- 1Hainan Provincial Key Laboratory of Carcinogenesis and Intervention, Hainan Medical College, Haikou, China
- 2Institution of Tumor, Hainan Medical College, Haikou, China
Alpha fetoprotein (AFP) plays a key role in stimulating the growth, metastasis and drug resistance of hepatocellular carcinoma (HCC). AFP is an important target molecule in the treatment of HCC. The application of AFP-derived peptides, AFP fragments and recombinant AFP (AFP-inhibiting fragments, AIFs) to inhibit the binding of AFP to intracellular proteins or its receptors is the basis of a new strategy for the treatment of HCC and other cancers. In addition, AIFs can be combined with drugs and delivery agents to target treatments to cancer. AIFs conjugated to anticancer drugs not only destroy cancer cells with these drugs but also activate immune cells to kill cancer cells. Furthermore, AIF delivery of drugs relieves immunosuppression and enhances chemotherapy effects. The synergism of immunotherapy and targeted chemotherapy is expected to play an important role in enhancing the treatment effect of patients with cancer. AIF delivery of drugs will be an available strategy for the targeted treatment of cancer in the future.
Introduction
Alpha fetoprotein (AFP) is an oncofetal protein that is highly expressed in fetal cells and in most patients with hepatocellular carcinoma (HCC), and it is a diagnostic marker of liver cancer (Bei and Mizejewski, 2011, 2020; Mizejewski, 2014, 2019; Bai et al., 2017; Kim et al., 2020; Mehta et al., 2020). Based on the origin, the types of AFP include natural AFP (nAFP), which is derived from fetal cells, and tumor-derived AFP (tAFP), which is highly expressed in HCC and other cancers. The forms of AFP are also categorized as secreted AFP (SeAFP) and cytoplasmic AFP (CyAFP) (Mizejewski, 2015a; Tcherkassova et al., 2017). Here, AFP mainly refers to tAFP, and CyAFP refers to cytoplasmic tAFP. The AFP amino acid sequence is highly homologous with albumin, and its structure is similar to that of albumin. However, the functions of AFP and albumin are different (Mizejewski, 2001, 2016). Albumin maintains stable plasma osmolality and delivers nutrients. AFP delivers nutrients, suppresses immunity and stimulates the growth of cancer cells. When the serum concentration of AFP is greater than 50 ng/ml in adult blood, it stimulates tissue regeneration or hematopoiesis, and it is also used by cancer cells to provide nutrients and stimulate growth (Mizejewski, 2002; He et al., 2014; Pak, 2018b).
Alpha fetoprotein regulates the expression of oncogenes, inhibits apoptosis, promotes cancer cell growth, enhances drug resistance, enhances the antitumor immune response, increases invasion, and increases metastasis, resulting in the malignant transformation of cancer, and these functions of AFP are referred to as AFP malignant behaviors (Meng et al., 2016; Lu et al., 2016; Suryatenggara et al., 2017; Komorowski et al., 2018; Mizejewski, 2019; Xue et al., 2020). AFP binds to its membrane receptor and cytoplasmic proteins to promote malignancy (Mizejewski, 2011b, 2014, 2015a, 2019; Pak, 2018b). Use of AFP-derived peptides, AFP fragments, and recombinant AFP (AFP-inhibiting fragments, AIFs) to prevent AFP from binding to signal transduction molecules or AFP receptors, thereby inhibiting the malignant behaviors mediated by AFP. Additionally, use of AIF conjugates with toxins or drugs to target its receptors to selectively destroy cancer cells (Mizejewski, 2011a, 2014; Pak, 2018b). Classically, AIF comprised of peptides and fragments which are derived from AFP domain-3. Here, we also categorize recombinant AFP (rhAFP) as an unique AIF because it can be used as a vector to deliver drugs to kill cancer cells. Peptide AIFs include AFP-derived growth inhibitory peptide (GIP) and its analogs. GIP does not bind the AFP receptor (AFPR), but it can enter the cells influence the enzyme activity of tumor cells (Mizejewski and Butterstein, 2006; Mizejewski et al., 2010). Other new peptides of AIF could be obtained from AFP domain-3, and these peptides can bind to AFPR or signal transduction molecules and serve as candidate decoy ligands to prevent malignancy mediated by AFP (Mizejewski, 2011b; Tcherkassova et al., 2017). Fragment AIFs include AFP-3BC, rAFP3D, and r3dAFP, which are fragments of protein derived from AFP domain-3, and they can deliver drugs and be endocytosed by cancer cells with high AFPR expression (Godovannyi et al., 2011; Posypanova et al., 2013; Yabbarov et al., 2013; Tcherkassova et al., 2017). The unique AIF, rhAFP is full-length AFP that is expressed in E. coli and in yeast cells (Arshad et al., 2015), and they can also be designed as candidate decoy ligands to deliver drugs to prevent AFP malignant behaviors. The reports on the use of AIFs for the treatment of cancer are shown in Table 1. In this review, we summarize the application of AIFs to deliver drugs for targeted cancer treatment.
Distribution and Function of the AFP Receptor in Cells
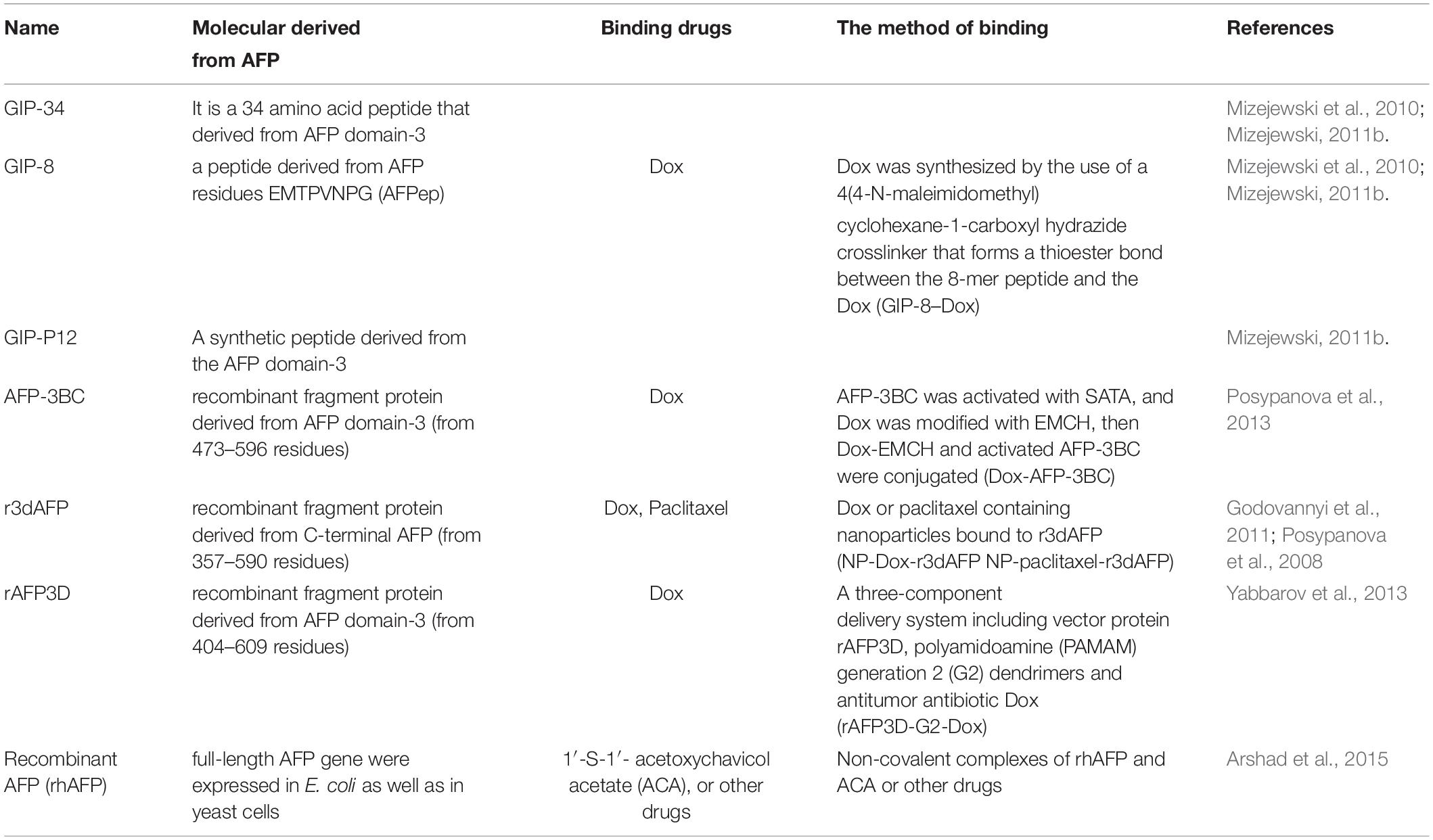
Alpha fetoprotein is a shuttle protein that is endocytosed mainly upon binding to its receptor. Previously, many researchers have suggested that AFP might bind to cellular membrane proteins (Naval et al., 1985; Biddle and Sarcione, 1987; Torres et al., 1989; Mizejewski, 1995, 2013, 2014, 2019), and further analysis indicated that these proteins are receptors of AFP (Suzuki et al., 1992; Mizejewski, 2014, 2019). The AFP receptor (AFPR) is expressed in myoblasts (fetal cells) (Lorenzo et al., 1996), NIH3T3 cells, and malignant cells (Laborda et al., 1987; Esteban et al., 1991; Torres et al., 1991; Li et al., 2002c; Mizejewski, 2011b), but it is not expressed in well-differentiated myotubes (adult-like cells) (Lorenzo et al., 1996). Recently, we detected the expression and location of AFPR in normal liver cells and HCC cells by immunohistochemistry and laser confocal microscopy. High expression of AFPR has been observed in the membrane of HCC cells (Li et al., 2013; Zhu et al., 2015b; Figure 1). Two subtypes of AFPR have been identified in NIH3T3 and HCC cells with different Kds (Li et al., 2002a,c), suggesting that AFPR exists in at least two subtypes. AFP binds with AFPR, which increases the concentrations of cAMP and Ca2+ in the cytoplasm and promotes the expression of some oncogenes (Li et al., 2002c, 2004). Activation of growth and signaling pathways are pivotal factors by which AFP promotes hepatocarcinogenesis (Wang et al., 2012; Mizejewski, 2015a; Zhang et al., 2015, 2016; Xue et al., 2020). These results suggest that AFPR in the cellular membrane has traits of G protein-coupled receptors (GPCRs) and that the signal transduction of AFPR follows the principles of GPCRs. Secreted AFP has many functions, such as immunosuppression, and it regulates the malignant behaviors of cancer cells through mediation by AFPR (Mizejewski, 2018, 2019).
