- 1Department of Paediatrics, University of Oxford, Oxford, United Kingdom
- 2MRC Molecular Haematology Unit, Weatherall Institute of Molecular Medicine, University of Oxford, Oxford, United Kingdom
- 3Department of Haematology, Great Ormond Street Hospital for Children, London, United Kingdom
Hematopoiesis is a finely orchestrated process, whereby hematopoietic stem cells give rise to all mature blood cells. Crucially, they maintain the ability to self-renew and/or differentiate to replenish downstream progeny. This process starts at an embryonic stage and continues throughout the human lifespan. Blood cancers such as leukemia occur when normal hematopoiesis is disrupted, leading to uncontrolled proliferation and a block in differentiation of progenitors of a particular lineage (myeloid or lymphoid). Although normal stem cell programs are crucial for tissue homeostasis, these can be co-opted in many cancers, including leukemia. Myeloid or lymphoid leukemias often display stem cell-like properties that not only allow proliferation and survival of leukemic blasts but also enable them to escape treatments currently employed to treat patients. In addition, some leukemias, especially in children, have a fetal stem cell profile, which may reflect the developmental origins of the disease. Aberrant fetal stem cell programs necessary for leukemia maintenance are particularly attractive therapeutic targets. Understanding how hijacked stem cell programs lead to aberrant gene expression in place and time, and drive the biology of leukemia, will help us develop the best treatment strategies for patients.
Introduction
Stem cells perform a complex balancing act between self-renewal and differentiation throughout ontogeny. To perform these functions, stem cells proliferate rapidly and repair DNA damage. However, these “stemness” properties present a vulnerability as, if hijacked, they provide cancer cells with the pathways required for growth and survival (Taipale and Beachy, 2001; Hanahan and Weinberg, 2011).
Hematopoietic stem cells
Human hematopoiesis is a dynamic process beginning at day 18 in the yolk sac (Palis and Yoder, 2001), with definitive hematopoietic stem cells (HSCs) originating in the aorta–gonad–mesonephros from 4 post-conception weeks (pcw) (Tavian and Peault, 2005; Ivanovs et al., 2011). HSCs subsequently migrate to fetal liver (FL), the main site of hematopoiesis until birth (Ivanovs et al., 2017), with contribution from fetal bone marrow (FBM) 10–12 pcw (Charbord et al., 1996; O’Byrne et al., 2019). After birth, BM becomes the sole site of hematopoiesis.
Fetal HSCs are molecularly and functionally distinct from postnatal HSCs
Fetal HSCs are more proliferative than their postnatal counterparts (Lansdorp et al., 1993; Muench et al., 1994; Bowie et al., 2006; Popescu et al., 2019; Roy et al., 2021) with higher self-renewal capacity (Copley and Eaves, 2013; Copley et al., 2013), better in vivo engraftment (Harrison et al., 1997; Holyoake et al., 1999), and a distinct metabolic profile (Manesia et al., 2015). Adult HSCs show a myeloid lineage bias (Benz et al., 2012); additionally, innate lymphoid cells such as B1a B-cells are derived exclusively from embryonic/fetal HSCs (Zhou et al., 2015; Montecino-Rodriguez et al., 2016). These functional differences may be a consequence of distinct developmental gene expression programs.
Stem cell programs in leukemia
In leukemia, stem cell programs may be inappropriately reactivated or retained and/or co-opted from fetal development. This may be a consequence of some leukemias originating in utero, especially in children (Greaves, 2005).
In this review, we discuss stem cell programs that are aberrantly active in the wrong cellular context (“place”) or stage of ontogeny (“time”) in pediatric leukemia and their potential applications in developing targeted therapies (Figure 1).
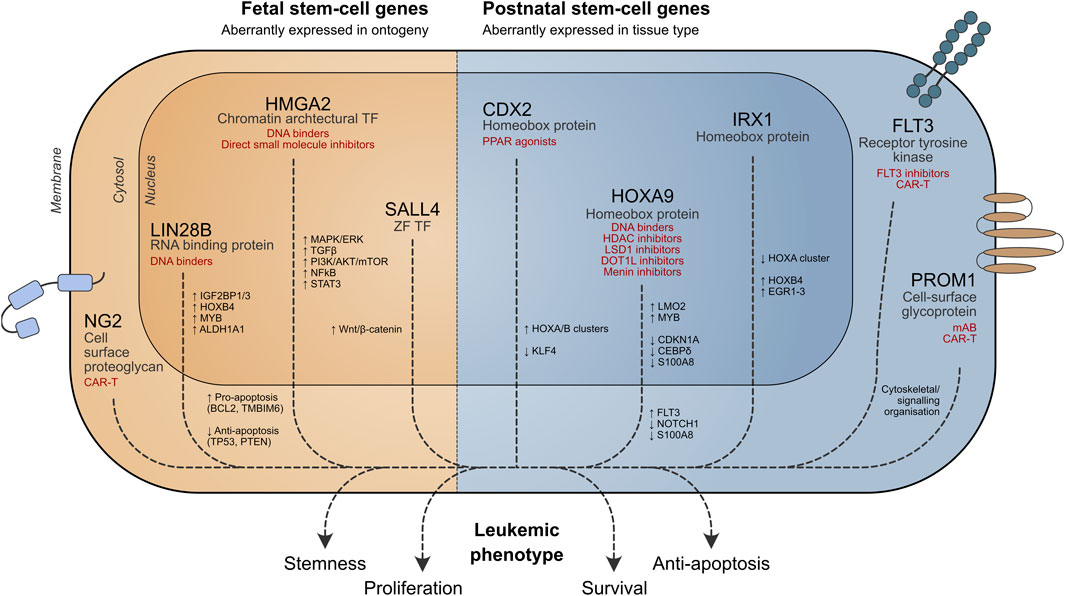
Figure 1. Role of stem cell genes in leukemia cell survival. Potential therapeutic strategies targeting aberrant stem cell-associated pathways are shown in red. TF = transcription factor, ZF = zinc finger.
Stem cell programs in the wrong place
Aberrant stem cell genes in leukemia
Leukemic blasts may exhibit stem cell properties, conferring a more aggressive phenotype. Several stem cell genes are important for leukemia biology (Table 1), and some key examples are discussed below.
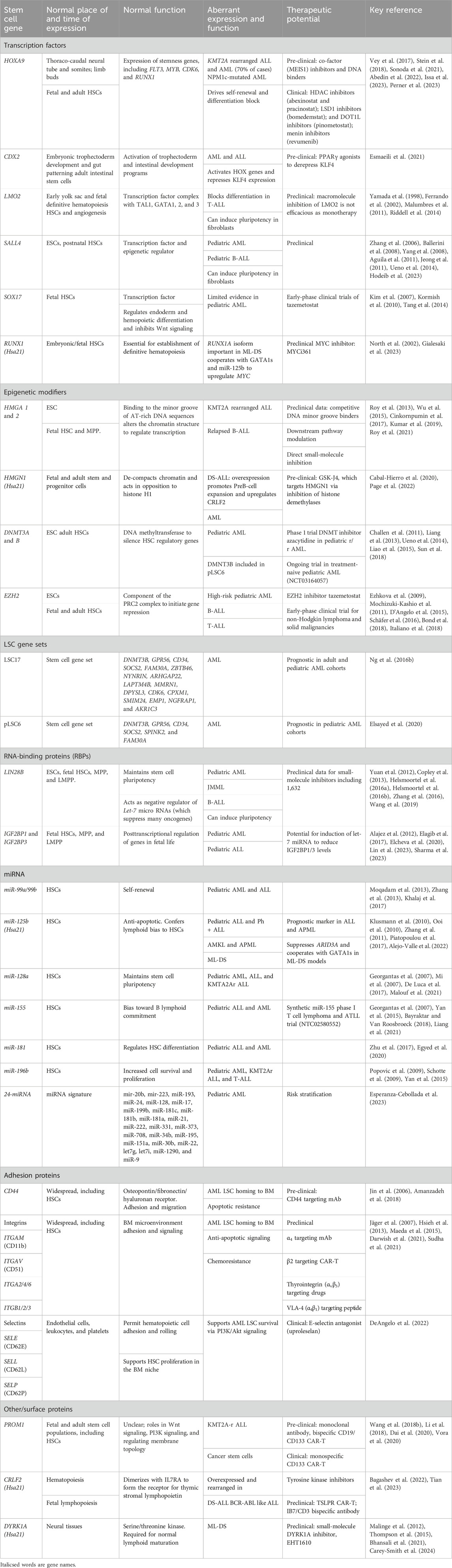
Table 1. Summary of developmental/stem cell genes involved in the pathophysiology of pediatric leukemia.
Homeobox genes
Homeobox genes form a group of transcription factors (TFs) with 235 functional genes in humans (Holland et al., 2007).
HOXA9
The largest class of homeobox genes, the HOX genes (Holland, 2013), play important roles in hematopoiesis, such as development and maintenance of HSCs (Magnusson et al., 2007; Ramos-Mejía et al., 2014). HOXA9 can reprogram tissues to a hematopoietic fate (Ng E. S. et al., 2016; Sugimura et al., 2017).
Over 50% of AML cases overexpress HOXA9, which correlates with poor survival (Drabkin et al., 2002; Andreeff et al., 2008; Tholouli et al., 2012). HOXA9 directly binds target genes along with PBX3/MEIS1 (Wong et al., 2007; Li et al., 2016), upregulating oncogenes such as FLT3, LMO2, and MYB(Huang et al., 2012; Collins et al., 2014). HOXA9 also suppresses apoptotic/differentiation factors, promoting leukemia cell survival and maintaining a more stem-like state (Agrawal-Singh et al., 2023).
HOXA9 plays a key role in KMT2A-rearranged (KMT2A-r) leukemia (Faber et al., 2009; Orlovsky et al., 2011), mediated by binding of the KMT2A fusion protein to HOX gene promoters (Milne et al., 2002; Milne et al., 2005) and recruitment of DOT1L H3K79 methyltransferases (Bernt et al., 2011; Kerry et al., 2017). Dysregulated HOXA9 expression is also seen in NPM1c-mutated AML (Brunetti et al., 2018; Uckelmann et al., 2023).
CDX2
CDX2 of the ParaHox family of homeobox genes (Brooke et al., 1998) is not normally expressed in hematopoietic cells and inhibits the hematopoietic potential of murine embryonic stem cells (ESCs) (McKinney-Freeman et al., 2008).
Over 90% of AML cases overexpress CDX2 (Chase et al., 1999; Scholl et al., 2007; Rawat et al., 2008), and ectopic expression of Cdx2 confers oncogenic properties to murine HSCs (Rawat et al., 2004; Scholl et al., 2007). CDX2 overexpression studies suggest upregulation of HOX genes as the oncogenic mechanism (Rawat et al., 2008), although at least one alternative pathway is through direct suppression of KLF4 (Faber et al., 2013).
Aberrant CDX2 expression is also frequently seen in ALL (Riedt et al., 2009; Thoene et al., 2009) and confers a poor prognosis (Yasuda et al., 2022).
IRX genes
The Iroquois genes (IRX1-6) belong to the TALE group of homeobox genes. IRX1 is required for normal development of kidney and neural tissues (Alarcón et al., 2008; Freese et al., 2014) but not expressed in most hematopoietic precursors (Nagel et al., 2022). IRX3 and IRX5 are not expressed in hematopoiesis.
In KMT2A-r infant ALL, IRX1 or HOXA9 expression defines two distinct subgroups (Stam et al., 2010; Symeonidou and Ottersbach, 2021). IRX1 prevents KMT2A::AFF1 from activating HOXA9 expression; instead, expression of HOXB4 causes persistence of HSC factors (Kuhn et al., 2016). IRX1+(HOXA-) KMT2A-r ALL has a poorer prognosis (Agraz-Doblas et al., 2019; Isobe et al., 2022).
In AML, IRX1/3/5 is aberrantly expressed (Nagel and Meyer, 2022). In AML cell lines, expressions of IRX1 and IRX3/5 are mutually exclusive with opposing effects on GATA1/2 activity (Somerville et al., 2018).
PROM1
Prominin-1 (PROM1/CD133) is a membrane pentaspan glycoprotein, identified in the mouse neuroepithelium (Weigmann et al., 1997) and human HSCs (Miraglia et al., 1997; Yin et al., 1997). Its function remains enigmatic; and its role in cytoskeletal organization is postulated. In solid organ malignancies, PROM1 regulates extracellular vesicle formation and release (Rappa et al., 2013; Kang et al., 2019; Zhao et al., 2020).
PROM1 is expressed in many cancers (Reya et al., 2001; Singh et al., 2004; Bao et al., 2006; Ferrandina et al., 2009), including leukemia, where the expression is associated with poor prognosis (Tolba et al., 2013). It is a common feature of KMT2A-r leukemias and essential for survival of KMT2A::AFF1 cell lines (Godfrey et al., 2021; Wang L.-l. et al., 2022).
Leukemic stem cells
In addition to inappropriate expression of stem cell genes, there may be very rare leukemic stem cells (LSCs) within a leukemia that can propagate disease in serial transplantation (Lapidot et al., 1994; Hope et al., 2004; Hong et al., 2008). This concept is well-established in AML (Dick, 2008), but less so in ALL (Cox et al., 2009). AML LSCs share the key feature of self-renewal with normal HSCs alongside similarities in gene expression (Sachs et al., 2020), immunophenotype (e.g., CD133+(Tolba et al., 2013), CD123+(Testa et al., 2002)), and metabolism (Manesia et al., 2015).
A stem cell signature (LSC17) has been used to risk stratify adult (Ng S. W. K. et al., 2016) and some pediatric AMLs (Duployez et al., 2019), where it confers a worse prognosis. A six-gene pediatric AML signature (pLSC6) (Elsayed et al., 2020) was derived from a cohort of 163 pediatric AMLs (Table 1). The adult and pediatric signatures share five genes (DNMT3B, GPR56, CD34, FAM30A/KIAA012, and SOCS2), although with different gene weightings, and pLSC6 includes a further unique gene, SPINK2.
