- 1Department of Biology and Geology, University of South Carolina Aiken, Aiken, SC, United States
- 2Department of Molecular Biology and Genetics, Cornell University, Ithaca, NY, United States
- 3Department of Crop and Soil Science, Institute of Plant Breeding, Genetics, and Genomics, University of Georgia, Athens, GA, United States
Introduction: Class II DNA transposable elements account for significant portions of eukaryotic genomes and contribute to genome evolution through their mobilization. To escape inactivating mutations and persist in the host genome over evolutionary time, these elements must be mobilized enough to result in additional copies. These elements utilize a “cut and paste” transposition mechanism that does not intrinsically include replication. However, elements such as the rice derived mPing element have been observed to increase in copy number over time.
Methods: We used yeast transposition assays to test several parameters that could affect the excision and insertion of mPing and its related elements. This included development of novel strategies for measuring element insertion and sequencing insertion sites.
Results: Increased transposase protein expression increased the mobilization frequency of a small (430 bp) element, while overexpression inhibition was observed for a larger (7,126 bp) element. Smaller element size increased both the frequency of excision and insertion of these elements. The effect of yeast ploidy on element excision, insertion, and copy number provided evidence that homology dependent repair allows for replicative transposition. These elements were found to preferentially insert into yeast rDNA repeat sequences.
Discussion: Identifying the parameters that influence transposition of these elements will facilitate their use for gene discovery and genome editing. These insights in to the behavior of these elements also provide important clues into how class II transposable elements have shaped eukaryotic genomes.
1 Introduction
Transposable elements (TEs) are mobile segments of DNA that can create new copies of themselves in the genome. When inserted within or near genes, TEs can disrupt gene expression, resulting in phenotypic changes. These mutations, coupled with the fact that TEs make up large portions of genomes, make these elements powerful drivers of genome evolution (Oliver et al., 2013). The observation that only recently replicated TEs are functional (Vitte and Bennetzen, 2006) indicates that movement and replication is crucial to TE survival on an evolutionary time scale. Thus, we must have a clear understanding of the mechanisms controlling TE mobility if we are to understand genome evolution.
Different classes of TEs rely on different transposition mechanisms. Class I TEs (retrotransposons) have a well described mechanism for replication where reverse transcriptase synthesizes a new copy of the element from an RNA template. In contrast, the transposition mechanism for eukaryotic class II (DNA) elements has been classified as “copy-in,” “copy-out/paste-in,” or “cut-out/paste-in” (Curcio and Derbyshire, 2003). Copy-in elements, such as Mu and Tn3, are hypothesized to only cleave and transfer a single strand of DNA and replication of the resulting “Shapiro” intermediate produces a second copy of the element (Shapiro, 1979). Copy-out/paste-in (IS3 family) elements form a circular plasmid like structure, which is replicated by the host before reinsertion into the genome (Sekine et al., 1999). Evidence for the Shapiro intermediates or circular elements only exists for the Maverick and Helitron super families in eukaryotes, respectively (Kapitonov and Jurka, 2001; Pritham et al., 2007). The majority of eukaryotic DNA TEs rely on the cut-out/paste-in mechanism, in which elements are excised from their genomic location by the activity of transposase proteins, then reinserted elsewhere in the genome (Yuan and Wessler, 2011; Craig, 2020). This “cut and paste” mechanism utilized by the majority of eukaryotic DNA TEs lacks an intrinsic replication mechanism and observations of element replication are rare. The finding that the Ac/Ds element transposes immediately after DNA replication, when the DNA is hemimethylated, suggests that the timing during the cell cycle may be an important factor (Ros and Kunze, 2001). However, relatively little is known about how the host genetic machinery is exploited to produce new copies of many elements.
The focus of this study is the PIF/Harbinger superfamily of TEs, which are found in almost all eukaryotes (Zhang et al., 2004; Han et al., 2015). These class II elements utilize the cut-out/paste-in mechanism catalyzed by two separate proteins referred to as ORF1 and Transposase [TPase] (Yang et al., 2007; Sinzelle et al., 2008; Johnson et al., 2021). The ORF1 protein contains a DNA binding domain that is hypothesized to bind to the terminal inverted repeat (TIR) sequences that delineate the ends of the elements (Sinzelle et al., 2008; Hancock et al., 2010). The TPase protein has a catalytic DDE domain for DNA cleavage (Yuan and Wessler, 2011) and interacts with ORF1 to form the functional transposition complex (Sinzelle et al., 2008; Velanis et al., 2020). The best studied PIF/Pong/Harbinger superfamily elements are the mPing, Ping, and Pong elements naturally found in the rice genome (Jiang et al., 2003; Kikuchi et al., 2003; Nakazaki et al., 2003). mPing is a Miniature Inverted-repeat Transposable Element (MITE) derived from the larger Ping element and is highly active in some rice cultivars (Naito et al., 2006; Naito et al., 2009). The mPing element was shown to increase in copy number during tissue culture treatment (Jiang et al., 2003), and naturally reached hundreds of genomic copies in some rice cultivars (Naito et al., 2006; Naito et al., 2009; Yasuda et al., 2013; Chen et al., 2019). As a non-autonomous element, mPing does not encode ORF1 and TPase proteins, but instead is mobilized by the ORF1 and TPase proteins encoded by the autonomous Ping or Pong elements (Yang et al., 2007; Hancock et al., 2010).
The activity of the mPing element can be studied using an established yeast transposition assay, in which mPing is inserted into the coding region of ADE2 to track the excision of mPing and subsequent repair of the excision site (Hancock et al., 2010). Expression of ORF1 and TPase proteins is induced by the presence of galactose and yeast that experience excision of mPing and precise repair of the excision site result in a colony on plates lacking adenine (Figure 1A). This assay has allowed for the discovery of several factors that regulate transposition of mPing and a synthetic mPong element. First, it was shown that increasing nuclear localization of the ORF1 and TPase proteins by strengthening a nuclear localization signal and deleting a nuclear export signal respectively results in a significant increase in element mobility (Hancock et al., 2010; Payero et al., 2016). Alteration of the TIR sequences away from the consensus sequence decreases the excision frequency (Chen et al., 2019; Redd et al., 2023). In addition, internal sequences are required for efficient transposition, and alteration can increase or decrease mobility (Redd et al., 2023). Yeast-based experiments also showed that repair of the ADE2 allele at mPing excision sites in haploid yeast is primarily a result of non-homologous end joining [NHEJ] (Gilbert et al., 2015). The goal of this study was to continue to describe factors that determine the mobility and retention of PIF/Pong/Harbinger elements in the genome.
