genes, which function together, tend to reside on fewer chromosomes than expected by chance. On the same chromosome, they are closer to each other than randomly chosen genes; on different chromosomes, they tend to be closer to each other in 3D space (Thevenin et al., 2014). Among the best documented inter-chromosomal interactions are those between the mouse X chromosomal gene, Xist, and the autosomal epigenetic factors mentioned above, that help silence the X chromosome from which the up-regulated Xist locus is being transcribed (Dekker and Misteli, 2015).
When extending her observations in mice to other mammals, Lyon suggested there was only a single active X, no matter the number of X’s in a cell (Lyon, 1962); however, the literature has persisted in labeling the mammalian process of X dosage compensation, X inactivation, which focuses us on the process of silencing the inactive X. Therefore, the salient question has been, “How does one choose the X chromosome that becomes inactive?” Because Xist RNA is able to silence any chromosome into which it is inserted (Jiang et al., 2013; Migeon et al., 1999), it is surprising that few ask the pertinent question, “What protects the single active X from silencing by its own Xist locus?” (Migeon, 2017; Migeon et al., 2017).
Further, it has not been easy to show how the mouse inactive X is chosen. Earlier studies suggested that an infrequent physical association (kissing) between the Xist loci of the two X chromosomes in mouse embryos determined the choice of inactive X (Xu et al., 2006; Augui et al., 2007), but more recent studies indicate that neither the expression of Xist nor Tsix, its antisense RNA, is affected by the interaction (Cheng et al., 2019; Pollex and Heard, 2019).
In addition, Inoue et al. (2018) and Harris et al. (2019) recently showed that in mice, the choice of active X is determined prenatally. Having been imprinted during oocyte differentiation [as predicted by Lyon and Rastan (1984)], the active X is always maternal in trophectoderm – the first tissue to undergo dosage compensation in the mouse embryo. Because X inactivation in the placenta occurs relatively early in mice, it is likely that the paternal X hasn’t had time to erase the inactivation imprint imposed during the early stages of spermatogenesis (Migeon, 2016). It remains to be seen if the rodent specific Tsix RNA, which is transcribed only from the maternal X in trophectoderm, protects the active X, regardless of its parental origin, from silencing by Xist in other mouse embryonic tissues.
With respect to human cells, we have learned that (1) human oocytes do not express PRC2 (which imprints the mouse oocyte) (Harris et al., 2019), (2) the human maternal X is not imprinted (Migeon, 2016), and (3) human TSIX is ineffective, having been truncated during human evolution (Migeon et al., 2001). Therefore, another means of repressing the XIST locus on the future active human X is needed to protect it from being silenced. Recent studies suggest that to prevent its heterochromatization by XIST, the future human active X needs to interact with human chromosome 19 (Migeon et al., 2017). They reveal a previously unsuspected eight MB region on the short arm of human chromosome 19 (19p13.3-13.2), which contains at least one dosage sensitive gene that is likely to play a role in silencing the XIST locus on one X chromosome in each cell (Migeon, 2017; Migeon et al., 2017) (Table 2). Candidate genes include satellite attachment factors SAFB and SAFB2, a cluster of zinc finger proteins that surround DNMT1 and its co-factor UHRF1, among many others. Although most of the zinc finger proteins clustered in the relevant region of human chromosome 19 arose after the split between rodents and humans, the other genes in this region can be found on mouse chromosomes 8, 9, and 17 – orthologous to human chromosome 19 (Table 2 and Figure 1B). Again, perhaps human 19 evolved to facilitate the interaction of genes that protect the future active X.
In the genomics era, many human geneticists tend not to specify which particular autosome encodes genes of interest; therefore, I was surprised to see that many of the proteins that interact with XIST to silence the X are encoded by human chromosome 1 (Migeon et al., 2017) (Table 1 and Figure 1A), and in the mouse, by the three orthologs of chromosome 1 (chromosomes 1, 3, and 4) (Table 1). In mice, these genes are bound to Xist at the same developmental stage (McHugh et al., 2015). To my knowledge, no one has examined the Xist-autosomal interactions by RNA FISH to determine if there is clustering of the three murine chromosome 1 orthologs. The positions of these genes on human chromosome 1 is of interest as some of the genes are present on opposing ends of the chromosome, which would require a large fold in the chromosome to facilitate any interaction (Figure 1A). Such intermingling and folding are frequently observed in the 3D nuclear space (Lieberman-Aiden et al., 2009).
