Abstract
Protein translation is an essential cellular process playing key roles in growth and development. Protein translation declines over the course of age in multiple animal species, including nematodes, fruit flies, mice, rats, and even humans. In all these species, protein translation transiently peaks in early adulthood with a subsequent drop over the course of age. Conversely, lifelong reductions in protein translation have been found to extend lifespan and healthspan in multiple animal models. These findings raise the protein synthesis paradox: age-related declines in protein synthesis should be detrimental, but life-long reductions in protein translation paradoxically slow down aging and prolong lifespan. This article discusses the nature of this paradox and complies an extensive body of work demonstrating protein translation as a modulator of lifespan and healthspan.
1 Introduction
The life of a protein begins with translation of mRNA into a nascent polypeptide chain [reviewed in (Hebert and Molinari 2007; Taylor and Dillin 2011; Balchin et al., 2016)]. During or after synthesis, a protein then adopts a higher-level stable three-dimensional structure to become biologically functional. This process is called co- or post-translational folding and occurs spontaneously; only the information contained in the amino acid sequence is required for folding of proteins into their native state. Especially, chemical forces created by specific amino acid sequences within the particular position of the polypeptide chain (e.g. hydrophobic interactions and hydrogen bonds) guide the proper folding. Although protein folding can occur spontaneously, there are several molecular chaperones that facilitate the folding by accelerating rate-limiting steps. Chaperones also assist refolding of unfolded or denatured proteins and prevent them from being aggregated. Once a protein is properly folded, it is chemically modified and trafficked to the correct cellular compartment. Finally, when proteins reach the end of their life or become damaged, they get degraded by the autophagy-lysosomal pathway or ubiquitin-proteasome system (UPS) (Figure 1).
FIGURE 1
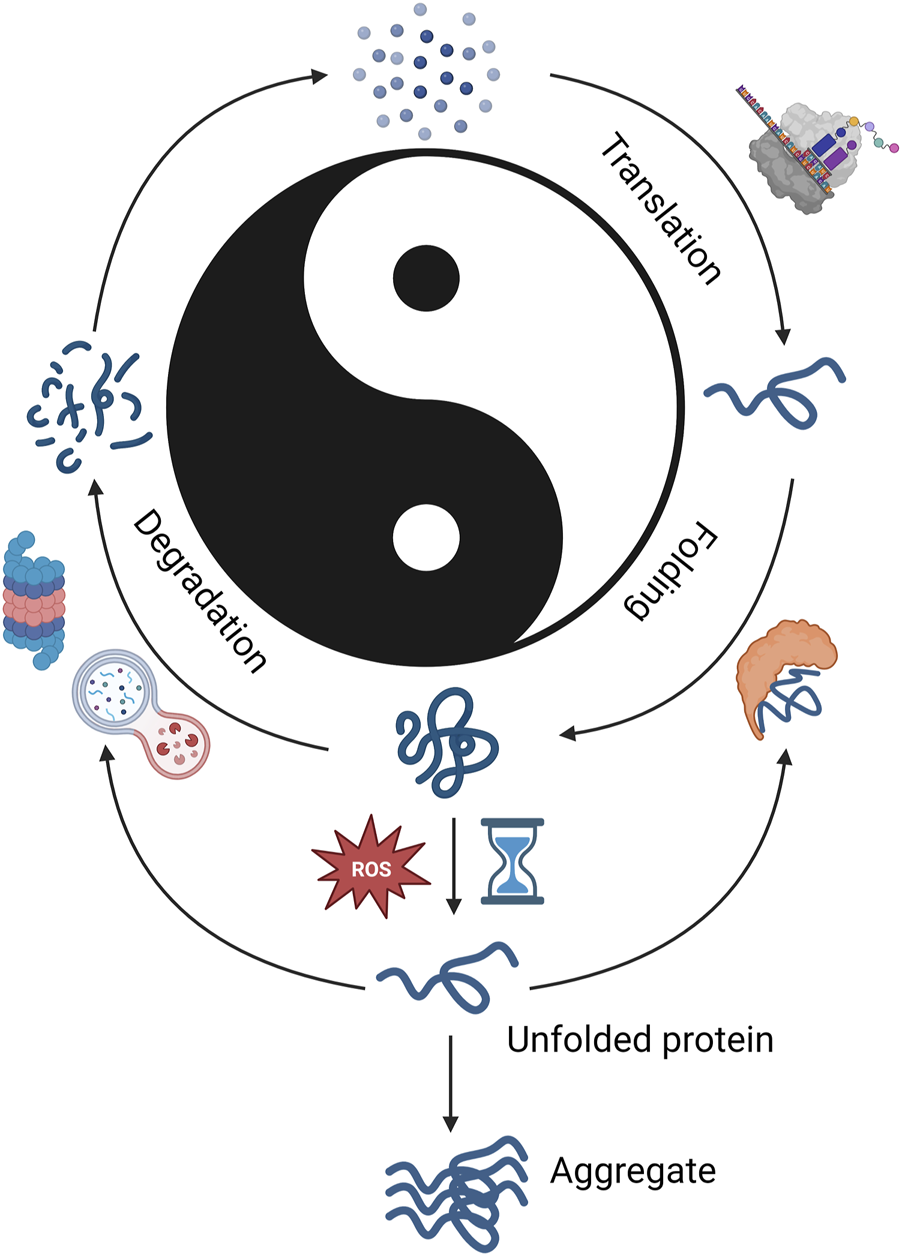
Protein lifecycle. Graphical representation of proteostatic cycle. Protein synthesis is shown on the right and protein degradation on the left. The Chinese philosophical concept Yin and Yang are denoted in the center with Yang denotating synthesis and Yin degradation. This is used to denote the importance of balance between synthesis and degradation. This figure shows that in balanced proteostasis, proteins are translated using the available amino acid pool (top right), folded by molecular chaperones (bottom right), degraded by proteasome or autophagial pathways (bottom left), and amino acids are recycled to form new proteins. Imbalance from insufficient molecular chaperone function, ROS, or insufficient degradation leads to unfolded proteins (bottom center), which may be refolded by molecular chaperones or removed by degradation pathways. If protein quality control does not occur, aggregates may form. Created with BioRender.com.
Disruption in any of the process mentioned above can disturb protein homeostasis (proteostasis), cause protein aggregations and cellular death, and ultimately contribute to the pathogenesis of several diseases, including neurodegenerative disorders (Hebert and Molinari 2007; Powers et al., 2009; Taylor and Dillin 2011).
To date, a large body of research has demonstrated dysfunction in many of aspects of the proteostatic network as a critical component of aging. The proteasome system was shown to deteriorate as a consequence of either age or disease; conversely, enhancing proteasome function can extend lifespan/healthspan and protect against age-related diseases (Munkacsy et al., 2019; Nguyen et al., 2019; Chocron et al., 2022). In addition, several studies demonstrated dysfunction of autophagial pathway with age and how augmentation of the autophagial system can extend lifespan (Hansen et al., 2018). Likewise, functions of molecular chaperones were shown to decline with age, whereas overexpression of such chaperones was able to extend lifespan (Soti and Csermely 2003). Despite extensive aging studies in degradation systems, there is a lack of studies in the other arm of the proteostatic control: protein synthesis. In this review, we extensively discuss how modulation of protein synthesis and lifetime protein translation dynamics regulate the onset of aging.
2 The process of eukaryotic protein translation
Protein translation occurs in three phases: 1) initiation of mRNA translation, 2) elongation of the polypeptide chain, and 3) termination of mRNA translation (Sonenberg and Hinnebusch 2009; Jackson et al., 2010) (Figure 2).
FIGURE 2
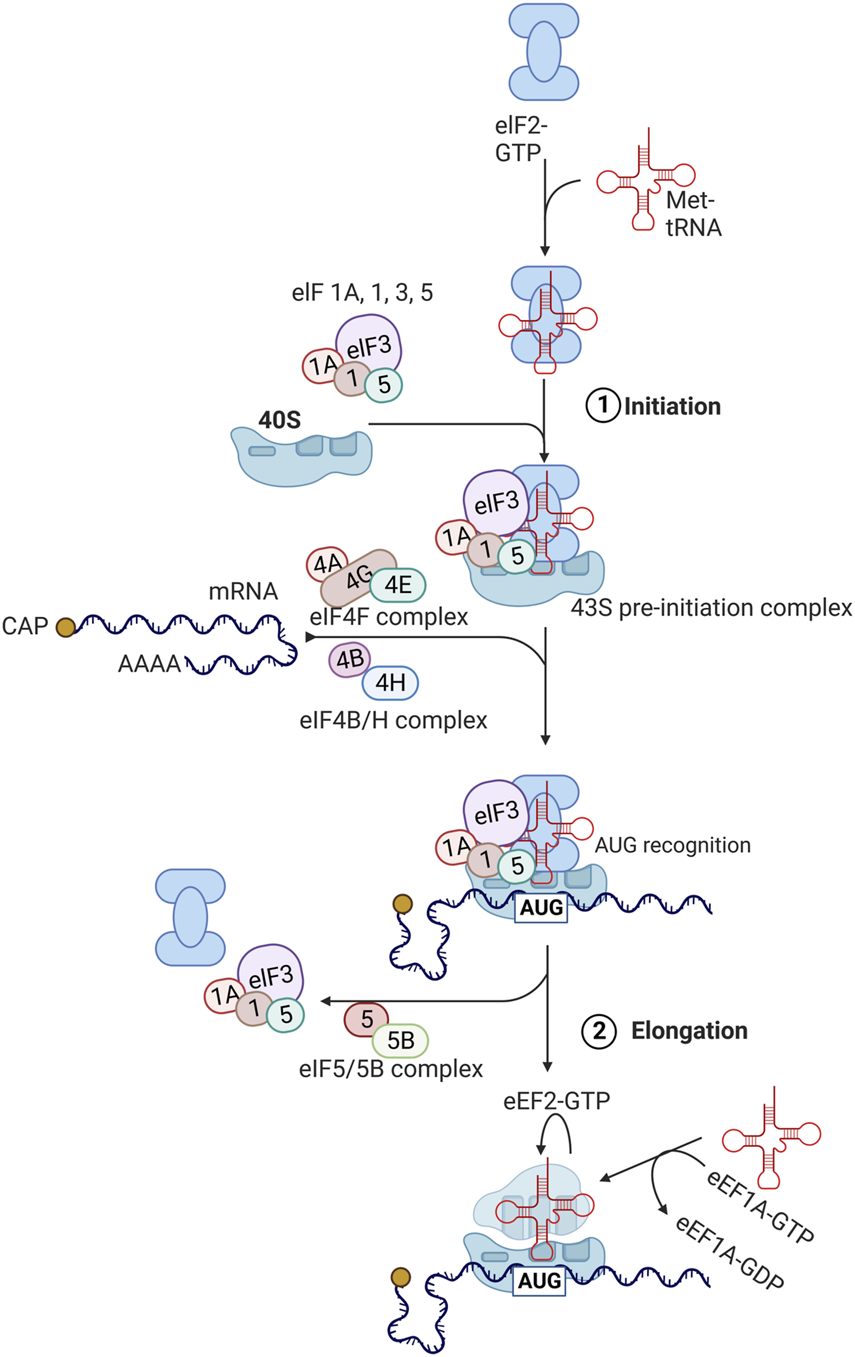
Protein translation pathways. The process of translation initiation and elongation in eukaryotic cells. Key eukaryotic initiation factors (eIFs) and eukaryotic elongation factors (eEFs) are highlighted. This process is described in detail in section 3 with involvement of highlighted factors in aging described in section 5. Created with BioRender.com.