Single-cell RNA-seq identified LSC clusters in pediatric AML; these LSC markers require functional validation (Zhang Y. et al., 2023). Fetal-specific genes (HMGA2) have been identified in a rare HSC-like fraction of KMT2A-r infant leukemia (Chen et al., 2022), and IGF2BP1 maintains LSC by regulating HOXB4, MYB, and ALDH1A1 in pediatric leukemia cell lines (Elcheva et al., 2020).
Aberrant stem cell gene expression in ontogeny
Fetal stem cell programs in pediatric leukemia
Pediatric leukemias [and solid malignancies such as neuroblastoma (Molenaar et al., 2012)] may exhibit properties of fetal HSCs. This may represent a fetal cell of origin or indicate reactivation of fetal programs (Monk and Holding, 2001; Sharma et al., 2022; Solé et al., 2022). Either way, the inappropriate expression of fetal genes is important for cancer biology. Leukemic blasts, especially LSCs (Somervaille et al., 2009), show fetal-specific gene expression profiles (Wu et al., 2015; Helsmoortel et al., 2016b; Elcheva et al., 2020; Bai et al., 2021; Tran et al., 2021). Down syndrome (DS)-associated leukemias and juvenile myelomonocytic leukemia (JMML) are two examples resulting from perturbation of fetal HSPCs.
Trisomy 21 causes global perturbation of fetal hematopoiesis, with increased phenotypic HSCs and megakaryocyte–erythroid progenitors (MEPs), as well as a marked skew to erythropoiesis with a concomitant decrease in B-lymphopoiesis (Chou et al., 2008; Roy et al., 2012; Jardine et al., 2021). DS fetal HSPCs and stromal cells also display increased inflammatory signatures (Jardine et al., 2021). Chromosome 21 (Hsa21) stem cell genes dysregulated/overexpressed in DS include transcription factor GABPA, which affects HSC maintenance/differentiation (Yu et al., 2011), and chromatin modifier HMGN1.
Children with DS have an increased risk of AML and ALL (Hasle et al., 2000). Mutations in the megakaryocyte–erythroid transcription factor GATA1 in fetal life lead to transient abnormal myelopoiesis (TAM) in the fetal/neonatal period (Roberts et al., 2013; Wagenblast et al., 2021). Additional mutations, most commonly in the cohesin complex genes (Labuhn et al., 2019), are required for myeloid leukemia of DS (Roberts and Izraeli, 2014). DS-ALL also likely stems from perturbed lymphopoiesis, which begins in utero, and is characterized by CRLF2/TSLPR overexpression in 50% and JAK2 mutations in 20% (Li et al., 2023). Key Hsa21 genes important for leukemogenesis are listed in Table 1.
JMML, an HSC-derived leukemia (Cooper et al., 2000; Louka et al., 2021), has a fetal molecular profile (Roy et al., 2021; Hartmann et al., 2023). As the oncogenic hit probably occurs in fetal HSCs, developmental stem cell programs are hijacked for leukemogenesis.
Fetal oncogenes relevant to stem cell activity and implicated in pediatric leukemia act via a diverse range of mechanisms and are discussed below (Figure 1).
Fetal genes with transcription factor activity implicated in pediatric leukemia
SCL/TAL1 was originally identified as overexpressed in T-ALL (Ferrando et al., 2002). Ablation of the gene causes embryonic death (Porcher et al., 2017), but it is dispensable for adult HSCs (Mikkola et al., 2003).
LMO2 is essential for fetal hematopoiesis (Yamada et al., 1998). LMO2 expression is seen in a subset of pediatric T-ALL (Ferrando et al., 2002) and B-ALL (Malumbres et al., 2011). In gene therapy trials for SCID, two patients developed T-ALL through off-target activation of LMO2 (Hacein-Bey-Abina et al., 2003).
SALL4 is expressed in ESCs (Zhang et al., 2006), but downregulated postnatally with only low-level expression in HSCs (Gao et al., 2013). It is aberrantly expressed in pediatric AML (Ballerini et al., 2008) and ALL (Den Boer et al., 2009; Ueno et al., 2014), with overexpression conferring poor prognosis (Harvey et al., 2010; Jeong et al., 2011).
Fetal genes important for post-transcriptional regulation
The bulk of post-transcriptional control is exerted by RNA-binding proteins (RBPs). Some RBPs such as LIN28B and insulin-like growth factor 2 mRNA-binding proteins (IGF2BP1/IGF2BP3) have a fetal expression pattern, a role in stem cell biology (Copley and Eaves, 2013; Degrauwe et al., 2016; Zhang et al., 2016) and pediatric leukemia.
LIN28B
LIN28B has wide-ranging physiological roles in fetal tissues; however, the expression after birth is limited to the placenta and testis (Uhlén et al., 2015). In hematopoiesis, FL HSCs have the highest expression of LIN28B (Roy et al., 2021), and expression in postnatal cells can reactivate fetal-like erythropoiesis (Lee et al., 2013; Basak et al., 2020) and B-lymphopoiesis (Yuan et al., 2012). LIN28B can reprogram somatic cells to induce pluripotency (Zhang et al., 2016); however, this can be oncogenic in other contexts (Wuputra et al., 2020).
The main action of LIN28B is to prevent maturation of let-7 miRNAs (Rybak et al., 2008; Viswanathan and Daley, 2010). LIN28B also directly stabilizes many mRNAs in conjunction with IGF2BP1/IGF2BP3. In murine B progenitors, this includes Pax5 and Arid3a, thereby driving fetal B-lymphopoiesis (Wang et al., 2019).
LIN28B is frequently aberrantly expressed in cancers, including leukemia (Viswanathan et al., 2009; Balzeau et al., 2017). Suppression of let-7 miRNAs by LIN28B leads to de-repression of oncogenes (MYC, RAS, MYB, and ARID3A) and epigenetic regulators, HMGA2 and CBX2 (Wang D. et al., 2022).
A meta-analysis showed that 7.5% of pediatric leukemias express LIN28B (Helsmoortel et al., 2016b). Aberrant LIN28B expression defines a poor prognosis subgroup in JMML (Helsmoortel et al., 2016a), where H19, a fetal oncogene (Matouk et al., 2014), is stabilized in the presence of LIN28B (Helsmoortel et al., 2016b). AML in children <3 years has higher levels of LIN28B (and IGF2BP1/3) expression than in children >3 years (Bolouri et al., 2021). Although LIN28B has predominantly been reported to have a pro-leukemic role in AML (Zhou et al., 2017), one study on a murine KMT2A::MLLT3 AML model suggests that LIN28B abrogates perinatal leukemia development (Eldeeb et al., 2023). Given >50% of human neonatal leukemias are of myeloid lineage, these findings seem counterintuitive, although it is possible that neonatal AML arises from LIN28B negative progenitors. Given the role of LIN28B in fetal B-lymphopoiesis, it may also be important for ALL initiation or maintenance.
IGF2BP1 and IGF2BP3
IGF2BP1 and IGF2BP3 are important for fetal organogenesis and are expressed in FL HSCs, but not in adult HSCs (Wang D. et al., 2022). Induction of IGF2BP3 in adult HSCs induces a fetal-type output (Palanichamy et al., 2016; Wang et al., 2019).
IGF2BP1 and IGF2BP3 have been linked to leukemia, as well as solid malignancies, and are often co-expressed with LIN28B (Elcheva et al., 2020; Tran et al., 2021). The mechanism of action for IGF2BP3 in oncogenesis is segregation of mRNA transcripts from the cytoplasmic RNA-induced silencing complex, including the let-7 miRNA family.
IGF2BP1 is linked to pediatric AML and ETV6/RUNX1 B-ALL, while IGF2BP3 is linked to AML, KMT2A::AFF1 ALL, and BCR::ABL1 ALL (Stoskus et al., 2011; Palanichamy et al., 2016; Elcheva et al., 2020; Zhang et al., 2022). IGF2BP1 supports an LSC phenotype in AML (Elcheva et al., 2020). In AML cell lines, knockdown of IGF2BP3 leads to reduced cell proliferation in an N6-methyladenosine (m6A)-dependent fashion (Zhang et al., 2022). Depletion of the murine paralog IGF2BP3 increases the latency of leukemia in murine models of KMT2A::AFF1 AML (Tran et al., 2021).
Fetal genes important for epigenetic regulation
HMGA1 and 2
HMGA1 and HMGA2 are fetal oncogenes affecting epigenetic regulation. The HMGA family encodes proteins with AT hooks which interact with DNA to alter the chromatin architecture. These genes have much lower expression in adult tissues than in the fetal counterparts (Kumar et al., 2019; Roy et al., 2021), and HMGA1 can promote a pluripotent state (Shah et al., 2012). HMGA2 is expressed mainly in fetal HSC/MPP and influences both differentiation and proliferation of stem cells (Battista et al., 2003; Li et al., 2007; Copley et al., 2013), as well as promoting long-term in vivo reconstitution by cord blood CD34+ cells (Kumar et al., 2019).
Reactivation of HMGA1 and HMGA2 has been demonstrated in a wide range of malignancies (Huso and Resar, 2014; Mansoori et al., 2021) including leukemia (Efanov et al., 2014). HMGA1 expression has been linked to risk of relapse in pediatric B-ALL (Roy et al., 2013). In pediatric and adult AML, high expression of HMGA2 is linked to poor prognosis, and knockdown of the gene has induced differentiation in primary blasts (Marquis et al., 2018; Tan et al., 2018). HMGA2 induces T-ALL in a Eμ-HMGA2 transgenic mouse (Efanov et al., 2014).
microRNAs in leukemia
Aberrant expression of microRNAs (miRNAs) specific to fetal life and stem cell compartment (O'Connell et al., 2010) is implicated in pediatric leukemia (Grobbelaar and Ford, 2019; Gaur et al., 2020) (Table1). Pediatric AML can be risk-stratified by a 24-miRNA signature (Esperanza-Cebollada et al., 2023). Eight of these have target genes within the pLSC6 signature and includes let-7 miRNAs (known repressors of oncogenes), with lower let-7g/let-7i expression in high-risk AML. One of the pLSC6 genes (FAM30A) is an lncRNA. Signatures based on lncRNA differentiate pediatric leukemia subtypes, but do not inform prognosis (Buono et al., 2022).
Targeting aberrant stem cell programs in leukemia for therapy
The inappropriate expression of stem cell genes, while conferring survival advantage to leukemic cells, can also render them dependent on specific proteins or pathways, and thus vulnerable to targeted disruption. Fetal stem cell genes are the most attractive targets as they are not expressed in healthy postnatal tissues, ameliorating concerns about off-target effects. Genes expressed in leukemic and healthy postnatal stem cells present more of a challenge. However, excessive leukemic reliance on the aberrant pathway, the so-called “oncogenic addiction,” can generate a therapeutic window, whereby leukemic cells can be killed while sparing normal stem cells. Potential targeting strategies are summarized in Table 1. Specific approaches relating to stem cell genes discussed in this review are explored below.
Small-molecule inhibitors
Many stem cell genes code for TFs or other DNA-binding proteins, considered “undruggable,” owing to their intrinsically disordered nature. Recent improvements in screening methods have identified HMGA2-binding compounds, including the antimicrobials sumarin and ciclopirox (Huang et al., 2019; Su et al., 2020) and MEIS1/2 inhibitors (Turan et al., 2020).
An alternative approach employs small molecules that bind the minor groove of the TF cognate sequence. DNA binders of this type can inhibit HOXA9 (Depauw et al., 2019), with in vitro activity against HOXA9-dependent cells lines (Sonoda et al., 2021). Similarly, netropsin and trabectedin demonstrate antitumor activity in HMGA2+ neoplasia. Treatment with both drugs shows a synergistic anti-proliferative effect in infant ALL cell lines (Wu et al., 2015). Other approaches to target HMGA2 include targeting downstream pathways such as G2M transition (Moison et al., 2022) and PI3K/Akt/mTOR (Tan et al., 2016).
TF function can also be impaired by preventing their expression. HOXA9 transcription is dependent on DOT1L-mediated H3K79 methylation in KMT2A-r leukemia. DOT1L has been successfully targeted with small molecules, pinometostat/EPZ5676 (Basavapathruni et al., 2014; Waters et al., 2016). Early-phase clinical trials suggest modest activity against KMT2A-r leukemia (Stein et al., 2018). Newer DOT1L inhibitors with oral availability and improved pharmacokinetics have been developed (Stauffer et al., 2019; Perner et al., 2020).
HOXA9 is also dependent on the scaffold protein menin for expression in KMT2A-r leukemias (Yokoyama et al., 2005) and NPM1c-mutated AML (Kühn et al., 2016). The small-molecule revumenib prevents the interaction of menin with its target proteins. Revumenib induces remission in 30% of relapsed/refractory leukemia patients (Issa et al., 2023), although mutations in MEN1 can lead to drug resistance (Perner et al., 2023).
Direct inhibition of CDX2 has not yet been possible; however, the observation that PPARγ signaling restores KLF4 expression offers a potential therapeutic route to partially opposing CDX2 activity. PPARγ agonists are toxic to CDX2 overexpressing leukemia cell lines in vitro (Faber et al., 2013; Esmaeili et al., 2021).
Targeting RNA-binding proteins
Like DNA-binding proteins, small-molecule inhibition of RBPs is difficult, although recent high-throughput approaches have generated candidates (Wu, 2020). The most promising LIN28(A/B) inhibitor is C1632, which targets LIN28B + cell lines both by disruption of LIN28B–let-7 interaction (Franses et al., 2020; Zhang Q. et al., 2023; Shahab et al., 2023) and in Ewing sarcoma by disruption of the interaction between EWS-FLI1 mRNA and LIN28B (Keskin et al., 2020). Other molecules such as LI71 bind the cold shock domain and have efficacy against LIN28B + cancer cell lines (Wang L. et al., 2018).
Small-molecule inhibitors of IGF2BP1 and IGF2BP3 are at the preclinical stage. BTYNB destabilizes oncogenic transcripts by disrupting the IGF2BP1–mRNA association (Müller et al., 2020; Jamal et al., 2023; Sharma et al., 2023). Combining menin inhibitors with depletion of IGF2BP3 impairs cell growth and increases differentiation of KMT2A::AFF1 leukemia (Lin et al., 2023).
Another potential strategy to boost let-7 miRNA expression, thus inhibiting several oncogenes (Cinkornpumin et al., 2017), has yet to be applied in leukemia.
