Abstract
Research involving autism spectrum disorder (ASD) most frequently focuses on its key diagnostic criteria: restricted interests and repetitive behaviors, altered sensory perception, and communication impairments. These core criteria, however, are often accompanied by numerous comorbidities, many of which result in severe negative impacts on quality of life, including seizures, epilepsy, sleep disturbance, hypotonia, and GI distress. While ASD is a clinically heterogeneous disorder, gastrointestinal (GI) distress is among the most prevalent co-occurring symptom complex, manifesting in upward of 70% of all individuals with ASD. Consistent with this high prevalence, over a dozen family foundations that represent genetically distinct, molecularly defined forms of ASD have identified GI symptoms as an understudied area with significant negative impacts on quality of life for both individuals and their caregivers. Moreover, GI symptoms are also correlated with more pronounced irritability, social withdrawal, stereotypy, hyperactivity, and sleep disturbances, suggesting that they may exacerbate the defining behavioral symptoms of ASD. Despite these facts (and to the detriment of the community), GI distress remains largely unaddressed by ASD research and is frequently regarded as a symptomatic outcome rather than a potential contributory factor to the behavioral symptoms. Allowing for examination of both ASD’s impact on the central nervous system (CNS) as well as its impact on the GI tract and the associated microbiome, the zebrafish has recently emerged as a powerful tool to study ASD. This is in no small part due to the advantages zebrafish present as a model system: their precocious development, their small transparent larval form, and their parallels with humans in genetics and physiology. While ASD research centered on the CNS has leveraged these advantages, there has been a critical lack of GI-centric ASD research in zebrafish models, making a holistic view of the gut-brain-microbiome axis incomplete. Similarly, high-throughput ASD drug screens have recently been developed but primarily focus on CNS and behavioral impacts while potential GI impacts have not been investigated. In this review, we aim to explore the great promise of the zebrafish model for elucidating the roles of the gut-brain-microbiome axis in ASD.
Introduction
The contribution of the gut-brain-microbiome axis to health and disease states is a relatively new field of research (Figure 1) with increasing interest from both public and scientific spheres (Drossman and Hasler, 2016). An understanding of this axis draws on a range of disciplines including neurobiology, gastroenterology, microbiology, endocrinology, and psychology (Liang et al., 2018; Neuhaus et al., 2018). This breadth of subjects relevant to the gut-brain-microbiome field speaks to the diversity of its potential applications. Specifically, its relevance to research on neurological disorders like autism spectrum disorder (ASD) is of particular interest, as such disorders often cause a wide range of symptoms involving multiple body systems. Furthermore, the interconnected aspects of the gut-brain-microbiome offer alternative causal explanations and treatment strategies for symptoms traditionally understood to be strictly caused by deficits within the central nervous system (CNS) (Neuhaus et al., 2018; Lefter et al., 2019; Srikantha and Mohajeri, 2019; Tye et al., 2019). While ASD is still diagnosed by deficits in social communication, repetitive behaviors, and/or restrictive interests, comorbidities (co-occurring symptoms) like seizures, epilepsy, sleep disturbance, hypotonia, and GI distress are also common with significant negative impacts on quality of life (Christensen et al., 2018; “IACC, 2019 Strategic Plan For Autism Spectrum Disorder 2018–2019 Update,” 2019; Leader et al., 2020). Here, we review how recent studies of the gut-brain-microbiome axis have changed our understanding of ASD related symptoms and highlight the important role the zebrafish model can play in future research.
FIGURE 1
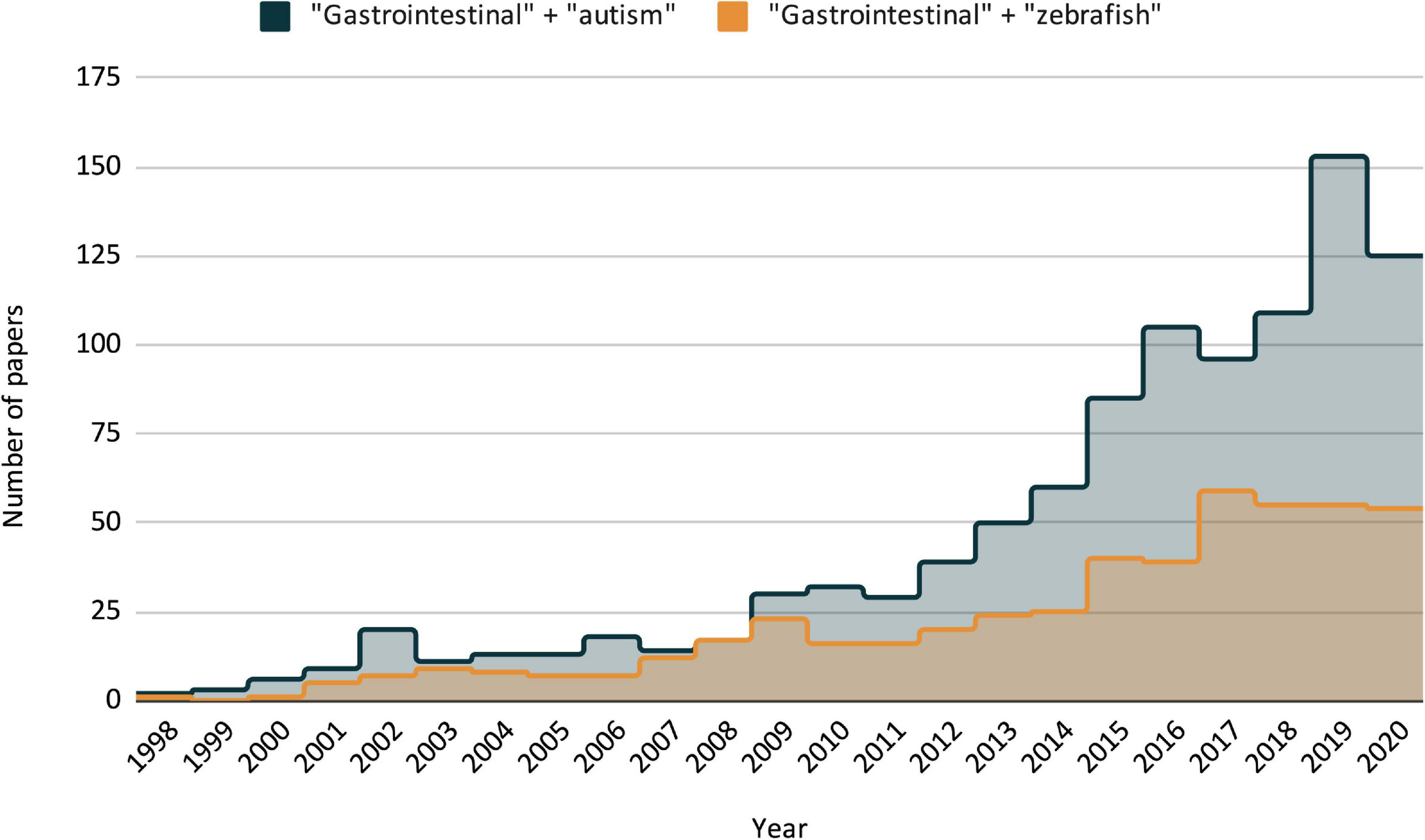
Publication trends as listed by PubMed/NLM over the last two decades, search criteria for each is “gastrointestinal + autism” and “gastrointestinal + zebrafish.” Including all three search terms (GI + ASD + zebrafish) only resulted in three publications, the earliest in 2014.
Since it was first described in a small subset of patients in 1943, the clinical definition of ASD has been subject to an ever-changing set of criteria in an attempt to capture a condition that is both common and heterogeneous. Likewise, estimates of the prevalence of comorbidities associated with ASD have changed (Geschwind, 2009; Chaidez et al., 2014; Bresnahan et al., 2015; De Rubeis and Buxbaum, 2015; Tye et al., 2019). This shift in criteria likely stems from diverse causal factors including hundreds of implicated genes, environmental, and gene-environment interactions that contribute to ASD prevalence (Chaste and Leboyer, 2012). Our current understanding is that ASD impacts more than 1% of the population and is both etiologically and clinically heterogeneous. Given this heterogeneity, addressing underlying mechanisms to develop treatment strategies has been difficult (Manoli and State, 2021). Moreover, the field would greatly benefit from determining how body systems work cooperatively and/or antagonistically to produce both core behavioral symptoms of ASD and the comorbid symptoms like gastrointestinal (GI) distress (McElhanon et al., 2014; Pellicano et al., 2014; Frye et al., 2015; Latorre et al., 2016; Rao and Gershon, 2016; Rose et al., 2017; Goodspeed et al., 2020). GI distress occurs at a disproportionately higher rate in individuals with ASD than the general population, and symptom severity ranges from relatively low-impact to severe (Bresnahan et al., 2015). As research into these GI symptoms has expanded, mounting evidence suggests that they also contribute to the behavioral symptoms associated with ASD, a finding well-recognized even in patients without ASD and explained by the biopsychosocial model of disorders of gut-brain interaction (Klarer et al., 2014; Mayer et al., 2014; Drossman and Hasler, 2016; Sharon et al., 2019). With this in mind, it becomes apparent why the gut-brain-microbiome axis is a critical focal point for studying both the pathophysiology of ASD-related GI dysfunction and ASD as a whole.
Addressing GI symptoms within neurological disorders is challenging because the regulation of GI function is complex and full of redundant feedback mechanisms involving multiple body systems (Holtmann and Talley, 2014). Adding to this complexity is the fact that the luminal space of the GI tract is technically “outside” of the body and not sterile, lending itself to microbial and chemical exposure which could influence regulatory mechanisms. Under normal conditions, communication between the GI tract and the CNS is modified by contributions from immune, microbial, hormonal, motor, and sensory inputs (Grundy et al., 2006; Vanner et al., 2016). The GI tract also exerts a large amount of autonomous control over its own functions, with the enteric nervous system (ENS) interfacing with various mechanosensory, chemosensory, endocrine, immune, and secretory cells, altering GI function as needed to deal with threats and maintain homeostasis (Holtmann and Talley, 2014). These typical functions may be altered in ASD (Hsiao, 2014), and recent findings from both clinical and rodent model studies have begun to frame the importance of the gut-brain-microbiome axis in ASD (Hsiao, 2014).
Viewing autism as a disorder of the brain, without consideration of gut/microbiome can have unintended negative consequences; a prime example is the use of antipsychotics to reduce aggressive behaviors, since these also suppress GI motility and thus are likely to increase GI distress (de Alvarenga et al., 2017). In this review, we contend that the zebrafish presents unique opportunities to approach to autism research holistically. In particular zebrafish ASD models are amenable to genetic modification, in vivo visualization of multiple organ systems, and high-throughput studies, providing an ideal model system to address a multidisciplinary gut-brain-microbiome approach to ASD research (Brugman, 2016; Kozol et al., 2016; Phelps et al., 2017; Kozol, 2018; James et al., 2019).
Gastrointestinal Issues and Their Link to Neurological Disorders
Gastrointestinal distress is a pervasive co-occurring ailment in a wide range of neurological disorders, including Parkinson’s (Mulak and Bonaz, 2015; Liddle, 2018; Brudek, 2019), schizophrenia (Severance et al., 2015, 2016; Dickerson et al., 2017), and Alzheimer’s (Hill et al., 2014; Jiang et al., 2017; Kowalski and Mulak, 2019; Goyal et al., 2020; He et al., 2020). Only in the last 5 years has GI distress been more widely recognized as an ASD-related comorbidity, and the potential causes have been the subject of considerable and ongoing debate. Although a comprehensive discussion on the clinical prevalence and significance of GI distress in ASD is outside of the scope of this review, we believe reviewing a few critical points are helpful in framing the current state of zebrafish-based research as it relates to ASD and GI comorbidities.