During the initiation phase, the eukaryotic initiation factor 2 (eIF2) forms a ternary complex with GTP and methionine-charged methionyl-tRNA (Met-tRNAiMet) (Sonenberg and Hinnebusch 2009; Jackson et al., 2010). The ternary complex then binds to the small 40S ribosomal subunit along with several eukaryotic initiation factors (eIF1, eIF1A, eIF3, and eIF5) to form the 43S pre-initiation complex. Subsequently, eIF4F complex, consisting of eIF4E, eIF4G, and eIF4A binds to the 5′ cap region of mRNA and unwinds this region in an ATP-dependent manner with eIF4B. This allows for the 43S pre-initiation complex to attach to the mRNA being translated. The 43S complex then scans the 5′untranslated region (5′ UTR) from the 5′ to 3′ direction to the initiation codon (AUG). After the AUG codon is recognized, eIF2-bound GTP is hydrolyzed by eIF5 and eIF5B, leading to dissociation of eIFs and joining of 60S ribosomal subunit to form the elongation-competent 80S ribosome.
Elongation of peptide chains by the 80S ribosome is assisted by eukaryotic translation elongation factors (eEFs) (Sonenberg and Hinnebusch 2009; Jackson et al., 2010). Once aminoacyl tRNA synthetase loads the tRNA with the amino acid corresponding to the codon, eEF1A-GTP complex brings this incoming aminoacyl-tRNA to the A site of the ribosome. If the correct aminoacyl-tRNA fits to the A site, GTP is hydrolyzed and eEF1A-GDP complex becomes dissociated. eEF1B catalyzes the exchange of bound GDP for GTP, enabling the next cycle to occur again. Then, peptidyltransferase in the 60S ribosomal subunit (28S ribosomal rRNA) catalyzes the transfer of peptide attached to the aminoacyl-tRNA in the P site to the amino group at the aminoacyl-tRNA in the A site, forming a peptide bond. Subsequently, eEF2-GTP complex facilitates the transfer of peptidyl-tRNA from the A site to the P site, which is called translocation.
Finally, when the A site encounters a stop codon (UAG, UAA, or UGA), releasing factor (eRF) binds GTP and stimulates the transfer of peptidyl group from the P-site tRNA to H2O instead of the A-site tRNA (Sonenberg and Hinnebusch 2009; Jackson et al., 2010). This results in the release of the peptide along with uncharged tRNA and eRF-GDP complex, followed by dissociation of 40S and 60 S ribosomal subunits.
3 Regulation of protein translation rate
The first initiation phase of protein translation is a rate limiting step, where most regulations are exerted (Gebauer and Hentze 2004; Sonenberg and Hinnebusch 2009; Jackson et al., 2010). The global rate of protein translation is mostly regulated by altering the activity and availability of eIFs. Two most heavily regulated eIFs to adjust the rate of protein translation are eIF2 and eIF4E.
eIF2 consists of three subunits: α, β, and γ (Gebauer and Hentze 2004; Sonenberg and Hinnebusch 2009; Jackson et al., 2010). GTP hydrolysis by the γ subunit of eIF2 facilitates the formation of ternary complex, which is the key step in translation initiation. Exchange of GDP for GTP is catalyzed by eIF2B and is necessary for regenerating active eIF2. Phosphorylation of the α subunit of eIF2 impedes the dissociation of eIF2B, blocks the GDP-GTP exchange, and thereby pronouncedly lowers the rate of protein translation (Donnelly et al., 2013). As such, eIF2α phosphorylation by eIF2α kinases is frequently used by cells to attenuate protein translation in response to physiological or environmental conditions. For example, when unfolded or misfolded proteins accumulate in the lumen of endoplasmic reticulum (ER), one of the eIF2α kinases called PKR-like ER kinase (PERK) phosphorylates eIF2α, reduces the protein translation rate, and thereby decreases the ER protein folding load (Walter and Ron 2011). This translational control by PERK is one of the branches of the so-called unfolded protein response (UPR) (Walter and Ron 2011). Another kind of eIF2α kinase called GCN2 is primarily activated by amino acid and glucose deprivation and lowers the rate of protein translation to adjust for nutrient availability (Zhang et al., 2002a; Donnelly et al., 2013).
The cap binding protein eIF4E facilitates the recruitment of the 43S pre-initiation complex to the 5′cap, which is regarded as the rate-limiting step in protein translation initiation (Jackson et al., 2010). The availability of active eIF4E is regulated by the eIF4E-binding protein (4E-BP) (Marcotrigiano et al., 1999; Musa et al., 2016). 4E-BP prevents eIF4E from interacting with eIF4G, thereby blocking the formation of eIF4F complex and halting protein translation initiation. The ability of 4E-BP to bind eIF4E is regulated by the phosphorylation of several serine and threonine residues of 4E-BP. Hypophosphorylated 4E-BP has a strong binding affinity to eIF4E, whereas hyperphosphorylated 4E-BP loses the binding affinity to eIF4E. Indeed, the deletion of 4E-BP significantly increases levels of active eIF4E and enhances the rate of global protein translation, whereas hyposphorylated forms of 4E-BP deplete active eIF4E and thereby inhibit protein translation (Oulhen et al., 2009; Carvalho et al., 2017).
Multiple signaling pathways regulate protein translation by modulating the activity of eIFs (Roux and Topisirovic 2012; Roux and Topisirovic 2018). Especially, mechanistic target of rapamycin (mTOR) is the major signaling pathway involved in regulating the global rate of protein synthesis (Wang and Proud 2006; Ma and Blenis 2009). mTOR is an evolutionarily conserved serine/threonine kinase that stimulates anabolic processes including protein translation, in response to high nutrient/oxygen availability, high energy status, and high levels of insulin/growth factors/growth hormones. mTOR complex 1 (mTORC1), consisting of mTOR, the scaffolding protein raptor, the GTPase β subunit-like protein (GβL), proline-rich AKT substrate of 40 kDa (PRAS40), and Deptor, can directly enhance the activity of translational machinery. For example, the activated mTORC1 hyperphosphorylates and inactivates 4E-BP, which normally attenuates protein translation by preventing eIF4E from interacting with eIF4G to form the eIF4F complex (Wang and Proud 2006; Ma and Blenis 2009).
In addition, mTORC1 can indirectly enhance protein translation by phosphorylating the Thr389 site of the S6 kinase (S6K) and thereby activating S6K (Wang and Proud 2006; Ma and Blenis 2009). S6K has been shown to promote protein translation by phosphorylating multiple components of the translational machinery. For instance, S6K stimulates ribosomal biogenesis and protein translation by phosphorylating ribosomal protein S6 (rpS6) of the 40S ribosomal subunit (Jastrzebski et al., 2007; Chauvin et al., 2014). S6K also disinhibits eEF2 by phosphorylating/inactivating the eEF2 kinase (eEF2K), a well-known negative regulator of eEF2 (Price et al., 1991; Wang et al., 2001). Since eEF2 is a GTPase facilitating the transfer of peptidyl-tRNA from the A site to the P site, S6K activation promotes the ribosomal translocation and ultimately protein translation by stimulating the eEF2 activity. Further, S6K activation upregulates the eIF4A activity by inactivating programmed cell death 4 (PDCD4) and stimulating eIF4B, leading to high protein translation rate (Dennis et al., 2012). PDCD4 is known to repress eIF4A activity by blocking eIF4G-eIF4A interactions (Yang et al., 2003). eIF4B has been shown to bolster the RNA-unwinding activity of eIF4A (Andreou et al., 2017).
Multiple upstream signaling pathways converge on mTOR signaling so that protein translation rate can be adjusted in response to external stimuli and internal cellular conditions (Wang and Proud 2006; Ma and Blenis 2009). Growth hormones and growth factors (e.g. insulin and insulin-like growth factor (IGF)) first activate phosphoinositide 3-kinase (PI3K) via receptor tyrosine kinases and associated adaptor proteins such as insulin receptor substrates (IRS). PI3K in turn activates protein kinase B (AKT), which feeds to the mTORC1 signaling via tuberous sclerosis complex 2 (TSC2). Growth factors can also stimulate the mTORC1 signaling via Ras GTPases and mitogen-activated protein kinase (MAPK) cascades, which also feed to TSC2. Information of the nutrient availability and energy status is conveyed to mTORC1 via Rag GTPases, AMPK serine/threonine kinase, and regulated in development and DNA damage responses 1 (REDD1). Via the crosstalk of mTOR signaling with other pathways, cells can timely promote protein translation when environmental conditions are favorable for growth.
