- 1Department of Pathology, Fudan University Shanghai Cancer Center, Shanghai, China
- 2Department of Medical Oncology, Shanghai Medical College, Fudan University, Shanghai, China
- 3Institute of Pathology, Fudan University, Shanghai, China
Long noncoding RNAs (lncRNAs), defined as ncRNAs no longer than 200 nucleotides, play an important role in cancer development. Accumulating research on lncRNAs offers a compelling new aspect of genome modulation, in which they are involved in chromatin remodeling, transcriptional and post-transcriptional regulation, and cross-talk with other nucleic acids. Increasing evidence suggests that lncRNAs reshape the tumor microenvironment (TME), which accounts for tumor development and progression. At the same time, the insightful findings on lncRNAs in immune recognition and evasion in tumor-infiltrating immune cells raise concerns with regard to immuno-oncology. In this review, we describe the essential characteristics of lncRNAs, elucidate functions of immune components engaged in tumor surveillance, and present some instructive examples in this new area.
Introduction
In recent years, in both basic and clinical medical research, a popular and widely appreciated topic is elucidating the role of noncoding RNAs in human physiology and diseases. Decades ago, the major part of the genome, which does not encode proteins, was recognized as the “junk” DNA (Pennisi, 2012; McNally, 2017). Owing to the development of high-throughput sequencing technology, the GENCODE project has helped researchers to understand that ncRNAs cover over 90% of the human genome, and has annotated various kinds of ncRNAs, which promoted a widespread interest in long noncoding RNAs (Harrow et al., 2012; Frankish et al., 2019). The extensive studies of lncRNA have revolutionized our understanding of genetic and cellular molecule regulation in physiological and pathological phenomena.
An increasing number of studies have emphasized the view that lncRNAs are frequently involved in the process of cancer development. Controversy remains regarding the regulatory actions and effects of some lncRNAs in tumorigenesis. While in clinic translation, research on lncRNAs as a genetic screening marker or therapeutic target is ongoing (Do and Kim, 2018; Slack and Chinnaiyan, 2019). Meanwhile, immuno-oncology is becoming one of the most active fields in academic discovery. The abundant signal networks of immune cells, the multi-layered mechanisms of innate and adaptive immunity, and the numerous compositions of the tumor microenvironment (TME) add complexity to the explication of cancer immunity (Mayes et al., 2018; Weiden et al., 2018; Jones et al., 2019). We propose that, for immuno-oncology, besides the recognized protein-coding oncogenes and suppressors, studies on lncRNAs provide an important way to broaden our horizons in cancer immune biology.
In this review, we intend to summarize the definition and classification of lncRNAs considering their functions, to reveal the connections of lncRNAs with cancer immunity, and to highlight the relative translational research.
Characteristics and Functions of lncRNA
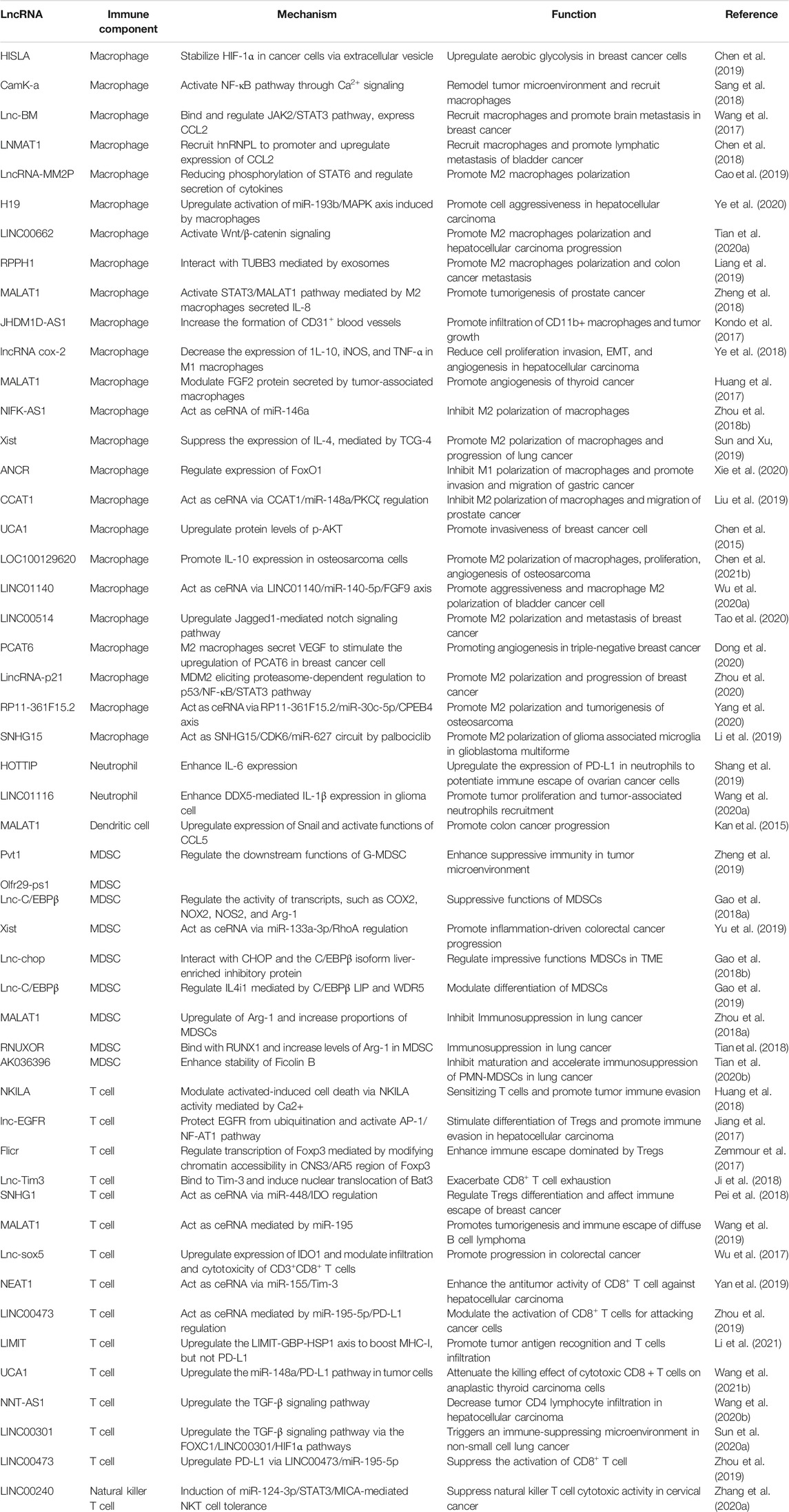
lncRNAs are defined as the noncoding RNAs with a length of at least 200 nucleotides (nt). Compared with conventional protein-coding RNAs, lncRNAs are generally characterized as having lower abundance, more frequent nuclear location, and less evolutionary conservation (Ulitsky and Bartel, 2013; Ulitsky, 2016). For molecular structure, the mature polynucleotide strand is often 5’ capped, tailed with poly adenosine (Poly A) and/or spliced (Gruber and Zavolan, 2019). The functions of lncRNAs rely on their interactions with biomolecules, such as DNA (enhancer, promoter), proteins (transcription factor, enzyme) and other types of RNA (mRNA, miRNA) (Figure 1). Given their interactions with DNA, RNA, and proteins, several key technologies and tools are commonly used to identify and annotate their roles, such as ChiRP (Chu et al., 2012), RIP (Gagliardi and Matarazzo, 2016), RNA-FISH (Mahadevaiah et al., 2009), and PAR-CLIP (Spitzer et al., 2014). Notably, new evidence has convinced scientists that most transcripts of the annotated lncRNAs do not have direct biological functions (Uszczynska-Ratajczak et al., 2018). The functional activity of some transcripts is not related to their abundance in cells and the specific-sequence, which might be consistent with the above features. Given the discovery of bi-directional transcription initiation in promoters and enhancers, lncRNAs represent the major group of RNAs that are initiated at the DNA elements, which lacks encoding functions. According to the relative location to the coding-gene and the direction of transcription, lncRNAs are broadly classified as antisense, intronic, intergenic, sense overlapping, and bidirectional lncRNAs (Figure 2) (Harrow et al., 2012).
