Abstract
Cancer has become the leading cause of death in recent years. As great advances in medical treatment, emerging therapies of various cancers have been developed. Current treatments include surgery, radiotherapy, chemotherapy, immunotherapy, and targeted therapy. Aptamers are synthetic ssDNA or RNA. They can bind tightly to target molecules due to their unique tertiary structure. It is easy for aptamers to be screened, synthesized, programmed, and chemically modified. Aptamers are emerging targeted drugs that hold great potentials, called therapeutic aptamers. There are few types of therapeutic aptamers that have already been approved by the US Food and Drug Administration (FDA) for disease treatment. Now more and more therapeutic aptamers are in the stage of preclinical research or clinical trials. This review summarized the screening and development of therapeutic aptamers against different types of cancer cells.
Introduction
Aptamers initially were described by researchers in 1990 (Ellington and Szostak, 1990). They are short-chain nucleotide sequences, generally DNA or RNA of 20 to 100 nucleotides. With a unique tertiary structure, aptamers specifically bind to target molecules (Röthlisberger and Hollenstein, 2018). Aptamers are reproducible and programmable. Enzymatic degradation is resisted by the chemical modification of aptamers. The chemical integrity and bioavailability of aptamers are ensured through optimization under the physiological condition (Mallikaratchy et al., 2006; Zhu and Chen, 2018). When aptamers are modified with a hydrophobic group, they have an excellent binding affinity (Dunn et al., 2017; Odeh et al., 2020). An outstanding method, Systematic Evolution of Ligands by Exponential Enrichment (SELEX), was used to screen aptamers (Dunn et al., 2017). The target of aptamers range from small molecules (Li and Liu) to biomacromolecules (Mallikaratchy et al., 2006), infected cells (Liu et al., 2019), stem cells (Hou et al., 2015; Gang et al., 2017), and cancer cells (Moosavian and Sahebkar, 2019; Kordasht and Hasanzadeh, 2020).
For most drugs, there are two problems in the treatment of tumors, the first one is weak therapeutic effects and another is strong off-target toxicity (Lammers et al., 2012). Therefore, improving the effectiveness of drugs and reducing side effects are major challenges faced by researchers (Moosavian and Sahebkar, 2019). Selecting specific ligands that target tumors is the most widely used strategy in targeted therapy. The specific ligand has almost no affinity for normal cells. Besides, it does not produce any toxic effects (Pérez-Herrero and Fernández-Medarde, 2015). Now various ligands, antibodies, antibody fragments, peptides, nucleic acids, and small molecules, have been used for this purpose (Dougan and Dougan, 2017). The conjugation of specific ligands and drugs enhances the internalization of encapsulated drugs and the accumulation of drugs in tumors (Alavi and Hamidi, 2019). Among these ligands, aptamers are considered the most suitable ligands for active tumor targeting. Due to high specificity, aptamers have become the subject of extensive research during the past decades as targeting ligands. Aptamers with great affinity and selectivity can be modified, including direct conjugation with drugs and drug-encapsulated nanoparticles. In addition, aptamer holds stable under a broad range of physical and chemical conditions. They can be released after the drug is transported to target cells (Soldevilla et al., 2018; Zhu and Chen, 2018).
Apart from being used as targeting ligands, aptamers also act as small molecular agents, which means that they inhibit the functions of target proteins (Gelinas et al., 2016; Adachi and Nakamura, 2019). So far, many therapeutic aptamers are in preclinical or clinical development (Adachi and Nakamura, 2019). Aptamers act as antagonists to impede the interaction of tumor-related targets (proteins or receptor-ligands) or act as agonists to activate the function of anti-cancer target receptors to achieve the purpose of cancer treatment. In the treatment of various diseases, including eye diseases, cardiovascular diseases, tumors, and inflammations, aptamers have entered clinical trials as therapeutic drugs. Pegaptanib is the only therapeutic aptamer approved by the FDA for the treatment of age-related macular degeneration (Ng et al.). The aptamers used in the treatment of cardiovascular diseases and inflammation are all in the first or second phase of clinical trials to evaluate the efficacy and safety without further updates (Maasch et al., 2008; Waters et al., 2009; Povsic et al., 2014). At present, there are three types of aptamers completed clinical trials for tumor treatment. The first one is AS1411, which binds to the external domain of the target (nucleolin) with high affinity. In various clinical cancer models, including lung cancer, breast cancer, kidney cancer and liver cancer, etc., AS1411 shows the ability to inhibit cell proliferation and to induce cell apoptosis (Yazdian-Robati et al., 2019). The second one is NOX-A12, which is an L-type RNA aptamer, so it can resist nuclease degradation. It is an antagonist of chemokine (C-X-C motif) ligand 12 (CXCL-12), which inhibits tumor cell proliferation, new blood vessel formation, and metastasis (Duda et al., 2011). Besides, AGRO100 is also an aptamer that binds to nucleolin to produce anti-proliferation effects in tumor cells. In some phase I clinical trials, AGRO100 showed high safety and anti-tumor proliferation effects (Laber et al., 2004). So far, aptamers have not been approved as clinical drugs for the treatment of tumors. Compared with antibodies, the development of therapeutic aptamers is relatively slow (Zhou and Rossi, 2017). There are still challenges in developing aptamers into tumor therapeutics. The success of aptamers as therapeutic drugs in the future requires overcoming the challenges and fully developing unique properties.
Properties of Aptamers
Due to the flexibility of short-chain nucleotides, aptamers have unique chemical properties. In general, aptamers fold into stem, loop, bugle, pseudoknot, G-quadruplex, and kissing hairpin structures, and further fold to form more complex tertiary structure. Complex folding allows aptamer to specifically recognize the target (Gelinas et al., 2016; Zhou and Rossi, 2017). A broad range of targets includes organic molecules (Umar and Kit, 2020), proteins (Umar and Kit, 2020), nucleotides (Yue et al., 2011), cancer cells (Guizhi and Xiaoyuan, 2018), bacteria (Alizadeh et al., 2018), toxins (Frohnmeyer et al., 2019), or viruses (Cesewski and Johnson, 2020). Compared with antibodies, aptamers with flexible structures and very small molecular weights can cross the blood-brain barrier (Cheng et al., 2013), a physiological barrier that antibodies cannot enter, and bind to target molecules with high affinity and specificity. Aptamers with unique properties are combined with targets to directly exert the efficacy of drugs to treat cancer. At the same time, aptamers act as carriers for drug delivery, combined with drugs to achieve targeted drug delivery, and play an important role in targeted tumor therapy.
Many therapeutic antibodies have been used clinically. Aptamers have many advantages over antibodies (Table 1). Aptamers are produced by chemical synthesis in vitro, which have great benefits including short synthesis time, low cost, high stability, and specificity (Zhang Y. et al., 2019). The small and flexible structure of aptamers allows them to bind with smaller targets or hide binding domains that some antibodies cannot access (Zhou and Rossi, 2017). However, disadvantages of aptamers are metabolic instability (Lakhin et al., 2013) and rapid kidney filtration (Guo, 2011). Therefore, methods of chemical modifications and conjugations have been developed to overcome the disadvantages. Despite these limitations, researchers have learned lessons from the clinical development and application of nucleic acids, and continue to pursue the development of therapeutic aptamers (Yoshihiro et al., 2018).
TABLE 1
| Aptamer | Antibodies | |
| Synthesis | In vitro, SELEX | Produced in vivo |
| Target potential | Can target any small molecules | Difficult to raise antibodies to toxins (not tolerated by an animal) or non-immunogenic targets |
| Stability | Stable at room temperature. | Must be refrigerated for storage and transport |
| Affinity | High and increased in multivalent aptamers. | Dependent on the number of epitopes on the antigen |
| Specificity | Single point mutations identifiable. | Different antibodies might bind the same antigen |
| Activity | Uniform activity regardless of batch synthesis | The activity of antibodies varies from batch to batch |
| Modifiability | Wide variety of chemical modifications to molecule for diverse functions | Limited modifications to molecule |
| Immunogenicity | No immunogenicity | Significant immunogenicity |
| Shelf life | Unlimited shelf life | Limited shelf life |
| Tissue uptake/kidney filtration | Fast | Slow |
Comparison of aptamers and antibodies.