Besides mTOR signaling, mRNA processing bodies (known as P-bodies) also play crucial roles in regulating the rate of protein translation in response to environmental stress (Decker and Parker 2012; Luo et al., 2018). P-bodies are cytoplasmic ribonucleoprotein granules known to sequester mRNA and facilitate the mRNA decay via deadenylation and decapping, which reduces the mRNA stability. In addition to promoting the mRNA decay, P-bodies can attenuate protein translation rate by trapping translation initiation factors, mainly eIF4E (Brengues and Parker 2007; Rieckher et al., 2018). The P-bodies-dependent halting of global protein translation is particularly beneficial in response to various cellular stresses since this can protect the proteome and divert energy sources into stress responses instead (Yamasaki and Anderson 2008; Rieckher et al., 2018).
4 Protein translation declines with age
Since the 1970s, many investigators have studied how aging impacts the rate of protein translation. In general, the rate of global protein translation is high during early-adulthood but thereafter drops with age (by up to 88%) in yeast, C. elegans, Drosophila, mice, rats, sheep, and humans (Hrachovec 1969; Young et al., 1975; Webster and Webster 1979; Dwyer et al., 1980; Ekstrom et al., 1980; Webster et al., 1981; Blazejowski and Webster 1983; Webster and Webster 1983; Bailey and Webster 1984; Ward and Richardson 1991; Sonntag et al., 1992; Rooyackers et al., 1996; Connors et al., 2008; Belozerov et al., 2014; Depuydt et al., 2016; Hu et al., 2018; Ravi et al., 2018). This age-related decline in protein translation was observed in a wide variety of cellular fractions (cytosol and mitochondria), tissues, and organs, including brain, lung, heart, thymus, muscle, liver, kidney, intestine, pancreas, etc and has almost universally shown a decline in PT with age this includes a review of >40 studies of protein translation in the liver of mice and rats using multiple measures of protein translation including cell free, liver slices, isolated hepatocytes, and perfused livers (Ward and Richardson 1991) (Figure 3). There is only one study to our knowledge showing PT to increase with age, comparing PT in heart tissue in 4 months–10 month old mice (Ravi et al., 2018). Most of studies examine an older age (22+ Months for the older time point), it is possible that PT rises early life and then subsequently falls. This is consistent with data in livers showing a rise in PT from month three–six then a subsequent decline (Ward and Richardson 1991).
FIGURE 3
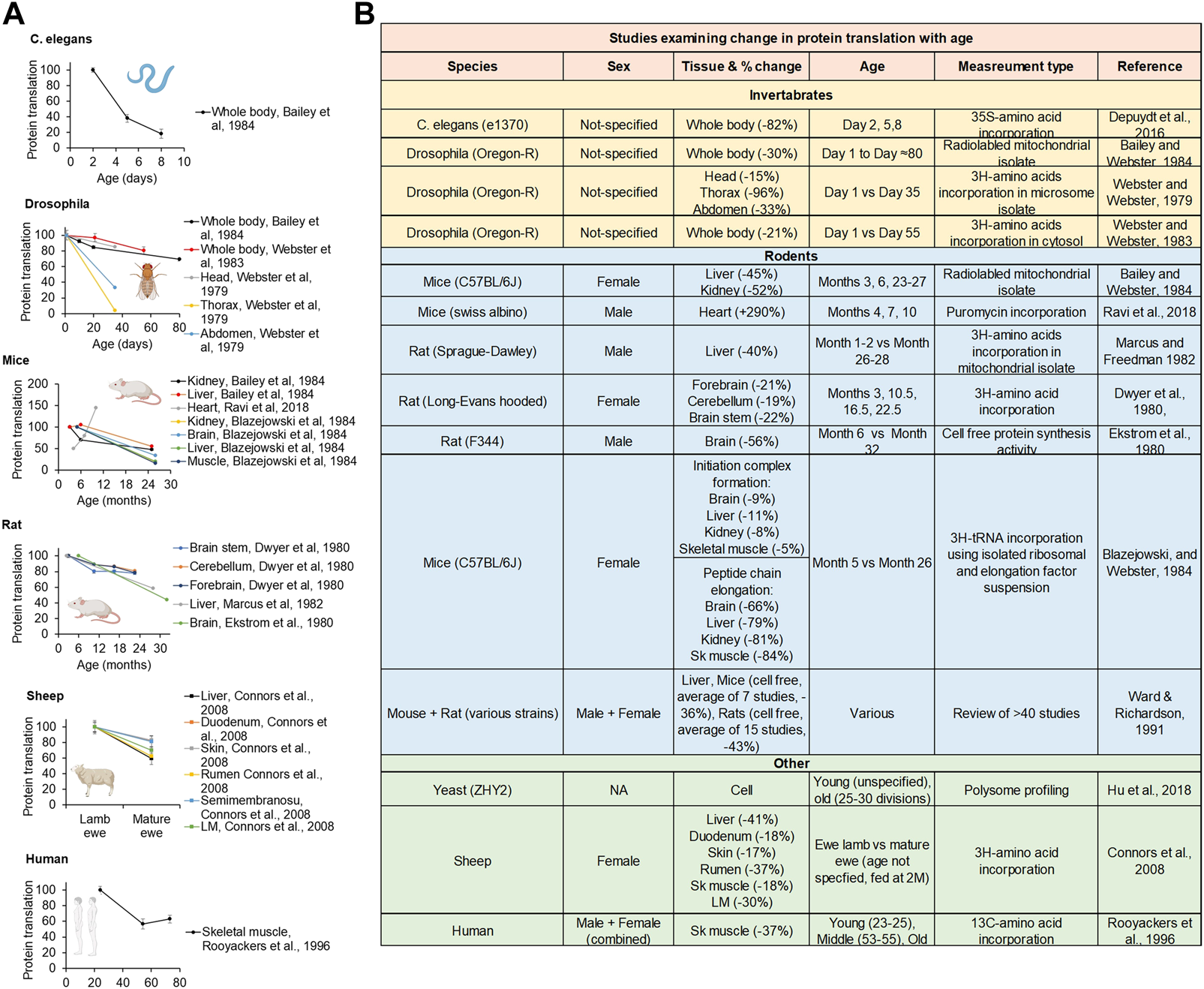
Lifetime protein translation dynamics. (A) Replotted protein translation rates across lifespan from studies in C.elegans, Drosophila, mice, rats, and humans. (B) Table of studies examining protein translation rates comparing young and old animals. Created with BioRender.com.
The exact molecular mechanism underlying this age-related decline in protein translation is still unknown. However, several studies suggest that reductions in the activity and levels of eIFs may impair the initiation step and thereby contribute to the age-dependent fall in protein translation. For example, the amount of eIF2 and the activity of eIF2 to promote ternary complex formation decline with age in multiple tissues of rats such as liver, spleen, kidney, lung, and brain (Vargas and Castaneda 1983; Cales et al., 1986; Kimball et al., 1992). Similarly, the amount and activity of eIF2B, which is essential for replenishing the eIF2 activity by facilitating the GDP-GTP exchange, declines with age in brains and livers of rats (Kimball et al., 1992). Interestingly, age-related reductions in levels of active eIF2 were strongly correlated with the decline in protein translation across the lifespan (Kimball et al., 1992). Besides eIF2, the amount of eIF5, which promotes the formation of the 43S pre-initiation complex, also declines with age in several brain areas of rats (Luchessi et al., 2008). Whether the activity and level of other eIFs such as eIF1, eIF3, eIF4A, eIF4E, eIF4G, etc. decline with age is still unknown.
The elongation step of protein translation is also compromised with age. For instance, the ribosomal half-transit time (the elongation time required for the synthesis of an average half-length of a nascent peptide) was increased by ∼60% in hepatocytes isolated from old rats (Coniglio et al., 1979). The later in vivo studies with yeast, C. elegans, mice, rats, and sheep also showed that the rate of peptide elongation significantly slows down with age (Blazejowski and Webster 1984; Merry and Holehan 1991; Connors et al., 2008; Stein et al., 2022). With age, as ribosomal elongation slows down, the frequency of ribosomal pausing increases as well, leading to increased ribosomal collisions and less efficient protein translation (Stein et al., 2022). Based on these observations, subsequent studies have investigated how the amount and function of elongation factors alter with age. The initial study done with Drosophila found that the protein level and activity of eEF1A declined with age by ∼60% (Webster and Webster 1983). The decline in eEF1A mRNA level occurred at the same time. Consistent with these data, the catalytic activity of eEF1A to bind aminoacyl tRNA to ribosomes was decreased by 60%–85% in liver, brain, kidney, and skeletal muscle of mice and rats (Moldave et al., 1979; Vargas and Castaneda 1981; Gabius et al., 1983; Webster and Webster 1983; Rattan et al., 1991). Unlike the eEF1 data, there have been some mixed reports on age-related alterations in eEF2. Some studies reported age-related decline in the amount and activity of eEF2 (Takahashi et al., 1985; Munoz et al., 2017), whereas others failed to observe such age-related changes (Moldave et al., 1979; Gabius et al., 1983).
Based on these results, in an attempt to prevent age-related decline in protein translation, Shepherd et al. created a Drosophila line that overexpresses the eEF1A gene (Shepherd et al., 1989). Interestingly, the extra copy of the eEF1A gene significantly prolonged the median and maximum lifespan of flies (∼41% increase in the median lifespan). However, the later study raised potential problems of the genetic construct and challenged interpretations of the data (Shikama et al., 1994). Shikama et al., just like the original study by Shepherd et al., did observe a robust lifespan extension in the fly line that overexpresses eEF1A. However, Shikama et al. found that the fly line that is supposed to overexpress eEF1A gene did not actually express more eEF1A mRNA or have more eEF1A protein levels. The lifespan extension was proposed to result from not the insertion of the transgene per se but the insertion of P-element reverting a life-shortening effect of the rosy gene mutation background. Since then, no studies have investigated how age-related alterations in eEF1A impact protein translation and lifespan.