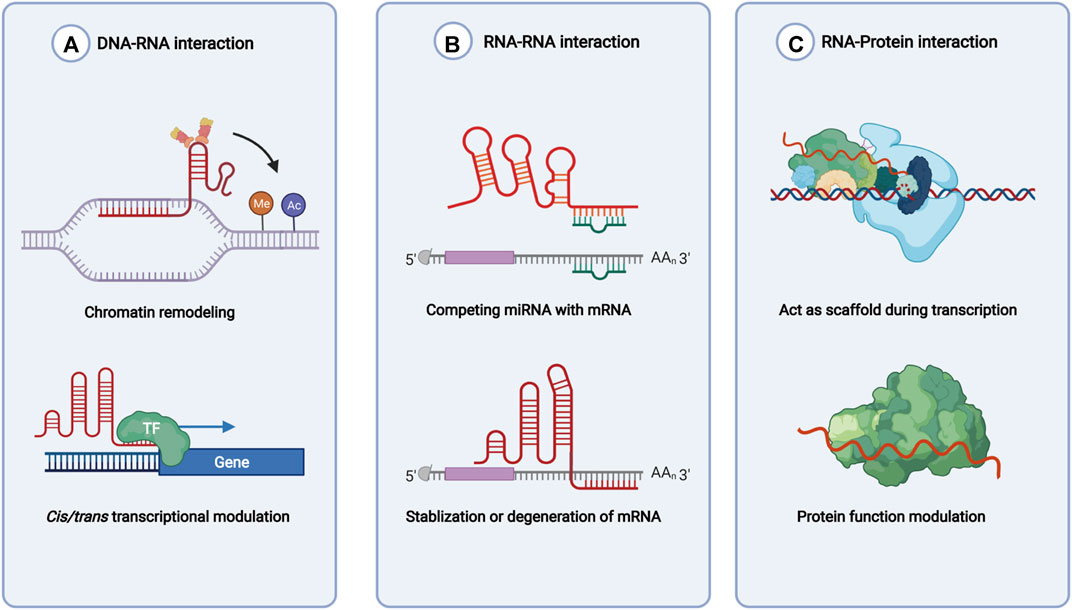
FIGURE 1. Functions of lncRNAs based on molecules interacted with. (A) lncRNAs recruit enzymes and transcription factors to involve in chromatin remodeling and transcription initiation. (B) lncRNAs regulate mRNA stability directly or indirectly. (C) lncRNAs bind with RNA binding protein and help with functions of the latter.
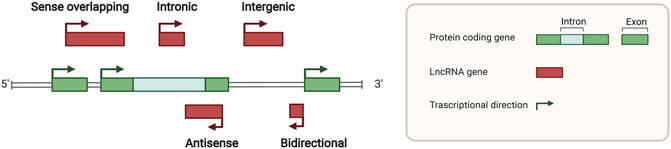
FIGURE 2. Locations of long noncoding RNAs in the genome. Green squares indicate protein coding genes in the genome, while red squares indicate noncoding RNA transcripts. Sense overlapping, antisense, intronic, intergenic, and bidirectional lncRNA loci was classified by different relative locations with protein coding genes.
Components and Events of Cancer Immune Recognition and Escape
The recent compelling achievements in cancer immune therapy are attributable to the persistent exploration of immunobiology during all stages of cancer development. It has been gradually appreciated that many, if not most, neoantigens from distinct cancer cells can be presented to T cells and set off a series of effects (Woo et al., 2015; Altorki et al., 2019). However, in observing individuals, the progression of cancer continues, arguing that the neoplasm finally escaped from the hunting of the immune system. This phenomenon involves the close cooperation of cells and molecules in the immune system.
The initial recognition of neoantigens starts with antigen exposure and uptake, which is accomplished by dendritic cells (DCs). The immigrated DCs present antigen peptides and promote the activation of adaptive immune cells, such as CD8+ T cells. The cascade activated CD8+ T cells flow back to the tumor microenvironment (TME), recognizing and attacking malignant cells. Simultaneously, other innate immune cells, also participate in the TME, including natural killer cells (NK cells), γδT cells, plasmacytoid DCs, macrophages, monocytes, and some myeloid cells (Ribas, 2015; Patel and Minn, 2018). Different kinds of innate immune cells comprise a great network to respond to the emergence of neoantigens.
Some macrophages are described as essential populations of cells in immune-oncology. The conventional recognized macrophages that are involved in the inflammatory response and act as antimicrobic and antitumor components of innate immunity are classified as M1 macrophages. M2 macrophages often have alternative activation, and function as immune modulators in Th2-related mechanisms and cancer development (Allavena et al., 2008; Mantovani and Locati, 2016). The term “tumor-associated macrophages” (TAMs) refers to macrophages existing in TME and share similar surface markers with M2 macrophages. Scientists have proposed that TAMs do not represent a subset of macrophages owing to their instability and dependence on tumors.
Myeloid-derived suppressor cells (MDSCs) comprise a group of myeloid cells generated in pathology, mainly cancer. The most remarkable proposed function of MDCSs is as a suppressor of T cell response. MDSCs are often closely involved in the progression of metastasis in different kinds of malignancies. Tumor cells secrete growth factors and chemokines to expand and recruit MDSCs, which play a role in protecting tumor cells from destruction by host immunity and physical injury, facilitate immune escape and support angiogenesis in neoplasms (Gabrilovich and Nagaraj, 2009; Talmadge and Gabrilovich, 2013; Veglia et al., 2018).
Latest Achievements in lncRNAs Modulation in Immune-Oncology
Here, we retrieved all relative original studies in PubMed (www.ncbi.nlm.nih.gov/pubmed). The Medical Subject Headings (MeSH) terms used were “RNA, Long Noncoding” AND “Neoplasms” AND cell types, containing “Macrophages” (51 results), “Dendritic Cells” (5 results), “Killer Cells, Natural” (4 results),” Neutrophils” (6 results), “Myeloid-Derived Suppressor Cells” (8 results), “T-Lymphocytes” (49 results), “B-Lymphocytes” (20 results) and “Lymphocytes, Tumor-Infiltrating” (14 results). Ultimately, all results were checked artificially. We only enrolled the lncRNA research referring to the immune effects confirmed by cell and/or animal oncological experiments. Studies on lymphomas caused by immune cell development disorders were eliminated (Table 1; Figure 3).