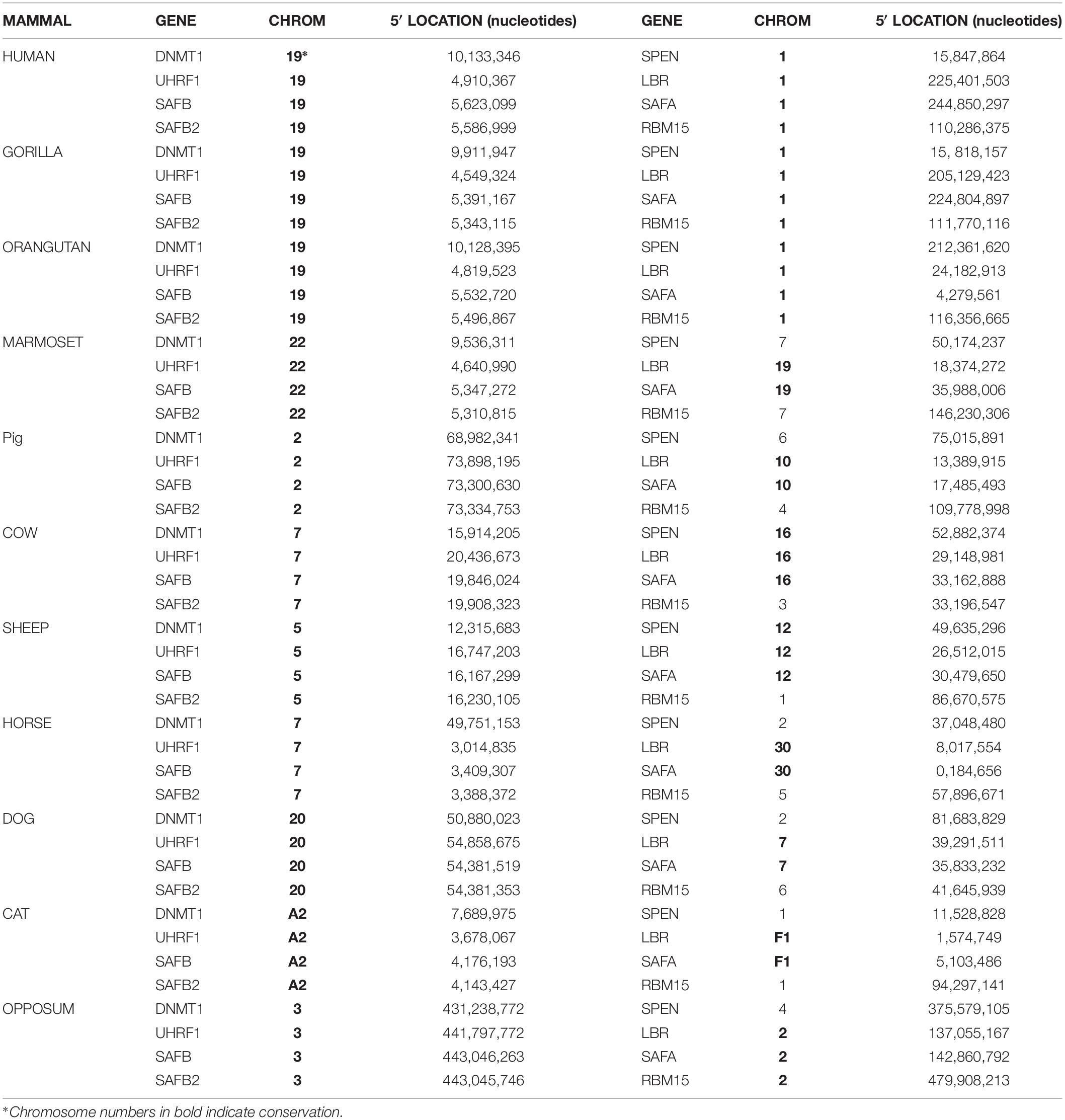
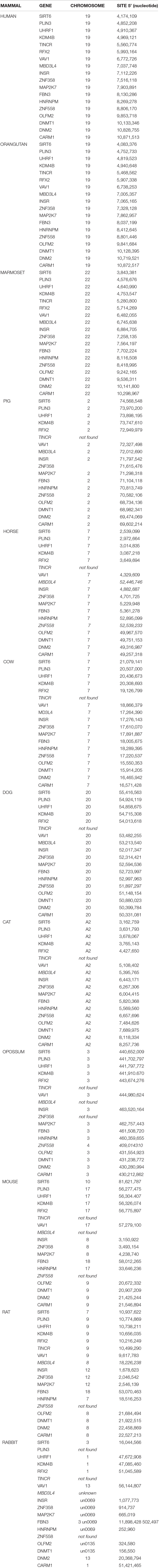
Table 3 presents conservation data obtained from the UCSC Genome Browser; it shows that of four relevant genes on chromosome 1 that aid Xist in silencing the inactive X, only SAFA and LBR have been on the chromosome since we evolved from marsupials. SPEN and RBM15 although on the same chromosome as SAFA and LBR in primates, are on other chromosomes in marmosets and non-primate mammals. In contrast, except in rodents (rat, mouse, and rabbit), the region on chromosome 19 that protects the active X is preserved in primates such as gorilla, orangutang, and marmoset, and other mammals such as cat, dog, pig, horse, cow, and opposum (Table 4). The exceptional genes that have left the group include the long noncoding RNA, TINCR, and the MD3L3-5, methyl CPG binding domain proteins, which are on chromosome 19 in primates and in marmoset but are not found in all mammals. The conserved cluster in pig, horse and cow is in the reverse orientation (Table 4). These differences interrupt what would otherwise be an exceptionally long synteny block, but the preservation of so many genes in this region, in spite of multiple evolutionary structural alterations, suggests that the local landscape may be important to function. That the chromosome 19 genes in rodents are not conserved as a group argues that their process of ensuring that one X will remain active differs from that of other mammals (Shevchenko et al., 2019), perhaps because only rodents have Tsix to protect the active X from silencing by Xist.
