- Australian Regenerative Medicine Institute, Monash University, Clayton, VIC, Australia
The Nuclear Receptor (NR) family of transcriptional regulators possess the ability to sense signalling molecules and directly couple that to a transcriptional response. While this large class of proteins are united by sequence and structural homology, individual NR functional output varies greatly depending on their expression, ligand selectivity and DNA binding sequence specificity. Many NRs have remained somewhat enigmatic, with the absence of a defined ligand categorising them as orphan nuclear receptors. One example is Nuclear Receptor subfamily 6 group A member 1 (Nr6a1), an orphan nuclear receptor that has no close evolutionary homologs and thus is alone in subfamily 6. Nonetheless, Nr6a1 has emerged as an important player in the regulation of key pluripotency and developmental genes, as functionally critical for mid-gestational developmental progression and as a possible molecular target for driving evolutionary change in animal body plan. Here, we review the current knowledge on this enigmatic nuclear receptor and how it impacts development and evolution.
Introduction
Nuclear receptors: an overview
The Nuclear Receptor superfamily of transcriptional regulators are generally known to be intracellular receptors whose conformational change in response to ligand binding leads to a direct effect on transcription (for general overview see Frigo et al., 2021). NRs are found throughout the animal kingdom, ranging from 2 NRs found in the sponge Amphimedon queenslandica (Bridgham et al., 2010), 48/49 NRs found in human/mouse respectively and upwards to 73/74 in teleosts such as zebrafish and tilapia (Zhao et al., 2015). Phylogenetic analysis has divided the NR superfamily into 7 structurally distinct groups (NR0-NR6; Bridgham et al., 2010; Holzer et al., 2017) that can be broadly clustered into 3 branches: steroid hormone-related, thyroid hormone-related and retinoid X receptor-related. This complex diversification of NRs across animal lineages has provided insight into their possible ancestral functions and raising questions as to whether the ancestral NR was even ligand regulated (reviewed in Holzer et al., 2017).
Some well characterised NR ligands include thyroid hormone, steroid hormones such as estrogen, progesterone and glucocorticoids, as well as Vitamin A and Vitamin D derivatives. For the most part, these small lipophilic molecules freely diffuse across cell membranes, except for thyroid hormone that requires receptor-mediated transport. Once internalised, these well characterised examples act as high affinity ligands for their cognate receptor. However, it is also clear that many NRs bind various metabolites and lipids with low affinity, increasing the complexity of NR-ligand interactions.
With such a diverse and notable list of ligands, it is not surprising that NR activity is critical throughout the course of animal life, including early embryonic growth and patterning, developmental transitions and metamorphosis, reproduction, metabolism, and adult homeostasis (McKenna et al., 1999; Escriva et al., 2000; Robinson-Rechavi et al., 2003). Moreover, the dysregulation of NR signalling in many human pathological states including diabetes, multiple cancers, cardiovascular diseases, asthma, and neurologic syndromes (Ranhotra, 2013; Oyekan, 2011; Lonard and O'Malley, 2012; Kadmiel and Cidlowski, 2013; Mazaira et al., 2018) has led to intense interest in targeting NR function therapeutically. Indeed, various estimates suggest 15%–20% of currently available therapeutic drugs modulate NR function. However, only about half of the human NRs have known ligands, those with uncharacterised ligands being termed as orphan NRs. This review will focus on the orphan nuclear receptor Nuclear receptor subfamily 6 group A member 1 (Nr6a1), identified in 1994 and originally called Germ cell nuclear factor (GCNF), Retinoid receptor-related testis-associated receptor (RTR) or Neuronal cell nuclear factor according to the varied contexts in which the same factor was identified (Chen et al., 1994; Hirose et al., 1995; Bauer et al., 1997).
Nr6a1: structural insight
The consensus NR structure is composed of i) a poorly conserved N-terminal domain (NTD) in terms of length and sequence, that usually harbours an activator function-1 (AF-1) region that interacts with transcriptional coregulator proteins, ii) a highly conserved DNA binding domain (DBD), iii) a discrete ligand-binding domain (LBD) that not only interacts with ligand(s) but also recruits transcriptional coregulator proteins, and iv) a hinge region that connects the DBD and the LBD (Figure 1) (Escriva et al., 2000; Khorasanizadeh and Rastinejad, 2001; Bain et al., 2007; Gallastegui et al., 2015). Nr6a1 occupies subfamily group 6 alone and, despite its earlier naming as retinoid receptor-related testis-associated receptor, is more closely related to proteins of the steroid hormone branch (Papageorgiou et al., 2021). Compared with other NRs, the LBD region of Nr6a1 lacks an activator function-2 (AF-2) domain though the corresponding region can facilitate co-repressor recruitment and dimerization (Greschik et al., 1999; Zechel, 2005). Collective studies based largely on classical gel mobility shift assays have shown the DNA binding region of Nr6a1 binds with higher affinity as a homodimer than monomer, to a direct repeat with zero spacing (DR0) of the consensus sequence AGGTCA or an extended half site TCAGGTCA (Chen et al., 1994; Borgmeyer, 1997; Yan et al., 1997; Cooney et al., 1998; Schmitz et al., 1999). At least in vitro, Nr6a1 does not dimerize with the retinoid X receptor (Borgmeyer, 1997). Gu et al. (2005) suggested that endogenous Nr6a1 forms an even larger oligomeric complex called transiently retinoid-induced factor (TRIF) that requires DNA to assemble. Certainly, the half-site sequence was corroborated as enriched in Nr6a1-bound regions using chromatin immunoprecipitation of a Flag-HA-tagged Nr6a1 protein expressed in mesenchymal stem cells (Gurtan et al., 2013), and the exact nature of what higher order protein complex Nr6a1 forms in vivo still requires clarification.
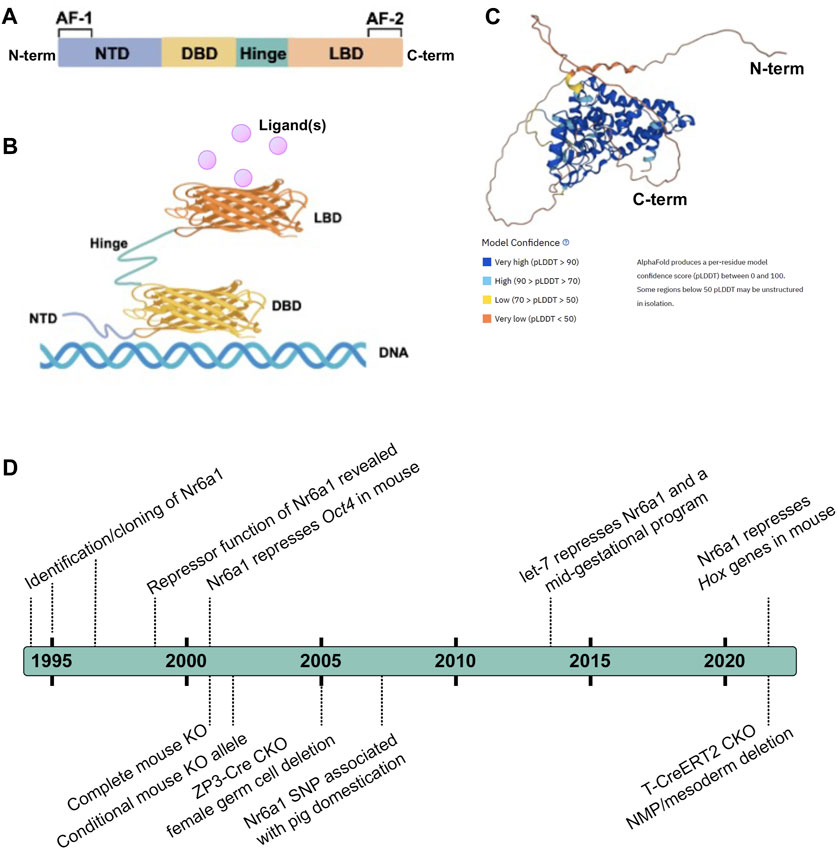
FIGURE 1. Nr6a1 Structural insight (A,B) Schematised structure of a canonical Nuclear Receptor. Nuclear receptors have a less conserved N-terminal domain (NTD) which harbours an activator function-1 (AF-1) region, a highly conserved DNA binding domain (DBD), a ligand-binding domain (LBD) that harbours an activator function-2 (AF2), and a hinge domain linking the DBD and LBD. (C) Predicted 3-Dimensional structure of human Nr6a1, generated by AlphaFold (Jumper et al., 2021; Varadi et al., 2022), model confidence indicated. (D) A timeline of key milestones in the identification and functional assessment of Nr6a1.