Broadly speaking, the link between psychological and gastrointestinal states has been acknowledged for centuries (Wolf, 1981), though this understanding has not been applied to neurodevelopmental disorders like ASD until recently. In fact, while research from the late 90s and early 2000s explored links between ASD and GI distress, no thorough categorization or treatment of ASD-related GI distress was attempted until 2010 (Buie et al., 2010). This interdisciplinary panel was unable to link a specific GI pathophysiology to individuals with ASD; nonetheless, they agreed that the prevalence of GI abnormalities was not completely understood, GI symptoms are frequently linked with negative behavioral manifestations, and that more research was required before coming to any definitive conclusions on evidence-based treatment recommendations. Current clinical research into the ASD-GI link is still hindered by many of the same obstacles that were identified over a decade ago: inconsistent or varying criteria used to define GI phenotypes, inconsistent or varying methodology (including differences in the reporting and measuring of GI phenotypes), and inconsistent criteria for patient participation and selection. This explains, in part, the wide variation in reported prevalence of ASD related GI symptoms, which ranges from 23% to 70% (Chaidez et al., 2014; McElhanon et al., 2014). Recent work has attempted to address these issues (Bresnahan et al., 2015). In a prospective population-based cohort study with well-defined methodology and participation criteria, Bresnahan et al. (2015) has shown that individuals with ASD are not only more likely to experience GI-related problems when compared to their typically developing counterparts, but that the type of GI distress varies with age. Encompassing a 10-year period, 95,278 mothers (with 114,516 children) from the Norwegian Mother and Child Cohort Study (MoBa) were recruited to participate, with “ongoing follow-up [including] health, behavioral, developmental, and nutritional questionnaires and collection of clinical and biological data” and maternal reports of GI symptoms. Additionally, by simplifying the categories of GI distress to only include constipation, food allergy, and diarrhea, the study focused on easily identifiable symptoms and limited the possibility of over or underreporting. This is a particularly important consideration when dealing with children who have communication deficits, or with non-verbal autistic individuals, irrespective of age. This study represents the first large-scale prospective cohort study on ASD-related GI symptoms that confirms GI distress existing at a higher rate within the ASD population. It also underscores the need for not only more GI-related ASD research, but for unified and consistent approaches to measuring GI distress (Margolis et al., 2019).
In addition to prospective studies, severe GI symptoms have been reported in at least eighteen molecularly identified forms of ASD (Table 1), many of which have corresponding zebrafish and/or rodent models that exhibit reduced intestinal tract motility (Figure 2). Because many of these molecularly identified forms of ASD are rare, caused by sporadic de novo genomic changes, clinical needs of these individuals can often go unmet (SHANK3; Figure 2B). Interestingly, gene expression analyses have shown that many of these ASD-linked genes are expressed in the intestine in both mammals (Sauer et al., 2019) and zebrafish (Lavergne et al., 2020; Wen et al., 2020; Willms et al., 2020) raising the possibility that GI distress is caused by gut-intrinsic mechanisms. Because GI homeostasis is maintained through a concert of influence from the CNS, the microbiome, and the GI tract itself, studies focusing on how the three interact stand to provide the most comprehensive explanations for why GI distress is prevalent within ASD populations.
TABLE 1
| Genetic locus | Foundation: Reported GI distress | Publications reporting GI distress case reportsCR |
| ADNP | ADNP Kids Research Foundation: GERD, reflux, cyclical vomiting, constipation, diarrhea, delayed digestion, stomach ulcers/scarring, IBS | Van Dijck et al. (2019) |
| CDKL5 | International Foundation for CDKL5 Research: Abdominal distension, constipation, diarrhea, reflux, slow gastric emptying, low motility, risk of life-threatening volvulus and intussusception | Amin et al. (2017) |
| CHD8 | SPARK: Gastrointestinal issues | Bernier et al. (2014) |
| CNTNAP2 | Pitt Hopkins Research Foundation: Syndrome 1 constipation | Gregor et al. (2011) |
| Dup Chr 15q | Dup 15 Q Alliance: Feeding issues in infancy, encopresis, acid reflux, some with G-tube | Shaaya et al. (2015) |
| FOXG1 | International FOXG1 Foundation: Constipation | McMahon et al. (2015) |
| FOXP1 | Siper et al., 2017 found that of 9 people with FOXP1 syndrome, 3 had feeding issues and 4 had constipation. | Frohlich et al. (2016, 2019), Siper et al. (2017) |
| KCNQ2 | KCNQ2 Cure Alliance: GI issues seen commonly | Inagaki et al. (2019) |
| MECP2 | Rett syndrome/Rett Syndrome Research Trust: 92% prevalence of GI dysmotility | Motil et al. (2012) |
| NRXN1 | Pitt Hopkins Research Foundation: Syndrome 2 constipation, reflux | Zweier et al. (2009); Harrison et al. (2011) |
| PTEN | PTEN Hamartoma Tumor Syndrome Foundation: Intestinal hamartomatous polyposis | Shaco-Levy et al. (2017) |
| SCN1A | Dravet Syndrome Foundation: Constipation, dysmotility | Villas et al. (2017) |
| SCN2A | Families SCN2A Foundation: Reflux and constipation | Tian et al. (2019) |
| SHANK3 | Phelan McDermid Syndrome Foundation: Constipation, reflux, some with G-tube | De Rubeis et al. (2018) |
| SYNGAP1 | Bridge the Gap SYNGAP1 ERF: Constipation, reflux, links btn GI and aggression, some with G-tube. | Parker et al. (2015); Prchalova et al. (2017) |
| TCF4 | Pitt Hopkins Research Foundation: Gastrointestinal issues | Peippo and Ignatius (2012) |
| TSC 1 and 2 | Tuberous Sclerosis Alliance: Rectal bleeding, papillomas in GI tract, constipation | Moulis et al. (1992) |
| UBE3A | Angelman Syndrome Foundation Inc.: Constipation/possibly due to low truncal tone, reflux/gagging | Williams et al. (2010) |
Molecularly-defined forms of ASD with GI symptoms.
FIGURE 2
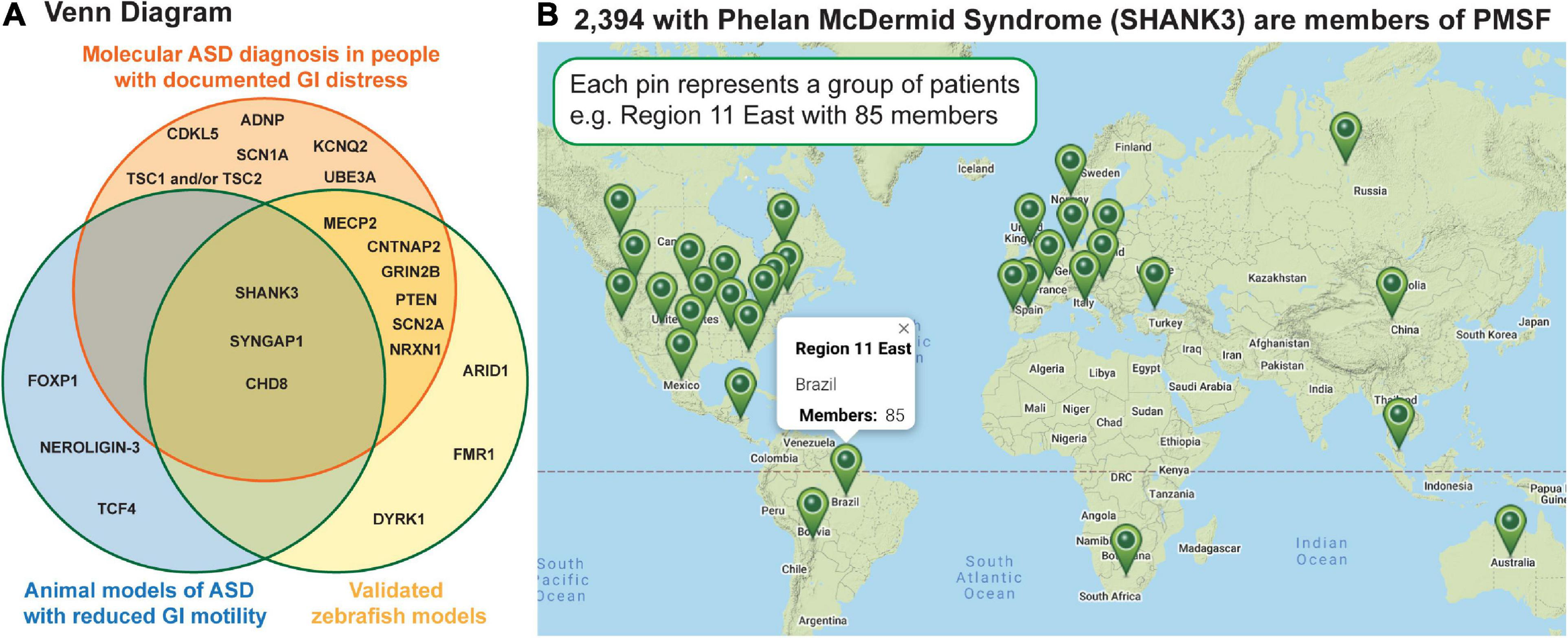
Human genetics has identified 100s of sporadic, de novo genetic changes that can cause ASD; shown are a subset of these that report GI distress as a major symptom. (A) The Venn diagram shows genes linked to GI distress in ASD in the orange circle, those which have extant zebrafish models in the blue circle, and those in which reduced GI motility has been reported in an animal model. (B) The map shows where families caring for individuals with Phelan McDermid Syndrome are scattered across the globe making a standard of care challenging. This Google map image was generated by the Phelan McDermid Syndrome Foundation and is reproduced above with their permission.
The Role of the CNS in the Gut-Brain-Microbiome Axis
Gastrointestinal function is regulated by crosstalk between the nervous system, the gut, and the microbiome (Stengel and Tache, 2010; Drossman and Hasler, 2016; Zhao and Pack, 2017; Ganz, 2018) and, as such, disruptions to this cross talk can contribute not only to GI distress (Bielefeldt et al., 2016) but also to core behaviors used to diagnose ASD (Chaidez et al., 2014; McCue et al., 2017; Penzol et al., 2019). The brain tracks gut luminal contents via sensory enteroendocrine cells (EECs) scattered throughout the gut lining. EECs signal using hormones like serotonin and cholecystokinin (CCK) released into the bloodstream; EECS also use both hormones and fast-acting neurotransmitters to regulate activity of the gut-intrinsic ENS and the gut-extrinsic the parasympathetic vagus and sympathetic dorsal root neurons. These nerves provide a physical, fast-acting conduit that modifies activity across the CNS (Bohorquez et al., 2015; Bellono et al., 2017; Kaelberer and Bohorquez, 2018). Therefore, visceral stimuli influence not only visceral function via gut and brainstem reflexes but also homeostasis, reward, affect, and executive function (Kaelberer et al., 2020). While genes linked to ASD have been extensively studied for their roles in brain and behavior (Kozol et al., 2016), their function along the gut-brain axis has received much less attention. Underscoring the importance of visceral signals, recent studies in zebrafish have been able to accurately predict behavior sequences by integrating environmental stimuli and internal state (Johnson et al., 2020; Marques et al., 2020). Below we discuss the brain regions most relevant to the Gut-Brain axis; we also describe opportunities in zebrafish to better understand the gut-brain axis as it relates to symptoms in ASD.