Immune effector cell therapy
Stem cell markers as targets for immunotherapy: The cell surface marker PROM1 is a highly attractive target for immunotherapy. A CD19/CD133 tandem CAR-T (Li et al., 2018) and CD19/133 bispecific CAR-iNKT (Ren et al., 2023) show efficacy in vivo against KMT2A-r cell lines. However, valid concerns about stem cell toxicity when targeting CD133 in patients have been raised (Bueno et al., 2019). Preclinical testing and early-phase trials using CD133-CAR-T in solid malignancies (Wang et al., 2018b; Dai et al., 2020; Vora et al., 2020) revealed no BM aplasia and only transient, reversible hematological toxicities. Longer-term follow-up and assessment will be required to confirm the safety of CD133 targeting. CAR-T directed against NG2 has shown promise in mobilizing leukemic blasts and rendering them more sensitive to chemotherapy in mouse models (Lopez-Millan et al., 2019). Anti-CD117 CAR-T therapy shows preclinical efficacy (Myburgh et al., 2020) but also eliminates healthy HSCs, necessitating novel approaches such as terminating “safety switches” (Magnani et al., 2023). Both CAR-T (Wang et al., 2018c) and CAR-NK (Mansour et al., 2023) cells have been used to target FLT3. Anti-CD123 CAR-T therapy has been used in early-stage clinical trials, appearing safe and potentially effective (Yao et al., 2019; Wermke et al., 2021).
Immunotherapy is particularly attractive in DS-associated leukemias where conventional treatments cause significant toxicities. In DS-ALL patients, CD19-, CD22-, and TSLPR-directed immunotherapies could yield promising results (Bagashev et al., 2022; Laetsch et al., 2023).
Discussion
Stem cells possess unique properties allowing the expansion, self-renewal, and differentiation required for tissue homeostasis. These programs are frequently co-opted by leukemias, where they provide growth and survival advantages. Although there is renewed interest in reprogramming adult HSCs to become more “fetal-like”, the potential of fetal stem cell genes to also promote oncogenesis must be considered.
Understanding stem cell programs in leukemia, including oncofetal genes, is vital to disentangling the biology of leukemias, including treatment resistance/relapse, and identifying mechanisms vulnerable to novel targeted therapies.
Author contributions
RL: conceptualization, writing–original draft, and writing–review and editing. JC: conceptualization, writing–original draft, and writing–review and editing. AR: conceptualization, supervision, and writing–review and editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. REL was supported by a CRUK Oxford DPhil in Cancer Sciences studentship (DCS-CRUK-CRTF20-RL). JWC was supported by a CCLG LPT Research Grant (CCLGA/2020/23/AR). AR was supported by the Wellcome Trust Clinical Research Career Development Fellowship (216632/Z/19/Z) and Medical Research Council Funding (MC_UU_00029/7).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abedin, S. M., Badar, T., Gauger, K., Michaelis, L. C., Runaas, L., Carlson, K.-S., et al. (2022). Safety and efficacy of pracinostat in combination with gemtuzumab ozogamicin (PraGO) in patients with relapsed/refractory acute myeloid leukemia. Leukemia Res. 123, 106984. doi:10.1016/j.leukres.2022.106984
Agrawal-Singh, S., Bagri, J., Giotopoulos, G., Azazi, D. M. A., Horton, S. J., Lopez, C. K., et al. (2023). HOXA9 forms a repressive complex with nuclear matrix-associated protein SAFB to maintain acute myeloid leukemia. Blood 141 (14), 1737–1754. doi:10.1182/blood.2022016528
Agraz-Doblas, A., Bueno, C., Bashford-Rogers, R., Roy, A., Schneider, P., Bardini, M., et al. (2019). Unraveling the cellular origin and clinical prognostic markers of infant B-cell acute lymphoblastic leukemia using genome-wide analysis. Haematologica 104 (6), 1176–1188. doi:10.3324/haematol.2018.206375
Aguila, J. R., Liao, W., Yang, J., Avila, C., Hagag, N., Senzel, L., et al. (2011). SALL4 is a robust stimulator for the expansion of hematopoietic stem cells. Blood 118 (3), 576–585. doi:10.1182/blood-2011-01-333641
Alajez, N. M., Shi, W., Wong, D., Lenarduzzi, M., Waldron, J., Weinreb, I., et al. (2012). Lin28b promotes head and neck cancer progression via modulation of the insulin-like growth factor survival pathway. Oncotarget 3 (12), 1641–1652. doi:10.18632/oncotarget.785
Alarcón, P., Rodríguez-Seguel, E., Fernández-González, A., Rubio, R., and Gómez-Skarmeta, J. L. (2008). A dual requirement for Iroquois genes during Xenopus kidney development. Dev. Camb. Engl. 135 (19), 3197–3207. doi:10.1242/dev.023697
Alejo-Valle, O., Weigert, K., Bhayadia, R., Ng, M., Issa, H., Beyer, C., et al. (2022). The megakaryocytic transcription factor ARID3A suppresses leukemia pathogenesis. Blood 139 (5), 651–665. doi:10.1182/blood.2021012231
Amanzadeh, A., Heidarnejad, F., Abdollahpour-Alitappeh, M., Molla-Kazemiha, V., Yari, S., Hadizadeh-Tasbiti, A., et al. (2018). Development of high-affinity monoclonal antibody using CD44 overexpressed cells as a candidate for targeted immunotherapy and diagnosis of acute myeloid leukemia. Hum. Antibodies 26 (1), 7–15. doi:10.3233/HAB-170315
Andreeff, M., Ruvolo, V., Gadgil, S., Zeng, C., Coombes, K., Chen, W., et al. (2008). HOX expression patterns identify a common signature for favorable AML. Leukemia 22 (11), 2041–2047. doi:10.1038/leu.2008.198
Bagashev, A., Loftus, J., Niswander, L., Ross, S., Falkenstein, C. D., Junco, J., et al. (2022). P1421: bimodal targeting of cytokine receptor-like factor 2 (crlf2) with jak inhibition and chimeric antigen receptor t cell immunotherapy in down syndrome acute lymphoblastic leukemia. HemaSphere 6, 1305–1306. doi:10.1097/01.Hs9.0000848544.25019.34
Bai, J., Yokomizo-Nakano, T., Kubota, S., Sun, Y., Kanai, A., Iimori, M., et al. (2021). Overexpression of Hmga2 activates Igf2bp2 and remodels transcriptional program of Tet2-deficient stem cells in myeloid transformation. Oncogene 40 (8), 1531–1541. doi:10.1038/s41388-020-01629-w
Ballerini, P., Boelle, P.-Y., Deswarte, C., Auvrignon, A., Couchy, G., Parker, J. L., et al. (2008). Prognostic significance of SALL4 expression levels in paediatric acute myeloid leukaemia (AML). Blood 112 (11), 2243. doi:10.1182/blood.V112.11.2243.2243
Balzeau, J., Menezes, M. R., Cao, S., and Hagan, J. P. (2017). The LIN28/let-7 pathway in cancer. Front. Genet. 8, 31. doi:10.3389/fgene.2017.00031
Bao, S., Wu, Q., McLendon, R. E., Hao, Y., Shi, Q., Hjelmeland, A. B., et al. (2006). Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature 444 (7120), 756–760. doi:10.1038/nature05236
Basak, A., Munschauer, M., Lareau, C. A., Montbleau, K. E., Ulirsch, J. C., Hartigan, C. R., et al. (2020). Control of human hemoglobin switching by LIN28B-mediated regulation of BCL11A translation. Nat. Genet. 52 (2), 138–145. doi:10.1038/s41588-019-0568-7
Basavapathruni, A., Olhava, E. J., Daigle, S. R., Therkelsen, C. A., Jin, L., Boriack-Sjodin, P. A., et al. (2014). Nonclinical pharmacokinetics and metabolism of EPZ-5676, a novel DOT1L histone methyltransferase inhibitor. Biopharm. Drug Dispos. 35 (4), 237–252. doi:10.1002/bdd.1889
Battista, S., Pentimalli, F., Baldassarre, G., Fedele, M., Fidanza, V., Croce, C. M., et al. (2003). Loss of Hmga1 gene function affects embryonic stem cell lympho-hematopoietic differentiation. FASEB J. 17 (11), 1496–1498. doi:10.1096/fj.02-0977fje
Bayraktar, R., and Van Roosbroeck, K. (2018). miR-155 in cancer drug resistance and as target for miRNA-based therapeutics. Cancer Metastasis Rev. 37, 33–44. doi:10.1007/s10555-017-9724-7
Benz, C., Copley, M. R., Kent, D. G., Wohrer, S., Cortes, A., Aghaeepour, N., et al. (2012). Hematopoietic stem cell subtypes expand differentially during development and display distinct lymphopoietic programs. Cell Stem Cell 10 (3), 273–283. doi:10.1016/j.stem.2012.02.007
Bernt, K. M., Zhu, N., Sinha, A. U., Vempati, S., Faber, J., Krivtsov, A. V., et al. (2011). MLL-rearranged leukemia is dependent on aberrant H3K79 methylation by DOT1L. Cancer Cell 20 (1), 66–78. doi:10.1016/j.ccr.2011.06.010
Bhansali, R. S., Rammohan, M., Lee, P., Laurent, A. P., Wen, Q., Suraneni, P., et al. (2021). DYRK1A regulates B cell acute lymphoblastic leukemia through phosphorylation of FOXO1 and STAT3. J. Clin. Invest. 131 (1), e135937. doi:10.1172/JCI135937
Bolouri, H., Ries, R., Pardo, L., Hylkema, T., Zhou, W., Smith, J. L., et al. (2021). A B-cell developmental gene regulatory network is activated in infant AML. PloS One 16 (11), e0259197. doi:10.1371/journal.pone.0259197
Bond, J., Labis, E., Marceau-Renaut, A., Duployez, N., Labopin, M., Hypolite, G., et al. (2018). Polycomb repressive complex 2 haploinsufficiency identifies a high-risk subgroup of pediatric acute myeloid leukemia. Leukemia 32 (8), 1878–1882. doi:10.1038/s41375-018-0187-9
Bowie, M. B., McKnight, K. D., Kent, D. G., McCaffrey, L., Hoodless, P. A., and Eaves, C. J. (2006). Hematopoietic stem cells proliferate until after birth and show a reversible phase-specific engraftment defect. J. Clin. Invest. 116 (10), 2808–2816. doi:10.1172/jci28310
Brooke, N. M., Garcia-Fernàndez, J., and Holland, P. W. (1998). The ParaHox gene cluster is an evolutionary sister of the Hox gene cluster. Nature 392 (6679), 920–922. doi:10.1038/31933
Brunetti, L., Gundry, M. C., Sorcini, D., Guzman, A. G., Huang, Y.-H., Ramabadran, R., et al. (2018). Mutant NPM1 maintains the leukemic state through HOX expression. Cancer Cell 34 (3), 499–512. doi:10.1016/j.ccell.2018.08.005
Bueno, C., Velasco-Hernandez, T., Gutiérrez-Agüera, F., Zanetti, S. R., Baroni, M. L., Sánchez-Martínez, D., et al. (2019). CD133-directed CAR T-cells for MLL leukemia: on-target, off-tumor myeloablative toxicity. Leukemia 33 (8), 2090–2125. doi:10.1038/s41375-019-0418-8
Buono, L., Iside, C., De Matteo, A., Stellato, P., Beneduce, G., de Vera d’Aragona, R. P., et al. (2022). Specific lncRNA signatures discriminate childhood acute leukaemias: a pilot study. Cancer Cell Int. 22 (1), 373. doi:10.1186/s12935-022-02789-3
Cabal-Hierro, L., van Galen, P., Prado, M. A., Higby, K. J., Togami, K., Mowery, C. T., et al. (2020). Chromatin accessibility promotes hematopoietic and leukemia stem cell activity. Nat. Commun. 11 (1), 1406. doi:10.1038/s41467-020-15221-z
Carey-Smith, S. L., Simad, M. H., Panchal, K., Aya-Bonilla, C., Smolders, H., Lin, S., et al. (2024). Efficacy of DYRK1A inhibitors in novel models of Down syndrome acute lymphoblastic leukemia. Haematologica. doi:10.3324/haematol.2023.284271
Challen, G. A., Sun, D., Jeong, M., Luo, M., Jelinek, J., Berg, J. S., et al. (2011). Dnmt3a is essential for hematopoietic stem cell differentiation. Nat. Genet. 44 (1), 23–31. doi:10.1038/ng.1009
Charbord, P., Tavian, M., Humeau, L., and Péault, B. (1996). Early ontogeny of the human marrow from long bones: an immunohistochemical study of hematopoiesis and its microenvironment [see comments]. Blood 87 (10), 4109–4119. doi:10.1182/blood.v87.10.4109.bloodjournal87104109