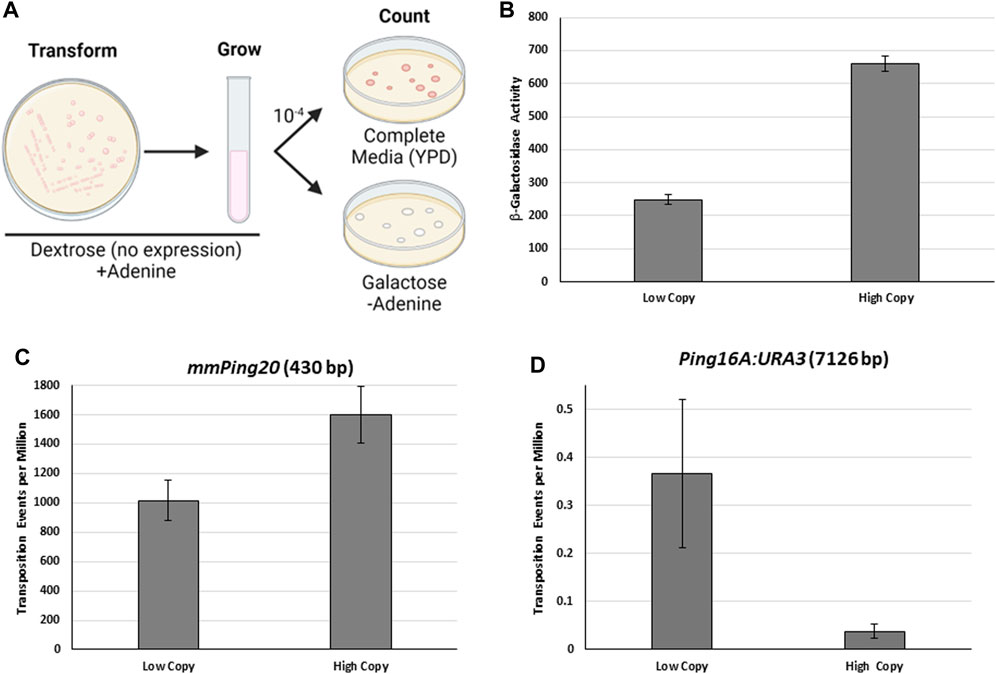
FIGURE 1. The effect of protein expression on mPing excision frequency. (A) Diagram of the yeast transposition assay used in this study. The number of colonies on the YPD and galactose plates is used to calculate the transposition frequency. The ADE2 revertant colonies on the galactose plates can be analyzed further to determine insertion frequency and location. (B) β-galactosidase assay comparing expression of LacZ from a low copy plasmid (pAG 413 GAL) and a high copy plasmid (pAG 423 GAL) in S. cerevisiae grown in 2% Galactose. The LacZ reporter provides an approximation for expression of ORF1 and TPase in (C,D). Error bars represent the standard error of three replicates. C&D: Yeast transposition assays using a genomic mmPing20:ADE2. (C) or Ping16A:URA3:ADE2. (D) reporter with expression of TPase:T2A:ORF1 from the same low and high copy plasmids. Error bars represent the standard error of six replicates.
2 Results
2.1 Altering transposase expression affects excision frequency
Many DNA TEs exhibit a phenomenon called overexpression inhibition, where an excess of transposase proteins results in reduced transposition (Heinlem et al., 1994; Geurts et al., 2003). Although previous experiments have shown that expression of ORF1 and TPase is required for mPing transposition, it was not known how expression level effected transposition frequency. To test the effect of protein expression, we used a previously developed genomic ADE2 reporter containing the hyperactive version of mPing called mmPing20 (Johnson et al., 2021). We altered the protein expression levels using low copy (pAG 413 GAL) and high copy (pAG 423 GAL) plasmids which are present at about 3 and 20 copies respectively (Karim et al., 2013). Figure 1B shows β-galactosidase assays for identical LacZ genes cloned into these plasmids. The high copy plasmid produces significantly (t-test, p < 0.001) higher levels of β-galactosidase protein than the low copy plasmid. When hyperactive Pong TPase and ORF1 proteins (Payero et al., 2016) fused by a T2A peptide (Kim et al., 2011) were expressed in these plasmids, we observed significantly higher (t-test, p < 0.05) excision of the 430 bp mmPing20 element by the high copy expression plasmid (Figure 1C). This result is consistent with increased protein concentration increasing the likelihood of forming an active transposition complex. In contrast, performing the same experiments with the much larger 7,126 bp Ping16A:Ura3 element produced the opposite effect (Figure 1D), with lower expression producing more transposition (t-test, p = 0.06). This suggests that overexpression inhibition is occurring for larger elements where the TIRs are farther apart.
2.2 Increased element copy number increases excision frequency
In addition to differences in the amount of protein expression, significant differences in mPing element copy number are found in different rice cultivars (Chen et al., 2019). We predicted that increased element abundance in the yeast genome would increase the probability of transposition complex formation. To test the effect of the element’s copy number, we compared the excision frequencies for mPing and mPong elements present as a single genomic copy to excision from multiple copies [2-5 copies per cell (Karim et al., 2013)] present on a low copy plasmid (Figure 2). We observed significantly more ADE2 revertant colonies from the plasmid encoded elements than the genomic copies (t-test, mPing p < 0.01, mPong p < 0.001), indicating that more excision and subsequent ADE2 repair occurred. This result supports a model in which excision frequency is correlated with the rate at which the ORF1 and TPase proteins form a complex with the element. Increasing the copy number appears to effectively increases its probability of interacting with the ORF1 and TPase proteins.
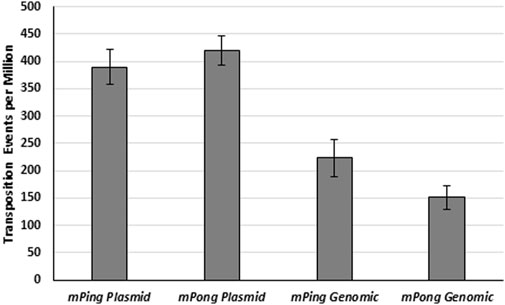
FIGURE 2. Transposition from plasmid and genome. Yeast transposition assay of mPing and mPong using pWL89a plasmid or genomic ADE2 reporters. A low copy plasmid (pAG 413 GAL) was used to express ORF1:T2A:TPase. Error bars represent the standard error of five to six replicates.
2.3 Smaller elements excise more frequently
The greater abundance of small elements, especially Tourist MITEs, has led to the hypothesis that small PIF/Pong/Harbinger elements transpose more frequently (Feschotte et al., 2002; Zhang et al., 2004). Previous yeast transposition assays were used to show that mPing (430 bp) transposes at significantly higher frequency than the larger Ping16A (5,341 bp) element (Redd et al., 2023). To further test the effect of size on element excision frequency, we performed transposition assays with synthetic mPong elements ranging from 430 bp to 5,166 bp in size (Figure 3). We observed a direct correlation (R2 = 0.97) between the element size and the number of ADE2 revertant colonies. This suggests that increasing the size of the elements decreases the ability of the ORF1 and TPase proteins to bind to the element and induce excision.
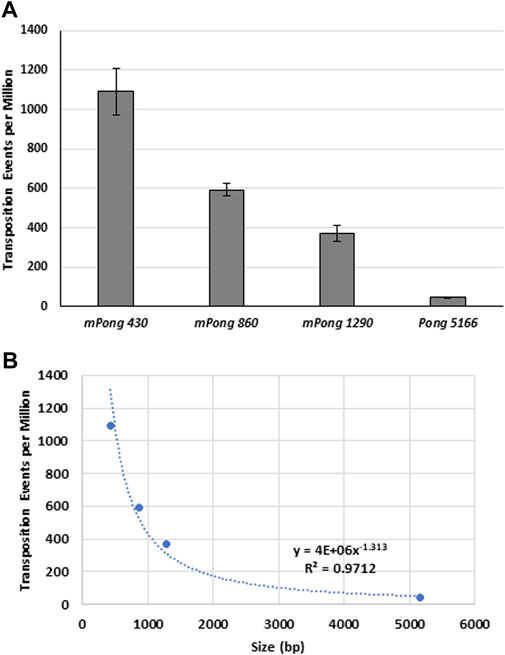
FIGURE 3. The effect of element size on excision frequency. (A) Yeast transposition assay with mPong elements of differing sizes on a pWL89a plasmid. Size in bp is indicated in the element name. ORF1 was expressed from pAG 413 GAL and TPase was expressed from pAG 425 GAL. Error bars represent the standard error of six replicates. (B) Graph showing the correlation between element size and transposition frequency.
