Abstract
As a byproduct of glycolysis, lactate functions as a signaling molecule, a substrate for energy metabolism, and a regulator of the tumor microenvironment (TME). It is involved in various biological processes, including energy shuttling, tumor growth and invasion, drug resistance, and immune evasion. Lactylation, a recently identified post-translational modification (PTM), acts as a bridge between gene regulation and cellular metabolism, thus playing a crucial role in tumor biology. Similar to other epigenetic modifications, lactylation influences the spatial conformation of chromatin, modulates DNA accessibility, and regulates gene expression. It intricately participates in TME-related processes by orchestrating immune state transitions and enhancing the malignant characteristics of tumors. This review summarizes lactylation-related genes in tumors, the role of lactylation in the TME, the interactions of the genes with other metabolic pathways, and the potential mechanisms underlying tumor progression as well as their clinical implications. Despite its nascent stage, research on the epigenetic regulation of tumor-related genes by lactylation holds promise. In this review, we highlighted unresolved challenges in this field and provided insights that may guide the development of novel targeted therapies for cancer.
1 Introduction
Glycolysis is a pathway of glucose breakdown that enables cells to generate energy under both normoxic and hypoxic conditions. In the tumor microenvironment (TME), glycolysis is remarkably active in cancer cells, even under high-oxygen concentrations. In the 1920s, Warburg et al. identified the characteristic “aerobic glycolysis” in tumors, now known as the Warburg effect (Warburg et al., 1927). Compared with surrounding tissues, tumors consume larger amounts of glucose. Through glycolysis, tumor cells generate adenosine triphosphate (ATP) and produce excessive lactate intracellularly, even in the presence of sufficient oxygen. Lactate, an oxidizable substrate, subsequently generates a substantial amount of ATP to promote cancer cell proliferation. Despite its lower efficiency than mitochondrial oxidative phosphorylation (OXPHOS), glycolysis-dependent ATP production allows tumor cells to obtain abundant metabolic intermediates. These intermediates are essential for synthesizing lipids, nucleotides, and other biomolecules required for tumor growth. In addition to energy production, glycolysis-derived lactate drives acidification of the extracellular matrix, impairing immune cell function (Li et al., 2022) and promoting tumor metastasis and invasion (Jiang et al., 2020). Lactate is transported between cells via monocarboxylate transporters (MCTs), particularly MCT1 (Brooks, 2018; Carneiro et al., 2017), or acts as a signaling molecule through specific receptors (Roland et al., 2014).
In 2019, Zhang et al. discovered that lactate could regulate gene expression by inducing histone lactylation, a process now termed lactylation (Zhang et al., 2019). Lactylation is a lactate-mediated post-translational modification (PTM) similar to phosphorylation, acetylation, methylation, and ubiquitination. Lactate-driven epigenetic alterations represent a critical link through which metabolic states influence gene expression. Increased levels of histone and non-histone lactylation have been observed in various malignancies and are closely associated with epigenetic dysregulation and metabolic dysfunction. These processes collectively regulate intracellular signaling, gene expression, and protein functionality (Chen H. et al., 2024; Zong et al., 2024; Chen et al., 2024b). Despite its significance, the mechanisms and functions of lactylation remain unclear. An in-depth understanding of the role of this novel epigenetic modification in tumor progression may provide insights into the molecular mechanisms underlying metabolic reprogramming in cancer and facilitate the development of therapeutic strategies targeting metabolism. This review provides a comprehensive overview of the research progress of lactylation in tumors.
2 Histone and non-histone lactylation in the TME
The TME comprises tumor cells, cancer-associated fibroblasts (CAFs), and tumor-associated immune cells, some of which are regulated by lactate (Han et al., 2023).
Both normoxic and hypoxic cellular populations are found in the TME. Hypoxic cells, inherently lacking the ability to oxidize lactate, are typically located in regions with the highest lactate levels. These cells rely on glucose for Warburg-like metabolism, generating a large amount of lactate, which is subsequently transported to adjacent oxygenated cells via MCT1. A metabolic symbiosis has been observed between normoxic and hypoxic cells, wherein some cells undergo Warburg-like metabolism, whereas others metabolize lactate through OXPHOS-dependent pathways (Hui et al., 2017; Faubert et al., 2017). This lactate shuttling facilitates the redistribution and efficient utilization of energy substrates, which are used by tumor cells to optimize energy production and meet the demands of rapid proliferation. This division of labor in energy metabolism serves as a crucial factor contributing to the formation of a TME (Brooks, 2018; Brooks, 2020). Over the past 5 years, studies have suggested that lactylation plays an essential role in shaping many pro-tumor characteristics of cells within the TME. In addition, lactylation serves as an important epigenetic mechanism in the TME (Jiao et al., 2024; Xiong et al., 2022). High lactate levels suppress immune cells and contribute to lactylation (Apostolova and Pearce, 2022; Elia et al., 2022) on both histone and non-histone proteins. This section systematically discusses the roles of histone and non-histone lactylation in the TME, providing a valuable reference for investigating how the TME is shaped by epigenetics.
2.1 Histone lactylation
Histones are proteins that form the core of nucleosomes wrapped by DNA, serving as the primary structural proteins of chromosomes. PTM of histones can regulate DNA replication, transcription, and repair by altering nucleosome interactions or recruiting non-histone proteins, which are essential for maintaining homeostasis (Millan-Zambrano et al., 2022). With the advancement of high-sensitivity mass spectrometry techniques in epigenetic research, the diversity of histone PTMs continues to expand (Sabari et al., 2017). Lysine acylation of histones is a common type of PTM, with hydrophobicity, charge, and hydrocarbon chain length being different across various acylation types (Hirschey and Zhao, 2015). This modification is regulated by specific enzymes that act as “writers” and “erasers”. For instance, histone acetyltransferase (HAT) p300 is a key “writer” of histone acetylation (Moreno-Yruela et al., 2022a), whereas histone deacetylases (HDACs) remove acetyl groups from the lysine residues of both histone and non-histone proteins. Effector proteins, termed “readers,” specifically interpret modified gene information and regulate downstream signaling pathways, contributing to the dynamic regulation of lysine acylation (Moreno-Yruela et al., 2022b; Rothbart et al., 2015).
Lysine lactylation (Kla) is a novel type of PTM that directly stimulates gene transcription (Zhang et al., 2019) (Figure 1). Zhao et al. detected a 72.021-Da mass shift at the lysine residues of histones in human tumor cells using tandem mass spectrometry (MS/MS) in conjunction with high-performance liquid chromatography (HPLC). This mass shift was hypothesized to result from the addition of a lactyl group to lysine residues, which validated lactate as a substrate for histone modification and established histone lactylation as a PTM in vivo (Zhang et al., 2019; Huang et al., 2014). Lactyl-CoA donates a lactyl group to lysine residues on histone tails via p300 (Zeng et al., 2023). HBO1, a HAT belonging to the MYST family, has been identified as another lactyltransferase mediating histone lactylation-dependent gene transcription (Niu et al., 2024). In addition, HDAC1–3 have been identified as “erasers” for histone lactylation at histone H3 lysine 1 (H3K1) (Moreno-Yruela et al., 2022b). However, the enzymes that produce lactyl-CoA, the enzymes that derive the lactyl group, and other enzymes that participate in the regulation of histone lactylation remain unclear (Hou et al., 2024; Yang et al., 2023a), necessitating further investigation.
FIGURE 1
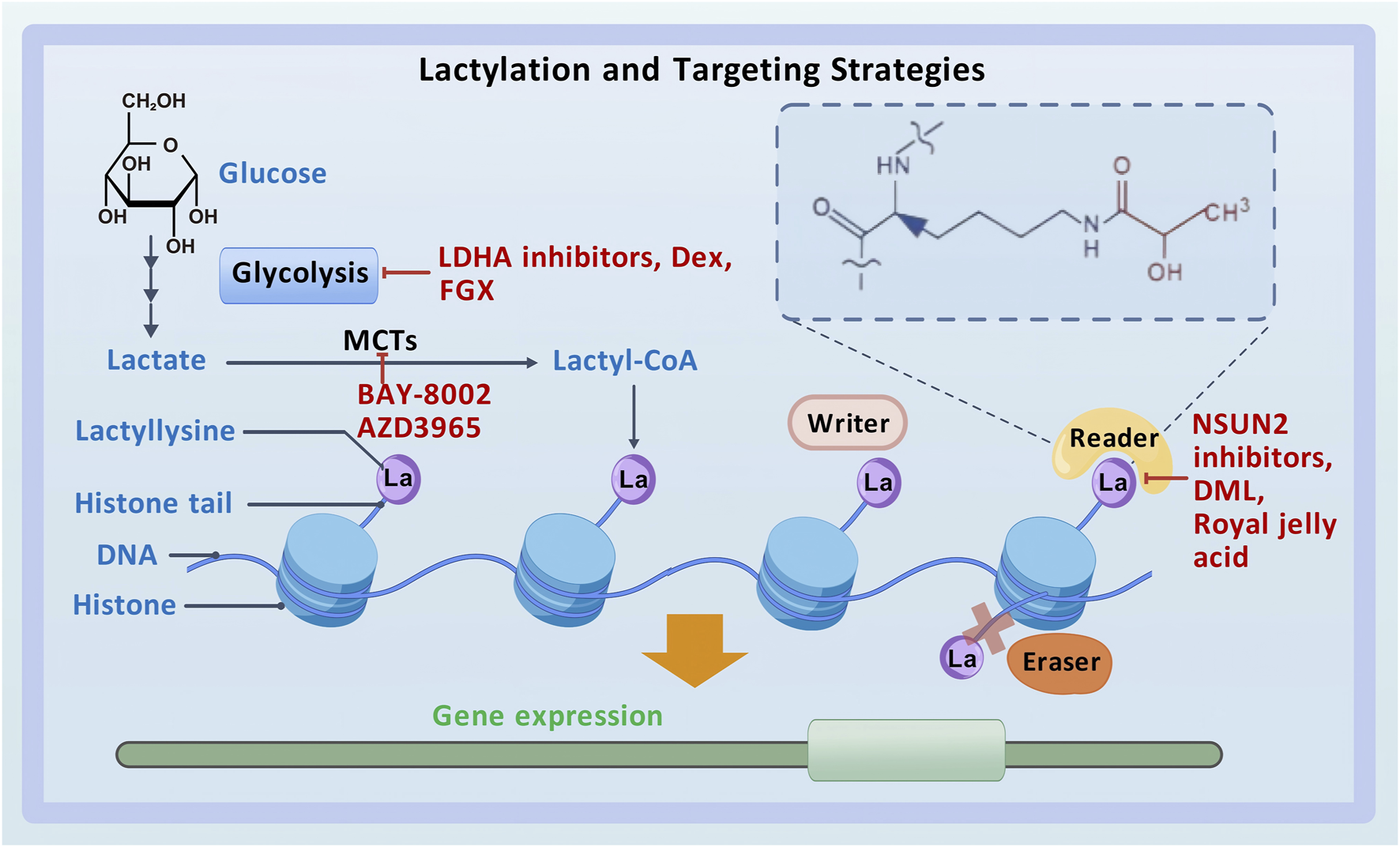
Lactylation and targeting strategies.
Studies have demonstrated that inhibition or promotion of glycolysis decreases or increases the total level of Kla, respectively, highlighting the sensitivity of histone lactylation to glycolysis-dependent lactate production (Jin et al., 2022; Irizarry-Caro et al., 2020). As an important component of the TME, lactate links metabolism, epigenetics, and transcription, thereby regulating gene expression. An immunosuppressive microenvironment is established and maintained in part by Kla. By directly binding to lactylation sites on the lysine residues of histones, lactate functions as an epigenetic regulator that directs the transcription of genes involved in damage repair, including arginase-1 (Arg1), and induces the polarization of macrophages toward the M2 phenotype through epigenetic reprogramming (Wang Z. H. et al., 2021). Pyruvate kinase M2 (PKM2) regulates glycolysis by directly interacting with lactate. In addition, lactylation at its K62 site enhances glycolytic activity, resulting in increased expression of Arg1. This regulation suppresses the inflammatory activity of M1 macrophages, promoting polarization toward the M2 phenotype (Wang et al., 2022a).
Histone lactylation exerts crucial epigenetic regulatory effects during tumorigenesis (Yu et al., 2021). Accumulation of lactate in the TME is a common feature of cancer that can induce histone lactylation, thus promoting the development of various types of tumors by disrupting the balance in gene transcription (Certo et al., 2021; Ippolito et al., 2019). Understanding the close relationship between histone lactylation and tumor development may offer novel avenues for developing more effective therapeutic strategies for cancer.
2.2 Non-histone lactylation
Lactylation can occur in non-histone proteins, affecting the structure and function of these proteins. Wan et al. found that peptides with lactylated lysine residues produced distinct immonium ions, which served as a marker for identifying lysine lactylation accurately. They discovered information regarding lactylation substrate proteins and modification sites by searching for these ions in existing human proteomic data resources. These findings suggest that lactylation plays an equally important regulatory role in non-histone proteins. Important cellular functions associated with both healthy and diseased states are mediated by these proteins (Wan et al., 2022). Identifying lactylation sites in non-histone proteins is another focus area of ongoing research in this field (Jing et al., 2024).
Lactylation can also occur in nuclear proteins. Studies have shown that the homologous recombination-associated protein MRE11 is lactylated at its K672 site via a p300-dependent mechanism. This modification leads to DNA end resection, promotes homologous recombination, and induces drug resistance in tumor cells (Chen et al., 2024b). Furthermore, the nuclear protein high-mobility group box-1 (HMGB1) can be lactylated (Yang K. et al., 2022) and subsequently secreted into the cytoplasm through exosomes. In the cytoplasm, lactylated HMGB1 regulates endothelial permeability, inflammation surrounding tumor cells, and stromal remodeling in the TME, thereby influencing the characteristics of the TME (Lan et al., 2020). Moreover, lactylation reduces the binding affinity of HMGB1 for immune cell surface receptors, impairing immune cell recognition and clearance of HMGB1, suppressing anti-tumor immune responses, and promoting cancer progression (Chen et al., 2024b). In a study, proteomic analysis of gastric cancer (GC) cells revealed HMGB1 among the 1,014 proteins with a total of 2,375 lactylation sites. The nucleus contained more than half of these lactylated proteins, whereas the remaining proteins were distributed across other cellular compartments. Lactylation is frequently found in spliceosomes and has been demonstrated to affect RNA splicing (Yang D. et al., 2022). Additionally, nuclear hypoxia-inducible factor 1-alpha (HIF1α) can undergo lactylation. In prostate cancer (PCa), the uptake of excess lactate via MCT1 results in lactylation of HIF1α, stabilizing the protein even under normoxic conditions. This stabilization activates downstream signaling, enhances transcriptional activity, and promotes angiogenesis (Luo et al., 2022). Alanyl-tRNA synthetase 1 (AARS1) has been identified as both a lactate sensor and a lactyltransferase that can lactylate p53 and promote tumorigenesis (Li H. et al., 2024; Ju et al., 2024). In a recent study, pan-cancer multi-omic analysis revealed overexpression of lactylation-related proteins such as glyoxalase 1 (GLO1), HDAC2, HDAC8, and HDAC1 in various tumor tissues (Wu Z. et al., 2024).
