Abstract
Long interspersed nuclear element-1 (LINE-1) retrotransposition is a major hallmark of cancer accompanied by global chromosomal instability, genomic instability, and genetic heterogeneity and has become one indicator for the occurrence, development, and poor prognosis of many diseases. LINE-1 also modulates the immune system and affects the immune microenvironment in a variety of ways. Aberrant expression of LINE-1 retrotransposon can provide strong stimuli for an innate immune response, activate the immune system, and induce autoimmunity and inflammation. Therefore, inhibition the activity of LINE-1 has become a potential treatment strategy for various diseases. In this review, we discussed the components and regulatory mechanisms involved with LINE-1, its correlations with disease and immunity, and multiple inhibitors of LINE-1, providing a new understanding of LINE-1.
Introduction
Long interspersed nuclear elements (LINEs) are the only autonomous and active retrotransposons, which include LINE-1, LINE-2, and LINE-3 (Cordaux and Batzer, 2009; de Koning et al., 2011). Also, 5–6% of LINE-2 and LINE-3 sequences in the human genome are as a truncated molecular fossil (Doxiadis et al., 2012; Ardeljan et al., 2017). LINE-1 retrotransposons are one of the most abundant and effective classes of mobile DNAs that account for 17% of the human genome (Lander et al., 2001; Hancks and Kazazian, 2016). Full-length LINE-1 is 6.0–7.0 kb and contains a 5′-untranslated region (5′-UTR) (Swergold, 1990), two open reading frames (ORF1 and ORF2), and a 3′-UTR punctuated with a poly-A tract (Babushok and Kazazian, 2007; Beck et al., 2011). Denli et al. (2015) revealed a new open reading frame, ORF0. It is located in the 5′-UTR of the LINE-1 transcript and on the strand opposite of the ORF1 and ORF2 structural genes. Antisense promotor (ASP) can initiate fusion transcripts and regulate ORF0 to enhance LINE-1 mobility (Roman-Gomez et al., 2005; Weber et al., 2010; Criscione et al., 2016).
Both ORFs are required for LINE-1 retrotransposition process. ORF1 encodes an RNA-binding protein named ORF1P that has nucleic acid chaperone activity, and ORF2 encodes a protein named ORF2P that has endonuclease and reverse-transcriptase activities (Mathias et al., 1991; Feng et al., 1996). The first step occurs when RNA polymerase II binds to the 5′-UTR promoter region of LINE-1 and mediates the transcription of full-length mRNA of LINE-1 (Lavie et al., 2004). The LINE-1 mRNA is exported to the cytoplasm where ORF1 and ORF2 are translated and combined to form a ribonucleoprotein (RNP) particle. The RNP is then incorporated into the nucleus, and the ORF2P endonuclease in the RNP identifies and cuts specific sequences on the bottom DNA strand at the consensus site 3AA/TTTT−5′. Subsequently, the free 3′ hydroxyl generated at the fracture is utilized by the ORF2P and LINE-1 mRNA in the RNP is used as the template for reverse transcription to produce the complementary DNA of the LINE-1 gene (Wei et al., 2001; Hancks and Kazazian, 2016; Wang and Jordan, 2018). The distribution of LINE-1 in the human genome is selective. LINE-1 endonuclease activity and DNA replication determine LINE-1 insertion preference (Flasch et al., 2019). For example, LINE-1 preferentially inserts into nucleosome-depleted DNA primarily as a result of its AT-rich sequences (Sultana et al., 2019). The direction of the DNA replication fork affects LINE-1 insertion preference because the cleaved strand is usually the lagging strand template.
LINE-1 elements play a crucial role in the course of species formation and evolution. On one hand, de-repressed LINE-1 functions as a driver of many diseases and even a diagnostic marker for some diseases (Pedersen and Zisoulis, 2016). On the other, it can affect the developmental processes and influence the behavior by generating multiple gene products and causing variable deleterious effects on the structure of the host genome through new insertions, deletions, and recombinations (Garcia-Perez et al., 2016). LINE-1 RNA and protein overexpression is related to apoptosis, DNA damage and repair, cellular plasticity, and stress responses and can even promote tumor progression (Morrish et al., 2002, 2007; Belgnaoui et al., 2006; Sinibaldi-Vallebona et al., 2006). DNA damage caused by genome-wide or intersperse repetitive sequences hypomethylation can induce inflammatory microenvironment (Lindqvist et al., 2017; Teerawattanapong et al., 2019). Here, we reviewed the correlation between LINE-1 and disease as well as immune system, meanwhile, conducted a new exploration in LINE-1 inhibitors by combining its regulation mechanisms.
Figure 1 shows the relative positions of the 5′ untranslated region (5′-UTR); the open reading frames ORF0, ORF1, and ORF2; the 3′ untranslated region (3′-UTR); and the Poly A tail. ORF2 encodes endonuclease (EN), reverse transcriptase (RT), and cysteine-rich domain (C). Full-length LINE-1 mRNA was generated using the sense promoter at 5′-UTR. The LINE-1 mRNA is exported to the cytoplasm where ORF1 and ORF2 are translated and combined to form a ribonucleoprotein (RNP) particle. The RNP is then incorporated into the nucleus, and the ORF2P endonuclease in the RNP identifies and cuts specific sequences on the bottom DNA strand at the consensus site 3AA/TTTT−5′. Subsequently, the free 3′ hydroxyl generated at the fracture is utilized by the ORF2P and LINE-1 mRNA in the RNP is used as the template for reverse transcription to produce the complementary DNA of the LINE-1 gene (Richardson et al., 2015; Kazazian and Moran, 2017).
FIGURE 1
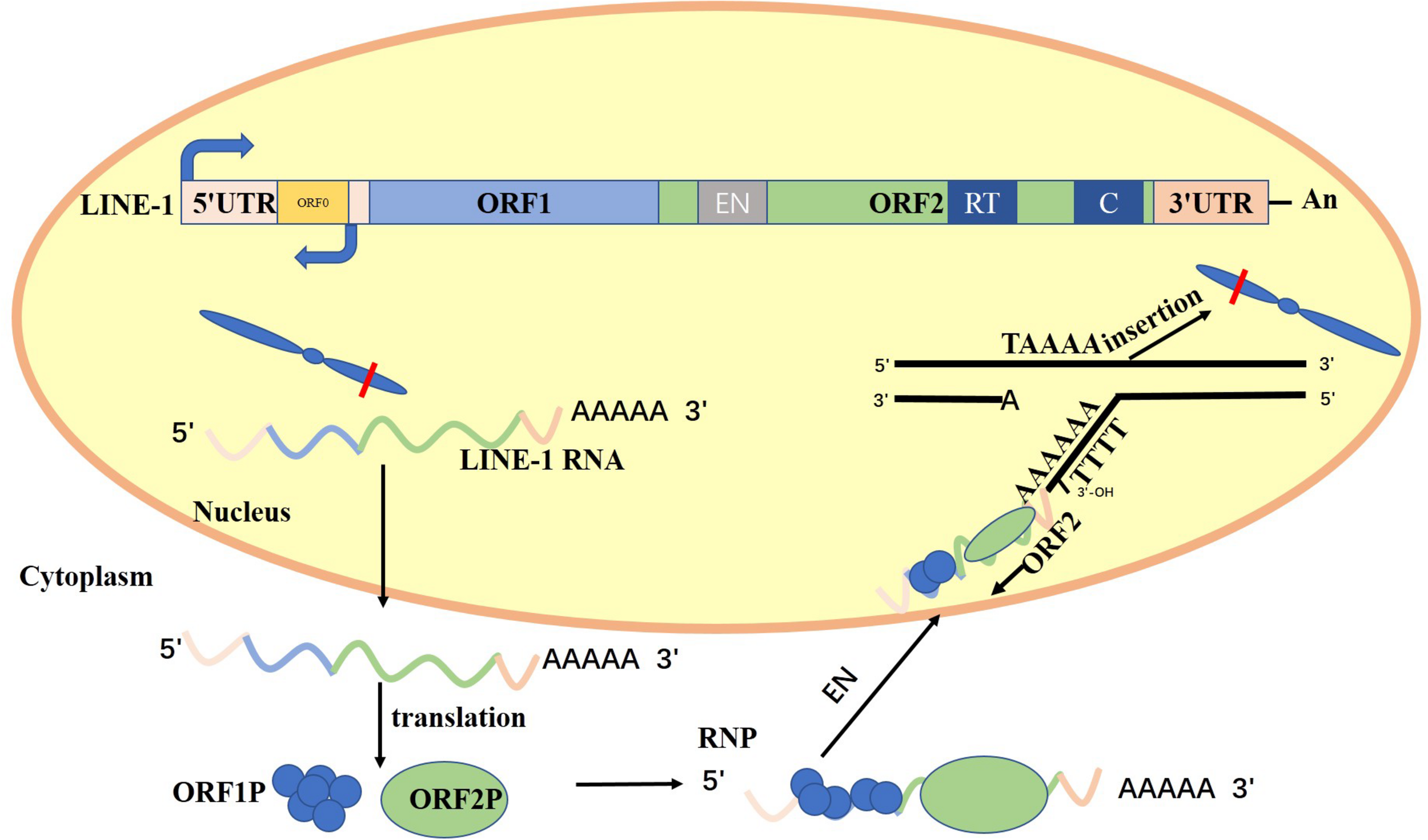
Structure of LINE-1 and LINE-1 retrotransposition cycle.