The ribosomal loading to mRNA is also impaired with age. For example, the proportion of ribosomes in polyribosome (polysome) fractions significantly decreases with age in multiple animal species (Wallach and Gershon 1974; Kurtz 1978; Webster et al., 1981; Pluskal et al., 1984; Lee et al., 1993; Hu et al., 2018). Conversely, there is an age-dependent increase in monomeric ribosomes (monosomes), implying that less ribosomes are actively associated with mRNA with increasing age. As a result, the ability of ribosomes to incorporate labeled amino acids into a polypeptide chain is severely compromised with age (Britton and Sherman 1975; Kurtz 1978; Mori et al., 1979; Ekstrom et al., 1980; Linnemann et al., 1993). The age-related reductions in the amount and functionality of various eIFs (discussed above) may contribute to the impairment in ribosomal attachment to the mRNA. Another contributor to the compromised ribosomal loading would be age-related decline in translational output of ribosomal protein mRNA and transcripts related to ribosomal biogenesis, potentially due to their increased methylation (D'Aquila et al., 2017; Hu et al., 2018; Anisimova et al., 2020). In fact, ribosome occupancy of translation-related genes drops with age by twofold or even more (Anisimova et al., 2020). At the same time, translational output of genes that downregulate protein translation is increased: e.g. ssd1, which sequesters mRNAs and translational machineries into P-bodies and Gcn2, which inactivates the eIF2-dependent ternary complex formation (Hu et al., 2018).
Recent studies suggest that not only reductions in ribosomes or translational machineries per se but also the disruption of the stoichiometry of translational machinery components may contribute to decline in protein translation with age. For example, the parallel analysis of Ribo-seq (ribosome profiling) data and RNA-seq data in young and old yeast cells showed that the abundance of transcripts increased with age whereas the translation of each transcript decreased with age (Hu et al., 2018; Tye et al., 2019). These data suggest that the imbalance between transcripts and translational machineries becomes exacerbated with age, which may cause inefficient loading of translational apparatus to mRNA. Similar divergent changes in transcripts and proteins with age were observed in mammals as well. For example, in African killifish, levels of transcripts encoding ribosomal proteins continuously increased with age whereas the abundance of ribosomal proteins decreased with age (Kelmer Sacramento et al., 2020). This age-related loss of stoichiometry of ribosomal proteins may impair the ribosomal assembly, leading to decreased efficiency of protein translation. The subsequently increased pool of orphan ribosomal proteins may increase the risk of protein misfolding and aggregation (Tye et al., 2019), which may further attenuate protein translation by activating the PERK-UPR axis (Walter and Ron 2011). Consistent with these results, the translational output of ribosomal proteins increased whereas that of other translational factors decreased in rat brains across ages (Ori et al., 2015). This proteomic imbalance between ribosomal proteins and translational factors may impair efficient assembly of pre-initiation complex and compromise the ribosomal loading to mRNA.
5 Life-long reduction in protein translation improves lifespan and healthspan
Protein translation is an essential cellular process, playing crucial roles in growth, development, and reproduction (Pan et al., 2007). As previously discussed, protein translation precipitously declines with age, staying at low basal levels throughout middle-old ages in multiple animal species, including humans. One would expect that lowering protein translation would be detrimental to health. However, a life-long reduction in protein translation slows down aging, prolongs lifespan, and ameliorates cellular senescence and several age age-related diseases (Scheuner et al., 2005; Pan et al., 2007; Syntichaki et al., 2007; Steffen et al., 2008; Selman et al., 2009; Tain et al., 2009; Blagden and Willis 2011; Rogers et al., 2011; Martin et al., 2014; Takauji et al., 2016; Ren et al., 2019) (Figure 4).
FIGURE 4
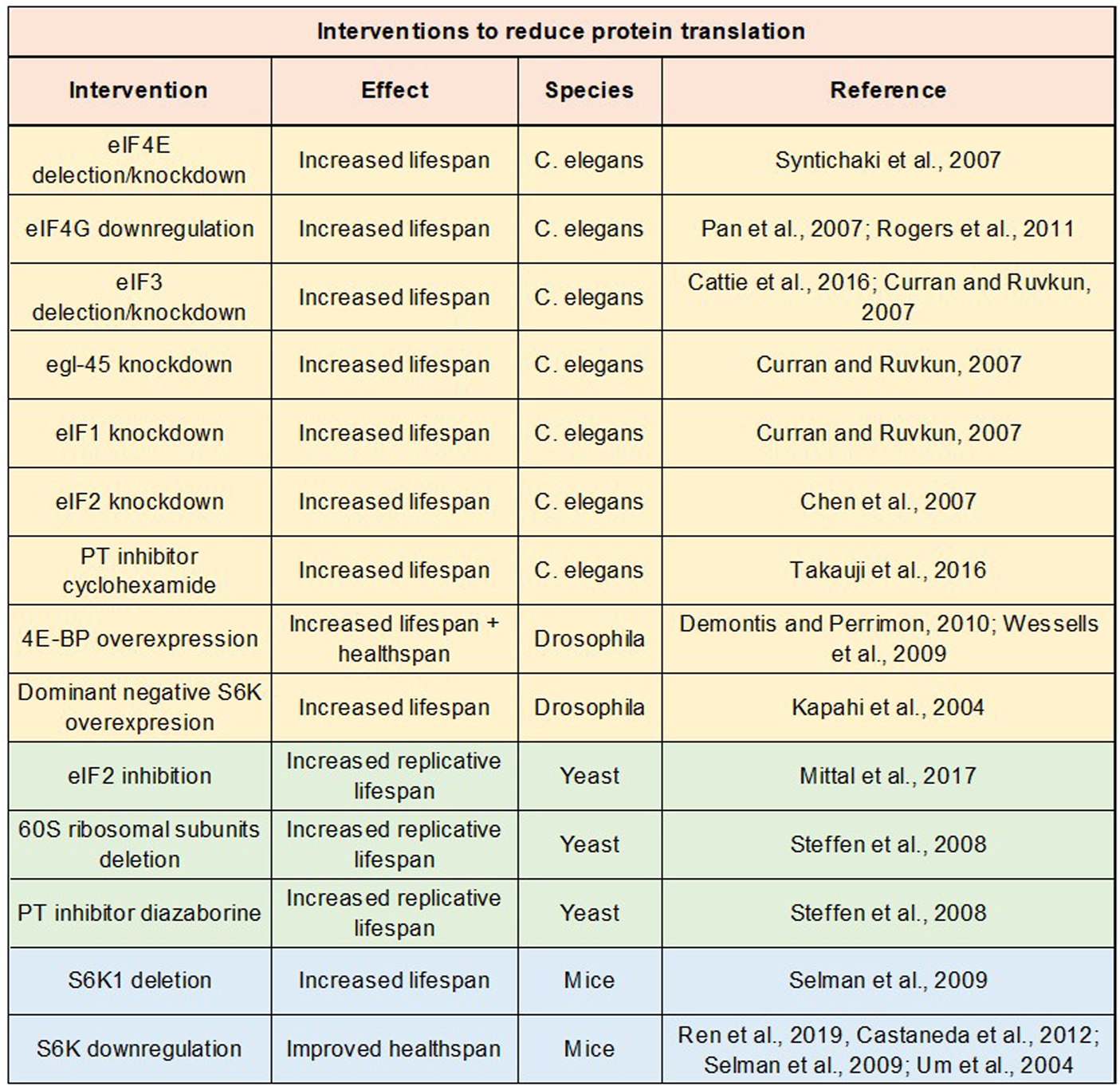
Impact of protein translation repression on lifespan and healthspan. Table of studies examining impact on lifespan or healthspan of genetic and drug interventions to repress protein translation.
From 1970s to 1990s, most of aging studies had focused on the effects of aging on protein synthesis. Since the early 2000s, investigators began to examine the reverse relationship: the effects of protein synthesis on the aging process. Syntichaki et al. were one of the first groups to study how perturbations in protein translation impact the animal lifespan (Syntichaki et al., 2007). Syntichaki et al. attenuated protein translation in C. elegans by deleting or knocking down the translation initiation factor eIF4E isoforms in somatic tissues. This manipulation substantially prolonged the nematode lifespan (∼55% extension in the median lifespan). Subsequent studies inhibited mRNA translation by mutating or knocking down other translation initiation factors and observed robust lifespan extension effects as well. For example, downregulation of eIF4G, which interacts with eIF4E to facilitate formation of the eIF4F complex, improved the nematode lifespan by more than 30% (Pan et al., 2007; Rogers et al., 2011). Similarly, deletion or RNAi of eIF3, which plays a critical role in formation of the 43S pre-initiation complex, led to ∼40% extension of the C. elegans lifespan (Curran and Ruvkun 2007; Cattie et al., 2016). Knocking down other translation initiation factors such as eIF1, eIF2, eIF2B, eIF4A, eIF5A, etc. also significantly improved the lifespan (Hamilton et al., 2005; Chen et al., 2007; Curran and Ruvkun 2007; Tohyama et al., 2008).
Later studies have targeted different translational machineries to inhibit protein synthesis in animal models other than C. elegans. Similar lifespan extension effects were observed. For example, overexpression of 4E-BP, which sequesters eIF4E and thereby attenuates protein translation, ameliorated age-related protein aggregation, improved functions of heart and skeletal muscle at old ages, and extended the Drosophila lifespan (Wessells et al., 2009; Demontis and Perrimon 2010). Moreover, reducing protein translation by inhibiting the eIF2 activity via Gcn4 extended the replicative lifespan of S. cerevisiae (Mittal et al., 2017). Further, deletions of or mutations in several ribosomal proteins significantly increased the lifespan of the budding yeast and filamentous fungi (Belcour et al., 1991; Steffen et al., 2008). Likewise, reducing the levels and activity of S6K, which promotes ribosomal biogenesis, robustly improved the lifespan of C. elegans, Drosophila, and rodents (Kapahi et al., 2004; Pan et al., 2007; Selman et al., 2009). For example, in fruit flies, dominant negative forms of S6K significantly increased the lifespan, whereas constitutively active forms of S6K caused accelerated aging and shortened the lifespan (Kapahi et al., 2004). Consistent with these results, inhibiting mTOR signaling, which enhances protein translation via S6K activation, promotes longevity in multiple animal models (Johnson et al., 2013).
Pharmacologically inhibiting protein translation has also been shown to slow down aging as well. For example, cyclohexamide, a well-known protein synthesis inhibitor targeting the elongation phase of translation, abolished senescent features in human cells and robustly improved the C. elegans lifespan (Takauji et al., 2016). In addition, diazaborine, which reduces levels of 60S ribosomal proteins and inhibits ribosomal biogenesis and maturation, significantly increased the replicative lifespan of the budding yeast (Steffen et al., 2008). Further, attenuating protein translation by minocycline enhanced proteostasis and promoted the longevity in C. elegans (Solis et al., 2018). Minocycline also had neuroprotective and anti-inflammatory effects and ameliorated AD-related cognitive deficits in rodents (Choi et al., 2005; Festoff et al., 2006; Seabrook et al., 2006; Choi et al., 2007; Noble et al., 2009). However, unlike cyclohexamide and diazaborine, minocycline is an antibiotic that does not specifically target protein translation and has the potential to impact other pathways.