With advancements in scientific technology, more lactylation sites will be identified and their roles and regulatory mechanisms will be comprehensively investigated, offering new avenues for therapeutic research. However, studies on non-histone lactylation remain limited, necessitating further investigation into the functions and mechanisms of this modification.
3 Interactions between lactylation and metabolic pathways in tumors
3.1 Glycolysis
In normal cells, energy is derived primarily from OXPHOS or glycolysis. However, aerobic glycolysis or the Warburg effect is considered the predominant mechanism of energy production in tumor cells (Icard et al., 2018). Glycolysis produces high amounts of lactate, and subsequent histone lactylation can regulate gene expression. During glycolysis, glucose undergoes incomplete oxidation, generating two molecules of pyruvate. Pyruvate is reduced to lactate under the action of lactate dehydrogenase. Subsequently, acyltransferases catalyze the conversion of lactate to lactyl-CoA, which modifies lysine residues on histone tails, completing lactylation (Zhang et al., 2019). Therefore, a key element in histone lactylation is the imbalance between glycolysis and the tricarboxylic acid cycle (TAC). Although the glycolytic intermediate lactylglutathione (LGSH) is the source of the lactyl group in histone lactylation, high LGSH levels are associated with an increase in histone lactylation levels of glycolytic enzymes and a notable reduction in glycolysis. Consequently, histone lactylation acts as a feedback loop during glycolysis, with higher blood glucose levels promoting the formation of LGSH and lactylation, which in turn inhibits glycolysis (Gaffney et al., 2020).
Hypoxia is a hallmark of the microenvironment of solid tumors. Under hypoxic conditions, the upregulated expression of serine hydroxymethyltransferase 2 (SHMT2) enhances the glycolytic activity of esophageal cancer (EC) cells. Additionally, glycolysis-derived lactate induces lactylation of SHMT2, stabilizing the protein. Mechanistically, SHMT2 interacts with mitochondrial monofunctional C1-tetrahydrofolate synthase (MTHFD1L), promoting malignant progression in EC (Qiao et al., 2024). A recent study showed that a positive feedback loop involving glycolysis, H3K18 lactylation (H3K18la), and the protein kinases TTK and BUB1B exacerbates dysfunction in pancreatic ductal adenocarcinoma (PDAC) (Li et al., 2024b). These findings indicate that histone lactylation not only occurs during glycolysis but also influences glycolysis, playing a regulatory role in pathological processes (Wang M. et al., 2023). Understanding histone lactylation may provide novel insights into the mechanisms through which the Warburg effect contributes to cancer progression and introduce promising avenues for investigating the interplay between glycolysis and epigenetic modifications in cancer.
3.2 Acetylation
Recent studies have revealed complex crosstalk between histone lactylation and acetylation, both independently and synergistically, providing insights into their roles in tumorigenesis and their potential as therapeutic targets. Both lactylation and acetylation occur on lysine residues. p300, a member of the HAT family, has been identified as the primary “writer” enzyme for lactylation (Dai X. et al., 2022; Chen et al., 2021). Despite their similarities and shared catalytic enzymes, histone lactylation exhibits slower dynamic equilibrium kinetics (24 h) than histone acetylation (6 h), which suggests that acetylation occurs more readily than lactylation under physiological conditions (Dai X. et al., 2022). During lactylation, lactyl-CoA donates lactyl groups to lysine residues (Dai S. K. et al., 2022). Because both lactyl-CoA and acetyl-CoA are derived from pyruvate, the dominance of these pathways can vary across cell types (Gaur et al., 2021). Cells generate lactate under hypoxic conditions; however, the presence of sufficient oxygen promotes the conversion of pyruvate to acetyl-CoA, driving TAC (Zhang et al., 2019). The interaction between acetylation and lactylation has been shown to influence tumor progression. Deactivation of the mitochondrial pyruvate dehydrogenase complex (PDC) is necessary for cancer cells to transition from OXPHOS to aerobic glycolysis. According to a recent study, p300 acetylates the E3-binding protein (E3BP) of PDC at Lys 488, interfering with the interaction of PDHX with dihydrolipoyl transacetylase and preventing the assembly and activation of PDC. The majority of glucose is converted to lactate as a result of this disruption, which promotes aerobic glycolysis and H3K56 lactylation-mediated gene expression, eventually accelerating the growth of malignant tumors (Jiang Z. et al., 2024). A competitive link between histone lactylation and acetylation has also been observed, which indicates that several genes that cannot undergo acetylation are susceptible to lactylation. The ratio of lactylation to acetylation reflects the pathway of pyruvate conversion and may serve as an indicator of a cell’s susceptibility to malignancy (Dai X. et al., 2022). This competitive interplay highlights the importance of understanding the metabolic and epigenetic regulatory processes involved in tumorigenesis.
3.3 Other metabolic pathways
Lipid metabolism and the pentose phosphate pathway (PPP) have been associated with lactylation. Chen et al. conducted a comprehensive lactyl-proteomic and metabolomic analysis on samples from patients with non-small cell lung cancer (NSCLC). The findings revealed that intracellular lactate promotes extracellular lipolysis through lactylation of apolipoprotein C-II (APOC2). Mechanistically, lactate enhances lactylation of APOC2, stabilizing the protein and resulting in the release of free fatty acids, accumulation of Tregs, resistance to immunotherapy, and tumor metastasis (Chen J. et al., 2024). Another study demonstrated that lactate dehydrogenase A (LDHA) is activated under nutrient deprivation by UNC-51-like kinase 1 (ULK1), which phosphorylates serine-196 and promotes lactate production. Subsequently, lactylation of the autophagy-related protein vacuolar protein sorting 34 (Vps34) enhances its binding to phosphoinositide-3-kinase class 3 (PIK3C3) and increases its lipid kinase activity. This process, which facilitates autophagy and lysosomal degradation, is closely associated with cancer progression (Jia et al., 2023). Furthermore, a recent study on cervical cancer found that lactate induces lactylation of discoidin, CUB, and LCCL domain-containing protein 1 (DCBLD1) at the K172 site. This modification stabilizes DCBLD1 expression, leading to an increase in the expression and enzymatic activity of glucose-6-phosphate dehydrogenase (G6PD), which stimulates the PPP and promotes the progression of cervical cancer (Meng et al., 2024). Additionally, lactylation of NMNAT1 can support the survival of pancreatic cancer cells by maintaining the nuclear NAD + salvage pathway (Huang H. et al., 2024).
These findings highlight the intricate crosstalk between lactylation and other metabolic pathways. However, research in this area remains limited. Understanding this crosstalk and the underlying mechanisms may provide novel avenues for preventing tumor progression, developing targeted therapeutic approaches, and optimizing existing treatment strategies.
4 Role of lactylation in tumor progression
4.1 Role in tumor initiation and growth
Studies have shown a strong correlation between lactylation and tumor initiation. In colorectal cancer (CRC), the overall level of Kla is negatively correlated with the prognosis. Among the lactylated proteins identified in CRC, the lysine acetyltransferase KAT8 mediates the lactylation of elongation factor 1 alpha (eEF1A2) at the K408 site, enhancing elongation during translation and protein synthesis, thus promoting tumorigenesis (Xie et al., 2024). In a study on cutaneous melanoma (CM), Yu et al. found that histone lactylation facilitated tumorigenesis by promoting the expression of YTH N6-methyladenosine RNA-binding protein 2 (YTHDF2) (Yu et al., 2021). Similarly, in CRC, histone lactylation increases the stability of potassium channel subfamily K member 6 (Kcnk6) in a YTHDF2-dependent manner, enhancing the activity of potassium channels, inducing the activation of the NOD-like receptor thermal protein domain-associated protein 3 (Nlrp3) inflammasome, and eventually driving inflammation-induced carcinogenesis (Yuan et al., 2024).
Furthermore, lactylation is involved in tumor cell proliferation (Fu et al., 2024; Zang et al., 2024). Miao et al. demonstrated that hypoxia promoted lactylation of β-catenin, enhancing its stability and driving CRC cell proliferation via the Wnt signaling pathway (Miao et al., 2023). Insulin-like growth factor 1 (IGF1) and its receptor (IGF1R) play crucial roles in regulating glycolysis in tumors. In lung cancer, IGF1R is highly expressed and its lactylation is associated with the malignant proliferation of tumor cells. Lactate enhances the stability of IGF1R protein, forming a feedback loop that promotes glycolysis and lactate production (Zhang R. et al., 2024). The histone H3 variant CENPA interacts with yin and yang 1 (YY1) to promote the production of neuropilin 2 (NRP2) and cyclin D1 (CCND1). Lactylation of CENPA at K124 promotes its activation, increasing the expression of its target genes and accelerating tumor development in hepatocellular carcinoma (HCC) (Liao et al., 2023). The transcription of serine/arginine-rich splicing factor 10 (SRSF10) is enhanced by increased aerobic glycolysis, which also increases histone lactylation levels at the c-Myc promoter. This phenomenon promotes alternative splicing of BCL2-like 1 (Bcl-x) and murine double minute 4 (MDM4), facilitating the proliferation of HCC cells (Pandkar et al., 2023; Yao and Yang, 2024). In anaplastic thyroid cancer (ATC), aerobic glycolysis increases cellular lactate utilization and overall protein lactylation. Specifically, H4K12la activates genes essential for the proliferation of ATC cells. The oncogenic mutation BRAFV600E further promotes glycolysis, re-inducing lactylation and leading to H4K12la-driven dysregulation of transcription and the cell cycle (Wang X. et al., 2023). Li et al. performed immunohistochemical analysis on 60 triple-negative breast cancer samples and found increased levels of total lactylated proteins and specific histone H4K12 lactylation in the samples. H4K12la may bind to the schlafen 5 (SLFN5) promoter region, suppressing the expression of SLFN5 and leading to altered gene profiles and reduced apoptosis (Li J. et al., 2024).
Tumor cells exhibit remarkable plasticity (Chiodi and Mondello, 2020). The acquisition of stem cell-like properties is a major factor contributing to the self-renewal and long-term proliferation of cancer cells (Li Q. et al., 2024; Han et al., 2020). Lactate serves as a crucial metabolite, signaling molecule, and energy source during the development of HCC. Intracellular lactylation is considered a contributing factor in the development of HCC. Compared with HCC cells, liver cancer stem cells (LCSCs), the source of phenotypic and functional heterogeneity in HCC, exhibit enhanced glycolysis, lactate accumulation, and lactylation. H3K56la is closely associated with tumor initiation and the stemness of LCSCs. In a study, a comprehensive analysis of the lactylome and proteome of LCSCs showed that lactylation of aldolase A (ALDOA) at K230/322 enhanced the function of DDX17 in maintaining stemness. However, the impact of Kla on HCC stem cells remains unclear. Targeting lactylated ALDOA represents a promising therapeutic strategy for HCC (Feng et al., 2024). In glioblastoma (GBM), lactylation of histone H3 increases the expression of LINC01127, which directs polymerase II (POLR2A) to the mitogen-activated protein kinase kinase kinase kinase 4 (MAP4K4) promoter region. This interaction activates the JNK pathway, regulating the self-renewal of GBM cells (Li et al., 2023).
4.2 Role in tumor invasion and migration
Invasion and migration are hallmarks of malignant tumors and significantly impact treatment outcomes and prognosis. Approximately 80% of cases of renal cell carcinoma (RCC), one of the three main cancers of the urinary system, are attributed to clear cell renal cell carcinoma (ccRCC). According to a bioinformatic study, FK506-binding protein 10 (FKBP10) is essential for the glycolytic pathway that leads to metastasis in ccRCC. FKBP10 directly interacts with LDHA via its C-terminal region, enhancing the phosphorylation of LDHA-Y10. This process leads to an overactive Warburg effect and histone lactylation (Liu R. et al., 2024). Lactylation at H3K18 is associated with several malignant features in tumors (Hou et al., 2024). Zhou et al. found that G protein-coupled receptor 37 (GPR37) was significantly overexpressed in human CRC specimens and was correlated with a poor prognosis. Mechanistically, GPR37 promotes LDHA expression and glycolysis by activating the Hippo pathway, which increases H3K18la levels, upregulates chemokine CXC motif ligand 1 (CXCL1) and CXCL5, and enhances tumor cell migration (Zhou et al., 2023). In GC, dynamic H3K18 lactylation activates the transcription of vascular cell adhesion molecule 1 (VCAM1), which stimulates downstream AKT–mTOR signaling, thereby promoting the proliferation and migration of GC cells. Therefore, H3K18 lactylation may serve as a therapeutic target for GC (Zhao Y. et al., 2024). In HCC, proline-5-carboxylate reductase 1 (PYCR1) modulates the transcription of insulin receptor substrate 1 (IRS1) by influencing H3K18 lactylation in its promoter region, thus driving metastasis and promoting other malignant characteristics of HCC cells (Wang H. et al., 2024).
Epithelial–mesenchymal transition (EMT) is a crucial mechanism through which tumor cells acquire metastatic potential. EMT decreases cell-to-cell interactions and promotes migration and motility (Dongre and Weinberg, 2019). This process is hijacked in cancer to enable morphological and motility-related changes that stimulate invasion and migration (Lu et al., 2019). Glucose transporter 3 (GLUT3) plays an important role in glycolysis in cancer. In a study on GC, single-cell sequencing revealed significant upregulation of GLUT3 expression in primary and metastatic tumor tissues. This increased expression of GLUT3 was positively correlated with the expression of LDHA and the activity of lactylation-associated pathways and was functionally associated with EMT (Yang et al., 2023a). Lysyl oxidase (LOX) secreted by CAFs controls EMT through TGFβ/IGF1 signaling and enhances PD-L1 expression through histone lactylation (Li Z. et al., 2024). In pancreatic cancer, Rho GTPase Rif (RHOF) promotes the lactylation and nuclear translocation of Snail 1 by enhancing PKM2-mediated glycolysis, thereby inducing EMT (Zhao R. et al., 2024). In HCC, increased lactylation at H3K9 and H3K56 activates the transcription of endothelial-specific molecule 1 (ESM1) to promote EMT (Zhao P. et al., 2024).
Angiogenesis is crucial for tumor cells to acquire nutrients and metastasize (Zheng et al., 2023; Bhat et al., 2021). Studies have shown that the lactation of H3K9 in Endothelial cells can regulate Vascular Endothelial Growth Factor (VEGF), thereby inducing angiogenesis (Fan et al., 2024). PCa is one of the most prevalent cancers in adult men worldwide. Recent studies have shown that the expression of the hyaluronan-binding protein KIAA1199 is correlated with tumor stage, HIF-1α overexpression, and angiogenic markers in PCa. Overexpression of KIAA1199 enhances the production of secretory vascular endothelial growth factor A (VEGFA), promoting angiogenesis and vasculogenic mimicry (VM). In a study, ChIP-PCR and dual-luciferase reporter assay showed that HIF-1α enhances the transcription of KIAA1199. Mechanistically, lactate imported via MCT1 stabilizes HIF-1α through lactylation, enhancing the transcription of KIAA1199 (Luo et al., 2022). HCC is characterized by abundant vasculature. Upon its activation by histone lactylation and c-Myc, Golgi phosphoprotein 73 (GP73) leads to the phosphorylation of signal transducer and activator of transcription 3 (STAT3), which in turn enhances the pro-angiogenic function of GP73 (Ye et al., 2024).
