- 1Department of Pharmacy, Xiangya Hospital, Central South University, Changsha, China
- 2Department of Pathology, Xiangya Hospital, Central South University, Changsha, China
- 3Key Laboratory of Molecular Radiation Oncology of Hunan Province, Center for Molecular Medicine, Xiangya Hospital, Central South University, Changsha, China
- 4National Clinical Research Center for Geriatric Disorders, Xiangya Hospital, Central South University, Changsha, China
Circadian rhythms are physical, behavioral and environmental cycles that respond primarily to light and dark, with a period of time of approximately 24 h. The most essential physiological functions of mammals are manifested in circadian rhythm patterns, including the sleep-wake cycle and nutrient and energy metabolism. Autophagy is a conserved biological process contributing to nutrient and cellular homeostasis. The factors affecting autophagy are numerous, such as diet, drugs, and aging. Recent studies have indicated that autophagy is activated rhythmically in a clock-dependent manner whether the organism is healthy or has certain diseases. In addition, autophagy can affect circadian rhythm by degrading circadian proteins. This review discusses the interaction and mechanisms between autophagy and circadian rhythm. Moreover, we introduce the molecules influencing both autophagy and circadian rhythm. We then discuss the drugs affecting the circadian rhythm of autophagy. Finally, we present the role of rhythmic autophagy in nutrient and energy metabolism and its significance in physiology and metabolic disease.
Introduction
Autophagy is an intracellular degradative procedure that targets cytosolic components to lysosomes for degradation to maintain cellular homeostasis and provide substrates for energy generation (Farias et al., 2019; Santin-Marquez et al., 2019; Yan et al., 2019). The activation of autophagy frequently occurs in the context of nutrient shortages and other stresses (Jacomin et al., 2020; Martel et al., 2020). Cytoplasmic materials are first surrounded by vesicles, which eventually form double membrane structures called autophagosomes (Klionsky et al., 2014; Gou et al., 2020). The autophagosomes then fuse with lysosomes and create autolysosomes where the cargo is degraded and released as critical nutrients, such as fatty acids, back into the cytosol (Gatica et al., 2018; Li et al., 2020; Patra et al., 2020). Autophagy is capable of regulating many0020biochemical processes, including embryogenesis, development, antigen presentation, metabolism and infection removal (Bonam et al., 2020; Farhan et al., 2020; Wu and Nagy, 2020). Aberrant autophagy leads to various diseases, such as neurodegeneration, cancer, aging process, autoimmunity, and others (Choi et al., 2013; Bonam et al., 2018; Brattas et al., 2020; Devis-Jauregui et al., 2020; Mizushima and Levine, 2020; Oeing et al., 2020). At present, of the three known basic types of autophagy, chaperone-mediated autophagy, microautophagy and macroautophagy, most of our current knowledge is focused on macroautophagy (hereafter referred to as autophagy), which is the center of this review (Kaushik and Cuervo, 2018; Lescat et al., 2020; Schafer et al., 2020; Zheng et al., 2020). Additionally, studies have shown that autophagy is a dynamic process in the biological circadian rhythm, which is related to the degradation of cellular components and is driven by a series of autophagy-related proteins (ATGs) (Sachdeva and Thompson, 2008; Wesselborg and Stork, 2015; He Y. et al., 2016).
Autophagy is an evolutionarily conserved catabolic process containing five unique phases: initiation, vesicle nucleation, vesicle elongation, vesicle fusion, and cargo degradation (Figure 1; Levy et al., 2017). The initiation of autophagy is caused by nutrient deprivation, infection, oxidative stress, and other factors (Pavlinov et al., 2020). Vesicle nucleation is mediated by the activation of the Unc-51-like autophagy activating kinase (ULK1) complex, which is composed of ULK1/2, ATG13, ATG 101, and focal adhesion kinase-family interacting protein of 200 kDa (FIP200), which is suppressed by activation of mechanistic target of rapamycin (mTOR) (Petherick et al., 2015; Vahsen et al., 2020). Next, Bcl-2-interacting protein (BECLIN)-1 is phosphorylated by ULK1 and acts as a scaffold to form the class III phosphatidylinositol-3 kinase (PI3K) complex (Fracchiolla et al., 2020). During this process, the ultraviolet radiation resistance-associated gene proteins ATG14 and p150 bind to BECLIN-1 to facilitate its interaction with vacuolar protein sorting protein 34 (VPS34) and phagophore formation (Boukhalfa et al., 2020; Wu M. Y. et al., 2020). Vesicle elongation is induced by two ubiquitin-like conjugating systems, ATG12-ATG5 and microtubule-associated protein 1A/1B-light chain 3-II (LC3-II), together with p62 and other molecules, resulting in the formation of a compartment called the autophagosome (Lystad et al., 2019; Yan et al., 2019). Finally, syntaxin 17 promotes the fusion of the autophagosome and the lysosome for autolysosome formation, with the degradation of cargo when the pH is lowered (Galluzzi et al., 2017; Levy et al., 2017). During the process of autophagy, many molecules, such as mTOR and ATGs, display robust circadian rhythms (Maiese, 2017; Kim et al., 2019).
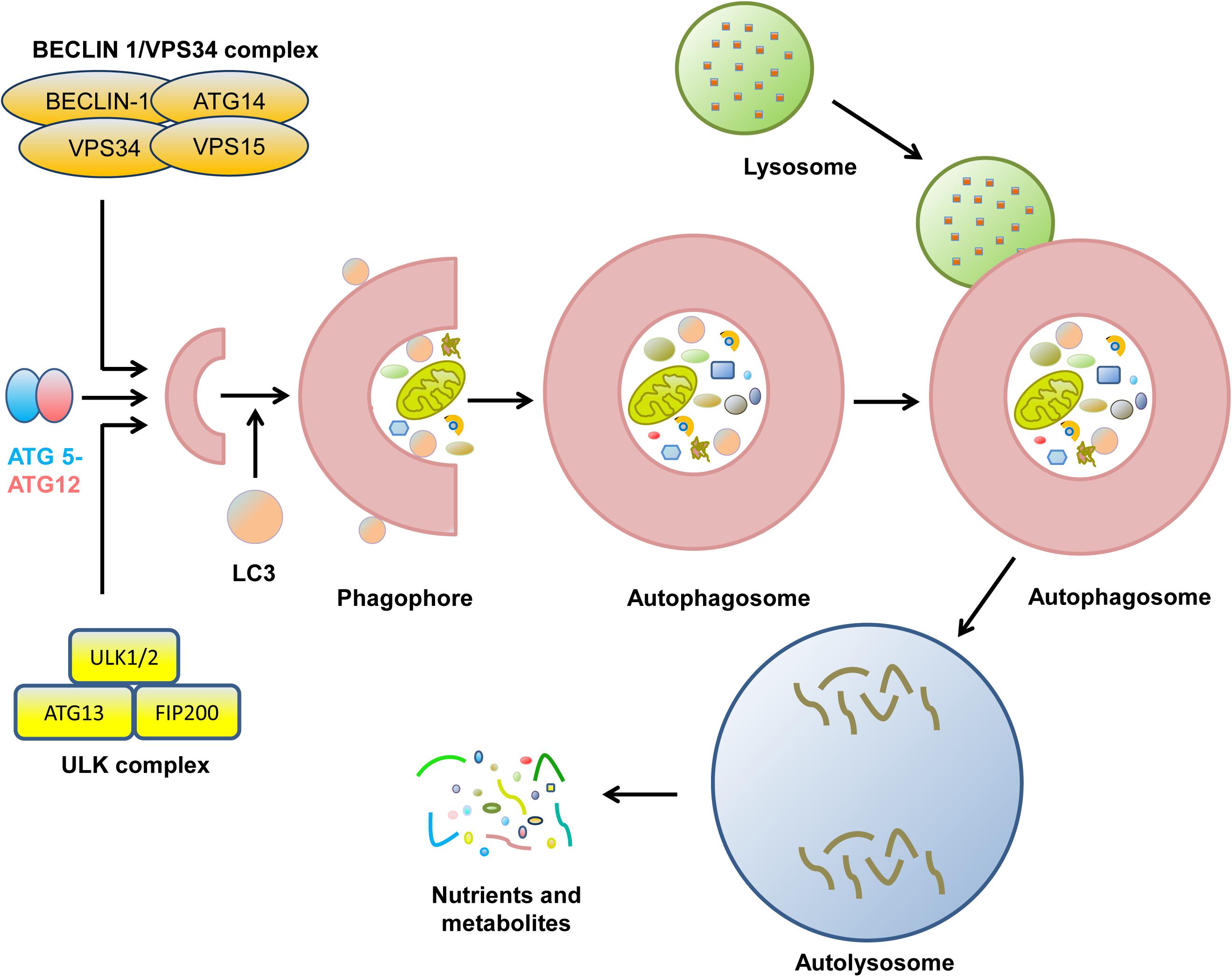
Figure 1. The biological process of cell autophagy. The initial stages of autophagy involve the nucleation, elongation, and maturation of a separation membrane, commonly known as a phagophore. Then, the formed phagophore unites to form the autophagosome. During the complex process, many protein complexes and autophagy-related proteins are sequentially involved. Eventually, autophagosome fuse with lysosome to form autolysosome, where the captured materials are eliminated. The final biomolecules are recycled back to the cytoplasm.
Circadian rhythms are daily predictable changes in physiology, behavior and environment with a period of time of approximately 24 h (Greco and Sassone-Corsi, 2020; Kim and Lazar, 2020). They exist on every biological scale, from macroscopic aspects such as sleep-wake cycles to microcosmic phenomena such as the rhythmic abundance of biomolecules (Ryzhikov et al., 2019b; Tan X. et al., 2019; Van Dyck and Casaer, 2019). In mammals, the daily changes are mediated by the hypothalamic master clock located in the suprachiasmatic nucleus (SCN) and by clock oscillations in peripheral tissues, which are synchronized by light and feeding time, respectively (Dreyer et al., 2019; Paul et al., 2020). The pacemakers are self-sustaining oscillators in the brain and peripheral tissues that synchronize their downstream transcriptional output (Carmo-Silva and Cavadas, 2017; Skene et al., 2018). A growing number of circadian-clock-controlled physiological processes exhibit daily oscillations, such as autophagy, whereas the dysfunction of the circadian system induced by shift work, for example, can increase the risk of many diseases, such as cancer (Rijo-Ferreira and Takahashi, 2019; Ryzhikov et al., 2019a).