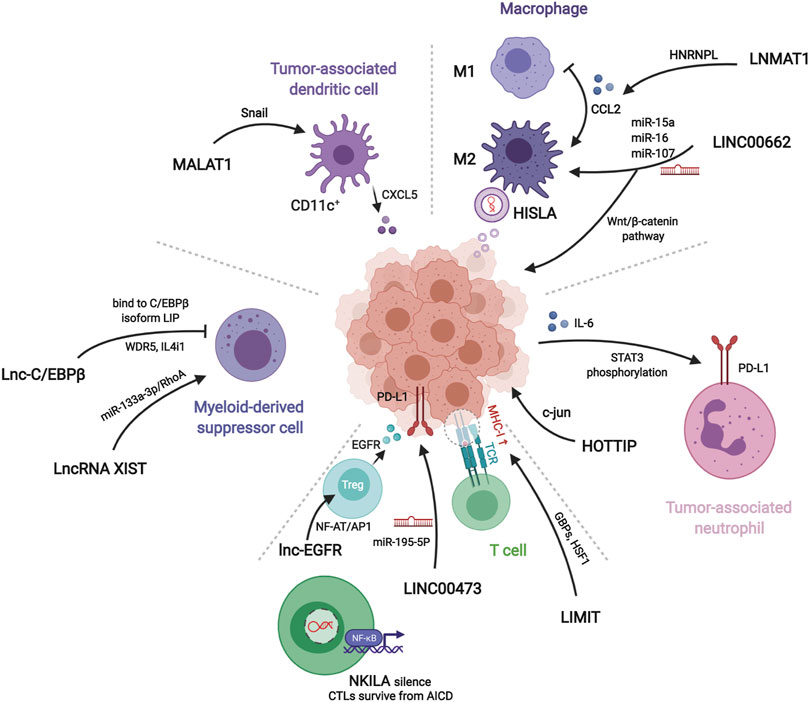
FIGURE 3. Mechanisms of lncRNAs related to different immune components in tumor microenvironment. LNMAT1, LINC00662, and HISLA regulate macrophages polariztion or tumor growth through chemokine, wnt pathway, or extracellular vesicles, respectively. HOTTIP stimulates tumor cells to secret IL-6 for inducing PD-L1 on the surface of tumor-associated neutrophils. LIMIT, LINC00473, NKILA, and lnc-EGFR modulate recognition and cytotoxicity of T cells to tumor cells. Myeloid-derived suppressor cells and tumor-associated dendritic cells assist tumor immune evasion by lnc-C/EBPβ, lncRNA XIST, and MALAT1.
Macrophages
In all articles retrieved, three classes of mechanisms of macrophages engaging in the immune modulation of tumor behavior were identified. Studies on macrophage polarization reported the most numerous RNA molecules. The epigenetic regulation implemented by lncRNAs influences the expression of CCL2, which is prominent in macrophage recruitment. The interaction of glucometabolism between TAM and cancer cells provides an interesting method for signaling communication via extracellular vesicles (EVs).
LINC00662 is identified as a tumor-promoting marker because of its involvement in patient’s prognosis in hepatocellular carcinoma (HCC). Its endogenous competing mechanism explains its behavior in cytoplasm. During competitive binding with miR-15a, miR-16, and miR-107, LINC00662 activates the expression and secretion of WNT3A. By the way of autocrine and paracrine, both in HCC cells and macrophages upregulate the Wnt/β-catenin pathway to promote cancer cell migration and macrophage polarization in TME, respectively (Tian et al., 2020a).
The lncRNA, LNMAT1, is highly expressed in tissue from lymph nodes of bladder cancer patients, which is significantly related to their prognosis and tumor metastasis. LNMAT1, as an intergenic gene, can dramatically enhance the transcription of CCL2, by recruiting hnRNPL to its promoter and mediatingH3K4 tri-methylation. The silenced cells demonstrate that CCL2 is the essential protein inducing LNMAT1-related lymph node metastasis. CCL2 enrolled macrophages can lead to TAM infiltration and lymph angiogenesis (Chen et al., 2018).
Lnc-BM also affects metastasis by CCL2 function of recruiting macrophages. A study of breast cancer brain metastases showed that the interaction between Lnc-BM and JAK activates the latter and results in phosphorylation of STAT3, of which the downstream proteins contain ICAM1 and CCL2. The consecutive action of Lnc-BM/JAK/STAT3 pathway prompts the vascular co-option and ultimately induces macrophages migration and penetration of the blood-brain barrier. Notably, the critical factors IL-6 and endostatin M, which trigger STAT3 phosphorylation, can in return be secreted by recruited macrophages. The mechanism shows a positive feedback loop in cytokines action (Wang et al., 2017).
HISLA is a myeloid-specific molecule, which is packaged in the EV secreted from TAMs of breast cancer cells. The extracellular vesicle has been reported as an efficient transporter in TME, carrying critical signaling molecules (Luga et al., 2012; Boelens et al., 2014). The transmission of HISLA from TAM to cancer cells restrains the hydroxylation via a reduction of the binding between PHD2 and HIF-1α, and stabilizes the latter (Semenza, 2012; Semenza, 2013). The relatively high level of the oxygen-sensing transcription factor, HIF-1α, enhances aerobic glycolysis and the capability of apoptosis resistance in breast cancer cells. Correspondingly, the accumulation of lactate up-regulates the abundance in cancer cells, which creates a positive feedback loop between interactions of TAMs and cancer cells, leading to the survival of cancer cells under stress and therapy. Finally, the resistance of chemotherapy was validated in a mouse model experiment (Chen et al., 2019).
Neutrophils
Tumor-associated neutrophils are cells in the TME that have gradually attracted attention, which can be recruited to regulate adaptive immunity through the PD-1/PD-L1 pathway (He et al., 2015). HOTTIP, a long noncoding RNA, promotes the secretion of IL-6 in ovarian cancer cells, maintaining high levels of PD-L1 on the surface of neutrophils. Consequently, the accumulating immune exhaustion inhibits the function of T cells and causing tumor cells to escape immune surveillance (Shang et al., 2019).
Dendritic Cells
MALAT-1 is a well-known lncRNA with a long history in epigenetic research (Ji et al., 2003). The latest studies report that MALAT1 expressed by tumor associated DCs is attributable to the epithelial-to-mesenchymal transition (EMT), invasion, and migration of tumor cells in colon cancer. Upregulation of MALAT-1 stimulates the expression of Snail, and subsequently activates the functions of CCL5. The downstream manner of CCL5 also refers to proliferation, angiogenesis, and chemotherapeutic resistance, as reported in other oncological studies (Kan et al., 2015; Aldinucci and Casagrande, 2018).
MDSCs
Studies have shown that MDSCs modulate immune responses in the TME. Lnc-C/EBPβ has been demonstrated as a suppressor in functional regulation of MDSCs. The functioning of lnc-C/EBPβ depends on a series of transcripts, including COX2, NOX2, NOS2, and Arg-1. The downstream targets of these molecules cover the secretion of IFN-γ, the distribution of CD4+ and CD8+ T cells, and the differentiation of MDSCs. Mechanically, the binding of lncRNA with C/EBPβ (located at the LIP isoform) obstructs the activation of the protein (Gao et al., 2018a).