Line-1 and Disease
LINE-1 and Cancer
When LINE-1 retrotransposition is out of control, it can lead to diseases. More than 1,000 articles focusing on LINE-1 and cancer are available in the PubMed archive (Rodic, 2018).
LINE-1 Hypomethylation and Cancer
The global hypomethylation of the genome promotes chromosomal instability, genomic instability, and genetic heterogeneity because specific changes in DNA methylation can affect a variety of genome sequences, especially the intergenic and intronic regions of the DNA, resulting in chromosome instability and mutations (Wilson et al., 2007). LINE-1 promoter hypomethylation is a biomarker for genome-wide DNA hypomethylation, which is itself a major hallmark of cancer. Thayer et al. (1993) first demonstrated the methylation status of LINE-1 in cancer cells. Since then, LINE-1 hypomethylation of tumors has attracted widespread attention (Thayer et al., 1993). LINE-1 hypomethylation was reported to be associated with poor survival in more than 200 cases of gastric cancer, suggesting its potential as a prognostic biomarker (Shigaki et al., 2013). This phenomenon was also subsequently found in lung cancer, liver cancer, esophageal cancer, prostate cancer, and endometrial cancer (Iwagami et al., 2013; Kawano et al., 2014; Lavasanifar et al., 2019). Ogino et al. (2008) analyzed 643 colon cancer samples from two independent prospective cohorts, demonstrating a linear correlation between LINE-1 hypomethylation and aggressive tumor behavior. It has been reported that global DNA hypomethylation promotes aggressive tumor behavior by amplifying oncogenes or through abnormal expression of microRNAs (Baba et al., 2014, 2018). In esophageal cancer with high mortality and poor endoscopic screening sensitivity, LINE-1 hypomethylation can serve as a good diagnostic biomarker, thereby improving 5-year survival (Shah et al., 2013). LINE-1 hypomethylation can also be seen in some precancerous lesions. For example, in colorectal cancer, LINE-1 hypomethylation had no significant difference between adenomas and cancerous tissues, but it was significantly lower in adenomas than in normal tissues (Dawwas, 2014). Therefore, LINE-1 hypomethylation also can be used as an early biomarker for cancer.
However, there was no significant difference in the hypomethylation of LINE-1 between the blood samples of patients with leukemia and those of normal subjects (Barchitta et al., 2014).
LINE-1 Integrations and Cancer
Many tumor tissues have been found to present a high level of LINE-1 activity that can rapidly increase their copy number through the “copy-and-paste” mechanism (Dunaeva et al., 2018). LINE-1 can be used as cis-regulatory elements to regulate the expression of host genes (Wanichnopparat et al., 2013). Pan-cancer Analysis of Whole Genomes analysis of 2,954 cancer genomes from 38 histological subtypes revealed that aberrant LINE-1 integrations could lead to gene rearrangement (Rodriguez-Martin et al., 2020). LINE-1-mediated rearrangement can trigger oncogene amplification. In breast cancer, Morse and colleagues first proposed that hypomethylation activates LINE-1 which can utilize the target primed reverse transcription pathway to insert into the oncogene MYC, causing tumor-specific rearrangement and amplification (Morse et al., 1988). LINE-1 was found to induce the amplification of CCND1 oncogene in esophageal tumor by inducing the breakage–fusion–bridge cycles (Rodriguez-Martin et al., 2020). LINE-1 can mediate the deletion of tumor suppressor genes. It may be through X inactivation mechanism that LINE-1 mRNA forms facultative heterochromatin in the inactivated region or LINE-1 mRNA and pre-mRNA form RISC complex to degrade complementary mRNA (Allen et al., 2003; Aporntewan et al., 2011). In colon cancer, Miki et al. reported that LINE-1 insertion disrupts the tumor suppressor gene APC, which can lead to gene inactivation (Miki et al., 1992). In lung squamous cell carcinoma, we found that LINE-1 insertion into tumor suppressor gene FGGY promotes cell proliferation and invasion in vitro, and facilitates tumorigenesis in vivo (Zhang et al., 2019).
High Expression of ORF1 and ORF2 of LINE-1 and Cancer
The activation of LINE-1 increases the translation of ORF1 and ORF2, which are not expressed in normal somatic tissues. ORF1 encodes an RNA-binding protein, and high expression level of ORF1 was proved to be more common in most of the cancers and therefore as a diagnostic marker. In breast cancer, high expression of nuclear ORF1 is associated with distant metastasis and poor prognosis (Harris et al., 2010). In high-grade ovarian carcinoma, the ORF1 level was high and correlated to the loss of TP53 (Rodic et al., 2014). The expression of both the LINE-1 ORF1 and c-Met protein was significantly increased and peaked in early stage in ovarian cancer, suggesting that LINE-1 ORF1 significantly activates c-Met (Ko et al., 2019). In tumor cell experiments, increased mRNA and protein expression of LINE1-ORF1 can result in significant enhancement in cell proliferation and colony formation (Tang et al., 2018). It is worth noting that the expression of ORF1 was heterogeneous and had histological specificity. Cancers originating in the endometrium, such as biliary tract, esophagus, bladder, head and neck, lung, and colon, exhibit ORF1 overexpression, whereas other cancers, such as renal, liver, and cervical cancer, show little expression of ORF1 (Ardeljan et al., 2017). Recent studies have shown that an ELISA method to measure ORF1 in serum can be better in prostate cancer detection (Hosseinnejad et al., 2018).
ORF2 encodes a protein with reverse transcriptase and endonuclease activities. High expression of endonuclease induces double-strand DNA breakage that can aggravate DNA damage repair and increase genomic instability (Kines et al., 2014). Reverse transcriptase activation can promote cell proliferation and differentiation and also alter the non-coding RNA transcription spectrum and other epigenetic phenotypes, resulting in alterations in cell regulatory networks, tumor development, and other important pathological processes (Rodic and Burns, 2013; Burns, 2017; Christian et al., 2017). ORF2 can express early in the tumorigenesis process, as it can be detected by a highly specific monoclonal antibody (mAb chA1-L1) in both transitional colon mucosa and prostate intraepithelial neoplasias (De Luca et al., 2016). However, studies have shown that chA1-L1 recognizes both ORF2p and the transcriptional regulator BCLAF1, so it is not specific (Briggs et al., 2019). But recently, tumor proteome profiling studies based on mass spectrometry have shown that ORF2p was difficult to be detected, and after affinity capture of ORF1p, ORF2p has not been detected in stem cell LINE-1 proteome analysis (Vuong et al., 2019; Ardeljan et al., 2020). Therefore, the detection and application of ORF2 in tumors are still worth exploring.
LINE-1 and Metabolic Disorders
New research has shown that LINE-1 is also associated with blood sugar and lipid levels (Turcot et al., 2012). LINE-1 methylation is associated with type 2 diabetes mellitus (T2DM). Studies showed that, compared with hypermethylation, LINE-1 hypomethylation was associated with a higher risk of worsening metabolic status, independent of other classic risk factors (Martin-Nunez et al., 2014). This discovery highlights the potential role for LINE-1 DNA methylation as a predictor of the risk of T2DM or other related metabolic disorders. LINE-1 DNA methylation is associated with increased LDL cholesterol and decreased HDL cholesterol levels, and these metabolic changes increase the risk of cardiovascular disease (Pearce et al., 2012). LINE-1 DNA methylation is also associated with many blood-based metabolic biomarkers. In fetal neural tissue with neural tube defects, it was found that the low methylation level of LINE-1 was associated with the significant reduction of vitamin B12 in maternal plasma, as well as lower folate levels and increased concentrations of homocysteine (Wang et al., 2010). Folic acid and other B vitamins play an important role in the biosynthesis of new purines and pyrimidines. Therefore, the methylation status of LINE-1 can be a predictor of some metabolic diseases. Current studies have shown that LINE-1 can also regulate metabolism by inserting metabolic genes. It was reported that LINE-1 insertions in the FGGY gene can upregulate cytochrome P450, arachidonic acid metabolism, and glycerolipid metabolism. These metabolic disorders can lead to the occurrence of a variety of diseases and poor prognosis (Zhang et al., 2019).