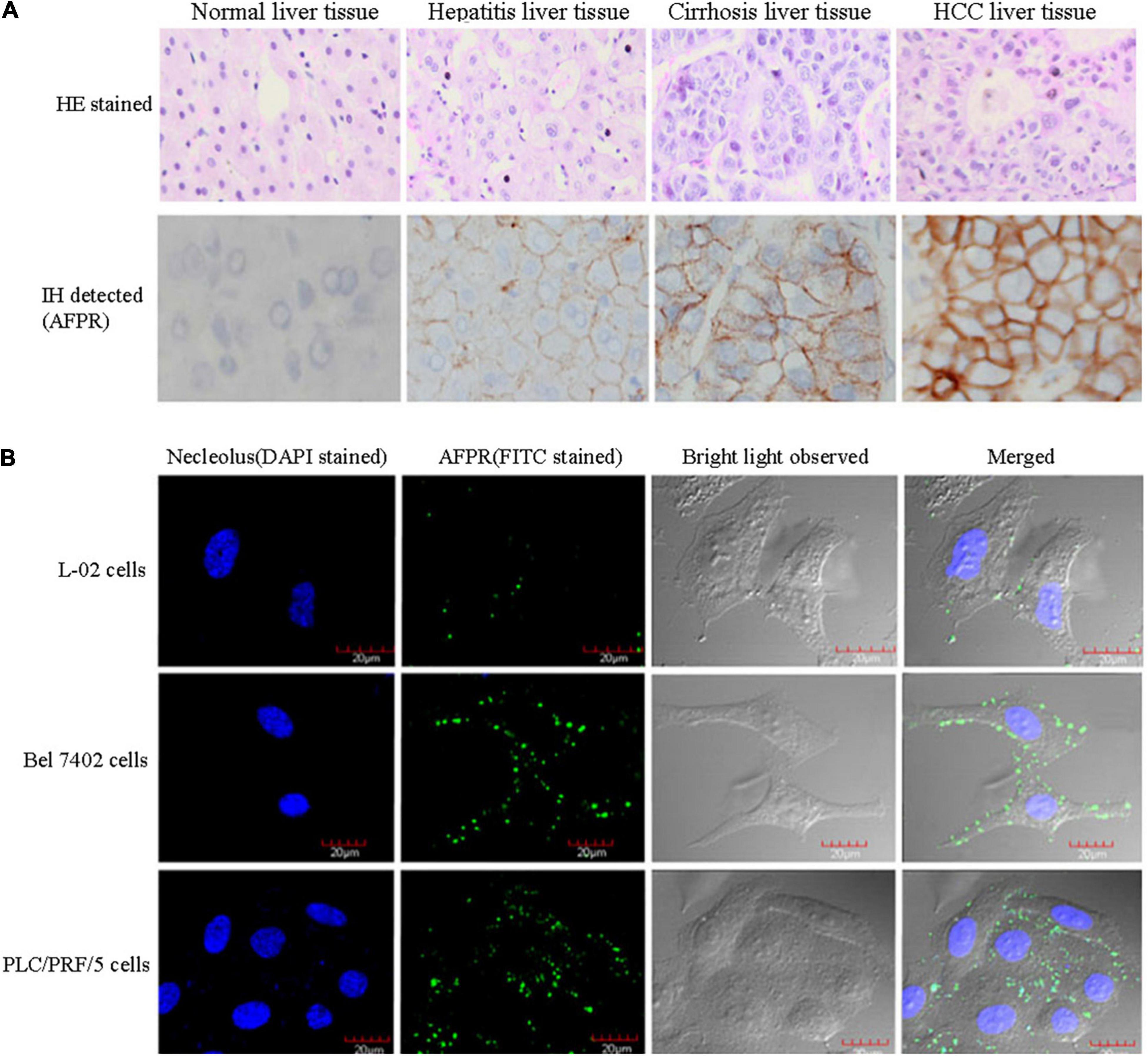
Figure 1. Expression of AFPR in clinical patient liver tissues and HCC cells. (A) Clinical liver tissue samples were collected after surgical hepatectomy, and the expression of AFPR in clinical patient liver tissues was detected by immunohistochemical assays. HE stained: hematoxylin-eosin stained; IH stained: immunohistochemical stained; red arrow indicates AFPR. (B) Expression and localization of AFPR (green fluorescence) in HCC cells were observed by laser confocal microscopy. Reprinted from Li et al. (2013) with permission from Elsevier.
AFP was initially found to be taken up by fetal cells (Sell et al., 1985; Iturralde et al., 1991; Alava et al., 1999; Nierhoff et al., 2005), and later studies found that muscle tumor cells also internalize exogenous AFP (Uriel et al., 1983; Lorenzo et al., 1996; Mizejewski, 2011b, 2018). In addition, AFP binds to cellular membrane receptors in pits in the membrane bilayer, thereby triggering their internalization by cells. AFP and its receptor are packaged in endosomal vesicles and transported to the trans-Golgi network distributed near the nucleus (Lorenzo et al., 1996; Torres et al., 1991; Mizejewski, 2011b, 2018). Finally, the vesicles release AFP and its receptor complex into the cytoplasm where they are translocated to cytoplasmic organelles undergoing lysosomal degradation or engage in signal transduction pathways. For example, AFP binds to PTEN in the cytoplasm and activates PI3K signaling pathways, thus stimulating the growth of many malignant cells (Wang et al., 2012; Zhang et al., 2015, 2016; Mizejewski, 2019).
Although AFP-binding receptors are critical for receptor-mediated endocytosis and the uptake of AFP into the cytoplasm, the complete AFP-binding receptor structure has not yet been elucidated. Many AFP-binding receptors have been reported, and they are mainly classified into two categories as follows: (a) the mucin (MUC) family and (b) the scavenger receptor (SR) family (Uriel et al., 1984; Mizejewski, 2013, 2014, 2015b, 2019). Although the details of the AFP-binding receptor structure are not known, many studies have shown that cancer cells take up AFP through AFP-binding receptors (Laborda et al., 1987; Esteban et al., 1991; Torres et al., 1991; Mizejewski, 2011b, 2013, 2014, 2019; Zhu et al., 2015b).
Because HCC and other cancer cells regain the ability to take up AFP via its receptor and exert malignant behaviors (Li et al., 2013; Zhu et al., 2015b), AFP delivery of cytotoxins is used to target and kill cancer cells. It has been demonstrated that AFP is effective for drug delivery, but AFP, especially tumor-derived AFP (tAFP), is also immunosuppressive and thus can stimulate immune escape of cancer cells. AFP may also promote initiation of cancer. Therefore, it is better to use AIFs to deliver drugs to target cancer therapeutics (Mizejewski, 2011a; Pak, 2014, 2018b). Experiments with radioactively labeled an AIF (AFP-3BC) have confirmed that they selectively accumulate in cancer cells and that AFP-3BC loaded with drugs binds to human breast MCF7 cells and ovarian adenocarcinoma SKOV3 cells, suppressing the proliferation of these cancer cells. Importantly, AFP-3BC do not bind to non-stimulated lymphocytes. These findings indicate that AFP-3BC can be a promising new vector for selectively targeting and inhibiting the malignant behaviors of cancer cells (Posypanova et al., 2008; Mizejewski, 2011a; Posypanova et al., 2013).
AFP Promotes the Malignant Behaviors of Cancer Cells
Because AFP is a growth-promoting factor, it mostly promotes the growth of cancer cells. AFP binding to its receptors activates the cAMP-PKA pathway and induces Ca2+ influx, which promotes the expression of the c-fos, c-jun and Ras oncogenes and stimulates the growth of hepatoma cells (Li et al., 2002a,c; Ma et al., 2010; Wang et al., 2012; Zhang et al., 2012, 2015, 2016). In addition, after binding to receptors, AFP not only triggers growth-promoting signals but also stimulates the endocytosis of AFP into cells (Torres et al., 1991; Mizejewski, 2011b; Kong et al., 2012). The endocytosed AFP is released (which becomes CyAFP) from its receptor and then binds with some cytoplasmic proteins, leading to the activation or inhibition of signaling pathways. For example, CyAFP binding to caspase-3 inhibits the apoptosis signaling pathway (Li et al., 2009a; Lin et al., 2017). CyAFP binding to caspase-3 is shown in Figure 2. Caspase-3, also called cysteine aspartyl proteinase 3, plays an important role in the apoptosis pathway of cancer cells (Riedl et al., 2001; Zhang et al., 2019; Jiao et al., 2020). Activated caspase-3 protein binds to its substrate and induces apoptosis through cascade amplification, indicating that caspase-3 is the main executor of apoptosis (Mittl et al., 1997; Rogers et al., 2017). The binding of CyAFP to caspase-3 prevents apoptotic signal transduction in HCC cells. CyAFP not only directly binds to caspase-3 and inhibits its activity but also affects the activation of caspase-3 through the mitochondrial apoptosis pathway. Yang et al. (2008, 2018) found that blocking the expression of AFP increases the ratio of Bax/Bcl-2 and releases cytochrome C from the mitochondria, thus activating caspase-3 to induce apoptosis. These results suggest that CyAFP inhibits apoptosis in HCC through the Bax/cytochrome C/caspase-3 signaling pathway and promotes the proliferation of hepatoma cells. In addition, CyAFP also binds to the all-trans retinoic acid (ATRA) receptor, RAR-β, and inhibits receptor entry into the nucleus, leading to increased expression of apoptosis-inhibiting proteins, such as survivin (Li et al., 2009b; Zhang et al., 2020).
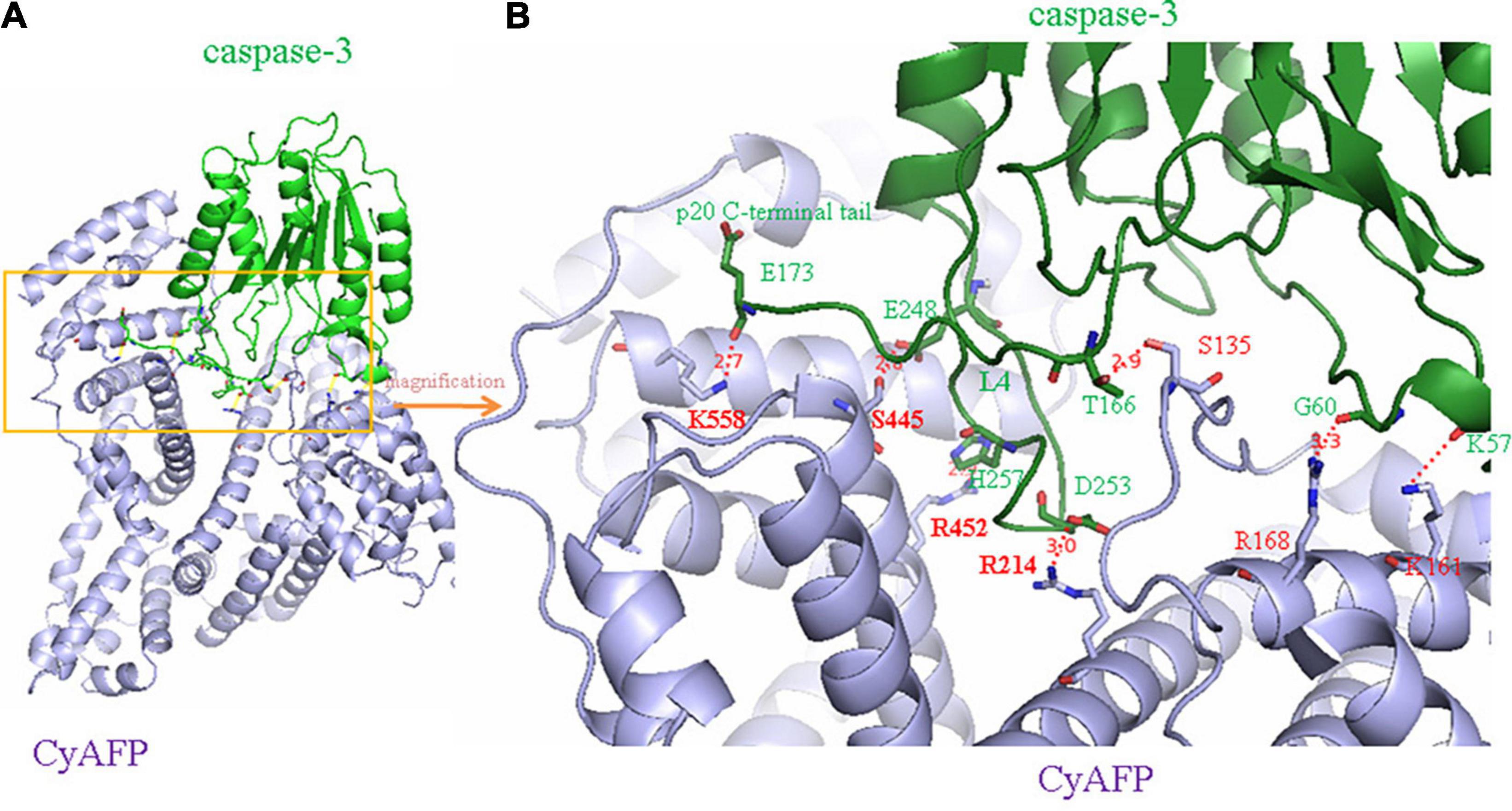
Figure 2. A molecular structure model of the interaction of CyAFP with caspase-3. (A) Overall structure of CyAFP in complex with caspase-3. CyAFP (blue) binds to caspase-3 (green) in the hydrophobic pocket. (B) Residues of CyAFP interact with caspase-3 (Lin et al., 2017).