Manipulations that lower protein translation improve not only lifespan but also healthspan and ameliorate several age-related diseases, including frailty, osteoporosis, metabolic disorders, cardiovascular disease, cancer, and neurodegenerative disorders. For example, diminishing protein translation rate by inhibiting ribosomal biogenesis alleviated physiological aging of human mesenchymal stem cells and counteracted the development of frailty and osteoarthritis in mice (Ren et al., 2019). Moreover, downregulating S6K signaling to attenuate protein translation in mice improved the locomotor activity, increased the population of naïve T cells, enhanced the bone volume and density, and protected against age- and diet-induced obesity/diabetes by improving insulin sensitivity and reducing adiposity (Um et al., 2004; Selman et al., 2009; Castaneda et al., 2012). Consistent with these studies, in rodent models of type II diabetes, S6K signaling was overactive and protein translation rates were elevated in islet β-cells (Hatanaka et al., 2014; Talaei et al., 2014). In addition, enhancing protein translation via overactivation of eIF2 impaired glucose tolerance and caused diabetic phenotypes in mice (Harding et al., 2001; Zhang et al., 2002b). Conversely, attenuating protein translation by lowering the eIF2 activity ameliorated the apoptosis of β cells and improved the glucose tolerance (Scheuner et al., 2005; Back et al., 2009).
Further, reduction in protein translation by depleting eIF4E with 4E-BP slowed down cardiac aging and decreased the frequency of cardiac failure at old ages (Wessells et al., 2009). Knocking down eIF4E ameliorated tumorigenesis as well, and consequently, drugs targeting eIF4E to inhibit mRNA translation have been heavily tested in clinical trials for hematologic malignancies (Rinker-Schaeffer et al., 1993; Blagden and Willis 2011). Overexpression of 4E-BP also prevented dopaminergic neuronal loss and ameliorated locomotor deficits in fly models of Parkinson’s disease, induced by parkin and pink1 mutations (Tain et al., 2009; Liu and Lu 2010). Likewise, in another fly model of Parkinson’s disease caused by the mutation in leucine-rich repeat kinase 2 (LRRK2), the most common genetic cause of both familial and sporadic Parkinson’s disease, global protein translation rate was elevated due to phosphorylation of ribosomal protein s15 (Martin et al., 2014). Introducing phosphodeficient s15 to attenuate protein translation ameliorated dopaminergic neuronal degeneration and age-related locomotor deficits in LRRK2 mutant flies (Martin et al., 2014).
A few studies suggest that overactive protein translation may contribute to the AD pathogenesis as well. For example, the pathogenic Aβ1-42 increased protein translation by activating the eIF4E activity (Ghosh et al., 2020). In addition, cytoplasmic FMR1-interacting protein (CYFIP), which represses protein translation by blocking eIF4E-eIF4G interactions, was downregulated in post-mortem brains of AD patients, and CYFIP reduction caused AD pathologies in mice (Tiwari et al., 2016; Ghosh et al., 2020). Further, tau K174 acetylation, which occurs in brains of AD patients/mice at early stages of the disease, enhanced protein translation by causing nucleolar expansion (Min et al., 2015; Portillo et al., 2021). Reduction in protein translation by downregulating S6K signaling improved spatial memory and synaptic plasticity in an AD mouse model (Caccamo et al., 2015).
Consistent with the data discussed above, long-lived strains of animals show low rates of protein translation. For example, long-lived C. elegans strains with reduced insulin/IGF signaling, reduced mTOR signaling, reduced germline signaling, or impaired mitochondrial electron transport chain functions, etc. showed smaller sized nucleoli, decreased ribosomal biogenesis, and significantly lower protein translation rates (Depuydt et al., 2013; Tiku et al., 2017). Likewise, long-lived growth hormone-deficient Snell dwarf mice exhibited decreased rates of protein translation in liver, heart, and skeletal muscle (Bates and Holder 1988; Thompson et al., 2016). Conversely, fibroblasts from patients with premature aging disorders showed nucleolar expansion, increased ribosomal biogenesis, and elevated protein translation rates (Buchwalter and Hetzer 2017). These data indirectly suggest that long-lived animals may achieve the longevity by sustaining low rates of protein translation.
6 Discussion and conclusion: Protein translation paradox
Extensive studies from the 1970s to 1990s have shown protein translation rates to decline over the course of age. These studies encompass a menagerie of organisms including yeast, C. elegans, Drosophila, mice, rats, sheep, and humans in a plethora of tissues including brain, lung, heart, thymus, muscle, liver, kidney, intestine, pancreas, etc. Thus, it has been postulated that reduction in protein translation would accelerate the aging process. However, research since the 2000s has shown that life-long reduction in protein translation actually robustly improves the lifespan and healthspan. These conflicting findings raise the so-called protein synthesis paradox (Blagosklonny et al., 2007). The paradox has not been resolved, which imposes a substantial barrier to fully understand how protein translation regulates aging.
The paradox may have mainly arisen from the assumption that age-related decline in protein translation is a passive byproduct of aging. Proteasomal and autophagial degradations decline with age, so in order to match with the reduced capacity of degradation machineries and thus maintain proteostasis, protein translation may have been repressed with age as an adaptive response. In this way, organisms can minimize proteostatic burden, reduce levels of damaged and aggregated proteins, and thus optimize the lifespan. This new theoretical framework can resolve inconsistently-seemed findings with regard to translational regulation of aging. To validate this idea, further studies should be done to investigate how the rise and fall in protein translation across lifespan regulate aging and proteostasis. Till now, all the aging studies have manipulated protein translation throughout the whole life rather than modifying it in a specific time window. With understanding of how lifetime protein translation dynamics regulate the onset of aging, we will be able to fully appreciate how protein translation modulates the aging process.
Statements
Author contributions
HK researched and wrote initial draft AP and HK reviewed and prepared final version together.
Funding
NIA/NIH R56 AG061051, Glenn Foundation for Medical Research, and AFAR Grants for Junior Faculty, Voelcker Young Investigator Award NIH/NIA RF1 AG065301, San Antonio Nathan Shock Center, San Antonio Pepper Center.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1
AndreouA. Z.HarmsU.KlostermeierD. (2017). eIF4B stimulates eIF4A ATPase and unwinding activities by direct interaction through its 7-repeats region. RNA Biol.14 (1), 113–123. 10.1080/15476286.2016.1259782
2
AnisimovaA. S.MeersonM. B.GerashchenkoM. V.KulakovskiyI. V.DmitrievS. E.GladyshevV. N. (2020). Multifaceted deregulation of gene expression and protein synthesis with age. Proc. Natl. Acad. Sci. U. S. A.117 (27), 15581–15590. 10.1073/pnas.2001788117
3
BackS. H.ScheunerD.HanJ.SongB.RibickM.WangJ.et al (2009). Translation attenuation through eIF2alpha phosphorylation prevents oxidative stress and maintains the differentiated state in beta cells. Cell Metab.10 (1), 13–26. 10.1016/j.cmet.2009.06.002
4
BaileyP. J.WebsterG. C. (1984). Lowered rates of protein synthesis by mitochondria isolated from organisms of increasing age. Mech. Ageing Dev.24 (2), 233–241. 10.1016/0047-6374(84)90074-5
5
BalchinD.Hayer-HartlM.HartlF. U. (2016). In vivo aspects of protein folding and quality control. Science353 (6294), aac4354. 10.1126/science.aac4354
6
BatesP. C.HolderA. T. (1988). The anabolic actions of growth hormone and thyroxine on protein metabolism in Snell dwarf and normal mice. J. Endocrinol.119 (1), 31–41. 10.1677/joe.0.1190031
7
BelozerovV. E.RatkovicS.McNeillH.HillikerA. J.McDermottJ. C. (2014). In vivo interaction proteomics reveal a novel p38 mitogen-activated protein kinase/Rack1 pathway regulating proteostasis in Drosophila muscle. Mol. Cell Biol.34 (3), 474–484. 10.1128/MCB.00824-13
8
BlagdenS. P.WillisA. E. (2011). The biological and therapeutic relevance of mRNA translation in cancer. Nat. Rev. Clin. Oncol.8 (5), 280–291. 10.1038/nrclinonc.2011.16