Production of Aptamers
To select high-affinity aptamers for targets, SELEX is a mature and widely used technology. Researchers have developed different SELEX methods to meet different experimental needs. These methods include capillary electrophoresis-SELEX (CE-SELEX) (Zhu et al., 2019), high-throughput (HT)-SELEX (Kato et al., 2020), microfluidic SELEX (Sinha et al., 2018), and cell SELEX (Harleen and Kaur, 2018).
The basic process of SELEX is a four-step repeated cycles of incubation, combination, distribution, and amplification (Figure 1). Firstly, a random oligonucleotide sequence library (starting library) is incubated with the target molecules. During the incubation process, some sequences bind to the target molecule, while others bind weakly or do not react with the target. Then, the bound sequence is separated from the weakly bound or unbound sequence. After that, Polymerase chain reaction (PCR) (DNA sequences) and reverse transcription PCR (RT-PCR) (RNA sequences) were adopted to amplifying eluted oligonucleotides to enrich the library. Various methods have been developed to generate ssDNA from the resulting double-stranded DNA (dsDNA), including asymmetric PCR (Citartan et al., 2012), denaturing urea-polyacrylamide gel (Ram et al., 2017), lambda exonuclease, and T7 gene 6 exonuclease digestion (Subramanian et al., 2003; Citartan et al., 2011), and magnetic separation by beads coated with streptavidin (Espelund et al., 1990). The obtained ssDNA is then used in the next round of selection (Bayat et al., 2018). The process of binding, separation, amplification, and pool conditioning needs to be repeated before the resulting nucleic acid pool is enriched enough with target-binding sequences (Komarova and Kuznetsov, 2019). In general, researchers get a rich pool through 5–15 rounds of selection. The enriched pool is sequenced and bioinformatics analysis is used to identify aptamer candidates from the sequencing results (Lewis et al., 2017). The selected aptamer is chemically synthesized and characterized (Wang et al., 2018).
FIGURE 1
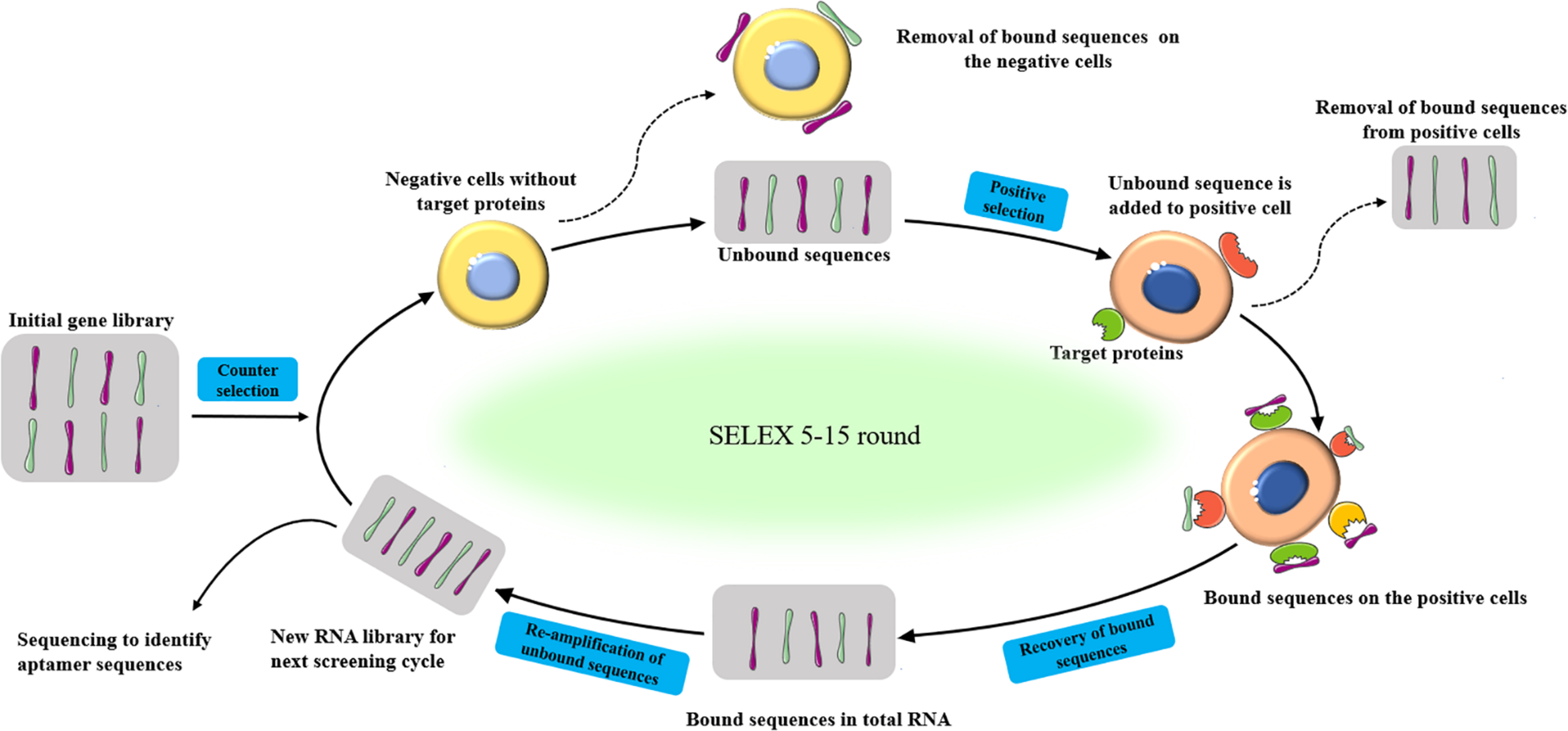
Schematic representation of the SELEX procedure to produced aptamers for specific target biomolecules.
Screening and Development of Therapeutic Aptamers Against Cancer Cells
In the past few decades, many specific aptamers have been developed for tumor treatment (Table 2), such as breast cancer (Liu et al., 2017), colorectal adenocarcinoma (Chen et al., 2017), lung cancer (2018), liver cancer (Zhang G.Q. et al., 2019), prostate cancer (Gray et al., 2018), leukemia (Tan et al., 2019), renal cell carcinoma (Zhang et al., 2018), oral cancer (Simmons et al., 2014), cervical cancer (Carvalho et al., 2019), bladder cancer (Yao et al., 2020), stomach cancer (Ding et al., 2015) and multiple myeloma (Waldschmidt et al., 2017).
