- 1College of Pharmaceutical Sciences, Zhejiang Chinese Medical University, Hangzhou, China
- 2Institute of Cancer and Basic Medicine, Chinese Academy of Sciences, Cancer Hospital of the University of Chinese Academy of Sciences, Zhejiang Cancer Hospital, Hangzhou, China
- 3School of Pharmacy, Naval Medical University, Shanghai, China
- 4Institute of Interdisciplinary Integrative Medicine Research, Shanghai University of Traditional Chinese Medicine, Shanghai, China
The p53 tumor suppressor protein and its major negative regulators MDM2 and MDMX oncoproteins form the MDM2/MDMX-p53 circuitry, which plays critical roles in regulating cancer cell growth, proliferation, cell cycle progression, apoptosis, senescence, angiogenesis, and immune response. Recent studies have shown that the stabilities of p53, MDM2, and MDMX are tightly controlled by the ubiquitin-proteasome system. Ubiquitin specific protease 7 (USP7), one of the most studied deubiquitinating enzymes plays a crucial role in protecting MDM2 and MDMX from ubiquitination-mediated proteasomal degradation. USP7 is overexpressed in human cancers and contributes to cancer initiation and progression. USP7 inhibition promotes the degradation of MDM2 and MDMX, activates the p53 signaling, and causes cell cycle arrest and apoptosis, making USP7 a potential target for cancer therapy. Several small-molecule inhibitors of USP7 have been developed and shown promising efficacy in preclinical settings. In the present review, we focus on recent advances in the understanding of the USP7-MDM2/MDMX-p53 network in human cancers as well as the discovery and development of USP7 inhibitors for cancer therapy.
Introduction
Ubiquitination refers to the cellular process by which ubiquitin specifically modifies a target protein under the cascade catalysis of the E1 ubiquitin-activating enzymes, the E2 ubiquitin-conjugating enzymes, and the E3 ubiquitin ligases (Zheng and Shabek, 2017; Senft et al., 2018). The polyubiquitinated proteins are further recognized and rapidly degraded into small peptides by the 26S proteasome (Zheng and Shabek, 2017; Senft et al., 2018). Protein ubiquitination is a common post-translational modification and plays a critical role in many cellular processes, such as apoptosis, cell cycle progression, DNA damage response, and membrane transport by causing the degradation of various regulatory proteins (Senft et al., 2018). Conversely, deubiquitination is the process that removes ubiquitin from the target proteins by the deubiquitinases (DUBs), which can specifically disassemble the ubiquitin-protein conjugates by cleaving the isopeptide bonds formed between the C-terminus of ubiquitin and the target proteins and protect them from degradation and inactivation (Harrigan et al., 2018; Rawat et al., 2019). Therefore, the ubiquitination-deubiquitination balance acts as a critical and powerful way to regulate protein turnover and functions and has been demonstrated as a promising target for developing therapeutic agents for several diseases, including cancer.
Ubiquitin-specific protease 7 (USP7), also named as herpesvirus-associated ubiquitin-specific protease (HAUSP) is one of the most investigated DUBs and has been associated with the initiation and progression of various cancers (Masuya et al., 2006; Su et al., 2018). USP7 has a broad range of substrates, such as the tumor suppressors p53 (Li et al., 2002) and PTEN (Phosphatase and tensin homolog) (Song et al., 2008) and the transcription factor FOXO4 (Forkhead box protein O4) (van der Horst et al., 2006), most of which are related to tumor suppression, DNA repair, and immune response (Pozhidaeva and Bezsonova, 2019). USP7 is also capable of deubiquitinating and stabilizing MDM2 (Murine double minute 2) and MDMX (Murine double minute X, also known as MDM4) oncoproteins, which are the major negative regulators of the p53 tumor suppressor (Sheng et al., 2006; Sarkari et al., 2010). Herein, targeting USP7 for destabilizing MDM2/MDMX and activating p53 has recently been proposed as a promising strategy for cancer therapy, and several USP7 inhibitors have been developed and shown promising efficacy in preclinical cancer models in vitro and in vivo (Zhou et al., 2018).
Due to the multifaceted nature of USP7 in various diseases, its structure, enzymatic activity, substrates, regulation, disease relevance, and inhibitors have been extensively reviewed recently (Bhattacharya et al., 2018; Wu et al., 2018; Zhou et al., 2018; Pozhidaeva and Bezsonova, 2019; Rawat et al., 2019). In the present review, we focus on the USP7-MDM2/MDMX-p53 network in human cancers as well as the current targeting strategies and small-molecule inhibitors. We also propose new targeting approaches that may lead to more specific and effective inhibitors for treating human cancers.
The USP7-MDM2/MDMX-p53 Network
The Protein Structures of USP7, MDM2, MDMX, and p53
The human USP7 gene is located on chromosome 16p13.2 and was initially identified as a new member of the ubiquitin-specific protease (USP) family in 1997 (Everett et al., 1997). The USP7 protein consists of 1102 amino acids that are distributed in three major domains, including the N-terminal tumor necrosis factor receptor-associated factor (TRAF) domain (amino acids 53–206), a central catalytic domain (amino acids 208–560), and the C-terminal tandem ubiquitin-like (Ubl) domain (UBL1-5, amino acids 560–1102) (Figure 1A). Among these domains, TRAF is critical for the binding of USP7 to its substrates, including MDM2, MDMX, and p53 via P/AxxS motifs (Hu et al., 2002, 2006; Saridakis et al., 2005; Sheng et al., 2006; Sarkari et al., 2010; Rouge et al., 2016). It has also been found that the nuclear localization of USP7 is partially dependent on the TRAF domain through the production of USP7 domain deletion mutants (Fernandez-Montalvan et al., 2007; Tavana and Gu, 2017).
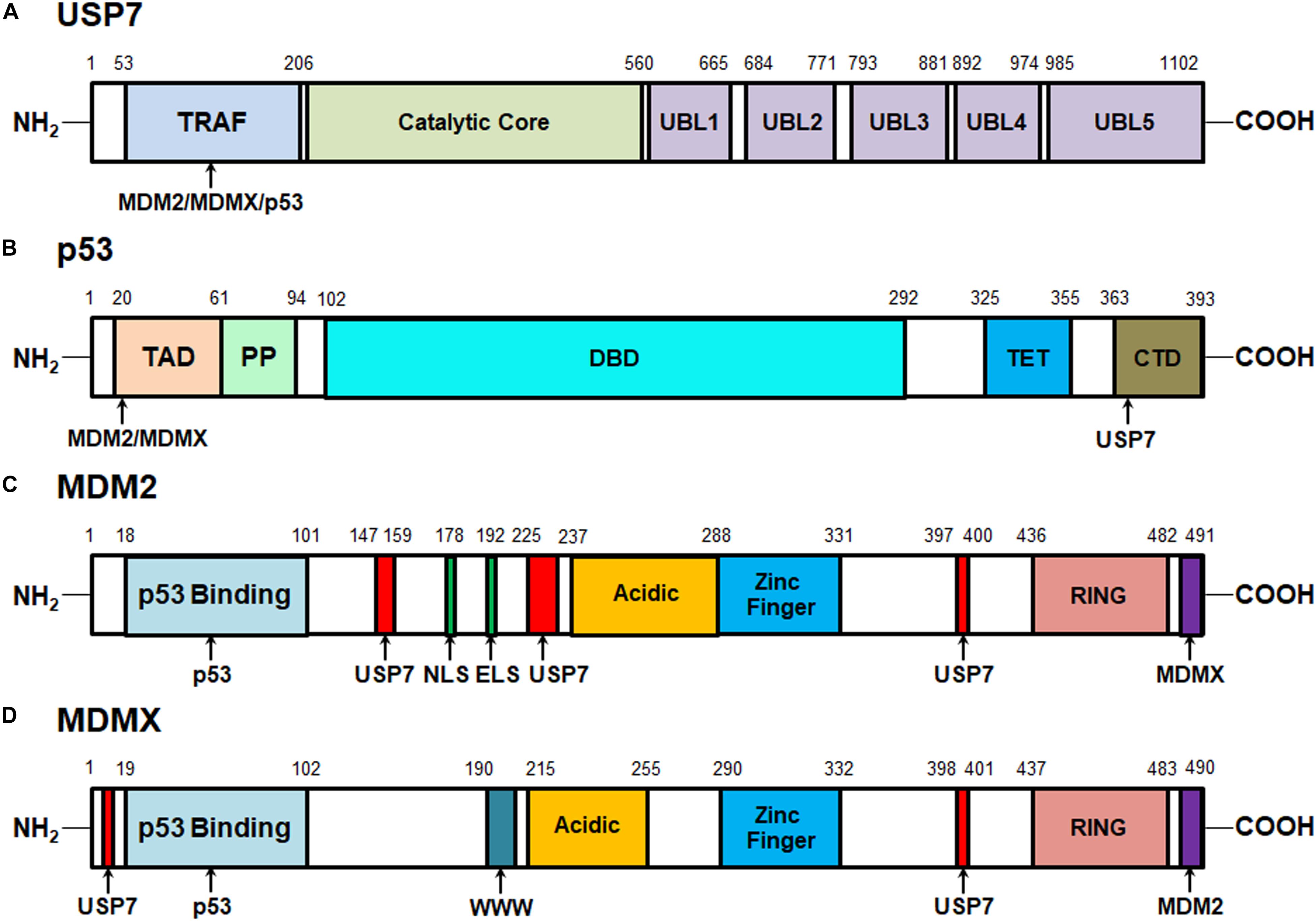
Figure 1. Structures of USP7, MDM2, MDMX, and p53 proteins and their interactive sites. (A) Structure of USP7 and the binding sites of MDM2, MDMX, and p53. (B) Structure of p53 and the MDM2-, MDMX-, and USP7-binding sites. (C) Structure of MDM2 and the p53-, MDMX-, and USP7-binding sites. (D) Structure of MDMX and the p53-, MDM2-, and USP7-binding sites.
The catalytic core of USP7 contains three distinct regions, Thumb, Palm, and Fingers, which form a unique binding pocket for ubiquitin. When ubiquitin binds, the catalytic core undergoes conformational changes, rearranging the catalytic ternary residues to adjacent positions and allowing ubiquitin catalysis (Hu et al., 2002). Mutations in the catalytic core region have been shown to significantly reduce USP7 activation (Faesen et al., 2011). Therefore, this catalytic domain is mainly responsible for the binding of USP7 with ubiquitin and the deubiquitination of the substrates (Hu et al., 2002). However, the weak catalytic activity of the separated catalytic core indicates that other regions are helpful to improve the efficiency of the ubiquitin catalytic reaction.