For studies of the gut-brain axis, visceral sensory and motor pathways in diverse taxa are marked by their expression of the Paired-like homeodomain Phox2b transcription factor (Pattyn et al., 1999; Bertucci and Arendt, 2013; Nomaksteinsky et al., 2013; Harrison et al., 2014). When Phox2b function is disrupted in rodents, motor neurons that would normally innervate the viscera, find muscle targets, indicating that Phox2b functions to make specific CNS nuclei attend to the viscera (D’Autreaux et al., 2011). While this marker is conserved, one pronounced difference between mammals and zebrafish is that zebrafish and their relatives taste with sensory cells on their skin and lips and, as such, the first CNS relay for visceral sensations, the solitary tract nucleus (nTS), is lobed to accommodate expanded vagal, glossopharyngeal and facial inputs; nonetheless these lobes are thought to be functionally homologous to the gustatory portion of the nTS in mammals (Coppola et al., 2012). Work using tract tracing in zebrafish has helped to map the connectivity of largely conserved fish visceral brain circuits (Yanez et al., 2017).
Setting the Stage
Even in advance of eating, sensations of external food stimuli as well as internal hunger or satiation states activate hypothalamic nuclei and play a highly conserved role in setting the stage (Sternson and Eiselt, 2017). Indeed hormonally regulated states of hunger, motivation to eat, satiety are similar in zebrafish and mammals (Jordi et al., 2015), though metabolic differences exist in leptin signaling associated with mammals being endotherms and fish being ectotherms (Gorissen and Flik, 2014). The ability to query the involvement of brain-wide circuits in zebrafish has been used to link behavioral states to brain activity (Randlett et al., 2015; Vanwalleghem et al., 2018). For example, seeing paramecia, a favored food of larval zebrafish, is sufficient to activate neural activity in the hypothalamus (Muto et al., 2017). Moreover, the transition between hunger and satiety can be mapped to activity in the ventromedial hypothalamus and lateral hypothalamus, respectively (Wee et al., 2019). In addition to hypothalamus, brainstem nuclei also respond to appetitive smell and taste in both fish and mammals (Vendrell-Llopis and Yaksi, 2015; Vincis and Fontanini, 2019) and sensorimotor integration during prey pursuit in zebrafish is modulated by feeding state (Filosa et al., 2016; Henriques et al., 2019). Due to the prevalence of eating disorders and sensory symptoms in ASD, an imbalance in sympathetic/parasympathetic tone is one of the hypotheses put forward to potentially explain these symptoms (Fenning et al., 2019). Currently, the physiological basis/es for eating difficulties in individuals with ASD is not well understood and is plagued by heterogeneity both in study design and how symptoms manifest across the spectrum (Margari et al., 2020). Recent studies show that while children with ASD are generally pickier about their food than neurotypical children (Babinska et al., 2020; Li C. et al., 2020), other symptoms may be unique to specific genetic forms of ASD. For example, in people with SYNGAP1 mutations, there is a correlation between eating and seizures (Vlaskamp et al., 2019).
Gut-Brain Connectivity
Innervating the gut, parasympathetic and sympathetic neurons link directly to the CNS and convey information about digestive and microbiome status as well as mechanical and/or chemical insult (Browning and Travagli, 2014; Niu et al., 2020). Vagus and sympathetic nerves have both sensory/afferent and motor/efferent components that carry out visceral reflexes as well as integrating and conveying information to and from widespread brain regions across the CNS.
Most of the recent gut-brain axis literature has focused on the vagus nerve. The cell bodies of the motor component of the vagal neurons reside in dorsal motor nucleus (DMV) in the caudal brainstem. Motor innervation of the viscera is denser in the anterior GI tract (esophagus, stomach, and proximal small intestine) and activity in these organs tends to promote regulation of GI secretions and motility appropriate to the phase of digestion (Tache et al., 2006; Browning et al., 2017). In zebrafish, islet1:GFP transgenics label all the cranial motor neurons including the vagal motor nucleus (Higashijima et al., 2000). The DMV is functionally distributed from rostral to caudal, with the neurons innervating the viscera enriched caudally (Barsh et al., 2017; Isabella et al., 2020). Sensory vagus neuron cell bodies reside outside the CNS in the Nodose ganglion, therefore, it is relatively straight-forward to monitor neuronal activity in these cells to identify salient gut stimuli (Bai et al., 2019; Tsang et al., 2020; Zhang W. et al., 2020) and this approach that has recently been used in zebrafish (Ye et al., 2020). Work in rodents supports a critical role for an intact vagus nerve in the ability of L. reuteri bacteria to rescue social deficits in a Shank3 ASD mouse model (Sgritta et al., 2019).
Sympathetic pre-ganglionic neurons when active during stress generally inhibit GI motility and secretion and also causes vasoconstriction that limits the blood supply to the viscera (Browning et al., 2017). Sensory sympathetic spinal afferents whose cell bodies reside in the dorsal root ganglia (DRG) also innervate the gut with denser innervation caudally (Muller et al., 2020). Sympathetic neurons are sensitive to digestion, injury, and microbes. While sympathetic innervation as it relates to digestion has not to our knowledge been studied in zebrafish, the DRG is accessible to electrophysiological recordings (Won et al., 2012) and both isl2b and ngn enhancers can drive expression of calcium sensors/light-gated channels in this cell type (Wright et al., 2010; Stil and Drapeau, 2016; Hall and Tropepe, 2018). Using these tools in zebrafish larvae could help elucidate GI stimuli and insults that activate sympathetic spinal afferents in vivo and how this activity is impacted zebrafish ASD models.
The brainstem/medulla oblongata is rich in nuclei that receive, process, and respond to sensory information from the GI tract. Vagal inputs directly innervate the Area Postrema (AP) and the Nucleus of the Solitary Tract (nTS) (Ma, 1997; Kaslin and Panula, 2001; McLean and Fetcho, 2004; Coppola et al., 2012). The AP is one of the few areas of the CNS that is not protected by the blood brain barrier and as such is responsive to factors/toxins in the bloodstream; neuronal activity in the AP is linked to the symptom of nausea (Zhang C. et al., 2020). Consistent with functions established in mammals, the zebrafish AP has been shown to be responsive to the pain-inducing Trp1A agonist AITC (Haney et al., 2020). The nTS serves as the first CNS relay to many other brain regions (Coppola et al., 2012; Yanez et al., 2017; Han et al., 2018) including the secondary gustatory nucleus aka parabrachial nucleus (PBN) as well as the DMV. Both nTS and PBN are marked by Phox2b expression with transgenic drivers available (Nechiporuk et al., 2007; Coppola et al., 2012). The zebrafish nTS is dorsal and sheetlike and, as such, is amenable to in vivo imaging studies (Vendrell-Llopis and Yaksi, 2015). Taste and visceral inputs map to different regions of the nTS in mammals (Vincis and Fontanini, 2019; Kaelberer et al., 2020) and in fishes, including zebrafish, visceral inputs map to the caudal part of the nTS in the adult brain (Kermen et al., 2013; Yanez et al., 2017). As a nucleus that integrates direct inputs from both viscera and CNS, the nTS holds promise for elucidating what aspects of the gut-microbiome-brain signaling may be altered in zebrafish ASD models.
Gut Feelings
In addition to visceral reflexes mediated at the level of hypothalamus and brainstem, widespread CNS nuclei mediating memory, emotion/affect, and motivation have been shown in rodents to also be responsive to gut stimuli (Kaelberer et al., 2020). Severing vagal afferents results in increased exploratory behaviors and risk-taking, heightened auditory-based fear conditioning, and altered neurotransmitters in the limbic system (Klarer et al., 2014, 2018). Stimulation of vagal afferents entering the brainstem on the right side engage a PBN to nucleus accumbens to dorsal striatum reward pathway and stimulating this pathway sufficient to elicit behaviors consistent with reward (Han et al., 2018). Another pathway activated through the vagus is the nTS to medial septum to hippocampus that when disrupted interferes with spatial memory (Suarez et al., 2018). Analogous zebrafish brain regions to those mediating memory, emotion/affect, and motivation in mammals are continuing to be elucidated in zebrafish, and anatomical studies indicate similar connectivity between visceral circuits and these brain regions in zebrafish (Yanez et al., 2017).
Elucidating the link between GI distress and negative behavioral symptoms could improve symptom management. Not only are GI symptoms common in ASD, but they correlate with more pronounced irritability, social withdrawal, stereotypy, hyperactivity, and sleep disturbances (De Rubeis and Buxbaum, 2015; McCue et al., 2017; Penzol et al., 2019). Such an intimate link between gut and brain symptoms is well-established in Parkinson’s disease where constipation often precedes the motor disturbances (Fasano et al., 2015; Mayer et al., 2015; Liddle, 2018; Ramprasad et al., 2018). The possibility that gut symptoms could contribute to the development of neurological symptoms has also been suggested in ASD (Mayer et al., 2014; Eshraghi et al., 2018) with recent studies providing empirical support (Sgritta et al., 2019).
Zebrafish as a Model for Microbial Studies and Their Potential Role in ASD
Humans are extensively colonized with microbial species, resulting in several distinct microbiomes determined by geographic distribution across the host’s body (Human Microbiome Project Consortium, 2012). Interactions between host and microbiome are complex, and our understanding of the bi-directional influence between the two is evolving as the scientific community adopts a less human-centric view (reviewed in Wiles and Guillemin, 2020). Nonetheless, the microbiome has long been understood to play an important role in host form and function. While the GI microbiome was traditionally thought to predominately interact with its host through nutrient processing, further study has shown that it is capable of influencing the host immune system and nervous system as well, and thus has direct implications in disorders like ASD.
The composition of the human GI microbiome is largely determined by functional, rather than taxonomic, qualification. The gut microbiome does not need any particular species profile to operate and maintain a commensal relationship with the host. Rather, it requires certain functions to be performed, which can be executed by any number of potential microbial species. In humans, the microbiome is composed of predominantly Firmicutes and Bacteroidetes species (Human Microbiome Project Consortium, 2012). Much like an ecosystem of macroorganisms, the human GI microbiome is influenced by its physical habitat (gut morphology), resource availability (host diet), and interspecies interaction (both between microorganisms and between the microbiome and host itself). Likewise, both the development and the function of the vertebrate immune and nervous system are affected by the GI microbiome. The gut microbiome is capable of exerting an effect on neurological functioning through several pathways, with metabolites able to travel through the host bloodstream or act locally upon the vagus (Schroeder and Backhed, 2016; Fulling et al., 2019). Immune responses elicited by microbes or their metabolites can also have implications on the brain and its function.
The GI microbiome also plays an important role in the development of the host immune system. Early-colonizing microbial species provide “training” to immune effectors, allowing them to become accustomed to commensal communities and to distinguish between them and pathogenic microorganisms. Severe negative consequences can occur when this process is interrupted or prevented. In germ-free mice, colonic epithelial cells are incapable of raising an immune response upon exposure to a bacterial pathogen (Lundin et al., 2008). Similar dysfunction can be seen in germ-free zebrafish, which display impaired differentiation of GI cell types such as goblet and enteroendocrine cells along with impaired nutrient uptake and death prior to adulthood if not conventionalized (Bates et al., 2006; Melancon et al., 2017). Human infants that avoid exposure to maternal microbiomes through cesarean delivery and/or formula feeding display increased inflammatory responses and autoimmune disorders (Toscano et al., 2017; Koch et al., 2018). As the microbiome shapes the immune system, the innate immune system of the host in turn shapes the native microbiome to one tailored to the individual’s metabolic needs (Thaiss et al., 2016).