TABLE 2
| Cancer types | Aptamers | Targets | Clinical applications | Cell lines | References |
| Breast cancer | PNDA-3 | Periostin | None | MCF7, MDA-MB-231cell | Lee et al., 2013 |
| HeA2-3 | HER2 | None | SKBR3, MDA-MB-231, SKOV3 cell | Gijs et al., 2016 | |
| ApMNK2F | MNK | None | MDA-MB-231, MCF7 cell | García-Recio et al., 2016 | |
| ApMNK3R | |||||
| Colorectal cancer | YJ-1 | CEA | None | LS174T,SW480 cell | Lee et al., 2012 |
| Aptamer | PDGF-BB | None | Caco-2, SW480 cell | Sae-Lim et al., 2019 | |
| MP7 | PD-1 | None | MC38 cell | Prodeus et al., 2015 | |
| Lung cancer | AptPD-L1 | PD-1 | None | CT26, LL/2 cell | Lai et al., 2016 |
| AP-74 M-545 | Gal-1 | None | LL/2 cell | Tsai et al., 2019 | |
| TBA535. | Hrombin | None | Calu-6 cell | Esposito et al., 2018 | |
| Liver cancer | AS1411 | HCC | None | Hepatocellular carcinoma cell | Cho et al., 2016 |
| CL-4RNV616 | EGFR | None | Huh-7 cell | Wang et al., 2019 | |
| Prostate cancer | Apt63 | Ecto-ATP5B | None | PC-3, RWPE-1, MCF10, 67NR, E0771 cell | Speransky et al., 2019 |
| A9g | PSMA | None | PC-3 cell | Dassie et al., 2014 | |
| AGRO100 | Nucleolin | I clinical trial | Prostate cancer cell | Laber et al., 2005 | |
| LeukemiaLeukemia | AS1411 | Nucleolin | None | MV4-11 cell | Soundararajan et al., 2009 |
| β-arr2As | β-arrestin | None | K562 cell | Kotula et al., 2014 | |
| Renal cell carcinoma | SW-4 | ccRCC | None | 786-O cell | Zhang et al., 2018 |
| AS1411 | Nucleolin | II clinical trial | Renal cell carcinoma | Rosenberg et al., 2014 | |
| Oral cancer | Aptamer | Heparanase | None | HSC-3 cell | Simmons et al., 2014 |
| Cervical cancer | A2 | E7 | None | SiHa, CaSki, HeLa cell | Clare et al., 2013 |
| Bladder cancer | Aptamer | β-catenin | None | SW780, 5637 cell | Xie et al., 2018 |
| Stomach cancer | Aptamer | HER2 | None | N87 cell | Mahlknecht et al., 2013 |
| Multiple myeloma | NOX-A12 | CXCL12 | I clinical trial | U266, NCI-H929 cell | Waldschmidt et al., 2017 |
Examples of aptamers with potential as targeted drugs.
Therapeutic Aptamers Against Breast Cancer Cells
Abnormal expression of human epidermal growth factor receptor 2 (HER2, also known as ErbB2) is present in 15–20% of all breast cancers (Waks and Winer, 2019). It is a molecular target with the potential for targeted cancer treatment. Gijs et al. screened the aptamer HeA2 through the SELEX method and identified two subtypes HeA2-1 and HeA2-3 by next-generation sequencing and bioinformatics methods. HeA2 binds to HER2 protein with high specificity. Interestingly, the authors’ results proved HeA2-3 was internalized into breast cancer cells and had an inhibitory effect on the growth and survival of breast cancer cells (Gijs et al., 2016). Periostin is a member of the extracellular matrix (ECM) protein. In tumor progression, its overexpression regulates the tumor microenvironment and affects the proliferation, invasion, and migration of tumor cells (Kudo, 2007). Lee et al. screened benzyl-d(U)TP modified DNA aptamers (PNDA-3) that are specifically bound to human periostin and characterized the function of the aptamers in breast cancer progression. PNDA-3 significantly inhibited the metastasis and growth of periostin-positive breast cancer. In the 4T1 orthotopic mouse model, the administration of PNDA-3 appreciably reduced the growth and distant metastasis of the primary tumor (Lee et al., 2013). MAP kinase-interacting kinase (MNK) is activated through the MAP kinase pathway and phosphorylates eukaryotic translation initiation factor 4E (eIF4E) at a single site. EIF4E and its phosphorylation are vital parts of cancer and tumorigenesis (Tian, 2017). García-Recio et al. (2016) screened two aptamers, called apMNK2F and apMNK3R. The selected aptamers were highly specific for MNK1. ApMNK2F and apMNK3R bind with MNK1 to produced significant translational inhibition, inhibiting tumor cell proliferation, migration, and colony formation in MDA-MB231 breast cancer cells (García-Recio et al., 2016).
Therapeutic Aptamers Against Colorectal Cancer Cells
Carcinoembryonic antigen (CEA, CEACAM5, or CD66e) is overexpressed in most cancers. It plays an important role in inducing tumor metastasis (Hammarström, 1999; Taheri et al., 2000). Liver metastasis is the main cause of death in colorectal cancer patients. Inhibiting liver metastasis is an effective way to improve the survival rate of patients with colorectal cancer (Tauriello et al., 2017). Lee et al. designed an RNA aptamer (YJ-1) that is specifically bound to CEA-positive cells. The homotype accumulation, migration, and invasion of CEA-positive cancer cells were inhibited by YJ-1. YJ-1 induced apoptosis of colon cancer cells by blocking the interaction between death receptor 5 and CEA. The transfer of human colon cancer cells to the liver was prevented by YJ-1 in the mouse experiments (Lee et al., 2012). Overexpression of platelet-derived growth factor-BB (PDGF-BB) is related to colorectal carcinogenesis. Sae-Lim et al. (2019) developed a DNA aptamer that is specifically bound to PDGF-BB. This DNA aptamer interfered with the binding of PDGF-BB and its receptor and inhibited the proliferation of colorectal cancer (CRC) cells by down-regulating the Ras/Raf/MEK/ERK signaling pathway (Sae-Lim et al., 2019). Programmed cell death-1 (PD-1, Pdcd1) negatively regulates antigen receptor signaling. It belongs to an immunoreceptor which is a family member of CD28/CTLA-4. The engagement of PD-1/PD-L1 suppresses T cell function and the axis is considered one of the major pathways involved in tumor immune evasion (Okazaki and Honjo, 2007). Proteus et al. synthesized a DNA aptamer (MP7), which mimicked an antibody that specifically bound to PD-1 and blocked the binding of PD-1 and PD-L1. The pegylated form of MP7 inhibited the growth of colon cancer in PD-L1-positive mice in vivo (Prodeus et al., 2015).
Therapeutic Aptamers Against Lung Cancer Cells
Immunotherapy has dramatically improved the survival rate of some lung cancer patients (Yu et al., 2016). Lai et al. (2016) developed a new type of PD-L1 antagonistic DNA aptamer (aptPD-L1). In CT26 and LL/2 murine syngeneic tumor models, aptPD-L1 had no direct cytotoxicity to cancer cells, but induced expression of IFNγ-inducible chemokines CXCL9/10 which regulated the tumor microenvironment. AptPD-L1 helped the recovery of T cell function and worked together with cytokines to resist tumor growth (Lai et al., 2016). Galectin-1 (Gal-1), which is related to several important biological processes in the development of tumors. Gal-1 has immunosuppressive effects on tumors by directly promoting T cell apoptosis or indirectly damaging the differentiation of tumor cells and T cells in their microenvironment (Pace et al., 1999). A DNA aptamer (AP-74 M-545) was developed for Galectin-1. The researchers developed a mouse lung cancer model to evaluate the characteristics, functions, and effects of AP-74 M-545. Aptamer blocked the interaction between Gal-1 and CD45 to inhibit T cells from apoptosis and restored T cell-mediated immunity. AP-74 M-545 exerted anti-tumor effect by restoring the immune function of mice (Tsai et al., 2019). The thrombin binding aptamer (TBA) has anticoagulant and antiproliferative effects. Esposito et al. (2018) chemically modified TBA to obtain two oligonucleotides, TBA353 and TBA535. Those two aptamers had significant anti-proliferative activity against lung cancer Calu-6 cells, while TBA535 inhibited cancer cell movement, indicating that it inhibited cell metastasis or tumor invasion (Esposito et al., 2018).