Recent studies have discovered that “clock genes” are essential for circadian rhythm generation (Figure 2; Kim and Lazar, 2020). In the clock gene family, members of the basic helix-loop-helix-PAS transcription factor family, BMAL1 and CLOCK, form heterodimers, which bind to E-boxes in the promoters of target genes to activate the expression of the Period (PER1, PER2, and PER3) and Cryptochrome (CRY1 and CRY2) genes (Jahanban-Esfahlan et al., 2018; Gabryelska et al., 2020). PER proteins are degraded in the cytoplasm by the proteasome, while PER:CRY heterodimers can translocate to the nucleus, preventing PER degradation, to suppress and block the activity of the BMAL1:CLOCK complex (Rabinovich-Nikitin et al., 2019). In a parallel arm of the transcriptional loop, the BMAL1:CLOCK heterodimer activates the transcription of orphan nuclear receptors REV-ERBs (REV-ERBα and REV-ERBβ) and RORs (RORα, RORβ, and RORγ) (Sulli et al., 2018). REV-ERBs and RORs can compete for binding to the Bmal1 promoter to activate and inhibit gene transcription effects, respectively, resulting in the circadian oscillation of Bmal1 (Maiese, 2017). In addition to regulating clock components, BMAL1:CLOCK also activates the transcription of genes involved in regulating autophagy, for example (Dong et al., 2016; Scotton et al., 2016; Maiese, 2017).
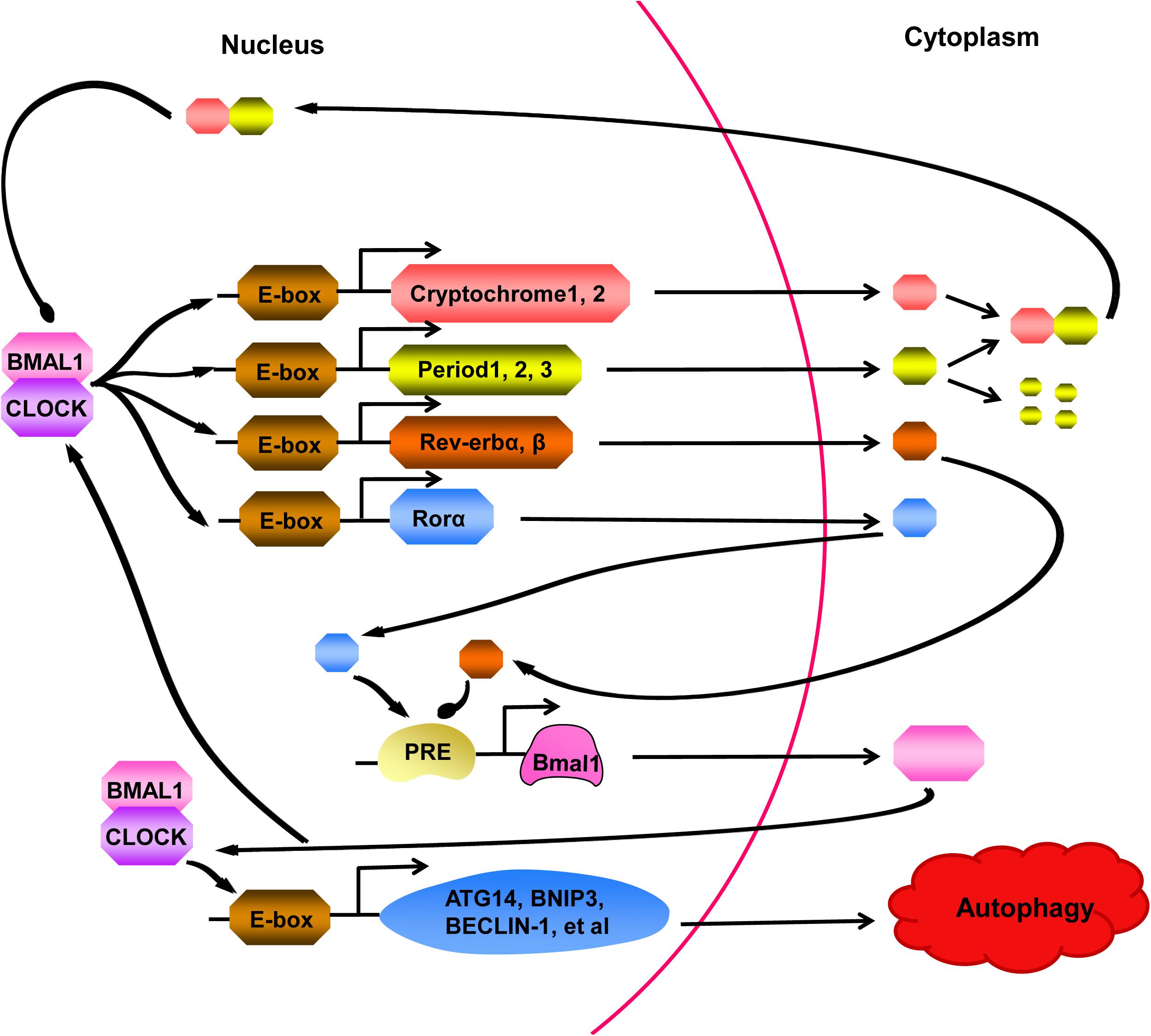
Figure 2. A series of cell processes are controlled by the circadian clock, which is activated by BMAL1:CLOCK heterodimer. The BMAL1:CLOCK complex binds to E-boxes in the promoters of target molecules, activating transcription of the Cryptochrome (CRY) genes, Period (PER) genes, Rev-erb genes, and Ror genes. PER proteins are degraded in the cytoplasm by the proteasome, however, binding to CRY prevents PER from degrading. The complex formed by PER and CRY can return to nucleus and suppressed activity of the BMAL1:CLOCK heterodimer. In a parallel arm of the transcriptional loop, Rev-erb and Ror proteins can compete for combining to the Bmal1 promoter, showing us another layer of regulation to cycling of the clock. Moreover, BMAL1:CLOCK complex can also activate the transcription of genes, participating in regulating many cell processes, such as ATG14, BNIP3, BECLIN-1. At present, studies have confirmed that autophagy can be regulated by the clock.
Here, we provide a general overview of the circadian rhythm of autophagy and discuss the potential mechanisms between circadian and autophagy. Moreover, we investigated the molecules and drugs affecting circadian rhythm and autophagy. Finally, we highlight the functions of the circadian rhythm of autophagy in biological processes and diseases.
Overview of the Circadian Rhythm of Autophagy
The first evidence for the connection between autophagy and circadian regulation was discovered in the early 1970s. A series of electron microscopy studies performed by Pfeifer and colleagues proved that the number of autophagic vacuoles varies on the basis of the time of day in several rat tissues, including hepatocytes, cardiomyocytes, proximal tubules of kidney, pancreatic acinar cells, and the inner segment of retina rod cells (Ma and Lin, 2012; Czaja et al., 2013; Li and Lin, 2015). In addition, the volume and numeric density of autophagic vacuoles in the heart were discovered to maintain a diurnal pattern, which peak at the late-night phase and decrease to lower levels in the early-dark period (Ma et al., 2011). Apart from heart tissue, recent work demonstrated that autophagy activity also presents a robust diurnal rhythm in skeletal muscle and liver through the combination of more specific autophagy markers and flux measurements (Ma et al., 2012). LC3-II, a molecular marker for autophagy induction, can be used to evaluate autophagy flux by its degradation rate. This measurement indicated that the rate of LC3-I to LC3-II conversion in the liver obviously peaks during the noon phase and later declines toward the dark phase (Czaja et al., 2013). In addition, autophagy undergoes rhythmic variation consistent with the circadian pattern of feeding in adult mammals. Studies have found that the number of autophagic vacuoles reaching the maximum before the initiation of feeding and the minimal number of autophagic vacuoles occurring after the animal has begun to feed (Mizushima and Murphy, 2020; Packer, 2020). These findings showed that autophagy, which is influenced by a series of factors, such as circulating hormones, may be inhibited under energy-rich conditions and be induced during starved conditions (Sachdeva and Thompson, 2008). Moreover, in the liver, the cyclic activation of autophagy flux is related to the rhythmic expression of autophagy genes (Li and Lin, 2015). In general, these studies confirmed that peripheral regulated circadian-dependent autophagy is manipulated in several organ systems and possibly plays a critical role in tissue and organ repair. This may also provide reasons for the theory that sleep hours are essential for daily physiological processes and tissue maintenance.
Additionally, the circadian regulation of autophagy was not only found in mammals but also discovered in eukaryotic cells, which contain several oscillating genes in the autophagy pathway (Kijak and Pyza, 2017). In yeast, microarray studies indicated that more than 50% of the yeast genome is loop-controlled in the period of metabolic restriction (Tu et al., 2005). Furthermore, the regulation of autophagy genes seems to follow a particular temporal expression pattern with a decrease in metabolic functions. This finding confirmed the point that cell homeostasis and cell death events take place simultaneously, which is greatly conserved even in a simple eukaryotic cell (Rabinovich-Nikitin et al., 2019).
Circadian Rhythm Regulation of Autophagy
The autophagy activities and the number of autophagic vacuoles have been found to vary during the day in many tissues. Several genes and pathways involved in autophagy are present in a rhythmical manner during diurnal variation (Table 1).
CCAAT/enhancer binding protein β (C/EBPβ) is a basic leucine zipper transcription factor that is critical for the activation of autophagy in response to starvation as well as during light/dark cycles (Xu et al., 2018). C/EBPβ can directly bind to the promoters of autophagy genes and induce their transcription (Barakat et al., 2016). At present, C/EBPβ is emerging as a significant factor involved in rhythmic autophagy gene expression, such as ULK1, microtubule-associated protein 1A/1B-light chain 3B (LC3B), BCL2/adenovirus E1B interacting protein 3 (BNIP3) and GABA (A) receptor-associated protein like 1 (GABARAPL1), which are expressed in an oscillating manner and are mediated by circadian and nutritional signals (Ma and Lin, 2012). In an in vivo study, Ma et al. found that adenoviral-mediated RNAi knockdown of C/EBPβ eliminated circadian-regulated autophagy. Furthermore, these authors also discovered altered C/EBPβ levels, disrupted rhythmic regulation of autophagy and decreased autophagic gene expression in liver-specific BMAL1 knockout mice (Ma et al., 2011). Interestingly, in this study, low autophagic flux occurred in line with feeding with the onset of the dark phase, while 24-h starvation did not influence the rhythmicity of autophagy gene expression. The results showed an association between the cyclic regulation of autophagy and the dependence of autophagic rhythm on nutritional signals. This finding indicated that the zeitgebers of the circadian rhythm can directly influence the temporal and spatial expression of autophagic genes.