2.4 Element size effects insertion frequency
Although element excision is a good indicator of overall transposition activity, excision is not directly linked to insertion back into the genome. Thus, it is important to also determine the rate at which various PIF/Pong/Harbinger elements insert into the genome after excision. We developed yeast strains containing genomic copies of the mPing, Ping16A and mPong elements with inserted URA3 selectable markers (mPing:URA3, Ping16A:URA3, mPong:URA3), allowing us to easily detect the elements. As expected, increasing the size by addition of the URA3 selectable marker reduced the number of ADE2 revertant colonies for all three elements, but the smaller mPing:URA3 and mPong:URA3 elements showed significantly higher excision than Ping16A:URA3 (Figure 4A). To measure element insertion, the ADE2 revertant colonies were isolated and tested for retention of the URA3 gene (Figure 4B). This result shows that the larger Ping16A:URA3 element (7,126 bp) was lost at a significantly higher frequency than the smaller mPing:URA3 and mPong:URA3 elements (2,258 bp). To confirm these results, we repeated these experiments with the natural mPing, Ping16A, and Pong elements encoded on a URA3 containing plasmid. The insertion rate was tested by isolating ADE2 revertant colonies, counter selecting against the URA3 gene to remove the original plasmids, then PCR screening to detect genomic copies of the element. Table 1 shows that the proportion of ADE2 revertant colonies that retained mPing was close to 100%, while the larger Pong and Ping16A elements were lost 25%–50% of the time.
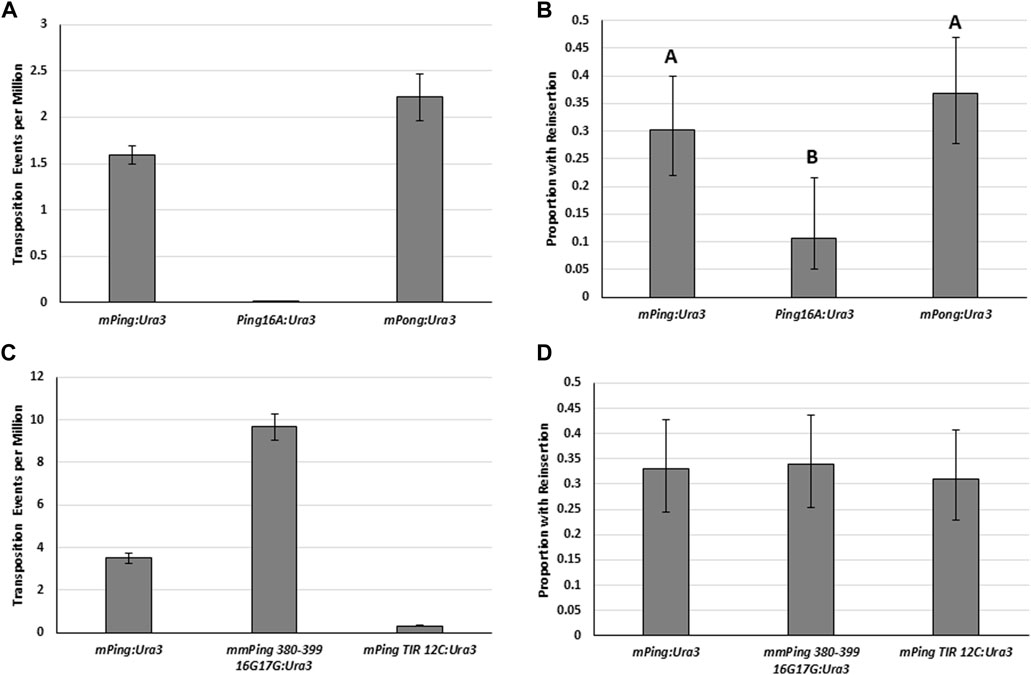
FIGURE 4. Excision and insertion frequency for URA3 containing elements. (A) Yeast transposition assay for URA3 containing versions of mPing (2,254 bp), Ping16A (7,126 bp), and mPong (2,254 bp) integrated into the genomic ADE2. A low copy plasmid (pAG 413 GAL) was used to express ORF1:T2A:TPase. Error bars represent the standard error of six replicates. (B) Proportion of ADE2 revertant colonies that retained URA3 as measured by the ability to grow on plates lacking uracil. Unique letter indicates statistical difference (p < 0.05, d.f. = 2) indicated by Chi2 analysis. (C) Yeast transposition assay for URA3 containing versions of mPing, mmPing 380–399 16G17G, and mPing TIR 12C (all 2,254 bp) integrated into the genomic ADE2. A low copy plasmid (pAG 413 GAL) was used to express ORF1:T2A:TPase. Error bars represent the standard error of six replicates. (D) Proportion of ADE2 revertant colonies that retained URA3 as measured by the ability to grow on plates lacking uracil.
We hypothesized that the observed differences in insertion ability could be due to differences in transposition complex stability or to the increased potential for self-insertion associated with longer DNA sequence. To address element stability, we tested the insertion frequency of URA3 versions of previously described hyperactive (mmPing 380–399 16G17G:URA3) and hypoactive (mPing TIR 12C:URA3) elements that have TIR mutations that are thought to affect protein binding (Redd et al., 2023). Although we observed the expected differences in excision frequency (Figure 4C), we observed no difference in insertion (Figure 4D). This result indicates that there is no direct correlation between excision and insertion efficiency and suggests that once the element has excised its insertion is controlled by other factors. To further explore the effect of size, we compared the insertion frequency of genomic copies of the mmPing20 (430 bp) and mmPong20:MET15 (2,361 bp) element using PCR analysis (Figure 5). These results supported the model that larger elements fail to reinsert into the genome as efficiently as smaller elements because they are likely to insert into themselves after excision.
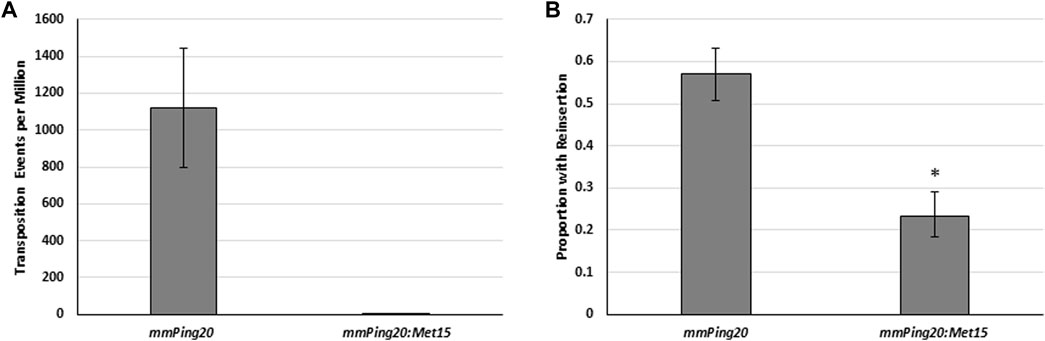
FIGURE 5. The effect of element size on insertion frequency. (A) Yeast transposition assay using genomic mmPing20 (430 bp) and mmPing20:MET15 (2,061 bp) ADE2 reporters. pAG 426 GAL was used to express ORF1:T2A:TPase. Error bars represent the standard error of six replicates. (B) Proportion of ADE2 revertant colonies that retained mmPing20 as measured by PCR. Asterix indicates statistical difference (p < 0.05, d.f. = 1) indicated by Chi2 analysis.
2.5 Excision site repair and insertion in diploid yeast
All the previously described experiments were performed in haploid yeast strains, but most of the life cycle of plants is in the diploid state. Thus, we performed yeast transposition assays in diploid yeast created from our haploid strains. Using a plasmid reporter, we see a drastic reduction in excision frequency for the diploid compared to the a and α parent strains (Figure 6). We attribute this reduction in ADE2 revertant colonies to differences in the repair of the mPing excision site. Diploid yeast is known to rely more on homology directed repair (HDR) instead of NHEJ for double strand break repair than haploids (Åström et al., 1999). Thus, we hypothesize that in diploid yeast the mPing excision sites are primarily being repaired using a homologous mPing:ADE2 plasmid, which prevents formation of a functional copy of ADE2.