9
BlazejowskiC. A.WebsterG. C. (1983). Decreased rates of protein synthesis by cell-free preparations from different organs of aging mice. Mech. Ageing Dev.21 (3-4), 345–356. 10.1016/0047-6374(83)90051-9
10
BlazejowskiC. A.WebsterG. C. (1984). Effect of age on peptide chain initiation and elongation in preparations from brain, liver, kidney and skeletal muscle of the C57B1/6J mouse. Mech. Ageing Dev.25 (3), 323–333. 10.1016/0047-6374(84)90005-8
11
BrenguesM.ParkerR. (2007). Accumulation of polyadenylated mRNA, Pab1p, eIF4E, and eIF4G with P-bodies in Saccharomyces cerevisiae. Mol. Biol. Cell18 (7), 2592–2602. 10.1091/mbc.e06-12-1149
12
BrittonG. W.ShermanF. G. (1975). Altered regulation of protein synthesis during aging as determined by in vitro ribosomal assays. Exp. Gerontol.10 (1), 67–77. 10.1016/0531-5565(75)90016-9
13
BuchwalterA.HetzerM. W. (2017). Nucleolar expansion and elevated protein translation in premature aging. Nat. Commun.8 (1), 328. 10.1038/s41467-017-00322-z
14
CaccamoA.BrancaC.TalboomJ. S.ShawD. M.TurnerD.MaL.et al (2015). Reducing ribosomal protein S6 kinase 1 expression improves spatial memory and synaptic plasticity in a mouse model of alzheimer's disease. J. Neurosci.35 (41), 14042–14056. 10.1523/JNEUROSCI.2781-15.2015
15
CalesC.FandoJ. L.AzuaraC.SalinasM. (1986). Developmental studies of the first step of the initiation of brain protein synthesis, role for initiation factor 2. Mech. Ageing Dev.33 (2), 147–156. 10.1016/0047-6374(86)90023-0
16
CarvalhoG. B.DragoI.HoxhaS.YamadaR.MahnevaO.BruceK. D.et al (2017). The 4E-BP growth pathway regulates the effect of ambient temperature on Drosophila metabolism and lifespan. Proc. Natl. Acad. Sci. U. S. A.114 (36), 9737–9742. 10.1073/pnas.1618994114
17
CastanedaT. R.AbplanalpW.UmS. H.PflugerP. T.SchrottB.BrownK.et al (2012). Metabolic control by S6 kinases depends on dietary lipids. PLoS One7 (3), e32631. 10.1371/journal.pone.0032631
18
CattieD. J.RichardsonC. E.ReddyK. C.Ness-CohnE. M.DrosteR.ThompsonM. K.et al (2016). Mutations in nonessential eIF3k and eIF3l genes confer lifespan extension and enhanced resistance to ER stress in Caenorhabditis elegans. PLoS Genet.12 (9), e1006326. 10.1371/journal.pgen.1006326
19
ChauvinC.KokaV.NouschiA.MieuletV.Hoareau-AveillaC.DreazenA.et al (2014). Ribosomal protein S6 kinase activity controls the ribosome biogenesis transcriptional program. Oncogene33 (4), 474–483. 10.1038/onc.2012.606
20
ChenD.PanK. Z.PalterJ. E.KapahiP. (2007). Longevity determined by developmental arrest genes in Caenorhabditis elegans. Aging Cell6 (4), 525–533. 10.1111/j.1474-9726.2007.00305.x
21
ChocronE. S.MunkacsyE.KimH. S.KarpowiczP.JiangN.Van SkikeC. E.et al (2022). Genetic and pharmacologic proteasome augmentation ameliorates Alzheimer's-like pathology in mouse and fly APP overexpression models. Sci. Adv.8 (23), eabk2252. 10.1126/sciadv.abk2252
22
ChoiS. H.LeeD. Y.ChungE. S.HongY. B.KimS. U.JinB. K. (2005). Inhibition of thrombin-induced microglial activation and NADPH oxidase by minocycline protects dopaminergic neurons in the substantia nigra in vivo. J. Neurochem.95 (6), 1755–1765. 10.1111/j.1471-4159.2005.03503.x
23
ChoiY.KimH. S.ShinK. Y.KimE. M.KimM.KimH. S.et al (2007). Minocycline attenuates neuronal cell death and improves cognitive impairment in Alzheimer's disease models. Neuropsychopharmacology32 (11), 2393–2404. 10.1038/sj.npp.1301377
24
ConiglioJ. J.LiuD. S.RichardsonA. (1979). A comparison of protein synthesis by liver parenchymal cells isolated from Fischer F344 rats of various ages. Mech. Ageing Dev.11 (2), 77–90. 10.1016/0047-6374(79)90026-5
25
ConnorsM. T.PoppiD. P.CantJ. P. (2008). Protein elongation rates in tissues of growing and adult sheep. J. Anim. Sci.86 (9), 2288–2295. 10.2527/jas.2007-0159
26
CurranS. P.RuvkunG. (2007). Lifespan regulation by evolutionarily conserved genes essential for viability. PLoS Genet.3 (4), e56. 10.1371/journal.pgen.0030056
27
D'AquilaP.MontesantoA.MandalaM.GarastoS.MariV.CorsonelloA.et al (2017). Methylation of the ribosomal RNA gene promoter is associated with aging and age-related decline. Aging Cell16 (5), 966–975. 10.1111/acel.12603
28
DeckerC. J.ParkerR. (2012). P-Bodies and stress granules: Possible roles in the control of translation and mRNA degradation. Cold Spring Harb. Perspect. Biol.4 (9), a012286. 10.1101/cshperspect.a012286
29
DemontisF.PerrimonN. (2010). FOXO/4E-BP signaling in Drosophila muscles regulates organism-wide proteostasis during aging. Cell143 (5), 813–825. 10.1016/j.cell.2010.10.007
30
DennisM. D.JeffersonL. S.KimballS. R. (2012). Role of p70S6K1-mediated phosphorylation of eIF4B and PDCD4 proteins in the regulation of protein synthesis. J. Biol. Chem.287 (51), 42890–42899. 10.1074/jbc.M112.404822
31
DepuydtG.ShanmugamN.RasulovaM.DhondtI.BraeckmanB. P. (2016). Increased protein stability and decreased protein turnover in the Caenorhabditis elegans ins/IGF-1 daf-2 mutant. J. Gerontol. A Biol. Sci. Med. Sci.71 (12), 1553–1559. 10.1093/gerona/glv221
32
DepuydtG.XieF.PetyukV. A.ShanmugamN.SmoldersA.DhondtI.et al (2013). Camp, 2ndReduced insulin/insulin-like growth factor-1 signaling and dietary restriction inhibit translation but preserve muscle mass in Caenorhabditis elegans. Mol. Cell Proteomics12 (12), 3624–3639. 10.1074/mcp.M113.027383
33
DonnellyN.GormanA. M.GuptaS.SamaliA. (2013). The eIF2α kinases: Their structures and functions. Cell Mol. Life Sci.70 (19), 3493–3511. 10.1007/s00018-012-1252-6
34
DwyerB. E.FandoJ. L.WasterlainC. G. (1980). Rat brain protein synthesis declines during postdevelopmental aging. J. Neurochem.35 (3), 746–749. 10.1111/j.1471-4159.1980.tb03717.x
35
EkstromR.LiuD. S.RichardsonA. (1980). Changes in brain protein synthesis during the life span of male Fischer rats. Gerontology26 (3), 121–128. 10.1159/000212405
36
FestoffB. W.AmeenuddinS.ArnoldP. M.WongA.SantacruzK. S.CitronB. A. (2006). Minocycline neuroprotects, reduces microgliosis, and inhibits caspase protease expression early after spinal cord injury. J. Neurochem.97 (5), 1314–1326. 10.1111/j.1471-4159.2006.03799.x
37
GabiusH. J.EngelhardtR.DeerbergF.CramerF. (1983). Age-related changes in different steps of protein synthesis of liver and kidney of rats. FEBS Lett.160 (1-2), 115–118. 10.1016/0014-5793(83)80948-x
38
GebauerF.HentzeM. W. (2004). Molecular mechanisms of translational control. Nat. Rev. Mol. Cell Biol.5 (10), 827–835. 10.1038/nrm1488
39
GhoshA.MizunoK.TiwariS. S.ProitsiP.Gomez Perez-NievasB.GlennonE.et al (2020). Alzheimer's disease-related dysregulation of mRNA translation causes key pathological features with ageing. Transl. Psychiatry10 (1), 192. 10.1038/s41398-020-00882-7
40
HamiltonB.DongY.ShindoM.LiuW.OdellI.RuvkunG.et al (2005). A systematic RNAi screen for longevity genes in C. elegans. Genes Dev.19 (13), 1544–1555. 10.1101/gad.1308205
41
HansenM.RubinszteinD. C.WalkerD. W. (2018). Autophagy as a promoter of longevity: Insights from model organisms. Nat. Rev. Mol. Cell Biol.19 (9), 579–593. 10.1038/s41580-018-0033-y
42
HardingH. P.ZengH.ZhangY.JungriesR.ChungP.PleskenH.et al (2001). Diabetes mellitus and exocrine pancreatic dysfunction in perk-/- mice reveals a role for translational control in secretory cell survival. Mol. Cell7 (6), 1153–1163. 10.1016/s1097-2765(01)00264-7
43
HatanakaM.MaierB.SimsE. K.TemplinA. T.KulkarniR. N.Evans-MolinaC.et al (2014). Palmitate induces mRNA translation and increases ER protein load in islet beta-cells via activation of the mammalian target of rapamycin pathway. Diabetes63 (10), 3404–3415. 10.2337/db14-0105
44
HebertD. N.MolinariM. (2007). In and out of the ER: Protein folding, quality control, degradation, and related human diseases. Physiol. Rev.87 (4), 1377–1408. 10.1152/physrev.00050.2006
45
HrachovecJ. P. (1969). Age changes in amino acid incorporation by rat liver microsomes. Gerontologia15 (1), 52–63. 10.1159/000211674
46
HuZ.XiaB.PostnikoffS. D.ShenZ. J.TomoiagaA. S.HarknessT. A.et al (2018). Ssd1 and Gcn2 suppress global translation efficiency in replicatively aged yeast while their activation extends lifespan. Elife7, e35551. 10.7554/eLife.35551
47
JacksonR. J.HellenC. U.PestovaT. V. (2010). The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell Biol.11 (2), 113–127. 10.1038/nrm2838
48
JastrzebskiK.HannanK. M.TchoubrievaE. B.HannanR. D.PearsonR. B. (2007). Coordinate regulation of ribosome biogenesis and function by the ribosomal protein S6 kinase, a key mediator of mTOR function. Growth factors.25 (4), 209–226. 10.1080/08977190701779101