Therapeutic Aptamers Against Liver Cancer Cells
AS1411 is a DNA aptamer with 26 nucleotide that forms a guanine quadruplex structure. AS1411 binding to nucleolin that is aberrantly expressed on the cell membrane of many tumors. Nucleolin is involved in cell adhesion, division, and migration. AS1411 binds with nucleolin and inhibits tumor cell growth (Trinh et al., 2015). Cho et al. (2016) evaluated the affinity and specificity of the modified AS1411 to hepatocellular carcinoma cells (HCC). Besides, the authors studied the therapeutic potential of the modified aptamer in liver cancer. Experimental data showed that the modified AS1411 significantly reduced the proliferation of HCC cells in vitro. Microarray analysis showed that the modified AS1411-aptamer inhibited the growth of HCC cells by up-regulating the expression of galectin-14 (Cho et al., 2016). Epidermal growth factor receptor (EGFR) is a member of the ErbB receptor family, in vivo EGFR expression is associated with the progression of a variety of cancers, including breast cancer, glioma, lung cancer, and liver cancer. Inhibition of EGFR limits the growth and proliferation of EGFR-positive cancers. Wang et al. (2019) investigated a 27mer aptamer with therapeutic potential. It is CL-4RNV616 that contains 2′-O-Methyl RNA and DNA nucleotides and specifically targets EGFR. The researchers found CL-4RNV616 not only effectively recognized and inhibited the proliferation of Huh-7 liver cancer, MDA-MB-231 breast cancer, and U87MG glioblastoma cells but also effectively induced cancer cell apoptosis (Wang et al., 2019).
Therapeutic Aptamers Against Prostate Cancer Cells
Speransky et al. (2019) designed an RNA aptamer (Apt63) to distinguish prostate cancer cell lines with high metastatic potential from low metastatic potential. Apt63 was not bound to non-metastatic cancer cells but stuck to the beta subunit of F1Fo ATP synthase (ATP5B), which existed in the plasma membrane of cancer cells. The binding of Apt63 and plasma membrane-located ATP synthase (Ecto-ATP5B) could destroy the basic survival mechanism of tumors with high metastatic potential and lead to rapid cell death (Speransky et al., 2019). Prostate-specific membrane antigen (PSMA) is a glycosylated type II membrane protein. In the process of malignant transformation, prostate cancer (PC) has an abnormally high level of PSMA on the cell surface. PSMA represents an ideal target for the diagnosis and treatment of PC (Haberkorn et al., 2016; Virgolini et al., 2018). Dassie et al. (2014) described the therapeutic potential of an RNA aptamer (A9g). A9g was specific for PC cells expressing PSMA and had a therapeutic effect on advanced PC by inhibiting the enzymatic activity of PSMA. AGRO100 was an oligonucleotide that bound to nucleolin. The combination of A9g and AGRO100 led to a strong anti-proliferative response in tumor cells. Laber et al. (2005) conducted a phase I clinical trial of the aptamer AGRO100. There are 17 patients with advanced cancer were enrolled, including prostate cancer patients. Patients who receive AGRO100 therapy were in stable condition and had no adverse immune reactions (Laber et al., 2005).
Therapeutic Aptamers Against Leukemia Cells
Compared with normal cells, nucleolin is an overexpressed protein in the cytoplasm and plasma membrane of some tumor cells. Soundararajan et al. (2009) found that nucleolin in MV4-11 cells is the functional receptor of AS1411. In clinical trials, AS1411 not only shows promising anti-tumor activity but also has low serious systemic toxicity (Soundararajan et al., 2009). β-arrestin is a cell scaffold protein that promotes tumorigenesis in various tumor models. Kotula et al. (2014) adopted an interesting delivery strategy. DNA “targeted aptamers” directly delivered RNA “therapeutic aptamers.” Nucleolin-specific DNA delivery aptamers were coupled with a therapeutic aptamer targeting β-arrestin2, and the therapeutic aptamer was delivered to leukemia cells. Inhibition of β-arrestin2 led to the obstruction of multiple β-arrestin-mediated signaling pathways required for the progression of chronic myeloid leukemia (CML) (Kotula et al., 2014).
Therapeutic Aptamers Against Renal Cell Carcinoma Cells
Zhang et al. identified the aptamer SW-4 targeting human renal clear cell adenocarcinoma cell 786-O from a library of known sequences. Further studies showed that aptamers were internalized into target cells in a temperature-dependent manner through caveolae-mediated endocytosis and accumulated at tumor sites. SW-4b inhibited the proliferation of 786-O cells by preventing the progression of the cell cycle in the S phase (Zhang et al., 2018). AS1411 has been completed in clinical trials. The safety of clinical treatment effect is excellent. However, the overall remission efficiency is low (Bates et al., 2017). It showed anti-tumor activity in a phase I study in patients with renal cell carcinoma, and no dose-limiting toxicity was observed (Miller et al., 2006; Stuart et al., 2009). Rosenberg et al. (2014) conducted a phase II study. The results were unsatisfactory. Continuous infusion of a dose of 40 mg/kg/day would not cause any serious side effects. AS1411 existed a lower cancer remission rate. No response was found in other patients, and the poor pharmacology and low potency of this unmodified DNA may limit its future development in unscreened cancer patients (Rosenberg et al., 2014).
Therapeutic Aptamers Against Oral Cancer Cells
Heparanase is a β-1,4-endoglycosidase, which regulates the degradation and remodeling of extracellular matrix (ECM) (Hulett et al., 1999). It has been found that elevated levels of heparanase mRNA and protein in cancer patients have significantly reduced the postoperative survival time. The expression level of heparanase in cells is related to the metastatic potential of tumors (Koliopanos et al., 2001). Simmons et al. (2014) proved that the heparinase-specific aptamers inhibited tissue invasion of cells, and at the same time, aptamer was stable under physiological conditions and had no cytotoxicity to play a role in the treatment of oral cancer.
Therapeutic Aptamers Against Cervical Cancer Cells
Human papillomavirus (HPV) is a DNA virus that is closely related to the cervical cancer. HPV16, a high-risk subtype, infects epithelial cells and leads to the development of cancer through viral oncogenes E6 and E7 (Bernard et al., 2010). Studies showed that E6 promoted the degradation of the tumor suppressor gene p53 (Lechner and Laimins, 1994), and E7 led to dysregulation of the S phase of the cell cycle (Nicol et al., 2011). Clare et al. (2013) discovered an RNA aptamer (A2) that had a high affinity for E7. A2 bound to the N-terminal residue of E7, which interacted with the cell cycle control protein pRb. In cervical cancer cell lines, A2 was able to induce apoptosis, and inhibit cell proliferation.
Therapeutic Aptamers Against Bladder Cancer Cells
Transcription factors (TFs) in the cell nucleus play a crucial role in the process of cell gene expression. During the development of cancer, TF regulated the expression levels of oncogenes, tumor suppressor genes, and cell cycle-related molecules, thereby affected tumor formation, evolution, and metastasis (Haberle and Stark, 2018; Lambert et al., 2018). Xie et al. (2018) linked TF-specific RNA aptamers with targeted oncogenic miRNAs to form artificial long non-coding RNA (alncRNA). The transcription activity of TF was inhibited by alncRNA, and the transcription of oncogenes was controlled. The experiment showed that alncRNA in bladder cancer cell lines (5637, SW780) reduced the expression of TF target genes, cell proliferation and migration, and cell apoptosis was increased (Xie et al., 2018). Compared with the CRISPR/Cas system, alncRNA had both transcriptional regulation and post-transcriptional regulation of bladder cancer cells, showing a higher inhibitory effect in terms of the cell phenotype (Yao et al., 2020).
Therapeutic Aptamers Against Stomach Cancer Cells
ErbB-2/HER2 has a high expression in gastric cancer, so it is used as a therapeutic target. For stomach cancer, Mahlknecht et al. (2013) screened an aptamer from the library. They found this aptamer was able to bind ErbB-2/HER2 specifically. Only when the aptamer formed trimers, it could play a therapeutic role. To be more specific, the complex contained trimeric aptamer and ErbB-2/HER2 entered the cytoplasm through endocytosis, then lysosomes hydrolyzed this complex. As a result, the growth of gastric cancer cells and the growth rate of tumors were blocked and reduced respectively (Mahlknecht et al., 2013).
