- Hubei Key Laboratory of Embryonic Stem Cell Research, Department of Physiology, Faculty of Basic Medical Sciences, Hubei University of Medicine, Shiyan, China
Satellite stem cell availability and high regenerative capacity have made them an ideal therapeutic approach for muscular dystrophies and neuromuscular diseases. Adult satellite stem cells remain in a quiescent state and become activated upon muscular injury. A series of molecular mechanisms succeed under the control of epigenetic regulation and various myogenic regulatory transcription factors myogenic regulatory factors, leading to their differentiation into skeletal muscles. The regulation of MRFs via various epigenetic factors, including DNA methylation, histone modification, and non-coding RNA, determine the fate of myogenesis. Furthermore, the development of histone deacetylation inhibitors (HDACi) has shown promising benefits in their use in clinical trials of muscular diseases. However, the complete application of using satellite stem cells in the clinic is still not achieved. While therapeutic advancements in the use of HDACi in clinical trials have emerged, histone methylation modulations and the long non-coding RNA (lncRNA) are still under study. A comprehensive understanding of these other significant epigenetic modulations is still incomplete. This review aims to discuss some of the current studies on these two significant epigenetic modulations, histone methylation and lncRNA, as potential epigenetic targets in skeletal muscle regeneration. Understanding the mechanisms that initiate myoblast differentiation from its proliferative state to generate new muscle fibres will provide valuable information to advance the field of regenerative medicine and stem cell transplant.
Introduction
Skeletal muscle cells were once understood to have locomotive function only, but advances in medicine and research have shown their essential role beyond just locomotive function. Various studies have documented their crucial role as an endocrine organ that secretes various proteins, like myokines, that regulate energy production and consumption (Iizuka et al., 2014; Giudice and Taylor, 2017). They are also believed to possess anti-tumour protective properties (Stölting et al., 2013). Also, there is a compelling association between various diseases and skeletal muscles, including HIV, neuromuscular diseases, cancer, heart failure, and chronic infectious diseases that induce muscle atrophy, known as cachexia. Muscle loss during cachexia is a malignant condition associated with increased morbidity and mortality (Powers et al., 2016). Finally, skeletal muscles have remarkable regenerative properties that make them especially intriguing to researchers in treating muscular injury and degenerative muscular diseases like Duchenne muscular dystrophy (DMD). Skeletal muscle’s high regenerative capacity is due to their large pool of stem cells called satellite stem cells, which reside beneath the basal lamina of muscle cells (Chang and Rudnicki, 2014). DMD is a genetic disorder caused by mutations in the gene encoding dystrophin characterised by progressive degeneration of muscle cells and weakness. Patridge et al. were the first to explore the restoration of dystrophin using myoblast transplantation in mdx mice (Partridge et al., 1989). Their promising results initiated the cascade of clinical trials in muscular dystrophies (Huard et al., 1991; Gussoni et al., 1992; Karpati et al., 1993; Mendell et al., 1995). However, there were still many limitations encountered with the allogeneic transplantation of satellite stem cells. Problems encountered included apoptosis of transplanted myoblast cells, poor migration with intramuscular injection, and host immune rejection. Inoculation with a high number of cells resolved the short-term survival rate and the low migration limitiation (Skuk et al., 1999). However, inoculation with a high number of stem cells increases the body’s stress response which activates a strong immune response and further causes a reduction in the oxygen and nutrition supply of the body (Kuang and Rudnicki, 2008). However, these failing limitations did not discourage the search into exploring more improved therapies in the use of satellite stem cell therapy. Various studies have managed to isolate quiescent satellite cells and successfully differentiate these cells into skeletal muscle cells (Collins et al., 2005; Montarras et al., 2005). Therefore, for satellite stem cell transplantation in regenerative medicine to have a successful clinical application, it is imperative to understand the underlying mechanisms that govern these cells from their quiescent state into fully matured myofiber cells.
The epigenetic control of gene expression is a major regulator of determining satellite stem cell fate via modifying the chromatin structure. This review revises two essential epigenetic regulators, histone methylation and long non-coding RNA (lncRNA) modulation. Histone modification and lncRNA modulation have shown progressive studies in the regulation of myogenesis and appear to be promising potential new targets in degenerative muscle diseases and muscular repair.
Overview of Skeletal Muscle Development
The initiation of skeletal muscle formation is established by the lineage commitment of satellite stem cells. These satellite stem cells remain quiescent, residing between the basal lamina and sarcolemma of myofibers until they are activated (Mauro, 1961). Upon muscle injury, satellite stem cells are activated and undergo a series of events, including myoblast proliferation, cell migration and differentiation, myotube fusion, and maturation of myofibers (Biressi et al., 2007). These events are regulated by myogenic regulatory transcription factors (MRFs), including MyoD, Myf5, myogenin (MyoG), and MRF4 (Sabourin and Rudnicki, 2000). The various expression markers characterise the different developmental stages of satellite and skeletal muscle cells. During embryogenesis, Pax3 and Pax7 characterise satellite cells, which are derived from embryonic progenitor cells (Relaix et al., 2005; Schienda et al., 2006). In the post-natal life, Pax3 is downregulated, and Pax7 assumes the dominant role of the quiescent adult satellite stem cells (Oustanina et al., 2004; Relaix et al., 2006). After muscle injury, these quiescent adult satellite stem cells dominantly expressing Pax7 are activated, and the cells start to express Myf5, which induces cell proliferation. Deletion of Pax7 resulted in decreased myogenic cells during cell culture due to defects in proliferation (Relaix et al., 2006). Therefore, Pax7 is believed to retain a significant role in the quiescent state of satellite stem cells and the proliferation state. Furthermore, Pax7+/Myf5+ cells are necessary to induce the myogenic marker, MyoD, which withdraws the cell from the cell cycle and initiates myotube cell differentiation (Wilson and Rotwein, 2006). MyoD and Myf5 further have the potential in converting non-myogenic cells, like fibroblast, into muscle cells (Davis et al., 1987; Choi et al., 1990).
MyoD is considered a master regulator of myogenesis for its functional role as an initiator of muscle cell differentiation. MyoD is responsible for activating essential muscle-specific genes of differentiation, such as MyHC, α-actin, troponin isoforms (Sabourin and Rudnicki, 2000), and MyoG (Bergstrom et al., 2002; Beylkin et al., 2006). MyoG initiates myotube fusion (Bergstrom et al., 2002), followed by the maturation of muscle fibre under the control of MRF4 (Megeney and Rudnicki, 1995). The role of MRF4 is not as direct and distinct as the other MRF members and seems to involve a more complex function. MRF4 appears to have a biphasic expression, whereby its expression is activated following the activation of Myf5 during myoblast proliferation and further reactivated during the differentiation phase (Zhu and Miller, 1997; Summerbell et al., 2002; Kassar-Duchossoy et al., 2004). Another study also found that MRF4 and MyoD likely have antagonistic roles during myogenesis (Jin et al., 2007). MRF4 was found to downregulate MyoD to allow for cell proliferation (Jin et al., 2007). However, forced expression of MyoD could inhibit MRF4 and induce partial myoblast differentiation (Jin et al., 2007). In addition, during myoblast differentiation, the overexpression of MRF4 could compensate for myogenin mutant cells (Zhu and Miller, 1997; Sumariwalla and Klein, 2001).
Finally, MRFs are believed to function synergistically with the myocyte enhancer factor-2 (MEF2) members to activate myotube formation (Molkentin et al., 1995; Black et al., 1998; Piasecka et al., 2021). Taken together, after muscle injury, Pax7 is downregulated, and the upstream factors, Myf5 and MyoD, determine cell lineage to initiate myogenic differentiation. After that, MyoG and MRF4 expression ensue that function in myotube formation and maturation. Finally, MEF2 synergistically functions together with MRFs to drive myogenesis during muscular regeneration. Figure 1 provides an overview of the development of skeletal muscles from a quiescent state to matured myofibers.
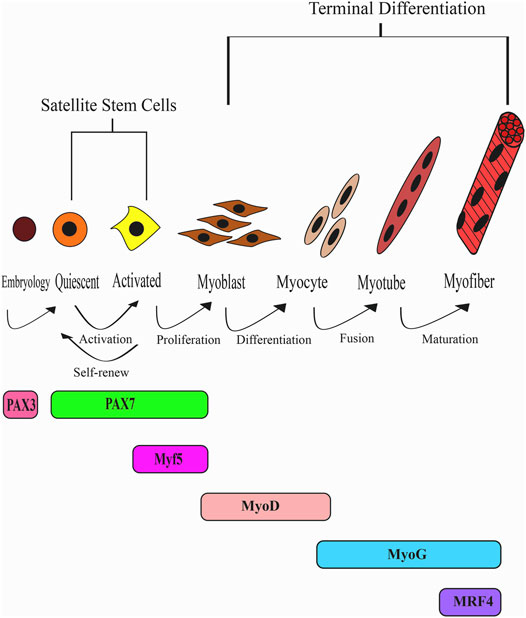
FIGURE 1. The myogenic regulatory factors (MRFs) pathway during skeletal muscle development and regeneration. Satellite stem cells remain in a quiescent state until activation upon muscle injury. Quiescent satellite stem cells expressing Pax7 are activated to committed progenitor cells expressing Myf5. However, a portion of the cells undergoesself-renewal to regenerate the stem cell pool. Proliferating myoblasts express Myf5 and MyoD while Pax7 is downregulated. MyoD is the master regulator of myoblast differentiation that initiates proliferating myoblast cells to exit the cell cycle. Differentiation and fusion precede to form myofibers under the regulation of MyoG and Mrf4.
Histone Modulation During Skeletal Muscle Cell Development
The eukaryotic chromatin is made up of nucleosomes composing of tightly wrapped DNA and histone proteins. Each nucleosome comprises an octamer of histones composed of two molecules of the canonical histone proteins, including H2A, H2B, H3, and H4 and about 150bp of DNA (Luger et al., 1997). The N-terminal histone tails are exposed on the surface of the nucleosome and are susceptible to post-translational modification via acetylation, methylation, phosphorylation, and ubiquitylation (Zhang et al., 2015). Post-translational histone modifications regulate the chromatin structure for proper gene regulation and transcription. Transcriptional regulators, including Nanog, Oct4, and Sox2 that maintain the pluripotency of stem cells, must be deactivated for cellular differentiation to occur (Wang et al., 2012; Chu et al., 2016). In addition to transcriptional regulators, induced pluripotent stem cells (iPCS) and embryonic stem cells (ESC) are poised by a bivalent chromatin, consisting of histone 3 lysine 4-trimethylation (H3K4me3) and histone 3 lysine 27-trimethylation (H3K27me3) (Bernstein et al., 2006; Mikkelsen et al., 2007). These bivalent marks are located on the same nucleosome in an asymmetric configuration regulating the chromatin structure (Bernstein et al., 2006). Generally, H3K4me3 is associated with gene activation, while H3K9me3 and H3K27me3 are associated with gene repression (Black et al., 2012). Upon receivingdifferentiation cues, the H3K27me3 repressive mark is lost, and H3K4me3 activity dominates the promoter region lineage-specific genes to activate gene transcription (Collinson et al., 2016). The bivalent gene is essential to regulate the transition between pluripotency and committed cells. Moreover, the H3K27me3 repressive mark of the bivalent gene maintains low expression levels of developmental genes in iPCS and ESC while allowing for their transcription upon differentiation. Furthermore, H3K27me3 protects the cell from aberrant gene activation by permitting only the target lineage-regulating genes to activate (Vastenhouw and Schier, 2012). H3K4me3 has vital significanceas a transcriptional activator. In addition to this main function, H3K4me3 may be essential to ensure that permanent gene silencing of developmental genes does not occur (Fouse et al., 2008; Vastenhouw and Schier, 2012).
During their quiescent state, satellite stem cells possess a similar bivalent chromatin (Pan et al., 2007; Liu et al., 2013; Akiyama et al., 2017). Interestingly, using ChiP-seq analysis, Liu et al. (Liu et al., 2013) found that the large number of genes containing H3K27me3 in quiescent satellite cells were non-myogenic genes and that the myogenic genes lacked H3K27me3. Furthermore, the knockout of the polycomb repressor complex 2 (PRC2)-Ezh2, responsible for the deposition of H3K27me3, in myogenic progenitors resulted in the activation of the non-myogenic gene lineages (Juan et al., 2011). This observation further highlights the relevance of H3K27me3 of the bivalent chromatin to suppress the activation of alternative lineage regulators, thus regulating cell fate.