Dynamic Nr6a1 expression
Initial characterisation of Nr6a1 expression in adult mouse and human tissues revealed exponentially higher levels of Nr6a1 in the testis compared to other organs, with low expression noted in ovary, kidney, and lung tissues (Chen et al., 1994). Cellular analysis by in-situ hybridization demonstrated Nr6a1 was localised to the germ cells of the male testis and the female ovary (Chen et al., 1994). In male germ cells, high expression was noted during the final stage of round spermatid development with dramatic reduction during elongation of the spermatid cells, suggesting Nr6a1 may act as a developmentally restricted post meiotic factor (Katz et al., 1997; Cooney et al., 1999; Yang et al., 2003). In developing oocytes however, Nr6a1 expression was detected before the completion of meiosis but not in the primordial follicles, supporting an earlier role for Nr6a1 in this context during the initiation of oogenesis (Katz et al., 1997).
During mouse embryonic development, Nr6a1 was detected as early as embryonic day (E) 6.5 in the ectoderm, with expression continuing throughout gastrulation stages in both anterior and posterior neuroepithelium as well nascent mesoderm emerging from the posterior primitive streak (Fuhrmann et al., 2001). At E8.5, expression appeared specific in the neural ectoderm and posterior growth zone (Chung et al., 2001), at least at the level of whole mount in situ hybridisation, with an absence of expression in the developing heart. Exploration of an E8.5 mouse single cell sequencing dataset (Pijuan-Sala et al., 2019) has confirmed widespread expression in cell types of all 3 germ layers, with high frequency across the caudolateral epiblast and bipotential neuromesodermal (NMP) progenitor populations (Chang et al., 2022), both key progenitor sources of the developing spinal cord and vertebral column (Henrique et al., 2015). By E9.5, expression remains strong in the anterior two-thirds of the embryo, including developing cranio-facial structures, limbs, neural tube and somites of the trunk, but is being visibly cleared from the posterior presomitic mesoderm and tailbud region (Chung et al., 2001; Chang et al., 2022). At E10.5, an overall reduction in Nr6a1 is observed, with cell-restricted expression in what appears to be trunk dorsal root ganglia and migrating cranial neural crest, though further characterisation is required. By E12.5, most Nr6a1 expression is extinguished, highlighting a tightly controlled and temporally-restricted mode of transcript regulation.
It should be noted that both initial and more recent in situ expression characterisation utilised a riboprobe detecting the 3′-UTR region of Nr6a1 transcript. With the vast wealth of transcript sequencing now available, it is clear the Nr6a1 genomic locus produces a multitude of transcript isoforms which can impact protein coding potential (Figure 2A). On the sense strand, there are at least 2 major alternate transcripts with coding potential and many additional transcripts (not depicted) where coding potential is not defined and thus are likely non-coding RNAs (ncRNAs). Moreover, on the antisense strand, two partially overlapping long ncRNA transcripts (lnc-Nr6a1-1 and lnc-Nr6a1-2) and a microRNA-encoding transcript (mir-181-a2 and mir-181-b2) are produced. Recent in vitro analysis has shown that Tgf-β induced epithelial-to-mesenchymal transition rapidly upregulates all antisense transcripts, with lnc-Nr6a1-1 and both pre-miRNAs being initially transcribed as a larger single unit (Polo-Generelo et al., 2022). At present, it is unclear if and how Nr6a1 protein functionality is controlled at the level of alternative transcript expression, nor whether there is coordinated regulation of sense and antisense transcripts in vivo—either positive or inverse correlation—as is often observed for sense-antisense pairs.
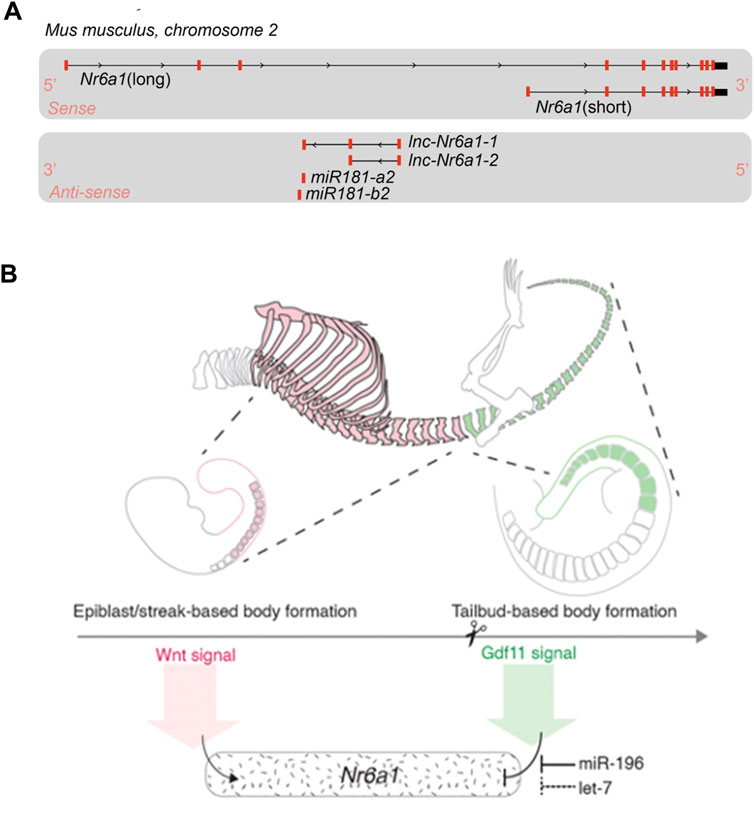
FIGURE 2. Nr6a1 genomic structure and transcript regulation (A) The Nr6a1 genomic locus of Mus musculus. Exons marked in red, not to scale. Multiple Nr6a1 transcripts with coding potential have been identified on the sense strand, while both long non-coding antisense and micro-RNA encoding transcripts are produced from the opposite strand. (B) Nr6a1 expression is defined by key developmental signals/regulators known to control axial elongation. Nr6a1 expression increases in response to Wnt signalling, while the synergistic actions of Gdf11 and miR-196, and potentially let-7 expression, terminate Nr6a1 expression at the trunk-to-tail transition.