LINE-1 and Neurological Disorders
LINE-1 can affect the developing brain at different stages of health and disease (Suarez et al., 2018). Ataxia telangiectasia (AT) is a progressive neurodegenerative disease caused by ataxia telangiectasia mutated (ATM) gene mutation. In 2011, researchers found that in nasopharyngeal carcinomas with ATM deficiency, LINE-1 retrotransposition increased, and ORF2 copy number increased in AT neurons, thus verifying the correlation between LINE-1 retrotransposition and ATM deficiency (Coufal et al., 2011). High expression of LINE-1 was found in Rett syndrome caused by mutation of methyl CpG binding protein 2 (MeCP2) in the X-linked gene, which was caused by the inclusion of LINE-1 5′-UTR sequence in the MeCP2 target, leading to methylation-dependent repression (Muotri et al., 2010). LINE-1 is involved in the aging process. In patients with frontotemporal lobe degeneration, LINE-1 transcripts were found to be elevated (Li et al., 2012). LINE-1 hypomethylation has been observed in most psychiatric studies. Increased copy numbers of LINE-1 as a result of LINE-1 hypomethylation were also found in patients with schizophrenia, bipolar disorder, and major depressive disorder (Liu et al., 2016; Li et al., 2018). The link between LINE-1 methylation levels and Alzheimer’s disease is still being studied.
LINE-1 and Genetic Disorders
LINE-1 is reported to be related to chromosome disorders. The first observation of LINE-1 insertion was in 1988, when Kazazian et al. observed a new exon of F8 LINE-1 insertion in the X-linked gene, which is a gene encoding coagulation factor VIII in a patient with hemophilia A (Kazazian et al., 1988). Then, a LINE-1 insertion was found in the CHM gene of a patient diagnosed with choroideremia. The reverse integration of a LINE-1 element into exon 6 resulted in aberrant splicing of the CHM mRNA (van den Hurk et al., 2003). Furthermore, LINE-1 can also promote mobilization of other RNAs in trans, Alu, and SVA, which can be trans-mobilized, leading to gene insertions (Kemp and Longworth, 2015). Retrotransposon insertions were found to account for up to 0.4% of all NF1 mutations (Wimmer et al., 2011). Neurofibromatosis type I is an autosomal dominant disorder caused by NF1 gene mutations (Messiaen et al., 2011). Alu insertion is located 44 bp upstream of NF1 exon 41, causing the exon 41 to skip and change the open reading frame (Payer and Burns, 2019). Only two cases were thought to be a result of independent SVA insertion in SUZ12P accompanied by 867-kb and 1-Mb deletions that encompassed the NF1 gene (Vogt et al., 2014). In autosomal recessive genetic disease, such as Fanconi anemia caused by SLX4FANCP deficiency and Aicardi–Goutieres syndrome (AGS) of three-prime repair exonuclease 1 mutations, LINE-1 expression was upregulated and pro-inflammatory cytokines were produced through the cGAS–STING pathway (Brégnard et al., 2016; Suarez et al., 2018).
Line-1 and Immune Regulation of Disease
LINE-1 and Autoimmune Disease
Hypomethylated and highly expressed LINE-1 has been found in autoimmune diseases such as systemic lupus erythematosus (SLE), Sjögren’s syndrome (SS), and psoriasis (Schulz et al., 2006; Yooyongsatit et al., 2015; Mavragani et al., 2016). LINE-1 RNA is characterized by viral RNA and exists as RNP particles, which can be recognized by RNA sensors and activate innate immune responses (Mavragani et al., 2016). Cell studies demonstrated that LINE-1 activates the production of IFNβ by RNA pathway (Zhao et al., 2018). When LINE-1 retrotransposition was inhibited by RT inhibitors, significant reductions were observed in IFNα, IFNβ, and IFNγ mRNA levels (Brégnard et al., 2016). LINE-1 transcripts and p40 protein (a 40−kDa RNA binding protein) that LINE-1 encodes have been detected in SLE and SS patients. It has been demonstrated that LINE-1 can induce the production of IFN-I in vitro by TLR-dependent and TLR-independent pathways (Mavragani et al., 2016). In MRL autoimmune lymphoproliferative syndrome, LINE-1 ORF2 encoding an RT and its products are associated with an MHC class I molecule on the cell membrane (Benihoud et al., 2002). In Fanconi anemia and AGS, LINE-1 was found to be associated with the activation of the autoimmune system. LINE-1 also regulates immunity by acting as a cis-regulatory element through the mechanism of LINE-1 mRNA and pre-mRNA forming RISC complex to degrade the complementary mRNA (Wanichnopparat et al., 2013).
LINE-1 and Tumor Immunity
In 112 TCGA cancer samples, the scientists measured the transcriptional activity of 1789 pathways and found that 49 of 176 immune pathways were significantly negatively correlated with LINE-1 (Jung et al., 2018). LINE-1 is inversely correlated with the expression of immunologic response genes. Less LINE-1 activity was found in tumors with high immune activity. In esophageal cancer tissues, scientists found that the LINE-1 methylation level in tumors was significantly positively associated with the peritumoral lymphocytic reaction (Kosumi et al., 2019). The activities of regulatory T cells and PD1 signaling as reported in cancer immune evasion and chronic inflammatory conditions also have negative correlations with LINE-1. It is reported that the negative correlation between LINE-1 and immune activity may be caused by the destruction of LINE-1 inhibition, but the specific mechanism is still unclear. LINE-1 may also mediate immune tolerance, which may change from immune stimulation mode to immunosuppression mode through continuous IFN signaling or directly affect lymphocyte signaling.
LINE-1 and Metabolism-Induced Immunity
LINE-1 is also associated with blood sugar and lipid levels. Abnormal glucose and lipid metabolism can lead to metabolic reprogramming in tumor cells. The most classic metabolism of tumor is Warburg effect, where a large amount of glucose is absorbed to fulfill the need for proliferation and produce lactic acid (Lunt and Vander Heiden, 2011). The acidic microenvironment caused by lactic acid leads to impaired T-cell activation and proliferation, prevents NK cell activation, stabilizes HIF1α to stimulate the polarization of anti-inflammatory M2 macrophages, and inhibits the production of IFN-γ in tumor-infiltrating T cells (Husain et al., 2013; Colegio et al., 2014; Brand et al., 2016). Abnormal lipid metabolism in tumor cells also can lead to local immunosuppression in the microenvironment (Hao et al., 2019). LINE-1 can affect local immune homeostasis by inserting elements into metabolism-related genes. FGGY is known to encode a protein that phosphorylates carbohydrates and is associated with obesity and sporadic amyotrophic lateral sclerosis (Zhang et al., 2011). LINE-1 retrotransposons suppress FGGY, leading to lipid metabolism disturbance and diet-induced obesity in mice (Taylor et al., 2018). Lung squamous cell carcinoma patients with L1-FGGY+ tissue have a poor prognosis, have low levels of CD3+ T cells, and have high levels of CD68+ macrophages and CD33+ myeloid-derived cells (Zhang et al., 2019). L1-FGGY+ also regulates the abnormal transcription of cytokines related to the immunosuppressive micromilieu.
Line-1 Inhibition
The correlation between LINE-1 and disease as well as immunity was analyzed (Figure 2). The life cycle of LINE-1 provides a plethora of ways to target and inhibit LINE-1 expression (Banuelos-Sanchez et al., 2019). The inhibition of LINE-1 has become a treatment strategy for some diseases.
FIGURE 2
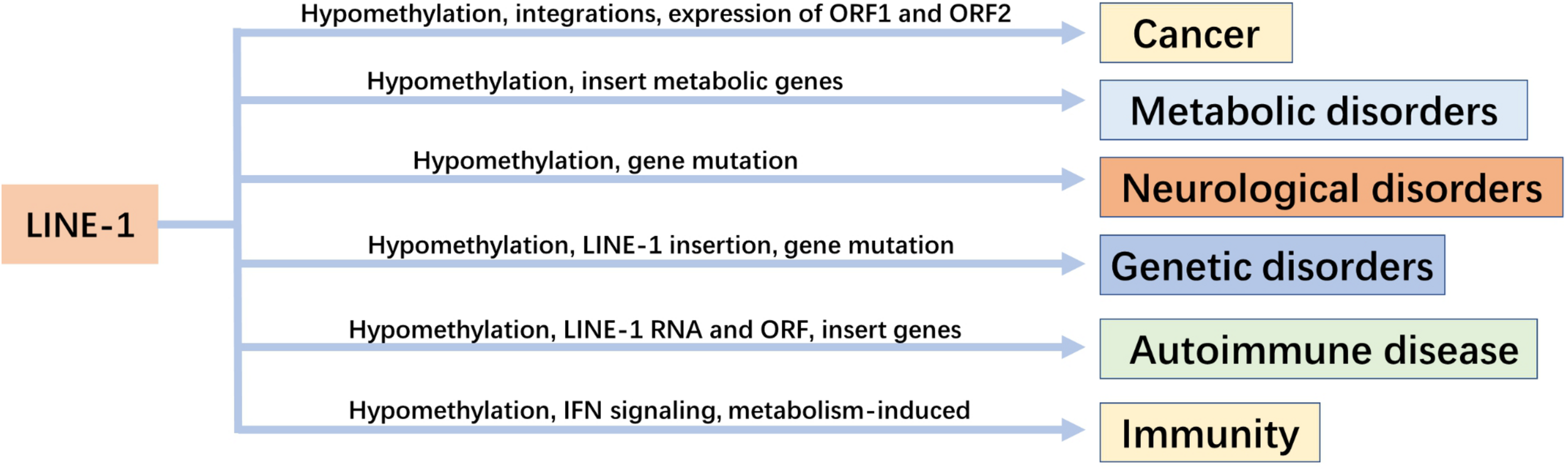
The relationship between LINE-1 and diseases and their regulatory mechanisms.