The C-terminal Ubl domains, which are ordered in a 2-1-2 manner (UBL1/2, UBL3, UBL4/5), contribute to the binding ability of USP7 to specific substrates and its deubiquitinating activity (Faesen et al., 2011; Kim et al., 2016). In vitro tests of mutants with different deletion regions have shown that the deletion of C-terminal strongly reduces the deubiquitination activity of USP7. Most of the catalytic activity of USP7 is reconstituted by adding UBL4/5 and 19 amino acid C-terminal tails, which suggests that they play an important role in this specific domain. Further mechanism analyses have found that UBL4/5 directly interacts and synergizes with the switch circuit in the catalytic field, which promotes the conformational change and thus increases the USP7 affinity to ubiquitin (Faesen et al., 2011). Therefore, all three domains are important for the recognition of the substrates by USP7 and the subsequent removal of ubiquitin from the target proteins (Tavana and Gu, 2017).
One of the most important USP7-interactive proteins is the p53 tumor suppressor (Saridakis et al., 2005). As shown in Figure 1B, p53 has 393 amino acid residues that can be subdivided into five domains: the N-terminal transactivation domains (TAD) that are responsible for its binding to the p53-binding sites on MDM2 and MDMX, a proline-rich region (PP) that contains five PxxP motifs and is essential for inducing apoptosis, a DNA-binding domain (DBD), a tetramerization domain (TET), and a C-terminal domain (CTD) that is critical for the binding of p53 to the TRAF domain of USP7 (Nag et al., 2013; Laptenko and Prives, 2017). As disclosed by biochemical and structural studies, the p53 CTD includes the amino acid residues 359PGGSRAHSS367 that harbor two distinct USP7-binding sites while Ser362 and Ser367 are essential for the USP7-p53 binding (Sheng et al., 2006).
The oncoproteins MDM2 and MDMX are not only the major negative regulators of p53 but also the well-characterized substrates of USP7 (Sarkari et al., 2010; Nag et al., 2013; Karni-Schmidt et al., 2016; Wang et al., 2019b). MDM2 and MDMX include 491 and 490 amino acid residues, respectively and share a great structural similarity (Nag et al., 2013; Karni-Schmidt et al., 2016). As depicted in Figures 1C,D, both MDM2 and MDMX proteins contain an N-terminal p53-binding domain, a central acidic domain that contributes to p53 ubiquitination, a zinc finger domain, and a C-terminal RING domain which is critical for MDM2’s E3 ligase activity and the formation of MDM2-MDMX heterodimers. Besides, MDM2 has the nuclear localization signal (NLS) and export localization signal (ELS) sequences but MDMX only has a unique WWW motif without NLS and ELS. The recent biochemical and structural analyses have characterized four USP7-binding sites on MDM2, including 147PSSSHLVSRPSTS159, 226AGVS229, and 397PSTS400, and Ser150 and Ser229 are critical for the MDM2-USP7 binding (Hu et al., 2006; Sheng et al., 2006; Sarkari et al., 2010). Also, two P/AxxS motifs of MDMX, 8AQCS11 and 398AHSS401 have been recognized and validated as the USP7-binding sites (Sarkari et al., 2010). These studies not only lead to a better understanding of the molecular recognition mechanisms of p53, MDM2, and MDMX by USP7 but also provide a viable option for developing new protein-protein interaction inhibitors.
The Regulation of MDM2/MDMX-p53 Circuitry by USP7
Although many USP7 substrates have been identified, the best-investigated function of USP7 is still the regulation of the MDM2/MDMX-p53 circuitry (Figure 2). As the major negative regulator of p53, MDM2 can directly bind to p53 and inhibit the transcription of its downstream target genes, including MDM2 itself, forming a negative feedback loop (Momand et al., 1992; Oliner et al., 1992; Barak et al., 1993; Perry et al., 1993). Importantly, MDM2 not only inhibits p53-mediated transactivation of its target genes but also promotes p53 ubiquitination and degradation by acting as an E3 ubiquitin ligase (Haupt et al., 1997; Honda et al., 1997; Kubbutat et al., 1997). Furthermore, the MDM2 homolog MDMX also binds to p53 and inhibits its transcriptional activity (Shvarts et al., 1996; Shvarts et al., 1997). However, MDMX does not affect the protein stability of p53 because it lacks the E3 ligase activity (Shvarts et al., 1996). MDM2 can also promote the ubiquitination and proteasomal degradation of MDMX and itself (Fang et al., 2000; de Graaf et al., 2003; Pan and Chen, 2003), while MDMX can bind to and stabilize MDM2 protein (Tanimura et al., 1999).
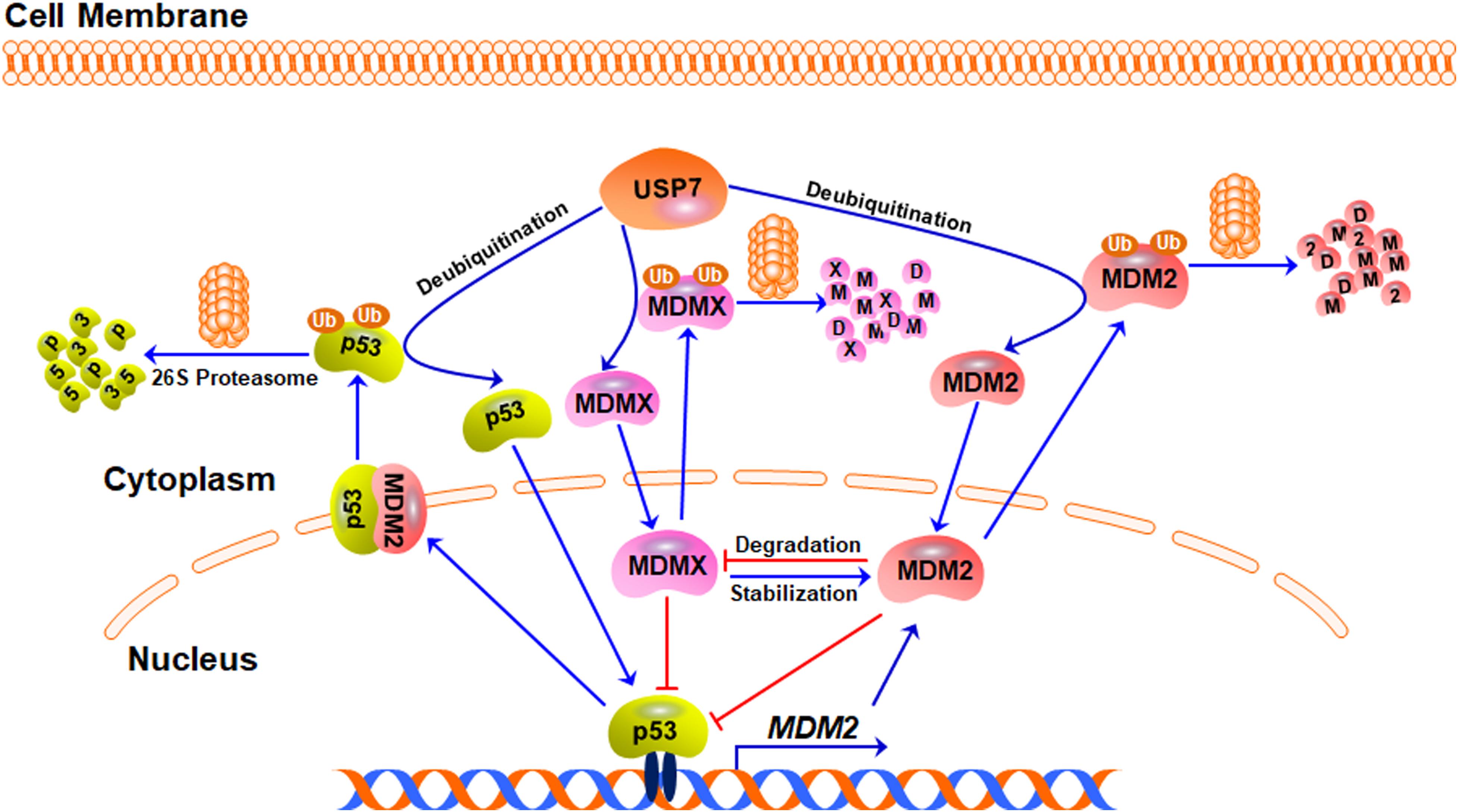
Figure 2. The USP7-MDM2/MDMX-p53 axis. Under stress-free conditions, MDM2, MDMX, and p53 form a negative feedback loop, in which p53 activates MDM2 and increases its expression level whereas MDM2 and MDMX directly bind to p53 and inhibit its transcriptional activity. MDM2 also acts as an E3 ubiquitin ligase and induces the ubiquitination and degradation of p53, MDMX, and MDM2 itself. USP7 directly binds to and stabilizes MDM2, MDMX, and p53 proteins through its deubiquitinating activity.
USP7 was initially identified as a p53-interactive protein by mass spectrometry analysis (Li et al., 2002). It has been found that the overexpression of USP7 stabilizes p53 even in the presence of excess MDM2 and the down-regulation of USP7 expression makes the endogenous p53 unstable (Tavana and Gu, 2017). Mechanistically, USP7 interacts with p53 and removes the ubiquitin from p53, causing p53 stabilization (Li et al., 2002). However, it has also been observed that the disruption of the USP7 gene stabilizes p53 (Cummins et al., 2004), which has been attributed to USP7-mediated MDM2 deubiquitination and stabilization (Li et al., 2004). Compared with p53, the binding between MDM2 and USP7 is much stronger (Hu et al., 2006; Sheng et al., 2006). USP7 binds to and stabilizes MDM2 under stress-free conditions, resulting in p53 turnover in an MDM2-dependent manner (Rawat et al., 2019). When the decrease of the USP7 level leads to MDM2 degradation, the available MDM2 pool will be not enough to exert the ubiquitin E3 ligase activity, which leads to the stabilization of p53 (Li et al., 2004). USP7 also deubiquitinates and stabilizes MDMX and the inhibition of USP7 causes MDMX degradation and the indirect activation of p53 (Meulmeester et al., 2005).
Taken together, USP7 is able to remove the ubiquitin from p53, MDM2, and MDMX and keep the stabilities of these proteins by rescuing them from degradation (Figure 2). However, due to the critical negative regulation of MDM2 and MDMX on p53, USP7-mediated stabilization of MDM2 and MDMX also causes p53 inactivation and degradation, which makes a paradoxical role of USP7 in regulating p53.