Therapeutic Aptamers Against Multiple Myeloma Cells
CXCL12 (Chemokine (C-X-C motif) ligand 12) is a chemokine, which is mainly related to cell trafficking and adhesion. Chemokines bind to specific G-protein-coupled seven-span transmembrane receptors. The binding of CXCL12 to its receptor CXC receptor 4 (CXCR4; CD184) will affect cell chemotaxis, cell survival, proliferation, and gene transcription (Teicher and Fricker, 2010). Waldschmidt et al. (2017) found that in multiple myeloma (MM), inhibition of cell adhesion-mediated drug resistance (CAM-DR) caused by bone marrow (BM) is the key to anti-myeloma treatment. NOX-A12, an L:-enantiomeric RNA oligonucleotide, is a specific inhibitor of CXCL12 (Sayyed et al., 2009). NOX-A12 functionally interfered with MM chemotaxis to the BM, caused multiple myeloma cells to be re-sensitized to therapeutic drugs. Aptamer NOX-A12 had anti-myeloma CAM-DR activity (Waldschmidt et al., 2017).
Conclusion
Aptamers have been widely applied to the treatment of diseases. They not only be used alone as therapeutics but also be combined with drugs covalently/non-covalently to achieve targeted drug delivery. With excellent binding affinity and specificity, aptamers have been more and more widely developed and applied to diagnosis, analytics, bio-imaging, and aptasensors of diseases (Ştefan et al., 2021). Among many application areas of aptamers, tumor therapy has aroused great interest. Researchers are committed to developing new aptamers as drugs to inhibit cancer progression. This article summarizes the aptamers screened for tumor cells in different cancer types. These aptamers require further animal experiments and clinical trials to confirm subsequent drug development.
Currently, the development of therapeutic aptamers is relatively slow, and there is no aptamer as a medicine to treat cancer clinically. Only a few aptamers had undergone clinical trials and they also have not been successfully used in the treatment of tumors. The underdevelopment may be mainly related to the following reasons. At the very beginning, aptamers are oligonucleotides that are susceptible to degradation by nucleases. Secondly, the aptamer has a small diameter and is easily filtered by the kidneys and excreted quickly. Thirdly, aptamers are artificially synthesized non-natural nucleotides, may cause chemical toxicity or immunogenicity. Last but not the least, when chemically synthesized in vitro aptamers are used in an in vivo environment, their conformation may be altered, affecting their affinity with targets or pharmacokinetic characteristics (Zhou and Rossi, 2017). For aptamers exist defects in affinity, specificity, and stability, a variety of strategies have been taken to overcome these difficulties and improve the effectiveness of aptamers in clinical disease treatment. For example, chemical modification of aptamers after SELEX and SELEX in vivo. Common chemical modifications include the introduction of 2′-O-methyl RNA bases, 2′-fluoro, 2′-thiol, 2′-hydroxymethyl, amino (2′-NH2) or 2′-azido, and sugar-modified nucleotide analogs such as unlocked nucleic acid or locked nucleic acid. At the same time, aptamers have spiegelmers, and L-nucleotides cannot be recognized by nuclease or the immune system. These chemical modification methods improve the nuclease resistance of the aptamer. The combination of aptamers and macromolecules effectively delays renal clearance. The introduced macromolecules include polyethyleneglycol, cholesterol 3′-biotin-streptavidinbioconjugates liposomes, proteins, dendrimers, and inorganic nanoparticles (Röthlisberger and Hollenstein, 2018). People use different chemical modifications to improve the deficiency of aptamers, which will facilitate further development of aptamers in disease treatment. Although aptamers have not been approved for clinical cancer treatment, as a unique and novel anti-cancer drug, it will have outstanding development. Furthermore, Aptamers provide a strong impetus for the development of cancer treatment.
Statements
Author contributions
CL supervised the whole project. ZL consulted the literature and wrote the manuscript. PZ revised the manuscript. JH, YH, XF, and XC provided the technical support and professional manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Natural Science Foundation Council of China (81922081 and 81700780).
Acknowledgments
We thank the administrative assistant (Ms. Yufang Zuo) for providing help and support.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Abbreviations
- BM
bone marrow
- CAM-DR
cell adhesion-mediated drug resistance
- ccRCC
clear cell renal cell carcinoma
- CEA
carcinoembryonic antigen
- CML
chronic myeloid leukemia
- CRC
colorectal cancer
- CXCL12
chemokine (C-X-C motif) ligand 12
- E7
viral oncogenes
- ECM
extracellular matrix
- Ecto-ATP5B
plasma membrane-located ATP synthase
- EGFR
epidermal growth factor receptor
- eIF4E
eukaryotic translation initiation factor 4E
- FDA
US Food and Drug Administration
- Gal-1
galectin-1
- HCC
hepatocellular carcinoma cells
- HER2
human epidermal growth factor receptor 2
- HPV
human papillomavirus
- MM
multiple myeloma
- MNK
MAP kinase-interacting kinase
- PC
prostate cancer
- PD-1
programmed cell death-1
- PDGF-BB
platelet-derived growth factor-BB
- PSMA
prostate-specific membrane antigen
- SELEX
systematic evolution of ligands by exponential enrichment
- TFs
transcription factors.
References
1
Adachi T. Nakamura Y. (2019). Aptamers: A Review of Their Chemical Properties and Modifications for Therapeutic Application.Molecules24:23.
2
Alavi M. Hamidi M. (2019). Passive and active targeting in cancer therapy by liposomes and lipid nanoparticles.Drug Metab. Pers. Ther.2019:34.
3
Alizadeh N. Memar M. Y. Mehramuz B. Abibiglou S. S. Hemmati F. Kafil H. S. (2018). Current advances in aptamer-assisted technologies for detecting bacterial and fungal toxins.J. Appl. Microbiol.124644–651. 10.1111/jam.13650
4
Bates P. J. Reyes-Reyes E. M. Malik M. T. Murphy E. M. O’toole M. G. Trent J. O. (2017). G-quadruplex oligonucleotide AS1411 as a cancer-targeting agent: Uses and mechanisms.Biochim. Biophys. Acta-Gen. Subj.18611414–1428. 10.1016/j.bbagen.2016.12.015
5
Bayat P. Nosrati R. Alibolandi M. Rafatpanah H. Abnous K. Khedri M. et al (2018). SELEX methods on the road to protein targeting with nucleic acid aptamers.Biochimie154132–155. 10.1016/j.biochi.2018.09.001
6
Bernard H. U. Burk R. D. Chen Z. Doorslaer K. V. Hausen H. Z. Villiers E. M. D. (2010). Classification of papillomaviruses (PVs) based on 189 PV types and proposal of taxonomic amendments.Virology40170–79. 10.1016/j.virol.2010.02.002
7
Carvalho J. Paiva A. Campello M. P. C. Paulo A. Mergny J.-L. Salgado G. F. et al (2019). Aptamer-based targeted Delivery of a G-quadruplex Ligand in Cervical Cancer Cells.Sci. Rep.91–12.
8
Cesewski E. Johnson B. N. (2020). Electrochemical biosensors for pathogen detection.Biosens. Bioelectr.2020:112214. 10.1016/j.bios.2020.112214
9
Chen C. Zhou S. Cai Y. Tang F. (2017). Nucleic acid aptamer application in diagnosis and therapy of colorectal cancer based on cell-SELEX technology.NPJ Precis. Oncol.1:37.
10
Cheng C. Chen Y. H. Lennox K. A. Behlke M. A. Davidson B. L. (2013). In vivo SELEX for Identification of Brain-penetrating Aptamers.Mole. Ther. Nucleic Acids2:e67. 10.1038/mtna.2012.59
11
Cho Y. Lee Y. B. Lee J.-H. Lee D. H. Cho E. J. Yu S. J. et al (2016). Modified AS1411 aptamer suppresses hepatocellular carcinoma by up-regulating galectin-14.PLoS One11:e0160822. 10.1371/journal.pone.0160822
12
Citartan M. Tang T. H. Tan S. C. Gopinath S. C. B. (2011). Conditions optimized for the preparation of single-stranded DNA (ssDNA) employing lambda exonuclease digestion in generating DNA aptamer.World J. Microb. Biotechnol.271167–1173. 10.1007/s11274-010-0563-8
13
Citartan M. Tang T. H. Tan S. C. Hoe C. H. Gopinath S. C. B. (2012). Asymmetric PCR for good quality ssDNA generation towards DNA aptamer production.Songklanakarin J. Technol.34125–131.