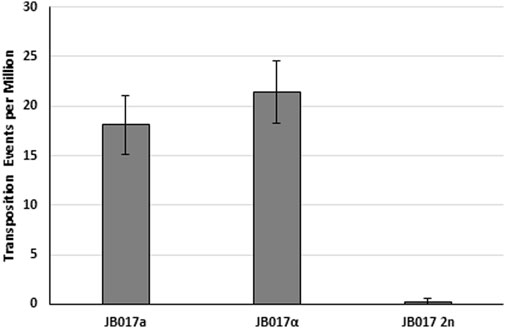
FIGURE 6. Excision frequency and ADE2 repair in haploid and diploid yeast. Yeast transposition assay of mPing using pWL89a plasmid reporters in haploid (JB017a and JB017α) and diploid (JB017 2n) yeast. A low copy plasmid (pAG 413 GAL) was used to express ORF1:T2A:TPase. Error bars represent the standard error of six replicates.
To test the effect of yeast ploidy on element reinsertion, we made a diploid yeast strain from haploid parents containing genomic copies of mmPing20:URA3 (a mating type) and mmPing20:Met15 (α mating type) inserted into the ADE2 gene. Transposition assays with the haploid parents and the diploid (Figure 7A) showed that 2,254 bp mmPing20:URA3 transposed at significantly higher frequency than the 2061 bp mmPing20:Met15 element. This is likely due to base composition of the added sequences which has previously been shown to affect mPing transposition efficiency (Johnson et al., 2021; Redd et al., 2023). We also observed the expected drop in ADE2 revertant colonies expected for the diploid. Analysis of the number of ADE2 revertant colonies showed no difference in element retention between the two haploid yeast strains but the diploid strain had a higher rate of element retention (Figure 7B). Because the elements were similar sizes, this result is likely from the difference in the amount of homologous repair. One possible explanation for this result is that in the diploid yeast, the ADE2 revertant colonies experienced multiple transposition events. If a transposition event was first repaired by homologous repair, followed by a second event repaired by NHEJ, it would provide an additional opportunity for the mmPing20 element to successfully insert into the genome.
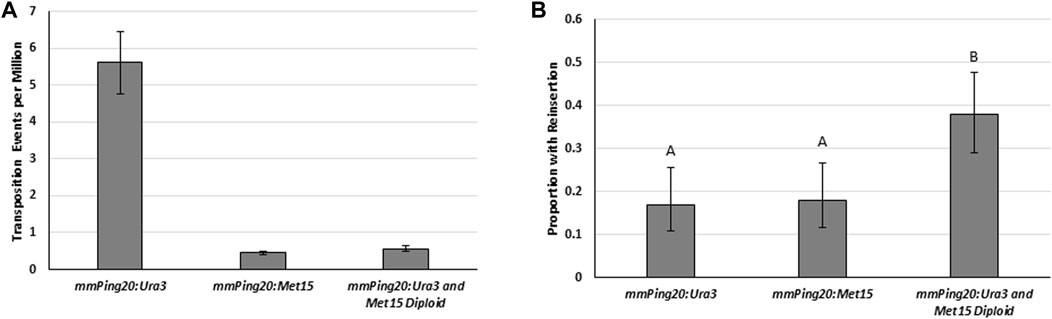
FIGURE 7. Excision and insertion frequency in haploid and diploid yeast. (A) Yeast transposition assay comparing haploid JB017a with genomic mmPing20:URA3 (2,254 bp) and JB017α with genomic mmPing20:MET15 (2061 bp) to the diploid JB017 2n made by mating the two haploids (contains both mmPing20:URA3 and mmPing20:MET15). A low copy plasmid (pAG 413 GAL) was used to express ORF1:T2A:TPase. Error bars represent the standard error of six replicates. (B) Proportion of ADE2 revertant colonies that retained the URA3 and MET15 containing elements as measured by the ability to grow on plates lacking uracil and methionine (diploid reinsertion indicates they retained both elements). Unique letter indicates statistical difference (p < 0.05, d.f. = 2) indicated by Chi2 analysis.
2.6 Insertion site preference and replicative transposition in yeast
The potential for HDR mediated gene conversion of the mPing excision sites in yeast provides an opportunity to test if HDR can act as the mechanisms for replication of class II TEs. In this replication model, there must be at least two matching chromosomes with identical class II elements at a particular locus. This could be from homologous chromosomes, or from sister chromatids after S-phase of the cell cycle. When one of these chromosomes has excision of one transposable element, the resulting excision site can be repaired by synthesis-dependent strand annealing or double-Holiday junction HDR (Ramakrishnan et al., 2018), potentially generating an additional copy of the element. To test if HDR-facilitated replication was occurring in our yeast, two diploid yeast strains containing the genomic ADE2:mmPing20 reporter were developed (Figure 8A). The first (1 Copy Diploid) contained a single copy of the ADE2:mmPing20 reporter construct on one chromosome and the other homologous chromosome had the ADE2 gene replaced by the hph gene. In this strain, gene conversion of the mmPing20 excision site does not produce a functional ADE2 allele, as the sequence including the hph gene acts as the HDR template for this strain. Reversion of ADE2 requires minimal disruption of the coding region, thus, the 1 Copy Diploid strain primarily indicates the number of excision events that are repaired by NHEJ. The second strain (2 Copy Diploid) contains an ADE2:mmPing20 reporter on both chromosomes. In this strain, HDR repair of the excision site with the homologous mmPing20 template could result in mmPing20 replication. However, the transposition assay measures ADE2 repair, so the number of colonies observed is still a measure of the excision events that are primarily repaired by NHEJ. Performing ADE2 reversion assays with these strains showed that similar to previous results, the ADE2 reversion frequency for both of the diploid strains was significantly lower than the haploid control (Figure 8B), consistent with them utilizing HDR repair to resolve the mmPing20 excision sites. The finding that the 2 Copy Diploid shows a slightly higher rate of ADE2 reversion than the 1 Copy Diploid is consistent with the increased excision observed when more element copies are present.
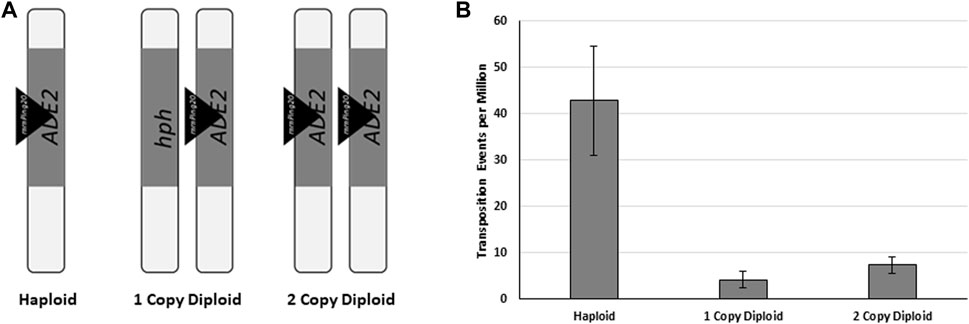
FIGURE 8. The effect of homologous templates. (A) Depiction of the ADE2 locus of the genomic mmPing20 yeast strains used. Vertical bars represent individual chromosomes, triangles represent inserted mmPing20 elements. (B) Yeast transposition assay with the strains shown in (A). ORF1 was expressed from pAG 413 GAL and TPase was expressed from pAG 425 GAL. Error bars represent the standard error of six replicates.
The mmPing20 insertion sites from these haploid and diploid yeast strains were identified by sequencing mPing20 containing amplicons from pooled genomic DNA from ADE2 revertant colonies (4 pools from each strain, 12 colonies per pool). We used a previously established protocol that randomly shears the genomic DNA, ligates the fragments to pGEM plasmid, and then amplifies the desired fragments using primers anchored in mmPing20 (Kanizay et al., 2015). This process resulted in the identification of 134 unique insertions that were mapped to the yeast genome (Supplementary Files S1). A significant percentage (haploid - 41.8%, 1 Copy diploid - 40.4%, and 2 Copy Diploid - 52%) of the mmPing20 elements were identified at various locations in the rDNA repeat region on Chromosome XII (about 150 tandem copies of a 9.1 Kb sequence). The number of mmPing20 insertions for each pool showed no statistically significant differences (Supplementary Table S1). All three yeast strains had at least one example of pools with more than the expected 12 copies of mmPing20. However, only Haploid and 1 Copy Diploid pools had examples with fewer mmPing20 copies than the expected 12. The highest number of insertions detected (14) was in a 2 Copy Diploid DNA pool (samples 37–48). While we cannot ensure that our sequencing results detected every copy of mmPing20, this result suggests that mmPing20 copies may be being lost at higher rates in the Haploid and 1 Copy Diploid strains.