In addition, a study based on laser confocal microscopy, immunoprecipitation, fluorescence energy resonance transfer, molecular simulation and site-directed mutagenesis has shown that CyAFP also binds to PTEN, which is an important tumor suppressor that negatively regulates the PI3K/Akt signaling pathway (Li et al., 2011; Wang et al., 2018). PTEN binds to PI3K subunits, inhibits the phosphorylation of PI3K and blocks signal transduction by PI3K/AKT (Lee et al., 2018). Specifically, the CyAFP interaction with PTEN activates the PI3K/AKT/mTOR pathway, inhibiting autophagy and promoting the malignant behavior of HCC by upregulating the expression of mTOR protein. After CyAFP binds to PTEN, the regulatory function of PTEN is lost, which leads to the continuous phosphorylation of PI3K and activation of the downstream molecule, AKT, thereby leading to the malignant transformation of liver cancer cells (Li et al., 2011; Wang et al., 2018). Activated AKT stimulates the mTOR transcription cofactor and the STAT3 and HIF-1α transcription factors, which regulates the expression of oncogenes, inhibits apoptosis and inhibits autophagy in hepatoma cells as well as promotes the growth of cancer cells (Missiaglia et al., 2010; Li et al., 2011; Lee et al., 2018; Wang et al., 2018).
Cytoplasmic AFP binding to PTEN not only promotes the growth of liver cancer cells but also enhances the drug resistance of cancer cells. For example, CyAFP plays an important role in promoting the drug resistance of HCC (Li et al., 2012, 2020). The binding of CyAFP to PTEN activates the PI3K/AKT signaling pathway and interferes with the activity of caspase-3 (Zhu et al., 2015a,c), which leads to the drug resistance of hepatoma cells. A high concentration of CyAFP in liver cancer cells not only promotes growth but also results in a loss of sensitivity to drugs in vivo (Cheng et al., 2013; Li et al., 2020). The 2018 EASL clinical practice guidelines suggest that AFP can be used as an indicator for the diagnosis and prognosis of advanced HCC (European Association for the Study of the Liver, 2018). Many HCC patients with elevated SeAFP or CyAFP expression may have drug resistance and a poor prognosis.
Design of AIFs From AFP Domain-3 and Applications for Targeting Deliver Drugs to Cancer Cells
Alpha fetoprotein has a molecular weight of 69 kDa and consists of a single peptide chain with 590∼609 amino acids and three domains. The N-terminal region of AFP, consisting of residues 1∼230, belongs to domain-1. The middle region of AFP, consisting of residues 230∼400, belongs to domain-2. The C-terminal region of AFP, consisting of residues 400∼609, belongs to domain-3. The overall AFP structure is V-shaped (see Figures 3A,B). Domain-1 (yellow) and domain-3 (red) are located on each side of the V-shape, and domain-2 (blue) is located at the bottom of the V-shape. A hydrophobic pocket is formed between domain-1 and domain-3, and it transports nutrients, such as fatty acids (Mizejewski, 2001, 2015b, 2016; Muehlemann et al., 2005).
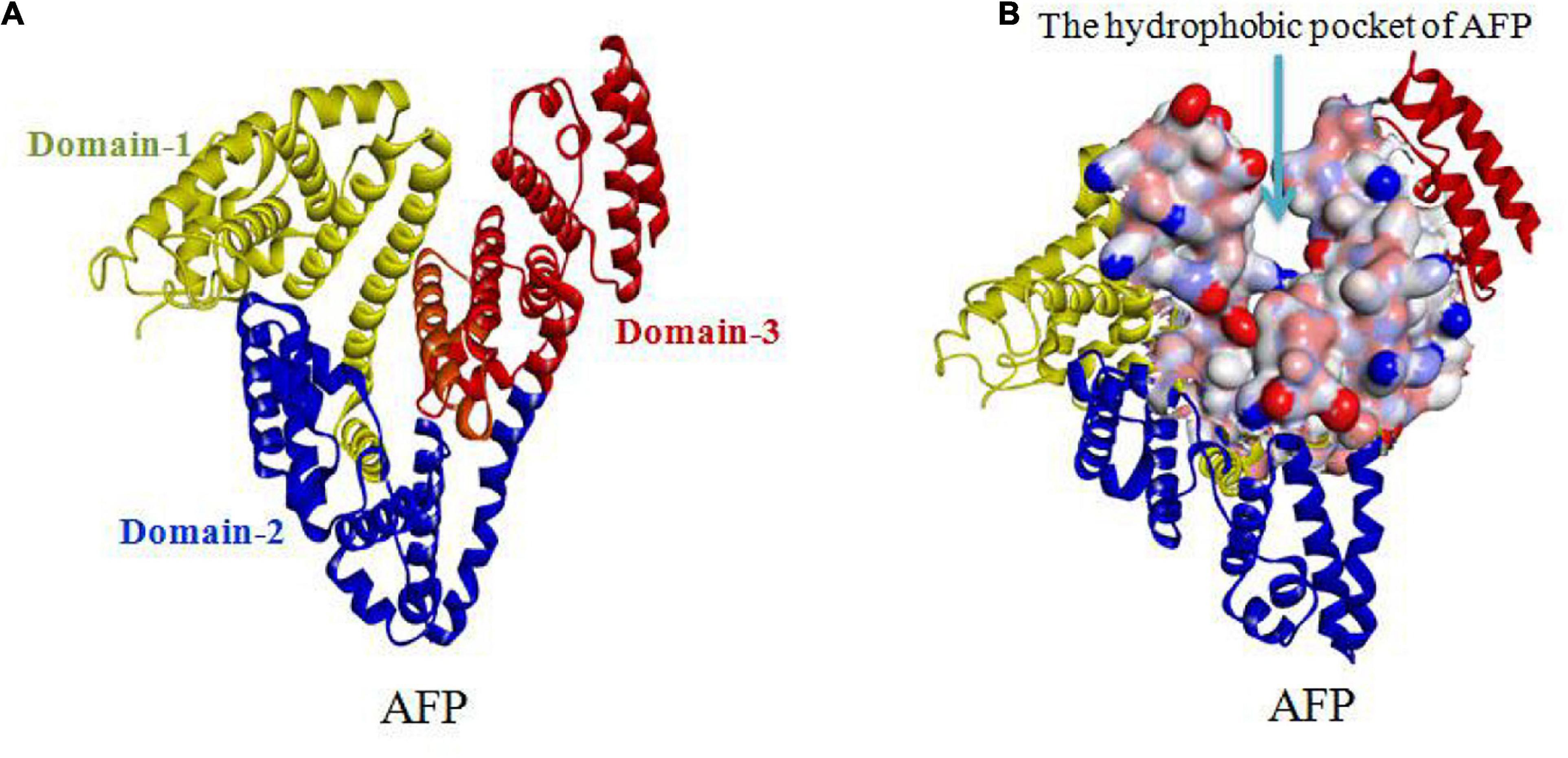
Figure 3. V-shaped AFP and its hydrophobic pocket. (A) Overall V-shaped structure of AFP comprised of three domains. Domain-1 (yellow) and domain-3 (red) are located on each side of the V-shape, and domain-2 (blue) is located at the bottom of the V-shape. (B) A hydrophobic pocket is formed between domain1 and domain-3 (Muehlemann et al., 2005; Pak, 2014; Mizejewski, 2015b, 2016).
Alpha fetoprotein domain-3 can be used to design peptide AIFs as vectors to deliver drugs to kill cancer cells. For example, GIP (Figures 4A,B) derived from domain-3 of the AFP sequence has the potential for treating cancer. GIP-34 and its analogs inhibit the migration and metastasis of cancer cells in both isograft and xenograft models. Additionally, GIP-34 and its analogs have been proposed to serve as vectors to deliver drugs to treat cancer, which will enhance the therapeutic effect (Mizejewski and Butterstein, 2006; Mizejewski et al., 2010; Mizejewski, 2011a). The anticancer mechanism of GIP may be explained as follows: GIP gains cell entry by: (1) direct cell membrane penetration; (2) channel formation; and/or (3) pore formation into the cell cytoplasm. And the cytoplasmic GIP influence enzyme activity which mediated by CyAFP during the growth and metastasis of cancer cells (Figure 4C). Such as GIP causes cancer cell growth suppression by inducing cell cycle arrest in the G1 to S-phase by preventing cell cycle p27 and p21 inhibitor degradations, thus halting cell cycle progression (Mizejewski and Butterstein, 2006; Mizejewski et al., 2010; Mizejewski, 2011a, 2013).
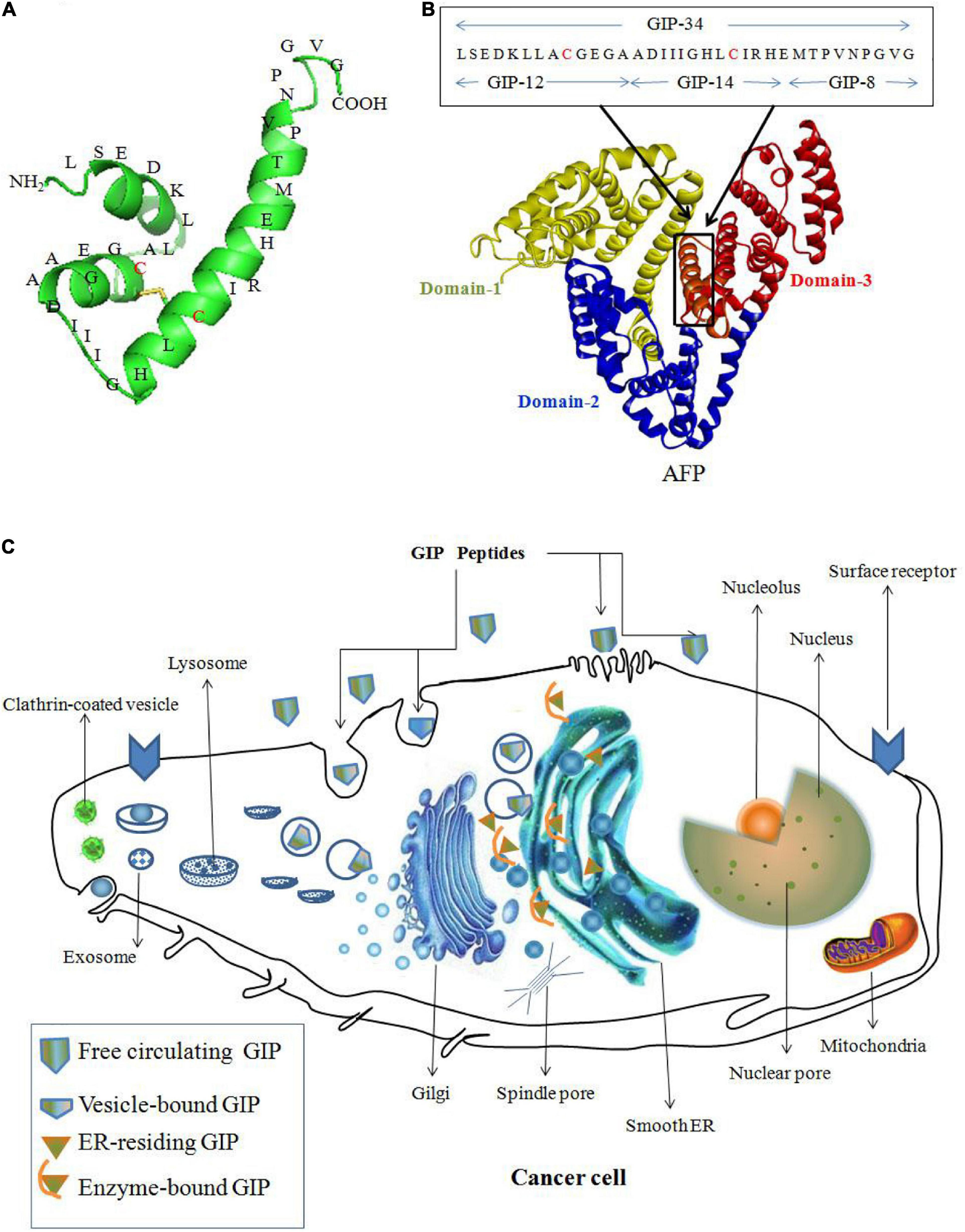
Figure 4. Peptide AIFs (GIP) derived from AFP domain-3 are taken up by cancer cells and prevent AFP-induced malignant functions. (A) Amino acid sequence of GIP-34. (B) Amino acid sequence of GIP analogs (GIP-8, 12, 14 and 34) derived from the AFP domain-3 sequence (Mizejewski et al., 2010). (C) The anticancer mechanism of GIP. GIP gains cell entry by: (1) direct cell membrane penetration; (2) channel formation; and/or (3) pore formation into the cell cytoplasm, and the cytoplasmic GIP target the smooth endoplasmic reticulum (ER) surrounding the nucleus and influence enzyme activity during the growth and metastasis of cancer cells (Mizejewski and Butterstein, 2006; Mizejewski et al., 2010; Mizejewski, 2011a).