Regulatory mechanisms controlling Nr6a1 expression
The timely induction and termination of Nr6a1 expression during development and differentiation is essential. The first clues as to regulatory factors capable of inducing Nr6a1 expression came following its identification as an early response target gene in embryonic carcinoma cells that were induced by retinoic acid (RA) towards a neural cell fate (Bauer et al., 1997; Heinzer et al., 1998). This finding was subsequently confirmed in both mouse (Sato et al., 2006) and human (Wang et al., 2016) embryonic stem cells treated with RA, and in vivo following exogenous application of RA to the Xenopus embryo (Barreto et al., 2003). Interestingly, some level of negative feedback exists between Nr6a1 and the key RA biosynthesis enzyme Aldh1a2, whereby complete loss of Nr6a1 in vivo results in a spatially discrete upregulation of Aldh1a2 in the tailbud (Chung et al., 2001), and conversely, Aldh1a2 is one of the most downregulated transcripts in the tailbud following in vivo Nr6a1 overexpression (Chang et al., 2022). Additional inducers of high level Nr6a1 expression have been identified in vitro, including the Fibroblast Growth Factor and the Wnt signalling pathways (Chang et al., 2022). During ESC differentiation to the bipotential NMP progenitor, the addition of FGF2 (inducing ESC to epiblast-like transition) resulted in an approx. 80-fold increase of Nr6a1, while subsequent addition of the Wnt pathway agonist CHIR99021 (inducing epiblast-like to NMP transition) led to a further increase to 175-fold relative to ESCs. In vivo spatio-temporal context of the relative contributions to induction remains to be delineated, however Wnt/Fgf-dependency correlates well with the strong in vivo posterior expression of Nr6a1 between E7.5-E8.5.
Slightly later in development, both in situ hybridisation and single cell RNAseq analysis have demonstrated a sharp clearance of Nr6a1 from the wildtype posterior growth zone at a key developmental transition known as the trunk-to-tail transition (approximately E9.5 in mouse). This transition marks the end of primary body elongation and is temporally regulated by both Gdf11 signalling (McPherron et al., 1999; Jurberg et al., 2013) and the miR-196 family of microRNAs (Wong et al., 2015), likely across vertebrate species (He et al., 2011; Matsubara et al., 2017). Our group has shown that addition of Gdf11 to in vitro-derived NMP cells almost completely abolishes Nr6a1 expression, back to the low levels seen in ESCs (Chang et al., 2022). In vivo, Gdf11 and miR-196 were shown to have individual and an additive role in the timely clearance of Nr6a1 at this site, with the clearance of Nr6a1 functionally mediating, at least in part, the role of Gdf11 at this critical transition point. Interestingly, the let-7 family of microRNAs are also predicted to extensively target both mouse and human Nr6a1 3′-UTRs (McGeary et al., 2019) and thus could be predicted to also participate in the timely clearance of Nr6a1. Let-7 has been experimentally validated to suppress Nr6a1 as part of a broad “mid-gestational” genetic signature in murine mesenchymal stem cells (Gurtan et al., 2013), and genetic reduction of let-7 paralogs in the mouse increased tail vertebral number by 5 elements (Robinton et al., 2019). Whether this latter phenotype is again in part due to a de-repression of Nr6a1 is not known, though perhaps unlikely given that transgenic overexpression of Nr6a1 in the posterior tailbud revealed the presence of Nr6a1 is detrimental to tail vertebrae morphology (Chang et al., 2022). Whether let-7 may act redundantly with Gdf11 and miR-196 at slightly more rostral locations however is possible and would require complex mouse genetics to dissect. In summary, Nr6a1 expression is tightly controlled during formation of the embryo by key developmental signals and post-transcriptional regulatory mechanisms, several of which are considered as regulators of developmental timing, prompting a detailed functional dissection in this context.
Nr6a1 is essential for embryonic survival
Complete genetic deletion of Nr6a1 in the mouse has revealed its indispensable role during mid-gestation development and in embryonic survival (Chung et al., 2001). The very early stages of Nr6a1−/− embryogenesis did not appear to be overtly compromised however, by E8.5, Nr6a1−/− embryos began to exhibit clear morphological defects including an open neural tube and a disorganisation of the primitive streak and posterior tissues that exacerbated over time. The allantois was larger than WT with defects in chorion attachment, while tissue of the posterior embryo proper began extending abnormally and were positioned outside of the yolk sac. By E9.5, Nr6a1−/− embryos had failed to rotate the posterior half of the main body axis as is normally observed by this stage in WT embryos and exhibited additional posterior alterations in hindgut and ventral body wall development. Somitogenesis was severely compromised, and overall embryo growth was stunted, though some regionalised expansion of the very anterior and posterior tissue was still observed at this stage. The latest time point where viable Nr6a1−/− embryos were recovered was E10.5, albeit at ratios lower than Mendelian expectations, with only resorbing Nr6a1−/− embryos observed at E11.5. The cause of lethality has not been precisely characterised but is likely due to altered chorioallantoic attachment and/or pericardial distention. The aetiology of this latter defect is not immediately obvious since Nr6a1 expression appears to be largely absent from the heart at E8.5-9.5, but whether Nr6a1 functions within very early mesodermal heart field(s) remains possible. An important molecular target of Nr6a1, the pluripotency factor Oct4/Pou5f1, was found to be ectopically expressed across much of the somatic tissue in E8.5 Nr6a1 null embryos (Fuhrmann et al., 2001), a time when this gene normally becomes highly restricted to the germline. These comprehensive studies, and the subsequent ubiquitous deletion of Nr6a1 DBD using Cre/LoxP technology that phenocopied early Nr6a1 null results (Lan et al., 2002), have provided the broad strokes for understanding Nr6a1 requirements during early development. However, the catastrophic defects resulting from global Nr6a1 deletion limited the ability to characterise later embryonic or adult requirements, or tissue-specific functions, of this important regulator.