The Role of USP7-MDM2/MDMX-p53 Network in Human Cancer and Immune Response
The p53 tumor suppressor plays a critical role in protecting normal cells from transformation and tumorigenesis through transactivation of the genes related to cell growth arrest, apoptosis, and senescence (Nag et al., 2013; Liu et al., 2019a). The MDM2 and MDMX oncogenes have been demonstrated to promote tumor growth and metastasis via both p53-dependent and p53-independent mechanisms (Karni-Schmidt et al., 2016; Wang et al., 2019b). The overexpression and amplification of MDM2 and MDMX and the loss-of-function mutation of p53 are the common and crucial events in the initiation, progression, and metastasis of human cancers, which have been comprehensively discussed recently (Walerych et al., 2015; Hou et al., 2019).
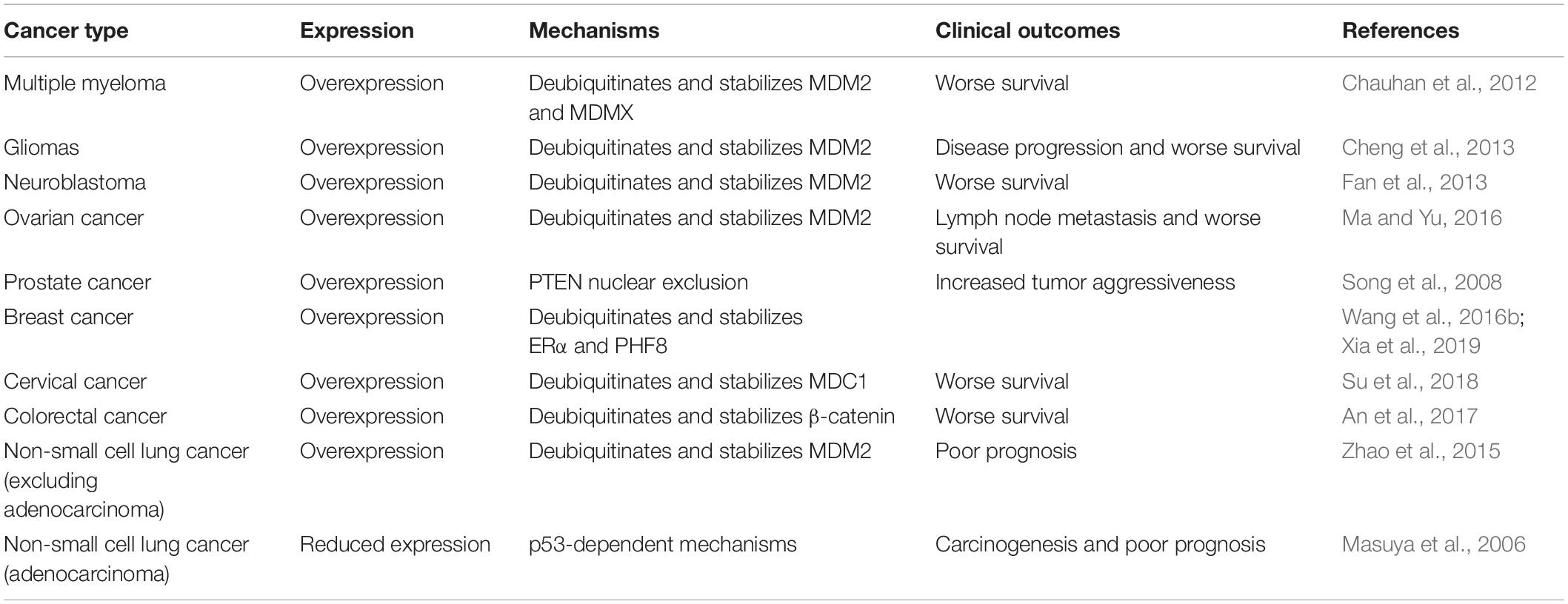
USP7, as a crucial controller of the MDM2/MDMX-p53 circuitry, has shown important functions in various human cancers and its expression and activation are associated with the progression and prognosis of these diseases (Table 1) (Bhattacharya et al., 2018; Zhou et al., 2018). It has been reported that both the protein and mRNA levels of USP7 are highly elevated in multiple myeloma (MM), and the MM patients with higher USP7 levels have significantly poorer survival (Chauhan et al., 2012). A similar role of USP7 has also been found in gliomas, neuroblastoma, and ovarian cancer, and USP7 overexpression is correlated with the disease progression and poor survival of patients (Cheng et al., 2013; Fan et al., 2013; Ma and Yu, 2016). Moreover, the carcinogenic role of USP7 in these types of cancer has been attributed to USP7-mediated deubiquitination and stabilization of MDM2 and MDMX, which has been validated by demonstrating the anticancer activity of USP7 inhibitors in vitro and in vivo (Chauhan et al., 2012; Fan et al., 2013).
USP7 overexpression has been observed in prostate cancer, which is correlated with the increased aggressiveness of the tumors. Moreover, the high expression level of USP7 is associated with the nuclear exclusion of the tumor suppressor PTEN (Song et al., 2008). USP7 is also overexpressed in breast, cervical, and colorectal cancer, which is positively associated with the worse survival of patients with these diseases (An et al., 2017; Su et al., 2018; Xia et al., 2019). In breast cancer cells, USP7 physically interacts with estrogen receptor α (ERα) and histone demethylase PHF8 and promotes their deubiquitination and stabilization (Wang et al., 2016b; Xia et al., 2019). In cervical cancer cells, USP7 directly binds to the MRN (MRE11-RAD50-NBS1)-MDC1 (mediator of DNA damage checkpoint protein 1) complex and deubiquitinates and stabilizes MDC1, which results in the maintenance of the DNA damage response (DDR) (Su et al., 2018). In colorectal cancer cells, USP7 may regulate the deubiquitination of β-catenin, although the detailed binding mechanisms need to be further investigated (An et al., 2017). Considering that MDM2 and MDMX are highly expressed in prostate, breast, cervical, and colorectal cancer (Cressey et al., 2006; Voruganti et al., 2015; Qin et al., 2016a, b, 2017; Stiasny et al., 2017; Wang et al., 2018a), USP7-mediated stabilization of MDM2 and MDMX may also contribute to the progression of these diseases.
Of note, USP7 is highly expressed in non-small cell lung cancer (NSCLC) and correlated with poor overall survival of patients (Zhao et al., 2015), excluding adenocarcinoma (Masuya et al., 2006). Conversely, NSCLC adenocarcinoma shows a lower expression level of USP7, which is negatively associated with the prognosis of this disease through p53-dependent mechanisms (Masuya et al., 2006), making a controversial and content-dependent role of USP7 in NSCLC. The contradictory effects of USP7 have also been found in the human colon carcinoma xenograft model (Becker et al., 2008). Both USP7 up- and down-regulation have been shown to stabilize and activate p53, thereby inhibiting the cancer cell growth and enhancing radiotherapy in vitro and in vivo. However, it has also been found that the down-regulation of USP7 reduces the camptothecin-induced mitochondrial translocation of p53, which further causes the resistance of cancer cells to stress-induced apoptosis (Becker et al., 2008).
USP7 has recently been found to promote tumor evasion of the immune system by deubiquitinating and stabilizing its substrates, e.g., Foxp3 (Forkhead box P3) and Tip60 (Tat-interactive protein-60 kDa) that play critical roles in enhancing the immunosuppressive functions of regulatory T cells (Treg) and suppressing tumoricidal effector T cells (Teff) (van Loosdregt et al., 2013; Wang et al., 2016a,2017a). Wang et al. (2016b) have demonstrated that USP7 deletion in Treg cells results in the expression alterations of 1,477 genes, notably causing the decreased expression of many transcription factors that play pivotal roles in Treg cell development and stability and the increased expression of genes associated with cell cycle, apoptosis, DNA damage response, and glycolysis pathways. Consequently, USP7 deletion significantly impairs the suppressive functions of Treg cells in vitro and in vivo. Therefore, USP7 has recently been proposed as a target of both tumor growth and immune evasion (Wu et al., 2018). Pharmacological modification of USP7 by its small-molecule inhibitors (including P5091 and P0217564) have been examined and shown promising efficacy in abrogating Treg cell functions and promoting antitumor immunity in murine Treg cells in vitro and in Treg-dependent syngeneic mouse models in vivo (Wang et al., 2016a; Fu et al., 2019). Wang and colleagues have also found that USP7 inhibitor treatment leads to the enhanced accumulation of CD8+ T cells in the tumor microenvironment and the reduced accumulation of Foxp3+ Treg cells, which cause the increased production of IFNγ and the break of immune tolerance. Therefore, the USP7 inhibitors could favorably target the Foxp3+ Treg cells and inhibit their functions, indicating that targeting USP7 may be a promising strategy to break immune tolerance and facilitate immunotherapy in cancer. However, it should be noted that the developmental deletion of USP7 in Foxp3+ Treg cells causes lethal systemic autoimmunity in mice within 4 weeks of birth (Wang et al., 2016a). It is still not clear whether USP7 inhibitor-mediated repression of Treg function impairs host immune responses and induces autoimmunity. Further investigation should be performed in more clinically relevant cancer models in vitro and in vivo.