Another bridge that functionally connects autophagy and circadian rhythm is the adenosine monophosphate-activated protein kinase (AMPK) pathway. AMPK is a well-known energy-sensing kinase that participates in various catabolic and anabolic processes, including oxidative metabolism, glucose uptake, and nutrient biosynthesis (Gonzalez et al., 2020). In addition, autophagy is negatively regulated by the mTOR signaling pathway and by a downstream cascade of Atgs, such as BECLIN-1, LC3, ATG1, and ATG5 (Mizushima et al., 2008). Recently, studies have shown that AMPK can induce autophagy through dual mechanisms, including the direct activation and the abolishment of inhibitory effects of mTOR complex 1 on ULK1 through activation of the hamartin (tuberous sclerosis 1)/tuberin (tuberous sclerosis 2) (TSC1/TSC2) complex (Kim et al., 2011; Shang et al., 2011). Interestingly, both AMPK and mTOR show rhythmic regulation. Lamia et al. found that AMPK activity and nuclear localization, which showed a cyclic rhythm, were negatively related to CRY1 nuclear expression in mouse hepatocytes (Lamia et al., 2009). In addition, activation of AMPK unbalanced CRY proteins and normal rhythmic oscillations. Destruction of AMPK pathways significantly impaired peripheral clock activity in mice (Lee and Kim, 2013). Thus, AMPK provides another direction for the rhythmic regulation of autophagy based on its ability to control the cyclic expression of mTOR through regulation of CRY1.
The peroxisome proliferator–activated receptor gamma coactivator 1 (PGC-1) family of transcriptional coactivators includes PGC-1α and PGC-1β (Piccinin et al., 2018). PGC-1α and PGC-1β can activate the expression of mitochondrial gene programs encoded by both nuclear and mitochondrial genomes in many cell types (Nierenberg et al., 2018). In addition, studies have verified that the expression of PGC-1α and PGC-1β presents robust circadian rhythms in the liver and skeletal muscle. For PGC-1α, the association with circadian gene expression indicated that PGC-1α might regulate mitochondrial turnover in a circadian rhythm manner (Sonoda et al., 2007). Moreover, mice lacking PGC-1α exhibit aberrant light/dark cycles (Liu et al., 2007). Therefore, the interaction between PGC-1α and the circadian system might suggest another connection between circadian rhythm and autophagy. PGC-1β was discovered to physically connect with C/EBPβ in transiently transfected 293T cells, which indicated that it might be a C/EBPβ transcriptional coactivator (Li and Lin, 2015). More importantly, adenoviral-mediated PGC-1β overexpression was shown to promote the expression of autophagy genes regulated by C/EBPβ in hepatocytes, including ULK1, BNIP3, FIP200, GABARAPL1, ATG2A, ATG16L1, and ATPase H+ transporting V0 subunit a2 (ATP6V0A2) (Li and Lin, 2015). These results illustrated that PGC-1β might function as a coactivator for C/EBPβ to facilitate the expression of autophagy and strengthen autophagy activity. Thus, it is possible that the rhythmic activation of PGC-1β and C/EBPβ might regulate circadian signaling to activate autophagy rhythmically.
The MiT-TFE transcription factors TFEB and TFE3 are the primary regulators of autophagy, lysosomal biogenesis and lysosomal exocytosis by activating the expression of many genes involved in these processes (Annunziata et al., 2019; Yang et al., 2020). Recent studies have found that TFEB and TFE3 are activated in a circadian manner and promote the expression of Rev-erbα (Nr1d1), a transcription inhibitor component of the main clock machinery that controls autophagy-related gene expression, such as BECLIN-1, BNIP3, ATG5, ATG7, and ULK1, however, their depletion damages Rev-erbα expression and oscillation (Pastore et al., 2019). In addition, TFEB/TFE3 and Rev-erbα bind the common promoter regions, indicating that they can induce the rhythmic expression of genes involved in autophagy (Pastore and Ballabio, 2019). Therefore, this study provides a novel mechanism by which the dynamic balance between TFEB/TFE3 and Rev-erbα can be used to regulate the rhythmic oscillation of autophagy.
Moreover, several other transcription factors, including forkhead transcription factor O3 (FoxO3), sterol regulatory element binding transcription factor 2 (SREBF2), heat shock factor 1 (HSF-1) and acyl-CoA-binding protein 3 (ACBP3), are involved in the circadian rhythm regulation of autophagy. FoxO3, a regulator of autophagy, has been shown to induce the expression of autophagy genes, including LC3B, BNIP3, BCL2 interacting protein 3 like (BNIP3L), and GABARAPL1 (Mammucari et al., 2007; Ruan et al., 2020). SREBF2 can increase autophagy in hepatocytes of high-fat diet-fed mice by directly activating the expression of the patatin-like phospholipase domain-containing enzyme (PNPLA8) gene, which associates with autophagosomes, in a circadian manner (Oishi et al., 2015; Kim et al., 2016). HSF-1 displays a circadian rhythm, inducing and inhibiting autophagy by acting on different autophagy-related genes under certain cellular conditions (Wang et al., 2015; Li et al., 2016; Barna et al., 2018). The overexpression of ACBP3 reinforces the degradation of ATG8 and obstructed autophagosome formation (Xiao and Chye, 2010). A previous study found that the circadian regulation of ACBP3, which is upregulated in darkness but suppressed by light, is mediated by the cis-responsive elements DNA-binding with one finger box (Dof) and GT-1 (Zheng et al., 2012). Furthermore, autophagy-related genes and mTOR also display robust circadian rhythms (Rotter and Rothermel, 2012). For example, the expression of the ATG14 gene presents a circadian rhythm, which is controlled by Clock/Bmal1, the core clock component (Xiong et al., 2012; Jenwitheesuk et al., 2014). Therefore, understanding these molecules, an important aspect of the autophagy rhythm, is essential for further exploring the mechanisms of circadian regulation of autophagy.
Regulation of Circadian Rhythm by Autophagy
Circadian rhythm can regulate autophagy and be regulated by autophagy. Jeong et al. (2015) first found evidence for the autophagic regulation of a core clock component. Pharmacological and molecular studies have shown the stabilization of BMAL1 by CLOCKΔ19 through the mechanism of attenuating the autophagic degradation of Bmal1 in Clk/ + mice. This investigation presented a novel style of clock regulation by autophagy. Another study showed that autophagy leads to the degradation of CRY1, which binds to LC3 via its LC3-interacting region (LIR) motifs, in the liver within a temporal window when rodents generally eat less and live on gluconeogenesis (Toledo et al., 2018). The livers were demonstrated to show enhanced CRY1 levels and reduced blood glucose levels in ATG7-deficient mice, which was rescued by reducing the hepatic CRY1 content. This phenomenon indicated that the autophagic degradation of CRY1 functions to maintain blood glucose levels during decreased feeding. This study also showed that PER2 might be a cargo adapter that promotes the interaction of CRY1 with LC3. Moreover, in a study on the mechanism of ferroptosis, Liu et al. discovered that autophagy facilitates ferroptotic cell death via the selective degradation of ARNTL/BMAL1 (aryl hydrocarbon receptor nuclear translocator-like), a vital circadian clock regulator, through the cargo receptor SQSTM1/p62 (Liu et al., 2019). Thus, these studies suggest that autophagy and circadian rhythm are reciprocally regulated.
Molecular Regulation of Both Autophagy and Circadian Rhythm
There are several molecules that regulate both autophagy and circadian rhythm (Table 2). For example, casein kinase 1α (CK1α), belonging to the CK1 family of proteins, exhibits dual functions in autophagy regulation (Cai et al., 2018; Hermanova and Carracedo, 2018). Suppression CK1α by D4476, a CK1 inhibitor, or siRNA-mediated knockdown of CK1α leaded to the inhibition of mTOR signaling, and activation of autophagy (Gao et al., 2011; Zhao et al., 2011). CK1α could suppress p53 downstream of MDM2-mediated autophagy in acute myeloid leukemia (Xu et al., 2020). However, the overexpression of CK1α has been shown to convectively activate autophagic flux in non-small cell lung cancer (NSCLC) through the PTEN/AKT/FOXO3A/ATG7 axis (Jiang et al., 2018). In addition, CK1α-mediated phosphorylation facilitates the degradation of PER1, indicating an effect on circadian rhythm (Lam et al., 2018). Melatonin, N-acetyl-5-methoxytryptamine, is produced by the pineal gland and various other tissues (Luo et al., 2019). The regulation of autophagy by melatonin is depending on physiological status and diseases. Under the state of aging, melatonin induces autophagy by decreasing the mitochondrial membrane rigidity, however, as for virus infection and high fructose consumption, melatonin functions as an inhibitor of autophagy via the antioxidant-mediated effect and ER stress-mediated effect (Garcia et al., 2011; San-Miguel et al., 2014; Xie et al., 2015; Lin C. et al., 2016; Bermejo-Millo et al., 2018). In addition, in some diseases, such as cancers, neurodegeneration and obesity, melatonin plays a dual role in autophagy through regulating the ROS/MST1 and PI3K/Akt/mTOR signaling pathways (Zheng et al., 2014; Yoo et al., 2016; Shen et al., 2018; Shi et al., 2018; Boga et al., 2019). Moreover, melatonin can upregulate the expression of CLOCK and PER2 proteins in human prostate cancer cells, showing an influence on circadian rhythm (Jung-Hynes et al., 2010). Silent mating type information regulation 2 homolog 1 (Saccharomyces cerevisiae) (SIRT1), a member of the sirtuin family, can promote autophagy by blocking mTOR or activating AMPK (Wu Y. et al., 2020). Furthermore, SIRT1 enables deacetylation of histone BMAL1, PER, and CRY and thus regulates the transcription of circadian proteins and hence the cycle (Hirayama et al., 2007; Chung et al., 2010; Maiese, 2018). HSF1 exhibits dual functions in autophagy, as mentioned above, and can also induce the synchronization of the circadian clock by regulating PER2 protein directly (Tamaru et al., 2011; Li et al., 2016; Watanabe et al., 2017). Therefore, these molecules play an essential role in the regulation of rhythmic autophagy and can be used as targets for the clinical treatment of diseases.