As mentioned previously, the AFP domain-3 can also be used to design fragment AIFs conjugated with toxins. For example, Yabbarov et al. applied a fragment AIF (rAFP3D, which is designed from AFP domain-3) as a vector molecule conjugated to doxorubicin (Dox) (shown in Figure 5A) and utilized the drug-sensitive human ovarian adenocarcinoma SKOV3 cell line and the drug-resistant human ovarian adenocarcinoma SKVLB cell line to observe rAFP3D-Dox in these cells (Yabbarov et al., 2013). These researchers found that in drug-sensitive SKOV3 cells, there was little difference in the accumulation of Dox in the cytoplasm and nucleus when treated with free Dox or rAFP3D-Dox, but in the drug-resistant SKVLB cells, there was a significant increase in the accumulation of Dox in the cytoplasm and nucleus when treated with the rAFP3D-Dox compared to the control free Dox (Figure 5B). These results show that rAFP3D conjugated with fluorescein or Dox can be taken up by cancer cells, indicating that AFPR mediates AFP-derived rAFP3D-fluorescein or rAFP3D-Dox endocytosis into cancer cells and that rAFP3D-Dox induces cytotoxicity, resulting in cancer cell destruction. Thus, these studies indicate that rAFP3D can be applied in cancer treatment (Yabbarov et al., 2013).
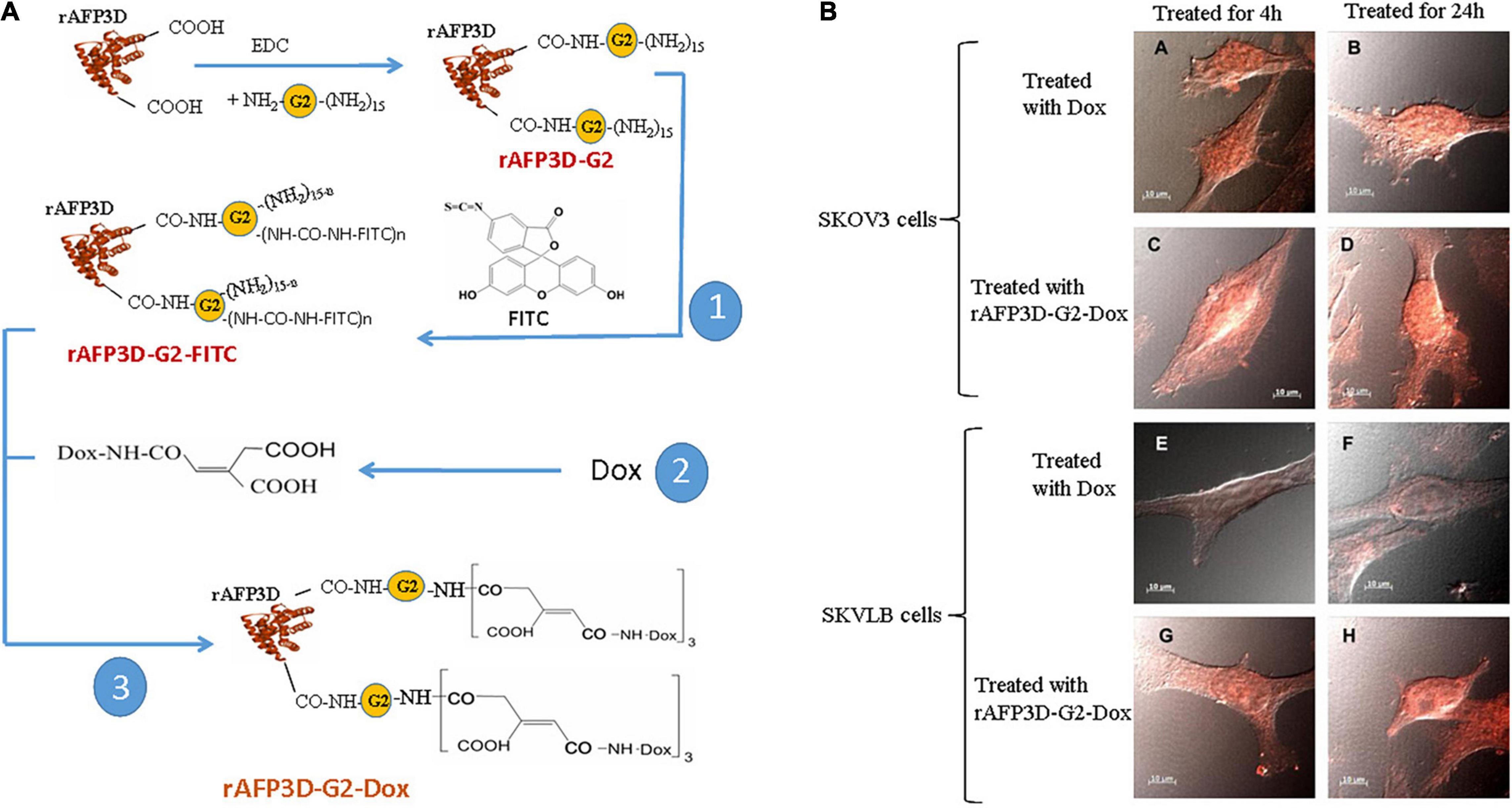
Figure 5. Scheme of conjugate synthesis of rAFP3D-G2-Dox and the accumulation of doxorubicin (Dox) in the drug-sensitive human ovarian adenocarcinoma SKOV3 cell line and the drug-resistant human ovarian adenocarcinoma SKVLB cell line. (A) rAFP3D conjugated to Dox (rAFP3D-G2-Dox). (B) Intracellular distribution of rAFP3D-G2-Dox and free Dox in (A∼D) the drug-sensitive human ovarian adenocarcinoma SKOV3 cell line and (E∼H) the drug-resistant human ovarian adenocarcinoma SKVLB cell line. Reprinted from Yabbarov et al. (2013) with permission from Elsevier.
Fragment AIFs can be used to prevent the AFP-mediated activation of proliferation-related signaling pathways to prevent drug resistance. Fragment AIFs can be conjugated with drugs to improve the sensitivity of cancer cells to agents (Godovannyi et al., 2011; Pak, 2018b). During cancer therapy, cancer cells may reduce their intake of anticancer drugs, such as methotrexate, paclitaxel, anthracyclines, platinum derivatives, 5-fluorouracil (5-FU), gemcitabine, capecitabine and sorafenib (Godovannyi et al., 2011; Pak, 2014), which may lead to decreased or inactivated drug sensitivity. However, these drugs can be conjugated to fragmented AIFs. Because fragment AIFs, such as rAFP3D, recognize and bind to AFP receptors on the membranes of cancer cells, they transport drugs into the cell through receptor-mediated endocytosis, which increases the intake of drugs and enhances the accumulation of drugs, allowing the drugs to exert their cytotoxic effects. For example, rAFP3D conjugated to Dox (rAFP3D-G2-Dox) increases the sensitivity of human ovarian carcinoma cells, breast cancer cells and other cancer cells to Dox (Godovannyi et al., 2011; Yabbarov et al., 2013; Tcherkassova et al., 2017).
There are several ways to conjugate AIFs with drugs. Figure 5A shows a method of rAFP3D conjugation to Dox (rAFP3D-G2-Dox) (Yabbarov et al., 2013; Posypanova et al., 2013). rAFP3D can also link nanoparticles and liposomes to increase the effectiveness of targeted therapy (Figures 6A,B; Godovannyi et al., 2011; Yabbarov et al., 2013). Other AIFs, such as peptide AIFs, can conjugate or synergize with drugs to treat cancer (Figures 6C,D). Recombinant AFP (a unique AIF) may be designed to retain the hydrophobic pockets of AFP, which may non-covalently bind to a variety of drugs and effectively release them inside cancer cells (Figure 6E) (Mizejewski et al., 2010; Arshad et al., 2015; Pak, 2018b).
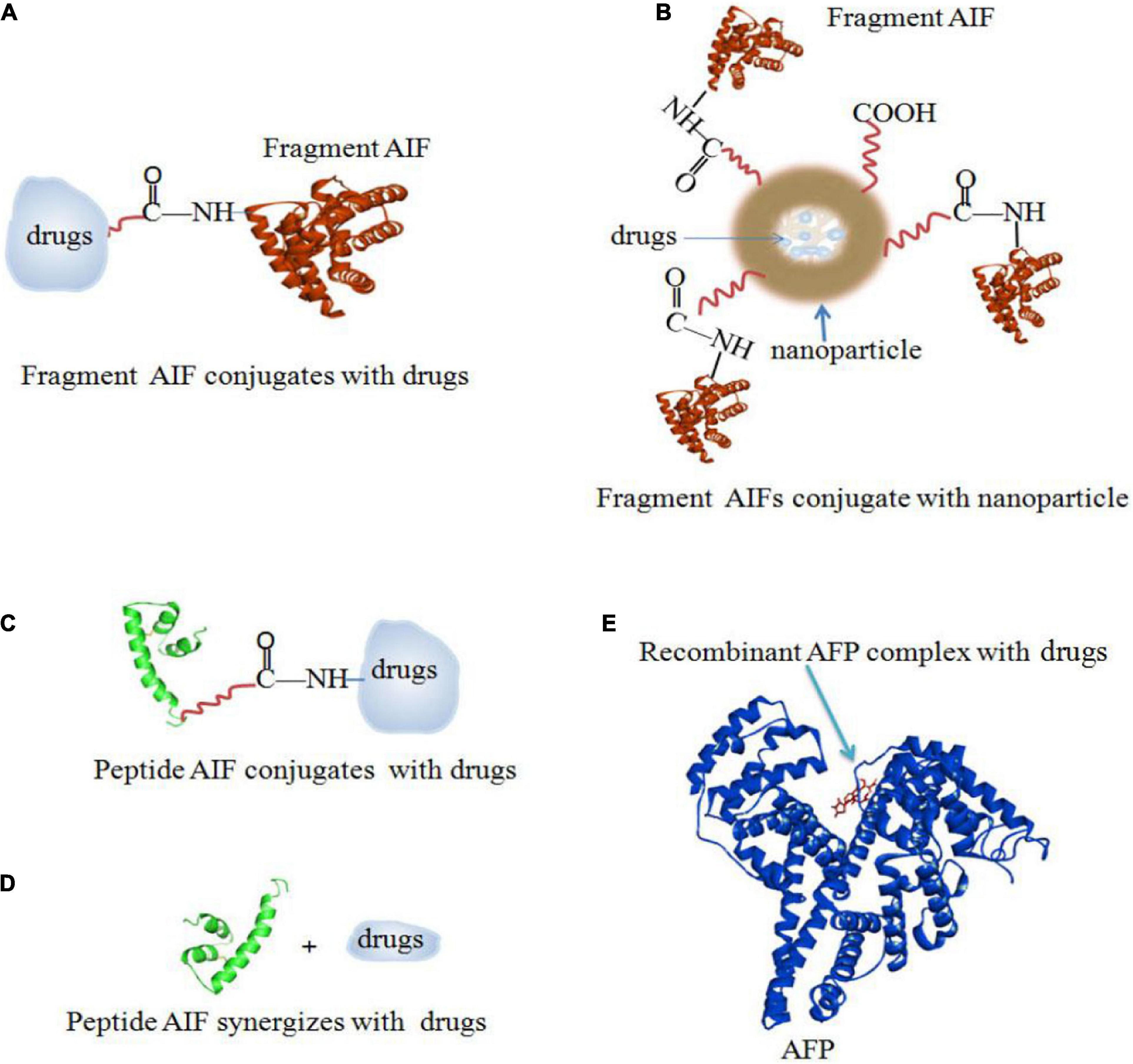
Figure 6. AIFs (fragment AIFs, peptide AIFs and recombinant AFP) conjugate, synergize and complex with drugs. (A) Fragment AIF conjugates with drugs (Posypanova et al., 2013; Yabbarov et al., 2013). (B) Fragment AIFs conjugate nanoparticles with drugs (Godovannyi et al., 2011; Yabbarov et al., 2013). (C) Peptide AIF conjugates with drugs (Mizejewski et al., 2010). (D) Peptide AIF synergizes with drugs (Mizejewski et al., 2010). (E) Recombinant AFP complex with drugs (Arshad et al., 2015; Pak, 2018b).