For differentiation of skeletal muscle cells to commence, three requirements need to be met: 1) Loss of repressive marks on the chromatin; 2) Activation of the permissive marks to promote gene expression; 3) Promotion of RNA PolII on the promoter of the chromatin.
Upon activation of satellite stem cells after muscle injury, they undergo one of the two fates, symmetrical cell division or asymmetric cell division. In symmetric cell division, satellite cells self-renew by giving rise to identical cells that re-enter the cell cycle to maintain the quiescent pool of satellite stem cells. Contrarily, asymmetrical cell division results in satellite stem cells giving rise to two daughter cells, one identical satellite stem cell and one committed cell that will undergo proliferation (Kuang et al., 2007). Demethylation of H3K27me3 is mediated by the Jummji C (JmjC) domain-containing demethylases UTX andJMJD3 (Agger et al., 2007; Shpargel et al., 2014). Interestingly, proceeding studies found that it is mainly the upregulation of JMJD3 and not UTX that results in H3K27me3 demethylation to drive the differentiation into committed myogenic cells (Akiyama et al., 2016). Upon the loss of H3K27me3, H3K4me3 activity is associated with euchromatin and promotes RNA polymerase II to the chromatin (Pekowska et al., 2011). This permits a less compacted chromatin structure that allows for gene transcriptional expression. These underlying mechanisms are discussed below:
Histone Modification in Muscle Development and Regeneration
H3K27 Modulation
The bivalent gene consisting of H3K27me3 and H3K4me3 (Bernstein et al., 2006) is associated with the heterochromatin because the H3K27me3 repressive mark in developmental genes maintains dominance. H3K27me3 is present in myoblasts and myotubes and regulates differentiation by silencing muscle-specific genes (Asp et al., 2011), including MyoD and MyoG (Dilworth and Blais, 2011). Upon satellite stem cell activation and lineage commitment, H3K27me3 silences differentiation muscle-specific genes, thus allowing cell proliferation. The Polycomb group proteins (PcG) consisting of the KMT6 family is responsible for depositing H3K27me3 on target promoter regions. The PcG protein Ezh2 is a subunit of the polycomb repressor complex 2 (PRC2) recruited on the promoter of regulatory genes and catalyses H3K27me3 resulting in gene inactivation (Palacios et al., 2010). The mechanism involves the phosphorylation of Ezh2 by MAPK signalling (p38), which permits YY1 to recruit the phosphorylated Ezh2 to promoter regions (Palacios et al., 2010). It is further believed that additional factors, such as the histone demethylase Jarid2, are also required to recruit Ezh2 to the promoter of target genes (Peng et al., 2009; Pasini et al., 2010).
Muscle cell differentiation requires the loss of H3K27me3 from promoter regions of differentiation regulating genes like MyoG. Various mechanisms have been identified in this process. The KDM6 family member, UTX, was reported to function in a two-step mechanism to demethylate H3K27me3 at target muscle genes (Seenundun et al., 2010). Firstly, the homeobox protein Six4 recruits UTX to regulatory regions of target genes, including MyoG, and the demethylation of H3K27me3 occurs at localised regions. Upon UTX association with a specific locus, H3K27me3 demethylation activity requires the elongation of RNA PolII to spread across the genome (Seenundun et al., 2010). Interestingly, this study also found that inhibition of the RNA Pol II elongation resulted in establishing the bivalent H3K27me3/H3K4me3 gene with increased Ezh2 and Suz12 expression and loss of UTX within the coding regions. The inhibition further resulted in the loss of MyoG expression.
Msk1 kinase has also been found to play a significant role in eliminating H3K27me3 from muscle-specific genes via the phosphorylation of histone three serine 28 (H3S28) (Stojic et al., 2011). This association depreciates Ezh2 interaction on promoter regions of target genes and instead favours binding of Ezh1, which possess weaker H3K27me3 activity (Margueron et al., 2008). Furthermore, the presence of Ezh1 is required for the recruitment of RNA Pol II to MyoG for transcription (Mousavi et al., 2012). Finally, UTX is then introduced to remove H3K27me3, including demethylating the weak marks established by Ezh1, thus ensuring complete loss of the repressive mark on the chromatin of muscle for gene activation (Seenundun et al., 2010).
The incorporation of histone variants H3.3 into differentiation-specific genes is necessary for gene activation (Harada et al., 2012). Furthermore, H3.3 and H3.1 appear to possess antagonistic effects on the state of H3K27me3 with knockdown of H3.3 decreasing H3K27me3 while expression of H3.1 increased H3K27me3 inhibiting myoblast differention (Harada et al., 2015). MEF2 causes a shift from H3.1 to H3.3 via the histone chaperon HIRA, thus eliminating H3K27me3 repression for gene activation (Yang et al., 2011).
Once the H3K27me3 repressive mark is removed, activation of TrxG complex (Ash2L) via Mef2d and Six1 is then allowed, which deposits trimethylation on H3K4, resulting in a euchromatin structure (Rampalli et al., 2007).
H3K9 Modulation
The Suv391H1 methyltransferase catalyses the di- and tri-methylation of H3K9 (Rea et al., 2000; Fritsch et al., 2010). Suv391H1 is recruited via interaction with MyoD to the promoter regions of muscle target genes resulting in H3K9me3 and gene repression (Mal and Harter, 2003). The mechanisms that modulate Suv391H1-MyoD interaction involves the phosphorylation of MyoD by p38γ MAPK (Mal, 2006). Various studies have confirmed that during the proliferation phase of myoblast cells, MyoD recruits H3K9me2 and H3K9me3 repressive marks on the promoter region of MyoG, hence, inhibiting myoblast differention (Zhang et al., 2002; Mal and Harter, 2003). Furthermore, in C2C12 cell lines, overexpressing Suv391H1 continues to inhibit differentiation-specific genes even in differentiation-inducing media (Mal, 2006). Deposition of H3K9me3 by Suv39h1 appear to repress the early muscle genes (Mal, 2006), while deposition of H3K27me3 by Ezh2 functions on repressing the late genes of undifferentiated cells (Caretti et al., 2004). In addition to maintaining myoblast cells in the proliferative phase, Suv39h1 also has a functional role in mediating muscle-specific genes involved during terminal differentiation (Ait-Si-Ali et al., 2004).
G9a is a member of the SET domain-containing Suv39 family (Tachibana et al., 2001) that catalyses the methylation of H3K9 (Tachibana et al., 2002; Yokochi et al., 2009; Ling et al., 2012). During myoblast proliferation, G9a deposits H3K9me2 on MyoD, repressing its transcriptional activity (Ling et al., 2012; Wang and Abate-Shen, 2012). This is achieved via a homeodomain repressor, Msk1, interaction with G9a on the Myod1 locus (Lee et al., 2004). Interaction of Msk1 with G9a leads to the deposition of the H3K9me2 repressive mark and inhibition of differention (Wang and Abate-Shen, 2012). However, the extensive function of G9a in skeletal muscles is still under investigation as one study reported on its redundant function, observing no phenotypic change after G9a knockout in mice skeletal muscles (Zhang et al., 2016).
Choi et al. (Choi et al., 2010) were the first group to report on the demethylation mechanisms of histones in myogenesis. MEF2 recruits the histone demethylase LSD1 that removes the H3K9me2 and H3K9me3 repressive marks from promoter regions of muscle-specific genes to promote muscle cell differentiation (Choi et al., 2010). Studies found that inhibition of LSD1 maintained the H3K9me2 repressive mark on the promoter regions of MyoG and MCK, inhibiting myoblast differentiation (Choi et al., 2010). JMJD2A (Kdm4a) is also required to remove the Suv39h1 mediated H3K9 methylation, and it is via JMJD2A mechanisms that LSD1 appears to be facilitated (Verrier et al., 2011). KDM4B is another H3K9 demethylation enzyme that acts on the promoter region of MyoD. However, unlike LSD1, which is essential for myogenic differentiation, KDM4B appears only to enhance myogenic differentiation as its depletion did not completely block myogenic differentiation but instead delayed it (Choi et al., 2015). Interestingly, H3K9 and H3K4 methylation appear to be mutually exclusive (Wang et al., 2001). Thus, to maintain the demethylation of H3K9, the Set7/9 methyltransferase enzymes must deposit H3K4me1 on MyoG to ensure that H3K9me3 is not re-introduced (Tao et al., 2011).
H3K4 Modulation
Myoblast differentiation requires the loss of repressive marks and the addition of permissive marks at promoter regions of muscle genes. There are four well-established permissive markers of myogenic differentiation, and these include H3K4me1, H3K27ac, p300, and RNA II polymerase (Blum et al., 2012). The active enhancer H3K4me1 deposition is regulated by H3-H4 histone methyltransferase Set7 (Tao et al., 2011). Studies found that Set7 directly interacts with Myod to regulate gene expression (Tao et al., 2011). As previously mentioned in the last section, the depletion of the H3K9me3 repressive mark appears to be mutually exclusive with the introduction of H3K4me1 by Set7/987. Indeed, inhibition of Set7 resulted in decreased H3K4me1 levels and MEF2 expression, impairing skeletal muscle differentiation (Tao et al., 2011). Furthermore, the inhibition of Set7 resulted in decreased levels of H3K4me1 with a loss of MyoD, MyHC, and MyoG expression (Wang et al., 2001; Nishioka et al., 2002; Tao et al., 2011). This ultimately resulted in a decreased number of myotube formation and impaired skeletal muscle differentiation (Tao et al., 2011).
At active promoters, H3K4me1 flanks with H3K4me3 in a bimodal pattern (Bae and Lesch, 2020). The H3K4me3 permissive mark in satellite stem cells is established via TrxG complexes (Ash2L/MLL2 methyltransferases) (McKinnell et al., 2008; Diao et al., 2012; Kawabe et al., 2012). The transcriptional factor Pax7 mediates the recruitment of Ash2L/MLL2 through binding upstream of the Myf5 transcriptional start site (TSS) (McKinnell et al., 2008; Soleimani et al., 2012). The methylation of arginine residues on the amino-terminus of Pax7 by arginine methyltransferase CARM1 is another factor that recruits TrxG complexes (McKinnell et al., 2008; Kawabe et al., 2012). By this methylation of Pax7 by CARM1, Pax7 can interact with Ash2L/MLL2 to target the Myf5 promoter region to deposit H3K4me3 and activate gene expression for myoblast proliferation (McKinnell et al., 2008; Diao et al., 2012; Kawabe et al., 2012). Similarly, Ash2L/MLL2 also targets the MyoG promoter region to deposit H3K4me3 and initiate proliferating myoblasts to commit to differentiation (Rampalli et al., 2007). Ash2L/MLL2 is recruited to the promoter region of MyoG via interactions with phosphorylated MEF2D by p38-α (Rampalli et al., 2007).
Furthermore, studies found that H3K4me3 co-localizes with the KDM5B demethylase enzyme at promoter regions of active genes (Kidder et al., 2014; Xue et al., 2020). Inhibition of KDM5B resulted in widespread H3K4 methylation into gene bodies, leading to defects in gene expression and impairment in the self-renewal and differentiation of embryonic cells (Kidder et al., 2014). Thus, it is proposed that KDM5B has significance in localising H3K4 methylation at the promoter and enhancer regions of relevant genes (Kidder et al., 2014; Xue et al., 2020). Whether similar regulatory mechanisms of KDM5B in muscle-regulated genes also exist is yet to be explored.
PARP1, a member of the Poly (ADP-ribose) polymerases family, has also been found to coordinate MyoD gene expression by regulating H3K4me3 (Matteini et al., 2020). In skeletal muscle cells, PARP1 impairs the accumulation of the permissive marker, H3K4me3, on the MyoD binding site, which inhibits myogenic gene expression. It is postulated that PARP1 directly interacts with KvDMR1 (Matteini et al., 2020), an imprinting control region necessary for MyoD regulation of muscle cell differentition (Busanello et al., 2012; Andresini et al., 2016). Interestingly, PARP1 appears to possess a cell-specific function in the regulation of H3K4me3. In its active state, PARP1 inhibits the H3K4me3 demethylase enzyme, KDM5B, and increases H3K4me3 in MCF-7 cells (Krishnakumar and Kraus, 2010). In contrast, in the HEK293T cell lines, the inactive state of PARP1 inhibited H3K4me3 in IL-6 by hindering methyltransferase MLL (Minotti et al., 2015).
