- 1Department of Obstetrics and Gynecology, Shengjing Hospital of China Medical University, Shenyang, China
- 2Department of Rheumatology and Immunology, Beijing Hospital, Chinese Academy of Medical Sciences, Beijing, China
- 3Department of Pediatrics, Clinical Medicine, China Medical University, Shenyang, China
- 4Department of Neurosurgery, Shengjing Hospital of China Medical University, Shenyang, China
Cancer is the primary cause of death worldwide. Cancer occurrence and progression are closely associated with DNA damage repair. DNA glycosylase were the first enzymes to initiate base excision repair, and the Nei-like DNA glycosylase (NEIL) family has attracted increasing attention as an important component of DNA glycosylases. Here, we introduce the role of the NEIL family in the malignant biological behaviors of cancer, including cell proliferation, chemoradiotherapy resistance, invasion and migration, apoptosis, and stemness. Mechanisms affecting the expression of the NEIL protein family range from the transcriptional level and mRNA editing to the level of post-translational modification. Additionally, we emphasize the different functions of the NEIL family in various malignancies and present useful information that supports the hypothesis that the NEIL family could be a potential target in the treatment and diagnosis of various cancers.
1 Introduction
Cellular DNA is persistently exposed to exogenous and endogenous damage (De Bont and van Larebeke, 2004; Hopkins et al., 2022; Bednarski and Sleckman, 2019). Exogenous sources of injury include ultraviolet light, environmental toxicants, and ionizing radiation (Hoeijmakers, 2001). Whereas, endogenous damage is predominantly caused by reactive oxygen species (ROS), including hydrogen peroxide (H2O2), the hydroxyl radical, ·OH, and the superoxide radical, O2−, and these compounds are important genotoxic substances (Hazra et al., 2002a). The accumulation of ROS can lead to oxidative damage and strand breaks in DNA, which are major threats to genome integrity (Sarker et al., 2021; Caliri et al., 2021; Seçme et al., 2023). Approximately 104–105 spontaneous DNA damage occurs in each cell every day (Yousefzadeh et al., 2021). Unrepaired damage may block DNA replication and transcription, and cause base mismatches, leading to aging and cancer (Chen et al., 2022; Yuan et al., 2023). Therefore, timely repair of DNA damage is vital for maintaining genome integrity.
Base excision repair (BER) is the primary pathway for repairing oxidative DNA damage (Hua and Sweasy, 2023). Specific DNA glycosylases that recognize and eliminate base damages initiate the BER pathway (Wiederhold et al., 2004). There are 11 DNA glycosylases in humans that are classified into three groups: Nei-like DNA glycosylases (NEILs, including NEIL1, NEIL2, and NEIL3), monofunctional enzymes and bifunctional glycosylases (Grundy and Parsons, 2020; Morland et al., 2002; Eckenroth et al., 2021; Hanna et al., 2021). After recognition, monofunctional enzymes catalyze the hydrolysis of the n-glycosidic bond, thereby removing damaged nitrogen bases and leaving an apurinic/apyrimidinic site. Then, apurinic/apyrimidinic nucleic acid endonuclease is recruited in order to hydrolyze the DNA backbone, forming a single strand break (SSB) with 5′ -deoxyribosephosphate and 3′-hydroxyl ends (Panigrahi et al., 2015; Gohil et al., 2023). In addition to cutting damage bases, bifunctional enzymes also exhibit innate apurinic/apyrimidinic cleavage enzyme activity, using β-elimination to generate 3′-αβ unsaturated aldehyde (Krokeide et al., 2013). In contrast, the NEIL family exhibits β,δ-elimination activity, creating a nucleotide gap with 3′-P and 5′-P termini (Dou et al., 2003). However, NEIL3 predominantly acts as a single functional enzyme using β-elimination in the presence of apurinic/apyrimidinic nucleic acid endonuclease, as a result of its weak apurinic/apyrimidinic lyase but vigorous glycosylase activity (Krokeide et al., 2013). DNA polymerase II repairs DNA, and ultimately, DNA ligase IIIa/X-ray repair cross complementing 1 completes the BER process (Tomkinson and Sallmyr, 2013).
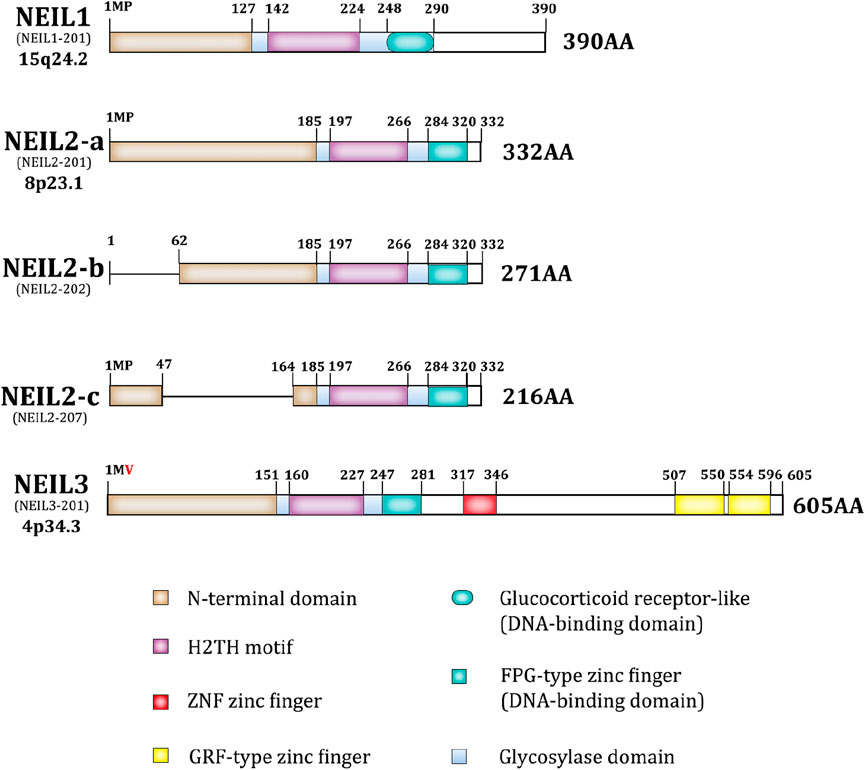
As previously reported, NEIL1/2/3, nth like DNA glycosylase 1 (NTH1), and 8-oxoguanine DNA glycosylase (OGG1) are bifunctional; however, the NEIL family differs from the others (Figure 1). It has a complete Fpg/Nei core protein structural domain at the N-terminus, including an N-terminal domain, a helix-2-turn-helix (H2TH) motif, and a zinc finger or zinc-like finger DNA-binding motif (Das et al., 2004; Hazra and Mitra, 2006; Kladova et al., 2019). The NEIL family proteins exhibit both shared and distinct features in their structural conformations and substrate specificities. NEIL1 and NEIL2 share a conserved N-terminal region with the Fpg/Nei family, characterized by the presence of the H2tH motif that forms an αG helix. NTH1 and OGG1 utilize an innate Lys residue as the active site nucleophile, while NEIL1, NEIL2 uses an N-terminal Pro (Hazra et al., 2002a; Hazra et al., 2002b). NEIL1 is primarily involved in DNA replication and the excision of various oxidative lesions, including thymine glycol, 2,6-diamino-4-hydroxy-5-formamidopyrimidine, 5-hydroxycytosine, 5-hydroxyuracil, and 4,6-diamino-5-formamidopyrimidine. In contrast, NEIL2 features a unique C-terminal zinc finger motif and demonstrates a preference for lesions in single-stranded DNA and bubble structures. Interestingly, in NEIL3, Val replaces the vital catalytic role of Pro in the N-terminal structural domain. NEIL3 is nearly twice as large as other Fpg/Nei family members (Liu et al., 2013) and includes a duplicated Glycine arginine phenylalanine GRF-zinc finger (GRF-ZF) motif and a RANbp-like zinc finger (ZNF) motif at the C-terminus, which makes NEIL3 distinct from NEIL1 and NEIL2 (Rodriguez et al., 2020; Ha et al., 2020; Huskova et al., 2022). Although NEIL3 is less studied than NEIL1 and NEIL2, this enzyme displays broad substrate specificity, targeting both single-stranded and double-stranded DNA.
The disordered C-terminal domain of NEIL1 may be associated with multiple NEIL1 interactions. Although it has no effect on apurinic/apyrimidinic lyase activity or lesion excision, it is required for efficient enzymatic activity (Hegde et al., 2013b; Hegde et al., 2012; Hegde et al., 2015). Typically, the catalytic core of BER enzymes is an inflexible mold for detecting distorted DNA, whereas the catalytic structure of NEIL2 is more specialized in that the two lobes of its catalytic core are not in a catalytically-active orientation, making NEIL2 inactive by default until the catalytic domains are clustered together in the correct orientation. This structure may confer more function to NEIL2 (Tsutakawa and Sarker, 2021; Zhdanova et al., 2022; Eckenroth et al., 2023). In general, the unique structures of the NEIL family lead to diverse substrates and excise ring-opened or damaged oxidized pyrimidines and purines (Chakraborty et al., 2015; Makasheva et al., 2020).
The unifying feature of the NEIL family is the Fpg/Nei-like core protein structural domain (Glycosoylase domain), including an N-terminal domain, a helix-2-turn-helix (H2TH) motif, and a DNA-binding domain. NEIL3 is unique in that it is twice as large as the other NEIL proteins, having a RANbp-like zinc finger motif and a duplicated Glycine arginine phenylalanine GRF-zinc finger (GRF-ZF) motif at the C-terminus. Besides, in NEIL3 valine replaces the central catalytic role of proline in the second amino acid. The ensembl transcript ID and the location of the gene is under each isoform name, and the protein length is at the end of each isoform, as shown in Figure 1.
NEIL1 and NEIL3 are mainly active in the S phase and are involved in the repair of damaged bases prior to replication, whereas NEIL2 expression is cell cycle-independent (Hazra et al., 2002a; Albelazi et al., 2019; Neurauter et al., 2012; Hegde et al., 2013a). As multifunctional DNA repair enzyme, NEILs are crucial in diverse cellular processes. NEIL2 participates in active DNA demethylation (Schomacher et al., 2016), inflammatory responses (Tapryal et al., 2021), transcription-coupled repair (Chakraborty et al., 2021) and maintenance of the mitochondrial genome (Sarker et al., 2021; Mandal et al., 2012). NEIL3 prefers ssDNA-derived base damage, maintenance of replication fork stability, repair of DNA damage in G-quadruplex structures, repair of DNA interstrand crosslinks (ICL) and repair of telomere damage (Chen et al., 2022; Li et al., 2022; Li et al., 2020; Wu et al., 2019). The NEIL family plays a critical role in DNA repair, and the overexpression and downregulation of NEILs may be associated with cancer, which we will elaborate in the following sections.
2 Common targets and cellular functions of NEILs in various cancer
Growing evidence suggests that the NEIL family is involved in several cellular events such as proliferation, chemoradiotherapy resistance, migration, invasion, cell death, and stemness.
2.1 Cell proliferation
According to previous studies, NEIL proteins act as tumor suppressors or oncogenes during cell proliferation under different circumstances. High NEIL1 expression, for example, may be associated with a poor prognosis in patients with gastric cancer. The DNA repair inhibitor, berzosertib, may inhibit the expression of NEIL1, thus limiting the proliferation of gastric cancer cells (Ni et al., 2019). However, this is not the case in breast cancer or multiple myeloma. One study has suggested that decreased NEIL1 expression predicts poor survival outcomes in patients with invasive breast cancer (Shinmura et al., 2016). In melphalan-resistant multiple myeloma, NEIL1 expression is downregulated, leading to a greater propensity of cells to repair toxic ICL, thereby reducing G2/M phase arrest (Sousa et al., 2013). NEIL3 expression is limited to cells with a high capacity for proliferation (Fleming et al., 2015), and is inhibited in non-dividing cells (Neurauter et al., 2012). The most interesting aspect of NEIL3 is that it can repair telomeres (Zhou et al., 2013; Zhou et al., 2015; Karlsen et al., 2022; Fouquerel et al., 2016). NEIL3 expression is upregulated in non-small-cell lung cancer (NSCLC) (Huang and Hua, 2022). In vitro, NEIL3 protects the genome by repairing oxidative damage to telomeres in the S/G2 phase, which plays a significant role in the proliferation of NSCLC cells (Zhou et al., 2017). In hepatocellular carcinoma (HCC), NEIL3 repairs oxidative damage to telomeres and prevents telomere shortening during mitosis (Zhao et al., 2021b). However, NEIL3 does not influence prostate cancer (PCa) cell proliferation in vitro (Wang et al., 2021b). Studies have suggested that NEIL3 promotes the progression of lung and liver cancers by regulating PI3K/Akt/mTOR signal transduction (Huang and Hua, 2022; Wang et al., 2021). These findings suggest that NEILs play a dual role in cancer that is specific to the cellular environment.
2.2 Chemotherapy resistance
Acquired chemotherapy resistance is a significant cause of death in patients with advanced malignant tumors; therefore, there is an urgent need to identify the mechanisms of chemotherapy resistance (Davodabadi et al., 2023). NEILs play a pivotal role in the resistance of PCa, HCC, and other tumors to chemotherapy.
During chemotherapy, NEIL3 confers resistance to cisplatin, which is one of the most commonly used chemotherapeutic drugs in the treatment of a wide range of solid tumors (Rottenberg et al., 2021). Cisplatin induces DNA ICL, which blocks DNA replication and transcription, and eventually triggers apoptosis, which is the main mechanism against tumors. NEIL3 is one of the main enzymes involved in ICL repair (Semlow and Walter, 2021; Yang et al., 2017; Chung et al., 2023; Imani Nejad et al., 2020).
Neuroendocrine prostate cancer, castration-resistant prostate cancer (CRPC), and metastatic PCa are all considered advanced PCa (Wang et al., 2018). NEIL3 knockdown markedly decreased the sensitivity of PCa cells to cisplatin in two ways (Wang et al., 2021c). On the one hand, the knockdown of NEIL3 reduces cell cycle arrest in the S phase, and on the other hand, it regulates ataxia-telangiectasia mutated (ATM) and ataxia-telangiectasia and Rad3 related (ATR) pathway activities. NEIL3 itself does not affect the levels of phosphorylated ATM (p-ATM) or phosphorylated ATR (p-ATR); however, upon exposure to docetaxel or cisplatin, the phosphorylation of ATM and ATR was significantly promoted by the loss of NEIL3, thereby triggering downstream pathways associated with DNA repair.