Chase, A., Reiter, A., Burci, L., Cazzaniga, G., Biondi, A., Pickard, J., et al. (1999). Fusion of ETV6 to the caudal-related homeobox gene CDX2 in acute myeloid leukemia with the t(12;13)(p13;q12). Blood 93 (3), 1025–1031. doi:10.1182/blood.V93.3.1025
Chen, C., Yu, W., Alikarami, F., Qiu, Q., Chen, C.-h., Flournoy, J., et al. (2022). Single-cell multiomics reveals increased plasticity, resistant populations, and stem-cell–like blasts in KMT2A-rearranged leukemia. Blood, J. Am. Soc. Hematol. 139 (14), 2198–2211. doi:10.1182/blood.2021013442
Chou, S. T., Opalinska, J. B., Yao, Y., Fernandes, M. A., Kalota, A., Brooks, J. S., et al. (2008). Trisomy 21 enhances human fetal erythro-megakaryocytic development. Blood 112 (12), 4503–4506. doi:10.1182/blood-2008-05-157859
Cinkornpumin, J., Roos, M., Nguyen, L., Liu, X., Gaeta, X., Lin, S., et al. (2017). A small molecule screen to identify regulators of let-7 targets. Sci. Rep. 7 (1), 15973. doi:10.1038/s41598-017-16258-9
Collins, C., Wang, J., Miao, H., Bronstein, J., Nawer, H., Xu, T., et al. (2014). C/EBPα is an essential collaborator in Hoxa9/Meis1-mediated leukemogenesis. Proc. Natl. Acad. Sci. U. S. A. 111 (27), 9899–9904. doi:10.1073/pnas.1402238111
Cooper, L. J., Shannon, K. M., Loken, M. R., Weaver, M., Stephens, K., and Sievers, E. L. (2000). Evidence that juvenile myelomonocytic leukemia can arise from a pluripotential stem cell. Blood, J. Am. Soc. Hematol. 96 (6), 2310–2313. doi:10.1182/blood.v96.6.2310
Copley, M. R., Babovic, S., Benz, C., Knapp, D. J., Beer, P. A., Kent, D. G., et al. (2013). The Lin28b-let-7-Hmga2 axis determines the higher self-renewal potential of fetal haematopoietic stem cells. Nat. Cell Biol. 15 (8), 916–925. doi:10.1038/ncb2783
Copley, M. R., and Eaves, C. J. (2013). Developmental changes in hematopoietic stem cell properties. Exp. Mol. Med. 45 (11), e55. doi:10.1038/emm.2013.98
Cox, C. V., Diamanti, P., Evely, R. S., Kearns, P. R., and Blair, A. (2009). Expression of CD133 on leukemia-initiating cells in childhood ALL. Blood 113 (14), 3287–3296. doi:10.1182/blood-2008-04-154187
Dai, H., Tong, C., Shi, D., Chen, M., Guo, Y., Chen, D., et al. (2020). Efficacy and biomarker analysis of CD133-directed CAR T cells in advanced hepatocellular carcinoma: a single-arm, open-label, phase II trial. Oncoimmunology 9 (1), 1846926. doi:10.1080/2162402X.2020.1846926
D'Angelo, V., Iannotta, A., Ramaglia, M., Lombardi, A., Zarone, M. R., Desiderio, V., et al. (2015). EZH2 is increased in paediatric T-cell acute lymphoblastic leukemia and is a suitable molecular target in combination treatment approaches. J. Exp. Clin. cancer Res. CR 34 (1), 83. doi:10.1186/s13046-015-0191-0
Darwish, N. H. E., Glinsky, G. V., Sudha, T., and Mousa, S. A. (2021). Targeting thyrointegrin αvβ3 using fluorobenzyl polyethylene glycol conjugated tetraiodothyroacetic acid (NP751) in acute myeloid leukemia. Front. Oncol. 11, 793810. doi:10.3389/fonc.2021.793810
DeAngelo, D. J., Jonas, B. A., Liesveld, J. L., Bixby, D. L., Advani, A. S., Marlton, P., et al. (2022). Phase 1/2 study of uproleselan added to chemotherapy in patients with relapsed or refractory acute myeloid leukemia. Blood 139 (8), 1135–1146. doi:10.1182/blood.2021010721
Degrauwe, N., Suvà, M. L., Janiszewska, M., Riggi, N., and Stamenkovic, I. (2016). IMPs: an RNA-binding protein family that provides a link between stem cell maintenance in normal development and cancer. Genes Dev. 30 (22), 2459–2474. doi:10.1101/gad.287540.116
De Luca, L., Trino, S., Laurenzana, I., Tagliaferri, D., Falco, G., Grieco, V., et al. (2017). Knockdown of miR-128a induces Lin28a expression and reverts myeloid differentiation blockage in acute myeloid leukemia. Cell death Dis. 8 (6), e2849. doi:10.1038/cddis.2017.253
Den Boer, M. L., van Slegtenhorst, M., De Menezes, R. X., Cheok, M. H., Buijs-Gladdines, J. G. C. A. M., Peters, S. T. C. J. M., et al. (2009). A subtype of childhood acute lymphoblastic leukaemia with poor treatment outcome: a genome-wide classification study. Lancet. Oncol. 10 (2), 125–134. doi:10.1016/S1470-2045(08)70339-5
Depauw, S., Lambert, M., Jambon, S., Paul, A., Peixoto, P., Nhili, R., et al. (2019). Heterocyclic diamidine DNA ligands as HOXA9 transcription factor inhibitors: design, molecular evaluation, and cellular consequences in a HOXA9-dependant leukemia cell model. J. Med. Chem. 62 (3), 1306–1329. doi:10.1021/acs.jmedchem.8b01448
Dick, J. E. (2008). Stem cell concepts renew cancer research. Blood 112 (13), 4793–4807. doi:10.1182/blood-2008-08-077941
Drabkin, H. A., Parsy, C., Ferguson, K., Guilhot, F., Lacotte, L., Roy, L., et al. (2002). Quantitative HOX expression in chromosomally defined subsets of acute myelogenous leukemia. Leukemia 16 (2), 186–195. doi:10.1038/sj.leu.2402354
Duployez, N., Marceau-Renaut, A., Villenet, C., Petit, A., Rousseau, A., Ng, S. W. K., et al. (2019). The stem cell-associated gene expression signature allows risk stratification in pediatric acute myeloid leukemia. Leukemia 33 (2), 348–357. doi:10.1038/s41375-018-0227-5
Efanov, A., Zanesi, N., Coppola, V., Nuovo, G., Bolon, B., Wernicle-Jameson, D., et al. (2014). Human HMGA2 protein overexpressed in mice induces precursor T-cell lymphoblastic leukemia. Blood Cancer J. 4 (7), e227. doi:10.1038/bcj.2014.46
Egyed, B., Kutszegi, N., Sági, J. C., Gézsi, A., Rzepiel, A., Visnovitz, T., et al. (2020). MicroRNA-181a as novel liquid biopsy marker of central nervous system involvement in pediatric acute lymphoblastic leukemia. J. Transl. Med. 18 (1), 250–312. doi:10.1186/s12967-020-02415-8
Elagib, K. E., Lu, C.-H., Mosoyan, G., Khalil, S., Zasadzińska, E., Foltz, D. R., et al. (2017). Neonatal expression of RNA-binding protein IGF2BP3 regulates the human fetal-adult megakaryocyte transition. J. Clin. investigation 127 (6), 2365–2377. doi:10.1172/JCI88936
Elcheva, I. A., Wood, T., Chiarolanzio, K., Chim, B., Wong, M., Singh, V., et al. (2020). RNA-binding protein IGF2BP1 maintains leukemia stem cell properties by regulating HOXB4, MYB, and ALDH1A1. Leukemia 34 (5), 1354–1363. doi:10.1038/s41375-019-0656-9
Eldeeb, M., Yuan, O., Guzzi, N., Thi Ngoc, P. C., Konturek-Ciesla, A., Kristiansen, T. A., et al. (2023). A fetal tumor suppressor axis abrogates MLL-fusion-driven acute myeloid leukemia. Cell Rep. 42 (2), 112099. doi:10.1016/j.celrep.2023.112099
Elsayed, A. H., Rafiee, R., Cao, X., Raimondi, S., Downing, J. R., Ribeiro, R., et al. (2020). A six-gene leukemic stem cell score identifies high risk pediatric acute myeloid leukemia. Leukemia 34 (3), 735–745. doi:10.1038/s41375-019-0604-8
Esmaeili, S., Salari, S., Kaveh, V., Ghaffari, S. H., and Bashash, D. (2021). Alteration of PPAR-GAMMA (PPARG; PPARγ) and PTEN gene expression in acute myeloid leukemia patients and the promising anticancer effects of PPARγ stimulation using pioglitazone on AML cells. Mol. Genet. Genomic Med. 9 (11), e1818. doi:10.1002/mgg3.1818
Esperanza-Cebollada, E., Gómez-González, S., Perez-Jaume, S., Vega-García, N., Vicente-Garcés, C., Richarte-Franqués, M., et al. (2023). A miRNA signature related to stemness identifies high-risk patients in paediatric acute myeloid leukaemia. Br. J. Haematol. 202 (1), 96–110. doi:10.1111/bjh.18746
Ezhkova, E., Pasolli, H. A., Parker, J. S., Stokes, N., Su, I. h., Hannon, G., et al. (2009). Ezh2 orchestrates gene expression for the stepwise differentiation of tissue-specific stem cells. Cell 136 (6), 1122–1135. doi:10.1016/j.cell.2008.12.043
Faber, J., Krivtsov, A. V., Stubbs, M. C., Wright, R., Davis, T. N., van den Heuvel-Eibrink, M., et al. (2009). HOXA9 is required for survival in human MLL-rearranged acute leukemias. Blood 113 (11), 2375–2385. doi:10.1182/blood-2007-09-113597
Faber, K., Bullinger, L., Ragu, C., Garding, A., Mertens, D., Miller, C., et al. (2013). CDX2-driven leukemogenesis involves KLF4 repression and deregulated PPARγ signaling. J. Clin. Investigation 123 (1), 299–314. doi:10.1172/JCI64745
Ferrandina, G., Petrillo, M., Bonanno, G., and Scambia, G. (2009). Targeting CD133 antigen in cancer. Expert Opin. Ther. Targets 13 (7), 823–837. doi:10.1517/14728220903005616
Ferrando, A. A., Neuberg, D. S., Staunton, J., Loh, M. L., Huard, C., Raimondi, S. C., et al. (2002). Gene expression signatures define novel oncogenic pathways in T cell acute lymphoblastic leukemia. Cancer Cell 1 (1), 75–87. doi:10.1016/s1535-6108(02)00018-1
Franses, J. W., Philipp, J., Missios, P., Bhan, I., Liu, A., Yashaswini, C., et al. (2020). Pancreatic circulating tumor cell profiling identifies LIN28B as a metastasis driver and drug target. Nat. Commun. 11 (1), 3303. doi:10.1038/s41467-020-17150-3
Freese, N. H., Lam, B. A., Staton, M., Scott, A., and Chapman, S. C. (2014). A novel gain-of-function mutation of the proneural IRX1 and IRX2 genes disrupts Axis elongation in the araucana rumpless chicken. PLOS ONE 9 (11), e112364. doi:10.1371/journal.pone.0112364
Gao, C., Kong, N. R., Li, A., Tatetu, H., Ueno, S., Yang, Y., et al. (2013). SALL4 is a key transcription regulator in normal human hematopoiesis. Transfusion 53 (5), 1037–1049. doi:10.1111/j.1537-2995.2012.03888.x
Gaur, V., Chaudhary, S., Tyagi, A., Agarwal, S., Sharawat, S. K., Sarkar, S., et al. (2020). Dysregulation of miRNA expression and their prognostic significance in paediatric cytogenetically normal acute myeloid leukaemia. Br. J. Haematol. 188 (6), e90–e94. doi:10.1111/bjh.16375
Georgantas, R. W., Hildreth, R., Morisot, S., Alder, J., Liu, C.-g., Heimfeld, S., et al. (2007). CD34+ hematopoietic stem-progenitor cell microRNA expression and function: a circuit diagram of differentiation control. Proc. Natl. Acad. Sci. 104 (8), 2750–2755. doi:10.1073/pnas.0610983104
Gialesaki, S., Brauer-Hartmann, D., Issa, H., Bhayadia, R., Alejo-Valle, O., Verboon, L., et al. (2023). RUNX1 isoform disequilibrium promotes the development of trisomy 21-associated myeloid leukemia. Blood 141 (10), 1105–1118. doi:10.1182/blood.2022017619
Godfrey, L., Crump, N. T., O’Byrne, S., Lau, I. J., Rice, S., Harman, J. R., et al. (2021). H3K79me2/3 controls enhancer–promoter interactions and activation of the pan-cancer stem cell marker PROM1/CD133 in MLL-AF4 leukemia cells. Leukemia 35 (1), 90–106. doi:10.1038/s41375-020-0808-y
Greaves, M. (2005). In utero origins of childhood leukaemia. Early Hum. Dev. 81 (1), 123–129. doi:10.1016/j.earlhumdev.2004.10.004
Grobbelaar, C., and Ford, A. M. (2019). The role of MicroRNA in paediatric acute lymphoblastic leukaemia: challenges for diagnosis and therapy. J. Oncol. 2019, 8941471. doi:10.1155/2019/8941471
Hacein-Bey-Abina, S., Von Kalle, C., Schmidt, M., McCormack, M. P., Wulffraat, N., Leboulch, P., et al. (2003). LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Sci. (New York, N.Y.) 302 (5644), 415–419. doi:10.1126/science.1088547
Hanahan, D., and Weinberg, R. A. (2011). Hallmarks of cancer: the next generation. Cell 144 (5), 646–674.
Harrison, D. E., Zhong, R. K., Jordan, C. T., Lemischka, I. R., and Astle, C. M. (1997). Relative to adult marrow, fetal liver repopulates nearly five times more effectively long-term than short-term. Exp. Hematol. 25 (4), 293–297.