Application of Aif to Enhance Immunotherapy of Cancer
Cancer cells can be produced in vivo at any time, but the immune system eliminates them quickly and maintains a healthy state. Mutations make cancer cells different, and immune cells can recognize cancer cells based on certain differences and attack them (Marx, 2008; Mohme et al., 2017). However, cancer cells can acquire skills to promote their own survival (DuPage et al., 2012; Ribas, 2015; McGray and Bramson, 2017). For example, cancer cells can mask proteins to prevent immune cells from recognizing them or produce proteins that suppress immunity to escape immunity without being attacked. Thus, cancer cells can survive and proliferate in vivo. As cancer cells proliferate and continue to evolve in vivo, it becomes increasingly difficult for the immune system to attack them.
Natural killer (NK) cells, which have a natural immune function, are the first-line defense system. NK cells can prevent cancer cell growth, invasion and metastasis as well as attack pathogens. NK cells constantly surveil and eliminate cells that pose a threat to health. Disruption of NK cell action can lead to diseases or carcinogenesis (Shimasaki et al., 2020). Similarly, immune T cells in vivo can monitor and eliminate cancer cells. However, cancer cells can disguise themselves through the production of myeloid-derived suppressor cells (MDSCs) and evade the surveillance of the immune system (Dumitru et al., 2012; Baniyash, 2016). MDSCs express proteins that bind with proteins on immune cells and signal them to “turn off” their immune functions (Bruno et al., 2019). MDSCs are advanced immunosuppressive cells that are produced from the bone marrow and transported to the primary lesion where they accumulate and suppress acquired immunity and innate immunity. In cancer, the immune system produces MDSCs from the bone marrow, and they proliferate in the blood and normal peripheral organs. Further, cancer cells develop immune tolerance.
Targeting MDSCs is a new approach to immunotherapy that can eliminate immune tolerance molecules, activate NK cells, activate T cells and engage the immune system in recognizing and destroying cancer cells through a positive response (Makkouk and Weiner, 2015; Pak, 2017). Many lines of evidence have indicated that AFP inhibits the immune response in patients with cancers (Um et al., 2004; Meng et al., 2016; Suryatenggara et al., 2017; Wang and Wang, 2018; Zheng et al., 2020). AFP inhibits the activity of NK cells and T cells by activating AFPR-positive MDSCs and promoting cancer development (Belyaev et al., 2018; Zamorina et al., 2018). Therefore, vaccines against AFP inhibit the growth of AFP receptor-positive cancer cells and prolong patient survival time (Lan et al., 2007). One study has reported that inoculation of the placental carcinoembryonic-derived proteins, AFP and AFPR, causes MDSCs to become exhausted, resulting in the elimination of maternal-fetal and host-tumor immune tolerance (Mizejewski, 2018; Pak, 2018b). Thus, an AFP vaccine promotes a longer survival of advanced patients with cancer. In some cancers, full-length glycosylated AFP has immunosuppressive effects by stimulating cancer growth and directly activating MDSCs (Pak, 2018a,b). Moreover, tAFP significantly inhibits dendritic cell (DC) differentiation, thereby playing a critical role in immunosuppression (Pardee et al., 2014; Li et al., 2019). Therefore, it is more suitable to use AIF than tAFP for manufacturing vaccines to prevent the initiation of cancer.
Currently, the main research direction of immunotherapy involves immune checkpoint inhibitors as represented by treatment with PD-1/PD-L1 inhibitors. PD-1/PD-L1 inhibitors prevent PD-L1 from binding to PD-1 on immune cells, relieving the inhibition of cancer immunosuppression and stimulating immune cells to attack cancer cells (Chen and Han, 2015; Postow et al., 2015; Sun et al., 2018; Andrews et al., 2019; Hayashi and Nakagawa, 2020). In patients with HCC, high expression of both PD-L1 and HIF-1α is significantly associated with high AFP levels (Dai et al., 2018; Liu G. M. et al., 2019), indicating that the expression of AFP is closely related to the expression of PD-L1 in HCC cells. The expression of PD-L1 is regulated by the HIF-1α transcription factor (Koh et al., 2015; Chen et al., 2016; Zerdes et al., 2018). Researchers have previously reported that AFP activates the PI3K/AKT signaling pathway to stimulate HIF-1α, which regulates the expression of some oncogenes (Zhu et al., 2015a,b), indicating that AFP has a biological role in stimulating the expression of PD-L1 in cancer cells. In HCC cells, the persistent expression of PD-L1 in HCC cells is a crucial factor for resisting immune checkpoint inhibitors (Liu Z. et al., 2019; Wu et al., 2019). We speculate that high AFP expression in HCC cells promotes HIF-1α to stimulate the expression of PD-L1, which plays a pivotal role in HCC cells resisting immune checkpoint inhibitors. Therefore, AIFs can be used to carry PD-1/PD-L1 inhibitors because AIFs do not induce the malignant behavior caused by tAFP. AIFs exhaust MDSCs and cancer cells, and PD-1/PD-L1 inhibitors reactivate the function of T cells, which leads to the activation of NK cells, restoring their normal function of recognizing cancer cells and destroying them.
In addition, synergistic immunotherapy with chemotherapy is better for treating cancer. AFP activates MDSCs and inhibits various non-specific immune reactions (Pardee et al., 2014; Belyaev et al., 2018). The structure of AFP-binding receptors is still unclear, but MDSCs and cancer cells have ‘scavenger’ receptors that are similar to AFP-binding receptors and are critical for receptor-mediated endocytosis. The receptors take up AFP and provide nutrients to cancer cells and bone marrow mesenchymal stem cells through shuttling; thus, AFP delivery of drugs instead of nutrients kills MDSCs and cancer cells. The use of AIFs combined with toxicity-inducing drugs is a new treatment that integrates chemotherapy and immunotherapy. AIFs can deplete MDSCs, and inhibiting fragments loaded with toxins can destroy cancer cells. For example, AIFs conjugated with paclitaxel, 5-Fu or other chemotherapeutic drugs not only are used as toxins to kill cancer cells but also serve as immunomodulators. AIFs conjugated with drugs selectively reduce the immunosuppression of MDSCs and destroy cancer cells to improve the treatment of cancers.
The conjugation of AIFs with drugs is a new type of treatment for cancer. It not only activates T cells and kills cancer cells by drugs but also depletes MDSCs, activates NK cells and destroys cancer stem and metastatic cells through AIFs. This combination also activates T cells through drug action. Conjugating AIFs and drugs is a new approach to immunotherapy and targeted chemotherapy, and this combination will play an important role in future cancer therapies (Mizejewski, 2011a; Pak, 2014, 2018b; Llovet et al., 2018; Pinter and Peck-Radosavljevic, 2018).
Forecasts of the Design and Application of AIFs in Targeting Therapeutics of Cancers
Cancer cells with multi-drug resistance (MDR) traits resist chemotherapy, and they express PD-L1 to suppress the immune response and escape immune surveillance, preventing the attack of immune cells (Ribas, 2015; Berraondo et al., 2016; Chen and Mellman, 2017; O’Donnell et al., 2019). tAFP is a crucial molecule for promoting the malignant behaviors of HCC cells, primarily by activating growth signaling pathways. Other cancer cells also take up AFP to activate malignant signaling pathways to acquire drug resistance, contributing to their survival in vivo. tAFP also has the capacity to impair immune cells (Bei et al., 1999; Pardee et al., 2014; Vujanovic et al., 2017; Santos et al., 2019) and stimulate the malignant behaviors of cancer cells. Therefore, it will better to design AIFs to prevent the malignant behaviors mediated by tAFP in cancer cells.
In previous studies, we found that the cytoplasmic tAFP (CyAFP) binds to caspase-3, ATRA, PTEN and other proteins (Li et al., 2002b, 2009a, 2011, 2012; Zhu et al., 2015c; Lin et al., 2017; Wang et al., 2018; Zhang et al., 2020) to affect the transduction of apoptosis- or proliferation-related signaling pathways. CyAFP binding to caspase-3 is shown in Figure 2. In particular, we found that the CyAFP domain-3 residues, K-558, S-445, R-452 and its adjacent residue R-214, directly interact with caspase-3 loop4 (L4) residues in the cytoplasm (Lin et al., 2017). Through these binding sites, a peptide or fragment AIF can be precisely designed as a ligand decoy, which will prevent the binding of CyAFP and caspase-3, thereby promoting cell apoptosis. Similarly, we found that the CyAFP domain-3 residues, M490 and D529, interact with PTEN (Zhu et al., 2015c). Based on the binding sites, relevant AFP-blocking peptides can be designed to inhibit AFP binding to PTEN, which will prevent the growth of cancer cells. In addition, cancer therapy realized by targeting AFP may overcome the problem of MDR. MDR is a major problem that vexes clinical oncologists. Although the MDR mechanism in cancer is complicated, studies have found that AFP is involved in MDR by inhibiting the function of PTEN and activating the PI3K/AKT signaling pathway, which leads to the inhibition of autophagy, induction of metabolic reprogramming of cancer stem cells, inhibition of the expression of apoptosis-related enzymes and resistance to tumor cell apoptosis, thereby enabling cancer cells to acquire a drug-resistant phenotype (Kang-Park et al., 2006; Fruman et al., 2017; Zhu et al., 2017; Janku et al., 2018; Hoxhaj and Manning, 2020).
Cytoplasmic tAFP domain-3 (CyAFP-3D) is a pivotal site for inhibiting PTEN and caspase-3 (Li et al., 2009a; Mizejewski, 2015b; Zhu et al., 2015c; Lin et al., 2017; Wang et al., 2018; Li et al., 2020), leading to MDR. Therefore, CyAFP-3D can be used to design AIF for interacting with signaling molecules that play crucial roles in inhibiting immune responses and cancer cell growth, drug resistance and metastasis. The designed AIF can bind to intracellular caspase-3, PTEN and other signaling molecules to prevent AFP from binding to them and activating malignant signaling pathways.
Cytoplasmic AFP domain-3 can also be used to design AIFs to deliver drugs to target cancer cells. Because some cancer cells and immune suppressive cells have high expression of AFPR vs normal cells, AIF will bind to AFPR and transport drugs into cancer cells and immune suppressive cells, thereby resulting in low cytotoxicity in normal cells. Therefore, a precisely designed AIF can be used to block AFP-stimulated malignant behavior and to carry anticancer drugs to selectively treat cancers (Godovannyi et al., 2011; Posypanova et al., 2013; Yabbarov et al., 2013; Tcherkassova et al., 2017).
Alpha fetoprotein-inhibiting fragments also competes with CyAFP in immune cells to decrease the immune suppression mediated by CyAFP. Additionally, AIF can be designed to block CyAFP from activating the transcriptional activity of HIF-1α, which regulates the expression of PD-L1 in cancer cells, thus contributing to immune cells attacking cancer cells.
Conclusion and Future Perspectives
Alpha fetoprotein-inhibiting fragments selectively deliver antineoplastic agents to cancer cells to inhibit the malignant behaviors mediated by CyAFP, representing a precise design for targeting and killing cancer cells. Moreover, blocking the immunosuppressive effect of CyAFP is a crucial issue for stimulating the immune response to cancer cells. CyAFP promotes the malignant behaviors of cancer cells and impairs the function of immune cells. Domain-3 of CyAFP can be applied to precisely design AIFs to carry anticancer drugs to selectively accumulate them in cancer or immunosuppressive cells. Precisely designed AIFs not only deliver drugs into cancer cells but also compete with CyAFP to bind to various signaling molecules, inhibiting the role of CyAFP in promoting the malignant behaviors of cancer cells and blocking its effect on immunosuppression. These AIFs can be combined with immunotherapy drugs to strengthen the therapeutic effect. In the future, computer simulation screening will be used to establish a database of AIFs that are effective in treating cancer and a database of drugs that can be conjugated with AIFs. Therefore, the application of AIFs will be a precise, readily available strategy for targeted treatment of cancers in the future.