To gain further insight, the individual ADE2 revertant colonies from two selected pools [2 Copy Diploid 37–48 (Figure 9A) and 1 Copy Diploid 1–12 (Figure 9B)] were analyzed by PCR with primers flanking insertions identified by sequencing (Supplementary Table S2). As expected, PCR of the ADE2 site showed that all 2-copy diploid isolates still contained one non-mobilized copy of mmPing20 at the ADE2 locus (Figure 9A). Using insertion site-specific primers with an mmPing20 specific primer (Supplementary Table S2) resulted in appropriately sized amplicons for 11 of the 14 insertions indicated by sequence analysis. Though intended to be insertion site specific, multiple insertion site primers amplified DNA from more than one sample. The non-specific binding of these primers is consistent with multiple mmPing20 insertions in the chromosome XII rDNA hotspot. Importantly, the PCR confirmation showed that three samples (44, 45, and 48) contained mmPing20 insertions at two new sites in addition to the original ADE2:mmPing20 (Figure 9A). This provides evidence that replicative transposition of mmPing20 can occur in the 2 Copy Diploid strain. This analysis also revealed that two of the samples (37 and 43) did not contain mobilized copies of mmPing20 despite evidence that it had excised from the ADE2 reporter (Figure 9A). This is consistent with our other results showing that mmPing20 does not always insert into the genome after excision. For the 1 Copy Diploid 1–12 pool (Figure 9B), ten of the 13 transposed mmPing20 copies indicated by sequencing were verified by PCR. Two of the ADE2 revertants did not contain any mmPing20 insertion, consistent with the previously described insertion failure. A single sample, clone 8, contained an additional copy of mmPing20, suggesting that mPing20 replication events are also possible in diploids that do not have homologous chromosomes with identical mPing insertions.
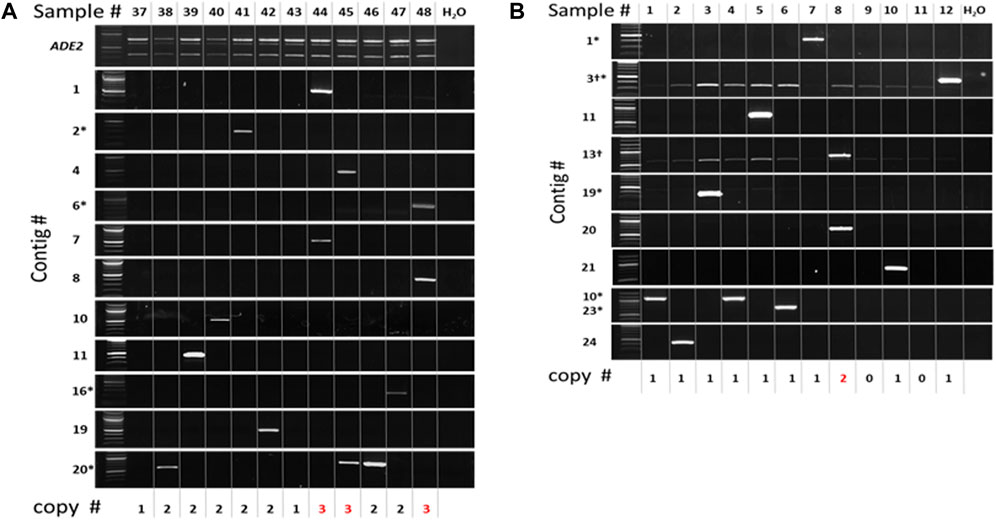
FIGURE 9. PCR Analysis of Selected ADE2 Revertant Clones. PCR verification of insertions identified by sequencing. Numbers at the bottom indicate the total number of mmPing20 elements detected by PCR. Red numbers indicate lines with evidence for replicative transposition. Asterisks indicate insertion in the Chromosome XII hotspot. (A) 2 Copy Diploid 37–48 pool. Top gel panel indicates the ADE2 gene, upper band is mmPing20 containing, lower band is ADE2 gene after excision. (B) 1 Copy Diploid 1–12 Pool.
3 Discussion
Our results indicate that like other class II TE superfamilies, increasing the expression of ORF1 and TPase protein in yeast reduced the excision frequency of the large Ping16A:URA3 element (Figure 1D). The presence of overexpression inhibition in both the Mariner/Tc1 and hAT superfamilies is well documented and is thought to prevent DNA elements from reaching extremely high copy number (Bouuaert et al., 2013). For example, Hsmar1 element transposition is inhibited at high TPase concentrations due to an allosteric effect controlled by a highly conserved linker motif between the N-terminal DNA-binding and C-terminal catalytic domain (Liu and Chalmers, 2014). Similarly, the Ac element TPase was found to oligomerize into a potentially catalytically inactive form at high concentrations (Heinlem et al., 1994). Using yeast transposition assays similar to those described in this paper, autoinhibition was detected when Ac TPase was expressed from the stronger GAL10 promoter, compared to expression from the weaker GALS promoter (Weil and Kunze, 2000). The absence of a detectable overproduction inhibition for the mPing20 element in our yeast assays supports a model in which smaller elements, especially MITEs, avoid the production of unproductive transposition complexes that only contain one TIR because of the close proximity of the two ends of the element. Thus, the activity of the smaller PIF/Pong/Harbinger element relies on alternative methods to prevent excessive transposition. Previous studies suggest that protein localization, terminal inverted repeat sequences, and internal sequences play a role in regulating the activity of these small elements (Hancock et al., 2010; Payero et al., 2016; Redd et al., 2023).
Our observation that transposition increases as element copy number increases (Figure 2) is consistent with the expected dynamics of enzyme substrate binding. However, we recognize that in situations with limited protein expression, flooding the system with too many copies of the element could inhibit transposition complex formation. Thus, under most cases in the cell, transposition activity would be somewhat self-limiting. However, if large amounts of transposase expression are allowed to go unchecked, it would be possible in theory to get a rapidly growing copy number of small TEs that would be detrimental to the cell.
The experiments testing the effect of element size on PIF/Pong/Harbinger element transposition (Figure 3) are consistent with what has been observed for other class II elements. For example, experiments with the Drosophila P-element transformation (Roberts, 1986) and in vitro Tc1 element transposition assays (Fischer et al., 1999) also indicated that small elements transpose more efficiently. Our results are also consistent with previous yeast transposition results showing that adding enhancer sequences to mPing resulted in significantly lower transposition (Johnson et al., 2021). We hypothesize that since both TIRs are part of the transposition complex, increasing the distance between the TIRs reduces the chances of interaction needed for transposition complex formation.
Our observations of PIF/Pong/Harbinger element insertion (Figure 4; Table 1) are consistent with what has been observed for other elements. The 627 bp miniDs element was reported to reinsert 57% of the time in yeast with either the wild-type or a hyperactive Ac Transposase protein (Lazarow et al., 2012). This is similar to the 50%–80% Ac/Ds insertion frequency reported for plants (Kunze et al., 1997). Similarly, the reinsertion frequency for the 2001 bp mini-piggyBac element in mouse cells with either a hyperactive or control transposase appears to be about 50% (Yusa et al., 2011). Additional experiments will be needed to confirm the role of self-insertion and determine if the effect of size on element insertion extends to other class II transposable elements.