Resistance to chemotherapy may be promoted by the activation of epithelial-mesenchymal transition (EMT), which enhances survival mechanisms and cell cycle progression (Marie-Egyptienne et al., 2013). In HCC, NEIL3 activates a key transcription factor, twist family bHLH transcription factor 1 (TWIST1), leading to an increase in the expression of the drug efflux gene and drug resistance in EMT (Lai et al., 2022).
In HeLa cells, NEIL3 interacts with WRN helicase interacting protein 1 (WRNIP1) through its C-terminal domain to target WRNIP1 to the proteasome, promotes WRNIP1 degradation and ICL repair, and induces cisplatin resistance (Aliyaskarova et al., 2023). WRNIP1 is a member of the AAA ATPase family and involved in the early stages of ICL repair (Socha et al., 2020). Its timely degradation at a later stage of repair can cause the ICL repair to proceed to the next step.
NEIL1 can also induce cisplatin resistance via a mechanism that is different from that of NEIL3. It has been suggested that BER maintains cisplatin cytotoxicity by decreasing ICL repair through competition with the cisplatin ICL DNA repair pathway (Kothandapani et al., 2011). In multiple myeloma cells, NEIL1 depletion contributes to melphalan resistance by downregulating the BER pathway, which facilitates the repair of the more toxic ICL (Sousa et al., 2013).
A defect in single-stranded DNA damage repair is considered a new factor that causes endocrine resistance in ER+ breast cancer. It has been reported that the loss of NEIL2 leads to endocrine resistance via disruption of the G1-S phase transition, but more specific mechanisms remain to be elucidated (Anurag et al., 2018). NEIL2 is involved in the regulation of cellular ROS concentrations in breast cancer stem cells. Its high expression repairs ROS-induced DNA damage, maintains ROS at low levels, and leads to resistance to doxorubicin and other chemotherapeutic drugs that produce ROS as the primary killing mechanism (Banerjee et al., 2020).
2.3 Other functions
Migratory and invasive cells have increased capacity during tumor development, which is closely related to metastasis at advanced stages of cancer. Various mechanisms are involved in the acquisition of these malignant features by tumor cells. EMT in hepatoma may be promoted by NEIL3 through activation of the BRAF/MEK/ERK/TWIST signaling pathway (Lai et al., 2022). NEIL3 directly induces BRAF transcription, which activates the downstream mitogen-activated protein kinase (MAPK) cascade of BRAF, phosphorylates ERK, and activates the downstream molecule, TWIST1, thereby promoting EMT. EMT is an important step in tumor metastasis (Zhu et al., 2016; Manfioletti and Fedele, 2022). Interestingly, NEIL3 promotes EMT independent of DNA repair, which was not observed for NEIL1 and NEIL2 in this study. NEIL3 can also partially promote the proliferation, invasion, and migration of NSCLC cells by regulating the classical PI3K/AKT/mTOR signaling pathway (Huang and Hua, 2022).
NEIL target genes also participate in apoptosis regulation. NEIL1 inhibits apoptosis by increasing the expression of the anti-apoptotic gene, BCL2 apoptosis regulator (Bcl-2), and decreasing the expression of pro-apoptotic genes (BCL2 associated X, apoptosis regulator (Bax) and caspase-9) in colorectal cancer (CRC) cells (Xue et al., 2020). NEIL2 expression level in cells is related to ROS levels. In breast cancer cells, low concentrations of ROS induce the upregulation of NEIL2 and enhance BER; however, high concentrations of ROS cause a decrease in NEIL2, which results in the activation of p53 and further activates the intrinsic apoptotic pathway (Banerjee et al., 2020; Chakraborti et al., 2017). NEIL3 also inhibits apoptosis by repairing gene damage (Wang et al., 2021c).
Maintenance of stemness is another malignant biological behavior of tumors. Breast cancer stem cells employ several molecular strategies to evade chemotherapy-induced death signals, and redox regulation is a key factor. High levels of NEIL2 expression can maintain low levels of ROS and stemness in breast cancer stem cells (Banerjee et al., 2020).
3 Common regulatory mechanisms for NEIL proteins in cancer
The NEIL protein family is regulated by diverse mechanisms, including transcriptional, post-transcriptional, and post-translational regulation (Figure 2).
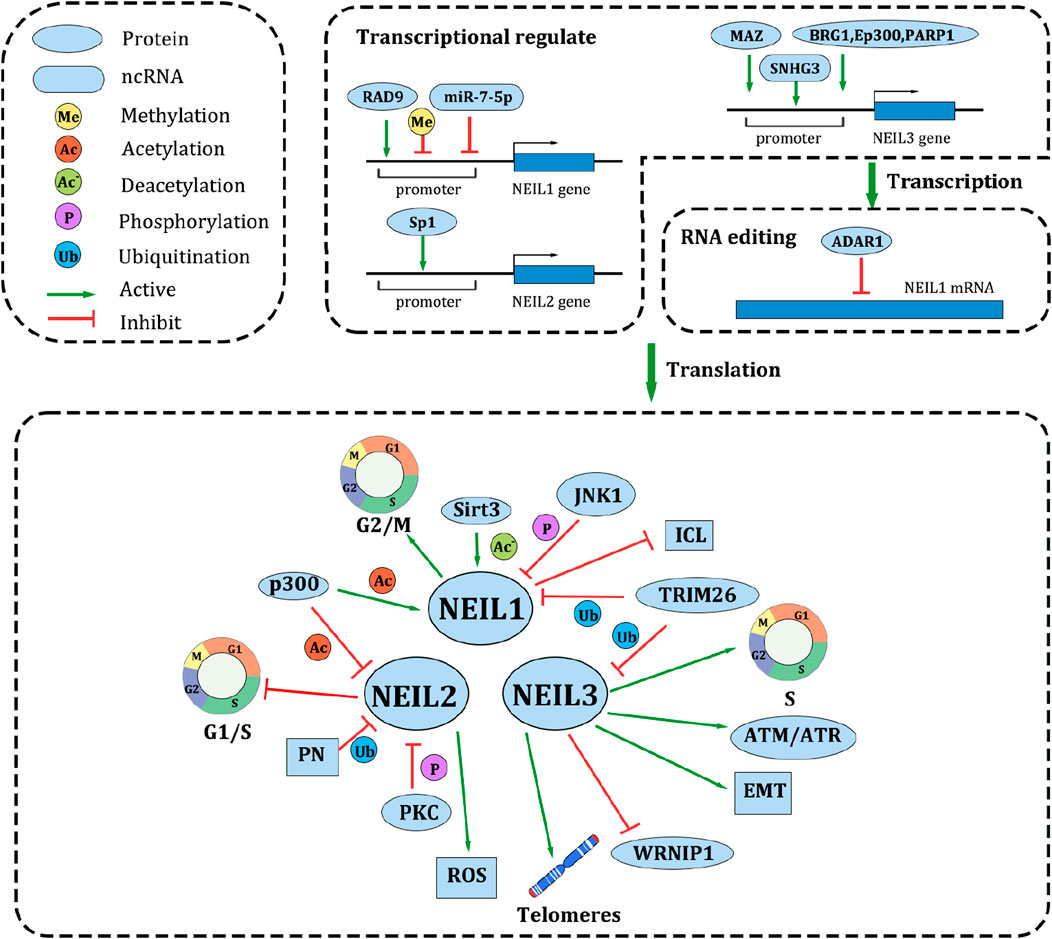
Figure 2. Common upstream regulatory mechanisms, targets and cellular functions of NEIL family. In the transcriptional level, promoter methylation, the combination of several proteins and ncRNAs may affect the NEIL family expression. Additionally, ADAR1 edits NEIL1 mRNA after transcription. After translation, a variety of protein modifications such as acetylation, deacetylation, phosphorylation, and ubiquitination affect the activity of NEIL protein. NEIL family also influence the malignant biological behaviors of cancer in several ways. Reducing cell cycle arrest and repairing telomeres are the main approaches to accelerate cell proliferation. In chemotherapy resistance, NEIL1 competes with ICL DNA repair pathway to maintains cisplatin cytotoxicity. NEIL2 promotes ROS repair, besides, loss of NEIL2 leads to disruption of the G1-S phase transition. NEIL3 promoting EMT pathway and ATM/ATR pathway, but inhibiting WRNIP. NEIL family also play a vital role in invasion and migration, apoptosis, and stemness.
3.1 Transcriptional and post-transcriptional regulation
The regulation of NEIL at the transcriptional level is primarily mediated by its promoter. A subset of factors modulates the expression of the NEIL family by directly binding to their promoters. Aberrant RAD9 checkpoint clamp component A (RAD9) expression is associated with various cancers. In PCa DU145 cells, RAD9 binds to the NEIL1 p53 consensus sequence in the promoter region, promoting NEIL1 transcription. RAD9 is crucial in maintaining genome stability, acting primarily as part of the 9-1-1 complex. It also acts as a transcription factor independent of 9-1-1, regulating DNA repair, cell cycle checkpoints, apoptosis, and telomere stability (Broustas et al., 2019; Lieberman et al., 2017; Panigrahi et al., 2015; McDonald et al., 2023; Guan et al., 2007). Oxidative stress can regulate the expression of NEIL2 (Kinslow et al., 2010). Binding of Sp1 to the NEIL2 promoter is boosted by low ROS levels, thereby promoting NEIL2 transcription and activating DNA damage repair (Chakraborti et al., 2017). Pyridinol reduces NEIL2 expression while inhibiting its association with RNA Pol II, thereby impeding NEIL2-mediated transcriptional coupling repair, promoting DNA damage, and initiating ROS production (Banerjee et al., 2020). Single nucleotide polymorphisms (SNPs) in the NEIL2 promoter may also influence its expression (Kinslow et al., 2008). The transcription factor, MYC associated zinc finger protein (MAZ), functions as an upstream regulator of NEIL3 to directly promote its transcription and this induces cisplatin resistance in lung adenocarcinoma (Wang et al., 2023).
Non-coding RNAs can also affect NEIL transcription. Small nucleolar RNA host gene 3 (SNHG3), a long non-coding RNA, recruits the transcription factor, E2F1, to the promoter region of NEIL3 and activates its transcription in HCC (Zhang et al., 2023). In colon cancer, miR-7-5p targets the 3′UTR of NEIL to suppress NEIL1 transcription, thereby increasing anti-apoptotic genes, Bcl-2, down-regulating pro-apoptotic genes (Bax and caspase-9), and inhibiting apoptosis (Xue et al., 2020).
Notably, the modification of the promoter region is an important way to regulate NEIL expression. NEIL1 expression is downregulated by the hypermethylation of the promoter region of NEIL1 in head and neck squamous cell carcinoma (Chaisaingmongkol et al., 2012). BRG1 is a member of the SWI/SNF chromatin-remodeling complex that acts as an activator of functionally connected genes and promotes DNA repair and mitotic cell division (Orlando et al., 2019; Takao et al., 2017). The BRG1-EP300 complex drives NEIL3 transcription in breast cancer. EP300 acetylates nucleosomes in the promoter region, and BRG1 evicts acetylated nucleosomes from the chromatin, thereby promoting transcription. PARP1 co-localizes with BRG1 on the highly acetylated promoter of NEIL3, ensuring an open chromatin structure (Sobczak et al., 2020; Sobczak et al., 2019).
RNA editing can also alter NEIL protein expression. The RNA editing enzyme, adenosine deaminase (ADAR1), converts adenosine to inosine (A-to-I) (Avesson and Barry, 2014). It has been reported that there may be different A-to-I editing levels at the NEIL1 RNA (lysine 242 AAA codon) site (Lee et al., 2017; Minko et al., 2020; Lotsof et al., 2022). Unedited NEIL1 repairs DNA much more quickly than edited NEIL1 (Yeo et al., 2021; Yeo et al., 2010). ADAR1 amplified lung cancer cell lines show a higher frequency of AAA-AIA and AAA-to-AII edits than ADAR1 normal cells (Anadón et al., 2016). NEIL1 is a vital and extensively edited ADAR1 target in multiple myeloma. The recoded NEIL1 protein shows a loss of oxidative damage repair capacity and an increased cell growth rate (Teoh et al., 2018). These data provide new insights into the molecular pathogenesis of multiple myeloma at the mRNA level.
3.2 Post-translational modifications
Post-translational modifications are currently a popular research topic. Studies have shown that the BER is strictly modified after translation. The addition of an acetyl group to a lysine (K) residue is a critical post-translational modification that modulates the function of a protein by altering its conformation, activity, stability, or ability to interact with other proteins (Bhakat et al., 2020; Shvedunova and Akhtar, 2022). Research has shown that NEIL1 is acetylated at multiple K residues (K296, K297, and K298) and that NEIL1 acetylation enhances its glycosylation enzyme activity in vitro. Acetylation of NEIL1 only combined with chromatin (Bacolla et al., 2021). In addition, acetylation-defective NEIL1 protein enhances cell sensitivity to DNA-damaging agents that produce SSBs or oxidized bases. At the same time, acetylation can affect the interaction of NEIL1 with other proteins. Compared to wild-type NEIL1, the 3KR mutant, which is acetylation-defective, forms less stable complexes with diverse chromatin proteins, such as BER/SSB repair partners and histone chaperones. The BER activity of the repair complex bound to the 3KR mutant was remarkably lower than that of wild-type NEIL1. In conclusion, the primary role of acetylated lysine residues in NEIL1 is to stabilize the chromatin-binding repair complex formation, thus protecting cells from oxidative stress (Sengupta et al., 2018). The mitochondrial protein, sirtuin 3 (Sirt3), participates in various metabolic regulatory processes (Onyango et al., 2002). Sirt3 not only controls metabolism at the transcriptional level but also directly regulates the activity of metabolic enzymes. In CRC, deacetylation of NEIL1 and NEIL2 is regulated by Sirt3. Meanwhile, Sirt3 directly engages with NEIL1 (Kabzinski et al., 2022; Kabziński et al., 2019). In vitro results showed that NEIL2 was acetylated by p300 at K49 and K153. Acetylation of K153 does not affect NEIL2 activity in vitro, whereas acetylation of K49 inhibits NEIL2 base excision and apurinic/apyrimidinic decomposition (Bhakat et al., 2004).