Nr6a1 is essential for male and female germ cell development
Using the original Nr6a1 null allele, it was shown that initial segregation of the germline cell lineage (primordial germ cells; PCGs) during early embryogenesis, and early PGC migration, do not require Nr6a1 function (Sabour et al., 2014). As Nr6a1 null embryos die soon after these stages, ex vivo knockdown of Nr6a1 (Nra61-KD) within testes-derived germline stem cells was performed, with transduced cells re-introduced into germ-cell depleted seminiferous tubules. Both control and Nr6a1-KD cells were able to re-colonise this environment, but by 3 months, Nr6a1-KD cells never produced functional sperm as compared to controls (Sabour et al., 2014). To dissect the role of Nr6a1 within maturing oocytes of an adult female mouse, a Zp3-Cre conditional knock-out model was employed (Lan et al., 2003). Phenotypically, homozygous conditional deletion did not affect germ cell number but led to reduced fertility owing to an extended diestrus of the estrus cycle, abnormal steroidogenesis, and double-oocyte follicles. The molecular aetiology of these largely non-cell autonomous consequences stemmed from the loss of repression specifically at diestrus of Bmp-15 and Gdf-9, two members of the TGF-b family of secreted ligands known to be critical for female reproduction (Carabatsos et al., 1998; Erickson and Shimasaki, 2001; Zhao et al., 2007). This led to a reduction of follicle stimulating hormone (FSH) and somatic cell-produced steroid hormones specifically in diestrus, underpinning the observed phenotypic consequences. In contrast, a separate conditional approach deleting the LBD of Nr6a1 from E10.5 using a ubiquitous by temporally controlled Cre deleter line observed a surprising lack of effect on the initiation of meiosis or early oogenesis (Okumura et al., 2013). A subsequent in vivo chimeric approach, using Nr6a1-null ESCs injected into a WT blastocyst stage embryo, revealed a reduced contribution to the germline in the absence of Nr6a1 activity (Sabour et al., 2014). These chimeric gonads were then implanted under the kidney capsule to develop further, with only degenerated oocytes present at 4 weeks post-transplant in Nr6a1-null chimeric gonads compared to normal oocyte development observed in the WT-chimeric controls.
Nr6a1 has a regionally-critical role in elongation of the main body axis
The vertebral column and spinal cord arise from progenitors of the posterior growth zone, with tissue being sequentially constructed over a series of days in an anterior-to-posterior (A-P; head-to-tail) direction. The abrupt termination of Nr6a1 expression from across the wildtype E9.5 posterior growth zone (Chung et al., 2001), supported by single cell RNAseq analysis of in vivo NMPs (Gouti et al., 2017), suggested that Nr6a1 function may be regionally-restricted during vertebrate axial elongation. To test this, conditional deletion of Nr6a1 using a tamoxifen-inducible Cre deleter line active in axial progenitors and early mesoderm (TCreERT2) was employed, circumventing the early embryonic lethality observed in Nr6a1 null embryos and allowing analysis of skeletal alterations in late stage embryos. Compared to the WT axial formulae of 7 cervical (C), 13 thoracic (T), 6 lumbar (L), 4 sacral (S) and 30-31 tail vertebrae, conditional knockout (CKO) of Nr6a1 activity from E7.5 resulted in a dose-dependent reduction in the number of trunk vertebral elements with increasing vertebra dysmorphology and rib fusions (Chang et al., 2022). CKO of one Nr6a1 allele reduced thoracic number by 2, while CKO of both Nr6a1 alleles resulted in 4 fewer thoracic elements. In each of these mutant scenarios, total vertebral number was reduced by 1 and 3 elements respectively, confirming the requirement for Nr6a1 in maintaining axial elongation. In parallel, while the entire lumbar region of TCreERT2; Nr6a1flx/flx embryos was not altered in terms of segment number, vertebral identity was transformed almost whole-sale to that of sacral elements based on characteristic lateral process morphology and fusion. This latter phenotype was particularly interesting since the developing hindlimbs, which normally align and ultimately articulate with sacral elements, were positioned normally in TCreERT2; Nr6a1flx/flx embryos, supporting a disassociation of the patterning events between hindlimb-forming lateral plate mesoderm and vertebral column-forming paraxial mesoderm. Finally, while the majority of thoraco-lumbar-sacral elements in TCreERT2; Nr6a1flx/flx embryos were highly dysmorphic, all post-sacral vertebral elements reverted back to WT morphology. This phenotype could be traced back to segmentation stages (E10.5) where a striking switch back to normal somite morphology could be observed immediately after the last sacral-forming somite (Chang et al., 2022).
In contrast to the above conditional loss-of-function scenario, Nr6a1 gain-of-function in the mouse posterior growth zone using a transient transgenic approach yielded almost mirror-image phenotypic alterations (Chang et al., 2022). These included an increased number of phenotypically normal thoraco-lumbar vertebrae by up to 5 elements, and a sharp switch this time to highly dysmorphic post-sacral elements and tail truncation. Together, these results not only delineated the critical requirement for Nr6a1 in axial elongation, but they also revealed quite unique phenotypes that will drive further research into lineage-specific patterning requirements and into axially-restricted gene regulatory networks that impact the seemingly uniform process of segmentation.
Molecular mechanisms and targets of Nr6a1
Nr6a1 has been characterised as a repressor of gene expression (Cooney et al., 1998; Yan and Jetten, 2000), with all evidence to date supporting this as its sole regulatory function. Early work revealed many examples of Nr6a1-dependent repression in germ cells that relied on the presence of a DR0 site, including protamine 1 and 2, mGDPH and ELP in male germ cells (Yan et al., 1997; Hummelke et al., 1998; Yang et al., 2003) and BMP-15 and Gdf-9 in female germ cells (Carabatsos et al., 1998; Erickson and Shimasaki, 2001; Zhao et al., 2007).
Certainly, one of the most notable target gene networks directly influenced by Nr6a1 activity is that of the pluripotency network. High expression levels of Pou5f1/Oct4 are part of a core network maintaining pluripotency within the in vivo mouse blastocyst and within in vitro blastocyst-derived ESCs. As differentiation proceeds in either system, the decrease and eventual termination of Oct4 expression was shown to inversely correlate with a rise in Nr6a1 expression levels (Figures 3A,B) (Fuhrmann et al., 2001; Gu et al., 2005), consistent with Nr6a1’s known repressive mode of action. A DR0 sequence was identified immediately upstream of the mouse Oct4 transcription start site (Fuhrmann et al., 2001), and subsequently across many vertebrate species (Wang et al., 2016), with direct binding to this site confirmed in mouse embryonal cells, mouse ESCs and human ESCs. In these various contexts, the Nr6a1 protein has been shown to directly interact with SMRT and N-Cor corepressor proteins in mouse embryonal cells (Fuhrmann et al., 2001), DNMT3b methyltransferase in human ESCs (Sato et al., 2006), and both Dnmt3A and methyl-CpG binding domain (MBD) proteins MBD-2 and MBD-3 in mouse ESCs (Gu et al., 2011), supporting histone deacetylation and de novo DNA methylation as ultimate mechanisms by which Nr6a1 reduces Oct4 target gene expression. LikeOct4, a second key pluripotency gene Nanog is directly repressed by Nr6a1, with indirect repressive consequences on the larger network including Sox2, Stella and Fgf4 (Gu et al., 2005).