Other Histone-Methyl Modulation
SUV4-20H1 is a dimethyltransferase that mediates the deposition of H4K20me3 (Boonsanay et al., 2016). Deposition of H4K20me2/3 has significance in maintaining satellite stem cell quiescent state (Boonsanay et al., 2016). One study demonstrated that SUV4-20H1 appears to deposit H4K20me2 on the MyoD locus and thus, inhibiting myoblast differentiation (Boonsanay et al., 2016). Inhibiting SUV4-20H1 decreases H4K20me2 expression on the MyoD distal regulatory region (DRR) and increases the permissive mark H3K4me3 (Boonsanay et al., 2016). This resulted in the activation of muscle stem cells with significant depletion of the quiescent stem cells impairing muscle cell regeneration capacity after muscular injury. Interestingly, the deletion of SUV4-20H1 led to the decreased expression levels of H3K20me2 and significantly decreased H3K27me3 levels (Boonsanay et al., 2016).
The SET domain containing 2 (Setd2) is an H3K36me3 that modulates the chromatin to an active state (Kouzarides, 2007; Edmunds et al., 2008). It was found that inhibition of Setd2 in the C2C12 cell line resulted in aberration in myotube formation with decreased expression of MyHC and MyoG (Yi et al., 2017). Interestingly, silencing of Setd2 did not appear to affect MyoD, Myf5, and MRF4 (Yi et al., 2017). Furthermore, silencing of Setd2 arrested the cell cycle and upregulated cyclin-dependent kinase inhibitor p21 resulting in decreased proliferation rate (Yi et al., 2017). The mechanisms involved are yet to be identified.
As the continual exploration into new epigenetic regulatory mechanisms is underway, these observations demonstrate the significant role that histone modifications play in muscle cell development and regeneration. From maintaining the quiescent stem cell pool through muscle cell activation and maturation of muscle cells, histone modifications may provide new novel target therapy in muscle dystrophies. Figure 2 provides a comprehensive summary of the current theories of histone methyltransferase modulations during skeletal muscle development.
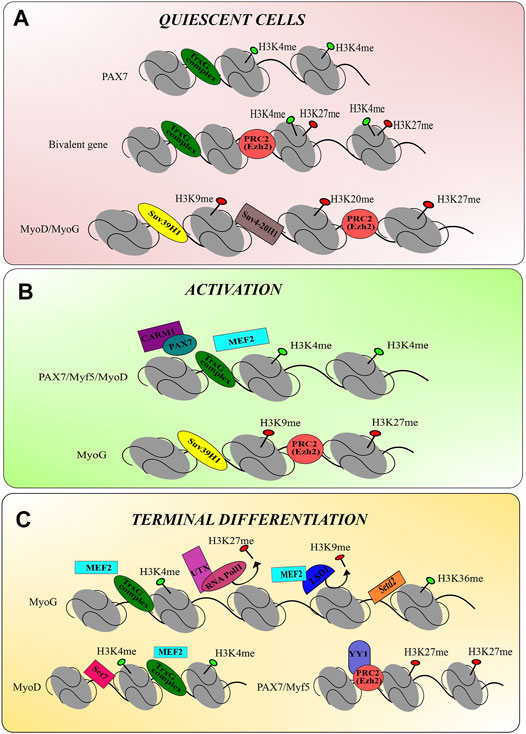
FIGURE 2. The epigenetic modulation of histone methyltransferases during skeletal muscle regeneration. (A) Epigenetic mechanisms of histone methyltransferases in quiescent stem cells. TrxG-mediated H3K4me catalyses the permissive chromatin of Pax7. MyoD and MyoG gene expression is repressed by H3K27me, H3K9me, and H3K20me via their respective enzymes. Interestingly, the bivalent gene consists of an activating H3K4me mark and the repressive H3K27me, however, H3K27me activity dominates to repress gene transcription during the quiescent state. (B) Epigenetic mechanisms of histone methyltransferases in activating progenitor cells. Upon the activation of stem cells, the permissive chromatin of Pax7, Myf5 and MyoD is catalysed by deposition of H3K4me via TrxG complexes. MEF2 and Carm-Pax7 complex recruits TrxG complexes to establish the permissive chromatin. MyoG remains with the repressive chromatin marks, H3K27me and H3K9me, catalysed by PRC2 and SUV39H1, respectively. (C) Epigenetic mechanisms of histone methyltransferases in during terminal differentiation. The MyoG chromatin permits gene transcription via H3K4me and H3K36me marks by TrxG complexes and Setd2, respectively. UTX and RNA PolII remove the repressive mark H3K27me, and MEF2 recruits LSD1 to remove the H3K9me repressive mark. MyoD gene transcription continues via the activating H3K4me. Pax7 and Myf5 are silenced by recruitment of PRC2 by YY1, which deposits the H3K27me repressive mark.
LncRNA in Myogenesis
Long non-coding-RNA (lncRNA) are a class of non-coding RNA molecules with more than 200 nucloetides (Engreitz et al., 2016). In epigenetics, lncRNAs have been reported with various functions. This includes: 1) mediating chromatin remodelling and modification (McHugh et al., 2015); 2) interacting with transcriptional factors for gene regulation (Atianand et al., 2016); 3) interacting with mRNAs to regulate the post-transcriptional processes of mRNAs (Tripathi et al., 2010; Wang et al., 2016). Recent studies have found that lncRNAs have significant roles in coordinating muscle-specific transcriptional factors during the different phases of myogenesis. LncRNAs direct epigenetic regulators to mediate MRFs during muscle cell development and regeneration (Dey et al., 2014; Wang et al., 2016; Chen et al., 2017; Xu et al., 2017). Furthermore, lncRNAs coordinate with muscle cell regeneration regulatory factors after contusion injury to regenerate muscle cells (Zheng et al., 2019). Amongst their diverse functions with other molecular factors, they also possess a cell-type-specific expression characteristic that makes them vital cell determination and commitment (Cesana et al., 2011; Wang et al., 2013). Below, we present lncRNAs that have been of recent interest in muscle cell development and regeneration.
H19
LncRNA H19 was one of the first lncRNAs identified with a crucial role in skeletal muscle. It promotes muscle cell differentiation and functions in skeletal muscle regeneration after injury (Dey et al., 2014). The mechanisms by which H19 works to promote skeletal muscle regeneration have been identified to involve inhibiting the myoblast inhibitory genes, Sirt1 and FoxO1, that leads to increased expression of MyoG and MyHC (Xu et al., 2017). Also, H19 is reported to encode two microRNAs (miRNA), miR-675-3p and miR-675-5p, which promote skeletal muscle cell differentiation (Dey et al., 2014). MiR-675-3p and miR-675-5p target the anti-differentiation factors, Smad proteins and Cdc6, respectively (Dey et al., 2014). MiR-675-3p and miR-675-5p are able to rescue abnormal muscle regeneration in H19 knockout (Dey et al., 2014). The significance of H19 during skeletal muscle regeneration was also evident after skeletal muscle injury in mice, where its expression was significantly increased during the early phase of muscle repair (Zheng et al., 2019). Further, the increase in H19 expression positively correlated with Myf5, MyoG and the angiogenic factors, HIF-1α and Angpt1 (Zheng et al., 2019). Interestingly, one study presented the discovery of a negative-feedback loop between H19-Igf2-MyoD (Borensztein et al., 2013). In this study, the knockdown of MyoD and Igf2 in mice models affected the development of muscles of the diaphragm (Borensztein et al., 2013). They reported that MyoD binds to the CS9 mesodermal enhancer of H19, thereby interacting with the promoter region of H19. This interaction increases the expression of H19, which results in trans repression of Igf2 (Wilkin et al., 2000). The repression of Igf2 further results in the negative regulation of MyoD via the suppression of the SRF transcription factor (Borensztein et al., 2013). Thus, H19 is essential for the development of muscle cells via regulation of key muscle development factors and pathways, including, Sirt1, FoxO1, microRNAs, and Igf2.
LncMyoD
During skeletal muscle differentiation, MyoD activates lncMyoD (Gong et al., 2015) and lnc-YY1 (Zhou et al., 2015) and promote skeletal muscle differentiation. Increased expression of LncMyoD is observed during muscle regeneration and development, and its knockdown negatively regulates a key number of muscle-related mRNAs (Gong et al., 2015). The mechanisms involve the binding of lncMyoD to IGF2-mRNA-binding protein 2 (IMP2), which negatively regulates the proliferation genes, N-Ras and c-Myc (Gong et al., 2015). The negative regulation of these genes then permits exit of the cell-cycle, thus, promoting myoblast differentiation (Gong et al., 2015). In addition to promoting myoblast differentiation, lncMyoD further has a regulatory role in the composition of muscle fibre-type (Zhang et al., 2021). Like most lncRNAs, lncMyoD also has the property of sponging miRNAs to activate transcription. Via the sponging of miR-370-3p, lncMyoD can regulate muscle fibre type composition (Zhang et al., 2021). LncMyoD promotes differentiation with an increased enhancement of fast muscle fibre while decreasing slow muscle fibre (Zhang et al., 2021). Correspondingly, in vitro studies in C2C12 cell lines revealed that the inhibition of lncMyoD upregulated miR-370-3p expression inhibiting myoblast differentiation, increasing slow muscle fibre formation, and decreasing fast muscle fibre formation. Furthermore, lncMyoD could rescue the inhibitory effects of miR-370-3p to promote myotube differentiation and switch to the formation of fast muscle fibres (Zhang et al., 2021). Conclusively, lncMyoD inhibits myoblast proliferation while promoting differentiation and regulates muscle fibre composition.
LncYY1
Lnc-YY1 is activated by MyoD and possess regulatory functions to activate myotube differentiation. Upon the activation of MyoD, lncYY1 destabilises the YY1/PRC2 repressive complex from the promoter region of target genes, which frees the chromatin and allows for gene activation (Zhou et al., 2015). During muscle regeneration, inhibition of lncYY1 decreased expression of Pax7, MyoD, MyoG, and MyHC (Zhou et al., 2015). Furthermore, together with lncRNA H19, lncYY1 shows a more prominent role during the early phase of skeletal muscle regeneration (Zheng et al., 2019). The expression pattern of lncYY1 during myogenesis display low expression in proliferating myotubes, peak expression during the early phase of myotube differentiation, and decreased expression during the late differentiation phase (Zhou et al., 2015). Studies found that inhibition of H19 or lncYY resulted in decreased expression of MyoD and MyoG, leading to the aberration of skeletal muscle repair post-injury (Cesana et al., 2011; Dey et al., 2014; Zhou et al., 2015). Therefore, lncYY1 may regulate significant key pathways necessary for myotube differentiation via MyoD and MyoG-related mechanisms.
Malat1
Metastasis-associated lung adenocarcinoma transcript 1 (Malat1) was initially identified in non-small cell lung cancer (NSCLC) as a prognostic marker (Ji et al., 2003). Watts et al. (Watts et al., 2013) were the first to report on the role of Malat1 in myogenesis. They found that Malat1 expression was upregulated during myoblast differentiation. Silencing of Malat1 resulted in decreased expression of MyoG and decreased myotube differentiation. The in vivo and in vitro studies further revealed that Malat1 was suppressed by myostatin, a negative regulator of myogenesis (Watts et al., 2013). Similarly to most lncRNAs, Malat1 functions as a sponge for miRNAs in the regulation of myogenesis. Malat1 consists of a miR-133 target region that targets serum response factor (SRF) to promote myoblast differentiation (Han et al., 2015). The inhibition of Malat1 resulted in decreased expression of SRF and inhibited differentiation during in vitro studies of C2C12 cells (Han et al., 2015). Further, inhibition of Malat1 arrested the cell-cycle in G0/G1 phase affecting myoblast proliferation and decreased MyoG expression levels (Watts et al., 2013). However, a contrasting study observed Malat1 acting as a repressor of myogenesis (Chen et al., 2017). In this study, Malat1 recruited Suv391h1 on the Myod1 loci during myotube proliferation. Differentiation cues degraded Malat1, destabilising the Suv391ha-complex resulting in the activation of MyoD. Furthermore, inhibition of Malat1 during in vivo studies of the mdx mouse model of muscular dystrophy enhanced muscle regeneration capacity, increasing muscle fibres (Chen et al., 2017). Similarly, during skeletal muscle repair after contusion injury, there was a parallel interaction between Malat1 expression with MyoD and MyoG (Zheng et al., 2019). These contrasting observations further make Malat1 an intriguing investigation because even with its abundant expression in all human tissues, its knockdown does not exhibit any obvious phenotypical aberration during development (Eißmann et al., 2012; Nakagawa et al., 2012; Zhang et al., 2012). Beyond myotube formation, Malat1 is further implicated in regulating the inflammatory response during skeletal muscle repair post-injury (Marques-Rocha et al., 2015; Zhao et al., 2016; Huang et al., 2017). There are indeed many inflammatory cytokines, including TNF-α, IL-6, and IL-10, that were found to positively correlate with Malat1 post-muscular injury (Zheng et al., 2019). Malat1 appears to regulate inflammatory activities via the recruitment of EZH2 (Yong et al., 2020). During sepsis of muscle tissue, Malat1 interacted with EZH2 to increase inflammatory cells and increase muscle cell apoptosis. In summary, Malat1 appears to have inconclusive yet a regulatory role during myogenesis with significant influence during the inflammatory response of skeletal muscle repair.