Most likely, the relevant genes on the same chromosome are co-regulated. The advantage of genes clustered in interphase is that they can be programmed for simultaneous transcription. To silence XIST on the future active X, some genes in the chromosome 19 cluster might be transcribed together, perhaps if they are close enough in 3D space, as a single transcript. The telomeric location of genes on primate chromosome 1 that participate in XIST silencing (Figure 1A) suggest that the two ends of the chromosome might physically interact at the time of transcription.
Several important questions remain unanswered: First, how do multiple genes in the inactivation pathway on human chromosome 1 (or in the activation pathway on chromosome 19) coordinately interact with each other? And then, how do autosomal genes encoding protein products, interact with the X chromosome?
Recent studies suggest that the intra-chromosomal gene interactions occur within the same topologically-associating-domain (TAD) (Nora et al., 2012; Galupa and Heard, 2018) and that TADS align with co-coordinately regulated gene clusters, fostering long-range contacts and preventing deleterious interactions between genes in different TADs (Galupa and Heard, 2018) One would like to examine the candidate genes on human chromosomes 1 and 19, at the appropriate time in development, to determine if they are located within the same TAD, or are otherwise coordinately regulated. It is unlikely that the occurrence of multiple silencers of the inactive X on human chromosome 1 and XIST repressors on human chromosome 19 is coincidental.
The question of how genes on an autosome interact with the genes on the X chromosome is especially challenging because in the human species either one or several X chromosomes can be silenced within a cell, the number dependent upon the number of X chromosomes in the genome. All but one X chromosome are silenced no matter how many are in the cell, nor the sex of the individual (Grumbach et al., 1963). Therefore, only one X chromosome resists silencing no matter the number of X chromosomes in the cell.