Ubiquitination is a key regulatory mechanism in this process (Cockram et al., 2021). Studies have discovered two enzymes that catalyze NEIL1 polyubiquitination, tripartite motif 26 (TRIM26) and Mcl-1 ubiquitin ligase E3 (Mule). These enzymes can polyubiquitinate NEIL1 in vitro, and both catalyze the ubiquitination of NEIL1 at the same C-terminal lysine residue. TRIM26 and Mule are important for sustaining steady-state levels of NEIL1 and are required for DNA damage responses. NEIL1 may be induced by ionizing radiation and may contribute to ionizing radiation resistance after TRIM26 silencing (Edmonds et al., 2017). TRIM26 tightly regulates NEIL1 and NEIL3 levels and regulates the oxidative stress response induced by hydrogen peroxide (Konis et al., 2022). Another study showed that Pyridoxine (PN) reduced the protein level of NEIL2 by enhancing its ubiquitination and degradation (Banerjee et al., 2020).
Phosphorylation is another method of regulating NEIL protein activity. The phosphorylation of three serine residues in NEIL1 was revealed by mass spectrometry: S207, S306, and S61. Phosphorylation did not influence the enzyme activity or DNA binding at the three serine sites. However, the mutation of another phosphorylation site, Y263, to E, produced a completely inactive enzyme. C-Jun N-terminal kinase 1 (JNK1) is involved in NEIL1 phosphorylation. As a member of the MAPK family, JNK1 interacts with NEIL1 in vitro and phosphorylates residues, S207, S306, and S61 (Prakash et al., 2016). Another study showed that two kinases, protein kinase C (PKC) and cyclin-dependent kinase 5 (CDK5), phosphorylate NEIL2 in human SH-SY5Y neuroblastoma cells. The two kinases regulate NEIL2 function in different ways. CDK5 does not directly influence NEIL2 activity in vitro, whereas PKC phosphorylation of NEIL2 results in a significant decrease in NEIL2 repair activity. Interestingly, NEIL2 is rapidly dephosphorylated in response to oxidative stress in SH-SY5Y cells, suggesting that phosphorylation is a critical regulator of NEIL2 function, especially under oxidative stress (Myrup Holst et al., 2023).
4 NEIL proteins in cancer
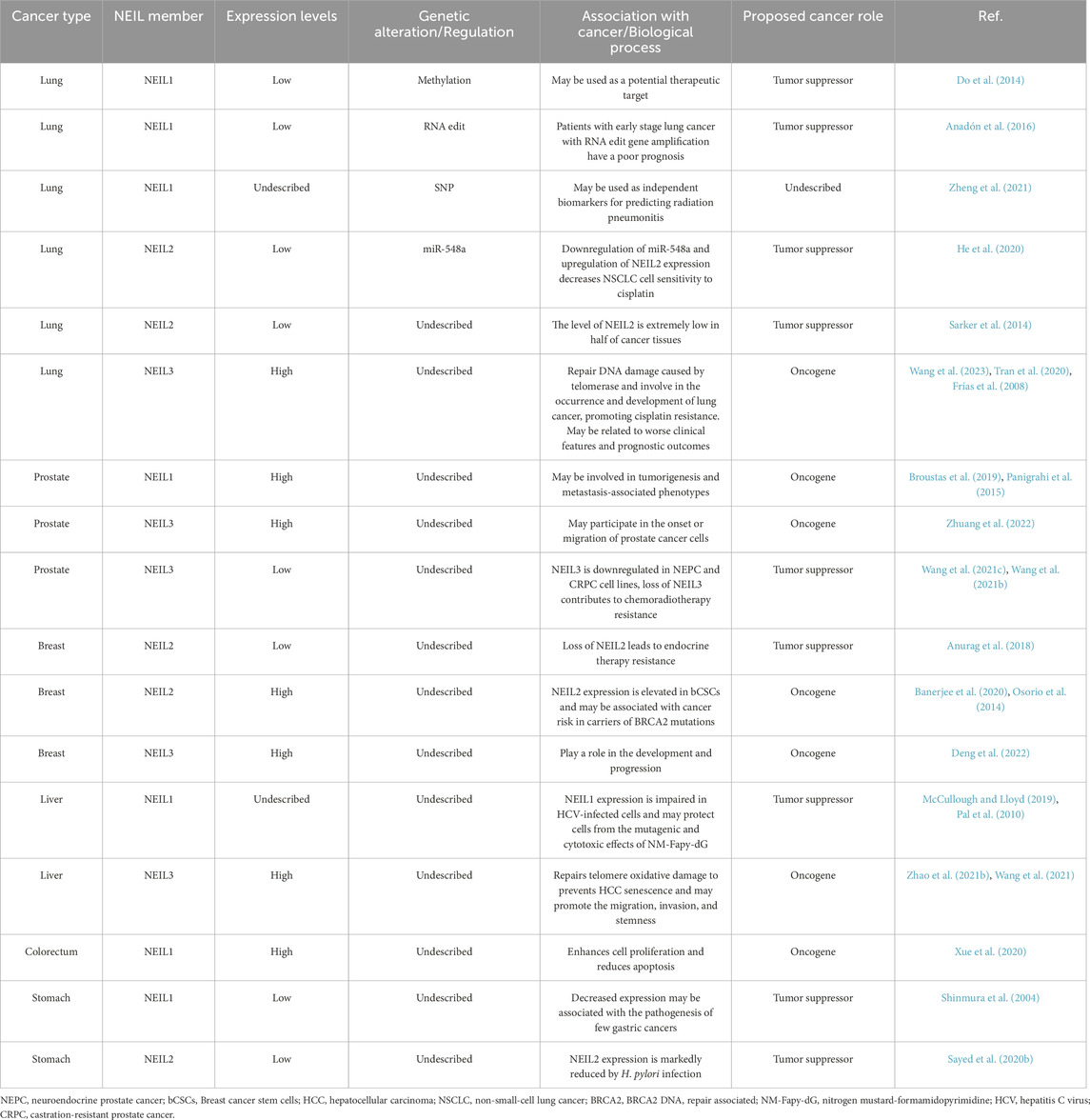
The NEIL family is modified in diverse cancer types (Table 1). These alterations include dysregulation of gene expression and SNPs. Our review focuses on the cancer types in which NEIL family members were involved and for which sufficient data was available.
4.1 NSCLC
With the second highest incidence of malignancy worldwide, NSCLC accounts for approximately half of all cancer-related deaths worldwide. Although considerable progress has been made in the diagnosis and treatment of NSCLC, the 5-year survival rate of patients with lung cancer remains less than 20%. Understanding the molecular mechanisms and potential therapeutic targets of NSCLC is urgently needed (Siegel et al., 2023; Herbst et al., 2018; Duma et al., 2019).
NEILs are crucial in the origin, progression, diagnosis, and treatment of NSCLC. The deletion, silencing, and abnormal expression of NEIL1 may play important roles in lung cancer pathogenesis. In one study, the incidence of lung tumors in Nth1−/−Neil1−/− mice was much higher than that in single knockout Nth1−/− or Neil1−/− mice (Chan et al., 2009). The NEIL1 promoter is frequently methylated in NSCLC, making it a potential therapeutic target (Do et al., 2014). The adeno-inosine-editing enzyme, ADAR1, is genetically amplified in NSCLC cell lines and primary tumors, and its overexpression enhances tumorigenic potential in cell culture and mouse models. Functionally, ADAR1 overexpression increases the editing frequency of lysine 242 AAA codons A-to-I of the target transcript of NEIL1. In the clinical setting, patients with early stage lung cancer with ADAR1 gene amplification have a poor prognosis (Anadón et al., 2016). Novel therapies targeting specific gene mutations in NSCLC are promising in terms of improving patient survival. Oxidative stress plays an important role in the progression of lung cancer. Smoking dramatically increases oxidative stress, and carotenoids are potent antioxidants. Lower doses of lycopene improve the levels of NEIL1, NEIL2, and NEIL3 in cigarette smoke-induced A549 human lung cancer epithelial cells. These findings can help elucidate the molecular mechanisms underlying the anti-lung cancer action (Cheng et al., 2020). Radiotherapy plays a vital role in the treatment of thoracic tumors; patients with lung cancer who receive radiotherapy may experience radiation-induced lung injury, which can cause radiation pneumonitis (Giuranno et al., 2019). A total of 174 lung cancer patients treated with radiotherapy were genotyped for the NEIL1 genetic variants, rs4462560 and rs7402844. NEIL1 mutations were related to the risk of radiation pneumonitis by regulation of NEIL1 expression and acted as independent biomarkers for predicting radiation pneumonitis in patients treated with thoracic radiotherapy (Zheng et al., 2021).
NEIL2 polymorphisms may affect the development and treatment sensitivity of lung cancer. It has been reported that in patients with advanced NSCLC, progression free survival is associated with rs8191670 in the Neil2 gene, which is a polymorphism (T/C). The potential molecular mechanism may be that miR-548a decreases the expression of NEIL2 through binding to its 3′UTR which contains rs8191670. Downregulation of miR-548a and upregulation of NEIL2 expression decreases NSCLC cell sensitivity to cisplatin (He et al., 2020). In addition, DNA from the R257L variant (rs8191664) shows a lower affinity for other repair proteins, especially Polβ, leading to reduced repair capacity and increased endogenous DNA damage, which can eventually lead to lung cancer (Dey et al., 2012). In lung cancer, NEIL2 acts as a tumor suppressor, and the level of NEIL2 is extremely low in half of cancer tissues. Smoking and exposure to secondhand smoke play vital roles in the development of lung cancer. Impaired NEIL2 expression in sidestream smoke-exposed nonsmokers may cause the accumulation of genomic DNA mutations, which could lead to sidestream smoke-induced lung cancer (Sarker et al., 2014).
Unlike NEIL1 and NEIL2, NEIL3 functions as an oncogenic factor in lung cancer. NEIL3 can repair DNA damage caused by telomerase in the S phase and reduce the destructive effects of ROS. NEIL3 is upregulated in NSCLC tissues, suggesting that NEIL3 is involved in the occurrence and development of lung cancer. Clinical correlation and prognostic analyses revealed that NEIL3 is related to worse clinical features and prognostic outcomes (Frías et al., 2008; Tran et al., 2020). Therefore, NEIL3 may be a potential therapeutic target and prognostic predictor. The transcription factor, MAZ, increases NEIL3 expression and inhibits DNA damage in lung adenocarcinoma cells, thereby promoting cisplatin resistance in the lung adenocarcinoma cells (Wang et al., 2023). In addition, NEIL3 partially activates the PI3K/AKT/mTOR signaling pathway. NEIL3 level positively correlate with chemosensitivity to cisplatin and paclitaxel (Huang and Hua, 2022). Several studies have established NEIL3-related survival and prognostic models, providing new diagnostic and treatment strategies for NSCLC (Zhao et al., 2022; Ali et al., 2022; Zhao et al., 2021a; Ömeroğlu Şimşek et al., 2020).
Taken together, NEIL1 and NEIL2 function as tumor suppressors and their abnormal expression can lead to the development of lung cancer and drug resistance. NEIL3 functions as an oncogenic factor and its high expression is associated with poor prognosis and chemoresistance.
4.2 Prostate cancer
PCa is a complex condition that affects a wide range of men globally. With early detection and treatment, patients with local disease and a low-to-moderate risk of recurrence generally have a good prognosis. Current research is aimed at improving the diagnosis and treatment of PCa and understanding of the basic biological characteristics of the disease at all stages (Rebello et al., 2021; Wasim et al., 2022; Lowrance et al., 2023).
RAD9 regulates BER by influencing NEIL1 levels, and RAD9A plays a vital role in prostate tumorigenesis and metastasis-associated phenotypes (Broustas et al., 2019; Panigrahi et al., 2015). Unlike NSCLC, NEIL3 plays multiple roles in PCa. It may participate in the onset or migration of PCa cells; however, its deletion can lead to resistance to chemoradiotherapy. NEIL3 is downregulated in neuroendocrine prostate cancer and CRPC cell lines, and NEIL3 is associated with a high Gleason score, but a good prognosis. NEIL3 modulates the cell cycle by negatively regulating ATR expression. Loss of NEIL3 contributes to chemoradiotherapy resistance in PCa, and may be a potential target for patients with chemoradiotherapy resistance (Wang et al., 2021c; Wang et al., 2021b). NEIL3 is a central gene involved in the inhibition of PCa progression by the combination of aspirin and lipitor (Wang et al., 2022b).
Fonofos, an organophosphate insecticide, the interaction between fonofos and NEIL3 rs1983132 significantly increases the risk of PCa in patients with a family history of PCa (Barry et al., 2011). Moreover, NEIL3 variants may be associated with PCa (Kim et al., 2016; Liu et al., 2020; Yadav et al., 2020). NEIL3 can also promote PCa metastasis. The regulation of NEIL3 by FOXM1 may be a potential pathway for promoting the migration of prostate cells while participating in anti-androgen resistance in PCa (Zhuang et al., 2022). According to bioinformatics analyses, NEIL3 is a potential biomarker for the prediction and prognosis of PCa (Teng et al., 2021).
In conclusion, NEIL proteins are related to the occurrence, metastasis, and sensitivity to chemoradiotherapy in PCa, and can be used as markers to predict efficacy and prognosis.
4.3 Breast cancer
Breast cancer is a complex disease involving both genetic and environmental factors. Diverse treatments for breast cancer have been developed; however, drug resistance remains a major problem. Breast cancer stem cells are major contributors to aggressiveness and drug resistance, posing a major challenge in cancer treatment. Thus, the detection and prognosis of breast cancer need to be improved (Barzaman et al., 2020; Tsang and Tse, 2020; Britt et al., 2020).