The microbiome has been shown to be capable of affecting or inducing multiple aberrant neurological phenotypes in various study systems. Some of these alterations have been traced to bacterial metabolites such as short chain fatty acids (SCFAs). Elevated levels of SCFAs have been directly detected in fecal samples from autistic patients (Wang et al., 2012). Although it was undetermined if those levels were mediated by the gut microbiome, other studies have likewise shown increased numbers of the Clostridia family (enterobacteria that are key producers of various SCFAs) in stool samples from individuals with autism. Studies in rats have recapitulated a behavioral phenotype resembling that of autistic patients by treatment with propionic acid, a short-chain fatty acid produced by Clostridia (MacFabe et al., 2007). Similar associations have been found in models of Parkinson’s disease, where bacterial SCFAs were sufficient to promote neuroinflammation in the mouse subjects (Sampson et al., 2016). The microbiome has also been shown to influence neurological conditions through modulation of the immune system (Benakis et al., 2016; Schroeder and Backhed, 2016). Microbial attenuation of inflammatory cytokines has been linked with reductions in anxiety (Cryan and O’Mahony, 2011) and antibiotic treatment in a stroke mouse model has been shown to confer neuroprotection post-ischemic injury through a reduction in intestinal immune effectors (Benakis et al., 2016). In addition to influencing immune activity, microbes in the gut are capable of altering hormone signaling as well. Spore-forming bacterial species in the gut have been shown to induce serotonin production by enterochromaffin cells through the release of several metabolites. This promotion of serotonin production was found to ameliorate the reduced GI motility seen in germ-free mice when the subjects were colonized with the spore-forming species (Yano et al., 2015).
Zebrafish are powerful model organisms for experiments involving microbiome contribution. As they are initially colonized by microbes via their environment, it is possible to raise them in sterilized conditions that result in germ-free individuals (Rawls et al., 2004). Once germ-free subjects are generated, experiments involving selective colonization, introduction of metabolites, and conventionalization effects are possible. The zebrafish gut is able to be imaged in vivo in the early life stages due to the optical transparency of larvae. This allows for examination of gut function like motility, and barrier function (Marjoram et al., 2015), as well as location and interspecies dynamics of fluorescently labeled bacterial species (Wiles and Guillemin, 2020). The gut is also easily dissectible, allowing for extraction of the microbiome for 16S sequencing.
Zebrafish as a Model for GI Form and Function, and Modeling ASD-Related GI Dysfunction
The use of zebrafish models for GI research has increased considerably over the last three decades (Figure 1), with publications exploring conserved and unique aspects of GI form and function, as well as more nuanced topics including specific disease models (reviewed in Zhao and Pack, 2017) and external impacts like microplastic exposure (Qiao et al., 2019) and chemotherapy treatment (Van Sebille et al., 2019). Like the animal model as a whole, the zebrafish GI system has many physiological similarities and differences with its mammalian model counterparts. There is conservation of key GI cell types in the zebrafish GI tract, with absorptive enterocytes, mucus producing goblet cells, chemo/mechano-sensitive enteroendocrine cells, and the innervation by the ENS (Roy-Carson et al., 2017) filling similar functional niches. The digestive tract and its accessory organs similarly form from a strip of endodermal tissue in early development (as early as 21 hpf in zebrafish) (Wallace and Pack, 2003). While there is some debate as to whether these organs develop individually or from the same interconnected endodermal strip (Ng et al., 2005), the differential expression of conserved genes key to GI development begins as early as 18hpf. Expression of these orthologs, such as pharyngeal endoderm associated gene axial, liver development gene hhex, pancreas development gene pdx, and esophageal gene gata-6, suggest that GI development in zebrafish differs slightly from mammals, with progenitors for the liver, pancreas, and pharynx existing before morphogenesis of the GI tract itself is complete. Similarly, small differences exist for the roles of morphogens like sonic-hedgehog (shh); required as a negative regulator for pancreatic development in mammals, in zebrafish it appears to be a positive regulator (Wallace and Pack, 2003). Like in mammals, the zebrafish GI tract has distinct functional layers, with an epithelial mucosal layer, and an underlying muscular layer innervated by the ENS. Unlike mammals, however, zebrafish lack a submucosal layer, villi are replaced by broad folds in the mucosal layer, the ENS is not organized into ganglia (acting instead as a nerve net), and the proliferative crypts of Lieberkuhn are absent (though proliferating cells still expand from stem cell niches at the base of the mucosal folds) (Wallace and Pack, 2003; Ng et al., 2005; Wallace et al., 2005; Uyttebroek et al., 2010). From a functional perspective, the zebrafish also has other simplifications when compared to mammalian models; they lack a true stomach (separated by sphincters and containing acid-producing Paneth cells) and instead have an intestinal bulb. This bulb likely acts as a reservoir for food, and motility patterns in this region are both anterograde and retrograde, acting to mix and break food down mechanically (Holmberg et al., 2004). These and other differences point to an overall simplification of the GI tract in zebrafish. Although components of the GI tract (including development, cell types, and molecular signaling) are conserved between zebrafish and mammals, there are important differences that need to be acknowledged when using zebrafish as a model for GI research.
From a GI-research prospective, zebrafish offer a key advantage over mammalian models: their external fertilization, development, and early transparency make studying GI function in vivo significantly easier. Measurements of digestive transit, peristaltic rate, and general ontogeny of GI motility can be made before the larvae begin feeding (with spontaneous motility developing before 5 dpf) (Holmberg et al., 2004), and do not require any complex or potentially variable-confounding surgical procedures. The measurement of motility in zebrafish also has its disadvantages; since this is a relatively new branch of zebrafish research, the approaches have not been standardized, with multiple labs (including our own) creating their own software for measurements of motility and transit (Field et al., 2009; Rich, 2009; Jordi et al., 2015; James et al., 2019). Although these different models share similar components and aims, the differences could present possible complications when comparing results. For instance, in our own model, while measuring transit and motility are relatively straightforward, determining the force of muscular contraction is difficult, and potentially confounding variables (such as food particles in the GI tract) mean that measuring GI motility specifically (and excluding the movement of particulates) becomes difficult. Additionally, as most motility software was developed for in-house use, the user-interface and technical aspects of each model present possible speedbumps for researchers unfamiliar with the software. The field, as a whole, would benefit from a unified method for GI motility measurement, especially when attempting to tackle questions on ASD-related GI distress.
While there is increasing interest in zebrafish-based GI research, and similarly, increasing interest in the relationship between ASD and GI dysfunction in non-zebrafish models (Figure 1), there has not been a coupling of the two. In fact, to our knowledge and excluding our own work, there have only been three publications that include data on ASD-related GI symptoms using zebrafish models, one of which is technically not ASD-specific (focusing on CHARGE syndrome, which has overlap with ASD but is distinct) (Bernier et al., 2014; van der Vaart et al., 2017; Cloney et al., 2018). This represents, in our view, a significant shortcoming that should be addressed in future studies. Previous work from our lab (James et al., 2019) attempts to address this shortcoming, and utilizes some of the aforementioned zebrafish GI assays to look at GI dysfunction in a monogenic ASD model. In this work, we focus on GI dysmotility found in CRISPR mutant of ASD related gene shank3ab. We found that while there was no difference in enteric neuron count, there was a significant decrease in the expression of serotonin-positive enteroendocrine cells in shank3ab mutants. We also note that RNA-seq data from a collaborating lab has found that shank3ab expression is detected in this population of cells (Wen et al., 2020). Interestingly, recent work in mouse models has found that enteroendocrine cells (specifically serotonin producing enterochromaffin cells) synapse directly with the ENS (Bellono et al., 2017). Taken together, this suggests that changes in GI physiology can have profound impacts on GI-CNS communication, and that there may be a non-CNS role for genes like shank3ab, which are largely understood and studied through their context in the CNS. To add further complexity to the picture, we also found increased goblet cell populations when comparing adult WT and shank3ab mutant fish, a difference not present in larval fish and indicative of possible age-related GI inflammation; we are currently exploring the microbial implications behind this finding. This work, coupled with the previously mentioned ASD-GI publications presents possible venues for establishing ASD-related GI pathophysiology, and it sets a foundation for future studies.
Discussion: Drawing Conclusion and Looking Forward
In this review, we have discussed some of the important research in the gut-brain-microbiome field, as well as presented the zebrafish as an ideal model for future multidisciplinary ASD studies. Moving forward, we hope research in zebrafish models increasingly integrates behavioral phenotypes and comorbidities of sleep, seizure, and GI function to obtain a more complete understanding of how genetic changes that can cause ASD impact the organism as a whole. To this end, there have been recent advances in the development of social assays that have helped characterize the neural circuits involved in social behavior (Dreosti et al., 2015; Stednitz et al., 2018). Additionally, adult social behaviors in a zebrafish ASD model have reduced shoaling behavior (Liu et al., 2018). While the bulk of current zebrafish ASD models are caused by indels that result in premature stop codons, recently developed CRISPR/Cas9 technologies in zebrafish are making tissue-specific mutagenesis and knockins that replicate patient-specific mis-sense mutations more broadly feasible (Albadri et al., 2017; Li J. et al., 2020). These technological advances continue to make the zebrafish an increasingly valuable model for ASD research.
It is also important to briefly highlight the role zebrafish are playing in high-throughput drug screening and drug discovery for treating ASD comorbidities. The zebrafish is becoming a well-established model for drug screening (Haesemeyer and Schier, 2015; Hoffman et al., 2016; Cassar et al., 2020), and is a particularly powerful model for high-throughput screens aimed at tailoring integrative care on a patient-symptom basis. The broad heterogeneity within ASD warrants an equally broad scope of research on different palliative drugs. It should be noted, however, that this heterogeneity also complicates the treatment of comorbidities along with the core behavioral phenotypes associated with ASD. Many current drugs are aimed at treating behavioral problems such as irritability and aggression (Stachnik and Gabay, 2010; Coleman et al., 2019), and in doing so frequently overlook downstream side-effects that might also be contributing to the initial behavioral issue. Risperidone, for example, is often used to treat problematic behavioral issues associated with ASD by inhibiting dopaminergic D2 receptors and serotonergic 5-HT2A receptors and can lead to constipation in human patients, which can in turn lead to a worsening of non-verbal behaviors such as agitation and anxiety. Similarly, common GI medications used to treat delays in gut transit (such as Metoclopramide) can lead to changes in behavior in zebrafish due to a compensatory mechanism following weakened dopamine signaling (Shontz et al., 2018). To further complicate the issue, the relationship between the GI microbiome and drug application is not well understood. As such, we believe the zebrafish also serves as an ideal model for ASD drug screening, not only for behavioral impacts, but for GI and microbiome impacts as well (Cassar et al., 2015). As their power in translational ASD research is already well established (Ijaz and Hoffman, 2016; Sakai et al., 2018), zebrafish could serve as an intersection between patient care and foundational exploratory research, with pre-clinical trials of newly discovered drugs helping to inform which treatments have the best benefit for multiple symptoms.
Autism spectrum disorder research has largely been compartmentalized, whether into behavioral, molecular, or microbial aspects. Given the interconnected regulatory pathways and feedback loops existing between the CNS, GI tract, and microbiome, we believe this compartmentalization acts to the detriment of a broader understanding of the disorder. Consequently, if the current lack of ASD-related GI research is filled, it could stand to provide critical components to both CNS and microbiome research, while also producing important foundational information on potential ASD-related GI pathophysiology.
Statements
Author contributions
All authors contributed to the research, writing, and editing of this review. DJ and JD conceived the scope of the review, coordinated efforts among authors, and wrote the gastrointestinal and CNS sections. ED wrote the bulk of the microbiome section. JY wrote the bulk of the drug screening section. BM aided in research and references to clinical studies.
Funding
This work was supported by grants from the National Institutes of Health, NICHD R21HD093021 to JD and BM, and from the Simon’s Foundation, SFARI Pilot Award 719401 to JD.
Acknowledgments
We would like to thank members (clinicians, researchers, and family members) of both the Phelan McDermid Syndrome Foundation and the Bridge the Gap SYNGAP Education and Research Foundation for their enthusiasm and support of our ongoing ASD-GI Research.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1
Albadri S. De Santis F. Di Donato V. Del Bene F. (2017). “CRISPR/Cas9-Mediated knockin and knockout in zebrafish,” in Genome Editing in Neurosciences, edsJaenischR.ZhangF.GageF. (Cham: Springer), 41–49.