These findings emphasize the crucial roles of lactylation in tumor invasion, migration, and angiogenesis, providing potential therapeutic targets for metastatic and vascularized tumors.
4.3 Role in immune regulation
Tumors form a complex ecosystem, with various immune cells being intricately involved. Studies have highlighted the important role of lactylation in immune regulation in tumors.
During tumor progression, cancer cells adopt diverse mechanisms to evade immune surveillance, such as downregulating antigen presentation pathways or inducing inhibitory immune checkpoints (Jhunjhunwal et al., 2021). Lactate-driven histone lactylation in KRAS-mutant tumor cells has been shown to activate the transcription of circATXN7, a circular RNA that interacts with NF-κB. circATXN7 sequesters p65 in the cytoplasm by binding to and masking the nuclear localization signal motif of the NF-κB p65 subunit, increasing the susceptibility of tumor-specific T cells to activation-induced cell death (AICD) and promoting immune evasion (Zhou et al., 2024). In NSCLC, H3K18la enhances immune evasion by activating the pore membrane protein 121 (POM121)/MYC/PD-L1 axis (Zhang C. et al., 2024). In GC, high lactylation levels are strongly correlated with poor overall survival and disease progression. Mechanistic experiments have shown that high lactylation levels are characterized by extensive macrophage infiltration, increased genomic instability, and a high risk of immune dysfunction in tumors (Yang et al., 2023b). In experimental models of PCa with PTEN/p53 abnormalities, activation of the Wnt/β-catenin pathway has been shown to result in lactylation at H3K18 in tumor cells and tumor-associated macrophages (TAMs), contributing to immune evasion (Chaudagar et al., 2023).
Cancer cells exploit immune cells, such as macrophages, CD8+ T cells and Tregs, to shape an immunosuppressive TME (Lei et al., 2020). Tumor-derived lactate is a critical driver of TAM polarization (Fang et al., 2023). For instance, Li et al. found that tumor-derived lactate promoted H3K18la, suppressing the transcription of RARγ in macrophages. This process enhanced IL-6 levels in the TME, activating STAT3 signaling in CRC cells and promoting the polarization of macrophages toward the M2 phenotype (Li X. M. et al., 2024). In neuroblastoma (NB), hexokinase-3 (HK3), a key enzyme in glucose metabolism, is correlated with M2 macrophage infiltration, increased lactate secretion by tumor cells, and histone lactylation (Wu X. et al., 2024). Proprotein convertase subtilisin/kexin type 9 (PCSK9) can inhibit M1 polarization by increasing lactate levels, promoting protein lactylation, and strengthening the activity of macrophage migration inhibitory factor (MIF) (Wang L. et al., 2022). In cervical cancer, lactate secreted by tumor cells increases H3K18la levels and promotes M2 polarization by upregulating glycerol-3-phosphate dehydrogenase 2 (GPD2), thereby supporting tumor progression (Huang C. et al., 2024). In advanced stages, monocyte-derived macrophages (MDMs) increase in number and lactate-driven histone lactylation increases IL-10 expression, forming an immunosuppressive TME by suppressing T-cell activity (De Leo et al., 2024).
Lactylation can influence the activity of T cells, one of the key components of the TME. Activation and functional differentiation of CD8+ T cells are regulated by lactate-associated metabolic pathways. Histone modifications such as H3K18la and H3K9la are enriched in CD8+ T cells, serving as transcriptional initiation factors for key regulatory genes (Raychaudhuri et al., 2024). SRSF10, which is upregulated in various tumors, enhances glycolysis-related enzymes, such as LDHA, increasing intracellular and extracellular lactate levels. This increase induces histone lactylation and amplifies SRSF10 expression. In addition, tumor-derived lactate can induce H3K18la in macrophages, activating transcription and enhancing cellular activity. M2 macrophages, in turn, inhibit CD8+ T cell infiltration, exacerbating the immunosuppressive TME (Cai et al., 2024).
Additionally, regulatory T cells (Tregs) are crucial for the development and maintenance of the suppressive TME (Li et al., 2020). A study showed that lactylation of lysine residues in membrane-organizing extension spike protein (MOESIN) increased the expression of forkhead box P3 (FOXP3) and enhanced the regulation of transforming growth factor-beta (TGF-β) signaling, thus sustaining Treg-mediated immunosuppression and promoting TME formation (Gu et al., 2022a). Lactylation at K72 in membrane proteins regulates Treg cell generation by enhancing their interactions with TGF-β receptors (Gu et al., 2022b). STAT5 expression promotes glycolysis in acute myeloid leukemia (AML) cells, leading to lactate accumulation. Excess lactate triggers the nuclear translocation and histone lactylation of E3BP, eventually inducing the transcription of PD-L1. Therefore, patients with AML characterized by STAT5-driven glycolysis and lactate accumulation may benefit from PD-1/PD-L1 blockade therapies (Huang et al., 2023) (Figure 2).
FIGURE 2
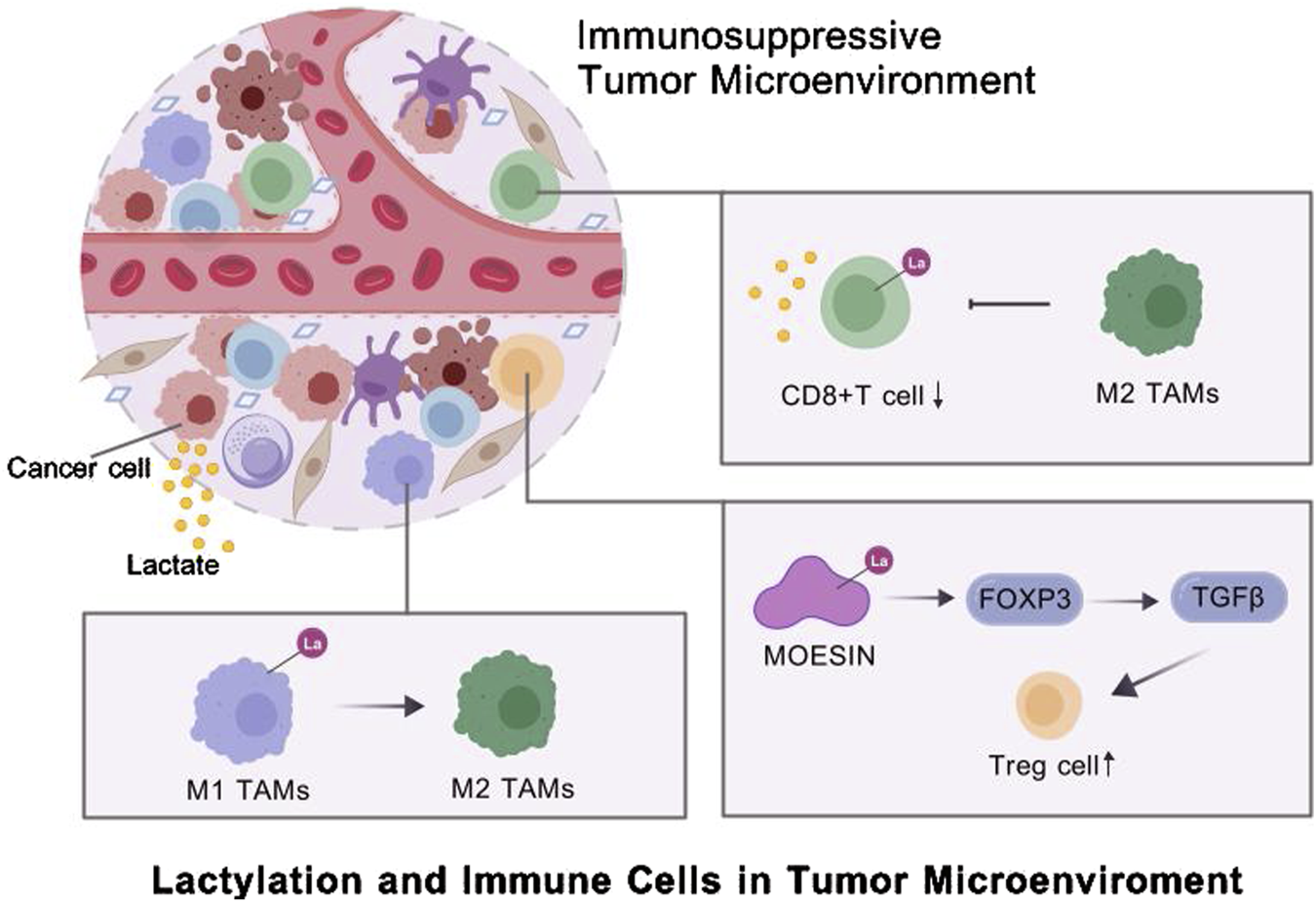
Lactylation and immune cells in tumor microenviroment.
Tumor immune regulation is a critical aspect of understanding tumorigenesis, with profound implications for developing novel treatments, elucidating mechanisms underlying drug resistance, and improving patient prognosis. Lactylation has demonstrated substantial research potential in this domain. Ongoing studies will likely uncover novel therapeutic targets, advancing the development of novel immunotherapies.
4.4 Role in the genome
As a PTM, lactylation inevitably exerts significant effects on the genome. TIMs, crucial players in tumor immune evasion, are regulated by various epigenetic mechanisms. Lactylation-induced activation of the m6A–YTHDF1 axis mediated by methyltransferase-like 3 (METTL3) improves the translation efficiency of JAK1 and phosphorylation of STAT3, supporting the immunosuppressive properties of TIMs (Xiong et al., 2022). N1-methyladenosine (m1A) RNA modification serves as a key regulator of RNA metabolism. Histone lactylation can upregulate AlkB homolog 3 (ALKBH3), which in turn suppresses m1A modification of speckled protein 100A (SP100A), weakening the formation of tumor-suppressive promyelocytic leukemia (PML) protein condensates and facilitating the malignant transformation of tumor cells (Gu X. et al., 2024). Therapeutic resistance and tumor development are significantly influenced by cellular plasticity. In a study, multi-omic analysis showed that the expression of zinc finger E-box-binding homeobox 1 (ZEB1) is indicative of an intermediate state that tumor cells go through during the progression of neuroendocrine prostate cancer (NEPC). EMT, stemness, and neuroendocrine traits are present in this stage. ZEB1 directs tumor cells toward glycolysis for energy production by controlling the transcription of numerous important glycolytic enzymes. Histone lactylation induced by lactate accumulation during this process improves cellular plasticity and chromatin accessibility (Wang D. et al., 2024).
These findings highlight the essential role of lactylation in modulating genomic and epigenomic landscapes, influencing tumor cell behavior, and promoting malignancy. Understanding these mechanisms may offer promising avenues for targeted therapeutic interventions in cancer.
4.5 Role in treatment resistance
Despite significant advancements in the treatment of cancer, therapeutic resistance associated with molecular and clinical relapse remains prevalent, resulting in a poor prognosis. Therefore, understanding the mechanisms underlying therapeutic resistance is necessary to improve the treatment of cancer. Recent studies have shown that lactylation may contribute to treatment resistance in cancer (Takata et al., 2024).
Resistance to traditional anti-cancer treatments has been closely associated with lactylation in various tumor types. Lactylation of Nijmegen breakage syndrome protein 1 (NBS1) at lysine 388 (K388) is essential for the formation of the MRE11–RAD50–NBS1 (MRN) complex and the accumulation of DNA repair proteins at DNA double-strand break sites. However, this process is associated with adverse outcomes of neoadjuvant chemotherapy (Yang et al., 2023b). In metastatic CRC, bevacizumab is an important drug in first- and second-line treatments. Studies have shown that patients with bevacizumab-resistant CRC exhibit increased levels of histone lactylation, which promotes the transcription of recombinant rubicon-like autophagy enhancer (RUBCNL) through interactions with beclin 1 (BECN1). This process drives the maturation of autophagosomes, recruitment of the class III phosphoinositide 3-kinase complex, and proliferation and survival of cancer cells under hypoxic conditions (Li W. et al., 2024). Patients with bladder cancer (BCa) often develop resistance to cisplatin. H3K18la drives the transcription of Y-box-binding protein 1 (YBX1) and YY1, which are closely associated with cisplatin resistance (Li et al., 2024h). Similarly, histone lactylation plays a role in the development of cisplatin resistance in oral squamous cell carcinoma (OSCC) (Huang G. et al., 2024). In a study on GBM, interactions between aldehyde dehydrogenase 1 family, member A3 (ALDH1A3) and PKM2 were found to enhance PKM2 tetramerization in glioblastoma stem cells (GSCs), promoting lactate accumulation. Proteomic analysis of these GSCs revealed lactylation of X-ray repair cross-complementing protein 1 (XRCC1) at K247. Lactylated XRCC1 enhanced DNA repair, conferring drug resistance in ALDH1A3-overexpressing GBM cells (Li et al., 2024). Temozolomide (TMZ) resistance is a major barrier to the successful treatment of GBM. Studies have shown that H3K9la significantly enriches the LUC7L2 promoter, activating its transcription and promoting TMZ resistance in GBM (Yue et al., 2024). Other studies on the role of lactylation in the acquisition of resistance to standard anti-cancer therapies are highlighted in Table 1 (Lu Y. et al., 2024; Chu et al., 2023; Duan et al., 2023; Sun X. et al., 2023).