Our finding that element insertion into the genome is correlated with element size has important implications to the replication and survival of PIF/Pong/Harbinger elements in the genome. It suggests that the abundance of smaller elements in the genome is not just due to increased excision frequency, but also due to increased genomic insertion. Thus, maintaining a small element size is a critical factor in overall TE replication strategy and may explain why most PIF/Harbinger elements do not carry excessive additional sequence outside of the coding regions. This finding also has implications for transposon tagging and genome editing applications, where addition of large cargos may lead to higher rates of element loss.
The results showing that the rDNA repeats are a common mPing insertion site is of interest as this region accounts for only 6.8%–12.7% of the genome (Kim et al., 2006). One possible explanation could be that this repetitive sequence can tolerate insertions better than other gene rich regions. However, a saturating mutagenesis of yeast using the MiniDs element showed that less than 7% of insertions mapped to the rDNA region (Michel et al., 2017). The insertions in this region were not site specific, suggesting it is not a sequence specific mechanism as observed for the Pokey element from Daphnia (Penton et al., 2002). Yeast rDNA sequences have previously been shown to be a hotspot for ectopic DNA integration (Tosato et al., 2005). However, our sequencing results indicated the presence of the normal target site duplications flanking the mPing insertion, suggesting that they were integrated by the normal catalytic mechanisms of the TPase protein. Analysis of the mPing insertion preference in rice (Naito et al., 2009) and soybean (Hancock et al., 2011) indicates that it primarily inserts upstream and downstream of genes and avoids GC rich sequences. However, no insertion hot spots have been observed in plants. The preference for yeast rDNA repeats suggests that these yeast sequences may have characteristics that mimic the normal insertion preference. Thus, this result warrants further study as it may provide more information about how the transposition complex identifies suitable insertion targets.
Though limited, our results provide some support for HDR mediated TE replication. Evidence for this gene conversion type replication mechanism has also been provided by experiments with the Drosophila P element where excision allowed for HDR of the double strand break, reverting the excision site to the sequence present on the homologous strand (Engels et al., 1990). This finding led the authors to predict that if chromosomes are homozygous for the TE insertion, excision and homologous repair of the excision site will result in the production of an additional copy of the element elsewhere in the genome. However, they had no mechanism to measure the copy number of the elements. Additional evidence for element replication by HDR comes from studies that found that deletion derivatives of the maize Mudr DNA element were derived from interrupted HDR (Plasterk, 1991; Hsia and Schnable, 1996). So, while there is support for this hypothesis, the experimental evidence is far from conclusive, and more information is needed about the frequency at which this phenomenon occurs.
4 Materials and methods
4.1 Yeast strains
All strains used in the study were derived from BY4741 (Baker Brachmann et al., 1998). JB017 (JIM17) yeast was described previously (Gilbert et al., 2015). To generate genomic reporter constructs, URA3 or MET15 containing elements with ADE2 tail sequences were constructed by bridge fusion PCR or synthesized as DNA fragments. These fragments were transformed into yeast with selection for the selectable marker gene. PCR amplicons of ADE2 were sequence verified to confirm proper insertion. To create versions lacking the internal URA3 gene, selection on FOA was used to induce recombination of homologous flanking sequences.
α mating type strains were made by transforming with pGAL HO and growing on CSM-URA galactose media to induce mating type switching. Individual clones were obtained on FOA to remove pGAL HO and yeast mating type assays were performed by patching on lawns of WS199 Mat α and GWS340 Mat a yeast. Genotype was verified using Mat a and Mat α specific primers (White and Haber, 1990). To create diploids, Mat a strains were transformed with pAG413 and Mat α strains were transformed with pAG 415 before mating, selection on CSM-His-Leu, and PCR verification.
4.2 Plasmid construction
Bridge fusion PCR with primers that contain the T2A sequence and attB sequences were used to generate the TPase:T2A:ORF1 and ORF1:T2A:TPase constructs. These were cloned into pDONR ZEO using a BP Clonase reaction. The LacZ gene was from pDONR223_LacZ, a gift from David Root [Addgene plasmid # 25893 (Yang et al., 2011)]. Genes were transferred to pAG 413 GAL-ccdb and pAG 423 GAL-ccdb, gifts from Susan Lindquist (Addgene plasmids # 14141 and # 14149) by performing a LR Clonase reaction.
4.3 Yeast transposition assays
Assays were performed as previously described (Redd et al., 2023). Briefly, liquid cultures were grown for 24 h before plating 100 μL directly on galactose media lacking adenine, and grown for 10 days at 30°C. For Figure 2, 100 μL of a 10−2 dilution was used to reduce the number of colonies to a countable range. To increase the number of ADE2 revertant colonies in Figures 4, 5, 7, a 5 mL culture was spun down and resuspended in 400 μL before plating on a 150 mm plate and allowed to incubate for 15 days. The quantity of yeast plated was determined by plating 100 μL of the 10−4diluted cultures onto YPD.
4.4 Determining element retention
ADE2 revertant colonies were patched out onto CSM-ADE plates to separate them from the yeast that did not experience transposition. For detection of URA3 and MET15 containing elements, clones were subsequently replica streaked onto CSM-URA and CSM-MET plates. For PCR detection, yeast was treated with Zymolyase prior to amplification with element specific primers.
4.5 DNA purification and sequencing
Yeast genomic DNA was purified from a fresh 3 mL liquid culture by washing in water and resuspending in 400 μL of lysis buffer (100 mM Tris pH 8.0, 50 mM EDTA, 1% SDS) before vortexing with acid washed glass beads. The liquid was transferred to a new tube, 250 μL of 7 M Ammonium Acetate pH 7.0 was added, incubated for 5 min at 65°C and then 5 min on ice before adding 500 μL of chloroform and centrifuging for 2 min. The resulting supernatant was transferred to a fresh tube, 950 μL of isopropanol was added, incubated for 5 min at room temperature, and centrifuged for 5 min to pellet the DNA. The pellet was washed with 70% ETOH, dried, and then resuspended in 50 μL of TE buffer. DNA was quantitated and pooled in groups of twelve before sequencing using a modified version of the procedure described previously (Kanizay et al., 2015). Sequence reads were de-multiplexed using Illumina BaseSpace (San Diego, CA). Trimmomatic (Bolger et al., 2014) was used to quality filter sequences. Sequences that contained the mPing target site duplication (TAA/TTA) were selected and the mmPing20 element sequence was removed. The remaining sequences were assembled into contigs using Cap3 (Huang and Madan, 1999). BLAST searching was used to map the resulting contigs to the Saccharomyces cerevisiae genome (Goffeau et al., 1997). The sequencing reads are available at NCBI (BioProject ID: PRJNA949205).
4.6 PCR verification of mmPing20 insertions
Sequencing data was used to design primers approximately 500 bp upstream or downstream of the target site duplication of each unique insertion site. These primers (Supplementary Table S2) were used with mPing primers (mPing 41 Rev or mPing 403 For) to screen DNA from individual ADE2 revertant colonies. Retention of an ADE2 inserted mPing copy was tested using the ADE2 CF and ADE2 CR primers (Hancock et al., 2010).
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA949205.
Author contributions
CH, DG, LP, CP, and PR contributed to conception and design of the study. CH, DG, PR, LP, SD, RK, and CP developed yeast strains and protein expression constructs. PR, CH, LP, DG, RK, DB, SD, and CP performed experiments. EM performed the insertion site sequencing. PR performed the statistical analysis. CH wrote the first draft of the manuscript and LP and CP wrote sections of the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by NSF Grant # 1651666 and #1444581, USCA INBRE supported by a grant from the National Institutes of Health National Institute of General Medical Sciences (P20GM13499-20), and Magellan Scholar Awards (University of South Carolina) to DG and LP.