Hartmann, M., Schoenung, M., Rajak, J., Maurer, V., Hai, L., Bauer, K., et al. (2023). Oncogenic RAS-pathway activation drives oncofetal reprogramming and creates therapeutic vulnerabilities in juvenile myelomonocytic leukemia. bioRxiv. Avail;able at: doi:10.1101/2023.10.27.563754
Harvey, R. C., Mullighan, C. G., Wang, X., Dobbin, K. K., Davidson, G. S., Bedrick, E. J., et al. (2010). Identification of novel cluster groups in pediatric high-risk B-precursor acute lymphoblastic leukemia with gene expression profiling: correlation with genome-wide DNA copy number alterations, clinical characteristics, and outcome. Blood 116 (23), 4874–4884. doi:10.1182/blood-2009-08-239681
Hasle, H., Clemmensen, I. H., and Mikkelsen, M. (2000). Risks of leukaemia and solid tumours in individuals with Down's syndrome. Lancet 355 (9199), 165–169. doi:10.1016/S0140-6736(99)05264-2
Helsmoortel, H. H., Bresolin, S., Lammens, T., Cavé, H., Noellke, P., Caye, A., et al. (2016a). LIN28B overexpression defines a novel fetal-like subgroup of juvenile myelomonocytic leukemia. Blood 127 (9), 1163–1172. doi:10.1182/blood-2015-09-667808
Helsmoortel, H. H., De Moerloose, B., Pieters, T., Ghazavi, F., Bresolin, S., Cavé, H., et al. (2016b). LIN28B is over-expressed in specific subtypes of pediatric leukemia and regulates lncRNA H19. Haematologica 101 (6), e240–e244. doi:10.3324/haematol.2016.143818
Hodeib, H., El Amrousy, D., Youssef, A., Khedr, R., Al-Asy, H., Shabana, A., et al. (2023). Acute lymphoblastic leukemia in children and SALL4 and BMI-1 gene expression. Pediatr. Res. 94 (4), 1510–1515. doi:10.1038/s41390-021-01854-3
Holland, P. W. H. (2013). Evolution of homeobox genes. Wiley Interdiscip. Rev. Dev. Biol. 2 (1), 31–45. doi:10.1002/wdev.78
Holland, P. W. H., Booth, H. A. F., and Bruford, E. A. (2007). Classification and nomenclature of all human homeobox genes. BMC Biol. 5 (1), 47. doi:10.1186/1741-7007-5-47
Holyoake, T. L., Nicolini, F. E., and Eaves, C. J. (1999). Functional differences between transplantable human hematopoietic stem cells from fetal liver, cord blood, and adult marrow. Exp. Hematol. 27 (9), 1418–1427. doi:10.1016/s0301-472x(99)00078-8
Hong, D., Gupta, R., Ancliff, P., Atzberger, A., Brown, J., Soneji, S., et al. (2008). Initiating and cancer-propagating cells in TEL-AML1-associated childhood leukemia. Science 319 (5861), 336–339. doi:10.1126/science.1150648
Hope, K. J., Jin, L., and Dick, J. E. (2004). Acute myeloid leukemia originates from a hierarchy of leukemic stem cell classes that differ in self-renewal capacity. Nat. Immunol. 5 (7), 738–743. doi:10.1038/ni1080
Hsieh, Y.-T., Gang, E. J., Geng, H., Park, E., Huantes, S., Chudziak, D., et al. (2013). Integrin alpha4 blockade sensitizes drug resistant pre-B acute lymphoblastic leukemia to chemotherapy. Blood 121 (10), 1814–1818. doi:10.1182/blood-2012-01-406272
Huang, Y., Sitwala, K., Bronstein, J., Sanders, D., Dandekar, M., Collins, C., et al. (2012). Identification and characterization of Hoxa9 binding sites in hematopoietic cells. Blood 119 (2), 388–398. doi:10.1182/blood-2011-03-341081
Huang, Y.-M., Cheng, C.-H., Pan, S.-L., Yang, P.-M., Lin, D.-Y., and Lee, K.-H. (2019). Gene expression signature-based approach identifies antifungal drug ciclopirox as a novel inhibitor of HMGA2 in colorectal cancer. Biomolecules 9 (11), 688. doi:10.3390/biom9110688
Huso, T. H., and Resar, L. M. S. (2014). The high mobility group A1 molecular switch: turning on cancer - can we turn it off? Expert Opin. Ther. Targets 18 (5), 541–553. doi:10.1517/14728222.2014.900045
Isobe, T., Takagi, M., Sato-Otsubo, A., Nishimura, A., Nagae, G., Yamagishi, C., et al. (2022). Multi-omics analysis defines highly refractory RAS burdened immature subgroup of infant acute lymphoblastic leukemia. Nat. Commun. 13 (1), 4501. doi:10.1038/s41467-022-32266-4
Issa, G. C., Aldoss, I., DiPersio, J., Cuglievan, B., Stone, R., Arellano, M., et al. (2023). The menin inhibitor revumenib in KMT2A-rearranged or NPM1-mutant leukaemia. Nature 615 (7954), 920–924. doi:10.1038/s41586-023-05812-3
Italiano, A., Soria, J.-C., Toulmonde, M., Michot, J.-M., Lucchesi, C., Varga, A., et al. (2018). Tazemetostat, an EZH2 inhibitor, in relapsed or refractory B-cell non-Hodgkin lymphoma and advanced solid tumours: a first-in-human, open-label, phase 1 study. Lancet. Oncol. 19 (5), 649–659. doi:10.1016/S1470-2045(18)30145-1
Ivanovs, A., Rybtsov, S., Ng, E. S., Stanley, E. G., Elefanty, A. G., and Medvinsky, A. (2017). Human haematopoietic stem cell development: from the embryo to the dish. Development 144 (13), 2323–2337. doi:10.1242/dev.134866
Ivanovs, A., Rybtsov, S., Welch, L., Anderson, R. A., Turner, M. L., and Medvinsky, A. (2011). Highly potent human hematopoietic stem cells first emerge in the intraembryonic aorta-gonad-mesonephros region. J. Exp. Med. 208 (12), 2417–2427. doi:10.1084/jem.20111688
Jäger, S., Jahnke, A., Wilmes, T., Adebahr, S., Vögtle, F. N., deLima-Hahn, E., et al. (2007). Leukemia targeting ligands isolated from phage display peptide libraries. Leukemia 21 (3), 411–420. doi:10.1038/sj.leu.2404548
Jamal, A., Hassan Dalhat, M., Jahan, S., Choudhry, H., and Imran Khan, M. (2023). BTYNB, an inhibitor of RNA binding protein IGF2BP1 reduces proliferation and induces differentiation of leukemic cancer cells. Saudi J. Biol. Sci. 30 (3), 103569. doi:10.1016/j.sjbs.2023.103569
Jardine, L., Webb, S., Goh, I., Quiroga Londono, M., Reynolds, G., Mather, M., et al. (2021). Blood and immune development in human fetal bone marrow and Down syndrome. Nature 598 (7880), 327–331. doi:10.1038/s41586-021-03929-x
Jeong, H.-W., Cui, W., Yang, Y., Lu, J., He, J., Li, A., et al. (2011). SALL4, a stem cell factor, affects the side population by regulation of the ATP-binding cassette drug transport genes. PloS One 6 (4), e18372. doi:10.1371/journal.pone.0018372
Jin, L., Hope, K. J., Zhai, Q., Smadja-Joffe, F., and Dick, J. E. (2006). Targeting of CD44 eradicates human acute myeloid leukemic stem cells. Nat. Med. 12 (10), 1167–1174. doi:10.1038/nm1483
Kang, M., Kim, S., and Ko, J. (2019). Roles of CD133 in microvesicle formation and oncoprotein trafficking in colon cancer. FASEB J. 33 (3), 4248–4260. doi:10.1096/fj.201802018R
Kerry, J., Godfrey, L., Repapi, E., Tapia, M., Blackledge, N. P., Ma, H., et al. (2017). MLL-AF4 spreading identifies binding sites that are distinct from super-enhancers and that govern sensitivity to DOT1L inhibition in leukemia. Cell Rep. 18 (2), 482–495. doi:10.1016/j.celrep.2016.12.054
Keskin, T., Bakaric, A., Waszyk, P., Boulay, G., Torsello, M., Cornaz-Buros, S., et al. (2020). LIN28B underlies the pathogenesis of a subclass of ewing sarcoma LIN28B control of EWS-FLI1 stability. Cell Rep. 30 (13), 4567–4583. doi:10.1016/j.celrep.2019.12.053
Khalaj, M., Woolthuis, C. M., Hu, W., Durham, B. H., Chu, S. H., Qamar, S., et al. (2017). miR-99 regulates normal and malignant hematopoietic stem cell self-renewal. J. Exp. Med. 214 (8), 2453–2470. doi:10.1084/jem.20161595
Kim, I., Saunders, T. L., and Morrison, S. J. (2007). Sox17 dependence distinguishes the transcriptional regulation of fetal from adult hematopoietic stem cells. Cell 130 (3), 470–483. doi:10.1016/j.cell.2007.06.011
Klusmann, J. H., Li, Z., Bohmer, K., Maroz, A., Koch, M. L., Emmrich, S., et al. (2010). miR-125b-2 is a potential oncomiR on human chromosome 21 in megakaryoblastic leukemia. Genes Dev. 24 (5), 478–490. doi:10.1101/gad.1856210
Kormish, J. D., Sinner, D., and Zorn, A. M. (2010). Interactions between SOX factors and Wnt/beta-catenin signaling in development and disease. Dev. Dyn. Official Publ. Am. Assoc. Anatomists 239 (1), 56–68. doi:10.1002/dvdy.22046
Kuhn, A., Loscher, D., and Marschalek, R. (2016). The IRX1/HOXA connection: insights into a novel t(4;11)- specific cancer mechanism. Oncotarget 7 (23), 35341–35352. doi:10.18632/oncotarget.9241
Kühn, M. W. M., Song, E., Feng, Z., Sinha, A., Chen, C.-W., Deshpande, A. J., et al. (2016). Targeting chromatin regulators inhibits leukemogenic gene expression in NPM1 mutant leukemia. Cancer Discov. 6 (10), 1166–1181. doi:10.1158/2159-8290.CD-16-0237
Kumar, P., Beck, D., Galeev, R., Thoms, J. A. I., Talkhoncheh, M. S., de Jong, I., et al. (2019). HMGA2 promotes long-term engraftment and myeloerythroid differentiation of human hematopoietic stem and progenitor cells. Blood Adv. 3 (4), 681–691. doi:10.1182/bloodadvances.2018023986
Labuhn, M., Perkins, K., Matzk, S., Varghese, L., Garnett, C., Papaemmanuil, E., et al. (2019). Mechanisms of progression of myeloid preleukemia to transformed myeloid leukemia in children with down syndrome. Cancer Cell 36 (3), 340. doi:10.1016/j.ccell.2019.08.014
Laetsch, T. W., Maude, S. L., Rives, S., Hiramatsu, H., Bittencourt, H., Bader, P., et al. (2023). Three-year update of tisagenlecleucel in pediatric and young adult patients with relapsed/refractory acute lymphoblastic leukemia in the ELIANA trial. J. Clin. Oncol. 41 (9), 1664–1669. doi:10.1200/JCO.22.00642
Lansdorp, P. M., Dragowska, W., and Mayani, H. (1993). Ontogeny-related changes in proliferative potential of human hematopoietic cells. J. Exp. Med. 178 (3), 787–791. doi:10.1084/jem.178.3.787
Lapidot, T., Sirard, C., Vormoor, J., Murdoch, B., Hoang, T., Caceres-Cortes, J., et al. (1994). A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature 367 (6464), 645–648. doi:10.1038/367645a0
Lee, Y. T., de Vasconcellos, J. F., Yuan, J., Byrnes, C., Noh, S. J., Meier, E. R., et al. (2013). LIN28B-mediated expression of fetal hemoglobin and production of fetal-like erythrocytes from adult human erythroblasts ex vivo. Blood 122 (6), 1034–1041. doi:10.1182/blood-2012-12-472308
Li, D., Hu, Y., Jin, Z., Zhai, Y., Tan, Y., Sun, Y., et al. (2018). TanCAR T cells targeting CD19 and CD133 efficiently eliminate MLL leukemic cells. Leukemia 32 (9), 2012–2016. doi:10.1038/s41375-018-0212-z
Li, O., Li, J., and Dröge, P. (2007). DNA architectural factor and proto-oncogene HMGA2 regulates key developmental genes in pluripotent human embryonic stem cells. FEBS Lett. 581 (18), 3533–3537. doi:10.1016/j.febslet.2007.06.072
Li, Z., Chang, T. C., Junco, J. J., Devidas, M., Li, Y., Yang, W., et al. (2023). Genomic landscape of Down syndrome-associated acute lymphoblastic leukemia. Blood 142 (2), 172–184. doi:10.1182/blood.2023019765
Li, Z., Chen, P., Su, R., Hu, C., Li, Y., Elkahloun, A. G., et al. (2016). PBX3 and MEIS1 cooperate in hematopoietic cells to drive acute myeloid leukemias characterized by a core transcriptome of the MLL-rearranged disease. Cancer Res. 76 (3), 619–629. doi:10.1158/0008-5472.CAN-15-1566
Liang, C., Li, Y., Wang, L.-N., Zhang, X.-L., Luo, J.-S., Peng, C.-J., et al. (2021). Up-regulated miR-155 is associated with poor prognosis in childhood acute lymphoblastic leukemia and promotes cell proliferation targeting ZNF238. Hematology 26 (1), 16–25. doi:10.1080/16078454.2020.1860187