2
Amin S. Majumdar A. Mallick A. A. Patel J. Scatchard R. Partridge C. A. et al (2017). Caregiver’s perception of epilepsy treatment, quality of life and comorbidities in an international cohort of CDKL5 patients.Hippokratia21130–135.
3
Babinska K. Celusakova H. Belica I. Szapuova Z. Waczulikova I. Nemcsicsova D. et al (2020). Gastrointestinal symptoms and feeding problems and their associations with dietary interventions, food supplement use, and behavioral characteristics in a sample of children and adolescents with autism spectrum disorders.Int. J. Environ. Res. Public Health17:6372. 10.3390/ijerph17176372
4
Bai L. Mesgarzadeh S. Ramesh K. S. Huey E. L. Liu Y. Gray L. A. et al (2019). Genetic identification of vagal sensory neurons that control feeding.Cell1791129.e23–1143.e23. 10.1016/j.cell.2019.10.031
5
Barsh G. R. Isabella A. J. Moens C. B. (2017). Vagus motor neuron topographic map determined by parallel mechanisms of hox5 expression and time of axon initiation.Curr. Biol.273812.e3–3825.e3. 10.1016/j.cub.2017.11.022
6
Bates J. M. Mittge E. Kuhlman J. Baden K. N. Cheesman S. E. Guillemin K. (2006). Distinct signals from the microbiota promote different aspects of zebrafish gut differentiation.Dev. Biol.297374–386. 10.1016/j.ydbio.2006.05.006
7
Bellono N. W. Bayrer J. R. Leitch D. B. Castro J. Zhang C. O’Donnell T. A. et al (2017). Enterochromaffin cells are gut chemosensors that couple to sensory neural pathways.Cell170185.e16–198.e16. 10.1016/j.cell.2017.05.034
8
Benakis C. Brea D. Caballero S. Faraco G. Moore J. Murphy M. et al (2016). Commensal microbiota affects ischemic stroke outcome by regulating intestinal gammadelta T cells.Nat. Med.22516–523. 10.1038/nm.4068
9
Bernier R. Golzio C. Xiong B. Stessman H. A. Coe B. P. Penn O. et al (2014). Disruptive CHD8 mutations define a subtype of autism early in development.Cell158263–276. 10.1016/j.cell.2014.06.017
10
Bertucci P. Arendt D. (2013). Somatic and visceral nervous systems - an ancient duality.BMC Biol.11:54. 10.1186/1741-7007-11-54
11
Bielefeldt K. Tuteja A. Nusrat S. (2016). Disorders of gastrointestinal hypomotility.F1000Res.5:F1000FacultyRev-1897. 10.12688/f1000research.8658.1
12
Bohorquez D. V. Shahid R. A. Erdmann A. Kreger A. M. Wang Y. Calakos N. et al (2015). Neuroepithelial circuit formed by innervation of sensory enteroendocrine cells.J. Clin. Invest.125782–786. 10.1172/JCI78361
13
Bresnahan M. Hornig M. Schultz A. F. Gunnes N. Hirtz D. Lie K. K. et al (2015). Association of maternal report of infant and toddler gastrointestinal symptoms with autism: evidence from a prospective birth cohort.JAMA Psychiatry72466–474. 10.1001/jamapsychiatry.2014.3034
14
Browning K. N. Travagli R. A. (2014). Central nervous system control of gastrointestinal motility and secretion and modulation of gastrointestinal functions.Compr. Physiol.41339–1368. 10.1002/cphy.c130055
15
Browning K. N. Verheijden S. Boeckxstaens G. E. (2017). The vagus nerve in appetite regulation, mood, and intestinal inflammation.Gastroenterology152730–744. 10.1053/j.gastro.2016.10.046
16
Brudek T. (2019). Inflammatory bowel diseases and Parkinson’s Disease.J. Parkinsons Dis.9(Suppl. 2), S331–S344. 10.3233/JPD-191729
17
Brugman S. (2016). The zebrafish as a model to study intestinal inflammation.Dev. Comp. Immunol.6482–92. 10.1016/j.dci.2016.02.020
18
Buie T. Campbell D. B. Fuchs G. J. III Furuta G. T. Levy J. Vandewater J. et al (2010). Evaluation, diagnosis, and treatment of gastrointestinal disorders in individuals with ASDs: a consensus report.Pediatrics125(Suppl. 1), S1–S18. 10.1542/peds.2009-1878C
19
Cassar S. Adatto I. Freeman J. L. Gamse J. T. Iturria I. Lawrence C. et al (2020). Use of zebrafish in drug discovery toxicology.Chem. Res. Toxicol.3395–118. 10.1021/acs.chemrestox.9b00335
20
Cassar S. Huang X. Cole T. (2015). A high-throughput method for predicting drug effects on gut transit time using larval zebrafish.J. Pharmacol. Toxicol. Methods7672–75. 10.1016/j.vascn.2015.08.156
21
Chaidez V. Hansen R. L. Hertz-Picciotto I. (2014). Gastrointestinal problems in children with autism, developmental delays or typical development.J. Autism Dev. Disord.441117–1127. 10.1007/s10803-013-1973-x
22
Chaste P. Leboyer M. (2012). Autism risk factors: genes, environment, and gene-environment interactions.Dialogues Clin. Neurosci.14281–292.
23
Christensen D. L. Braun K. V. N. Baio J. Bilder D. Charles J. Constantino J. N. et al (2018). Prevalence and characteristics of autism spectrum disorder among children aged 8 years - autism and developmental disabilities monitoring network, 11 Sites. United States, 2012.MMWR Surveill. Summ.651–23. 10.15585/mmwr.ss6513a1
24
Cloney K. Steele S. L. Stoyek M. R. Croll R. P. Smith F. M. Prykhozhij S. V. et al (2018). Etiology and functional validation of gastrointestinal motility dysfunction in a zebrafish model of CHARGE syndrome.FEBS J.2852125–2140. 10.1111/febs.14473
25
Coleman D. M. Adams J. B. Anderson A. L. Frye R. E. (2019). Rating of the effectiveness of 26 psychiatric and seizure medications for autism spectrum disorder: results of a national survey.J. Child Adolesc. Psychopharmacol.29107–123. 10.1089/cap.2018.0121
26
Coppola E. D’Autreaux F. Nomaksteinsky M. Brunet J. F. (2012). Phox2b expression in the taste centers of fish.J. Comp. Neurol.5203633–3649. 10.1002/cne.23117
27
Cryan J. F. O’Mahony S. M. (2011). The microbiome-gut-brain axis: from bowel to behavior.Neurogastroenterol. Motil.23187–192. 10.1111/j.1365-2982.2010.01664.x
28
D’Autreaux F. Coppola E. Hirsch M. R. Birchmeier C. Brunet J. F. (2011). Homeoprotein Phox2b commands a somatic-to-visceral switch in cranial sensory pathways.Proc. Natl. Acad. Sci. U.S.A.10820018–20023. 10.1073/pnas.1110416108
29
de Alvarenga K. A. F. Sacramento E. K. Rosa D. V. Souza B. R. de Rezende V. B. Romano-Silva M. A. (2017). Effects of antipsychotics on intestinal motility in zebrafish larvae.Neurogastroenterol. Motil.29:e13006. 10.1111/nmo.13006
30
De Rubeis S. Buxbaum J. D. (2015). Recent advances in the genetics of autism spectrum disorder.Curr. Neurol. Neurosci. Rep.15:36. 10.1007/s11910-015-0553-1
31
De Rubeis S. Siper P. M. Durkin A. Weissman J. Muratet F. Halpern D. et al (2018). Delineation of the genetic and clinical spectrum of Phelan-McDermid syndrome caused by SHANK3 point mutations.Mol. Autism9:31. 10.1186/s13229-018-0205-9
32
Dickerson F. Severance E. Yolken R. (2017). The microbiome, immunity, and schizophrenia and bipolar disorder.Brain Behav. Immun.6246–52. 10.1016/j.bbi.2016.12.010
33
Dreosti E. Lopes G. Kampff A. R. Wilson S. W. (2015). Development of social behavior in young zebrafish.Front. Neural Circuits9:39. 10.3389/fncir.2015.00039
34
Drossman D. A. Hasler W. L. (2016). Rome IV-Functional GI disorders: disorders of gut-brain interaction.Gastroenterology1501257–1261. 10.1053/j.gastro.2016.03.035
35
Eshraghi R. S. Deth R. C. Mittal R. Aranke M. Kay S. S. Moshiree B. et al (2018). Early disruption of the microbiome leading to decreased antioxidant capacity and epigenetic changes: implications for the rise in autism.Front. Cell Neurosci.12:256. 10.3389/fncel.2018.00256
36
Fasano A. Visanji N. P. Liu L. W. Lang A. E. Pfeiffer R. F. (2015). Gastrointestinal dysfunction in Parkinson’s disease.Lancet Neurol.14625–639. 10.1016/S1474-4422(15)00007-1
37
Fenning R. M. Erath S. A. Baker J. K. Messinger D. S. Moffitt J. Baucom B. R. et al (2019). Sympathetic-parasympathetic interaction and externalizing problems in children with autism spectrum disorder.Autism Res.121805–1816. 10.1002/aur.2187
38
Field H. A. Kelley K. A. Martell L. Goldstein A. M. Serluca F. C. (2009). Analysis of gastrointestinal physiology using a novel intestinal transit assay in zebrafish.Neurogastroenterol. Motil.21304–312. 10.1111/j.1365-2982.2008.01234.x
39
Filosa A. Barker A. J. Dal Maschio M. Baier H. (2016). Feeding state modulates behavioral choice and processing of prey stimuli in the zebrafish tectum.Neuron90596–608. 10.1016/j.neuron.2016.03.014
40
Frohlich E. E. Farzi A. Mayerhofer R. Reichmann F. Jacan A. Wagner B. et al (2016). Cognitive impairment by antibiotic-induced gut dysbiosis: analysis of gut microbiota-brain communication.Brain Behav. Immun.56140–155. 10.1016/j.bbi.2016.02.020
41
Frohlich H. Kollmeyer M. L. Linz V. C. Stuhlinger M. Groneberg D. Reigl A. et al (2019). Gastrointestinal dysfunction in autism displayed by altered motility and achalasia in Foxp1 (+/-) mice.Proc. Natl. Acad. Sci. U.S.A.11622237–22245. 10.1073/pnas.1911429116
42
Frye R. E. Rose S. Slattery J. MacFabe D. F. (2015). Gastrointestinal dysfunction in autism spectrum disorder: the role of the mitochondria and the enteric microbiome.Microb. Ecol. Health Dis.26:27458. 10.3402/mehd.v26.27458
43
Fulling C. Dinan T. G. Cryan J. F. (2019). Gut microbe to brain signaling: what happens in vagus.Neuron101998–1002. 10.1016/j.neuron.2019.02.008
44
Ganz J. (2018). Gut feelings: studying enteric nervous system development, function, and disease in the zebrafish model system.Dev. Dyn.247268–278. 10.1002/dvdy.24597
45
Geschwind D. H. (2009). Advances in autism.Annu. Rev. Med.60367–380. 10.1146/annurev.med.60.053107.121225
46
Goodspeed K. Bliss G. Linnehan D. (2020). Bringing everyone to the table - findings from the 2018 phelan-mcdermid syndrome foundation international conference.Orphanet. J. Rare Dis.15:152. 10.1186/s13023-020-01389-6
47
Gorissen M. Flik G. (2014). Leptin in teleostean fish, towards the origins of leptin physiology.J. Chem. Neuroanat.61-62200–206. 10.1016/j.jchemneu.2014.06.005
48
Goyal D. Ali S. A. Singh R. K. (2020). Emerging role of gut microbiota in modulation of neuroinflammation and neurodegeneration with emphasis on Alzheimer’s disease.Prog. Neuropsychopharmacol. Biol. Psychiatry106:110112. 10.1016/j.pnpbp.2020.110112
49
Gregor A. Albrecht B. Bader I. Bijlsma E. K. Ekici A. B. Engels H. et al (2011). Expanding the clinical spectrum associated with defects in CNTNAP2 and NRXN1.BMC Med. Genet.12:106. 10.1186/1471-2350-12-106
50
Grundy D. Al-Chaer E. D. Aziz Q. Collins S. M. Ke M. Tache Y. et al (2006). Fundamentals of neurogastroenterology: basic science.Gastroenterology1301391–1411. 10.1053/j.gastro.2005.11.060
51
Haesemeyer M. Schier A. F. (2015). The study of psychiatric disease genes and drugs in zebrafish.Curr. Opin. Neurobiol.30122–130. 10.1016/j.conb.2014.12.002
52
Hall Z. J. Tropepe V. (2018). Movement maintains forebrain neurogenesis via peripheral neural feedback in larval zebrafish.eLife7:e31045. 10.7554/eLife.31045
53
Han W. Tellez L. A. Perkins M. H. Perez I. O. Qu T. Ferreira J. et al (2018). A neural circuit for gut-induced reward.Cell175887–888. 10.1016/j.cell.2018.10.018
54
Haney W. A. Moussaoui B. Strother J. A. (2020). Prolonged exposure to stressors suppresses exploratory behavior in zebrafish larvae.J. Exp. Biol.223(Pt 22):jeb224964. 10.1242/jeb.224964
55
Harrison C. Wabbersen T. Shepherd I. T. (2014). In vivo visualization of the development of the enteric nervous system using a Tg(-8.3bphox2b:Kaede) transgenic zebrafish.Genesis52985–990. 10.1002/dvg.22826
56
Harrison V. Connell L. Hayesmoore J. McParland J. Pike M. G. Blair E. (2011). Compound heterozygous deletion of NRXN1 causing severe developmental delay with early onset epilepsy in two sisters.Am. J. Med. Genet. A155A2826–2831. 10.1002/ajmg.a.34255
57
He Y. Li B. Sun D. Chen S. (2020). Gut microbiota: implications in Alzheimer’s Disease.J. Clin. Med.9:2042. 10.3390/jcm9072042
58
Henriques P. M. Rahman N. Jackson S. E. Bianco I. H. (2019). Nucleus isthmi is required to sustain target pursuit during visually guided prey-catching.Curr. Biol.291771.e5–1786.e5. 10.1016/j.cub.2019.04.064
59
Higashijima S. Hotta Y. Okamoto H. (2000). Visualization of cranial motor neurons in live transgenic zebrafish expressing green fluorescent protein under the control of the islet-1 promoter/enhancer.J. Neurosci.20206–218.