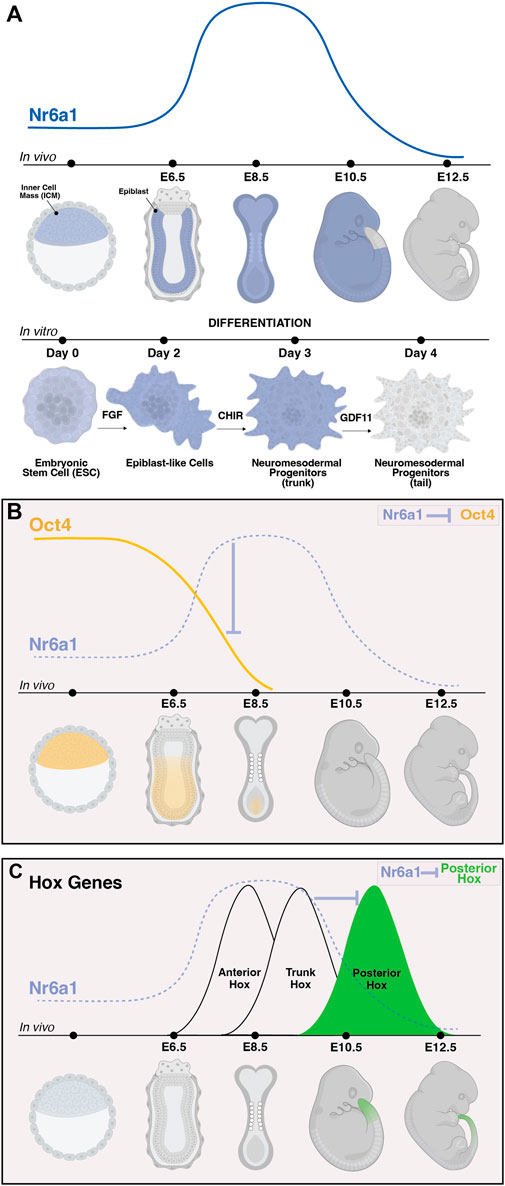
FIGURE 3. The dynamic expression of Nr6a1 controls key developmental genes and transitions. The expression of Nr6a1 over time in vivo and during in vitro differentiation (A). In vivo, Nr6a1 expression is detected within the inner cell mass (ICM) at very early stage of mouse development, broadly within the epiblast (epi) at embryonic day (E)6.5, across most tissues and germ layers at E8.5, with a gradual clearing of expression beginning with the posterior tail bud from E9.5 and expression largely cleared from the embryo by E12.5. A similar expression dynamic is seen during in vitro ESC-to-NMP differentiation: Nr6a1 expression within ESCs increases following exposure to Fgf2, and further increases following activation of Wnt signalling (CHIR). The transition in vitro from a trunk NMP to a tail NMP following exposure to Gdf11 downregulates Nr6a1 to low/basal levels. The dynamic in vivo expression of Nr6a1 overlaid with key target genes Oct4 and posterior Hox genes (B,C). The rise of Nr6a1 within the epiblast directly repress Oct4 levels, leading to broadly complementary patterns of expression between early and mid-gestation (B). Conversely, the rise of Nr6a1 prevents precocious expression of posterior Hox genes, leading to broadly complementary patterns of expression between mid and late-gestation (C). Whether posterior Hox repression by Nr6a1 is via direct mechanisms is currently unclear. Images created in Biorender.
During later tailbud stages in the mouse, global expression changes elicited by in vivo ectopic Nr6a1 activity revealed the maintenance of a core trunk gene regulatory network (Chang et al., 2022) longer than wildtype, consistent with the Nr6a1-dependent “mid-gestation program” identified in mesenchymal stem cells (Gurtan et al., 2013). Of particular importance was the identification that Nr6a1 activity significantly impacts the timing of gene activation across all 4 Hox clusters, central regulators of body plan formation across bilateria (reviewed in Hubert and Wellik, 2023). Ectopic expression of Nr6a1 stalled progression of Hox gene activation at a trunk (Hox5-9) code with a concomitant downregulation (or likely delayed activation) of posterior/terminal (Hox11-13) genes. Conversely, loss of Nr6a1 activity, both in vitro and in vivo, led to a speeding up of Hox cluster progression and precocious activation of posterior/terminal Hox genes (Figure 3C) (Chang et al., 2022). In parallel, Nr6a1 was shown to impact the balance of neural vs. Mesodermal gene signatures in the tailbud. Further work is needed to understand the direct vs. Indirect regulatory nature of each of these interactions and whether changes in histone marks and DNA methylation are involved.
Nr6a1 and the evolution of animal body plan
The body plan and ensuing axial formulae of a given vertebrate species is remarkably robust, particularly for isogenic mouse strains such as C57Bl6 that are used in many genetic studies. In contrast, the diversity of body plans across the vertebrate species can be extreme, and the molecular mechanisms driving such changes are of intense interest yet still largely unknown. As the precise expression level of Nr6a1 has now been shown to control total vertebral number in the mouse, both positively and negatively, this work raises the possibility that Nr6a1 may be a molecular target for evolutionary change. In support of this view, numerous studies have identified a genetic association between Nr6a1 and the increase in vertebral count of domesticated animals, a trait possibly selected for in the livestock industry due to its advantage in boosting meat yield. In 2005, Mikawa et al. (2005) conducted a quantitative trait loci (QTL) analysis comparing domesticated pigs and wild boar populations, pinpointing regions on Sus scrofa chromosomes 1 and 7 as associated with an increase of more than two presacral vertebrae. Subsequent fine mapping of the chromosome 1 loci identified a C>T single nucleotide polymorphism (SNP) at nucleotide 748 of Nr6a1 which segregated with the phenotype (Mikawa et al., 2007). This SNP led to a non-conservative amino acid substitution of proline to leucine at amino acid 192 within the protein’s hinge region which, using a yeast-two-hybrid assay, led to enhanced binding between Nr6a1 and the corepressors NCOR1 and RAP80. The predicted molecular gain-of-function aligns well with phenotypic outcomes of in vivo Nr6a1 gain-of-function studies in increasing thoraco-lumbar number (Chang et al., 2022). Subsequent studies by Zhang et al. (2019) have discovered an A>C SNP within exon 8 of Nr6a1 that potentially influences the number of lumbar vertebrae in sheep. Similarly, Fang et al. (2019) identified a 13 bp deletion within intron 1 of NR6A1 in various donkey breeds, linking it to body size attributes such as height and length. The exact molecular consequences of these later polymorphisms are still to be delineated but support the possibility that subtle changes in Nr6a1 regulation and/or function may have been an important event in the evolution of intra-species variation. In this light, further analysis of Nr6a1 sequence and function in vertebrate animals with extreme body plans, such as the snake, would be of major interest.
Conclusion
Nr6a1 is a critical developmental regulator with emerging roles in disease. It’s connection to key signalling pathways (RA, Wnt, and Fgf) and its ability to repress fundamental developmental molecules (Oct4, Nanog, posterior Hox genes) have been characterised in disparate contexts and a clearer consensus of the similarities and difference in how Nr6a1 functions across time and space is needed. What can be speculated however given the notable list of target genes this protein represses, and the quantitative manner in which Nr6a1 levels affect these targets (both positively and negatively), is that Nr6a1 could be utilised to guide positional identity and/or cell identity in 3D in vitro cell-based models of development either in its native form or as an engineered transcription factor. For example, manipulating the precise level of Nr6a1 is likely to “speed up” or “slow down” the Hox clock in 3D models of axial elongation such as gastruloids (Beccari et al., 2018) or somitoids (Sanaki-Matsumiya et al., 2022; Yamanaka et al., 2023). Along similar lines, the current inability of many cellular or organoid platforms to transition to a mature state, the often-lengthy (thus costly) protocols required, and the exhaustion of progenitor pools may all be enhanced by direct manipulation of positional identity via Nr6a1. Alternatively, Nr6a1 expression may provide a robust exit from pluripotency in blastoid models (Rivron et al., 2018; Liu et al., 2021; Yu et al., 2021) of early embryogenesis.