Other researchers describe different effects of Lnc-C/EBPβ on the subset transformation of CD11b+Ly6ChiLy6G− monocytic MDSC (Mo-MDSC) and CD11b+Ly6Clow/negLy6G+ polymorphonuclear MDSC (PMN-MDSC). In vitro and in vivo experiments demonstrate that Lnc-C/EBPβ induces bias of PMN-promotion and Mo-suppression during development. This tendency occurs by a two-way method: One is the aforementioned bounding to LIP, and the other is interaction with WDR5 to stop the activation of H3K4me3 in the promoter of IL4i1. The distinct functions of the two subsets of MDSC lead to different influences on neighboring tumor cells (Gao et al., 2019).
Transgenic mouse model experiments showed that LncRNA XIST acts as a regulator of the CXCL12/CXCR4 axis. Higher levels of LncRNA XIST bind with miR-133a-3p and functioning as molecule sponge, liberating the block of RhoA by miRNA. These actions promote the EMT, recruitment of MDSCs and macrophages, and accumulation of antigen-presenting cells, contributing to tumorigenesis under a continuous inflammatory environment (Yu et al., 2019).
T Cells
In cancer immunosurveillance, cytotoxic T lymphocytes are dominant in the identification and destruction of cancer cells. However, for the existence of regulation factors, such as TNF, FasL, and TRAIL, activated T cells in TME often suffer immunological elimination, called activation-induced cell death (AICD) (Schietinger and Greenberg, 2014). NKILA, an lncRNA translated in T cells, sets off the activating threshold of AICD. Under the status of T cell activation, the Ca2+ related signal removes the deacetylase in area of NKILA promoter and up-regulates the transcription mediated by STAT1. Treatment of patient-derived xenografts (PDXs) of breast cancer with NKILA-CTLs injection has shown compelling efficiency in suppressing tumor growth. This study indicates the stirring potential strategies of immunotherapy of lncRNAs engineering (Huang et al., 2018).
The promotion of differentiation of regulatory T cells (Tregs) is also a subject of T cell studies in cancer immunity. Lnc-EGFR can combine with the target transcript EGFR in a sequence-specific manner and protect it from ubiquitination mediated by c-CBL. Stabilization of EGFR sets up the AP-1/NF-AT1 pathway, which consists of positive feedback enhancing the expression of EGFR. The modulation of lnc-EGFR was confirmed in both PDX and humans with HCC, and results in Treg differentiation, CTL suppression, and tumor growth (Jiang et al., 2017).
The co-stimulation signals between activated T cells and tumor cells have been proposed as a convincing target of immune therapy. LINC00473 has been identified as a regulator of programmed death-ligand-1 (PD-L1) in pancreatic cancer. The RNA molecule works as an ceRNA, buffering the levels of miR-195-5p and mRNA of PD-L1. The results demonstrated that silencing of LINC00473 contributes to enhanced expression of IFN-γ, Bax, and IL-4, and, simultaneously, decreases the levels of MMP-2, MMP-9, IL-10, and Bcl-2. Thus, LINC00473 induces a reduction of apoptosis and enhanced proliferation, invasion, and migration of cancer cells (Zhou et al., 2019).
Major histocompatibility complex-I (MHC-I) represents a type of molecule that presents antigens to CD8+ T cells, involving tumor antigen recognition. The lncRNA, inducing MHC-I and immunogenicity of tumor (LIMIT) acts as an accelerator in MHC-I generation. LIMIT can be stimulated by IFN-γ and cis-activate the guanylate-binding protein (GBP) gene cluster. GBPs can disrupt the interaction between HSP90 and HSF1, followed by transcriptional activation of MHC-I, but not PD-L1. This finding indicates a potential pan-cancer epigenetic target of immune therapy (Li et al., 2021).
Clinical Translational Research
Exploring the mechanisms of lncRNAs provides new insights into applications of the molecules as drugs and biomarkers. As this is still an emerging field, only a handful of lncRNA drugs have been approved, but there is accumulated experience with RNA drugs. Small interfering RNAs (siRNAs), antisense oligonucleotides (ASOs), and the CRISPR/Cas9 system are commonly used in drug design (Matsui and Corey, 2017; Chen et al., 2021a). Malat1, a well-studied lncRNA, was systemically knocked down using ASOs in a mouse mammary carcinoma model, and showed lower tumor burden and significant reduction in metastasis. Given the roles of the drugs acting on cancer immunity have not been well elucidated, and their efficacy and safety determined through lnRNAs primary studies will stimulate confidence for them to be tested in clinical trials (Arun et al., 2016). In the meantime, bioinformatics is used in an attempt to filter out the vital lncRNAs in cancer immunity of different kinds of malignancies (Wang et al., 2018; Sun J. et al., 2020; Wu Y. et al., 2020; Zhang et al., 2020b; Wang J. et al., 2021; Mao et al., 2021; Zhou et al., 2021), and the findings may help with biomarker construction or further mechanism research. In a clinical trial of efficacy biomarkers based on the phase 2 IMvigor210 cohort, the authors reported a novel lncRNA-based immune classification in cancer immunotherapy and recommended immunotherapy for the immune-active class.
Conclusion and Perspective
Research on the immunological modulation of lncRNAs in affecting cancer manners is an emerging area enriched by interactive innovation in multiple disciplines. As detailed in the review, lncRNAs engage in many critical events in the immunological balance of elimination or escape in tumor locations. For oncologists, much more attention should be paid to the interactions between cells rather than focusing on single cancer cells and doubling the difficulty. The major challenge to study on lncRNAs is that despite their importance, the current technologies are used to reveal their molecular mechanisms are quite technically difficult. Explaining the mechanism of lncRNA regulation requires the excellent ability to perform epigenetic and molecular biologic experiments. New technologies, for example, new sequencing methods that can directly sequence DNA, RNA, and RNA modifications as well as proteomics will shed light on their role in the transcription processes, immune modulation, and cancer progression. All this technological progress may enhance our understanding of lncRNAs in cancer and will give a better view of disease etiology and will help guide future diagnosis and ultimately therapeutic options. Nevertheless, the possibility of enhancing our understanding of immune surveillance and finding new therapeutic targets for cancer offers continuous motion of relative discovery.
Author Contributions
Xi W conceived the idea of the review and wrote the paper. Xu W contributed to revision. MX and WS are responsible for ensuring that the descriptions are accurate and agreed by all authors.