60
Hill J. M. Bhattacharjee S. Pogue A. I. Lukiw W. J. (2014). The gastrointestinal tract microbiome and potential link to Alzheimer’s disease.Front. Neurol.5:43. 10.3389/fneur.2014.00043
61
Hoffman E. J. Turner K. J. Fernandez J. M. Cifuentes D. Ghosh M. Ijaz S. et al (2016). Estrogens suppress a behavioral phenotype in zebrafish mutants of the autism risk gene. CNTNAP2. Neuron89725–733. 10.1016/j.neuron.2015.12.039
62
Holmberg A. Schwerte T. Pelster B. Holmgren S. (2004). Ontogeny of the gut motility control system in zebrafish Danio rerio embryos and larvae.J. Exp. Biol.207(Pt 23), 4085–4094. 10.1242/jeb.01260
63
Holtmann G. Talley N. J. (2014). The stomach-brain axis.Best Pract. Res. Clin. Gastroenterol.28967–979. 10.1016/j.bpg.2014.10.001
64
Hsiao E. Y. (2014). Gastrointestinal issues in autism spectrum disorder.Harv. Rev. Psychiatry22104–111. 10.1097/HRP.0000000000000029
65
Human Microbiome Project Consortium (2012). A framework for human microbiome research.Nature486215–221. 10.1038/nature11209
66
IACC (2019). IACC Strategic Plan For Autism Spectrum Disorder 2018-2019 Update. Avaliable at: https://iacc.hhs.gov/publications/strategic-plan/2019/(accessed February 8, 2021).
67
Ijaz S. Hoffman E. J. (2016). Zebrafish: a translational model system for studying neuropsychiatric disorders.J. Am. Acad. Child Adolesc. Psychiatry55746–748. 10.1016/j.jaac.2016.06.008
68
Inagaki A. Hayashi M. Andharia N. Matsuda H. (2019). Involvement of butyrate in electrogenic K(+) secretion in rat rectal colon.Pflugers Arch.471313–327. 10.1007/s00424-018-2208-y
69
Isabella A. J. Barsh G. R. Stonick J. A. Dubrulle J. Moens C. B. (2020). Retinoic acid organizes the zebrafish vagus motor topographic map via spatiotemporal coordination of Hgf/Met Signaling.Dev. Cell53344.e5–357.e5. 10.1016/j.devcel.2020.03.017
70
James D. M. Kozol R. A. Kajiwara Y. Wahl A. L. Storrs E. C. Buxbaum J. D. et al (2019). Intestinal dysmotility in a zebrafish (Danio rerio) shank3a;shank3b mutant model of autism.Mol. Autism10:3. 10.1186/s13229-018-0250-4
71
Jiang C. Li G. Huang P. Liu Z. Zhao B. (2017). The gut microbiota and Alzheimer’s Disease.J. Alzheimers Dis.581–15. 10.3233/JAD-161141
72
Johnson R. E. Linderman S. Panier T. Wee C. L. Song E. Herrera K. J. et al (2020). Probabilistic models of larval zebrafish behavior reveal structure on many scales.Curr. Biol.3070.e4–82.e4. 10.1016/j.cub.2019.11.026
73
Jordi J. Guggiana-Nilo D. Soucy E. Song E. Y. Lei Wee C. Engert F. (2015). A high-throughput assay for quantifying appetite and digestive dynamics.Am. J. Physiol. Regul. Integr. Comp. Physiol.309R345–R357. 10.1152/ajpregu.00225.2015
74
Kaelberer M. M. Bohorquez D. V. (2018). The now and then of gut-brain signaling.Brain Res.1693(Pt B), 192–196. 10.1016/j.brainres.2018.03.027
75
Kaelberer M. M. Rupprecht L. E. Liu W. W. Weng P. Bohorquez D. V. (2020). Neuropod cells: the emerging biology of gut-brain sensory transduction.Annu. Rev. Neurosci.43337–353. 10.1146/annurev-neuro-091619-022657
76
Kaslin J. Panula P. (2001). Comparative anatomy of the histaminergic and other aminergic systems in zebrafish (Danio rerio).J. Comp. Neurol.440342–377. 10.1002/cne.1390
77
Kermen F. Franco L. M. Wyatt C. Yaksi E. (2013). Neural circuits mediating olfactory-driven behavior in fish.Front. Neural Circuits7:62. 10.3389/fncir.2013.00062
78
Klarer M. Arnold M. Gunther L. Winter C. Langhans W. Meyer U. (2014). Gut vagal afferents differentially modulate innate anxiety and learned fear.J. Neurosci.347067–7076. 10.1523/JNEUROSCI.0252-14.2014
79
Klarer M. Krieger J. P. Richetto J. Weber-Stadlbauer U. Gunther L. Winter C. et al (2018). Abdominal vagal afferents modulate the brain transcriptome and behaviors relevant to schizophrenia.J. Neurosci.381634–1647. 10.1523/JNEUROSCI.0813-17.2017
80
Koch B. E. V. Yang S. Lamers G. Stougaard J. Spaink H. P. (2018). Intestinal microbiome adjusts the innate immune setpoint during colonization through negative regulation of MyD88.Nat. Commun.9:4099. 10.1038/s41467-018-06658-4
81
Kowalski K. Mulak A. (2019). Brain-Gut-microbiota axis in Alzheimer’s Disease.J. Neurogastroenterol. Motil.2548–60. 10.5056/jnm18087
82
Kozol R. A. (2018). Prenatal neuropathologies in autism spectrum disorder and intellectual disability: the gestation of a comprehensive zebrafish model.J. Dev. Biol.6:29. 10.3390/jdb6040029
83
Kozol R. A. Abrams A. J. James D. M. Buglo E. Yan Q. Dallman J. E. (2016). Function over form: modeling groups of inherited neurological conditions in zebrafish.Front. Mol. Neurosci.9:55. 10.3389/fnmol.2016.00055
84
Latorre R. Sternini C. De Giorgio R. Greenwood-Van Meerveld B. (2016). Enteroendocrine cells: a review of their role in brain-gut communication.Neurogastroenterol. Motil.28620–630. 10.1111/nmo.12754
85
Lavergne A. Tarifeno-Saldivia E. Pirson J. Reuter A. S. Flasse L. Manfroid I. et al (2020). Pancreatic and intestinal endocrine cells in zebrafish share common transcriptomic signatures and regulatory programmes.BMC Biol.18:109. 10.1186/s12915-020-00840-1
86
Leader G. Tuohy E. Chen J. L. Mannion A. Gilroy S. P. (2020). Feeding problems, gastrointestinal symptoms, challenging behavior and sensory issues in children and adolescents with autism spectrum disorder.J. Autism Dev. Disord.501401–1410. 10.1007/s10803-019-04357-7
87
Lefter R. Ciobica A. Timofte D. Stanciu C. Trifan A. (2019). A descriptive review on the prevalence of gastrointestinal disturbances and their multiple associations in autism spectrum disorder.Medicina56:11. 10.3390/medicina56010011
88
Li C. Liu Y. Fang H. Chen Y. Weng J. Zhai M. et al (2020). Study on aberrant eating behaviors, food intolerance, and stereotyped behaviors in autism spectrum disorder.Front. Psychiatry11:493695. 10.3389/fpsyt.2020.493695
89
Li J. Li H. Y. Gu S. Y. Zi H. X. Jiang L. Du J. L. (2020). One-step generation of zebrafish carrying a conditional knockout-knockin visible switch via CRISPR/Cas9-mediated intron targeting.Sci. China Life Sci.6359–67. 10.1007/s11427-019-1607-9
90
Liang S. Wu X. Jin F. (2018). Gut-brain psychology: rethinking psychology from the microbiota–gut–brain axis.Front. Integr. Neurosci.12:33. 10.3389/fnint.2018.00033
91
Liddle R. A. (2018). Parkinson’s disease from the gut.Brain Res.1693(Pt B), 201–206. 10.1016/j.brainres.2018.01.010
92
Liu C. X. Li C. Y. Hu C. C. Wang Y. Lin J. Jiang Y. H. et al (2018). CRISPR/Cas9-induced shank3b mutant zebrafish display autism-like behaviors.Mol. Autism9:23. 10.1186/s13229-018-0204-x
93
Lundin A. Bok C. M. Aronsson L. Bjorkholm B. Gustafsson J. A. Pott S. et al (2008). Gut flora, Toll-like receptors and nuclear receptors: a tripartite communication that tunes innate immunity in large intestine.Cell Microbiol.101093–1103. 10.1111/j.1462-5822.2007.01108.x
94
Ma P. M. (1997). Catecholaminergic systems in the zebrafish. III. Organization and projection pattern of medullary dopaminergic and noradrenergic neurons.J. Comp. Neurol.381411–427.