49
JohnsonS. C.RabinovitchP. S.KaeberleinM. (2013). mTOR is a key modulator of ageing and age-related disease. Nature493 (7432), 338–345. 10.1038/nature11861
50
KapahiP.ZidB. M.HarperT.KosloverD.SapinV.BenzerS. (2004). Regulation of lifespan in Drosophila by modulation of genes in the TOR signaling pathway. Curr. Biol.14 (10), 885–890. 10.1016/j.cub.2004.03.059
51
Kelmer SacramentoE.KirkpatrickJ. M.MazzettoM.BaumgartM.BartolomeA.Di SanzoS.et al (2020). Reduced proteasome activity in the aging brain results in ribosome stoichiometry loss and aggregation. Mol. Syst. Biol.16 (6), e9596. 10.15252/msb.20209596
52
KimballS. R.VaryT. C.JeffersonL. S. (1992). Age-dependent decrease in the amount of eukaryotic initiation factor 2 in various rat tissues. Biochem. J.286 (1), 263–268. 10.1042/bj2860263
53
KurtzD. I. (1978). A decrease in the number of active mouse liver ribosomes during aging. Exp. Gerontol.13 (6), 397–402. 10.1016/0531-5565(78)90050-5
54
LeeD. Y.HayesJ. J.PrussD.WolffeA. P. (1993). A positive role for histone acetylation in transcription factor access to nucleosomal DNA. Cell72 (1), 73–84. 10.1016/0092-8674(93)90051-q
55
LinnemannD.GaardsvollH.OlsenM.BockE. (1993). Expression of NCAM mRNA and polypeptides in aging rat brain. Int. J. Dev. Neurosci.11 (1), 71–81. 10.1016/0736-5748(93)90036-d
56
LiuS.LuB. (2010). Reduction of protein translation and activation of autophagy protect against PINK1 pathogenesis in Drosophila melanogaster. PLoS Genet.6 (12), e1001237. 10.1371/journal.pgen.1001237
57
LuchessiA. D.CambiaghiT. D.AlvesA. S.ParreirasE. S. L. T.BrittoL. R.Costa-NetoC. M.et al (2008). Insights on eukaryotic translation initiation factor 5A (eIF5A) in the brain and aging. Brain Res.1228, 6–13. 10.1016/j.brainres.2008.06.057
58
LuoY.NaZ.SlavoffS. A. (2018). P-Bodies: Composition, properties, and functions. Biochemistry57 (17), 2424–2431. 10.1021/acs.biochem.7b01162
59
MaX. M.BlenisJ. (2009). Molecular mechanisms of mTOR-mediated translational control. Nat. Rev. Mol. Cell Biol.10 (5), 307–318. 10.1038/nrm2672
60
MarcotrigianoJ.GingrasA. C.SonenbergN.BurleyS. K. (1999). Cap-dependent translation initiation in eukaryotes is regulated by a molecular mimic of eIF4G. Mol. Cell3 (6), 707–716. 10.1016/s1097-2765(01)80003-4
61
MartinI.KimJ. W.LeeB. D.KangH. C.XuJ. C.JiaH.et al (2014). Ribosomal protein s15 phosphorylation mediates LRRK2 neurodegeneration in Parkinson's disease. Cell157 (2), 472–485. 10.1016/j.cell.2014.01.064
62
MerryB. J.HolehanA. M. (1991). Effect of age and restricted feeding on polypeptide chain assembly kinetics in liver protein synthesis in vivo. Mech. Ageing Dev.58 (2-3), 139–150. 10.1016/0047-6374(91)90088-h
63
MinS. W.ChenX.TracyT. E.LiY.ZhouY.WangC.et al (2015). Critical role of acetylation in tau-mediated neurodegeneration and cognitive deficits. Nat. Med.21 (10), 1154–1162. 10.1038/nm.3951
64
MittalN.GuimaraesJ. C.GrossT.SchmidtA.Vina-VilasecaA.NedialkovaD. D.et al (2017). The Gcn4 transcription factor reduces protein synthesis capacity and extends yeast lifespan. Nat. Commun.8 (1), 457. 10.1038/s41467-017-00539-y
65
MoldaveK.HarrisJ.SaboW.SadnikI. (1979). Protein synthesis and aging: Studies with cell-free mammalian systems. Fed. Proc.38 (6), 1979–1983.
66
MoriN.MizunoD.GotoS. (1979). Conservation of ribosomal fidelity during ageing. Mech. Ageing Dev.10 (6), 379–398. 10.1016/0047-6374(79)90020-4
67
MunkacsyE.ChocronE. S.QuintanillaL.GendronC. M.PletcherS. D.PickeringA. M. (2019). Neuronal-specific proteasome augmentation via Prosβ5 overexpression extends lifespan and reduces age-related cognitive decline. Aging Cell18 (5), e13005. 10.1111/acel.13005
68
MunozM. F.ArguellesS.CanoM.MarottaF.AyalaA. (2017). Aging and oxidative stress decrease pineal elongation factor 2: In vivo protective effect of melatonin in young rats treated with cumene hydroperoxide. J. Cell Biochem.118 (1), 182–190. 10.1002/jcb.25624
69
MusaJ.OrthM. F.DallmayerM.BaldaufM.PardoC.RotblatB.et al (2016). Eukaryotic initiation factor 4E-binding protein 1 (4E-BP1): A master regulator of mRNA translation involved in tumorigenesis. Oncogene35 (36), 4675–4688. 10.1038/onc.2015.515
70
NguyenN. N.RanaA.GoldmanC.MooreR.TaiJ.HongY.et al (2019). Proteasome β5 subunit overexpression improves proteostasis during aging and extends lifespan in Drosophila melanogaster. Sci. Rep.9 (1), 3170. 10.1038/s41598-019-39508-4
71
NobleW.GarwoodC.StephensonJ.KinseyA. M.HangerD. P.AndertonB. H. (2009). Minocycline reduces the development of abnormal tau species in models of Alzheimer's disease. FASEB J.23 (3), 739–750. 10.1096/fj.08-113795
72
OriA.ToyamaB. H.HarrisM. S.BockT.IskarM.BorkP.et al (2015). Integrated transcriptome and proteome analyses reveal organ-specific proteome deterioration in old rats. Cell Syst.1 (3), 224–237. 10.1016/j.cels.2015.08.012
73
OulhenN.BoulbenS.BidinostiM.MoralesJ.CormierP.CossonB. (2009). A variant mimicking hyperphosphorylated 4E-BP inhibits protein synthesis in a sea urchin cell-free, cap-dependent translation system. PLoS One4 (3), e5070. 10.1371/journal.pone.0005070
74
PanK. Z.PalterJ. E.RogersA. N.OlsenA.ChenD.LithgowG. J.et al (2007). Inhibition of mRNA translation extends lifespan in Caenorhabditis elegans. Aging Cell6 (1), 111–119. 10.1111/j.1474-9726.2006.00266.x
75
PluskalM. G.MoreyraM.BuriniR. C.YoungV. R. (1984). Protein synthesis studies in skeletal muscle of aging rats. I. Alterations in nitrogen composition and protein synthesis using a crude polyribosome and pH 5 enzyme system. J. Gerontol.39 (4), 385–391. 10.1093/geronj/39.4.385
76
PortilloM.EremenkoE.KaluskiS.Garcia-VenzorA.OnnL.SteinD.et al (2021). SIRT6-CBP-dependent nuclear Tau accumulation and its role in protein synthesis. Cell Rep.35 (4), 109035. 10.1016/j.celrep.2021.109035
77
PowersE. T.MorimotoR. I.DillinA.KellyJ. W.BalchW. E. (2009). Biological and chemical approaches to diseases of proteostasis deficiency. Annu. Rev. Biochem.78, 959–991. 10.1146/annurev.biochem.052308.114844
78
PriceN. T.RedpathN. T.SeverinovK. V.CampbellD. G.RussellJ. M.ProudC. G. (1991). Identification of the phosphorylation sites in elongation factor-2 from rabbit reticulocytes. FEBS Lett.282 (2), 253–258. 10.1016/0014-5793(91)80489-p
79
RattanS. I.WardW. F.GlentingM.SvendsenL.RiisB.ClarkB. F. (1991). Dietary calorie restriction does not affect the levels of protein elongation factors in rat livers during ageing. Mech. Ageing Dev.58 (1), 85–91. 10.1016/0047-6374(91)90122-g
80
RaviV.JainA.AhamedF.FathmaN.DesinguP. A.SundaresanN. R. (2018). Systematic evaluation of the adaptability of the non-radioactive SUnSET assay to measure cardiac protein synthesis. Sci. Rep.8 (1), 4587. 10.1038/s41598-018-22903-8
81
RenX.HuB.SongM.DingZ.DangY.LiuZ.et al (2019). Maintenance of nucleolar homeostasis by CBX4 alleviates senescence and osteoarthritis. Cell Rep.26 (13), 3643–3656. e3647. 10.1016/j.celrep.2019.02.088
82
RieckherM.MarkakiM.PrinczA.SchumacherB.TavernarakisN. (2018). Maintenance of proteostasis by P body-mediated regulation of eIF4E availability during aging in Caenorhabditis elegans. Cell Rep.25 (1), 199–211. 10.1016/j.celrep.2018.09.009
83
Rinker-SchaefferC. W.GraffJ. R.De BenedettiA.ZimmerS. G.RhoadsR. E. (1993). Decreasing the level of translation initiation factor 4E with antisense RNA causes reversal of ras-mediated transformation and tumorigenesis of cloned rat embryo fibroblasts. Int. J. Cancer55 (5), 841–847. 10.1002/ijc.2910550525
84
RogersA. N.ChenD.McCollG.CzerwieniecG.FelkeyK.GibsonB. W.et al (2011). Life span extension via eIF4G inhibition is mediated by posttranscriptional remodeling of stress response gene expression in C. elegans. Cell Metab.14 (1), 55–66. 10.1016/j.cmet.2011.05.010
85
RooyackersO. E.AdeyD. B.AdesP. A.NairK. S. (1996). Effect of age on in vivo rates of mitochondrial protein synthesis in human skeletal muscle. Proc. Natl. Acad. Sci. U. S. A.93 (26), 15364–15369. 10.1073/pnas.93.26.15364
86
RouxP. P.TopisirovicI. (2012). Regulation of mRNA translation by signaling pathways. Cold Spring Harb. Perspect. Biol.4 (11), a012252. 10.1101/cshperspect.a012252
87
RouxP. P.TopisirovicI. (2018). Signaling pathways involved in the regulation of mRNA translation. Mol. Cell Biol.38 (12), e00070-18. 10.1128/MCB.00070-18
88
ScheunerD.Vander MierdeD.SongB.FlamezD.CreemersJ. W.TsukamotoK.et al (2005). Control of mRNA translation preserves endoplasmic reticulum function in beta cells and maintains glucose homeostasis. Nat. Med.11 (7), 757–764. 10.1038/nm1259