Clearly, suppressing the XIST locus on the future active X is easier for males than females. We know this because of the specific loss of females who reduplicate the essential chromosome 19 gene(s), presumably because reduplication enables both X’s to be active – a known lethal event in diploid cells. At least five percent more pre-implantation human females are miscarried than are males (Migeon et al., 2017). If males reduplicate the XIST repressor, it has little consequence, but females who by chance inactivate both XIST loci, die before they implant into the uterus. This suggests that not only when this region of chromosome 19 is duplicated, but even, when the chromosome is normal, the required interaction is a difficult one, as either too little or too much XIST repressor would lead to a lethal event (too many active X’s or no active X). The former does not occur as often in males who have only one X chromosome: too much repressor is not lethal, although too little might be.
And there is the question of gene dosage. How in a diploid cell do two autosomes cooperate to make an inhibitor for a single X chromosome? In the case of more than two X chromosomes, how is the right dosage of gene product from chromosome 1 achieved? On one hand Lyon (1971) and more recently Nguyen et al. (2019) suggest that the two autosomes might pair to synthesize a single product. One such product might be a dimeric protein, there is also the possibility of competitive inhibition. Once, a molecule of gene product arrives on one X chromosome then the other(s) are unable to be hit. On the other hand, perhaps, not all attempts to activate or inactivate the chromosome are successful, and so the process is stochastic. That many errors occur while repressing XIST on the future active X might explain a significant loss of pre-implantation females, even in absence of gene reduplication.
To answer these questions one needs to identify genome interactions during the pre-implantation development of the human embryo, at the time of X inactivation. One can use chromosome capture such as Hi-C, 3D RNA-FISH (Shiura and Abe, 2019) (to see if nascent transcripts are transcribed together). Single-cell RNA-Seq as has been recently described in the mouse (Cheng et al., 2019), examining the candidate genes. The best human model would be the beginning of cleavage to embryonic day 10. The inability to study available human embryos is a decided disadvantage for American investigators, but I hope that my colleagues in other countries will carry out such studies. For the human X: 19 interaction, embryonic day 4–7 would probably be appropriate, whereas human embryonic day 6–9 should capture the chromosome 1: X interaction.
Author Contributions
BM conceived the study, obtained the data, and wrote the manuscript.
Conflict of Interest Statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The author is most grateful to Drs. Hans Bjornsonn and Teresa Luperchio for incredibly insightful discussions, and to Drs. Haig Kazazian and Roger Reeves for their helpful comments about the manuscript. The author is deeply indebted to Dr. Sarah Wheelan for her contribution to the gene conservation analysis. The author appreciates the suggestions of the reviewers.
References
Augui, S., Filion, G. J., Huart, S., Nora, E., Guggiari, M., Maresca, M., et al. (2007). Sensing X chromosome pairs before X inactivation via a novel X-pairing region of the Xic. Science 318, 1632–1636. doi: 10.1126/science.1149420
Brockdorff, N., Ashworth, A., Kay, G., McCabe, V., Norris, D. P., Cooper, P., et al. (1992). The product of the mouse Xist gene is a 15 kb inactive X-specific transcript containing no conserved ORF and located in the nucleus. Cell 71, 515–526. doi: 10.1016/0092-8674(92)90519-i
Brown, C. J., Hendrich, B. D., Rupert, J. L., Lafreniere, R. G., Xing, Y., Lawrence, J., et al. (1992). The human XIST gene: analysis of a 17 kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell 71, 527–542. doi: 10.1016/0092-8674(92)90520-m
Chen, C. K., Blanco, M., Jackson, C., Aznauryan, E., Ollikainen, N., Surka, C., et al. (2016). Xist recruits the X chromosome to the nuclear lamina to enable chromosome-wide silencing. Science 354, 468–472. doi: 10.1126/science.aae0047
Cheng, S., Pei, Y., He, L., Peng, G., Reinius, B., Tam, P. P. L., et al. (2019). Single-cell RNA-seq reveals cellular heterogeneity of pluripotency transition and x chromosome dynamics during early mouse development. Cell Rep. 26, 2593–2607.e3. doi: 10.1016/j.celrep.2019.02.031
Dekker, J., and Misteli, T. (2015). Long-range chromatin interactions. Cold Spring Harb. Perspect. Biol. 7:a019356. doi: 10.1101/cshperspect.a019356
Fraser, P., and Bickmore, W. (2007). Nuclear organization of the genome and the potential for gene regulation. Nature 447, 413–417. doi: 10.1038/nature05916
Galupa, R., and Heard, E. (2018). X-chromosome inactivation: a crossroads between chromosome architecture and gene regulation. Annu. Rev. Genet. 52, 535–566. doi: 10.1146/annurev-genet-120116-024611
Grant, J., Mahadevaiah, S. K., Khil, P., Sangrithi, M. N., Royo, H., Duckworth, J., et al. (2012). Rsx is a metatherian RNA with Xist-like properties in X-chromosome inactivation. Nature 487, 254–258. doi: 10.1038/nature11171
Grumbach, M. M., Morishima, A., and Taylor, J. H. (1963). Human sex chromosome abnormalities in relation to DNA replication and heterochromatization. Proc. Natl. Acad. Sci. U.S.A. 69, 3138–3141.