Myparr
The lncRNA Myparr is transcribed by the MyoG promoter region (Hitachi et al., 2019). Myparr functions to regulate myogenesis by two mechanisms. Firstly, Myparr activates the myogenic regulatory miRNAs to withdraw the myoblast cell from the cell-cycle and initiate myoblast differentiation (Hitachi et al., 2019). Secondly, Myparr regulates MyoG to promote myoblast differentiation (Hitachi et al., 2019). Studies found that silencing of Myparr resulted in decreased occupancy of PolII and a significant reduction in the permissive marks, H3K4me3 and H3K27ac, on the MyoG locus. The underlying mechanisms involve regulating the interaction of Ddx17, a coactivator of MyoD, with PCAF proteins (Hitachi et al., 2019). Interestingly, there also appears to exist a positive-feedback between Myparr and MRFs. During the inhibition of MyoG, compensatory mechanisms enhanced the expression of Myparr, together with Myf5 and MRF4 (Hitachi et al., 2019).
As summarised in Figure 3, there are several emerging evidence of the role of lncRNAs during myoblast differentiation into matured muscle cells. LncRNAs possesses great potential as effective targets during muscular regeneration. It is worth exploring further for their upstream regulations during myogenesis for a comprehensive understanding of their potential in clinical application.
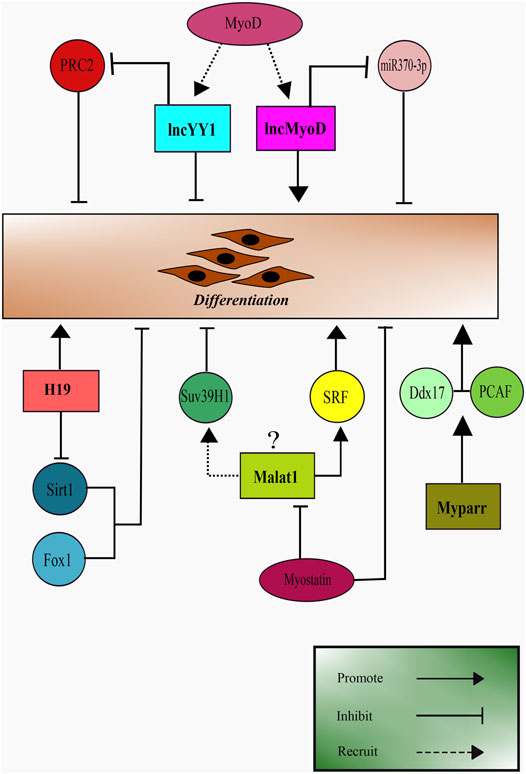
FIGURE 3. Schematic representation of LncRNAs role in skeletal muscle differentiation. LncRNA H19, lncMyoD, lncYY1, and Myparr promote myoblast differentiation. Malat1 appears to have contrasting results being a promoter of differentiation and recruiting SUV39H1, an inhibitor of differentiation.
Long Non-coding RNAs Interaction With Histone Methylation Proteins
More than just their interaction with MRFs, lncRNAs further have underlying mechanisms with epigenetic proteins, including histone methylation enzymes to regulate gene expression. Their functional roles include recruitment of histone methylation proteins and acting further as a molecular scaffold to coordinate the localisation of these epigenetic complexes (Hanly and Esteller, 2018). One of the well-documented roles of lncRNAs regulation of epigenetic modulators is their mediation of gene suppression via interaction with PRC2 (Hanly and Esteller, 2018). LncRNAs function as a molecular scaffold to coordinate the localisation and binding of PRC2 on promoter regions of target genes to deposit H3K27me3 (Rinn et al., 2007; Tsai et al., 2010; Yap et al., 2010). Furthermore, an individual lncRNA has the unique ability to harness different histone enzymes on their distinct domains. Tsai et al. (Tsai et al., 2010) found that while the 5′-end of lncRNA HOTAIR binds with the PRC2 complex, its 3′-end binds with the H3K4 histone demethylase, LSD1, to coordinate gene repression. LncRNAs were also found to recruit G9a, responsible for H3K9me3 deposition, to silence target genes in mouse placental tissue (Nagano et al., 2008) and to coordinate lineage-specific transcriptional silencing of target genes (Pandey et al., 2008). Whether the aim is to turn genes off via H3K27me3, H3K9me3, or demethylate H3K4me3, there is expanding evidence of the significant role of lncRNAs coordination gene suppressive effects. An interesting observation about the target mechanisms of lncRNAs appears to be highly cell type-specific. In relation to cancer cells, lncRNA ANRIL mediates the repression of tumour suppressor proteins via the recruitment of PRC1 and PRC2 (Aguilo et al., 2011). However, in smooth muscle cells, the levels of H3K27me3 were unaffected by the inhibition of ANRIL during the phenotypic switching of aortic smooth muscle cells (Zhang et al., 2020). Instead, ANRIL acted as a molecular scaffold for the WD-40 repeat-containing protein 5 (WDR5) and histone deacetylase HDAC3 complex to activate gene transcription via H3K4me3 (Zhang et al., 2020). In developmental genes, lncRNA HOTTIP is present at the bivalent gene, and its transcription resulted in increased H3K4me3 levels and decreased H3K27me3 (Wang et al., 2011). Similarly toANRIL, HOTTIP mediated gene activation by modulating the localisation and occupancy of WDR5 to deposit H3K4me3 (Wang et al., 2011). Furthermore, the loss of HOTTIP led to the diffusespread of MLL and WDR5 across the gene body and decreased H3K4me3 levels at the promoter region of the HOXA gene (Wang et al., 2011). These studies provide compelling evidence of the specificity of lncRNAs to modulate epigenetic events via the methylation of histone proteins. Thus, these discoveries about lncRNAs make them particularly intriguing in stem cell therapy to mediate epigenetic events during cellular differentiation.
Epigenetic Therapy in Muscle Diseases and Muscle Regeneration
The evidence above provides a strong indicator that epigenetic modulations like histone methylation and lncRNA alter gene expression. The dysregulation of any of these processes can lead to aberrations in muscular development and regeneration. Therefore, a comprehensive understanding of these epigenetic changes can advance the development of epigenetic drugs to treat muscular dystrophy diseases and enhance satellite stem cell therapy. Thus far, histone deacetylase inhibitors (HDACi) are the only epigenetic drugs that have been well studied and used in clinical trials to treat muscular dystrophies. HDACi inhibits myostatin activity, resulting in increased formation of myotubes to treat DMD (Myostatin, 2008). Givinostat is the first HDACi used in the treatment of muscle diseases. It showed a successful increase in myofiber mass and size in mdx mice (Minetti et al., 2006) and showed increased muscle tissue accompanied with decreased fibrotic tissue and necrosis in clinical trials of DMD patients (Bettica et al., 2016). However, a major challenge of using epigenetic drugs is their non-enzyme specificity effects. The coordination between methylation substrates and acetylation of several histone lysines often results in off-target enzyme activation with the use of HDACi (Huang et al., 2011; Zentner and Henikoff, 2013). Therefore, as successful as these studies are, HDACi has systematic effects that influence many gene expressions. Thus, they are often accompanied by major side effects, including nausea, neutropenia, thrombocytopenia, or ventricular arrhythmia (Suraweera et al., 2018). In contrast, lncRNAs function to regulate gene expression in a locus and allele-specific way. They appear to function in a more tissue-type specific manner (Parasramka et al., 2016).
Because of these various histone cross-talks, exploring combinatorial drug therapies between HDACi and histone methyltransferase drugs may augment therapeutic effects than serving an individual drug target. Indeed, there is a growing interest in the innovation of methyltransferase inhibitors in the clinic. EZH2 is a catalytic enzyme for polycomb repressive complex 2 (PRC2) responsible for the repressive H2K27me3 mark (Marchesi et al., 2014). At the initiation of skeletal muscle differentiation, EZH2 is displaced from promoter regions of muscle-specific genes, and KDM6a, a histone demethylase enzyme, is recruited (Seenundun et al., 2010). Further, the overexpression of EZH2 leads to impaired muscle differentiation (Caretti et al., 2004). It is thus essential to destabilise this component from target genes for myogenesis to occur. EZH2 inhibitor drugs demonstrate a high potency against EZH2 and are highly selective (Morera et al., 2016). They also exhibit tolerable side effects, including nausea, asthenia, anorexia, dyspnea, and anorexia that were of low grade (Knutson et al., 2014). Synthetic histone demethylase-mimicking drugs are also a great potential for therapeutic exploration.
In contrast, lncRNAs regulate gene expression in a locus and allele-specific way that is cell-type specific (Parasramka et al., 2016). The specificity of lncRNAs to target different epigenetic modulators at different developmental phases may be a resourceful therapeuticapproach to explore. Furthermore, the current epigenetic drug mechanisms aim to target the overexpressed aberrations of epigenetic enzymes. Because of their role to recruit and activate epigenetic enzymes, lncRNAs may possess therapeutic effects to upregulate poorly expressed epigenetic enzymes. Finally, an individual lncRNA may target only one or a small group of related proteins (Khorkova et al., 2015). Thus this makes them an exceptional approach to avert redundant gene activation.
During muscular diseases including neurogenic atrophy, ALS, spinal muscular atrophy and any loss of nerve supply, MyoG is usually upregulated to promote proteolyisis (Moresi et al., 2010; Bricceno et al., 2012; Galbiati et al., 2012; Mielcarek et al., 2015). Studies found that inhibiting Myparr in denervated muscles resulted in decreased expression of MyoG and increased muscle weight (Hitachi et al., 2019). Hence, Myparr and many other lncRNAs astherapeutic targets in skeletal muscle differentiation may further be an effective novel therapy in muscular disorders.
Future
Understanding the mechanisms that control and regulate muscle cell development and differentiation enables the research community to improve and enhance the effectiveness of satellite stem cell therapy in muscular disorders. In this review, we outlined the mechanisms of muscle development under the epigenetic control of histone modification and lncRNAs. MRFs reveal to be an excellent factors target to enhance stem cell differentiation into matured muscle fibre for muscle regeneration. Therefore, modulating MRFs via epigenetic mechanisms is showing to be a promising therapeutic approach for muscle differentiation. Histone methylation plays a vital role in modifying the chromatin to switch target genes on or off. Thus, it is a major determining factor for the expression state of genes. Further, emerging studies into lncRNAs provide cogent evidence of their regulatory roles in significant muscle-related signalling pathways. LncRNAs further have an advantageous property of their ability to control myogenesis at specific phases. Hence, they have a high specificity to relevant MRFs. Epigenetic factors do not work in isolation, and lncRNAs have been shown to have significant cross-talks with DNA methyltransferases and histone modifiers. While much progress has been made to explore many of these epigenetic factors during myogenesis, there are still limitations to employ them in clinical use. Firstly, the regulation of histone methylation and lncRNA during myogenesis have only been studied in isolation. Further in vivo and in vitro investigations are needed for the interplay between lncRNAs modulation with histone modifiers to translate to clinical use. Secondly, harnessing lncRNAs for therapeutic use is still an area that requires much exploration in order for effective clinical use. Finally, augmenting the current trial therapies of HDACi with lncRNAs and histone methylation-mimicking drugs may be a potential therapeutic approach to enhance satellite stem cell implantation and decrease the side effects. Further exploration to advance the knowledge of epigenetic modulation during stem cell activation and muscle development will ultimately progress the regenerative field of muscle cell diseases and muscle cell injury.