Funding
This work was supported by National Natural Science Foundation of China (81972249, 81802367, 81802361), Shanghai Clinical science and technology innovation project of municipal hospital (SHDC12020102), Shanghai Clinical Research Plan of SHDC (SHDC2020CR4068), Shanghai Nova Young Medical Talents Funding Program, Shanghai Health Personnel (2020 No. 087), Fudan University's 2019 "Double First-class" Original Research Personalized Support Project (XM03190634), Shanghai Science and Technology Development Fund (18ZR1408000), Shanghai Science and technology development fund 19MC1911000), Clinical Research Project of Shanghai Municipal Health Committee (20194Y0348), and Shanghai Anticancer Association EYAS project (SACA-CY19B10).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Aldinucci, D., and Casagrande, N. (2018). Inhibition of the CCL5/CCR5 Axis against the Progression of Gastric Cancer. Int. J. Mol. Sci. 19 (5), 1477. doi:10.3390/ijms19051477
Allavena, P., Sica, A., Garlanda, C., and Mantovani, A. (2008). The Yin-Yang of Tumor-Associated Macrophages in Neoplastic Progression and Immune Surveillance. Immunol. Rev. 222, 155–161. doi:10.1111/j.1600-065X.2008.00607.x
Altorki, N. K., Markowitz, G. J., Gao, D., Port, J. L., Saxena, A., Stiles, B., et al. (2019). The Lung Microenvironment: an Important Regulator of Tumour Growth and Metastasis. Nat. Rev. Cancer 19 (1), 9–31. doi:10.1038/s41568-018-0081-9
Arun, G., Diermeier, S., Akerman, M., Chang, K.-C., Wilkinson, J. E., Hearn, S., et al. (2016). Differentiation of Mammary Tumors and Reduction in Metastasis uponMalat1lncRNA Loss. Genes Dev. 30 (1), 34–51. doi:10.1101/gad.270959.115
Boelens, M. C., Wu, T. J., Nabet, B. Y., Xu, B., Qiu, Y., Yoon, T., et al. (2014). Exosome Transfer from Stromal to Breast Cancer Cells Regulates Therapy Resistance Pathways. Cell 159 (3), 499–513. doi:10.1016/j.cell.2014.09.051
Cao, J., Dong, R., Jiang, L., Gong, Y., Yuan, M., You, J., et al. (2019). LncRNA-MM2P Identified as a Modulator of Macrophage M2 Polarization. Cancer Immunol. Res. 7 (2), 292–305. doi:10.1158/2326-6066.CIR-18-0145
Chen, C., He, W., Huang, J., Wang, B., Li, H., Cai, Q., et al. (2018). LNMAT1 Promotes Lymphatic Metastasis of Bladder Cancer via CCL2 Dependent Macrophage Recruitment. Nat. Commun. 9 (1), 3826. doi:10.1038/s41467-018-06152-x
Chen, F., Chen, J., Yang, L., Liu, J., Zhang, X., Zhang, Y., et al. (2019). Extracellular Vesicle-Packaged HIF-1α-Stabilizing lncRNA from Tumour-Associated Macrophages Regulates Aerobic Glycolysis of Breast Cancer Cells. Nat. Cel Biol 21 (4), 498–510. doi:10.1038/s41556-019-0299-0
Chen, S., Shao, C., Xu, M., Ji, J., Xie, Y., Lei, Y., et al. (2015). Macrophage Infiltration Promotes Invasiveness of Breast Cancer Cells via Activating Long Non-coding RNA UCA1. Int. J. Clin. Exp. Pathol. 8 (8), 9052–9061.
Chen, Y., Li, Z., Chen, X., and Zhang, S. (2021a). Long Non-coding RNAs: From Disease Code to Drug Role. Acta Pharmaceutica Sinica B 11 (2), 340–354. doi:10.1016/j.apsb.2020.10.001
Chen, Y., Tang, G., Qian, H., Chen, J., Cheng, B., Zhou, C., et al. (2021b). LncRNA LOC100129620 Promotes Osteosarcoma Progression through Regulating CDK6 Expression, Tumor Angiogenesis, and Macrophage Polarization. Aging 13 (10), 14258–14276. doi:10.18632/aging.203042
Chu, C., Quinn, J., and Chang, H. Y. (2012). Chromatin Isolation by RNA Purification (ChIRP). J. Vis. Exp. 25 (61), 3912. doi:10.3791/3912
Do, H., and Kim, W. (2018). Roles of Oncogenic Long Non-coding RNAs in Cancer Development. Genomics Inform. 16 (4), e18. doi:10.5808/GI.2018.16.4.e18
Dong, F., Ruan, S., Wang, J., Xia, Y., Le, K., Xiao, X., et al. (2020). M2 Macrophage-Induced lncRNA PCAT6 Facilitates Tumorigenesis and Angiogenesis of Triple-Negative Breast Cancer through Modulation of VEGFR2. Cell Death Dis 11 (9), 728. doi:10.1038/s41419-020-02926-8
Frankish, A., Diekhans, M., Ferreira, A.-M., Johnson, R., Jungreis, I., Loveland, J., et al. (2019). GENCODE Reference Annotation for the Human and Mouse Genomes. Nucleic Acids Res. 47 (D1), D766–D773. doi:10.1093/nar/gky955
Gabrilovich, D. I., and Nagaraj, S. (2009). Myeloid-derived Suppressor Cells as Regulators of the Immune System. Nat. Rev. Immunol. 9 (3), 162–174. doi:10.1038/nri2506
Gagliardi, M., and Matarazzo, M. R. (2016). RIP: RNA Immunoprecipitation. Methods Mol. Biol. 1480, 73–86. doi:10.1007/978-1-4939-6380-5_7
Gao, Y., Shang, W., Zhang, D., Zhang, S., Zhang, X., Zhang, Y., et al. (2019). Lnc-C/Ebpβ Modulates Differentiation of MDSCs through Downregulating IL4i1 with C/EBPβ LIP and WDR5. Front. Immunol. 10 (1661), 1661. doi:10.3389/fimmu.2019.01661
Gao, Y., Sun, W., Shang, W., Li, Y., Zhang, D., Wang, T., et al. (2018a). Lnc-C/Ebpβ Negatively Regulates the Suppressive Function of Myeloid-Derived Suppressor Cells. Cancer Immunol. Res. 6 (11), 1352–1363. doi:10.1158/2326-6066.CIR-18-0108
Gao, Y., Wang, T., Li, Y., Zhang, Y., and Yang, R. (2018b). Lnc-chop Promotes Immunosuppressive Function of Myeloid-Derived Suppressor Cells in Tumor and Inflammatory Environments. J. Immunol. 200 (8), 2603–2614. doi:10.4049/jimmunol.1701721
Gruber, A. J., and Zavolan, M. (2019). Alternative Cleavage and Polyadenylation in Health and Disease. Nat. Rev. Genet. 20 (10), 599–614. doi:10.1038/s41576-019-0145-z
Harrow, J., Frankish, A., Gonzalez, J. M., Tapanari, E., Diekhans, M., Kokocinski, F., et al. (2012). GENCODE: the Reference Human Genome Annotation for the ENCODE Project. Genome Res. 22 (9), 1760–1774. doi:10.1101/gr.135350.111