Drugs Affecting the Circadian Rhythm of Autophagy
In addition to certain molecules influencing rhythmic autophagy, some drugs can also exert similar effects (Table 3). For example, rapamycin, already used in clinical practice, is a potent regulator of autophagy (Alvers et al., 2009; Pulakat and Chen, 2020). Chloroquine and Lys05 are clinically relevant lysosomotropic agents that inhibit autophagy (Baquero et al., 2019; Erkisa et al., 2020). The compound SR8278 is the first REV-ERB antagonist, although its pharmacokinetic properties limit its pharmacological uses (Kojetin et al., 2011). A recent study found that the compound ARN5187 has a dual inhibitory effect on both autophagy and REV-ERB in BT-474 cells (De Mei et al., 2015). ARN5187 was demonstrated to have the ability to block lysosomal function, disrupt the autophagy process in the late stage and decrease breast cancer cell viability, which showed that it is an autophagy suppressor. ARN5187 was also shown to repress REV-ERB-mediated transcription regulation. Moreover, ARN5187 was demonstrated to be more cytotoxic than chloroquine, and REV-ERB inhibitors were shown to be able to enhance the cytotoxicity of chloroquine in BT-474 cells. In addition, research has showed that folic acid deficiency can enhance the activity of autophagy in HT-22 hippocampal neuron cells in vitro, accompanied by the induction of the expression of autophagy- and circadian-related genes, such as ATG12, ATG13, and PER2, through the glucocorticoid receptor-mediated pathway (Sun et al., 2016). This research suggested that folic acid might be involved in the regulation of rhythmic autophagy. Apart from these results glucocorticoids have been reported to decrease autophagic activity in the placenta and bone, whereas they are activated in muscle and lymphocytes (Harr et al., 2010; Troncoso et al., 2014; He B. et al., 2016; Lin N. Y. et al., 2016). Morphine was shown to increase the mRNA expression levels of autophagy-related genes, including ATG3, ATG5, ATG7, and ATG12, in C6 cells (Feng et al., 2013). Autophagy can be regulated during doxorubicin-induced cardiotoxicity (Kobayashi et al., 2010; Sishi et al., 2013). In summary, these studies demonstrated the significance of rhythmic autophagy in organisms and suggest that this process could be used as a target for the clinical therapy of diseases.
The Applications of Circadian Rhythm of Autophagy in Organisms and Diseases
Rhythmic autophagy induction enables the maintenance of energy and nutrient homeostasis, the implementation of temporal compartmentalization of tissue metabolism and remodeling of proteomes and organelles throughout the light/dark and feeding cycles (Solanas et al., 2017; Mazzoccoli et al., 2018; Stockman et al., 2018). The concentrations of plasma amino acids and metabolites show intense circadian rhythms, which are mediated by autophagy in part. In addition, these nutrients can be used for the biosynthesis of essential macromolecules in nutrient-limited periods and can also enter systemic circulation for energy homeostasis in organisms (Ma and Lin, 2012). The expression of genes participating in glucose metabolism, cholesterol biosynthesis, de novo lipogenesis and fatty acid β-oxidation greatly oscillate in the liver, indicating that the coordination of circadian patterns of metabolic cycles with rhythmic autophagy has the ability to optimize nutrient storage and fuel oxidation (Tarquini and Mazzoccoli, 2017; Roohbakhsh et al., 2018).
Moreover, a comprehensive understanding of the associations between autophagy and circadian rhythm has significant impacts on human diseases (Kondratova and Kondratov, 2012; Esterline et al., 2018). For example, autophagy is involved in the regulation of tumor development and progression at both the cellular and organismal levels, a condition also impacted by circadian rhythm (Czaja et al., 2013; Chok et al., 2019). Autophagy has been shown to be involved in aging, a process related to circadian oscillation, based on the finding that the amassing of neural aggregates observed in aging is associated with a decrease in the autophagy pathway (Blagosklonny et al., 2010; Jirakkakul et al., 2018). The dysfunction of circadian rhythm is also connected to age-related neurodegenerative disorders, such as Alzheimer’s disease, in which autophagy plays a part in the pathogenesis and progression of disease (Chen et al., 2015; Doktor et al., 2019). The absence of the PER1 protein in the hippocampus might exacerbate the pathology of cerebral ischemia due to depressed autophagy (Maiese, 2017). Additionally, other diseases, such as osteoporosis, obesity, diabetes and cardiovascular disease, are also influenced by both autophagy and circadian rhythm (Blaney Davidson et al., 2017; McGinnis et al., 2017).
Discussion and Conclusion
Interference with the circadian rhythm of autophagy has been demonstrated to result in many disorders in organisms (Rozman, 2018; Sarker and Franks, 2018; Dong et al., 2019). Therefore, the reasons for the diseases induced by rhythmic autophagy dysfunction are discussed here. Rhythmic autophagy is capable of removing damaged materials from cells regularly, and a deficiency in this process can lead to the accumulation of abnormal cellular components, which finally induce cells and whole organisms to undergo pathological conditions (Li et al., 2020; Wu and Nagy, 2020). Autophagy might be activated to eliminate damaged organelles, proteins and lipids after oxidative phases of metabolism (Pacheco et al., 2020). The dysfunction of rhythmic autophagy might cause misfolded proteins to gather in cells, which can lead to alteration in membrane permeability, the generation of reactive oxygen species, and the disruption of mitochondria and DNA, facilitating metabolic diseases, neurodegeneration and aging (Sachdeva and Thompson, 2008; Abdrakhmanov et al., 2020; Gu et al., 2020). The circadian rhythm of autophagy regulation is considered to limit the accumulation of dysfunctional cell components, which are the core constituents of the pathogenesis of each of these disorders.
In addition, autophagy plays an important role in the development, organization and functions of the immune system, including cell survival, cell-autonomous defense and regulation of complex multicellular immune responses (Keller et al., 2020). For example, autophagy is necessary for keeping T cell survival, development and functional integrity under the circumstance of activating, however, autophagy deficiency in Treg cells will result in defective Treg function (Wei et al., 2016; Tan P. et al., 2019). As mention above, autophagy present circadian rhythm in many tissues and light/dark phase can influence its activity. Therefore, we speculate that light phase and less dark phase or vice-versa may have effects on autoimmunity by affecting autophagy, although it has not been reported yet. Above all, maintain the circadian rhythm of autophagy is essential for the construction of immune system.
Thus, a comprehensive study of the relationship between autophagy and circadian rhythm will allow us to better understand multiple disease processes, including cancer, neurodegeneration, aging, and metabolic disorders, all of which have been associated either directly or indirectly with both autophagy and circadian-controlled genes. The understanding of the mechanisms of the interaction between autophagy and circadian rhythm provides insights into the clinical treatment of diseases.
Author Contributions
ZG, YY, XW, and YC collected the related manuscript. ZX, XW, and XR drafted and wrote the manuscript. ZX, XW, SZ, BP and XR revised the manuscript. All authors have read and approved the final manuscript.
Funding
The study was supported by the grants from Natural Science Foundation of Hunan Province (2020JJ5934, 2019JJ50932), National Natural Science Foundation of China (81703036, 81803035), China Postdoctoral Science Foundation (2020M672521, 2017M610510), The Postdoctoral Science Foundation of Central South University (248485), and Fundamental Research Funds for the Central Universities of Central South University (2019zzts345, 2019zzts800).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Abbreviations
ATGs, Autophagy-related proteins; ULK1, Unc-51-like autophagy activating kinase; Mtor, Mechanistic target of rapamycin; PI3K, Phosphatidylinositol-3 kinase; FIP200, Focal adhesion kinase-family interacting protein of 200 kDa; VPS34, Vacuolar protein sorting protein 34; LC3-II, Microtubule-associated protein 1A/1B-light chain 3-II; LC3B, Microtubule-associated protein 1A/1B-light chain 3B; BNIP3, BCL2/adenovirus E1B interacting protein 3; GABARAPL1, GABA (A) receptor-associated protein like 1; ATP6V0A2, ATPase H+ transporting V0 subunit a2; SCN, Suprachiasmatic nucleus; PER, Period; CRY, Cryptochrome; C/EBP β, CCAAT/enhancer binding protein β; AMPK, Adenosine monophosphate-activated protein kinase; BNIP3L, BCL2 interacting protein 3 like; PGC-1, Peroxisome proliferator–activated receptor gamma coactivator 1; FoxO3, Forkhead transcription factor O3; SREBF2, Sterol regulatory element binding transcription factor 2; HSF-1, Heat shock factor 1; ACBP3, Acyl-CoA-binding protein 3; PNPLA8, Patatin-like phospholipase domain-containing enzyme; Dof, DNA-binding with one finger box; LIR, LC3-interacting region; CK1 α, Casein kinase 1 α; NSCLC, Non-small cell lung cancer; SIRT1, Silent mating type information regulation 2 homolog 1.