Author Contributions
MNM drafted and wrote the manuscript, J-xZ and J-mT revised the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the Scientific and Technological Project of Shiyan City of Hubei Province (2021K63).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Agger, K., Cloos, P. A. C., Christensen, J., Pasini, D., Rose, S., Rappsilber, J., et al. (2007). UTX and JMJD3 Are Histone H3K27 Demethylases Involved in HOX Gene Regulation and Development. Nature 449 (7163), 731–734. doi:10.1038/nature06145
Aguilo, F., Zhou, M.-M., and Walsh, M. J. (2011). Long Noncoding RNA, Polycomb, and the Ghosts Haunting INK4b-ARF-INK4a Expression: Figure 1. Cancer Res. 71 (16), 5365–5369. doi:10.1158/0008-5472.CAN-10-4379
Ait-Si-Ali, S., Guasconi, V., Fritsch, L., Yahi, H., Sekhri, R., Naguibneva, I., et al. (2004). A Suv39h-dependent Mechanism for Silencing S-phase Genes in Differentiating but Not in Cycling Cells. Embo J. 23, 605–615. doi:10.1038/sj.emboj.7600074
Akiyama, T., Wakabayashi, S., Soma, A., Sato, S., Nakatake, Y., Oda, M., et al. (2017). Epigenetic Manipulation Facilitates the Generation of Skeletal Muscle Cells from Pluripotent Stem Cells. Stem Cell Int. 2017, 1–8. doi:10.1155/2017/7215010
Akiyama, T., Wakabayashi, S., Soma, A., Sato, S., Nakatake, Y., Oda, M., et al. (2016). Transient Ectopic Expression of the Histone Demethylase JMJD3 Accelerates the Differentiation of Human Pluripotent Stem Cells. Dev 143 (20), 3674–3685. doi:10.1242/dev.139360
Andresini, O., Ciotti, A., Rossi, M. N., Battistelli, C., Carbone, M., and Maione, R. (2016). A Cross-Talk between DNA Methylation and H3 Lysine 9 Dimethylation at the KvDMR1 Region Controls the Induction of Cdkn1c in Muscle Cells. Epigenetics 11, 791–803. doi:10.1080/15592294.2016.1230576
Asp, P., Blum, R., Vethantham, V., Parisi, F., Micsinai, M., Cheng, J., et al. (2011). Genome-wide Remodeling of the Epigenetic Landscape during Myogenic Differentiation. Proc. Natl. Acad. Sci. 108, E149–E158. doi:10.1073/pnas.1102223108
Atianand, M. K., Hu, W., Satpathy, A. T., Shen, Y., Ricci, E. P., Alvarez-Dominguez, J. R., et al. (2016). A Long Noncoding RNA lincRNA-EPS Acts as a Transcriptional Brake to Restrain Inflammation. Cell 165, 1672–1685. doi:10.1016/j.cell.2016.05.075
Bae, S., and Lesch, B. J. (2020). H3K4me1 Distribution Predicts Transcription State and Poising at Promoters. Front. Cel Dev. Biol. 8. doi:10.3389/fcell.2020.00289
Bergstrom, D. A., Penn, B. H., Strand, A., Perry, R. L. S., Rudnicki, M. A., and Tapscott, S. J. (2002). Promoter-specific Regulation of MyoD Binding and Signal Transduction Cooperate to Pattern Gene Expression. Mol. Cel. 9, 587–600. doi:10.1016/S1097-2765(02)00481-1
Bernstein, B. E., Mikkelsen, T. S., Xie, X., Kamal, M., Huebert, D. J., Cuff, J., et al. (2006). A Bivalent Chromatin Structure Marks Key Developmental Genes in Embryonic Stem Cells. Cell 125, 315–326. doi:10.1016/j.cell.2006.02.041
Bettica, P., Petrini, S., D'Oria, V., D'Amico, A., Catteruccia, M., Pane, M., et al. (2016). Histological Effects of Givinostat in Boys with Duchenne Muscular Dystrophy. Neuromuscul. Disord. 26, 643–649. doi:10.1016/j.nmd.2016.07.002
Beylkin, D. H., Allen, D. L., and Leinwand, L. A. (2006). MyoD, Myf5, and the Calcineurin Pathway Activate the Developmental Myosin Heavy Chain Genes. Develop. Biol. 294, 541–553. doi:10.1016/j.ydbio.2006.02.049
Biressi, S., Molinaro, M., and Cossu, G. (2007). Cellular Heterogeneity during Vertebrate Skeletal Muscle Development. Develop. Biol. 308, 281–293. doi:10.1016/j.ydbio.2007.06.006
Black, B. L., Molkentin, J. D., and Olson, E. N. (1998). Multiple Roles for the MyoD Basic Region in Transmission of Transcriptional Activation Signals and Interaction with MEF2. Mol. Cel Biol 18, 69–77. doi:10.1128/mcb.18.1.69
Black, J. C., Van Rechem, C., and Whetstine, J. R. (2012). Histone Lysine Methylation Dynamics: Establishment, Regulation, and Biological Impact. Mol. Cel 48, 491–507. doi:10.1016/j.molcel.2012.11.006
Blum, R., Vethantham, V., Bowman, C., Rudnicki, M., and Dynlacht, B. D. (2012). Genome-wide Identification of Enhancers in Skeletal Muscle: The Role of MyoD1. Genes Dev. 26, 2763–2779. doi:10.1101/gad.200113.112
Boonsanay, V., Zhang, T., Georgieva, A., Kostin, S., Qi, H., Yuan, X., et al. (2016). Regulation of Skeletal Muscle Stem Cell Quiescence by Suv4-20h1-dependent Facultative Heterochromatin Formation. Cell Stem Cell 18, 229–242. doi:10.1016/j.stem.2015.11.002
Borensztein, M., Monnier, P., Court, F., Louault, Y., Ripoche, M.-A., Tiret, L., et al. (2013). Myod and H19-Igf2 Locus Interactions Are Required for Diaphragm Formation in the Mouse. Dev 140 (6), 1231–1239. doi:10.1242/dev.084665
Bricceno, K. V., Sampognaro, P. J., Van meerbeke, J. P., Sumner, C. J., Fischbeck, K. H., and Burnett, B. G. (2012). Histone Deacetylase Inhibition Suppresses Myogenin-dependent Atrogene Activation in Spinal Muscular Atrophy Mice. Hum. Mol. Genet. 21, 4448–4459. doi:10.1093/hmg/dds286
Busanello, A., Battistelli, C., Carbone, M., Mostocotto, C., and Maione, R. (2012). MyoD Regulates P57 Kip2 Expression by Interacting with a Distant Cis -element and Modifying a Higher Order Chromatin Structure. Nucleic Acids Res. 40, 8266–8275. doi:10.1093/nar/gks619
Caretti, G., Di Padova, M., Micales, B., Lyons, G. E., and Sartorelli, V. (2004). The Polycomb Ezh2 Methyltransferase Regulates Muscle Gene Expression and Skeletal Muscle Differentiation. Genes Dev. 18, 2627–2638. doi:10.1101/gad.1241904
Cesana, M., Cacchiarelli, D., Legnini, I., Santini, T., Sthandier, O., Chinappi, M., et al. (2011). A Long Noncoding RNA Controls Muscle Differentiation by Functioning as a Competing Endogenous RNA. Cell 147, 358–369. doi:10.1016/j.cell.2011.09.028
Chang, N. C., and Rudnicki, M. A. (2014). “Satellite Cells,” in Current Topics in Developmental Biology. (Amsterdam: Elsevier Inc), 161–181. doi:10.1016/B978-0-12-416022-4.00006-8
Chen, X., He, L., Zhao, Y., Li, Y., Zhang, S., Sun, K., et al. (2017). Malat1 Regulates Myogenic Differentiation and Muscle Regeneration through Modulating MyoD Transcriptional Activity. Cell Discov 3, 1–23. doi:10.1038/celldisc.2017.2
Choi, J., Costa, M. L., Mermelstein, C. S., Chagas, C., Holtzer, S., and Holtzer, H. (1990). MyoD Converts Primary Dermal Fibroblasts, Chondroblasts, Smooth Muscle, and Retinal Pigmented Epithelial Cells into Striated Mononucleated Myoblasts and Multinucleated Myotubes. Proc. Natl. Acad. Sci. 87, 7988–7992. doi:10.1073/pnas.87.20.7988
Choi, J. H., Song, Y. J., and Lee, H. (2015). The Histone Demethylase KDM4B Interacts with MyoD to Regulate Myogenic Differentiation in C2C12 Myoblast Cells. Biochem. Biophysical Res. Commun. 456, 872–878. doi:10.1016/j.bbrc.2014.12.061
Choi, J., Jang, H., Kim, H., Kim, S.-T., Cho, E.-J., and Youn, H.-D. (2010). Histone Demethylase LSD1 Is Required to Induce Skeletal Muscle Differentiation by Regulating Myogenic Factors. Biochem. Biophysical Res. Commun. 401 (3), 327–332. doi:10.1016/j.bbrc.2010.09.014
Chu, L.-F., Leng, N., Zhang, J., Hou, Z., Mamott, D., Vereide, D. T., et al. (2016). Single-cell RNA-Seq Reveals Novel Regulators of Human Embryonic Stem Cell Differentiation to Definitive Endoderm. Genome Biol. 17 (1). doi:10.1186/s13059-016-1033-x
Collins, C. A., Olsen, I., Zammit, P. S., Heslop, L., Petrie, A., Partridge, T. A., et al. (2005). Stem Cell Function, Self-Renewal, and Behavioral Heterogeneity of Cells from the Adult Muscle Satellite Cell Niche. Cell 122, 289–301. doi:10.1016/j.cell.2005.05.010
Collinson, A., Collier, A. J., Morgan, N. P., Sienerth, A. R., Chandra, T., Andrews, S., et al. (2016). Deletion of the Polycomb-Group Protein EZH2 Leads to Compromised Self-Renewal and Differentiation Defects in Human Embryonic Stem Cells. Cel Rep. 17 (10), 2700–2714. doi:10.1016/j.celrep.2016.11.032
Davis, R. L., Weintraub, H., and Lassar, A. B. (1987). Expression of a Single Transfected cDNA Converts Fibroblasts to Myoblasts. Cell. 51, 987–1000. doi:10.1016/0092-8674(87)90585-X
Dey, B. K., Pfeifer, K., and Dutta, A. (2014). The H19 Long Noncoding RNA Gives Rise to microRNAs miR-675-3p and miR-675-5p to Promote Skeletal Muscle Differentiation and Regeneration. Genes Dev. 28, 491–501. doi:10.1101/gad.234419.113
Diao, Y., Guo, X., Li, Y., Sun, K., Lu, L., Jiang, L., et al. (2012). Pax3/7BP Is a Pax7- and Pax3-Binding Protein that Regulates the Proliferation of Muscle Precursor Cells by an Epigenetic Mechanism. Cell Stem Cell 11, 231–241. doi:10.1016/j.stem.2012.05.022
Dilworth, F. J., and Blais, A. (2011). Epigenetic Regulation of Satellite Cell Activation during Muscle Regeneration. Stem Cel Res. Ther. 2 (2), 18–8. doi:10.1186/scrt59
Edmunds, J. W., Mahadevan, L. C., and Clayton, A. L. (2008). Dynamic Histone H3 Methylation during Gene Induction: HYPB/Setd2 Mediates All H3K36 Trimethylation. Embo J. 27, 406–420. doi:10.1038/sj.emboj.7601967
Eißmann, M., Gutschner, T., Hämmerle, M., Günther, S., Caudron-Herger, M., Groß, M., et al. (2012). Loss of the Abundant Nuclear Non-coding RNAMALAT1is Compatible with Life and Development. RNA Biol. 9, 1076–1087. doi:10.4161/rna.21089
Engreitz, J. M., Ollikainen, N., and Guttman, M. (2016). Long Non-coding RNAs: Spatial Amplifiers that Control Nuclear Structure and Gene Expression. Nat. Rev. Mol. Cel Biol 17, 756–770. doi:10.1038/nrm.2016.126
Fouse, S. D., Shen, Y., Pellegrini, M., Cole, S., Meissner, A., Van Neste, L., et al. (2008). Promoter CpG Methylation Contributes to ES Cell Gene Regulation in Parallel with Oct4/Nanog, PcG Complex, and Histone H3 K4/K27 Trimethylation. Cell Stem Cell 2 (2), 160–169. doi:10.1016/j.stem.2007.12.011
Fritsch, L., Robin, P., Mathieu, J. R. R., Souidi, M., Hinaux, H., Rougeulle, C., et al. (2010). A Subset of the Histone H3 Lysine 9 Methyltransferases Suv39h1, G9a, GLP, and SETDB1 Participate in a Multimeric Complex. Mol. Cel 37, 46–56. doi:10.1016/j.molcel.2009.12.017
Galbiati, M., Onesto, E., Zito, A., Crippa, V., Rusmini, P., Mariotti, R., et al. (2012). The Anabolic/androgenic Steroid Nandrolone Exacerbates Gene Expression Modifications Induced by Mutant SOD1 in Muscles of Mice Models of Amyotrophic Lateral Sclerosis. Pharmacol. Res. 65, 221–230. doi:10.1016/j.phrs.2011.12.001
Giudice, J., and Taylor, J. M. (2017). Muscle as a Paracrine and Endocrine Organ. Curr. Opin. Pharmacol. 34, 49–55. doi:10.1016/j.coph.2017.05.005
Gong, C., Li, Z., Ramanujan, K., Clay, I., Zhang, Y., Lemire-Brachat, S., et al. (2015). A Long Non-coding RNA, LncMyoD, Regulates Skeletal Muscle Differentiation by Blocking IMP2-Mediated mRNA Translation. Develop. Cel 34, 181–191. doi:10.1016/j.devcel.2015.05.009