TABLE 1
| Malignant characteristics | Tumor type | Protein | Modification site | Mechanism | References |
|---|---|---|---|---|---|
| Tumorigenesis | Colorectal cancer | non-histone | K408 of eEF1A2 | Enhance translation elongation and protein synthesis | Xie et al. (2024) |
| histone | not clear | Increase the stability of Kcnk6 in a YTHDF2-dependent manner, inducing Nlrp3 activation | Yuan et al. (2024) | ||
| Cutaneous melanoma | histone | H3K18 | Promote the expression of YTHDF2 | Yu et al. (2021) | |
| Cell proliferation | Colorectal cancer | non-histone | β-catenin | Activate the Wnt signaling pathway | Miao et al. (2023) |
| Lung cancer | non-histone | IGF1R | Form a feedback loop that promotes glycolysis and lactate production | Zhang et al. (2024a) | |
| Hepatocellular carcinoma | histone | K124 of histone H3 variant CENPA | Work with YY1 to promote the production of NRP2 and CCND1 | Liao et al. (2023) | |
| histone | H3K18 | Promote alternative splicing of Bcl-x and MDM4 | Pandkar et al. (2023) | ||
| Anaplastic thyroid cancer | histone | H4K12 | Activate BRAFV600E, further promoting glycolysis, reconstructing the lactylation environment and leading to cell cycle dysregulation | Wang et al. (2024a) | |
| Triple-negative breast cancer | histone | H4K12 | Suppress SLFN5 expression and lead to altered gene profiles and reduced apoptosis | Li et al. (2024c) | |
| Generation of stem cell like characteristics | Hepatocellular carcinoma | non-histone | K230/322 of ALDOA | Enhance the regulatory function of DDX17 | Feng et al. (2024) |
| Glioblastoma | histone | H3K18 | Increase the expression of LINC01127, which directs POLR2A to the MAP4K4 promoter region. This interaction activates the JNK pathway | Li et al. (2023) | |
| Invasion and migration | Clear cell renal cell carcinoma | histone | Not clear | Not clear | Liu et al. (2024a) |
| Colorectal cancer | histone | H3K18 | Upregulate CXCL1 and CXCL5 | Zhou et al. (2023) | |
| Gastric cancer | histone | H3K18 | Activate the transcription of VCAM1, which subsequently stimulates downstream AKT-mTOR signaling | Zhao et al. (2024a) | |
| Hepatocellular carcinoma | histone | H3K18 | PYCR1 modulates IRS1 transcription by influencing H3K18la in its promoter region | Wang et al. (2024a) | |
| EMT | Gastric cancer | histone | H3K9, H3K18, H3K56, H4K8, H4K12 | Not clear | Yang et al. (2023a) |
| histone | H3K18 | CAF-secreted LOX controls EMT via TGFβ/IGF1 signaling and enhances PD-L1 expression through histone lactylation | Li et al. (2024e) | ||
| Pancreatic cancer | non-histone | Snail | RHOF enhances glycolysis mediated by PKM2, promoting lactylation and nuclear translocation of Snail 1, thereby inducing EMT | Zhao et al. (2024b) | |
| Hepatocellular carcinoma | histone | H3K9, H3K56 | Activate the transcription of ESM1 | Zhao et al. (2024c) | |
| Tumor angiogenesis | Prostate cancer | non-histone | HIF-1α | Enhance KIAA1199 transcription, improving the production of secretory VEGFA | Luo et al. (2022) |
| Hepatocellular carcinoma | histone | H3K18, H4K5 | Activate GP73, causing STAT3 to become phosphorylated which further amplifies GP73-mediated pro-angiogenic functions | Ye et al. (2024) | |
| Immune escape | Prostate cancer | histone | H3K18 | Activate the transcription of circATXN7 sequestering p65 in the cytoplasm that make tumor-specific T cells more susceptible to AICD | Zhou et al. (2024) |
| Non-small cell lung cancer | histone | H3K18 | Activate the POM121/MYC/PD-L1 axis | Zhang et al. (2024b) | |
| Prostate cancer | histone | H3K18 | Activation of the Wnt/β-catenin pathway results in H3K18la and TAMs with PTEN/p53 abnormalities | Chaudagar et al. (2023) | |
| Immune suppression | Colorectal cancer | histone | H3K18 | Suppress the transcription of RARγ in macrophages which enhances IL-6 levels in the TME, activating STAT3 signaling and fostering M2-like macrophage polarization | Li et al. (2024f) |
| not clear | not clear | Inhibit M1 macrophage polarization | Wang et al. (2022b) | ||
| Neuroblastoma | histone | not clear | M2 macrophage infiltration | Wu et al. (2024b) | |
| Cervical cancer | histone | H3K18 | Promote M2 macrophage polarization | Huang et al. (2024b) | |
| Glioblastoma | histone | not clear | Boost IL-10 expression forming an immunosuppressive TME by suppressing T cell activity | De Leo et al. (2024) | |
| Hepatocellular carcinoma | histone | K72 | Regulate Treg cell generation by enhancing interactions with TGF-β receptors | Gu et al. (2022b) | |
| Acute myeloid leukemia | histone | H3K18 | Triggers E3BP nuclear translocation and induce PD-L1 transcription | Huang et al. (2023) | |
| Not clear | histone | H3K18, H3K9 | Serve as transcriptional initiation factors for key regulatory genes | Raychaudhuri et al. (2024) | |
| Hepatocellular carcinoma | histone | H3K18 | Amplify SRSF10 expression, activate transcription and boost TAM activity | Cai et al. (2024) | |
| Impacts on genome | Colorectal cancer | non-histone | METTL3 | Lactylation-induced activation of the m6A-YTHDF1 axis mediated by METTL3 improves JAK1 protein translation efficiency and STAT3 phosphorylation, supporting the immunosuppressive properties of TIMs | Xiong et al. (2022) |
| Acute myeloid leukemia | histone | H3K18 | Upregulate ALKBH3 expression, then reduce m1A methylation on SP100A weakening the formation of tumor-suppressive PML protein condensates and facilitating malignant cancer transformation | Gu et al. (2024a) | |
| Prostate cancer | histone | H3K18 | Improve cellular plasticity and chromatin accessibility | Wang et al. (2024b) | |
| Therapy resistance | Gastric cancer | non-histone | NBS1 | Be essential for the formation of the MRN complex and the accumulation of DNA repair proteins | Yang et al. (2023b) |
| Colorectal cancer | histone | H3K18 | Promote the transcription of RUBCNL through interaction with BECN1 which drives autophagosome maturation, recruitment of the class III phosphoinositide 3-kinase complex, and cancer cell proliferation and survival under hypoxic conditions | Li et al. (2024g) | |
| histone | not clear | Activate the ALDOB/PDK1/lactate/CEACAM6 axis | Chu et al. (2023) | ||
| histone | H4K12 | Increase ABC transporter | Sun et al. (2023a) | ||
| Bladder cancer | histone | H3K18 | Drive the transcription of key transcription factors YBX1 and YY1 | Li et al. (2024h) | |
| Oral squamous cell cancer | histone | H3K18, H3K27 | Induce BCAM expression | Huang et al. (2024c) | |
| Glioblastoma | non-histone | XRCC1 | Enhance DNA repair conferring therapy resistance in ALDH1A3-overexpressing | Li et al. (2024) | |
| histone | H3K9 | Enrich the LUC7L2 promoter activating its transcription and promoting TMZ resistance | Yue et al. (2024) | ||
| Hepatocellular carcinoma | non-histone | IGF2BP3 | By maintaining high levels of PCK2 and NRF2, the lactylated IGF2BP3-PCK2-SAM-m6A loop strengthens the antioxidant system | Lu et al. (2024a) | |
| Lung cancer-derived brain metastasis | histone | H4K12 | Activate the transcription of CCNB1 and accelerate the DNA replication and cell cycle | Duan et al. (2023) | |
| Lung squamous cell carcinoma | histone | not clear | Be linked to high SLC2A1 expression which has a negative correlation with TIGIT, CTLA4, LAG3, PD-1 | Hao et al. (2024) | |
| non-histone | APOC 2 | Resulte in FFA release | Chen et al. (2024c) | ||
| Head and neck squamous cell cancer | histone | H3K9 | Control downstream IL-11 through the JAK2/STAT3 pathway that activates immunological checkpoint genes in CD8+ T cells | Wang et al. (2024c) | |
| Microorganism | Colorectal liver metastasis | non-histone | RIG-I | E. coli inhibits NF-κB recruitment to the Nlrp3 promoter in macrophages by inducing retinoic RIG-I lactylation, which reduces Nlrp3 transcription and mediates M2 macrophage polarization | Gu et al. (2024b) |
| Colorectal cancer | histone | H4K8 H4K5 H3K18 |
Increase production of LINC00152 | Wang et al. (2022c) | |
| HBV-related hepatocellular carcinoma | non-histone | 9,256 sites | Lactylation-dependent metabolic adaption pathways | Y et al. (2023) | |
| Cellular senescence | Lung cancer | histone | H4K8 H4K16 | Control the production of TERT which lowers telomerase activity and causes telomere dysfunction, offering fresh information on the function of LKB1-mediated senescence | Liu et al. (2024b) |
| Autophagy | Cervical cancer | non-histone | TFEB | Enhance autophagic flux | Huang et al. (2024d) |
| Colorectal cancer | histone | H3K18 | Encourage autophagosome maturation, which is essential for hypoxic cancer cells to proliferate and survive | Li et al. (2024g) | |
| Lung cancer, Gastric cancer | non-histone | VPS34 | Enhance the association of VPS34 with BECN1, ATG14, and UVRAG, promoting autophagy and lysosomal degradation pathways | Sun et al. (2023b) |
Summary of lactylation’s functions in the malignant characteristics of cancers.
Immune checkpoint blockade (ICB) therapies have shown remarkable efficacy across multiple tumor types. However, resistance to these therapies is increasingly being reported. The expression of solute carrier family two member 1 (SLC2A1), which encodes the GLUT1 protein, is different between lung squamous cell carcinoma (LUSC) tissues and normal lung tissues. In a study, GLUT1 was identified as an independent prognostic factor for LUSC. According to LASSO analysis, high protein lactylation levels were associated with high SLC2A1 expression. Furthermore, SLC2A1 expression was found to have a negative correlation with the expression of TIGIT, CTLA4, LAG3, PD-1, and other common immune checkpoints (Hao et al., 2024). Increased levels of histone lactylation have been associated with a poor response to immunotherapy in head and neck squamous cell cancer (HNSCC). H3K9la can control downstream interleukin-11 (IL-11), which activates the transcription of various immune checkpoint genes in CD8+ T cells through the JAK2/STAT3 pathway. In addition, H3K9la is strongly correlated with IL-11 expression and a poor response to immunotherapy in patients with HNSCC in clinical settings (Wang R. et al., 2024). APOC2 can also promote resistance to immunotherapy in NSCLC (Chen J. et al., 2024).
Understanding the mechanisms through which lactylation contributes to drug resistance is essential for improving treatment outcomes in patients with cancer.
4.6 Microorganisms participate in lactation modification
The microbiome may play an important role in tumor initiation, progression, and metastasis. Microorganisms can influence cancer-related processes by modulating immune responses, promoting inflammation, and producing carcinogenic substances. Numerous intratumoral bacteria, such as E. coli, have been found to reside within tumors and contribute to liver metastasis of colorectal cancer (CRLM). E. coli inhibits the recruitment of NF-κB to the Nlrp3 promoter in macrophages by inducing lactylation of retinoic acid-inducible gene 1 (RIG-I), which suppresses the transcription of Nlrp3 and mediates M2 polarization. The absence of Nlrp3 impacts the immunosuppressive activity of Tregs and the anti-tumor responses of CD8+ T cells (Gu J. et al., 2024). In a prospective study, Yang et al. investigated the lactylome of patients with HCC with hepatitis B virus (HBV) infection through integrated genomic and proteomic analysis of the tumor and surrounding liver tissues. The results revealed 9,275 lactylation sites; of which, 9,256 sites were found on non-histone proteins, indicating that Kla is a ubiquitous alteration that affects both histone and non-histone proteins and performs other functions in addition to transcriptional regulation. Furthermore, analysis of the lactylome revealed lactylation-dependent metabolic adaption pathways in HBV-related HCC (Y et al., 2023). Gut microorganisms can play a role in disease development by interacting with the host genome through epigenetic factors, such as long non-coding RNAs (lncRNAs). Wang et al. measured transcriptomic alterations in a human colon cell line infected with the common gut pathogen Salmonella typhimurium SL1344. The results showed that bacterial lipopolysaccharides (LPSs) led to increased production of LINC00152, which was controlled by histone lactylation. The expression of LINC00152 was higher in CRC tissues than in adjacent normal tissues (Wang et al., 2022c).
4.7 Other mechanisms
In addition to its well-known tumor-suppressive function in lung cancer, liver kinase B1 (LKB1) is reported to be involved in cellular senescence. In LKB1-deficient A549 cells, overexpression of LKB1 increases H4K8 and H4K16 levels and causes lactate generation. By controlling the production of telomerase reverse transcriptase (TERT), this inhibition reduces telomerase activity and causes telomere dysfunction, providing novel insights into the function of LKB1-mediated cellular senescence in lung cancer (Liu M. et al., 2024).
Autophagy is a lysosome-mediated self-degradation process that recycles damaged organelles and long-lived proteins, maintaining cellular homeostasis under metabolic stresses, such as starvation and hypoxia. Lactate has been shown to regulate autophagy. Transcription factor EB (TFEB), a key regulator of lysosomal biogenesis and autophagy, undergoes covalent lactylation, leading to its stabilization and increased activity, which enhances autophagic flux (Huang Y. et al., 2024). Furthermore, histone lactylation promotes autophagosome maturation, which is essential for the proliferation and survival of hypoxic cancer cells (Li W. et al., 2024). Lactylation of PIK3C3/VPS34 at K356 and K781 through lysine acetyltransferase 5 (KAT5) can enhance the association of VPS34 with BECN1, ATG14, and UVRAG, promoting autophagy and other lysosomal degradation pathways (Sun W. et al., 2023) (Figure 3).
FIGURE 3
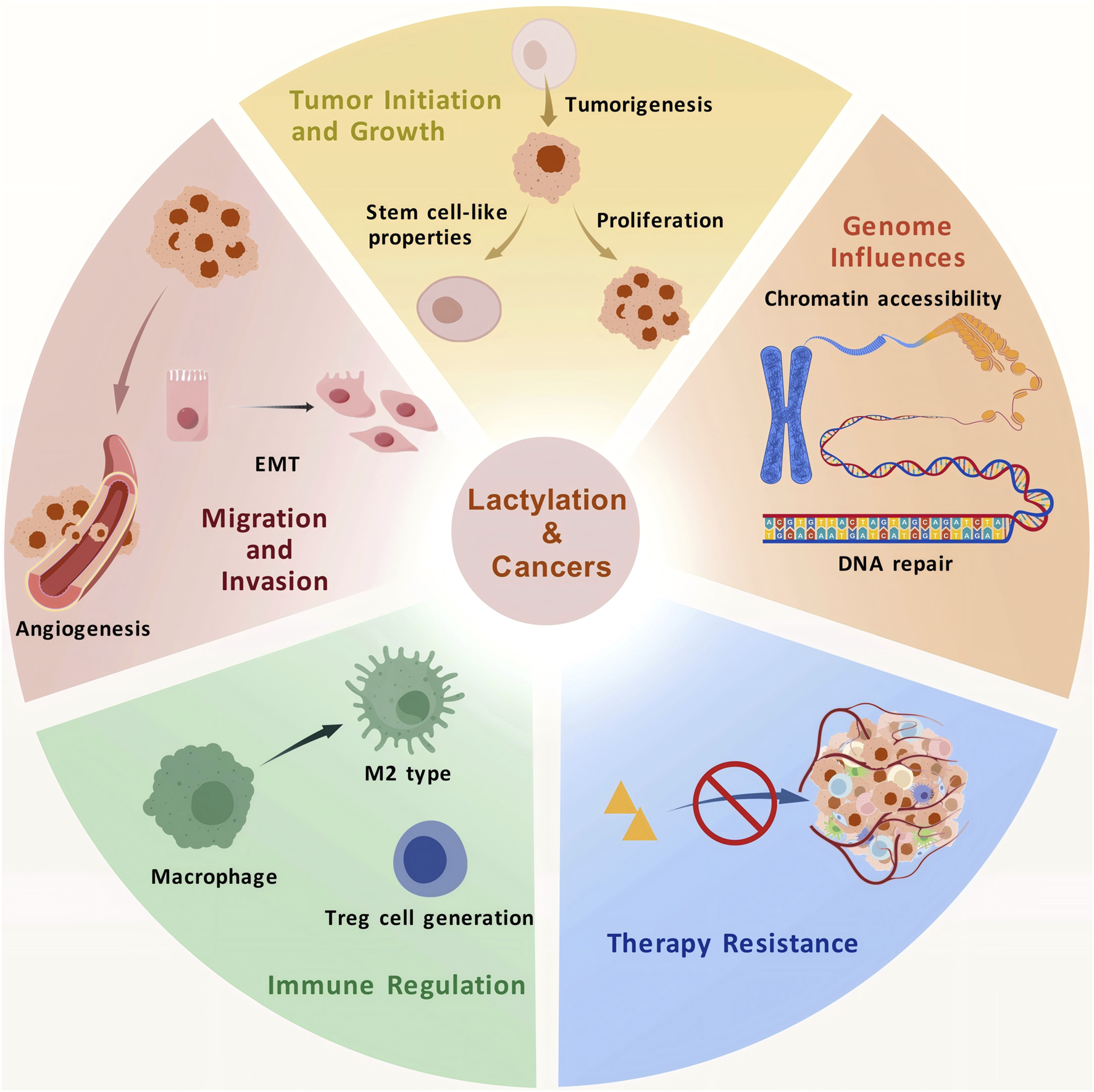
The roles of lactylation in tumor progression.