In future research, the dynamic transcriptional output of the genomic loci encompassing Nr6a1, Nr6a1 antisense transcripts and the two miR-181 microRNAs requires careful in vivo dissection, first to understand any co-regulation or anti-regulation that may shape Nr6a1’s spatio-temporal functional output and second, to determine if antisense transcripts have independent functional roles of as has been shown in vitro (Polo-Generelo et al., 2022). Once expressed, how is Nr6a1 protein subcellular localisation controlled and what, if any, ligand(s) trigger a downstream transcriptional response? What is the full complement of direct genomics targets of Nr6a1 in vivo, particularly in more recent areas of focus such as axial elongation, and how is lineage-restricted target gene regulation achieved? What higher order protein complexes is Nr6a1 guiding to the chromatin in vivo and is this lineage- or cell-restricted? And finally, how have the array of Nr6a1 SNPs identified across vertebrate species altered its molecular function and were these changes an important driver of phenotypic change across evolution.
Author contributions
JL: Writing–original draft, Writing–review and editing. PM: Writing–original draft, Writing–review and editing. EM: Writing–original draft, Writing–review and editing, Funding acquisition.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by Australian Research Council Discovery Project DP180102157 to EM. The Australian Regenerative Medicine Institute is supported by grants from the State Government of Victoria and the Australian Government.
Acknowledgments
The authors thank Yi-Cheng Chang for critical reading of the manuscript and for contributing Figure 2B.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Bain, D. L., Heneghan, A. F., Connaghan-Jones, K. D., and Miura, M. T. (2007). Nuclear receptor structure: implications for function. Annu. Rev. Physiol. 69, 201–220. doi:10.1146/annurev.physiol.69.031905.160308
Barreto, G., Borgmeyer, U., and Dreyer, C. (2003). The germ cell nuclear factor is required for retinoic acid signaling during Xenopus development. Mech. Dev. 120, 415–428. doi:10.1016/s0925-4773(03)00018-2
Bauer, U. M., Schneider-Hirsch, S., Reinhardt, S., Pauly, T., Maus, A., Wang, F., et al. (1997). Neuronal cell nuclear factor--a nuclear receptor possibly involved in the control of neurogenesis and neuronal differentiation. Eur. J. Biochem. 249, 826–837. doi:10.1111/j.1432-1033.1997.t01-1-00826.x
Beccari, L., Moris, N., Girgin, M., Turner, D. A., Baillie-Johnson, P., Cossy, A. C., et al. (2018). Multi-axial self-organization properties of mouse embryonic stem cells into gastruloids. Nature 562, 272–276. doi:10.1038/s41586-018-0578-0
Borgmeyer, U. (1997). Dimeric binding of the mouse germ cell nuclear factor. Eur. J. Biochem. 244, 120–127. doi:10.1111/j.1432-1033.1997.00120.x
Bridgham, J. T., Eick, G. N., Larroux, C., Deshpande, K., Harms, M. J., Gauthier, M. E., et al. (2010). Protein evolution by molecular tinkering: diversification of the nuclear receptor superfamily from a ligand-dependent ancestor. PLoS Biol. 8, e1000497. doi:10.1371/journal.pbio.1000497
Carabatsos, M. J., Elvin, J., Matzuk, M. M., and Albertini, D. F. (1998). Characterization of oocyte and follicle development in growth differentiation factor-9-deficient mice. Dev. Biol. 204, 373–384. doi:10.1006/dbio.1998.9087
Chang, Y. C., Manent, J., Schroeder, J., Wong, S. F. L., Hauswirth, G. M., Shylo, N. A., et al. (2022). Nr6a1 controls Hox expression dynamics and is a master regulator of vertebrate trunk development. Nat. Commun. 13, 7766. doi:10.1038/s41467-022-35303-4
Chen, F., Cooney, A. J., Wang, Y., Law, S. W., and O'malley, B. W. (1994). Cloning of a novel orphan receptor (GCNF) expressed during germ cell development. Mol. Endocrinol. 8, 1434–1444. doi:10.1210/mend.8.10.7854358
Chung, A. C., Katz, D., Pereira, F. A., Jackson, K. J., Demayo, F. J., Cooney, A. J., et al. (2001). Loss of orphan receptor germ cell nuclear factor function results in ectopic development of the tail bud and a novel posterior truncation. Mol. Cell Biol. 21, 663–677. doi:10.1128/MCB.21.2.663-677.2001
Cooney, A. J., Hummelke, G. C., Herman, T., Chen, F., and Jackson, K. J. (1998). Germ cell nuclear factor is a response element-specific repressor of transcription. Biochem. Biophys. Res. Commun. 245, 94–100. doi:10.1006/bbrc.1998.8391
Cooney, A. J., Katz, D., Hummelke, G. C., and Jackson, K. J. (1999). Germ cell nuclear factor: an orphan receptor in search of a function. Am. zoologist 39, 796–806. doi:10.1093/icb/39.4.796
Erickson, G. F., and Shimasaki, S. (2001). The physiology of folliculogenesis: the role of novel growth factors. Fertil. Steril. 76, 943–949. doi:10.1016/s0015-0282(01)02859-x
Escriva, H., Delaunay, F., and Laudet, V. (2000). Ligand binding and nuclear receptor evolution. Bioessays 22, 717–727. doi:10.1002/1521-1878(200008)22:8<717::AID-BIES5>3.0.CO;2-I
Fang, X., Lai, Z., Liu, J., Zhang, C., Li, S., Wu, F., et al. (2019). A novel 13 bp deletion within the NR6A1 gene is significantly associated with growth traits in donkeys. Anim. (Basel) 9, 681. doi:10.3390/ani9090681
Frigo, D. E., Bondesson, M., and Williams, C. (2021). Nuclear receptors: from molecular mechanisms to therapeutics. Essays Biochem. 65, 847–856. doi:10.1042/EBC20210020
Fuhrmann, G., Chung, A. C., Jackson, K. J., Hummelke, G., Baniahmad, A., Sutter, J., et al. (2001). Mouse germline restriction of Oct4 expression by germ cell nuclear factor. Dev. Cell 1, 377–387. doi:10.1016/s1534-5807(01)00038-7
Gallastegui, N., Mackinnon, J. A., Fletterick, R. J., and Estébanez-Perpiñá, E. (2015). Advances in our structural understanding of orphan nuclear receptors. Trends Biochem. Sci. 40, 25–35. doi:10.1016/j.tibs.2014.11.002
Gouti, M., Delile, J., Stamataki, D., Wymeersch, F. J., Huang, Y., Kleinjung, J., et al. (2017). A gene regulatory network balances neural and mesoderm specification during vertebrate trunk development. Dev. Cell 41, 243–261. doi:10.1016/j.devcel.2017.04.002