Author Contributions
BL, XD, and QW gathered the related literature, prepared the figures, and drafted the manuscript. WL, MZ, and ML participated in the design of the review and drafted the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (Nos. 82060514, 81960519, 81660463, 81560450, and 31560243), The Natural Science Foundation of Hainan Province (Nos. 820RC634, 2019CXTD406, 2019CR204, and 20168263), and Hainan Provincial Association for Science and Technology Program of Youth Science Talent and Academic Innovation (No. QCXM 201922).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Abbreviations
AFP, alpha fetoprotein; AFP-3BC, recombinant fragment protein derived from AFP domain-3 (from 473 ∼ 596 residues); AFPR, AFP receptor; AIFs, AFP-inhibiting fragments; ATRA, all-transretinoic acid; CyAFP, cytoplasmic AFP; DC, dendritic cell; Dox, doxorubicin; GIP, AFP-derived growth inhibitory peptide (synthetic growth inhibitory peptide derived from AFP domain-3); GPCRs, G protein-coupled receptors; HCC, hepatocellular carcinoma; HIF-1 α, hypoxia-induced factor (HIF)-1 α; Kds, dissociation constants; MDR, multi-drug resistance; MDSCs, myeloid-derived suppressor cells; nAFP, natural AFP (derived from fetal cells); NK, natural killer; PTEN, phosphatase and tensin homolog gene deleted on chromosome 10; r3dAFP, recombinant fragment protein derived from C-terminal AFP (from 357 ∼ 590 residues); rAFP3D, recombinant fragment protein derived from AFP domain-3 (from 404 ∼ 609 residues); RAR, retinoic acid receptor; rhAFP, recombinant AFP (full-length AFP gene was expressed in E. coli as well as in yeast cells, which had biological properties related to but not identical to native human AFP); SeAFP, secreted AFP; tAFP, tumor-derived AFP.
References
Alava, M. A., Iturralde, M., Gonzalez, B., and Piñeiro, A. (1999). Fatty acid desaturation: effect of alpha fetoprotein on alpha-linolenic acid conversion by fetal rat hepatocytes. Prostaglandins Leukot Essent Fatty Acids 60, 209–215. doi: 10.1054/plef.1999.0026
Andrews, L. P., Yano, H., and Vignali, D. A. A. (2019). Inhibitory receptors and ligands beyond PD-1, PD-L1 and CTLA-4: breakthroughs or backups. Nat. Immunol. 20, 1425–1434. doi: 10.1038/s41590-019-0512-0
Arshad, N. M., In, L. L., Soh, T. L., Azmi, M. N., Ibrahim, H., Awang, K., et al. (2015). Recombinant human alpha fetoprotein synergistically potentiates the anti-cancer effects of 1’-S-1’-acetoxychavicol acetate when used as a complex against human tumours harbouring AFP-receptors. Oncotarget 6, 16151–16167. doi: 10.18632/oncotarget.3951
Bai, D. S., Zhang, C., Chen, P., Jin, S. J., and Jiang, G. Q. (2017). The prognostic correlation of AFP level at diagnosis with pathological grade, progression, and survival of patients with hepatocellular carcinoma. Sci. Rep. 7:12870.
Baniyash, M. (2016). Myeloid-derived suppressor cells as intruders and targets: clinical implications in cancer therapy. Cancer Immunol. Immunother. 65, 857–867. doi: 10.1007/s00262-016-1849-y
Bei, R., Budillon, A., Reale, M. G., Capuano, G., Pomponi, D., Budillon, G., et al. (1999). Cryptic epitopes on alpha-fetoprotein induce spontaneous immune responses in hepatocellular carcinoma, liver cirrhosis, and chronic hepatitis patients. Cancer Res. 59, 5471–5474.
Bei, R., and Mizejewski, G. J. (2011). Alpha fetoprotein is more than a hepatocellular cancer biomarker: from spontaneous immune response in cancer patients to the development of an AFP-based cancer vaccine. Curr. Mol. Med. 11, 564–581. doi: 10.2174/156652411800615162
Bei, R., and Mizejewski, G. J. (2020). Alpha-fetoprotein is an autoantigen in hepatocellular carcinoma and juvenile Batten disease. Front. Biosci. 25:912–929. doi: 10.2741/4840
Belyaev, N. N., Abdolla, N., Perfilyeva, Y. V., Ostapchuk, Y. O., Krasnoshtanov, V. K., Kali, A., et al. (2018). Daunorubicin conjugated with alpha-fetoprotein selectively eliminates myeloid-derived suppressor cells (MDSCs) and inhibits experimental tumor growth. Cancer Immunol. Immunother. 67, 101–111. doi: 10.1007/s00262-017-2067-y
Berraondo, P., Minute, L., Ajona, D., Corrales, L., Melero, I., and Pio, R. (2016). Innate immune mediators in cancer: between defense and resistance. Immunol. Rev. 274, 290–306. doi: 10.1111/imr.12464
Biddle, W., and Sarcione, E. J. (1987). Specific cytoplasmic alpha-fetoprotein binding protein in MCF-7 human breast cancer cells and primary breast cancer tissue. Breast Cancer Res. Treat. 10, 279–286. doi: 10.1007/bf01805765
Bruno, A., Mortara, L., Baci, D., Noonan, D. M., and Albini, A. (2019). Myeloid derived suppressor cells interactions with Natural Killer cells and pro-angiogenic activities: roles in tumor progression. Front. Immunol. 10:771.
Chen, D. S., and Mellman, I. (2017). Elements of cancer immunity and the cancer-immune set point. Nature 541, 321–330. doi: 10.1038/nature21349
Chen, J., Jiang, C. C., Jin, L., and Zhang, X. D. (2016). Regulation of PD-L1: a novel role of pro-survival signalling in cancer. Ann. Oncol. 27, 409–416. doi: 10.1093/annonc/mdv615
Chen, L., and Han, X. (2015). Anti-PD-1/PD-L1 therapy of human cancer: past, present, and future. J. Clin. Invest. 125, 3384–3391. doi: 10.1172/jci80011
Cheng, L., Luo, S., Jin, C., Ma, H., Zhou, H., and Jia, L. (2013). FUT family mediates the multidrug resistance of human hepatocellular carcinoma via the PI3K/Akt signaling pathway. Cell Death Dis. 4:e923. doi: 10.1038/cddis.2013.450
Dai, X., Pi, G., Yang, S. L., Chen, G. G., Liu, L. P., and Dong, H. H. (2018). Association of PD-L1 and HIF-1α coexpression with poor prognosis in hepatocellular carcinoma. Transl. Oncol. 11, 559–566. doi: 10.1016/j.tranon.2018.02.014
Dumitru, C. A., Moses, K., Trellakis, S., Lang, S., and Brandau, S. (2012). Neutrophils and granulocytic myeloid-derived suppressor cells: immunophenotyping, cell biology and clinical relevance in human oncology. Cancer Immunol. Immunother. 61, 1155–1167. doi: 10.1007/s00262-012-1294-5
DuPage, M., Mazumdar, C., Schmidt, L. M., Cheung, A. F., and Jacks, T. (2012). Expression of tumour-specific antigens underlies cancer immunoediting. Nature 482, 405–409. doi: 10.1038/nature10803
Esteban, C., Geuskens, M., and Uriel, J. (1991). Activation of an alpha-fetoprotein (AFP)/receptor autocrine loop in HT-29 human colon carcinoma cells. Int. J. Cancer 49, 425–430. doi: 10.1002/ijc.2910490320
European Association for the Study of the Liver, (2018). EASL clinical practice guidelines: management of hepatocellular carcinoma. J. Hepatol. 69, 182–136.
Fruman, D. A., Chiu, H., Hopkins, B. D., Bagrodia, S., Cantley, L. C., and Abraham, R. T. (2017). The PI3K pathway in human disease. Cell 170, 605–635. doi: 10.1016/j.cell.2017.07.029
Godovannyi, A. V., Vorontsov, E. A., Gukasova, N. V., Pozdnyakova, N. V., Vasilenko, E. A., Yabbarov, N. G., et al. (2011). Targeted delivery of paclitaxel-loaded recombinant α-fetoprotein fragment-conjugated nanoparticles to tumor cells. Dokl. Biochem. Biophys. 439, 158–160. doi: 10.1134/s160767291104003x
Hayashi, H., and Nakagawa, K. (2020). Combination therapy with PD-1 or PD-L1 inhibitors for cancer. Int. J. Clin. Oncol. 25, 818–830. doi: 10.1007/s10147-019-01548-1
He, Y., Hong, Y., and Mizejewski, G. J. (2014). Engineering alpha-fetoprotein-based gene vaccines to prevent and treat hepatocellular carcinoma: review and future prospects. Immunotherapy 6, 725–736. doi: 10.2217/imt.14.46
Hoxhaj, G., and Manning, B. D. (2020). The PI3K-AKT network at the interface of oncogenic signalling and cancer metabolism. Nat. Rev. Cancer 20, 74–88. doi: 10.1038/s41568-019-0216-7
Iturralde, M., Alava, M. A., González, B., Anel, A., and Piñeiro, A. (1991). Effect of alpha-fetoprotein and albumin on the uptake of polyunsaturated fatty acids by rat hepatoma cells and fetal rat hepatocytes. Biochim. Biophys. Acta 1086, 81–88. doi: 10.1016/0005-2760(91)90157-d
Janku, F., Yap, T. A., and Meric-Bernstam, F. (2018). Targeting the PI3K pathway in cancer: are we making headway? Nat. Rev. Clin. Oncol. 15, 273–291. doi: 10.1038/nrclinonc.2018.28
Jiao, C., Chen, W., Tan, X., Liang, H., Li, J., Yun, H., et al. (2020). Ganoderma lucidum spore oil induces apoptosis of breast cancer cells in vitro and in vivo by activating caspase-3 and caspase-9. J. Ethnopharmacol. 30:112256. doi: 10.1016/j.jep.2019.112256
Kang-Park, S., Im, J. H., Lee, J. H., and Lee, Y. I. (2006). PTEN modulates hepatitis B virus-X protein induced survival signaling in Chang liver cells. Virus Res. 122, 53–60. doi: 10.1016/j.virusres.2006.06.010
Kim, H., Lee, S. J., and Yoon, M. (2020). Alpha-fetoprotein is correlated with intrahepatic recurrence of hepatocellular carcinoma after a hepatectomy. Ann. Surg. Treat. Res. 98, 168–176. doi: 10.4174/astr.2020.98.4.168
Koh, J., Jang, J. Y., Keam, B., Kim, S., Kim, M. Y., Go, H., et al. (2015). EML4-ALK enhances programmed cell death-ligand1expression in pulmonary adenocarcinoma via hypoxia-inducible factor (HIF)-1α and STAT3. Oncoimmunology 5:e1108514. doi: 10.1080/2162402x.2015.1108514
Komorowski, A. L., Hsu, C. C., Julka, K. D., Vasavada, B., Lin, C. C., Wang, C. C., et al. (2018). AFP role in predicting recurence of hepatocellular carcinoma after living donor liver transplantation in HCV patients. Neoplasma 65, 455–460. doi: 10.4149/neo_2018_170315n184
Kong, M., Tian, S., Shi, H., Zhao, J., Feng, X., Zheng, S., et al. (2012). The effect of alpha-fetoprotein on the activation and phagocytosis of granulocytes and monocytes. Hepatogastroenterology 59, 2385–2388.