Gussoni, E., Pavlath, G. K., Lanctot, A. M., Sharma, K. R., Miller, R. G., Steinman, L., et al. (1992). Normal Dystrophin Transcripts Detected in Duchenne Muscular Dystrophy Patients after Myoblast Transplantation. Nature 356, 435–438. doi:10.1038/356435a0
Han, X., Yang, F., Cao, H., and Liang, Z. (2015). Malat1 Regulates Serum Response Factor through miR‐133 as a Competing Endogenous RNA in Myogenesis. FASEB j. 29 (7), 3054–3064. doi:10.1096/fj.14-259952
Hanly, D. J., and Esteller, M. (2018). Interplay between Long Non-coding RNAs and Epigenetic Machinery : Emerging Targets in Cancer ? Philos. Trans. R. Soc. Lond. B Biol. Sci. 373, 20170074. doi:10.1098/rstb.2017.0074
Harada, A., Maehara, K., Sato, Y., Konno, D., Tachibana, T., Kimura, H., et al. (2015). Incorporation of Histone H3.1 Suppresses the Lineage Potential of Skeletal Muscle. Nucleic Acids Res. 43, 775–786. doi:10.1093/nar/gku1346
Harada, A., Okada, S., Konno, D., Odawara, J., Yoshimi, T., Yoshimura, S., et al. (2012). Chd2 Interacts with H3.3 to Determine Myogenic Cell Fate. EMBO J. 31, 2994–3007. doi:10.1038/emboj.2012.136
Hitachi, K., Nakatani, M., Takasaki, A., Ouchi, Y., Uezumi, A., Ageta, H., et al. (2019). Myogenin Promoter‐associated Lnc RNA Myoparr Is Essential for Myogenic Differentiation. EMBO Rep. 20 (3), 1–22. doi:10.15252/embr.201847468
Huang, J. k., Ma, L., Song, W. h., Lu, B. y., Huang, Y. b., Dong, H. m., et al. (2017). LncRNA‐MALAT1 Promotes Angiogenesis of Thyroid Cancer by Modulating Tumor‐Associated Macrophage FGF2 Protein Secretion. J. Cel. Biochem. 118, 4821–4830. doi:10.1002/jcb.26153
Huang, P. H., Plass, C., and Chen, C. S. (2011). Effects of Histone Deacetylase Inhibitors on Modulating H3K4 Methylation marks - A Novel Cross-Talk Mechanism between Histone-Modifying Enzymes. Mol. Cel Pharmacol 3 (2), 39–43. doi:10.4255/mcpharmacol.11.06
Huard, J., Bouchard, J.-P., Roy, R., Labrecque, C., Dansereau, G., Lemieux, B., et al. (1991). Correspondence Section: Myoblast Transplantation Produced Dystrophin-Positive Muscle Fibres in a 16-Year-Old Patient with Duchenne Muscular Dystrophy. Clin. Sci. 81, 287–288. doi:10.1042/cs0810287
Iizuka, K., Machida, T., and Hirafuji, M. (2014). Skeletal Muscle Is an Endocrine Organ. J. Pharmacol. Sci. 125, 125–131. doi:10.1254/jphs.14R02CP
Ji, P., Diederichs, S., Wang, W., Böing, S., Metzger, R., Schneider, P. M., et al. (2003). MALAT-1, a Novel Noncoding RNA, and Thymosin β4 Predict Metastasis and Survival in Early-Stage Non-small Cell Lung Cancer. Oncogene 22 (39), 8031–8041. doi:10.1038/sj.onc.1206928
Jin, X., Kim, J.-G., Oh, M.-J., Oh, H.-Y., Sohn, Y.-W., Pian, X., et al. (2007). Opposite Roles of MRF4 and MyoD in Cell Proliferation and Myogenic Differentiation. Biochem. Biophysical Res. Commun. 364 (3), 476–482. doi:10.1016/j.bbrc.2007.10.042
Juan, A. H., Derfoul, A., Feng, X., Ryall, J. G., Dell'Orso, S., Pasut, A., et al. (2011). Polycomb EZH2 Controls Self-Renewal and Safeguards the Transcriptional Identity of Skeletal Muscle Stem Cells. Genes Dev. 25 (8), 789–794. doi:10.1101/gad.2027911
Karpati, G., Ajdukovic, D., Arnold, D., Gledhill, R. B., Guttmann, R., Holland, P., et al. (1993). Myoblast Transfer in Duchenne Muscular Dystrophy. Ann. Neurol. 34, 8–17. doi:10.1002/ana.410340105
Kassar-Duchossoy, L., Gayraud-Morel, B., Gomès, D., Rocancourt, D., Buckingham, M., Shinin, V., et al. (2004). Mrf4 Determines Skeletal Muscle Identity in Myf5:Myod Double-Mutant Mice. Nature 431, 466–471. doi:10.1038/nature02876
Kawabe, Y.-i., Wang, Y. X., McKinnell, I. W., Bedford, M. T., and Rudnicki, M. A. (2012). Carm1 Regulates Pax7 Transcriptional Activity through MLL1/2 Recruitment during Asymmetric Satellite Stem Cell Divisions. Cell Stem Cell 11, 333–345. doi:10.1016/j.stem.2012.07.001
Khorkova, O., Hsiao, J., and Wahlestedt, C. (2015). Basic Biology and Therapeutic Implications of lncRNA. Adv. Drug Deliv. Rev. 87, 15–24. doi:10.1016/j.addr.2015.05.012
Kidder, B. L., Hu, G., and Zhao, K. (2014). KDM5B Focuses H3K4 Methylation Near Promoters and Enhancers during Embryonic Stem Cell Self-Renewal and Differentiation. Genome Biol. 15, R32. doi:10.1186/gb-2014-15-2-r32
Knutson, S. K., Kawano, S., Minoshima, Y., Warholic, N. M., Huang, K.-C., Xiao, Y., et al. (2014). Selective Inhibition of EZH2 by EPZ-6438 Leads to Potent Antitumor Activity in EZH2-Mutant Non-hodgkin Lymphoma. Mol. Cancer Ther. 13, 842–854. doi:10.1158/1535-7163.MCT-13-0773
Kouzarides, T. (2007). Chromatin Modifications and Their Function. Cell 128, 693–705. doi:10.1016/j.cell.2007.02.005
Krishnakumar, R., and Kraus, W. L. (2010). PARP-1 Regulates Chromatin Structure and Transcription through a KDM5B-dependent Pathway. Mol. Cel 39 (5), 736–749. doi:10.1016/j.molcel.2010.08.014
Kuang, S., Kuroda, K., Le Grand, F., and Rudnicki, M. A. (2007). Asymmetric Self-Renewal and Commitment of Satellite Stem Cells in Muscle. Cell 129, 999–1010. doi:10.1016/j.cell.2007.03.044
Kuang, S., and Rudnicki, M. A. (2008). The Emerging Biology of Satellite Cells and Their Therapeutic Potential. Trends Mol. Med. 14 (January), 82–91. doi:10.1016/j.molmed.2007.12.004
Lee, H., Habas, R., and Abate-Shen, C. (2004). Msx1 Cooperates with Histone H1b for Inhibition of Transcription and Myogenesis. Science 304, 1675–1678. doi:10.1126/science.1098096
Ling, B. M. T., Bharathy, N., Chung, T.-K., Kok, W. K., Li, S., Tan, Y. H., et al. (2012). Lysine Methyltransferase G9a Methylates the Transcription Factor MyoD and Regulates Skeletal Muscle Differentiation. Proc. Natl. Acad. Sci. 109, 841–846. doi:10.1073/pnas.1111628109
Liu, L., Cheung, T. H., Charville, G. W., Hurgo, B. M. C., Leavitt, T., Shih, J., et al. (2013). Chromatin Modifications as Determinants of Muscle Stem Cell Quiescence and Chronological Aging. Cel Rep. 4 (1), 189–204. doi:10.1016/j.celrep.2013.05.043
Luger, K., Mäder, A. W., Richmond, R. K., Sargent, D. F., and Richmond, T. J. (1997). Crystal Structure of the Nucleosome Core Particle at 2.8 Å Resolution. Nature 389 (6648), 251–260. doi:10.1038/38444
Mal, A., and Harter, M. L. (2003). MyoD Is Functionally Linked to the Silencing of a Muscle-specific Regulatory Gene Prior to Skeletal Myogenesis. Proc. Natl. Acad. Sci. 100, 1735–1739. doi:10.1073/pnas.0437843100
Mal, A. K. (2006). Histone Methyltransferase Suv39h1 Represses MyoD-Stimulated Myogenic Differentiation. Embo J. 25, 3323–3334. doi:10.1038/sj.emboj.7601229
Marchesi, I., Giordano, A., and Bagella, L. (2014). Roles of Enhancer of Zeste Homolog 2: From Skeletal Muscle Differentiation to Rhabdomyosarcoma Carcinogenesis. Cell Cycle 13, 516–527. doi:10.4161/cc.27921
Margueron, R., Li, G., Sarma, K., Blais, A., Zavadil, J., Woodcock, C. L., et al. (2008). Ezh1 and Ezh2 Maintain Repressive Chromatin through Different Mechanisms. Mol. Cel 32, 503–518. doi:10.1016/j.molcel.2008.11.004
Marques-Rocha, J. L., Samblas, M., Milagro, F. I., Bressan, J., Martínez, J. A., and Marti, A. (2015). Noncoding RNAs, Cytokines, and Inflammation-Related Diseases. FASEB J. 29, 3595–3611. doi:10.1096/fj.14-260323
Matteini, F., Andresini, O., Petrai, S., Battistelli, C., Rossi, M. N., and Maione, R. (2020). Poly(ADP-ribose) Polymerase 1 (PARP1) Restrains MyoD-dependent Gene Expression during Muscle Differentiation. Sci. Rep. 15, 15086. doi:10.1038/s41598-020-72155-8
Mauro, A. (1961). Satellite Cell of Skeletal Muscle Fibers. J. Biophys. Biochem. Cytol. 9, 493–495. doi:10.1083/jcb.9.2.493
McHugh, C. A., Chen, C.-K., Chow, A., Surka, C. F., Tran, C., McDonel, P., et al. (2015). The Xist lncRNA Interacts Directly with SHARP to Silence Transcription through HDAC3. Nature 521, 232–236. doi:10.1038/nature14443
McKinnell, I. W., Ishibashi, J., Le Grand, F., Punch, V. G. J., Addicks, G. C., Greenblatt, J. F., et al. (2008). Pax7 Activates Myogenic Genes by Recruitment of a Histone Methyltransferase Complex. Nat. Cel Biol 10, 77–84. doi:10.1038/ncb1671
Megeney, L. A., and Rudnicki, M. A. (1995). Determination versus Differentiation and the MyoD Family of Transcription Factors. Biochem. Cel Biol. 73, 723–732. doi:10.1139/o95-080
Mendell, J. R., Kissel, J. T., Amato, A. A., King, W., Signore, L., Prior, T. W., et al. (1995). Myoblast Transfer in the Treatment of Duchenne's Muscular Dystrophy. N. Engl. J. Med. 333, 832–838. doi:10.1056/nejm199509283331303
Mielcarek, M., Toczek, M., Smeets, C. J. L. M., Franklin, S. A., Bondulich, M. K., Jolinon, N., et al. (2015). HDAC4-Myogenin Axis as an Important Marker of HD-Related Skeletal Muscle Atrophy. Plos Genet. 11, e1005021. doi:10.1371/journal.pgen.1005021
Mikkelsen, T. S., Ku, M., Jaffe, D. B., Issac, B., Lieberman, E., Giannoukos, G., et al. (2007). Genome-wide Maps of Chromatin State in Pluripotent and Lineage-Committed Cells. Nature 448 (7153), 553–560. doi:10.1038/nature06008
Minetti, G. C., Colussi, C., Adami, R., Serra, C., Mozzetta, C., Parente, V., et al. (2006). Functional and Morphological Recovery of Dystrophic Muscles in Mice Treated with Deacetylase Inhibitors. Nat. Med. 12, 1147–1150. doi:10.1038/nm1479
Minotti, R., Andersson, A., and Hottiger, M. O. (2015). ARTD1 Suppresses Interleukin 6 Expression by Repressing MLL1-dependent Histone H3 Trimethylation. Mol. Cel. Biol. 35, 00196–215. doi:10.1128/mcb.00196-15
Molkentin, J. D., Black, B. L., Martin, J. F., and Olson, E. N. (1995). Cooperative Activation of Muscle Gene Expression by MEF2 and Myogenic bHLH Proteins. Cell. 83, 1125–1136. doi:10.1016/0092-8674(95)90139-6
Montarras, D., Morgan, J., and Collins, C. (2005). Direct Isolation of Satellite Cells for Skeletal Muscle Regeneration. Mol. Cel Biol. 309, 2064–2067. doi:10.1126/science.1114758
Morera, L., Lübbert, M., and Jung, M. (2016). Targeting Histone Methyltransferases and Demethylases in Clinical Trials for Cancer Therapy. Clin. Epigenet 8, 1–16. doi:10.1186/s13148-016-0223-4
Moresi, V., Williams, A. H., Meadows, E., Flynn, J. M., Potthoff, M. J., McAnally, J., et al. (2010). Myogenin and Class II HDACs Control Neurogenic Muscle Atrophy by Inducing E3 Ubiquitin Ligases. Cell 143, 35–45. doi:10.1016/j.cell.2010.09.004
Mousavi, K., Zare, H., Wang, A. H., and Sartorelli, V. (2012). Polycomb Protein Ezh1 Promotes RNA Polymerase II Elongation. Mol. Cel 45, 255–262. doi:10.1016/j.molcel.2011.11.019
Myostatin, K. T. (2008). Inhibition by a Follistatin-Derived Peptide Ameliorates the Pathophysiology of Muscular Dystrophy Model Mice. Acta Myol. 27, 14–18. Available at: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2859604/.