Liang, D.-C., Liu, H.-C., Yang, C.-P., Jaing, T.-H., Hung, I.-J., Yeh, T.-C., et al. (2013). Cooperating gene mutations in childhood acute myeloid leukemia with special reference on mutations of ASXL1, TET2, IDH1, IDH2, and DNMT3A. Blood 121 (15), 2988–2995. doi:10.1182/blood-2012-06-436782
Liao, J., Karnik, R., Gu, H., Ziller, M. J., Clement, K., Tsankov, A. M., et al. (2015). Targeted disruption of DNMT1, DNMT3A and DNMT3B in human embryonic stem cells. Nat. Genet. 47 (5), 469–478. doi:10.1038/ng.3258
Lin, T. L., Jaiswal, A. K., Ritter, A. J., Reppas, J., Tran, T. M., Neeb, Z. T., et al. (2023). Targeting IGF2BP3 enhances antileukemic effects of menin-MLL inhibition in MLL-AF4 leukemia. Blood Adv. 8, 261–275. doi:10.1182/bloodadvances.2023011132
Lopez-Millan, B., Sanchéz-Martínez, D., Roca-Ho, H., Gutiérrez-Agüera, F., Molina, O., Diaz de la Guardia, R., et al. (2019). NG2 antigen is a therapeutic target for MLL-rearranged B-cell acute lymphoblastic leukemia. Leukemia 33 (7), 1557–1569. doi:10.1038/s41375-018-0353-0
Louka, E., Povinelli, B., Rodriguez-Meira, A., Buck, G., Wen, W. X., Wang, G., et al. (2021). Heterogeneous disease-propagating stem cells in juvenile myelomonocytic leukemia. J. Exp. Med. 218 (2), e20180853. doi:10.1084/jem.20180853
Maeda, N., Ohashi, T., Chagan-Yasutan, H., Hattori, T., Takahashi, Y., Harigae, H., et al. (2015). Osteopontin-integrin interaction as a novel molecular target for antibody-mediated immunotherapy in adult T-cell leukemia. Retrovirology 12 (1), 99. doi:10.1186/s12977-015-0225-x
Magnani, C. F., Myburgh, R., Brunn, S., Chambovey, M., Ponzo, M., Volta, L., et al. (2023). Anti-CD117 CAR T cells incorporating a safety switch eradicate human acute myeloid leukemia and hematopoietic stem cells. Mol. Ther. Oncolytics 30, 56–71. doi:10.1016/j.omto.2023.07.003
Magnusson, M., Brun, A. C. M., Lawrence, H. J., and Karlsson, S. (2007). Hoxa9/hoxb3/hoxb4 compound null mice display severe hematopoietic defects. Exp. Hematol. 35 (9), 1421–1428. doi:10.1016/j.exphem.2007.05.011
Malinge, S., Bliss-Moreau, M., Kirsammer, G., Diebold, L., Chlon, T., Gurbuxani, S., et al. (2012). Increased dosage of the chromosome 21 ortholog Dyrk1a promotes megakaryoblastic leukemia in a murine model of Down syndrome. J. Clin. Invest. 122 (3), 948–962. doi:10.1172/JCI60455
Malouf, C., Antunes, E. T. B., O’Dwyer, M., Jakobczyk, H., Sahm, F., Landua, S.-L., et al. (2021). miR-130b and miR-128a are essential lineage-specific codrivers of t(4;11) MLL-AF4 acute leukemia. Blood 138 (21), 2066–2092. doi:10.1182/blood.2020006610
Malumbres, R., Fresquet, V., Roman-Gomez, J., Bobadilla, M., Robles, E. F., Altobelli, G. G., et al. (2011). LMO2 expression reflects the different stages of blast maturation and genetic features in B-cell acute lymphoblastic leukemia and predicts clinical outcome. Haematologica 96 (7), 980–986. doi:10.3324/haematol.2011.040568
Manesia, J. K., Xu, Z., Broekaert, D., Boon, R., van Vliet, A., Eelen, G., et al. (2015). Highly proliferative primitive fetal liver hematopoietic stem cells are fueled by oxidative metabolic pathways. Stem Cell Res. 15 (3), 715–721. doi:10.1016/j.scr.2015.11.001
Mansoori, B., Mohammadi, A., Ditzel, H. J., Duijf, P. H. G., Khaze, V., Gjerstorff, M. F., et al. (2021). HMGA2 as a critical regulator in cancer development. Genes (Basel) 12 (2), 269. doi:10.3390/genes12020269
Mansour, A. G., Teng, K.-Y., Li, Z., Zhu, Z., Chen, H., Tian, L., et al. (2023). Off-the-shelf CAR–engineered natural killer cells targeting FLT3 enhance killing of acute myeloid leukemia. Blood Adv. 7 (20), 6225–6239. doi:10.1182/bloodadvances.2022007405
Marquis, M., Beaubois, C., Lavallée, V. P., Abrahamowicz, M., Danieli, C., Lemieux, S., et al. (2018). High expression of HMGA2 independently predicts poor clinical outcomes in acute myeloid leukemia. Blood Cancer J. 8 (8), 68. doi:10.1038/s41408-018-0103-6
Matouk, I. J., Raveh, E., Abu-lail, R., Mezan, S., Gilon, M., Gershtain, E., et al. (2014). Oncofetal H19 RNA promotes tumor metastasis. Biochim. Biophys. Acta 1843 (7), 1414–1426. doi:10.1016/j.bbamcr.2014.03.023
McKinney-Freeman, S. L., Lengerke, C., Jang, I.-H., Schmitt, S., Wang, Y., Philitas, M., et al. (2008). Modulation of murine embryonic stem cell-derived CD41+c-kit+ hematopoietic progenitors by ectopic expression of Cdx genes. Blood 111 (10), 4944–4953. doi:10.1182/blood-2007-11-124644
Mi, S., Lu, J., Sun, M., Li, Z., Zhang, H., Neilly, M. B., et al. (2007). MicroRNA expression signatures accurately discriminate acute lymphoblastic leukemia from acute myeloid leukemia. Proc. Natl. Acad. Sci. 104 (50), 19971–19976. doi:10.1073/pnas.0709313104
Mikkola, H. K., Klintman, J., Yang, H., Hock, H., Schlaeger, T. M., Fujiwara, Y., et al. (2003). Haematopoietic stem cells retain long-term repopulating activity and multipotency in the absence of stem-cell leukaemia SCL/tal-1 gene. Nature 421 (6922), 547–551. doi:10.1038/nature01345
Milne, T. A., Briggs, S. D., Brock, H. W., Martin, M. E., Gibbs, D., Allis, C. D., et al. (2002). MLL targets SET domain methyltransferase activity to Hox gene promoters. Mol. Cell 10 (5), 1107–1117. doi:10.1016/s1097-2765(02)00741-4
Milne, T. A., Martin, M. E., Brock, H. W., Slany, R. K., and Hess, J. L. (2005). Leukemogenic MLL fusion proteins bind across a broad region of the Hox a9 locus, promoting transcription and multiple histone modifications. Cancer Res. 65 (24), 11367–11374. doi:10.1158/0008-5472.CAN-05-1041
Miraglia, S., Godfrey, W., Yin, A. H., Atkins, K., Warnke, R., Holden, J. T., et al. (1997). A novel five-transmembrane hematopoietic stem cell antigen: isolation, characterization, and molecular cloning. Blood 90 (12), 5013–5021. doi:10.1182/blood.v90.12.5013
Mochizuki-Kashio, M., Mishima, Y., Miyagi, S., Negishi, M., Saraya, A., Konuma, T., et al. (2011). Dependency on the polycomb gene Ezh2 distinguishes fetal from adult hematopoietic stem cells. Blood 118 (25), 6553–6561. doi:10.1182/blood-2011-03-340554
Moison, C., Spinella, J.-F., Chagraoui, J., Lavallée, V.-P., Lehnertz, B., Thiollier, C., et al. (2022). HMGA2 expression defines a subset of human AML with immature transcriptional signature and vulnerability to G2/M inhibition. Blood Adv. 6 (16), 4793–4806. doi:10.1182/bloodadvances.2021005828
Molenaar, J. J., Domingo-Fernández, R., Ebus, M. E., Lindner, S., Koster, J., Drabek, K., et al. (2012). LIN28B induces neuroblastoma and enhances MYCN levels via let-7 suppression. Nat. Genet. 44 (11), 1199–1206. doi:10.1038/ng.2436
Monk, M., and Holding, C. (2001). Human embryonic genes re-expressed in cancer cells. Oncogene 20 (56), 8085–8091. doi:10.1038/sj.onc.1205088
Montecino-Rodriguez, E., Fice, M., Casero, D., Berent-Maoz, B., Barber, C. L., and Dorshkind, K. (2016). Distinct genetic networks orchestrate the emergence of specific waves of fetal and adult B-1 and B-2 development. Immunity 45 (3), 527–539. doi:10.1016/j.immuni.2016.07.012
Moqadam, F. A., Lange-Turenhout, E., Aries, I., Pieters, R., and Den Boer, M. (2013). MiR-125b, miR-100 and miR-99a co-regulate vincristine resistance in childhood acute lymphoblastic leukemia. Leukemia Res. 37 (10), 1315–1321. doi:10.1016/j.leukres.2013.06.027
Muench, M. O., Cupp, J., Polakoff, J., and Roncarolo, M. G. (1994). Expression of CD33, CD38, and HLA-DR on CD34+ human fetal liver progenitors with a high proliferative potential. Blood 83 (11), 3170–3181. doi:10.1182/blood.v83.11.3170.3170
Müller, S., Bley, N., Busch, B., Glaß, M., Lederer, M., Misiak, C., et al. (2020). The oncofetal RNA-binding protein IGF2BP1 is a druggable, post-transcriptional super-enhancer of E2F-driven gene expression in cancer. Nucleic Acids Res. 48 (15), 8576–8590. doi:10.1093/nar/gkaa653
Myburgh, R., Kiefer, J. D., Russkamp, N. F., Magnani, C. F., Nuñez, N., Simonis, A., et al. (2020). Anti-human CD117 CAR T-cells efficiently eliminate healthy and malignant CD117-expressing hematopoietic cells. Leukemia 34 (10), 2688–2703. doi:10.1038/s41375-020-0818-9
Nagel, S., and Meyer, C. (2022). Normal and aberrant TALE-class homeobox gene activities in pro-B-cells and B-cell precursor acute lymphoblastic leukemia. Int. J. Mol. Sci. 23 (19), 11874. doi:10.3390/ijms231911874
Nagel, S., Pommerenke, C., Meyer, C., and MacLeod, R. A. F. (2022). The hematopoietic TALE-code shows normal activity of IRX1 in myeloid progenitors and reveals ectopic expression of IRX3 and IRX5 in acute myeloid leukemia. Int. J. Mol. Sci. 23 (6), 3192. doi:10.3390/ijms23063192
Ng, E. S., Azzola, L., Bruveris, F. F., Calvanese, V., Phipson, B., Vlahos, K., et al. (2016a). Differentiation of human embryonic stem cells to HOXA+ hemogenic vasculature that resembles the aorta-gonad-mesonephros. Nat. Biotechnol. 34 (11), 1168–1179. doi:10.1038/nbt.3702
Ng, S. W. K., Mitchell, A., Kennedy, J. A., Chen, W. C., McLeod, J., Ibrahimova, N., et al. (2016b). A 17-gene stemness score for rapid determination of risk in acute leukaemia. Nature 540 (7633), 433–437. doi:10.1038/nature20598
North, T. E., de Bruijn, M. F., Stacy, T., Talebian, L., Lind, E., Robin, C., et al. (2002). Runx1 expression marks long-term repopulating hematopoietic stem cells in the midgestation mouse embryo. Immunity 16 (5), 661–672. doi:10.1016/s1074-7613(02)00296-0
O’Byrne, S., Elliott, N., Rice, S., Buck, G., Fordham, N., Garnett, C., et al. (2019). Discovery of a CD10-negative B-progenitor in human fetal life identifies unique ontogeny-related developmental programs. Blood, J. Am. Soc. Hematol. 134 (13), 1059–1071. doi:10.1182/blood.2019001289
O'Connell, R. M., Chaudhuri, A. A., Rao, D. S., Gibson, W. S., Balazs, A. B., and Baltimore, D. (2010). MicroRNAs enriched in hematopoietic stem cells differentially regulate long-term hematopoietic output. Proc. Natl. Acad. Sci. 107 (32), 14235–14240. doi:10.1073/pnas.1009798107
Ooi, A. L., Sahoo, D., Adorno, M., Wang, Y., Weissman, I. L., and Park, C. Y. (2010). MicroRNA-125b expands hematopoietic stem cells and enriches for the lymphoid-balanced and lymphoid-biased subsets. Proc. Natl. Acad. Sci. 107 (50), 21505–21510. doi:10.1073/pnas.1016218107
Orlovsky, K., Kalinkovich, A., Rozovskaia, T., Shezen, E., Itkin, T., Alder, H., et al. (2011). Down-regulation of homeobox genes MEIS1 and HOXA in MLL-rearranged acute leukemia impairs engraftment and reduces proliferation. Proc. Natl. Acad. Sci. U. S. A. 108 (19), 7956–7961. doi:10.1073/pnas.1103154108
Page, E. C., Heatley, S. L., Eadie, L. N., McClure, B. J., de Bock, C. E., Omari, S., et al. (2022). HMGN1 plays a significant role in CRLF2 driven Down Syndrome leukemia and provides a potential therapeutic target in this high-risk cohort. Oncogene 41 (6), 797–808. doi:10.1038/s41388-021-02126-4
Palanichamy, J. K., Tran, T. M., Howard, J. M., Contreras, J. R., Fernando, T. R., Sterne-Weiler, T., et al. (2016). RNA-binding protein IGF2BP3 targeting of oncogenic transcripts promotes hematopoietic progenitor proliferation. J. Clin. Invest. 126 (4), 1495–1511. doi:10.1172/jci80046