NEIL2 and NEIL3 play a role in the development and progression of breast cancer. NEIL2 rs1466785 and rs804271 are associated with cancer risk in carriers of BRCA2 mutations (Osorio et al., 2014; Benítez-Buelga et al., 2017). Furthermore, increased NEIL2 expression enhances the sensitivity of breast cancer cells to double-strand breaks and apolipoprotein B mRNA editing enzyme catalytic subunit 3 (APOBEC3) deaminase-mediated mutations by interfering with BER (Shen et al., 2020). The BRG1-EP300 complex drives NEIL3 transcription in breast cancer. EP300 acetylates nucleosomes in the promoter region, and BRG1 evicts acetylated nucleosomes from the chromatin, thereby promoting transcription. PARP1 co-localizes with BRG1 on the highly acetylated promoter of NEIL3, ensuring an open chromatin structure (Sobczak et al., 2020; Sobczak et al., 2019). NEIL3 may also be involved in the tumorigenesis induced by estrogen and progestin therapy in breast cancer (Deng et al., 2022).
Therapeutically, the curcumin analog, 3,5-bis (4-hydroxy-3-methoxy benzylidene) -N-methyl-4-piperidone (PAC), induces apoptosis by upregulating Bax expression and downregulating Bcl-2 expression in triple-negative breast cancer cell lines. PAC also upregulates the expression of NEIL2. This opens up a new perspective for triple-negative breast cancer treatment (Almalki et al., 2023). Regulation of cellular redox status may also be a potential way to treat drug-resistant breast cancer. Peg-functionalized zno nanoparticles can generate ROS and exert anti-cancer effects. In breast cancer, the binding of Sp1 to the NEIL2 promoter is boosted by low ROS, thereby promoting NEIL2 transcription and enhancing BER. However, high concentrations of ROS lead to a decrease in NEIL2, which results in the activation of p53 and further activates the intrinsic apoptotic pathway (Chakraborti et al., 2017). Another study showed that PN enhances the chemotherapy responsiveness of breast cancer stem cells through redox modulation. NEIL2 expression is elevated in breast cancer stem cells, leading to doxorubicin resistance. However, vitamin B6 and PN, inhibit NEIL2-mediated transcriptional coupling repair processes by reducing NEIL2 expression and inhibiting its association with RNA Pol II, thereby stimulating DNA damage and triggering ROS production (Banerjee et al., 2020).
Research on the mechanisms underlying resistance to endocrine therapy has also progressed. Loss of NEIL2 leads to endocrine therapy resistance via dysregulation of the G1-S transition, and miRNA regulation of NEIL2 may mediate the prognosis of hormone-treated breast cancer (Anurag et al., 2018). The SNPs of miRNA binding sites (miRSNPs) in the 3′-untranslated region of NEIL2 may affect the binding affinity of miRNA. For example, patients with genotype of NEIL2 rs6997097 who received only hormone therapy had significantly shorter disease-free survival and overall survival (Cumova et al., 2021). In addition, NEILs may serve as risk indicators of breast cancer (Matta et al., 2013).
In conclusion, NEIL2 may provide new perspectives for breast cancer treatment, and NEIL3 may have a role in the onset and progression of breast cancer.
4.4 Hepatocellular carcinoma
HCC is one of the most common cancers and a major global healthcare challenge (Vogel et al., 2022; Yang et al., 2019; Zhou and Song, 2021). The NEIL family is associated with liver cancer caused by hepatitis virus and aflatoxins (Li et al., 2017). Dietary exposure to aflatoxi (AFB1) and subsequent DNA damage are important promoters. Currently, there are several treatment options for HCC, and there is an urgent need to identify predictive biomarkers to inform treatment selection.
NEIL1 expression is impaired, whereas NEIL2 expression is unaffected in HCV-infected cells (Pal et al., 2010). Alkylation damage of DNA bases can be caused by diverse drugs, such as AFB1 and chemotherapeutic nitrogen mustard (NM). NEIL1 effectively recognizes and excises the highly mutagenic AFB1-deoxyguanosine adduct in mice, and NEIL1−/− mice have increased sensitivity to AFB1-induced HCC. Both NEIL1 and NEIL3 may protect cells from the mutagenic and cytotoxic effects of NM-formamidopyrimidine; however, NEIL1 may play an important role in initiating BER of AFB1-deoxyguanosine adduct (McCullough and Lloyd, 2019; Vartanian et al., 2017; Minko et al., 2019a).
Additionally, NEIL1 SNP variants are associated with an elevated risk of early onset HCC; in sub-Saharan Africa, patients with the NEIL1 I182M variant are at potential risk of early onset HCC (Zuckerman et al., 2023). The P68H variant showed a slight decrease in efficiency among residents of Qidong County, China, but the A51V and G245R variants showed almost the same activity as the wide type. However, A51V is highly sensitive to temperature, suggesting that its biological activity will be greatly reduced (Minko et al., 2019b).
Similar to NEIL1, NEIL3 is overexpressed in HCC and associated with poor survival. We showed that SNHG3 increases the binding of E2F1 to the promoter region of NEIL3, thereby activating the transcriptional signature of NEIL3 (Zhang et al., 2023). NEIL3 repairs telomere oxidative damage during mitosis and prevents HCC senescence (Zhao et al., 2021b). NEIL3 may promote the migration, invasion, and stemness of HCC cells by activating the BRAF/MEK/ERK/TWIST pathway or by regulating the PI3K/Akt/mTOR signaling pathway (Lai et al., 2022; Wang et al., 2021). A peptide vaccine cocktail derived from NEIL3 has shown initial success in phase I studies of advanced HCC (Ikeda et al., 2021). Several other studies have established prognostic models for HCC involving NEIL3 (Wang et al., 2022a; Hu et al., 2022; Ding et al., 2022; Wu et al., 2022; Wu et al., 2021; Wang et al., 2021a; Lu et al., 2023).
4.5 Other cancers
The roles of the NEIL family in various other cancers have been previously reported. CRC is one of the most common cancers worldwide (Dekker et al., 2019; Biller and Schrag, 2021); in CRC, an increase in NEIL1 enhances cell proliferation and reduces apoptosis (Xue et al., 2020); NEIL1 can act as a substrate for the enzymatic deacetylation activity of Sirt3, which may result in the regulation of CRC risk (Kabzinski et al., 2022; Kabziński et al., 2019). However, it remains unclear whether variants in the NEIL family contribute to CRC susceptibility (Dallosso et al., 2008; Broderick et al., 2006). NEIL2 may act as an important marker for predicting radiosensitivity in patients with cancer and is related to overall survival (Kobunai et al., 2011; Pardini et al., 2013; Sayed et al., 2020a; Corral et al., 2013). NEIL3 may also be a prognostic factor for CRC (Jiraskova et al., 2018).
Furthermore, the low activity of NEIL1 caused by mutations and its decreased expression in gastric cancer may be associated with the pathogenesis of few gastric cancers (Goto et al., 2010; Shinmura et al., 2004). Helicobacter pylori infection is closely associated with gastric cancer and NEIL2 expression is markedly reduced by Helicobacter pylori infection (Sayed et al., 2020b). In addition, the NEIL2 SNP is potentially associated with gastric cancer, esophageal adenocarcinoma, and Barrett’s esophagus risk (Mou et al., 2015; Elingarami et al., 2015; Ali et al., 2022).
The role of the NEIL family in astrocytoma, myeloma, and glioblastoma (Jin et al., 2013; Klattenhoff et al., 2017) and renal clear-cell carcinoma has also been reported (Sousa et al., 2019; Sousa et al., 2013; de Sousa et al., 2017; Sun and Liu, 2023; Peng et al., 2022).
5 Concluding remarks/future perspectives
Cellular DNA is constantly exposed to exogenous and endogenous damage, and the accumulation of this damage may result in oxidative DNA damage and strand breaks, thereby affecting genomic integrity and leading to cancer. BER is the primary method used to repair oxidative DNA damage (Iyama and Wilson, 2013). This pathway is initiated by damage-specific DNA glycosylases that recognize and clear damaged bases (Rosenquist et al., 2003; Bj Rås et al., 2017). As a special class of human DNA glycosylases, the NEIL family has a unique structure and a wide range of substrates (Grin and Zharkov, 2011). There is mounting evidence linking members of the NEIL family to cancer occurrence in humans.
As expected given the important role of DNA glycosylases in gene repair, all NEILs directly or indirectly affected cancer characteristics, including cell proliferation, chemoradiotherapy resistance, apoptosis, metastasis, and stemness. This review integrates the mechanisms by which the NEIL family influences the malignant behavior of cancer. Variants in the NEIL family affect cell growth, mainly by affecting the cell cycle. In addition, the abnormal expression of the NEIL family can affect DNA integrity and cause cancer cells to acquire drug resistance and stemness. Notably, NEIL3 can repair telomeres and may play a crucial role in promoting cancer progression.
The expression of members of the NEIL family is regulated by upstream molecules such as RAD9, EP300, and ADAR1. Moreover, post-translational modifications, especially acetylation, significantly influence NEIL expression. ROS levels in cells may also influence the expression of the NEIL family (Fleming et al., 2019).
Possible roles for the NEIL family in many common cancers have been reported. For example, multiple NEIL variants may be associated with cancer susceptibility (Kakhkharova et al., 2022; Zhai et al., 2008; Chae et al., 2016; Wei et al., 2012; Galick et al., 2017; Roy et al., 2007; Prakash et al., 2014). Many studies have listed the NEIL family as potential targets for cancer treatment or as prognostic prediction markers, which may provide suggestions for the precise classification of various cancers and the treatment of drug-resistant cancers (Liao et al., 2022). However, the role of the NEIL family varies in different cancers. Therefore, it is important to identify the pathways involved in the role of the NEIL family in specific cancers. Further comprehensive mechanistic studies are required to confirm these findings.
Author contributions
YC: Data curation, Writing – original draft. MM: Data curation, Writing – original draft. AZ: Data curation, Writing – original draft. XW: Supervision, Writing – review and editing, Conceptualization. WD: Conceptualization, Writing – review and editing, Supervision, Funding acquisition, Project administration.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This research was supported by grant from Natural Science Foundation of Liaoning Province (2023-BS-094).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Albelazi, M. S., Martin, P. R., Mohammed, S., Mutti, L., Parsons, J. L., and Elder, R. H. (2019). The biochemical role of the human NEIL1 and NEIL3 DNA glycosylases on model DNA replication forks. Genes (Basel) 10, 315. doi:10.3390/genes10040315
Ali, M. W., Chen, J., Yan, L., Wang, X., Dai, J. Y., Vaughan, T. L., et al. (2022). A risk variant for Barrett's esophagus and esophageal adenocarcinoma at chr8p23.1 affects enhancer activity and implicates multiple gene targets. Hum. Mol. Genet. 31, 3975–3986. doi:10.1093/hmg/ddac141
Aliyaskarova, U., Baiken, Y., Renaud, F., Couve, S., Kisselev, A. F., Saparbaev, M., et al. (2023). NEIL3-mediated proteasomal degradation facilitates the repair of cisplatin-induced DNA damage in human cells. Sci. Rep. 13, 5174. doi:10.1038/s41598-023-32186-3
Almalki, E., Al-Amri, A., Alrashed, R., Al-Zharani, M., and Semlali, A. (2023). The curcumin analog PAC is a potential solution for the treatment of triple-negative breast cancer by modulating the gene expression of DNA repair pathways. Int. J. Mol. Sci. 24, 9649. doi:10.3390/ijms24119649
AnadóN, C., Guil, S., Simó-Riudalbas, L., Moutinho, C., Setien, F., MartíNEZ-CardúS, A., et al. (2016). Gene amplification-associated overexpression of the RNA editing enzyme ADAR1 enhances human lung tumorigenesis. Oncogene 35, 4407–4413. doi:10.1038/onc.2015.469