He, G., Zhang, H., Zhou, J., Wang, B., Chen, Y., Kong, Y., et al. (2015). Peritumoural Neutrophils Negatively Regulate Adaptive Immunity via the PD-L1/pd-1 Signalling Pathway in Hepatocellular Carcinoma. J. Exp. Clin. Cancer Res. 34, 141. doi:10.1186/s13046-015-0256-0
Huang, D., Chen, J., Yang, L., Ouyang, Q., Li, J., Lao, L., et al. (2018). NKILA lncRNA Promotes Tumor Immune Evasion by Sensitizing T Cells to Activation-Induced Cell Death. Nat. Immunol. 19 (10), 1112–1125. doi:10.1038/s41590-018-0207-y
Huang, J. k., Ma, L., Song, W. h., Lu, B. y., Huang, Y. b., Dong, H. m., et al. (2017). LncRNA‐MALAT1 Promotes Angiogenesis of Thyroid Cancer by Modulating Tumor‐Associated Macrophage FGF2 Protein Secretion. J. Cel. Biochem. 118 (12), 4821–4830. doi:10.1002/jcb.26153
Ji, J., Yin, Y., Ju, H., Xu, X., Liu, W., Fu, Q., et al. (2018). Long Non-coding RNA Lnc-Tim3 Exacerbates CD8 T Cell Exhaustion via Binding to Tim-3 and Inducing Nuclear Translocation of Bat3 in HCC. Cel Death Dis 9 (5), 478. doi:10.1038/s41419-018-0528-7
Ji, P., Diederichs, S., Wang, W., Böing, S., Metzger, R., Schneider, P. M., et al. (2003). MALAT-1, a Novel Noncoding RNA, and Thymosin β4 Predict Metastasis and Survival in Early-Stage Non-small Cell Lung Cancer. Oncogene 22 (39), 8031–8041. doi:10.1038/sj.onc.1206928
Jiang, R., Tang, J., Chen, Y., Deng, L., Ji, J., Xie, Y., et al. (2017). The Long Noncoding RNA Lnc-EGFR Stimulates T-Regulatory Cells Differentiation Thus Promoting Hepatocellular Carcinoma Immune Evasion. Nat. Commun. 8 (1), 15129. doi:10.1038/ncomms15129
Jones, P. A., Ohtani, H., Chakravarthy, A., and De Carvalho, D. D. (2019). Epigenetic Therapy in Immune-Oncology. Nat. Rev. Cancer 19 (3), 151–161. doi:10.1038/s41568-019-0109-9
Kan, J.-Y., Wu, D.-C., Yu, F.-J., Wu, C.-Y., Ho, Y.-W., Chiu, Y.-J., et al. (2015). Chemokine (C-C Motif) Ligand 5 Is Involved in Tumor-Associated Dendritic Cell-Mediated Colon Cancer Progression through Non-coding RNA MALAT-1. J. Cel. Physiol. 230 (8), 1883–1894. doi:10.1002/jcp.24918
Kondo, A., Nonaka, A., Shimamura, T., Yamamoto, S., Yoshida, T., Kodama, T., et al. (2017). Long Noncoding RNA JHDM1D-AS1 Promotes Tumor Growth by Regulating Angiogenesis in Response to Nutrient Starvation. Mol. Cel Biol 37 (18). doi:10.1128/MCB.00125-17
Li, G., Kryczek, I., Nam, J., Li, X., Li, S., Li, J., et al. (2021). LIMIT Is an Immunogenic lncRNA in Cancer Immunity and Immunotherapy. Nat. Cel Biol 23 (5), 526–537. doi:10.1038/s41556-021-00672-3
Li, Z., Zhang, J., Zheng, H., Li, C., Xiong, J., Wang, W., et al. (2019). Modulating lncRNA SNHG15/CDK6/miR-627 Circuit by Palbociclib, Overcomes Temozolomide Resistance and Reduces M2-Polarization of Glioma Associated Microglia in Glioblastoma Multiforme. J. Exp. Clin. Cancer Res. 38 (1), 380. doi:10.1186/s13046-019-1371-0
Liang, Z.-x., Liu, H.-s., Wang, F.-w., Xiong, L., Zhou, C., Hu, T., et al. (2019). LncRNA RPPH1 Promotes Colorectal Cancer Metastasis by Interacting with TUBB3 and by Promoting Exosomes-Mediated Macrophage M2 Polarization. Cel Death Dis 10 (11), 829. doi:10.1038/s41419-019-2077-0
Liu, J., Ding, D., Jiang, Z., Du, T., Liu, J., and Kong, Z. (2019). Long Non-coding RNA CCAT1/miR-148a/PKCζ Prevents Cell Migration of Prostate Cancer by Altering Macrophage Polarization. Prostate 79 (1), 105–112. doi:10.1002/pros.23716
Luga, V., Zhang, L., Viloria-Petit, A. M., Ogunjimi, A. A., Inanlou, M. R., Chiu, E., et al. (2012). Exosomes Mediate Stromal Mobilization of Autocrine Wnt-PCP Signaling in Breast Cancer Cell Migration. Cell 151 (7), 1542–1556. doi:10.1016/j.cell.2012.11.024
Mahadevaiah, S. K., Costa, Y., and Turner, J. M. A. (2009). Using RNA FISH to Study Gene Expression during Mammalian Meiosis. Methods Mol. Biol. 558, 433–444. doi:10.1007/978-1-60761-103-5_25
Mantovani, A., and Locati, M. (2016). Macrophage Metabolism Shapes Angiogenesis in Tumors. Cel Metab. 24 (6), 887–888. doi:10.1016/j.cmet.2016.11.007
Mao, X., Chen, S., and Li, G. (2021). Identification of a Ten-Long Noncoding RNA Signature for Predicting the Survival and Immune Status of Patients with Bladder Urothelial Carcinoma Based on the GEO Database: a superior Machine Learning Model. Aging 13 (5), 6957–6981. doi:10.18632/aging.202553
Matsui, M., and Corey, D. R. (2017). Non-coding RNAs as Drug Targets. Nat. Rev. Drug Discov. 16 (3), 167–179. doi:10.1038/nrd.2016.117
Mayes, P. A., Hance, K. W., and Hoos, A. (2018). The Promise and Challenges of Immune Agonist Antibody Development in Cancer. Nat. Rev. Drug Discov. 17 (7), 509–527. doi:10.1038/nrd.2018.75
McNally, F. J. (2017). Competing Chromosomes Explain Junk DNA. Science 358 (6363), 594–595. doi:10.1126/science.aaq0200
Patel, S. A., and Minn, A. J. (2018). Combination Cancer Therapy with Immune Checkpoint Blockade: Mechanisms and Strategies. Immunity 48 (3), 417–433. doi:10.1016/j.immuni.2018.03.007
Pei, X., Wang, X., and Li, H. (2018). LncRNA SNHG1 Regulates the Differentiation of Treg Cells and Affects the Immune Escape of Breast Cancer via Regulating miR-448/Ido. Int. J. Biol. Macromolecules 118 (Pt A), 24–30. doi:10.1016/j.ijbiomac.2018.06.033
Pennisi, E. (2012). ENCODE Project Writes Eulogy for Junk DNA. Science 337 (6099), 1159–1161. doi:10.1126/science.337.6099.1159
Ribas, A. (2015). Adaptive Immune Resistance: How Cancer Protects from Immune Attack. Cancer Discov. 5 (9), 915–919. doi:10.1158/2159-8290.CD-15-0563
Sang, L.-j., Ju, H.-q., Liu, G.-p., Tian, T., Ma, G.-l., Lu, Y.-x., et al. (2018). LncRNA CamK-A Regulates Ca2+-Signaling-Mediated Tumor Microenvironment Remodeling. Mol. Cel 72 (1), 71–83. doi:10.1016/j.molcel.2018.08.014
Schietinger, A., and Greenberg, P. D. (2014). Tolerance and Exhaustion: Defining Mechanisms of T Cell Dysfunction. Trends Immunol. 35 (2), 51–60. doi:10.1016/j.it.2013.10.001