Targeting MDM2/MDMX-p53 Circuitry by USP7 Inhibitors
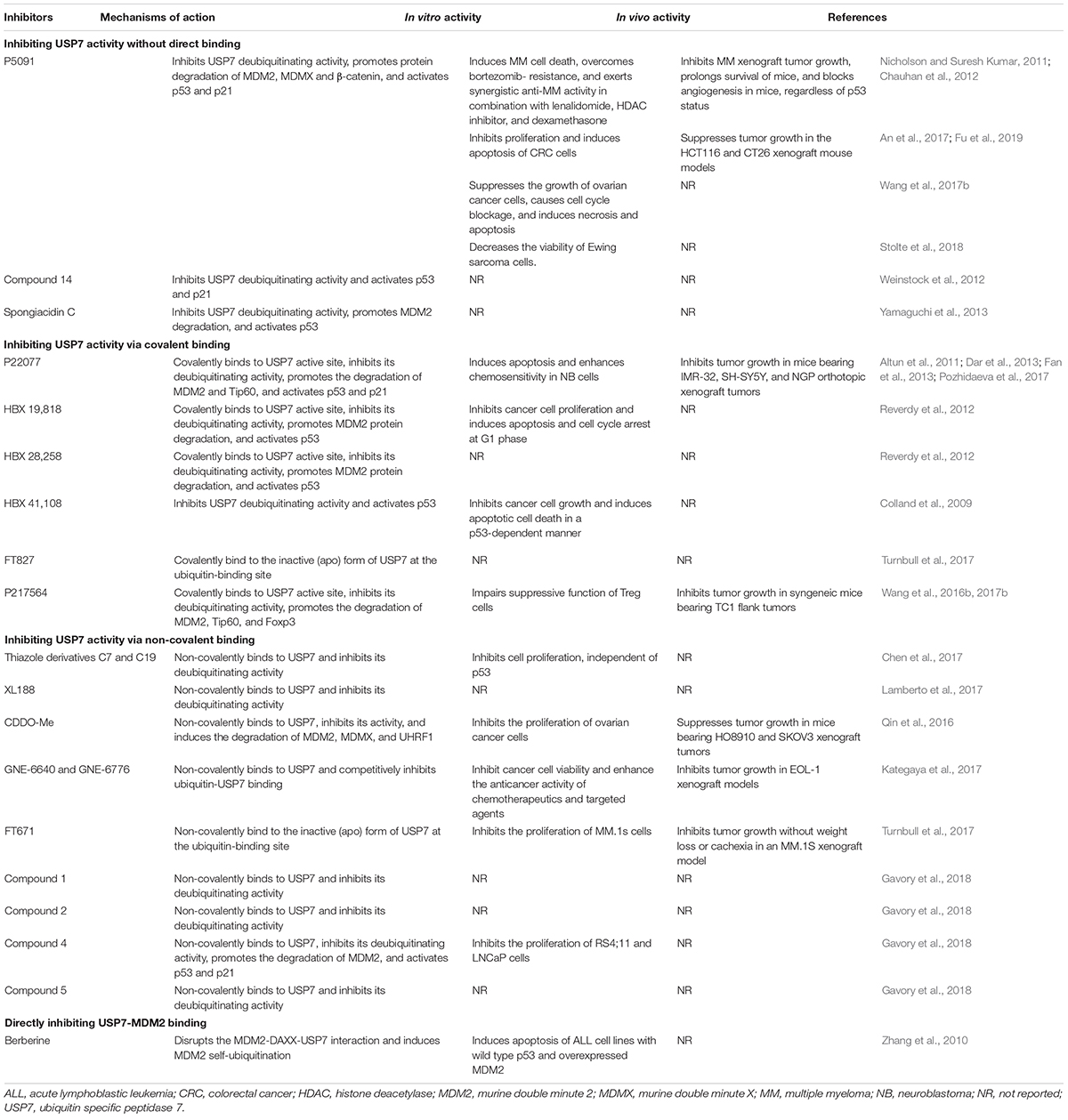
Considering the pivotal role of USP7 in regulating the MDM2/MDMX-p53 pathway in cancer cells, many small-molecule inhibitors have been developed for treating human cancers. The majority of USP7 inhibitors have been found to inhibit its deubiquitinating activity, promote the degradation of MDM2 and MDMX, and activate p53 signaling (Table 2 and Figure 3). All the deubiquitinating activity inhibitors can be classified into three categories based on their molecular mechanisms, i.e., inhibiting USP7 activity (1) without direct binding, (2) via covalent binding, or (3) via non-covalent binding. It has also been reported that direct inhibition of the USP7-MDM2 binding can promote MDM2 ubiquitination and degradation (strategy 4 in Figure 3). Accordingly, direct inhibition of the USP7-MDMX interaction may cause MDMX degradation, but there is no specific inhibitor reported yet (strategy 5 in Figure 3).
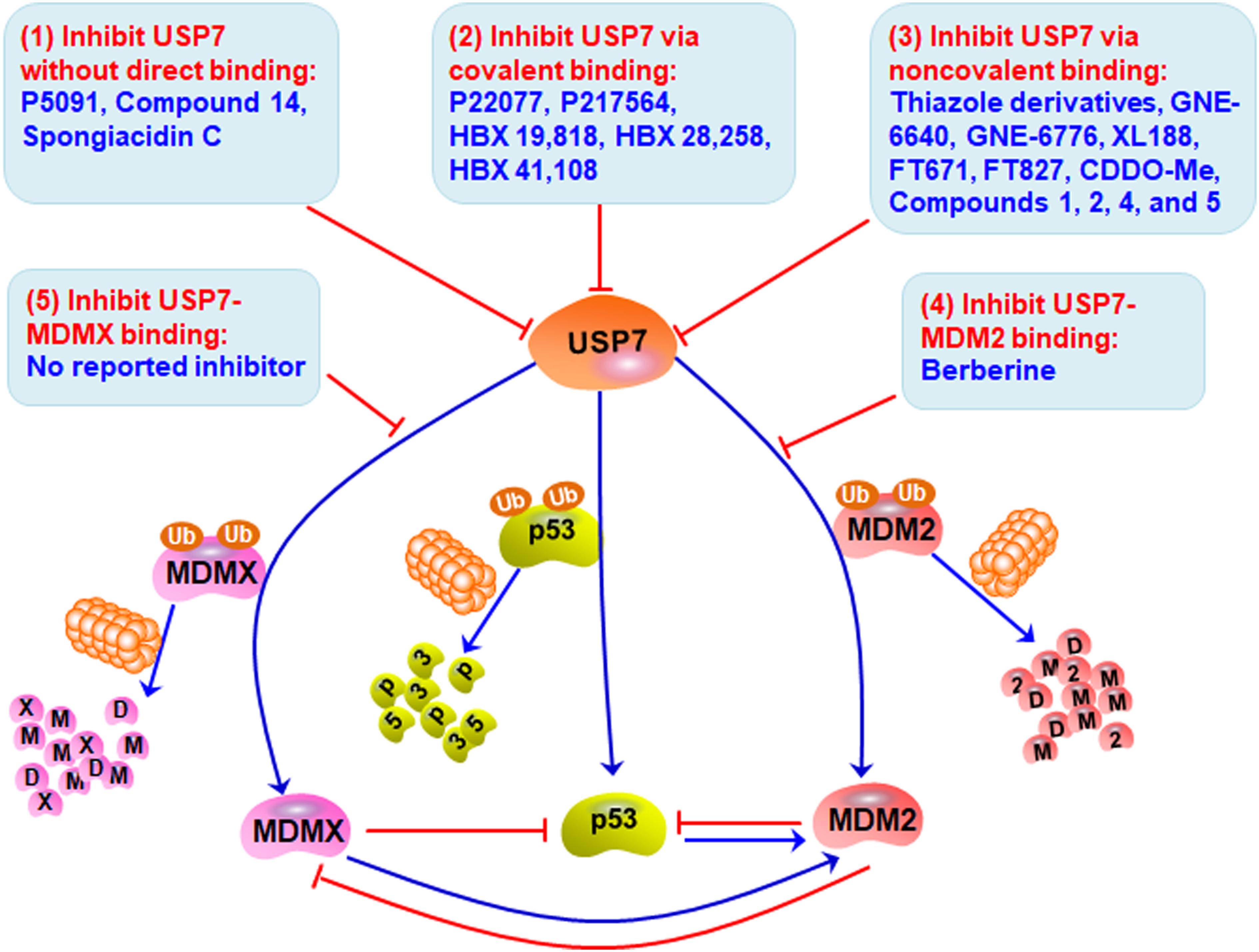
Figure 3. Targeting USP7-MDM2/MDMX-p53 network for cancer therapy. Several small-molecule inhibitors have been developed to broadly inhibit the deubiquitinating activity of USP7 without direct binding (Strategy 1), via covalent binding (Strategy 2), or via non-covalent binding (Strategy 3), therefore inducing the degradation of MDM2 and MDMX and stabilizing and activating p53. These compounds have shown anticancer efficacy in vitro and in vivo. Directly inhibiting the bindings of USP7 to MDM2 (Strategy 4) and MDMX (Strategy 5) may be examined to develop more safe and effective inhibitors for cancer therapy.
Inhibiting USP7 Activity Without Direct Binding
P5091 is the first identified specific USP7 inhibitor through a high-throughput screening from a small-molecule library using a ubiquitin-phospholipase A2 enzyme reporter assay (Nicholson and Suresh Kumar, 2011; Chauhan et al., 2012). It has been found that P5091 potently and selectively inhibits the deubiquitinating activity of USP7 with an IC50 value of 4.2 μM, without affecting other DUBs at 100 μM (Chauhan et al., 2012). As expected, P5091 induces the polyubiquitination of MDM2 and MDMX and accelerates their proteasomal degradation in MM cells, thereby activating p53 and its downstream target p21 (Chauhan et al., 2012). It has further been observed that P5091 induces cancer cell death and overcomes chemoresistance in MM cells in vitro and in MM xenograft models in vivo. Moreover, P5091 has been shown to inhibit cell proliferation and induce cell cycle arrest and apoptosis in colorectal cancer (An et al., 2017; Fu et al., 2019), ovarian cancer (Wang et al., 2017b), and Ewing sarcoma (Stolte et al., 2018) cells in vitro and in vivo. Besides, P5091 inhibits the Wnt signaling by enhancing the ubiquitination and degradation of β-catenin, suggesting that β-catenin is a potential substrate of USP7 (An et al., 2017).
Considering the important role of USP7 in Treg cell functions, Wang et al. (2016a) have recently demonstrated that P5091 promotes the antitumor immune response by impairing the immunosuppressive activity of Treg cells. Fu et al. (2019) have further evaluated the efficacy and possible molecular mechanisms of P5091 in the colon cancer CT26 xenograft model. The USP7 inhibitor has comparable efficacy versus anti-PD-1 antibody in inhibiting the growth of CT26 xenograft tumors in mice. Mechanism studies have shown that P5091 treatment down-regulates IL-10 expression and upregulates the expression of IFN-γ and TNF-α in tumor tissue at the mRNA level (Fu et al., 2019). Similar changes have been observed in serum from tumor-bearing mice at the protein level. Importantly, P5091 treatment increases IFN-γ expression in both CD4+ and CD8+ T cells from the spleen of tumor-bearing mice, which suggests that this compound boosts the antitumor immune response (Fu et al., 2019). It has been further found that P5091 down-regulates the expression of Foxp3 in CD4+ T cells and decreases the proportion of Treg cells in vivo, suggesting an impaired immune tolerance in the tumor microenvironment.
In addition, a new class of P5091 derivatives has recently been designed and synthesized, and a lead compound termed compound 14 has been discovered to inhibit the deubiquitinating activities of both USP7 and USP47 with IC50 values of 0.42 and 1.0 μM, respectively (Weinstock et al., 2012). Compound 14 also activates p53 and p21 in HCT116 human colon cancer cells (Weinstock et al., 2012). A pyrrole alkaloid, named spongiacidin C has recently been isolated from the marine sponge Stylissa massa and identified as a specific USP7 inhibitor with an IC50 of 3.8 μM (Yamaguchi et al., 2013). However, neither compound 14 nor spongiacidin C are evaluated for their anticancer activity yet.