Anurag, M., Punturi, N., Hoog, J., Bainbridge, M. N., Ellis, M. J., and Haricharan, S. (2018). Comprehensive profiling of DNA repair defects in breast cancer identifies a novel class of endocrine therapy resistance drivers. Clin. Cancer Res. 24, 4887–4899. doi:10.1158/1078-0432.Ccr-17-3702
Avesson, L., and Barry, G. (2014). The emerging role of RNA and DNA editing in cancer. Biochim. Biophys. Acta 1845, 308–316. doi:10.1016/j.bbcan.2014.03.001
Bacolla, A., Sengupta, S., Ye, Z., Yang, C., Mitra, J., De-Paula, R. B., et al. (2021). Heritable pattern of oxidized DNA base repair coincides with pre-targeting of repair complexes to open chromatin. Nucleic Acids Res. 49, 221–243. doi:10.1093/nar/gkaa1120
Banerjee, S., Mukherjee, S., Bhattacharya, A., Basak, U., Chakraborty, S., Paul, S., et al. (2020). Pyridoxine enhances chemo-responsiveness of breast cancer stem cells via redox reconditioning. Free Radic. Biol. Med. 152, 152–165. doi:10.1016/j.freeradbiomed.2020.02.031
Barry, K. H., Koutros, S., Berndt, S. I., Andreotti, G., Hoppin, J. A., Sandler, D. P., et al. (2011). Genetic variation in base excision repair pathway genes, pesticide exposure, and prostate cancer risk. Environ. Health Perspect. 119, 1726–1732. doi:10.1289/ehp.1103454
Barzaman, K., Karami, J., Zarei, Z., Hosseinzadeh, A., Kazemi, M. H., Moradi-Kalbolandi, S., et al. (2020). Breast cancer: biology, biomarkers, and treatments. Int. Immunopharmacol. 84, 106535. doi:10.1016/j.intimp.2020.106535
Bednarski, J. J., and Sleckman, B. P. (2019). At the intersection of DNA damage and immune responses. Nat. Rev. Immunol. 19, 231–242. doi:10.1038/s41577-019-0135-6
Benítez-Buelga, C., Baquero, J. M., Vaclova, T., Fernández, V., Martín, P., Inglada-Perez, L., et al. (2017). Genetic variation in the NEIL2 DNA glycosylase gene is associated with oxidative DNA damage in BRCA2 mutation carriers. Oncotarget 8, 114626–114636. doi:10.18632/oncotarget.22638
Bhakat, K. K., Hazra, T. K., and Mitra, S. (2004). Acetylation of the human DNA glycosylase NEIL2 and inhibition of its activity. Nucleic Acids Res. 32, 3033–3039. doi:10.1093/nar/gkh632
Bhakat, K. K., Sengupta, S., and Mitra, S. (2020). Fine-tuning of DNA base excision/strand break repair via acetylation. DNA Repair (Amst) 93, 102931. doi:10.1016/j.dnarep.2020.102931
Biller, L. H., and Schrag, D. (2021). Diagnosis and treatment of metastatic colorectal cancer: a review. Jama 325, 669–685. doi:10.1001/jama.2021.0106
Bj Rås, K., Sousa, M. M. L., Sharma, A., Fonseca, D. M., Ck, S. G., Bj Rås, M., et al. (2017). Monitoring of the spatial and temporal dynamics of BER/SSBR pathway proteins, including MYH, UNG2, MPG, NTH1 and NEIL1-3, during DNA replication. Nucleic Acids Res. 45, 8291–8301. doi:10.1093/nar/gkx476
Britt, K. L., Cuzick, J., and Phillips, K. A. (2020). Key steps for effective breast cancer prevention. Nat. Rev. Cancer 20, 417–436. doi:10.1038/s41568-020-0266-x
Broderick, P., Bagratuni, T., Vijayakrishnan, J., Lubbe, S., Chandler, I., and Houlston, R. S. (2006). Evaluation of NTHL1, NEIL1, NEIL2, MPG, TDG, UNG and SMUG1 genes in familial colorectal cancer predisposition. BMC Cancer 6, 243. doi:10.1186/1471-2407-6-243
Broustas, C. G., Hopkins, K. M., Panigrahi, S. K., Wang, L., Virk, R. K., and Lieberman, H. B. (2019). RAD9A promotes metastatic phenotypes through transcriptional regulation of anterior gradient 2 (AGR2). Carcinogenesis 40, 164–172. doi:10.1093/carcin/bgy131
Caliri, A. W., Tommasi, S., and Besaratinia, A. (2021). Relationships among smoking, oxidative stress, inflammation, macromolecular damage, and cancer. Mutat. Res. Rev. Mutat. Res. 787, 108365. doi:10.1016/j.mrrev.2021.108365
Chae, Y. K., Anker, J. F., Carneiro, B. A., Chandra, S., Kaplan, J., Kalyan, A., et al. (2016). Genomic landscape of DNA repair genes in cancer. Oncotarget 7, 23312–23321. doi:10.18632/oncotarget.8196
Chaisaingmongkol, J., Popanda, O., Warta, R., Dyckhoff, G., Herpel, E., Geiselhart, L., et al. (2012). Epigenetic screen of human DNA repair genes identifies aberrant promoter methylation of NEIL1 in head and neck squamous cell carcinoma. Oncogene 31, 5108–5116. doi:10.1038/onc.2011.660
Chakraborti, S., Chakraborty, S., Saha, S., Manna, A., Banerjee, S., Adhikary, A., et al. (2017). PEG-functionalized zinc oxide nanoparticles induce apoptosis in breast cancer cells through reactive oxygen species-dependent impairment of DNA damage repair enzyme NEIL2. Free Radic. Biol. Med. 103, 35–47. doi:10.1016/j.freeradbiomed.2016.11.048
Chakraborty, A., Tapryal, N., Islam, A., Mitra, S., and Hazra, T. (2021). Transcription coupled base excision repair in mammalian cells: so little is known and so much to uncover. DNA Repair (Amst) 107, 103204. doi:10.1016/j.dnarep.2021.103204
Chakraborty, A., Wakamiya, M., Venkova-Canova, T., Pandita, R. K., Aguilera-Aguirre, L., Sarker, A. H., et al. (2015). Neil2-null mice accumulate oxidized DNA bases in the transcriptionally active sequences of the genome and are susceptible to innate inflammation. J. Biol. Chem. 290, 24636–24648. doi:10.1074/jbc.M115.658146
Chan, M. K., Ocampo-Hafalla, M. T., Vartanian, V., Jaruga, P., Kirkali, G., Koenig, K. L., et al. (2009). Targeted deletion of the genes encoding NTH1 and NEIL1 DNA N-glycosylases reveals the existence of novel carcinogenic oxidative damage to DNA. DNA Repair (Amst) 8, 786–794. doi:10.1016/j.dnarep.2009.03.001
Cheng, J., Miller, B., Balbuena, E., and Eroglu, A. (2020). Lycopene protects against smoking-induced lung cancer by inducing base excision repair. Antioxidants (Basel) 9, 643. doi:10.3390/antiox9070643
Chen, L., Huan, X., Gao, X. D., Yu, W. H., Xiao, G. H., Li, T. F., et al. (2022). Biological functions of the DNA glycosylase NEIL3 and its role in disease progression including cancer. Cancers (Basel) 14, 5722. doi:10.3390/cancers14235722
Chung, H. J., Lee, J. R., Kim, T. M., Kim, S., Park, K., Kim, M. J., et al. (2023). ZNF212 promotes genomic integrity through direct interaction with TRAIP. Nucleic Acids Res. 51, 631–649. doi:10.1093/nar/gkac1226
Cockram, P. E., Kist, M., Prakash, S., Chen, S. H., Wertz, I. E., and Vucic, D. (2021). Ubiquitination in the regulation of inflammatory cell death and cancer. Cell Death Differ. 28, 591–605. doi:10.1038/s41418-020-00708-5
Corral, R., Lewinger, J. P., Joshi, A. D., Levine, A. J., Vandenberg, D. J., Haile, R. W., et al. (2013). Genetic variation in the base excision repair pathway, environmental risk factors, and colorectal adenoma risk. PLoS One 8, e71211. doi:10.1371/journal.pone.0071211
Cumova, A., Vymetalkova, V., Opattova, A., Bouskova, V., Pardini, B., Kopeckova, K., et al. (2021). Genetic variations in 3'UTRs of SMUG1 and NEIL2 genes modulate breast cancer risk, survival and therapy response. Mutagenesis 36, 269–279. doi:10.1093/mutage/geab017
Dallosso, A. R., Dolwani, S., Jones, N., Jones, S., Colley, J., Maynard, J., et al. (2008). Inherited predisposition to colorectal adenomas caused by multiple rare alleles of MUTYH but not OGG1, NUDT1, NTH1 or NEIL 1, 2 or 3. Gut 57, 1252–1255. doi:10.1136/gut.2007.145748
Das, A., Rajagopalan, L., Mathura, V. S., Rigby, S. J., Mitra, S., and Hazra, T. K. (2004). Identification of a zinc finger domain in the human NEIL2 (Nei-like-2) protein. J. Biol. Chem. 279, 47132–47138. doi:10.1074/jbc.M406224200
Davodabadi, F., Sajjadi, S. F., Sarhadi, M., Mirghasemi, S., Nadali Hezaveh, M., Khosravi, S., et al. (2023). Cancer chemotherapy resistance: mechanisms and recent breakthrough in targeted drug delivery. Eur. J. Pharmacol. 958, 176013. doi:10.1016/j.ejphar.2023.176013
De Bont, R., and Van Larebeke, N. (2004). Endogenous DNA damage in humans: a review of quantitative data. Mutagenesis 19, 169–185. doi:10.1093/mutage/geh025
Dekker, E., Tanis, P. J., Vleugels, J. L. A., Kasi, P. M., and Wallace, M. B. (2019). Colorectal cancer. Lancet 394, 1467–1480. doi:10.1016/s0140-6736(19)32319-0
Deng, Y., Huang, H., Shi, J., and Jin, H. (2022). Identification of candidate genes in breast cancer induced by estrogen plus progestogens using bioinformatic analysis. Int. J. Mol. Sci. 23, 11892. doi:10.3390/ijms231911892
De Sousa, J. F., Torrieri, R., Serafim, R. B., Di Cristofaro, L. F., Escanfella, F. D., Ribeiro, R., et al. (2017). Expression signatures of DNA repair genes correlate with survival prognosis of astrocytoma patients. Tumour Biol. 39, 1010428317694552. doi:10.1177/1010428317694552
Dey, S., Maiti, A. K., Hegde, M. L., Hegde, P. M., Boldogh, I., Sarkar, P. S., et al. (2012). Increased risk of lung cancer associated with a functionally impaired polymorphic variant of the human DNA glycosylase NEIL2. DNA Repair (Amst) 11, 570–578. doi:10.1016/j.dnarep.2012.03.005
Ding, J., He, X., Luo, W., Zhou, W., Chen, R., Cao, G., et al. (2022). Development and validation of a pyroptosis-related signature for predicting prognosis in hepatocellular carcinoma. Front. Genet. 13, 801419. doi:10.3389/fgene.2022.801419
Do, H., Wong, N. C., Murone, C., John, T., Solomon, B., Mitchell, P. L., et al. (2014). A critical re-assessment of DNA repair gene promoter methylation in non-small cell lung carcinoma. Sci. Rep. 4, 4186. doi:10.1038/srep04186
Dou, H., Mitra, S., and Hazra, T. K. (2003). Repair of oxidized bases in DNA bubble structures by human DNA glycosylases NEIL1 and NEIL2. J. Biol. Chem. 278, 49679–49684. doi:10.1074/jbc.M308658200
Duma, N., Santana-Davila, R., and Molina, J. R. (2019). Non-small cell lung cancer: epidemiology, screening, diagnosis, and treatment. Mayo Clin. Proc. 94, 1623–1640. doi:10.1016/j.mayocp.2019.01.013
Eckenroth, B. E., Bumgarner, J. D., Matsumoto-Elliott, O., David, S. S., and Doublié, S. (2023). Structural and biochemical insights into NEIL2's preference for abasic sites. Nucleic Acids Res. 51, 12508–12521. doi:10.1093/nar/gkad1075
Eckenroth, B. E., Cao, V. B., Averill, A. M., Dragon, J. A., and Doublié, S. (2021). Unique structural features of mammalian NEIL2 DNA glycosylase prime its activity for diverse DNA substrates and environments. Structure 29, 29–42.e4. doi:10.1016/j.str.2020.08.001
Edmonds, M. J., Carter, R. J., Nickson, C. M., Williams, S. C., and Parsons, J. L. (2017). Ubiquitylation-dependent regulation of NEIL1 by Mule and TRIM26 is required for the cellular DNA damage response. Nucleic Acids Res. 45, 726–738. doi:10.1093/nar/gkw959
Elingarami, S., Liu, H., Kalinjuma, A. V., Hu, W., Li, S., and He, N. (2015). Polymorphisms in NEIL-2, APE-1, CYP2E1 and MDM2 genes are independent predictors of gastric cancer risk in a Northern Jiangsu population (China). J. Nanosci. Nanotechnol. 15, 4815–4828. doi:10.1166/jnn.2015.10028
Fleming, A. M., Zhou, J., Wallace, S. S., and Burrows, C. J. (2015). A role for the fifth G-track in G-quadruplex forming oncogene promoter sequences during oxidative stress: do these “spare tires” have an evolved function? ACS Cent. Sci. 1, 226–233. doi:10.1021/acscentsci.5b00202
Fleming, A. M., Zhu, J., Howpay Manage, S. A., and Burrows, C. J. (2019). Human NEIL3 gene expression regulated by epigenetic-like oxidative DNA modification. J. Am. Chem. Soc. 141, 11036–11049. doi:10.1021/jacs.9b01847
Fouquerel, E., Parikh, D., and Opresko, P. (2016). DNA damage processing at telomeres: the ends justify the means. DNA Repair (Amst) 44, 159–168. doi:10.1016/j.dnarep.2016.05.022
Frías, C., García-Aranda, C., De Juan, C., Morán, A., Ortega, P., Gómez, A., et al. (2008). Telomere shortening is associated with poor prognosis and telomerase activity correlates with DNA repair impairment in non-small cell lung cancer. Lung Cancer 60, 416–425. doi:10.1016/j.lungcan.2007.11.001
Galick, H. A., Marsden, C. G., Kathe, S., Dragon, J. A., Volk, L., Nemec, A. A., et al. (2017). The NEIL1 G83D germline DNA glycosylase variant induces genomic instability and cellular transformation. Oncotarget 8, 85883–85895. doi:10.18632/oncotarget.20716