Laborda, J., Naval, J., Allouche, M., Calvo, M., Georgoulias, V., Mishal, Z., et al. (1987). Specific uptake of alpha-fetoprotein by malignant human lymphoid cells. Int. J. Cancer 40, 314–318. doi: 10.1002/ijc.2910400306
Lan, Y. H., Li, Y. G., Liang, Z. W., Chen, M., Peng, M. L., Tang, L., et al. (2007). A DNA vaccine against chimeric AFP enhanced by HSP70 suppresses growth of hepatocellular carcinoma. Cancer Immunol. Immunother. 56, 1009–1016. doi: 10.1007/s00262-006-0254-3
Lee, Y. R., Chen, M., and Pandolfi, P. P. (2018). The functions and regulation of the PTEN tumour suppressor: new modes and prospects. Nat. Rev. Mol. Cell Biol. 19, 547–562. doi: 10.1038/s41580-018-0015-0
Li, C., Song, B., Santos, P. M., and Butterfield, L. H. (2019). Hepatocellular cancer-derived alpha fetoprotein uptake reduces CD1 molecules on monocyte-derived dendritic cells. Cell. Immunol. 335, 59–67. doi: 10.1016/j.cellimm.2018.10.011
Li, C., Wang, S., Jiang, W., Li, H., Liu, Z., Zhang, C., et al. (2012). Impact of intracellular alpha fetoprotein on retinoic acid receptors-mediated expression of GADD153 in human hepatoma cell lines. Int. J. Cancer 130, 754–764. doi: 10.1002/ijc.26025
Li, M., Li, H., Li, C., Guo, L., Liu, H., Zhou, S., et al. (2009b). Cytoplasmic alpha-fetoprotein functions as a co-repressor in RA-RAR signaling to promote the growth of human hepatoma Bel 7402 cells. Cancer Lett. 285, 190–199. doi: 10.1016/j.canlet.2009.05.014
Li, M., Li, H., Li, C., Wang, S., Jiang, W., Liu, Z., et al. (2011). Alpha-fetoprotein: a new member of intracellular signal molecules in regulation of the PI3K/AKT signaling in human hepatoma cell lines. Int. J. Cancer 128, 524–532. doi: 10.1002/ijc.25373
Li, M., Li, H., Li, C., Zhou, S., Guo, L., Liu, H., et al. (2009a). Alpha fetoprotein is a novel protein-binding partner forcaspase-3 and blocks the apoptotic signaling pathway in human hepatoma cells. Int. J. Cancer 124, 2845–2854. doi: 10.1002/ijc.24272
Li, M., Li, P. F., Chen, Q., Du, G. G., and Li, G. (2004). Alpha-fetoprotein stimulated the expression of some oncogenes in human hepatocellular carcinoma Bel 7402 cells. World J. Gastroenterol. 10, 819–824. doi: 10.3748/wjg.v10.i6.819
Li, M., Li, P. F., He, S. P., Du, G. G., and Li, G. (2002a). The promoting molecular mechanism of alpha-fetoprotein on the growth of human hepatoma Bel7402 cell line. World J. Gastroenterol. 8, 469–475. doi: 10.3748/wjg.v8.i3.469
Li, M., Li, P. F., Li, G., and Du, G. G. (2002b). Enhancement of proliferation of HeLa cells by the alpha-fetoprotein. Sheng Wu Hua Xue Yu Sheng Wu Wu Li Xue Bao 34, 769–774.
Li, M., Li, P. F., Yang, F. Y., He, S. P., Du, G. G., and Li, G. (2002c). The intracellular mechanism of alpha-fetoprotein promoting the proliferation of NIH 3T3 cells. Cell Res. 12, 151–156. doi: 10.1038/sj.cr.7290121
Li, M., Zhu, M., Li, W., Lu, Y., Xie, X., Wu, Y., et al. (2013). Alpha-fetoprotein receptor as an early indicator of HBx-driven hepatocarcinogenesis and its applications in tracing cancer cell metastasis. Cancer Lett. 330, 170–180. doi: 10.1016/j.canlet.2012.11.042
Li, W., Liu, K., Chen, Y., Zhu, M., and Li, M. (2020). Role of alpha-fetoprotein in hepatocellular carcinoma drug resistance. Curr. Med. Chem. [Epub ahead of print]. doi: 10.2174/0929867327999200729151247
Lin, B., Zhu, M., Wang, W., Li, W., Dong, X., Chen, Y., et al. (2017). Structural basis for alpha fetoprotein-mediated inhibition of caspase-3 activity in hepatocellular carcinoma cells. Int. J. Cancer 141, 1413–1421. doi: 10.1002/ijc.30850
Liu, G. M., Li, X. G., and Zhang, Y. M. (2019). Prognostic role of PD-L1 for HCC patients after potentially curative resection: a meta-analysis. Cancer Cell Int. 19:22.
Liu, Z., Lin, Y., Zhang, J., Zhang, Y., Li, Y., Liu, Z., et al. (2019). Molecular targeted and immune checkpoint therapy for advanced hepatocellular carcinoma. J. Exp. Clin. Cancer Res. 38:447.
Llovet, J. M., Montal, R., Sia, D., and Finn, R. S. (2018). Molecular therapies and precision medicine for hepatocellular carcinoma. Nat. Rev. Clin. Oncol. 15, 599–616. doi: 10.1038/s41571-018-0073-4
Lorenzo, H. C., Geuskens, M., Macho, A., Lachkar, S., Verdiere-Sahuque, M., Pineiro, A., et al. (1996). Alpha-fetoprotein binding and uptake by primary cultures of human skeletal muscle. Tumour Biol. 17, 251–260. doi: 10.1159/000217986
Lu, Y., Zhu, M., Li, W., Lin, B., Dong, X., Chen, Y., et al. (2016). Alpha fetoprotein plays a critical role in promoting metastasis of hepatocellular carcinoma cells. J. Cell Mol. Med. 20, 549–558. doi: 10.1111/jcmm.12745
Ma, S. H., Chen, G. G., Yip, J., and Lai, P. B. (2010). Therapeutic effect of alpha-fetoprotein promoter-mediated tBid and chemotherapeutic agents on orthotopic liver tumor in mice. Gene Ther. 17, 905–912. doi: 10.1038/gt.2010.34
Makkouk, A., and Weiner, G. J. (2015). Cancer immunotherapy and breaking immune tolerance: new approaches to an old challenge. Cancer Res. 75, 5–10. doi: 10.1158/0008-5472.can-14-2538
Marx, J. (2008). Cancer immunology. Cancer’s bulwark against immune attack: MDS cells. Science 319, 154–156. doi: 10.1126/science.319.5860.154
McGray, A. J. R., and Bramson, J. (2017). Adaptive resistance to cancer immunotherapy. Adv. Exp. Med. Biol. 1036, 213–227. doi: 10.1007/978-3-319-67577-0_14
Mehta, N., Dodge, J. L., Grab, J. D., and Yao, F. Y. (2020). National experience on down-staging of hepatocellular carcinoma before liver transplant: Influence of tumor burden, alpha-fetoprotein, and wait time. Hepatology 71, 943–954. doi: 10.1002/hep.30879
Meng, W., Bai, B., Bai, Z., Li, Y., Yue, P., Li, X., et al. (2016). The immunosuppression role of alpha-fetoprotein in human hepatocellular carcinoma. Discov. Med. 21, 489–494.
Missiaglia, E., Dalai, I., Barbi, S., Beghelli, S., Falconi, M., della Peruta, M., et al. (2010). Pancreatic endocrine tumors: expression profiling evidences a role for AKT-mTOR pathway. J. Clin. Oncol. 28, 245–255. doi: 10.1200/jco.2008.21.5988
Mittl, P. R., Di Marco, S., Krebs, J. F., Bai, X., Karanewsky, D. S., Priestle, J. P., et al. (1997). Structure of recombinant human CPP32 in complex with the tetrapeptide acetyl-Asp-Val-Ala-Asp fluoromethyl ketone. J. Biol. Chem. 272, 6539–6547. doi: 10.1074/jbc.272.10.6539
Mizejewski, G. J. (1995). Alpha-fetoprotein binding proteins: implications for transmembrane passage and subcellular localization. Life Sci. 56, 1–9. doi: 10.1016/0024-3205(94)00401-d
Mizejewski, G. J. (2001). Alpha-fetoprotein structure and function: relevance to isoforms, epitopes, and conformational variants. Exp. Biol. Med. 226, 377–408. doi: 10.1177/153537020122600503
Mizejewski, G. J. (2002). Biological role of alpha-fetoprotein in cancer: prospects for anticancer therapy. Expert Rev. Anticancer Ther. 2, 709–735. doi: 10.1586/14737140.2.6.709
Mizejewski, G. J. (2011a). Mechanism of cancer growth suppression of alpha-fetoprotein derived growth inhibitory peptides (GIP): comparison of GIP-34 versus GIP-8 (AFPep). Updates and Prospects. Cancers 3, 2709–2733. doi: 10.3390/cancers3022709
Mizejewski, G. J. (2011b). Review of the putative cell-surface receptors foralpha-fetoprotein: identification of a candidate receptor protein family. Tumour Biol. 32, 241–258. doi: 10.1007/s13277-010-0134-5
Mizejewski, G. J. (2013). Review of the adenocarcinoma cell surface receptor for human alpha-fetoprotein; proposed identification of a widespread mucin as the tumor cell receptor. Tumour Biol. 34, 1317–1336. doi: 10.1007/s13277-013-0704-4
Mizejewski, G. J. (2014). The adenocarcinoma cell surface mucin receptor for alpha-fetoprotein: is the same receptor present on circulating monocytes and macrophages? A commentary. Tumour Biol. 35, 7397–7402. doi: 10.1007/s13277-014-2183-7
Mizejewski, G. J. (2015a). Nonsecreted cytoplasmic alpha-fetoprotein: a newly discovered role in intracellular signaling and regulation. An update and commentary. Tumour Biol. 36, 9857–9864. doi: 10.1007/s13277-015-3736-0
Mizejewski, G. J. (2015b). The alpha-fetoprotein third domain receptor binding fragment: in search of scavenger and associated receptor targets. J. Drug Target. 23, 538–551. doi: 10.3109/1061186x.2015.1015538
Mizejewski, G. J. (2016). The alpha-fetoprotein (AFP) third domain: a search for AFP interaction sites of cell cycle proteins. Tumour Biol. 37, 12697–12711. doi: 10.1007/s13277-016-5131-x
Mizejewski, G. J. (2018). Alpha-fetoprotein uptake and cytoplasmic trafficking in cancer and immune-associated cells: relevance to adaptive immunity. EC Clin. Exp. Anat. 1, 71–77.
Mizejewski, G. J. (2019). Protein binding and interactions with alpha-fetoprotein (AFP): a review of multiple AFP cell surface receptors, intracytoplasmic binding, and inter- molecular complexing proteins. J. Mol. Cell. Biol. Forecast. 2:1016.
Mizejewski, G. J., and Butterstein, G. (2006). Survey of functional activities of alpha-fetoprotein derived growth inhibitory peptides: review, and prospects. Curr. Protein Peptide Sci. 7, 73–100. doi: 10.2174/138920306775474130
Mizejewski, G. J., Mirowski, M., Garnuszek, P., Maurin, M., Cohen, B. D., Poiesz, B. J., et al. (2010). Targeted delivery of anti-cancer growth inhibitory peptides derived from human alpha-fetoprotein: review of an International Multi-Center Collaborative Study. J. Drug Target. 18, 575–588. doi: 10.3109/10611861003587243
Mohme, M., Riethdorf, S., and Pantel, K. (2017). Circulating and disseminated tumour cells- mechanisms ofimmune surveillance and escape. Nat. Rev. Clin. Oncol. 14, 155–167. doi: 10.1038/nrclinonc.2016.144
Muehlemann, M., Miller, K. D., Dauphinee, M., and Mizejewski, G. J. (2005). Review of growth inhibitory peptide as a biotherapeutic agent for tumor growth, adhesion, and metastasis. Cancer Metastasis Rev. 24, 441–467. doi: 10.1007/s10555-005-5135-2
Naval, J., Villacampa, M. J., Goguel, A. F., and Uriel, J. (1985). Cell-type-specific receptors for alpha-fetoprotein in a mouse T-lymphoma cell line. Proc. Natl. Acad. Sci. U.S.A. 82, 3301–3305. doi: 10.1073/pnas.82.10.3301
Nierhoff, D., Ogawa, A., Oertel, M., Chen, Y. Q., and Shafritz, D. A. (2005). Purification and characterization of mouse fetal livere pithelial cells with high in vivo repopulation capacity. Hepatology 42, 130–139. doi: 10.1002/hep.20735
O’Donnell, J. S., Teng, M. W. L., and Smyth, M. J. (2019). Cancer immunoediting and resistance to T cell-based immunotherapy. Nat. Rev. Clin. Oncol. 16, 151–167. doi: 10.1038/s41571-018-0142-8
Pak, V. (2014). The use of α-fetoprotein for the delivery of cytotoxic payloads to cancer cells. Ther. Deliv. 5, 885–892. doi: 10.4155/tde.14.59
Pak, V. N. (2018a). Selective targeting of myeloid-derived suppressor cells in cancer patients through AFP-binding receptors. Future Sci. OA 5:FSO321.