Palis, J., and Yoder, M. C. (2001). Yolk-sac hematopoiesis: the first blood cells of mouse and man. Exp. Hematol. 29 (8), 927–936. doi:10.1016/s0301-472x(01)00669-5
Perner, F., Gadrey, J. Y., Xiong, Y., Hatton, C., Eschle, B. K., Weiss, A., et al. (2020). Novel inhibitors of the histone methyltransferase DOT1L show potent antileukemic activity in patient-derived xenografts. Blood 136 (17), 1983–1988. doi:10.1182/blood.2020006113
Perner, F., Stein, E. M., Wenge, D. V., Singh, S., Kim, J., Apazidis, A., et al. (2023). MEN1 mutations mediate clinical resistance to menin inhibition. Nature 615 (7954), 913–919. doi:10.1038/s41586-023-05755-9
Piatopoulou, D., Avgeris, M., Marmarinos, A., Xagorari, M., Baka, M., Doganis, D., et al. (2017). miR-125b predicts childhood acute lymphoblastic leukaemia poor response to BFM chemotherapy treatment. Br. J. cancer 117 (6), 801–812. doi:10.1038/bjc.2017.256
Popescu, D.-M., Botting, R. A., Stephenson, E., Green, K., Webb, S., Jardine, L., et al. (2019). Decoding human fetal liver haematopoiesis. Nature 574 (7778), 365–371. doi:10.1038/s41586-019-1652-y
Popovic, R., Riesbeck, L. E., Velu, C. S., Chaubey, A., Zhang, J., Achille, N. J., et al. (2009). Regulation of mir-196b by MLL and its overexpression by MLL fusions contributes to immortalization. Blood, J. Am. Soc. Hematol. 113 (14), 3314–3322. doi:10.1182/blood-2008-04-154310
Porcher, C., Chagraoui, H., and Kristiansen, M. S. (2017). SCL/TAL1: a multifaceted regulator from blood development to disease. Blood 129 (15), 2051–2060. doi:10.1182/blood-2016-12-754051
Ramos-Mejía, V., Navarro-Montero, O., Ayllón, V., Bueno, C., Romero, T., Real, P. J., et al. (2014). HOXA9 promotes hematopoietic commitment of human embryonic stem cells. Blood 124 (20), 3065–3075. doi:10.1182/blood-2014-03-558825
Rappa, G., Mercapide, J., Anzanello, F., Pope, R. M., and Lorico, A. (2013). Biochemical and biological characterization of exosomes containing prominin-1/CD133. Mol. Cancer 12, 62. doi:10.1186/1476-4598-12-62
Rawat, V. P. S., Cusan, M., Deshpande, A., Hiddemann, W., Quintanilla-Martinez, L., Humphries, R. K., et al. (2004). Ectopic expression of the homeobox gene Cdx2 is the transforming event in a mouse model of t(12;13)(p13;q12) acute myeloid leukemia. Proc. Natl. Acad. Sci. U. S. A. 101 (3), 817–822. doi:10.1073/pnas.0305555101
Rawat, V. P. S., Thoene, S., Naidu, V. M., Arseni, N., Heilmeier, B., Metzeler, K., et al. (2008). Overexpression of CDX2 perturbs HOX gene expression in murine progenitors depending on its N-terminal domain and is closely correlated with deregulated HOX gene expression in human acute myeloid leukemia. Blood 111 (1), 309–319. doi:10.1182/blood-2007-04-085407
Ren, H., Elliott, N., Lye, B., Cross, J., Field, L., Ponnusamy, K., et al. (2023). Bispecific CAR-iNKT immunotherapy for high risk MLL-rearranged acute lymphoblastic leukemia. Blood 142 (1), 766. doi:10.1182/blood-2023-186442
Reya, T., Morrison, S. J., Clarke, M. F., and Weissman, I. L. (2001). Stem cells, cancer, and cancer stem cells. Nature 414 (6859), 105–111. doi:10.1038/35102167
Riddell, J., Gazit, R., Garrison, B. S., Guo, G., Saadatpour, A., Mandal, P. K., et al. (2014). Reprogramming committed murine blood cells to induced hematopoietic stem cells with defined factors. Cell 157 (3), 549–564. doi:10.1016/j.cell.2014.04.006
Riedt, T., Ebinger, M., Salih, H. R., Tomiuk, J., Handgretinger, R., Kanz, L., et al. (2009). Aberrant expression of the homeobox gene CDX2 in pediatric acute lymphoblastic leukemia. Blood 113 (17), 4049–4051. doi:10.1182/blood-2008-12-196634
Roberts, I., Alford, K., Hall, G., Juban, G., Richmond, H., Norton, A., et al. (2013). GATA1-mutant clones are frequent and often unsuspected in babies with Down syndrome: identification of a population at risk of leukemia. Blood 122 (24), 3908–3917. doi:10.1182/blood-2013-07-515148
Roberts, I., and Izraeli, S. (2014). Haematopoietic development and leukaemia in Down syndrome. Br. J. Haematol. 167 (5), 587–599. doi:10.1111/bjh.13096
Roy, A., Cowan, G., Mead, A. J., Filippi, S., Bohn, G., Chaidos, A., et al. (2012). Perturbation of fetal liver hematopoietic stem and progenitor cell development by trisomy 21. Proc. Natl. Acad. Sci. 109 (43), 17579–17584. doi:10.1073/pnas.1211405109
Roy, A., Wang, G., Iskander, D., O’Byrne, S., Elliott, N., O’Sullivan, J., et al. (2021). Transitions in lineage specification and gene regulatory networks in hematopoietic stem/progenitor cells over human development. Cell Rep. 36 (11), 109698. doi:10.1016/j.celrep.2021.109698
Roy, S., Di Cello, F., Kowalski, J., Hristov, A. C., Tsai, H.-L., Bhojwani, D., et al. (2013). HMGA1 overexpression correlates with relapse in childhood B-lineage acute lymphoblastic leukemia. Leukemia Lymphoma 54 (11), 2565–2567. doi:10.3109/10428194.2013.782610
Rybak, A., Fuchs, H., Smirnova, L., Brandt, C., Pohl, E. E., Nitsch, R., et al. (2008). A feedback loop comprising lin-28 and let-7 controls pre-let-7 maturation during neural stem-cell commitment. Nat. Cell Biol. 10 (8), 987–993. doi:10.1038/ncb1759
Sachs, K., Sarver, A. L., Noble-Orcutt, K. E., LaRue, R. S., Antony, M. L., Chang, D., et al. (2020). Single-cell gene expression analyses reveal distinct self-renewing and proliferating subsets in the leukemia stem cell compartment in acute myeloid leukemia. Cancer Res. 80 (3), 458–470. doi:10.1158/0008-5472.Can-18-2932
Schäfer, V., Ernst, J., Rinke, J., Winkelmann, N., Beck, J. F., Hochhaus, A., et al. (2016). EZH2 mutations and promoter hypermethylation in childhood acute lymphoblastic leukemia. J. Cancer Res. Clin. Oncol. 142 (7), 1641–1650. doi:10.1007/s00432-016-2174-8
Scholl, C., Bansal, D., Döhner, K., Eiwen, K., Huntly, B. J. P., Lee, B. H., et al. (2007). The homeobox gene CDX2 is aberrantly expressed in most cases of acute myeloid leukemia and promotes leukemogenesis. J. Clin. Investigation 117 (4), 1037–1048. doi:10.1172/JCI30182
Schotte, D., Chau, J. C. K., Sylvester, G., Liu, G., Chen, C., van der Velden, V. H. J., et al. (2009). Identification of new microRNA genes and aberrant microRNA profiles in childhood acute lymphoblastic leukemia. Leukemia 23 (2), 313–322. doi:10.1038/leu.2008.286
Shah, S. N., Kerr, C., Cope, L., Zambidis, E., Liu, C., Hillion, J., et al. (2012). HMGA1 reprograms somatic cells into pluripotent stem cells by inducing stem cell transcriptional networks. PLOS ONE 7 (11), e48533. doi:10.1371/journal.pone.0048533
Shahab, S. W., Roggeveen, C. M., Sun, J., Kunhiraman, H., McSwain, L. F., Juraschka, K., et al. (2023). The LIN28B-let-7-PBK pathway is essential for group 3 medulloblastoma tumor growth and survival. Mol. Oncol. 17 (9), 1784–1802. doi:10.1002/1878-0261.13477
Sharma, A., Blériot, C., Currenti, J., and Ginhoux, F. (2022). Oncofetal reprogramming in tumour development and progression. Nat. Rev. Cancer 22 (10), 593–602. doi:10.1038/s41568-022-00497-8
Sharma, G., Tran, T. M., Bansal, I., Beg, M. S., Bhardwaj, R., Bassi, J., et al. (2023). RNA binding protein IGF2BP1 synergizes with ETV6-RUNX1 to drive oncogenic signaling in B-cell Acute Lymphoblastic Leukemia. J. Exp. Clin. cancer Res. CR 42 (1), 231. doi:10.1186/s13046-023-02810-1
Singh, S. K., Hawkins, C., Clarke, I. D., Squire, J. A., Bayani, J., Hide, T., et al. (2004). Identification of human brain tumour initiating cells. Nature 432 (7015), 396–401. doi:10.1038/nature03128
Solé, L., Lobo-Jarne, T., Álvarez-Villanueva, D., Alonso-Marañón, J., Guillén, Y., Guix, M., et al. (2022). p53 wild-type colorectal cancer cells that express a fetal gene signature are associated with metastasis and poor prognosis. Nat. Commun. 13 (1), 2866. doi:10.1038/s41467-022-30382-9
Somervaille, T. C., Matheny, C. J., Spencer, G. J., Iwasaki, M., Rinn, J. L., Witten, D. M., et al. (2009). Hierarchical maintenance of MLL myeloid leukemia stem cells employs a transcriptional program shared with embryonic rather than adult stem cells. Cell stem Cell 4 (2), 129–140. doi:10.1016/j.stem.2008.11.015
Somerville, T. D. D., Simeoni, F., Chadwick, J. A., Williams, E. L., Spencer, G. J., Boros, K., et al. (2018). Derepression of the Iroquois homeodomain transcription factor gene IRX3 confers differentiation block in acute leukemia. Cell Rep. 22 (3), 638–652. doi:10.1016/j.celrep.2017.12.063
Sonoda, Y., Itoh, M., and Tohda, S. (2021). Effects of HOXA9 inhibitor DB818 on the growth of acute myeloid leukaemia cells. Anticancer Res. 41 (4), 1841–1847. doi:10.21873/anticanres.14950
Stam, R. W., Schneider, P., Hagelstein, J. A. P., van der Linden, M. H., Stumpel, D. J. P. M., de Menezes, R. X., et al. (2010). Gene expression profiling-based dissection of MLL translocated and MLL germline acute lymphoblastic leukemia in infants. Blood 115 (14), 2835–2844. doi:10.1182/blood-2009-07-233049
Stauffer, F., Weiss, A., Scheufler, C., Möbitz, H., Ragot, C., Beyer, K. S., et al. (2019). New potent DOT1L inhibitors for in vivo evaluation in mouse. ACS Med. Chem. Lett. 10 (12), 1655–1660. doi:10.1021/acsmedchemlett.9b00452
Stein, E. M., Garcia-Manero, G., Rizzieri, D. A., Tibes, R., Berdeja, J. G., Savona, M. R., et al. (2018). The DOT1L inhibitor pinometostat reduces H3K79 methylation and has modest clinical activity in adult acute leukemia. Blood 131 (24), 2661–2669. doi:10.1182/blood-2017-12-818948
Stoskus, M., Gineikiene, E., Valceckiene, V., Valatkaite, B., Pileckyte, R., and Griskevicius, L. (2011). Identification of characteristic IGF2BP expression patterns in distinct B-ALL entities. Blood Cells, Mol. Dis. 46 (4), 321–326. doi:10.1016/j.bcmd.2011.02.005
Su, L., Bryan, N., Battista, S., Freitas, J., Garabedian, A., D’Alessio, F., et al. (2020). Identification of HMGA2 inhibitors by AlphaScreen-based ultra-high-throughput screening assays. Sci. Rep. 10 (1), 18850. doi:10.1038/s41598-020-75890-0
Sudha, T., Godugu, K., Darwish, N. H. E., Nazeer, T., and Mousa, S. A. (2021). Novel polyethylene glycol-conjugated triazole derivative with high thyrointegrin αvβ3 affinity in acute myeloid leukemia management. Cancers 13 (16), 4070. doi:10.3390/cancers13164070
Sugimura, R., Jha, D. K., Han, A., Soria-Valles, C., da Rocha, E. L., Lu, Y.-F., et al. (2017). Haematopoietic stem and progenitor cells from human pluripotent stem cells. Nature 545 (7655), 432–438. doi:10.1038/nature22370
Sun, W., Triche, T., Malvar, J., Gaynon, P., Sposto, R., Yang, X., et al. (2018). A phase 1 study of azacitidine combined with chemotherapy in childhood leukemia: a report from the TACL consortium. Blood 131 (10), 1145–1148. doi:10.1182/blood-2017-09-803809
Symeonidou, V., and Ottersbach, K. (2021). HOXA9/IRX1 expression pattern defines two subgroups of infant MLL-AF4-driven acute lymphoblastic leukemia. Exp. Hematol. 93, 38–43.e5. doi:10.1016/j.exphem.2020.10.002
Taipale, J., and Beachy, P. A. (2001). The Hedgehog and Wnt signalling pathways in cancer. Nature 411 (6835), 349–354.