Giuranno, L., Ient, J., De Ruysscher, D., and Vooijs, M. A. (2019). Radiation-induced lung injury (RILI), Front. Oncol. 9, 877. doi:10.3389/fonc.2019.00877
Gohil, D., Sarker, A. H., and Roy, R. (2023). Base excision repair: mechanisms and impact in biology, disease, and medicine. Int. J. Mol. Sci. 24, 14186. doi:10.3390/ijms241814186
Goto, M., Shinmura, K., Tao, H., Tsugane, S., and Sugimura, H. (2010). Three novel NEIL1 promoter polymorphisms in gastric cancer patients. World J. Gastrointest. Oncol. 2, 117–120. doi:10.4251/wjgo.v2.i2.117
Grin, I. R., and Zharkov, D. O. (2011). Eukaryotic endonuclease VIII-like proteins: new components of the base excision DNA repair system. Biochem. (Mosc) 76, 80–93. doi:10.1134/s000629791101010x
Grundy, G. J., and Parsons, J. L. (2020). Base excision repair and its implications to cancer therapy. Essays Biochem. 64, 831–843. doi:10.1042/ebc20200013
Guan, X., Bai, H., Shi, G., Theriot, C. A., Hazra, T. K., Mitra, S., et al. (2007). The human checkpoint sensor Rad9-Rad1-Hus1 interacts with and stimulates NEIL1 glycosylase. Nucleic Acids Res. 35, 2463–2472. doi:10.1093/nar/gkm075
Ha, A., Lin, Y., and Yan, S. (2020). A non-canonical role for the DNA glycosylase NEIL3 in suppressing APE1 endonuclease-mediated ssDNA damage. J. Biol. Chem. 295, 14222–14235. doi:10.1074/jbc.RA120.014228
Hanna, B. M. F., Michel, M., Helleday, T., and Mortusewicz, O. (2021). NEIL1 and NEIL2 are recruited as potential backup for OGG1 upon OGG1 depletion or inhibition by TH5487. Int. J. Mol. Sci. 22, 4542. doi:10.3390/ijms22094542
Hazra, T. K., Izumi, T., Boldogh, I., Imhoff, B., Kow, Y. W., Jaruga, P., et al. (2002a). Identification and characterization of a human DNA glycosylase for repair of modified bases in oxidatively damaged DNA. Proc. Natl. Acad. Sci. U. S. A. 99, 3523–3528. doi:10.1073/pnas.062053799
Hazra, T. K., Kow, Y. W., Hatahet, Z., Imhoff, B., Boldogh, I., Mokkapati, S. K., et al. (2002b). Identification and characterization of a novel human DNA glycosylase for repair of cytosine-derived lesions. J. Biol. Chem. 277, 30417–30420. doi:10.1074/jbc.C200355200
Hazra, T. K., and Mitra, S. (2006). Purification and characterization of NEIL1 and NEIL2, members of a distinct family of mammalian DNA glycosylases for repair of oxidized bases. Methods Enzymol. 408, 33–48. doi:10.1016/s0076-6879(06)08003-7
Hegde, M. L., Hegde, P. M., Arijit, D., Boldogh, I., and Mitra, S. (2012). Human DNA glycosylase NEIL1's interactions with downstream repair proteins is critical for efficient repair of oxidized DNA base damage and enhanced cell survival. Biomolecules 2, 564–578. doi:10.3390/biom2040564
Hegde, M. L., Hegde, P. M., Bellot, L. J., Mandal, S. M., Hazra, T. K., Li, G. M., et al. (2013a). Prereplicative repair of oxidized bases in the human genome is mediated by NEIL1 DNA glycosylase together with replication proteins. Proc. Natl. Acad. Sci. U. S. A. 110, E3090–E3099. doi:10.1073/pnas.1304231110
Hegde, M. L., Tsutakawa, S. E., Hegde, P. M., Holthauzen, L. M., Li, J., Oezguen, N., et al. (2013b). The disordered C-terminal domain of human DNA glycosylase NEIL1 contributes to its stability via intramolecular interactions. J. Mol. Biol. 425, 2359–2371. doi:10.1016/j.jmb.2013.03.030
Hegde, P. M., Dutta, A., Sengupta, S., Mitra, J., Adhikari, S., Tomkinson, A. E., et al. (2015). The C-terminal domain (CTD) of human DNA glycosylase NEIL1 is required for forming BERosome repair complex with DNA replication proteins at the replicating genome: dominant negative function of the ctd. J. Biol. Chem. 290, 20919–20933. doi:10.1074/jbc.M115.642918
Herbst, R. S., Morgensztern, D., and Boshoff, C. (2018). The biology and management of non-small cell lung cancer. Nature 553, 446–454. doi:10.1038/nature25183
He, W., Pang, L., Gong, S., Wang, X., and Hou, L. (2020). Nei endonuclease VIII-like 2 gene rs8191670 polymorphism affects the sensitivity of non-small cell lung cancer to cisplatin by binding with MiR-548a. J. Cancer 11, 4801–4809. doi:10.7150/jca.47495
Hoeijmakers, J. H. (2001). Genome maintenance mechanisms for preventing cancer. Nature 411, 366–374. doi:10.1038/35077232
Hopkins, J. L., Lan, L., and Zou, L. (2022). DNA repair defects in cancer and therapeutic opportunities. Genes Dev. 36, 278–293. doi:10.1101/gad.349431.122
Hua, A. B., and Sweasy, J. B. (2023). Functional roles and cancer variants of the bifunctional glycosylase NEIL2. Environ. Mol. Mutagen 65 (Suppl. 1), 40–56. doi:10.1002/em.22555
Huang, H., and Hua, Q. (2022). NEIL3 mediates lung cancer progression and modulates PI3K/AKT/mTOR signaling: a potential therapeutic target. Int. J. Genomics 2022, 8348499. doi:10.1155/2022/8348499
Hu, B., Qu, C., Qi, W. J., Liu, C. H., and Xiu, D. R. (2022). Development and verification of the glycolysis-associated and immune-related prognosis signature for hepatocellular carcinoma. Front. Genet. 13, 955673. doi:10.3389/fgene.2022.955673
Huskova, A., Dinesh, D. C., Srb, P., Boura, E., Veverka, V., and Silhan, J. (2022). Model of abasic site DNA cross-link repair; from the architecture of NEIL3 DNA binding domains to the X-structure model. Nucleic Acids Res. 50, 10436–10448. doi:10.1093/nar/gkac793
Ikeda, M., Okusaka, T., Ohno, I., Mitsunaga, S., Kondo, S., Ueno, H., et al. (2021). Phase I studies of peptide vaccine cocktails derived from GPC3, WDRPUH and NEIL3 for advanced hepatocellular carcinoma. Immunotherapy 13, 371–385. doi:10.2217/imt-2020-0278
Imani Nejad, M., Housh, K., Rodriguez, A. A., Haldar, T., Kathe, S., Wallace, S. S., et al. (2020). Unhooking of an interstrand cross-link at DNA fork structures by the DNA glycosylase NEIL3. DNA Repair (Amst) 86, 102752. doi:10.1016/j.dnarep.2019.102752
Iyama, T., and Wilson, D. M. (2013). DNA repair mechanisms in dividing and non-dividing cells. DNA Repair (Amst) 12, 620–636. doi:10.1016/j.dnarep.2013.04.015
Jin, T. B., Li, X. L., Yang, H., Jiri, M., Shi, X. G., Yuan, D. Y., et al. (2013). Association of polymorphisms in FLT3, EGFR, ALOX5, and NEIL3 with glioblastoma in the Han Chinese population. Med. Oncol. 30, 718. doi:10.1007/s12032-013-0718-1
Jiraskova, K., Hughes, D. J., Brezina, S., Gumpenberger, T., Veskrnova, V., Buchler, T., et al. (2018). Functional polymorphisms in DNA repair genes are associated with sporadic colorectal cancer susceptibility and clinical outcome. Int. J. Mol. Sci. 20, 97. doi:10.3390/ijms20010097
Kabzinski, J., Walczak, A., and Majsterek, I. (2022). Sirt3 regulates response to oxidative stress by interacting with BER proteins in colorectal cancer. Genet. Res. (Camb) 2022, 7299555. doi:10.1155/2022/7299555
Kabziński, J., Walczak, A., Mik, M., and Majsterek, I. (2019). Sirt3 regulates the level of mitochondrial DNA repair activity through deacetylation of NEIL1, NEIL2, OGG1, MUTYH, APE1 and LIG3 in colorectal cancer. Pol. Przegl Chir. 92, 1–4. doi:10.5604/01.3001.0013.5539
Kakhkharova, Z. I., Zharkov, D. O., and Grin, I. R. (2022). A low-activity polymorphic variant of human NEIL2 DNA glycosylase. Int. J. Mol. Sci. 23, 2212. doi:10.3390/ijms23042212
Karlsen, T. R., Olsen, M. B., Kong, X. Y., Yang, K., Quiles-Jiménez, A., Kroustallaki, P., et al. (2022). NEIL3-deficient bone marrow displays decreased hematopoietic capacity and reduced telomere length. Biochem. Biophys. Rep. 29, 101211. doi:10.1016/j.bbrep.2022.101211
Kim, Y. S., Kim, Y., Choi, J. W., Oh, H. E., and Lee, J. H. (2016). Genetic variants and risk of prostate cancer using pathway analysis of a genome-wide association study. Neoplasma 63, 629–634. doi:10.4149/neo_2016_418
Kinslow, C. J., El-Zein, R. A., Hill, C. E., Wickliffe, J. K., and Abdel-Rahman, S. Z. (2008). Single nucleotide polymorphisms 5' upstream the coding region of the NEIL2 gene influence gene transcription levels and alter levels of genetic damage. Genes Chromosom. Cancer 47, 923–932. doi:10.1002/gcc.20594
Kinslow, C. J., El-Zein, R. A., Rondelli, C. M., Hill, C. E., Wickliffe, J. K., and Abdel-Rahman, S. Z. (2010). Regulatory regions responsive to oxidative stress in the promoter of the human DNA glycosylase gene NEIL2. Mutagenesis 25, 171–177. doi:10.1093/mutage/gep058
Kladova, O. A., Grin, I. R., Fedorova, O. S., Kuznetsov, N. A., and Zharkov, D. O. (2019). Conformational dynamics of damage processing by human DNA glycosylase NEIL1. J. Mol. Biol. 431, 1098–1112. doi:10.1016/j.jmb.2019.01.030
Klattenhoff, A. W., Thakur, M., Chu, C. S., Ray, D., Habib, S. L., and Kidane, D. (2017). Loss of NEIL3 DNA glycosylase markedly increases replication associated double strand breaks and enhances sensitivity to ATR inhibitor in glioblastoma cells. Oncotarget 8, 112942–112958. doi:10.18632/oncotarget.22896
Kobunai, T., Watanabe, T., and Fukusato, T. (2011). REG4, NEIL2, and BIRC5 gene expression correlates with gamma-radiation sensitivity in patients with rectal cancer receiving radiotherapy. Anticancer Res. 31, 4147–4153. Available online at: https://ar.iiarjournals.org/content/31/12/4147.long
Konis, S. M. R., Hughes, J. R., and Parsons, J. L. (2022). TRIM26 maintains cell survival in response to oxidative stress through regulating DNA glycosylase stability. Int. J. Mol. Sci. 23, 11613. doi:10.3390/ijms231911613
Kothandapani, A., Dangeti, V. S., Brown, A. R., Banze, L. A., Wang, X. H., Sobol, R. W., et al. (2011). Novel role of base excision repair in mediating cisplatin cytotoxicity. J. Biol. Chem. 286, 14564–14574. doi:10.1074/jbc.M111.225375
Krokeide, S. Z., Laerdahl, J. K., Salah, M., Luna, L., Cederkvist, F. H., Fleming, A. M., et al. (2013). Human NEIL3 is mainly a monofunctional DNA glycosylase removing spiroimindiohydantoin and guanidinohydantoin. DNA Repair (Amst) 12, 1159–1164. doi:10.1016/j.dnarep.2013.04.026
Lai, H. H., Hung, L. Y., Yen, C. J., Hung, H. C., Chen, R. Y., Ku, Y. C., et al. (2022). NEIL3 promotes hepatoma epithelial-mesenchymal transition by activating the BRAF/MEK/ERK/TWIST signaling pathway. J. Pathol. 258, 339–352. doi:10.1002/path.6001
Lee, S. H., Kim, H. P., Kang, J. K., Song, S. H., Han, S. W., and Kim, T. Y. (2017). Identification of diverse adenosine-to-inosine RNA editing subtypes in colorectal cancer. Cancer Res. Treat. 49, 1077–1087. doi:10.4143/crt.2016.301
Liao, W., Huang, S., Li, L., Wang, J., Li, J., Chen, Y., et al. (2022). Pan-cancer landscape of NEIL3 in tumor microenvironment: a promising predictor for chemotherapy and immunotherapy. Cancers (Basel) 15, 109. doi:10.3390/cancers15010109
Lieberman, H. B., Panigrahi, S. K., Hopkins, K. M., Wang, L., and Broustas, C. G. (2017). p53 and RAD9, the DNA damage response, and regulation of transcription networks. Radiat. Res. 187, 424–432. doi:10.1667/rr003cc.1
Li, N., Wang, J., Wallace, S. S., Chen, J., Zhou, J., and D'Andrea, A. D. (2020). Cooperation of the NEIL3 and Fanconi anemia/BRCA pathways in interstrand crosslink repair. Nucleic Acids Res. 48, 3014–3028. doi:10.1093/nar/gkaa038
Li, N., Xu, Y., Chen, H., Chen, L., Zhang, Y., Yu, T., et al. (2022). NEIL3 contributes to the Fanconi anemia/BRCA pathway by promoting the downstream double-strand break repair step. Cell Rep. 41, 111600. doi:10.1016/j.celrep.2022.111600
Liu, M., Doublié, S., and Wallace, S. S. (2013). Neil3, the final frontier for the DNA glycosylases that recognize oxidative damage. Mutat. Res. 743-744, 4–11. doi:10.1016/j.mrfmmm.2012.12.003
Liu, W., Zheng, S. L., Na, R., Wei, L., Sun, J., Gallagher, J., et al. (2020). Distinct genomic alterations in prostate tumors derived from African American men. Mol. Cancer Res. 18, 1815–1824. doi:10.1158/1541-7786.Mcr-20-0648
Li, Y., Xia, Y., Han, M., Chen, G., Zhang, D., Thasler, W. E., et al. (2017). IFN-α-mediated base excision repair pathway correlates with antiviral response against hepatitis B virus infection. Sci. Rep. 7, 12715. doi:10.1038/s41598-017-13082-z
Lotsof, E. R., Krajewski, A. E., Anderson-Steele, B., Rogers, J., Zhang, L., Yeo, J., et al. (2022). NEIL1 recoding due to RNA editing impacts lesion-specific recognition and excision. J. Am. Chem. Soc. 144, 14578–14589. doi:10.1021/jacs.2c03625