5 Predicting clinical prognosis using lactylation-related genes
With the emergence of new bioinformatic technologies, such as machine learning, and publicly available databases, clinicians can analyze gene expression profiles to predict patient prognosis in cancer (Yin et al., 2024; Zheng et al., 2024; Pan et al., 2024; Cai et al., 2023). Studies have shown that lactylation is closely associated with the clinical outcomes of patients with cancer (Chen et al., 2024d; Lu X. et al., 2024; Liu J. et al., 2024). Feng et al. used machine learning algorithms to construct a risk model for CM based on lactylation-related genes using data from the TCGA and GEO databases. Eight genes, namely, TMSB4X, CRABP2, WAS, LAP3, SATB1, GATAD2A, CALML5, and MDC1 were identified as candidate lactylation-related genes for constructing the model. Patients in the TCGA-CM cohort were divided into high- and low-risk groups based on the median risk score. In the high-risk group, the expression levels of CRABP2, GATAD2A, and MDC1 were substantially high, whereas those of LAP3, SATB1, and WAS were low. In both TCGA and GEO cohorts, survival analysis revealed a negative correlation between the risk scores and overall survival (OS) in patients with CM (Feng et al., 2023). In another study, machine learning revealed SMAD4, KRAS, TTN, CDKN2A, and TP53 as the top five mutated genes in PDAC. In addition, the researchers investigated mutation trends in different PDAC subgroups based on lactylation scores. The tumor mutation burden (TMB) was positively correlated with lactylation scores, with high-scoring clusters having a greater TMB than low-scoring clusters. Compared with patients with low TMB and lactylation scores, those with high TMB and lactylation scores had notably shorter OS (Peng et al., 2024). In a study on cervical cancer, lactylation-related genes (KRGs) were used to classify tumors into two subtypes, C1 and C2. The C2 cluster exhibited suppressed glycolysis and enhanced OXPHOS, which corresponded to higher survival rates. Additionally, a prognostic model based on two lactate signaling genes, ISY1 and PPP1R14B, was developed. A negative correlation was observed between PPP1R14B expression and CD8+ T cell infiltration, which was associated with low survival rates. The prognostic model was validated in two independent cohorts of patients with cervical cancer, one treated in Uganda (n = 106) and the other in Seoul (n = 300). Survival analysis revealed considerably lower OS or disease-free survival (DFS) rates among high-risk patients (He et al., 2024). Chao et al. investigated the relationship between lactylation and OS or progression-free survival (PFS) in ovarian cancer. Higher H3K18la levels were associated with worse PFS (p < 0.001) and OS (p = 0.028). Multivariate Cox regression analysis showed that H3K18la was an independent risk factor for PFS (p = 0.001). Moreover, H3K18la levels were correlated with the tumor stage (p = 0.037). Altogether, high H3K18la levels were associated with a poor prognosis in epithelial ovarian cancer (Chao et al., 2024).
Understanding the role of lactylation in cancer prognosis can assist clinicians in devising personalized treatment plans, evaluating therapeutic efficacy, and monitoring adverse reactions (Hao et al., 2024; Yu et al., 2024). As mentioned earlier, lactylation is found in various tumor types; however, studies investigating its clinical prognostic value remain limited. Future research in this area is essential to broaden the understanding and application of lactylation-related biomarkers in cancer.
6 Therapeutic strategies targeting lactylation
Lactylation plays an important role in the development and progression of various tumors, making it a promising therapeutic target for cancer (Li X. M. et al., 2024; Sun L. et al., 2023). With the deepening of research on lactylation in recent years, numerous targeted therapies have been developed for cancer, primarily encompassing the following three categories: inhibitors of lactate metabolism, lactate transporter proteins, and lactylation sites (Figure 1).
LDHA inhibitors, dexmedetomidine (Dex), and fargesin (FGS) are inhibitors of lactate metabolism. In patients with HCC, the combination of anti-PD-1 therapy and LDHA inhibitors has demonstrated stronger anti-tumor effects than monotherapy with anti-PD-1 antibodies (Gu et al., 2022b). In GBM, the LDHA inhibitor oxamate can enhance the efficacy of CAR-T cell therapy by inhibiting the lactylation of exonucleases and C-C motif chemokine receptor 8 (CCR8) (Sun T. et al., 2023). In addition, Dex can suppress glycolysis and reduce lactylated c-Myc levels, thereby decreasing the stability of c-Myc protein and inhibiting the migratory and invasive abilities and glycolytic activity of GBM cells (Zhu and Zhang, 2024). In NSCLC, FGS interacts with PKM2 to regulate glycolysis and pyruvate metabolism, suppressing lactate production and LDHA expression. Moreover, it inhibits tumor-associated histone lactylation, thereby exerting anti-tumor effects (Guo et al., 2024).
As mentioned earlier, lactate is transported between cells via MCTs. MCT4, located on glycolytic tumor cells, transports the lactate produced by these cells to the intercellular space. This lactate is subsequently internalized by oxidative tumor cells via MCT1. BAY-8002 and AZD3965 are two potent inhibitors of MCT1 that can alter its open conformation and inhibit lactate transport in the TME (Wang N. et al., 2021).
Inhibitors of lactylation sites directly target lactylation to achieve anti-cancer effects. Accumulation of lactate in CRC cells activates the transcription of the m5C methyltransferase NSUN2 via H3K18la. Lactylation of NSUN2 at K356 contributes to its oncogenic role. Therefore, combining NSUN2 inhibitors with immunotherapy represents a promising therapeutic strategy for CRC (Chen B. et al., 2024). Cancer stem cells can reduce the effectiveness of anti-cancer medications by driving tumor development, progression, and recurrence. A triterpenoid anti-cancer substance called dihydrocelastrol (DML) prevents carcinogenesis in liver cancer by interfering with histone lactylation induced by metabolic stress (Pan et al., 2022). Royal jelly acid, a component of traditional Chinese medicine, exerts inhibitory effects on lactylation. For instance, it suppresses H3K9la and H3K14la to reduce tumorigenicity in HCC (Xu et al., 2023).
Effective therapeutic strategies targeting lactylation are lacking at present. Therefore, developing broad-spectrum inhibitors of lactylation is necessary. Lactylation plays an oncogenic role in various tumor types, thus serving as a promising therapeutic target for cancer. Further studies are warranted to explore and optimize lactylation-targeted strategies for the treatment of cancer in clinical settings.
7 Discussion
While lactylation research is still in its early stages, emerging evidence highlights lactylation’s potential as both a diagnostic/prognostic biomarker and a therapeutic target across diverse cancer types. With continued investigation into its regulatory enzymes, context-specific roles, and clinical translation, lactylation-based strategies may pave the way for more precise, metabolism-driven cancer therapies that can enhance patient outcomes and overcome resistance mechanisms.
Despite the growing interest in targeting lactylation as a cancer therapeutic strategy, several critical knowledge gaps and shortcomings hinder its clinical translation. First, the enzymatic machinery that governs histone lactylation remains incompletely characterized. Although p300 and HBO1 have been identified as potential lactyltransferases, the full complement of “writers,” “erasers,” and “readers” of lactylation is largely unknown, including the enzymes responsible for generating and regulating lactyl-CoA. This limited understanding impairs our ability to design specific modulators of lactylation. Second, most of the compounds described—such as LDHA inhibitors, oxamate, and dexmedetomidine—impact broad metabolic or epigenetic pathways rather than selectively targeting lactylation, raising concerns about off-target effects and therapeutic specificity.
Another significant gap lies in the tumor-type specificity and context dependency of lactylation. While lactylation appears to promote tumor progression in various cancers, it is not yet clear why certain tumor types rely more heavily on lactylation or how tumor microenvironmental differences shape lactylation dynamics. This lack of precision complicates the development of broadly effective therapies. Moreover, there is a noticeable absence of clinically validated biomarkers or non-invasive tools to measure lactylation activity in patients, making it difficult to stratify patients or monitor therapeutic responses.
Importantly, although some studies suggest lactylation plays a role in modulating the immune microenvironment—such as through macrophage polarization and Treg activation—detailed mechanistic insights into how lactylation-targeted therapies influence immune cell function are missing. Lastly, while initial combinations with immunotherapies have shown promise, systematic evaluations of combination strategies and potential resistance mechanisms to lactylation-targeted agents are lacking. Addressing these gaps through mechanistic, translational, and clinical research will be essential for realizing the full therapeutic potential of lactylation in cancer.
8 Conclusion
Lactylation, a PTM first identified in 2019, has received increased attention from researchers over the past 5 years for its important role in cancer. It not only participates in tumor pathogenesis but also serves as a potential prognostic biomarker and therapeutic target. Studies have demonstrated that lactylation impacts key malignant features, such as tumor initiation, growth, metastasis, immune regulation, and therapeutic resistance. Furthermore, genes associated with lactylation are closely related to clinical outcomes, highlighting the potential of lactylation as a therapeutic target. However, using lactylation as a diagnostic biomarker or therapeutic target is associated with several challenges. As discussed earlier, lactylation sites associated biological processes, and the regulatory molecules or pathways involved in lactylation vary across tumor types, complicating the development of lactylation as a universal therapeutic target for cancer. Moreover, due to the lack of universal and efficient detection methods, research on the prediction of patient clinical prognosis related to lactylation-modified genes is still insufficient in many tumor types.In addition, effective and selective lactylation inhibitors are still lacking. During the development of new inhibitors, the possibility of off-target effects, the challenges of tissue-specific drug delivery, and the need for reliable biomarkers to monitor treatment responses should also be taken into account. Despite these challenges, lactylation holds promise as a highly effective diagnostic/prognostic biomarker and therapeutic target for cancer owing to its unique role in disease progression.
Existing studies have introduced valuable avenues for investigating the pathophysiological, diagnostic, prognostic, and therapeutic roles of lactylation in tumors. Future research directions may focus on high-throughput screening to identify lactylation regulators, in vivo studies to test therapeutic effects, development of tissue-specific delivery systems, or the use of advanced techniques such as ChIP-seq or single-cell RNA-seq to describe lactylation dynamics in the tumor microenvironment. Further in-depth and innovative research will improve the understanding of the role of lactylation in tumor progression, laying a robust theoretical foundation for the clinical application of lactylation-targeted strategies. This progress may significantly benefit patients that alleviating the disease burdens caused by malignant tumors.
Statements
Author contributions
ZG: Writing – review and editing, Methodology, Software, Conceptualization, Writing – original draft, Investigation. QS: Validation, Investigation, Writing – review and editing. JL: Writing – review and editing, Investigation, Supervision, Funding acquisition.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by the Financial Department of Jilin Province (grant number 2024WSZY-D01).