Tan, L., Wei, X., Zheng, L., Zeng, J., Liu, H., Yang, S., et al. (2016). Amplified HMGA2 promotes cell growth by regulating Akt pathway in AML. J. Cancer Res. Clin. Oncol. 142 (2), 389–399. doi:10.1007/s00432-015-2036-9
Tan, L., Xu, H., Chen, G., Wei, X., Yu, B., Ye, J., et al. (2018). Silencing of HMGA2 reverses retardance of cell differentiation in human myeloid leukaemia. Br. J. Cancer 118 (3), 405–415. doi:10.1038/bjc.2017.403
Tang, C.-Y., Lin, J., Qian, W., Yang, J., Ma, J.-C., Deng, Z.-Q., et al. (2014). Low SOX17 expression: prognostic significance in de novo acute myeloid leukemia with normal cytogenetics. Clin. Chem. Laboratory Med. 52 (12), 1843–1850. doi:10.1515/cclm-2014-0487
Tavian, M., and Peault, B. (2005). Embryonic development of the human hematopoietic system. Int. J. Dev. Biol. 49 (2-3), 243–250. doi:10.1387/ijdb.041957mt
Testa, U., Riccioni, R., Militi, S., Coccia, E., Stellacci, E., Samoggia, P., et al. (2002). Elevated expression of IL-3Ralpha in acute myelogenous leukemia is associated with enhanced blast proliferation, increased cellularity, and poor prognosis. Blood, J. Am. Soc. Hematol. 100 (8), 2980–2988. doi:10.1182/blood-2002-03-0852
Thoene, S., Rawat, V. P. S., Heilmeier, B., Hoster, E., Metzeler, K. H., Herold, T., et al. (2009). The homeobox gene CDX2 is aberrantly expressed and associated with an inferior prognosis in patients with acute lymphoblastic leukemia. Leukemia 23 (4), 649–655. doi:10.1038/leu.2008.355
Tholouli, E., MacDermott, S., Hoyland, J., Yin, J. L., and Byers, R. (2012). Quantitative multiplex quantum dot in-situ hybridisation based gene expression profiling in tissue microarrays identifies prognostic genes in acute myeloid leukaemia. Biochem. Biophysical Res. Commun. 425 (2), 333–339. doi:10.1016/j.bbrc.2012.07.092
Thompson, B. J., Bhansali, R., Diebold, L., Cook, D. E., Stolzenburg, L., Casagrande, A. S., et al. (2015). DYRK1A controls the transition from proliferation to quiescence during lymphoid development by destabilizing Cyclin D3. J. Exp. Med. 212 (6), 953–970. doi:10.1084/jem.20150002
Tian, Z., Shi, C., Yang, G., Allen, J. K., Shi, Q., Al-Shami, A., et al. (2023). Preclinical development of 1B7/CD3, a novel anti-TSLPR bispecific antibody that targets CRLF2-rearranged Ph-like B-ALL. Leukemia 37 (10), 2006–2016. doi:10.1038/s41375-023-02010-y
Tolba, F. M., Foda, M. E., kamal, H. M., and Elshabrawy, D. A. (2013). Expression of CD133 in acute leukemia. Med. Oncol. 30 (2), 527. doi:10.1007/s12032-013-0527-6
Tran, T. M., Philipp, J., Bassi, J. S., Nibber, N., Draper, J. M., Lin, T. L., et al. (2021). The RNA-binding protein IGF2BP3 is critical for MLL-AF4-mediated leukemogenesis. Leukemia 36, 68–79. doi:10.1038/s41375-021-01346-7
Turan, R. D., Albayrak, E., Uslu, M., Siyah, P., Alyazici, L. Y., Kalkan, B. M., et al. (2020). Development of small molecule MEIS inhibitors that modulate HSC activity. Sci. Rep. 10 (1), 7994. doi:10.1038/s41598-020-64888-3
Uckelmann, H. J., Haarer, E. L., Takeda, R., Wong, E. M., Hatton, C., Marinaccio, C., et al. (2023). Mutant NPM1 directly regulates oncogenic transcription in acute myeloid leukemia. Cancer Discov. 13 (3), 746–765. doi:10.1158/2159-8290.CD-22-0366
Ueno, S., Lu, J., He, J., Li, A., Zhang, X., Ritz, J., et al. (2014). Aberrant expression of SALL4 in acute B cell lymphoblastic leukemia: mechanism, function, and implication for a potential novel therapeutic target. Exp. Hematol. 42 (4), 307–316. doi:10.1016/j.exphem.2014.01.005
Uhlén, M., Fagerberg, L., Hallström, B. M., Lindskog, C., Oksvold, P., Mardinoglu, A., et al. (2015). Proteomics. Tissue-based map of the human proteome. Sci. (New York, N.Y.) 347 (6220), 1260419. doi:10.1126/science.1260419
Vey, N., Prebet, T., Thalamas, C., Charbonnier, A., Rey, J., Kloos, I., et al. (2017). Phase 1 dose-escalation study of oral abexinostat for the treatment of patients with relapsed/refractory higher-risk myelodysplastic syndromes, acute myeloid leukemia, or acute lymphoblastic leukemia. Leukemia Lymphoma 58 (8), 1880–1886. doi:10.1080/10428194.2016.1263843
Viswanathan, S. R., and Daley, G. Q. (2010). Lin28: a MicroRNA regulator with a macro role. Cell 140 (4), 445–449. doi:10.1016/j.cell.2010.02.007
Viswanathan, S. R., Powers, J. T., Einhorn, W., Hoshida, Y., Ng, T. L., Toffanin, S., et al. (2009). Lin28 promotes transformation and is associated with advanced human malignancies. Nat. Genet. 41 (7), 843–848. doi:10.1038/ng.392
Vora, P., Venugopal, C., Salim, S. K., Tatari, N., Bakhshinyan, D., Singh, M., et al. (2020). The rational development of cd133-targeting immunotherapies for glioblastoma. Cell Stem Cell 26 (6), 832–844. doi:10.1016/j.stem.2020.04.008
Wagenblast, E., Araujo, J., Gan, O. I., Cutting, S. K., Murison, A., Krivdova, G., et al. (2021). Mapping the cellular origin and early evolution of leukemia in Down syndrome. Science 373 (6551), eabf6202. doi:10.1126/science.abf6202
Wang, D., Tanaka-Yano, M., Meader, E., Kinney, M. A., Morris, V., Lummertz da Rocha, E., et al. (2022a). Developmental maturation of the hematopoietic system controlled by a Lin28b-let-7-Cbx2 axis. Cell Rep. 39 (1), 110587. doi:10.1016/j.celrep.2022.110587
Wang, L., Rowe, R. G., Jaimes, A., Yu, C., Nam, Y., Pearson, D. S., et al. (2018a). Small-molecule inhibitors disrupt let-7 oligouridylation and release the selective blockade of let-7 processing by LIN28. Cell Rep. 23 (10), 3091–3101. doi:10.1016/j.celrep.2018.04.116
Wang, L.-l., Tang, X., Zhou, G., Liu, S., Wang, Y., Chen, F., et al. (2022b). PROM1 and CTGF expression in childhood MLL-rearrangement acute lymphoblastic leukemia. J. Oncol. 2022, 5896022. doi:10.1155/2022/5896022
Wang, S., Chim, B., Su, Y., Khil, P., Wong, M., Wang, X., et al. (2019). Enhancement of LIN28B-induced hematopoietic reprogramming by IGF2BP3. Genes and Dev. 33 (15-16), 1048–1068. doi:10.1101/gad.325100.119
Wang, Y., Chen, M., Wu, Z., Tong, C., Dai, H., Guo, Y., et al. (2018b). CD133-directed CAR T cells for advanced metastasis malignancies: A phase I trial. Oncoimmunology 7 (7), e1440169. doi:10.1080/2162402X.2018.1440169
Wang, Y., Xu, Y., Li, S., Liu, J., Xing, Y., Xing, H., et al. (2018c). Targeting FLT3 in acute myeloid leukemia using ligand-based chimeric antigen receptor-engineered T cells. J. Hematol. Oncol. 11 (1), 60. doi:10.1186/s13045-018-0603-7
Waters, N. J., Smith, S. A., Olhava, E. J., Duncan, K. W., Burton, R. D., O'Neill, J., et al. (2016). Metabolism and disposition of the DOT1L inhibitor, pinometostat (EPZ-5676), in rat, dog and human. Cancer Chemother. Pharmacol. 77 (1), 43–62. doi:10.1007/s00280-015-2929-y
Weigmann, A., Corbeil, D., Hellwig, A., and Huttner, W. B. (1997). Prominin, a novel microvilli-specific polytopic membrane protein of the apical surface of epithelial cells, is targeted to plasmalemmal protrusions of non-epithelial cells. Proc. Natl. Acad. Sci. U. S. A. 94 (23), 12425–12430. doi:10.1073/pnas.94.23.12425
Wermke, M., Kraus, S., Ehninger, A., Bargou, R. C., Goebeler, M.-E., Middeke, J. M., et al. (2021). Proof of concept for a rapidly switchable universal CAR-T platform with UniCAR-T-CD123 in relapsed/refractory AML. Blood 137 (22), 3145–3148. doi:10.1182/blood.2020009759
Wong, P., Iwasaki, M., Somervaille, T. C. P., So, C. W. E., and Cleary, M. L. (2007). Meis1 is an essential and rate-limiting regulator of MLL leukemia stem cell potential. Genes and Dev. 21 (21), 2762–2774. doi:10.1101/gad.1602107
Wu, P. (2020). Inhibition of RNA-binding proteins with small molecules. Nat. Rev. Chem. 4 (9), 441–458. doi:10.1038/s41570-020-0201-4
Wu, Z., Eguchi-Ishimae, M., Yagi, C., Iwabuki, H., Gao, W., Tauchi, H., et al. (2015). HMGA2 as a potential molecular target in KMT2A-AFF1-positive infant acute lymphoblastic leukaemia. Br. J. Haematol. 171 (5), 818–829. doi:10.1111/bjh.13763
Wuputra, K., Ku, C.-C., Wu, D.-C., Lin, Y.-C., Saito, S., and Yokoyama, K. K. (2020). Prevention of tumor risk associated with the reprogramming of human pluripotent stem cells. J. Exp. Clin. Cancer Res. 39 (1), 100–124. doi:10.1186/s13046-020-01584-0
Yamada, Y., Warren, A. J., Dobson, C., Forster, A., Pannell, R., and Rabbitts, T. H. (1998). The T cell leukemia LIM protein Lmo2 is necessary for adult mouse hematopoiesis. Proc. Natl. Acad. Sci. U. S. A. 95 (7), 3890–3895. doi:10.1073/pnas.95.7.3890
Yan, W., Xu, L., Sun, Z., Lin, Y., Zhang, W., Chen, J., et al. (2015). MicroRNA biomarker identification for pediatric acute myeloid leukemia based on a novel bioinformatics model. Oncotarget 6 (28), 26424–26436. doi:10.18632/oncotarget.4459
Yang, J., Chai, L., Gao, C., Fowles, T. C., Alipio, Z., Dang, H., et al. (2008). SALL4 is a key regulator of survival and apoptosis in human leukemic cells. Blood 112 (3), 805–813. doi:10.1182/blood-2007-11-126326
Yao, S., Jianlin, C., Yarong, L., Botao, L., Qinghan, W., Hongliang, F., et al. (2019). Donor-derived cd123-targeted CAR T cell serves as a RIC regimen for haploidentical transplantation in a patient with FUS-ERG+ AML. Front. Oncol. 9, 1358. doi:10.3389/fonc.2019.01358
Yasuda, T., Sanada, M., Kawazu, M., Kojima, S., Tsuzuki, S., Ueno, H., et al. (2022). Two novel high-risk adult B-cell acute lymphoblastic leukemia subtypes with high expression of CDX2 and IDH1/2 mutations. Blood 139 (12), 1850–1862. doi:10.1182/blood.2021011921
Yin, A. H., Miraglia, S., Zanjani, E. D., Almeida-Porada, G., Ogawa, M., Leary, A. G., et al. (1997). AC133, a novel marker for human hematopoietic stem and progenitor cells. Blood 90 (12), 5002–5012. doi:10.1182/blood.v90.12.5002.5002_5002_5012
Yokoyama, A., Somervaille, T. C. P., Smith, K. S., Rozenblatt-Rosen, O., Meyerson, M., and Cleary, M. L. (2005). The menin tumor suppressor protein is an essential oncogenic cofactor for MLL-associated leukemogenesis. Cell 123 (2), 207–218. doi:10.1016/j.cell.2005.09.025
Yu, S., Cui, K., Jothi, R., Zhao, D. M., Jing, X., Zhao, K., et al. (2011). GABP controls a critical transcription regulatory module that is essential for maintenance and differentiation of hematopoietic stem/progenitor cells. Blood 117 (7), 2166–2178. doi:10.1182/blood-2010-09-306563
Yuan, J., Nguyen, C. K., Liu, X., Kanellopoulou, C., and Muljo, S. A. (2012). Lin28b reprograms adult bone marrow hematopoietic progenitors to mediate fetal-like lymphopoiesis. Science 335 (6073), 1195–1200. doi:10.1126/science.1216557
Zhang, H., Luo, X.-Q., Feng, D.-D., Zhang, X.-J., Wu, J., Zheng, Y.-S., et al. (2011). Upregulation of microRNA-125b contributes to leukemogenesis and increases drug resistance in pediatric acute promyelocytic leukemia. Mol. cancer 10, 108–113. doi:10.1186/1476-4598-10-108
Zhang, J., Ratanasirintrawoot, S., Chandrasekaran, S., Wu, Z., Ficarro, S. B., Yu, C., et al. (2016). LIN28 regulates stem cell metabolism and conversion to primed pluripotency. Cell Stem Cell 19 (1), 66–80. doi:10.1016/j.stem.2016.05.009
Zhang, J., Tam, W.-L., Tong, G. Q., Wu, Q., Chan, H.-Y., Soh, B.-S., et al. (2006). Sall4 modulates embryonic stem cell pluripotency and early embryonic development by the transcriptional regulation of Pou5f1. Nat. Cell Biol. 8 (10), 1114–1123. doi:10.1038/ncb1481
Zhang, L., Li, X., Ke, Z., Huang, L., Liang, Y., Wu, J., et al. (2013). MiR-99a may serve as a potential oncogene in pediatric myeloid leukemia. Cancer Cell Int. 13 (1), 110–111. doi:10.1186/1475-2867-13-110
Zhang, N., Shen, Y., Li, H., Chen, Y., Zhang, P., Lou, S., et al. (2022). The m6A reader IGF2BP3 promotes acute myeloid leukemia progression by enhancing RCC2 stability. Exp. Mol. Med. 54 (2), 194–205. doi:10.1038/s12276-022-00735-x
Zhang, Q., Shi, M., Zheng, R., Han, H., Zhang, X., and Lin, F. (2023a). C1632 inhibits ovarian cancer cell growth and migration by inhibiting LIN28 B/let-7/FAK signaling pathway and FAK phosphorylation. Eur. J. Pharmacol. 956, 175935. doi:10.1016/j.ejphar.2023.175935
Zhang, Y., Jiang, S., He, F., Tian, Y., Hu, H., Gao, L., et al. (2023b). Single-cell transcriptomics reveals multiple chemoresistant properties in leukemic stem and progenitor cells in pediatric AML. Genome Biol. 24 (1), 199. doi:10.1186/s13059-023-03031-7
Zhao, H., Chen, S., and Fu, Q. (2020). Exosomes from CD133+ cells carrying circ-ABCC1 mediate cell stemness and metastasis in colorectal cancer. J. Cell. Biochem. 121 (5-6), 3286–3297. doi:10.1002/jcb.29600
Zhou, J., Bi, C., Ching, Y. Q., Chooi, J.-Y., Lu, X., Quah, J. Y., et al. (2017). Inhibition of LIN28B impairs leukemia cell growth and metabolism in acute myeloid leukemia. J. Hematol. Oncol. 10 (1), 138. doi:10.1186/s13045-017-0507-y
Zhou, Y., Li, Y. S., Bandi, S. R., Tang, L., Shinton, S. A., Hayakawa, K., et al. (2015). Lin28b promotes fetal B lymphopoiesis through the transcription factor Arid3a. J. Exp. Med. 212 (4), 569–580. doi:10.1084/jem.20141510
Keywords: leukemia, stem cells, development genes, gene regulation, fetal oncogenes
Citation: Ling RE, Cross JW and Roy A (2024) Aberrant stem cell and developmental programs in pediatric leukemia. Front. Cell Dev. Biol. 12:1372899. doi: 10.3389/fcell.2024.1372899
Received: 18 January 2024; Accepted: 11 March 2024;
Published: 27 March 2024.
Edited by:
Antonella Lettieri, University of Milan, ItalyReviewed by:
Anilkumar Gopalakrishnapillai, Alfred I. duPont Hospital for Children, United StatesCopyright © 2024 Ling, Cross and Roy. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Anindita Roy, anindita.roy@paediatrics.ox.ac.uk
†These authors have contributed equally to this work
 Rebecca E. Ling
Rebecca E. Ling