Targeting LINE-1 Methylation
Full-length LINE-1 transcription is driven by a CpG dinucleotide-rich internal promoter. Hypomethylation of LINE-1 causes the activation of LINE-1, which causes retroelement transposition and chromosomal alteration (Saito et al., 2010). The hypomethylation of LINE-1 has become an important factor in the occurrence and development of diseases, so maintaining the state of LINE-1 methylation has become a key method for the treatment of diseases. Soy isoflavone supplementation can regulate the level of LINE-1 methylation in head and neck squamous cell carcinoma (HNSCC). In a clinical trial of 39 patients with HNSCC who took a soy isoflavone supplement (300 mg/day) orally for 3 weeks before surgery, a positive correlation was found between LINE-1 methylation level and daily isoflavone intake (Rozek et al., 2019). Some cell-based studies and clinical data have shown that LINE-1 dysregulation is associated with tumor drug resistance (Zhu et al., 2015; Lavasanifar et al., 2019). It was found in breast cancer cells treated with paclitaxel that DNMT3a, a member of the DNA methyltransferase family, could enhance the methylation level in the gene by binding to the inner region of the LINE-1 gene, and then upregulate the expression level of LINE-1. Downregulating the expression of DNMT3a can effectively inhibit the expression of LINE-1 (Wang et al., 2020). LINE-1 retrotransposon silenced also through histone modifications. Histone demethylase KDM4B may enhance the LINE-1 retrotransposition efficacy, whereas depletion of KDM4B reduced it in breast cancer (Xiang et al., 2019). Elevated LINE-1 expression was found in PC9 drug-tolerant persister (DTP) cancer cells treated with the EGFR inhibitor erlotinib. HDAC inhibitors can suppress LINE-1 in DTP cancer cells (Guler et al., 2017). Currently, DNA methyltransferase inhibitors and histone deacetylase inhibitors have entered clinical trials (Gaillard et al., 2019).
Targeting RT Activity
LINE-1 elements harbor ORF1 and ORF2, which has reverse transcriptase (RT) activity, and RT inhibition may be a novel, non-cytotoxic anticancer therapy strategy (Sciamanna et al., 2018). RT is a key player in retrotransposition and functions by transcribing LINE-1 mRNA or other RNAs to cDNA at the integration sites (Khalid et al., 2018). Specific reverse transcription inhibitors, including nevirapine (NVR) and efavirenz (EFV), which target the HIV-1-encoded RT and are currently used in AIDS therapy, reduce cell proliferation and promotes differentiation of a variety of cancer cell lines of unrelated histological origin (Mangiacasale et al., 2003; Landriscina et al., 2005; Sciamanna et al., 2005). In vivo assays using murine models inoculated with various human cancer cell lines revealed that daily treatment of animals with EFV significantly delayed the progression of tumors (Oricchio et al., 2007). NVR and EFV dramatically countered L1-FGGY abundance, inhibited tumor growth, attenuated metabolism dysfunction, and improved the local immune evasion in lung squamous cell carcinomas (Zhang et al., 2019). EFV has recently undergone a phase II clinical trial in patients with metastatic prostate cancer (Houédé et al., 2014). Another RT inhibitor, F2-DABOs, has shown anti-proliferative activity in nude mice, helping to promote cell differentiation and inhibit tumor growth (Sbardella et al., 2011). Later, the nucleoside reverse transcription inhibitor abacavir was also shown to inhibit cell growth, migration, and invasion (Carlini et al., 2010). Capsaicin is the main chemical component of Asiasari radix and Capsicum annuum, as well as the major component of a Chinese traditional herbal medicine, Sho-seiryu-to (Friedman et al., 2018). Capsaicin suppresses LINE-1 by inhibiting the RT activity of LINE-1 ORF2P, which is the LINE-1-encoded RT responsible for LINE-1 activity (Nishikawa et al., 2018). A recent study revealed that azidothymidine (AZT) inhibits the RT activity of ORF2P in a fetal oocyte attrition model. Experiments showed that AZT-treated oocytes have a reduction of LINE-1 ORF1 ssDNA compared with untreated oocytes (Tharp et al., 2020). It is important to note that RT inhibitors do not eliminate the tumor but only control its progression. Therefore, in addition to the anti-AIDS drugs approved by the FDA, the combination of Chinese and western medicine can be regarded as an emerging treatment.
Combined Immunotherapy
Recent studies suggest that LINE-1 hypomethylation may be a positive indicator of immunotherapy. DNA methyltransferase (DNMT) is an important epigenetic molecule that catalyzed DNA methylation and can induce the development of various tumors. Downregulating the expression of DNMT can effectively inhibit the expression of LINE-1 (Wang et al., 2020). So DNA methyltransferase inhibitors (DNMTis) play an important role in the anti-tumor process. DNMTI can improve tumor immunogenicity, promote NK cells and CD8+ T cells to play a cell-mediated cytotoxic role, and promote immune response to participate in antigen commission by regulating immunosuppressive cells (Chiappinelli et al., 2015). DNMTi can enhance the expression of cancer-testis (CT) antigen, making the tumor more susceptible to CT antigen vaccine. The combination of decitabine, a DNA methyltransferase inhibitor, and cancer-testis/cancer-germline antigen NY-ESO-1 vaccine has a good therapeutic effect in the primary treatment of human recurrent epithelial ovarian cancer (Odunsi et al., 2014). A clinical trial has shown that combination therapy with carboplatin and anti-programmed death-1 has a good therapeutic effect in lung cancer because carboplatin can induce LINE-1 expression (Langer et al., 2016). Therefore, LINE-1 can be used as a target of combined immunotherapy in tumor therapy.
Other Inhibitors
Recently, a number of other regulatory approaches have been reported. In somatic cells, microRNAs (miRNAs or miRs) also regulate the activity of LINE-1 (Idica et al., 2017). MiR-128 regulates LINE-1 activity in somatic cells by targeting the nuclear import factor transportin-1 (TNPO1) 3′-UTR, which mediates nuclear import and requires RanGTP for cargo delivery into the nucleus (Twyffels et al., 2014). MiR-128 inhibits the expression of TNPO1 mRNA and protein, and TNPO1 deficiency suppresses LINE-1 mobilization by inhibiting nuclear import of LINE-1–RNP (Idica et al., 2017). MiR-128 also guides the miRNA-induced silencing complex to bind directly to a target site residing in the ORF2 RNA of LINE-1 (Hamdorf et al., 2015). At present, a novel target of miR-128 has been identified as heterogeneous nuclear ribonucleoprotein A1 (hnRNPA1), which is required for LINE-1 retrotransposition (Goodier et al., 2013; Fung et al., 2019). MiR-128 represses hnRNPA1 mRNA and protein by targeting the CDS of hnRNPA1, which interacts with LINE-1 ORF1p via RNA bridge to promote LINE-1 mobilization (Goodier et al., 2013). This interaction results in translational repression of the LINE-1 retrotransposition, thereby reducing the risk of LINE-1-mediated mutagenesis (Pedersen and Zisoulis, 2016). Therefore, microRNAs can be a target for LINE-1 inhibition.
Aryl hydrocarbon receptor (AHR) is a ligand-activated transcription factor that activates LINE-1 expression (Teneng et al., 2007). AHR is overexpressed in breast and thyroid cancers, suggesting that these tumors also overexpress LINE-1 (Powell et al., 2013). Lai et al. found that biseugenol, a novel AHR inhibitor, impeded cancer growth and inhibited EMT in gastric cancer cells (Lai et al., 2014). These findings suggest that targeting AHR with small molecule inhibitors may be a novel therapeutic approach. ORF1P phosphorylation by protein kinase A is also required for LINE-1. Kinase inhibitors specifically designed to target LINE-1 ORF1P phosphorylation may be associated with inhibition of LINE-1 (Bojang et al., 2013). Therefore, there is room for drug development research focusing on targeting and inhibiting LINE-1 ORF1P phosphorylation.
Conclusion
The activation of LINE-1 retrotransposon is associated with a variety of human diseases and is involved in the occurrence and progression of disease through retrotransposition-dependent and retrotransposition-independent mechanisms. Currently, it has even become a marker of tumorigenesis and prognosis and is related to immune regulation. The effective inhibition of LINE-1 activation has become a treatment for some diseases. The inhibition of LINE-1 in animal experiments can inhibit the occurrence and development of tumors, so the clinical application of LINE-1 inhibitors is imminent. In addition to exploring some known inhibitors, other mechanisms of LINE-1 inhibition should also be explored. We summarized the relationship between LINE-1 and disease-related immunity, and proposed that LINE-1 may affect the immune status of the body by regulating metabolism, leading to poor prognosis. Metabolic substances can affect the immune microenvironment, for example, lactic acid can lead to immunosuppressive microenvironment, leading to poor prognosis of tumors. The dysregulation of LINE-1 can lead to the disorder of glucose and lipid metabolism, and the inhibition of glucose and lipid metabolism may reverse the disease progression caused by LINE-1. Now the anti-tumor effect of regulating the body’s metabolism has entered clinical trials, such as the significant effect of metformin in the treatment of tumors. Therefore, the metabolic status of diseases caused by LINE-1 can be checked. Metabolic therapy combined with LINE-1 inhibitors may inhibit the progression of LINE-1 and may improve the immune microenvironment to achieve the optimal therapeutic effect.