References
Abdrakhmanov, A., Gogvadze, V., and Zhivotovsky, B. (2020). To eat or to die: deciphering selective forms of autophagy. Trends Biochem. Sci. 45, 347–364. doi: 10.1016/j.tibs.2019.11.006
Alvers, A. L., Fishwick, L. K., Wood, M. S., Hu, D., Chung, H. S., and Dunn, W. A. Jr., et al. (2009). Autophagy and amino acid homeostasis are required for chronological longevity in Saccharomyces cerevisiae. Aging Cell 8, 353–369. doi: 10.1111/j.1474-9726.2009.00469.x
Annunziata, I., van de Vlekkert, D., Wolf, E., Finkelstein, D., Neale, G., Machado, E., et al. (2019). MYC competes with MiT/TFE in regulating lysosomal biogenesis and autophagy through an epigenetic rheostat. Nat. Commun. 10:3623. doi: 10.1038/s41467-019-11568-0
Baquero, P., Dawson, A., Mukhopadhyay, A., Kuntz, E. M., Mitchell, R., Olivares, O., et al. (2019). Targeting quiescent leukemic stem cells using second generation autophagy inhibitors. Leukemia 33, 981–994. doi: 10.1038/s41375-018-0252-4
Barakat, D. J., Mendonca, J., Barberi, T., Zhang, J., Kachhap, S. K., Paz-Priel, I., et al. (2016). C/EBPbeta regulates sensitivity to bortezomib in prostate cancer cells by inducing REDD1 and autophagosome-lysosome fusion. Cancer Lett. 375, 152–161. doi: 10.1016/j.canlet.2016.03.005
Barna, J., Csermely, P., and Vellai, T. (2018). Roles of heat shock factor 1 beyond the heat shock response. Cell. Mol. Life Sci. 75, 2897–2916. doi: 10.1007/s00018-018-2836-6
Bermejo-Millo, J. C., Guimaraes, M. R. M., de Luxan-Delgado, B., Potes, Y., Perez-Martinez, Z., Diaz-Luis, A., et al. (2018). High-fructose consumption impairs the redox system and protein quality control in the brain of syrian hamsters: therapeutic effects of melatonin. Mol. Neurobiol. 55, 7973–7986. doi: 10.1007/s12035-018-0967-2
Blagosklonny, M. V., Campisi, J., Sinclair, D. A., Bartke, A., Blasco, M. A., Bonner, W. M., et al. (2010). Impact papers on aging in 2009. Aging 2, 111–121. doi: 10.18632/aging.100132
Blaney Davidson, E. N., van Caam, A. P., and van der Kraan, P. M. (2017). Osteoarthritis year in review 2016: biology. Osteoarthr. Cartil. 25, 175–180. doi: 10.1016/j.joca.2016.09.024
Boga, J. A., Caballero, B., Potes, Y., Perez-Martinez, Z., Reiter, R. J., Vega-Naredo, I., et al. (2019). Therapeutic potential of melatonin related to its role as an autophagy regulator: a review. J. Pineal Res. 66:e12534. doi: 10.1111/jpi.12534
Bonam, S. R., Bayry, J., Tschan, M. P., and Muller, S. (2020). Progress and challenges in the use of MAP1LC3 as a legitimate marker for measuring dynamic autophagy In Vivo. Cells 9:1321. doi: 10.3390/cells9051321
Bonam, S. R., Wang, F., and Muller, S. (2018). Autophagy: A new concept in autoimmunity regulation and a novel therapeutic option. J. Autoimm. 94, 16–32. doi: 10.1016/j.jaut.2018.08.009
Boukhalfa, A., Nascimbeni, A. C., Ramel, D., Dupont, N., Hirsch, E., Gayral, S., et al. (2020). PI3KC2alpha-dependent and VPS34-independent generation of PI3P controls primary cilium-mediated autophagy in response to shear stress. Nat. Commun. 11:294. doi: 10.1038/s41467-019-14086-1
Brattas, P. L., Hersbach, B. A., Madsen, S., Petri, R., Jakobsson, J., and Pircs, K. (2020). Impact of differential and time-dependent autophagy activation on therapeutic efficacy in a model of Huntington disease. Autophagy 6, 1–14. doi: 10.1080/15548627.2020.1760014
Cai, J., Li, R., Xu, X., Zhang, L., Lian, R., Fang, L., et al. (2018). CK1alpha suppresses lung tumour growth by stabilizing PTEN and inducing autophagy. Nat. Cell Biol. 20, 465–478. doi: 10.1038/s41556-018-0065-8
Carmo-Silva, S., and Cavadas, C. (2017). Hypothalamic dysfunction in obesity and metabolic disorders. Adv. Neurobiol. 19, 73–116. doi: 10.1007/978-3-319-63260-5_4
Chen, X., Kondo, K., Motoki, K., Homma, H., and Okazawa, H. (2015). Fasting activates macroautophagy in neurons of Alzheimer’s disease mouse model but is insufficient to degrade amyloid-beta. Sci. Rep. 5:12115. doi: 10.1038/srep12115
Choi, A. M., Ryter, S. W., and Levine, B. (2013). Autophagy in human health and disease. New Engl. J. Med. 368, 651–662. doi: 10.1056/NEJMra1205406
Chok, K. C., Ng, C. H., Koh, R. Y., Ng, K. Y., and Chye, S. M. (2019). The potential therapeutic actions of melatonin in colorectal cancer. Hormone Mol. Biol. Clin. Investigat. 39, 1–13. doi: 10.1515/hmbci-2019-0001
Chung, S., Yao, H., Caito, S., Hwang, J. W., Arunachalam, G., and Rahman, I. (2010). Regulation of SIRT1 in cellular functions: role of polyphenols. Arch. Biochem. Biophys. 501, 79–90. doi: 10.1016/j.abb.2010.05.003
Czaja, M. J., Ding, W. X., Donohue, T. M. Jr., Friedman, S. L., Kim, J. S., Komatsu, M., et al. (2013). Functions of autophagy in normal and diseased liver. Autophagy 9, 1131–1158. doi: 10.4161/auto.25063
De Mei, C., Ercolani, L., Parodi, C., Veronesi, M., Lo Vecchio, C., Bottegoni, G., et al. (2015). Dual inhibition of REV-ERBbeta and autophagy as a novel pharmacological approach to induce cytotoxicity in cancer cells. Oncogene 34, 2597–2608. doi: 10.1038/onc.2014.203
Devis-Jauregui, L., Eritja, N., Davis, M. L., Matias-Guiu, X., and Llobet-Navas, D. (2020). Autophagy in the physiological endometrium and cancer. Autophagy 13, 1–19. doi: 10.1080/15548627.2020.1752548
Doktor, B., Damulewicz, M., and Pyza, E. (2019). Effects of MUL1 and PARKIN on the circadian clock, brain and behaviour in Drosophila Parkinson’s disease models. BMC Neurosci. 20:24. doi: 10.1186/s12868-019-0506-8
Dong, C., Gongora, R., Sosulski, M. L., Luo, F., and Sanchez, C. G. (2016). Regulation of transforming growth factor-beta1 (TGF-beta1)-induced pro-fibrotic activities by circadian clock gene BMAL1. Respir. Res. 17:4. doi: 10.1186/s12931-016-0320-0
Dong, Y., Digman, M. A., and Brewer, G. J. (2019). Age- and AD-related redox state of NADH in subcellular compartments by fluorescence lifetime imaging microscopy. GeroScience 41, 51–67. doi: 10.1007/s11357-019-00052-8
Dreyer, A. P., Martin, M. M., Fulgham, C. V., Jabr, D. A., Bai, L., Beshel, J., et al. (2019). A circadian output center controlling feeding:fasting rhythms in Drosophila. PLoS Genet. 15:e1008478. doi: 10.1371/journal.pgen.1008478
Erkisa, M., Aydinlik, S., Cevatemre, B., Aztopal, N., Akar, R. O., Celikler, S., et al. (2020). A promising therapeutic combination for metastatic prostate cancer: chloroquine as autophagy inhibitor and palladium(II) barbiturate complex. Biochimie 175, 159–172. doi: 10.1016/j.biochi.2020.05.010
Esterline, R. L., Vaag, A., Oscarsson, J., and Vora, J. (2018). Mechanisms in endocrinology: SGLT2 inhibitors: clinical benefits by restoration of normal diurnal metabolism? Eur. J. Endocrinol. 178, R113–R125. doi: 10.1530/EJE-17-0832
Farhan, M., Silva, M., Li, S., Yan, F., Fang, J., Peng, T., et al. (2020). The role of FOXOs and autophagy in cancer and metastasis-Implications in therapeutic development. Med. Res. Rev. 40, 2089–2113. doi: 10.1002/med.21695
Farias, Q. G. E., Mao, K., Hu, Z., Novaj, A., Cui, M. H., Gulinello, M., et al. (2019). Central IGF-1 protects against features of cognitive and sensorimotor decline with aging in male mice. GeroScience 41, 185–208. doi: 10.1007/s11357-019-00065-3
Feng, Y. M., Jia, Y. F., Su, L. Y., Wang, D., Lv, L., Xu, L., et al. (2013). Decreased mitochondrial DNA copy number in the hippocampus and peripheral blood during opiate addiction is mediated by autophagy and can be salvaged by melatonin. Autophagy 9, 1395–1406. doi: 10.4161/auto.25468
Fracchiolla, D., Chang, C., Hurley, J. H., and Martens, S. A. (2020). PI3K-WIPI2 positive feedback loop allosterically activates LC3 lipidation in autophagy. J. Cell Biol. 219:e201912098. doi: 10.1083/jcb.201912098
Gabryelska, A., Sochal, M., Turkiewicz, S., and Bialasiewicz, P. (2020). Relationship between HIF-1 and circadian clock proteins in obstructive sleep apnea patients-preliminary study. J. Clin. Med. 9:1599. doi: 10.3390/jcm9051599
Galluzzi, L., Bravo-San Pedro, J. M., Levine, B., Green, D. R., and Kroemer, G. (2017). Pharmacological modulation of autophagy: therapeutic potential and persisting obstacles. Nat. Rev. Drug Discov. 16, 487–511. doi: 10.1038/nrd.2017.22
Gao, D., Inuzuka, H., Tan, M. K., Fukushima, H., Locasale, J. W., Liu, P., et al. (2011). mTOR drives its own activation via SCF(betaTrCP)-dependent degradation of the mTOR inhibitor DEPTOR. Mol. Cell 44, 290–303. doi: 10.1016/j.molcel.2011.08.030
Garcia, J. J., Pinol-Ripoll, G., Martinez-Ballarin, E., Fuentes-Broto, L., Miana-Mena, F. J., Venegas, C., et al. (2011). Melatonin reduces membrane rigidity and oxidative damage in the brain of SAMP8 mice. Neurobiol. Aging 32, 2045–2054. doi: 10.1016/j.neurobiolaging.2009.12.013
Gatica, D., Lahiri, V., and Klionsky, D. J. (2018). Cargo recognition and degradation by selective autophagy. Nat. Cell Biol. 20, 233–242. doi: 10.1038/s41556-018-0037-z
Gonzalez, A., Hall, M. N., Lin, S. C., and Hardie, D. G. (2020). AMPK and TOR: the yin and yang of cellular nutrient sensing and growth control. Cell Metab. 31, 472–492. doi: 10.1016/j.cmet.2020.01.015