Nagano, T., Mitchell, J. A., Sanz, L. A., Pauler, F. M., Ferguson-Smith, A. C., Feil, R., et al. (2008). The Air Noncoding RNA Epigenetically Silences Transcription by Targeting G9a to Chromatin. Science 322 (5908), 1717–1720. doi:10.1126/science.1163802
Nakagawa, S., Ip, J. Y., Shioi, G., Tripathi, V., Zong, X., Hirose, T., et al. (2012). Malat1 Is Not an Essential Component of Nuclear Speckles in Mice. Rna 18, 1487–1499. doi:10.1261/rna.033217.112
Nishioka, K., Chuikov, S., Sarma, K., Erdjument-Bromage, H., Allis, C. D., Tempst, P., et al. (2002). Set9, a Novel Histone H3 Methyltransferase that Facilitates Transcription by Precluding Histone Tail Modifications Required for Heterochromatin Formation. Genes Dev. 16, 479–489. doi:10.1101/gad.967202
Oustanina, S., Hause, G., and Braun, T. (2004). Pax7 Directs Postnatal Renewal and Propagation of Myogenic Satellite Cells but Not Their Specification. Embo J. 23, 3430–3439. doi:10.1038/sj.emboj.7600346
Palacios, D., Mozzetta, C., Consalvi, S., Caretti, G., Saccone, V., Proserpio, V., et al. (2010). TNF/p38α/Polycomb Signaling to Pax7 Locus in Satellite Cells Links Inflammation to the Epigenetic Control of Muscle Regeneration. Cell Stem Cell 7, 455–469. doi:10.1016/j.stem.2010.08.013
Pan, G., Tian, S., Nie, J., Yang, C., Ruotti, V., Wei, H., et al. (2007). Whole-Genome Analysis of Histone H3 Lysine 4 and Lysine 27 Methylation in Human Embryonic Stem Cells. Cell Stem Cell 1 (3), 299–312. doi:10.1016/j.stem.2007.08.003
Pandey, R. R., Mondal, T., Mohammad, F., Enroth, S., Redrup, L., Komorowski, J., et al. (2008). Kcnq1ot1 Antisense Noncoding RNA Mediates Lineage-specific Transcriptional Silencing through Chromatin-Level Regulation. Mol. Cel 32 (2), 232–246. doi:10.1016/j.molcel.2008.08.022
Parasramka, M. A., Maji, S., Matsuda, A., Yan, I. K., and Patel, T. (2016). Long Non-coding RNAs as Novel Targets for Therapy in Hepatocellular Carcinoma. Pharmacol. Ther. 161, 67–78. doi:10.1016/j.pharmthera.2016.03.004
Partridge, T. A., Morgan, J. E., Coulton, G. R., Hoffman, E. P., and Kunkel, L. M. (1989). Conversion of Mdx Myofibres from Dystrophin-Negative to -positive by Injection of normal Myoblasts. Nature 337, 176–179. doi:10.1038/337176a0
Pasini, D., Cloos, P. A. C., Walfridsson, J., Olsson, L., Bukowski, J.-P., Johansen, J. V., et al. (2010). JARID2 Regulates Binding of the Polycomb Repressive Complex 2 to Target Genes in ES Cells. Nature 464, 306–310. doi:10.1038/nature08788
Pekowska, A., Benoukraf, T., Zacarias-Cabeza, J., Belhocine, M., Koch, F., Holota, H., et al. (2011). H3K4 Tri-methylation Provides an Epigenetic Signature of Active Enhancers. EMBO J. 30, 4198–4210. doi:10.1038/emboj.2011.295
Peng, J. C., Valouev, A., Swigut, T., Zhang, J., Zhao, Y., Sidow, A., et al. (2009). Jarid2/Jumonji Coordinates Control of PRC2 Enzymatic Activity and Target Gene Occupancy in Pluripotent Cells. Cell 139, 1290–1302. doi:10.1016/j.cell.2009.12.002
Piasecka, A., Sekrecki, M., Szcześniak, M. W., and Sobczak, K. (2021). MEF2C Shapes the Microtranscriptome during Differentiation of Skeletal Muscles. Sci. Rep. 11, 3476. doi:10.1038/s41598-021-82706-2
Powers, S. K., Lynch, G. S., Murphy, K. T., Reid, M. B., and Zijdewind, I. (2016). Disease-induced Skeletal Muscle Atrophy and Fatigue. Med. Sci. Sports Exerc. 48, 2307–2319. doi:10.1249/MSS.0000000000000975
Rampalli, S., Li, L., Mak, E., Ge, K., Brand, M., Tapscott, S. J., et al. (2007). p38 MAPK Signaling Regulates Recruitment of Ash2L-Containing Methyltransferase Complexes to Specific Genes during Differentiation. Nat. Struct. Mol. Biol. 14, 1150–1156. doi:10.1038/nsmb1316
Rea, S., Eisenhaber, F., O'Carroll, D., Strahl, B. D., Sun, Z.-W., Schmid, M., et al. (2000). Regulation of Chromatin Structure by Site-specific Histone H3 Methyltransferases. Nature 406, 593–599. doi:10.1038/35020506
Relaix, F., Montarras, D., Zaffran, S., Gayraud-Morel, B., Rocancourt, D., Tajbakhsh, S., et al. (2006). Pax3 and Pax7 Have Distinct and Overlapping Functions in Adult Muscle Progenitor Cells. J. Cel Biol. 172, 91–102. doi:10.1083/jcb.200508044
Relaix, F., Rocancourt, D., Mansouri, A., and Buckingham, M. (2005). A Pax3/Pax7-dependent Population of Skeletal Muscle Progenitor Cells. Nature 435, 948–953. doi:10.1038/nature03594
Rinn, J. L., Kertesz, M., Wang, J. K., Squazzo, S. L., Xu, X., Brugmann, S. A., et al. (2007). Functional Demarcation of Active and Silent Chromatin Domains in Human HOX Loci by Noncoding RNAs. Cell 129 (7), 1311–1323. doi:10.1016/j.cell.2007.05.022
Sabourin, L. A., and Rudnicki, M. A. (2000). The Molecular Regulation of Myogenesis. Clin. Genet. 57, 16–25. doi:10.1034/j.1399-0004.2000.570103.x
Schienda, J., Engleka, K. A., Jun, S., Hansen, M. S., Epstein, J. A., Tabin, C. J., et al. (2006). Somitic Origin of Limb Muscle Satellite and Side Population Cells. Proc. Natl. Acad. Sci. 103, 945–950. doi:10.1073/pnas.0510164103
Seenundun, S., Rampalli, S., Liu, Q.-C., Aziz, A., Palii, C., Hong, S., et al. (2010). UTX Mediates Demethylation of H3K27me3 at Muscle-specific Genes during Myogenesis. EMBO J. 29 (8), 1401–1411. doi:10.1038/emboj.2010.37
Shpargel, K. B., Starmer, J., Yee, D., Pohlers, M., and Magnuson, T. (2014). KDM6 Demethylase Independent Loss of Histone H3 Lysine 27 Trimethylation during Early Embryonic Development. Plos Genet. 10 (8), e1004507. doi:10.1371/journal.pgen.1004507
Skuk, D., Roy, B., Goulet, M., and Tremblay, J. P. (1999). Successful Myoblast Transplantation in Primates Depends on Appropriate Cell Delivery and Induction of Regeneration in the Host Muscle. Exp. Neurol. 155, 22–30. doi:10.1006/exnr.1998.6973
Soleimani, V. D., Punch, V. G., Kawabe, Y.-i., Jones, A. E., Palidwor, G. A., Porter, C. J., et al. (2012). Transcriptional Dominance of Pax7 in Adult Myogenesis Is Due to High-Affinity Recognition of Homeodomain Motifs. Develop. Cel 22, 1208–1220. doi:10.1016/j.devcel.2012.03.014
Stojic, L., Jasencakova, Z., Prezioso, C., Stützer, A., Bodega, B., Pasini, D., et al. (2011). Chromatin Regulated Interchange between Polycomb Repressive Complex 2 (PRC2)-Ezh2 and PRC2-Ezh1 Complexes Controls Myogenin Activation in Skeletal Muscle Cells. Epigenetics & Chromatin 4. doi:10.1186/1756-8935-4-16
Stölting, M. N. L., Ferrari, S., Handschin, C., Becskei, A., Provenzano, M., Sulser, T., et al. (2013). Myoblasts Inhibit Prostate Cancer Growth by Paracrine Secretion of Tumor Necrosis Factor-α. J. Urol. 189, 1952–1959. doi:10.1016/j.juro.2012.10.071
Sumariwalla, V. M., and Klein, W. H. (2001). Similar Myogenic Functions for Myogenin and MRF4 but Not MyoD in Differentiated Murine Embryonic Stem Cells. genesis 30, 239–249. doi:10.1002/gene.1070
Summerbell, D., Halai, C., and Pwj, Rigby. (2002). Expression of the Myogenic Regulatory Factor Mrf4 Precedes or Is Contemporaneous with that of Myf5 in the Somitic Bud. Mech. Dev. 117, 331–335. doi:10.1016/S0925-4773(02)00208-3
Suraweera, A., O’Byrne, K. J., and Richard, D. J. (2018). Combination Therapy with Histone Deacetylase Inhibitors (HDACi) for the Treatment of Cancer: Achieving the Full Therapeutic Potential of HDACi. Front. Oncol. 8, 92. doi:10.3389/fonc.2018.00092
Tachibana, M., Sugimoto, K., Nozaki, M., Ueda, J., Ohta, T., Ohki, M., et al. (2002). G9a Histone Methyltransferase Plays a Dominant Role in Euchromatic Histone H3 Lysine 9 Methylation and Is Essential for Early Embryogenesis. Genes Dev. 16, 1779–1791. doi:10.1101/gad.989402
Tachibana, M., Sugimoto, K., Fukushima, T., and Shinkai, Y. (2001). SET Domain-Containing Protein, G9a, Is a Novel Lysine-Preferring Mammalian Histone Methyltransferase with Hyperactivity and Specific Selectivity to Lysines 9 and 27 of Histone H3. J. Biol. Chem. 276, 25309–25317. doi:10.1074/jbc.M101914200
Tao, Y., Neppl, R. L., Huang, Z.-P., Chen, J., Tang, R.-H., Cao, R., et al. (2011). The Histone Methyltransferase Set7/9 Promotes Myoblast Differentiation and Myofibril Assembly. J. Cel Biol. 194, 551–565. doi:10.1083/jcb.201010090