Acknowledgments
We used BioGDP (BioGDP.com) to create our figures (Jiang S. et al., 2024).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Glossary
- TME
tumor microenvironment
- PTM
post-translational modification
- OXPHOS
oxidative phosphorylation
- MCTs
monocarboxylate transporters
- CAFs
cancer-associated fibroblasts
- HAT
histone acetyltransferase
- HDACs
histone deacetylases
- Kla
lysine lactylation
- MS/MS
mass spectrometry
- HPLC
high-performance liquid chromatography
- H3K1
histone H3 lysine 1
- Arg1
arginase-1
- PKM2Pyruvate kinase M2TIMs
Pyruvate kinase M2TIMstumor-infiltrating myeloid cells
- Treg
regulatory T cells
- MOESIN
membrane-organizing extension spike protein
- TGF-β
transforming growth factor-β
- HMGB1
high mobility group box-1
- GC
gastric cancer
- HIF1α
hypoxia-inducible factor 1α
- PCa
prostate cancer
- AARS1
Alanyl-tRNA synthetase 1
- GLO1
glyoxalase 1
- TAC
tricarboxylic acid cycle
- LGSH
lactylglutathione
- SHMT2
serine hydroxymethyltransferase 2
- EC
esophageal cancer
- MTHFD1L
mitochondrial monofunctional C1-tetrahydrofolate synthase
- H3K18la
H3K18 lactylation
- PDAC
pancreatic ductal adenocarcinoma
- E3BP
E3 binding protein
- PDC
dehydrogenase complex
- PPP
pentose phosphate pathway
- NSCLC
non-small cell lung cancer
- APOC2
apolipoprotein C-II
- LDHA
lactate dehydrogenase A
- ULK1
UNC-51-like kinase 1
- PIK3C3
phosphoinositide-3-kinase Class 3
- Vps34
vacuolar protein sorting 34
- DCBLD1
Discoidin
- CUB
and LCCL domain-containing protein 1
- G6PD
glucose-6-phosphate dehydrogenase
- CRC
colorectal cancer
- eEF1A2
Elongation Factor 1 Alpha
- CM
cutaneous melanoma
- YTHDF2
YTH N6-methyladenosine RNA-binding protein 2
- Kcnk6
Potassium Channel Subfamily K Member 6
- Nlrp3
NOD-like receptor thermal protein domain-associated protein 3
- IGF1
Insulin-like growth factor 1
- YY1
Yin and Yang 1 protein
- CCND1
Cyclin D1
- NRP2
Neuropilin 2
- HCC
hepatocellular carcinoma
- SRSF10
Serine/Arginine-Rich Splicing Factor 10
- Bcl-x
BCL2-like 1
- MDM4
Murine Double Minute 4
- ATC
anaplastic thyroid cancer
- SLFN5
Schlafen 5
- LCSCs
Liver cancer stem cells
- ALDOA
Aldolase A
- GBM
glioblastoma
- POLR2A
Polymerase II
- MAP4K4
Mitogen-Activated Protein Kinase Kinase Kinase Kinase 4
- RCC
Renal cell carcinoma
- ccRCC
clear cell renal cell carcinoma
- FKBP10
FK506 Binding Protein 10
- GPR37
G Protein Coupled Receptor 37
- CXCL1
chemokine CXC motif ligand 1
- VCAM1
Vascular Cell Adhesion Molecule 1
- PYCR1
Proline-5-Carboxylate Reductase 1
- IRS1
Insulin Receptor Substrate 1
- EMTEpithelial-mesenchymal transitionGLUT3
Epithelial-mesenchymal transitionGLUT3Glucose Transporter 3
- LOX
lysyl oxidase;
- RHOF
Rho GTPase Rif
- ESM1
Endothelial-Specific Molecule 1
- VEGFA
Vascular Endothelial Growth Factor A
- VM
vasculogenic mimicry
- GP73
Golgi Phosphoprotein 73
- STAT3
Signal Transducer and Activator of Transcription 3
- AICD
activation-induced cell death
- POM121Pore Membrane Protein 121TAM
Pore Membrane Protein 121TAMstumor-associated macrophages
- NB
neuroblastoma
- HK3
Hexokinase-3
- PCSK9
Proprotein Convertase Subtilisin/Kexin Type 9
- MIF
macrophage migration inhibitory factor
- GPD2
Glycerol-3-Phosphate Dehydrogenase 2
- MDMs
monocyte-derived macrophages
- AML
acute myeloid leukemia
- METTL3
methyltransferase-like 3
- ALKBH3
AlkB homolog 3
- SP100A
Speckled Protein 100A
- PML
promyelocytic leukemia
- NEPC
neuroendocrine prostate cancer;
- ZEB1
Zinc Finger E-Box Binding Homeobox 1
- m1A
N1-methyladenosine
- NBS1
Nijmegen Breakage Syndrome Protein 1
- K388
lysine 388
- MRN
MRE11-RAD50-NBS1
- BECN1
Beclin 1
- BCa
Bladder cancer
- RUBCNL
Recombinant Rubicon-Like Autophagy Enhancer Protein
- YBX1
Y-box binding protein 1
- OSCC
oral squamous cell carcinoma
- ALDH1A3
Aldehyde Dehydrogenase 1 Family, Member A3
- GSCs
glioblastoma stem cells
- XRCC1
X-ray Repair Cross-Complementing Protein 1
- TMZ
Temozolomide
- ICB
Immune checkpoint blockade
- LUSC
lung squamous cell carcinoma
- SLC2A1
solute carrier family 2 member 1
- HNSCC
head and neck squamous cell carcinoma
- IL-11
interleukin-11
- CRLM
colorectal liver metastasis
- RIG-I
retinoic acid-inducible gene 1
- HBV
hepatitis B virus
- lncRNAslong non-coding RNAsLPS
long non-coding RNAsLPSlipopolysaccharides
- LKB1
liver kinase B1
- TERT
telomerase reverse transcriptase
- TFEB
Transcription factor EB
- KAT5
Lysine Acetyltransferase 5
- OS
overall survival
- TMB
Tumor mutation burden
- KRGs
lactylation-related genes
- DFS
disease-free survival
- PFS
progression-free survival
- CCR8
C-C Motif Chemokine Receptor 8
- Dex
dexmedetomidine
- K356
lysine 356
- DML
Dihydrocelastrol
- FGS
Folylpolyglutamate synthase
References
1
Apostolova P. Pearce E. L. (2022). Lactic acid and lactate: revisiting the physiological roles in the tumor microenvironment. Trends Immunol.43 (12), 969–977. 10.1016/j.it.2022.10.005
2
Bhat S. M. Badiger V. A. Vasishta S. Chakraborty J. Prasad S. Ghosh S. et al (2021). 3D tumor angiogenesis models: recent advances and challenges. J. Cancer Res. Clin. Oncol.147 (12), 3477–3494. 10.1007/s00432-021-03814-0
3
Brooks G. A. (2018). The science and translation of lactate Shuttle theory. Cell Metab.27 (4), 757–785. 10.1016/j.cmet.2018.03.008
4
Brooks G. A. (2020). Lactate as a fulcrum of metabolism. Redox Biol.35, 101454. 10.1016/j.redox.2020.101454
5
Cai D. Yuan X. Cai D. Q. Li A. Yang S. Yang W. et al (2023). Integrative analysis of lactylation-related genes and establishment of a novel prognostic signature for hepatocellular carcinoma. J. Cancer Res. Clin. Oncol.149 (13), 11517–11530. 10.1007/s00432-023-04947-0
6
Cai J. Song L. Zhang F. Wu S. Zhu G. Zhang P. et al (2024). Targeting SRSF10 might inhibit M2 macrophage polarization and potentiate anti-PD-1 therapy in hepatocellular carcinoma. Cancer Commun. (Lond)44, 1231–1260. 10.1002/cac2.12607
7
Carneiro L. Asrih M. Repond C. Sempoux C. Stehle J. C. Leloup C. et al (2017). AMPK activation caused by reduced liver lactate metabolism protects against hepatic steatosis in MCT1 haploinsufficient mice. Mol. Metab.6 (12), 1625–1633. 10.1016/j.molmet.2017.10.005
8
Certo M. Tsai C. H. Pucino V. Ho P. C. Mauro C. (2021). Lactate modulation of immune responses in inflammatory versus tumour microenvironments. Nat. Rev. Immunol.21 (3), 151–161. 10.1038/s41577-020-0406-2
9
Chao J. Chen G. d. Huang S. T. Gu H. Liu Y. Y. Luo Y. et al (2024). High histone H3K18 lactylation level is correlated with poor prognosis in epithelial ovarian cancer. Neoplasma71 (4), 319–332. 10.4149/neo_2024_240127N41
10
Chaudagar K. Hieromnimon H. M. Kelley A. Labadie B. Shafran J. Rameshbabu S. et al (2023). Suppression of tumor cell lactate-generating signaling pathways Eradicates murine PTEN/p53-deficient Aggressive-variant prostate cancer via macrophage Phagocytosis. Clin. Cancer Res.29 (23), 4930–4940. 10.1158/1078-0432.CCR-23-1441
11
Chen A. N. Luo Y. Yang Y. H. Fu J. T. Geng X. M. Shi J. P. et al (2021). Lactylation, a novel metabolic reprogramming Code: Current Status and prospects. Front. Immunol.12, 688910. 10.3389/fimmu.2021.688910
12
Chen B. Deng Y. Hong Y. Fan L. Zhai X. Hu H. et al (2024e). Metabolic Recoding of NSUN2-mediated m(5)C modification promotes the progression of colorectal cancer via the NSUN2/YBX1/m(5)C-ENO1 positive feedback loop. Adv. Sci. (Weinh)11 (28), e2309840. 10.1002/advs.202309840
13
Chen H. Li Y. Li H. Chen X. Fu H. Mao D. et al (2024a). NBS1 lactylation is required for efficient DNA repair and chemotherapy resistance. Nature631 (8021), 663–669. 10.1038/s41586-024-07620-9
14
Chen J. Zhao D. Wang Y. Liu M. Zhang Y. Feng T. et al (2024c). Lactylated apolipoprotein C-II induces immunotherapy resistance by promoting extracellular lipolysis. Adv. Sci. (Weinh)11 (38), e2406333. 10.1002/advs.202406333
15
Chen Y. Chang L. Hu L. Yan C. Dai L. Shelat V. G. et al (2024d). Identification of a lactylation-related gene signature to characterize subtypes of hepatocellular carcinoma using bulk sequencing data. J. Gastrointest. Oncol.15 (4), 1636–1646. 10.21037/jgo-24-405
16
Chen Y. Wu J. Zhai L. Zhang T. Yin H. Gao H. et al (2024b). Metabolic regulation of homologous recombination repair by MRE11 lactylation. Cell187 (2), 294–311 e21. 10.1016/j.cell.2023.11.022
17
Chiodi I. Mondello C. (2020). Life style factors, tumor cell plasticity and cancer stem cells. Mutat. Res. Rev. Mutat. Res.784, 108308. 10.1016/j.mrrev.2020.108308
18
Chu Y. D. Cheng L. C. Lim S. N. Lai M. W. Yeh C. T. Lin W. R. (2023). Aldolase B-driven lactagenesis and CEACAM6 activation promote cell renewal and chemoresistance in colorectal cancer through the Warburg effect. Cell Death Dis.14 (10), 660. 10.1038/s41419-023-06187-z
19
Dai S. K. Liu P. P. Li X. Jiao L. F. Teng Z. Q. Liu C. M. (2022b). Dynamic profiling and functional interpretation of histone lysine crotonylation and lactylation during neural development. Development149 (14), dev200049. 10.1242/dev.200049
20
Dai X. Lv X. Thompson E. W. Ostrikov K. K. (2022a). Histone lactylation: epigenetic mark of glycolytic switch. Trends Genet.38 (2), 124–127. 10.1016/j.tig.2021.09.009
21
De Leo A. Ugolini A. Yu X. Scirocchi F. Scocozza D. Peixoto B. et al (2024). Glucose-driven histone lactylation promotes the immunosuppressive activity of monocyte-derived macrophages in glioblastoma. Immunity57 (5), 1105–1123 e8. 10.1016/j.immuni.2024.04.006
22
Dongre A. Weinberg R. A. (2019). New insights into the mechanisms of epithelial-mesenchymal transition and implications for cancer. Nat. Rev. Mol. Cell Biol.20 (2), 69–84. 10.1038/s41580-018-0080-4
23
Duan W. Liu W. Xia S. Zhou Y. Tang M. Xu M. et al (2023). Warburg effect enhanced by AKR1B10 promotes acquired resistance to pemetrexed in lung cancer-derived brain metastasis. J. Transl. Med.21 (1), 547. 10.1186/s12967-023-04403-0
24
Elia I. Rowe J. H. Johnson S. Joshi S. Notarangelo G. Kurmi K. et al (2022). Tumor cells dictate anti-tumor immune responses by altering pyruvate utilization and succinate signaling in CD8(+) T cells. Cell Metab.34 (8), 1137–1150 e6. 10.1016/j.cmet.2022.06.008
25
Fan W. Zeng S. Wang X. Wang G. Liao D. Li R. et al (2024). A feedback loop driven by H3K9 lactylation and HDAC2 in endothelial cells regulates VEGF-induced angiogenesis. Genome Biol.25 (1), 165. 10.1186/s13059-024-03308-5
26
Fang X. Zhao P. Gao S. Liu D. Zhang S. Shan M. et al (2023). Lactate induces tumor-associated macrophage polarization independent of mitochondrial pyruvate carrier-mediated metabolism. Int. J. Biol. Macromol.237, 123810. 10.1016/j.ijbiomac.2023.123810
27
Faubert B. Li K. Y. Cai L. Hensley C. T. Kim J. Zacharias L. G. et al (2017). Lactate metabolism in human lung tumors. Cell171 (2), 358–371 e9. 10.1016/j.cell.2017.09.019
28
Feng F. Wu J. Chi Q. Wang S. Liu W. Yang L. et al (2024). Lactylome analysis Unveils lactylation-dependent mechanisms of stemness remodeling in the liver cancer stem cells. Adv. Sci. (Weinh)11 (38), e2405975. 10.1002/advs.202405975
29
Feng H. Chen W. Zhang C. (2023). Identification of lactylation gene CALML5 and its correlated lncRNAs in cutaneous melanoma by machine learning. Med. Baltim.102 (47), e35999. 10.1097/MD.0000000000035999
30
Fu C. Jiang W. Wang C. Song S. J. Tao H. Zhang X. G. et al (2024). AP001885.4 promotes the proliferation of esophageal squamous cell carcinoma cells by histone lactylation- and NF-κB (p65)-dependent transcription activation and METTL3-mediated mRNA stability of c-myc. Anim. Cells Syst. Seoul.28 (1), 536–550. 10.1080/19768354.2024.2417458
31
Gaffney D. O. Jennings E. Q. Anderson C. C. Marentette J. O. Shi T. Schou Oxvig A. M. et al (2020). Non-enzymatic lysine Lactoylation of glycolytic enzymes. Cell Chem. Biol.27 (2), 206–213 e6. 10.1016/j.chembiol.2019.11.005
32
Gaur P. Prasad S. Kumar B. Sharma S. K. Vats P. (2021). High-altitude hypoxia induced reactive oxygen species generation, signaling, and mitigation approaches. Int. J. Biometeorol.65 (4), 601–615. 10.1007/s00484-020-02037-1
33
Gu J. Xu X. Li X. Yue L. Zhu X. Chen Q. et al (2024b). Tumor-resident microbiota contributes to colorectal cancer liver metastasis by lactylation and immune modulation. Oncogene43 (31), 2389–2404. 10.1038/s41388-024-03080-7
34
Gu J. Zhou J. Chen Q. Xu X. Gao J. Li X. et al (2022a). Tumor metabolite lactate promotes tumorigenesis by modulating MOESIN lactylation and enhancing TGF-beta signaling in regulatory T cells. Cell Rep.39 (12), 110986. 10.1016/j.celrep.2022.110986
35
Gu J. Zhou J. Chen Q. Xu X. Gao J. Li X. et al (2022b). Tumor metabolite lactate promotes tumorigenesis by modulating MOESIN lactylation and enhancing TGF-beta signaling in regulatory T cells. Cell Rep.40 (3), 111122. 10.1016/j.celrep.2022.111122
36
Gu X. Zhuang A. Yu J. Yang L. Ge S. Ruan J. et al (2024a). Histone lactylation-boosted ALKBH3 potentiates tumor progression and diminished promyelocytic leukemia protein nuclear condensates by m1A demethylation of SP100A. Nucleic Acids Res.52 (5), 2273–2289. 10.1093/nar/gkad1193
37
Guo Z. Tang Y. Wang S. Huang Y. Chi Q. Xu K. et al (2024). Natural product fargesin interferes with H3 histone lactylation via targeting PKM2 to inhibit non-small cell lung cancer tumorigenesis. Biofactors50 (3), 592–607. 10.1002/biof.2031
38