Statements
Author contributions
XZ wrote the article. RZ and JY reviewed and revised the article. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China (Grant Nos. 81702280 and 81872143), National Science and Technology Support Program of China (Grant No. 2018ZX09201015), and Project of Science and Technology of Tianjin (Grant No. 18JCQNJC82700).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1
AllenE.HorvathS.TongF.KraftP.SpiteriE.RiggsA. D.et al (2003). High concentrations of long interspersed nuclear element sequence distinguish monoallelically expressed genes.Proc. Natl. Acad. Sci. U.S.A.1009940–9945. 10.1073/pnas.1737401100
2
AporntewanC.PhokaewC.PiriyapongsaJ.NgamphiwC.IttiwutC.TongsimaS.et al (2011). Hypomethylation of intragenic LINE-1 represses transcription in cancer cells through AGO2.PLoS One6:e17934. 10.1371/journal.pone.0017934
3
ArdeljanD.TaylorM. S.TingD. T.BurnsK. H. (2017). The human long interspersed element-1 retrotransposon: an emerging biomarker of neoplasia.Clin. Chem.63816–822. 10.1373/clinchem.2016.257444
4
ArdeljanD.WangX.OghbaieM.TaylorM. S.HusbandD.DeshpandeV.et al (2020). LINE-1 ORF2p expression is nearly imperceptible in human cancers.Mob. DNA11:1. 10.1186/s13100-019-0191-2
5
BabaY.WatanabeM.MurataA.ShigakiH.MiyakeK.IshimotoT.et al (2014). LINE-1 hypomethylation, DNA copy number alterations, and CDK6 amplification in esophageal squamous cell carcinoma.Clin. Cancer Res.201114–1124. 10.1158/1078-0432.CCR-13-1645
6
BabaY.YagiT.SawayamaH.HiyoshiY.IshimotoT.IwatsukiM.et al (2018). Long interspersed element-1 methylation level as a prognostic biomarker in gastrointestinal cancers.Digestion9726–30. 10.1159/000484104
7
BabushokD. V.KazazianH. H.Jr. (2007). Progress in understanding the biology of the human mutagen LINE-1.Hum. Mutat.28527–539. 10.1002/humu.20486
8
Banuelos-SanchezG.SanchezL.Benitez-GuijarroM.Sanchez-CarnereroV.Salvador-PalomequeC.Tristan-RamosP.et al (2019). Synthesis and characterization of specific reverse transcriptase inhibitors for mammalian LINE-1 retrotransposons.Cell Chem. Biol.261095–1109.e14. 10.1016/j.chembiol.2019.04.010
9
BarchittaM.QuattrocchiA.MaugeriA.VinciguerraM.AgodiA. (2014). LINE-1 hypomethylation in blood and tissue samples as an epigenetic marker for cancer risk: a systematic review and meta-analysis.PLoS One9:e109478. 10.1371/journal.pone.0109478
10
BeckC. R.Garcia-PerezJ. L.BadgeR. M.MoranJ. V. (2011). LINE-1 elements in structural variation and disease.Annu. Rev. Genom. Hum. Genet.12187–215. 10.1146/annurev-genom-082509-141802
11
BelgnaouiS. M.GosdenR. G.SemmesO. J.HaoudiA. (2006). Human LINE-1 retrotransposon induces DNA damage and apoptosis in cancer cells.Cancer Cell Int.6:13. 10.1186/1475-2867-6-13
12
BenihoudK.BonardelleD.Soual-HoebekeE.Durand-GasselinI.EmilieD.KigerN.et al (2002). Unusual expression of LINE-1 transposable element in the MRL autoimmune lymphoproliferative syndrome-prone strain.Oncogene215593–5600. 10.1038/sj.onc.1205730
13
BojangP.Jr.RobertsR. A.AndertonM. J.RamosK. S. (2013). Reprogramming of the HepG2 genome by long interspersed nuclear element-1.Mol. Oncol.7812–825. 10.1016/j.molonc.2013.04.003
14
BrandA.SingerK.KoehlG. E.KolitzusM.SchoenhammerG.ThielA.et al (2016). LDHA-associated lactic acid production blunts tumor immunosurveillance by T and NK cells.Cell Metab.24657–671. 10.1016/j.cmet.2016.08.011
15
BrégnardC.GuerraJ.DéjardinS.PassalacquaF.BenkiraneM.LaguetteN. (2016). Upregulated LINE-1 activity in the fanconi anemia cancer susceptibility syndrome leads to spontaneous pro-inflammatory cytokine production.EBioMedicine8184–194. 10.1016/j.ebiom.2016.05.005
16
BriggsE. M.SpadaforaC.LoganS. K. (2019). A re-evaluation of LINE-1 ORF2 expression in LNCaP prostate cancer cells.Mob. DNA10:51. 10.1186/s13100-019-0196-x
17
BurnsK. H. (2017). Transposable elements in cancer.Nat. Rev. Cancer17415–424. 10.1038/nrc.2017.35
18
CarliniF.RidolfiB.MolinariA.ParisiC.BozzutoG.ToccacieliL.et al (2010). The reverse transcription inhibitor abacavir shows anticancer activity in prostate cancer cell lines.PLoS One5:e14221. 10.1371/journal.pone.0014221
19
ChiappinelliK. B.StrisselP. L.DesrichardA.LiH.HenkeC.AkmanB.et al (2015). Inhibiting DNA methylation causes an interferon response in cancer via dsRNA including endogenous retroviruses.Cell162974–986. 10.1016/j.cell.2015.07.011
20
ChristianC. M.SokolowskiM.deHaroD.KinesK. J.BelancioV. P. (2017). Involvement of conserved amino acids in the C-terminal region of LINE-1 ORF2p in retrotransposition.Genetics2051139–1149. 10.1534/genetics.116.191403
21
ColegioO. R.ChuN. Q.SzaboA. L.ChuT.RhebergenA. M.JairamV.et al (2014). Functional polarization of tumour-associated macrophages by tumour-derived lactic acid.Nature513559–563. 10.1038/nature13490
22
CordauxR.BatzerM. A. (2009). The impact of retrotransposons on human genome evolution.Nat. Rev. Genet.10691–703. 10.1038/nrg2640
23
CoufalN. G.Garcia-PerezJ. L.PengG. E.MarchettoM. C.MuotriA. R.MuY.et al (2011). Ataxia telangiectasia mutated (ATM) modulates long interspersed element-1 (L1) retrotransposition in human neural stem cells.Proc. Natl. Acad. Sci. U.S.A.10820382–20387. 10.1073/pnas.1100273108
24
CriscioneS. W.TheodosakisN.MicevicG.CornishT. C.BurnsK. H.NerettiN.et al (2016). Genome-wide characterization of human L1 antisense promoter-driven transcripts.BMC Genomics17:463. 10.1186/s12864-016-2800-5
25
DawwasM. F. (2014). Adenoma detection rate and risk of colorectal cancer and death.N. Engl. J. Med.3702539–2540. 10.1056/NEJMc1405329
26
de KoningA. P.GuW.CastoeT. A.BatzerM. A.PollockD. D. (2011). Repetitive elements may comprise over two-thirds of the human genome.PLoS Genet.7:e1002384. 10.1371/journal.pgen.1002384
27
De LucaC.GuadagniF.Sinibaldi-VallebonaP.SentinelliS.GallucciM.HoffmannA.et al (2016). Enhanced expression of LINE-1-encoded ORF2 protein in early stages of colon and prostate transformation.Oncotarget74048–4061. 10.18632/oncotarget.6767
28
DenliA. M.NarvaizaI.KermanB. E.PenaM.BennerC.MarchettoM. C. N.et al (2015). Primate-specific ORF0 contributes to retrotransposon-mediated diversity.Cell163583–593. 10.1016/j.cell.2015.09.025
29
DoxiadisG. G.HoofI.de GrootN.BontropR. E. (2012). Evolution of HLA-DRB genes.Mol. Biol. Evol.293843–3853. 10.1093/molbev/mss186
30
DunaevaM.DerksenM.PruijnG. J. M. (2018). LINE-1 hypermethylation in serum cell-free DNA of relapsing remitting multiple sclerosis patients.Mol. Neurobiol.554681–4688. 10.1007/s12035-017-0679-z
31
FengQ.MoranJ. V.KazazianH. H.Jr.BoekeJ. D. (1996). Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition.Cell87905–916. 10.1016/S0092-8674(00)81997-2
32
FlaschD. A.MaciaÁSánchezL.LjungmanM.HerasS. R.García-PérezJ. L.et al (2019). Genome-wide de novo L1 retrotransposition connects endonuclease activity with replication.Cell177837–851.e28. 10.1016/j.cell.2019.02.050
33
FriedmanJ. R.NolanN. A.BrownK. C.MilesS. L.AkersA. T.ColcloughK. W.et al (2018). Anticancer activity of natural and synthetic capsaicin analogs.J. Pharmacol. Exp. Ther.364462–473. 10.1124/jpet.117.243691
34
FungL.GuzmanH.SevrioukovE.IdicaA.ParkE.BochnakianA.et al (2019). miR-128 restriction of LINE-1 (L1) retrotransposition is dependent on targeting hnRNPA1 mRNA.Int. J. Mol. Sci.201955. 10.3390/ijms20081955
35
GaillardS. L.ZahurakM.SharmaA.DurhamJ. N.ReissK. A.Sartorius-MergenthalerS.et al (2019). A phase 1 trial of the oral DNA methyltransferase inhibitor CC-486 and the histone deacetylase inhibitor romidepsin in advanced solid tumors.Cancer1252837–2845. 10.1002/cncr.32138
36
Garcia-PerezJ. L.WidmannT. J.AdamsI. R. (2016). The impact of transposable elements on mammalian development.Development (Cambridge, England)1434101–4114. 10.1242/dev.132639
37
GoodierJ. L.CheungL. E.KazazianH. H.Jr. (2013). Mapping the LINE1 ORF1 protein interactome reveals associated inhibitors of human retrotransposition.Nucleic Acids Res.417401–7419. 10.1093/nar/gkt512
38