95
MacFabe D. F. Cain D. P. Rodriguez-Capote K. Franklin A. E. Hoffman J. E. Boon F. et al (2007). Neurobiological effects of intraventricular propionic acid in rats: possible role of short chain fatty acids on the pathogenesis and characteristics of autism spectrum disorders.Behav. Brain Res.176149–169. 10.1016/j.bbr.2006.07.025
96
Manoli D. S. State M. W. (2021). Autism spectrum disorder genetics and the search for pathological mechanisms.Am. J. Psychiatry17830–38. 10.1176/appi.ajp.2020.20111608
97
Margari L. Marzulli L. Gabellone A. de Giambattista C. (2020). Eating and mealtime behaviors in patients with autism spectrum disorder: current perspectives.Neuropsychiatr. Dis. Treat.162083–2102. 10.2147/NDT.S224779
98
Margolis K. G. Buie T. M. Turner J. B. Silberman A. E. Feldman J. F. Murray K. F. et al (2019). Development of a brief parent-report screen for common gastrointestinal disorders in autism spectrum disorder.J. Autism Dev. Disord.49349–362. 10.1007/s10803-018-3767-7
99
Marjoram L. Alvers A. Deerhake M. E. Bagwell J. Mankiewicz J. Cocchiaro J. L. et al (2015). Epigenetic control of intestinal barrier function and inflammation in zebrafish.Proc. Natl. Acad. Sci. U.S.A.1122770–2775. 10.1073/pnas.1424089112
100
Marques J. C. Li M. Schaak D. Robson D. N. Li J. M. (2020). Internal state dynamics shape brainwide activity and foraging behaviour.Nature577239–243. 10.1038/s41586-019-1858-z
101
Mayer E. A. Padua D. Tillisch K. (2014). Altered brain-gut axis in autism: comorbidity or causative mechanisms?Bioessays36933–939. 10.1002/bies.201400075
102
Mayer E. A. Tillisch K. Gupta A. (2015). Gut/brain axis and the microbiota.J. Clin. Invest.125926–938. 10.1172/JCI76304
103
McCue L. M. Flick L. H. Twyman K. A. Xian H. (2017). Gastrointestinal dysfunctions as a risk factor for sleep disorders in children with idiopathic autism spectrum disorder: a retrospective cohort study.Autism211010–1020. 10.1177/1362361316667061
104
McElhanon B. O. McCracken C. Karpen S. Sharp W. G. (2014). Gastrointestinal symptoms in autism spectrum disorder: a meta-analysis.Pediatrics133872–883. 10.1542/peds.2013-3995
105
McLean D. L. Fetcho J. R. (2004). Relationship of tyrosine hydroxylase and serotonin immunoreactivity to sensorimotor circuitry in larval zebrafish.J. Comp. Neurol.48057–71. 10.1002/cne.20281
106
McMahon K. Q. Papandreou A. Ma M. Barry B. J. Mirzaa G. M. Dobyns W. B. et al (2015). Familial recurrences of FOXG1-related disorder: evidence for mosaicism.Am. J. Med. Genet. A167A3096–3102. 10.1002/ajmg.a.37353
107
Melancon E. Gomez De La Torre Canny S. Sichel S. Kelly M. Wiles T. J. Rawls J. F. et al (2017). Best practices for germ-free derivation and gnotobiotic zebrafish husbandry.Methods Cell Biol.13861–100. 10.1016/bs.mcb.2016.11.005
108
Motil K. J. Caeg E. Barrish J. O. Geerts S. Lane J. B. Percy A. K. et al (2012). Gastrointestinal and nutritional problems occur frequently throughout life in girls and women with Rett syndrome.J. Pediatr. Gastroenterol. Nutr.55292–298. 10.1097/MPG.0b013e31824b6159
109
Moulis H. Garsten J. J. Marano A. R. Elser J. M. (1992). Tuberous sclerosis complex: review of the gastrointestinal manifestations and report of an unusual case.Am. J. Gastroenterol.87914–918.
110
Mulak A. Bonaz B. (2015). Brain-gut-microbiota axis in Parkinson’s disease.World J. Gastroenterol.2110609–10620. 10.3748/wjg.v21.i37.10609
111
Muller P. A. Schneeberger M. Matheis F. Wang P. Kerner Z. Ilanges A. et al (2020). Microbiota modulate sympathetic neurons via a gut-brain circuit.Nature583441–446. 10.1038/s41586-020-2474-7
112
Muto A. Lal P. Ailani D. Abe G. Itoh M. Kawakami K. (2017). Activation of the hypothalamic feeding centre upon visual prey detection.Nat. Commun.8:15029. 10.1038/ncomms15029
113
Nechiporuk A. Linbo T. Poss K. D. Raible D. W. (2007). Specification of epibranchial placodes in zebrafish.Development134611–623. 10.1242/dev.02749
114
Neuhaus E. Bernier R. A. Tham S. W. Webb S. J. (2018). Gastrointestinal and psychiatric symptoms among children and adolescents with autism spectrum disorder.Front. Psychiatry9:515. 10.3389/fpsyt.2018.00515
115
Ng A. N. de Jong-Curtain T. A. Mawdsley D. J. White S. J. Shin J. Appel B. et al (2005). Formation of the digestive system in zebrafish: III. Intestinal epithelium morphogenesis.Dev. Biol.286114–135. 10.1016/j.ydbio.2005.07.013
116
Niu X. Liu L. Wang T. Chuan X. Yu Q. Du M. et al (2020). Mapping of extrinsic innervation of the gastrointestinal tract in the mouse embryo.J. Neurosci.406691–6708. 10.1523/JNEUROSCI.0309-20.2020
117
Nomaksteinsky M. Kassabov S. Chettouh Z. Stoekle H. C. Bonnaud L. Fortin G. et al (2013). Ancient origin of somatic and visceral neurons.BMC Biol.11:53. 10.1186/1741-7007-11-53
118
Parker M. J. Fryer A. E. Shears D. J. Lachlan K. L. McKee S. A. Magee A. C. et al (2015). De novo, heterozygous, loss-of-function mutations in SYNGAP1 cause a syndromic form of intellectual disability.Am. J. Med. Genet. A167A2231–2237. 10.1002/ajmg.a.37189
119
Pattyn A. Morin X. Cremer H. Goridis C. Brunet J. F. (1999). The homeobox gene Phox2b is essential for the development of autonomic neural crest derivatives.Nature399366–370. 10.1038/20700
120
Peippo M. Ignatius J. (2012). Pitt-hopkins syndrome.Mol. Syndromol.2171–180. 10.1159/000335287
121
Pellicano E. Dinsmore A. Charman T. (2014). What should autism research focus upon? Community views and priorities from the United Kingdom.Autism18756–770. 10.1177/1362361314529627
122
Penzol M. J. Salazar de Pablo G. Llorente C. Moreno C. Hernandez P. Dorado M. L. et al (2019). Functional gastrointestinal disease in autism spectrum disorder: a retrospective descriptive study in a clinical sample.Front. Psychiatry10:179. 10.3389/fpsyt.2019.00179
123
Phelps D. Brinkman N. E. Keely S. P. Anneken E. M. Catron T. R. Betancourt D. et al (2017). Microbial colonization is required for normal neurobehavioral development in zebrafish.Sci. Rep.7:11244. 10.1038/s41598-017-10517-5
124
Prchalova D. Havlovicova M. Sterbova K. Stranecky V. Hancarova M. Sedlacek Z. (2017). Analysis of 31-year-old patient with SYNGAP1 gene defect points to importance of variants in broader splice regions and reveals developmental trajectory of SYNGAP1-associated phenotype: case report.BMC Med. Genet.18:62. 10.1186/s12881-017-0425-4
125
Qiao R. Sheng C. Lu Y. Zhang Y. Ren H. Lemos B. (2019). Microplastics induce intestinal inflammation, oxidative stress, and disorders of metabolome and microbiome in zebrafish.Sci. Total Environ.662246–253. 10.1016/j.scitotenv.2019.01.245
126
Ramprasad C. Douglas J. Y. Moshiree B. (2018). Parkinson’s Disease and current treatments for its gastrointestinal neurogastromotility effects.Curr. Treat. Opt. Gastroenterol.16489–510. 10.1007/s11938-018-0201-3
127
Randlett O. Wee C. L. Naumann E. A. Nnaemeka O. Schoppik D. Fitzgerald J. E. et al (2015). Whole-brain activity mapping onto a zebrafish brain atlas.Nat. Methods121039–1046. 10.1038/nmeth.3581
128
Rao M. Gershon M. D. (2016). The bowel and beyond: the enteric nervous system in neurological disorders.Nat. Rev. Gastroenterol. Hepatol.13517–528. 10.1038/nrgastro.2016.107
129
Rawls J. F. Samuel B. S. Gordon J. I. (2004). Gnotobiotic zebrafish reveal evolutionarily conserved responses to the gut microbiota.Proc. Natl. Acad. Sci. U.S.A.1014596–4601. 10.1073/pnas.0400706101
130
Rich A. (2009). A new high-content model system for studies of gastrointestinal transit: the zebrafish.Neurogastroenterol. Motil.21225–228. 10.1111/j.1365-2982.2008.01251.x
131
Rose S. Bennuri S. C. Murray K. F. Buie T. Winter H. Frye R. E. (2017). Mitochondrial dysfunction in the gastrointestinal mucosa of children with autism: a blinded case-control study.PLoS One12:e0186377. 10.1371/journal.pone.0186377
132
Roy-Carson S. Natukunda K. Chou H. C. Pal N. Farris C. Schneider S. Q. et al (2017). Defining the transcriptomic landscape of the developing enteric nervous system and its cellular environment.BMC Genomics18:290. 10.1186/s12864-017-3653-2
133
Sakai C. Ijaz S. Hoffman E. J. (2018). Zebrafish models of neurodevelopmental disorders: past, present, and future.Front. Mol. Neurosci.11:294. 10.3389/fnmol.2018.00294
134
Sampson T. R. Debelius J. W. Thron T. Janssen S. Shastri G. G. Ilhan Z. E. et al (2016). Gut microbiota regulate motor deficits and neuroinflammation in a model of Parkinson’s Disease.Cell1671469.e12–1480.e12. 10.1016/j.cell.2016.11.018
135
Sauer A. K. Bockmann J. Steinestel K. Boeckers T. M. Grabrucker A. M. (2019). Altered intestinal morphology and microbiota composition in the autism spectrum disorders associated SHANK3 mouse model.Int. J. Mol. Sci.20:2134. 10.3390/ijms20092134
136
Schroeder B. O. Backhed F. (2016). Signals from the gut microbiota to distant organs in physiology and disease.Nat. Med.221079–1089. 10.1038/nm.4185
137
Severance E. G. Prandovszky E. Castiglione J. Yolken R. H. (2015). Gastroenterology issues in schizophrenia: why the gut matters.Curr. Psychiatry Rep.17:27. 10.1007/s11920-015-0574-0
138
Severance E. G. Yolken R. H. Eaton W. W. (2016). Autoimmune diseases, gastrointestinal disorders and the microbiome in schizophrenia: more than a gut feeling.Schizophr. Res.17623–35. 10.1016/j.schres.2014.06.027
139
Sgritta M. Dooling S. W. Buffington S. A. Momin E. N. Francis M. B. Britton R. A. et al (2019). Mechanisms underlying microbial-mediated changes in social behavior in mouse models of autism spectrum disorder.Neuron101246.e6–259.e6. 10.1016/j.neuron.2018.11.018
140
Shaaya E. A. Pollack S. F. Boronat S. Davis-Cooper S. Zella G. C. Thibert R. L. (2015). Gastrointestinal problems in 15q duplication syndrome.Eur. J. Med. Genet.58191–193. 10.1016/j.ejmg.2014.12.012
141