Han J. Won M. Kim J. H. Jung E. Min K. Jangili P. et al (2020). Cancer stem cell-targeted bio-imaging and chemotherapeutic perspective. Chem. Soc. Rev.49 (22), 7856–7878. 10.1039/d0cs00379d
39
Han S. Bao X. Zou Y. Wang L. Li Y. Yang L. et al (2023). d-lactate modulates M2 tumor-associated macrophages and remodels immunosuppressive tumor microenvironment for hepatocellular carcinoma. Sci. Adv.9 (29), eadg2697. 10.1126/sciadv.adg2697
40
Hao B. Dong H. Xiong R. Song C. Xu C. Li N. et al (2024). Identification of SLC2A1 as a predictive biomarker for survival and response to immunotherapy in lung squamous cell carcinoma. Comput. Biol. Med.171, 108183. 10.1016/j.compbiomed.2024.108183
41
He C. Zhang J. Bai X. Lu C. Zhang K. (2024). Lysine lactylation-based insight to understanding the characterization of cervical cancer. Biochim. Biophys. Acta Mol. Basis Dis.1870 (7), 167356. 10.1016/j.bbadis.2024.167356
42
Hirschey M. D. Zhao Y. (2015). Metabolic regulation by lysine Malonylation, Succinylation, and Glutarylation. Mol. Cell Proteomics14 (9), 2308–2315. 10.1074/mcp.R114.046664
43
Hou X. Ouyang J. Tang L. Wu P. Deng X. Yan Q. et al (2024). KCNK1 promotes proliferation and metastasis of breast cancer cells by activating lactate dehydrogenase A (LDHA) and up-regulating H3K18 lactylation. PLoS Biol.22 (6), e3002666. 10.1371/journal.pbio.3002666
44
Huang C. Xue L. Lin X. Shen Y. Wang X. (2024b). Histone lactylation-driven GPD2 mediates M2 macrophage polarization to promote malignant transformation of cervical cancer progression. DNA Cell Biol.43, 605–618. 10.1089/dna.2024.0122
45
Huang G. Chen S. He J. Li H. Ma Z. Lubamba G. P. et al (2024c). Histone lysine lactylation (Kla)-induced BCAM promotes OSCC progression and cis-Platinum resistance. Oral Dis.31, 1116–1132. 10.1111/odi.15179
46
Huang H. Sabari B. R. Garcia B. A. Allis C. D. Zhao Y. (2014). SnapShot: histone modifications. Cell159 (2), 458–458 e1. 10.1016/j.cell.2014.09.037
47
Huang H. Wang S. Xia H. Zhao X. Chen K. Jin G. et al (2024a). Lactate enhances NMNAT1 lactylation to sustain nuclear NAD(+) salvage pathway and promote survival of pancreatic adenocarcinoma cells under glucose-deprived conditions. Cancer Lett.588, 216806. 10.1016/j.canlet.2024.216806
48
Huang Y. Luo G. Peng K. Song Y. Wang Y. Zhang H. et al (2024d). Lactylation stabilizes TFEB to elevate autophagy and lysosomal activity. J. Cell Biol.223 (11), e202308099. 10.1083/jcb.202308099
49
Huang Z. W. Zhang X. N. Zhang L. Liu L. L. Zhang J. W. Sun Y. X. et al (2023). STAT5 promotes PD-L1 expression by facilitating histone lactylation to drive immunosuppression in acute myeloid leukemia. Signal Transduct. Target Ther.8 (1), 391. 10.1038/s41392-023-01605-2
50
Hui S. Ghergurovich J. M. Morscher R. J. Jang C. Teng X. Lu W. et al (2017). Glucose feeds the TCA cycle via circulating lactate. Nature551 (7678), 115–118. 10.1038/nature24057
51
Icard P. Shulman S. Farhat D. Steyaert J. M. Alifano M. Lincet H. (2018). How the Warburg effect supports aggressiveness and drug resistance of cancer cells?Drug Resist Updat38, 1–11. 10.1016/j.drup.2018.03.001
52
Ippolito L. Morandi A. Giannoni E. Chiarugi P. (2019). Lactate: a metabolic driver in the tumour landscape. Trends Biochem. Sci.44 (2), 153–166. 10.1016/j.tibs.2018.10.011
53
Irizarry-Caro R. A. McDaniel M. M. Overcast G. R. Jain V. G. Troutman T. D. Pasare C. (2020). TLR signaling adapter BCAP regulates inflammatory to reparatory macrophage transition by promoting histone lactylation. Proc. Natl. Acad. Sci. U. S. A.117 (48), 30628–30638. 10.1073/pnas.2009778117
54
Jhunjhunwala S. Hammer C. Delamarre L. (2021). Antigen presentation in cancer: insights into tumour immunogenicity and immune evasion. Nat. Rev. Cancer21 (5), 298–312. 10.1038/s41568-021-00339-z
55
Jia M. Yue X. Sun W. Zhou Q. Chang C. Gong W. et al (2023). ULK1-mediated metabolic reprogramming regulates Vps34 lipid kinase activity by its lactylation. Sci. Adv.9 (22), eadg4993. 10.1126/sciadv.adg4993
56
Jiang J. Zheng Q. Zhu W. Chen X. Lu H. Chen D. et al (2020). Alterations in glycolytic/cholesterogenic gene expression in hepatocellular carcinoma. Aging (Albany NY)12 (11), 10300–10316. 10.18632/aging.103254
57
Jiang S. Li H. Zhang L. Mu W. Zhang Y. Chen T. et al (2024b). Generic Diagramming Platform (GDP): a comprehensive database of high-quality biomedical graphics. Nucleic Acids Res.53, D1670–D1676. 10.1093/nar/gkae973
58
Jiang Z. Xiong N. Yan R. Li S. T. Liu H. Mao Q. et al (2024a). PDHX acetylation facilitates tumor progression by disrupting PDC assembly and activating lactylation mediated gene expression. Protein Cell16, 49–63. 10.1093/procel/pwae052
59
Jiao Y. Ji F. Hou L. Lv Y. Zhang J. (2024). Lactylation-related gene signature for prognostic prediction and immune infiltration analysis in breast cancer. Heliyon10 (3), e24777. 10.1016/j.heliyon.2024.e24777
60
Jin M. Cao W. Chen B. Xiong M. Cao G. (2022). Tumor-derived lactate creates a Favorable Niche for tumor via Supplying energy source for tumor and modulating the tumor microenvironment. Front. Cell Dev. Biol.10, 808859. 10.3389/fcell.2022.808859
61
Jing F. Zhu L. Zhang J. Zhou X. Bai J. Li X. et al (2024). Multi-omics reveals lactylation-driven regulatory mechanisms promoting tumor progression in oral squamous cell carcinoma. Genome Biol.25 (1), 272. 10.1186/s13059-024-03383-8
62
Ju J. Zhang H. Lin M. Yan Z. An L. Cao Z. et al (2024). The alanyl-tRNA synthetase AARS1 moonlights as a lactyltransferase to promote YAP signaling in gastric cancer. J. Clin. Invest134 (10), e174587. 10.1172/JCI174587
63
Lan J. Luo H. Wu R. Wang J. Zhou B. Zhang Y. et al (2020). Internalization of HMGB1 (high mobility group box 1) promotes angiogenesis in endothelial cells. Arterioscler. Thromb. Vasc. Biol.40 (12), 2922–2940. 10.1161/ATVBAHA.120.315151
64
Lei X. Lei Y. Li J. K. Du W. X. Li R. G. Yang J. et al (2020). Immune cells within the tumor microenvironment: biological functions and roles in cancer immunotherapy. Cancer Lett.470, 126–133. 10.1016/j.canlet.2019.11.009
65
Li C. Jiang P. Wei S. Xu X. Wang J. (2020). Regulatory T cells in tumor microenvironment: new mechanisms, potential therapeutic strategies and future prospects. Mol. Cancer19 (1), 116. 10.1186/s12943-020-01234-1
66
Li F. Si W. Xia L. Yin D. Wei T. Tao M. et al (2024b). Positive feedback regulation between glycolysis and histone lactylation drives oncogenesis in pancreatic ductal adenocarcinoma. Mol. Cancer23 (1), 90. 10.1186/s12943-024-02008-9
67
Li F. Zhang H. Huang Y. Li D. Zheng Z. Xie K. et al (2024h). Single-cell transcriptome analysis reveals the association between histone lactylation and cisplatin resistance in bladder cancer. Drug Resist Updat73, 101059. 10.1016/j.drup.2024.101059
68
Li G. Wang D. Zhai Y. Pan C. Zhang J. Wang C. et al (2024). Glycometabolic reprogramming-induced XRCC1 lactylation confers therapeutic resistance in ALDH1A3-overexpressing glioblastoma. Cell Metab.36 (8), 1696–1710 e10. 10.1016/j.cmet.2024.07.011
69
Li H. Liu C. Li R. Zhou L. Ran Y. Yang Q. et al (2024a). AARS1 and AARS2 sense L-lactate to regulate cGAS as global lysine lactyltransferases. Nature634 (8036), 1229–1237. 10.1038/s41586-024-07992-y
70
Li J. Chen Z. Jin M. Gu X. Wang Y. Huang G. et al (2024c). Histone H4K12 lactylation promotes malignancy progression in triple-negative breast cancer through SLFN5 downregulation. Cell Signal124, 111468. 10.1016/j.cellsig.2024.111468
71
Li L. Li Z. Meng X. Wang X. Song D. Liu Y. et al (2023). Histone lactylation-derived LINC01127 promotes the self-renewal of glioblastoma stem cells via the cis-regulating the MAP4K4 to activate JNK pathway. Cancer Lett.579, 216467. 10.1016/j.canlet.2023.216467
72
Li Q. Lin G. Zhang K. Liu X. Li Z. Bing X. et al (2024d). Hypoxia exposure induces lactylation of Axin1 protein to promote glycolysis of esophageal carcinoma cells. Biochem. Pharmacol.226, 116415. 10.1016/j.bcp.2024.116415
73
Li W. Zhou C. Yu L. Hou Z. Liu H. Kong L. et al (2024g). Tumor-derived lactate promotes resistance to bevacizumab treatment by facilitating autophagy enhancer protein RUBCNL expression through histone H3 lysine 18 lactylation (H3K18la) in colorectal cancer. Autophagy20 (1), 114–130. 10.1080/15548627.2023.2249762
74
Li X. Yang Y. Zhang B. Lin X. Fu X. An Y. et al (2022). Lactate metabolism in human health and disease. Signal Transduct. Target Ther.7 (1), 305. 10.1038/s41392-022-01151-3
75
Li X. M. Yang Y. Jiang F. Q. Hu G. Wan S. Yan W. Y. et al (2024f). Histone lactylation inhibits RARγ expression in macrophages to promote colorectal tumorigenesis through activation of TRAF6-IL-6-STAT3 signaling. Cell Rep.43 (2), 113688. 10.1016/j.celrep.2024.113688
76
Li Z. Liang P. Chen Z. Chen Z. Jin T. He F. et al (2024e). CAF-secreted LOX promotes PD-L1 expression via histone Lactylation and regulates tumor EMT through TGFβ/IGF1 signaling in gastric Cancer. Cell Signal124, 111462. 10.1016/j.cellsig.2024.111462
77
Liao J. Chen Z. Chang R. Yuan T. Li G. Zhu C. et al (2023). CENPA functions as a transcriptional regulator to promote hepatocellular carcinoma progression via cooperating with YY1. Int. J. Biol. Sci.19 (16), 5218–5232. 10.7150/ijbs.85656
78
Liu J. Chen P. Zhou J. Li H. Pan Z. (2024c). Prognostic impact of lactylation-associated gene modifications in clear cell renal cell carcinoma: insights into molecular landscape and therapeutic opportunities. Environ. Toxicol.39 (3), 1360–1373. 10.1002/tox.24040
79
Liu M. Gu L. Zhang Y. Li Y. Zhang L. Xin Y. et al (2024b). LKB1 inhibits telomerase activity resulting in cellular senescence through histone lactylation in lung adenocarcinoma. Cancer Lett.595, 217025. 10.1016/j.canlet.2024.217025
80
Liu R. Zou Z. Chen L. Feng Y. Ye J. Deng Y. et al (2024a). FKBP10 promotes clear cell renal cell carcinoma progression and regulates sensitivity to the HIF2α blockade by facilitating LDHA phosphorylation. Cell Death Dis.15 (1), 64. 10.1038/s41419-024-06450-x
81
Lu W. Kang Y. (2019). Epithelial-mesenchymal plasticity in cancer progression and metastasis. Dev. Cell49 (3), 361–374. 10.1016/j.devcel.2019.04.010
82
Lu X. Zhou Z. Qiu P. Xin T. (2024b). Integrated single-cell and bulk RNA-sequencing data reveal molecular subtypes based on lactylation-related genes and prognosis and therapeutic response in glioma. Heliyon10 (9), e30726. 10.1016/j.heliyon.2024.e30726
83
Lu Y. Zhu J. Zhang Y. Li W. Xiong Y. Fan Y. et al (2024a). Lactylation-driven IGF2BP3-mediated serine metabolism reprogramming and RNA m6A-modification promotes Lenvatinib resistance in HCC. Adv. Sci. (Weinh)11, e2401399. 10.1002/advs.202401399
84
Luo Y. Yang Z. Yu Y. Zhang P. (2022). HIF1α lactylation enhances KIAA1199 transcription to promote angiogenesis and vasculogenic mimicry in prostate cancer. Int. J. Biol. Macromol.222 (Pt B), 2225–2243. 10.1016/j.ijbiomac.2022.10.014
85
Meng Q. Sun H. Zhang Y. Yang X. Hao S. Liu B. et al (2024). Lactylation stabilizes DCBLD1 activating the pentose phosphate pathway to promote cervical cancer progression. J. Exp. Clin. Cancer Res.43 (1), 36. 10.1186/s13046-024-02943-x
86
Miao Z. Zhao X. Liu X. (2023). Hypoxia induced beta-catenin lactylation promotes the cell proliferation and stemness of colorectal cancer through the wnt signaling pathway. Exp. Cell Res.422 (1), 113439. 10.1016/j.yexcr.2022.113439
87
Millan-Zambrano G. Burton A. Bannister A. J. Schneider R. (2022). Histone post-translational modifications - cause and consequence of genome function. Nat. Rev. Genet.23 (9), 563–580. 10.1038/s41576-022-00468-7
88
Moreno-Yruela C. Bæk M. Monda F. Olsen C. A. (2022a). Chiral Posttranslational modification to lysine epsilon-Amino groups. Acc. Chem. Res.55 (10), 1456–1466. 10.1021/acs.accounts.2c00115
89
Moreno-Yruela C. Zhang D. Wei W. Bæk M. Liu W. Gao J. et al (2022b). Class I histone deacetylases (HDAC1-3) are histone lysine delactylases. Sci. Adv.8 (3), eabi6696. 10.1126/sciadv.abi6696
90
Niu Z. Chen C. Wang S. Lu C. Wu Z. Wang A. et al (2024). HBO1 catalyzes lysine lactylation and mediates histone H3K9la to regulate gene transcription. Nat. Commun.15 (1), 3561. 10.1038/s41467-024-47900-6
91
Pan J. Zhang J. Lin J. Cai Y. Zhao Z. (2024). Constructing lactylation-related genes prognostic model to effectively predict the disease-free survival and treatment responsiveness in prostate cancer based on machine learning. Front. Genet.15, 1343140. 10.3389/fgene.2024.1343140
92
Pan L. Feng F. Wu J. Fan S. Han J. Wang S. et al (2022). Demethylzeylasteral targets lactate by inhibiting histone lactylation to suppress the tumorigenicity of liver cancer stem cells. Pharmacol. Res.181, 106270. 10.1016/j.phrs.2022.106270
93
Pandkar M. R. Sinha S. Samaiya A. Shukla S. (2023). Oncometabolite lactate enhances breast cancer progression by orchestrating histone lactylation-dependent c-Myc expression. Transl. Oncol.37, 101758. 10.1016/j.tranon.2023.101758
94
Peng T. Sun F. Yang J. C. Cai M. H. Huai M. X. Pan J. X. et al (2024). Novel lactylation-related signature to predict prognosis for pancreatic adenocarcinoma. World J. Gastroenterol.30 (19), 2575–2602. 10.3748/wjg.v30.i19.2575
95