Gou, Q., Jiang, Y., Zhang, R., Xu, Y., Xu, H., Zhang, W., et al. (2020). PPARdelta is a regulator of autophagy by its phosphorylation. Oncogene 39, 4844–4853. doi: 10.1038/s41388-020-1329-x
Greco, C. M., and Sassone-Corsi, P. (2020). Personalized medicine and circadian rhythms: opportunities for modern society. J. Exp. Med. 217:e20200702. doi: 10.1084/jem.20200702
Gu, Y., Han, J., Jiang, C., and Zhang, Y. (2020). Biomarkers, oxidative stress and autophagy in skin aging. Ageing Res. Rev. 59:101036. doi: 10.1016/j.arr.2020.101036
Harr, M. W., McColl, K. S., Zhong, F., Molitoris, J. K., and Distelhorst, C. W. (2010). Glucocorticoids downregulate Fyn and inhibit IP(3)-mediated calcium signaling to promote autophagy in T lymphocytes. Autophagy 6, 912–921. doi: 10.4161/auto.6.7.13290
He, B., Zhang, N., and Zhao, R. (2016). Dexamethasone downregulates SLC7A5 expression and promotes cell cycle arrest, autophagy and apoptosis in BeWo cells. J. Cell. Physiol. 231, 233–242. doi: 10.1002/jcp.25076
He, Y., Cornelissen-Guillaume, G. G., He, J., Kastin, A. J., Harrison, L. M., and Pan, W. (2016). Circadian rhythm of autophagy proteins in hippocampus is blunted by sleep fragmentation. Chronobiol. Int. 33, 553–560. doi: 10.3109/07420528.2015.1137581
Hermanova, I., and Carracedo, A. (2018). CK1alpha promotes tumour suppressive autophagy. Nat. Cell Biol. 20, 369–371. doi: 10.1038/s41556-018-0079-2
Hirayama, J., Sahar, S., Grimaldi, B., Tamaru, T., Takamatsu, K., Nakahata, Y., et al. (2007). CLOCK-mediated acetylation of BMAL1 controls circadian function. Nature 450, 1086–1090. doi: 10.1038/nature06394
Jacomin, A. C., Petridi, S., Di Monaco, M., Bhujabal, Z., Jain, A., Mulakkal, N. C., et al. (2020). Regulation of Expression of autophagy genes by Atg8a-interacting partners sequoia, YL-1, and Sir2 in Drosophila. Cell Rep. 31:107695. doi: 10.1016/j.celrep.2020.107695
Jahanban-Esfahlan, R., Mehrzadi, S., Reiter, R. J., Seidi, K., Majidinia, M., Baghi, H. B., et al. (2018). Melatonin in regulation of inflammatory pathways in rheumatoid arthritis and osteoarthritis: involvement of circadian clock genes. Br. J. Pharmacol. 175, 3230–3238. doi: 10.1111/bph.13898
Jenwitheesuk, A., Nopparat, C., Mukda, S., Wongchitrat, P., and Govitrapong, P. (2014). Melatonin regulates aging and neurodegeneration through energy metabolism, epigenetics, autophagy and circadian rhythm pathways. Int. J. Mol. Sci. 15, 16848–16884. doi: 10.3390/ijms150916848
Jeong, K., He, B., Nohara, K., Park, N., Shin, Y., Kim, S., et al. (2015). Dual attenuation of proteasomal and autophagic BMAL1 degradation in Clock Delta19/+ mice contributes to improved glucose homeostasis. Sci. Rep. 5:12801. doi: 10.1038/srep12801
Jiang, S., Zhang, M., Sun, J., and Yang, X. (2018). Casein kinase 1alpha: biological mechanisms and theranostic potential. Cell Commun. Signal. 16:23. doi: 10.1186/s12964-018-0236-z
Jirakkakul, J., Roytrakul, S., Srisuksam, C., Swangmaneecharern, P., Kittisenachai, S., Jaresitthikunchai, J., et al. (2018). Culture degeneration in conidia of Beauveria bassiana and virulence determinants by proteomics. Fungal Biol. 122, 156–171. doi: 10.1016/j.funbio.2017.12.010
Jung-Hynes, B., Huang, W., Reiter, R. J., and Ahmad, N. (2010). Melatonin resynchronizes dysregulated circadian rhythm circuitry in human prostate cancer cells. J. Pineal Res. 49, 60–68. doi: 10.1111/j.1600-079X.2010.00767.x
Kaushik, S., and Cuervo, A. M. (2018). The coming of age of chaperone-mediated autophagy. Nat. Rev. Mol. Cell Biol. 19, 365–381. doi: 10.1038/s41580-018-0001-6
Keller, C. W., Munz, C., and Lunemann, J. D. (2020). Autophagy pathways in CNS myeloid cell immune functions. Trends Neurosci. 43, 1–10. doi: 10.1016/j.tins.2020.09.003
Kijak, E., and Pyza, E. (2017). TOR signaling pathway and autophagy are involved in the regulation of circadian rhythms in behavior and plasticity of L2 interneurons in the brain of Drosophila melanogaster. PLoS One 12:e0171848. doi: 10.1371/journal.pone.0171848
Kim, H., Williams, D., Qiu, Y., Song, Z., Yang, Z., Kimler, V., et al. (2019). Regulation of hepatic autophagy by stress-sensing transcription factor CREBH. FASEB J. 33, 7896–7914. doi: 10.1096/fj.201802528R
Kim, J., Kundu, M., Viollet, B., and Guan, K. L. (2011). AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat. Cell Biol. 13, 132–141. doi: 10.1038/ncb2152
Kim, K. Y., Jang, H. J., Yang, Y. R., Park, K. I., Seo, J., Shin, I. W., et al. (2016). SREBP-2/PNPLA8 axis improves non-alcoholic fatty liver disease through activation of autophagy. Sci. Rep. 6:35732. doi: 10.1038/srep35732
Kim, Y. H., and Lazar, M. A. (2020). Transcriptional control of circadian rhythms and metabolism: a matter of time and space. Endocr. Rev. 41:bnaa014. doi: 10.1210/endrev/bnaa014
Klionsky, D. J., Eskelinen, E. L., and Deretic, V. (2014). Autophagosomes, phagosomes, autolysosomes, phagolysosomes, autophagolysosomes wait, I’m confused. Autophagy 10, 549–551. doi: 10.4161/auto.28448
Kobayashi, S., Volden, P., Timm, D., Mao, K., Xu, X., and Liang, Q. (2010). Transcription factor GATA4 inhibits doxorubicin-induced autophagy and cardiomyocyte death. J. Biol. Chem. 285, 793–804. doi: 10.1074/jbc.M109.070037
Kojetin, D., Wang, Y., Kamenecka, T. M., and Burris, T. P. (2011). Identification of SR8278, a synthetic antagonist of the nuclear heme receptor REV-ERB. ACS Chem. Biol. 6, 131–134. doi: 10.1021/cb1002575
Kondratova, A. A., and Kondratov, R. V. (2012). The circadian clock and pathology of the ageing brain. Nat. Rev. Neurosci. 13, 325–335. doi: 10.1038/nrn3208
Lam, V. H., Li, Y. H., Liu, X., Murphy, K. A., Diehl, J. S., Kwok, R. S., et al. (2018). CK1alpha collaborates with doubletime to regulate period function in the Drosophila circadian clock. J. Neurosci. 38, 10631–10643. doi: 10.1523/JNEUROSCI.0871-18.2018
Lamia, K. A., Sachdeva, U. M., DiTacchio, L., Williams, E. C., Alvarez, J. G., Egan, D. F., et al. (2009). AMPK regulates the circadian clock by cryptochrome phosphorylation and degradation. Science 326, 437–440. doi: 10.1126/science.1172156
Lee, Y., and Kim, E. K. (2013). AMP-activated protein kinase as a key molecular link between metabolism and clockwork. Exp. Mol. Med. 45:e33. doi: 10.1038/emm.2013.65
Lescat, L., Veron, V., Mourot, B., Peron, S., Chenais, N., Dias, K., et al. (2020). Chaperone-mediated autophagy in the light of evolution: insight from fish. Mol. Biol. Evol. 37, 2887–2899. doi: 10.1093/molbev/msaa127
Levy, J. M. M., Towers, C. G., and Thorburn, A. (2017). Targeting autophagy in cancer. Nat. Rev. Cancer 17, 528–542. doi: 10.1038/nrc.2017.53
Li, F., Zhang, C., Tang, Z., Zhang, L., Dai, Z., Lyu, S., et al. (2020). A plant RNA virus activates selective autophagy in a UPR-dependent manner to promote virus infection. New Phytol. 288, 622–639. doi: 10.1111/nph.16716
Li, J., Chauve, L., Phelps, G., Brielmann, R. M., and Morimoto, R. I. (2016). E2F coregulates an essential HSF developmental program that is distinct from the heat-shock response. Genes Dev. 30, 2062–2075. doi: 10.1101/gad.283317.116
Li, S., and Lin, J. D. (2015). Transcriptional control of circadian metabolic rhythms in the liver. Diabetes Obesity Metab. 17(Suppl. 1), 33–38. doi: 10.1111/dom.12520
Lin, C., Chao, H., Li, Z., Xu, X., Liu, Y., Hou, L., et al. (2016). Melatonin attenuates traumatic brain injury-induced inflammation: a possible role for mitophagy. J. Pineal Res. 61, 177–186. doi: 10.1111/jpi.12337
Lin, N. Y., Chen, C. W., Kagwiria, R., Liang, R., Beyer, C., Distler, A., et al. (2016). Inactivation of autophagy ameliorates glucocorticoid-induced and ovariectomy-induced bone loss. Ann. Rheum. Dis. 75, 1203–1210. doi: 10.1136/annrheumdis-2015-207240
Liu, C., Li, S., Liu, T., Borjigin, J., and Lin, J. D. (2007). Transcriptional coactivator PGC-1alpha integrates the mammalian clock and energy metabolism. Nature 447, 477–481. doi: 10.1038/nature05767
Liu, J., Yang, M., Kang, R., Klionsky, D. J., and Tang, D. (2019). Autophagic degradation of the circadian clock regulator promotes ferroptosis. Autophagy 15, 2033–2035. doi: 10.1080/15548627.2019.1659623
Luo, C., Yang, Q., Liu, Y., Zhou, S., Jiang, J., Reiter, R. J., et al. (2019). The multiple protective roles and molecular mechanisms of melatonin and its precursor N-acetylserotonin in targeting brain injury and liver damage and in maintaining bone health. Free Radical Biol. Med. 130, 215–233. doi: 10.1016/j.freeradbiomed.2018.10.402
Lystad, A. H., Carlsson, S. R., and Simonsen, A. (2019). Toward the function of mammalian ATG12-ATG5-ATG16L1 complex in autophagy and related processes. Autophagy 15, 1485–1486. doi: 10.1080/15548627.2019.1618100
Ma, D., Li, S., Molusky, M. M., and Lin, J. D. (2012). Circadian autophagy rhythm: a link between clock and metabolism? Trends Endocrinol. Metab. 23, 319–325. doi: 10.1016/j.tem.2012.03.004
Ma, D., and Lin, J. D. (2012). Circadian regulation of autophagy rhythm through transcription factor C/EBPbeta. Autophagy 8, 124–125. doi: 10.4161/auto.8.1.18081
Ma, D., Panda, S., and Lin, J. D. (2011). Temporal orchestration of circadian autophagy rhythm by C/EBPbeta. EMBO J. 30, 4642–4651. doi: 10.1038/emboj.2011.322