Tripathi, V., Ellis, J. D., Shen, Z., Song, D. Y., Pan, Q., Watt, A. T., et al. (2010). The Nuclear-Retained Noncoding RNA MALAT1 Regulates Alternative Splicing by Modulating SR Splicing Factor Phosphorylation. Mol. Cel 39, 925–938. doi:10.1016/j.molcel.2010.08.011
Tsai, M.-C., Manor, O., Wan, Y., Mosammaparast, N., Wang, J. K., Lan, F., et al. (2010). Long Noncoding RNA as Modular Scaffold of Histone Modification Complexes. Science 329 (5992), 689–693. doi:10.1126/science.1192002
Vastenhouw, N. L., and Schier, A. F. (2012). Bivalent Histone Modifications in Early Embryogenesis. Curr. Opin. Cel Biol. 24 (3), 374–386. doi:10.1016/j.ceb.2012.03.009
Verrier, L., Escaffit, F., Chailleux, C., Trouche, D., and Vandromme, M. (2011). A New Isoform of the Histone Demethylase JMJD2A/KDM4A Is Required for Skeletal Muscle Differentiation. Plos Genet. 7, e1001390. doi:10.1371/journal.pgen.1001390
Wang, G.-q., Wang, Y., Xiong, Y., Chen, X.-C., Ma, M.-l., Cai, R., et al. (2016). Sirt1 AS lncRNA Interacts with its mRNA to Inhibit Muscle Formation by Attenuating Function of miR-34a. Sci. Rep. 6. doi:10.1038/srep21865
Wang, H., Cao, R., Xia, L., Erdjument-Bromage, H., Borchers, C., Tempst, P., et al. (2001). Purification and Functional Characterisation of a Histone H3-Lysine 4-specific Methyltransferase. Mol. Cel. 8, 1207–1217. doi:10.1016/S1097-2765(01)00405-1
Wang, J., and Abate-Shen, C. (2012). The Msx1 Homeoprotein Recruits G9a Methyltransferase to Repressed Target Genes in Myoblast Cells. PLoS One 7, e37647. doi:10.1371/journal.pone.0037647
Wang, K. C., Yang, Y. W., Liu, B., Sanyal, A., Corces-Zimmerman, R., Chen, Y., et al. (2011). A Long Noncoding RNA Maintains Active Chromatin to Coordinate Homeotic Gene Expression. Nature 472 (7341), 120–124. doi:10.1038/nature09819
Wang, Y., Xu, Z., Jiang, J., Xu, C., Kang, J., Xiao, L., et al. (2013). Endogenous miRNA Sponge lincRNA-RoR Regulates Oct4, Nanog, and Sox2 in Human Embryonic Stem Cell Self-Renewal. Develop. Cel 25, 69–80. doi:10.1016/j.devcel.2013.03.002
Wang, Z., Oron, E., Nelson, B., Razis, S., and Ivanova, N. (2012). Distinct Lineage Specification Roles for NANOG, OCT4, and SOX2 in Human Embryonic Stem Cells. Cell Stem Cell 10 (4), 440–454. doi:10.1016/j.stem.2012.02.016
Watts, R., Johnsen, V. L., Shearer, J., and Hittel, D. S. (2013). Myostatin-induced Inhibition of the Long Noncoding RNA Malat1 Is Associated with Decreased Myogenesis. Am. J. Physiology-Cell Physiol. 304, C995–C1001. doi:10.1152/ajpcell.00392.2012
Wilkin, F., Paquette, J., Ledru, E., Mamelin, C., Pollak, M., and Deal, C. L. (2000). H19sense and Antisense Transgenes Modify Insulin-like Growth Factor-II mRNA Levels. Eur. J. Biochem. 267, 4020–4027. doi:10.1046/j.1432-1327.2000.01438.x
Wilson, E. M., and Rotwein, P. (2006). Control of MyoD Function during Initiation of Muscle Differentiation by an Autocrine Signaling Pathway Activated by Insulin-like Growth Factor-II. J. Biol. Chem. 281, 29962–29971. doi:10.1074/jbc.M605445200
Xu, X., Ji, S., Li, W., Yi, B., Li, H., Zhang, H., et al. (2017). LncRNA H19 Promotes the Differentiation of Bovine Skeletal Muscle Satellite Cells by Suppressing Sirt1/FoxO1. Cell Mol Biol Lett 22. doi:10.1186/s11658-017-0040-6
Xue, S., Lam, Y. M., He, Z., Zheng, Y., Li, L., Zhang, Y., et al. (2020). Histone Lysine Demethylase KDM5B Maintains Chronic Myeloid Leukemia via Multiple Epigenetic Actions. Exp. Hematol. 82, 53–65. doi:10.1016/j.exphem.2020.01.006
Yang, J.-H., Song, Y., Seol, J.-H., Park, J. Y., Yang, Y.-J., Han, J.-W., et al. (2011). Myogenic Transcriptional Activation of MyoD Mediated by Replication-independent Histone Deposition. Proc. Natl. Acad. Sci. USA 108, 85–90. doi:10.1073/pnas.1009830108
Yap, K. L., Li, S., Muñoz-Cabello, A. M., Raguz, S., Zeng, L., Mujtaba, S., et al. (2010). Molecular Interplay of the Noncoding RNA ANRIL and Methylated Histone H3 Lysine 27 by Polycomb CBX7 in Transcriptional Silencing of INK4a. Mol. Cel 38 (5), 662–674. doi:10.1016/j.molcel.2010.03.021
Yi, X., Tao, Y., Lin, X., Dai, Y., Yang, T., Yue, X., et al. (2017). Histone Methyltransferase Setd2 Is Critical for the Proliferation and Differentiation of Myoblasts. Biochim. Biophys. Acta (Bba) - Mol. Cel Res. 1864 (4), 697–707. doi:10.1016/j.bbamcr.2017.01.012
Yokochi, T., Poduch, K., Ryba, T., Lu, J., Hiratani, I., Tachibana, M., et al. (2009). G9a Selectively Represses a Class of Late-Replicating Genes at the Nuclear Periphery. Proc. Natl. Acad. Sci. 106, 19363–19368. doi:10.1073/pnas.0906142106
Yong, H., Wu, G., Chen, J., Liu, X., Bai, Y., Tang, N., et al. (2020). lncRNA MALAT1 Accelerates Skeletal Muscle Cell Apoptosis and Inflammatory Response in Sepsis by Decreasing BRCA1 Expression by Recruiting EZH2. Mol. Ther. - Nucleic Acids 19 (25), 97–108. doi:10.1016/j.omtn.2019.10.028
Zentner, G. E., and Henikoff, S. (2013). Regulation of Nucleosome Dynamics by Histone Modifications. Nat. Struct. Mol. Biol. 20, 259–266. doi:10.1038/nsmb.2470
Zhang, B., Arun, G., Mao, Y. S., Lazar, Z., Hung, G., Bhattacharjee, G., et al. (2012). The lncRNA Malat1 Is Dispensable for Mouse Development but its Transcription Plays a Cis-Regulatory Role in the Adult. Cel Rep. 2, 111–123. doi:10.1016/j.celrep.2012.06.003
Zhang, C., Ge, S., Gong, W., Xu, J., Guo, Z., Liu, Z., et al. (2020). LncRNA ANRIL Acts as a Modular Scaffold of WDR5 and HDAC3 Complexes and Promotes Alteration of the Vascular Smooth Muscle Cell Phenotype. Cell Death Dis 11 (6), 435. doi:10.1038/s41419-020-2645-3
Zhang, C. L., McKinsey, T. A., and Olson, E. N. (2002). Association of Class II Histone Deacetylases with Heterochromatin Protein 1: Potential Role for Histone Methylation in Control of Muscle Differentiation. Mol. Cel Biol 22, 7302–7312. doi:10.1128/mcb.22.20.7302-7312.2002
Zhang, P., Du, J., Guo, X., Wu, S., He, J., Li, X., et al. (2021). LncMyoD Promotes Skeletal Myogenesis and Regulates Skeletal Muscle Fiber-type Composition by Sponging miR-370-3p. Genes 12, 589. doi:10.3390/genes12040589
Zhang, R.-H., Judson, R. N., Liu, D. Y., Kast, J., and Rossi, F. M. V. (2016). The Lysine Methyltransferase Ehmt2/G9a Is Dispensable for Skeletal Muscle Development and Regeneration. Skeletal Muscle 6. doi:10.1186/s13395-016-0093-7
Zhang, T., Cooper, S., and Brockdorff, N. (2015). The Interplay of Histone Modifications - Writers that Read. EMBO Rep. 16 (11), 1467–1481. doi:10.15252/embr.201540945
Zhao, G., Su, Z., Song, D., Mao, Y., and Mao, X. (2016). The Long Noncoding RNA MALAT1 Regulates the Lipopolysaccharide-Induced Inflammatory Response through its Interaction with NF-Κb. FEBS Lett. 590, 2884–2895. doi:10.1002/1873-3468.12315
Zheng, L., Liu, X., Chen, P., and Xiao, W. (2019). Expression and Role of lncRNAs in the Regeneration of Skeletal Muscle Following Contusion Injury. Exp. Ther. Med. 18, 2617–2627. doi:10.3892/etm.2019.7871
Zhou, L., Sun, K., Zhao, Y., Zhang, S., Wang, X., Li, Y., et al. (2015). Linc-YY1 Promotes Myogenic Differentiation and Muscle Regeneration through an Interaction with the Transcription Factor YY1. Nat. Commun. 6. doi:10.1038/ncomms10026
Keywords: epigenetics, skeletal muscle, skeletal muscle regeneration, histone methylation, lncRNAs, satellite stem cells
Citation: Mbadhi MN, Tang J-m and Zhang J-x (2021) Histone Lysine Methylation and Long Non-Coding RNA: The New Target Players in Skeletal Muscle Cell Regeneration. Front. Cell Dev. Biol. 9:759237. doi: 10.3389/fcell.2021.759237
Received: 16 August 2021; Accepted: 11 November 2021;
Published: 03 December 2021.
Edited by:
Masatoshi Suzuki, University of Wisconsin-Madison, United StatesReviewed by:
Liliana Burlibasa, University of Bucharest, RomaniaKoji Hayakawa, Okayama University of Science, Japan
Copyright © 2021 Mbadhi, Tang and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jing-xuan Zhang, emhhbmdqeEBoYm11LmVkdS5jbg==
 Magdaleena Naemi Mbadhi
Magdaleena Naemi Mbadhi Jun-ming Tang
Jun-ming Tang Jing-xuan Zhang
Jing-xuan Zhang