89
SeabrookT. J.JiangL.MaierM.LemereC. A. (2006). Minocycline affects microglia activation, Abeta deposition, and behavior in APP-tg mice. Glia53 (7), 776–782. 10.1002/glia.20338
90
SelmanC.TulletJ. M.WieserD.IrvineE.LingardS. J.ChoudhuryA. I.et al (2009). Ribosomal protein S6 kinase 1 signaling regulates mammalian life span. Science326 (5949), 140–144. 10.1126/science.1177221
91
ShepherdJ. C.WalldorfU.HugP.GehringW. J. (1989). Fruit flies with additional expression of the elongation factor EF-1 alpha live longer. Proc. Natl. Acad. Sci. U. S. A.86 (19), 7520–7521. 10.1073/pnas.86.19.7520
92
ShikamaN.AckermannR.BrackC. (1994). Protein synthesis elongation factor EF-1 alpha expression and longevity in Drosophila melanogaster. Proc. Natl. Acad. Sci. U. S. A.91 (10), 4199–4203. 10.1073/pnas.91.10.4199
93
SolisG. M.KardakarisR.ValentineE. R.Bar-PeledL.ChenA. L.BlewettM. M.et al (2018). Translation attenuation by minocycline enhances longevity and proteostasis in old post-stress-responsive organisms. Elife7, e40314. 10.7554/eLife.40314
94
SonenbergN.HinnebuschA. G. (2009). Regulation of translation initiation in eukaryotes: Mechanisms and biological targets. Cell136 (4), 731–745. 10.1016/j.cell.2009.01.042
95
SonntagW. E.LenhamJ. E.IngramR. L. (1992). Effects of aging and dietary restriction on tissue protein synthesis: Relationship to plasma insulin-like growth factor-1. J. Gerontol.47 (5), B159–B163. 10.1093/geronj/47.5.b159
96
SotiC.CsermelyP. (2003). Aging and molecular chaperones. Exp. Gerontol.38 (10), 1037–1040. 10.1016/s0531-5565(03)00185-2
97
SteffenK. K.MacKayV. L.KerrE. O.TsuchiyaM.HuD.FoxL. A.et al (2008). Yeast life span extension by depletion of 60s ribosomal subunits is mediated by Gcn4. Cell133 (2), 292–302. 10.1016/j.cell.2008.02.037
98
SteinK. C.Morales-PolancoF.van der LiendenJ.RainboltT. K.FrydmanJ. (2022). Ageing exacerbates ribosome pausing to disrupt cotranslational proteostasis. Nature601 (7894), 637–642. 10.1038/s41586-021-04295-4
99
SyntichakiP.TroulinakiK.TavernarakisN. (2007). eIF4E function in somatic cells modulates ageing in Caenorhabditis elegans. Nature445 (7130), 922–926. 10.1038/nature05603
100
TainL. S.MortiboysH.TaoR. N.ZivianiE.BandmannO.WhitworthA. J. (2009). Rapamycin activation of 4E-BP prevents parkinsonian dopaminergic neuron loss. Nat. Neurosci.12 (9), 1129–1135. 10.1038/nn.2372
101
TakahashiR.MoriN.GotoS. (1985). Accumulation of heat-labile elongation factor 2 in the liver of mice and rats. Exp. Gerontol.20 (6), 325–331. 10.1016/0531-5565(85)90012-9
102
TakaujiY.WadaT.TakedaA.KudoI.MikiK.FujiiM.et al (2016). Restriction of protein synthesis abolishes senescence features at cellular and organismal levels. Sci. Rep.6, 18722. 10.1038/srep18722
103
TalaeiF.Van PraagV. M.ShishavanM. H.LandheerS. W.BuikemaH.HenningR. H. (2014). Increased protein aggregation in zucker diabetic fatty rat brain: Identification of key mechanistic targets and the therapeutic application of hydrogen sulfide. BMC Cell Biol.15, 1. 10.1186/1471-2121-15-1
104
TaylorR. C.DillinA. (2011). Aging as an event of proteostasis collapse. Cold Spring Harb. Perspect. Biol.3 (5), a004440. 10.1101/cshperspect.a004440
105
ThompsonA. C.BrussM. D.PriceJ. C.KhambattaC. F.HolmesW. E.ColangeloM.et al (2016). Reduced in vivo hepatic proteome replacement rates but not cell proliferation rates predict maximum lifespan extension in mice. Aging Cell15 (1), 118–127. 10.1111/acel.12414
106
TikuV.JainC.RazY.NakamuraS.HeestandB.LiuW.et al (2017). Small nucleoli are a cellular hallmark of longevity. Nat. Commun.8, 16083. 10.1038/ncomms16083
107
TiwariS. S.MizunoK.GhoshA.AzizW.TroakesC.DaoudJ.et al (2016). Alzheimer-related decrease in CYFIP2 links amyloid production to tau hyperphosphorylation and memory loss. Brain139 (10), 2751–2765. 10.1093/brain/aww205
108
TohyamaD.YamaguchiA.YamashitaT. (2008). Inhibition of a eukaryotic initiation factor (eIF2Bdelta/F11A3.2) during adulthood extends lifespan in Caenorhabditis elegans. FASEB J.22 (12), 4327–4337. 10.1096/fj.08-112953
109
TyeB. W.ComminsN.RyazanovaL. V.WuhrM.SpringerM.PincusD.et al (2019). Proteotoxicity from aberrant ribosome biogenesis compromises cell fitness. Elife8, e43002. 10.7554/eLife.43002
110
UmS. H.FrigerioF.WatanabeM.PicardF.JoaquinM.StickerM.et al (2004). Absence of S6K1 protects against age- and diet-induced obesity while enhancing insulin sensitivity. Nature431 (7005), 200–205. 10.1038/nature02866
111
VargasR.CastanedaM. (1983). Age-dependent decrease in the activity of protein-synthesis initiation factors in rat brain. Mech. Ageing Dev.21 (2), 183–191. 10.1016/0047-6374(83)90073-8
112
VargasR.CastanedaM. (1981). Role of elongation factor 1 in the translational control of rodent brain protein synthesis. J. Neurochem.37 (3), 687–694. 10.1111/j.1471-4159.1982.tb12542.x
113
WallachZ.GershonD. (1974). Altered ribosomal particles in senescent nematodes. Mech. Ageing Dev.3 (3-4), 225–234. 10.1016/0047-6374(74)90018-9
114
WalterP.RonD. (2011). The unfolded protein response: From stress pathway to homeostatic regulation. Science334 (6059), 1081–1086. 10.1126/science.1209038
115
WangX.LiW.WilliamsM.TeradaN.AlessiD. R.ProudC. G. (2001). Regulation of elongation factor 2 kinase by p90(RSK1) and p70 S6 kinase. EMBO J.20 (16), 4370–4379. 10.1093/emboj/20.16.4370
116
WangX.ProudC. G. (2006). The mTOR pathway in the control of protein synthesis. Physiol. (Bethesda)21, 362–369. 10.1152/physiol.00024.2006
117
WardW.RichardsonA. (1991). Effect of age on liver protein synthesis and degradation. Hepatology14 (5), 935–948. 10.1002/hep.1840140529
118
WebsterG. C.WebsterS. L. (1983). Decline in synthesis of elongation factor one (EF-1) precedes the decreased synthesis of total protein in aging Drosophila melanogaster. Mech. Ageing Dev.22 (2), 121–128. 10.1016/0047-6374(83)90105-7
119
WebsterG. C.WebsterS. L. (1979). Decreased protein synthesis by microsomes from aging Drosophila melanogaster. Exp. Gerontol.14 (6), 343–348. 10.1016/0531-5565(79)90047-0
120
WebsterG. C.WebsterS. L.LandisW. A. (1981). The effect of age on the initiation of protein synthesis in Drosophila melanogaster. Mech. Ageing Dev.16 (1), 71–79. 10.1016/0047-6374(81)90034-8
121
WessellsR.FitzgeraldE.PiazzaN.OcorrK.MorleyS.DaviesC.et al (2009). d4eBP acts downstream of both dTOR and dFoxo to modulate cardiac functional aging in Drosophila. Aging Cell8 (5), 542–552. 10.1111/j.1474-9726.2009.00504.x
122
YamasakiS.AndersonP. (2008). Reprogramming mRNA translation during stress. Curr. Opin. Cell Biol.20 (2), 222–226. 10.1016/j.ceb.2008.01.013
123
YangH. S.JansenA. P.KomarA. A.ZhengX.MerrickW. C.CostesS.et al (2003). The transformation suppressor Pdcd4 is a novel eukaryotic translation initiation factor 4A binding protein that inhibits translation. Mol. Cell Biol.23 (1), 26–37. 10.1128/mcb.23.1.26-37.2003
124
YoungV. R.SteffeeW. P.PencharzP. B.WintererJ. C.ScrimshawN. S. (1975). Total human body protein synthesis in relation to protein requirements at various ages. Nature253 (5488), 192–194. 10.1038/253192a0
125
ZhangP.McGrathB. C.ReinertJ.OlsenD. S.LeiL.GillS.et al (2002b). The GCN2 eIF2alpha kinase is required for adaptation to amino acid deprivation in mice. Mol. Cell Biol.22 (19), 6681–6688. 10.1128/mcb.22.19.6681-6688.2002
126
ZhangP.McGrathB.LiS.FrankA.ZambitoF.ReinertJ.et al (2002a). The PERK eukaryotic initiation factor 2 alpha kinase is required for the development of the skeletal system, postnatal growth, and the function and viability of the pancreas. Mol. Cell Biol.22 (11), 3864–3874. 10.1128/mcb.22.11.3864-3874.2002
Summary
Keywords
protein translation, aging, ageing, lifespan, hallmarks of aging, sk6, eIF, theories of aging
Citation
Kim HS and Pickering AM (2023) Protein translation paradox: Implications in translational regulation of aging. Front. Cell Dev. Biol. 11:1129281. doi: 10.3389/fcell.2023.1129281
Received
21 December 2022
Accepted
02 January 2023
Published
13 January 2023
Volume
11 - 2023
Edited by
Fu-Hui Xiao, Kunming Institute of Zoology (CAS), China
Reviewed by
Qin Yu, Kunming Institute of Zoology (CAS), China
Junying Qin, Beijing Institute of Genomics (CAS), China
Updates
Copyright
© 2023 Kim and Pickering.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Andrew M. Pickering, ampick@uab.edu
This article was submitted to Cellular Biochemistry, a section of the journal Frontiers in Cell and Developmental Biology
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.