Greschik, H., Wurtz, J. M., Hublitz, P., Köhler, F., Moras, D., and Schüle, R. (1999). Characterization of the DNA-binding and dimerization properties of the nuclear orphan receptor germ cell nuclear factor. Mol. Cell Biol. 19, 690–703. doi:10.1128/mcb.19.1.690
Gu, P., Lemenuet, D., Chung, A. C., Mancini, M., Wheeler, D. A., and Cooney, A. J. (2005). Orphan nuclear receptor GCNF is required for the repression of pluripotency genes during retinoic acid-induced embryonic stem cell differentiation. Mol. Cell Biol. 25, 8507–8519. doi:10.1128/MCB.25.19.8507-8519.2005
Gu, P., Xu, X., Le Menuet, D., Chung, A. C., and Cooney, A. J. (2011). Differential recruitment of methyl CpG-binding domain factors and DNA methyltransferases by the orphan receptor germ cell nuclear factor initiates the repression and silencing of Oct4. Stem Cells 29, 1041–1051. doi:10.1002/stem.652
Gurtan, A. M., Ravi, A., Rahl, P. B., Bosson, A. D., Jnbaptiste, C. K., Bhutkar, A., et al. (2013). Let-7 represses Nr6a1 and a mid-gestation developmental program in adult fibroblasts. Genes Dev. 27, 941–954. doi:10.1101/gad.215376.113
He, X., Yan, Y. L., Eberhart, J. K., Herpin, A., Wagner, T. U., Schartl, M., et al. (2011). miR-196 regulates axial patterning and pectoral appendage initiation. Dev. Biol. 357, 463–477. doi:10.1016/j.ydbio.2011.07.014
Heinzer, C., Süsens, U., Schmitz, T. P., and Borgmeyer, U. (1998). Retinoids induce differential expression and DNA binding of the mouse germ cell nuclear factor in P19 embryonal carcinoma cells. Biol. Chem. 379, 349–359. doi:10.1515/bchm.1998.379.3.349
Henrique, D., Abranches, E., Verrier, L., and Storey, K. G. (2015). Neuromesodermal progenitors and the making of the spinal cord. Development 142, 2864–2875. doi:10.1242/dev.119768
Hirose, T., O'brien, D. A., and Jetten, A. M. (1995). RTR: a new member of the nuclear receptor superfamily that is highly expressed in murine testis. Gene 152, 247–251. doi:10.1016/0378-1119(94)00656-d
Holzer, G., Markov, G. V., and Laudet, V. (2017). Evolution of nuclear receptors and ligand signaling: toward a soft key-lock model? Curr. Top. Dev. Biol. 125, 1–38. doi:10.1016/bs.ctdb.2017.02.003
Hubert, K. A., and Wellik, D. M. (2023). Hox genes in development and beyond. Development 150, dev192476. doi:10.1242/dev.192476
Hummelke, G. C., Meistrich, M. L., and Cooney, A. J. (1998). Mouse protamine genes are candidate targets for the novel orphan nuclear receptor, germ cell nuclear factor. Mol. Reprod. Dev. 50, 396–405. doi:10.1002/(SICI)1098-2795(199808)50:4<396::AID-MRD3>3.0.CO;2-D
Jumper, J., Evans, R., Pritzel, A., Green, T., Figurnov, M., Ronneberger, O., et al. (2021). Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589. doi:10.1038/s41586-021-03819-2
Jurberg, A. D., Aires, R., Varela-Lasheras, I., Nóvoa, A., and Mallo, M. (2013). Switching axial progenitors from producing trunk to tail tissues in vertebrate embryos. Dev. Cell 25, 451–462. doi:10.1016/j.devcel.2013.05.009
Kadmiel, M., and Cidlowski, J. A. (2013). Glucocorticoid receptor signaling in health and disease. Trends Pharmacol. Sci. 34, 518–530. doi:10.1016/j.tips.2013.07.003
Katz, D., Niederberger, C., Slaughter, G. R., and Cooney, A. J. (1997). Characterization of germ cell-specific expression of the orphan nuclear receptor, germ cell nuclear factor. Endocrinology 138, 4364–4372. doi:10.1210/endo.138.10.5444
Khorasanizadeh, S., and Rastinejad, F. (2001). Nuclear-receptor interactions on DNA-response elements. Trends Biochem. Sci. 26, 384–390. doi:10.1016/s0968-0004(01)01800-x
Lan, Z. J., Chung, A. C., Xu, X., Demayo, F. J., and Cooney, A. J. (2002). The embryonic function of germ cell nuclear factor is dependent on the DNA binding domain. J. Biol. Chem. 277, 50660–50667. doi:10.1074/jbc.M209586200
Lan, Z. J., Gu, P., Xu, X., Jackson, K. J., Demayo, F. J., O'malley, B. W., et al. (2003). GCNF-dependent repression of BMP-15 and GDF-9 mediates gamete regulation of female fertility. Embo J. 22, 4070–4081. doi:10.1093/emboj/cdg405
Liu, X., Tan, J. P., Schroder, J., Aberkane, A., Ouyang, J. F., Mohenska, M., et al. (2021). Modelling human blastocysts by reprogramming fibroblasts into iBlastoids. Nature 591, 627–632. doi:10.1038/s41586-021-03372-y
Lonard, D. M., and O'malley, B. W. (2012). Nuclear receptor coregulators: modulators of pathology and therapeutic targets. Nat. Rev. Endocrinol. 8, 598–604. doi:10.1038/nrendo.2012.100
Matsubara, Y., Hirasawa, T., Egawa, S., Hattori, A., Suganuma, T., Kohara, Y., et al. (2017). Anatomical integration of the sacral-hindlimb unit coordinated by GDF11 underlies variation in hindlimb positioning in tetrapods. Nat. Ecol. Evol. 1, 1392–1399. doi:10.1038/s41559-017-0247-y
Mazaira, G. I., Zgajnar, N. R., Lotufo, C. M., Daneri-Becerra, C., Sivils, J. C., Soto, O. B., et al. (2018). The nuclear receptor field: a historical overview and future challenges. Nucl. Recept. Res. 5, 101320. doi:10.11131/2018/101320
Mcgeary, S. E., Lin, K. S., Shi, C. Y., Pham, T. M., Bisaria, N., Kelley, G. M., et al. (2019). The biochemical basis of microRNA targeting efficacy. Science 366, eaav1741. doi:10.1126/science.aav1741
Mckenna, N. J., Lanz, R. B., and O'malley, B. W. (1999). Nuclear receptor coregulators: cellular and molecular biology. Endocr. Rev. 20, 321–344. doi:10.1210/edrv.20.3.0366
Mcpherron, A. C., Lawler, A. M., and Lee, S. J. (1999). Regulation of anterior/posterior patterning of the axial skeleton by growth/differentiation factor 11. Nat. Genet. 22, 260–264. doi:10.1038/10320
Mikawa, S., Hayashi, T., Nii, M., Shimanuki, S., Morozumi, T., and Awata, T. (2005). Two quantitative trait loci on Sus scrofa chromosomes 1 and 7 affecting the number of vertebrae. J. Anim. Sci. 83, 2247–2254. doi:10.2527/2005.83102247x
Mikawa, S., Morozumi, T., Shimanuki, S., Hayashi, T., Uenishi, H., Domukai, M., et al. (2007). Fine mapping of a swine quantitative trait locus for number of vertebrae and analysis of an orphan nuclear receptor, germ cell nuclear factor (NR6A1). Genome Res. 17, 586–593. doi:10.1101/gr.6085507