Inhibiting USP7 Activity via Covalent Binding
The medicinal chemistry optimization of P5091 leads to the identification of a USP7/USP47 dual inhibitor termed P22077 (Altun et al., 2011). In comparison to P5091, P22077 shows low inhibitory effects on the deubiquitinating activity with effective concentrations at 15–45 μM (Altun et al., 2011). The compound has further been shown to promote MDM2 degradation and activate p53 and p21 (Altun et al., 2011). Pozhidaeva et al. (2017) have recently demonstrated that P22077 covalently modifies cysteine 223 (Cys223) in the catalytic core of USP7 and induces conformational changes in the active site, resulting in the inhibition of its enzymatic activity. P22077 also induces apoptosis and enhances chemosensitivity in neuroblastoma (NB) cells in vitro (Fan et al., 2013). Its in vivo efficacy has been demonstrated in mice bearing IMR-32, SH-SY5Y, and NGP orthotopic xenograft tumors (Fan et al., 2013). Based on the observation that P22077 promotes the ubiquitination and degradation of Tip60, this protein has been identified as a substrate of USP7 and has been found to contribute to P22077-mediated cytotoxicity (Dar et al., 2013).
The USP7 inhibitors P22077 and compound 14 have shown unexpected inhibition on USP47 (Altun et al., 2011; Weinstock et al., 2012). To identify more specific USP7 inhibitors, Reverdy et al. (2012) have developed a high-throughput screening assay using an optimized USP substrate ubiquitin C-terminal 7-amido-4-methylcoumarin, which has led to the discovery of a novel class of USP7 inhibitors from a chemically diverse library of small molecules. The lead compounds HBX 19,818 and HBX 28,258 can specifically and selectively inhibit USP7 activity with IC50 values of 28.1 and 22.6 μM, respectively, without apparent effects on other tested deubiquitinating enzymes (Reverdy et al., 2012). Importantly, this class of compounds can covalently form a bond with the Cys223 located in the catalytic core of USP7, thereby selectively causing its inactivation as well as MDM2 degradation and p53 stabilization and activation (Reverdy et al., 2012). HBX 19,818 has further been shown to inhibit cell proliferation and induce G1 arrest and apoptosis in HCT116 cells. It has also been found that HBX 19,818 inhibits cancer cell viability in a p53-independent manner, which emphasizes the critical role of the p53-independent functions of MDM2 and MDMX in cancer cells (Qin et al., 2018; Wang et al., 2019b). However, both HBX 19,818 and HBX 28,258 have not been evaluated in any animal models, and further investigation for their in vivo efficacy and safety should be performed.
Further optimization of HBX 19,818 and HBX 28,258 has led to a more potent and specific inhibitor named HBX41,108, which inhibits USP7 activity with an IC50 of 424 nM (Colland et al., 2009). This new USP7 inhibitor has been found to significantly reverse the USP7-mediated MDM2 stabilization and increase the expression of p53 and p21 in a dose-dependent manner. The molecular modeling and docking studies have been performed and the results suggest that HBX41,108 may covalently bind to the ubiquitin-binding sites on USP7 and allosterically inhibit its catalytic activity (Colland et al., 2009). Importantly, HBX41,108 can significantly inhibit cell proliferation and induce apoptosis in HCT116 cells at submicromolar concentrations (Colland et al., 2009). Another high-throughput screening of a library containing ∼500K compounds at FORMA Therapeutics has recently been performed by using a ubiquitin-rhodamine assay, and a covalent inhibitor FT827 has been identified (Turnbull et al., 2017). Similar to HBX 19,818 and HBX 28,258, FT827 binds to the catalytic domain of USP7 (KD = 7.8 μM) by covalently modifying Cys223 of this domain and inhibits the enzymatic activity (Turnbull et al., 2017). However, the in vivo efficacy, toxicology, and pharmacokinetics of HBX41,108 and FT827 are still unknown yet, which are critical for further developing these compounds as anticancer agents.
Another P5091 derivative, termed P217564 has been generated and shown to impair the suppressive functions of Foxp3+ Treg cells in a Tip60-dependent way, thereby enhancing the antitumor immune response in vivo (Wang et al., 2016a). Molecular mechanism studies have indicated that P217564 treatment promotes the ubiquitination and degradation of Foxp3 and Tip60, which play essential roles in maintaining the functions of Treg cells. P217564 has also been shown to inhibit tumor growth in syngeneic mice by improving the host antitumor immune responses (Wang et al., 2016b). Of note, the Teff cells from P217564-treated mice have exhibited resistance to the suppression by Treg cells, which suggests that the USP7 inhibitor does not abrogate the functions of Teff cells in vivo (Wang et al., 2016a). Further studies have indicated that P217564 is a USP7/USP47 dual inhibitor with IC50 values of 0.48 and 0.66 μM, respectively (Wang et al., 2017a). Different from P5091, P217564 directly targets the catalytic core domain of USP7 and forms a covalent adduct with Cys223 on USP7, consequently inducing the inactivation of USP7 and the degradation of USP7 substrates, including MDM2, Foxp3, and Tip60 (Wang et al., 2017a).
Inhibiting USP7 Activity via Non-covalent Binding
Thanks to the advances in understanding the crystal structures of USP7, USP7-ligand complexes, and the functional domains, especially the catalytic core (Hu et al., 2002), several small-molecule inhibitors have been designed and developed to non-covalently bind to USP7 and selectively inhibit its deubiquitinating activity, including but not limited to the thiazole derivatives C7 and C19 (Chen et al., 2017), XL188 (Lamberto et al., 2017), FT671 (Turnbull et al., 2017), CDDO-Me (Qin et al., 2016), GNE-6640 (Kategaya et al., 2017), GNE-6776 (Kategaya et al., 2017), and compounds 1, 2, 4, and 5 (Gavory et al., 2018).
Some of these inhibitors have been designed to target the catalytic domain of USP7 via non-covalent binding and inhibit its deubiquitinating activity. Chen et al. (2017) have designed and synthesized a new class of thiazole derivatives based on the chemical structure of P22077 and the crystal structure of USP7. The lead compounds C7 and C19 have been found to directly bind to the ubiquitin-binding site of USP7 and inhibit its activity with IC50 values of 0.67 and 1.35 μM, respectively (Chen et al., 2017). Both compounds have exhibited anti-proliferation activity in cancer cells in vitro, independent of p53 (Chen et al., 2017). Lamberto et al. (2017) have performed a structure-based design and developed a non-covalent-binding USP7 inhibitor termed XL188, which selectively binds to the S4–S5 pocket of USP7 (KD = 104 nM) and potently inhibits its enzymatic activity (IC50 = 90 nM). XL188 has also been found to accelerate MDM2 protein degradation and increase the accumulation of p53 and p21 (Lamberto et al., 2017). However, the in vivo efficacy and safety profiles of these inhibitors have not been examined yet.
Turnbull et al. (2017) have recently characterized a non-covalent inhibitor FT671, which binds to the USP7 catalytic domain with a KD of 65 nM and inhibits USP7 activity with an IC50 of 69 nM. FT671 has also been found to decrease the MDM2 level and increase the p53 level in a concentration-dependent manner, thereby blocking the proliferation of MM.1S cells (IC50 = 33 nM) in vitro and inhibiting the growth of MM.1S xenograft tumors in vivo (Turnbull et al., 2017). In addition, a synthetic triterpenoid CDDO-Me has recently been characterized as a USP7 inhibitor using the Ub-AMC protease assay (Qin et al., 2016). Qin et al. (2016) have demonstrated that CDDO-Me directly binds to USP7, markedly inhibits its activity (IC50 = 14.08 μM), and induces the degradation of MDM2, MDMX, and UHRF1. CDDO-Me has also shown anticancer activity in ovarian cancer cells in vitro and in HO8910 and SKOV3 xenograft models in vivo.
Several non-covalent-binding inhibitors have been designed to attenuate the USP7-ubiquitin binding for inhibiting its deubiquitinating activity. Kategaya et al. (2017) have recently developed two selective USP7 inhibitors GNE-6640 and GNE-6776 using nuclear magnetic resonance-based screening and structure-based design. Both compounds non-covalently interact with and cleave ubiquitin moieties harboring free K48 side chains, thereby competitively inhibiting the binding of ubiquitin to USP7 and destabilizing its substrates, especially MDM2 (Kategaya et al., 2017). GNE-6640 and GNE-6776 have also been shown to inhibit the viability of cancer cells and enhance the anticancer activity of chemotherapeutics and targeted agents in vitro and delay the EOL-1 xenograft tumor growth in vivo (Kategaya et al., 2017).
Gavory et al. (2018) have recently identified a new class of non-covalent and reversible USP7 inhibitors. By employing fragment-based methods as well as combining structural features of published USP7 inhibitors, Gavory et al. (2018) have designed and synthesized a chemically stable and reversible USP7 binder, compound 1 (IC50 = 13.1 μM) with high water solubility and without redox liabilities. By introducing a chiral methyl group (R stereochemistry) at the benzylic position of the phenethylamide chain, they have further obtained a sub-micromolar USP7 inhibitor, compound 2 (IC50 = 0.3 μM) (Gavory et al., 2018). The non-covalent binding of compound 2 to USP7 has been confirmed by the X-ray crystal structure of their complex. The key amino acid residues with conformational changes, including Phe409, Gly458, Gly463, and His461 have been disclosed, indicating a novel binding site that is situated 5.5 Å from the previously reported catalytic cysteine. Further optimization of compound 2 has led to the discovery of highly potent and selective allosteric USP7 inhibitors, compound 4 (IC50 = 6.0 nM) and compound 5 (IC50 = 22.0 nM) (Gavory et al., 2018). The co-crystal structure of compound 5 and USP7 complex is highly similar to the USP7-compound 2 complex. Besides, compound 4 has been found to tightly bind to USP7 (KD = 2.0 nM) and non-competitively inhibit its activity. Compound 4 has also been shown to decrease the MDM2 level and increase the levels of p53 and p21 in vitro. In comparison to the clinically relevant MDM2 inhibitors, compound 4 has equal or superior anticancer activity in acute lymphoblastic leukemia cell line RS4;11 and prostate cancer cell line LNCaP, which are hypersensitive to USP7 inhibition (Gavory et al., 2018).
Directly Inhibiting USP7-MDM2 Binding
Zhang et al. (2010) have recently characterized berberine, a natural product isolated from Chinese herbal medicine as a novel USP7 inhibitor with a distinct molecular mechanism. Berberine has been shown to disrupt the MDM2-DAXX-USP7 complex and promote MDM2 self-ubiquitination and degradation. It has also been observed that berberine induces cytotoxicity and apoptosis in acute lymphoblastic leukemia (ALL) cells containing wild-type p53 and overexpressed MDM2 (Zhang et al., 2010). Considering that USP7 binds to MDM2 via the TRAF domain instead of the catalytic core domain, berberine may block the USP7-MDM2 binding via interacting with the TRAF domain. However, the binding mechanisms should be further investigated and may be used to develop specific USP7 inhibitors, whose molecular mechanisms are distinct from the current USP7 inhibitors.