Harris, C., Cloutier, M., Trotter, M., Hinten, M., Gayen, S., Du, Z., et al. (2019). Conversion of random X-inactivation to imprinted X-inactivation by maternal PRC2. eLife 8:e44258. doi: 10.7554/eLife.44258
Inoue, A., Chen, Z., Yin, Q., and Zhang, Y. (2018). Maternal Eed knockout causes loss of H3K27me3 imprinting and random X inactivation in the extraembryonic cells. Genes Dev. 32, 1525–1536. doi: 10.1101/gad.318675.118
Jegu, T., Blum, R., Cochrane, J. C., Yang, L., Wang, C. Y., Gilles, M. E., et al. (2019). Xist RNA antagonizes the SWI/SNF chromatin remodeler BRG1 on the inactive X chromosome. Nat. Struct. Mol. Biol. 26, 96–109. doi: 10.1038/s41594-018-0176-8
Jiang, J., Jing, Y., Cost, G. J., Chiang, J. C., Kolpa, H. J., Cotton, A. M., et al. (2013). Translating dosage compensation to trisomy 21. Nature 500, 296–300. doi: 10.1038/nature12394
Lee, J. T., and Lu, N. (1999). Targeted mutagenesis of Tsix leads to nonrandom inactivation. Cell 99, 47–57. doi: 10.1016/s0092-8674(00)80061-6
Lieberman-Aiden, E., van Berkum, N. L., Williams, L., Imakaev, M., Ragoczy, T., Telling, A., et al. (2009). Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326, 289–293. doi: 10.1126/science.1181369
Lyon, M. F. (1971). Possible mechanisms of X chromosome inactivation. Nat. New Biol. 232, 229–232. doi: 10.1038/newbio232229a0
Lyon, M. F. (1962). Sex chromatin and gene action in the mammalian X-chromosome. Am. J. Hum. Genet. 1962, 135–148.
Lyon, M. F., and Rastan, S. (1984). Paternal source of chromosome imprinting and its relevance for X chromosome inactivation. Differentiation 26, 63–67. doi: 10.1111/j.1432-0436.1984.tb01375.x
McHugh, C. A., Chen, C. K., Chow, A., Surka, C. F., Tran, C., McDonel, P., et al. (2015). The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature 521, 232–236. doi: 10.1038/nature14443
Migeon, B. R. (2016). An overview of X inactivation based on species differences. Semin. Cell Dev. Biol. 56, 111–116. doi: 10.1016/j.semcdb.2016.01.024
Migeon, B. R. (2017). Choosing the active X: the human version of X inactivation. Trends Genet. 33, 899–909. doi: 10.1016/j.tig.2017.09.005
Migeon, B. R., Beer, M. A., and Bjornsson, H. T. (2017). Embryonic loss of human females with partial trisomy 19 identifies region critcal for the single active X. PLoS One 12:e0170403. doi: 10.1371/journal.pone.0170403
Migeon, B. R., Chowdhury, A. K., Dunston, J. A., and McIntosh, I. (2001). Identification of TSIX encoding an RNA antisense to human XIST, reveals differences from its murine counterpart: implications for X inactivation. Am. J. Hum. Genet. 69, 951–960. doi: 10.1086/324022
Migeon, B. R., Kazi, E., Haisley-Royster, C., Hu, J., Reeves, R. H., Call, L., et al. (1999). Human X inactivation center induces random X inactivation in male transgenic mice. Genomics 59, 113–121. doi: 10.1006/geno.1999.5861
Moindrot, B., and Brockdorff, N. (2016). RNA binding proteins implicated in Xist-mediated chromosome silencing. Semin. Cell Dev. Biol. 56, 58–70. doi: 10.1016/j.semcdb.2016.01.029
Nora, E. P., Lajoie, B. R., Schulz, E. G., Giorgetti, L., Okamoto, I., Servant, N., et al. (2012). Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature 485, 381–385. doi: 10.1038/nature11049
Patil, D. P., Chen, C. K., Pickering, B. F., Chow, A., Jackson, C., Guttman, M., et al. (2016). m(6)A RNA methylation promotes XIST-mediated transcriptional repression. Nature 537, 369–373. doi: 10.1038/nature19342
Percec, I., Thorvaldsen, J. L., Plenge, R. M., Krapp, C. J., Nadeau, J. H., Willard, H. F., et al. (2003). An N-Ethyl-N-Nitrosourea mutagenesis screen for epigenetic mutations in the mouse. Genetics 164, 1481–1494.