14
Clare N. Zlem C. Sophie F. Belyaeva T. A. Bunka D. H. J. Eric B. G. et al (2013). An RNA Aptamer Provides a Novel Approach for the Induction of Apoptosis by Targeting the HPV16 E7 Oncoprotein.PLoS One8:e64781. 10.1371/journal.pone.0064781
15
Dassie J. P. Hernandez L. I. Thomas G. S. Long M. E. Rockey W. M. Howell C. A. et al (2014). Targeted inhibition of prostate cancer metastases with an RNA aptamer to prostate specific membrane antigen (PSMA).Mol. Ther.221910–1922.
16
Ding F. Guo S. Xie M. Luo W. Yuan C. Huang W. et al (2015). Diagnostic applications of gastric carcinoma cell aptamers in vitro and in vivo.Talanta13430–36. 10.1016/j.talanta.2014.09.036
17
Dougan M. Dougan S. K. (2017). Targeting Immunotherapy to the Tumor Microenvironment.J. Cell. Biochem.1183049–3054.
18
Duda D. G. Kozin S. V. Kirkpatrick N. D. Xu L. Fukumura D. Jain R. K. (2011). CXCL12 (SDF1α)-CXCR4/CXCR7 pathway inhibition: an emerging sensitizer for anticancer therapies?Clin. Res.172074–2080. 10.1158/1078-0432.ccr-10-2636
19
Dunn M. R. Jimenez R. M. Chaput J. C. (2017). Analysis of aptamer discovery and technology.Nat. Rev. Chem.1:0076.
20
Ellington A. D. Szostak J. W. (1990). In vitro selection of RNA molecules that bind specific ligands.Nature346818–822. 10.1038/346818a0
21
Espelund M. Prentice S. R. A. Jakobsen K. S. (1990). A simple method for generating single-stranded DNA probes labeled to high activities.Nuclc Acids Res.186157–6158. 10.1093/nar/18.20.6157
22
Esposito V. Russo A. Vellecco V. Bucci M. Russo G. Mayol L. et al (2018). Thrombin binding aptamer analogues containing inversion of polarity sites endowed with antiproliferative and anti-motility properties against Calu-6 cells.Biochim. Biophys. Acta (BBA)-Gen. Subj.18622645–2650. 10.1016/j.bbagen.2018.07.031
23
Frohnmeyer E. Tuschel N. Sitz T. Hermann C. Dahl G. T. Schulz F. et al (2019). Aptamer lateral flow assays for rapid and sensitive detection of cholera toxin.Analyst1441840–1849. 10.1039/c8an01616j
24
Gang Z. Olivier L. Mary B. Lionel H. Wei D. Christopher L. et al (2017). Aptamer-Based Therapeutic Approaches to Target Cancer Stem Cells.Theranostics73948–3961. 10.7150/thno.20725
25
García-Recio E. M. Pinto-Díez C. Pérez-Morgado M. I. García-Hernández M. Fernández G. Martín M. E. et al (2016). Characterization of MNK1b DNA aptamers that inhibit proliferation in MDA-MB231 breast cancer cells.Mole. Ther. Nucleic Acids5:e275. 10.1038/mtna.2015.50
26
Gelinas A. D. Davies D. R. Janjic N. (2016). Embracing proteins: structural themes in aptamer–protein complexes.Curr. Opin. Struct. Biol.36:122. 10.1016/j.sbi.2016.01.009
27
Gijs M. Penner G. Blackler G. B. Impens N. R. Baatout S. Luxen A. et al (2016). Improved aptamers for the diagnosis and potential treatment of HER2-positive cancer.Pharmaceuticals9:29. 10.3390/ph9020029
28
Gray B. P. Kelly L. Ahrens D. P. Barry A. P. Sullenger B. A. (2018). Tunable cytotoxic aptamer–drug conjugates for the treatment of prostate cancer.Proc. Natl. Acad.115:201717705.
29
Guizhi Z. Xiaoyuan C. (2018). Aptamer-based targeted therapy. Adv. Drug. Deliv. Rev.134, 65–78. 10.1016/j.addr.2018.08.005
30
Guo P. (2011). The emerging field of RNA nanotechnology.Chem.5833–842. 10.1038/nnano.2010.231
31
Haberkorn U. Eder M. Kopka K. Babich J. W. Eisenhut M. (2016). New Strategies in Prostate Cancer: Prostate-Specific Membrane Antigen (PSMA) Ligands for Diagnosis and Therapy.Clin. Cancer Res.229–15. 10.1158/1078-0432.ccr-15-0820
32
Haberle V. Stark A. (2018). Eukaryotic core promoters and the functional basis of transcription initiation.Nat. Rev. Mole. Cell Biol.19621–637. 10.1038/s41580-018-0028-8
33
Hammarström S. (1999). The carcinoembryonic antigen (CEA) family: structures, suggested functions and expression in normal and malignant tissues.Semin. Cancer Biol.967–81. 10.1006/scbi.1998.0119
34
Hou Z. Meyer S. Propson N. Nie J. Jiang P. Stewart R. et al (2015). Characterization and target identification of a DNA aptamer that labels pluripotent stem cells.Cell Res.25390–393. 10.1038/cr.2015.7
35
Hulett M. D. Freeman C. Hamdorf B. J. Baker R. T. Harris M. J. Parish C. R. (1999). Cloning of mammalian heparanase, an important enzyme in tumor invasion and metastasis.Nat. Med.5803–809. 10.1038/10525
36
Kato S. Ono T. Minagawa H. Horii K. Aoki T. (2020). FSBC: fast string-based clustering for HT-SELEX data.BMC Bioinformatics21:3607. 10.1186/s12859-020-03607-1
37
Kaur H. (2018). Recent developments in cell-SELEX technology for aptamer selection.Biochim. Biophys. Acta Gen. Subj.18622323–2329. 10.1016/j.bbagen.2018.07.029
38
Koliopanos A. Friess H. Kleeff J. Shi X. Büchler M. W. (2001). Heparanase expression in primary and metastatic pancreatic cancer.Cancer Res.614655–4659.
39
Komarova N. Kuznetsov A. (2019). Inside the Black Box: What Makes SELEX Better?Molecules24:19.
40
Kordasht H. K. Hasanzadeh M. (2020). Aptamer based recognition of cancer cells: Recent progress and challenges in bioanalysis.Talanta220:121436. 10.1016/j.talanta.2020.121436
41
Kotula J. W. Sun J. Li M. Pratico E. D. Fereshteh M. P. Ahrens D. P. et al (2014). Targeted disruption of β-arrestin 2-mediated signaling pathways by aptamer chimeras leads to inhibition of leukemic cell growth.PLoS One9:e93441. 10.1371/journal.pone.0093441
42
Kudo Y. (2007). Periostin: novel diagnostic and therapeutic target for cancer.Histol. Histopathol.221167–1174.
43
Laber D. Choudry M. Taft B. Bhupalam L. Sharma V. Hendler F. et al (2004). A phase I study of AGRO100 in advanced cancer.J. Clin. Oncol.223112–3112. 10.1200/jco.2004.22.14_suppl.3112
44
Laber D. Sharma V. Bhupalam L. Taft B. Hendler F. Barnhart K. (2005). Update on the first phase I study of AGRO100 in advanced cancer.J. Clin. Oncol.233064–3064. 10.1200/jco.2005.23.16_suppl.3064
45
Lai W.-Y. Huang B.-T. Wang J.-W. Lin P.-Y. Yang P.-C. (2016). A novel PD-L1-targeting antagonistic DNA aptamer with antitumor effects.Mole. Ther. Nucleic Acids5:e397. 10.1038/mtna.2016.102
46
Lakhin A. V. Tarantul V. Z. Gening L. V. (2013). Aptamers: Problems, Solutions and Prospects.Acta Nat.534–43. 10.32607/20758251-2013-5-4-34-43
47
Lambert S. A. Jolma A. Campitelli L. F. Das P. K. Yin Y. Albu M. et al (2018). The Human Transcription Factors.Cell172650–665.