GulerG. D.TindellC. A.PittiR.WilsonC.NicholsK.KaiWai CheungT.et al (2017). Repression of stress-induced LINE-1 expression protects cancer cell subpopulations from lethal drug exposure.Cancer Cell32221–237.e13. 10.1016/j.ccell.2017.07.002
39
HamdorfM.IdicaA.ZisoulisD. G.GamelinL.MartinC.SandersK. J.et al (2015). miR-128 represses L1 retrotransposition by binding directly to L1 RNA.Nat. Struct. Mol. Biol.22824–831. 10.1038/nsmb.3090
40
HancksD. C.KazazianH. H.Jr. (2016). Roles for retrotransposon insertions in human disease.Mob. DNA7:9. 10.1186/s13100-016-0065-9
41
HaoY.LiD.XuY.OuyangJ.WangY.ZhangY.et al (2019). Investigation of lipid metabolism dysregulation and the effects on immune microenvironments in pan-cancer using multiple omics data.BMC Bioinformatics20:195. 10.1186/s12859-019-2734-4
42
HarrisC. R.NormartR.YangQ.StevensonE.HafftyB. G.GanesanS.et al (2010). Association of nuclear localization of a long interspersed nuclear element-1 protein in breast tumors with poor prognostic outcomes.Genes Cancer1115–124. 10.1177/1947601909360812
43
HosseinnejadK.YinT.GaskinsJ. T.BailenJ. L.JortaniS. A. (2018). Discovery of the long interspersed nuclear element-1 activation product [Open Reading Frame-1 (ORF1) protein] in human blood.Clin. Chim. Acta487228–232. 10.1016/j.cca.2018.09.040
44
HouédéN.PulidoM.MoureyL.JolyF.FerreroJ.-M.BelleraC.et al (2014). A phase II trial evaluating the efficacy and safety of efavirenz in metastatic castration-resistant prostate cancer.Oncologist191227–1228. 10.1634/theoncologist.2014-0345
45
HusainZ.HuangY.SethP.SukhatmeV. P. (2013). Tumor-derived lactate modifies antitumor immune response: effect on myeloid-derived suppressor cells and NK cells.J. Immunol.1911486–1495. 10.4049/jimmunol.1202702
46
IdicaA.SevrioukovE. A.ZisoulisD. G.HamdorfM.DaugaardI.KadandaleP.et al (2017). MicroRNA miR-128 represses LINE-1 (L1) retrotransposition by down-regulating the nuclear import factor TNPO1.J. Biol. Chem.29220494–20508. 10.1074/jbc.M117.807677
47
IwagamiS.BabaY.WatanabeM.ShigakiH.MiyakeK.IshimotoT.et al (2013). LINE-1 hypomethylation is associated with a poor prognosis among patients with curatively resected esophageal squamous cell carcinoma.Ann. Surg.257449–455. 10.1097/SLA.0b013e31826d8602
48
JungH.ChoiJ. K.LeeE. A. (2018). Immune signatures correlate with L1 retrotransposition in gastrointestinal cancers.Genome Res.281136–1146. 10.1101/gr.231837.117
49
KawanoH.SaekiH.KitaoH.TsudaY.OtsuH.AndoK.et al (2014). Chromosomal instability associated with global DNA hypomethylation is associated with the initiation and progression of esophageal squamous cell carcinoma.Ann. Surg. Oncol.21(Suppl. 4)S696–S702. 10.1245/s10434-014-3818-z
50
KazazianH. H.Jr.MoranJ. V. (2017). Mobile DNA in health and disease. N Engl. J. Med.377, 361–370. 10.1056/NEJMra1510092
51
KazazianH. H.Jr.WongC.YoussoufianH.ScottA. F.PhillipsD. G.AntonarakisS. E. (1988). Haemophilia A resulting from de novo insertion of L1 sequences represents a novel mechanism for mutation in man.Nature332164–166. 10.1038/332164a0
52
KempJ. R.LongworthM. S. (2015). Crossing the LINE toward genomic instability: LINE-1 retrotransposition in cancer.Front. Chem.3:68. 10.3389/fchem.2015.00068
53
KhalidM.BojangP.Jr.HassaninA. A. I.BowersE. C.Reyes-ReyesE. M.IRamosN.et al (2018). Line-1: Implications in the etiology of cancer, clinical applications, and pharmacologic targets.Mutat. Res.77851–60. 10.1016/j.mrrev.2018.09.003
54
KinesK. J.SokolowskiM.deHaroD. L.ChristianC. M.BelancioV. P. (2014). Potential for genomic instability associated with retrotranspositionally-incompetent L1 loci.Nucleic Acids Res.4210488–10502. 10.1093/nar/gku687
55
KoE.-J.OhY. L.KimH. Y.EoW. K.KimH.OckM. S.et al (2019). Correlation of long interspersed element-1 open reading frame 1 and c-Met proto-oncogene protein expression in ovarian cancer.Genes Genomics411293–1299. 10.1007/s13258-019-00858-y
56
KosumiK.BabaY.OkadomeK.YagiT.KiyozumiY.YoshidaN.et al (2019). Tumor long-interspersed nucleotide element-1 methylation level and immune response to esophageal cancer.Ann. Surg.10.1097/SLA.0000000000003264
57
LaiD.-W.LiuS.-H.KarlssonA. I.LeeW.-J.WangK.-B.ChenY.-C.et al (2014). The novel Aryl hydrocarbon receptor inhibitor biseugenol inhibits gastric tumor growth and peritoneal dissemination.Oncotarget57788–7804. 10.18632/oncotarget.2307
58
LanderE. S.LintonL. M.BirrenB.NusbaumC.ZodyM. C.BaldwinJ.et al (2001). Initial sequencing and analysis of the human genome.Nature409860–921. 10.1038/35057062
59
LandriscinaM.FabianoA.AltamuraS.BagalàC.PiscazziA.CassanoA.et al (2005). Reverse transcriptase inhibitors down-regulate cell proliferation in vitro and in vivo and restore thyrotropin signaling and iodine uptake in human thyroid anaplastic carcinoma.J. Clin. Endocrinol. Metab.905663–5671. 10.1210/jc.2005-0367
60
LangerC. J.GadgeelS. M.BorghaeiH.PapadimitrakopoulouV. A.PatnaikA.PowellS. F.et al (2016). Carboplatin and pemetrexed with or without pembrolizumab for advanced, non-squamous non-small-cell lung cancer: a randomised, phase 2 cohort of the open-label KEYNOTE-021 study.Lancet Oncol.171497–1508. 10.1016/S1470-2045(16)30498-3
61
LavasanifarA.SharpC. N.KorteE. A.YinT.HosseinnejadK.JortaniS. A. (2019). Long interspersed nuclear element-1 mobilization as a target in cancer diagnostics, prognostics and therapeutics.Clin. Chim. Acta49352–62. 10.1016/j.cca.2019.02.015
62
LavieL.MaldenerE.BrouhaB.MeeseE. U.MayerJ. (2004). The human L1 promoter: variable transcription initiation sites and a major impact of upstream flanking sequence on promoter activity.Genome Res.142253–2260. 10.1101/gr.2745804
63
LiS.YangQ.HouY.JiangT.ZongL.WangZ.et al (2018). Hypomethylation of LINE-1 elements in schizophrenia and bipolar disorder.J. Psychiatr. Res.10768–72. 10.1016/j.jpsychires.2018.10.009
64
LiW.JinY.PrazakL.HammellM.DubnauJ. (2012). Transposable elements in TDP-43-mediated neurodegenerative disorders.PLoS One7:e44099. 10.1371/journal.pone.0044099
65
LindqvistD.DhabharF. S.JamesS. J.HoughC. M.JainF. A.BersaniF. S.et al (2017). Oxidative stress, inflammation and treatment response in major depression.Psychoneuroendocrinology76197–205. 10.1016/j.psyneuen.2016.11.031
66
LiuS.DuT.LiuZ.ShenY.XiuJ.XuQ. (2016). Inverse changes in L1 retrotransposons between blood and brain in major depressive disorder.Sci. Rep.6:37530. 10.1038/srep37530
67
LuntS. Y.Vander HeidenM. G. (2011). Aerobic glycolysis: meeting the metabolic requirements of cell proliferation.Annu. Rev. Cell Dev. Biol.27441–464. 10.1146/annurev-cellbio-092910-154237
68
MangiacasaleR.PittoggiC.SciamannaI.CaredduA.MatteiE.LorenziniR.et al (2003). Exposure of normal and transformed cells to nevirapine, a reverse transcriptase inhibitor, reduces cell growth and promotes differentiation.Oncogene222750–2761. 10.1038/sj.onc.1206354
69
Martin-NunezG. M.Rubio-MartinE.Cabrera-MuleroR.Rojo-MartinezG.OlveiraG.ValdesS.et al (2014). Type 2 diabetes mellitus in relation to global LINE-1 DNA methylation in peripheral blood: a cohort study.Epigenetics91322–1328. 10.4161/15592294.2014.969617
70
MathiasS. L.ScottA. F.KazazianH. H.Jr.BoekeJ. D.GabrielA. (1991). Reverse transcriptase encoded by a human transposable element.Science2541808–1810. 10.1126/science.1722352
71
MavraganiC. P.SagalovskiyI.GuoQ.NezosA.KapsogeorgouE. K.LuP.et al (2016). Expression of long interspersed nuclear element 1 retroelements and induction of type I interferon in patients with systemic autoimmune disease.Arthritis Rheum.682686–2696. 10.1002/art.39795
72
MessiaenL.VogtJ.BengesserK.FuC.MikhailF.SerraE.et al (2011). Mosaic type-1 NF1 microdeletions as a cause of both generalized and segmental neurofibromatosis type-1 (NF1).Hum. Mutat.32213–219. 10.1002/humu.21418
73
MikiY.NishishoI.HoriiA.MiyoshiY.UtsunomiyaJ.KinzlerK. W.et al (1992). Disruption of the APC gene by a retrotransposal insertion of L1 sequence in a colon cancer.Cancer Res.52643–645.