Maiese, K. (2017). Moving to the rhythm with clock (Circadian) genes, autophagy, mTOR, and SIRT1 in degenerative disease and cancer. Curr. Neurovas. Res. 14, 299–304. doi: 10.2174/1567202614666170718092010
Maiese, K. (2018). Novel treatment strategies for the nervous system: circadian clock genes, non-coding RNAs, and Forkhead transcription factors. Curr. Neurovas. Res. 15, 81–91. doi: 10.2174/1567202615666180319151244
Mammucari, C., Milan, G., Romanello, V., Masiero, E., Rudolf, R., Del Piccolo, P., et al. (2007). FoxO3 controls autophagy in skeletal muscle in vivo. Cell Metab. 6, 458–471. doi: 10.1016/j.cmet.2007.11.001
Martel, J., Ojcius, D. M., Ko, Y. F., and Young, J. D. (2020). Phytochemicals as prebiotics and biological stress inducers. Trends Biochem. Sci. 45, 462–471. doi: 10.1016/j.tibs.2020.02.008
Mazzoccoli, G., De Cosmo, S., and Mazza, T. (2018). The biological clock: a pivotal hub in non-alcoholic fatty liver disease pathogenesis. Front. Physiol. 9:193. doi: 10.3389/fphys.2018.00193
McGinnis, G. R., Tang, Y., Brewer, R. A., Brahma, M. K., Stanley, H. L., Shanmugam, G., et al. (2017). Genetic disruption of the cardiomyocyte circadian clock differentially influences insulin-mediated processes in the heart. J. Mol. Cell. Cardiol. 110, 80–95. doi: 10.1016/j.yjmcc.2017.07.005
Mizushima, N., and Levine, B. (2020). Autophagy in human diseases. New Engl. J. Med. 383, 1564–1576. doi: 10.1056/NEJMra2022774
Mizushima, N., Levine, B., Cuervo, A. M., and Klionsky, D. J. (2008). Autophagy fights disease through cellular self-digestion. Nature 451, 1069–1075. doi: 10.1038/nature06639
Mizushima, N., and Murphy, L. O. (2020). Autophagy assays for biological discovery and therapeutic development. Trends Biochem. Sci. 45, 1080–1093. doi: 10.1016/j.tibs.2020.07.006
Nierenberg, A. A., Ghaznavi, S. A., Sande Mathias, I., Ellard, K. K., Janos, J. A., and Sylvia, L. G. (2018). Peroxisome proliferator-activated receptor gamma coactivator-1 alpha as a novel target for bipolar disorder and other neuropsychiatric disorders. Biol. Psychiatry 83, 761–769. doi: 10.1016/j.biopsych.2017.12.014
Oeing, C. U., Nakamura, T., Pan, S., Mishra, S., Dunkerly-Eyring, B. L., Kokkonen-Simon, K. M., et al. (2020). PKG1alpha cysteine-42 redox state controls mTORC1 activation in pathological cardiac hypertrophy. Circulat. Res. 127, 522–533. doi: 10.1161/CIRCRESAHA.119.315714
Oishi, K., Yamamoto, S., Itoh, N., Nakao, R., Yasumoto, Y., Tanaka, K., et al. (2015). Wheat alkylresorcinols suppress high-fat, high-sucrose diet-induced obesity and glucose intolerance by increasing insulin sensitivity and cholesterol excretion in male mice. J. Nutr. 145, 199–206. doi: 10.3945/jn.114.202754
Pacheco, Y., Lim, C. X., Weichhart, T., Valeyre, D., Bentaher, A., and Calender, A. (2020). Sarcoidosis and the mTOR, Rac1, and autophagy triad. Trends Immunol. 41, 286–299. doi: 10.1016/j.it.2020.01.007
Packer, M. (2020). Cardioprotective effects of sirtuin-1 and Its downstream effectors: potential role in mediating the heart failure benefits of SGLT2 (Sodium-Glucose Cotransporter 2) inhibitors. Circulat. Heart Fail. 13:e007197. doi: 10.1161/CIRCHEARTFAILURE.120.007197
Pastore, N., and Ballabio, A. (2019). Keeping the autophagy tempo. Autophagy 15, 1854–1856. doi: 10.1080/15548627.2019.1645545
Pastore, N., Vainshtein, A., Herz, N. J., Huynh, T., Brunetti, L., Klisch, T. J., et al. (2019). Nutrient-sensitive transcription factors TFEB and TFE3 couple autophagy and metabolism to the peripheral clock. EMBO J. 38:e101347. doi: 10.15252/embj.2018101347
Patra, S., Mishra, S. R., Behera, B. P., Mahapatra, K. K., Panigrahi, D. P., Bhol, C. S., et al. (2020). Autophagy-modulating phytochemicals in cancer therapeutics: current evidences and future perspectives. Semin. Cancer Biol. 62, 1–13. doi: 10.1016/j.semcancer.2020.05.008
Paul, S., Hanna, L., Harding, C., Hayter, E. A., Walmsley, L., Bechtold, D. A., et al. (2020). Output from VIP cells of the mammalian central clock regulates daily physiological rhythms. Nat. Commun. 11:1453. doi: 10.1038/s41467-020-15277-x
Pavlinov, I., Salkovski, M., and Aldrich, L. N. (2020). Beclin 1-ATG14L protein-protein interaction inhibitor selectively inhibits autophagy through disruption of VPS34 complex I. J. Am. Chem. Soc. 142, 8174–8182. doi: 10.1021/jacs.9b12705
Petherick, K. J., Conway, O. J., Mpamhanga, C., Osborne, S. A., Kamal, A., Saxty, B., et al. (2015). Pharmacological inhibition of ULK1 kinase blocks mammalian target of rapamycin (mTOR)-dependent autophagy. J. Biol. Chem. 290, 11376–11383. doi: 10.1074/jbc.C114.627778
Piccinin, E., Peres, C., Bellafante, E., Ducheix, S., Pinto, C., Villani, G., et al. (2018). Hepatic peroxisome proliferator-activated receptor gamma coactivator 1beta drives mitochondrial and anabolic signatures that contribute to hepatocellular carcinoma progression in mice. Hepatology 67, 884–898. doi: 10.1002/hep.29484
Pulakat, L., and Chen, H. H. (2020). Pro-senescence and anti-senescence mechanisms of cardiovascular aging: cardiac MicroRNA regulation of longevity drug-induced autophagy. Front. Pharmacol. 11:774. doi: 10.3389/fphar.2020.00774
Rabinovich-Nikitin, I., Lieberman, B., Martino, T. A., and Kirshenbaum, L. A. (2019). Circadian-regulated Cell death in cardiovascular diseases. Circulation 139, 965–980. doi: 10.1161/CIRCULATIONAHA.118.036550
Rijo-Ferreira, F., and Takahashi, J. S. (2019). Genomics of circadian rhythms in health and disease. Genome Med. 11:82. doi: 10.1186/s13073-019-0704-0
Roohbakhsh, A., Shamsizadeh, A., Hayes, A. W., Reiter, R. J., and Karimi, G. (2018). Melatonin as an endogenous regulator of diseases: the role of autophagy. Pharmacol. Res. 133, 265–276. doi: 10.1016/j.phrs.2018.01.022
Rotter, D., and Rothermel, B. A. (2012). Targets, trafficking, and timing of cardiac autophagy. Pharmacol. Res. 66, 494–504. doi: 10.1016/j.phrs.2012.10.001
Rozman, P. (2018). The potential of non-myeloablative heterochronous autologous hematopoietic stem cell transplantation for extending a healthy life span. GeroScience 40, 221–242. doi: 10.1007/s11357-018-0027-x
Ruan, C., Wang, C., Gong, X., Zhang, Y., Deng, W., Zhou, J., et al. (2020). An integrative multi-omics approach uncovers the regulatory role of CDK7 and CDK4 in autophagy activation induced by silica nanoparticles. Autophagy 24, 1–22. doi: 10.1080/15548627.2020.1763019
Ryzhikov, M., Ehlers, A., and Haspel, J. A. (2019a). Adventures in spacetime: circadian rhythms and the dynamics of protein catabolism. Autophagy 15, 1115–1116. doi: 10.1080/15548627.2019.1596498
Ryzhikov, M., Eubanks, A., and Haspel, J. A. (2019b). Measuring diurnal rhythms in autophagic and proteasomal flux. J. Visual. Exp. 151, 1–6. doi: 10.3791/60133
Sachdeva, U. M., and Thompson, C. B. (2008). Diurnal rhythms of autophagy: implications for cell biology and human disease. Autophagy 4, 581–589. doi: 10.4161/auto.6141
San-Miguel, B., Crespo, I., Vallejo, D., Alvarez, M., Prieto, J., Gonzalez-Gallego, J., et al. (2014). Melatonin modulates the autophagic response in acute liver failure induced by the rabbit hemorrhagic disease virus. J. Pineal Res. 56, 313–321. doi: 10.1111/jpi.12124
Santin-Marquez, R., Alarcon-Aguilar, A., Lopez-Diazguerrero, N. E., Chondrogianni, N., and Konigsberg, M. (2019). Sulforaphane – role in aging and neurodegeneration. GeroScience 41, 655–670. doi: 10.1007/s11357-019-00061-7
Sarker, M. R., and Franks, S. F. (2018). Efficacy of curcumin for age-associated cognitive decline: a narrative review of preclinical and clinical studies. GeroScience 40, 73–95. doi: 10.1007/s11357-018-0017-z
Schafer, J. A., Schessner, J. P., Bircham, P. W., Tsuji, T., Funaya, C., Pajonk, O., et al. (2020). ESCRT machinery mediates selective microautophagy of endoplasmic reticulum in yeast. EMBO J. 39:e102586. doi: 10.15252/embj.2019102586
Scotton, C., Bovolenta, M., Schwartz, E., Falzarano, M. S., Martoni, E., Passarelli, C., et al. (2016). Deep RNA profiling identified CLOCK and molecular clock genes as pathophysiological signatures in collagen VI myopathy. J. Cell Sci. 129, 1671–1684. doi: 10.1242/jcs.175927
Shang, L., Chen, S., Du, F., Li, S., Zhao, L., and Wang, X. (2011). Nutrient starvation elicits an acute autophagic response mediated by Ulk1 dephosphorylation and its subsequent dissociation from AMPK. Proc. Natl. Acad. Sci. U.S.A. 108, 4788–4793. doi: 10.1073/pnas.1100844108
Shen, Y. Q., Guerra-Librero, A., Fernandez-Gil, B. I., Florido, J., Garcia-Lopez, S., Martinez-Ruiz, L., et al. (2018). Combination of melatonin and rapamycin for head and neck cancer therapy: suppression of AKT/mTOR pathway activation, and activation of mitophagy and apoptosis via mitochondrial function regulation. J. Pineal Res. 64:e12461. doi: 10.1111/jpi.12461
Shi, L., Liang, F., Zheng, J., Zhou, K., Chen, S., Yu, J., et al. (2018). Melatonin regulates apoptosis and autophagy Via ROS-MST1 pathway in subarachnoid hemorrhage. Front. Mol. Neurosci. 11:93. doi: 10.3389/fnmol.2018.00093
Sishi, B. J., Loos, B., van Rooyen, J., and Engelbrecht, A. M. (2013). Autophagy upregulation promotes survival and attenuates doxorubicin-induced cardiotoxicity. Biochem. Pharmacol. 85, 124–134. doi: 10.1016/j.bcp.2012.10.005