Pak, V. N. (2018b). The use of alpha-fetoprotein for the treatment of autoimmune diseases and cancer. Ther. Deliv. 9, 37–46. doi: 10.4155/tde-2017-0073
Pardee, A. D., Shi, J., and Butterfield, L. H. (2014). Tumor-derived α-fetoprotein impairs the differentiation and T cell stimulatory activity of human dendritic cells. J. Immunol. 193, 5723–5732. doi: 10.4049/jimmunol.1400725
Pinter, M., and Peck-Radosavljevic, M. (2018). Review article: systemic treatment of hepatocellular carcinoma. Aliment. Pharmacol. Ther. 48, 598–609. doi: 10.1111/apt.14913
Postow, M. A., Callahan, M. K., and Wolchok, J. D. (2015). Immune checkpoint blockade in cancer therapy. J. Clin. Oncol. 33, 1974–1982.
Posypanova, G. A., Gorokhovets, N. V., Makarov, V. A., Savvateeva, L. V., Kireeva, N. N., Severin, S. E., et al. (2008). Recombinant alpha-fetoprotein C-terminal fragment: the new recombinant vector for targeted delivery. J. Drug Target 16, 321–328. doi: 10.1080/10611860801927721
Posypanova, G. A., Makarov, V. A., Savvateeva, M. V., Bereznikova, A. V., and Severin, E. S. (2013). The receptor binding fragment of alpha-fetoprotein is a promising new vector for the selective delivery of antineoplastic agents. J. Drug Target 21, 458–465. doi: 10.3109/1061186x.2013.765441
Ribas, A. (2015). Adaptive immune resistance: how cancer protects from immune attack. Cancer Discov. 5, 915–919. doi: 10.1158/2159-8290.cd-15-0563
Riedl, S. J., Renatus, M., Schwarzenbacher, R., Zhou, Q., Sun, C., Fesik, S. W., et al. (2001). Structural basis for the inhibition of caspase-3 by XIAP. Cell 104, 791–800. doi: 10.1016/s0092-8674(01)00274-4
Rogers, C., Fernandes-Alnemri, T., Mayes, L., Alnemri, D., and Cingolani, G. (2017). Alnemri ES Cleavage of DFNA5 by caspase-3 during apoptosis mediates progression to secondary necrotic/pyroptotic cell death. Nat. Commun. 3:14128.
Santos, P. M., Menk, A. V., Shi, J., Tsung, A., Delgoffe, G. M., and Butterfield, L. H. (2019). Tumor-derived α-fetoprotein suppresses fatty acid metabolism and oxidative phosphorylation in dendritic cells. Cancer Immunol. Res. 7, 1001–1012. doi: 10.1158/2326-6066.cir-18-0513
Sell, S., Longley, M. A., and Boulter, J. (1985). Alpha-Fetoprotein and albumin gene expression in brain and other tissues of fetal and adult rats. Brain Res. 354, 49–53. doi: 10.1016/0165-3806(85)90067-7
Shimasaki, N., Jain, A., and Campana, D. (2020). NK cells for cancer immunotherapy. Nat. Rev. Drug Discov. 19, 200–218.
Sun, C., Mezzadra, R., and Schumacher, T. N. (2018). Regulation and function of the PD-L1 checkpoint. Immunity 48, 434–452. doi: 10.1016/j.immuni.2018.03.014
Suryatenggara, J., Wibowo, H., Atmodjo, W. L., and Mathew, G. (2017). Characterization of alpha-fetoprotein effects on dendritic cell and its function as effector immune response activator. J. Hepatocell. Carcinoma 4, 139–151. doi: 10.2147/jhc.s139070
Suzuki, Y., Zeng, C. Q., and Alpert, E. (1992). Isolation and partial characterization of a specific alpha-fetoprotein receptor on human monocytes. J. Clin. Invest. 90, 1530–1536. doi: 10.1172/jci116021
Tcherkassova, J., Tsurkan, S., Smirnova, G., Borisova, J., Moro, R., and Treshalina, H. (2017). Binding characterization of the targeting drug AIMPILA to AFP receptors in human tumor xenografts. Tumor Biol. 39:1010428317734815.
Torres, J. M., Geuskens, M., and Uriel, J. (1991). Receptor-mediated endocytosis and recycling of alpha-fetoprotein in human B-lymphoma and T-leukemia cells. Int. J. Cancer 47, 110–117. doi: 10.1002/ijc.2910470120
Torres, J. M., Laborda, J., Naval, J., Darracq, N., Calvo, M., Mishal, Z., et al. (1989). Expression of alpha-fetoprotein receptors by human T-lymphocytes during blastic transformation. Mol. Immunol. 26, 851–857. doi: 10.1016/0161-5890(89)90141-7
Um, S. H., Mulhall, C., Alisa, A., Ives, A. R., Karani, J., Williams, R., et al. (2004). Alpha-fetoprotein impairs APC function and induces their apoptosis. J. Immunol. 173, 1772–1778. doi: 10.4049/jimmunol.173.3.1772
Uriel, J., Failly-Crepin, C., Villacampa, M. J., Pineiro, A., and Geuskens, M. (1984). Incorporation of alpha fetoprotein by the MCF-7 human breast cancer cell line. Tumour Biol. 5, 41–51.
Uriel, J., Poupon, M. F., and Geuskens, M. (1983). Alpha foetoprotein uptake by cloned cell lines derived from a nickel-induced rat rhabdomyosarcoma. Br. J. Cancer 48, 261–269. doi: 10.1038/bjc.1983.181
Vujanovic, L., Stahl, E. C., Pardee, A. D., Geller, D. A., Tsung, A., Watkins, S. C., et al. (2017). Tumor-derivedα-fetoprotein directly drives human natural killer-cell activation and subsequent cell death. Cancer Immunol. Res. 5, 493–502. doi: 10.1158/2326-6066.cir-16-0216
Wang, S., Jiang, W., Chen, X., Zhang, C., Li, H., Hou, W., et al. (2012). Alpha-fetoprotein acts as a novel signal molecule and mediates transcription of Fn14 in human hepatocellular carcinoma. J. Hepatol. 57, 322–329. doi: 10.1016/j.jhep.2012.03.029
Wang, S., Zhu, M., Wang, Q., Hou, Y., Li, L., Weng, H., et al. (2018). Alpha-fetoprotein inhibits autophagy to promote malignant behaviour in hepatocellular carcinoma cells by activating PI3K/AKT/mTOR signalling. Cell Death Dis. 9:1027.
Wang, X., and Wang, Q. (2018). Alpha-fetoprotein and hepatocellular carcinoma immunity. Can. J. Gastroenterol. Hepatol. 2018:9049252.
Wu, Q., Zhou, W., Yin, S., Zhou, Y., Chen, T., Qian, J., et al. (2019). Blocking triggering receptor expressed on Myeloid cells-1-positive tumor-associated macrophages induced by hypoxia reverses immunosuppression and anti-programmed cell death ligand 1 resistancein liver cancer. Hepatology 70, 198–214. doi: 10.1002/hep.30593
Xue, J., Cao, Z., Cheng, Y., Wang, J., Liu, Y., Yang, R., et al. (2020). Acetylation of alpha-fetoprotein promotes hepatocellular carcinoma progression. Cancer Lett. 471, 12–26. doi: 10.1016/j.canlet.2019.11.043
Yabbarov, N. G., Posypanova, G. A., Vorontsov, E. A., Obydenny, S. I., and Severin, E. S. (2013). A new system for targeted delivery of doxorubicin into tumor cells. J. Control. Release 168, 135–141. doi: 10.1016/j.jconrel.2013.03.007
Yang, X., Chen, L., Liang, Y., Si, R., Jiang, Z., Ma, B., et al. (2018). Knockdown of alpha-fetoprotein expression inhibits HepG2 cell growth and induces apoptosis. J. Cancer Res. Ther. 14(Suppl.), S634–S643.
Yang, X., Zhang, Y., Zhang, L., Zhang, L., and Mao, J. (2008). Silencing alpha-fetoprotein expression induces growth arrest and apoptosis in human hepatocellular cancer cell. Cancer Lett. 271, 281–293. doi: 10.1016/j.canlet.2008.06.017
Zamorina, S. A., Timganova, V. P., Bochkova, M. S., Khramtsov, P. V., Fomicheva, K. A., Rayev, M. B., et al. (2018). α-Fetoprotein influence on the conversion of Naïve T-Helpers into Memory T-cell effector subpopulations. Dokl. Biol. Sci. 482, 210–213. doi: 10.1134/s0012496618050113
Zerdes, I., Matikas, A., Bergh, J., Rassidakis, G. Z., and Foukakis, T. (2018). Genetic, transcriptional and post-translational regulation of the programmed death protein ligand 1 in cancer: biology and clinical correlations. Oncogene 37, 4639–4661. doi: 10.1038/s41388-018-0303-3
Zhang, C., Chen, X., Liu, H., Li, H., Jiang, W., Hou, W., et al. (2015). Alpha fetoprotein mediates HBx induced carcinogenesis in the hepatocyte cytoplasm. Int. J. Cancer 37, 1818–1829. doi: 10.1002/ijc.29548
Zhang, C., Li, H., Jiang, W., Zhang, X., and Li, G. (2016). Icaritin inhibits the expression of alpha-fetoprotein in hepatitis B virus-infected hepatoma cell lines through post-transcriptional regulation. Oncotarget 7, 83755–83766. doi: 10.18632/oncotarget.13194
Zhang, C., Zhang, J., Wang, J., Yan, Y., and Zhang, C. (2020). Alpha-fetoprotein accelerates the progression of hepatocellular carcinoma by promoting Bcl-2 gene expression through an RA-RAR signalling pathway. J. Cell Mol. Med. 24, 13804–1381. doi: 10.1111/jcmm.15962
Zhang, C. C., Li, C. G., Wang, Y. F., Xu, L. H., He, X. H., Zeng, Q. Z., et al. (2019). Chemotherapeutic paclitaxel and cisplatin differentially induce pyroptosis in A549 lung cancer cells via caspase-3/GSDME activation. Apoptosis 24, 312–325. doi: 10.1007/s10495-019-01515-1
Zhang, L., He, T., Cui, H., Wang, Y., Huang, C., and Han, F. (2012). Effects of AFP gene silencing on apoptosis and proliferation of a hepatocellular carcinoma cell line. Discov. Med. 14, 115–124.
Zheng, Y., Zhu, M., and Li, M. (2020). Effects of alpha-fetoproteinon the occurrence and progression of hepatocellular carcinoma. J. Cancer Res. Clin. Oncol. 146, 2439–2446. doi: 10.1007/s00432-020-03331-6
Zhu, M., Guo, J., Li, W., Lu, Y., Fu, S., Xie, X., et al. (2015a). Hepatitis B virus X protein induces expression of alpha-fetoprotein and activates PI3K/mTOR signaling pathway in liver cells. Oncotarget 6, 12196–12208. doi: 10.18632/oncotarget.2906
Zhu, M., Guo, J., Li, W., Xia, H., Lu, Y., Dong, X., et al. (2015b). HBx induced AFP receptor expressed to activatePI3K/AKT signal to promote expression of Src in liver cells and hepatoma cells. BMC Cancer 15:362.
Zhu, M., Li, W., Lu, Y., Dong, X., Lin, B., Chen, Y., et al. (2017). HBx drives alpha fetoprotein expression to promote initiation of liver cancer stem cells through activating PI3K/AKT signal pathway. Int. J. Cancer 140, 1346–1355. doi: 10.1002/ijc.30553
Keywords: AFP molecular structure, AFP inhibiting fragments, drug delivery, targeted cancer therapy, drug design
Citation: Lin B, Dong X, Wang Q, Li W, Zhu M and Li M (2021) AFP-Inhibiting Fragments for Drug Delivery: The Promise and Challenges of Targeting Therapeutics to Cancers. Front. Cell Dev. Biol. 9:635476. doi: 10.3389/fcell.2021.635476
Received: 30 November 2020; Accepted: 12 March 2021;
Published: 08 April 2021.
Edited by:
Lucio Miele, Louisiana State University, United StatesReviewed by:
Gerald J. Mizejewski, New York State Department of Health, United StatesGang Li, Peking University Health Science Centre, China
Copyright © 2021 Lin, Dong, Wang, Li, Zhu and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author (s) and the copyright owner (s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mingyue Zhu, bWluZ3l1ZXpodTIwMDJAMTYzLmNvbQ==; Mengsen Li, bWVuZ3NlbmxpQDE2My5jb20=
†These authors have contributed equally to this work and share first authorship
 Bo Lin
Bo Lin Xu Dong
Xu Dong Qiujiao Wang
Qiujiao Wang Wei Li1
Wei Li1 Mingyue Zhu
Mingyue Zhu Mengsen Li
Mengsen Li