74
MorrishT. A.Garcia-PerezJ. L.StamatoT. D.TaccioliG. E.SekiguchiJ.MoranJ. V. (2007). Endonuclease-independent LINE-1 retrotransposition at mammalian telomeres.Nature446208–212. 10.1038/nature05560
75
MorrishT. A.GilbertN.MyersJ. S.VincentB. J.StamatoT. D.TaccioliG. E.et al (2002). DNA repair mediated by endonuclease-independent LINE-1 retrotransposition.Nat. Genet.31159–165. 10.1038/ng898
76
MorseB.RothergP. G.SouthV. J.SpandorferJ. M.AstrinS. M. (1988). Insertional mutagenesis of the myc locus by a LINE-1 sequence in a human breast carcinoma.Nature33387–90. 10.1038/333087a0
77
MuotriA. R.MarchettoM. C.CoufalN. G.OefnerR.YeoG.NakashimaK.et al (2010). L1 retrotransposition in neurons is modulated by MeCP2.Nature468443–446. 10.1038/nature09544
78
NishikawaY.NakayamaR.ObikaS.OhsakiE.UedaK.HondaT. (2018). Inhibition of LINE-1 retrotransposition by capsaicin.Int. J. Mol. Sci.193243. 10.3390/ijms19103243
79
OdunsiK.MatsuzakiJ.JamesS. R.Mhawech-FaucegliaP.TsujiT.MillerA.et al (2014). Epigenetic potentiation of NY-ESO-1 vaccine therapy in human ovarian cancer.Cancer Immunol. Res.237–49. 10.1158/2326-6066.CIR-13-0126
80
OginoS.NoshoK.KirknerG. J.KawasakiT.ChanA. T.SchernhammerE. S.et al (2008). A cohort study of tumoral LINE-1 hypomethylation and prognosis in colon cancer.J. Natl. Cancer Inst.1001734–1738. 10.1093/jnci/djn359
81
OricchioE.SciamannaI.BeraldiR.TolstonogG. V.SchumannG. G.SpadaforaC. (2007). Distinct roles for LINE-1 and HERV-K retroelements in cell proliferation, differentiation and tumor progression.Oncogene264226–4233. 10.1038/sj.onc.1210214
82
PayerL. M.BurnsK. H. (2019). Transposable elements in human genetic disease.Nat. Rev. Genet.20760–772. 10.1038/s41576-019-0165-8
83
PearceM. S.McConnellJ. C.PotterC.BarrettL. M.ParkerL.MathersJ. C.et al (2012). Global LINE-1 DNA methylation is associated with blood glycaemic and lipid profiles.Int. J. Epidemiol.41210–217. 10.1093/ije/dys020
84
PedersenI. M.ZisoulisD. G. (2016). Transposable elements and miRNA: regulation of genomic stability and plasticity.Mob. Genet. Elements6:e1175537. 10.1080/2159256X.2016.1175537
85
PowellJ. B.GoodeG. D.EltomS. E. (2013). The Aryl hydrocarbon receptor: a target for breast cancer therapy.J. Cancer Ther.41177–1186. 10.4236/jct.2013.47137
86
RichardsonS. R.DoucetA. J.KoperaH. C.MoldovanJ. B.Garcia-PerezJ. L.MoranJ. V. (2015). The influence of LINE-1 and SINE retrotransposons on mammalian genomes. Microbiol. Spectr.3:MDNA3-2014. 10.1128/microbiolspec.MDNA3-0061-2014
87
RodicN. (2018). LINE-1 activity and regulation in cancer.Front. Biosci.23:1680–1686. 10.2741/4666
88
RodicN.BurnsK. H. (2013). Long interspersed element-1 (LINE-1): passenger or driver in human neoplasms?PLoS Genet9:e1003402. 10.1371/journal.pgen.1003402
89
RodicN.SharmaR.SharmaR.ZampellaJ.DaiL.TaylorM. S.et al (2014). Long interspersed element-1 protein expression is a hallmark of many human cancers.Am. J. Pathol.1841280–1286. 10.1016/j.ajpath.2014.01.007
90
Rodriguez-MartinB.AlvarezE. G.Baez-OrtegaA.ZamoraJ.SupekF.DemeulemeesterJ.et al (2020). Pan-cancer analysis of whole genomes identifies driver rearrangements promoted by LINE-1 retrotransposition.Nat. Genet.52306–319. 10.1038/s41588-019-0562-0
91
Roman-GomezJ.Jimenez-VelascoA.AgirreX.CervantesF.SanchezJ.GarateL.et al (2005). Promoter hypomethylation of the LINE-1 retrotransposable elements activates sense/antisense transcription and marks the progression of chronic myeloid leukemia.Oncogene247213–7223. 10.1038/sj.onc.1208866
92
RozekL. S.ViraniS.BellileE. L.TaylorJ. M. G.SartorM. A.ZarinsK. R.et al (2019). Soy isoflavone supplementation increases long interspersed nucleotide element-1 (LINE-1) methylation in head and neck squamous cell carcinoma.Nutr. Cancer71772–780. 10.1080/01635581.2019.1577981
93
SaitoK.KawakamiK.MatsumotoI.OdaM.WatanabeG.MinamotoT. (2010). Long interspersed nuclear element 1 hypomethylation is a marker of poor prognosis in stage IA non-small cell lung cancer.Clin. Cancer Res.162418–2426. 10.1158/1078-0432.CCR-09-2819
94
SbardellaG.MaiA.BartoliniS.CastellanoS.CirilliR.RotiliD.et al (2011). Modulation of cell differentiation, proliferation, and tumor growth by dihydrobenzyloxopyrimidine non-nucleoside reverse transcriptase inhibitors.J. Med. Chem.545927–5936. 10.1021/jm200734j
95
SchulzW. A.SteinhoffC.FlorlA. R. (2006). Methylation of endogenous human retroelements in health and disease.Curr. Top. Microbiol. Immunol.310211–250. 10.1007/3-540-31181-5_11
96
SciamannaI.LandriscinaM.PittoggiC.QuirinoM.MearelliC.BeraldiR.et al (2005). Inhibition of endogenous reverse transcriptase antagonizes human tumor growth.Oncogene243923–3931. 10.1038/sj.onc.1208562
97
SciamannaI.Sinibaldi-VallebonaP.SerafinoA.SpadaforaC. (2018). LINE-1-encoded reverse transcriptase as a target in cancer therapy.Front. Biosci.23:1360–1369. 10.2741/4648
98
ShahA. K.SaundersN. A.BarbourA. P.HillM. M. (2013). Early diagnostic biomarkers for esophageal adenocarcinoma–the current state of play.Cancer Epidemiol. Biomark. Prevent.221185–1209. 10.1158/1055-9965.EPI-12-1415
99
ShigakiH.BabaY.WatanabeM.MurataA.IwagamiS.MiyakeK.et al (2013). LINE-1 hypomethylation in gastric cancer, detected by bisulfite pyrosequencing, is associated with poor prognosis.Gastric Cancer16480–487. 10.1007/s10120-012-0209-7
100
Sinibaldi-VallebonaP.LaviaP.GaraciE.SpadaforaC. (2006). A role for endogenous reverse transcriptase in tumorigenesis and as a target in differentiating cancer therapy.Genes Chrom. Cancer451–10. 10.1002/gcc.20266
101
SuarezN. A.MaciaA.MuotriA. R. (2018). LINE-1 retrotransposons in healthy and diseased human brain.Dev. Neurobiol.78434–455. 10.1002/dneu.22567
102
SultanaT.van EssenD.SiolO.Bailly-BechetM.PhilippeC.El AabidineA. Z.et al (2019). The landscape of L1 retrotransposons in the human genome is shaped by pre-insertion sequence biases and post-insertion selection.Mol. Cell74555–570.e7. 10.1016/j.molcel.2019.02.036
103
SwergoldG. D. (1990). Identification, characterization, and cell specificity of a human LINE-1 promoter.Mol. Cell. Biol.106718–6729. 10.1128/MCB.10.12.6718
104
TangM. L.XiaoP.ZouJ. Z.CaoD. D.LiY. Y.ChangH. B. (2018). [Effect of LINE1-ORF1p overexpression on the proliferation of nephroblastoma WT_CLS1 cells].Chin. J. Contemp. Pediatr.20501–507.