Conclusion and Future Directions
The MDM2/MDMX-p53 circuitry plays a pivotal role in cancer cell proliferation, cell cycle progression, apoptosis, and senescence (Karni-Schmidt et al., 2016; Wang et al., 2019b), while USP7 is a critical regulator of this circuitry and tightly controls the stabilities of these proteins, thereby contributing to cancer initiation, progression, and metastasis (Bhattacharya et al., 2018; Rawat et al., 2019). Many MDM2/MDMX inhibitors have been developed for cancer therapy (Qin et al., 2015; Wang et al., 2018a, b, 2019b; de Oliveira Ribeiro et al., 2020), and some of them are in clinical trials (Liu et al., 2019b; Rafal et al., 2019). Most of the MDM2/MDMX inhibitors are designed to target the MDM2/p53 and MDMX/p53 interactions, and wild-type p53 is critical for their anticancer activity (Liu et al., 2019b; Rafal et al., 2019). However, p53 mutation and inactivation and MDM2/MDMX overexpression and amplification are frequently observed in various types of human cancer, and the current MDM2/MDMX inhibitors have low or no efficacy in patients with these cancers (Bohlman and Manfredi, 2014; Wang et al., 2014a, b). Therefore, it is urgently needed to find a new strategy for developing effective and safe therapeutics for such types of cancer.
USP7 directly interacts with MDM2 and MDMX, regardless of p53 status. The current USP7 inhibitors have been shown to promote the ubiquitination and degradation of MDM2 and MDMX, independent of p53. Consistently, these USP7 inhibitors have shown anticancer activity in cancer cells with various p53 statuses in vitro and in vivo. Therefore, targeting the USP7-MDM2/MDMX-p53 network is a promising strategy to develop novel targeted therapy for human cancers, especially for those with mutant p53 and overexpressed MDM2 and MDMX. Currently, almost all the USP7 inhibitors have been developed to directly inhibit its deubiquitinating activity with or without the binding to its catalytic core. However, some of the USP7 substrates function as tumor suppressors, such as p53 and PTEN. Directly inhibiting the enzymatic activity of USP7 with or without binding to the catalytic domain by small-molecule inhibitors may cause the degradation of all the substrates of USP7, regardless of tumor suppressors (e.g., p53 and PTEN) and oncoproteins (e.g., MDM2 and MDMX), which may cause adverse effects in cancer patients.
Inhibiting the expression of USP7 has been considered as a strategy to develop USP7 inhibitors. However, it has been reported that the down-regulation of USP7 expression also leads to the degradation of the p53 tumor suppressor (Tavana and Gu, 2017). Also, small molecules developed to inhibit the expression of molecular targets have been frequently found to have low specificity and selectivity (Qin et al., 2019a, b; Dong et al., 2020). The characterization of berberine as a USP7-MDM2 binding inhibitor provides novel strategies to develop specific inhibitors that selectively promote the degradation of oncoproteins. It is speculated that berberine binds to the TRAF domain of USP7 and blocks the USP7-MDM2 binding, consequently inhibiting USP7-mediated MDM2 deubiquitination and promoting its proteasomal degradation. However, the detailed binding mechanism is still not clear yet. Importantly, it has been reported that the MDM2-USP7 binding is much stronger than the p53-USP7 binding (Hu et al., 2006; Sheng et al., 2006), which provides a rationale for developing selective inhibitors of MDM2-USP7 interaction without affecting the deubiquitinating effects of USP7 on p53. Mechanistically, the small molecules that mimic the P/AxxS motifs and competitively block the bindings of USP7 to MDM2/MDMX may specifically and selectively destabilize MDM2 and MDMX, without affecting other proteins. Nevertheless, this new targeting strategy should be extensively investigated in the future.
Author Contributions
J-JQ, X-DC, and W-DZ conceptualized the manuscript. S-MQ, GC, X-DC, ZX, BX, and J-JQ collected the literature, wrote the manuscript, and made the figures. J-JQ edited and made significant revisions to the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by National Natural Science Foundation of China (81903842), Program of Zhejiang Provincial TCM Sci-tech Plan (2020ZZ005), and Zhejiang Chinese Medical University Startup Funding (111100E014).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the current and former members of our laboratories and collaborators for their contributions to the publications cited in this review article. The research field in USP7 is rapidly growing, and we apologize for not being able to cite all the recent publications, due to space limitation.
References
Altun, M., Kramer, H. B., Willems, L. I., Mcdermott, J. L., Leach, C. A., Goldenberg, S. J., et al. (2011). Activity-based chemical proteomics accelerates inhibitor development for deubiquitylating enzymes. Chem. Biol. 18, 1401–1412. doi: 10.1016/j.chembiol.2011.08.018
An, T., Gong, Y., Li, X., Kong, L., Ma, P., Gong, L., et al. (2017). USP7 inhibitor P5091 inhibits Wnt signaling and colorectal tumor growth. Biochem. Pharmacol. 131, 29–39. doi: 10.1016/j.bcp.2017.02.011
Barak, Y., Juven, T., Haffner, R., and Oren, M. (1993). mdm2 expression is induced by wild type p53 activity. EMBO J. 12, 461–468.
Becker, K., Marchenko, N. D., Palacios, G., and Moll, U. M. (2008). A role of HAUSP in tumor suppression in a human colon carcinoma xenograft model. Cell Cycle 7, 1205–1213.
Bhattacharya, S., Chakraborty, D., Basu, M., and Ghosh, M. K. (2018). Emerging insights into HAUSP (USP7) in physiology, cancer and other diseases. Signal. Transd. Target Ther. 3:17. doi: 10.1038/s41392-018-0012-y
Bohlman, S., and Manfredi, J. J. (2014). p53-independent effects of Mdm2. Subcell. Biochem. 85, 235–246. doi: 10.1007/978-94-017-9211-0_13
Chauhan, D., Tian, Z., Nicholson, B., Kumar, K. G., Zhou, B., Carrasco, R., et al. (2012). A small molecule inhibitor of ubiquitin-specific protease-7 induces apoptosis in multiple myeloma cells and overcomes bortezomib resistance. Cancer Cell 22, 345–358. doi: 10.1016/j.ccr.2012.08.007
Chen, C., Song, J., Wang, J., Xu, C., Chen, C., Gu, W., et al. (2017). Synthesis and biological evaluation of thiazole derivatives as novel USP7 inhibitors. Bioorg. Med. Chem. Lett. 27, 845–849. doi: 10.1016/j.bmcl.2017.01.018
Cheng, C., Niu, C., Yang, Y., Wang, Y., and Lu, M. (2013). Expression of HAUSP in gliomas correlates with disease progression and survival of patients. Oncol. Rep. 29, 1730–1736.
Colland, F., Formstecher, E., Jacq, X., Reverdy, C., Planquette, C., Conrath, S., et al. (2009). Small-molecule inhibitor of USP7/HAUSP ubiquitin protease stabilizes and activates p53 in cells. Mol. Cancer Ther. 8, 2286–2295. doi: 10.1158/1535-7163.MCT-09-0097
Cressey, R., Pimpa, S., Tontrong, W., Watananupong, O., and Leartprasertsuke, N. (2006). Expression of cyclooxygenase-2 in colorectal adenocarcinoma is associated with p53 accumulation and hdm2 overexpression. Cancer Lett. 233, 232–239.
Cummins, J. M., Rago, C., Kohli, M., Kinzler, K. W., Lengauer, C., and Vogelstein, B. (2004). Tumour suppression: disruption of HAUSP gene stabilizes p53. Nature 428:486.
Dar, A., Shibata, E., and Dutta, A. (2013). Deubiquitination of Tip60 by USP7 determines the activity of the p53-dependent apoptotic pathway. Mol. Cell Biol. 33, 3309–3320. doi: 10.1128/MCB.00358-13
de Graaf, P., Little, N. A., Ramos, Y. F., Meulmeester, E., Letteboer, S. J., and Jochemsen, A. G. (2003). Hdmx protein stability is regulated by the ubiquitin ligase activity of Mdm2. J. Biol. Chem. 278, 38315–38324.
de Oliveira Ribeiro, H., Pereira Cortez, A., De Avila, R. I., Silva, A., De Carvalho, F. S., Menegatti, R., et al. (2020). Small-molecule MDM2 inhibitor LQFM030-induced apoptosis in p53-null K562 chronic myeloid leukemia cells. Fundam Clin. Pharmacol. doi: 10.1111/fcp.12540 [Epub ahead of print].
Dong, J., Qin, Z., Zhang, W. D., Cheng, G., Yehuda, A. G., Ashby, C. R., et al. (2020). Medicinal chemistry strategies to discover P-glycoprotein inhibitors: an update. Drug Resist. Updat. 49:100681. doi: 10.1016/j.drup.2020.100681
Everett, R. D., Meredith, M., Orr, A., Cross, A., Kathoria, M., and Parkinson, J. (1997). A novel ubiquitin-specific protease is dynamically associated with the PML nuclear domain and binds to a herpesvirus regulatory protein. EMBO J. 16, 1519–1530.
Faesen, A. C., Dirac, A. M., Shanmugham, A., Ovaa, H., Perrakis, A., and Sixma, T. K. (2011). Mechanism of USP7/HAUSP activation by its C-terminal ubiquitin-like domain and allosteric regulation by GMP-synthetase. Mol. Cell 44, 147–159. doi: 10.1016/j.molcel.2011.06.034
Fan, Y. H., Cheng, J., Vasudevan, S. A., Dou, J., Zhang, H., Patel, R. H., et al. (2013). USP7 inhibitor P22077 inhibits neuroblastoma growth via inducing p53-mediated apoptosis. Cell Death Dis. 4:e867. doi: 10.1038/cddis.2013.400
Fang, S., Jensen, J. P., Ludwig, R. L., Vousden, K. H., and Weissman, A. M. (2000). Mdm2 is a RING finger-dependent ubiquitin protein ligase for itself and p53. J. Biol. Chem. 275, 8945–8951.
Fernandez-Montalvan, A., Bouwmeester, T., Joberty, G., Mader, R., Mahnke, M., Pierrat, B., et al. (2007). Biochemical characterization of USP7 reveals post-translational modification sites and structural requirements for substrate processing and subcellular localization. FEBS J. 274, 4256–4270.