Semenza, G. L. (2013). HIF-1 Mediates Metabolic Responses to Intratumoral Hypoxia and Oncogenic Mutations. J. Clin. Invest. 123 (9), 3664–3671. doi:10.1172/JCI67230
Semenza, G. L. (2012). Hypoxia-inducible Factors: Mediators of Cancer Progression and Targets for Cancer Therapy. Trends Pharmacol. Sci. 33 (4), 207–214. doi:10.1016/j.tips.2012.01.005
Shang, A., Wang, W., Gu, C., Chen, C., Zeng, B., Yang, Y., et al. (2019). Long Non-coding RNA HOTTIP Enhances IL-6 Expression to Potentiate Immune Escape of Ovarian Cancer Cells by Upregulating the Expression of PD-L1 in Neutrophils. J. Exp. Clin. Cancer Res. 38 (1), 411. doi:10.1186/s13046-019-1394-6
Slack, F. J., and Chinnaiyan, A. M. (2019). The Role of Non-coding RNAs in Oncology. Cell 179 (5), 1033–1055. doi:10.1016/j.cell.2019.10.017
Spitzer, J., Hafner, M., Landthaler, M., Ascano, M., Farazi, T., Wardle, G., et al. (2014). PAR-CLIP (Photoactivatable Ribonucleoside-Enhanced Crosslinking and Immunoprecipitation). Methods Enzymol. 539, 113–161. doi:10.1016/b978-0-12-420120-0.00008-6
Sun, C.-C., Zhu, W., Li, S.-J., Hu, W., Zhang, J., Zhuo, Y., et al. (2020a). FOXC1-mediated LINC00301 Facilitates Tumor Progression and Triggers an Immune-Suppressing Microenvironment in Non-small Cell Lung Cancer by Regulating the HIF1α Pathway. Genome Med. 12 (1), 77. doi:10.1186/s13073-020-00773-y
Sun, J., Zhang, Z., Bao, S., Yan, C., Hou, P., Wu, N., et al. (2020b). Identification of Tumor Immune Infiltration-Associated lncRNAs for Improving Prognosis and Immunotherapy Response of Patients with Non-small Cell Lung Cancer. J. Immunother. Cancer 8 (1), e000110. doi:10.1136/jitc-2019-000110
Sun, Y., and Xu, J. (2019). TCF-4 Regulated lncRNA-XIST Promotes M2 Polarization of Macrophages and Is Associated with Lung Cancer. OncoTargets Ther. 2, 8055–8062. doi:10.2147/OTT.S210952
Talmadge, J. E., and Gabrilovich, D. I. (2013). History of Myeloid-Derived Suppressor Cells. Nat. Rev. Cancer 13 (10), 739–752. doi:10.1038/nrc3581
Tao, S., Chen, Q., Lin, C., and Dong, H. (2020). Linc00514 Promotes Breast Cancer Metastasis and M2 Polarization of Tumor-Associated Macrophages via Jagged1-Mediated Notch Signaling Pathway. J. Exp. Clin. Cancer Res. 39 (1), 191. doi:10.1186/s13046-020-01676-x
Tian, X., Ma, J., Wang, T., Tian, J., Zheng, Y., Peng, R., et al. (2018). Long Non-coding RNA RUNXOR Accelerates MDSC-Mediated Immunosuppression in Lung Cancer. BMC Cancer 18 (1), 660. doi:10.1186/s12885-018-4564-6
Tian, X., Wu, Y., Yang, Y., Wang, J., Niu, M., Gao, S., et al. (2020a). Long Noncoding RNA LINC00662 Promotes M2 Macrophage Polarization and Hepatocellular Carcinoma Progression via Activating Wnt/β‐catenin Signaling. Mol. Oncol. 14 (2), 462–483. doi:10.1002/1878-0261.12606
Tian, X., Zheng, Y., Yin, K., Ma, J., Tian, J., Zhang, Y., et al. (2020b). LncRNAAK036396Inhibits Maturation and Accelerates Immunosuppression of Polymorphonuclear Myeloid-Derived Suppressor Cells by Enhancing the Stability of Ficolin B. Cancer Immunol. Res. 8 (4), 565–577. doi:10.1158/2326-6066.CIR-19-0595
Ulitsky, I., and Bartel, D. P. (2013). lincRNAs: Genomics, Evolution, and Mechanisms. Cell 154 (1), 26–46. doi:10.1016/j.cell.2013.06.020
Ulitsky, I. (2016). Evolution to the rescue: Using Comparative Genomics to Understand Long Non-coding RNAs. Nat. Rev. Genet. 17 (10), 601–614. doi:10.1038/nrg.2016.85
Uszczynska-Ratajczak, B., Lagarde, J., Frankish, A., Guigó, R., and Johnson, R. (2018). Towards a Complete Map of the Human Long Non-coding RNA Transcriptome. Nat. Rev. Genet. 19 (9), 535–548. doi:10.1038/s41576-018-0017-y
Veglia, F., Perego, M., and Gabrilovich, D. (2018). Myeloid-derived Suppressor Cells Coming of Age. Nat. Immunol. 19 (2), 108–119. doi:10.1038/s41590-017-0022-x
Wang, J., Shen, C., Dong, D., Zhong, X., Wang, Y., and Yang, X. (2021a). Identification and Verification of an Immune-Related lncRNA Signature for Predicting the Prognosis of Patients with Bladder Cancer. Int. Immunopharmacology 90, 107146. doi:10.1016/j.intimp.2020.107146
Wang, L., Felts, S. J., Van Keulen, V. P., Scheid, A. D., Block, M. S., Markovic, S. N., et al. (2018). Integrative Genome-wide Analysis of Long Noncoding RNAs in Diverse Immune Cell Types of Melanoma Patients. Cancer Res. 78 (15), 4411–4423. doi:10.1158/0008-5472.CAN-18-0529
Wang, Q.-M., Lian, G.-Y., Song, Y., Huang, Y.-F., and Gong, Y. (2019). LncRNA MALAT1 Promotes Tumorigenesis and Immune Escape of Diffuse Large B Cell Lymphoma by Sponging miR-195. Life Sci. 231, 116335. doi:10.1016/j.lfs.2019.03.040
Wang, S., Liang, K., Hu, Q., Li, P., Song, J., Yang, Y., et al. (2017). JAK2-binding Long Noncoding RNA Promotes Breast Cancer Brain Metastasis. J. Clin. Invest. 127 (12), 4498–4515. doi:10.1172/JCI91553
Wang, T., Cao, L., Dong, X., Wu, F., De, W., Huang, L., et al. (2020a). LINC01116 Promotes Tumor Proliferation and Neutrophil Recruitment via DDX5-Mediated Regulation of IL-1β in Glioma Cell. Cel Death Dis 11 (5), 302. doi:10.1038/s41419-020-2506-0
Wang, X., Zhang, Y., Zheng, J., Yao, C., and Lu, X. (2021b). LncRNA UCA1 Attenuated the Killing Effect of Cytotoxic CD8 + T Cells on Anaplastic Thyroid Carcinoma via miR-148a/PD-L1 Pathway. Cancer Immunol. Immunother. 70 (8), 2235–2245. doi:10.1007/s00262-020-02753-y
Wang, Y., Yang, L., Dong, X., Yang, X., Zhang, X., Liu, Z., et al. (2020b). Overexpression of NNT-AS1 Activates TGF-β Signaling to Decrease Tumor CD4 Lymphocyte Infiltration in Hepatocellular Carcinoma. Biomed. Res. Int. 2020, 1–11. doi:10.1155/2020/8216541
Weiden, J., Tel, J., and Figdor, C. G. (2018). Synthetic Immune Niches for Cancer Immunotherapy. Nat. Rev. Immunol. 18 (3), 212–219. doi:10.1038/nri.2017.89