105
TaylorJ. A.ShiodaK.MitsunagaS.YawataS.AngleB. M.NagelS. C.et al (2018). Prenatal exposure to bisphenol a disrupts naturally occurring bimodal dna methylation at proximal promoter of fggy, an obesity-relevant gene encoding a carbohydrate kinase, in gonadal white adipose tissues of CD-1 mice.Endocrinology159779–794. 10.1210/en.2017-00711
106
TeerawattanapongN.UdomsinprasertW.NgarmukosS.TanavaleeA.HonsawekS. (2019). Blood leukocyte LINE-1 hypomethylation and oxidative stress in knee osteoarthritis.Heliyon5:e01774. 10.1016/j.heliyon.2019.e01774
107
TenengI.StribinskisV.RamosK. S. (2007). Context-specific regulation of LINE-1.Genes Cells121101–1110. 10.1111/j.1365-2443.2007.01117.x
108
TharpM. E.MalkiS.BortvinA. (2020). Maximizing the ovarian reserve in mice by evading LINE-1 genotoxicity.Nat. Commun.11:330. 10.1038/s41467-019-14055-8
109
ThayerR. E.SingerM. F.FanningT. G. (1993). Undermethylation of specific LINE-1 sequences in human cells producing a LINE-1-encoded protein.Gene133273–277. 10.1016/0378-1119(93)90651-I
110
TurcotV.TchernofA.DeshaiesY.PerusseL.BelisleA.MarceauS.et al (2012). LINE-1 methylation in visceral adipose tissue of severely obese individuals is associated with metabolic syndrome status and related phenotypes.Clin. Epigenet.4:10. 10.1186/1868-7083-4-10
111
TwyffelsL.GueydanC.KruysV. (2014). Transportin-1 and Transportin-2: protein nuclear import and beyond.FEBS Lett.5881857–1868. 10.1016/j.febslet.2014.04.023
112
van den HurkJ. A.van de PolD. J.WissingerB.van DrielM. A.HoefslootL. H.Ide WijsJ.et al (2003). Novel types of mutation in the choroideremia (CHM) gene: a full-length L1 insertion and an intronic mutation activating a cryptic exon.Hum. Genet.113268–275. 10.1007/s00439-003-0970-0
113
VogtJ.BengesserK.ClaesK. B. M.WimmerK.MautnerV.-F.van MinkelenR.et al (2014). SVA retrotransposon insertion-associated deletion represents a novel mutational mechanism underlying large genomic copy number changes with non-recurrent breakpoints.Genome Biol.15R80–R80. 10.1186/gb-2014-15-6-r80
114
VuongL. M.PanS.DonovanP. J. (2019). Proteome profile of endogenous retrotransposon-associated complexes in human embryonic stem cells.Proteomics19:e1900169. 10.1002/pmic.201900169
115
WangL.JordanI. K. (2018). Transposable element activity, genome regulation and human health.Curr. Opin. Genet. Dev.4925–33. 10.1016/j.gde.2018.02.006
116
WangL.WangF.GuanJ.LeJ.WuL.ZouJ.et al (2010). Relation between hypomethylation of long interspersed nucleotide elements and risk of neural tube defects.Am. J. Clin. Nutr.911359–1367. 10.3945/ajcn.2009.28858
117
WangX. Y.ZhangY.YangN.ChengH.SunY. J. (2020). DNMT3a mediates paclitaxel-induced abnormal expression of LINE-1 by increasing the intragenic methylation.Hereditas42100–111.
118
WanichnopparatW.SuwanwongseK.Pin-OnP.AporntewanC.MutiranguraA. (2013). Genes associated with the cis-regulatory functions of intragenic LINE-1 elements.BMC Genomics14:205. 10.1186/1471-2164-14-205
119
WeberB.KimhiS.HowardG.EdenA.LykoF. (2010). Demethylation of a LINE-1 antisense promoter in the cMet locus impairs Met signalling through induction of illegitimate transcription.Oncogene295775–5784. 10.1038/onc.2010.227
120
WeiW.GilbertN.OoiS. L.LawlerJ. F.OstertagE. M.KazazianH. H.et al (2001). Human L1 retrotransposition: cis preference versus trans complementation.Mol. Cell. Biol.211429–1439. 10.1128/MCB.21.4.1429-1439.2001
121
WilsonA. S.PowerB. E.MolloyP. L. (2007). DNA hypomethylation and human diseases.Biochim. Biophys. Acta1775138–162. 10.1016/j.bbcan.2006.08.007
122
WimmerK.CallensT.WernstedtA.MessiaenL. (2011). The NF1 gene contains hotspots for L1 endonuclease-dependent de novo insertion.PLoS Genet.7:e1002371. 10.1371/journal.pgen.1002371
123
XiangY.YanK.ZhengQ.KeH.ChengJ.XiongW.et al (2019). Histone demethylase KDM4B promotes DNA damage by activating long interspersed nuclear element-1.Cancer Res.7986–98. 10.1158/0008-5472.CAN-18-1310
124
YooyongsatitS.RuchusatsawatK.NoppakunN.HirankarnN.MutiranguraA.WongpiyabovornJ. (2015). Patterns and functional roles of LINE-1 and Alu methylation in the keratinocyte from patients with psoriasis vulgaris.J. Hum. Genet.60349–355. 10.1038/jhg.2015.33
125
ZhangR.ZhangF.SunZ.LiuP.ZhangX.YeY.et al (2019). LINE-1 retrotransposition promotes the development and progression of lung squamous cell carcinoma by disrupting the tumor-suppressor gene FGGY.Cancer Res.794453–4465. 10.1158/0008-5472.CAN-19-0076
126
ZhangY.ZagnitkoO.RodionovaI.OstermanA.GodzikA. (2011). The FGGY carbohydrate kinase family: insights into the evolution of functional specificities.PLoS Comput. Biol.7:e1002318. 10.1371/journal.pcbi.1002318
127
ZhaoK.DuJ.PengY.LiP.WangS.WangY.et al (2018). LINE1 contributes to autoimmunity through both RIG-I- and MDA5-mediated RNA sensing pathways.J. Autoimmun.90105–115. 10.1016/j.jaut.2018.02.007
128
ZhuJ.LingY.XuY.LuM.-Z.LiuY.-P.ZhangC.-S. (2015). Elevated expression of MDR1 associated with Line-1 hypomethylation in esophageal squamous cell carcinoma.Int. J. Clin. Exp. Pathol.814392–14400.
Summary
Keywords
retrotransposons, LINE-1, regulatory mechanisms, cancer, immune, inhibitor
Citation
Zhang X, Zhang R and Yu J (2020) New Understanding of the Relevant Role of LINE-1 Retrotransposition in Human Disease and Immune Modulation. Front. Cell Dev. Biol. 8:657. doi: 10.3389/fcell.2020.00657
Received
15 April 2020
Accepted
01 July 2020
Published
07 August 2020
Volume
8 - 2020
Edited by
Trygve Tollefsbol, The University of Alabama at Birmingham, United States
Reviewed by
Sandra Rose Richardson, The University of Queensland, Australia; Apiwat Mutirangura, Chulalongkorn University, Thailand; John LaCava, The Rockefeller University, United States
Updates
Copyright
© 2020 Zhang, Zhang and Yu.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jinpu Yu, yujinpu@tjmuch.com
This article was submitted to Epigenomics and Epigenetics, a section of the journal Frontiers in Cell and Developmental Biology
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.