Fu, C., Zhu, X., Xu, P., and Li, Y. (2019). Pharmacological inhibition of USP7 promotes antitumor immunity and contributes to colon cancer therapy. Onco Targets Ther. 12, 609–617. doi: 10.2147/OTT.S182806
Gavory, G., O’dowd, C. R., Helm, M. D., Flasz, J., Arkoudis, E., Dossang, A., et al. (2018). Discovery and characterization of highly potent and selective allosteric USP7 inhibitors. Nat. Chem. Biol. 14, 118–125. doi: 10.1038/nchembio.2528
Harrigan, J. A., Jacq, X., Martin, N. M., and Jackson, S. P. (2018). Deubiquitylating enzymes and drug discovery: emerging opportunities. Nat. Rev. Drug Discov. 17, 57–78. doi: 10.1038/nrd.2017.152
Haupt, Y., Maya, R., Kazaz, A., and Oren, M. (1997). Mdm2 promotes the rapid degradation of p53. Nature 387, 296–299.
Honda, R., Tanaka, H., and Yasuda, H. (1997). Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett. 420, 25–27.
Hou, H., Sun, D., and Zhang, X. (2019). The role of MDM2 amplification and overexpression in therapeutic resistance of malignant tumors. Cancer Cell Int. 19:216. doi: 10.1186/s12935-019-0937-4
Hu, M., Gu, L., Li, M., Jeffrey, P. D., Gu, W., and Shi, Y. (2006). Structural basis of competitive recognition of p53 and MDM2 by HAUSP/USP7: implications for the regulation of the p53-MDM2 pathway. PLoS Biol. 4:e27. doi: 10.1371/journal.pbio.0040027
Hu, M., Li, P., Li, M., Li, W., Yao, T., Wu, J. W., et al. (2002). Crystal structure of a UBP-family deubiquitinating enzyme in isolation and in complex with ubiquitin aldehyde. Cell 111, 1041–1054.
Karni-Schmidt, O., Lokshin, M., and Prives, C. (2016). The roles of MDM2 and MDMX in cancer. Annu. Rev. Pathol. 11, 617–644. doi: 10.1146/annurev-pathol-012414-040349
Kategaya, L., Di Lello, P., Rouge, L., Pastor, R., Clark, K. R., Drummond, J., et al. (2017). USP7 small-molecule inhibitors interfere with ubiquitin binding. Nature 550, 534–538. doi: 10.1038/nature24006
Kim, R. Q., Van Dijk, W. J., and Sixma, T. K. (2016). Structure of USP7 catalytic domain and three Ubl-domains reveals a connector alpha-helix with regulatory role. J. Struct. Biol. 195, 11–18. doi: 10.1016/j.jsb.2016.05.005
Kubbutat, M. H., Jones, S. N., and Vousden, K. H. (1997). Regulation of p53 stability by Mdm2. Nature 387, 299–303.
Lamberto, I., Liu, X., Seo, H. S., Schauer, N. J., Iacob, R. E., Hu, W., et al. (2017). Structure-guided development of a potent and selective non-covalent active-site inhibitor of USP7. Cell Chem. Biol. 24, 1490.e11–1500.e11. doi: 10.1016/j.chembiol.2017.09.003
Laptenko, O., and Prives, C. (2017). p53: master of life, death, and the epigenome. Genes Dev. 31, 955–956. doi: 10.1101/gad.302364.117
Li, M., Brooks, C. L., Kon, N., and Gu, W. (2004). A dynamic role of HAUSP in the p53-Mdm2 pathway. Mol. Cell 13, 879–886.
Li, M., Chen, D., Shiloh, A., Luo, J., Nikolaev, A. Y., Qin, J., et al. (2002). Deubiquitination of p53 by HAUSP is an important pathway for p53 stabilization. Nature 416, 648–653.
Liu, Y., Tavana, O., and Gu, W. (2019a). p53 modifications: exquisite decorations of the powerful guardian. J. Mol. Cell Biol. 11, 564–577. doi: 10.1093/jmcb/mjz060
Liu, Y., Wang, X., Wang, G., Yang, Y., Yuan, Y., and Ouyang, L. (2019b). The past, present and future of potential small-molecule drugs targeting p53-MDM2/MDMX for cancer therapy. Eur. J. Med. Chem. 176, 92–104. doi: 10.1016/j.ejmech.2019.05.018
Ma, M., and Yu, N. (2016). Ubiquitin-specific protease 7 expression is a prognostic factor in epithelial ovarian cancer and correlates with lymph node metastasis. Onco Targets Ther. 9, 1559–1569. doi: 10.2147/OTT.S100050
Masuya, D., Huang, C., Liu, D., Nakashima, T., Yokomise, H., Ueno, M., et al. (2006). The HAUSP gene plays an important role in non-small cell lung carcinogenesis through p53-dependent pathways. J. Pathol. 208, 724–732.
Meulmeester, E., Maurice, M. M., Boutell, C., Teunisse, A. F., Ovaa, H., Abraham, T. E., et al. (2005). Loss of HAUSP-mediated deubiquitination contributes to DNA damage-induced destabilization of Hdmx and Hdm2. Mol. Cell. 18, 565–576.
Momand, J., Zambetti, G. P., Olson, D. C., George, D., and Levine, A. J. (1992). The mdm-2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell 69, 1237–1245.
Nag, S., Qin, J., Srivenugopal, K. S., Wang, M., and Zhang, R. (2013). The MDM2-p53 pathway revisited. J. Biomed. Res. 27, 254–271. doi: 10.7555/JBR.27.20130030
Nicholson, B., and Suresh Kumar, K. G. (2011). The multifaceted roles of USP7: new therapeutic opportunities. Cell Biochem. Biophys. 60, 61–68. doi: 10.1007/s12013-011-9185-5
Oliner, J. D., Kinzler, K. W., Meltzer, P. S., George, D. L., and Vogelstein, B. (1992). Amplification of a gene encoding a p53-associated protein in human sarcomas. Nature 358, 80–83.
Pan, Y., and Chen, J. (2003). MDM2 promotes ubiquitination and degradation of MDMX. Mol. Cell. Biol. 23, 5113–5121.
Perry, M. E., Piette, J., Zawadzki, J. A., Harvey, D., and Levine, A. J. (1993). The mdm-2 gene is induced in response to UV light in a p53-dependent manner. Proc. Natl. Acad. Sci. U.S.A. 90, 11623–11627.
Pozhidaeva, A., and Bezsonova, I. (2019). USP7: structure, substrate specificity, and inhibition. DNA Repair 76, 30–39.
Pozhidaeva, A., Valles, G., Wang, F., Wu, J., Sterner, D. E., Nguyen, P., et al. (2017). USP7-specific inhibitors target and modify the enzyme’s active site via distinct chemical mechanisms. Cell Chem. Biol. 24, 1501.e5–1512.e5. doi: 10.1016/j.chembiol.2017.09.004
Qin, D., Wang, W., Lei, H., Luo, H., Cai, H., Tang, C., et al. (2016). CDDO-Me reveals USP7 as a novel target in ovarian cancer cells. Oncotarget 7, 77096–77109. doi: 10.18632/oncotarget.12801
Qin, J. J., Cheng, X. D., Zhang, J., and Zhang, W. D. (2019a). Dual roles and therapeutic potential of Keap1-Nrf2 pathway in pancreatic cancer: a systematic review. Cell Commun. Signal. 17:121. doi: 10.1186/s12964-019-0435-2
Qin, J. J., Li, X., Hunt, C., Wang, W., Wang, H., and Zhang, R. (2018). Natural products targeting the p53-MDM2 pathway and mutant p53: recent advances and implications in cancer medicine. Genes Dis. 5, 204–219. doi: 10.1016/j.gendis.2018.07.002
Qin, J. J., Li, X., Wang, W., Zi, X., and Zhang, R. (2017). Targeting the NFAT1-MDM2-MDMX network inhibits the proliferation and invasion of prostate cancer cells, independent of p53 and androgen. Front. Pharmacol. 8:917. doi: 10.3389/fphar.2017.00917
Qin, J. J., Wang, W., Sarkar, S., and Zhang, R. (2016b). Oral delivery of anti-MDM2 inhibitor SP141-loaded FcRn-targeted nanoparticles to treat breast cancer and metastasis. J. Control Release. 237, 101–114. doi: 10.1016/j.jconrel.2016.07.008
Qin, J. J., Wang, W., Sarkar, S., Voruganti, S., Agarwal, R., and Zhang, R. (2016a). Inulanolide A as a new dual inhibitor of NFAT1-MDM2 pathway for breast cancer therapy. Oncotarget 7, 32566–32578. doi: 10.18632/oncotarget.8873
Qin, J. J., Wang, W., Voruganti, S., Wang, H., Zhang, W. D., and Zhang, R. (2015). Identification of a new class of natural product MDM2 inhibitor: in vitro and in vivo anti-breast cancer activities and target validation. Oncotarget 6, 2623–2640.
Qin, J. J., Yan, L., Zhang, J., and Zhang, W. D. (2019b). STAT3 as a potential therapeutic target in triple negative breast cancer: a systematic review. J. Exp. Clin. Cancer Res. 38:195. doi: 10.1186/s13046-019-1206-z
Rafal, R., Jakub, W., and Joanna, J. A. (2019). MDM2-p53 Interaction Inhibitors: current State of Art and Updated Patent Review (2010-present). Recent Pat. Anticancer Drug Discov. 14, 324–369.
Rawat, R., Starczynowski, D. T., and Ntziachristos, P. (2019). Nuclear deubiquitination in the spotlight: the multifaceted nature of USP7 biology in disease. Curr. Opin. Cell Biol. 58, 85–94. doi: 10.1016/j.ceb.2019.02.008
Reverdy, C., Conrath, S., Lopez, R., Planquette, C., Atmanene, C., Collura, V., et al. (2012). Discovery of specific inhibitors of human USP7/HAUSP deubiquitinating enzyme. Chem. Biol. 19, 467–477. doi: 10.1016/j.chembiol.2012.02.007
Rouge, L., Bainbridge, T. W., Kwok, M., Tong, R., Di Lello, P., Wertz, I. E., et al. (2016). Molecular understanding of USP7 substrate recognition and C-terminal activation. Structure 24, 1335–1345. doi: 10.1016/j.str.2016.05.020
Saridakis, V., Sheng, Y., Sarkari, F., Holowaty, M. N., Shire, K., Nguyen, T., et al. (2005). Structure of the p53 binding domain of HAUSP/USP7 bound to Epstein-Barr nuclear antigen 1 implications for EBV-mediated immortalization. Mol. Cell. 18, 25–36.
Sarkari, F., La Delfa, A., Arrowsmith, C. H., Frappier, L., Sheng, Y., and Saridakis, V. (2010). Further insight into substrate recognition by USP7: structural and biochemical analysis of the HdmX and Hdm2 interactions with USP7. J. Mol. Biol. 402, 825–837. doi: 10.1016/j.jmb.2010.08.017
Senft, D., Qi, J., and Ronai, Z. A. (2018). Ubiquitin ligases in oncogenic transformation and cancer therapy. Nat. Rev. Cancer 18, 69–88.