Woo, S.-R., Corrales, L., and Gajewski, T. F. (2015). Innate Immune Recognition of Cancer. Annu. Rev. Immunol. 33 (1), 445–474. doi:10.1146/annurev-immunol-032414-112043
Wu, K., Zhao, Z., Liu, K., Zhang, J., Li, G., and Wang, L. (2017). Long Noncoding RNA Lnc-Sox5 Modulates CRC Tumorigenesis by Unbalancing Tumor Microenvironment. Cell Cycle 16 (13), 1295–1301. doi:10.1080/15384101.2017.1317416
Wu, S., Xu, R., Zhu, X., He, H., Zhang, J., Zeng, Q., et al. (2020a). The Long Noncoding RNA LINC01140/miR-140-5p/FGF9 axis Modulates Bladder Cancer Cell Aggressiveness and Macrophage M2 Polarization. Aging 12 (24), 25845–25864. doi:10.18632/aging.202147
Wu, Y., Zhang, L., He, S., Guan, B., He, A., Yang, K., et al. (2020b). Identification of Immune-Related LncRNA for Predicting Prognosis and Immunotherapeutic Response in Bladder Cancer. aging 12 (22), 23306–23325. doi:10.18632/aging.104115
Xie, C., Guo, Y., and Lou, S. (2020). LncRNA ANCR Promotes Invasion and Migration of Gastric Cancer by Regulating FoxO1 Expression to Inhibit Macrophage M1 Polarization. Dig. Dis. Sci. 65 (10), 2863–2872. doi:10.1007/s10620-019-06019-1
Yan, K., Fu, Y., Zhu, N., Wang, Z., Hong, J.-l., Li, Y., et al. (2019). Repression of lncRNA NEAT1 Enhances the Antitumor Activity of CD8+T Cells against Hepatocellular Carcinoma via Regulating miR-155/Tim-3. Int. J. Biochem. Cel Biol. 110, 1–8. doi:10.1016/j.biocel.2019.01.019
Yang, D., Liu, K., Fan, L., Liang, W., Xu, T., Jiang, W., et al. (2020). LncRNA RP11-361F15.2 Promotes Osteosarcoma Tumorigenesis by Inhibiting M2-like Polarization of Tumor-Associated Macrophages of CPEB4. Cancer Lett. 473, 33–49. doi:10.1016/j.canlet.2019.12.041
Ye, Y., Guo, J., Xiao, P., Ning, J., Zhang, R., Liu, P., et al. (2020). Macrophages-induced Long Noncoding RNA H19 Up-Regulation Triggers and Activates the miR-193b/MAPK1 axis and Promotes Cell Aggressiveness in Hepatocellular Carcinoma. Cancer Lett. 469, 310–322. doi:10.1016/j.canlet.2019.11.001
Ye, Y., Xu, Y., Lai, Y., He, W., Li, Y., Wang, R., et al. (2018). Long Non‐coding RNA Cox‐2 Prevents Immune Evasion and Metastasis of Hepatocellular Carcinoma by Altering M1/M2 Macrophage Polarization. J. Cel. Biochem. 119 (3), 2951–2963. doi:10.1002/jcb.26509
Yu, X., Wang, D., Wang, X., Sun, S., Zhang, Y., Wang, S., et al. (2019). CXCL12/CXCR4 Promotes Inflammation-Driven Colorectal Cancer Progression through Activation of RhoA Signaling by Sponging miR-133a-3p. J. Exp. Clin. Cancer Res. 38 (1), 32. doi:10.1186/s13046-018-1014-x
Zemmour, D., Pratama, A., Loughhead, S. M., Mathis, D., and Benoist, C. (2017). Flicr, a Long Noncoding RNA, Modulates Foxp3 Expression and Autoimmunity. Proc. Natl. Acad. Sci. USA 114 (17), E3472–E3480. doi:10.1073/pnas.1700946114
Zhang, Y., Li, X., Zhang, J., and Liang, H. (2020a). Natural Killer T Cell Cytotoxic Activity in Cervical Cancer Is Facilitated by the LINC00240/microRNA-124-3p/STAT3/MICA axis. Cancer Lett. 474, 63–73. doi:10.1016/j.canlet.2019.12.038
Zhang, Y., Zhang, L., Xu, Y., Wu, X., Zhou, Y., and Mo, J. (2020b). Immune‐related Long Noncoding RNA Signature for Predicting Survival and Immune Checkpoint Blockade in Hepatocellular Carcinoma. J. Cel Physiol 235 (12), 9304–9316. doi:10.1002/jcp.29730
Zheng, T., Ma, G., Tang, M., Li, Z., and Xu, R. (2018). IL-8 Secreted from M2 Macrophages Promoted Prostate Tumorigenesis via STAT3/MALAT1 Pathway. Int. J. Mol. Sci. 20 (1), 98. doi:10.3390/ijms20010098
Zheng, Y., Tian, X., Wang, T., Xia, X., Cao, F., Tian, J., et al. (2019). Long Noncoding RNA Pvt1 Regulates the Immunosuppression Activity of Granulocytic Myeloid-Derived Suppressor Cells in Tumor-Bearing Mice. Mol. Cancer 18 (1), 61. doi:10.1186/s12943-019-0978-2
Zhou, L., Tian, Y., Guo, F., Yu, B., Li, J., Xu, H., et al. (2020). LincRNA-p21 Knockdown Reversed Tumor-Associated Macrophages Function by Promoting MDM2 to Antagonize* P53 Activation and Alleviate Breast Cancer Development. Cancer Immunol. Immunother. 69 (5), 835–846. doi:10.1007/s00262-020-02511-0
Zhou, M., Zhang, Z., Bao, S., Hou, P., Yan, C., Su, J., et al. (2021). Computational Recognition of lncRNA Signature of Tumor-Infiltrating B Lymphocytes with Potential Implications in Prognosis and Immunotherapy of Bladder Cancer. Brief Bioinform 22 (3). doi:10.1093/bib/bbaa047
Zhou, Q., Tang, X., Tian, X., Tian, J., Zhang, Y., Ma, J., et al. (2018a). LncRNA MALAT1 Negatively Regulates MDSCs in Patients with Lung Cancer. J. Cancer 9 (14), 2436–2442. doi:10.7150/jca.24796
Zhou, W. Y., Zhang, M. M., Liu, C., Kang, Y., Wang, J. O., and Yang, X. H. (2019). Long Noncoding RNA LINC00473 Drives the Progression of Pancreatic Cancer via Upregulating Programmed Death‐ligand 1 by Sponging microRNA‐195‐5p. J. Cel Physiol 234 (12), 23176–23189. doi:10.1002/jcp.28884
Keywords: lncRNA, cancer, tumor environment, immune surveillance, immuno-oncology
Citation: Wang X, Wang X, Xu M and Sheng W (2021) Emerging Roles of Long Noncoding RNAs in Immuno-Oncology. Front. Cell Dev. Biol. 9:722904. doi: 10.3389/fcell.2021.722904
Received: 09 June 2021; Accepted: 01 November 2021;
Published: 25 November 2021.
Edited by:
Zong Sheng Guo, Roswell Park Comprehensive Cancer Center, United StatesReviewed by:
Ma Del Rocio Banos-Lara, Universidad Popular Autónoma del Estado de Puebla, MexicoGuillaume Chemin, University of Limoges, France
Copyright © 2021 Wang, Wang, Xu and Sheng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Weiqi Sheng, c2hlbmd3ZWlxaTIwMDZAMTYzLmNvbQ==; Midie Xu, eHVtZDI3MjAyMDAzQHNpbmEuY29t
 Xin Wang1,2
Xin Wang1,2 Xu Wang
Xu Wang