48
Lammers T. Kiessling F. Hennink W. E. Storm G. (2012). Drug targeting to tumors: Principles, pitfalls and (pre-) clinical progress.J. Controlled Release161175–187. 10.1016/j.jconrel.2011.09.063
49
Lechner M. S. Laimins L. A. (1994). Inhibition of p53 DNA binding by human papillomavirus E6 proteins.J. Virol.68:4262. 10.1128/jvi.68.7.4262-4273.1994
50
Lee Y. J. Han S. R. Kim N. Y. Lee S. H. Jeong J. S. Lee S. W. (2012). An RNA aptamer that binds carcinoembryonic antigen inhibits hepatic metastasis of colon cancer cells in mice.Gastroenterology143155.–165.
51
Lee Y. J. Kim I. S. Park S.-A. Kim Y. Lee J. E. Noh D.-Y. et al (2013). Periostin-binding DNA aptamer inhibits breast cancer growth and metastasis.Mole. Ther.211004–1013. 10.1038/mt.2013.30
52
Lewis F. Liang S. Simon S. Julian T. (2017). Aptamer Bioinformatics.Internatl. J. Mole. Sci.18:2516. 10.3390/ijms18122516
53
Liu M. Yu X. Chen Z. Yang T. Yang D. Liu Q. et al (2017). Aptamer selection and applications for breast cancer diagnostics and therapy.J. Nanobiotechnol.15:81.
54
Liu Y. Jiang W. Yang S. Hu J. Zu Y. (2019). Rapid Detection of Mycoplasma-Infected Cells by an ssDNA Aptamer Probe.ACS Sensors4:8.
55
Maasch C. Buchner K. Eulberg D. Vonhoff S. Klussmann S. (2008). Physicochemical stability of NOX-E36, a 40mer L-RNA (Spiegelmer) for therapeutic applications.Nucleic Acids Symp. Series200861–62. 10.1093/nass/nrn031
56
Mahlknecht G. Maron R. Mancini M. Schechter B. Sela M. Yarden Y. (2013). Aptamer to ErbB-2/HER2 enhances degradation of the target and inhibits tumorigenic growth.Proc. Natl. Acad. Sci.1108170–8175. 10.1073/pnas.1302594110
57
Mallikaratchy P. Stahelin R. Cao Z. Cho W. Tan W. (2006). Selection of DNA ligands for protein kinase C-δ.Chem. Comm.3229–3231. 10.1039/b604778e
58
Miller D. M. Laber D. A. Bates P. J. Trent J. O. Kloecker G. H. (2006). Extended phase i study of AS1411 in renal and non-small cell lung cancers.Ann. Oncol.17147–148.
59
Moosavian S. A. Sahebkar A. (2019). Aptamer-functionalized liposomes for targeted cancer therapy.Can. Lett.448144–154. 10.1016/j.canlet.2019.01.045
60
Nicol C. Bunka D. H. J. Blair G. E. Stonehouse N. J. (2011). Effects of single nucleotide changes on the binding and activity of RNA aptamers to human papillomavirus 16 E7 oncoprotein.Biochem. Biophys. Res. Commun.405417–421. 10.1016/j.bbrc.2011.01.044
61
Odeh F. Nsairat H. Alshaer W. Ismail M. A. Ismail S. I. (2020). Aptamers Chemistry: Chemical Modifications and Conjugation Strategies.Molecules25:3. 10.3390/molecules25010003
62
Okazaki T. Honjo T. (2007). PD-1 and PD-1 ligands: from discovery to clinical application.Internat. Immunol.19813–824. 10.1093/intimm/dxm057
63
Pace K. E. Lee C. Stewart P. L. Baum L. G. (1999). Restricted receptor segregation into membrane microdomains occurs on human T cells during apoptosis induced by galectin-1.J. Immunol.1633801–3811.
64
Pérez-Herrero E. Fernández-Medarde A. (2015). Advanced targeted therapies in cancer: Drug nanocarriers, the future of chemotherapy.Eur. J. Pharm. Biopharm.9352–79. 10.1016/j.ejpb.2015.03.018
65
Povsic T. J. Vavalle J. P. Alexander J. H. Aberle L. H. Mehran R. (2014). Use of the REG1 anticoagulation system in patients with acute coronary syndromes undergoing percutaneous coronary intervention: results from the phase II RADAR-PCI study.Eur. J. Eur. Collab. Intervent. Cardiol. Eur. Soc. Cardiol.10431–438. 10.4244/eijy14m06_01
66
Prodeus A. Abdul-Wahid A. Fischer N. W. Huang E. H. Cydzik M. Gariépy J. (2015). Targeting the PD-1/PD-L1 immune evasion axis with DNA aptamers as a novel therapeutic strategy for the treatment of disseminated cancers.Mole. Ther. Nucleic Acids4:e237. 10.1038/mtna.2015.11
67
Ram D. T. Ellington A. D. Allen P. B. (2017). Purification of single-stranded DNA by co-polymerization with acrylamide and electrophoresis.Biotechniques62275–282.
68
Rosenberg J. E. Bambury R. M. Van Allen E. M. Drabkin H. A. Lara P. N. Harzstark A. L. et al (2014). A phase II trial of AS1411 (a novel nucleolin-targeted DNA aptamer) in metastatic renal cell carcinoma.Investigat. New Drugs32178–187. 10.1007/s10637-013-0045-6
69
Röthlisberger P. Hollenstein M. (2018). Aptamer chemistry.Advanced drug delivery reviews1343–21.
70
Sae-Lim S. Soontornworajit B. Pichayanoot P. (2019). Inhibition of Colorectal Cancer Cell Proliferation by Regulating Platelet-Derived Growth Factor B Signaling with a DNA Aptamer.Asian Pacific J. Cancer Preven. APJCP20:487. 10.31557/apjcp.2019.20.2.487
71
Sayyed S. G. Hele H. Kulkarni O. P. Endlich K. Segerer S. Eulberg D. et al (2009). Podocytes produce homeostatic chemokine stromal cell-derived factor-1/CXCL12, which contributes to glomerulosclerosis, podocyte loss and albuminuria in a mouse model of type 2 diabetes.Diabetologia522445–2454. 10.1007/s00125-009-1493-6
72
Simmons S. C. Jämsä H. Silva D. Cortez C. M. McKenzie E. A. Bitu C. C. et al (2014). Anti-heparanase aptamers as potential diagnostic and therapeutic agents for oral cancer.PLoS One9:e96846. 10.1371/journal.pone.0096846
73
Sinha A. Gopinathan P. Chung Y-D. Lin Y-D. Lic K-H. Ma H-P. et al (2018). An integrated microfluidic platform to perform uninterrupted SELEX cycles to screen affinity reagents specific to cardiovascular biomarkers.Biosens. Bioelectr.122104–112. 10.1016/j.bios.2018.09.040
74
Soldevilla M. Daniel M. C. D. C. Menon A. Pastor F. (2018). Aptamer-iRNAs as Therapeutics for Cancer Treatment.Pharmaceuticals11:4.