Acknowledgments
The WS199 Mat α and GWS340 Mat a, mating type tester strains were provided by Walter Schmidt at the University of Georgia. We would also like to thank the Georgia Genomics and Bioinformatics Core as well as the Georgia Advanced Computing Resource Center for sequencing advice and bioinformatics support.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2023.1184046/full#supplementary-material
References
Åström, S. U., Okamura, S. M., and Rine, J. (1999). Yeast cell-type regulation of DNA repair. Nature 397 (6717), 310. doi:10.1038/16833
Baker Brachmann, C., Davies, A., Cost, G. J., Caputo, E., Li, J., Hieter, P., et al. (1998). Designer deletion strains derived from Saccharomyces cerevisiae S288C: A useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14 (2), 115–132. doi:10.1002/(SICI)1097-0061(19980130)14:2<115:AID-YEA204>3.0.CO;2-2
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30 (15), 2114–2120. doi:10.1093/bioinformatics/btu170
Bouuaert, C. C., Lipkow, K., Andrews, S. S., Liu, D., and Chalmers, R. (2013). The autoregulation of a eukaryotic DNA transposon. Elife 2, e00668. doi:10.7554/eLife.00668
Chen, J., Lu, L., Benjamin, J., Diaz, S., Hancock, C. N., Stajich, J. E., et al. (2019). Tracking the origin of two genetic components associated with transposable element bursts in domesticated rice. Nat. Commun. 10 (1), 641–710. doi:10.1038/s41467-019-08451-3
Curcio, M. J., and Derbyshire, K. M. (2003). The outs and ins of transposition: From mu to kangaroo. Nat. Rev. Mol. Cell Biol. 4 (11), 865–877. doi:10.1038/nrm1241
Engels, W. R., Johnson-Schlitz, D. M., Eggleston, W. B., and Sved, J. (1990). High-frequency P element loss in Drosophila is homolog dependent. Cell 62 (3), 515–525. doi:10.1016/0092-8674(90)90016-8
Feschotte, C., Zhang, X. Y., and Wessler, S. R. (2002). “Miniature Inverted-repeat Transposable Elements (MITEs) and their relationship with established DNA transposons,” in Mobile DNA II. Editors N. L. Craig, R. Craige, M. Gellert, and A. Lambowitz (Washington D.C. American Society of Microbiology Press).
Fischer, S. E. J., van Luenen, H. G. A. M., and Plasterk, R. H. A. (1999). Cis requirements for transposition of Tc1-like transposons in C. elegans. Mol. General Genet. 262 (2), 268–274. doi:10.1007/pl00008641
Geurts, A. M., Yang, Y., Clark, K. J., Liu, G., Cui, Z., Dupuy, A. J., et al. (2003). Gene transfer into genomes of human cells by the Sleeping Beauty transposon system. Mol. Ther. 8 (1), 108–117. doi:10.1016/s1525-0016(03)00099-6
Gilbert, D. M., Bridges, M. C., Strother, A. E., Burckhalter, C. E., Burnette, J. M., and Hancock, C. N. (2015). Precise repair of mPing excision sites is facilitated by target site duplication derived microhomology. Mob. DNA 6, 15. doi:10.1186/s13100-015-0046-4
Goffeau, A., Aert, R., Agostini-Carbone, M., Ahmed, A., Aigle, M., Alberghina, L., et al. (1997). The yeast genome directory. Nature 387 (6632), 5–6. doi:10.1038/387s005
Han, M.-J., Xiong, C.-L., Zhang, H.-B., Zhang, M.-Q., Zhang, H.-H., and Zhang, Z. (2015). The diversification of PHIS transposon superfamily in eukaryotes. Mob. DNA 6 (1), 12–11. doi:10.1186/s13100-015-0043-7
Hancock, C. N., Zhang, F., Floyd, K., Richardson, A. O., LaFayette, P., Tucker, D., et al. (2011). The rice miniature inverted repeat transposable element mPing is an effective insertional mutagen in soybean. Plant Physiol. 157 (2), 552–562. doi:10.1104/pp.111.181206
Hancock, C. N., Zhang, F., and Wessler, S. (2010). Transposition of the Tourist-MITE mPing in yeast: An assay that retains key features of catalysis by the Class 2 PIF/Harbinger superfamily. Mob. DNA 1 (5), 5. doi:10.1186/1759-8753-1-5
Heinlem, M., Brattig, T., and Kunze, R. (1994). In vivo aggregation of maize Activator (Ac) transposase in nuclei of maize endosperm and Petunia protoplasts. Plant J. 5 (5), 705–714. doi:10.1111/j.1365-313x.1994.00705.x
Hsia, A.-P., and Schnable, P. S. (1996). DNA sequence analyses support the role of interrupted gap repair in the origin of internal deletions of the maize transposon, MuDR. MuDR. Genet. 142 (2), 603–618. doi:10.1093/genetics/142.2.603
Huang, X., and Madan, A. (1999). CAP3: A DNA sequence assembly program. Genome Res. 9 (9), 868–877. doi:10.1101/gr.9.9.868
Jiang, N., Bao, Z. R., Zhang, X. Y., Hirochika, H., Eddy, S. R., McCouch, S. R., et al. (2003). An active DNA transposon family in rice. Nature 421 (6919), 163–167. doi:10.1038/nature01214
Johnson, A., McAssey, E., Diaz, S., Reagin, J., Redd, P. S., Parrilla, D. R., et al. (2021). Development of mPing-based activation tags for crop insertional mutagenesis. Plant Direct 5 (1), e00300. doi:10.1002/pld3.300
Kanizay, L. B., Jacobs, T. B., Gillespie, K., Newsome, J. A., Spaid, B. N., and Parrott, W. A. (2015). HtStuf: High-throughput sequencing to locate unknown DNA junction fragments. Plant Genome 8 (1), eplantgenome2014.10.0070. doi:10.3835/plantgenome2014.10.0070
Kapitonov, V. V., and Jurka, J. (2001). Rolling-circle transposons in eukaryotes. Proc. Natl. Acad. Sci. 98 (15), 8714–8719. doi:10.1073/pnas.151269298
Karim, A. S., Curran, K. A., and Alper, H. S. (2013). Characterization of plasmid burden and copy number in Saccharomyces cerevisiae for optimization of metabolic engineering applications. FEMS yeast Res. 13 (1), 107–116. doi:10.1111/1567-1364.12016
Kikuchi, K., Terauchi, K., Wada, M., and Hirano, H. Y. (2003). The plant MITE mPing is mobilized in anther culture. Nature 421 (6919), 167–170. doi:10.1038/nature01218
Kim, J. H., Lee, S.-R., Li, L.-H., Park, H.-J., Park, J.-H., Lee, K. Y., et al. (2011). High cleavage efficiency of a 2A peptide derived from porcine teschovirus-1 in human cell lines, zebrafish and mice. PloS one 6 (4), e18556. doi:10.1371/journal.pone.0018556
Kim, Y.-H., Ishikawa, D., Ha, H. P., Sugiyama, M., Kaneko, Y., and Harashima, S. (2006). Chromosome XII context is important for rDNA function in yeast. Nucleic acids Res. 34 (10), 2914–2924. doi:10.1093/nar/gkl293
Kunze, R., Saedler, H., and Lönnig, W.-E. (1997). “Plant transposable elements,” in Advances in botanical research (Elsevier), 331–470.