Qiao Z. Li Y. Li S. Liu S. Cheng Y. (2024). Hypoxia-induced SHMT2 protein lactylation facilitates glycolysis and stemness of esophageal cancer cells. Mol. Cell Biochem.479 (11), 3063–3076. 10.1007/s11010-023-04913-x
96
Raychaudhuri D. Singh P. Chakraborty B. Hennessey M. Tannir A. J. Byregowda S. et al (2024). Histone lactylation drives CD8(+) T cell metabolism and function. Nat. Immunol.25 (11), 2140–2151. 10.1038/s41590-024-01985-9
97
Roland C. L. Arumugam T. Deng D. Liu S. H. Philip B. Gomez S. et al (2014). Cell surface lactate receptor GPR81 is crucial for cancer cell survival. Cancer Res.74 (18), 5301–5310. 10.1158/0008-5472.CAN-14-0319
98
Rothbart S. B. Dickson B. M. Strahl B. D. (2015). From histones to ribosomes: a chromatin regulator tangoes with translation. Cancer Discov.5 (3), 228–230. 10.1158/2159-8290.CD-15-0073
99
Sabari B. R. Zhang D. Allis C. D. Zhao Y. (2017). Metabolic regulation of gene expression through histone acylations. Nat. Rev. Mol. Cell Biol.18 (2), 90–101. 10.1038/nrm.2016.140
100
Sun L. Zhang Y. Yang B. Sun S. Zhang P. Luo Z. et al (2023c). Lactylation of METTL16 promotes cuproptosis via m(6)A-modification on FDX1 mRNA in gastric cancer. Nat. Commun.14 (1), 6523. 10.1038/s41467-023-42025-8
101
Sun T. Liu B. Li Y. Wu J. Cao Y. Yang S. et al (2023d). Oxamate enhances the efficacy of CAR-T therapy against glioblastoma via suppressing ectonucleotidases and CCR8 lactylation. J. Exp. Clin. Cancer Res.42 (1), 253. 10.1186/s13046-023-02815-w
102
Sun W. Jia M. Feng Y. Cheng X. (2023b). Lactate is a bridge linking glycolysis and autophagy through lactylation. Autophagy19 (12), 3240–3241. 10.1080/15548627.2023.2246356
103
Sun X. He L. Liu H. Thorne R. F. Zeng T. Liu L. et al (2023a). The diapause-like colorectal cancer cells induced by SMC4 attenuation are characterized by low proliferation and chemotherapy insensitivity. Cell Metab.35 (9), 1563–1579 e8. 10.1016/j.cmet.2023.07.005
104
Takata T. Nakamura A. Yasuda H. Miyake H. Sogame Y. Sawai Y. et al (2024). Pathophysiological implications of protein lactylation in pancreatic epithelial tumors. Acta Histochem Cytochem57 (2), 57–66. 10.1267/ahc.24-00010
105
Wan N. Wang N. Yu S. Zhang H. Tang S. Wang D. et al (2022). Cyclic immonium ion of lactyllysine reveals widespread lactylation in the human proteome. Nat. Methods19 (7), 854–864. 10.1038/s41592-022-01523-1
106
Wang D. Du G. Chen X. Wang J. Liu K. Zhao H. et al (2024b). Zeb1-controlled metabolic plasticity enables remodeling of chromatin accessibility in the development of neuroendocrine prostate cancer. Cell Death Differ.31 (6), 779–791. 10.1038/s41418-024-01295-5
107
Wang H. Xu M. Zhang T. Pan J. Li C. Pan B. et al (2024a). PYCR1 promotes liver cancer cell growth and metastasis by regulating IRS1 expression through lactylation modification. Clin. Transl. Med.14 (10), e70045. 10.1002/ctm2.70045
108
Wang J. Liu Z. Xu Y. Wang Y. Wang F. Zhang Q. et al (2022c). Enterobacterial LPS-inducible LINC00152 is regulated by histone lactylation and promotes cancer cells invasion and migration. Front. Cell Infect. Microbiol.12, 913815. 10.3389/fcimb.2022.913815
109
Wang J. Yang P. Yu T. Gao M. Liu D. Zhang J. et al (2022a). Lactylation of PKM2 suppresses inflammatory metabolic adaptation in pro-inflammatory macrophages. Int. J. Biol. Sci.18 (16), 6210–6225. 10.7150/ijbs.75434
110
Wang L. Li S. Luo H. Lu Q. Yu S. (2022b). PCSK9 promotes the progression and metastasis of colon cancer cells through regulation of EMT and PI3K/AKT signaling in tumor cells and phenotypic polarization of macrophages. J. Exp. Clin. Cancer Res.41 (1), 303. 10.1186/s13046-022-02477-0
111
Wang M. He T. Meng D. Lv W. Ye J. Cheng L. et al (2023a). BZW2 modulates lung adenocarcinoma progression through glycolysis-mediated IDH3G lactylation modification. J. Proteome Res.22 (12), 3854–3865. 10.1021/acs.jproteome.3c00518
112
Wang N. Jiang X. Zhang S. Zhu A. Yuan Y. Xu H. et al (2021b). Structural basis of human monocarboxylate transporter 1 inhibition by anti-cancer drug candidates. Cell184 (2), 370–383 e13. 10.1016/j.cell.2020.11.043
113
Wang R. Li C. Cheng Z. Li M. Shi J. Zhang Z. et al (2024c). H3K9 lactylation in malignant cells facilitates CD8(+) T cell dysfunction and poor immunotherapy response. Cell Rep.43 (9), 114686. 10.1016/j.celrep.2024.114686
114
Wang X. Ying T. Yuan J. Wang Y. Su X. Chen S. et al (2023b). BRAFV600E restructures cellular lactylation to promote anaplastic thyroid cancer proliferation. Endocr. Relat. Cancer30 (8), e220344. 10.1530/ERC-22-0344
115
Wang Z. H. Peng W. B. Zhang P. Yang X. P. Zhou Q. (2021a). Lactate in the tumour microenvironment: from immune modulation to therapy. EBioMedicine73, 103627. 10.1016/j.ebiom.2021.103627
116
Warburg O. Wind F. Negelein E. (1927). The metabolism of tumors in the Body. J. Gen. Physiol.8 (6), 519–530. 10.1085/jgp.8.6.519
117
Wu X. Mi T. Jin L. Ren C. Wang J. Zhang Z. et al (2024b). Dual roles of HK3 in regulating the network between tumor cells and tumor-associated macrophages in neuroblastoma. Cancer Immunol. Immunother.73 (7), 122. 10.1007/s00262-024-03702-9
118
Wu Z. Wu H. Dai Y. Wang Z. Han H. Shen Y. et al (2024a). A pan-cancer multi-omics analysis of lactylation genes associated with tumor microenvironment and cancer development. Heliyon10 (5), e27465. 10.1016/j.heliyon.2024.e27465
119
Xie B. Zhang M. Li J. Cui J. Zhang P. Liu F. et al (2024). KAT8-catalyzed lactylation promotes eEF1A2-mediated protein synthesis and colorectal carcinogenesis. Proc. Natl. Acad. Sci. U. S. A.121 (8), e2314128121. 10.1073/pnas.2314128121
120
Xiong J. He J. Zhu J. Pan J. Liao W. Ye H. et al (2022). Lactylation-driven METTL3-mediated RNA m(6)A modification promotes immunosuppression of tumor-infiltrating myeloid cells. Mol. Cell82 (9), 1660–1677 e10. 10.1016/j.molcel.2022.02.033
121
Xu H. Li L. Wang S. Wang Z. Qu L. Wang C. et al (2023). Royal jelly acid suppresses hepatocellular carcinoma tumorigenicity by inhibiting H3 histone lactylation at H3K9la and H3K14la sites. Phytomedicine118, 154940. 10.1016/j.phymed.2023.154940
122
Yang D. Yin J. Shan L. Yi X. Zhang W. Ding Y. (2022b). Identification of lysine-lactylated substrates in gastric cancer cells. iScience25 (7), 104630. 10.1016/j.isci.2022.104630
123
Yang H. Yang S. He J. Li W. Zhang A. Li N. et al (2023a). Glucose transporter 3 (GLUT3) promotes lactylation modifications by regulating lactate dehydrogenase A (LDHA) in gastric cancer. Cancer Cell Int.23 (1), 303. 10.1186/s12935-023-03162-8
124
Yang H. Zou X. Yang S. Zhang A. Ma Z. (2023b). Identification of lactylation related model to predict prognostic, tumor infiltrating immunocytes and response of immunotherapy in gastric cancer. Front. Immunol.14, 1149989. 10.3389/fimmu.2023.1149989
125
Yang K. Fan M. Wang X. Xu J. Wang Y. Tu F. et al (2022a). Lactate promotes macrophage HMGB1 lactylation, acetylation, and exosomal release in polymicrobial sepsis. Cell Death Differ.29 (1), 133–146. 10.1038/s41418-021-00841-9
126
Yang Z. Yan C. Ma J. Peng P. Ren X. Cai S. et al (2023). Lactylome analysis suggests lactylation-dependent mechanisms of metabolic adaptation in hepatocellular carcinoma. Nat. Metab.5 (1), 61–79. 10.1038/s42255-022-00710-w
127
Yao G. Yang Z. (2024). Glypican-3 knockdown inhibits the cell growth, stemness, and glycolysis development of hepatocellular carcinoma cells under hypoxic microenvironment through lactylation. Arch. Physiol. Biochem.130 (5), 546–554. 10.1080/13813455.2023.2206982
128
Ye J. Gao X. Huang X. Huang S. Zeng D. Luo W. et al (2024). Integrating single-cell and spatial transcriptomics to uncover and elucidate GP73-mediated pro-angiogenic regulatory Networks in hepatocellular carcinoma. Research (Wash D C)7, 0387. 10.34133/research.0387
129
Yin X. Xing W. Yi N. Zhou Y. Chen Y. Jiang Z. et al (2024). Comprehensive analysis of lactylation-related gene sets and mitochondrial functions in gastric adenocarcinoma: implications for prognosis and therapeutic strategies. Front. Immunol.15, 1451725. 10.3389/fimmu.2024.1451725
130
Yu J. Chai P. Xie M. Ge S. Ruan J. Fan X. et al (2021). Histone lactylation drives oncogenesis by facilitating m(6)A reader protein YTHDF2 expression in ocular melanoma. Genome Biol.22 (1), 85. 10.1186/s13059-021-02308-z
131
Yu L. Jing C. Zhuang S. Ji L. Jiang L. (2024). A novel lactylation-related gene signature for effectively distinguishing and predicting the prognosis of ovarian cancer. Transl. Cancer Res.13 (5), 2497–2508. 10.21037/tcr-24-319
132
Yuan X. Wang Q. Zhao J. Xie H. Pu Z. (2024). The m6A methyltransferase METTL3 modifies Kcnk6 promoting on inflammation associated carcinogenesis is essential for colon homeostasis and defense system through histone lactylation dependent YTHDF2 binding. Int. Rev. Immunol.44, 1–16. 10.1080/08830185.2024.2401358
133
Yue Q. Wang Z. Shen Y. Lan Y. Zhong X. Luo X. et al (2024). Histone H3K9 lactylation confers Temozolomide resistance in glioblastoma via LUC7L2-mediated MLH1 Intron Retention. Adv. Sci. (Weinh)11 (19), e2309290. 10.1002/advs.202309290
134
Zang Y. Wang A. Zhang J. Xia M. Jiang Z. Jia B. et al (2024). Hypoxia promotes histone H3K9 lactylation to enhance LAMC2 transcription in esophageal squamous cell carcinoma. iScience27 (7), 110188. 10.1016/j.isci.2024.110188
135
Zeng Q. Wang K. Zhao Y. Ma Q. Chen Z. Huang W. (2023). Effects of the acetyltransferase p300 on tumour regulation from the novel perspective of Posttranslational protein modification. Biomolecules13 (3), 417. 10.3390/biom13030417
136
Zhang C. Zhou L. Zhang M. Du Y. Li C. Ren H. et al (2024b). H3K18 lactylation potentiates immune Escape of non-small cell lung cancer. Cancer Res.84 (21), 3589–3601. 10.1158/0008-5472.CAN-23-3513
137
Zhang D. Tang Z. Huang H. Zhou G. Cui C. Weng Y. et al (2019). Metabolic regulation of gene expression by histone lactylation. Nature574 (7779), 575–580. 10.1038/s41586-019-1678-1
138
Zhang R. Li L. Yu J. (2024a). Lactate-induced IGF1R protein lactylation promotes proliferation and metabolic reprogramming of lung cancer cells. Open Life Sci.19 (1), 20220874. 10.1515/biol-2022-0874
139
Zhao P. Qiao C. Wang J. Zhou Y. Zhang C. (2024c). Histone lactylation facilitates hepatocellular carcinoma progression by upregulating endothelial cell-specific molecule 1 expression. Mol. Carcinog.63 (11), 2078–2089. 10.1002/mc.23794
140
Zhao R. Yi Y. Liu H. Xu J. Chen S. Wu D. et al (2024b). RHOF promotes Snail1 lactylation by enhancing PKM2-mediated glycolysis to induce pancreatic cancer cell endothelial-mesenchymal transition. Cancer Metab.12 (1), 32. 10.1186/s40170-024-00362-2
141
Zhao Y. Jiang J. Zhou P. Deng K. Liu Z. Yang M. et al (2024a). H3K18 lactylation-mediated VCAM1 expression promotes gastric cancer progression and metastasis via AKT-mTOR-CXCL1 axis. Biochem. Pharmacol.222, 116120. 10.1016/j.bcp.2024.116120
142
Zheng P. Mao Z. Luo M. Zhou L. Wang L. Liu H. et al (2023). Comprehensive bioinformatics analysis of the solute carrier family and preliminary exploration of SLC25A29 in lung adenocarcinoma. Cancer Cell Int.23 (1), 222. 10.1186/s12935-023-03082-7
143
Zheng Y. Yang Y. Xiong Q. Ma Y. Zhu Q. (2024). Establishment and Verification of a novel gene signature connecting hypoxia and lactylation for predicting prognosis and immunotherapy of pancreatic ductal adenocarcinoma patients by integrating multi-machine learning and single-cell analysis. Int. J. Mol. Sci.25 (20), 11143. 10.3390/ijms252011143
144
Zhou C. Li W. Liang Z. Wu X. Cheng S. Peng J. et al (2024). Mutant KRAS-activated circATXN7 fosters tumor immunoescape by sensitizing tumor-specific T cells to activation-induced cell death. Nat. Commun.15 (1), 499. 10.1038/s41467-024-44779-1
145
Zhou J. Xu W. Wu Y. Wang M. Zhang N. Wang L. et al (2023). GPR37 promotes colorectal cancer liver metastases by enhancing the glycolysis and histone lactylation via Hippo pathway. Oncogene42 (45), 3319–3330. 10.1038/s41388-023-02841-0
146
Zhu J. Zhang Y. (2024). Dexmedetomidine inhibits the migration, invasion, and glycolysis of glioblastoma cells by lactylation of c-myc. Neurol. Res.46, 1105–1112. 10.1080/01616412.2024.2395069
147
Zong Z. Xie F. Wang S. Wu X. Zhang Z. Yang B. et al (2024). Alanyl-tRNA synthetase, AARS1, is a lactate sensor and lactyltransferase that lactylates p53 and contributes to tumorigenesis. Cell187 (10), 2375–2392 e33. 10.1016/j.cell.2024.04.002
Summary
Keywords
lactylation, post-translational modification, tumor progression, prognostic biomarkers, therapeutic target
Citation
Gou Z, Sun Q and Li J (2025) Exploring lactylation and cancer biology: insights from pathogenesis to clinical applications. Front. Cell Dev. Biol. 13:1598232. doi: 10.3389/fcell.2025.1598232
Received
22 March 2025
Accepted
02 June 2025
Published
13 June 2025
Volume
13 - 2025
Edited by
Jessica Lilian Bell, Children’s Cancer Institute Australia, Australia
Reviewed by
Rani Ojha, Post Graduate Institute of Medical Education and Research (PGIMER), India
Suchandrima Saha, Stony Brook Medicine, United States
Updates
Copyright
© 2025 Gou, Sun and Li.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jiannan Li, jnli@jlu.edu.cn, jnli@ciac.ac.cn
†These authors have contributed equally to this work and share first authorship
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.