Skene, D. J., Skornyakov, E., Chowdhury, N. R., Gajula, R. P., Middleton, B., Satterfield, B. C., et al. (2018). Separation of circadian- and behavior-driven metabolite rhythms in humans provides a window on peripheral oscillators and metabolism. Proc. Natl. Acad. Sci. U.S.A. 115, 7825–7830. doi: 10.1073/pnas.1801183115
Solanas, G., Peixoto, F. O., Perdiguero, E., Jardi, M., Ruiz-Bonilla, V., Datta, D., et al. (2017). Aged stem cells reprogram their daily rhythmic functions to adapt to stress. Cell 170, 678–692.e20. doi: 10.1016/j.cell.2017.07.035
Sonoda, J., Mehl, I. R., Chong, L. W., Nofsinger, R. R., and Evans, R. M. (2007). PGC-1beta controls mitochondrial metabolism to modulate circadian activity, adaptive thermogenesis, and hepatic steatosis. Proc. Natl. Acad. Sci. U.S.A. 104, 5223–5228. doi: 10.1073/pnas.0611623104
Stockman, M. C., Thomas, D., Burke, J., and Apovian, C. M. (2018). Intermittent fasting: is the wait worth the weight? Curr. Obesity Rep. 7, 172–185. doi: 10.1007/s13679-018-0308-9
Sulli, G., Rommel, A., Wang, X., Kolar, M. J., Puca, F., Saghatelian, A., et al. (2018). Pharmacological activation of REV-ERBs is lethal in cancer and oncogene-induced senescence. Nature 553, 351–355. doi: 10.1038/nature25170
Sun, Q., Yang, Y., Li, X., He, B., Jia, Y., Zhang, N., et al. (2016). Folate deprivation modulates the expression of autophagy- and circadian-related genes in HT-22 hippocampal neuron cells through GR-mediated pathway. Steroids 112, 12–19. doi: 10.1016/j.steroids.2016.04.010
Tamaru, T., Hattori, M., Honda, K., Benjamin, I., Ozawa, T., and Takamatsu, K. (2011). Synchronization of circadian Per2 rhythms and HSF1-BMAL1:CLOCK interaction in mouse fibroblasts after short-term heat shock pulse. PLoS One 6:e24521. doi: 10.1371/journal.pone.0024521
Tan, P., Ye, Y., Mao, J., and He, L. (2019). Autophagy and immune-related diseases. Adv. Exp. Med. Biol. 1209, 167–179. doi: 10.1007/978-981-15-0606-2_10
Tan, X., Ye, J., Liu, W., Zhao, B., Shi, X., Zhang, C., et al. (2019). Acrylamide aggravates cognitive deficits at night period via the gut-brain axis by reprogramming the brain circadian clock. Arch. Toxicol. 93, 467–486. doi: 10.1007/s00204-018-2340-7
Tarquini, R., and Mazzoccoli, G. (2017). Clock genes, metabolism, and cardiovascular risk. Heart Fail. Clin. 13, 645–655. doi: 10.1016/j.hfc.2017.05.001
Toledo, M., Batista-Gonzalez, A., Merheb, E., Aoun, M. L., Tarabra, E., Feng, D., et al. (2018). Autophagy regulates the liver clock and glucose metabolism by degrading CRY1. Cell Metab. 28, 268–281.e4. doi: 10.1016/j.cmet.2018.05.023
Troncoso, R., Paredes, F., Parra, V., Gatica, D., Vasquez-Trincado, C., Quiroga, C., et al. (2014). Dexamethasone-induced autophagy mediates muscle atrophy through mitochondrial clearance. Cell Cycle 13, 2281–2295. doi: 10.4161/cc.29272
Tu, B. P., Kudlicki, A., Rowicka, M., and McKnight, S. L. (2005). Logic of the yeast metabolic cycle: temporal compartmentalization of cellular processes. Science 310, 1152–1158. doi: 10.1126/science.1120499
Vahsen, B. F., Ribas, V. T., Sundermeyer, J., Boecker, A., Dambeck, V., Lenz, C., et al. (2020). Inhibition of the autophagic protein ULK1 attenuates axonal degeneration in vitro and in vivo, enhances translation, and modulates splicing. Cell Death Different. 27, 2810–2827. doi: 10.1038/s41418-020-0543-y
Van Dyck, L., and Casaer, M. P. (2019). Intermittent or continuous feeding: any difference during the first week? Curr. Opin. Crit. Care 25, 356–362. doi: 10.1097/MCC.0000000000000617
Wang, Y., Cai, S., Yin, L., Shi, K., Xia, X., Zhou, Y., et al. (2015). Tomato HsfA1a plays a critical role in plant drought tolerance by activating ATG genes and inducing autophagy. Autophagy 11, 2033–2047. doi: 10.1080/15548627.2015.1098798
Watanabe, Y., Tsujimura, A., Taguchi, K., and Tanaka, M. (2017). HSF1 stress response pathway regulates autophagy receptor SQSTM1/p62-associated proteostasis. Autophagy 13, 133–148. doi: 10.1080/15548627.2016.1248018
Wei, J., Long, L., Yang, K., Guy, C., Shrestha, S., Chen, Z., et al. (2016). Autophagy enforces functional integrity of regulatory T cells by coupling environmental cues and metabolic homeostasis. Nat. Immunol. 17, 277–285. doi: 10.1038/ni.3365
Wesselborg, S., and Stork, B. (2015). Autophagy signal transduction by ATG proteins: from hierarchies to networks. Cell. Mol. Life Sci. 72, 4721–4757. doi: 10.1007/s00018-015-2034-8
Wu, M. Y., Liu, L., Wang, E. J., Xiao, H. T., Cai, C. Z., Wang, J., et al. (2020). PI3KC3 complex subunit NRBF2 is required for apoptotic cell clearance to restrict intestinal inflammation. Autophagy 16, 1–16. doi: 10.1080/15548627.2020.1741332
Wu, X., and Nagy, L. E. (2020). MLKL contributes to Western diet-induced liver injury through inhibiting autophagy. Autophagy 16, 1351–1352. doi: 10.1080/15548627.2020.1760624
Wu, Y., Li, W., Hu, Y., Liu, Y., and Sun, X. (2020). Suppression of sirtuin 1 alleviates airway inflammation through mTORmediated autophagy. Mol. Med. Rep. 22, 2219–2226. doi: 10.3892/mmr.2020.11338
Xiao, S., and Chye, M. L. (2010). The Arabidopsis thaliana ACBP3 regulates leaf senescence by modulating phospholipid metabolism and ATG8 stability. Autophagy 6, 802–804. doi: 10.1105/tpc.110.075333
Xie, S., Deng, Y., Pan, Y. Y., Wang, Z. H., Ren, J., Guo, X. L., et al. (2015). Melatonin protects against chronic intermittent hypoxia-induced cardiac hypertrophy by modulating autophagy through the 5’ adenosine monophosphate-activated protein kinase pathway. Biochem. Biophys. Res. Commun. 464, 975–981. doi: 10.1016/j.bbrc.2015.06.149
Xiong, X., Tao, R., DePinho, R. A., and Dong, X. C. (2012). The autophagy-related gene 14 (Atg14) is regulated by forkhead box O transcription factors and circadian rhythms and plays a critical role in hepatic autophagy and lipid metabolism. J. Biol. Chem. 287, 39107–39114. doi: 10.1074/jbc.M112.412569
Xu, W., Huang, Z., Gan, Y., Chen, R., Huang, Y., Xue, B., et al. (2020). Casein kinase 1alpha inhibits p53 downstream of MDM2mediated autophagy and apoptosis in acute myeloid leukemia. Oncol. Rep. 44, 1895–1904. doi: 10.3892/or.2020.7760
Xu, X., Huang, E., Luo, B., Cai, D., Zhao, X., Luo, Q., et al. (2018). Methamphetamine exposure triggers apoptosis and autophagy in neuronal cells by activating the C/EBPbeta-related signaling pathway. FASEB J. 32, 6737–6759. doi: 10.1096/fj.201701460RRR
Yan, Y., Chen, X., Wang, X., Zhao, Z., Hu, W., Zeng, S., et al. (2019). The effects and the mechanisms of autophagy on the cancer-associated fibroblasts in cancer. J. Exp. Clin. Cancer Res. 38:171. doi: 10.1186/s13046-019-1172-5
Yang, C., Zhu, Z., Tong, B. C., Iyaswamy, A., Xie, W. J., Zhu, Y., et al. (2020). A stress response p38 MAP kinase inhibitor SB202190 promoted TFEB/TFE3-dependent autophagy and lysosomal biogenesis independent of p38. Redox Biol. 32:101445. doi: 10.1016/j.redox.2020.101445
Yoo, Y. M., Han, T. Y., and Kim, H. S. (2016). Melatonin suppresses autophagy induced by clinostat in preosteoblast MC3T3-E1 cells. Int. J. Mol. Sci. 17:526. doi: 10.3390/ijms17040526
Zhao, Y., Xiong, X., and Sun, Y. (2011). DEPTOR, an mTOR inhibitor, is a physiological substrate of SCF(betaTrCP) E3 ubiquitin ligase and regulates survival and autophagy. Mol. Cell 44, 304–316. doi: 10.1016/j.molcel.2011.08.029
Zheng, S. X., Xiao, S., and Chye, M. L. (2012). The gene encoding Arabidopsis acyl-CoA-binding protein 3 is pathogen inducible and subject to circadian regulation. J. Exp. Bot. 63, 2985–3000. doi: 10.1093/jxb/ers009
Zheng, Y., Hou, J., Liu, J., Yao, M., Li, L., Zhang, B., et al. (2014). Inhibition of autophagy contributes to melatonin-mediated neuroprotection against transient focal cerebral ischemia in rats. J. Pharmacol. Sci. 124, 354–364. doi: 10.1254/jphs.13220fp
Zheng, Z. G., Zhu, S. T., Cheng, H. M., Zhang, X., Cheng, G., Thu, P. M., et al. (2020). Discovery of a potent SCAP degrader that ameliorates HFD-induced obesity, hyperlipidemia and insulin resistance via an autophagy-independent lysosomal pathway. Autophagy 20, 1–22. doi: 10.1080/15548627.2020.1757955
Keywords: autophagy, circadian rhythm, mTOR, AMPK, diseases
Citation: Wang X, Xu Z, Cai Y, Zeng S, Peng B, Ren X, Yan Y and Gong Z (2020) Rheostatic Balance of Circadian Rhythm and Autophagy in Metabolism and Disease. Front. Cell Dev. Biol. 8:616434. doi: 10.3389/fcell.2020.616434
Received: 12 October 2020; Accepted: 04 November 2020;
Published: 24 November 2020.
Edited by:
Bechan Sharma, Allahabad University, IndiaReviewed by:
Srinivasa Reddy Bonam, Institut National de la Santé et de la Recherche Médicale (INSERM), FranceBrijesh Kumar Singh, Duke-NUS Medical School, Singapore
Copyright © 2020 Wang, Xu, Cai, Zeng, Peng, Ren, Yan and Gong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhicheng Gong, Z29uZ3poaWNoZW5nQGNzdS5lZHUuY24=; Yuanliang Yan, eWFueXVhbmxpYW5nQGNzdS5lZHUuY24=
 Xiang Wang
Xiang Wang Zhijie Xu
Zhijie Xu Yuan Cai2
Yuan Cai2 Yuanliang Yan
Yuanliang Yan Zhicheng Gong
Zhicheng Gong