Lazarow, K., Du, M.-L., Weimer, R., and Kunze, R. (2012). A hyperactive transposase of the maize transposable element Activator (Ac). Genetics 191 (3), 747–756. doi:10.1534/genetics.112.139642
Liu, D., and Chalmers, R. (2014). Hyperactive mariner transposons are created by mutations that disrupt allosterism and increase the rate of transposon end synapsis. Nucleic Acids Res. 42 (4), 2637–2645. doi:10.1093/nar/gkt1218
Michel, A. H., Hatakeyama, R., Kimmig, P., Arter, M., Peter, M., Matos, J., et al. (2017). Functional mapping of yeast genomes by saturated transposition. Elife 6, e23570. doi:10.7554/eLife.23570
Naito, K., Cho, E., Yang, G. J., Campbell, M. A., Yano, K., Okumoto, Y., et al. (2006). Dramatic amplification of a rice transposable element during recent domestication. Proc. Natl. Acad. Sci. U. S. A. 103 (47), 17620–17625. doi:10.1073/pnas.0605421103
Naito, K., Zhang, F., Tsukiyama, T., Saito, H., Hancock, C. N., Richardson, A. O., et al. (2009). Unexpected consequences of a sudden and massive transposon amplification on rice gene expression. Nature 461 (7267), 1130–1134. doi:10.1038/nature08479
Nakazaki, T., Okumoto, Y., Horibata, A., Yamahira, S., Teraishi, M., Nishida, H., et al. (2003). Mobilization of a transposon in the rice genome. Nature 421 (6919), 170–172. doi:10.1038/nature01219
Oliver, K. R., McComb, J. A., and Greene, W. K. (2013). Transposable elements: Powerful contributors to angiosperm evolution and diversity. Genome Biol. Evol. 5 (10), 1886–1901. doi:10.1093/gbe/evt141
Payero, L., Outten, G., Burckhalter, C., and Hancock, C. N. (2016). Alteration of the Ping and Pong ORF1 proteins allows for hyperactive transposition of mPing. J. S. C. Acad. Sci. 14 (2), 3.
Penton, E. H., Sullender, B. W., and Crease, T. J. (2002). Pokey, a new DNA transposon in Daphnia (cladocera: Crustacea). J. Mol. Evol. 55, 664–673. doi:10.1007/s00239-002-2362-9
Plasterk, R. H. (1991). The origin of footprints of the Tc1 transposon of Caenorhabditis elegans. EMBO J. 10 (7), 1919–1925. doi:10.1002/j.1460-2075.1991.tb07718.x
Pritham, E. J., Putliwala, T., and Feschotte, C. (2007). Mavericks, a novel class of giant transposable elements widespread in eukaryotes and related to DNA viruses. Gene 390 (1-2), 3–17. doi:10.1016/j.gene.2006.08.008
Ramakrishnan, S., Kockler, Z., Evans, R., Downing, B. D., and Malkova, A. (2018). Single-strand annealing between inverted DNA repeats: Pathway choice, participating proteins, and genome destabilizing consequences. PLoS Genet. 14 (8), e1007543. doi:10.1371/journal.pgen.1007543
Redd, P. S., Diaz, S., Weidner, D., Benjamin, J., and Hancock, C. N. (2023). Mobility of mPing and its associated elements is regulated by both internal and terminal sequences. Mob. DNA 14 (1), 1. doi:10.1186/s13100-023-00289-3
Ros, F., and Kunze, R. (2001). Regulation of Activator/Dissociation transposition by replication and DNA methylation. Genetics 157 (4), 1723–1733. doi:10.1093/genetics/157.4.1723
Sekine, Y., Aihara, K., and Ohtsubo, E. (1999). Linearization and transposition of circular molecules of insertion sequence IS3. J. Mol. Biol. 294 (1), 21–34. doi:10.1006/jmbi.1999.3181
Shapiro, J. A. (1979). Molecular model for the transposition and replication of bacteriophage Mu and other transposable elements. Proc. Natl. Acad. Sci. 76 (4), 1933–1937. doi:10.1073/pnas.76.4.1933
Sinzelle, L., Kapitonov, V. V., Grzela, D. P., Jursch, T., Jurka, J., Izsvak, Z., et al. (2008). Transposition of a reconstructed Harbinger element in human cells and functional homology with two transposon-derived cellular genes. Proc. Natl. Acad. Sci. U. S. A. 105 (12), 4715–4720. doi:10.1073/pnas.0707746105
Tosato, V., Waghmare, S. K., and Bruschi, C. V. (2005). Non-reciprocal chromosomal bridge-induced translocation (BIT) by targeted DNA integration in yeast. Chromosoma 114, 15–27. doi:10.1007/s00412-005-0332-x
Velanis, C. N., Perera, P., Thomson, B., de Leau, E., Liang, S. C., Hartwig, B., et al. (2020). The domesticated transposase ALP2 mediates formation of a novel Polycomb protein complex by direct interaction with MSI1, a core subunit of Polycomb Repressive Complex 2 (PRC2). PLoS Genet. 16 (5), e1008681. doi:10.1371/journal.pgen.1008681
Vitte, C., and Bennetzen, J. L. (2006). Analysis of retrotransposon structural diversity uncovers properties and propensities in angiosperm genome evolution. Proc. Natl. Acad. Sci. U. S. A. 103 (47), 17638–17643. doi:10.1073/pnas.0605618103
Weil, C. F., and Kunze, R. (2000). Transposition of maize Ac/Ds transposable elements in the yeast Saccharomyces cerevisiae. Nat. Genet. 26 (2), 187–190. doi:10.1038/82827
White, C., and Haber, J. (1990). Intermediates of recombination during mating type switching in Saccharomyces cerevisiae. EMBO J. 9 (3), 663–673. doi:10.1002/j.1460-2075.1990.tb08158.x
Yang, G. J., Zhang, F., Hancock, C. N., and Wessler, S. R. (2007). Transposition of the rice miniature inverted repeat transposable element mPing in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U. S. A. 104 (26), 10962–10967. doi:10.1073/pnas.0702080104
Yang, X., Boehm, J. S., Yang, X., Salehi-Ashtiani, K., Hao, T., Shen, Y., et al. (2011). A public genome-scale lentiviral expression library of human ORFs. Nat. methods 8 (8), 659–661. doi:10.1038/nmeth.1638
Yasuda, K., Ito, M., Sugita, T., Tsukiyama, T., Saito, H., Naito, K., et al. (2013). Utilization of transposable element mPing as a novel genetic tool for modification of the stress response in rice. Mol. Breed. 32, 505–516. doi:10.1007/s11032-013-9885-1
Yuan, Y.-W., and Wessler, S. R. (2011). The catalytic domain of all eukaryotic cut-and-paste transposase superfamilies. Proc. Natl. Acad. Sci. 108 (19), 7884–7889. doi:10.1073/pnas.1104208108
Yusa, K., Zhou, L., Li, M. A., Bradley, A., and Craig, N. L. (2011). A hyperactive piggyBac transposase for mammalian applications. Proc. Natl. Acad. Sci. 108 (4), 1531–1536. doi:10.1073/pnas.1008322108
Keywords: transposable elements, excision, insertion, replication, rDNA
Citation: Redd PS, Payero L, Gilbert DM, Page CA, King R, McAssey EV, Bodie D, Diaz S and Hancock CN (2023) Transposase expression, element abundance, element size, and DNA repair determine the mobility and heritability of PIF/Pong/Harbinger transposable elements. Front. Cell Dev. Biol. 11:1184046. doi: 10.3389/fcell.2023.1184046
Received: 10 March 2023; Accepted: 22 May 2023;
Published: 09 June 2023.
Edited by:
Jinfeng Chen, Institute of Zoology (CAS), ChinaReviewed by:
Valentina Tosato, University of Trieste, ItalyMichael Tellier, University of Leicester, United Kingdom
Copyright © 2023 Redd, Payero, Gilbert, Page, King, McAssey, Bodie, Diaz and Hancock. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: C. Nathan Hancock, bmF0aGFuaEB1c2NhLmVkdQ==
†Present addresses: Clinton A. Page, USDA-Agricultural Research Service, Raleigh, NC, United States Edward V. McAssey, Department of Ecology and Evolutionary Biology, University of Connecticut, Storrs, CT, United States; Stephanie Diaz, Cardiovascular Disease Initiative, Bayer and Broad Institute of MIT and Harvard, Cambridge, MA, United States
 Priscilla S. Redd1
Priscilla S. Redd1 Edward V. McAssey
Edward V. McAssey C. Nathan Hancock
C. Nathan Hancock