Shaco-Levy R. Jasperson K. W. Martin K. Samadder N. J. Burt R. W. Ying J. et al (2017). Gastrointestinal polyposis in cowden syndrome.J. Clin. Gastroenterol.51e60–e67. 10.1097/MCG.0000000000000703
142
Sharon G. Cruz N. J. Kang D. W. Gandal M. J. Wang B. Kim Y. M. et al (2019). Human gut microbiota from autism spectrum disorder promote behavioral symptoms in mice.Cell1771600.e17–1618.e17. 10.1016/j.cell.2019.05.004
143
Shontz E. C. Souders C. L. II Schmidt J. T. Martyniuk C. J. (2018). Domperidone upregulates dopamine receptor expression and stimulates locomotor activity in larval zebrafish (Danio rerio).Genes Brain Behav.17:e12460. 10.1111/gbb.12460
144
Siper P. M. De Rubeis S. Trelles M. D. P. Durkin A. Di Marino D. Muratet F. et al (2017). Prospective investigation of FOXP1 syndrome.Mol. Autism8:57. 10.1186/s13229-017-0172-6
145
Srikantha P. Mohajeri M. H. (2019). The possible role of the microbiota-gut-brain-axis in autism spectrum disorder.Int. J. Mol. Sci.20:2115. 10.3390/ijms20092115
146
Stachnik J. Gabay M. (2010). Emerging role of aripiprazole for treatment of irritability associated with autistic disorder in children and adolescents.Adolesc. Health Med. Ther.1105–114. 10.2147/AHMT.S9819
147
Stednitz S. J. McDermott E. M. Ncube D. Tallafuss A. Eisen J. S. Washbourne P. (2018). Forebrain control of behaviorally driven social orienting in zebrafish.Curr. Biol.282445.e3–2451.e3. 10.1016/j.cub.2018.06.016
148
Stengel A. Tache Y. (2010). Corticotropin-releasing factor signaling and visceral response to stress.Exp. Biol. Med.2351168–1178. 10.1258/ebm.2010.009347
149
Sternson S. M. Eiselt A. K. (2017). Three Pillars for the neural control of appetite.Annu. Rev. Physiol.79401–423. 10.1146/annurev-physiol-021115-104948
150
Stil A. Drapeau P. (2016). Neuronal labeling patterns in the spinal cord of adult transgenic Zebrafish.Dev. Neurobiol.76642–660. 10.1002/dneu.22350
151
Suarez A. N. Hsu T. M. Liu C. M. Noble E. E. Cortella A. M. Nakamoto E. M. et al (2018). Gut vagal sensory signaling regulates hippocampus function through multi-order pathways.Nat. Commun.9:2181. 10.1038/s41467-018-04639-1
152
Tache Y. Yang H. Miampamba M. Martinez V. Yuan P. Q. (2006). Role of brainstem TRH/TRH-R1 receptors in the vagal gastric cholinergic response to various stimuli including sham-feeding.Auton. Neurosci.12542–52. 10.1016/j.autneu.2006.01.014
153
Thaiss C. A. Zmora N. Levy M. Elinav E. (2016). The microbiome and innate immunity.Nature53565–74. 10.1038/nature18847
154
Tian X. Chen J. Zhang J. Yang X. Ji T. Zhang Y. et al (2019). The efficacy of ketogenic diet in 60 chinese patients with dravet syndrome.Front. Neurol.10:625. 10.3389/fneur.2019.00625
155
Toscano M. De Grandi R. Grossi E. Drago L. (2017). Role of the human breast milk-associated microbiota on the newborns’ immune system: a mini review.Front. Microbiol.8:2100. 10.3389/fmicb.2017.02100
156
Tsang A. H. Nuzzaci D. Darwish T. Samudrala H. Blouet C. (2020). Nutrient sensing in the nucleus of the solitary tract mediates non-aversive suppression of feeding via inhibition of AgRP neurons.Mol. Metab.42:101070. 10.1016/j.molmet.2020.101070
157
Tye C. Runicles A. K. Whitehouse A. J. O. Alvares G. A. (2019). Characterizing the interplay between autism spectrum disorder and comorbid medical conditions: an integrative review.Front. Psychiatry9:751. 10.3389/fpsyt.2018.00751
158
Uyttebroek L. Shepherd I. T. Harrisson F. Hubens G. Blust R. Timmermans J. P. et al (2010). Neurochemical coding of enteric neurons in adult and embryonic zebrafish (Danio rerio).J. Comp. Neurol.5184419–4438. 10.1002/cne.22464
159
van der Vaart M. Svoboda O. Weijts B. G. Espin-Palazon R. Sapp V. Pietri T. et al (2017). Mecp2 regulates tnfa during zebrafish embryonic development and acute inflammation.Dis. Model Mech.101439–1451. 10.1242/dmm.026922
160
Van Dijck A. Vulto-van Silfhout A. T. Cappuyns E. van der Werf I. M. Mancini G. M. Tzschach A. et al (2019). Clinical presentation of a complex neurodevelopmental disorder caused by mutations in ADNP.Biol. Psychiatry85287–297. 10.1016/j.biopsych.2018.02.1173
161
Van Sebille Y. Z. Gibson R. J. Wardill H. R. Carney T. J. Bowen J. M. (2019). Use of zebrafish to model chemotherapy and targeted therapy gastrointestinal toxicity.Exp. Biol. Med.2441178–1185. 10.1177/1535370219855334
162
Vanner S. Greenwood-Van Meerveld B. Mawe G. Shea-Donohue T. Verdu E. F. Wood J. et al (2016). Fundamentals of neurogastroenterology: basic science.Gastroenterology1501280–1291. 10.1053/j.gastro.2016.02.018
163
Vanwalleghem G. C. Ahrens M. B. Scott E. K. (2018). Integrative whole-brain neuroscience in larval zebrafish.Curr. Opin. Neurobiol.50136–145. 10.1016/j.conb.2018.02.004
164
Vendrell-Llopis N. Yaksi E. (2015). Evolutionary conserved brainstem circuits encode category, concentration and mixtures of taste.Sci. Rep.5:17825. 10.1038/srep17825
165
Villas N. Meskis M. A. Goodliffe S. (2017). Dravet syndrome: characteristics, comorbidities, and caregiver concerns.Epilepsy Behav.7481–86. 10.1016/j.yebeh.2017.06.031
166
Vincis R. Fontanini A. (2019). Central taste anatomy and physiology.Handb. Clin. Neurol.164187–204. 10.1016/B978-0-444-63855-7.00012-5
167
Vlaskamp D. R. M. Shaw B. J. Burgess R. Mei D. Montomoli M. Xie H. et al (2019). SYNGAP1 encephalopathy: a distinctive generalized developmental and epileptic encephalopathy.Neurology92e96–e107. 10.1212/WNL.0000000000006729
168
Wallace K. N. Akhter S. Smith E. M. Lorent K. Pack M. (2005). Intestinal growth and differentiation in zebrafish.Mech. Dev.122157–173. 10.1016/j.mod.2004.10.009
169
Wallace K. N. Pack M. (2003). Unique and conserved aspects of gut development in zebrafish.Dev. Biol.25512–29. 10.1016/s0012-1606(02)00034-9
170
Wang L. Christophersen C. T. Sorich M. J. Gerber J. P. Angley M. T. Conlon M. A. (2012). Elevated fecal short chain fatty acid and ammonia concentrations in children with autism spectrum disorder.Dig Dis. Sci.572096–2102. 10.1007/s10620-012-2167-7
171
Wee C. L. Song E. Y. Johnson R. E. Ailani D. Randlett O. Kim J. Y. et al (2019). A bidirectional network for appetite control in larval zebrafish.eLife8:e43775. 10.7554/eLife.43775
172
Wen J. Mercado G. P. Volland A. Doden H. L. Lickwar C. R. Crooks T. et al (2020). Fxr signaling and microbial metabolism of bile salts in the zebrafish intestine.bioRxiv [Preprint]. 10.1101/2020.12.13.422569
173
Wiles T. J. Guillemin K. (2020). Patterns of partnership: surveillance and mimicry in host-microbiota mutualisms.Curr. Opin. Microbiol.5487–94. 10.1016/j.mib.2020.01.012
174
Williams C. A. Driscoll D. J. Dagli A. I. (2010). Clinical and genetic aspects of angelman syndrome.Genet. Med.12385–395. 10.1097/GIM.0b013e3181def138
175
Willms R. J. Hocking J. C. Foley E. (2020). A cell atlas of microbe-responsive processes in the zebrafish intestine.bioRxiv [Preprint]. 10.1101/2020.11.06.371609
176
Wolf S. (1981). The psyche and the stomach. A historical vignette.Gastroenterology80605–614.
177
Won Y. J. Ono F. Ikeda S. R. (2012). Characterization of Na+ and Ca2+ channels in zebrafish dorsal root ganglion neurons.PLoS One7:e42602. 10.1371/journal.pone.0042602
178
Wright M. A. Mo W. Nicolson T. Ribera A. B. (2010). In vivo evidence for transdifferentiation of peripheral neurons.Development1373047–3056. 10.1242/dev.052696
179
Yanez J. Souto Y. Pineiro L. Folgueira M. Anadon R. (2017). Gustatory and general visceral centers and their connections in the brain of adult zebrafish: a carbocyanine dye tract-tracing study.J. Comp. Neurol.525333–362. 10.1002/cne.24068
180
Yano J. M. Yu K. Donaldson G. P. Shastri G. G. Ann P. Ma L. et al (2015). Indigenous bacteria from the gut microbiota regulate host serotonin biosynthesis.Cell161264–276. 10.1016/j.cell.2015.02.047
181
Ye L. Bae M. Cassilly C. D. Jabba S. V. Thorpe D. W. Martin A. M. et al (2020). Enteroendocrine cells sense bacterial tryptophan catabolites to activate enteric and vagal neuronal pathways.Cell Host Microbe29179.e9–196.e9. 10.1016/j.chom.2020.11.011
182
Zhang C. Kaye J. A. Cai Z. Wang Y. Prescott S. L. Liberles S. D. (2020). Area postrema cell types that mediate nausea-associated behaviors.Neuron109461.e5–472.e5. 10.1016/j.neuron.2020.11.010
183
Zhang W. Waise T. M. Z. Toshinai K. Tsuchimochi W. Naznin F. Islam M. N. et al (2020). Functional interaction between Ghrelin and GLP-1 regulates feeding through the vagal afferent system.Sci. Rep.10:18415. 10.1038/s41598-020-75621-5
184
Zhao X. Pack M. (2017). Modeling intestinal disorders using zebrafish.Methods Cell Biol.138241–270. 10.1016/bs.mcb.2016.11.006
185
Zweier C. de Jong E. K. Zweier M. Orrico A. Ousager L. B. Collins A. L. et al (2009). CNTNAP2 and NRXN1 are mutated in autosomal-recessive Pitt-Hopkins-like mental retardation and determine the level of a common synaptic protein in Drosophila.Am. J. Hum. Genet.85655–666. 10.1016/j.ajhg.2009.10.004
Summary
Keywords
gastrointestinal, microbiome, comorbidities, ASD, GI, CNS
Citation
James DM, Davidson EA, Yanes J, Moshiree B and Dallman JE (2021) The Gut-Brain-Microbiome Axis and Its Link to Autism: Emerging Insights and the Potential of Zebrafish Models. Front. Cell Dev. Biol. 9:662916. doi: 10.3389/fcell.2021.662916
Received
01 February 2021
Accepted
15 March 2021
Published
15 April 2021
Volume
9 - 2021
Edited by
Yasuhito Shimada, Mie University, Japan
Reviewed by
Arlene Mannion, National University of Ireland Galway, Ireland; Youliang Wang, Beijing Institute of Technology, China
Updates
Copyright
© 2021 James, Davidson, Yanes, Moshiree and Dallman.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Julia E. Dallman, j.dallman@miami.edu
This article was submitted to Molecular Medicine, a section of the journal Frontiers in Cell and Developmental Biology
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.