75
Soundararajan S. Wang L. Sridharan V. Chen W. Courtenay-Luck N. Jones D. et al (2009). Plasma membrane nucleolin is a receptor for the anticancer aptamer AS1411 in MV4-11 leukemia cells.Mole. Pharm.76984–991. 10.1124/mol.109.055947
76
Speransky S. Serafini P. Caroli J. Bicciato S. Lippman M. Bishopric N. (2019). A novel RNA aptamer identifies plasma membrane ATP synthase beta subunit as an early marker and therapeutic target in aggressive cancer.Breast Cancer Res. Treat.176271–289. 10.1007/s10549-019-05174-3
77
Ştefan G. Hosu O. De Wael K. Lobo-Castañón M. J. Cristea C. (2021). Aptamers in biomedicine: Selection strategies and recent advances.Electrochimica Acta376:137994. 10.1016/j.electacta.2021.137994
78
Stuart R. Stockerl-Goldstein K. Cooper M. Devetten M. Herzig R. Medeiros B. et al (2009). Randomized phase II trial of the nucleolin targeting aptamer AS1411 combined with high-dose cytarabine in relapsed/refractory acute myeloid leukemia (AML).J. Clin. Oncol.277019–7019. 10.1200/jco.2009.27.15_suppl.7019
79
Subramanian K. Rutvisuttinunt W. Scott W. Myers R. S. (2003). The enzymatic basis of processivity in lambda exonuclease.Nuclc Acids Res.311585–1596. 10.1093/nar/gkg266
80
Taheri M. Saragovi U. Fuks A. Makkerh J. Mort J. Stanners C. P. (2000). Self recognition in the Ig superfamily: Identification of precise sub domains in CEA required for intercellular adhesion.J. Biol. Chem.275:35. 10.1201/9781482283471-9
81
Tan Y. Li Y. Tang F. (2019). Nucleic Acid Aptamer: A Novel Potential Diagnostic and Therapeutic Tool for Leukemia.OncoTargets Ther.12:10597. 10.2147/ott.s223946
82
Tauriello D. V. Calon A. Lonardo E. Batlle E. (2017). Determinants of metastatic competency in colorectal cancer.Mole. Oncol.1197–119. 10.1002/1878-0261.12018
83
Teicher B. A. Fricker S. P. (2010). CXCL12 (SDF-1)/CXCR4 Pathway in Cancer.Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res.162927–2931. 10.1158/1078-0432.ccr-09-2329
84
Tian S. (2017). Novel Roles of the MAP Kinase-interacting Kinases. Ph.D. thesis, University of Adelaide, School of Biological Sciences.
85
Trinh T. L. Zhu G. Xiao X. Puszyk W. Sefah K. Wu Q. et al (2015). A Synthetic Aptamer-Drug Adduct for Targeted Liver Cancer Therapy.PLoS One10:e0136673. 10.1371/journal.pone.0136673
86
Tsai Y.-T. Liang C.-H. Yu J.-H. Huang K.-C. Tung C.-H. Wu J.-E. et al (2019). A DNA Aptamer targeting galectin-1 as a novel immunotherapeutic strategy for lung cancer.Mole. Ther. Nucleic Acids18991–998. 10.1016/j.omtn.2019.10.029
87
Umar M. I. Kit K. C. (2020). Specific suppression of D-RNA G-quadruplex–protein interaction with an L-RNA aptamer.Nucleic Acids Res.18:18.
88
Virgolini I. Decristoforo C. Haug A. Fanti S. Uprimny C. (2018). Current status of theranostics in prostate cancer.Eur. J. Nuclear Med. Mole. Imag.45 (Suppl. 3), 471–495.
89
Waks A. G. Winer E. P. (2019). Breast cancer treatment: a review.JAMA321288–300.
90
Waldschmidt J. M. Simon A. Wider D. Müller S. Follo M. Ihorst G. et al (2017). CXCL12 and CXCR7 are relevant targets to reverse cell adhesion-mediated drug resistance in multiple myeloma.Br. J. Haematol.17936–49. 10.1111/bjh.14807
91
Wang T. Chen C. Larcher L. M. Barrero R. A. Veedu R. N. et al (2018). Three decades of nucleic acid aptamer technologies: Lessons learned, progress and opportunities on aptamer development.Biotechnol. Adv.3728–50. 10.1016/j.biotechadv.2018.11.001
92
Wang T. Philippovich S. Mao J. Veedu R. N. (2019). Efficient epidermal growth factor receptor targeting oligonucleotide as a potential molecule for targeted cancer therapy.Internat. J. Mole. Sci.20:4700. 10.3390/ijms20194700
93
Waters E. Richardson J. Schaub R. Kurz J. (2009). Effect of NU172 and bivalirudin on ecarin clotting time in human plasma and whole blood-Posters.J. Thromb. Haemos.2009:7.
94
Xie H. Zhan H. Gao Q. Li J. Zhou Q. Chen Z. et al (2018). Synthetic artificial “long non-coding RNAs” targeting oncogenic microRNAs and transcriptional factors inhibit malignant phenotypes of bladder cancer cells.Cancer Lett.42294–106. 10.1016/j.canlet.2018.02.038
95
Yao L. Zhang Q. Li A. Ma B. Zhang Z. Liu J. et al (2020). Synthetic Artificial Long Non-coding RNA Shows Higher Efficiency in Specific Malignant Phenotype Inhibition Compared to the CRISPR/Cas Systems.Front. Mole. Biosci.7:387.
96
Yazdian-Robati R. Bayat P. Oroojalian F. Zargari M. Ramezani M. Taghdisi S. M. et al (2019). Therapeutic applications of AS1411 aptamer, an update review.Internat. J. Biol. Macromole.1551420–1431. 10.1016/j.ijbiomac.2019.11.118
97
Yoshihiro M. Macall L. Hiroyasu K. David V. Takemi T. (2018). Aptamer Therapeutics in Cancer: Current and Future.Cancers10:80. 10.3390/cancers10030080
98
Yu H. Boyle T. A. Zhou C. Rimm D. L. Hirsch F. R. (2016). PD-L1 expression in lung cancer.J. Thorac. Oncol.11964–975.
99
Yue H. Wang Z. G. Tang H. W. Pang D. W. (2011). Low background signal platform for the detection of ATP: When a molecular aptamer beacon meets graphene oxide.Biosens. Bioelectr.2976–81. 10.1016/j.bios.2011.07.069
100
Zhang G. Q. Zhong L. P. Yang N. Zhao Y. X. (2019). Screening of aptamers and their potential application in targeted diagnosis and therapy of liver cancer.World J. Gastroenterol.253359–3369. 10.3748/wjg.v25.i26.3359
101
Zhang H. Wang Z. Xie L. Zhang Y. Deng T. Li J. et al (2018). Molecular recognition and in-vitro-targeted inhibition of renal cell carcinoma using a DNA aptamer.Mole. Ther. Nucleic Acids12758–768. 10.1016/j.omtn.2018.07.015
102
Zhang Y. Lai B. Juhas M. (2019). Recent Advances in Aptamer Discovery and Applications.Molecules24:5.
103
Zhou J. Rossi J. (2017). Aptamers as targeted therapeutics: current potential and challenges.Nat. Rev. Drug Discov.16181–202. 10.1038/nrd.2016.199
104
Zhu C. Yang G. Ghulam M. Li L. Qu F. (2019). Evolution of multi-functional capillary electrophoresis for high-efficiency selection of aptamers.Biotechnol. Adv.37:107432. 10.1016/j.biotechadv.2019.107432
105
Zhu G. Chen X. (2018). Aptamer-based targeted therapy.Adv. Drug Deliv. Rev.13465–78.
Summary
Keywords
aptamer, cancer cells, targeted therapy, SELEX, clinical application
Citation
Li Z, Fu X, Huang J, Zeng P, Huang Y, Chen X and Liang C (2021) Advances in Screening and Development of Therapeutic Aptamers Against Cancer Cells. Front. Cell Dev. Biol. 9:662791. doi: 10.3389/fcell.2021.662791
Received
01 February 2021
Accepted
21 April 2021
Published
19 May 2021
Volume
9 - 2021
Edited by
Yuanyuan Yu, Hong Kong Baptist University, Hong Kong
Reviewed by
Zexian Liu, Sun Yat-sen University Cancer Center (SYSUCC), China; Jing Nie, People’s Liberation Army General Hospital, China
Updates
Copyright
© 2021 Li, Fu, Huang, Zeng, Huang, Chen and Liang.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chao Liang, liangc@sustech.edu.cn
This article was submitted to Molecular Medicine, a section of the journal Frontiers in Cell and Developmental Biology
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.