Lowrance, W., Dreicer, R., Jarrard, D. F., Scarpato, K. R., Kim, S. K., Kirkby, E., et al. (2023). Updates to advanced prostate cancer: AUA/SUO guideline (2023). J. Urol. 209, 1082–1090. doi:10.1097/ju.0000000000003452
Lu, Y., Wang, S., Chi, T., Zhao, Y., Guo, H., Wang, H., et al. (2023). DNA damage repair-related gene signature for identifying the immune status and predicting the prognosis of hepatocellular carcinoma. Sci. Rep. 13, 18978. doi:10.1038/s41598-023-45999-z
Makasheva, K. A., Endutkin, A. V., and Zharkov, D. O. (2020). Requirements for DNA bubble structure for efficient cleavage by helix-two-turn-helix DNA glycosylases. Mutagenesis 35, 119–128. doi:10.1093/mutage/gez047
Mandal, S. M., Hegde, M. L., Chatterjee, A., Hegde, P. M., Szczesny, B., Banerjee, D., et al. (2012). Role of human DNA glycosylase Nei-like 2 (NEIL2) and single strand break repair protein polynucleotide kinase 3'-phosphatase in maintenance of mitochondrial genome. J. Biol. Chem. 287, 2819–2829. doi:10.1074/jbc.M111.272179
Manfioletti, G., and Fedele, M. (2022). Epithelial-mesenchymal transition (EMT) 2021. Int. J. Mol. Sci. 23, 5848. doi:10.3390/ijms23105848
Marie-Egyptienne, D. T., Lohse, I., and Hill, R. P. (2013). Cancer stem cells, the epithelial to mesenchymal transition (EMT) and radioresistance: potential role of hypoxia. Cancer Lett. 341, 63–72. doi:10.1016/j.canlet.2012.11.019
Matta, J., Morales, L., Dutil, J., Bayona, M., Alvarez, C., and Suarez, E. (2013). Differential expression of DNA repair genes in Hispanic women with breast cancer. Mol. Cancer Biol. 1, 54. doi:10.9777/mcb.2013.10006
Mccullough, A. K., and Lloyd, R. S. (2019). Mechanisms underlying aflatoxin-associated mutagenesis - implications in carcinogenesis. DNA Repair (Amst) 77, 76–86. doi:10.1016/j.dnarep.2019.03.004
Mcdonald, D. T., Wang, P. S., Moitoza Johnson, J., and Tsai, M. S. (2023). Using affinity pulldown assays to study protein-protein interactions of human NEIL1 glycosylase and the checkpoint protein RAD9-RAD1-HUS1 (9-1-1) complex. Methods Mol. Biol. 2701, 199–207. doi:10.1007/978-1-0716-3373-1_13
Minko, I. G., Christov, P. P., Li, L., Stone, M. P., Mccullough, A. K., and Lloyd, R. S. (2019a). Processing of N(5)-substituted formamidopyrimidine DNA adducts by DNA glycosylases NEIL1 and NEIL3. DNA Repair (Amst) 73, 49–54. doi:10.1016/j.dnarep.2018.11.001
Minko, I. G., Vartanian, V. L., Tozaki, N. N., Coskun, E., Coskun, S. H., Jaruga, P., et al. (2020). Recognition of DNA adducts by edited and unedited forms of DNA glycosylase NEIL1. DNA Repair (Amst) 85, 102741. doi:10.1016/j.dnarep.2019.102741
Minko, I. G., Vartanian, V. L., Tozaki, N. N., Linde, O. K., Jaruga, P., Coskun, S. H., et al. (2019b). Characterization of rare NEIL1 variants found in East Asian populations. DNA Repair (Amst) 79, 32–39. doi:10.1016/j.dnarep.2019.05.001
Morland, I., Rolseth, V., Luna, L., Rognes, T., BjøRåS, M., and Seeberg, E. (2002). Human DNA glycosylases of the bacterial Fpg/MutM superfamily: an alternative pathway for the repair of 8-oxoguanine and other oxidation products in DNA. Nucleic Acids Res. 30, 4926–4936. doi:10.1093/nar/gkf618
Mou, X., Li, T., Wang, J., Ali, Z., Zhang, Y., Chen, Z., et al. (2015). Genetic variation of BCL2 (rs2279115), NEIL2 (rs804270), LTA (rs909253), PSCA (rs2294008) and PLCE1 (rs3765524, rs10509670) genes and their correlation to gastric cancer risk based on universal tagged arrays and Fe3O4 magnetic nanoparticles. J. Biomed. Nanotechnol. 11, 2057–2066. doi:10.1166/jbn.2015.2113
Myrup Holst, C., Brøndum Andersen, N., Thinggaard, V., Tilken, M., Lautrup, S., Tesauro, C., et al. (2023). Phosphorylation of the human DNA glycosylase NEIL2 is affected by oxidative stress and modulates its activity. Antioxidants (Basel) 12, 355. doi:10.3390/antiox12020355
Neurauter, C. G., Luna, L., and BjøRåS, M. (2012). Release from quiescence stimulates the expression of human NEIL3 under the control of the Ras dependent ERK-MAP kinase pathway. DNA Repair (Amst) 11, 401–409. doi:10.1016/j.dnarep.2012.01.007
Ni, F., Tang, H., Wang, C., Wang, Z., Yu, F., Chen, B., et al. (2019). Berzosertib (VE-822) inhibits gastric cancer cell proliferation via base excision repair system. Cancer Manag. Res. 11, 8391–8405. doi:10.2147/cmar.S217375
Ömeroğlu Şimşek, G., Ağababaoğlu, İ., Dursun, D., Özekinci, S., ErçETIN, P., Ellidokuz, H., et al. (2020). Evaluation of gene expression levels in the diagnosis of lung adenocarcinoma and malignant pleural mesothelioma. Turk Gogus Kalp Damar Cerrahisi Derg. 28, 188–196. doi:10.5606/tgkdc.dergisi.2020.17279
Onyango, P., Celic, I., Mccaffery, J. M., Boeke, J. D., and Feinberg, A. P. (2002). SIRT3, a human SIR2 homologue, is an NAD-dependent deacetylase localized to mitochondria. Proc. Natl. Acad. Sci. U. S. A. 99, 13653–13658. doi:10.1073/pnas.222538099
Orlando, K. A., Nguyen, V., Raab, J. R., Walhart, T., and Weissman, B. E. (2019). Remodeling the cancer epigenome: mutations in the SWI/SNF complex offer new therapeutic opportunities. Expert Rev. Anticancer Ther. 19, 375–391. doi:10.1080/14737140.2019.1605905
Osorio, A., Milne, R. L., Kuchenbaecker, K., Vaclová, T., Pita, G., Alonso, R., et al. (2014). DNA glycosylases involved in base excision repair may be associated with cancer risk in BRCA1 and BRCA2 mutation carriers. PLoS Genet. 10, e1004256. doi:10.1371/journal.pgen.1004256
Pal, S., Polyak, S. J., Bano, N., Qiu, W. C., Carithers, R. L., Shuhart, M., et al. (2010). Hepatitis C virus induces oxidative stress, DNA damage and modulates the DNA repair enzyme NEIL1. J. Gastroenterol. Hepatol. 25, 627–634. doi:10.1111/j.1440-1746.2009.06128.x
Panigrahi, S. K., Hopkins, K. M., and Lieberman, H. B. (2015). Regulation of NEIL1 protein abundance by RAD9 is important for efficient base excision repair. Nucleic Acids Res. 43, 4531–4546. doi:10.1093/nar/gkv327
Pardini, B., Rosa, F., Barone, E., Di Gaetano, C., Slyskova, J., Novotny, J., et al. (2013). Variation within 3'-UTRs of base excision repair genes and response to therapy in colorectal cancer patients: a potential modulation of microRNAs binding. Clin. Cancer Res. 19, 6044–6056. doi:10.1158/1078-0432.Ccr-13-0314
Peng, L., Liang, J., Wang, Q., and Chen, G. (2022). A DNA damage repair gene signature associated with immunotherapy response and clinical prognosis in clear cell renal cell carcinoma. Front. Genet. 13, 798846. doi:10.3389/fgene.2022.798846
Prakash, A., Cao, V. B., and Doublié, S. (2016). Phosphorylation sites identified in the NEIL1 DNA glycosylase are potential targets for the JNK1 kinase. PLoS One 11, e0157860. doi:10.1371/journal.pone.0157860
Prakash, A., Carroll, B. L., Sweasy, J. B., Wallace, S. S., and Doublié, S. (2014). Genome and cancer single nucleotide polymorphisms of the human NEIL1 DNA glycosylase: activity, structure, and the effect of editing. DNA Repair (Amst) 14, 17–26. doi:10.1016/j.dnarep.2013.12.003
Rebello, R. J., Oing, C., Knudsen, K. E., Loeb, S., Johnson, D. C., Reiter, R. E., et al. (2021). Prostate cancer. Nat. Rev. Dis. Prim. 7, 9. doi:10.1038/s41572-020-00243-0
Rodriguez, A. A., Wojtaszek, J. L., Greer, B. H., Haldar, T., Gates, K. S., Williams, R. S., et al. (2020). An autoinhibitory role for the GRF zinc finger domain of DNA glycosylase NEIL3. J. Biol. Chem. 295, 15566–15575. doi:10.1074/jbc.RA120.015541
Rosenquist, T. A., Zaika, E., Fernandes, A. S., Zharkov, D. O., Miller, H., and Grollman, A. P. (2003). The novel DNA glycosylase, NEIL1, protects mammalian cells from radiation-mediated cell death. DNA Repair (Amst) 2, 581–591. doi:10.1016/s1568-7864(03)00025-9
Rottenberg, S., Disler, C., and Perego, P. (2021). The rediscovery of platinum-based cancer therapy. Nat. Rev. Cancer 21, 37–50. doi:10.1038/s41568-020-00308-y
Roy, L. M., Jaruga, P., Wood, T. G., Mccullough, A. K., Dizdaroglu, M., and Lloyd, R. S. (2007). Human polymorphic variants of the NEIL1 DNA glycosylase. J. Biol. Chem. 282, 15790–15798. doi:10.1074/jbc.M610626200
Sarker, A. H., Chatterjee, A., Williams, M., Lin, S., Havel, C., Jacob, P., et al. (2014). NEIL2 protects against oxidative DNA damage induced by sidestream smoke in human cells. PLoS One 9, e90261. doi:10.1371/journal.pone.0090261
Sarker, A. H., Cooper, P. K., and Hazra, T. K. (2021). DNA glycosylase NEIL2 functions in multiple cellular processes. Prog. Biophys. Mol. Biol. 164, 72–80. doi:10.1016/j.pbiomolbio.2021.03.003
Sayed, I. M., Chakraborty, A., Abd El-Hafeez, A. A., Sharma, A., Sahan, A. Z., Huang, W. J. M., et al. (2020a). The DNA glycosylase NEIL2 suppresses fusobacterium-infection-induced inflammation and DNA damage in colonic epithelial cells. Cells 9, 1980. doi:10.3390/cells9091980
Sayed, I. M., Sahan, A. Z., Venkova, T., Chakraborty, A., Mukhopadhyay, D., Bimczok, D., et al. (2020b). Helicobacter pylori infection downregulates the DNA glycosylase NEIL2, resulting in increased genome damage and inflammation in gastric epithelial cells. J. Biol. Chem. 295, 11082–11098. doi:10.1074/jbc.RA119.009981
Schomacher, L., Han, D., Musheev, M. U., Arab, K., KienhöFER, S., Von Seggern, A., et al. (2016). Neil DNA glycosylases promote substrate turnover by Tdg during DNA demethylation. Nat. Struct. Mol. Biol. 23, 116–124. doi:10.1038/nsmb.3151
Seçme, M., Urgancı, A. B. E., Üzen, R., Aslan, A., and TıRAŞ, F. (2023). Determination of the effects of fusaric acid, a mycotoxin, on cytotoxicity, gamma-H2AX, 8-hydroxy-2 deoxyguanosine and DNA repair gene expressions in pancreatic cancer cells. Toxicon 231, 107179. doi:10.1016/j.toxicon.2023.107179
Semlow, D. R., and Walter, J. C. (2021). Mechanisms of vertebrate DNA interstrand cross-link repair. Annu. Rev. Biochem. 90, 107–135. doi:10.1146/annurev-biochem-080320-112510
Sengupta, S., Yang, C., Hegde, M. L., Hegde, P. M., Mitra, J., Pandey, A., et al. (2018). Acetylation of oxidized base repair-initiating NEIL1 DNA glycosylase required for chromatin-bound repair complex formation in the human genome increases cellular resistance to oxidative stress. DNA Repair (Amst) 66-67, 1–10. doi:10.1016/j.dnarep.2018.04.001
Shen, B., Chapman, J. H., Custance, M. F., Tricola, G. M., Jones, C. E., and Furano, A. V. (2020). Perturbation of base excision repair sensitizes breast cancer cells to APOBEC3 deaminase-mediated mutations. Elife 9, e51605. doi:10.7554/eLife.51605
Shinmura, K., Kato, H., Kawanishi, Y., Igarashi, H., Goto, M., Tao, H., et al. (2016). Abnormal expressions of DNA glycosylase genes NEIL1, NEIL2, and NEIL3 are associated with somatic mutation loads in human cancer. Oxid. Med. Cell Longev. 2016, 1546392. doi:10.1155/2016/1546392
Shinmura, K., Tao, H., Goto, M., Igarashi, H., Taniguchi, T., Maekawa, M., et al. (2004). Inactivating mutations of the human base excision repair gene NEIL1 in gastric cancer. Carcinogenesis 25, 2311–2317. doi:10.1093/carcin/bgh267
Shvedunova, M., and Akhtar, A. (2022). Modulation of cellular processes by histone and non-histone protein acetylation. Nat. Rev. Mol. Cell Biol. 23, 329–349. doi:10.1038/s41580-021-00441-y
Siegel, R. L., Miller, K. D., Wagle, N. S., and Jemal, A. (2023). Cancer statistics, 2023. CA Cancer J. Clin. 73, 17–48. doi:10.3322/caac.21763
Sobczak, M., Pietrzak, J., Płoszaj, T., and Robaszkiewicz, A. (2020). BRG1 activates proliferation and transcription of cell cycle-dependent genes in breast cancer cells. Cancers (Basel) 12, 349. doi:10.3390/cancers12020349
Sobczak, M., Pitt, A. R., Spickett, C. M., and Robaszkiewicz, A. (2019). PARP1 Co-regulates EP300-BRG1-dependent transcription of genes involved in breast cancer cell proliferation and DNA repair. Cancers (Basel) 11, 1539. doi:10.3390/cancers11101539
Socha, A., Yang, D., Bulsiewicz, A., Yaprianto, K., Kupculak, M., Liang, C. C., et al. (2020). WRNIP1 is recruited to DNA interstrand crosslinks and promotes repair. Cell Rep. 32, 107850. doi:10.1016/j.celrep.2020.107850