Pollex, T., and Heard, E. (2019). Nuclear positioning and pairing of X-chromosome inactivation centers are not primary determinants during initiation of random X-inactivation. Nat. Genet. 51, 285–295. doi: 10.1038/s41588-018-0305-7
Rieder, D., Ploner, C., Krogsdam, A. M., Stocker, G., Fischer, M., Scheideler, M., et al. (2014). Co-expressed genes prepositioned in spatial neighborhoods stochastically associate with SC35 speckles and RNA polymerase II factories. Cell Mol. Life Sci. 71, 1741–1759. doi: 10.1007/s00018-013-1465-3
Shevchenko, A. I., Dementyeva, E. V., Zakharova, I. S., and Zakian, S. M. (2019). Diverse developmental strategies of X chromosome dosage compensation in eutherian mammals. Int. J. Dev. Biol. 63, 223–233. doi: 10.1387/ijdb.180376as
Shiura, H., and Abe, K. (2019). Xist/Tsix expression dynamics during mouse peri-implantation development revealed by whole-mount 3D RNA-FISH. Sci. Rep. 9:3637. doi: 10.1038/s41598-019-38807-0
Sunwoo, H., Colognori, D., Froberg, J. E., Jeon, Y., and Lee, J. T. (2017). Repeat E anchors Xist RNA to the inactive X chromosomal compartment through CDKN1A-interacting protein (CIZ1). Proc. Natl. Acad. Sci. U.S.A. 114, 10654–10659. doi: 10.1073/pnas.1711206114
Thevenin, A., Ein-Dor, L., Ozery-Flato, M., and Shamir, R. (2014). Functional gene groups are concentrated within chromosomes, among chromosomes and in the nuclear space of the human genome. Nucleic Acids Res. 42, 9854–9861. doi: 10.1093/nar/gku667
Vallot, C., Patrat, C., Collier, A. J., Huret, C., Casanova, M., Liyakat Ali, T. M., et al. (2017). XACT noncoding RNA competes with XIST in the control of X chromosome activity during human early development. Cell Stem Cell 20, 102–111. doi: 10.1016/j.stem.2016.10.014
Keywords: single active X, intra-chromosomal interaction, inter-chromosomal interaction, X-chromosome dosage compensation, autosomes in X inactivation, evolutionary conservation, clustered gene interactions
Citation: Migeon BR (2019) The Non-random Location of Autosomal Genes That Participate in X Inactivation. Front. Cell Dev. Biol. 7:144. doi: 10.3389/fcell.2019.00144
Received: 30 May 2019; Accepted: 11 July 2019;
Published: 06 August 2019.
Edited by:
Bernhard Payer, Centre for Genomic Regulation (CRG), SpainReviewed by:
Sha Sun, University of California, Irvine, United StatesMichael Cowley, North Carolina State University, United States
Copyright © 2019 Migeon. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Barbara R. Migeon, Ym1pZ2VvbkBqaG1pLmVkdQ==
 Barbara R. Migeon
Barbara R. Migeon