Sheng, Y., Saridakis, V., Sarkari, F., Duan, S., Wu, T., Arrowsmith, C. H., et al. (2006). Molecular recognition of p53 and MDM2 by USP7/HAUSP. Nat. Struct. Mol. Biol. 13, 285–291.
Shvarts, A., Bazuine, M., Dekker, P., Ramos, Y. F., Steegenga, W. T., Merckx, G., et al. (1997). Isolation and identification of the human homolog of a new p53-binding protein, Mdmx. Genomics 43, 34–42.
Shvarts, A., Steegenga, W. T., Riteco, N., Van Laar, T., Dekker, P., Bazuine, M., et al. (1996). MDMX: a novel p53-binding protein with some functional properties of MDM2. EMBO J. 15, 5349–5357.
Song, M. S., Salmena, L., Carracedo, A., Egia, A., Lo-Coco, F., Teruya-Feldstein, J., et al. (2008). The deubiquitinylation and localization of PTEN are regulated by a HAUSP-PML network. Nature 455, 813–817. doi: 10.1038/nature07290
Stiasny, A., Freier, C. P., Kuhn, C., Schulze, S., Mayr, D., Alexiou, C., et al. (2017). The involvement of E6, p53, p16, MDM2 and Gal-3 in the clinical outcome of patients with cervical cancer. Oncol. Lett. 14, 4467–4476. doi: 10.3892/ol.2017.6752
Stolte, B., Iniguez, A. B., Dharia, N. V., Robichaud, A. L., Conway, A. S., Morgan, A. M., et al. (2018). Genome-scale CRISPR-Cas9 screen identifies druggable dependencies in TP53 wild-type Ewing sarcoma. J. Exp. Med. 215, 2137–2155. doi: 10.1084/jem.20171066
Su, D., Ma, S., Shan, L., Wang, Y., Wang, Y., Cao, C., et al. (2018). Ubiquitin-specific protease 7 sustains DNA damage response and promotes cervical carcinogenesis. J. Clin. Invest. 128, 4280–4296. doi: 10.1172/JCI120518
Tanimura, S., Ohtsuka, S., Mitsui, K., Shirouzu, K., Yoshimura, A., and Ohtsubo, M. (1999). MDM2 interacts with MDMX through their RING finger domains. FEBS Lett. 447, 5–9.
Tavana, O., and Gu, W. (2017). Modulation of the p53/MDM2 interplay by HAUSP inhibitors. J. Mol. Cell Biol. 9, 45–52. doi: 10.1093/jmcb/mjw049
Turnbull, A. P., Ioannidis, S., Krajewski, W. W., Pinto-Fernandez, A., Heride, C., Martin, A. C. L., et al. (2017). Molecular basis of USP7 inhibition by selective small-molecule inhibitors. Nature 550, 481–486. doi: 10.1038/nature24451
van der Horst, A., De Vries-Smits, A. M., Brenkman, A. B., Van Triest, M. H., Van Den Broek, N., Colland, F., et al. (2006). FOXO4 transcriptional activity is regulated by monoubiquitination and USP7/HAUSP. Nat. Cell. Biol. 8, 1064–1073.
van Loosdregt, J., Fleskens, V., Fu, J., Brenkman, A. B., Bekker, C. P., Pals, C. E., et al. (2013). Stabilization of the transcription factor Foxp3 by the deubiquitinase USP7 increases Treg-cell-suppressive capacity. Immunity 39, 259–271. doi: 10.1016/j.immuni.2013.05.018
Voruganti, S., Qin, J. J., Sarkar, S., Nag, S., Walbi, I. A., Wang, S., et al. (2015). Oral nano-delivery of anticancer ginsenoside 25-OCH3-PPD, a natural inhibitor of the MDM2 oncogene: nanoparticle preparation, characterization, in vitro and in vivo anti-prostate cancer activity, and mechanisms of action. Oncotarget 6, 21379–21394.
Walerych, D., Lisek, K., and Del Sal, G. (2015). Mutant p53: one, no one, and one hundred thousand. Front. Oncol. 5:289. doi: 10.3389/fonc.2015.00289
Wang, F., Wang, L., Wu, J., Sokirniy, I., Nguyen, P., Bregnard, T., et al. (2017a). Active site-targeted covalent irreversible inhibitors of USP7 impair the functions of Foxp3+ T-regulatory cells by promoting ubiquitination of Tip60. PLoS One 12:e0189744. doi: 10.1371/journal.pone.0189744
Wang, M., Zhang, Y., Wang, T., Zhang, J., Zhou, Z., Sun, Y., et al. (2017b). The USP7 Inhibitor P5091 Induces Cell Death in Ovarian Cancers with Different P53 Status. Cell Physiol. Biochem. 43, 1755–1766. doi: 10.1159/000484062
Wang, L., Kumar, S., Dahiya, S., Wang, F., Wu, J., Newick, K., et al. (2016a). Ubiquitin-specific Protease-7 Inhibition Impairs Tip60-dependent Foxp3+ T-regulatory Cell Function and Promotes Antitumor Immunity. EBioMedicine 13, 99–112. doi: 10.1016/j.ebiom.2016.10.018
Wang, Q., Ma, S., Song, N., Li, X., Liu, L., Yang, S., et al. (2016b). Stabilization of histone demethylase PHF8 by USP7 promotes breast carcinogenesis. J. Clin. Invest. 126, 2205–2220. doi: 10.1172/JCI85747
Wang, W., Cheng, J. W., Qin, J. J., Hu, B., Li, X., Nijampatnam, B., et al. (2019a). MDM2-NFAT1 dual inhibitor, MA242: effective against hepatocellular carcinoma, independent of p53. Cancer Lett. 459, 156–167. doi: 10.1016/j.canlet.2019.114429
Wang, W., Qin, J. J., Li, X., Tao, G., Wang, Q., Wu, X., et al. (2018a). Prevention of prostate cancer by natural product MDM2 inhibitor GS25: in vitro and in vivo activities and molecular mechanisms. Carcinogenesis 39, 1026–1036. doi: 10.1093/carcin/bgy063
Wang, W., Qin, J. J., Rajaei, M., Li, X., Yu, X., Hunt, C., et al. (2019b). Targeting MDM2 for novel molecular therapy: beyond oncology. Med. Res. Rev. doi: 10.1002/med.21637 [Epub ahead of print].
Wang, W., Qin, J. J., Voruganti, S., Nijampatnam, B., Velu, S. E., Ruan, K. H., et al. (2018b). Discovery and characterization of dual inhibitors of MDM2 and NFAT1 for pancreatic cancer therapy. Cancer Res. 78, 5656–5667. doi: 10.1158/0008-5472.CAN-17-3939
Wang, W., Qin, J. J., Voruganti, S., Srivenugopal, K. S., Nag, S., Patil, S., et al. (2014a). The pyrido[b]indole MDM2 inhibitor SP-141 exerts potent therapeutic effects in breast cancer models. Nat. Commun. 5:5086.
Wang, W., Qin, J. J., Voruganti, S., Wang, M. H., Sharma, H., Patil, S., et al. (2014b). Identification of a new class of MDM2 inhibitor that inhibits growth of orthotopic pancreatic tumors in mice. Gastroenterology 147, 893.e2–902.e2. doi: 10.1053/j.gastro.2014.07.001
Weinstock, J., Wu, J., Cao, P., Kingsbury, W. D., Mcdermott, J. L., Kodrasov, M. P., et al. (2012). Selective dual inhibitors of the cancer-related deubiquitylating proteases USP7 and USP47. ACS Med. Chem. Lett. 3, 789–792. doi: 10.1021/ml200276j
Wu, J., Kumar, S., Wang, F., Wang, H., Chen, L., Arsenault, P., et al. (2018). Chemical approaches to intervening in ubiquitin specific protease 7 (USP7) function for oncology and immune oncology therapies. J. Med. Chem. 61, 422–443. doi: 10.1021/acs.jmedchem.7b00498
Xia, X., Liao, Y., Huang, C., Liu, Y., He, J., Shao, Z., et al. (2019). Deubiquitination and stabilization of estrogen receptor alpha by ubiquitin-specific protease 7 promotes breast tumorigenesis. Cancer Lett. 465, 118–128.
Yamaguchi, M., Miyazaki, M., Kodrasov, M. P., Rotinsulu, H., Losung, F., Mangindaan, R. E., et al. (2013). Spongiacidin C, a pyrrole alkaloid from the marine sponge Stylissa massa, functions as a USP7 inhibitor. Bioorg. Med. Chem. Lett. 23, 3884–3886.
Zhang, X., Gu, L., Li, J., Shah, N., He, J., Yang, L., et al. (2010). Degradation of MDM2 by the interaction between berberine and DAXX leads to potent apoptosis in MDM2-overexpressing cancer cells. Cancer Res. 70, 9895– 9904.
Zhao, G. Y., Lin, Z. W., Lu, C. L., Gu, J., Yuan, Y. F., Xu, F. K., et al. (2015). USP7 overexpression predicts a poor prognosis in lung squamous cell carcinoma and large cell carcinoma. Tumour Biol. 36, 1721–1729.
Zheng, N., and Shabek, N. (2017). Ubiquitin ligases: structure, function, and regulation. Annu. Rev. Biochem. 86, 129–157.
Keywords: USP7, deubiquitination, p53, MDM2, MDMX, small-molecule inhibitor, cancer
Citation: Qi S-M, Cheng G, Cheng X-D, Xu Z, Xu B, Zhang W-D and Qin J-J (2020) Targeting USP7-Mediated Deubiquitination of MDM2/MDMX-p53 Pathway for Cancer Therapy: Are We There Yet? Front. Cell Dev. Biol. 8:233. doi: 10.3389/fcell.2020.00233
Received: 28 October 2019; Accepted: 19 March 2020;
Published: 02 April 2020.
Edited by:
Luisa Lanfrancone, European Institute of Oncology (IEO), ItalyCopyright © 2020 Qi, Cheng, Cheng, Xu, Xu, Zhang and Qin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiang-Dong Cheng, Y2hlbmd4ZDUxNkAxMjYuY29t; Wei-Dong Zhang, d2R6aGFuZ3lAaG90bWFpbC5jb20=; Jiang-Jiang Qin, anFpbkB6Y211LmVkdS5jbg==; enlseXNqdHVAaG90bWFpbC5jb20=
 Si-Min Qi
Si-Min Qi Gang Cheng1
Gang Cheng1 Wei-Dong Zhang
Wei-Dong Zhang Jiang-Jiang Qin
Jiang-Jiang Qin