Sousa, J. F., Serafim, R. B., Freitas, L. M., Fontana, C. R., and Valente, V. (2019). DNA repair genes in astrocytoma tumorigenesis, progression and therapy resistance. Genet. Mol. Biol. 43, e20190066. doi:10.1590/1678-4685-gmb-2019-0066
Sousa, M. M., Zub, K. A., Aas, P. A., Hanssen-Bauer, A., Demirovic, A., Sarno, A., et al. (2013). An inverse switch in DNA base excision and strand break repair contributes to melphalan resistance in multiple myeloma cells. PLoS One 8, e55493. doi:10.1371/journal.pone.0055493
Sun, X., and Liu, P. (2023). Prognostic biomarker NEIL3 and its association with immune infiltration in renal clear cell carcinoma. Front. Oncol. 13, 1073941. doi:10.3389/fonc.2023.1073941
Takao, C., Morikawa, A., Ohkubo, H., Kito, Y., Saigo, C., Sakuratani, T., et al. (2017). Downregulation of ARID1A, a component of the SWI/SNF chromatin remodeling complex, in breast cancer. J. Cancer 8, 1–8. doi:10.7150/jca.16602
Tapryal, N., Shahabi, S., Chakraborty, A., Hosoki, K., Wakamiya, M., Sarkar, G., et al. (2021). Intrapulmonary administration of purified NEIL2 abrogates NF-κB-mediated inflammation. J. Biol. Chem. 296, 100723. doi:10.1016/j.jbc.2021.100723
Teng, P. C., Huang, S. P., Liu, C. H., Lin, T. Y., Cho, Y. C., Lai, Y. L., et al. (2021). Identification of DNA damage repair-associated prognostic biomarkers for prostate cancer using transcriptomic data analysis. Int. J. Mol. Sci. 22, 11771. doi:10.3390/ijms222111771
Teoh, P. J., An, O., Chung, T. H., Chooi, J. Y., Toh, S. H. M., Fan, S., et al. (2018). Aberrant hyperediting of the myeloma transcriptome by ADAR1 confers oncogenicity and is a marker of poor prognosis. Blood 132, 1304–1317. doi:10.1182/blood-2018-02-832576
Tomkinson, A. E., and Sallmyr, A. (2013). Structure and function of the DNA ligases encoded by the mammalian LIG3 gene. Gene 531, 150–157. doi:10.1016/j.gene.2013.08.061
Tran, O. T., Tadesse, S., Chu, C., and Kidane, D. (2020). Overexpression of NEIL3 associated with altered genome and poor survival in selected types of human cancer. Tumour Biol. 42, 1010428320918404. doi:10.1177/1010428320918404
Tsang, J. Y. S., and Tse, G. M. (2020). Molecular classification of breast cancer. Adv. Anat. Pathol. 27, 27–35. doi:10.1097/pap.0000000000000232
Tsutakawa, S. E., and Sarker, A. H. (2021). Breaking the rules: protein sculpting in NEIL2 regulation. Structure 29, 1–2. doi:10.1016/j.str.2020.12.011
Vartanian, V., Minko, I. G., Chawanthayatham, S., Egner, P. A., Lin, Y. C., Earley, L. F., et al. (2017). NEIL1 protects against aflatoxin-induced hepatocellular carcinoma in mice. Proc. Natl. Acad. Sci. U. S. A. 114, 4207–4212. doi:10.1073/pnas.1620932114
Vogel, A., Meyer, T., Sapisochin, G., Salem, R., and Saborowski, A. (2022). Hepatocellular carcinoma. Lancet 400, 1345–1362. doi:10.1016/s0140-6736(22)01200-4
Wang, G., Zhao, D., Spring, D. J., and Depinho, R. A. (2018). Genetics and biology of prostate cancer. Genes Dev. 32, 1105–1140. doi:10.1101/gad.315739.118
Wang, L., Zhang, W., Yang, T., He, L., Liao, Y., and Lu, J. (2021a). Construction and comprehensive analysis of a stratification system based on AGTRAP in patients with hepatocellular carcinoma. Dis. Markers 2021, 6144476. doi:10.1155/2021/6144476
Wang, Q., Li, Z., Yang, J., Peng, S., Zhou, Q., Yao, K., et al. (2021b). Loss of NEIL3 activates radiotherapy resistance in the progression of prostate cancer. Cancer Biol. Med. 19, 1193–1210. doi:10.20892/j.issn.2095-3941.2020.0550
Wang, T., Dai, L., Shen, S., Yang, Y., Yang, M., Yang, X., et al. (2022a). Comprehensive molecular analyses of a macrophage-related gene signature with regard to prognosis, immune features, and biomarkers for immunotherapy in hepatocellular carcinoma based on WGCNA and the LASSO algorithm. Front. Immunol. 13, 843408. doi:10.3389/fimmu.2022.843408
Wang, T., Zhu, X., Wang, K., Li, J., Hu, X., Lin, P., et al. (2023). Transcriptional factor MAZ promotes cisplatin-induced DNA damage repair in lung adenocarcinoma by regulating NEIL3. Pulm. Pharmacol. Ther. 80, 102217. doi:10.1016/j.pupt.2023.102217
Wang, W., Yin, Q., Guo, S., and Wang, J. (2021). NEIL3 contributes toward the carcinogenesis of liver cancer and regulates PI3K/Akt/mTOR signaling. Exp. Ther. Med. 22, 1053. doi:10.3892/etm.2021.10487
Wang, X., Wu, Y., Liu, J., Xu, X., Sheng, Z., Liu, W., et al. (2022b). Identification of target and pathway of aspirin combined with Lipitor treatment in prostate cancer through integrated bioinformatics analysis. Toxicol. Appl. Pharmacol. 452, 116169. doi:10.1016/j.taap.2022.116169
Wang, Y., Xu, L., Shi, S., Wu, S., Meng, R., Chen, H., et al. (2021c). Deficiency of NEIL3 enhances the chemotherapy resistance of prostate cancer. Int. J. Mol. Sci. 22, 4098. doi:10.3390/ijms22084098
Wasim, S., Lee, S. Y., and Kim, J. (2022). Complexities of prostate cancer. Int. J. Mol. Sci. 23, 14257. doi:10.3390/ijms232214257
Wei, H., Kamat, A., Chen, M., Ke, H. L., Chang, D. W., Yin, J., et al. (2012). Association of polymorphisms in oxidative stress genes with clinical outcomes for bladder cancer treated with Bacillus Calmette-Guérin. PLoS One 7, e38533. doi:10.1371/journal.pone.0038533
Wiederhold, L., Leppard, J. B., Kedar, P., Karimi-Busheri, F., Rasouli-Nia, A., Weinfeld, M., et al. (2004). AP endonuclease-independent DNA base excision repair in human cells. Mol. Cell 15, 209–220. doi:10.1016/j.molcel.2004.06.003
Wu, R. A., Semlow, D. R., Kamimae-Lanning, A. N., Kochenova, O. V., Chistol, G., Hodskinson, M. R., et al. (2019). TRAIP is a master regulator of DNA interstrand crosslink repair. Nature 567, 267–272. doi:10.1038/s41586-019-1002-0
Wu, C., Luo, Y., Chen, Y., Qu, H., Zheng, L., and Yao, J. (2022). Development of a prognostic gene signature for hepatocellular carcinoma. Cancer Treat. Res. Commun. 31, 100511. doi:10.1016/j.ctarc.2022.100511
Wu, D., Zhang, G., Ma, J., Wu, H., Xiong, J., Huang, X., et al. (2021). Upregulation of Nei-like DNA glycosylase 3 predicts poor prognosis in hepatocellular carcinoma. J. Oncol. 2021, 1301671. doi:10.1155/2021/1301671
Xue, W., Liu, Y., Xin, N., Miao, J., Du, J., Wang, Y., et al. (2020). Nei endonuclease VIII-Like1 (NEIL1) inhibits apoptosis of human colorectal cancer cells. Biomed. Res. Int. 2020, 5053975. doi:10.1155/2020/5053975
Yadav, S., Anbalagan, M., Baddoo, M., Chellamuthu, V. K., Mukhopadhyay, S., Woods, C., et al. (2020). Somatic mutations in the DNA repairome in prostate cancers in African Americans and Caucasians. Oncogene 39, 4299–4311. doi:10.1038/s41388-020-1280-x
Yang, J. D., Hainaut, P., Gores, G. J., Amadou, A., Plymoth, A., and Roberts, L. R. (2019). A global view of hepatocellular carcinoma: trends, risk, prevention and management. Nat. Rev. Gastroenterol. Hepatol. 16, 589–604. doi:10.1038/s41575-019-0186-y
Yang, Z., Nejad, M. I., Varela, J. G., Price, N. E., Wang, Y., and Gates, K. S. (2017). A role for the base excision repair enzyme NEIL3 in replication-dependent repair of interstrand DNA cross-links derived from psoralen and abasic sites. DNA Repair (Amst) 52, 1–11. doi:10.1016/j.dnarep.2017.02.011
Yeo, J., Goodman, R. A., Schirle, N. T., David, S. S., and Beal, P. A. (2010). RNA editing changes the lesion specificity for the DNA repair enzyme NEIL1. Proc. Natl. Acad. Sci. U. S. A. 107, 20715–20719. doi:10.1073/pnas.1009231107
Yeo, J., Lotsof, E. R., Anderson-Steele, B. M., and David, S. S. (2021). RNA editing of the human DNA glycosylase NEIL1 alters its removal of 5-hydroxyuracil lesions in DNA. Biochemistry 60, 1485–1497. doi:10.1021/acs.biochem.1c00062
Yousefzadeh, M., Henpita, C., Vyas, R., Soto-Palma, C., Robbins, P., and Niedernhofer, L. (2021). DNA damage-how and why we age? Elife 10, e62852. doi:10.7554/eLife.62852
Yuan, H., Qing, T., Zhu, S., Yang, X., Wu, W., Xu, K., et al. (2023). The effects of altered DNA damage repair genes on mutational processes and immune cell infiltration in esophageal squamous cell carcinoma. Cancer Med. 12, 10077–10090. doi:10.1002/cam4.5663
Zhai, X., Zhao, H., Liu, Z., Wang, L. E., El-Naggar, A. K., Sturgis, E. M., et al. (2008). Functional variants of the NEIL1 and NEIL2 genes and risk and progression of squamous cell carcinoma of the oral cavity and oropharynx. Clin. Cancer Res. 14, 4345–4352. doi:10.1158/1078-0432.Ccr-07-5282
Zhang, F., Lu, J., Yang, J., Dai, Q., Du, X., Xu, Y., et al. (2023). SNHG3 regulates NEIL3 via transcription factor E2F1 to mediate malignant proliferation of hepatocellular carcinoma. Immunogenetics 75, 39–51. doi:10.1007/s00251-022-01277-2
Zhao, C., Liu, J., Zhou, H., Qian, X., Sun, H., Chen, X., et al. (2021a). NEIL3 may act as a potential prognostic biomarker for lung adenocarcinoma. Cancer Cell Int. 21, 228. doi:10.1186/s12935-021-01938-4
Zhao, Y., Feng, H. M., Yan, W. J., and Qin, Y. (2022). Identification of the signature genes and network of reactive oxygen species related genes and DNA repair genes in lung adenocarcinoma. Front. Med. (Lausanne) 9, 833829. doi:10.3389/fmed.2022.833829
Zhao, Z., Gad, H., Benitez-Buelga, C., Sanjiv, K., Xiangwei, H., Kang, H., et al. (2021b). NEIL3 prevents senescence in hepatocellular carcinoma by repairing oxidative lesions at telomeres during mitosis. Cancer Res. 81, 4079–4093. doi:10.1158/0008-5472.Can-20-1028
Zhdanova, P. V., Ishchenko, A. A., Chernonosov, A. A., Zharkov, D. O., and Koval, V. V. (2022). Dynamics and conformational changes in human NEIL2 DNA glycosylase analyzed by hydrogen/deuterium exchange mass spectrometry. J. Mol. Biol. 434, 167334. doi:10.1016/j.jmb.2021.167334
Zheng, Y., Zheng, L., Yu, J., Jiang, M., Zhang, S., Cai, X., et al. (2021). Genetic variations in DNA repair gene NEIL1 associated with radiation pneumonitis risk in lung cancer patients. Mol. Genet. Genomic Med. 9, e1698. doi:10.1002/mgg3.1698
Zhou, H., and Song, T. (2021). Conversion therapy and maintenance therapy for primary hepatocellular carcinoma. Biosci. Trends 15, 155–160. doi:10.5582/bst.2021.01091
Zhou, J., Chan, J., Lambelé, M., Yusufzai, T., Stumpff, J., Opresko, P. L., et al. (2017). NEIL3 repairs telomere damage during S phase to secure chromosome segregation at mitosis. Cell Rep. 20, 2044–2056. doi:10.1016/j.celrep.2017.08.020
Zhou, J., Fleming, A. M., Averill, A. M., Burrows, C. J., and Wallace, S. S. (2015). The NEIL glycosylases remove oxidized guanine lesions from telomeric and promoter quadruplex DNA structures. Nucleic Acids Res. 43, 4039–4054. doi:10.1093/nar/gkv252
Zhou, J., Liu, M., Fleming, A. M., Burrows, C. J., and Wallace, S. S. (2013). Neil3 and NEIL1 DNA glycosylases remove oxidative damages from quadruplex DNA and exhibit preferences for lesions in the telomeric sequence context. J. Biol. Chem. 288, 27263–27272. doi:10.1074/jbc.M113.479055
Zhu, Q. Q., Ma, C., Wang, Q., Song, Y., and Lv, T. (2016). The role of TWIST1 in epithelial-mesenchymal transition and cancers. Tumour Biol. 37, 185–197. doi:10.1007/s13277-015-4450-7
Zhuang, J., Li, M., Zhang, X., Xian, S., Zhang, J., Yin, H., et al. (2022). Construction of bone metastasis-specific regulation network based on prognostic stemness-related signatures in prostate cancer. Dis. Markers 2022, 8495923. doi:10.1155/2022/8495923
Keywords: NEIL, cancer, DNA, glycosylases, malignancy, treatment
Citation: Chen Y, Ma M, Zou A, Wang X and Dong W (2025) Research progress on the role of the NEIL family in cancer. Front. Cell Dev. Biol. 13:1612329. doi: 10.3389/fcell.2025.1612329
Received: 15 April 2025; Accepted: 10 July 2025;
Published: 21 July 2025.
Edited by:
Sneha Saxena, Mass General Brigham, United StatesReviewed by:
Dipika Gupta, New York University, United StatesUmeshkumar Vekariya, Temple University, United States
Copyright © 2025 Chen, Ma, Zou, Wang and Dong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Weiwei Dong, ZG9uZ3d3QHNqLWhvc3BpdGFsLm9yZw==; Xinjia Wang, d2FuZ3hqOTcwNDAxMUAxNjMuY29tJiN4MDIwMGE7
†These authors have contributed equally to this work
 Yinghan Chen1†
Yinghan Chen1† Weiwei Dong
Weiwei Dong