Okumura, L. M., Lesch, B. J., and Page, D. C. (2013). The ligand binding domain of GCNF is not required for repression of pluripotency genes in mouse fetal ovarian germ cells. PLoS One 8, e66062. doi:10.1371/journal.pone.0066062
Oyekan, A. (2011). PPARs and their effects on the cardiovascular system. Clin. Exp. Hypertens. 33, 287–293. doi:10.3109/10641963.2010.531845
Papageorgiou, L., Shalzi, L., Pierouli, K., Papakonstantinou, E., Manias, S., Dragoumani, K., et al. (2021). An updated evolutionary study of the nuclear receptor protein family. World Acad. Sci. Journa 3, 51–58. doi:10.3892/wasj.2021.122
Pijuan-Sala, B., Griffiths, J. A., Guibentif, C., Hiscock, T. W., Jawaid, W., Calero-Nieto, F. J., et al. (2019). A single-cell molecular map of mouse gastrulation and early organogenesis. Nature 566, 490–495. doi:10.1038/s41586-019-0933-9
Polo-Generelo, S., Torres, B., Guerrero-Martinez, J. A., Camafeita, E., Vazquez, J., Reyes, J. C., et al. (2022). TGF-β-Upregulated lnc-nr6a1 acts as a reservoir of miR-181 and mediates assembly of a glycolytic complex. Noncoding RNA 8, 62. doi:10.3390/ncrna8050062
Ranhotra, H. S. (2013). The orphan nuclear receptors in cancer and diabetes. J. Recept Signal Transduct. Res. 33, 207–212. doi:10.3109/10799893.2013.781624
Rivron, N. C., Frias-Aldeguer, J., Vrij, E. J., Boisset, J. C., Korving, J., Vivie, J., et al. (2018). Blastocyst-like structures generated solely from stem cells. Nature 557, 106–111. doi:10.1038/s41586-018-0051-0
Robinson-Rechavi, M., Escriva Garcia, H., and Laudet, V. (2003). The nuclear receptor superfamily. J. Cell Sci. 116, 585–586. doi:10.1242/jcs.00247
Robinton, D. A., Chal, J., Lummertz Da Rocha, E., Han, A., Yermalovich, A. V., Oginuma, M., et al. (2019). The lin28/let-7 pathway regulates the mammalian caudal body Axis elongation program. Dev. Cell 48, 396–405. doi:10.1016/j.devcel.2018.12.016
Sabour, D., Xu, X., Chung, A. C., Le Menuet, D., Ko, K., Tapia, N., et al. (2014). Germ cell nuclear factor regulates gametogenesis in developing gonads. PLoS One 9, e103985. doi:10.1371/journal.pone.0103985
Sanaki-Matsumiya, M., Matsuda, M., Gritti, N., Nakaki, F., Sharpe, J., Trivedi, V., et al. (2022). Periodic formation of epithelial somites from human pluripotent stem cells. Nat. Commun. 13, 2325. doi:10.1038/s41467-022-29967-1
Sato, N., Kondo, M., and Arai, K. (2006). The orphan nuclear receptor GCNF recruits DNA methyltransferase for Oct-3/4 silencing. Biochem. Biophys. Res. Commun. 344, 845–851. doi:10.1016/j.bbrc.2006.04.007
Schmitz, T. P., Susens, U., and Borgmeyer, U. (1999). DNA binding, protein interaction and differential expression of the human germ cell nuclear factor. Biochim. Biophys. Acta 1446, 173–180. doi:10.1016/s0167-4781(99)00079-2
Varadi, M., Anyango, S., Deshpande, M., Nair, S., Natassia, C., Yordanova, G., et al. (2022). AlphaFold Protein Structure Database: massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 50, D439–d444. doi:10.1093/nar/gkab1061
Wang, H., Wang, X., Xu, X., Kyba, M., and Cooney, A. J. (2016). Germ cell nuclear factor (GCNF) represses Oct4 expression and globally modulates gene expression in human embryonic stem (hES) cells. J. Biol. Chem. 291, 8644–8652. doi:10.1074/jbc.M115.694208
Wong, S. F., Agarwal, V., Mansfield, J. H., Denans, N., Schwartz, M. G., Prosser, H. M., et al. (2015). Independent regulation of vertebral number and vertebral identity by microRNA-196 paralogs. Proc. Natl. Acad. Sci. U. S. A. 112, E4884–E4893. doi:10.1073/pnas.1512655112
Yamanaka, Y., Hamidi, S., Yoshioka-Kobayashi, K., Munira, S., Sunadome, K., Zhang, Y., et al. (2023). Reconstituting human somitogenesis in vitro. Nature 614, 509–520. doi:10.1038/s41586-022-05649-2
Yan, Z., and Jetten, A. M. (2000). Characterization of the repressor function of the nuclear orphan receptor retinoid receptor-related testis-associated receptor/germ cell nuclear factor. J. Biol. Chem. 275, 35077–35085. doi:10.1074/jbc.M005566200
Yan, Z. H., Medvedev, A., Hirose, T., Gotoh, H., and Jetten, A. M. (1997). Characterization of the response element and DNA binding properties of the nuclear orphan receptor germ cell nuclear factor/retinoid receptor-related testis-associated receptor. J. Biol. Chem. 272, 10565–10572. doi:10.1074/jbc.272.16.10565
Yang, G., Zhang, Y. L., Buchold, G. M., Jetten, A. M., and O'brien, D. A. (2003). Analysis of germ cell nuclear factor transcripts and protein expression during spermatogenesis. Biol. Reprod. 68, 1620–1630. doi:10.1095/biolreprod.102.012013
Yu, L., Wei, Y., Duan, J., Schmitz, D. A., Sakurai, M., Wang, L., et al. (2021). Blastocyst-like structures generated from human pluripotent stem cells. Nature 591, 620–626. doi:10.1038/s41586-021-03356-y
Zechel, C. (2005). The germ cell nuclear factor (GCNF). Mol. Reprod. Dev. 72, 550–556. doi:10.1002/mrd.20377
Zhang, X., Li, C., Li, X., Liu, Z., Ni, W., Cao, Y., et al. (2019). Association analysis of polymorphism in the NR6A1 gene with the lumbar vertebrae number traits in sheep. Genes Genomics 41, 1165–1171. doi:10.1007/s13258-019-00843-5
Zhao, H., Li, Z., Cooney, A. J., and Lan, Z. J. (2007). Orphan nuclear receptor function in the ovary. Front. Biosci. 12, 3398–3405. doi:10.2741/2321
Keywords: Nr6a1, GCNF, orphan nuclear receptor, axial elongation, Oct4, Hox genes, retinoic acid
Citation: Li J, Mascarinas P and McGlinn E (2024) The expanding roles of Nr6a1 in development and evolution. Front. Cell Dev. Biol. 12:1357968. doi: 10.3389/fcell.2024.1357968
Received: 19 December 2023; Accepted: 31 January 2024;
Published: 19 February 2024.
Edited by:
Jose F. de Celis, Spanish National Research Council (CSIC), SpainReviewed by:
Yogesh Srivastava, University of Texas MD Anderson Cancer Center, United StatesAlessandro Fiorenzano, Lund University, Sweden
Copyright © 2024 Li, Mascarinas and McGlinn. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Edwina McGlinn, ZWR3aW5hLm1jZ2xpbm5AbW9uYXNoLmVkdQ==
†Lead contact
 Jingxuan Li
Jingxuan Li Pauline Mascarinas
Pauline Mascarinas Edwina McGlinn
Edwina McGlinn